Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
|
|
|
- Marian Holmes
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri mutations, and cpm/pri compound heterozygous animals. The latter show the failure of the two alleles to complement one another to form marginal zone B cells. Columns are arithmetic means, error bars standard error of the mean, and dots individual mice. Results of three independent experiments. MZP MZ ns * * ** * ns ** ns Supplementary Figure 2. Marginal zone B cell defect not corrected by overexpression of BAFF. Absence of marginal zone B cells is not corrected in bone marrow chimeras constructed with bone marrow and irradiated BAFF-transgenic mice. Note that the overexpression of BAFF increases the marginal zone subset in chimeras with wildtype bone marrow. Columns are arithmetic means, error bars standard error of the mean, and dots individual mice. Statistical analysis by unpaired, two-tailed t test with Welch s correction, ns, not significant;*,p<0.05, **,P<0.01. Experiment performed once.
2 Supplementary Figure 3. Mutation maps to chromosome 19. Key recombinant haplotypes, SNP markers, chromosomal locations, and corresponding phenotypes of informative (B6xCBA)F 2 intercross cpm animals.
3 B220+ B cells CD4+ CD8+ T cells thymus Supplementary Figure 4. DOCK8 is highly expressed in cells of the immune system. Relative expression of DOCK8 mrna in the indicated mouse tissues, as determined by GNF SYMATLAS Affymetrix microarray profiling data (Su AI et al 2002 Proc Natl Acad Sci USA 99: ;
4 d2.5 d3.5 d4.5 +/+ * Supplementary Figure 5. Location of SW HEL B cells day after adoptive transfer. Histochemical staining for SW HEL B cells of the indicated genotypes in spleen on day 2.5, 3.5 or 4.5 after adoptive transfer and immunization with HEL 2x - SRBC or unconjugated SRBC (*). Blue, HEL binding; Brown, IgD.
5 +/+ * srbc Supplementary Figure 6. Adoptive transfer spleen or lymph node SW HEL cells. The primurus mutation results in a comparable decrease in SW HEL B cell accumulation in GCs on day 4.5 after HEL 2X -SRBC immunization, when either spleen or lymph node cells are adoptively transferred.
6 wildtype SWHEL GC B cells: average # mut silent mutations Mutations in CDR2: r 31 total r s # sequences Total mutations CDR2 D V Q L Q E S G P S L V K P S Q T L S L T C S V T G D S I T S D Y W S W I R K F P G N R L E Y M G Y V S Y S G S T Y Y N P S L K S R I S I T R D T S K N Q Y Y L D L N S V T T E D T A T Y Y C A N W D G D Y W G Q G T L V T V S A Single cell product total r s WT3-C R N S C WT3-C F WT3-C F F T D WT3-E N N WT3-E WT4-A F C N F A n WT2-B A D l H I WT2-B P WT2-B Y q S WT2-B C N N I WT2-E C WT4-A R - WT4A WT4-B g s I F C WT1-D WT1-D s F a WT1-D K r F N k WT1-D e N F A WT1-D WT1-E WT1-E N WT2-E N S G H t y WT2-E WT2-E WT2-E WT2-E D R y WT3-B WT3-B N D T C P WT3-D T D D S WT3-D L R WT3-D s C WT3-D K e H D WT3-D T T S g G Y WT4-A WT4-D N P WT4-E N WT4-E Y WT4-E Sums: # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # Mutations per residue: r s C 82B 82A SWHEL GC B cells: total r s s 3 # sequences Total mutations average # mut CDR2 D V Q L Q E S G P S L V K P S Q T L S L T C S V T G D S I T S D Y W S W I R K F P G N R L E Y M G Y V S Y S G S T Y Y N P S L K S R I S I T R D T S K N Q Y Y L D L N S V T T E D T A T Y Y C A N W D G D Y W G Q G T L V T V S A Single cell product total r s P4-A P4-A P4-A P4-C P3-A s P3-A P3-A P3-C P3-C P3-E P N F P1-D V i F g N y t L P1-D s D s V P1-E p S P1-F P2-B2 * P2-B g P2-B V t I I C P2-C s Y P1-A v T S P1-B G R H t P1-B s N T p S y H P1-B F P1-B P2-G D P2-F H I N s M A P2-F F C S Y P2-F P2-E R C s t P2-E P2-D s F P3-E P3-E L g N N S P3-E I v F F S P3-G P3-G P4-D C e v R T H l G P4-D D T P4-E i P4-E N N S P4-E P4-E A N R T P4-G P P4-G V v Y Y P1-G Sums # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # Mutations per residue: r s C 82B 82A Mutations in CDR2: r 24 s 10 Supplementary Figure 7. Heavy chain VDJ-region mutations in flow-sorted single germinal center wildtype or SWHEL B cells. SWHEL GC cells were sorted on day 9 after HEL2x-SRBC immunization from four different recipients of wildtype SWHEL cells (prefix WT1 - WT4) and four different recipients of SWHEL cells (prefix P1-P4). The SWHEL heavy chain V region was amplified by nested PCR from single cells, and the PCR products directly sequenced without cloning. Silent (s) and replacement (r ) mutations are shown. Heavy chain residues 50, 52, 53, 54, 56 and 58 make contacts with HEL on or next to D101. At this stage of the response the recurrent Y53D mutation observed on day 15 has yet to become dominant in wildtype SWHEL GC cells. Recurrent Y58F mutations are observed prior to and independently of Y53D in normal SWHEL cells (R Brink et al unpublished data), and these are evident in six wildtype SWHEL GC cells at day 9. X-ray structural comparisons show that the Y58F substitution increases the buried apolar surface between antibody and antigen, explaining increased HEL-affinity in the related antibody HyHEL8 compared to HyHEL10 SwHEL (Li Y et al Nat Struct Biol :482. X-ray snapshots of the maturation of an antibody response to a protein antigen).
7 +/ Counts Stimulus: Anti-IgM (!g/ml) IC10 (!g/ml)! !!! 10 Supplementary Figure 8. Assessment of proliferation by DNA synthesis. DNA synthesis by cultured lymph node cells from four individual wild-type and four homozygous animals. The cells from each mouse were cultured in triplicate wells, either unstimulated (US), or with the indicated concentrations of antibody to IgM and/or antibody to CD40 (1C10). Each culture was pulsed with 3H-thymidine after 36 hours, harvested, and the mean and standard error of incorporated radioactivity (counts) in triplicates from individual animals expressed by columns and error bars, respectively.
8 Stimulus: Anti-IgM (!g/ml): cpm/cpm cpm/cpm LPS CFSE 1.0 Supplementary Figure 9. Proliferation assessed by CFSE. Mutant or cpm/cpm cells (red histograms) were CFSE labelled and cultured with wild-type, congenically cells (black histograms) in the same wells to control for experimental variation. Each panel shows an independent pair of mutant and wild-type samples co-cultured. Each independent sample was from separate mice, and typically pooled from two or more animals. In 6 of the 8 samples the CFSE dilution profiles of mutant and wild-type were superimposable, and in 2 there was a subtle decrease in CFSE dilution in mutant cells.
9 Supplementary Figure 10. Expression of activation markers. Median and s.e. of cell surface fluorescence staining for activation markers CD25, CD69 and CD86 on lymph node B cells cultured from four individual wild-type and four homozygous animals. The cells from each mouse were cultured either unstimulated (US), or with the indicated concentrations of antibody to IgM and CD40 (1C10). Each culture was harvested after 18 hr, stained for B220 and the indicated markers, analyzed by flow cytometry, and median fluorescence intensity calculated. There was no statistical difference in median fluorescence between mutant and wildtype mice for any stimulus condition. Supplementary Figure 11. Assessment of B:T conjugation. Enumeration of B cell: T cell conjugates formed in vitro after addition of separately cultured OT-II T cells (with and without peptide stimulation) and B cells (stimulated with LPS or CD40L and IL-4). Results show decreased conjugation formation with SAP KO OT-II T cells (as expected), while conjugation formation with B cells was comparable to +/+ B cells. Columns are arithmetic means, error bars standard error of the mean, and dots individual results. Representative of two independent experiments.
10 CD45.2 (%) Spleen pln Supplementary Figure 12. Assessment of LN B cells in bone marrow chimeras. Mutant B cells competitively accumulate in peripheral lymph nodes in mixed bone marrow chimeras reconstituted with wild-type CD45.1 bone marrow and CD45.2 marrow. Each black bar shows the percentage contribution of CD45.2 cells in individual mice (numbered 1-4) to the follicular B cell subset in the spleen and the peripheral lymph nodes (pln). Note that there is no consistent difference in the percentage of B cells that are derived from CD45.2 in pln compared to the percentage of these cells in the spleen (P=0.83, unpaired, two-tailed t-test with Welch s correction). Similar results are found in cpm/cpm mixed bone marrow chimeras (data not shown). Stimulus: nil S1P (mm) CXCL13 CXCL /+ cpm/cpm Migrating cells (% input) Supplementary Figure 13. B cell chemotaxis. Migration of wild-type and mutant CD19 + B cells to S1P, CXCL12 (0.3 µg/ml) and CXCL13 (1 µ g/ml) in vitro. Data from 3 independent experiments. Columns are arithmetic means, bars s.e.m, and dots individual mice.
11 Primers for amplification of DOCK8 cdna cdna Primers Primer sequence Spanning exons 1F GGTGGAAATGCGGAAGTTT 1-5 1R AGCACGTTTAAGGGATGACG 2F ACCTTGGAGTGCAGTGAACC R TTGGCAGAAGGATTCAGCTT 3F AAAGAAAGCGATGGTGGAAA R TGGGGGTATACGTACAGAAGG 4F GCGTCCACACAAGGAGATTT R CGGTTTGAAGACGTTCGTTTA 5F CAGCTGTCAGCAGAAGCAAG R GTAGGAGGCCAGAAGGCAGT 6F GCATCATCTGCCTCAACTCC R GGCAAGGCTGATGTTGATCT 7F GGTCACCTCGGAGATAGCAG R TCATGATCCACAGGAAGCAA 8F TGAAGACCAGTGGAGCAATG R CTCCTCGAACAGCAGGTCTC 9F ACCAGAGCACCACCTACCTG R CTGCAGACTCCTCAAGCACA 10F ATGCTGGAGGACCACAGCTA R CACCCGGTCTTTCATCTCAT 11F TGTTTCGGTGCAGAGTTTGT R GATTTTCCGCTCAATCATGG 12F CAAGCTGAGGTTGTGCTTCA R TGCTGGACAGTGTTCCACTC Primers for amplification of genomic DOCK8 DNA spanning introns Genomic Primers Primer sequence D8-Intron1F CTGTTTCTTCCGTCCACACC D8-Intron1R GAGTGGCAAAGGGTGAAGAA D8-Intron2F CTTCCTCCACCTGGTGCTT D8-Intron2R AGTACATCTGCCTGCCTGCT D8-Intron3F AGCAGGTGGGTAGGCAGTC D8-Intron3R CTTTCGGGATAGCATTTGAA D8-Intron4F TGTGCATCTCCAGTTTAAAGG D8-Intron4R AGGGCCCACAATGGAGTAG D8-Intron5F TAGGGTTGCAGACCCCTTC D8-Intron5R CTAATTCATGGCATCGCTCA D8-Intron6F GCAAGTAAAGATGCTTGTCTCCA D8-Intron6R GTGTACGTGCGTGCCTGTAT D8-Intron7F AGCATCTGGCCTCTGCATAC D8-Intron7R AGCCAACTTTGTGCCTGTTT D8-Intron8F AGAATGGGATGAGCTGATGG D8-Intron8R CATGCCTGGCTTACACCTTT D8-Intron9F CCCAGTTTTCAGCTGAGTTT D8-Intron9R TTCCTCAGGTCCTTGCTGTT Supplementary Table 1: Primer sequences spanning DOCK8 cdna and introns 19-21
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
FSC-W FSC-H CD4 CD62-L
 Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1
 Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Table S1. Sequence of primers used in RT-qPCR
 Table S1. Sequence of primers used in RT-qPCR Primer Name P16Ink4a-F P16Ink4a-R P15Ink4b-F P15Ink4b-R P19Arf-F P19Arf-R P53-F P53-R P21cip1-F P21cip1-R P27kip1-F P27kip1-R P18Ink4c-F P18Ink4c-R P19Ink4d-F
Table S1. Sequence of primers used in RT-qPCR Primer Name P16Ink4a-F P16Ink4a-R P15Ink4b-F P15Ink4b-R P19Arf-F P19Arf-R P53-F P53-R P21cip1-F P21cip1-R P27kip1-F P27kip1-R P18Ink4c-F P18Ink4c-R P19Ink4d-F
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. NAD + -related genes expression in mouse tissues and in cells overexpressing NRK1. mrna (a) and protein (b) levels of NRK1, NRK2 and other enzymes involved
Supplementary Figure 1 Supplementary Figure 1. NAD + -related genes expression in mouse tissues and in cells overexpressing NRK1. mrna (a) and protein (b) levels of NRK1, NRK2 and other enzymes involved
Actinobacteria Relative abundance (%) Co-housed CD300f WT. CD300f KO. Colon length (cm) Day 9. Microscopic inflammation score
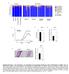 y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
Table S1 List of primers used for genotyping and qrt-pcr.
 Table S1 List of primers used for genotyping and qrt-pcr. genotyping! allele! ligomer*! 5'-sequence-3'! rice! d10-2! F! TTGGCTTTGCCTCGTTTC!!! R! AGCCTCCACTTGTACTGTG! Arabidopsis! max2-3, max2-4! F! ACTCTCTCCGACCTCCCTGACG!!!
Table S1 List of primers used for genotyping and qrt-pcr. genotyping! allele! ligomer*! 5'-sequence-3'! rice! d10-2! F! TTGGCTTTGCCTCGTTTC!!! R! AGCCTCCACTTGTACTGTG! Arabidopsis! max2-3, max2-4! F! ACTCTCTCCGACCTCCCTGACG!!!
Solutions to Problem Set 4
 Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
CELL REPLICATION. Fluorescent light microscopy showing mitosis, especially immunolabelled cytoskeleton and tubulin
 CELL REPLICATION Fluorescent light microscopy showing mitosis, especially immunolabelled cytoskeleton and tubulin Cell REPLICATION PROLIFERATION MUTIPLICATION DIVISION CELL REPLICATION Fluorescent light
CELL REPLICATION Fluorescent light microscopy showing mitosis, especially immunolabelled cytoskeleton and tubulin Cell REPLICATION PROLIFERATION MUTIPLICATION DIVISION CELL REPLICATION Fluorescent light
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
RANK. Alternative names. Discovery. Structure. William J. Boyle* SUMMARY BACKGROUND
 RANK William J. Boyle* Department of Cell Biology, Amgen, Inc., One Amgen Center Drive, Thousand Oaks, CA 91320-1799, USA * corresponding author tel: 805-447-4304, fax: 805-447-1982, e-mail: bboyle@amgen.com
RANK William J. Boyle* Department of Cell Biology, Amgen, Inc., One Amgen Center Drive, Thousand Oaks, CA 91320-1799, USA * corresponding author tel: 805-447-4304, fax: 805-447-1982, e-mail: bboyle@amgen.com
The phenotype of this worm is wild type. When both genes are mutant: The phenotype of this worm is double mutant Dpy and Unc phenotype.
 Series 2: Cross Diagrams - Complementation There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
Series 2: Cross Diagrams - Complementation There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
Unit 3 - Molecular Biology & Genetics - Review Packet
 Name Date Hour Unit 3 - Molecular Biology & Genetics - Review Packet True / False Questions - Indicate True or False for the following statements. 1. Eye color, hair color and the shape of your ears can
Name Date Hour Unit 3 - Molecular Biology & Genetics - Review Packet True / False Questions - Indicate True or False for the following statements. 1. Eye color, hair color and the shape of your ears can
mrna Codon Table Mutant Dinosaur Name: Period:
 Mutant Dinosaur Name: Period: Intro Your dinosaur is born with a new genetic mutation. Your job is to map out the genes that are influenced by the mutation and to discover how the new dinosaurs interact
Mutant Dinosaur Name: Period: Intro Your dinosaur is born with a new genetic mutation. Your job is to map out the genes that are influenced by the mutation and to discover how the new dinosaurs interact
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09414 Supplementary Figure 1 FACS-isolated 8c hepatocytes are highly pure. a, Gating strategy for identifying hepatocyte populations based on DNA content. b, Detection of mchry and mchr9
doi:10.1038/nature09414 Supplementary Figure 1 FACS-isolated 8c hepatocytes are highly pure. a, Gating strategy for identifying hepatocyte populations based on DNA content. b, Detection of mchry and mchr9
Reading: Chapter 5, pp ; Reference chapter D, pp Problem set F
 Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants. GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
 SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
Supplementary Figure 1. Nature Genetics: doi: /ng.3848
 Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Biological Roles of Cytokinins
 Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Chromosome Chr Duplica Duplic t a ion Pixley
 Chromosome Duplication Pixley Figure 4-6 Molecular Biology of the Cell ( Garland Science 2008) Figure 4-72 Molecular Biology of the Cell ( Garland Science 2008) Interphase During mitosis (cell division),
Chromosome Duplication Pixley Figure 4-6 Molecular Biology of the Cell ( Garland Science 2008) Figure 4-72 Molecular Biology of the Cell ( Garland Science 2008) Interphase During mitosis (cell division),
The E-M Algorithm in Genetics. Biostatistics 666 Lecture 8
 The E-M Algorithm in Genetics Biostatistics 666 Lecture 8 Maximum Likelihood Estimation of Allele Frequencies Find parameter estimates which make observed data most likely General approach, as long as
The E-M Algorithm in Genetics Biostatistics 666 Lecture 8 Maximum Likelihood Estimation of Allele Frequencies Find parameter estimates which make observed data most likely General approach, as long as
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
** LCA LCN PCA
 % of wild type value % of wild type value a 12 1 8 2 b 12 1 8 2 LCA LCN PCA Col- sod3-1 Supplementary Figure 1 sod3-1 influences cell proliferation. (a) Fifth leaf cell area (LCA) and leaf cell number
% of wild type value % of wild type value a 12 1 8 2 b 12 1 8 2 LCA LCN PCA Col- sod3-1 Supplementary Figure 1 sod3-1 influences cell proliferation. (a) Fifth leaf cell area (LCA) and leaf cell number
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Supplementary Methods
 Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Designer Genes C Test
 Northern Regional: January 19 th, 2019 Designer Genes C Test Name(s): Team Name: School Name: Team Number: Rank: Score: Directions: You will have 50 minutes to complete the test. You may not write on the
Northern Regional: January 19 th, 2019 Designer Genes C Test Name(s): Team Name: School Name: Team Number: Rank: Score: Directions: You will have 50 minutes to complete the test. You may not write on the
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/243/ra68/dc1 Supplementary Materials for Superbinder SH2 Domains Act as Antagonists of Cell Signaling Tomonori Kaneko, Haiming Huang, Xuan Cao, Xing Li, Chengjun
www.sciencesignaling.org/cgi/content/full/5/243/ra68/dc1 Supplementary Materials for Superbinder SH2 Domains Act as Antagonists of Cell Signaling Tomonori Kaneko, Haiming Huang, Xuan Cao, Xing Li, Chengjun
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX.
 Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Curriculum Links. AQA GCE Biology. AS level
 Curriculum Links AQA GCE Biology Unit 2 BIOL2 The variety of living organisms 3.2.1 Living organisms vary and this variation is influenced by genetic and environmental factors Causes of variation 3.2.2
Curriculum Links AQA GCE Biology Unit 2 BIOL2 The variety of living organisms 3.2.1 Living organisms vary and this variation is influenced by genetic and environmental factors Causes of variation 3.2.2
Principles of Genetics
 Principles of Genetics Snustad, D ISBN-13: 9780470903599 Table of Contents C H A P T E R 1 The Science of Genetics 1 An Invitation 2 Three Great Milestones in Genetics 2 DNA as the Genetic Material 6 Genetics
Principles of Genetics Snustad, D ISBN-13: 9780470903599 Table of Contents C H A P T E R 1 The Science of Genetics 1 An Invitation 2 Three Great Milestones in Genetics 2 DNA as the Genetic Material 6 Genetics
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Biology I Level - 2nd Semester Final Review
 Biology I Level - 2nd Semester Final Review The 2 nd Semester Final encompasses all material that was discussed during second semester. It s important that you review ALL notes and worksheets from the
Biology I Level - 2nd Semester Final Review The 2 nd Semester Final encompasses all material that was discussed during second semester. It s important that you review ALL notes and worksheets from the
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Regulation of Phosphate Homeostasis by microrna in Plants
 Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
Supplementary Figure 1.
 Supplementary Figure 1. Immunodominant epitope profiles of human S-Ag protein for HLA-DR4 alleles. Spleen cells from S-Ag immunized mice were used in an in-vitro lymphocyte proliferation assay in the presence
Supplementary Figure 1. Immunodominant epitope profiles of human S-Ag protein for HLA-DR4 alleles. Spleen cells from S-Ag immunized mice were used in an in-vitro lymphocyte proliferation assay in the presence
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field.
 Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
RNA Synthesis and Processing
 RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
ESR1 Allele Frequency. PIK3CA Allele Frequency
 Supplementary Figure 1 Distribution of mutant allele frequencies ESR1 Allele Frequency Median allele frequency 0.45% Inset graph 10 20 30 40 50 PIK3CA Allele Frequency Median allele frequency 3.6% Inset
Supplementary Figure 1 Distribution of mutant allele frequencies ESR1 Allele Frequency Median allele frequency 0.45% Inset graph 10 20 30 40 50 PIK3CA Allele Frequency Median allele frequency 3.6% Inset
Cells and Their Processes. 1. What element do organic compounds have that inorganic compounds do not?
 Name: Date: Cells and Their Processes 1. What element do organic compounds have that inorganic compounds do not? 2. List the four types of organic compounds, describe the function of each AND list a food
Name: Date: Cells and Their Processes 1. What element do organic compounds have that inorganic compounds do not? 2. List the four types of organic compounds, describe the function of each AND list a food
 www.lessonplansinc.com Topic: Dinosaur Evolution Project Summary: Students pretend to evolve two dinosaurs using genetics and watch how the dinosaurs adapt to an environmental change. This is a very comprehensive
www.lessonplansinc.com Topic: Dinosaur Evolution Project Summary: Students pretend to evolve two dinosaurs using genetics and watch how the dinosaurs adapt to an environmental change. This is a very comprehensive
Q2 (4.6) Put the following in order from biggest to smallest: Gene DNA Cell Chromosome Nucleus. Q8 (Biology) (4.6)
 Q1 (4.6) What is variation? Q2 (4.6) Put the following in order from biggest to smallest: Gene DNA Cell Chromosome Nucleus Q3 (4.6) What are genes? Q4 (4.6) What sort of reproduction produces genetically
Q1 (4.6) What is variation? Q2 (4.6) Put the following in order from biggest to smallest: Gene DNA Cell Chromosome Nucleus Q3 (4.6) What are genes? Q4 (4.6) What sort of reproduction produces genetically
Chapter 17. From Gene to Protein. Biology Kevin Dees
 Chapter 17 From Gene to Protein DNA The information molecule Sequences of bases is a code DNA organized in to chromosomes Chromosomes are organized into genes What do the genes actually say??? Reflecting
Chapter 17 From Gene to Protein DNA The information molecule Sequences of bases is a code DNA organized in to chromosomes Chromosomes are organized into genes What do the genes actually say??? Reflecting
Multiple Choice Review- Eukaryotic Gene Expression
 Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
SUPPLEMENTARY INFORMATION
 reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
NB-DNJ/GCase-pH 7.4 NB-DNJ+/GCase-pH 7.4 NB-DNJ+/GCase-pH 4.5
 SUPPLEMENTARY TABLES Suppl. Table 1. Protonation states at ph 7.4 and 4.5. Protonation states of titratable residues in GCase at ph 7.4 and 4.5. Histidine: HID, H at δ-nitrogen; HIE, H at ε-nitrogen; HIP,
SUPPLEMENTARY TABLES Suppl. Table 1. Protonation states at ph 7.4 and 4.5. Protonation states of titratable residues in GCase at ph 7.4 and 4.5. Histidine: HID, H at δ-nitrogen; HIE, H at ε-nitrogen; HIP,
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated
 Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
TE content correlates positively with genome size
 TE content correlates positively with genome size Mb 3000 Genomic DNA 2500 2000 1500 1000 TE DNA Protein-coding DNA 500 0 Feschotte & Pritham 2006 Transposable elements. Variation in gene numbers cannot
TE content correlates positively with genome size Mb 3000 Genomic DNA 2500 2000 1500 1000 TE DNA Protein-coding DNA 500 0 Feschotte & Pritham 2006 Transposable elements. Variation in gene numbers cannot
Nature Methods: doi: /nmeth Supplementary Figure 1. In vitro screening of recombinant R-CaMP2 variants.
 Supplementary Figure 1 In vitro screening of recombinant R-CaMP2 variants. Baseline fluorescence compared to R-CaMP1.07 at nominally zero calcium plotted versus dynamic range ( F/F) for 150 recombinant
Supplementary Figure 1 In vitro screening of recombinant R-CaMP2 variants. Baseline fluorescence compared to R-CaMP1.07 at nominally zero calcium plotted versus dynamic range ( F/F) for 150 recombinant
What is the structure of DNA?
 NAME Biology Final Review Sem. II Genetics 1. Define: a. allele b. phenotype c. genotype d. recessive e. dominant f. heterozygous g. homozygous h. autosomes i. sex chromosomes j. Punnett square k. pedigree
NAME Biology Final Review Sem. II Genetics 1. Define: a. allele b. phenotype c. genotype d. recessive e. dominant f. heterozygous g. homozygous h. autosomes i. sex chromosomes j. Punnett square k. pedigree
A complementation test would be done by crossing the haploid strains and scoring the phenotype in the diploids.
 Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication
 SUPPORTING ONLINE MATERIALS Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication Bin Cong, Luz Barrero, & Steven Tanksley 1 SUPPORTING
SUPPORTING ONLINE MATERIALS Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication Bin Cong, Luz Barrero, & Steven Tanksley 1 SUPPORTING
FW 1 CDR 1 FW 2 CDR 2
 Supplementary Figure 1 Supplementary Figure 1: Interface of the E9:Fas structure. The two interfaces formed by V H and V L of E9 with Fas are shown in stereo. The Fas receptor is represented as a surface
Supplementary Figure 1 Supplementary Figure 1: Interface of the E9:Fas structure. The two interfaces formed by V H and V L of E9 with Fas are shown in stereo. The Fas receptor is represented as a surface
1.5.1 ESTIMATION OF HAPLOTYPE FREQUENCIES:
 .5. ESTIMATION OF HAPLOTYPE FREQUENCIES: Chapter - 8 For SNPs, alleles A j,b j at locus j there are 4 haplotypes: A A, A B, B A and B B frequencies q,q,q 3,q 4. Assume HWE at haplotype level. Only the
.5. ESTIMATION OF HAPLOTYPE FREQUENCIES: Chapter - 8 For SNPs, alleles A j,b j at locus j there are 4 haplotypes: A A, A B, B A and B B frequencies q,q,q 3,q 4. Assume HWE at haplotype level. Only the
Development 143: doi: /dev : Supplementary information
 Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
When one gene is wild type and the other mutant:
 Series 2: Cross Diagrams Linkage Analysis There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
Series 2: Cross Diagrams Linkage Analysis There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
m1 m2 m3 m4 m5 m6 m7 m8 wt m m m m m m m m8 - + wt +
 otherwise, you couldn't grow them!) You perform pairwise infections with each of your mutant bacteriophage strains and get the following results: (+) = pair of phages lysed host cells, (-) = pair of phages
otherwise, you couldn't grow them!) You perform pairwise infections with each of your mutant bacteriophage strains and get the following results: (+) = pair of phages lysed host cells, (-) = pair of phages
Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures
 Nature Methods Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures Yannick Doyon, Thuy D Vo, Matthew C Mendel, Shon G Greenberg, Jianbin Wang, Danny F Xia, Jeffrey
Nature Methods Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures Yannick Doyon, Thuy D Vo, Matthew C Mendel, Shon G Greenberg, Jianbin Wang, Danny F Xia, Jeffrey
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Different crystal forms obtained for Sky
 Supplementary Figure 1 Different crystal forms obtained for Sky 1 353. (a) Crystal form 1 obtained in the presence of 20% PEG 3350 and 0.2 M ammonium citrate tribasic ph 7.0. (b) Crystal form 1 of the
Supplementary Figure 1 Different crystal forms obtained for Sky 1 353. (a) Crystal form 1 obtained in the presence of 20% PEG 3350 and 0.2 M ammonium citrate tribasic ph 7.0. (b) Crystal form 1 of the
EST1 Homology Domain. 100 aa. hest1a / SMG6 PIN TPR TPR. Est1-like DBD? hest1b / SMG5. TPR-like TPR. a helical. hest1c / SMG7.
 hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
Supporting Information
 Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B-NS3 protease
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
Supplemental Data. Yamamoto et al. (2016). Plant Cell /tpc WT + HCO 3. slac1-4/δnc #5 + HCO 3 WT + ABA. slac1-4/δnc #5 + ABA
 I (pa) I (pa) A WT + HCO 3 B 2 V (mv) 15 1 5 5 2 slac14/δnc #5 + HCO 3 4 6 WT (n = 6) 8 WT + HCO 3 (n = 7) slac14/δnc #5 (n = 6) slac14/δnc #5 + HCO 3 (n = 1) C WT + ABA D 2 V (mv) 15 1 5 5 2 slac14/δnc
I (pa) I (pa) A WT + HCO 3 B 2 V (mv) 15 1 5 5 2 slac14/δnc #5 + HCO 3 4 6 WT (n = 6) 8 WT + HCO 3 (n = 7) slac14/δnc #5 (n = 6) slac14/δnc #5 + HCO 3 (n = 1) C WT + ABA D 2 V (mv) 15 1 5 5 2 slac14/δnc
Michigan State University Diagnostic Center for Population and Animal Health, Lansing MI USA. QIAGEN Leipzig GmbH, Leipzig, Germany
 Detection of Bovine Viral Diarrhea (BVD) Virus in Ear Skin Tissue Samples: Evaluation of a Combination of QIAGEN MagAttract 96 cador Pathogen Extraction and virotype BVDV RT-PCR-based Amplification. Roger
Detection of Bovine Viral Diarrhea (BVD) Virus in Ear Skin Tissue Samples: Evaluation of a Combination of QIAGEN MagAttract 96 cador Pathogen Extraction and virotype BVDV RT-PCR-based Amplification. Roger
Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1
 Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1 (a) Fold-change in UCP-1 mrna abundance in white adipocytes upon β-adrenergic stimulation
Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1 (a) Fold-change in UCP-1 mrna abundance in white adipocytes upon β-adrenergic stimulation
A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae.
 Genetics: Published Articles Ahead of Print, published on July 30, 2012 as 10.1534/genetics.112.143818 A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces
Genetics: Published Articles Ahead of Print, published on July 30, 2012 as 10.1534/genetics.112.143818 A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces
Outline. Leaf Development. Leaf Structure - Morphology. Leaf Structure - Morphology
 Outline 1. Leaf Structure: Morphology & Anatomy 2. Leaf Development A. Anatomy B. Sector analysis C. Leaf Development Leaf Structure - Morphology Leaf Structure - Morphology 1 Leaf Structure - Morphology
Outline 1. Leaf Structure: Morphology & Anatomy 2. Leaf Development A. Anatomy B. Sector analysis C. Leaf Development Leaf Structure - Morphology Leaf Structure - Morphology 1 Leaf Structure - Morphology
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/331/6019/876/dc1 Supporting Online Material for Synthetic Clonal Reproduction Through Seeds Mohan P. A. Marimuthu, Sylvie Jolivet, Maruthachalam Ravi, Lucie Pereira,
www.sciencemag.org/cgi/content/full/331/6019/876/dc1 Supporting Online Material for Synthetic Clonal Reproduction Through Seeds Mohan P. A. Marimuthu, Sylvie Jolivet, Maruthachalam Ravi, Lucie Pereira,
Molecular Evolution & the Origin of Variation
 Molecular Evolution & the Origin of Variation What Is Molecular Evolution? Molecular evolution differs from phenotypic evolution in that mutations and genetic drift are much more important determinants
Molecular Evolution & the Origin of Variation What Is Molecular Evolution? Molecular evolution differs from phenotypic evolution in that mutations and genetic drift are much more important determinants
Molecular Evolution & the Origin of Variation
 Molecular Evolution & the Origin of Variation What Is Molecular Evolution? Molecular evolution differs from phenotypic evolution in that mutations and genetic drift are much more important determinants
Molecular Evolution & the Origin of Variation What Is Molecular Evolution? Molecular evolution differs from phenotypic evolution in that mutations and genetic drift are much more important determinants
Functional expression of PEPT2 and its regulation in alveolar epithelial cells
 Workshop on Drug Transporters in the Lungs TCD, Dublin, IRL, Sept. 22-23, 216 Functional expression of PEPT2 and its regulation in alveolar epithelial cells Mikihisa Takano, Ph.D. Graduate School of Biomedical
Workshop on Drug Transporters in the Lungs TCD, Dublin, IRL, Sept. 22-23, 216 Functional expression of PEPT2 and its regulation in alveolar epithelial cells Mikihisa Takano, Ph.D. Graduate School of Biomedical
A Mathematical Study of Germinal Center Formation
 A Mathematical Study of Germinal Center Formation Samantha Erwin Adviser: Dr. Stanca Ciupe Virginia Tech October 1, 2014 Samantha Erwin Modeling Germinal Center Formation 1/19 1 Biology 2 The Model 3 Results
A Mathematical Study of Germinal Center Formation Samantha Erwin Adviser: Dr. Stanca Ciupe Virginia Tech October 1, 2014 Samantha Erwin Modeling Germinal Center Formation 1/19 1 Biology 2 The Model 3 Results
. Supplementary Information
 . Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
. Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
Allele interactions: Terms used to specify interactions between alleles of the same gene: Dominant/recessive incompletely dominant codominant
 Biol 321 Feb 3, 2010 Allele interactions: Terms used to specify interactions between alleles of the same gene: Dominant/recessive incompletely dominant codominant Gene interactions: the collaborative efforts
Biol 321 Feb 3, 2010 Allele interactions: Terms used to specify interactions between alleles of the same gene: Dominant/recessive incompletely dominant codominant Gene interactions: the collaborative efforts
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
Inferring Genetic Architecture of Complex Biological Processes
 Inferring Genetic Architecture of Complex Biological Processes BioPharmaceutical Technology Center Institute (BTCI) Brian S. Yandell University of Wisconsin-Madison http://www.stat.wisc.edu/~yandell/statgen
Inferring Genetic Architecture of Complex Biological Processes BioPharmaceutical Technology Center Institute (BTCI) Brian S. Yandell University of Wisconsin-Madison http://www.stat.wisc.edu/~yandell/statgen
Heterozygous BMN lines
 Optical density at 80 hours 0.8 0.6 0.4 0.2 0.8 0.6 0.4 0.2 0.8 0.6 0.4 0.2 0.8 0.6 0.4 0.2 a YPD b YPD + 1µM nystatin c YPD + 2µM nystatin d YPD + 4µM nystatin 1 3 5 6 9 13 16 20 21 22 23 25 28 29 30
Optical density at 80 hours 0.8 0.6 0.4 0.2 0.8 0.6 0.4 0.2 0.8 0.6 0.4 0.2 0.8 0.6 0.4 0.2 a YPD b YPD + 1µM nystatin c YPD + 2µM nystatin d YPD + 4µM nystatin 1 3 5 6 9 13 16 20 21 22 23 25 28 29 30
Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr.
 Molecular Cell, Volume 30 Supplemental Data Coupling of Ribosomal L1 Stalk and trna Dynamics during Translation Elongation Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr. Supplemental
Molecular Cell, Volume 30 Supplemental Data Coupling of Ribosomal L1 Stalk and trna Dynamics during Translation Elongation Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr. Supplemental
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Information #
 Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Details of PRF Methodology In the Poisson Random Field PRF) model, it is assumed that non-synonymous mutations at a given gene are either
Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Details of PRF Methodology In the Poisson Random Field PRF) model, it is assumed that non-synonymous mutations at a given gene are either
Nature Genetics: doi: /ng Supplementary Figure 1. The FIN and FAB genes act separately from the meristem maturation pathway.
 Supplementary Figure 1 The FIN and FAB genes act separately from the meristem maturation pathway. (a) Representative inflorescence from the compound inflorescence (s, defective in the homolog of Arabidopsis
Supplementary Figure 1 The FIN and FAB genes act separately from the meristem maturation pathway. (a) Representative inflorescence from the compound inflorescence (s, defective in the homolog of Arabidopsis
CJ LI LRRK2 MOUSE COMPARISON STUDY. Phenotyping Data Results
 CJ LI MOUSE COMPARISON STUDY Phenotyping Data Results NOTE FOR USE This comparison study was run as an opportunistic look at the phenotype of multiple CJ Li-generated mouse lines. The primary purpose of
CJ LI MOUSE COMPARISON STUDY Phenotyping Data Results NOTE FOR USE This comparison study was run as an opportunistic look at the phenotype of multiple CJ Li-generated mouse lines. The primary purpose of
Case-Control Association Testing. Case-Control Association Testing
 Introduction Association mapping is now routinely being used to identify loci that are involved with complex traits. Technological advances have made it feasible to perform case-control association studies
Introduction Association mapping is now routinely being used to identify loci that are involved with complex traits. Technological advances have made it feasible to perform case-control association studies
4º ESO BIOLOGY & GEOLOGY SUMMER REINFORCEMENT: CONTENTS & ACTIVITIES
 COLEGIO INTERNACIONAL SEK ALBORÁN 4º ESO BIOLOGY & GEOLOGY SUMMER REINFORCEMENT: CONTENTS & ACTIVITIES 1 ST EVALUATION UNIT 4: CELLS 1. Levels of biological organization 2. Cell theory 3. Basic unit of
COLEGIO INTERNACIONAL SEK ALBORÁN 4º ESO BIOLOGY & GEOLOGY SUMMER REINFORCEMENT: CONTENTS & ACTIVITIES 1 ST EVALUATION UNIT 4: CELLS 1. Levels of biological organization 2. Cell theory 3. Basic unit of
