Review Article Putting Markov Chains Back into Markov Chain Monte Carlo
|
|
|
- Jean Lyons
- 5 years ago
- Views:
Transcription
1 Hindawi Publishing Corporation Journal of Applied Mathematics and Decision Sciences Volume 2007, Article ID 98086, 13 pages doi: /2007/98086 Review Article Putting Markov Chains Back into Markov Chain Monte Carlo Richard J. Barker and Matthew R. Schofield Received 7 May 2007; Accepted 8 August 2007 Recommended by Graeme Charles Wake Markov chain theory plays an important role in statistical inference both in the formulation of models for data and in the construction of efficient algorithms for inference. The use of Markov chains in modeling data has a long history, however the use of Markov chain theory in developing algorithms for statistical inference has only become popular recently. Using mark-recapture models as an illustration, we show how Markov chains can be used for developing demographic models and also in developing efficient algorithms for inference. We anticipate that a major area of future research involving mark-recapture data will be the development of hierarchical models that lead to better demographic models that account for all uncertainties in the analysis. A key issue is determining when the chains produced by Markov chain Monte Carlo sampling have converged. Copyright 2007 R. J. Barker and M. R. Schofield. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. 1. Introduction Markov chains, and related stochastic models, have long played an important role in helping ecologists understand population dynamics. In the main, this has been through the application of probability models to the problem of predicting the realized dynamics of plant and animal populations over time. In this context, the challenge is to construct models that are relatively simple, in terms of the numbers of parameters and the relationships between (and among) parameters and state variables, and yet that are able to provide a reasonable approximation to the sorts of dynamics typically observed in nature.
2 2 Journal of Applied Mathematics and Decision Sciences The development of these models has been driven by ecologists motivated by an interest in underlying theory, and ecologists with applications in mind. These applications are generally based on inference about the long-term dynamics of the study population. Examples include predicting the time to possible extinction for threatened populations or establishing safe levels of harvest for exploited populations. Complementary to the problem of developing theoretical models that predict the temporal dynamics of populations has been the development of a body of theory for estimating population parameters. This estimation theory is the main emphasis in the remainder of this contribution. Almost universally, these models treat quantities such as population size at time t, N t, or the numbers of individuals born between t and t + Δt, as fixed quantities to be predicted (here we use prediction in the sense of predicting the value of an unobserved realization of a random variable as well as future realizations). In contrast, the population models discussed above treat these same quantities as random variables whose behavior we are interested in describing. A particularly important class of estimation models for population dynamics is the mark-recapture model [1]. Mark-recapture data comprises repeated measures on individual animals in the population obtained through samples of animals drawn from the population (usually) at discrete sample times t 1,...,t k. Because the capture process is imperfect, not all animals in the population at time t j are captured, and a model is required to describe this process of repeated imperfect captures. In the simplest case, the population is regarded as closed to births and deaths and so the population size at time t j is the same for all k sample occasions. Studies of the dynamics of the population can then be based on repeated experiments generating a sequence of abundance estimates. A more interesting class of models is the open population models in which individuals may enter (through birth or immigration) or leave (through death or emigration) during the interval (t 1,t k ). Here the challenge is to model the sequence of captures of animals in terms of parameters of demographic interest. These are usually survival probabilities S j, and parameters that describe the birth process. Quantities such as N j, the abundance of animals at the time of sample j, andb j, the number of animals born between samples j and j + 1, are then predicted from the model. Traditionally, inference for mark-recapture models has been based on maximum likelihood. Importantly, demographic models for {N t } have played no role in these estimation models. Inference about the {N t } process has instead been based on ad hoc methods for summarizing sequences of estimates such as { N t }. Recently there have been many developments that apply Bayesian inference methods to mark recapture models based on Markov chain Monte Carlo (MCMC). Here the utility of Markov chain theory appears in a fundamentally different context to that described above for population modeling; it arises as a tool for inference. An exciting feature of Bayesian inference methods based on MCMC is that fitting complicated hierarchical models has become feasible. Hierarchical models provide a link between the demographic models and estimation models in a way that should lead to better and more relevant inference. It is recent developments in Markov chain theory, in particular Gibbs sampling [2] and reversible jump Markov chain Monte Carlo [3], that allow this link to take place.
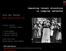
3 R.J.BarkerandM.R.Schofield 3 X i1 X i2 X i3 X ik Y i1 Y i2 Y i3 Y ik Figure 2.1. Directed acyclic graph for the hidden Markov model for mark-recapture data. 2. Open population capture-recapture models Define ν as the total number of animals alive on at least one of the sampling occasions t j (j = 1,...,k). Let Y ij be the indicator that animal i (i = 1,...,ν) is caught in sample j, which takes place at time t j. Any animal that is caught for the first time in a particular sample is marked with some form of unique tag and then released, with any recaptures noted. The data are of the form y ={y ij } for i = 1,...,u. andj = 1,...,k, whereu j is the number of unmarked animals caught in sample j and u. = j u j is the total number of animals caught at least once. For animal i, the set of values y i ={y i1,..., y ik } is called the capture history and these provide censored information on the time of birth and death. We know that the time of birth occurred before the sample time corresponding to the first nonzero value of y i and the time of death occurred sometime after the sample time corresponding to the last nonzero value of y i Hidden Markov model. The problem with capture-recapture methods is that not all animals in the population are caught; this causes the censoring of the birth and death intervals. Define X ij as the indicator of the event that animal i is alive and in the study population at time t j.ifx ={x ij }, i = 1,...,ν, j = 1,...,k, were observed for an animal population in which ν animals were ever alive and available for capture on at least one of the k sample occasions, then inference about the parameters {S j } and {β j } would be straight-forward. It would also be straight-forward to calculate observed values of the random variables N j = i x ij. Instead of observing x ij,weobservey ij with the joint distribution of the sequence of pairs {X ij,y ij } described by a hidden Markov model (Figure 2.1). Importantly, y does not include the history given by the vector of k zeros associated with animals that were available for capture at sometime during the experiment but never caught. Conditional on the event that animal i is alive (and available for capture) on at least one of the capture occasions t j, the sequence (X i1,...,x ik ) can then be modeled as a threestate Markov chain. X ij = 0 corresponds to the event that animal i has not yet entered the population. X ij = 1 corresponds to the event that animal i is in the population and alive, and X ij = 2 corresponds to animal i being dead. The possible transitions are 0 0, 0 1,
4 4 Journal of Applied Mathematics and Decision Sciences 1 1, 1 2, and 2 2. Define: β 0 as the probability that an animal is born before the start of the experiment, given that it is alive at some time during the experiment, β j as the probability that an animal is born between the times of sample j and j + 1, given that it was available for capture at sometime during the experiment, S j as the probability that an animal alive at the time of sample j is still alive at the start of sample j +1. Conditioning on the ν individuals that are alive and available for capture at sometime during the experiment means that β 0 + β β k 1 = 1. If is used to denote the event that animal i is alive and available for capture at sometime during the experiment, then Pr ( X i1 = x i1 ) β 0, x i1 = 1, = 1 β 0, x i1 = 0, (2.1) and for j = 2,...,k 1, 1, x ij = 2,x ij 1 = 2, 0, x ij = 1,x ij 1 = 2, 0, x ij = 0,x ij 1 = 2, Pr ( X ij = x ij x ij 1, ) 1 S j, x ij = 2,x ij 1 = 1, = S j, x ij = 1,x ij 1 = 1, 0, x ij = 0,x ij 1 = 1, 0, x ij = 2,x ij 1 = 1, β j 1, x ij = 1,x ij 1 = 0, 1 β j 1, x ij = 0,x ij 1 = 0, (2.2) where β j = β j / k 1 h=j β h. Note that we exclude from the study population individuals that are born and then die during (t j,t j+1 ). Clearly these individuals are invisible to the markrecapture experiment Observed data likelihood. A common assumption in mark recapture models is that y ij x ij is the outcome of a Bernoulli trial with probability p j if x ij = 1 or 0 otherwise. That is, only those animals alive and in the population at the time of sample j areatrisk of being caught, which happens with probability p j. The standard approach to fitting this model is to derive an observed data likelihood from the marginal distribution with pdf [y ν,s,β, p] which is described by summing up across all possible sample paths for X that are compatible with the data (here we use the notation [y x] to denote the pdf of the distribution for the random variable Y, evaluated at y, conditional on X, evaluated at x). Using the notation y ={y ij } (i = 1,...,u.; j = 1,...,k) andu = (u 1,...,u k ) the likelihood can be expressed as [y ν,s,β, p] = [u. ν][u u.,β,s][y u,s, p], (2.3)
5 R.J.BarkerandM.R.Schofield 5 where [u. ν] describes a binomial distribution with index ν and parameter π 0 and [u u.] a multinomial distribution with index u.andparametervectorξ. Both π 0 and ξ are complicated functions of the parameters S and β that account for the censoring of the birth times in y. Ifweletψ j denote the probability that an animal is available for capture in sample j but has not yet been caught, then ψ j+1 = ψ j ( 1 p j ) Sj + β j (2.4) for j = 2,...,k 1, with ψ 1 = β 0. Such an animal is first caught in sample j with probability ψ j p j hence π 0 = 1 t ψ j p j, j=1 ξ j = ψ j p j 1 π 0. (2.5) The term [y u,s, p] describes the celebrated Cormack-Jolly-Seber model (see [4 6]). If we index the first sample in which an animal i was caught by r i and the last sample in which animal i was seen by l i, then we can write u. [y u,s, p] = i=1 l i p yij j j=r i ( 1 p j ) 1 yij χ li, (2.6) where the term χ j, which accounts for the censoring of time of death, can be defined recursively as 1, j = k, χ j = ( ) 1 S j + S j 1 p j+1 χj+1, j = 1,...,k 1. (2.7) A nice feature of the observed data likelihood (2.3) isthatitisstraightforwardtofind maximum likelihood estimators. [7] showed that the parameter ν is not identifiable, however the partial likelihood [u u.,β,s][y u,s, p] obtained by conditioning on u. contains all practically useful information on the identifiable parameters in the model. Closedform solutions to the ML equations exist for all identifiable parameters in this partiallikelihood. The model can also be easily generalized to allow parameters to be individual-specific, by introducing covariates, or to allow captures to depend on the earlier capture history of the animal. The model (2.3) has also been extended by reparameterizing the model in terms of per capita birth rate f j [7, 8] and an index to population growth rate [8] λ j = S j + f j = E[ N j+1 N j,ν ] E [ N j ν ]. (2.8)
6 6 Journal of Applied Mathematics and Decision Sciences The complete model (2.3), which we refer to as the Crosbie-Manly-Arnason-Schwarz model, was first described in [9] building on earlier work of [10]. It differs from the wellknown Jolly-Seber (JS) model ([5, 6]) in the way that captures of unmarked animals are modeled. In the JS model, the terms [u. ν][u u.] (2.9) are replaced by [u U] (2.10) and the elements of U = (U 1,...,U k ),whereu j is the number of unmarked animals in the population at the time of sample j, are treated as unknown parameters to be estimated. While historically important, this approach does not allow the reparameterizations in terms of f j and λ j described above as f j and λ j cannot be written as deterministic functions of the parameters S j and β j.also,theu j parameters are of little demographic interest although predictors of N j and B j exist Complete data likelihood. An alternative to using the observed data likelihood is to describe the model in terms of the complete data likelihood (CDL) [11]. The CDL is the likelihood of all data, both missing and observed. The observed data are (i) the values y ij for all u. individuals that we observed at least once, (ii) the censored information about X that we obtain from y. The missing data are (i) the unknown number of individuals that were available for capture but not caught (we include this by specifying ν as a parameter), (ii) the realized but unknown values of X ij, x ij. Including both the observed and missing data gives the CDL that we can express as [x, y S,β, p,ν] = [y p,x,ν][x S,β,ν]. (2.11) The CDL has been naturally factored into a term that describes how the data were corrupted, [y p,x,ν], and a term that models the underlying birth and death processes of interest, [x S,β,ν]. As for the observed data likelihood section, we model the corruption by assuming that an individual that was alive in sample j (x ij = 1) was observed in sample j (y ij = 1) with probability p j. We model the birth and death process as in the Markov chain model for X described in Section 2.1. Even though the CDL provides a natural approach to looking at the problem, we must still integrate over the missing data in order to obtain valid inference. Computational techniques such as Markov chain Monte Carlo (MCMC, discussed below) can be used that iteratively integrate out all missing data, allowing models to be specified in terms of the CDL. This means that in each iteration of the MCMC chain we need values for the missing quantities x and ν (and all other parameters) that were obtained from the
7 R.J.BarkerandM.R.Schofield 7 posterior distribution of all unknowns. One such MCMC algorithm we can use is Gibbs sampling, described in the following section Markov chain Monte Carlo. The Gibbs sampler, also known as alternating conditional sampling, is a remarkable algorithm for efficiently constructing a Markov chain for complex joint probability distributions [Z 1,...,Z k θ] by sampling from the full conditional distributions [Z i {Z j } j i,θ] of each component. The stationary distribution of this Markov chain has the target density [Z 1,...,Z k θ]. A particularly useful feature of the Gibbs sampler is that the Markov chain can be constructed even though the target joint probability density may only be known up to the normalizing constant. This has led to a resurgence of interest in Bayesian inference which, historically, has been held back by the need for high-dimension integration needed to normalize posterior probability densities. Here, we outline the construction of a Gibbs sampler whose target density is the CDL described in the previous section. Once we have collected our mark-recapture data we have data y, that is known; and the unknown are any parameters and any unknown realized values of random variables of interest. For the Crosbie-Manly-Arnason-Schwarz model (2.3), the parameters are p 1, p 2,..., p k ; S 1,...,S k 1, β 0,β 1,...,β k 1,andν. In addition unknown realized random variables of interest (might) include N 1,...,N k, B 0,B 1,..., B k 1. Starting with (2.3) and specifying independent beta Be(α p,γ p ) prior distributions for the parameters p j, we can show that the full conditional distribution for p j is a beta Be(n j + α p,n j n j + γ p )forj = 1,...,k, andwheren j is the total number of individuals caught at time of sample j. If we specify independent beta Be(α S,γ S ) prior distributions for the parameters S j,we obtain beta Be(N j D j + α S,D j + γ S ) full conditional distributions for j = 1,...,k 1. If we specify independent beta Be(α β,γ β ) prior distributions for the parameters β j,we obtain beta Be(B j + α β,n j h=0 B h + γ β ) full conditional distributions for j = 1,...,k 2 (note that β k 1 = 1). If desired, we can transform the generated values of β j to any other birth parameter, such as β j or η j.forexample,β j, j = 0,...,k 1, is obtained by taking β j, j = 0, β j = j 1 β ( ) (2.12) j 1 β h, j = 1,...,k 2. h=0 We also need to calculate the full conditional probability of the missing values of x. This can be done a number of ways, but we choose to represent the information in x by matrices that give the interval censored times of birth and death, denoted by b and d, respectively. The value b ij = 1 means individual i was born between sample j and j + 1withb ij = 0 otherwise. The value b i0 = 1 means the individual was born before the study began. The value d ij = 1 means that individual i died between sample j and j +1 with d ij = 0 otherwise. The value d ik = 1 means the individual was still alive at the end of the study. The assumptions about the underlying birth and death processes impose
8 8 Journal of Applied Mathematics and Decision Sciences restrictions on the values of b and d, k 1 b ij = 1, j=0 k d ij = 1, d ij b ij 0 i, j. (2.13) j=1 The matrices b and d are censored. For the u. individuals that were ever observed we know that they were not born after first capture, b ij = 0, j = r i,...,k,wherer i indexes the sample in which animal i first appeared. Likewise we know that they did not die before thelastcapture,d ij = 0, j = 1,...,l i,wherel i indexes the sample in which animal i last appeared. For the ν u. individuals that were never observed we have no information about either b or d. The full conditional distribution of the unknown values of b for individual i is a multinomial distribution with probability vector γ b,where γ b j = ζb j λ b i 1 h=0 ζb h, (2.14) λb ζ b i 1 ( ) j =β j S h 1 ph, h=j and λ b i is the time of first capture r i for all individuals observed, and it is the last period in which the individual was alive (obtained from d) for individuals i = u. +1,...,ν that were not observed. The full conditional distribution of the unknown values of d for individual i is a multinomial distribution with probability vector γ d,where γ d j = ζ d j = ζd j t h=λ ζ d d i h j 1 S h, j h=λ d i h=λ d i +1 j 1 j S h h=λ d i h=λ d i +1 ( 1 ph )( 1 Sj ), j<k, ( 1 ph ), j = k, (2.15) and λ d i is the time of last capture l i for all individuals observed, and it is the first period in which the individual was alive (obtained from b) for individuals i = u. +1,...,ν that were not observed. For the parameter ν we specify a discrete uniform prior distribution DU(0,κ ν ). Obtaining a sample from the full conditional distribution for ν has two problems: (1) the full conditional distribution is only known to a proportionality constant, (2) the value of the parameter changes the dimension of other unknowns in the model.
9 R.J.BarkerandM.R.Schofield 9 Table 2.1. Summary statistics for fitting the Crosbie-Manly-Arnason-Schwarz model to the meadow vole data. The statistic u j is the number of individuals caught in sample j, R j is the number of individuals released following sample j, q j is the number of the R j that were ever recaptured, and m j is the number of marked animals caught in sample j. Month u j = R j q j m j Table 2.2. Maximum likelihood estimates of identifiable parameters in the Crosbie-Manly-Arnason- Schwarz model fitted to the meadow vole data. 95% C. I. Parameter Estimate SE lower upper S S S S p p p p f f f Toovercomethefirstproblemwecanuseasamplingscheme,suchastheMetropolis- Hastings algorithm, that allows us to sample from a distribution that we only know up to the proportionality constant. The second problem requires an extension of the Metropolis-Hastings algorithm called reversible jump Markov chain Monte Carlo [3] where there is dimension matching to ensure reversibility of the Markov chain. Details of the reversible-jump sampler are given in [11] Example. We illustrate the use of the MCMC algorithm described in the previous section by fitting the model (2.3) to meadow vole (Microtus pensylvannicus) datacollected at the Patuxent Wildlife Research Center, Laurel, Md, USA [12]. The meadow vole population was trapped at one-month intervals; untagged animals were tagged and released, tagged animals had their identity recorded and were then released. The model (2.3) can be fitted using sufficient statistics (Table 2.1) and closed-form solutions to the ML equations [7]. The ML estimates (Table 2.2) and the posterior summaries from fitting the model using the Gibbs sample algorithm (Table 2.3) of the previous section are
10 10 Journal of Applied Mathematics and Decision Sciences Table 2.3. Posterior distribution summary statistics for the Gibbs sampler posterior simulations and for the identifiable parameters in the Crosbie-Manly-Arnason-Schwarz model fitted to the meadow vole data. Included are predictions of the abundances N j at the time of each sample and B j,the number of individuals born in (t j,t j+1 ). Quantile Parameters Mean Median S S S S f f f p p p p Realized random variables N N N N B B B in close agreement. For the Gibbs sampler we used beta Be(1,1) priors for S i, p i,andβ i with the f i parameters found deterministically as a function of the β i and S i parameters [7]. We used a discrete uniform DU(200000) prior for ν. A nice feature of the MCMC approach is that it is relatively simple to obtain a posterior distribution for unobserved random variables of interest such as N i and B i,alsoreported in Table 2.3. Predictions for these in a likelihood analysis must be based on method of moment-type estimators as the N i and B i parameters do not explicitly appear in the likelihood function. Obtaining predictions using MCMC is straightforward with values sampled from the posterior predictive distribution. In practice, we simply use the set of b and d values obtained in the Markov chain and for each iteration compute the current value of the variable of interest. For example, [11] show that N j = ν i=1 ( j 1 b ih d ih ). (2.16) h=0 Progress of the Markov chains is instructive (Figure 2.2). Convergence of the Markov chains for the identifiable parameters S 1,...,S k 2 ; p 2,..., p k 1 ; f 2,..., f k 2 is rapid and the j 1 h=1
11 R.J.BarkerandM.R.Schofield S p S 1 S p 2 p ν ν ν Figure 2.2. Gibbs sampler Markov chains for the first 1000 iterations (column of plots on left-hand side), for iterations following a discarded burn-in of iterations (middle column of plots) and posterior density plots (right-hand column of plots) for the identifiable parameters S 1 and p 1 and the nonidentifiable parameter ν, for the Gibbs sampler posterior simulations from fitting the Crosbie-Manly-Arnason-Schwarz model fitted to the meadow vole data. chains appear well-mixed after a few thousand iterations. Even chains for nonidentifiable parameters such as ν appear well mixed after a few thousand iterations. 3. Discussion Markov chain theory plays an important role in statistical inference both in the formulation of models for data and in the construction of efficient algorithms for inference. The ability to compactly represent stochastic processes that evolve over time has meant that Markov chain theory has played an important role in describing animal and plant population dynamics. The fact that each generation gives rise to the next makes the Markov property a natural starting point in developing population models. The role of the Markov chain in statistical inference is a much more recent development. Although the Metropolis-Hastings algorithm first appeared in 1953 [13], with roots back to the Manhattan project, and the Gibbs sampler in 1984 [2] MCMCentered into widespread use in statistics only in the last 15 years. This followed [14] who illustrated the application of Gibbs sampling to a variety of statistical problems. MCMC is particularly useful for fitting hierarchical models using Bayesian inference methods. A simple example of a hierarchical model is one where we have two components
12 12 Journal of Applied Mathematics and Decision Sciences to the model [Y θ][θ γ]. Here, conditional on parameters θ,therandomvariabley has density [Y θ]. The parameters θ are themselves modeled as random variables and have distribution with pdf [θ γ]. In order to fit this model by a method such as maximum likelihood (ML), we must find the marginal likelihood: (θ y] = θ[y θ][θ γ]dθ. In practice this integral is very difficult, if not practically impossible, in most cases of interest. The equivalent Bayesian problem is to find the posterior marginal density [γ y] [Y θ][θ γ][γ] which involves essentially the same integral. However, MCMC allows us to approximate a sample from the required density but we must specify a prior distribution from γ. Thus, Bayesian inference methods based on MCMC allows inference for the model [Y θ][θ γ] butthesameproblemmaybedifficult or impossible using standard ML theory. In the context of mark-recapture modeling, the advent of MCMC has meant that it is now possible to fit complex hierarchical models. Starting with a model such as (2.3), it is relatively easy to add hierarchical structure. In the context of the hidden Markov model described in Figure 2.1, we mean that it is relativelystraightforwardto add in components that allow us to further model [X β,s,ν]. These issues have been explored by [7, 11]. [7] showed how the model (2.3) could be used to explore relationships among parameters and in particular considered a hierarchical model in which it was assumed that survival probabilities were related to per capita birth rates (functions of the β parameters). Such a model would make sense when there were common environmental influences on both survival probabilities and birth rates. Such an analysis is virtually impossible using ML methods. Similarly, [11] showed how the CDL discussed above could be used in hierarchical modeling, and in particular fit models in which there is feedback between the unknown random variables N j and survival S j or per capita birth rates f j.thepractical benefit of hierarchical modeling is that a much richer class of models is available for the ecologist to explore, and the advent of MCMC means that methods of inference are fully able to incorporate all different sources of uncertainty in the analysis. A particularly important technical issue with MCMC is that of convergence. A typical problem in Markov chain theory is the determination of a stationary distribution given the transition kernel. In MCMC, the problem is reversed, and is one of constructing a transition kernel that has, as its stationary distribution, the target distribution of interest. Under mild conditions, the Metropolis-Hastings algorithm and the Gibbs sampler as a special case are methods of constructing transition kernels that have the required property. An important practical problem is the development of rules for helping determine when the chain has converged. Experience indicates that for some problems, convergence can be rapid and in others it can be slow. Rapid convergence appears to be an associated with likelihood functions that are well behaved. Methods for assessing convergence are ad hoc, and generally based on informal graphical assessment (Figure 2.2) or computation of simple statistics, such as the Brooks-Gelman-Rubin diagnostic statistic. There is surprisingly little underlying theory about the rate at which Markov chains constructed in MCMC should converge to the stationary distribution that can be applied to the practical problems of MCMC. Clearly, time to stationarity will be a property of the model. Again, observation indicates that problems for which the likelihood function are well behaved tendtoconvergerapidly.theworkin[15, 16] is important in this regard.
13 R.J.BarkerandM.R.Schofield 13 References [1] G.A.F.Seber,The Estimation of Animal Abundance and Related Parameters, Macmillan,New York, NY, USA, 2nd edition, [2] S. Geman and D. Geman, Stochastic relaxation, Gibbs distributions and the Bayesian restoration of images, IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 6, no. 6, pp , [3] P. J. Green, Reversible jump Markov chain Monte Carlo computation and Bayesian model determination, Biometrika, vol. 82, no. 4, pp , [4] R. M. Cormack, Estimates of survival from the sighting of marked animals, Biometrika, vol. 51, pp , [5] G. M. Jolly, Explicit estimates from capture-recapture data with both death and immigrationstochastic model, Biometrika, vol. 52, pp , [6] G. A. F. Seber, A note on the multiple-recapture census, Biometrika, vol. 52, pp , [7] W. A. Link and R. J. Barker, Modeling association among demographic parameters in analysis of open population capture-recapture data, Biometrics, vol. 61, no. 1, pp , [8] R. Pradel, Utilization of capture-mark-recapture for the study of recruitment and population growth rate, Biometrics, vol. 52, no. 2, pp , [9] C. J. Schwarz and A. N. Arnason, A general methodology for the analysis of capture-recapture experiments in open populations, Biometrics, vol. 52, no. 3, pp , [10] S. F. Crosbie and B. F. J. Manly, Parsimonious modelling of capture-mark-recapture studies, Biometrics, vol. 41, no. 2, pp , [11] M. R. Schofield and R. J. Barker, A unified capture-recapture framework, to appear in Journal of Agricultural, Biological and Ecological Statistics. [12] B. K. Williams, J. D. Nichols, and M. J. Conroy, Analysis and Management of Animal Populations, Academic Press, London, UK, [13] N. Metropolis, A. W. Rosenbluth, M. N. Rosenbluth, A. H. Teller, and E. Teller, Equations of state calculations by fast computing machines, The Journal of Chemical Physics, vol. 21, no. 6, pp , [14] A. E. Gelfand and A. F. M. Smith, Sampling-based approaches to calculating marginal densities, Journal of the American Statistical Association, vol. 85, no. 410, pp , [15] J. J. Hunter, Mixing times with applications to perturbed Markov chains, Linear Algebra and Its Applications, vol. 417, no. 1, pp , [16] J. J. Hunter, Coupling and mixing times in a Markov chains, Research Letters in Information and Mathematical Science, vol. 11, pp. 1 22, Richard J. Barker: Department of Mathematics and Statistics, University of Otago, P.O. Box 56, Dunedin 9010, New Zealand address: rbarker@maths.otago.ac.nz Matthew R. Schofield: Department of Mathematics and Statistics, University of Otago, P.O. Box 56, Dunedin 9010, New Zealand address: mschofield@maths.otago.ac.nz
Introduction to capture-markrecapture
 E-iNET Workshop, University of Kent, December 2014 Introduction to capture-markrecapture models Rachel McCrea Overview Introduction Lincoln-Petersen estimate Maximum-likelihood theory* Capture-mark-recapture
E-iNET Workshop, University of Kent, December 2014 Introduction to capture-markrecapture models Rachel McCrea Overview Introduction Lincoln-Petersen estimate Maximum-likelihood theory* Capture-mark-recapture
Markov Chain Monte Carlo methods
 Markov Chain Monte Carlo methods By Oleg Makhnin 1 Introduction a b c M = d e f g h i 0 f(x)dx 1.1 Motivation 1.1.1 Just here Supresses numbering 1.1.2 After this 1.2 Literature 2 Method 2.1 New math As
Markov Chain Monte Carlo methods By Oleg Makhnin 1 Introduction a b c M = d e f g h i 0 f(x)dx 1.1 Motivation 1.1.1 Just here Supresses numbering 1.1.2 After this 1.2 Literature 2 Method 2.1 New math As
An Extension of the Cormack Jolly Seber Model for Continuous Covariates with Application to Microtus pennsylvanicus
 Biometrics 62, 142 149 March 2006 DOI: 10.1111/j.1541-0420.2005.00399.x An Extension of the Cormack Jolly Seber Model for Continuous Covariates with Application to Microtus pennsylvanicus S. J. Bonner
Biometrics 62, 142 149 March 2006 DOI: 10.1111/j.1541-0420.2005.00399.x An Extension of the Cormack Jolly Seber Model for Continuous Covariates with Application to Microtus pennsylvanicus S. J. Bonner
SUPPLEMENT TO EXTENDING THE LATENT MULTINOMIAL MODEL WITH COMPLEX ERROR PROCESSES AND DYNAMIC MARKOV BASES
 SUPPLEMENT TO EXTENDING THE LATENT MULTINOMIAL MODEL WITH COMPLEX ERROR PROCESSES AND DYNAMIC MARKOV BASES By Simon J Bonner, Matthew R Schofield, Patrik Noren Steven J Price University of Western Ontario,
SUPPLEMENT TO EXTENDING THE LATENT MULTINOMIAL MODEL WITH COMPLEX ERROR PROCESSES AND DYNAMIC MARKOV BASES By Simon J Bonner, Matthew R Schofield, Patrik Noren Steven J Price University of Western Ontario,
Markov Chain Monte Carlo methods
 Markov Chain Monte Carlo methods Tomas McKelvey and Lennart Svensson Signal Processing Group Department of Signals and Systems Chalmers University of Technology, Sweden November 26, 2012 Today s learning
Markov Chain Monte Carlo methods Tomas McKelvey and Lennart Svensson Signal Processing Group Department of Signals and Systems Chalmers University of Technology, Sweden November 26, 2012 Today s learning
Session 3A: Markov chain Monte Carlo (MCMC)
 Session 3A: Markov chain Monte Carlo (MCMC) John Geweke Bayesian Econometrics and its Applications August 15, 2012 ohn Geweke Bayesian Econometrics and its Session Applications 3A: Markov () chain Monte
Session 3A: Markov chain Monte Carlo (MCMC) John Geweke Bayesian Econometrics and its Applications August 15, 2012 ohn Geweke Bayesian Econometrics and its Session Applications 3A: Markov () chain Monte
Bayesian Methods for Machine Learning
 Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Markov chain Monte Carlo methods for visual tracking
 Markov chain Monte Carlo methods for visual tracking Ray Luo rluo@cory.eecs.berkeley.edu Department of Electrical Engineering and Computer Sciences University of California, Berkeley Berkeley, CA 94720
Markov chain Monte Carlo methods for visual tracking Ray Luo rluo@cory.eecs.berkeley.edu Department of Electrical Engineering and Computer Sciences University of California, Berkeley Berkeley, CA 94720
STAT 425: Introduction to Bayesian Analysis
 STAT 425: Introduction to Bayesian Analysis Marina Vannucci Rice University, USA Fall 2017 Marina Vannucci (Rice University, USA) Bayesian Analysis (Part 2) Fall 2017 1 / 19 Part 2: Markov chain Monte
STAT 425: Introduction to Bayesian Analysis Marina Vannucci Rice University, USA Fall 2017 Marina Vannucci (Rice University, USA) Bayesian Analysis (Part 2) Fall 2017 1 / 19 Part 2: Markov chain Monte
Cormack-Jolly-Seber Models
 Cormack-Jolly-Seber Models Estimating Apparent Survival from Mark-Resight Data & Open-Population Models Ch. 17 of WNC, especially sections 17.1 & 17.2 For these models, animals are captured on k occasions
Cormack-Jolly-Seber Models Estimating Apparent Survival from Mark-Resight Data & Open-Population Models Ch. 17 of WNC, especially sections 17.1 & 17.2 For these models, animals are captured on k occasions
Statistical Inference for Stochastic Epidemic Models
 Statistical Inference for Stochastic Epidemic Models George Streftaris 1 and Gavin J. Gibson 1 1 Department of Actuarial Mathematics & Statistics, Heriot-Watt University, Riccarton, Edinburgh EH14 4AS,
Statistical Inference for Stochastic Epidemic Models George Streftaris 1 and Gavin J. Gibson 1 1 Department of Actuarial Mathematics & Statistics, Heriot-Watt University, Riccarton, Edinburgh EH14 4AS,
Principles of Bayesian Inference
 Principles of Bayesian Inference Sudipto Banerjee University of Minnesota July 20th, 2008 1 Bayesian Principles Classical statistics: model parameters are fixed and unknown. A Bayesian thinks of parameters
Principles of Bayesian Inference Sudipto Banerjee University of Minnesota July 20th, 2008 1 Bayesian Principles Classical statistics: model parameters are fixed and unknown. A Bayesian thinks of parameters
(5) Multi-parameter models - Gibbs sampling. ST440/540: Applied Bayesian Analysis
 Summarizing a posterior Given the data and prior the posterior is determined Summarizing the posterior gives parameter estimates, intervals, and hypothesis tests Most of these computations are integrals
Summarizing a posterior Given the data and prior the posterior is determined Summarizing the posterior gives parameter estimates, intervals, and hypothesis tests Most of these computations are integrals
Online appendix to On the stability of the excess sensitivity of aggregate consumption growth in the US
 Online appendix to On the stability of the excess sensitivity of aggregate consumption growth in the US Gerdie Everaert 1, Lorenzo Pozzi 2, and Ruben Schoonackers 3 1 Ghent University & SHERPPA 2 Erasmus
Online appendix to On the stability of the excess sensitivity of aggregate consumption growth in the US Gerdie Everaert 1, Lorenzo Pozzi 2, and Ruben Schoonackers 3 1 Ghent University & SHERPPA 2 Erasmus
Bayesian model selection in graphs by using BDgraph package
 Bayesian model selection in graphs by using BDgraph package A. Mohammadi and E. Wit March 26, 2013 MOTIVATION Flow cytometry data with 11 proteins from Sachs et al. (2005) RESULT FOR CELL SIGNALING DATA
Bayesian model selection in graphs by using BDgraph package A. Mohammadi and E. Wit March 26, 2013 MOTIVATION Flow cytometry data with 11 proteins from Sachs et al. (2005) RESULT FOR CELL SIGNALING DATA
Introduction to Machine Learning CMU-10701
 Introduction to Machine Learning CMU-10701 Markov Chain Monte Carlo Methods Barnabás Póczos & Aarti Singh Contents Markov Chain Monte Carlo Methods Goal & Motivation Sampling Rejection Importance Markov
Introduction to Machine Learning CMU-10701 Markov Chain Monte Carlo Methods Barnabás Póczos & Aarti Singh Contents Markov Chain Monte Carlo Methods Goal & Motivation Sampling Rejection Importance Markov
MH I. Metropolis-Hastings (MH) algorithm is the most popular method of getting dependent samples from a probability distribution
 MH I Metropolis-Hastings (MH) algorithm is the most popular method of getting dependent samples from a probability distribution a lot of Bayesian mehods rely on the use of MH algorithm and it s famous
MH I Metropolis-Hastings (MH) algorithm is the most popular method of getting dependent samples from a probability distribution a lot of Bayesian mehods rely on the use of MH algorithm and it s famous
Webinar Session 1. Introduction to Modern Methods for Analyzing Capture- Recapture Data: Closed Populations 1
 Webinar Session 1. Introduction to Modern Methods for Analyzing Capture- Recapture Data: Closed Populations 1 b y Bryan F.J. Manly Western Ecosystems Technology Inc. Cheyenne, Wyoming bmanly@west-inc.com
Webinar Session 1. Introduction to Modern Methods for Analyzing Capture- Recapture Data: Closed Populations 1 b y Bryan F.J. Manly Western Ecosystems Technology Inc. Cheyenne, Wyoming bmanly@west-inc.com
Monte Carlo in Bayesian Statistics
 Monte Carlo in Bayesian Statistics Matthew Thomas SAMBa - University of Bath m.l.thomas@bath.ac.uk December 4, 2014 Matthew Thomas (SAMBa) Monte Carlo in Bayesian Statistics December 4, 2014 1 / 16 Overview
Monte Carlo in Bayesian Statistics Matthew Thomas SAMBa - University of Bath m.l.thomas@bath.ac.uk December 4, 2014 Matthew Thomas (SAMBa) Monte Carlo in Bayesian Statistics December 4, 2014 1 / 16 Overview
eqr094: Hierarchical MCMC for Bayesian System Reliability
 eqr094: Hierarchical MCMC for Bayesian System Reliability Alyson G. Wilson Statistical Sciences Group, Los Alamos National Laboratory P.O. Box 1663, MS F600 Los Alamos, NM 87545 USA Phone: 505-667-9167
eqr094: Hierarchical MCMC for Bayesian System Reliability Alyson G. Wilson Statistical Sciences Group, Los Alamos National Laboratory P.O. Box 1663, MS F600 Los Alamos, NM 87545 USA Phone: 505-667-9167
Maximum likelihood estimation of mark-recapture-recovery models in the presence of continuous covariates
 Maximum likelihood estimation of mark-recapture-recovery models in the presence of continuous covariates & Ruth King University of St Andrews........ 1 Outline of the problem 2 MRR data in the absence
Maximum likelihood estimation of mark-recapture-recovery models in the presence of continuous covariates & Ruth King University of St Andrews........ 1 Outline of the problem 2 MRR data in the absence
Stat 535 C - Statistical Computing & Monte Carlo Methods. Lecture 18-16th March Arnaud Doucet
 Stat 535 C - Statistical Computing & Monte Carlo Methods Lecture 18-16th March 2006 Arnaud Doucet Email: arnaud@cs.ubc.ca 1 1.1 Outline Trans-dimensional Markov chain Monte Carlo. Bayesian model for autoregressions.
Stat 535 C - Statistical Computing & Monte Carlo Methods Lecture 18-16th March 2006 Arnaud Doucet Email: arnaud@cs.ubc.ca 1 1.1 Outline Trans-dimensional Markov chain Monte Carlo. Bayesian model for autoregressions.
Capture-recapture experiments
 4 Inference in finite populations Binomial capture model Two-stage capture-recapture Open population Accept-Reject methods Arnason Schwarz s Model 236/459 Inference in finite populations Inference in finite
4 Inference in finite populations Binomial capture model Two-stage capture-recapture Open population Accept-Reject methods Arnason Schwarz s Model 236/459 Inference in finite populations Inference in finite
Continuous Covariates in Mark-Recapture-Recovery Analysis: A Comparison. of Methods
 Biometrics 000, 000 000 DOI: 000 000 0000 Continuous Covariates in Mark-Recapture-Recovery Analysis: A Comparison of Methods Simon J. Bonner 1, Byron J. T. Morgan 2, and Ruth King 3 1 Department of Statistics,
Biometrics 000, 000 000 DOI: 000 000 0000 Continuous Covariates in Mark-Recapture-Recovery Analysis: A Comparison of Methods Simon J. Bonner 1, Byron J. T. Morgan 2, and Ruth King 3 1 Department of Statistics,
Markov Chain Monte Carlo
 Markov Chain Monte Carlo Recall: To compute the expectation E ( h(y ) ) we use the approximation E(h(Y )) 1 n n h(y ) t=1 with Y (1),..., Y (n) h(y). Thus our aim is to sample Y (1),..., Y (n) from f(y).
Markov Chain Monte Carlo Recall: To compute the expectation E ( h(y ) ) we use the approximation E(h(Y )) 1 n n h(y ) t=1 with Y (1),..., Y (n) h(y). Thus our aim is to sample Y (1),..., Y (n) from f(y).
Theory of Stochastic Processes 8. Markov chain Monte Carlo
 Theory of Stochastic Processes 8. Markov chain Monte Carlo Tomonari Sei sei@mist.i.u-tokyo.ac.jp Department of Mathematical Informatics, University of Tokyo June 8, 2017 http://www.stat.t.u-tokyo.ac.jp/~sei/lec.html
Theory of Stochastic Processes 8. Markov chain Monte Carlo Tomonari Sei sei@mist.i.u-tokyo.ac.jp Department of Mathematical Informatics, University of Tokyo June 8, 2017 http://www.stat.t.u-tokyo.ac.jp/~sei/lec.html
LECTURE 15 Markov chain Monte Carlo
 LECTURE 15 Markov chain Monte Carlo There are many settings when posterior computation is a challenge in that one does not have a closed form expression for the posterior distribution. Markov chain Monte
LECTURE 15 Markov chain Monte Carlo There are many settings when posterior computation is a challenge in that one does not have a closed form expression for the posterior distribution. Markov chain Monte
Bayesian Statistical Methods. Jeff Gill. Department of Political Science, University of Florida
 Bayesian Statistical Methods Jeff Gill Department of Political Science, University of Florida 234 Anderson Hall, PO Box 117325, Gainesville, FL 32611-7325 Voice: 352-392-0262x272, Fax: 352-392-8127, Email:
Bayesian Statistical Methods Jeff Gill Department of Political Science, University of Florida 234 Anderson Hall, PO Box 117325, Gainesville, FL 32611-7325 Voice: 352-392-0262x272, Fax: 352-392-8127, Email:
ST 740: Markov Chain Monte Carlo
 ST 740: Markov Chain Monte Carlo Alyson Wilson Department of Statistics North Carolina State University October 14, 2012 A. Wilson (NCSU Stsatistics) MCMC October 14, 2012 1 / 20 Convergence Diagnostics:
ST 740: Markov Chain Monte Carlo Alyson Wilson Department of Statistics North Carolina State University October 14, 2012 A. Wilson (NCSU Stsatistics) MCMC October 14, 2012 1 / 20 Convergence Diagnostics:
MCMC algorithms for fitting Bayesian models
 MCMC algorithms for fitting Bayesian models p. 1/1 MCMC algorithms for fitting Bayesian models Sudipto Banerjee sudiptob@biostat.umn.edu University of Minnesota MCMC algorithms for fitting Bayesian models
MCMC algorithms for fitting Bayesian models p. 1/1 MCMC algorithms for fitting Bayesian models Sudipto Banerjee sudiptob@biostat.umn.edu University of Minnesota MCMC algorithms for fitting Bayesian models
MCMC: Markov Chain Monte Carlo
 I529: Machine Learning in Bioinformatics (Spring 2013) MCMC: Markov Chain Monte Carlo Yuzhen Ye School of Informatics and Computing Indiana University, Bloomington Spring 2013 Contents Review of Markov
I529: Machine Learning in Bioinformatics (Spring 2013) MCMC: Markov Chain Monte Carlo Yuzhen Ye School of Informatics and Computing Indiana University, Bloomington Spring 2013 Contents Review of Markov
Rank Regression with Normal Residuals using the Gibbs Sampler
 Rank Regression with Normal Residuals using the Gibbs Sampler Stephen P Smith email: hucklebird@aol.com, 2018 Abstract Yu (2000) described the use of the Gibbs sampler to estimate regression parameters
Rank Regression with Normal Residuals using the Gibbs Sampler Stephen P Smith email: hucklebird@aol.com, 2018 Abstract Yu (2000) described the use of the Gibbs sampler to estimate regression parameters
CPSC 540: Machine Learning
 CPSC 540: Machine Learning MCMC and Non-Parametric Bayes Mark Schmidt University of British Columbia Winter 2016 Admin I went through project proposals: Some of you got a message on Piazza. No news is
CPSC 540: Machine Learning MCMC and Non-Parametric Bayes Mark Schmidt University of British Columbia Winter 2016 Admin I went through project proposals: Some of you got a message on Piazza. No news is
Mixture modelling of recurrent event times with long-term survivors: Analysis of Hutterite birth intervals. John W. Mac McDonald & Alessandro Rosina
 Mixture modelling of recurrent event times with long-term survivors: Analysis of Hutterite birth intervals John W. Mac McDonald & Alessandro Rosina Quantitative Methods in the Social Sciences Seminar -
Mixture modelling of recurrent event times with long-term survivors: Analysis of Hutterite birth intervals John W. Mac McDonald & Alessandro Rosina Quantitative Methods in the Social Sciences Seminar -
Markov Chain Monte Carlo
 Markov Chain Monte Carlo (and Bayesian Mixture Models) David M. Blei Columbia University October 14, 2014 We have discussed probabilistic modeling, and have seen how the posterior distribution is the critical
Markov Chain Monte Carlo (and Bayesian Mixture Models) David M. Blei Columbia University October 14, 2014 We have discussed probabilistic modeling, and have seen how the posterior distribution is the critical
Bayesian Inference in GLMs. Frequentists typically base inferences on MLEs, asymptotic confidence
 Bayesian Inference in GLMs Frequentists typically base inferences on MLEs, asymptotic confidence limits, and log-likelihood ratio tests Bayesians base inferences on the posterior distribution of the unknowns
Bayesian Inference in GLMs Frequentists typically base inferences on MLEs, asymptotic confidence limits, and log-likelihood ratio tests Bayesians base inferences on the posterior distribution of the unknowns
Stat 451 Lecture Notes Markov Chain Monte Carlo. Ryan Martin UIC
 Stat 451 Lecture Notes 07 12 Markov Chain Monte Carlo Ryan Martin UIC www.math.uic.edu/~rgmartin 1 Based on Chapters 8 9 in Givens & Hoeting, Chapters 25 27 in Lange 2 Updated: April 4, 2016 1 / 42 Outline
Stat 451 Lecture Notes 07 12 Markov Chain Monte Carlo Ryan Martin UIC www.math.uic.edu/~rgmartin 1 Based on Chapters 8 9 in Givens & Hoeting, Chapters 25 27 in Lange 2 Updated: April 4, 2016 1 / 42 Outline
Supplement to A Hierarchical Approach for Fitting Curves to Response Time Measurements
 Supplement to A Hierarchical Approach for Fitting Curves to Response Time Measurements Jeffrey N. Rouder Francis Tuerlinckx Paul L. Speckman Jun Lu & Pablo Gomez May 4 008 1 The Weibull regression model
Supplement to A Hierarchical Approach for Fitting Curves to Response Time Measurements Jeffrey N. Rouder Francis Tuerlinckx Paul L. Speckman Jun Lu & Pablo Gomez May 4 008 1 The Weibull regression model
ABC methods for phase-type distributions with applications in insurance risk problems
 ABC methods for phase-type with applications problems Concepcion Ausin, Department of Statistics, Universidad Carlos III de Madrid Joint work with: Pedro Galeano, Universidad Carlos III de Madrid Simon
ABC methods for phase-type with applications problems Concepcion Ausin, Department of Statistics, Universidad Carlos III de Madrid Joint work with: Pedro Galeano, Universidad Carlos III de Madrid Simon
Continuous, Individual, Time-Dependent Covariates in the Cormack-Jolly-Seber Model
 Continuous, Individual, Time-Dependent Covariates in the Cormack-Jolly-Seber Model by Simon J. Bonner B.Sc. Hons., McGill University, 2001 a Project submitted in partial fulfillment of the requirements
Continuous, Individual, Time-Dependent Covariates in the Cormack-Jolly-Seber Model by Simon J. Bonner B.Sc. Hons., McGill University, 2001 a Project submitted in partial fulfillment of the requirements
Control Variates for Markov Chain Monte Carlo
 Control Variates for Markov Chain Monte Carlo Dellaportas, P., Kontoyiannis, I., and Tsourti, Z. Dept of Statistics, AUEB Dept of Informatics, AUEB 1st Greek Stochastics Meeting Monte Carlo: Probability
Control Variates for Markov Chain Monte Carlo Dellaportas, P., Kontoyiannis, I., and Tsourti, Z. Dept of Statistics, AUEB Dept of Informatics, AUEB 1st Greek Stochastics Meeting Monte Carlo: Probability
Bagging During Markov Chain Monte Carlo for Smoother Predictions
 Bagging During Markov Chain Monte Carlo for Smoother Predictions Herbert K. H. Lee University of California, Santa Cruz Abstract: Making good predictions from noisy data is a challenging problem. Methods
Bagging During Markov Chain Monte Carlo for Smoother Predictions Herbert K. H. Lee University of California, Santa Cruz Abstract: Making good predictions from noisy data is a challenging problem. Methods
Brief introduction to Markov Chain Monte Carlo
 Brief introduction to Department of Probability and Mathematical Statistics seminar Stochastic modeling in economics and finance November 7, 2011 Brief introduction to Content 1 and motivation Classical
Brief introduction to Department of Probability and Mathematical Statistics seminar Stochastic modeling in economics and finance November 7, 2011 Brief introduction to Content 1 and motivation Classical
Markov chain Monte Carlo methods in atmospheric remote sensing
 1 / 45 Markov chain Monte Carlo methods in atmospheric remote sensing Johanna Tamminen johanna.tamminen@fmi.fi ESA Summer School on Earth System Monitoring and Modeling July 3 Aug 11, 212, Frascati July,
1 / 45 Markov chain Monte Carlo methods in atmospheric remote sensing Johanna Tamminen johanna.tamminen@fmi.fi ESA Summer School on Earth System Monitoring and Modeling July 3 Aug 11, 212, Frascati July,
An ABC interpretation of the multiple auxiliary variable method
 School of Mathematical and Physical Sciences Department of Mathematics and Statistics Preprint MPS-2016-07 27 April 2016 An ABC interpretation of the multiple auxiliary variable method by Dennis Prangle
School of Mathematical and Physical Sciences Department of Mathematics and Statistics Preprint MPS-2016-07 27 April 2016 An ABC interpretation of the multiple auxiliary variable method by Dennis Prangle
CS242: Probabilistic Graphical Models Lecture 7B: Markov Chain Monte Carlo & Gibbs Sampling
 CS242: Probabilistic Graphical Models Lecture 7B: Markov Chain Monte Carlo & Gibbs Sampling Professor Erik Sudderth Brown University Computer Science October 27, 2016 Some figures and materials courtesy
CS242: Probabilistic Graphical Models Lecture 7B: Markov Chain Monte Carlo & Gibbs Sampling Professor Erik Sudderth Brown University Computer Science October 27, 2016 Some figures and materials courtesy
Likelihood-free MCMC
 Bayesian inference for stable distributions with applications in finance Department of Mathematics University of Leicester September 2, 2011 MSc project final presentation Outline 1 2 3 4 Classical Monte
Bayesian inference for stable distributions with applications in finance Department of Mathematics University of Leicester September 2, 2011 MSc project final presentation Outline 1 2 3 4 Classical Monte
Reconstruction of individual patient data for meta analysis via Bayesian approach
 Reconstruction of individual patient data for meta analysis via Bayesian approach Yusuke Yamaguchi, Wataru Sakamoto and Shingo Shirahata Graduate School of Engineering Science, Osaka University Masashi
Reconstruction of individual patient data for meta analysis via Bayesian approach Yusuke Yamaguchi, Wataru Sakamoto and Shingo Shirahata Graduate School of Engineering Science, Osaka University Masashi
MONTE CARLO METHODS. Hedibert Freitas Lopes
 MONTE CARLO METHODS Hedibert Freitas Lopes The University of Chicago Booth School of Business 5807 South Woodlawn Avenue, Chicago, IL 60637 http://faculty.chicagobooth.edu/hedibert.lopes hlopes@chicagobooth.edu
MONTE CARLO METHODS Hedibert Freitas Lopes The University of Chicago Booth School of Business 5807 South Woodlawn Avenue, Chicago, IL 60637 http://faculty.chicagobooth.edu/hedibert.lopes hlopes@chicagobooth.edu
Bayesian Inference and MCMC
 Bayesian Inference and MCMC Aryan Arbabi Partly based on MCMC slides from CSC412 Fall 2018 1 / 18 Bayesian Inference - Motivation Consider we have a data set D = {x 1,..., x n }. E.g each x i can be the
Bayesian Inference and MCMC Aryan Arbabi Partly based on MCMC slides from CSC412 Fall 2018 1 / 18 Bayesian Inference - Motivation Consider we have a data set D = {x 1,..., x n }. E.g each x i can be the
Capture-Recapture Analyses of the Frog Leiopelma pakeka on Motuara Island
 Capture-Recapture Analyses of the Frog Leiopelma pakeka on Motuara Island Shirley Pledger School of Mathematical and Computing Sciences Victoria University of Wellington P.O.Box 600, Wellington, New Zealand
Capture-Recapture Analyses of the Frog Leiopelma pakeka on Motuara Island Shirley Pledger School of Mathematical and Computing Sciences Victoria University of Wellington P.O.Box 600, Wellington, New Zealand
A Single Series from the Gibbs Sampler Provides a False Sense of Security
 A Single Series from the Gibbs Sampler Provides a False Sense of Security Andrew Gelman Department of Statistics University of California Berkeley, CA 9472 Donald B. Rubin Department of Statistics Harvard
A Single Series from the Gibbs Sampler Provides a False Sense of Security Andrew Gelman Department of Statistics University of California Berkeley, CA 9472 Donald B. Rubin Department of Statistics Harvard
MCMC and Gibbs Sampling. Kayhan Batmanghelich
 MCMC and Gibbs Sampling Kayhan Batmanghelich 1 Approaches to inference l Exact inference algorithms l l l The elimination algorithm Message-passing algorithm (sum-product, belief propagation) The junction
MCMC and Gibbs Sampling Kayhan Batmanghelich 1 Approaches to inference l Exact inference algorithms l l l The elimination algorithm Message-passing algorithm (sum-product, belief propagation) The junction
Factorization of Seperable and Patterned Covariance Matrices for Gibbs Sampling
 Monte Carlo Methods Appl, Vol 6, No 3 (2000), pp 205 210 c VSP 2000 Factorization of Seperable and Patterned Covariance Matrices for Gibbs Sampling Daniel B Rowe H & SS, 228-77 California Institute of
Monte Carlo Methods Appl, Vol 6, No 3 (2000), pp 205 210 c VSP 2000 Factorization of Seperable and Patterned Covariance Matrices for Gibbs Sampling Daniel B Rowe H & SS, 228-77 California Institute of
BAYESIAN METHODS FOR VARIABLE SELECTION WITH APPLICATIONS TO HIGH-DIMENSIONAL DATA
 BAYESIAN METHODS FOR VARIABLE SELECTION WITH APPLICATIONS TO HIGH-DIMENSIONAL DATA Intro: Course Outline and Brief Intro to Marina Vannucci Rice University, USA PASI-CIMAT 04/28-30/2010 Marina Vannucci
BAYESIAN METHODS FOR VARIABLE SELECTION WITH APPLICATIONS TO HIGH-DIMENSIONAL DATA Intro: Course Outline and Brief Intro to Marina Vannucci Rice University, USA PASI-CIMAT 04/28-30/2010 Marina Vannucci
Markov Chain Monte Carlo
 Chapter 5 Markov Chain Monte Carlo MCMC is a kind of improvement of the Monte Carlo method By sampling from a Markov chain whose stationary distribution is the desired sampling distributuion, it is possible
Chapter 5 Markov Chain Monte Carlo MCMC is a kind of improvement of the Monte Carlo method By sampling from a Markov chain whose stationary distribution is the desired sampling distributuion, it is possible
Research Article A Nonparametric Two-Sample Wald Test of Equality of Variances
 Advances in Decision Sciences Volume 211, Article ID 74858, 8 pages doi:1.1155/211/74858 Research Article A Nonparametric Two-Sample Wald Test of Equality of Variances David Allingham 1 andj.c.w.rayner
Advances in Decision Sciences Volume 211, Article ID 74858, 8 pages doi:1.1155/211/74858 Research Article A Nonparametric Two-Sample Wald Test of Equality of Variances David Allingham 1 andj.c.w.rayner
Parameter Estimation. William H. Jefferys University of Texas at Austin Parameter Estimation 7/26/05 1
 Parameter Estimation William H. Jefferys University of Texas at Austin bill@bayesrules.net Parameter Estimation 7/26/05 1 Elements of Inference Inference problems contain two indispensable elements: Data
Parameter Estimation William H. Jefferys University of Texas at Austin bill@bayesrules.net Parameter Estimation 7/26/05 1 Elements of Inference Inference problems contain two indispensable elements: Data
Represent processes and observations that span multiple levels (aka multi level models) R 2
 Hierarchical models Hierarchical models Represent processes and observations that span multiple levels (aka multi level models) R 1 R 2 R 3 N 1 N 2 N 3 N 4 N 5 N 6 N 7 N 8 N 9 N i = true abundance on a
Hierarchical models Hierarchical models Represent processes and observations that span multiple levels (aka multi level models) R 1 R 2 R 3 N 1 N 2 N 3 N 4 N 5 N 6 N 7 N 8 N 9 N i = true abundance on a
Probabilistic Graphical Models
 2016 Robert Nowak Probabilistic Graphical Models 1 Introduction We have focused mainly on linear models for signals, in particular the subspace model x = Uθ, where U is a n k matrix and θ R k is a vector
2016 Robert Nowak Probabilistic Graphical Models 1 Introduction We have focused mainly on linear models for signals, in particular the subspace model x = Uθ, where U is a n k matrix and θ R k is a vector
Metropolis-Hastings Algorithm
 Strength of the Gibbs sampler Metropolis-Hastings Algorithm Easy algorithm to think about. Exploits the factorization properties of the joint probability distribution. No difficult choices to be made to
Strength of the Gibbs sampler Metropolis-Hastings Algorithm Easy algorithm to think about. Exploits the factorization properties of the joint probability distribution. No difficult choices to be made to
Multilevel Statistical Models: 3 rd edition, 2003 Contents
 Multilevel Statistical Models: 3 rd edition, 2003 Contents Preface Acknowledgements Notation Two and three level models. A general classification notation and diagram Glossary Chapter 1 An introduction
Multilevel Statistical Models: 3 rd edition, 2003 Contents Preface Acknowledgements Notation Two and three level models. A general classification notation and diagram Glossary Chapter 1 An introduction
Computational statistics
 Computational statistics Markov Chain Monte Carlo methods Thierry Denœux March 2017 Thierry Denœux Computational statistics March 2017 1 / 71 Contents of this chapter When a target density f can be evaluated
Computational statistics Markov Chain Monte Carlo methods Thierry Denœux March 2017 Thierry Denœux Computational statistics March 2017 1 / 71 Contents of this chapter When a target density f can be evaluated
Basic math for biology
 Basic math for biology Lei Li Florida State University, Feb 6, 2002 The EM algorithm: setup Parametric models: {P θ }. Data: full data (Y, X); partial data Y. Missing data: X. Likelihood and maximum likelihood
Basic math for biology Lei Li Florida State University, Feb 6, 2002 The EM algorithm: setup Parametric models: {P θ }. Data: full data (Y, X); partial data Y. Missing data: X. Likelihood and maximum likelihood
Bayesian Estimation of Input Output Tables for Russia
 Bayesian Estimation of Input Output Tables for Russia Oleg Lugovoy (EDF, RANE) Andrey Polbin (RANE) Vladimir Potashnikov (RANE) WIOD Conference April 24, 2012 Groningen Outline Motivation Objectives Bayesian
Bayesian Estimation of Input Output Tables for Russia Oleg Lugovoy (EDF, RANE) Andrey Polbin (RANE) Vladimir Potashnikov (RANE) WIOD Conference April 24, 2012 Groningen Outline Motivation Objectives Bayesian
Computer Vision Group Prof. Daniel Cremers. 10a. Markov Chain Monte Carlo
 Group Prof. Daniel Cremers 10a. Markov Chain Monte Carlo Markov Chain Monte Carlo In high-dimensional spaces, rejection sampling and importance sampling are very inefficient An alternative is Markov Chain
Group Prof. Daniel Cremers 10a. Markov Chain Monte Carlo Markov Chain Monte Carlo In high-dimensional spaces, rejection sampling and importance sampling are very inefficient An alternative is Markov Chain
\ fwf The Institute for Integrating Statistics in Decision Sciences
 # \ fwf The Institute for Integrating Statistics in Decision Sciences Technical Report TR-2007-8 May 22, 2007 Advances in Bayesian Software Reliability Modelling Fabrizio Ruggeri CNR IMATI Milano, Italy
# \ fwf The Institute for Integrating Statistics in Decision Sciences Technical Report TR-2007-8 May 22, 2007 Advances in Bayesian Software Reliability Modelling Fabrizio Ruggeri CNR IMATI Milano, Italy
18 : Advanced topics in MCMC. 1 Gibbs Sampling (Continued from the last lecture)
 10-708: Probabilistic Graphical Models 10-708, Spring 2014 18 : Advanced topics in MCMC Lecturer: Eric P. Xing Scribes: Jessica Chemali, Seungwhan Moon 1 Gibbs Sampling (Continued from the last lecture)
10-708: Probabilistic Graphical Models 10-708, Spring 2014 18 : Advanced topics in MCMC Lecturer: Eric P. Xing Scribes: Jessica Chemali, Seungwhan Moon 1 Gibbs Sampling (Continued from the last lecture)
Down by the Bayes, where the Watermelons Grow
 Down by the Bayes, where the Watermelons Grow A Bayesian example using SAS SUAVe: Victoria SAS User Group Meeting November 21, 2017 Peter K. Ott, M.Sc., P.Stat. Strategic Analysis 1 Outline 1. Motivating
Down by the Bayes, where the Watermelons Grow A Bayesian example using SAS SUAVe: Victoria SAS User Group Meeting November 21, 2017 Peter K. Ott, M.Sc., P.Stat. Strategic Analysis 1 Outline 1. Motivating
Mixture models. Mixture models MCMC approaches Label switching MCMC for variable dimension models. 5 Mixture models
 5 MCMC approaches Label switching MCMC for variable dimension models 291/459 Missing variable models Complexity of a model may originate from the fact that some piece of information is missing Example
5 MCMC approaches Label switching MCMC for variable dimension models 291/459 Missing variable models Complexity of a model may originate from the fact that some piece of information is missing Example
State Space and Hidden Markov Models
 State Space and Hidden Markov Models Kunsch H.R. State Space and Hidden Markov Models. ETH- Zurich Zurich; Aliaksandr Hubin Oslo 2014 Contents 1. Introduction 2. Markov Chains 3. Hidden Markov and State
State Space and Hidden Markov Models Kunsch H.R. State Space and Hidden Markov Models. ETH- Zurich Zurich; Aliaksandr Hubin Oslo 2014 Contents 1. Introduction 2. Markov Chains 3. Hidden Markov and State
Four aspects of a sampling strategy necessary to make accurate and precise inferences about populations are:
 Why Sample? Often researchers are interested in answering questions about a particular population. They might be interested in the density, species richness, or specific life history parameters such as
Why Sample? Often researchers are interested in answering questions about a particular population. They might be interested in the density, species richness, or specific life history parameters such as
Ages of stellar populations from color-magnitude diagrams. Paul Baines. September 30, 2008
 Ages of stellar populations from color-magnitude diagrams Paul Baines Department of Statistics Harvard University September 30, 2008 Context & Example Welcome! Today we will look at using hierarchical
Ages of stellar populations from color-magnitude diagrams Paul Baines Department of Statistics Harvard University September 30, 2008 Context & Example Welcome! Today we will look at using hierarchical
FW Laboratory Exercise. Program MARK: Joint Live Recapture and Dead Recovery Data and Pradel Model
 FW663 -- Laboratory Exercise Program MARK: Joint Live Recapture and Dead Recovery Data and Pradel Model Today s exercise explores parameter estimation using both live recaptures and dead recoveries. We
FW663 -- Laboratory Exercise Program MARK: Joint Live Recapture and Dead Recovery Data and Pradel Model Today s exercise explores parameter estimation using both live recaptures and dead recoveries. We
Default Priors and Effcient Posterior Computation in Bayesian
 Default Priors and Effcient Posterior Computation in Bayesian Factor Analysis January 16, 2010 Presented by Eric Wang, Duke University Background and Motivation A Brief Review of Parameter Expansion Literature
Default Priors and Effcient Posterior Computation in Bayesian Factor Analysis January 16, 2010 Presented by Eric Wang, Duke University Background and Motivation A Brief Review of Parameter Expansion Literature
A generalization of the Multiple-try Metropolis algorithm for Bayesian estimation and model selection
 A generalization of the Multiple-try Metropolis algorithm for Bayesian estimation and model selection Silvia Pandolfi Francesco Bartolucci Nial Friel University of Perugia, IT University of Perugia, IT
A generalization of the Multiple-try Metropolis algorithm for Bayesian estimation and model selection Silvia Pandolfi Francesco Bartolucci Nial Friel University of Perugia, IT University of Perugia, IT
The Mixture Approach for Simulating New Families of Bivariate Distributions with Specified Correlations
 The Mixture Approach for Simulating New Families of Bivariate Distributions with Specified Correlations John R. Michael, Significance, Inc. and William R. Schucany, Southern Methodist University The mixture
The Mixture Approach for Simulating New Families of Bivariate Distributions with Specified Correlations John R. Michael, Significance, Inc. and William R. Schucany, Southern Methodist University The mixture
Efficient adaptive covariate modelling for extremes
 Efficient adaptive covariate modelling for extremes Slides at www.lancs.ac.uk/ jonathan Matthew Jones, David Randell, Emma Ross, Elena Zanini, Philip Jonathan Copyright of Shell December 218 1 / 23 Structural
Efficient adaptive covariate modelling for extremes Slides at www.lancs.ac.uk/ jonathan Matthew Jones, David Randell, Emma Ross, Elena Zanini, Philip Jonathan Copyright of Shell December 218 1 / 23 Structural
Learning latent structure in complex networks
 Learning latent structure in complex networks Lars Kai Hansen www.imm.dtu.dk/~lkh Current network research issues: Social Media Neuroinformatics Machine learning Joint work with Morten Mørup, Sune Lehmann
Learning latent structure in complex networks Lars Kai Hansen www.imm.dtu.dk/~lkh Current network research issues: Social Media Neuroinformatics Machine learning Joint work with Morten Mørup, Sune Lehmann
Probabilistic Graphical Networks: Definitions and Basic Results
 This document gives a cursory overview of Probabilistic Graphical Networks. The material has been gleaned from different sources. I make no claim to original authorship of this material. Bayesian Graphical
This document gives a cursory overview of Probabilistic Graphical Networks. The material has been gleaned from different sources. I make no claim to original authorship of this material. Bayesian Graphical
Bayesian Linear Regression
 Bayesian Linear Regression Sudipto Banerjee 1 Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, U.S.A. September 15, 2010 1 Linear regression models: a Bayesian perspective
Bayesian Linear Regression Sudipto Banerjee 1 Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, U.S.A. September 15, 2010 1 Linear regression models: a Bayesian perspective
A nonparametric two-sample wald test of equality of variances
 University of Wollongong Research Online Faculty of Informatics - Papers (Archive) Faculty of Engineering and Information Sciences 211 A nonparametric two-sample wald test of equality of variances David
University of Wollongong Research Online Faculty of Informatics - Papers (Archive) Faculty of Engineering and Information Sciences 211 A nonparametric two-sample wald test of equality of variances David
Principles of Bayesian Inference
 Principles of Bayesian Inference Sudipto Banerjee and Andrew O. Finley 2 Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, U.S.A. 2 Department of Forestry & Department
Principles of Bayesian Inference Sudipto Banerjee and Andrew O. Finley 2 Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, U.S.A. 2 Department of Forestry & Department
Approximate Bayesian computation: methods and applications for complex systems
 Approximate Bayesian computation: methods and applications for complex systems Mark A. Beaumont, School of Biological Sciences, The University of Bristol, Bristol, UK 11 November 2015 Structure of Talk
Approximate Bayesian computation: methods and applications for complex systems Mark A. Beaumont, School of Biological Sciences, The University of Bristol, Bristol, UK 11 November 2015 Structure of Talk
Stat 535 C - Statistical Computing & Monte Carlo Methods. Arnaud Doucet.
 Stat 535 C - Statistical Computing & Monte Carlo Methods Arnaud Doucet Email: arnaud@cs.ubc.ca 1 1.1 Outline Introduction to Markov chain Monte Carlo The Gibbs Sampler Examples Overview of the Lecture
Stat 535 C - Statistical Computing & Monte Carlo Methods Arnaud Doucet Email: arnaud@cs.ubc.ca 1 1.1 Outline Introduction to Markov chain Monte Carlo The Gibbs Sampler Examples Overview of the Lecture
Learning the hyper-parameters. Luca Martino
 Learning the hyper-parameters Luca Martino 2017 2017 1 / 28 Parameters and hyper-parameters 1. All the described methods depend on some choice of hyper-parameters... 2. For instance, do you recall λ (bandwidth
Learning the hyper-parameters Luca Martino 2017 2017 1 / 28 Parameters and hyper-parameters 1. All the described methods depend on some choice of hyper-parameters... 2. For instance, do you recall λ (bandwidth
Markov Chain Monte Carlo (MCMC)
 Markov Chain Monte Carlo (MCMC Dependent Sampling Suppose we wish to sample from a density π, and we can evaluate π as a function but have no means to directly generate a sample. Rejection sampling can
Markov Chain Monte Carlo (MCMC Dependent Sampling Suppose we wish to sample from a density π, and we can evaluate π as a function but have no means to directly generate a sample. Rejection sampling can
Bayesian nonparametric estimation of finite population quantities in absence of design information on nonsampled units
 Bayesian nonparametric estimation of finite population quantities in absence of design information on nonsampled units Sahar Z Zangeneh Robert W. Keener Roderick J.A. Little Abstract In Probability proportional
Bayesian nonparametric estimation of finite population quantities in absence of design information on nonsampled units Sahar Z Zangeneh Robert W. Keener Roderick J.A. Little Abstract In Probability proportional
Mark-Recapture. Mark-Recapture. Useful in estimating: Lincoln-Petersen Estimate. Lincoln-Petersen Estimate. Lincoln-Petersen Estimate
 Mark-Recapture Mark-Recapture Modern use dates from work by C. G. J. Petersen (Danish fisheries biologist, 1896) and F. C. Lincoln (U. S. Fish and Wildlife Service, 1930) Useful in estimating: a. Size
Mark-Recapture Mark-Recapture Modern use dates from work by C. G. J. Petersen (Danish fisheries biologist, 1896) and F. C. Lincoln (U. S. Fish and Wildlife Service, 1930) Useful in estimating: a. Size
Principles of Bayesian Inference
 Principles of Bayesian Inference Sudipto Banerjee 1 and Andrew O. Finley 2 1 Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, U.S.A. 2 Department of Forestry & Department
Principles of Bayesian Inference Sudipto Banerjee 1 and Andrew O. Finley 2 1 Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, U.S.A. 2 Department of Forestry & Department
Ecological applications of hidden Markov models and related doubly stochastic processes
 . Ecological applications of hidden Markov models and related doubly stochastic processes Roland Langrock School of Mathematics and Statistics & CREEM Motivating example HMM machinery Some ecological applications
. Ecological applications of hidden Markov models and related doubly stochastic processes Roland Langrock School of Mathematics and Statistics & CREEM Motivating example HMM machinery Some ecological applications
Application of Algebraic Statistics to Mark-Recapture Models with Misidentificaton
 Application of Algebraic Statistics to Mark-Recapture Models with Misidentificaton Dr. Simon Bonner 1, Dr. Matthew Schofield 2, Patrik Noren 3, and Dr. Ruriko Yoshida 1 1 Department of Statistics, University
Application of Algebraic Statistics to Mark-Recapture Models with Misidentificaton Dr. Simon Bonner 1, Dr. Matthew Schofield 2, Patrik Noren 3, and Dr. Ruriko Yoshida 1 1 Department of Statistics, University
SAMPLING ALGORITHMS. In general. Inference in Bayesian models
 SAMPLING ALGORITHMS SAMPLING ALGORITHMS In general A sampling algorithm is an algorithm that outputs samples x 1, x 2,... from a given distribution P or density p. Sampling algorithms can for example be
SAMPLING ALGORITHMS SAMPLING ALGORITHMS In general A sampling algorithm is an algorithm that outputs samples x 1, x 2,... from a given distribution P or density p. Sampling algorithms can for example be
Bayesian Networks in Educational Assessment
 Bayesian Networks in Educational Assessment Estimating Parameters with MCMC Bayesian Inference: Expanding Our Context Roy Levy Arizona State University Roy.Levy@asu.edu 2017 Roy Levy MCMC 1 MCMC 2 Posterior
Bayesian Networks in Educational Assessment Estimating Parameters with MCMC Bayesian Inference: Expanding Our Context Roy Levy Arizona State University Roy.Levy@asu.edu 2017 Roy Levy MCMC 1 MCMC 2 Posterior
Improving the Computational Efficiency in Bayesian Fitting of Cormack-Jolly-Seber Models with Individual, Continuous, Time-Varying Covariates
 University of Kentucky UKnowledge Theses and Dissertations--Statistics Statistics 2017 Improving the Computational Efficiency in Bayesian Fitting of Cormack-Jolly-Seber Models with Individual, Continuous,
University of Kentucky UKnowledge Theses and Dissertations--Statistics Statistics 2017 Improving the Computational Efficiency in Bayesian Fitting of Cormack-Jolly-Seber Models with Individual, Continuous,
Text Mining for Economics and Finance Latent Dirichlet Allocation
 Text Mining for Economics and Finance Latent Dirichlet Allocation Stephen Hansen Text Mining Lecture 5 1 / 45 Introduction Recall we are interested in mixed-membership modeling, but that the plsi model
Text Mining for Economics and Finance Latent Dirichlet Allocation Stephen Hansen Text Mining Lecture 5 1 / 45 Introduction Recall we are interested in mixed-membership modeling, but that the plsi model
Hastings-within-Gibbs Algorithm: Introduction and Application on Hierarchical Model
 UNIVERSITY OF TEXAS AT SAN ANTONIO Hastings-within-Gibbs Algorithm: Introduction and Application on Hierarchical Model Liang Jing April 2010 1 1 ABSTRACT In this paper, common MCMC algorithms are introduced
UNIVERSITY OF TEXAS AT SAN ANTONIO Hastings-within-Gibbs Algorithm: Introduction and Application on Hierarchical Model Liang Jing April 2010 1 1 ABSTRACT In this paper, common MCMC algorithms are introduced
A Search and Jump Algorithm for Markov Chain Monte Carlo Sampling. Christopher Jennison. Adriana Ibrahim. Seminar at University of Kuwait
 A Search and Jump Algorithm for Markov Chain Monte Carlo Sampling Christopher Jennison Department of Mathematical Sciences, University of Bath, UK http://people.bath.ac.uk/mascj Adriana Ibrahim Institute
A Search and Jump Algorithm for Markov Chain Monte Carlo Sampling Christopher Jennison Department of Mathematical Sciences, University of Bath, UK http://people.bath.ac.uk/mascj Adriana Ibrahim Institute
Dynamic models. Dependent data The AR(p) model The MA(q) model Hidden Markov models. 6 Dynamic models
 6 Dependent data The AR(p) model The MA(q) model Hidden Markov models Dependent data Dependent data Huge portion of real-life data involving dependent datapoints Example (Capture-recapture) capture histories
6 Dependent data The AR(p) model The MA(q) model Hidden Markov models Dependent data Dependent data Huge portion of real-life data involving dependent datapoints Example (Capture-recapture) capture histories
Statistical Methods in Particle Physics Lecture 1: Bayesian methods
 Statistical Methods in Particle Physics Lecture 1: Bayesian methods SUSSP65 St Andrews 16 29 August 2009 Glen Cowan Physics Department Royal Holloway, University of London g.cowan@rhul.ac.uk www.pp.rhul.ac.uk/~cowan
Statistical Methods in Particle Physics Lecture 1: Bayesian methods SUSSP65 St Andrews 16 29 August 2009 Glen Cowan Physics Department Royal Holloway, University of London g.cowan@rhul.ac.uk www.pp.rhul.ac.uk/~cowan
