Slice Sampling Mixture Models
|
|
|
- Esmond Byrd
- 5 years ago
- Views:
Transcription
1 Slice Sampling Mixture Models Maria Kalli, Jim E. Griffin & Stephen G. Walker Centre for Health Services Studies, University of Kent Institute of Mathematics, Statistics & Actuarial Science, University of Kent Abstract We propose a more efficient version of the slice sampler for Dirichlet process mixture models described by Walker (27). This sampler allows the fitting of infinite mixture models with a wide range of prior specification. To illustrate this flexiblity we develop a new nonparametric prior for mixture models by normalizing an infinite sequence of independent positive random variables and show how the slice sampler can be applied to make inference in this model. Two submodels are studied in detail. The first one assumes that the positive random variables are Gamma distributed and the second assumes that they are inverse Gaussian distributed. Both priors have two hyperparameters and we consider their effect on the prior distribution of the number of occupied clusters in a sample. Extensive computational comparisons with alternative conditional simulation techniques for mixture models using the standard Dirichlet process prior and our new prior are made. The properties of the new prior are illustrated on a density estimation problem. Keywords: Dirichlet process; Markov chain Monte Carlo; Mixture model; Normalized Weights; Slice sampler. Corresponding author: Jim E. Griffin, Institute of Mathematics, Statistics & Actuarial Science, University of Kent, Canterbury, U. K. Tel.: ; Fax: ; jeg28@kent.ac.uk 1
2 1 Introduction The well known and widely used mixture of Dirichlet process (MDP) model was first introduced by Lo (1984). The MDP model, with Gaussian kernel, is given by f P (y) = K(y; φ) dp(φ) with K(y; φ) being a normal kernel and P D(M, P ). We write P D(M, P ) to denote that P is a Dirichlet process (Ferguson, 1973) with parameters M >, the scale parameter, and P, a distribution on the real line and φ = (µ, σ 2 ) with µ to represent the mean and σ 2 the variance of the normal component. Since the advent of Markov chain Monte Carlo methods within the mainstream statistics literature (Smith and Roberts, 1993), and the specific application to the MDP model (Escobar, 1988; Escobar, 1994; Escobar and West, 1995), the model has become one of the most popular in Bayesian nonparametrics since it is possible to integrate P from the posterior defined by this model. Variations of the original algorithm of Escobar (1988) have been numerous; for example, MacEachern (1994); Müller and MacEachern (1998); Neal (2). All of these algorithms rely on integrating out the random distribution function from the model, removing the infinite dimensional problem. These are usually referred to as marginal methods. Recent ideas have left the infinite dimensional distribution in the model and found ways of sampling a sufficient but finite number of variables at each iteration of a Markov chain with the correct stationary distribution. See Papaspiliopoulos and Roberts (28) and Walker (27); the latter paper using slice sampling ideas. These define so called conditional methods. There has recently been interest in defining nonparametric priors for P that move beyond the Dirichlet process (see e.g. Lijoi et al (27)) in infinite mixture models. These alternative priors allow more control over the prior cluster structure than would be possible with the Dirichlet process. The availability of computational methods for posterior inference,that do not integrate out P, allows us to implement these priors. The purpose of this paper is two fold: 1) to develop an efficient version of the slice sampling algorithm for MDP models proposed by Walker (27) and to extend it to more general nonparametric priors such as general stick breaking processes and normalised weights priors and 2) to develop a new class of nonparametric prior for infinite mixture models by normalizing an infinite sequence of positive random variables, which will be termed a Normalized Weights prior. The lay out of the paper is 2
3 as follows. In Section 2 we describe the slice efficient sampler for the MDP model. Section 3 describes the normalized weights prior and discusses constructing a slice sampler for infinite mixture models with this prior. Section 4 discusses an application of the normalized weights prior to modelling the hazard in survival analysis and Section 5 contains numerical illustrations and an application of the normalized weight prior to density estimation. Finally, Section 6 contains conclusions and a discussion. 2 The slice efficient sampler for the MDP It is well known that P D(M, P ) has a stick breaking representation (Sethuraman, 1994) given by P = w j δ φj, where the {φ j } are independent and identically distributed from P and w 1 = z 1, w j = z j (1 z l ) j=1 with the {z j } being independent and identically distributed from beta(1, M). It is possible to integrate P from the posterior defined by the MDP model. However, the stick breaking representation is essential to estimation via the non marginal methods of Papaspiliopoulos and Roberts (28) and Walker (27). The idea is that we can write f z,φ (y) = l<j w j K(y; φ j ) j=1 and the key is to find exactly which (finite number of) variables need to be sampled to produce a valid Markov chain with correct stationary distribution. The details of the slice sampler algorithm are given in Walker (27), but we briefly describe the basis for the algorithm here and note an improvement, also noticed by Papaspiliopoulos (28). The joint density f z,φ (y, u) = 1(u < w j ) K(y; φ j ) j=1 is the starting point. Given the latent variable u, the number of mixtures is finite, the indices being A u = {k : w k > u}. One has f z,φ (y u) = Nu 1 K(y; φ j ), j A u 3
4 and the size of A u is N u = j=1 1(w j > u). One can then introduce a further latent variable, d, which indicates which of these finite number of mixtures provides the observation to give the joint density f z,φ (y, u, d) = 1(u < w d ) K(y; φ d ). Hence, a complete likelihood function for (z, φ) is available as a simple product of terms and crucially d is finite. Without u, d can take an infinite number of values which would make the implementation of a Markov chain Monte Carlo algorithm problematic. We briefly describe the simulation algorithm, but only provide the sampling procedure without derivation since this has appeared elsewhere (Walker, 27). However, as mentioned earlier, we do sample one of the full conditionals in a different and more efficient manner. We sample π(z, u ) as a block and this involves sampling π(z exclude u) and then π(u z, ), where π(z exclude u) is obtained by integrating out u from π(z, u ). The distribution π(z exclude u) will be the standard full conditional for a stick breaking process (see Ishwaran and James (21)). Standard MCMC theory on blocking suggests that this should lead to a more efficient sampler. Recall that we have the model f(y) = w j K(y; φ j ), j=1 where the {φ j } are independent and identically distributed from P, the {w j } have a stick breaking process based on the Dirichlet process, described earlier in this section. The variables that need to be sampled at each sweep of a Gibbs sampler are 1. π(φ j ) p (φ j ) d i =j K(y i; φ j ). {(φ j, z j ), j = 1, 2,...; (d i, u i ), i = 1,...,n}. 2. π(z j exclude u) beta(z j ; a j, b j ), where and a j = 1 + b j = M + n 1(d i = j) i=1 n 1(d i > j). i=1 4
5 3. π(u i ) 1( < u i < w di ). 4. P(d i = k ) 1(k : w k > u i ) K(y i ; φ k ). Obviously, we can not sample all of the (φ j, z j ). But it is not required to in order to proceed with the chain. We only need to sample up to the integer N for which we have found all the appropriate w k in order to do step 4 exactly. Since the weights sum to 1 if we find N i such that N i k=1 w k > 1 u i then it is not possible for any of the w k, for k > N i, to be greater than u i. There are some important points to make here. First, it is a trivial extension to consider more general stick breaking processes for which z j beta(α j, β j ) independently. Then, in this case, we would have a j = α j + n 1(d i = j) i=1 and n b j = β j + 1(d i > j). i=1 This easy extension to more general priors is not a feature of alternative, marginal sampling algorithms. Secondly, the algorithm is remarkably simple to implement; all full conditionals are standard. Later, for the illustrations and comparison, we will consider two types of slice sampler. The slice efficient which is the one described above and the slice which is the original algorithm appearing in Walker (27) and is noted by the fact that the v is sampled conditional on u in this case. The retrospective sampler (Papaspiliopoulos and Roberts 28) is an alternative, conditional method. The following argument gives some understanding for the difference between retrospective sampling (which uses Metropolis sampling) and slice sampling. Suppose we wish to sample from f(x) l(x)π(x) using Metropolis sampling and use π(x) as the proposal density. Let x c be the current sample and x π(x) and u Un(, 1), so the new sample x n is x if u < l(x )/l(x c ) or else is x c. On the other hand, the slice sampler would work by considering f(x, u) 1(u < l(x))π(x) and so a move from x c to x n would work by sampling x n from π(x) restricted to {x : l(x)/l(x c ) > u} where u Un(, 1). So the two sampling strategies are using the same variables but in a fundamentally different way, which allows the slice sampling version to always move. 5
6 This illustration is obviously demonstrated on a simple level, but we believe the principle applies to the difference between the retrospective sampler and the slice sampler for the mixture of Dirichlet process model. 3 Mixtures Based on Normalized Weights 3.1 Definition and Properties The slice sampling idea can be extended to mixture models with weights obtained via normalization. The Dirichlet process has been the dominant prior in nonparametrics but the definition of alternative nonparametric priors has been a recent area of interest. For example, Lijoi et al (27) define nonparametric priors through the normalization of the generalized Gamma process. We discuss an alternative form of normalization. We consider f(y) = w j K(y; φ j ) j=1 where w j = λ j /Λ and Λ = j=1 λ j. We will also use Λ m = j=m+1 λ j. Here the {λ j } are positive and will be assigned independent prior distributions, say λ j π j (λ j ). These must be constructed so as to ensure that j=1 λ j < + a.s. We suggest defining specific priors by defining E[λ j ] = ξq j where ξ > and q j = P(X = j) where X is a random variable whose distribution is discrete on the positive integers. For example, we could assume that X = Y + 1 where Y follows a geometric distribution. Then q j = (1 θ)θ j 1. The parameter θ controls the rate at which E[λ 1 ], E[λ 2 ], E[λ 2 ],... tends to zero. We have defined a nonparametric prior with two parameters θ and ξ. As we will see in the following examples, the choice of the distributions π 1, π 2, π 3,... controls the properties of the process. Many other families of nonparametric prior distribution can be generated by different choices of X. For example, we could assume that X = Y +1 where Y follows a Poisson distribution. Example 1: Gamma distribution. Here we take the {λ j } to be independent gamma distributions, say λ j Ga(γ j, 1). To ensure that Λ < + a.s. we take j=1 γ j < +. Clearly, w j has expectation 6
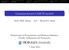
7 ξ =.1 ξ = 1 ξ = θ = θ = θ = Figure 1: Prior distribution of the number of clusters from 3 observations with the infinite Dirichlet prior q j and variance q j (1 q j )/(ξ + 1) and we can interpret ξ as a mass parameter. We will refer to this model as an infinite Dirichlet prior since if we have a finite number of unnormalized weights λ 1, λ 2,...,λ N then w 1, w 2,...,w N would be Dirichlet distributed. In infinite mixture models, the prior distribution on the number of clusters from n observations is important. Figure 1 shows this distribution for n = 3. Larger values of θ for fixed ξ place more mass on larger numbers of clusters (as we would expect since the weights decay increasingly slowly with larger θ). The mass parameter ξ also plays an important role. Larger values of ξ lead to more dispersed distributions with a larger median value. Stick breaking priors were introduced to Bayesian nonparametrics by Ishwaran and James (21). They are defined by two infinite vectors of parameters. Clearly, there is a need to develop priors within this class that have a few hyperparameters to allow easy prior specification. The Dirichlet process and Poisson-Dirichlet process are two such priors and the infinite Dirichlet prior represents another. The( stick-breaking representation of the infinite Dirichlet prior takes α j = ξq j and β j = ξ 1 ) j i=1 q i. 7
8 Example 2: Inverse Gaussian distribution The inverse Gaussian distribution, IG(γ, η), has a density function given by π(λ) = γ { λ 3/2 exp 1 ( ) } γ 2 2π 2 λ + η2 λ + ηγ, where γ and η can be interpreted as a shape and a scale parameter, respectively. We take λ j to follow independent IG(γ j, 1) distributions. Then Λ m = j=m+1 λ j is disξ =.1 ξ = 1 ξ = θ = θ = θ = Figure 2: Prior distribution of the number of clusters for infinite normalized inverse Gaussian prior tributed as IG( j=m+1 γ j, 1) and the normalization is well defined if j=1 γ j < + which implies that Λ is almost surely finite. The finite dimensional normalized distribution (λ 1 /Λ, λ 2 /Λ,..., λ m /Λ) has been studied by Lijoi et al. (25) as the normalized inverse Gaussian distribution. We again define γ j = ξq j and it follows directly from their results that w i has expectation q i and variance q i (1 q i )ξ 2 exp{ξ}γ( 2, ξ). This prior will be referred to as the infinite normalized inverse Gaussian prior. Figure 2 shows the prior distribution of the number of clusters in 3 observations. The effects of ξ and θ follow the same pattern as the infinite Dirichlet case discussed above. However, the effect of ξ is less marked for small ξ. In the infinite Dirichlet case for ξ =.1, the distributions are almost indistinguishable for different values of θ but in 8
9 this case it is clear that the location of the distribution is increasing with θ. This allows easier prior specification for the infinite normalized inverse Gaussian prior 3.2 Slice sampler The model can be fitted using an extension of the slice sampler developed in section 2. We will assume that the distribution of Λ m has a known form for all m, which we will denote by π m (Λ m). The introduction of a normalizing constant, Λ, makes MCMC trickier. Simpler updating is possible when we introduce the additional latent variable v, and consider the joint density Clearly the marginal density f(y, v, u, d) = exp( vλ)1(u < λ d ) K(y; φ d ). f(y, d) = λ d Λ K(y; φ d), as required. The likelihood function based on a sample of size n is given by n exp( v i Λ)1(u i < λ di ) K(y i ; φ di ). i=1 We will only consider those conditional distributions which are not immediately trivial; those that are completely trivial being u i, v i and φ j. The distribution of d i is trivial but as before we need to find the number of λ j s (and also φ j s) to be sampled in order to implement the sampling of d i. Hence, the non trivial aspect to the algorithm is the sampling of the sufficient number of {λ j } and Λ. We will, as before, work on the conditional distribution of the ({λ j }, Λ) excluding the {u i }. We simulate λ 1,..., λ m, Λ m (where m is the number of atoms given in the previous iteration) in a block from their full conditional distribution which is proportional to exp{ V Λ m }π m (Λ m) m j=1 exp{ V λ j }λ n j j π j(λ j ), where n j = n i=1 1(d i = j) and V = n i=1 v i. We need to find the smallest value of m for which Λ m < min i {u i } so that we can evaluate the full conditional distribution of d i. This value can be found by sequentially simulating [λ j, Λ j Λ j 1 ] for j = m + 1,...,m. The conditional distribution of [λ j = x, Λ j = Λ j 1 x Λ j 1 ] is given by f(x) π j (x)π j (Λ j 1 x), < x < Λ j 1. 9
10 In some cases simulation from the distribution will be straightforward. If not, generic univariate simulation methods such as Adaptive Rejection Metropolis Sampling (Gilks et al. 1995) can be employed. We now consider a couple of examples. Example 1: Gamma distribution It is easy to see that ( π(λ 1 /Λ,...,λ m /Λ Λ,, exclude u) = Dir γ 1 + n 1,...,γ m + n m, l=m+1 γ l ) and ( ) Λ m Ga j=m+1 γ j, 1 + V. The conditional distribution of λ j /Λ j is Be(γ j, i=j+1 γ i). This prior can also be represented as a stick breaking prior. Example 2: Inverse Gaussian distribution The full conditional distribution of λ j is given by { π(λ j ) λ n j 3/2 j exp 1 ( γ 2 )} j + (1 + 2V )λ j, 2 λ j where n j is the number of observations allocated to component j. The full conditional distribution of Λ m is proportional to { Λm 3/2 exp 1 ( ( i=m+1 γ i) 2 )} + (1 + 2V )λ j. 2 λ j These are both generalized inverse Gaussian distributions which can be simulated directly; see e.g. Devroye (1986). We can simulate from [λ j+1, Λ j+1 Λ j ] by defining λ j+1 = x j+1 Λ j and Λ j+1 = (1 x j+1 )Λ j where the density of x j+1 is given by { [ g(x j+1 ) x 3/2 j+1 (1 x j+1) 3/2 exp 1 γ 2 j + ( i=j+1 γ ]} i) 2. 2 Λ j x j+1 Λ j (1 x j+1 ) Unlike the gamma case, this conditional distribution depends on Λ m. The distribution of x j+1 /(1 x j+1 ) can be identified as a two mixture of generalized inverse Gaussian distributions and hence can be sampled easily (details are given in the Appendix). 1
11 4 Hazard Functions The normalized procedure can also be applied to the modeling of random hazard functions. Suppose we model the unknown hazard function h(t), for t >, using a set of known functions {h k (t)} k=1, via h(t) = λ k h k (t). k=1 Here the {λ k > } are the model parameters and can be assigned independent gamma prior distributions; say λ k Ga(a k, b k ). Obviously we will need to select (a k, b k ) to ensure that h(t) < + a.s. for all t < +. The corresponding density function is given by { } f(t) = λ k h k (t) exp λ k H k (t), k=1 k=1 where H k is the cumulative hazard corresponding to h k. So with observations {t i } n i=1, the likelihood function is given by [ n { }] l(λ t) λ k h k (t i ) exp λ k H k (t i ). i=1 k=1 k=1 Our approach is based on the introduction of a latent variable, say u, so that we consider the joint density with t given by { } f(t, u) = 1(u < λ k ) h k (t) exp λ k H k (t). k=1 k=1 A further latent variable d picks out the mixture component from which (t, u) come, { } f(t, u, d) = 1(u < λ d ) h d (t) exp λ k H k (t). We will now introduce the key latent variables, one for each observation, and label them (u i, d i ), into the likelihood, which is given by { } n l(λ t, u, d) 1(u i < λ di ) h di (t i ) exp λ k H k (t i ). k=1 i=1 k=1 The point is that the choice of d i is finite. It is now clear that the sampling algorithm for this model is basically the same now as for the normalized case. We could take 11
12 the λ j to be gamma with parameters a j + d i =j 1 and b j + d i =j H j(t i ) and we would first sample up to M = max i d i. Then the u i are from Un(, λ di ). In order to sample the d i we need to find all the λ j greater than u i. We can do this by sampling Λ M = j>m λ j as a gamma distribution and then sampling [λ M+1,...λ Ni ] Λ M so that N i is the smallest integer for which N i j=m+1 λ j > Λ M u i. Finally, once we have found all the λ j > u i, we can sample d i from Pr(d i = j) 1(λ j > u i )h j (t i ). 5 Illustration and Comparisons In this section we carry out a comparison of the slice sampling algorithm with the retrospective sampler using the Dirichlet process and the normalized weights prior. The algorithms are compared using the normal kernel K(y φ) with components φ = (µ, ζ), and P (µ, ζ) = N(µ µ, ξ 2 ) G(ζ γ, β). Here G(γ, β) denotes the gamma distribution. We also consider inference for the commonly used galaxy data set with the infinite Dirichlet and infinite normalized inverse Gaussian priors. For comparison purposes we consider two real data sets and two simulated data sets. The real data sets are: 1. Galaxy data set which consists of the velocities of 82 distant galaxies diverging from our own galaxy. This is the most commonly used data set in density estimation studies, due to its mulimodality. We will also use it to illustrate the effect of the prior choice on the posterior density in Section S & P 5 data set which consist of 223 daily index returns. This is yet another commonly used data set in density estimation and volatility studies of financial asset returns; see, Jacquier, Polson, and Rossi (1994, 24). This data set is unimodal, not necessarily symmetric, around zero, and it is characterized by heavy tails. We chose these data sets because of their size, as we would like to study the performance of the algorithms on both small and large data sets. The simulated data sets are based on the models used in Green and Richardson (21) and consist of 1 draws from a bimodal and a leptokurtic mixture. 1. The bimodal mixture:.5n( 1,.5 2 ) +.5N(1,.5 2 ). 2. The leptokurtic mixture:.67n(, 1) +.33N(.3,.25 2 ). 12
13 Both of these simulated data sets were used in the algorithm comparison study carried out in Papaspiliopoulos and Roberts (28); since we are comparing our slice sampler with the retrospective sampler, we decided to use these simulated data sets. The parameters for our MDP mixture are also set according to Green and Richardson (21). If R is the range of the data; then we take µ = R/2, ξ = R, γ = 2, and β =.2R 2. The precision parameter of the Dirichlet Process is set at M = 1. In the comparison of the estimates of the statistics used, we took the Monte Carlo sample size to be S = 25, for each algorithm, with the initial 1, used as a burn in period. Density estimates using the retrospective and slice efficient samplers are shown in figure 3 for the Dirichlet process mixture model. Bi modal Leptokurtic S & P 5 (a) (b) Figure 3: Predictive densities: (a) retrospective and (b) slice efficient 5.1 Algorithmic performance To monitor the performance of the algorithms we look at the convergence of two quantities: The number of clusters: at each iteration there are j = 1,..., N clusters of the i = 1,...,n data points with m j being the size of the j cluster, so that N j=1 m j = n. The deviance, D, of the estimated density, calculated as ( ) n m j D = 2 log n K(y i φ j ). i=1 j 13
14 These variables have been used in the previous comparison studies of Papaspiliopoulos and Roberts (28), Green and Richardson (21) and Neal (2). Here D is one of the most common functionals used in comparing algorithms, because it is seen as a global function of all model parameters. Although we produce this variable and study its algorithmic performance we are also concerned with the convergence of the number of clusters. The efficiency of the algorithms is summarized by computing an estimate τ of the integrated autocorrelation time, τ, for each of the variables. Integrated autocorrelation time is defined in Sokal (1997) as τ = ρ l. where ρ l is the autocorrelation at lag l. An estimate of τ has been used in Papaspiliopoulos and Roberts (28), Green and Richardson (21) and Neal (2). Integrated autocorrelation time is of interest as it controls the statistical error in Monte Carlo measurements of a desired function f. To clarify this point, consider the Monte Carlo sample mean, f 1 S l=1 S f j, where S is the number of iterations. The variance of f according to Sokal (1997) is l=1 Var( f) 1 2τ V, S where V is the marginal variance. Sokal (1997) concludes that Var( f) is a factor 2τ larger than what it would be if the {f j } were statistically independent. In other words, τ determines the statistical error of the Monte Carlo measurements of f once equilibrium has been attained. Therefore a run of S iterations contains only S/(2τ) effectively independent data points. This means that the algorithm with the smallest estimated value of τ will be the most efficient. The problem with the calculation of τ lies in accurately estimating the covariance between the states, which in turn is used to calculate the autocorrelation ρ l. It must be noted that in MCMC the covariance and the autocorrelation are not single values but random variables. Based on Sokal (1997) the estimator for τ is given by τ = 1 C l=1 ρ l.
15 where ρ l is the estimated autocorrelation at lag l (obtained via MatLab) and C is a cut off point, normally set by the researcher. In our comparisons we define, as is commonly done, { C = min l : ρ l < 2/ } S. Then C is the smallest lag for which we would not reject the null hypothesis H : ρ l =. A similar approach has also been used in Papaspiliopoulos (28). The issue here, see Sokal (1997), is the cut off point C; it introduces bias equal to Bias( τ) = 1 ( ) 1 ρ l + o. 2 S l >C On the other hand, the variance of τ can be computed using 2(2C 1) Var( τ) τ 2. S The choice of C will be a trade off between the bias and the variance of ˆτ, which means that we really cannot say how good an algorithm is since the choice of C point is left to the researcher. According to Sokal (1997), this approach works well when a sufficient quantity of data is available which we can control by running the sampler for a sufficient number of iterations. 5.2 Results The following tables compare the estimated integrated autocorrelation time τ of the two variables of interest; the number of clusters and the deviance Dirichlet process Looking at the estimates of τ for the real data sets we come to the following conclusions: For the galaxy data set there is little difference between the two samplers. Even though the retrospective sampler performs marginally better, the slice efficient sampler is easier to use as simulating the z and k is carried out in an easy way, as opposed to the complexity of the set up of the retrospective sampling steps. For the S&P data set which is large, unimodal, asymmetric and heavy tailed, it is the slice efficient sampler that outperforms the retrospective sampler, in terms of τ for the number of clusters; τ for the slice efficient sampler is about half that of the retrospective sampler. 15
16 Galaxy data Leptokurtic data τ for clust τ for D τ for clust τ for D Slice Slice efficient Retrospective Bimodal data S&P 5 data τ for clust τ for D τ for clust τ for D Slice Slice efficient Retrospective Table 1: Estimates of the integrated autocorrelation times for the deviance (D) and for the number of clusters with four data sets with the Dirichlet process mixture model Galaxy Bimodal Leptokurtic S & P 5 clust Dev Figure 4: Autocorrelation of MCMC output for: slice sampler (red), efficient slice sampler (blue) and retrospective sampler (green) Mixtures based on normalized weights We reject the slice sampler in favour of the slice efficient sampler. We use the infinite Dirichlet and infinite normalized inverse Gaussian mixtures models with ξ = 1 and θ =.5 on the four data sets. We find similar performance for the normalized weights prior as for the Dirichlet process prior. The retrospective sampler is usually more efficient than the slice sampler with a two times relative improvement on average. 16
17 Galaxy data Leptokurtic data τ for clust τ for D τ for clust τ for D Slice efficient Retrospective Bimodal data S&P 5 data τ for clust τ for D τ for clust τ for D Slice efficient Retrospective Table 2: Estimates of the integrated autocorrelation times for the deviance (D) and for the number of clusters with four data sets with the infinite Dirichlet distribution mixture model The improvement is typically larger for the simulated rather than the real data sets. The effect is also more pronounced for the infinite Dirichlet distribution prior rather than the infinite normalized inverse Gaussian prior. Galaxy data Leptokurtic data τ for clust τ for D τ for clust τ for D Slice efficient Retrospective Bimodal data S&P 5 data τ for clust τ for D τ for clust τ for D Slice efficient Retrospective Table 3: Estimates of the integrated autocorrelation times for the deviance (D) and for the number of clusters with four data sets with the infinite normalized inverse Gaussian distribution mixture model These results are not surprising. The slice sampler introduces auxiliary variables to help simulation which will slow convergence through over conditioning. The slice efficient sampler reduces this effect by jointly updating u and λ (or V in the Dirichlet 17
18 process case) in a block. The retrospective sampler will mix slowly when the proposal distribution is a poor approximation to the full conditional distribution. Therefore it is usually difficult to be sure about the ranking of the methods. In these illustrations, we have seen examples where the slice efficient sampler is more efficient than the retrospective sampler. 5.3 Inference for the Normalized Weights Priors The galaxy data has been a popular data set in Bayesian nonparametric modelling and we will illustrate the infinite Dirichlet and infinite normalized inverse Gaussian priors on it. The posterior mean density estimates are shown in figure 5 for the infinite Dirichlet prior and figure 6 for the infinite normalized inverse Gaussian prior. The hyperparameters of the prior distributions have a clear effect on the posterior.25 ξ =.1 ξ = 1 ξ = θ = θ = θ = Figure 5: Posterior mean density estimates for the galaxy data using the infinite Dirichlet prior with different values of M and θ mean estimates. Prior distributions that places more mass on a small number of components tend to find estimates with three clear modes. As the prior mean number of components increases so do the number of modes in the estimate from 4 to 5 for 18
19 the prior within each class that places most mass on a large number of components (ξ = 1 and φ =.9). However, there are some clear differences between the two.25 ξ =.1 ξ = 1 ξ = θ = θ = θ = Figure 6: Posterior mean density estimates for the galaxy data using the infinite normalized inverse Gaussian prior with different values of ξ and θ classes of prior. The effects of the two hyperparameters on the prior distribution of the number of non empty components were more clearly distinguishable in the infinite normalized inverse Gaussian prior than the infinite Dirichlet prior. In the infinite normalized inverse Gaussian prior θ controls the mean number of non empty components whereas ξ controls the dispersion around the mean. This property is carried forward to the posterior mean density and the number of modes in the posterior mean increases with θ. For example, when ξ =.1, there are three modes in the posterior mean if θ =.4 whereas there are 4 when θ =.9. Similarly, larger values of ξ are associated with larger variability in the prior mean and favour distributions which uses a larger number of components. This suggests that infinite normalized inverse Gaussian distribution may be a more easily specified prior distribution than the infinite Dirichlet prior. 19
20 6 Conclusions and Discussion This paper has shown how mixture models based on random probability measures, of either the stick breaking or normalized types, can be easily handled via the introduction of a key latent variable which makes finite the number of mixtures. The more complicated of the two is the normalized type, which requires particular distributions of the unnormalized weights in order to be able to make the simulation algorithm work. Nevertheless, such distributions based on the gamma and inverse Gaussian distributions are popular choices anyway. Further ideas which need to be worked out include the case when we can generate weights which are decreasing. This for example would make the search for those w j > u are far simpler exercise and would lead to more efficient algorithms. In conclusion, concerning performance of slice efficient and retrospective samplers, we note that once running, both samplers are approximately the same in terms of efficiency and performance. In terms of time efficiency we have found that for large data sets, like the S&P 5 the slice efficient sampler is more efficient than the retrospective sampler, it takes approximately half the time to run than the retrospective sampler. The most notable savings of the slice efficient sampler are in the pre running work where setting up a slice sampler is far easier than setting up a retrospective sampler. The slice sampler allows the Gibbs sampling step for a finite mixture model to be used at each iteration and introduce a method for updating the truncation point in each iteration. This allows standard methods for finite mixture models to be used directly. For example, Van Gael et al (28) fit an infinite hidden Markov model using the forward backward sampler for finite hidden Markov model using the slice sampling idea. This would be difficult to implement in a retrospective framework since the truncation point changes when updating the allocations. 2
21 References Devroye, L. (1986): Non Uniform Random Variate Generation, Springer Verlag: New York. Escobar, M.D Estimating the means of several normal populations by nonparametric estimation of the distribution of the means. Unpublished Ph.D. dissertation, Department of Statistics, Yale University. Escobar, M.D. (1994). Estimating normal means with a Dirichlet process prior. Journal of the American Statistical Association 89, Escobar, M.D. and West, M. (1995). Bayesian density estimation and inference using mixtures. Journal of the American Statistical Association 9, Ferguson, T.S. (1973). A Bayesian analysis of some nonparametric problems. Annals of Statistics 1, Gilks, W.R., Best, N.G. and Tan, K.K.C. (1995). Adaptive rejection Metropolis sampling within Gibbs sampling. Applied Statistics 44, Green, P. J. and Richardson, S. (21). Modelling heterogeneity with and without the Dirichlet process. Scandinavian Journal of Statistics 28, Ishwaran, H. and James, L.F. (21). Gibbs sampling methods for stick breaking priors. Journal of the American Statistical Association 96, Jacquier, E., Polson, N., and Rossi P.E. (1994). Bayesian Analysis of stochastic volatility models. Journal of Business and Economic Statistics 12, Jacquier, E., Polson, N., and Rossi P.E. (24). Bayesian Analysis of stochastic volatility models with fat tails and correlated errors. Journal of Econometrics 122, Lijoi, A., Mena, R. H. and Prünster, I. (25). Hierarchical Mixture Modeling with Normalized Inverse Gaussian Priors. Journal of the American Statistical Association 1, Lijoi, A., Mena, R. H. and Prüenster, I (27): Controlling the reinforcement in Bayesian nonparametric mixture models, Journal of the Royal Statistical Society B, 69,
22 Lo, A.Y. (1984). On a class of Bayesian nonparametric estimates I. Density estimates. Annals of Statistics 12, MacEachern, S.N. (1994). Estimating normal means with a conjugate style Dirichlet process prior. Communications in Statistics: Simulation and Computation 23, MacEachern, S.N. and Müller, P. (1998). Estimating mixtures of Dirichlet process models. Journal of Computational and Graphical Statistics 7, Neal, R. (2). Markov chain sampling methods for Dirichlet process mixture models. Journal of Computational and Graphical Statistics 9, Papaspiliopoulos, O. and Roberts, G.O. (28). Retrospective Markov chain Monte Carlo methods for Dirichlet process hierarchical models. Biometrika 95, Papaspiliopoulos, O. (28). A note on posterior sampling from Dirichlet mixture models. Preprint. Sethuraman, J. (1994). A constructive definition of Dirichlet priors. Statistica Sinica 4, Sokal, A. (1997). Monte Carlo Methods in Statistical Mechanics: Foundations and New Algorithms. Functional Integration (Cargése, 1996) 361of NATO Adv. Sci. Inst. Ser. B Phys., New York: Plenum, Smith, A.F.M. and Roberts, G.O. (1993). Bayesian computations via the Gibbs sampler and related Markov chain Monte Carlo methods. Journal of the Royal Statistical Society, Series B 55, Van Gael, J., Saatchi, Y., Teh, Y.W., and Ghahramani, Z. (28). Beam Sampling for the Infinite Hidden Markov Model. Technical Report : Engineering Department, University of Cambridge. Walker, S.G. (27). Sampling the Dirichlet mixture model with slices. Communications in Statistics: Simulation and Computation 36,
23 Appendix Simulation for the Inverse Gaussian model. We wish to simulate from the density g(x j+1 ) g(x j+1 ) x 3/2 j+1 (1 x j+1) 3/2 exp The transformation y j+1 = x j+1 1 x j+1 has the density g(y j+1 ) y 3/2 j+1 (1 + y j+1) exp { 1 2 { [ 1 γ 2 j + ( i=j+1 γ ]} i) 2. 2 Λ j x j+1 Λ j (1 x j+1 ) [ γ 2 j + ( i=j+1 γ ]} i) 2 y j+1. Λ j y j+1 Λ j which can be expressed as a mixture of two generalized inverse Gaussian distributions ( ) ( ) w GIG 1/2, γ j /Λ j, γ i /Λ j + (1 w) GIG 1/2, γ j /Λ j, γ i /Λ j i=j+1 i=j+1 where w = γ j i=j+1 γ i and GIG(p, a, b) denotes a distribution with density (b/a) ( p/2 ab )x (p 1) exp{ (a/x + bx)/2} 2K p where K ν denotes the modified Bessel function of the third kind with index ν. 23
A Nonparametric Model for Stationary Time Series
 A Nonparametric Model for Stationary Time Series Isadora Antoniano-Villalobos Bocconi University, Milan, Italy. isadora.antoniano@unibocconi.it Stephen G. Walker University of Texas at Austin, USA. s.g.walker@math.utexas.edu
A Nonparametric Model for Stationary Time Series Isadora Antoniano-Villalobos Bocconi University, Milan, Italy. isadora.antoniano@unibocconi.it Stephen G. Walker University of Texas at Austin, USA. s.g.walker@math.utexas.edu
Slice sampling σ stable Poisson Kingman mixture models
 ISSN 2279-9362 Slice sampling σ stable Poisson Kingman mixture models Stefano Favaro S.G. Walker No. 324 December 2013 www.carloalberto.org/research/working-papers 2013 by Stefano Favaro and S.G. Walker.
ISSN 2279-9362 Slice sampling σ stable Poisson Kingman mixture models Stefano Favaro S.G. Walker No. 324 December 2013 www.carloalberto.org/research/working-papers 2013 by Stefano Favaro and S.G. Walker.
Bayesian Statistics. Debdeep Pati Florida State University. April 3, 2017
 Bayesian Statistics Debdeep Pati Florida State University April 3, 2017 Finite mixture model The finite mixture of normals can be equivalently expressed as y i N(µ Si ; τ 1 S i ), S i k π h δ h h=1 δ h
Bayesian Statistics Debdeep Pati Florida State University April 3, 2017 Finite mixture model The finite mixture of normals can be equivalently expressed as y i N(µ Si ; τ 1 S i ), S i k π h δ h h=1 δ h
Default priors for density estimation with mixture models
 Bayesian Analysis ) 5, Number, pp. 45 64 Default priors for density estimation with mixture models J.E. Griffin Abstract. The infinite mixture of normals model has become a popular method for density estimation
Bayesian Analysis ) 5, Number, pp. 45 64 Default priors for density estimation with mixture models J.E. Griffin Abstract. The infinite mixture of normals model has become a popular method for density estimation
Bayesian Methods for Machine Learning
 Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
CSC 2541: Bayesian Methods for Machine Learning
 CSC 2541: Bayesian Methods for Machine Learning Radford M. Neal, University of Toronto, 2011 Lecture 3 More Markov Chain Monte Carlo Methods The Metropolis algorithm isn t the only way to do MCMC. We ll
CSC 2541: Bayesian Methods for Machine Learning Radford M. Neal, University of Toronto, 2011 Lecture 3 More Markov Chain Monte Carlo Methods The Metropolis algorithm isn t the only way to do MCMC. We ll
27 : Distributed Monte Carlo Markov Chain. 1 Recap of MCMC and Naive Parallel Gibbs Sampling
 10-708: Probabilistic Graphical Models 10-708, Spring 2014 27 : Distributed Monte Carlo Markov Chain Lecturer: Eric P. Xing Scribes: Pengtao Xie, Khoa Luu In this scribe, we are going to review the Parallel
10-708: Probabilistic Graphical Models 10-708, Spring 2014 27 : Distributed Monte Carlo Markov Chain Lecturer: Eric P. Xing Scribes: Pengtao Xie, Khoa Luu In this scribe, we are going to review the Parallel
A marginal sampler for σ-stable Poisson-Kingman mixture models
 A marginal sampler for σ-stable Poisson-Kingman mixture models joint work with Yee Whye Teh and Stefano Favaro María Lomelí Gatsby Unit, University College London Talk at the BNP 10 Raleigh, North Carolina
A marginal sampler for σ-stable Poisson-Kingman mixture models joint work with Yee Whye Teh and Stefano Favaro María Lomelí Gatsby Unit, University College London Talk at the BNP 10 Raleigh, North Carolina
Computational statistics
 Computational statistics Markov Chain Monte Carlo methods Thierry Denœux March 2017 Thierry Denœux Computational statistics March 2017 1 / 71 Contents of this chapter When a target density f can be evaluated
Computational statistics Markov Chain Monte Carlo methods Thierry Denœux March 2017 Thierry Denœux Computational statistics March 2017 1 / 71 Contents of this chapter When a target density f can be evaluated
Markov Chain Monte Carlo methods
 Markov Chain Monte Carlo methods Tomas McKelvey and Lennart Svensson Signal Processing Group Department of Signals and Systems Chalmers University of Technology, Sweden November 26, 2012 Today s learning
Markov Chain Monte Carlo methods Tomas McKelvey and Lennart Svensson Signal Processing Group Department of Signals and Systems Chalmers University of Technology, Sweden November 26, 2012 Today s learning
18 : Advanced topics in MCMC. 1 Gibbs Sampling (Continued from the last lecture)
 10-708: Probabilistic Graphical Models 10-708, Spring 2014 18 : Advanced topics in MCMC Lecturer: Eric P. Xing Scribes: Jessica Chemali, Seungwhan Moon 1 Gibbs Sampling (Continued from the last lecture)
10-708: Probabilistic Graphical Models 10-708, Spring 2014 18 : Advanced topics in MCMC Lecturer: Eric P. Xing Scribes: Jessica Chemali, Seungwhan Moon 1 Gibbs Sampling (Continued from the last lecture)
Markov Chain Monte Carlo
 Markov Chain Monte Carlo Recall: To compute the expectation E ( h(y ) ) we use the approximation E(h(Y )) 1 n n h(y ) t=1 with Y (1),..., Y (n) h(y). Thus our aim is to sample Y (1),..., Y (n) from f(y).
Markov Chain Monte Carlo Recall: To compute the expectation E ( h(y ) ) we use the approximation E(h(Y )) 1 n n h(y ) t=1 with Y (1),..., Y (n) h(y). Thus our aim is to sample Y (1),..., Y (n) from f(y).
Gibbs Sampling for (Coupled) Infinite Mixture Models in the Stick Breaking Representation
 Gibbs Sampling for (Coupled) Infinite Mixture Models in the Stick Breaking Representation Ian Porteous, Alex Ihler, Padhraic Smyth, Max Welling Department of Computer Science UC Irvine, Irvine CA 92697-3425
Gibbs Sampling for (Coupled) Infinite Mixture Models in the Stick Breaking Representation Ian Porteous, Alex Ihler, Padhraic Smyth, Max Welling Department of Computer Science UC Irvine, Irvine CA 92697-3425
On Reparametrization and the Gibbs Sampler
 On Reparametrization and the Gibbs Sampler Jorge Carlos Román Department of Mathematics Vanderbilt University James P. Hobert Department of Statistics University of Florida March 2014 Brett Presnell Department
On Reparametrization and the Gibbs Sampler Jorge Carlos Román Department of Mathematics Vanderbilt University James P. Hobert Department of Statistics University of Florida March 2014 Brett Presnell Department
CPSC 540: Machine Learning
 CPSC 540: Machine Learning MCMC and Non-Parametric Bayes Mark Schmidt University of British Columbia Winter 2016 Admin I went through project proposals: Some of you got a message on Piazza. No news is
CPSC 540: Machine Learning MCMC and Non-Parametric Bayes Mark Schmidt University of British Columbia Winter 2016 Admin I went through project proposals: Some of you got a message on Piazza. No news is
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistical Sciences! rsalakhu@cs.toronto.edu! h0p://www.cs.utoronto.ca/~rsalakhu/ Lecture 7 Approximate
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistical Sciences! rsalakhu@cs.toronto.edu! h0p://www.cs.utoronto.ca/~rsalakhu/ Lecture 7 Approximate
Infinite-State Markov-switching for Dynamic. Volatility Models : Web Appendix
 Infinite-State Markov-switching for Dynamic Volatility Models : Web Appendix Arnaud Dufays 1 Centre de Recherche en Economie et Statistique March 19, 2014 1 Comparison of the two MS-GARCH approximations
Infinite-State Markov-switching for Dynamic Volatility Models : Web Appendix Arnaud Dufays 1 Centre de Recherche en Economie et Statistique March 19, 2014 1 Comparison of the two MS-GARCH approximations
On the Fisher Bingham Distribution
 On the Fisher Bingham Distribution BY A. Kume and S.G Walker Institute of Mathematics, Statistics and Actuarial Science, University of Kent Canterbury, CT2 7NF,UK A.Kume@kent.ac.uk and S.G.Walker@kent.ac.uk
On the Fisher Bingham Distribution BY A. Kume and S.G Walker Institute of Mathematics, Statistics and Actuarial Science, University of Kent Canterbury, CT2 7NF,UK A.Kume@kent.ac.uk and S.G.Walker@kent.ac.uk
Hastings-within-Gibbs Algorithm: Introduction and Application on Hierarchical Model
 UNIVERSITY OF TEXAS AT SAN ANTONIO Hastings-within-Gibbs Algorithm: Introduction and Application on Hierarchical Model Liang Jing April 2010 1 1 ABSTRACT In this paper, common MCMC algorithms are introduced
UNIVERSITY OF TEXAS AT SAN ANTONIO Hastings-within-Gibbs Algorithm: Introduction and Application on Hierarchical Model Liang Jing April 2010 1 1 ABSTRACT In this paper, common MCMC algorithms are introduced
17 : Markov Chain Monte Carlo
 10-708: Probabilistic Graphical Models, Spring 2015 17 : Markov Chain Monte Carlo Lecturer: Eric P. Xing Scribes: Heran Lin, Bin Deng, Yun Huang 1 Review of Monte Carlo Methods 1.1 Overview Monte Carlo
10-708: Probabilistic Graphical Models, Spring 2015 17 : Markov Chain Monte Carlo Lecturer: Eric P. Xing Scribes: Heran Lin, Bin Deng, Yun Huang 1 Review of Monte Carlo Methods 1.1 Overview Monte Carlo
Bayesian Nonparametrics: Dirichlet Process
 Bayesian Nonparametrics: Dirichlet Process Yee Whye Teh Gatsby Computational Neuroscience Unit, UCL http://www.gatsby.ucl.ac.uk/~ywteh/teaching/npbayes2012 Dirichlet Process Cornerstone of modern Bayesian
Bayesian Nonparametrics: Dirichlet Process Yee Whye Teh Gatsby Computational Neuroscience Unit, UCL http://www.gatsby.ucl.ac.uk/~ywteh/teaching/npbayes2012 Dirichlet Process Cornerstone of modern Bayesian
Variational Bayesian Dirichlet-Multinomial Allocation for Exponential Family Mixtures
 17th Europ. Conf. on Machine Learning, Berlin, Germany, 2006. Variational Bayesian Dirichlet-Multinomial Allocation for Exponential Family Mixtures Shipeng Yu 1,2, Kai Yu 2, Volker Tresp 2, and Hans-Peter
17th Europ. Conf. on Machine Learning, Berlin, Germany, 2006. Variational Bayesian Dirichlet-Multinomial Allocation for Exponential Family Mixtures Shipeng Yu 1,2, Kai Yu 2, Volker Tresp 2, and Hans-Peter
Advances and Applications in Perfect Sampling
 and Applications in Perfect Sampling Ph.D. Dissertation Defense Ulrike Schneider advisor: Jem Corcoran May 8, 2003 Department of Applied Mathematics University of Colorado Outline Introduction (1) MCMC
and Applications in Perfect Sampling Ph.D. Dissertation Defense Ulrike Schneider advisor: Jem Corcoran May 8, 2003 Department of Applied Mathematics University of Colorado Outline Introduction (1) MCMC
Outline. Binomial, Multinomial, Normal, Beta, Dirichlet. Posterior mean, MAP, credible interval, posterior distribution
 Outline A short review on Bayesian analysis. Binomial, Multinomial, Normal, Beta, Dirichlet Posterior mean, MAP, credible interval, posterior distribution Gibbs sampling Revisit the Gaussian mixture model
Outline A short review on Bayesian analysis. Binomial, Multinomial, Normal, Beta, Dirichlet Posterior mean, MAP, credible interval, posterior distribution Gibbs sampling Revisit the Gaussian mixture model
On Simulations form the Two-Parameter. Poisson-Dirichlet Process and the Normalized. Inverse-Gaussian Process
 On Simulations form the Two-Parameter arxiv:1209.5359v1 [stat.co] 24 Sep 2012 Poisson-Dirichlet Process and the Normalized Inverse-Gaussian Process Luai Al Labadi and Mahmoud Zarepour May 8, 2018 ABSTRACT
On Simulations form the Two-Parameter arxiv:1209.5359v1 [stat.co] 24 Sep 2012 Poisson-Dirichlet Process and the Normalized Inverse-Gaussian Process Luai Al Labadi and Mahmoud Zarepour May 8, 2018 ABSTRACT
Non-Parametric Bayes
 Non-Parametric Bayes Mark Schmidt UBC Machine Learning Reading Group January 2016 Current Hot Topics in Machine Learning Bayesian learning includes: Gaussian processes. Approximate inference. Bayesian
Non-Parametric Bayes Mark Schmidt UBC Machine Learning Reading Group January 2016 Current Hot Topics in Machine Learning Bayesian learning includes: Gaussian processes. Approximate inference. Bayesian
Normalized kernel-weighted random measures
 Normalized kernel-weighted random measures Jim Griffin University of Kent 1 August 27 Outline 1 Introduction 2 Ornstein-Uhlenbeck DP 3 Generalisations Bayesian Density Regression We observe data (x 1,
Normalized kernel-weighted random measures Jim Griffin University of Kent 1 August 27 Outline 1 Introduction 2 Ornstein-Uhlenbeck DP 3 Generalisations Bayesian Density Regression We observe data (x 1,
16 : Approximate Inference: Markov Chain Monte Carlo
 10-708: Probabilistic Graphical Models 10-708, Spring 2017 16 : Approximate Inference: Markov Chain Monte Carlo Lecturer: Eric P. Xing Scribes: Yuan Yang, Chao-Ming Yen 1 Introduction As the target distribution
10-708: Probabilistic Graphical Models 10-708, Spring 2017 16 : Approximate Inference: Markov Chain Monte Carlo Lecturer: Eric P. Xing Scribes: Yuan Yang, Chao-Ming Yen 1 Introduction As the target distribution
STA 294: Stochastic Processes & Bayesian Nonparametrics
 MARKOV CHAINS AND CONVERGENCE CONCEPTS Markov chains are among the simplest stochastic processes, just one step beyond iid sequences of random variables. Traditionally they ve been used in modelling a
MARKOV CHAINS AND CONVERGENCE CONCEPTS Markov chains are among the simplest stochastic processes, just one step beyond iid sequences of random variables. Traditionally they ve been used in modelling a
ABC methods for phase-type distributions with applications in insurance risk problems
 ABC methods for phase-type with applications problems Concepcion Ausin, Department of Statistics, Universidad Carlos III de Madrid Joint work with: Pedro Galeano, Universidad Carlos III de Madrid Simon
ABC methods for phase-type with applications problems Concepcion Ausin, Department of Statistics, Universidad Carlos III de Madrid Joint work with: Pedro Galeano, Universidad Carlos III de Madrid Simon
Bayesian Inference for Discretely Sampled Diffusion Processes: A New MCMC Based Approach to Inference
 Bayesian Inference for Discretely Sampled Diffusion Processes: A New MCMC Based Approach to Inference Osnat Stramer 1 and Matthew Bognar 1 Department of Statistics and Actuarial Science, University of
Bayesian Inference for Discretely Sampled Diffusion Processes: A New MCMC Based Approach to Inference Osnat Stramer 1 and Matthew Bognar 1 Department of Statistics and Actuarial Science, University of
Statistical Inference for Stochastic Epidemic Models
 Statistical Inference for Stochastic Epidemic Models George Streftaris 1 and Gavin J. Gibson 1 1 Department of Actuarial Mathematics & Statistics, Heriot-Watt University, Riccarton, Edinburgh EH14 4AS,
Statistical Inference for Stochastic Epidemic Models George Streftaris 1 and Gavin J. Gibson 1 1 Department of Actuarial Mathematics & Statistics, Heriot-Watt University, Riccarton, Edinburgh EH14 4AS,
David B. Dahl. Department of Statistics, and Department of Biostatistics & Medical Informatics University of Wisconsin Madison
 AN IMPROVED MERGE-SPLIT SAMPLER FOR CONJUGATE DIRICHLET PROCESS MIXTURE MODELS David B. Dahl dbdahl@stat.wisc.edu Department of Statistics, and Department of Biostatistics & Medical Informatics University
AN IMPROVED MERGE-SPLIT SAMPLER FOR CONJUGATE DIRICHLET PROCESS MIXTURE MODELS David B. Dahl dbdahl@stat.wisc.edu Department of Statistics, and Department of Biostatistics & Medical Informatics University
Bayesian nonparametric models for bipartite graphs
 Bayesian nonparametric models for bipartite graphs François Caron Department of Statistics, Oxford Statistics Colloquium, Harvard University November 11, 2013 F. Caron 1 / 27 Bipartite networks Readers/Customers
Bayesian nonparametric models for bipartite graphs François Caron Department of Statistics, Oxford Statistics Colloquium, Harvard University November 11, 2013 F. Caron 1 / 27 Bipartite networks Readers/Customers
Bayesian Inference in GLMs. Frequentists typically base inferences on MLEs, asymptotic confidence
 Bayesian Inference in GLMs Frequentists typically base inferences on MLEs, asymptotic confidence limits, and log-likelihood ratio tests Bayesians base inferences on the posterior distribution of the unknowns
Bayesian Inference in GLMs Frequentists typically base inferences on MLEs, asymptotic confidence limits, and log-likelihood ratio tests Bayesians base inferences on the posterior distribution of the unknowns
On the posterior structure of NRMI
 On the posterior structure of NRMI Igor Prünster University of Turin, Collegio Carlo Alberto and ICER Joint work with L.F. James and A. Lijoi Isaac Newton Institute, BNR Programme, 8th August 2007 Outline
On the posterior structure of NRMI Igor Prünster University of Turin, Collegio Carlo Alberto and ICER Joint work with L.F. James and A. Lijoi Isaac Newton Institute, BNR Programme, 8th August 2007 Outline
Bayesian Nonparametric Regression for Diabetes Deaths
 Bayesian Nonparametric Regression for Diabetes Deaths Brian M. Hartman PhD Student, 2010 Texas A&M University College Station, TX, USA David B. Dahl Assistant Professor Texas A&M University College Station,
Bayesian Nonparametric Regression for Diabetes Deaths Brian M. Hartman PhD Student, 2010 Texas A&M University College Station, TX, USA David B. Dahl Assistant Professor Texas A&M University College Station,
7. Estimation and hypothesis testing. Objective. Recommended reading
 7. Estimation and hypothesis testing Objective In this chapter, we show how the election of estimators can be represented as a decision problem. Secondly, we consider the problem of hypothesis testing
7. Estimation and hypothesis testing Objective In this chapter, we show how the election of estimators can be represented as a decision problem. Secondly, we consider the problem of hypothesis testing
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 7 Approximate
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 7 Approximate
An ABC interpretation of the multiple auxiliary variable method
 School of Mathematical and Physical Sciences Department of Mathematics and Statistics Preprint MPS-2016-07 27 April 2016 An ABC interpretation of the multiple auxiliary variable method by Dennis Prangle
School of Mathematical and Physical Sciences Department of Mathematics and Statistics Preprint MPS-2016-07 27 April 2016 An ABC interpretation of the multiple auxiliary variable method by Dennis Prangle
Nonparametric Bayesian Methods - Lecture I
 Nonparametric Bayesian Methods - Lecture I Harry van Zanten Korteweg-de Vries Institute for Mathematics CRiSM Masterclass, April 4-6, 2016 Overview of the lectures I Intro to nonparametric Bayesian statistics
Nonparametric Bayesian Methods - Lecture I Harry van Zanten Korteweg-de Vries Institute for Mathematics CRiSM Masterclass, April 4-6, 2016 Overview of the lectures I Intro to nonparametric Bayesian statistics
Graphical Models for Query-driven Analysis of Multimodal Data
 Graphical Models for Query-driven Analysis of Multimodal Data John Fisher Sensing, Learning, & Inference Group Computer Science & Artificial Intelligence Laboratory Massachusetts Institute of Technology
Graphical Models for Query-driven Analysis of Multimodal Data John Fisher Sensing, Learning, & Inference Group Computer Science & Artificial Intelligence Laboratory Massachusetts Institute of Technology
Control Variates for Markov Chain Monte Carlo
 Control Variates for Markov Chain Monte Carlo Dellaportas, P., Kontoyiannis, I., and Tsourti, Z. Dept of Statistics, AUEB Dept of Informatics, AUEB 1st Greek Stochastics Meeting Monte Carlo: Probability
Control Variates for Markov Chain Monte Carlo Dellaportas, P., Kontoyiannis, I., and Tsourti, Z. Dept of Statistics, AUEB Dept of Informatics, AUEB 1st Greek Stochastics Meeting Monte Carlo: Probability
SAMPLING ALGORITHMS. In general. Inference in Bayesian models
 SAMPLING ALGORITHMS SAMPLING ALGORITHMS In general A sampling algorithm is an algorithm that outputs samples x 1, x 2,... from a given distribution P or density p. Sampling algorithms can for example be
SAMPLING ALGORITHMS SAMPLING ALGORITHMS In general A sampling algorithm is an algorithm that outputs samples x 1, x 2,... from a given distribution P or density p. Sampling algorithms can for example be
Marginal Specifications and a Gaussian Copula Estimation
 Marginal Specifications and a Gaussian Copula Estimation Kazim Azam Abstract Multivariate analysis involving random variables of different type like count, continuous or mixture of both is frequently required
Marginal Specifications and a Gaussian Copula Estimation Kazim Azam Abstract Multivariate analysis involving random variables of different type like count, continuous or mixture of both is frequently required
Hyperparameter estimation in Dirichlet process mixture models
 Hyperparameter estimation in Dirichlet process mixture models By MIKE WEST Institute of Statistics and Decision Sciences Duke University, Durham NC 27706, USA. SUMMARY In Bayesian density estimation and
Hyperparameter estimation in Dirichlet process mixture models By MIKE WEST Institute of Statistics and Decision Sciences Duke University, Durham NC 27706, USA. SUMMARY In Bayesian density estimation and
Supplementary Appendix to Dynamic Asset Price Jumps: the Performance of High Frequency Tests and Measures, and the Robustness of Inference
 Supplementary Appendix to Dynamic Asset Price Jumps: the Performance of High Frequency Tests and Measures, and the Robustness of Inference Worapree Maneesoonthorn, Gael M. Martin, Catherine S. Forbes August
Supplementary Appendix to Dynamic Asset Price Jumps: the Performance of High Frequency Tests and Measures, and the Robustness of Inference Worapree Maneesoonthorn, Gael M. Martin, Catherine S. Forbes August
Bayesian non-parametric model to longitudinally predict churn
 Bayesian non-parametric model to longitudinally predict churn Bruno Scarpa Università di Padova Conference of European Statistics Stakeholders Methodologists, Producers and Users of European Statistics
Bayesian non-parametric model to longitudinally predict churn Bruno Scarpa Università di Padova Conference of European Statistics Stakeholders Methodologists, Producers and Users of European Statistics
The simple slice sampler is a specialised type of MCMC auxiliary variable method (Swendsen and Wang, 1987; Edwards and Sokal, 1988; Besag and Green, 1
 Recent progress on computable bounds and the simple slice sampler by Gareth O. Roberts* and Jerey S. Rosenthal** (May, 1999.) This paper discusses general quantitative bounds on the convergence rates of
Recent progress on computable bounds and the simple slice sampler by Gareth O. Roberts* and Jerey S. Rosenthal** (May, 1999.) This paper discusses general quantitative bounds on the convergence rates of
Bayesian Nonparametric Models
 Bayesian Nonparametric Models David M. Blei Columbia University December 15, 2015 Introduction We have been looking at models that posit latent structure in high dimensional data. We use the posterior
Bayesian Nonparametric Models David M. Blei Columbia University December 15, 2015 Introduction We have been looking at models that posit latent structure in high dimensional data. We use the posterior
Dirichlet Enhanced Latent Semantic Analysis
 Dirichlet Enhanced Latent Semantic Analysis Kai Yu Siemens Corporate Technology D-81730 Munich, Germany Kai.Yu@siemens.com Shipeng Yu Institute for Computer Science University of Munich D-80538 Munich,
Dirichlet Enhanced Latent Semantic Analysis Kai Yu Siemens Corporate Technology D-81730 Munich, Germany Kai.Yu@siemens.com Shipeng Yu Institute for Computer Science University of Munich D-80538 Munich,
Markov Chain Monte Carlo (MCMC)
 Markov Chain Monte Carlo (MCMC Dependent Sampling Suppose we wish to sample from a density π, and we can evaluate π as a function but have no means to directly generate a sample. Rejection sampling can
Markov Chain Monte Carlo (MCMC Dependent Sampling Suppose we wish to sample from a density π, and we can evaluate π as a function but have no means to directly generate a sample. Rejection sampling can
Lecture 3a: Dirichlet processes
 Lecture 3a: Dirichlet processes Cédric Archambeau Centre for Computational Statistics and Machine Learning Department of Computer Science University College London c.archambeau@cs.ucl.ac.uk Advanced Topics
Lecture 3a: Dirichlet processes Cédric Archambeau Centre for Computational Statistics and Machine Learning Department of Computer Science University College London c.archambeau@cs.ucl.ac.uk Advanced Topics
CMPS 242: Project Report
 CMPS 242: Project Report RadhaKrishna Vuppala Univ. of California, Santa Cruz vrk@soe.ucsc.edu Abstract The classification procedures impose certain models on the data and when the assumption match the
CMPS 242: Project Report RadhaKrishna Vuppala Univ. of California, Santa Cruz vrk@soe.ucsc.edu Abstract The classification procedures impose certain models on the data and when the assumption match the
A Nonparametric Approach Using Dirichlet Process for Hierarchical Generalized Linear Mixed Models
 Journal of Data Science 8(2010), 43-59 A Nonparametric Approach Using Dirichlet Process for Hierarchical Generalized Linear Mixed Models Jing Wang Louisiana State University Abstract: In this paper, we
Journal of Data Science 8(2010), 43-59 A Nonparametric Approach Using Dirichlet Process for Hierarchical Generalized Linear Mixed Models Jing Wang Louisiana State University Abstract: In this paper, we
Probabilistic Graphical Models Lecture 17: Markov chain Monte Carlo
 Probabilistic Graphical Models Lecture 17: Markov chain Monte Carlo Andrew Gordon Wilson www.cs.cmu.edu/~andrewgw Carnegie Mellon University March 18, 2015 1 / 45 Resources and Attribution Image credits,
Probabilistic Graphical Models Lecture 17: Markov chain Monte Carlo Andrew Gordon Wilson www.cs.cmu.edu/~andrewgw Carnegie Mellon University March 18, 2015 1 / 45 Resources and Attribution Image credits,
Likelihood-free MCMC
 Bayesian inference for stable distributions with applications in finance Department of Mathematics University of Leicester September 2, 2011 MSc project final presentation Outline 1 2 3 4 Classical Monte
Bayesian inference for stable distributions with applications in finance Department of Mathematics University of Leicester September 2, 2011 MSc project final presentation Outline 1 2 3 4 Classical Monte
Introduction to Bayesian methods in inverse problems
 Introduction to Bayesian methods in inverse problems Ville Kolehmainen 1 1 Department of Applied Physics, University of Eastern Finland, Kuopio, Finland March 4 2013 Manchester, UK. Contents Introduction
Introduction to Bayesian methods in inverse problems Ville Kolehmainen 1 1 Department of Applied Physics, University of Eastern Finland, Kuopio, Finland March 4 2013 Manchester, UK. Contents Introduction
Hmms with variable dimension structures and extensions
 Hmm days/enst/january 21, 2002 1 Hmms with variable dimension structures and extensions Christian P. Robert Université Paris Dauphine www.ceremade.dauphine.fr/ xian Hmm days/enst/january 21, 2002 2 1 Estimating
Hmm days/enst/january 21, 2002 1 Hmms with variable dimension structures and extensions Christian P. Robert Université Paris Dauphine www.ceremade.dauphine.fr/ xian Hmm days/enst/january 21, 2002 2 1 Estimating
A Simple Proof of the Stick-Breaking Construction of the Dirichlet Process
 A Simple Proof of the Stick-Breaking Construction of the Dirichlet Process John Paisley Department of Computer Science Princeton University, Princeton, NJ jpaisley@princeton.edu Abstract We give a simple
A Simple Proof of the Stick-Breaking Construction of the Dirichlet Process John Paisley Department of Computer Science Princeton University, Princeton, NJ jpaisley@princeton.edu Abstract We give a simple
Bayesian Hidden Markov Models and Extensions
 Bayesian Hidden Markov Models and Extensions Zoubin Ghahramani Department of Engineering University of Cambridge joint work with Matt Beal, Jurgen van Gael, Yunus Saatci, Tom Stepleton, Yee Whye Teh Modeling
Bayesian Hidden Markov Models and Extensions Zoubin Ghahramani Department of Engineering University of Cambridge joint work with Matt Beal, Jurgen van Gael, Yunus Saatci, Tom Stepleton, Yee Whye Teh Modeling
Large-scale Ordinal Collaborative Filtering
 Large-scale Ordinal Collaborative Filtering Ulrich Paquet, Blaise Thomson, and Ole Winther Microsoft Research Cambridge, University of Cambridge, Technical University of Denmark ulripa@microsoft.com,brmt2@cam.ac.uk,owi@imm.dtu.dk
Large-scale Ordinal Collaborative Filtering Ulrich Paquet, Blaise Thomson, and Ole Winther Microsoft Research Cambridge, University of Cambridge, Technical University of Denmark ulripa@microsoft.com,brmt2@cam.ac.uk,owi@imm.dtu.dk
Principles of Bayesian Inference
 Principles of Bayesian Inference Sudipto Banerjee University of Minnesota July 20th, 2008 1 Bayesian Principles Classical statistics: model parameters are fixed and unknown. A Bayesian thinks of parameters
Principles of Bayesian Inference Sudipto Banerjee University of Minnesota July 20th, 2008 1 Bayesian Principles Classical statistics: model parameters are fixed and unknown. A Bayesian thinks of parameters
Bayesian nonparametrics
 Bayesian nonparametrics 1 Some preliminaries 1.1 de Finetti s theorem We will start our discussion with this foundational theorem. We will assume throughout all variables are defined on the probability
Bayesian nonparametrics 1 Some preliminaries 1.1 de Finetti s theorem We will start our discussion with this foundational theorem. We will assume throughout all variables are defined on the probability
MCMC and Gibbs Sampling. Sargur Srihari
 MCMC and Gibbs Sampling Sargur srihari@cedar.buffalo.edu 1 Topics 1. Markov Chain Monte Carlo 2. Markov Chains 3. Gibbs Sampling 4. Basic Metropolis Algorithm 5. Metropolis-Hastings Algorithm 6. Slice
MCMC and Gibbs Sampling Sargur srihari@cedar.buffalo.edu 1 Topics 1. Markov Chain Monte Carlo 2. Markov Chains 3. Gibbs Sampling 4. Basic Metropolis Algorithm 5. Metropolis-Hastings Algorithm 6. Slice
Lecture 4: Dynamic models
 linear s Lecture 4: s Hedibert Freitas Lopes The University of Chicago Booth School of Business 5807 South Woodlawn Avenue, Chicago, IL 60637 http://faculty.chicagobooth.edu/hedibert.lopes hlopes@chicagobooth.edu
linear s Lecture 4: s Hedibert Freitas Lopes The University of Chicago Booth School of Business 5807 South Woodlawn Avenue, Chicago, IL 60637 http://faculty.chicagobooth.edu/hedibert.lopes hlopes@chicagobooth.edu
A Review of Pseudo-Marginal Markov Chain Monte Carlo
 A Review of Pseudo-Marginal Markov Chain Monte Carlo Discussed by: Yizhe Zhang October 21, 2016 Outline 1 Overview 2 Paper review 3 experiment 4 conclusion Motivation & overview Notation: θ denotes the
A Review of Pseudo-Marginal Markov Chain Monte Carlo Discussed by: Yizhe Zhang October 21, 2016 Outline 1 Overview 2 Paper review 3 experiment 4 conclusion Motivation & overview Notation: θ denotes the
An adaptive truncation method for inference in Bayesian nonparametric models
 An adaptive truncation method for inference in Bayesian nonparametric models arxiv:1308.045v [stat.co] 1 May 014 J.E. Griffin School of Mathematics, Statistics and Actuarial Science, University of Kent,
An adaptive truncation method for inference in Bayesian nonparametric models arxiv:1308.045v [stat.co] 1 May 014 J.E. Griffin School of Mathematics, Statistics and Actuarial Science, University of Kent,
eqr094: Hierarchical MCMC for Bayesian System Reliability
 eqr094: Hierarchical MCMC for Bayesian System Reliability Alyson G. Wilson Statistical Sciences Group, Los Alamos National Laboratory P.O. Box 1663, MS F600 Los Alamos, NM 87545 USA Phone: 505-667-9167
eqr094: Hierarchical MCMC for Bayesian System Reliability Alyson G. Wilson Statistical Sciences Group, Los Alamos National Laboratory P.O. Box 1663, MS F600 Los Alamos, NM 87545 USA Phone: 505-667-9167
Web Appendix for Hierarchical Adaptive Regression Kernels for Regression with Functional Predictors by D. B. Woodard, C. Crainiceanu, and D.
 Web Appendix for Hierarchical Adaptive Regression Kernels for Regression with Functional Predictors by D. B. Woodard, C. Crainiceanu, and D. Ruppert A. EMPIRICAL ESTIMATE OF THE KERNEL MIXTURE Here we
Web Appendix for Hierarchical Adaptive Regression Kernels for Regression with Functional Predictors by D. B. Woodard, C. Crainiceanu, and D. Ruppert A. EMPIRICAL ESTIMATE OF THE KERNEL MIXTURE Here we
(5) Multi-parameter models - Gibbs sampling. ST440/540: Applied Bayesian Analysis
 Summarizing a posterior Given the data and prior the posterior is determined Summarizing the posterior gives parameter estimates, intervals, and hypothesis tests Most of these computations are integrals
Summarizing a posterior Given the data and prior the posterior is determined Summarizing the posterior gives parameter estimates, intervals, and hypothesis tests Most of these computations are integrals
Reminder of some Markov Chain properties:
 Reminder of some Markov Chain properties: 1. a transition from one state to another occurs probabilistically 2. only state that matters is where you currently are (i.e. given present, future is independent
Reminder of some Markov Chain properties: 1. a transition from one state to another occurs probabilistically 2. only state that matters is where you currently are (i.e. given present, future is independent
A Process over all Stationary Covariance Kernels
 A Process over all Stationary Covariance Kernels Andrew Gordon Wilson June 9, 0 Abstract I define a process over all stationary covariance kernels. I show how one might be able to perform inference that
A Process over all Stationary Covariance Kernels Andrew Gordon Wilson June 9, 0 Abstract I define a process over all stationary covariance kernels. I show how one might be able to perform inference that
Bayesian finite mixtures with an unknown number of. components: the allocation sampler
 Bayesian finite mixtures with an unknown number of components: the allocation sampler Agostino Nobile and Alastair Fearnside University of Glasgow, UK 2 nd June 2005 Abstract A new Markov chain Monte Carlo
Bayesian finite mixtures with an unknown number of components: the allocation sampler Agostino Nobile and Alastair Fearnside University of Glasgow, UK 2 nd June 2005 Abstract A new Markov chain Monte Carlo
Markov Chain Monte Carlo Using the Ratio-of-Uniforms Transformation. Luke Tierney Department of Statistics & Actuarial Science University of Iowa
 Markov Chain Monte Carlo Using the Ratio-of-Uniforms Transformation Luke Tierney Department of Statistics & Actuarial Science University of Iowa Basic Ratio of Uniforms Method Introduced by Kinderman and
Markov Chain Monte Carlo Using the Ratio-of-Uniforms Transformation Luke Tierney Department of Statistics & Actuarial Science University of Iowa Basic Ratio of Uniforms Method Introduced by Kinderman and
Markov Chain Monte Carlo methods
 Markov Chain Monte Carlo methods By Oleg Makhnin 1 Introduction a b c M = d e f g h i 0 f(x)dx 1.1 Motivation 1.1.1 Just here Supresses numbering 1.1.2 After this 1.2 Literature 2 Method 2.1 New math As
Markov Chain Monte Carlo methods By Oleg Makhnin 1 Introduction a b c M = d e f g h i 0 f(x)dx 1.1 Motivation 1.1.1 Just here Supresses numbering 1.1.2 After this 1.2 Literature 2 Method 2.1 New math As
Graphical Models and Kernel Methods
 Graphical Models and Kernel Methods Jerry Zhu Department of Computer Sciences University of Wisconsin Madison, USA MLSS June 17, 2014 1 / 123 Outline Graphical Models Probabilistic Inference Directed vs.
Graphical Models and Kernel Methods Jerry Zhu Department of Computer Sciences University of Wisconsin Madison, USA MLSS June 17, 2014 1 / 123 Outline Graphical Models Probabilistic Inference Directed vs.
Computer Practical: Metropolis-Hastings-based MCMC
 Computer Practical: Metropolis-Hastings-based MCMC Andrea Arnold and Franz Hamilton North Carolina State University July 30, 2016 A. Arnold / F. Hamilton (NCSU) MH-based MCMC July 30, 2016 1 / 19 Markov
Computer Practical: Metropolis-Hastings-based MCMC Andrea Arnold and Franz Hamilton North Carolina State University July 30, 2016 A. Arnold / F. Hamilton (NCSU) MH-based MCMC July 30, 2016 1 / 19 Markov
Gaussian kernel GARCH models
 Gaussian kernel GARCH models Xibin (Bill) Zhang and Maxwell L. King Department of Econometrics and Business Statistics Faculty of Business and Economics 7 June 2013 Motivation A regression model is often
Gaussian kernel GARCH models Xibin (Bill) Zhang and Maxwell L. King Department of Econometrics and Business Statistics Faculty of Business and Economics 7 June 2013 Motivation A regression model is often
Bayesian Nonparametric Models for Ranking Data
 Bayesian Nonparametric Models for Ranking Data François Caron 1, Yee Whye Teh 1 and Brendan Murphy 2 1 Dept of Statistics, University of Oxford, UK 2 School of Mathematical Sciences, University College
Bayesian Nonparametric Models for Ranking Data François Caron 1, Yee Whye Teh 1 and Brendan Murphy 2 1 Dept of Statistics, University of Oxford, UK 2 School of Mathematical Sciences, University College
Lecture 7 and 8: Markov Chain Monte Carlo
 Lecture 7 and 8: Markov Chain Monte Carlo 4F13: Machine Learning Zoubin Ghahramani and Carl Edward Rasmussen Department of Engineering University of Cambridge http://mlg.eng.cam.ac.uk/teaching/4f13/ Ghahramani
Lecture 7 and 8: Markov Chain Monte Carlo 4F13: Machine Learning Zoubin Ghahramani and Carl Edward Rasmussen Department of Engineering University of Cambridge http://mlg.eng.cam.ac.uk/teaching/4f13/ Ghahramani
Computer Vision Group Prof. Daniel Cremers. 10a. Markov Chain Monte Carlo
 Group Prof. Daniel Cremers 10a. Markov Chain Monte Carlo Markov Chain Monte Carlo In high-dimensional spaces, rejection sampling and importance sampling are very inefficient An alternative is Markov Chain
Group Prof. Daniel Cremers 10a. Markov Chain Monte Carlo Markov Chain Monte Carlo In high-dimensional spaces, rejection sampling and importance sampling are very inefficient An alternative is Markov Chain
Adaptive Rejection Sampling with fixed number of nodes
 Adaptive Rejection Sampling with fixed number of nodes L. Martino, F. Louzada Institute of Mathematical Sciences and Computing, Universidade de São Paulo, Brazil. Abstract The adaptive rejection sampling
Adaptive Rejection Sampling with fixed number of nodes L. Martino, F. Louzada Institute of Mathematical Sciences and Computing, Universidade de São Paulo, Brazil. Abstract The adaptive rejection sampling
MCMC for non-linear state space models using ensembles of latent sequences
 MCMC for non-linear state space models using ensembles of latent sequences Alexander Y. Shestopaloff Department of Statistical Sciences University of Toronto alexander@utstat.utoronto.ca Radford M. Neal
MCMC for non-linear state space models using ensembles of latent sequences Alexander Y. Shestopaloff Department of Statistical Sciences University of Toronto alexander@utstat.utoronto.ca Radford M. Neal
Rank Regression with Normal Residuals using the Gibbs Sampler
 Rank Regression with Normal Residuals using the Gibbs Sampler Stephen P Smith email: hucklebird@aol.com, 2018 Abstract Yu (2000) described the use of the Gibbs sampler to estimate regression parameters
Rank Regression with Normal Residuals using the Gibbs Sampler Stephen P Smith email: hucklebird@aol.com, 2018 Abstract Yu (2000) described the use of the Gibbs sampler to estimate regression parameters
STAT Advanced Bayesian Inference
 1 / 32 STAT 625 - Advanced Bayesian Inference Meng Li Department of Statistics Jan 23, 218 The Dirichlet distribution 2 / 32 θ Dirichlet(a 1,...,a k ) with density p(θ 1,θ 2,...,θ k ) = k j=1 Γ(a j) Γ(
1 / 32 STAT 625 - Advanced Bayesian Inference Meng Li Department of Statistics Jan 23, 218 The Dirichlet distribution 2 / 32 θ Dirichlet(a 1,...,a k ) with density p(θ 1,θ 2,...,θ k ) = k j=1 Γ(a j) Γ(
MCMC algorithms for fitting Bayesian models
 MCMC algorithms for fitting Bayesian models p. 1/1 MCMC algorithms for fitting Bayesian models Sudipto Banerjee sudiptob@biostat.umn.edu University of Minnesota MCMC algorithms for fitting Bayesian models
MCMC algorithms for fitting Bayesian models p. 1/1 MCMC algorithms for fitting Bayesian models Sudipto Banerjee sudiptob@biostat.umn.edu University of Minnesota MCMC algorithms for fitting Bayesian models
Bayesian Nonparametric Autoregressive Models via Latent Variable Representation
 Bayesian Nonparametric Autoregressive Models via Latent Variable Representation Maria De Iorio Yale-NUS College Dept of Statistical Science, University College London Collaborators: Lifeng Ye (UCL, London,
Bayesian Nonparametric Autoregressive Models via Latent Variable Representation Maria De Iorio Yale-NUS College Dept of Statistical Science, University College London Collaborators: Lifeng Ye (UCL, London,
Metropolis-Hastings Algorithm
 Strength of the Gibbs sampler Metropolis-Hastings Algorithm Easy algorithm to think about. Exploits the factorization properties of the joint probability distribution. No difficult choices to be made to
Strength of the Gibbs sampler Metropolis-Hastings Algorithm Easy algorithm to think about. Exploits the factorization properties of the joint probability distribution. No difficult choices to be made to
A quick introduction to Markov chains and Markov chain Monte Carlo (revised version)
 A quick introduction to Markov chains and Markov chain Monte Carlo (revised version) Rasmus Waagepetersen Institute of Mathematical Sciences Aalborg University 1 Introduction These notes are intended to
A quick introduction to Markov chains and Markov chain Monte Carlo (revised version) Rasmus Waagepetersen Institute of Mathematical Sciences Aalborg University 1 Introduction These notes are intended to
The Recycling Gibbs Sampler for Efficient Learning
 The Recycling Gibbs Sampler for Efficient Learning L. Martino, V. Elvira, G. Camps-Valls Universidade de São Paulo, São Carlos (Brazil). Télécom ParisTech, Université Paris-Saclay. (France), Universidad
The Recycling Gibbs Sampler for Efficient Learning L. Martino, V. Elvira, G. Camps-Valls Universidade de São Paulo, São Carlos (Brazil). Télécom ParisTech, Université Paris-Saclay. (France), Universidad
Adaptive Rejection Sampling with fixed number of nodes
 Adaptive Rejection Sampling with fixed number of nodes L. Martino, F. Louzada Institute of Mathematical Sciences and Computing, Universidade de São Paulo, São Carlos (São Paulo). Abstract The adaptive
Adaptive Rejection Sampling with fixed number of nodes L. Martino, F. Louzada Institute of Mathematical Sciences and Computing, Universidade de São Paulo, São Carlos (São Paulo). Abstract The adaptive
Bayesian model selection in graphs by using BDgraph package
 Bayesian model selection in graphs by using BDgraph package A. Mohammadi and E. Wit March 26, 2013 MOTIVATION Flow cytometry data with 11 proteins from Sachs et al. (2005) RESULT FOR CELL SIGNALING DATA
Bayesian model selection in graphs by using BDgraph package A. Mohammadi and E. Wit March 26, 2013 MOTIVATION Flow cytometry data with 11 proteins from Sachs et al. (2005) RESULT FOR CELL SIGNALING DATA
A nonparametric Bayesian approach to inference for non-homogeneous. Poisson processes. Athanasios Kottas 1. (REVISED VERSION August 23, 2006)
 A nonparametric Bayesian approach to inference for non-homogeneous Poisson processes Athanasios Kottas 1 Department of Applied Mathematics and Statistics, Baskin School of Engineering, University of California,
A nonparametric Bayesian approach to inference for non-homogeneous Poisson processes Athanasios Kottas 1 Department of Applied Mathematics and Statistics, Baskin School of Engineering, University of California,
Bayesian Nonparametric Modelling with the Dirichlet Process Regression Smoother
 Bayesian Nonparametric Modelling with the Dirichlet Process Regression Smoother J. E. Griffin and M. F. J. Steel University of Warwick Bayesian Nonparametric Modelling with the Dirichlet Process Regression
Bayesian Nonparametric Modelling with the Dirichlet Process Regression Smoother J. E. Griffin and M. F. J. Steel University of Warwick Bayesian Nonparametric Modelling with the Dirichlet Process Regression
STAT 518 Intro Student Presentation
 STAT 518 Intro Student Presentation Wen Wei Loh April 11, 2013 Title of paper Radford M. Neal [1999] Bayesian Statistics, 6: 475-501, 1999 What the paper is about Regression and Classification Flexible
STAT 518 Intro Student Presentation Wen Wei Loh April 11, 2013 Title of paper Radford M. Neal [1999] Bayesian Statistics, 6: 475-501, 1999 What the paper is about Regression and Classification Flexible
Slice Sampling with Adaptive Multivariate Steps: The Shrinking-Rank Method
 Slice Sampling with Adaptive Multivariate Steps: The Shrinking-Rank Method Madeleine B. Thompson Radford M. Neal Abstract The shrinking rank method is a variation of slice sampling that is efficient at
Slice Sampling with Adaptive Multivariate Steps: The Shrinking-Rank Method Madeleine B. Thompson Radford M. Neal Abstract The shrinking rank method is a variation of slice sampling that is efficient at
Bayesian Semiparametric GARCH Models
 Bayesian Semiparametric GARCH Models Xibin (Bill) Zhang and Maxwell L. King Department of Econometrics and Business Statistics Faculty of Business and Economics xibin.zhang@monash.edu Quantitative Methods
Bayesian Semiparametric GARCH Models Xibin (Bill) Zhang and Maxwell L. King Department of Econometrics and Business Statistics Faculty of Business and Economics xibin.zhang@monash.edu Quantitative Methods
Predictivist Bayes density estimation
 Predictivist Bayes density estimation P. Richard Hahn Abstract: This paper develops a novel computational approach for Bayesian density estimation, using a kernel density representation of the Bayesian
Predictivist Bayes density estimation P. Richard Hahn Abstract: This paper develops a novel computational approach for Bayesian density estimation, using a kernel density representation of the Bayesian
The Mixture Approach for Simulating New Families of Bivariate Distributions with Specified Correlations
 The Mixture Approach for Simulating New Families of Bivariate Distributions with Specified Correlations John R. Michael, Significance, Inc. and William R. Schucany, Southern Methodist University The mixture
The Mixture Approach for Simulating New Families of Bivariate Distributions with Specified Correlations John R. Michael, Significance, Inc. and William R. Schucany, Southern Methodist University The mixture
