Supervised classification for structured data: Applications in bio- and chemoinformatics
|
|
|
- Randell Rodgers
- 5 years ago
- Views:
Transcription
1 Supervised classification for structured data: Applications in bio- and chemoinformatics Jean-Philippe Vert Mines ParisTech / Curie Institute / Inserm The third school on The Analysis of Patterns, Pula Science Park, Italy, June 1, 2009 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 1 / 114
2 Virtual screening for drug discovery active inactive inactive active inactive active NCI AIDS screen results (from Jean-Philippe Vert (ParisTech) The Analysis of Patterns 2 / 114
3 Image retrieval and classification From Harchaoui and Bach (2007). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 3 / 114
4 Cancer diagnosis N Glycan biosynthesis Glycolysis / Gluconeogenesis Porphyrin and chlorophyll metabolism Protein kinases Sulfur metabolism Nitrogen, asparagine metabolism - Riboflavin metabolism Folate biosynthesis DNA and RNA polymerase subunits Biosynthesis of steroids, ergosterol metabolism Lysine biosynthesis Oxidative phosphorylation, TCA cycle Phenylalanine, tyrosine and tryptophan biosynthesis Jean-Philippe Vert (ParisTech) Purine metabolism The Analysis of Patterns 4 / 114
5 Cancer prognosis Jean-Philippe Vert (ParisTech) The Analysis of Patterns 5 / 114
6 Pattern recognition, aka supervised classification active 4 inactive 2 0 inactive 2 active inactive active Jean-Philippe Vert (ParisTech) 4 The Analysis of Patterns 6 / 114
7 Pattern recognition, aka supervised classification active 4 inactive 2 0 inactive 2 active inactive active Jean-Philippe Vert (ParisTech) 4 The Analysis of Patterns 6 / 114
8 Pattern recognition, aka supervised classification active 4 inactive 2 0 inactive 2 active inactive active Jean-Philippe Vert (ParisTech) 4 The Analysis of Patterns 6 / 114
9 Pattern recognition, aka supervised classification active 4 inactive 2 0 inactive 2 active inactive active Jean-Philippe Vert (ParisTech) 4 The Analysis of Patterns 6 / 114
10 Pattern recognition, aka supervised classification Challenges High dimension Few samples Structured data Heterogeneous data Prior knowledge Fast and scalable implementations Interpretable models Jean-Philippe Vert (ParisTech) The Analysis of Patterns 7 / 114
11 Formalization The problem Given a set of training instances (x 1, y 1 ),..., (x n, y n ), where x i X are data and y i Y are continuous or discrete variables of interest, Estimate a function y = f (x) where x is any new data to be labeled. f should be accurate and intepretable. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 8 / 114
12 Linear classifiers The model Each sample x X is represented by a vector of features (or descriptors, or patterns): Φ(x) = (Φ 1 (x),..., Φ p (x)) Based on the training set we estimate a linear function: f β (x) = p β i Φ i (x) = β Φ(x). i=1 Two (related) questions How to design the features Φ(x)? How to estimate the model β? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 9 / 114
13 Linear classifiers The model Each sample x X is represented by a vector of features (or descriptors, or patterns): Φ(x) = (Φ 1 (x),..., Φ p (x)) Based on the training set we estimate a linear function: f β (x) = p β i Φ i (x) = β Φ(x). i=1 Two (related) questions How to design the features Φ(x)? How to estimate the model β? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 9 / 114
14 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 10 / 114
15 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 11 / 114
16 Motivation active inactive inactive active inactive active NCI AIDS screen results (from Jean-Philippe Vert (ParisTech) The Analysis of Patterns 12 / 114
17 The approach 1 Represent explicitly each graph x by a vector of fixed dimension Φ(x) R p. 2 Use an algorithm for regression or pattern recognition in R p. X Jean-Philippe Vert (ParisTech) The Analysis of Patterns 13 / 114
18 The approach 1 Represent explicitly each graph x by a vector of fixed dimension Φ(x) R p. 2 Use an algorithm for regression or pattern recognition in R p. X φ H Jean-Philippe Vert (ParisTech) The Analysis of Patterns 13 / 114
19 The approach 1 Represent explicitly each graph x by a vector of fixed dimension Φ(x) R p. 2 Use an algorithm for regression or pattern recognition in R p. X φ H Jean-Philippe Vert (ParisTech) The Analysis of Patterns 13 / 114
20 Example 2D structural keys in chemoinformatics Index a molecule by a binary fingerprint defined by a limited set of pre-defined stuctures O O O N N N O O O N O Use a machine learning algorithms such as SVM, NN, PLS, decision tree,... O Jean-Philippe Vert (ParisTech) The Analysis of Patterns 14 / 114
21 Challenge: which descriptors (patterns)? O O O N N N O O O N O O Expressiveness: they should retain as much information as possible from the graph Computation : they should be fast to compute Large dimension of the vector representation: memory storage, speed, statistical issues Jean-Philippe Vert (ParisTech) The Analysis of Patterns 15 / 114
22 Indexing by substructures O O O N N N O O O N O O Often we believe that the presence substructures are important predictive patterns Hence it makes sense to represent a graph by features that indicate the presence (or the number of occurrences) of particular substructures However, detecting the presence of particular substructures may be computationally challenging... Jean-Philippe Vert (ParisTech) The Analysis of Patterns 16 / 114
23 Subgraphs Definition A subgraph of a graph (V, E) is a connected graph (V, E ) with V V and E E. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 17 / 114
24 Indexing by all subgraphs? Theorem Computing all subgraph occurrences is NP-hard. Proof. The linear graph of size n is a subgraph of a graph X with n vertices iff X has an Hamiltonian path The decision problem whether a graph has a Hamiltonian path is NP-complete. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 18 / 114
25 Indexing by all subgraphs? Theorem Computing all subgraph occurrences is NP-hard. Proof. The linear graph of size n is a subgraph of a graph X with n vertices iff X has an Hamiltonian path The decision problem whether a graph has a Hamiltonian path is NP-complete. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 18 / 114
26 Indexing by all subgraphs? Theorem Computing all subgraph occurrences is NP-hard. Proof. The linear graph of size n is a subgraph of a graph X with n vertices iff X has an Hamiltonian path The decision problem whether a graph has a Hamiltonian path is NP-complete. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 18 / 114
27 Paths Definition A path of a graph (V, E) is sequence of distinct vertices v 1,..., v n V (i j = v i v j ) such that (v i, v i+1 ) E for i = 1,..., n 1. Equivalently the paths are the linear subgraphs. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 19 / 114
28 Indexing by all paths? A B A A B (0,...,0,1,0,...,0,1,0,...) A A A B A Theorem Computing all path occurrences is NP-hard. Proof. Same as for subgraphs. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 20 / 114
29 Indexing by all paths? A B A A B (0,...,0,1,0,...,0,1,0,...) A A A B A Theorem Computing all path occurrences is NP-hard. Proof. Same as for subgraphs. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 20 / 114
30 Indexing by all paths? A B A A B (0,...,0,1,0,...,0,1,0,...) A A A B A Theorem Computing all path occurrences is NP-hard. Proof. Same as for subgraphs. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 20 / 114
31 Indexing by what? Substructure selection We can imagine more limited sets of substuctures that lead to more computationnally efficient indexing (non-exhaustive list) substructures selected by domain knowledge (MDL fingerprint) all path up to length k (Openeye fingerprint, Nicholls 2005) all shortest paths (Borgwardt and Kriegel, 2005) all subgraphs up to k vertices (graphlet kernel, Sherashidze et al., 2009) all frequent subgraphs in the database (Helma et al., 2004) Jean-Philippe Vert (ParisTech) The Analysis of Patterns 21 / 114
32 Example : Indexing by all shortest paths A A B A B A B A A B (0,...,0,2,0,...,0,1,0,...) A A A A A B Properties (Borgwardt and Kriegel, 2005) There are O(n 2 ) shortest paths. The vector of counts can be computed in O(n 4 ) with the Floyd-Warshall algorithm. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 22 / 114
33 Example : Indexing by all shortest paths A A B A B A B A A B (0,...,0,2,0,...,0,1,0,...) A A A A A B Properties (Borgwardt and Kriegel, 2005) There are O(n 2 ) shortest paths. The vector of counts can be computed in O(n 4 ) with the Floyd-Warshall algorithm. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 22 / 114
34 Example : Indexing by all subgraphs up to k vertices Properties (Shervashidze et al., 2009) Naive enumeration scales as O(n k ). Enumeration of connected graphlets in O(nd k 1 ) for graphs with degree d and k 5. Randomly sample subgraphs if enumeration is infeasible. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 23 / 114
35 Example : Indexing by all subgraphs up to k vertices Properties (Shervashidze et al., 2009) Naive enumeration scales as O(n k ). Enumeration of connected graphlets in O(nd k 1 ) for graphs with degree d and k 5. Randomly sample subgraphs if enumeration is infeasible. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 23 / 114
36 Summary Explicit computation of substructure occurrences can be computationnally prohibitive (subgraph, paths) Several ideas to reduce the set of substructures considered In practice, NP-hardness may not be so prohibitive (e.g., graphs with small degrees), the strategy followed should depend on the data considered. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 24 / 114
37 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 25 / 114
38 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 26 / 114
39 Positive definite kernels Definition Let Φ(x) be a vector representation of the data x The kernel between two graphs is defined by: K (x, x ) = Φ(x) Φ(x ). X φ F Jean-Philippe Vert (ParisTech) The Analysis of Patterns 27 / 114
40 The kernel trick The trick Many linear algorithms for regression or pattern recognition can be expressed only in terms of inner products between vectors Computing the kernel is often more efficient than computing Φ(x), especially in high or infinite dimensions! Perhaps we can consider more features with kernels than with explicit feature computation? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 28 / 114
41 Learning linear classifiers with kernels Training the model Minimize an empirical risk on the training samples: min β R p+1 R emp (β) = 1 n... subject to a constraint on β: n l(β Φ(x i ), y i ), i=1 β C. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 29 / 114
42 Making kernels Two important strategies (not the only ones!) Feature design : K (x, x ) = Φ(x) Φ(x ). We illustrate this idea with graph kernels. Regularization design : β C. We illustrate this idea with kernels for microarray data. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 30 / 114
43 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 31 / 114
44 The idea 1 Represent implicitly each graph x by a vector Φ(x) H through the kernel K (x, x ) = Φ(x) Φ(x ). 2 Use a kernel method for classification in H. X Jean-Philippe Vert (ParisTech) The Analysis of Patterns 32 / 114
45 The idea 1 Represent implicitly each graph x by a vector Φ(x) H through the kernel K (x, x ) = Φ(x) Φ(x ). 2 Use a kernel method for classification in H. X φ H Jean-Philippe Vert (ParisTech) The Analysis of Patterns 32 / 114
46 The idea 1 Represent implicitly each graph x by a vector Φ(x) H through the kernel K (x, x ) = Φ(x) Φ(x ). 2 Use a kernel method for classification in H. X φ H Jean-Philippe Vert (ParisTech) The Analysis of Patterns 32 / 114
47 Expressiveness vs Complexity Definition: Complete graph kernels A graph kernel is complete if it separates non-isomorphic graphs, i.e.: G 1, G 2 X, d K (G 1, G 2 ) = 0 = G 1 G 2. Equivalently, Φ(G 1 ) Φ(G 1 ) if G 1 and G 2 are not isomorphic. Expressiveness vs Complexity trade-off If a graph kernel is not complete, then there is no hope to learn all possible functions over X : the kernel is not expressive enough. On the other hand, kernel computation must be tractable, i.e., no more than polynomial (with small degree) for practical applications. Can we define tractable and expressive graph kernels? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 33 / 114
48 Expressiveness vs Complexity Definition: Complete graph kernels A graph kernel is complete if it separates non-isomorphic graphs, i.e.: G 1, G 2 X, d K (G 1, G 2 ) = 0 = G 1 G 2. Equivalently, Φ(G 1 ) Φ(G 1 ) if G 1 and G 2 are not isomorphic. Expressiveness vs Complexity trade-off If a graph kernel is not complete, then there is no hope to learn all possible functions over X : the kernel is not expressive enough. On the other hand, kernel computation must be tractable, i.e., no more than polynomial (with small degree) for practical applications. Can we define tractable and expressive graph kernels? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 33 / 114
49 Complexity of complete kernels Proposition (Gärtner et al., 2003) Computing any complete graph kernel is at least as hard as the graph isomorphism problem. Proof For any kernel K the complexity of computing d K is the same as the complexity of computing K, because: d K (G 1, G 2 ) 2 = K (G 1, G 1 ) + K (G 2, G 2 ) 2K (G 1, G 2 ). If K is a complete graph kernel, then computing d K solves the graph isomorphism problem (d K (G 1, G 2 ) = 0 iff G 1 G 2 ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 34 / 114
50 Complexity of complete kernels Proposition (Gärtner et al., 2003) Computing any complete graph kernel is at least as hard as the graph isomorphism problem. Proof For any kernel K the complexity of computing d K is the same as the complexity of computing K, because: d K (G 1, G 2 ) 2 = K (G 1, G 1 ) + K (G 2, G 2 ) 2K (G 1, G 2 ). If K is a complete graph kernel, then computing d K solves the graph isomorphism problem (d K (G 1, G 2 ) = 0 iff G 1 G 2 ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 34 / 114
51 Subgraph kernel Definition Let (λ G ) G X a set or nonnegative real-valued weights For any graph G X, let H X, Φ H (G) = { G is a subgraph of G : G H }. The subgraph kernel between any two graphs G 1 and G 2 X is defined by: K subgraph (G 1, G 2 ) = H X λ H Φ H (G 1 )Φ H (G 2 ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 35 / 114
52 Subgraph kernel complexity Proposition (Gärtner et al., 2003) Computing the subgraph kernel is NP-hard. Proof (1/2) Let P n be the path graph with n vertices. Subgraphs of P n are path graphs: Φ(P n ) = ne P1 + (n 1)e P e Pn. The vectors Φ(P 1 ),..., Φ(P n ) are linearly independent, therefore: e Pn = n α i Φ(P i ), i=1 where the coefficients α i can be found in polynomial time (solving a n n triangular system). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 36 / 114
53 Subgraph kernel complexity Proposition (Gärtner et al., 2003) Computing the subgraph kernel is NP-hard. Proof (1/2) Let P n be the path graph with n vertices. Subgraphs of P n are path graphs: Φ(P n ) = ne P1 + (n 1)e P e Pn. The vectors Φ(P 1 ),..., Φ(P n ) are linearly independent, therefore: e Pn = n α i Φ(P i ), i=1 where the coefficients α i can be found in polynomial time (solving a n n triangular system). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 36 / 114
54 Subgraph kernel complexity Proposition (Gärtner et al., 2003) Computing the subgraph kernel is NP-hard. Proof (2/2) If G is a graph with n vertices, then it has a path that visits each node exactly once (Hamiltonian path) if and only if Φ(G) e n > 0, i.e., ( n ) n Φ(G) α i Φ(P i ) = α i K subgraph (G, P i ) > 0. i=1 The decision problem whether a graph has a Hamiltonian path is NP-complete. i=1 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 37 / 114
55 Path kernel A B A A B (0,...,0,1,0,...,0,1,0,...) A A A B A Definition The path kernel is the subgraph kernel restricted to paths, i.e., K path (G 1, G 2 ) = H P λ H Φ H (G 1 )Φ H (G 2 ), where P X is the set of path graphs. Proposition (Gärtner et al., 2003) Computing the path kernel is NP-hard. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 38 / 114
56 Path kernel A B A A B (0,...,0,1,0,...,0,1,0,...) A A A B A Definition The path kernel is the subgraph kernel restricted to paths, i.e., K path (G 1, G 2 ) = H P λ H Φ H (G 1 )Φ H (G 2 ), where P X is the set of path graphs. Proposition (Gärtner et al., 2003) Computing the path kernel is NP-hard. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 38 / 114
57 Summary Expressiveness vs Complexity trade-off It is intractable to compute complete graph kernels. It is intractable to compute the subgraph kernels. Restricting subgraphs to be linear does not help: it is also intractable to compute the path kernel. One approach to define polynomial time computable graph kernels is to have the feature space be made up of graphs homomorphic to subgraphs, e.g., to consider walks instead of paths. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 39 / 114
58 Walks Definition A walk of a graph (V, E) is sequence of v 1,..., v n V such that (v i, v i+1 ) E for i = 1,..., n 1. We note W n (G) the set of walks with n vertices of the graph G, and W(G) the set of all walks. etc... Jean-Philippe Vert (ParisTech) The Analysis of Patterns 40 / 114
59 Walks paths Jean-Philippe Vert (ParisTech) The Analysis of Patterns 41 / 114
60 Walk kernel Definition Let S n denote the set of all possible label sequences of walks of length n (including vertices and edges labels), and S = n 1 S n. For any graph X let a weight λ G (w) be associated to each walk w W(G). Let the feature vector Φ(G) = (Φ s (G)) s S be defined by: Φ s (G) = w W(G) λ G (w)1 (s is the label sequence of w). A walk kernel is a graph kernel defined by: K walk (G 1, G 2 ) = s S Φ s (G 1 )Φ s (G 2 ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 42 / 114
61 Walk kernel Definition Let S n denote the set of all possible label sequences of walks of length n (including vertices and edges labels), and S = n 1 S n. For any graph X let a weight λ G (w) be associated to each walk w W(G). Let the feature vector Φ(G) = (Φ s (G)) s S be defined by: Φ s (G) = w W(G) λ G (w)1 (s is the label sequence of w). A walk kernel is a graph kernel defined by: K walk (G 1, G 2 ) = s S Φ s (G 1 )Φ s (G 2 ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 42 / 114
62 Walk kernel examples Examples The nth-order walk kernel is the walk kernel with λ G (w) = 1 if the length of w is n, 0 otherwise. It compares two graphs through their common walks of length n. The random walk kernel is obtained with λ G (w) = P G (w), where P G is a Markov random walk on G. In that case we have: K (G 1, G 2 ) = P(label(W 1 ) = label(w 2 )), where W 1 and W 2 are two independant random walks on G 1 and G 2, respectively (Kashima et al., 2003). The geometric walk kernel is obtained (when it converges) with λ G (w) = β length(w), for β > 0. In that case the feature space is of infinite dimension (Gärtner et al., 2003). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 43 / 114
63 Walk kernel examples Examples The nth-order walk kernel is the walk kernel with λ G (w) = 1 if the length of w is n, 0 otherwise. It compares two graphs through their common walks of length n. The random walk kernel is obtained with λ G (w) = P G (w), where P G is a Markov random walk on G. In that case we have: K (G 1, G 2 ) = P(label(W 1 ) = label(w 2 )), where W 1 and W 2 are two independant random walks on G 1 and G 2, respectively (Kashima et al., 2003). The geometric walk kernel is obtained (when it converges) with λ G (w) = β length(w), for β > 0. In that case the feature space is of infinite dimension (Gärtner et al., 2003). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 43 / 114
64 Walk kernel examples Examples The nth-order walk kernel is the walk kernel with λ G (w) = 1 if the length of w is n, 0 otherwise. It compares two graphs through their common walks of length n. The random walk kernel is obtained with λ G (w) = P G (w), where P G is a Markov random walk on G. In that case we have: K (G 1, G 2 ) = P(label(W 1 ) = label(w 2 )), where W 1 and W 2 are two independant random walks on G 1 and G 2, respectively (Kashima et al., 2003). The geometric walk kernel is obtained (when it converges) with λ G (w) = β length(w), for β > 0. In that case the feature space is of infinite dimension (Gärtner et al., 2003). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 43 / 114
65 Computation of walk kernels Proposition These three kernels (nth-order, random and geometric walk kernels) can be computed efficiently in polynomial time. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 44 / 114
66 Product graph Definition Let G 1 = (V 1, E 1 ) and G 2 = (V 2, E 2 ) be two graphs with labeled vertices. The product graph G = G 1 G 2 is the graph G = (V, E) with: 1 V = {(v 1, v 2 ) V 1 V 2 : v 1 and v 2 have the same label}, 2 E {( = (v1, v 2 ), (v 1, v 2 )) V V : (v 1, v 1 ) E 1 and (v 2, v 2 ) E } 2. 1 a b 1b 2a 1d 2 c 1a 2b 3c 2d 3e 3 4 d e 4c 4e G1 G2 G1 x G2 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 45 / 114
67 Walk kernel and product graph Lemma There is a bijection between: 1 The pairs of walks w 1 W n (G 1 ) and w 2 W n (G 2 ) with the same label sequences, 2 The walks on the product graph w W n (G 1 G 2 ). Corollary K walk (G 1, G 2 ) = Φ s (G 1 )Φ s (G 2 ) s S = λ G1 (w 1 )λ G2 (w 2 )1(l(w 1 ) = l(w 2 )) = (w 1,w 2 ) W(G 1 ) W(G 1 ) w W(G 1 G 2 ) λ G1 G 2 (w). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 46 / 114
68 Walk kernel and product graph Lemma There is a bijection between: 1 The pairs of walks w 1 W n (G 1 ) and w 2 W n (G 2 ) with the same label sequences, 2 The walks on the product graph w W n (G 1 G 2 ). Corollary K walk (G 1, G 2 ) = Φ s (G 1 )Φ s (G 2 ) s S = λ G1 (w 1 )λ G2 (w 2 )1(l(w 1 ) = l(w 2 )) = (w 1,w 2 ) W(G 1 ) W(G 1 ) w W(G 1 G 2 ) λ G1 G 2 (w). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 46 / 114
69 Computation of the nth-order walk kernel For the nth-order walk kernel we have λ G1 G 2 (w) = 1 if the length of w is n, 0 otherwise. Therefore: K nth order (G 1, G 2 ) = w W n(g 1 G 2 ) Let A be the adjacency matrix of G 1 G 2. Then we get: 1. K nth order (G 1, G 2 ) = i,j [A n ] i,j = 1 A n 1. Computation in O(n G 1 G 2 d 1 d 2 ), where d i is the maximum degree of G i. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 47 / 114
70 Computation of random and geometric walk kernels In both cases λ G (w) for a walk w = v 1... v n can be decomposed as: n λ G (v 1... v n ) = λ i (v 1 ) λ t (v i 1, v i ). Let Λ i be the vector of λ i (v) and Λ t be the matrix of λ t (v, v ): n K walk (G 1, G 2 ) = λ i (v 1 ) λ t (v i 1, v i ) = i=2 n=1 w W n(g 1 G 2 ) Λ i Λ n t 1 n=0 = Λ i (I Λ t ) 1 1 Computation in O( G 1 3 G 2 3 ) i=2 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 48 / 114
71 Extensions 1: label enrichment Atom relabebling with the Morgan index N 1 O N 3 O N 5 O 3 No Morgan Indices O 1 Order 1 indices O 1 Order 2 indices O 3 Compromise between fingerprints and structural keys features. Other relabeling schemes are possible (graph coloring). Faster computation with more labels (less matches implies a smaller product graph). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 49 / 114
72 Extension 2: Non-tottering walk kernel Tottering walks A tottering walk is a walk w = v 1... v n with v i = v i+2 for some i. Non tottering Tottering Tottering walks seem irrelevant for many applications Focusing on non-tottering walks is a way to get closer to the path kernel (e.g., equivalent on trees). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 50 / 114
73 Computation of the non-tottering walk kernel (Mahé et al., 2005) Second-order Markov random walk to prevent tottering walks Written as a first-order Markov random walk on an augmented graph Normal walk kernel on the augmented graph (which is always a directed graph). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 51 / 114
74 Extension 3: Subtree kernels Jean-Philippe Vert (ParisTech) The Analysis of Patterns 52 / 114
75 Example: Tree-like fragments of molecules O N C N C C... C N C N N N C O C N O N C C C Jean-Philippe Vert (ParisTech) The Analysis of Patterns 53 / 114
76 Computation of the subtree kernel Like the walk kernel, amounts to compute the (weighted) number of subtrees in the product graph. Recursion: if T (v, n) denotes the weighted number of subtrees of depth n rooted at the vertex v, then: T (v, n + 1) = λ t (v, v )T (v, n), R N (v) v R where N (v) is the set of neighbors of v. Can be combined with the non-tottering graph transformation as preprocessing to obtain the non-tottering subtree kernel. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 54 / 114
77 Application in chemoinformatics (Mahé et al., 2004) MUTAG dataset aromatic/hetero-aromatic compounds high mutagenic activity /no mutagenic activity, assayed in Salmonella typhimurium. Results 188 compouunds: / fold cross-validation accuracy Method Accuracy Progol1 81.4% 2D kernel 91.2% Jean-Philippe Vert (ParisTech) The Analysis of Patterns 55 / 114
78 2D Subtree vs walk kernels AUC Walks Subtrees CCRF CEM HL 60(TB) K 562 MOLT 4 RPMI 8226 SR A549/ATCC EKVX HOP 62 HOP 92 NCI H226 NCI H23 NCI H322M NCI H460 NCI H522 COLO_205 HCC 2998 HCT 116 HCT 15 HT29 KM12 SW 620 SF 268 SF 295 SF 539 SNB 19 SNB 75 U251 LOX_IMVI MALME 3M M14 SK MEL 2 SK MEL 28 SK MEL 5 UACC 257 UACC 62 IGR OV1 OVCAR 3 OVCAR 4 OVCAR 5 OVCAR 8 SK OV A498 ACHN CAKI 1 RXF_393 SN12C TK 10 UO 31 PC 3 DU 145 MCF7 NCI/ADR RES MDA MB 231/ATCC HS_578T MDA MB 435 MDA N BT 549 T 47D Screening of inhibitors for 60 cancer cell lines. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 56 / 114
79 Image classification (Harchaoui and Bach, 2007) COREL14 dataset 1400 natural images in 14 classes Compare kernel between histograms (H), walk kernel (W), subtree kernel (TW), weighted subtree kernel (wtw), and a combination (M). Performance comparison on Corel Test error H W TW wtw M Kernels Jean-Philippe Vert (ParisTech) The Analysis of Patterns 57 / 114
80 Summary: graph kernels What we saw Kernels do not allow to overcome the NP-hardness of subgraph patterns They allow to work with approximate subgraphs (walks, subtrees), in infinite dimension, thanks to the kernel trick However: using kernels makes it difficult to come back to patterns after the learning stage Jean-Philippe Vert (ParisTech) The Analysis of Patterns 58 / 114
81 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 59 / 114
82 Microarrays measure gene expression Jean-Philippe Vert (ParisTech) The Analysis of Patterns 60 / 114
83 Cancer classification from microarray data Jean-Philippe Vert (ParisTech) The Analysis of Patterns 61 / 114
84 Gene networks N Glycan biosynthesis Glycolysis / Gluconeogenesis Porphyrin and chlorophyll metabolism Riboflavin metabolism Sulfur metabolism - Protein kinases Nitrogen, asparagine metabolism Folate biosynthesis Biosynthesis of steroids, ergosterol metabolism DNA and RNA polymerase subunits Lysine biosynthesis Phenylalanine, tyrosine and tryptophan biosynthesis Oxidative phosphorylation, TCA cycle Purine metabolism Jean-Philippe Vert (ParisTech) The Analysis of Patterns 62 / 114
85 Gene networks and expression data Motivation Basic biological functions usually involve the coordinated action of several proteins: Formation of protein complexes Activation of metabolic, signalling or regulatory pathways Many pathways and protein-protein interactions are already known Hypothesis: the weights of the classifier should be coherent with respect to this prior knowledge Jean-Philippe Vert (ParisTech) The Analysis of Patterns 63 / 114
86 An idea 1 Use the gene network to extract the important information in gene expression profiles by Fourier analysis on the graph 2 Learn a linear classifier on the smooth components Jean-Philippe Vert (ParisTech) The Analysis of Patterns 64 / 114
87 Graph Laplacian Definition The Laplacian of the graph is the matrix L = D A. 1 2 L = D A = Jean-Philippe Vert (ParisTech) The Analysis of Patterns 65 / 114
88 Fourier basis L is positive semidefinite The eigenvectors e 1,..., e n of L with eigenvalues 0 = λ 1... λ n form a basis called Fourier basis For any f : V R, the Fourier transform of f is the vector ˆf R n defined by: ˆfi = f e i, i = 1,..., n. The inverse Fourier formula holds: f = n ˆfi e i. i=1 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 66 / 114
89 Fourier basis λ=0 λ= 0.5 λ= 1 λ= 2.3 λ= 4.2 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 67 / 114
90 Fourier basis Jean-Philippe Vert (ParisTech) The Analysis of Patterns 68 / 114
91 Smoothing operator Definition Let φ : R + R + be non-increasing. A smoothing operator S φ transform a function f : V R into a smoothed version: S φ (f ) = n ˆfi φ(λ i )e i. i=1 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 69 / 114
92 Smoothing operators Examples Identity operator (S φ (f ) = f ): φ(λ) = 1, λ Low-pass filter: φ(λ) = { 1 if λ λ, 0 otherwise. Attenuation of high frequencies: φ(λ) = exp( βλ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 70 / 114
93 Smoothing operators Examples Identity operator (S φ (f ) = f ): φ(λ) = 1, λ Low-pass filter: φ(λ) = { 1 if λ λ, 0 otherwise. Attenuation of high frequencies: φ(λ) = exp( βλ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 70 / 114
94 Smoothing operators Examples Identity operator (S φ (f ) = f ): φ(λ) = 1, λ Low-pass filter: φ(λ) = { 1 if λ λ, 0 otherwise. Attenuation of high frequencies: φ(λ) = exp( βλ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 70 / 114
95 Supervised classification and regression Working with smoothed profiles Classical methods for linear classification and regression with a ridge penalty solve: 1 min β R p n n l i=1 (β f i, y i ) + λβ β. Applying these algorithms on the smooth profiles means solving: 1 min β R p n n l i=1 (β S φ (f i ), y i ) + λβ β. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 71 / 114
96 Link with shrinkage estimator Lemma This is equivalent to: 1 min v R p n n l i=1 (v f i, y i ) + λ hence the linear classifier v is smooth. Proof Let v = n i=1 φ(λ i)e i e i β, then β S φ (f i ) = β Then ˆv i = φ(λ i ) ˆβ i and β β = n i=1 p i=1 ˆv 2 i φ(λ i ), n ˆfi φ(λ i )e i = f v. i=1 ˆv 2 i φ(λ i ) 2. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 72 / 114
97 Link with shrinkage estimator Lemma This is equivalent to: 1 min v R p n n l i=1 (v f i, y i ) + λ hence the linear classifier v is smooth. Proof Let v = n i=1 φ(λ i)e i e i β, then β S φ (f i ) = β Then ˆv i = φ(λ i ) ˆβ i and β β = n i=1 p i=1 ˆv 2 i φ(λ i ), n ˆfi φ(λ i )e i = f v. i=1 ˆv 2 i φ(λ i ) 2. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 72 / 114
98 Kernel methods Smoothing kernel Kernel methods (SVM, kernel ridge regression..) only need the inner product between smooth profiles: K (f, g) = S φ (f ) S φ (g) n = ˆfi ĝ i φ(λ i ) 2 i=1 ( n = f φ(λ i ) 2 e i ei i=1 = f K φ g, ) g (1) with K φ = n φ(λ i ) 2 e i ei. i=1 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 73 / 114
99 Examples For φ(λ) = exp( tλ), we recover the diffusion kernel: K φ = exp M ( 2tL). For φ(λ) = 1/ 1 + λ, we obtain and the penalization is: n i=1 K φ = (L + I) 1, ˆv i 2 φ(λ i ) = v (L + I) v = v (v i v j ) 2. i j Jean-Philippe Vert (ParisTech) The Analysis of Patterns 74 / 114
100 Examples For φ(λ) = exp( tλ), we recover the diffusion kernel: K φ = exp M ( 2tL). For φ(λ) = 1/ 1 + λ, we obtain and the penalization is: n i=1 K φ = (L + I) 1, ˆv i 2 φ(λ i ) = v (L + I) v = v (v i v j ) 2. i j Jean-Philippe Vert (ParisTech) The Analysis of Patterns 74 / 114
101 Data Expression Graph Study the effect of low irradiation doses on the yeast 12 non irradiated vs 6 irradiated Which pathways are involved in the response at the transcriptomic level? KEGG database of metabolic pathways Two genes are connected is they code for enzymes that catalyze successive reactions in a pathway (metabolic gene network). 737 genes, 4694 vertices. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 75 / 114
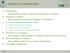
102 a) b) Classification performance Glycerophospholipid metabolism Alanine and aspartate metabolism riboflavin metabolism Fig. 2. PCA plots of the initial expression profiles (a) and the transformed profiles using network topology (80% of the eigenvalues removed) (b). The green squares are non-irradiated samples and the red rhombuses are irradiated samples. Individual sample labels are shown together with GO and KEGG annotations associated with each principal component. LOO hinge loss / error Error Hinge LOO hinge loss / error Error Hinge Beta Percentage of eigenvalues removed Fig. 3. Performance of the supervised classification when changing the metric with the function φexp(λ) = exp( βλ) for different values of β (left picture), or the function φ thres(λ) = 1(λ < λ0) for different values of λ0 (i.e., keeping only a fraction of the smallest eigenvalues, right picture). The performance is estimated from the number of misclassifications in a leave-one-out error. shift). Jean-Philippe The reconstruction Vert (ParisTech) of this from our data with no prior Representing KEGG as a large The Analysis networkof helps Patterns keep the bio- 76 / 114
103 Fig. Jean-Philippe 4. Global connection Vert (ParisTech) map of KEGG with mapped coefficients of the decision function obtainedthe by applying Analysisa customary of Patterns linear SVM 77 / 114 Classifier Rapaport et al N Glycan biosynthesis Glycolysis / Gluconeogenesis Porphyrin and chlorophyll metabolism Riboflavin metabolism Sulfur metabolism - Protein kinases Nitrogen, asparagine metabolism Folate biosynthesis Biosynthesis of steroids, ergosterol metabolism DNA and RNA polymerase subunits Lysine biosynthesis Phenylalanine, tyrosine and tryptophan biosynthesis Oxidative phosphorylation, TCA cycle Purine metabolism
104 Classifier Spectral analysis of gene expression profiles using gene networks a) b) Fig. 5. The glycolysis/gluconeogenesis pathways of KEGG with mapped coefficients of the decision function obtained by applying a customary linear SVM (a) and using high-frequency eigenvalue attenuation (b). The pathways are mutually exclusive in a cell, as clearly highlighted by Jean-Philippe Vert (ParisTech) The Analysis of Patterns 78 / 114
105 Summary With kernels we are able to soft constrain the shape of the classifier through regularization, e.g.: min R emp(v) + λ v R p p i=1 ˆv 2 i φ(λ i ), This is related to priors in Bayesian learning The resulting classifier is interpretable, even without selection of a specific list of features. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 79 / 114
106 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 80 / 114
107 Linear classifiers Training the model Minimize an empirical risk on the training samples: min β R p+1 R emp (β) = 1 n n l(f β (x i ), y i ), i=1... subject to some constraint on β, e.g.: Ω(β) C. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 81 / 114
108 Linear classifiers Training the model Minimize an empirical risk on the training samples: min β R p+1 R emp (β) = 1 n n l(f β (x i ), y i ), i=1... subject to some constraint on β, e.g.: Ω(β) C. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 81 / 114
109 Example : Norm Constraints The approach A common method in statistics to learn with few samples in high dimension is to constrain the Euclidean norm of β Ω ridge (β) = β 2 2 = p βi 2, i=1 (ridge regression, support vector machines, kernel methods...) Pros Good performance in classification Cons Limited interpretation (small weights) No prior biological knowledge Jean-Philippe Vert (ParisTech) The Analysis of Patterns 82 / 114
110 Example : Feature Selection The approach Constrain most weights to be 0, i.e., select a few genes whose expression are sufficient for classification. Ω Best subset selection (β) = β 0 = p 1(β i > 0). i=1 This is usually a NP-hard problem, many greedy variants have been proposed (filter methods, wrapper methods) Pros Good performance Biomarker selection Interpretability Cons NP-hard Gene selection not robust No use of prior knowledge Jean-Philippe Vert (ParisTech) The Analysis of Patterns 83 / 114
111 Example : Sparsity inducing convex priors The approach Constrain most weights to be 0 through a convex non-differentiable penalty: p Ω LASSO (β) = β 1 = β i. Several variants exist, e.g., elastic net penalty ( β 1 + β 2 ),... ) i=1 Pros Good performance Biomarker selection Interpretability Cons Gene selection not robust No use of prior knowledge Jean-Philippe Vert (ParisTech) The Analysis of Patterns 84 / 114
112 Why LASSO leads to sparse solutions Jean-Philippe Vert (ParisTech) The Analysis of Patterns 85 / 114
113 Efficienty computation of the regularization path min R n (f β ) = β R p+1 n (f β (x i ) y i ) 2 + λ i=1 p β i (2) i=1 No explicit solution, but this is just a quadratic program. LARS (Efron et al., 2004) provides a fast algorithm to compute the solution for all λ s simultaneously (regularization path) Jean-Philippe Vert (ParisTech) The Analysis of Patterns 86 / 114
114 Incorporating prior knowledge The idea If we have a specific prior knowledge about the correct weights, it can be included in Ω in the contraint: Minimize R emp (β) subject to Ω(β) C. If we design a convex function Ω, then the algorithm boils down to a convex optimization problem (usually easy to solve). Similar to priors in Bayesian statistics Jean-Philippe Vert (ParisTech) The Analysis of Patterns 87 / 114
115 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 88 / 114
116 Motivation Indexing by all subgraphs is appealing but intractable in practice (both explicitly and with the kernel trick) Can we work implicitly with this representation using sparse learning, e.g., LASSO regression or boosting? This may lead to both accurate predictive model and the identification of discriminative patterns. The iterations of LARS or boosting amount to an optimization problem over subgraphs, which may be solved efficiently using graph mining technique... Jean-Philippe Vert (ParisTech) The Analysis of Patterns 89 / 114
117 Boosting over subgraph indexation (Kudo et al., 2004) Weak learner = decision stump indexed by subgraph H and α = ±1: h α,h (G) = αφ H (G) Boosting: at each iteration, for a given distribution d d n = 1 over the training points (G i, y i ), select a weak learner (subgraph H) which maximizes the gain gain(h, α) = n y i h α,h (G i ). i=1 This can be done "efficiently" by branch-and-bound over a DFS code tree (Yan and Han, 2002). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 90 / 114
118 The DFS code tree Jean-Philippe Vert (ParisTech) The Analysis of Patterns 91 / 114
119 Graph LASSO regularization path (Tsuda, 2007) Jean-Philippe Vert (ParisTech) The Analysis of Patterns 92 / 114
120 Summary Sparse learning is practically feasible in the space of graphs indexed by all subgraphs Leads to subgraph selection Several extensions LASSO regularization path (Tsuda, 2007) gboost (Saigo et al., 2009) A beautiful and promising marriage between machine learning and data mining Jean-Philippe Vert (ParisTech) The Analysis of Patterns 93 / 114
121 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 94 / 114
122 Chromosomic aberrations in cancer Jean-Philippe Vert (ParisTech) The Analysis of Patterns 95 / 114
123 Comparative Genomic Hybridization (CGH) Motivation Comparative genomic hybridization (CGH) data measure the DNA copy number along the genome Very useful, in particular in cancer research Can we classify CGH arrays for diagnosis or prognosis purpose? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 96 / 114
124 Aggressive vs non-aggressive melanoma Jean-Philippe Vert (ParisTech) The Analysis of Patterns 97 / 114
125 Classification of array CGH Prior knowledge Let x be a CGH profile We focus on linear classifiers, i.e., the sign of : f (x) = x β. We expect β to be sparse : only a few positions should be discriminative piecewise constant : within a region, all probes should contribute equally Jean-Philippe Vert (ParisTech) The Analysis of Patterns 98 / 114
126 A penalty for CGH array classification The fused LASSO penalty (Tibshirani et al., 2005) Ω fusedlasso (β) = i β i + i j β i β j. First term leads to sparse solutions Second term leads to piecewise constant solutions Combined with a hinge loss leads to a fused SVM (Rapaport et al., 2008); Jean-Philippe Vert (ParisTech) The Analysis of Patterns 99 / 114
127 Application: metastasis prognosis in melanoma Weight BAC Weight BAC Jean-Philippe Vert (ParisTech) The Analysis of Patterns 100 / 114
128 Outline 1 Explicit computation of features : the case of graph features 2 Using kernels Introduction to kernels Graph kernels Kernels for gene expression data using gene networks 3 Using sparsity-inducing shrinkage estimators Feature selection for all subgraph indexation Classification of array CGH data with piecewise-linear models Structured gene selection for microarray classification 4 Conclusion Jean-Philippe Vert (ParisTech) The Analysis of Patterns 101 / 114
129 How to select jointly genes belonging to the same pathways? Jean-Philippe Vert (ParisTech) The Analysis of Patterns 102 / 114
130 Selecting pre-defined groups of variables Group lasso (Yuan & Lin, 2006) If groups of covariates are likely to be selected together, the l 1 /l 2 -norm induces sparse solutions at the group level: Ω group (w) = g w g 2 Ω(w 1, w 2, w 3 ) = (w 1, w 2 ) 2 + w 3 2 Jean-Philippe Vert (ParisTech) The Analysis of Patterns 103 / 114
131 What if a gene belongs to several groups? Issue of using the group-lasso Ω group (w) = g w g 2 sets groups to 0. One variable is selected all the groups to which it belongs are selected. w g1 2 = w g3 2 =0 IGF selection selection of unwanted groups Removal of any group containing a gene the weight of the gene is 0. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 104 / 114
132 Overlap norm (Jacob et al., 2009) An idea Introduce latent variables v g : min L(w) + λ v g 2 w,v g G w = g G v g supp ( ) v g g. Properties Resulting support is a union of groups in G. Possible to select one variable without selecting all the groups containing it. Setting one v g to 0 doesn t necessarily set to 0 all its variables in w. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 105 / 114
133 A new norm Overlap norm min L(w) + λ v g 2 w,v g G with = min L(w) + λω overlap (w) w w = g G v g supp ( ) v g g. min v g 2 v Ω overlap (w) = g G w = g G v g supp ( ) v g g. ( ) Property Ω overlap (w) is a norm of w. Ω overlap (.) associates to w a specific (not necessarily unique) decomposition (v g ) g G which is the argmin of ( ). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 106 / 114
134 Overlap and group unity balls Balls for Ω G group ( ) (middle) and Ω G overlap ( ) (right) for the groups G = {{1, 2}, {2, 3}} where w 2 is represented as the vertical coordinate. Left: group-lasso (G = {{1, 2}, {3}}), for comparison. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 107 / 114
135 Theoretical results Consistency in group support (Jacob et al., 2009) Then Let w be the true parameter vector. Assume that there exists a unique decomposition v g such that w = g v g and Ω G overlap ( w) = v g 2. Consider the regularized empirical risk minimization problem L(w) + λω G overlap (w). under appropriate mutual incoherence conditions on X, as n, with very high probability, the optimal solution ŵ admits a unique decomposition (ˆv g ) g G such that { g G ˆvg 0 } = { g G v g 0 }. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 108 / 114
136 Theoretical results Consistency in group support (Jacob et al., 2009) Then Let w be the true parameter vector. Assume that there exists a unique decomposition v g such that w = g v g and Ω G overlap ( w) = v g 2. Consider the regularized empirical risk minimization problem L(w) + λω G overlap (w). under appropriate mutual incoherence conditions on X, as n, with very high probability, the optimal solution ŵ admits a unique decomposition (ˆv g ) g G such that { g G ˆvg 0 } = { g G v g 0 }. Jean-Philippe Vert (ParisTech) The Analysis of Patterns 108 / 114
137 Experiments Synthetic data: overlapping groups 10 groups of 10 variables with 2 variables of overlap between two successive groups :{1,..., 10}, {9,..., 18},..., {73,..., 82}. Support: union of 4th and 5th groups. Learn from 100 training points. Frequency of selection of each variable with the lasso (left) and Ω G overlap (.) (middle), comparison of the RMSE of both methods (right). Jean-Philippe Vert (ParisTech) The Analysis of Patterns 109 / 114
Group lasso for genomic data
 Group lasso for genomic data Jean-Philippe Vert Mines ParisTech / Curie Institute / Inserm Machine learning: Theory and Computation workshop, IMA, Minneapolis, March 26-3, 22 J.P Vert (ParisTech) Group
Group lasso for genomic data Jean-Philippe Vert Mines ParisTech / Curie Institute / Inserm Machine learning: Theory and Computation workshop, IMA, Minneapolis, March 26-3, 22 J.P Vert (ParisTech) Group
Machine learning for ligand-based virtual screening and chemogenomics!
 Machine learning for ligand-based virtual screening and chemogenomics! Jean-Philippe Vert Institut Curie - INSERM U900 - Mines ParisTech In silico discovery of molecular probes and drug-like compounds:
Machine learning for ligand-based virtual screening and chemogenomics! Jean-Philippe Vert Institut Curie - INSERM U900 - Mines ParisTech In silico discovery of molecular probes and drug-like compounds:
S1324: Support Vector Machines
 S1324: Support Vector Machines Chloé-Agathe Azencott chloe-agathe.azencott@mines-paristech.fr based on slides by J.-P. Vert 2015-05-22 Overview Large margin linear classification Support vector machines
S1324: Support Vector Machines Chloé-Agathe Azencott chloe-agathe.azencott@mines-paristech.fr based on slides by J.-P. Vert 2015-05-22 Overview Large margin linear classification Support vector machines
Statistical learning on graphs
 Statistical learning on graphs Jean-Philippe Vert Jean-Philippe.Vert@ensmp.fr ParisTech, Ecole des Mines de Paris Institut Curie INSERM U900 Seminar of probabilities, Institut Joseph Fourier, Grenoble,
Statistical learning on graphs Jean-Philippe Vert Jean-Philippe.Vert@ensmp.fr ParisTech, Ecole des Mines de Paris Institut Curie INSERM U900 Seminar of probabilities, Institut Joseph Fourier, Grenoble,
Graph Wavelets to Analyze Genomic Data with Biological Networks
 Graph Wavelets to Analyze Genomic Data with Biological Networks Yunlong Jiao and Jean-Philippe Vert "Emerging Topics in Biological Networks and Systems Biology" symposium, Swedish Collegium for Advanced
Graph Wavelets to Analyze Genomic Data with Biological Networks Yunlong Jiao and Jean-Philippe Vert "Emerging Topics in Biological Networks and Systems Biology" symposium, Swedish Collegium for Advanced
Statistical learning theory, Support vector machines, and Bioinformatics
 1 Statistical learning theory, Support vector machines, and Bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group ENS Paris, november 25, 2003. 2 Overview 1.
1 Statistical learning theory, Support vector machines, and Bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group ENS Paris, november 25, 2003. 2 Overview 1.
Machine learning in molecular classification
 Machine learning in molecular classification Hongyu Su Department of Computer Science PO Box 68, FI-00014 University of Helsinki, Finland firstname.lastname@cs.helsinki.fi Abstract. Chemical and biological
Machine learning in molecular classification Hongyu Su Department of Computer Science PO Box 68, FI-00014 University of Helsinki, Finland firstname.lastname@cs.helsinki.fi Abstract. Chemical and biological
Metabolic networks: Activity detection and Inference
 1 Metabolic networks: Activity detection and Inference Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Advanced microarray analysis course, Elsinore, Denmark, May 21th,
1 Metabolic networks: Activity detection and Inference Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Advanced microarray analysis course, Elsinore, Denmark, May 21th,
Support Vector Machines (SVM) in bioinformatics. Day 1: Introduction to SVM
 1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
Convex relaxation for Combinatorial Penalties
 Convex relaxation for Combinatorial Penalties Guillaume Obozinski Equipe Imagine Laboratoire d Informatique Gaspard Monge Ecole des Ponts - ParisTech Joint work with Francis Bach Fête Parisienne in Computation,
Convex relaxation for Combinatorial Penalties Guillaume Obozinski Equipe Imagine Laboratoire d Informatique Gaspard Monge Ecole des Ponts - ParisTech Joint work with Francis Bach Fête Parisienne in Computation,
Support Vector Machine (continued)
 Support Vector Machine continued) Overlapping class distribution: In practice the class-conditional distributions may overlap, so that the training data points are no longer linearly separable. We need
Support Vector Machine continued) Overlapping class distribution: In practice the class-conditional distributions may overlap, so that the training data points are no longer linearly separable. We need
6. Regularized linear regression
 Foundations of Machine Learning École Centrale Paris Fall 2015 6. Regularized linear regression Chloé-Agathe Azencot Centre for Computational Biology, Mines ParisTech chloe agathe.azencott@mines paristech.fr
Foundations of Machine Learning École Centrale Paris Fall 2015 6. Regularized linear regression Chloé-Agathe Azencot Centre for Computational Biology, Mines ParisTech chloe agathe.azencott@mines paristech.fr
Linear & nonlinear classifiers
 Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1394 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1394 1 / 34 Table
Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1394 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1394 1 / 34 Table
Linear & nonlinear classifiers
 Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1396 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1396 1 / 44 Table
Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1396 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1396 1 / 44 Table
Support Vector Machine (SVM) and Kernel Methods
 Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2014 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2014 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support vector machines, Kernel methods, and Applications in bioinformatics
 1 Support vector machines, Kernel methods, and Applications in bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Machine Learning in Bioinformatics conference,
1 Support vector machines, Kernel methods, and Applications in bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Machine Learning in Bioinformatics conference,
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie
 A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
Support Vector Machines for Classification: A Statistical Portrait
 Support Vector Machines for Classification: A Statistical Portrait Yoonkyung Lee Department of Statistics The Ohio State University May 27, 2011 The Spring Conference of Korean Statistical Society KAIST,
Support Vector Machines for Classification: A Statistical Portrait Yoonkyung Lee Department of Statistics The Ohio State University May 27, 2011 The Spring Conference of Korean Statistical Society KAIST,
Support Vector Machines
 Support Vector Machines Reading: Ben-Hur & Weston, A User s Guide to Support Vector Machines (linked from class web page) Notation Assume a binary classification problem. Instances are represented by vector
Support Vector Machines Reading: Ben-Hur & Weston, A User s Guide to Support Vector Machines (linked from class web page) Notation Assume a binary classification problem. Instances are represented by vector
Support Vector Machine
 Support Vector Machine Fabrice Rossi SAMM Université Paris 1 Panthéon Sorbonne 2018 Outline Linear Support Vector Machine Kernelized SVM Kernels 2 From ERM to RLM Empirical Risk Minimization in the binary
Support Vector Machine Fabrice Rossi SAMM Université Paris 1 Panthéon Sorbonne 2018 Outline Linear Support Vector Machine Kernelized SVM Kernels 2 From ERM to RLM Empirical Risk Minimization in the binary
DATA MINING AND MACHINE LEARNING
 DATA MINING AND MACHINE LEARNING Lecture 5: Regularization and loss functions Lecturer: Simone Scardapane Academic Year 2016/2017 Table of contents Loss functions Loss functions for regression problems
DATA MINING AND MACHINE LEARNING Lecture 5: Regularization and loss functions Lecturer: Simone Scardapane Academic Year 2016/2017 Table of contents Loss functions Loss functions for regression problems
Introduction to Machine Learning Midterm, Tues April 8
 Introduction to Machine Learning 10-701 Midterm, Tues April 8 [1 point] Name: Andrew ID: Instructions: You are allowed a (two-sided) sheet of notes. Exam ends at 2:45pm Take a deep breath and don t spend
Introduction to Machine Learning 10-701 Midterm, Tues April 8 [1 point] Name: Andrew ID: Instructions: You are allowed a (two-sided) sheet of notes. Exam ends at 2:45pm Take a deep breath and don t spend
Structure-Activity Modeling - QSAR. Uwe Koch
 Structure-Activity Modeling - QSAR Uwe Koch QSAR Assumption: QSAR attempts to quantify the relationship between activity and molecular strcucture by correlating descriptors with properties Biological activity
Structure-Activity Modeling - QSAR Uwe Koch QSAR Assumption: QSAR attempts to quantify the relationship between activity and molecular strcucture by correlating descriptors with properties Biological activity
Chapter 9. Support Vector Machine. Yongdai Kim Seoul National University
 Chapter 9. Support Vector Machine Yongdai Kim Seoul National University 1. Introduction Support Vector Machine (SVM) is a classification method developed by Vapnik (1996). It is thought that SVM improved
Chapter 9. Support Vector Machine Yongdai Kim Seoul National University 1. Introduction Support Vector Machine (SVM) is a classification method developed by Vapnik (1996). It is thought that SVM improved
Support Vector Machine (SVM) and Kernel Methods
 Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2015 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2015 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2013
 UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2013 Exam policy: This exam allows two one-page, two-sided cheat sheets; No other materials. Time: 2 hours. Be sure to write your name and
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2013 Exam policy: This exam allows two one-page, two-sided cheat sheets; No other materials. Time: 2 hours. Be sure to write your name and
Regression. Goal: Learn a mapping from observations (features) to continuous labels given a training set (supervised learning)
 Linear Regression Regression Goal: Learn a mapping from observations (features) to continuous labels given a training set (supervised learning) Example: Height, Gender, Weight Shoe Size Audio features
Linear Regression Regression Goal: Learn a mapping from observations (features) to continuous labels given a training set (supervised learning) Example: Height, Gender, Weight Shoe Size Audio features
Regression. Goal: Learn a mapping from observations (features) to continuous labels given a training set (supervised learning)
 Linear Regression Regression Goal: Learn a mapping from observations (features) to continuous labels given a training set (supervised learning) Example: Height, Gender, Weight Shoe Size Audio features
Linear Regression Regression Goal: Learn a mapping from observations (features) to continuous labels given a training set (supervised learning) Example: Height, Gender, Weight Shoe Size Audio features
Mark your answers ON THE EXAM ITSELF. If you are not sure of your answer you may wish to provide a brief explanation.
 CS 189 Spring 2015 Introduction to Machine Learning Midterm You have 80 minutes for the exam. The exam is closed book, closed notes except your one-page crib sheet. No calculators or electronic items.
CS 189 Spring 2015 Introduction to Machine Learning Midterm You have 80 minutes for the exam. The exam is closed book, closed notes except your one-page crib sheet. No calculators or electronic items.
Regularization Paths
 December 2005 Trevor Hastie, Stanford Statistics 1 Regularization Paths Trevor Hastie Stanford University drawing on collaborations with Brad Efron, Saharon Rosset, Ji Zhu, Hui Zhou, Rob Tibshirani and
December 2005 Trevor Hastie, Stanford Statistics 1 Regularization Paths Trevor Hastie Stanford University drawing on collaborations with Brad Efron, Saharon Rosset, Ji Zhu, Hui Zhou, Rob Tibshirani and
Applied Machine Learning Annalisa Marsico
 Applied Machine Learning Annalisa Marsico OWL RNA Bionformatics group Max Planck Institute for Molecular Genetics Free University of Berlin 29 April, SoSe 2015 Support Vector Machines (SVMs) 1. One of
Applied Machine Learning Annalisa Marsico OWL RNA Bionformatics group Max Planck Institute for Molecular Genetics Free University of Berlin 29 April, SoSe 2015 Support Vector Machines (SVMs) 1. One of
Final Overview. Introduction to ML. Marek Petrik 4/25/2017
 Final Overview Introduction to ML Marek Petrik 4/25/2017 This Course: Introduction to Machine Learning Build a foundation for practice and research in ML Basic machine learning concepts: max likelihood,
Final Overview Introduction to ML Marek Petrik 4/25/2017 This Course: Introduction to Machine Learning Build a foundation for practice and research in ML Basic machine learning concepts: max likelihood,
A Sparse Solution Approach to Gene Selection for Cancer Diagnosis Using Microarray Data
 A Sparse Solution Approach to Gene Selection for Cancer Diagnosis Using Microarray Data Yoonkyung Lee Department of Statistics The Ohio State University http://www.stat.ohio-state.edu/ yklee May 13, 2005
A Sparse Solution Approach to Gene Selection for Cancer Diagnosis Using Microarray Data Yoonkyung Lee Department of Statistics The Ohio State University http://www.stat.ohio-state.edu/ yklee May 13, 2005
Fast Regularization Paths via Coordinate Descent
 August 2008 Trevor Hastie, Stanford Statistics 1 Fast Regularization Paths via Coordinate Descent Trevor Hastie Stanford University joint work with Jerry Friedman and Rob Tibshirani. August 2008 Trevor
August 2008 Trevor Hastie, Stanford Statistics 1 Fast Regularization Paths via Coordinate Descent Trevor Hastie Stanford University joint work with Jerry Friedman and Rob Tibshirani. August 2008 Trevor
Infinite Ensemble Learning with Support Vector Machinery
 Infinite Ensemble Learning with Support Vector Machinery Hsuan-Tien Lin and Ling Li Learning Systems Group, California Institute of Technology ECML/PKDD, October 4, 2005 H.-T. Lin and L. Li (Learning Systems
Infinite Ensemble Learning with Support Vector Machinery Hsuan-Tien Lin and Ling Li Learning Systems Group, California Institute of Technology ECML/PKDD, October 4, 2005 H.-T. Lin and L. Li (Learning Systems
hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference
 CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
Machine Learning for OR & FE
 Machine Learning for OR & FE Regression II: Regularization and Shrinkage Methods Martin Haugh Department of Industrial Engineering and Operations Research Columbia University Email: martin.b.haugh@gmail.com
Machine Learning for OR & FE Regression II: Regularization and Shrinkage Methods Martin Haugh Department of Industrial Engineering and Operations Research Columbia University Email: martin.b.haugh@gmail.com
Machine learning methods to infer drug-target interaction network
 Machine learning methods to infer drug-target interaction network Yoshihiro Yamanishi Medical Institute of Bioregulation Kyushu University Outline n Background Drug-target interaction network Chemical,
Machine learning methods to infer drug-target interaction network Yoshihiro Yamanishi Medical Institute of Bioregulation Kyushu University Outline n Background Drug-target interaction network Chemical,
Kernel Methods and Support Vector Machines
 Kernel Methods and Support Vector Machines Oliver Schulte - CMPT 726 Bishop PRML Ch. 6 Support Vector Machines Defining Characteristics Like logistic regression, good for continuous input features, discrete
Kernel Methods and Support Vector Machines Oliver Schulte - CMPT 726 Bishop PRML Ch. 6 Support Vector Machines Defining Characteristics Like logistic regression, good for continuous input features, discrete
Chapter 6: Classification
 Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
ESL Chap3. Some extensions of lasso
 ESL Chap3 Some extensions of lasso 1 Outline Consistency of lasso for model selection Adaptive lasso Elastic net Group lasso 2 Consistency of lasso for model selection A number of authors have studied
ESL Chap3 Some extensions of lasso 1 Outline Consistency of lasso for model selection Adaptive lasso Elastic net Group lasso 2 Consistency of lasso for model selection A number of authors have studied
Kernel Methods. Jean-Philippe Vert Last update: Jan Jean-Philippe Vert (Mines ParisTech) 1 / 444
 Kernel Methods Jean-Philippe Vert Jean-Philippe.Vert@mines.org Last update: Jan 2015 Jean-Philippe Vert (Mines ParisTech) 1 / 444 What we know how to solve Jean-Philippe Vert (Mines ParisTech) 2 / 444
Kernel Methods Jean-Philippe Vert Jean-Philippe.Vert@mines.org Last update: Jan 2015 Jean-Philippe Vert (Mines ParisTech) 1 / 444 What we know how to solve Jean-Philippe Vert (Mines ParisTech) 2 / 444
Introduction to Machine Learning. PCA and Spectral Clustering. Introduction to Machine Learning, Slides: Eran Halperin
 1 Introduction to Machine Learning PCA and Spectral Clustering Introduction to Machine Learning, 2013-14 Slides: Eran Halperin Singular Value Decomposition (SVD) The singular value decomposition (SVD)
1 Introduction to Machine Learning PCA and Spectral Clustering Introduction to Machine Learning, 2013-14 Slides: Eran Halperin Singular Value Decomposition (SVD) The singular value decomposition (SVD)
CIS 520: Machine Learning Oct 09, Kernel Methods
 CIS 520: Machine Learning Oct 09, 207 Kernel Methods Lecturer: Shivani Agarwal Disclaimer: These notes are designed to be a supplement to the lecture They may or may not cover all the material discussed
CIS 520: Machine Learning Oct 09, 207 Kernel Methods Lecturer: Shivani Agarwal Disclaimer: These notes are designed to be a supplement to the lecture They may or may not cover all the material discussed
Statistical Learning with the Lasso, spring The Lasso
 Statistical Learning with the Lasso, spring 2017 1 Yeast: understanding basic life functions p=11,904 gene values n number of experiments ~ 10 Blomberg et al. 2003, 2010 The Lasso fmri brain scans function
Statistical Learning with the Lasso, spring 2017 1 Yeast: understanding basic life functions p=11,904 gene values n number of experiments ~ 10 Blomberg et al. 2003, 2010 The Lasso fmri brain scans function
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2014
 UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2014 Exam policy: This exam allows two one-page, two-sided cheat sheets (i.e. 4 sides); No other materials. Time: 2 hours. Be sure to write
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2014 Exam policy: This exam allows two one-page, two-sided cheat sheets (i.e. 4 sides); No other materials. Time: 2 hours. Be sure to write
Discriminative Models
 No.5 Discriminative Models Hui Jiang Department of Electrical Engineering and Computer Science Lassonde School of Engineering York University, Toronto, Canada Outline Generative vs. Discriminative models
No.5 Discriminative Models Hui Jiang Department of Electrical Engineering and Computer Science Lassonde School of Engineering York University, Toronto, Canada Outline Generative vs. Discriminative models
A Submodular Framework for Graph Comparison
 A Submodular Framework for Graph Comparison Pinar Yanardag Department of Computer Science Purdue University West Lafayette, IN, 47906, USA ypinar@purdue.edu S.V.N. Vishwanathan Department of Computer Science
A Submodular Framework for Graph Comparison Pinar Yanardag Department of Computer Science Purdue University West Lafayette, IN, 47906, USA ypinar@purdue.edu S.V.N. Vishwanathan Department of Computer Science
Machine Learning Concepts in Chemoinformatics
 Machine Learning Concepts in Chemoinformatics Martin Vogt B-IT Life Science Informatics Rheinische Friedrich-Wilhelms-Universität Bonn BigChem Winter School 2017 25. October Data Mining in Chemoinformatics
Machine Learning Concepts in Chemoinformatics Martin Vogt B-IT Life Science Informatics Rheinische Friedrich-Wilhelms-Universität Bonn BigChem Winter School 2017 25. October Data Mining in Chemoinformatics
Sparse Approximation and Variable Selection
 Sparse Approximation and Variable Selection Lorenzo Rosasco 9.520 Class 07 February 26, 2007 About this class Goal To introduce the problem of variable selection, discuss its connection to sparse approximation
Sparse Approximation and Variable Selection Lorenzo Rosasco 9.520 Class 07 February 26, 2007 About this class Goal To introduce the problem of variable selection, discuss its connection to sparse approximation
SVMs: Non-Separable Data, Convex Surrogate Loss, Multi-Class Classification, Kernels
 SVMs: Non-Separable Data, Convex Surrogate Loss, Multi-Class Classification, Kernels Karl Stratos June 21, 2018 1 / 33 Tangent: Some Loose Ends in Logistic Regression Polynomial feature expansion in logistic
SVMs: Non-Separable Data, Convex Surrogate Loss, Multi-Class Classification, Kernels Karl Stratos June 21, 2018 1 / 33 Tangent: Some Loose Ends in Logistic Regression Polynomial feature expansion in logistic
CSE 417T: Introduction to Machine Learning. Final Review. Henry Chai 12/4/18
 CSE 417T: Introduction to Machine Learning Final Review Henry Chai 12/4/18 Overfitting Overfitting is fitting the training data more than is warranted Fitting noise rather than signal 2 Estimating! "#$
CSE 417T: Introduction to Machine Learning Final Review Henry Chai 12/4/18 Overfitting Overfitting is fitting the training data more than is warranted Fitting noise rather than signal 2 Estimating! "#$
The lasso: some novel algorithms and applications
 1 The lasso: some novel algorithms and applications Newton Institute, June 25, 2008 Robert Tibshirani Stanford University Collaborations with Trevor Hastie, Jerome Friedman, Holger Hoefling, Gen Nowak,
1 The lasso: some novel algorithms and applications Newton Institute, June 25, 2008 Robert Tibshirani Stanford University Collaborations with Trevor Hastie, Jerome Friedman, Holger Hoefling, Gen Nowak,
Linear regression methods
 Linear regression methods Most of our intuition about statistical methods stem from linear regression. For observations i = 1,..., n, the model is Y i = p X ij β j + ε i, j=1 where Y i is the response
Linear regression methods Most of our intuition about statistical methods stem from linear regression. For observations i = 1,..., n, the model is Y i = p X ij β j + ε i, j=1 where Y i is the response
Support Vector Machines.
 Support Vector Machines www.cs.wisc.edu/~dpage 1 Goals for the lecture you should understand the following concepts the margin slack variables the linear support vector machine nonlinear SVMs the kernel
Support Vector Machines www.cs.wisc.edu/~dpage 1 Goals for the lecture you should understand the following concepts the margin slack variables the linear support vector machine nonlinear SVMs the kernel
A Survey of L 1. Regression. Céline Cunen, 20/10/2014. Vidaurre, Bielza and Larranaga (2013)
 A Survey of L 1 Regression Vidaurre, Bielza and Larranaga (2013) Céline Cunen, 20/10/2014 Outline of article 1.Introduction 2.The Lasso for Linear Regression a) Notation and Main Concepts b) Statistical
A Survey of L 1 Regression Vidaurre, Bielza and Larranaga (2013) Céline Cunen, 20/10/2014 Outline of article 1.Introduction 2.The Lasso for Linear Regression a) Notation and Main Concepts b) Statistical
Mining Classification Knowledge
 Mining Classification Knowledge Remarks on NonSymbolic Methods JERZY STEFANOWSKI Institute of Computing Sciences, Poznań University of Technology COST Doctoral School, Troina 2008 Outline 1. Bayesian classification
Mining Classification Knowledge Remarks on NonSymbolic Methods JERZY STEFANOWSKI Institute of Computing Sciences, Poznań University of Technology COST Doctoral School, Troina 2008 Outline 1. Bayesian classification
6.036 midterm review. Wednesday, March 18, 15
 6.036 midterm review 1 Topics covered supervised learning labels available unsupervised learning no labels available semi-supervised learning some labels available - what algorithms have you learned that
6.036 midterm review 1 Topics covered supervised learning labels available unsupervised learning no labels available semi-supervised learning some labels available - what algorithms have you learned that
Midterm Review CS 6375: Machine Learning. Vibhav Gogate The University of Texas at Dallas
 Midterm Review CS 6375: Machine Learning Vibhav Gogate The University of Texas at Dallas Machine Learning Supervised Learning Unsupervised Learning Reinforcement Learning Parametric Y Continuous Non-parametric
Midterm Review CS 6375: Machine Learning Vibhav Gogate The University of Texas at Dallas Machine Learning Supervised Learning Unsupervised Learning Reinforcement Learning Parametric Y Continuous Non-parametric
Discriminative Models
 No.5 Discriminative Models Hui Jiang Department of Electrical Engineering and Computer Science Lassonde School of Engineering York University, Toronto, Canada Outline Generative vs. Discriminative models
No.5 Discriminative Models Hui Jiang Department of Electrical Engineering and Computer Science Lassonde School of Engineering York University, Toronto, Canada Outline Generative vs. Discriminative models
CSCE 478/878 Lecture 6: Bayesian Learning
 Bayesian Methods Not all hypotheses are created equal (even if they are all consistent with the training data) Outline CSCE 478/878 Lecture 6: Bayesian Learning Stephen D. Scott (Adapted from Tom Mitchell
Bayesian Methods Not all hypotheses are created equal (even if they are all consistent with the training data) Outline CSCE 478/878 Lecture 6: Bayesian Learning Stephen D. Scott (Adapted from Tom Mitchell
Support Vector Machine (SVM) & Kernel CE-717: Machine Learning Sharif University of Technology. M. Soleymani Fall 2012
 Support Vector Machine (SVM) & Kernel CE-717: Machine Learning Sharif University of Technology M. Soleymani Fall 2012 Linear classifier Which classifier? x 2 x 1 2 Linear classifier Margin concept x 2
Support Vector Machine (SVM) & Kernel CE-717: Machine Learning Sharif University of Technology M. Soleymani Fall 2012 Linear classifier Which classifier? x 2 x 1 2 Linear classifier Margin concept x 2
Introduction to Machine Learning
 1, DATA11002 Introduction to Machine Learning Lecturer: Teemu Roos TAs: Ville Hyvönen and Janne Leppä-aho Department of Computer Science University of Helsinki (based in part on material by Patrik Hoyer
1, DATA11002 Introduction to Machine Learning Lecturer: Teemu Roos TAs: Ville Hyvönen and Janne Leppä-aho Department of Computer Science University of Helsinki (based in part on material by Patrik Hoyer
Effective Linear Discriminant Analysis for High Dimensional, Low Sample Size Data
 Effective Linear Discriant Analysis for High Dimensional, Low Sample Size Data Zhihua Qiao, Lan Zhou and Jianhua Z. Huang Abstract In the so-called high dimensional, low sample size (HDLSS) settings, LDA
Effective Linear Discriant Analysis for High Dimensional, Low Sample Size Data Zhihua Qiao, Lan Zhou and Jianhua Z. Huang Abstract In the so-called high dimensional, low sample size (HDLSS) settings, LDA
Nearest Neighbors Methods for Support Vector Machines
 Nearest Neighbors Methods for Support Vector Machines A. J. Quiroz, Dpto. de Matemáticas. Universidad de Los Andes joint work with María González-Lima, Universidad Simón Boĺıvar and Sergio A. Camelo, Universidad
Nearest Neighbors Methods for Support Vector Machines A. J. Quiroz, Dpto. de Matemáticas. Universidad de Los Andes joint work with María González-Lima, Universidad Simón Boĺıvar and Sergio A. Camelo, Universidad
PAC-learning, VC Dimension and Margin-based Bounds
 More details: General: http://www.learning-with-kernels.org/ Example of more complex bounds: http://www.research.ibm.com/people/t/tzhang/papers/jmlr02_cover.ps.gz PAC-learning, VC Dimension and Margin-based
More details: General: http://www.learning-with-kernels.org/ Example of more complex bounds: http://www.research.ibm.com/people/t/tzhang/papers/jmlr02_cover.ps.gz PAC-learning, VC Dimension and Margin-based
EE613 Machine Learning for Engineers. Kernel methods Support Vector Machines. jean-marc odobez 2015
 EE613 Machine Learning for Engineers Kernel methods Support Vector Machines jean-marc odobez 2015 overview Kernel methods introductions and main elements defining kernels Kernelization of k-nn, K-Means,
EE613 Machine Learning for Engineers Kernel methods Support Vector Machines jean-marc odobez 2015 overview Kernel methods introductions and main elements defining kernels Kernelization of k-nn, K-Means,
Midterm Review CS 7301: Advanced Machine Learning. Vibhav Gogate The University of Texas at Dallas
 Midterm Review CS 7301: Advanced Machine Learning Vibhav Gogate The University of Texas at Dallas Supervised Learning Issues in supervised learning What makes learning hard Point Estimation: MLE vs Bayesian
Midterm Review CS 7301: Advanced Machine Learning Vibhav Gogate The University of Texas at Dallas Supervised Learning Issues in supervised learning What makes learning hard Point Estimation: MLE vs Bayesian
Support'Vector'Machines. Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan
 Support'Vector'Machines Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan kasthuri.kannan@nyumc.org Overview Support Vector Machines for Classification Linear Discrimination Nonlinear Discrimination
Support'Vector'Machines Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan kasthuri.kannan@nyumc.org Overview Support Vector Machines for Classification Linear Discrimination Nonlinear Discrimination
Mining Molecular Fragments: Finding Relevant Substructures of Molecules
 Mining Molecular Fragments: Finding Relevant Substructures of Molecules Christian Borgelt, Michael R. Berthold Proc. IEEE International Conference on Data Mining, 2002. ICDM 2002. Lecturers: Carlo Cagli
Mining Molecular Fragments: Finding Relevant Substructures of Molecules Christian Borgelt, Michael R. Berthold Proc. IEEE International Conference on Data Mining, 2002. ICDM 2002. Lecturers: Carlo Cagli
Regularization Path Algorithms for Detecting Gene Interactions
 Regularization Path Algorithms for Detecting Gene Interactions Mee Young Park Trevor Hastie July 16, 2006 Abstract In this study, we consider several regularization path algorithms with grouped variable
Regularization Path Algorithms for Detecting Gene Interactions Mee Young Park Trevor Hastie July 16, 2006 Abstract In this study, we consider several regularization path algorithms with grouped variable
Statistical Data Mining and Machine Learning Hilary Term 2016
 Statistical Data Mining and Machine Learning Hilary Term 2016 Dino Sejdinovic Department of Statistics Oxford Slides and other materials available at: http://www.stats.ox.ac.uk/~sejdinov/sdmml Naïve Bayes
Statistical Data Mining and Machine Learning Hilary Term 2016 Dino Sejdinovic Department of Statistics Oxford Slides and other materials available at: http://www.stats.ox.ac.uk/~sejdinov/sdmml Naïve Bayes
Machine Learning, Midterm Exam
 10-601 Machine Learning, Midterm Exam Instructors: Tom Mitchell, Ziv Bar-Joseph Wednesday 12 th December, 2012 There are 9 questions, for a total of 100 points. This exam has 20 pages, make sure you have
10-601 Machine Learning, Midterm Exam Instructors: Tom Mitchell, Ziv Bar-Joseph Wednesday 12 th December, 2012 There are 9 questions, for a total of 100 points. This exam has 20 pages, make sure you have
Statistical Pattern Recognition
 Statistical Pattern Recognition Support Vector Machine (SVM) Hamid R. Rabiee Hadi Asheri, Jafar Muhammadi, Nima Pourdamghani Spring 2013 http://ce.sharif.edu/courses/91-92/2/ce725-1/ Agenda Introduction
Statistical Pattern Recognition Support Vector Machine (SVM) Hamid R. Rabiee Hadi Asheri, Jafar Muhammadi, Nima Pourdamghani Spring 2013 http://ce.sharif.edu/courses/91-92/2/ce725-1/ Agenda Introduction
L5 Support Vector Classification
 L5 Support Vector Classification Support Vector Machine Problem definition Geometrical picture Optimization problem Optimization Problem Hard margin Convexity Dual problem Soft margin problem Alexander
L5 Support Vector Classification Support Vector Machine Problem definition Geometrical picture Optimization problem Optimization Problem Hard margin Convexity Dual problem Soft margin problem Alexander
Decision Tree Learning Lecture 2
 Machine Learning Coms-4771 Decision Tree Learning Lecture 2 January 28, 2008 Two Types of Supervised Learning Problems (recap) Feature (input) space X, label (output) space Y. Unknown distribution D over
Machine Learning Coms-4771 Decision Tree Learning Lecture 2 January 28, 2008 Two Types of Supervised Learning Problems (recap) Feature (input) space X, label (output) space Y. Unknown distribution D over
Regularization Paths. Theme
 June 00 Trevor Hastie, Stanford Statistics June 00 Trevor Hastie, Stanford Statistics Theme Regularization Paths Trevor Hastie Stanford University drawing on collaborations with Brad Efron, Mee-Young Park,
June 00 Trevor Hastie, Stanford Statistics June 00 Trevor Hastie, Stanford Statistics Theme Regularization Paths Trevor Hastie Stanford University drawing on collaborations with Brad Efron, Mee-Young Park,
Sparse Additive Functional and kernel CCA
 Sparse Additive Functional and kernel CCA Sivaraman Balakrishnan* Kriti Puniyani* John Lafferty *Carnegie Mellon University University of Chicago Presented by Miao Liu 5/3/2013 Canonical correlation analysis
Sparse Additive Functional and kernel CCA Sivaraman Balakrishnan* Kriti Puniyani* John Lafferty *Carnegie Mellon University University of Chicago Presented by Miao Liu 5/3/2013 Canonical correlation analysis
Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function.
 Bayesian learning: Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function. Let y be the true label and y be the predicted
Bayesian learning: Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function. Let y be the true label and y be the predicted
Learning Eigenfunctions: Links with Spectral Clustering and Kernel PCA
 Learning Eigenfunctions: Links with Spectral Clustering and Kernel PCA Yoshua Bengio Pascal Vincent Jean-François Paiement University of Montreal April 2, Snowbird Learning 2003 Learning Modal Structures
Learning Eigenfunctions: Links with Spectral Clustering and Kernel PCA Yoshua Bengio Pascal Vincent Jean-François Paiement University of Montreal April 2, Snowbird Learning 2003 Learning Modal Structures
MULTIPLEKERNELLEARNING CSE902
 MULTIPLEKERNELLEARNING CSE902 Multiple Kernel Learning -keywords Heterogeneous information fusion Feature selection Max-margin classification Multiple kernel learning MKL Convex optimization Kernel classification
MULTIPLEKERNELLEARNING CSE902 Multiple Kernel Learning -keywords Heterogeneous information fusion Feature selection Max-margin classification Multiple kernel learning MKL Convex optimization Kernel classification
Plan. Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics.
 Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Lecture 3: Statistical Decision Theory (Part II)
 Lecture 3: Statistical Decision Theory (Part II) Hao Helen Zhang Hao Helen Zhang Lecture 3: Statistical Decision Theory (Part II) 1 / 27 Outline of This Note Part I: Statistics Decision Theory (Classical
Lecture 3: Statistical Decision Theory (Part II) Hao Helen Zhang Hao Helen Zhang Lecture 3: Statistical Decision Theory (Part II) 1 / 27 Outline of This Note Part I: Statistics Decision Theory (Classical
PAC-learning, VC Dimension and Margin-based Bounds
 More details: General: http://www.learning-with-kernels.org/ Example of more complex bounds: http://www.research.ibm.com/people/t/tzhang/papers/jmlr02_cover.ps.gz PAC-learning, VC Dimension and Margin-based
More details: General: http://www.learning-with-kernels.org/ Example of more complex bounds: http://www.research.ibm.com/people/t/tzhang/papers/jmlr02_cover.ps.gz PAC-learning, VC Dimension and Margin-based
Stat542 (F11) Statistical Learning. First consider the scenario where the two classes of points are separable.
 Linear SVM (separable case) First consider the scenario where the two classes of points are separable. It s desirable to have the width (called margin) between the two dashed lines to be large, i.e., have
Linear SVM (separable case) First consider the scenario where the two classes of points are separable. It s desirable to have the width (called margin) between the two dashed lines to be large, i.e., have
CSCI-567: Machine Learning (Spring 2019)
 CSCI-567: Machine Learning (Spring 2019) Prof. Victor Adamchik U of Southern California Mar. 19, 2019 March 19, 2019 1 / 43 Administration March 19, 2019 2 / 43 Administration TA3 is due this week March
CSCI-567: Machine Learning (Spring 2019) Prof. Victor Adamchik U of Southern California Mar. 19, 2019 March 19, 2019 1 / 43 Administration March 19, 2019 2 / 43 Administration TA3 is due this week March
Nonlinear Dimensionality Reduction. Jose A. Costa
 Nonlinear Dimensionality Reduction Jose A. Costa Mathematics of Information Seminar, Dec. Motivation Many useful of signals such as: Image databases; Gene expression microarrays; Internet traffic time
Nonlinear Dimensionality Reduction Jose A. Costa Mathematics of Information Seminar, Dec. Motivation Many useful of signals such as: Image databases; Gene expression microarrays; Internet traffic time
Support Vector Machine (SVM) and Kernel Methods
 Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2016 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2016 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Unsupervised dimensionality reduction
 Unsupervised dimensionality reduction Guillaume Obozinski Ecole des Ponts - ParisTech SOCN course 2014 Guillaume Obozinski Unsupervised dimensionality reduction 1/30 Outline 1 PCA 2 Kernel PCA 3 Multidimensional
Unsupervised dimensionality reduction Guillaume Obozinski Ecole des Ponts - ParisTech SOCN course 2014 Guillaume Obozinski Unsupervised dimensionality reduction 1/30 Outline 1 PCA 2 Kernel PCA 3 Multidimensional
Linear Classifiers (Kernels)
 Universität Potsdam Institut für Informatik Lehrstuhl Linear Classifiers (Kernels) Blaine Nelson, Christoph Sawade, Tobias Scheffer Exam Dates & Course Conclusion There are 2 Exam dates: Feb 20 th March
Universität Potsdam Institut für Informatik Lehrstuhl Linear Classifiers (Kernels) Blaine Nelson, Christoph Sawade, Tobias Scheffer Exam Dates & Course Conclusion There are 2 Exam dates: Feb 20 th March
Midterm exam CS 189/289, Fall 2015
 Midterm exam CS 189/289, Fall 2015 You have 80 minutes for the exam. Total 100 points: 1. True/False: 36 points (18 questions, 2 points each). 2. Multiple-choice questions: 24 points (8 questions, 3 points
Midterm exam CS 189/289, Fall 2015 You have 80 minutes for the exam. Total 100 points: 1. True/False: 36 points (18 questions, 2 points each). 2. Multiple-choice questions: 24 points (8 questions, 3 points
10-701/ Machine Learning - Midterm Exam, Fall 2010
 10-701/15-781 Machine Learning - Midterm Exam, Fall 2010 Aarti Singh Carnegie Mellon University 1. Personal info: Name: Andrew account: E-mail address: 2. There should be 15 numbered pages in this exam
10-701/15-781 Machine Learning - Midterm Exam, Fall 2010 Aarti Singh Carnegie Mellon University 1. Personal info: Name: Andrew account: E-mail address: 2. There should be 15 numbered pages in this exam
Genetic Networks. Korbinian Strimmer. Seminar: Statistical Analysis of RNA-Seq Data 19 June IMISE, Universität Leipzig
 Genetic Networks Korbinian Strimmer IMISE, Universität Leipzig Seminar: Statistical Analysis of RNA-Seq Data 19 June 2012 Korbinian Strimmer, RNA-Seq Networks, 19/6/2012 1 Paper G. I. Allen and Z. Liu.
Genetic Networks Korbinian Strimmer IMISE, Universität Leipzig Seminar: Statistical Analysis of RNA-Seq Data 19 June 2012 Korbinian Strimmer, RNA-Seq Networks, 19/6/2012 1 Paper G. I. Allen and Z. Liu.
Is the test error unbiased for these programs? 2017 Kevin Jamieson
 Is the test error unbiased for these programs? 2017 Kevin Jamieson 1 Is the test error unbiased for this program? 2017 Kevin Jamieson 2 Simple Variable Selection LASSO: Sparse Regression Machine Learning
Is the test error unbiased for these programs? 2017 Kevin Jamieson 1 Is the test error unbiased for this program? 2017 Kevin Jamieson 2 Simple Variable Selection LASSO: Sparse Regression Machine Learning
Max Margin-Classifier
 Max Margin-Classifier Oliver Schulte - CMPT 726 Bishop PRML Ch. 7 Outline Maximum Margin Criterion Math Maximizing the Margin Non-Separable Data Kernels and Non-linear Mappings Where does the maximization
Max Margin-Classifier Oliver Schulte - CMPT 726 Bishop PRML Ch. 7 Outline Maximum Margin Criterion Math Maximizing the Margin Non-Separable Data Kernels and Non-linear Mappings Where does the maximization
Kernels for small molecules
 Kernels for small molecules Günter Klambauer June 23, 2015 Contents 1 Citation and Reference 1 2 Graph kernels 2 2.1 Implementation............................ 2 2.2 The Spectrum Kernel........................
Kernels for small molecules Günter Klambauer June 23, 2015 Contents 1 Citation and Reference 1 2 Graph kernels 2 2.1 Implementation............................ 2 2.2 The Spectrum Kernel........................
Support Vector Machines: Maximum Margin Classifiers
 Support Vector Machines: Maximum Margin Classifiers Machine Learning and Pattern Recognition: September 16, 2008 Piotr Mirowski Based on slides by Sumit Chopra and Fu-Jie Huang 1 Outline What is behind
Support Vector Machines: Maximum Margin Classifiers Machine Learning and Pattern Recognition: September 16, 2008 Piotr Mirowski Based on slides by Sumit Chopra and Fu-Jie Huang 1 Outline What is behind
STA414/2104 Statistical Methods for Machine Learning II
 STA414/2104 Statistical Methods for Machine Learning II Murat A. Erdogdu & David Duvenaud Department of Computer Science Department of Statistical Sciences Lecture 3 Slide credits: Russ Salakhutdinov Announcements
STA414/2104 Statistical Methods for Machine Learning II Murat A. Erdogdu & David Duvenaud Department of Computer Science Department of Statistical Sciences Lecture 3 Slide credits: Russ Salakhutdinov Announcements
Kernel Methods. Foundations of Data Analysis. Torsten Möller. Möller/Mori 1
 Kernel Methods Foundations of Data Analysis Torsten Möller Möller/Mori 1 Reading Chapter 6 of Pattern Recognition and Machine Learning by Bishop Chapter 12 of The Elements of Statistical Learning by Hastie,
Kernel Methods Foundations of Data Analysis Torsten Möller Möller/Mori 1 Reading Chapter 6 of Pattern Recognition and Machine Learning by Bishop Chapter 12 of The Elements of Statistical Learning by Hastie,
ECE G: Special Topics in Signal Processing: Sparsity, Structure, and Inference
 ECE 18-898G: Special Topics in Signal Processing: Sparsity, Structure, and Inference Sparse Recovery using L1 minimization - algorithms Yuejie Chi Department of Electrical and Computer Engineering Spring
ECE 18-898G: Special Topics in Signal Processing: Sparsity, Structure, and Inference Sparse Recovery using L1 minimization - algorithms Yuejie Chi Department of Electrical and Computer Engineering Spring
