Draft document version 0.6; ClustalX version 2.1(PC), (Mac); NJplot version 2.3; 3/26/2012
|
|
|
- Philomena Sanders
- 6 years ago
- Views:
Transcription
1 Comparing DNA Sequences to Determine Evolutionary Relationships of Molluscs This activity serves as a supplement to the online activity Biodiversity and Evolutionary Trees: An Activity on Biological Classification at Overview In the online activity Biodiversity and Evolutionary Trees: An Activity on Biological Classification, you generated a phylogenetic tree of molluscs using only shell morphology information. In this exercise, you will revisit that classification and reconstruct the phylogenetic tree using ClustalX, software that aligns and compares DNA sequences. You will use a simple viewer program called NJplot to view the tree. Software and Files Install ClustalX, which is available from (For Windows, download clustalx-2.1-win.msi; for Mac OS, clustalx-2.1-macosx.dmg). Next, install NJplot, which is available from Then download the zip file containing all the DNA sequence files used in this exercise, which is found under the Biodiversity and Evolutionary Trees section at Format of DNA Sequence Information There are different formats for representing DNA sequences. Shown below is a partial sequence from the dog s cytochrome oxidase subunit I (COI) gene in FASTA format. FASTA format starts with a >, followed by information about the file to the end of the first line, followed by the DNA sequence. >gi gb JN Canis lupus familiaris isolate dog_3 cytochrome oxidase subunit I (COI) gene, partial cds; mitochondrial TACTTTATACTTACTATTTGGAGCATGAGCCGGTATAGTAGGCACTGCCTTGAGCCTCCTCATCCGAGCC GAACTAGGTCAGCCCGGTACTTTACTAGGTGACGATCAAATTTATAATGTCATYGTAACCGCCCATGCTT A file containing FASTA format sequence information may contain multiple sequences one after another. For example: 2012 Howard Hughes Medical Institute 1
2 What Sequences Do We Choose to Compare? In modern taxonomic practice, scientists routinely analyze the DNA from specimens they collect to obtain a DNA barcode, a short DNA sequence unique to a particular organism, which is used to identify the species it belongs to. For animals and many other eukaryotes, the mitochondrial cytochrome c oxidase subunit I (COI) gene, that encodes an important enzyme for cellular respiration, has been used to generate such barcodes. As a result, COI sequences are available from a wide range of species, making it possible to use this gene sequence to explore phylogenetic relationships. COI is a good choice for DNA barcoding, because, in general, there is little variation in COI sequences of organisms within the same species, while there is significant variation in COI sequences of organisms from different species. Therefore, a COI sequence provides a unique sequence signature for a particular species. For the same reasons, COI gene is suitable for comparing phylogenetic relationships between species. Because the COI sequences are so similar within the same species, the COI gene is not a good choice for studying variations within the same species, or even among species that have recently speciated. COI sequences also have a low mutation rate among many species of plants, and cannot be used for DNA barcoding or phylogenetic comparisons of those species Howard Hughes Medical Institute 2
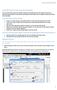
3 Exercise 1: An Overview of ClustalX Let s use ClustalX to compare DNA sequences. For this exercise, use the test sequence file test.txt, which contains the three short DNA sequences (test1, test2, and test3) shown below: >test1 AAGGAAGGAAGGAAGGAAGGAAGG >test2 AAGGAAGGAATGGAAGGAAGGAAGG >test3 AAGGAACGGAATGGTAGGAAGGAAGG Load these sequences into ClustalX by choosing from the menu, File -> Load Sequences, and then selecting test.txt. ClustalX shows these sequences as shown in the illustration on the right (PC version shown). Before you can compare sequences, you have to align them, which means lining up the sequences and sliding them past one another until the best matching pattern is found. Alignment allows you to examine differences between related sequences; such differences reflect evolutionary relationships. From the menu, choose Alignment -> Do Complete Alignment. When prompted for output file names, use the default names given and click OK. The screen changes to looks like the second illustration on the right. Notice that it s a lot easier to see differences among DNA sequences after alignment. You can figure out what kinds of mutations have occurred in each sequence by how it compares to the others (as shown in the third illustration). The number of differences among sequences determines how closely or distantly related the corresponding organisms are. Deletion Insertion Substitution Based on this information alone, which two sequences do you think are more closely related? 2012 Howard Hughes Medical Institute 3
4 To check whether your answer was accurate, we can use ClustalX to generate a phylogenetic tree. From the menu, choose Trees -> Draw Tree. This creates a phylogenetic tree file called test1.ph, which can be opened using NJplot.exe. Launch NJplot, then from its menu, choose File -> Open, and select test1.ph. The result shows that test1 and test2 are on the same branch of the tree, indicating they are more closely related to each other than to test Howard Hughes Medical Institute 4
5 Exercise 2: Phylogeny of Neogastropods, Tonnoids, and Cowries In the online seashell sorting activity, Biodiversity and Evolutionary Trees: An Activity on Biological Classification, one of the questions you had to answer was which two groups of gastropods among neogastropods, tonnoids, and cowries are more closely related. Based on the information in that activity, the correct answer was tonnoids and cowries. Let s confirm that conclusion using molecular data. Load the file molluscs1.txt into ClustalX. (For this exercise, the DNA sequences used were obtained from representative species from each group as follows: neogastropods = Conus magus; tonnoids = Bursa granularis; and cowries = Cypraea tigris) Align the sequences by choosing from the menu, Alignment -> Do Complete Alignment Then choose from the menu, Trees -> Draw Tree Howard Hughes Medical Institute 5
6 The result is shown below; it confirms that the tonnoids and the cowries are more closely related to each other than to the neogastropods Howard Hughes Medical Institute 6
7 Exercise 3: Phylogeny of Molluscs The final phylogeny obtained from the online seashell sorting activity is shown below Conus magus 2. Neritina communis 3. Bursa ignobilis 4. Conus capitaneus 5. Cypraea annulus 6. Conus marmoreus Distorsio anus 9. Conus omaria 10. Cypraea isabella 11. Pecten pallium 12. Cypraea tigris 13. Conus ebraeus Conus chaldeus Imbricaria conularis 18. Conus circumcisus 19. Cypraea moneta 20. Turris babylonia We will compare the phylogenetic tree above, obtained using morphological information, with a tree obtained using DNA sequence data. The file molluscs2.txt contains COI DNA sequences from all of the shells in this tree*. Load that file into ClustalX Howard Hughes Medical Institute 7
8 Align the sequences and make a phylogenetic tree. Examine the resulting tree in NJplot and compare it to the one above. Confirm that the patterns of the various groupings are the same. Even though the order of species in the tree on the left is not the same as the tree above, the two trees are the same topologically. That means, if you imagine the tree on the left is like a mobile hanging from a ceiling, you could rotate each branch point in the tree to make it the same as the tree above. However, in the ClustalX tree individual cone snails and cowries have their own branches. Cone Snails Tonnoids Cowries * Because COI gene sequences for five of the species used in the online seashell activity were not available, we used substitute sequences from closely related species. Substitutions were as follows: Neritina pulligera was substituted for Neritina communis; Distorsio reticularis for Distorsio anus; Bursa granularis for Bursa ignobilis; Mitra lens for Imbricaria conularis; Pecten jacobaeus for Pecten pallium Howard Hughes Medical Institute 8
9 Exercise 4: Inquiry-Based Activity Comparing Sequences of Your Choice In this exercise, you will find the DNA sequences you want to compare. To find DNA sequences, go to the NCBI s nucleotide resource page at NCBI website images are current as of March Search Box In the search box at the top of the page, enter the species you are interested in, and coi (for cytochrome oxidase I gene). Your search will be more effective if you use the scientific species name. For example, instead of dog, use canis lupus familiaris. FASTA link Look for something about 600 bases listed as cytochrome oxidase subunit I (COI) gene, and partial cds; mitochondrial. In the screenshot above, both search result 1 and 2 are DNA sequences from dog and would be good choices for this activity. But you have to be careful in selecting which file to use, as sequences from different species or sequences of different genes may be present. For example, the third result is for Dirofilaria, a common parasite of dogs Howard Hughes Medical Institute 9
10 Once you know which sequence you want to use, click on the FASTA file link, then select all the text, copy it, and paste it into a new document as a plain text file. Remember that in FASTA format, the first line contains the information about the sequence and starts with > and ends at the end of the line. >gi gb JN Canis lupus familiaris isolate dog_3 cytochrome oxidase subunit I (COI) gene, partial cds; mitochondrial ClustalX uses the letters after > until the first space as the label for the file. In this case, the label is gi gb JN The label will be used onscreen and in the tree diagram, so it would help if you edited it to something simpler for this exercise. For example, you could insert Dog after the > so that it reads: >Dog gi gb JN Canis lupus familiaris isolate dog_3 cytochrome oxidase subunit I (COI) gene, partial cds; mitochondrial Once you have the DNA sequence information from all the organisms you want to compare, assemble them all into a single text file. Be sure to save your file as a plain text (.txt) file. If you want to see what that looks like, you can look at the molluscs2.txt as a model. Once you have all the sequences loaded, the procedure for aligning them and drawing a tree and looking at it is the same as before. Be creative, and explore obscure species that you only read about in books or websites! You may also want to repeat this exercise using different genes. For phylogenetic comparisons to work, the gene should be conserved among different species you are interested in, similar enough to be compared, yet different enough to give enough variability. You could try genes like actin, myosin, or ubiquitin. If you restrict the range of species, even genes like hemoglobin may work Howard Hughes Medical Institute 10
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST
 Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES
 INTRODUCTION CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES This worksheet complements the Click and Learn developed in conjunction with the 2011 Holiday Lectures on Science, Bones, Stones, and Genes:
INTRODUCTION CREATING PHYLOGENETIC TREES FROM DNA SEQUENCES This worksheet complements the Click and Learn developed in conjunction with the 2011 Holiday Lectures on Science, Bones, Stones, and Genes:
BIOINFORMATICS LAB AP BIOLOGY
 BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
Using Bioinformatics to Study Evolutionary Relationships Instructions
 3 Using Bioinformatics to Study Evolutionary Relationships Instructions Student Researcher Background: Making and Using Multiple Sequence Alignments One of the primary tasks of genetic researchers is comparing
3 Using Bioinformatics to Study Evolutionary Relationships Instructions Student Researcher Background: Making and Using Multiple Sequence Alignments One of the primary tasks of genetic researchers is comparing
Open a Word document to record answers to any italicized questions. You will the final document to me at
 Molecular Evidence for Evolution Open a Word document to record answers to any italicized questions. You will email the final document to me at tchnsci@yahoo.com Pre Lab Activity: Genes code for amino
Molecular Evidence for Evolution Open a Word document to record answers to any italicized questions. You will email the final document to me at tchnsci@yahoo.com Pre Lab Activity: Genes code for amino
Phylogenetic analyses. Kirsi Kostamo
 Phylogenetic analyses Kirsi Kostamo The aim: To construct a visual representation (a tree) to describe the assumed evolution occurring between and among different groups (individuals, populations, species,
Phylogenetic analyses Kirsi Kostamo The aim: To construct a visual representation (a tree) to describe the assumed evolution occurring between and among different groups (individuals, populations, species,
BIOINFORMATICS: An Introduction
 BIOINFORMATICS: An Introduction What is Bioinformatics? The term was first coined in 1988 by Dr. Hwa Lim The original definition was : a collective term for data compilation, organisation, analysis and
BIOINFORMATICS: An Introduction What is Bioinformatics? The term was first coined in 1988 by Dr. Hwa Lim The original definition was : a collective term for data compilation, organisation, analysis and
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
08/21/2017 BLAST. Multiple Sequence Alignments: Clustal Omega
 BLAST Multiple Sequence Alignments: Clustal Omega What does basic BLAST do (e.g. what is input sequence and how does BLAST look for matches?) Susan Parrish McDaniel College Multiple Sequence Alignments
BLAST Multiple Sequence Alignments: Clustal Omega What does basic BLAST do (e.g. what is input sequence and how does BLAST look for matches?) Susan Parrish McDaniel College Multiple Sequence Alignments
Bioinformatics Exercises
 Bioinformatics Exercises AP Biology Teachers Workshop Susan Cates, Ph.D. Evolution of Species Phylogenetic Trees show the relatedness of organisms Common Ancestor (Root of the tree) 1 Rooted vs. Unrooted
Bioinformatics Exercises AP Biology Teachers Workshop Susan Cates, Ph.D. Evolution of Species Phylogenetic Trees show the relatedness of organisms Common Ancestor (Root of the tree) 1 Rooted vs. Unrooted
Chapter 19: Taxonomy, Systematics, and Phylogeny
 Chapter 19: Taxonomy, Systematics, and Phylogeny AP Curriculum Alignment Chapter 19 expands on the topics of phylogenies and cladograms, which are important to Big Idea 1. In order for students to understand
Chapter 19: Taxonomy, Systematics, and Phylogeny AP Curriculum Alignment Chapter 19 expands on the topics of phylogenies and cladograms, which are important to Big Idea 1. In order for students to understand
Emily Blanton Phylogeny Lab Report May 2009
 Introduction It is suggested through scientific research that all living organisms are connected- that we all share a common ancestor and that, through time, we have all evolved from the same starting
Introduction It is suggested through scientific research that all living organisms are connected- that we all share a common ancestor and that, through time, we have all evolved from the same starting
USING BLAST TO IDENTIFY PROTEINS THAT ARE EVOLUTIONARILY RELATED ACROSS SPECIES
 USING BLAST TO IDENTIFY PROTEINS THAT ARE EVOLUTIONARILY RELATED ACROSS SPECIES HOW CAN BIOINFORMATICS BE USED AS A TOOL TO DETERMINE EVOLUTIONARY RELATIONSHPS AND TO BETTER UNDERSTAND PROTEIN HERITAGE?
USING BLAST TO IDENTIFY PROTEINS THAT ARE EVOLUTIONARILY RELATED ACROSS SPECIES HOW CAN BIOINFORMATICS BE USED AS A TOOL TO DETERMINE EVOLUTIONARY RELATIONSHPS AND TO BETTER UNDERSTAND PROTEIN HERITAGE?
Gridded Ambient Air Pollutant Concentrations for Southern California, User Notes authored by Beau MacDonald, 11/28/2017
 Gridded Ambient Air Pollutant Concentrations for Southern California, 1995-2014 User Notes authored by Beau, 11/28/2017 METADATA: Each raster file contains data for one pollutant (NO2, O3, PM2.5, and PM10)
Gridded Ambient Air Pollutant Concentrations for Southern California, 1995-2014 User Notes authored by Beau, 11/28/2017 METADATA: Each raster file contains data for one pollutant (NO2, O3, PM2.5, and PM10)
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution
 Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction. Lesser Tenrec (Echinops telfairi)
 Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction Lesser Tenrec (Echinops telfairi) Goals: 1. Use phylogenetic experimental design theory to select optimal taxa to
Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction Lesser Tenrec (Echinops telfairi) Goals: 1. Use phylogenetic experimental design theory to select optimal taxa to
Skulls & Evolution. Procedure In this lab, groups at the same table will work together.
 Skulls & Evolution Objectives To illustrate trends in the evolution of humans. To demonstrate what you can learn from bones & fossils. To show the adaptations of various mammals to different habitats and
Skulls & Evolution Objectives To illustrate trends in the evolution of humans. To demonstrate what you can learn from bones & fossils. To show the adaptations of various mammals to different habitats and
Phylogenetic Trees. How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species?
 Why? Phylogenetic Trees How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species? The saying Don t judge a book by its cover. could be applied
Why? Phylogenetic Trees How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species? The saying Don t judge a book by its cover. could be applied
Tree Building Activity
 Tree Building Activity Introduction In this activity, you will construct phylogenetic trees using a phenotypic similarity (cartoon microbe pictures) and genotypic similarity (real microbe sequences). For
Tree Building Activity Introduction In this activity, you will construct phylogenetic trees using a phenotypic similarity (cartoon microbe pictures) and genotypic similarity (real microbe sequences). For
LAB 4: PHYLOGENIES & MAPPING TRAITS
 LAB 4: PHYLOGENIES & MAPPING TRAITS *This is a good day to check your Physcomitrella (protonema, buds, gametophores?) and Ceratopteris cultures (embryos, young sporophytes?)* Phylogeny Introduction The
LAB 4: PHYLOGENIES & MAPPING TRAITS *This is a good day to check your Physcomitrella (protonema, buds, gametophores?) and Ceratopteris cultures (embryos, young sporophytes?)* Phylogeny Introduction The
CHAPTERS 24-25: Evidence for Evolution and Phylogeny
 CHAPTERS 24-25: Evidence for Evolution and Phylogeny 1. For each of the following, indicate how it is used as evidence of evolution by natural selection or shown as an evolutionary trend: a. Paleontology
CHAPTERS 24-25: Evidence for Evolution and Phylogeny 1. For each of the following, indicate how it is used as evidence of evolution by natural selection or shown as an evolutionary trend: a. Paleontology
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week:
 Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week: Course general information About the course Course objectives Comparative methods: An overview R as language: uses and
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week: Course general information About the course Course objectives Comparative methods: An overview R as language: uses and
PHYLOGENY AND SYSTEMATICS
 AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 11 Chapter 26 Activity #15 NAME DATE PERIOD PHYLOGENY AND SYSTEMATICS PHYLOGENY Evolutionary history of species or group of related species SYSTEMATICS Study
AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 11 Chapter 26 Activity #15 NAME DATE PERIOD PHYLOGENY AND SYSTEMATICS PHYLOGENY Evolutionary history of species or group of related species SYSTEMATICS Study
let s examine pupation rates. With the conclusion of that data collection, we will go on to explore the rate at which new adults appear, a process
 Population Dynamics and Initial Population Size (Module website: http://web.as.uky.edu/biology/faculty/cooper/population%20dynamics%20examples%20 with%20fruit%20flies/theamericanbiologyteacher-populationdynamicswebpage.html
Population Dynamics and Initial Population Size (Module website: http://web.as.uky.edu/biology/faculty/cooper/population%20dynamics%20examples%20 with%20fruit%20flies/theamericanbiologyteacher-populationdynamicswebpage.html
Characteristics of Life
 UNIT 2 BIODIVERSITY Chapter 4- Patterns of Life Biology 2201 Characteristics of Life All living things share some basic characteristics: 1) living things are organized systems made up of one or more cells
UNIT 2 BIODIVERSITY Chapter 4- Patterns of Life Biology 2201 Characteristics of Life All living things share some basic characteristics: 1) living things are organized systems made up of one or more cells
module, with the exception that the vials are larger and you only use one initial population size.
 Population Dynamics and Space Availability (http://web.as.uky.edu/biology/faculty/cooper/population%20dynamics%20examples%2 0with%20fruit%20flies/TheAmericanBiologyTeacher- PopulationDynamicsWebpage.html
Population Dynamics and Space Availability (http://web.as.uky.edu/biology/faculty/cooper/population%20dynamics%20examples%2 0with%20fruit%20flies/TheAmericanBiologyTeacher- PopulationDynamicsWebpage.html
Date: Summer Stem Section:
 Page 1 of 7 Name: Date: Summer Stem Section: Summer assignment: Build a Molecule Computer Simulation Learning Goals: 1. Students can describe the difference between a molecule name and chemical formula.
Page 1 of 7 Name: Date: Summer Stem Section: Summer assignment: Build a Molecule Computer Simulation Learning Goals: 1. Students can describe the difference between a molecule name and chemical formula.
Plant Names and Classification
 Plant Names and Classification Science of Taxonomy Identification (necessary!!) Classification (order out of chaos!) Nomenclature (why not use common names?) Reasons NOT to use common names Theophrastus
Plant Names and Classification Science of Taxonomy Identification (necessary!!) Classification (order out of chaos!) Nomenclature (why not use common names?) Reasons NOT to use common names Theophrastus
RELATIONSHIPS BETWEEN GENES/PROTEINS HOMOLOGUES
 Molecular Biology-2018 1 Definitions: RELATIONSHIPS BETWEEN GENES/PROTEINS HOMOLOGUES Heterologues: Genes or proteins that possess different sequences and activities. Homologues: Genes or proteins that
Molecular Biology-2018 1 Definitions: RELATIONSHIPS BETWEEN GENES/PROTEINS HOMOLOGUES Heterologues: Genes or proteins that possess different sequences and activities. Homologues: Genes or proteins that
Foundations of Computation
 The Australian National University Semester 2, 2018 Research School of Computer Science Tutorial 1 Dirk Pattinson Foundations of Computation The tutorial contains a number of exercises designed for the
The Australian National University Semester 2, 2018 Research School of Computer Science Tutorial 1 Dirk Pattinson Foundations of Computation The tutorial contains a number of exercises designed for the
Overview Objectives Materials Background Activity Procedure
 Overview In this activity, students will explore how to determine how to assemble information about plant distributions, and relate those distributional to the environment by assembling information on
Overview In this activity, students will explore how to determine how to assemble information about plant distributions, and relate those distributional to the environment by assembling information on
Bio 1B Lecture Outline (please print and bring along) Fall, 2007
 Bio 1B Lecture Outline (please print and bring along) Fall, 2007 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #5 -- Molecular genetics and molecular evolution
Bio 1B Lecture Outline (please print and bring along) Fall, 2007 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #5 -- Molecular genetics and molecular evolution
Presenting Tree Inventory. Tomislav Sapic GIS Technologist Faculty of Natural Resources Management Lakehead University
 Presenting Tree Inventory Tomislav Sapic GIS Technologist Faculty of Natural Resources Management Lakehead University Suggested Options 1. Print out a Google Maps satellite image of the inventoried block
Presenting Tree Inventory Tomislav Sapic GIS Technologist Faculty of Natural Resources Management Lakehead University Suggested Options 1. Print out a Google Maps satellite image of the inventoried block
Biology 160 Cell Lab. Name Lab Section: 1:00pm 3:00 pm. Student Learning Outcomes:
 Biology 160 Cell Lab Name Lab Section: 1:00pm 3:00 pm Student Learning Outcomes: Upon completion of today s lab you will be able to do the following: Properly use a compound light microscope Discuss the
Biology 160 Cell Lab Name Lab Section: 1:00pm 3:00 pm Student Learning Outcomes: Upon completion of today s lab you will be able to do the following: Properly use a compound light microscope Discuss the
Evolutionary Tree Analysis. Overview
 CSI/BINF 5330 Evolutionary Tree Analysis Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Distance-Based Evolutionary Tree Reconstruction Character-Based
CSI/BINF 5330 Evolutionary Tree Analysis Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Distance-Based Evolutionary Tree Reconstruction Character-Based
Bioinformatics tools for phylogeny and visualization. Yanbin Yin
 Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
The Tree of Life. Chapter 17
 The Tree of Life Chapter 17 1 17.1 Taxonomy The science of naming and classifying organisms 2000 years ago Aristotle Grouped plants and animals Based on structural similarities Greeks and Romans included
The Tree of Life Chapter 17 1 17.1 Taxonomy The science of naming and classifying organisms 2000 years ago Aristotle Grouped plants and animals Based on structural similarities Greeks and Romans included
GENERAL BIOLOGY LABORATORY EXERCISE Amino Acid Sequence Analysis of Cytochrome C in Bacteria and Eukarya Using Bioinformatics
 GENERAL BIOLOGY LABORATORY EXERCISE Amino Acid Sequence Analysis of Cytochrome C in Bacteria and Eukarya Using Bioinformatics INTRODUCTION: All life forms undergo metabolic processes to obtain energy.
GENERAL BIOLOGY LABORATORY EXERCISE Amino Acid Sequence Analysis of Cytochrome C in Bacteria and Eukarya Using Bioinformatics INTRODUCTION: All life forms undergo metabolic processes to obtain energy.
Welcome to Evolution 101 Reading Guide
 Name: Welcome to Evolution 101 Reading Guide http://evolution.berkeley.edu/evolibrary/article/evo_01 Read the information on the website. Click on the next arrow at the bottom of each page to move to the
Name: Welcome to Evolution 101 Reading Guide http://evolution.berkeley.edu/evolibrary/article/evo_01 Read the information on the website. Click on the next arrow at the bottom of each page to move to the
SPECIATION. REPRODUCTIVE BARRIERS PREZYGOTIC: Barriers that prevent fertilization. Habitat isolation Populations can t get together
 SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
BASIC TECHNOLOGY Pre K starts and shuts down computer, monitor, and printer E E D D P P P P P P P P P P
 BASIC TECHNOLOGY Pre K 1 2 3 4 5 6 7 8 9 10 11 12 starts and shuts down computer, monitor, and printer P P P P P P practices responsible use and care of technology devices P P P P P P opens and quits an
BASIC TECHNOLOGY Pre K 1 2 3 4 5 6 7 8 9 10 11 12 starts and shuts down computer, monitor, and printer P P P P P P practices responsible use and care of technology devices P P P P P P opens and quits an
Molecular Evolution and DNA systematics
 Biology 4505 - Biogeography & Systematics Dr. Carr Molecular Evolution and DNA systematics Ultimately, the source of all organismal variation that we have examined in this course is the genome, written
Biology 4505 - Biogeography & Systematics Dr. Carr Molecular Evolution and DNA systematics Ultimately, the source of all organismal variation that we have examined in this course is the genome, written
Investigating Evolutionary Questions Using Online Molecular Databases *
 Investigating Evolutionary Questions Using Online Molecular Databases * Adapted from Puterbaugh and Burleigh, and The American Biology Teacher Lesson Background and Overview [Student Information Handout]
Investigating Evolutionary Questions Using Online Molecular Databases * Adapted from Puterbaugh and Burleigh, and The American Biology Teacher Lesson Background and Overview [Student Information Handout]
v Prerequisite Tutorials GSSHA WMS Basics Watershed Delineation using DEMs and 2D Grid Generation Time minutes
 v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
Companion to Cells, Heredity & Classification Student Resources
 Companion to Cells, Heredity & Classification Student Resources The 21st Century Center for Research and Development in Cognition and Science Instruction The CaSEbook Companion: Student Resource Book
Companion to Cells, Heredity & Classification Student Resources The 21st Century Center for Research and Development in Cognition and Science Instruction The CaSEbook Companion: Student Resource Book
Organizing Life s Diversity
 17 Organizing Life s Diversity section 2 Modern Classification Classification systems have changed over time as information has increased. What You ll Learn species concepts methods to reveal phylogeny
17 Organizing Life s Diversity section 2 Modern Classification Classification systems have changed over time as information has increased. What You ll Learn species concepts methods to reveal phylogeny
Assignment #0 Using Stellarium
 Name: Class: Date: Assignment #0 Using Stellarium The purpose of this exercise is to familiarize yourself with the Stellarium program and its many capabilities and features. Stellarium is a visually beautiful
Name: Class: Date: Assignment #0 Using Stellarium The purpose of this exercise is to familiarize yourself with the Stellarium program and its many capabilities and features. Stellarium is a visually beautiful
Organizing Life on Earth
 Organizing Life on Earth Inquire: Organizing Life on Earth Overview Scientists continually obtain new information that helps to understand the evolutionary history of life on Earth. Each group of organisms
Organizing Life on Earth Inquire: Organizing Life on Earth Overview Scientists continually obtain new information that helps to understand the evolutionary history of life on Earth. Each group of organisms
Using SkyTools to log Texas 45 list objects
 Houston Astronomical Society Using SkyTools to log Texas 45 list objects You can use SkyTools to keep track of objects observed in Columbus and copy the output into the Texas 45 observation log. Preliminary
Houston Astronomical Society Using SkyTools to log Texas 45 list objects You can use SkyTools to keep track of objects observed in Columbus and copy the output into the Texas 45 observation log. Preliminary
(THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE)
 PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
Name: Class: Date: ID: A
 Class: _ Date: _ Ch 17 Practice test 1. A segment of DNA that stores genetic information is called a(n) a. amino acid. b. gene. c. protein. d. intron. 2. In which of the following processes does change
Class: _ Date: _ Ch 17 Practice test 1. A segment of DNA that stores genetic information is called a(n) a. amino acid. b. gene. c. protein. d. intron. 2. In which of the following processes does change
Protein Bioinformatics Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski
 Protein Bioinformatics 260.655 Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski Goals: Approx. Time [1] Use the Protein Data Bank PDB website. 10 minutes [2] Use the WebMol Viewer.
Protein Bioinformatics 260.655 Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski Goals: Approx. Time [1] Use the Protein Data Bank PDB website. 10 minutes [2] Use the WebMol Viewer.
Lab 9: Maximum Likelihood and Modeltest
 Integrative Biology 200A University of California, Berkeley "PRINCIPLES OF PHYLOGENETICS" Spring 2010 Updated by Nick Matzke Lab 9: Maximum Likelihood and Modeltest In this lab we re going to use PAUP*
Integrative Biology 200A University of California, Berkeley "PRINCIPLES OF PHYLOGENETICS" Spring 2010 Updated by Nick Matzke Lab 9: Maximum Likelihood and Modeltest In this lab we re going to use PAUP*
Interacting Galaxies
 Interacting Galaxies Contents Introduction... 1 Downloads... 1 Selecting Interacting Galaxies to Observe... 2 Measuring the sizes of the Galaxies... 5 Making a Colour Image in IRIS... 8 External Resources...
Interacting Galaxies Contents Introduction... 1 Downloads... 1 Selecting Interacting Galaxies to Observe... 2 Measuring the sizes of the Galaxies... 5 Making a Colour Image in IRIS... 8 External Resources...
THEORY. Based on sequence Length According to the length of sequence being compared it is of following two types
 Exp 11- THEORY Sequence Alignment is a process of aligning two sequences to achieve maximum levels of identity between them. This help to derive functional, structural and evolutionary relationships between
Exp 11- THEORY Sequence Alignment is a process of aligning two sequences to achieve maximum levels of identity between them. This help to derive functional, structural and evolutionary relationships between
Richter Scale and Logarithms
 activity 7.1 Richter Scale and Logarithms In this activity, you will investigate earthquake data and explore the Richter scale as a measure of the intensity of an earthquake. You will consider how numbers
activity 7.1 Richter Scale and Logarithms In this activity, you will investigate earthquake data and explore the Richter scale as a measure of the intensity of an earthquake. You will consider how numbers
PROJECT LEADER TIP #6 HOW TO ADD SAMPLING UNITS
 PROJECT LEADER TIP #6 HOW TO ADD SAMPLING UNITS Before beginning, it is important to understand that sampling locations are set-up hierarchically. For example, Pacific Flyway Shorebird Survey the hierarchy
PROJECT LEADER TIP #6 HOW TO ADD SAMPLING UNITS Before beginning, it is important to understand that sampling locations are set-up hierarchically. For example, Pacific Flyway Shorebird Survey the hierarchy
Figure 2.1 The Inclined Plane
 PHYS-101 LAB-02 One and Two Dimensional Motion 1. Objectives The objectives of this experiment are: to measure the acceleration due to gravity using one-dimensional motion, i.e. the motion of an object
PHYS-101 LAB-02 One and Two Dimensional Motion 1. Objectives The objectives of this experiment are: to measure the acceleration due to gravity using one-dimensional motion, i.e. the motion of an object
Geomap Downloading and changes in New Version
 1 Geomap Downloading and changes in New Version 1.9.10.0 14 June 2014 William O'Connor and Udo Kaschner about-us.html 2 Published by: William O'Connor PO Box 2207 Clarkson Western Australia http://www.gold-prospecting-wa.com
1 Geomap Downloading and changes in New Version 1.9.10.0 14 June 2014 William O'Connor and Udo Kaschner about-us.html 2 Published by: William O'Connor PO Box 2207 Clarkson Western Australia http://www.gold-prospecting-wa.com
Chapter 26. Phylogeny and the Tree of Life. Lecture Presentations by Nicole Tunbridge and Kathleen Fitzpatrick Pearson Education, Inc.
 Chapter 26 Phylogeny and the Tree of Life Lecture Presentations by Nicole Tunbridge and Kathleen Fitzpatrick Investigating the Tree of Life Phylogeny is the evolutionary history of a species or group of
Chapter 26 Phylogeny and the Tree of Life Lecture Presentations by Nicole Tunbridge and Kathleen Fitzpatrick Investigating the Tree of Life Phylogeny is the evolutionary history of a species or group of
HR Diagram of Globular Cluster Messier 80 Using Hubble Space Telescope Data
 Jason Kendall, William Paterson University, Department of Physics HR Diagram of Globular Cluster Messier 80 Using Hubble Space Telescope Data Background Purpose: HR Diagrams are central to understanding
Jason Kendall, William Paterson University, Department of Physics HR Diagram of Globular Cluster Messier 80 Using Hubble Space Telescope Data Background Purpose: HR Diagrams are central to understanding
Pogil Phylogenetic Trees Answer Key Ap Biology
 POGIL PHYLOGENETIC TREES ANSWER KEY AP BIOLOGY PDF - Are you looking for pogil phylogenetic trees answer key ap biology Books? Now, you will be happy that at this time pogil phylogenetic trees answer key
POGIL PHYLOGENETIC TREES ANSWER KEY AP BIOLOGY PDF - Are you looking for pogil phylogenetic trees answer key ap biology Books? Now, you will be happy that at this time pogil phylogenetic trees answer key
Prokaryotes Bacteria And Archaea Study Guide Answers READ ONLINE
 Prokaryotes Bacteria And Archaea Study Guide Answers READ ONLINE If you are searched for a ebook Prokaryotes bacteria and archaea study guide answers in pdf format, then you have come on to the correct
Prokaryotes Bacteria And Archaea Study Guide Answers READ ONLINE If you are searched for a ebook Prokaryotes bacteria and archaea study guide answers in pdf format, then you have come on to the correct
Biology Classification Unit 11. CLASSIFICATION: process of dividing organisms into groups with similar characteristics
 Biology Classification Unit 11 11:1 Classification and Taxonomy CLASSIFICATION: process of dividing organisms into groups with similar characteristics TAXONOMY: the science of classifying living things
Biology Classification Unit 11 11:1 Classification and Taxonomy CLASSIFICATION: process of dividing organisms into groups with similar characteristics TAXONOMY: the science of classifying living things
Homework Assignment, Evolutionary Systems Biology, Spring Homework Part I: Phylogenetics:
 Homework Assignment, Evolutionary Systems Biology, Spring 2009. Homework Part I: Phylogenetics: Introduction. The objective of this assignment is to understand the basics of phylogenetic relationships
Homework Assignment, Evolutionary Systems Biology, Spring 2009. Homework Part I: Phylogenetics: Introduction. The objective of this assignment is to understand the basics of phylogenetic relationships
1 Abstract. 2 Introduction. 3 Requirements. 4 Procedure
 1 Abstract None 2 Introduction The archaeal core set is used in testing the completeness of the archaeal draft genomes. The core set comprises of conserved single copy genes from 25 genomes. Coverage statistic
1 Abstract None 2 Introduction The archaeal core set is used in testing the completeness of the archaeal draft genomes. The core set comprises of conserved single copy genes from 25 genomes. Coverage statistic
CLASSIFICATION OF LIVING THINGS
 CLASSIFICATION OF LIVING THINGS 1. Taxonomy The branch of biology that deals with the classification of living organisms About 1.8 million species of plants and animals have been identified. Some scientists
CLASSIFICATION OF LIVING THINGS 1. Taxonomy The branch of biology that deals with the classification of living organisms About 1.8 million species of plants and animals have been identified. Some scientists
OECD QSAR Toolbox v.3.4. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
UoN, CAS, DBSC BIOL102 lecture notes by: Dr. Mustafa A. Mansi. The Phylogenetic Systematics (Phylogeny and Systematics)
 - Phylogeny? - Systematics? The Phylogenetic Systematics (Phylogeny and Systematics) - Phylogenetic systematics? Connection between phylogeny and classification. - Phylogenetic systematics informs the
- Phylogeny? - Systematics? The Phylogenetic Systematics (Phylogeny and Systematics) - Phylogenetic systematics? Connection between phylogeny and classification. - Phylogenetic systematics informs the
CLASSIFICATION UNIT GUIDE DUE WEDNESDAY 3/1
 CLASSIFICATION UNIT GUIDE DUE WEDNESDAY 3/1 MONDAY TUESDAY WEDNESDAY THURSDAY FRIDAY 2/13 2/14 - B 2/15 2/16 - B 2/17 2/20 Intro to Viruses Viruses VS Cells 2/21 - B Virus Reproduction Q 1-2 2/22 2/23
CLASSIFICATION UNIT GUIDE DUE WEDNESDAY 3/1 MONDAY TUESDAY WEDNESDAY THURSDAY FRIDAY 2/13 2/14 - B 2/15 2/16 - B 2/17 2/20 Intro to Viruses Viruses VS Cells 2/21 - B Virus Reproduction Q 1-2 2/22 2/23
Synteny Portal Documentation
 Synteny Portal Documentation Synteny Portal is a web application portal for visualizing, browsing, searching and building synteny blocks. Synteny Portal provides four main web applications: SynCircos,
Synteny Portal Documentation Synteny Portal is a web application portal for visualizing, browsing, searching and building synteny blocks. Synteny Portal provides four main web applications: SynCircos,
Session 5: Phylogenomics
 Session 5: Phylogenomics B.- Phylogeny based orthology assignment REMINDER: Gene tree reconstruction is divided in three steps: homology search, multiple sequence alignment and model selection plus tree
Session 5: Phylogenomics B.- Phylogeny based orthology assignment REMINDER: Gene tree reconstruction is divided in three steps: homology search, multiple sequence alignment and model selection plus tree
Introduction to Bioinformatics Online Course: IBT
 Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec1 Building a Multiple Sequence Alignment Learning Outcomes 1- Understanding Why multiple
Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec1 Building a Multiple Sequence Alignment Learning Outcomes 1- Understanding Why multiple
Moving into the information age: From records to Google Earth
 Moving into the information age: From records to Google Earth David R. R. Smith Psychology, School of Life Sciences, University of Hull e-mail: davidsmith.butterflies@gmail.com Introduction Many of us
Moving into the information age: From records to Google Earth David R. R. Smith Psychology, School of Life Sciences, University of Hull e-mail: davidsmith.butterflies@gmail.com Introduction Many of us
Homology and Information Gathering and Domain Annotation for Proteins
 Homology and Information Gathering and Domain Annotation for Proteins Outline Homology Information Gathering for Proteins Domain Annotation for Proteins Examples and exercises The concept of homology The
Homology and Information Gathering and Domain Annotation for Proteins Outline Homology Information Gathering for Proteins Domain Annotation for Proteins Examples and exercises The concept of homology The
Distance From the Sun
 Distance From the Sun Computer 32 Have you ever thought about what it would be like if you were on another planet looking back at the sun? In this activity, you will use the Light Probe to get an idea
Distance From the Sun Computer 32 Have you ever thought about what it would be like if you were on another planet looking back at the sun? In this activity, you will use the Light Probe to get an idea
Hands-On Nine The PAX6 Gene and Protein
 Hands-On Nine The PAX6 Gene and Protein Main Purpose of Hands-On Activity: Using bioinformatics tools to examine the sequences, homology, and disease relevance of the Pax6: a master gene of eye formation.
Hands-On Nine The PAX6 Gene and Protein Main Purpose of Hands-On Activity: Using bioinformatics tools to examine the sequences, homology, and disease relevance of the Pax6: a master gene of eye formation.
BCMB/CHEM 8190 Lab Exercise Using Maple for NMR Data Processing and Pulse Sequence Design March 2012
 BCMB/CHEM 8190 Lab Exercise Using Maple for NMR Data Processing and Pulse Sequence Design March 2012 Introduction Maple is a powerful collection of routines to aid in the solution of mathematical problems
BCMB/CHEM 8190 Lab Exercise Using Maple for NMR Data Processing and Pulse Sequence Design March 2012 Introduction Maple is a powerful collection of routines to aid in the solution of mathematical problems
Project 3: Molecular Orbital Calculations of Diatomic Molecules. This project is worth 30 points and is due on Wednesday, May 2, 2018.
 Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
TRANSCRIPTION VS TRANSLATION FILE
 23 April, 2018 TRANSCRIPTION VS TRANSLATION FILE Document Filetype: PDF 352.85 KB 0 TRANSCRIPTION VS TRANSLATION FILE Get an answer for 'Compare and contrast transcription and translation in Prokaryotes
23 April, 2018 TRANSCRIPTION VS TRANSLATION FILE Document Filetype: PDF 352.85 KB 0 TRANSCRIPTION VS TRANSLATION FILE Get an answer for 'Compare and contrast transcription and translation in Prokaryotes
Simulating Future Climate Change Using A Global Climate Model
 Simulating Future Climate Change Using A Global Climate Model Introduction: (EzGCM: Web-based Version) The objective of this abridged EzGCM exercise is for you to become familiar with the steps involved
Simulating Future Climate Change Using A Global Climate Model Introduction: (EzGCM: Web-based Version) The objective of this abridged EzGCM exercise is for you to become familiar with the steps involved
Evolution: An Introduction By Rolf F. Hoekstra, Stephen C. Stearns
 Evolution: An Introduction By Rolf F. Hoekstra, Stephen C. Stearns Evolution Webquest - Evolution Webquest In this webquest you will be exploring evolution and the mechanisms that A. Click on Evolution
Evolution: An Introduction By Rolf F. Hoekstra, Stephen C. Stearns Evolution Webquest - Evolution Webquest In this webquest you will be exploring evolution and the mechanisms that A. Click on Evolution
Creating Empirical Calibrations
 030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
TIphysics.com. Physics. Bell Ringer: Determining the Relationship Between Displacement, Velocity, and Acceleration ID: 13308
 Bell Ringer: Determining the Relationship Between Displacement, Velocity, and Acceleration ID: 13308 Time required 15 minutes Topic: Kinematics Construct and interpret graphs of displacement, velocity,
Bell Ringer: Determining the Relationship Between Displacement, Velocity, and Acceleration ID: 13308 Time required 15 minutes Topic: Kinematics Construct and interpret graphs of displacement, velocity,
Phylogenetic Collection Lab
 Phylogenetic Collection Lab Objectives To connect the diversity of organisms described in class with the real world. To connect particular phyla of organisms with their characteristic habitats. To compare
Phylogenetic Collection Lab Objectives To connect the diversity of organisms described in class with the real world. To connect particular phyla of organisms with their characteristic habitats. To compare
A Brief Introduction To. GRTensor. On MAPLE Platform. A write-up for the presentation delivered on the same topic as a part of the course PHYS 601
 A Brief Introduction To GRTensor On MAPLE Platform A write-up for the presentation delivered on the same topic as a part of the course PHYS 601 March 2012 BY: ARSHDEEP SINGH BHATIA arshdeepsb@gmail.com
A Brief Introduction To GRTensor On MAPLE Platform A write-up for the presentation delivered on the same topic as a part of the course PHYS 601 March 2012 BY: ARSHDEEP SINGH BHATIA arshdeepsb@gmail.com
Avida-ED Lab Book Summer 2018 Workshop Version Avidians Competing
 Avida-ED Lab Book Summer 2018 Workshop Version Avidians Competing Avida-ED Project Curriculum Development Team Michigan State University NSF BEACON Center for the Study of Evolution in Action http://avida-ed.msu.edu
Avida-ED Lab Book Summer 2018 Workshop Version Avidians Competing Avida-ED Project Curriculum Development Team Michigan State University NSF BEACON Center for the Study of Evolution in Action http://avida-ed.msu.edu
Taxonomy and Biodiversity
 Chapter 25/26 Taxonomy and Biodiversity Evolutionary biology The major goal of evolutionary biology is to reconstruct the history of life on earth Process: a- natural selection b- mechanisms that change
Chapter 25/26 Taxonomy and Biodiversity Evolutionary biology The major goal of evolutionary biology is to reconstruct the history of life on earth Process: a- natural selection b- mechanisms that change
CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis
 CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis Purpose: The purpose of this laboratory is to introduce some of the basic visualization and modeling tools for viewing
CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis Purpose: The purpose of this laboratory is to introduce some of the basic visualization and modeling tools for viewing
Senior astrophysics Lab 2: Evolution of a 1 M star
 Senior astrophysics Lab 2: Evolution of a 1 M star Name: Checkpoints due: Friday 13 April 2018 1 Introduction This is the rst of two computer labs using existing software to investigate the internal structure
Senior astrophysics Lab 2: Evolution of a 1 M star Name: Checkpoints due: Friday 13 April 2018 1 Introduction This is the rst of two computer labs using existing software to investigate the internal structure
The practice of naming and classifying organisms is called taxonomy.
 Chapter 18 Key Idea: Biologists use taxonomic systems to organize their knowledge of organisms. These systems attempt to provide consistent ways to name and categorize organisms. The practice of naming
Chapter 18 Key Idea: Biologists use taxonomic systems to organize their knowledge of organisms. These systems attempt to provide consistent ways to name and categorize organisms. The practice of naming
Using Microsoft Excel
 Using Microsoft Excel Objective: Students will gain familiarity with using Excel to record data, display data properly, use built-in formulae to do calculations, and plot and fit data with linear functions.
Using Microsoft Excel Objective: Students will gain familiarity with using Excel to record data, display data properly, use built-in formulae to do calculations, and plot and fit data with linear functions.
Introduction. Pre-Lab Questions: Physics 1CL PERIODIC MOTION - PART II Fall 2009
 Introduction This is the second of two labs on simple harmonic motion (SHM). In the first lab you studied elastic forces and elastic energy, and you measured the net force on a pendulum bob held at an
Introduction This is the second of two labs on simple harmonic motion (SHM). In the first lab you studied elastic forces and elastic energy, and you measured the net force on a pendulum bob held at an
Lab #10 Atomic Radius Rubric o Missing 1 out of 4 o Missing 2 out of 4 o Missing 3 out of 4
 Name: Date: Chemistry ~ Ms. Hart Class: Anions or Cations 4.7 Relationships Among Elements Lab #10 Background Information The periodic table is a wonderful source of information about all of the elements
Name: Date: Chemistry ~ Ms. Hart Class: Anions or Cations 4.7 Relationships Among Elements Lab #10 Background Information The periodic table is a wonderful source of information about all of the elements
Kingdom: What s missing? List the organisms now missing from the above list..
 Life Science 7 Chapter 9-1 p 222-227 Classification: Sorting It All Out Objectives Explain why and how organisms are classified. List the eight levels of classification. Explain scientific names. Describe
Life Science 7 Chapter 9-1 p 222-227 Classification: Sorting It All Out Objectives Explain why and how organisms are classified. List the eight levels of classification. Explain scientific names. Describe
Physics 212E Spring 2004 Classical and Modern Physics. Computer Exercise #2
 Physics 212E Spring 2004 Classical and Modern Physics Chowdary Computer Exercise #2 Launch Mathematica by clicking on the Start menu (lower left hand corner of the screen); from there go up to Science
Physics 212E Spring 2004 Classical and Modern Physics Chowdary Computer Exercise #2 Launch Mathematica by clicking on the Start menu (lower left hand corner of the screen); from there go up to Science
RGP finder: prediction of Genomic Islands
 Training courses on MicroScope platform RGP finder: prediction of Genomic Islands Dynamics of bacterial genomes Gene gain Horizontal gene transfer Gene loss Deletion of one or several genes Duplication
Training courses on MicroScope platform RGP finder: prediction of Genomic Islands Dynamics of bacterial genomes Gene gain Horizontal gene transfer Gene loss Deletion of one or several genes Duplication
Journal of Proteomics & Bioinformatics - Open Access
 Abstract Methodology for Phylogenetic Tree Construction Kudipudi Srinivas 2, Allam Appa Rao 1, GR Sridhar 3, Srinubabu Gedela 1* 1 International Center for Bioinformatics & Center for Biotechnology, Andhra
Abstract Methodology for Phylogenetic Tree Construction Kudipudi Srinivas 2, Allam Appa Rao 1, GR Sridhar 3, Srinubabu Gedela 1* 1 International Center for Bioinformatics & Center for Biotechnology, Andhra
