Learning Concave Conditional Likelihood Models for Improved Analysis of Tandem Mass Spectra
|
|
|
- Gordon Lucas
- 5 years ago
- Views:
Transcription
1 Learning Concave Conditional Likelihood Models for Improved Analysis of Tandem Mass Spectra John T. Halloran Department of Public Health Sciences University of California, Davis David M. Rocke Department of Public Health Sciences University of California, Davis Abstract The most widely used technology to identify the proteins present in a complex biological sample is tandem mass spectrometry, which quickly produces a large collection of spectra representative of the peptides (i.e., protein subsequences) present in the original sample. In this work, we greatly expand the parameter learning capabilities of a dynamic Bayesian network (DBN) peptide-scoring algorithm, Didea [25], by deriving emission distributions for which its conditional log-likelihood scoring function remains concave. We show that this class of emission distributions, called Convex Virtual Emissions (CVEs), naturally generalizes the log-sum-exp function while rendering both maximum likelihood estimation and conditional maximum likelihood estimation concave for a wide range of Bayesian networks. Utilizing CVEs in Didea allows efficient learning of a large number of parameters while ensuring global convergence, in stark contrast to Didea s previous parameter learning framework (which could only learn a single parameter using a costly grid search) and other trainable models [12, 13, 14] (which only ensure convergence to local optima). The newly trained scoring function substantially outperforms the state-of-the-art in both scoring function accuracy and downstream Fisher kernel analysis. Furthermore, we significantly improve Didea s runtime performance through successive optimizations to its message passing schedule and derive explicit connections between Didea s new concave score and related MS/MS scoring functions. 1 Introduction A fundamental task in medicine and biology is identifying the proteins present in a complex biological sample, such as a drop of blood. The most widely used technology to accomplish this task is tandem mass spectrometry (MS/MS), which quickly produces a large collection of spectra representative of the peptides (i.e., protein subsequences) present in the original sample. A critical problem in MS/MS analysis, then, is the accurate identification of the peptide generating each observed spectrum. The most accurate methods which solve this problem search a database of peptides derived from the mapped organism of interest. Such database-search algorithms score peptides from the database and return the top-ranking peptide per spectrum. The pair consisting of an observed spectrum and scored peptide are typically referred to as a peptide-spectrum match (PSM). Many scoring functions have been proposed, ranging from simple dot-products [2, 28], to cross-correlation based [8], to p-value based [, 19, 16]. Recently, dynamic Bayesian networks (DBNs) have been shown to achieve the state-of-the-art in both PSM identification accuracy and post-search discriminative analysis, owing to their temporal modeling capabilities, parameter learning capabilities, and generative nature. The first such DBN-based scoring function, Didea [25], used sum-product inference to efficiently compute a log-posterior score for highly accurate PSM identification. However, Didea utilized a 32nd Conference on Neural Information Processing Systems (NeurIPS 2018), Montréal, Canada.
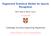
2 Figure 1: Example tandem mass spectrum with precursor charge c s = 2 and generating peptide x = TGPSPQPESQGSFYQR. Plotted in red and blue are, respectively, b- and y-ion peaks (discussed in Section 2.1), while unidentified peaks are colored gray. complicated emission distribution for which only a single parameter could be learned through a costly grid search. Subsequently, a DBN for Rapid Identification of Peptides (DRIP) [13, 11], was shown in [12] to outperform Didea due to its ability to generatively learn a large number of model parameters. Most recently, DRIP s generative nature was further exploited to derive log-likelihood gradients detailing the manner in which peptides align with observed spectra [14]. Combining these gradients with a discriminative postprocessor [17], the resulting DRIP Fisher kernel substantially improved upon all state-of-the-art methods for downstream analysis on a large number of datasets. However, while DRIP significantly improves several facets of MS/MS analysis due to its parameter learning capabilities, these improvements come at high runtime cost. In practice, DRIP inference is slow due to its large model complexity (the state-space grows exponentially in the lengths of both the observed spectrum and peptide). For instance, DRIP search required an order of magnitude longer than the slowest implementation of Didea for the timing tests in Section 5.2. Herein, we greatly improve upon all the analysis strengths provided by DRIP using the much faster Didea model. Furthermore, we optimize Didea s message passing schedule for a 64.2% speed improvement, leading to runtimes two orders of magnitude faster than DRIP and comparable to less accurate(but widely used) methods. Thus, the work described herein not only improves upon state-of-the-art DBN analysis for effective parameter learning, scoring function accuracy, and downstream Fisher kernel recalibration, but also renders such analysis practical by significantly decreasing state-of-the-art DBN inference time. In this work, we begin by discussing relevant MS/MS background and previous work. We then greatly expand the parameter learning capabilities of Didea by deriving a class of Bayesian network (BN) emission distributions for which both maximum likelihood learning and, most importantly, conditional maximum likelihood learning are concave. Called Convex Virtual Emissions (CVEs), we show that this class of emission distributions generalizes the widely used log-sum-exp function and naturally arises from the solution of a nonlinear differential equation representing convex conditions for general BN emissions. We incorporate CVEs into Didea to quickly and efficiently learn a substantial number of model parameters, considerably improving upon the previous learning framework. The newly trained model drastically improves PSM identification accuracy, outperforming all state-of-the-art methods over the presented datasets; at a strict FDR of 1% and averaged over the presented datasets, the trained Didea scoring function identifies 16% more spectra than DRIP and 17.4% more spectra than the highly accurate and widely used MS-GF+ [19]. Under the newly parameterized model, we then derive a bound explicitly relating Didea s score to the popular XCorr scoring function, thus providing potential avenues to train XCorr using the presented parameter learning work. With efficient parameter learning in place, we next utilize the new Didea model to improve MS/MS recalibration performance. We use gradient information derived from Didea s conditional loglikelihood in the feature-space of a kernel-based classifier [17]. Training the resulting conditional Fisher kernel substantially improves upon the state-of-the-art recalibration performance previously achieved by DRIP; at a strict FDR of 1%, discriminative recalibration using Didea s conditional Fisher kernel results in an average 11.3% more identifications than using the DRIP Fisher kernel. Finally, we conclude with a discussion of several avenues for future work. 2
3 2 Tandem mass spectrometry With a complex sample as input, a typical MS/MS experiment begins by cleaving the proteins of the sample into peptides using a digesting enzyme, such as trypsin. The digested peptides are then separated via liquid chromatography and undergo two rounds of mass spectrometry. The first round of mass spectrometry measures the mass and charge of the intact peptide, referred to as the precursor mass and precursor charge, respectively. Peptides are then fragmented into prefix and suffix ions. The mass-to-charge (m/z) ratios of the resulting fragment ions are measured in the second round of mass spectrometry, producing an observed spectrum of m/z versus intensity values representative of the fragmented peptide. The output of this overall process is a large collection of spectra (often numbering in the hundreds-of-thousands), each of which is representative of a peptide from the original complex sample and requires identification. The x-axis of such observed spectra denotes m/z, measured in thomsons (Th), and y-axis measures the intensity at a particular m/z value. A sample such observed spectrum is illustrated in Figure Database search and theoretical spectra Let s S be an observed spectrum with precursor m/z m s and precursor charge c s, where S is the universe of tandem mass spectra. The generating peptide of s is identified by searching a database of peptides, as follows. Let P be the universe of all peptides and x P be an arbitrary peptide of length l. x = x 1... x l is a string comprised of characters called amino acids, the dictionary size of which are 20. We denote peptide substrings as x i:j = x i,..., x j, where i > 0, j l, i < j, and the mass of x as m(x). Given a peptide database D P, the set of peptides considered is constrained to those within a precursor mass tolerance window w of m s. The set of candidate peptides to be scored is thus D(s, D, w) = {x : x D, m(x) c m s w}. Using a scoring function ψ : P S R, a database search outputs the top-scoring s PSM, x = argmax x D ψ(x, s). In order to score a PSM, the idealized fragment ions of x are first collected into a theoretical spectrum. The most commonly encountered fragment ions are called b-ions and y-ions. B- and y-ions correspond to prefix and suffix mass pairs, respectively, such that the precursor charge c s is divided amongst the pair. For b-ion charge c b c s, the kth b-ion and the accompanying y-ion are then, respectively, [ k ] ] b(m(x 1:k ), c b ) = m(x 1:k) + c b i=1 m(xi) + c b i=k+1 m(xi) c y =, y(m(x k+1:l ), c y) =, c b c b c y where c y is the y-ion charge, the b-ion offset corresponds to a c b charged hydrogen atom, and the y-ion offset corresponds to a c y charged hydrogen atom plus a water molecule. For singly charged spectra c s = 1, only singly charged fragment ions are detectable, so that c b = c y = 1. For higher precursor charge states c s 2, the total charge is split between each b- and y-ion pair, so that 0 < c b < c s and c y = c s c b. The annotated b- and y-ions of an identified observed spectrum are illustrated in Figure 1. 3 Previous work Many database search scoring algorithms have been proposed, each of which is characterized by the scoring function they employ. These scoring functions have ranged from dot-products (X!Tandem [2] and Morpheus [28]), to cross-correlation based (XCorr [8]), to exact p-values computed over linear scores [19, 16]. Recently, DBNs have been used to substantially improve upon the accuracy of previous approaches. In the first such DBN, Didea [25], the time series being modeled is the sequence of a peptide s amino acids (i.e., an amino acid is observed in each frame) and the quantized observed spectrum is observed in each frame. In successive frames, the sequence of b- and y-ions are computed and used as indices into the observed spectrum via virtual evidence [24]. A hidden variable in the first frame, corresponding to the amount to shift the observed spectrum by, is then marginalized in order to compute a conditional log-likehood probability consisting of a foreground score minus a background score, similar in form to XCorr (described in Section 4.2.1). The resulting scoring function outperformed the most accurate scoring algorithms at the time (including MS-GF+, then called MS-GFDB) on a majority of datasets. However, parameter learning in the model was severely limited and inefficient; a single hyperparameter controlling the reweighting of peak intensities was learned via an expensive grid search, requiring repeated database searches over a dataset. [ l 3
4 Subsequent work saw the introduction of DRIP [12, 13], a DBN with substantial parameter learning capabilities. In DRIP, the time series being modeled is the sequence of observed spectrum peaks (i.e., each frame in DRIP corresponds to an observed peak) and two types of prevalent phenomena are explicitly modeled via sequences of random variables: spurious observed peaks (called insertions) and absent theoretical peaks (called deletions). A large collection of Gaussians parameterizing the m/z axis are generatively learned, via expectation-maximization (EM) [4], and used to score observed peaks. DRIP then uses max-product inference to calculate the most probable sequences of insertions and deletions in order to score PSMs. In practice, the majority of PSM scoring functions discussed are typically poorly calibrated, i.e., it is often difficult to compare the PSM scores across different spectra. In order to combat such poor calibration, postprocessors are commonly employed to recalibrate PSM scores [17, 26, 27]. In recent work, DRIP s generative framework was further exploited to calculate highly accurate features based on the log-likelihood gradients of its learnable parameters. Combining these new gradient-based features with a popular kernel-based classifier for recalibrating PSM scores [17], the resulting Fisher kernel was shown to significantly improve postprocessing accuracy [14]. 4 Didea Figure 2: Graph of Didea. Unshaded nodes are hidden, shaded nodes are observed, and edges denote deterministic functions of parent variables. We now derive Didea s scoring function in detail. The graph of Didea is displayed in Figure 2. Shaded variables are observed and unshaded variables are hidden (random). Groups of variables are collected into time instances called frames, where the first frame is called the prologue, the final frame is called the epilogue, and the chunk dynamically expands to fill all frames in between. Let 0 t l be an arbitrary frame. The amino acids of a peptide are observed in each frame after the prologue. The variable M t successively accumulates the prefix masses of the peptide such that p(m 0 = 0) = 1 and p(m t = M t 1 + m(x t ) M t 1, x t ) = 1, while the variable M t successively accumulates the suffix masses of the peptide such that p(m l = 0) = 1 and p(m t = M t+1 + m(x t+1 ) M t+1, x t+1 ) = 1. Denoting the maximum spectra shift as L, the shift variable τ 0 [ L, L] is hidden, uniform, and deterministically copied by its descendents in successive frames, such that p(τ t = τ τ 0 = τ) = 1 for t > 1. Let s Rō+1 be the binned observed spectrum, i.e., a vector of length ō + 1 whose ith element is s(i), where ō is the maximum observable discretized m/z value. Shifted versions of the tth b- and y-ion pair (where the shift is denoted by subscript) are deterministic functions of the shift variable as well as prefix and suffix masses, i.e., p(b t = b τt (M t, 1) M t, τ t ) = p(b t = max(min(b(m t, 1) τ t, 0), ō) M t, τ t ), p(y t = y τt (m(x t+1:l ), 1) M t, τ t ) = p(y t = max(min(y(m(x t+1:l ), 1) τ, 0), ō) M t, τ t ), respectively. ξ b and ξ y are virtual evidence children, i.e., leaf nodes whose conditional distribution need not be normalized (only non-negative) to compute posterior probabilities in the DBN. A comprehensive overview of virtual evidence is available in [15]. ξ b and ξ y compare the b- and y-ions, respectively, to the observed spectrum, such that p(ξ b B t ) = f(s(b τt (m(x 1:t ), 1))), p(ξ y Y t ) = f(s(y τt (m(x t+1:l ), 1))), where f is a non-negative emission function. 4
5 Let 1 { } denote the indicator function. Didea s log-likelihood is then log p(τ 0 = τ, x, s) = log p(τ 0 = τ)p(m 0)p(M 0 M 1, x 1)p(M l )p(m l M l 1, x 1)+ l log [p(τ t τ t 1)p(M t M t 1, x t)p(m t M t+1, x t+1)p(b t M t, τ t)p(y t M t, τ t)p(ξ b B t)p(ξ y Y t)] t=1 l 1 = log p(τ 0 = τ) + log t=1 ( 1{τt = τ M t =m(x 1:t) M l 1 = log p(τ 0 = τ) + log p(ξ b b τ (M t, 1))p(ξ y y τ (M t, 1))] t=1 t =m(x t+1:l)}p(b t M t, τ t)p(y t M t, τ t)p(ξ b B t)p(ξ y Y t) ) l 1 ( = log p(τ 0 = τ) + log f(s τ (b(m(x 1:t), 1))) + log f(s τ (y(m(x t+1:l ), 1))) ). t=1 In order to score PSMs, Didea computes the conditional log-likelihood ψ(s, x) = log p(τ 0 = 0 x, s) = log p(τ 0 = 0, x, s) log = log p(τ 0 = 0, x, s) log 1 τ 0 L τ= L L τ= L p(τ 0 = τ)p(x, s τ τ 0 = τ) p(x, s τ τ 0 = τ). (1) As previously mentioned, ψ(s, x) is a foreground score minus a background score, where the background score consists of averaging over τ 0 shifted versions of the foreground score, much like the XCorr scoring function. Thus, Didea may be thought of as a probabilistic analogue of XCorr. 4.1 Convex Virtual Emissions for Bayesian networks Consider an arbitrary Bayesian network where the observed variables are leaf nodes, as is common in a large number of time-series models such as hidden Markov models (HMMs), hierarchical HMMs [22], DBNs for speech recognition [5], hybrid HMMs/DBNs [3], as well as DRIP and Didea. Let E be the set of observed random variables, H be the hypothesis space composed of the cross-product of the n hidden discrete random variables in the network, and h H be an arbitrary hypothesis (i.e., an instantiation of the hidden variables). As is the case in Didea, often desired is the log-posterior probability log p(h E) = log p(h,e) p(h,e) p(e) = log h H p( h,e) = log p(h, E) log h H p(h)p(e h). Under general assumptions, we d like to find emission functions for which log p(h E) is concave. Assume p(h) and p(e h) are non-negative, that the emission density p(e h) is parameterized by θ (which we d like to learn), and that there is a parameter θ h to be learned for every hypothesis of latent variables (though if we have fewer parameters, parameter estimation becomes strictly easier). We make this parameterization explicit by denoting the emission distributions of interest as p θh (E h). Assume that p θh (E h) is smooth on R for all h H. Applying virtual evidence for such models, p θh (E h) need not be normalized for posterior inference (as well as Viterbi inference and comparative inference between sets of observations; an extensive review of virtual evidence may be found in [15]). Given the factorization of the joint distribution described by the BN, the quantity p θh (h, E) = p(h)p θh (E h) may often be efficiently computed for any given h. Thus, the computationally difficult portion of log p(h E) is the calculation of the log-likelihood in the denominator, wherein all hidden variables are marginalized over. We therefore first seek emission functions for which the loglikelihood log p(e) = log h H p(h)p θ h (E h) is convex. For such emission functions, we have the following theorem. Theorem 1. The unique convex functions of the form log h H p(h)p θ h (E h), such that (p θ h (E h)) 2 p θ h (E h)p θh (E h) = 0, are log h H p(h)p θ h (E h) = log h H α he β hθ h, where α h = p(h)a h and a h, β h are hyperparameters. The full proof of Theorem 1 is given in [15]. The nonlinear differential equation (p θ h (E h)) 2 p θ h (E h)p θh (E h) = 0 describes the curvature of the desired emission functions and arises from the necessary and sufficient conditions for twice differentiable convex functions (i.e., the Hessian 5
6 must be p.s.d.) and the Cauchy-Schwarz inequality. Particular values of the hyperparameters a h and β h correspond to unique initial conditions for this nonlinear differential equation. Note that when α h = 1, p θh (E h) = e θ h, we have the well-known log-sum-exp (LSE) convex function. Thus, this result generalizes the LSE function to a broader class of convex functions. We call the unique class of convex functions which arise from solving the nonlinear differential in Theorem 1, p θh (E h) = a h e β hθ h, Convex Virtual Emissions (CVEs). Note that utilizing CVEs, maximimum likelihood estimation (i.e., argmax θ log h H p(h)p θ h (E h)) is thus concave and guaranteed to converge to a global optimum. Furthermore, and most importantly for Didea, we have the following result for the conditional log-likelihood (the full proof of which is in [15]). Corollary 1.1. For convex log p(e) = log h H p(h)p θ h (E h) such that (p θ h (E h)) 2 p θ h (E h)p θh (E h) = 0, the log-posterior log p θ (h E) is concave in θ. Thus, utilizing CVEs, conditional maximum likelihood estimation is also rendered concave. 4.2 CVEs in Didea In [25], the virtual evidence emission function to score peak intensities was f λ (s(i)) = 1 λe λ + λe λ(1 s(i)). Under this function, Didea was shown to perform well on a variety of datasets. However, this function is non-convex and does not permit efficient parameter learning; although only a single model parameter, λ, was trained, learning required a grid search wherein each step consisted of a database search over a dataset and subsequent target-decoy analysis to assess each new parameter value. While this training scheme is already costly and impractical, it quickly becomes infeasible when looking to learn more than a single model parameter. We use CVEs to render Didea s conditional log-likelihood concave given a large number of parameters. To efficiently learn a distinct observation weight θ τ for each spectral shift τ [ L, L], we thus utilize the emission function f θτ (s(i)) = e θτ s(i). Denote the set of parameters per spectra shift as θ = {θ L,..., θ L }. Due to the concavity of Equation 1 using f θτ under Corollary 1.1, given n PSM training pairs (s 1, x 1 ),..., (s n, x n ), the learned parameters θ n = argmax θ i=1 ψ θ(s i, x i ) are guaranteed to converge to a global optimum. Further analysis of Didea s scoring function under this new emission function may be found in [15], including the derivation of the new model s gradients (i.e., conditional Fisher scores) Relating Didea s conditional log-likelihood to XCorr using CVEs XCorr [8], the very first database search scoring function for peptide identification, remains one of the most widely used tools in the field today. Owing to its prominence, XCorr remains an active subject of analysis and continuous development [20, 23, 7, 6, 16, 21, 9, 14]. As previously noted, the scoring functions of XCorr and Didea share several similarities in form, where, in fact, the former served as the motivating example in [25] for both the design of the Didea model and its posterior-based scoring function. While cosmetic similarities have thus far been noted, the reparameterization of Didea s conditional log-likelihood using CVEs permits the derivation of an explicit relationship between the two. Let u be the theoretical spectrum of peptide x. As with Didea, let L be the maximum spectra shift considered and, for shift τ, denote a vector shift via subscript, such that s τ is the vector of observed spectrum elements shifted by τ units. In order to compare u and s, XCorr is thus computed as XCorr(s, x) = u T s 1 L 2L+1 τ= L ut s τ. Intuitively, the cross-correlation background term is meant to penalize overfitting of the theoretical spectrum. Under the newly parameterized Didea conditional log-likelihood described herein, we have the following theorem explicitly relating the XCorr and Didea scoring functions. Theorem 2. Assume the PSM scoring function ψ(s, x) is that of Didea (i.e., Equation 1) where the emission function f θτ (s(i)) has uniform weights θ i = θ j, for i, j [ L, L]. Then ψ(s, x) O(XCorr(s, x)). The full proof of Theorem 2 may be found in [15]. Thus, Didea s scoring function effectively serves to lower bound XCorr. This opens possible avenues for extending the learning results detailed herein to the widely used XCorr function. For instance, a natural extension is to use a variational Bayesian 6
7 inference approach and learn XCorr parameters through iterative maximization of the Didea lower bound, made efficient by the concavity of new Didea model derived in this work. 4.3 Faster Didea sum-product inference We successively improved Didea s inference time when conducting a database search using the intensive charge-varying model (discussed in [15]). Firstly, we removed the need for a backward pass by keeping track of the foreground log-likelihood during the forward pass (which computes the background score, i.e., the probability of evidence in the model). Next, by exploiting the symmetry of the spectral shifts, we cut the effective cardinality of τ in half during inference. While this requires twice as much memory in practice, this is not close to being prohibitive on modern machines. Finally, a large portion of the speedup was achieved by offsetting the virtual evidence vector by τ and pre/post buffering with zeros and offsetting each computed b- and y-ion by τ. Under this construction, the scores do not change, but, during inference, we are able to shift each computed b- and y-ion by ±τ without requiring any bound checking. Hashing virtual evidence bin values by b-/y-ion value and τ was also pursued, but did not offer any runtime benefit over the aforementioned speedups (due to the cost of constructing the hash table per spectrum). 5 Results In practice, assessing peptide identification accuracy is made difficult by the lack of ground-truth encountered in real-world data. Thus, it is most common to estimate the false discovery rate (FDR) [1] by searching a decoy database of peptides which are unlikely to occur in nature, typically generated by shuffling entries in the target database [18]. For a particular score threshold, t, the FDR is calculated as the proportion of decoys scoring better than t to the number of targets scoring better than t. Once the target and decoy PSM scores are calculated, a curve displaying the FDR threshold versus the number of correctly identified targets at each given threshold may be calculated. In place of FDR along the x-axis, we use the [18], defined to be the minimum FDR threshold at which a given score is deemed to be significant. As many applications require a search algorithm perform well at low thresholds, we only plot q [0, 0.1]. The benchmark datasets and search settings used to recently evaluate the DRIP Fisher kernel in [14] are adapted in this work. The charge-varying Didea model (which integrates over multiple charge states, further described in [15]) with concave emissions (described in Section 4.2) was used to score and rank database peptides. Concave Didea parameters were learned using the high-quality PSMs used to generatively train the DRIP model in [13] and gradient ascent. Didea s newly trained database-search scoring function is benchmarked against the Didea model from [25] trained using a costly grid search for a single parameter (denoted as Didea-0 ) and four other state-of-the-art scoring algorithms: DRIP, MS-GF+, XCorr p-values, and XCorr. DRIP searches were conducted using the DRIP Toolkit and the generatively trained parameters described in [12, 13]. MS-GF+, one of the most accurate search algorithms in wide-spread use, was run using version 9980, with PSMs ranked by E-value. XCorr and XCorr p-value scores were collected using Crux v All database searches were run using a ±3.0Th mass tolerance, XCorr flanking peaks not allowed in Crux searches, and all search algorithm settings otherwise left to their defaults. Peptides were derived from the protein databases using trypsin cleavage rules without suppression of proline and a single fixed carbamidomethyl modification was included. The resulting database-search accuracy plots are displayed in Figure 3. The trained Didea model outperforms all competitors across all presented datasets; compared to highly accurate scoring algorithms DRIP and MS-GF+, the trained Didea scoring function identifies 16% more spectra than DRIP and 17.4% more spectra than MS-GF+, at a strict FDR of 1% averaged over the presented datasets. This high-level performance is attributable to the expanded and efficient parameter learning framework, which greatly improves upon the limited parameter learning capabilities of the original Didea model, identifying 9.8% more spectra than Didea-0 at a strict FDR of 1% averaged over the presented datasets. 7
8 Didea Didea-0 DRIP MS-GF+ XCorr p-value XCorr (d) Worm (b) Worm-1 (e) Worm (c) Worm-2 (f) Yeast (g) Yeast-2 (h) Yeast-3 (i) Yeast-4 Figure 3: Database search accuracy plots measured by versus number of spectra identified for worm (C. elegans) and yeast (Saccharomyces cerevisiae) datasets. All methods are run with as equivalent settings as possible. The Didea charge-varying model (described in [15]) was used to score PSMs, with Didea denoting the model trained per charge state using the concave framework described in Section 4.2 and Didea-0 denoting the model from [25] trained using a grid search. DRIP, another DBN-based scoring function, was run using the generatively learned parameters described in [13]. 5.1 Conditional Fisher kernel for improved discriminative analysis Facilitated by Didea s effective parameter learning framework, we look to leverage gradient-based PSM information to aid in discriminative postprocessing analysis. We utilize the same set of features as the DRIP Fisher kernel [14]. However, in order to measure the relative utility of the gradients under study, we replace the DRIP log-likelihood gradients with Didea gradient information (derived in [15]). These features are used to train an SVM classifier, Percolator [17], which recalibrates PSM scores based on the learned decision boundary between input targets and decoys. Didea s resulting conditional Fisher kernel is benchmarked against the DRIP Fisher kernel and the previously benchmarked scoring algorithms using their respective standard Percolator features sets. DRIP Kernel features were computed using the customized version of the DRIP Toolkit from [14]. MS-GF+ Percolator features were collected using msgf2pin and XCorr/XCorr p-value features collected using Crux. For the resulting postprocessing results, the trained Didea scoring function outperforms all competitors, identifying 12.3% more spectra than DRIP and 13.4% more spectra than MS-GF+ at a strict FDR of 1% and averaged over the presented datasets. The full panel of results is displayed in [15]. Compared to DRIP s log-likelihood gradient features, the conditional 8
9 log-likelihood gradients of Didea contain much richer PSM information, thus allowing Percolator to better distinguish target from decoy PSMs for much greater recalibration performance. 5.2 Optimized exact sum-product inference for improved Didea runtime Implementing the speedups to exact Didea sum-product inference described in Section 4.3, we benchmark the optimized search algorithm using 1, 000 randomly sampled spectra (with charges varying from 1+ to 3+) from the Worm-1 dataset and averaged database-search times (reported in wall clock time) over runs. The resulting runtimes are listed in Table 1. DRIP was run using the DRIP Toolkit and XCorr p-values were collected using Crux v All benchmarked search algorithms were run on the same machine with an Intel Xeon E and 64GB RAM. The described optimizations result in a 64.2% runtime improvement, and brings search time closer to less accurate, but faster, search algorithms. Didea-0 Didea Opt. XCorr p-values DRIP runtime Table 1: Database search runtimes per spectrum, in seconds, searching 1,000 worm spectra randomly sampled from the Worm-1 dataset. Didea-0 is the implementation of Didea used in [25] and Didea Opt is the speed-optimized implementation described herein. All reported search algorithm runtimes were averaged over runs. 6 Conclusions and future work In this work, we ve derived a widely applicable class of Bayesian network emission distributions, CVEs, which naturally generalize the convex log-sum-exp function and carry important theoretical properties for parameter learning. Using CVEs, we ve substantially improved the parameter learning capabilities of the DBN scoring algorithm, Didea, by rendering its conditional log-likelihood concave with respect to a large set of learnable parameters. Unlike previous DBN parameter learning solutions, which only guarantee convergence to a local optimum, the new learning framework thus guarantees global convergence. Didea s newly trained database-search scoring function significantly outperforms all state-of-the-art scoring algorithms on the presented datasets. With efficient parameter learning in hand, we derived the gradients of Didea s conditional log-likelihood and used this gradient information in the feature space of a kernel-based discriminative postprocessor. The resulting conditional Fisher kernel once again outperforms the state-of-the-art on all presented datasets, including a highly accurate, recently proposed Fisher kernel. Furthermore, we successively optimized Didea s message passing schedule, leading to DBN analysis times two orders of magnitude faster than other leading DBN tools for MS/MS analysis. Thus, the presented results improve upon all aspects of state-of-theart DBN analysis for MS/MS. Finally, using the new learning framework, we ve proven that Didea is proportionally lower bounds the widely used XCorr scoring function. There are a number of exciting avenues for future work. Considering the large amount of PSM information held in the gradient space of Didea s conditional log-likelihood, we plan on pursuing kernel-based approaches to peptide identification using the Hessian of the scoring function. This is especially exciting given the high degree of recalibration accuracy provided by Percolator, a kernel-based post-processor. Using a variational approach, we also plan on investigating parameter learning options for XCorr given the Didea lower bound and the concavity of Didea s parameterized scoring function. Finally, in perhaps the most ambitious plan for future work, we plan to further build upon Didea s parameter learning framework by learning distance matrices between the theoretical and observed spectra. Such matrices naturally generalize the class of CVEs derived herein. Acknowledgments: This work was supported by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, through grant UL1 TR References [1] Y. Benjamini and Y. Hochberg. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B, 57: ,
10 [2] R. Craig and R. C. Beavis. Tandem: matching proteins with tandem mass spectra. Bioinformatics, 20: , [3] George E Dahl, Dong Yu, Li Deng, and Alex Acero. Context-dependent pre-trained deep neural networks for large-vocabulary speech recognition. IEEE Transactions on audio, speech, and language processing, 20(1):30 42, [4] A. P. Dempster, N. M. Laird, and D. B. Rubin. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society. Series B (Methodological), 39:1 22, [5] Li Deng. Dynamic speech models: theory, algorithms, and applications. Synthesis Lectures on Speech and Audio Processing, 2(1):1 118, [6] Benjamin J. Diament and William Stafford Noble. Faster SEQUEST searching for peptide identification from tandem mass spectra. Journal of Proteome Research, (9): , [7] J. K. Eng, B. Fischer, J. Grossman, and M. J. MacCoss. A fast SEQUEST cross correlation algorithm. Journal of Proteome Research, 7(): , [8] J. K. Eng, A. L. McCormack, and J. R. Yates, III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry, 5: , [9] Jimmy K Eng, Tahmina A Jahan, and Michael R Hoopmann. Comet: An open-source ms/ms sequence database search tool. Proteomics, 13(1):22 24, [] Lewis Y. Geer, Sanford P. Markey, Jeffrey A. Kowalak, Lukas Wagner, Ming Xu, Dawn M. Maynard, Xiaoyu Yang, Wenyao Shi, and Stephen H. Bryant. Open mass spectrometry search algorithm. Journal of Proteome Research, 3(5): , [11] John T Halloran. Analyzing tandem mass spectra using the drip toolkit: Training, searching, and post-processing. In Data Mining for Systems Biology, pages Springer, [12] John T. Halloran, Jeff A. Bilmes, and William S. Noble. Learning peptide-spectrum alignment models for tandem mass spectrometry. In Uncertainty in Artificial Intelligence (UAI), Quebec City, Quebec Canada, July AUAI. [13] John T Halloran, Jeff A Bilmes, and William S Noble. Dynamic bayesian network for accurate detection of peptides from tandem mass spectra. Journal of Proteome Research, 15(8): , [14] John T Halloran and David M Rocke. Gradients of generative models for improved discriminative analysis of tandem mass spectra. In Advances in Neural Information Processing Systems, pages , [15] John T. Halloran and David M. Rocke. Learning Concave Conditional Likelihood Models for Improved Analysis of Tandem Mass Spectra: Supplementary Material. In Advances in Neural Information Processing Systems, [16] J Jeffry Howbert and William S Noble. Computing exact p-values for a cross-correlation shotgun proteomics score function. Molecular & Cellular Proteomics, pages mcp O113, [17] L. Käll, J. Canterbury, J. Weston, W. S. Noble, and M. J. MacCoss. A semi-supervised machine learning technique for peptide identification from shotgun proteomics datasets. Nature Methods, 4:923 25, [18] Uri Keich, Attila Kertesz-Farkas, and William Stafford Noble. Improved false discovery rate estimation procedure for shotgun proteomics. Journal of proteome research, 14(8): , [19] Sangtae Kim and Pavel A Pevzner. Ms-gf+ makes progress towards a universal database search tool for proteomics. Nature communications, 5, 2014.
11 [20] A. A. Klammer, C. Y. Park, and W. S. Noble. Statistical calibration of the SEQUEST XCorr function. Journal of Proteome Research, 8(4): , PMC [21] Sean McIlwain, Kaipo Tamura, Attila Kertesz-Farkas, Charles E Grant, Benjamin Diament, Barbara Frewen, J Jeffry Howbert, Michael R Hoopmann, Lukas Käll, Jimmy K Eng, et al. Crux: rapid open source protein tandem mass spectrometry analysis. Journal of proteome research, [22] Kevin P Murphy and Mark A Paskin. Linear-time inference in hierarchical hmms. In Advances in neural information processing systems, pages , [23] C. Y. Park, A. A. Klammer, L. Käll, M. P. MacCoss, and W. S. Noble. Rapid and accurate peptide identification from tandem mass spectra. Journal of Proteome Research, 7(7): , [24] J. Pearl. Probabilistic Reasoning in Intelligent Systems : Networks of Plausible Inference. Morgan Kaufmann, [25] Ajit P. Singh, John Halloran, Jeff A. Bilmes, Katrin Kirchoff, and William S. Noble. Spectrum identification using a dynamic bayesian network model of tandem mass spectra. In Uncertainty in Artificial Intelligence (UAI), Catalina Island, USA, July AUAI. [26] M. Spivak, J. Weston, L. Bottou, L. Käll, and W. S. Noble. Improvements to the Percolator algorithm for peptide identification from shotgun proteomics data sets. Journal of Proteome Research, 8(7): , PMC [27] M. Spivak, J. Weston, D. Tomazela, M. J. MacCoss, and W. S. Noble. Direct maximization of protein identifications from tandem mass spectra. Molecular and Cellular Proteomics, 11(2):M , PMC [28] C. D. Wenger and J. J. Coon. A proteomics search algorithm specifically designed for highresolution tandem mass spectra. Journal of proteome research,
Learning Peptide-Spectrum Alignment Models for Tandem Mass Spectrometry
 Learning Peptide-Spectrum Alignment Models for Tandem Mass Spectrometry John T. Halloran Dept. of Electrical Engineering University of Washington Seattle, WA 99, USA Jeff A. Bilmes Dept. of Electrical
Learning Peptide-Spectrum Alignment Models for Tandem Mass Spectrometry John T. Halloran Dept. of Electrical Engineering University of Washington Seattle, WA 99, USA Jeff A. Bilmes Dept. of Electrical
Tandem Mass Spectrometry: Generating function, alignment and assembly
 Tandem Mass Spectrometry: Generating function, alignment and assembly With slides from Sangtae Kim and from Jones & Pevzner 2004 Determining reliability of identifications Can we use Target/Decoy to estimate
Tandem Mass Spectrometry: Generating function, alignment and assembly With slides from Sangtae Kim and from Jones & Pevzner 2004 Determining reliability of identifications Can we use Target/Decoy to estimate
Sequence labeling. Taking collective a set of interrelated instances x 1,, x T and jointly labeling them
 HMM, MEMM and CRF 40-957 Special opics in Artificial Intelligence: Probabilistic Graphical Models Sharif University of echnology Soleymani Spring 2014 Sequence labeling aking collective a set of interrelated
HMM, MEMM and CRF 40-957 Special opics in Artificial Intelligence: Probabilistic Graphical Models Sharif University of echnology Soleymani Spring 2014 Sequence labeling aking collective a set of interrelated
Learning Score Function Parameters for Improved Spectrum Identification in Tandem Mass Spectrometry Experiments
 pubs.acs.org/jpr Learning Score Function Parameters for Improved Spectrum Identification in Tandem Mass Spectrometry Experiments Marina Spivak, Michael S. Bereman, Michael J. MacCoss, and William Stafford
pubs.acs.org/jpr Learning Score Function Parameters for Improved Spectrum Identification in Tandem Mass Spectrometry Experiments Marina Spivak, Michael S. Bereman, Michael J. MacCoss, and William Stafford
Bayesian Networks BY: MOHAMAD ALSABBAGH
 Bayesian Networks BY: MOHAMAD ALSABBAGH Outlines Introduction Bayes Rule Bayesian Networks (BN) Representation Size of a Bayesian Network Inference via BN BN Learning Dynamic BN Introduction Conditional
Bayesian Networks BY: MOHAMAD ALSABBAGH Outlines Introduction Bayes Rule Bayesian Networks (BN) Representation Size of a Bayesian Network Inference via BN BN Learning Dynamic BN Introduction Conditional
O 3 O 4 O 5. q 3. q 4. Transition
 Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
An Introduction to Bioinformatics Algorithms Hidden Markov Models
 Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Hidden Markov Models
 Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 11 Project
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 11 Project
Nature Methods: doi: /nmeth Supplementary Figure 1. Fragment indexing allows efficient spectra similarity comparisons.
 Supplementary Figure 1 Fragment indexing allows efficient spectra similarity comparisons. The cost and efficiency of spectra similarity calculations can be approximated by the number of fragment comparisons
Supplementary Figure 1 Fragment indexing allows efficient spectra similarity comparisons. The cost and efficiency of spectra similarity calculations can be approximated by the number of fragment comparisons
A NESTED MIXTURE MODEL FOR PROTEIN IDENTIFICATION USING MASS SPECTROMETRY
 Submitted to the Annals of Applied Statistics A NESTED MIXTURE MODEL FOR PROTEIN IDENTIFICATION USING MASS SPECTROMETRY By Qunhua Li,, Michael MacCoss and Matthew Stephens University of Washington and
Submitted to the Annals of Applied Statistics A NESTED MIXTURE MODEL FOR PROTEIN IDENTIFICATION USING MASS SPECTROMETRY By Qunhua Li,, Michael MacCoss and Matthew Stephens University of Washington and
Workflow concept. Data goes through the workflow. A Node contains an operation An edge represents data flow The results are brought together in tables
 PROTEOME DISCOVERER Workflow concept Data goes through the workflow Spectra Peptides Quantitation A Node contains an operation An edge represents data flow The results are brought together in tables Protein
PROTEOME DISCOVERER Workflow concept Data goes through the workflow Spectra Peptides Quantitation A Node contains an operation An edge represents data flow The results are brought together in tables Protein
Dynamic Data Modeling, Recognition, and Synthesis. Rui Zhao Thesis Defense Advisor: Professor Qiang Ji
 Dynamic Data Modeling, Recognition, and Synthesis Rui Zhao Thesis Defense Advisor: Professor Qiang Ji Contents Introduction Related Work Dynamic Data Modeling & Analysis Temporal localization Insufficient
Dynamic Data Modeling, Recognition, and Synthesis Rui Zhao Thesis Defense Advisor: Professor Qiang Ji Contents Introduction Related Work Dynamic Data Modeling & Analysis Temporal localization Insufficient
Modeling Mass Spectrometry-Based Protein Analysis
 Chapter 8 Jan Eriksson and David Fenyö Abstract The success of mass spectrometry based proteomics depends on efficient methods for data analysis. These methods require a detailed understanding of the information
Chapter 8 Jan Eriksson and David Fenyö Abstract The success of mass spectrometry based proteomics depends on efficient methods for data analysis. These methods require a detailed understanding of the information
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2014
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Probabilistic Time Series Classification
 Probabilistic Time Series Classification Y. Cem Sübakan Boğaziçi University 25.06.2013 Y. Cem Sübakan (Boğaziçi University) M.Sc. Thesis Defense 25.06.2013 1 / 54 Problem Statement The goal is to assign
Probabilistic Time Series Classification Y. Cem Sübakan Boğaziçi University 25.06.2013 Y. Cem Sübakan (Boğaziçi University) M.Sc. Thesis Defense 25.06.2013 1 / 54 Problem Statement The goal is to assign
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2016
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2016 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2016 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Sequence Modelling with Features: Linear-Chain Conditional Random Fields. COMP-599 Oct 6, 2015
 Sequence Modelling with Features: Linear-Chain Conditional Random Fields COMP-599 Oct 6, 2015 Announcement A2 is out. Due Oct 20 at 1pm. 2 Outline Hidden Markov models: shortcomings Generative vs. discriminative
Sequence Modelling with Features: Linear-Chain Conditional Random Fields COMP-599 Oct 6, 2015 Announcement A2 is out. Due Oct 20 at 1pm. 2 Outline Hidden Markov models: shortcomings Generative vs. discriminative
Dynamic Approaches: The Hidden Markov Model
 Dynamic Approaches: The Hidden Markov Model Davide Bacciu Dipartimento di Informatica Università di Pisa bacciu@di.unipi.it Machine Learning: Neural Networks and Advanced Models (AA2) Inference as Message
Dynamic Approaches: The Hidden Markov Model Davide Bacciu Dipartimento di Informatica Università di Pisa bacciu@di.unipi.it Machine Learning: Neural Networks and Advanced Models (AA2) Inference as Message
STA 414/2104: Machine Learning
 STA 414/2104: Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistics! rsalakhu@cs.toronto.edu! http://www.cs.toronto.edu/~rsalakhu/ Lecture 9 Sequential Data So far
STA 414/2104: Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistics! rsalakhu@cs.toronto.edu! http://www.cs.toronto.edu/~rsalakhu/ Lecture 9 Sequential Data So far
Variational Inference (11/04/13)
 STA561: Probabilistic machine learning Variational Inference (11/04/13) Lecturer: Barbara Engelhardt Scribes: Matt Dickenson, Alireza Samany, Tracy Schifeling 1 Introduction In this lecture we will further
STA561: Probabilistic machine learning Variational Inference (11/04/13) Lecturer: Barbara Engelhardt Scribes: Matt Dickenson, Alireza Samany, Tracy Schifeling 1 Introduction In this lecture we will further
Experiments with a Gaussian Merging-Splitting Algorithm for HMM Training for Speech Recognition
 Experiments with a Gaussian Merging-Splitting Algorithm for HMM Training for Speech Recognition ABSTRACT It is well known that the expectation-maximization (EM) algorithm, commonly used to estimate hidden
Experiments with a Gaussian Merging-Splitting Algorithm for HMM Training for Speech Recognition ABSTRACT It is well known that the expectation-maximization (EM) algorithm, commonly used to estimate hidden
Part of Speech Tagging: Viterbi, Forward, Backward, Forward- Backward, Baum-Welch. COMP-599 Oct 1, 2015
 Part of Speech Tagging: Viterbi, Forward, Backward, Forward- Backward, Baum-Welch COMP-599 Oct 1, 2015 Announcements Research skills workshop today 3pm-4:30pm Schulich Library room 313 Start thinking about
Part of Speech Tagging: Viterbi, Forward, Backward, Forward- Backward, Baum-Welch COMP-599 Oct 1, 2015 Announcements Research skills workshop today 3pm-4:30pm Schulich Library room 313 Start thinking about
A Gentle Tutorial of the EM Algorithm and its Application to Parameter Estimation for Gaussian Mixture and Hidden Markov Models
 A Gentle Tutorial of the EM Algorithm and its Application to Parameter Estimation for Gaussian Mixture and Hidden Markov Models Jeff A. Bilmes (bilmes@cs.berkeley.edu) International Computer Science Institute
A Gentle Tutorial of the EM Algorithm and its Application to Parameter Estimation for Gaussian Mixture and Hidden Markov Models Jeff A. Bilmes (bilmes@cs.berkeley.edu) International Computer Science Institute
Fast and Accurate Causal Inference from Time Series Data
 Fast and Accurate Causal Inference from Time Series Data Yuxiao Huang and Samantha Kleinberg Stevens Institute of Technology Hoboken, NJ {yuxiao.huang, samantha.kleinberg}@stevens.edu Abstract Causal inference
Fast and Accurate Causal Inference from Time Series Data Yuxiao Huang and Samantha Kleinberg Stevens Institute of Technology Hoboken, NJ {yuxiao.huang, samantha.kleinberg}@stevens.edu Abstract Causal inference
Conditional Random Field
 Introduction Linear-Chain General Specific Implementations Conclusions Corso di Elaborazione del Linguaggio Naturale Pisa, May, 2011 Introduction Linear-Chain General Specific Implementations Conclusions
Introduction Linear-Chain General Specific Implementations Conclusions Corso di Elaborazione del Linguaggio Naturale Pisa, May, 2011 Introduction Linear-Chain General Specific Implementations Conclusions
Variational Autoencoders
 Variational Autoencoders Recap: Story so far A classification MLP actually comprises two components A feature extraction network that converts the inputs into linearly separable features Or nearly linearly
Variational Autoencoders Recap: Story so far A classification MLP actually comprises two components A feature extraction network that converts the inputs into linearly separable features Or nearly linearly
Lecture : Probabilistic Machine Learning
 Lecture : Probabilistic Machine Learning Riashat Islam Reasoning and Learning Lab McGill University September 11, 2018 ML : Many Methods with Many Links Modelling Views of Machine Learning Machine Learning
Lecture : Probabilistic Machine Learning Riashat Islam Reasoning and Learning Lab McGill University September 11, 2018 ML : Many Methods with Many Links Modelling Views of Machine Learning Machine Learning
Log-Linear Models, MEMMs, and CRFs
 Log-Linear Models, MEMMs, and CRFs Michael Collins 1 Notation Throughout this note I ll use underline to denote vectors. For example, w R d will be a vector with components w 1, w 2,... w d. We use expx
Log-Linear Models, MEMMs, and CRFs Michael Collins 1 Notation Throughout this note I ll use underline to denote vectors. For example, w R d will be a vector with components w 1, w 2,... w d. We use expx
Performance Comparison of K-Means and Expectation Maximization with Gaussian Mixture Models for Clustering EE6540 Final Project
 Performance Comparison of K-Means and Expectation Maximization with Gaussian Mixture Models for Clustering EE6540 Final Project Devin Cornell & Sushruth Sastry May 2015 1 Abstract In this article, we explore
Performance Comparison of K-Means and Expectation Maximization with Gaussian Mixture Models for Clustering EE6540 Final Project Devin Cornell & Sushruth Sastry May 2015 1 Abstract In this article, we explore
Extensions of Bayesian Networks. Outline. Bayesian Network. Reasoning under Uncertainty. Features of Bayesian Networks.
 Extensions of Bayesian Networks Outline Ethan Howe, James Lenfestey, Tom Temple Intro to Dynamic Bayesian Nets (Tom Exact inference in DBNs with demo (Ethan Approximate inference and learning (Tom Probabilistic
Extensions of Bayesian Networks Outline Ethan Howe, James Lenfestey, Tom Temple Intro to Dynamic Bayesian Nets (Tom Exact inference in DBNs with demo (Ethan Approximate inference and learning (Tom Probabilistic
Recent Advances in Bayesian Inference Techniques
 Recent Advances in Bayesian Inference Techniques Christopher M. Bishop Microsoft Research, Cambridge, U.K. research.microsoft.com/~cmbishop SIAM Conference on Data Mining, April 2004 Abstract Bayesian
Recent Advances in Bayesian Inference Techniques Christopher M. Bishop Microsoft Research, Cambridge, U.K. research.microsoft.com/~cmbishop SIAM Conference on Data Mining, April 2004 Abstract Bayesian
Hidden Markov models
 Hidden Markov models Charles Elkan November 26, 2012 Important: These lecture notes are based on notes written by Lawrence Saul. Also, these typeset notes lack illustrations. See the classroom lectures
Hidden Markov models Charles Elkan November 26, 2012 Important: These lecture notes are based on notes written by Lawrence Saul. Also, these typeset notes lack illustrations. See the classroom lectures
NIH Public Access Author Manuscript IEEE/ACM Trans Comput Biol Bioinform. Author manuscript; available in PMC 2013 May 01.
 NIH Public Access Author Manuscript Published in final edited form as: IEEE/ACM Trans Comput Biol Bioinform. 2012 ; 9(3): 809 817. doi:10.1109/tcbb.2012.26. Faster Mass Spectrometry-based Protein Inference:
NIH Public Access Author Manuscript Published in final edited form as: IEEE/ACM Trans Comput Biol Bioinform. 2012 ; 9(3): 809 817. doi:10.1109/tcbb.2012.26. Faster Mass Spectrometry-based Protein Inference:
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, etworks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, etworks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Data Mining in Bioinformatics HMM
 Data Mining in Bioinformatics HMM Microarray Problem: Major Objective n Major Objective: Discover a comprehensive theory of life s organization at the molecular level 2 1 Data Mining in Bioinformatics
Data Mining in Bioinformatics HMM Microarray Problem: Major Objective n Major Objective: Discover a comprehensive theory of life s organization at the molecular level 2 1 Data Mining in Bioinformatics
hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference
 CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
Introduction to Machine Learning Midterm, Tues April 8
 Introduction to Machine Learning 10-701 Midterm, Tues April 8 [1 point] Name: Andrew ID: Instructions: You are allowed a (two-sided) sheet of notes. Exam ends at 2:45pm Take a deep breath and don t spend
Introduction to Machine Learning 10-701 Midterm, Tues April 8 [1 point] Name: Andrew ID: Instructions: You are allowed a (two-sided) sheet of notes. Exam ends at 2:45pm Take a deep breath and don t spend
Computational Methods for Mass Spectrometry Proteomics
 Computational Methods for Mass Spectrometry Proteomics Eidhammer, Ingvar ISBN-13: 9780470512975 Table of Contents Preface. Acknowledgements. 1 Protein, Proteome, and Proteomics. 1.1 Primary goals for studying
Computational Methods for Mass Spectrometry Proteomics Eidhammer, Ingvar ISBN-13: 9780470512975 Table of Contents Preface. Acknowledgements. 1 Protein, Proteome, and Proteomics. 1.1 Primary goals for studying
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra. Andrew Keller
 PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Outline Need to validate peptide assignments to MS/MS spectra Statistical approach to validation Running PeptideProphet
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Outline Need to validate peptide assignments to MS/MS spectra Statistical approach to validation Running PeptideProphet
Augmented Statistical Models for Speech Recognition
 Augmented Statistical Models for Speech Recognition Mark Gales & Martin Layton 31 August 2005 Trajectory Models For Speech Processing Workshop Overview Dependency Modelling in Speech Recognition: latent
Augmented Statistical Models for Speech Recognition Mark Gales & Martin Layton 31 August 2005 Trajectory Models For Speech Processing Workshop Overview Dependency Modelling in Speech Recognition: latent
Gaussian processes. Chuong B. Do (updated by Honglak Lee) November 22, 2008
 Gaussian processes Chuong B Do (updated by Honglak Lee) November 22, 2008 Many of the classical machine learning algorithms that we talked about during the first half of this course fit the following pattern:
Gaussian processes Chuong B Do (updated by Honglak Lee) November 22, 2008 Many of the classical machine learning algorithms that we talked about during the first half of this course fit the following pattern:
Recurrent Latent Variable Networks for Session-Based Recommendation
 Recurrent Latent Variable Networks for Session-Based Recommendation Panayiotis Christodoulou Cyprus University of Technology paa.christodoulou@edu.cut.ac.cy 27/8/2017 Panayiotis Christodoulou (C.U.T.)
Recurrent Latent Variable Networks for Session-Based Recommendation Panayiotis Christodoulou Cyprus University of Technology paa.christodoulou@edu.cut.ac.cy 27/8/2017 Panayiotis Christodoulou (C.U.T.)
Undirected Graphical Models
 Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Properties Properties 3 Generative vs. Conditional
Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Properties Properties 3 Generative vs. Conditional
Clustering K-means. Machine Learning CSE546. Sham Kakade University of Washington. November 15, Review: PCA Start: unsupervised learning
 Clustering K-means Machine Learning CSE546 Sham Kakade University of Washington November 15, 2016 1 Announcements: Project Milestones due date passed. HW3 due on Monday It ll be collaborative HW2 grades
Clustering K-means Machine Learning CSE546 Sham Kakade University of Washington November 15, 2016 1 Announcements: Project Milestones due date passed. HW3 due on Monday It ll be collaborative HW2 grades
General comments, TODOs, etc.
 7 9 7 9 7 9 7 9 7 9 General comments, TODOs, etc. -Think up a catchy title. -Decide which references to anonymize to maintain double-blindness. -Write the abstract -Fill in remaining [CITE], which are
7 9 7 9 7 9 7 9 7 9 General comments, TODOs, etc. -Think up a catchy title. -Decide which references to anonymize to maintain double-blindness. -Write the abstract -Fill in remaining [CITE], which are
Pattern Recognition and Machine Learning
 Christopher M. Bishop Pattern Recognition and Machine Learning ÖSpri inger Contents Preface Mathematical notation Contents vii xi xiii 1 Introduction 1 1.1 Example: Polynomial Curve Fitting 4 1.2 Probability
Christopher M. Bishop Pattern Recognition and Machine Learning ÖSpri inger Contents Preface Mathematical notation Contents vii xi xiii 1 Introduction 1 1.1 Example: Polynomial Curve Fitting 4 1.2 Probability
Learning Gaussian Process Models from Uncertain Data
 Learning Gaussian Process Models from Uncertain Data Patrick Dallaire, Camille Besse, and Brahim Chaib-draa DAMAS Laboratory, Computer Science & Software Engineering Department, Laval University, Canada
Learning Gaussian Process Models from Uncertain Data Patrick Dallaire, Camille Besse, and Brahim Chaib-draa DAMAS Laboratory, Computer Science & Software Engineering Department, Laval University, Canada
Linear Dynamical Systems
 Linear Dynamical Systems Sargur N. srihari@cedar.buffalo.edu Machine Learning Course: http://www.cedar.buffalo.edu/~srihari/cse574/index.html Two Models Described by Same Graph Latent variables Observations
Linear Dynamical Systems Sargur N. srihari@cedar.buffalo.edu Machine Learning Course: http://www.cedar.buffalo.edu/~srihari/cse574/index.html Two Models Described by Same Graph Latent variables Observations
Key questions of proteomics. Bioinformatics 2. Proteomics. Foundation of proteomics. What proteins are there? Protein digestion
 s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture 2 roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture 2 roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
Lecture 21: Spectral Learning for Graphical Models
 10-708: Probabilistic Graphical Models 10-708, Spring 2016 Lecture 21: Spectral Learning for Graphical Models Lecturer: Eric P. Xing Scribes: Maruan Al-Shedivat, Wei-Cheng Chang, Frederick Liu 1 Motivation
10-708: Probabilistic Graphical Models 10-708, Spring 2016 Lecture 21: Spectral Learning for Graphical Models Lecturer: Eric P. Xing Scribes: Maruan Al-Shedivat, Wei-Cheng Chang, Frederick Liu 1 Motivation
Expectation Maximization (EM)
 Expectation Maximization (EM) The EM algorithm is used to train models involving latent variables using training data in which the latent variables are not observed (unlabeled data). This is to be contrasted
Expectation Maximization (EM) The EM algorithm is used to train models involving latent variables using training data in which the latent variables are not observed (unlabeled data). This is to be contrasted
MS-MS Analysis Programs
 MS-MS Analysis Programs Basic Process Genome - Gives AA sequences of proteins Use this to predict spectra Compare data to prediction Determine degree of correctness Make assignment Did we see the protein?
MS-MS Analysis Programs Basic Process Genome - Gives AA sequences of proteins Use this to predict spectra Compare data to prediction Determine degree of correctness Make assignment Did we see the protein?
Today s Lecture: HMMs
 Today s Lecture: HMMs Definitions Examples Probability calculations WDAG Dynamic programming algorithms: Forward Viterbi Parameter estimation Viterbi training 1 Hidden Markov Models Probability models
Today s Lecture: HMMs Definitions Examples Probability calculations WDAG Dynamic programming algorithms: Forward Viterbi Parameter estimation Viterbi training 1 Hidden Markov Models Probability models
Final Examination CS 540-2: Introduction to Artificial Intelligence
 Final Examination CS 540-2: Introduction to Artificial Intelligence May 7, 2017 LAST NAME: SOLUTIONS FIRST NAME: Problem Score Max Score 1 14 2 10 3 6 4 10 5 11 6 9 7 8 9 10 8 12 12 8 Total 100 1 of 11
Final Examination CS 540-2: Introduction to Artificial Intelligence May 7, 2017 LAST NAME: SOLUTIONS FIRST NAME: Problem Score Max Score 1 14 2 10 3 6 4 10 5 11 6 9 7 8 9 10 8 12 12 8 Total 100 1 of 11
Design and Implementation of Speech Recognition Systems
 Design and Implementation of Speech Recognition Systems Spring 2013 Class 7: Templates to HMMs 13 Feb 2013 1 Recap Thus far, we have looked at dynamic programming for string matching, And derived DTW from
Design and Implementation of Speech Recognition Systems Spring 2013 Class 7: Templates to HMMs 13 Feb 2013 1 Recap Thus far, we have looked at dynamic programming for string matching, And derived DTW from
Using Expectation-Maximization for Reinforcement Learning
 NOTE Communicated by Andrew Barto and Michael Jordan Using Expectation-Maximization for Reinforcement Learning Peter Dayan Department of Brain and Cognitive Sciences, Center for Biological and Computational
NOTE Communicated by Andrew Barto and Michael Jordan Using Expectation-Maximization for Reinforcement Learning Peter Dayan Department of Brain and Cognitive Sciences, Center for Biological and Computational
Machine Learning and Bayesian Inference. Unsupervised learning. Can we find regularity in data without the aid of labels?
 Machine Learning and Bayesian Inference Dr Sean Holden Computer Laboratory, Room FC6 Telephone extension 6372 Email: sbh11@cl.cam.ac.uk www.cl.cam.ac.uk/ sbh11/ Unsupervised learning Can we find regularity
Machine Learning and Bayesian Inference Dr Sean Holden Computer Laboratory, Room FC6 Telephone extension 6372 Email: sbh11@cl.cam.ac.uk www.cl.cam.ac.uk/ sbh11/ Unsupervised learning Can we find regularity
Learning in Bayesian Networks
 Learning in Bayesian Networks Florian Markowetz Max-Planck-Institute for Molecular Genetics Computational Molecular Biology Berlin Berlin: 20.06.2002 1 Overview 1. Bayesian Networks Stochastic Networks
Learning in Bayesian Networks Florian Markowetz Max-Planck-Institute for Molecular Genetics Computational Molecular Biology Berlin Berlin: 20.06.2002 1 Overview 1. Bayesian Networks Stochastic Networks
Intelligent Systems (AI-2)
 Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 24, 2016 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 24, 2016 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
13: Variational inference II
 10-708: Probabilistic Graphical Models, Spring 2015 13: Variational inference II Lecturer: Eric P. Xing Scribes: Ronghuo Zheng, Zhiting Hu, Yuntian Deng 1 Introduction We started to talk about variational
10-708: Probabilistic Graphical Models, Spring 2015 13: Variational inference II Lecturer: Eric P. Xing Scribes: Ronghuo Zheng, Zhiting Hu, Yuntian Deng 1 Introduction We started to talk about variational
Introduction to Machine Learning
 Introduction to Machine Learning Brown University CSCI 1950-F, Spring 2012 Prof. Erik Sudderth Lecture 25: Markov Chain Monte Carlo (MCMC) Course Review and Advanced Topics Many figures courtesy Kevin
Introduction to Machine Learning Brown University CSCI 1950-F, Spring 2012 Prof. Erik Sudderth Lecture 25: Markov Chain Monte Carlo (MCMC) Course Review and Advanced Topics Many figures courtesy Kevin
1 Kernel methods & optimization
 Machine Learning Class Notes 9-26-13 Prof. David Sontag 1 Kernel methods & optimization One eample of a kernel that is frequently used in practice and which allows for highly non-linear discriminant functions
Machine Learning Class Notes 9-26-13 Prof. David Sontag 1 Kernel methods & optimization One eample of a kernel that is frequently used in practice and which allows for highly non-linear discriminant functions
Weighted Finite-State Transducers in Computational Biology
 Weighted Finite-State Transducers in Computational Biology Mehryar Mohri Courant Institute of Mathematical Sciences mohri@cims.nyu.edu Joint work with Corinna Cortes (Google Research). 1 This Tutorial
Weighted Finite-State Transducers in Computational Biology Mehryar Mohri Courant Institute of Mathematical Sciences mohri@cims.nyu.edu Joint work with Corinna Cortes (Google Research). 1 This Tutorial
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra
 PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Day 2 October 17, 2006 Andrew Keller Rosetta Bioinformatics, Seattle Outline Need to validate peptide assignments to MS/MS
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Day 2 October 17, 2006 Andrew Keller Rosetta Bioinformatics, Seattle Outline Need to validate peptide assignments to MS/MS
Chapter 4 Dynamic Bayesian Networks Fall Jin Gu, Michael Zhang
 Chapter 4 Dynamic Bayesian Networks 2016 Fall Jin Gu, Michael Zhang Reviews: BN Representation Basic steps for BN representations Define variables Define the preliminary relations between variables Check
Chapter 4 Dynamic Bayesian Networks 2016 Fall Jin Gu, Michael Zhang Reviews: BN Representation Basic steps for BN representations Define variables Define the preliminary relations between variables Check
Overview - MS Proteomics in One Slide. MS masses of peptides. MS/MS fragments of a peptide. Results! Match to sequence database
 Overview - MS Proteomics in One Slide Obtain protein Digest into peptides Acquire spectra in mass spectrometer MS masses of peptides MS/MS fragments of a peptide Results! Match to sequence database 2 But
Overview - MS Proteomics in One Slide Obtain protein Digest into peptides Acquire spectra in mass spectrometer MS masses of peptides MS/MS fragments of a peptide Results! Match to sequence database 2 But
Intelligent Systems (AI-2)
 Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 23, 2015 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 23, 2015 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Math 350: An exploration of HMMs through doodles.
 Math 350: An exploration of HMMs through doodles. Joshua Little (407673) 19 December 2012 1 Background 1.1 Hidden Markov models. Markov chains (MCs) work well for modelling discrete-time processes, or
Math 350: An exploration of HMMs through doodles. Joshua Little (407673) 19 December 2012 1 Background 1.1 Hidden Markov models. Markov chains (MCs) work well for modelling discrete-time processes, or
Brief Introduction of Machine Learning Techniques for Content Analysis
 1 Brief Introduction of Machine Learning Techniques for Content Analysis Wei-Ta Chu 2008/11/20 Outline 2 Overview Gaussian Mixture Model (GMM) Hidden Markov Model (HMM) Support Vector Machine (SVM) Overview
1 Brief Introduction of Machine Learning Techniques for Content Analysis Wei-Ta Chu 2008/11/20 Outline 2 Overview Gaussian Mixture Model (GMM) Hidden Markov Model (HMM) Support Vector Machine (SVM) Overview
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Comparison of Modern Stochastic Optimization Algorithms
 Comparison of Modern Stochastic Optimization Algorithms George Papamakarios December 214 Abstract Gradient-based optimization methods are popular in machine learning applications. In large-scale problems,
Comparison of Modern Stochastic Optimization Algorithms George Papamakarios December 214 Abstract Gradient-based optimization methods are popular in machine learning applications. In large-scale problems,
Machine Learning for Signal Processing Bayes Classification and Regression
 Machine Learning for Signal Processing Bayes Classification and Regression Instructor: Bhiksha Raj 11755/18797 1 Recap: KNN A very effective and simple way of performing classification Simple model: For
Machine Learning for Signal Processing Bayes Classification and Regression Instructor: Bhiksha Raj 11755/18797 1 Recap: KNN A very effective and simple way of performing classification Simple model: For
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
CSEP 573: Artificial Intelligence
 CSEP 573: Artificial Intelligence Hidden Markov Models Luke Zettlemoyer Many slides over the course adapted from either Dan Klein, Stuart Russell, Andrew Moore, Ali Farhadi, or Dan Weld 1 Outline Probabilistic
CSEP 573: Artificial Intelligence Hidden Markov Models Luke Zettlemoyer Many slides over the course adapted from either Dan Klein, Stuart Russell, Andrew Moore, Ali Farhadi, or Dan Weld 1 Outline Probabilistic
Machine Learning Summer School
 Machine Learning Summer School Lecture 3: Learning parameters and structure Zoubin Ghahramani zoubin@eng.cam.ac.uk http://learning.eng.cam.ac.uk/zoubin/ Department of Engineering University of Cambridge,
Machine Learning Summer School Lecture 3: Learning parameters and structure Zoubin Ghahramani zoubin@eng.cam.ac.uk http://learning.eng.cam.ac.uk/zoubin/ Department of Engineering University of Cambridge,
Supplementary Material for: Clustering Millions of Tandem Mass Spectra
 Supplementary Material for: Clustering Millions of Tandem Mass Spectra Ari M. Frank 1 Nuno Bandeira 1 Zhouxin Shen 2 Stephen Tanner 3 Steven P. Briggs 2 Richard D. Smith 4 Pavel A. Pevzner 1 October 4,
Supplementary Material for: Clustering Millions of Tandem Mass Spectra Ari M. Frank 1 Nuno Bandeira 1 Zhouxin Shen 2 Stephen Tanner 3 Steven P. Briggs 2 Richard D. Smith 4 Pavel A. Pevzner 1 October 4,
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistical Sciences! rsalakhu@cs.toronto.edu! h0p://www.cs.utoronto.ca/~rsalakhu/ Lecture 7 Approximate
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistical Sciences! rsalakhu@cs.toronto.edu! h0p://www.cs.utoronto.ca/~rsalakhu/ Lecture 7 Approximate
p(d θ ) l(θ ) 1.2 x x x
 p(d θ ).2 x 0-7 0.8 x 0-7 0.4 x 0-7 l(θ ) -20-40 -60-80 -00 2 3 4 5 6 7 θ ˆ 2 3 4 5 6 7 θ ˆ 2 3 4 5 6 7 θ θ x FIGURE 3.. The top graph shows several training points in one dimension, known or assumed to
p(d θ ).2 x 0-7 0.8 x 0-7 0.4 x 0-7 l(θ ) -20-40 -60-80 -00 2 3 4 5 6 7 θ ˆ 2 3 4 5 6 7 θ ˆ 2 3 4 5 6 7 θ θ x FIGURE 3.. The top graph shows several training points in one dimension, known or assumed to
Dynamic Time-Alignment Kernel in Support Vector Machine
 Dynamic Time-Alignment Kernel in Support Vector Machine Hiroshi Shimodaira School of Information Science, Japan Advanced Institute of Science and Technology sim@jaist.ac.jp Mitsuru Nakai School of Information
Dynamic Time-Alignment Kernel in Support Vector Machine Hiroshi Shimodaira School of Information Science, Japan Advanced Institute of Science and Technology sim@jaist.ac.jp Mitsuru Nakai School of Information
Clustering K-means. Clustering images. Machine Learning CSE546 Carlos Guestrin University of Washington. November 4, 2014.
 Clustering K-means Machine Learning CSE546 Carlos Guestrin University of Washington November 4, 2014 1 Clustering images Set of Images [Goldberger et al.] 2 1 K-means Randomly initialize k centers µ (0)
Clustering K-means Machine Learning CSE546 Carlos Guestrin University of Washington November 4, 2014 1 Clustering images Set of Images [Goldberger et al.] 2 1 K-means Randomly initialize k centers µ (0)
STA414/2104. Lecture 11: Gaussian Processes. Department of Statistics
 STA414/2104 Lecture 11: Gaussian Processes Department of Statistics www.utstat.utoronto.ca Delivered by Mark Ebden with thanks to Russ Salakhutdinov Outline Gaussian Processes Exam review Course evaluations
STA414/2104 Lecture 11: Gaussian Processes Department of Statistics www.utstat.utoronto.ca Delivered by Mark Ebden with thanks to Russ Salakhutdinov Outline Gaussian Processes Exam review Course evaluations
Sum-Product Networks. STAT946 Deep Learning Guest Lecture by Pascal Poupart University of Waterloo October 17, 2017
 Sum-Product Networks STAT946 Deep Learning Guest Lecture by Pascal Poupart University of Waterloo October 17, 2017 Introduction Outline What is a Sum-Product Network? Inference Applications In more depth
Sum-Product Networks STAT946 Deep Learning Guest Lecture by Pascal Poupart University of Waterloo October 17, 2017 Introduction Outline What is a Sum-Product Network? Inference Applications In more depth
Lecture 3: Pattern Classification
 EE E6820: Speech & Audio Processing & Recognition Lecture 3: Pattern Classification 1 2 3 4 5 The problem of classification Linear and nonlinear classifiers Probabilistic classification Gaussians, mixtures
EE E6820: Speech & Audio Processing & Recognition Lecture 3: Pattern Classification 1 2 3 4 5 The problem of classification Linear and nonlinear classifiers Probabilistic classification Gaussians, mixtures
Lecture 6: Graphical Models: Learning
 Lecture 6: Graphical Models: Learning 4F13: Machine Learning Zoubin Ghahramani and Carl Edward Rasmussen Department of Engineering, University of Cambridge February 3rd, 2010 Ghahramani & Rasmussen (CUED)
Lecture 6: Graphical Models: Learning 4F13: Machine Learning Zoubin Ghahramani and Carl Edward Rasmussen Department of Engineering, University of Cambridge February 3rd, 2010 Ghahramani & Rasmussen (CUED)
Final Exam, Machine Learning, Spring 2009
 Name: Andrew ID: Final Exam, 10701 Machine Learning, Spring 2009 - The exam is open-book, open-notes, no electronics other than calculators. - The maximum possible score on this exam is 100. You have 3
Name: Andrew ID: Final Exam, 10701 Machine Learning, Spring 2009 - The exam is open-book, open-notes, no electronics other than calculators. - The maximum possible score on this exam is 100. You have 3
Outline of Today s Lecture
 University of Washington Department of Electrical Engineering Computer Speech Processing EE516 Winter 2005 Jeff A. Bilmes Lecture 12 Slides Feb 23 rd, 2005 Outline of Today s
University of Washington Department of Electrical Engineering Computer Speech Processing EE516 Winter 2005 Jeff A. Bilmes Lecture 12 Slides Feb 23 rd, 2005 Outline of Today s
CSCI-567: Machine Learning (Spring 2019)
 CSCI-567: Machine Learning (Spring 2019) Prof. Victor Adamchik U of Southern California Mar. 19, 2019 March 19, 2019 1 / 43 Administration March 19, 2019 2 / 43 Administration TA3 is due this week March
CSCI-567: Machine Learning (Spring 2019) Prof. Victor Adamchik U of Southern California Mar. 19, 2019 March 19, 2019 1 / 43 Administration March 19, 2019 2 / 43 Administration TA3 is due this week March
Probabilistic Graphical Models: MRFs and CRFs. CSE628: Natural Language Processing Guest Lecturer: Veselin Stoyanov
 Probabilistic Graphical Models: MRFs and CRFs CSE628: Natural Language Processing Guest Lecturer: Veselin Stoyanov Why PGMs? PGMs can model joint probabilities of many events. many techniques commonly
Probabilistic Graphical Models: MRFs and CRFs CSE628: Natural Language Processing Guest Lecturer: Veselin Stoyanov Why PGMs? PGMs can model joint probabilities of many events. many techniques commonly
Expectation Maximization (EM)
 Expectation Maximization (EM) The Expectation Maximization (EM) algorithm is one approach to unsupervised, semi-supervised, or lightly supervised learning. In this kind of learning either no labels are
Expectation Maximization (EM) The Expectation Maximization (EM) algorithm is one approach to unsupervised, semi-supervised, or lightly supervised learning. In this kind of learning either no labels are
Algorithmisches Lernen/Machine Learning
 Algorithmisches Lernen/Machine Learning Part 1: Stefan Wermter Introduction Connectionist Learning (e.g. Neural Networks) Decision-Trees, Genetic Algorithms Part 2: Norman Hendrich Support-Vector Machines
Algorithmisches Lernen/Machine Learning Part 1: Stefan Wermter Introduction Connectionist Learning (e.g. Neural Networks) Decision-Trees, Genetic Algorithms Part 2: Norman Hendrich Support-Vector Machines
Hidden Markov Models in Language Processing
 Hidden Markov Models in Language Processing Dustin Hillard Lecture notes courtesy of Prof. Mari Ostendorf Outline Review of Markov models What is an HMM? Examples General idea of hidden variables: implications
Hidden Markov Models in Language Processing Dustin Hillard Lecture notes courtesy of Prof. Mari Ostendorf Outline Review of Markov models What is an HMM? Examples General idea of hidden variables: implications
Efficient Marginalization to Compute Protein Posterior Probabilities from Shotgun Mass Spectrometry Data
 Efficient Marginalization to Compute Protein Posterior Probabilities from Shotgun Mass Spectrometry Data Oliver Serang Department of Genome Sciences, University of Washington, Seattle, Washington Michael
Efficient Marginalization to Compute Protein Posterior Probabilities from Shotgun Mass Spectrometry Data Oliver Serang Department of Genome Sciences, University of Washington, Seattle, Washington Michael
STATS 306B: Unsupervised Learning Spring Lecture 5 April 14
 STATS 306B: Unsupervised Learning Spring 2014 Lecture 5 April 14 Lecturer: Lester Mackey Scribe: Brian Do and Robin Jia 5.1 Discrete Hidden Markov Models 5.1.1 Recap In the last lecture, we introduced
STATS 306B: Unsupervised Learning Spring 2014 Lecture 5 April 14 Lecturer: Lester Mackey Scribe: Brian Do and Robin Jia 5.1 Discrete Hidden Markov Models 5.1.1 Recap In the last lecture, we introduced
Lecture 4: Hidden Markov Models: An Introduction to Dynamic Decision Making. November 11, 2010
 Hidden Lecture 4: Hidden : An Introduction to Dynamic Decision Making November 11, 2010 Special Meeting 1/26 Markov Model Hidden When a dynamical system is probabilistic it may be determined by the transition
Hidden Lecture 4: Hidden : An Introduction to Dynamic Decision Making November 11, 2010 Special Meeting 1/26 Markov Model Hidden When a dynamical system is probabilistic it may be determined by the transition
Approximating the Partition Function by Deleting and then Correcting for Model Edges (Extended Abstract)
 Approximating the Partition Function by Deleting and then Correcting for Model Edges (Extended Abstract) Arthur Choi and Adnan Darwiche Computer Science Department University of California, Los Angeles
Approximating the Partition Function by Deleting and then Correcting for Model Edges (Extended Abstract) Arthur Choi and Adnan Darwiche Computer Science Department University of California, Los Angeles
Learning from Sequential and Time-Series Data
 Learning from Sequential and Time-Series Data Sridhar Mahadevan mahadeva@cs.umass.edu University of Massachusetts Sridhar Mahadevan: CMPSCI 689 p. 1/? Sequential and Time-Series Data Many real-world applications
Learning from Sequential and Time-Series Data Sridhar Mahadevan mahadeva@cs.umass.edu University of Massachusetts Sridhar Mahadevan: CMPSCI 689 p. 1/? Sequential and Time-Series Data Many real-world applications
STATISTICAL METHODS FOR THE ANALYSIS OF MASS SPECTROMETRY- BASED PROTEOMICS DATA. A Dissertation XUAN WANG
 STATISTICAL METHODS FOR THE ANALYSIS OF MASS SPECTROMETRY- BASED PROTEOMICS DATA A Dissertation by XUAN WANG Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of
STATISTICAL METHODS FOR THE ANALYSIS OF MASS SPECTROMETRY- BASED PROTEOMICS DATA A Dissertation by XUAN WANG Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of
MaSS-Simulator: A highly configurable MS/MS simulator for generating test datasets for big data algorithms.
 MaSS-Simulator: A highly configurable MS/MS simulator for generating test datasets for big data algorithms. Muaaz Gul Awan 1 and Fahad Saeed 1 1 Department of Computer Science, Western Michigan University,
MaSS-Simulator: A highly configurable MS/MS simulator for generating test datasets for big data algorithms. Muaaz Gul Awan 1 and Fahad Saeed 1 1 Department of Computer Science, Western Michigan University,
Spectrum-to-Spectrum Searching Using a. Proteome-wide Spectral Library
 MCP Papers in Press. Published on April 30, 2011 as Manuscript M111.007666 Spectrum-to-Spectrum Searching Using a Proteome-wide Spectral Library Chia-Yu Yen, Stephane Houel, Natalie G. Ahn, and William
MCP Papers in Press. Published on April 30, 2011 as Manuscript M111.007666 Spectrum-to-Spectrum Searching Using a Proteome-wide Spectral Library Chia-Yu Yen, Stephane Houel, Natalie G. Ahn, and William
