Formation and specification of a Drosophila dopaminergic precursor cell
|
|
|
- Kelly Atkins
- 5 years ago
- Views:
Transcription
1 3316 RESEARCH ARTICLE AND STEM CELLS Development 139, (2012) doi: /dev Published by The Company of Biologists Ltd Formation and specification of a Drosophila dopaminergic precursor cell Joseph D. Watson and Stephen T. Crews* SUMMARY Dopaminergic neurons play important roles in animal behavior, including motivation, reward and locomotion. The Drosophila dopaminergic H-cell interneuron is an attractive system for studying the genetics of neural development because analysis is focused on a single neuronal cell type. Here we provide a mechanistic understanding of how MP3, the precursor to the H-cell, forms and acquires its identity. We show that the gooseberry/gooseberry-neuro (gsb/gsb-n) transcription factor genes act to specify MP3 cell fate. It is proposed that single-minded commits neuroectodermal cells to a midline fate, followed by a series of signaling events that result in the formation of a single gsb + /gsb-n + MP3 cell per segment. The wingless signaling pathway establishes a midline anterior domain by activating expression of the forkhead transcription factors sloppy paired 1 and sloppy paired 2. This is followed by hedgehog signaling that activates gsb/gsb-n expression in a subgroup of anterior cells. Finally, Notch signaling results in the selection of a single MP3, with the remaining cells becoming midline glia. In MP3, gsb/gsb-n direct H-cell development, in large part by activating expression of the lethal of scute and tailup H-cell regulatory genes. Thus, a series of signaling and transcriptional events result in the specification of a unique dopaminergic precursor cell. Additional genetic experiments indicate that the molecular mechanisms that govern MP3/H-cell development might also direct the development of non-midline dopaminergic neurons. KEY WORDS: Cell fate, CNS midline, Dopamine, Drosophila, gooseberry, Neuron INTRODUCTION A key aspect of neurogenesis concerns how neural precursors are generated and acquire specific fates. The simplified view is that patterning proteins, consisting of intercellular signaling pathway components and transcription factors, activate downstream transcription factors that promote neural precursor formation and direct specific precursor fates (Skeath and Thor, 2003). The combined action of these proteins activates additional factors that control neuron-specific differentiation. Despite conceptual understanding of the factors involved, there are few in vivo examples in which the developmental progression of individual neurons has been comprehensively followed from the undifferentiated neuroectoderm to the differentiated neuron. Yet, studies of individual neurons and their precursors are particularly valuable for their detailed, mechanistic insights. An attractive system for the systematic study of neuronal development is provided by Drosophila CNS midline cells (Fig. 1) (Wheeler et al., 2006). These cells reside between the two hemiganglia of the Drosophila ventral nerve cord (VNC). Initially consisting of 16 ectodermal cells per segment (referred to as mesectoderm ), these cells express single-minded (sim), which acts as a master regulator of midline cell development (Nambu et al., 1991). From this seemingly uniform set of precursor cells emerge an array of diverse interneurons, motoneurons, neurosecretory neurons, axon-ensheathing anterior midline glia (AMG) and non-ensheathing posterior midline glia (PMG) (Wheeler et al., 2006). Five of the 16 mesectodermal cells give rise Department of Biochemistry and Biophysics, Program in Molecular Biology and Biotechnology, The University of North Carolina at Chapel Hill, Chapel Hill, NC , USA. *Author for correspondence (steve_crews@unc.edu) Accepted 21 June 2012 to midline precursors (MPs; MP1, MP3, MP4, MP5 and MP6) that divide only once to generate two neurons (Wheeler et al., 2008). MP1 divides symmetrically to generate two identical MP1 peptidergic motoneurons, MP3 divides asymmetrically into the dopaminergic (DA) H-cell and glutamatergic H-cell sib interneurons, and MP4-6 each divide asymmetrically to yield a GABAergic ivum interneuron and a glutamatergic/ octopaminergic mvum motoneuron. One additional midline neural precursor, the median neuroblast (MNB), is a neural stem cell that divides throughout embryonic and postembryonic development. Analysis of midline cell development has been greatly facilitated by large-scale identification of midline-expressed genes and the ability to identify each cell type at all stages of embryonic development (Kearney et al., 2004; Wheeler et al., 2006; Wheeler et al., 2008; Wheeler et al., 2009). Recent work has focused on the development of the midline DA H-cell neuron (Stagg et al., 2011; Wheeler et al., 2008). The MP3 precursor to the H-cell emerges during stage 11 from the mesectodermal cells (Fig. 1), rotates its spindle perpendicular to the longitudinal axis and divides asymmetrically into a basal H-cell and an apical H-cell sib. Notch signaling is required for H-cell sib fate, but the asymmetric localization of Numb in the H-cell blocks Notch signaling, leading to its divergent fate (Wheeler et al., 2008). Both the Lethal of scute [L(1)sc] and Tailup (Tup) transcription factors are present in the H-cell (Stagg et al., 2011; Thor and Thomas, 1997). Embryos mutant for l(1)sc fail to express any of the genes required for H-cell-specific differentiation, whereas tup mutants lack expression of a subset of H-cell differentiation genes, including pale (ple; tyrosine hydroxylase), Dopamine transporter (DAT) and Dopa decarboxylase (Ddc). l(1)sc is also required for the formation of MP4-6 and controls mvum differentiation (Stagg et al., 2011). Since l(1)sc function leads in one case (H-cell) to a DA fate and in the other case (mvum) to a motoneuron fate, this suggests that the distinct identities and properties of their precursors (MP3 and MP4-6) lead to alternative neuronal fates.
2 DA precursor formation and specification RESEARCH ARTICLE 3317 (Patel et al., 1989), rabbit anti-gfp (1:100; Abcam), rabbit anti-hb (1:100) (Tran and Doe, 2008), guinea pig and rat anti-l(1)sc (1:250 with TSA) (Stagg et al., 2011), guinea pig anti-lim3 (1:250) (Broihier and Skeath, 2002), guinea pig anti-runt (1:400 with TSA) (Kosman et al., 1998), mouse anti-tau (1:100; Tau-2, Sigma) and guinea pig anti-zfh1 (1:250) (Vogler and Urban, 2008). Stained embryos were imaged on Zeiss LSM- PASCAL, LSM-510 and LSM-710 confocal microscopes. Fig. 1. Summary of Drosophila midline neurogenesis. During stages 10 to 11, 16 mesectodermal cells (gray) develop into six midline neural precursors [MP1, MP3, MP4, MP5, MP6 and the median neuroblast (MNB)] and two groups of midline glia [anterior midline glia (AMG) and posterior midline glia (PMG)]. Each MP divides once to generate two neurons. This proposition raises several related issues: (1) what is the molecular basis of MP3 formation and MP3 identity specification; (2) how do these genes regulate l(1)sc and tup to ultimately control H-cell differentiation; (3) are midline cells pre-patterned into domains permissive and non-permissive for MP3 specification; and (4) are the genes that specify MP3 development also used to control development of other Drosophila DA neuronal lineages? Here, we address the genetic mechanisms involved in MP3 formation and specification. MATERIALS AND METHODS Drosophila strains and genetic analysis Drosophila mutant strains included: Df(2R)gsb (Nusslein-Volhard et al., 1984), Df(2R)Kr10 (Gutjahr et al., 1993), hh AC (Lee et al., 1992), ptc 7 wg 1-12 (Nusslein-Volhard et al., 1984), ptc 9 (Nusslein-Volhard et al., 1984), slp 34B (Grossniklaus et al., 1992), slp1 2 (Nusslein-Volhard et al., 1984) and wg 1-8 (Nusslein-Volhard et al., 1984). Mutant strains were obtained from the Bloomington Drosophila Stock Center. Gal4 and UAS lines employed were: sim-gal4 (Xiao et al., 1996), prd-gal4 (Xiao et al., 1996), UAS-ci.VP16 (Larsen et al., 2003), UAS-en (Guillén et al., 1995), UAS-gsb (Marie et al., 2010), UAS-gsb-n (Colomb et al., 2008), UAS-hh (Porter et al., 1996), UAS-slp1 (Sato and Tomlinson, 2007) and UAS-tau-GFP (Brand, 1995). The ptc 7 wg 1-12 strain was grown at 29 C to reduce wg function. Wild-type, mutant and misexpression embryos contained sim-gal4 UAStau-GFP in the background to assist in visualizing midline cells. Homozygous mutant embryos were identified by either: (1) staining for the absence of marked balancer TM3 P[ftz-lacZ] expression; (2) staining by in situ hybridization for lack of gene expression in deletion mutants; or (3) assaying for characteristic alterations in gene expression. In situ hybridization, immunostaining and microscopy Embryo collection, in situ hybridization and immunostaining were performed as previously described (Kearney et al., 2004). Embryos were commonly hybridized to two RNA probes, one labeled with digoxygenin and another with biotin, along with immunostaining with two antibodies (see under Information>protocols). RNA probes for in situ hybridization were generated from the Drosophila Gene Collection (Open Biosystems) (en, Gad1, gsb-n, odd, ple, slp1, slp2 and VGlut) or PCR amplified from genomic DNA (gsb, hh, tup). Primary antibodies used were: rat anti-elav (1:3; Developmental Studies Hybridoma Bank), mouse anti-en (1:25) RESULTS MP3 gives rise to both the H-cell and H-cell sib, and genes were sought that affected the development of MP3 and its progeny. Since MPs arise in defined positions along the anterior-posterior axis (Bate and Grunewald, 1981; Wheeler et al., 2008), we hypothesized that anterior-posterior patterning genes [hedgehog (hh), patched (ptc), wingless (wg), slp1, slp2, gsb, gsb-n, engrailed (en)] might play a role in directing MP cell fate. Eight of these genes were assayed for effects on MP3 lineage development using mutant and misexpression/overexpression approaches. gsb and gsb-n transcription factor genes specify MP3 fate The gsb and gsb-n genes are related in sequence, reside within 9.7 kb, have similar patterns of embryonic expression and are partially redundant (Duman-Scheel et al., 1997). At stages 10-11, both gsb and gsb-n are expressed in neuroectodermal stripes that are collinear with, and include, MP3 (Fig. 2A,C; supplementary material Fig. S1A,E) (Bossing and Brand, 2006). MP3 is the only MP in which either gene is expressed. Both genes are also expressed in the H-cell and H-cell sib at stage 11 (Fig. 2B,D), but are absent from all other midline neurons and MPs. Expression of both genes is absent in midline cells by late stage 12. The highly specific expression of gsb and gsb-n in MP3 and its progeny suggested that one or both of these genes play important roles in MP3 cell fate. This was initially tested using a gsb gsb-n double-mutant strain [Df(2R)gsb]. In these experiments and throughout, midline cell identity was based on position, morphology and the use of cell-specific markers in multiply stained embryos (for examples, see Fig. 2I-N, Fig. 5B,B,E,I,J, Fig. 6H,N,P). Cell fate changes were assessed by analyzing embryos at stages for the following midline neuronal differentiation markers: H-cell (ple, tup), H-cell sib (high levels of VGlut), MP1 (Lim3, odd, Runt), ivums (En, Gad1) and mvums (Zfh1, low VGlut) (Fig. 2E-H) (Stagg et al., 2011; Wheeler et al., 2006; Wheeler et al., 2008); these data are quantitated in supplementary material Figs S2-S5. The Df(2R)gsb embryos showed an absence of cells expressing ple (H-cell) or high VGlut (H-cell sib) and a corresponding doubling of Lim3 + MP1 neurons was often observed (29% of segments scored) (Fig. 2I-K). Zfh1 + mvum numbers also increased in 50% of Df(2R)gsb segments, most often from three to four cells (Fig. 2L). The interpretation of these findings is that gsb/gsb-n are required for MP3 cell fate, and in their absence MP3 is transformed into either an MP1 or MP4-6. Since in Df(2R)gsb embryos MP3 fate is transformed rather than fails to appear, this indicates that gsb/gsb-n are not required for MP3 formation or cell division. However, we note that the timing of division, as revealed by the appearance of some Tau-dense dividing cells adjacent to MP1 neurons, is delayed compared with the normal timing of the MP3 division (Fig. 2W). The respective roles of gsb and gsb-n were analyzed by examining embryos homozygous mutant for only gsb. Since singlegene mutants for gsb and gsb-n are not available, we analyzed
3 Development 139 (18) Fig. 2. gsb and gsb-n direct MP3 cell fate. All panels are sagittal views of single segments; anterior is to the left and internal up. Embryos carry sim-gal4 UAS-tau-GFP and are stained with anti-tau to illustrate midline cells (green), with in situ hybridization or immunostaining as indicated. (A) At stage 10, gsb RNA is present in MP3 (yellow arrowhead) and surrounding MG (asterisks), but not in MP4 (white arrowhead). (B) At stage 11, gsb is present in the H-cell and H-cell sib (yellow arrows) but is absent in MP1 (blue arrowhead) and mvum4 and ivum4 (white arrows). (C) At stage 10, gsb-n is expressed only in MP3 (yellow arrowhead), not MP1 (blue arrowhead) and MP4 (white arrowhead). (D) At stage 11, gsb-n is expressed in the H-cell and H-cell sib (yellow arrows), but is absent from MP1 (blue arrowhead) and mvum4 and ivum4 (white arrows). (E-P) Stage embryos. (F,J,N ) Only VGlut expression is shown (compare with F,J,N). (E-H) Wild-type expression of (E) ple in Hcell (arrow), (F,F ) high levels of VGlut in H-cell sib (white arrow) and low levels in mvums (yellow arrows), (G) Lim3 in two MP1 neurons (arrows) and (H) Zfh1, which is strong in the three mvums (white arrows) and weak in the tup+ H-cell (yellow arrow). (IL) Df(2R)gsb embryos. (I) ple expression is absent. (J,J ) High VGlut expression is absent; yellow arrows indicate low VGlut mvums. (K) Four Lim3+ MP1 neurons are present (arrows). (L) Three Zfh1+ mvums are present (arrows); the additional Zfh1+ cell (asterisk) might be a fourth mvum. (M-P) sim-gal4 UAS-gsb-n embryos. Two ple+ cells (M) and two high VGlut cells (N,N ) are present (arrows). (O) Lim3 protein is absent. (P) Only one strong Zfh1+ mvum (arrow) is present. There are two weak Zfh1+ cells (asterisks) that are ple+ H-cells. (Q-V) Embryos at (Q-S) stages 13-14, (T) mid stage 11 (sm11), (U) early stage 11 (se11) and (V) late stage 11 (sl11). (Q) Wild-type expression of tup in H-cell (arrow). (R) Expression of tup is absent in Df(2R)gsb embryos. (S) Misexpression of gsb-n results in two tup+ cells (arrows). (T) Wild-type embryo shows L(1)sc protein in H-cell and H-cell sib (yellow arrows), but its absence in MP1 neurons (blue arrows) and AMG (white asterisks). L(1)sc is also present in MP5, MP6, MNB and PMG (yellow asterisks). (U) Misexpression of gsb-n results in activation of L(1)sc in MP1 (blue arrowhead) and AMG (white asterisks). H-cell and H-cell sib are indicated by yellow arrows and PMG by yellow asterisks. (V) In older gsb-n misexpression embryos, L(1)sc was present in MP1 neurons (blue arrow; only one neuron shown), AMG (white asterisks) and H-cell and H-cell sib (yellow arrows). (W-Y) Stage 11 Df(2R)gsb embryo stained for (W) Runt and (X,Y) L(1)sc. Shown is a Runt+ MP1 neuron (blue arrow), dividing MP3 (yellow arrowhead) and mvum4 and ivum4 (white arrows). L(1)sc is present in posterior cells but not in MP3/H-cell [compare with wild-type H-cell and H-cell sib L(1)sc staining in T; yellow arrows]. Note that the timing of the dividing MP3 is delayed compared with wild type (Wheeler et al., 2008). transheterozygous embryos that were heterozygous for gsb-n but lacked both copies of gsb [Df(2R)Kr10/Df(2R)gsb]. Expression of gsb-n was significantly reduced in the lateral CNS (supplementary material Fig. S1G, compare with S1A), as observed previously (Gutjahr et al., 1993). In the midline, gsb-n was present in some segments, but absent in others (supplementary material Fig. S1G). Examination of embryos for ple and high VGlut expression indicated an absence of ple expression in 85% of segments (supplementary material Fig. S1H, Fig. S2) and an absence of high VGlut in 54% of segments (supplementary material Fig. S1I, Fig. S3). Thus, mutant analysis indicates that gsb plays a role in activating gsb-n expression in MP3 and is important for MP RESEARCH ARTICLE
4 DA precursor formation and specification RESEARCH ARTICLE 3319 lineage development. However, as Df(2R)Kr10/Df(2R)gsb embryos do not show as severe a phenotype as Df(2R)gsb embryos (supplementary material Figs S2, S3), both gsb and gsb-n are required for MP3 development. In a complementary experiment, sim-gal4 UAS-gsb-n embryos, in which gsb-n is expressed in all midline cells at stages 10-11, showed an increase in ple + H-cells (Fig. 2M) and high VGlut H- cell sibs (Fig. 2N,N ) from one to two cells per segment. Accordingly, Lim3 expression was absent (Fig. 2O), suggesting that MP1 was transformed to MP3. There was also a general decrease in VUM neurons from six to two cells, as assayed by zfh1 expression (Fig. 2P). However, these cells were not transformed to additional H-cells and H-cell sibs, so their fate is unclear. In contrast to UAS-gsb-n, misexpression of gsb (sim-gal4 UAS-gsb) did not show an obvious effect on midline neuron cell fate (not shown). In summary, the gsb/gsb-n mutant and misexpression data are consistent with gsb-n and gsb driving MP3 cell fate, but not its formation. Mechanistically, gsb/gsb-n normally repress MP1 and MP4 fate in MP3, while also promoting MP3 fate. The l(1)sc and tup transcription factor genes are both expressed in the H-cell and control H-cell differentiation and gene expression (Fig. 2Q,T) (Stagg et al., 2011). We addressed whether their expression was controlled by gsb/gsb-n. H-cell expression of tup was absent in Df(2R)gsb (Fig. 2R) and misexpression of gsb-n most often led to the appearance of two or more tup + H-cells in 28/38 segments scored (Fig. 2S). Similarly, l(1)sc expression was absent from the H-cell in Df(2R)gsb mutants (Fig. 2W-Y), and gsbn misexpression resulted in a strong increase in l(1)sc expression in MP1 neurons and AMG (Fig. 2U,V). These results indicate that gsb/gsb-n (directly or indirectly) regulate the expression of tup and l(1)sc. slp1/2 establish a permissive anterior midline domain for MP3 and MP1 cell fates Having established that gsb/gsb-n control MP3 cell fate, the next issue concerns how gsb/gsb-n expression is activated in the midline cells. The first developmental event involves establishing an anterior midline domain, compatible with the formation of MP3. This hypothesis is based on the observation that ectopic gsb-n most often generates a single additional MP3 at the expense of the more anterior MP1 or adjacent MP4; it does not generally convert more posterior MPs (MP5 and MP6) and the MNB to MP3. We analyzed the role of slp1 and slp2, which are closely related in sequence, reside within 9.7 kb, have similar expression patterns and genetically have largely redundant segmentation phenotypes (Cadigan et al., 1994). At stage 9, both genes are expressed in mesectodermal rows D-F, which are adjacent to, but do not overlap with, the en expression domain (rows G,H) (Wheeler et al., 2006). Rows D-F are likely to give rise to MP3. At stage 10, both genes are expressed in anterior midline cells, including MP1, MP3 and Fig. 3. slp1/2 establish an MP3/MP1 permissive region in anterior midline cells. (A,C) At stage 10, slp1 and slp2 are expressed in MP1 (blue arrowhead), MP3 (yellow arrowhead), AMG (asterisks), but not MP4 (white arrowhead). (B,D) At stage 11, slp1 and slp2 expression is present in MP1 (blue arrowhead) and some AMG (asterisks), but absent in H-cell and H-cell sib (yellow arrows) and ivum4 and mvum4 (white arrows). (E-H) In slp 34B embryos, H-cell (gsb-n, ple), H-cell sib (high VGlut) and MP1 (odd) gene expression is absent. (M) En is present in multiple anterior midline neurons (white arrows) in addition to the three ivums (yellow arrows). (N,O) Additional Zfh1 + (N, asterisks) and low VGlut (O, asterisks) neurons are present in slp 34B in addition to the three mvums (also asterisks). (P) At stage 12, En expands into most midline cells in slp 34B embryos. In this segment, two glia (asterisks) and four neurons (not shown) were En. (I-K) slp1 mutant (slp1 2 ) had reductions in gsb-n, ple and VGlut expression. (L) Lim3 protein was weakly present (arrows) in slp1 2. (Q,R) There is an extra En + cell (Q, white arrow) and Zfh1 + cell (R, white arrow) in anterior midline cells in slp1 2 embryos at the position of MP1 neurons. Yellow arrows mark (Q) ivums and (R) mvums. (S-U) Stage 15 prd-gal4 UAS-slp1 embryos were stained for (S) ple, (T) Zfh1 and (U) En. Multiple ple + cells (yellow arrows) were observed posterior to the H-cell (white arrow), whereas no Zfh1 + mvums or En + ivums were observed. (U) The En + cell (asterisk) is either MNB progeny or PMG.
5 3320 RESEARCH ARTICLE Development 139 (18) AMG (Fig. 3A,C; supplementary material Fig. S1B,F). However, after MP3 divides during stage 11, expression of both slp1 and slp2 is absent from the H-cell and H-cell sib (Fig. 3B,D). Thus, slp1 and slp2 overlap in expression in the midline cells from which MP3 will form. The potential role of slp1 and slp2 in midline cell development was tested by genetic analysis using an slp1 slp2 double-null strain (slp 34B ) and an slp1 null mutant strain (slp1 2 ). In slp 34B embryos, MP3 and H-cell gsb-n expression was absent at stages (Fig. 3E), and H-cell ple and tup expression and H-cell sib high VGlut expression (Fig. 3F,G; tup not shown) were absent at later stages. MP1 neuronal odd expression was also absent in slp 34B embryos at stages 12 and 13 (Fig. 3H). Thus, MP3 and MP1 progeny were absent. There was a corresponding increase in MP4-6 VUM progeny, as indicated by additional En + ivums and Zfh1 + low VGlut mvums (Fig. 3M-O). Examination of earlier, stage 12 mutant embryos indicated that En expanded throughout most of the segment (Fig. 3P). This result reinforces the view that slp1/2 repress posterior gene expression in anterior cells. Similar results were observed for the slp1 2 single mutant, except that the effects were weaker (Fig. 3I-L,Q,R), suggesting that slp1 and slp2 act redundantly. Together, these results indicate that slp1/2 are required for MP1 and MP3 fates and repress MP4-6 fates in both MP3 and MP1. The slp1/2 mutant results suggested that misexpressing slp1/2 in all midline cells might convert MP4-6 to MP3 or MP1. Whereas sim-gal4 UAS-slp1 embryos were unaffected in midline cell fates (data not shown), when we expressed slp1 earlier using prd-gal4 we observed an increase in the number of ple + cells at the expense of Zfh1 + mvums and En + ivums (Fig. 3S-U). These results reinforce the slp1/2 mutant results and indicate that slp1/2 influence MP3 and MP1 fate by establishing a permissive anterior midline environment at stages 9-10 for specification of MP1 and MP3 identity. slp1/2 might accomplish this by repressing genes, including en, that, if expressed in these cells, would shift them toward posterior midline MP4-6 fates. Mutants in wg activate slp1/2 expression and MP3 fate The next issue concerns how the slp1/2 anterior domain is established. Previous work has shown that wg can induce slp1/2 expression in other cell types (Bhat et al., 2000) and wg can influence midline gene expression (Bossing and Brand, 2006). Consequently, we addressed whether wg signaling influences slp1/2 expression and MP3 development. wg encodes a secreted signaling protein, and at stage 9 it is expressed in a stripe, including midline cell rows E and F, that spans the neuroectoderm (supplementary material Fig. S1C) (Wheeler et al., 2006; Xiao et al., 1996). Expression of wg is absent from the midline at stages 10-11, but remains in the lateral stripe adjacent to the midline. Analysis of wg null mutant (wg 1-8 ) embryos indicates a strong reduction of gsb-n, ple, tup and of high VGlut expression (Fig. 4A- D), consistent with a loss of MP3. There was a mild reduction in the number of MP1 neurons, with 25% of segments possessing no MP1 neurons (Fig. 4E) and 75% of segments with the wild-type number of two neurons. VUMs were relatively unaffected, although one to two additional Zfh1 + cells were observed in 61% of segments (Fig. 4F). Of particular note is that slp1 expression was absent from the midline in wg mutant embryos (Fig. 4G). In summary, these data suggest a model in which wg activates slp1/2 in an anterior midline domain, thus allowing MP1 and MP3 to form. Ectopic en alters MP3 and MP1 fates One potential role of slp1/2 is to repress posterior gene expression in the anterior midline cells. A strong candidate is en, the posterior expression of which is adjacent to, but does not overlap with, slp1/2 expression (supplementary material Fig. S1B,F). At stage 9, en is expressed in a stripe that includes midline rows G and H and is collinear with the lateral ectodermal stripe (Kearney et al., 2004). Expression of en at stage 10 is present in two MG that lie between the MP3 and MP4 neural precursors (Fig. 5A) (Watson et al., 2011). At stage 11, en expands into MP4, MP5, MP6, MNB and all PMG (Fig. 5B) (Wheeler et al., 2006). Most importantly, en is not expressed in MP1, MP3, H-cell or H-cell sib (Fig. 5B ). Later, at stage 15, En is prominently expressed in the three ivum neurons (Fig. 5C). Analysis of slp1/2 mutant and slp1 misexpression embryos indicates that they repress en (Fig. 3M,P,Q,U), and possibly other genes, in anterior midline cells. This suggests that if en is present in MP3 its fate might be altered. This was addressed by misexpressing en in MP3 and MP1 in sim-gal4 UAS-en embryos. The expression of gsb-n in MP3 was not significantly affected (Fig. 5D), indicating that the presence of en did not block activation of gsb-n. However, ectopic en resulted in a decrease of ple, tup (both H-cell), high VGlut (H-cell sib) and Lim3 (MP1) expression (Fig. 5E-H). There was a small increase in Zfh1 + mvums (3.4±0.5; Fig. 5I) and Gad1 + ivums (3.6±0.6; Fig. 5J) compared with wild type (3.0 cells). The additional VUM cells were usually the anteriormost neurons at the position of MP1 neurons. By contrast, neurons at the position of H-cell and H-cell sib (Fig. 5I,J, asterisks) generally did not express any midline neuron marker, although occasionally low levels of ple were present. This suggests that the presence of en in the H-cell and H-cell sib alters their neuronal identity, but does not Fig. 4. wg signaling influences slp1/2 expression. (A-E) In wg 1-8 embryos, expression of gsb-n, ple, tup, high VGlut and Lim3 was strongly reduced. The weak midline VGlut staining (D, arrows) is from mvums. (F) Additional Zfh1 + cells (white arrows) were often observed in wg 1-8 embryos (mvums, yellow arrows). (G) Expression of slp1 was absent in wg 1-8.
6 DA precursor formation and specification RESEARCH ARTICLE 3321 Fig. 5. Ectopic en alters MP1 and MP3 cell fate. (A) At stage 10, En is present in only two MG cells (arrow; only one cell is shown). En is absent from all neural precursors, including MP1 (blue arrowhead), MP3 (yellow arrowhead) and MP4 (white arrowhead). (B,B ) At stage 11, En defines the posterior midline cells. The same segment is shown stained for (B) En and (B ) gsb-n. (B) En is present in VUM4 progeny (white arrows) and MP5, MP6, MNB and PMG (asterisks). En is absent from H-cell and H-cell sib (yellow arrows), which are both gsb-n + (B ), and En is also absent in MP1 (blue arrowheads). (C) At stage 15, En is present in the three ivum4-6 neurons (arrows). (D-H) In stage 11 sim-gal4 UAS-en embryos, gsb-n expression in H-cell and H-cell sib (arrows) was unaffected (D) and expression of ple, tup, high VGlut and lim3 was greatly reduced (E-H). Low levels of ple could be detected in a single cell (asterisk), indicating that it is likely to be an H-cell. (I,J) Additional Zfh1 + and Gad1 + cells are present in the position of MP1 neurons (white arrows) in sim-gal4 UAS-en embryos. Yellow arrows indicate (I) mvums and (J) ivums, and probable H-cell and H- cell sib are indicated by asterisks (see E). (K) Absence of slp1 midline expression in a stage 11 sim-gal4 UAS-en embryo. transform these cells into VUMs; MP1s might be more permissive to VUM transformation by en. Ectopic en also resulted in an absence of slp1 from midline cells (Fig. 5K). Since slp1/2 repress en, these results indicate that en and slp1/2 mutually repress each other. If en is expressed in MP1 and MP3 it can alter their fates and thus it is important to restrict its expression, which is a function of slp1/2. These results also indicate that misexpressed en has the ability to drive MP1 cells into an MP4-6 fate or that en interferes with the establishment of MP1 fate leading to a default state resembling MP4-6. MP3 fate is dependent on hh signaling slp1/2, activated by wg, establish an anterior domain that is necessary for the expression of gsb-n in MP3. However, we propose that wg and slp1/2 commit anterior midline cells to an MP1 fate and that an additional factor is required to commit a group of those cells to an MP3 fate. An attractive candidate for this factor is Hh, a prominent secreted signaling protein that directs alternative MG fates (Watson et al., 2011). At stages 10-11, hh is largely absent from the midline, but is expressed as a stripe in the lateral neuroectoderm (supplementary material Fig. S1D). We addressed whether hh signaling plays a role in MP3 cell fate. Initially, elav expression was examined at stages in hh mutants to assess whether hh affects midline neuronal cell number (Robinow and White, 1991). Segments contained 4.2±1.1 Elav + cells, whereas wild-type embryos contained 12.0 neurons at these stages (Fig. 6A,B; supplementary material Fig. S6A). However, 72% of embryos contained either four (46%) or six (26%) neurons. These data indicate that hh is required for the production of many midline neurons. Staining with neuron-specific markers indicated that when four or six neurons were present in hh null mutant (hh AC ) embryos, they comprised two MP1 neurons (the progeny of a single MP1 precursor), one to two mvums and one to two ivums (Fig. 6G,I,K; supplementary material Fig. S6B-D; stages 12-14). By contrast, H-cell (Fig. 6M), H-cell sib (Fig. 6O), one to two ivums, one to two mvums, and the MNB and its progeny were absent. Consistent with the absence of H-cell and H-cell sib gene expression, gsb-n expression was absent at stages (Fig. 6Q) in the midline of hh AC embryos. As expected, tup was also absent (Fig. 6S) at stage 11 and later, and L(1)sc staining was absent in the midline of hh AC stage 11 embryos (Fig. 6U). The presence of only four to six neurons in hh AC embryos indicates that, generally, one or two of three MP4-6 cells and the MNB require hh signaling for formation. Surprisingly, the slp1 expression domain is increased in some segments in hh mutant embryos at stages 10-11, although levels are reduced compared with wild type (Fig. 6C-F). This suggests that hh signaling helps in maintaining a sharp boundary of slp1/2 expression. Despite this expansion of slp1/2, hh mutants fail to generate an MP3 or additional MP1s. Thus, we hypothesize that hh signaling plays three roles in midline neuronal development: (1) a proneural role for MPs and the MNB; (2) a role in specifying MP3 cell fate; and (3) a role in restricting slp1/2 expression to anterior midline cells. Consistent with the hh MP3 cell fate mutant phenotype, overactivation of hh signaling in all midline cells resulted in an increase in MP3s. ptc encodes an Hh receptor that, in the absence of hh signaling, inhibits the ability of Smoothened (Smo) to activate the hh signaling pathway (Ingham and McMahon, 2001). When activated by Hh, Smo is released from Ptc inhibition. Thus, ptc expression is present in cells responding to hh signaling, and ptc mutants act as constitutive activators of hh signaling. At stage 10, ptc is expressed in most midline cells, including all MPs, with the exception of two en + MG that lie between MP3 and MP4 (Watson et al., 2011). Most striking are the high levels of ptc in
7 3322 RESEARCH ARTICLE Development 139 (18) Fig. 6. hh signaling is required for MP3 formation and identity. Analyses of sim-gal4 UAStau-GFP embryos are shown. (A) This wild-type embryo had 11 Elav+ neurons (eight are shown). (B) This hhac embryo had six Elav+ neurons (asterisks marksfour). (C,D) slp1 expression was expanded in the epidermis of hhac mutants [compare the width of the white bar in wild-type (C) and mutant (D)]. (E,F) slp1 expression was expanded (white bar) in midline cells in hhac. (G) In hhac mutants, two Runt+ MP1 neurons (arrows) were present, as in wild type. (I,K) hhac embryos possessed only (I) one Zfh1+ mvum and (K) one En+ ivum (arrows). (M,O,Q,S,U) In hhac mutants, there was an absence of (M) ple, (O) high VGlut, (Q) gsb-n, (S) tup and (U) l(1)sc expression. (H,J,L) ptc9 mutants had an absence of Lim3 (H), but a wild-type number of three mvums (J) and three ivums (L) (arrows). (N,P) ptc9 mutant had additional (N) ple+ and (P) high VGlut cells (arrows); two segments are shown. (R) Stage 11 ptc9 embryo had four gsb-n+ cells (arrows). (T) tup is ectopically expressed in a ptc9 mutant in three cells (arrows). (V) In ptc9, L(1)sc is ectopically present in MP1 neurons (blue arrow; only one MP1 neuron is shown), as well as in the H-cell and H-cell sib (yellow arrows) and AMG (white asterisks). material Fig. S7A,B,D,G), whereas zfh1 mvum expression and en ivum expression were expanded (supplementary material Fig. S7C,E,F). Thus, wg is required for gsb-n MP3 expression. Similarly, the ability of activated hh in anterior midline cells to generate additional gsb-n+ MP3s via sim-gal4 UAS-Ci.VP16 indicates that hh can activate gsb-n expression. In summary, the hh mutant and misexpression experiments demonstrate that hh signaling emanating from outside the midline is directly responsible for MP3 cell fate. When overexpressed, hh signaling has the ability to convert MP1 into MP3, indicating that MP1 and MP3 derive from a developmentally similar ground state. Genes controlling MP3 development are also required for the development of other Drosophila dopaminergic neurons We have demonstrated that gsb/gsb-n, hh/ptc, wg and slp1/2 are required for MP3/H-cell development. Do these genes also control the development of DA neurons in other lineages? In the embryonic VNC, two additional DA neurons exist per hemisegment in addition to the H-cell (Lundell and Hirsh, 1994). These are the paramedial DA neurons and dorsal lateral DA neurons (Fig. 7A). Interestingly, most of these neurons are collinear with the H-cell suggesting that the same segmentation genes that control H-cell development also play a role in paramedial and dorsal lateral neuron development. Consistent with this view, recent work (Tio et al., 2011) has indicated that both DA neurons are wg+, which overlaps in lateral CNS expression with gsb and slp1/2 MP3 (Watson et al., 2011). In a ptc null mutant (ptc9) there were commonly two ple+ H-cells (Fig. 6N) and two high VGlut H-cell sibs (Fig. 6P). MP1 marker gene expression was absent (Fig. 6H), whereas ivum and mvum marker expression resembled that of wild type (Fig. 6J,L). There was also an increase in gsb-n+ neurons (H-cell and H-cell sib) in ptc9 (Fig. 6R), further indicating that hh activates gsb/gsb-n expression in MP3. Consistent with this result, an increase in L(1)sc levels and tup+ cells also occurred in ptc mutant embryos (Fig. 6T,V). Similar results to the ptc9 mutant were observed for sim-gal4 UAS-hh embryos, which overexpress hh in midline cells (supplementary material Figs S2-S5). One issue regarding the hh and ptc mutant and hh overexpression experiments is whether the effects of hh are due to alterations in ectodermal patterning and are not autonomous to midline cells. This was addressed by examining sim-gal4 UASci.VP16, in which the hh signaling pathway is only active in midline cells (ci encodes the transcriptional effector of the hh signaling pathway). These experiments also showed an increase in H-cell and H-cell sib at the expense of MP1 (supplementary material Figs S2-S4). Since the hh and wg signaling pathways are known to regulate each other (Hatini and DiNardo, 2001) it is possible that the wg MP3/H-cell phenotype only reflects a reduction in hh signaling and is not due to a direct effect of wg. To address this issue, ptc wg double-mutant embryos were examined, as they lack wg function but have constitutively active hh signaling. The results showed that expression of gsb-n, odd, ple and slp1 was absent (supplementary
8 DA precursor formation and specification RESEARCH ARTICLE 3323 Fig. 7. Genes controlling H-cell development also affect non-midline DA neuronal development. Horizontal views of the CNS of stage sim-gal4 UAS-tau-GFP embryos stained for ple expression. (A) Wild-type embryo showing the ventral nerve cord (VNC) of dopaminergic (DA) neurons: H-cell (H), paramedial DA neurons (M) and dorsal lateral DA neurons (DL). (B-D) DA neuron ple expression was absent from Df(2R)gsb (B) and slp 34B (C) but expanded in ptc 9 (D). The ple + staining in B is in epidermal stripes. White bars indicate the midline. (supplementary material Fig. S1A-C,E,F). We looked at ple expression in the paramedial DA and dorsal lateral DA neurons in Df(2R)gsb, ptc 9 and slp 34B embryos at stage 16. In Df(2R)gsb and slp 34B embryos (Fig. 7B,C) ple expression was absent in all lateral DA neurons. In ptc 9 embryos (Fig. 7D) there were additional ple + dorsal lateral neurons in 57% of hemisegments scored (n 42) and additional paramedial DA neurons in 52% of hemisegments (n 50). These results are similar to the effects seen on the H-cell and provide an initial indication that different Drosophila DA neurons might share a common set of genes for their development. DISCUSSION The results presented here and in the literature (Stagg et al., 2011; Wheeler et al., 2008) have identified key regulators of Drosophila MP3/H-cell development, as summarized in Fig. 8. sim and the functional role of master regulatory genes The Drosophila sim gene is a master regulator of CNS midline cell development. sim mutants fail to develop midline neuronal and glial precursors, and midline transcription of almost all genes normally expressed in the midline is absent (Nambu et al., 1990). Similarly, ectopic expression of sim in the neuroectoderm transforms the entire CNS into midline cells (Nambu et al., 1991). However, here we suggest a further refinement of sim function and propose that sim commits cells to an MP4 neural precursor fate that is followed by a series of signaling events that act on these cells to generate a diverse group of midline neuronal precursors and glia. This concept extends the notion of master regulator to posit a specific function for sim in initiating MP4 fate while subsequently working combinatorially with other transcription factors to control midline cell type-specific gene expression (Ma et al., 2000). wg, slp1/2 and hh establish a midline anterior neural precursor domain The midline cells initially appear morphologically and molecularly uniform, as characterized by the expression of sim in all mesectodermal cells. In our model, sim initially commits mesectodermal cells to an MP4 neural precursor fate (Fig. 8A). This is followed by wg signaling that establishes an anterior domain in which cells are committed to an MP1 fate (Fig. 8B). This is mediated by activation of slp1/2 expression in anterior cells. Signaling by hh also maintains a distinct slp1/2 anterior-posterior boundary. One important aspect of slp1/2 function is the repression of en in the anterior region, as experimentally inducing en in wildtype anterior cells disrupts MP3 and MP1 neuronal development. In this sense, slp1/2 play a role in midline neural precursor development that is conceptually similar to that of runt in MG development (Watson et al., 2011). The major function of runt in MG is to repress en expression in ensheathing glia (AMG) and ensure that AMG do not become en + non-ensheathing glia (PMG). hh signaling specifies MP3 identity Both hh mutant and misexpression/overexpression experiments indicate that hh signaling is required for MP3 identity (Fig. 8C). The influence of hh on MP3 identity occurs largely, if not completely, by hh activation of gsb/gsb-n expression. hh is also required for expression of the bhlh factor L(1)sc in MPs, and both hh and l(1)sc mutants have similar proneural phenotypes with regard to the formation of MP4-6 and the MNB (Stagg et al., 2011). However, l(1)sc does not play a proneural role in MP3, even though it is expressed in MP3 (Stagg et al., 2011). There are two interpretations of the hh mutant results. In one scenario, MP3 fails to form and divide in an hh mutant, and thus hh plays an MP3 proneural role. Since MP3 formation is unaffected in gsb/gsb-n and l(1)sc mutants, the proneural function of hh may act through direct activation of proneural target genes by the hh pathway transcriptional effector Ci, or it could be through indirect Ci activation of additional transcription factors. Another interpretation is that MP3 is transformed in an hh mutant into an MP4-6-like cell, and MP4-6 fail to form. In this case, hh would not be acting as an MP3 proneural gene. It is important to note that hh signaling is postulated to convert a group of about five cells to an MP3 fate (Fig. 8C). The selection of the single MP3 found in each segment is through Notch signaling (Fig. 8D) (Wheeler et al., 2008). In this manner, Delta- Notch lateral inhibition results in the appearance of a single MP3, while the remaining cells become AMG and PMG. However, the division of MP3 is dependent on hh, and not Notch, signaling, as both MP3 and MP1 divide and differentiate in Delta mutant embryos (Wheeler et al., 2008). When hh signaling is activated in all midline cells in either ptc mutants or by hh pathway gene overexpression, cells destined to become MP1 instead become MP3. This suggests that, in wild-type embryos, the Hh morphogen is insufficiently active to direct the anteriormost cells to become MP3 even though these cells have the intrinsic ability to become MP3 if hh signaling is activated. Future studies will address the pathway by which hh controls MP3 formation, how MP1 is specified, and how hh signaling is inhibited in the cells that give rise to MP1. gsb/gsb-n direct MP3 cell fate gsb and gsb-n are targets of hh signaling and act to specify MP3 cell fate. Although not required for MP3 delamination or division, gsb/gsb-n mutant embryos did show a delay in the timing of MP3 division, which can be considered an aspect of cell fate. Both genes
9 3324 RESEARCH ARTICLE Development 139 (18) Fig. 8. Major steps in MP3 and H-cell development. A single segment is shown. (A) sim commits mesectodermal cells to an MP4 fate. (B) wg signaling activates slp1/2 expression in the anterior region, which represses en and commits cells to an MP1 fate (pink). (C) hh signaling directs a group of anterior cells to an MP3 fate (green) by activating expression of gsb/gsb-n. (D) Notch signaling selects one cell in the MP3 cluster to become MP3. (E) H-cell and H-cell sib fates are determined by Notch signaling and asymmetric localization of Numb. gsb/gsb-n activate expression of l(1)sc and tup in the H-cell. (F) l(1)sc and tup control H-cell-specific gene expression and differentiation. Genes expressed in common between the H-cell and H-cell sib are regulated by a different pathway. are expressed in MP3 and each plays a role in MP3 cell fate specification. Embryos homozygous mutant for gsb show defects in MP3 development, but these are less severe than in gsb gsb-n double-mutant embryos, indicating functional roles for both genes. One function of gsb is to activate expression of gsb-n in MP3, indicating that these genes might function in a hierarchical manner. The role of gsb-n was reinforced from misexpression experiments, in which MP1 was transformed into MP3. Upon division of MP3, the two progeny, i.e. the H-cell and H- cells sib, acquire their distinct identities due to Numb asymmetric localization and Notch signaling (Fig. 8E) (Wheeler et al., 2008). Whereas H-cell sib differentiation is dependent on Notch signaling, H-cell differentiation is largely dependent on the L(1)sc and Tup transcription factors (Fig. 8F). Genetically, gsb/gsb-n function is required for expression of l(1)sc and tup, linking cell fate to differentiation. Interestingly, l(1)sc also controls mvum-specific gene expression in addition to H-cell-specific gene expression. This raises the question of how the same transcription factor, L(1)sc, controls two distinct developmental programs. Since the H-cell and mvums differ in their MP precursors, one possibility is that Gsb/Gsb-n combinatorially interact with L(1)sc to control H-cell transcription and differentiation, whereas L(1)sc interacts with an unknown MP4-6 cell fate factor to control mvum transcription and differentiation. Acknowledgements We thank Chris Doe, Florence Maschat, Bruno Marie, Jim Skeath and Andrew Tomlinson for antibodies and Drosophila strains; Scott Wheeler for advice; Tony Perdue for assistance with microscopy; Frank Conlon for comments on the manuscript; the Developmental Studies Hybridoma Bank for monoclonal antibodies; and the Bloomington Drosophila Stock Center for Drosophila strains. Funding This work was supported by National Institutes of Health (NIH) grants [R01 NS64264 (NINDS) and R37 RD25251 (NICHD) to S.T.C.]; a University of North Carolina Developmental Biology NIH training grant fellowship to J.D.W.; and a National Research Service Award (NRSA) postdoctoral fellowship to J.D.W. Deposited in PMC for release after 12 months. Competing interests statement The authors declare no competing financial interests. Supplementary material Supplementary material available online at References Bate, C. M. and Grunewald, E. B. (1981). Embryogenesis of an insect nervous system II: a second class of neuron precursor cells and the origin of the intersegmental connectives. J. Embryol. Exp. Morphol. 61, Bhat, K. M., van Beers, E. H. and Bhat, P. (2000). Sloppy paired acts as the downstream target of wingless in the Drosophila CNS and interaction between sloppy paired and gooseberry inhibits sloppy paired during neurogenesis. Development 127, Bossing, T. and Brand, A. H. (2006). Determination of cell fate along the anteroposterior axis of the Drosophila ventral midline. Development 133, Brand, A. (1995). GFP in Drosophila. Trends Genet. 11, Broihier, H. T. and Skeath, J. B. (2002). Drosophila homeodomain protein dhb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron 35, Cadigan, K. M., Grossniklaus, U. and Gehring, W. J. (1994). Functional redundancy: the respective roles of the two sloppy paired genes in Drosophila segmentation. Proc. Natl. Acad. Sci. USA 91, Colomb, S., Joly, W., Bonneaud, N. and Maschat, F. (2008). A concerted action of Engrailed and Gooseberry-Neuro in neuroblast 6-4 is triggering the formation of embryonic posterior commissure bundles. PLoS ONE 3, e2197. Duman-Scheel, M., Li, X., Orlov, I., Noll, M. and Patel, N. H. (1997). Genetic separation of the neural and cuticular patterning functions of gooseberry. Development 124, Grossniklaus, U., Pearson, R. K. and Gehring, W. J. (1992). The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 6, Guillén, I., Mullor, J. L., Capdevila, J., Sánchez-Herrero, E., Morata, G. and Guerrero, I. (1995). The function of engrailed and the specification of Drosophila wing pattern. Development 121, Gutjahr, T., Patel, N. H., Li, X., Goodman, C. S. and Noll, M. (1993). Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and activates gooseberry neuro. Development 118, Hatini, V. and DiNardo, S. (2001). Divide and conquer: pattern formation in Drosophila embryonic epidermis. Trends Genet. 17, Ingham, P. W. and McMahon, A. P. (2001). Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, Kearney, J. B., Wheeler, S. R., Estes, P., Parente, B. and Crews, S. T. (2004). Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 275, Kosman, D., Small, S. and Reinitz, J. (1998). Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208, Larsen, C. W., Hirst, E., Alexandre, C. and Vincent, J. P. (2003). Segment boundary formation in Drosophila embryos. Development 130,
10 DA precursor formation and specification RESEARCH ARTICLE 3325 Lee, J. J., von Kessler, D. P., Parks, S. and Beachy, P. A. (1992). Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71, Lundell, M. J. and Hirsh, J. (1994). Temporal and spatial development of serotonin and dopamine neurons in the Drosophila CNS. Dev. Biol. 165, Ma, Y., Certel, K., Gao, Y., Niemitz, E., Mosher, J., Mukherjee, A., Mutsuddi, M., Huseinovic, N., Crews, S. T., Johnson, W. A. et al. (2000). Functional interactions between Drosophila bhlh/pas, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J. Neurosci. 20, Marie, B., Pym, E., Bergquist, S. and Davis, G. W. (2010). Synaptic homeostasis is consolidated by the cell fate gene gooseberry, a Drosophila pax3/7 homolog. J. Neurosci. 30, Nambu, J. R., Franks, R. G., Hu, S. and Crews, S. T. (1990). The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63, Nambu, J. R., Lewis, J. O., Wharton, K. A., Jr and Crews, S. T. (1991). The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67, Nusslein-Volhard, C., Wieschaus, E. and Kluding, H. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III. Zygotic loci on the second chromosome. Dev. Genes Evol. 193, Patel, N. H., Martin-Blanco, E., Coleman, K. G., Poole, S. J., Ellis, M. C., Kornberg, T. B. and Goodman, C. S. (1989). Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, Porter, J. A., Ekker, S. C., Park, W. J., von Kessler, D. P., Young, K. E., Chen, C. H., Ma, Y., Woods, A. S., Cotter, R. J., Koonin, E. V. et al. (1996). Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxyterminal autoprocessing domain. Cell 86, Robinow, S. and White, K. (1991). Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, Sato, A. and Tomlinson, A. (2007). Dorsal-ventral midline signaling in the developing Drosophila eye. Development 134, Skeath, J. B. and Thor, S. (2003). Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13, Stagg, S. B., Guardiola, A. R. and Crews, S. T. (2011). Dual role for Drosophila lethal of scute in CNS midline precursor formation and dopaminergic neuron and motoneuron cell fate. Development 138, Thor, S. and Thomas, J. B. (1997). The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron 18, Tio, M., Toh, J., Fang, W., Blanco, J. and Udolph, G. (2011). Asymmetric cell division and Notch signaling specify dopaminergic neurons in Drosophila. PLoS ONE 6, e Tran, K. D. and Doe, C. Q. (2008). Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development 135, Vogler, G. and Urban, J. (2008). The transcription factor Zfh1 is involved in the regulation of neuropeptide expression and growth of larval neuromuscular junctions in Drosophila melanogaster. Dev. Biol. 319, Watson, J. D., Wheeler, S. R., Stagg, S. B. and Crews, S. T. (2011). Drosophila hedgehog signaling and engrailed-runt mutual repression direct midline glia to alternative ensheathing and non-ensheathing fates. Development 138, Wheeler, S. R., Kearney, J. B., Guardiola, A. R. and Crews, S. T. (2006). Singlecell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev. Biol. 294, Wheeler, S. R., Stagg, S. B. and Crews, S. T. (2008). Multiple Notch signaling events control Drosophila CNS midline neurogenesis, gliogenesis and neuronal identity. Development 135, Wheeler, S. R., Stagg, S. B. and Crews, S. T. (2009). MidExDB: a database of Drosophila CNS midline cell gene expression. BMC Dev. Biol. 9, 56. Xiao, H., Hrdlicka, L. A. and Nambu, J. R. (1996). Alternate functions of the single-minded and rhomboid genes in development of the Drosophila ventral neuroectoderm. Mech. Dev. 58,
11 Fig. S1. CNS midline and ectodermal expression of Drosophila segmentation genes. Horizontal views are shown with anterior to the left. Short white lines mark the location of the midline in some panels. (A) At stage 10, gsb-n is expressed in MP3 (arrowheads) and in broader collinear neuroectodermal stripes. Three segments are shown. Expression of gsb is similar but broader (data not shown). (B) slp1 is expressed in anterior midline cells at stage 10. (C) Expression of wg at stage 9 is largely absent from midline cells, but is present in a stripe of cells perpendicular to the midline cells. wg overlaps in expression with the posteriormost slp1 + cells. (D) The hh gene is expressed in a stripe perpendicular to the midline cells at stage 10, but is absent from the midline cells. The hh stripe overlaps the En stripe. (E-F0) Horizontal views of stage 11 sim-gal4 UAS-tau-GFP embryos stained for (E) gsb-n RNA or (F) slp1 RNA in addition to anti-en and anti-tau (blue). Three different focal planes reveal the epidermis, lateral CNS, and midline H-cell and MP1 neurons. (E-E0) gsb-n expression in the lateral CNS is broader than midline expression, which is restricted to H-cell and H-cell sib (arrow). (F-F0) slp1 is expressed in anterior midline cells (above the white line) and is adjacent to, but does not overlap, En. (G) Df(2R)Kr10/Df(2R)gsb embryos are homozygous mutant for gsb and lack one copy of gsb-n. Lateral CNS expression of gsb-n (arrows) is reduced compared with wild type (A). Midline expression of gsb-n is present in some segments (arrowheads) and absent in others (asterisks). (H,I) Both ple and VGlut expression are frequently absent in Df(2R)Kr10/Df(2R)gsb embryos.
12 % segments with # ple+ neurons cells 1 cell 2 cells 3 cells Fig. S2. Summary of genetic experiments analyzing H-cell development. The number of ple + midline cells observed in each segment was counted and expressed as a percentage. Segments scored for each mutant or misexpression genotype 100 % segments with # high VGlut+ neurons cells 1 cell 2 cells 3 cells Fig. S3. Summary of genetic experiments analyzing H-cell sib development. The number of high VGlut cells observed in each segment was counted and expressed as a percentage. Segments scored for each mutant or misexpression genotype ranged between nine and 22.
13 % segments with # Lim3+ or odd+ neurons cells 1 cell 2 cells 3 cells 4 cells Fig. S4. Summary of genetic experiments analyzing MP1 neuron development. The number of Lim3 + or odd + cells observed in each segment was counted and expressed as a percentage. Segments scored for each mutant or misexpression genotype ranged between eight and % segments with # Zfh1+ neurons cells 1 cell 2 cells 3 cells 4 cells 5 cells Fig. S5. Summary of genetic experiments analyzing mvum neuron development. The number of Zfh1 + cells observed in each segment was counted and expressed as a percentage. Segments scored for each mutant or misexpression genotype ranged between eight and 44.
14 Fig. S6. Number and identity of midline neurons in hh mutant embryos. (A) The number of Elav + neurons observed in each segment of wild-type (n=22 segments) and hh AC mutant (n=39 segments) embryos was counted and expressed as a percentage. Most mutant segments had either four or six neurons, whereas wild-type segments had 11 or more. (B-D) Horizontal views of a single segment of a stage 15 hh AC sim-gal4 UAS-tau-GFP mutant embryo shown at three focal planes (difference in distance is indicated on the left). The embryo was stained for anti-tau (green), Zfh1, odd and Elav (all magenta). Imaging throughout the segment revealed that only six Tau-GFP + cells were Elav +. Two of these were Zfh1 + (mvums) and two were odd + (MP1s). The other two are likely to be ivums.
15 Fig. S7. Both wg and hh are required for activation of gsb-n in MP3. (A-C) In the same ptc 7 wg 1-12 double-mutant embryo, expression of (A) ple (H-cell) and (B) odd (MP1 neurons) was absent, and (C) Zfh1 (mvums) was expanded. (D) slp1 was absent in ptc 7 wg 1-12 embryos. (E,F) In another embryo, both (E) en and (F) Zfh1 were expanded. In the midline cells shown (which might constitute more than one fused segment), there were 20 En + cells and ten Zfh1 + cells (not all can be seen in this focal plane). Together, they constitute all of the midline cells present. (G) gsb-n expression was absent in ptc 7 wg 1-12.
Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells
 Developmental Biology 294 (2006) 509 524 www.elsevier.com/locate/ydbio Genomes & Developmental Control Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells
Developmental Biology 294 (2006) 509 524 www.elsevier.com/locate/ydbio Genomes & Developmental Control Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells
Nirupama Deshpande, Rainer Dittrich, Gerhard M. Technau and Joachim Urban* SUMMARY
 Development 128, 3253-3261 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV8801 3253 Successive specification of Drosophila neuroblasts NB 6-4 and NB 7-3 depends on interaction
Development 128, 3253-3261 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV8801 3253 Successive specification of Drosophila neuroblasts NB 6-4 and NB 7-3 depends on interaction
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Segment boundary formation in Drosophila embryos
 Segment boundary formation in Drosophila embryos Development 130, August 2003 Camilla W. Larsen, Elizabeth Hirst, Cyrille Alexandre and Jean Paul Vincent 1. Introduction: - Segment boundary formation:
Segment boundary formation in Drosophila embryos Development 130, August 2003 Camilla W. Larsen, Elizabeth Hirst, Cyrille Alexandre and Jean Paul Vincent 1. Introduction: - Segment boundary formation:
The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
 The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord Mary Ann Manavalan, Ivana Gaziova, Krishna Moorthi Bhat* Department of
The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord Mary Ann Manavalan, Ivana Gaziova, Krishna Moorthi Bhat* Department of
Lecture 7. Development of the Fruit Fly Drosophila
 BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
Axis determination in flies. Sem 9.3.B.5 Animal Science
 Axis determination in flies Sem 9.3.B.5 Animal Science All embryos are in lateral view (anterior to the left). Endoderm, midgut; mesoderm; central nervous system; foregut, hindgut and pole cells in yellow.
Axis determination in flies Sem 9.3.B.5 Animal Science All embryos are in lateral view (anterior to the left). Endoderm, midgut; mesoderm; central nervous system; foregut, hindgut and pole cells in yellow.
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
10/2/2015. Chapter 4. Determination and Differentiation. Neuroanatomical Diversity
 Chapter 4 Determination and Differentiation Neuroanatomical Diversity 1 Neurochemical diversity: another important aspect of neuronal fate Neurotransmitters and their receptors Excitatory Glutamate Acetylcholine
Chapter 4 Determination and Differentiation Neuroanatomical Diversity 1 Neurochemical diversity: another important aspect of neuronal fate Neurotransmitters and their receptors Excitatory Glutamate Acetylcholine
Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and activates gooseberry neuro
 Development 118, 21-31 (1993) Printed in Great Britain The Company of Biologists Limited 1993 21 Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and
Development 118, 21-31 (1993) Printed in Great Britain The Company of Biologists Limited 1993 21 Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and
Midterm 1. Average score: 74.4 Median score: 77
 Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Cell division genes promote asymmetric interaction between Numb and Notch in the Drosophila CNS
 Development 126, 2759-2770 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV5300 2759 Cell division genes promote asymmetric interaction between Numb and Notch in the Drosophila
Development 126, 2759-2770 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV5300 2759 Cell division genes promote asymmetric interaction between Numb and Notch in the Drosophila
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila
 Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
178 Part 3.2 SUMMARY INTRODUCTION
 178 Part 3.2 Chapter # DYNAMIC FILTRATION OF VARIABILITY WITHIN EXPRESSION PATTERNS OF ZYGOTIC SEGMENTATION GENES IN DROSOPHILA Surkova S.Yu. *, Samsonova M.G. St. Petersburg State Polytechnical University,
178 Part 3.2 Chapter # DYNAMIC FILTRATION OF VARIABILITY WITHIN EXPRESSION PATTERNS OF ZYGOTIC SEGMENTATION GENES IN DROSOPHILA Surkova S.Yu. *, Samsonova M.G. St. Petersburg State Polytechnical University,
Why Flies? stages of embryogenesis. The Fly in History
 The Fly in History 1859 Darwin 1866 Mendel c. 1890 Driesch, Roux (experimental embryology) 1900 rediscovery of Mendel (birth of genetics) 1910 first mutant (white) (Morgan) 1913 first genetic map (Sturtevant
The Fly in History 1859 Darwin 1866 Mendel c. 1890 Driesch, Roux (experimental embryology) 1900 rediscovery of Mendel (birth of genetics) 1910 first mutant (white) (Morgan) 1913 first genetic map (Sturtevant
Cellular Neurobiology BIPN 140 Fall 2016 Problem Set #8
 Cellular Neurobiology BIPN 140 Fall 2016 Problem Set #8 1. Inductive signaling is a hallmark of vertebrate and mammalian development. In early neural development, there are multiple signaling pathways
Cellular Neurobiology BIPN 140 Fall 2016 Problem Set #8 1. Inductive signaling is a hallmark of vertebrate and mammalian development. In early neural development, there are multiple signaling pathways
Drosophila homeodomain protein Nkx6 coordinates motoneuron
 Research article 5233 Drosophila homeodomain protein Nkx6 coordinates motoneuron subtype identity and axonogenesis Heather T. Broihier 1, *,,, Alexander Kuzin 2,, Yi Zhu 1, Ward Odenwald 2 and James B.
Research article 5233 Drosophila homeodomain protein Nkx6 coordinates motoneuron subtype identity and axonogenesis Heather T. Broihier 1, *,,, Alexander Kuzin 2,, Yi Zhu 1, Ward Odenwald 2 and James B.
purpose of this Chapter is to highlight some problems that will likely provide new
 119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
BILD7: Problem Set. 2. What did Chargaff discover and why was this important?
 BILD7: Problem Set 1. What is the general structure of DNA? 2. What did Chargaff discover and why was this important? 3. What was the major contribution of Rosalind Franklin? 4. How did solving the structure
BILD7: Problem Set 1. What is the general structure of DNA? 2. What did Chargaff discover and why was this important? 3. What was the major contribution of Rosalind Franklin? 4. How did solving the structure
Neuronal cell death in grasshopper embryos: variable patterns in different species, clutches, and clones
 J. Embryol. exp. Morph. 78, 169-182 (1983) Printed in Great Britain The Company of Biologists Limited 1983 Neuronal cell death in grasshopper embryos: variable patterns in different species, clutches,
J. Embryol. exp. Morph. 78, 169-182 (1983) Printed in Great Britain The Company of Biologists Limited 1983 Neuronal cell death in grasshopper embryos: variable patterns in different species, clutches,
Drosophila melanogaster- Morphogen Gradient
 NPTEL Biotechnology - Systems Biology Drosophila melanogaster- Morphogen Gradient Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by
NPTEL Biotechnology - Systems Biology Drosophila melanogaster- Morphogen Gradient Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by
9/4/2015 INDUCTION CHAPTER 1. Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology. Fig 1.
 INDUCTION CHAPTER 1 Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology Fig 1.1 1 EVOLUTION OF METAZOAN BRAINS GASTRULATION MAKING THE 3 RD GERM LAYER
INDUCTION CHAPTER 1 Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology Fig 1.1 1 EVOLUTION OF METAZOAN BRAINS GASTRULATION MAKING THE 3 RD GERM LAYER
Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS
 Development 128, 4103-4112 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV3509 4103 Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in
Development 128, 4103-4112 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV3509 4103 Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in
PRACTICE EXAM. 20 pts: 1. With the aid of a diagram, indicate how initial dorsal-ventral polarity is created in fruit fly and frog embryos.
 PRACTICE EXAM 20 pts: 1. With the aid of a diagram, indicate how initial dorsal-ventral polarity is created in fruit fly and frog embryos. No Low [] Fly Embryo Embryo Non-neural Genes Neuroectoderm Genes
PRACTICE EXAM 20 pts: 1. With the aid of a diagram, indicate how initial dorsal-ventral polarity is created in fruit fly and frog embryos. No Low [] Fly Embryo Embryo Non-neural Genes Neuroectoderm Genes
MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION
 MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION Drosophila is the best understood of all developmental systems, especially at the genetic level, and although it is an invertebrate it has had an enormous
MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION Drosophila is the best understood of all developmental systems, especially at the genetic level, and although it is an invertebrate it has had an enormous
Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea
 Development 129, 4941-4951 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV7966 4941 Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila
Development 129, 4941-4951 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV7966 4941 Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila
Sonic hedgehog (Shh) signalling in the rabbit embryo
 Sonic hedgehog (Shh) signalling in the rabbit embryo In the first part of this thesis work the physical properties of cilia-driven leftward flow were characterised in the rabbit embryo. Since its discovery
Sonic hedgehog (Shh) signalling in the rabbit embryo In the first part of this thesis work the physical properties of cilia-driven leftward flow were characterised in the rabbit embryo. Since its discovery
Unicellular: Cells change function in response to a temporal plan, such as the cell cycle.
 Spatial organization is a key difference between unicellular organisms and metazoans Unicellular: Cells change function in response to a temporal plan, such as the cell cycle. Cells differentiate as a
Spatial organization is a key difference between unicellular organisms and metazoans Unicellular: Cells change function in response to a temporal plan, such as the cell cycle. Cells differentiate as a
Positional Cues in the Drosophila Nerve Cord: Semaphorins Pattern the Dorso-Ventral Axis
 Positional Cues in the Drosophila Nerve Cord: Semaphorins Pattern the Dorso-Ventral Axis Marta Zlatic 1,2,3 *, Feng Li 1, Maura Strigini 4, Wesley Grueber 2, Michael Bate 1 * 1 Department of Zoology, University
Positional Cues in the Drosophila Nerve Cord: Semaphorins Pattern the Dorso-Ventral Axis Marta Zlatic 1,2,3 *, Feng Li 1, Maura Strigini 4, Wesley Grueber 2, Michael Bate 1 * 1 Department of Zoology, University
Neural development its all connected
 Neural development its all connected How do you build a complex nervous system? How do you build a complex nervous system? 1. Learn how tissue is instructed to become nervous system. Neural induction 2.
Neural development its all connected How do you build a complex nervous system? How do you build a complex nervous system? 1. Learn how tissue is instructed to become nervous system. Neural induction 2.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Systematic Screening of Drosophila Deficiency Mutations for Embryonic Phenotypes and Orphan Receptor Ligands
 Systematic Screening of Drosophila Deficiency Mutations for Embryonic Phenotypes and Orphan Receptor Ligands Ashley P. Wright 1, A. Nicole Fox 1, Karl G. Johnson 2, Kai Zinn 1 * 1 Division of Biology,
Systematic Screening of Drosophila Deficiency Mutations for Embryonic Phenotypes and Orphan Receptor Ligands Ashley P. Wright 1, A. Nicole Fox 1, Karl G. Johnson 2, Kai Zinn 1 * 1 Division of Biology,
Segment-specific prevention of pioneer neuron apoptosis by cellautonomous, postmitotic Hox gene activity
 First posted online on 10 November 2004 as 10.1242/dev.01521 Access Development the most recent epress version at online http://dev.biologists.org/lookup/doi/10.1242/dev.01521 publication date November
First posted online on 10 November 2004 as 10.1242/dev.01521 Access Development the most recent epress version at online http://dev.biologists.org/lookup/doi/10.1242/dev.01521 publication date November
5/4/05 Biol 473 lecture
 5/4/05 Biol 473 lecture animals shown: anomalocaris and hallucigenia 1 The Cambrian Explosion - 550 MYA THE BIG BANG OF ANIMAL EVOLUTION Cambrian explosion was characterized by the sudden and roughly simultaneous
5/4/05 Biol 473 lecture animals shown: anomalocaris and hallucigenia 1 The Cambrian Explosion - 550 MYA THE BIG BANG OF ANIMAL EVOLUTION Cambrian explosion was characterized by the sudden and roughly simultaneous
Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during CNS development in Drosophila
 Development 129, 4219-4228 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV7977 4219 Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during
Development 129, 4219-4228 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV7977 4219 Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
The jing Zn-finger transcription factor is a mediator of cellular differentiation in the Drosophila CNS midline and trachea
 Development 129, 2591-2606 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV5932 2591 The jing Zn-finger transcription factor is a mediator of cellular differentiation in the Drosophila
Development 129, 2591-2606 (2002) Printed in Great Britain The Company of Biologists Limited 2002 DEV5932 2591 The jing Zn-finger transcription factor is a mediator of cellular differentiation in the Drosophila
ATranscriptionFactorNetworkCoordinates Attraction, Repulsion, and Adhesion Combinatorially to Control Motor Axon Pathway Selection
 Article ATranscriptionFactorNetworkCoordinates Attraction, Repulsion, and Adhesion Combinatorially to Control Motor Axon Pathway Selection Aref Arzan Zarin, 1,2 Jamshid Asadzadeh, 1,2 Karsten Hokamp, 1
Article ATranscriptionFactorNetworkCoordinates Attraction, Repulsion, and Adhesion Combinatorially to Control Motor Axon Pathway Selection Aref Arzan Zarin, 1,2 Jamshid Asadzadeh, 1,2 Karsten Hokamp, 1
Supplementary Figures
 Supplementary Figures Supplementary Fig. S1: Normal development and organization of the embryonic ventral nerve cord in Platynereis. (A) Life cycle of Platynereis dumerilii. (B-F) Axonal scaffolds and
Supplementary Figures Supplementary Fig. S1: Normal development and organization of the embryonic ventral nerve cord in Platynereis. (A) Life cycle of Platynereis dumerilii. (B-F) Axonal scaffolds and
Gonadal mesoderm and fat body initially follow a common developmental path in Drosophila
 Development 125, 837-844 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV7627 837 Gonadal and fat body initially follow a common developmental path in Drosophila Lisa A. Moore
Development 125, 837-844 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV7627 837 Gonadal and fat body initially follow a common developmental path in Drosophila Lisa A. Moore
UNIVERSITY OF YORK BIOLOGY. Developmental Biology
 Examination Candidate Number: UNIVERSITY OF YORK BSc Stage 2 Degree Examinations 2017-18 Department: BIOLOGY Title of Exam: Developmental Biology Desk Number: Time allowed: 1 hour and 30 minutes Total
Examination Candidate Number: UNIVERSITY OF YORK BSc Stage 2 Degree Examinations 2017-18 Department: BIOLOGY Title of Exam: Developmental Biology Desk Number: Time allowed: 1 hour and 30 minutes Total
Building the brain (1): Evolutionary insights
 Building the brain (1): Evolutionary insights Historical considerations! Initial insight into the general role of the brain in human behaviour was already attained in antiquity and formulated by Hippocrates
Building the brain (1): Evolutionary insights Historical considerations! Initial insight into the general role of the brain in human behaviour was already attained in antiquity and formulated by Hippocrates
Report. Functional Diversity of Robo Receptor Immunoglobulin Domains Promotes Distinct Axon Guidance Decisions
 Current Biology 20, 567 572, March 23, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.02.021 Functional Diversity of Robo Receptor Immunoglobulin Domains Promotes Distinct Axon Guidance
Current Biology 20, 567 572, March 23, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.02.021 Functional Diversity of Robo Receptor Immunoglobulin Domains Promotes Distinct Axon Guidance
Morphogens in biological development: Drosophila example
 LSM5194 Morphogens in biological development: Drosophila example Lecture 29 The concept of morphogen gradients The concept of morphogens was proposed by L. Wolpert as a part of the positional information
LSM5194 Morphogens in biological development: Drosophila example Lecture 29 The concept of morphogen gradients The concept of morphogens was proposed by L. Wolpert as a part of the positional information
Embryogenesis of an insect nervous system II: A second class of neuron precursor cells and the origin of the intersegmental connectives
 J. Embryol. exp. Morph. Vol. 61,pp. 317-330, 1981 3J7 Printed in Great Britain @ Company of Biologists Limited 1981 Embryogenesis of an insect nervous system II: A second class of neuron precursor cells
J. Embryol. exp. Morph. Vol. 61,pp. 317-330, 1981 3J7 Printed in Great Britain @ Company of Biologists Limited 1981 Embryogenesis of an insect nervous system II: A second class of neuron precursor cells
The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm
 The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm Brian Biehs, Vincent Francois, 1 and Ethan Bier 2 Department of Biology and Center
The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm Brian Biehs, Vincent Francois, 1 and Ethan Bier 2 Department of Biology and Center
Reading: Chapter 5, pp ; Reference chapter D, pp Problem set F
 Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Current address: Whitney Laboratory for Marine Bioscience. University of Florida, St. Augustine, FL 32080
 *Manuscript The eneuro atlas: a modifiable, searchable 3D atlas that individually identifies the majority of interneurons in the Drosophila embryonic CNS and provides tools for functional access Ellie
*Manuscript The eneuro atlas: a modifiable, searchable 3D atlas that individually identifies the majority of interneurons in the Drosophila embryonic CNS and provides tools for functional access Ellie
Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system
 Development 116, 855-863 (1992) Printed in Great Britain The Company of Biologists Limited 1992 855 Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous
Development 116, 855-863 (1992) Printed in Great Britain The Company of Biologists Limited 1992 855 Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous
Hedgehog and Wingless stabilize but do not induce cell fate during Drosophila dorsal embryonic epidermal patterning
 Development epress online publication date 9 July 2008 RESEARCH ARTICLE 2767 Development 135, 2767-2775 (2008) doi:10.1242/dev.017814 Hedgehog and Wingless stabilize but do not induce cell fate during
Development epress online publication date 9 July 2008 RESEARCH ARTICLE 2767 Development 135, 2767-2775 (2008) doi:10.1242/dev.017814 Hedgehog and Wingless stabilize but do not induce cell fate during
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Chapter 4. DROSOPHILA bhlh-pas DEVELOPMENTAL REGULATORY PROTEINS 1. INTRODUCTION
 Chapter 4 DROSOPHILA bhlh-pas DEVELOPMENTAL REGULATORY PROTEINS Stephen T. Crews The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 1. INTRODUCTION Drosophila bhlh-pas proteins play
Chapter 4 DROSOPHILA bhlh-pas DEVELOPMENTAL REGULATORY PROTEINS Stephen T. Crews The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 1. INTRODUCTION Drosophila bhlh-pas proteins play
Wingless and Hedgehog pattern Drosophila denticle belts by regulating the production of short-range signals
 Development 126, 5689-5698 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV7768 5689 Wingless and Hedgehog pattern Drosophila denticle belts by regulating the production of short-range
Development 126, 5689-5698 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV7768 5689 Wingless and Hedgehog pattern Drosophila denticle belts by regulating the production of short-range
Developmental Biology 3230 Midterm Exam 1 March 2006
 Name Developmental Biology 3230 Midterm Exam 1 March 2006 1. (20pts) Regeneration occurs to some degree to most metazoans. When you remove the head of a hydra a new one regenerates. Graph the inhibitor
Name Developmental Biology 3230 Midterm Exam 1 March 2006 1. (20pts) Regeneration occurs to some degree to most metazoans. When you remove the head of a hydra a new one regenerates. Graph the inhibitor
Drosophila Homeodomain Protein dhb9 Directs Neuronal Fate via Crossrepressive and Cell-Nonautonomous Mechanisms
 Neuron, Vol. 35, 39 50, July 3, 2002, Copyright 2002 by Cell Press Drosophila Homeodomain Protein dhb9 Directs Neuronal Fate via Crossrepressive and Cell-Nonautonomous Mechanisms Heather T. Broihier and
Neuron, Vol. 35, 39 50, July 3, 2002, Copyright 2002 by Cell Press Drosophila Homeodomain Protein dhb9 Directs Neuronal Fate via Crossrepressive and Cell-Nonautonomous Mechanisms Heather T. Broihier and
MBios 401/501: Lecture 14.2 Cell Differentiation I. Slide #1. Cell Differentiation
 MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
Segmental patterning of heart precursors in Drosophila
 Development 121, 4303-4308 (1995) Printed in Great Britain The Company of Biologists Limited 1995 DEV5025 4303 Segmental patterning of heart precursors in Drosophila Peter A. Lawrence 1, *, Rolf Bodmer
Development 121, 4303-4308 (1995) Printed in Great Britain The Company of Biologists Limited 1995 DEV5025 4303 Segmental patterning of heart precursors in Drosophila Peter A. Lawrence 1, *, Rolf Bodmer
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
NN-A29932: Supplementary Data 1 Diversity and Wiring Variability of Olfactory Local Interneurons in the Drosophila Antennal Lobe
 NN-A29932: Supplementary Data 1 Diversity and Wiring Variability of Olfactory Local Interneurons in the Drosophila Antennal Lobe Ya-Hui Chou, Maria L. Spletter, Emre Yaksi, Jonathan C. S. Leong, Rachel
NN-A29932: Supplementary Data 1 Diversity and Wiring Variability of Olfactory Local Interneurons in the Drosophila Antennal Lobe Ya-Hui Chou, Maria L. Spletter, Emre Yaksi, Jonathan C. S. Leong, Rachel
Drosophila. Cubitus interruptus-independent transduction of the Hedgehog signal in
 Development 127, 5509-5522 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV7818 5509 Cubitus interruptus-independent transduction of the Hedgehog signal in Drosophila Armel Gallet
Development 127, 5509-5522 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV7818 5509 Cubitus interruptus-independent transduction of the Hedgehog signal in Drosophila Armel Gallet
Synaptic Homeostasis Is Consolidated by the Cell Fate Gene gooseberry,adrosophila pax3/7 Homolog
 The Journal of Neuroscience, June 16, 2010 30(24):8071 8082 8071 Development/Plasticity/Repair Synaptic Homeostasis Is Consolidated by the Cell Fate Gene gooseberry,adrosophila pax3/7 Homolog Bruno Marie,
The Journal of Neuroscience, June 16, 2010 30(24):8071 8082 8071 Development/Plasticity/Repair Synaptic Homeostasis Is Consolidated by the Cell Fate Gene gooseberry,adrosophila pax3/7 Homolog Bruno Marie,
Developmental genetics: finding the genes that regulate development
 Developmental Biology BY1101 P. Murphy Lecture 9 Developmental genetics: finding the genes that regulate development Introduction The application of genetic analysis and DNA technology to the study of
Developmental Biology BY1101 P. Murphy Lecture 9 Developmental genetics: finding the genes that regulate development Introduction The application of genetic analysis and DNA technology to the study of
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo
 Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Boolean Modeling of Genetic Regulatory Networks
 Boolean Modeling of Genetic Regulatory Networks Réka Albert Department of Physics, Pennsylvania State University, University Park, PA 16802, USA Abstract. Biological systems form complex networks of interaction
Boolean Modeling of Genetic Regulatory Networks Réka Albert Department of Physics, Pennsylvania State University, University Park, PA 16802, USA Abstract. Biological systems form complex networks of interaction
Conservation of arthropod midline netrin accumulation revealed with a cross-reactive antibody provides evidence for midline cell homology
 EVOLUTION & DEVELOPMENT 11:3, 260 268 (2009) DOI: 10.1111/j.1525-142X.2009.00328.x Conservation of arthropod midline netrin accumulation revealed with a cross-reactive antibody provides evidence for midline
EVOLUTION & DEVELOPMENT 11:3, 260 268 (2009) DOI: 10.1111/j.1525-142X.2009.00328.x Conservation of arthropod midline netrin accumulation revealed with a cross-reactive antibody provides evidence for midline
Pattern formation: Wingless on the move Robert Howes and Sarah Bray
 R222 Dispatch Pattern formation: Wingless on the move Robert Howes and Sarah Bray Wingless is a key morphogen in Drosophila. Although it is evident that Wingless acts at a distance from its site of synthesis,
R222 Dispatch Pattern formation: Wingless on the move Robert Howes and Sarah Bray Wingless is a key morphogen in Drosophila. Although it is evident that Wingless acts at a distance from its site of synthesis,
Homeotic genes in flies. Sem 9.3.B.6 Animal Science
 Homeotic genes in flies Sem 9.3.B.6 Animal Science So far We have seen that identities of each segment is determined by various regulators of segment polarity genes In arthopods, and in flies, each segment
Homeotic genes in flies Sem 9.3.B.6 Animal Science So far We have seen that identities of each segment is determined by various regulators of segment polarity genes In arthopods, and in flies, each segment
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Neuronal Subtype Specification within a Lineage by Opposing Temporal Feed-Forward Loops
 Neuronal Subtype Specification within a Lineage by Opposing Temporal Feed-Forward Loops Magnus Baumgardt, 1 Daniel Karlsson, 1 Javier Terriente, 2,3 Fernando J. Díaz-Benjumea, 2 and Stefan Thor 1, 1 Department
Neuronal Subtype Specification within a Lineage by Opposing Temporal Feed-Forward Loops Magnus Baumgardt, 1 Daniel Karlsson, 1 Javier Terriente, 2,3 Fernando J. Díaz-Benjumea, 2 and Stefan Thor 1, 1 Department
Drosophila Somatic Anterior-Posterior Axis (A-P Axis) Formation
 Home Biol 4241 Luria-Delbruck 1943 Hershey-Chase 1952 Meselson-Stahl 1958 Garapin et al. 1978 McClintock 1953 King-Wilson 1975 Sanger et al. 1977 Rothberg et al. 2011 Jeffreys et al. 1985 Bacterial Genetics
Home Biol 4241 Luria-Delbruck 1943 Hershey-Chase 1952 Meselson-Stahl 1958 Garapin et al. 1978 McClintock 1953 King-Wilson 1975 Sanger et al. 1977 Rothberg et al. 2011 Jeffreys et al. 1985 Bacterial Genetics
Trans-regulatory functions in the Abdominal-B gene of the bithorax complex
 Development 101, 117-122 (1987) Printed in Great Britain The Company of Biologists Limited 1987 117 Trans-regulatory functions in the Abdominal-B gene of the bithorax complex JORDI CASANOVA 1 and ROBERT
Development 101, 117-122 (1987) Printed in Great Britain The Company of Biologists Limited 1987 117 Trans-regulatory functions in the Abdominal-B gene of the bithorax complex JORDI CASANOVA 1 and ROBERT
Reading. Lecture VI. Making Connections 9/17/12. Bio 3411 Lecture VI. Making Connections. Bio 3411 Monday September 17, 2012
 Lecture VI. Making Connections Bio 3411 Monday September 17, 2012!! 1! Reading NEUROSCIENCE: 5 th ed, pp!507?536! 4 th ed, pp 577-609 Bentley, D., & Caudy, M. (1983). Nature, 304(5921), 62-65. Dickson,
Lecture VI. Making Connections Bio 3411 Monday September 17, 2012!! 1! Reading NEUROSCIENCE: 5 th ed, pp!507?536! 4 th ed, pp 577-609 Bentley, D., & Caudy, M. (1983). Nature, 304(5921), 62-65. Dickson,
Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene
 Development 103, 733-741 (1988) Printed in Great Britain The Company of Biologists Limited 1988 733 Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos
Development 103, 733-741 (1988) Printed in Great Britain The Company of Biologists Limited 1988 733 Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos
18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis
 18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis An organism arises from a fertilized egg cell as the result of three interrelated processes: cell division, cell
18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis An organism arises from a fertilized egg cell as the result of three interrelated processes: cell division, cell
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila July 9, 2008 Drosophila Development Overview Fertilization Cleavage Gastrulation Drosophila body plan Oocyte formation Genetic control
Developmental Biology Biology 4361 Axis Specification in Drosophila July 9, 2008 Drosophila Development Overview Fertilization Cleavage Gastrulation Drosophila body plan Oocyte formation Genetic control
Biology 218, practise Exam 2, 2011
 Figure 3 The long-range effect of Sqt does not depend on the induction of the endogenous cyc or sqt genes. a, Design and predictions for the experiments shown in b-e. b-e, Single-cell injection of 4 pg
Figure 3 The long-range effect of Sqt does not depend on the induction of the endogenous cyc or sqt genes. a, Design and predictions for the experiments shown in b-e. b-e, Single-cell injection of 4 pg
Conclusions. The experimental studies presented in this thesis provide the first molecular insights
 C h a p t e r 5 Conclusions 5.1 Summary The experimental studies presented in this thesis provide the first molecular insights into the cellular processes of assembly, and aggregation of neural crest and
C h a p t e r 5 Conclusions 5.1 Summary The experimental studies presented in this thesis provide the first molecular insights into the cellular processes of assembly, and aggregation of neural crest and
Studying the effect of cell division on expression patterns of the segment polarity genes
 Studying the effect of cell division on expression patterns of the segment polarity genes Madalena Chaves and Réka Albert Abstract The segment polarity gene family, and its gene regulatory network, is
Studying the effect of cell division on expression patterns of the segment polarity genes Madalena Chaves and Réka Albert Abstract The segment polarity gene family, and its gene regulatory network, is
Axon guidance I. Paul Garrity March 15, /9.013
 Axon guidance I Paul Garrity March 15, 2004 7.68/9.013 Neuronal Wiring: Functional Framework of the Nervous System Stretch reflex circuit Early theories of axonogenesis Schwann: many neurons link to form
Axon guidance I Paul Garrity March 15, 2004 7.68/9.013 Neuronal Wiring: Functional Framework of the Nervous System Stretch reflex circuit Early theories of axonogenesis Schwann: many neurons link to form
Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors
 Research article 5429 Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors Sarah J. Certel and Stefan Thor*, Department of Neurobiology, Harvard Medical
Research article 5429 Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors Sarah J. Certel and Stefan Thor*, Department of Neurobiology, Harvard Medical
A glial cell arises from an additional division within the mechanosensory lineage during development of the microchaete on the Drosophila notum
 Development 126, 4617-4622 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV8615 4617 A glial cell arises from an additional division within the mechanosensory lineage during development
Development 126, 4617-4622 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV8615 4617 A glial cell arises from an additional division within the mechanosensory lineage during development
Figure 1. Automated 4D detection and tracking of endosomes and polarity markers.
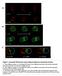 A t = 0 z t = 90 B t = 0 z t = 90 C t = 0-90 t = 0-180 t = 0-300 Figure 1. Automated 4D detection and tracking of endosomes and polarity markers. A: Three different z planes of a dividing SOP shown at
A t = 0 z t = 90 B t = 0 z t = 90 C t = 0-90 t = 0-180 t = 0-300 Figure 1. Automated 4D detection and tracking of endosomes and polarity markers. A: Three different z planes of a dividing SOP shown at
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
split ends encodes large nuclear proteins that regulate neuronal cell fate and
 Development 127, 1517-1529 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV8693 1517 split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension
Development 127, 1517-1529 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV8693 1517 split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension
Roles of wingless in patterning the larval epidermis of Drosophila
 Development 113, 471-485 (1991) Printed in Great Britain (E) The Company of Biologists Limited 1991 471 Roles of wingless in patterning the larval epidermis of Drosophila AMY BEJSOVEC* and ALFONSO MARTINEZ
Development 113, 471-485 (1991) Printed in Great Britain (E) The Company of Biologists Limited 1991 471 Roles of wingless in patterning the larval epidermis of Drosophila AMY BEJSOVEC* and ALFONSO MARTINEZ
Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290
 Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290 Question (from Introduction): How does svb control the
Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290 Question (from Introduction): How does svb control the
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental processes Differential gene expression Introduction to determination The model organisms used to study developmental processes
 Date Title Topic(s) Learning Outcomes: Sept 28 Oct 3 1. What is developmental biology and why should we care? 2. What is so special about stem cells and gametes? Developmental processes Differential gene
Date Title Topic(s) Learning Outcomes: Sept 28 Oct 3 1. What is developmental biology and why should we care? 2. What is so special about stem cells and gametes? Developmental processes Differential gene
IN the Drosophila ventral nerve cord, there are 20
 Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.107.075085 Regulation of Axon Guidance by Slit and Netrin Signaling in the Drosophila Ventral Nerve Cord Krishna Moorthi Bhat,
Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.107.075085 Regulation of Axon Guidance by Slit and Netrin Signaling in the Drosophila Ventral Nerve Cord Krishna Moorthi Bhat,
Role of Organizer Chages in Late Frog Embryos
 Ectoderm Germ Layer Frog Fate Map Frog Fate Map Role of Organizer Chages in Late Frog Embryos Organizer forms three distinct regions Notochord formation in chick Beta-catenin localization How does beta-catenin
Ectoderm Germ Layer Frog Fate Map Frog Fate Map Role of Organizer Chages in Late Frog Embryos Organizer forms three distinct regions Notochord formation in chick Beta-catenin localization How does beta-catenin
Drosophila Life Cycle
 Drosophila Life Cycle 1 Early Drosophila Cleavage Nuclei migrate to periphery after 10 nuclear divisions. Cellularization occurs when plasma membrane folds in to divide nuclei into cells. Drosophila Superficial
Drosophila Life Cycle 1 Early Drosophila Cleavage Nuclei migrate to periphery after 10 nuclear divisions. Cellularization occurs when plasma membrane folds in to divide nuclei into cells. Drosophila Superficial
The function of engrailed and the specification of Drosophila wing pattern
 Development 121, 3447-3456 (1995) Printed in Great Britain The Company of Biologists Limited 1995 3447 The function of engrailed and the specification of Drosophila wing pattern Isabel Guillén 1, *, Jose
Development 121, 3447-3456 (1995) Printed in Great Britain The Company of Biologists Limited 1995 3447 The function of engrailed and the specification of Drosophila wing pattern Isabel Guillén 1, *, Jose
Follow this and additional works at: Part of the Medical Sciences Commons
 Bucknell University Bucknell Digital Commons Master s Theses Student Theses 2010 The overexpression of homeotic complex gene Ultrabithorax in the post-embryonic neuronal lineages of the ventral nervous
Bucknell University Bucknell Digital Commons Master s Theses Student Theses 2010 The overexpression of homeotic complex gene Ultrabithorax in the post-embryonic neuronal lineages of the ventral nervous
Development of Drosophila
 Development of Drosophila Hand-out CBT Chapter 2 Wolpert, 5 th edition March 2018 Introduction 6. Introduction Drosophila melanogaster, the fruit fly, is found in all warm countries. In cooler regions,
Development of Drosophila Hand-out CBT Chapter 2 Wolpert, 5 th edition March 2018 Introduction 6. Introduction Drosophila melanogaster, the fruit fly, is found in all warm countries. In cooler regions,
Expression of a set of glial cell-specific markers in the Drosophila embryonic central nervous system
 BMB Reports BMB Rep. 2014; 47(6): 354-359 www.bmbreports.org Expression of a set of glial cell-specific markers in the Drosophila embryonic central nervous system Hui Jeong Ahn 1, Sang-Hak Jeon 2 & Sang
BMB Reports BMB Rep. 2014; 47(6): 354-359 www.bmbreports.org Expression of a set of glial cell-specific markers in the Drosophila embryonic central nervous system Hui Jeong Ahn 1, Sang-Hak Jeon 2 & Sang
Question Set # 4 Answer Key 7.22 Nov. 2002
 Question Set # 4 Answer Key 7.22 Nov. 2002 1) A variety of reagents and approaches are frequently used by developmental biologists to understand the tissue interactions and molecular signaling pathways
Question Set # 4 Answer Key 7.22 Nov. 2002 1) A variety of reagents and approaches are frequently used by developmental biologists to understand the tissue interactions and molecular signaling pathways
MCN. Complex Genetic Interactions among Four Receptor Tyrosine Phosphatases Regulate Axon Guidance in Drosophila
 MCN Molecular and Cellular Neuroscience 17, 274 291 (2001) doi:10.1006/mcne.2000.0939, available online at http://www.idealibrary.com on Complex Genetic Interactions among Four Receptor Tyrosine Phosphatases
MCN Molecular and Cellular Neuroscience 17, 274 291 (2001) doi:10.1006/mcne.2000.0939, available online at http://www.idealibrary.com on Complex Genetic Interactions among Four Receptor Tyrosine Phosphatases
