Supplementary information
|
|
|
- Julian Beasley
- 6 years ago
- Views:
Transcription
1 Supplementary information Superoxide dismutase 1 is positively selected in great apes to minimize protein misfolding Pouria Dasmeh 1, and Kasper P. Kepp* 2 1 Harvard University, Department of Chemistry and Chemical Biology, Cambridge, MA, USA 2 Technical University of Denmark, DTU Chemistry, DK-2800 Kongens Lyngby, Denmark. Current address: Department of Biochemistry and Cedergren Center for Bioinformatics and Genomics, Faculty of Medicine, University of Montreal, 2900 Edouard-Montpetit, Montreal, Quebec H3T 1J4, Canada. * Corresponding Author kpj@kemi.dtu.dk Phone: S1
2 Figure S1. Multiple sequence alignment (Clustal Omega) of SOD1 in species studied in this work. Charged amino acids are marked in red and blue, respectively. Consensus amino acids are shown at the bottom of the alignment. S2
3 S3
4 Figure S2. Complete multiple sequence alignment of SOD1 in rodents and primates. Identical and similar positions are shown in blue and pink, respectively (made using the software msa from R[1]). Consensus amino acids are shown at the bottom of the alignment. S4
5 Figure S3. Tree from DataMonkey used to conduct relaxation tests. S5
6 Table S1. Codons detected to be under positive selection using different methods as explained in the main text. Datamonkey PAML SLAC FEL REL FUBAR MEME M8_rel Branchsite test[2] Table S2. Branches detected to be under positive selection. Branch leading to Corrected p-value a ω+ b Pr [ω=ω+] c Pig Beaver Great apes Opossum a: p-values corrected for multiple sampling using the Holm-Bonferooni method[3]. b: dn/ds for positively selected sites, c: fraction of sequence evolving with ω+. Table S3. Logarithm of likelihood function for null and alternative model in which relaxation is ignored and assumed. Branch set a ω1 b ω2 ω3 log L c LR (p-value) d Reference 0.00 (65%) (23%) 3.65 (12%) Test 0.00 (65%) (23%) 3.65 (12%) Unclassified (80%) (17%) 54.1 (2.8%) Reference (79%) (14%) 6.66 (6.6%) 6.20 (p = 0.013) Test (79%) (14%) 2.85 (6.6%) Unclassified (80%) (17%) 75.3 (2.8%) a: Branches refer to test (i.e., primates), alternative (i.e., rodents) and unclassified (i.e., rest) branch sets. b: different ωs refer to different site classes assumed for residues with the fraction of residues written in parenthesis. c: logarithm of likelihood function. d: Twice the log-likelihood (i.e., selection criteria) and the p-value associated with it in parenthesis. S6
7 Computed ΔΔG (kcal/mol) Computed ΔΔG (kcal/mol) Popmusic 2.1 y = 1.48x R² = Experimental normalized ΔΔG (Byström et al.) I-Mutant 2.0 y = 1.25x R² = Experimental normalized ΔΔG (Byström et al.) Figure S4. Correlation between experimental G (normalized G(norm) from Ref. [4]) and computed G using I-Mutant 2.0 and Popmusic 2.1. S7
8 Table S4. Data used for benchmarking Popmusic 2.1 and I-Mutant 2.0 (Figure S4). The normalized free energy changes G(norm) are taken from Ref. [4] Mutant G(norm) I-Mutant G(norm) Popmusic A4V V7E G37R L38V G41D G41S H43R H46R H48Q D76V D76Y L84V G85R N86D N86K N86S D90A D90V G93A G93D G93R G93S G93V E100G E100K D101G D101N I104F S105L L106V I113T G114A D124V D125H S134N N139D N139K L144F L144S V148G S8
9 # mutations ΔΔG (kcal/mol) Figure S5. The number of mutations with a ΔΔG in a certain range (in kcal/mol) for all possible mutations in SOD1 computed with Popmusic; most introduced mutations are destabilizing, and the distribution agrees well with the general behavior of such distributions[5]. S9
10 Table S5. Substitutions occurring in all branches of the studied phylogeny and associated changes in thermodynamic stability computed with I-Mutant 2.0 and Popmusic 2.1, and solvent accessibility of the substituted site s(sasa in %). Notice that the two methods have reverse sign conventions for G, so that stabilizing effects corresponds to positive for I-Mutant and negative for Popmusic. Details: "Calculation" describes whether the G value was obtained as the reverse substitution (with inversed sign of the G value) in the 2C9V structure, or as a composite substitution, requiring two substitutions to be added. For example, 1 T > A is "reverse" because A is the wild type residue in the 2C9V structure, so the mutation A to T was computed and the sign inversed. Similarly, if neither the start X or end Y residue was present in the structure but rather a third residue Z, the site substitution of interest was computed as G (XZ) + G (ZY) (referred to as "composite"). Summary of changes along branches. IMUTANT SASA POPMUSIC Calculation Branch 1: C9V 1 T > A # reverse S > N reverse K > E T > S composite K > T composite S > T reverse E > L reverse E > N composite S > A reverse N > E reverse K > T reverse SUM Branch 2: L > M composite E > V composite D > E composite E > Q A > P reverse P > E composite P > S reverse K > Q reverse Branch 3: SUM Branch 4: T > R composite E > Q SUM Branch 5: M > T reverse A > E S10
11 SUM Branch 6: Branch 7: Branch 8: T > M G > S reverse N > G G > E R > Q composite F > Y L > H K > A S > R E > Q SUM Branch 9: (Mouse) 12 N > V > L E > I > SUM Branch 10: (Rat) 12 N > T > V composite Q > E composite 57 E > I > S > T T > A SUM Branch 11: (Rabbit) D > G reverse N > G > S Q > E H > D composite A > N > T S > K composite Q > L reverse S11
12 54 57 E > I > K > S D > N N > D reverse S > L E > D reverse H > M I > V M > L reverse E > D Q > P SUM Branch 12: G > N composite V > L SUM Branch 13: (Squirrel) 12 N > A > K composite N > K composite N > A G > N P > K T > A L > Q K > T T > I G > D reverse E > Q composite E > P composite SUM Branch 14: (Beaver) N > P > S I > V N > G G > S > R composite Q > L reverse E > I > K > L S12
13 77 80 E > Q G > N composite N > D reverse S > N E > D A > T SUM Branch 15: S > K Q > E reverse K > T V > L E > A H > N SUM Branch 16: (Naked_Mole_rat) 12 N > Q > H K > Q A > T > A composite T > N Q > K composite 57 E > I > N > E composite S > F A > P composite SUM Branch 17: (Guinea_pig) 12 N > T > I reverse 25 A > G > A composite T > V Q > K composite 57 E > I > K > Q T > A composite E > P composite SUM S13
14 Branch 18: Branch 19: (Tree_Shrew) 2 M > L composite 12 N > G > E composite V > L composite S > T composite T > M composite 57 E > I > L > E K > S E > Q T > I N > D S > V E > A composite K > R SUM Branch 20: Branch 21: (Galago) 1 A > T N > P > A K > Q A > V > M composite S > K composite T > A Q > D composite 57 E > I > L > Q D > N V > E N > I composite V > M S > G SUM Branch 22: H > N A > E S14
15 25 26 G > S V > K S > W R > S Q > L K > R N > K composite E > D M > L SUM Branch 23: T > A K > E SUM Branch 24: (Marmoset) N > E > I > K > S composite D > V SUM Branch 25: (Capuchin) 12 N > E > I > Branch 26: G > S SUM Branch 27: K > Q I > F T > K S > G SUM Branch 28: (Baboon) N > N > S E > I > K > N composite S15
16 SUM Branch 29: (Macaque) 12 N > E > I > Branch 30: T > I G > D SUM Branch 31: (Gibbon) 12 N > W > Y S > R E > I > SUM Branch 32: M > T T > K Q > E Q > A K > D destab 114 S > C exp SUM Branch 33: (Orangutan) Q > K S > R K > E A > V E > I > D > S SUM Branch 34: S > G SUM Branch 35: (Gorilla) 12 N > E > S16
17 58 I > S > F SUM Branch 36: (Human) 12 N > E > I > Branch 37: (Bat) 1 A > T M > T reverse 3 K > R N > H > R composite K > E A > N > T V > K reverse S > F E > K Q > E reverse 57 E > I > S > R K > T K > Q G > E composite K > E D > N N > E composite I > L E > K V > Q E > A composite I > V A > R K > R E > D T > K SUM Branch 38: Branch 39: A > E S17
18 11 11 D > Q Q > E E > V V > L S > A E > D reverse A > R K > Q Q > K SUM Branch 40: E > Q SUM Branch 41: G > D K > Q reverse SUM Branch 42: (Dog) 12 N > A > N > S G > E N > X E > I > N > I composite H > Y SUM Branch 43: (Fox) 1 1 E > T composite 2 2 M > T reverse Q > D reverse N > P > K E > Q reverse V > E reverse A > G > A composite P > L E > D composite E > I > S18
19 68 71 S > G K > T Q > E reverse V > M N > H composite S > H S > A L > M composite R > P composite D > E composite E > D T > K Q > K reverse SUM Branch 44: (Cat) 12 N > E > I > I > M SUM Branch 45: (Giant_panda) 12 N > A > K composite G > E composite N > G E > I > N > T composite I > L SUM Branch 46: G > E composite N > G E > D composite S > D A > P SUM Branch 47: (Horse) 2 2 M > L composite N > Q > H T > V composite S19
20 23 24 K > Q A > Q composite V > L S > K composite T > F composite T > E composite E > K Q > E reverse E > I > S > T P > A G > D reverse K > E D > N V > K N > D reverse I > M E > K E > K composite P > Q composite Q > P SUM Branch 48: M > T reverse P > T S > T N > T K > R SUM Branch 49: (Pig) N > H > Y composite Q > L A > G G > K G > V > L T > K reverse T > A E > I > L > E E > Q S20
21 D > Y S > A E > D A > T SUM Branch 50: Q > A G > D T > S reverse G > D reverse D > N T > I composite E > V S > P V > L H > Y Q > P SUM Branch 51: (Goat) 12 N > A > E > T > K E > I > T > K R > C SUM Branch 52: Branch 53: (Red_deer) 12 N > H > R composite 25 A > E > D > H composite 57 E > I > I > K composite P > S reverse Y > H reverse G > R S > N S21
22 P > Q SUM Branch 54: Branch 55: (Bovine) 12 N > A > E > E > I > P > K composite SUM Branch 56: (Sheep) 12 N > H > R composite 25 A > E > T > K E > I > T > K S > G SUM Branch 57: (Opossum) 1 A > V N > H > F composite A > Q composite G > V composite N > G G > E V > L T > S reverse T > K reverse T > A E > I > L > H K > T G > N composite N > T composite E > K V > H S22
23 S > E H > M E > A T > E SUM Branch 58: (Chicken) 8 8 L > M N > G > A Q > E T > V composite E > Q K > Q A > E > D Q > N reverse E > I > S > G K > Q E > A E > D G > D > G S > E S > T S > C reverse E > A K > R N > D K > L SUM S23
24 References [1] U. Bodenhofer, E. Bonatesta, C. Horejš-Kainrath, S. Hochreiter, msa: an R package for multiple sequence alignment, Bioinformatics. (2015) btv494. [2] J. Zhang, R. Nielsen, Z. Yang, Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level, Mol. Biol. Evol. 22 (2005) [3] S. Holm, A simple sequentially rejective multiple test procedure, Scand. J. Stat. 6 (1979) [4] R. Byström, P.M. Andersen, G. Gröbner, M. Oliveberg, SOD1 mutations targeting surface hydrogen bonds promote amyotrophic lateral sclerosis without reducing apo-state stability., J. Biol. Chem. 285 (2010) doi: /jbc.m [5] N. Tokuriki, F. Stricher, J. Schymkowitz, L. Serrano, D.S. Tawfik, The stability effects of protein mutations appear to be universally distributed., J. Mol. Biol. 369 (2007) doi: /j.jmb S24
7. Tests for selection
 Sequence analysis and genomics 7. Tests for selection Dr. Katja Nowick Group leader TFome and Transcriptome Evolution Bioinformatics group Paul-Flechsig-Institute for Brain Research www. nowicklab.info
Sequence analysis and genomics 7. Tests for selection Dr. Katja Nowick Group leader TFome and Transcriptome Evolution Bioinformatics group Paul-Flechsig-Institute for Brain Research www. nowicklab.info
Orthologous loci for phylogenomics from raw NGS data
 Orthologous loci for phylogenomics from raw NS data Rachel Schwartz The Biodesign Institute Arizona State University Rachel.Schwartz@asu.edu May 2, 205 Big data for phylogenetics Phylogenomics requires
Orthologous loci for phylogenomics from raw NS data Rachel Schwartz The Biodesign Institute Arizona State University Rachel.Schwartz@asu.edu May 2, 205 Big data for phylogenetics Phylogenomics requires
Emily Blanton Phylogeny Lab Report May 2009
 Introduction It is suggested through scientific research that all living organisms are connected- that we all share a common ancestor and that, through time, we have all evolved from the same starting
Introduction It is suggested through scientific research that all living organisms are connected- that we all share a common ancestor and that, through time, we have all evolved from the same starting
Evolution of the Sry gene within the African pygmy mice Nannomys
 Evolution of the Sry gene within the African pygmy mice Nannomys Subgenus of the genus Mus Widespread in Sub Saharan Africa ~ 20 species Mus minutoides Very high proportion (> 75%) of fertile sex reversed
Evolution of the Sry gene within the African pygmy mice Nannomys Subgenus of the genus Mus Widespread in Sub Saharan Africa ~ 20 species Mus minutoides Very high proportion (> 75%) of fertile sex reversed
Positively Selected Sites in Cetacean Myoglobins Contribute to Protein Stability
 Contribute to Protein Stability The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Published Version Accessed Citable
Contribute to Protein Stability The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Published Version Accessed Citable
Cladistics and Bioinformatics Questions 2013
 AP Biology Name Cladistics and Bioinformatics Questions 2013 1. The following table shows the percentage similarity in sequences of nucleotides from a homologous gene derived from five different species
AP Biology Name Cladistics and Bioinformatics Questions 2013 1. The following table shows the percentage similarity in sequences of nucleotides from a homologous gene derived from five different species
Estimation of species divergence dates with a sloppy molecular clock
 Estimation of species divergence dates with a sloppy molecular clock Ziheng Yang Department of Biology University College London Date estimation with a clock is easy. t 2 = 13my t 3 t 1 t 4 t 5 Node Distance
Estimation of species divergence dates with a sloppy molecular clock Ziheng Yang Department of Biology University College London Date estimation with a clock is easy. t 2 = 13my t 3 t 1 t 4 t 5 Node Distance
Fixation of Deleterious Mutations at Critical Positions in Human Proteins
 Fixation of Deleterious Mutations at Critical Positions in Human Proteins Author Sankarasubramanian, Sankar Published 2011 Journal Title Molecular Biology and Evolution DOI https://doi.org/10.1093/molbev/msr097
Fixation of Deleterious Mutations at Critical Positions in Human Proteins Author Sankarasubramanian, Sankar Published 2011 Journal Title Molecular Biology and Evolution DOI https://doi.org/10.1093/molbev/msr097
Introduction to Bioinformatics Online Course: IBT
 Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec1 Building a Multiple Sequence Alignment Learning Outcomes 1- Understanding Why multiple
Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec1 Building a Multiple Sequence Alignment Learning Outcomes 1- Understanding Why multiple
Sequence motif analysis
 Sequence motif analysis Alan Moses Associate Professor and Canada Research Chair in Computational Biology Departments of Cell & Systems Biology, Computer Science, and Ecology & Evolutionary Biology Director,
Sequence motif analysis Alan Moses Associate Professor and Canada Research Chair in Computational Biology Departments of Cell & Systems Biology, Computer Science, and Ecology & Evolutionary Biology Director,
AP Biology. Evolution is "so overwhelmingly established that it has become irrational to call it a theory." Evidence of Evolution by Natural Selection
 Evidence of Evolution by Natural Selection Evolution is "so overwhelmingly established that it has become irrational to call it a theory." -- Ernst Mayr What Evolution Is 2001 Professor Emeritus, Evolutionary
Evidence of Evolution by Natural Selection Evolution is "so overwhelmingly established that it has become irrational to call it a theory." -- Ernst Mayr What Evolution Is 2001 Professor Emeritus, Evolutionary
Plan: Evolutionary trees, characters. Perfect phylogeny Methods: NJ, parsimony, max likelihood, Quartet method
 Phylogeny 1 Plan: Phylogeny is an important subject. We have 2.5 hours. So I will teach all the concepts via one example of a chain letter evolution. The concepts we will discuss include: Evolutionary
Phylogeny 1 Plan: Phylogeny is an important subject. We have 2.5 hours. So I will teach all the concepts via one example of a chain letter evolution. The concepts we will discuss include: Evolutionary
Combined Isothermal Titration and Differential Scanning Calorimetry Define. Three-State Thermodynamics of fals-associated Mutant Apo SOD1 Dimers
 Supporting Information for: Combined Isothermal Titration and Differential Scanning Calorimetry Define Three-State Thermodynamics of fals-associated Mutant Apo SOD1 Dimers and an Increased Population of
Supporting Information for: Combined Isothermal Titration and Differential Scanning Calorimetry Define Three-State Thermodynamics of fals-associated Mutant Apo SOD1 Dimers and an Increased Population of
Probabilistic modeling and molecular phylogeny
 Probabilistic modeling and molecular phylogeny Anders Gorm Pedersen Molecular Evolution Group Center for Biological Sequence Analysis Technical University of Denmark (DTU) What is a model? Mathematical
Probabilistic modeling and molecular phylogeny Anders Gorm Pedersen Molecular Evolution Group Center for Biological Sequence Analysis Technical University of Denmark (DTU) What is a model? Mathematical
DO NOT WRITE ON THIS. Evidence from Evolution Activity. The Fossilization Process. Types of Fossils
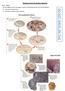 Evidence from Evolution Activity Part 1 - Fossils Use the diagrams on the next page to answer the following questions IN YOUR NOTEBOOK. 1. Describe how fossils form. 2. Describe the different types of
Evidence from Evolution Activity Part 1 - Fossils Use the diagrams on the next page to answer the following questions IN YOUR NOTEBOOK. 1. Describe how fossils form. 2. Describe the different types of
Quantifying sequence similarity
 Quantifying sequence similarity Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, February 16 th 2016 After this lecture, you can define homology, similarity, and identity
Quantifying sequence similarity Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, February 16 th 2016 After this lecture, you can define homology, similarity, and identity
Evidence of Evolution by Natural Selection. Dodo bird
 Evidence of Evolution by Natural Selection Dodo bird 2007-2008 Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development
Evidence of Evolution by Natural Selection Dodo bird 2007-2008 Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11510 Supplementary Table 1. Indel Index Removal Gene Number of Starting Sequences Number of Final Sequences Percentage of Sequences Removed based on the Indel
SUPPLEMENTARY INFORMATION doi:10.1038/nature11510 Supplementary Table 1. Indel Index Removal Gene Number of Starting Sequences Number of Final Sequences Percentage of Sequences Removed based on the Indel
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST
 Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
Likelihood Ratio Tests for Detecting Positive Selection and Application to Primate Lysozyme Evolution
 Likelihood Ratio Tests for Detecting Positive Selection and Application to Primate Lysozyme Evolution Ziheng Yang Department of Biology, University College, London An excess of nonsynonymous substitutions
Likelihood Ratio Tests for Detecting Positive Selection and Application to Primate Lysozyme Evolution Ziheng Yang Department of Biology, University College, London An excess of nonsynonymous substitutions
Mechanisms of Evolution Darwinian Evolution
 Mechanisms of Evolution Darwinian Evolution Descent with modification by means of natural selection All life has descended from a common ancestor The mechanism of modification is natural selection Concept
Mechanisms of Evolution Darwinian Evolution Descent with modification by means of natural selection All life has descended from a common ancestor The mechanism of modification is natural selection Concept
POPULATION GENETICS Biology 107/207L Winter 2005 Lab 5. Testing for positive Darwinian selection
 POPULATION GENETICS Biology 107/207L Winter 2005 Lab 5. Testing for positive Darwinian selection A growing number of statistical approaches have been developed to detect natural selection at the DNA sequence
POPULATION GENETICS Biology 107/207L Winter 2005 Lab 5. Testing for positive Darwinian selection A growing number of statistical approaches have been developed to detect natural selection at the DNA sequence
NB-DNJ/GCase-pH 7.4 NB-DNJ+/GCase-pH 7.4 NB-DNJ+/GCase-pH 4.5
 SUPPLEMENTARY TABLES Suppl. Table 1. Protonation states at ph 7.4 and 4.5. Protonation states of titratable residues in GCase at ph 7.4 and 4.5. Histidine: HID, H at δ-nitrogen; HIE, H at ε-nitrogen; HIP,
SUPPLEMENTARY TABLES Suppl. Table 1. Protonation states at ph 7.4 and 4.5. Protonation states of titratable residues in GCase at ph 7.4 and 4.5. Histidine: HID, H at δ-nitrogen; HIE, H at ε-nitrogen; HIP,
Proximal point algorithm in Hadamard spaces
 Proximal point algorithm in Hadamard spaces Miroslav Bacak Télécom ParisTech Optimisation Géométrique sur les Variétés - Paris, 21 novembre 2014 Contents of the talk 1 Basic facts on Hadamard spaces 2
Proximal point algorithm in Hadamard spaces Miroslav Bacak Télécom ParisTech Optimisation Géométrique sur les Variétés - Paris, 21 novembre 2014 Contents of the talk 1 Basic facts on Hadamard spaces 2
Estimating the Distribution of Selection Coefficients from Phylogenetic Data with Applications to Mitochondrial and Viral DNA
 Estimating the Distribution of Selection Coefficients from Phylogenetic Data with Applications to Mitochondrial and Viral DNA Rasmus Nielsen* and Ziheng Yang *Department of Biometrics, Cornell University;
Estimating the Distribution of Selection Coefficients from Phylogenetic Data with Applications to Mitochondrial and Viral DNA Rasmus Nielsen* and Ziheng Yang *Department of Biometrics, Cornell University;
Week 6: Restriction sites, RAPDs, microsatellites, likelihood, hidden Markov models
 Week 6: Restriction sites, RAPDs, microsatellites, likelihood, hidden Markov models Genome 570 February, 2012 Week 6: Restriction sites, RAPDs, microsatellites, likelihood, hidden Markov models p.1/63
Week 6: Restriction sites, RAPDs, microsatellites, likelihood, hidden Markov models Genome 570 February, 2012 Week 6: Restriction sites, RAPDs, microsatellites, likelihood, hidden Markov models p.1/63
Bioinformatics tools for phylogeny and visualization. Yanbin Yin
 Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
THEORY. Based on sequence Length According to the length of sequence being compared it is of following two types
 Exp 11- THEORY Sequence Alignment is a process of aligning two sequences to achieve maximum levels of identity between them. This help to derive functional, structural and evolutionary relationships between
Exp 11- THEORY Sequence Alignment is a process of aligning two sequences to achieve maximum levels of identity between them. This help to derive functional, structural and evolutionary relationships between
Bioinformatics Exercises
 Bioinformatics Exercises AP Biology Teachers Workshop Susan Cates, Ph.D. Evolution of Species Phylogenetic Trees show the relatedness of organisms Common Ancestor (Root of the tree) 1 Rooted vs. Unrooted
Bioinformatics Exercises AP Biology Teachers Workshop Susan Cates, Ph.D. Evolution of Species Phylogenetic Trees show the relatedness of organisms Common Ancestor (Root of the tree) 1 Rooted vs. Unrooted
Lecture 16: Again on Regression
 Lecture 16: Again on Regression S. Massa, Department of Statistics, University of Oxford 10 February 2016 The Normality Assumption Body weights (Kg) and brain weights (Kg) of 62 mammals. Species Body weight
Lecture 16: Again on Regression S. Massa, Department of Statistics, University of Oxford 10 February 2016 The Normality Assumption Body weights (Kg) and brain weights (Kg) of 62 mammals. Species Body weight
Statistical Machine Learning Methods for Bioinformatics II. Hidden Markov Model for Biological Sequences
 Statistical Machine Learning Methods for Bioinformatics II. Hidden Markov Model for Biological Sequences Jianlin Cheng, PhD Department of Computer Science University of Missouri 2008 Free for Academic
Statistical Machine Learning Methods for Bioinformatics II. Hidden Markov Model for Biological Sequences Jianlin Cheng, PhD Department of Computer Science University of Missouri 2008 Free for Academic
Natural selection on the molecular level
 Natural selection on the molecular level Fundamentals of molecular evolution How DNA and protein sequences evolve? Genetic variability in evolution } Mutations } forming novel alleles } Inversions } change
Natural selection on the molecular level Fundamentals of molecular evolution How DNA and protein sequences evolve? Genetic variability in evolution } Mutations } forming novel alleles } Inversions } change
Using phylogenetics to estimate species divergence times... Basics and basic issues for Bayesian inference of divergence times (plus some digression)
 Using phylogenetics to estimate species divergence times... More accurately... Basics and basic issues for Bayesian inference of divergence times (plus some digression) "A comparison of the structures
Using phylogenetics to estimate species divergence times... More accurately... Basics and basic issues for Bayesian inference of divergence times (plus some digression) "A comparison of the structures
part 4: phenomenological load and biological inference. phenomenological load review types of models. Gαβ = 8π Tαβ. Newton.
 2017-07-29 part 4: and biological inference review types of models phenomenological Newton F= Gm1m2 r2 mechanistic Einstein Gαβ = 8π Tαβ 1 molecular evolution is process and pattern process pattern MutSel
2017-07-29 part 4: and biological inference review types of models phenomenological Newton F= Gm1m2 r2 mechanistic Einstein Gαβ = 8π Tαβ 1 molecular evolution is process and pattern process pattern MutSel
Accuracy and Power of the Likelihood Ratio Test in Detecting Adaptive Molecular Evolution
 Accuracy and Power of the Likelihood Ratio Test in Detecting Adaptive Molecular Evolution Maria Anisimova, Joseph P. Bielawski, and Ziheng Yang Department of Biology, Galton Laboratory, University College
Accuracy and Power of the Likelihood Ratio Test in Detecting Adaptive Molecular Evolution Maria Anisimova, Joseph P. Bielawski, and Ziheng Yang Department of Biology, Galton Laboratory, University College
The Evolutionary Origins of Protein Sequence Variation
 Temple University Structural Bioinformatics II The Evolutionary Origins of Protein Sequence Variation Protein Evolution (tour of concepts & current ideas) Protein Fitness, Marginal Stability, Compensatory
Temple University Structural Bioinformatics II The Evolutionary Origins of Protein Sequence Variation Protein Evolution (tour of concepts & current ideas) Protein Fitness, Marginal Stability, Compensatory
Effects of Gap Open and Gap Extension Penalties
 Brigham Young University BYU ScholarsArchive All Faculty Publications 200-10-01 Effects of Gap Open and Gap Extension Penalties Hyrum Carroll hyrumcarroll@gmail.com Mark J. Clement clement@cs.byu.edu See
Brigham Young University BYU ScholarsArchive All Faculty Publications 200-10-01 Effects of Gap Open and Gap Extension Penalties Hyrum Carroll hyrumcarroll@gmail.com Mark J. Clement clement@cs.byu.edu See
RELATING PHYSICOCHEMMICAL PROPERTIES OF AMINO ACIDS TO VARIABLE NUCLEOTIDE SUBSTITUTION PATTERNS AMONG SITES ZIHENG YANG
 RELATING PHYSICOCHEMMICAL PROPERTIES OF AMINO ACIDS TO VARIABLE NUCLEOTIDE SUBSTITUTION PATTERNS AMONG SITES ZIHENG YANG Department of Biology (Galton Laboratory), University College London, 4 Stephenson
RELATING PHYSICOCHEMMICAL PROPERTIES OF AMINO ACIDS TO VARIABLE NUCLEOTIDE SUBSTITUTION PATTERNS AMONG SITES ZIHENG YANG Department of Biology (Galton Laboratory), University College London, 4 Stephenson
Proceedings of the SMBE Tri-National Young Investigators Workshop 2005
 Proceedings of the SMBE Tri-National Young Investigators Workshop 25 Control of the False Discovery Rate Applied to the Detection of Positively Selected Amino Acid Sites Stéphane Guindon,* Mik Black,*à
Proceedings of the SMBE Tri-National Young Investigators Workshop 25 Control of the False Discovery Rate Applied to the Detection of Positively Selected Amino Acid Sites Stéphane Guindon,* Mik Black,*à
Phylogenomics, Multiple Sequence Alignment, and Metagenomics. Tandy Warnow University of Illinois at Urbana-Champaign
 Phylogenomics, Multiple Sequence Alignment, and Metagenomics Tandy Warnow University of Illinois at Urbana-Champaign Phylogeny (evolutionary tree) Orangutan Gorilla Chimpanzee Human From the Tree of the
Phylogenomics, Multiple Sequence Alignment, and Metagenomics Tandy Warnow University of Illinois at Urbana-Champaign Phylogeny (evolutionary tree) Orangutan Gorilla Chimpanzee Human From the Tree of the
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION
 THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
CONSTRUCTION OF PHYLOGENETIC TREE FROM MULTIPLE GENE TREES USING PRINCIPAL COMPONENT ANALYSIS
 INTERNATIONAL JOURNAL OF ELECTRONICS AND COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) Proceedings of the International Conference on Emerging Trends in Engineering and Management (ICETEM14) ISSN 0976
INTERNATIONAL JOURNAL OF ELECTRONICS AND COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) Proceedings of the International Conference on Emerging Trends in Engineering and Management (ICETEM14) ISSN 0976
Temporal Trails of Natural Selection in Human Mitogenomes. Author. Published. Journal Title DOI. Copyright Statement.
 Temporal Trails of Natural Selection in Human Mitogenomes Author Sankarasubramanian, Sankar Published 2009 Journal Title Molecular Biology and Evolution DOI https://doi.org/10.1093/molbev/msp005 Copyright
Temporal Trails of Natural Selection in Human Mitogenomes Author Sankarasubramanian, Sankar Published 2009 Journal Title Molecular Biology and Evolution DOI https://doi.org/10.1093/molbev/msp005 Copyright
Bayesian Models for Phylogenetic Trees
 Bayesian Models for Phylogenetic Trees Clarence Leung* 1 1 McGill Centre for Bioinformatics, McGill University, Montreal, Quebec, Canada ABSTRACT Introduction: Inferring genetic ancestry of different species
Bayesian Models for Phylogenetic Trees Clarence Leung* 1 1 McGill Centre for Bioinformatics, McGill University, Montreal, Quebec, Canada ABSTRACT Introduction: Inferring genetic ancestry of different species
Drosophila melanogaster and D. simulans, two fruit fly species that are nearly
 Comparative Genomics: Human versus chimpanzee 1. Introduction The chimpanzee is the closest living relative to humans. The two species are nearly identical in DNA sequence (>98% identity), yet vastly different
Comparative Genomics: Human versus chimpanzee 1. Introduction The chimpanzee is the closest living relative to humans. The two species are nearly identical in DNA sequence (>98% identity), yet vastly different
Supplementary text and figures: Comparative assessment of methods for aligning multiple genome sequences
 Supplementary text and figures: Comparative assessment of methods for aligning multiple genome sequences Xiaoyu Chen Martin Tompa Department of Computer Science and Engineering Department of Genome Sciences
Supplementary text and figures: Comparative assessment of methods for aligning multiple genome sequences Xiaoyu Chen Martin Tompa Department of Computer Science and Engineering Department of Genome Sciences
Microscopic analysis of protein oxidative damage: effect of. carbonylation on structure, dynamics and aggregability of.
 Microscopic analysis of protein oxidative damage: effect of carbonylation on structure, dynamics and aggregability of villin headpiece Drazen etrov 1,2,3 and Bojan Zagrovic 1,2,3,* Supplementary Information
Microscopic analysis of protein oxidative damage: effect of carbonylation on structure, dynamics and aggregability of villin headpiece Drazen etrov 1,2,3 and Bojan Zagrovic 1,2,3,* Supplementary Information
"Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky
 MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
InDel 3-5. InDel 8-9. InDel 3-5. InDel 8-9. InDel InDel 8-9
 Lecture 5 Alignment I. Introduction. For sequence data, the process of generating an alignment establishes positional homologies; that is, alignment provides the identification of homologous phylogenetic
Lecture 5 Alignment I. Introduction. For sequence data, the process of generating an alignment establishes positional homologies; that is, alignment provides the identification of homologous phylogenetic
SUPPLEMENTARY INFORMATION
 Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
BIOINFORMATICS: An Introduction
 BIOINFORMATICS: An Introduction What is Bioinformatics? The term was first coined in 1988 by Dr. Hwa Lim The original definition was : a collective term for data compilation, organisation, analysis and
BIOINFORMATICS: An Introduction What is Bioinformatics? The term was first coined in 1988 by Dr. Hwa Lim The original definition was : a collective term for data compilation, organisation, analysis and
08/21/2017 BLAST. Multiple Sequence Alignments: Clustal Omega
 BLAST Multiple Sequence Alignments: Clustal Omega What does basic BLAST do (e.g. what is input sequence and how does BLAST look for matches?) Susan Parrish McDaniel College Multiple Sequence Alignments
BLAST Multiple Sequence Alignments: Clustal Omega What does basic BLAST do (e.g. what is input sequence and how does BLAST look for matches?) Susan Parrish McDaniel College Multiple Sequence Alignments
Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition
 Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition David D. Pollock* and William J. Bruno* *Theoretical Biology and Biophysics, Los Alamos National
Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition David D. Pollock* and William J. Bruno* *Theoretical Biology and Biophysics, Los Alamos National
METHODS FOR DETERMINING PHYLOGENY. In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task.
 Chapter 12 (Strikberger) Molecular Phylogenies and Evolution METHODS FOR DETERMINING PHYLOGENY In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task. Modern
Chapter 12 (Strikberger) Molecular Phylogenies and Evolution METHODS FOR DETERMINING PHYLOGENY In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task. Modern
Nucleotides containing variously modified sugars: energetics, structure, and mechanical properties
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 ELECTRONIC SUPPLEMENTARY INFORMATION Nucleotides containing variously modified
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 ELECTRONIC SUPPLEMENTARY INFORMATION Nucleotides containing variously modified
Molecular dynamics simulations of anti-aggregation effect of ibuprofen. Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov
 Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Taming the Beast Workshop
 Workshop and Chi Zhang June 28, 2016 1 / 19 Species tree Species tree the phylogeny representing the relationships among a group of species Figure adapted from [Rogers and Gibbs, 2014] Gene tree the phylogeny
Workshop and Chi Zhang June 28, 2016 1 / 19 Species tree Species tree the phylogeny representing the relationships among a group of species Figure adapted from [Rogers and Gibbs, 2014] Gene tree the phylogeny
Detecting the correlated mutations based on selection pressure with CorMut
 Detecting the correlated mutations based on selection pressure with CorMut Zhenpeng Li April 30, 2018 Contents 1 Introduction 1 2 Methods 2 3 Implementation 3 1 Introduction In genetics, the Ka/Ks ratio
Detecting the correlated mutations based on selection pressure with CorMut Zhenpeng Li April 30, 2018 Contents 1 Introduction 1 2 Methods 2 3 Implementation 3 1 Introduction In genetics, the Ka/Ks ratio
7.36/7.91 recitation CB Lecture #4
 7.36/7.91 recitation 2-19-2014 CB Lecture #4 1 Announcements / Reminders Homework: - PS#1 due Feb. 20th at noon. - Late policy: ½ credit if received within 24 hrs of due date, otherwise no credit - Answer
7.36/7.91 recitation 2-19-2014 CB Lecture #4 1 Announcements / Reminders Homework: - PS#1 due Feb. 20th at noon. - Late policy: ½ credit if received within 24 hrs of due date, otherwise no credit - Answer
T h e C S E T I P r o j e c t
 T h e P r o j e c t T H E P R O J E C T T A B L E O F C O N T E N T S A r t i c l e P a g e C o m p r e h e n s i v e A s s es s m e n t o f t h e U F O / E T I P h e n o m e n o n M a y 1 9 9 1 1 E T
T h e P r o j e c t T H E P R O J E C T T A B L E O F C O N T E N T S A r t i c l e P a g e C o m p r e h e n s i v e A s s es s m e n t o f t h e U F O / E T I P h e n o m e n o n M a y 1 9 9 1 1 E T
Molecular phylogeny How to infer phylogenetic trees using molecular sequences
 Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 2009 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 2009 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Nature Structural & Molecular Biology: doi: /nsmb.3194
 Supplementary Figure 1 Mass spectrometry and solution NMR data for -syn samples used in this study. (a) Matrix-assisted laser-desorption and ionization time-of-flight (MALDI-TOF) mass spectrum of uniformly-
Supplementary Figure 1 Mass spectrometry and solution NMR data for -syn samples used in this study. (a) Matrix-assisted laser-desorption and ionization time-of-flight (MALDI-TOF) mass spectrum of uniformly-
Phylogenetic inference
 Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Edward Susko Department of Mathematics and Statistics, Dalhousie University. Introduction. Installation
 1 dist est: Estimation of Rates-Across-Sites Distributions in Phylogenetic Subsititution Models Version 1.0 Edward Susko Department of Mathematics and Statistics, Dalhousie University Introduction The
1 dist est: Estimation of Rates-Across-Sites Distributions in Phylogenetic Subsititution Models Version 1.0 Edward Susko Department of Mathematics and Statistics, Dalhousie University Introduction The
SUPPLEMENTARY INFORMATION
 Supplementary information S1 (box). Supplementary Methods description. Prokaryotic Genome Database Archaeal and bacterial genome sequences were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/)
Supplementary information S1 (box). Supplementary Methods description. Prokaryotic Genome Database Archaeal and bacterial genome sequences were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/)
A Method for Aligning RNA Secondary Structures
 Method for ligning RN Secondary Structures Jason T. L. Wang New Jersey Institute of Technology J Liu, JTL Wang, J Hu and B Tian, BM Bioinformatics, 2005 1 Outline Introduction Structural alignment of RN
Method for ligning RN Secondary Structures Jason T. L. Wang New Jersey Institute of Technology J Liu, JTL Wang, J Hu and B Tian, BM Bioinformatics, 2005 1 Outline Introduction Structural alignment of RN
A Model of Proteostatic Energy Cost and Its Use in Analysis of Proteome Trends and Sequence Evolution.
 Downloaded from orbit.u.dk on: Dec 18, 2017 A Model of Proteostatic Energy Cost and Its Use in Analysis of Proteome Trends and Sequence Evolution. Kepp, Kasper Planeta Published in: PLoS ONE Link to article,
Downloaded from orbit.u.dk on: Dec 18, 2017 A Model of Proteostatic Energy Cost and Its Use in Analysis of Proteome Trends and Sequence Evolution. Kepp, Kasper Planeta Published in: PLoS ONE Link to article,
Reconstructing the History of Large-scale Genomic Changes. Jian Ma
 Reconstructing the History of Large-scale Genomic Changes Jian Ma The Human Genome: the blueprint of our body Initial sequencing and analysis of the human genome International Human Genome Sequencing Consortium*
Reconstructing the History of Large-scale Genomic Changes Jian Ma The Human Genome: the blueprint of our body Initial sequencing and analysis of the human genome International Human Genome Sequencing Consortium*
CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES
 INTRODUCTION CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES This worksheet complements the Click and Learn developed in conjunction with the 2011 Holiday Lectures on Science, Bones, Stones, and Genes:
INTRODUCTION CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES This worksheet complements the Click and Learn developed in conjunction with the 2011 Holiday Lectures on Science, Bones, Stones, and Genes:
Concepts and Methods in Molecular Divergence Time Estimation
 Concepts and Methods in Molecular Divergence Time Estimation 26 November 2012 Prashant P. Sharma American Museum of Natural History Overview 1. Why do we date trees? 2. The molecular clock 3. Local clocks
Concepts and Methods in Molecular Divergence Time Estimation 26 November 2012 Prashant P. Sharma American Museum of Natural History Overview 1. Why do we date trees? 2. The molecular clock 3. Local clocks
Protein evolutionary rates that is, the rate by which protein
 Slow protein evolutionary rates are dictated by surface core association Ágnes Tóth-Petróczy and Dan S. Tawfik 1 Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel
Slow protein evolutionary rates are dictated by surface core association Ágnes Tóth-Petróczy and Dan S. Tawfik 1 Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel
Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection
 Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection Ziheng Yang,* Wendy S.W. Wong, and Rasmus Nielsen à *Department of Biology, University College London, London, United Kingdom;
Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection Ziheng Yang,* Wendy S.W. Wong, and Rasmus Nielsen à *Department of Biology, University College London, London, United Kingdom;
Molecular phylogeny How to infer phylogenetic trees using molecular sequences
 Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 200 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 200 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Investigating Evolutionary Relationships between Species through the Light of Graph Theory based on the Multiplet Structure of the Genetic Code
 07 IEEE 7th International Advance Computing Conference Investigating Evolutionary Relationships between Species through the Light of Graph Theory based on the Multiplet Structure of the Genetic Code Antara
07 IEEE 7th International Advance Computing Conference Investigating Evolutionary Relationships between Species through the Light of Graph Theory based on the Multiplet Structure of the Genetic Code Antara
Journal of Molecular Evolution Springer-Verlag New York Inc. 1994
 J Mol Evol (1994) 39:306-314 Journal of Molecular Evolution Springer-Verlag New York Inc. 1994 Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate
J Mol Evol (1994) 39:306-314 Journal of Molecular Evolution Springer-Verlag New York Inc. 1994 Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate
Phylogenetic Trees. How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species?
 Why? Phylogenetic Trees How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species? The saying Don t judge a book by its cover. could be applied
Why? Phylogenetic Trees How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species? The saying Don t judge a book by its cover. could be applied
Multiple sequence alignment
 Multiple sequence alignment Multiple sequence alignment: today s goals to define what a multiple sequence alignment is and how it is generated; to describe profile HMMs to introduce databases of multiple
Multiple sequence alignment Multiple sequence alignment: today s goals to define what a multiple sequence alignment is and how it is generated; to describe profile HMMs to introduce databases of multiple
Determining the Null Model for Detecting Adaptive Convergence from Genomic Data: A Case Study using Echolocating Mammals
 Determining the Null Model for Detecting Adaptive Convergence from Genomic Data: A Case Study using Echolocating Mammals Gregg W.C. Thomas 1 and Matthew W. Hahn*,1,2 1 School of Informatics and Computing,
Determining the Null Model for Detecting Adaptive Convergence from Genomic Data: A Case Study using Echolocating Mammals Gregg W.C. Thomas 1 and Matthew W. Hahn*,1,2 1 School of Informatics and Computing,
Erasing Errors Due to Alignment Ambiguity When Estimating Positive Selection
 Article (Methods) Erasing Errors Due to Alignment Ambiguity When Estimating Positive Selection Authors Benjamin Redelings 1,2 1 Biology Department, Duke University 2 National Evolutionary Synthesis Center
Article (Methods) Erasing Errors Due to Alignment Ambiguity When Estimating Positive Selection Authors Benjamin Redelings 1,2 1 Biology Department, Duke University 2 National Evolutionary Synthesis Center
Constructing Evolutionary/Phylogenetic Trees
 Constructing Evolutionary/Phylogenetic Trees 2 broad categories: Distance-based methods Ultrametric Additive: UPGMA Transformed Distance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
Constructing Evolutionary/Phylogenetic Trees 2 broad categories: Distance-based methods Ultrametric Additive: UPGMA Transformed Distance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
Application of new distance matrix to phylogenetic tree construction
 Application of new distance matrix to phylogenetic tree construction P.V.Lakshmi Computer Science & Engg Dept GITAM Institute of Technology GITAM University Andhra Pradesh India Allam Appa Rao Jawaharlal
Application of new distance matrix to phylogenetic tree construction P.V.Lakshmi Computer Science & Engg Dept GITAM Institute of Technology GITAM University Andhra Pradesh India Allam Appa Rao Jawaharlal
Software GASP: Gapped Ancestral Sequence Prediction for proteins Richard J Edwards* and Denis C Shields
 BMC Bioinformatics BioMed Central Software GASP: Gapped Ancestral Sequence Prediction for proteins Richard J Edwards* and Denis C Shields Open Access Address: Bioinformatics Core, Clinical Pharmacology,
BMC Bioinformatics BioMed Central Software GASP: Gapped Ancestral Sequence Prediction for proteins Richard J Edwards* and Denis C Shields Open Access Address: Bioinformatics Core, Clinical Pharmacology,
Motifs and Logos. Six Introduction to Bioinformatics. Importance and Abundance of Motifs. Getting the CDS. From DNA to Protein 6.1.
 Motifs and Logos Six Discovering Genomics, Proteomics, and Bioinformatics by A. Malcolm Campbell and Laurie J. Heyer Chapter 2 Genome Sequence Acquisition and Analysis Sami Khuri Department of Computer
Motifs and Logos Six Discovering Genomics, Proteomics, and Bioinformatics by A. Malcolm Campbell and Laurie J. Heyer Chapter 2 Genome Sequence Acquisition and Analysis Sami Khuri Department of Computer
Structural Perspectives on Drug Resistance
 Structural Perspectives on Drug Resistance Irene Weber Departments of Biology and Chemistry Molecular Basis of Disease Program Georgia State University Atlanta, GA, USA What have we learned from 20 years
Structural Perspectives on Drug Resistance Irene Weber Departments of Biology and Chemistry Molecular Basis of Disease Program Georgia State University Atlanta, GA, USA What have we learned from 20 years
Simple Methods for Testing the Molecular Evolutionary Clock Hypothesis
 Copyright 0 1998 by the Genetics Society of America Simple s for Testing the Molecular Evolutionary Clock Hypothesis Fumio Tajima Department of Population Genetics, National Institute of Genetics, Mishima,
Copyright 0 1998 by the Genetics Society of America Simple s for Testing the Molecular Evolutionary Clock Hypothesis Fumio Tajima Department of Population Genetics, National Institute of Genetics, Mishima,
Potts Models and Protein Covariation. Allan Haldane Ron Levy Group
 Temple University Structural Bioinformatics II Potts Models and Protein Covariation Allan Haldane Ron Levy Group Outline The Evolutionary Origins of Protein Sequence Variation Contents of the Pfam database
Temple University Structural Bioinformatics II Potts Models and Protein Covariation Allan Haldane Ron Levy Group Outline The Evolutionary Origins of Protein Sequence Variation Contents of the Pfam database
Objectives. Comparison and Analysis of Heat Shock Proteins in Organisms of the Kingdom Viridiplantae. Emily Germain 1,2 Mentor Dr.
 Comparison and Analysis of Heat Shock Proteins in Organisms of the Kingdom Viridiplantae Emily Germain 1,2 Mentor Dr. Hugh Nicholas 3 1 Bioengineering & Bioinformatics Summer Institute, Department of Computational
Comparison and Analysis of Heat Shock Proteins in Organisms of the Kingdom Viridiplantae Emily Germain 1,2 Mentor Dr. Hugh Nicholas 3 1 Bioengineering & Bioinformatics Summer Institute, Department of Computational
Molecular evolution. Joe Felsenstein. GENOME 453, Autumn Molecular evolution p.1/49
 Molecular evolution Joe Felsenstein GENOME 453, utumn 2009 Molecular evolution p.1/49 data example for phylogeny inference Five DN sequences, for some gene in an imaginary group of species whose names
Molecular evolution Joe Felsenstein GENOME 453, utumn 2009 Molecular evolution p.1/49 data example for phylogeny inference Five DN sequences, for some gene in an imaginary group of species whose names
Supplementary Information for Hurst et al.: Causes of trends of amino acid gain and loss
 Supplementary Information for Hurst et al.: Causes of trends of amino acid gain and loss Methods Identification of orthologues, alignment and evolutionary distances A preliminary set of orthologues was
Supplementary Information for Hurst et al.: Causes of trends of amino acid gain and loss Methods Identification of orthologues, alignment and evolutionary distances A preliminary set of orthologues was
Hidden Markov Models for Unaligned DNA Sequence Comparison. James Cook University, Townsville, QLD 4811, Australia.
 Hidden Markov Models for Unaligned DNA Sequence Comparison TUAN D. PHAM 1, DOMINIK BECK 2, and DENIS I. CRANE 3 1 School of Information Technology James Cook University, Townsville, QLD 4811, Australia.
Hidden Markov Models for Unaligned DNA Sequence Comparison TUAN D. PHAM 1, DOMINIK BECK 2, and DENIS I. CRANE 3 1 School of Information Technology James Cook University, Townsville, QLD 4811, Australia.
A bioinformatics approach to the structural and functional analysis of the glycogen phosphorylase protein family
 A bioinformatics approach to the structural and functional analysis of the glycogen phosphorylase protein family Jieming Shen 1,2 and Hugh B. Nicholas, Jr. 3 1 Bioengineering and Bioinformatics Summer
A bioinformatics approach to the structural and functional analysis of the glycogen phosphorylase protein family Jieming Shen 1,2 and Hugh B. Nicholas, Jr. 3 1 Bioengineering and Bioinformatics Summer
Week 8: Testing trees, Bootstraps, jackknifes, gene frequencies
 Week 8: Testing trees, ootstraps, jackknifes, gene frequencies Genome 570 ebruary, 2016 Week 8: Testing trees, ootstraps, jackknifes, gene frequencies p.1/69 density e log (density) Normal distribution:
Week 8: Testing trees, ootstraps, jackknifes, gene frequencies Genome 570 ebruary, 2016 Week 8: Testing trees, ootstraps, jackknifes, gene frequencies p.1/69 density e log (density) Normal distribution:
Supplementary Figure 1 Crystal packing of ClR and electron density maps. Crystal packing of type A crystal (a) and type B crystal (b).
 Supplementary Figure 1 Crystal packing of ClR and electron density maps. Crystal packing of type A crystal (a) and type B crystal (b). Crystal contacts at B-C loop are magnified and stereo view of A-weighted
Supplementary Figure 1 Crystal packing of ClR and electron density maps. Crystal packing of type A crystal (a) and type B crystal (b). Crystal contacts at B-C loop are magnified and stereo view of A-weighted
98 Algorithms in Bioinformatics I, WS 06, ZBIT, D. Huson, December 6, 2006
 98 Algorithms in Bioinformatics I, WS 06, ZBIT, D. Huson, December 6, 2006 8.3.1 Simple energy minimization Maximizing the number of base pairs as described above does not lead to good structure predictions.
98 Algorithms in Bioinformatics I, WS 06, ZBIT, D. Huson, December 6, 2006 8.3.1 Simple energy minimization Maximizing the number of base pairs as described above does not lead to good structure predictions.
Week 6: Protein sequence models, likelihood, hidden Markov models
 Week 6: Protein sequence models, likelihood, hidden Markov models Genome 570 February, 2016 Week 6: Protein sequence models, likelihood, hidden Markov models p.1/57 Variation of rates of evolution across
Week 6: Protein sequence models, likelihood, hidden Markov models Genome 570 February, 2016 Week 6: Protein sequence models, likelihood, hidden Markov models p.1/57 Variation of rates of evolution across
Achievement of Protein Thermostability by Amino Acid Substitution. 2018/6/30 M1 Majima Sohei
 Achievement of Protein Thermostability by Amino Acid Substitution 2018/6/30 M1 Majima Sohei Contents of Today s seminar Introduction Study on hyperthermophilic enzymes to understand the origin of thermostability
Achievement of Protein Thermostability by Amino Acid Substitution 2018/6/30 M1 Majima Sohei Contents of Today s seminar Introduction Study on hyperthermophilic enzymes to understand the origin of thermostability
Monomeric Clavularia CFP July 19, 2006
 SUPPLEMENTARY MATERIAL Table 1S Rationale for design of the synthetic gene library. Residue number Mutation Codon a Rationale His42 Leu44 Gln66 (residue of chromophore) His, Asn, Gln, Lys Leu, Val, Ala,
SUPPLEMENTARY MATERIAL Table 1S Rationale for design of the synthetic gene library. Residue number Mutation Codon a Rationale His42 Leu44 Gln66 (residue of chromophore) His, Asn, Gln, Lys Leu, Val, Ala,
Position-specific scoring matrices (PSSM)
 Regulatory Sequence nalysis Position-specific scoring matrices (PSSM) Jacques van Helden Jacques.van-Helden@univ-amu.fr Université d ix-marseille, France Technological dvances for Genomics and Clinics
Regulatory Sequence nalysis Position-specific scoring matrices (PSSM) Jacques van Helden Jacques.van-Helden@univ-amu.fr Université d ix-marseille, France Technological dvances for Genomics and Clinics
High-throughput identification of protein mutant stability computed from a double mutant fitness landscape
 FOR THE RECORD High-throughput identification of protein mutant stability computed from a double mutant fitness landscape Nicholas C. Wu, 1,2,3 C. Anders Olson, 1 and Ren Sun 1 * 1 Department of Molecular
FOR THE RECORD High-throughput identification of protein mutant stability computed from a double mutant fitness landscape Nicholas C. Wu, 1,2,3 C. Anders Olson, 1 and Ren Sun 1 * 1 Department of Molecular
Local Alignment Statistics
 Local Alignment Statistics Stephen Altschul National Center for Biotechnology Information National Library of Medicine National Institutes of Health Bethesda, MD Central Issues in Biological Sequence Comparison
Local Alignment Statistics Stephen Altschul National Center for Biotechnology Information National Library of Medicine National Institutes of Health Bethesda, MD Central Issues in Biological Sequence Comparison
