TreeShrink: efficient detection of outlier tree leaves
|
|
|
- Julian Osborne
- 5 years ago
- Views:
Transcription
1 TreeShrink: efficient detection of outlier tree leaves Uyen Mai Siavash Mirarab University of California at San Diego 1
2 Lesser Hedgehog Tenrec Long branches are suspect Sphagnum lesc Amborella trichopoda Nuphar advena Monilophytes Flowering plants Gnetum montanum Ephedra sinica Gymnosperms Tarsier Guinea Pig Kangaroo Rat Squirrel Galagos Mouse Lemur Tree Shrew Selaginella moellendorffii 1kp Selaginella moellendorffii genome Chaetosphaeridium globosum Klebsormidium subtile Entransia fimbriata Coleochaete irregularis Monomastix opisthostigma Netrium digitus Pyramimonas parkeae Roya obtusa Cosmarium ochthodes Nephroselmis pyriformis Coleochaete scutata Penium margaritaceum Chlorokybus atmophyticus Mougeotia sp Cylindrocystis brebissonii Mesotaenium endlicherianum Cylindrocystis cushleckae Spirogyra sp Chara vulgaris Pseudolycopodiella caroliniana Dendrolycopodium obscurum Huperzia squarrosa Nothoceros aenigmaticus Nothoceros vincentianus Mosses Liverworts 0.6 A gene tree from the1kp plant dataset (Wicket et al, PNAS, 2014) Rat Pika Rabbit Mouse Macaque Chimp Human Orangutan Gorilla Marmoset Alpaca
3 Lesser Hedgehog Tenrec Long branches are suspect Sphagnum lesc Idea: find errors in the data by building a phylogeny and detecting long branches Amborella trichopoda Nuphar advena Monilophytes Flowering plants Gnetum montanum Ephedra sinica Gymnosperms Tarsier Guinea Pig Kangaroo Rat Squirrel Galagos Mouse Lemur Tree Shrew Selaginella moellendorffii 1kp Selaginella moellendorffii genome Chaetosphaeridium globosum Klebsormidium subtile Entransia fimbriata Coleochaete irregularis Monomastix opisthostigma Netrium digitus Pyramimonas parkeae Roya obtusa Cosmarium ochthodes Nephroselmis pyriformis Coleochaete scutata Penium margaritaceum Chlorokybus atmophyticus Mougeotia sp Cylindrocystis brebissonii Mesotaenium endlicherianum Cylindrocystis cushleckae Spirogyra sp Chara vulgaris Pseudolycopodiella caroliniana Dendrolycopodium obscurum Huperzia squarrosa Nothoceros aenigmaticus Nothoceros vincentianus Mosses Liverworts 0.6 A gene tree from the1kp plant dataset (Wicket et al, PNAS, 2014) Rat Pika Rabbit Mouse Macaque Chimp Human Orangutan Gorilla Marmoset Alpaca
4 For unrooted trees? Diameter: the longest path between any two species A gene tree from the1kp plant dataset (Wicket et al, PNAS, 2014) 3 0.2
5 For unrooted trees? Diameter: the longest path between any two species A gene tree from the1kp plant dataset (Wicket et al, PNAS, 2014)
6 An optimization problem The k-shrink problem: Given: a tree with n leaves and branch lengths some 1 k n 4
7 An optimization problem The k-shrink problem: Given: a tree with n leaves and branch lengths some 1 k n Find: for every 1 i k: the set of i leaves that should be removed to reduce the tree diameter maximally 4
8 An optimization problem The k-shrink problem: Given: a tree with n leaves and branch lengths some 1 k n Find: We have a polynomial for every 1 i k: time solution the set of i leaves that should be removed to reduce the tree diameter maximally 4
9 Running Time k-shrink can be solved in O(k 2 h+n) where h = the tree height by default, we set k=o(n 0.5 ) 5
10 Running Time k-shrink can be solved in O(k 2 h+n) where h = the tree height by default, we set k=o(n 0.5 ) Fast enough: processes a tree of n=203,452 leaves with k=2255 in 28 mins 5
11 How many do we remove? How do we decide how many things to remove? We have the optimal removals for 1 i k. What i should we use? 6
12 How many do we remove? How do we decide how many things to remove? We have the optimal removals for 1 i k. What i should we use? Find an i where the corresponding reduction in the diameter is unexpectedly high needs statistical tests to find outliers 6
13 What to remove? 0.2 7
14 What to remove? the diameter after i-1 removals Let ν i = the diameter after i removals 0.2 7
15 What to remove? the diameter after i-1 removals Let ν i = the diameter after i removals 5 4 νi ratio removal 7
16 What to remove? 5 4 ratio νi removal 8
17 Signature of each species Signature of x = max log(ν i ) among all i that remove x Pyramimonas parkeae Anomodon attenuatus Nephroselmis pyriformis ratio (ν) Species Signatures: Smilax bona-nox log(3.43) Pyramimonas parkeae log(1.12) Equisetum diffusum log(3.43) Anomodon attenuatus log(1.80) Klebsormidium subtile log(3.43) Nephroselmis pyriformis log(1.12) Smilax bona-nox Klebsormidium subtile Equisetum diffusum removal Optimal removing sets: i= Anomodon attenuatus i= Equisetum diffusum, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Pyramimonas parkeae, Anomodon attenuatus
18 Signature of each species Signature of x = max log(ν i ) among all i that remove x Pyramimonas parkeae Anomodon attenuatus Nephroselmis pyriformis ratio (ν) Species Signatures: Smilax bona-nox log(3.43) Pyramimonas parkeae log(1.12) Equisetum diffusum log(3.43) Anomodon attenuatus log(1.80) Klebsormidium subtile log(3.43) Nephroselmis pyriformis log(1.12) Smilax bona-nox Klebsormidium subtile Equisetum diffusum removal Optimal removing sets: i= Anomodon attenuatus i= Equisetum diffusum, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Pyramimonas parkeae, Anomodon attenuatus
19 Signature of each species Signature of x = max log(ν i ) among all i that remove x Pyramimonas parkeae Anomodon attenuatus Nephroselmis pyriformis ratio (ν) Species Signatures: Smilax bona-nox log(3.43) Pyramimonas parkeae log(1.12) Equisetum diffusum log(3.43) Anomodon attenuatus log(1.80) Klebsormidium subtile log(3.43) Nephroselmis pyriformis log(1.12) Smilax bona-nox Klebsormidium subtile Equisetum diffusum removal Optimal removing sets: i= Anomodon attenuatus i= Equisetum diffusum, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Pyramimonas parkeae, Anomodon attenuatus
20 Signature of each species Signature of x = max log(ν i ) among all i that remove x Pyramimonas parkeae Anomodon attenuatus Nephroselmis pyriformis ratio (ν) Species Signatures: Smilax bona-nox log(3.43) Pyramimonas parkeae log(1.12) Equisetum diffusum log(3.43) Anomodon attenuatus log(1.80) Klebsormidium subtile log(3.43) Nephroselmis pyriformis log(1.12) Smilax bona-nox Klebsormidium subtile Equisetum diffusum removal Optimal removing sets: i= Anomodon attenuatus i= Equisetum diffusum, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Anomodon attenuatus i= Equisetum diffusum, Smilax bona-nox, Klebsormidium subtile, Nephroselmis pyriformis, Pyramimonas parkeae, Anomodon attenuatus
21 Three statistical tests of TreeShrink The per-gene test: requires only a single tree The all-gene test: requires a collection of gene trees The per-species test: requires a collection of gene trees 10
22 Statistical tests The per-gene test (input: a single tree) Fit a log-normal distribution to the signatures Remove taxa with outlier signatures Outlier: CDF above 1 α fore a given α (false positive tolerance) The all-gene test The per-species test 11
23 Statistical tests The per-gene test The all-gene test (input: a collection of gene trees) Combine all signature values across all genes Compute a kernel density over the empirical distribution Remove the taxa of the outlier signatures Outlier: CDF above 1 α fore a given α The per-species test 12
24 Statistical tests The per-gene test The all-gene test The per-species test (input: a collection of gene trees) Compute a kernel density function for each species over its signatures across genes Remove the taxa of the outlier signatures Outlier: CDF above 1 α for a given α 13
25 Methods The three tests of TreeShrink Alternative filtering methods RootedFiltering: root gene trees and remove taxa X standard deviations more distant to the root than average RogueNarok: rogue taxon removal based; finds unstable nodes based on bootstrap replicates RandomFiltering: randomly choose what to remove. 14
26 Measurements Effects of filtering on taxon occupancy Proportion of data retained for each species Effects of filtering on gene tree discordance Reduction in pairwise MS distance of gene trees on controlled amount of filtering 15
27 Datasets Genes Species Plants phylogenomic datasets Gene number: Species number: Insects Mammals Frogs Metazoa- Cannon Metazoa- Rouse
28 Results: outgroup removal 30 Cannon Frogs Insects Percent of the data removed for α=0.05 for All species 20 Percent removed Percent removed Mammals Plants Rouse Outgroups all gene per gene per species all gene per gene per species all gene per gene per species all gene per gene per species All All taxa Outgroups Outgroups 17 Mammalian dataset
29 Results: outgroup removal 30 Cannon Frogs Insects Percent of the data removed for α=0.05 for All species Outgroups Percent removed Mammals Plants Rouse all gene per gene per species all gene per gene per species all gene per gene per species All taxa Outgroups 17
30 Impact of filtering on discordance Random_pruning 20 TreeShrink_all_gene TreeShrink_per_gene TreeShrink_per_species Delta MS Proportion of taxa retained Plant dataset 18
31 TreeShrink versus alternative methods (discordance) (b) Plants Random_pruning Delta MS Delta MS Proportion of taxa retained Random_pruning RogueNarok Rooted_pruning TreeShrink RogueNarok Rooted_pruning TreeShrink
32 TreeShrink versus alternative methods (b) Plants (discordance) (a) Insects Random_pruning Delta MS Delta MS Delta MS Proportion of taxa retained 20 RogueNarok Random_pruning RogueNarok Rooted_pruning Rooted_pruning TreeShrink TreeShrink
33 Results: TreeShrink versus Alternative Methods (e) Mammals (f) Frogs Random_pruning RogueNarok 50 Rooted_pruning TreeShrink Delta MS 5 Delta MS Proportion of taxa retained Proportion of taxa retained 21
34 TreeShrink versus alternative methods (occupancy) Arabidopsis_thaliana Eschscholzia_californica Amborella_trichopoda SOriginalhum_bicolor Catharanthus_roseus Brachypodium_distachyon Oryza_sativa Pinus_taeda Nuphar_advena Liriodendron_tulipifera Vitis_vinifera Carica_papaya Persea_americana Diospyros_malabarica Prumnopitys_andina Allamanda_cathartica Saruma_henryi Ipomoea_purpurea Acorus_americanus Hibiscus_cannabinus Boehmeria_nivea Tanacetum_parthenium Dioscorea_villosa Podophyllum_peltatum Kochia_scoparia Sabal_bermudana Sciadopitys_verticillata Yucca_filamentosa Sarcandra_glabra Hedwigia_ciliata Rosmarinus_officinalis Zea_mays Smilax_bona nox Cedrus_libani Rhynchostegium_serrulatum Ricciocarpos_natans Cunninghamia_lanceolata Sphagnum_lescurii Ginkgo_biloba Leucodon_brachypus Taxus_baccata Gnetum_montanum Thuidium_delicatulum Marchantia_polymorpha Selaginella_moellendorffii_genome Juniperus_scopulorum Anomodon_attenuatus Larrea_tridentata Pteridium_aquilinum Ceratodon_purpureus Polytrichum_commune Pseudolycopodiella_caroliniana Cycas_micholitzii Inula_helenium Sphaerocarpos_texanus Equisetum_diffusum Cylindrocystis_cushleckae Ophioglossum_petiolatum Rosulabryum_cf_capillare Dendrolycopodium_obscurum Kadsura_heteroclita Huperzia_squarrosa Nothoceros_aenigmaticus Metzgeria_crassipilis Coleochaete_scutata Bryum_argenteum Cylindrocystis_brebissonii Houttuynia_cordata Bazzania_trilobata Coleochaete_irregularis Psilotum_nudum Selaginella_moellendorffii_1kp Mesotaenium_endlicherianum Entransia_fimbriata Aquilegia_formosa Medicago_truncatula Mougeotia_sp Angiopteris_evecta Physcomitrella_patens Nothoceros_vincentianus Populus_trichocarpa Marchantia_emarginata Alsophila_spinulosa Colchicum_autumnale Zamia_vazquezii Chlorokybus_atmophyticus Roya_obtusa Klebsormidium_subtile Ephedra_sinica Netrium_digitus Pyramimonas_parkeae Chaetosphaeridium_globosum Nephroselmis_pyriformis Cosmarium_ochthodes Spirogyra_sp Monomastix_opisthostigma Chara_vulgaris Spirotaenia_minuta Mesostigma_viride Cycas_rumphii Penium_margaritaceum Welwitschia_mirabilis Uronema_sp Occupancy (# genes) Original RogueNarok Rooted_pruning TreeShrink Original RogueNarok Rooted_pruning TreeShrink 22
35 {} ab h a d f g e c on-diameter b removed
36 remove b {} ab h a {b} ac d f g e c on-diameter b removed
37 remove b {} ab remove a h a {b} ac {a} db d f g e c on-diameter b removed
38 remove b {} ab remove a h a remove c {b} ac {a} db d {b,c} ae f g e c on-diameter b removed
39 remove b {} ab remove a h a remove c {b} ac remove a {a} db remove b d {b,c} ae {a,b} dc f g e c on-diameter b removed
40 remove b {} ab remove a h a remove c {b} ac remove a {a} db remove b remove d d {b,c} ae {a,b} dc {a,d} fb f g e c on-diameter b removed
41 remove b {} ab remove a h a remove c {b} ac remove a {a} db remove b remove d d remove e {b,c} ae {a,b} dc remove a remove c remove d remove b {a,d} fb f remove f {b,c,e} ag {b,c,a} de {a,b,d} fc {a,d,f} hb g e c on-diameter b removed
42 Solution space i = 0 remove b {} ab remove a i = 1 {b} ac {a} db k = 3 remove c remove a remove b remove d i = 2 remove e {b,c} ae {a,b} dc remove a remove c remove d remove b {a,d} fb remove f i = 3 {b,c,e} ag {b,c,a} de {a,b,d} fc {a,d,f} hb
43 The TreeShrink tool is publicly available Uyen Mai 31
44 A single HIV tree 648 HIV-1 partial pol sequences 639 subtype B 7 non-subtype B 2 unassigned TreeShrink RogueNarok TreeShrink and RogueNarok Unassigned Subtype 32
45 Results: TreeShrink versus Alternative Methods (c) Metazoa - Cannon (d) Metazoa - Rouse Random_pruning RogueNarok Delta MS Delta MS 5.0 Rooted_pruning TreeShrink Proportion of taxa retained Proportion of taxa retained 33
46 Results: The 3 Tests of TreeShrink (c) Metazoa - Cannon (d) Metazoa - Rouse 9 40 Delta MS Delta MS Proportion of taxa retained Proportion of taxa retained 34
47 Results: The 3 Tests of TreeShrink (e) Mammals (f) Frogs Delta MS Delta MS Proportion of taxa retained Proportion of taxa retained 35
48 Can be done in other ways too (e.g., O(n.k+k 2 logk)), but harder to implement
49 Can be just outgroups Sphagnum lesc Chicken Lesser Hedgehog Tenrec Platypus Wallaby Opossum Tarsier Guinea Pig Kangaroo Rat Squirrel Galagos Mouse Lemur Tree Shrew Hyrax Elephant Pika Rabbit Sloth Armadillos Rat Mouse Horse Megabat Macaque Chimp Human Orangutan Gorilla Marmoset Alpaca Dolphin Cow Microbat Cat Dog Shrew Hedgehog 0.05 b) Hard case: a gene tree in the mammalian dataset 37 Pig
Orthologous loci for phylogenomics from raw NGS data
 Orthologous loci for phylogenomics from raw NS data Rachel Schwartz The Biodesign Institute Arizona State University Rachel.Schwartz@asu.edu May 2, 205 Big data for phylogenetics Phylogenomics requires
Orthologous loci for phylogenomics from raw NS data Rachel Schwartz The Biodesign Institute Arizona State University Rachel.Schwartz@asu.edu May 2, 205 Big data for phylogenetics Phylogenomics requires
Complex evolutionary history of the vertebrate sweet/umami taste receptor genes
 Article SPECIAL ISSUE Adaptive Evolution and Conservation Ecology of Wild Animals doi: 10.1007/s11434-013-5811-5 Complex evolutionary history of the vertebrate sweet/umami taste receptor genes FENG Ping
Article SPECIAL ISSUE Adaptive Evolution and Conservation Ecology of Wild Animals doi: 10.1007/s11434-013-5811-5 Complex evolutionary history of the vertebrate sweet/umami taste receptor genes FENG Ping
Sequence motif analysis
 Sequence motif analysis Alan Moses Associate Professor and Canada Research Chair in Computational Biology Departments of Cell & Systems Biology, Computer Science, and Ecology & Evolutionary Biology Director,
Sequence motif analysis Alan Moses Associate Professor and Canada Research Chair in Computational Biology Departments of Cell & Systems Biology, Computer Science, and Ecology & Evolutionary Biology Director,
Fast coalescent-based branch support using local quartet frequencies
 Fast coalescent-based branch support using local quartet frequencies Molecular Biology and Evolution (2016) 33 (7): 1654 68 Erfan Sayyari, Siavash Mirarab University of California, San Diego (ECE) anzee
Fast coalescent-based branch support using local quartet frequencies Molecular Biology and Evolution (2016) 33 (7): 1654 68 Erfan Sayyari, Siavash Mirarab University of California, San Diego (ECE) anzee
Stat 529 (Winter 2011) A simple linear regression (SLR) case study. Mammals brain weights and body weights
 Stat 529 (Winter 2011) A simple linear regression (SLR) case study Reading: Sections 8.1 8.4, 8.6, 8.7 Mammals brain weights and body weights Questions of interest Scatterplots of the data Log transforming
Stat 529 (Winter 2011) A simple linear regression (SLR) case study Reading: Sections 8.1 8.4, 8.6, 8.7 Mammals brain weights and body weights Questions of interest Scatterplots of the data Log transforming
Reconstruction of species trees from gene trees using ASTRAL. Siavash Mirarab University of California, San Diego (ECE)
 Reconstruction of species trees from gene trees using ASTRAL Siavash Mirarab University of California, San Diego (ECE) Phylogenomics Orangutan Chimpanzee gene 1 gene 2 gene 999 gene 1000 Gorilla Human
Reconstruction of species trees from gene trees using ASTRAL Siavash Mirarab University of California, San Diego (ECE) Phylogenomics Orangutan Chimpanzee gene 1 gene 2 gene 999 gene 1000 Gorilla Human
Supplementary text and figures: Comparative assessment of methods for aligning multiple genome sequences
 Supplementary text and figures: Comparative assessment of methods for aligning multiple genome sequences Xiaoyu Chen Martin Tompa Department of Computer Science and Engineering Department of Genome Sciences
Supplementary text and figures: Comparative assessment of methods for aligning multiple genome sequences Xiaoyu Chen Martin Tompa Department of Computer Science and Engineering Department of Genome Sciences
Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals
 Project Report Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals By Shubhra Gupta CBS 598 Phylogenetic Biology and Analysis Life Science
Project Report Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals By Shubhra Gupta CBS 598 Phylogenetic Biology and Analysis Life Science
ASTRAL: Fast coalescent-based computation of the species tree topology, branch lengths, and local branch support
 ASTRAL: Fast coalescent-based computation of the species tree topology, branch lengths, and local branch support Siavash Mirarab University of California, San Diego Joint work with Tandy Warnow Erfan Sayyari
ASTRAL: Fast coalescent-based computation of the species tree topology, branch lengths, and local branch support Siavash Mirarab University of California, San Diego Joint work with Tandy Warnow Erfan Sayyari
Molecules consolidate the placental mammal tree.
 Molecules consolidate the placental mammal tree. The morphological concensus mammal tree Two decades of molecular phylogeny Rooting the placental mammal tree Parallel adaptative radiations among placental
Molecules consolidate the placental mammal tree. The morphological concensus mammal tree Two decades of molecular phylogeny Rooting the placental mammal tree Parallel adaptative radiations among placental
Phylogenetic Tree Reconstruction
 I519 Introduction to Bioinformatics, 2011 Phylogenetic Tree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Evolution theory Speciation Evolution of new organisms is driven
I519 Introduction to Bioinformatics, 2011 Phylogenetic Tree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Evolution theory Speciation Evolution of new organisms is driven
Mechanisms of Evolution Darwinian Evolution
 Mechanisms of Evolution Darwinian Evolution Descent with modification by means of natural selection All life has descended from a common ancestor The mechanism of modification is natural selection Concept
Mechanisms of Evolution Darwinian Evolution Descent with modification by means of natural selection All life has descended from a common ancestor The mechanism of modification is natural selection Concept
Reconstructing the History of Large-scale Genomic Changes. Jian Ma
 Reconstructing the History of Large-scale Genomic Changes Jian Ma The Human Genome: the blueprint of our body Initial sequencing and analysis of the human genome International Human Genome Sequencing Consortium*
Reconstructing the History of Large-scale Genomic Changes Jian Ma The Human Genome: the blueprint of our body Initial sequencing and analysis of the human genome International Human Genome Sequencing Consortium*
Realism and Instrumentalism. in models of. molecular evolution
 Galileo Realism and Instrumentalism in models of molecular evolution David Penny Montpellier, June 08 Overview sites free to vary summing sources of error rates of molecular evolution estimates of time
Galileo Realism and Instrumentalism in models of molecular evolution David Penny Montpellier, June 08 Overview sites free to vary summing sources of error rates of molecular evolution estimates of time
Proximal point algorithm in Hadamard spaces
 Proximal point algorithm in Hadamard spaces Miroslav Bacak Télécom ParisTech Optimisation Géométrique sur les Variétés - Paris, 21 novembre 2014 Contents of the talk 1 Basic facts on Hadamard spaces 2
Proximal point algorithm in Hadamard spaces Miroslav Bacak Télécom ParisTech Optimisation Géométrique sur les Variétés - Paris, 21 novembre 2014 Contents of the talk 1 Basic facts on Hadamard spaces 2
1 ATGGGTCTC 2 ATGAGTCTC
 We need an optimality criterion to choose a best estimate (tree) Other optimality criteria used to choose a best estimate (tree) Parsimony: begins with the assumption that the simplest hypothesis that
We need an optimality criterion to choose a best estimate (tree) Other optimality criteria used to choose a best estimate (tree) Parsimony: begins with the assumption that the simplest hypothesis that
Changes in the composition of the RNA virome mark evolutionary transitions in green plants
 Mushegian et al. BMC Biology (2016) 14:68 DOI 10.1186/s12915-016-0288-8 RESEARCH ARTICLE Open Access Changes in the composition of the RNA virome mark evolutionary transitions in green plants Arcady Mushegian
Mushegian et al. BMC Biology (2016) 14:68 DOI 10.1186/s12915-016-0288-8 RESEARCH ARTICLE Open Access Changes in the composition of the RNA virome mark evolutionary transitions in green plants Arcady Mushegian
Workshop III: Evolutionary Genomics
 Identifying Species Trees from Gene Trees Elizabeth S. Allman University of Alaska IPAM Los Angeles, CA November 17, 2011 Workshop III: Evolutionary Genomics Collaborators The work in today s talk is joint
Identifying Species Trees from Gene Trees Elizabeth S. Allman University of Alaska IPAM Los Angeles, CA November 17, 2011 Workshop III: Evolutionary Genomics Collaborators The work in today s talk is joint
Supplementary information
 Supplementary information Superoxide dismutase 1 is positively selected in great apes to minimize protein misfolding Pouria Dasmeh 1, and Kasper P. Kepp* 2 1 Harvard University, Department of Chemistry
Supplementary information Superoxide dismutase 1 is positively selected in great apes to minimize protein misfolding Pouria Dasmeh 1, and Kasper P. Kepp* 2 1 Harvard University, Department of Chemistry
part 4: phenomenological load and biological inference. phenomenological load review types of models. Gαβ = 8π Tαβ. Newton.
 2017-07-29 part 4: and biological inference review types of models phenomenological Newton F= Gm1m2 r2 mechanistic Einstein Gαβ = 8π Tαβ 1 molecular evolution is process and pattern process pattern MutSel
2017-07-29 part 4: and biological inference review types of models phenomenological Newton F= Gm1m2 r2 mechanistic Einstein Gαβ = 8π Tαβ 1 molecular evolution is process and pattern process pattern MutSel
Phylogeny: traditional and Bayesian approaches
 Phylogeny: traditional and Bayesian approaches 5-Feb-2014 DEKM book Notes from Dr. B. John Holder and Lewis, Nature Reviews Genetics 4, 275-284, 2003 1 Phylogeny A graph depicting the ancestor-descendent
Phylogeny: traditional and Bayesian approaches 5-Feb-2014 DEKM book Notes from Dr. B. John Holder and Lewis, Nature Reviews Genetics 4, 275-284, 2003 1 Phylogeny A graph depicting the ancestor-descendent
9/30/11. Evolution theory. Phylogenetic Tree Reconstruction. Phylogenetic trees (binary trees) Phylogeny (phylogenetic tree)
 I9 Introduction to Bioinformatics, 0 Phylogenetic ree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & omputing, IUB Evolution theory Speciation Evolution of new organisms is driven by
I9 Introduction to Bioinformatics, 0 Phylogenetic ree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & omputing, IUB Evolution theory Speciation Evolution of new organisms is driven by
Phylogenetic inference
 Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Algorithms in Bioinformatics
 Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Distance Methods Character Methods
Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Distance Methods Character Methods
Phylogenetics in the Age of Genomics: Prospects and Challenges
 Phylogenetics in the Age of Genomics: Prospects and Challenges Antonis Rokas Department of Biological Sciences, Vanderbilt University http://as.vanderbilt.edu/rokaslab http://pubmed2wordle.appspot.com/
Phylogenetics in the Age of Genomics: Prospects and Challenges Antonis Rokas Department of Biological Sciences, Vanderbilt University http://as.vanderbilt.edu/rokaslab http://pubmed2wordle.appspot.com/
Chapter 26 Phylogeny and the Tree of Life
 Chapter 26 Phylogeny and the Tree of Life Biologists estimate that there are about 5 to 100 million species of organisms living on Earth today. Evidence from morphological, biochemical, and gene sequence
Chapter 26 Phylogeny and the Tree of Life Biologists estimate that there are about 5 to 100 million species of organisms living on Earth today. Evidence from morphological, biochemical, and gene sequence
Plan: Evolutionary trees, characters. Perfect phylogeny Methods: NJ, parsimony, max likelihood, Quartet method
 Phylogeny 1 Plan: Phylogeny is an important subject. We have 2.5 hours. So I will teach all the concepts via one example of a chain letter evolution. The concepts we will discuss include: Evolutionary
Phylogeny 1 Plan: Phylogeny is an important subject. We have 2.5 hours. So I will teach all the concepts via one example of a chain letter evolution. The concepts we will discuss include: Evolutionary
Cladistics and Bioinformatics Questions 2013
 AP Biology Name Cladistics and Bioinformatics Questions 2013 1. The following table shows the percentage similarity in sequences of nucleotides from a homologous gene derived from five different species
AP Biology Name Cladistics and Bioinformatics Questions 2013 1. The following table shows the percentage similarity in sequences of nucleotides from a homologous gene derived from five different species
Constructing Evolutionary Trees
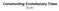 Constructing Evolutionary Trees 0-0 HIV Evolutionary Tree SIVs (monkeys)! HIV (human)! human infection! human HIV/M human HIV/M chimpanzee SIV chimpanzee SIV human HIV/N human HIV/N chimpanzee SIV chimpanzee
Constructing Evolutionary Trees 0-0 HIV Evolutionary Tree SIVs (monkeys)! HIV (human)! human infection! human HIV/M human HIV/M chimpanzee SIV chimpanzee SIV human HIV/N human HIV/N chimpanzee SIV chimpanzee
Emily Blanton Phylogeny Lab Report May 2009
 Introduction It is suggested through scientific research that all living organisms are connected- that we all share a common ancestor and that, through time, we have all evolved from the same starting
Introduction It is suggested through scientific research that all living organisms are connected- that we all share a common ancestor and that, through time, we have all evolved from the same starting
Bayesian Phylogenetics
 Bayesian Phylogenetics 30 July 2014 Workshop on Molecular Evolution Woods Hole Paul O. Lewis Department of Ecology & Evolutionary Biology Paul O. Lewis (2014 Woods Hole Molecular Evolution Workshop) 1
Bayesian Phylogenetics 30 July 2014 Workshop on Molecular Evolution Woods Hole Paul O. Lewis Department of Ecology & Evolutionary Biology Paul O. Lewis (2014 Woods Hole Molecular Evolution Workshop) 1
Week 8: Testing trees, Bootstraps, jackknifes, gene frequencies
 Week 8: Testing trees, ootstraps, jackknifes, gene frequencies Genome 570 ebruary, 2016 Week 8: Testing trees, ootstraps, jackknifes, gene frequencies p.1/69 density e log (density) Normal distribution:
Week 8: Testing trees, ootstraps, jackknifes, gene frequencies Genome 570 ebruary, 2016 Week 8: Testing trees, ootstraps, jackknifes, gene frequencies p.1/69 density e log (density) Normal distribution:
Estimation of species divergence dates with a sloppy molecular clock
 Estimation of species divergence dates with a sloppy molecular clock Ziheng Yang Department of Biology University College London Date estimation with a clock is easy. t 2 = 13my t 3 t 1 t 4 t 5 Node Distance
Estimation of species divergence dates with a sloppy molecular clock Ziheng Yang Department of Biology University College London Date estimation with a clock is easy. t 2 = 13my t 3 t 1 t 4 t 5 Node Distance
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST
 Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
Phylogeny. Properties of Trees. Properties of Trees. Trees represent the order of branching only. Phylogeny: Taxon: a unit of classification
 Multiple sequence alignment global local Evolutionary tree reconstruction Pairwise sequence alignment (global and local) Substitution matrices Gene Finding Protein structure prediction N structure prediction
Multiple sequence alignment global local Evolutionary tree reconstruction Pairwise sequence alignment (global and local) Substitution matrices Gene Finding Protein structure prediction N structure prediction
Anatomy of a species tree
 Anatomy of a species tree T 1 Size of current and ancestral Populations (N) N Confidence in branches of species tree t/2n = 1 coalescent unit T 2 Branch lengths and divergence times of species & populations
Anatomy of a species tree T 1 Size of current and ancestral Populations (N) N Confidence in branches of species tree t/2n = 1 coalescent unit T 2 Branch lengths and divergence times of species & populations
Phylogeny and Evolution. Gina Cannarozzi ETH Zurich Institute of Computational Science
 Phylogeny and Evolution Gina Cannarozzi ETH Zurich Institute of Computational Science History Aristotle (384-322 BC) classified animals. He found that dolphins do not belong to the fish but to the mammals.
Phylogeny and Evolution Gina Cannarozzi ETH Zurich Institute of Computational Science History Aristotle (384-322 BC) classified animals. He found that dolphins do not belong to the fish but to the mammals.
C3020 Molecular Evolution. Exercises #3: Phylogenetics
 C3020 Molecular Evolution Exercises #3: Phylogenetics Consider the following sequences for five taxa 1-5 and the known outgroup O, which has the ancestral states (note that sequence 3 has changed from
C3020 Molecular Evolution Exercises #3: Phylogenetics Consider the following sequences for five taxa 1-5 and the known outgroup O, which has the ancestral states (note that sequence 3 has changed from
An Introduction to Bayesian Phylogenetics
 An Introduction to Bayesian Phylogenetics Workshop on Molecular Evolution Woods Hole, Mass. 22 July 2015 Paul O. Lewis Department of Ecology & Evolutionary Biology Paul O. Lewis (2015 Woods Hole Molecular
An Introduction to Bayesian Phylogenetics Workshop on Molecular Evolution Woods Hole, Mass. 22 July 2015 Paul O. Lewis Department of Ecology & Evolutionary Biology Paul O. Lewis (2015 Woods Hole Molecular
Evidence of Evolution by Natural Selection. Dodo bird
 Evidence of Evolution by Natural Selection Dodo bird 2007-2008 Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development
Evidence of Evolution by Natural Selection Dodo bird 2007-2008 Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development
Phylogeny. November 7, 2017
 Phylogeny November 7, 2017 Phylogenetics Phylon = tribe/race, genetikos = relative to birth Phylogenetics: study of evolutionary relationships among organisms, sequences, or anything in between Related
Phylogeny November 7, 2017 Phylogenetics Phylon = tribe/race, genetikos = relative to birth Phylogenetics: study of evolutionary relationships among organisms, sequences, or anything in between Related
Phylogenetic Trees. Phylogenetic Trees Five. Phylogeny: Inference Tool. Phylogeny Terminology. Picture of Last Quagga. Importance of Phylogeny 5.
 Five Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu v Distance Methods v Character Methods v Molecular Clock v UPGMA v Maximum Parsimony
Five Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu v Distance Methods v Character Methods v Molecular Clock v UPGMA v Maximum Parsimony
Supplemental Information: Origin of land plants revisited in the light of sequence
 Supplemental Information: Origin of land plants revisited in the light of sequence contamination and missing data Simon Laurin Lemay, Henner Brinkmann and Hervé Philippe Figure S1. Comparison of the trees
Supplemental Information: Origin of land plants revisited in the light of sequence contamination and missing data Simon Laurin Lemay, Henner Brinkmann and Hervé Philippe Figure S1. Comparison of the trees
Lecture 16: Again on Regression
 Lecture 16: Again on Regression S. Massa, Department of Statistics, University of Oxford 10 February 2016 The Normality Assumption Body weights (Kg) and brain weights (Kg) of 62 mammals. Species Body weight
Lecture 16: Again on Regression S. Massa, Department of Statistics, University of Oxford 10 February 2016 The Normality Assumption Body weights (Kg) and brain weights (Kg) of 62 mammals. Species Body weight
How to read and make phylogenetic trees Zuzana Starostová
 How to read and make phylogenetic trees Zuzana Starostová How to make phylogenetic trees? Workflow: obtain DNA sequence quality check sequence alignment calculating genetic distances phylogeny estimation
How to read and make phylogenetic trees Zuzana Starostová How to make phylogenetic trees? Workflow: obtain DNA sequence quality check sequence alignment calculating genetic distances phylogeny estimation
Copyright notice. Molecular Phylogeny and Evolution. Goals of the lecture. Introduction. Introduction. December 15, 2008
 opyright notice Molecular Phylogeny and volution ecember 5, 008 ioinformatics J. Pevsner pevsner@kennedykrieger.org Many of the images in this powerpoint presentation are from ioinformatics and Functional
opyright notice Molecular Phylogeny and volution ecember 5, 008 ioinformatics J. Pevsner pevsner@kennedykrieger.org Many of the images in this powerpoint presentation are from ioinformatics and Functional
Many of the slides that I ll use have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks!
 Many of the slides that I ll use have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks! Paul has many great tools for teaching phylogenetics at his web site: http://hydrodictyon.eeb.uconn.edu/people/plewis
Many of the slides that I ll use have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks! Paul has many great tools for teaching phylogenetics at his web site: http://hydrodictyon.eeb.uconn.edu/people/plewis
DO NOT WRITE ON THIS. Evidence from Evolution Activity. The Fossilization Process. Types of Fossils
 Evidence from Evolution Activity Part 1 - Fossils Use the diagrams on the next page to answer the following questions IN YOUR NOTEBOOK. 1. Describe how fossils form. 2. Describe the different types of
Evidence from Evolution Activity Part 1 - Fossils Use the diagrams on the next page to answer the following questions IN YOUR NOTEBOOK. 1. Describe how fossils form. 2. Describe the different types of
Lecture 6 Phylogenetic Inference
 Lecture 6 Phylogenetic Inference From Darwin s notebook in 1837 Charles Darwin Willi Hennig From The Origin in 1859 Cladistics Phylogenetic inference Willi Hennig, Cladistics 1. Clade, Monophyletic group,
Lecture 6 Phylogenetic Inference From Darwin s notebook in 1837 Charles Darwin Willi Hennig From The Origin in 1859 Cladistics Phylogenetic inference Willi Hennig, Cladistics 1. Clade, Monophyletic group,
Evidence of Evolution by Natural Selection (Ch. 16.4) Dodo bird
 Evidence of Evolution by Natural Selection (Ch. 16.4) Dodo bird Evidence supporting evolution Fossil record Anatomical record Molecular record Artificial selection Fossil record Layers of sedimentary rock
Evidence of Evolution by Natural Selection (Ch. 16.4) Dodo bird Evidence supporting evolution Fossil record Anatomical record Molecular record Artificial selection Fossil record Layers of sedimentary rock
Molecular phylogeny How to infer phylogenetic trees using molecular sequences
 Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 2009 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 2009 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
X X (2) X Pr(X = x θ) (3)
 Notes for 848 lecture 6: A ML basis for compatibility and parsimony Notation θ Θ (1) Θ is the space of all possible trees (and model parameters) θ is a point in the parameter space = a particular tree
Notes for 848 lecture 6: A ML basis for compatibility and parsimony Notation θ Θ (1) Θ is the space of all possible trees (and model parameters) θ is a point in the parameter space = a particular tree
Phylogenetic Analysis. Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center
 Phylogenetic Analysis Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center Outline Basic Concepts Tree Construction Methods Distance-based methods
Phylogenetic Analysis Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center Outline Basic Concepts Tree Construction Methods Distance-based methods
CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES
 INTRODUCTION CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES This worksheet complements the Click and Learn developed in conjunction with the 2011 Holiday Lectures on Science, Bones, Stones, and Genes:
INTRODUCTION CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES This worksheet complements the Click and Learn developed in conjunction with the 2011 Holiday Lectures on Science, Bones, Stones, and Genes:
Lab 22: Classification of Species
 Name: Period: Lab 22: Classification of Species Instructions: 1. Pick up a bag of plastic toy animals (note: these are a choking hazard ) 2. At the top of this page, note which bag number you have 3. Set
Name: Period: Lab 22: Classification of Species Instructions: 1. Pick up a bag of plastic toy animals (note: these are a choking hazard ) 2. At the top of this page, note which bag number you have 3. Set
Molecular phylogeny How to infer phylogenetic trees using molecular sequences
 Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 200 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 200 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Multiple Sequence Alignment. Sequences
 Multiple Sequence Alignment Sequences > YOR020c mstllksaksivplmdrvlvqrikaqaktasglylpe knveklnqaevvavgpgftdangnkvvpqvkvgdqvl ipqfggstiklgnddevilfrdaeilakiakd > crassa mattvrsvksliplldrvlvqrvkaeaktasgiflpe
Multiple Sequence Alignment Sequences > YOR020c mstllksaksivplmdrvlvqrikaqaktasglylpe knveklnqaevvavgpgftdangnkvvpqvkvgdqvl ipqfggstiklgnddevilfrdaeilakiakd > crassa mattvrsvksliplldrvlvqrvkaeaktasgiflpe
AP Biology. Evolution is "so overwhelmingly established that it has become irrational to call it a theory." Evidence of Evolution by Natural Selection
 Evidence of Evolution by Natural Selection Evolution is "so overwhelmingly established that it has become irrational to call it a theory." -- Ernst Mayr What Evolution Is 2001 Professor Emeritus, Evolutionary
Evidence of Evolution by Natural Selection Evolution is "so overwhelmingly established that it has become irrational to call it a theory." -- Ernst Mayr What Evolution Is 2001 Professor Emeritus, Evolutionary
Evolution by duplication
 6.095/6.895 - Computational Biology: Genomes, Networks, Evolution Lecture 18 Nov 10, 2005 Evolution by duplication Somewhere, something went wrong Challenges in Computational Biology 4 Genome Assembly
6.095/6.895 - Computational Biology: Genomes, Networks, Evolution Lecture 18 Nov 10, 2005 Evolution by duplication Somewhere, something went wrong Challenges in Computational Biology 4 Genome Assembly
Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation
 Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any later version published
Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any later version published
Tree thinking pretest
 Page 1 Tree thinking pretest This quiz is in three sections. Questions 1-10 assess your basic understanding of phylogenetic trees. Questions 11-15 assess whether you are equipped to accurately extract
Page 1 Tree thinking pretest This quiz is in three sections. Questions 1-10 assess your basic understanding of phylogenetic trees. Questions 11-15 assess whether you are equipped to accurately extract
Introduction to Biological Anthropology: Notes 11 What is a primate, and why do we study them? Copyright Bruce Owen 2011
 Why study non-human primates? Introduction to Biological Anthropology: Notes 11 What is a primate, and why do we study them? Copyright Bruce Owen 2011 They give us clues about human nature and the nature
Why study non-human primates? Introduction to Biological Anthropology: Notes 11 What is a primate, and why do we study them? Copyright Bruce Owen 2011 They give us clues about human nature and the nature
Bootstraps and testing trees. Alog-likelihoodcurveanditsconfidenceinterval
 ootstraps and testing trees Joe elsenstein epts. of Genome Sciences and of iology, University of Washington ootstraps and testing trees p.1/20 log-likelihoodcurveanditsconfidenceinterval 2620 2625 ln L
ootstraps and testing trees Joe elsenstein epts. of Genome Sciences and of iology, University of Washington ootstraps and testing trees p.1/20 log-likelihoodcurveanditsconfidenceinterval 2620 2625 ln L
User s Manual for. Continuous. (copyright M. Pagel) Mark Pagel School of Animal and Microbial Sciences University of Reading Reading RG6 6AJ UK
 User s Manual for Continuous (copyright M. Pagel) Mark Pagel School of Animal and Microbial Sciences University of Reading Reading RG6 6AJ UK email: m.pagel@rdg.ac.uk (www.ams.reading.ac.uk/zoology/pagel/)
User s Manual for Continuous (copyright M. Pagel) Mark Pagel School of Animal and Microbial Sciences University of Reading Reading RG6 6AJ UK email: m.pagel@rdg.ac.uk (www.ams.reading.ac.uk/zoology/pagel/)
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics
 POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
Darwin's Theory. Use Target Reading Skills. Darwin's Observations. Changes Over Time Guided Reading and Study
 Darwin's Theory This section discusses Charles Darwin and his theories ofevolution, which are based on what he saw during his trip around the world. Use Target Reading Skills In the graphic organizer,
Darwin's Theory This section discusses Charles Darwin and his theories ofevolution, which are based on what he saw during his trip around the world. Use Target Reading Skills In the graphic organizer,
Elements of Bioinformatics 14F01 TP5 -Phylogenetic analysis
 Elements of Bioinformatics 14F01 TP5 -Phylogenetic analysis 10 December 2012 - Corrections - Exercise 1 Non-vertebrate chordates generally possess 2 homologs, vertebrates 3 or more gene copies; a Drosophila
Elements of Bioinformatics 14F01 TP5 -Phylogenetic analysis 10 December 2012 - Corrections - Exercise 1 Non-vertebrate chordates generally possess 2 homologs, vertebrates 3 or more gene copies; a Drosophila
Microbial Diversity and Assessment (II) Spring, 2007 Guangyi Wang, Ph.D. POST103B
 Microbial Diversity and Assessment (II) Spring, 007 Guangyi Wang, Ph.D. POST03B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm General introduction and overview Taxonomy [Greek
Microbial Diversity and Assessment (II) Spring, 007 Guangyi Wang, Ph.D. POST03B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm General introduction and overview Taxonomy [Greek
Lecture 11 Friday, October 21, 2011
 Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Evolution and divergence of the mammalian SAMD9/SAMD9L gene family
 Lemos de Matos et al. BMC Evolutionary Biology 2013, 13:121 RESEARCH ARTICLE Evolution and divergence of the mammalian SAMD9/SAMD9L gene family Ana Lemos de Matos 1,2,3, Jia Liu 3, Grant McFadden 3 and
Lemos de Matos et al. BMC Evolutionary Biology 2013, 13:121 RESEARCH ARTICLE Evolution and divergence of the mammalian SAMD9/SAMD9L gene family Ana Lemos de Matos 1,2,3, Jia Liu 3, Grant McFadden 3 and
Evidence for Evolution by Natural Selection. Raven Chapters 1 & 22
 Evidence for Evolution by Natural Selection Raven Chapters 1 & 22 2006-2007 Science happens within a culture What was the doctrine of the time? TINTORETTO The Creation of the Animals 1550 Then along comes
Evidence for Evolution by Natural Selection Raven Chapters 1 & 22 2006-2007 Science happens within a culture What was the doctrine of the time? TINTORETTO The Creation of the Animals 1550 Then along comes
Mul$ple Sequence Alignment Methods. Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu
 Mul$ple Sequence Alignment Methods Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu Species Tree Orangutan Gorilla Chimpanzee Human From the Tree of the Life
Mul$ple Sequence Alignment Methods Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu Species Tree Orangutan Gorilla Chimpanzee Human From the Tree of the Life
Lecture 27. Phylogeny methods, part 7 (Bootstraps, etc.) p.1/30
 Lecture 27. Phylogeny methods, part 7 (Bootstraps, etc.) Joe Felsenstein Department of Genome Sciences and Department of Biology Lecture 27. Phylogeny methods, part 7 (Bootstraps, etc.) p.1/30 A non-phylogeny
Lecture 27. Phylogeny methods, part 7 (Bootstraps, etc.) Joe Felsenstein Department of Genome Sciences and Department of Biology Lecture 27. Phylogeny methods, part 7 (Bootstraps, etc.) p.1/30 A non-phylogeny
Primate phylogeny: molecular evidence for a pongid clade excluding humans and a prosimian clade containing tarsiers
 Huang, 1 Primate phylogeny: molecular evidence for a pongid clade excluding humans and a prosimian clade containing tarsiers Shi Huang State Key Laboratory of Medical Genetics Xiangya Medical School Central
Huang, 1 Primate phylogeny: molecular evidence for a pongid clade excluding humans and a prosimian clade containing tarsiers Shi Huang State Key Laboratory of Medical Genetics Xiangya Medical School Central
Being Bayesian About Network Structure:
 Being Bayesian About Network Structure: A Bayesian Approach to Structure Discovery in Bayesian Networks Nir Friedman and Daphne Koller Machine Learning, 2003 Presented by XianXing Zhang Duke University
Being Bayesian About Network Structure: A Bayesian Approach to Structure Discovery in Bayesian Networks Nir Friedman and Daphne Koller Machine Learning, 2003 Presented by XianXing Zhang Duke University
Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction. Lesser Tenrec (Echinops telfairi)
 Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction Lesser Tenrec (Echinops telfairi) Goals: 1. Use phylogenetic experimental design theory to select optimal taxa to
Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction Lesser Tenrec (Echinops telfairi) Goals: 1. Use phylogenetic experimental design theory to select optimal taxa to
Determining the Null Model for Detecting Adaptive Convergence from Genomic Data: A Case Study using Echolocating Mammals
 Determining the Null Model for Detecting Adaptive Convergence from Genomic Data: A Case Study using Echolocating Mammals Gregg W.C. Thomas 1 and Matthew W. Hahn*,1,2 1 School of Informatics and Computing,
Determining the Null Model for Detecting Adaptive Convergence from Genomic Data: A Case Study using Echolocating Mammals Gregg W.C. Thomas 1 and Matthew W. Hahn*,1,2 1 School of Informatics and Computing,
Anthro 101: Human Biological Evolution. Lecture 7: Taxonomy/Primate Adaptations. Prof. Kenneth Feldmeier
 Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the PLAN Listen to this lecture and read about Taxonomy in the text I will ask you a question(s)
Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the PLAN Listen to this lecture and read about Taxonomy in the text I will ask you a question(s)
BINF6201/8201. Molecular phylogenetic methods
 BINF60/80 Molecular phylogenetic methods 0-7-06 Phylogenetics Ø According to the evolutionary theory, all life forms on this planet are related to one another by descent. Ø Traditionally, phylogenetics
BINF60/80 Molecular phylogenetic methods 0-7-06 Phylogenetics Ø According to the evolutionary theory, all life forms on this planet are related to one another by descent. Ø Traditionally, phylogenetics
Station A: #3. If two organisms belong to the same order, they must also belong to the same
 Station A: #1. Write your mnemonic for remembering the order of the taxa (from the broadest, most generic taxon to the most specific). Out to the side of each, write the name of each taxon the mnemonic
Station A: #1. Write your mnemonic for remembering the order of the taxa (from the broadest, most generic taxon to the most specific). Out to the side of each, write the name of each taxon the mnemonic
Week 7: Bayesian inference, Testing trees, Bootstraps
 Week 7: ayesian inference, Testing trees, ootstraps Genome 570 May, 2008 Week 7: ayesian inference, Testing trees, ootstraps p.1/54 ayes Theorem onditional probability of hypothesis given data is: Prob
Week 7: ayesian inference, Testing trees, ootstraps Genome 570 May, 2008 Week 7: ayesian inference, Testing trees, ootstraps p.1/54 ayes Theorem onditional probability of hypothesis given data is: Prob
Anthro 101: Human Biological Evolution. Lecture 7: Taxonomy/Primate Adaptations. Prof. Kenneth Feldmeier
 Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the deal, read though the lecture and hopefully the audio works on youtube Classifying species
Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the deal, read though the lecture and hopefully the audio works on youtube Classifying species
Bio94 Discussion Activity week 3: Chapter 27 Phylogenies and the History of Life
 Bio94 Discussion Activity week 3: Chapter 27 Phylogenies and the History of Life 1. Constructing a phylogenetic tree using a cladistic approach Construct a phylogenetic tree using the following table:
Bio94 Discussion Activity week 3: Chapter 27 Phylogenies and the History of Life 1. Constructing a phylogenetic tree using a cladistic approach Construct a phylogenetic tree using the following table:
ELE4120 Bioinformatics Tutorial 8
 ELE4120 ioinformatics Tutorial 8 ontent lassifying Organisms Systematics and Speciation Taxonomy and phylogenetics Phenetics versus cladistics Phylogenetic trees iological classification Goal: To develop
ELE4120 ioinformatics Tutorial 8 ontent lassifying Organisms Systematics and Speciation Taxonomy and phylogenetics Phenetics versus cladistics Phylogenetic trees iological classification Goal: To develop
CHAPTER 26 PHYLOGENY AND THE TREE OF LIFE Connecting Classification to Phylogeny
 CHAPTER 26 PHYLOGENY AND THE TREE OF LIFE Connecting Classification to Phylogeny To trace phylogeny or the evolutionary history of life, biologists use evidence from paleontology, molecular data, comparative
CHAPTER 26 PHYLOGENY AND THE TREE OF LIFE Connecting Classification to Phylogeny To trace phylogeny or the evolutionary history of life, biologists use evidence from paleontology, molecular data, comparative
Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis
 Method Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis John E. McCormack, 1,8 Brant C. Faircloth, 2 Nicholas G. Crawford,
Method Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis John E. McCormack, 1,8 Brant C. Faircloth, 2 Nicholas G. Crawford,
Evolution. Darwin s Voyage
 Evolution Darwin s Voyage Charles Darwin Explorer on an observation trip to the Galapagos Islands. He set sail on the HMS Beagle in 1858 from England on a 5 year trip. He was a naturalist (a person who
Evolution Darwin s Voyage Charles Darwin Explorer on an observation trip to the Galapagos Islands. He set sail on the HMS Beagle in 1858 from England on a 5 year trip. He was a naturalist (a person who
Multidimensional Vector Space Representation for Convergent Evolution and Molecular Phylogeny
 MBE Advance Access published November 17, 2004 Multidimensional Vector Space Representation for Convergent Evolution and Molecular Phylogeny Yasuhiro Kitazoe*, Hirohisa Kishino, Takahisa Okabayashi*, Teruaki
MBE Advance Access published November 17, 2004 Multidimensional Vector Space Representation for Convergent Evolution and Molecular Phylogeny Yasuhiro Kitazoe*, Hirohisa Kishino, Takahisa Okabayashi*, Teruaki
Introduction to Biological Anthropology: Notes 9 What is a primate, and why do we study them? Copyright Bruce Owen 2008
 Why study non-human primates? Introduction to Biological Anthropology: Notes 9 What is a primate, and why do we study them? Copyright Bruce Owen 2008 They give us clues about human nature and the nature
Why study non-human primates? Introduction to Biological Anthropology: Notes 9 What is a primate, and why do we study them? Copyright Bruce Owen 2008 They give us clues about human nature and the nature
Minimum Regularized Covariance Determinant Estimator
 Minimum Regularized Covariance Determinant Estimator Honey, we shrunk the data and the covariance matrix Kris Boudt (joint with: P. Rousseeuw, S. Vanduffel and T. Verdonck) Vrije Universiteit Brussel/Amsterdam
Minimum Regularized Covariance Determinant Estimator Honey, we shrunk the data and the covariance matrix Kris Boudt (joint with: P. Rousseeuw, S. Vanduffel and T. Verdonck) Vrije Universiteit Brussel/Amsterdam
Biology Keystone (PA Core) Quiz Theory of Evolution - (BIO.B ) Theory Of Evolution, (BIO.B ) Scientific Terms
 Biology Keystone (PA Core) Quiz Theory of Evolution - (BIO.B.3.2.1 ) Theory Of Evolution, (BIO.B.3.3.1 ) Scientific Terms Student Name: Teacher Name: Jared George Date: Score: 1) Evidence for evolution
Biology Keystone (PA Core) Quiz Theory of Evolution - (BIO.B.3.2.1 ) Theory Of Evolution, (BIO.B.3.3.1 ) Scientific Terms Student Name: Teacher Name: Jared George Date: Score: 1) Evidence for evolution
Biology 211 (2) Week 1 KEY!
 Biology 211 (2) Week 1 KEY Chapter 1 KEY FIGURES: 1.2, 1.3, 1.4, 1.5, 1.6, 1.7 VOCABULARY: Adaptation: a trait that increases the fitness Cells: a developed, system bound with a thin outer layer made of
Biology 211 (2) Week 1 KEY Chapter 1 KEY FIGURES: 1.2, 1.3, 1.4, 1.5, 1.6, 1.7 VOCABULARY: Adaptation: a trait that increases the fitness Cells: a developed, system bound with a thin outer layer made of
Basic Tree Thinking Assessment David A. Baum, Stacey DeWitt Smith, Samuel S. Donovan
 Basic Tree Thinking Assessment David A. Baum, Stacey DeWitt Smith, Samuel S. Donovan This quiz includes a number of multiple-choice questions you can use to test yourself on your ability to accurately
Basic Tree Thinking Assessment David A. Baum, Stacey DeWitt Smith, Samuel S. Donovan This quiz includes a number of multiple-choice questions you can use to test yourself on your ability to accurately
0 Mya - Humans Goodbye Big Dinosaurs Mammals EXPLODE First flowers 100 Mya- First 200 Mya-
 0 Mya - Humans Goodbye Big Dinosaurs Mammals EXPLODE First flowers 100 Mya- First 200 Mya- 300 Mya- Dinosaurs First First Coal lforests Pennsylvanian Mississippian 400 Mya- 500 Mya- First Forests First
0 Mya - Humans Goodbye Big Dinosaurs Mammals EXPLODE First flowers 100 Mya- First 200 Mya- 300 Mya- Dinosaurs First First Coal lforests Pennsylvanian Mississippian 400 Mya- 500 Mya- First Forests First
Bootstrapping and Tree reliability. Biol4230 Tues, March 13, 2018 Bill Pearson Pinn 6-057
 Bootstrapping and Tree reliability Biol4230 Tues, March 13, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Rooting trees (outgroups) Bootstrapping given a set of sequences sample positions randomly,
Bootstrapping and Tree reliability Biol4230 Tues, March 13, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Rooting trees (outgroups) Bootstrapping given a set of sequences sample positions randomly,
Are Guinea Pigs Rodents? The Importance of Adequate Models in Molecular Phylogenetics
 Journal of Mammalian Evolution, Vol. 4, No. 2, 1997 Are Guinea Pigs Rodents? The Importance of Adequate Models in Molecular Phylogenetics Jack Sullivan1'2 and David L. Swofford1 The monophyly of Rodentia
Journal of Mammalian Evolution, Vol. 4, No. 2, 1997 Are Guinea Pigs Rodents? The Importance of Adequate Models in Molecular Phylogenetics Jack Sullivan1'2 and David L. Swofford1 The monophyly of Rodentia
Origins of Life. Fundamental Properties of Life. Conditions on Early Earth. Evolution of Cells. The Tree of Life
 The Tree of Life Chapter 26 Origins of Life The Earth formed as a hot mass of molten rock about 4.5 billion years ago (BYA) -As it cooled, chemically-rich oceans were formed from water condensation Life
The Tree of Life Chapter 26 Origins of Life The Earth formed as a hot mass of molten rock about 4.5 billion years ago (BYA) -As it cooled, chemically-rich oceans were formed from water condensation Life
CLASSIFICATION OF LIVING THINGS. Chapter 18
 CLASSIFICATION OF LIVING THINGS Chapter 18 How many species are there? About 1.8 million species have been given scientific names Nearly 2/3 of which are insects 99% of all known animal species are smaller
CLASSIFICATION OF LIVING THINGS Chapter 18 How many species are there? About 1.8 million species have been given scientific names Nearly 2/3 of which are insects 99% of all known animal species are smaller
 Tree of Life iological Sequence nalysis Chapter http://tolweb.org/tree/ Phylogenetic Prediction ll organisms on Earth have a common ancestor. ll species are related. The relationship is called a phylogeny
Tree of Life iological Sequence nalysis Chapter http://tolweb.org/tree/ Phylogenetic Prediction ll organisms on Earth have a common ancestor. ll species are related. The relationship is called a phylogeny
Master Biomedizin ) UCSC & UniProt 2) Homology 3) MSA 4) Phylogeny. Pablo Mier
 Master Biomedizin 2018 1) UCSC & UniProt 2) Homology 3) MSA 4) 1 12 a. All of the sequences in file1.fasta (https://cbdm.uni-mainz.de/mb18/) are homologs. How many groups of orthologs would you say there
Master Biomedizin 2018 1) UCSC & UniProt 2) Homology 3) MSA 4) 1 12 a. All of the sequences in file1.fasta (https://cbdm.uni-mainz.de/mb18/) are homologs. How many groups of orthologs would you say there
