Using MnovaScreen to Process, Analyze and Report Ligand-Protein Binding Spectra for Fragment-based Lead Design
|
|
|
- Sandra Henry
- 5 years ago
- Views:
Transcription
1 September 2014 Using MnovaScreen to Process, Analyze and Report Ligand-Protein Binding Spectra for Fragment-based Lead Design Dr Manuel Perez Senior VP - Mestrelab
2 1996: A research project in University of Santiago de Compostela, Spain, developed free MestReC software for NMR processing 2004: Mestrelab Research incorporated in Santiago de Compostela 2004: New MestReNova (Mnova) platform and NMR plugin released 2006: NMRPredict Desktop for NMR prediction 2009: MS plugin for LC/GC/MS data analysis 2009: Global Spectral Deconvolution (GSD) algorithm released for NMR 2011: DB plugin for Database Management of NMR and MS 2012: Verify plugin for auto structure verification 2012: qnmr plugin for quantitative NMR analysis 2013: Reaction Monitor plugin for NMR-based reaction kinetics studies 2013: Screen plugin for high-throughput ligand-protein binding analysis An R&D company with >20 people and >80,000 registered users MESTRELAB INTERNAL USE ONLY - CONFIDENTIAL
3 Mnova products and applications NMR Mnova Verify Mnova Screen Mnova DB Creating databases, Storing and searching structures, NMR, LC/GC/MS raw data and analysis results, Texts etc. LC/MS GC/MS Arrayed 2D 1D Mnova NMR Quick processing, analysis, reporting, and verifying structures Chemists Mnova NMRPredict Desktop Visualization and peak integration, molecular verification, elemental composition determination, Reporting, etc. Detailed str. verification & elucidation, peak assign. & deconvolution, spin simulation, quantitation etc. Analysts Mnova qnmr Arrayed spectra: relaxation studies, diffusion studies, reaction monitoring, ligand-protein binding screening (FBDD), metabolomics studies. etc. Specialists Batch processing, analysis and reporting, quantitation, etc. Mnova R. Monitor Mnova Scripting Service Users Mnova MS
4 Fragment-based Drug Design Using NMR Lead generation from a compound library with weak binding (K d ~um mm) in early stage of drug discovery Ligand-based screening: 1D 1 H or 19 F of mixtures of protein & ~8 ligands Saturation transfer difference (STD) Relaxation editing : T1ρ CPMG WaterLOGSY Target-based screening: 1 H- 15 N HSQC of target: Profiling, and titration using ligand ( SAR by NMR ) Analysis is a bottleneck S.B. Shuker, P.J, Hajduk, R. P. Meadows, S. W. Fesik, Discovering high-affinity ligands for proteins: SAR by NMR., Science 274 (5292): , 1996 J.Fejzo, C.A. Lepre, J.W. Peng, G.W. Bemis., M.A Ajay, J.M. Moore.; Chem. Biol. 6(10):755-69, 1999 C. Dalvit,, P. Pevarello, M. Tato, M. Veronesi, A. Vulpetti, M. Sundstrom, J. Biomol. NMR, 18(1), 65-68, 2000 M. Coles, M. Heller, H. Kessler, Drug Discovery Today 8(17): , 2003
5 The challenges of automated data analysis A lot of compounds and spectra: how to manage them? Batch processing and collating spectra: no consistent way of organizing data Peak picking: overlap, different S/N, a consistent threshold is impossible Peak misalignment: systematic and local How to detect the peak changes? How to report the results? How to facilitate the inspection of the results?
6 What Mnova can help you* Library compound: spectral processing, verification of structure, assignment of peaks Mnova NMR Mnova NMRPredict Desktop Mnova Verify Compound solubility/concentration determination Mnova qnmr Library compound/spectral management: databasing Mnova DB Plate design: auto choosing compounds with minimum peak overlap from a group of 1D spectra Mnova Scripting Batch processing and analysis of STD, waterlogsy, T1ρ and similar spectra Mnova Screen Inspection and editing results Mnova NMR Batch processing and binning 2D HSQC Mnova NMR
7 The common NMR experiments and what we do with them Experiment Sample contents What to observe? How to analyze? Reference spectra Single library compound The reference peaks of the library compounds Process the spectra, pick ref. peaks by GSD Saturation transfer Detection (STD) Mixture of N library compounds and protein. Blank (no protein) and/or competition (with protein and inhibitor) may be used for comparison The on-resonance spectral peaks are expected to decrease if that compound is binding with the protein. If the STD difference spectra are used, then the peaks are picked and matched to ref peaks. If the on/off-resonance spectral pairs are used, then the ref peaks are mapped to the STD pair. The change of peak intensities within the mapped regions are calculated and averaged. If available, blank and competition data will be used to detect false positive and nonspecific binding if available. T1ρ (and CPMG) Mixture of N library compounds and protein. Blank (no protein) and/or competition (with protein and inhibitor) may be used for comparison The peaks from the long spin-lock time spectrum are expected to decrease compared with the short spin-lock one, if that compound is binding with the protein. If inhibitor is used, the peaks are expected to restore to the original height if it is a specific binding The ref peaks are mapped to the T1ρ pair. The change of peak intensities within the mapped regions are calculated and averaged. If available, blank and competition data will be used to detect false positive and nonspecific binding if available. WaterLOGSY Mixture of N library compounds without protein (blank), and with protein. Competition sample (with protein and inhibitor) may be used for comparison Negative are expected in the blank. Positive peaks are expected for a compound that binds with the protein. If inhibitor is used, such peaks are expected to restore to negative if it is a specific binding The ref peaks are mapped to the wlogsy spectra. The change of peak intensities within the mapped regions are calculated and averaged. If available, blank and competition data will be used to detect false positive and nonspecific binding if available.
8 What types of data can Mnova Screen handle? Mnova Screen handles any combination of the experimental types: Reference : H-1 of the pure library compounds* Scout: H-1 of the mixture, with or without protein STD: without protein (blank), with protein, and with protein+inhibitor, in the form of difference spectra or on/off resonance pairs** T1ρ (and CPMG): without protein (blank), with protein, and with protein+inhibitor, in the form of short/long spin-lock time pairs*** WaterLOGSY: without protein (blank), with protein, and with protein+inhibitor, all as single spectra 19 F data The spectral data can be raw data or processed data from various vendors (Bruker, Varian/Agilent, JEOL, JCAMP etc.) Note: *The use of ref spec is optional. If you don t use ref spec, you can use the Scout as the reference, and Mnova Screen will report the maximum and average peak intensity changes for all the Scout peaks. **The use of on/off resonance pairs is preferred, as their signal to noise ratio is higher than the difference spectra, and it is possible to calculate the percentage of intensity changes relative to the original spectra. *** Mnova Screen can also handle if only 3 experiments are used: Blank with short spin-lock time, Protein with long spin-lock time, and Protein+Inhibitor with long spin-lock time.
9 How to organize your dataset? There have been no standard ways to organize screening data. Mnova Screen was designed to accept as general ways of data organization as possible: Mixture data and reference data are saved in separate folders with matching folder names Mixture data and reference data are saved in the same folder for each sample, but with distinguished experimental names Mixture data and reference data are matched by a definition file Several examples are illustrated in the subsequent slides If necessary, you can write a shell script to reorganize your data in the similar ways so that Mnova Screen can import them correctly
10 Data org. example I: Ref + STD In this example the ref. spectra for each mixture are saved under a folder <mix_name>, where <mix_name> is the folder name of the corresponding mixture. The number of ref spectra can be different for each mixture. The mixture spectra (STD in this case) are saved under a separate folder. Each mixture, <mix_name>, has several experiments (scouts and STD difference spectra, and some others - to be ignored here) : 10: To be ignored 11: STD difference spectra 12: Scout 13: To be ignored Note: This format is not limited to STD only. You can have other types of experiments for each mixture. You just need to name the experimental folders in a consistent way.
11 Data org. example II: Ref + STD + T1r In this example the ref spectra for each mixture are saved under a separate folders, such as Mix005, Mix009, etc. The mixture spectra are saved under separate folders with names matching those for the ref spectra. The experiment names have a consistent pattern defined as follows: 1[nnn]: STD difference spectra 2[nnn]: Scout 3[nnn]: T1r with short spin-lock time 4[nnn]: T1r with long spin-lock time
12 Data org. example III: Ref + STD + T1r This example is similar to Example II, but the mixture spectra are all saved under the same folder; hence there are rigorous requirements their experiment folder names match the ref folder names The ref spectra for each mixture are saved under a folder P1Mix[nnn], where [nnn] corresponds to the mixture #. The mixture spectra are all saved under one folder, with a naming convention of [e][nnn], where [e] is the experimental type # and nnn matches the ref spectral folder names: 1[nnn]: STD difference spectra 2[nnn]: Scout 3[nnn]: T1r with short spin-lock time 4[nnn]: T1r with long spin-lock time
13 Data org. example IV: Ref + T1ρ In this example all the T1ρ and ref. spectra for the same mixture are saved under the same folder, with single digit names [m] for T1ρ, and double digit names [rr] for the 8 reference spectra. 3 T1ρ s are used for each mixture: 1: T1ρ blank 2: T1ρ with protein 3: T1ρ with protein and inhibitor 8 ref. spectra are saved in the same folder (11-18)
14 Data org. example V: Ref + STD + T1r + wlogsy In this example all mixture blank spectra [m][e] are saved under one folder (e=0-4): [m]0: Scout [m]1: T1ρ with short spin-lock [m]2: T1ρ with long spin-lock [m]3: WaterLOGSY [m]4 : STD pair (on/off resonance) All mixture spectra [m][e] with protein are saved under another folder with exactly the same names as for the blanks. * *Note: The same number of folders with the exactly same names are expected in either folders. Each mixture must have exactly the same number (5) of experiments.
15 Data org. example V: Ref + STD + T1r + wlogsy (continued) The names of the library compounds for each mixture are saved in the Comment parameter (read from the title file) of each experiment* The reference spectra are saved in other folders. The correspondence between the lib compound names and the reference spectra are defined in an Excel file** Note: *The number of library compounds can be fewer than a maximum (8 in this case). If the spectrum of a lib compound cannot be found, it is ignored without interrupting the program. **Mnova Screen does not read Excel file, so the original Excel file should be exported to a.csv file.
16 To install Mnova Screen Mnova Screen is a separately licensed plugin to the Mnova system. To run it, you need to install Mnova NMR and Screen, and have licenses to both plugins After installation of Mnova Screen you will see two commands on the Advanced menu: Mnova Screen: for processing and analyzing FBS data View Mnova Screen Results: for inspecting and refining the results from previous runs Choose Help > License Manager and make sure you have properly activated both the NMR and Mnova Screen plugins** To download the installer of Mnova Suite (including NMR and NMRPredict Desktop): To download the installer of Mnova Screen:
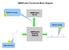
17 Workflow of Mnova Screen Processing template(s) STD/T1ρ/wL spectra Library spectra Batch process and group spectra (STD,T1ρ, wlogsy, reference spectra) Deconvolute spectra and match peaks User inspects selected experiments Stack spectra and generate reports Mnova file for each sample Overall results in a spreadsheet
18 Before you start Make sure your data files are organized as described in the previous slides, and the incomplete experiments are moved away from the folders Open a few spectra data in Mnova and see how the auto processing works for your data. If not satisfactory, you choose Process > Processing Template, change the parameters until you are satisfied, and then save a processing template for later use. Stack a few sets of ref. spectra and mix. spectra together to see how well the peaks align. Find a peak (e.g. DSS) that can be used as reference and the chemical shift offset for aligning them. Also observe the local misalignment of individual peaks across the spectra, so that you have an idea about the tolerance for matching ref. peaks and mix. peaks. For T1ρ and WaterLOGSY, identify the peak (e.g. DMSO or DSS) that can be used as reference to normalize the spectral intensity, or make sure your experimental and acquisition conditions are identical Decide the spectral region(s) that you are interested in (ROI). Make sure solvent and buffer peaks are not included. The subsequent slides shows an example of how to figure out these settings.
19 Before you start: Check the processing of spectra Open all the mix. and ref. spectra of the first sample in Mnova, and stack them together. Use the Cut tool to show only the aromatic and DSS regions. In this case, the auto processing gives good phasing. So no processing template is needed.* Ref. spectra Mix. spectra Note: *Mnova Screen will automatically apply a baseline correction (3 rd order Bernstein Polynomial) to a spectrum if no processing template is specified for it.
20 Before you start: Check the Y-scales It s observed that the spectra have very different Y scales so normalization is needed to make their peaks visible in the stack. The DSS peaks can be used for the ref spectra (e.g. setting their height to be 25,000) The STD diff spectra does not have a visible DSS peak, so we can normalize them by setting the highest peak within the aromatic region to 25,000 DSS peaks Ref. spectra Scout spectrum STD difference
21 Before you start: Check the normalization The T1ρ pair can be normalized based on DMSO peak s areas within ppm. By setting the integral to a proper number (e.g. 250,000), it will also make the Y scale of T1ρ similar to that of the other spectra, so that their peaks are visible in the stacked spectra. Choose Processing > Normalize, and set the parameters as above
22 Before you start: Check the spectral alignment In this case, the ref. spectra are not properly aligned, and they are not wellaligned with mix. spectra either. By trial and error, it s found that the misalignment can be offset by setting the DSS peaks of the ref spectra to ppm Check the ref. spectra in the Stacked Spectra table so only they will be changed. Choose the Reference tool, and click on the DSS peak of the active spectrum Turn on the Auto Tuning option and enter the values as shown above.
23 Before you start: Check the spectral alignment Most of the peaks are well-aligned now, though local shift of some peaks is inevitable. A typical local peak mis-alignment that cannot be offset by changing chemical shift reference. Hence a tolerance for peak matching is always needed. See next page for details
24 Before you start: Estimate peak match tolerance Shift = 3.0 Hz Peak width = 1.6 Hz Tolerance= ± 4 Hz Tip: To measure the distance between two peaks, click C key for Cross-hair Mode. Next click on a peak top and drag to another one. The distance will be reported in an Info View dialog. A tolerance of ±2.5 x peak width (=8 Hz for this peak) will be used for matching peaks, or integrating peaks for measuring the change of peak intensity
25 Setup parameters for Mnova Screen Mnova Screen allows you to run against a specific mixture, a group of mixtures, or all of your mixtures. It is better to start with one or a few mixtures to optimize your parameters before you start to process all mixtures Choose Scripts > Mnova Screen, and fill in the relevant info in the Project Info tab: Tip: Click the question buttons to see specific instructions
26 Define your datasets Parent folder for the mixture data This will read all folders whose name starts with 1, 2, 3 or 4, and ends with 3 other characters.* The file name is limited to 3 characters ( fid or ser ) The matched mix. data (and ref. data when defined) are summarized here The matched data files for the first mixture is listed here. You click on the Type cells to define their experimental Types and sample Contents** Parent folder for the reference data This will read all folders under each folder that that starts with P1Mix and ends with a matching number with the mix. folder. fid file is searched. The actual file paths for the ref. spectra are listed here. Note: * You can limit the samples you want to run, e.g. use [1-4]005/??? for one sample, or [1-4]??5/??? for all samples ending with 5, and so on. ** Click on a Type cell to define it as Scout, STD, T1ρ, or wlogsy. Click on a Contents cell to define it as Blank, Protein, or Protein+Inhibitor.
27 Define processing and analysis parameters These parameters are defined based on the previous inspection of the sample spectra. See details in the subsequent slides
28 Processing settings for Scout spectra In this case, the scout spectra are used to identify whether a ref compound presents in the mixture or not. Click the Edit button next to open it: Scout spectra are normalized by setting the highest peak within ROIs to 25,000. This is for visualization only. No alignment is done for scouts Scout are processed using the original processing parameters, except zerofilling to 64K, and applying Bernstein trinomial baseline correction No extra processing script is applied Peaks with S/N Ratio less than 10 will be discarded to reduce noise/impurity peaks.
29 Processing settings for STD diff spectra In this case, STD diff spectra are used to identify primary hits Screen matches ref peaks with the STD diff peaks and identify primary hits based on the percentage of matched ref peaks STD diff spectra are normalized by setting the highest peak within ROIs to 25,000 (This is for visualization only) No alignment is done for STD STD are processed using the original processing parameters, except zerofilling to 64K, and applying Bernstein trinomial baseline correction No extra processing script is applied STD peaks with height less than 10% of the max. peak in the ROIs are discarded to reduce noise/impurity peaks.
30 Processing settings for T1ρ spectra In this case, T1ρ spectral pairs are used to identify primary hits: Ref peaks are mapped to the T1ρ spectra and the decrease of peak intensity from the short spin-lock T1ρ to the long spin-lock T1ρ is used to identify primary hits T1ρ spectra are normalized by setting the spectral area within ppm to 2500,000. Note this affects both the analysis results and visualization. No alignment is done for STD T1ρ are processed using the original processing parameters, except zerofilling to 64K, and applying Bernstein trinomial baseline correction No extra processing script is applied This threshold is used to avoid false positives caused by comparing noise and impurity peaks. See details in the next slide
31 To avoid false positives caused by local peak shift In this case, local peak shift makes Ref Peak B map to a region that contains mostly base points in either T1ρ spectrum (shown as the pink box, with +/- 2.5* ref. peak width). The resulting intensity change can be random and easily causes a false positive. Our solution is to get the highest intensity within the region for each T1r. Next compare it with the highest intensity within the ROIs. The ratio from at least one T1r must exceed the threshold (e.g. 5%). Otherwise we ignore this ref peak. Note that the T1r with long spinlock may have a peak totally disappears, so we can only require one of them to meet this category. The absolute values are used above to cover wlogsy, as peaks can be positive and negative in it. Integration region = 5.0Hz Ref. peak width = 1.0Hz Blue curve: Ref spec. Brown and green curves: T1r spec A B C D E
32 Processing settings for reference spectra It is critical to align ref spectra with mixture ones, as the ref peaks (picked using GSD) are used to match mixture peaks. Ref spectra are normalized by setting the spectral area within ppm to 25,000. This is only for visualization. Ref spectra are aligned with the other mixture spectra by setting their highest (TSS) peak within 0 +/ ppm to ppm Ref spec are processed using the original processing parameters, except zero-filling to 64K, and applying Bernstein trinomial baseline correction No extra processing script is applied Ref peaks with S/N Ratio less than 10 will be discarded to reduce noise/impurity peaks.
33 Other settings for analysis It is critical to align ref spectra with mixture ones, as the ref peaks (picked using GSD) are used to match mixture peaks. A fragment is considered a primary hit if at least 20% of its ref peaks match STD diff peaks A fragment is considered a primary hit if the average T1ρ intensity decrease is 30% or more Options to display difference spectra for T1ρ spectral pairs A fragment is considered present (in the mixture) if at least 20% of the ref peaks match Scout peaks Tolerance for match a ref peak with STD diff peak or T1ρ integration region: +/- 2.5xRef peak width Which ref spectra to display in Stacked Spectral Table Region of Interets (ROIs) is set to 6-9 ppm
34 Save, load, or reset your parameters All the parameters can be saved into a.txt file for later use You can load a parameters file to reuse the previous settings Some times Mnova Screen gets confused during the parameters setup. Click Reset to remove all the settings.
35 Results from Mnova Screen After the completion of a run, Mnova Screen summarizes the results in a dialog Upon clicking OK, Screen displays a spreadsheet showing an overview of the results Use the Reference Display Text box to choose the details for each type of data Missing: not enough ref peaks matched the scout peaks, hence the compound is considered missing from the mixture. Present: The compound presents, but none of the intensity changes meets the criteria Non-selective hit: At least one of the intensity changes meets the criteria, but has not passed the competition test (if Protein+Inhibitor spec are used). Specific hit: At least one of the intensity changes meets the criteria, and also passed the competition test, if any.
36 Display stacked spectra in Mnova Double click on the row header to open the corresponding.mnova document in Mnova You can double click on other cells to change the status, or edit the comments On the stacked spectra in Mnova, you can also double click on a ref. spectrum to change its status Double click on a cell or on the spectrum here to change its status manually Ref. spectra Mix. spectra
37 Tools for inspecting the results Mnova provides many tools for visualizing stacked spectra conveniently Click to open and dock the Stacked Spectra Table to the right of the Mnova window. Use it to choose which ones to display Use the Stacked Tool to switch to Stacked, Superimposed or Active Only mode to compare spectra in different ways The log.txt file generated in the Results folder logs many details of the processing and analysis for every spectra. Tip: Mnova Screen cuts the display of the stacked spectra so that only the region of interest is visible. To restore the display of the cut regions, click Restore All in the Cut options.
38 Full View: The whole spectrum and zoom-in region. Drag the blue box to see other parts of the spectra. (Choose View Full View to open Full View if not yet) Stacked Mode: choose to display the spectra as Stacked/Superim posed/active Only Pages View: scroll to different pages to see the processing and pick picking results for each spectrum Stacked Spectra Table: Check/uncheck the boxes here to choose which spectra to display
39 Details of the results: The missing ones Choose to display the Average Scout/Blank Match Proportion results to see why one of them is taken as missing (red): This one is missing as the only peak within ROI does not matches any Scout peaks This one is missing as there is no ref peaks within the ROI
40 Details of the results: Hits from STD Choose to display the STD Protein Match Proportion results. Fragment 5 of Experiment 005 are taken as a hit because 87.5% of its ref peaks matched with the STD difference peaks (> the threshold of 20%): Double click on Comment cell to open the Comments dialog, and you can manually edit the texts here
41 Details of the results: Hits from STD Use the display tools to check the STD difference and the ref spec #5 Ref spectrum #5 STD difference spec Tip: Scroll to the pages containing the single ref. or STD diff spectra to see the GSD peak picking results. In the Superimposed or Active Only mode, you can also see the peak labels of the active spectrum in the stacked spectra. The details of peak matching analysis can be found in the log.txt file in the result folder.
42 Details of the results: Hits from T1ρ Choose to display the T1rho Protein Average Intensity Change results. Fragment 5 of Experiment 005 are taken as a hit because its peaks have an average intensity changes of 45.4% ( > threshold of 30%):
43 Details of the results: Hits from T1ρ Use the display tools to check the T1ρ pair and the Ref #5 T1ρ with short spin lock time T1ρ with long spin lock time Ref spec #5 Tip: In the Superimposed or Active Only mode, you can also see the peak labels of the active spectrum (as shown above). The details of peak matching analysis can be found in the log.txt file in the result folder.
44 Log.txt: Details of the analysis The log.txt file logs all the details of spectral processing and analysis. Use a text editor such as Notepad++ to view it. Details of the GSD peaks from the ref spec 5_5 Details of the match of ref peaks with the scout peaks Details of the match of the ref peaks with the STD diff peaks Details of the calculation of peak intensity changes in T1ρ. For example, for ref peak at ppm, sumintegration is done within ppm, and the decrease of spectral integration is %
45 Export results to a spreadsheet After examining and revising the results, you can export the results to a spreadsheet by clicking the button, and then choosing the results that you want to export:
46 Save and reload the results After examining and revising the results, you can click the Save button to save the results before closing the Editor. If you want to edit the previous results again, choose Advanced > View Mnova Screen Results, and open the editor.csv file saved in the Results folder.
47 Thank you for your time!
Mnova Software Tools for Fragment-Based Drug Discovery
 Mnova Software Tools for Fragment-Based Drug Discovery Chen Peng, PhD, VP of Business Development, US & China Mestrelab Research SL San Diego, CA chen.peng@mestrelab.com 858.736.4563 Agenda Brief intro
Mnova Software Tools for Fragment-Based Drug Discovery Chen Peng, PhD, VP of Business Development, US & China Mestrelab Research SL San Diego, CA chen.peng@mestrelab.com 858.736.4563 Agenda Brief intro
Mnova Software for Analyzing Reaction Monitoring NMR Spectra
 Mnova Software for Analyzing Reaction Monitoring NMR Spectra Version 10 Chen Peng, PhD, VP of Business Development, US & China Mestrelab Research SL San Diego, CA, USA chen.peng@mestrelab.com 858.736.4563
Mnova Software for Analyzing Reaction Monitoring NMR Spectra Version 10 Chen Peng, PhD, VP of Business Development, US & China Mestrelab Research SL San Diego, CA, USA chen.peng@mestrelab.com 858.736.4563
Implementation of novel tools to facilitate fragment-based drug discovery by NMR:
 Implementation of novel tools to facilitate fragment-based drug discovery by NMR: Automated analysis of large sets of ligand-observed NMR binding data and 19 F methods Andreas Lingel Global Discovery Chemistry
Implementation of novel tools to facilitate fragment-based drug discovery by NMR: Automated analysis of large sets of ligand-observed NMR binding data and 19 F methods Andreas Lingel Global Discovery Chemistry
NMR Solutions for drug discovery
 NMR Solutions for drug discovery Dr. Matteo Pennestri London, UK Bruker Users Meeting Innovation with Integrity The principle of Fragment Based Screening from efficient fragments to Drug candidates Fragment
NMR Solutions for drug discovery Dr. Matteo Pennestri London, UK Bruker Users Meeting Innovation with Integrity The principle of Fragment Based Screening from efficient fragments to Drug candidates Fragment
NMR Predictor. Introduction
 NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
ProMass Deconvolution User Training. Novatia LLC January, 2013
 ProMass Deconvolution User Training Novatia LLC January, 2013 Overview General info about ProMass Features Basics of how ProMass Deconvolution works Example Spectra Manual Deconvolution with ProMass Deconvolution
ProMass Deconvolution User Training Novatia LLC January, 2013 Overview General info about ProMass Features Basics of how ProMass Deconvolution works Example Spectra Manual Deconvolution with ProMass Deconvolution
Login -the operator screen should be in view when you first sit down at the spectrometer console:
 Lab #2 1D 1 H Double Resonance (Selective Decoupling) operation of the 400 MHz instrument using automated sample insertion (robot) and automated locking and shimming collection of 1D 1 H spectra retrieving
Lab #2 1D 1 H Double Resonance (Selective Decoupling) operation of the 400 MHz instrument using automated sample insertion (robot) and automated locking and shimming collection of 1D 1 H spectra retrieving
Fragment-based drug discovery
 Fragment-based drug discovery Dr. Till Kühn VP Applications Development MRS, Bruker BioSpion User s meeting, Brussels, November 2016 Innovation with Integrity The principle of Fragment Based Screening
Fragment-based drug discovery Dr. Till Kühn VP Applications Development MRS, Bruker BioSpion User s meeting, Brussels, November 2016 Innovation with Integrity The principle of Fragment Based Screening
MassHunter TOF/QTOF Users Meeting
 MassHunter TOF/QTOF Users Meeting 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks
MassHunter TOF/QTOF Users Meeting 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
MassHunter Software Overview
 MassHunter Software Overview 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks A
MassHunter Software Overview 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks A
Handling Human Interpreted Analytical Data. Workflows for Pharmaceutical R&D. Presented by Peter Russell
 Handling Human Interpreted Analytical Data Workflows for Pharmaceutical R&D Presented by Peter Russell 2011 Survey 88% of R&D organizations lack adequate systems to automatically collect data for reporting,
Handling Human Interpreted Analytical Data Workflows for Pharmaceutical R&D Presented by Peter Russell 2011 Survey 88% of R&D organizations lack adequate systems to automatically collect data for reporting,
NMR Data workup using NUTS
 omework 1 Chem 636, Fall 2008 due at the beginning of the 2 nd week lab (week of Sept 9) NMR Data workup using NUTS This laboratory and homework introduces the basic processing of one dimensional NMR data
omework 1 Chem 636, Fall 2008 due at the beginning of the 2 nd week lab (week of Sept 9) NMR Data workup using NUTS This laboratory and homework introduces the basic processing of one dimensional NMR data
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Application Note 12: Fully Automated Compound Screening and Verification Using Spinsolve and MestReNova
 Application Note : Fully Automated Compound Screening and Verification Using Spinsolve and MestReNova Paul Bowyer, Magritek, Inc. and Mark Dixon, Mestrelab Sample screening to verify the identity or integrity
Application Note : Fully Automated Compound Screening and Verification Using Spinsolve and MestReNova Paul Bowyer, Magritek, Inc. and Mark Dixon, Mestrelab Sample screening to verify the identity or integrity
Chemistry Department
 Chemistry Department NMR/Instrumentation Facility Users Guide - VNMRJ Prepared by Leila Maurmann The following procedures should be used to acquire one-dimensional proton and carbon NMR data on the 400MHz
Chemistry Department NMR/Instrumentation Facility Users Guide - VNMRJ Prepared by Leila Maurmann The following procedures should be used to acquire one-dimensional proton and carbon NMR data on the 400MHz
2D NMR: HMBC Assignments and Publishing NMR Data Using MNova
 Homework 10 Chem 636, Fall 2014 due at the beginning of lab Nov 18-20 updated 10 Nov 2014 (cgf) 2D NMR: HMBC Assignments and Publishing NMR Data Using MNova Use Artemis (Av-400) or Callisto (Av-500) for
Homework 10 Chem 636, Fall 2014 due at the beginning of lab Nov 18-20 updated 10 Nov 2014 (cgf) 2D NMR: HMBC Assignments and Publishing NMR Data Using MNova Use Artemis (Av-400) or Callisto (Av-500) for
Cerno Application Note Extending the Limits of Mass Spectrometry
 Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Quantification of JEOL XPS Spectra from SpecSurf
 Quantification of JEOL XPS Spectra from SpecSurf The quantification procedure used by the JEOL SpecSurf software involves modifying the Scofield cross-sections to account for both an energy dependency
Quantification of JEOL XPS Spectra from SpecSurf The quantification procedure used by the JEOL SpecSurf software involves modifying the Scofield cross-sections to account for both an energy dependency
Ligand Scout Tutorials
 Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
ncounter PlexSet Data Analysis Guidelines
 ncounter PlexSet Data Analysis Guidelines NanoString Technologies, Inc. 530 airview Ave North Seattle, Washington 98109 USA Telephone: 206.378.6266 888.358.6266 E-mail: info@nanostring.com Molecules That
ncounter PlexSet Data Analysis Guidelines NanoString Technologies, Inc. 530 airview Ave North Seattle, Washington 98109 USA Telephone: 206.378.6266 888.358.6266 E-mail: info@nanostring.com Molecules That
La RMN quantitative appliquée aux petites molécules
 La RMN quantitative appliquée aux petites molécules Fabrice Moriaud - Applications Development - Fällanden 30ème Réunion d Utilisateurs RMN Bruker December 9, 2016 1 Covered in this presentation Quantification
La RMN quantitative appliquée aux petites molécules Fabrice Moriaud - Applications Development - Fällanden 30ème Réunion d Utilisateurs RMN Bruker December 9, 2016 1 Covered in this presentation Quantification
Tutorial. Getting started. Sample to Insight. March 31, 2016
 Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
OECD QSAR Toolbox v.4.1. Tutorial illustrating new options for grouping with metabolism
 OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
Building Inflation Tables and CER Libraries
 Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Protein Deconvolution Version 2.0
 Thermo Protein Deconvolution Version 2.0 User Guide XCALI-97414 Revision A August 2012 2012 Thermo Fisher Scientific Inc. All rights reserved. ReSpect is a trademark of Positive Probability Ltd. Xcalibur
Thermo Protein Deconvolution Version 2.0 User Guide XCALI-97414 Revision A August 2012 2012 Thermo Fisher Scientific Inc. All rights reserved. ReSpect is a trademark of Positive Probability Ltd. Xcalibur
Viewing and Analyzing Proteins, Ligands and their Complexes 2
 2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
Last updated: Copyright
 Last updated: 2012-08-20 Copyright 2004-2012 plabel (v2.4) User s Manual by Bioinformatics Group, Institute of Computing Technology, Chinese Academy of Sciences Tel: 86-10-62601016 Email: zhangkun01@ict.ac.cn,
Last updated: 2012-08-20 Copyright 2004-2012 plabel (v2.4) User s Manual by Bioinformatics Group, Institute of Computing Technology, Chinese Academy of Sciences Tel: 86-10-62601016 Email: zhangkun01@ict.ac.cn,
Operation of the Bruker 400 JB Stothers NMR Facility Department of Chemistry Western University
 Operation of the Bruker 400 JB Stothers NMR Facility Department of Chemistry Western University 1. INTRODUCTION...3 1.1. Overview of the Bruker 400 NMR Spectrometer...3 1.2. Overview of Software... 3 1.2.1.
Operation of the Bruker 400 JB Stothers NMR Facility Department of Chemistry Western University 1. INTRODUCTION...3 1.1. Overview of the Bruker 400 NMR Spectrometer...3 1.2. Overview of Software... 3 1.2.1.
SeeSAR 7.1 Beginners Guide. June 2017
 SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
Simulation of Second Order Spectra Using SpinWorks. CHEM/BCMB 8190 Biomolecular NMR UGA, Spring, 2005
 Simulation of Second Order Spectra Using SpinWorks CHEM/BCMB 8190 Biomolecular NMR UGA, Spring, 2005 Introduction Although we frequently assume that scalar couplings are small compared to the differences
Simulation of Second Order Spectra Using SpinWorks CHEM/BCMB 8190 Biomolecular NMR UGA, Spring, 2005 Introduction Although we frequently assume that scalar couplings are small compared to the differences
Intelligent NMR productivity tools
 Intelligent NMR productivity tools Till Kühn VP Applications Development Pittsburgh April 2016 Innovation with Integrity A week in the life of Brian Brian Works in a hypothetical pharma company / university
Intelligent NMR productivity tools Till Kühn VP Applications Development Pittsburgh April 2016 Innovation with Integrity A week in the life of Brian Brian Works in a hypothetical pharma company / university
Data Structures & Database Queries in GIS
 Data Structures & Database Queries in GIS Objective In this lab we will show you how to use ArcGIS for analysis of digital elevation models (DEM s), in relationship to Rocky Mountain bighorn sheep (Ovis
Data Structures & Database Queries in GIS Objective In this lab we will show you how to use ArcGIS for analysis of digital elevation models (DEM s), in relationship to Rocky Mountain bighorn sheep (Ovis
Sample Alignment (2D detector) Part
 Sample Alignment (2D detector) Part Contents Contents 1 How to set Part conditions...1 1.1 Setting conditions... 1 1.2 Customizing scan conditions and slit conditions... 6 2 Sample alignment sequence...13
Sample Alignment (2D detector) Part Contents Contents 1 How to set Part conditions...1 1.1 Setting conditions... 1 1.2 Customizing scan conditions and slit conditions... 6 2 Sample alignment sequence...13
Determining C-H Connectivity: ghmqc and ghmbc (VnmrJ-2.2D Version: For use with the new Software)
 Determining C-H Connectivity: ghmqc and ghmbc (VnmrJ-2.2D Version: For use with the new Software) Heteronuclear Multiple Quantum Coherence (HMQC) and Heteronuclear Multiple Bond Coherence (HMBC) are 2-dimensional
Determining C-H Connectivity: ghmqc and ghmbc (VnmrJ-2.2D Version: For use with the new Software) Heteronuclear Multiple Quantum Coherence (HMQC) and Heteronuclear Multiple Bond Coherence (HMBC) are 2-dimensional
VCell Tutorial. Building a Rule-Based Model
 VCell Tutorial Building a Rule-Based Model We will demonstrate how to create a rule-based model of EGFR receptor interaction with two adapter proteins Grb2 and Shc. A Receptor-monomer reversibly binds
VCell Tutorial Building a Rule-Based Model We will demonstrate how to create a rule-based model of EGFR receptor interaction with two adapter proteins Grb2 and Shc. A Receptor-monomer reversibly binds
Performing Map Cartography. using Esri Production Mapping
 AGENDA Performing Map Cartography Presentation Title using Esri Production Mapping Name of Speaker Company Name Kannan Jayaraman Agenda Introduction What s New in ArcGIS 10.1 ESRI Production Mapping Mapping
AGENDA Performing Map Cartography Presentation Title using Esri Production Mapping Name of Speaker Company Name Kannan Jayaraman Agenda Introduction What s New in ArcGIS 10.1 ESRI Production Mapping Mapping
Creating Empirical Calibrations
 030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
Sample Alignment Part
 Sample Alignment Part Contents Contents 1. How to set Part conditions...1 1.1 Setting conditions... 1 1.2 Customizing scan conditions and slit conditions... 6 2. Sample alignment sequence...13 2.1 Direct
Sample Alignment Part Contents Contents 1. How to set Part conditions...1 1.1 Setting conditions... 1 1.2 Customizing scan conditions and slit conditions... 6 2. Sample alignment sequence...13 2.1 Direct
Agilent MassHunter Quantitative Data Analysis
 Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch and Method Setup Outliers, Data Review, Reporting 1 MassHunter Quantitative Analysis
Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch and Method Setup Outliers, Data Review, Reporting 1 MassHunter Quantitative Analysis
All Ions MS/MS: Targeted Screening and Quantitation Using Agilent TOF and Q-TOF LC/MS Systems
 All Ions MS/MS: Targeted Screening and Quantitation Using Agilent TOF and Q-TOF LC/MS Systems Technical Overview Introduction All Ions MS/MS is a technique that is available for Agilent high resolution
All Ions MS/MS: Targeted Screening and Quantitation Using Agilent TOF and Q-TOF LC/MS Systems Technical Overview Introduction All Ions MS/MS is a technique that is available for Agilent high resolution
Agilent MassHunter Quantitative Data Analysis
 Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch Table, Compound Information Setup, Calibration Curve and Globals Settings 1 MassHunter
Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch Table, Compound Information Setup, Calibration Curve and Globals Settings 1 MassHunter
ST-Links. SpatialKit. Version 3.0.x. For ArcMap. ArcMap Extension for Directly Connecting to Spatial Databases. ST-Links Corporation.
 ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
Vector Analysis: Farm Land Suitability Analysis in Groton, MA
 Vector Analysis: Farm Land Suitability Analysis in Groton, MA Written by Adrienne Goldsberry, revised by Carolyn Talmadge 10/9/2018 Introduction In this assignment, you will help to identify potentially
Vector Analysis: Farm Land Suitability Analysis in Groton, MA Written by Adrienne Goldsberry, revised by Carolyn Talmadge 10/9/2018 Introduction In this assignment, you will help to identify potentially
Agilent All Ions MS/MS
 Agilent All Ions MS/MS Workflow Overview A Determine fragment ions for LC/MS Quant method B Develop final Quant method Develop LC/MS Qualitative Analysis method Process data with Find by Formula Build
Agilent All Ions MS/MS Workflow Overview A Determine fragment ions for LC/MS Quant method B Develop final Quant method Develop LC/MS Qualitative Analysis method Process data with Find by Formula Build
mylab: Chemical Safety Module Last Updated: January 19, 2018
 : Chemical Safety Module Contents Introduction... 1 Getting started... 1 Login... 1 Receiving Items from MMP Order... 3 Inventory... 4 Show me Chemicals where... 4 Items Received on... 5 All Items... 5
: Chemical Safety Module Contents Introduction... 1 Getting started... 1 Login... 1 Receiving Items from MMP Order... 3 Inventory... 4 Show me Chemicals where... 4 Items Received on... 5 All Items... 5
Skin Damage Visualizer TiVi60 User Manual
 Skin Damage Visualizer TiVi60 User Manual PIONEERS IN TISSUE VIABILITY IMAGING User Manual 3.2 Version 3.2 October 2013 Dear Valued Customer! TiVi60 Skin Damage Visualizer Welcome to the WheelsBridge Skin
Skin Damage Visualizer TiVi60 User Manual PIONEERS IN TISSUE VIABILITY IMAGING User Manual 3.2 Version 3.2 October 2013 Dear Valued Customer! TiVi60 Skin Damage Visualizer Welcome to the WheelsBridge Skin
This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus.
 PCI GEOMATICS GEOMATICA QUICKSTART 1. Introduction This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus. All data used in
PCI GEOMATICS GEOMATICA QUICKSTART 1. Introduction This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus. All data used in
Automated, High- Throughput Data Processing & Quantification: Illustrated by a series of Non-Steroidal Anti-Inflammatory Drugs (NSAIDs)
 Automated, High- Throughput Data Processing & Quantification: Illustrated by a series of Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) Automatic Data Processing Mnova offers a number of listeners that
Automated, High- Throughput Data Processing & Quantification: Illustrated by a series of Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) Automatic Data Processing Mnova offers a number of listeners that
SIERRA ANALYTICS, INC. Version Polymerix Software User Manual
 SIERRA ANALYTICS, INC. Version 3.0.0 Polymerix Software User Manual V E R S I O N 3.0.0 M A R C H 2013 Polymerix Software User Manual Copyright 2010 to 2013 Sierra Analytics, Inc. 5815 Stoddard Road, Suite
SIERRA ANALYTICS, INC. Version 3.0.0 Polymerix Software User Manual V E R S I O N 3.0.0 M A R C H 2013 Polymerix Software User Manual Copyright 2010 to 2013 Sierra Analytics, Inc. 5815 Stoddard Road, Suite
Skyline Small Molecule Targets
 Skyline Small Molecule Targets The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline documents. Originally developed
Skyline Small Molecule Targets The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline documents. Originally developed
Introduction to Computer Tools and Uncertainties
 Experiment 1 Introduction to Computer Tools and Uncertainties 1.1 Objectives To become familiar with the computer programs and utilities that will be used throughout the semester. To become familiar with
Experiment 1 Introduction to Computer Tools and Uncertainties 1.1 Objectives To become familiar with the computer programs and utilities that will be used throughout the semester. To become familiar with
OECD QSAR Toolbox v.4.1. Tutorial on how to predict Skin sensitization potential taking into account alert performance
 OECD QSAR Toolbox v.4.1 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.1 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.0. Tutorial on how to predict Skin sensitization potential taking into account alert performance
 OECD QSAR Toolbox v.4.0 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.0 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
How to Create Stream Networks using DEM and TauDEM
 How to Create Stream Networks using DEM and TauDEM Take note: These procedures do not describe all steps. Knowledge of ArcGIS, DEMs, and TauDEM is required. TauDEM software ( http://hydrology.neng.usu.edu/taudem/
How to Create Stream Networks using DEM and TauDEM Take note: These procedures do not describe all steps. Knowledge of ArcGIS, DEMs, and TauDEM is required. TauDEM software ( http://hydrology.neng.usu.edu/taudem/
Downloading GPS Waypoints
 Downloading Data with DNR- GPS & Importing to ArcMap and Google Earth Written by Patrick Florance & Carolyn Talmadge, updated on 4/10/17 DOWNLOADING GPS WAYPOINTS... 1 VIEWING YOUR POINTS IN GOOGLE EARTH...
Downloading Data with DNR- GPS & Importing to ArcMap and Google Earth Written by Patrick Florance & Carolyn Talmadge, updated on 4/10/17 DOWNLOADING GPS WAYPOINTS... 1 VIEWING YOUR POINTS IN GOOGLE EARTH...
Advanced Forecast. For MAX TM. Users Manual
 Advanced Forecast For MAX TM Users Manual www.maxtoolkit.com Revised: June 24, 2014 Contents Purpose:... 3 Installation... 3 Requirements:... 3 Installer:... 3 Setup: spreadsheet... 4 Setup: External Forecast
Advanced Forecast For MAX TM Users Manual www.maxtoolkit.com Revised: June 24, 2014 Contents Purpose:... 3 Installation... 3 Requirements:... 3 Installer:... 3 Setup: spreadsheet... 4 Setup: External Forecast
protein interaction analysis bulletin 6300
 protein interaction analysis bulletin 6300 Guide to SPR Data Analysis on the ProteOn XPR36 System Ruben Luo, Bio-Rad Laboratories, Inc., 2000 Alfred Nobel Drive, Hercules, CA 94547 Kinetic Analysis To
protein interaction analysis bulletin 6300 Guide to SPR Data Analysis on the ProteOn XPR36 System Ruben Luo, Bio-Rad Laboratories, Inc., 2000 Alfred Nobel Drive, Hercules, CA 94547 Kinetic Analysis To
You w i ll f ol l ow these st eps : Before opening files, the S c e n e panel is active.
 You w i ll f ol l ow these st eps : A. O pen a n i m a g e s t a c k. B. Tr a c e t h e d e n d r i t e w i t h t h e user-guided m ode. C. D e t e c t t h e s p i n e s a u t o m a t i c a l l y. D. C
You w i ll f ol l ow these st eps : A. O pen a n i m a g e s t a c k. B. Tr a c e t h e d e n d r i t e w i t h t h e user-guided m ode. C. D e t e c t t h e s p i n e s a u t o m a t i c a l l y. D. C
ACD/Labs Software Impurity Resolution Management. Presented by Peter Russell
 ACD/Labs Software Impurity Resolution Management Presented by Peter Russell Impurity Resolution Process Chemists Method Development Specialists Toxicology Groups Stability Groups Analytical Chemists 7/8/2016
ACD/Labs Software Impurity Resolution Management Presented by Peter Russell Impurity Resolution Process Chemists Method Development Specialists Toxicology Groups Stability Groups Analytical Chemists 7/8/2016
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
85. Geo Processing Mineral Liberation Data
 Research Center, Pori / Pertti Lamberg 15023-ORC-J 1 (23) 85. Geo Processing Mineral Liberation Data 85.1. Introduction The Mineral Liberation Analyzer, MLA, is an automated mineral analysis system that
Research Center, Pori / Pertti Lamberg 15023-ORC-J 1 (23) 85. Geo Processing Mineral Liberation Data 85.1. Introduction The Mineral Liberation Analyzer, MLA, is an automated mineral analysis system that
Using SkyTools to log Texas 45 list objects
 Houston Astronomical Society Using SkyTools to log Texas 45 list objects You can use SkyTools to keep track of objects observed in Columbus and copy the output into the Texas 45 observation log. Preliminary
Houston Astronomical Society Using SkyTools to log Texas 45 list objects You can use SkyTools to keep track of objects observed in Columbus and copy the output into the Texas 45 observation log. Preliminary
MAGNETITE OXIDATION EXAMPLE
 HSC Chemistry 7.0 1 MAGNETITE OXIDATION EXAMPLE Pelletized magnetite (Fe 3 O 4 ) ore may be oxidized to hematite (Fe 2 O 3 ) in shaft furnace. Typical magnetite content in ore is some 95%. Oxidation is
HSC Chemistry 7.0 1 MAGNETITE OXIDATION EXAMPLE Pelletized magnetite (Fe 3 O 4 ) ore may be oxidized to hematite (Fe 2 O 3 ) in shaft furnace. Typical magnetite content in ore is some 95%. Oxidation is
Import Digital Spatial Data into OneStop
 Import Digital Spatial Data into OneStop» Intended User: Pipeline Licence or Amendments Applicants Overview To apply for a pipeline or pipeline installation licence, applicants must upload the proposed
Import Digital Spatial Data into OneStop» Intended User: Pipeline Licence or Amendments Applicants Overview To apply for a pipeline or pipeline installation licence, applicants must upload the proposed
HOWTO, example workflow and data files. (Version )
 HOWTO, example workflow and data files. (Version 20 09 2017) 1 Introduction: SugarQb is a collection of software tools (Nodes) which enable the automated identification of intact glycopeptides from HCD
HOWTO, example workflow and data files. (Version 20 09 2017) 1 Introduction: SugarQb is a collection of software tools (Nodes) which enable the automated identification of intact glycopeptides from HCD
85. Geo Processing Mineral Liberation Data
 Research Center, Pori / Pertti Lamberg 14024-ORC-J 1 (23) 85. Geo Processing Mineral Liberation Data 85.1. Introduction The Mineral Liberation Analyzer, MLA, is an automated mineral analysis system that
Research Center, Pori / Pertti Lamberg 14024-ORC-J 1 (23) 85. Geo Processing Mineral Liberation Data 85.1. Introduction The Mineral Liberation Analyzer, MLA, is an automated mineral analysis system that
(THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE)
 PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
Preparing a PDB File
 Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Integrated Cheminformatics to Guide Drug Discovery
 Integrated Cheminformatics to Guide Drug Discovery Matthew Segall, Ed Champness, Peter Hunt, Tamsin Mansley CINF Drug Discovery Cheminformatics Approaches August 23 rd 2017 Optibrium, StarDrop, Auto-Modeller,
Integrated Cheminformatics to Guide Drug Discovery Matthew Segall, Ed Champness, Peter Hunt, Tamsin Mansley CINF Drug Discovery Cheminformatics Approaches August 23 rd 2017 Optibrium, StarDrop, Auto-Modeller,
Tutorial 2: Analysis of DIA data in Skyline
 Tutorial 2: Analysis of DIA data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantitation using
Tutorial 2: Analysis of DIA data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantitation using
Information Dependent Acquisition (IDA) 1
 Information Dependent Acquisition (IDA) Information Dependent Acquisition (IDA) enables on the fly acquisition of MS/MS spectra during a chromatographic run. Analyst Software IDA is optimized to generate
Information Dependent Acquisition (IDA) Information Dependent Acquisition (IDA) enables on the fly acquisition of MS/MS spectra during a chromatographic run. Analyst Software IDA is optimized to generate
1H 1D-NOE Difference Spectra and Spin-Saturation Transfer Experiments on the GN500
 UGN526 VVM-21JUN88CD VVM-31OCT91UD 1H 1D-NOE Difference Spectra and Spin-Saturation Transfer Experiments on the GN500 Double-resonance experiments are techniques which use a second irradiating field (B
UGN526 VVM-21JUN88CD VVM-31OCT91UD 1H 1D-NOE Difference Spectra and Spin-Saturation Transfer Experiments on the GN500 Double-resonance experiments are techniques which use a second irradiating field (B
Physical Chemistry II Laboratory
 Kuwata Spring 2003 Physical Chemistry II Laboratory The Rovibrational Spectra of H 35 Cl and H 37 Cl Using FTIR Write-Up Due Date: Thursday, April 17 (You may record spectra and write your reports in teams
Kuwata Spring 2003 Physical Chemistry II Laboratory The Rovibrational Spectra of H 35 Cl and H 37 Cl Using FTIR Write-Up Due Date: Thursday, April 17 (You may record spectra and write your reports in teams
Using a GIS to Calculate Area of Occupancy. Part 1: Creating a Shapefile Grid
 Using a GIS to Calculate Area of Occupancy Part 1: Creating a Shapefile Grid By Ryan Elliott California Natural Diversity Database, A NatureServe network program December 2008 This document describes the
Using a GIS to Calculate Area of Occupancy Part 1: Creating a Shapefile Grid By Ryan Elliott California Natural Diversity Database, A NatureServe network program December 2008 This document describes the
ICM-Chemist-Pro How-To Guide. Version 3.6-1h Last Updated 12/29/2009
 ICM-Chemist-Pro How-To Guide Version 3.6-1h Last Updated 12/29/2009 ICM-Chemist-Pro ICM 3D LIGAND EDITOR: SETUP 1. Read in a ligand molecule or PDB file. How to setup the ligand in the ICM 3D Ligand Editor.
ICM-Chemist-Pro How-To Guide Version 3.6-1h Last Updated 12/29/2009 ICM-Chemist-Pro ICM 3D LIGAND EDITOR: SETUP 1. Read in a ligand molecule or PDB file. How to setup the ligand in the ICM 3D Ligand Editor.
The OptiSage module. Use the OptiSage module for the assessment of Gibbs energy data. Table of contents
 The module Use the module for the assessment of Gibbs energy data. Various types of experimental data can be utilized in order to generate optimized parameters for the Gibbs energies of stoichiometric
The module Use the module for the assessment of Gibbs energy data. Various types of experimental data can be utilized in order to generate optimized parameters for the Gibbs energies of stoichiometric
Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis
 Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis Technical Overview Introduction Metabolomics studies measure the relative abundance of metabolites
Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis Technical Overview Introduction Metabolomics studies measure the relative abundance of metabolites
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build a userdefined
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build a userdefined QSAR Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build a userdefined QSAR Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed
New Approaches to the Development of GC/MS Selected Ion Monitoring Acquisition and Quantitation Methods Technique/Technology
 New Approaches to the Development of GC/MS Selected Ion Monitoring Acquisition and Quantitation Methods Technique/Technology Gas Chromatography/Mass Spectrometry Author Harry Prest 1601 California Avenue
New Approaches to the Development of GC/MS Selected Ion Monitoring Acquisition and Quantitation Methods Technique/Technology Gas Chromatography/Mass Spectrometry Author Harry Prest 1601 California Avenue
SRM assay generation and data analysis in Skyline
 in Skyline Preparation 1. Download the example data from www.srmcourse.ch/eupa.html (3 raw files, 1 csv file, 1 sptxt file). 2. The number formats of your computer have to be set to English (United States).
in Skyline Preparation 1. Download the example data from www.srmcourse.ch/eupa.html (3 raw files, 1 csv file, 1 sptxt file). 2. The number formats of your computer have to be set to English (United States).
OECD QSAR Toolbox v.4.1. Tutorial of how to use Automated workflow for ecotoxicological prediction
 OECD QSAR Toolbox v.4.1 Tutorial of how to use Automated workflow for ecotoxicological prediction Outlook Aim Automated workflow The exercise Report The OECD QSAR Toolbox for Grouping Chemicals into Categories
OECD QSAR Toolbox v.4.1 Tutorial of how to use Automated workflow for ecotoxicological prediction Outlook Aim Automated workflow The exercise Report The OECD QSAR Toolbox for Grouping Chemicals into Categories
ICM-Chemist How-To Guide. Version 3.6-1g Last Updated 12/01/2009
 ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
ISIS/Draw "Quick Start"
 ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
NMRPredict Functional Block Diagram
 NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
Pymol Practial Guide
 Pymol Practial Guide Pymol is a powerful visualizor very convenient to work with protein molecules. Its interface may seem complex at first, but you will see that with a little practice is simple and powerful
Pymol Practial Guide Pymol is a powerful visualizor very convenient to work with protein molecules. Its interface may seem complex at first, but you will see that with a little practice is simple and powerful
LED Lighting Facts: Product Submission Guide
 LED Lighting Facts: Product Submission Guide NOVEMBER 2017 1 P a g e L E D L i g h t i n g F a c t s : M a n u f a c t u r e r P r o d u c t S u b m i s s i o n G u i d e TABLE OF CONTENTS Section 1) Accessing
LED Lighting Facts: Product Submission Guide NOVEMBER 2017 1 P a g e L E D L i g h t i n g F a c t s : M a n u f a c t u r e r P r o d u c t S u b m i s s i o n G u i d e TABLE OF CONTENTS Section 1) Accessing
ON SITE SYSTEMS Chemical Safety Assistant
 ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
Titrator 3.0 Tutorial: Calcite precipitation
 Titrator 3.0 Tutorial: Calcite precipitation November 2008 Steve Cabaniss A. Introduction This brief tutorial is intended to acquaint you with some of the features of the program Titrator. It assumes that
Titrator 3.0 Tutorial: Calcite precipitation November 2008 Steve Cabaniss A. Introduction This brief tutorial is intended to acquaint you with some of the features of the program Titrator. It assumes that
An Introduction to the Cycle 4 ALMA Observing Tool How to turn that great idea into ALMA data...
 An Introduction to the Cycle 4 ALMA Observing Tool How to turn that great idea into ALMA data... Atacama Large Millimeter/submillimeter Array Expanded Very Large Array Robert C. Byrd Green Bank Telescope
An Introduction to the Cycle 4 ALMA Observing Tool How to turn that great idea into ALMA data... Atacama Large Millimeter/submillimeter Array Expanded Very Large Array Robert C. Byrd Green Bank Telescope
In order to save time, the following data files have already been preloaded to the computer (most likely under c:\odmc2012\data\)
 ODMC2012 QGIS Ex1 Schools and Public Transport In this exercise, users will learn how to a) map the location of secondary schools in and around the Southampton area; b) overlay the school location map
ODMC2012 QGIS Ex1 Schools and Public Transport In this exercise, users will learn how to a) map the location of secondary schools in and around the Southampton area; b) overlay the school location map
Athena Visual Software, Inc. 1
 Athena Visual Studio Visual Kinetics Tutorial VisualKinetics is an integrated tool within the Athena Visual Studio software environment, which allows scientists and engineers to simulate the dynamic behavior
Athena Visual Studio Visual Kinetics Tutorial VisualKinetics is an integrated tool within the Athena Visual Studio software environment, which allows scientists and engineers to simulate the dynamic behavior
Creating Questions in Word Importing Exporting Respondus 4.0. Importing Respondus 4.0 Formatting Questions
 1 Respondus Creating Questions in Word Importing Exporting Respondus 4.0 Importing Respondus 4.0 Formatting Questions Creating the Questions in Word 1. Each question must be numbered and the answers must
1 Respondus Creating Questions in Word Importing Exporting Respondus 4.0 Importing Respondus 4.0 Formatting Questions Creating the Questions in Word 1. Each question must be numbered and the answers must
Moving into the information age: From records to Google Earth
 Moving into the information age: From records to Google Earth David R. R. Smith Psychology, School of Life Sciences, University of Hull e-mail: davidsmith.butterflies@gmail.com Introduction Many of us
Moving into the information age: From records to Google Earth David R. R. Smith Psychology, School of Life Sciences, University of Hull e-mail: davidsmith.butterflies@gmail.com Introduction Many of us
Automatic Watershed Delineation using ArcSWAT/Arc GIS
 Automatic Watershed Delineation using ArcSWAT/Arc GIS By: - Endager G. and Yalelet.F 1. Watershed Delineation This tool allows the user to delineate sub watersheds based on an automatic procedure using
Automatic Watershed Delineation using ArcSWAT/Arc GIS By: - Endager G. and Yalelet.F 1. Watershed Delineation This tool allows the user to delineate sub watersheds based on an automatic procedure using
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Measuring ph with Smart Cuvettes
 1 Measuring ph with Smart Cuvettes Hardware Setup for ph Measurements ph is measured in Absorbance mode. Configure your spectroscopy components for Absorbance measurements. Sample Absorbance setups for
1 Measuring ph with Smart Cuvettes Hardware Setup for ph Measurements ph is measured in Absorbance mode. Configure your spectroscopy components for Absorbance measurements. Sample Absorbance setups for
GPS Mapping with Esri s Collector App. What We ll Cover
 GPS Mapping with Esri s Collector App Part 1: Overview What We ll Cover Part 1: Overview and requirements Part 2: Preparing the data in ArcGIS for Desktop Part 3: Build a web map in ArcGIS Online Part
GPS Mapping with Esri s Collector App Part 1: Overview What We ll Cover Part 1: Overview and requirements Part 2: Preparing the data in ArcGIS for Desktop Part 3: Build a web map in ArcGIS Online Part
OECD QSAR Toolbox v.4.1. Step-by-step example for predicting skin sensitization accounting for abiotic activation of chemicals
 OECD QSAR Toolbox v.4.1 Step-by-step example for predicting skin sensitization accounting for abiotic activation of chemicals Background Outlook Objectives The exercise Workflow 2 Background This is a
OECD QSAR Toolbox v.4.1 Step-by-step example for predicting skin sensitization accounting for abiotic activation of chemicals Background Outlook Objectives The exercise Workflow 2 Background This is a
