A: Up regulated proteins B: Down regulated proteins. Susceptible Resistant Susceptible Resistant Resistant Susceptible
|
|
|
- Janis McKenzie
- 5 years ago
- Views:
Transcription
1 Supplementary Materials: Identification of Biomarkers for Resistance to Fusarium oxysporum f. sp. cubense Infection and in Silico Studies in Musa paradisiaca Cultivar Puttabale through Proteomic Approach Venkatesh Ramu, Krishna Venkatarangaiah, Pradeepa Krishnappa, Santosh Kumar Shimoga Rajanna, Nagaraja Deeplanaik, Anup Chandra Pal and Kukkundoor Ramachandra Kini A: Up regulated proteins B: Down regulated proteins Susceptible Resistant Susceptible Resistant Resistant Susceptible Figure S1. 3D images of resolved differentially expressed proteins. (A) Up regulated protein spots; (B) Down regulated protein spots Pathogenesis-related protein; 1401 Ring fyve phd zinc finger protein; 2401 Dehydroascorbate reductase; 2403 Salicylate o-methyltransferase-like; 3004 Sucrose synthase; 3103 Dynein heavy chain; 3205 Pathogenesis-related protein; 4205 Cadmium/zinctransporting atpase; s proteasome non-atpase regulatory subunit; 4403 Ras-related protein rabb1c-like; 4503 Alcohol dehydrogenase 1; 4505 Polyphosphoinositide binding protein ssh2p; 5007 Albumin-1 D; 5302 Peptide methionine sulfoxide reductase chlo; 6001 Disease resistance rpp13-like protein 1-like; 5303 Subtilisin-like protease; 4601 Protein odr-4; 7704 Lrr repeats and ubiquitinlike domain-containing protein; 4404 Auxin-responsive protein; s acidic ribosomal protein. Proteomes 2016, 4, 9; doi: /proteomes
2 S2 of S22 a b Figure S2. (a) Melting curve and (b) Amplification plot Specificity of the PCR reactions of PR, PBPssh2p, PMSRc, IAA, DRrpp13 gene. Figure S3. The specific amplicon size of Tubulin, PR, PMSRc, PBPssh2p, IAA, DRrpp13 gene on 3.5% agarose gel.
3 S3 of S22 Figure S4. PR (Musa sps.) sequence with homologues Birch Pollen Allergen Bet V 1 (PDB-1FM4_A) shared 41% of identity. Conserved residues are represented by asterisk (*), gaps are represented by hyphen ( ), weak conserved amino acids are denoted by (.), respectively. Figure S5. Alignment of PMSRc (Musa sps.) sequence with homologues Peptide Methionine Sulfoxide Reductase Msrb (Bacillus Subtilis; PDB-2KZN_A) shared 51% of identity. Conserved residues are represented by asterisk (*), gaps are represented by hyphen ( ), weak conserved amino acids are denoted by (.), respectively.
4 S4 of S22 Figure S6. Alignment of PBPssh2p (Musa sps.) sequence with homologues Functional Phosphatidylinositol Transfer Protein from A Pseudo-Sec14 Scaffold (PDB-3Q8G_A) By Directed Evolution shared 35% of identity. Conserved residues are represented by asterisk (*), gaps are represented by hyphen ( ), weak conserved amino acids are denoted by (.), respectively. Figure S7. Alignment of IAA (Musa sps.) sequence with homologues Aux/iaa Transcription Factor Ps-iaa4 From Pea (Pisum sativum, PDB-2M1M_A) shared 57% of identity. Conserved residues are represented by asterisk (*), gaps are represented by hyphen ( ), weak conserved amino acids are denoted by (.), respectively.
5 S5 of S22 Figure S8. The Homology modeling and structure validation of PR protein. (a) Homology modeling; (b) Ramachandra plot; (c) The prosa surface energy z score plot; (d) profile of prosa plot.
6 S6 of S22 Figure S9. The Homology modeling and structure validation of PMSRc protein. (a) Homology modeling; (b) Ramachandra plot; (c) The prosa surface energy z score plot; (d) profile of prosa plot.
7 S7 of S22 Figure S10. The Homology modeling and structure validation of PBPssh2p protein. (a) Homology modeling; (b) Ramachandra plot; (c) The prosa surface energy z score plot; (d) profile of prosa plot.
8 S8 of S22 Figure S11. The Homology modeling and structure validation of IAA protein. (a) Homology modeling; (b) Ramachandra plot; (c) The prosa surface energy z score plot; (d) profile of prosa plot.
9 S9 of S22 a b c Figure S12. Molecular dynamic simulation of PR protein. (a) RMSD plot at 10 ns timescale; (b) Radius of gyration plot for compactness of structure; (c) RMSF plot for fluctuation in backbone residues. a b c Figure S13. Molecular dynamic simulation of PMSRc protein. (a) RMSD plot at 10 ns timescale; (b) Radius of gyration plot for compactness of structure; (c) RMSF plot for fluctuation in backbone residues.
10 S10 of S22 a b c Figure S14. Molecular dynamic simulation of PBPssh2p protein. (a) RMSD plot at 10 ns timescale; (b) Radius of gyration plot for compactness of structure; (c) RMSF plot for fluctuation in backbone residues. a b c Figure S15. Molecular dynamic simulation of IAA protein. (a) RMSD plot at 10 ns timescale; (b) Radius of gyration plot for compactness of structure; (c) RMSF plot for fluctuation in backbone residues.
11 S11 of S22 a b a b c d c d Figure S16. Total potential energy of the modeled proteins viz., (a) PR; (b) PMSRc; (c) PBPssh2p; (d) IAA.
12 S12 of S22 Figure S17. The protein-protein docking of PR protein to PG molecule using Gramm-X server. The PG molecule highlighted with tv_red, active pocket colored with yellow color and PR proetin are highlighted with hotpink color. (a) Cartoon view; (b) Molecular surface view. Figure S18. Protein-protein docking of PMSRc protein with PG molecule. The PG molecule highlighted with tv_red, active pocket colored with yellow and the PMSRc protein highlighted with lightmagenta color. (a) Cartoon view; (b) Molecular surface view. Figure S19. Protein-protein docking of PBPssh2p protein to PG molecule. The PG molecule highlighted with tv_red, active pocket colored with yellow and the PBPssh2p protein highlighted with cyans color. (a) Cartoon view; (b) Molecular surface view.
13 S13 of S22 Figure S20. Protein-protein docking of IAA protein to PG molecule. The PG molecule highlighted with tv_red, active pocket colored with yellow and IAA protein highlighted with blue color. (a) Cartoon view; (b) Molecular surface view. Figure S21. Docking of PR protein with Glucan compound. (A) Ribbon structure of PR and Glucan compound; (B) Molecular surface view of PR and Glucan compound; (C) Glucan compound compactly inserted into the active pocket of PR protein. Figure S22. Docking of PMSRc protein with Glucan compound. (A) Ribbon structure of PMSRc and Glucan compound; (B) Molecular surface view of PMSRc and Glucan compound; (C) Glucan compound compactly inserted into the active pocket of PMSRc protein.
14 S14 of S22 a b c Figure S23. Docking of PBPssh2p protein with Glucan compound. (a) Ribbon structure of PBPssh2p and Glucan compound; (b) Molecular surface view of PBPssh2p and Glucan compound; (c) Glucan compound compactly inserted into the active pocket of PBPssh2p protein. Figure S24. Docking of IAA protein with Glucan compound. (A) Ribbon structure of IAA and Glucan compound; (B) Molecular surface view of PBPssh2p and Glucan compound; (C) Glucan compound compactly inserted into the active pocket of IAA protein. Figure S25. Docking of PR protein with chitin substrate. (A) Molecular surface view of PR and chitin substrate; (B) Ribbon structure of PR and Glucan substrate; (C) Chitin substrate compactly inserted into the active pocket of PR protein.
15 S15 of S22 Figure S26. Docking of PMSRc protein with chitin substrate. (A) Molecular surface view of PMSRc and chitin substrate; (B) Ribbon structure of PMSRc and Glucan substrate; (C) Chitin substrate compactly inserted into the active pocket of PMSRc protein. Figure S27. Docking of PBPssh2p protein with chitin substrate. (A) Ribbon structure of PBPssh2p and Glucan substrate; (B) Molecular surface view of PBPssh2p and chitin substrate; (C) Chitin substrate compactly inserted into the active pocket of PBPssh2p protein. Figure S28. Docking of IAA protein with chitin substrate. (A) Ribbon structure of IAA and Glucan substrate; (B) Molecular surface view of IAA and chitin substrate; (C) Chitin substrate compactly inserted into the active pocket of IAA protein.
16 S16 of S22 Table S1. The PDquest software analysis of differentially accumulated proteins in Foc infected leaves sample. SL. NO. SSP Mr pi co Ratio tr Ratio , , , ,
17 S17 of S22 Table 1. Cont. SL. NO. SSP Mr pi co Ratio tr Ratio , Highlighted proteins spots are used for MALDI TOF analysis. Table S2. Significance level correction by Benjamini-Hochberg FDR method. Sl. No. SSP No. p q* p < q* TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE The Benjamini-Hochberg s FDR (q*) for the p-value of a t-test were calculated as [(i/m) q], where m is the number of p-values of the given dataset. The hypothesis for a given test is considered truly significant, i.e., significantly rejects the null hypothesis only if its p-value is equal or less than that of Benjamini- Hochberg s correction (q*), i.e., p < q*. The null hypothesis is that there is no significant difference in the protein abundance for a protein spot between control and treated samples.
18 Proteomes 2016, 4, x; doi: / S18 of S22 Sl. No. Surface Area, Å 2 Buried Area, Å 2 Table S3. Analysis of the Interface of protein-protein docking of PR with PG using GRAMM X server. ΔG int kcal/m ΔG diss kcal/m No. of HB and HBE No. of SB No. of HI within 5 Å EE vwe TSE II within 6 Å 01 19, / , / , / , / , / , / , / , / , / , / ΔG int : solvation Free energy gain upon formation of the assembly; ΔG diss : Free energy of assembly dissociation; HB: Hydrogen bonds across the interface; HBE: Hydrogen Bond ; SB: Salt bridges across the interface; HI: Hydrophobic Interactions; EE: Electrostatic ; vwe: van der Waals ; TSE: Total Stabilizing ; II: Ionic Interactions. p-value Sl. No. Surface Area, Å 2 Table S4. Analysis of the Interface of protein-protein docking of PMSRc with PG using GRAMM X server. Buried Area, Å 2 ΔG int kcal/m ΔG diss kcal/m No. of HB and HBE No. of SB No. of HI within 5 Å EE vwe TSE II within 6 Å 01 25, / , / , / , / , / , / , / , / , / , / ΔG int : solvation Free energy gain upon formation of the assembly; ΔG diss : Free energy of assembly dissociation; HB: Hydrogen bonds across the interface; HBE: Hydrogen Bond ; SB: Salt bridges across the interface; HI: Hydrophobic Interactions; EE: Electrostatic ; vwe: van der Waals ; TSE: Total Stabilizing ; II: Ionic Interactions. p-value
19 Proteomes 2016, 4, x; doi: / S19 of S22 Sl. No. Surface Area, Å 2 Table S5. Analysis of the Interface of protein-protein docking of PBPssh2p with PG using GRAMM X server. Buried Area, Å 2 ΔG int kcal/m ΔG diss kcal/m No. of HB and HBE No. of SB No. of HI within 5 Å EE vwe TSE II within 6 Å 01 42, / , / , / , / , / , / , / , / , / , / ΔG int : solvation Free energy gain upon formation of the assembly; ΔG diss : Free energy of assembly dissociation; HB: Hydrogen bonds across the interface; HBE: Hydrogen Bond ; SB: Salt bridges across the interface; HI: Hydrophobic Interactions; EE: Electrostatic ; vwe: van der Waals ; TSE: Total Stabilizing ; II: Ionic Interactions. p-value Sl. No. Surface Area, Å 2 Table S6. Analysis of the Interface of protein-protein docking of IAA with PG using GRAMM X server. Buried Area, Å 2 ΔG int kcal/m ΔG diss kcal/m No. of HB and HBE No. of SB No. of HI within 5 Å EE vwe TSE II within 6 Å 01 30, / , / , / , / , / , / , / , / , / , / ΔG int : solvation Free energy gain upon formation of the assembly; ΔG diss : Free energy of assembly dissociation; HB: Hydrogen bonds across the interface; HBE: Hydrogen Bond ; SB: Salt bridges across the interface; HI: Hydrophobic Interactions; EE: Electrostatic ; vwe: van der Waals ; TSE: Total Stabilizing ; II: Ionic Interactions. p-value
20 Proteomes 2016, 4, x; doi: / S20 of S22 Sl. No. Binding Table S7. Molecular docking of PR protein to β 1, 3- glucan. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed Sl. No. Binding Table S8. Molecular docking of PMSRc protein to β 1, 3- glucan. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed x x e x x Sl. No. Binding Table S9. Molecular docking of PBPssh2p protein with β 1, 3- glucan. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed x x x Sl. No. Binding Table S10. Molecular docking of IAA protein to β 1, 3- glucan. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed x x x x x e x
21 Proteomes 2016, 4, x; doi: / S21 of S22 Sl. No. Binding Table S11. Molecular docking of PR protein with chitin molecule. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed Sl. No. Binding Table S12. Molecular docking of PMSRc protein with chitin molecule. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed Sl. No. Table S13. Molecular docking of PBPssh2p protein with chitin molecule. Binding Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed Sl. No. Binding Table S14. Molecular docking of IAA protein with chitin molecule. Docking Ligand Efficiency Inhibition Constant Inter Molecular Internal Hydrogen Bond Formed
22 Proteomes 2016, 4, x; doi: / S22 of S22 Table S15. List of primer sequences used for qrt-pcr expression studies. Sl. No 1 Gene Indoleacetic acid-induced-like protein (IAA) 2 Pathogenesis-related protein (PR) Polyphosphoinositide binding protein ssh2p (PBPssh2p) Peptide methionine sulfoxide reductase chloroplastic-like (PMSRc) Disease resistance rpp13-like protein 1-like (DRrpp13) 6 Tubulin PRIMERS Primer Tm GC Content F- ACCTCTCGATTCCCTTCTCTCT 59.9 C 45.5% R- CTTCTCCTTGCTTCCTTTCTCA 60.4 C 50.0% F- CTTCTCACCAGCGTTACCATT 60.1 C 45.5% R- CGGCTTGGAACTTGAAATGA 61.1 C 45.5% F- TTCCTGGTGAGTTTAGTGCTGA 59.9 C 50.0% R- CAACTTGCTTAACCCTCTGCTC 60.1 C 45.5% F- GAAACTTATCGCACCAGAAAGG 60.1 C 45.5% R- ATGAGGCAGGATTGACATTAGC 60.5 C 45.5% F- ATGAAGCCAGCCATCTATCTC 59.7 C 45.5% R- TGATACCTCCTCCGTTCAAAGT 60.0 C 45.5% F- GGCTTGTTTCTCAGGTCATTTC 60.1 C 45.5% R- GAGAGTTGCTCGTGGTAGGC 60.0 C 60.0% F: Forward primer; R: Reverse primer. Product Size (bp)
Supplementary Information
 Supplementary Information Resveratrol Serves as a Protein-Substrate Interaction Stabilizer in Human SIRT1 Activation Xuben Hou,, David Rooklin, Hao Fang *,,, Yingkai Zhang Department of Medicinal Chemistry
Supplementary Information Resveratrol Serves as a Protein-Substrate Interaction Stabilizer in Human SIRT1 Activation Xuben Hou,, David Rooklin, Hao Fang *,,, Yingkai Zhang Department of Medicinal Chemistry
Nature Structural & Molecular Biology doi: /nsmb Supplementary Figure 1. CRBN binding assay with thalidomide enantiomers.
 Supplementary Figure 1 CRBN binding assay with thalidomide enantiomers. (a) Competitive elution assay using thalidomide-immobilized beads coupled with racemic thalidomide. Beads were washed three times
Supplementary Figure 1 CRBN binding assay with thalidomide enantiomers. (a) Competitive elution assay using thalidomide-immobilized beads coupled with racemic thalidomide. Beads were washed three times
REVIEW 1: BIOCHEMISTRY UNIT. A. Top 10 If you learned anything from this unit, you should have learned:
 Period Date REVIEW 1: BIOCHEMISTRY UNIT A. Top 10 If you learned anything from this unit, you should have learned: 1. All living matter made up of CHONPS 2. Bonds a. covalent bonds are strong b. hydrogen
Period Date REVIEW 1: BIOCHEMISTRY UNIT A. Top 10 If you learned anything from this unit, you should have learned: 1. All living matter made up of CHONPS 2. Bonds a. covalent bonds are strong b. hydrogen
Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine. JAK2 Selective Mechanism Combined Molecular Dynamics Simulation
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supporting Information Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supporting Information Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine
Docking. GBCB 5874: Problem Solving in GBCB
 Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Cryo-EM structure and model of the C. thermophilum 90S preribosome. a, Gold standard FSC curve showing the average resolution of the 90S preribosome masked and unmasked (left). FSC
Supplementary Figure 1 Cryo-EM structure and model of the C. thermophilum 90S preribosome. a, Gold standard FSC curve showing the average resolution of the 90S preribosome masked and unmasked (left). FSC
PDBe TUTORIAL. PDBePISA (Protein Interfaces, Surfaces and Assemblies)
 PDBe TUTORIAL PDBePISA (Protein Interfaces, Surfaces and Assemblies) http://pdbe.org/pisa/ This tutorial introduces the PDBePISA (PISA for short) service, which is a webbased interactive tool offered by
PDBe TUTORIAL PDBePISA (Protein Interfaces, Surfaces and Assemblies) http://pdbe.org/pisa/ This tutorial introduces the PDBePISA (PISA for short) service, which is a webbased interactive tool offered by
Chapter 1. Topic: Overview of basic principles
 Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides Zaixing
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides Zaixing
Insights into the Biotransformation of 2,4,6- Trinitrotoluene by the Old Yellow Enzyme Family of Flavoproteins. A Computational Study
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2019 Supporting Information for Insights into the Biotransformation of 2,4,6- Trinitrotoluene
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2019 Supporting Information for Insights into the Biotransformation of 2,4,6- Trinitrotoluene
Structure Investigation of Fam20C, a Golgi Casein Kinase
 Structure Investigation of Fam20C, a Golgi Casein Kinase Sharon Grubner National Taiwan University, Dr. Jung-Hsin Lin University of California San Diego, Dr. Rommie Amaro Abstract This research project
Structure Investigation of Fam20C, a Golgi Casein Kinase Sharon Grubner National Taiwan University, Dr. Jung-Hsin Lin University of California San Diego, Dr. Rommie Amaro Abstract This research project
Supporting Information
 Supporting Information Ottmann et al. 10.1073/pnas.0907587106 Fig. S1. Primary structure alignment of SBT3 with C5 peptidase from Streptococcus pyogenes. The Matchmaker tool in UCSF Chimera (http:// www.cgl.ucsf.edu/chimera)
Supporting Information Ottmann et al. 10.1073/pnas.0907587106 Fig. S1. Primary structure alignment of SBT3 with C5 peptidase from Streptococcus pyogenes. The Matchmaker tool in UCSF Chimera (http:// www.cgl.ucsf.edu/chimera)
Tu 1,*, , Sweden
 Supplementary Material Computational studiess of the binding profile of phosphoinositide PtdIns(,4,5)P with the pleckstrin homology domain d of an oomycetee cellulose synthase Guanglin Kuang 1, Vincent
Supplementary Material Computational studiess of the binding profile of phosphoinositide PtdIns(,4,5)P with the pleckstrin homology domain d of an oomycetee cellulose synthase Guanglin Kuang 1, Vincent
Bioengineering & Bioinformatics Summer Institute, Dept. Computational Biology, University of Pittsburgh, PGH, PA
 Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Computational engineering of cellulase Cel9A-68 functional motions through mutations in its linker region. WT 1TF4 (crystal) -90 ERRAT PROVE VERIFY3D
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 218 Supplementary Material: Computational engineering of cellulase Cel9-68 functional
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 218 Supplementary Material: Computational engineering of cellulase Cel9-68 functional
Visualization of Macromolecular Structures
 Visualization of Macromolecular Structures Present by: Qihang Li orig. author: O Donoghue, et al. Structural biology is rapidly accumulating a wealth of detailed information. Over 60,000 high-resolution
Visualization of Macromolecular Structures Present by: Qihang Li orig. author: O Donoghue, et al. Structural biology is rapidly accumulating a wealth of detailed information. Over 60,000 high-resolution
Week 10: Homology Modelling (II) - HHpred
 Week 10: Homology Modelling (II) - HHpred Course: Tools for Structural Biology Fabian Glaser BKU - Technion 1 2 Identify and align related structures by sequence methods is not an easy task All comparative
Week 10: Homology Modelling (II) - HHpred Course: Tools for Structural Biology Fabian Glaser BKU - Technion 1 2 Identify and align related structures by sequence methods is not an easy task All comparative
SUPPLEMENTARY INFORMATION
 Data collection Supplementary Table 1 Statistics of data collection, phasing and refinement Native Se-MAD Space group P2 1 2 1 2 1 P2 1 2 1 2 1 Cell dimensions a, b, c (Å) 50.4, 94.2, 115.4 49.8, 94.2,
Data collection Supplementary Table 1 Statistics of data collection, phasing and refinement Native Se-MAD Space group P2 1 2 1 2 1 P2 1 2 1 2 1 Cell dimensions a, b, c (Å) 50.4, 94.2, 115.4 49.8, 94.2,
Supplementary information
 Supplementary information The structural basis of modularity in ECF-type ABC transporters Guus B. Erkens 1,2, Ronnie P-A. Berntsson 1,2, Faizah Fulyani 1,2, Maria Majsnerowska 1,2, Andreja Vujičić-Žagar
Supplementary information The structural basis of modularity in ECF-type ABC transporters Guus B. Erkens 1,2, Ronnie P-A. Berntsson 1,2, Faizah Fulyani 1,2, Maria Majsnerowska 1,2, Andreja Vujičić-Žagar
DISCRETE TUTORIAL. Agustí Emperador. Institute for Research in Biomedicine, Barcelona APPLICATION OF DISCRETE TO FLEXIBLE PROTEIN-PROTEIN DOCKING:
 DISCRETE TUTORIAL Agustí Emperador Institute for Research in Biomedicine, Barcelona APPLICATION OF DISCRETE TO FLEXIBLE PROTEIN-PROTEIN DOCKING: STRUCTURAL REFINEMENT OF DOCKING CONFORMATIONS Emperador
DISCRETE TUTORIAL Agustí Emperador Institute for Research in Biomedicine, Barcelona APPLICATION OF DISCRETE TO FLEXIBLE PROTEIN-PROTEIN DOCKING: STRUCTURAL REFINEMENT OF DOCKING CONFORMATIONS Emperador
Lec.1 Chemistry Of Water
 Lec.1 Chemistry Of Water Biochemistry & Medicine Biochemistry can be defined as the science concerned with the chemical basis of life. Biochemistry can be described as the science concerned with the chemical
Lec.1 Chemistry Of Water Biochemistry & Medicine Biochemistry can be defined as the science concerned with the chemical basis of life. Biochemistry can be described as the science concerned with the chemical
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 Å resolution: comparison with the Azotobacter vinelandii MoFe protein
 Acta Cryst. (2015). D71, 274-282, doi:10.1107/s1399004714025243 Supporting information Volume 71 (2015) Supporting information for article: Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08
Acta Cryst. (2015). D71, 274-282, doi:10.1107/s1399004714025243 Supporting information Volume 71 (2015) Supporting information for article: Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08
Supplementary Figures
 1 Supplementary Figures Supplementary Figure 1 Type I FGFR1 inhibitors (a) Chemical structures of a pyrazolylaminopyrimidine inhibitor (henceforth referred to as PAPI; PDB-code of the FGFR1-PAPI complex:
1 Supplementary Figures Supplementary Figure 1 Type I FGFR1 inhibitors (a) Chemical structures of a pyrazolylaminopyrimidine inhibitor (henceforth referred to as PAPI; PDB-code of the FGFR1-PAPI complex:
Transmembrane Domains (TMDs) of ABC transporters
 Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Electronic Supplementary Information Effective lead optimization targeted for displacing bridging water molecule
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Electronic Supplementary Information Effective lead optimization targeted for displacing
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Electronic Supplementary Information Effective lead optimization targeted for displacing
Molecular Mechanics, Dynamics & Docking
 Molecular Mechanics, Dynamics & Docking Lawrence Hunter, Ph.D. Director, Computational Bioscience Program University of Colorado School of Medicine Larry.Hunter@uchsc.edu http://compbio.uchsc.edu/hunter
Molecular Mechanics, Dynamics & Docking Lawrence Hunter, Ph.D. Director, Computational Bioscience Program University of Colorado School of Medicine Larry.Hunter@uchsc.edu http://compbio.uchsc.edu/hunter
Bioinformatics. Macromolecular structure
 Bioinformatics Macromolecular structure Contents Determination of protein structure Structure databases Secondary structure elements (SSE) Tertiary structure Structure analysis Structure alignment Domain
Bioinformatics Macromolecular structure Contents Determination of protein structure Structure databases Secondary structure elements (SSE) Tertiary structure Structure analysis Structure alignment Domain
CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis
 CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis Purpose: The purpose of this laboratory is to introduce some of the basic visualization and modeling tools for viewing
CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis Purpose: The purpose of this laboratory is to introduce some of the basic visualization and modeling tools for viewing
The protein folding problem consists of two parts:
 Energetics and kinetics of protein folding The protein folding problem consists of two parts: 1)Creating a stable, well-defined structure that is significantly more stable than all other possible structures.
Energetics and kinetics of protein folding The protein folding problem consists of two parts: 1)Creating a stable, well-defined structure that is significantly more stable than all other possible structures.
Supplementary Figure 1. Aligned sequences of yeast IDH1 (top) and IDH2 (bottom) with isocitrate
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Aligned sequences of yeast IDH1 (top) and IDH2 (bottom) with isocitrate dehydrogenase from Escherichia coli [ICD, pdb 1PB1, Mesecar, A. D., and Koshland,
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Aligned sequences of yeast IDH1 (top) and IDH2 (bottom) with isocitrate dehydrogenase from Escherichia coli [ICD, pdb 1PB1, Mesecar, A. D., and Koshland,
Homology Modeling (Comparative Structure Modeling) GBCB 5874: Problem Solving in GBCB
 Homology Modeling (Comparative Structure Modeling) Aims of Structural Genomics High-throughput 3D structure determination and analysis To determine or predict the 3D structures of all the proteins encoded
Homology Modeling (Comparative Structure Modeling) Aims of Structural Genomics High-throughput 3D structure determination and analysis To determine or predict the 3D structures of all the proteins encoded
Time-dependence of key H-bond/electrostatic interaction distances in the sirna5-hago2 complexes... Page S14
 Supporting Information Probing the Binding Interactions between Chemically Modified sirnas and Human Argonaute 2 Using Microsecond Molecular Dynamics Simulations S. Harikrishna* and P. I. Pradeepkumar*
Supporting Information Probing the Binding Interactions between Chemically Modified sirnas and Human Argonaute 2 Using Microsecond Molecular Dynamics Simulations S. Harikrishna* and P. I. Pradeepkumar*
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis Sergio de Cima, Luis M. Polo, Carmen Díez-Fernández, Ana I. Martínez, Javier
SUPPLEMENTARY INFORMATION Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis Sergio de Cima, Luis M. Polo, Carmen Díez-Fernández, Ana I. Martínez, Javier
From Amino Acids to Proteins - in 4 Easy Steps
 From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
Protein Structure Prediction, Engineering & Design CHEM 430
 Protein Structure Prediction, Engineering & Design CHEM 430 Eero Saarinen The free energy surface of a protein Protein Structure Prediction & Design Full Protein Structure from Sequence - High Alignment
Protein Structure Prediction, Engineering & Design CHEM 430 Eero Saarinen The free energy surface of a protein Protein Structure Prediction & Design Full Protein Structure from Sequence - High Alignment
Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R)
 Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R) Shown in cyan and green are two adjacent tetramers from the crystallographic lattice of COP, forming the only unique inter-tetramer
Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R) Shown in cyan and green are two adjacent tetramers from the crystallographic lattice of COP, forming the only unique inter-tetramer
Tracking Protein Allostery in Evolution
 Tracking Protein Allostery in Evolution Glycogen phosphorylase frees sugars to provide energy GP orthologs diverged 600,000,000 years can respond to transcription controls, metabolite concentrations and
Tracking Protein Allostery in Evolution Glycogen phosphorylase frees sugars to provide energy GP orthologs diverged 600,000,000 years can respond to transcription controls, metabolite concentrations and
SUPPLEMENTARY INFORMATION
 Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
Protein Dynamics. The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron.
 Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Supplemental Data SUPPLEMENTAL FIGURES
 Supplemental Data CRYSTAL STRUCTURE OF THE MG.ADP-INHIBITED STATE OF THE YEAST F 1 C 10 ATP SYNTHASE Alain Dautant*, Jean Velours and Marie-France Giraud* From Université Bordeaux 2, CNRS; Institut de
Supplemental Data CRYSTAL STRUCTURE OF THE MG.ADP-INHIBITED STATE OF THE YEAST F 1 C 10 ATP SYNTHASE Alain Dautant*, Jean Velours and Marie-France Giraud* From Université Bordeaux 2, CNRS; Institut de
Targeting protein-protein interactions: A hot topic in drug discovery
 Michal Kamenicky; Maria Bräuer; Katrin Volk; Kamil Ödner; Christian Klein; Norbert Müller Targeting protein-protein interactions: A hot topic in drug discovery 104 Biomedizin Innovativ patientinnenfokussierte,
Michal Kamenicky; Maria Bräuer; Katrin Volk; Kamil Ödner; Christian Klein; Norbert Müller Targeting protein-protein interactions: A hot topic in drug discovery 104 Biomedizin Innovativ patientinnenfokussierte,
SUPPLEMENTARY INFORMATION
 Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Molecular dynamics simulations of anti-aggregation effect of ibuprofen. Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov
 Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Supplementary Figure 1
 Supplementary Figure 1 The correlation of n-score cutoff and FDR in both CID-only and CID-ETD fragmentation strategies. A bar diagram of different n-score thresholds applied in the search, plotted against
Supplementary Figure 1 The correlation of n-score cutoff and FDR in both CID-only and CID-ETD fragmentation strategies. A bar diagram of different n-score thresholds applied in the search, plotted against
Progress of Compound Library Design Using In-silico Approach for Collaborative Drug Discovery
 21 th /June/2018@CUGM Progress of Compound Library Design Using In-silico Approach for Collaborative Drug Discovery Kaz Ikeda, Ph.D. Keio University Self Introduction Keio University, Tokyo, Japan (Established
21 th /June/2018@CUGM Progress of Compound Library Design Using In-silico Approach for Collaborative Drug Discovery Kaz Ikeda, Ph.D. Keio University Self Introduction Keio University, Tokyo, Japan (Established
The MOLECULES of LIFE
 The MLEULE of LIFE Physical and hemical Principles olutions Manual Prepared by James Fraser and amuel Leachman hapter 1 From enes to RA and Proteins Problems and olutions and Multiple hoice 1. When two
The MLEULE of LIFE Physical and hemical Principles olutions Manual Prepared by James Fraser and amuel Leachman hapter 1 From enes to RA and Proteins Problems and olutions and Multiple hoice 1. When two
Structural insights into Aspergillus fumigatus lectin specificity - AFL binding sites are functionally non-equivalent
 Acta Cryst. (2015). D71, doi:10.1107/s1399004714026595 Supporting information Volume 71 (2015) Supporting information for article: Structural insights into Aspergillus fumigatus lectin specificity - AFL
Acta Cryst. (2015). D71, doi:10.1107/s1399004714026595 Supporting information Volume 71 (2015) Supporting information for article: Structural insights into Aspergillus fumigatus lectin specificity - AFL
SUPPLEMENTARY INFORMATION
 Supplementary Table 1: Amplitudes of three current levels. Level 0 (pa) Level 1 (pa) Level 2 (pa) TrkA- TrkH WT 200 K 0.01 ± 0.01 9.5 ± 0.01 18.7 ± 0.03 200 Na * 0.001 ± 0.01 3.9 ± 0.01 12.5 ± 0.03 200
Supplementary Table 1: Amplitudes of three current levels. Level 0 (pa) Level 1 (pa) Level 2 (pa) TrkA- TrkH WT 200 K 0.01 ± 0.01 9.5 ± 0.01 18.7 ± 0.03 200 Na * 0.001 ± 0.01 3.9 ± 0.01 12.5 ± 0.03 200
Supplementary Information: Table S1. Potential energy and essential dynamics (ED) analysis of the MD simulations of the Bcl-2 complexes under study.
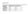 Supplementary Information: Table S1. Potential energy and essential dynamics (ED) analysis of the MD simulations of the Bcl-2 complexes under study. Bcl2 and complex Potential energy (kj/mol) ED 2D projection
Supplementary Information: Table S1. Potential energy and essential dynamics (ED) analysis of the MD simulations of the Bcl-2 complexes under study. Bcl2 and complex Potential energy (kj/mol) ED 2D projection
Modeling for 3D structure prediction
 Modeling for 3D structure prediction What is a predicted structure? A structure that is constructed using as the sole source of information data obtained from computer based data-mining. However, mixing
Modeling for 3D structure prediction What is a predicted structure? A structure that is constructed using as the sole source of information data obtained from computer based data-mining. However, mixing
Supplement information
 Electronic Supplementary Material (ESI) for Physil Chemistry Chemil Physics. This journal is the Owner Societies 216 Supplement information Fullerenol C 6 (OH) 16 prevents amyloid fibrillization of Aβ
Electronic Supplementary Material (ESI) for Physil Chemistry Chemil Physics. This journal is the Owner Societies 216 Supplement information Fullerenol C 6 (OH) 16 prevents amyloid fibrillization of Aβ
Supplementary Figure 3 a. Structural comparison between the two determined structures for the IL 23:MA12 complex. The overall RMSD between the two
 Supplementary Figure 1. Biopanningg and clone enrichment of Alphabody binders against human IL 23. Positive clones in i phage ELISA with optical density (OD) 3 times higher than background are shown for
Supplementary Figure 1. Biopanningg and clone enrichment of Alphabody binders against human IL 23. Positive clones in i phage ELISA with optical density (OD) 3 times higher than background are shown for
Ranjit P. Bahadur Assistant Professor Department of Biotechnology Indian Institute of Technology Kharagpur, India. 1 st November, 2013
 Hydration of protein-rna recognition sites Ranjit P. Bahadur Assistant Professor Department of Biotechnology Indian Institute of Technology Kharagpur, India 1 st November, 2013 Central Dogma of life DNA
Hydration of protein-rna recognition sites Ranjit P. Bahadur Assistant Professor Department of Biotechnology Indian Institute of Technology Kharagpur, India 1 st November, 2013 Central Dogma of life DNA
CSCE555 Bioinformatics. Protein Function Annotation
 CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
Structural basis of PROTAC cooperative recognition for selective protein degradation
 SUPPLEMENTARY INFORMATION Structural basis of PROTAC cooperative recognition for selective protein degradation Morgan S. Gadd 1, Andrea Testa 1, Xavier Lucas 1, Kwok-Ho Chan, Wenzhang Chen, Douglas J.
SUPPLEMENTARY INFORMATION Structural basis of PROTAC cooperative recognition for selective protein degradation Morgan S. Gadd 1, Andrea Testa 1, Xavier Lucas 1, Kwok-Ho Chan, Wenzhang Chen, Douglas J.
Papers listed: Cell2. This weeks papers. Chapt 4. Protein structure and function. The importance of proteins
 1 Papers listed: Cell2 During the semester I will speak of information from several papers. For many of them you will not be required to read these papers, however, you can do so for the fun of it (and
1 Papers listed: Cell2 During the semester I will speak of information from several papers. For many of them you will not be required to read these papers, however, you can do so for the fun of it (and
Protein Structures. 11/19/2002 Lecture 24 1
 Protein Structures 11/19/2002 Lecture 24 1 All 3 figures are cartoons of an amino acid residue. 11/19/2002 Lecture 24 2 Peptide bonds in chains of residues 11/19/2002 Lecture 24 3 Angles φ and ψ in the
Protein Structures 11/19/2002 Lecture 24 1 All 3 figures are cartoons of an amino acid residue. 11/19/2002 Lecture 24 2 Peptide bonds in chains of residues 11/19/2002 Lecture 24 3 Angles φ and ψ in the
Unfolding CspB by means of biased molecular dynamics
 Chapter 4 Unfolding CspB by means of biased molecular dynamics 4.1 Introduction Understanding the mechanism of protein folding has been a major challenge for the last twenty years, as pointed out in the
Chapter 4 Unfolding CspB by means of biased molecular dynamics 4.1 Introduction Understanding the mechanism of protein folding has been a major challenge for the last twenty years, as pointed out in the
Structural Bioinformatics (C3210) Molecular Docking
 Structural Bioinformatics (C3210) Molecular Docking Molecular Recognition, Molecular Docking Molecular recognition is the ability of biomolecules to recognize other biomolecules and selectively interact
Structural Bioinformatics (C3210) Molecular Docking Molecular Recognition, Molecular Docking Molecular recognition is the ability of biomolecules to recognize other biomolecules and selectively interact
HOMOLOGY MODELING. The sequence alignment and template structure are then used to produce a structural model of the target.
 HOMOLOGY MODELING Homology modeling, also known as comparative modeling of protein refers to constructing an atomic-resolution model of the "target" protein from its amino acid sequence and an experimental
HOMOLOGY MODELING Homology modeling, also known as comparative modeling of protein refers to constructing an atomic-resolution model of the "target" protein from its amino acid sequence and an experimental
Supplemental Information
 Supplemental Information Combinatorial Readout of Unmodified H3R2 and Acetylated H3K14 by the Tandem PHD Finger of MOZ Reveals a Regulatory Mechanism for HOXA9 Transcription Yu Qiu 1, Lei Liu 1, Chen Zhao
Supplemental Information Combinatorial Readout of Unmodified H3R2 and Acetylated H3K14 by the Tandem PHD Finger of MOZ Reveals a Regulatory Mechanism for HOXA9 Transcription Yu Qiu 1, Lei Liu 1, Chen Zhao
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability
 Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
User Guide for LeDock
 User Guide for LeDock Hongtao Zhao, PhD Email: htzhao@lephar.com Website: www.lephar.com Copyright 2017 Hongtao Zhao. All rights reserved. Introduction LeDock is flexible small-molecule docking software,
User Guide for LeDock Hongtao Zhao, PhD Email: htzhao@lephar.com Website: www.lephar.com Copyright 2017 Hongtao Zhao. All rights reserved. Introduction LeDock is flexible small-molecule docking software,
Rex-Family Repressor/NADH Complex
 Kasey Royer Michelle Lukosi Rex-Family Repressor/NADH Complex Part A The biological sensing protein that we selected is the Rex-family repressor/nadh complex. We chose this sensor because it is a calcium
Kasey Royer Michelle Lukosi Rex-Family Repressor/NADH Complex Part A The biological sensing protein that we selected is the Rex-family repressor/nadh complex. We chose this sensor because it is a calcium
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
β 2 -microglobulin fibrillation?
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Can small hydrophobic gold nanoparticles inhibit β 2 -microglobulin fibrillation? G. Brancolini,,
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Can small hydrophobic gold nanoparticles inhibit β 2 -microglobulin fibrillation? G. Brancolini,,
Protein Structures. Sequences of amino acid residues 20 different amino acids. Quaternary. Primary. Tertiary. Secondary. 10/8/2002 Lecture 12 1
 Protein Structures Sequences of amino acid residues 20 different amino acids Primary Secondary Tertiary Quaternary 10/8/2002 Lecture 12 1 Angles φ and ψ in the polypeptide chain 10/8/2002 Lecture 12 2
Protein Structures Sequences of amino acid residues 20 different amino acids Primary Secondary Tertiary Quaternary 10/8/2002 Lecture 12 1 Angles φ and ψ in the polypeptide chain 10/8/2002 Lecture 12 2
Equilibrated atomic models of outward-facing P-glycoprotein and effect of ATP binding on structural dynamics (Supplementary Information)
 Equilibrated atomic models of outward-facing P-glycoprotein and effect of ATP binding on structural dynamics (Supplementary Information) Lurong Pan 1 and Stephen G. Aller 2 * 1,2 Department of Pharmacology
Equilibrated atomic models of outward-facing P-glycoprotein and effect of ATP binding on structural dynamics (Supplementary Information) Lurong Pan 1 and Stephen G. Aller 2 * 1,2 Department of Pharmacology
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
2015 AP Biology Unit 2 PRETEST- Introduction to the Cell and Biochemistry
 Name: Class: _ Date: _ 2015 AP Biology Unit 2 PRETEST- Introduction to the Cell and Biochemistry Multiple Choice Identify the choice that best completes the statement or answers the question. 1) In what
Name: Class: _ Date: _ 2015 AP Biology Unit 2 PRETEST- Introduction to the Cell and Biochemistry Multiple Choice Identify the choice that best completes the statement or answers the question. 1) In what
Table 1. Crystallographic data collection, phasing and refinement statistics. Native Hg soaked Mn soaked 1 Mn soaked 2
 Table 1. Crystallographic data collection, phasing and refinement statistics Native Hg soaked Mn soaked 1 Mn soaked 2 Data collection Space group P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 Cell
Table 1. Crystallographic data collection, phasing and refinement statistics Native Hg soaked Mn soaked 1 Mn soaked 2 Data collection Space group P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 Cell
Programme Last week s quiz results + Summary Fold recognition Break Exercise: Modelling remote homologues
 Programme 8.00-8.20 Last week s quiz results + Summary 8.20-9.00 Fold recognition 9.00-9.15 Break 9.15-11.20 Exercise: Modelling remote homologues 11.20-11.40 Summary & discussion 11.40-12.00 Quiz 1 Feedback
Programme 8.00-8.20 Last week s quiz results + Summary 8.20-9.00 Fold recognition 9.00-9.15 Break 9.15-11.20 Exercise: Modelling remote homologues 11.20-11.40 Summary & discussion 11.40-12.00 Quiz 1 Feedback
Homology Modeling. Roberto Lins EPFL - summer semester 2005
 Homology Modeling Roberto Lins EPFL - summer semester 2005 Disclaimer: course material is mainly taken from: P.E. Bourne & H Weissig, Structural Bioinformatics; C.A. Orengo, D.T. Jones & J.M. Thornton,
Homology Modeling Roberto Lins EPFL - summer semester 2005 Disclaimer: course material is mainly taken from: P.E. Bourne & H Weissig, Structural Bioinformatics; C.A. Orengo, D.T. Jones & J.M. Thornton,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11744 Supplementary Table 1. Crystallographic data collection and refinement statistics. Wild-type Se-Met-BcsA-B SmCl 3 -soaked EMTS-soaked Data collection Space
SUPPLEMENTARY INFORMATION doi:10.1038/nature11744 Supplementary Table 1. Crystallographic data collection and refinement statistics. Wild-type Se-Met-BcsA-B SmCl 3 -soaked EMTS-soaked Data collection Space
Structural insight into the binding of C60-derivatives with enoyl-pyruvate transferase from Helicobacter pylori
 www.bioinformation.net Volume 13(6) Hypothesis Structural insight into the binding of C60-derivatives with enoyl-pyruvate transferase from Helicobacter pylori Mohammad Teimouri 1,*, Muhammad Junaid 2,
www.bioinformation.net Volume 13(6) Hypothesis Structural insight into the binding of C60-derivatives with enoyl-pyruvate transferase from Helicobacter pylori Mohammad Teimouri 1,*, Muhammad Junaid 2,
The Molecular Dynamics Method
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
Francisco Melo, Damien Devos, Eric Depiereux and Ernest Feytmans
 From: ISMB-97 Proceedings. Copyright 1997, AAAI (www.aaai.org). All rights reserved. ANOLEA: A www Server to Assess Protein Structures Francisco Melo, Damien Devos, Eric Depiereux and Ernest Feytmans Facultés
From: ISMB-97 Proceedings. Copyright 1997, AAAI (www.aaai.org). All rights reserved. ANOLEA: A www Server to Assess Protein Structures Francisco Melo, Damien Devos, Eric Depiereux and Ernest Feytmans Facultés
Tutorial: Structural Analysis of a Protein-Protein Complex
 Molecular Modeling Section (MMS) Department of Pharmaceutical and Pharmacological Sciences University of Padova Via Marzolo 5-35131 Padova (IT) @contact: stefano.moro@unipd.it Tutorial: Structural Analysis
Molecular Modeling Section (MMS) Department of Pharmaceutical and Pharmacological Sciences University of Padova Via Marzolo 5-35131 Padova (IT) @contact: stefano.moro@unipd.it Tutorial: Structural Analysis
A primer on pharmacology pharmacodynamics
 A primer on pharmacology pharmacodynamics Drug binding & effect Universidade do Algarve Faro 2017 by Ferdi Engels, Ph.D. 1 Pharmacodynamics Relation with pharmacokinetics? dosage plasma concentration site
A primer on pharmacology pharmacodynamics Drug binding & effect Universidade do Algarve Faro 2017 by Ferdi Engels, Ph.D. 1 Pharmacodynamics Relation with pharmacokinetics? dosage plasma concentration site
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Examples of Protein Modeling. Protein Modeling. Primary Structure. Protein Structure Description. Protein Sequence Sources. Importing Sequences to MOE
 Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
Nature Structural and Molecular Biology: doi: /nsmb.2938
 Supplementary Figure 1 Characterization of designed leucine-rich-repeat proteins. (a) Water-mediate hydrogen-bond network is frequently visible in the convex region of LRR crystal structures. Examples
Supplementary Figure 1 Characterization of designed leucine-rich-repeat proteins. (a) Water-mediate hydrogen-bond network is frequently visible in the convex region of LRR crystal structures. Examples
Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits. AhpC and AhpF from Escherichia coli
 Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits AhpC and AhpF from Escherichia coli Phat Vinh Dip 1,#, Neelagandan Kamariah 2,#, Malathy Sony Subramanian Manimekalai
Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits AhpC and AhpF from Escherichia coli Phat Vinh Dip 1,#, Neelagandan Kamariah 2,#, Malathy Sony Subramanian Manimekalai
Introduction to Comparative Protein Modeling. Chapter 4 Part I
 Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
What is the central dogma of biology?
 Bellringer What is the central dogma of biology? A. RNA DNA Protein B. DNA Protein Gene C. DNA Gene RNA D. DNA RNA Protein Review of DNA processes Replication (7.1) Transcription(7.2) Translation(7.3)
Bellringer What is the central dogma of biology? A. RNA DNA Protein B. DNA Protein Gene C. DNA Gene RNA D. DNA RNA Protein Review of DNA processes Replication (7.1) Transcription(7.2) Translation(7.3)
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12045 Supplementary Table 1 Data collection and refinement statistics. Native Pt-SAD X-ray source SSRF BL17U SPring-8 BL41XU Wavelength (Å) 0.97947 1.07171 Space group P2 1 2 1 2 1 P2
doi:10.1038/nature12045 Supplementary Table 1 Data collection and refinement statistics. Native Pt-SAD X-ray source SSRF BL17U SPring-8 BL41XU Wavelength (Å) 0.97947 1.07171 Space group P2 1 2 1 2 1 P2
Advanced Certificate in Principles in Protein Structure. You will be given a start time with your exam instructions
 BIRKBECK COLLEGE (University of London) Advanced Certificate in Principles in Protein Structure MSc Structural Molecular Biology Date: Thursday, 1st September 2011 Time: 3 hours You will be given a start
BIRKBECK COLLEGE (University of London) Advanced Certificate in Principles in Protein Structure MSc Structural Molecular Biology Date: Thursday, 1st September 2011 Time: 3 hours You will be given a start
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1 Protein sequence alignment of Vibrionaceae with either a 40-residue insertion or a 44-residue insertion. Identical residues are indicated by red background.
SUPPLEMENTARY FIGURES Supplementary Figure 1 Protein sequence alignment of Vibrionaceae with either a 40-residue insertion or a 44-residue insertion. Identical residues are indicated by red background.
Structure-Function Relationship of Cytoplasmic and Nuclear IkB Proteins: An In Silico Analysis
 Structure-Function Relationship of Cytoplasmic and Nuclear IkB Proteins: An In Silico Analysis Balachandran Manavalan 1, Shaherin Basith 1, Yong-Min Choi 1, Gwang Lee 1,2, Sangdun Choi 1 * 1 Department
Structure-Function Relationship of Cytoplasmic and Nuclear IkB Proteins: An In Silico Analysis Balachandran Manavalan 1, Shaherin Basith 1, Yong-Min Choi 1, Gwang Lee 1,2, Sangdun Choi 1 * 1 Department
schematic diagram; EGF binding, dimerization, phosphorylation, Grb2 binding, etc.
 Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
BSc and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes Marking
Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes Marking
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase Cwc27
 Acta Cryst. (2014). D70, doi:10.1107/s1399004714021695 Supporting information Volume 70 (2014) Supporting information for article: Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase
Acta Cryst. (2014). D70, doi:10.1107/s1399004714021695 Supporting information Volume 70 (2014) Supporting information for article: Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase
The structure of vanadium nitrogenase reveals an unusual bridging ligand
 SUPPLEMENTARY INFORMATION The structure of vanadium nitrogenase reveals an unusual bridging ligand Daniel Sippel and Oliver Einsle Lehrstuhl Biochemie, Institut für Biochemie, Albert-Ludwigs-Universität
SUPPLEMENTARY INFORMATION The structure of vanadium nitrogenase reveals an unusual bridging ligand Daniel Sippel and Oliver Einsle Lehrstuhl Biochemie, Institut für Biochemie, Albert-Ludwigs-Universität
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
CAP 5510 Lecture 3 Protein Structures
 CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
