Structural Analysis of Expanding Metabolic Networks
|
|
|
- Lucas Pearson
- 6 years ago
- Views:
Transcription
1 Genome Informatics 15(1): (24) 35 Structural Analysis of Expanding Metabolic Networks Oliver Ebenhöh Reinhart Heinrich Thomas Handorf Theoretical Biophysics, Institute of Biology, Humboldt University Berlin, Germany Abstract Methods are developed for structural analysis of metabolic networks expanding in size. Expansion proceeds in consecutive generations in which new reactions are attached to the network produced in the previous stage. Different rules are applied resulting in various modes of expansion. Expansion is performed on the set of glycolytic reactions as well as on a very large set of reactions taken from the KEGG database. It is shown that reactions and compounds strongly differ in the generation in which they are attached to the network allowing conclusions for the temporal order of the acquisition during network evolution. The expansion provides efficient tools for detecting new structural characteristics such as substrate-product relationships over long distances. Keywords: metabolic network, evolution, stoichiometry, optimisation, scope, KEGG database 1 Introduction Metabolic networks in living cells are characterised by several quantities. Variables such as the concentrations of metabolites or fluxes may change on a short time scale whereas system parameters such as stoichiometric numbers defining the structure of the networks or kinetic constants of enzymatic reactions can be considered to remain constant throughout the life span of a single organism. Traditionally, mathematical modelling of metabolic networks aims at simulating stationary states or time courses for the system variables with the system parameters used as input quantities. Recently, structural analyses gained much interest since in contrast to enzyme kinetic parameters the stoichiometric properties are often much better known [5]. The development of metabolic databases such as KEGG [3, 4] allows for studying the structure of metabolism on a large scale. In previous work we performed structural analyses on subnetworks by applying evolutionary optimisation using a limited set of generic reactions. We could show that the stoichiometry of the central energy metabolism, in particular the ordering of and NADH producing and consuming reactions, can be explained under the premise that contemporary networks ensure a high production rate [1, 6]. Furthermore we introduced the concept of stoichiometric robustness characterising the effect of structural changes on the function of metabolic networks [2]. The goal of the present work is to reconstruct extant metabolic systems on a much larger scale by taking into account as many specific reactions as possible. We perform this reconstruction by simulating evolutionary processes starting from a small set of reactions and compounds. During this procedure new reactions are repeatedly attached according to certain rules leading to networks of increasing size. Each individual process we call network expansion. We show that the mutual dependencies of reactions by their substrates and products allow for conclusions on the temporal order of the emergence of enzymatic reactions and subpathways. Such studies will help to elucidate the principles under which network structures evolved towards increasing complexity. In this work we develop methods for this kind of investigation which are tested first on the limited set of reactions of the glycolytic pathway. Application to the large set leads to the definition of new structural characteristics such as the scope of compounds.
2 36 Ebenhöh et al. 2 Basic Assumptions Any metabolic network depends on substrates which have to be provided by the environment. Through a series of biochemical reactions these substrates get converted into end products which are released into the environment. Such substrates and products are called external compounds of the given network. Under steady-state conditions, the production and consumption of all other (internal) metabolites is balanced resulting in time independent concentrations. Let C = (X, S) T denote the vector of the concentrations of the metabolites such that X comprises all internal and S all external compounds. The dynamic behaviour of the network is governed by a set of differential equations for the metabolite concentrations having the form dc = N V, (1) dt where N is the stoichiometric matrix which can be decomposed into two submatrices N X and N S for the internal and external compounds, respectively, and V is the vector of the rates of the biochemical reactions. In principle, all reactions are reversible allowing for positive and negative values of V. Under physiological conditions, however, many reactions proceed only in one direction leading to sign restrictions for the corresponding components of V. In the following, we consider all reactions to be reversible. The steady-state of the system is determined by the equation N X V =. (2) In the framework of stoichiometric analysis, Eq. (2) is treated as a linear equation system for feasible fluxes V. In general, the solution of the system is not unique with the general solution being a superposition of a set of fundamental solutions. We distinguish between two different types of solutions, one where the fluxes result in a net conversion of external metabolites, N S V, and one where the fluxes represent only internal cycles, N S V =. In the present work we investigate expanding network structures leading to sequences of stoichiometric matrices of increasing size. This analysis is performed in view of evolution where metabolic networks have acquired new metabolic capabilities in form of additional enzymatic reactions. On shorter time scales, changes in network size could be mediated by a gene transfer as can be observed between bacteria. For deriving rules for network expansion, we assume that a newly acquired biochemical reaction may be especially useful to an organism if it makes it less dependent from external resources or provides it with new resources. This can be fulfilled by reactions allowing to treat one or more compounds to be internal which were external before the acquisition. If the acquired reaction depends on metabolites not present in the network before, these compounds must be considered external. However, for some chemicals which were always highly abundant in the environment there was no need for an organism to balance their production and consumption. In our analysis we consider such compounds to remain external during the whole process of network expansion. We investigate several scenarios with different sets of these highly abundant compounds. The computational analysis of network expansion is performed by an algorithm which can be summarised as follows: 1. The process starts with a set of one or more biochemical compounds acting as a seed of the expanding network. 2. From the set of all available reactions a set of candidate reactions is determined. Candidate reactions are defined by the property that they can in principle be attached to the current network. Candidate reactions must fulfil one of the two expansion conditions: At least one of its compounds must already be present in the current network (weak expansion condition C1);
3 Structural Analysis of Expanding Metabolic Networks 37 all of its substrates or all of its products must already be present (strong expansion condition C2). 3. From the set of candidate reactions, one or more are attached to the current network according to specific selection rules. In this work, the selection rules utilise Eqs. 2 and 1, for example for deciding which reactions perform net conversions under steady state conditions (see Section 3.1). Steps 2 and 3 are repeated continuously until no candidate reaction can be found. Each loop leads to a new network generation. 3 Expansion on the Set of Glycolytic Reactions In this section we investigate network expansion on a limited set of reactions. Candidate reactions are chosen from the thirteen reactions of the glycolytic pathway (see Figure 1). Since this pathway supplies energy in form of, consumption of by other processes (ases) is also taken into account (the reaction placed on top of the first glycolytic reaction at the left end of Figure 1). PEP Pyr Lac 1,3-BPG 3-PG 2-PG H+ H+ NADH NAD+ NAD+ NADH Figure 1: Base set of reactions comprising the glycolytic pathway. Abbreviations for the compounds are: - glucose, - glucose-6-phosphate, - fructose-6-phosphate, - fructose-1,6- bisphosphate, - dihydroxyacetone phosphate, - glyceraldehyde-3-phosphate, 1,3-BPG - 1,3-bisphosphoglycerate, - 2,3-bisphosphoglycerate, 3-PG - 3-phosphoglycerate, 2-PG - 2- phosphoglycerate, PEP - phosphoenolpyruvate, Pyr - pyruvate, Lac - lactate. 3.1 Selection Rule We analyse network expansion on this set of reactions under the following conditions. First, we assume that the network in each stage depends on as few as possible external metabolites. Second, we assume that the network produces as many as possible internal metabolites since these compounds may serve as building blocks for other metabolic processes. Third, we consider it disadvantageous for a network to possess reactions that are not required for conversions under steady-state conditions. These reactions are characterised by the fact that the corresponding entries in all solution vectors V of Eq. (2) vanish. The selection rule can be formalised as follows: From the candidate reactions, those reactions are chosen maximising a selection value for which we use the expression f = αx βs γz, (3) where x and s are the numbers of internal and external metabolites, respectively, and z is the number of reactions that are not required in steady-state and α, β, and γ are weight factors. The presented simulations have been performed for α = β and γ < β. The first choice ensures that the value of f remains unchanged if the acquisition of a new reaction involves the incorporation of a new external compound into the network and simultaneously an external compound becomes internal. The second choice entails that an additional reaction which is not required for conversions is less disadvantageous in terms of the selection value f than an additional external compound.
4 38 Ebenhöh et al. Additional criteria to be fulfilled during the process are: a) an internal compound cannot become external in later generations; b) whenever a reaction is attached to one or more external compounds, at least one of these compounds must become internal; c) a maximal set of external compounds becomes internal under the condition that there exists a steady-state solution with a net conversion between the remaining external metabolites (N S V ). In the set of glycolytic reactions there are certain compounds whose production and consumption are strictly coupled. These are and as well as NAD +, NADH and H +. Therefore, the stoichiometric matrix is reduced by considering only one compound from each group. We choose to omit the rows corresponding to the compounds, NADH and H Model Scenarios Using the algorithm described in section 2, we analyse three scenarios. All of them are based on the weak expansion condition C1, using glucose as the only initial compound and considering water as highly abundant. The three scenarios differ in the choice of compounds which, in addition to water, are forced to be external and cannot become internal during the expansion. These compounds are selected from the metabolites, NAD + and inorganic phosphate. Case 1:, NAD + and Inorganic Phosphate as Forced External Compounds In the first step of the algorithm, there is only one candidate reaction (hexokinase HK) which can be attached to glucose. In this stage the networks contains three new compounds, all of which are external. Two of them ( and ) are forced to be external whereas can become internal later (see generation 1 in Figure 2). Like in the first step, for each of the following three steps there 1: 4: 5: 7: 12: 1,3-BPG H+ NAD+ NADH 1,3-BPG 3-PG 2-PG H+ NAD+ NADH PEP Pyr Lac H+ NADH NAD+ 5 : 13: 1,3-BPG H+ NAD+ NADH 1,3-BPG 3-PG 2-PG H+ NAD+ NADH PEP Pyr Lac H+ NADH NAD+ Figure 2: Expansion process on the limited set of the glycolytic reactions (case 1). It is assumed that water,, NAD +, and inorganic phosphate are highly abundant in the environment and forced to be external. These compounds are marked in grey., NADH, and H + are strictly coupled to other compounds and also do not contribute to the selection value f and are therefore marked in grey as well. Compounds which are external in one generation but may become internal in a later generation are underlined. The calculations were performed with the parameters α = β = 1 and γ = β/2. exists only one candidate reaction. This leads to the generation 4 which includes the reactions HK, glucosephosphate isomerase (PGI), phosphofructokinase (PFK) and aldolase (Ald). In the fifth step there are two candidate reactions, triose phosphate isomerase (TIM) and glyceraldehyde phosphate dehydrogenase (DH). Incorporation of TIM leads to generation 5 whereas incorporation of DH leads to a network depicted by 5 in Figure 2. These two possibilities differ in their relations between
5 Structural Analysis of Expanding Metabolic Networks 39 the numbers of external and internal compounds. Whereas both networks contain four internal compounds, networks 5 and 5 include two and three external compounds, respectively. Therefore, the expansion proceeds with network 5. In step 6 there is only one candidate reaction, leading to the attachment of DH. In step 7 there are two candidate reactions, namely bisphosphoglycerate mutase (BPGM) and phosphoglycerate kinase (PGK). Both resulting networks have the properties s = 2, x = 6, and z = resulting in the same selection value f = 4. The algorithm randomly chooses the first possibility. In step 8, from the two candidate reactions PGK and BPGase, the latter is selected since in this case the network expands increasing the number of internal as well as external compounds by one, whereas in the other case only one external compound is added. In the subsequent steps 9 12, the metabolic chain is expanded until the incorporation of lactate dehydrogenase (LDH). In this stage only two reactions are not yet incorporated (PGK and ase). Since in this scenario ase has only forced external compounds, its attachment cannot result in compounds becoming internal and therefore this reaction is not one of the candidate reactions. Consequently, the last reaction added is PGK leading to generation 13 representing the full glycolytic pathway. Case 2: Inorganic Phosphate as Forced External Compound As in case 1, the first step represents the incorporation of HK. Since in this scenario can become an internal compound, the expansion continues in a different fashion compared to case 1. In fact, there are five candidate reactions in step 2, four of them proceed with turnover and are therefore suited to make an internal substrate when combined HK. The fifth candidate reaction is PGI. The algorithm selects ase since no new external compounds are incorporated (note that in this scenario inorganic phosphate does not contribute to the selection value f). From this step the expansion proceeds in the same way as in case 1. Case 3: No Additional Forced External Compound In this scenario in principle all compounds except water can become internal during the expansion. The first six steps proceed in the same way as in case 2 resulting in the network 6 in Figure 3. This networks converts glucose into two molecules and consumes two molecules of which are produced by ase working in the opposite direction by the expense of inorganic phosphate which in this stage is external. 6: 1: 3-PG 2-PG 7: 12: PEP Pyr H+ NAD+ NADH 3-PG 1,3-BPG 3-PG 2-PG Figure 3: Expansion on the glycolytic pathway (case 3). Conditions, labels, and parameter values are the same as for Figure 2 with the exception that only water is considered to be highly abundant. From this step on, the expansion differs dramatically from both previously considered cases. In step 7 the sugar converting chain is not extended by further metabolising but rather the reaction BPGase is attached allowing inorganic phosphate to become an internal compound. PEP Pyr
6 4 Ebenhöh et al. The expansion proceeds by prolongation of the chain from 3PG to pyruvate. This leads in step 1 to a pathway consuming besides glucose also with a net consumption of one. ase still works in the opposite direction where the rate is halved compared to network 6. In steps 11 and 12 the two sugar converting chains are connected by incorporation of BPGM and DH. This network transforms glucose into pyruvate with no net consumption of. Therefore, ase occurs with a zero rate resulting in a value z = 1. In step 13 LDH is attached allowing for the internalisation of NAD + and pyruvate which were external in step 12. Only with the incorporation of PGK in the last step, the glycolytic pathway achieves the ability to produce. Consequently, ase now operates in the forward direction. We have chosen the well known glycolytic pathway to demonstrate in detail the procedure of network expansion. Despite its simplicity the expansion may proceed in many modes differing in the order of attachment of the reactions. Expansion conditions and selection rules restrict the number of possibilities significantly. We could demonstrate that applying a small number of such rules is sufficient to ensure that intermediary generations do not represent arbitrary collections of reactions but rather networks with preliminary capabilities. Interestingly, the same criteria did not lead to a premature end but the expansion evolved until all reactions of the glycolytic pathway were incorporated. 4 Network Expansion on a Large Scale We will now apply the expansion methods on a set of 5161 reactions and 445 compounds as provided by the KEGG database. Whereas in section 3 we examined specific expansion modes, we now characterise the processes by general properties. 4.1 Strong Expansion Condition without Selection Rules First, we investigate the expansion on this global set of reactions according to the following rules: a) Several randomly chosen metabolites serve as initial compounds; b) the expansion condition C2 is used which is more restricted than condition C1; c) all candidate reactions are attached when proceeding from one generation to the next. number of compounds/reactions compounds reactions network generation Figure 4: Evolution of the number of compounds and reactions during network expansion. The process has been started with 2 randomly chosen compounds. Figure 4 shows the development of the total numbers of compounds and reactions during one particular expansion. The curve is characterised by a strong initial increase and saturation in later generations. Interestingly, the algorithm stops after 5 generations resulting in a network of about 21 compounds which is considerably smaller than the global network. This behaviour is due to the fact that at this stage no candidate reaction can be found fulfilling the strong expansion condition C2.
7 Structural Analysis of Expanding Metabolic Networks 41 Similar results were obtained for other sets of initial compounds leading to final networks of different sizes. The strong initial increase of the network size results from the fact that in this phase with each additional reaction a large number of new compounds are recruited which in turn increase the number of candidate reactions for the next generation. Another interesting feature is the tendency of the curve to level off in intermediate stages followed by another strong increase. We explain this by the existence of bottlenecks in the global network where only few reactions link highly connected domains. An intriguing question is in which generation a certain compound is acquired. Naturally, the particular value is highly dependent on the choice of initial compounds. We have therefore run a large number of simulations, each starting from a random initial set of 2 compounds. Figure 5 represents the results for the two compounds NAD + and α-d-glucose. The diagram shows the frequency of acquisition in a certain generation. It is observed that glucose is generally acquired earlier during the expansion than NAD +. The mean generations of acquisition for NAD + and glucose are 12.4 and 6.8, the standard deviations are 3.3 and 3.8, respectively. 6 6 frequency of acquisition NAD+ frequency of acquisition alpha-d-glucose generation of acquisition generation of acquisition Figure 5: Histograms of the network generations at which the compounds glucose and NAD + are acquired during the expansion. We have calculated corresponding histograms and their mean values for all compounds. Figure 6 shows the number of compounds having a certain mean generation of acquisition. number of compounds mean generation of acquisition Figure 6: Histogram of mean generations of acquisition. Simulations have been performed for all compounds as explained for NAD + and glucose in the legend of Figure 5. The diagram includes the values of those compounds which were acquired in at least 5% of the simulations. In Table 1, left part, are listed those ten compounds having the lowest mean generation of acquisition. For each compound the number of reactions is listed in which it participates as substrate or product (connectivity). In the right part we list those ten compounds having the highest connectivity.
8 42 Ebenhöh et al. Table 1: The ten compounds with the lowest mean generation of acquisition (left side) and the ten compounds with the highest connectivity (right side). compound generation conn. compound conn. generation Water 184 Water 184 Oxygen 1, H ,11 H + 2,11 14 Oxygen 648 1,83 CO 2 2,2 333 NAD ,41 H 2 O 2 2, NADH ,44 NH 3 2, N ,46 Orthophosphate 3,3 329 NH ,39 O 2 3, ,4 Carbonic acid 3,42 3 CO ,2 Pyruvate 3, Orthophosphate 329 3,3 number of compounds scope size number of compounds scope size Figure 7: Numbers of compounds leading to a scope of a certain size. A section of the histogram for low sizes is shown on the right. The calculations were performed under the assumption that water is highly abundant. It can be seen that five compounds appear on both sides meaning that compounds with a high connectivity tend to be acquired early in the expansion. Interestingly, there is no one to one correspondence between these two tables, an observation which will be discussed below. 4.2 Scope of Compounds Network expansion can also start with a single initial compound (A). Again we apply the strong expansion condition C2 resulting in a final network containing besides A all possible compounds which can be synthesised from A. This set of compounds we denote by Σ(A) and call it the scope of A. If a compound B lies in the scope of A, then the scope of B is a subset of the scope of A: B Σ(A) Σ(B) Σ(A). (4) There exist compounds which can be interconverted. For two such compounds A and B, the following implication holds true due to relation (4): B Σ(A) and A Σ(B) Σ(A) = Σ(B). (5) Figure 7 shows a histogram of the scope sizes. It can be observed that most scopes are of a size smaller than 5. There are only a few scopes of very large size. Not all scope sizes occur leading to gaps in the histogram. Scopes exist with almost all sizes below 5. Of special interest are the scopes of large sizes. Closer inspection shows that the largest set of 242 compounds represents one scope that can be reached from four initial compounds which are
9 Structural Analysis of Expanding Metabolic Networks phosphoadenosine 5 -phosphosulfate (PAPS), adenosine 5 -phosphosulfate (APS), dephospho-coa, and UDP-6-sulfoquinovose. From the property of scopes expressed by Eq. (5) it follows that these four compounds are interconvertible. Similarly, the set of 1545 compounds represents a single scope which can be reached by 15 initial compounds. Among these are central cofactors such as, GTP, NADH, NH as well as the compound UDPglucose. Further investigation reveals that the latter scope is a subset of the largest scope. This means, for example, that can be synthesised from PAPS whereas the opposite process is not possible. 4.3 Expansion with Selection Rules So far we expanded the global network by adding at the transition from one generation to the next all candidate reactions allowed by the expansion condition C2. Now we perform expansion on the global network in a very similar way as it was done in section 3 on the glycolytic pathway. Again, we start from glucose, apply the weak expansion condition C1, and attach in each generation the one reaction which maximises the selection value f given in Eq. (3). We assume that during expansion all compounds can in principle be internalised. We compare this process to the expansion where in each generation one randomly chosen candidate reaction is attached. In each case we characterise the expansion by the development of the numbers of external and internal compounds as well as the numbers of independent steady state fluxes and internal cycles. For the first 3 generations, the results are shown in Figure 8. number of compounds selected, ext selected, int random, ext random, int number of independent fluxes selected, fluxes selected, cycles random, fluxes random, cycles network generation network generation Figure 8: Initial phase of the expansion on the global network. Left: development of the numbers of external (squares) and internal (triangles) compounds for random (dashed lines) and selected (solid lines) expansion. Right: development of the numbers of independent steady state fluxes (squares) and internal cycles (triangles). Dashed lines are used for random and solid lines for selected expansion. Figure 8 demonstrates that the two expansions proceed in significantly different ways. With the selection rule, the number of external compounds always remains below the number of internal compounds. For the random expansion the opposite is true. It can be seen that for the random expansion there exist a higher number of fluxes which can be explained by the greater amount of external compounds. The number of internal compounds remains so low that in the observed phase of the random expansion no cycles appear. Interestingly, the networks appearing during the expansion with selection rule possess a small number of independent fluxes but a comparably high number of internal cycles. This means that these networks are rather independent from external compounds and additionally can interconvert them in a very flexible manner. 5 Discussion The main results of our analysis can be summarised as follows. 1) Single metabolic pathways as well as large parts of the global network of cellular metabolism can be reconstructed by simulating evolu-
10 44 Ebenhöh et al. tionary processes resulting in series of networks of increasing size. 2) Depending on the availability of resources, different modes of expansion lead to the glycolytic pathway. 3) Expansion on the global network yields a ranking of the metabolites according to the mean generation in which they were acquired when starting from randomly selected initial metabolites. All compounds which are on average acquired early in the expansion are well known core components of the metabolism and moreover are rather simple in their chemical structure. 4) Compounds of a network may be characterised by their scope defining the set of substances that can be synthesised through the available reactions without further use of other resources. Compounds with a large scope are found to be composed of a rather large number of chemical groups. 5) Expansion processes obeying selection rules proceed significantly different to random expansion. Selected expansion results in networks being able to perform relatively few interconversions but flexibly via various different internal routes whereas random expansion results in networks which can perform many interconversions but are highly dependent on external compounds. We observed that the compounds with a low mean generation of acquisition are not necessarily those participating in many reactions. Carbonic acid, for example, is acquired very early in the expansion but has an extremely low connectivity (see Table 1). This can be understood by the fact that it can be produced from water and carbon dioxide both having a very high connectivity and are acquired very early. Moreover, metabolites with high connectivities are not necessarily acquired early. Examples are NAD + and other cofactors. This can be explained by the fact that the synthesis of these compounds require a large number of steps. The present investigation was performed by considering all reactions as reversible. For some reactions it is well known that they proceed only in one direction under physiological conditions. This information could easily be taken into account by applying a sign restrictions for the corresponding elements of the feasible solution vectors of Eq. (2). We refrained from including this information since the results would be dependent on the concentrations of external metabolites which do not represent a structural property. We do not claim that the calculated expansion modes reflect the details of the evolutionary development during natural selection of metabolic pathways. This particularly concerns the results obtained using expansion condition C1 in which many intermediate generations depend on the abundance of external metabolites. This problem is less severe when using expansion condition C2 since networks get expanded only by such reactions which further convert metabolites made available by already existing reactions of the network. Our studies use only structural information about the biochemical reactions. However, it is conceivable to include further biological information. For example, using the results from comparative genomics, one can identify enzymes which are closely related to enzymes catalysing a biochemical reaction which is already present in the network. Such additional information could be incorporated into the specific selection rules. Further refinement of the methods presented here may contribute to reveal whether the history of network evolution is encoded in their present design. References [1] Ebenhöh, O. and Heinrich, R., Evolutionary optimization of metabolic pathways. Theoretical reconstruction of the stoichiometry of and NADH producing systems, Bull. Math. Biol., 63:21 55, 21. [2] Ebenhöh, O. and Heinrich, R., Stoichiometric design of metabolic networks: Multifunctionality, clusters, optimization, weak and strong robustness, Bull. Math. Biol., 65: , 21. [3] Kanehisa, M., A database for post-genome analysis, Trends Genet., 13: , 1997.
11 Structural Analysis of Expanding Metabolic Networks 45 [4] Kanehisa, M. and Goto, S., KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res., 28:27 3, 2. [5] Schilling, C.H., Schuster, S., Palsson, B.O., and Heinrich, R., Metabolic pathway analysis: Basic concepts and scientific applications in the post-genomic era, Biotechnol. Prog., 15:296 33, [6] Stephani, A., Nuño, J.C., and Heinrich, R., Optimal stoichiometric design of -producing systems as determined by an evolutionary algorithm, J. Theor. Biol., 199:45 61, 1999.
Expanding Metabolic Networks: Scopes of Compounds, Robustness and Evolution
 1 Expanding Metabolic Networks: Scopes of Compounds, Robustness and Evolution by Thomas Handorf, Oliver Ebenhöh, Reinhart Heinrich published in Journal of Molecular Evolution Address: Humboldt University
1 Expanding Metabolic Networks: Scopes of Compounds, Robustness and Evolution by Thomas Handorf, Oliver Ebenhöh, Reinhart Heinrich published in Journal of Molecular Evolution Address: Humboldt University
Evolutionary Optimization of Metabolic Pathways. Theoretical Reconstruction of the Stoichiometry of ATP and NADH Producing Systems
 Bulletin of Mathematical Biology (2001) 63, 21 55 doi:10.1006/bulm.2000.0197 Available online at http://www.idealibrary.com on Evolutionary Optimization of Metabolic Pathways. Theoretical Reconstruction
Bulletin of Mathematical Biology (2001) 63, 21 55 doi:10.1006/bulm.2000.0197 Available online at http://www.idealibrary.com on Evolutionary Optimization of Metabolic Pathways. Theoretical Reconstruction
Description of the algorithm for computing elementary flux modes
 Description of the algorithm for computing elementary flux modes by Stefan Schuster, Thomas Dandekar,, and David Fell Department of Bioinformatics, Max Delbrück Centre for Molecular Medicine D-9 Berlin-Buch,
Description of the algorithm for computing elementary flux modes by Stefan Schuster, Thomas Dandekar,, and David Fell Department of Bioinformatics, Max Delbrück Centre for Molecular Medicine D-9 Berlin-Buch,
V19 Metabolic Networks - Overview
 V19 Metabolic Networks - Overview There exist different levels of computational methods for describing metabolic networks: - stoichiometry/kinetics of classical biochemical pathways (glycolysis, TCA cycle,...
V19 Metabolic Networks - Overview There exist different levels of computational methods for describing metabolic networks: - stoichiometry/kinetics of classical biochemical pathways (glycolysis, TCA cycle,...
BBS2710 Microbial Physiology. Module 5 - Energy and Metabolism
 BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
V14 extreme pathways
 V14 extreme pathways A torch is directed at an open door and shines into a dark room... What area is lighted? Instead of marking all lighted points individually, it would be sufficient to characterize
V14 extreme pathways A torch is directed at an open door and shines into a dark room... What area is lighted? Instead of marking all lighted points individually, it would be sufficient to characterize
Chapter 15 part 2. Biochemistry I Introduction to Metabolism Bioenergetics: Thermodynamics in Biochemistry. ATP 4- + H 2 O ADP 3- + P i + H +
 Biochemistry I Introduction to Metabolism Bioenergetics: Thermodynamics in Biochemistry ATP 4- + 2 ADP 3- + P i 2- + + Chapter 15 part 2 Dr. Ray 1 Energy flow in biological systems: Energy Transformations
Biochemistry I Introduction to Metabolism Bioenergetics: Thermodynamics in Biochemistry ATP 4- + 2 ADP 3- + P i 2- + + Chapter 15 part 2 Dr. Ray 1 Energy flow in biological systems: Energy Transformations
Chapter 7: Metabolic Networks
 Chapter 7: Metabolic Networks 7.1 Introduction Prof. Yechiam Yemini (YY) Computer Science epartment Columbia University Introduction Metabolic flux analysis Applications Overview 2 1 Introduction 3 Metabolism:
Chapter 7: Metabolic Networks 7.1 Introduction Prof. Yechiam Yemini (YY) Computer Science epartment Columbia University Introduction Metabolic flux analysis Applications Overview 2 1 Introduction 3 Metabolism:
Predicting rice (Oryza sativa) metabolism
 Predicting rice (Oryza sativa) metabolism Sudip Kundu Department of Biophysics, Molecular Biology & Bioinformatics, University of Calcutta, WB, India. skbmbg@caluniv.ac.in Collaborators: Mark G Poolman
Predicting rice (Oryza sativa) metabolism Sudip Kundu Department of Biophysics, Molecular Biology & Bioinformatics, University of Calcutta, WB, India. skbmbg@caluniv.ac.in Collaborators: Mark G Poolman
Giving you the energy you need!
 Giving you the energy you need! Use your dominant hand Open and close the pin (with your thumb and forefinger) as many times as you can for 20 seconds while holding the other fingers straight out! Repeat
Giving you the energy you need! Use your dominant hand Open and close the pin (with your thumb and forefinger) as many times as you can for 20 seconds while holding the other fingers straight out! Repeat
Exam 3 Review (4/12/2011) Lecture note excerpt covering lectures (Exam 3 topics: Chapters 8, 12, 14 & 15)
 Exam 3 Review (4/12/2011) Lecture note excerpt covering lectures 17-23 (Exam 3 topics: Chapters 8, 12, 14 & 15) Enzyme Kinetics, Inhibition, and Regulation Chapter 12 Enzyme Kinetics When the concentration
Exam 3 Review (4/12/2011) Lecture note excerpt covering lectures 17-23 (Exam 3 topics: Chapters 8, 12, 14 & 15) Enzyme Kinetics, Inhibition, and Regulation Chapter 12 Enzyme Kinetics When the concentration
Cell population modelling of yeast glycolytic oscillations
 Biochem. J. (2002) 368, 433 446 (Printed in Great Britain) 433 Cell population modelling of yeast glycolytic oscillations Michael A. HENSON* 1, Dirk MU LLER and Matthias REUSS *Department of Chemical Engineering,
Biochem. J. (2002) 368, 433 446 (Printed in Great Britain) 433 Cell population modelling of yeast glycolytic oscillations Michael A. HENSON* 1, Dirk MU LLER and Matthias REUSS *Department of Chemical Engineering,
System Reduction of Nonlinear Positive Systems by Linearization and Truncation
 System Reduction of Nonlinear Positive Systems by Linearization and Truncation Hanna M. Härdin 1 and Jan H. van Schuppen 2 1 Department of Molecular Cell Physiology, Vrije Universiteit, De Boelelaan 1085,
System Reduction of Nonlinear Positive Systems by Linearization and Truncation Hanna M. Härdin 1 and Jan H. van Schuppen 2 1 Department of Molecular Cell Physiology, Vrije Universiteit, De Boelelaan 1085,
Biochemical Pathways
 Biochemical Pathways Living organisms can be divided into two large groups according to the chemical form in which they obtain carbon from the environment. Autotrophs can use carbon dioxide from the atmosphere
Biochemical Pathways Living organisms can be divided into two large groups according to the chemical form in which they obtain carbon from the environment. Autotrophs can use carbon dioxide from the atmosphere
Review Questions - Lecture 5: Metabolism, Part 1
 Review Questions - Lecture 5: Metabolism, Part 1 Questions: 1. What is metabolism? 2. What does it mean to say that a cell has emergent properties? 3. Define metabolic pathway. 4. What is the difference
Review Questions - Lecture 5: Metabolism, Part 1 Questions: 1. What is metabolism? 2. What does it mean to say that a cell has emergent properties? 3. Define metabolic pathway. 4. What is the difference
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Outer Glycolysis mitochondrial membrane Glucose ATP
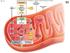 Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
V15 Flux Balance Analysis Extreme Pathways
 V15 Flux Balance Analysis Extreme Pathways Stoichiometric matrix S: m n matrix with stochiometries of the n reactions as columns and participations of m metabolites as rows. The stochiometric matrix is
V15 Flux Balance Analysis Extreme Pathways Stoichiometric matrix S: m n matrix with stochiometries of the n reactions as columns and participations of m metabolites as rows. The stochiometric matrix is
Exam 4 April 15, 2005 CHEM 3511 Print Name: KEY Signature
 1) (8 pts) General Properties of Enzymes. Give four properties of enzymaticallycatalyzed reactions. The answers should indicate how enzymatic reactions differ from non-enzymatic reactions. Write four only
1) (8 pts) General Properties of Enzymes. Give four properties of enzymaticallycatalyzed reactions. The answers should indicate how enzymatic reactions differ from non-enzymatic reactions. Write four only
Biochemistry 3300 Problems (and Solutions) Metabolism I
 (1) Provide a reasonable systematic name for an enzyme that catalyzes the following reaction: fructose + ATP > fructose-1 phosphate + ADP (2) The IUBMB has a developed a set of rules for classifying enzymes
(1) Provide a reasonable systematic name for an enzyme that catalyzes the following reaction: fructose + ATP > fructose-1 phosphate + ADP (2) The IUBMB has a developed a set of rules for classifying enzymes
Gene Regulatory Network Identification
 Gene Regulatory Network Identification Korkut Uygun and Yinlun Huang Department of Chemical Engineering and Materials Science Wayne State University AIChE Annual National Meeting San Francisco November
Gene Regulatory Network Identification Korkut Uygun and Yinlun Huang Department of Chemical Engineering and Materials Science Wayne State University AIChE Annual National Meeting San Francisco November
Glycolysis and Fermentation. Chapter 8
 Glycolysis and Fermentation Chapter 8 Cellular Respiration and Photosynthesis 0 Things to know in these chapters 0 Names and order of the processes 0 Reactants and products of each process 0 How do they
Glycolysis and Fermentation Chapter 8 Cellular Respiration and Photosynthesis 0 Things to know in these chapters 0 Names and order of the processes 0 Reactants and products of each process 0 How do they
Oxidative Phosphorylation versus. Photophosphorylation
 Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
AHL Topic 8 IB Biology Miss Werba
 CELL RESPIRATION & PHOTOSYNTHESIS AHL Topic 8 IB Biology Miss Werba TOPIC 8 CELL RESPIRATION & PHOTOSYNTHESIS 8.1 CELL RESPIRATION 1. STATE that oxidation involves the loss of electrons from an element,
CELL RESPIRATION & PHOTOSYNTHESIS AHL Topic 8 IB Biology Miss Werba TOPIC 8 CELL RESPIRATION & PHOTOSYNTHESIS 8.1 CELL RESPIRATION 1. STATE that oxidation involves the loss of electrons from an element,
Stoichiometric analysis of metabolic networks
 1 SGN-6156 Computational systems biology II Antti Larjo antti.larjo@tut.fi Computational Systems Biology group 30.4.2008 30.4.2008 2 Contents Networks in cells Metabolism Metabolic networks and pathways
1 SGN-6156 Computational systems biology II Antti Larjo antti.larjo@tut.fi Computational Systems Biology group 30.4.2008 30.4.2008 2 Contents Networks in cells Metabolism Metabolic networks and pathways
Enzymes I. Dr. Mamoun Ahram Summer semester,
 Enzymes I Dr. Mamoun Ahram Summer semester, 2017-2018 Resources Mark's Basic Medical Biochemistry Other resources NCBI Bookshelf: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=books The Medical Biochemistry
Enzymes I Dr. Mamoun Ahram Summer semester, 2017-2018 Resources Mark's Basic Medical Biochemistry Other resources NCBI Bookshelf: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=books The Medical Biochemistry
Quantitative, scalable discrete event simulation of metabolic pathways
 From: ISMB-99 Proceedings. Copyright 1999, AAAI (www.aaai.org). All rights reserved. Quantitative, scalable discrete event simulation of metabolic pathways Peter A. Meric Basser Department of Computer
From: ISMB-99 Proceedings. Copyright 1999, AAAI (www.aaai.org). All rights reserved. Quantitative, scalable discrete event simulation of metabolic pathways Peter A. Meric Basser Department of Computer
Table 1. ODEs used in the model based on mass balances.
 Table. ODEs used in the model based on mass balances. ODE s _ = _ 6 = + _ 6 = + + = +! 3 = +2!!$ = +!$ %&' ( = _ + %&' ) *+ = + _, - +2 ),$.(/0 = +,!%!-.(023 = + - $!. 56..(023_ = +!. 56. 2,3 8.3(20 =
Table. ODEs used in the model based on mass balances. ODE s _ = _ 6 = + _ 6 = + + = +! 3 = +2!!$ = +!$ %&' ( = _ + %&' ) *+ = + _, - +2 ),$.(/0 = +,!%!-.(023 = + - $!. 56..(023_ = +!. 56. 2,3 8.3(20 =
RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES. Bio 107 Week 6
 RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES Bio 107 Week 6 Procedure 7.2 Label test tubes well, including group name 1) Add solutions listed to small test tubes 2)
RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES Bio 107 Week 6 Procedure 7.2 Label test tubes well, including group name 1) Add solutions listed to small test tubes 2)
build stuff!! Stomates Warm-up Remember what it means to be a Photosynthesis: The Calvin Cycle Life from Air Autotrophs Light reactions
 Warm-up Objective: Describe how the chemical products of the light-trapping reactions couple to the synthesis of carbohydrates. Warm-up: What is the advantage of the light reaction producing H and ATP
Warm-up Objective: Describe how the chemical products of the light-trapping reactions couple to the synthesis of carbohydrates. Warm-up: What is the advantage of the light reaction producing H and ATP
' Institute for Biology, Humboldt University Berlin, Germany
 METABOLIC SYNERGY: INCREASING BIOSYNTHETIC CAPABILITIES BY NETWORK COOPERATION NILS CHRISTIAN' nils.christianqphysik.fu-berlin.de THOMAS HANDORF' thomas.handorfc3physik.hu-berlin.de OLIVER EBENHOH~ ebenhoehompimp-golm.mpg.de
METABOLIC SYNERGY: INCREASING BIOSYNTHETIC CAPABILITIES BY NETWORK COOPERATION NILS CHRISTIAN' nils.christianqphysik.fu-berlin.de THOMAS HANDORF' thomas.handorfc3physik.hu-berlin.de OLIVER EBENHOH~ ebenhoehompimp-golm.mpg.de
Biological Pathways Representation by Petri Nets and extension
 Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Energy in Chemical and Biochemical Reactions
 Energy in Chemical and Biochemical Reactions Reaction Progress Diagram for Exothermic Reaction Reactants activated complex Products ENERGY A + B Reactants E a C + D Products Δ rxn Reaction coordinate The
Energy in Chemical and Biochemical Reactions Reaction Progress Diagram for Exothermic Reaction Reactants activated complex Products ENERGY A + B Reactants E a C + D Products Δ rxn Reaction coordinate The
CHAPTER 15 Metabolism: Basic Concepts and Design
 CHAPTER 15 Metabolism: Basic Concepts and Design Chapter 15 An overview of Metabolism Metabolism is the sum of cellular reactions - Metabolism the entire network of chemical reactions carried out by living
CHAPTER 15 Metabolism: Basic Concepts and Design Chapter 15 An overview of Metabolism Metabolism is the sum of cellular reactions - Metabolism the entire network of chemical reactions carried out by living
Stoichiometric network analysis
 Stoichiometric network analysis In stoichiometric analysis of metabolic networks, one concerns the effect of the network structure on the behaviour and capabilities of metabolism. Questions that can be
Stoichiometric network analysis In stoichiometric analysis of metabolic networks, one concerns the effect of the network structure on the behaviour and capabilities of metabolism. Questions that can be
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy
 Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of October
 Name: Class: _ Date: _ 2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of 19-23 October Multiple Choice Identify the choice that best completes the statement or answers the question. 1) Which
Name: Class: _ Date: _ 2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of 19-23 October Multiple Choice Identify the choice that best completes the statement or answers the question. 1) Which
BIOCHEMISTRY. František Vácha. JKU, Linz.
 BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
BCH Graduate Survey of Biochemistry
 BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Marsha Durosier Lecture 55 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html
BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Marsha Durosier Lecture 55 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html
Metabolic modelling. Metabolic networks, reconstruction and analysis. Esa Pitkänen Computational Methods for Systems Biology 1 December 2009
 Metabolic modelling Metabolic networks, reconstruction and analysis Esa Pitkänen Computational Methods for Systems Biology 1 December 2009 Department of Computer Science, University of Helsinki Metabolic
Metabolic modelling Metabolic networks, reconstruction and analysis Esa Pitkänen Computational Methods for Systems Biology 1 December 2009 Department of Computer Science, University of Helsinki Metabolic
New Results on Energy Balance Analysis of Metabolic Networks
 New Results on Energy Balance Analysis of Metabolic Networks Qinghua Zhou 1, Simon C.K. Shiu 2,SankarK.Pal 3,andYanLi 1 1 College of Mathematics and Computer Science, Hebei University, Baoding City 071002,
New Results on Energy Balance Analysis of Metabolic Networks Qinghua Zhou 1, Simon C.K. Shiu 2,SankarK.Pal 3,andYanLi 1 1 College of Mathematics and Computer Science, Hebei University, Baoding City 071002,
Photosynthesis in Higher Plants
 Photosynthesis in Higher Plants Very Short Answers Questions: 1. Name the processes which take place in the grana and stroma regions of chloroplasts? A: Grana Light reactions. Trapping light energy, synthesizing
Photosynthesis in Higher Plants Very Short Answers Questions: 1. Name the processes which take place in the grana and stroma regions of chloroplasts? A: Grana Light reactions. Trapping light energy, synthesizing
Mechanism of CO 2 Fixation: Rubisco Step 1
 dark means light-indendent! The Dark Reaction: Carbon Fixation Requires Rubisco: Ribulose-1,5-bisphosphate carboxylase RuBPi + C 6 x 3PG MW = 550, 000 g/mol Subunits eight small (14,000 g/mol) eight large
dark means light-indendent! The Dark Reaction: Carbon Fixation Requires Rubisco: Ribulose-1,5-bisphosphate carboxylase RuBPi + C 6 x 3PG MW = 550, 000 g/mol Subunits eight small (14,000 g/mol) eight large
Chapter 6- An Introduction to Metabolism*
 Chapter 6- An Introduction to Metabolism* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Energy of Life
Chapter 6- An Introduction to Metabolism* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Energy of Life
Energy for biological processes
 1 Energy transfer When you have finished revising this topic, you should: be able to explain the difference between catabolic and anabolic reactions be able to describe the part played by in cell metabolism
1 Energy transfer When you have finished revising this topic, you should: be able to explain the difference between catabolic and anabolic reactions be able to describe the part played by in cell metabolism
13.3A: The general mechanism for an aldol reaction
 Ashley Robison My Preferences Site Tools FAQ Sign Out If you like us, please share us on social media. The latest UCD Hyperlibrary newsletter is now complete, check it out. ChemWiki BioWiki GeoWiki StatWiki
Ashley Robison My Preferences Site Tools FAQ Sign Out If you like us, please share us on social media. The latest UCD Hyperlibrary newsletter is now complete, check it out. ChemWiki BioWiki GeoWiki StatWiki
NOTES: CH 10, part 3 Calvin Cycle (10.3) & Alternative Mechanisms of C-Fixation (10.4)
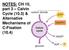 NOTES: CH 10, part 3 Calvin Cycle (10.3) & Alternative Mechanisms of C-Fixation (10.4) 10.3 - The Calvin cycle uses ATP and NADPH to convert CO 2 to sugar The Calvin cycle, like the citric acid cycle,
NOTES: CH 10, part 3 Calvin Cycle (10.3) & Alternative Mechanisms of C-Fixation (10.4) 10.3 - The Calvin cycle uses ATP and NADPH to convert CO 2 to sugar The Calvin cycle, like the citric acid cycle,
Integrated Knowledge-based Reverse Engineering of Metabolic Pathways
 Integrated Knowledge-based Reverse Engineering of Metabolic Pathways Shuo-Huan Hsu, Priyan R. Patkar, Santhoi Katare, John A. Morgan and Venkat Venkatasubramanian School of Chemical Engineering, Purdue
Integrated Knowledge-based Reverse Engineering of Metabolic Pathways Shuo-Huan Hsu, Priyan R. Patkar, Santhoi Katare, John A. Morgan and Venkat Venkatasubramanian School of Chemical Engineering, Purdue
Prediction of Enzyme Kinetic Parameters Based on Statistical Learning
 80 Genome Informatics 17(1): 80 87 (2006) Prediction of Enzyme Kinetic Parameters Based on Statistical Learning Simon Borger Wolfram Liebermeister Edda Klipp borger@molgen.mpg.de lieberme@molgen.mpg.de
80 Genome Informatics 17(1): 80 87 (2006) Prediction of Enzyme Kinetic Parameters Based on Statistical Learning Simon Borger Wolfram Liebermeister Edda Klipp borger@molgen.mpg.de lieberme@molgen.mpg.de
BIOLOGY 10/11/2014. An Introduction to Metabolism. Outline. Overview: The Energy of Life
 8 An Introduction to Metabolism CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Forms of Energy II. Laws of Thermodynamics III. Energy and metabolism IV. ATP V. Enzymes
8 An Introduction to Metabolism CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Forms of Energy II. Laws of Thermodynamics III. Energy and metabolism IV. ATP V. Enzymes
Applications of Free Energy. NC State University
 Chemistry 433 Lecture 15 Applications of Free Energy NC State University Thermodynamics of glycolysis Reaction kj/mol D-glucose + ATP D-glucose-6-phosphate + ADP ΔG o = -16.7 D-glucose-6-phosphate p D-fructose-6-phosphate
Chemistry 433 Lecture 15 Applications of Free Energy NC State University Thermodynamics of glycolysis Reaction kj/mol D-glucose + ATP D-glucose-6-phosphate + ADP ΔG o = -16.7 D-glucose-6-phosphate p D-fructose-6-phosphate
Section A2: The Pathways of Photosynthesis
 CHAPTER 10 PHOTOSYNTHESIS Section A2: The Pathways of Photosynthesis 4. The Calvin cycle uses ATP and NADPH to convert CO2 to sugar: a closer look 5. Alternative mechanisms of carbon fixation have evolved
CHAPTER 10 PHOTOSYNTHESIS Section A2: The Pathways of Photosynthesis 4. The Calvin cycle uses ATP and NADPH to convert CO2 to sugar: a closer look 5. Alternative mechanisms of carbon fixation have evolved
Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013
 Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013 Lecture 10. Biochemical Transformations II. Phosphoryl transfer and the kinetics and thermodynamics of energy currency in the cell: ATP and GTP.
Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013 Lecture 10. Biochemical Transformations II. Phosphoryl transfer and the kinetics and thermodynamics of energy currency in the cell: ATP and GTP.
Analyze the roles of enzymes in biochemical reactions
 ENZYMES and METABOLISM Elements: Cell Biology (Enzymes) Estimated Time: 6 7 hours By the end of this course, students will have an understanding of the role of enzymes in biochemical reactions. Vocabulary
ENZYMES and METABOLISM Elements: Cell Biology (Enzymes) Estimated Time: 6 7 hours By the end of this course, students will have an understanding of the role of enzymes in biochemical reactions. Vocabulary
V14 Graph connectivity Metabolic networks
 V14 Graph connectivity Metabolic networks In the first half of this lecture section, we use the theory of network flows to give constructive proofs of Menger s theorem. These proofs lead directly to algorithms
V14 Graph connectivity Metabolic networks In the first half of this lecture section, we use the theory of network flows to give constructive proofs of Menger s theorem. These proofs lead directly to algorithms
Stoichiometric vector and matrix
 Stoichiometric vector and matrix The stoichiometric coefficients of a reaction are collected to a vector s r In s r there is a one position for each metabolite in the metabolic system, and the stoichiometric
Stoichiometric vector and matrix The stoichiometric coefficients of a reaction are collected to a vector s r In s r there is a one position for each metabolite in the metabolic system, and the stoichiometric
STUDYING ENZYME KINETICS BY MEANS OF PROGRESS-CURVE ANALYSIS
 99 ESCEC, Oct. 5 th - 8 th 2003, Rüdesheim, Germany STUDYING ENZYME KINETICS BY MEANS OF PROGRESS-CURVE ANALYSIS HERMANN-GEORG HOLZHÜTTER Humboldt-University Berlin, Medical School (Charité), Institute
99 ESCEC, Oct. 5 th - 8 th 2003, Rüdesheim, Germany STUDYING ENZYME KINETICS BY MEANS OF PROGRESS-CURVE ANALYSIS HERMANN-GEORG HOLZHÜTTER Humboldt-University Berlin, Medical School (Charité), Institute
Constraint-Based Workshops
 Constraint-Based Workshops 2. Reconstruction Databases November 29 th, 2007 Defining Metabolic Reactions ydbh hslj ldha 1st level: Primary metabolites LAC 2nd level: Neutral Formulas C 3 H 6 O 3 Charged
Constraint-Based Workshops 2. Reconstruction Databases November 29 th, 2007 Defining Metabolic Reactions ydbh hslj ldha 1st level: Primary metabolites LAC 2nd level: Neutral Formulas C 3 H 6 O 3 Charged
Cellular Automata Approaches to Enzymatic Reaction Networks
 Cellular Automata Approaches to Enzymatic Reaction Networks Jörg R. Weimar Institute of Scientific Computing, Technical University Braunschweig, D-38092 Braunschweig, Germany J.Weimar@tu-bs.de, http://www.jweimar.de
Cellular Automata Approaches to Enzymatic Reaction Networks Jörg R. Weimar Institute of Scientific Computing, Technical University Braunschweig, D-38092 Braunschweig, Germany J.Weimar@tu-bs.de, http://www.jweimar.de
Assimilation of Carbon in Plants
 Module 0210101: Molecular Biology and Biochemistry of the Cell Lecture 18 Assimilation of Carbon in Plants Dale Sanders 17 March 2009 Objectives By the end of the lecture you should understand: 1. How
Module 0210101: Molecular Biology and Biochemistry of the Cell Lecture 18 Assimilation of Carbon in Plants Dale Sanders 17 March 2009 Objectives By the end of the lecture you should understand: 1. How
Introduction to Systems Biology. James Gomes KSBS, IITD
 Introduction to Systems Biology James Gomes KSBS, IITD Inner Life of a Cell Cells Exhibit a High Degree of Complexity What are the different types of cells? Is Life an emergent property? What is Systems
Introduction to Systems Biology James Gomes KSBS, IITD Inner Life of a Cell Cells Exhibit a High Degree of Complexity What are the different types of cells? Is Life an emergent property? What is Systems
2. In regards to the fluid mosaic model, which of the following is TRUE?
 General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
Supplementary Figure 3
 Supplementary Figure 3 a 1 (i) (ii) (iii) (iv) (v) log P gene Q group, % ~ ε nominal 2 1 1 8 6 5 A B C D D' G J L M P R U + + ε~ A C B D D G JL M P R U -1 1 ε~ (vi) Z group 2 1 1 (vii) (viii) Z module
Supplementary Figure 3 a 1 (i) (ii) (iii) (iv) (v) log P gene Q group, % ~ ε nominal 2 1 1 8 6 5 A B C D D' G J L M P R U + + ε~ A C B D D G JL M P R U -1 1 ε~ (vi) Z group 2 1 1 (vii) (viii) Z module
Photosynthetic autotrophs use the energy of sunlight to convert low-g CO 2 and H 2 O into energy-rich complex sugar molecules.
 Chapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
Chapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
Unit 1C Practice Exam (v.2: KEY)
 Unit 1C Practice Exam (v.2: KEY) 1. Which of the following statements concerning photosynthetic pigments (chlorophylls a and b, carotenes, and xanthophylls) is correct? (PT1-12) a. The R f values obtained
Unit 1C Practice Exam (v.2: KEY) 1. Which of the following statements concerning photosynthetic pigments (chlorophylls a and b, carotenes, and xanthophylls) is correct? (PT1-12) a. The R f values obtained
Welcome to Class 8! Introductory Biochemistry! Announcements / Reminders! Midterm TA led Review Sessions!
 Announcements / Reminders Midterm TA led Review Sessions Welcome to Class 8 Sunday, February 23 from 8-10pm Location: Science Center Main Room (315) Office Hours Prof Salomon: SFH 270 on Thursday Feb 20,
Announcements / Reminders Midterm TA led Review Sessions Welcome to Class 8 Sunday, February 23 from 8-10pm Location: Science Center Main Room (315) Office Hours Prof Salomon: SFH 270 on Thursday Feb 20,
This is an example of cellular respiration, which can be used to make beer and wine using different metabolic pathways For these reasons we call this
 Chapter 6 Carvings from ancient Egypt show barley being crushed and mixed with water (left) and then put into closed vessels (centre) where airless conditions are suitable for the production of alcohol
Chapter 6 Carvings from ancient Egypt show barley being crushed and mixed with water (left) and then put into closed vessels (centre) where airless conditions are suitable for the production of alcohol
Overview of Kinetics
 Overview of Kinetics [P] t = ν = k[s] Velocity of reaction Conc. of reactant(s) Rate of reaction M/sec Rate constant sec -1, M -1 sec -1 1 st order reaction-rate depends on concentration of one reactant
Overview of Kinetics [P] t = ν = k[s] Velocity of reaction Conc. of reactant(s) Rate of reaction M/sec Rate constant sec -1, M -1 sec -1 1 st order reaction-rate depends on concentration of one reactant
37 Modelling and experimental evidence for two separate steady states in the photosynthetic Calvin cycle
 37 Modelling and experimental evidence for two separate steady states in the photosynthetic Calvin cycle M.G. Poolman and D.A. Fell School of Biology and Molecular Science, Oxford Brookes University, Headington,
37 Modelling and experimental evidence for two separate steady states in the photosynthetic Calvin cycle M.G. Poolman and D.A. Fell School of Biology and Molecular Science, Oxford Brookes University, Headington,
Chapter 8: An Introduction to Metabolism. 1. Energy & Chemical Reactions 2. ATP 3. Enzymes & Metabolic Pathways
 Chapter 8: An Introduction to Metabolism 1. Energy & Chemical Reactions 2. ATP 3. Enzymes & Metabolic Pathways 1. Energy & Chemical Reactions 2 Basic Forms of Energy Kinetic Energy (KE) energy in motion
Chapter 8: An Introduction to Metabolism 1. Energy & Chemical Reactions 2. ATP 3. Enzymes & Metabolic Pathways 1. Energy & Chemical Reactions 2 Basic Forms of Energy Kinetic Energy (KE) energy in motion
Flux Balance Analysis
 Lecture 10: Flux Balance Analysis Tue 7 March 2006 with the collaboration of Luna De Ferrari 1 Images and text from: E. Klipp, Systems Biology in Practice, Wiley-VCH, 2005 Ch 5 Edwards JS, Palsson BO,
Lecture 10: Flux Balance Analysis Tue 7 March 2006 with the collaboration of Luna De Ferrari 1 Images and text from: E. Klipp, Systems Biology in Practice, Wiley-VCH, 2005 Ch 5 Edwards JS, Palsson BO,
Name Date Class. Photosynthesis and Respiration
 Concept Mapping Photosynthesis and Respiration Complete the Venn diagram about photosynthesis and respiration. These terms may be used more than once: absorbs, Calvin cycle, chlorophyll, CO 2, H 2 O, Krebs
Concept Mapping Photosynthesis and Respiration Complete the Venn diagram about photosynthesis and respiration. These terms may be used more than once: absorbs, Calvin cycle, chlorophyll, CO 2, H 2 O, Krebs
Lower glycolysis carries a higher flux than any biochemically possible alternative Supporting Information
 Lower glycolysis carries a higher flux than any biochemically possible alternative Supporting Information Steven J. Court, Bart lomiej Waclaw, and Rosalind J. Allen I. THE TRUNK PATHWAY OF GLYCOLYSIS AND
Lower glycolysis carries a higher flux than any biochemically possible alternative Supporting Information Steven J. Court, Bart lomiej Waclaw, and Rosalind J. Allen I. THE TRUNK PATHWAY OF GLYCOLYSIS AND
METABOLIC PATHWAY PREDICTION/ALIGNMENT
 COMPUTATIONAL SYSTEMIC BIOLOGY METABOLIC PATHWAY PREDICTION/ALIGNMENT Hofestaedt R*, Chen M Bioinformatics / Medical Informatics, Technische Fakultaet, Universitaet Bielefeld Postfach 10 01 31, D-33501
COMPUTATIONAL SYSTEMIC BIOLOGY METABOLIC PATHWAY PREDICTION/ALIGNMENT Hofestaedt R*, Chen M Bioinformatics / Medical Informatics, Technische Fakultaet, Universitaet Bielefeld Postfach 10 01 31, D-33501
BIOLOGICAL SCIENCE. Lecture Presentation by Cindy S. Malone, PhD, California State University Northridge. FIFTH EDITION Freeman Quillin Allison
 BIOLOGICAL SCIENCE FIFTH EDITION Freeman Quillin Allison 8 Lecture Presentation by Cindy S. Malone, PhD, California State University Northridge Roadmap 8 In this chapter you will learn how Enzymes use
BIOLOGICAL SCIENCE FIFTH EDITION Freeman Quillin Allison 8 Lecture Presentation by Cindy S. Malone, PhD, California State University Northridge Roadmap 8 In this chapter you will learn how Enzymes use
Program for the rest of the course
 Program for the rest of the course 16.4 Enzyme kinetics 17.4 Metabolic Control Analysis 19.4. Exercise session 5 23.4. Metabolic Control Analysis, cont. 24.4 Recap 27.4 Exercise session 6 etabolic Modelling
Program for the rest of the course 16.4 Enzyme kinetics 17.4 Metabolic Control Analysis 19.4. Exercise session 5 23.4. Metabolic Control Analysis, cont. 24.4 Recap 27.4 Exercise session 6 etabolic Modelling
Dynamic Stability of Signal Transduction Networks Depending on Downstream and Upstream Specificity of Protein Kinases
 Dynamic Stability of Signal Transduction Networks Depending on Downstream and Upstream Specificity of Protein Kinases Bernd Binder and Reinhart Heinrich Theoretical Biophysics, Institute of Biology, Math.-Nat.
Dynamic Stability of Signal Transduction Networks Depending on Downstream and Upstream Specificity of Protein Kinases Bernd Binder and Reinhart Heinrich Theoretical Biophysics, Institute of Biology, Math.-Nat.
Photosynthesis and Cellular Respiration
 Photosynthesis and Cellular Respiration Photosynthesis and Cellular Respiration All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs:
Photosynthesis and Cellular Respiration Photosynthesis and Cellular Respiration All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs:
Aerobic Cellular Respiration
 Aerobic Cellular Respiration Under aerobic conditions (oxygen gas is available), cells will undergo aerobic cellular respiration. The end products of aerobic cellular respiration are carbon dioxide gas,
Aerobic Cellular Respiration Under aerobic conditions (oxygen gas is available), cells will undergo aerobic cellular respiration. The end products of aerobic cellular respiration are carbon dioxide gas,
9/25/2011. Outline. Overview: The Energy of Life. I. Forms of Energy II. Laws of Thermodynamics III. Energy and metabolism IV. ATP V.
 Chapter 8 Introduction to Metabolism Outline I. Forms of Energy II. Laws of Thermodynamics III. Energy and metabolism IV. ATP V. Enzymes Overview: The Energy of Life Figure 8.1 The living cell is a miniature
Chapter 8 Introduction to Metabolism Outline I. Forms of Energy II. Laws of Thermodynamics III. Energy and metabolism IV. ATP V. Enzymes Overview: The Energy of Life Figure 8.1 The living cell is a miniature
Cellular Respiration: Harvesting Chemical Energy. 9.1 Catabolic pathways yield energy by oxidizing organic fuels
 Cellular Respiration: Harvesting Chemical Energy 9.1 Catabolic pathways yield energy by oxidizing organic fuels 9.2 Glycolysis harvests chemical energy by oxidizing glucose to pyruvate 9.3 The citric acid
Cellular Respiration: Harvesting Chemical Energy 9.1 Catabolic pathways yield energy by oxidizing organic fuels 9.2 Glycolysis harvests chemical energy by oxidizing glucose to pyruvate 9.3 The citric acid
What Is Energy? Energy is the capacity to do work. First Law of Thermodynamics. Types of energy
 What Is Energy? Energy is the capacity to do work. Synthesizing molecules Moving objects Generating heat and light Types of Kinetic: of movement otential: stored First Law of Thermodynamics Energy cannot
What Is Energy? Energy is the capacity to do work. Synthesizing molecules Moving objects Generating heat and light Types of Kinetic: of movement otential: stored First Law of Thermodynamics Energy cannot
Chapter 5. The Chloroplast. 5.1 Matter and Energy Pathways in Living Systems. Photosynthesis & Cellular Respiration
 Chapter 5 Photosynthesis & Cellular Respiration 5.1 Matter and Energy Pathways in Living Systems Both cellular respiration and photosynthesis are examples of biological processes that involve matter &
Chapter 5 Photosynthesis & Cellular Respiration 5.1 Matter and Energy Pathways in Living Systems Both cellular respiration and photosynthesis are examples of biological processes that involve matter &
An introduction to biochemical reaction networks
 Chapter 1 An introduction to biochemical reaction networks (This part is discussed in the slides). Biochemical reaction network distinguish themselves from general chemical reaction networks in that the
Chapter 1 An introduction to biochemical reaction networks (This part is discussed in the slides). Biochemical reaction network distinguish themselves from general chemical reaction networks in that the
All organisms require a constant expenditure of energy to maintain the living state - "LIFE".
 CELLULAR RESPIRATION All organisms require a constant expenditure of energy to maintain the living state - "LIFE". Where does the energy come from and how is it made available for life? With rare exception,
CELLULAR RESPIRATION All organisms require a constant expenditure of energy to maintain the living state - "LIFE". Where does the energy come from and how is it made available for life? With rare exception,
QUALITATIVE PATH ANALYSIS OF METABOLIC PATHWAYS USING PETRI NETS FOR GENERIC MODELLING
 BRANDENBURG UNIVERSITY OF TECHNOLOGY AT COTTBUS Faculty of Mathematics, Natural Sciences and Computer Science Institute of Computer Science COMPUTER SCIENCE REPORTS Report 03/04 August 2004 QUALITATIVE
BRANDENBURG UNIVERSITY OF TECHNOLOGY AT COTTBUS Faculty of Mathematics, Natural Sciences and Computer Science Institute of Computer Science COMPUTER SCIENCE REPORTS Report 03/04 August 2004 QUALITATIVE
Flux balance analysis in metabolic networks
 Flux balance analysis in metabolic networks Lecture notes by Eran Eden, https://systemsbiology.usu.edu/documents/flux%20balance%20analysis%20overview%20-%202016.pdf additional slides by B. Palsson from
Flux balance analysis in metabolic networks Lecture notes by Eran Eden, https://systemsbiology.usu.edu/documents/flux%20balance%20analysis%20overview%20-%202016.pdf additional slides by B. Palsson from
Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013
 Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013 Lecture 9 Biochemical Transformations I. Carbon-carbon bond forming and cleaving reactions in Biology (see the Lexicon). Enzymes catalyze a limited
Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013 Lecture 9 Biochemical Transformations I. Carbon-carbon bond forming and cleaving reactions in Biology (see the Lexicon). Enzymes catalyze a limited
The products have more enthalpy and are more ordered than the reactants.
 hapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
hapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
Winter School in Mathematical & Computational Biology
 1 Network reconstruction, topology and feasible solution space From component to systems biology Component biology Systems biology Component view Systems view Needed homeostasis Function S+E X E+P Reaction
1 Network reconstruction, topology and feasible solution space From component to systems biology Component biology Systems biology Component view Systems view Needed homeostasis Function S+E X E+P Reaction
Photosynthesis in Detail. 3/19/2014 Averett
 Photosynthesis in Detail 1 In photosynthesis many chemical reactions, enzymes and ions work together in a precise order. Enzymes Biological catalyst Substance that initiates or speeds up the rate of a
Photosynthesis in Detail 1 In photosynthesis many chemical reactions, enzymes and ions work together in a precise order. Enzymes Biological catalyst Substance that initiates or speeds up the rate of a
Biology Reading Assignment: Chapter 9 in textbook
 Biology 205 5.10.06 Reading Assignment: Chapter 9 in textbook HTTP://WUNMR.WUSTL.EDU/EDUDEV/LABTUTORIALS/CYTOCHROMES/CYTOCHROMES.HTML What does a cell need to do? propagate itself (and its genetic program)
Biology 205 5.10.06 Reading Assignment: Chapter 9 in textbook HTTP://WUNMR.WUSTL.EDU/EDUDEV/LABTUTORIALS/CYTOCHROMES/CYTOCHROMES.HTML What does a cell need to do? propagate itself (and its genetic program)
Sabrina Hoffmann, Andreas Hoppe and Hermann-Georg Holzhutter
 Beilstein-Institut ESCEC, March 19 th 23 rd, 2006, Rüdesheim/Rhein, Germany 91 Decomposition into Minimal Flux Modes: A Novel Flux-Balance Approach to Predict Flux Changes in Metabolic Networks from Changes
Beilstein-Institut ESCEC, March 19 th 23 rd, 2006, Rüdesheim/Rhein, Germany 91 Decomposition into Minimal Flux Modes: A Novel Flux-Balance Approach to Predict Flux Changes in Metabolic Networks from Changes
Computational Systems Biology Exam
 Computational Systems Biology Exam Dr. Jürgen Pahle Aleksandr Andreychenko, M.Sc. 31 July, 2012 Name Matriculation Number Do not open this exam booklet before we ask you to. Do read this page carefully.
Computational Systems Biology Exam Dr. Jürgen Pahle Aleksandr Andreychenko, M.Sc. 31 July, 2012 Name Matriculation Number Do not open this exam booklet before we ask you to. Do read this page carefully.
1. Plants and other autotrophs are the producers of the biosphere
 1. Plants and other autotrophs are the producers of the biosphere Photosynthesis nourishes almost all of the living world directly or indirectly. All organisms require organic compounds for energy and
1. Plants and other autotrophs are the producers of the biosphere Photosynthesis nourishes almost all of the living world directly or indirectly. All organisms require organic compounds for energy and
1. Plants and other autotrophs are the producers of the biosphere
 1. Plants and other autotrophs are the producers of the biosphere Photosynthesis nourishes almost all of the living world directly or indirectly. All organisms require organic compounds for energy and
1. Plants and other autotrophs are the producers of the biosphere Photosynthesis nourishes almost all of the living world directly or indirectly. All organisms require organic compounds for energy and
Cellular Energy: Respiration. Goals: Anaerobic respiration
 Cellular Energy: Respiration Anaerobic respiration Goals: Define and describe the 3 sets of chemical reactions that comprise aerobic cellular respiration Describe the types of anaerobic respiration Compare
Cellular Energy: Respiration Anaerobic respiration Goals: Define and describe the 3 sets of chemical reactions that comprise aerobic cellular respiration Describe the types of anaerobic respiration Compare
Located in the thylakoid membranes. Chlorophyll have Mg + in the center. Chlorophyll pigments harvest energy (photons) by absorbing certain
 a review Located in the thylakoid membranes. Chlorophyll have Mg + in the center. Chlorophyll pigments harvest energy (photons) by absorbing certain wavelengths (blue-420 nm and red-660 nm are most important).
a review Located in the thylakoid membranes. Chlorophyll have Mg + in the center. Chlorophyll pigments harvest energy (photons) by absorbing certain wavelengths (blue-420 nm and red-660 nm are most important).
METABOLISM CHAPTER 04 BIO 211: ANATOMY & PHYSIOLOGY I. Dr. Lawrence G. Altman Some illustrations are courtesy of McGraw-Hill.
 BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. CELLULAR METABOLISM Dr. Lawrence G. Altman
BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. CELLULAR METABOLISM Dr. Lawrence G. Altman
CELL METABOLISM OVERVIEW Keep the big picture in mind as we discuss the particulars!
 BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 CELLULAR METABOLISM 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. Dr. Lawrence G. Altman
BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 CELLULAR METABOLISM 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. Dr. Lawrence G. Altman
Metabolic Network Analysis
 Metabolic Network Analysis Christoph Flamm Institute for Theoretical Chemistry University Vienna Harvest Network Course, Brno, April 1-6, 2011 The Systems Biology Modelling Space Network Models Level of
Metabolic Network Analysis Christoph Flamm Institute for Theoretical Chemistry University Vienna Harvest Network Course, Brno, April 1-6, 2011 The Systems Biology Modelling Space Network Models Level of
