Sequence context effect on the structure of nitrous acid induced DNA interstrand cross-links
|
|
|
- Laurel Wells
- 5 years ago
- Views:
Transcription
1 Published online May 20, 2004 Sequence context effect on the structure of nitrous acid induced DNA interstrand cross-links N. B. Fredrik Edfeldt*, Eric A. Harwood, Snorri Th. Sigurdsson, Paul B. Hopkins and Brian R. Reid Department of Chemistry, University of Washington, Seattle, WA 98195, USA Nucleic Acids Research, 2004, Vol. 32, No ±2801 DOI: /nar/gkh607 Received January 7, 2004; Revised and Accepted April 22, 2004 ABSTRACT In the preceding paper in this journal, we described the solution structure of the nitrous acid crosslinked dodecamer duplex [d(gcatccggatgc)] 2 (the cross-linked guanines are underlined). The structure revealed that the cross-linked guanines form a nearly planar covalently linked `G:G base pair', with the complementary partner cytidines ipped out of the helix. Here we explore the anking sequence context effect on the structure of nitrous acid cross-links in [d(cg)] 2 and the factors allowing the extrahelical cytidines to adopt such xed positions in the minor groove. We have used NMR spectroscopy to determine the solution structure of a second cross-linked dodecamer duplex, [d(cgctacgtagcg)] 2, which shows that the identity of the anking base pairs signi cantly alters the stacking patterns and phosphate backbone conformations. The cross-linked guanines are now stacked well on adenines preceding the extrahelical cytidines, illustrating the importance of purine± purine base stacking. Observation of an imino proton resonance at 15.6 p.p.m. provides evidence for hydrogen bonding between the two cross-linked guanines. Preliminary structural studies on the cross-linked duplex [d(cgcgacgtcgcg)] 2 show that the extrahelical cytidines are very mobile in this sequence context. We suggest that favorable van der Waals interactions between the cytidine and the adenine 2 bp away from the cross-link localize the cytidines in the previous cross-linked structures. INTRODUCTION In the preceding paper in this journal (1), we described the solution structure of the cross-linked dodecamer duplex [d(gcatccggatgc)] 2 (the cross-linked guanines are underlined), henceforth referred to as CCGG. We found that the cross-linked guanines form a nearly planar, covalently linked `G:G base pair', stacked on the 3 side guanines of the spatially adjacent G:C base pairs. The observed planar geometry of the G:G base pair is consistent with a single H1±N1 hydrogen bond, although this could not be established experimentally as the cross-linked guanine imino protons were absent from the NMR spectra. The cytidines, which normally would base pair with the cross-linked guanines, were found to be ipped out of the helix, adopting well de ned extrahelical positions in the minor groove. The phosphate backbone was found to be in the highly unusual e(g ± ) z(g + ) a(g + ) b(t) g(t) conformation on the 5 side of the extrahelical cytidine, causing a local strand reversal and directing the base out of the helix. Somewhat surprisingly, more modest deviations from idealized B-DNA dihedral angles were observed for the 3 side, all within the typical e(t) z(g ± ) a(g ± ) b(t) g(g + ) conformational domains, resulting in normal strand continuation. Extrahelical cytidines are well documented phenomena, and have previously been observed in cytosine bulges (2,3), C:C mismatches (4,5), a cisplatin-dna interstrand cross-link (6,7), and in DNA bound to proteins such as bacterial cytosine-speci c methyl transferase (8) and bacterial methylase (9). However, the particular location and lack of exibility of the cytosines in the minor groove, and the unusual phosphate backbone conformation are unique to this system. This paper describes a more detailed investigation of nitrous acid cross-linked DNA. First, it was of interest to determine if the planar G:G base pair and the extrahelical cytidines are general features of cross-links in [d(cg)] 2, and to investigate the anking sequence context effect on the structure. Secondly, we hoped to con rm experimentally the presence of a hydrogen bond between the two guanines within the cross-link. Thirdly, we wanted to learn more about the factors allowing the extrahelical cytidines to adopt such xed positions in the minor groove. Therefore, we decided to determine the structure of the cross-linked dodecamer duplex [d(cgctacgtagcg)] 2 (the cross-linked guanines are underlined), henceforth referred to as ACGT, with the crosslink in a [d(acgt)] 2 sequence and compare it to the structure of CCGG. The CCGG and ACGT sequences are the most susceptible to cross-linking in vitro (10). In addition, we performed preliminary structural characterization of the crosslinked duplex [d(cgcgacgtcgcg)] 2, which is analogous to ACGT but with anking G:C base pairs instead of T:A base *To whom correspondence should be addressed at present address: AstraZeneca Structural Chemistry Laboratory, AstraZeneca R&D MoÈlndal, , MoÈlndal, Sweden. Tel: ; Fax: ; fredrik.edfeldt@astrazeneca.com Present address: Eric A. Harwood, Chiron Corporation, 201 Elliott Avenue West, Suite 150, Seattle, WA 98119, USA Nucleic Acids Research, Vol. 32 No. 9 ã Oxford University Press 2004; all rights reserved
2 2796 Nucleic Acids Research, 2004, Vol. 32, No. 9 pairs (the T to G and A to C changes are underlined), henceforth referred to as GACGTC. The results from the study of GACGTC indicates that the second anking base pairs affect the mobility of the extrahelical cytidines. Shown in Figure 1 are the schematic representations of all three duplexes used in these studies. MATERIALS AND METHODS The cross-linked duplexes ACGT and GACGTC were synthesized as described previously (1,11). NMR samples were prepared, and NMR experiments were conducted, as in the previous paper in this journal (1). The solution structure of ACGT was determined using the distance geometry, restrained molecular dynamics and iterative NOE re nement protocol used for CCGG (1). Brie y, distance restraints for nonexchangeable protons were derived from 2 H 2 O-NOESY spectra collected at 25 C with 60, 120, 180, 240 and 360 ms mixing times, qualitative exchangeable proton restraints, including hydrogen bonding restraints, were derived from the 1 H 2 O-NOESY spectrum collected at 0 C, and phosphate backbone restraints were derived from the 2 H 2 O-NOESY, DQF-COSY and 1 H- 31 P HETCOR spectra (see Results). A total of 647 distance restraints were used, of which 215 were intraresidue, 432 interresidue, 52 hydrogen bonding, 42 derived from the 1 H 2 O-NOESY spectrum, and 81 were repulsive. 104 phosphate backbone dihedral angle restraints and 72 chiral constraints were also used. The phosphate backbone was restrained to: non-trans ( ) for a and z; trans ( ) for b and e (except C6 b, A5e and C6 e, which were not restrained); and gauche + ( ) for g [except C6 g, which was restrained to gauche ± (± )]. In each cycle the lowest energy structure was subjected to distance geometry/simulated annealing (DGII, Biosym/MSI), restrained molecular dynamics and energy minimization (DISCOVER, Biosym/MSI) resulting in a family of structures. The NOESY spectra of the resulting lowest energy structure were subsequently back-calculated using the NOESY simulation program BIRDER (12) with an empirically determined correlation time of 4.0 ns. The distance restraints were adjusted and the procedure repeated until the back-calculated spectra matched the experimental spectra, resulting in an R NOE factor of The nal set of 12 structures converged with pair wise RMS deviation values of AÊ, and low total and restraint violation energies. The cross-link was modeled with one guanine in the keto form with a single N1- imino proton and the other in the enol form, as was done previously for CCGG (1). As a test, in later stages of re nement, the cross-link was modeled with two N1-imino protons. However, these protons were consistently forced closer than their combined van der Waals radii (as we observed for CCGG). The presence of two G7/G7 N1-imino protons is clearly inconsistent with the experimental data. RESULTS Exchangeable proton studies of ACGT In the imino proton spectrum of ACGT shown in Figure 2A, there are ve peaks corresponding to the hydrogen bonded residues G2, T4, T8, G10 and G12 (unambiguously assigned Figure 1. Schematic representation of the three cross-linked self-complementary dodecamer duplexes with their abbreviated designations and residue numbering schemes. Although the duplexes are symmetrical, the residues of one of the strands are designated with a prime ( ) for convenience when describing interstrand interactions. Note, in particular, the crosslinked guanines (G7 and G7 ), and the cytosines preceding the cross-link, C6 and C6, which would normally base-pair with G7 and G7, respectively. using the 1 H 2 O-NOESY spectrum; data not shown). In addition, there is a broad peak at 15.6 p.p.m., which is only observed at very low temperature (0 C). This is most likely the G7 N1-imino proton resonance, although an unambiguous assignment could not be made in the 1 H 2 O-NOESY spectrum. Rapid solvent exchange is the probable cause of such a broad peak, and it could explain why this resonance did not give any crosspeaks other than a large solvent exchange peak at the water resonance in the 0 C 1 H 2 O-NOESY spectrum (data not shown). The chemical shift is highly anomalous for a guanine imino proton and ~2.5 p.p.m. further down eld than that of a typical hydrogen bonded guanine imino proton. However, it is clearly more consistent with a hydrogen bond than a lack thereof. Furthermore, this peak integrates to less than half of the others, which is expected for a single shared N1-imino proton. We suggest that this result is consistent with the G7 H1±G7 N1 hydrogen bond we observed in the solution structure of CCGG (1). Such a hydrogen bond requires that one of the guanines adopt the unusual enol tautomer, or more likely that each guanine alternates between the keto and enol forms as shown in Figure 2B. For normal guanines in aqueous solutions, the keto tautomer dominates over the enol tautomer due to more favorable hydration energy in spite of nearly equal intrinsic stability (13). However, a less hydrophilic environment, such as the interior of a protein binding pocket, could shift the equilibrium to favor the enol form (14). In this case, it could be that since the two guanines are covalently linked they are less susceptible to the typical `breathing' motions. This could make the microenvironment less hydrophilic which would indeed favor the presence of the enol form. Furthermore, since the guanine enol tautomer is fully aromatic, each guanine would be partially aromatic, and when bridged by the N2 lone pair, a large delocalized p electron cloud involving both purine rings would be created. We predict that this would result in a large deshielding ring current that could account for the large down eld shift of the G7 N1-imino proton. This type of keto-enol interconversion could also explain why, in spite of the apparent hydrogen bonding, the imino resonance is quite broad. If the imino proton is rst transferred to the spatially adjacent carbonyl
3 Nucleic Acids Research, 2004, Vol. 32, No Figure 2. (A) The down eld region (7±17 p.p.m.) of the 1D proton 750 MHz NMR spectrum of ACGT, collected in 90% 1 H 2 O/10% 2 H 2 Oat0 C. Note the broad G7 imino proton resonance. (B) The proposed interconversion between the enol and keto forms for each of the cross-linked guanines. The two guanines are planar, with a single shared imino proton and an H1±N1 hydrogen bond. oxygen (O6) as the guanine is converted to the enol form, it could subsequently be exchanged readily with the solvent from the hydroxyl position. The xed geometry of the crosslink should facilitate this process, in which the O6 would act as an intrinsic exchange catalyst (15). While this explanation accounts for the appearance of the G7 imino proton resonance in ACGT, it does not explain why the G7 resonance was not observed in CCGG. We conclude that for some unknown reason the exchange process is simply more ef cient in that duplex. Non-exchangeable proton studies of ACGT There are several features in H6/H8±H1 /H5 H6/H8±H2 1/ H2 2 regions of the 2 H 2 O-NOESY spectrum of ACGT, shown in Figure 3, that indicate an overall structural similarity with CCGG. The intensities of the intraresidue aromatic to H1 Figure 3. The 750 MHz 2 H 2 O-NOESY spectrum of ACGT, collected at 25 C with a mixing time of 360 ms. (A) The H6/H8±H1 /H5 region, showing the sequential aromatic to H1 interresidue walk is indicated with lines and the intraresidue H6/H8±H1 connectivities are labeled with the corresponding residue name and number. The absence of the C6 H1 ±G7 H8 and A5 H8±C6 H5 connectivities are marked X, and the C6 H5±A9 H2, A5 H1 ±G7 H8 and A5 H2±G7 H1 connectivities are labeled and indicated with arrows. (B) The H6/H8±H2 1/H2 2 region, showing the intraresidue aromatic to H2 1/H2 2 connectivities which are labeled and connected with lines. The sequential aromatic to H2 1/H2 2 interresidue walk for residues A5-G7 is indicated with dashed lines. The A5 H2 1±G7 H8 and A5 H2 2±G7 H8 connectivities are in a box. crosspeaks establish that all c torsion angles are in the typical anti conformation. The relatively weak intrasugar H6/H8±H3, H1 ±H4 and H2 2±H4 crosspeaks in the NOESY spectrum, and the strong intrasugar H1 ±H2 1 ( 3 J H1 ±H2 1 ) but weak H2 2±H3 ( 3 J H2 ±2H3 ) and H3 ±H4 ( 3 J H3 ±H4 ) crosspeaks in the DQF-COSY spectrum, suggest that all residues adopt typical C2 -endo type sugar conformations (data not shown). The cross-linked guanines appear to form a head to head
4 2798 Nucleic Acids Research, 2004, Vol. 32, No. 9 G:G base pair that is well stacked in the helix. The normal G7 H1 ±T8 H6, G7 H2 1±T8 H6, G7 H2 2±T8 H6 and G7 H8±T8 H6 connectivities (Fig. 3) indicate that the cross-linked G7 is stacked on the adjacent T8 base. As was the case in CCGG, the C6±G7 base±base stacking is disrupted, which is con rmed by the absence of C6 H1 ±G7 H8 and C6 H6±G7 H8 connectivities, and the weak C6 H2 1±G7 H8 and C6 H2 2±G7 H8 connectivities. The A5±C6 base±base stacking is also disrupted, as evidenced by the weak A5 H2 1±C6 H6 and A5 H2 2±C6 H6 connectivities, and the lack of A5 H8±C6 H5, A5 H8±C6 H6, and A5 H3 ±C6 H6 connectivities. The unusual interstrand C6 H5±A9 H2 and C6 H6±A9 H2 connectivities, previously seen in CCGG, con rm that the C6 base is ipped out of the helix and located in the minor groove. In spite of the numerous similarities with CCGG, a signi cant structural difference is evidenced by the unusual A5 H1 ±G7 H8, A5 H2±G7 H1, A5H2 1±G7 H8 and A5 H2 2±G7 H8 connectivities. These surprising crosspeaks are indicative of A5±G7 base±base stacking, which is in contrast to CCGG where no C5±G7 base stacking was observed. The phosphate backbone appears to adopt several unusual torsion angles, particularly in the A5±C6 step, and also exhibits some interesting differences compared to CCGG. For instance, the unusual C6 H5 1±G7 H8 and C6 H5 2±G7 H8 connectivities seen in CCGG are not observed, and unlike in CCGG, the intraresidue C6 H6±H5 1 connectivity is stronger than the intraresidue C6 H6±H5 2 connectivity (data not shown). The 1 H- 31 P HETCOR (see Supplementary Figure S1) and DQF-COSY (see Supplementary Figure S2) spectra are very useful in deriving backbone torsion angles. In particular, the b and g angles can be obtained from the intraresidue P±H4 crosspeak, the relative H4 ±H5 1 and H4 ±H5 2 crosspeaks, and the relative intraresidue P±H5 1 and P±H5 2 crosspeaks (16±19). The C6 P±H4 crosspeak is absent, indicating that one or both of the C6 b and g torsion angles adopt unusual conformations. Based on the strong H4 ±H5 1 crosspeak and absence of a H4 ±H5 2 crosspeak in the DQF-COSY spectrum (Supplementary Figure S2), C6 g adopts the unusual gauche ± conformation, which is also consistent with the relative intensities of the H3 ±H5 1, H3 ±H5 2, H4 ±H5 2 and C6 H4 ±H5 1 2 H 2 O-NOESY crosspeaks (data not shown). The relative intensities of the intraresidue P±H5 1 and P±H5 2 crosspeaks (P±H5 2 is more intense than P±H5 1; Supplementary Figure S1) are consistent with a C6 b angle in either the low end of the trans (120±150 ) or the gauche ± (±30 to ±60 ) conformation. The z and a angles are correlated to the phosphorus chemical shift. The C6 phosphorus chemical shift (±3.87) is at the low end of the normal range of ±3.8 to ±4.8 p.p.m. This appears to be inconsistent with unusual A5 z(t) and C6 a(t) torsion angles, as they would probably lead to a down eld shift of the C6 phosphorus resonance (20). Structure determination of ACGT The iterative relaxation matrix and back-calculation re nement process described in Materials and Methods yielded a set of 12 independently generated nal structures. The skeletal stereo view of the lowest energy structure is shown in Figure 4A. Overall the structure is very similar to that of CCGG, with the bases of residues C6 and C6 ipped out of the helix and residing in the minor groove, pointing towards the 5 end of the strand and with its hydrophobic (H5±H6) Figure 4. (A) The skeletal stereo view of the lowest energy structure of the nal set of re ned structures of ACGT, with the cross-linked guanines (G7 and G7 ) colored blue and the extrahelical cytidine residues (C6 and C6 ) in yellow. This view is looking into the narrow major groove at the cross-link, showing the nearly planar covalently linked G7:G7 base pair. The ribbon backbone trace is shown to emphasize the unusual major and minor groove widths. The 5 -ends of the two strands are top-right and bottom-left, respectively. (B) Detailed representation of the cross-link region of ACGT showing the A5:T8 base pair in red, the extrahelical C6 and C6 in yellow, the G7:G7 base pair in blue, and the T8:A5 base pair in orange. A top view (top) and a view into the minor groove (bottom) are shown. The G7:G7 base pair is well stacked on the spatially adjacent A5:T8 and T8:A5 base pairs. edge towards the core of the helix. The cross-linked guanines form a nearly planar covalently linked G7:G7 base pair with only minor propeller twisting, and they are stacked well on the spatially adjacent A5:T8 and T8:A5 base pairs. The minor groove is widened to accommodate the extrahelical cytidines, reaching a maximum of 7.2 AÊ, while the major groove is narrowed to just 7.0 AÊ at the cross-link as the two strands are
5 Nucleic Acids Research, 2004, Vol. 32, No Table 1. Backbone torsion angles (a±z) and glycosidic torsion angles (c) for the cross-linked ACGT duplex Residue a b g d e z c C1 ± ± ± ± ± ± G2 ± ± ± C3 ± ± ± ± T4 ± ± ± ± A5 ± ± ± C6 ± ± ± ± ± G7 ± ± ± T8 ± ± ± ± A9 ± ± ± ± G10 ± ± ± ± C11 ± ± ± ± G12 ± ± ± ± ± The angles are given in degrees ( ) and averaged over the nal 12 re ned structures. Unusual torsion angles are in bold. forced together by the short covalent linkage. A closer look at the cross-link region, shown in Figure 4B, reveals that the intrastrand base±base stacking is particularly good between the spatially adjacent purine rings of A5 and G7 (and between A5 and G7 ). There is a large helical twist of 50 between the spatially adjacent A5:T8 and G7:G7 base pairs (and between G7:G7 and T8:A5 ), due to the intervening backbone segment of the C6 residue (and C6 ). As a result, the duplex is not overall underwound as one might expect. The two cross-linked guanines exhibit a relatively minor propeller twist of ±24. The rise between the G7:G7 base pair and the adjacent A5:T8 and T8:A5 base pairs is 3.1 AÊ, which is typical for well stacked adjacent base pairs. All residues adopt C2 -endo type sugar conformations and anti glycosidic torsion angles, although the C6 c value is somewhat unusual (±71 ) compared to B-DNA. Shown in Table 1 are the torsion angles for the duplex. Non-exchangeable proton studies of GACGTC In both CCGG and ACGT the anking T:A base pairs are in contact with the extrahelical cytidines, resulting in a number of unusual intra- and interstrand NOEs. We wanted to determine if the identity of these anking base pairs is of importance to the location and exibility of the extrahelical cytidines by studying GACGTC, which has anking G:C base pairs. Shown in Figure 5 is the H6/H8±H1 /H5 region of the 2 H 2 O-NOESY spectrum of GACGTC. The diagnostic A5 H1 ±G7 H8 and A5 H2±G7 H1 connectivities, which were also observed in ACGT, are present, indicating that there is A5±G7 base±base stacking and that the C6 base is extrahelical in this duplex as well. However, the connectivities involving C6 H5 and H6 are all weak and quite broad, particularly the intraresidue C6 H6±H5 connectivity, which is much weaker than the other cytosine H5±H6 connectivities and the intraresidue C6 H6±H1 connectivity. This indicates that the extrahelical cytidines are more exible in this sequence context, and subject to conformational exchange which is intermediate on the proton chemical shift timescale. The relatively slow dynamics, in turn, suggest a signi cant structural rearrangement. This C6-speci c effect is observed both at higher and lower temperatures. We did not observe C6-speci c line broadening in the CCGG and ACGT duplexes at any temperature, which suggests that there is an intrinsic difference in the interaction of the extrahelical cytidines with the minor groove in GACGTC. The observed line broadening and exibility makes GACGTC unsuitable for structure determination. DISCUSSION Structural comparison between ACGT and CCGG Although the structures of ACGT and CCGG are similar overall, there are some very interesting structural differences. Listed in Table 2 are several of the unusual interproton distances observed in CCGG and ACGT, and in Table 3 the unusual phosphate backbone conformations. These features are also shown in Figure 6, which depicts the cross-link region of the two duplexes. Most notably, the importance of purine± purine base stacking is apparent. In CCGG the cross-linked guanines are stacked well on the 3 guanines of the adjacent G:C base pairs. In ACGT the stacking is still on the purine bases of the adjacent base pairs, but in this case that means the adenines preceding the extrahelical cytidines. Another notable difference lies in the phosphate backbone which adopts different conformations that, none the less, both result in an extrahelical cytidine. In ACGT the backbone adopts the unusual e(g ± ) z(t) a(g ± ) b(t) g(g ± ) conformation in the A5±C6 step, whereas the C5±C6 step in CCGG adopts the unusual e(g ± ) z(g + ) a(g + ) b(t) g(t) conformation. Interestingly, none of the dihedral angles in the C6±G7 step differ by more than 15 between ACGT and CCGG. They are all within the normal e(t) z(g ± ) a(g ± ) b(t) g (g + ) conformational domains, yet direct the next base (G7) back into the helix. The unusual A5/C5 e and z angles, which direct the backbone out of the helix, are essentially the same in both duplexes, although A5 z (ACGT) is in the low trans domain and C5 z (CCGG) in the high gauche ± domain. The fact that A5 a is in the very low end of the trans domain (136 ) might explain why the z(g ± ) a(t) conformation leads to such a minor phosphorus down eld shift in the spectrum of ACGT, although the C6 phosphorus chemical shift is indeed the furthest down eld of the phosphorus resonances. In both duplexes, the C6 a, b and g angles turn the C6 base into the minor groove. In ACGT, only the g angle is in an unusual conformation (gauche ± ), whereas in CCGG, both a and g are unusual (gauche + and trans, respectively). This difference manifests itself in the orientation of C6 H5 1 and H5 2: in ACGT they are pointing away
6 2800 Nucleic Acids Research, 2004, Vol. 32, No. 9 Figure 5. The H6/H8±H1 /H5 region of the 2 H 2 O-NOESY spectrum of GACGTC, collected at 25 C with a mixing time of 360 ms. The sequential aromatic to H1 interresidue walk is indicated with lines and the intraresidue H6/H8±H1 connectivities are labeled with the corresponding residue name and number. The A5 H1 ±G7 H8, A5 H2±G7 H1 and A5 H1 ±C6 H6 connectivities are labeled and in boxes. The absence of a C6 H1 ±G7 H8 connectivity is marked X. Table 2. A list of unusual interproton distances in ACGT and CCGG, compared with idealized B-DNA Proton pair ACGT CCGG Idealized B-DNA G7 H8 (A)±C6 H5 2 (B) 6.7 AÊ 4.2 AÊ >7 AÊ G7 H8 (A)±C6 H5 1 (C) 5.9 AÊ 4.9 AÊ >7 AÊ G7 H8 (A)±C5/A5 H1 (D) 3.8 AÊ 5.5 AÊ >7 AÊ G7 H8 (A)±C5/A5 H2 2 (E) 4.1 AÊ 6.5 AÊ >7 AÊ C6 H6 (F)±C6 H5 2 (B) 5.5 AÊ 3.3 AÊ ~4 AÊ C6 H6 (F)±C6 H5 1 (C) 4.1 AÊ 4.8 AÊ ~3.5 AÊ C6 H6 (F)±A9 H2 (H) 4.5 AÊ 4.7 AÊ >7 AÊ C6 H5 (G)±A9 H2 (H) 3.4 AÊ 3.2 AÊ >7 AÊ C6 H6 (F)±T4 H4 (I) 4.3 AÊ 3.4 AÊ >5.5 AÊ C6 H5 (G)±T4 H4 (I) 5.1 AÊ 3.8 AÊ >6 AÊ These protons are shown in Figure 6. from G7 H8; in CCGG they are pointing toward G7 H8. This also results in very different C6 H6±H5 2 distances. The b angle is normal in both duplexes, but the low end of trans (162 ) observed for ACGT is consistent with the J P±H5 1 and J P±H5 2 coupling data. An additional difference is that the C6 Figure 6. Comparison of the cross-link regions in ACGT (top) and CCGG (bottom), showing residues C3±G7, A9 and G10 of ACGT, and residues A3±G7, A9 and T10 of CCGG. Labeled in white are several protons giving rise to diagnostic NOEs: G7 H8 (A), C6 H5 1 (B), C6 H5 2 (C), C5/A5 H1 (D), C5/A5 H2 2 (E), C6 H6 (F), C6 H5 (G), A9 H2 (H) and C5/A5 H4 (I). Several of the corresponding interproton distances are shown in Table 2. Also labeled are the C5/A5 e and z dihedral angels (orange), and the C6 a and b angles (yellow), the values of which are shown in Table 3. base is pushed further down into the minor groove in ACGT, toward the C3:G10 base pair, although there are no NOE contacts with either of these residues. This does, however, lead to greater C6 H6±T4 H4 and C6 H5±T4 H4 interproton distances in ACGT. The particular ACGT backbone conformation effectively decreases the distance between the A5 and G7 bases allowing them to stack well on each other, which results in G7 H8±A5 H1 and G7 H8±A5 H2 2 distances that are much shorter than the corresponding G7 H8±C5 H1 and G7 H8±C5 H2 2 distances in CCGG. In both duplexes, the diagnostic interstrand C6 H5±A9 H2 and C6 H6±A9 H2 distances are virtually the same. Thus, this is an elegant Table 3. A list of the unusual dihedral angles and the conformational domains they adopt in ACGT and CCGG, compared with idealized B-DNA Dihedral angle ACGT CCGG Idealized B-DNA C5/A5 e Gauche ± (±86 ) Gauche ± (±75 ) Trans C5/A5 z High gauche + /low trans (136 ) High gauche + /low trans (110 ) Gauche ± C6 a Gauche ± (±83 ) Gauche + (65 ) Gauche ± C6 g Gauche ± (±35 ) Trans (165 ) Gauche + These dihedral angles are shown in Figure 6.
7 Nucleic Acids Research, 2004, Vol. 32, No example of how the phosphate backbone conformation is altered in order to accommodate the purine±purine stacking. Effects of anking sequence on mobility of extrahelical cytidines In both ACGT and CCGG, the extrahelical C6 base appears to be xed in the minor groove and lacking exibility, as evidenced by narrow spectral line widths and an apparent correlation time similar to that of the other cytidine bases (~4 ns). In CCGG the C6 amino group is close enough in space to the carbonyl oxygen of T4 to form an intrastrand hydrogen bond. However, such a hydrogen bond would form with less than ideal geometry, making it very weak at best. Furthermore, there is no experimental evidence for this hydrogen bond since the C6 amino protons are exchanging too rapidly with solvent to be observed (1). A hydrogen bonding cytidine amino proton should be protected from exchange. In ACGT the C6 amino group is not even within hydrogen bonding distance of the T4 base, and again the amino protons are not observed (data not shown). Thus, it seems that the stability of the cytidine in the minor groove must be derived from other interactions. For example, the adenine H2 in the minor groove could create a small hydrophobic patch into which the cytosine H5 and H6 can t snugly, resulting in favorable van der Waals interactions. As evidence for this hypothesis, we refer to GACGTC, which has a G4:C9 base pair instead of a T4:A9 base pair, and in which the extrahelical cytidines are quite mobile. It seems unlikely that this is a steric effect since the cytosine (C9 ) is smaller than the adenine (A9 ). On the other hand, this cytosine (C9 ) has a hydrophilic carbonyl oxygen roughly in the same location as the adenine (A9 ) H2. This would presumably prevent favorable van der Waals interactions with the hydrophobic edge of the extrahelical cytidine, and prevent it from tting snugly in the groove. Furthermore, although it intuitively appears that the xed location of the extrahelical cytidine would be entropically unfavorable, we suggest that the opposite might be true. If the C6 base were more exible and exposed to the solvent, a network of water molecules would most likely surround it, which could result in an even more unfavorable entropic contribution. Other possible factors explaining the xed location of the extrahelical cytidine could be that the geometry of the phosphate backbone simply will not allow this residues to move, and that the C6 c angle rotation is sterically hindered by the sides of the minor groove (the two backbones). These latter factors, however, would not explain the exibility observed in GACGTC. SUPPLEMENTARY MATERIAL Supplementary Material is available at NAR Online. ACKNOWLEDGEMENTS This work was supported in part by NIH grants GM to B.R.R. and GM to P.B.H. The coordinates have been deposited in the RSCB Protein Data Bank (Piscataway, NJ) with the accession no. 1S9O. REFERENCES 1. Edfeldt,N.B.F., Harwood,E.A., Sigurdsson,S.T., Hopkins,P.B. and Reid,B.R. (2004) Solution structure of a nitrous acid induced DNA interstrand cross-link. Nucleic Acids Res., 32, 2. Morden,K.M. and Maskos,K. (1993) NMR Studies of an extrahelical cytosine in an A. T rich region of a deoxyribodecanucleotide. Biopolymers, 33, 27± Moe,J.G., Reddy,G.R., Lawrence,J.M. and Stone,M.P. (1994) 1 H NMR Characterization of a duplex oligonucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG) 3 frameshift hotspot of Salmonella typhimuium hisd3052. Chem. Res. Toxicol., 7, 319± Gao,X., Huang,X., Smith,G.K., Zheng,M. and Liu,H. (1995) A new antiparallel duplex motif of DNA CCG repeats that is stabilized by extrahelical bases symmetrically located in the minor groove. J. Am. Chem. Soc., 117, 8883± Edfeldt,N.B.F. (2001) 2D NMR Studies of Unusual DNA Structures in Solution. PhD Thesis, University of Washington, Seattle, USA. 6. Huang,H., Zhu,L., Reid,B.R., Drobny,G.P. and Hopkins,P.B. (1995) Solution structure of a cisplatin-induced DNA interstrand cross-link. Science, 270, 1842± Coste,F., Malinge,J.-M., Serre,L., Shepard,W., Roth,M., Leng,M. and Zelwer C. (1999) Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 AÊ resolution: hydration at the platinated site. Nucleic Acids Res., 27, 1837± O'Gara,M., Horton,J.R., Roberts,R.J. and Cheng X. (1998) Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nature Struct. Biol., 5, 872± Reinisch,K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143± Kirchner,J.J. and Hopkins,P.B. (1991) Nitrous acid cross-links duplex DNA fragments through deoxyguanosine residues at the sequence 5 -CG. J. Am. Chem. Soc., 113, 4681± Harwood,E.A., Sigurdsson,S.T., Edfeldt,N.B.F., Reid,B.R. and Hopkins,P.B. (1999) Chemical synthesis and preliminary structural characterization of a nitrous acid interstrand cross-linked duplex DNA. J. Am. Chem. Soc., 121, 5081± Zhu,L. and Reid,B.R. (1995) An improved NOESY simulation program for partially relaxed spectra: BIRDER. J. Magn. Reson. Ser. B, 106, 227± Poltev,V.I., Malenkov,G.G., Gonzalez,E.J., Teplukhin,A.V., Rein,R., Shibata,M. and Miller,J.H. (1996) Modeling DNA hydration: comparision of calculated and experimental hydration properties of nucleic acid bases. J. Biomol. Struct. Dyn., 13, 717± Poltev,V.I., Gonzalez,E.J. and Teplukhin,A.V. (1995) The possible role of rare tautomers of DNA bases in mutagenesis: study of the effect of hydration on tautomeric equilibrium bt the Monte-Carlo method. Molekuliarnaia Biol., 29, 365± GueÂron,M. and Leroy,J.-L. (1992) Base-pair opening in double-stranded nucleic acids. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Heidelberg, Vol. 6, pp. 1± Sarma,R.H., Mynott,R.J., Wood,D.J. and Hruska,F.E. (1973) Determination of the preferred conformations constrained along the C-4 -C-5 and C-5 -O-5 bonds of b-5 -nucleotides in solution. J. Am. Chem. Soc., 95, 6457± Altona,C. (1982) Conformational analysis of nucleic acids. Determination of backbone geometry of single-helical RNA and DNA in aqueous solution. Recl. Trav. Chim. Pays-Bas, 101, 413± Kim,S.-G., Lin,L.-J. and Reid,B.R. (1992) Determination of nucleic acid backbone conformation by 1 H NMR. Biochemistry, 31, 3564± Chou,S.-H., Zhu,L., Gao,Z., Cheng,J.-W. and Reid,B.R. (1996) Hairpin loops consisting of single adenine residues closed by sheared A. A and G. G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(gtacaaagtac) hairpin. J. Mol. Biol., 264, 981± Gorenstein,D.G., Schroeder,S.A., Fu,J.M., Metz,J.T., Roongta,V. and Jones,C.L. (1988) Assignment of 31 P resonances in oligodeoxyribonucleotides: origin of sequence-speci c variations in the deoxyribose phosphate backbone conformation and the 31 P chemical shifts of double-helical nucleic acids. Biochemistry, 27, 7223±7237.
The wonderful world of NUCLEIC ACID NMR!
 Lecture 12 M230 Feigon Sequential resonance assignments in DNA (and RNA): homonuclear method 2 structure determination Reading resources Evans Chap 9 The wonderful world of NUCLEIC ACID NMR! Catalytically
Lecture 12 M230 Feigon Sequential resonance assignments in DNA (and RNA): homonuclear method 2 structure determination Reading resources Evans Chap 9 The wonderful world of NUCLEIC ACID NMR! Catalytically
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Resonance assignment and NMR spectra for hairpin and duplex A 6 constructs. (a) 2D HSQC spectra of hairpin construct (hp-a 6 -RNA) with labeled assignments. (b) 2D HSQC or SOFAST-HMQC
Supplementary Figure 1 Resonance assignment and NMR spectra for hairpin and duplex A 6 constructs. (a) 2D HSQC spectra of hairpin construct (hp-a 6 -RNA) with labeled assignments. (b) 2D HSQC or SOFAST-HMQC
NMR parameters intensity chemical shift coupling constants 1D 1 H spectra of nucleic acids and proteins
 Lecture #2 M230 NMR parameters intensity chemical shift coupling constants Juli Feigon 1D 1 H spectra of nucleic acids and proteins NMR Parameters A. Intensity (area) 1D NMR spectrum: integrated intensity
Lecture #2 M230 NMR parameters intensity chemical shift coupling constants Juli Feigon 1D 1 H spectra of nucleic acids and proteins NMR Parameters A. Intensity (area) 1D NMR spectrum: integrated intensity
NUCLEIC ACIDS. Basic terms and notions. Presentation by Eva Fadrná adapted by Radovan Fiala
 UCLEIC ACIDS Basic terms and notions Presentation by Eva Fadrná adapted by Radovan Fiala RA vs DA Single strand A-RA B-DA duplex Length of A Total length of DA in a human cell 1 m (1000 km) DA in typical
UCLEIC ACIDS Basic terms and notions Presentation by Eva Fadrná adapted by Radovan Fiala RA vs DA Single strand A-RA B-DA duplex Length of A Total length of DA in a human cell 1 m (1000 km) DA in typical
DNA Structure. Voet & Voet: Chapter 29 Pages Slide 1
 DNA Structure Voet & Voet: Chapter 29 Pages 1107-1122 Slide 1 Review The four DNA bases and their atom names The four common -D-ribose conformations All B-DNA ribose adopt the C2' endo conformation All
DNA Structure Voet & Voet: Chapter 29 Pages 1107-1122 Slide 1 Review The four DNA bases and their atom names The four common -D-ribose conformations All B-DNA ribose adopt the C2' endo conformation All
NMR of Nucleic Acids. K.V.R. Chary Workshop on NMR and it s applications in Biological Systems November 26, 2009
 MR of ucleic Acids K.V.R. Chary chary@tifr.res.in Workshop on MR and it s applications in Biological Systems TIFR ovember 26, 2009 ucleic Acids are Polymers of ucleotides Each nucleotide consists of a
MR of ucleic Acids K.V.R. Chary chary@tifr.res.in Workshop on MR and it s applications in Biological Systems TIFR ovember 26, 2009 ucleic Acids are Polymers of ucleotides Each nucleotide consists of a
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
SUPPLEMENTARY INFORMATION
 Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
I690/B680 Structural Bioinformatics Spring Protein Structure Determination by NMR Spectroscopy
 I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
Lecture 2-3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2-3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions Van der Waals Interactions
Lecture 2-3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions Van der Waals Interactions
SUPPLEMENTARY INFORMATION
 Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Dihedral Angles. Homayoun Valafar. Department of Computer Science and Engineering, USC 02/03/10 CSCE 769
 Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Ranjit P. Bahadur Assistant Professor Department of Biotechnology Indian Institute of Technology Kharagpur, India. 1 st November, 2013
 Hydration of protein-rna recognition sites Ranjit P. Bahadur Assistant Professor Department of Biotechnology Indian Institute of Technology Kharagpur, India 1 st November, 2013 Central Dogma of life DNA
Hydration of protein-rna recognition sites Ranjit P. Bahadur Assistant Professor Department of Biotechnology Indian Institute of Technology Kharagpur, India 1 st November, 2013 Central Dogma of life DNA
Nature Structural & Molecular Biology: doi: /nsmb
 Supplementary Table 1 Mean inclination angles a of wild-type and mutant K10 RNAs Base pair K10-wt K10-au-up K10-2gc-up K10-2gc-low K10-A-low 1-44 2.8 ± 0.7 4.8 ± 0.8 3.2 ± 0.6 4.3 ± 1.9 12.5 ± 0.4 2 43-2.5
Supplementary Table 1 Mean inclination angles a of wild-type and mutant K10 RNAs Base pair K10-wt K10-au-up K10-2gc-up K10-2gc-low K10-A-low 1-44 2.8 ± 0.7 4.8 ± 0.8 3.2 ± 0.6 4.3 ± 1.9 12.5 ± 0.4 2 43-2.5
Physiochemical Properties of Residues
 Physiochemical Properties of Residues Various Sources C N Cα R Slide 1 Conformational Propensities Conformational Propensity is the frequency in which a residue adopts a given conformation (in a polypeptide)
Physiochemical Properties of Residues Various Sources C N Cα R Slide 1 Conformational Propensities Conformational Propensity is the frequency in which a residue adopts a given conformation (in a polypeptide)
Introduction to Polymer Physics
 Introduction to Polymer Physics Enrico Carlon, KU Leuven, Belgium February-May, 2016 Enrico Carlon, KU Leuven, Belgium Introduction to Polymer Physics February-May, 2016 1 / 28 Polymers in Chemistry and
Introduction to Polymer Physics Enrico Carlon, KU Leuven, Belgium February-May, 2016 Enrico Carlon, KU Leuven, Belgium Introduction to Polymer Physics February-May, 2016 1 / 28 Polymers in Chemistry and
Introduction to Comparative Protein Modeling. Chapter 4 Part I
 Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Sequential Assignment Strategies in Proteins
 Sequential Assignment Strategies in Proteins NMR assignments in order to determine a structure by traditional, NOE-based 1 H- 1 H distance-based methods, the chemical shifts of the individual 1 H nuclei
Sequential Assignment Strategies in Proteins NMR assignments in order to determine a structure by traditional, NOE-based 1 H- 1 H distance-based methods, the chemical shifts of the individual 1 H nuclei
Microbiology with Diseases by Taxonomy, 5e (Bauman) Chapter 2 The Chemistry of Microbiology. 2.1 Multiple Choice Questions
 Microbiology with Diseases by Taxonomy, 5e (Bauman) Chapter 2 The Chemistry of Microbiology 2.1 Multiple Choice Questions 1) Which of the following does not contribute significantly to the mass of an atom?
Microbiology with Diseases by Taxonomy, 5e (Bauman) Chapter 2 The Chemistry of Microbiology 2.1 Multiple Choice Questions 1) Which of the following does not contribute significantly to the mass of an atom?
Model Worksheet Teacher Key
 Introduction Despite the complexity of life on Earth, the most important large molecules found in all living things (biomolecules) can be classified into only four main categories: carbohydrates, lipids,
Introduction Despite the complexity of life on Earth, the most important large molecules found in all living things (biomolecules) can be classified into only four main categories: carbohydrates, lipids,
Timescales of Protein Dynamics
 Timescales of Protein Dynamics From Henzler-Wildman and Kern, Nature 2007 Dynamics from NMR Show spies Amide Nitrogen Spies Report On Conformational Dynamics Amide Hydrogen Transverse Relaxation Ensemble
Timescales of Protein Dynamics From Henzler-Wildman and Kern, Nature 2007 Dynamics from NMR Show spies Amide Nitrogen Spies Report On Conformational Dynamics Amide Hydrogen Transverse Relaxation Ensemble
Section 10/5/06. Junaid Malek, M.D.
 Section 10/5/06 Junaid Malek, M.D. Equilibrium Occurs when two interconverting states are stable This is represented using stacked arrows: State A State B, where State A and State B can represent any number
Section 10/5/06 Junaid Malek, M.D. Equilibrium Occurs when two interconverting states are stable This is represented using stacked arrows: State A State B, where State A and State B can represent any number
2: CHEMICAL COMPOSITION OF THE BODY
 1 2: CHEMICAL COMPOSITION OF THE BODY Although most students of human physiology have had at least some chemistry, this chapter serves very well as a review and as a glossary of chemical terms. In particular,
1 2: CHEMICAL COMPOSITION OF THE BODY Although most students of human physiology have had at least some chemistry, this chapter serves very well as a review and as a glossary of chemical terms. In particular,
Timescales of Protein Dynamics
 Timescales of Protein Dynamics From Henzler-Wildman and Kern, Nature 2007 Summary of 1D Experiment time domain data Fourier Transform (FT) frequency domain data or Transverse Relaxation Ensemble of Nuclear
Timescales of Protein Dynamics From Henzler-Wildman and Kern, Nature 2007 Summary of 1D Experiment time domain data Fourier Transform (FT) frequency domain data or Transverse Relaxation Ensemble of Nuclear
Chapter 1. Topic: Overview of basic principles
 Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
Solution Structure of a Pyrimidine Purine Pyrimidine Triplex Containing the Sequence-specific Intercalating Non-natural Base D 3
 J. Mol. Biol. (1996) 257, 1052 1069 Solution Structure of a Pyrimidine Purine Pyrimidine Triplex Containing the Sequence-specific Intercalating Non-natural Base D 3 Edmond Wang 1, Karl M. Koshlap 1, Paul
J. Mol. Biol. (1996) 257, 1052 1069 Solution Structure of a Pyrimidine Purine Pyrimidine Triplex Containing the Sequence-specific Intercalating Non-natural Base D 3 Edmond Wang 1, Karl M. Koshlap 1, Paul
Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination
 Lecture 9 M230 Feigon Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination Reading resources v Roberts NMR of Macromolecules, Chap 4 by Christina Redfield
Lecture 9 M230 Feigon Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination Reading resources v Roberts NMR of Macromolecules, Chap 4 by Christina Redfield
BIOCHEMISTRY GUIDED NOTES - AP BIOLOGY-
 BIOCHEMISTRY GUIDED NOTES - AP BIOLOGY- ELEMENTS AND COMPOUNDS - anything that has mass and takes up space. - cannot be broken down to other substances. - substance containing two or more different elements
BIOCHEMISTRY GUIDED NOTES - AP BIOLOGY- ELEMENTS AND COMPOUNDS - anything that has mass and takes up space. - cannot be broken down to other substances. - substance containing two or more different elements
1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI )
 Uses of NMR: 1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI ) 3) NMR is used as a method for determining of protein, DNA,
Uses of NMR: 1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI ) 3) NMR is used as a method for determining of protein, DNA,
1. (5) Draw a diagram of an isomeric molecule to demonstrate a structural, geometric, and an enantiomer organization.
 Organic Chemistry Assignment Score. Name Sec.. Date. Working by yourself or in a group, answer the following questions about the Organic Chemistry material. This assignment is worth 35 points with the
Organic Chemistry Assignment Score. Name Sec.. Date. Working by yourself or in a group, answer the following questions about the Organic Chemistry material. This assignment is worth 35 points with the
Molecular Modeling lecture 2
 Molecular Modeling 2018 -- lecture 2 Topics 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction 4. Where do protein structures come from? X-ray crystallography
Molecular Modeling 2018 -- lecture 2 Topics 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction 4. Where do protein structures come from? X-ray crystallography
Introduction solution NMR
 2 NMR journey Introduction solution NMR Alexandre Bonvin Bijvoet Center for Biomolecular Research with thanks to Dr. Klaartje Houben EMBO Global Exchange course, IHEP, Beijing April 28 - May 5, 20 3 Topics
2 NMR journey Introduction solution NMR Alexandre Bonvin Bijvoet Center for Biomolecular Research with thanks to Dr. Klaartje Houben EMBO Global Exchange course, IHEP, Beijing April 28 - May 5, 20 3 Topics
Supersecondary Structures (structural motifs)
 Supersecondary Structures (structural motifs) Various Sources Slide 1 Supersecondary Structures (Motifs) Supersecondary Structures (Motifs): : Combinations of secondary structures in specific geometric
Supersecondary Structures (structural motifs) Various Sources Slide 1 Supersecondary Structures (Motifs) Supersecondary Structures (Motifs): : Combinations of secondary structures in specific geometric
ELECTRONIC SUPPLEMENTARY INFORMATION
 This journal is The Royal Society of Chemistry 1 ELECTRONIC SUPPLEMENTARY INFORMATION More than one non-canonical phosphodiester bond into the G-tract: formation of unusual parallel G-quadruplex structures
This journal is The Royal Society of Chemistry 1 ELECTRONIC SUPPLEMENTARY INFORMATION More than one non-canonical phosphodiester bond into the G-tract: formation of unusual parallel G-quadruplex structures
Why Is Molecular Interaction Important in Our Life
 Why Is Molecular Interaction Important in ur Life QuLiS and Graduate School of Science iroshima University http://www.nabit.hiroshima-u.ac.jp/iwatasue/indexe.htm Suehiro Iwata Sept. 29, 2007 Department
Why Is Molecular Interaction Important in ur Life QuLiS and Graduate School of Science iroshima University http://www.nabit.hiroshima-u.ac.jp/iwatasue/indexe.htm Suehiro Iwata Sept. 29, 2007 Department
Principles of Physical Biochemistry
 Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Useful background reading
 Overview of lecture * General comment on peptide bond * Discussion of backbone dihedral angles * Discussion of Ramachandran plots * Description of helix types. * Description of structures * NMR patterns
Overview of lecture * General comment on peptide bond * Discussion of backbone dihedral angles * Discussion of Ramachandran plots * Description of helix types. * Description of structures * NMR patterns
Structure Determination by NMR Spectroscopy #3-1
 Structure Determination by NMR Spectroscopy #3-1 3. Computational methods in NMR-spectroscopy 3.1 Nomenclature of proteins and nucleic acids 3.1.1. Classification of structural relationships between molecules
Structure Determination by NMR Spectroscopy #3-1 3. Computational methods in NMR-spectroscopy 3.1 Nomenclature of proteins and nucleic acids 3.1.1. Classification of structural relationships between molecules
Chapter 25 Organic and Biological Chemistry
 Chapter 25 Organic and Biological Chemistry Organic Chemistry The chemistry of carbon compounds. Carbon has the ability to form long chains. Without this property, large biomolecules such as proteins,
Chapter 25 Organic and Biological Chemistry Organic Chemistry The chemistry of carbon compounds. Carbon has the ability to form long chains. Without this property, large biomolecules such as proteins,
Protein Structure Determination Using NMR Restraints BCMB/CHEM 8190
 Protein Structure Determination Using NMR Restraints BCMB/CHEM 8190 Programs for NMR Based Structure Determination CNS - Brünger, A. T.; Adams, P. D.; Clore, G. M.; DeLano, W. L.; Gros, P.; Grosse-Kunstleve,
Protein Structure Determination Using NMR Restraints BCMB/CHEM 8190 Programs for NMR Based Structure Determination CNS - Brünger, A. T.; Adams, P. D.; Clore, G. M.; DeLano, W. L.; Gros, P.; Grosse-Kunstleve,
Introduction to" Protein Structure
 Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10657 Supplementary Text Introduction. All retroviruses contain three genes: namely, gag, pol and env, which code for structural, enzymatic and glycoprotein receptor proteins, respectively.
doi:10.1038/nature10657 Supplementary Text Introduction. All retroviruses contain three genes: namely, gag, pol and env, which code for structural, enzymatic and glycoprotein receptor proteins, respectively.
NMR, X-ray Diffraction, Protein Structure, and RasMol
 NMR, X-ray Diffraction, Protein Structure, and RasMol Introduction So far we have been mostly concerned with the proteins themselves. The techniques (NMR or X-ray diffraction) used to determine a structure
NMR, X-ray Diffraction, Protein Structure, and RasMol Introduction So far we have been mostly concerned with the proteins themselves. The techniques (NMR or X-ray diffraction) used to determine a structure
2) Matter composed of a single type of atom is known as a(n) 2) A) element. B) mineral. C) electron. D) compound. E) molecule.
 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Which of the following is a particle found in the nucleus of an atom and that has no electrical
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Which of the following is a particle found in the nucleus of an atom and that has no electrical
Structures of the Molecular Components in DNA and RNA with Bond Lengths Interpreted as Sums of Atomic Covalent Radii
 The Open Structural Biology Journal, 2008, 2, 1-7 1 Structures of the Molecular Components in DNA and RNA with Bond Lengths Interpreted as Sums of Atomic Covalent Radii Raji Heyrovska * Institute of Biophysics
The Open Structural Biology Journal, 2008, 2, 1-7 1 Structures of the Molecular Components in DNA and RNA with Bond Lengths Interpreted as Sums of Atomic Covalent Radii Raji Heyrovska * Institute of Biophysics
in Halogen-Bonded Complexes
 9 Resonance Assistance and Cooperativity in Halogen-Bonded Complexes Previously appeared as Covalency in Resonance-Assisted Halogen Bonds Demonstrated with Cooperativity in N-Halo-Guanine Quartets L. P.
9 Resonance Assistance and Cooperativity in Halogen-Bonded Complexes Previously appeared as Covalency in Resonance-Assisted Halogen Bonds Demonstrated with Cooperativity in N-Halo-Guanine Quartets L. P.
 There are two types of polysaccharides in cell: glycogen and starch Starch and glycogen are polysaccharides that function to store energy Glycogen Glucose obtained from primary sources either remains soluble
There are two types of polysaccharides in cell: glycogen and starch Starch and glycogen are polysaccharides that function to store energy Glycogen Glucose obtained from primary sources either remains soluble
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Resonance assignments in proteins. Christina Redfield
 Resonance assignments in proteins Christina Redfield 1. Introduction The assignment of resonances in the complex NMR spectrum of a protein is the first step in any study of protein structure, function
Resonance assignments in proteins Christina Redfield 1. Introduction The assignment of resonances in the complex NMR spectrum of a protein is the first step in any study of protein structure, function
Secondary and sidechain structures
 Lecture 2 Secondary and sidechain structures James Chou BCMP201 Spring 2008 Images from Petsko & Ringe, Protein Structure and Function. Branden & Tooze, Introduction to Protein Structure. Richardson, J.
Lecture 2 Secondary and sidechain structures James Chou BCMP201 Spring 2008 Images from Petsko & Ringe, Protein Structure and Function. Branden & Tooze, Introduction to Protein Structure. Richardson, J.
F. Piazza Center for Molecular Biophysics and University of Orléans, France. Selected topic in Physical Biology. Lecture 1
 Zhou Pei-Yuan Centre for Applied Mathematics, Tsinghua University November 2013 F. Piazza Center for Molecular Biophysics and University of Orléans, France Selected topic in Physical Biology Lecture 1
Zhou Pei-Yuan Centre for Applied Mathematics, Tsinghua University November 2013 F. Piazza Center for Molecular Biophysics and University of Orléans, France Selected topic in Physical Biology Lecture 1
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability
 Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Hydrogen and hydration of DNA and RNA oligonucleotides
 Biophysical Chemistry 95 (2002) 273 282 Hydrogen and hydration of DNA and RNA oligonucleotides Muttaiya Sundaralingam*, Baocheng Pan Biological Macromolecular Structure Center, Departments of Chemistry,
Biophysical Chemistry 95 (2002) 273 282 Hydrogen and hydration of DNA and RNA oligonucleotides Muttaiya Sundaralingam*, Baocheng Pan Biological Macromolecular Structure Center, Departments of Chemistry,
NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B-NS3 protease
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
J. Isaksson, E. Zamaratski, T. V. Maltseva, P. Agback, A. Kumar, and J. Chattopadhyaya*
 Journal of Biomolecular Structure & Dynamics, ISSN 0739-1102 Volume 18, Issue Number 6, (2001) Adenine Press (2001) The First Example of a Hoogsteen Basepaired DNA Duplex in Dynamic Equilibrium with a
Journal of Biomolecular Structure & Dynamics, ISSN 0739-1102 Volume 18, Issue Number 6, (2001) Adenine Press (2001) The First Example of a Hoogsteen Basepaired DNA Duplex in Dynamic Equilibrium with a
Biomolecules: lecture 10
 Biomolecules: lecture 10 - understanding in detail how protein 3D structures form - realize that protein molecules are not static wire models but instead dynamic, where in principle every atom moves (yet
Biomolecules: lecture 10 - understanding in detail how protein 3D structures form - realize that protein molecules are not static wire models but instead dynamic, where in principle every atom moves (yet
A Dimeric DNA Interface Stabilized by Stacked A(GGGG)A Hexads and Coordinated Monovalent Cations
 doi:10.1006/jmbi.2000.3524 available online at http://www.idealibrary.com on J. Mol. Biol. (2000) 297, 627±644 A Dimeric DNA Interface Stabilized by Stacked A(GGGG)A Hexads and Coordinated Monovalent Cations
doi:10.1006/jmbi.2000.3524 available online at http://www.idealibrary.com on J. Mol. Biol. (2000) 297, 627±644 A Dimeric DNA Interface Stabilized by Stacked A(GGGG)A Hexads and Coordinated Monovalent Cations
Conformational Geometry of Peptides and Proteins:
 Conformational Geometry of Peptides and Proteins: Before discussing secondary structure, it is important to appreciate the conformational plasticity of proteins. Each residue in a polypeptide has three
Conformational Geometry of Peptides and Proteins: Before discussing secondary structure, it is important to appreciate the conformational plasticity of proteins. Each residue in a polypeptide has three
Conformational Analysis
 Conformational Analysis C01 3 C C 3 is the most stable by 0.9 kcal/mole C02 K eq = K 1-1 * K 2 = 0.45-1 * 0.048 = 0.11 C04 The intermediate in the reaction of 2 has an unfavorable syn-pentane interaction,
Conformational Analysis C01 3 C C 3 is the most stable by 0.9 kcal/mole C02 K eq = K 1-1 * K 2 = 0.45-1 * 0.048 = 0.11 C04 The intermediate in the reaction of 2 has an unfavorable syn-pentane interaction,
PROTEIN'STRUCTURE'DETERMINATION'
 PROTEIN'STRUCTURE'DETERMINATION' USING'NMR'RESTRAINTS' BCMB/CHEM'8190' Programs for NMR Based Structure Determination CNS - Brünger, A. T.; Adams, P. D.; Clore, G. M.; DeLano, W. L.; Gros, P.; Grosse-Kunstleve,
PROTEIN'STRUCTURE'DETERMINATION' USING'NMR'RESTRAINTS' BCMB/CHEM'8190' Programs for NMR Based Structure Determination CNS - Brünger, A. T.; Adams, P. D.; Clore, G. M.; DeLano, W. L.; Gros, P.; Grosse-Kunstleve,
A prevalent intraresidue hydrogen bond stabilizes proteins
 Supplementary Information A prevalent intraresidue hydrogen bond stabilizes proteins Robert W. Newberry 1 & Ronald T. Raines 1,2 * 1 Department of Chemistry and 2 Department of Biochemistry, University
Supplementary Information A prevalent intraresidue hydrogen bond stabilizes proteins Robert W. Newberry 1 & Ronald T. Raines 1,2 * 1 Department of Chemistry and 2 Department of Biochemistry, University
The Molecules of Life Chapter 2
 The Molecules of Life Chapter 2 Core concepts 1.The atom is the fundamental unit of matter. 2.Atoms can combine to form molecules linked by chemical bonds. 3.Water is essential for life. 4.Carbon is the
The Molecules of Life Chapter 2 Core concepts 1.The atom is the fundamental unit of matter. 2.Atoms can combine to form molecules linked by chemical bonds. 3.Water is essential for life. 4.Carbon is the
BMB/Bi/Ch 173 Winter 2018
 BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4:00-5:00pm and SFL 229, Monday 3/5 4:00-5:30pm.
BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4:00-5:00pm and SFL 229, Monday 3/5 4:00-5:30pm.
Basic One- and Two-Dimensional NMR Spectroscopy
 Horst Friebolin Basic One- and Two-Dimensional NMR Spectroscopy Third Revised Edition Translated by Jack K. Becconsall WILEY-VCH Weinheim New York Chichester Brisbane Singapore Toronto Contents XV 1 The
Horst Friebolin Basic One- and Two-Dimensional NMR Spectroscopy Third Revised Edition Translated by Jack K. Becconsall WILEY-VCH Weinheim New York Chichester Brisbane Singapore Toronto Contents XV 1 The
Supporting Information
 Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Supplementary information Silver (I) as DNA glue: Ag + - mediated guanine pairing revealed by removing Watson- Crick constraints
 Supplementary information Silver (I) as DNA glue: Ag + - mediated guanine pairing revealed by removing Watson- Crick constraints Steven M. Swasey [b], Leonardo Espinosa Leal [c], Olga Lopez- Acevedo [c],
Supplementary information Silver (I) as DNA glue: Ag + - mediated guanine pairing revealed by removing Watson- Crick constraints Steven M. Swasey [b], Leonardo Espinosa Leal [c], Olga Lopez- Acevedo [c],
Chem. 27 Section 1 Conformational Analysis Week of Feb. 6, TF: Walter E. Kowtoniuk Mallinckrodt 303 Liu Laboratory
 Chem. 27 Section 1 Conformational Analysis TF: Walter E. Kowtoniuk wekowton@fas.harvard.edu Mallinckrodt 303 Liu Laboratory ffice hours are: Monday and Wednesday 3:00-4:00pm in Mallinckrodt 303 Course
Chem. 27 Section 1 Conformational Analysis TF: Walter E. Kowtoniuk wekowton@fas.harvard.edu Mallinckrodt 303 Liu Laboratory ffice hours are: Monday and Wednesday 3:00-4:00pm in Mallinckrodt 303 Course
Biophysics II. Hydrophobic Bio-molecules. Key points to be covered. Molecular Interactions in Bio-molecular Structures - van der Waals Interaction
 Biophysics II Key points to be covered By A/Prof. Xiang Yang Liu Biophysics & Micro/nanostructures Lab Department of Physics, NUS 1. van der Waals Interaction 2. Hydrogen bond 3. Hydrophilic vs hydrophobic
Biophysics II Key points to be covered By A/Prof. Xiang Yang Liu Biophysics & Micro/nanostructures Lab Department of Physics, NUS 1. van der Waals Interaction 2. Hydrogen bond 3. Hydrophilic vs hydrophobic
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1 Protein sequence alignment of Vibrionaceae with either a 40-residue insertion or a 44-residue insertion. Identical residues are indicated by red background.
SUPPLEMENTARY FIGURES Supplementary Figure 1 Protein sequence alignment of Vibrionaceae with either a 40-residue insertion or a 44-residue insertion. Identical residues are indicated by red background.
Protein Structure. W. M. Grogan, Ph.D. OBJECTIVES
 Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Sample Question Solutions for the Chemistry of Life Topic Test
 Sample Question Solutions for the Chemistry of Life Topic Test 1. Enzymes play a crucial role in biology by serving as biological catalysts, increasing the rates of biochemical reactions by decreasing
Sample Question Solutions for the Chemistry of Life Topic Test 1. Enzymes play a crucial role in biology by serving as biological catalysts, increasing the rates of biochemical reactions by decreasing
Focus on PNA Flexibility and RNA Binding using Molecular Dynamics and Metadynamics
 SUPPLEMENTARY INFORMATION Focus on PNA Flexibility and RNA Binding using Molecular Dynamics and Metadynamics Massimiliano Donato Verona 1, Vincenzo Verdolino 2,3,*, Ferruccio Palazzesi 2,3, and Roberto
SUPPLEMENTARY INFORMATION Focus on PNA Flexibility and RNA Binding using Molecular Dynamics and Metadynamics Massimiliano Donato Verona 1, Vincenzo Verdolino 2,3,*, Ferruccio Palazzesi 2,3, and Roberto
1/23/2012. Atoms. Atoms Atoms - Electron Shells. Chapter 2 Outline. Planetary Models of Elements Chemical Bonds
 Chapter 2 Outline Atoms Chemical Bonds Acids, Bases and the p Scale Organic Molecules Carbohydrates Lipids Proteins Nucleic Acids Are smallest units of the chemical elements Composed of protons, neutrons
Chapter 2 Outline Atoms Chemical Bonds Acids, Bases and the p Scale Organic Molecules Carbohydrates Lipids Proteins Nucleic Acids Are smallest units of the chemical elements Composed of protons, neutrons
titin, has 35,213 amino acid residues (the human version of titin is smaller, with only 34,350 residues in the full length protein).
 Introduction to Protein Structure Proteins are large heteropolymers usually comprised of 50 2500 monomer units, although larger proteins are observed 8. The monomer units of proteins are amino acids. Proteins
Introduction to Protein Structure Proteins are large heteropolymers usually comprised of 50 2500 monomer units, although larger proteins are observed 8. The monomer units of proteins are amino acids. Proteins
Full file at
 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Which of the following is an uncharged particle found in the nucleus of 1) an atom and which has
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Which of the following is an uncharged particle found in the nucleus of 1) an atom and which has
Chapter 9 DNA recognition by eukaryotic transcription factors
 Chapter 9 DNA recognition by eukaryotic transcription factors TRANSCRIPTION 101 Eukaryotic RNA polymerases RNA polymerase RNA polymerase I RNA polymerase II RNA polymerase III RNA polymerase IV Function
Chapter 9 DNA recognition by eukaryotic transcription factors TRANSCRIPTION 101 Eukaryotic RNA polymerases RNA polymerase RNA polymerase I RNA polymerase II RNA polymerase III RNA polymerase IV Function
Name: Date: Period: Biology Notes: Biochemistry Directions: Fill this out as we cover the following topics in class
 Name: Date: Period: Biology Notes: Biochemistry Directions: Fill this out as we cover the following topics in class Part I. Water Water Basics Polar: part of a molecule is slightly, while another part
Name: Date: Period: Biology Notes: Biochemistry Directions: Fill this out as we cover the following topics in class Part I. Water Water Basics Polar: part of a molecule is slightly, while another part
CHAPTER 29 HW: AMINO ACIDS + PROTEINS
 CAPTER 29 W: AMI ACIDS + PRTEIS For all problems, consult the table of 20 Amino Acids provided in lecture if an amino acid structure is needed; these will be given on exams. Use natural amino acids (L)
CAPTER 29 W: AMI ACIDS + PRTEIS For all problems, consult the table of 20 Amino Acids provided in lecture if an amino acid structure is needed; these will be given on exams. Use natural amino acids (L)
Dana Alsulaibi. Jaleel G.Sweis. Mamoon Ahram
 15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
Organic and Biochemical Molecules. 1. Compounds composed of carbon and hydrogen are called hydrocarbons.
 Organic and Biochemical Molecules 1. Compounds composed of carbon and hydrogen are called hydrocarbons. 2. A compound is said to be saturated if it contains only singly bonded carbons. Such hydrocarbons
Organic and Biochemical Molecules 1. Compounds composed of carbon and hydrogen are called hydrocarbons. 2. A compound is said to be saturated if it contains only singly bonded carbons. Such hydrocarbons
DSHIFT: a web server for predicting DNA chemical shifts
 Nucleic Acids Research, 007, Vol. 35, Web Server issue W713 W717 doi:10.1093/nar/gkm30 DSHIF: a web server for predicting DNA chemical shifts Sik Lok Lam* Department of Chemistry, he Chinese University
Nucleic Acids Research, 007, Vol. 35, Web Server issue W713 W717 doi:10.1093/nar/gkm30 DSHIF: a web server for predicting DNA chemical shifts Sik Lok Lam* Department of Chemistry, he Chinese University
CHAPTER 23 HW: ENOLS + ENOLATES
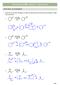 CAPTER 23 W: ENLS + ENLATES KET-ENL TAUTMERSM 1. Draw the curved arrow mechanism to show the interconversion of the keto and enol form in either trace acid or base. trace - 2 trace 3 + 2 + E1 2 c. trace
CAPTER 23 W: ENLS + ENLATES KET-ENL TAUTMERSM 1. Draw the curved arrow mechanism to show the interconversion of the keto and enol form in either trace acid or base. trace - 2 trace 3 + 2 + E1 2 c. trace
Supporting Information. Copyright Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 2009
 Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2009 Helical Hairpin Structure of a potent Antimicrobial Peptide MSI-594 in Lipopolysaccharide Micelles by NMR Anirban
Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2009 Helical Hairpin Structure of a potent Antimicrobial Peptide MSI-594 in Lipopolysaccharide Micelles by NMR Anirban
Computational Protein Design
 11 Computational Protein Design This chapter introduces the automated protein design and experimental validation of a novel designed sequence, as described in Dahiyat and Mayo [1]. 11.1 Introduction Given
11 Computational Protein Design This chapter introduces the automated protein design and experimental validation of a novel designed sequence, as described in Dahiyat and Mayo [1]. 11.1 Introduction Given
Saba Al Fayoumi. Tamer Barakat. Dr. Mamoun Ahram + Dr. Diala Abu-Hassan
 1 Saba Al Fayoumi Tamer Barakat Dr. Mamoun Ahram + Dr. Diala Abu-Hassan What is BIOCHEMISTRY??? Biochemistry = understanding life Chemical reactions are what makes an organism (An organism is simply atoms
1 Saba Al Fayoumi Tamer Barakat Dr. Mamoun Ahram + Dr. Diala Abu-Hassan What is BIOCHEMISTRY??? Biochemistry = understanding life Chemical reactions are what makes an organism (An organism is simply atoms
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor
 LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
Ch 3: Chemistry of Life. Chemistry Water Macromolecules Enzymes
 Ch 3: Chemistry of Life Chemistry Water Macromolecules Enzymes Chemistry Atom = smallest unit of matter that cannot be broken down by chemical means Element = substances that have similar properties and
Ch 3: Chemistry of Life Chemistry Water Macromolecules Enzymes Chemistry Atom = smallest unit of matter that cannot be broken down by chemical means Element = substances that have similar properties and
The biomolecules of terrestrial life
 Functional groups in biomolecules Groups of atoms that are responsible for the chemical properties of biomolecules The biomolecules of terrestrial life Planets and Astrobiology (2017-2018) G. Vladilo 1
Functional groups in biomolecules Groups of atoms that are responsible for the chemical properties of biomolecules The biomolecules of terrestrial life Planets and Astrobiology (2017-2018) G. Vladilo 1
Chapter Two: The Chemistry of Biology. The molecules of life make up the structure of cells Chemistry of biological molecule
 Chapter Two: The Chemistry of Biology The molecules of life make up the structure of cells Chemistry of biological molecule Atoms and Elements: Atoms: The basic units of all matter, containing three major
Chapter Two: The Chemistry of Biology The molecules of life make up the structure of cells Chemistry of biological molecule Atoms and Elements: Atoms: The basic units of all matter, containing three major
Chapter 002 The Chemistry of Biology
 Chapter 002 The Chemistry of Biology Multiple Choice Questions 1. Anything that occupies space and has mass is called A. Atomic B. Living C. Matter D. Energy E. Space 2. The electrons of an atom are A.
Chapter 002 The Chemistry of Biology Multiple Choice Questions 1. Anything that occupies space and has mass is called A. Atomic B. Living C. Matter D. Energy E. Space 2. The electrons of an atom are A.
New insight into DNA damage by cisplatin at the atomic scale
 1 New insight into DNA damage by cisplatin at the atomic scale Raji Heyrovska Institute of Biophysics, Academy of Sciences of the Czech Republic, 135 Kralovopolska, 612 65 Brno, Czech Republic. Email:
1 New insight into DNA damage by cisplatin at the atomic scale Raji Heyrovska Institute of Biophysics, Academy of Sciences of the Czech Republic, 135 Kralovopolska, 612 65 Brno, Czech Republic. Email:
Lecture 26: Polymers: DNA Packing and Protein folding 26.1 Problem Set 4 due today. Reading for Lectures 22 24: PKT Chapter 8 [ ].
![Lecture 26: Polymers: DNA Packing and Protein folding 26.1 Problem Set 4 due today. Reading for Lectures 22 24: PKT Chapter 8 [ ]. Lecture 26: Polymers: DNA Packing and Protein folding 26.1 Problem Set 4 due today. Reading for Lectures 22 24: PKT Chapter 8 [ ].](/thumbs/84/90810734.jpg) Lecture 26: Polymers: DA Packing and Protein folding 26.1 Problem Set 4 due today. eading for Lectures 22 24: PKT hapter 8 DA Packing for Eukaryotes: The packing problem for the larger eukaryotic genomes
Lecture 26: Polymers: DA Packing and Protein folding 26.1 Problem Set 4 due today. eading for Lectures 22 24: PKT hapter 8 DA Packing for Eukaryotes: The packing problem for the larger eukaryotic genomes
Identification of Two Antiparallel-sheet Structure of Cobrotoxin in Aqueous Solution by'hnmr
 188 Bulletin of Magnetic Resonance Identification of Two Antiparallel-sheet Structure of Cobrotoxin in Aqueous Solution by'hnmr Chang-Shin Lee and Chin Yu* Department of Chemistry, National Tsing Hua University
188 Bulletin of Magnetic Resonance Identification of Two Antiparallel-sheet Structure of Cobrotoxin in Aqueous Solution by'hnmr Chang-Shin Lee and Chin Yu* Department of Chemistry, National Tsing Hua University
NMR in Structural Biology
 NMR in Structural Biology Exercise session 2 1. a. List 3 NMR observables that report on structure. b. Also indicate whether the information they give is short/medium or long-range, or perhaps all three?
NMR in Structural Biology Exercise session 2 1. a. List 3 NMR observables that report on structure. b. Also indicate whether the information they give is short/medium or long-range, or perhaps all three?
Molecular Mechanics. I. Quantum mechanical treatment of molecular systems
 Molecular Mechanics I. Quantum mechanical treatment of molecular systems The first principle approach for describing the properties of molecules, including proteins, involves quantum mechanics. For example,
Molecular Mechanics I. Quantum mechanical treatment of molecular systems The first principle approach for describing the properties of molecules, including proteins, involves quantum mechanics. For example,
Comparing crystal structure of M.HhaI with and without DNA1, 2 (PDBID:1hmy and PDBID:2hmy),
 Supporting Information 1. Constructing the starting structure Comparing crystal structure of M.HhaI with and without DNA1, 2 (PDBID:1hmy and PDBID:2hmy), we find that: the RMSD of overall structure and
Supporting Information 1. Constructing the starting structure Comparing crystal structure of M.HhaI with and without DNA1, 2 (PDBID:1hmy and PDBID:2hmy), we find that: the RMSD of overall structure and
Crystal Structures of the Two Isomorphous A-DNA Decamers d(gtacgcgtac) and d(ggccgcggcc)
 568 Bull. Korean Chem. Soc. 2006, Vol. 27, No. 4 Taegyun Kim et al. Crystal Structures of the Two Isomorphous A-DNA Decamers d(gtacgcgtac) and d(ggccgcggcc) Taegyun Kim, Taek Hun Kwon, Hyesun Jung, Ja
568 Bull. Korean Chem. Soc. 2006, Vol. 27, No. 4 Taegyun Kim et al. Crystal Structures of the Two Isomorphous A-DNA Decamers d(gtacgcgtac) and d(ggccgcggcc) Taegyun Kim, Taek Hun Kwon, Hyesun Jung, Ja
Magnetic Resonance Lectures for Chem 341 James Aramini, PhD. CABM 014A
 Magnetic Resonance Lectures for Chem 341 James Aramini, PhD. CABM 014A jma@cabm.rutgers.edu " J.A. 12/11/13 Dec. 4 Dec. 9 Dec. 11" " Outline" " 1. Introduction / Spectroscopy Overview 2. NMR Spectroscopy
Magnetic Resonance Lectures for Chem 341 James Aramini, PhD. CABM 014A jma@cabm.rutgers.edu " J.A. 12/11/13 Dec. 4 Dec. 9 Dec. 11" " Outline" " 1. Introduction / Spectroscopy Overview 2. NMR Spectroscopy
Computational Studies of the Photoreceptor Rhodopsin. Scott E. Feller Wabash College
 Computational Studies of the Photoreceptor Rhodopsin Scott E. Feller Wabash College Rhodopsin Photocycle Dark-adapted Rhodopsin hn Isomerize retinal Photorhodopsin ~200 fs Bathorhodopsin Meta-II ms timescale
Computational Studies of the Photoreceptor Rhodopsin Scott E. Feller Wabash College Rhodopsin Photocycle Dark-adapted Rhodopsin hn Isomerize retinal Photorhodopsin ~200 fs Bathorhodopsin Meta-II ms timescale
Chapter 25. Organic and Biological Chemistry. Organic and
 Chapter 25 Calculate grade: (Add exam1 - exam 4 scores)x1.5 Add 7 best quizzes (each quiz is worth 29) Gives you the # points you have so far. Final (worth 200 points) Grades: 800-4 750-3.5 700-3 650-2.5
Chapter 25 Calculate grade: (Add exam1 - exam 4 scores)x1.5 Add 7 best quizzes (each quiz is worth 29) Gives you the # points you have so far. Final (worth 200 points) Grades: 800-4 750-3.5 700-3 650-2.5
