A differential equation model for functional mapping of a virus-cell dynamic system
|
|
|
- Bryce Stokes
- 6 years ago
- Views:
Transcription
1 J. Math. Biol. DOI /s Mathematical Biology A differential equation model for functional mapping of a virus-cell dynamic system Jiangtao Luo William W. Hager Rongling Wu Received: 23 January 2009 / Revised: 13 July 2009 Springer-Verlag 2009 Abstract The dynamic pattern of viral load in a patient s body critically depends on the host s genes. For this reason, the identification of those genes responsible for virus dynamics, although difficult, is of fundamental importance to design an optimal drug therapy based on patients genetic makeup. Here, we present a differential equation (DE) model for characterizing specific genes or quantitative trait loci (QTLs) that affect viral load trajectories within the framework of a dynamic system. The model is formulated with the principle of functional mapping, originally derived to map dynamic QTLs, and implemented with a Markov chain process. The DE-integrated model enhances the mathematical robustness of functional mapping, its quantitative prediction about the temporal pattern of genetic expression, and therefore its practical utilization and effectiveness for gene discovery in clinical settings. The model was used to analyze simulated data for viral dynamics, aimed to investigate its statistical properties and validate its usefulness. With an increasing availability of genetic polymorphic data, the model will have great implications for probing the molecular genetic mechanism of virus dynamics and disease progression. Keywords Differential equation Functional mapping Quantitative trait loci Viral dynamics Mathematics Subject Classification (2000) 62 Statistics J. Luo W. W. Hager Department of Mathematics, University of Florida, Gainesville, FL 32611, USA R. Wu (B) Departments of Public Health Sciences and Statistics, Pennsylvania State University College of Medicine, Hershey, PA 17033, USA rwu@hes.hmc.psu.edu
2 J. Luo et al. 1 Introduction Several serious human diseases, such as AIDS, hepatitis B, influenza, and rabies, are caused by viruses. To control these diseases, antiviral drugs have been developed to prevent infection of new viral cells or stop already-infected cells from producing infectious virus particles by inhibiting specific viral enzymes. This process constitutes a complex dynamic system, in which different types of viral cells, including uninfected cells, infected cells, and free virus particles, interact with each other to determine the pattern of viral change in response to drugs (Ho et al. 1999; Wei et al. 2004; Perelson et al. 1997, 1996; Sedaghat et al. 2008). A major challenge that faces drug development and delivery for controlling viral diseases is to develop a quantitative model for analyzing and predicting the dynamics of decline in virus load during drug therapy and further providing estimates of the rate of emergence of resistant virus. The development of such a model can now be made possible with recent advances in two seemingly unrelated areas. First, the combination between novel instruments and an increasing understanding of molecular genetics has led to the birth of highthroughput genotyping assays for single nucleotide polymorphisms (SNPs). Through the construction of a haplotype map (HapMap) with SNP data (The International HapMap Consortium 2003), we are able to characterize concrete nucleotides or their combinations that encode a complex phenotype, and ultimately document, map and understand the structure and patterns of the human genome linked to drug response. Second, the past two decades have witnessed a tremendous growth of interest in deriving sophisticated mathematical models for characterizing virus dynamics from molecular and cellular mechanisms of interactions between virus and drug (Ho et al. 1999; Wei et al. 2004; Perelson et al. 1997, 1996; Sedaghat et al. 2008; Bonhoeffer et al. 1997; Bonhoeffer et al. 1999; Nowak and May 2000). These models mostly built with differential equations (DE) have been instrumental for studying the function of virus and the origins and properties of virus dynamics. These two advances can be integrated to identify specific genes or quantitative trait loci (QTLs) that regulate a dynamic system of viral infection through a new statistical model called functional mapping (Ma et al. 2002; Wang and Wu 2004; Wu et al. 2004a,b,c; Wu and Lin 2006). The basic idea of functional mapping is to map dynamic QTL for the pattern of developmental changes in time course. The purpose of this article is to propose a statistical strategy for implementing a system of DE into the functional mapping framework, ultimately to map QTLs from the host genome that determine the dynamic pattern of virus load in patients bodies. The new strategy is founded on a set of random samples drawn from a natural population at Hardy Weinberg equilibrium (HWE). We integrate the Markov chain properties of dynamic data into the model to facilitate the estimation of parameters that define virus dynamics. Simulation studies were performed to investigate statistical properties of the model and validate its usefulness and utilization.
3 Functional mapping of a virus-cell dynamic system 2 Dynamic models of virus load 2.1 Differential equations A basic model for describing short-term virus dynamics was provided by many researchers (Bonhoeffer et al. 1997; Bonhoeffer et al. 1999; Nowak and May 2000). This model includes three variables: uninfected cells, x, infected cells, y, and free virus particles, v. These three types of cells interact with each other to determine the dynamic changes of virus in a host s body, which can be described by a system of ordinary differential equations (ODE): dx dt dy dt dv dt = λ dx βxv = βxv ay (1) = ky uv, where uninfected cells are yielded at a constant rate, λ, and die at the rate dx; free virus infects uninfected cells to yield infected cells at rate βxv; infected cells die at rate ay; and new virus is yielded from infected cells at rate ky and dies at rate uv (Bonhoeffer et al. 1999). The system (1) is defined by six parameters {λ, d,β,a, k, u} and the initial conditions for x, y, and v. The dynamic pattern of this system can be determined and predicted by the change of these parameters and the initial conditions of x, y, and v. There are some practical problems in the real application. First, we can only observe the data for x, y, and v at discrete time points, and it is difficult to get the continuous dx dv, and dt terms. Second, any biological development is related to genes, but the model does not involve any genetic components. Third, the dynamic change of the virus is accompanied by noise which cannot be neglected in the dynamic modeling. It should be noted that the model (1) used to explain our idea in this article is a basic sculpture of real virus infection as it ignores the dynamics of immune responses and virus mutations. Let 0 = t 0 < t 1 < < t N = T denote a mesh on the time interval [0, T ] and define t k = t k+1 t k. The Euler approximation to the continuous differential equations (1)is dt, dy dt x(t k+1 ) x(t k ) = λ dx(t k ) βx(t k )v(t k ) t k y(t k+1 ) y(t k ) = βx(t k )v(t k ) ay(t k ) (2) t k v(t k+1 ) v(t k ) = ky(t k ) uv(t k ), t k
4 J. Luo et al. or equivalently, x(t k+1 ) = x(t k ) + λ t k dx(t k ) t k βx(t k )v(t k ) t k y(t k+1 ) = y(t k ) + βx(t k )v(t k ) t k ay(t k ) t k (3) v(t k+1 ) = v(t k ) + ky(t k ) t k uv(t k ) t k. 2.2 Markov properties Suppose there is a random sample with n patients from a population carrying a certain virus. Each patient is measured for uninfected cells, x, infected cells, y, and free virus particles, v, at a series of time points, (t 0, t 1,...,t N ). Thus, three sets of serial measurements are expressed as x i =[x i (t 0 ),...,x i (t N )], y i =[y i (t 0 ),..., y i (t N )], and v i =[v i (t 0 ),...,v i (t N )], where the subscript i corresponds to the patient and t j, 0 j N, are the measurement times. A transitional Markov model is used to describe the random process of the system by x i (t k+1 ) = x i (t k ) + λ t k dx i (t k ) t k βx i (t k )v i (t k ) t k + ɛ xi (t k ) y i (t k+1 ) = y i (t k ) + βx i (t k )v i (t k ) t k ay i (t k ) t k + ɛ yi (t k ) (4) v i (t k+1 ) = v i (t k ) + ky i (t k ) t k uv i (t k ) t k + ɛ vi (t k ), where ɛ xi (t k ) N(0,σx 2), ɛ y i (t k ) N(0,σy 2), and ɛ v i (t k ) N(0,σv 2 ) are the innovation errors for three variables, x, y, and v, respectively, each of which is assumed to be iid and time-independent. To simplify our line of analysis, we assume that these three variables are independent of each other, although this assumption can be relaxed. For simplicity, we use x ik, y ik, and v ik to stand for x i (t k ), y i (t k ), and v i (t k ), respectively. For a conditional density function, f (..), we derive the Markov properties of the dynamic system (1) asfollows: Theorem 1 All the future values of uninfected cells, infected cells, and free virus particles depend statistically only on their present values. That is, f (x ik+1, y ik+1,v ik+1 (x i1, y i1,v i1 ),...,(x ik, y ik,v ik )) = f (x ik+1, y ik+1,v ik+1 (x ik, y ik,v ik )), f (x ik+1 (x i1, y i1,v i1 ),...,(x ik, y ik,v ik )) = f (x ik+1 (x ik, y ik,v ik )), f (y ik+1 (x i1, y i1,v i1 ),...,(x ik, y ik,v ik )) = f (y ik+1 (x ik, y ik,v ik )), f (v ik+1 (x i1, y i1,v i1 ),...,(x ik, y ik,v ik )) = f (v ik+1 (x ik, y ik,v ik )). The proof follows directly from (4) and the definitions of ɛ xi (t k ), ɛ yi (t k ), and ɛ zi (t k ). From this theorem, we have the following results. Corollary 2.1 Conditional on (x ik, y ik,v ik ), (x ik 1, y ik 1,v ik 1 ) and (x ik+1, y ik+1, v ik+1 ) are statistically independent.
5 Functional mapping of a virus-cell dynamic system Corollary 2.2 Conditional on (x ik, y ik,v ik ),x ik 1 and x ik+1 are statistically independent. Corollary 2.3 Conditional on (x ik, y ik,v ik ), y ik 1 and y ik+1 are statistically independent. Corollary 2.4 Conditional on (x ik, y ik,v ik ), v ik 1 and v ik+1 are statistically independent. Since ((x ik+1, y ik+1,v ik+1 ) (x ik, y ik,v ik ), (x ik 1, y ik 1,v ik 1 )) = f ((x ik+1, y ik+1,v ik+1 ) (x ik, y ik,v ik )), conditional on (x ik, y ik,v ik ), (x ik 1, y ik 1,v ik 1 ) and (x ik+1, y ik+1,v ik+1 ) are statistically independent (Bremaud 1999). Hence, Corollary 2.1 holds. The proofs of Corollaries can be made in a similar way. Now, we get the following theorems: Corollary 2.5 Conditional on (x ik, y ik,v ik ), (x ij, y ij,v ij ) for j = 0, 1,...,k 1 and (x ik+1, y ik+1,v ik+1 ) are statistically independent. Corollary 2.6 Conditional on (x ik, y ik,v ik ), {x i1,...,x ik 1 }, and x ik+1 are statistically independent. Corollary 2.7 Conditional on (x ik, y ik,v ik ), {y i1,...,y ik 1 }, and y ik+1 are statistically independent. Corollary 2.8 Conditional on (x ik, y ik,v ik ), {v i1,...,v ik 1 }, and v ik+1 are statistically independent. The Theorem and all these corollaries will be used to derive computing algorithms for solving a system of differential equations (4) embedded in functional mapping. 3 Functional mapping 3.1 Genetic design Genetic mapping of QTLs can be based on linkage analysis for a pedigree (Lander and Bostein 1989) or linkage disequilibrium analysis for a natural population (Wang and Wu 2004). In this article, we assume that the population used to map human QTLs for viral load trajectories is composed of n patients randomly sampled from a natural population at HWE. A panel of SNP markers are genotyped for all patients, aimed at the identification of QTLs affecting virus dynamics. Suppose there is a functional QTL of alleles A and a for virus dynamics. Let q and 1 q denote the allele frequencies of A and a. The QTL forms three possible genotypes, AA, Aa, and aa. We assume that this QTL is associated with a SNP marker of alleles M (in a frequency of p) and m
6 J. Luo et al. (in a frequency of 1 p). The detection of significant linkage disequilibrium between the marker and QTL implies that the QTL may be linked with and,therefore, can be genetically manipulated by the marker. The four haplotypes for the marker and QTL are MA, Ma, ma and ma, with respective frequencies expressed as p 11 = pq + D, p 10 = p(1 q) D, p 01 = (1 p)q D, and p 00 = (1 p)(1 q) + D, where D is the linkage disequilibrium between the marker and QTL. Thus, the population genetic parameters ( p, q, and D) can be estimated by solving a group of regular equations if we can estimate the four haplotype frequencies = (p 11, p 10, p 01, p 00 ). Joint marker-qtl diplotype frequencies can be expressed as a product of the corresponding haplotype frequencies under the HWE assumption, from which joint marker-qtl genotype frequencies are derived. Because the marker is observed, an unknown genotype of the QTL can be inferred from the conditional probability of the QTL genotype given a marker genotype. Each sampled patient is measured for three different traits, uninfected cells, x, infected cells, y, and free virus particles, v, at a series of time points, (t i1,...,t iti ). 3.2 Likelihood For a given QTL genotype j ( j =2forAA,1forAa, or0foraa), the parameters describing virus dynamics are denoted by j ={λ j, d j,β j, a j, k j, u j }. The comparisons of these parameters between the three different QTL genotypes can determine whether and how this QTL affects the pattern of virus dynamics. The likelihood of longitudinal viral data (x i, y i, v i ) ={x i (t k ), y i (t k ), v i (t k )} N k=0 and marker information M i for patient i is formulated by the mixture transitional Markov model, expressed as L(x, y, v; M) = n 2 ω j i f j (x i, y i, v i ; j, ), (5) i=1 j=0 where ω j i is a mixture proportion, that is, the conditional probability of QTL genotype j given the marker genotype of subject i, which can be expressed as a function of haplotype frequencies (Table 1), and f j (x i, y i, v i ; j, ) is a multivariate normal distribution with QTL genotype-specific mean vector specified by ODE parameters ( j ) and covariance matrix specified by parametric, non-parametric, or semiparametric models ( )(Ma et al. 2002; Wu and Lin 2006). Based on the Corollaries given above, the multivariate distribution can be specified by the following transition model f j (x i, y i, v i ; j, ) N 1 = f j (x i1, y i1,v i1 j, ) k=0 f j (x ik+1, y ik+1,v ik+1 x ik, y ik,v ik ; j, ) (6)
7 Functional mapping of a virus-cell dynamic system Table 1 Joint genotype frequencies at the marker and QTL in terms of gametic haplotype frequencies, from which the conditional probabilities of QTL genotypes given marker genotypes can be calculated according to Bayes theorem Genotype Diplotype AA Aa aa Observations A A A a + a A a a MM M M p p 11 p 10 p 2 10 N 1 Mm M m 2p 11 p 01 2p 11 p p 10 p 01 2p 10 p 00 N 2 mm m m p p 01 p 00 p 2 00 N 3 where f j (x ik+1, y ik+1,v ik+1 x ik, y ik,v ik ; j, ) = f j (x ik+1 x ik, y ik,v ik ; j, ) f j (y ik+1 x ik, y ik,v ik ; j, ) f j (v ik+1 x ik, y ik,v ik ; j, ), [ f j (x ik+1 x ik, y ik,v ik ; j,σx 2 ) = 1 exp 1 2πσ 2 x 2σx 2 f j (y ik+1 x ik, y ik,v ik ; j,σy 2 ) = 1 2πσy 2 [ exp 1 2σy 2 [ f j (v ik+1 x ik, y ik,v ik ; j,σv 2 ) = 1 exp 1 2πσ 2 v 2σv 2 with = (σx 2,σ2 y,σ2 v ), and ( xik+1 g j (x ik+1 ) ) ] 2, ( yik+1 h j (y ik+1 ) ) ] 2, ( vik+1 l j (v ik+1 ) ) ] 2, g j (x ik+1 ) = x ik + λ j t k d j x ik t k β j x ik v ik t k h j (y ik+1 ) = y ik + β j x ik v ik t k a j y ik t k (7) l j (v ik+1 ) = v ik + k j y ik t k u j v ik t k. 3.3 Estimation and algorithm The EM algorithm (Dempster et al. 1977; Little and Rubin 2002) is implemented to get the maximum likelihood estimates (MLE) of all unknown parameters. The gradient of the log-likelihood function n 2 log L(x, y, v; M) = log ω j i f j (x i, y i, v i ; j, ), (8) i=1 j=0
8 J. Luo et al. is given by j log L(x, y, v; M) = n 2 i=1 j=0 log L(x, y, v; M) = n 2 i=1 j=0 ω j i f j (x i, y i, v i ; j, ) 2j =0 ω j i f j (x i, y i, v i ; j, ) j log f j (x i, y i, v i ; j, ), ω j i f j (x i, y i, v i ; j, ) 2j =0 ω j i f j (x i, y i, v i ; j, ) log f j (x i, y i, v i ; j, ), and ω j i log L(x, y, v; M) = n 2 i=1 j=0 ω j i f j (x i, y i, v i ; j, ) 2j =0 ω j i f j (x i, y i, v i ; j, ) ω j i log(ω j i ). A iterative loop for the EM algorithm is formulated as follows. In the E step, the posterior probability with which a patient i carries a specific QTL genotype j based on the marker and phenotypic data is calculated by j i = ω j i f j (x i, y i, v i ; j, ) 2j =0 ω j i f j (x i, y i, v i ; j, ). (9) In the M step, the parameters are estimated by solving the following log-likelihood equations: j log L(x, y, v; M) = 0, (10) log L(x, y, v; M) = 0, (11) ω j i log L(x, y, v; M) = 0. (12) Wang and Wu (2004) derived a closed algorithmic form to obtain the MLEs of haplotype frequencies p 11, p 10, p 01 and p 00 and, therefore, allele frequencies of the marker (p) and QTL (q) and their linkage disequilibrium (D). Genotype-specific mathematical parameters for viral dynamics and variances for the three types of viruses are calculated by implementing the Newton algorithm with the Armijo search (Bertsekas 2003).
9 Functional mapping of a virus-cell dynamic system 4 Hypothesis testing 4.1 The significance of QTL Whether there is a specific QTL responsible for viral dynamics described by a system of differential equations 1 can be tested by using the following hypotheses: H 0 : j, (j = 2, 1, 0) H 1 : At least one of the equalities above does not hold, (13) The likelihoods under the null (L 0 ) and alternative hypotheses (L 1 ) are calculated, from which a log-likelihood ratio test statistic is computed by [ ] LR = 2 log L 0 (, x, y, z) log L 1 ( ˆ, ˆ j, ˆ x, y, z, M), where the tildes and hats present the MLE under the null and alternative hypotheses, respectively. Because of violation of the regularity assumption, the LR may not asymptotically follow a χ 2 -distribution with the degrees of freedom equal to the difference of parameter numbers between the two hypotheses (13). For this reason, the threshold for claiming the existence of a significant QTL is determined from empirical permutation tests (Churchill and Doerge 1994) because this approach does not rely on the distribution of LR values. After a significant QTL is claimed, its significant association with the marker considered can be tested by the following hypotheses: H 0 : D = 0vs. H 1 : D = 0, (14) whose log-likelihood ratio test statistic asymptotically follow the χ 2 -distribution with one degree of freedom. 4.2 Genetic mechanisms The model allows the test of whether the QTL triggers a pleiotropic effect on three different types of cells. To do so, three null hypotheses for uninfected cells, infected cells, and free virus particles are formulated as follows: H 0 : (λ j, d j,β j ) (λ, d,β), (15) H 0 : (β j, a j ) (β, a), (16) H 0 : (k j, u j ) (k, u), (17) for j = 2, 1, 0. If all the null hypotheses are rejected, then this means that the QTL pleiotropically affect these three different aspects of viral dynamics. The pleiotropic effect of the QTL on any pair of three types of cells can also be tested accordingly. An empirical approach for determining the critical threshold is based on simulation studies.
10 J. Luo et al. 4.3 Physiological control of QTL Several physiological important parameters define the dynamic system (1)(Bonhoeffer et al. 1999), including 1. The average life-times, 1/d, 1/a, and 1/u, of uninfected cells, infected cells, and free virus, respectively, 2. The average number of virus particles or the burst size, k/a, yielded over the lifetime of a single infected cell, 3. Basic reproductive ratio, R 0 = βλk/(adu), i.e., the average number of newly infected cells that arise from any one infected cell when almost all cells are uninfected. How a QTL affects these physiological aspects of viral dynamics separately or jointly can be tested. 5 Application to simulated data Monte Carlo simulation was performed to examine the statistical properties of the model for genetic mapping of viral dynamics. Also, the use of the model to analyze simulated data will validate its practical usefulness and utilization. We randomly choose 100 subjects from an HWE population. Consider one of the markers genotyped for all subjects. This marker of two alleles M and m is used to infer a QTL of two alleles A and a for viral dynamics based on the non-random association between the marker and QTL. The allele frequencies are assumed as p = 0.6 for allele M 0.4 for allele m as well as q = 0.6 for allele A and 0.4 for allele a. A positive value of linkage disequilibrium (D = 0.08) between alleles M and A is assumed, suggesting that these two more common alleles are in coupling phase. The three QTL genotypes, AA, Aa, and aa, are each hypothesized to have different response systems for uninfected cells, x, infected cells, y, and free virus particles, v, constructed by Eqs. (1). Six curve parameters {λ j, d j,β j, a j, k j, u j } that define QTL genotype-specific systems were chosen from their spaces of biological relevance (Bonhoeffer et al. 1999). The phenotypic values of these three variables are expressed as the sum of the genotype-specific means and innovation errors assumed to follow a multivariate normal distribution. The phenotypic data were simulated for a practically reasonable number of equally spaced time points (say 22) under two different levels of heritability, low (0.1) and high (0.4). The genetic variance due to the QTL for virus response at a middle measurement point was used to define the heritability. The residual variances for each of the three virus traits were then calculated under different heritabilities. To assure the homoscedasticity of variances, the transforms-both-sides (TBS) model was used to simulate innovation errors. The TBS model can preserve biological means of parameters in original DE and also avoid negative phenotypic values (Wu et al. 2004b). The DE-incorporated functional mapping model was used to analyze the simulated data, with the results suggesting that the QTL responsible for the dynamic system of viral infection can be detected using a molecular marker in association with the
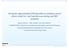
11 Functional mapping of a virus-cell dynamic system QTL. As expected, population genetic parameters about QTL segregation in a population can well be estimated with a closed form of the EM algorithm derived in (Wang and Wu 2004). The curve parameters for virus responses of each QTL genotype can be estimated accurately and precisely with a modest sample size (100) even for a low heritability of viral loads (Tables 2, 3). The precision of all parameters can increase with increasing heritability level. By drawing the curves of viral trajectories with six parameters, the dynamic behavior of the system can be visualized. Figure 1 illustrates QTL genotype-specific curves of uninfected cells, infected cells, and free virus particles in a dynamic system from a random run of simulation. It is found that the shapes of the estimated curves are broadly consistent with the those of the true curves, suggesting that the system can be reasonably estimated with the new model. Simulation studies showed that the new model displays reasonably high power, 0.75 for a modest heritability (0.1) and 0.99 for a high heritability (0.4), to detect a significant QTL responsible for a dynamic system of viral infection. Hypothesis tests described in Sects. 4.2 and 4.3 provide a general platform for addressing the genetic control machinery of viral dynamics. For a given set of simulation data, it appears that these tests can be reasonably made. For example, the power for detecting a pleiotropic QTL for three types of viral cell dynamics is adequately high ( 0.7) for a modest sample size and heritability level. On the other hand, under this circumstance, type I error rates for detecting a significant QTL despite its absence is reasonably low Table 2 The MLEs of parameters that define virus-host dynamics for three different QTL genotypes, and the association between the marker and QTL in a natural population, assuming that the heritability of the simulated QTL is H 2 = 0.1 AA Aa aa Given MLE Given MLE Given MLE Virus-host parameters λ (0.0137) (0.0115) (0.0142) d (0.0017) (0.0014) (0.0017) β (0.0001) (0.0001) (0.0031) a (0.0008) (0.0006) (0.0008) k (0.0010) (0.0008) (0.0012) u (0.0025) (0.0023) (0.0033) Given MLE Genetic parameters and variances p (0.0312) q (0.0680) D (0.0108) σx (0.0107) σy (0.0105) σv (0.0239) The numbers in the parentheses are the square roots of the mean square errors of the MLEs
12 J. Luo et al. Table 3 The MLEs of parameters that define virus-host dynamics for three different QTL genotypes, and the association between the marker and QTL in a natural population, assuming that the heritability of the simulated QTL is H 2 = 0.4 AA Aa aa Given MLE Given MLE Given MLE Rhythmic parameters λ (0.0069) (0.0057) (0.0100) d (0.0009) (0.0007) (0.0012) β (0.0000) (0.0000) (0.0000) a (0.0003) (0.0003) (0.0005) k (0.0004) (0.0003) (0.0006) u (0.0011) (0.0009) (0.0017) Given MLE Genetic parameters p (0.0336) q (0.0407) D (0.0047) σx (0.0020) σy (0.0031) σv (0.0045) The numbers in the parentheses are the square roots of the mean square errors of the MLEs ( 0.1). These results suggest that our model will be practically useful in statistical analysis of the genetic control of viral dynamics. 6 Discussion A combination of functional mapping (Ma et al. 2002; Wang and Wu 2004; Wu et al. 2004a,b,c; Wu and Lin 2006) and mathematical models (Bonhoeffer et al. 1997; Bonhoeffer et al. 1999; Nowak and May 2000) provides new insights into the genetic control of virus population dynamics. In this article, we have proposed a statistical model for mapping QTLs that affect the dynamic pattern of viral infection. One of the meritorious advantages of the new model, as compared to existing functional mapping models, lies in the organization of multiple correlated aspects of viral infection into a dynamic system through a group of ODE and the implementation of such a viral dynamic system into the framework of functional mapping. To our best knowledge, the work presented here is a first model of genetic mapping which treats multiple complex traits as a complex system. The current model is not a simple extension of functional mapping for multiple traits (Zhao et al. 2005). The previous multi-trait models do not take into account the relationships of genotypic values of different traits, although they model across-trait correlations in residual errors. The new model views multiple traits as a whole in which different traits coordinate each other to determine the dynamic behavior of the
13 Functional mapping of a virus-cell dynamic system 10 Heritability Heritability 0.4 Uninfected Cells Aa aa AA Uninfected Cells AA Aa aa Time Heritability Time Heritability 0.4 Infected Cells AA aa Aa Infected Cells AA aa Aa Time Heritability Time Heritability Virus Particles AA aa Aa Virus Particles AA aa Aa Time Time Fig. 1 Estimated and true curves for a system of viral infection including uninfected cells, x, infected cells, y, and free virus particles, v for three genotypes at a simulated QTL, AA, Aa,andaa, under different heritability levels, 0.1 (right panel) and 0.4 (left panel). The broad consistency between the estimated and true curves suggests that the model can provide a reasonably good estimate of the dynamic system system. Thus, by altering one variable or trait, other variables will change, leading to the change of the entire system. The genetic mapping of genes for a dynamic system will provide a powerful means for understanding the genetic architecture of a biological process. The mathematical strength of the new model is the deployment of a system of DE in a genetic mapping context. The solution of multiple DE, especially high-dimensional
14 J. Luo et al. ones, is computationally challenging. In this article, we apply a Newton algorithm with the EM setting to provide numerical estimates of the parameters that define the dynamic system. With the corollaries derived from several assumptions of independence, the algorithm is shown from simulation studies to be computationally efficient and provides precise estimates of the parameters, even when the sample size used is modest (Tables 2, 3). As a demonstration of the new model, we assume that a dynamic system is controlled by a single QTL, although this assumption is too simple in real world. The genome-wide modeling of multiple QTLs throughout the genome can be incorporated into the current model setting, allowing the characterization of epistatic interactions among different QTLs (Wang et al. 2005; Wu et al. 2006). A multi-locus linkage disequilibrium model has been available to specify high-order non-random associations among multiple loci in a natural population (Li and Wu 2009). Although more parameters are involved in a multi-locus model, the closed forms derived for the EM algorithm (Li and Wu 2009) facilitates the estimation of many parameters at the same time. Also, a multi-locus model allows the test of the role of genetic interference in recombination events between adjacent intervals. Although linkage disequilibrium mapping has proven to be powerful for the high-resolution of QTLs, it often gives spurious results due to population structure and other evolutionary forces. A new genetic design that samples a set of random families, each composed of parents and their offspring, can overcome this limitation of linkage disequilibrium mapping (Li and Wu 2009; Wu et al. 2002). This design allows the simultaneous estimation of the linkage and linkage disequilibrium between different genes, thus making it possible to construct a genome-wide linkage disequilibrium map for gene discovery. Our model focuses on the identification of genes for a dynamic system of viral changes in a host s body before the administration of an anti-viral drug. When the patients are treated with a drug, the equilibrium state of the system will be violated, from which a new equilibrium will be generated. Bonhoeffer et al. (1999) described a series of DE that specify the dynamic change of the system after drug treatment. The current model can be readily extended to model the genetic control of viral declines in a response to the anti-viral drug and half-lives of infected cells in the body. Perhaps, the most promising aspect of the new model is that it, when incorporated with the dynamics of virus drug resistance, can provide scientific guidance for drug delivery and development by characterizing genes for drug resistance. The emergence of drug-resistant virus presents a main problem with antiviral therapy. A system of DE that captures the essential dynamics of resistance is given in the literature (Ribeiro and Bonhoeffer 2000; Wodarz and Nowak 2000). With the idea presented in this article, they can be readily incorporated into the functional mapping model, in a hope to achieve the maximum prevention of virus resistance to drugs by determining an optimal administration dose and time for individual patients based on their genetic makeups. Acknowledgments We thank two anonymous referees for their constructive comments on the manuscript which led to a better presentation of it. This work is partially supported by NSF Grants and and NSF/NIH Joint grant DMS/NIGMS
15 Functional mapping of a virus-cell dynamic system References Bertsekas DP (2003) Nonlinear programming, 2nd edn. Athena Scientific, Belmont Bonhoeffer S, Coffin JM, Nowak MA (1997) Human immuodeficiency virus drug therapy and virus load. J Virol 71: Bonhoeffer S, May RM, Shaw GM, Nowak MA (1999) Virus dynamics and drug therapy. Proc Natl Acad Sci USA 94: Bremaud P (1999) Markov chains: Gibbs fields, Monte Carlo simulation and queues. Springer, New York Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via EM algorithm. J R Stat Soc B 39:1 38 Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M (1999) Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373: 126 Lander ES, Bostein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: Little RJA, Rubin DB (2002) Statistical analysis with missing data, 2nd edn. Wiley, New York Li Q, Wu RL (2009) A multilocus odel for constructing a linkage disequilibrium map in human populations. Stat Appl Mol Genet Biol 8:Article 18 Ma CX, Casella G, Wu RL (2002) Functional mapping of quantitative trait loci underlying the character process: a theoretical framework. Genetics 161: Nowak MA, May RM (2000) Virus dynamics. Oxford University, New York Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD (1996) HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271: Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD (1997) Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387: Ribeiro RM, Bonhoeffer S (2000) Production of resistant HIV mutants during antiretroviral therapy. Proc Natl Acad Sci USA 97: Sedaghat AR, Dinoso JB, Shen L, Wilke CO, Siliciano RF (2008) Decay dynamics of HIV-1 depend on the inhibited stages of the viral life cycle. Proc Natl Acad Sci 105: The International HapMap Consortium (2003) The International HapMap Project. Nature 426: Wang ZH, Wu RL (2004) A statistical model for high-resolution mapping of quantitative trait loci determining HIV dynamics. Stat Med 23: Wang ZH, Hou W, Wu RL (2005) A statistical model to analyze quantitative trait locus interactions for HIV dynamics from the virus and human genomes. Stat Med 25: Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, Saag MS, Shaw GM (2004) Viral dynamics in human immunodeficiency virus type 1 infection. Nature 23: Wodarz D, Nowak MA (2000) HIV therapy: managing resistance. Proc Natl Acad Sci USA 97: Wu RL, Lin M (2006) Functional mapping how to study the genetic architecture of dynamic complex traits. Nat Rev Genet 7: Wu RL, Ma C-X, Casella G (2002) Joint linkage and linkage disequilibrium mapping of quantitative trait loci in natural populations. Genetics 160: Wu RL, Ma CX, Lin M, Casella G (2004a) A general framework for analyzing the genetic architecture of developmental characteristics. Genetics 166: Wu RL, Ma CX, Lin M, Wang ZH, Casella G (2004b) Functional mapping of growth quantitative trait loci using a transform-both-sides logistic model. Biometrics 60: Wu RL, Wang ZH, Zhao W, Cheverud JM (2004c) A mechanistic model for genetic machinery of ontogenetic growth. Genetics 168: Wu S, Yang J, Wu RL (2006) Multilocus linkage disequilibrium mapping of quantitative trait loci that affect HIV dynamics: a simulation approach. Stat Med 25: Zhao W, Hou W, Littell RC, Wu RL (2005) Structured antedependence models for functional mapping of multivariate longitudinal quantitative traits. Stat Appl Mol Genet Biol 4:Article 33
1 Springer. Nan M. Laird Christoph Lange. The Fundamentals of Modern Statistical Genetics
 1 Springer Nan M. Laird Christoph Lange The Fundamentals of Modern Statistical Genetics 1 Introduction to Statistical Genetics and Background in Molecular Genetics 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 Springer Nan M. Laird Christoph Lange The Fundamentals of Modern Statistical Genetics 1 Introduction to Statistical Genetics and Background in Molecular Genetics 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
DNA polymorphisms such as SNP and familial effects (additive genetic, common environment) to
 1 1 1 1 1 1 1 1 0 SUPPLEMENTARY MATERIALS, B. BIVARIATE PEDIGREE-BASED ASSOCIATION ANALYSIS Introduction We propose here a statistical method of bivariate genetic analysis, designed to evaluate contribution
1 1 1 1 1 1 1 1 0 SUPPLEMENTARY MATERIALS, B. BIVARIATE PEDIGREE-BASED ASSOCIATION ANALYSIS Introduction We propose here a statistical method of bivariate genetic analysis, designed to evaluate contribution
A State Space Model Approach for HIV Infection Dynamics
 A State Space Model Approach for HIV Infection Dynamics Jiabin Wang a, Hua Liang b, and Rong Chen a Mathematical models have been proposed and developed to model HIV dynamics over the past few decades,
A State Space Model Approach for HIV Infection Dynamics Jiabin Wang a, Hua Liang b, and Rong Chen a Mathematical models have been proposed and developed to model HIV dynamics over the past few decades,
Introduction to QTL mapping in model organisms
 Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics and Medical Informatics University of Wisconsin Madison www.biostat.wisc.edu/~kbroman [ Teaching Miscellaneous lectures]
Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics and Medical Informatics University of Wisconsin Madison www.biostat.wisc.edu/~kbroman [ Teaching Miscellaneous lectures]
Gene mapping in model organisms
 Gene mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman Goal Identify genes that contribute to common human diseases. 2
Gene mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman Goal Identify genes that contribute to common human diseases. 2
Introduction to QTL mapping in model organisms
 Introduction to QTL mapping in model organisms Karl Broman Biostatistics and Medical Informatics University of Wisconsin Madison kbroman.org github.com/kbroman @kwbroman Backcross P 1 P 2 P 1 F 1 BC 4
Introduction to QTL mapping in model organisms Karl Broman Biostatistics and Medical Informatics University of Wisconsin Madison kbroman.org github.com/kbroman @kwbroman Backcross P 1 P 2 P 1 F 1 BC 4
2. Map genetic distance between markers
 Chapter 5. Linkage Analysis Linkage is an important tool for the mapping of genetic loci and a method for mapping disease loci. With the availability of numerous DNA markers throughout the human genome,
Chapter 5. Linkage Analysis Linkage is an important tool for the mapping of genetic loci and a method for mapping disease loci. With the availability of numerous DNA markers throughout the human genome,
Use of hidden Markov models for QTL mapping
 Use of hidden Markov models for QTL mapping Karl W Broman Department of Biostatistics, Johns Hopkins University December 5, 2006 An important aspect of the QTL mapping problem is the treatment of missing
Use of hidden Markov models for QTL mapping Karl W Broman Department of Biostatistics, Johns Hopkins University December 5, 2006 An important aspect of the QTL mapping problem is the treatment of missing
Introduction to QTL mapping in model organisms
 Human vs mouse Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University www.biostat.jhsph.edu/~kbroman [ Teaching Miscellaneous lectures] www.daviddeen.com
Human vs mouse Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University www.biostat.jhsph.edu/~kbroman [ Teaching Miscellaneous lectures] www.daviddeen.com
Preprint núm May Stability of a stochastic model for HIV-1 dynamics within a host. L. Shaikhet, A. Korobeinikov
 Preprint núm. 91 May 2014 Stability of a stochastic model for HIV-1 dynamics within a host L. Shaikhet, A. Korobeinikov STABILITY OF A STOCHASTIC MODEL FOR HIV-1 DYNAMICS WITHIN A HOST LEONID SHAIKHET
Preprint núm. 91 May 2014 Stability of a stochastic model for HIV-1 dynamics within a host L. Shaikhet, A. Korobeinikov STABILITY OF A STOCHASTIC MODEL FOR HIV-1 DYNAMICS WITHIN A HOST LEONID SHAIKHET
Binomial Mixture Model-based Association Tests under Genetic Heterogeneity
 Binomial Mixture Model-based Association Tests under Genetic Heterogeneity Hui Zhou, Wei Pan Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455 April 30,
Binomial Mixture Model-based Association Tests under Genetic Heterogeneity Hui Zhou, Wei Pan Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455 April 30,
Bayesian Inference of Interactions and Associations
 Bayesian Inference of Interactions and Associations Jun Liu Department of Statistics Harvard University http://www.fas.harvard.edu/~junliu Based on collaborations with Yu Zhang, Jing Zhang, Yuan Yuan,
Bayesian Inference of Interactions and Associations Jun Liu Department of Statistics Harvard University http://www.fas.harvard.edu/~junliu Based on collaborations with Yu Zhang, Jing Zhang, Yuan Yuan,
Evolution of phenotypic traits
 Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
A A A A B B1
 LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
Enduring understanding 1.A: Change in the genetic makeup of a population over time is evolution.
 The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
AP Curriculum Framework with Learning Objectives
 Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Case-Control Association Testing. Case-Control Association Testing
 Introduction Association mapping is now routinely being used to identify loci that are involved with complex traits. Technological advances have made it feasible to perform case-control association studies
Introduction Association mapping is now routinely being used to identify loci that are involved with complex traits. Technological advances have made it feasible to perform case-control association studies
Linear Regression (1/1/17)
 STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
Methods for Cryptic Structure. Methods for Cryptic Structure
 Case-Control Association Testing Review Consider testing for association between a disease and a genetic marker Idea is to look for an association by comparing allele/genotype frequencies between the cases
Case-Control Association Testing Review Consider testing for association between a disease and a genetic marker Idea is to look for an association by comparing allele/genotype frequencies between the cases
Big Idea 1: The process of evolution drives the diversity and unity of life.
 Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Overview. Background
 Overview Implementation of robust methods for locating quantitative trait loci in R Introduction to QTL mapping Andreas Baierl and Andreas Futschik Institute of Statistics and Decision Support Systems
Overview Implementation of robust methods for locating quantitative trait loci in R Introduction to QTL mapping Andreas Baierl and Andreas Futschik Institute of Statistics and Decision Support Systems
Introduction to QTL mapping in model organisms
 Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University kbroman@jhsph.edu www.biostat.jhsph.edu/ kbroman Outline Experiments and data Models ANOVA
Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University kbroman@jhsph.edu www.biostat.jhsph.edu/ kbroman Outline Experiments and data Models ANOVA
On the limiting distribution of the likelihood ratio test in nucleotide mapping of complex disease
 On the limiting distribution of the likelihood ratio test in nucleotide mapping of complex disease Yuehua Cui 1 and Dong-Yun Kim 2 1 Department of Statistics and Probability, Michigan State University,
On the limiting distribution of the likelihood ratio test in nucleotide mapping of complex disease Yuehua Cui 1 and Dong-Yun Kim 2 1 Department of Statistics and Probability, Michigan State University,
Bayesian Methods with Monte Carlo Markov Chains II
 Bayesian Methods with Monte Carlo Markov Chains II Henry Horng-Shing Lu Institute of Statistics National Chiao Tung University hslu@stat.nctu.edu.tw http://tigpbp.iis.sinica.edu.tw/courses.htm 1 Part 3
Bayesian Methods with Monte Carlo Markov Chains II Henry Horng-Shing Lu Institute of Statistics National Chiao Tung University hslu@stat.nctu.edu.tw http://tigpbp.iis.sinica.edu.tw/courses.htm 1 Part 3
Lecture 11: Multiple trait models for QTL analysis
 Lecture 11: Multiple trait models for QTL analysis Julius van der Werf Multiple trait mapping of QTL...99 Increased power of QTL detection...99 Testing for linked QTL vs pleiotropic QTL...100 Multiple
Lecture 11: Multiple trait models for QTL analysis Julius van der Werf Multiple trait mapping of QTL...99 Increased power of QTL detection...99 Testing for linked QTL vs pleiotropic QTL...100 Multiple
Lecture 9. QTL Mapping 2: Outbred Populations
 Lecture 9 QTL Mapping 2: Outbred Populations Bruce Walsh. Aug 2004. Royal Veterinary and Agricultural University, Denmark The major difference between QTL analysis using inbred-line crosses vs. outbred
Lecture 9 QTL Mapping 2: Outbred Populations Bruce Walsh. Aug 2004. Royal Veterinary and Agricultural University, Denmark The major difference between QTL analysis using inbred-line crosses vs. outbred
Alternative implementations of Monte Carlo EM algorithms for likelihood inferences
 Genet. Sel. Evol. 33 001) 443 45 443 INRA, EDP Sciences, 001 Alternative implementations of Monte Carlo EM algorithms for likelihood inferences Louis Alberto GARCÍA-CORTÉS a, Daniel SORENSEN b, Note a
Genet. Sel. Evol. 33 001) 443 45 443 INRA, EDP Sciences, 001 Alternative implementations of Monte Carlo EM algorithms for likelihood inferences Louis Alberto GARCÍA-CORTÉS a, Daniel SORENSEN b, Note a
The Admixture Model in Linkage Analysis
 The Admixture Model in Linkage Analysis Jie Peng D. Siegmund Department of Statistics, Stanford University, Stanford, CA 94305 SUMMARY We study an appropriate version of the score statistic to test the
The Admixture Model in Linkage Analysis Jie Peng D. Siegmund Department of Statistics, Stanford University, Stanford, CA 94305 SUMMARY We study an appropriate version of the score statistic to test the
Introduction to QTL mapping in model organisms
 Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University kbroman@jhsph.edu www.biostat.jhsph.edu/ kbroman Outline Experiments and data Models ANOVA
Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University kbroman@jhsph.edu www.biostat.jhsph.edu/ kbroman Outline Experiments and data Models ANOVA
Outline of lectures 3-6
 GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 007 Population genetics Outline of lectures 3-6 1. We want to know what theory says about the reproduction of genotypes in a population. This results
GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 007 Population genetics Outline of lectures 3-6 1. We want to know what theory says about the reproduction of genotypes in a population. This results
Multiple QTL mapping
 Multiple QTL mapping Karl W Broman Department of Biostatistics Johns Hopkins University www.biostat.jhsph.edu/~kbroman [ Teaching Miscellaneous lectures] 1 Why? Reduce residual variation = increased power
Multiple QTL mapping Karl W Broman Department of Biostatistics Johns Hopkins University www.biostat.jhsph.edu/~kbroman [ Teaching Miscellaneous lectures] 1 Why? Reduce residual variation = increased power
QTL model selection: key players
 Bayesian Interval Mapping. Bayesian strategy -9. Markov chain sampling 0-7. sampling genetic architectures 8-5 4. criteria for model selection 6-44 QTL : Bayes Seattle SISG: Yandell 008 QTL model selection:
Bayesian Interval Mapping. Bayesian strategy -9. Markov chain sampling 0-7. sampling genetic architectures 8-5 4. criteria for model selection 6-44 QTL : Bayes Seattle SISG: Yandell 008 QTL model selection:
Combining dependent tests for linkage or association across multiple phenotypic traits
 Biostatistics (2003), 4, 2,pp. 223 229 Printed in Great Britain Combining dependent tests for linkage or association across multiple phenotypic traits XIN XU Program for Population Genetics, Harvard School
Biostatistics (2003), 4, 2,pp. 223 229 Printed in Great Britain Combining dependent tests for linkage or association across multiple phenotypic traits XIN XU Program for Population Genetics, Harvard School
Lecture WS Evolutionary Genetics Part I 1
 Quantitative genetics Quantitative genetics is the study of the inheritance of quantitative/continuous phenotypic traits, like human height and body size, grain colour in winter wheat or beak depth in
Quantitative genetics Quantitative genetics is the study of the inheritance of quantitative/continuous phenotypic traits, like human height and body size, grain colour in winter wheat or beak depth in
Nonparametric Modeling of Longitudinal Covariance Structure in Functional Mapping of Quantitative Trait Loci
 Nonparametric Modeling of Longitudinal Covariance Structure in Functional Mapping of Quantitative Trait Loci John Stephen Yap 1, Jianqing Fan 2 and Rongling Wu 1 1 Department of Statistics, University
Nonparametric Modeling of Longitudinal Covariance Structure in Functional Mapping of Quantitative Trait Loci John Stephen Yap 1, Jianqing Fan 2 and Rongling Wu 1 1 Department of Statistics, University
MODEL-FREE LINKAGE AND ASSOCIATION MAPPING OF COMPLEX TRAITS USING QUANTITATIVE ENDOPHENOTYPES
 MODEL-FREE LINKAGE AND ASSOCIATION MAPPING OF COMPLEX TRAITS USING QUANTITATIVE ENDOPHENOTYPES Saurabh Ghosh Human Genetics Unit Indian Statistical Institute, Kolkata Most common diseases are caused by
MODEL-FREE LINKAGE AND ASSOCIATION MAPPING OF COMPLEX TRAITS USING QUANTITATIVE ENDOPHENOTYPES Saurabh Ghosh Human Genetics Unit Indian Statistical Institute, Kolkata Most common diseases are caused by
Calculation of IBD probabilities
 Calculation of IBD probabilities David Evans and Stacey Cherny University of Oxford Wellcome Trust Centre for Human Genetics This Session IBD vs IBS Why is IBD important? Calculating IBD probabilities
Calculation of IBD probabilities David Evans and Stacey Cherny University of Oxford Wellcome Trust Centre for Human Genetics This Session IBD vs IBS Why is IBD important? Calculating IBD probabilities
I Have the Power in QTL linkage: single and multilocus analysis
 I Have the Power in QTL linkage: single and multilocus analysis Benjamin Neale 1, Sir Shaun Purcell 2 & Pak Sham 13 1 SGDP, IoP, London, UK 2 Harvard School of Public Health, Cambridge, MA, USA 3 Department
I Have the Power in QTL linkage: single and multilocus analysis Benjamin Neale 1, Sir Shaun Purcell 2 & Pak Sham 13 1 SGDP, IoP, London, UK 2 Harvard School of Public Health, Cambridge, MA, USA 3 Department
Genotype Imputation. Biostatistics 666
 Genotype Imputation Biostatistics 666 Previously Hidden Markov Models for Relative Pairs Linkage analysis using affected sibling pairs Estimation of pairwise relationships Identity-by-Descent Relatives
Genotype Imputation Biostatistics 666 Previously Hidden Markov Models for Relative Pairs Linkage analysis using affected sibling pairs Estimation of pairwise relationships Identity-by-Descent Relatives
Association studies and regression
 Association studies and regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Association studies and regression 1 / 104 Administration
Association studies and regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Association studies and regression 1 / 104 Administration
CHAPTER 23 THE EVOLUTIONS OF POPULATIONS. Section C: Genetic Variation, the Substrate for Natural Selection
 CHAPTER 23 THE EVOLUTIONS OF POPULATIONS Section C: Genetic Variation, the Substrate for Natural Selection 1. Genetic variation occurs within and between populations 2. Mutation and sexual recombination
CHAPTER 23 THE EVOLUTIONS OF POPULATIONS Section C: Genetic Variation, the Substrate for Natural Selection 1. Genetic variation occurs within and between populations 2. Mutation and sexual recombination
Processes of Evolution
 15 Processes of Evolution Forces of Evolution Concept 15.4 Selection Can Be Stabilizing, Directional, or Disruptive Natural selection can act on quantitative traits in three ways: Stabilizing selection
15 Processes of Evolution Forces of Evolution Concept 15.4 Selection Can Be Stabilizing, Directional, or Disruptive Natural selection can act on quantitative traits in three ways: Stabilizing selection
(Genome-wide) association analysis
 (Genome-wide) association analysis 1 Key concepts Mapping QTL by association relies on linkage disequilibrium in the population; LD can be caused by close linkage between a QTL and marker (= good) or by
(Genome-wide) association analysis 1 Key concepts Mapping QTL by association relies on linkage disequilibrium in the population; LD can be caused by close linkage between a QTL and marker (= good) or by
Outline of lectures 3-6
 GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 009 Population genetics Outline of lectures 3-6 1. We want to know what theory says about the reproduction of genotypes in a population. This results
GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 009 Population genetics Outline of lectures 3-6 1. We want to know what theory says about the reproduction of genotypes in a population. This results
Association Testing with Quantitative Traits: Common and Rare Variants. Summer Institute in Statistical Genetics 2014 Module 10 Lecture 5
 Association Testing with Quantitative Traits: Common and Rare Variants Timothy Thornton and Katie Kerr Summer Institute in Statistical Genetics 2014 Module 10 Lecture 5 1 / 41 Introduction to Quantitative
Association Testing with Quantitative Traits: Common and Rare Variants Timothy Thornton and Katie Kerr Summer Institute in Statistical Genetics 2014 Module 10 Lecture 5 1 / 41 Introduction to Quantitative
Major Genes, Polygenes, and
 Major Genes, Polygenes, and QTLs Major genes --- genes that have a significant effect on the phenotype Polygenes --- a general term of the genes of small effect that influence a trait QTL, quantitative
Major Genes, Polygenes, and QTLs Major genes --- genes that have a significant effect on the phenotype Polygenes --- a general term of the genes of small effect that influence a trait QTL, quantitative
Population Genetics. with implications for Linkage Disequilibrium. Chiara Sabatti, Human Genetics 6357a Gonda
 1 Population Genetics with implications for Linkage Disequilibrium Chiara Sabatti, Human Genetics 6357a Gonda csabatti@mednet.ucla.edu 2 Hardy-Weinberg Hypotheses: infinite populations; no inbreeding;
1 Population Genetics with implications for Linkage Disequilibrium Chiara Sabatti, Human Genetics 6357a Gonda csabatti@mednet.ucla.edu 2 Hardy-Weinberg Hypotheses: infinite populations; no inbreeding;
Supplementary Materials for Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control
 Supplementary Materials for Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control Xiaoquan Wen Department of Biostatistics, University of Michigan A Model
Supplementary Materials for Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control Xiaoquan Wen Department of Biostatistics, University of Michigan A Model
Map of AP-Aligned Bio-Rad Kits with Learning Objectives
 Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Linkage and Linkage Disequilibrium
 Linkage and Linkage Disequilibrium Summer Institute in Statistical Genetics 2014 Module 10 Topic 3 Linkage in a simple genetic cross Linkage In the early 1900 s Bateson and Punnet conducted genetic studies
Linkage and Linkage Disequilibrium Summer Institute in Statistical Genetics 2014 Module 10 Topic 3 Linkage in a simple genetic cross Linkage In the early 1900 s Bateson and Punnet conducted genetic studies
MIXED MODELS THE GENERAL MIXED MODEL
 MIXED MODELS This chapter introduces best linear unbiased prediction (BLUP), a general method for predicting random effects, while Chapter 27 is concerned with the estimation of variances by restricted
MIXED MODELS This chapter introduces best linear unbiased prediction (BLUP), a general method for predicting random effects, while Chapter 27 is concerned with the estimation of variances by restricted
Proportional Variance Explained by QLT and Statistical Power. Proportional Variance Explained by QTL and Statistical Power
 Proportional Variance Explained by QTL and Statistical Power Partitioning the Genetic Variance We previously focused on obtaining variance components of a quantitative trait to determine the proportion
Proportional Variance Explained by QTL and Statistical Power Partitioning the Genetic Variance We previously focused on obtaining variance components of a quantitative trait to determine the proportion
Stationary Distribution of the Linkage Disequilibrium Coefficient r 2
 Stationary Distribution of the Linkage Disequilibrium Coefficient r 2 Wei Zhang, Jing Liu, Rachel Fewster and Jesse Goodman Department of Statistics, The University of Auckland December 1, 2015 Overview
Stationary Distribution of the Linkage Disequilibrium Coefficient r 2 Wei Zhang, Jing Liu, Rachel Fewster and Jesse Goodman Department of Statistics, The University of Auckland December 1, 2015 Overview
Lecture 2: Genetic Association Testing with Quantitative Traits. Summer Institute in Statistical Genetics 2017
 Lecture 2: Genetic Association Testing with Quantitative Traits Instructors: Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2017 1 / 29 Introduction to Quantitative Trait Mapping
Lecture 2: Genetic Association Testing with Quantitative Traits Instructors: Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2017 1 / 29 Introduction to Quantitative Trait Mapping
A simple genetic model with non-equilibrium dynamics
 J. Math. Biol. (1998) 36: 550 556 A simple genetic model with non-equilibrium dynamics Michael Doebeli, Gerdien de Jong Zoology Institute, University of Basel, Rheinsprung 9, CH-4051 Basel, Switzerland
J. Math. Biol. (1998) 36: 550 556 A simple genetic model with non-equilibrium dynamics Michael Doebeli, Gerdien de Jong Zoology Institute, University of Basel, Rheinsprung 9, CH-4051 Basel, Switzerland
Lecture 28: BLUP and Genomic Selection. Bruce Walsh lecture notes Synbreed course version 11 July 2013
 Lecture 28: BLUP and Genomic Selection Bruce Walsh lecture notes Synbreed course version 11 July 2013 1 BLUP Selection The idea behind BLUP selection is very straightforward: An appropriate mixed-model
Lecture 28: BLUP and Genomic Selection Bruce Walsh lecture notes Synbreed course version 11 July 2013 1 BLUP Selection The idea behind BLUP selection is very straightforward: An appropriate mixed-model
Stochastic approximation EM algorithm in nonlinear mixed effects model for viral load decrease during anti-hiv treatment
 Stochastic approximation EM algorithm in nonlinear mixed effects model for viral load decrease during anti-hiv treatment Adeline Samson 1, Marc Lavielle and France Mentré 1 1 INSERM E0357, Department of
Stochastic approximation EM algorithm in nonlinear mixed effects model for viral load decrease during anti-hiv treatment Adeline Samson 1, Marc Lavielle and France Mentré 1 1 INSERM E0357, Department of
Methodology Report EM Algorithm for Mapping Quantitative Trait Loci in Multivalent Tetraploids
 International Journal of Plant Genomics Volume 00, Article ID 7, 0 pages doi:0./00/7 Methodology Report EM Algorithm for Mapping Quantitative Trait Loci in Multivalent Tetraploids Jiahan Li, Kiranmoy Das,
International Journal of Plant Genomics Volume 00, Article ID 7, 0 pages doi:0./00/7 Methodology Report EM Algorithm for Mapping Quantitative Trait Loci in Multivalent Tetraploids Jiahan Li, Kiranmoy Das,
Evolutionary quantitative genetics and one-locus population genetics
 Evolutionary quantitative genetics and one-locus population genetics READING: Hedrick pp. 57 63, 587 596 Most evolutionary problems involve questions about phenotypic means Goal: determine how selection
Evolutionary quantitative genetics and one-locus population genetics READING: Hedrick pp. 57 63, 587 596 Most evolutionary problems involve questions about phenotypic means Goal: determine how selection
Principles of QTL Mapping. M.Imtiaz
 Principles of QTL Mapping M.Imtiaz Introduction Definitions of terminology Reasons for QTL mapping Principles of QTL mapping Requirements For QTL Mapping Demonstration with experimental data Merit of QTL
Principles of QTL Mapping M.Imtiaz Introduction Definitions of terminology Reasons for QTL mapping Principles of QTL mapping Requirements For QTL Mapping Demonstration with experimental data Merit of QTL
Calculation of IBD probabilities
 Calculation of IBD probabilities David Evans University of Bristol This Session Identity by Descent (IBD) vs Identity by state (IBS) Why is IBD important? Calculating IBD probabilities Lander-Green Algorithm
Calculation of IBD probabilities David Evans University of Bristol This Session Identity by Descent (IBD) vs Identity by state (IBS) Why is IBD important? Calculating IBD probabilities Lander-Green Algorithm
Approximate Bayesian Computation
 Approximate Bayesian Computation Michael Gutmann https://sites.google.com/site/michaelgutmann University of Helsinki and Aalto University 1st December 2015 Content Two parts: 1. The basics of approximate
Approximate Bayesian Computation Michael Gutmann https://sites.google.com/site/michaelgutmann University of Helsinki and Aalto University 1st December 2015 Content Two parts: 1. The basics of approximate
Model-Free Knockoffs: High-Dimensional Variable Selection that Controls the False Discovery Rate
 Model-Free Knockoffs: High-Dimensional Variable Selection that Controls the False Discovery Rate Lucas Janson, Stanford Department of Statistics WADAPT Workshop, NIPS, December 2016 Collaborators: Emmanuel
Model-Free Knockoffs: High-Dimensional Variable Selection that Controls the False Discovery Rate Lucas Janson, Stanford Department of Statistics WADAPT Workshop, NIPS, December 2016 Collaborators: Emmanuel
1.5.1 ESTIMATION OF HAPLOTYPE FREQUENCIES:
 .5. ESTIMATION OF HAPLOTYPE FREQUENCIES: Chapter - 8 For SNPs, alleles A j,b j at locus j there are 4 haplotypes: A A, A B, B A and B B frequencies q,q,q 3,q 4. Assume HWE at haplotype level. Only the
.5. ESTIMATION OF HAPLOTYPE FREQUENCIES: Chapter - 8 For SNPs, alleles A j,b j at locus j there are 4 haplotypes: A A, A B, B A and B B frequencies q,q,q 3,q 4. Assume HWE at haplotype level. Only the
Expression QTLs and Mapping of Complex Trait Loci. Paul Schliekelman Statistics Department University of Georgia
 Expression QTLs and Mapping of Complex Trait Loci Paul Schliekelman Statistics Department University of Georgia Definitions: Genes, Loci and Alleles A gene codes for a protein. Proteins due everything.
Expression QTLs and Mapping of Complex Trait Loci Paul Schliekelman Statistics Department University of Georgia Definitions: Genes, Loci and Alleles A gene codes for a protein. Proteins due everything.
Friday Harbor From Genetics to GWAS (Genome-wide Association Study) Sept David Fardo
 Friday Harbor 2017 From Genetics to GWAS (Genome-wide Association Study) Sept 7 2017 David Fardo Purpose: prepare for tomorrow s tutorial Genetic Variants Quality Control Imputation Association Visualization
Friday Harbor 2017 From Genetics to GWAS (Genome-wide Association Study) Sept 7 2017 David Fardo Purpose: prepare for tomorrow s tutorial Genetic Variants Quality Control Imputation Association Visualization
Sensitivity and Stability Analysis of Hepatitis B Virus Model with Non-Cytolytic Cure Process and Logistic Hepatocyte Growth
 Global Journal of Pure and Applied Mathematics. ISSN 0973-1768 Volume 12, Number 3 2016), pp. 2297 2312 Research India Publications http://www.ripublication.com/gjpam.htm Sensitivity and Stability Analysis
Global Journal of Pure and Applied Mathematics. ISSN 0973-1768 Volume 12, Number 3 2016), pp. 2297 2312 Research India Publications http://www.ripublication.com/gjpam.htm Sensitivity and Stability Analysis
A Robust Test for Two-Stage Design in Genome-Wide Association Studies
 Biometrics Supplementary Materials A Robust Test for Two-Stage Design in Genome-Wide Association Studies Minjung Kwak, Jungnam Joo and Gang Zheng Appendix A: Calculations of the thresholds D 1 and D The
Biometrics Supplementary Materials A Robust Test for Two-Stage Design in Genome-Wide Association Studies Minjung Kwak, Jungnam Joo and Gang Zheng Appendix A: Calculations of the thresholds D 1 and D The
Major questions of evolutionary genetics. Experimental tools of evolutionary genetics. Theoretical population genetics.
 Evolutionary Genetics (for Encyclopedia of Biodiversity) Sergey Gavrilets Departments of Ecology and Evolutionary Biology and Mathematics, University of Tennessee, Knoxville, TN 37996-6 USA Evolutionary
Evolutionary Genetics (for Encyclopedia of Biodiversity) Sergey Gavrilets Departments of Ecology and Evolutionary Biology and Mathematics, University of Tennessee, Knoxville, TN 37996-6 USA Evolutionary
MA/ST 810 Mathematical-Statistical Modeling and Analysis of Complex Systems
 MA/ST 810 Mathematical-Statistical Modeling and Analysis of Complex Systems Integrating Mathematical and Statistical Models Recap of mathematical models Models and data Statistical models and sources of
MA/ST 810 Mathematical-Statistical Modeling and Analysis of Complex Systems Integrating Mathematical and Statistical Models Recap of mathematical models Models and data Statistical models and sources of
1. Understand the methods for analyzing population structure in genomes
 MSCBIO 2070/02-710: Computational Genomics, Spring 2016 HW3: Population Genetics Due: 24:00 EST, April 4, 2016 by autolab Your goals in this assignment are to 1. Understand the methods for analyzing population
MSCBIO 2070/02-710: Computational Genomics, Spring 2016 HW3: Population Genetics Due: 24:00 EST, April 4, 2016 by autolab Your goals in this assignment are to 1. Understand the methods for analyzing population
p(d g A,g B )p(g B ), g B
 Supplementary Note Marginal effects for two-locus models Here we derive the marginal effect size of the three models given in Figure 1 of the main text. For each model we assume the two loci (A and B)
Supplementary Note Marginal effects for two-locus models Here we derive the marginal effect size of the three models given in Figure 1 of the main text. For each model we assume the two loci (A and B)
Global Analysis of a HCV Model with CTL, Antibody Responses and Therapy
 Applied Mathematical Sciences Vol 9 205 no 8 3997-4008 HIKARI Ltd wwwm-hikaricom http://dxdoiorg/02988/ams20554334 Global Analysis of a HCV Model with CTL Antibody Responses and Therapy Adil Meskaf Department
Applied Mathematical Sciences Vol 9 205 no 8 3997-4008 HIKARI Ltd wwwm-hikaricom http://dxdoiorg/02988/ams20554334 Global Analysis of a HCV Model with CTL Antibody Responses and Therapy Adil Meskaf Department
Methods for QTL analysis
 Methods for QTL analysis Julius van der Werf METHODS FOR QTL ANALYSIS... 44 SINGLE VERSUS MULTIPLE MARKERS... 45 DETERMINING ASSOCIATIONS BETWEEN GENETIC MARKERS AND QTL WITH TWO MARKERS... 45 INTERVAL
Methods for QTL analysis Julius van der Werf METHODS FOR QTL ANALYSIS... 44 SINGLE VERSUS MULTIPLE MARKERS... 45 DETERMINING ASSOCIATIONS BETWEEN GENETIC MARKERS AND QTL WITH TWO MARKERS... 45 INTERVAL
Global Properties for Virus Dynamics Model with Beddington-DeAngelis Functional Response
 Global Properties for Virus Dynamics Model with Beddington-DeAngelis Functional Response Gang Huang 1,2, Wanbiao Ma 2, Yasuhiro Takeuchi 1 1,Graduate School of Science and Technology, Shizuoka University,
Global Properties for Virus Dynamics Model with Beddington-DeAngelis Functional Response Gang Huang 1,2, Wanbiao Ma 2, Yasuhiro Takeuchi 1 1,Graduate School of Science and Technology, Shizuoka University,
Lecture 8. QTL Mapping 1: Overview and Using Inbred Lines
 Lecture 8 QTL Mapping 1: Overview and Using Inbred Lines Bruce Walsh. jbwalsh@u.arizona.edu. University of Arizona. Notes from a short course taught Jan-Feb 2012 at University of Uppsala While the machinery
Lecture 8 QTL Mapping 1: Overview and Using Inbred Lines Bruce Walsh. jbwalsh@u.arizona.edu. University of Arizona. Notes from a short course taught Jan-Feb 2012 at University of Uppsala While the machinery
Population Genetics: a tutorial
 : a tutorial Institute for Science and Technology Austria ThRaSh 2014 provides the basic mathematical foundation of evolutionary theory allows a better understanding of experiments allows the development
: a tutorial Institute for Science and Technology Austria ThRaSh 2014 provides the basic mathematical foundation of evolutionary theory allows a better understanding of experiments allows the development
Introduction to Linkage Disequilibrium
 Introduction to September 10, 2014 Suppose we have two genes on a single chromosome gene A and gene B such that each gene has only two alleles Aalleles : A 1 and A 2 Balleles : B 1 and B 2 Suppose we have
Introduction to September 10, 2014 Suppose we have two genes on a single chromosome gene A and gene B such that each gene has only two alleles Aalleles : A 1 and A 2 Balleles : B 1 and B 2 Suppose we have
Confidence Intervals of the Simple Difference between the Proportions of a Primary Infection and a Secondary Infection, Given the Primary Infection
 Biometrical Journal 42 (2000) 1, 59±69 Confidence Intervals of the Simple Difference between the Proportions of a Primary Infection and a Secondary Infection, Given the Primary Infection Kung-Jong Lui
Biometrical Journal 42 (2000) 1, 59±69 Confidence Intervals of the Simple Difference between the Proportions of a Primary Infection and a Secondary Infection, Given the Primary Infection Kung-Jong Lui
Lecture 1: Case-Control Association Testing. Summer Institute in Statistical Genetics 2015
 Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2015 1 / 1 Introduction Association mapping is now routinely being used to identify loci that are involved with complex traits.
Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2015 1 / 1 Introduction Association mapping is now routinely being used to identify loci that are involved with complex traits.
Outline of lectures 3-6
 GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 013 Population genetics Outline of lectures 3-6 1. We ant to kno hat theory says about the reproduction of genotypes in a population. This results
GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 013 Population genetics Outline of lectures 3-6 1. We ant to kno hat theory says about the reproduction of genotypes in a population. This results
Biology II : Embedded Inquiry
 Biology II : Embedded Inquiry Conceptual Strand Understandings about scientific inquiry and the ability to conduct inquiry are essential for living in the 21 st century. Guiding Question What tools, skills,
Biology II : Embedded Inquiry Conceptual Strand Understandings about scientific inquiry and the ability to conduct inquiry are essential for living in the 21 st century. Guiding Question What tools, skills,
Pairwise rank based likelihood for estimating the relationship between two homogeneous populations and their mixture proportion
 Pairwise rank based likelihood for estimating the relationship between two homogeneous populations and their mixture proportion Glenn Heller and Jing Qin Department of Epidemiology and Biostatistics Memorial
Pairwise rank based likelihood for estimating the relationship between two homogeneous populations and their mixture proportion Glenn Heller and Jing Qin Department of Epidemiology and Biostatistics Memorial
Tutorial Session 2. MCMC for the analysis of genetic data on pedigrees:
 MCMC for the analysis of genetic data on pedigrees: Tutorial Session 2 Elizabeth Thompson University of Washington Genetic mapping and linkage lod scores Monte Carlo likelihood and likelihood ratio estimation
MCMC for the analysis of genetic data on pedigrees: Tutorial Session 2 Elizabeth Thompson University of Washington Genetic mapping and linkage lod scores Monte Carlo likelihood and likelihood ratio estimation
Affected Sibling Pairs. Biostatistics 666
 Affected Sibling airs Biostatistics 666 Today Discussion of linkage analysis using affected sibling pairs Our exploration will include several components we have seen before: A simple disease model IBD
Affected Sibling airs Biostatistics 666 Today Discussion of linkage analysis using affected sibling pairs Our exploration will include several components we have seen before: A simple disease model IBD
Modeling IBD for Pairs of Relatives. Biostatistics 666 Lecture 17
 Modeling IBD for Pairs of Relatives Biostatistics 666 Lecture 7 Previously Linkage Analysis of Relative Pairs IBS Methods Compare observed and expected sharing IBD Methods Account for frequency of shared
Modeling IBD for Pairs of Relatives Biostatistics 666 Lecture 7 Previously Linkage Analysis of Relative Pairs IBS Methods Compare observed and expected sharing IBD Methods Account for frequency of shared
Statistical issues in QTL mapping in mice
 Statistical issues in QTL mapping in mice Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman Outline Overview of QTL mapping The X chromosome Mapping
Statistical issues in QTL mapping in mice Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman Outline Overview of QTL mapping The X chromosome Mapping
Homework Assignment, Evolutionary Systems Biology, Spring Homework Part I: Phylogenetics:
 Homework Assignment, Evolutionary Systems Biology, Spring 2009. Homework Part I: Phylogenetics: Introduction. The objective of this assignment is to understand the basics of phylogenetic relationships
Homework Assignment, Evolutionary Systems Biology, Spring 2009. Homework Part I: Phylogenetics: Introduction. The objective of this assignment is to understand the basics of phylogenetic relationships
Big Idea 3: Living systems store, retrieve, transmit, and respond to information essential to life processes.
 Big Idea 3: Living systems store, retrieve, transmit, and respond to information essential to life processes. Enduring understanding 3.A: Heritable information provides for continuity of life. Essential
Big Idea 3: Living systems store, retrieve, transmit, and respond to information essential to life processes. Enduring understanding 3.A: Heritable information provides for continuity of life. Essential
25 : Graphical induced structured input/output models
 10-708: Probabilistic Graphical Models 10-708, Spring 2016 25 : Graphical induced structured input/output models Lecturer: Eric P. Xing Scribes: Raied Aljadaany, Shi Zong, Chenchen Zhu Disclaimer: A large
10-708: Probabilistic Graphical Models 10-708, Spring 2016 25 : Graphical induced structured input/output models Lecturer: Eric P. Xing Scribes: Raied Aljadaany, Shi Zong, Chenchen Zhu Disclaimer: A large
Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination)
 12/5/14 Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination) Linkage Disequilibrium Genealogical Interpretation of LD Association Mapping 1 Linkage and Recombination v linkage equilibrium ²
12/5/14 Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination) Linkage Disequilibrium Genealogical Interpretation of LD Association Mapping 1 Linkage and Recombination v linkage equilibrium ²
Asymptotic properties of the likelihood ratio test statistics with the possible triangle constraint in Affected-Sib-Pair analysis
 The Canadian Journal of Statistics Vol.?, No.?, 2006, Pages???-??? La revue canadienne de statistique Asymptotic properties of the likelihood ratio test statistics with the possible triangle constraint
The Canadian Journal of Statistics Vol.?, No.?, 2006, Pages???-??? La revue canadienne de statistique Asymptotic properties of the likelihood ratio test statistics with the possible triangle constraint
1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms.
 Practicing Biology BIG IDEA 3.A 1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms. 2. Using at least 2 well-known experiments, describe which features of DNA and RNA
Practicing Biology BIG IDEA 3.A 1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms. 2. Using at least 2 well-known experiments, describe which features of DNA and RNA
Chapters AP Biology Objectives. Objectives: You should know...
 Objectives: You should know... Notes 1. Scientific evidence supports the idea that evolution has occurred in all species. 2. Scientific evidence supports the idea that evolution continues to occur. 3.
Objectives: You should know... Notes 1. Scientific evidence supports the idea that evolution has occurred in all species. 2. Scientific evidence supports the idea that evolution continues to occur. 3.
Lecture 2: Linear Models. Bruce Walsh lecture notes Seattle SISG -Mixed Model Course version 23 June 2011
 Lecture 2: Linear Models Bruce Walsh lecture notes Seattle SISG -Mixed Model Course version 23 June 2011 1 Quick Review of the Major Points The general linear model can be written as y = X! + e y = vector
Lecture 2: Linear Models Bruce Walsh lecture notes Seattle SISG -Mixed Model Course version 23 June 2011 1 Quick Review of the Major Points The general linear model can be written as y = X! + e y = vector
Computational Aspects of Aggregation in Biological Systems
 Computational Aspects of Aggregation in Biological Systems Vladik Kreinovich and Max Shpak University of Texas at El Paso, El Paso, TX 79968, USA vladik@utep.edu, mshpak@utep.edu Summary. Many biologically
Computational Aspects of Aggregation in Biological Systems Vladik Kreinovich and Max Shpak University of Texas at El Paso, El Paso, TX 79968, USA vladik@utep.edu, mshpak@utep.edu Summary. Many biologically
Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Information #
 Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Details of PRF Methodology In the Poisson Random Field PRF) model, it is assumed that non-synonymous mutations at a given gene are either
Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Details of PRF Methodology In the Poisson Random Field PRF) model, it is assumed that non-synonymous mutations at a given gene are either
The Mechanisms of Evolution
 The Mechanisms of Evolution Figure.1 Darwin and the Voyage of the Beagle (Part 1) 2/8/2006 Dr. Michod Intro Biology 182 (PP 3) 4 The Mechanisms of Evolution Charles Darwin s Theory of Evolution Genetic
The Mechanisms of Evolution Figure.1 Darwin and the Voyage of the Beagle (Part 1) 2/8/2006 Dr. Michod Intro Biology 182 (PP 3) 4 The Mechanisms of Evolution Charles Darwin s Theory of Evolution Genetic
Introduction to Quantitative Genetics. Introduction to Quantitative Genetics
 Introduction to Quantitative Genetics Historical Background Quantitative genetics is the study of continuous or quantitative traits and their underlying mechanisms. The main principals of quantitative
Introduction to Quantitative Genetics Historical Background Quantitative genetics is the study of continuous or quantitative traits and their underlying mechanisms. The main principals of quantitative
Restricted Maximum Likelihood in Linear Regression and Linear Mixed-Effects Model
 Restricted Maximum Likelihood in Linear Regression and Linear Mixed-Effects Model Xiuming Zhang zhangxiuming@u.nus.edu A*STAR-NUS Clinical Imaging Research Center October, 015 Summary This report derives
Restricted Maximum Likelihood in Linear Regression and Linear Mixed-Effects Model Xiuming Zhang zhangxiuming@u.nus.edu A*STAR-NUS Clinical Imaging Research Center October, 015 Summary This report derives
