Searching CrossFire Databases-
|
|
|
- Doreen Gilbert
- 6 years ago
- Views:
Transcription
1 Searching CrossFire Databases- based on CrossFire Beilstein Training Guide CrossFire Commander Version 7.1
2
3 Searching CrossFire Databases- based on CrossFire Beilstein CrossFire Commander Version 7.1 Training Guide CrossFire Software Copyright , Elsevier Information Systems GmbH. CrossFire Beilstein Database: Copyright , Elsevier Information Systems GmbH. Gmelin Database: Copyright , Gesellschaft Deutscher Chemiker, licensed to Elsevier Information Systems GmbH; , Gmelin Institut fuer Anorganische Chemie und Grenzgebiete der Max-Planck-Gesellschaft zur Förderung der Wissenschaften. All rights reserved. This work is subject to copyright. All rights are reserved, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilm or in other ways, and storage in data banks. Duplication of this publication or parts thereof is only permitted under the provisions of the German Copyright Law of September 9, 1965, in its version of June 24, 1985, and a copyright fee must always be paid. Violations fall under the prosecution act of the German Copyright Law. The use of registered names, trademarks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and are therefore free for general use. Furthermore the Elsevier CrossFire system is subject to the license agreement and the terms and conditions for the CrossFire Beilstein data and database system. Training Guide: May 2008, for Elsevier CrossFire Commander Version 7.1
4
5 TABLE OF CONTENTS INTRODUCTION Introduction Module objectives Connect to the CrossFire server password access Connect to the CrossFire server IP access Select the database Beilstein database structure What is a Beilstein compound? Elements included in Beilstein Layout of the interface Structure Editors DATA SEARCHING Data searching Scenario # 1 General information Search All Text area Text Search context selection Hitset Family relationship Results tab toolbar Finding general information Data operators Modify the original query Scenario # 2 Search for data Datastructure Tree Clearing a query Chemical Name search Check the occurrence in the database CAS number search Use the Predefined Search Forms Scenario # 3 Retrieving specifics Search Fields area Enter the data query Relational operators Enlarge the search grid Multiple data parameter search results Scenario # 4 Search for reactions Formulate the query Reaction data search results Exercise descriptions Exercise 1: conduct a data search to retrieve the solubility Exercise 2: conduct a search to retrieve compounds isolated from digitalis purpurea Exercise 3: conduct a search to retrieve demethylation reactions
6 BASIC STRUCTURE SEARCHING Basic structure searching Scenario # 5 Basic structure searches Select Structure Editor CrossFire Commander structure editor Conduct an Exact Match search Search Status Report AutoSearch progression Search results Grid view Search results Details view Link to boiling point data Substructure search Search Structure Query Options View substructure search results Scenario # 6 Modifying the search Eliminating additional rings Extended Structure Query Options Saving a query Limiting the number of fragments List manipulation create search logic NOT list results Saving and deleting a hitset Exercise descriptions Exercise 1: conduct a structure search Exercise 2: conduct a substructure search Exercise 3: conduct a tautomer search REFINED SUBSTRUCTURE SEARCHING Redefined substructure searching Scenario # 7 Generalize the structure Predefined generics Implicit versus explicit substitution Create the query Create the query Alkyl substituted search results Scenario # 8 Bond variation Allowing bond variation Bond topology Draw the query Structural portion of the query Adding the data constraint View the results Scenario # 9 Fragment search Combination query
7 Enter the Search Fields parameter Combination search results View specific information Link to the Citation View the detailed reaction information Hyperlink to the aromatic product Hitset History Browser Exercise descriptions Exercise 1: conduct a substructure search for 4-amino-indole Exercise 2: conduct a substructure search with predefined generic groups Exercise 3: conduct a combined structure and data search USER-DEFINED AND NESTED GENERIC GROUPS User-defined and nested generic groups Scenario # 10 User-defined generics Create the user-defined generic group Set the Attachment point User-defined generic group results Scenario # 11 Specifying frequency Draw the query Setting the frequency Search results frequency of Scenario # 12 Modify the frequency Search results frequency of Intersection of hitsets List manipulation results Scenario # 13 Synthesis of imidazoles What is a nested generic group? Acceptable nested queries Nested search results View solubility data Search results Scenario # 14 Allowing ring expansion Creating an atom list Combined query Search results Exercise descriptions Exercise 1: retrieve disubstituted indoles Exercise 2: create a use-defined generic group with an atom list Exercise 3: retrieve benzoic acids STEREOCHEMISTRY Stereochemistry Scenario #15 Stereochemical search (E or Z) Stereochemical settings Stereochemical bonds
8 Stereochemical search options Relative search results Retrieve a list of citations for a hitset Absolute stereochemical search Absolute search results Convert a hit to a list of citations Exercise descriptions Exercise 1: retrieve penicillanic acid and esters REACTION SEARCHING Reaction searching Possible reaction queries Scenario # 16 Partial reaction search Define the role in the reaction Launching the reaction search Partial reaction search results Mark reaction centers Final partial search results Scenario # 17 Search with fragments Retrosynthetic pathway Scenario # 18 Mapping the reaction Analyzing the results Mapping a reaction query Results of mapping a query Find similar reactions InfoChem Class Codes Reaction similarity choices Query parser interpretation Reaction similarity search results Sort/Group hitsets Viewing grouped results Scenario # 19 Reaction stereochemistry Stereo configuration settings Stereochemistry search results Scenario # 20 Mapping a reaction Apply atom maps Search results Exercise descriptions Exercise 1: conduct a partial reaction search Exercise 2: retrieve examples of Knorr Pyrrol synthesis Exercise 3: synthesis search for quinolines with a sulphuric acid reagent Exercise 4: search for inversion reactions MANIPULATING RESULTS Manipulating results Copy selected data
9 Viewing copied data Create a report Create a report in Word Create a report in the Commander Export Predefined export choices Predefined export process Create a new export file Export Wizard Define Format Export formats Export target Export Wizard Define Content Selecting data fields Save the User View Changing field names Inclusion/exclusion of data Export Wizard Finish Save the export Exported data Specify Print Options
10
11 Introduction 1-1 Searching CrossFire Databasesbased on CrossFire Beilstein Welcome to the Searching CrossFire Databases module using CrossFire Commander 7.1. In this module, we will search only the Beilstein database using factual, structural, and reaction queries. Searching techniques covered in this module can also be applied to the Gmelin database and the Patent Chemistry Database.
12 1-2 Searching CrossFire Databases- based on CrossFire Beilstein Module objectives Access functional aspects of the CrossFire Commander Conduct structural searches: Substances as structure exact match and substructure as reactant as product Reactions partial reaction full reaction Specify stereochemistry Use predefined generic groups Use user-defined generic groups Conduct data searches Search All Text keyword searches Search Fields select data fields from the datastructure use Predefined Search Forms Search for Citations Manipulate results Export Create reports In this module, you will learn the qualifications used to define a Beilstein compound. You will learn how to conduct data searches using keywords, predefined search forms, and data fields. You will learn how to use query features applied to atoms and bonds. You will learn how to conduct reaction searches, how to mark the reacting centers, and apply atom-atom maps. You will learn how to manipulate your retrieved hitsets.
13 Introduction 1-3 Connect to the CrossFire server password access Desktop icon Your Server Check to save your user id and password The CrossFire Commander is the desktop portion of the software. CrossFire is the server portion of the software. The Connect button starts the login script. Enter your User ID and Password. Your organization can subscribe to CrossFire in-house or through the CrossFire Direct server. These configurations allow you to perform an unlimited number of searches. When you check the Save user id and password box, the system automatically stores your entry in a server profile.
14 1-4 Searching CrossFire Databases- based on CrossFire Beilstein Connect to the CrossFire server IP access Desktop icon Click Connect to be logged in as an anonymous user. After connecting, click the Login button Click here to register for a User ID and Password. CrossFire also accomodates site-wide IP access. If your organization has IP access, you will be logged in as an anonymous user when you hit the connect button. In order to perform certain functions, such as saving Hitsets and defining alerts, you must register for a user name and password. To register, click the login button. Then click Register.
15 Introduction 1-5 Select the database Once connected, the Connect button is replaced with the Select Database button. If more than one database is selected, only the search fields common to all databases will be available. After connection to the CrossFire server has been established, click the Select Database button to open a list box of the available databases. The database list box varies based on the databases licensed at your site. The choices are the Beilstein, Gmelin, and Patent Chemistry databases, but can also include a customized database for your site. You can select one or more databases. If you select more than one, the available search fields will change to include only the fields common to all databases.
16 1-6 Searching CrossFire Databases- based on CrossFire Beilstein Beilstein database structure Beilstein(2008/01) Basic Indexes All BRNs All Chemical names All Chemical name segments Substance Basic Index Reaction Basic Index Citation Basic Index EcoPharm Basic Index Bibliographic Information Substance Identification Chemical Properties Physical Properties Pharmacological and Ecological Data This is the structure of the Beilstein database. Every compound in this database has structure and identification data. Basic Indexes are available to conduct keyword searches. There are seven indexes: All BRNs, All Chemical names, All Chemical name segments, Substance Basic Index, Reaction Basic Index, Citation Basic Index, and the EcoPharm Basic Index.
17 Introduction 1-7 What is a Beilstein compound? Organic compounds that have: A completely defined structure Chemical or physical data available An acceptable state of purity The Beilstein database contains structures, synthesis, and property data for compounds that meet the criteria shown. The restrictions are for the structural components in the database. Reagents and catalysts are included in reaction records. The reagents and catalysts are not structural components in the database. They are typically indexed by chemical name. Peptides are registered if they contain 12 or fewer amino acids. Multi-component systems, such as salts or mixtures, must have at least one organic component. Polymers are defined by their names, not by their structures.
18 1-8 Searching CrossFire Databases- based on CrossFire Beilstein Elements included in Beilstein H B Literature In addition to carbon, all elements of the periodic table are allowed. Literature abstracts > All carbon compounds which contain those elements highlighted in the periodic table above are allowed. From 1771 to 1979 all carbon-containing compounds are allowed. Since 1980, only carbon-containing compounds that also contain B, Si, N, P, As, O, S, Se, Te, F, Cl, Br, I, H, Li, Na, K, Rb, Cs, Mg, Ca, Sr, Ba are allowed. The elements in green, in combination with carbon, are contained in the Gmelin database. The Beilstein and Gmelin databases are complementary. Information in Beilstein is derived from The Beilstein Handbook, journals, and patents (prior to 1980.) Patents after 1979 are covered in the Patent Chemistry Database.
19 Introduction 1-9 Layout of the interface Help files Tabbed forms Datastructure Tree Structure/Reaction query builder Factual query builder Sets search context Launches search The CrossFire Commander uses one window with tabbed forms to access all the functionality. The Query tab is used to construct the query that is posed to the database. The Results tab displays the information retrieved from the query posed. The Reports tab displays a custom report created from data selected on the Results form. The Alerts tab is used to create a periodic alert for a specific query on updates of the database. Help can be found on the menu bar and also by clicking the blue question mark (?) in various locations throughout the application.
20 1-10 Searching CrossFire Databases- based on CrossFire Beilstein Structure Editors Options > Select Structure Editor The CrossFire Structure Editor and MDL ISIS/Draw are included in the Commander 7.1 Information about MDL ISIS/Draw is included at the end of this module. A license for MDL Draw can be obtained from Symyx. Other structure editors may also be used. Please consult the CrossFire Administration Guide for details. Choose Options > Select Structure Editor to make your selection The CrossFire Structure Editor (default). is recommended to avoid possible query feature discrepancies between the Commander and other structure editors.
21 Data Searching 2-1 Data searching In this section, you will learn how to: Conduct data searches to retrieve substances, reactions, and citations Use all factual query areas in the user application In this section, you will learn how to conduct data searches to retrieve substances, reactions, and citations. You will use the All Text field and the Search Fields area to create factual queries. You will review the Advanced Search and Guided Search modes. You will check for the occurrence of a fact in the database prior to conducting the search.
22 2-2 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 1 General information You are interested in retrieving general information on epibatidine; a natural product obtained from the Ecuadorian frog used as an analgesic. You do not know the structure of this compound. Modify the original search to retrieve information on the synthesis of this compound. You are interested in retrieving information on the analgesic epibatidine. You do not know the structure. Therefore, you will conduct a factual search to obtain information on this substance. You will use the All Text field to enter keywords to search over the Beilstein database. You will modify the original search to retrieve information on the synthesis of this compound.
23 Data Searching 2-3 Search All Text area Allows you to combine a text search with a structure or factual query using the and, or, or not operators. Allows you to search for a keyword or expression in the Basic Indexes. You can enter a phrase or keyword into the All Text field. The Commander default is to perform a search in the basic index of the selected database to enable you to get a full overview of the specific topic. Generalized text searches can be combined with structure or factual queries using the and, or, or not operators.
24 2-4 Searching CrossFire Databases- based on CrossFire Beilstein Text Search context selection Query interpretation The Search Status Report dialog box will display the analysis of your query. The number of hits per context (Substance, Reaction, or Citation) is displayed along with the query. You can select 1 or more hitsets to view.
25 Data Searching 2-5 Hitset Family relationship One search generated three lists creating a Hitset Family. Multiple searches creates a cascade of windows. Results can be individually selected to view. The Hitset Family tracks your search results, from a single query, and ties them together for future analysis. You will generate a Hitset Family if you conduct a search over multiple contexts, or you have selected more than one database. You will obtain a window for each context. Each window functions independently from the others.
26 2-6 Searching CrossFire Databases- based on CrossFire Beilstein Results tab toolbar Goto first hit Goto previous hit Goto next hit Goto last hit Previous hyperlink Next hyperlink View History Sort/Group Define the view of the data Grid view Details view Switch structure display on and off Copy structure to structure editor Get additional information Export selected data Copy data to a report Print a hitset The Results tab has a series of tools to make viewing your records easier. The first set of commands allows you to navigate through a hitset. You can scroll through a list to the first, previous, next, or the last record. The Previous and Next hyperlink buttons are used to move between hyperlinked information. The next set of commands allows you to control the display of the data. The last set of commands provides additional tools for getting related information, and copying, and exporting results.
27 Data Searching 2-7 Finding general information Substance context Reaction context Citation context The initial query retrieved hits from all three contexts selected. Select View> Highlight Hit Terms to see your query highlighted in the results. This selection will become part of your profile until you change it. Reviewing the records, some references deal with information outside the scope of our original intent. We will modify our query to reduce the number of pertinent hits.
28 2-8 Searching CrossFire Databases- based on CrossFire Beilstein Data operators Operators And Or Not Proximity Next Near Use to retrieve records that: Satisfy both criteria Satisfy either criterion Do not satisfy the criterion Satisfy both criteria in the same subrecord Contain the preceding word and following word as a phrase in that order in a text field Contain the preceding word and following word as a phrase in either order in a text field Wildcards Use in text string to represent * Any number of characters (including zero)? One character?? Two characters Operators are required to connect multiple data fields. The operators are listed in the slide. The two wildcards are the asterisk (*) and the question mark (?). Both can be used at the beginning, in the middle, or at the end of an entry. The asterisk (*) allows any number of characters, including zero. The question mark (?) requires a single character.
29 Data Searching 2-9 Modify the original query Create a more specific query. Select a data operator from the drop-down menu We modified the text entry to include the keyword synthesis. When additional keywords are entered, the Commander changes the interpretation of the parsed query. If more than one word is entered, the Commander will run a combined query. We also entered the NEAR operator to indicate the relationship between the two keywords. This operator retrieves hit lists in which the individual records contain both of the search terms and the search terms are adjacent to each other.
30 2-10 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 2 Search for data You are interested in retrieving data on the compound ibuprofen, a well known non-steroidal anti-inflammatory. You are unsure of the chemical structure. Conduct a search using the chemical name and then conduct a second search using the CAS number. Conduct both searches two ways: using the Search Fields grid and then using the Predefined Search forms. You are interested in retrieving data on the non-steroidal anti-inflammatory, ibuprofen. You do not know the chemical structure but can conduct a search for information using the chemical name or the CAS number (if known). First, conduct the searches using the search fields grid. Then conduct the same searches using the Predefined Search Forms.
31 Data Searching 2-11 Datastructure Tree Database data fields used to formulate your query. Predefined search forms used to formulate your query. Browse or use current, saved, or previous queries. Browse or use current or saved hitsets. The Datastructure Tree is composed of four tabs and has multiple functions. The Search Fields tab allows you to access all fields in the datastructure. The Predefined Search Forms tab has a series of forms that can be accessed to create a search query. The Queries tab allows you to access queries that were previously launched or saved. The Hitsets tab allows you to access hitsets that were previously retrieved or saved and allows you to do a subset search.
32 2-12 Searching CrossFire Databases- based on CrossFire Beilstein Clearing a query Clears structure and text/data queries and resets Structure Search Options and additional options ( More ) Clearing queries is important or they can be included in subsequent searches. Use the Clear Structure button to remove structures from the Structure/Reaction query builder. Use the Clear Table button to remove entries from specific cells, specific rows, or the entire grid. Use the Clear Query button to remove structures and remove text/field entries. The Clear Query button also resets the options under Allow and resets the extended options (found by clicking the More button).
33 Data Searching 2-13 Chemical Name search Single-click opens the Field Help Double-click adds field to the Search Fields grid There are two ways to enter the current query. We will do both. First we will use the Search Fields query builder in conjunction with the Search Fields tab in the Datastructure Tree. Use the Find Field or Form button to find an instance of the data field in the database datastructure (and in the Field Help). Clicking Find Next will take you to subsequent instances of that field name. Double-click the Field Name to add it to the Search Fields Grid.
34 2-14 Searching CrossFire Databases- based on CrossFire Beilstein Check the occurrence in the database Once the data field is added to the Search Fields grid, you can enter relational operators and Field content to complete the query. To the right of the Field content column is a column labeled List. Click the arrow to open the Expand <data field> dialog box. This dialog box is used to confirm the presence of the entry in the database. Each field has a controlled vocabulary, so every entry for ibuprofen is listed.
35 Data Searching 2-15 CAS number search CAS CAS numbers are searched in the same manner you used to search for the chemical name. Clear the previous query using the Clear Table button before continuing. Use the Search Fields tab in the datastructure tree to obtain the correct data field.
36 2-16 Searching CrossFire Databases- based on CrossFire Beilstein Use the Predefined Search Forms This query form provides search criteria specific to fields related to substance identification data. Click list to obtain the Expand Field dialog box. Clear the previous query. You will now use the Substance Identification form to perform the same searches. Predefined Search Forms are specific to the datastructure of the database you are using. Predefined Search Forms are dialog boxes used for easy query formulation in the factual portion of a CrossFire database. It is not necessary to know any field codes or to know how to use operators and truncations. The Substance Identification predefined search form provides search criteria specific to fields related to substance identification data.
37 Data Searching 2-17 Scenario # 3 Retrieving specifics You are interested in retrieving papers discussing information on the pheromones of the bark beetle published after You want to create a search using more specific data parameters. In this example, you are interested in obtaining information on the pheromones of the bark beetle published after 1995.
38 2-18 Searching CrossFire Databases- based on CrossFire Beilstein Search Fields area Operators and, or, or not used to connect a structure or text search. Guided Search mode Use the Tree pane (or the List drop-down menu) and the List database index entry. Advanced Search mode Manually enter (type) search syntax. You can enter more than one data field into the Search Fields grid. CrossFire Commander has two modes for creating queries using the Search Fields query builder: the Guided Search mode and an Advanced Search mode. The Guided Search mode allows you to select a data field and a data operator and then enter the field content. The Advanced Search mode allows direct input of a CrossFire query if all the various field codes and operators are known.
39 Data Searching 2-19 Enter the data query Select a field from the Tree Pane or from the List Drop-down menu. List Dropdown menu List Database Index Tree Pane Addition of the wildcard (*)helps retrieve a long list of selections. In addition to using the Search Fields tab and the Predefined Search Forms tab in the Tree Pane, you can also select a field from a drop-down menu of recently-used fields. When entering more than one data field into the Search Fields grid, you can use field operators to connect the search criteria. The field operators are AND, OR, NOT, NEAR, NEXT, and PROXIMITY. You can also use wild cards to enhance your search.
40 2-20 Searching CrossFire Databases- based on CrossFire Beilstein Relational operators Numeric Operators Use to retrieve records that: = satisfy the exact entry < are less than the specified entry <= are less than or equal to the specified entry > are greater than the specified entry >= are greater than or equal to the specified entry Text Operators Is Starts with Ends with Contains Use to retrieve records that: have the exact entry begin with the entry end with the entry have the term contained within When using numeric fields, the following operators are available: Equal to (=), less than (<), less than or equal to (<=), greater than (>), and greater than or equal to (>=). When using text values, the following operators are available: Is, Starts with, Ends with, and Contains. The selection Is will search for the exact term. The selection Starts with will set right truncation. The selection Ends with will set left truncation. The selection Contains will set left and right truncation.
41 Data Searching 2-21 Enlarge the search grid Query: bicit= pheromon* and bicit= bark* next bicit= beetle* and py> 1995 The Search Fields grid can accommodate up to 99 field entries. The grid on the Query tab displays eight data fields, but will add more fields when needed. To view greater than eight data entries at a time, select View> Enlarge the Field Table.
42 2-22 Searching CrossFire Databases- based on CrossFire Beilstein Multiple data parameter search results View> Hit Only To ensure that the term bark beetle was found together, you used the NEXT operator. As already stated, the NEXT operator requires that the text remain in the order specified in the query. Examining the results, you will see all the terms in this abstract with the requirement that bark and beetle be next to each other in the order they were entered. You will also see the publication date greater than Select View> Hit Only to display only relevant information.
43 Data Searching 2-23 Scenario # 4 Search for reactions You are interested in retrieving all reactions where Raney Nickel has been used as a reagent and/or catalyst. In this example, you are interested in retrieving hydrogenation reactions that use Raney-Ni as a reagent or a catalyst.
44 2-24 Searching CrossFire Databases- based on CrossFire Beilstein Formulate the query Include all reagent occurrences and then include all catalyst occurrences. Look for alternative spellings Due to authors inconsistencies in reporting various reagents and catalysts in the literature, catalyst information can be stored in the catalyst and/or reagent field. Search both fields for completeness. The substance, Raney Ni, occurs in the database using different entries. Use the Expand <Data field> dialog box to include all occurrences of the catalyst and the reagent in the query. Include various spellings, such as ranay, ranaey. Alternate spellings will automatically be interpreted using the Or operator.
45 Data Searching 2-25 Reaction data search results View> Hit Only Viewing the results, you can see that we retrieved a list of reactions that use Raney nickel as a reagent or catalyst. Select View> Hit Only to display only reactions using Raney Ni.
46 2-26 Searching CrossFire Databases- based on CrossFire Beilstein
47 Data searching 2-27 Exercise descriptions The following descriptions explain the goal of each exercise. If you like to figure things out on your own, use the descriptions to conduct the exercises. If you prefer step-bystep instructions, go to the page listed below the description. Exercise 1 Conduct a combination data search. Conduct a search to retrieve the solubility of benzocaine in the solvent ethanol. View the solubility data. For a step-by-step solution, see page Exercise 2 Conduct a search to retrieve compounds that have been isolated from digitalis purpurea. Use the isolation field in the database and make sure digitalis and purpurea come from the same subrecord. Browse the hitset and review the pharmacological data. For a step-by-step solution, see page Exercise 3 Conduct a search to retrieve demethylation reactions of anisole. View the various reactions. For a step-by-step solution, see page 2-35.
48 2-28 Searching CrossFire Databases- based on CrossFire Beilstein Conduct a data search to retrieve the solubility Exercise 1 Conduct a search to retrieve the solubility of benzocaine in the solvent ethanol. View the solubility data. Prepare for a new search 1. Click the Query tab. Search for the desired field 2. Click the Search Field Tab on the Tree Pane. Click the Find Field or Form button. Type chemical name into the text box. Click OK. 3. Click the Find Next button until you open the Substance Identification portion of the datastructure tree. 4. Double-click the Chemical Name field. 5. In the Field content box, type benzocaine.
49 Data searching 2-29 Check for the occurrence in the database 6. Click the List button to check for the occurrence in the database. 7. Click Cancel. 8. In the second row of the Search Fields grid, verify that the and operator is selected. Locate the solubility field 9. Click the Search Field Tab on the Tree Pane. Click the Find Field or Form button. Type solvent into the text box. Click OK. 10. Double-click the Solvent field under the Solubility (MCS) category.
50 2-30 Searching CrossFire Databases- based on CrossFire Beilstein 11. In the Field content box, type ethanol. 12. Click the List button to check for the occurrence in the database. Click Cancel. Set the search Context 13. For the Search Context, select Substances. 14. Click the Start Search button. 15. Click View in the Search Status Report. 16. Click the Details button.
51 Data searching Click the View all button and select Hit only. View the results.
52 2-32 Searching CrossFire Databases- based on CrossFire Beilstein Conduct a search to retrieve compounds isolated from digitalis purpurea Exercise 2 Conduct a search to retrieve compounds that have been isolated from digitalis purpurea. Use the isolation field in the database and make sure digitalis and purpurea come from the same subrecord. Browse the hitset and review the pharmacological data. Prepare for a new search 1. Click the Query tab. 2. Click the Clear Query button. Locate the Isolation from Natural Product field 3. Click the Search Field Tab on the Tree Pane. Click the Find Field or Form button. Type isolation into the text box. Click OK. 4. Double-click the Isolation from Natural Product field. Enter the search data 5. In the Field content box, type digitalis. Check for the occurrence in the database 6. Click the List button to check for the occurrence in the database.
53 Data searching Click Cancel. Add the proximity operator 8. In the second row of the Search Fields grid, select the proximity operator. 9. In the Field name box, type inp. 10. In the Field content box, type purpurea. Check for the occurrence in the database 11. Click the List button to check for the occurrence of purpurea in the database. 12. Click Cancel. 13. Click the Start Search button. 14. Click View in the Search Status Report.
54 2-34 Searching CrossFire Databases- based on CrossFire Beilstein 15. Click the Details button. 16. Click the View All button. Select Hit Only. 17. Browse the results. 18. Choose View > All Fields. Click the PHARM link 19. Under Field Availability, click the PHARM link in the Code column. Review the pharmacological data.
55 Data searching 2-35 Conduct a search to retrieve demethylation reactions Exercise 3 Conduct a search to retrieve demethylation reactions of anisole. Prepare for a new search 1. Click the Query tab. 2. Click the Clear Query button. Locate the title field 3. Click the Search Field Tab on the Tree Pane. Click the Find Field or Form button. Type title into the text box. Click OK. 4. Click Find again to locate the Abstract Title field. 5. Double-click the Title field. 6. In the Field contents box, type demethylation.
56 2-36 Searching CrossFire Databases- based on CrossFire Beilstein Check for the occurrence in the database 7. Click the List button to check for the occurrence. 8. In the second row of the Search Fields grid, verify that the and operator is selected. 9. In the Field name box, type ti. Use wildcards or operators to retrieve information 10. Press Tab and type *anisole* in the Field content box. (Alternatively, you could select the contains operator found in the Relation column just prior to the anisole entry.) 11. Click the gray boxes in the appropriate columns to add parentheses before the Title (TI) Field name and after the *anisole* term, as shown.
57 Data searching In the third row of the Search Fields grid, choose the or operator. Locate the abstract field 13. Confirm that the Search Field Tab on the Tree Pane is selected. Click the Find Field or Form button. Type abstract into the text box. Click OK. 14. Double-click the Abstract field. 15. In the Field content box, type demethylation. 16. In the fourth row of the Search Fields grid, type ab in the Field name box. 17. Press Tab and type *anisole*. (Alternatively, you could select the contains operator found in the Relation column just prior to the anisole entry.) 18. Click the gray boxes in the appropriate columns to add parentheses around the Abstract (AB) Field entries.
58 2-38 Searching CrossFire Databases- based on CrossFire Beilstein 19. For the Search Context, choose Citations. 20. Click the Start Search button. 21. Click View in the Search Status Report. 22. Click the Details button. 23. If necessary, click the View all button and select Hit only. Browse the results. 24. Click the View hit button and select All Fields.
59 Basic structure searching 3-1 Basic structure searching In this section, you will learn how to: Create an exact match search Modify the Results view Use the tools on the Results tab Use internal links to view specific data Conduct a substructure search Set the Search Structure Query options Set the Extended Structure Query options Use list manipulation In this section, you will learn how to conduct an exact structure search and a substructure search. You will learn how the AutoSearch feature conducts structural searches. You will use internal links to view specific data. You will use the Structure Search Options and the Extended Query options (the More button)to modify your search. You will use list manipulation to eliminate records from one list by subtracting out another list.
60 3-2 Searching CrossFire Databases- based on CrossFire Beilstein Scenario #5 Basic structure searches Retrieve the following spiro hydrocarbon compound based on cyclopropane and indene. Review the boiling point information using internal links. Use the same structure to conduct a substructure search for all compounds containing this basic core structure. In this example, you are interested in retrieving spiro hydrocarbons based on cyclopropane and indene. You will then use the same core structure to conduct a substructure search.
61 Basic structure searching 3-3 Select Structure Editor Options> Select Structure Editor Default Structure Editor Double click to open Structure Editor To view structure editor choices, choose Options > Select Structure Editors. We will use the CrossFire Structure Editor. This is the default structure editor. Double-click in the Structure Query builder window to open the structure editor.
62 3-4 Searching CrossFire Databases- based on CrossFire Beilstein CrossFire Commander structure editor Atoms Bonds Template tools Edit Select Reaction and Template Lasso Structure modes file Eraser Rotate Define atom list To Commander Define generic group Enlarge display Reduce display 1. Click the Benzene template tool. 2. Click the cyclopentadiene template tool. 3. Click the dot in the middle of a double bond on the cyclopentadiene and drag bond to merge it with the benzene ring. 4. Click the cyclopropyl tool. Click an atom in the cyclopropyl and drag to the appropriate position on the indene structure. The CrossFire Structure Editor has a series of atom tools, bond tools, and predrawn templates that can be used to draw your compounds. Use the instructions above to draw the compound. Any information in the structure window will be transferred when the To Commander button is clicked.
63 Basic structure searching 3-5 Conduct an Exact Match search Enable as structure only. Click Start Search An Exact Match search will retrieve the structure you have drawn without any variations.
64 3-6 Searching CrossFire Databases- based on CrossFire Beilstein Search Status Report Hitset Family tracks your search results and ties them together for future analysis. Sends information to a report Displays results The Search Status Report window provides detailed information about the search results. Structure search query options are noted above the structure. Any additional structure search options are provided in the last column. The Hitset Family status is reported in this area. To copy the Search Status Report window to a report, click the To Report button. To display the results of the search, click the View button.
65 Basic structure searching 3-7 AutoSearch progression AutoSearch ON If this option is enabled, CrossFire Commander will do appropriate variations on the original query to retrieve results closely related to the original request. Stop AutoSearches after 1st result If this option is enabled, AutoSearch will stop when the first set of results closest to the original query are retrieved. The AutoSearch feature performs a tolerant search, starting with the original query, to retrieve results that are closely related. This function is designed to avoid searches resulting in zero hits. The AutoSearch feature is on by default. To disable this option, uncheck it by selecting Query > AutoSearch ON. The AutoSearch feature will proceed to the next search variation if no results are found for the primary search query. This happens only when the Query > Stop AutoSearches after 1st result command is turned off.
66 3-8 Searching CrossFire Databases- based on CrossFire Beilstein Search results Grid view Displays information on the status of the application By default, structures are displayed on the Results tab in Grid View. The Grid View displays many pieces of data. A match to the structure query is highlighted in red. By default, the highlighting is turned on. The status of the system is displayed at the bottom of the screen.
67 Basic structure searching 3-9 Search results Details view Field Availability displays the populated data fields and the number of occurrences for the record. To review a complete listing of all data retrieved for this record, use the DetailsView. You can switch to DetailsView by clicking the Details button or by doubleclicking a structure. Specific data can be viewed by using the internal links. The Field Availability summarizes which fields have data for the current record and the number of occurrences.
68 3-10 Searching CrossFire Databases- based on CrossFire Beilstein Link to boiling point data Internal links allow you to quickly jump to items of interest. Click the Home icon to return to the top of the page. Clicking an internal link takes you to the location in the record where that information is summarized. Alternatively, you can choose View > Field Availability in separate window or F5 to access a separate window displaying the same information. Whenever you use an internal link, such as the data field code, you can quickly return to the top of the page by clicking the home icon.
69 Basic structure searching 3-11 Substructure search SSS Rules: the core structure must be embedded in all hits retrieved substitution can occur at any open valence To retrieve compounds that contain the same core structure, conduct a substructure search. To conduct a substructure search, you must enable the Implicit Free Sites query feature. Implicit Free Sites allows, but does not require, substitution at any open valence. The rules of a substructure search are: the core structure must be embedded in all hits retrieved, and substitution can occur at any open valence.
70 3-12 Searching CrossFire Databases- based on CrossFire Beilstein Search Structure Query Options hetero atoms sets maximum free sites on all heteroatoms all atoms sets maximum free sites on all atoms Stereo Search set to: off absolute relative racemic Check the appropriate box to allow salts, compounds with ring closures between atoms or groups, isotopes, charged compounds, radicals, and mixtures. as structure searches a structure as drawn as reactant searches a structure as a reactant as product searches a structure as the product as reagent/as catalyst/as solvent not available with this database The Structure query options area, next to the Structure Query box, allows you to set most of the common structure and reaction query options at run time. This easy access option setting is very intuitive and allows you to focus on the structure, rather than on where to find the options.
71 Basic structure searching 3-13 View substructure search results Previous results 1 Substance Current results 2062 Substances The substructure search retrieved more hits than the previous search. This is expected since you are opening the possibility of substitution at all the atom sites.
72 3-14 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 6 Modifying the search Use the following structure to conduct a substructure search eliminating compounds with additional rings. Use the Extended Structure Query Options to limit the number of structural fragments retrieved to 1. Create a query, using the generated lists, to display only those results where the core substructure exists and has multiple fragments and no additional rings. A substructure search can retrieve very general results. You can modify the search, using query options, to obtain a more focused list of results. In the next few steps, you will be shown how to eliminate compounds having additional rings on the core structure, as well as limiting the structural fragments retrieved to 1. Finally, you will use your results to create a list of multi-fragment compounds containing the core structure with no additional rings.
73 Basic structure searching 3-15 Eliminating additional rings Eliminates additional rings only on the core structure when conducting a substructure search. To eliminate additional rings from the core structure, uncheck the addl. rings option under Allow. This will only eliminate additional rings on the core structure. Rings can be retrieved at the open sites where substitution occurs.
74 3-16 Searching CrossFire Databases- based on CrossFire Beilstein Extended Structure Query Options Allow Tautomers allows retrieval of tautomers of the query structure Keep Fragments separate requires that isolated fragments in the structure query are retrieved as separate components Ignore Atom Mappings requires that defined mapping on a reaction be ignored in the search. Number of Atoms, Fragments, Ring Closures sets the total number of atoms, fragments, or rings to be found in the retrieved structure Click the More button to open the Extended Structure Query Options dialog box. The CrossFire Commander has a query feature called Number of Fragments. This feature sets the total number of fragments to be found in the retrieved structure (for example, salts and addition compounds). Setting the minimum and maximum to 1 eliminates multiple fragments being found in the retrieved list.
75 Basic structure searching 3-17 Saving a query Click the Save Query button OR Choose File > Save Query as Queries are saved locally on your desktop. Saved queries can be opened at any time to use or to modify. Generally, it is better to save a query rather than a hitset. Saving a query allows you to conduct the same search over an updated database. This ensures that you are working with a complete hitset. To save a query, click the Save Query button or choose File > Save Query.
76 3-18 Searching CrossFire Databases- based on CrossFire Beilstein Limiting the number of fragments Multi-fragment compounds are eliminated. Conducting the search using these two additional parameters eliminates additional rings from appearing on the core structure and limits the fragments retrieved to 1.
77 Basic structure searching 3-19 List manipulation create search logic Double-click the first modified Hitset. Scroll to the logical operator. Double-click the second modified Hitset. Key Points You can conduct a Boolean operation on your retrieved lists. Use list logic to view the difference between what we retrieved in the original substructure search and the search with limitations. Use the Hitsets tab to access all permanent and temporary lists. Double-click a list to add it to the Search Fields grid. You want to view the records that are in one list and not in the other, so you will select the NOT operator. Click Start Search.
78 3-20 Searching CrossFire Databases- based on CrossFire Beilstein NOT list results These are the records eliminated from the original substructure search list. You can see that all have more than one component, which we eliminated in the second search.
79 Basic structure searching 3-21 Saving and deleting a hitset Choose File > Save Hitset as Choose File > Delete Hitset Enter a file name and click OK. Select the hitset and click OK. Click Yes at the confirmation prompt. Choose File > Save Hitset as from the Results tab to save the current hitset to the CrossFire server for use in another session. Enter a file name for your hitset. Any existing file names from permanently saved hitsets are shown in the main window. You are limited to naming conventions. You can only use the letters a-z and the digits 0-9 along with the underscore "_". File names on the server are not case sensitive.
80 3-22 Searching CrossFire Databases- based on CrossFire Beilstein
81 Basic structure searching 3-23 Exercise descriptions Exercise 1 The following descriptions explain the goal of each exercise. If you like to figure things out on your own, use the descriptions to conduct the exercises. If you prefer step-bystep instructions using the CrossFire Structure Editor, go to the page listed below the description. Conduct a structure search. Conduct an exact match structure search for tryptamine. N N Save the query as an.xfque file. View the hitst. For a step-by-step solution, see page Exercise 2 Conduct a substructure search. Conduct a search for the tryptamine structure with implicit free sites enabled. Use the Details button to view specific data. Switch to the Grid display to view all of the records at once. Repeat the search preventing implicit ring closures. For a step-by-step solution, see page Exercise 3 Conduct a tautomer search. Draw the structure shown.
82 3-24 Searching CrossFire Databases- based on CrossFire Beilstein O N N Set query options to include tautomers and exclude multicomponent compounds. Browse the hitset. View dissociation exponent data. Use hyperlinks to view citations. For a step-by-step solution, see page 3-30.
83 Basic structure searching 3-25 Conduct a structure search for tryptamine Exercise 1 Conduct an exact match structure search to retrieve the compound tryptamine. Save the query as a.xfque file. View the hitset. 1. Click the Query tab. Click Clear Query. Launch the structure editor 2. Double-click the Structure/Reaction Search box. Draw tryptamine 3. Click the Benzene template tool. CrossFire Structure Editor instructions 4. Click the Cyclopentadiene template tool. Click the dot on the cyclopentadiene bond and drag to fuse the two rings. Click and drag to connect 5. Click the Edit tool. Click the Single Bond tool. Draw a 3- carbon chain on the 3-position. 6. Click the N atom tool. Shift-click the two nitrogen atom positions.
84 3-26 Searching CrossFire Databases- based on CrossFire Beilstein N N 7. Click the to Commander button. 8. If necessary, uncheck all of the boxes under Allow. Save the query as a.xfque file 9. Click the Save Query button. 10. Type tryptamine in the File name box and click Save. 11. Click Start Search. 12. Click the View button.
85 Basic structure searching 3-27
86 3-28 Searching CrossFire Databases- based on CrossFire Beilstein Search for tryptamine with implicit free sites Exercise 2 Conduct a substructure search for the tryptamine structure with implicit frees sites enabled. Repeat the search with no implicit ring closures. View the hitset. Modify the query 1. Click the Query tab. 2. In the Structure Search Options area under Free Sites, check the all atoms box. By default, salts, additional rings, isotopes, charges, and radicals are allowed in the search. Conduct the search 3. Click Start Search. 4. Click the View button. 5. Browse through the hitset. Notice that many of the hits have additional fused rings. 6. Click the Query tab. 7. In the Structure Search Options area under Allow, uncheck addl. rings. 8. Click Start Search.
87 Basic structure searching Click the View button. 10. Click the Details button. 11. Browse through the hitset. 12. Click the Grid button to return to grid view.
88 3-30 Searching CrossFire Databases- based on CrossFire Beilstein Conduct a tautomer search Exercise 3 Search for tautomers of the following structure while excluding multicomponent compounds. Browse the hitset and view the dissociation exponent. Use hyperlinks to view the citations. O N N Prepare for a new search 1. Click the Query tab. 2. Click Clear Query. 3. Double-click the Structure/Reaction Search box. Draw the query 4. Click the Cyclohexane tool. 5. Click the Edit tool. Click the Double Bond tool. Hold down the Shift key and click a single bond to change it into a double bond. Repeat for the other double bond. 6. Click to select the oxygen tool. Click the carbon in the ring and drag to add the oxo substituent.
89 Basic structure searching Click to select the Nitrogen. Hold down the shift key and click the 2 carbons in the ring where you want to place nitrogen atoms. 8. Click the To Commander button. 9. In the Structure Search Options area under Free Sites, uncheck the all atoms box. 10. Click the More button. 11. Click to check Allow Tautomers. 12. Use the micro-scrolls to set the minimum and maximum values for the Number of Fragments to Click OK.
90 3-32 Searching CrossFire Databases- based on CrossFire Beilstein Conduct the search 14. Click Start Search. 15. Click View. 16. On the Results tab, click the Details button. Link to the Dissociation Exponent 17. Under Field Availability click the DE (Dissociation Exponent) link. Link to the citation 18. Click the citation hyperlink for the first Dissociation Exponent. 19. View the data.
91 Refined substructure searching 4-1 Refined substructure searching In this section, you will learn how to: Expand your structure search by applying query features to atoms and bonds Apply predefined generic groups Use structural fragments Use data entries to enhance or restrict the query Use hyperlinks to view data In this section, you will learn how to apply query features for atoms and bonds. You will learn how to apply predefined generic groups. You will learn how to formulate a query using structural fragments. You will use data entries to enhance or restrict the query.
92 4-2 Searching CrossFire Databases- based on CrossFire Beilstein Scenario #7 Generalize the structure You want to create a generalized structure query that will retrieve any alkyl-substituted oxaziridine compounds. ALK N O C * In this example, you will conduct a generalized substructure search using a structural fragment. Refining a query can be as simple as removing unnecessary structural components. Beilstein generic atoms and Beilstein generic groups are query options that can be used to generalize your search query.
93 Refined substructure searching 4-3 Predefined generics In the CrossFire Structure Editor: Options> Define Atom The Generic groups ending with H also allow for Hydrogen as a substituent ACY, ACH Acyclic G, GH AnyGroup Generics G*, GH* Any Group with ring closure CYC, CYH Cyclic ABC, ABH Carbacyclic AHC, AHH Heteroacyclic CHC, CHH Heterocyclic CBC, CBH Carbocyclic CXX, CXH Cyclic, no carbon AOX, AOH Alkoxy HAR, HAH Heteroaryl CEL represents a CYCLOALLENYL group; CEH represents either a CYCLOALLENYL group or a hydrogen. ALK, ALH Alkyl AEL, AEH Alkenyl AYL, AYH Alkynyl ARY, ARH Aryl CAL, CAH Cycloallyl CEL, CEH Cycloallenyl The atom generics and predefined generic groups are shown in this slide. The atom generics are: A Any atom except hydrogen Q Any atom except H and C M Any metal X Any halogen G* and GH* represent any group with ring closure. Any generic atom/group ending in H will allow for either the generic atom/group or a hydrogen to be retrieved as a substituent in the specified location on the molecule.
94 4-4 Searching CrossFire Databases- based on CrossFire Beilstein Implicit versus explicit substitution Implicit free sites Substitution applied to the entire structure Explicit free sites Substitution applied to specified atoms Specify the number of free sites N O ALK ALK N N C O * Max free sites (only limited by the valence of the atom) O C *1 1 free site O N O ALK N C O *2 2 free sites (up to the this number of substituents) Implicit Free Sites allows substitution at any open valence. Explicit Free Sites allows substitution only at the sites you specify. Free Sites allows any substituent, any bond order, and ring closure. When you specify MAX Free Sites at an atom, the number of additional substituents is limited only by its valence. For example a carbon with one substituent and MAX Free Sites could have 0, 1, 2, or 3 additional substituents. The chosen number implies that up to that number of substituents is possible.
95 Refined substructure searching 4-5 Create the query Edit Draw the core structure. 2. Use the Edit tool tool to select the methyl. 3. Scroll to select Generics in the Atom Attributes box. 4. Click ALK. Click OK twice. 5. Use the Edit tool to select the carbon. 6. Choose MAX Free Sites. Click OK Draw the structure above. Add the predefined generic group and use the Maximum free sites feature. Click the To Commander button.
96 4-6 Searching CrossFire Databases- based on CrossFire Beilstein Create the query Predefined generic group ALK N O C * MAX free sites substitution feature Manually reset Generalized alkyl substituted oxaziridine Set your Structure Search Options to retrieve salts, additional rings, isotopes, charges, and radicals. Click Start Search.
97 Refined substructure searching 4-7 Alkyl substituted search results All compounds retrieved contain the oxaziridine substructure and any alkyl group. Generalizing the query returned compounds that could not be found using a fully defined structural query.
98 4-8 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 8 Bond variation You are interested in retrieving cyclic sulfoxides that are represented in an isolated ring system. You also want to prevent the retrieval of isotopes. Finally, isolate the list to those substances with a melting point between 100 o C and 120 o C. You have seen that combining multiple search parameters is a convenient way to limit the number of records retrieved in a hitset to only the most relevant. In this example, you want to retrieve cyclic sulfoxides in an isolated ring system eliminating isotopes. You also want to limit the list to products that have a melting point between 100 o C and 120 o C.
99 Refined substructure searching 4-9 Allowing bond variation Bond Type and Topology allow for bond variation on your query. CrossFire Structure Editor The query bond features are: - Any retrieves structures containing any type of bond where applied. - Single/Double retrieves structures containing either single or double bonds where applied. - Double/Triple retrieves structures containing either double or triple bonds where applied. Use the bond topology to specify whether a bond is part of a ring, chain, or either.
100 4-10 Searching CrossFire Databases- based on CrossFire Beilstein Bond topology O S O Rn S Rn Bonds must be part of a ring. O S O O O Bonds must be part of a chain. O S O Ch S Ch S S O When the topology is set to Ring, the bond can only be part of a ring structure. The size and composition of the ring is not defined. When the topology is set to Chain, the bond can only be part of an acyclic system.
101 Refined substructure searching 4-11 Draw the query Edit Select 1 2 (IV) 7 1. Draw the core structure. 2. Use the Select tool tool to select the 2 carbons and the 2 bonds attached to them. 3. Select Options> Define Atom. 4. Choose MAX Free Sites. Click OK. 5. Select Options> Define Bond. 6. Choose Ring. Click OK. 7. Use the Edit tool to select the S. 8. Select 4 for Valency. Click OK Draw the structure above. Specify bond topology and set maximum free sites. Specify a valence of 4 on the S.
102 4-12 Searching CrossFire Databases- based on CrossFire Beilstein Structural portion of the query Forces bond to be part of a ring system Limits valence on the S Eliminates isotopes Allows substitution Controls the number of ring systems Applying ring topology ensures retrieval of cyclic systems. Applying Max free sites ensures that substitution will occur at these sites. Setting the Number of Ring Closures to 1 for the maximum and the minimum retrieves an isolated ring system. Click the More button to access this option. To eliminate isotopes, we unchecked the Allow isotopes feature in the Structure Query Options Area. To eliminate sulfones, we specified the valence on the S.
103 Refined substructure searching 4-13 Adding the data constraint This query form provides search criteria specific to fields related to physical data. The last portion of the proposed query is to consider only those compound records where the melting point of the sulfoxide is between 100 o C and 120 o C. The Physical Data predefined search form provides search criteria specific to fields related to physical data. The top portion of the form allows you to perform an Exists search. This is a search for the existence of facts in a specific field. Simply click the checkbox next to the area of interest. The lower portion of the form allows you to perform a detailed search for a specified data point or range of data points within a field.
104 4-14 Searching CrossFire Databases- based on CrossFire Beilstein View the results Click the button to display the structures. Click the button and select Hit only. With inclusion of the data constraint, the list of results goes from > 1700 to approximately 80.
105 Refined substructure searching 4-15 Scenario # 9 Fragment search Chloramphenicol, a very potent antibiotic, is one of the rare chloro-nitro-compounds. You are interested in retrieving any other chloro-nitro-compounds that have been recently isolated from natural products. O C Cl C N * * O Use hyperlinks to investigate the results of the search. You are interested in retrieving any chloro-nitro compounds that have been recently isolated from natural products. This example shows you how the CrossFire Commander searches using random multiple fragments.
106 4-16 Searching CrossFire Databases- based on CrossFire Beilstein Combination query Query: Structure entry AND Search Fields entry Draw the fragments shown above. We want to retrieve all compounds which have a nitro group attached to a carbon and also a chlorine attached to a carbon atom. The carbons can be the same, but it is not required. Following the search rules of the CrossFire Commander, both groups must be present in all the compounds retrieved, but we are not defining how they must attach. We also want to include the data constraint where the compound retrieved must be isolated from natural products. (See next page).
107 Refined substructure searching 4-17 Enter the Search Fields parameter isolation Click Find Field or Form. Enter a keyword and click OK to access the Search Fields and the Field Help. Folder Page Folders can be searched without values. Pages must have values. Some field codes are both folders and pages; search them with or without Click Find Field or Form. Type isolation to find the Isolation from Natural Products data field folder. Click once on the folder to view the Help file. Notice that the field code is INP. Close the Help file. Click Find Next to find the next instance of isolation in the tree (the Isolation from Natural Products data field page). Click once on the page to view the Help file. Notice the field code is INP. Close the Help file. Double click either the folder or the page to select it. Click Search. Commander assumes an Exists relation operator.
108 4-18 Searching CrossFire Databases- based on CrossFire Beilstein Combination search results Click the Details button. Click the View All button and select Hit Only. The hitset contains information about compounds such as pyrrolnitrin (BRN ) and chloramphenicol (BRN ). You also have a record for the compound tetracycline (BRN ).
109 Refined substructure searching 4-19 View specific information In Grid View, click the checkboxes to select specific hits. Hit the View Selected Hits only button. Double-click structure to view details Click the Grid View button. In Grid view, you can click to select specific compounds to view. We chose the pyrrolnitrin, chloramphenicol, and tetracycline compounds. To select the compounds that you are interested in, click in the checkbox next to each compound and choose View > Selected Hits only. Double-click BRN to switch to the Details view. Tetracycline is described as Dactylocycline B in the record retrieved.
110 4-20 Searching CrossFire Databases- based on CrossFire Beilstein Link to the Citation Click the View Details button to view information about other compounds in the article. You are interested in other compounds described in the paper. Click the View Details button in the Substance record to link to the Citation record. Examination of the Field Availability tells us that there are 9 substances and 2 reactions reported in this literature reference.
111 Refined substructure searching 4-21 View the detailed reaction information Click the Reaction (RX) hyperlink to examine the two reactions. You can see that you have a dehydration reaction and a hydrochlorination reaction. We are interested in following the dehydration reaction, so we will use the Reaction ID hyperlink ( ) to obtain more details about this reaction. From this window, we can now use the Product BRN hyperlink ( ) to obtain more details about the aromatic product.
112 4-22 Searching CrossFire Databases- based on CrossFire Beilstein Hyperlink to the aromatic product The Results Tree maintains a history of the session hitsets (Q:) including those created by hyperlinking (L:). We see that the same reaction is indexed in this record, as well as in the last. Seven of the 9 compounds are dactylocyclines and unique to the citation. There are 2 hydroxylamine derivatives which are even more unusual than nitro compounds in the natural product field. In the Tree view, the display keeps a history of all hyperlinks opened since our initial search for fragments with a data constraint. Session hitsets are designated using a Q: descriptor, and hyperlinks are designated using an L: descriptor.
113 Refined substructure searching 4-23 Hitset History Browser Choose View > Show Hitset History Goes to the selected hitset and closes the browser. Goes to the preceding hit. Copies an ACSII text version of the tree to the clipboard. You can also monitor the use of hyperlinks through the Hitset History Browser. Choose View > Hitset History to open the dialog box and scan the hits.
114 4-24 Searching CrossFire Databases- based on CrossFire Beilstein
115 Refined substructure searching 4-25 Exercise descriptions The following descriptions explain the goal of each exercise. If you like to figure things out on your own, use the descriptions to conduct the exercises. If you prefer step-bystep instructions using the CrossFire Structure Editor, go to the page listed below the description. Exercise 1 Conduct a substructure search for 4-amino-indole. Draw 4-amino-indole. Conduct a substructure search. Hyperlink to one of the reactions. For a step-by-step solution, see page Exercise 2 Conduct a substructure search with predefined generic groups. Conduct a search for quinoline derivatives using predefined generic groups. Allow tautomers. Save the query as an.xfque file. O AOX N AOX For a step-by-step solution, see page 4-30.
116 4-26 Searching CrossFire Databases- based on CrossFire Beilstein Exercise 3 Conduct a combined structure and data search. Search for benzodiazepine derivatives. Use predefined generic groups and an X halogen atom to generalize the query. Add a data constraint to retrieve compounds where the melting point is greater than 150 o C. G N O X N ARY For a step-by-step solution, see page 4-33.
117 Refined substructure searching 4-27 Conduct a search for 4-amino-indole Exercise 1 Conduct a substructure search for 4-amino-indole. Use hyperlinks to view one of the reactions. Find similar reactions to the reaction chosen. 1. Click the Query tab. 2. Click Clear Query. Launch the Structure Editor 3. Double-click the Structure/Reaction Search box. CrossFire Structure Editor instructions 4. Click the Benzene template tool. Click the Cyclopentadiene template tool. Click the middle of one of the double bonds on the cyclopentadiene ring and drag to connect it to the benzene ring. Click and drag to connect 5. Click the Edit tool. Single Bond tool. Click the Carbon atom and draw the single bond.
118 4-28 Searching CrossFire Databases- based on CrossFire Beilstein 6. Click to select the Nitrogen. Hold down the Shift key and click the atoms where you want to place the nitrogens. N N 7. Click the To Commander button. Reset the Extended Query Options Set the set parameters 8. Click the More button. Click Reset and then click OK. 9. In the Structure Search Options area under Free Sites, check the all atoms box. By default, salts, additional rings, isotopes, charges, and radicals are allowed in the search. 10. Click Start Search. View the hitset 11. Click the View button.
119 Refined substructure searching In Grid View, double-click the following structure, or browse to a substance of your choice. Hyperlink to a reaction 13. Under Field Availability, click RX in the Code column. 14. Click the Reaction ID hyperlink to view the reaction and the reaction details. 15. Choose View > Structures included. 16. Click the Previous screen hyperlink button to return to the original record. Search for quinoline derivatives
120 4-30 Searching CrossFire Databases- based on CrossFire Beilstein Exercise 2 Conduct a search for quinoline derivatives using predefined generic groups. Allow tautomers. Save the query. O AOX N AOX Prepare for a new search 18. Click the Query tab. 19. Click the Clear Query button. Launch the Structure Editor 20. Double-click the Structure/Reaction Search box. CrossFire Structure Editor instructions 21. Click the Cyclohexane template tool. Click the Cyclohexane template tool again. Click the middle of a bond on the second cyclohexane and drag to fuse it to the first ring. 22. Click the Single Bond tool. Click an atom and drag to draw the bond. Click the Double Bond tool. Hold down the shift key and click on a single bond to change it to a double bond.
121 Refined substructure searching Click the Oxygen. Click the Double Bond tool. Click the atom where you would like to place the oxo group and drag to draw the oxo group. 24. Click the Nitrogen. Hold down the shift key and click the atom where you would like to place the nitrogen atom. 25. Click a carbon on one of the methyl groups. Click Generics in the drop-down menu. Click AOX. Click OK. Check the box for Set to Current. Click OK. Hold down the shift key and click the other carbon to change it to AOX. AOX O AOX N
122 4-32 Searching CrossFire Databases- based on CrossFire Beilstein 26. Click the To Commander button. 27. In the Structure Search Options area under Free Sites, uncheck the all atoms box. 28. Click the More button. 29. Check the box for Allow Tautomers. Click OK. 30. Click the Save Query button. 31. Type quinolin and click Save. 32. Click Start Search. 33. Click View.
123 Refined substructure searching 4-33 Retrieve benzodiazepine derivatives Exercise 3 Search for benzodiazepine derivatives. Use predefined generic groups and an X halogen atom to generalize the query. G is the abbreviation for any group. ARY is the abbreviation for aryl groups. Add a data constraint to retrieve compounds where the melting point is greater than 150 o C. G N O X ARY N Prepare for a new search 20. Click the Query tab. 21. Click the Clear Query button. Launch the Structure Editor 22. Double click the Structure/Reaction Search box. 23. Click the Template tool. Select Cyc_Alk.bsd. Doubleclick the cycloheptane ring. CrossFire Structure Editor instructions 24. Click the Benzene template tool. Click the middle of a bond in the benzene and drag it to a bond on the cycloheptane ring to draw a fused ring.
124 4-34 Searching CrossFire Databases- based on CrossFire Beilstein 25. Click the Edit tool. Click the Single Bond tool. Click an atom and drag to draw a bond. Click the Double Bond tool. Hold down the shift key and click a single bond to change it to a double bond. 26. Click the Oxygen. Click the atom where you want to place an oxo group and drag to draw the oxo group. Click the Nitrogen. Hold down the shift key and click the carbon atoms that you want to change to nitrogen atoms. 27. Click a carbon on one of the methyl groups. Click Generics in the drop-down menu. Click ARY, X, or G in
125 Refined substructure searching 4-35 the appropriate location. Click OK. Click OK. Repeat with the remaining 2 carbon atoms. 28. Click the To Commander button. 29. Click the More button. 30. Uncheck the box for Allow Tautomers. Click OK. Enter the melting point constraint 31. Click the Predefined Search Forms tab. 32. Double-click Physical Data. 33. Under Melting Point change the range operator to greater than >. 34. In the Melting Point text box enter 150 and click OK.
126 4-36 Searching CrossFire Databases- based on CrossFire Beilstein 35. Click Start Search. 36. Click View. 37. Click the Details button. 38. Click View all and choose Hit only.
127 User-defined and nested generic groups 5-1 User-defined and nested generic groups In this section, you will learn how to: Create a user-defined generic group Specify the frequency of user-defined generic groups Create nested user-defined generic groups Create a user-defined atom list In this section, you will create a userdefined generic group to allow only the specified substituents at a particular site. You will create a user-defined generic group to allow ring expansion. You will specify the frequency of userdefined generic groups to retrieve multifunctional compounds. You will create nested user-defined generic groups. You will create a user-defined atom list.
128 5-2 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 10 User-defined generics Create one query to retrieve para-substituted nitrobenzenes while limiting the substituents to amines, halogens, or nitriles. You want to retrieve para-substituted nitrobenzenes while limiting the substituents to amines, halogens, and nitriles. A predefined generic group is too restrictive and a free site is too general. Create a user-defined generic group to allow only the substituents specified.
129 User-defined and nested generic groups 5-3 Create the user-defined generic group Edit 1. Draw the core structure and fragment(s). 2. Use the Edit tool tool to select the methyl. 3. Select Generics from the Atom Attributes box. 4. Select G1 from the Generics box. Select Atom List 2 1 G Group Create a user-defined generic group to allow only the specified substituents at a particular site. A user-defined generic group allows the possible substituents to be described precisely. G group labels can be found in the Generics dialog box Select an atom using the Edit tool. Scroll down under Symbol in the Atom Attributes dialog box and select Generics. The CrossFire Structure Editor can define 49 different G groups.
130 5-4 Searching CrossFire Databases- based on CrossFire Beilstein Set the Attachment point Edit Select Use the Select tool to select the fragments. 2. Click the G group button to assign the fragments to the G group. 3. Use the Edit tool. Click the carbon on the cyano and click the Attachment button. Atom List 2 G Group 3 Assign the fragments to the G group label by selecting the fragments and clicking the Gn button. You must set attachment points for fragments containing more than one atom. Attachment points define where the fragment will attach to the core structure. Using the Edit tool, click the carbon on the cyano fragment. Click the Attachments button in the Atom Attributes dialog box. Choose 1 because this is the first (and in this fragment s case, the only) point of attachment to the main structure.
131 User-defined and nested generic groups 5-5 User-defined generic group results amine nitrile halogen Return to the Commander and conduct an Exact search for substances. Your results consist of a list of nitrobenzenes substituted in the paraposition with an amine, halogen, or nitrile.
132 5-6 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 11 Specifying frequency Quinoline is a naturally occurring, persistent molecule found in creosote, coal tar, and other fossil fuel derivatives. Create a query to retrieve up to four halogenations on quinoline. View the di-, tri-, and tetra-substituted compounds. In the next example, we want to create a single query to retrieve mono-, di-, triand tetra-halogenated quinolines. We then want to modify the view to only those substances that have two, three, or four halogenations. You can define generic queries where a specific number of substituents can be present at different positions in the molecule. This is done by defining frequencies for the generic group. Specifying the frequency of a userdefined generic group is a convenient method for retrieving multi-functional compounds
133 User-defined and nested generic groups Draw the query 1 1. Draw the core structure and fragment shown left. 2. Use the Edit tool tool to select the methyl. 3. Select Generics from the Atom Attributes box. 4. Select G1 from the Generics box. 5. Check the box for Set to Current. 6. Continue drawing to add G1 to other locations on the core structure You may want your results to include your specified G group fragment(s) in more than one location on the molecule. After choosing your first G1 label, click Set to Current in the Atom Attributes dialog box. Then continue drawing to add G1 to additional locations on the molecule.
134 5-8 Searching CrossFire Databases- based on CrossFire Beilstein Setting the frequency Use the Select tool to select the X. 2. Click the G group button to assign the fragment to the G group. 3. Select Query>Generic Frequency and choose 4 for the frequency. You can limit the number of G group fragments in your results. After drawing your G group query, Select Query> Generic Frequency. The number you choose will limit the number of G group fragments on a structure to up to the chosen number. In our example, the results can contain 1, 2, 3, or 4 halogens at the G1 sites. MDL ISIS/Draw instructions can be found in the addendum.
135 User-defined and nested generic groups 5-9 Search results frequency of 4 tri tetra mono di Retrieves molecules with up to four G group substitutions on any of the possible sites. Results show the retrieval of mono-, di-, tri- and tetra-halogenated quinolines. Remember, once we obtain these results we will want to view only the compounds with two, three, and four halogenations.
136 5-10 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 12 Modify the frequency Modify the previous query to retrieve only single halogenations of quinoline. Choose Query > Generic Frequency Our last query was the first step in the process of retrieving the two, three, or four halogenated compounds. We must now modify our query to represent a frequency of 1 to retrieve the monohalogenations. In the CrossFire Structure Editor, choose Query> Generic Frequency, and select one for the frequency.
137 User-defined and nested generic groups 5-11 Search results frequency of 1 Retrieves molecules with one G group substitution on any of the four possible sites. This query retrieves only the monosubstituted compounds.
138 5-12 Searching CrossFire Databases- based on CrossFire Beilstein Intersection of hitsets Saved hitsets Current session hitsets Double-click to add to the query Final query Use the not operator Use the Search/Text Field grid to perform list manipulation. Click the Hitsets tab to view the available hitsets. Right-click on a hitset to view the query from which it originated. This will help you choose the correct Hitset. Double click the Hitsets, as shown above, to add them to the Search/Text Grid. The Exists relation will automatically appear. The NOT operator will retrieve the records contained in the first hitset and not in the second hitset.
139 User-defined and nested generic groups 5-13 List manipulation results tri tetra di This query retrieves only the two-, threeand four halogenated quinolines. All mono-halogenated substances have been removed through list manipulation. If you select AND, the operation will retrieve the records that are in both the first and second hitsets. If you select OR, the operation will retrieve the records that are contained in either the first or the second hitset.
140 5-14 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 13 Synthesis of imidazoles Create one reaction query, using nested user-defined generic groups, that would retrieve both the reaction where the intermediate is characterized and where it is not characterized. O O N O N N O O Overall reaction query O N N O O O N O Query if intermediate is NOT characterized N N O O O N O N O N N Query if intermediate IS characterized O View the solubility data for the compounds reported. You are interested in the synthesis of imidazoles. Reactions are indexed as single steps in the Beilstein Database. In the case of a multi-step reaction, each step is indexed as an individual reaction if the intermediates are characterized. If the intermediate product was not characterized, the second reaction would be an appropriate query; if the intermediate is characterized, the third reaction would be an appropriate query" You will create a single query with userdefined generic groups that can retrieve both possible reactions. You will also retrieve solubility data for the reactants and products.
141 User-defined and nested generic groups 5-15 What is a nested generic group? A nested generic group is a query where a user-defined generic group appears within a user-defined generic group. This slide shows an example of a nested generic group. A nested generic group is a query where a user-defined generic group appears within a user-defined generic group.
142 5-16 Searching CrossFire Databases- based on CrossFire Beilstein Acceptable nested queries Substance query Reaction query You can search for substances and reactions with user-defined and nested user-defined generic groups. Create the reaction query shown above. Please note: Reactions with userdefined generic groups created with ISIS/Draw or MDL Draw cannot be used with the CrossFire Commander.
143 User-defined and nested generic groups 5-17 Nested search results The hitset contains examples of synthesis where the intermediate product is characterized and examples where the intermediate product is not characterized.
144 5-18 Searching CrossFire Databases- based on CrossFire Beilstein View solubility data View the solubility data for the imidazole compounds reported. Combined query You want to view solubility data for the reactants and products in these reactions. Solubility is not available in reaction context. Change context and create a combined query to retrieve this data. Create a query using the hitset from the previous search combined with an exists search for solubility data. Select the Hitsets tab and double click the hitset to select it. Select the Predefined Search Forms tab. Double click to select the Solubility Data form and click to select solubility data. Click OK.
145 User-defined and nested generic groups 5-19 Search results Use the View Hit Only command to isolate the facts associated with solubility data. If you were planning a synthesis, you could verify that the solvent selected is appropriate.
146 5-20 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 14 Allowing ring expansion Create a user-defined atom list to retrieve five and six membered spiro-fused thioacetals. Isolate the list to those compounds with NMR data reported. You will create a user-defined atom list and a user-defined generic group to retrieve five and six membered thioacetals.
147 User-defined and nested generic groups Atom List Creating an atom list 1 1. Draw the core structure. Assign fragments to the G group, assign 2 attachment points to the ethyl fragment (allows for ring expansion). See pages 3-4 for general instructions. 2. Use the Edit tool to select carbon 7 and select Generics from the Atom Attributes box. 3. Choose the A1 label from the Generics box. Click OK. 4. Select the An button, click OK, and select atoms from the Periodic Table You can create user-defined Atom lists. To create an atom list, choose the A1 label from the Generics dialog box. Click the An button and select the atoms from the Periodic Table. The structure shows an atom list that allows the retrieval of nitrogen, oxygen, or sulfur in a specific position on the core structure.
148 5-22 Searching CrossFire Databases- based on CrossFire Beilstein Combined query In addition to a structure search, we are interested in NMR data associated with retrieved compounds. To find the nmr field, click the Find Field or Form button and type nmr into box. Double-click to select the (+) NMR category. When you select a category of data, a Field Availability search is performed. If you want a more specific search, you must search on a subcategory of data within NMR. Alternatively, you can use the Spectroscopic Data predefined search form and check the box next to NMR.
149 User-defined and nested generic groups 5-23 Search results Viewing the results shows five and six membered spiro-fused compounds for which NMR data is reported. The user-defined atom allowed us to vary the atoms at a specific site. The user-defined generic group allowed us to define the ring expansion.
150 5-24 Searching CrossFire Databases- based on CrossFire Beilstein
151 User-defined and nested generic groups 5-25 Exercise descriptions Exercise 1 The following descriptions explain the goal of each exercise. If you like to figure things out on your own, use the descriptions to conduct the exercises. If you prefer step-bystep instructions using the CrossFire Structure Editor, go to the page listed below the description. Retrieve disubstituted indoles. Allow halogens or amines. Construct the user-defined generic group shown. Specify a generic frequency of 2. Conduct the search. Modify the query to specify a generic frequency of 1. Intersect the hitsets to view only disubstituted compounds. The first query is shown below. For a step-by-step solution, see page Exercise 2 Create a user-defined generic group with an atom list. Retrieve 2,4-substituted cyclopentadienes, where one substituent is an alkyl group and the other is a phenyl ring, a halogen, a methyl, or an ethyl group. The phenyl ring may have any substituent at the 4-position. The heteroatom in the 5-membered ring can be oxygen, sulfur, selenium, tellurium, or nitrogen. The query is shown below.
152 5-26 Searching CrossFire Databases- based on CrossFire Beilstein For a step-by-step solution, see page Exercise 3 Retrieve all 1-, 2-, and 3-carbon esters of benzoic, benzenesulfonic, and benzenephosphonic acids. Construct a nested user-defined generic group to minimize drawing. Retrieve the hitset in citation context and view the citations in short display. The query is shown below. For a step-by-step solution, see page 5-33.
153 User-defined and nested generic groups 5-27 Retrieve disubstituted indoles Exercise 1 Construct the user-defined generic group shown to retrieve disubstituted indoles. Allow halogens or amines as the substituents. Specify a generic frequency of 2. Conduct the search. Modify the query to specify a generic frequency of 1. Intersect the hitsets to view only disubstituted compounds. Prepare for a new query 1. Click the To Commander button to return to the CrossFire Commander. 2. Clear the previous query. Launch the Structure Editor 3. Double click the Structure/Reaction Search box. CrossFire Structure Editor instructions 5. Draw the core structure and the generic group members. 6. To label the G1 sites, click the Edit tool. Click an atom that you want to label. Click the dropdown list under Symbol in the Atom Attributes box and scroll to the bottom. Click Generics. Select G1 in the Generics dialog box. Click OK. Click Set to Current in the Atom Attributes box. Click OK. Hold down the shift key and click the remaining atoms to apply the G1 label.
154 5-28 Searching CrossFire Databases- based on CrossFire Beilstein Alternatively, you can click the Select tool. Click an atom that you want to label. Hold down the ctrl key and click the remaining atoms to select them. Then click the dropdown menu in the toolbar and select Generics. Select G1. Click OK. Define the G1group 7. Click the Select tool. Select the generic group members. Click the Gn tool. Select G1. Click OK. Define the frequency 8. Select Query> Generic Frequency. Choose 2. Click OK
155 User-defined and nested generic groups 5-29 Return to Commander 9. Click the To Commander icon. Use the following settings: 10. Click the Start Search button. 11. Click View. 12. View the results. Note the number of hits retrieved. Beilstein (2008/01) = 145 Modify your query 13. Click the Query tab. 14. Double-click the structure box to return to the CrossFire Structure Editor. Define the frequency 15. Select Query> Generic Frequency. Select 1. Click OK. 16. Click the To Commander icon. 17. Click the Start Search button.
156 5-30 Searching CrossFire Databases- based on CrossFire Beilstein Note the hitset number 18. Click the View button. Note the number of hits retrieved. Beilstein (2008/01) = Click the Query tab. 20. Click the Clear Query button. Intersect the hitsets 21. Click the Hitsets tab. 22. Double-click the hitset where generic frequency was equal to Choose not in the Operator column. 24. Double-click the hitset where the generic frequency was equal to Click the Start Search button. 26. Click View. 27. Examine the hitset. Beilstein (2008/01) = 80.
157 User-defined and nested generic groups 5-31 Retrieve 2,4-substituted heterocyclic 5-membered rings Exercise 2 Retrieve 2,4-substituted heterocyclic 5-membered rings, where one substituent is an alkyl group and the other is a phenyl ring, a halogen, a methyl, or an ethyl group. The phenyl ring can have any substituent at the 4-position. The heteroatom in the 5-membered ring can be oxygen, sulfur, selenium, tellurium, or nitrogen. Prepare for a new query 1. Click the Query tab. 2. Click the Clear Query button. Launch the Structure Editor 3. Double click the Structure/Reaction Search box. 4. Draw the query structure and generic group members CrossFire Structure Editor instructions C X 5. To label the ALK, A1, and G1 sites, click the Edit tool, click the site, type the desired atom label under Symbol in the Atom Attributes box, and then click OK. Create Atom List 6. To create an Atom List, click the An tool. Click OK. Select the atoms for your atom list: O, S, Se, Te, and N. Click OK.
158 5-32 Searching CrossFire Databases- based on CrossFire Beilstein Define generic group 7. To allow substitution on a carbon in the benzene ring, use the Edit tool to click a carbon. Select MAX from the Atom Attributes box. Click OK. Use the Select tool to select the G1 fragments. Click the Gn button. Click OK. Set Attachment points 8. Use the Edit tool and click an atom on the ethyl fragment. Click the Attachments button in the Atom Attributes box and select 1. Click OK. Repeat with the carbon in the benzene that is para to the carbon set to MAX Free Sites.. 9. Click the To Commander icon. Check all boxes under Allow. Conduct the search 10. Click the Start Search button. 11. Click View. Beilstein (2008/01) = 94.
159 User-defined and nested generic groups 5-33 Retrieve benzoic acids Exercise 3 Retrieve all 1-, 2-, and 3-carbon esters of benzoic, benzenesulfonic, and benzenephosphonic acids. Construct a nested user-defined generic group to minimize drawing. Retrieve the hitset in citation context and view the citations in short display. Prepare for a new query 1. Click the Query tab. 2. Click the Clear Query button. Launch the Structure Editor 3. Double click the Structure/Reaction Search box. 4. Draw the core structure and the generic group members. See previous exercises for instructions on adding the G group labels. Define the G groups 5. Click the Select tool. Select the members of G1. Click the Gn button, select 1, and click OK. Select the members of G2. Click the Gn button, select 2, and click OK.
160 5-34 Searching CrossFire Databases- based on CrossFire Beilstein Add attachment points 6. Click the Edit tool. For each fragment, do the following: -Click the atom in the fragment that will be the attachment point. -Click the Attachment button in the Atom Attributes box. -Click 1. Click OK. Return to the Commander 7. Click the To Commander icon. 8. Click the Start Search button. 9. Click the View button. Beilstein (2008/01) = 134. Retrieve the hitset in citation context 10. In the Tree View, click the hitset corresponding to the last search, and choose Get > Get All Related Citations. 11. Check the radio button for all Substances in the current hitset Q##.
161 User-defined and nested generic groups Click OK. 13. Click View. 14. Choose Options > Define Rows/Columns in Grid View. Enter 12 for rows and 1 for columns. Click OK.
162 5-36 Searching CrossFire Databases- based on CrossFire Beilstein
163 Stereochemistry 6-1 Stereochemistry In this section, you will learn how to: Draw structures specifying the stereochemistry Conduct a structure search specifying stereochemistry Retrieve a list of citations for the current record and a list of records In this section, you will draw and conduct a search for structures and reactions that have stereochemistry specified. You will also retrieve citations for the current record and a list of records.
164 6-2 Searching CrossFire Databases- based on CrossFire Beilstein Scenario # 15 Stereochemical search Retrieve compounds having the same relative stereochemistry as the following compound. Retrieve compounds with the absolute stereochemistry of the following compound. Convert the hitset to a list of citations. You will conduct a search to retrieve compounds that have relative and absolute stereochemistry of the structure drawn. Before we begin, let s take a closer look at stereochemical features for double and single bonds.
165 Stereochemistry 6-3 (E or Z) Stereochemical settings Choose absolute or relative in the structure Query Builder Click Double Steric in the CrossFire Structure Editor Defines a double bond where the configuration (E or Z) is considered. Search for Trans-2-butene Stereo Off = (Z) unspecified (E) Absolute Relative = (E) You can specify stereochemical settings in the Bond Attributes dialog box in the Structure Editor. If you select Double Steric, a double bond with either an E or Z configuration is considered. To retrieve the Absolute or Relative configuration, set the drop-down menu in the Query Builder to either Absolute or Relative.
166 6-4 Searching CrossFire Databases- based on CrossFire Beilstein Stereochemical bonds Apply Up and Down stereo bonds Defines a stereochemical bond in the up direction You can specify stereochemical settings in the Structure Editor using either the drop-down menu or the Bond Attributes box. The Up bond defines a stereochemically directed bond in the up direction. The Down bond defines a stereochemically directed bond in the down direction. The Steric Unknown bond defines any stereo configuration at the site where it is placed. Create a query using the structure above, including appropriate stereochemical settings.
167 Stereochemistry 6-5 Stereochemical search options Off retrieves all configurations of the compound. Absolute retrieves compounds which have the identical, absolute configuration as the query structure. Relative retrieves compounds with the absolute configuration, the mirror image, and the racemate of the query. Racemic retrieves only the racemate of the query. Stereochemistry is considered only if the proper Stereo option has been selected. To perform a stereo search, activate the appropriate selection. The choices are: - Off, the stereo search option is not in use - Absolute, retrieves compounds having the identical absolute configuration as the query structure - Relative, retrieves compounds and their enantiomers - Racemic, retrieves only the racemate of the query structure Conduct a structure search using your structure query and the settings above.
168 6-6 Searching CrossFire Databases- based on CrossFire Beilstein Relative search results The search results include the exact stereoconfiguration we drew as well as the enantiomers.
169 Stereochemistry 6-7 Retrieve a list of citations for a hitset Use a structure query to retrieve a list of citations Results Let s retrieve the list of citations from the previous search. Use the previous query, but change the context to Citations using the drop-down menu next to to As result I want to get. CrossFire Commander 7.1 defines the recommended search context as the one that matches the context of the query. Since our search context differs from the recommended search context, the Select Search Context dialog box appears. Choose Citations as the Search Context.
170 6-8 Searching CrossFire Databases- based on CrossFire Beilstein Absolute stereochemical search Now we will retrieve compounds with the same Absolute stereochemistry shown.
171 Stereochemistry 6-9 Absolute search results Fewer compounds are retrieved for this search than with the relative stereochemical search.
172 6-10 Searching CrossFire Databases- based on CrossFire Beilstein Convert a hit to a list of citations Choose: Get > Get All Related Citations OR Right-click the hitset When you have retrieved a list of results, you can convert them from one context to another. Click the Get button and choose Substances, Reactions, or Citations to perform the operation. You can also right-click on a Hitset in the Tree to perform the same operation. The same can be applied to a single hit. When converting from Substances to Reactions context, you can choose whether the substance occurs as a reactant, product, or either.
173 Stereochemistry 6-11 Exercise descriptions The following descriptions explain the goal of each exercise. If you like to figure things out on your own, use the descriptions to conduct the exercises. If you prefer step-bystep instructions using the CrossFire Structure Editor, go to the page listed below the description. Exercise 1 Retrieve penicillanic acid and esters. Search with absolute stereo enabled. Repeat the search for reactions where penicillanic acid is a reactant. For a step-by-step solution, see page 6-12.
174 6-12 Searching CrossFire Databases- based on CrossFire Beilstein Search for penicillanic acid Exercise 1 Retrieve penicillanic acid and esters. Search with absolute stereo enabled. Repeat the search for reactions where penicillanic acid is a reactant. If you are using the CrossFire Structure Editor, you can use a template to draw it. Be sure to set two explicit Free Sites. Prepare for a new query 4. Click the To Commander button to return to the CrossFire Commander. 5. Clear the previous query. Launch the Structure Editor 6. Double-click the Structure/Reaction Search box. CrossFire Structure Editor instructions 7. Draw the query structure. 8. Click the To Commander icon. Enable absolute stereo 9. Under stereo, choose Absolute. Conduct the search 10. Click the Start Search button.
175 Stereochemistry Click View. Beilstein (2008/01) = Click the Details button. Browse the hitset. 13. Click the Query tab. Define as a reactant 14. Under Search, in the Structure Query Options area, select as reactant. Conduct the search 15. Click the Start Search button. 16. Click View. Beilstein (2008/01) = Hyperlink to a citation.
176 6-14 Searching CrossFire Databases- based on CrossFire Beilstein
177 Reaction searching 7-1 Reaction searching In this section, you will learn how to: Conduct a partial reaction search Conduct a full reaction search Mark reacting centers, specify atom-atom mapping, and use stereochemistry Use classification codes to retrieve similar reactions Use the Sort/Group feature Incorporate data constraints In this section, you will learn how to conduct searches to retrieve reaction queries. You will learn how to apply reacting centers and atom-atom mapping. You will learn how to search for similar reactions using classcodes. You will continue to incorporate data constraints to focus your search query. You will learn how to use the Sort/Group feature.
178 7-2 Searching CrossFire Databases- based on CrossFire Beilstein Possible reaction queries Partial reaction S reactant(s) product(s) Full reaction reactant(s) product(s) C * C ALK * + C X C * * ALK When you search for reactions, you specify the role of each molecule. You can have one reactant and one product, or you can have multiple reactants and products. You can also search using a partial reaction. A partial reaction is defined as only a reactant or only a product. Query features can be used on structures of complete or partial reactions.
179 Reaction searching 7-3 Scenario # 16 Partial reaction search You want to retrieve compounds where all ring closures produce benzo[b]thiophenes and the new bonds are made into an existing thiophene ring in the same preparation step. S You want to retrieve compounds where all ring closures produce benzo[b]thiophenes and the new bonds are made into an existing thiophene ring in the same preparation. Since there is no mechanistic implication associated with this query, we will use reaction centers to assign one.
180 7-4 Searching CrossFire Databases- based on CrossFire Beilstein Define the role in the reaction as reactant as product Partial reaction queries can be created using the Commander Query Builder or by using the Structure editor. Reactions can be created quickly using the Structure Search Query Options area. You can define the role of the molecule to be a reactant or a product. When creating a partial reaction this way, you can only have one component. (Note: You may want to add features from the Structure Editor to your query. However, when creating a partial reaction this way, certain features within the Structure Editor may not be available.) Reactions can also be created using the structure editor. (An arrow will appear in front of the product.)
181 Reaction searching 7-5 Launching the reaction search When the context does not match the query, the Select Search Context dialog box opens. In this example, you will draw a structure that will retrieve all benzothiophenes, including dibenzocompounds, and define it as a product. You will use a partial reaction query. The context is set to Substances, but the CrossFire Commander understands that you are trying to search for a reaction since you set the Search feature to As Product. When the query does not match the context, a Select Search Context box appears giving you a choice of context.
182 7-6 Searching CrossFire Databases- based on CrossFire Beilstein Partial reaction search results The partial reaction retrieved a large number of extraneous results. A larger number of reactions are retrieved when the reacting centers are left undefined. You will retrieve a variety of extraneous reactions. To answer the query proposed in our scenario, we have to define which bonds will be made during the course of the reaction. To retrieve specific transformations, mark the reaction centers.
183 Reaction searching 7-7 Mark reaction centers To apply reaction centers, your reaction query must be created using a structure editor. To define which bonds get made in the course of the reaction, we will use the Mark Reaction Center feature. A reacting center can be defined on an atom or a bond. The terminology and choices vary based on the structure editor you are using. You can specify that a bond is made or broken, changes bond order, or is not changed in the reaction. Reaction centers can only be applied to reactions created with a structure editor. Partial reactions created using the Structure Search Query Options cannot have reacting centers applied.
184 7-8 Searching CrossFire Databases- based on CrossFire Beilstein Final partial search results Results without reaction centers 26,428 reactions. Results with reaction centers 23 reactions. This slide displays the difference between the number of reactions retrieved with and without reacting centers defined. This indicates that a huge number of extraneous reactions, not related to the scenario, where retrieved in the first search.
185 Reaction searching 7-9 Scenario # 17 Search with fragments You have a compound that contains, among other groups, an amine and an oxime. You want to react the amine with a sulfinyl chloride, but you would like to know what happens to the oxime during this reaction. Conduct a partial reaction search that will retrieve any type of reaction of an oxime with this sulfinyl chloride. Cl C * N O + C S * O Trace the synthetic pathway for the sulfinyl chloride. In this example, you have a compound that contains, among other groups, an amine and oxime. You want to react the amine with a sulfinyl chloride, but you would like to know what happens to the oxime during the reaction. To do this, you will conduct a search using a partial reaction query that specifies the oxime and sulfinyl chloride using structural fragments. You will use query features to introduce flexibility into the query and once the search is complete, you will trace the synthetic pathway for the sulfinyl chloride.
186 7-10 Searching CrossFire Databases- based on CrossFire Beilstein Retrosynthetic pathway N O + O S Cl O S N O O S Cl Reaction ID: Cl Reactant BRN: Cl Cl Cl S Cl Cl S Cl O S Cl S Cl S Cl S Reaction ID: Reactant BRN: Reaction ID: S S Reactant BRN: You can trace the links using the Hitset History. The substitution feature allows an atom site to accept various undefined structural elements. One of the reactions retrieved has a 1,2 shift of the oxygen from the nitrogen to the sulfur. The oxime N-O bond is completely eliminated (this answers the question we posed as to what happens to the oxime in the reaction). There has been a thermal homolysis (free radicals) and recombination of the radical pair that takes place immediately after the formation of the N-O-S bond. The final portion of our original query is to trace the synthetic pathway for the sulfinyl chloride. Use the CrossFire Commander s internal links to access this information.
187 Reaction searching 7-11 Scenario # 18 Mapping the reaction You are interested in studying Friedel-Crafts alkylations of naphthalenes. Conduct a full reaction search to retrieve these types of reactions. C * C ALK * + C X C * * ALK Utilize the atom-atom mapping to remove ambiguous records from the list. Select a search of interest and find similar reactions. In the last example of reaction searching, you will create a full reaction query to retrieve reactions that show the alkylation of naphthalenes. In the process, you will use predefined atoms, predefined generic groups, and atom-atom mapping.
188 7-12 Searching CrossFire Databases- based on CrossFire Beilstein Analyzing the results + + Cl Cl When you analyze the results, note that most conform to the criteria specified in the query. H O + I Some results do not fit the criteria specified in the query. Se + O I H Although you created a searchable query, the application can only do a broad interpretation of what you presented and will retrieve some results that do not satisfy exactly what you were trying to retrieve. Despite the symmetry of the naphthalenes, the assumption was made that the application would understand that certain atoms on the left side of the equation would correspond to certain atoms on the right side of the equation. You can see that this is not true by looking at the bottom two examples on the slide.
189 Reaction searching 7-13 Mapping a reaction query To apply the atom-atom mapping, press and drag from the corresponding atom on the reactant to the corresponding atom on the product. Mapping generics can only be done using the CrossFire Structure Editor. To be more precise when you are searching for reactions, you can specify atom-atom mapping to require that a specific atom in the reactant correspond to a specific atom in the product. Not all of the reactions in the Beilstein database are mapped. When you search with atom mapping, you are limiting your search to about 80% of the database. The method of applying atom-atom mapping varies based on the structure editor you are using, the search results are the same. When mapping a generic, you must use the CrossFire Structure editor.
190 7-14 Searching CrossFire Databases- based on CrossFire Beilstein Results of mapping a query Scenario Results Reaction search unmapped 48 Reaction search carbon mapped 19 Reaction search carbon and alkyl group mapped 15 It is possible to retrieve ambiguous records when you do not map a reaction. If you do not use atom-atom mapping, you can retrieve ambiguous hits. When mapping, it is recommended that you map as few atoms as possible. If you map too many atoms, you increase the chance that the reactions in the database will not be mapped the same way, and therefore will not be retrieved.
191 Reaction searching 7-15 Find similar reactions When viewing reactions, either from reaction searches or hyperlinking, you will notice that there is a link below the reaction data called Find similar reactions. When you use the click here link a dialog box opens displaying various similar reaction searching parameters. This link is only accessible in Details view.
192 7-16 Searching CrossFire Databases- based on CrossFire Beilstein InfoChem ClassCodes Reaction Similarity Broad Only reaction centers are compared. Reaction Similarity Medium Reaction centers and the alpha atoms and bonds are compared. Reaction Similarity Narrow Reaction centers and the alpha and beta atoms and bonds are compared Reactions in the CrossFire databases are indexed using InfoChem ClassCodes. Similar reactions have the same ClassCodes in these databases. In the Reaction Similarity Search dialog box, you can create a search for similar reactions using broad, medium, or narrow classcodes. The broad classification retrieves reactions that are similar at the reaction sites only. The medium classification retrieves reactions that are similar at the reaction sites and the alpha atoms. The narrow classification retrieves reactions that are similar at the reaction sites and the alpha and beta atoms. All Choices conducts a similarity search using all three classcodes and returns three separate hitsets.
193 Reaction searching 7-17 Reaction similarity choices Select Beilstein from the drop-down menu. You are shown a preview of the number of hits found for each of the 3 classification codes. The Find All link covers all three. Click the Find All link to begin the search.
194 7-18 Searching CrossFire Databases- based on CrossFire Beilstein Query parser interpretation RX.BCODE Broad search for reaction sites only RX.MCODE Medium search for reactions sites and alpha atoms RX.NCODE Narrow search for reaction sites and alpha and beta atoms You selected All Choices so the application searched and created a hitset of each of the three classcodes. The Search Status Report box indicates which classcode was used in the Query parser.
195 Reaction searching 7-19 Reaction similarity search results Narrow search results Original query The results shown are from the list generated using the narrow classcodes (reaction sites and the alpha and beta atom sites).
196 7-20 Searching CrossFire Databases- based on CrossFire Beilstein Sort/Group hitsets Click to sort or group Group Sort Preview the results and their occurance Click to ungroup Select (no_sort) to remove sorting All hitsets can be sorted and/or grouped. The hitsets can be sorted/grouped only by the fields in the drop-down list. The selection of fields is database dependent. Certain fields display an analysis of the results and their occurrence. To ungroup, click the Ungroup button. To remove the sorting from a hitset, choose No Sort in the first drop-down list. Click the Sort/Group button. Choose Group. Select Solvent from the dropdown menu.
197 Reaction searching 7-21 Viewing grouped results In Grid View, the grouped hitsets will display the group number, the number of members, and the grouping criteria. The first group member is displayed. The grouping criterion appears in the tree after the hitset name and additionally in the status bar.
198 7-22 Searching CrossFire Databases- based on CrossFire Beilstein Scenario #19 Reaction stereochemistry Retrieve all the preparations of -amino-aldehydes from the corresponding alcohols. What are the results of searching with the absolute stereo configuration versus the relative stereo specification? You will retrieve all the preparations of alpha-amino-aldehydes from the corresponding alcohol. You will look at the results of an Absolute stereo search and a Relative stereo search.
199 Reaction searching 7-23 Stereo configuration settings 1. Select Reaction Mode. 2. Click the Select tool. Select the product structure and click the Product button. Select the reactant structure and click the Reactant button. 3. Use the Edit tool and click the stereo center of the structure. 4. Select Retain and click OK In reactions, you can ignore stereochemistry or specify that bonds invert or retain their configuration. In the CrossFire Structure Editor, apply the Retain label by clicking the stereo center of the structure using the Edit tool. The Atom Attributes in Reaction box will appear. Start your search
200 7-24 Searching CrossFire Databases- based on CrossFire Beilstein Stereochemistry search results Relative Absolute When the Relative designation is applied to the query, 594 reactions are retrieved. Note that the racemic mixture is retrieved. When the Absolute designation is applied to the query, 475 reactions are retrieved. When no designation is applied, 599 reactions are retrieved.
201 Reaction searching 7-25 Scenario # 20 Mapping a reaction Refine the previous query to include atom-atom mapping specifications on two atoms in the reaction. How does this change the results? We will modify the previous reaction and apply atom-atom maps to two atoms. When you search for reactions, you can specify atom-atom mapping to require that a specific atom in the reactant correspond to a specific atom in the product. You can map atoms from the reactant to the product. Not all of the reactions in the Beilstein database are mapped. When you search with atom mapping, you are limiting your search to about 80% of the database.
202 7-26 Searching CrossFire Databases- based on CrossFire Beilstein Apply atom maps 1. Select Reaction Mode. 2. Click the Select tool. Select the product structure and click the Product button. Select the reactant structure and click the Reactant button. 3. Use the Edit tool and click to draw a line from an atom in the Reactant to an atom in the Product In the example, the nitrogen and oxygen atoms in the reactant are mapped to the product, and the nitrogen-oxygen bond in the product is made during the reaction. To map a reaction in the CrossFire Structure Editor, select Reaction Mode. Using the Edit tool, click an atom in the reactant and draw a line to the corresponding atom in the product. To remove atom mapping, use the Delete All Atom Mappings button.
203 Reaction searching 7-27 Search results Our search results include the specific transformation of alpha-aminoaldehydes from the corresponding alcohol.
204 7-28 Searching CrossFire Databases- based on CrossFire Beilstein
205 Reaction Searching 7-29 Exercise descriptions Exercise 1 The following descriptions explain the goal of each exercise. If you like to figure things out on your own, use the descriptions to conduct the exercises. If you prefer step-bystep instructions using the CrossFire Structure Editor, go to the page listed below the description. Conduct a partial reaction search. Retrieve methods for the synthesis of 6-nitro-nicotinic acid. O N N O O O Create the partial reaction query using the Structure query builder. Retrieve the list of citations for a hit with multiple citations. For a step-by-step solution, see page Exercise 2 Retrieve examples of Knorr Pyrrol synthesis. Draw the query shown. Use predefined generic groups to generalize the structure query. Mark the reacting centers. Group the hitset by solvent. For a step-by-step solution, see page 7-34.
206 7-30 Searching CrossFire Databases- based on CrossFire Beilstein Exercise 3 Search for synthesis of quinolines with anilines and glycerol as the starting materials. Draw the reaction, assign molecule roles in the reaction, draw atom-atom mapping, and mark the reacting center. Include a data query for sulphuric acid reagents. Be sure your query will retrieve different spellings and forms of sulphuric acid. For a step-by-step solution, see page Exercise 4 Search for inversion reactions of 1-monosubstituted propan-2- ols. Specify stereo inversion and search with absolute stereo enabled. Browse your hitset and hyperlink to a reactant. For a step-by-step solution, see page 7-43.
207 Reaction Searching 7-31 Retrieve synthetic methods for 6-nitro-nicotinic acid Exercise 1 Retrieve methods for synthesis of 6-nitro-nicotinic acid. Use the Convert option to retrieve the list of citations for a hit with multiple citations. O N N O O O Prepare for a new search 34. Click the Query tab. 35. Click the Clear Query button. Launch the Structure Editor 36. Double-click the Structure/Reaction Search box. Draw the compound 37. Draw the 6-nitro-nicotinic acid structure. 38. Click the To Commander button. 39. In the Structure Search Options area under Search, select as product.
208 7-32 Searching CrossFire Databases- based on CrossFire Beilstein 40. Under Allow, check the boxes for addl. rings, isotopes, charges, and radicals. 41. Under Search Context, choose Reactions. 42. Click Start Search. 43. Click View. Get all related citations 44. Choose Get > Get All Related Citations. 45. Click the radio button for all Reactions in the current hitset Q##. Click OK.
209 46. Click the View button. Reaction Searching 7-33
210 7-34 Searching CrossFire Databases- based on CrossFire Beilstein Retrieve examples of Knorr Pyrrol synthesis Exercise 2 Retrieve examples of Knorr Pyrrol synthesis by conducting a structural search. Use predefined generic groups to generalize the structure query. Use Grid display to view multiple reactions at the same time. Mark the reacting centers. Group the hitset by solvent. Prepare for a new search 1. Click the Query tab. 2. Click Clear Query. Launch the Structure Editor 3. Double-click the Structure/Reaction Search box. 4. Draw the reactants and product. 5. Click the Reaction Mode button. CrossFire Structure Editor instructions 6. Select one of the 2 reactants and click the Reactant button. Select the other reactant and click the Reactant button.
211 Reaction Searching Select the product and click the Product button. Map the Reaction 8. Click the Edit tool. Click the N in the reactant and draw a line to the N in the product. 9. Click the To Commander button. 10. Under Allow, check the boxes for addl. rings, isotopes, charges, and radicals.
212 7-36 Searching CrossFire Databases- based on CrossFire Beilstein 11. Click Start Search. 12. Click View and browse the hitset. 13. In Grid View, choose Options > Define Rows/Columns in Grid View. Specify 1 Rows and 1 Columns, and then click OK. 14. Choose Options > Define Structure/Reaction View. 15. Under Reaction Attributes, check Reaction Center and click OK. 16. View the reaction center markings on the results.
213 Reaction Searching 7-37 Group the hitset 17. Click the Sort/Group button. 18. Click the Group tab. 19. Use the drop-down list to select Solvent (RX.SOL).
214 7-38 Searching CrossFire Databases- based on CrossFire Beilstein 20. Notice the preview of the different groups of solvents. Accept the default Ascending order. Click OK. 21. Navigate the groups using the Tree View.
215 Reaction Searching 7-39 Search for synthesis of quinolines with a sulphuric acid reagent Exercise 3 Search for synthesis of quinolines with anilines and glycerol as the starting materials. Map the reaction. Mark the reacting centers. Create a data query to find sulphuric acid reagents. Make sure that your query will find different spellings and forms of sulphuric acid. Prepare for a new search 1. Click the Query tab. 2. Click the Clear Query button. Launch the Structure Editor 3. Double-click the Structure/Reaction Search box. 4. Draw the reactants and the product. 5. Using the Lasso tool, select the 4 carbons shown in the reactant below. Choose Options > Define Atom and choose Max for Free Sites. Repeat, using the 4 carbons in the product. CrossFire Structure Editor instructions
216 7-40 Searching CrossFire Databases- based on CrossFire Beilstein 6. Select Reaction Mode. Select the first reactant and click the Reactant button. Select the second reactant and click the Reactant button. Select the product and click the Product button. Map the reaction 7. Click the Edit tool, and draw lines to map the nitrogens and carbons as shown below. 8. Using the Edit tool, click the bond shown below and select Make/Break Bond from the Bond Change by Reaction dialog box.
217 Reaction Searching Click the To Commander button. Enter the data query 10. On the Search Fields tab, click to expand the Basic Indexes category. Double-click the Reaction Basic Index field. 11. In the Field content box, type sulphuric acid*. (Alternatively, you could set the operator to Starts with and remove the wildcard placed after acid.) 12. In the second row, choose or for the Operator. Press tab and type birea. Press Tab and type sulfuric acid* in the Field content box.
218 7-42 Searching CrossFire Databases- based on CrossFire Beilstein 13. In the third row, choose or for the Operator. Press tab and type birea. Press Tab and type h2so4* in the Field content box. Select a bracket before the first entry and after the last entry. 14. Click Start Search. 15. Click View.
219 Reaction Searching 7-43 Search for inversion reactions Exercise 4 Search for inversion reactions of 1-monosubstituted propan-2- ols. Specify stereo inversion and search with absolute stereo enabled. Browse your hitset and hyperlink to a reactant. Prepare for a new query 1. Click the Query tab. 2. Clear the previous query Launch the Structure Editor 3. Double-click the Structure/Reaction Search box. CrossFire Structure Editor instructions 4. Draw the structures. Set stereo inversion 5. Click to switch to Reaction Mode. Using the Select tool, select the reactant. Click the Reactant button. Select the product. Click the Product button. 6. Using the Edit tool, click the center carbon in the reactant. Select Invert in the Atom Attributes for Reaction box. Click OK. Repeat for the product.
220 7-44 Searching CrossFire Databases- based on CrossFire Beilstein 7. Click the To Commander icon. Enable absolute stereo 8. Under Stereo, choose absolute. 9. Choose Reactions next to As result I want to get. 10. Click the Start Search button. 11. Click View. Beilstein (2008/01) = 19. Click the Details button. 12. Click a hyperlink to a reactant.
221 Manipulating results 8-1 Manipulating results In this section, you will learn how to: Copy selected information from the Results tab Create a report using retrieved information Export retrieved information using the predefined export features Export retrieved information using the Export Wizard Print retrieved information from the Results tab In this section, you will learn how to copy selected information from the Results tab and view it in an external application. You will learn how to create a report, in Microsoft Excel or Microsoft Word, using specific retrieved facts. You will learn how to export using the predefined export settings. You will also learn how to export using the Export Wizard. You will learn how to print retrieved information.
222 8-2 Searching CrossFire Databases- based on CrossFire Beilstein Copy selected data Copy the Selected Facts button Check the box to select the fact. Click the Copy the Selected Facts button to open the menu. To copy a structure to an external application such as PowerPoint, select the structure and choose Edit> Copy. Copying data and creating a report is done in the Details view of the Results Tab. You can copy data and structures into WORD or an xml, html, or ASCII file by clicking the Copy the Selected Facts button. You can copy text to the clipboard by selecting the text and choosing Edit> Copy. You can copy a structure to the clipboard by selecting the structure that appears in a separate window (controlled by the Show button on the button bar) and choosing Edit> Copy.
223 Manipulating results 8-3 Viewing copied data To perform a direct copy into Microsoft Word, click the button shown above, and select Copy Selection to Word. When the Copy Selection to Word choice is selected, Microsoft Word automatically launches and displays the selected information in a formatted table.
224 8-4 Searching CrossFire Databases- based on CrossFire Beilstein Create a report Creating a report uses features similar to those used to copy data. Copies the selected facts to a new or existing xml file The citation for that fact is copied to a text file Change settings for report files, xml files, and citation files Opens MS Word and copies the selected facts Selects facts, starting with the current fact selected Copies a new selection to a previously defined HTML report Includes structures or reactions in your HTML Document (use bitmaps for MS Word, use molfiles for HTML) Creating a report uses features similar to those used to copy data. Choose Define Settings. prior to creating your report to specify the name and location of the report. Choosing the Commander> Reports folder will allow you to view your report from the Reports Tab within the Commander. You must select which facts to include. To do this, check the Select facts for printing or copying box in the appropriate data section. Then click the Copy the selected facts button.
225 Manipulating results 8-5 Create a report in Word Substance data, NMR, and IR facts selected to create a report in MS Word. Here is an example of a Microsoft Word report where we specified Substance data, NMR data, and IR data. When selected, the structure will appear below the initial substance fact table. Hyperlinks are active. When you click on the hyperlink for the BRN, the Commander will open and display the record for that BRN.
226 8-6 Searching CrossFire Databases- based on CrossFire Beilstein Create a report in the Commander From the Results tab, click Copy to Report. Switch to the Reports tab to view the report. NMR.html Choose Define Settings. prior to creating your report to specify the name and location of the report. Choosing the Commander> Reports folder will allow you to view your report from the Reports Tab within the Commander. Click the Copy the Selected Facts button or the Copy to button on the button bar to copy the selected facts to the.html. Click the Report tab to view the report created.
227 Manipulating results 8-7 Export Decide which records to include prior to starting the export Export record options: 1. Export without selecting hits 2. Export after selecting hits 3. Export after selecting hits and creating an export list To Create an Export List, select hits and click the View Selected hits Only button Another method for viewing and manipulating data is to export it to an external application. Decide which records you would like to export prior to starting your export. During the export you will be further prompted to select (depending on your selection prior to exporting): All of the records, the current record, selected hits only, or a range of records. For this example, view a substance hitset and select several substances. Click the View Selected Hits Only button.
228 8-8 Searching CrossFire Databases- based on CrossFire Beilstein Predefined export choices Click the Export Hits button Or Choose File > Export Hits Substance Context Reaction Context Citation Context A list of predefined export settings is available after clicking the Export Hits button, or choosing File > Export Hits. Predefined export settings depend on the available databases. They are also dependent on the current search context (Substances or Citations or Reactions). The export function enables you to save results as HTML documents. Structures within these HTML documents are displayed. Hyperlinks (Full text, BRN, GRN) are retained live during export.
229 Manipulating results 8-9 Predefined export process 1 File name: Compound ID to Table Select File Select Format Name the file 2 File description: Identification to Table 3 Name the newly exported File Exporting information using the predefined export settings can be done in 3 easy steps. First, click the Export Hits button, select an export setting, and click the Start Export button. Second, select a format, specify which records to include in the export, and click the Start button. Third, name the export file and click the Save button.
230 8-10 Searching CrossFire Databases- based on CrossFire Beilstein Create a new export file Click the Export Hits button Or Choose File > Export Hits Click New You don t always have to use the predefined export settings; you can create your own setting. Select File > Export Hits, or click the Export Hits button. Instead of choosing a setting in the list, click the New button. The Export Wizard will open and guide you through the process of defining a new export setting.
231 Manipulating results 8-11 Export Wizard Define Format The Define Format step allows you to choose the format in which to export. Allows you to start a program after the export is complete. You must select an Export Target. Selection will change according to the format chosen (above) In the Define Content step, you must define both the Export Format and the Export Target. You can also choose to start a program after the export is complete. The export formats define how the information is stored. The export target is the location where the information is stored. Program Starting Options allows you to start a program after the export is completed.
232 8-12 Searching CrossFire Databases- based on CrossFire Beilstein Export formats Export Format Structures Facts MS Word (as HTML file) Yes Yes Browser (HTML) Yes Yes MS Excel (as HTML file) Yes Yes ASCII No Yes Literature Management Systems No Yes BSD File Yes Yes SD File Yes Yes RD File Yes Yes Information is exported as shown in the slide. Please note that structures are not exported with Literature Management Systems or with ASCII files.
233 Manipulating results 8-13 Export target To File - Ask for Name: Causes Display Hits to present a file dialog box before the export begins. To File - Autocreate: Maintains the same file name originally entered, incrementing the extension automatically. To File: Allows user to define a specific file name for each export. Append: If checked, appends exported data to an existing export file. If unchecked, the file is overwritten. The Export Target must be defined. You can export to the clipboard only, to a file only, or to a file and the clipboard. To Clipboard only exports to the clipboard only. To export to a file, select To File and click the Change Settings button. The selections are shown in the slide.
234 8-14 Searching CrossFire Databases- based on CrossFire Beilstein Export Wizard Define Content Available to export MDL SD, MDL RD, or BSD defined formats. To export data, you must define the data fields. In this dialog box, you will define the User View. The user view is the data you want to export from a list of results. Check the appropriate structure box to export the structures in the desired format. If the chosen format does not allow storing structures, such as an ACSII file, then this check box will be grayed out. The Facts check box is used to export data. Before data can be exported, you need to define the fields from which the data should be taken. The list of fields to be exported is called an Export View.
235 Manipulating results 8-15 Selecting data fields Searches using keywords Copies selected fields Removes selected fields User views depend on the context of the query and the database for which the view is defined. Some predefined user views are provided. To view predefined user views, click the Select Facts to View button. Click a predefined user view and then click the Edit button. The fact fields selected for that user view will appear on the right side of the dialog box. You can add or delete fields and click OK to edit that user view or click Cancel to return to the previous dialog box. To create a new user view, click the Select Facts to View button. Click the New button, select fields, and click OK.
236 8-16 Searching CrossFire Databases- based on CrossFire Beilstein Save the User View Enter a file name and click Save. Select a file and click Apply. You are prompted to save the User View. Enter a file name and click Save. Choose the appropriate User View, select it, and click Apply.
237 Manipulating results 8-17 Changing field names Select the display. Type in a new name under My Field Name if you would like to change it. Many of the original field codes do not adequately describe the type of data they represent. To change the identification label of the exported data, click the Define Field Names button. Select the desired display for your label: Short, Long, or My Field Name. You can type in a new data label under My Field Name. Click OK.
238 8-18 Searching CrossFire Databases- based on CrossFire Beilstein Inclusion/exclusion of data Controls the number of multiple data occurrences. Check these boxes to include information in the export. Click to copy to a single file or multiple files. It is very difficult to predict how many times a fact is added to a given structure or reaction. For example, some structures might only have one melting point referenced, others 100. If you want to ensure that all facts are exported, choose All in the list box next to At most. It is recommended that you limit this to 10 or 15, or choose HitOnly, if a factual search was performed. Additional data can be exported without selecting the data fields. Field Availability and References are fields of the original database, which cannot be selected in the User View dialog box. If you want this data to be exported, you must check the appropriate box.
239 Manipulating results 8-19 Export Wizard Finish Summarizes your settings. Click Finish to confirm. Save the setting for future use. In this dialog box, you will view a summary of the settings that you defined. You can add a comment to describe the export setting. This comment will be visible when you select the export setting later. Click the Finish button. A dialog box opens that allows you to save the export settings. If an error occurred during the definition process, or if information is missing, then this will be displayed in red in the final wizard dialog box.
240 8-20 Searching CrossFire Databases- based on CrossFire Beilstein Start the export Select the newly-created setting The final step in the Export Wizard is to start the export. To start the export, click the name of the setting, and then click the Start button. You can choose to export all of the records in the current list, the current record, selected hits only, or a range of hits.
241 Manipulating results 8-21 Exported data We performed an HTML export. The data selected in the Define Export View dialog box appears. The structure display in an HTML document requires the MDL Chime application. This is included with the CrossFire Commander Installer, both on the CD-ROM and on the Commander download site.
242 8-22 Searching CrossFire Databases- based on CrossFire Beilstein Specify Print Options Click the Print button Or Choose File > Print Hits Defines which records to print. Defines what to include in the printout. To print, choose File > Print Hits, or click the Print Hits button. You can choose to print the given structures, all referenced structures, no structures, or the Field Availability list. For multiple records, mark the check box to start a new page for each hit. Click the Setup button to select the destination printer and its connection. Margins can be defined in cm or inch at the left, right, top, and bottom margin. The default is 2 cm. You can define a header and/or footer
DiscoveryGate SM Version 1.4 Participant s Guide
 Citation Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Citation Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Elsevier MDL 14600
Citation Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Citation Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Elsevier MDL 14600
Structure Searching in CrossFire Beilstein. DiscoveryGate SM Version 1.4 Participant s Guide
 Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Elsevier MDL 14600
Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Elsevier MDL 14600
Searching CrossFire Beilstein Using DiscoveryGate. DiscoveryGate Version 2.2 Participant s Guide
 Searching CrossFire Beilstein Using DiscoveryGate DiscoveryGate Version 2.2 Participant s Guide Searching CrossFire Beilstein using DiscoveryGate DiscoveryGate Version 2.2 Participant s Guide Elsevier
Searching CrossFire Beilstein Using DiscoveryGate DiscoveryGate Version 2.2 Participant s Guide Searching CrossFire Beilstein using DiscoveryGate DiscoveryGate Version 2.2 Participant s Guide Elsevier
ISIS/Draw "Quick Start"
 ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
How to Create a Substance Answer Set
 How to Create a Substance Answer Set Select among five search techniques to find substances Since substances can be described by multiple names or other characteristics, SciFinder gives you the flexibility
How to Create a Substance Answer Set Select among five search techniques to find substances Since substances can be described by multiple names or other characteristics, SciFinder gives you the flexibility
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Table of Contents. Scope of the Database 3 Searching by Structure 3. Searching by Substructure 4. Searching by Text 11
 Searrcchiing fforr Subssttanccess and Reaccttiionss iin Beiillsstteiin and Gmelliin 1 Table of Contents Scope of the Database 3 Searching by Structure 3 Introduction to the Structure Editor 3 Searching
Searrcchiing fforr Subssttanccess and Reaccttiionss iin Beiillsstteiin and Gmelliin 1 Table of Contents Scope of the Database 3 Searching by Structure 3 Introduction to the Structure Editor 3 Searching
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Reaxys Pipeline Pilot Components Installation and User Guide
 1 1 Reaxys Pipeline Pilot components for Pipeline Pilot 9.5 Reaxys Pipeline Pilot Components Installation and User Guide Version 1.0 2 Introduction The Reaxys and Reaxys Medicinal Chemistry Application
1 1 Reaxys Pipeline Pilot components for Pipeline Pilot 9.5 Reaxys Pipeline Pilot Components Installation and User Guide Version 1.0 2 Introduction The Reaxys and Reaxys Medicinal Chemistry Application
Searching Substances in Reaxys
 Searching Substances in Reaxys Learning Objectives Understand that substances in Reaxys have different sources (e.g., Reaxys, PubChem) and can be found in Document, Reaction and Substance Records Recognize
Searching Substances in Reaxys Learning Objectives Understand that substances in Reaxys have different sources (e.g., Reaxys, PubChem) and can be found in Document, Reaction and Substance Records Recognize
Searching CrossFire Gmelin
 Searching CrossFire Gmelin Training Guide CrossFire Commander Version 7.1 Searching CrossFire Gmelin CrossFire Commander Version 7.1 Training Guide CrossFire Software Copyright 1995-2008, Elsevier Information
Searching CrossFire Gmelin Training Guide CrossFire Commander Version 7.1 Searching CrossFire Gmelin CrossFire Commander Version 7.1 Training Guide CrossFire Software Copyright 1995-2008, Elsevier Information
E-EROS TUTORIAL Encyclopedia of Reagents for Organic Synthesis at the UIUC Chemistry Library
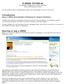 E-EROS TUTORIAL Encyclopedia of Reagents for Organic Synthesis at the UIUC Chemistry Library For any questions about e-eros please contact Tina Chrzastowski or call (217) 333-3737. Introduction About e-eros:
E-EROS TUTORIAL Encyclopedia of Reagents for Organic Synthesis at the UIUC Chemistry Library For any questions about e-eros please contact Tina Chrzastowski or call (217) 333-3737. Introduction About e-eros:
Basic Techniques in Structure and Substructure
 Truncating Molecules Basic Techniques in Structure and Substructure Searching for Information Professionals Judith Currano Head, Chemistry Library University of Pennsylvania currano@pobox.upenn.edu Acknowledgements
Truncating Molecules Basic Techniques in Structure and Substructure Searching for Information Professionals Judith Currano Head, Chemistry Library University of Pennsylvania currano@pobox.upenn.edu Acknowledgements
M E R C E R W I N WA L K T H R O U G H
 H E A L T H W E A L T H C A R E E R WA L K T H R O U G H C L I E N T S O L U T I O N S T E A M T A B L E O F C O N T E N T 1. Login to the Tool 2 2. Published reports... 7 3. Select Results Criteria...
H E A L T H W E A L T H C A R E E R WA L K T H R O U G H C L I E N T S O L U T I O N S T E A M T A B L E O F C O N T E N T 1. Login to the Tool 2 2. Published reports... 7 3. Select Results Criteria...
ON SITE SYSTEMS Chemical Safety Assistant
 ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
NMR Predictor. Introduction
 NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
ArcGIS 9 ArcGIS StreetMap Tutorial
 ArcGIS 9 ArcGIS StreetMap Tutorial Copyright 2001 2008 ESRI All Rights Reserved. Printed in the United States of America. The information contained in this document is the exclusive property of ESRI. This
ArcGIS 9 ArcGIS StreetMap Tutorial Copyright 2001 2008 ESRI All Rights Reserved. Printed in the United States of America. The information contained in this document is the exclusive property of ESRI. This
DECEMBER 2014 REAXYS R201 ADVANCED STRUCTURE SEARCHING
 DECEMBER 2014 REAXYS R201 ADVANCED STRUCTURE SEARCHING 1 NOTES ON REAXYS R201 THIS PRESENTATION COMMENTS AND SUMMARY Outlines how to: a. Perform Substructure and Similarity searches b. Use the functions
DECEMBER 2014 REAXYS R201 ADVANCED STRUCTURE SEARCHING 1 NOTES ON REAXYS R201 THIS PRESENTATION COMMENTS AND SUMMARY Outlines how to: a. Perform Substructure and Similarity searches b. Use the functions
ICM-Chemist How-To Guide. Version 3.6-1g Last Updated 12/01/2009
 ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
OECD QSAR Toolbox v.3.2. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.2 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.2 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
NOVEMBER 2014 REAXYS R101 BASIC REACTION QUERIES
 NOVEMBER 2014 REAXYS R101 BASIC REACTION QUERIES 1 NOTES ON REAXYS R101 BASIC REACTION QUERIES THIS PRESENTATION: REAXYS R101 BASIC REACTION QUERIES REAXYS R201 ADVANCED REACTION QUERIES Outlines the four
NOVEMBER 2014 REAXYS R101 BASIC REACTION QUERIES 1 NOTES ON REAXYS R101 BASIC REACTION QUERIES THIS PRESENTATION: REAXYS R101 BASIC REACTION QUERIES REAXYS R201 ADVANCED REACTION QUERIES Outlines the four
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
OECD QSAR Toolbox v.3.4. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.4.1. Step-by-step example for building QSAR model
 OECD QSAR Toolbox v.4.1 Step-by-step example for building QSAR model Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed to take
OECD QSAR Toolbox v.4.1 Step-by-step example for building QSAR model Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed to take
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
RADIATION PROCEDURES MANUAL Procedure Cover Sheet
 RADIATION PROCEDURES MANUAL Procedure Cover Sheet Procedure Title: Radioactive Material Inventory Procedure Number: TSO-09-16-REV 0 Effective Date: June 18, 2009 Approved By: Date: 11 August, 2009 Technical
RADIATION PROCEDURES MANUAL Procedure Cover Sheet Procedure Title: Radioactive Material Inventory Procedure Number: TSO-09-16-REV 0 Effective Date: June 18, 2009 Approved By: Date: 11 August, 2009 Technical
General Chemistry Lab Molecular Modeling
 PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
The Geodatabase Working with Spatial Analyst. Calculating Elevation and Slope Values for Forested Roads, Streams, and Stands.
 GIS LAB 7 The Geodatabase Working with Spatial Analyst. Calculating Elevation and Slope Values for Forested Roads, Streams, and Stands. This lab will ask you to work with the Spatial Analyst extension.
GIS LAB 7 The Geodatabase Working with Spatial Analyst. Calculating Elevation and Slope Values for Forested Roads, Streams, and Stands. This lab will ask you to work with the Spatial Analyst extension.
(THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE)
 PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
Defense Acquisition University
 STEP 1 STEP 2 STEP 3 STEP 4 STEP 5 STEP 6 Search For Training Take DAU Orientation Take Online Training Take Instructor-Led Training Edit Your User Record View Your Course Completions Defense Acquisition
STEP 1 STEP 2 STEP 3 STEP 4 STEP 5 STEP 6 Search For Training Take DAU Orientation Take Online Training Take Instructor-Led Training Edit Your User Record View Your Course Completions Defense Acquisition
SciFinder Scholar Guide to Getting Started
 SciFinder Scholar Guide to Getting Started What is SciFinder Scholar? SciFinder Scholar is the online version of Chemical Abstracts, covering chemistry publications worldwide from 1907 to the present.
SciFinder Scholar Guide to Getting Started What is SciFinder Scholar? SciFinder Scholar is the online version of Chemical Abstracts, covering chemistry publications worldwide from 1907 to the present.
Reaxys Medicinal Chemistry Fact Sheet
 R&D SOLUTIONS FOR PHARMA & LIFE SCIENCES Reaxys Medicinal Chemistry Fact Sheet Essential data for lead identification and optimization Reaxys Medicinal Chemistry empowers early discovery in drug development
R&D SOLUTIONS FOR PHARMA & LIFE SCIENCES Reaxys Medicinal Chemistry Fact Sheet Essential data for lead identification and optimization Reaxys Medicinal Chemistry empowers early discovery in drug development
Search for Substance Data
 Search for Substance Data Virtual class structure Part 1 Find property data for a substance Retrieve chemical supplier information for a substance Part 2 Import a structure and conduct an exact structure
Search for Substance Data Virtual class structure Part 1 Find property data for a substance Retrieve chemical supplier information for a substance Part 2 Import a structure and conduct an exact structure
Building Inflation Tables and CER Libraries
 Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Preparing a PDB File
 Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Elsevier R&D Solutions. Tool Sheet. Exploring a chemical reaction
 Elsevier R&D Solutions Exploring a chemical reaction Exploring a chemical reaction Use Ask Reaxys to explore reactions, substances and concepts Learning objectives Use Ask Reaxys as an entry point to browse
Elsevier R&D Solutions Exploring a chemical reaction Exploring a chemical reaction Use Ask Reaxys to explore reactions, substances and concepts Learning objectives Use Ask Reaxys as an entry point to browse
OECD QSAR Toolbox v.4.1. Tutorial on how to predict Skin sensitization potential taking into account alert performance
 OECD QSAR Toolbox v.4.1 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.1 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
Introduction to Spark
 1 As you become familiar or continue to explore the Cresset technology and software applications, we encourage you to look through the user manual. This is accessible from the Help menu. However, don t
1 As you become familiar or continue to explore the Cresset technology and software applications, we encourage you to look through the user manual. This is accessible from the Help menu. However, don t
Getting started with BatchReactor Example : Simulation of the Chlorotoluene chlorination
 Getting started with BatchReactor Example : Simulation of the Chlorotoluene chlorination 2011 ProSim S.A. All rights reserved. Introduction This document presents the different steps to follow in order
Getting started with BatchReactor Example : Simulation of the Chlorotoluene chlorination 2011 ProSim S.A. All rights reserved. Introduction This document presents the different steps to follow in order
OECD QSAR Toolbox v.4.0. Tutorial on how to predict Skin sensitization potential taking into account alert performance
 OECD QSAR Toolbox v.4.0 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.0 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
Planning Softproviding Meat User Documentation
 Great ideas are always simple Softproviding simply makes them happen. Planning Softproviding Meat User Documentation Version: 1.00 Date: 24 August 2017 Release: v5.50 Softproviding AG Riehenring 175 CH-4058
Great ideas are always simple Softproviding simply makes them happen. Planning Softproviding Meat User Documentation Version: 1.00 Date: 24 August 2017 Release: v5.50 Softproviding AG Riehenring 175 CH-4058
CHEMICAL INVENTORY ENTRY GUIDE
 CHEMICAL INVENTORY ENTRY GUIDE Version Date Comments 1 October 2013 Initial A. SUMMARY All chemicals located in research and instructional laboratories at George Mason University are required to be input
CHEMICAL INVENTORY ENTRY GUIDE Version Date Comments 1 October 2013 Initial A. SUMMARY All chemicals located in research and instructional laboratories at George Mason University are required to be input
Space Objects. Section. When you finish this section, you should understand the following:
 GOLDMC02_132283433X 8/24/06 2:21 PM Page 97 Section 2 Space Objects When you finish this section, you should understand the following: How to create a 2D Space Object and label it with a Space Tag. How
GOLDMC02_132283433X 8/24/06 2:21 PM Page 97 Section 2 Space Objects When you finish this section, you should understand the following: How to create a 2D Space Object and label it with a Space Tag. How
ST-Links. SpatialKit. Version 3.0.x. For ArcMap. ArcMap Extension for Directly Connecting to Spatial Databases. ST-Links Corporation.
 ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
2010 Autodesk, Inc. All rights reserved. NOT FOR DISTRIBUTION.
 Wastewater Profiles 2010 Autodesk, Inc. All rights reserved. NOT FOR DISTRIBUTION. The contents of this guide were created for Autodesk Topobase 2011. The contents of this guide are not intended for other
Wastewater Profiles 2010 Autodesk, Inc. All rights reserved. NOT FOR DISTRIBUTION. The contents of this guide were created for Autodesk Topobase 2011. The contents of this guide are not intended for other
Using SkyTools to log Texas 45 list objects
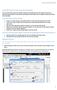 Houston Astronomical Society Using SkyTools to log Texas 45 list objects You can use SkyTools to keep track of objects observed in Columbus and copy the output into the Texas 45 observation log. Preliminary
Houston Astronomical Society Using SkyTools to log Texas 45 list objects You can use SkyTools to keep track of objects observed in Columbus and copy the output into the Texas 45 observation log. Preliminary
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build a userdefined
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build a userdefined QSAR Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build a userdefined QSAR Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed
Chem 253. Tutorial for Materials Studio
 Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
LAB 2 - ONE DIMENSIONAL MOTION
 Name Date Partners L02-1 LAB 2 - ONE DIMENSIONAL MOTION OBJECTIVES Slow and steady wins the race. Aesop s fable: The Hare and the Tortoise To learn how to use a motion detector and gain more familiarity
Name Date Partners L02-1 LAB 2 - ONE DIMENSIONAL MOTION OBJECTIVES Slow and steady wins the race. Aesop s fable: The Hare and the Tortoise To learn how to use a motion detector and gain more familiarity
SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide
 SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide This page is intentionally left blank. SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide The ACTiSys IR Programmer and SuperCELL
SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide This page is intentionally left blank. SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide The ACTiSys IR Programmer and SuperCELL
SeeSAR 7.1 Beginners Guide. June 2017
 SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SciFinder Scholar Guide to Getting Started
 SciFinder Scholar Guide to Getting Started What is SciFinder Scholar? SciFinder Scholar is the online version of Chemical Abstracts, covering chemistry publications worldwide from 1907 to the present.
SciFinder Scholar Guide to Getting Started What is SciFinder Scholar? SciFinder Scholar is the online version of Chemical Abstracts, covering chemistry publications worldwide from 1907 to the present.
Science of Synthesis Guided Examples
 Science of Synthesis Guided Examples 1 Science of Synthesis Guided Examples Compiled by: Dr. Thomas Krimmer Table of Contents 1 Exact Structure Search...2 2 Houben-Weyl...5 3 Reaction Search...6 4 Outbound
Science of Synthesis Guided Examples 1 Science of Synthesis Guided Examples Compiled by: Dr. Thomas Krimmer Table of Contents 1 Exact Structure Search...2 2 Houben-Weyl...5 3 Reaction Search...6 4 Outbound
Location Intelligence Infrastructure Asset Management. Confirm. Confirm Mapping Link to ArcMap Version v18.00b.am
 Location Intelligence Infrastructure Asset Management Confirm Confirm Mapping Link to ArcMap Version v18.00b.am Information in this document is subject to change without notice and does not represent a
Location Intelligence Infrastructure Asset Management Confirm Confirm Mapping Link to ArcMap Version v18.00b.am Information in this document is subject to change without notice and does not represent a
REPLACE DAMAGED OR MISSING TEXTBOOK BARCODE LABEL
 Destiny Textbook Manager allows users to create and print replacement barcode labels for textbooks. In this tutorial you will learn how to: Replace damaged textbook barcode label(s) Replace missing textbook
Destiny Textbook Manager allows users to create and print replacement barcode labels for textbooks. In this tutorial you will learn how to: Replace damaged textbook barcode label(s) Replace missing textbook
v Prerequisite Tutorials GSSHA WMS Basics Watershed Delineation using DEMs and 2D Grid Generation Time minutes
 v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
mylab: Chemical Safety Module Last Updated: January 19, 2018
 : Chemical Safety Module Contents Introduction... 1 Getting started... 1 Login... 1 Receiving Items from MMP Order... 3 Inventory... 4 Show me Chemicals where... 4 Items Received on... 5 All Items... 5
: Chemical Safety Module Contents Introduction... 1 Getting started... 1 Login... 1 Receiving Items from MMP Order... 3 Inventory... 4 Show me Chemicals where... 4 Items Received on... 5 All Items... 5
PDF-4+ Tools and Searches
 PDF-4+ Tools and Searches PDF-4+ 2018 The PDF-4+ 2018 database is powered by our integrated search display software. PDF-4+ 2018 boasts 72 search selections coupled with 125 display fields resulting in
PDF-4+ Tools and Searches PDF-4+ 2018 The PDF-4+ 2018 database is powered by our integrated search display software. PDF-4+ 2018 boasts 72 search selections coupled with 125 display fields resulting in
Lightcloud Application
 Controlling Your Lightcloud System Lightcloud Application Lightcloud Application Navigating the Application Devices Device Settings Organize Control Energy Scenes Schedules Demand Response Power Up State
Controlling Your Lightcloud System Lightcloud Application Lightcloud Application Navigating the Application Devices Device Settings Organize Control Energy Scenes Schedules Demand Response Power Up State
Chemistry 14CL. Worksheet for the Molecular Modeling Workshop. (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell)
 Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Version 1.2 October 2017 CSD v5.39
 Mogul Geometry Check Table of Contents Introduction... 2 Example 1. Using Mogul to assess intramolecular geometry... 3 Example 2. Using Mogul to explain activity data... 5 Conclusions... 8 Further Exercises...
Mogul Geometry Check Table of Contents Introduction... 2 Example 1. Using Mogul to assess intramolecular geometry... 3 Example 2. Using Mogul to explain activity data... 5 Conclusions... 8 Further Exercises...
Introduction to the Problem-Solving Methodology (PSM)
 Problem Name: Problem Description: Date: P01 Simple Reactor Introduction to the Problem-Solving Methodology (PSM) Your Name: Problem Session Objectives To learn the five stages of the problem solving methodology.
Problem Name: Problem Description: Date: P01 Simple Reactor Introduction to the Problem-Solving Methodology (PSM) Your Name: Problem Session Objectives To learn the five stages of the problem solving methodology.
Working with ArcGIS: Classification
 Working with ArcGIS: Classification 2 Abbreviations D-click R-click TOC Double Click Right Click Table of Content Introduction The benefit from the use of geographic information system (GIS) software is
Working with ArcGIS: Classification 2 Abbreviations D-click R-click TOC Double Click Right Click Table of Content Introduction The benefit from the use of geographic information system (GIS) software is
OECD QSAR Toolbox v.3.3. Predicting acute aquatic toxicity to fish of Dodecanenitrile (CAS ) taking into account tautomerism
 OECD QSAR Toolbox v.3.3 Predicting acute aquatic toxicity to fish of Dodecanenitrile (CAS 2437-25-4) taking into account tautomerism Outlook Background Objectives The exercise Workflow Save prediction
OECD QSAR Toolbox v.3.3 Predicting acute aquatic toxicity to fish of Dodecanenitrile (CAS 2437-25-4) taking into account tautomerism Outlook Background Objectives The exercise Workflow Save prediction
Cerno Application Note Extending the Limits of Mass Spectrometry
 Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Chemically Intelligent Experiment Data Management
 Chemically Intelligent Experiment Data Management Offering tools specifically designed to optimize the workflow of synthetic, medicinal, process and analytical chemists, the E-WorkBook Suite delivers a
Chemically Intelligent Experiment Data Management Offering tools specifically designed to optimize the workflow of synthetic, medicinal, process and analytical chemists, the E-WorkBook Suite delivers a
Ligand Scout Tutorials
 Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
PDF-4+ Tools and Searches
 PDF-4+ Tools and Searches PDF-4+ 2019 The PDF-4+ 2019 database is powered by our integrated search display software. PDF-4+ 2019 boasts 74 search selections coupled with 126 display fields resulting in
PDF-4+ Tools and Searches PDF-4+ 2019 The PDF-4+ 2019 database is powered by our integrated search display software. PDF-4+ 2019 boasts 74 search selections coupled with 126 display fields resulting in
CREATING CUSTOMIZED DATE RANGE COLLECTIONS IN PRESENTATION STUDIO
 CREATING CUSTOMIZED DATE RANGE COLLECTIONS IN PRESENTATION STUDIO Date range collections are pre-defined reporting periods for performance data. You have two options: Dynamic date ranges automatically
CREATING CUSTOMIZED DATE RANGE COLLECTIONS IN PRESENTATION STUDIO Date range collections are pre-defined reporting periods for performance data. You have two options: Dynamic date ranges automatically
SteelSmart System Cold Formed Steel Design Software Download & Installation Instructions
 Step 1 - Login or Create an Account at the ASI Portal: Login: https://portal.appliedscienceint.com/account/login Create Account: https://portal.appliedscienceint.com/account/register 2 0 1 7 A p p l i
Step 1 - Login or Create an Account at the ASI Portal: Login: https://portal.appliedscienceint.com/account/login Create Account: https://portal.appliedscienceint.com/account/register 2 0 1 7 A p p l i
Accountability. User Guide
 Accountability User Guide The information in this document is subject to change without notice and does not represent a commitment on the part of Horizon. The software described in this document is furnished
Accountability User Guide The information in this document is subject to change without notice and does not represent a commitment on the part of Horizon. The software described in this document is furnished
OECD QSAR Toolbox v.4.1
 OECD QSAR Toolbox v.4.1 Step-by-step example on how to predict the skin sensitisation potential approach of a chemical by read-across based on an analogue approach Outlook Background Objectives Specific
OECD QSAR Toolbox v.4.1 Step-by-step example on how to predict the skin sensitisation potential approach of a chemical by read-across based on an analogue approach Outlook Background Objectives Specific
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
DRUG DISCOVERY TODAY ELN ELN. Chemistry. Biology. Known ligands. DBs. Generate chemistry ideas. Check chemical feasibility In-house.
 DRUG DISCOVERY TODAY Known ligands Chemistry ELN DBs Knowledge survey Therapeutic target Generate chemistry ideas Check chemical feasibility In-house Analyze SAR Synthesize or buy Report Test Journals
DRUG DISCOVERY TODAY Known ligands Chemistry ELN DBs Knowledge survey Therapeutic target Generate chemistry ideas Check chemical feasibility In-house Analyze SAR Synthesize or buy Report Test Journals
GIS Workshop UCLS_Fall Forum 2014 Sowmya Selvarajan, PhD TABLE OF CONTENTS
 TABLE OF CONTENTS TITLE PAGE NO. 1. ArcGIS Basics I 2 a. Open and Save a Map Document 2 b. Work with Map Layers 2 c. Navigate in a Map Document 4 d. Measure Distances 4 2. ArcGIS Basics II 5 a. Work with
TABLE OF CONTENTS TITLE PAGE NO. 1. ArcGIS Basics I 2 a. Open and Save a Map Document 2 b. Work with Map Layers 2 c. Navigate in a Map Document 4 d. Measure Distances 4 2. ArcGIS Basics II 5 a. Work with
PDF-2 Tools and Searches
 PDF-2 Tools and Searches PDF-2 2019 The PDF-2 2019 database is powered by our integrated search display software. PDF-2 2019 boasts 69 search selections coupled with 53 display fields resulting in a nearly
PDF-2 Tools and Searches PDF-2 2019 The PDF-2 2019 database is powered by our integrated search display software. PDF-2 2019 boasts 69 search selections coupled with 53 display fields resulting in a nearly
ncounter PlexSet Data Analysis Guidelines
 ncounter PlexSet Data Analysis Guidelines NanoString Technologies, Inc. 530 airview Ave North Seattle, Washington 98109 USA Telephone: 206.378.6266 888.358.6266 E-mail: info@nanostring.com Molecules That
ncounter PlexSet Data Analysis Guidelines NanoString Technologies, Inc. 530 airview Ave North Seattle, Washington 98109 USA Telephone: 206.378.6266 888.358.6266 E-mail: info@nanostring.com Molecules That
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
OECD QSAR Toolbox v.3.0
 OECD QSAR Toolbox v.3.0 Step-by-step example of how to categorize an inventory by mechanistic behaviour of the chemicals which it consists Background Objectives Specific Aims Trend analysis The exercise
OECD QSAR Toolbox v.3.0 Step-by-step example of how to categorize an inventory by mechanistic behaviour of the chemicals which it consists Background Objectives Specific Aims Trend analysis The exercise
University of Colorado Denver Anschutz Medical Campus Online Chemical Inventory System User s Manual
 University of Colorado Denver Anschutz Medical Campus Online Chemical Inventory System User s Manual Hazardous Materials Division 303-724-0345 chemical.inventory@ucdenver.edu May, 2017 Table of Contents
University of Colorado Denver Anschutz Medical Campus Online Chemical Inventory System User s Manual Hazardous Materials Division 303-724-0345 chemical.inventory@ucdenver.edu May, 2017 Table of Contents
Information Dependent Acquisition (IDA) 1
 Information Dependent Acquisition (IDA) Information Dependent Acquisition (IDA) enables on the fly acquisition of MS/MS spectra during a chromatographic run. Analyst Software IDA is optimized to generate
Information Dependent Acquisition (IDA) Information Dependent Acquisition (IDA) enables on the fly acquisition of MS/MS spectra during a chromatographic run. Analyst Software IDA is optimized to generate
Computer simulation of radioactive decay
 Computer simulation of radioactive decay y now you should have worked your way through the introduction to Maple, as well as the introduction to data analysis using Excel Now we will explore radioactive
Computer simulation of radioactive decay y now you should have worked your way through the introduction to Maple, as well as the introduction to data analysis using Excel Now we will explore radioactive
OECD QSAR Toolbox v.3.3
 OECD QSAR Toolbox v.3.3 Step-by-step example on how to predict the skin sensitisation potential of a chemical by read-across based on an analogue approach Outlook Background Objectives Specific Aims Read
OECD QSAR Toolbox v.3.3 Step-by-step example on how to predict the skin sensitisation potential of a chemical by read-across based on an analogue approach Outlook Background Objectives Specific Aims Read
OECD QSAR Toolbox v.4.1. Tutorial of how to use Automated workflow for ecotoxicological prediction
 OECD QSAR Toolbox v.4.1 Tutorial of how to use Automated workflow for ecotoxicological prediction Outlook Aim Automated workflow The exercise Report The OECD QSAR Toolbox for Grouping Chemicals into Categories
OECD QSAR Toolbox v.4.1 Tutorial of how to use Automated workflow for ecotoxicological prediction Outlook Aim Automated workflow The exercise Report The OECD QSAR Toolbox for Grouping Chemicals into Categories
Software BioScout-Calibrator June 2013
 SARAD GmbH BioScout -Calibrator 1 Manual Software BioScout-Calibrator June 2013 SARAD GmbH Tel.: ++49 (0)351 / 6580712 Wiesbadener Straße 10 FAX: ++49 (0)351 / 6580718 D-01159 Dresden email: support@sarad.de
SARAD GmbH BioScout -Calibrator 1 Manual Software BioScout-Calibrator June 2013 SARAD GmbH Tel.: ++49 (0)351 / 6580712 Wiesbadener Straße 10 FAX: ++49 (0)351 / 6580718 D-01159 Dresden email: support@sarad.de
How do I do that in Quantum GIS: illustrating classic GIS tasks Edited by: Arthur J. Lembo, Jr.; Salisbury University
 How do I do that in Quantum GIS: illustrating classic GIS tasks Edited by: Arthur J. Lembo, Jr.; Salisbury University How do I do that in Quantum GIS Page 1 Introduction from the editor:... 4 Database
How do I do that in Quantum GIS: illustrating classic GIS tasks Edited by: Arthur J. Lembo, Jr.; Salisbury University How do I do that in Quantum GIS Page 1 Introduction from the editor:... 4 Database
CHEMDRAW ULTRA ITEC107 - Introduction to Computing for Pharmacy. ITEC107 - Introduction to Computing for Pharmacy 1
 CHEMDRAW ULTRA 12.0 ITEC107 - Introduction to Computing for Pharmacy 1 Objectives Basic drawing skills with ChemDraw Bonds, captions, hotkeys, chains, arrows Checking and cleaning up structures Chemical
CHEMDRAW ULTRA 12.0 ITEC107 - Introduction to Computing for Pharmacy 1 Objectives Basic drawing skills with ChemDraw Bonds, captions, hotkeys, chains, arrows Checking and cleaning up structures Chemical
NINE CHOICE SERIAL REACTION TIME TASK
 instrumentation and software for research NINE CHOICE SERIAL REACTION TIME TASK MED-STATE NOTATION PROCEDURE SOF-700RA-8 USER S MANUAL DOC-025 Rev. 1.3 Copyright 2013 All Rights Reserved MED Associates
instrumentation and software for research NINE CHOICE SERIAL REACTION TIME TASK MED-STATE NOTATION PROCEDURE SOF-700RA-8 USER S MANUAL DOC-025 Rev. 1.3 Copyright 2013 All Rights Reserved MED Associates
OECD QSAR Toolbox v.4.1. Tutorial illustrating new options for grouping with metabolism
 OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
OECD QSAR Toolbox v.3.4
 OECD QSAR Toolbox v.3.4 Step-by-step example on how to predict the skin sensitisation potential approach of a chemical by read-across based on an analogue approach Outlook Background Objectives Specific
OECD QSAR Toolbox v.3.4 Step-by-step example on how to predict the skin sensitisation potential approach of a chemical by read-across based on an analogue approach Outlook Background Objectives Specific
Molecular Modeling and Conformational Analysis with PC Spartan
 Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Data Structures & Database Queries in GIS
 Data Structures & Database Queries in GIS Objective In this lab we will show you how to use ArcGIS for analysis of digital elevation models (DEM s), in relationship to Rocky Mountain bighorn sheep (Ovis
Data Structures & Database Queries in GIS Objective In this lab we will show you how to use ArcGIS for analysis of digital elevation models (DEM s), in relationship to Rocky Mountain bighorn sheep (Ovis
Preparing Spatial Data
 13 CHAPTER 2 Preparing Spatial Data Assessing Your Spatial Data Needs 13 Assessing Your Attribute Data 13 Determining Your Spatial Data Requirements 14 Locating a Source of Spatial Data 14 Performing Common
13 CHAPTER 2 Preparing Spatial Data Assessing Your Spatial Data Needs 13 Assessing Your Attribute Data 13 Determining Your Spatial Data Requirements 14 Locating a Source of Spatial Data 14 Performing Common
Learning ArcGIS: Introduction to ArcCatalog 10.1
 Learning ArcGIS: Introduction to ArcCatalog 10.1 Estimated Time: 1 Hour Information systems help us to manage what we know by making it easier to organize, access, manipulate, and apply knowledge to the
Learning ArcGIS: Introduction to ArcCatalog 10.1 Estimated Time: 1 Hour Information systems help us to manage what we know by making it easier to organize, access, manipulate, and apply knowledge to the
A powerful site for all chemists CHOICE CRC Handbook of Chemistry and Physics
 Chemical Databases Online A powerful site for all chemists CHOICE CRC Handbook of Chemistry and Physics Combined Chemical Dictionary Dictionary of Natural Products Dictionary of Organic Dictionary of Drugs
Chemical Databases Online A powerful site for all chemists CHOICE CRC Handbook of Chemistry and Physics Combined Chemical Dictionary Dictionary of Natural Products Dictionary of Organic Dictionary of Drugs
Introduction to ArcMap
 Introduction to ArcMap ArcMap ArcMap is a Map-centric GUI tool used to perform map-based tasks Mapping Create maps by working geographically and interactively Display and present Export or print Publish
Introduction to ArcMap ArcMap ArcMap is a Map-centric GUI tool used to perform map-based tasks Mapping Create maps by working geographically and interactively Display and present Export or print Publish
OECD QSAR Toolbox v.3.4
 OECD QSAR Toolbox v.3.4 Predicting developmental and reproductive toxicity of Diuron (CAS 330-54-1) based on DART categorization tool and DART SAR model Outlook Background Objectives The exercise Workflow
OECD QSAR Toolbox v.3.4 Predicting developmental and reproductive toxicity of Diuron (CAS 330-54-1) based on DART categorization tool and DART SAR model Outlook Background Objectives The exercise Workflow
From BASIS DD to Barista Application in Five Easy Steps
 Y The steps are: From BASIS DD to Barista Application in Five Easy Steps By Jim Douglas our current BASIS Data Dictionary is perfect raw material for your first Barista-brewed application. Barista facilitates
Y The steps are: From BASIS DD to Barista Application in Five Easy Steps By Jim Douglas our current BASIS Data Dictionary is perfect raw material for your first Barista-brewed application. Barista facilitates
Structure Drawing. March Use the New Features. Keyboard Shortcuts and Paste from ChemDraw
 Use the New Features March 2010 SciFinder offers many new structure drawing, reaction search and display, answer set manipulation, and results postprocessing features and enhancements. Improved performance
Use the New Features March 2010 SciFinder offers many new structure drawing, reaction search and display, answer set manipulation, and results postprocessing features and enhancements. Improved performance
