A. J. BORDNER, G. A. HUBER Department of Bioengineering, University of California, San Diego, 9500 Gilman Drive, San Diego, California
|
|
|
- Solomon Stafford
- 5 years ago
- Views:
Transcription
1 Boundary Element Solution of the Linear Poisson Boltzmann Equation and a Multipole Method for the Rapid Calculation of Forces on Macromolecules in Solution A. J. BORDNER, G. A. HUBER Department of Bioengineering, University of California, San Diego, 9500 Gilman Drive, San Diego, California Received 7 May 2002; Accepted 23 August 2002 Abstract: The Poisson Boltzmann equation is widely used to describe the electrostatic potential of molecules in an ionic solution that is treated as a continuous dielectric medium. The linearized form of this equation, applicable to many biologic macromolecules, may be solved using the boundary element method. A single-layer formulation of the boundary element method, which yields simpler integral equations than the direct formulations previously discussed in the literature, is given. It is shown that the electrostatic force and torque on a molecule may be calculated using its boundary element representation and also the polarization charge for two rigid molecules may be rapidly calculated using a noniterative scheme. An algorithm based on a fast adaptive multipole method is introduced to further increase the speed of the calculation. This method is particularly suited for Brownian dynamics or molecular dynamics simulations of large molecules, in which the electrostatic forces must be calculated for many different relative positions and orientations of the molecules. It has been implemented as a set of programs in C, which are used to study the accuracy and speed of this method for two actin monomers Wiley Periodicals, Inc. J Comput Chem 24: , 2003 Key words: Poisson Boltzmann equation; boundary element method; cell multipole method; Brownian dynamics; molecular dynamics Introduction Electrostatic interactions are important in determining the structure, thermodynamic properties, and reaction kinetics of macromolecules in solution. In particular, for noncovalent protein association reactions Monte Carlo calculations of the thermodynamic quantities and Brownian dynamics calculations of reaction rates require evaluating the electrostatic interaction energy and force, respectively, many times during the calculation. The water may be represented in atomic detail, as in molecular dynamics simulations; however, this is computationally intensive. Alternatively, treating the water as a continuum medium with macroscopic properties, such as a bulk dielectric constant, allows a faster computation of the electrostatic and hydrodynamic effects. 3 Methods for evaluating the electrostatic potential in continuum solvation models include the generalized Born method, 4,5 the effective charge method of ref. 6, and various numerical methods for direct solution of the Poisson Boltzmann equation. In the latter case the charge density of the molecules is usually approximately represented by point charges located at the atomic centers. The interior of the molecule, delimited by the solvent-excluded surface, has a low dielectric constant (2 4), whereas the exterior has a high dielectric constant (80). We will be primarily interested in simulations of proteins under physiologic conditions in which ions are present and are included in the electrostatic calculation by assuming that the electric potential satisfies the Poisson Boltzmann equation. When the electrostatic energy of the ions is much less than their thermal energy, i.e. q/kt, the nonlinear Poisson Boltzmann equation may be linearized. This approximation is reasonably accurate for molecules with a relatively low charge density in solutions at physiologic ion concentrations (0. M). 7 The boundary element method (BEM) for solving the linear Poisson Boltzmann equation has several advantages over two Correspondence to: A. J. Bordner, The Scripps Research Institute, 0550 North Torrey Pines Rd., TPC-28, San Diego, CA 92037; bordner@scripps.edu Contract/grant sponsor: National Institutes of Health (to A.J.B.) 2003 Wiley Periodicals, Inc.
2 354 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry other numerical methods: the finite difference and finite element methods. One is that infinite domains may be simply treated without introducing a large artificial boundary. Another advantage is that the equations are defined over the two-dimensional molecular surface rather than a three-dimensional domain so that a smaller number of finite elements, resulting from discretizing these equations on a mesh, are required. Also, there is no need to distribute the partial atomic charges over nearby node points, as in the other two methods. Finally, because the electrostatic potential is represented by charge distributions on the molecular surfaces it may be rapidly calculated for a large number of different relative configurations by calculating the perturbation of the charge density for an isolated molecule due to the electric field of the other molecule. This is because, unlike the other two numerical methods, the discretization mesh moves with the molecules and is not fixed in space. This property will be the basis for our method. The BEM was first applied to the case of zero ionic strength, in which the potential is a solution of the Poisson equation. 8 Studies that include ions in solution may be found in refs Several recent articles have used multipole or adaptive grid methods to reduce the size of the matrix resulting from the BEM for the Poisson equation and thus reducing storage requirements and solution time. References 7 9 used methods based on the cell multipole algorithm of refs. 20 and 2, whereas ref. 22 used a multilevel grid on the surface to reduce the size of the system of linear equations. In this article we will first describe a BEM for solving the linear Poisson Boltzmann equation for two proteins in an ionic solution based on a single-layer formulation of the equations. This gives a simpler set of equations on the boundary and hence a more efficient starting point for solving them than the direct formulation of the boundary integral equations based on Green s theorem used in previous studies. More importantly, this will allow us to use a method based on the cell multipole algorithm to rapidly evaluate the force and torque on one molecule due to the electric potential of the other. It should be noted that the single-layer formulation of the Poisson equation yields matrix equations of half the size of those presented in this article and, hence, are preferable in the case of zero ionic strength.,23 Our cell multipole method is applicable to rigid molecules because the calculation of the fields of the isolated molecules needs to be done only once before the simulation begins. The relatively slow calculations, namely that of the surface charge distribution on an isolated molecule s surface using the BEM and this distribution s multipole moments, up to quadrupole order, within a multilevel set of cubic cells are done at leisure before the simulation begins. The calculation of the total force and torque on a molecule are then done rapidly by using an adaptive cell multipole algorithm. Basically, the potential at one molecule due to the charges of the other are calculated using the cell multipole algorithm and a local Taylor expansion about the center of a cell in the first molecule. The contributions to the force and torque due to this quadratic approximation to the electric field within the cells on the first molecule are then summed. Actually, linear combinations of the multipole and Taylor coefficients, that are coefficients in an expansion in orthogonal polynomials within each cell, are used. The next section describes the boundary element solution of the Poisson Boltzmann equation and some associated numerical methods. Then we discuss the adaptive multipole algorithm for fast calculation of the force and torque on a rigid molecule. The error and computational speed are then tested on a system of two actin monomers, and finally, the results and possible areas of further investigation are discussed. Boundary Element Solution of the Poisson Boltzmann Equation Boundary Integral Equations The first step in the BEM is to formulate the solution of a boundary value problem for a partial differential equation in a region in terms of integral equations on its boundary, called the boundary integral equations (BIE). In the literature, previous applications of the boundary element method to solve the linear Poisson Boltzmann equation for the electric potential of macromolecules in an ionic solution used the direct formulation of the BIE, in which the BIE are derived using Green s theorem. However the single-layer formulation of a boundary integral equation, when it exists, always yields simpler equations, and hence, a more efficient computational scheme. We consider the case of two molecules in an ionic solution which occupy regions and 2, delimited by the solvent-excluded, or Connolly, surfaces of the respective molecules. The solvent-excluded surface is formed by the boundary of the region in which a probe sphere, typically the size of a water oxygen Van der Waals radius of around.4 Å, is excluded from the Van der Waals volume of the molecule. 24 Outside of these regions, i.e., in the solution, the potential satisfies the linear Poisson Boltzmann equation without sources where the inverse Debye length is 2 x 2 x 0 (.) 8n 2 e2 out kt (.2) for a neutral solution of monovalent ions of concentration n. e is the fundamental electronic charge, out is the dielectric constant in the solution, k is the Boltzmann constant and T is the absolute temperature. Inside each region the potential satisfies the Poisson equation 2 x 4 x 4 in j N C in k q k j x x k j, x j, j, 2 (.3) where the partial atomic charges q k ( j) are located at the atomic centers x k ( j) within j and in is the dielectric constant inside the molecules. The fundamental solution or Green s function E P ( x) for the Poisson equation is
3 Boundary Element Solution of Linear Poisson Boltzmann Equation 355 and satisfies Next define and E P x 4x (.4) 2 E P x x. (.5) u j 4 dxe P x xx, inj j N C in k q k j x x k (.6) The potential in the exterior region x c 2 c may be described by a single-layer representation in terms of charge densities h j ( x) as x dxe PB, x xh j x j,2j 4 dx exx x x h j x (.4) j,2j The solutions of the Poisson Boltzmann equation for the inside regions [eq. (.)], and the outside region [eq. (.4)], must satisfy the following boundary conditions on j for y j lim x lim x, (.5) x3y x3y x j x j c j( x) then satisfies the Laplace equation inside j, because j x x u j x. (.7) 2 jx 0 (.8) 2 u j x 4 in x. (.9) j( x) inside j may then be represented by a single-layer representation in terms of a surface charge density f j ( x) as 23 lim x3y in nˆ x lim out nˆ x (.6) x3y x j x j c with nˆ the outward pointing unit length normal vector to j. The integrals in the expressions for the potential in each region also satisfy the following jump conditions at the boundaries j for y j 23 lim dxe P x x fx dxe P y x fx, (.7) x3y x jj j j x j dxe P x x f j x (.0) lim dxe PB x x fx dxe PB y x fx, (.8) x3y x cj j j so that the potential inside j is x j N C j q in k x x j k k f j x dx x x. (.) 4j Because there are no charges in the exterior region the potential there satisfies the Poisson Boltzmann equation without sources [eq. (.)]. The fundamental solution E PB (, x) for this equation is which satisfies E PB, x ex x (.2) 2 2 E PB, x x. (.3) lim dxnˆ x E P x x fx x3y x jj dxnˆ y E P y x fx fy, (.9) 2 j lim dxnˆ x E PB x x fx x3y x cj j dxnˆ y E PB y x fx fy. (.20) 2 j Using these jump conditions eq. (.5) becomes
4 356 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry N, , N 2, 2 2, N 3, , N 4, 2 4 2, N 5, , Figure. Mapping x(, 2 ) from local coordinates to a triangular quadratic surface element. j N C j q k in x x k k dx 4j and eq. (.6) becomes k f j x x x 4 dx exx x x h kx, k,2k j N C in q j k nˆx x x x k x j dxf j xnˆx x x x 2 f jx 4j out in e 4 xx dxh k xnˆx x x x k,2k (.2) 2 h jx, x j. (.22) Surface Discretization and Boundary Element Equations The BIE of eqs. (.2) and (.22) are then solved for f,2 ( x) and h,2 ( x) by first discretizing both the boundary surfaces and the function space in terms of finite elements. We choose to use quadratic surface elements and linear function elements. The solvent-excluded surfaces of the molecules,2 are approximated by a closed mesh of triangles with quadratic curves as edges. Local coordinates (, 2 ) for each triangle T m ( j) on the mesh for surface j, such that they map the standard triangle, with vertices at (0, 0), (, 0), and (0, ) onto the triangular surface element defined by the six vertices {v,...,v 6 } are (see Fig. ) N 6, (.24) The function N j (, 2 ) is at vertex v j and 0 at the other vertices. A function defined on the surface h j (x), x j is approximated by linear function elements {H k (j) (x), k,...,n V }as j N V h x h j k H j k x (.25) k where N V ( j) is the number of vertices in the triangulation of j.a component of the vector h j,(h j ) k, is the value of h j ( x) at vertex k. H k ( j) ( x) is linear in the local coordinates and 2, at vertex k and 0 at the other vertices so that only the vertices of triangles containing x contribute to the sum in eq. (.25). On surface element T m H k ( x Tm (, 2 )) is one of the functions S j (, 2 ), j, 2, 3 defined by S, 2 2, S 2, 2, S 3, 2 2 (.26) depending on whether vertex k is at (0, 0), (, 0), or (0, ), ( respectively, in the local coordinates on T j) m. We denote the ( appropriate function for H j) ( k ( x) ont j) ( m as S j) k,m (, 2 ). The four BIE of eqs. (.2) and (.22) are then solved using the method of collocation in which the discrete approximation to the solution is assumed to be exact at a finite number of node points, which in this case are the corner vertices of the triangles. This then yields a system of linear equations in terms of the coefficients in the expansion of the surface charge densities f,2 ( x) and h,2 ( x) in terms of the finite elements. Define the following integrals I j l x, 2 2 m l jt m j m l j0 dx exx x x H l j x, d 0 d 2 x T m j 6 x Tm, 2 N j, 2 v j (.23) j j, 2 exxtm x x j Tm, 2 S l,m j, 2, with the shape functions N j (, 2 ) defined as I j 2l x, nˆx x I j l x, (.27)
5 Boundary Element Solution of Linear Poisson Boltzmann Equation 357 where l ( j) is the set of indices for surface elements that contain vertex l on the mesh for j. The Jacobian factor in the integral x j T m x j Tm, 2 2 x j T m, 2. (.28) may be calculated from eq. (.23). The integrals in eqs. (.27) are calculated using two-dimensional Gaussian quadrature as described in Appendix A. Next we define the following matrix elements in terms of the integrals in eq. (.27) A j mn I j n v j m,0, j, 2 B jk mn I k n v j m,, j, k, 2 C j mn I j 2n v j m,0 mn, j, 2 Iterative Solution of Boundary Element Equations Another approach to solving eq. (.3) is a perturbative method in which the surface charge densities for two molecules in isolation are first calculated and then the relatively small change in these densities due to the charges of the other molecule are determined. This is a fast method to calculate the charge densities for a large number of different relative configurations of the two rigid molecules. Although we will use a faster, but approximate, noniterative method that is described later to solve for the polarization charge in the fast multipole version of the BEM, we first review an iterative method of this type, developed by Zhou in ref. 4, that may be adapted to solve eq. (.3). The results of this method will be used for comparison with our one-step algorithm. In the iterative scheme the surface charge densities for an isolated molecule in solution are first given by solving the equations f j 0 A j B jj h j s j, D jj mn out I j 2n v j m, mn, j, 2 in where h j 0 U j t j C j A j s j (.32) D jk mn out I k 2n v j m,, j k, 2 (.29) in and the vector elements s k j 2 in t k j 2 in j N C m j N C m q m j v k j x m j q m j nˆv k j v k j x m j v k j x m j 3 (.30) where v m ( j) is the coordinate of vertex m on surface j and x m ( j) is the coordinate of atomic charge q m ( j). Furthermore, we define vectors of coefficients f and f 2 in the expansion of the surface charge densities f ( x) and f 2 ( x), respectively, in terms of the finite function elements according to eq. (.25). Likewise, vectors h and h 2 are defined for the surface charge densities h ( x) and h 2 ( x), respectively. Equations (.2) and (.22) then become v A f B h B 2 h 2, v 2 A 2 f 2 B 2 h B 22 h 2, w C f D h D 2 h 2, w 2 C 2 f 2 D 2 h D 22 h 2. (.3) These linear equations may then be solved for the charge density vectors f,2 and h,2 using a suitable iterative method with preconditioner. U j D jj C j A j B jj. (.33) Then substituting f j f j 0 f j and h j h j 0 h j in eq. (.3) gives the following equations for the changes in the surface charge densities due to the electric field of the other molecule, h j and f j,as h U D 2 C A B 2 h 2 h 2, f A B h B 2 h 2 h 2 (.34) and likewise for f 2 and h 2. These equations may be solved to the desired accuracy by starting with f j h j 0 and iteratively substituting the previous values for f j and h j into eq. (.34) to obtain the value for the next iteration. One-Step Solution of Boundary Element Equations Next, we will derive a one-step calculation to approximate h j and f j as a faster alternative to the iterative solution of eq. (.34). The method is to approximate the change in the charge density of molecule, after introducing molecule 2, by solving the Poisson Boltzmann boundary value problem for molecule without any internal point charges, i.e. simply a dielectric cavity, and in the presence of an externally applied potential due only to molecule 2. Of course, the resulting change in charge density on one molecule, in turn, induces a correction to the charge density on the other one and these corrections must be evaluated iteratively to obtain an exact result. However, for values of the dielectric constants relevant to protein molecules in water, each succeeding correction to the charge density is much smaller than the previous one, and hence, the first term is a reasonably accurate approximation. The boundary value problem for the dielectric cavity of molecule in the presence of an external field is again solved using the
6 358 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry boundary element method. There is a surface charge density on the surface due to an externally applied electric field with potential ext ( x), no free charges inside, and a constant dielectric coefficient in inside of and out outside of,asinthe previous derivation. Also, as before, ions are present outside of but excluded from the inside. The boundary integral equations for each region are then where U is defined in eq. (.33). Substituting the potential due to an isolated molecule 2 for the external potential gives the charge on molecule due to presence of molecule 2 as h j 2 out in k U jk x ext x dxe P x xf x, x, nˆx x2 dx exx x x h 2xxxk (.42) x ext x dxe PB x xh x, x c (.35) with E P ( x x) and E PB ( x x) defined in eq. (.4) and eq. (.2), respectively. Matching the solutions in the two regions at the boundary using eq. (.5) and (.6) and using the jump conditions of eq. (.7) (.20) gives the boundary integral equations dxf x x x dxh x exx x x, 4 out in dxf xnˆ x x x x 2 f x dxh xnˆx x out 4 x (.36) e xx x x 2 x h in nˆx ext x, x. (.37) After discretizing the BIE, as before, and defining the components of vector u as u k 2 out in nˆx k ext x k (.38) where the x k are the nodal coordinates for the mesh of and using the matrices A, B, C, and D, defined in eq. (.29), gives the linear equations A f B h, (.39) C f D h u. (.40) Only h is needed, with its value given by solving the preceding equations to yield h U u, (.4) which, upon substituting the discretized form of the integral, gives h in U D 2 h 2. (.43) out The corresponding equation for h 2 may be obtained by exchanging indices 7 2. Force and Torque Calculation We next describe a simple method to calculate the force and torque on one molecule due to the electric field of the other using the surface charge densities of the boundary element representation. Consider the force, F 2, on molecule due to the field of molecule 2. This may be calculated by integrating the normal component of the electromagnetic stress tensor T jk x out 4 E jxe k x 8 E x 2 jk kt i n i x jk out 4 E j xe k x 8 E x 2 jk 2nqx (.44) over a surface S surrounding molecule. The second equation is obtained by substituting the value of the ionic concentrations of the two species of oppositely charged ions implicit in the linear Poisson Boltzmann equation n ( x) n ( x) 2n q( x)/kt. Because the last term for the contribution of the ionic pressure is negligible for the range of ionic concentrations considered under physiologic conditions we use only the electric field contribution 25 T jk E x out 4 E j xe k x 8 E x 2 jk (.45) and the force on molecule is then F 2 S dxt E x nˆx (.46) where nˆ ( x) is the unit normal to the surface at x. For our purposes, the important fact about this expression is that the force depends only on the value of the electric field on the surface S. Thus, any other charge distributions that generates the same values of E ( x)
7 Boundary Element Solution of Linear Poisson Boltzmann Equation 359 on S will experience the same force. Because the boundary integral representation of the potential in the region exterior to the molecular surfaces, eq. (.4) is the same as that for surface charge densities j x out 4 h jx, x j (.47) on the respective molecular surfaces in a surrounding medium with dielectric constant out and Debye parameter the force is simply F 2 dx xe 2 x. (.48) Note that j ( x) from the boundary integral representation is not the same as the physical charge density that exists at the dielectric interface. Substituting E 2 x 2 x 42 gives the expression for the total force dxh 2 x x e xx x x (.49) F 2 out e xx 6 2 dxh xh 2 x x x x dx2. (.50) Using a similar argument, the torque on molecule due to the charges on molecule 2 is T 2 out e xx 6 2 dxh xh 2 xx x 0 x x x dx2. (.5) Fast Force Evaluation Using an Adaptive Multipole Algorithm Next we describe a method that uses a variation of the cell multipole method along with the BEM for solving the linear Poisson Boltzmann equation shown earlier to rapidly calculate the force and torque on the molecules due to electrostatic interactions. As will be shown in the next section, the polarization corrections to the charge densities of each molecule due to the electric field of the other only adds a small correction to the calculated force and torque. Because it would be computationally expensive, relative to their magnitude, to calculate these corrections only the charge densities h j 0 from the BEM solution of the isolated molecules are used in the multipole method. Also, because this method is designed to be part of a Brownian dynamics or molecular dynamics simulation, as many quantities as possible are precomputed before the simulation begins. The speed of these computations are therefore considered unimportant. The cell multipole method is a fast, efficient algorithm to calculate the electric potential due to a set of particles that interact according to a pairwise potential. 20,2 In this method, a cubic cell is defined such that it contains all of the particles of interest. This cell is then equally subdivided into eight smaller cubic cells, and this procedure iterated until no cell at the finest level contains too many particles. The grouping of the cells is represented by an octatree data structure in which any given cell is linked to its eight daughter cells. Contributions to the potential are then calculated using a multipole expansion about the center of a cell containing the source charges and a Taylor expansion about the center of a cell containing the field point. The cells are chosen such that the source cells and field cells each form a set that covers the largest cubic cell and the sizes of the cells are such that the approximation error is less than a specified value. It is useful to define a set of unique indices for each cubic cell in the octatree structure with N L levels. The cells will be denoted by C j,k, where j 0,..., N L is the level number, and the cells within a level are indexed by a vector k with integer components and 0 (k) l 2 j. The cell C j,k is then defined by the region x R 3, a2 j x c j,k m a2 j, m, 2, 3 (2.52) with the center c j,k a2 j k 2 2j,,. (2.53) This is a convenient notation because whether a cubic region at the next higher level is contained in a given cubic region may be determined simply from the index as C j,l C j, k N l m 2 k m, m, 2, 3 (2.54) where [ x] denotes the integer part of x. Each cell at level j N L 2 has exactly eight daughter cells at level j contained within it. The unique level 0 cell is centered at the origin, i.e., c 0,0 0, and has sides of length a. The set of polynomials m j,k ( x) for the multipole expansions within each cell are chosen to be mutually orthonormal over the cell C j,k so that dx m j, k x n j, k x m,n (2.55) Cj, k A set of polynomials up to quadratic order that are orthonormal in the cubic region centered at the origin with sides of length, C 0,, are given in Table. The polynomials for cell C j,k are then defined in terms of these by with m j, k x 8 j/2 m 2 j x c j, k (2.56) m x a 3/ 2 P m a x (2.57)
8 360 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry Table. Polynomials That Are Orthonormal on the Unit Cube Centered at the Origin. P 0 P 23x P 2 23y P 3 23z P 2 65x P 22 65y P 23 65z P 24 2xy P 25 2yz 2xz P 26 with 0 8 m A m,n n b j,dk, n b j,k A m,n dx C0, m n x,d0, x 0 (2.60) C0, 0 dx m x n2x 4 2, 2 2, 2 where d(k, ) is the vector index of the th daughter cell (2.6) where c j,k is the center of the cell C j,k and P m ( x) is one of the polynomials in Table. It will sometimes be convenient to refer to the indices m, j, and k for the function m j,k ( x) collectively by a single index, denoted by a capital letter, i.e., I ( x) orbythe corresponding cell C j,k and the index m. Precomputation All of the calculations in this section are repeated for each molecule in isolation before the simulation begins. The molecules are also assumed to be rigid so that these solutions remain valid throughout the simulation. The molecule and its associated highest level cell C 0,0 should be centered at the origin. First a smooth representation of the solvent-excluded surface j of molecule j ( j, 2) is triangulated and the surface charge densities at each node, h j 0, are calculated with the BEM using eq. (.32). Next, a cubic region that contains the molecule is found and is recursively divided into eight daughter cells until the cells at the finest level have a size that is at least about five times the average length of the triangles sides. These cubes are contained within an octatree data structure described in the previous section. Next, the multipole coefficients of the surface charge density at the highest (finest) level in the octatree structure are calculated using m b NL,k dxh 0 m x NL,k x (2.58) where h 0( x) is the boundary element solution of an isolated molecule calculated using h 0 j in eq. (.32) and the approximation by linear function element in eq. (.25), i.e., h 0 x h 0 j l H l x. (2.59) lxlcnl,k where the sum is over the indices for vertices of the j mesh that are in cell C NL,k. The multipole coefficients of the surface charge density h 0( x) for the lower levels are then calculated by recursively applying dk, 2k 4, 2, (2.62) and k [/k], k, 2, 4 is the value of the appropriate bit in the binary representation of. The sum in eq. (2.60) is over the multipole coefficients for the eight cells contained within cell C j,k. As an aside we note that the orthogonal polynomials may be interpreted as scaling functions in a multiresolution analysis and that except for the fact that only the scaling function coefficients, not the wavelet coefficients are computed, this calculation of the lower level scaling coefficients is the same as the fast wavelet transform algorithm. 26 Force and Torque Evaluation during the Simulation During the simulation, the multipole coefficients calculated beforehand will be used for fast evaluation of the electrostatic force and torque on a molecule. For simplicity, it is assumed here that only molecule 2 moves and molecule remains at the origin, i.e., only relative motion is considered. Let b be the displacement of molecule 2 relative to the origin and A be the 3 3 matrix that effects a rotation of the molecule relative to its initial orientation. A point x on the molecule when it is initially situated at the origin is then transformed as x 3 Ax b. The expansion of h 0( x) for molecule 2 at the origin h 0 x b 2 I I x (2.63) I with b I the coefficients calculated using eqs. (2.58) and (2.60) becomes h 0 x b 2 I I A x b (2.64) I for molecule 2 in the new position and orientation. The coefficients b I (2) in the latter expansion are the same as those in the former. To calculate the force on molecule at the origin due to molecule 2 at the position and orientation specified by b and A using the multipole representation of the charge densities the integral kernel in eq. (.4) must be calculated by
9 Boundary Element Solution of Linear Poisson Boltzmann Equation 36 K I,J dx dx Ix x e xx dx dx Ix x x x JA x b e xaxb x Ax b Jx. (2.65) Using the definition of eq. (2.56) for the polynomials this becomes K I,J sj3cj, k dxcj, k2dxp m sj x c j,k e xaxb P m2 sj x c j,k x x Ax b (2.66) where the polynomial indices are I (m, j, k ) and J (m 2, j, k 2 ) and s( j) 2 j a is the length of a side of a cell at level j. Because only matrix elements between cells at the same level are needed, the level indices are the same. Defining y s ( x c j,k ) and y s ( x c j,k 2 ) the previous equation becomes F 2 out 6 2 C,CS 0 m,m b C,m 2 b C,m K C,m,C,m (2.7) where the sum is over pairs of cells belonging to the set S. We next describe a method to find the pairs of cells in S such that the approximation error resulting from the sparse representation of the matrix K is less than a specified value. Throughout this article we make the assumption that the cells and corresponding octatree structures for each molecule are the same. This simplifies the discussion; however, it is straightforward to use different sets of cells for each molecule. A choice of indices consistent with the desired error bound is to include matrix elements K (C,m),(C,m) between cells that are as large as possible, subject to the condition that d(c, C) for a specified. d(c, C) is the upper bound on the approximation error described in appendix B. If this condition is not satisfied for nearby cells, even at the finest level, then the matrix element is calculated directly using the original boundary element representation. We denote the octatree structures for molecule and molecule 2 as list and list 2, respectively. A recursive algorithm for choosing such pairs of cells for which their corresponding indices are summed over in eq. (2.7) is K I,J e s js jr yay dx sj sjc0, dxc0, R y Ay P m yp m2 y 0 0 (2.67) with R c j,k Ac j,k 2 b. Defining e R xax K, R, x, x x R x Ax and its associated matrix elements at level 0 (2.68). Let j be the lowest level (largest cells) for which cells exist in both list and list 2 2. For each pair of nonempty cells (C (), C (2) ) at level j, one from each of list and list 2 3. If d(c (), C (2) ) for these cells calculate and store the matrix elements {K (C (),m ),(C (2),m 2 ), m, m 2,..., 0} 4. Else If j 0 Mark for direct calculation and stop Else For pairs of nonempty daughter cells, one from each of C () and C (2) Repeat step 3 K m,m 2, R C0, 0 dxc0, 0 dxk, R, x, xp m xp m2 x gives the relation between matrix elements of K and K (2.69) K I,J sjk m,m 2 sj, sj R. (2.70) This implies that the matrix elements of the integral kernel K ( x, x) defined in eq. (2.65) for any choice of indices are related to the level 0 coefficients of K m,m 2, m, m 2,..., 0. Analytical approximations for these latter coefficients are given in Appendix B. According to eq. (.50) the force on molecule due to the electric field of molecule 2 may be calculated using the multipole coefficients as The resulting matrix K will be sparse with nonzero elements appearing in the sum in eq. (2.7). Because the charge distribution is two-dimensional, many cells at the finer levels are empty. The condition that both cells corresponding to the matrix element are nonempty is checked before calculating the approximation error. The large number of empty cells also contributes to the sparsity of K. The terms for the pairs of cells that are marked for direct calculation are calculated using the boundary element representation of the charge density on mesh triangles within the corresponding cells. Because the matrix is sparse the calculation of the force using eq. (2.7) will be fast, particularly if the molecules are widely separated. In fact, if the molecules are sufficiently separated only the matrix elements corresponding to the unique lowest level cell for each molecule will be used. This choice of cells is the same as would be used in the cell multipole method if the approximation error controlling the choice of cells were d(c (), C (2) ) for both the multipole expansion and local Taylor expansion about the field evaluation point.
10 362 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry The same method used to calculate F 2 may also be used to calculate the torque T 2. A new integral kernel L ( x, x) with multipole coefficients L I,J dx dx Ixx x e xx dx dx Ixx x x x J A x b e xaxb x Ax b J x. (2.72) is required. As before, these coefficients may be calculated using only the level 0 coefficients L m,m 2, R C0,0 of the kernel by using dxc0,0 dxl, R, x, xp m xp m2 x (2.73) e R xax L, R, x, x x x R x Ax (2.74) L I,J sjl m,m 2 sj, sj R. (2.75) Analytical approximations of the coefficients L m,m 2, R are given in Appendix B. The torque about x 0 may then be calculated from T 2 out 6 2 C,CS K C,m,C,m out 6 2 C,CS 0 m,m 0 m,m b C,m b C,m 2 b C,m 2 b C,m L C,m,C,m c x 0 L C,m,C,m c K C,m,C,m x 0 F 2. (2.76) The indices to sum over are chosen using the same algorithm described above with the same value of the parameter. Sample Calculations The method described in this article for fast calculation of the electrostatic forces on large molecules in solution has been implemented as a program package in C. In this section we give the results of some calculations of the electrostatic force between two proteins, both actin monomers (G-actin), as a demonstration of the method. G-actin was chosen because it is sufficiently large, containing 5837 atoms with a maximum separation of 76 Å between their centers, and because of our interest in studying actin polymerization using Brownian dynamics simulations. The atomic coordinates were those used in ref. 27 with the bound Ca ion and ADP removed. AMBER force-field values were used for the partial atomic charges and Van der Waals radii. 28 A triangulation of the solvent-excluded surface with probe radius.5 Å was generated using the program MSMS. 29 Because this triangulation contained many triangles with a large vertex angle (near 80 degrees), which lead to large interpolation errors in the boundary element calculation, it was repeatedly refined until 4634 corner vertices and 9272 faces remained. 23 All calculations were done on a 500- MHz Pentium workstation. Dielectric constants in 4.0 and out 80.0 were used as typical values for a protein molecule in water. All calculations were repeated for Debye parameter 0.0 and Å corresponding to pure water and 0. M monovalent ions at 300 K, the approximate physiologic concentration of Na ions in solution, respectively. The center of mass separations of the two molecules was set to 50, 75, 00, and 200 Å. A 50-Å center of mass separation corresponds to a separation of only 8.0 Å for the nearest vertices on each molecular surface. First, the accuracy and speed of the fast noniterative calculation of the polarization correction to the total force, F step 2,is compared with the value, F iter 2, calculated from an iterative solution of h j, j, 2 using eq. (.34). Equation (.34) was iterated until (h, h 2 ) The average computation time for the noniterative method was 390 s, which is about 36% of the time required for the iterative method. The relative errors in the total force F 2 for different separations are given in Table 2. The relative error in the polarization force increases for larger separation distances for 0.0 but decreases for However, the relative error in the force remains small (0.3%). Next, iterative solutions of the boundary element equations were used to compare the fraction of the force resulting from the perturbation of the charge density on one molecule due to the electric field of the other, F polariz 2. The results are also shown in Table 2. As expected, polarization effects are largest when the molecules are near one another but the error remains below 0%, thus justifying the neglect of these corrections in the multipole version of the force calculation. The force was then calculated using the fast multipole algorithm with 4 3 cells at the finest level for each molecule. Twentyfour of the cells were empty and the remaining ones contained an average of 6 vertices per cell. The results for 0.0 and Å are shown in Tables 3 and 4, respectively. Although a single value for the error bound for different relative configurations would be used in practice, smaller values of were chosen at larger separation distances to demonstrate results for a range of different numbers of nonzero matrix elements for a given separation distance. Also, smaller values of were chosen for Å compared to 0.0 to achieve a similar sparsity of matrix K because the electric field decreases much faster in the former case. For the same reason, the relative error for the same distance and matrix sparsity is
11 Boundary Element Solution of Linear Poisson Boltzmann Equation 363 Table 2. Fraction of Force from Polarization in the Electric Field of the Other Molecule and Error in the Force Calculated Using the Fast Noniterative Method of Eq. (.43) Relative to That Calculated Using an Iterative Method of Eq. (.34) r (Å) F polariz 2 F 2 iter F step 2 F iter 2 F iter 2 F polariz 2 F 2 iter F step 2 F iter 2 F iter r is the center of mass separation distance. larger for Å than for 0.0; however, for simulations, one is usually interested in the absolute error, which remains small. Conclusion We have given a detailed description of a calculational scheme, which is a combination of the BEM and the cell multipole method, which allows a fast calculation of the electrostatic force and torque on a macromolecule for use in Brownian dynamics or molecular dynamics simulations. Because the molecules are assumed to be rigid, the relatively slow calculation of the surface charge density on an isolated molecule is done before the simulation and the force evaluation during the simulation is done quickly using the multipole representation of this charge density. When the separation of the molecular surfaces is much closer than the 8 Å tested in the previous section it is expected that the continuum approximation of the ionic solution implicit in the Poisson Boltzmann equation breaks down and that the method should be modified. This is true for any method that uses a continuum representation of the solvent. Numerical errors also become important when the separation of the molecular surfaces is on the order of the size of the surface elements. One solution to these problems is to merge the molecular surfaces near their interface. Another possibility is to replace the solvent layer separating the molecules by explicit molecules whose conformations Table 3. Errors and Execution Times for Calculating the Force on Two Actin Monomers in Pure Water ( 0) Whose Centers of Mass Are Separated by r Å. r (Å) Error bound # Matrix elements # Direct elements F mult 2 F step 2 (0 6 dynes) F mult 2 F step 2 F 2 step tf mult 2 tf 2 step Each molecular surface of the monomer has 9272 triangular surface elements. is the error bound used in the multipole algorithm described in the text. The number of nonzero elements in the matrix K and the number of elements calculated directly using the boundary elements are given. F mult 2 and F step 2 are the forces calculated using the multipole algorithm and the direct one-step BEM with the charge density of eq. (.43), respectively. Execution times for the multipole method, t(f mult 2 ), are given as a fraction of the time for the same calculation using the boundary element method without multipoles, t(f step 2 ). t(f step 2 ) is approximately 390 s on a 500-MHz Pentium workstation.
12 364 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry Table 4. Errors and Execution Times for Calculating the Force on Two Actin Monomers in 0. M Solution of Monovalent Ions ( Å ). r (Å) Error bound # Matrix elements # Direct elements F mult 2 F step 2 (0 6 dynes) F mult 2 F step 2 F 2 step tf mult 2 tf 2 step The notation is the same as in Table 3. are sampled from an equilibrium ensemble using either a Monte Carlo or a molecular dynamics simulation. Although quadratic orthogonal polynomials were used for the multipole expansion, it would be more efficient to use spherical harmonics up to a given order if polynomials of greater than quadratic order are used. This is because the error bound estimate, at least for the Coulomb case, may be formulated in terms of spherical harmonics, and there are fewer of these than independent polynomials of the same order. 20 This was not done here because the number of spherical harmonics up to quadratic order (9) is not sufficiently smaller than the number of independent quadratic polynomials (0) to justify the added complication. There are several ways in which the method may be made faster. First, a higher order multipole approximation would be expected to improve performance because the computation time is currently dominated by direct boundary element contributions. This would require using spherical harmonics, as described above. Two approximations that would improve the speed of the boundary element calculation are using a lower order Gaussian quadrature formula for the off-diagonal elements and using larger surface elements such as described in ref. 30. Although these two approximations would lower the accuracy of the direct boundary element contribution to the force, this is acceptable if the multipole error bound is comparable. It is also possible to apply the methods described here for solving the linear Poisson Boltzmann equation to more highly charged systems. Reference 7 describes a method for scaling the solution of the linear Poisson Boltzmann equation so that it agrees fairly well with the nonlinear Poisson Boltzmann equation solution. The authors of this reference also found that the correction was unnecessary for systems with low charge density at physiologic ionic strengths, where the solutions are virtually identical. Finally, it would be useful to apply some of the methods used in this article to solve for the surface charge density of an isolated molecule as well. Although this would not speed up the calculations during the simulation, it would reduce time and memory constraints on the initial step and allow the use of larger surface meshes. Acknowledgements We thank D. Sept for the actin coordinates and D. Steiger for many useful discussions. Numerical Integration Over Boundary Elements The integrals over the standard triangles, with vertices (0, 0), (, 0), and (0, ) in eq. (.27) are calculated using Gaussian quadrature. The two-dimensional integrals are evaluated using a nonproduct form of the Gaussian quadrature scheme, i.e., not simply using a one-dimensional form in each dimension. Although it is more difficult to calculate the coefficients, the nonproduct form requires less function evaluations, in general. The integral of a function w( x, y) f( x, y) over a region T in this approximation is evaluated using NP w x, y f x, y dx dy w j f x j, y j. (.77) T j The w j, x j, and y j depend on both the region T and the weight function w( x, y). These coefficients are chosen such that this equation is exact for f( x, y) x m y n, m n P. Because the integrands for the diagonal matrix elements in eq. (.29) have a /r singularity, the weight function is chosen to be w( x, y) /x 2 y 2. The weight function w( x, y) for the integrals in the off-diagonal elements. A third-order formula due to Radon is used, with the coefficients for w( x, y) taken from ref. 3 for the off-diagonal matrix elements and a fifth-order Radon formula, with the coefficients for w( x, y) /x 2 y 2 calculated using Macsyma, is used for the diagonal elements. 32 The coefficients for the latter are given in Table 5. Approximate Integral Kernel Matrix Elements and Error Bounds All of the matrix elements of the integral kernels K and L may be calculated from the level 0 matrix elements of the kernel K using
13 Boundary Element Solution of Linear Poisson Boltzmann Equation 365 Table 5. Weights w j and Evaluation Points ( x j, y j ) for Fifth-Order Gaussian Quadrature Over a Triangular Region with Vertices (0, 0), (, 0), and (0, ) and with Weight Function w( x, y) /x 2 y 2. w x y eq. (2.70) and the matrix elements of the kernel L using eq. (2.75), respectively. A closed form expression for these matrix elements may be obtained by using the Taylor series expansion of the kernel to ( x j m x k n ), m n 2 and using the orthonormality of the polynomials P( x) in eqs. (2.69) and (2.73). Defining R F R 3, F 2 2 R 2 3R 3 R 5, F 3 3 R R 2 5R 5 R 7, (2.78) the matrix elements for the integral kernel K (, R, x, x) are K 0,0 l e R F 2 2R l, K j,0 l er 23 F jl F 2 R j R l, K 0,j l er 23 F 2RA j R l F Al, j, K j,k l er 24 F 2 jl RA k kl RA j A jk R l A kj R l A lj R k A lk R j ) F 3 R j RA k R l RA j R k R l ), K 2j,0 l er 25 F 3R j 2 R l F 2 2 jl R j R l, j, 2, 3 K 0,2j l er 25 F 3RA j 2 R j F 2 A2Al, jra j R l )), j, 2, 3 K 24,0 l er 2 F 3R R 2 R l F 2 l R 2 l2 R, K 25,0 l er 2 F 3R 2 R 3 R l F 2 l2 R 3 l3 R 2, K 26,0 l er 2 F 3R R 3 R l F 2 l R 3 l3 R, K 0,24 l er 2 F 3RA RA 2 R l F 2 Al, RA 2 RA Al, 2)), K 0,25 l er 2 F 3RA 2 RA 3 R l F 2 Al, 2RA 3 RA 2 Al, 3)), K 0,26 l er 2 F 3RA RA 3 R l F 2 Al, RA 3 RA Al, 3)), K j,2k l K 2k,j l 0, j, 2, 3; k,...,6 K 2j,2k l 0, j, k,..., 6. (2.79) The matrix elements for the integral kernel L (, R, x, x) are L j,0 er 3 23 F ljk R k, k L j,k l er 3 2 ljm F 2 RA k R m F Am, k, m L 2j,0 er 3 65 F 2R j ljm R m, j, 2, 3 m
14 366 Bordner and Huber Vol. 24, No. 3 Journal of Computational Chemistry charge, at the closest pair of points. The error bound for the terms 0 () (2) m,m b (C,m) b (C,m) K (C,m),(C,m) is denoted by d(c, C) and is dc, C max xc, xc K R, x x K 2 R, x x (2.8) where K 2 (R, x x) is the Taylor series expansion up to quadratic order in x x 3 K 2 R, x x d 0 R d k x x k (2.82) k Figure 2. The recursive algorithm for choosing pairs of cells is illustrated by showing one step. The error bound d(c, C) for two cells in (a) is larger than the desired value so the error bounds for each pair of daughter cells are compared with. If the bound for these cells is found to be less than, as in (b), the recursion terminates. The algorithm then continues checking other pairs of cells at the coarser level. L 24,0 er 2 F 2R R 3, R 2 R 3, R 2 R 2 2, L 25,0 er 2 F 2R 2 2 R 3 2, R R 2, R R 3, L 26,0 er 2 F 2R R 2, R 3 2 R 2, R 2 R 3, L 0,0 l 0, L 0,j l 0, L 0,2j l 0, j,...,6 L j,2k l L 2k,j l 0, j, 2, 3; k,...,6 L 2j,2k l 0, j, k,..., 6. (2.80) Because many matrix elements of K and L are zero, sparse matrix multiplication is used in the program implementing the method. We next derive a bound on the approximation error of each term in the sum of eq. (2.7) for F 2 resulting from truncating the Taylor series expansion of the kernel K to obtain the matrix elements given above. This bound is used to determine which matrix elements in the compressed form of the orthonormal polynomial basis of K (R, x x) are nonzero. Because truncating the Taylor series expansion for K to ( x j m x k n ), m n 2 corresponds to the same approximation for K we examine the error in the latter. We also know that the maximum approximation error for each term in eq. (.50) occurs when the charge distribution in the cells corresponding to each index is a delta function, i.e., point 3 d kl 2 x x k x x l. k,l Defining y x x the kernel function is y K R, y e R R y 3 R y 2R y. (2.83) Next, we assume that the maximum in eq. (2.8) occurs when R y R y. Defining R R, r y, and Rˆ R /R yields Rr K R, r e R r 2 (2.84) R rrˆ and the scalar function K(R, r) may be defined by and likewise for K 2 (R, r) to give K 2 R, r er R 2 K R, r KR, rrˆ (2.85) R 2 R 2 2R 2 R 3 R R 2 6R 6 2 R 2 r 2. (2.86) The error bound of eq. (2.8) then becomes dc, C max 0r3s j KR, r K 2 R, r (2.87) with s( j) the length of one side of the cells C and C and the 3 factor coming from the relative configuration of the cells with the largest error in which the line connecting their centers also passes through a corner of each cell. This expression is the remainder term for the Taylor series, whose value may be found from the Lagrange form R 3 r, c 3! 3 KR, c r 3 r 3, 0 c r. (2.88) r
Notes: Most of the material presented in this chapter is taken from Jackson, Chap. 2, 3, and 4, and Di Bartolo, Chap. 2. 2π nx i a. ( ) = G n.
 Chapter. Electrostatic II Notes: Most of the material presented in this chapter is taken from Jackson, Chap.,, and 4, and Di Bartolo, Chap... Mathematical Considerations.. The Fourier series and the Fourier
Chapter. Electrostatic II Notes: Most of the material presented in this chapter is taken from Jackson, Chap.,, and 4, and Di Bartolo, Chap... Mathematical Considerations.. The Fourier series and the Fourier
Dielectrics. Lecture 20: Electromagnetic Theory. Professor D. K. Ghosh, Physics Department, I.I.T., Bombay
 What are dielectrics? Dielectrics Lecture 20: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay So far we have been discussing electrostatics in either vacuum or in a conductor.
What are dielectrics? Dielectrics Lecture 20: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay So far we have been discussing electrostatics in either vacuum or in a conductor.
lim = F F = F x x + F y y + F z
 Physics 361 Summary of Results from Lecture Physics 361 Derivatives of Scalar and Vector Fields The gradient of a scalar field f( r) is given by g = f. coordinates f g = ê x x + ê f y y + ê f z z Expressed
Physics 361 Summary of Results from Lecture Physics 361 Derivatives of Scalar and Vector Fields The gradient of a scalar field f( r) is given by g = f. coordinates f g = ê x x + ê f y y + ê f z z Expressed
SUMMARY PHYSICS 707 Electrostatics. E(x) = 4πρ(x) and E(x) = 0 (1)
 SUMMARY PHYSICS 707 Electrostatics The basic differential equations of electrostatics are E(x) = 4πρ(x) and E(x) = 0 (1) where E(x) is the electric field and ρ(x) is the electric charge density. The field
SUMMARY PHYSICS 707 Electrostatics The basic differential equations of electrostatics are E(x) = 4πρ(x) and E(x) = 0 (1) where E(x) is the electric field and ρ(x) is the electric charge density. The field
Computer simulation methods (2) Dr. Vania Calandrini
 Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
3. Numerical integration
 3. Numerical integration... 3. One-dimensional quadratures... 3. Two- and three-dimensional quadratures... 3.3 Exact Integrals for Straight Sided Triangles... 5 3.4 Reduced and Selected Integration...
3. Numerical integration... 3. One-dimensional quadratures... 3. Two- and three-dimensional quadratures... 3.3 Exact Integrals for Straight Sided Triangles... 5 3.4 Reduced and Selected Integration...
LECTURE NOTES ELEMENTARY NUMERICAL METHODS. Eusebius Doedel
 LECTURE NOTES on ELEMENTARY NUMERICAL METHODS Eusebius Doedel TABLE OF CONTENTS Vector and Matrix Norms 1 Banach Lemma 20 The Numerical Solution of Linear Systems 25 Gauss Elimination 25 Operation Count
LECTURE NOTES on ELEMENTARY NUMERICAL METHODS Eusebius Doedel TABLE OF CONTENTS Vector and Matrix Norms 1 Banach Lemma 20 The Numerical Solution of Linear Systems 25 Gauss Elimination 25 Operation Count
G pol = 1 2 where ρ( x) is the charge density at position x and the reaction electrostatic potential
 EFFICIENT AND ACCURATE HIGHER-ORDER FAST MULTIPOLE BOUNDARY ELEMENT METHOD FOR POISSON BOLTZMANN ELECTROSTATICS CHANDRAJIT BAJAJ SHUN-CHUAN CHEN Abstract. The Poisson-Boltzmann equation is a partial differential
EFFICIENT AND ACCURATE HIGHER-ORDER FAST MULTIPOLE BOUNDARY ELEMENT METHOD FOR POISSON BOLTZMANN ELECTROSTATICS CHANDRAJIT BAJAJ SHUN-CHUAN CHEN Abstract. The Poisson-Boltzmann equation is a partial differential
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany
 Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
l=0 The expansion coefficients can be determined, for example, by finding the potential on the z-axis and expanding that result in z.
 Electrodynamics I Exam - Part A - Closed Book KSU 15/11/6 Name Electrodynamic Score = 14 / 14 points Instructions: Use SI units. Where appropriate, define all variables or symbols you use, in words. Try
Electrodynamics I Exam - Part A - Closed Book KSU 15/11/6 Name Electrodynamic Score = 14 / 14 points Instructions: Use SI units. Where appropriate, define all variables or symbols you use, in words. Try
APPENDIX A. Background Mathematics. A.1 Linear Algebra. Vector algebra. Let x denote the n-dimensional column vector with components x 1 x 2.
 APPENDIX A Background Mathematics A. Linear Algebra A.. Vector algebra Let x denote the n-dimensional column vector with components 0 x x 2 B C @. A x n Definition 6 (scalar product). The scalar product
APPENDIX A Background Mathematics A. Linear Algebra A.. Vector algebra Let x denote the n-dimensional column vector with components 0 x x 2 B C @. A x n Definition 6 (scalar product). The scalar product
On the adaptive finite element analysis of the Kohn-Sham equations
 On the adaptive finite element analysis of the Kohn-Sham equations Denis Davydov, Toby Young, Paul Steinmann Denis Davydov, LTM, Erlangen, Germany August 2015 Denis Davydov, LTM, Erlangen, Germany College
On the adaptive finite element analysis of the Kohn-Sham equations Denis Davydov, Toby Young, Paul Steinmann Denis Davydov, LTM, Erlangen, Germany August 2015 Denis Davydov, LTM, Erlangen, Germany College
On the positivity of linear weights in WENO approximations. Abstract
 On the positivity of linear weights in WENO approximations Yuanyuan Liu, Chi-Wang Shu and Mengping Zhang 3 Abstract High order accurate weighted essentially non-oscillatory (WENO) schemes have been used
On the positivity of linear weights in WENO approximations Yuanyuan Liu, Chi-Wang Shu and Mengping Zhang 3 Abstract High order accurate weighted essentially non-oscillatory (WENO) schemes have been used
Molecular Modeling -- Lecture 15 Surfaces and electrostatics
 Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Interpolation Theory
 Numerical Analysis Massoud Malek Interpolation Theory The concept of interpolation is to select a function P (x) from a given class of functions in such a way that the graph of y P (x) passes through the
Numerical Analysis Massoud Malek Interpolation Theory The concept of interpolation is to select a function P (x) from a given class of functions in such a way that the graph of y P (x) passes through the
Department of Structural, Faculty of Civil Engineering, Architecture and Urban Design, State University of Campinas, Brazil
 Blucher Mechanical Engineering Proceedings May 2014, vol. 1, num. 1 www.proceedings.blucher.com.br/evento/10wccm A SIMPLIFIED FORMULATION FOR STRESS AND TRACTION BOUNDARY IN- TEGRAL EQUATIONS USING THE
Blucher Mechanical Engineering Proceedings May 2014, vol. 1, num. 1 www.proceedings.blucher.com.br/evento/10wccm A SIMPLIFIED FORMULATION FOR STRESS AND TRACTION BOUNDARY IN- TEGRAL EQUATIONS USING THE
Lecture 11: Potential Energy Functions
 Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Non-bonded interactions
 speeding up the number-crunching continued Marcus Elstner and Tomáš Kubař December 3, 2013 why care? number of individual pair-wise interactions bonded interactions proportional to N: O(N) non-bonded interactions
speeding up the number-crunching continued Marcus Elstner and Tomáš Kubař December 3, 2013 why care? number of individual pair-wise interactions bonded interactions proportional to N: O(N) non-bonded interactions
CE 530 Molecular Simulation
 1 CE 530 Molecular Simulation Lecture 14 Molecular Models David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Review Monte Carlo ensemble averaging, no dynamics easy
1 CE 530 Molecular Simulation Lecture 14 Molecular Models David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Review Monte Carlo ensemble averaging, no dynamics easy
Lectures notes. Rheology and Fluid Dynamics
 ÉC O L E P O L Y T E C H N IQ U E FÉ DÉR A L E D E L A U S A N N E Christophe Ancey Laboratoire hydraulique environnementale (LHE) École Polytechnique Fédérale de Lausanne Écublens CH-05 Lausanne Lectures
ÉC O L E P O L Y T E C H N IQ U E FÉ DÉR A L E D E L A U S A N N E Christophe Ancey Laboratoire hydraulique environnementale (LHE) École Polytechnique Fédérale de Lausanne Écublens CH-05 Lausanne Lectures
Chapter Two: Numerical Methods for Elliptic PDEs. 1 Finite Difference Methods for Elliptic PDEs
 Chapter Two: Numerical Methods for Elliptic PDEs Finite Difference Methods for Elliptic PDEs.. Finite difference scheme. We consider a simple example u := subject to Dirichlet boundary conditions ( ) u
Chapter Two: Numerical Methods for Elliptic PDEs Finite Difference Methods for Elliptic PDEs.. Finite difference scheme. We consider a simple example u := subject to Dirichlet boundary conditions ( ) u
Lecture 4: Numerical solution of ordinary differential equations
 Lecture 4: Numerical solution of ordinary differential equations Department of Mathematics, ETH Zürich General explicit one-step method: Consistency; Stability; Convergence. High-order methods: Taylor
Lecture 4: Numerical solution of ordinary differential equations Department of Mathematics, ETH Zürich General explicit one-step method: Consistency; Stability; Convergence. High-order methods: Taylor
18.02 Multivariable Calculus Fall 2007
 MIT OpenCourseWare http://ocw.mit.edu 18.02 Multivariable Calculus Fall 2007 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. V9. Surface Integrals Surface
MIT OpenCourseWare http://ocw.mit.edu 18.02 Multivariable Calculus Fall 2007 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. V9. Surface Integrals Surface
CS 273 Prof. Serafim Batzoglou Prof. Jean-Claude Latombe Spring Lecture 12 : Energy maintenance (1) Lecturer: Prof. J.C.
 CS 273 Prof. Serafim Batzoglou Prof. Jean-Claude Latombe Spring 2006 Lecture 12 : Energy maintenance (1) Lecturer: Prof. J.C. Latombe Scribe: Neda Nategh How do you update the energy function during the
CS 273 Prof. Serafim Batzoglou Prof. Jean-Claude Latombe Spring 2006 Lecture 12 : Energy maintenance (1) Lecturer: Prof. J.C. Latombe Scribe: Neda Nategh How do you update the energy function during the
Poisson Solver, Pseudopotentials, Atomic Forces in the BigDFT code
 CECAM Tutorial on Wavelets in DFT, CECAM - LYON,, in the BigDFT code Kernel Luigi Genovese L_Sim - CEA Grenoble 28 November 2007 Outline, Kernel 1 The with Interpolating Scaling Functions in DFT for Interpolating
CECAM Tutorial on Wavelets in DFT, CECAM - LYON,, in the BigDFT code Kernel Luigi Genovese L_Sim - CEA Grenoble 28 November 2007 Outline, Kernel 1 The with Interpolating Scaling Functions in DFT for Interpolating
Scientific Computing
 2301678 Scientific Computing Chapter 2 Interpolation and Approximation Paisan Nakmahachalasint Paisan.N@chula.ac.th Chapter 2 Interpolation and Approximation p. 1/66 Contents 1. Polynomial interpolation
2301678 Scientific Computing Chapter 2 Interpolation and Approximation Paisan Nakmahachalasint Paisan.N@chula.ac.th Chapter 2 Interpolation and Approximation p. 1/66 Contents 1. Polynomial interpolation
Math Methods for Polymer Physics Lecture 1: Series Representations of Functions
 Math Methods for Polymer Physics ecture 1: Series Representations of Functions Series analysis is an essential tool in polymer physics and physical sciences, in general. Though other broadly speaking,
Math Methods for Polymer Physics ecture 1: Series Representations of Functions Series analysis is an essential tool in polymer physics and physical sciences, in general. Though other broadly speaking,
d 1 µ 2 Θ = 0. (4.1) consider first the case of m = 0 where there is no azimuthal dependence on the angle φ.
 4 Legendre Functions In order to investigate the solutions of Legendre s differential equation d ( µ ) dθ ] ] + l(l + ) m dµ dµ µ Θ = 0. (4.) consider first the case of m = 0 where there is no azimuthal
4 Legendre Functions In order to investigate the solutions of Legendre s differential equation d ( µ ) dθ ] ] + l(l + ) m dµ dµ µ Θ = 0. (4.) consider first the case of m = 0 where there is no azimuthal
Jim Lambers MAT 610 Summer Session Lecture 2 Notes
 Jim Lambers MAT 610 Summer Session 2009-10 Lecture 2 Notes These notes correspond to Sections 2.2-2.4 in the text. Vector Norms Given vectors x and y of length one, which are simply scalars x and y, the
Jim Lambers MAT 610 Summer Session 2009-10 Lecture 2 Notes These notes correspond to Sections 2.2-2.4 in the text. Vector Norms Given vectors x and y of length one, which are simply scalars x and y, the
Non-bonded interactions
 speeding up the number-crunching Marcus Elstner and Tomáš Kubař May 8, 2015 why care? key to understand biomolecular structure and function binding of a ligand efficiency of a reaction color of a chromophore
speeding up the number-crunching Marcus Elstner and Tomáš Kubař May 8, 2015 why care? key to understand biomolecular structure and function binding of a ligand efficiency of a reaction color of a chromophore
A Fast N-Body Solver for the Poisson(-Boltzmann) Equation
 A Fast N-Body Solver for the Poisson(-Boltzmann) Equation Robert D. Skeel Departments of Computer Science (and Mathematics) Purdue University http://bionum.cs.purdue.edu/2008december.pdf 1 Thesis Poisson(-Boltzmann)
A Fast N-Body Solver for the Poisson(-Boltzmann) Equation Robert D. Skeel Departments of Computer Science (and Mathematics) Purdue University http://bionum.cs.purdue.edu/2008december.pdf 1 Thesis Poisson(-Boltzmann)
Transactions on Modelling and Simulation vol 18, 1997 WIT Press, ISSN X
 Application of the fast multipole method to the 3-D BEM analysis of electron guns T. Nishida and K. Hayami Department of Mathematical Engineering and Information Physics, School of Engineering, University
Application of the fast multipole method to the 3-D BEM analysis of electron guns T. Nishida and K. Hayami Department of Mathematical Engineering and Information Physics, School of Engineering, University
%#, for x = a & ' 0, for x $ a. but with the restriction that the area it encloses equals 1. Therefore, whenever it is used it is implied that
 Chapter 1. Electrostatics I Notes: Most of the material presented in this chapter is taken from Jackson, Chap. 1. Units from the ystème International (I will be used in this chapter. 1.1 Mathematical Considerations
Chapter 1. Electrostatics I Notes: Most of the material presented in this chapter is taken from Jackson, Chap. 1. Units from the ystème International (I will be used in this chapter. 1.1 Mathematical Considerations
Solvation and Macromolecular Structure. The structure and dynamics of biological macromolecules are strongly influenced by water:
 Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
CHAPTER 3 POTENTIALS 10/13/2016. Outlines. 1. Laplace s equation. 2. The Method of Images. 3. Separation of Variables. 4. Multipole Expansion
 CHAPTER 3 POTENTIALS Lee Chow Department of Physics University of Central Florida Orlando, FL 32816 Outlines 1. Laplace s equation 2. The Method of Images 3. Separation of Variables 4. Multipole Expansion
CHAPTER 3 POTENTIALS Lee Chow Department of Physics University of Central Florida Orlando, FL 32816 Outlines 1. Laplace s equation 2. The Method of Images 3. Separation of Variables 4. Multipole Expansion
Multipole moments. November 9, 2015
 Multipole moments November 9, 5 The far field expansion Suppose we have a localized charge distribution, confined to a region near the origin with r < R. Then for values of r > R, the electric field must
Multipole moments November 9, 5 The far field expansion Suppose we have a localized charge distribution, confined to a region near the origin with r < R. Then for values of r > R, the electric field must
Preliminary Examination in Numerical Analysis
 Department of Applied Mathematics Preliminary Examination in Numerical Analysis August 7, 06, 0 am pm. Submit solutions to four (and no more) of the following six problems. Show all your work, and justify
Department of Applied Mathematics Preliminary Examination in Numerical Analysis August 7, 06, 0 am pm. Submit solutions to four (and no more) of the following six problems. Show all your work, and justify
Scientific Computing: An Introductory Survey
 Scientific Computing: An Introductory Survey Chapter 5 Nonlinear Equations Prof. Michael T. Heath Department of Computer Science University of Illinois at Urbana-Champaign Copyright c 2002. Reproduction
Scientific Computing: An Introductory Survey Chapter 5 Nonlinear Equations Prof. Michael T. Heath Department of Computer Science University of Illinois at Urbana-Champaign Copyright c 2002. Reproduction
Chapter 4. Electric Fields in Matter
 Chapter 4. Electric Fields in Matter 4.1.2 Induced Dipoles What happens to a neutral atom when it is placed in an electric field E? The atom now has a tiny dipole moment p, in the same direction as E.
Chapter 4. Electric Fields in Matter 4.1.2 Induced Dipoles What happens to a neutral atom when it is placed in an electric field E? The atom now has a tiny dipole moment p, in the same direction as E.
Practical Linear Algebra: A Geometry Toolbox
 Practical Linear Algebra: A Geometry Toolbox Third edition Chapter 12: Gauss for Linear Systems Gerald Farin & Dianne Hansford CRC Press, Taylor & Francis Group, An A K Peters Book www.farinhansford.com/books/pla
Practical Linear Algebra: A Geometry Toolbox Third edition Chapter 12: Gauss for Linear Systems Gerald Farin & Dianne Hansford CRC Press, Taylor & Francis Group, An A K Peters Book www.farinhansford.com/books/pla
Finite Element Method (FEM)
 Finite Element Method (FEM) The finite element method (FEM) is the oldest numerical technique applied to engineering problems. FEM itself is not rigorous, but when combined with integral equation techniques
Finite Element Method (FEM) The finite element method (FEM) is the oldest numerical technique applied to engineering problems. FEM itself is not rigorous, but when combined with integral equation techniques
Practical in Numerical Astronomy, SS 2012 LECTURE 9
 Practical in Numerical Astronomy, SS 01 Elliptic partial differential equations. Poisson solvers. LECTURE 9 1. Gravity force and the equations of hydrodynamics. Poisson equation versus Poisson integral.
Practical in Numerical Astronomy, SS 01 Elliptic partial differential equations. Poisson solvers. LECTURE 9 1. Gravity force and the equations of hydrodynamics. Poisson equation versus Poisson integral.
CS 450 Numerical Analysis. Chapter 8: Numerical Integration and Differentiation
 Lecture slides based on the textbook Scientific Computing: An Introductory Survey by Michael T. Heath, copyright c 2018 by the Society for Industrial and Applied Mathematics. http://www.siam.org/books/cl80
Lecture slides based on the textbook Scientific Computing: An Introductory Survey by Michael T. Heath, copyright c 2018 by the Society for Industrial and Applied Mathematics. http://www.siam.org/books/cl80
C. Show your answer in part B agrees with your answer in part A in the limit that the constant c 0.
 Problem #1 A. A projectile of mass m is shot vertically in the gravitational field. Its initial velocity is v o. Assuming there is no air resistance, how high does m go? B. Now assume the projectile is
Problem #1 A. A projectile of mass m is shot vertically in the gravitational field. Its initial velocity is v o. Assuming there is no air resistance, how high does m go? B. Now assume the projectile is
What is Classical Molecular Dynamics?
 What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
INTERPOLATION. and y i = cos x i, i = 0, 1, 2 This gives us the three points. Now find a quadratic polynomial. p(x) = a 0 + a 1 x + a 2 x 2.
 INTERPOLATION Interpolation is a process of finding a formula (often a polynomial) whose graph will pass through a given set of points (x, y). As an example, consider defining and x 0 = 0, x 1 = π/4, x
INTERPOLATION Interpolation is a process of finding a formula (often a polynomial) whose graph will pass through a given set of points (x, y). As an example, consider defining and x 0 = 0, x 1 = π/4, x
Fixed point iteration and root finding
 Fixed point iteration and root finding The sign function is defined as x > 0 sign(x) = 0 x = 0 x < 0. It can be evaluated via an iteration which is useful for some problems. One such iteration is given
Fixed point iteration and root finding The sign function is defined as x > 0 sign(x) = 0 x = 0 x < 0. It can be evaluated via an iteration which is useful for some problems. One such iteration is given
Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects
 Statistical Thermodynamics Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects Dr. Ronald M. Levy ronlevy@temple.edu Bare Molecular Mechanics Atomistic Force Fields: torsion
Statistical Thermodynamics Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects Dr. Ronald M. Levy ronlevy@temple.edu Bare Molecular Mechanics Atomistic Force Fields: torsion
Foundations of Matrix Analysis
 1 Foundations of Matrix Analysis In this chapter we recall the basic elements of linear algebra which will be employed in the remainder of the text For most of the proofs as well as for the details, the
1 Foundations of Matrix Analysis In this chapter we recall the basic elements of linear algebra which will be employed in the remainder of the text For most of the proofs as well as for the details, the
Outline. Scientific Computing: An Introductory Survey. Nonlinear Equations. Nonlinear Equations. Examples: Nonlinear Equations
 Methods for Systems of Methods for Systems of Outline Scientific Computing: An Introductory Survey Chapter 5 1 Prof. Michael T. Heath Department of Computer Science University of Illinois at Urbana-Champaign
Methods for Systems of Methods for Systems of Outline Scientific Computing: An Introductory Survey Chapter 5 1 Prof. Michael T. Heath Department of Computer Science University of Illinois at Urbana-Champaign
you expect to encounter difficulties when trying to solve A x = b? 4. A composite quadrature rule has error associated with it in the following form
 Qualifying exam for numerical analysis (Spring 2017) Show your work for full credit. If you are unable to solve some part, attempt the subsequent parts. 1. Consider the following finite difference: f (0)
Qualifying exam for numerical analysis (Spring 2017) Show your work for full credit. If you are unable to solve some part, attempt the subsequent parts. 1. Consider the following finite difference: f (0)
The change in free energy on transferring an ion from a medium of low dielectric constantε1 to one of high dielectric constant ε2:
 The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
CHAPTER 3 Further properties of splines and B-splines
 CHAPTER 3 Further properties of splines and B-splines In Chapter 2 we established some of the most elementary properties of B-splines. In this chapter our focus is on the question What kind of functions
CHAPTER 3 Further properties of splines and B-splines In Chapter 2 we established some of the most elementary properties of B-splines. In this chapter our focus is on the question What kind of functions
Electric fields in matter
 Electric fields in matter November 2, 25 Suppose we apply a constant electric field to a block of material. Then the charges that make up the matter are no longer in equilibrium: the electrons tend to
Electric fields in matter November 2, 25 Suppose we apply a constant electric field to a block of material. Then the charges that make up the matter are no longer in equilibrium: the electrons tend to
Lecture 9 Approximations of Laplace s Equation, Finite Element Method. Mathématiques appliquées (MATH0504-1) B. Dewals, C.
 Lecture 9 Approximations of Laplace s Equation, Finite Element Method Mathématiques appliquées (MATH54-1) B. Dewals, C. Geuzaine V1.2 23/11/218 1 Learning objectives of this lecture Apply the finite difference
Lecture 9 Approximations of Laplace s Equation, Finite Element Method Mathématiques appliquées (MATH54-1) B. Dewals, C. Geuzaine V1.2 23/11/218 1 Learning objectives of this lecture Apply the finite difference
Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum
 THE JOURNAL OF CHEMICAL PHYSICS 126, 124114 2007 Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum Michael J. Schnieders Department of Biomedical Engineering, Washington University
THE JOURNAL OF CHEMICAL PHYSICS 126, 124114 2007 Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum Michael J. Schnieders Department of Biomedical Engineering, Washington University
Discrete Projection Methods for Integral Equations
 SUB Gttttingen 7 208 427 244 98 A 5141 Discrete Projection Methods for Integral Equations M.A. Golberg & C.S. Chen TM Computational Mechanics Publications Southampton UK and Boston USA Contents Sources
SUB Gttttingen 7 208 427 244 98 A 5141 Discrete Projection Methods for Integral Equations M.A. Golberg & C.S. Chen TM Computational Mechanics Publications Southampton UK and Boston USA Contents Sources
Properties of the stress tensor
 Appendix C Properties of the stress tensor Some of the basic properties of the stress tensor and traction vector are reviewed in the following. C.1 The traction vector Let us assume that the state of stress
Appendix C Properties of the stress tensor Some of the basic properties of the stress tensor and traction vector are reviewed in the following. C.1 The traction vector Let us assume that the state of stress
CS 450 Numerical Analysis. Chapter 5: Nonlinear Equations
 Lecture slides based on the textbook Scientific Computing: An Introductory Survey by Michael T. Heath, copyright c 2018 by the Society for Industrial and Applied Mathematics. http://www.siam.org/books/cl80
Lecture slides based on the textbook Scientific Computing: An Introductory Survey by Michael T. Heath, copyright c 2018 by the Society for Industrial and Applied Mathematics. http://www.siam.org/books/cl80
In practice one often meets a situation where the function of interest, f(x), is only represented by a discrete set of tabulated points,
 1 Interpolation 11 Introduction In practice one often meets a situation where the function of interest, f(x), is only represented by a discrete set of tabulated points, {x i, y i = f(x i ) i = 1 n, obtained,
1 Interpolation 11 Introduction In practice one often meets a situation where the function of interest, f(x), is only represented by a discrete set of tabulated points, {x i, y i = f(x i ) i = 1 n, obtained,
Linear Algebra. Session 12
 Linear Algebra. Session 12 Dr. Marco A Roque Sol 08/01/2017 Example 12.1 Find the constant function that is the least squares fit to the following data x 0 1 2 3 f(x) 1 0 1 2 Solution c = 1 c = 0 f (x)
Linear Algebra. Session 12 Dr. Marco A Roque Sol 08/01/2017 Example 12.1 Find the constant function that is the least squares fit to the following data x 0 1 2 3 f(x) 1 0 1 2 Solution c = 1 c = 0 f (x)
Lecture Notes to Accompany. Scientific Computing An Introductory Survey. by Michael T. Heath. Chapter 5. Nonlinear Equations
 Lecture Notes to Accompany Scientific Computing An Introductory Survey Second Edition by Michael T Heath Chapter 5 Nonlinear Equations Copyright c 2001 Reproduction permitted only for noncommercial, educational
Lecture Notes to Accompany Scientific Computing An Introductory Survey Second Edition by Michael T Heath Chapter 5 Nonlinear Equations Copyright c 2001 Reproduction permitted only for noncommercial, educational
Notes for Lecture 10
 February 2, 26 Notes for Lecture Introduction to grid-based methods for solving Poisson and Laplace Equations Finite difference methods The basis for most grid-based finite difference methods is the Taylor
February 2, 26 Notes for Lecture Introduction to grid-based methods for solving Poisson and Laplace Equations Finite difference methods The basis for most grid-based finite difference methods is the Taylor
In manycomputationaleconomicapplications, one must compute thede nite n
 Chapter 6 Numerical Integration In manycomputationaleconomicapplications, one must compute thede nite n integral of a real-valued function f de ned on some interval I of
Chapter 6 Numerical Integration In manycomputationaleconomicapplications, one must compute thede nite n integral of a real-valued function f de ned on some interval I of
Chapter 5 HIGH ACCURACY CUBIC SPLINE APPROXIMATION FOR TWO DIMENSIONAL QUASI-LINEAR ELLIPTIC BOUNDARY VALUE PROBLEMS
 Chapter 5 HIGH ACCURACY CUBIC SPLINE APPROXIMATION FOR TWO DIMENSIONAL QUASI-LINEAR ELLIPTIC BOUNDARY VALUE PROBLEMS 5.1 Introduction When a physical system depends on more than one variable a general
Chapter 5 HIGH ACCURACY CUBIC SPLINE APPROXIMATION FOR TWO DIMENSIONAL QUASI-LINEAR ELLIPTIC BOUNDARY VALUE PROBLEMS 5.1 Introduction When a physical system depends on more than one variable a general
Molecular Driving Forces
 Molecular Driving Forces Statistical Thermodynamics in Chemistry and Biology SUBGfittingen 7 At 216 513 073 / / Ken A. Dill Sarina Bromberg With the assistance of Dirk Stigter on the Electrostatics chapters
Molecular Driving Forces Statistical Thermodynamics in Chemistry and Biology SUBGfittingen 7 At 216 513 073 / / Ken A. Dill Sarina Bromberg With the assistance of Dirk Stigter on the Electrostatics chapters
Jim Lambers MAT 460/560 Fall Semester Practice Final Exam
 Jim Lambers MAT 460/560 Fall Semester 2009-10 Practice Final Exam 1. Let f(x) = sin 2x + cos 2x. (a) Write down the 2nd Taylor polynomial P 2 (x) of f(x) centered around x 0 = 0. (b) Write down the corresponding
Jim Lambers MAT 460/560 Fall Semester 2009-10 Practice Final Exam 1. Let f(x) = sin 2x + cos 2x. (a) Write down the 2nd Taylor polynomial P 2 (x) of f(x) centered around x 0 = 0. (b) Write down the corresponding
Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte
 THE JOURNAL OF CHEMICAL PHYSICS 122, 044903 2005 Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte Zhi-Jie Tan and Shi-Jie Chen a) Department of Physics and
THE JOURNAL OF CHEMICAL PHYSICS 122, 044903 2005 Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte Zhi-Jie Tan and Shi-Jie Chen a) Department of Physics and
To make this point clear, let us rewrite the integral in a way that emphasizes its dependence on the momentum variable p:
 3.2 Construction of the path integral 101 To make this point clear, let us rewrite the integral in a way that emphasizes its dependence on the momentum variable p: q f e iĥt/ q i = Dq e i t 0 dt V (q)
3.2 Construction of the path integral 101 To make this point clear, let us rewrite the integral in a way that emphasizes its dependence on the momentum variable p: q f e iĥt/ q i = Dq e i t 0 dt V (q)
FORMULA SHEET FOR QUIZ 2 Exam Date: November 8, 2017
 MASSACHUSETTS INSTITUTE OF TECHNOLOGY Physics Department Physics 8.07: Electromagnetism II November 5, 207 Prof. Alan Guth FORMULA SHEET FOR QUIZ 2 Exam Date: November 8, 207 A few items below are marked
MASSACHUSETTS INSTITUTE OF TECHNOLOGY Physics Department Physics 8.07: Electromagnetism II November 5, 207 Prof. Alan Guth FORMULA SHEET FOR QUIZ 2 Exam Date: November 8, 207 A few items below are marked
INTERMOLECULAR AND SURFACE FORCES
 INTERMOLECULAR AND SURFACE FORCES SECOND EDITION JACOB N. ISRAELACHVILI Department of Chemical & Nuclear Engineering and Materials Department University of California, Santa Barbara California, USA ACADEMIC
INTERMOLECULAR AND SURFACE FORCES SECOND EDITION JACOB N. ISRAELACHVILI Department of Chemical & Nuclear Engineering and Materials Department University of California, Santa Barbara California, USA ACADEMIC
Topics for the Qualifying Examination
 Topics for the Qualifying Examination Quantum Mechanics I and II 1. Quantum kinematics and dynamics 1.1 Postulates of Quantum Mechanics. 1.2 Configuration space vs. Hilbert space, wave function vs. state
Topics for the Qualifying Examination Quantum Mechanics I and II 1. Quantum kinematics and dynamics 1.1 Postulates of Quantum Mechanics. 1.2 Configuration space vs. Hilbert space, wave function vs. state
Multivariable Calculus
 2 Multivariable Calculus 2.1 Limits and Continuity Problem 2.1.1 (Fa94) Let the function f : R n R n satisfy the following two conditions: (i) f (K ) is compact whenever K is a compact subset of R n. (ii)
2 Multivariable Calculus 2.1 Limits and Continuity Problem 2.1.1 (Fa94) Let the function f : R n R n satisfy the following two conditions: (i) f (K ) is compact whenever K is a compact subset of R n. (ii)
Fast and accurate methods for the discretization of singular integral operators given on surfaces
 Fast and accurate methods for the discretization of singular integral operators given on surfaces James Bremer University of California, Davis March 15, 2018 This is joint work with Zydrunas Gimbutas (NIST
Fast and accurate methods for the discretization of singular integral operators given on surfaces James Bremer University of California, Davis March 15, 2018 This is joint work with Zydrunas Gimbutas (NIST
 Title Theory of solutions in the energy r of the molecular flexibility Author(s) Matubayasi, N; Nakahara, M Citation JOURNAL OF CHEMICAL PHYSICS (2003), 9702 Issue Date 2003-11-08 URL http://hdl.handle.net/2433/50354
Title Theory of solutions in the energy r of the molecular flexibility Author(s) Matubayasi, N; Nakahara, M Citation JOURNAL OF CHEMICAL PHYSICS (2003), 9702 Issue Date 2003-11-08 URL http://hdl.handle.net/2433/50354
Minimal Element Interpolation in Functions of High-Dimension
 Minimal Element Interpolation in Functions of High-Dimension J. D. Jakeman, A. Narayan, D. Xiu Department of Mathematics Purdue University West Lafayette, Indiana Random Model as a function We can consider
Minimal Element Interpolation in Functions of High-Dimension J. D. Jakeman, A. Narayan, D. Xiu Department of Mathematics Purdue University West Lafayette, Indiana Random Model as a function We can consider
INTRODUCTION TO FINITE ELEMENT METHODS ON ELLIPTIC EQUATIONS LONG CHEN
 INTROUCTION TO FINITE ELEMENT METHOS ON ELLIPTIC EQUATIONS LONG CHEN CONTENTS 1. Poisson Equation 1 2. Outline of Topics 3 2.1. Finite ifference Method 3 2.2. Finite Element Method 3 2.3. Finite Volume
INTROUCTION TO FINITE ELEMENT METHOS ON ELLIPTIC EQUATIONS LONG CHEN CONTENTS 1. Poisson Equation 1 2. Outline of Topics 3 2.1. Finite ifference Method 3 2.2. Finite Element Method 3 2.3. Finite Volume
A Tutorial on Wavelets and their Applications. Martin J. Mohlenkamp
 A Tutorial on Wavelets and their Applications Martin J. Mohlenkamp University of Colorado at Boulder Department of Applied Mathematics mjm@colorado.edu This tutorial is designed for people with little
A Tutorial on Wavelets and their Applications Martin J. Mohlenkamp University of Colorado at Boulder Department of Applied Mathematics mjm@colorado.edu This tutorial is designed for people with little
Section Taylor and Maclaurin Series
 Section.0 Taylor and Maclaurin Series Ruipeng Shen Feb 5 Taylor and Maclaurin Series Main Goal: How to find a power series representation for a smooth function us assume that a smooth function has a power
Section.0 Taylor and Maclaurin Series Ruipeng Shen Feb 5 Taylor and Maclaurin Series Main Goal: How to find a power series representation for a smooth function us assume that a smooth function has a power
Numerical Integration for Multivariable. October Abstract. We consider the numerical integration of functions with point singularities over
 Numerical Integration for Multivariable Functions with Point Singularities Yaun Yang and Kendall E. Atkinson y October 199 Abstract We consider the numerical integration of functions with point singularities
Numerical Integration for Multivariable Functions with Point Singularities Yaun Yang and Kendall E. Atkinson y October 199 Abstract We consider the numerical integration of functions with point singularities
NONLINEAR EQUATIONS AND TAYLOR S THEOREM
 APPENDIX C NONLINEAR EQUATIONS AND TAYLOR S THEOREM C.1 INTRODUCTION In adjustment computations it is frequently necessary to deal with nonlinear equations. For example, some observation equations relate
APPENDIX C NONLINEAR EQUATIONS AND TAYLOR S THEOREM C.1 INTRODUCTION In adjustment computations it is frequently necessary to deal with nonlinear equations. For example, some observation equations relate
4 / Gaussian Ensembles. Level Density
 4 / Gaussian Ensembles. Level Density In this short chapter we reproduce a statistical mechanical argument of Wigner to "derive" the level density for the Gaussian ensembles. The joint probability density
4 / Gaussian Ensembles. Level Density In this short chapter we reproduce a statistical mechanical argument of Wigner to "derive" the level density for the Gaussian ensembles. The joint probability density
Performance Comparison of Generalized Born and Poisson Methods in the Calculation of Electrostatic Solvation Energies for Protein Structures
 Performance Comparison of Generalized Born and Poisson Methods in the Calculation of Electrostatic Solvation Energies for Protein Structures MICHAEL FEIG, 1 ALEXEY ONUFRIEV, 1 MICHAEL S. LEE, 2 WONPIL
Performance Comparison of Generalized Born and Poisson Methods in the Calculation of Electrostatic Solvation Energies for Protein Structures MICHAEL FEIG, 1 ALEXEY ONUFRIEV, 1 MICHAEL S. LEE, 2 WONPIL
CIV-E1060 Engineering Computation and Simulation Examination, December 12, 2017 / Niiranen
 CIV-E16 Engineering Computation and Simulation Examination, December 12, 217 / Niiranen This examination consists of 3 problems rated by the standard scale 1...6. Problem 1 Let us consider a long and tall
CIV-E16 Engineering Computation and Simulation Examination, December 12, 217 / Niiranen This examination consists of 3 problems rated by the standard scale 1...6. Problem 1 Let us consider a long and tall
Expansion of 1/r potential in Legendre polynomials
 Expansion of 1/r potential in Legendre polynomials In electrostatics and gravitation, we see scalar potentials of the form V = K d Take d = R r = R 2 2Rr cos θ + r 2 = R 1 2 r R cos θ + r R )2 Use h =
Expansion of 1/r potential in Legendre polynomials In electrostatics and gravitation, we see scalar potentials of the form V = K d Take d = R r = R 2 2Rr cos θ + r 2 = R 1 2 r R cos θ + r R )2 Use h =
(f(x) P 3 (x)) dx. (a) The Lagrange formula for the error is given by
 1. QUESTION (a) Given a nth degree Taylor polynomial P n (x) of a function f(x), expanded about x = x 0, write down the Lagrange formula for the truncation error, carefully defining all its elements. How
1. QUESTION (a) Given a nth degree Taylor polynomial P n (x) of a function f(x), expanded about x = x 0, write down the Lagrange formula for the truncation error, carefully defining all its elements. How
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian. which is given in spherical coordinates by (2)
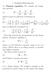 1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
V. Electrostatics Lecture 24: Diffuse Charge in Electrolytes
 V. Electrostatics Lecture 24: Diffuse Charge in Electrolytes MIT Student 1. Poisson-Nernst-Planck Equations The Nernst-Planck Equation is a conservation of mass equation that describes the influence of
V. Electrostatics Lecture 24: Diffuse Charge in Electrolytes MIT Student 1. Poisson-Nernst-Planck Equations The Nernst-Planck Equation is a conservation of mass equation that describes the influence of
NUMERICAL METHODS. x n+1 = 2x n x 2 n. In particular: which of them gives faster convergence, and why? [Work to four decimal places.
 NUMERICAL METHODS 1. Rearranging the equation x 3 =.5 gives the iterative formula x n+1 = g(x n ), where g(x) = (2x 2 ) 1. (a) Starting with x = 1, compute the x n up to n = 6, and describe what is happening.
NUMERICAL METHODS 1. Rearranging the equation x 3 =.5 gives the iterative formula x n+1 = g(x n ), where g(x) = (2x 2 ) 1. (a) Starting with x = 1, compute the x n up to n = 6, and describe what is happening.
Chapter 4. Electrostatic Fields in Matter
 Chapter 4. Electrostatic Fields in Matter 4.1. Polarization 4.2. The Field of a Polarized Object 4.3. The Electric Displacement 4.4. Linear Dielectrics 4.5. Energy in dielectric systems 4.6. Forces on
Chapter 4. Electrostatic Fields in Matter 4.1. Polarization 4.2. The Field of a Polarized Object 4.3. The Electric Displacement 4.4. Linear Dielectrics 4.5. Energy in dielectric systems 4.6. Forces on
MATH 590: Meshfree Methods
 MATH 590: Meshfree Methods Chapter 34: Improving the Condition Number of the Interpolation Matrix Greg Fasshauer Department of Applied Mathematics Illinois Institute of Technology Fall 2010 fasshauer@iit.edu
MATH 590: Meshfree Methods Chapter 34: Improving the Condition Number of the Interpolation Matrix Greg Fasshauer Department of Applied Mathematics Illinois Institute of Technology Fall 2010 fasshauer@iit.edu
Integration, differentiation, and root finding. Phys 420/580 Lecture 7
 Integration, differentiation, and root finding Phys 420/580 Lecture 7 Numerical integration Compute an approximation to the definite integral I = b Find area under the curve in the interval Trapezoid Rule:
Integration, differentiation, and root finding Phys 420/580 Lecture 7 Numerical integration Compute an approximation to the definite integral I = b Find area under the curve in the interval Trapezoid Rule:
Electronic properties of CdSe nanocrystals in the absence and presence of a dielectric medium
 JOURNAL OF CHEMICAL PHYSICS VOLUME 110, NUMBER 11 15 MARCH 1999 Electronic properties of CdSe nanocrystals in the absence and presence of a dielectric medium Eran Rabani, Balázs Hetényi, and B. J. Berne
JOURNAL OF CHEMICAL PHYSICS VOLUME 110, NUMBER 11 15 MARCH 1999 Electronic properties of CdSe nanocrystals in the absence and presence of a dielectric medium Eran Rabani, Balázs Hetényi, and B. J. Berne
MY PUTNAM PROBLEMS. log(1 + x) dx = π2
 MY PUTNAM PROBLEMS These are the problems I proposed when I was on the Putnam Problem Committee for the 984 86 Putnam Exams. Problems intended to be A or B (and therefore relatively easy) are marked accordingly.
MY PUTNAM PROBLEMS These are the problems I proposed when I was on the Putnam Problem Committee for the 984 86 Putnam Exams. Problems intended to be A or B (and therefore relatively easy) are marked accordingly.
PEAT SEISMOLOGY Lecture 2: Continuum mechanics
 PEAT8002 - SEISMOLOGY Lecture 2: Continuum mechanics Nick Rawlinson Research School of Earth Sciences Australian National University Strain Strain is the formal description of the change in shape of a
PEAT8002 - SEISMOLOGY Lecture 2: Continuum mechanics Nick Rawlinson Research School of Earth Sciences Australian National University Strain Strain is the formal description of the change in shape of a
Scientific Computing II
 Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
Intermolecular Forces and Monte-Carlo Integration 열역학특수연구
 Intermolecular Forces and Monte-Carlo Integration 열역학특수연구 2003.3.28 Source of the lecture note. J.M.Prausnitz and others, Molecular Thermodynamics of Fluid Phase Equiliria Atkins, Physical Chemistry Lecture
Intermolecular Forces and Monte-Carlo Integration 열역학특수연구 2003.3.28 Source of the lecture note. J.M.Prausnitz and others, Molecular Thermodynamics of Fluid Phase Equiliria Atkins, Physical Chemistry Lecture
4. Numerical Quadrature. Where analytical abilities end... continued
 4. Numerical Quadrature Where analytical abilities end... continued Where analytical abilities end... continued, November 30, 22 1 4.3. Extrapolation Increasing the Order Using Linear Combinations Once
4. Numerical Quadrature Where analytical abilities end... continued Where analytical abilities end... continued, November 30, 22 1 4.3. Extrapolation Increasing the Order Using Linear Combinations Once
