Interhead distance measurements in myosin VI via SHRImP support a simplified
|
|
|
- Hugo Flynn
- 5 years ago
- Views:
Transcription
1 Biophys J BioFAST, published on April 29, 2005 as doi: /biophysj Interhead distance measurements in myosin VI via SHRImP support a simplified hand-over-hand model Hamza Balci 1, Taekjip Ha 1, H. Lee Sweeney 2, Paul R. Selvin 1* 1 Physics and Biophysics Depts. Department, University of Illinois, Urbana-Champaign, and 2 University of Pennsylvania, Penn. Muscle Institute. *To whom correspondence should be addressed: Loomis Lab of Physics 1110 W. Green St. Univ. of Illinois Urbana, IL selvin@uiuc.edu (217) (tel); (217) (fax) Copyright 2005 by The Biophysical Society.
2 Myosin-VI walks in a hand-over-hand fashion with an average step size of 30 nm, which is much larger than its 10 nm lever arm. Recent experiments suggest that myosin VI structure has an unfolded and flexible region in the proximal tail which makes such a large step possible. In addition, cryo-em images of actomyosin-vi show the two heads bound to the actin monomers with a broad distribution of distances, including some as close as a few nanometers. This observation when combined with the existence of a flexible region in the structure, which takes part in stepping, challenged the hand-over-hand model. In the handover-hand model the lever arm is considered to be rigid and the inter-head separation should not be very different from 30 nm. We considered an alternative model in which myosin-vi heads sequentially take 60 nm steps while the inter-head separation alternates between a large and small value (x and 60-x, where x<30). In order to clarify these issues, we used a new technique, SHRImP, to measure the inter-head distance of nearly rigor myosin-vi molecules. Our data shows a single peak at 29.3±0.7 nm, in agreement with the straightforward hand-over-hand model.
3 Myosin VI, similar to the other 17 members of the myosin superfamily (1), has three domains in its structure: a motor domain (N terminal) that binds to actin and shows ATPase activity, a neck domain that binds light chains or calmodulin, and a tail domain (C terminal) that binds to cargo. It was first discovered in Drosophila (2), and later was identified in many higher organisms ranging from Caenorhabditis elegans to humans (3-5). The gene for myosin VI is defective in the deaf mouse, Snell s Waltzer, suggesting that myosin VI is involved in the function of sensory hair cells (5,6). Myosin VI is unique in the myosin family in that it is the only member of the family that walks towards the pointed (-) end of actin (7). It is also observed that myosin VI colocalizes with clathrin coated pits (8), which when combined with its directionality, suggests that it is involved in endocytosis (9). Recently myosin VI has attracted attention due to two of its unusual properties - processivity and step-size. Biochemical characterization and electron microscopy studies on myosin VI have shown that expressed full-length myosin VI is a monomer and is a nonprocessive motor (10). However, it has also been shown that dimerized myosin VI can take many steps before detaching from actin (11-14), and hence acts as a processive motor. It is not clear at this point how myosin VI functions within the cell: as a monomer, a dimer, or both. However, extensive optical trap experiments on myosin VI imply a possible explanation by attributing multi funtionality, which might reguire both monomers and dimers of this motor (15). The molecules that we use in this study are dimerized constructs of myosin VI. The second unusual property of myosin VI, which is the topic of this article, was realized when motility experiments on dimerized constructs of myosin VI measured a
4 step size that is not compatible with the assumed structure of this motor using the well established lever arm mechanism (11-14). Hand-over-hand model of walking using a lever arm mechanism has been well established for processive motors in the myosin and kinesin families (16-24). The step of these molecular motors consists of two parts: a power stroke and a diffusive search that follows the power stroke (22). The size of the power stroke is comparable to the size of the light chain domain which acts as the lever arm. For example for myosin V, the step is formed from a ~23 nm (size of the light chain domain) power stroke and a ~12 nm diffusive search (22). However, in the case of myosin VI the situation is quite different since the light chain domain (~10 nm) is much shorter than the measured step size (~30 nm) (13,14). Two possible scenarios were considered: there is a rigid extension to the lever arm which would enable a longer power stroke, or there is a flexible compliant region which enables the motor to go through a longer diffusive search. Several experimental observations (13,25) favor the proximal tail to act as a flexible compliant region rather than a rigid extension to the lever arm. The emerging picture is that, after the power stroke of the motor (~12 nm), the head domain goes through a biased diffusive search on the actin monomer (~20 nm) until it finds the next binding site. The complications in myosin VI motility were further increased with cryo-electron microscopy images of myosin VI. These images showed that for some of the motors, the two heads of the dimer were bound to adjacent actin monomers with a separation much less than 30 nm (12). These images, when considered with the additional degree of freedom
5 provided by the flexible region in the proximal tail domain, challenged the hand-over-hand model of walking. Fig. 1b shows an alternative model where the heads alternately take 60 nm steps and the distance between the heads is either x nm or 60-x nm, depending on which head took the last step, where x could have any value between ~5 nm (since the two heads can not be on the same actin monomer) and 30 nm (see Fig.1-b). This model will be referred to as asymmetric hand-over-hand model in the rest of the manuscript. In order to clarify this issue we measured the separation between the myosin VI heads, rather than the step size. In the hand-over-hand model, we would expect the inter-head separation to show a single peak at 30 nm, whereas the asymmetric hand-over-hand model should show two peaks: one at x nm and another at 60-x nm. We used our recently introduced Singlemolecule High Resolution Imaging with Photobleaching (SHRImP) to determine the distance between egfp molecules on the heads of myosin VI with nanometer precision (26,27). Materials and Methods Imaging. Olympus (Melville, NY) IX-71 microscope, Olympus Apo 100, 1.65 NA oil objective, and high refractive index cover slips (also from Olympus) were used in the objective type TIR setup. The egfp molecules were excited using Melles Griot (Carlsbad, CA) ion laser at 488 nm wavelength. Images were captured by using a slow-scan backthinned Andor (South Windsor, CT) CCD camera, allowing continuous imaging with no interframe dead time. The image capture frequency was 2 Hz (13).
6 Sample preparation. The samples were prepared as described in (13). The sapphire cover slips and glass slides were sonicated in acetone for 20 minutes, rinsed with doubly deionized water, sonicated in 1 M KOH for another 20 minutes, and finally rinsed again with doubly deionized water. The sample chamber was prepared by flowing 10 mm BSA-biotin, waiting for 10 min, washing with M5BufBH (20 mm HEPES ph 7.6, 2 mm MgCl2, 25 mm KCl, 1 mm EGTA), followed by 0.5 mg/ml streptavidin, waiting for 5 min, followed by 0.2 µm 1:10 biotinylated/unbiotinylated phalloidin-stabilized F-actin, followed by M5+ (M5BufBH plus 10 mm DTT and 100 mg/ml calmodulin) wash, followed by myosin VI sample solution. Myosin VI sample solution consists of 50 pm egfp-myosin VI, 100 mm DTT, 100 mg/ml calmodulin, and 1 2 µm ATP, in M5BufBH. Data Analysis We have used SHRImP (26) to measure the distance between the myosin VI heads in the presence of very low concentration of ATP. SHRImP takes advantage of quantal photobleaching of fluorescent molecules to resolve two fluorophores which lie on the same plane and which have overlapping point spread functions (PSF). In this case, there would be a two-step photobleaching event, and each of the steps corresponds to the photobleaching of one of the dyes. The images both before and after photobleaching of one of the dyes are fit to two dimensional Gaussian functions. The image after one of the dyes photobleaches is used to determine the PSF of one of the dyes and the difference between this image and the one before the first photobleaching is used to determine the PSF of the other dye. Hence, both dyes can be simultaneously localized by using this simple idea.
7 Results and Discussion We used SHRImP to measure the distance between the myosin VI heads. The fluorophores are two enhanced green fluorescent proteins (egfp), located at the N-terminus, or head region, of the myosin VI dimer. The data is taken in the presence of very low concentration of ATP in order to make sure the myosin heads are bound to the actins in a configuration relevant for stepping (1 µm which is about 3 % of that used for motility experiments). However, the concentration of ATP is low enough that almost all of the dyes are stationary. Among the thousands of molecules we imaged in the course of this study, we noticed only a few which moved 1-2 steps. These were not included in the analysis. This is a necessary condition in order to be able to measure the separation between the two heads accurately and avoid any confusion that could result from the moving of one or both heads during the measurement. Since the myosin molecules do not move, it is important to make sure that they land on actins rather than non-specifically bind to the surface. In order to test if any non-specific binding occurs, the actins were decorated with a high concentration of myosin VI molecules. We include an image of the actin filaments decorated with myosin VI molecules as Supplementary Online Material. This image clearly shows that the myosin molecules bind only to the actins and not to the surface. In addition, the actin concentration is low enough and the length of the actin where multiple actins cross-over is far shorter than the full-length of the actin filaments to assure us that a myosin VI molecule would bind to only one actin filament. Fig. 2a shows an example of two-step photobleaching. The intensity on the y-axis corresponds to the maximum of the PSF although the same two-step photobleaching can be
8 observed in the total integrated area under the PSF as well. Fig. 2b shows the corresponding PSF s before and after photobleaching. The difference between the first two graphs is shown in the third graph, which is the PSF of the egfp that photobleached first. The details of the data analysis using SHRImP are given in Ref. (26). The frequency of the two-step photobleaching events was sensitively dependent on the experimental conditions used in preparing the sample. However, 1-2 % was the typical frequency that showed clear two-step photobleaching. In the best cases, this frequency was as high as 5%. Most of the dyes showed a single step photobleaching. It is not clear at this point why higher frequency of two-step photobleaching could not be observed, since in gel filtration experiments our samples showed a single band of doubly-labeled myosin VI (data not shown). However, there are several reasons which would reduce the frequency of observing clear two-step photobleaching events. The relatively long integration time we use in acquiring the images is one of these reasons. The images were taken every 0.5 seconds, therefore if the dyes photobleached one after another within 1-2 seconds, we would not be able to clearly distinguish this event from a single step photobleaching event. In addition, it is possible that one of the dyes photobleached within the first 1-2 seconds after the laser is turned on. This case would not be counted as a two-step photobleaching event since the time is too short to get reliable statistics. In a few rare cases we observed broad, rather than single step, photobleaching events which indicates that the myosins formed aggregates. We submit an image as Supplementary Online Material showing the concentration of myosin VI molecules we used in the experiments. As is clear in this image, the concentration of myosin VI molecules is low enough to image single molecules.
9 Fig. 3 shows a histogram of the separation between the myosin VI heads determined from the two-step photobleaching events and various fits made to this histogram. There are 96 points in the histogram. The average inter-head separation of the 96 molecules is 31.0 nm, and the standard deviation is 15.4 nm. Each point reflects an average distance determined from the best Gaussian fits to several pre-and post-photobleach events. Typically 4 frames were used to compute the average distance. However, in some cases only 2 frames yielded good enough fits that could be used, and in some cases as many as 9 frames were used to find the average distance. The center of a Gaussian for each frame was typically determined within an error of 5-6 nm. The best resolution (in localizing the center of the Gaussian) obtained was 3 nm, and the minimum acceptable resolution (to be included in the histogram) was 10 nm. The curves in Fig. 3 are three different fits to the data. Different fits were used in order to model the alternative walking styles presented in Fig. 1. In the case of symmetric handover-hand model, we would expect the interhead separation to peak at 30 nm, and hence a single Gaussian should be able to represent the histogram. A Gaussian fit of the form y=y 0 +a exp[-0.5*([x-x(0)]/b) 2 ] to the histogram results in x(0) = 29.2 ± 0.8 nm, which is in very good agreement with the symmetric hand-over-hand model. The other parameters generated by the fit are y 0 =0.9 ± 0.6 nm, a= 19.9±1.4, and b=9.7±0.9 nm. The full-width at half maximum of the Gaussian is 23.8 nm. However, the broadness of the peak we observe in the histogram and in the corresponding Gaussian fit suggests that two closely spaced Gaussians may be able to describe the data as well. In order to study this possibility further,
10 we fit the data with a function of the form y=y 0 +a exp[-0.5*([x-x 1 (0)]/b) 2 ] + c exp[-0.5*([xx 2 (0)]/d) 2 ]. This fit is shown as the black curve in Fig.2, and the corresponding parameters are y 0 =0.9 ± 0.3 nm, a=21.7±0.9, x 1 (0)=27.3±0.5 nm, b=7.2±0.5 nm, c=8.2±1.3, x 2 (0)=43.9±1.0 nm, and d=4.2±0.7 nm. This function corresponds to a model in which the separation between the heads alternate between 27.3 nm and 43.9 nm. This would suggest that even if the asymmetry between the interhead separations is as large as =16.6 nm we would not be able to distinguish it from the symmetric case presented earlier. Even though the data seems to be very well described by such a fitting function (Reduced χ 2 = 1.0), the amplitudes of the Gaussians which is a measure of the frequency of observing the corresponding interhead separation do not represent the experimental conditions we used. The amplitude of the Gaussian for 27.3 nm interhead separation is 21.7 and the amplitude for 43.9 nm separation is 8.2, which means that 73 % of the time we observe the myosins when the interhead separation is 27.3 nm. However, there is no reason for such a strong bias in the statistics to happen towards one of the interhead separation, and on average we should observe the two interhead separations with equal frequency. Another problem with this fit is that 27.3 nm and 43.9 nm result in a step size of 35.6 nm, which is significantly larger than the expected 30 nm step. A more realistic fit which consists of two Gaussians would be one in which the amplitudes of the Gaussians are equal to each other (a=c in the previous function), and the sum of the two separations equals 60 nm (the red curve in Fig.3). Such a fit, y=y 0 +a exp[-0.5*([x-x 1 (0)]/b) 2 ] + a exp[-0.5*([x-(60-x 1 (0))]/d) 2 ], is shown as the red line in Fig. 2. The parameters of the function are determined to be y 0 =0.6 ± 0.5 nm, a= 12.0±0.8, x 1 (0)=34.3±0.9 nm, b=11.6±1.6 nm, and d=5.5±0.7 nm, and the reduced χ 2 = 1.5. According to this fit, the peaks of the Gaussians are at 34.3 nm and 25.7
11 nm which result in an asymmetry of 8.6 nm. Hence, the resolution of our data in separating two peaks from one would be somewhere between 8.6 nm and 16.6 nm. A non-rigorous and conservative estimate for resolution would be 14 nm. Conclusion Our results are the first application of SHRImP on motor proteins, and they represent the unique capabilities of this technique in measuring distances between 10 nm to 100 nm. We show that in the presence of very low concentration of ATP the separation between the heads of myosin VI is 29.3 ± 0.7 nm, in agreement with the straightforward hand-overhand model. Although the distribution in the measured head-to-head separation does not allow us to rule out a small asymmetry, we can set an upper limit of 14 nm for this asymmetry in the interhead separation (i.e. the interhead separation can not be more asymmetric than x 1 (0)=23 nm and x 1 (0)=37 nm). Since such large separations between the heads are structurally not possible by considering only the light chain domains (the maximum possible separation a 10 nm long lever arm can provide is 10+10=20 nm), the flexible region which enables the 60 nm step should also be at least partially open in between steps as well. We thank Matthew Gordon for sharing his SHRImP software with us, and Hyokeun Park and Ahmet Yildiz for their instructions regarding the sample preparation. We thank Anna Li and Damien Garbett for protein preparation. This work was supported by National Institutes of Health Grants AR44420, AR48931 and GM68625 and National Science Foundation Grants DBI and
12 References 1. Berg, J. S., B. C. Powell, and R. E. Cheney A millennial myosin census. Mol Biol Cell 12: Kellerman, K. A. and K. G. Miller An unconventional myosin heavy chain gene from Drosophila melanogaster. J Cell Biol 119: Hasson, T. and M. S. Mooseker Porcine myosin-vi: characterization of a new mammalian unconventional myosin. J Cell Biol 127: Avraham, K. B., T. Hasson, T. Sobe, B. Balsara, J. R. Testa, A. B. Skvorak, C. C. Morton, N. G. Copeland, and N. A. Jenkins Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice. Hum Mol Genet 6: Avraham, K. B., T. Hasson, K. P. Steel, D. M. Kingsley, L. B. Russell, M. S. Mooseker, N. G. Copeland, and N. A. Jenkins The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 11: Hasson, T., P. G. Gillespie, J. A. Garcia, R. B. MacDonald, Y. Zhao, A. G. Yee, M. S. Mooseker, and D. P. Corey Unconventional myosins in inner-ear sensory epithelia. J Cell Biol 137: Wells, A. L., A. W. Lin, L. Q. Chen, D. Safer, S. M. Cain, T. Hasson, B. O. Carragher, R. A. Milligan, and H. L. Sweeney Myosin VI is an actin-based motor that moves backwards. Nature 401: Buss, F., S. D. Arden, M. Lindsay, J. P. Luzio, and J. Kendrick-Jones Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. Embo J 20: Aschenbrenner, L., T. Lee, and T. Hasson Myo6 facilitates the translocation of endocytic vesicles from cell peripheries. Mol Biol Cell 14: Lister, I., S. Schmitz, M. Walker, J. Trinick, F. Buss, C. Veigel, and J. Kendrick- Jones A monomeric myosin VI with a large working stroke. Embo J 23: Rock, R. S., S. E. Rice, A. L. Wells, T. J. Purcell, J. A. Spudich, and H. L. Sweeney Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci U S A 98:
13 12. Nishikawa, S., K. Homma, Y. Komori, M. Iwaki, T. Wazawa, A. Hikikoshi Iwane, J. Saito, R. Ikebe, E. Katayama, T. Yanagida, and M. Ikebe Class VI myosin moves processively along actin filaments backward with large steps. Biochem Biophys Res Commun 290: Yildiz, A., H. Park, D. Safer, Z. Yang, L. Q. Chen, P. R. Selvin, and H. L. Sweeney Myosin VI Steps via a Hand-over-Hand Mechanism with Its Lever Arm Undergoing Fluctuations when Attached to Actin. J Biol Chem 279: Okten, Z., L. S. Churchman, R. S. Rock, and J. A. Spudich Myosin VI walks hand-over-hand along actin. Nat Struct Mol Biol 11(9):: Altman, D., H. L. Sweeney, and J. A. Spudich The mechanism of myosin VI translocation and its load-induced anchoring. Cell 116: Mehta, A. D., R. S. Rock, M. Rief, J. A. Spudich, M. S. Mooseker, and R. E. Cheney Myosin-V is a processive actin-based motor. Nature 400: Yildiz, A., M. Tomishige, R. D. Vale, and P. R. Selvin Kinesin Walks Hand-Over-Hand. Science 303: Yildiz, A., J. N. Forkey, S. A. McKinney, T. Ha, Y. E. Goldman, and P. R. Selvin Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300: Sakamoto, T., I. Amitani, E. Yokota, and T. Ando Direct observation of processive movement by individual myosin V molecules. Biochem Biophys Res Commun 272: Purcell, T. J., C. Morris, J. A. Spudich, and H. L. Sweeney Role of the lever arm in the processive stepping of myosin V. Proc Natl Acad Sci U S A 99: Mehta, A Myosin learns to walk. J Cell Sci 114: Veigel, C., F. Wang, M. L. Bartoo, J. R. Sellers, and J. E. Molloy The gated gait of the processive molecular motor, myosin V. Nat Cell Biol 4: Forkey, J. N., M. E. Quinlan, M. A. Shaw, J. E. Corrie, and Y. E. Goldman Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 422: Warshaw, D. M., G. Kennedy, S. Work, E. Krementsova, S. Beck, and K. Trybus Differential Labeling of Myosin V Heads with Quantum Dots Allows Direct Visualization of Hand-Over-Hand Processivity. Biophys J 88:L30-L32.
14 25. Rock, R. S., B. Ramamurthy, A. R. Dunn, S. Beccafico, B. R. Rami, C. Morris, B. J. Spink, C. Franzini-Armstrong, J. A. Spudich, and H. L. Sweeney A flexible domain is essential for the large step size and processivity of myosin VI. Mol Cell 17: Gordon, M. P., T. Ha, and P. R. Selvin Single-molecule high-resolution imaging with photobleaching. Proc Natl Acad Sci U S A 101: Qu, X., D. Wu, L. Mets, and N. F. Scherer Nanometer-localized multiple single-molecule fluorescence microscopy. PNAS:
15 Figure Legends Fig. 1. (a) Hand-over-hand mechanism of walking. (b) An alternative model of motility in which the separation between the myosin heads alternates between x and 60-x nm, while each head takes 60 nm steps sequentially. Cryo-electron microscopy images suggest that x could be much less than 30 nm. Fig. 2. (a) Peak intensity versus time for a myosin VI dimer labeled with two egfp s. (b) Pre and post photobleach of one of the egfp s. The last graph shows the difference between the first two, which is the emission from the egfp that photobleached first. Fig.3. Histogram of measured separations between the myosin VI heads and three Gaussian fits to the data. The blue curve is a single Gaussian fit representing the hand-over-hand model, the black curve is an unconstraint fit of two Gaussians, and the red curve is fit of two Gaussians with constraints.
16 Figure 1
17 Figure 2 a Time (sec) b Y Pre-photobleach Y Post-photobleach 8 X 3600 Intensity Intensity Y Difference 8 X 6000 Intensity Counts 5400
18 Figure Data single Gaussian two Gaussians two Gaussians with constraints Counts Distance (nm)
A new model for myosin dimeric motors incorporating Brownian ratchet and powerstroke mechanisms
 Invited Paper A new model for myosin dimeric motors incorporating Brownian ratchet and powerstroke mechanisms Brian Geislinger and Ryoichi Kawai Department of Physics, University of Alabama at Birmingham,
Invited Paper A new model for myosin dimeric motors incorporating Brownian ratchet and powerstroke mechanisms Brian Geislinger and Ryoichi Kawai Department of Physics, University of Alabama at Birmingham,
Introduction: actin and myosin
 Introduction: actin and myosin Actin Myosin Myosin V and actin 375 residues Found in all eukaryotes Polymeric Forms track for myosin Many other cellular functions 36 nm pseudo-helical repeat Catalytic
Introduction: actin and myosin Actin Myosin Myosin V and actin 375 residues Found in all eukaryotes Polymeric Forms track for myosin Many other cellular functions 36 nm pseudo-helical repeat Catalytic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
Myosin V Passing over Arp2/3 Junctions: Branching Ratio Calculated from the Elastic Lever Arm Model
 Biophysical Journal Volume 94 May 28 345 342 345 Myosin V Passing over Arp2/3 Junctions: Branching Ratio Calculated from the Elastic Lever Arm Model Andrej Vilfan J. Stefan Institute, Ljubljana, Slovenia
Biophysical Journal Volume 94 May 28 345 342 345 Myosin V Passing over Arp2/3 Junctions: Branching Ratio Calculated from the Elastic Lever Arm Model Andrej Vilfan J. Stefan Institute, Ljubljana, Slovenia
arxiv: v1 [physics.bio-ph] 9 Aug 2011
![arxiv: v1 [physics.bio-ph] 9 Aug 2011 arxiv: v1 [physics.bio-ph] 9 Aug 2011](/thumbs/95/124074436.jpg) Phenomenological analysis of ATP dependence of motor protein Yunxin Zhang Laboratory of Mathematics for Nonlinear Science, Centre for Computational System Biology, School of Mathematical Sciences, Fudan
Phenomenological analysis of ATP dependence of motor protein Yunxin Zhang Laboratory of Mathematics for Nonlinear Science, Centre for Computational System Biology, School of Mathematical Sciences, Fudan
Single-Molecule Methods I - in vitro
 Single-Molecule Methods I - in vitro Bo Huang Macromolecules 2014.03.10 F 1 -ATPase: a case study Membrane ADP ATP Rotation of the axle when hydrolyzing ATP Kinosita group, 1997-2005 Single Molecule Methods
Single-Molecule Methods I - in vitro Bo Huang Macromolecules 2014.03.10 F 1 -ATPase: a case study Membrane ADP ATP Rotation of the axle when hydrolyzing ATP Kinosita group, 1997-2005 Single Molecule Methods
Supplementary Information
 Supplementary Information Switching of myosin-v motion between the lever-arm swing and Brownian search-and-catch Keisuke Fujita 1*, Mitsuhiro Iwaki 2,3*, Atsuko H. Iwane 1, Lorenzo Marcucci 1 & Toshio
Supplementary Information Switching of myosin-v motion between the lever-arm swing and Brownian search-and-catch Keisuke Fujita 1*, Mitsuhiro Iwaki 2,3*, Atsuko H. Iwane 1, Lorenzo Marcucci 1 & Toshio
Coarse-Grained Structural Modeling of Molecular Motors Using Multibody Dynamics
 Cellular and Molecular Bioengineering, Vol. 2, No. 3, September 2009 (Ó 2009) pp. 366 374 DOI: 10.1007/s12195-009-0084-4 Coarse-Grained Structural Modeling of Molecular Motors Using Multibody Dynamics
Cellular and Molecular Bioengineering, Vol. 2, No. 3, September 2009 (Ó 2009) pp. 366 374 DOI: 10.1007/s12195-009-0084-4 Coarse-Grained Structural Modeling of Molecular Motors Using Multibody Dynamics
NIH Public Access Author Manuscript Cell Mol Bioeng. Author manuscript; available in PMC 2010 April 27.
 NIH Public Access Author Manuscript Published in final edited form as: Cell Mol Bioeng. 2009 September 1; 2(3): 366 374. doi:10.1007/s12195-009-0084-4. Coarse-Grained Structural Modeling of Molecular Motors
NIH Public Access Author Manuscript Published in final edited form as: Cell Mol Bioeng. 2009 September 1; 2(3): 366 374. doi:10.1007/s12195-009-0084-4. Coarse-Grained Structural Modeling of Molecular Motors
Mechanochemical coupling of two substeps in a single myosin V motor
 Mechanochemical coupling of two substeps in a single myosin V motor Sotaro Uemura 1, Hideo Higuchi 2,3, Adrian O Olivares 4, Enrique M De La Cruz 4 & Shin ichi Ishiwata 1,5 Myosin V is a double-headed
Mechanochemical coupling of two substeps in a single myosin V motor Sotaro Uemura 1, Hideo Higuchi 2,3, Adrian O Olivares 4, Enrique M De La Cruz 4 & Shin ichi Ishiwata 1,5 Myosin V is a double-headed
Five models for myosin V
 Five models for myosin V Andrej Vilfan J. Stefan Institute, Jamova 39, 1000 Ljubljana, Slovenia Myosin V was the first discovered processive motor from the myosin family. It has therefore been subject
Five models for myosin V Andrej Vilfan J. Stefan Institute, Jamova 39, 1000 Ljubljana, Slovenia Myosin V was the first discovered processive motor from the myosin family. It has therefore been subject
A Simple Kinetic Model Describes the Processivity of Myosin-V
 1642 Biophysical Journal Volume 84 March 2003 1642 1650 A Simple Kinetic Model Describes the Processivity of Myosin-V Anatoly B. Kolomeisky* and Michael E. Fisher y *Department of Chemistry, Rice University,
1642 Biophysical Journal Volume 84 March 2003 1642 1650 A Simple Kinetic Model Describes the Processivity of Myosin-V Anatoly B. Kolomeisky* and Michael E. Fisher y *Department of Chemistry, Rice University,
Anatoly B. Kolomeisky. Department of Chemistry CAN WE UNDERSTAND THE COMPLEX DYNAMICS OF MOTOR PROTEINS USING SIMPLE STOCHASTIC MODELS?
 Anatoly B. Kolomeisky Department of Chemistry CAN WE UNDERSTAND THE COMPLEX DYNAMICS OF MOTOR PROTEINS USING SIMPLE STOCHASTIC MODELS? Motor Proteins Enzymes that convert the chemical energy into mechanical
Anatoly B. Kolomeisky Department of Chemistry CAN WE UNDERSTAND THE COMPLEX DYNAMICS OF MOTOR PROTEINS USING SIMPLE STOCHASTIC MODELS? Motor Proteins Enzymes that convert the chemical energy into mechanical
Motor Proteins as Nanomachines: The Roles of Thermal Fluctuations in Generating Force and Motion
 Séminaire Poincaré XII (2009) 33 44 Séminaire Poincaré Motor Proteins as Nanomachines: The Roles of Thermal Fluctuations in Generating Force and Motion Jonathon Howard Max Planck Institute of Molecular
Séminaire Poincaré XII (2009) 33 44 Séminaire Poincaré Motor Proteins as Nanomachines: The Roles of Thermal Fluctuations in Generating Force and Motion Jonathon Howard Max Planck Institute of Molecular
Dynamics of Myosin-V Processivity
 Biophysical Journal Volume 88 February 2005 999 1008 999 Dynamics of Myosin-V Processivity Ganhui Lan* and Sean X. Sun* y *Department of Mechanical Engineering, and y Whitaker Institute of Biomedical Engineering,
Biophysical Journal Volume 88 February 2005 999 1008 999 Dynamics of Myosin-V Processivity Ganhui Lan* and Sean X. Sun* y *Department of Mechanical Engineering, and y Whitaker Institute of Biomedical Engineering,
arxiv:cond-mat/ v2 [cond-mat.stat-mech] 8 Jun 2006
![arxiv:cond-mat/ v2 [cond-mat.stat-mech] 8 Jun 2006 arxiv:cond-mat/ v2 [cond-mat.stat-mech] 8 Jun 2006](/thumbs/93/113637182.jpg) Brownian molecular motors driven by rotation-translation coupling Brian Geislinger and Ryoichi Kawai Department of Physics, University of Alabama at Birmingham, Birmingham, AL 35294 arxiv:cond-mat/6487v2
Brownian molecular motors driven by rotation-translation coupling Brian Geislinger and Ryoichi Kawai Department of Physics, University of Alabama at Birmingham, Birmingham, AL 35294 arxiv:cond-mat/6487v2
Are motor proteins power strokers, Brownian motors or both?
 Invited Paper Are motor proteins power strokers, Brownian motors or both? Brian Geislinger a,erindarnell b, Kimberly Farris c,andryoichikawai a a Dept. of Physics, Univ. of Alabama at Birmingham, 153 3rd
Invited Paper Are motor proteins power strokers, Brownian motors or both? Brian Geislinger a,erindarnell b, Kimberly Farris c,andryoichikawai a a Dept. of Physics, Univ. of Alabama at Birmingham, 153 3rd
Transport of single molecules along the periodic parallel lattices with coupling
 THE JOURNAL OF CHEMICAL PHYSICS 124 204901 2006 Transport of single molecules along the periodic parallel lattices with coupling Evgeny B. Stukalin The James Franck Institute The University of Chicago
THE JOURNAL OF CHEMICAL PHYSICS 124 204901 2006 Transport of single molecules along the periodic parallel lattices with coupling Evgeny B. Stukalin The James Franck Institute The University of Chicago
Biophysik der Moleküle!
 Biophysik der Moleküle!!"#$%&'()*+,-$./0()'$12$34!4! Molecular Motors:! - linear motors" 6. Dec. 2010! Muscle Motors and Cargo Transporting Motors! There are striking structural similarities but functional
Biophysik der Moleküle!!"#$%&'()*+,-$./0()'$12$34!4! Molecular Motors:! - linear motors" 6. Dec. 2010! Muscle Motors and Cargo Transporting Motors! There are striking structural similarities but functional
Acto-myosin: from muscles to single molecules. Justin Molloy MRC National Institute for Medical Research LONDON
 Acto-myosin: from muscles to single molecules. Justin Molloy MRC National Institute for Medical Research LONDON Energy in Biological systems: 1 Photon = 400 pn.nm 1 ATP = 100 pn.nm 1 Ion moving across
Acto-myosin: from muscles to single molecules. Justin Molloy MRC National Institute for Medical Research LONDON Energy in Biological systems: 1 Photon = 400 pn.nm 1 ATP = 100 pn.nm 1 Ion moving across
Detailed Tuning of Structure and Intramolecular Communication Are Dispensable for Processive Motion of Myosin VI
 430 Biophysical Journal Volume 100 January 2011 430 439 Detailed Tuning of Structure and Intramolecular Communication Are Dispensable for Processive Motion of Myosin VI Mary Williard Elting, Zev Bryant,
430 Biophysical Journal Volume 100 January 2011 430 439 Detailed Tuning of Structure and Intramolecular Communication Are Dispensable for Processive Motion of Myosin VI Mary Williard Elting, Zev Bryant,
Drosophila myosin VIIA is a high duty ratio motor with a unique kinetic mechanism
 JBC Papers in Press. Published on January 16, 2006 as Manuscript M511592200 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m511592200 Drosophila myosin VIIA is a high duty ratio motor
JBC Papers in Press. Published on January 16, 2006 as Manuscript M511592200 The latest version is at http://www.jbc.org/cgi/doi/10.1074/jbc.m511592200 Drosophila myosin VIIA is a high duty ratio motor
The Photon Counting Histogram: Statistical Analysis of Single Molecule Populations
 The Photon Counting Histogram: Statistical Analysis of Single Molecule Populations E. Gratton Laboratory for Fluorescence Dynamics University of California, Irvine Transition from FCS The Autocorrelation
The Photon Counting Histogram: Statistical Analysis of Single Molecule Populations E. Gratton Laboratory for Fluorescence Dynamics University of California, Irvine Transition from FCS The Autocorrelation
Tilting and Wobble of Myosin V by High-Speed Single-Molecule Polarized Fluorescence Microscopy
 University of Pennsylvania ScholarlyCommons Department of Physics Papers Department of Physics 3-2013 Tilting and Wobble of Myosin V by High-Speed Single-Molecule Polarized Fluorescence Microscopy John
University of Pennsylvania ScholarlyCommons Department of Physics Papers Department of Physics 3-2013 Tilting and Wobble of Myosin V by High-Speed Single-Molecule Polarized Fluorescence Microscopy John
Polymerization/depolymerization motors
 Polymerization/depolymerization motors Movement formation Kuo Lab, J.H.U. http://www.nature.com/nature/journal/v407/n6807/extref/40 71026a0_S3.mov http://www.bme.jhu.edu/~skuo/movies/macrophchase.mov http://www.bme.jhu.edu/~skuo/movies/gc_filo.mov
Polymerization/depolymerization motors Movement formation Kuo Lab, J.H.U. http://www.nature.com/nature/journal/v407/n6807/extref/40 71026a0_S3.mov http://www.bme.jhu.edu/~skuo/movies/macrophchase.mov http://www.bme.jhu.edu/~skuo/movies/gc_filo.mov
JBC Papers in Press. Published on June 16, 2005 as Manuscript M
 JBC Papers in Press. Published on June 16, 2005 as Manuscript M504779200 1 MYOSIN X IS A HIGH DUTY RATIO MOTOR Kazuaki Homma & Mitsuo Ikebe* Department of Physiology, University of Massachusetts Medical
JBC Papers in Press. Published on June 16, 2005 as Manuscript M504779200 1 MYOSIN X IS A HIGH DUTY RATIO MOTOR Kazuaki Homma & Mitsuo Ikebe* Department of Physiology, University of Massachusetts Medical
Neurite formation & neuronal polarization
 Neurite formation & neuronal polarization Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 An immature neuron in cell culture first sprouts
Neurite formation & neuronal polarization Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 An immature neuron in cell culture first sprouts
Neurite formation & neuronal polarization. The cytoskeletal components of neurons have characteristic distributions and associations
 Mechanisms of neuronal migration & Neurite formation & neuronal polarization Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 The cytoskeletal
Mechanisms of neuronal migration & Neurite formation & neuronal polarization Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 The cytoskeletal
Chapter 16. Cellular Movement: Motility and Contractility. Lectures by Kathleen Fitzpatrick Simon Fraser University Pearson Education, Inc.
 Chapter 16 Cellular Movement: Motility and Contractility Lectures by Kathleen Fitzpatrick Simon Fraser University Two eukaryotic motility systems 1. Interactions between motor proteins and microtubules
Chapter 16 Cellular Movement: Motility and Contractility Lectures by Kathleen Fitzpatrick Simon Fraser University Two eukaryotic motility systems 1. Interactions between motor proteins and microtubules
Elastic Lever-Arm Model for Myosin V
 3792 Biophysical Journal Volume 88 June 2005 3792 3805 Elastic Lever-Arm Model for Myosin V Andrej Vilfan J. Stefan Institute, Ljubljana, Slovenia ABSTRACT We present a mechanochemical model for myosin
3792 Biophysical Journal Volume 88 June 2005 3792 3805 Elastic Lever-Arm Model for Myosin V Andrej Vilfan J. Stefan Institute, Ljubljana, Slovenia ABSTRACT We present a mechanochemical model for myosin
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
Nuclear import of DNA: genetic modification of plants
 Nuclear import of DNA: genetic modification of plants gene delivery by Agrobacterium tumefaciens T. Tzfira & V. Citovsky. 2001. Trends in Cell Biol. 12: 121-129 VirE2 binds ssdna in vitro forms helical
Nuclear import of DNA: genetic modification of plants gene delivery by Agrobacterium tumefaciens T. Tzfira & V. Citovsky. 2001. Trends in Cell Biol. 12: 121-129 VirE2 binds ssdna in vitro forms helical
In biomolecular systems, free energy frequently needs to be
 Kinetic equilibrium of forces and molecular events in muscle contraction Erwin W. Becker* Institut für Mikrostrukturtechnik, Forschungszentrum, and Universität Karlsruhe, 76021 Karlsruhe, P.O. Box 3640,
Kinetic equilibrium of forces and molecular events in muscle contraction Erwin W. Becker* Institut für Mikrostrukturtechnik, Forschungszentrum, and Universität Karlsruhe, 76021 Karlsruhe, P.O. Box 3640,
Research Article Flexural Stiffness of Myosin Va Subdomains as Measured from Tethered Particle Motion
 Journal of Biophysics Volume 2015, Article ID 465693, 9 pages http://dx.doi.org/10.1155/2015/465693 Research Article Flexural Stiffness of Myosin Va Subdomains as Measured from Tethered Particle Motion
Journal of Biophysics Volume 2015, Article ID 465693, 9 pages http://dx.doi.org/10.1155/2015/465693 Research Article Flexural Stiffness of Myosin Va Subdomains as Measured from Tethered Particle Motion
LAB 3: Confocal Microscope Imaging of single-emitter fluorescence. LAB 4: Hanbury Brown and Twiss setup. Photon antibunching. Roshita Ramkhalawon
 LAB 3: Confocal Microscope Imaging of single-emitter fluorescence LAB 4: Hanbury Brown and Twiss setup. Photon antibunching Roshita Ramkhalawon PHY 434 Department of Physics & Astronomy University of Rochester
LAB 3: Confocal Microscope Imaging of single-emitter fluorescence LAB 4: Hanbury Brown and Twiss setup. Photon antibunching Roshita Ramkhalawon PHY 434 Department of Physics & Astronomy University of Rochester
ATP Synthase. Proteins as nanomachines. ATP Synthase. Protein physics, Lecture 11. Create proton gradient across. Thermal motion and small objects
 Proteins as nanomachines Protein physics, Lecture 11 ATP Synthase ATP synthase, a molecular motor Thermal motion and small objects Brownian motors and ratchets Actin and Myosin using random motion to go
Proteins as nanomachines Protein physics, Lecture 11 ATP Synthase ATP synthase, a molecular motor Thermal motion and small objects Brownian motors and ratchets Actin and Myosin using random motion to go
The Photon Counting Histogram (PCH)
 The Photon Counting Histogram (PCH) While Fluorescence Correlation Spectroscopy (FCS) can measure the diffusion coefficient and concentration of fluorophores, a complementary method, a photon counting
The Photon Counting Histogram (PCH) While Fluorescence Correlation Spectroscopy (FCS) can measure the diffusion coefficient and concentration of fluorophores, a complementary method, a photon counting
The neuron as a secretory cell
 The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
Importance of Hydrodynamic Interactions in the Stepping Kinetics of Kinesin
 pubs.acs.org/jpcb Importance of Hydrodynamic Interactions in the Stepping Kinetics of Kinesin Yonathan Goldtzvik, Zhechun Zhang, and D. Thirumalai*,, Biophysics Program, Institute for Physical Science
pubs.acs.org/jpcb Importance of Hydrodynamic Interactions in the Stepping Kinetics of Kinesin Yonathan Goldtzvik, Zhechun Zhang, and D. Thirumalai*,, Biophysics Program, Institute for Physical Science
Linear Motors. Nanostrukturphysik II, Manuel Bastuck
 Molecular l Motors I: Linear Motors Nanostrukturphysik II, Manuel Bastuck Why can he run so fast? Usain Bolt 100 m / 9,58 s Usain Bolt: http://www.wallpaperdev.com/stock/fantesty-usain-bolt.jpg muscle:
Molecular l Motors I: Linear Motors Nanostrukturphysik II, Manuel Bastuck Why can he run so fast? Usain Bolt 100 m / 9,58 s Usain Bolt: http://www.wallpaperdev.com/stock/fantesty-usain-bolt.jpg muscle:
Laboratory 3: Confocal Microscopy Imaging of Single Emitter Fluorescence and Hanbury Brown, and Twiss Setup for Photon Antibunching
 Laboratory 3: Confocal Microscopy Imaging of Single Emitter Fluorescence and Hanbury Brown, and Twiss Setup for Photon Antibunching Jonathan Papa 1, * 1 Institute of Optics University of Rochester, Rochester,
Laboratory 3: Confocal Microscopy Imaging of Single Emitter Fluorescence and Hanbury Brown, and Twiss Setup for Photon Antibunching Jonathan Papa 1, * 1 Institute of Optics University of Rochester, Rochester,
Actin and myosin constitute thin and thick filaments, respectively,
 Conformational change of the actomyosin complex drives the multiple stepping movement Tomoki P. Terada, Masaki Sasai, and Tetsuya Yomo Graduate School of Human Informatics, Nagoya University, Nagoya 464-860,
Conformational change of the actomyosin complex drives the multiple stepping movement Tomoki P. Terada, Masaki Sasai, and Tetsuya Yomo Graduate School of Human Informatics, Nagoya University, Nagoya 464-860,
According to the diagram, which of the following is NOT true?
 Instructions: Review Chapter 44 on muscular-skeletal systems and locomotion, and then complete the following Blackboard activity. This activity will introduce topics that will be covered in the next few
Instructions: Review Chapter 44 on muscular-skeletal systems and locomotion, and then complete the following Blackboard activity. This activity will introduce topics that will be covered in the next few
Intracellular transport
 Transport in cells Intracellular transport introduction: transport in cells, molecular players etc. cooperation of motors, forces good and bad transport regulation, traffic issues, Stefan Klumpp image
Transport in cells Intracellular transport introduction: transport in cells, molecular players etc. cooperation of motors, forces good and bad transport regulation, traffic issues, Stefan Klumpp image
ParM filament images were extracted and from the electron micrographs and
 Supplemental methods Outline of the EM reconstruction: ParM filament images were extracted and from the electron micrographs and straightened. The digitized images were corrected for the phase of the Contrast
Supplemental methods Outline of the EM reconstruction: ParM filament images were extracted and from the electron micrographs and straightened. The digitized images were corrected for the phase of the Contrast
Lecture 7 : Molecular Motors. Dr Eileen Nugent
 Lecture 7 : Molecular Motors Dr Eileen Nugent Molecular Motors Energy Sources: Protonmotive Force, ATP Single Molecule Biophysical Techniques : Optical Tweezers, Atomic Force Microscopy, Single Molecule
Lecture 7 : Molecular Motors Dr Eileen Nugent Molecular Motors Energy Sources: Protonmotive Force, ATP Single Molecule Biophysical Techniques : Optical Tweezers, Atomic Force Microscopy, Single Molecule
Supporting Information Converter domain mutations in myosin alter structural kinetics and motor function. Hershey, PA, MN 55455, USA
 Supporting Information Converter domain mutations in myosin alter structural kinetics and motor function Laura K. Gunther 1, John A. Rohde 2, Wanjian Tang 1, Shane D. Walton 1, William C. Unrath 1, Darshan
Supporting Information Converter domain mutations in myosin alter structural kinetics and motor function Laura K. Gunther 1, John A. Rohde 2, Wanjian Tang 1, Shane D. Walton 1, William C. Unrath 1, Darshan
For slowly varying probabilities, the continuum form of these equations is. = (r + d)p T (x) (u + l)p D (x) ar x p T(x, t) + a2 r
 3.2 Molecular Motors A variety of cellular processes requiring mechanical work, such as movement, transport and packaging material, are performed with the aid of protein motors. These molecules consume
3.2 Molecular Motors A variety of cellular processes requiring mechanical work, such as movement, transport and packaging material, are performed with the aid of protein motors. These molecules consume
Fluorescence Resonance Energy Transfer (FRET) Microscopy
 Fluorescence Resonance Energy Transfer () Microscopy Mike Lorenz Optical Technology Development mlorenz@mpi-cbg.de -FLM course, May 2009 What is fluorescence? Stoke s shift Fluorescence light is always
Fluorescence Resonance Energy Transfer () Microscopy Mike Lorenz Optical Technology Development mlorenz@mpi-cbg.de -FLM course, May 2009 What is fluorescence? Stoke s shift Fluorescence light is always
The biological motors
 Motor proteins The definition of motor proteins Miklós Nyitrai, November 30, 2016 Molecular machines key to understand biological processes machines in the micro/nano-world (unidirectional steps): nm,
Motor proteins The definition of motor proteins Miklós Nyitrai, November 30, 2016 Molecular machines key to understand biological processes machines in the micro/nano-world (unidirectional steps): nm,
Confocal Microscopy Imaging of Single Emitter Fluorescence and Hanbury Brown and Twiss Photon Antibunching Setup
 1 Confocal Microscopy Imaging of Single Emitter Fluorescence and Hanbury Brown and Twiss Photon Antibunching Setup Abstract Jacob Begis The purpose of this lab was to prove that a source of light can be
1 Confocal Microscopy Imaging of Single Emitter Fluorescence and Hanbury Brown and Twiss Photon Antibunching Setup Abstract Jacob Begis The purpose of this lab was to prove that a source of light can be
The mechanical events responsible for muscle contraction are
 Repriming the actomyosin crossbridge cycle Walter Steffen and John Sleep* Randall Centre, King s College, London SE1 1UL, United Kingdom Edited by Edwin W. Taylor, University of Chicago, Chicago, IL, and
Repriming the actomyosin crossbridge cycle Walter Steffen and John Sleep* Randall Centre, King s College, London SE1 1UL, United Kingdom Edited by Edwin W. Taylor, University of Chicago, Chicago, IL, and
Single molecule biochemistry using optical tweezers
 FEBS 20359 FEBS Letters 430 (1998) 23^27 Minireview Single molecule biochemistry using optical tweezers Amit D. Mehta*, Katherine A. Pullen, James A. Spudich Department of Biochemistry, Stanford University
FEBS 20359 FEBS Letters 430 (1998) 23^27 Minireview Single molecule biochemistry using optical tweezers Amit D. Mehta*, Katherine A. Pullen, James A. Spudich Department of Biochemistry, Stanford University
SINGLE-MOLECULE PHYSIOLOGY
 SINGLE-MOLECULE PHYSIOLOGY Kazuhiko Kinosita, Jr. Center for Integrative Bioscience, Okazaki National Research Institutes Higashiyama 5-1, Myodaiji, Okazaki 444-8585, Japan Single-Molecule Physiology under
SINGLE-MOLECULE PHYSIOLOGY Kazuhiko Kinosita, Jr. Center for Integrative Bioscience, Okazaki National Research Institutes Higashiyama 5-1, Myodaiji, Okazaki 444-8585, Japan Single-Molecule Physiology under
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Supplementary Information
 Supplementary Information Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism Duyoung Min, Kipom Kim, Changbong Hyeon, Yong Hoon Cho, Yeon-Kyun
Supplementary Information Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism Duyoung Min, Kipom Kim, Changbong Hyeon, Yong Hoon Cho, Yeon-Kyun
Conventional kinesin is a processive motor protein that walks
 Kinesin crouches to sprint but resists pushing Michael E. Fisher* and Young C. Kim Institute for Physical Science and Technology, University of Maryland, College Park, MD 20742 Contributed by Michael E.
Kinesin crouches to sprint but resists pushing Michael E. Fisher* and Young C. Kim Institute for Physical Science and Technology, University of Maryland, College Park, MD 20742 Contributed by Michael E.
SUPPLEMENTARY INFORMATION. microscopy of membrane proteins
 SUPPLEMENTARY INFORMATION Glass is a viable substrate for precision force microscopy of membrane proteins Nagaraju Chada 1, Krishna P. Sigdel 1, Raghavendar Reddy Sanganna Gari 1, Tina Rezaie Matin 1,
SUPPLEMENTARY INFORMATION Glass is a viable substrate for precision force microscopy of membrane proteins Nagaraju Chada 1, Krishna P. Sigdel 1, Raghavendar Reddy Sanganna Gari 1, Tina Rezaie Matin 1,
Molecular Motors. Structural and Mechanistic Overview! Kimberly Nguyen - December 6, 2013! MOLECULAR MOTORS - KIMBERLY NGUYEN
 Molecular Motors Structural and Mechanistic Overview!! Kimberly Nguyen - December 6, 2013!! 1 Molecular Motors: A Structure and Mechanism Overview! Introduction! Molecular motors are fundamental agents
Molecular Motors Structural and Mechanistic Overview!! Kimberly Nguyen - December 6, 2013!! 1 Molecular Motors: A Structure and Mechanism Overview! Introduction! Molecular motors are fundamental agents
Extension of a Three-Helix Bundle Domain of Myosin VI and Key Role of Calmodulins
 2964 Biophysical Journal Volume 100 June 2011 2964 2973 Extension of a Three-Helix Bundle Domain of Myosin VI and Key Role of Calmodulins Yanxin Liu, 6 Jen Hsin, 6 HyeongJun Kim, Paul R. Selvin, * and
2964 Biophysical Journal Volume 100 June 2011 2964 2973 Extension of a Three-Helix Bundle Domain of Myosin VI and Key Role of Calmodulins Yanxin Liu, 6 Jen Hsin, 6 HyeongJun Kim, Paul R. Selvin, * and
(i.e. what you should be able to answer at end of lecture)
 Today s Announcements 1. Test given back next Wednesday 2. HW assigned next Wednesday. 3. Next Monday 1 st discussion about Individual Projects. Today s take-home lessons (i.e. what you should be able
Today s Announcements 1. Test given back next Wednesday 2. HW assigned next Wednesday. 3. Next Monday 1 st discussion about Individual Projects. Today s take-home lessons (i.e. what you should be able
Nature Methods: doi: /nmeth Supplementary Figure 1
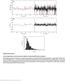 Supplementary Figure 1 Control experiments on the fluorescence stability of single Atto647N-biotin complexes. a, Exemplary signal trace (black line) in counts per second (cps) of a single Atto647N molecule
Supplementary Figure 1 Control experiments on the fluorescence stability of single Atto647N-biotin complexes. a, Exemplary signal trace (black line) in counts per second (cps) of a single Atto647N molecule
Use of Stable Analogs of Myosin ATPase Intermediates for Kinetic Studies of the Weak Binding of Myosin Heads to F-Actin
 Biochemistry (Moscow), Vol. 64, No. 8, 1999, pp. 875-882. Translated from Biokhimiya, Vol. 64, No. 8, 1999, pp. 1043-1051. Original Russian Text Copyright 1999 by Rostkova, Moiseeva, Teplova, Nikolaeva,
Biochemistry (Moscow), Vol. 64, No. 8, 1999, pp. 875-882. Translated from Biokhimiya, Vol. 64, No. 8, 1999, pp. 1043-1051. Original Russian Text Copyright 1999 by Rostkova, Moiseeva, Teplova, Nikolaeva,
SUPPLEMENTARY FIGURE 1. Force dependence of the unbinding rate: (a) Force-dependence
 (b) BSA-coated beads double exponential low force exponential high force exponential 1 unbinding time tb [sec] (a) unbinding time tb [sec] SUPPLEMENTARY FIGURES BSA-coated beads without BSA.2.1 5 1 load
(b) BSA-coated beads double exponential low force exponential high force exponential 1 unbinding time tb [sec] (a) unbinding time tb [sec] SUPPLEMENTARY FIGURES BSA-coated beads without BSA.2.1 5 1 load
Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding
 Supplementary information Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding Marcus Braun* 1,2,3, Zdenek Lansky* 1,2,3, Agata Szuba 1,2, Friedrich W. Schwarz 1,2, Aniruddha
Supplementary information Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding Marcus Braun* 1,2,3, Zdenek Lansky* 1,2,3, Agata Szuba 1,2, Friedrich W. Schwarz 1,2, Aniruddha
PHYSIOLOGY CHAPTER 9 MUSCLE TISSUE Fall 2016
 PHYSIOLOGY CHAPTER 9 MUSCLE TISSUE Fall 2016 2 Chapter 9 Muscles and Muscle Tissue Overview of Muscle Tissue types of muscle: are all prefixes for muscle Contractility all muscles cells can Smooth & skeletal
PHYSIOLOGY CHAPTER 9 MUSCLE TISSUE Fall 2016 2 Chapter 9 Muscles and Muscle Tissue Overview of Muscle Tissue types of muscle: are all prefixes for muscle Contractility all muscles cells can Smooth & skeletal
Solution set for EXAM IN TFY4265/FY8906 Biophysical microtechniques
 ENGLISH NORWEGIAN UNIVERSITY OF SCIENCE AND TECHNOLOGY DEPARTMENT OF PHYSICS Contact during exam: Magnus Borstad Lilledahl Telefon: 73591873 (office) 92851014 (mobile) Solution set for EXAM IN TFY4265/FY8906
ENGLISH NORWEGIAN UNIVERSITY OF SCIENCE AND TECHNOLOGY DEPARTMENT OF PHYSICS Contact during exam: Magnus Borstad Lilledahl Telefon: 73591873 (office) 92851014 (mobile) Solution set for EXAM IN TFY4265/FY8906
Muscle tissue. Types. Functions. Cardiac, Smooth, and Skeletal
 Types Cardiac, Smooth, and Skeletal Functions movements posture and body position Support soft tissues Guard openings body temperature nutrient reserves Muscle tissue Special Characteristics of Muscle
Types Cardiac, Smooth, and Skeletal Functions movements posture and body position Support soft tissues Guard openings body temperature nutrient reserves Muscle tissue Special Characteristics of Muscle
Muscle regulation and Actin Topics: Tropomyosin and Troponin, Actin Assembly, Actin-dependent Movement
 1 Muscle regulation and Actin Topics: Tropomyosin and Troponin, Actin Assembly, Actin-dependent Movement In the last lecture, we saw that a repeating alternation between chemical (ATP hydrolysis) and vectorial
1 Muscle regulation and Actin Topics: Tropomyosin and Troponin, Actin Assembly, Actin-dependent Movement In the last lecture, we saw that a repeating alternation between chemical (ATP hydrolysis) and vectorial
Velocity, processivity and individual steps of single myosin V molecules in live cells
 Biophysical Journal, Volume 96 Supporting Material Velocity, processivity and individual steps of single myosin V molecules in live cells Paolo Pierobon, Sarra Achouri, Sébastien Courty, Alexander R. Dunn,
Biophysical Journal, Volume 96 Supporting Material Velocity, processivity and individual steps of single myosin V molecules in live cells Paolo Pierobon, Sarra Achouri, Sébastien Courty, Alexander R. Dunn,
Lecture 13, 05 October 2004 Chapter 10, Muscle. Vertebrate Physiology ECOL 437 University of Arizona Fall instr: Kevin Bonine t.a.
 Lecture 13, 05 October 2004 Chapter 10, Muscle Vertebrate Physiology ECOL 437 University of Arizona Fall 2004 instr: Kevin Bonine t.a.: Nate Swenson Vertebrate Physiology 437 18 1. Muscle A. Sarcomere
Lecture 13, 05 October 2004 Chapter 10, Muscle Vertebrate Physiology ECOL 437 University of Arizona Fall 2004 instr: Kevin Bonine t.a.: Nate Swenson Vertebrate Physiology 437 18 1. Muscle A. Sarcomere
Dynamic 3D behavior of dynein using DXT
 MERIT Self-directed Joint Research Dynamic 3D behavior of dynein using DXT Yoshimi KINOSHITA1 and Yufuku MATSUSHITA2 1 Department of Physics, Graduate School of Science, UTokyo 2 Department of Advanced
MERIT Self-directed Joint Research Dynamic 3D behavior of dynein using DXT Yoshimi KINOSHITA1 and Yufuku MATSUSHITA2 1 Department of Physics, Graduate School of Science, UTokyo 2 Department of Advanced
BMB Class 17, November 30, Single Molecule Biophysics (II)
 BMB 178 2018 Class 17, November 30, 2018 15. Single Molecule Biophysics (II) New Advances in Single Molecule Techniques Atomic Force Microscopy Single Molecule Manipulation - optical traps and tweezers
BMB 178 2018 Class 17, November 30, 2018 15. Single Molecule Biophysics (II) New Advances in Single Molecule Techniques Atomic Force Microscopy Single Molecule Manipulation - optical traps and tweezers
Biological motors 18.S995 - L10
 Biological motors 18.S995 - L1 Reynolds numbers Re = UL µ = UL m the organism is mo E.coli (non-tumbling HCB 437) Drescher, Dunkel, Ganguly, Cisneros, Goldstein (211) PNAS Bacterial motors movie: V. Kantsler
Biological motors 18.S995 - L1 Reynolds numbers Re = UL µ = UL m the organism is mo E.coli (non-tumbling HCB 437) Drescher, Dunkel, Ganguly, Cisneros, Goldstein (211) PNAS Bacterial motors movie: V. Kantsler
EXPERIMENTAL PROCEDURES
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 284, NO. 4, pp. 2138 2149, January 23, 2009 Printed in the U.S.A. Switch 1 Mutation S217A Converts Myosin V into a Low Duty Ratio Motor * Received for publication,
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 284, NO. 4, pp. 2138 2149, January 23, 2009 Printed in the U.S.A. Switch 1 Mutation S217A Converts Myosin V into a Low Duty Ratio Motor * Received for publication,
c 2006 by Prasanth Sankar. All rights reserved.
 c 2006 by Prasanth Sankar. All rights reserved. PHENOMENOLOGICAL MODELS OF MOTOR PROTEINS BY PRASANTH SANKAR M. S., University of Illinois at Urbana-Champaign, 2000 DISSERTATION Submitted in partial fulfillment
c 2006 by Prasanth Sankar. All rights reserved. PHENOMENOLOGICAL MODELS OF MOTOR PROTEINS BY PRASANTH SANKAR M. S., University of Illinois at Urbana-Champaign, 2000 DISSERTATION Submitted in partial fulfillment
arxiv: v1 [physics.bio-ph] 27 Sep 2014
![arxiv: v1 [physics.bio-ph] 27 Sep 2014 arxiv: v1 [physics.bio-ph] 27 Sep 2014](/thumbs/95/122806024.jpg) Ensemble velocity of non-processive molecular motors with multiple chemical states Andrej Vilfan J. Stefan Institute, Jamova 39, Ljubljana, Slovenia and Faculty of Mathematics and Physics, University of
Ensemble velocity of non-processive molecular motors with multiple chemical states Andrej Vilfan J. Stefan Institute, Jamova 39, Ljubljana, Slovenia and Faculty of Mathematics and Physics, University of
Cytokinesis in fission yeast: Modeling the assembly of the contractile ring
 Cytokinesis in fission yeast: Modeling the assembly of the contractile ring Nikola Ojkic, Dimitrios Vavylonis Department of Physics, Lehigh University Damien Laporte, Jian-Qiu Wu Department of Molecular
Cytokinesis in fission yeast: Modeling the assembly of the contractile ring Nikola Ojkic, Dimitrios Vavylonis Department of Physics, Lehigh University Damien Laporte, Jian-Qiu Wu Department of Molecular
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell
 FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
Laboratory 3&4: Confocal Microscopy Imaging of Single-Emitter Fluorescence and Hanbury Brown and Twiss setup for Photon Antibunching
 Laboratory 3&4: Confocal Microscopy Imaging of Single-Emitter Fluorescence and Hanbury Brown and Twiss setup for Photon Antibunching Jose Alejandro Graniel Institute of Optics University of Rochester,
Laboratory 3&4: Confocal Microscopy Imaging of Single-Emitter Fluorescence and Hanbury Brown and Twiss setup for Photon Antibunching Jose Alejandro Graniel Institute of Optics University of Rochester,
Fluorescence polarisation, anisotropy FRAP
 Fluorescence polarisation, anisotropy FRAP Reminder: fluorescence spectra Definitions! a. Emission sp. b. Excitation sp. Stokes-shift The difference (measured in nm) between the peak of the excitation
Fluorescence polarisation, anisotropy FRAP Reminder: fluorescence spectra Definitions! a. Emission sp. b. Excitation sp. Stokes-shift The difference (measured in nm) between the peak of the excitation
Fitting Force-dependent Kinetic Models to Single-Molecule Dwell Time Records
 Fitting Force-dependent Kinetic Models to Single-Molecule Dwell Time Records Abstract Micromanipulation techniques such as optical tweezers atomic force microscopy allow us to identify characterize force-dependent
Fitting Force-dependent Kinetic Models to Single-Molecule Dwell Time Records Abstract Micromanipulation techniques such as optical tweezers atomic force microscopy allow us to identify characterize force-dependent
Bidirectional Power Stroke by Ncd Kinesin
 Biophysical Journal Volume 99 December 1 39 391 39 Bidirectional Power Stroke by Ncd Kinesin Anthony E. Butterfield, * ussell J. Stewart, Christoph F. Schmidt, and Mikhail Skliar * Department of Chemical
Biophysical Journal Volume 99 December 1 39 391 39 Bidirectional Power Stroke by Ncd Kinesin Anthony E. Butterfield, * ussell J. Stewart, Christoph F. Schmidt, and Mikhail Skliar * Department of Chemical
Chapter 4 Bidirectional membrane tubes driven by nonprocessive motors
 Chapter 4 Bidirectional membrane tubes driven by nonprocessive motors In cells, membrane tubes are extracted by molecular motors. Although individual motors cannot provide enough force to pull a tube,
Chapter 4 Bidirectional membrane tubes driven by nonprocessive motors In cells, membrane tubes are extracted by molecular motors. Although individual motors cannot provide enough force to pull a tube,
Does the S2 Rod of Myosin II Uncoil upon Two-Headed Binding to Actin? A Leucine-Zippered HMM Study
 12886 Biochemistry 2003, 42, 12886-12892 Does the S2 Rod of Myosin II Uncoil upon Two-Headed Binding to Actin? A Leucine-Zippered HMM Study Tania Chakrabarty,, Chris Yengo, Corry Baldacchino, Li-Qiong
12886 Biochemistry 2003, 42, 12886-12892 Does the S2 Rod of Myosin II Uncoil upon Two-Headed Binding to Actin? A Leucine-Zippered HMM Study Tania Chakrabarty,, Chris Yengo, Corry Baldacchino, Li-Qiong
Speed-detachment tradeoff and its effect. on track bound transport of single motor. protein
 Speed-detachment tradeoff and its effect on track bound transport of single motor protein arxiv:1811.11414v1 [q-bio.sc] 28 Nov 2018 Mohd Suhail Rizvi Department of Biological Sciences and Bioengineering,
Speed-detachment tradeoff and its effect on track bound transport of single motor protein arxiv:1811.11414v1 [q-bio.sc] 28 Nov 2018 Mohd Suhail Rizvi Department of Biological Sciences and Bioengineering,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
Visualize and Measure Nanoparticle Size and Concentration
 NTA : Nanoparticle Tracking Analysis Visualize and Measure Nanoparticle Size and Concentration 30 Apr 2015 NanoSight product range LM 10 series NS300 series NS500 series Dec 13 34 www.nanosight.com NanoSight
NTA : Nanoparticle Tracking Analysis Visualize and Measure Nanoparticle Size and Concentration 30 Apr 2015 NanoSight product range LM 10 series NS300 series NS500 series Dec 13 34 www.nanosight.com NanoSight
Neurite initiation. Neurite formation begins with a bud that sprouts from the cell body. One or several neurites can sprout at a time.
 Neurite initiation. Neuronal maturation initiation f-actin polarization and maturation tubulin stage 1: "spherical" neuron stage 2: neurons extend several neurites stage 3: one neurite accelerates its
Neurite initiation. Neuronal maturation initiation f-actin polarization and maturation tubulin stage 1: "spherical" neuron stage 2: neurons extend several neurites stage 3: one neurite accelerates its
Supplementary Figure 1 Level structure of a doubly charged QDM (a) PL bias map acquired under 90 nw non-resonant excitation at 860 nm.
 Supplementary Figure 1 Level structure of a doubly charged QDM (a) PL bias map acquired under 90 nw non-resonant excitation at 860 nm. Charging steps are labeled by the vertical dashed lines. Intensity
Supplementary Figure 1 Level structure of a doubly charged QDM (a) PL bias map acquired under 90 nw non-resonant excitation at 860 nm. Charging steps are labeled by the vertical dashed lines. Intensity
Fig. 1. Stereo images showing (A) the best fit of the atomic model for F actin and the F actin map obtained by cryo-em and image analysis, and (B) goo
 Fig. 1. Stereo images showing (A) the best fit of the atomic model for F actin and the F actin map obtained by cryo-em and image analysis, and (B) good correspondence between the location of Cys374 and
Fig. 1. Stereo images showing (A) the best fit of the atomic model for F actin and the F actin map obtained by cryo-em and image analysis, and (B) good correspondence between the location of Cys374 and
Manipulating and Probing Enzymatic Conformational Fluctuations and Enzyme-Substrate Interactions by Single-Molecule FRET- Magnetic Tweezers Microscopy
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2014 Supporting Information (SI) Manipulating and Probing Enzymatic Conformational Fluctuations
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2014 Supporting Information (SI) Manipulating and Probing Enzymatic Conformational Fluctuations
type GroEL-GroES complex. Crystals were grown in buffer D (100 mm HEPES, ph 7.5,
 Supplementary Material Supplementary Materials and Methods Structure Determination of SR1-GroES-ADP AlF x SR1-GroES-ADP AlF x was purified as described in Materials and Methods for the wild type GroEL-GroES
Supplementary Material Supplementary Materials and Methods Structure Determination of SR1-GroES-ADP AlF x SR1-GroES-ADP AlF x was purified as described in Materials and Methods for the wild type GroEL-GroES
Supplementary Information Effects of asymmetric nanostructures on the extinction. difference properties of actin biomolecules and filaments
 Supplementary Information Effects of asymmetric nanostructures on the extinction difference properties of actin biomolecules and filaments 1 E. H. Khoo, 2 Eunice S. P. Leong, 1 W. K. Phua, 2 S. J. Wu,
Supplementary Information Effects of asymmetric nanostructures on the extinction difference properties of actin biomolecules and filaments 1 E. H. Khoo, 2 Eunice S. P. Leong, 1 W. K. Phua, 2 S. J. Wu,
Stress Effects on Myosin Mutant Root Length in Arabidopsis thaliana
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
Single Molecule Spectroscopy and Imaging
 Single Molecule Spectroscopy and Imaging Ingo Gregor, Thomas Dertinger, Iris von der Hocht, Jan Sykora, Luru Dai, Jörg Enderlein Institute for Biological Information Processing 1 Forschungszentrum Jülich
Single Molecule Spectroscopy and Imaging Ingo Gregor, Thomas Dertinger, Iris von der Hocht, Jan Sykora, Luru Dai, Jörg Enderlein Institute for Biological Information Processing 1 Forschungszentrum Jülich
Kinematics of the lever arm swing in myosin VI
 Kinematics of the lever arm swing in myosin VI Mauro L. Mugnai a,b,1,2 and D. Thirumalai a,b,1,2 PNAS PLUS a Biophysics Program, Institute for Physical Science and Technology, University of Maryland, College
Kinematics of the lever arm swing in myosin VI Mauro L. Mugnai a,b,1,2 and D. Thirumalai a,b,1,2 PNAS PLUS a Biophysics Program, Institute for Physical Science and Technology, University of Maryland, College
NIH Public Access Author Manuscript J Phys Condens Matter. Author manuscript; available in PMC 2014 November 20.
 NIH Public Access Author Manuscript Published in final edited form as: J Phys Condens Matter. 2013 November 20; 25(46):. doi:10.1088/0953-8984/25/46/463101. Motor Proteins and Molecular Motors: How to
NIH Public Access Author Manuscript Published in final edited form as: J Phys Condens Matter. 2013 November 20; 25(46):. doi:10.1088/0953-8984/25/46/463101. Motor Proteins and Molecular Motors: How to
TitleElastically coupled two-dimensional. Citation PHYSICAL REVIEW E (2005), 71(6)
 TitleElastically coupled two-dimensional Author(s) Goko, H; Igarashi, A Citation PHYSICAL REVIEW E (2005), 71(6) Issue Date 2005-06 URL http://hdl.handle.net/2433/50298 RightCopyright 2005 American Physical
TitleElastically coupled two-dimensional Author(s) Goko, H; Igarashi, A Citation PHYSICAL REVIEW E (2005), 71(6) Issue Date 2005-06 URL http://hdl.handle.net/2433/50298 RightCopyright 2005 American Physical
Interactions and Dynamics within the Troponin Complex
 Interactions and Dynamics within the Troponin Complex Tharin Blumenschein Steve Matthews Lab - Imperial College London (formerly Brian Sykes Lab, Canada) Striated muscle Thin filament proteins - regulation
Interactions and Dynamics within the Troponin Complex Tharin Blumenschein Steve Matthews Lab - Imperial College London (formerly Brian Sykes Lab, Canada) Striated muscle Thin filament proteins - regulation
