NTChem. ntprep. ntprep
|
|
|
- Karen Watts
- 6 years ago
- Views:
Transcription
1 NTChem NTChem.inp ) bash ) csh RCCS)) ntprep ntprep /opt/aics/ntchem/scripts RCCS) ccpg: /local/apl/pg/ntchem/scripts ccuv: /local/apl/uv/ntchem/scripts Focus: /home1/share/ntchem/ntchem /scripts ntprep 1) xmol xyz 2).bash_profile ntprep.bash_profile $PATH ntprep * # User specific environment and startup programs $ PATH=$PATH: /opt/aics/ntchem/scripts $ export PATH 3) ntprep NTChem ntprep ntprep.basis 4) ntprep cd ntprep Enter ntprep $ cd ~/my_ntchem_wrkdir
2 $ ntprep 5) Enter geometry file name: xmol xyz Enter : h2o.xyz h2o.xyz [Enter] 6) NTChem Enter input file name NTChem Enter : NTChem h2o_dft_opt.inp h2o_dft_opt [Enter] Enter 3) 3) h2o.xyz h2o.inp 7) Select the type of task 1 Enter Enter : 3 [Enter] 8) Select the quantum chemistry method DFT Enter Enter : HF 1 [Enter] 9) Select how to assign the basis set Enter 2 Enter, Select the basis set for
3 Def2-SVP Enter Enter, 0 Enter Select basis set card Enter : mybasis.basis mybasis.basis [Enter] 10) Select the scalar relativistic Hamiltonian Enter Enter : DK 2 [Enter] - Select the spin-orbit relativistic Hamiltonian - - Enter - Enter : - DK3 - DK1 2[Enter] 2[Enter] 11) Define the total charge 0) Enter Enter : -1-1 [Enter]
4 12) Define the spin multiplicity 1 2) Enter Enter : 3 3 [Enter] 13) SCF Select the type of SCF SCF SCF 1 Restricted 1 Unrestricted) Enter SCF SCF Enter : 3 SCF Restricted-Open 2 [Enter] 14) DFT 7) DFT Select the DFT exchange-correlation functional DFT wb97xd Enter Enter : DFT B3LYP 3 [Enter] 15) SCF Fock 2 Coulomb Select the method for evaluation of Coulomb contribution in SCF SCF Fock 2 Coulomb 2 Enter RI 2 Enter 16) SCF Select the SCF initial guess NDDO Enter Enter : NTChem 3 [Enter]
5 .MO 17) TD-DFT TD-HF) 6)1 7) HF DFT ) Calculate the excitation energy by TD-DFTTD-HF) TD-DFT TD-HF) Enter 2 Enter Select the TD approximation TD-DFT TD-HF) RPA Enter TDA 2 Enter Define the number of excited states Enter Enter SCF RHF Select target state, 1 Enter 3 2 Enter 18) Mulliken Calculate the Mulliken populations Mulliken Mulliken Enter Mulliken 2 Enter 19) Calculate the electrostatic potential Enter 2 Enter 20) Perform the parallel calculations Enter 2 Enter Define number of nodes used for parallel calculations Enter : [Enter] 21)
6 Define the job time limit in hh:mm:ss 1 Enter hh:mm:ss Enter : :30:00 [Enter]
7 NTChem NTChem ntprep inp.bash pjsub : ntprep NTChem h2o_dft_opt.bash $ pjsub h2o_dft_opt.bash NTChem NTChem ntprep inp.cshjsub : ntprep NTChem h2o_dft_opt.csh ccpg $ jsub q PF h2o_dft_opt.csh NTChem.out
8 ab initio Molecular Dynamics) gradient RATTLE Nosé Hoover Tully ) Geom Velo Velo Velo Vx1 Vy1 Vz1 VxN VyN VzN End Vx Vy Vz x y z &AIMD VelUnit = 'AngFs' Angstrom/Femto second NormMod Geom Classical Quantum &AIMD VibSample Classical Quantum ) 0 ) &AIMD RotSample =.TRUE. 1 1/2kT k:
9 T : &AIMD Kelvin )) Molecule1 Molecule2 Geom Molecule1 Molecule2 Geom!--- Molecule 1 Symbol1 X1 Y1 Z1 SymbolN XN YN ZN!--- Molecule 2 SymbolN+1) XN+1) YN+1) ZN+1) SymbolN+M) XN+M) YN+M) ZN+M) End Symbol X Y Z x y z Molecule1 Molecule2 &AIMD NAtom1=N NAtom2=M 2 &AIMD CollSample =.TRUE. Molecule1 Molecule2 &AIMD Distance 2 &AIMD ImpParam 2kT T &AIMD Kelvin Molecule1 Molecule2 Z &AIMD OriSample =.TRUE. Molecule1 Molecule2 &AIMD VibSample RotSample NormMod Geom Molecule1 Molecul2 NormMod Prefix Name &AIMD Name1 = 'Name1' Name2 = 'Name2'
10 &AIMD Kelvin MD AIMD AIMD AIMD &AIMD Restart =.TRUE. Nosé Hoover chain &AIMD Ensemble &AIMD Kelvin RATTLE MD H X ) RATTLE &AIMD RATTLE =.TRUE. RATTLE RATTLE Num1 NumN End Num2 NumM Num Geom ) NYI) Tully &AIMD SurfHopt =.TRUE. Gradient StateNum
11 Molecular Mechanics) Tinker &Tinker2NT Param TinXYZ TinXYZ Tinker XYZ TinXYZ 1 Symbol X1 Y1 Z1 Param1 Bond1-1 Bond1-2 Bond1-3 N Symbol XN YN ZN ParamN BondN-1 BondN-2 BondN-3 End Symbol X Y Z x y z Param MM Bond Geom TinXYZ Key Prefix Name
12 QM/MM Quantum Mechanics QM) Molecular Mechanics MM) 1 QM MM van der Waals Molecular Mechanics MM) QM MM QM/MM 3 Name.inp Name.QM.inp Name.MM.inp QM MM QM/MM Geom &QMMM NameQM NameMM QM MM Prefix Name QM MM van der Waals TinXYZ &QMMM Param van der Waals TinXYZ Tinker XYZ TinXYZ 1 Symbol1 X1 Y1 Z1 Param1 Bond1-1 Bond1-2 Bond1-3 N SymbolN XN YN ZN ParamN BondN-1 BondN-2 BondN-3 N+1 SymbolN+1) XN+1) YN+1) ZN+1) ParamN+1) BondN+1)-1 BondN+1)-2 BondN+1)-3 N+M SymbolN+M) XN+M) YN+M) ZN+M) ParamN+M) BondN+M)-1 BondN+M)-2 BondN+M)-3 End Symbol X Y Z x y z Param MM Bond Geom TinXYZ QM QM Geom Geom_ONIOM T
13 QM MM Ch &MDInt1 CalChg =.TRUE. QM Geom_ONIOM!--- QM Symbol1 X1 Y1 Z1 T SymbolN XN YN ZN T!--- MM Ch XN+1) YN+1) ZN+1) ChargeN+1) Ch XN+M) YN+M) ZN+M) ChargeN+M) End X Y Z CX CY CZ Charge MM MM Geom Geom_ONIOM T MM TinXYZ MM MM Geom_ONIOM!--- QM Symbol1 X1 Y1 Z1 SymbolN XN YN ZN!--- MM SymbolN+1) XN+1) YN+1) ZN+1) T SymbolN+M) XN+M) YN+M) ZN+M) T End TinXYZ!--- QM
14 ! 1 Symbol1 X1 Y1 Z1 Param1 Bond1-1 Bond1-2 Bond1-3! N SymbolN XN YN ZN ParamN BondN-1 BondN-2 BondN-3!--- MM N+1 SymbolN+1) XN+1) YN+1) ZN+1) ParamN+1) BondN+1)-1 BondN+1)-2 BondN+1)-3 N+M SymbolN+M) XN+M) YN+M) ZN+M) ParamN+M) BondN+M)-1 BondN+M)-2 BondN+M)-3 End
15 ONIOM NTChem 2-layer ONIOM Molecular Mechanics ONIOM 4 Name.inp NameLR.inp NameLM.inp NameHM.inp Low-Real Low-Model High-Model ONIOM Geom &ONIOM 3 Prefix Name NameLR = NameLR NameLM = NameLM NameHM = NameHM Low-Real Prefix Name Low-Model Prefix Name High-Real Prefix Name Low-Real Low-Real ONIOM Geom Low-Model Low-Model Geom Geom_ONIOM T Model Model Geom T Geom_ONIOM Symbol1 X1 Y1 Z1 SymbolN XN YN ZN
16 !--- Model part SymbolN+1) XN+1) YN+1) ZN+1) T CapN+1) ScaleN+1) SymbolN+M) XN+M) YN+M) ZN+M) T CapN+M) ScaleN+M) End Molecular Mechanics TinXYZ Model C 5 H 8 2 Model 2 Geom_ONIOM C T C H T C H H H H C H C T H H T End TinXYZ 1 C ! 2 C H ! 4 C ! 5 H ! 6 H
17 ! 7 H ! 8 H ! 9 C !10 H C !12 H H ! Cap atom 2 H H End High-Model High-Model Low-Model
18 DL-Find Nudged Elastic Band Dimer Geom &DLFind Coord.xyz Cartesian FixGeom CoordType = 'Internal') FixGeom FixGeom!--- Fix bond length atom1 - atom2 atom1 atom2!--- Fix bond angle atom3 - atom4 - atom5 atom3 atom4 atom5!--- Fix dihedral angle atom6 - atom7 - atom8 - atom9 atom6 atom7 atom8 atom9 End Cartesian FixXYZ Cartesian Cartesian CoordType = 'XYZ') FixXYZ "X" "Y" "Z" FixXYZ!--- Fix X coordinate of atom1 atom1 X!--- Fix X and Y coordinate of atom2 atom2 XY!--- Fix X, Y and Z coordinate of atom3
19 atom3 End XYZ Hessian gradient Hessian Nudged Elastic Band NEB) NEB NEB 2 Name.inp Neme.2nd.inp Name.inp Geom &DLFind NEB =.TRUE. Prefix Name OtherGeom = 'Name.2nd' Geom NEB nebpath.xyz NEB nebinfo Image XYZ XYZ Image &DLFind NEBPath NInitImg NInitImg NImage - 1 Image Image Image Image Image Dimer Dimer &DLFind Dimer =.TRUE.
20 &DLFind Dump dlf_*.chk) &DLFind Restart =.TRUE.
21 Tin2QMMM ' ' version2015/3/2 Tin2QMMM Tin2QMMM.exe ' QM/MM,ONIOM NTChem TinkerXMolXYZ MD QM NTChem QM/MM
22 QM/MM QM/MM ' =QM+MM QM MM MM QM QM MM QM MM NTChemQM/MM QM/MM link'atom ONIOM ONIOM ' linkatom Real Model NTChemlinkatom C C link'atom 1
23 NTChem MMTinker' version6.2 Tinker2NT.exe Tinker ' Tinker' Tinker.xyz.Mn!!1362! "!!!!!1!!!!C!!!!!' !!!! !!!! !!!!!174!!!!!!!!2!!!!!3!!!!!4!!!!!!2!!!!HC!!!!' !!!! !!!! !!!!!194!!!!!!!!1!!!!!!3!!!!O!!!!!' !!!! !!!' !!!!!175!!!!!!!!1!!!!!!4!!!!N!!!!!! !!!! !!!! !!!!!176!!!!!!!!1!!!!!5!!!!!6!!!!!!5!!!!H!!!!!! !!!' !!!' !!!!!179!!!!!!!!4!!!!!!6!!!!H!!!!!! !!!! !!!! !!!!!179!!!!!!!!4!!!!!!7!!!!OW!!!!' !!!' !!!' !!!!!!53!!!!!!!!8!!!!!9!!!!!!8!!!!HW!!!!' !!!' !!!' !!!!!!54!!!!!!!!7!!!!!!9!!!!HW!!!!' !!!' !!!' !!!!!!54!!!!!!!!7!! )")"""X)"""""Y)""""""Z)""" )""")!!!!!1362!!!!HW!!!!! !!!! !!!! !!!!!!54!!!!!1360! Tinker' Tinker.prm
24 ' XMolXYZfileformat.xyz!!1362! " )"!C!!!!!!!' !!!! !!!! !!H!!!!!!!' !!!! !!!! !!O!!!!!!!' !!!! !!!' !!N!!!!!!!! !!!! !!!! !!H!!!!!!!! !!!' !!!' !!H!!!!!!!! !!!! !!!! !!O!!!!!!!' !!!' !!!' !!H!!!!!!!' !!!' !!!' !!H!!!!!!!' !!!' !!!' !!O!!!!!!!' !!!' !!!' !!:! )"""X)""""""Y)"""""Z)"!:!!H!!!!!!!! !!!! !!!! QM/MM ' ${Name}.inp ${Name}.QM.inp QM ${Name}.MM.inp MM 3 ONIOMQM/MM) ${Name}.inp ${Name}.LR.inp LowVReal ${Name}.LM.inp LowVModel ${Name}.HM.inp HighVModel 4
25 NTPrep Tin2QMMM ' &Control Name= alice,ncoreperio=8, / &QMMM' 'NameQM= alice.qm,' 'NameMM= alice.mm,' /' Geom' ) : : End' &Control Name= alice.qm,/ &SCF' )' /' Basis' ) End Geom_ONIOM' ) : : End' ' &Control Name= alice.mm,/ &Tinker' )' /' Geom_ONIOM' ) : : End' TinXYZ' ) End Tinker' Tinker ' NTPrep Tinker Tin2QMMM ' Tin2QMMM.exe
26 1 QM/MM ' MM MM QM QM/MM 1) ' Tinker formamide+water.mn XMol formamide+water.xmol.xyz MM oplsaa.prm
27 QM/MM 2) ' Tin2QMMM.exe Tin2QMMM.exe QM/MM 2' ' Tinker' 1362! ) *) " 16QM "! QM! "! >$!${NTChem}/util/Tin2QMMM.exe" Select!calculation!type![!1:ONIOMQM/MM)!2:QM/MM!]!>>!2" Enter!tinker!param!file!>>!oplsaa.prm" Enter!tinker!input!file!>>!formamide+water.tin" NAtom!=!1362!!!!!! NFrag!=!453!!!!!!!>>!Press!enter!to!continue!Press!Y!to!show!fragment!information)! Enter )"!Input!QM!atom!list:!!!!!Enter!''I!J'!to!set!sequential!atoms!from!I!to!J!!!!!Enter!'0'!to!terminate!input! E1"6"0!!!!!!!!Accept!this!QM/MM!division!?!Y:!Accept!/!Q:!Quit!/!other:!Redo)! y!!!!!!! Enter!output!base!name!>>!exam01!!!!!! >$!ls" DLFindFixXYZ.txt!exam01.QMMM.MM"exam01.QMMM.QM"exam01.QMMM.QM.xyz! formamide+water.tin!formamide+water.xmol.xyz!oplsaa.prm" " *) 1'2'3'4 ' I J TI'J '0 QM/MM 3) ' exam01.qmmm.mm" exam01.qmmm.qm" exam01.qmmm.qm.xyz" DLFindFixXYZ.txt" '
28 2 ONIOMQM/MM) PDB 1L2Y)' Model Real ONIOMQM/MM) 1) ' Tinker TrCage_1L2Y.Mn XMol TrCage_1L2Y.xmol.xyz MM oplsaa.prm
29 ONIOMQM/MM) 2) ' Tin2QMMM.exe Tin2QMMM.exe >$!${NTChem}/util/Tin2QMMM.exe" ONIOMQM/MM) 1 Select!calculation!type![!1:ONIOMQM/MM)!2:QM/MM!]!>>!1" Enter!tinker!param!file!>>!oplsaa.prm" Tinker Enter!tinker!input!file!>>!TrCage_1L2Y.tin" 304! NAtom!=!304! 1) NFrag!=!!!1!!!!!!!!!!!!>>!Press!enter!to!continue!Press!Y!to!show!fragment!information)! Enter )"!Input!QM!atom!list:!!!!!Enter!''I!J'!to!set!sequential!atoms!from!I!to!J!!!!!Enter!'0'!to!terminate!input! 99116QM! E99"116"0!!!!!!!!!*!Caution!!Fragment!!!!!!!!!!!!1!!has!partial!QM!attribute!!!!!!Fragment!!!!!!!!!!!!1! Fragment1)" 99! QM'MM!connect!!!!!99[CT!!!!]!'!!!!!!94[CT!!!!]! 94!!Accept!this!QM/MM!division!?!Y:!Accept!/!Q:!Quit!/!other:!Redo)! QM! y!!!!!!!! Enter!output!base!name!>>!exam02!!!!! link!atom!!notice!:!please!change!hydrogen!atom!type!manually!!! >$!ls" DLFindFixXYZ.txt"TrCage_1L2Y.tin!!TrCage_1L2Y.xmol.xyz!exam02.ONIOM.Gl" exam02.oniom.hm"exam02.oniom.hm.model.xyz"exam02.oniom.lm"exam02.oniom.lr! oplsaa.prm!!! " ONIOMQM/MM)6) ' exam02.oniom.gl" exam02.oniom.hm" exam02.oniom.hm.model.xyz" exam02.oniom.lm" ChangeToH exam02.oniom.lr" DLFindFixXYZ.txt" '
Hints on Using the Orca Program
 Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Hints on Using the Orca Program
Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Hints on Using the Orca Program
Basic introduction of NWChem software
 Basic introduction of NWChem software Background NWChem is part of the Molecular Science Software Suite Designed and developed to be a highly efficient and portable Massively Parallel computational chemistry
Basic introduction of NWChem software Background NWChem is part of the Molecular Science Software Suite Designed and developed to be a highly efficient and portable Massively Parallel computational chemistry
IFM Chemistry Computational Chemistry 2010, 7.5 hp LAB2. Computer laboratory exercise 1 (LAB2): Quantum chemical calculations
 Computer laboratory exercise 1 (LAB2): Quantum chemical calculations Introduction: The objective of the second computer laboratory exercise is to get acquainted with a program for performing quantum chemical
Computer laboratory exercise 1 (LAB2): Quantum chemical calculations Introduction: The objective of the second computer laboratory exercise is to get acquainted with a program for performing quantum chemical
NWChem: Molecular Dynamics and QM/MM
 NWChem: Molecular Dynamics and QM/MM Molecular Dynamics Functionality Target systems: General features: biomolecules (proteins, DNA/RNA, biomembranes) energy evaluation (SP) energy minimization (EM) molecular
NWChem: Molecular Dynamics and QM/MM Molecular Dynamics Functionality Target systems: General features: biomolecules (proteins, DNA/RNA, biomembranes) energy evaluation (SP) energy minimization (EM) molecular
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland
 Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Basic introduction of NWChem software
 Basic introduction of NWChem software Background! NWChem is part of the Molecular Science Software Suite! Designed and developed to be a highly efficient and portable Massively Parallel computational chemistry
Basic introduction of NWChem software Background! NWChem is part of the Molecular Science Software Suite! Designed and developed to be a highly efficient and portable Massively Parallel computational chemistry
Development and Application of Combined Quantum Mechanical and Molecular Mechanical Methods
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Student Research Projects, Dissertations, and Theses - Chemistry Department Chemistry, Department of Fall 12-2014 Development
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Student Research Projects, Dissertations, and Theses - Chemistry Department Chemistry, Department of Fall 12-2014 Development
Chemistry 4560/5560 Molecular Modeling Fall 2014
 Final Exam Name:. User s guide: 1. Read questions carefully and make sure you understand them before answering (if not, ask). 2. Answer only the question that is asked, not a different question. 3. Unless
Final Exam Name:. User s guide: 1. Read questions carefully and make sure you understand them before answering (if not, ask). 2. Answer only the question that is asked, not a different question. 3. Unless
Fragmentation methods
 Fragmentation methods Scaling of QM Methods HF, DFT scale as N 4 MP2 scales as N 5 CC methods scale as N 7 What if we could freeze the value of N regardless of the size of the system? Then each method
Fragmentation methods Scaling of QM Methods HF, DFT scale as N 4 MP2 scales as N 5 CC methods scale as N 7 What if we could freeze the value of N regardless of the size of the system? Then each method
Jack Smith. Center for Environmental, Geotechnical and Applied Science. Marshall University
 Jack Smith Center for Environmental, Geotechnical and Applied Science Marshall University -- Division of Science and Research WV Higher Education Policy Commission WVU HPC Summer Institute June 20, 2014
Jack Smith Center for Environmental, Geotechnical and Applied Science Marshall University -- Division of Science and Research WV Higher Education Policy Commission WVU HPC Summer Institute June 20, 2014
Tools for QM studies of large systems
 Tools for QM studies of large systems Automated, hessian-free saddle point search & characterization QM/MM implementation for zeolites Shaama Mallikarjun Sharada Advisors: Prof. Alexis T Bell, Prof. Martin
Tools for QM studies of large systems Automated, hessian-free saddle point search & characterization QM/MM implementation for zeolites Shaama Mallikarjun Sharada Advisors: Prof. Alexis T Bell, Prof. Martin
Modeling Ultrafast Deactivation in Oligothiophenes via Nonadiabatic Dynamics
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 Supplementary Data for Modeling Ultrafast Deactivation in Oligothiophenes via Nonadiabatic
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2015 Supplementary Data for Modeling Ultrafast Deactivation in Oligothiophenes via Nonadiabatic
DFT calculations of NMR indirect spin spin coupling constants
 DFT calculations of NMR indirect spin spin coupling constants Dalton program system Program capabilities Density functional theory Kohn Sham theory LDA, GGA and hybrid theories Indirect NMR spin spin coupling
DFT calculations of NMR indirect spin spin coupling constants Dalton program system Program capabilities Density functional theory Kohn Sham theory LDA, GGA and hybrid theories Indirect NMR spin spin coupling
Introduction to Computational Chemistry for Experimental Chemists... (Part 2/2)
 12 th PhD seminar, Garching, October 31 st 2008 Introduction to Computational Chemistry for Experimental Chemists... (Part 2/2) Dr. Markus Drees, TU München Universität Regensburg Universität Augsburg
12 th PhD seminar, Garching, October 31 st 2008 Introduction to Computational Chemistry for Experimental Chemists... (Part 2/2) Dr. Markus Drees, TU München Universität Regensburg Universität Augsburg
Computational Chemistry Using the University of Alaska WebMO Site
 2/7/2017 1 Computational Chemistry Using the University of Alaska WebMO Site John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Intro and operation of WebMO and MOPAC Basic
2/7/2017 1 Computational Chemistry Using the University of Alaska WebMO Site John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Intro and operation of WebMO and MOPAC Basic
Electric properties of molecules
 Electric properties of molecules For a molecule in a uniform electric fielde the Hamiltonian has the form: Ĥ(E) = Ĥ + E ˆµ x where we assume that the field is directed along the x axis and ˆµ x is the
Electric properties of molecules For a molecule in a uniform electric fielde the Hamiltonian has the form: Ĥ(E) = Ĥ + E ˆµ x where we assume that the field is directed along the x axis and ˆµ x is the
Multiscale Materials Modeling
 Multiscale Materials Modeling Lecture 09 Quantum Mechanics/Molecular Mechanics (QM/MM) Techniques Fundamentals of Sustainable Technology These notes created by David Keffer, University of Tennessee, Knoxville,
Multiscale Materials Modeling Lecture 09 Quantum Mechanics/Molecular Mechanics (QM/MM) Techniques Fundamentals of Sustainable Technology These notes created by David Keffer, University of Tennessee, Knoxville,
Exercise 1: Structure and dipole moment of a small molecule
 Introduction to computational chemistry Exercise 1: Structure and dipole moment of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the dipole moment of a small
Introduction to computational chemistry Exercise 1: Structure and dipole moment of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the dipole moment of a small
Session 1. Introduction to Computational Chemistry. Computational (chemistry education) and/or (Computational chemistry) education
 Session 1 Introduction to Computational Chemistry 1 Introduction to Computational Chemistry Computational (chemistry education) and/or (Computational chemistry) education First one: Use computational tools
Session 1 Introduction to Computational Chemistry 1 Introduction to Computational Chemistry Computational (chemistry education) and/or (Computational chemistry) education First one: Use computational tools
Using Web-Based Computations in Organic Chemistry
 10/30/2017 1 Using Web-Based Computations in Organic Chemistry John Keller UAF Department of Chemistry & Biochemistry The UAF WebMO site Practical aspects of computational chemistry theory and nomenclature
10/30/2017 1 Using Web-Based Computations in Organic Chemistry John Keller UAF Department of Chemistry & Biochemistry The UAF WebMO site Practical aspects of computational chemistry theory and nomenclature
XYZ file format Protein Data Bank (pdb) file format Z - matrix
 Chemistry block (exercise 1) In this exercise, students will be introduced how to preform simple quantum chemical calculations. Input files for Gaussian09. Output file structure. Geometry optimization,
Chemistry block (exercise 1) In this exercise, students will be introduced how to preform simple quantum chemical calculations. Input files for Gaussian09. Output file structure. Geometry optimization,
LUMO + 1 LUMO. Tómas Arnar Guðmundsson Report 2 Reikniefnafræði G
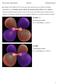 Q1: Display all the MOs for N2 in your report and classify each one of them as bonding, antibonding or non-bonding, and say whether the symmetry of the orbital is σ or π. Sketch a molecular orbital diagram
Q1: Display all the MOs for N2 in your report and classify each one of them as bonding, antibonding or non-bonding, and say whether the symmetry of the orbital is σ or π. Sketch a molecular orbital diagram
QUANTUM CHEMISTRY FOR TRANSITION METALS
 QUANTUM CHEMISTRY FOR TRANSITION METALS Outline I Introduction II Correlation Static correlation effects MC methods DFT III Relativity Generalities From 4 to 1 components Effective core potential Outline
QUANTUM CHEMISTRY FOR TRANSITION METALS Outline I Introduction II Correlation Static correlation effects MC methods DFT III Relativity Generalities From 4 to 1 components Effective core potential Outline
Electronic Supplementary Material (ESI) for Chemical Science This journal is The Royal Society of Chemistry 2012
 Fig. S1 CASSCF (13,10) active space orbitals with Ru-Ru distance of 2.4 Å. Occupation numbers are on the left and energies in Hartrees are on the right of each orbital. The δ orbital is also included here,
Fig. S1 CASSCF (13,10) active space orbitals with Ru-Ru distance of 2.4 Å. Occupation numbers are on the left and energies in Hartrees are on the right of each orbital. The δ orbital is also included here,
Study of Ozone in Tribhuvan University, Kathmandu, Nepal. Prof. S. Gurung Central Department of Physics, Tribhuvan University, Kathmandu, Nepal
 Study of Ozone in Tribhuvan University, Kathmandu, Nepal Prof. S. Gurung Central Department of Physics, Tribhuvan University, Kathmandu, Nepal 1 Country of the Mt Everest 2 View of the Mt Everest 3 4 5
Study of Ozone in Tribhuvan University, Kathmandu, Nepal Prof. S. Gurung Central Department of Physics, Tribhuvan University, Kathmandu, Nepal 1 Country of the Mt Everest 2 View of the Mt Everest 3 4 5
计算物理作业二. Excercise 1: Illustration of the convergence of the dissociation energy for H 2 toward HF limit.
 计算物理作业二 Excercise 1: Illustration of the convergence of the dissociation energy for H 2 toward HF limit. In this exercise, basis indicates one of the following basis sets: STO-3G, cc-pvdz, cc-pvtz, cc-pvqz
计算物理作业二 Excercise 1: Illustration of the convergence of the dissociation energy for H 2 toward HF limit. In this exercise, basis indicates one of the following basis sets: STO-3G, cc-pvdz, cc-pvtz, cc-pvqz
CHEM6085: Density Functional Theory Lecture 10
 CHEM6085: Density Functional Theory Lecture 10 1) Spin-polarised calculations 2) Geometry optimisation C.-K. Skylaris 1 Unpaired electrons So far we have developed Kohn-Sham DFT for the case of paired
CHEM6085: Density Functional Theory Lecture 10 1) Spin-polarised calculations 2) Geometry optimisation C.-K. Skylaris 1 Unpaired electrons So far we have developed Kohn-Sham DFT for the case of paired
Exchange Correlation Functional Investigation of RT-TDDFT on a Sodium Chloride. Dimer. Philip Straughn
 Exchange Correlation Functional Investigation of RT-TDDFT on a Sodium Chloride Dimer Philip Straughn Abstract Charge transfer between Na and Cl ions is an important problem in physical chemistry. However,
Exchange Correlation Functional Investigation of RT-TDDFT on a Sodium Chloride Dimer Philip Straughn Abstract Charge transfer between Na and Cl ions is an important problem in physical chemistry. However,
Oslo node. Highly accurate calculations benchmarking and extrapolations
 Oslo node Highly accurate calculations benchmarking and extrapolations Torgeir Ruden, with A. Halkier, P. Jørgensen, J. Olsen, W. Klopper, J. Gauss, P. Taylor Explicitly correlated methods Pål Dahle, collaboration
Oslo node Highly accurate calculations benchmarking and extrapolations Torgeir Ruden, with A. Halkier, P. Jørgensen, J. Olsen, W. Klopper, J. Gauss, P. Taylor Explicitly correlated methods Pål Dahle, collaboration
The Basics of Theoretical and Computational Chemistry
 Bernd M. Rode, Thomas S. Hofer, and Michael D. Kugler The Basics of Theoretical and Computational Chemistry BICENTENNIA BICENTBNN I AL. WILEY-VCH Verlag GmbH & Co. KGaA V Contents Preface IX 1 Introduction
Bernd M. Rode, Thomas S. Hofer, and Michael D. Kugler The Basics of Theoretical and Computational Chemistry BICENTENNIA BICENTBNN I AL. WILEY-VCH Verlag GmbH & Co. KGaA V Contents Preface IX 1 Introduction
Figure 1: Transition State, Saddle Point, Reaction Pathway
 Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Potential Energy Surfaces
Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Potential Energy Surfaces
List of Figures Page Figure No. Figure Caption No. Figure 1.1.
 List of Figures Figure No. Figure Caption Page No. Figure 1.1. Cation- interactions and their modulations. 4 Figure 1.2. Three conformations of benzene dimer, S is not a minimum on the potential energy
List of Figures Figure No. Figure Caption Page No. Figure 1.1. Cation- interactions and their modulations. 4 Figure 1.2. Three conformations of benzene dimer, S is not a minimum on the potential energy
Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations
 Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations Alexandre V. Morozov, Tanja Kortemme, Kiril Tsemekhman, David Baker
Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations Alexandre V. Morozov, Tanja Kortemme, Kiril Tsemekhman, David Baker
Lec20 Fri 3mar17
 564-17 Lec20 Fri 3mar17 [PDF]GAUSSIAN 09W TUTORIAL www.molcalx.com.cn/wp-content/uploads/2015/01/gaussian09w_tutorial.pdf by A Tomberg - Cited by 8 - Related articles GAUSSIAN 09W TUTORIAL. AN INTRODUCTION
564-17 Lec20 Fri 3mar17 [PDF]GAUSSIAN 09W TUTORIAL www.molcalx.com.cn/wp-content/uploads/2015/01/gaussian09w_tutorial.pdf by A Tomberg - Cited by 8 - Related articles GAUSSIAN 09W TUTORIAL. AN INTRODUCTION
Q-Chem 5: Facilitating Worldwide Scientific Breakthroughs
 Q-Chem 5: Facilitating Worldwide Scientific Breakthroughs Founded in 1993, Q-Chem strives to bring its customers state-ofthe-art methods and algorithms for performing quantum chemistry calculations. Cutting-edge
Q-Chem 5: Facilitating Worldwide Scientific Breakthroughs Founded in 1993, Q-Chem strives to bring its customers state-ofthe-art methods and algorithms for performing quantum chemistry calculations. Cutting-edge
Computational Modeling of Protein-Ligand Interactions
 Computational Modeling of Protein-Ligand Interactions Steven R. Gwaltney Department of Chemistry Mississippi State University Mississippi State, MS 39762 Auguste Comte, 1830 Every attempt to refer chemical
Computational Modeling of Protein-Ligand Interactions Steven R. Gwaltney Department of Chemistry Mississippi State University Mississippi State, MS 39762 Auguste Comte, 1830 Every attempt to refer chemical
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Free Energy Simulation Methods
 Free Energy Simulation Methods Free energy simulation methods Many methods have been developed to compute (relative) free energies on the basis of statistical mechanics Free energy perturbation Thermodynamic
Free Energy Simulation Methods Free energy simulation methods Many methods have been developed to compute (relative) free energies on the basis of statistical mechanics Free energy perturbation Thermodynamic
One-sided Communication Implementation in FMO Method
 One-sided Communication mplementation in FMO Method J. Maki, Y. nadomi, T. Takami, R. Susukita, H. Honda, J. Ooba, T. obayashi, R. Nogita,. noue and M. Aoyagi Computing and Communications Center, yushu
One-sided Communication mplementation in FMO Method J. Maki, Y. nadomi, T. Takami, R. Susukita, H. Honda, J. Ooba, T. obayashi, R. Nogita,. noue and M. Aoyagi Computing and Communications Center, yushu
NH 3 inversion: Potential energy surfaces and transition states CH342L March 28, 2016
 N 3 inversion: Potential energy surfaces and transition states C342L March 28, 2016 Last week, we used the IR spectrum of ammonia to determine the splitting of energy levels due to inversion of the umbrella
N 3 inversion: Potential energy surfaces and transition states C342L March 28, 2016 Last week, we used the IR spectrum of ammonia to determine the splitting of energy levels due to inversion of the umbrella
Electronic structure theory: Fundamentals to frontiers. 1. Hartree-Fock theory
 Electronic structure theory: Fundamentals to frontiers. 1. Hartree-Fock theory MARTIN HEAD-GORDON, Department of Chemistry, University of California, and Chemical Sciences Division, Lawrence Berkeley National
Electronic structure theory: Fundamentals to frontiers. 1. Hartree-Fock theory MARTIN HEAD-GORDON, Department of Chemistry, University of California, and Chemical Sciences Division, Lawrence Berkeley National
Computational Chemistry I
 Computational Chemistry I Text book Cramer: Essentials of Quantum Chemistry, Wiley (2 ed.) Chapter 3. Post Hartree-Fock methods (Cramer: chapter 7) There are many ways to improve the HF method. Most of
Computational Chemistry I Text book Cramer: Essentials of Quantum Chemistry, Wiley (2 ed.) Chapter 3. Post Hartree-Fock methods (Cramer: chapter 7) There are many ways to improve the HF method. Most of
Same idea for polyatomics, keep track of identical atom e.g. NH 3 consider only valence electrons F(2s,2p) H(1s)
 XIII 63 Polyatomic bonding -09 -mod, Notes (13) Engel 16-17 Balance: nuclear repulsion, positive e-n attraction, neg. united atom AO ε i applies to all bonding, just more nuclei repulsion biggest at low
XIII 63 Polyatomic bonding -09 -mod, Notes (13) Engel 16-17 Balance: nuclear repulsion, positive e-n attraction, neg. united atom AO ε i applies to all bonding, just more nuclei repulsion biggest at low
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/NCHEM.1677 Entangled quantum electronic wavefunctions of the Mn 4 CaO 5 cluster in photosystem II Yuki Kurashige 1 *, Garnet Kin-Lic Chan 2, Takeshi Yanai 1 1 Department of Theoretical and
DOI: 10.1038/NCHEM.1677 Entangled quantum electronic wavefunctions of the Mn 4 CaO 5 cluster in photosystem II Yuki Kurashige 1 *, Garnet Kin-Lic Chan 2, Takeshi Yanai 1 1 Department of Theoretical and
Computational Methods. Chem 561
 Computational Methods Chem 561 Lecture Outline 1. Ab initio methods a) HF SCF b) Post-HF methods 2. Density Functional Theory 3. Semiempirical methods 4. Molecular Mechanics Computational Chemistry " Computational
Computational Methods Chem 561 Lecture Outline 1. Ab initio methods a) HF SCF b) Post-HF methods 2. Density Functional Theory 3. Semiempirical methods 4. Molecular Mechanics Computational Chemistry " Computational
Calculating Vibrational Spectra from Molecular Dynamics
 Calculating Vibrational Spectra from Molecular Dynamics A Simulating a Trajectory with Wannier Centers To calculate IR spectra from Molecular Dynamics, it is necessary to have dipole information for the
Calculating Vibrational Spectra from Molecular Dynamics A Simulating a Trajectory with Wannier Centers To calculate IR spectra from Molecular Dynamics, it is necessary to have dipole information for the
Tutorial I: IQ MOL and Basic DFT and MP2 Calculations 1 / 30
 Tutorial I: IQ MOL and Basic DFT and MP2 Calculations Q-Chem User Workshop, Denver March 21, 2015 1 / 30 2 / 30 Introduction to IQMOL DFT and MP2 Calculations 3 / 30 IQMOL and Q-CHEM IQMOL is an open-source
Tutorial I: IQ MOL and Basic DFT and MP2 Calculations Q-Chem User Workshop, Denver March 21, 2015 1 / 30 2 / 30 Introduction to IQMOL DFT and MP2 Calculations 3 / 30 IQMOL and Q-CHEM IQMOL is an open-source
John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks
 10/15/2016 1 WebMO & Gaussian John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Corrections and updates 9-5-2017 SCHEDULE 9-10 Intro and basic operation of WebMO and MOPAC
10/15/2016 1 WebMO & Gaussian John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Corrections and updates 9-5-2017 SCHEDULE 9-10 Intro and basic operation of WebMO and MOPAC
Introduction to Hartree-Fock calculations using ORCA and Chemcraft
 Introduction to Hartree-Fock calculations using ORCA and Chemcraft In this exercise, you will get to use software for carrying out calculations of wavefunctions for molecules, the ORCA program. While ORCA
Introduction to Hartree-Fock calculations using ORCA and Chemcraft In this exercise, you will get to use software for carrying out calculations of wavefunctions for molecules, the ORCA program. While ORCA
Answers Quantum Chemistry NWI-MOL406 G. C. Groenenboom and G. A. de Wijs, HG00.307, 8:30-11:30, 21 jan 2014
 Answers Quantum Chemistry NWI-MOL406 G. C. Groenenboom and G. A. de Wijs, HG00.307, 8:30-11:30, 21 jan 2014 Question 1: Basis sets Consider the split valence SV3-21G one electron basis set for formaldehyde
Answers Quantum Chemistry NWI-MOL406 G. C. Groenenboom and G. A. de Wijs, HG00.307, 8:30-11:30, 21 jan 2014 Question 1: Basis sets Consider the split valence SV3-21G one electron basis set for formaldehyde
Neural Network Potentials
 Neural Network Potentials ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost Smith, Isayev and Roitberg Sabri Eyuboglu February 6, 2018 What are Neural Network
Neural Network Potentials ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost Smith, Isayev and Roitberg Sabri Eyuboglu February 6, 2018 What are Neural Network
Chapter: 22. Visualization: Making INPUT File and Processing of Output Results
 Chapter: 22 Visualization: Making INPUT File and Processing of Output Results Keywords: visualization, input and output structure, molecular orbital, electron density. In the previous chapters, we have
Chapter: 22 Visualization: Making INPUT File and Processing of Output Results Keywords: visualization, input and output structure, molecular orbital, electron density. In the previous chapters, we have
Introduction to computational chemistry Exercise I: Structure and electronic energy of a small molecule. Vesa Hänninen
 Introduction to computational chemistry Exercise I: Structure and electronic energy of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the electronic energy
Introduction to computational chemistry Exercise I: Structure and electronic energy of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the electronic energy
74 these states cannot be reliably obtained from experiments. In addition, the barriers between the local minima can also not be obtained reliably fro
 73 Chapter 5 Development of Adiabatic Force Field for Polyvinyl Chloride (PVC) and Chlorinated PVC (CPVC) 5.1 Introduction Chlorinated polyvinyl chloride has become an important specialty polymer due to
73 Chapter 5 Development of Adiabatic Force Field for Polyvinyl Chloride (PVC) and Chlorinated PVC (CPVC) 5.1 Introduction Chlorinated polyvinyl chloride has become an important specialty polymer due to
Density Functional Theory
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard October 28, 2015 Density Functional Theory What is a Functional? A functional is a general mathematical quantity that represents a rule to convert a function
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard October 28, 2015 Density Functional Theory What is a Functional? A functional is a general mathematical quantity that represents a rule to convert a function
MO Calculation for a Diatomic Molecule. /4 0 ) i=1 j>i (1/r ij )
 MO Calculation for a Diatomic Molecule Introduction The properties of any molecular system can in principle be found by looking at the solutions to the corresponding time independent Schrodinger equation
MO Calculation for a Diatomic Molecule Introduction The properties of any molecular system can in principle be found by looking at the solutions to the corresponding time independent Schrodinger equation
Quantum Chemistry. NC State University. Lecture 5. The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy
 Quantum Chemistry Lecture 5 The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy NC State University 3.5 Selective absorption and emission by atmospheric gases (source:
Quantum Chemistry Lecture 5 The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy NC State University 3.5 Selective absorption and emission by atmospheric gases (source:
Due: since the calculation takes longer than before, we ll make it due on 02/05/2016, Friday
 Homework 3 Due: since the calculation takes longer than before, we ll make it due on 02/05/2016, Friday Email to: jqian@caltech.edu Introduction In this assignment, you will be using a commercial periodic
Homework 3 Due: since the calculation takes longer than before, we ll make it due on 02/05/2016, Friday Email to: jqian@caltech.edu Introduction In this assignment, you will be using a commercial periodic
3: Many electrons. Orbital symmetries. l =2 1. m l
 3: Many electrons Orbital symmetries Atomic orbitals are labelled according to the principal quantum number, n, and the orbital angular momentum quantum number, l. Electrons in a diatomic molecule experience
3: Many electrons Orbital symmetries Atomic orbitals are labelled according to the principal quantum number, n, and the orbital angular momentum quantum number, l. Electrons in a diatomic molecule experience
XMVB 3.0 A Right Way To Do Valence Bond Calculations. Zhenhua Chen Xiamen University
 XMVB 3.0 A Right Way To Do Valence Bond Calculations Zhenhua Chen Xiamen University XMVB 3.0 Xiamen Valence Bond An Ab Initio Non-orthogonal Valence Bond Program Version 3.0 Lingchun Song, Zhenhua Chen,
XMVB 3.0 A Right Way To Do Valence Bond Calculations Zhenhua Chen Xiamen University XMVB 3.0 Xiamen Valence Bond An Ab Initio Non-orthogonal Valence Bond Program Version 3.0 Lingchun Song, Zhenhua Chen,
Introduction to Ab Initio Quantum Chemical Computation
 c:\374-17\computation\computation17.doc for 9mar17 Prof. Patrik Callis 8mar17 Introduction to Ab Initio Quantum Chemical Computation Purpose: 1. To become acquainted with basic concepts of ab initio quantum
c:\374-17\computation\computation17.doc for 9mar17 Prof. Patrik Callis 8mar17 Introduction to Ab Initio Quantum Chemical Computation Purpose: 1. To become acquainted with basic concepts of ab initio quantum
Q-Chem Workshop Tasks
 Marek Freindorf Q-Chem Workshop Tasks Washington DC August 2009 Basic Calculations Carbon Dioxide, Example 1A 1. Calculate an optimal geometry of carbon dioxide using the B3LYP/6-31+G* level of theory
Marek Freindorf Q-Chem Workshop Tasks Washington DC August 2009 Basic Calculations Carbon Dioxide, Example 1A 1. Calculate an optimal geometry of carbon dioxide using the B3LYP/6-31+G* level of theory
Electron Correlation Methods
 Electron Correlation Methods HF method: electron-electron interaction is replaced by an average interaction E HF c = E 0 E HF E 0 exact ground state energy E HF HF energy for a given basis set HF E c
Electron Correlation Methods HF method: electron-electron interaction is replaced by an average interaction E HF c = E 0 E HF E 0 exact ground state energy E HF HF energy for a given basis set HF E c
CHEM3023: Spins, Atoms and Molecules
 CHEM3023: Spins, Atoms and Molecules Lecture 5 The Hartree-Fock method C.-K. Skylaris Learning outcomes Be able to use the variational principle in quantum calculations Be able to construct Fock operators
CHEM3023: Spins, Atoms and Molecules Lecture 5 The Hartree-Fock method C.-K. Skylaris Learning outcomes Be able to use the variational principle in quantum calculations Be able to construct Fock operators
Solid State Theory: Band Structure Methods
 Solid State Theory: Band Structure Methods Lilia Boeri Wed., 11:15-12:45 HS P3 (PH02112) http://itp.tugraz.at/lv/boeri/ele/ Plan of the Lecture: DFT1+2: Hohenberg-Kohn Theorem and Kohn and Sham equations.
Solid State Theory: Band Structure Methods Lilia Boeri Wed., 11:15-12:45 HS P3 (PH02112) http://itp.tugraz.at/lv/boeri/ele/ Plan of the Lecture: DFT1+2: Hohenberg-Kohn Theorem and Kohn and Sham equations.
Appendix D Simulating Spectroscopic Bands Using Gaussian and PGopher
 429 Appendix D Simulating Spectroscopic Bands Using Gaussian and PGopher This appendix contains methods for using Gaussian 09 121 and PGopher 120 to simulate vibrational and electronic bands of molecules.
429 Appendix D Simulating Spectroscopic Bands Using Gaussian and PGopher This appendix contains methods for using Gaussian 09 121 and PGopher 120 to simulate vibrational and electronic bands of molecules.
From Electrons to Materials Properties
 From Electrons to Materials Properties: DFT for Engineers and Materials Scientists Funk Fachgebiet Werkstoffe des Bauwesens und Bauchemie Mar-17, 2016 From Electrons to Materials Properties Density Functional
From Electrons to Materials Properties: DFT for Engineers and Materials Scientists Funk Fachgebiet Werkstoffe des Bauwesens und Bauchemie Mar-17, 2016 From Electrons to Materials Properties Density Functional
Q-Chem Workshop. Doubletree Hotel 2085 S. Harbor Boulevard Anaheim, CA March 26, Schedule
 Q-Chem Workshop Doubletree Hotel 2085 S. Harbor Boulevard Anaheim, CA 92802 March 26, 2011 1 8:30 Schedule Welcome remarks, Prof. Peter Gill, Australian National Univ and President of Q-Chem 8:45-9:15
Q-Chem Workshop Doubletree Hotel 2085 S. Harbor Boulevard Anaheim, CA 92802 March 26, 2011 1 8:30 Schedule Welcome remarks, Prof. Peter Gill, Australian National Univ and President of Q-Chem 8:45-9:15
Gustavus Adolphus College. Lab #5: Computational Chemistry
 CHE 372 Gustavus Adolphus College Lab #5: Computational Chemistry Introduction In this investigation we will apply the techniques of computational chemistry to several of the molecular systems that we
CHE 372 Gustavus Adolphus College Lab #5: Computational Chemistry Introduction In this investigation we will apply the techniques of computational chemistry to several of the molecular systems that we
Density Func,onal Theory (Chapter 6, Jensen)
 Chem 580: DFT Density Func,onal Theory (Chapter 6, Jensen) Hohenberg- Kohn Theorem (Phys. Rev., 136,B864 (1964)): For molecules with a non degenerate ground state, the ground state molecular energy and
Chem 580: DFT Density Func,onal Theory (Chapter 6, Jensen) Hohenberg- Kohn Theorem (Phys. Rev., 136,B864 (1964)): For molecules with a non degenerate ground state, the ground state molecular energy and
C:\Users\Leonardo\Desktop\README_ELMO_NOTES.txt Montag, 7. Juli :48
 *********** * Input * *********** Besides the normal GAMESS-UK input directives, to perform an ELMO calculation one needs to specify the elmo keyword and to provide additional instructions that are contained
*********** * Input * *********** Besides the normal GAMESS-UK input directives, to perform an ELMO calculation one needs to specify the elmo keyword and to provide additional instructions that are contained
Q-Chem Workshop Examples Part 2
 Marek Freindorf Q-Chem Workshop Examples Part 2 Louisville, KY March 2010 CO, MP2 Calculations Open Avogadro using the "Build" button of QUI Page 2 CO, MP2 Calculations Using Avogadro to create coordinates
Marek Freindorf Q-Chem Workshop Examples Part 2 Louisville, KY March 2010 CO, MP2 Calculations Open Avogadro using the "Build" button of QUI Page 2 CO, MP2 Calculations Using Avogadro to create coordinates
CONTROLLED BY QUANTUM COMPUTING LOGIC GATES
 Quantum QUANTUM Mechanical MECHANICAL Modeling of Self-Reproducible MODELING OF Living SELF-REPRODUCIBLE PNA Chip Immersed in the LIVING Lipid PNA Bilayer CHIP Vesicle IMMERSED Controlled IN by Quantum
Quantum QUANTUM Mechanical MECHANICAL Modeling of Self-Reproducible MODELING OF Living SELF-REPRODUCIBLE PNA Chip Immersed in the LIVING Lipid PNA Bilayer CHIP Vesicle IMMERSED Controlled IN by Quantum
Electronic structure calculations: fundamentals George C. Schatz Northwestern University
 Electronic structure calculations: fundamentals George C. Schatz Northwestern University Electronic Structure (often called Quantum Chemistry) calculations use quantum mechanics to determine the wavefunctions
Electronic structure calculations: fundamentals George C. Schatz Northwestern University Electronic Structure (often called Quantum Chemistry) calculations use quantum mechanics to determine the wavefunctions
Benzene Dimer: dispersion forces and electronic correlation
 Benzene Dimer: dispersion forces and electronic correlation Introduction The Benzene dimer is an ideal example of a system bound by π-π interaction, which is in several cases present in many biologically
Benzene Dimer: dispersion forces and electronic correlation Introduction The Benzene dimer is an ideal example of a system bound by π-π interaction, which is in several cases present in many biologically
 3/23/2010 More basics of DFT Kieron Burke and friends UC Irvine Physics and Chemistry References for ground-state DFT ABC of DFT, by KB and Rudy Magyar, http://dft.uci.edu A Primer in Density Functional
3/23/2010 More basics of DFT Kieron Burke and friends UC Irvine Physics and Chemistry References for ground-state DFT ABC of DFT, by KB and Rudy Magyar, http://dft.uci.edu A Primer in Density Functional
Joint ICTP-IAEA Workshop on Fusion Plasma Modelling using Atomic and Molecular Data January 2012
 2327-3 Joint ICTP-IAEA Workshop on Fusion Plasma Modelling using Atomic and Molecular Data 23-27 January 2012 Qunatum Methods for Plasma-Facing Materials Alain ALLOUCHE Univ.de Provence, Lab.de la Phys.
2327-3 Joint ICTP-IAEA Workshop on Fusion Plasma Modelling using Atomic and Molecular Data 23-27 January 2012 Qunatum Methods for Plasma-Facing Materials Alain ALLOUCHE Univ.de Provence, Lab.de la Phys.
Electron Correlation
 Electron Correlation Levels of QM Theory HΨ=EΨ Born-Oppenheimer approximation Nuclear equation: H n Ψ n =E n Ψ n Electronic equation: H e Ψ e =E e Ψ e Single determinant SCF Semi-empirical methods Correlation
Electron Correlation Levels of QM Theory HΨ=EΨ Born-Oppenheimer approximation Nuclear equation: H n Ψ n =E n Ψ n Electronic equation: H e Ψ e =E e Ψ e Single determinant SCF Semi-empirical methods Correlation
Jaguar DFT Optimizations and Transition State Searches
 Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Symmetry: Translation and Rotation
 Symmetry: Translation and Rotation The sixth column of the C 2v character table indicates the symmetry species for translation along (T) and rotation about (R) the Cartesian axes. y y y C 2 F v (x) T x
Symmetry: Translation and Rotation The sixth column of the C 2v character table indicates the symmetry species for translation along (T) and rotation about (R) the Cartesian axes. y y y C 2 F v (x) T x
9.33 Ab initio Molecular Dynamics Simulations
 9.33 Ab initio Molecular Dynamics Simulations 1 9.33 Ab initio Molecular Dynamics Simulations Recently, it was decided to include an ab initio molecular dynamics (AIMD) module in ORCA. 1 As a plethora
9.33 Ab initio Molecular Dynamics Simulations 1 9.33 Ab initio Molecular Dynamics Simulations Recently, it was decided to include an ab initio molecular dynamics (AIMD) module in ORCA. 1 As a plethora
Semiempirical, empirical and hybrid methods
 Semiempirical, empirical and hybrid methods Gérald MONARD Théorie - Modélisation - Simulation UMR 7565 CNRS - Université de Lorraine Faculté des Sciences - B.P. 7239 5456 Vandœuvre-les-Nancy Cedex - FRANCE
Semiempirical, empirical and hybrid methods Gérald MONARD Théorie - Modélisation - Simulation UMR 7565 CNRS - Université de Lorraine Faculté des Sciences - B.P. 7239 5456 Vandœuvre-les-Nancy Cedex - FRANCE
TECHNIQUES TO LOCATE A TRANSITION STATE
 32 III. TECHNIQUES TO LOCATE A TRANSITION STATE In addition to the location of minima, the calculation of transition states and reaction pathways is an interesting task for Quantum Chemistry (QC). The
32 III. TECHNIQUES TO LOCATE A TRANSITION STATE In addition to the location of minima, the calculation of transition states and reaction pathways is an interesting task for Quantum Chemistry (QC). The
1 Density functional theory (DFT)
 1 Density functional theory (DFT) 1.1 Introduction Density functional theory is an alternative to ab initio methods for solving the nonrelativistic, time-independent Schrödinger equation H Φ = E Φ. The
1 Density functional theory (DFT) 1.1 Introduction Density functional theory is an alternative to ab initio methods for solving the nonrelativistic, time-independent Schrödinger equation H Φ = E Φ. The
ABC of DFT: Hands-on session 1 Introduction into calculations on molecules
 ABC of DFT: Hands-on session 1 Introduction into calculations on molecules Tutor: Alexej Bagrets Wann? 09.11.2012, 11:30-13:00 Wo? KIT Campus Nord, Flachbau Physik, Geb. 30.22, Computerpool, Raum FE-6
ABC of DFT: Hands-on session 1 Introduction into calculations on molecules Tutor: Alexej Bagrets Wann? 09.11.2012, 11:30-13:00 Wo? KIT Campus Nord, Flachbau Physik, Geb. 30.22, Computerpool, Raum FE-6
Example: H 2 O (the car file)
 Example: H 2 O (the car file) As a practical example of DFT methods we calculate the energy and electronic properties of the water molecule. In order to carry out the DFT calculation you will need a set
Example: H 2 O (the car file) As a practical example of DFT methods we calculate the energy and electronic properties of the water molecule. In order to carry out the DFT calculation you will need a set
Putting a spin on time dependent electronic structure theory! Joshua Goings! General Exam! Thursday, May 14, 10:00am CHB 339
 Putting a spin on time dependent electronic structure theory Joshua Goings General Exam Thursday, May 14, 10:00am CHB 339 Thank you to General Exam Committee Xiaosong Li (chair) Jim Pfaendtner (GSR) Matthew
Putting a spin on time dependent electronic structure theory Joshua Goings General Exam Thursday, May 14, 10:00am CHB 339 Thank you to General Exam Committee Xiaosong Li (chair) Jim Pfaendtner (GSR) Matthew
ABIN: AB INitio Molecular Dynamics (and much more)
 ABIN: AB INitio Molecular Dynamics (and much more) version 1.0 comming soon(ish) Daniel Hollas FIRST PHOTOX seminar, ICT Prague 17.10.2014 Daniel Hollas (ICT Prague) ABIN 17.10.2014 1 / 30 (Really) Brief
ABIN: AB INitio Molecular Dynamics (and much more) version 1.0 comming soon(ish) Daniel Hollas FIRST PHOTOX seminar, ICT Prague 17.10.2014 Daniel Hollas (ICT Prague) ABIN 17.10.2014 1 / 30 (Really) Brief
Walter Kohn was awarded with the Nobel Prize in Chemistry in 1998 for his development of the density functional theory.
 Walter Kohn was awarded with the Nobel Prize in Chemistry in 1998 for his development of the density functional theory. Walter Kohn receiving his Nobel Prize from His Majesty the King at the Stockholm
Walter Kohn was awarded with the Nobel Prize in Chemistry in 1998 for his development of the density functional theory. Walter Kohn receiving his Nobel Prize from His Majesty the King at the Stockholm
Molecular Simulation I
 Molecular Simulation I Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences
Molecular Simulation I Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences
Semi-Empirical MO Methods
 Semi-Empirical MO Methods the high cost of ab initio MO calculations is largely due to the many integrals that need to be calculated (esp. two electron integrals) semi-empirical MO methods start with the
Semi-Empirical MO Methods the high cost of ab initio MO calculations is largely due to the many integrals that need to be calculated (esp. two electron integrals) semi-empirical MO methods start with the
Speed-up of ATK compared to
 What s new @ Speed-up of ATK 2008.10 compared to 2008.02 System Speed-up Memory reduction Azafulleroid (molecule, 97 atoms) 1.1 15% 6x6x6 MgO (bulk, 432 atoms, Gamma point) 3.5 38% 6x6x6 MgO (k-point sampling
What s new @ Speed-up of ATK 2008.10 compared to 2008.02 System Speed-up Memory reduction Azafulleroid (molecule, 97 atoms) 1.1 15% 6x6x6 MgO (bulk, 432 atoms, Gamma point) 3.5 38% 6x6x6 MgO (k-point sampling
Density Functional Theory
 Density Functional Theory March 26, 2009 ? DENSITY FUNCTIONAL THEORY is a method to successfully describe the behavior of atomic and molecular systems and is used for instance for: structural prediction
Density Functional Theory March 26, 2009 ? DENSITY FUNCTIONAL THEORY is a method to successfully describe the behavior of atomic and molecular systems and is used for instance for: structural prediction
Gaussian: Basic Tutorial
 Input file: # hf sto-g pop=full Water - Single Point Energy 0 H.0 H.0 H 04.5 Route Section Start with # Contains the keywords Gaussian: Basic Tutorial Route Section Title Section Charge-Multiplicity Molecule
Input file: # hf sto-g pop=full Water - Single Point Energy 0 H.0 H.0 H 04.5 Route Section Start with # Contains the keywords Gaussian: Basic Tutorial Route Section Title Section Charge-Multiplicity Molecule
Figure 1. Molecules geometries of 5021 and Each neutral group in CHARMM topology was grouped in dash circle.
 Project I Chemistry 8021, Spring 2005/2/23 This document was turned in by a student as a homework paper. 1. Methods First, the cartesian coordinates of 5021 and 8021 molecules (Fig. 1) are generated, in
Project I Chemistry 8021, Spring 2005/2/23 This document was turned in by a student as a homework paper. 1. Methods First, the cartesian coordinates of 5021 and 8021 molecules (Fig. 1) are generated, in
X-ray absorption spectroscopy.
 X-ray absorption spectroscopy www.anorg.chem.uu.nl/people/staff/frankdegroot/ X-ray absorption spectroscopy www.anorg.chem.uu.nl/people/staff/frankdegroot/ Frank de Groot PhD: solid state chemistry U Nijmegen
X-ray absorption spectroscopy www.anorg.chem.uu.nl/people/staff/frankdegroot/ X-ray absorption spectroscopy www.anorg.chem.uu.nl/people/staff/frankdegroot/ Frank de Groot PhD: solid state chemistry U Nijmegen
Hartree, Hartree-Fock and post-hf methods
 Hartree, Hartree-Fock and post-hf methods MSE697 fall 2015 Nicolas Onofrio School of Materials Engineering DLR 428 Purdue University nonofrio@purdue.edu 1 The curse of dimensionality Let s consider a multi
Hartree, Hartree-Fock and post-hf methods MSE697 fall 2015 Nicolas Onofrio School of Materials Engineering DLR 428 Purdue University nonofrio@purdue.edu 1 The curse of dimensionality Let s consider a multi
DENSITY FUNCTIONAL THEORY FOR NON-THEORISTS JOHN P. PERDEW DEPARTMENTS OF PHYSICS AND CHEMISTRY TEMPLE UNIVERSITY
 DENSITY FUNCTIONAL THEORY FOR NON-THEORISTS JOHN P. PERDEW DEPARTMENTS OF PHYSICS AND CHEMISTRY TEMPLE UNIVERSITY A TUTORIAL FOR PHYSICAL SCIENTISTS WHO MAY OR MAY NOT HATE EQUATIONS AND PROOFS REFERENCES
DENSITY FUNCTIONAL THEORY FOR NON-THEORISTS JOHN P. PERDEW DEPARTMENTS OF PHYSICS AND CHEMISTRY TEMPLE UNIVERSITY A TUTORIAL FOR PHYSICAL SCIENTISTS WHO MAY OR MAY NOT HATE EQUATIONS AND PROOFS REFERENCES
Basis Set for Molecular Orbital Theory
 Basis Set for Molecular Orbital Theory! Different Types of Basis Functions! Different Types of Atom Center Basis Functions! Classifications of Gaussian Basis Sets! Pseudopotentials! Molecular Properties
Basis Set for Molecular Orbital Theory! Different Types of Basis Functions! Different Types of Atom Center Basis Functions! Classifications of Gaussian Basis Sets! Pseudopotentials! Molecular Properties
GEM4 Summer School OpenCourseWare
 GEM4 Summer School OpenCourseWare http://gem4.educommons.net/ http://www.gem4.org/ Lecture: Molecular Mechanics by Ju Li. Given August 9, 2006 during the GEM4 session at MIT in Cambridge, MA. Please use
GEM4 Summer School OpenCourseWare http://gem4.educommons.net/ http://www.gem4.org/ Lecture: Molecular Mechanics by Ju Li. Given August 9, 2006 during the GEM4 session at MIT in Cambridge, MA. Please use
