Table of contents Appendix Figure S1 Appendix Figure S2 Appendix Figure S3 Appendix Table S1 Appendix Table S2
|
|
|
- Shana Goodwin
- 5 years ago
- Views:
Transcription
1 Table of contents Appendix Figure S1 Appendix Figure S2 Appendix Figure S3 Appendix Table S1 Appendix Table S2 1
2 Appendix Figure S1 Fig. S1. Proteome analysis of the CPE-derived EVs and the choroid plexus in the presence and absence of LPS. (A) Schematic representation of the EV proteomics workflow. Mice were injected i.p. with PBS or LPS and 6 h later CSF was isolated using the cisterna magna puncture technique. Fifty microliters of CSF was pooled and EVs were isolated using the Total Exosome Isolation reagent (Life Technologies). Isolated EVs were separated by SDS-PAGE, stained with Coomassie brilliant blue, and digested with trypsin. After pre-fractionation using RP-HPLC, peptides were analyzed by LC-MS/MS (Q-Exactive; Thermo Fisher Scientific). Proteins were identified by using the Mascot Daemon software and quantified (label-free quantification) by MaxLFQ (MaxQuant). Finally, the data were analyzed using DAVID GO enrichment. (B) Scatter plots displaying LFQ intensities of the different 2
3 proteome samples. The calculated correlation coefficient (Pearson correlation) for each pair of samples is indicated in blue at the left corner of the graphs. -LPS1 and -LPS2 are biological replicates derived from EVs purified from CSF of PBS-injected mice. +LPS1 and +LPS2 are biological replicates derived from EVs purified from CSF of LPS-injected mice. (C-D) Overlap of proteins identified in EVs isolated from two independent pooled CSF samples (total volume 50 µl) of control (C; blue) and LPS-injected mice (D; yellow). (E) Schematic representation of the choroid plexus proteomics workflow. Mice were injected i.p. with PBS or LPS and 6 h later the choroid plexus (CP) was isolated. Choroid plexus tissue was lysed by mechanical disruption in Laemmli buffer followed by high speed centrifugation to remove the debris. Supernatant was collected, separated by SDS-PAGE, stained with Coomassie brilliant blue, and excised protein bands were digested with trypsin. Next, the peptide mixture was labeled with freshly prepared 12C (light; PBS) or 13C (heavy; LPS) N-hydroxy succinimide propionate, excess labeling was quenched, and light and heavy labeled samples were mixed in a 1:1 ratio. After pre-fractionation using RP-HPLC, the peptide samples were analyzed by LC- MS/MS (Q-Exactive; Thermo fisher scientific). Proteins were identified by using Mascot Daemon software, and the data were analyzed using DAVID. 3
4 Appendix Figure S2 Fig. S2. Analysis of fluorescent labeling of the EVs. (A-B) Double-labelling of CSF-isolated EVs with the membrane label PKH26 (A; red) and RNA label Ribogreen (B; green). 4
5 Appendix Figure S3 Fig. S3. Analysis of TLR4 dependency of the EV effect on recipient cells. QPCR gene expression analysis of inflammatory genes in wild type (black) and TLR4 -/- (grey) derived primary mixed cortical cultures incubated for 24 h with EVs derived from LPS-injected mice (n=3). 5
6 Appendix Table S1 Primer Anxa5_Fw Anxa5_Rev Calm2_Fw Calm2_Rev Cd63_Forward Cd63_Reverse Cd81_Fw Cd81_Rev Cd9_Fw Cd9_Rev Dicer1_Fw Dicer1_Rev Dnmt3a_Fw Dnmt3a_Rev Gapdh_Fw Gapdh_Rev Hprt_Fw Hprt_Rev Hspa1a_Fw Hspa1a_Rev Nfkbia_Fw Nfkbia_Rev Il1b_Fw Il1b_Rev Il6_Fw Il6_Rev Nos2_Fw Nos2_Rev Irak1_Fw Irak1_Rev Mapk3_Fw Mapk3_Rev Notch1_Fw Notch1_Rev Smad2_Fw Smad2_Rev Smad5_Fw Smad5_Rev Sox2_Fw Sox2_Rev Tab2_Fw Sequence (5'-3') ATCCTGAACCTGTTGACATCCC AGTCGTGAGGGCTTCATCATA ACGGGGATGGGACAATAACAA TGCTGCACTAATATAGCCATTGC GAAGCAGGCCATTACCCATGA TGACTTCACCTGGTCTCTAAACA GTGGAGGGCTGCACCAAAT GACGCAACCACAGAGCTACA ATGCCGGTCAAAGGAGGTAG GCCATAGTCCAATAGCAAGCA GGTCCTTTCTTTGGACTGCCA GCGATGAACGTCTTCCCTGA GAGGGAACTGAGACCCCAC CTGGAAGGTGAGTCTTGGCA TGAAGCAGGCATCTGAGGG CGAAGGTGGAAGAGTGGGAG AGTGTTGGATACAGGCCAGAC CGTGATTCAAATCCCTGAAGT TCTCGGCACCACCTACTCC CTACGCCCGATCAGACGTTT CTCACGGAGGACGGAGACTC CTCTTCGTGGATGATTGCCA CACCTCACAAGCAGAGCACAAG GCATTAGAAACAGTCCAGCCCATAC GCCTAAGCATATCAGTTTGTGGAC AGTGTCCCAACATTCATATTGTCAG CAGCTGGGCTGTACAAACCTT CATTGGAAGTGAAGCGTTTCG AGCCGAGGTCTGCATTACATT TGGCAGTCTGGATAACTGATGA TCCGCCATGAGAATGTTATAGGC GGTGGTGTTGATAAGCAGATTGG CCCTTGCTCTGCCTAACGC GGAGTCCTGGCATCGTTGG ATGTCGTCCATCTTGCCATTC AACCGTCCTGTTTTCTTTAGCTT TTGTTCAGAGTAGGAACTGCAAC GAAGCTGAGCAAACTCCTGAT GCGGAGTGGAAACTTTTGTCC CGGGAAGCGTGTACTTATCCTT CATGACCTGCGACAAAAATTCC 6
7 Tab2_Rev Tnf_Fw Tnf_Rev Ubc_Fw Ubc_Rev TGATTGCGTAGACCAGAAATTCC ACCCTGGTATGAGCCCATATAC ACACCCATTCCCTTCACAGAG AGGTCAAACAGGAAGACAGACGTA TCACACCCAAGAACAAGCACA Appendix Table S1. Forward (Fw) and reversed (Rev) primer sequences for qpcr. 7
8 Appendix Table S2 Figure panel comparison p-value n-value Figure 1 B 2 h vs 0 h 0,03170 n = 5 4 h vs 0 h 0,00790 n = 5 6 h vs 0 h 0,00790 n = 5 D 0,02860 n = 4 F 1 h vs 0 h 0,02860 n = 4 6 h vs 0 h 0,02860 n = 4 Figure 2 A 0,02680 n = 5 C 0,03250 n = 3 D 0,02100 n = 3 and n =4 E 0,01640 n = 4 G 0,00970 n = 3 H 0,00030 n = 3 I 0,00160 n = 3 Figure 3 B 0,02290 n = 3 D 0,02250 n = 3 E 0,03630 n = 3 F 0,00950 n = 3 Figure 4 D 3 h vs 0 h 0,02830 n = 21 and n = 20 4 h vs 0 h < 0,0001 n = 13 and n = 20 6 h vs 0 h 0,00960 n = 23 and n = 20 E 3 h vs 0 h < 0,0001 n = 21 and n = 20 4 h vs 0 h 0,00260 n = 13 and n = 20 6 h vs 0 h < 0,0001 n = 23 and n = 20 F 3 h vs 0 h 0,0002 n = 21 and n = 20 4 h vs 0 h < 0,0001 n = 13 and n = 20 6 h vs 0 h 0,0004 n = 23 and n = 20 G 1 h vs 0 h 0,00840 n = 5 and n = 4 6 h vs 0 h 0,00450 n = 5 and n = 4 24 h vs 0 h 0,03680 n = 3 and n = 4 H 1 h vs 0 h 0,00170 n = 3 24 h vs 0 h 0,00120 n = 3 I 6 h vs 0 h 0,01780 n = 5 and n = 4 24 h vs 0 h 0,01140 n = 3 and n = 4 J 1 h vs 0 h 0,00830 n = 5 and n = 4 6 h vs 0 h 0,00030 n = 5 and n = 4 24 h vs 0 h 0,00050 n = 3 and n = 4 K 0,04400 n = 8 and n = 10 L mr-9 0,04090 n = 4 mir-146a 0,03719 n = 4 mir-155 0,03032 n = 4 M 0,00660 n = 7 Figure 5 C 0,00210 n = 3 and n = 4 D mir-9 0,04427 n = 6 and n = 5 mir-155 0,00061 n = 5 and n = 7 E mir-9 0,02126 n = 5 and n = 4 mir-155 0,03627 n = 5 and n = 4 8
9 F 0,01710 n = 5 and n = 4 G mir-9 0,00820 n = 5 and n = 7 mir-146a 0,00574 n = 6 mir-155 0,00000 n = 7 H mir-146a 0,02993 n = 3 mir-155 0,04198 n = 3 Figure 8 A Mapk3 0,00280 n = 3 Notch1 0,02234 n = 3 Dicer1 0,03244 n = 3 Tab2 0,00666 n = 3 Sox2 0,00729 n = 3 Smad2 0,01496 n = 3 Smad5 0,00178 n = 3 Irak1 0,03244 n = 3 B Notch1 0,03290 n = 3 Dicer1 0,04111 n = 3 Smad2 0,04737 n = 3 Smad5 0,04486 n = 3 Dnmt3a 0,00884 n = 3 C in vitro Il1b 0,00181 n = 3 Tnf 0,00588 n = 3 Il6 0,00395 n = 3 Nos2 0,00258 n = 3 Nfkbia 0,00215 n = 3 C in vivo Il1b 0,00149 n = 3 Tnf 0,01980 n = 3 Il6 0,00262 n = 3 Nos2 0,02425 n = 3 Nfkbia 0,00771 n = 3 D in vitro IL-6 0,01131 n = 3 IL-1β 0,00009 n = 3 TNF 0,00747 n = 3 D in vivo IL-6 0,01346 n = 3 and n= 5 IL-1β 0,01190 n = 3 and n= 8 TNF 0,00304 n = 3 and n= 8 E Nfkbia 0,00686 n = 3 F Dnmt3a 0,00505 n = 3 G Dicer1 0,04714 n = 6 and n = 7 Tab2 0,03036 n = 6 and n = 8 Sox2 0,00344 n = 6 and n = 7 Dnmt3a 0,04355 n = 6 and n = 8 Irak1 0,00053 n = 6 and n = 8 H Il1b 0,03524 n = 6 and n = 7 Tnf 0,00294 n = 6 and n = 8 Nos2 0,02709 n = 6 Nfkbia 0,00035 n = 3 Figure EV1 E 2 h vs 0 h 0,00090 n = 6 3 h vs 0 h 0,00060 n = 6 4 h vs 0 h 0,00010 n = 6 9
10 5 h vs 0 h < 0,0001 n = 6 6 h vs 0 h < 0,0001 n = 6 7 h vs 0 h < 0,0001 n = 6 8 h vs 0 h < 0,0001 n = 6 9 h vs 0 h < 0,0001 n = 6 10 h vs 0 h < 0,0001 n = 6 Figure EV4 A 0,01180 n = 3 and n = 4 C 0,00210 n = 3 D 0,02280 n = 3 E 0,02350 n = 3 and n = 4 Figure EV5 B 0,04230 n = 3 and n = 5 Appendix Table S2. Overview of the exact p- and n-values of the significant results. 10
NPTEL VIDEO COURSE PROTEOMICS PROF. SANJEEVA SRIVASTAVA
 LECTURE-25 Quantitative proteomics: itraq and TMT TRANSCRIPT Welcome to the proteomics course. Today we will talk about quantitative proteomics and discuss about itraq and TMT techniques. The quantitative
LECTURE-25 Quantitative proteomics: itraq and TMT TRANSCRIPT Welcome to the proteomics course. Today we will talk about quantitative proteomics and discuss about itraq and TMT techniques. The quantitative
MS-based proteomics to investigate proteins and their modifications
 MS-based proteomics to investigate proteins and their modifications Francis Impens VIB Proteomics Core October th 217 Overview Mass spectrometry-based proteomics: general workflow Identification of protein
MS-based proteomics to investigate proteins and their modifications Francis Impens VIB Proteomics Core October th 217 Overview Mass spectrometry-based proteomics: general workflow Identification of protein
Chemical Labeling Strategy for Generation of Internal Standards for Targeted Quantitative Proteomics
 Chemical Labeling Strategy for Generation of Internal Standards for Targeted Quantitative Proteomics mtraq Reagents Triplex Christie Hunter, Brian Williamson, Marjorie Minkoff AB SCIEX, USA The utility
Chemical Labeling Strategy for Generation of Internal Standards for Targeted Quantitative Proteomics mtraq Reagents Triplex Christie Hunter, Brian Williamson, Marjorie Minkoff AB SCIEX, USA The utility
Michigan State University Diagnostic Center for Population and Animal Health, Lansing MI USA. QIAGEN Leipzig GmbH, Leipzig, Germany
 Detection of Bovine Viral Diarrhea (BVD) Virus in Ear Skin Tissue Samples: Evaluation of a Combination of QIAGEN MagAttract 96 cador Pathogen Extraction and virotype BVDV RT-PCR-based Amplification. Roger
Detection of Bovine Viral Diarrhea (BVD) Virus in Ear Skin Tissue Samples: Evaluation of a Combination of QIAGEN MagAttract 96 cador Pathogen Extraction and virotype BVDV RT-PCR-based Amplification. Roger
Improved 6- Plex TMT Quantification Throughput Using a Linear Ion Trap HCD MS 3 Scan Jane M. Liu, 1,2 * Michael J. Sweredoski, 2 Sonja Hess 2 *
 Improved 6- Plex TMT Quantification Throughput Using a Linear Ion Trap HCD MS 3 Scan Jane M. Liu, 1,2 * Michael J. Sweredoski, 2 Sonja Hess 2 * 1 Department of Chemistry, Pomona College, Claremont, California
Improved 6- Plex TMT Quantification Throughput Using a Linear Ion Trap HCD MS 3 Scan Jane M. Liu, 1,2 * Michael J. Sweredoski, 2 Sonja Hess 2 * 1 Department of Chemistry, Pomona College, Claremont, California
Chapter 4. strategies for protein quantitation Ⅱ
 Proteomics Chapter 4. strategies for protein quantitation Ⅱ 1 Multiplexed proteomics Multiplexed proteomics is the use of fluorescent stains or probes with different excitation and emission spectra to
Proteomics Chapter 4. strategies for protein quantitation Ⅱ 1 Multiplexed proteomics Multiplexed proteomics is the use of fluorescent stains or probes with different excitation and emission spectra to
Methods for proteome analysis of obesity (Adipose tissue)
 Methods for proteome analysis of obesity (Adipose tissue) I. Sample preparation and liquid chromatography-tandem mass spectrometric analysis Instruments, softwares, and materials AB SCIEX Triple TOF 5600
Methods for proteome analysis of obesity (Adipose tissue) I. Sample preparation and liquid chromatography-tandem mass spectrometric analysis Instruments, softwares, and materials AB SCIEX Triple TOF 5600
SERVA ICPL Kit (Cat.-No )
 INSTRUCTION MANUAL SERVA ICPL Kit (Cat.-No. 39230.01) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 Heidelberg Phone +49-6221-138400, Fax +49-6221-1384010 e-mail: info@serva.de http://www.serva.de
INSTRUCTION MANUAL SERVA ICPL Kit (Cat.-No. 39230.01) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 Heidelberg Phone +49-6221-138400, Fax +49-6221-1384010 e-mail: info@serva.de http://www.serva.de
PC235: 2008 Lecture 5: Quantitation. Arnold Falick
 PC235: 2008 Lecture 5: Quantitation Arnold Falick falickam@berkeley.edu Summary What you will learn from this lecture: There are many methods to perform quantitation using mass spectrometry (any method
PC235: 2008 Lecture 5: Quantitation Arnold Falick falickam@berkeley.edu Summary What you will learn from this lecture: There are many methods to perform quantitation using mass spectrometry (any method
SILAC and TMT. IDeA National Resource for Proteomics Workshop for Graduate Students and Post-docs Renny Lan 5/18/2017
 SILAC and TMT IDeA National Resource for Proteomics Workshop for Graduate Students and Post-docs Renny Lan 5/18/2017 UHPLC peak chosen at 26.47 min LC Mass at 571.36 chosen for MS/MS MS/MS MS This is a
SILAC and TMT IDeA National Resource for Proteomics Workshop for Graduate Students and Post-docs Renny Lan 5/18/2017 UHPLC peak chosen at 26.47 min LC Mass at 571.36 chosen for MS/MS MS/MS MS This is a
Reagents. Affinity Tag (Biotin) Acid Cleavage Site. Figure 1. Cleavable ICAT Reagent Structure.
 DATA SHEET Protein Expression Analysis Reagents Background The ultimate goal of proteomics is to identify and quantify proteins that are relevant to a given biological state; and to unearth networks of
DATA SHEET Protein Expression Analysis Reagents Background The ultimate goal of proteomics is to identify and quantify proteins that are relevant to a given biological state; and to unearth networks of
Quantitation of a target protein in crude samples using targeted peptide quantification by Mass Spectrometry
 Quantitation of a target protein in crude samples using targeted peptide quantification by Mass Spectrometry Jon Hao, Rong Ye, and Mason Tao Poochon Scientific, Frederick, Maryland 21701 Abstract Background:
Quantitation of a target protein in crude samples using targeted peptide quantification by Mass Spectrometry Jon Hao, Rong Ye, and Mason Tao Poochon Scientific, Frederick, Maryland 21701 Abstract Background:
Colorimetric GAPDH Assay Cat. No. 8148, 100 tests
 Colorimetric GAPDH Assay Cat. No. 8148, 1 tests Introduction Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) is a tetrameric enzyme that catalyzes glycolysis and thus serves to break down glucose for
Colorimetric GAPDH Assay Cat. No. 8148, 1 tests Introduction Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) is a tetrameric enzyme that catalyzes glycolysis and thus serves to break down glucose for
Proteome-wide label-free quantification with MaxQuant. Jürgen Cox Max Planck Institute of Biochemistry July 2011
 Proteome-wide label-free quantification with MaxQuant Jürgen Cox Max Planck Institute of Biochemistry July 2011 MaxQuant MaxQuant Feature detection Data acquisition Initial Andromeda search Statistics
Proteome-wide label-free quantification with MaxQuant Jürgen Cox Max Planck Institute of Biochemistry July 2011 MaxQuant MaxQuant Feature detection Data acquisition Initial Andromeda search Statistics
Improved Throughput and Reproducibility for Targeted Protein Quantification Using a New High-Performance Triple Quadrupole Mass Spectrometer
 Improved Throughput and Reproducibility for Targeted Protein Quantification Using a New High-Performance Triple Quadrupole Mass Spectrometer Reiko Kiyonami, Mary Blackburn, Andreas FR Hühme: Thermo Fisher
Improved Throughput and Reproducibility for Targeted Protein Quantification Using a New High-Performance Triple Quadrupole Mass Spectrometer Reiko Kiyonami, Mary Blackburn, Andreas FR Hühme: Thermo Fisher
DEVELOP YOUR LAB, YOUR WAY
 Technical brochure DEVELOP YOUR LAB, YOUR WAY The FLOW Solution: Expand your potential today Redesign your lab with the FLOW Solution The FLOW Solution is a highly flexible modular, semi-automated data
Technical brochure DEVELOP YOUR LAB, YOUR WAY The FLOW Solution: Expand your potential today Redesign your lab with the FLOW Solution The FLOW Solution is a highly flexible modular, semi-automated data
Quantitative Proteomics
 Quantitative Proteomics Quantitation AND Mass Spectrometry Condition A Condition B Identify and quantify differently expressed proteins resulting from a change in the environment (stimulus, disease) Lyse
Quantitative Proteomics Quantitation AND Mass Spectrometry Condition A Condition B Identify and quantify differently expressed proteins resulting from a change in the environment (stimulus, disease) Lyse
Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea nd Semester
 Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea 2018. 2 nd Semester Absorbance Assay (280 nm) Considerations for use Absorbance assays are fast and
Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea 2018. 2 nd Semester Absorbance Assay (280 nm) Considerations for use Absorbance assays are fast and
In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE
 In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE (Lamond Lab / April 2008)! Perform all the pipetting steps in a laminar flow hood. We routinely do our digestions in our TC room hoods.
In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE (Lamond Lab / April 2008)! Perform all the pipetting steps in a laminar flow hood. We routinely do our digestions in our TC room hoods.
U.S. Patent No. 9,051,563 and other pending patents. Ver
 INSTRUCTION MANUAL Direct-zol 96 RNA Catalog Nos. R2054, R2055, R2056 & R2057 Highlights Quick, 96-well purification of high-quality (DNA-free) total RNA directly from TRIzol, TRI Reagent and all other
INSTRUCTION MANUAL Direct-zol 96 RNA Catalog Nos. R2054, R2055, R2056 & R2057 Highlights Quick, 96-well purification of high-quality (DNA-free) total RNA directly from TRIzol, TRI Reagent and all other
Enrichment Media and Kits
 Enrichment Media and Kits Innovative Biotin/Streptavidin-free Tools for Capturing Azide- or Alkyne-tagged Proteins Enrichment Media and Kits Protein Enrichment Kits Far superior digest purity compared
Enrichment Media and Kits Innovative Biotin/Streptavidin-free Tools for Capturing Azide- or Alkyne-tagged Proteins Enrichment Media and Kits Protein Enrichment Kits Far superior digest purity compared
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
Comprehensive support for quantitation
 Comprehensive support for quantitation One of the major new features in the current release of Mascot is support for quantitation. This is still work in progress. Our goal is to support all of the popular
Comprehensive support for quantitation One of the major new features in the current release of Mascot is support for quantitation. This is still work in progress. Our goal is to support all of the popular
Computational Methods for Mass Spectrometry Proteomics
 Computational Methods for Mass Spectrometry Proteomics Eidhammer, Ingvar ISBN-13: 9780470512975 Table of Contents Preface. Acknowledgements. 1 Protein, Proteome, and Proteomics. 1.1 Primary goals for studying
Computational Methods for Mass Spectrometry Proteomics Eidhammer, Ingvar ISBN-13: 9780470512975 Table of Contents Preface. Acknowledgements. 1 Protein, Proteome, and Proteomics. 1.1 Primary goals for studying
Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ* S
 Author s Choice Technological Innovation and Resources 2014 by The American Society for Biochemistry and Molecular Biology, Inc. This paper is available on line at http://www.mcponline.org Accurate Proteome-wide
Author s Choice Technological Innovation and Resources 2014 by The American Society for Biochemistry and Molecular Biology, Inc. This paper is available on line at http://www.mcponline.org Accurate Proteome-wide
Increasing the Multiplexing of Protein Quantitation from 6- to 10-Plex with Reporter Ion Isotopologues
 Increasing the Multiplexing of Protein Quantitation from 6- to 1-Plex with Reporter Ion Isotopologues Rosa Viner, 1 Ryan Bomgarden, 2 Michael Blank, 1 John Rogers 2 1 Thermo Fisher Scientific, San Jose,
Increasing the Multiplexing of Protein Quantitation from 6- to 1-Plex with Reporter Ion Isotopologues Rosa Viner, 1 Ryan Bomgarden, 2 Michael Blank, 1 John Rogers 2 1 Thermo Fisher Scientific, San Jose,
Protein Catalog Number Quantity Myoglobin Sigma M ug
 Dear Colleagues, November 6, 2014 Please find enclosed the ABRF 2015 PSRG samples that you requested from the ABRF Protein Sequence Research Group. This is the 27 th study in an annual series designed
Dear Colleagues, November 6, 2014 Please find enclosed the ABRF 2015 PSRG samples that you requested from the ABRF Protein Sequence Research Group. This is the 27 th study in an annual series designed
Aplicació de la proteòmica a la cerca de Biomarcadors proteics Barcelona, 08 de Juny 2010
 Aplicació de la proteòmica a la cerca de Biomarcadors proteics Barcelona, 8 de Juny 21 Eliandre de Oliveira Plataforma de Proteòmica Parc Científic de Barcelona Protein Chemistry Proteomics Hypothesis-free
Aplicació de la proteòmica a la cerca de Biomarcadors proteics Barcelona, 8 de Juny 21 Eliandre de Oliveira Plataforma de Proteòmica Parc Científic de Barcelona Protein Chemistry Proteomics Hypothesis-free
TrioMol Isolation Reagent
 TrioMol Isolation Reagent Technical Manual No. 0242 Version 06142007 I Description... 1 II Key Features... 1 III Storage..... 1 IV General Protocol Using Triomol Isolation Reagent 1 V Troubleshooting.
TrioMol Isolation Reagent Technical Manual No. 0242 Version 06142007 I Description... 1 II Key Features... 1 III Storage..... 1 IV General Protocol Using Triomol Isolation Reagent 1 V Troubleshooting.
TrioMol Isolation Reagent
 TrioMol Isolation Reagent Technical Manual No. 0242 Version 06142007 I Description... 1 II Key Features... 1 III Storage..... 1 IV General Protocol Using Triomol Isolation Reagent 1 V Troubleshooting.
TrioMol Isolation Reagent Technical Manual No. 0242 Version 06142007 I Description... 1 II Key Features... 1 III Storage..... 1 IV General Protocol Using Triomol Isolation Reagent 1 V Troubleshooting.
Workflow concept. Data goes through the workflow. A Node contains an operation An edge represents data flow The results are brought together in tables
 PROTEOME DISCOVERER Workflow concept Data goes through the workflow Spectra Peptides Quantitation A Node contains an operation An edge represents data flow The results are brought together in tables Protein
PROTEOME DISCOVERER Workflow concept Data goes through the workflow Spectra Peptides Quantitation A Node contains an operation An edge represents data flow The results are brought together in tables Protein
Appendix II- Bioanalytical Method Development and Validation
 A2. Bioanalytical method development 1. Optimization of chromatographic conditions Method development and optimization of chromatographic parameters is of utmost important for validating a method in biological
A2. Bioanalytical method development 1. Optimization of chromatographic conditions Method development and optimization of chromatographic parameters is of utmost important for validating a method in biological
Purdue-UAB Botanicals Center for Age- Related Disease
 Purdue-UAB Botanicals Center for Age- Related Disease MALDI-TOF Mass Spectrometry Fingerprinting Technique Landon Wilson MALDI-TOF mass spectrometry is an advanced technique for rapid protein identification
Purdue-UAB Botanicals Center for Age- Related Disease MALDI-TOF Mass Spectrometry Fingerprinting Technique Landon Wilson MALDI-TOF mass spectrometry is an advanced technique for rapid protein identification
Supplemental Information
 Supplemental Information Figure S. Regions recognized by the specific antibodies. The amino acid sequence of the major p3-alc species, p3-alc α 35, p3-alc β 37 and p3-alc γ 3, are indicated as red letters.
Supplemental Information Figure S. Regions recognized by the specific antibodies. The amino acid sequence of the major p3-alc species, p3-alc α 35, p3-alc β 37 and p3-alc γ 3, are indicated as red letters.
Site-specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells
 Supplementary Information Site-specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells Tong Zhou 1, 2, Ying-hua Chung 1, 2, Jianji Chen 1, Yue Chen 1 1. Department of Biochemistry,
Supplementary Information Site-specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells Tong Zhou 1, 2, Ying-hua Chung 1, 2, Jianji Chen 1, Yue Chen 1 1. Department of Biochemistry,
Overview - MS Proteomics in One Slide. MS masses of peptides. MS/MS fragments of a peptide. Results! Match to sequence database
 Overview - MS Proteomics in One Slide Obtain protein Digest into peptides Acquire spectra in mass spectrometer MS masses of peptides MS/MS fragments of a peptide Results! Match to sequence database 2 But
Overview - MS Proteomics in One Slide Obtain protein Digest into peptides Acquire spectra in mass spectrometer MS masses of peptides MS/MS fragments of a peptide Results! Match to sequence database 2 But
Supporting Information for. PNA FRET Pair Miniprobes for Quantitative. Fluorescent in Situ Hybridization to Telomeric DNA in Cells and Tissue
 Supporting Information for PNA FRET Pair Miniprobes for Quantitative Fluorescent in Situ Hybridization to Telomeric DNA in Cells and Tissue Orenstein, et al Page S2... PNA synthesis procedure Page S3...
Supporting Information for PNA FRET Pair Miniprobes for Quantitative Fluorescent in Situ Hybridization to Telomeric DNA in Cells and Tissue Orenstein, et al Page S2... PNA synthesis procedure Page S3...
Isotopic-Labeling and Mass Spectrometry-Based Quantitative Proteomics
 Isotopic-Labeling and Mass Spectrometry-Based Quantitative Proteomics Xiao-jun Li, Ph.D. Current address: Homestead Clinical Day 4 October 19, 2006 Protein Quantification LC-MS/MS Data XLink mzxml file
Isotopic-Labeling and Mass Spectrometry-Based Quantitative Proteomics Xiao-jun Li, Ph.D. Current address: Homestead Clinical Day 4 October 19, 2006 Protein Quantification LC-MS/MS Data XLink mzxml file
Protein Quantitation II: Multiple Reaction Monitoring. Kelly Ruggles New York University
 Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot RPPA Immunohistochemistry
Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot RPPA Immunohistochemistry
Effective desalting and concentration of in-gel digest samples with Vivapure C18 Micro spin columns prior to MALDI-TOF analysis.
 Introduction The identification of proteins plays an important role in today s pharmaceutical and proteomics research. Commonly used methods for separating proteins from complex samples are 1D or 2D gels.
Introduction The identification of proteins plays an important role in today s pharmaceutical and proteomics research. Commonly used methods for separating proteins from complex samples are 1D or 2D gels.
Protein assay. Absorbance Fluorescence Emission Colorimetric detection BIO/MDT 325. Absorbance
 Protein assay Absorbance Fluorescence Emission Colorimetric detection BIO/MDT 325 Absorbance Using A280 to Determine Protein Concentration Determination of protein concentration by measuring absorbance
Protein assay Absorbance Fluorescence Emission Colorimetric detection BIO/MDT 325 Absorbance Using A280 to Determine Protein Concentration Determination of protein concentration by measuring absorbance
SUPPLEMENTARY INFORMATION
 5 N 4 8 20 22 24 2 28 4 8 20 22 24 2 28 a b 0 9 8 7 H c (kda) 95 0 57 4 28 2 5.5 Precipitate before NMR expt. Supernatant before NMR expt. Precipitate after hrs NMR expt. Supernatant after hrs NMR expt.
5 N 4 8 20 22 24 2 28 4 8 20 22 24 2 28 a b 0 9 8 7 H c (kda) 95 0 57 4 28 2 5.5 Precipitate before NMR expt. Supernatant before NMR expt. Precipitate after hrs NMR expt. Supernatant after hrs NMR expt.
Tandem mass spectra were extracted from the Xcalibur data system format. (.RAW) and charge state assignment was performed using in house software
 Supplementary Methods Software Interpretation of Tandem mass spectra Tandem mass spectra were extracted from the Xcalibur data system format (.RAW) and charge state assignment was performed using in house
Supplementary Methods Software Interpretation of Tandem mass spectra Tandem mass spectra were extracted from the Xcalibur data system format (.RAW) and charge state assignment was performed using in house
Supporting Information. Chemo-enzymatic Synthesis of Isotopically Labeled Nicotinamide Ribose
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Chemo-enzymatic Synthesis of Isotopically Labeled
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supporting Information Chemo-enzymatic Synthesis of Isotopically Labeled
Quantitative Proteomics
 BSPR workshop 16 th July 2010 Quantitative Proteomics Kathryn Lilley Cambridge Centre for Proteomics Department of Biochemistry University of Cambridge k.s.lilley@bioc.cam.ac.uk www.bio.cam.ac.uk/proteomics/
BSPR workshop 16 th July 2010 Quantitative Proteomics Kathryn Lilley Cambridge Centre for Proteomics Department of Biochemistry University of Cambridge k.s.lilley@bioc.cam.ac.uk www.bio.cam.ac.uk/proteomics/
Workshop: SILAC and Alternative Labeling Strategies in Quantitative Proteomics
 Workshop: SILAC and Alternative Labeling Strategies in Quantitative Proteomics SILAC and Stable Isotope Dimethyl-Labeling Approaches in Quantitative Proteomics Ho-Tak Lau, Hyong-Won Suh, Shao-En Ong UW
Workshop: SILAC and Alternative Labeling Strategies in Quantitative Proteomics SILAC and Stable Isotope Dimethyl-Labeling Approaches in Quantitative Proteomics Ho-Tak Lau, Hyong-Won Suh, Shao-En Ong UW
Protocol for 2D-E. Protein Extraction
 Protocol for 2D-E Protein Extraction Reagent 1 inside the ReadyPrep TM Sequential Extraction kit (in powder form) 50ml of deionized water is used to dissolve all the Reagent 1. The solution is known as
Protocol for 2D-E Protein Extraction Reagent 1 inside the ReadyPrep TM Sequential Extraction kit (in powder form) 50ml of deionized water is used to dissolve all the Reagent 1. The solution is known as
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Peptide Targeted Quantification By High Resolution Mass Spectrometry A Paradigm Shift? Zhiqi Hao Thermo Fisher Scientific San Jose, CA
 Peptide Targeted Quantification By High Resolution Mass Spectrometry A Paradigm Shift? Zhiqi Hao Thermo Fisher Scientific San Jose, CA Proteomics is Turning Quantitative Hmmm.. Which ones are my targets?
Peptide Targeted Quantification By High Resolution Mass Spectrometry A Paradigm Shift? Zhiqi Hao Thermo Fisher Scientific San Jose, CA Proteomics is Turning Quantitative Hmmm.. Which ones are my targets?
OMX-S basic Kit Instruction Manual March/09
 OMX-S for rapid in-gel digestion OMX-S basic Kit Instruction Manual March/09 Table of Contents Part No.: 17002 1. Introduction...2 Content...2 Kit components...2 Storage conditions...2 Additionally required
OMX-S for rapid in-gel digestion OMX-S basic Kit Instruction Manual March/09 Table of Contents Part No.: 17002 1. Introduction...2 Content...2 Kit components...2 Storage conditions...2 Additionally required
Protocol. Product Use & Liability. Contact us: InfoLine: Order per fax: www:
 Protocol SpikeTides Sets SpikeTides Sets_L heavy SpikeMix SpikeMix_L heavy Peptide Sets for relative quantification of Proteins in Mass Spectrometry Based Assays Contact us: InfoLine: +49-30-6392-7878
Protocol SpikeTides Sets SpikeTides Sets_L heavy SpikeMix SpikeMix_L heavy Peptide Sets for relative quantification of Proteins in Mass Spectrometry Based Assays Contact us: InfoLine: +49-30-6392-7878
M e a n. F l u o r e s c e n c e I n t e n s i t y P r o t e i n ( p g / m L )
 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0. 0. 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 P r o t e i n ( p g / m L ) 0. 0. 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 P r o t e i n ( p g / m L ) Figure. Representative
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0. 0. 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 P r o t e i n ( p g / m L ) 0. 0. 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 P r o t e i n ( p g / m L ) Figure. Representative
U.S. Patent No. 9,051,563 and other pending patents. Ver
 INSTRUCTION MANUAL Direct-zol RNA MiniPrep Catalog Nos. R050, R05, R05, & R053 Highlights Quick, spin column purification of high-quality (DNA-free) total RNA directly from TRIzol, TRI Reagent and other
INSTRUCTION MANUAL Direct-zol RNA MiniPrep Catalog Nos. R050, R05, R05, & R053 Highlights Quick, spin column purification of high-quality (DNA-free) total RNA directly from TRIzol, TRI Reagent and other
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
High-Throughput Protein Quantitation Using Multiple Reaction Monitoring
 High-Throughput Protein Quantitation Using Multiple Reaction Monitoring Application Note Authors Ning Tang, Christine Miller, Joe Roark, Norton Kitagawa and Keith Waddell Agilent Technologies, Inc. Santa
High-Throughput Protein Quantitation Using Multiple Reaction Monitoring Application Note Authors Ning Tang, Christine Miller, Joe Roark, Norton Kitagawa and Keith Waddell Agilent Technologies, Inc. Santa
HPLC Winter Webinars Part 2: Sample Preparation for HPLC
 HPLC Winter Webinars Part 2: Sample Preparation for HPLC Jon Bardsley, Application Chemist Thermo Fisher Scientific, Runcorn/UK The world leader in serving science What am I Going to Talk About? What do
HPLC Winter Webinars Part 2: Sample Preparation for HPLC Jon Bardsley, Application Chemist Thermo Fisher Scientific, Runcorn/UK The world leader in serving science What am I Going to Talk About? What do
Protein Quantitation II: Multiple Reaction Monitoring. Kelly Ruggles New York University
 Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot Immunohistochemistry
Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot Immunohistochemistry
Analysis of Labeled and Non-Labeled Proteomic Data Using Progenesis QI for Proteomics
 Analysis of Labeled and Non-Labeled Proteomic Data Using Progenesis QI for Proteomics Lee Gethings, Gushinder Atwal, Martin Palmer, Chris Hughes, Hans Vissers, and James Langridge Waters Corporation, Wilmslow,
Analysis of Labeled and Non-Labeled Proteomic Data Using Progenesis QI for Proteomics Lee Gethings, Gushinder Atwal, Martin Palmer, Chris Hughes, Hans Vissers, and James Langridge Waters Corporation, Wilmslow,
Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1
 Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1 (a) Fold-change in UCP-1 mrna abundance in white adipocytes upon β-adrenergic stimulation
Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1 (a) Fold-change in UCP-1 mrna abundance in white adipocytes upon β-adrenergic stimulation
Supporting Information
 Supporting Information Multifunctional Albumin-MnO 2 Nanoparticles Modulate Solid Tumor Microenvironment by Attenuating Hypoxia, Acidosis, Vascular Endothelial Growth Factor and Enhance Radiation Response
Supporting Information Multifunctional Albumin-MnO 2 Nanoparticles Modulate Solid Tumor Microenvironment by Attenuating Hypoxia, Acidosis, Vascular Endothelial Growth Factor and Enhance Radiation Response
SRM assay generation and data analysis in Skyline
 in Skyline Preparation 1. Download the example data from www.srmcourse.ch/eupa.html (3 raw files, 1 csv file, 1 sptxt file). 2. The number formats of your computer have to be set to English (United States).
in Skyline Preparation 1. Download the example data from www.srmcourse.ch/eupa.html (3 raw files, 1 csv file, 1 sptxt file). 2. The number formats of your computer have to be set to English (United States).
According to the manufacture s direction (Pierce), RNA and DNA
 Supplementary method Electrophoretic Mobility-shift assay (EMSA) According to the manufacture s direction (Pierce), RNA and DNA oligonuleotides were firstly labeled by biotin. TAVb (1pM) was incubated
Supplementary method Electrophoretic Mobility-shift assay (EMSA) According to the manufacture s direction (Pierce), RNA and DNA oligonuleotides were firstly labeled by biotin. TAVb (1pM) was incubated
Data Sheet. Azide Cy5 RNA T7 Transcription Kit
 Cat. No. Size 1. Description PP-501-Cy5 10 reactions à 40 µl For in vitro use only Quality guaranteed for 12 months Store all components at -20 C. Avoid freeze and thaw cycles. DBCO-Sulfo-Cy5 must be stored
Cat. No. Size 1. Description PP-501-Cy5 10 reactions à 40 µl For in vitro use only Quality guaranteed for 12 months Store all components at -20 C. Avoid freeze and thaw cycles. DBCO-Sulfo-Cy5 must be stored
RayBio Protease Activity Assay Kit
 RayBio Protease Activity Assay Kit User Manual Version 1.0 Mar 25, 2013 RayBio Protease Activity Assay (Cat#: 68AT-Protease-S100) RayBiotech, Inc. We Provide You With Excellent Support And Service Tel:(Toll
RayBio Protease Activity Assay Kit User Manual Version 1.0 Mar 25, 2013 RayBio Protease Activity Assay (Cat#: 68AT-Protease-S100) RayBiotech, Inc. We Provide You With Excellent Support And Service Tel:(Toll
Actinobacteria Relative abundance (%) Co-housed CD300f WT. CD300f KO. Colon length (cm) Day 9. Microscopic inflammation score
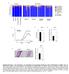 y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
Quantification of Protein Half-Lives in the Budding Yeast Proteome
 Supporting Methods Quantification of Protein Half-Lives in the Budding Yeast Proteome 1 Cell Growth and Cycloheximide Treatment Three parallel cultures (17 ml) of each TAP-tagged strain were grown in separate
Supporting Methods Quantification of Protein Half-Lives in the Budding Yeast Proteome 1 Cell Growth and Cycloheximide Treatment Three parallel cultures (17 ml) of each TAP-tagged strain were grown in separate
A pentose bisphosphate pathway for nucleoside degradation in Archaea. Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto , Japan.
 SUPPLEMENTARY INFORMATION A pentose bisphosphate pathway for nucleoside degradation in Archaea Riku Aono 1,, Takaaki Sato 1,, Tadayuki Imanaka, and Haruyuki Atomi 1, * 7 8 9 10 11 1 Department of Synthetic
SUPPLEMENTARY INFORMATION A pentose bisphosphate pathway for nucleoside degradation in Archaea Riku Aono 1,, Takaaki Sato 1,, Tadayuki Imanaka, and Haruyuki Atomi 1, * 7 8 9 10 11 1 Department of Synthetic
Plasma-free Metanephrines Quantitation with Automated Online Sample Preparation and a Liquid Chromatography-Tandem Mass Spectrometry Method
 Plasma-free Metanephrines Quantitation with Automated Online Sample Preparation and a Liquid Chromatography-Tandem Mass Spectrometry Method Xiang He and Marta Kozak ThermoFisher Scientific, San Jose, CA,
Plasma-free Metanephrines Quantitation with Automated Online Sample Preparation and a Liquid Chromatography-Tandem Mass Spectrometry Method Xiang He and Marta Kozak ThermoFisher Scientific, San Jose, CA,
OMX-S pro Instruction Manual* Oct/07
 OMX-S Instruction Manual OMX-S for rapid in-gel digestion OMX-S pro Instruction Manual* Oct/07 Part No.: 17001 Table of Contents 1. Introduction...2 2. Content and precautions...2 2.1. Content...2 2.2.
OMX-S Instruction Manual OMX-S for rapid in-gel digestion OMX-S pro Instruction Manual* Oct/07 Part No.: 17001 Table of Contents 1. Introduction...2 2. Content and precautions...2 2.1. Content...2 2.2.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
Designed for Accuracy. Innovation with Integrity. High resolution quantitative proteomics LC-MS
 Designed for Accuracy High resolution quantitative proteomics Innovation with Integrity LC-MS Setting New Standards in Accuracy The development of mass spectrometry based proteomics approaches has dramatically
Designed for Accuracy High resolution quantitative proteomics Innovation with Integrity LC-MS Setting New Standards in Accuracy The development of mass spectrometry based proteomics approaches has dramatically
NAD + /NADH Assay [Colorimetric]
![NAD + /NADH Assay [Colorimetric] NAD + /NADH Assay [Colorimetric]](/thumbs/91/105496846.jpg) G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name NAD + /NADH Assay [Colorimetric] (Cat. #786 1539, 786 1540) think proteins! think G-Biosciences
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name NAD + /NADH Assay [Colorimetric] (Cat. #786 1539, 786 1540) think proteins! think G-Biosciences
FSC-W FSC-H CD4 CD62-L
 Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Gene expression in prokaryotic and eukaryotic cells, Plasmids: types, maintenance and functions. Mitesh Shrestha
 Gene expression in prokaryotic and eukaryotic cells, Plasmids: types, maintenance and functions. Mitesh Shrestha Plasmids 1. Extrachromosomal DNA, usually circular-parasite 2. Usually encode ancillary
Gene expression in prokaryotic and eukaryotic cells, Plasmids: types, maintenance and functions. Mitesh Shrestha Plasmids 1. Extrachromosomal DNA, usually circular-parasite 2. Usually encode ancillary
Identification of TARP Phosphorylation Sites and Their Significance in the Brain
 Identification of TARP Phosphorylation Sites and Their Significance in the Brain Susumu Tomita @ SHM BE17 Department of Cellular and Molecular Physiology NIDA Neuroproteomics Center, December 13, 2008
Identification of TARP Phosphorylation Sites and Their Significance in the Brain Susumu Tomita @ SHM BE17 Department of Cellular and Molecular Physiology NIDA Neuroproteomics Center, December 13, 2008
Nitric Oxide Synthase Assay Kit
 Nitric Oxide Synthase Assay Kit Catalog Number KA1634 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay...
Nitric Oxide Synthase Assay Kit Catalog Number KA1634 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay...
MALDI-HDMS E : A Novel Data Independent Acquisition Method for the Enhanced Analysis of 2D-Gel Tryptic Peptide Digests
 -HDMS E : A Novel Data Independent Acquisition Method for the Enhanced Analysis of 2D-Gel Tryptic Peptide Digests Emmanuelle Claude, 1 Mark Towers, 1 and Rachel Craven 2 1 Waters Corporation, Manchester,
-HDMS E : A Novel Data Independent Acquisition Method for the Enhanced Analysis of 2D-Gel Tryptic Peptide Digests Emmanuelle Claude, 1 Mark Towers, 1 and Rachel Craven 2 1 Waters Corporation, Manchester,
Scope: Materials: Labeled 1.5 ml or 2 ml Eppendorf tubes Appropriate pipette tips (see Table 1) Labeled LC vials
 Scope: This SOP applies to sample protein precipitation for global metabolomics analysis by reverse phase or HILIC- HPLC-MS. Samples include but are not limited to tissue, cells, plasma, serum, and stool.
Scope: This SOP applies to sample protein precipitation for global metabolomics analysis by reverse phase or HILIC- HPLC-MS. Samples include but are not limited to tissue, cells, plasma, serum, and stool.
Quantum Dot-Peptide-Fullerene Bioconjugates for Visualization of In Vitro and In Vivo Cellular Membrane Potential
 Quantum Dot-Peptide-Fullerene Bioconjugates for Visualization of In Vitro and In Vivo Cellular Membrane Potential Okhil K. Nag, Michael H. Stewart, Jeffrey R. Deschamps, Kimihiro Susumu, Eunkeu Oh, Vassiliy
Quantum Dot-Peptide-Fullerene Bioconjugates for Visualization of In Vitro and In Vivo Cellular Membrane Potential Okhil K. Nag, Michael H. Stewart, Jeffrey R. Deschamps, Kimihiro Susumu, Eunkeu Oh, Vassiliy
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Developing Algorithms for the Determination of Relative Abundances of Peptides from LC/MS Data
 Developing Algorithms for the Determination of Relative Abundances of Peptides from LC/MS Data RIPS Team Jake Marcus (Project Manager) Anne Eaton Melanie Kanter Aru Ray Faculty Mentors Shawn Cokus Matteo
Developing Algorithms for the Determination of Relative Abundances of Peptides from LC/MS Data RIPS Team Jake Marcus (Project Manager) Anne Eaton Melanie Kanter Aru Ray Faculty Mentors Shawn Cokus Matteo
Data sheet. UV-Tracer TM Biotin-Maleimide. For Labeling of Thiol-groups with UV-detectable Biotin CLK-B Description
 Cat. No. CLK-B105-10 CLK-B105-100 Amount 10 mg 100 mg For in vitro use only! Quality guaranteed for 12 months Store at -20 C 1. Description UV-Tracer TM Biotin Maleimide for biotinylation reactions of
Cat. No. CLK-B105-10 CLK-B105-100 Amount 10 mg 100 mg For in vitro use only! Quality guaranteed for 12 months Store at -20 C 1. Description UV-Tracer TM Biotin Maleimide for biotinylation reactions of
Mass Spectrometry and Proteomics - Lecture 5 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 5 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Proteomics Sample prep 144 Lecture 5 Quantitation techniques Search Algorithms Proteomics
Mass Spectrometry and Proteomics - Lecture 5 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Proteomics Sample prep 144 Lecture 5 Quantitation techniques Search Algorithms Proteomics
NOVABEADS FOOD 1 DNA KIT
 NOVABEADS FOOD 1 DNA KIT NOVABEADS FOOD DNA KIT is the new generation tool in molecular biology techniques and allows DNA isolations from highly processed food products. The method is based on the use
NOVABEADS FOOD 1 DNA KIT NOVABEADS FOOD DNA KIT is the new generation tool in molecular biology techniques and allows DNA isolations from highly processed food products. The method is based on the use
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
Protein and antibody functionalization using continuous flow microreactor technology. Table of Contents
 1 Protein and antibody functionalization using continuous flow microreactor technology Meaghan M. Sebeika, Nicholas G. Gedeon, Sara Sadler, Nicholas L. Kern, Devan J. Wilkins, David E. Bell and Graham
1 Protein and antibody functionalization using continuous flow microreactor technology Meaghan M. Sebeika, Nicholas G. Gedeon, Sara Sadler, Nicholas L. Kern, Devan J. Wilkins, David E. Bell and Graham
Elastic CNT-polyurethane nanocomposite: Synthesis, performance and assessment of fragments released during use Online Supporting Information
 Elastic CNT-polyurethane nanocomposite: Synthesis, performance and assessment of fragments released during use Online Supporting Information Methods of lifecycle simulation testing a b Fig. SI_1 Setups
Elastic CNT-polyurethane nanocomposite: Synthesis, performance and assessment of fragments released during use Online Supporting Information Methods of lifecycle simulation testing a b Fig. SI_1 Setups
Chem 250 Unit 1 Proteomics by Mass Spectrometry
 Chem 250 Unit 1 Proteomics by Mass Spectrometry Article #1 Quantitative MS for proteomics: teaching a new dog old tricks. MacCoss MJ, Matthews DE., Anal Chem. 2005 Aug 1;77(15):294A-302A. 1. Synopsis 1.1.
Chem 250 Unit 1 Proteomics by Mass Spectrometry Article #1 Quantitative MS for proteomics: teaching a new dog old tricks. MacCoss MJ, Matthews DE., Anal Chem. 2005 Aug 1;77(15):294A-302A. 1. Synopsis 1.1.
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1
 Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
SERVA ICPL Quadruplex PLUS Kit
 INSTRUCTION MANUAL SERVA ICPL Quadruplex PLUS Kit (Cat. No. 39233.01) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 Heidelberg Phone +49-6221-138400, Fax +49-6221-1384010 e-mail: info@serva.de http://www.serva.de
INSTRUCTION MANUAL SERVA ICPL Quadruplex PLUS Kit (Cat. No. 39233.01) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 Heidelberg Phone +49-6221-138400, Fax +49-6221-1384010 e-mail: info@serva.de http://www.serva.de
Using an environmentally-relevant panel of Gramnegative bacteria to assess the toxicity of polyallylamine hydrochloride-wrapped gold nanoparticles
 Electronic Supplementary Material (ESI) for Environmental Science: Nano. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Using an environmentally-relevant panel
Electronic Supplementary Material (ESI) for Environmental Science: Nano. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Using an environmentally-relevant panel
Statistical mass spectrometry-based proteomics
 1 Statistical mass spectrometry-based proteomics Olga Vitek www.stat.purdue.edu Outline What is proteomics? Biological questions and technologies Protein quantification in label-free workflows Joint analysis
1 Statistical mass spectrometry-based proteomics Olga Vitek www.stat.purdue.edu Outline What is proteomics? Biological questions and technologies Protein quantification in label-free workflows Joint analysis
Key questions of proteomics. Bioinformatics 2. Proteomics. Foundation of proteomics. What proteins are there? Protein digestion
 s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture 2 roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture 2 roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
Peptide & Protein Quantification Kit, with RED EpicoccoStab for the accurate and sensitive determination of peptides, as well proteins
 Peptide & Protein Quantification Kit, with RED EpicoccoStab for the accurate and sensitive determination of peptides, as well proteins Description Catalog #: CH4191, 1 kit up to 2000 assays Name: Protein&Peptide
Peptide & Protein Quantification Kit, with RED EpicoccoStab for the accurate and sensitive determination of peptides, as well proteins Description Catalog #: CH4191, 1 kit up to 2000 assays Name: Protein&Peptide
Strategies for the Analysis of Therapeutic Peptides in Biofluids by LC-MS/MS. Lee Goodwin
 Strategies for the Analysis of Therapeutic Peptides in Biofluids by LC-MS/MS Lee Goodwin Sample Preparation Chromatography Detection General Strategies Examples New Approaches Summary Outline ABUNDANCE
Strategies for the Analysis of Therapeutic Peptides in Biofluids by LC-MS/MS Lee Goodwin Sample Preparation Chromatography Detection General Strategies Examples New Approaches Summary Outline ABUNDANCE
Protocol. Product Use & Liability. Contact us: InfoLine: Order per fax: www:
 Protocol SpikeTides Set TAA - light SpikeTides Set TAA_L - heavy Peptide Sets for relative quantification of Tumor Associated Antigens (TAAs) in SRM and MRM Assays Contact us: InfoLine: +49-30-6392-7878
Protocol SpikeTides Set TAA - light SpikeTides Set TAA_L - heavy Peptide Sets for relative quantification of Tumor Associated Antigens (TAAs) in SRM and MRM Assays Contact us: InfoLine: +49-30-6392-7878
ICP-MS based methods for the quantitative analysis of nanoparticles in biological samples
 ICP-MS based methods for the quantitative analysis of nanoparticles in biological samples Norbert Jakubowski Heike Traub Janina Kneipp, HU Daniela Drescher, HU norbert.jakubowski@bam.de BAM Federal Institute
ICP-MS based methods for the quantitative analysis of nanoparticles in biological samples Norbert Jakubowski Heike Traub Janina Kneipp, HU Daniela Drescher, HU norbert.jakubowski@bam.de BAM Federal Institute
Human papillomavirus,hpv ELISA Kit
 Human papillomavirus,hpv ELISA Kit Catalog No: E0787h 96 Tests Operating instructions www.eiaab.com FOR RESEARCH USE ONLY; NOT FOR THERAPEUTIC OR DIAGNOSTIC APPLICATIONS! PLEASE READ THROUGH ENTIRE PROCEDURE
Human papillomavirus,hpv ELISA Kit Catalog No: E0787h 96 Tests Operating instructions www.eiaab.com FOR RESEARCH USE ONLY; NOT FOR THERAPEUTIC OR DIAGNOSTIC APPLICATIONS! PLEASE READ THROUGH ENTIRE PROCEDURE
Supporting Information
 Copyright WILEY-VCH Verlag GmbH & Co. KGaA, 69469 Weinheim, Germany, 2013. Supporting Information for Adv. Mater., DOI: 10.1002/adma.201301472 Reconfigurable Infrared Camouflage Coatings from a Cephalopod
Copyright WILEY-VCH Verlag GmbH & Co. KGaA, 69469 Weinheim, Germany, 2013. Supporting Information for Adv. Mater., DOI: 10.1002/adma.201301472 Reconfigurable Infrared Camouflage Coatings from a Cephalopod
