J. Hasenauer a J. Heinrich b M. Doszczak c P. Scheurich c D. Weiskopf b F. Allgöwer a
|
|
|
- Chad Mason
- 6 years ago
- Views:
Transcription
1 J. Hasenauer a J. Heinrich b M. Doszczak c P. Scheurich c D. Weiskopf b F. Allgöwer a Visualization methods and support vector machines as tools for determining markers in models of heterogeneous populations: Proapoptotic signaling as a case study Stuttgart, June 2011 a Institute of Systems Theory and Automatic Control, University of Stuttgart, Pfaffenwaldring 9, Stuttgart/ Germany {jan.hasenauer,frank.allgower}@ist.uni-stuttgart.de b Visualization Research Center University of Stuttgart, Allmandring 19, Stuttgart/ Germany {julian.heinrich, daniel.weiskopf}@visus.uni-stuttgart.de c Institute of Cell Biology and Immunology, University of Stuttgart, Allmandring 31, Stuttgart/ Germany {peter.scheurich,malgorzata.doszczak}@izi.uni-stuttgart.de This work has been presented at the 8th Workshop for Computational Systems Biology (WCSB 2011), 6-8 July, Please cite this article as: J. Hasenauer, J. Heinrich, M. Doszczak, P. Scheurich, D. Weiskopf, and F. Allgöwer. Visualization methods and support vector machines as tools for determining markers in models of heterogeneous populations: Proapoptotic signaling as a case study. In Proc. of 8th Workshop for Computational Systems Biology (WCSB 2011), Zürich, Switzerland, pages 61 64, Abstract In recent years, cell population models have become very common, as they allow for the study of population heterogeneity. Unfortunately, the complexity of population models so far has prevented the development of analysis tools permitting an in-depth study of the source of heterogeneity, like genetics and epigenetics. In this paper we propose an explorative analysis combining parallel-coordinate plots and nonlinear support vector machines to determine the main sources of cell-to-cell variability within decision processes in heterogeneous cell populations. The approach is applied to analyze proapoptotic signal transduction in cancer cell populations and to determine decision markers. Keywords visual analytics parallel coordinates support vector machines cell population Postprint Series Issue No Stuttgart Research Centre for Simulation Technology (SRC SimTech) SimTech Cluster of Excellence Pfaffenwaldring 7a Stuttgart publications@simtech.uni-stuttgart.de
2 2 J. Hasenauer et al. 1 INTRODUCTION Models of intracellular signaling pathways become increasingly complex. Most of the commonly used quantitative models have tens, sometimes hundreds of states and parameters. Due to this complexity, understanding the models and the important elements of the pathway is challenging. This is a problem particularly in situations where, apart from single cell dynamics, cell-to-cell variability has to be considered as well [1,2]. In such cases, the complexity of the model often prevents the application of classical analysis tools for dynamical systems. One of these cases is the model-driven identification of markers for decision processes in heterogeneous cell populations. In this work, we address the question: Which parameters cause the heterogeneity of the population s response?. We propose the application of two methods widely used in data analysis: Parallel-coordinate plots and nonlinear support vector machines (SVMs). The former method provides an easy tool to obtain qualitative understanding, whereas the latter allows for assessing the performance of decision marker combinations quantitatively. Good decision markers are thereby parameters that allow for a good prediction of the decision outcome of an individual cell. The paper is structured as follows: In Section 2 the considered system class and problem is described in mathematical terms. The general idea is discussed in Section 3, and the application of parallel-coordinate plots and SVMs is outlined. Section 4 provides the example application of the proposed method to a model of the caspase cascade. The results are summarized in Section 5. 2 PROBLEM DESCRIPTION In this paper, heterogeneous cell populations are studied. The population dynamics are described using a cell ensemble model [1,2], which is a collection of N cells, Σ pop = { Σ(θ (i) ) i = {1,..., N}, θ (i) Θ }. (1) The signaling process in each individual cell in Σ pop is described by ordinary differential equations, Σ(θ (i) ) : ẋ (i) = f (x (i), θ (i) ), x (i) (0) = x 0 (θ (i) ), (2) with state vector x (i) (t) R n + and parameter vector θ (i) R q +. The index i specifies individual cells within the population. The vector field f : R n + R q + R n describing the cell dynamics is locally Lipschitz and the mapping x 0 : R q + R n + is continuously differentiable. The parameters θ (i) may be kinetic constants, e.g. synthesis, degradation, or reaction rates. Heterogeneity within the cell ensemble is modeled by differential parameter values θ (i) among individual cells. The density of parameters θ (i) is given by a probability density function Θ : R q + R +. Thus, the probability of obtaining θ (i) [θ, θ+dθ] is Θ(θ)dθ. This modeling approach is employed in several publications (e.g. [1,2]) and has been proven to be useful to study short-term dynamic processes, e.g. cellular apoptosis. In the following, discrete decision processes are considered. Therefore, the functional d : l 1 { 1, +1} is introduced, which maps the single cell trajectory x (i) ( ) to a discrete decision δ (i) { 1, +1}, δ (i) = d(x (i) ( )). (3) This functional is used to evaluate the outcome of the decision process in each cell. Example: To exemplify the decision functional, we considered the process of cell death. There are two possible decisions a cell can make: Either it stays alive, δ (i) = +1, or it dies δ (i) = 1. In many cases, the cell is assumed to die if a certain protein concentration x j exceeds a threshold x j,th [2,3]. This would yield the decision functional { +1 if max d(x (i) x (i) ( )) := t j (t) x j,th (4) 1 otherwise. Note that the response x (i) ( ) of a cell implicitly depends on the cell s parameters θ (i). Furthermore, the parameters are the only difference between two cells. Thus, the decision of a single cell can be viewed as a function of the parameters, δ (i) = δ (i) (θ (i) ). To understand the heterogeneity within the population response, it is necessary to understand how the decision depends on these parameters. In particular, the question arises which parameters θ m := [θ m1,..., θ mr ] T, j : m j {1,..., q} (5)
3 A maximum likelihood estimator for parameter distributions in heterogeneous cell populations 3 cause and explain most of the heterogeneity. If parameters θ m can be determined, they can be used (i) to predict the outcome of the decision process and (ii) as markers to distinguish between individual cells with different responses. 3 METHODS 3.1 Basic idea In this contribution, two methods are combined to determine decision markers, θ m, for models of heterogeneous cell populations. The proposed methods are well-known, but almost exclusively applied to study highdimensional sets of measurement data. To exploit the methods in the context of population model analysis, in a first step the cell ensemble is simulated for N 1, yielding many pairs of parameters and trajectories, ( θ (i), x (i) ( ) ), i = 1,..., N. (6) Given these pairs, a sample of cell decisions is constructed, S = {( θ (1), δ (1)),..., ( θ (N), δ (N))}, with δ (i) = d(x (i) ( )). This sample contains much information about the dependency of the decision δ on the parameters θ. In the following, the sample S is considered as a dataset and the dependency δ = δ(θ) is analyzed using visualization techniques and SVMs. The integration of interactive visualization with automated methods (such as SVMs) thereby closely follows the visual analytics paradigm [4] for finding insights from complex data. In this paper, parallel coordinates [5] are employed to visually select the most promising dimensions which are then used to train a SVM. 3.2 Parallel coordinates for marker selection Parallel coordinates [5] are constructed by placing axes in parallel within a 2-D Cartesian embedding. An N- dimensional data point is then represented by a polyline intersecting the axes at the respective values. While parallel coordinates are widely used to identify patterns or trends in high-dimensional data, they greatly suffer from overplotting if many lines have to be drawn. Instead of rendering opaque lines, continuous parallel coordinates [6] estimate the (line-)density of the resulting image from the sample S. In this work, a pointwise density-estimate is obtained using additive alpha-blending. To indicate class-membership of a sample member in parallel coordinates, we use different colors for each class. Combining density estimate and colors, good markers θ m can visually be determined. They correspond to coordinate axes on which the different colors (classes) are well separated. 3.3 SVMs to quantitative marker properties Given a qualitative understanding of the importance of the parameters and a selection of potential markers θ m, a quantitative assessment of the classification power of θ m is necessary. To obtain this the sample S is used to learn a nonlinear SVM. This is a two step process illustrated in Figure 1. First, a mapping Φ : R r R r is constructed that transforms the input space into a feature space of higher dimension (r > r). Secondly, a linear separation of the data is performed in the feature space [7]. Therefore, the optimization problem min w,b,ξ 1 2 wt w + C N i=1 ξ i s.t. δ (i) ( w T Φ(θ (i) m ) + b ) 1 ξ i, i = 1,..., N, ξ i 0, i = 1,..., N, is solved, in which w and b denote the normal vector of the separating hyperplane and its offset, respectively. The objective function combines a misclassification penalty, S i=1 ξ i, and a margin maximization, 1 2 wt w. For (7)
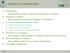
4 4 J. Hasenauer et al. input space feature space feature space Step 1 Step 2 θ 2 Φ 2(θ) Φ 2(θ) θ 1 Φ 1(θ) Φ 1(θ) Fig. 1 Visualization of the SVM approach for separating cells with δ (i) = +1 (+ ) and δ (i) = 1 ( ). Left: Distributed data in the input space. Middle: Sample transformed in the feature space which allows for better separation. Right: Separation result for separating hyperplane with normal vector w ( ). As a perfect separation is in general not possible, misclassifications ( ) exist. k 8 k8 k 9 k9 k4 IAP k7 stimulus (e.g. TNF or TRAIL) C8 k 3 IAP k3 C3a k2 C3a k6 k5 C8a k1 cell death C8a CARP k11 k 11 C3 k13 CARP k 10 k10 k 12 k12 Fig. 2 Illustration of proapoptotic signaling pathway [3]. Normal arrows ( ) refer to conversion reactions, dashed arrows ( ) indicate enzymatic activity, and thick arrows ( ) illustrate inputs and outputs of the system. a detailed introduction to SMVs, we refer to [7]. An application for the study of dynamical systems can be found in [8]. Given the solution of (7), the percentage of true positive classifications, TP m, and false positive classifications, FP m, can be evaluated. This is done for a second sample, S, to avoid overfitting. TP m and FP m provide information about predictability of the outcome for θ (i) using solely θ m (i). Thus, the marker quality can be assessed via TP m and FP m. If a low-dimensional θ m exists that provides TP m 1 and FP m 0, the parameters θ m dominate the decision process and are good markers. For a quantification of this effect, the classification performance can be analyzed in ROC space. For details we refer to [9]. Summing up, parallel-coordinate plots allow for an intuitive visual assessment of the dependency of the decision on the parameters and the selection of potential markers. A quantitative evaluation of the marker quality is possible using SVMs. By combining both methods, the combinatorial explosion related to checking all possible marker combinations using SVMs can be avoided, resulting in a tremendously reduced computational complexity. 4 ANALYSIS OF PROAPOPTOTIC SIGNALING To illustrate what insight can be gained using the proposed methods, proapoptotic signaling is analyzed. Proapoptotic signaling is involved in the process of apoptosis (programmed cell death). Apoptosis is an important physiological process to remove infected, malfunctioning, or no longer needed cells from a multicellular organism. The apoptotic signaling pathways converge at the caspase cascade. In [3], a mathematical model for the signal transduction in a single cell has been proposed. This model is also studied in this paper, and depicted in Figure 2. For details about the model we refer to [3]. The process of apoptosis induction is known to be heterogeneous [2,3]. Therefore, the single cell model is extended by introducing differences in parameters: 1) The amount of caspase 8 (C8), caspase 3 (C3), caspase 8- and 10-associated RING protein (CARP), and inhibitor of apoptosis protein (IAP) is known to be different among cells. To account for this, the synthesis rates (k 8, k 9, k 10, and k 12 ) in individual cells are assumed to be different. The distribution of k 8, k 9, k 10, and k 12 within the population is modeled as log-normal distribution, with mean as published in [3] and a
5 A maximum likelihood estimator for parameter distributions in heterogeneous cell populations 5 log(θ j) E[log(θ j)] C8a(0) k 8 k 9 k 10 k 12 Fig. 3 Parallel coordinate density-plot in which each polyline represents the parameter of a single cell, θ (i). The color of a polyline encodes whether the cell survived ( ) or died ( ). After estimating line-density with additive alpha-blending, a logarithmic colormap is applied. The coordinates k 8 and k 10 show the best separation of colors and hence correspond to potential markers. IAP synthesis, k C3 synthesis, k 10 (a) Classification employing C3 synthesis, k 10, and IAP synthesis, k 8, as markers. For the classification of cell survival: TP = 0.77, FP = init. active C8, C8a(0) CARP synthesis, k 12 (b) Classification employing initial amount of C8a, C8a(0), and C8 synthesis, k 8, as markers. For the classification of cell survival: TP = 0.68, FP = True positive False positive C8a(0) k 8 k 9 k 10 k 12 C8a(0) and k 8 C8a(0) and k 9 C8a(0) and k 10 C8a(0) and k 12 k 8 and k 9 k 8 and k 10 k 8 and k 12 k 9 and k 10 k 9 and k 12 k 10 and k 12 k 8, k 10 and k 12 (c) Classification performance for different marker combinations m in ROC space. The performance of all individual markers, all marker pairs, and the best marker triplet is shown. Note that an optimal classifier would be in the upper left corner. Fig. 4 Illustration of achieved classification (prediction) performance using different marker combinations. In plot (b) and (a) the prediction ( = alive; = dead) is shown of two marker combinations, as well as a test sample ( = alive; = dead). Plot (c) depicts the classification performance of different marker combinations in ROC space. coefficient of variation of 0.4 (own unpublished data). The initial conditions of C8, C3, CARP, and IAP are set to their steady state values. 2) Similar to [3], the activation of the caspase cascade is modeled by a non-zero initial condition of active caspase 8, C8a(0). In the population, C8a(0) is log-normally distributed with a mean of 4,000 molecules per cells and a coefficient of variation of 0.4. The variation of C8a(0) accounts for variability up-stream of the caspase cascade. The binding affinities and kinetic rates are the same for all cells. The precise values can be found in [3]. Given this heterogeneous population, it is analyzed which cells undergo apoptosis during the first 12 hours. As indicator for this, the amount of active caspase 3 (C3a) is used. If more than 5,000 copies of active caspase 3 are present in a cell, this cell is assumed to undergo apoptosis, yielding the decision functional similar to (4). Hence, the question we address is which low-dimensional subset of the parameters, θ = [C8a(0), k 8, k 9, k 10, k 12 ] T, are good markers for cell death and survival, respectively. Parallel coordinates: In Figure 3, a sample S with 100, 000 members is visualized in parallel coordinates. The second and fourth parameters (θ m = [k 8, k 10 ] T ) indicate a good separation between the classes (orange = dead, blue = alive). Most of the surviving cells have high values of k 8 and low values of k 10, which corresponds to a high IAP expression and a low C3 expression, respectively. Although the other parameters also influence the process, their influence seems to be minor. SVM: Given the results from the visual analysis, we select θ m = [k 8, k 10 ] T and compute the classification quality using SVMs (with Gaussian kernels). As visible in Figure 4(a), we obtain a good separation (TP = 0.77, FP = 0.11). For comparison, all other combinations of two markers are evaluated and depicted in
6 6 J. Hasenauer et al. Figure 4(c). The marker θ m = [k 8, k 10 ] T outperforms all other combinations considering the norm distance to the optimal classifier. Some other combinations result in more than 50 % of false positive classifications (see e.g. Figure 4(b)). Of course, extending the marker vector e.g. by adding k 12 results in further improvement. This case study shows that parallel coordinate plots are a proper tools to easily determine markers. The predictive power of the markers can then be quantified using SVM. In this example, the markers found agree well with those found in the literature. In particular, the important role of IAP is outlined in several publications. This study suggests that the amount of available C3 is even more important than expected. 5 CONCLUSION In this paper, a first and novel explorative approach has been presented to determine markers for decision processes in heterogeneous populations. It has been shown that methods used for data analysis can also be employed to gain insight into complex models. Especially the potential of parallel coordinate plots and SVMs has been illustrated. The proposed visual analytics approach has been applied to a cell population model for the tumor necrosis factor induced proapoptotic signaling. The markers found are the same as those mentioned in the literature. This provides an additional and so far missing validation of the model and thus proves the usefulness of our approach. 6 ACKNOWLEDGMENTS The authors acknowledge financial support from the German Research Foundation within the Cluster of Excellence in Simulation Technology (EXC 310/1) at the University of Stuttgart, from the German Federal Ministry of Education and Research (BMBF) within the FORSYS-Partner program (grant nr A and D), and from Center Systems Biology at the University of Stuttgart. References 1. J. Hasenauer, S. Waldherr, N. Radde, M. Doszczak, P. Scheurich, and F. Allgöwer, A maximum likelihood estimator for parameter distributions in heterogeneous cell populations, Procedia Computer Science, vol. 1, no. 1, pp , S. Spencer, S. Gaudet, J. Albeck, J. Burke, and P. Sorger, Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis, Nature, vol. 459, no. 7245, pp , T. Eissing, H. Conzelmann, E. Gilles, F. Allgöwer, E. Bullinger, and P. Scheurich, Bistability analyses of a caspase activation model for receptor-induced apoptosis, Journal of Biological Chemistry, vol. 279, no. 35, pp , J. J. Thomas and K. A. Cook, A Visual Analytics Agenda., IEEE Computer Graphics and Applications, vol. 26, no. 1, pp. 10 3, A. Inselberg and B. Dimsdale, Parallel coordinates: a tool for visualizing multi-dimensional geometry, in Proc. of IEEE Visualization, 1990, pp J. Heinrich and D. Weiskopf, Continuous parallel coordinates, IEEE Transactions of Visual Computer Graphics, vol. 15, no. 6, pp , O. Ivanciuc, Reviews in computational chemistry, vol. 23, chapter Applications of support vector machines in chemistry, pp , Wiley-VCH, Weinheim, J. Hasenauer, C. Breindl, S. Waldherr, and F. Allgöwer, Approximative classification of regions in parameter spaces of nonlinear ODEs yielding different qualitative behavior, in Proc. IEEE Conference on Decision and Control (CDC 2010), Atlanta, USA, 2010, pp M. Zweig and G. Campbell, Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine, Clinical Chemistry, vol. 39, no. 8, pp , 1993.
Density-based modeling and identification of biochemical networks in cell populations
 Density-based modeling and identification of biochemical networks in cell populations J. Hasenauer 1,, S. Waldherr 1, M. Doszczak 2, P. Scheurich 2, and F. Allgöwer 1 arxiv:1002.4599v1 [q-bio.mn] 24 Feb
Density-based modeling and identification of biochemical networks in cell populations J. Hasenauer 1,, S. Waldherr 1, M. Doszczak 2, P. Scheurich 2, and F. Allgöwer 1 arxiv:1002.4599v1 [q-bio.mn] 24 Feb
Approximative Classification of Regions in Parameter Spaces of Nonlinear ODEs Yielding Different Qualitative Behavior
 J. Hasenauer C. Breindl S. Waldherr F. Allgöwer Approximative Classification of Regions in Parameter Spaces of Nonlinear ODEs Yielding Different Qualitative Behavior Stuttgart, December 2010 Institute
J. Hasenauer C. Breindl S. Waldherr F. Allgöwer Approximative Classification of Regions in Parameter Spaces of Nonlinear ODEs Yielding Different Qualitative Behavior Stuttgart, December 2010 Institute
Death wins against life in a spatially extended model of the caspase-3/8 feedback loop
 M. Daub a S. Waldherr b F. Allgöwer b P. Scheurich c G. Schneider a Death wins against life in a spatially extended model of the caspase-3/8 feedback loop Stuttgart, Januar 212 a Institute of Analysis,
M. Daub a S. Waldherr b F. Allgöwer b P. Scheurich c G. Schneider a Death wins against life in a spatially extended model of the caspase-3/8 feedback loop Stuttgart, Januar 212 a Institute of Analysis,
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie
 A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
Introduction to Support Vector Machines
 Introduction to Support Vector Machines Hsuan-Tien Lin Learning Systems Group, California Institute of Technology Talk in NTU EE/CS Speech Lab, November 16, 2005 H.-T. Lin (Learning Systems Group) Introduction
Introduction to Support Vector Machines Hsuan-Tien Lin Learning Systems Group, California Institute of Technology Talk in NTU EE/CS Speech Lab, November 16, 2005 H.-T. Lin (Learning Systems Group) Introduction
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Analysis and Simulation of Biological Systems
 Analysis and Simulation of Biological Systems Dr. Carlo Cosentino School of Computer and Biomedical Engineering Department of Experimental and Clinical Medicine Università degli Studi Magna Graecia Catanzaro,
Analysis and Simulation of Biological Systems Dr. Carlo Cosentino School of Computer and Biomedical Engineering Department of Experimental and Clinical Medicine Università degli Studi Magna Graecia Catanzaro,
Support'Vector'Machines. Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan
 Support'Vector'Machines Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan kasthuri.kannan@nyumc.org Overview Support Vector Machines for Classification Linear Discrimination Nonlinear Discrimination
Support'Vector'Machines Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan kasthuri.kannan@nyumc.org Overview Support Vector Machines for Classification Linear Discrimination Nonlinear Discrimination
Machine Learning Linear Classification. Prof. Matteo Matteucci
 Machine Learning Linear Classification Prof. Matteo Matteucci Recall from the first lecture 2 X R p Regression Y R Continuous Output X R p Y {Ω 0, Ω 1,, Ω K } Classification Discrete Output X R p Y (X)
Machine Learning Linear Classification Prof. Matteo Matteucci Recall from the first lecture 2 X R p Regression Y R Continuous Output X R p Y {Ω 0, Ω 1,, Ω K } Classification Discrete Output X R p Y (X)
Plan. Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics.
 Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Bayesian Decision Theory
 Introduction to Pattern Recognition [ Part 4 ] Mahdi Vasighi Remarks It is quite common to assume that the data in each class are adequately described by a Gaussian distribution. Bayesian classifier is
Introduction to Pattern Recognition [ Part 4 ] Mahdi Vasighi Remarks It is quite common to assume that the data in each class are adequately described by a Gaussian distribution. Bayesian classifier is
HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH
 HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH Hoang Trang 1, Tran Hoang Loc 1 1 Ho Chi Minh City University of Technology-VNU HCM, Ho Chi
HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH Hoang Trang 1, Tran Hoang Loc 1 1 Ho Chi Minh City University of Technology-VNU HCM, Ho Chi
Support Vector Machines. Introduction to Data Mining, 2 nd Edition by Tan, Steinbach, Karpatne, Kumar
 Data Mining Support Vector Machines Introduction to Data Mining, 2 nd Edition by Tan, Steinbach, Karpatne, Kumar 02/03/2018 Introduction to Data Mining 1 Support Vector Machines Find a linear hyperplane
Data Mining Support Vector Machines Introduction to Data Mining, 2 nd Edition by Tan, Steinbach, Karpatne, Kumar 02/03/2018 Introduction to Data Mining 1 Support Vector Machines Find a linear hyperplane
PATTERN RECOGNITION AND MACHINE LEARNING
 PATTERN RECOGNITION AND MACHINE LEARNING Chapter 1. Introduction Shuai Huang April 21, 2014 Outline 1 What is Machine Learning? 2 Curve Fitting 3 Probability Theory 4 Model Selection 5 The curse of dimensionality
PATTERN RECOGNITION AND MACHINE LEARNING Chapter 1. Introduction Shuai Huang April 21, 2014 Outline 1 What is Machine Learning? 2 Curve Fitting 3 Probability Theory 4 Model Selection 5 The curse of dimensionality
Non-Bayesian Classifiers Part II: Linear Discriminants and Support Vector Machines
 Non-Bayesian Classifiers Part II: Linear Discriminants and Support Vector Machines Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Fall 2018 CS 551, Fall
Non-Bayesian Classifiers Part II: Linear Discriminants and Support Vector Machines Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Fall 2018 CS 551, Fall
Support Vector Machines, Kernel SVM
 Support Vector Machines, Kernel SVM Professor Ameet Talwalkar Professor Ameet Talwalkar CS260 Machine Learning Algorithms February 27, 2017 1 / 40 Outline 1 Administration 2 Review of last lecture 3 SVM
Support Vector Machines, Kernel SVM Professor Ameet Talwalkar Professor Ameet Talwalkar CS260 Machine Learning Algorithms February 27, 2017 1 / 40 Outline 1 Administration 2 Review of last lecture 3 SVM
Lecture 4 Discriminant Analysis, k-nearest Neighbors
 Lecture 4 Discriminant Analysis, k-nearest Neighbors Fredrik Lindsten Division of Systems and Control Department of Information Technology Uppsala University. Email: fredrik.lindsten@it.uu.se fredrik.lindsten@it.uu.se
Lecture 4 Discriminant Analysis, k-nearest Neighbors Fredrik Lindsten Division of Systems and Control Department of Information Technology Uppsala University. Email: fredrik.lindsten@it.uu.se fredrik.lindsten@it.uu.se
L5 Support Vector Classification
 L5 Support Vector Classification Support Vector Machine Problem definition Geometrical picture Optimization problem Optimization Problem Hard margin Convexity Dual problem Soft margin problem Alexander
L5 Support Vector Classification Support Vector Machine Problem definition Geometrical picture Optimization problem Optimization Problem Hard margin Convexity Dual problem Soft margin problem Alexander
Prediction and Classif ication of Human G-protein Coupled Receptors Based on Support Vector Machines
 Article Prediction and Classif ication of Human G-protein Coupled Receptors Based on Support Vector Machines Yun-Fei Wang, Huan Chen, and Yan-Hong Zhou* Hubei Bioinformatics and Molecular Imaging Key Laboratory,
Article Prediction and Classif ication of Human G-protein Coupled Receptors Based on Support Vector Machines Yun-Fei Wang, Huan Chen, and Yan-Hong Zhou* Hubei Bioinformatics and Molecular Imaging Key Laboratory,
Applied Machine Learning Annalisa Marsico
 Applied Machine Learning Annalisa Marsico OWL RNA Bionformatics group Max Planck Institute for Molecular Genetics Free University of Berlin 22 April, SoSe 2015 Goals Feature Selection rather than Feature
Applied Machine Learning Annalisa Marsico OWL RNA Bionformatics group Max Planck Institute for Molecular Genetics Free University of Berlin 22 April, SoSe 2015 Goals Feature Selection rather than Feature
Regulation of Apoptosis via the NFκB Pathway: Modeling and Analysis
 Regulation of Apoptosis via the NFκB Pathway: Modeling and Analysis Madalena Chaves, Thomas Eissing, 2 and Frank Allgöwer 3 COMORE, INRIA, 24 Route des Lucioles, BP 93, 692 Sophia-Antipolis, France; mchaves@sophia.inria.fr
Regulation of Apoptosis via the NFκB Pathway: Modeling and Analysis Madalena Chaves, Thomas Eissing, 2 and Frank Allgöwer 3 COMORE, INRIA, 24 Route des Lucioles, BP 93, 692 Sophia-Antipolis, France; mchaves@sophia.inria.fr
Machine Learning in Action
 Machine Learning in Action Tatyana Goldberg (goldberg@rostlab.org) August 16, 2016 @ Machine Learning in Biology Beijing Genomics Institute in Shenzhen, China June 2014 GenBank 1 173,353,076 DNA sequences
Machine Learning in Action Tatyana Goldberg (goldberg@rostlab.org) August 16, 2016 @ Machine Learning in Biology Beijing Genomics Institute in Shenzhen, China June 2014 GenBank 1 173,353,076 DNA sequences
Statistical Pattern Recognition
 Statistical Pattern Recognition Support Vector Machine (SVM) Hamid R. Rabiee Hadi Asheri, Jafar Muhammadi, Nima Pourdamghani Spring 2013 http://ce.sharif.edu/courses/91-92/2/ce725-1/ Agenda Introduction
Statistical Pattern Recognition Support Vector Machine (SVM) Hamid R. Rabiee Hadi Asheri, Jafar Muhammadi, Nima Pourdamghani Spring 2013 http://ce.sharif.edu/courses/91-92/2/ce725-1/ Agenda Introduction
18.9 SUPPORT VECTOR MACHINES
 744 Chapter 8. Learning from Examples is the fact that each regression problem will be easier to solve, because it involves only the examples with nonzero weight the examples whose kernels overlap the
744 Chapter 8. Learning from Examples is the fact that each regression problem will be easier to solve, because it involves only the examples with nonzero weight the examples whose kernels overlap the
15-388/688 - Practical Data Science: Nonlinear modeling, cross-validation, regularization, and evaluation
 15-388/688 - Practical Data Science: Nonlinear modeling, cross-validation, regularization, and evaluation J. Zico Kolter Carnegie Mellon University Fall 2016 1 Outline Example: return to peak demand prediction
15-388/688 - Practical Data Science: Nonlinear modeling, cross-validation, regularization, and evaluation J. Zico Kolter Carnegie Mellon University Fall 2016 1 Outline Example: return to peak demand prediction
BISTABILITY is a recurrent motif in biology, and there
 PREPRINT ACCEPTED TO SPECIAL ISSUE IEEE TAC TCAS-I 1 Bistable biological systems: a characterization through local compact input-to-state stability Madalena Chaves Thomas Eissing and Frank Allgöwer Member
PREPRINT ACCEPTED TO SPECIAL ISSUE IEEE TAC TCAS-I 1 Bistable biological systems: a characterization through local compact input-to-state stability Madalena Chaves Thomas Eissing and Frank Allgöwer Member
Support Vector Machine (SVM) and Kernel Methods
 Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2016 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2016 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods
 Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2015 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2015 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Pattern Recognition and Machine Learning. Perceptrons and Support Vector machines
 Pattern Recognition and Machine Learning James L. Crowley ENSIMAG 3 - MMIS Fall Semester 2016 Lessons 6 10 Jan 2017 Outline Perceptrons and Support Vector machines Notation... 2 Perceptrons... 3 History...3
Pattern Recognition and Machine Learning James L. Crowley ENSIMAG 3 - MMIS Fall Semester 2016 Lessons 6 10 Jan 2017 Outline Perceptrons and Support Vector machines Notation... 2 Perceptrons... 3 History...3
Jeff Howbert Introduction to Machine Learning Winter
 Classification / Regression Support Vector Machines Jeff Howbert Introduction to Machine Learning Winter 2012 1 Topics SVM classifiers for linearly separable classes SVM classifiers for non-linearly separable
Classification / Regression Support Vector Machines Jeff Howbert Introduction to Machine Learning Winter 2012 1 Topics SVM classifiers for linearly separable classes SVM classifiers for non-linearly separable
Data Mining: Concepts and Techniques. (3 rd ed.) Chapter 8. Chapter 8. Classification: Basic Concepts
 Data Mining: Concepts and Techniques (3 rd ed.) Chapter 8 1 Chapter 8. Classification: Basic Concepts Classification: Basic Concepts Decision Tree Induction Bayes Classification Methods Rule-Based Classification
Data Mining: Concepts and Techniques (3 rd ed.) Chapter 8 1 Chapter 8. Classification: Basic Concepts Classification: Basic Concepts Decision Tree Induction Bayes Classification Methods Rule-Based Classification
Sig2GRN: A Software Tool Linking Signaling Pathway with Gene Regulatory Network for Dynamic Simulation
 Sig2GRN: A Software Tool Linking Signaling Pathway with Gene Regulatory Network for Dynamic Simulation Authors: Fan Zhang, Runsheng Liu and Jie Zheng Presented by: Fan Wu School of Computer Science and
Sig2GRN: A Software Tool Linking Signaling Pathway with Gene Regulatory Network for Dynamic Simulation Authors: Fan Zhang, Runsheng Liu and Jie Zheng Presented by: Fan Wu School of Computer Science and
Pointwise Exact Bootstrap Distributions of Cost Curves
 Pointwise Exact Bootstrap Distributions of Cost Curves Charles Dugas and David Gadoury University of Montréal 25th ICML Helsinki July 2008 Dugas, Gadoury (U Montréal) Cost curves July 8, 2008 1 / 24 Outline
Pointwise Exact Bootstrap Distributions of Cost Curves Charles Dugas and David Gadoury University of Montréal 25th ICML Helsinki July 2008 Dugas, Gadoury (U Montréal) Cost curves July 8, 2008 1 / 24 Outline
Support Vector Machines
 Support Vector Machines Here we approach the two-class classification problem in a direct way: We try and find a plane that separates the classes in feature space. If we cannot, we get creative in two
Support Vector Machines Here we approach the two-class classification problem in a direct way: We try and find a plane that separates the classes in feature space. If we cannot, we get creative in two
Apoptosis in Mammalian Cells
 Apoptosis in Mammalian Cells 7.16 2-10-05 Apoptosis is an important factor in many human diseases Cancer malignant cells evade death by suppressing apoptosis (too little apoptosis) Stroke damaged neurons
Apoptosis in Mammalian Cells 7.16 2-10-05 Apoptosis is an important factor in many human diseases Cancer malignant cells evade death by suppressing apoptosis (too little apoptosis) Stroke damaged neurons
Introduction to Support Vector Machines
 Introduction to Support Vector Machines Shivani Agarwal Support Vector Machines (SVMs) Algorithm for learning linear classifiers Motivated by idea of maximizing margin Efficient extension to non-linear
Introduction to Support Vector Machines Shivani Agarwal Support Vector Machines (SVMs) Algorithm for learning linear classifiers Motivated by idea of maximizing margin Efficient extension to non-linear
6.036 midterm review. Wednesday, March 18, 15
 6.036 midterm review 1 Topics covered supervised learning labels available unsupervised learning no labels available semi-supervised learning some labels available - what algorithms have you learned that
6.036 midterm review 1 Topics covered supervised learning labels available unsupervised learning no labels available semi-supervised learning some labels available - what algorithms have you learned that
Outline. Basic concepts: SVM and kernels SVM primal/dual problems. Chih-Jen Lin (National Taiwan Univ.) 1 / 22
 Outline Basic concepts: SVM and kernels SVM primal/dual problems Chih-Jen Lin (National Taiwan Univ.) 1 / 22 Outline Basic concepts: SVM and kernels Basic concepts: SVM and kernels SVM primal/dual problems
Outline Basic concepts: SVM and kernels SVM primal/dual problems Chih-Jen Lin (National Taiwan Univ.) 1 / 22 Outline Basic concepts: SVM and kernels Basic concepts: SVM and kernels SVM primal/dual problems
Chapter 6: Classification
 Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
c 4, < y 2, 1 0, otherwise,
 Fundamentals of Big Data Analytics Univ.-Prof. Dr. rer. nat. Rudolf Mathar Problem. Probability theory: The outcome of an experiment is described by three events A, B and C. The probabilities Pr(A) =,
Fundamentals of Big Data Analytics Univ.-Prof. Dr. rer. nat. Rudolf Mathar Problem. Probability theory: The outcome of an experiment is described by three events A, B and C. The probabilities Pr(A) =,
Decision-Tree Based Model. for Efficient Identification of Parameter Relations Leading. to Different Signaling States
 Decision-Tree Based Model Analysis for Efficient Identification of Parameter Relations Leading to Different Signaling States The Harvard community has made this article openly available. Please share how
Decision-Tree Based Model Analysis for Efficient Identification of Parameter Relations Leading to Different Signaling States The Harvard community has made this article openly available. Please share how
INTRODUCTION TO PATTERN RECOGNITION
 INTRODUCTION TO PATTERN RECOGNITION INSTRUCTOR: WEI DING 1 Pattern Recognition Automatic discovery of regularities in data through the use of computer algorithms With the use of these regularities to take
INTRODUCTION TO PATTERN RECOGNITION INSTRUCTOR: WEI DING 1 Pattern Recognition Automatic discovery of regularities in data through the use of computer algorithms With the use of these regularities to take
Control structure and limitations of biochemical networks
 21 American Control Conference Marriott Waterfront, Baltimore, MD, USA June 3-July 2, 21 FrC11.6 Control structure and limitations of biochemical networs F. López-Caamal, D. A. Oyarzún, J. A. Moreno, D.
21 American Control Conference Marriott Waterfront, Baltimore, MD, USA June 3-July 2, 21 FrC11.6 Control structure and limitations of biochemical networs F. López-Caamal, D. A. Oyarzún, J. A. Moreno, D.
Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function.
 Bayesian learning: Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function. Let y be the true label and y be the predicted
Bayesian learning: Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function. Let y be the true label and y be the predicted
Support Vector Machine (SVM) & Kernel CE-717: Machine Learning Sharif University of Technology. M. Soleymani Fall 2012
 Support Vector Machine (SVM) & Kernel CE-717: Machine Learning Sharif University of Technology M. Soleymani Fall 2012 Linear classifier Which classifier? x 2 x 1 2 Linear classifier Margin concept x 2
Support Vector Machine (SVM) & Kernel CE-717: Machine Learning Sharif University of Technology M. Soleymani Fall 2012 Linear classifier Which classifier? x 2 x 1 2 Linear classifier Margin concept x 2
The exam is closed book, closed notes except your one-page (two sides) or two-page (one side) crib sheet.
 CS 189 Spring 013 Introduction to Machine Learning Final You have 3 hours for the exam. The exam is closed book, closed notes except your one-page (two sides) or two-page (one side) crib sheet. Please
CS 189 Spring 013 Introduction to Machine Learning Final You have 3 hours for the exam. The exam is closed book, closed notes except your one-page (two sides) or two-page (one side) crib sheet. Please
Machine Learning. Support Vector Machines. Fabio Vandin November 20, 2017
 Machine Learning Support Vector Machines Fabio Vandin November 20, 2017 1 Classification and Margin Consider a classification problem with two classes: instance set X = R d label set Y = { 1, 1}. Training
Machine Learning Support Vector Machines Fabio Vandin November 20, 2017 1 Classification and Margin Consider a classification problem with two classes: instance set X = R d label set Y = { 1, 1}. Training
Unraveling Apoptosis Signalling using Linear Control Methods: Linking the Loop Gain to Reverting the Decision to Undergo Apoptosis
 Preprints of the 9th International Symposium on Advanced Control of Chemical Processes The International Federation of Automatic Control TuPoster.3 Unraveling Apoptosis Signalling using Linear Control
Preprints of the 9th International Symposium on Advanced Control of Chemical Processes The International Federation of Automatic Control TuPoster.3 Unraveling Apoptosis Signalling using Linear Control
Introduction to SVM and RVM
 Introduction to SVM and RVM Machine Learning Seminar HUS HVL UIB Yushu Li, UIB Overview Support vector machine SVM First introduced by Vapnik, et al. 1992 Several literature and wide applications Relevance
Introduction to SVM and RVM Machine Learning Seminar HUS HVL UIB Yushu Li, UIB Overview Support vector machine SVM First introduced by Vapnik, et al. 1992 Several literature and wide applications Relevance
Performance Evaluation and Comparison
 Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Cross Validation and Resampling 3 Interval Estimation
Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Cross Validation and Resampling 3 Interval Estimation
Modeling heterogeneous responsiveness of intrinsic apoptosis pathway
 Ooi and Ma BMC Systems Biology 213, 7:65 http://www.biomedcentral.com/1752-59/7/65 RESEARCH ARTICLE OpenAccess Modeling heterogeneous responsiveness of intrinsic apoptosis pathway Hsu Kiang Ooi and Lan
Ooi and Ma BMC Systems Biology 213, 7:65 http://www.biomedcentral.com/1752-59/7/65 RESEARCH ARTICLE OpenAccess Modeling heterogeneous responsiveness of intrinsic apoptosis pathway Hsu Kiang Ooi and Lan
Analysis of Feedback Mechanisms in Cell-biological Systems
 Proceedings of the 17th World Congress The International Federation of Automatic Control Analysis of Feedback Mechanisms in Cell-biological Systems Steffen Waldherr, Thomas Eissing, and Frank Allgöwer
Proceedings of the 17th World Congress The International Federation of Automatic Control Analysis of Feedback Mechanisms in Cell-biological Systems Steffen Waldherr, Thomas Eissing, and Frank Allgöwer
The definitions and notation are those introduced in the lectures slides. R Ex D [h
 Mehryar Mohri Foundations of Machine Learning Courant Institute of Mathematical Sciences Homework assignment 2 October 04, 2016 Due: October 18, 2016 A. Rademacher complexity The definitions and notation
Mehryar Mohri Foundations of Machine Learning Courant Institute of Mathematical Sciences Homework assignment 2 October 04, 2016 Due: October 18, 2016 A. Rademacher complexity The definitions and notation
Support Vector Machine (SVM) and Kernel Methods
 Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2014 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Support Vector Machine (SVM) and Kernel Methods CE-717: Machine Learning Sharif University of Technology Fall 2014 Soleymani Outline Margin concept Hard-Margin SVM Soft-Margin SVM Dual Problems of Hard-Margin
Machine Learning Concepts in Chemoinformatics
 Machine Learning Concepts in Chemoinformatics Martin Vogt B-IT Life Science Informatics Rheinische Friedrich-Wilhelms-Universität Bonn BigChem Winter School 2017 25. October Data Mining in Chemoinformatics
Machine Learning Concepts in Chemoinformatics Martin Vogt B-IT Life Science Informatics Rheinische Friedrich-Wilhelms-Universität Bonn BigChem Winter School 2017 25. October Data Mining in Chemoinformatics
Linear classifiers selecting hyperplane maximizing separation margin between classes (large margin classifiers)
 Support vector machines In a nutshell Linear classifiers selecting hyperplane maximizing separation margin between classes (large margin classifiers) Solution only depends on a small subset of training
Support vector machines In a nutshell Linear classifiers selecting hyperplane maximizing separation margin between classes (large margin classifiers) Solution only depends on a small subset of training
A Sparse Solution Approach to Gene Selection for Cancer Diagnosis Using Microarray Data
 A Sparse Solution Approach to Gene Selection for Cancer Diagnosis Using Microarray Data Yoonkyung Lee Department of Statistics The Ohio State University http://www.stat.ohio-state.edu/ yklee May 13, 2005
A Sparse Solution Approach to Gene Selection for Cancer Diagnosis Using Microarray Data Yoonkyung Lee Department of Statistics The Ohio State University http://www.stat.ohio-state.edu/ yklee May 13, 2005
Class 4: Classification. Quaid Morris February 11 th, 2011 ML4Bio
 Class 4: Classification Quaid Morris February 11 th, 211 ML4Bio Overview Basic concepts in classification: overfitting, cross-validation, evaluation. Linear Discriminant Analysis and Quadratic Discriminant
Class 4: Classification Quaid Morris February 11 th, 211 ML4Bio Overview Basic concepts in classification: overfitting, cross-validation, evaluation. Linear Discriminant Analysis and Quadratic Discriminant
Statistical Methods for SVM
 Statistical Methods for SVM Support Vector Machines Here we approach the two-class classification problem in a direct way: We try and find a plane that separates the classes in feature space. If we cannot,
Statistical Methods for SVM Support Vector Machines Here we approach the two-class classification problem in a direct way: We try and find a plane that separates the classes in feature space. If we cannot,
ECE662: Pattern Recognition and Decision Making Processes: HW TWO
 ECE662: Pattern Recognition and Decision Making Processes: HW TWO Purdue University Department of Electrical and Computer Engineering West Lafayette, INDIANA, USA Abstract. In this report experiments are
ECE662: Pattern Recognition and Decision Making Processes: HW TWO Purdue University Department of Electrical and Computer Engineering West Lafayette, INDIANA, USA Abstract. In this report experiments are
Learning Kernel Parameters by using Class Separability Measure
 Learning Kernel Parameters by using Class Separability Measure Lei Wang, Kap Luk Chan School of Electrical and Electronic Engineering Nanyang Technological University Singapore, 3979 E-mail: P 3733@ntu.edu.sg,eklchan@ntu.edu.sg
Learning Kernel Parameters by using Class Separability Measure Lei Wang, Kap Luk Chan School of Electrical and Electronic Engineering Nanyang Technological University Singapore, 3979 E-mail: P 3733@ntu.edu.sg,eklchan@ntu.edu.sg
Machine Learning and Data Mining. Support Vector Machines. Kalev Kask
 Machine Learning and Data Mining Support Vector Machines Kalev Kask Linear classifiers Which decision boundary is better? Both have zero training error (perfect training accuracy) But, one of them seems
Machine Learning and Data Mining Support Vector Machines Kalev Kask Linear classifiers Which decision boundary is better? Both have zero training error (perfect training accuracy) But, one of them seems
natural development from this collection of knowledge: it is more reliable to predict the property
 1 Chapter 1 Introduction As the basis of all life phenomena, the interaction of biomolecules has been under the scrutiny of scientists and cataloged meticulously [2]. The recent advent of systems biology
1 Chapter 1 Introduction As the basis of all life phenomena, the interaction of biomolecules has been under the scrutiny of scientists and cataloged meticulously [2]. The recent advent of systems biology
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Machine Learning, Fall 2009: Midterm
 10-601 Machine Learning, Fall 009: Midterm Monday, November nd hours 1. Personal info: Name: Andrew account: E-mail address:. You are permitted two pages of notes and a calculator. Please turn off all
10-601 Machine Learning, Fall 009: Midterm Monday, November nd hours 1. Personal info: Name: Andrew account: E-mail address:. You are permitted two pages of notes and a calculator. Please turn off all
Probabilistic Machine Learning. Industrial AI Lab.
 Probabilistic Machine Learning Industrial AI Lab. Probabilistic Linear Regression Outline Probabilistic Classification Probabilistic Clustering Probabilistic Dimension Reduction 2 Probabilistic Linear
Probabilistic Machine Learning Industrial AI Lab. Probabilistic Linear Regression Outline Probabilistic Classification Probabilistic Clustering Probabilistic Dimension Reduction 2 Probabilistic Linear
FIND A FUNCTION TO CLASSIFY HIGH VALUE CUSTOMERS
 LINEAR CLASSIFIER 1 FIND A FUNCTION TO CLASSIFY HIGH VALUE CUSTOMERS x f y High Value Customers Salary Task: Find Nb Orders 150 70 300 100 200 80 120 100 Low Value Customers Salary Nb Orders 40 80 220
LINEAR CLASSIFIER 1 FIND A FUNCTION TO CLASSIFY HIGH VALUE CUSTOMERS x f y High Value Customers Salary Task: Find Nb Orders 150 70 300 100 200 80 120 100 Low Value Customers Salary Nb Orders 40 80 220
Navigation and Obstacle Avoidance via Backstepping for Mechanical Systems with Drift in the Closed Loop
 Navigation and Obstacle Avoidance via Backstepping for Mechanical Systems with Drift in the Closed Loop Jan Maximilian Montenbruck, Mathias Bürger, Frank Allgöwer Abstract We study backstepping controllers
Navigation and Obstacle Avoidance via Backstepping for Mechanical Systems with Drift in the Closed Loop Jan Maximilian Montenbruck, Mathias Bürger, Frank Allgöwer Abstract We study backstepping controllers
BAYESIAN CLASSIFICATION OF HIGH DIMENSIONAL DATA WITH GAUSSIAN PROCESS USING DIFFERENT KERNELS
 BAYESIAN CLASSIFICATION OF HIGH DIMENSIONAL DATA WITH GAUSSIAN PROCESS USING DIFFERENT KERNELS Oloyede I. Department of Statistics, University of Ilorin, Ilorin, Nigeria Corresponding Author: Oloyede I.,
BAYESIAN CLASSIFICATION OF HIGH DIMENSIONAL DATA WITH GAUSSIAN PROCESS USING DIFFERENT KERNELS Oloyede I. Department of Statistics, University of Ilorin, Ilorin, Nigeria Corresponding Author: Oloyede I.,
CMU-Q Lecture 24:
 CMU-Q 15-381 Lecture 24: Supervised Learning 2 Teacher: Gianni A. Di Caro SUPERVISED LEARNING Hypotheses space Hypothesis function Labeled Given Errors Performance criteria Given a collection of input
CMU-Q 15-381 Lecture 24: Supervised Learning 2 Teacher: Gianni A. Di Caro SUPERVISED LEARNING Hypotheses space Hypothesis function Labeled Given Errors Performance criteria Given a collection of input
Multisurface Proximal Support Vector Machine Classification via Generalized Eigenvalues
 Multisurface Proximal Support Vector Machine Classification via Generalized Eigenvalues O. L. Mangasarian and E. W. Wild Presented by: Jun Fang Multisurface Proximal Support Vector Machine Classification
Multisurface Proximal Support Vector Machine Classification via Generalized Eigenvalues O. L. Mangasarian and E. W. Wild Presented by: Jun Fang Multisurface Proximal Support Vector Machine Classification
Programmed Cell Death
 Programmed Cell Death Dewajani Purnomosari Department of Histology and Cell Biology Faculty of Medicine Universitas Gadjah Mada d.purnomosari@ugm.ac.id What is apoptosis? a normal component of the development
Programmed Cell Death Dewajani Purnomosari Department of Histology and Cell Biology Faculty of Medicine Universitas Gadjah Mada d.purnomosari@ugm.ac.id What is apoptosis? a normal component of the development
Analysis of heterogeneous cell populations: A density-based modeling and identification framework
 Analysis of heterogeneous cell populations: A density-based modeling and identification framework Jan Hasenauer a, Steffen Waldherr a, Malgorzata Doszczak b, Peter Scheurich b, Nicole Radde a, Frank Allgöwer
Analysis of heterogeneous cell populations: A density-based modeling and identification framework Jan Hasenauer a, Steffen Waldherr a, Malgorzata Doszczak b, Peter Scheurich b, Nicole Radde a, Frank Allgöwer
Linear, threshold units. Linear Discriminant Functions and Support Vector Machines. Biometrics CSE 190 Lecture 11. X i : inputs W i : weights
 Linear Discriminant Functions and Support Vector Machines Linear, threshold units CSE19, Winter 11 Biometrics CSE 19 Lecture 11 1 X i : inputs W i : weights θ : threshold 3 4 5 1 6 7 Courtesy of University
Linear Discriminant Functions and Support Vector Machines Linear, threshold units CSE19, Winter 11 Biometrics CSE 19 Lecture 11 1 X i : inputs W i : weights θ : threshold 3 4 5 1 6 7 Courtesy of University
Computation of the posterior entropy in a Bayesian framework for parameter estimation in biological networks
 Andrei Kramer Jan Hasenauer Frank Allgöwer Nicole Radde Computation of the posterior entropy in a Bayesian framework for parameter estimation in biological networks Stuttgart, March Institute for Systems
Andrei Kramer Jan Hasenauer Frank Allgöwer Nicole Radde Computation of the posterior entropy in a Bayesian framework for parameter estimation in biological networks Stuttgart, March Institute for Systems
Notes of Dr. Anil Mishra at 1
 Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Infinite Ensemble Learning with Support Vector Machinery
 Infinite Ensemble Learning with Support Vector Machinery Hsuan-Tien Lin and Ling Li Learning Systems Group, California Institute of Technology ECML/PKDD, October 4, 2005 H.-T. Lin and L. Li (Learning Systems
Infinite Ensemble Learning with Support Vector Machinery Hsuan-Tien Lin and Ling Li Learning Systems Group, California Institute of Technology ECML/PKDD, October 4, 2005 H.-T. Lin and L. Li (Learning Systems
Mark your answers ON THE EXAM ITSELF. If you are not sure of your answer you may wish to provide a brief explanation.
 CS 189 Spring 2015 Introduction to Machine Learning Midterm You have 80 minutes for the exam. The exam is closed book, closed notes except your one-page crib sheet. No calculators or electronic items.
CS 189 Spring 2015 Introduction to Machine Learning Midterm You have 80 minutes for the exam. The exam is closed book, closed notes except your one-page crib sheet. No calculators or electronic items.
Course plan Academic Year Qualification MSc on Bioinformatics for Health Sciences. Subject name: Computational Systems Biology Code: 30180
 Course plan 201-201 Academic Year Qualification MSc on Bioinformatics for Health Sciences 1. Description of the subject Subject name: Code: 30180 Total credits: 5 Workload: 125 hours Year: 1st Term: 3
Course plan 201-201 Academic Year Qualification MSc on Bioinformatics for Health Sciences 1. Description of the subject Subject name: Code: 30180 Total credits: 5 Workload: 125 hours Year: 1st Term: 3
Biological Pathways Representation by Petri Nets and extension
 Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Linear & nonlinear classifiers
 Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1396 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1396 1 / 44 Table
Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1396 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1396 1 / 44 Table
Machine Learning : Support Vector Machines
 Machine Learning Support Vector Machines 05/01/2014 Machine Learning : Support Vector Machines Linear Classifiers (recap) A building block for almost all a mapping, a partitioning of the input space into
Machine Learning Support Vector Machines 05/01/2014 Machine Learning : Support Vector Machines Linear Classifiers (recap) A building block for almost all a mapping, a partitioning of the input space into
Mathematically modeling the sequential application of a cytotoxic nanoparticle and a PI3K-inhibitor enhances anti-tumor efficacy
 Supplementary information Mathematically modeling the sequential application of a cytotoxic nanoparticle and a PI3K-inhibitor enhances anti-tumor efficacy Ambarish Pandey 1, 5, Ashish Kulkarni 1, Bhaskar
Supplementary information Mathematically modeling the sequential application of a cytotoxic nanoparticle and a PI3K-inhibitor enhances anti-tumor efficacy Ambarish Pandey 1, 5, Ashish Kulkarni 1, Bhaskar
Perceptron Revisited: Linear Separators. Support Vector Machines
 Support Vector Machines Perceptron Revisited: Linear Separators Binary classification can be viewed as the task of separating classes in feature space: w T x + b > 0 w T x + b = 0 w T x + b < 0 Department
Support Vector Machines Perceptron Revisited: Linear Separators Binary classification can be viewed as the task of separating classes in feature space: w T x + b > 0 w T x + b = 0 w T x + b < 0 Department
BAYESIAN DECISION THEORY
 Last updated: September 17, 2012 BAYESIAN DECISION THEORY Problems 2 The following problems from the textbook are relevant: 2.1 2.9, 2.11, 2.17 For this week, please at least solve Problem 2.3. We will
Last updated: September 17, 2012 BAYESIAN DECISION THEORY Problems 2 The following problems from the textbook are relevant: 2.1 2.9, 2.11, 2.17 For this week, please at least solve Problem 2.3. We will
Unsupervised Learning with Permuted Data
 Unsupervised Learning with Permuted Data Sergey Kirshner skirshne@ics.uci.edu Sridevi Parise sparise@ics.uci.edu Padhraic Smyth smyth@ics.uci.edu School of Information and Computer Science, University
Unsupervised Learning with Permuted Data Sergey Kirshner skirshne@ics.uci.edu Sridevi Parise sparise@ics.uci.edu Padhraic Smyth smyth@ics.uci.edu School of Information and Computer Science, University
Characterization of Jet Charge at the LHC
 Characterization of Jet Charge at the LHC Thomas Dylan Rueter, Krishna Soni Abstract The Large Hadron Collider (LHC) produces a staggering amount of data - about 30 petabytes annually. One of the largest
Characterization of Jet Charge at the LHC Thomas Dylan Rueter, Krishna Soni Abstract The Large Hadron Collider (LHC) produces a staggering amount of data - about 30 petabytes annually. One of the largest
Proceedings of the FOSBE 2007 Stuttgart, Germany, September 9-12, 2007
 Proceedings of the FOSBE 2007 Stuttgart, Germany, September 9-12, 2007 A FEEDBACK APPROACH TO BIFURCATION ANALYSIS IN BIOCHEMICAL NETWORKS WITH MANY PARAMETERS Steffen Waldherr 1 and Frank Allgöwer Institute
Proceedings of the FOSBE 2007 Stuttgart, Germany, September 9-12, 2007 A FEEDBACK APPROACH TO BIFURCATION ANALYSIS IN BIOCHEMICAL NETWORKS WITH MANY PARAMETERS Steffen Waldherr 1 and Frank Allgöwer Institute
Networks in systems biology
 Networks in systems biology Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Spring 2017 M. Macauley (Clemson) Networks in systems
Networks in systems biology Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Spring 2017 M. Macauley (Clemson) Networks in systems
Speaker Verification Using Accumulative Vectors with Support Vector Machines
 Speaker Verification Using Accumulative Vectors with Support Vector Machines Manuel Aguado Martínez, Gabriel Hernández-Sierra, and José Ramón Calvo de Lara Advanced Technologies Application Center, Havana,
Speaker Verification Using Accumulative Vectors with Support Vector Machines Manuel Aguado Martínez, Gabriel Hernández-Sierra, and José Ramón Calvo de Lara Advanced Technologies Application Center, Havana,
Using an Ensemble of One-Class SVM Classifiers to Harden Payload-based Anomaly Detection Systems
 Using an Ensemble of One-Class SVM Classifiers to Harden Payload-based Anomaly Detection Systems Roberto Perdisci,, Guofei Gu,WenkeLee College of Computing, Georgia Institute of Technology, Atlanta, GA
Using an Ensemble of One-Class SVM Classifiers to Harden Payload-based Anomaly Detection Systems Roberto Perdisci,, Guofei Gu,WenkeLee College of Computing, Georgia Institute of Technology, Atlanta, GA
Support Vector Machine for Classification and Regression
 Support Vector Machine for Classification and Regression Ahlame Douzal AMA-LIG, Université Joseph Fourier Master 2R - MOSIG (2013) November 25, 2013 Loss function, Separating Hyperplanes, Canonical Hyperplan
Support Vector Machine for Classification and Regression Ahlame Douzal AMA-LIG, Université Joseph Fourier Master 2R - MOSIG (2013) November 25, 2013 Loss function, Separating Hyperplanes, Canonical Hyperplan
Linear & nonlinear classifiers
 Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1394 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1394 1 / 34 Table
Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1394 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1394 1 / 34 Table
Predictive analysis on Multivariate, Time Series datasets using Shapelets
 1 Predictive analysis on Multivariate, Time Series datasets using Shapelets Hemal Thakkar Department of Computer Science, Stanford University hemal@stanford.edu hemal.tt@gmail.com Abstract Multivariate,
1 Predictive analysis on Multivariate, Time Series datasets using Shapelets Hemal Thakkar Department of Computer Science, Stanford University hemal@stanford.edu hemal.tt@gmail.com Abstract Multivariate,
Analysis of N-terminal Acetylation data with Kernel-Based Clustering
 Analysis of N-terminal Acetylation data with Kernel-Based Clustering Ying Liu Department of Computational Biology, School of Medicine University of Pittsburgh yil43@pitt.edu 1 Introduction N-terminal acetylation
Analysis of N-terminal Acetylation data with Kernel-Based Clustering Ying Liu Department of Computational Biology, School of Medicine University of Pittsburgh yil43@pitt.edu 1 Introduction N-terminal acetylation
Support Vector Machines (SVM) in bioinformatics. Day 1: Introduction to SVM
 1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
Support Vector Machines and Speaker Verification
 1 Support Vector Machines and Speaker Verification David Cinciruk March 6, 2013 2 Table of Contents Review of Speaker Verification Introduction to Support Vector Machines Derivation of SVM Equations Soft
1 Support Vector Machines and Speaker Verification David Cinciruk March 6, 2013 2 Table of Contents Review of Speaker Verification Introduction to Support Vector Machines Derivation of SVM Equations Soft
Incorporating detractors into SVM classification
 Incorporating detractors into SVM classification AGH University of Science and Technology 1 2 3 4 5 (SVM) SVM - are a set of supervised learning methods used for classification and regression SVM maximal
Incorporating detractors into SVM classification AGH University of Science and Technology 1 2 3 4 5 (SVM) SVM - are a set of supervised learning methods used for classification and regression SVM maximal
Neural Networks. Prof. Dr. Rudolf Kruse. Computational Intelligence Group Faculty for Computer Science
 Neural Networks Prof. Dr. Rudolf Kruse Computational Intelligence Group Faculty for Computer Science kruse@iws.cs.uni-magdeburg.de Rudolf Kruse Neural Networks 1 Supervised Learning / Support Vector Machines
Neural Networks Prof. Dr. Rudolf Kruse Computational Intelligence Group Faculty for Computer Science kruse@iws.cs.uni-magdeburg.de Rudolf Kruse Neural Networks 1 Supervised Learning / Support Vector Machines
Chapter 6 Classification and Prediction (2)
 Chapter 6 Classification and Prediction (2) Outline Classification and Prediction Decision Tree Naïve Bayes Classifier Support Vector Machines (SVM) K-nearest Neighbors Accuracy and Error Measures Feature
Chapter 6 Classification and Prediction (2) Outline Classification and Prediction Decision Tree Naïve Bayes Classifier Support Vector Machines (SVM) K-nearest Neighbors Accuracy and Error Measures Feature
