mixmod Statistical Documentation
|
|
|
- Reginald Greene
- 5 years ago
- Views:
Transcription
1 mixmod Statistical Documentation February 10, 2016 Contents 1 Introduction 3 2 Mixture model Density estimation from a mixture model Clustering with mixture model The mixture approach The classification approach Discriminant Analysis Algorithms in mixmod EM algorithm SEM algorithm CEM algorithm Using mixmod Stopping rules Initialization strategies Criteria to select a model The Bayesian Information Criterion (BIC) The Integrated Completed Likelihood (ICL) The Normalized Entropy Criterion (NEC) The cross-validation criterion (CV) The double cross-validation criterion (DCV) Partial labeling of individuals Weighting the units
2 5 The Gaussian mixture model Definition Fourteen Gaussian models Eigenvalue decomposition of variance matrices The general family The diagonal family The spherical family M step for each of the 14 models The general family The diagonal family The spherical family Mixture of Factor Analyzers The general high dimensional model Sub-models of model [a kj b k D k δ k ] The MAP step Estimation of the model parameters The multinomial mixture model Definition Five multinomial models M step for each of the five models The Gaussian-multinomial mixture model Definition Thirty combined Gaussian-multinomial models M step for the 30 models Selected references 38 2
3 1 Introduction Finite mixture models is a powerful tool for density estimation, cluster analysis and discriminant analysis. mixmod is a software for mixture modelling which considered those three different aspects of mixtures and gives great place to the multivariate context. In its present version, mixmod is dealing with multivariate Gaussian mixture models for quantitative data, with multivariate multinomial mixture models for categorical data and with combined multivariate Gaussian-multinomial mixture models for mixed quantitative and categorical data. Basing cluster or discriminant analysis on Gaussian mixture models is a classical and powerful approach since Gaussian models are useful both for understanding and suggesting powerful clustering criteria. One of the originality of mixmod is to consider a parameterization of the variance matrix of a cluster through its eigenvalue decomposition leading to many meaningful models for clustering and classification. In the same manner, different more or less parsimonious parameterizations are entering in the multinomial mixture models. Combining both the mixed case quantitative/categorical beneficits from both advantages. This documentation is organized as follows. In Section 2, the general setting of finite mixture modelling is sketched. In Section 3, the different available algorithms in mixmod for estimating mixture parameters are presented. In Section 4, the possible strategies for using mixmod algorithms and for initiating them are described. Moreover criteria to select a model are presented. Section 5 is devoted to the detailed presentation of the Gaussian mixture models considered in mixmod and to a mixture of factor analyser models useful to treat high dimensional supervised classification problems. Section 6 is devoted to the detailed presentation of multivariate multinomial mixture models. Section 7 is devoted to the detailed presentation of multivariate combined Gaussian-multivariate mixture models. 2 Mixture model Let x = {x 1,...,x n } be n independent vectors in X such that each x i arises from a probability distribution with density f(x i θ) = K p k h(x i λ k ) (1) 3
4 where the p k s are the mixing proportions (0 < p k < 1 for all k = 1,...,K and p p K = 1), h( λ k ) denotes a d-dimensional distribution parameterized by λ k. As we will see in Section 5, h is for instance the density of a Gaussian distribution with X = R d, mean µ and variance matrix Σ and thus, λ = (µ,σ). It is worth noting that for a mixture distribution, a sample of indicator vectors or labels z = {z 1,...,z n }, with z i = (z i1,...,z ik ), z ik = 1 or 0, according to the fact that x i is arising from the kth mixture component or not, is associated to the observed data x. The sample z can be known in which case we are in a discriminant analysis context where the problem is essentially to predict an indicator vector z n+1 from a new observed data vector x n+1. But the sample z can be unknown in which case we are in a density estimation context or cluster analysis context if the estimation of the z i s are of primary interest. In each case, the vector parameter to be estimated is θ = (p 1,...,p K,λ 1,...,λ K ). 2.1 Density estimation from a mixture model Mixture modelling can be regarded as a flexible way to represent a probability density function, and thus providing a semi parametric tool for density estimation. When the labels z are unknown, maximum likelihood estimation of mixture models can be performed in mixmod via the EM algorithm of Dempster, Laird and Rubin (1977) or by a stochastic version of EM called SEM(seeMcLachlanandPeel, 2000). Ineachcase, theparameterθ ischosen to maximize the observed log-likelihood ( n K ) L(θ x 1,...,x n ) = ln p k h(x i,λ k ). (2) i=1 2.2 Clustering with mixture model Cluster analysis is concerned with discovering a group structure in a n by d data matrix x = {x 1,...,x n } where x i is an individual in X. Consequently, the result provided by clustering is typically a partition of x into K groups defined with the labels z = { z 1,..., z n }, with z i = ( z i1,..., z ik ), z ik = 1 or 0 according to x i is assigned to the kth group or not. Many authors have considered non hierarchical clustering methods in which a mixture of distributions is used as a statistical model. In this con- 4
5 text, two commonly used maximum likelihood (m.l.) approaches have been proposed: the mixture approach and the classification approach. Loosely speaking, the mixture approach is aimed to maximize the likelihood over the mixture parameters, whereas the classification approach is aimed to maximize the likelihood over the mixture parameters and over the mixture component labels The mixture approach In this approach, a partition of the data can directly be derived from the m.l. estimates ˆθ of the mixture parameters obtained, for instance, by the EM or the SEM algorithm described hereafter, by assigning each x i to the component providing the largest conditional probability that x i arises from it using a MAP (Maximum A Posteriori) principle. Denoting z i the label of x i, the MAP principle is as follows { 1 if k = argmaxl=1...,k t z ik = l (x i ˆθ) 0 if not (3) where t k (x i ˆθ) = ˆp k h(x i ˆλ k ) K l=1 ˆp lh(x i ˆλ l ) The classification approach The second approach available in mixmod is the classification approach. In this approach, the indicator vectors z = {z 1,...,z n }, identifying the mixture component origin, are treated as unknown parameters. The Classification Maximum Likelihood (c.m.l.) method is used to estimate both the parameters θ and z. The classification likelihood criterion is defined by CL(θ,z 1,...,z n x 1,...,x n ) = n K z ik ln[p k h(x i λ k )]. (4) i=1 The CL criterion can be maximized by making use of a classification version of the EM algorithm, the so-called CEM algorithm (Celeux and Govaert 1992) which includes a classification step (C-step) between the E and M steps. 5
6 2.3 Discriminant Analysis When the labels z are known, we are concerned with discriminant analysis: in discriminant analysis, data are composed by n observations x = (x 1,...,x n } (x i R d ) and a partition of x into K groups defined with the labels z = {z 1,...,z n }. The aim is to estimate the group z n+1 of any new individual x n+1 of R d with unknown label. Inthiscontext, thencouples(x i,z i ),..., (x n,z n )arerealizationsofnidentically and independently distributed random vectors (X i,z i ),...,(X n,z n ). The distribution of each (X i,z i ) (1 i n) is f(x i,z i θ) = K p z ik k [h(x i λ k )] z ik, (5) where p k is the prior probability of the kth group (the mixing proportion), h(x i λ k )isaprobabilitydensitywithparametersλ k andthewholeparameter is θ = (p 1,...,p K,λ 1,...,λ K ). An estimate ˆθ of θ is obtained by the m.l. method ˆθ = argmaxl(θ x, z) (6) θ where the log-likelihood function L(θ x, z) is defined by L(θ x,z) = n K z ik ln(p k h(x i λ k )). (7) i=1 This estimate ˆθ allows to assign any new point x n+1 with unknown membership in one of the K groups by the maximum a posteriori (MAP) procedure. Computing the conditional probability t k (x n+1 ˆθ) that x n+1 arises from the kth group ˆp k h(x n+1 ˆλ k ) t k (x n+1 ˆθ) = K l=1 ˆp lh(x n+1 ˆλ l ), (8) the MAP procedure consists of assigning x n+1 to the group maximizing this conditional probability, i.e. { 1 if k = argmaxl=1...,k t ẑ n+1 k = l (x n+1 ˆθ) 0 if not. (9) 6
7 3 Algorithms in mixmod 3.1 EM algorithm Starting from an initial arbitrary parameter θ 0, the mth iteration of the EM algorithm consists of repeating the following E and M steps. E step: The current conditional probabilities that z ik = 1 for i = 1,...,n and k = 1,...,K are computed using the current value θ m 1 of the parameter: t m ik = t m k (x i θ m 1 ) = p m 1 k K l=1 pm 1 l h(x i λ m 1 k ) h(x i λ m 1 l ). (10) M step: The m.l. estimate θ m of θ is updated using the conditional probabilities t m ik as conditional mixing weights. It leads to maximize F(θ x 1,...,x n,t m ) = n K t m ikln[p k Φ(x i λ k )], (11) i=1 where t m = (t m ik,i = 1,...,n,k = 1,...,K). Updated expression of mixture proportions are, for k = 1,...,K, p m k = n i=1 tm ik n. (12) Detailed formula for the updating of the λ k s are depending of the component parameterization λ and cannot be detailed here. 3.2 SEM algorithm The SEM algorithm is a stochastic version of EM incorporating between the EandMstepsarestorationoftheunknowncomponentlabelsz i,i = 1,...,n, by drawing them at random from their current conditional distribution. Starting from an initial parameter θ 0, an iteration of SEM consists of three steps. E step: The conditional probabilities t m ik (1 i n,1 k K) are computed for the current value of θ as done in the E step of EM. 7
8 S step: A partition P m = (P m 1,...,P m K ) of x 1,...,x n is designed by assigning each point x i at random to one of the mixture components according to the multinomial distribution with parameter (t m ik,1 k K). M step: The m.l. estimate of θ is updated using the cluster P m k as sub-sample (1 k K) of the kth mixture component. This step leads generally to simple formula. For instance, p m k = card(pm k ). (13) n SEM does not converge pointwise. It generates a Markov chain whose stationary distribution is more or less concentrated around the m.l. parameter estimator. A natural parameter estimate from a SEM sequence (θ r ) r=1,...,r is the mean R r=b+1 θr /(R b) of the iterates values where the first b burnin iterates have been discarded when computing this mean. An alternative estimate is to consider the parameter value leading to the highest likelihood in a SEM sequence. A remark is to be made. When several observations are associated to the same vector, they are assigned to the same mixture component in the S step. This choice can make a difference when concerned with categorical data. It is expected to give a larger influence to the random assignments. 3.3 CEM algorithm This algorithm incorporates a classification step between the E and M steps of EM. Starting from an initial parameter θ 0, an iteration of CEM consists of three steps. E step: The conditional probabilities t m ik (1 i n,1 k K) are computed for the current value of θ as done in the E step of EM. C step: A partition P m = (P1 m,...,pk m) of x 1,...,x n is designed by assigning each point x i to the component maximizing the conditional probability (t m ik,1 k K). M step: The m.l. estimates (ˆp k,λ k ) are computed using the cluster P m k as sub-sample (1 k K) of the kth mixture component as done in the M step of SEM. 8
9 CEM is a K-means-like algorithm and contrary to EM, it converges in a finite number of iterations. CEM is not maximizing the observed log-likelihood L (2) but is maximizing in θ and z 1,...,z n the complete data log-likelihood CL (4) where the missing component indicator vector z i of each sample point is included in the data set. As a consequence, CEM is not expected to converge to the m.l. estimate of θ and yields inconsistent estimates of the parameters especially when the mixture components are overlapping or are in disparate proportions (see McLachlan and Peel 2000, Section 2.21). 4 Using mixmod 4.1 Stopping rules In mixmod they are three ways to stop an algorithm. An algorithm can be stopped after a pre-defined number of iterations (100 by default in mixmod). This possibility is available for EM, SEM and CEM. An algorithm can be stopped using a threshold for the relative change of the criterion at hand (the likelihood L or the classification likelihood CL). This possibility is available with EM and CEM. It is not recommended since EM can encounter slow convergence situations and CEM is converging in a finite number of iterations. An algorithm can be stopped at stationarity. Obviously, this possibility is only available for CEM. 4.2 Initialization strategies The solution provided by EM can highly depend on its starting position especially in a multivariate context. Thus, it is important to have sensible ways for initiating EM to get a sensible optimum of the likelihood. Obviously, in some cases it is possible to start from a particular partition of the data or from a pre-defined θ 0 and those initializations of EM are possible in mixmod. But there is the need to have more general strategies. In mixmod, it is possible to easily link the algorithms EM, SEM and CEM in all imaginable ways. Thus, in Biernacki et al. (2003), we have experimented an efficient three step Search/Run/Select (S/R/S) strategy for maximizing the likelihood: 9
10 1. Build a search method for generating p initial positions. This could be based on random starts or the output from an algorithm like a Classification EM (CEM) algorithm, a Stochastic EM (SEM) algorithm or short runs of the standard EM algorithm. The parameter p is depending on an allotment of iterations. 2. Run the EM algorithm a set number of times at each initial position with a fixed number of iterations. 3. Select the solution providing best likelihood among the p trials, say θ. This three-step strategy can be compounded by repeating the three steps x times and using the θ 1,...,θ x as the starting positions in step 1. By compounding, one increases starting position variation, but one must decrease the length of the EM runs possible within the steps in order to fix the total number of steps. Possible variants of this strategy are now described. Random initialization Usually this random initial position is obtained by drawing at random component means in the data set. Since this is probably the most employed way of initiating EM, it can be regarded as a reference strategy. An extension of this simple strategy consists of repeating it x times from different random positions and selecting the solution maximizing the likelihood among those x runs. This xem strategy is the basic S/R/S algorithm. Using the CEM algorithm Runs of CEM from random positions followed by EM from the position providing the highest complete data loglikelihood obtained with CEM. And, x repetitions of the previous strategy give rise to an additional strategy denoted xcem-em. Using short runs of EM By a short run of EM, we mean that we do not wait for convergence and that we stop the algorithm as soon as L m L m 1 L m L , (14) L m denoting the observed log-likelihood at mth iteration. Here 10 2 represents a threshold value which has to be chosen on a pragmatic ground. It 10
11 leads to the following strategies : several short runs of EM from random positions followed by a long run of EM from the solution maximizing the observed log-likelihood. And, x repetitions of the previous strategy lead to the so called xem-em strategy. Using Stochastic EM The stochastic EM algorithm generates an ergodic Markov chain. Thus a sequence of parameter estimates via SEM is expected to visit the whole parameter space with long sojourns in the neighborhood of sensible maxima of likelihood functions. This characteristic of SEM leads to the following strategies. SEMmean-EM : A run of SEM, followed by a run of EM from the solution obtained by computing the mean values of the sequence of parameter estimates provided by SEM after a burn-in period. The idea underlying this strategy is that SEM is expected to spend most of the time near sensible likelihood maxima with a large attractive neighborhood. SEMmax-EM : The same run of SEM followed by a run of EM from the position leading to the highest maximum likelihood value reached by SEM. Here, the idea is that a SEM sequence is expected to enter rapidly in the neighborhood of the global maximum of the likelihood function. It is difficult to recommend a particular strategy among the ones presented above. However, the strategy xem-em gives generally good performances and is the default strategy in mixmod. 4.3 Criteria to select a model It is of high interest to automatically select a model and the number K of mixture components. However, choosing a sensible mixture model is highly dependent of the modelling purpose. In mixmod, two criteria are proposed in a supervised setting: BIC and cross-validation. In an unsupervised setting, three criteria are available: BIC, ICL and NEC. In a density estimation perspective, BIC must be preferred. But in a cluster analysis perspective, ICL and NEC can provide more parsimonious answers. Nevertheless, NEC is essentially devoted to choose the number of mixture components K, rather than the model parameterization. 11
12 Before describing those criteria, it can be noted that if no information on K is available, it is recommended to vary it between 1 and the smallest integer larger than n 0.3 (see Bozdogan 1993) The Bayesian Information Criterion (BIC) A finite mixture model is characterized by the number of components K and the vector parameter θ = (p 1,...,p K,λ 1,...,λ K ). A classical way of choosing a model is to select this one maximizing the integrated likelihood, (ˆm, ˆK) = argmaxf(x m,k) (15) m,k where the integrated likelihood is f(x m,k) = f(x m,k,θ)π(θ m,k)dθ, (16) Θ m,k with the likelihood f(x m,k,θ) = n f(x i m,k,θ), (17) i=1 and Θ m,k being the parameter space of the model m with K components and π(θ m,k) a non informative or a weakly informative prior distribution on θ for this model. An asymptotic approximation of the integrated likelihood, valid under regularity conditions, has been proposed by Schwarz (1978) logf(x m,k) logf(x m,k,ˆθ) ν m,k 2 where ˆθ is the m.l. estimate of θ log(n), (18) ˆθ = argmaxf(x m,k,θ) (19) θ and ν m,k is the number of free parameters in the model m with K components. It leads to minimize the so-called BIC criterion BIC m,k = 2L m,k +ν m,k lnn, (20) where L m,k = logf(x m,k,ˆθ) is the maximum log-likelihood for m and K. Despite the fact that those regularity conditions are not fulfilled for mixtures, it has been proved that the criterion BIC is consistent (Keribin 2000) and has been proved to be efficient on a practical ground (see for instance Fraley and Raftery 1998). 12
13 4.3.2 The Integrated Completed Likelihood (ICL) The use of the integrated likelihood(16) does not take into account the ability of the mixture model to give evidence for a clustering structure of the data. An alternative is to consider the integrated likelihood of the complete data (x, z) (or integrated completed likelihood) (Biernacki et al. 2000) f(x,z m,k) = f(x,z m,k,θ)π(θ m,k)dθ, (21) Θ m,k where with f(x,z m,k,θ) = f(x i,z i m,k,θ) = n f(x i,z i m,k,θ) (22) i=1 K p z ik k [h(x i λ k )] z ik. (23) This integrated completed likelihood can be approximated from a BIC-like approximation. That is where logf(x,z m,k) logf(x,z m,k,ˆθ ) ν m,k 2 log n (24) ˆθ = argmaxf(x,z m,k,θ). (25) θ But z is unknown. It means that the objective functions to be maximized in (21) and (25) are not available and so is ˆθ. However, for n large enough, ˆθ can be approximated by the m.l. estimator ˆθ. Moreover, the missing data z can be replaced using the MAP principle: z = MAP(ˆθ). It leads finally to the ICL criterion to be minimized (Biernacki et al. 2000) ICL m,k = 2logf(x, z m,k,ˆθ)+ν m,k logn, (26) that we can also write as a BIC criterion penalized by an entropy term: ICL m,k = BIC m,k 2 n i=1 K z ik lnt ik. (27) 13
14 4.3.3 The Normalized Entropy Criterion (NEC) This entropy criterion measures the ability of a mixture model to provide well-separated clusters and is derived from a relation highlighting the differences between the maximum likelihood (m.l.) approach and the classification maximum likelihood (c.m.l.) approach to the mixture problem. Recall that NEC is essentially devoted to choose the number of mixture components K, not the model m. We note ˆθ the m.l. estimator of θ and t ik = t k (x i ˆθ) = ˆp k h(x i ˆλ k ) K k =1 ˆp k h(x i ˆλ k ) (28) the associated conditional probability that x i arises from to the kth mixture component. Direct calculations show that with L K the maximum log-likelihood, and C K = L K = C K +E K, (29) K E K = n t ik ln [ˆp k h(x i ˆλ k )], (30) i=1 K n t ik ln t ik 0. (31) i=1 This relation with the fact that the entropy term E K measures the overlap of the mixture components (If the mixture components are well-separated E K 0. But if the mixture components are poorly separated, E K has a large value.) leads to the normalized entropy criterion (Celeux and Soromenho 1996) NEC K = E K L K L 1 (32) as a criterion to be minimized for assessing the number of clusters arising from a mixture. Note that NEC 1 is not defined. Biernacki et al. (1999) proposed the following efficient rule to deal with this problem. Let K be the value minimizing NEC K, (2 K Ksup), Ksup being an upper bound for the number of mixture components. We choose K clusters if NEC K 1, otherwise we declare no clustering structure in the data. 14
15 4.3.4 The cross-validation criterion (CV) This criterion is valid only in the discriminant analysis (supervised) context. In this situation, note that only the model m has to be selected. Cross validation is a resampling method which can be summarised as follows: Let S be the whole dataset. Consider random splits of S into V independent datasets S 1,...,S V of approximatively equal sizes n 1,...,n V. (If n/v is an integer h, we have n 1 =... = n V = h.) The CV criterion is defined by CV m = 1 n V v=1 i S v δ(ẑ (v) i,z i ) (33) with δ the 0-1 cost and ẑ (v) i denotes the group to which x i is assigned when designing the assignment rule from the entire data set (x,z) without S v. When V = 1 the cross validation is known as the leave one out method, and, in this case, fast estimation of the n discriminant rules is implemented in the Gaussian situation (Biernacki and Govaert 1999). In mixmod, the default value for the cross validation criterion is V = The double cross-validation criterion (DCV) The CV error rate described above gives an optimistic estimate of the actual error rate because the method includes the selection of one model among several ones. Thus, there is a need to assess the actual error rate from an independent sample. This is the purpose of the DCV criterion, implemented in mixmod version 1.7. The double cross-validated error rate is computed in mixmod as follows: Repeat the three following steps for v in 1,...,V with S v = S \ S v build the models using the S v dataset select the best model regarding the CV criterion: m v estimate the error rate e v of m v using S v e v = 1 n v δ(ẑ m v i S v i,z i ). (34) 15
16 The DCV error rate (ē) is finally obtained by averaging the e 1,...,e V. The empirical standard error of the error rate is given by σ e ( ) 1/2 ē = 1 V 1 V e v, σ e = (e v ē) 2. (35) V V 1 v=1 v=1 Recall that in mixmod, the default value of V is Partial labeling of individuals MIXMOD allows partial labeling. Recall that in density estimation or clustering context, observed data are x = {x 1,...,x n }, the corresponding labels z = {z 1,...,z n } being unknown. On the contrary, in the discriminant analysis context, all the labels z are available to estimate the mixture parameter θ. In some cases, the following intermediate situation may occur: the set x of individuals is divided into two sets x = (x l,x u ) where x l = {x 1,...,x m } (1 m n) are units with known labels z l = {z 1,...,z m }, and x u = {x m+1,...,x n } units with unknown labels z u = {z m+1,...,z n }. The m.l. mixture parameter estimate is derived by maximizing the following log-likelihood ( m K n K ) L(θ x,z l ) = z ik ln[p k h(x i λ k )]+ ln p k h(x i λ k ). (36) i=1 i=m+1 In a clustering context using the classification approach, the c.m.l. method, is maximizing the following completed log-likelihood CL(θ,z u x,z l ) = m i=1 K z ik ln[p k h(x i λ k )]+ n i=m+1 K z ik ln[p k h(x i λ k )]. (37) In practice, the modifications of the algorithms are straightforward. It is simply necessary to replace t ik by z ik for all k and i = 1,...,m in the M step of EM, and to fix z ik to constant known values for all k and i = 1,...,m in the M step of SEM and CEM. 4.5 Weighting the units In some cases, it arises that some units are duplicated. Typically, it happens when the number of possible values for the units is low in regard to the 16
17 sample size. To avoid entering unnecessarily large lists of units, mixmod allows to specify a weight w i for each unit y i (i = 1,...,r). The set y w = {(y 1,w 1 ),...,(y r,w r )} is strictly equivalent to the set with eventual replications x = {x 1,...,x n }, so we have the relation n = w w r. All formula are easily adapted to take account of this weighting scheme. For instance, the log-likelihood L becomes ( r K ) L(θ x) = L(θ y w ) = w i ln p k h(y i λ k ), (38) i=1 and the proportion estimation equation at the mth iteration becomes p m k = r i=1 w it m ik. (39) n 5 The Gaussian mixture model 5.1 Definition In the Gaussian mixture model, X = R d and each x i is assumed to arise independently from a mixture with density f(x i θ) = K p k h(x i µ k,σ k ) (40) where p k is the mixing proportion (0 < p k < 1 for all k = 1,...,K and p p K = 1) of the kth component and h( µ k,σ k ) denotes the d- dimensional Gaussian density with mean µ k and variance matrix Σ k, { h(x i µ k,σ k ) = (2π) d/2 Σ k 1/2 exp 1 } 2 (x i µ k ) Σ 1 k (x i µ k ), (41) and θ = (p 1,...,p K,µ 1,...,µ K,Σ 1,...,Σ K ) is the vector of the mixture parameters. Thus, clusters associated to the mixture components are ellipsoidal, centered at the means µ k and variance matrices Σ k determine their geometric characteristics. 17
18 5.2 Fourteen Gaussian models Eigenvalue decomposition of variance matrices Following Banfield and Raftery (1993) and Celeux and Govaert (1995), we consider a parameterization of the variance matrices of the mixture components consisting of expressing the variance matrix Σ k in terms of its eigenvalue decomposition Σ k = λ k D k A k D k (42) whereλ k = Σ k 1/d,D k isthematrixofeigenvectorsofσ k anda k isadiagonal matrix, such that A k = 1, with the normalized eigenvalues of Σ k on the diagonal in a decreasing order. The parameter λ k determines the volume of thekthcluster,d k itsorientation anda k itsshape. Byallowingsomebutnot all of these quantities to vary between clusters, we obtain parsimonious and easily interpreted models which are appropriate to describe various clustering situations The general family First, we can allow the volumes, the shapes and the orientations of clusters to vary or to be equal between clusters. Variations on assumptions on the parameters λ k,d k and A k (1 k K) lead to 8 general models of interest. For instance, we can assume different volumes and keep the shapes and orientations equal by requiring that A k = A (A unknown) and D k = D (D unknown) for k = 1,...,K. We denote this model [λ k DAD ]. With this convention, writing [λd k AD k ] means that we consider the mixture model with equal volumes, equal shapes and different orientations The diagonal family Another family of interest consists of assuming that the variance matrices Σ k are diagonal. In the parameterization (42), it means that the orientation matrices D k are permutation matrices. We write Σ k = λ k B k where B k is a diagonal matrix with B k = 1. This particular parameterization gives rise to 4 models: [λb], [λ k B], [λb k ] and [λ k B k ]. 18
19 5.2.4 The spherical family Thelastfamily ofmodelsconsistsofassumingsphericalshapes, namelya k = I, I denoting the identity matrix. In such a case, two parsimonious models are in competition: [λi] and [λ k I]. Finally, we get 14 different models (see Table 1). Those 14 Gaussian mixture models are implemented, specifying different clustering situations from the eigenvalue decomposition of the variance matrices of the mixture components. The main advantage of variance matrices eigenvalue decomposition is the simple geometric interpretation of the models. To stress this point, Figure 1 shows a contour plot for each model, for K = 2 groups with dimension d = 2, consisting of a single ellipse of isodensity per group. model number of parameters M step inertia criteria [λdad ] α+β CF W [λ k DAD ] α+β +K 1 IP - [λda k D ] α+β +(K 1)(d 1) IP - [λ k DA k D ] α+β +(K 1)d IP - [λd k AD k ] α+kβ (K 1)d CF Σ kω k [λ k D k AD k ] α+kβ (K 1)(d 1) IP - [λd k A k D k ] α+kβ (K 1) CF Σ k W k 1 d [λ k D k A k D k ] α+kβ CF Σ kn k ln( W k n k ) [λb] α + d CF diag(w) [λ k B] α+d+k 1 IP - [λb k ] α+kd K +1 CF Σ k diag(w k ) 1 d [λ k B k ] α+kd CF Σ k n k ln( diag(w k) n k ) [λi] α+1 CF tr(w) [λ k I] α+k CF Σ k n k lntr( W k n k ) Table 1: Some characteristics of the 14 models. We have α = Kd+K 1 in the case of free proportions and α = Kd in the case of equal proportions, and β = d(d+1) ; CF means that the M step is closed form, IP means that the 2 M step needs an iterative procedure. The last column gives the inertia type criterion to be minimized in the case of equal proportions for each model. Exact definition of W and W k are given in (46) and (47). 19
20 [LI] [lkbk] [LDkADk ] [LkI] [LDAD ] [LkDkADk ] [LB] [LkDAD ] [LDkAkDk ] [LkB] [LDAkD ] [LkDkAkDk ] [LBk] [LkDAkD ] Figure 1: For two groups in two dimensions, this graphic displays the typical ellipse of isodensity per group for each of the 14 Gaussian models. 20
21 5.3 M step for each of the 14 models The M step has to be detailed for each of the 14 models. It is obviously present in the EM algorithm and its variants (SEM, CEM), but it is also useful for the discrimination purpose since maximizing the likelihood with complete data (x,z) can be performed with a single iteration of the M step. To unify the presentation, we make use of a classification matrix c = (c ik,i = 1,...,n;k = 1,...,K) with 0 c ik 1 and K c ik = 1, with the constraint c ik {0,1} when c defines a partition as in the classification approach. With this convention, in both the mixture and the classification approaches, the M step consists of maximizing in θ the function: F(θ x 1,...,x n,c) = n i=1 K c ik ln[p k h(x i µ k,σ k )] (43) for fixed c and x 1,...,x n. When we are concerned with the EM algorithm, c defines a fuzzy classification and we have c ik = t ik for 1 i n and 1 k K. When we are concerned with the CEM algorithm, c defines a partition and we have c ik = 1 if x i belongs to the group k and 0 otherwise (1 i n, 1 k K). Thus, for both approaches and for each of the considered models, the updating formulas for the proportions and the mean vectors of the mixture are, for 1 k K, n i=1 ˆµ k = x k = c ikx i (44) n k where n k = n c ik. (45) i=1 Remark that when c defines a partition n k = card(p k ). Moreover, we note W the within cluster scattering matrix W = K n c ik (x i x k )(x i x k ) (46) i=1 andw k thescatteringmatrixofacluster(orfuzzycluster),fork = 1,...,K, W k = n c ik (x i x k )(x i x k ). (47) i=1 21
22 The updating formulas for the variance matrices depend on the considered mixture model and are presented in the next subsections. Table 1 summarizes some features of the 14 models. In this table, the first column specifies the model. The second column gives the number of parameters to be estimated. The third column indicates if the M step can be achievedwithclosedformformulas(cf)orifthereisaneedtomakeuseofan iterative procedure (IP). The last column displays the inertia type criterion to be minimized for the case of equal proportions when the M step is closed form. These criteria can be derived from standard algebraic calculations. Some of them corresponds to standard criteria that was proposed without any reference to a statistical model. For instance, in clustering, tr(w) is the K-means criterion of Ward (1963), W was suggested by Friedman and Rubin (1967) and Σ k n k lntr( W k n k ) was proposed by Scott and Symons (1971). Indiscrimination,models[λC]and[λ k C k ]withequalproportionsrespectively correspond to classical linear and quadratic allocation rules (see for instance McLachlan 1982) The general family From Table 1, it can be seen that the inertia type criteria derived from the models [λdad ],[λd k A k D k ] and [λ kd k A k D k ] are classical clustering criteria (see Scott and Symons 1971, Maronna, Jacovkis 1974). On the contrary, the unusual models [λ k DAD ],[λ k DA k D ] and [λ k D k AD k ] which allow different volumes for the clusters do not lead to any inertia type criteria. Moreover, it is worth noting that the 8 models of the general family are invariant under any linear transformation of the data. We now detail the m.l. estimations of the variance matrices from a classification matrix c for the 8 situations. Model [λdad ] In this well-known situation, the common variance matrix Σ is estimated by ˆΣ = W n. (48) Model [λ k DAD ] In this situation, it is convenient to write Σ k = λ k C with C = DAD. M-step consists of two steps to minimize K tr(w kc 1 )/λ k + 22
23 d K n kln(λ k ) Step 1 (C fixed): λ k = tr(w kc 1 ) (49) dn k Step 2 (λ k s fixed): C = K 1 λ k W k K 1 λ k W k 1 d. (50) Model [λda k D ] In this situation and in the next one, there is no interest to assume that the terms of the diagonal matrices A k are in decreasing order. Thus for the models [λda k D ] and [λ k DA k D ] we do not assume that the diagonal terms of A k are in decreasing order. First, direct calculation of λ is λ = K tr(da 1 k D W k ). (51) nd Then,M-stepperformsiterativelytwostepstominimize K tr(da 1 k D W k ) Step 1 (D fixed): A k = diag(d W k D) diag(d W k D) 1 d (52) Step 2 (A k s fixed): see Flury and Gautschi (1986). (53) Model [λ k DA k D ] In this situation, there is no need to isolate the volume and it is convenient to write Σ k = DA k D where A k = Σ k. M-step consists of two steps to minimize K [ tr(da 1 k D W k )+n k dln A k ] Step 1 (D fixed): A k = diag(d W k D) (54) Step 2 (A k s fixed): see Flury and Gautschi (1986). (55) Model [λd k AD k ] Considering for k = 1,...,K the eigenvalue decomposition W k = L k Ω k L k of the symmetric definite positive matrix W k with the eigenvalues in the diagonal matrix Ω k in decreasing order, we have D k = L k, A = K Ω k K Ω k 1 d, λ = K Ω k 1 d. (56) n 23
24 Model [λ k D k AD k ] UsingagaintheeigenvaluedecompositionW k = L k Ω k L k, M-step consists of three steps to minimize K tr(w kd k A 1 D k )/λ k+ d K n kln(λ k ) Step 1 (D k s, A fixed): λ k = tr(w kd k A 1 D k ) (57) dn k Step 2 (λ k s, A fixed): D k = L k (58) Step 3 (λ k s, D k s fixed): A = K 1 λ k Ω k K 1 λ k Ω k 1 d. (59) Model [λd k A k D k ] In this situation, it is convenient to write Σ k = λc k where C k = D k A k D k. Direct calculation shows that C k = W K k, λ = W k 1 d. (60) W k 1 d n Model [λ k D k A k D k ] This is the most general situation and we have ˆΣ k = 1 n k W k. (61) The diagonal family For this more parsimonious family of models, the eigenvectors of Σ k (1 k K) are the vectors generating the basis associated to the d variables (D k = J k ). If the J k are equal, the variables are independent. If the J k are different, the variables are independent conditionally to the z i (1 i n). In this situation, Gaussian mixture with diagonal variance matrices can be viewed as an elegant model for weighting variables in a cluster analysis context. It leads to adaptive weighting algorithms assuming same weights for each cluster if the J k s are assumed equal and different weights for each cluster if the J k s are assumed different. We considered four models of interest. The main features of these four models are summarized in Table 1. The three inertia type criteria for the models [λb],[λb k ] and [λ k B k ] are simple adaptations of the corresponding criteria of the general family. The interesting model [λ k B] does not lead to an inertia type criterion. Moreover, it is worth noting that the 4 models of the diagonal family are invariant under any scaling of the variables but not under any linear transformation. We now derive the m.l. 24
25 estimation of the variance matrices from a classification matrix c for each of the four situations. Model [λb] We have B = diag(w), λ = diag(w) 1 d. (62) diag(w) 1 d n Model [λ k B] M-step consists of two steps to minimize K tr(w kb 1 )+ d K n kln(λ k ) Step 1 (B fixed): λ k = tr(w kb 1 ) (63) dn ( k K ) 1 diag λ k W k Step 2 (λ k s fixed): B = Model [λb k ] We have diag ( K ) 1 λ k W k 1 d. (64) Model [λ k B k ] We get B k = diag(w k) diag(w k ) 1 d, λ = K diag(w k) 1 d. (65) n The spherical family B k = diag(w k), λ diag(w k ) 1 k = diag(w k) 1 d. (66) d n k We consider here very parsimonious models for which the variance matrices are spherical. Two situations have to be considered: Σ k = λi and Σ k = λ k I, I denoting the(d d) identity matrix. The inertia type criterion tr(w) of the model [λi] is certainly the oldest and the most employed clustering criterion. On the contrary, as far as we know, the criterion K n k ln tr(w k) n k (67) 25
26 has been proposed for the first time by Banfield and Raftery (1993). Note that the 2 models of the spherical family are invariant under any isometric transformation. We derive the m.l. estimations of the volumes of the clusters for these models. Model [λi] We get λ = tr(w) dn. (68) Model [λ k I] We get λ k = tr(w k) dn k. (69) Formally models [λi] and [λ k I] do not seem to be very different and the increase of the number of parameters when considering model [λ k I] instead of model [λi] is small (see Table 1). In fact, these two models can lead to very different clustering structures. 5.4 Mixture of Factor Analyzers In order to deal with high dimensional data, mixture of factor analyzers have been considered by several authors including Bouveyron et al. (2007), McNicholas and Murphy (2008), McLachlan and Peel (2000), Chapter 8, Tipping and Bishop (1999). In mixmod, a family of eight Gaussian mixture models introduced by Bouveyron et al. (2007) have been implemented for discriminant analysis in high dimensional spaces. They are denoted as HD (for High Dimensional) models in the following The general high dimensional model The same eigenvalue decomposition of the mixture component variance matrices Σ k, k = 1,...,K, is considered: Σ k = D k k D t k, where D k is the orthogonal matrix of the eigenvectors of Σ k and k is a diagonal matrix containing the eigenvalues of Σ k. It is further assume that 26
27 k has the following form (Note that in this section, there is no need to isolate the volume of the variance matrices.): a k δ k 0 a kδk k = 0 b k b k (d δ k) where a kj b k, for j = 1,...,δ k and δ k < d. The class-specific subspace generated by the δ k first eigenvectors corresponding to the eigenvalues a kj and containing the mean µ k is denoted E k. In the orthogonal of E k, the component variance is characterized with a single parameter b k. The projectors on E k and E k are denoted P k and Pk. Figure 2 summarizes the model which is referred as [a kj b k D k δ k ] in the following. Figure 2: The parameters of the model [a kj b k D k δ k ] in the case of two classes Sub-models of model [a kj b k D k δ k ] Starting from the general model [a kj b k D k δ k ] and allowing the elements of the model to vary or to be equal between classes, leads to 28 different models 27
28 related to different types of regularization. In mixmod eight useful models have been selected: Two models with free dimension δ k : the model [a kj b k D k δ k ] the model [a k b k D k δ k ] Six models with fixed dimension δ: the model [a kj b k D k δ] the model [a j b k D k δ] the model [a kj bd k δ] the model [a j bd k δ] the model [a k b k D k δ] the model [a k bd k δ] Their main features are summarized in Table 2. The second column of this table gives the number of parameters to be estimated. The third column provides the asymptotic order of the number of parameters to be estimated (with the assumption K δ k d). The last column gives this number in the particular case K = 4, d = 100 and k, δ k = 10. These values are also given for the standard classification methods QDA and LDA. It is worthwhile to note that, in this cases, all HD models are more parsimonious than both QDA and LDA. Some particular situations lead to standard discriminant methods. For example, if δ k = (d 1), for k = 1,...,K, the model reduces to QDA. Moreover, if a kj = a j, b k = b and D k = D, for i = 1,...,k, it reduces to LDA The MAP step The MAP decision rule for model [a kj b k D k δ k ] yields to classify x in class C k if k = argmin,...,k {Γ k (x)} with Γ k (x) = µ k P k (x) 2 A k + 1 b k x P k (x) 2 + δ k j=1 log(a kj )+(d δ k )log(b k ) 2log(π k )+plog(2π), 28
29 Model Number of Asymptotic Values of n for K = 4, parameters n order p = 100 and d = 10 [a kj b k D k δ k ] ρ+ τ +2K +D Kdδ 4231 [a k b k D k δ k ] ρ+ τ +3K Kdδ 4195 [a kj b k D k d] ρ+k(τ +δ +1)+1 Kdδ 4228 [a j b k D k d] ρ+k(τ +1)+δ +1 Kdδ 4198 [a kj bd k d] ρ+k(τ +δ)+2 Kdδ 4225 [a j bd k d] ρ+kτ +δ +2 Kdδ 4195 [a k b k D k d] ρ+k(τ +2)+1 Kdδ 4192 [a k bd k d] ρ+k(τ +1)+2 Kdδ 4189 QDA ρ+kd(δ +1)/2 Kp 2 / LDA ρ+δ(δ +1)/2 p 2 / Table 2: Features of the HD models: ρ = Kd + K 1 is the number of parameters required for the estimation of means and proportions, τ = K δ k[p (δ k + 1)/2] and τ = δ[d (δ + 1)/2] are the number of parameters required for the estimation of Dk and D, and D = K δ k. For asymptotic order, it is assumed that K δ d.. Ak being a norm on E k such that x 2 A k = x t A k x with A k = D k 1 t k D k. This decision rule is based on two distances: the distance between the observation and the subspace E k, and the distance between the projection of x on E k and the mean of the class. It also depends on the variances a kj and b k and on prior probabilities π k. Figure 3 depicts the decision rule. It illustratesthefacttheprojectionone k isnotrequired,reducingdramatically the number of parameters to be estimated and avoiding numerical difficulties Estimation of the model parameters The parameters of the mixture of factor analyzers models are estimated through the maximum likelihood approach. Estimation of parameters π k and µ k of class C k are ˆπ k = n k n, ˆµ k = 1 n k x k C k x j. In what follows, we make use of W k = x j C k (x j ˆµ k ) t (x j ˆµ k ), n k = card(c k ), W = K i=k ˆπ kw k, λ kj which denotes the jth largest eigenvalue of 29
30 Figure 3: The subspaces E k and E k of the class C k. 30
31 W k and λ j the jth largest eigenvalue of W, to define the m.l. estimate of the mixture component variance that are now presented. Details can be found in Bouveyron et al. (2007). Models with free δ k Assuming that dimensions δ k are known for k = 1,..., K, the following closed form estimators of model parameters are derived. Subspace E k The δ k first columns of D k are estimated by the eigenvectors associated with the δ k largest eigenvalues λ kj of W k. Model [a kj b k D k δ k ] The estimators of a kj are the δ k largest eigenvalues λ kj of W k divided by n k and ( ) 1 δ k ˆbk = trace(w k ) λ kj. (70) n k (d δ k ) Model [a k b k D k δ k ] The estimator of b k is given by (70) and â k = 1 n k δ k δ k j=1 j=1 λ kj, (71) Models with common δ k Assumingthatparameterδ isknown, weobtain the following closed form estimators for the parameters of the models with common δ k, equal to δ. Subspace E k The δ first columns of D k are estimated by the eigenvectors associated with the δ largest eigenvalues λ kj of W k. Model [a kj b k D k d] The estimators of a kj are the δ largest eigenvalues λ kj of W k divided by n k and ( ) 1 d ˆbk = trace(w k ) λ kj. (72) n k (d δ) j=1 31
32 Model [a j b k D k d] The estimator of b k is given by (72) and â j = 1 n K ˆπ k λ kj. (73) Model [a kj bd k d] The estimators of a kj are the δ largest eigenvalues λ kj of W k divided by n k and ( ) 1 K d ˆb = trace(w) ˆπ k λ kj. (74) n(d δ) Model [a j bd k d] The estimators of a j are given by (73) and the estimator of b is given by (74). Model [a k b k D k d] The estimator of b k is given by (72) and â k = 1 nd j=1 d λ kj, (75) Model [a k bd k d] The estimator of a k is given by (75) and the estimator of b is given by (74). Estimation of intrinsic dimensions The last parameters to be estimated are the intrinsic dimensions δ k of the K classes. It is not possible to estimate the dimensions δ k using the maximum likelihood approach and minimizing the cross validated error rate is considered. However, this minimization technique is not implemented in the present version of mixmod and the user has to provide all the intrinsic dimensions δ k. 6 The multinomial mixture model 6.1 Definition We consider now that data are n objects described by d categorical variables, with respective number of categories m 1,...,m d, so X = {1,...,m 1 }... {1,...,m d }. The data can be represented by n binary vectors x i = (x jh i ;j = 32 j=1
33 1,...,d;h = 1,...,m j ) (i = 1,...,n) where x jh i = 1 if the object i belongs to the category h of the variable j and 0 otherwise. Denoting m = d j=1 m j the total number of categories, the data are defined by the matrix x = (x 1,...,x n ) with n rows and m columns. Binary data can be seen as a particular case of categorical data with d dichotomous variables, i.e. m j = 2 for any j = 1,...,d. The latent class model assumes that the d categorical variables are independent given the latent variable. Formulated in mixture terms (Everitt 1984), each x i arises independently from a mixture of multivariate multinomial distributions defined by f(x i θ) = K p k h(x i α k ) (76) where p k is the mixing proportion (0 < p k < 1 for all k = 1,...,K and p p K = 1) of the kth component and where, for k = 1,...,K, h(x i α k ) = m d j (α jh k )xjh j=1 h=1 i (77) with α k = (α jh k ;j = 1,...,d;h = 1,...,m j). In (77), we recognize the product of d conditionally independent multinomial distributions of parameters α j k. The mixture parameters is denoted by θ = (p 1,...,p K 1,α 1,...,α K ). This model may present problems of identifiability(see for instance Goodman 1974) but most situations of interest are identified. 6.2 Five multinomial models In order to propose more parsimonious models than the previous one, we present the following extension of the parameterization of Bernoulli distributions used by Celeux and Govaert (1991) for clustering and also by Aitchison and Aitken (1976) for kernel discriminant analysis. The basic idea is to impose the vector α j k = (αj1 k,...,αjm j k ) to take the form (β j k,...,βj k,γj k,βj k,...,βj k ) with γj k > βj k. Since m j h=1 αjh k = 1, we have (m j 1)β j k +γj k = 1 and, consequently, βj k = (1 γj k )/(m j 1). The constraint γ j k > βj k becomes finally γj k > 1/m j. Then, the vector α j k can be broken up into the two following parameters: 33
34 a j k = (aj1 k,...,ajm j k ) where a jh k = 1 if h corresponds to the rank of γ j k (in the following, this rank will be noted h(k,j)), 0 otherwise; ε j k = 1 γj k whichcorrespondstotheprobabilitythatthedatax i arising from the kth component are such that x jh(k,j) i 1. In other words, the multinomial distribution associated to the jth variable of the kth component is reparameterized by a center a j k and the dispersion ε j k around this center. Thus, it allows us to give an interpretation similar to the center and the variance matrix used for continuous data in the Gaussian mixture context. Since, the relationship between the initial parameterization and the new one is given by: { 1 ε j k = k if h = h(k,j) ε j k /(m j 1) otherwise, α jh (78) Equation (77) can be rewritten with a k = (a j k ;j = 1,...,d) and ε k = (ε j k ;j = 1,...,d) h(x i α k ) = h(x i a k,ε k ) = m d j j=1 h=1 ) x jh jk ((1 ε )ajh k (ε j k /(m j 1)) 1 ajh i k. (79) In the following, this model will be denoted by [ε j k ]. In this context, three other models can be easily deduced. We note [ε k ] the model where ε j k is independent of the variable j, [ε j ] the model where ε j k is independent of the component k and, finally, [ε] the model where ε j k is independent of both the variable j and the component k. In order to maintain some unity in the notation, we will denote also [ε jh k ] the most general model introduced at the previous section. The number of free parameters associated to each models is given in Table M step for each of the five models The M step has to be detailed for each of the five models presented above. Using notation already defined in the Gaussian mixture context, the M step consists of maximizing in θ the function: F(θ x,c) = n i=1 K c ik ln[p k h(x i α k )] (80) 34
Model-based cluster analysis: a Defence. Gilles Celeux Inria Futurs
 Model-based cluster analysis: a Defence Gilles Celeux Inria Futurs Model-based cluster analysis Model-based clustering (MBC) consists of assuming that the data come from a source with several subpopulations.
Model-based cluster analysis: a Defence Gilles Celeux Inria Futurs Model-based cluster analysis Model-based clustering (MBC) consists of assuming that the data come from a source with several subpopulations.
Finite Mixture Models and Clustering
 Finite Mixture Models and Clustering Mohamed Nadif LIPADE, Université Paris Descartes, France Nadif (LIPADE) EPAT, May, 2010 Course 3 1 / 40 Introduction Outline 1 Introduction Mixture Approach 2 Finite
Finite Mixture Models and Clustering Mohamed Nadif LIPADE, Université Paris Descartes, France Nadif (LIPADE) EPAT, May, 2010 Course 3 1 / 40 Introduction Outline 1 Introduction Mixture Approach 2 Finite
INRIA Rh^one-Alpes. Abstract. Friedman (1989) has proposed a regularization technique (RDA) of discriminant analysis
 Regularized Gaussian Discriminant Analysis through Eigenvalue Decomposition Halima Bensmail Universite Paris 6 Gilles Celeux INRIA Rh^one-Alpes Abstract Friedman (1989) has proposed a regularization technique
Regularized Gaussian Discriminant Analysis through Eigenvalue Decomposition Halima Bensmail Universite Paris 6 Gilles Celeux INRIA Rh^one-Alpes Abstract Friedman (1989) has proposed a regularization technique
Different points of view for selecting a latent structure model
 Different points of view for selecting a latent structure model Gilles Celeux Inria Saclay-Île-de-France, Université Paris-Sud Latent structure models: two different point of views Density estimation LSM
Different points of view for selecting a latent structure model Gilles Celeux Inria Saclay-Île-de-France, Université Paris-Sud Latent structure models: two different point of views Density estimation LSM
High Dimensional Discriminant Analysis
 High Dimensional Discriminant Analysis Charles Bouveyron 1,2, Stéphane Girard 1, and Cordelia Schmid 2 1 LMC IMAG, BP 53, Université Grenoble 1, 38041 Grenoble cedex 9 France (e-mail: charles.bouveyron@imag.fr,
High Dimensional Discriminant Analysis Charles Bouveyron 1,2, Stéphane Girard 1, and Cordelia Schmid 2 1 LMC IMAG, BP 53, Université Grenoble 1, 38041 Grenoble cedex 9 France (e-mail: charles.bouveyron@imag.fr,
Model selection criteria in Classification contexts. Gilles Celeux INRIA Futurs (orsay)
 Model selection criteria in Classification contexts Gilles Celeux INRIA Futurs (orsay) Cluster analysis Exploratory data analysis tools which aim is to find clusters in a large set of data (many observations
Model selection criteria in Classification contexts Gilles Celeux INRIA Futurs (orsay) Cluster analysis Exploratory data analysis tools which aim is to find clusters in a large set of data (many observations
High Dimensional Discriminant Analysis
 High Dimensional Discriminant Analysis Charles Bouveyron LMC-IMAG & INRIA Rhône-Alpes Joint work with S. Girard and C. Schmid High Dimensional Discriminant Analysis - Lear seminar p.1/43 Introduction High
High Dimensional Discriminant Analysis Charles Bouveyron LMC-IMAG & INRIA Rhône-Alpes Joint work with S. Girard and C. Schmid High Dimensional Discriminant Analysis - Lear seminar p.1/43 Introduction High
Journal of Statistical Software
 JSS Journal of Statistical Software January 2012, Volume 46, Issue 6. http://www.jstatsoft.org/ HDclassif: An R Package for Model-Based Clustering and Discriminant Analysis of High-Dimensional Data Laurent
JSS Journal of Statistical Software January 2012, Volume 46, Issue 6. http://www.jstatsoft.org/ HDclassif: An R Package for Model-Based Clustering and Discriminant Analysis of High-Dimensional Data Laurent
Classification of high dimensional data: High Dimensional Discriminant Analysis
 Classification of high dimensional data: High Dimensional Discriminant Analysis Charles Bouveyron, Stephane Girard, Cordelia Schmid To cite this version: Charles Bouveyron, Stephane Girard, Cordelia Schmid.
Classification of high dimensional data: High Dimensional Discriminant Analysis Charles Bouveyron, Stephane Girard, Cordelia Schmid To cite this version: Charles Bouveyron, Stephane Girard, Cordelia Schmid.
Variable selection for model-based clustering
 Variable selection for model-based clustering Matthieu Marbac (Ensai - Crest) Joint works with: M. Sedki (Univ. Paris-sud) and V. Vandewalle (Univ. Lille 2) The problem Objective: Estimation of a partition
Variable selection for model-based clustering Matthieu Marbac (Ensai - Crest) Joint works with: M. Sedki (Univ. Paris-sud) and V. Vandewalle (Univ. Lille 2) The problem Objective: Estimation of a partition
Model-based clustering of high-dimensional data: an overview and some recent advances
 Model-based clustering of high-dimensional data: an overview and some recent advances Charles BOUVEYRON Laboratoire SAMM, EA 4543 Université Paris 1 Panthéon-Sorbonne This presentation is based on several
Model-based clustering of high-dimensional data: an overview and some recent advances Charles BOUVEYRON Laboratoire SAMM, EA 4543 Université Paris 1 Panthéon-Sorbonne This presentation is based on several
High Dimensional Discriminant Analysis
 High Dimensional Discriminant Analysis Charles Bouveyron LMC-IMAG & INRIA Rhône-Alpes Joint work with S. Girard and C. Schmid ASMDA Brest May 2005 Introduction Modern data are high dimensional: Imagery:
High Dimensional Discriminant Analysis Charles Bouveyron LMC-IMAG & INRIA Rhône-Alpes Joint work with S. Girard and C. Schmid ASMDA Brest May 2005 Introduction Modern data are high dimensional: Imagery:
Choosing a model in a Classification purpose. Guillaume Bouchard, Gilles Celeux
 Choosing a model in a Classification purpose Guillaume Bouchard, Gilles Celeux Abstract: We advocate the usefulness of taking into account the modelling purpose when selecting a model. Two situations are
Choosing a model in a Classification purpose Guillaume Bouchard, Gilles Celeux Abstract: We advocate the usefulness of taking into account the modelling purpose when selecting a model. Two situations are
Unsupervised machine learning
 Chapter 9 Unsupervised machine learning Unsupervised machine learning (a.k.a. cluster analysis) is a set of methods to assign objects into clusters under a predefined distance measure when class labels
Chapter 9 Unsupervised machine learning Unsupervised machine learning (a.k.a. cluster analysis) is a set of methods to assign objects into clusters under a predefined distance measure when class labels
An Introduction to mixture models
 An Introduction to mixture models by Franck Picard Research Report No. 7 March 2007 Statistics for Systems Biology Group Jouy-en-Josas/Paris/Evry, France http://genome.jouy.inra.fr/ssb/ An introduction
An Introduction to mixture models by Franck Picard Research Report No. 7 March 2007 Statistics for Systems Biology Group Jouy-en-Josas/Paris/Evry, France http://genome.jouy.inra.fr/ssb/ An introduction
Adaptive Mixture Discriminant Analysis for. Supervised Learning with Unobserved Classes
 Adaptive Mixture Discriminant Analysis for Supervised Learning with Unobserved Classes Charles Bouveyron SAMOS-MATISSE, CES, UMR CNRS 8174 Université Paris 1 (Panthéon-Sorbonne), Paris, France Abstract
Adaptive Mixture Discriminant Analysis for Supervised Learning with Unobserved Classes Charles Bouveyron SAMOS-MATISSE, CES, UMR CNRS 8174 Université Paris 1 (Panthéon-Sorbonne), Paris, France Abstract
Model-Based Clustering of High-Dimensional Data: A review
 Model-Based Clustering of High-Dimensional Data: A review Charles Bouveyron, Camille Brunet To cite this version: Charles Bouveyron, Camille Brunet. Model-Based Clustering of High-Dimensional Data: A review.
Model-Based Clustering of High-Dimensional Data: A review Charles Bouveyron, Camille Brunet To cite this version: Charles Bouveyron, Camille Brunet. Model-Based Clustering of High-Dimensional Data: A review.
Gaussian Mixtures and the EM algorithm
 Gaussian Mixtures and the EM algorithm 1 sigma=1.0 sigma=1.0 Responsibilities 0.0 0.2 0.4 0.6 0.8 1.0 sigma=0.2 sigma=0.2 Responsibilities 0.0 0.2 0.4 0.6 0.8 1.0 2 Details of figure Left panels: two Gaussian
Gaussian Mixtures and the EM algorithm 1 sigma=1.0 sigma=1.0 Responsibilities 0.0 0.2 0.4 0.6 0.8 1.0 sigma=0.2 sigma=0.2 Responsibilities 0.0 0.2 0.4 0.6 0.8 1.0 2 Details of figure Left panels: two Gaussian
Mixtures and Hidden Markov Models for analyzing genomic data
 Mixtures and Hidden Markov Models for analyzing genomic data Marie-Laure Martin-Magniette UMR AgroParisTech/INRA Mathématique et Informatique Appliquées, Paris UMR INRA/UEVE ERL CNRS Unité de Recherche
Mixtures and Hidden Markov Models for analyzing genomic data Marie-Laure Martin-Magniette UMR AgroParisTech/INRA Mathématique et Informatique Appliquées, Paris UMR INRA/UEVE ERL CNRS Unité de Recherche
Performance Comparison of K-Means and Expectation Maximization with Gaussian Mixture Models for Clustering EE6540 Final Project
 Performance Comparison of K-Means and Expectation Maximization with Gaussian Mixture Models for Clustering EE6540 Final Project Devin Cornell & Sushruth Sastry May 2015 1 Abstract In this article, we explore
Performance Comparison of K-Means and Expectation Maximization with Gaussian Mixture Models for Clustering EE6540 Final Project Devin Cornell & Sushruth Sastry May 2015 1 Abstract In this article, we explore
Finite Singular Multivariate Gaussian Mixture
 21/06/2016 Plan 1 Basic definitions Singular Multivariate Normal Distribution 2 3 Plan Singular Multivariate Normal Distribution 1 Basic definitions Singular Multivariate Normal Distribution 2 3 Multivariate
21/06/2016 Plan 1 Basic definitions Singular Multivariate Normal Distribution 2 3 Plan Singular Multivariate Normal Distribution 1 Basic definitions Singular Multivariate Normal Distribution 2 3 Multivariate
Latent Variable Models and EM Algorithm
 SC4/SM8 Advanced Topics in Statistical Machine Learning Latent Variable Models and EM Algorithm Dino Sejdinovic Department of Statistics Oxford Slides and other materials available at: http://www.stats.ox.ac.uk/~sejdinov/atsml/
SC4/SM8 Advanced Topics in Statistical Machine Learning Latent Variable Models and EM Algorithm Dino Sejdinovic Department of Statistics Oxford Slides and other materials available at: http://www.stats.ox.ac.uk/~sejdinov/atsml/
Clustering by Mixture Models. General background on clustering Example method: k-means Mixture model based clustering Model estimation
 Clustering by Mixture Models General bacground on clustering Example method: -means Mixture model based clustering Model estimation 1 Clustering A basic tool in data mining/pattern recognition: Divide
Clustering by Mixture Models General bacground on clustering Example method: -means Mixture model based clustering Model estimation 1 Clustering A basic tool in data mining/pattern recognition: Divide
The Expectation-Maximization Algorithm
 1/29 EM & Latent Variable Models Gaussian Mixture Models EM Theory The Expectation-Maximization Algorithm Mihaela van der Schaar Department of Engineering Science University of Oxford MLE for Latent Variable
1/29 EM & Latent Variable Models Gaussian Mixture Models EM Theory The Expectation-Maximization Algorithm Mihaela van der Schaar Department of Engineering Science University of Oxford MLE for Latent Variable
Exact and Monte Carlo Calculations of Integrated Likelihoods for the Latent Class Model
 Exact and Monte Carlo Calculations of Integrated Likelihoods for the Latent Class Model C. Biernacki a,, G. Celeux b, G. Govaert c a CNRS & Université de Lille 1, Villeneuve d Ascq, France b INRIA, Orsay,
Exact and Monte Carlo Calculations of Integrated Likelihoods for the Latent Class Model C. Biernacki a,, G. Celeux b, G. Govaert c a CNRS & Université de Lille 1, Villeneuve d Ascq, France b INRIA, Orsay,
L11: Pattern recognition principles
 L11: Pattern recognition principles Bayesian decision theory Statistical classifiers Dimensionality reduction Clustering This lecture is partly based on [Huang, Acero and Hon, 2001, ch. 4] Introduction
L11: Pattern recognition principles Bayesian decision theory Statistical classifiers Dimensionality reduction Clustering This lecture is partly based on [Huang, Acero and Hon, 2001, ch. 4] Introduction
Discriminant analysis and supervised classification
 Discriminant analysis and supervised classification Angela Montanari 1 Linear discriminant analysis Linear discriminant analysis (LDA) also known as Fisher s linear discriminant analysis or as Canonical
Discriminant analysis and supervised classification Angela Montanari 1 Linear discriminant analysis Linear discriminant analysis (LDA) also known as Fisher s linear discriminant analysis or as Canonical
STATS 306B: Unsupervised Learning Spring Lecture 2 April 2
 STATS 306B: Unsupervised Learning Spring 2014 Lecture 2 April 2 Lecturer: Lester Mackey Scribe: Junyang Qian, Minzhe Wang 2.1 Recap In the last lecture, we formulated our working definition of unsupervised
STATS 306B: Unsupervised Learning Spring 2014 Lecture 2 April 2 Lecturer: Lester Mackey Scribe: Junyang Qian, Minzhe Wang 2.1 Recap In the last lecture, we formulated our working definition of unsupervised
Clustering VS Classification
 MCQ Clustering VS Classification 1. What is the relation between the distance between clusters and the corresponding class discriminability? a. proportional b. inversely-proportional c. no-relation Ans:
MCQ Clustering VS Classification 1. What is the relation between the distance between clusters and the corresponding class discriminability? a. proportional b. inversely-proportional c. no-relation Ans:
Families of Parsimonious Finite Mixtures of Regression Models arxiv: v1 [stat.me] 2 Dec 2013
![Families of Parsimonious Finite Mixtures of Regression Models arxiv: v1 [stat.me] 2 Dec 2013 Families of Parsimonious Finite Mixtures of Regression Models arxiv: v1 [stat.me] 2 Dec 2013](/thumbs/86/94023882.jpg) Families of Parsimonious Finite Mixtures of Regression Models arxiv:1312.0518v1 [stat.me] 2 Dec 2013 Utkarsh J. Dang and Paul D. McNicholas Department of Mathematics & Statistics, University of Guelph
Families of Parsimonious Finite Mixtures of Regression Models arxiv:1312.0518v1 [stat.me] 2 Dec 2013 Utkarsh J. Dang and Paul D. McNicholas Department of Mathematics & Statistics, University of Guelph
Probabilistic & Unsupervised Learning
 Probabilistic & Unsupervised Learning Week 2: Latent Variable Models Maneesh Sahani maneesh@gatsby.ucl.ac.uk Gatsby Computational Neuroscience Unit, and MSc ML/CSML, Dept Computer Science University College
Probabilistic & Unsupervised Learning Week 2: Latent Variable Models Maneesh Sahani maneesh@gatsby.ucl.ac.uk Gatsby Computational Neuroscience Unit, and MSc ML/CSML, Dept Computer Science University College
Testing for a Global Maximum of the Likelihood
 Testing for a Global Maximum of the Likelihood Christophe Biernacki When several roots to the likelihood equation exist, the root corresponding to the global maximizer of the likelihood is generally retained
Testing for a Global Maximum of the Likelihood Christophe Biernacki When several roots to the likelihood equation exist, the root corresponding to the global maximizer of the likelihood is generally retained
arxiv: v1 [stat.me] 7 Aug 2015
![arxiv: v1 [stat.me] 7 Aug 2015 arxiv: v1 [stat.me] 7 Aug 2015](/thumbs/85/92929102.jpg) Dimension reduction for model-based clustering Luca Scrucca Università degli Studi di Perugia August 0, 05 arxiv:508.07v [stat.me] 7 Aug 05 Abstract We introduce a dimension reduction method for visualizing
Dimension reduction for model-based clustering Luca Scrucca Università degli Studi di Perugia August 0, 05 arxiv:508.07v [stat.me] 7 Aug 05 Abstract We introduce a dimension reduction method for visualizing
MODEL BASED CLUSTERING FOR COUNT DATA
 MODEL BASED CLUSTERING FOR COUNT DATA Dimitris Karlis Department of Statistics Athens University of Economics and Business, Athens April OUTLINE Clustering methods Model based clustering!"the general model!"algorithmic
MODEL BASED CLUSTERING FOR COUNT DATA Dimitris Karlis Department of Statistics Athens University of Economics and Business, Athens April OUTLINE Clustering methods Model based clustering!"the general model!"algorithmic
CMSC858P Supervised Learning Methods
 CMSC858P Supervised Learning Methods Hector Corrada Bravo March, 2010 Introduction Today we discuss the classification setting in detail. Our setting is that we observe for each subject i a set of p predictors
CMSC858P Supervised Learning Methods Hector Corrada Bravo March, 2010 Introduction Today we discuss the classification setting in detail. Our setting is that we observe for each subject i a set of p predictors
New Global Optimization Algorithms for Model-Based Clustering
 New Global Optimization Algorithms for Model-Based Clustering Jeffrey W. Heath Department of Mathematics University of Maryland, College Park, MD 7, jheath@math.umd.edu Michael C. Fu Robert H. Smith School
New Global Optimization Algorithms for Model-Based Clustering Jeffrey W. Heath Department of Mathematics University of Maryland, College Park, MD 7, jheath@math.umd.edu Michael C. Fu Robert H. Smith School
More on Unsupervised Learning
 More on Unsupervised Learning Two types of problems are to find association rules for occurrences in common in observations (market basket analysis), and finding the groups of values of observational data
More on Unsupervised Learning Two types of problems are to find association rules for occurrences in common in observations (market basket analysis), and finding the groups of values of observational data
Parsimonious Gaussian Mixture Models
 Parsimonious Gaussian Mixture Models Brendan Murphy Department of Statistics, Trinity College Dublin, Ireland. East Liguria West Liguria Umbria North Apulia Coast Sardina Inland Sardinia South Apulia Calabria
Parsimonious Gaussian Mixture Models Brendan Murphy Department of Statistics, Trinity College Dublin, Ireland. East Liguria West Liguria Umbria North Apulia Coast Sardina Inland Sardinia South Apulia Calabria
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
Adaptive Mixture Discriminant Analysis for Supervised Learning with Unobserved Classes
 Adaptive Mixture Discriminant Analysis for Supervised Learning with Unobserved Classes Charles Bouveyron To cite this version: Charles Bouveyron. Adaptive Mixture Discriminant Analysis for Supervised Learning
Adaptive Mixture Discriminant Analysis for Supervised Learning with Unobserved Classes Charles Bouveyron To cite this version: Charles Bouveyron. Adaptive Mixture Discriminant Analysis for Supervised Learning
Machine Learning. Gaussian Mixture Models. Zhiyao Duan & Bryan Pardo, Machine Learning: EECS 349 Fall
 Machine Learning Gaussian Mixture Models Zhiyao Duan & Bryan Pardo, Machine Learning: EECS 349 Fall 2012 1 The Generative Model POV We think of the data as being generated from some process. We assume
Machine Learning Gaussian Mixture Models Zhiyao Duan & Bryan Pardo, Machine Learning: EECS 349 Fall 2012 1 The Generative Model POV We think of the data as being generated from some process. We assume
Regularized Discriminant Analysis and Reduced-Rank LDA
 Regularized Discriminant Analysis and Reduced-Rank LDA Department of Statistics The Pennsylvania State University Email: jiali@stat.psu.edu Regularized Discriminant Analysis A compromise between LDA and
Regularized Discriminant Analysis and Reduced-Rank LDA Department of Statistics The Pennsylvania State University Email: jiali@stat.psu.edu Regularized Discriminant Analysis A compromise between LDA and
An Extended BIC for Model Selection
 An Extended BIC for Model Selection at the JSM meeting 2007 - Salt Lake City Surajit Ray Boston University (Dept of Mathematics and Statistics) Joint work with James Berger, Duke University; Susie Bayarri,
An Extended BIC for Model Selection at the JSM meeting 2007 - Salt Lake City Surajit Ray Boston University (Dept of Mathematics and Statistics) Joint work with James Berger, Duke University; Susie Bayarri,
LECTURE NOTE #10 PROF. ALAN YUILLE
 LECTURE NOTE #10 PROF. ALAN YUILLE 1. Principle Component Analysis (PCA) One way to deal with the curse of dimensionality is to project data down onto a space of low dimensions, see figure (1). Figure
LECTURE NOTE #10 PROF. ALAN YUILLE 1. Principle Component Analysis (PCA) One way to deal with the curse of dimensionality is to project data down onto a space of low dimensions, see figure (1). Figure
Nonstationary spatial process modeling Part II Paul D. Sampson --- Catherine Calder Univ of Washington --- Ohio State University
 Nonstationary spatial process modeling Part II Paul D. Sampson --- Catherine Calder Univ of Washington --- Ohio State University this presentation derived from that presented at the Pan-American Advanced
Nonstationary spatial process modeling Part II Paul D. Sampson --- Catherine Calder Univ of Washington --- Ohio State University this presentation derived from that presented at the Pan-American Advanced
Introduction to Machine Learning
 1, DATA11002 Introduction to Machine Learning Lecturer: Teemu Roos TAs: Ville Hyvönen and Janne Leppä-aho Department of Computer Science University of Helsinki (based in part on material by Patrik Hoyer
1, DATA11002 Introduction to Machine Learning Lecturer: Teemu Roos TAs: Ville Hyvönen and Janne Leppä-aho Department of Computer Science University of Helsinki (based in part on material by Patrik Hoyer
Web Appendix for Hierarchical Adaptive Regression Kernels for Regression with Functional Predictors by D. B. Woodard, C. Crainiceanu, and D.
 Web Appendix for Hierarchical Adaptive Regression Kernels for Regression with Functional Predictors by D. B. Woodard, C. Crainiceanu, and D. Ruppert A. EMPIRICAL ESTIMATE OF THE KERNEL MIXTURE Here we
Web Appendix for Hierarchical Adaptive Regression Kernels for Regression with Functional Predictors by D. B. Woodard, C. Crainiceanu, and D. Ruppert A. EMPIRICAL ESTIMATE OF THE KERNEL MIXTURE Here we
Probabilistic Fisher Discriminant Analysis
 Probabilistic Fisher Discriminant Analysis Charles Bouveyron 1 and Camille Brunet 2 1- University Paris 1 Panthéon-Sorbonne Laboratoire SAMM, EA 4543 90 rue de Tolbiac 75013 PARIS - FRANCE 2- University
Probabilistic Fisher Discriminant Analysis Charles Bouveyron 1 and Camille Brunet 2 1- University Paris 1 Panthéon-Sorbonne Laboratoire SAMM, EA 4543 90 rue de Tolbiac 75013 PARIS - FRANCE 2- University
COM336: Neural Computing
 COM336: Neural Computing http://www.dcs.shef.ac.uk/ sjr/com336/ Lecture 2: Density Estimation Steve Renals Department of Computer Science University of Sheffield Sheffield S1 4DP UK email: s.renals@dcs.shef.ac.uk
COM336: Neural Computing http://www.dcs.shef.ac.uk/ sjr/com336/ Lecture 2: Density Estimation Steve Renals Department of Computer Science University of Sheffield Sheffield S1 4DP UK email: s.renals@dcs.shef.ac.uk
The Bayes classifier
 The Bayes classifier Consider where is a random vector in is a random variable (depending on ) Let be a classifier with probability of error/risk given by The Bayes classifier (denoted ) is the optimal
The Bayes classifier Consider where is a random vector in is a random variable (depending on ) Let be a classifier with probability of error/risk given by The Bayes classifier (denoted ) is the optimal
Clustering using Mixture Models
 Clustering using Mixture Models The full posterior of the Gaussian Mixture Model is p(x, Z, µ,, ) =p(x Z, µ, )p(z )p( )p(µ, ) data likelihood (Gaussian) correspondence prob. (Multinomial) mixture prior
Clustering using Mixture Models The full posterior of the Gaussian Mixture Model is p(x, Z, µ,, ) =p(x Z, µ, )p(z )p( )p(µ, ) data likelihood (Gaussian) correspondence prob. (Multinomial) mixture prior
Notes on Noise Contrastive Estimation (NCE)
 Notes on Noise Contrastive Estimation NCE) David Meyer dmm@{-4-5.net,uoregon.edu,...} March 0, 207 Introduction In this note we follow the notation used in [2]. Suppose X x, x 2,, x Td ) is a sample of
Notes on Noise Contrastive Estimation NCE) David Meyer dmm@{-4-5.net,uoregon.edu,...} March 0, 207 Introduction In this note we follow the notation used in [2]. Suppose X x, x 2,, x Td ) is a sample of
Bayesian Methods for Machine Learning
 Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Classification Methods II: Linear and Quadratic Discrimminant Analysis
 Classification Methods II: Linear and Quadratic Discrimminant Analysis Rebecca C. Steorts, Duke University STA 325, Chapter 4 ISL Agenda Linear Discrimminant Analysis (LDA) Classification Recall that linear
Classification Methods II: Linear and Quadratic Discrimminant Analysis Rebecca C. Steorts, Duke University STA 325, Chapter 4 ISL Agenda Linear Discrimminant Analysis (LDA) Classification Recall that linear
Data Preprocessing. Cluster Similarity
 1 Cluster Similarity Similarity is most often measured with the help of a distance function. The smaller the distance, the more similar the data objects (points). A function d: M M R is a distance on M
1 Cluster Similarity Similarity is most often measured with the help of a distance function. The smaller the distance, the more similar the data objects (points). A function d: M M R is a distance on M
Regularized Parameter Estimation in High-Dimensional Gaussian Mixture Models
 LETTER Communicated by Clifford Lam Regularized Parameter Estimation in High-Dimensional Gaussian Mixture Models Lingyan Ruan lruan@gatech.edu Ming Yuan ming.yuan@isye.gatech.edu School of Industrial and
LETTER Communicated by Clifford Lam Regularized Parameter Estimation in High-Dimensional Gaussian Mixture Models Lingyan Ruan lruan@gatech.edu Ming Yuan ming.yuan@isye.gatech.edu School of Industrial and
MACHINE LEARNING ADVANCED MACHINE LEARNING
 MACHINE LEARNING ADVANCED MACHINE LEARNING Recap of Important Notions on Estimation of Probability Density Functions 22 MACHINE LEARNING Discrete Probabilities Consider two variables and y taking discrete
MACHINE LEARNING ADVANCED MACHINE LEARNING Recap of Important Notions on Estimation of Probability Density Functions 22 MACHINE LEARNING Discrete Probabilities Consider two variables and y taking discrete
Mixture Models & EM. Nicholas Ruozzi University of Texas at Dallas. based on the slides of Vibhav Gogate
 Mixture Models & EM icholas Ruozzi University of Texas at Dallas based on the slides of Vibhav Gogate Previously We looed at -means and hierarchical clustering as mechanisms for unsupervised learning -means
Mixture Models & EM icholas Ruozzi University of Texas at Dallas based on the slides of Vibhav Gogate Previously We looed at -means and hierarchical clustering as mechanisms for unsupervised learning -means
ECE521 week 3: 23/26 January 2017
 ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
Communities, Spectral Clustering, and Random Walks
 Communities, Spectral Clustering, and Random Walks David Bindel Department of Computer Science Cornell University 26 Sep 2011 20 21 19 16 22 28 17 18 29 26 27 30 23 1 25 5 8 24 2 4 14 3 9 13 15 11 10 12
Communities, Spectral Clustering, and Random Walks David Bindel Department of Computer Science Cornell University 26 Sep 2011 20 21 19 16 22 28 17 18 29 26 27 30 23 1 25 5 8 24 2 4 14 3 9 13 15 11 10 12
Determining the number of components in mixture models for hierarchical data
 Determining the number of components in mixture models for hierarchical data Olga Lukočienė 1 and Jeroen K. Vermunt 2 1 Department of Methodology and Statistics, Tilburg University, P.O. Box 90153, 5000
Determining the number of components in mixture models for hierarchical data Olga Lukočienė 1 and Jeroen K. Vermunt 2 1 Department of Methodology and Statistics, Tilburg University, P.O. Box 90153, 5000
Data Mining Techniques
 Data Mining Techniques CS 6220 - Section 2 - Spring 2017 Lecture 6 Jan-Willem van de Meent (credit: Yijun Zhao, Chris Bishop, Andrew Moore, Hastie et al.) Project Project Deadlines 3 Feb: Form teams of
Data Mining Techniques CS 6220 - Section 2 - Spring 2017 Lecture 6 Jan-Willem van de Meent (credit: Yijun Zhao, Chris Bishop, Andrew Moore, Hastie et al.) Project Project Deadlines 3 Feb: Form teams of
MACHINE LEARNING ADVANCED MACHINE LEARNING
 MACHINE LEARNING ADVANCED MACHINE LEARNING Recap of Important Notions on Estimation of Probability Density Functions 2 2 MACHINE LEARNING Overview Definition pdf Definition joint, condition, marginal,
MACHINE LEARNING ADVANCED MACHINE LEARNING Recap of Important Notions on Estimation of Probability Density Functions 2 2 MACHINE LEARNING Overview Definition pdf Definition joint, condition, marginal,
12 Discriminant Analysis
 12 Discriminant Analysis Discriminant analysis is used in situations where the clusters are known a priori. The aim of discriminant analysis is to classify an observation, or several observations, into
12 Discriminant Analysis Discriminant analysis is used in situations where the clusters are known a priori. The aim of discriminant analysis is to classify an observation, or several observations, into
Latent Variable Models and Expectation Maximization
 Latent Variable Models and Expectation Maximization Oliver Schulte - CMPT 726 Bishop PRML Ch. 9 2 4 6 8 1 12 14 16 18 2 4 6 8 1 12 14 16 18 5 1 15 2 25 5 1 15 2 25 2 4 6 8 1 12 14 2 4 6 8 1 12 14 5 1 15
Latent Variable Models and Expectation Maximization Oliver Schulte - CMPT 726 Bishop PRML Ch. 9 2 4 6 8 1 12 14 16 18 2 4 6 8 1 12 14 16 18 5 1 15 2 25 5 1 15 2 25 2 4 6 8 1 12 14 2 4 6 8 1 12 14 5 1 15
Mixture Models & EM. Nicholas Ruozzi University of Texas at Dallas. based on the slides of Vibhav Gogate
 Mixture Models & EM icholas Ruozzi University of Texas at Dallas based on the slides of Vibhav Gogate Previously We looed at -means and hierarchical clustering as mechanisms for unsupervised learning -means
Mixture Models & EM icholas Ruozzi University of Texas at Dallas based on the slides of Vibhav Gogate Previously We looed at -means and hierarchical clustering as mechanisms for unsupervised learning -means
Lecture 4: Probabilistic Learning
 DD2431 Autumn, 2015 1 Maximum Likelihood Methods Maximum A Posteriori Methods Bayesian methods 2 Classification vs Clustering Heuristic Example: K-means Expectation Maximization 3 Maximum Likelihood Methods
DD2431 Autumn, 2015 1 Maximum Likelihood Methods Maximum A Posteriori Methods Bayesian methods 2 Classification vs Clustering Heuristic Example: K-means Expectation Maximization 3 Maximum Likelihood Methods
Unsupervised Learning
 2018 EE448, Big Data Mining, Lecture 7 Unsupervised Learning Weinan Zhang Shanghai Jiao Tong University http://wnzhang.net http://wnzhang.net/teaching/ee448/index.html ML Problem Setting First build and
2018 EE448, Big Data Mining, Lecture 7 Unsupervised Learning Weinan Zhang Shanghai Jiao Tong University http://wnzhang.net http://wnzhang.net/teaching/ee448/index.html ML Problem Setting First build and
Randomized Algorithms
 Randomized Algorithms Saniv Kumar, Google Research, NY EECS-6898, Columbia University - Fall, 010 Saniv Kumar 9/13/010 EECS6898 Large Scale Machine Learning 1 Curse of Dimensionality Gaussian Mixture Models
Randomized Algorithms Saniv Kumar, Google Research, NY EECS-6898, Columbia University - Fall, 010 Saniv Kumar 9/13/010 EECS6898 Large Scale Machine Learning 1 Curse of Dimensionality Gaussian Mixture Models
STATS306B STATS306B. Discriminant Analysis. Jonathan Taylor Department of Statistics Stanford University. June 3, 2010
 STATS306B Discriminant Analysis Jonathan Taylor Department of Statistics Stanford University June 3, 2010 Spring 2010 Classification Given K classes in R p, represented as densities f i (x), 1 i K classify
STATS306B Discriminant Analysis Jonathan Taylor Department of Statistics Stanford University June 3, 2010 Spring 2010 Classification Given K classes in R p, represented as densities f i (x), 1 i K classify
STATISTICAL LEARNING SYSTEMS
 STATISTICAL LEARNING SYSTEMS LECTURE 8: UNSUPERVISED LEARNING: FINDING STRUCTURE IN DATA Institute of Computer Science, Polish Academy of Sciences Ph. D. Program 2013/2014 Principal Component Analysis
STATISTICAL LEARNING SYSTEMS LECTURE 8: UNSUPERVISED LEARNING: FINDING STRUCTURE IN DATA Institute of Computer Science, Polish Academy of Sciences Ph. D. Program 2013/2014 Principal Component Analysis
Unsupervised Learning Techniques Class 07, 1 March 2006 Andrea Caponnetto
 Unsupervised Learning Techniques 9.520 Class 07, 1 March 2006 Andrea Caponnetto About this class Goal To introduce some methods for unsupervised learning: Gaussian Mixtures, K-Means, ISOMAP, HLLE, Laplacian
Unsupervised Learning Techniques 9.520 Class 07, 1 March 2006 Andrea Caponnetto About this class Goal To introduce some methods for unsupervised learning: Gaussian Mixtures, K-Means, ISOMAP, HLLE, Laplacian
CS534 Machine Learning - Spring Final Exam
 CS534 Machine Learning - Spring 2013 Final Exam Name: You have 110 minutes. There are 6 questions (8 pages including cover page). If you get stuck on one question, move on to others and come back to the
CS534 Machine Learning - Spring 2013 Final Exam Name: You have 110 minutes. There are 6 questions (8 pages including cover page). If you get stuck on one question, move on to others and come back to the
ECE 521. Lecture 11 (not on midterm material) 13 February K-means clustering, Dimensionality reduction
 ECE 521 Lecture 11 (not on midterm material) 13 February 2017 K-means clustering, Dimensionality reduction With thanks to Ruslan Salakhutdinov for an earlier version of the slides Overview K-means clustering
ECE 521 Lecture 11 (not on midterm material) 13 February 2017 K-means clustering, Dimensionality reduction With thanks to Ruslan Salakhutdinov for an earlier version of the slides Overview K-means clustering
Chapter 10. Semi-Supervised Learning
 Chapter 10. Semi-Supervised Learning Wei Pan Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455 Email: weip@biostat.umn.edu PubH 7475/8475 c Wei Pan Outline
Chapter 10. Semi-Supervised Learning Wei Pan Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455 Email: weip@biostat.umn.edu PubH 7475/8475 c Wei Pan Outline
Latent Variable Models and EM algorithm
 Latent Variable Models and EM algorithm SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic 3.1 Clustering and Mixture Modelling K-means and hierarchical clustering are non-probabilistic
Latent Variable Models and EM algorithm SC4/SM4 Data Mining and Machine Learning, Hilary Term 2017 Dino Sejdinovic 3.1 Clustering and Mixture Modelling K-means and hierarchical clustering are non-probabilistic
Label Switching and Its Simple Solutions for Frequentist Mixture Models
 Label Switching and Its Simple Solutions for Frequentist Mixture Models Weixin Yao Department of Statistics, Kansas State University, Manhattan, Kansas 66506, U.S.A. wxyao@ksu.edu Abstract The label switching
Label Switching and Its Simple Solutions for Frequentist Mixture Models Weixin Yao Department of Statistics, Kansas State University, Manhattan, Kansas 66506, U.S.A. wxyao@ksu.edu Abstract The label switching
Curves clustering with approximation of the density of functional random variables
 Curves clustering with approximation of the density of functional random variables Julien Jacques and Cristian Preda Laboratoire Paul Painlevé, UMR CNRS 8524, University Lille I, Lille, France INRIA Lille-Nord
Curves clustering with approximation of the density of functional random variables Julien Jacques and Cristian Preda Laboratoire Paul Painlevé, UMR CNRS 8524, University Lille I, Lille, France INRIA Lille-Nord
Lecture 4: Probabilistic Learning. Estimation Theory. Classification with Probability Distributions
 DD2431 Autumn, 2014 1 2 3 Classification with Probability Distributions Estimation Theory Classification in the last lecture we assumed we new: P(y) Prior P(x y) Lielihood x2 x features y {ω 1,..., ω K
DD2431 Autumn, 2014 1 2 3 Classification with Probability Distributions Estimation Theory Classification in the last lecture we assumed we new: P(y) Prior P(x y) Lielihood x2 x features y {ω 1,..., ω K
Introduction to Machine Learning
 Introduction to Machine Learning Brown University CSCI 1950-F, Spring 2012 Prof. Erik Sudderth Lecture 20: Expectation Maximization Algorithm EM for Mixture Models Many figures courtesy Kevin Murphy s
Introduction to Machine Learning Brown University CSCI 1950-F, Spring 2012 Prof. Erik Sudderth Lecture 20: Expectation Maximization Algorithm EM for Mixture Models Many figures courtesy Kevin Murphy s
Regression Clustering
 Regression Clustering In regression clustering, we assume a model of the form y = f g (x, θ g ) + ɛ g for observations y and x in the g th group. Usually, of course, we assume linear models of the form
Regression Clustering In regression clustering, we assume a model of the form y = f g (x, θ g ) + ɛ g for observations y and x in the g th group. Usually, of course, we assume linear models of the form
Neural Network Training
 Neural Network Training Sargur Srihari Topics in Network Training 0. Neural network parameters Probabilistic problem formulation Specifying the activation and error functions for Regression Binary classification
Neural Network Training Sargur Srihari Topics in Network Training 0. Neural network parameters Probabilistic problem formulation Specifying the activation and error functions for Regression Binary classification
CS6220: DATA MINING TECHNIQUES
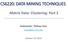 CS6220: DATA MINING TECHNIQUES Matrix Data: Clustering: Part 2 Instructor: Yizhou Sun yzsun@ccs.neu.edu October 19, 2014 Methods to Learn Matrix Data Set Data Sequence Data Time Series Graph & Network
CS6220: DATA MINING TECHNIQUES Matrix Data: Clustering: Part 2 Instructor: Yizhou Sun yzsun@ccs.neu.edu October 19, 2014 Methods to Learn Matrix Data Set Data Sequence Data Time Series Graph & Network
Latent Variable Models and Expectation Maximization
 Latent Variable Models and Expectation Maximization Oliver Schulte - CMPT 726 Bishop PRML Ch. 9 2 4 6 8 1 12 14 16 18 2 4 6 8 1 12 14 16 18 5 1 15 2 25 5 1 15 2 25 2 4 6 8 1 12 14 2 4 6 8 1 12 14 5 1 15
Latent Variable Models and Expectation Maximization Oliver Schulte - CMPT 726 Bishop PRML Ch. 9 2 4 6 8 1 12 14 16 18 2 4 6 8 1 12 14 16 18 5 1 15 2 25 5 1 15 2 25 2 4 6 8 1 12 14 2 4 6 8 1 12 14 5 1 15
arxiv: v1 [stat.ml] 22 Jun 2012
![arxiv: v1 [stat.ml] 22 Jun 2012 arxiv: v1 [stat.ml] 22 Jun 2012](/thumbs/75/71627085.jpg) Hidden Markov Models with mixtures as emission distributions Stevenn Volant 1,2, Caroline Bérard 1,2, Marie-Laure Martin Magniette 1,2,3,4,5 and Stéphane Robin 1,2 arxiv:1206.5102v1 [stat.ml] 22 Jun 2012
Hidden Markov Models with mixtures as emission distributions Stevenn Volant 1,2, Caroline Bérard 1,2, Marie-Laure Martin Magniette 1,2,3,4,5 and Stéphane Robin 1,2 arxiv:1206.5102v1 [stat.ml] 22 Jun 2012
Hidden Markov Models and Gaussian Mixture Models
 Hidden Markov Models and Gaussian Mixture Models Hiroshi Shimodaira and Steve Renals Automatic Speech Recognition ASR Lectures 4&5 23&27 January 2014 ASR Lectures 4&5 Hidden Markov Models and Gaussian
Hidden Markov Models and Gaussian Mixture Models Hiroshi Shimodaira and Steve Renals Automatic Speech Recognition ASR Lectures 4&5 23&27 January 2014 ASR Lectures 4&5 Hidden Markov Models and Gaussian
Communications in Statistics - Simulation and Computation. Comparison of EM and SEM Algorithms in Poisson Regression Models: a simulation study
 Comparison of EM and SEM Algorithms in Poisson Regression Models: a simulation study Journal: Manuscript ID: LSSP-00-0.R Manuscript Type: Original Paper Date Submitted by the Author: -May-0 Complete List
Comparison of EM and SEM Algorithms in Poisson Regression Models: a simulation study Journal: Manuscript ID: LSSP-00-0.R Manuscript Type: Original Paper Date Submitted by the Author: -May-0 Complete List
EXAM IN STATISTICAL MACHINE LEARNING STATISTISK MASKININLÄRNING
 EXAM IN STATISTICAL MACHINE LEARNING STATISTISK MASKININLÄRNING DATE AND TIME: August 30, 2018, 14.00 19.00 RESPONSIBLE TEACHER: Niklas Wahlström NUMBER OF PROBLEMS: 5 AIDING MATERIAL: Calculator, mathematical
EXAM IN STATISTICAL MACHINE LEARNING STATISTISK MASKININLÄRNING DATE AND TIME: August 30, 2018, 14.00 19.00 RESPONSIBLE TEACHER: Niklas Wahlström NUMBER OF PROBLEMS: 5 AIDING MATERIAL: Calculator, mathematical
applications Rome, 9 February Università di Roma La Sapienza Robust model based clustering: methods and applications Francesco Dotto Introduction
 model : fuzzy model : Università di Roma La Sapienza Rome, 9 February Outline of the presentation model : fuzzy 1 General motivation 2 algorithm on trimming and reweigthing. 3 algorithm on trimming and
model : fuzzy model : Università di Roma La Sapienza Rome, 9 February Outline of the presentation model : fuzzy 1 General motivation 2 algorithm on trimming and reweigthing. 3 algorithm on trimming and
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistical Sciences! rsalakhu@cs.toronto.edu! h0p://www.cs.utoronto.ca/~rsalakhu/ Lecture 7 Approximate
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistical Sciences! rsalakhu@cs.toronto.edu! h0p://www.cs.utoronto.ca/~rsalakhu/ Lecture 7 Approximate
Model-Based Co-Clustering of Multivariate Functional Data
 Model-Based Co-Clustering of Multivariate Functional Data Faicel Chamroukhi, Christophe Biernacki To cite this version: Faicel Chamroukhi, Christophe Biernacki. Model-Based Co-Clustering of Multivariate
Model-Based Co-Clustering of Multivariate Functional Data Faicel Chamroukhi, Christophe Biernacki To cite this version: Faicel Chamroukhi, Christophe Biernacki. Model-Based Co-Clustering of Multivariate
Hmms with variable dimension structures and extensions
 Hmm days/enst/january 21, 2002 1 Hmms with variable dimension structures and extensions Christian P. Robert Université Paris Dauphine www.ceremade.dauphine.fr/ xian Hmm days/enst/january 21, 2002 2 1 Estimating
Hmm days/enst/january 21, 2002 1 Hmms with variable dimension structures and extensions Christian P. Robert Université Paris Dauphine www.ceremade.dauphine.fr/ xian Hmm days/enst/january 21, 2002 2 1 Estimating
Introduction. Chapter 1
 Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
Factor Analysis (10/2/13)
 STA561: Probabilistic machine learning Factor Analysis (10/2/13) Lecturer: Barbara Engelhardt Scribes: Li Zhu, Fan Li, Ni Guan Factor Analysis Factor analysis is related to the mixture models we have studied.
STA561: Probabilistic machine learning Factor Analysis (10/2/13) Lecturer: Barbara Engelhardt Scribes: Li Zhu, Fan Li, Ni Guan Factor Analysis Factor analysis is related to the mixture models we have studied.
STA414/2104. Lecture 11: Gaussian Processes. Department of Statistics
 STA414/2104 Lecture 11: Gaussian Processes Department of Statistics www.utstat.utoronto.ca Delivered by Mark Ebden with thanks to Russ Salakhutdinov Outline Gaussian Processes Exam review Course evaluations
STA414/2104 Lecture 11: Gaussian Processes Department of Statistics www.utstat.utoronto.ca Delivered by Mark Ebden with thanks to Russ Salakhutdinov Outline Gaussian Processes Exam review Course evaluations
Lecture 6: Methods for high-dimensional problems
 Lecture 6: Methods for high-dimensional problems Hector Corrada Bravo and Rafael A. Irizarry March, 2010 In this Section we will discuss methods where data lies on high-dimensional spaces. In particular,
Lecture 6: Methods for high-dimensional problems Hector Corrada Bravo and Rafael A. Irizarry March, 2010 In this Section we will discuss methods where data lies on high-dimensional spaces. In particular,
Gaussian Mixture Models with Component Means Constrained in Pre-selected Subspaces
 Gaussian Mixture Models with Component Means Constrained in Pre-selected Subspaces Mu Qiao and Jia Li Abstract We investigate a Gaussian mixture model (GMM) with component means constrained in a pre-selected
Gaussian Mixture Models with Component Means Constrained in Pre-selected Subspaces Mu Qiao and Jia Li Abstract We investigate a Gaussian mixture model (GMM) with component means constrained in a pre-selected
Model-based clustering for conditionally correlated categorical data
 Model-based clustering for conditionally correlated categorical data arxiv:1401.5684v3 [stat.co] 10 Jul 2014 Matthieu Marbac Inria & DGA Christophe Biernacki University Lille 1 & CNRS & Inria Vincent Vandewalle
Model-based clustering for conditionally correlated categorical data arxiv:1401.5684v3 [stat.co] 10 Jul 2014 Matthieu Marbac Inria & DGA Christophe Biernacki University Lille 1 & CNRS & Inria Vincent Vandewalle
MSA220 Statistical Learning for Big Data
 MSA220 Statistical Learning for Big Data Lecture 4 Rebecka Jörnsten Mathematical Sciences University of Gothenburg and Chalmers University of Technology More on Discriminant analysis More on Discriminant
MSA220 Statistical Learning for Big Data Lecture 4 Rebecka Jörnsten Mathematical Sciences University of Gothenburg and Chalmers University of Technology More on Discriminant analysis More on Discriminant
Lecture 5. Gaussian Models - Part 1. Luigi Freda. ALCOR Lab DIAG University of Rome La Sapienza. November 29, 2016
 Lecture 5 Gaussian Models - Part 1 Luigi Freda ALCOR Lab DIAG University of Rome La Sapienza November 29, 2016 Luigi Freda ( La Sapienza University) Lecture 5 November 29, 2016 1 / 42 Outline 1 Basics
Lecture 5 Gaussian Models - Part 1 Luigi Freda ALCOR Lab DIAG University of Rome La Sapienza November 29, 2016 Luigi Freda ( La Sapienza University) Lecture 5 November 29, 2016 1 / 42 Outline 1 Basics
