Amphibian populations around the world have been experiencing
|
|
|
- Marian Charles
- 5 years ago
- Views:
Transcription
1 Global gene expression profiles for life stages of the deadly amphibian pathogen Batrachochytrium dendrobatidis Erica Bree Rosenblum a,b,1, Jason E. Stajich c, Nicole Maddox a, and Michael B. Eisen a,d Departments of a Molecular and Cell Biology, and c Plant and Microbial Biology, and d Howard Hughes Medical Institute, University of California, Berkeley, CA 94720; and b Department of Biological Sciences, University of Idaho, Moscow, ID Edited by David B. Wake, University of California, Berkeley, CA, and accepted by the Editorial Board September 8, 2008 (received for review April 30, 2008) Amphibians around the world are being threatened by an emerging pathogen, the chytrid fungus Batrachochytrium dendrobatidis (Bd). Despite intensive ecological study in the decade since Bd was discovered, little is known about the mechanism by which Bd kills frogs. Here, we compare patterns of global gene expression in controlled laboratory conditions for the two phases of the life cycle of Bd: the free-living zoospore and the substrate-embedded sporangia. We find zoospores to be transcriptionally less complex than sporangia. Several transcripts more abundant in zoospores provide clues about how this motile life stage interacts with its environment. Genes with higher levels of expression in sporangia provide new hypotheses about the molecular pathways involved in metabolic, flagellar function, and pathogenicity in Bd. We highlight expression patterns for a group of fungalysin metallopeptidase genes, a gene family thought to be involved in pathogenicity in another group of fungal pathogens that similarly cause cutaneous infection of vertebrates. Finally we discuss the challenges inherent in developing a molecular toolkit for chytrids, a basal fungal lineage separated by vast phylogenetic distance from other well characterized fungi. amphibian population decline chytrid fungal pathogenicity genomics Amphibian populations around the world have been experiencing massive declines for several decades, with both local and global extinctions reported (1). Although these declines have been precipitated by a number of interacting factors (2), the recently discovered fungal pathogen Batrachochytrium dendrobatidis (Bd) (3) is responsible for many of the observed catastrophic die-offs. Bd has been documented in hundreds of amphibian species worldwide (4), has decimated many local frog populations (5, 6), and remains a devastating threat. Bd infects keratinized amphibian skin, and there are distinct morphological and physiological changes observed in frogs suffering from Bd-induced chytridiomycosis (7, 8). However, because most studies of Bd in the decade since its discovery have focused on ecology and population genetics (9, 10), very little is known about the molecular and cellular mechanism underlying the lethality of Bd. The challenges in studying the molecular biology of Bd are compounded by its position in the poorly characterized Chytridiomycota. Chytrids are basal fungi, separated by a vast phylogenetic distance from any well characterized relatives (11). Approximately 1.0 to 1.5 billion years of branch length lies between this pathogen and other fungi with fully sequenced genomes (12). The complete genome of Bd has recently been sequenced (J.E.S., E.B.R., M.B.E., and Joint Genome Institute, unpublished data), enabling experimental genomics in this species for the first time. Genomic data have been used with some success to begin understanding the genetic basis of pathogenicity in other pathogens of vertebrates (14, 15). Given the speed at which Bd is decimating host populations, whole-genome assays promise a relatively rapid way to gain mechanistic insight into Bd disease processes. Here, we initiate a functional genomics approach to understanding the molecular biology of Bd and conduct whole-genome expression assays (i.e., quantification of RNA abundance) to genetically characterize Bd life stages. The life cycle of Bd is divided into two broad categories: substrate-independent and substrate-dependent (Fig. 1). Bd zoospores are free-living, flagellated, and substrate-independent. Zoospores have a relatively short period and travel relatively short distances (16). However, in nature they are critical in initiating the infection of amphibian tissue. Zoospores in Bd exhibit chemotaxis (17), so they likely play an active role in finding appropriate substrates to colonize. Once a zoospore encysts, the substrate-dependent portion of the life stage begins. Germlings develop into zoosporangia, which produce additional zoospores. Mature zoospores are released from sporangia and can reinfect the same substrate or return to the surrounding aquatic environment (18). In nature, sporangia are of particular interest because they grow and reproduce in host tissue, and are responsible for increased pathogen loads because they release additional zoospores. Although we are ultimately interested in the interaction between Bd and its amphibian hosts in natural systems, here we use controlled laboratory culturing conditions to (i) obtain sufficient genetic material for whole-genome assays, (ii) compare zoospores and sporangia under identical conditions, and (iii) describe genetic differences between Bd life stages in the absence of an amphibian host. These whole-genome data provide a necessary baseline for all future studies that endeavor to document host-specific or condition-specific patterns of Bd gene expression. Results We used the complete genomic sequence of Bd to generate a species-specific, whole-genome array. We then compared gene expression profiles for substrate-independent (i.e., zoospore) and substrate-dependent (i.e., sporangia) samples grown under standard laboratory conditions. Particularly for reporting zoospore results, we refer to RNA abundance rather than gene expression because zoospores may contained stored transcripts (as described later). Because Bd is phylogenetically distant from other fungi with well characterized genomes, determining the exact function of Bd genes is often difficult. Therefore, patterns of expression are generally more robustly described for functional classes of genes rather than for individual genes. Herein Author contributions: E.B.R. and M.B.E. designed research; E.B.R. and N.M. performed research; E.B.R. and J.E.S. analyzed data; and E.B.R. wrote the paper. The authors declare no conflict of interest. This article is a PNAS Direct Submission. Freely available online through the PNAS open access option. 1 To whom correspondence should be addressed. rosenblum@uidaho.edu by The National Academy of Sciences of the USA PNAS November 4, 2008 vol. 105 no cgi doi pnas
2 Substrate-independent Encysted zoospore Developing sporangia Zoospores Substrate-dependent Sporangia releasing zoospores Sporangia forming zoospores Fig. 1. The life cycle of Bd. In the substrate-independent portion of the life stage, flagellated zoospores are motile and free-living. In the substratedependent portion of the life cycle, zoospores encyst and develop into zoosporangia, which produce and release new zoospores. we present results from two types of analyses: (i) analysis of enrichment patterns by using broad-scale functional classifications in the Gene Ontology (GO) database (19), and (ii) analysis of particular protein families and protein domains by using fine-scale functional classifications in the InterPro database (20). Summary. The overall expression profiles of Bd life stages were strikingly different; more than half the genes in the genome exhibited differential expression between zoospore and sporangia samples. There are 9,000 genes in the Bd genome, 8,255 for which we designed probes. Of these 8,255 genes, 4,538 (55% of genes in the genome) satisfied our criteria of containing multiple statistically significant probes (at the P 0.05 level after correction for multiple tests), all with differential expression in the same direction. In addition to differentially expressed genes, we recorded 1,522 invariant genes those genes without any differentially expressed probes. Of the 4,538 genes with differential expression between life stages, 3,179 showed higher levels of expression in sporangia (39% of genes in the genome) and 1,358 showed increased RNA abundances in zoospores (16% of genes in the genome). Although we were able to functionally annotate the majority of genes in the Bd genome, it is important to note that a large number of Bd genes in our categories of interest currently have no GO or InterPro database numbers assigned (16% in the zoospore set, 14% in the sporangia set, and 28% in the invariant set). Additional genes of interest may therefore come to light as we learn more about the functions of currently unclassified genes. Broad-Scale Patterns of Functional Enrichment. The GO database contains three different ontologies to describe the biological role of particular genes: biological process, cellular component, and molecular function. We searched for enrichment of functional categories in each of the three ontologies for each of the three gene categories of interest (i.e., higher expression in sporangia, increased abundance in zoospores, consistent representation in both samples). The GO database is organized as a network of nested functional categories, with terminal nodes being the most specific functional classification available. We define unique terminal nodes as those specific functional terms enriched in only one of our three gene lists. Results for the molecular function ontology (Fig. 2) were generated by using our most stringently chosen gene set ( 100 genes per life-stage category). In this ontology, many terminal nodes were unique to each sample. Both the sporangia sample and the invariant sample showed enrichment for peptidase : metalloexopeptidases in the sporangia sample and serine peptidases in the invariant sample. If the less stringent gene set ( 1,000 genes per life-stage category) was used for the molecular function ontology, several additional unique nodes were observed: ligase and helicase in the zoospore sample and transmembrane transporter in the sporangia sample. There was no consistent signature of enrichment for the biological process ontology by using our most stringently chosen gene set, however, many unique nodes were recovered by using our less stringent gene set (Fig. 2). In this ontology, genes with higher levels of expression in the sporangia sample were enriched for many aspects of carbohydrate and alcohol metabolism. Transcripts with greater abundance in the zoospore sample were enriched for phosphorylation and posttranslational protein modification. Invariant genes were involved in aspects of DNA metabolic process (e.g., DNA integration and modification), regulation of gene expression, and autophagy. There was less resolution for the cellular component ontology for both the stringent and expanded gene sets. The only organelle with a life stage specific enrichment pattern was the endoplasmic reticulum for the sporangia sample. Fine Scale Patterns of Differential Expression. In addition to evaluating patterns of enrichment for broad GO functional categories, we also examined genes and gene families with specific InterPro functional domains of interest. There were more than EVOLUTION Higher Expression in Sporangia Sample biological process Higher Expression in Zoospore Sample biological process Invariant Expression in Both Samples biological process metabolic process celluluar process metabolic process celluluar process metabolic process celluluar process biological regulation gene expression alcohol, carbohydrate, monosaccharide, hexose, glucose catabolic process phosphorylation post-translational protein modification DNA modification, alykylation, methylation, integration p<0.01 transcription, regulation of gene expression p<0.01 molecular function molecular function molecular function binding catalytic binding catalytic catalytic nucleotide binding p=0.010 metalloexopeptidase p=0.024 zinc ion binding p=0.017 phosphoric ester hydrolase p=0.003 serine-type peptidase Fig. 2. Enrichment for GO terms in the biological process and molecular function ontologies. Gray boxes represent unique nodes for each sample. Rosenblum et al. PNAS November 4, 2008 vol. 105 no
3 A GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP B average expression zoospore sporangia GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP GLEANP Aspergillus fumigatus outgroup Rhizopus oryzae outgroup average expression zoospore sporangia Fig. 3. Relationship and gene expression patterns for Bd fungalysin metallopeptidase genes. (A) Genes in bold font exhibited higher levels of expression in sporangia. RNA abundance for gene (boxed) was greater in zoospores. Gray gene names indicate Bd genes with no expression differences. (B) RT-PCR validation of patterns at genes and showing average relative expression for the two samples. 1,000 unique InterPro identifiers in our set of differentially expressed genes. Here, we restrict our discussion to those functional categories of particular relevance to Bd biology. Below we refer to specific Bd genes by number, which can be cross-referenced to the Joint Genome Institute s Bd portal ( First we examined the expression profiles of putative pathogenicity genes. Several gene families have been hypothesized to play a role in pathogenicity in other disease-causing fungi, and we were particularly interested in candidate genes for pathogen interaction with vertebrate skin cells. We highlight expression data from two peptidase families, both inferred to have undergone expansions in the chytrid lineage (relative to the inferred common ancestor of all fungal species with full genomes available). The fungalysin metallopeptidases (peptidase M36) may play a role in keratin degradation by dermatophytes, fungi that infect vertebrate skin (21). We observed a dramatic expansion of the fungalysin metallopeptidase gene family in Bd; there are 25 genes in the Bd genome with a peptidase M36 domain and probes on our array. Of these 25 genes, 19 (76%) show differential expression between life stages (Fig. 3). Nearly all of these (n 18) exhibited higher levels of expression in the sporangia. However, there was one gene (046850) with a robust zoosporespecific pattern (31 of 40 probes had significant differences). By using RT-PCR, we confirmed patterns for this zoospore-specific fungalysin and one sporangia-specific fungalysin. With three biological replicates, we recorded fivefold higher levels of RNA abundance in the zoospore sample for gene and 10-fold higher levels of RNA abundance in the sporangia sample for gene (Fig. 3). The serine-type peptidases (peptidase S41) also exhibit an expansion in Bd and similarly could be involved in host substrate or peptide degradation. Peptide degradation may be important for Bd because it is confronted with host antimicrobial peptides that are known to inhibit Bd growth (22). We found 29 genes in the serine peptidase family in the Bd genome, and of these, 41% showed differential expression between life stages. Most of these (n 12) showed higher levels of expression in the sporangia sample (012080, , , , , , , , , , , and ), many with quite robust patterns ( 30 probes per gene and 75% of probes giving a strong consistent signal). Only two peptidase S41 genes ( and ) displayed increased RNA abundances in the zoospore sample, and the patterns at these two genes were less robust (i.e., fewer probes with statistically significant differences in expression). An additional 41% of serine protease genes had invariant expression patterns. Apart from genes with peptidase M36 or S41 domains, we did not identify other putative fungal pathogenicity factors in the Bd genome. However, there were several differentially expressed genes with sequence similarity to genes thought to be involved in viral or bacterial evasion of vertebrate immune systems [e.g., ITAM, interleukin-1 (002700, )] (23, 24). Second, we were interested in genes that could be involved in Bd flagella because flagellar function is critical to Bd zoospores. Flagella likely were present in the fungal-animal (opisthokont) ancestor, with subsequent losses of flagella on branches leading to the majority of extant fungal taxa (11). Therefore, we identified genes that are shared between Bd and Metazoa and are known to play a role in flagellar formation or function in other taxa. We identified two radial spoke genes ( and ) in the Bd genome with orthologs that are important in eukaryotic flagella; both showed higher expression in the sporangia sample. All four genes (028800, , , and ) with membrane occupation and recognition nexus (MORN) motifs, which are involved in a number of cellular processes, including cgi doi pnas Rosenblum et al.
4 in animal flagella (25), exhibited higher levels of expression in the sporangia sample. In addition, several Bd genes with sequence similarity to genes implicated in mammalian sperm maturation and motility (e.g., Tcp11, Tpx-1) showed differential expression between life stages. However, these mammalian sperm function genes have fungal orthologs involved in different cellular processes, so it is premature to conclude a role for them in Bd flagellar function. Third, a number of genes potentially involved in structural aspects of Bd cells displayed differential expression. Most notably, six of eight chitin synthases and three of six chitin binding genes showed differential expression between life stages, some with greater RNA abundances in zoospores (076580, , , , , and ) and some in sporangia (072590, , and ). These genes each contained a large number of probes (n 22 66), and patterns of differential expression were robust. Additionally, several genes involved in metazoan cell adhesion showed differential expression; two of two vinculin genes ( and ), two of two fibronectin genes ( and ), and one of four genes with a FAS1 domain (069890) exhibited higher expression in the sporangia sample. Fourth, several genes involved in cell growth and cell death showed higher levels of expression in the sporangia sample. Three of seven cullin genes (029560, , ), which are ubiquitin ligase subunits involved in controlling yeast cell division (26), and one TAP42-like protein (047320), which regulates a signaling pathway for cell growth in response to nutrients (27), showed higher expression in sporangia. The TAP42-like protein is the only gene in our set with a GO annotation of response to biotic stimulus (GO: ). Several apoptosis regulatory proteins also exhibited higher levels of expression in sporangia ( and ). Four of five universal stress protein genes (019030, , , and ) showed robust patterns of increased expression in the sporangia sample. Few genes displayed zoospore-specific patterns, but four of six genes in a particular class of cell division/gtp-binding proteins (010000, , , and ) had greater RNA abundances in the zoospore sample, with a large number of probes in each gene supporting this result. Fifth, genes involved in cellular signaling and transport were of interest given the importance of the interaction of Bd with its environment. The G- -protein genes, which play a role in cellular signaling (28), exhibit an expansion in Bd. In this family, transcripts from four genes were more abundant in sporangia (001760, , , ), three were more abundant in zoospores (001570, , ). Most G- genes with differential expression contained at least 30 probes per gene and had very robust patterns (up to 97% of probes with statistical significance). Additionally, two of two ( and ) peptidase A22 genes (presenilin) revealed zoospore-specific patterns. Although there are a large number of peptidase genes with aspartic catalytic in the Bd genome, the presenilins are thought to be important in signaling pathways (29). Last, in the Bd genome there are 22 clathrin- and clathrin adaptor related genes (involved in cellular transport), and nearly 70% of them showed higher expression in the sporangia. Discussion Here, we provide a molecular profile of different life stages in the pathogenic chytrid fungus Bd. We compare global gene expression patterns for two portions of the Bd life cycle cultured under controlled laboratory conditions: the substrateindependent zoospore stage and the substrate-dependent sporangia stage. The two life stages exhibit strikingly different RNA abundance profiles, with 55% of genes in the genome showing differential expression. In the following paragraphs we discuss the development of a molecular toolkit for Bd, the genetic differences between Bd life stages, and some of the first hypotheses about pathogenicity factors in this species. Gene Expression Differences Between Bd Life Stages. Global patterns of gene expression indicate that zoospores are transcriptionally less complex than sporangia, with RNAs from fewer than half as many genes showing increased abundances in the zoospore compared with the sporangia sample. Previous research in another chytrid, Blastocladiella emersonii, suggests that chytrid zoospores contain stored mrnas and exhibit reduced RNA and protein synthesis (30). Our finding that genes involved in posttranslational protein modification are enriched in zoospores is interesting in light of possible reduced transcriptional/ translational in this life stage. Preliminary trials exposing Bd zoospores to the transcriptional inhibitor actinomycin-d did not compromise zoospore survival or motility and suggest that transcriptional provisioning may be adequate for basic zoospore function. Because most genes in the Bd genome do not exhibit zoosporespecific patterns, genes with increased transcript representation in the zoospore sample are of particular interest because they provide a window into the pool of stored and newly transcribed mrnas necessary for zoospore function. First, zoospores exhibit greater RNA abundances for several putative pathogenicity genes, a pattern discussed later. Second, we observed higher RNA abundances in the zoospore sample for several genes that may play a role in signaling. Several presenilin and G- genes may provide clues for understanding how Bd zoospores interact with their environment. Third, we saw greater RNA abundances for several chitin synthases and chitin binding genes in the zoospore sample. Although chitin synthases with higher expression in the sporangia may be involved in cell wall remodeling during development, the function of these gene products in the zoospore remains to be determined. Finally, and perhaps surprisingly, we did not observe any putative flagellar genes with increased transcript representation in the zoospore sample. This is likely explained by the fact that zoospores develop within zoosporangia; all putative flagellar genes showed higher levels of expression in the sporangia sample. In contrast to the zoospores, sporangia are transcriptionally and metabolically active and complex. The sporangia sample exhibited more than twice as many genes with higher levels of expression than the zoospore sample. The large number of genes involved in sporangia function is not surprising given that our substrate-dependent sample contained life-stage diversity not found in the zoospore sample from encysted zoospores to mature sporangia. Our gene expression data capture this developmental variation and suggest some molecular mechanisms for the functional and structural complexity of sporangia. As expected, we saw higher levels of expression in our sporangia sample for genes involved in cell growth, cell death, and various aspects of alcohol and carbohydrate metabolism. A persistent gap in our knowledge of Bd biology is understanding what vertebrate cellular components are directly used by Bd. Although Bd attacks keratinized amphibian tissue, it is not clear that keratin is a primary nutrient source for this pathogen (16). Therefore, describing the metabolic pathways used by the substrate-dependent portion of the life cycle is important for gaining insight into the particular vertebrate cellular components necessary for Bd growth in host tissue. By identifying genes involved in sporangia metabolism, subsequent studies can evaluate the behavior of these genes under different nutrient and stress regimens. Also of interest in the sporangia sample are a number of genes that have clear orthologs with metazoans. First, several genes (vinculin, fibronectin, fasciclin) involved in cell adhesion show higher levels of expression in the sporangia sample. Although Bd does not exhibit true multicellularity, sporangia can grow in a EVOLUTION Rosenblum et al. PNAS November 4, 2008 vol. 105 no
5 colonial fashion and can appear in clusters. This group effect, whereby Bd cells survive better in colonies, has been observed in laboratory conditions (3) and merits further dissection. Second, several genes involved in flagellar biogenesis in other taxa were identified in the sporangia sample. Radial spoke like genes showed higher levels of expression in sporangia. These genes are well conserved in eukaryotes and play a role in flagellar movement of such diverse taxa as the green algae Chlamydomonas reinhardtii and the sea squirt Ciona intestinalis (31). Similarly, a number of genes with MORN motifs had higher levels of expression in sporangia. MORN motifs are found along many deep branches of the tree of life and have many hypothesized functions, including flagellar biogenesis in animal sperm. These motifs are found in proteins localized to sperm basal bodies and flagellum in the sea squirt C. intestinalis and the carp Cyprinus carpio (32). Zoospore development occurs entirely inside of zoosporangia, and flagellar genes expressed in developing sporangia are likely to provide more clues about the molecular basis for Bd flagellar development than pure zoospore samples. Synchronizing Bd cultures to examine expression profiles sliced more finely through the life cycle was outside the scope of this study but could provide additional insight into genes involved in zoospore development. Although it is difficult to obtain adequate amounts of RNA from synchronized cultures for wholegenome assays, it will now be feasible to analyze patterns throughout development at specific genetic targets identified here. Gene Expression Patterns at Putative Pathogenicity Genes. We have identified two gene families with intriguing patterns of gene expression that are thought to specifically play a role in fungal pathogenicity. Serine protease and fungalysin metallopeptidase families both show large expansions in Bd and represent our most promising leads for understanding life stage specific mechanisms of pathogenicity in Bd. Of the putative pathogenicity genes identified in this study, of greatest interest are the fungalysin metallopeptidase genes (peptidase M36). Not only did we observe an expansion of this gene family in Bd, but the vast majority (76%) exhibited differential expression between life stages. Nearly all differentially expressed fungalysins showed higher levels of expression in sporangia, the life stage associated in nature with keratinized host tissue. One fungalysin gene showed greater RNA abundance in the zoospore sample, and this gene may be an interesting target for understanding the initial stages of zoospore colonization of amphibian skin and entry into host cells. The serine protease family (peptidase S41) also requires further study in Bd; some genes in this family showed higher levels of expression in the sporangia sample, and others showed consistently high in both life stages. Both fungalysin metallopeptidases and serine proteases have been implicated in pathogenesis in a variety of fungal pathogens that infect diverse animal hosts from nematodes to insects to humans (33, 34). Extracellular metallopeptidases are also expressed by a number of pathogenic fungi that specifically parasitize vertebrates (35). For example, multiplication of fungalysin metallopeptidases has been observed in dermatophytes, fungal pathogens causing cutaneous infections in vertebrates (21). Bd attacks keratinized amphibian tissue, and appears to exhibit chemotaxis toward soluble keratin in laboratory assays (17). However, it is not clear that Bd directly metabolizes keratin. Growth assays have suggested that Bd grows poorly and does not produce keratinases on pure industrial keratin (16), so it is important to consider the possibility that fungalysin metallopeptidases may be involved in degrading host cellular components other than keratin. The methods used in the current study are not appropriate for conclusively demonstrating a functional role for particular peptidases in Bd pathogenicity or virulence. Rather, the purpose of our approach is to describe promising targets for further study. By identifying particular peptidase M36 and S41 genes with life stage specific expression patterns in laboratory conditions, targeted follow-up studies can now be conducted to understand the role of these specific genes in the presence of host tissue. In addition to factors thought to play a role specifically in fungal pathogenicity, several genes and gene families known to be involved in pathogenicity of more distantly related taxa showed differential expression between life stages in our study. We observed differential expression of a large number of clathrin-related genes, a gene family used by some pathogenic viruses and bacteria to enter host cells by mimicking ligands to the host s clathrin-dependent signaling system (36). Several genes that are involved in viral immune evasion by downregulating host immune response also showed differential expression in our study. Although mechanisms of pathogenicity in viral, bacterial, protist, and fungal systems are likely to be quite divergent, motifs associated with ITAM, interleukin-1, and clathrin genes are conserved in vertebrates. It is possible that fungal pathogens could similarly mimic vertebrate host proteins to evade the immune system or make use of the host intracellular signaling machinery to gain entry into host cells. These possibilities are speculative and further work is necessary to determine whether the association of these genes with different Bd life stages is meaningful. Genomics of Uncharacterized Fungi. The challenges of interpreting molecular profiles for uncharacterized organisms are augmented in Bd as a result of the difficulty in assigning orthology between Bd genes and those in phylogenetically distant taxa. Of genes lacking functional annotation, 58% displayed differential expression between life stages. Further, there is a large intersection between genes that appear to be unique to Bd and genes that showed increased RNA abundance in the zoospore sample. Understanding the molecular mechanisms that drive zoospore function in particular will likely require substantial new data acquisition for currently un-annotated genes in the chytrid genome. As we learn more about the genomes of basal fungi, our understanding of the functional classifications of Bd genes will become more refined and will provide additional insights into the molecular biology of this emerging pathogen. Materials and Methods Sample Preparation. Three Bd strains, isolated from different frog species, were used as biological replicates. These isolates were from a natural population of Rana muscosa from California (JAM81), a natural population of Phyllomedusa lemur from Panama (JEL423), and a captive population of Xenopus tropicalis from the University of California at Berkeley (JP005). Cultures were grown at room temperature on 1% tryptone, 1% agar plates for 2 weeks and then separated into two samples: (i) the substrate-independent sample contained free-swimming zoospores and (ii) the substrate-dependent sample contained all remaining Bd growth once zoospores were collected. To separate life stages, plates were flooded with water and incubated for 1 h so mature zoospores would release from zoosporangia. The water was then passed through a 20- m Spectramesh filter (Spectrum Medical Instruments) to isolate zoospores. Additional water was added to plates and remaining growth was collected as the substrate-dependent sample. The two samples were pelleted and flash-frozen. It is important to note that the substratedependent sample was not homogenous: newly encysted, developing, mature, and empty sporangia were all captured in this sample. We designed our experiment to pool stages of sporangia growth because the comparison between free-living and substrate-embedded portions of the Bd life cycle is biologically relevant from a pathogenicity perspective. Additionally, our whole-genome approach required large quantities of RNA and precluded analyses of more finely sliced life stages. Experimental Design. We designed a custom chip for Bd based on the complete genome of strain JAM81. The array contained 377,075 species-specific oligonucleotides, or 1 oligonucleotide every 100 base pairs throughout the genome. The two samples from each strain were labeled with different cgi doi pnas Rosenblum et al.
6 cyanine dyes and hybridized to a common chip to provide a direct comparison between life stages. Five chips were used for biological and technical replication. Samples for three chips were biological replicates (JAM81, JEL423, and JP005) and were prepared as a batch to ensure identical treatment. The remaining chips were technical replicates (one a direct comparison for JEL423 and one a dye-swap comparison for JAM81) and were treated individually to control for variation in technical conditions. Molecular Methods. RNA was extracted with an RNeasy Mini Kit (Qiagen) with the standard protocol and a DNase digestion. Double-stranded cdnas were synthesized by using the SuperScript cdna Synthesis Kit (Invitrogen). cdna samples were fluorescently labeled with custom Cy3- and Cy5-labeled oligonucleotides from TRILink BioTechnologies. A 16-hour hybridization was conducted at 42 in a Hybex microarray incubation system (SciGene). Chips were washed and then scanned on a GenePix Professional 4200 Scanner (Axon). We confirmed chip-based gene expression patterns for a subset of genes with RT-PCR by using SYBR green (Qiagen) and an ABI 7300 system (Applied Biosystems). We quantified gene expression of two target genes of interest relative to reference genes, which had high levels of expression in both the zoospore and sporangia samples. Data Analysis. NimbleScan software (NimbleGen) was used to align a chipspecific grid to control features and extract probe-by-probe intensity data, and the LIMMA package in the R Project for Statistical Computing was used for statistical analysis. Global Loess normalization was used to normalize within arrays. Normalization among arrays was not necessary because mean and variance in intensities were similar across chips. A linear model was fit to the data, and a Bayesian t test was used to identify probes with significantly differential expression. The Benjamini and Hochberg method was used to control for the expected false discovery rate given multiple tests (37). Because there were multiple probes per gene, we used the following criteria to draw inference at the gene level. Genes were considered differentially expressed if (i) multiple probes in the gene showed significant differences in expression between life stages at the P 0.05 level (after correction for multiple tests) and (ii) all probes in the gene with significant differences in expression showed higher expression in the same sample. In addition to identifying genes with differential expression between life stages, we identified invariant genes those for which zero probes showed significant differences between zoospore and sporangia samples. We then conducted two related analyses. First, we looked for significant enrichment in GO terms by using GO Term Finder (38), a hyper-geometric test for enrichment at nested nodes throughout the GO ontology. Because so many genes showed differential expression, we modified our stringency criteria to reduce gene lists to 100 and 1,000 for each life stage for the GO analysis. For the sporangia sample, P value cutoff points of and gave us 126 and 745 genes, respectively. For the zoospore sample, P value cutoff values of and gave us 81 and 810 genes, respectively. Second, we used InterPro identifiers to describe specific gene families and functional categories with differential expression between life stages. Finally, we identified several expansions in Bd gene families by evaluating the distribution of Pfam domains (39) across the currently available fungal genomes. Gene trees were constructed for the Bd expansions based on hidden Markov model guided alignments to the Pfam domain and by using MrBayes (40) and RAxML (13). ACKNOWLEDGMENTS. We thank John Taylor, David Wake, Jamie Voyles, Craig Moritz, and two anonymous reviewers for comments on the manuscript, Karen Vranizan for consultation about data analysis, and Joyce Longcore (University of Maine, Orono, ME) for providing JEL423 cultures. This work was supported by a postdoctoral fellowship in Bioinformatics from the National Science Foundation (to E.B.R.) and a postdoctoral fellowship from the Miller Institute for Basic Research (to J.E.S.). 1. Loetters S, La Marca E, Stuart S, Gagliardo R, Veith M (2004) A new dimension of current biodiversity loss? Herpetotropicos 1: Blaustein AR, Hatch AC, Beldon LK, Kiesecker J (2004) Multiple causes for declining amphibian populations. Experimental approaches to conservation biology, ed Gordon MS, Bartol SM (Univ of California, Berkeley), pp Longcore JE, Pessier AP, Nichols DK (1999) Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibians. Mycologia 91: Skerratt LF, et al. (2007) Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth 4: Lips KR, et al. (2006) Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci USA Rachowicz LJ, et al. (2006) Emerging infectious disease as a proximate cause of amphibian mass mortality. Ecology 87: Parker JM, Mikaelian I, Hahn N, Diggs HE (2002) Clinical diagnosis and treatment of epidermal chytridiomycosis in African clawed frogs (Xenopus tropicalis). Comp Med 52: Voyles J, et al. (2007) Electrolyte depletion and osmotic imbalance in amphibians with chytridiomycosis. Dis Aquat Org 77: Morgan JAT, et al. (2007) Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc Natl Acad Sci 104: Morehouse EA, et al. (2003) Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Mol Ecol 12: James TY, et al. (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: Taylor JW, Berbee ML (2006) Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia 98: Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: Dunning H, Julie C, Lin M, et al. (2006) Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2: Nierman WC, et al. (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438: Piotrowski JS, Annis SL, Longcore JE (2004) Physiology of Batrachochytrium dendrobatidis, a chytrid pathogen of amphibians. Mycologia 96: Moss AS, Reddy NS, Dortaj IM, San Francisco MJ (2008) Chemotaxis of the amphibian pathogen Batrachochytrium dendrobatidis and its response to a variety of attractants. Mycologia 100: Berger L, Hyatt AD, Speare R, Longcore JE (2005) Life cycle stages of the amphibian chytrid Batrachchytrium dendrobatidis. Dis Aquat Org 68: The Gene Ontology Consortium (2000) Gene ontology: Tool for the unification of biology. Nat Genet 25: Mulder NJ, et al. (2007) New developments in the InterPro database. Nucleic Acids Res 35:D224 D Jousson O, et al. (2004) Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology 150: Rollins-Smith LA, et al. (2006) Antimicrobial peptide defenses of the mountain yellowlegged frog (Rana muscosa). Dev Comp Immunol 30: Smith GL, Chan YS (1991) 2 Vaccinia virus proteins structurally related to the interleukin-1 receptor and the immunoglobulin superfamily. J Gen Virol 72: Geimonen E, et al. (2003) Hantavirus pulmonary syndrome-associated hantaviruses contain conserved and functional ITAM signaling elements. J Virol 77: Inaba K (2007) Molecular basis of sperm flagellar axonemes: Structural and evolutionary aspects. Reprod Biomech 1101: Michel JJ, McCarville JF, Xiong Y (2003) A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J Biol Chem 278: Beck T, Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: Sandhya K, Vemuri MC (1997) Regulation of cellular signals by G-proteins. J Biosci 22: Parks AL, Curtis D (2007) Presenilin diversifies its portfolio. Trends Genet 23: Johnson SA, Lovett JS (1984) Gene expression during development of Blastocladiella emersonii. Exp Mycol 8: Satouh Y, et al. (2005) Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol Biol Cell 16: Ju TK, Huang FL (2004) MSAP, the Meichroacidin homolog of carp (Cyprinus carpio), differs from the rodent counterpart in germline expression and involves flagellar differentiation. Biol Reprod 71: Xu J, Baldwin D, Kindrachuk C, Hegedus DD (2006) Serine proteases and metalloproteases associated with pathogenesis but not host specificity in the Entomophthoralean fungus Zoophthora radicans. Can J Microbiol 52: Huang XW, Zhao NH, Zhang KQ (2004) Extracellular enzymes serving as virulence factors in nematophagous fungi involved in infection of the host. Res Microbiol 155: da Silva BA, Santos ALS, Barreto-Bergter E, Pinto MR (2006) Extracellular peptidase in the fungal pathogen Pseudallescheria boydii. Curr Microbiol 53: Bonazzi M, Cossart P (2006) Bacterial entry into cells: A role for the endocytic machinery. FEBS Lett 580: Benjamini Y, Hockberg Y (1995) Controlling the false discovery rate a practical and powerful approach to multiple testing. J R Statistics Soc Ser B 57: Boyle EI, et al. (2004) GO::TermFinder - open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: Finn RD, et al. (2006) Pfam: Clans, web tools and services. Nucleic Acids Res 34:D247 D Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: EVOLUTION Rosenblum et al. PNAS November 4, 2008 vol. 105 no
Evolution and pathogenicity in the deadly chytrid pathogen of amphibians. Erica Bree Rosenblum UC Berkeley
 Evolution and pathogenicity in the deadly chytrid pathogen of amphibians Erica Bree Rosenblum UC Berkeley Emerging infectious disease Emerging infectious disease EID events have risen significantly over
Evolution and pathogenicity in the deadly chytrid pathogen of amphibians Erica Bree Rosenblum UC Berkeley Emerging infectious disease Emerging infectious disease EID events have risen significantly over
[Byline: Erica Bree Rosenblum1*, Jamie Voyles2, Thomas J. Poorten1, Jason E. Stajich3,4
 2010-02-03-35 Chytrid fungus, frogs - worldwide: review article To: (05) Zoonoses, general; (23) Veterinary education ************************************************* CHYTRID FUNGUS, FROGS - WORLDWIDE:
2010-02-03-35 Chytrid fungus, frogs - worldwide: review article To: (05) Zoonoses, general; (23) Veterinary education ************************************************* CHYTRID FUNGUS, FROGS - WORLDWIDE:
SPECIES OF ARCHAEA ARE MORE CLOSELY RELATED TO EUKARYOTES THAN ARE SPECIES OF PROKARYOTES.
 THE TERMS RUN AND TUMBLE ARE GENERALLY ASSOCIATED WITH A) cell wall fluidity. B) cell membrane structures. C) taxic movements of the cell. D) clustering properties of certain rod-shaped bacteria. A MAJOR
THE TERMS RUN AND TUMBLE ARE GENERALLY ASSOCIATED WITH A) cell wall fluidity. B) cell membrane structures. C) taxic movements of the cell. D) clustering properties of certain rod-shaped bacteria. A MAJOR
This is a repository copy of Microbiology: Mind the gaps in cellular evolution.
 This is a repository copy of Microbiology: Mind the gaps in cellular evolution. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/114978/ Version: Accepted Version Article:
This is a repository copy of Microbiology: Mind the gaps in cellular evolution. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/114978/ Version: Accepted Version Article:
Chapters AP Biology Objectives. Objectives: You should know...
 Objectives: You should know... Notes 1. Scientific evidence supports the idea that evolution has occurred in all species. 2. Scientific evidence supports the idea that evolution continues to occur. 3.
Objectives: You should know... Notes 1. Scientific evidence supports the idea that evolution has occurred in all species. 2. Scientific evidence supports the idea that evolution continues to occur. 3.
VCE BIOLOGY Relationship between the key knowledge and key skills of the Study Design and the Study Design
 VCE BIOLOGY 2006 2014 Relationship between the key knowledge and key skills of the 2000 2005 Study Design and the 2006 2014 Study Design The following table provides a comparison of the key knowledge (and
VCE BIOLOGY 2006 2014 Relationship between the key knowledge and key skills of the 2000 2005 Study Design and the 2006 2014 Study Design The following table provides a comparison of the key knowledge (and
AP Biology Exam #7 (PRACTICE) Subunit #7: Diversity of Life
 AP Biology Exam #7 (PRACTICE) Subunit #7: Diversity of Life Multiple Choice Questions: Choose the best answer then bubble your answer on your scantron sheet. 1. Armadillos and spiny anteaters are not related.
AP Biology Exam #7 (PRACTICE) Subunit #7: Diversity of Life Multiple Choice Questions: Choose the best answer then bubble your answer on your scantron sheet. 1. Armadillos and spiny anteaters are not related.
California Subject Examinations for Teachers
 California Subject Examinations for Teachers TEST GUIDE SCIENCE SUBTEST II: LIFE SCIENCES Subtest Description This document contains the Life Sciences subject matter requirements arranged according to
California Subject Examinations for Teachers TEST GUIDE SCIENCE SUBTEST II: LIFE SCIENCES Subtest Description This document contains the Life Sciences subject matter requirements arranged according to
Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis
 Title Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis Author list Yu Han 1, Huihua Wan 1, Tangren Cheng 1, Jia Wang 1, Weiru Yang 1, Huitang Pan 1* & Qixiang
Title Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis Author list Yu Han 1, Huihua Wan 1, Tangren Cheng 1, Jia Wang 1, Weiru Yang 1, Huitang Pan 1* & Qixiang
The Prokaryotic World
 The Prokaryotic World A. An overview of prokaryotic life There is no doubt that prokaryotes are everywhere. By everywhere, I mean living in every geographic region, in extremes of environmental conditions,
The Prokaryotic World A. An overview of prokaryotic life There is no doubt that prokaryotes are everywhere. By everywhere, I mean living in every geographic region, in extremes of environmental conditions,
STAAR Biology Assessment
 STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
A. Correct! Taxonomy is the science of classification. B. Incorrect! Taxonomy is the science of classification.
 DAT - Problem Drill 07: Diversity of Life Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as 1. What is taxonomy? Question #01 (A) Taxonomy
DAT - Problem Drill 07: Diversity of Life Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as 1. What is taxonomy? Question #01 (A) Taxonomy
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
NORTHERN ILLINOIS UNIVERSITY. Screening of Chemical Libraries in Search of Inhibitors of Aflatoxin Biosynthesis. A Thesis Submitted to the
 NORTHERN ILLINOIS UNIVERSITY Screening of Chemical Libraries in Search of Inhibitors of Aflatoxin Biosynthesis A Thesis Submitted to the University Honors Program In Partial Fulfillment of the Requirements
NORTHERN ILLINOIS UNIVERSITY Screening of Chemical Libraries in Search of Inhibitors of Aflatoxin Biosynthesis A Thesis Submitted to the University Honors Program In Partial Fulfillment of the Requirements
Biology Assessment. Eligible Texas Essential Knowledge and Skills
 Biology Assessment Eligible Texas Essential Knowledge and Skills STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules
Biology Assessment Eligible Texas Essential Knowledge and Skills STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules
BIOLOGY STANDARDS BASED RUBRIC
 BIOLOGY STANDARDS BASED RUBRIC STUDENTS WILL UNDERSTAND THAT THE FUNDAMENTAL PROCESSES OF ALL LIVING THINGS DEPEND ON A VARIETY OF SPECIALIZED CELL STRUCTURES AND CHEMICAL PROCESSES. First Semester Benchmarks:
BIOLOGY STANDARDS BASED RUBRIC STUDENTS WILL UNDERSTAND THAT THE FUNDAMENTAL PROCESSES OF ALL LIVING THINGS DEPEND ON A VARIETY OF SPECIALIZED CELL STRUCTURES AND CHEMICAL PROCESSES. First Semester Benchmarks:
AP Curriculum Framework with Learning Objectives
 Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Map of AP-Aligned Bio-Rad Kits with Learning Objectives
 Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Variation in the genetic response to high temperature in Montastraea faveolata from the Florida Keys & Mexico
 Variation in the genetic response to high temperature in Montastraea faveolata from the Florida Keys & Mexico Nicholas R. Polato 1, Christian R. Voolstra 2, Julia Schnetzer 3, Michael K. DeSalvo 4, Carly
Variation in the genetic response to high temperature in Montastraea faveolata from the Florida Keys & Mexico Nicholas R. Polato 1, Christian R. Voolstra 2, Julia Schnetzer 3, Michael K. DeSalvo 4, Carly
Curriculum Map. Biology, Quarter 1 Big Ideas: From Molecules to Organisms: Structures and Processes (BIO1.LS1)
 1 Biology, Quarter 1 Big Ideas: From Molecules to Organisms: Structures and Processes (BIO1.LS1) Focus Standards BIO1.LS1.2 Evaluate comparative models of various cell types with a focus on organic molecules
1 Biology, Quarter 1 Big Ideas: From Molecules to Organisms: Structures and Processes (BIO1.LS1) Focus Standards BIO1.LS1.2 Evaluate comparative models of various cell types with a focus on organic molecules
A A A A B B1
 LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
Introduction to Bioinformatics
 CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
TEST SUMMARY AND FRAMEWORK TEST SUMMARY
 Washington Educator Skills Tests Endorsements (WEST E) TEST SUMMARY AND FRAMEWORK TEST SUMMARY BIOLOGY Copyright 2014 by the Washington Professional Educator Standards Board 1 Washington Educator Skills
Washington Educator Skills Tests Endorsements (WEST E) TEST SUMMARY AND FRAMEWORK TEST SUMMARY BIOLOGY Copyright 2014 by the Washington Professional Educator Standards Board 1 Washington Educator Skills
Big Idea 1: The process of evolution drives the diversity and unity of life.
 Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Chapter Chemical Uniqueness 1/23/2009. The Uses of Principles. Zoology: the Study of Animal Life. Fig. 1.1
 Fig. 1.1 Chapter 1 Life: Biological Principles and the Science of Zoology BIO 2402 General Zoology Copyright The McGraw Hill Companies, Inc. Permission required for reproduction or display. The Uses of
Fig. 1.1 Chapter 1 Life: Biological Principles and the Science of Zoology BIO 2402 General Zoology Copyright The McGraw Hill Companies, Inc. Permission required for reproduction or display. The Uses of
1. The basic structural and physiological unit of all living organisms is the A) aggregate. B) organelle. C) organism. D) membrane. E) cell.
 Name: Date: Test File Questions 1. The basic structural and physiological unit of all living organisms is the A) aggregate. B) organelle. C) organism. D) membrane. E) cell. 2. A cell A) can be composed
Name: Date: Test File Questions 1. The basic structural and physiological unit of all living organisms is the A) aggregate. B) organelle. C) organism. D) membrane. E) cell. 2. A cell A) can be composed
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Enduring understanding 1.A: Change in the genetic makeup of a population over time is evolution.
 The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
SCOTCAT Credits: 20 SCQF Level 7 Semester 1 Academic year: 2018/ am, Practical classes one per week pm Mon, Tue, or Wed
 Biology (BL) modules BL1101 Biology 1 SCOTCAT Credits: 20 SCQF Level 7 Semester 1 10.00 am; Practical classes one per week 2.00-5.00 pm Mon, Tue, or Wed This module is an introduction to molecular and
Biology (BL) modules BL1101 Biology 1 SCOTCAT Credits: 20 SCQF Level 7 Semester 1 10.00 am; Practical classes one per week 2.00-5.00 pm Mon, Tue, or Wed This module is an introduction to molecular and
16 The Cell Cycle. Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization
 The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
Microbial Diversity and Assessment (II) Spring, 2007 Guangyi Wang, Ph.D. POST103B
 Microbial Diversity and Assessment (II) Spring, 007 Guangyi Wang, Ph.D. POST03B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm General introduction and overview Taxonomy [Greek
Microbial Diversity and Assessment (II) Spring, 007 Guangyi Wang, Ph.D. POST03B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm General introduction and overview Taxonomy [Greek
Biology Teach Yourself Series Topic 2: Cells
 Biology Teach Yourself Series Topic 2: Cells A: Level 14, 474 Flinders Street Melbourne VIC 3000 T: 1300 134 518 W: tssm.com.au E: info@tssm.com.au TSSM 2013 Page 1 of 14 Contents Cells... 3 Prokaryotic
Biology Teach Yourself Series Topic 2: Cells A: Level 14, 474 Flinders Street Melbourne VIC 3000 T: 1300 134 518 W: tssm.com.au E: info@tssm.com.au TSSM 2013 Page 1 of 14 Contents Cells... 3 Prokaryotic
CELL PRACTICE TEST
 Name: Date: 1. As a human red blood cell matures, it loses its nucleus. As a result of this loss, a mature red blood cell lacks the ability to (1) take in material from the blood (2) release hormones to
Name: Date: 1. As a human red blood cell matures, it loses its nucleus. As a result of this loss, a mature red blood cell lacks the ability to (1) take in material from the blood (2) release hormones to
ADVANCED PLACEMENT BIOLOGY
 ADVANCED PLACEMENT BIOLOGY Description Advanced Placement Biology is designed to be the equivalent of a two-semester college introductory course for Biology majors. The course meets seven periods per week
ADVANCED PLACEMENT BIOLOGY Description Advanced Placement Biology is designed to be the equivalent of a two-semester college introductory course for Biology majors. The course meets seven periods per week
Evidence for dynamically organized modularity in the yeast protein-protein interaction network
 Evidence for dynamically organized modularity in the yeast protein-protein interaction network Sari Bombino Helsinki 27.3.2007 UNIVERSITY OF HELSINKI Department of Computer Science Seminar on Computational
Evidence for dynamically organized modularity in the yeast protein-protein interaction network Sari Bombino Helsinki 27.3.2007 UNIVERSITY OF HELSINKI Department of Computer Science Seminar on Computational
Biology Release Notes 2016
 Biology Release Notes 2016 Publish Date: March 30, 2016 Revision Number: BM-2013-002(06/16)-BW Page Count Difference: This edition of Biology, has a 47 page count variation when compared to the last edition.
Biology Release Notes 2016 Publish Date: March 30, 2016 Revision Number: BM-2013-002(06/16)-BW Page Count Difference: This edition of Biology, has a 47 page count variation when compared to the last edition.
Virginia Western Community College BIO 101 General Biology I
 BIO 101 General Biology I Prerequisites Successful completion of MTE 1, 2, 3, 4, and 5; and a placement recommendation for ENG 111, co-enrollment in ENF 3/ENG 111, or successful completion of all developmental
BIO 101 General Biology I Prerequisites Successful completion of MTE 1, 2, 3, 4, and 5; and a placement recommendation for ENG 111, co-enrollment in ENF 3/ENG 111, or successful completion of all developmental
no.1 Raya Ayman Anas Abu-Humaidan
 no.1 Raya Ayman Anas Abu-Humaidan Introduction to microbiology Let's start! As you might have concluded, microbiology is the study of all organisms that are too small to be seen with the naked eye, Ex:
no.1 Raya Ayman Anas Abu-Humaidan Introduction to microbiology Let's start! As you might have concluded, microbiology is the study of all organisms that are too small to be seen with the naked eye, Ex:
Text of objective. Investigate and describe the structure and functions of cells including: Cell organelles
 This document is designed to help North Carolina educators teach the s (Standard Course of Study). NCDPI staff are continually updating and improving these tools to better serve teachers. Biology 2009-to-2004
This document is designed to help North Carolina educators teach the s (Standard Course of Study). NCDPI staff are continually updating and improving these tools to better serve teachers. Biology 2009-to-2004
Introduction to Bioinformatics Integrated Science, 11/9/05
 1 Introduction to Bioinformatics Integrated Science, 11/9/05 Morris Levy Biological Sciences Research: Evolutionary Ecology, Plant- Fungal Pathogen Interactions Coordinator: BIOL 495S/CS490B/STAT490B Introduction
1 Introduction to Bioinformatics Integrated Science, 11/9/05 Morris Levy Biological Sciences Research: Evolutionary Ecology, Plant- Fungal Pathogen Interactions Coordinator: BIOL 495S/CS490B/STAT490B Introduction
Valley Central School District 944 State Route 17K Montgomery, NY Telephone Number: (845) ext Fax Number: (845)
 Valley Central School District 944 State Route 17K Montgomery, NY 12549 Telephone Number: (845)457-2400 ext. 18121 Fax Number: (845)457-4254 Advance Placement Biology Presented to the Board of Education
Valley Central School District 944 State Route 17K Montgomery, NY 12549 Telephone Number: (845)457-2400 ext. 18121 Fax Number: (845)457-4254 Advance Placement Biology Presented to the Board of Education
Principles of Cellular Biology
 Principles of Cellular Biology آشنایی با مبانی اولیه سلول Biologists are interested in objects ranging in size from small molecules to the tallest trees: Cell Basic building blocks of life Understanding
Principles of Cellular Biology آشنایی با مبانی اولیه سلول Biologists are interested in objects ranging in size from small molecules to the tallest trees: Cell Basic building blocks of life Understanding
2012 Univ Aguilera Lecture. Introduction to Molecular and Cell Biology
 2012 Univ. 1301 Aguilera Lecture Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the
2012 Univ. 1301 Aguilera Lecture Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the
Biology II : Embedded Inquiry
 Biology II : Embedded Inquiry Conceptual Strand Understandings about scientific inquiry and the ability to conduct inquiry are essential for living in the 21 st century. Guiding Question What tools, skills,
Biology II : Embedded Inquiry Conceptual Strand Understandings about scientific inquiry and the ability to conduct inquiry are essential for living in the 21 st century. Guiding Question What tools, skills,
Prereq: Concurrent 3 CH
 0201107 0201101 General Biology (1) General Biology (1) is an introductory course which covers the basics of cell biology in a traditional order, from the structure and function of molecules to the structure
0201107 0201101 General Biology (1) General Biology (1) is an introductory course which covers the basics of cell biology in a traditional order, from the structure and function of molecules to the structure
Ledyard Public Schools Science Curriculum. Biology. Level-2. Instructional Council Approval June 1, 2005
 Ledyard Public Schools Science Curriculum Biology Level-2 1422 Instructional Council Approval June 1, 2005 Suggested Time: Approximately 9 weeks Essential Question Cells & Cell Processes 1. What compounds
Ledyard Public Schools Science Curriculum Biology Level-2 1422 Instructional Council Approval June 1, 2005 Suggested Time: Approximately 9 weeks Essential Question Cells & Cell Processes 1. What compounds
13.1 Biological Classification - Kingdoms and Domains Modern species are divided into three large groups, or domains. Bacteria Archaea Eukarya
 Chapter 13 Prospecting for Biological Gold Biodiversity and Classification 13.1 Biological Classification- How Many Species Exist? Biodiversity is the variety within and among living species Number of
Chapter 13 Prospecting for Biological Gold Biodiversity and Classification 13.1 Biological Classification- How Many Species Exist? Biodiversity is the variety within and among living species Number of
Rapid Learning Center Chemistry :: Biology :: Physics :: Math
 Rapid Learning Center Chemistry :: Biology :: Physics :: Math Rapid Learning Center Presents Teach Yourself AP Biology in 24 Hours 1/37 *AP is a registered trademark of the College Board, which does not
Rapid Learning Center Chemistry :: Biology :: Physics :: Math Rapid Learning Center Presents Teach Yourself AP Biology in 24 Hours 1/37 *AP is a registered trademark of the College Board, which does not
The facts about cells
 The facts about cells By Regina Bailey, ThoughtCo.com on 10.18.17 Word Count 867 Level MAX An illustration of cells. Photo from Pixabay. Cells are the fundamental units of life. Whether they be unicellular
The facts about cells By Regina Bailey, ThoughtCo.com on 10.18.17 Word Count 867 Level MAX An illustration of cells. Photo from Pixabay. Cells are the fundamental units of life. Whether they be unicellular
Slide 1 / Describe the setup of Stanley Miller s experiment and the results. What was the significance of his results?
 Slide 1 / 57 1 Describe the setup of Stanley Miller s experiment and the results. What was the significance of his results? Slide 2 / 57 2 Explain how dehydration synthesis and hydrolysis are related.
Slide 1 / 57 1 Describe the setup of Stanley Miller s experiment and the results. What was the significance of his results? Slide 2 / 57 2 Explain how dehydration synthesis and hydrolysis are related.
AP Biology Essential Knowledge Cards BIG IDEA 1
 AP Biology Essential Knowledge Cards BIG IDEA 1 Essential knowledge 1.A.1: Natural selection is a major mechanism of evolution. Essential knowledge 1.A.4: Biological evolution is supported by scientific
AP Biology Essential Knowledge Cards BIG IDEA 1 Essential knowledge 1.A.1: Natural selection is a major mechanism of evolution. Essential knowledge 1.A.4: Biological evolution is supported by scientific
Bioinformatics. Dept. of Computational Biology & Bioinformatics
 Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Cell (Learning Objectives)
 Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Biology Scope & Sequence
 Process Standards: Tools to Know: B.1(A) demonstrate safe practices during laboratory and field investigations B.1(B) demonstrate an understanding of the use and conservation of resources and the proper
Process Standards: Tools to Know: B.1(A) demonstrate safe practices during laboratory and field investigations B.1(B) demonstrate an understanding of the use and conservation of resources and the proper
Course Descriptions Biology
 Course Descriptions Biology BIOL 1010 (F/S) Human Anatomy and Physiology I. An introductory study of the structure and function of the human organ systems including the nervous, sensory, muscular, skeletal,
Course Descriptions Biology BIOL 1010 (F/S) Human Anatomy and Physiology I. An introductory study of the structure and function of the human organ systems including the nervous, sensory, muscular, skeletal,
Introduction to Microbiology BIOL 220 Summer Session I, 1996 Exam # 1
 Name I. Multiple Choice (1 point each) Introduction to Microbiology BIOL 220 Summer Session I, 1996 Exam # 1 B 1. Which is possessed by eukaryotes but not by prokaryotes? A. Cell wall B. Distinct nucleus
Name I. Multiple Choice (1 point each) Introduction to Microbiology BIOL 220 Summer Session I, 1996 Exam # 1 B 1. Which is possessed by eukaryotes but not by prokaryotes? A. Cell wall B. Distinct nucleus
Microbiota: Its Evolution and Essence. Hsin-Jung Joyce Wu "Microbiota and man: the story about us
 Microbiota: Its Evolution and Essence Overview q Define microbiota q Learn the tool q Ecological and evolutionary forces in shaping gut microbiota q Gut microbiota versus free-living microbe communities
Microbiota: Its Evolution and Essence Overview q Define microbiota q Learn the tool q Ecological and evolutionary forces in shaping gut microbiota q Gut microbiota versus free-living microbe communities
Bio/Life: Cell Biology
 Bio/Life: Cell Biology 1a The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism's cells. As a basis for understanding
Bio/Life: Cell Biology 1a The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism's cells. As a basis for understanding
Overview of Cells. Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory
 Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Genomes and Their Evolution
 Chapter 21 Genomes and Their Evolution PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Chapter 21 Genomes and Their Evolution PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Ap Biology Chapter 17 Packet Answers
 AP BIOLOGY CHAPTER 17 PACKET ANSWERS PDF - Are you looking for ap biology chapter 17 packet answers Books? Now, you will be happy that at this time ap biology chapter 17 packet answers PDF is available
AP BIOLOGY CHAPTER 17 PACKET ANSWERS PDF - Are you looking for ap biology chapter 17 packet answers Books? Now, you will be happy that at this time ap biology chapter 17 packet answers PDF is available
Introduction to Molecular and Cell Biology
 Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the molecular basis of disease? What
Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the molecular basis of disease? What
METHODS FOR DETERMINING PHYLOGENY. In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task.
 Chapter 12 (Strikberger) Molecular Phylogenies and Evolution METHODS FOR DETERMINING PHYLOGENY In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task. Modern
Chapter 12 (Strikberger) Molecular Phylogenies and Evolution METHODS FOR DETERMINING PHYLOGENY In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task. Modern
8/23/2014. Phylogeny and the Tree of Life
 Phylogeny and the Tree of Life Chapter 26 Objectives Explain the following characteristics of the Linnaean system of classification: a. binomial nomenclature b. hierarchical classification List the major
Phylogeny and the Tree of Life Chapter 26 Objectives Explain the following characteristics of the Linnaean system of classification: a. binomial nomenclature b. hierarchical classification List the major
Honors Biology summer assignment. Review the notes and study them. There will be a test on this information the 1 st week of class
 Honors Biology summer assignment Review the notes and study them. There will be a test on this information the 1 st week of class Biomolecules Molecules that make up living things. There are 4 molecules
Honors Biology summer assignment Review the notes and study them. There will be a test on this information the 1 st week of class Biomolecules Molecules that make up living things. There are 4 molecules
Biochemistry: A Review and Introduction
 Biochemistry: A Review and Introduction CHAPTER 1 Chem 40/ Chem 35/ Fundamentals of 1 Outline: I. Essence of Biochemistry II. Essential Elements for Living Systems III. Classes of Organic Compounds IV.
Biochemistry: A Review and Introduction CHAPTER 1 Chem 40/ Chem 35/ Fundamentals of 1 Outline: I. Essence of Biochemistry II. Essential Elements for Living Systems III. Classes of Organic Compounds IV.
SPRINGFIELD TECHNICAL COMMUNITY COLLEGE ACADEMIC AFFAIRS
 SPRINGFIELD TECHNICAL COMMUNITY COLLEGE ACADEMIC AFFAIRS Course Number: BIOL 102 Department: Biological Sciences Course Title: Principles of Biology 1 Semester: Spring Year: 1997 Objectives/ 1. Summarize
SPRINGFIELD TECHNICAL COMMUNITY COLLEGE ACADEMIC AFFAIRS Course Number: BIOL 102 Department: Biological Sciences Course Title: Principles of Biology 1 Semester: Spring Year: 1997 Objectives/ 1. Summarize
Big Idea 1: Does the process of evolution drive the diversity and unit of life?
 AP Biology Syllabus 2016-2017 Course Overview: AP Biology is equivalent to an introductory college level biology program in order to develop student led inquiry into science. The class is designed to go
AP Biology Syllabus 2016-2017 Course Overview: AP Biology is equivalent to an introductory college level biology program in order to develop student led inquiry into science. The class is designed to go
Grade Level: AP Biology may be taken in grades 11 or 12.
 ADVANCEMENT PLACEMENT BIOLOGY COURSE SYLLABUS MRS. ANGELA FARRONATO Grade Level: AP Biology may be taken in grades 11 or 12. Course Overview: This course is designed to cover all of the material included
ADVANCEMENT PLACEMENT BIOLOGY COURSE SYLLABUS MRS. ANGELA FARRONATO Grade Level: AP Biology may be taken in grades 11 or 12. Course Overview: This course is designed to cover all of the material included
A.P. Biology Lecture Notes Unit 1A - Themes of Life
 A.P. Biology Lecture Notes Unit 1A - Themes of Life I. Why study biology? A. Life is attractive, diverse, and interesting. B. The study of biology is enormous in scope C. Organisms span size scales from
A.P. Biology Lecture Notes Unit 1A - Themes of Life I. Why study biology? A. Life is attractive, diverse, and interesting. B. The study of biology is enormous in scope C. Organisms span size scales from
Comparative Virology as Context. Loy Volkman Professor Emerita Plant & Microbial Biology University of California, Berkeley
 Comparative Virology as Context Loy Volkman Professor Emerita Plant & Microbial Biology University of California, Berkeley LEFT FIELD Oxygen Accumulation and Evolutionary History of Life on Earth PHOTOSYNTHESIS
Comparative Virology as Context Loy Volkman Professor Emerita Plant & Microbial Biology University of California, Berkeley LEFT FIELD Oxygen Accumulation and Evolutionary History of Life on Earth PHOTOSYNTHESIS
Understanding Science Through the Lens of Computation. Richard M. Karp Nov. 3, 2007
 Understanding Science Through the Lens of Computation Richard M. Karp Nov. 3, 2007 The Computational Lens Exposes the computational nature of natural processes and provides a language for their description.
Understanding Science Through the Lens of Computation Richard M. Karp Nov. 3, 2007 The Computational Lens Exposes the computational nature of natural processes and provides a language for their description.
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/8/e1500527/dc1 Supplementary Materials for A phylogenomic data-driven exploration of viral origins and evolution The PDF file includes: Arshan Nasir and Gustavo
advances.sciencemag.org/cgi/content/full/1/8/e1500527/dc1 Supplementary Materials for A phylogenomic data-driven exploration of viral origins and evolution The PDF file includes: Arshan Nasir and Gustavo
MICROORGANISMS AND AMPHIBIANS
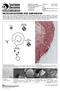 The life cycle of the chytrid fungus Batrachochytrium dendrobatidis begins with a motile zoospore, which is the infective stage of this pathogen. During the course of infection, chytrid zoospores enter
The life cycle of the chytrid fungus Batrachochytrium dendrobatidis begins with a motile zoospore, which is the infective stage of this pathogen. During the course of infection, chytrid zoospores enter
Components of a functional cell. Boundary-membrane Cytoplasm: Cytosol (soluble components) & particulates DNA-information Ribosomes-protein synthesis
 Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Computational Genomics. Reconstructing dynamic regulatory networks in multiple species
 02-710 Computational Genomics Reconstructing dynamic regulatory networks in multiple species Methods for reconstructing networks in cells CRH1 SLT2 SLR3 YPS3 YPS1 Amit et al Science 2009 Pe er et al Recomb
02-710 Computational Genomics Reconstructing dynamic regulatory networks in multiple species Methods for reconstructing networks in cells CRH1 SLT2 SLR3 YPS3 YPS1 Amit et al Science 2009 Pe er et al Recomb
Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes. - Supplementary Information -
 Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes - Supplementary Information - Martin Bartl a, Martin Kötzing a,b, Stefan Schuster c, Pu Li a, Christoph Kaleta b a
Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes - Supplementary Information - Martin Bartl a, Martin Kötzing a,b, Stefan Schuster c, Pu Li a, Christoph Kaleta b a
Inferring Transcriptional Regulatory Networks from Gene Expression Data II
 Inferring Transcriptional Regulatory Networks from Gene Expression Data II Lectures 9 Oct 26, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday
Inferring Transcriptional Regulatory Networks from Gene Expression Data II Lectures 9 Oct 26, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday
COMPETENCY GOAL 1: The learner will develop abilities necessary to do and understand scientific inquiry.
 North Carolina Draft Standard Course of Study and Grade Level Competencies, Biology BIOLOGY COMPETENCY GOAL 1: The learner will develop abilities necessary to do and understand scientific inquiry. 1.01
North Carolina Draft Standard Course of Study and Grade Level Competencies, Biology BIOLOGY COMPETENCY GOAL 1: The learner will develop abilities necessary to do and understand scientific inquiry. 1.01
BACTERIAL PHYSIOLOGY SMALL GROUP. Monday, August 25, :00pm. Faculty: Adam Driks, Ph.D. Alan Wolfe, Ph.D.
 BACTERIAL PHYSIOLOGY SMALL GROUP Monday, August 25, 2014 1:00pm Faculty: Adam Driks, Ph.D. Alan Wolfe, Ph.D. Learning Goal To understand how bacterial physiology applies to the diagnosis and treatment
BACTERIAL PHYSIOLOGY SMALL GROUP Monday, August 25, 2014 1:00pm Faculty: Adam Driks, Ph.D. Alan Wolfe, Ph.D. Learning Goal To understand how bacterial physiology applies to the diagnosis and treatment
CSCE555 Bioinformatics. Protein Function Annotation
 CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
Chapter 15 Active Reading Guide Regulation of Gene Expression
 Name: AP Biology Mr. Croft Chapter 15 Active Reading Guide Regulation of Gene Expression The overview for Chapter 15 introduces the idea that while all cells of an organism have all genes in the genome,
Name: AP Biology Mr. Croft Chapter 15 Active Reading Guide Regulation of Gene Expression The overview for Chapter 15 introduces the idea that while all cells of an organism have all genes in the genome,
Only skin deep: shared genetic response to the deadly chytrid fungus in susceptible frog species
 Molecular Ecology (2012) 21, 3110 3120 doi: 10.1111/j.1365-294X.2012.05481.x FROM THE COVER Only skin deep: shared genetic response to the deadly chytrid fungus in susceptible frog species ERICA BREE ROSENBLUM,*
Molecular Ecology (2012) 21, 3110 3120 doi: 10.1111/j.1365-294X.2012.05481.x FROM THE COVER Only skin deep: shared genetic response to the deadly chytrid fungus in susceptible frog species ERICA BREE ROSENBLUM,*
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
BIOLOGY Grades Summer Units: 10 high school credits UC Requirement Category: d. General Description:
 Summer 2015 Units: 10 high school credits UC Requirement Category: d General Description: BIOLOGY Grades 9-12 Summer session biology will be an intense, fast paced course. Students will gain an understanding
Summer 2015 Units: 10 high school credits UC Requirement Category: d General Description: BIOLOGY Grades 9-12 Summer session biology will be an intense, fast paced course. Students will gain an understanding
Bundle at a Glance Biology 2015/16
 Introduction: Scientific Investigation and Reasoning Skills (3 A/B days) Biology Process TEKS: 1A demonstrate safe practices during laboratory and field investigations. 1B demonstrate an understanding
Introduction: Scientific Investigation and Reasoning Skills (3 A/B days) Biology Process TEKS: 1A demonstrate safe practices during laboratory and field investigations. 1B demonstrate an understanding
SPRING GROVE AREA SCHOOL DISTRICT. Course Description. Instructional Strategies, Learning Practices, Activities, and Experiences.
 SPRING GROVE AREA SCHOOL DISTRICT PLANNED COURSE OVERVIEW Course Title: Advanced Placement Biology Grade Level(s): 12 Units of Credit: 1.50 Classification: Elective Length of Course: 30 cycles Periods
SPRING GROVE AREA SCHOOL DISTRICT PLANNED COURSE OVERVIEW Course Title: Advanced Placement Biology Grade Level(s): 12 Units of Credit: 1.50 Classification: Elective Length of Course: 30 cycles Periods
Biology 112 Practice Midterm Questions
 Biology 112 Practice Midterm Questions 1. Identify which statement is true or false I. Bacterial cell walls prevent osmotic lysis II. All bacterial cell walls contain an LPS layer III. In a Gram stain,
Biology 112 Practice Midterm Questions 1. Identify which statement is true or false I. Bacterial cell walls prevent osmotic lysis II. All bacterial cell walls contain an LPS layer III. In a Gram stain,
Evaluate evidence provided by data from many scientific disciplines to support biological evolution. [LO 1.9, SP 5.3]
![Evaluate evidence provided by data from many scientific disciplines to support biological evolution. [LO 1.9, SP 5.3] Evaluate evidence provided by data from many scientific disciplines to support biological evolution. [LO 1.9, SP 5.3]](/thumbs/75/72965665.jpg) Learning Objectives Evaluate evidence provided by data from many scientific disciplines to support biological evolution. [LO 1.9, SP 5.3] Refine evidence based on data from many scientific disciplines
Learning Objectives Evaluate evidence provided by data from many scientific disciplines to support biological evolution. [LO 1.9, SP 5.3] Refine evidence based on data from many scientific disciplines
Exhaustive search. CS 466 Saurabh Sinha
 Exhaustive search CS 466 Saurabh Sinha Agenda Two different problems Restriction mapping Motif finding Common theme: exhaustive search of solution space Reading: Chapter 4. Restriction Mapping Restriction
Exhaustive search CS 466 Saurabh Sinha Agenda Two different problems Restriction mapping Motif finding Common theme: exhaustive search of solution space Reading: Chapter 4. Restriction Mapping Restriction
Biology: Life on Earth
 Biology: Life on Earth Eighth Edition Lecture for Chapter 11 The Continuity of Life: Cellular Reproduction Cellular Reproduction Intracellular activity between one cell division to the next is the cell
Biology: Life on Earth Eighth Edition Lecture for Chapter 11 The Continuity of Life: Cellular Reproduction Cellular Reproduction Intracellular activity between one cell division to the next is the cell
AP Biology Curriculum Framework
 AP Biology Curriculum Framework This chart correlates the College Board s Advanced Placement Biology Curriculum Framework to the corresponding chapters and Key Concept numbers in Campbell BIOLOGY IN FOCUS,
AP Biology Curriculum Framework This chart correlates the College Board s Advanced Placement Biology Curriculum Framework to the corresponding chapters and Key Concept numbers in Campbell BIOLOGY IN FOCUS,
Class Work 31. Describe the function of the Golgi apparatus? 32. How do proteins travel from the E.R. to the Golgi apparatus? 33. After proteins are m
 Eukaryotes Class Work 1. What does the word eukaryote mean? 2. What is the one major difference between eukaryotes and prokaryotes? 3. List the different kingdoms of the eukaryote domain in the order in
Eukaryotes Class Work 1. What does the word eukaryote mean? 2. What is the one major difference between eukaryotes and prokaryotes? 3. List the different kingdoms of the eukaryote domain in the order in
Introduction. Gene expression is the combined process of :
 1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
Dr. Amira A. AL-Hosary
 Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
 1 of 13 8/11/2014 10:32 AM Units: Teacher: APBiology, CORE Course: APBiology Year: 2012-13 Chemistry of Life Chapters 1-4 Big Idea 1, 2 & 4 Change in the genetic population over time is feedback mechanisms
1 of 13 8/11/2014 10:32 AM Units: Teacher: APBiology, CORE Course: APBiology Year: 2012-13 Chemistry of Life Chapters 1-4 Big Idea 1, 2 & 4 Change in the genetic population over time is feedback mechanisms
The most widely used biological classification system has six kingdoms within three domains.
 Section 3: The most widely used biological classification system has six kingdoms within three domains. K What I Know W What I Want to Find Out L What I Learned Essential Questions What are the major characteristics
Section 3: The most widely used biological classification system has six kingdoms within three domains. K What I Know W What I Want to Find Out L What I Learned Essential Questions What are the major characteristics
Gene Ontology and Functional Enrichment. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Supplementary Information
 Supplementary Information Supplementary Figure 1. Schematic pipeline for single-cell genome assembly, cleaning and annotation. a. The assembly process was optimized to account for multiple cells putatively
Supplementary Information Supplementary Figure 1. Schematic pipeline for single-cell genome assembly, cleaning and annotation. a. The assembly process was optimized to account for multiple cells putatively
Computational Biology Course Descriptions 12-14
 Computational Biology Course Descriptions 12-14 Course Number and Title INTRODUCTORY COURSES BIO 311C: Introductory Biology I BIO 311D: Introductory Biology II BIO 325: Genetics CH 301: Principles of Chemistry
Computational Biology Course Descriptions 12-14 Course Number and Title INTRODUCTORY COURSES BIO 311C: Introductory Biology I BIO 311D: Introductory Biology II BIO 325: Genetics CH 301: Principles of Chemistry
