Metadynamics-biased ab initio Molecular Dynamics Study of
|
|
|
- Anthony Golden
- 5 years ago
- Views:
Transcription
1 Metadynamics-biased ab initio Molecular Dynamics Study of Heterogeneous CO 2 Reduction via Surface Frustrated Lewis Pairs Mireille Ghoussoub 1, Shwetank Yadav 1, Kulbir Kaur Ghuman 1, Geoffrey A. Ozin 2, and Chandra Veer Singh* 1. [*] 1 Department of Materials Science and Engineering, University of Toronto, 184 College St., Toronto, Ontario M5S 3E4, Canada, chandraveer.singh@utoronto.ca 2 Department of Chemistry, University of Toronto, 80 St. George St., Toronto, Ontario, M5S 3H6, Canada, gozin@chem.utoronto.ca
2 1. Background into Metadynamics In order to investigate the reaction mechanism at finite temperature, we performed ab initio molecular dynamics (AIMD) on the Born Oppenheimer surface. The computationally expensive nature of these simulations restricts them to explore the system s dynamics over short durations only, and therefore AIMD alone is unable to capture full reactions of interest. This limitation is evaded by employing metadynamics (MetaD), a recently developed approach that applies a periodic bias to the system to encourage efficient, self-avoiding exploration of the free energy landscape 25,26. MetaD has proven to be a useful tool for overcoming the sampling limitations of traditional molecular dynamics on complex molecular processes In this method, a historydependent potential is added intermittently throughout the simulation to encourage efficient sampling of the associated free energy surface. The latter is defined over a set of collective variables (CVs), carefully selected to provide a complete description of the slow degrees of freedom of the system. The Helmholtz free energy surface corresponds to simulations carried out in the canonical ensemble (N,V,T) and therefore accounts for entropic contributions to the displacement of ions. The history-dependent bias takes the form of Gaussian functions to allow the system to escape beyond high energy barriers, thus allowing for faster sampling of the reaction surface. The sum of the deposited bias potentials may be used to reconstruct the free energy surface as shown here. The free energy surface F(s) can be written as =, (1) where T is the temperature of the system and, =, is the histogram of the set of CVs s corresponding to the unbiased simulation. Here we employ well-tempered MetaD which includes an additional tuning parameter to represent the effective sampling temperature of the CV space. 25 In this method, the added Gaussian potentials are rescaled at each step to allow 2
3 for the net bias to converge more smoothly, and thus prevent the simulation from overfilling the free energy surface. The bias potential at time t takes the form:, = 1+, (2) where V(s,t) is the bias potential defined over the set of CV coordinates s, where T is a tuning parameter and ω is the initial bias deposition rate. The dynamics of the CVs approaches the thermodynamic equilibrium as the simulation proceeds such that their probability distribution in the long time limit (t ) becomes, (3) and the bias potential converges to, = + (4) where k B is the Boltzmann constant and C is a nonphysical constant. 25 Introducing the tuning parameter T limits the exploration of the free energy surface to physically interesting regions on the order of T+ T, avoids the risk of overfilling the free energy surface, and guarantees convergence in the long time limit, making well-tempered MetaD a more effective method, operating between the limiting cases of standard MetaD and nonbiased sampling Simulation Details 3
4 In order to capture the full reaction using a feasible amount of computational resources, separate simulations were carried out for each step of the mechanism; ie. H 2 dissociation, CO 2 reduction, and H 2 O desorption. In the parameters described below, Sigma and Height refer to the width and height of the deposited gaussian potentials, and collective variables (CVs) refers to the defined quantities (ie. atomic distances, angles) used to define the free energy surface and/or the constraints on in the MetaD-biased AIMD simulations. The column MetaD refers to the CV(s) that were used to define the free energy surface. Constraints describe the upper and lower walls applied to the simulation, where ARG is the quantity subject to constraint, and KAPPA is the value of the force constant on the walls. 2.1 H 2 Dissocation Table S1. Parameters for PLUMED plugin used in test simulations Test # Temp. Timeste p K 1.25 fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE ATOMS=3, K 1.25 fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE ATOMS=1, K 0.8 fs d1: DISTANCE ATOMS=1,2 c1: CENTER ATOMS=1,2 WEIGHTS=1,1 c2: CENTER ATOMS=3,32 WEIGHTS=1,1 d2: DISTANCE ATOMS=c1,c2 CVs MetaD Sigma Height Constraints d ev UPPER_WALLS ARG=d2 AT=1.1 KAPPA=150.0 d1,d2 0.02, ev UPPER_WALLS ARG=d1 AT=2.888 KAPPA=150.0 d1, d2 0.1, ev Run 1: LOWER_WALLS ARG=d1 AT=0.74 KAPPA=400 UPPER_WALLS ARG=d2 AT=1 KAPPA=400 Run 2: LOWER_WALLS ARG=d1 AT=0.7 KAPPA=400 UPPER_WALLS ARG=d1 AT=0.76 KAPPA=400 LOWER_WALLS ARG=d2 AT=1.3 KAPPA=400 4
5 4 450 K 0.8 fs d1: DISTANCE ATOMS=1,2 c1: CENTER ATOMS=1,2 WEIGHTS=1,1 c2: CENTER ATOMS=3,32 WEIGHTS=1,1 d2: DISTANCE ATOMS=c1,c2 d1, d2 0.1, ev Run 1: LOWER_WALLS ARG=d1 AT=0.74 KAPPA=400 UPPER_WALLS ARG=d2 AT=1 KAPPA=400 Run 2: LOWER_WALLS ARG=d1 AT=0.7 KAPPA=400 UPPER_WALLS ARG=d1 AT=0.76 KAPPA=400 LOWER_WALLS ARG=d2 AT=1.3 KAPPA=400 After exploring different CV and PLUMED parameter choices (see Table S1), the following variables were selected for simulating the dissociation of H 2 over the In 2 O 3-x (OH) y surface. The CVs used to define the free energy surface are shown in Figure S1; CV1 is defined as the distance from the In (In28) to the nearest H on the adsorbate H 2 molecule (H1), while CV2 is the distance from O on the surface hydroxyl group (O1) to H (H2) on the adsorbate H 2 molecule. No constraints were placed on the system in the first simulation, and bias potentials 0.1 ev in height were applied intermittently over the course of 640 fs. Based on the area of the free energy surface explored in the first simulation, three constraints were introduced to ensure exploration of the region relevant to the reaction: an upper wall limiting the distance from O1 to H2 to a maximum of 2 Å, a lower wall limiting the distance from H2 to In28 to a minimum of 1.8 Å, and another lower wall limiting the distance from H1 to H2 to a minimum of 0.74 Å. Gaussian bias potentials of height 0.05 ev were deposited over the course of 640 fs for the latter, at which point dissociation was visibly achieved. The free energy surface was generated by two separate simulations, both employing a tuning parameter of 7000, a bias factor of 6, Gaussian widths of 0.1, and bias potentials placed every 30 iterations. MetaD-biased AIMD simulations 5
6 were performed at 20 C and 180 C; plots of the simulation temperature over time are shown in Figure S2. Figure S1: CVs used to define the free energy surface for H 2 dissociation over the In 2 O 3-x (OH) y surface. Figure S2: Simulation Temperature over time for of H 2 dissociation over the In 2 O 3-x (OH) y surface. Selecting the In-H1 distance, as well as the O-H2 distance allowed us to capture the H-H bond separation in a more reasonable amount of simulation time by guiding the reaction to the CV space most relevant to the adsorbate-surface interaction. The various H-H distances are still captured by the selected CVs whereas they ensure the molecule will interact with the surface and 6
7 adsorb to atoms which have been suggested to very likely participate in any dissociation from previous studies 19 as well as informed conjectures. It should be noted that selecting one set of CVS does not neglect any other possible CVs, some portion of the domain of almost all CVs is explored in the process of exploring other CVs' domain (as ultimately almost all CVs can be reduced to relationships based on atomic positions) and for such a simple variable as H-H distance most of the relevant distances would certainly be sampled. We provide further justification for our choice of CVs by performing the same calculation by taking the H-H distance as CV1, and the distance from the midpoint of the H1 and H2 atoms to the midpoint of the surface In and the O on surface bound hydroxyl group as CV2. PLUMED plugin parameters are provided in Table S1 and correspond to test simulations #3 and #4, for 20 C and 180 C, respectively. The collective variable definitions definitions are shown in Figure S3. Figure S3: CV1 and CV2 used in MetaD-biased AIMD test simulations #3 and #4. 7
8 Figure S4: Free Energy surface for hydrogen dissociation over In 2 O 3-x (OH) y at 20 C (top) and 180 C (bottom) generated from MetaD-biased AIMD test simulations #3 and #4. A TS B Figure S5: Atomic configurations corresponding to states A, TS, and B shown in the Free Energy surface in Figure S4. 8
9 The free energy surfaces are shown in Figure S4, with the reactant, transition, and product states indicated by points A, TS, and B, respectively; the atomic configurations corresponding to these states are shown in Figure S5. We find that this choice of collective variables instead favours exploration of the region of the Free Energy surface corresponding to adsorption of the H 2 molecule immediately prior to splitting. The H 2 molecule begins at a distance relatively far from the surface site, and then approaches the surface almost independently of the H1-H2 bond distance. The product state of the reaction, in which the In-H bond is 1.89 Å appears to coincide with the reactant state of the simulation carried out with our original choice of collective variables, in which H2 is already interacting with the surface In atom (In-H bond is 1.85 Å). These new results offer confirmation of our original choice of collective variables, as the O-H bond distance appears to be a significant quantity in driving the splitting of the H 2 molecule. 2.2 CO 2 Adsorption and Dissociation Table S2: Parameters for PLUMED plugin used in test simulations Test # K 1.25 fs c1: CENTER ATOMS=35,67 WEIGHTS=1,1 d1: DISTANCE ATOMS=c1,3 Temp. Timestep CVs MetaD Sigma Height Constraints d ev LOWER_WALLS ARG=d1 AT=1.1 KAPPA= K fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE ATOMS=3, K fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE ATOMS=3, K 1.25 fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE ATOMS=3, K 0.8 fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE d ev UPPER_WALLS ARG=d2 AT=1.1 KAPPA=150.0 d ev UPPER_WALLS ARG=d1 AT=2.888 KAPPA=150.0 d1,d2 0.02, 0.02 d1,d2 0.02, ev UPPER_WALLS ARG=d2 AT=1.0 KAPPA= ev UPPER_WALLS ARG=d2 AT=1.0 KAPPA=
10 ATOMS=3, K 0.8 fs d1: DISTANCE ATOMS=2,3 d2: DISTANCE ATOMS=3,4 d1,d2 0.02, ev UPPER_WALLS ARG=d1 AT=2.888 KAPPA=150.0 After exploring different CVs and PLUMED parameter choices (see Table S2), the following variables were selected for simulating the dissociation of CO 2 adsorption and dissociation over the In 2 O 3-x (OH) y surface. The CVs used to define the free energy surface are shown in Figure S6. CV1 is defined as the angle formed by O1, C1, and O2, and CV2 is the distance from C1 to the midpoint between In28 and In60. In the first simulation, bias potentials of height 0.3 ev were deposited with the distances from O2 to H1 constrained to a maximum of 1.2 Å. In the second, bias potentials of height 0.1 ev were deposited with an additional constraint forcing the distance from O2 to the surface to a maximum of 3.2 Å. In the third and fourth simulations, Gaussian bias potentials of height 0.1 ev were also deposited; however, the constraint on the distance from O2 to the surface was decreased to 2.8 Å. A final run was performed with no constraints placed on the system using 0.1 ev bias potentials. The combination of the various simulations modeled the reaction over a total of 1520 fs at both 20 C and 180 C. Bias potentials were placed every 30 iterations for all runs. Gaussian widths of 0.1, a bias factor of 6.0, and a tuning parameter of 10,000 were employed for all simulations. MetaDbiased AIMD simulations were performed at 20 C and 180 C; plots of the simulation temperature over time are shown in Figure S7. 10
11 Figure S6: CVs used to define the free energy surface of the adsorption and reduction of CO 2 over the In 2 O 3-x (OH) y surface. Figure S7: Simulation Temperature over time for the adsorption and reduction of CO 2 over the In 2 O 3-x (OH) y surface. 11
12 2.3 Dissociative adsorption of H 2 O Table S3: Parameters for PLUMED plugin used in test simulation Test # Temp. Timeste p K 1.75 fs d1: DISTANCE ATOMS=67,6 CVs MetaD Sigma Height Constraints d LOWER_WALLS ARG=d1 AT=2.25 KAPPA=150.0 After exploring different CVs and PLUMED parameter choices (see Table S3), the following variables were selected for simulating the dissociation of CO 2 adsorption and dissociation over the In 2 O 3-x (OH) y surface. The CVs used to define the free energy surface are shown in Figure S8. CV1 is defined as the distance from In60 on the surface to O2, while CV2 is the distance from In28 to O1. Four simulations were performed to capture the diffusive behavior of H 2 O at the surface. In the first simulation, Gaussian bias potentials of height 0.3 ev were deposited, with no force constraints applied to the system. In the second, forces limiting the distance from O1 to In28 to 2.08 Å, and the distance from O1 to H2 to 1.22 Å, were introduced. In the third and fourth simulations, the height of the Gaussian potentials was reduced to 0.1 ev, and the constraint on the distance between O1 and H2 was increased to 2.75 Å. The combination of the simulations modeled the reaction over the course of 2075 fs at both 20 C and 180 C. Gaussian bias potentials of width 0.1 were deposited every 30 iterations and a bias factor of 6 and a tuning parameter of 10,000 were applied in all simulations. MetaD-biased AIMD simulations were performed at 20 C and 180 C; plots of the simulation temperature over time are shown in Figure S9. 12
13 Figure S8: CVs used to define the free energy surface. Figure S9: Simulation Temperature over time for the dissociative adsorption of H 2 O over the In 2 O 3- x(oh) y surface. 13
14 3. Atomic Coordinates 3.1 Input atomic coordinates for MetaD-biased AIMD simulations of H 2 dissociation (for simulations at both 20 C and 180 C): CELL_PARAMETERS (alat= ) ATOMIC_POSITIONS angstrom H H O H In In In In In In In In In In In In In In In In In In In In In In In
15 In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In
16 In In In O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
17 O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
18 O O O O O O O O O O O O O O O O O O O O O O Input coordinates for MetaD-biased AIMD simulations of CO 2 reduction (for simulations at both 20 C and 180 C) CELL_PARAMETERS (alat= ) ATOMIC_POSITIONS angstrom O C O H H O
19 H In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In
20 In In In In In In In In In In In In In In In In In In In In In In In In In In In O O O O O O O O O O O
21 O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
22 O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
23 O O O O O O O O Input coordinates for MetaD-biased AIMD simulations for the motion of product H 2 O (for simulations at both 20 C and 180 C): CELL_PARAMETERS (alat= ) ATOMIC_POSITIONS angstrom O H H O H In In In In In In In In In In In In In In In
24 In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In In
25 In In In In In In In In In In In O O O O O O O O O O O O O O O O O O O O O O O O O O O
26 O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
27 O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
Au-C Au-Au. g(r) r/a. Supplementary Figures
 g(r) Supplementary Figures 60 50 40 30 20 10 0 Au-C Au-Au 2 4 r/a 6 8 Supplementary Figure 1 Radial bond distributions for Au-C and Au-Au bond. The zero density regime between the first two peaks in g
g(r) Supplementary Figures 60 50 40 30 20 10 0 Au-C Au-Au 2 4 r/a 6 8 Supplementary Figure 1 Radial bond distributions for Au-C and Au-Au bond. The zero density regime between the first two peaks in g
Ab initio molecular dynamics
 Ab initio molecular dynamics Kari Laasonen, Physical Chemistry, Aalto University, Espoo, Finland (Atte Sillanpää, Jaakko Saukkoriipi, Giorgio Lanzani, University of Oulu) Computational chemistry is a field
Ab initio molecular dynamics Kari Laasonen, Physical Chemistry, Aalto University, Espoo, Finland (Atte Sillanpää, Jaakko Saukkoriipi, Giorgio Lanzani, University of Oulu) Computational chemistry is a field
Density Functional Theory: from theory to Applications
 Density Functional Theory: from theory to Applications Uni Mainz May 27, 2012 Large barrier-activated processes time-dependent bias potential extended Lagrangian formalism Basic idea: during the MD dynamics
Density Functional Theory: from theory to Applications Uni Mainz May 27, 2012 Large barrier-activated processes time-dependent bias potential extended Lagrangian formalism Basic idea: during the MD dynamics
Performing Metadynamics Simulations Using NAMD
 Performing Metadynamics Simulations Using NAMD Author: Zhaleh Ghaemi Contents 1 Introduction 2 1.1 Goals of this tutorial.......................... 2 2 Standard metadynamics simulations 3 2.1 Free energy
Performing Metadynamics Simulations Using NAMD Author: Zhaleh Ghaemi Contents 1 Introduction 2 1.1 Goals of this tutorial.......................... 2 2 Standard metadynamics simulations 3 2.1 Free energy
MD Thermodynamics. Lecture 12 3/26/18. Harvard SEAS AP 275 Atomistic Modeling of Materials Boris Kozinsky
 MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
Consequences of Surface Oxophilicity of Ni, Ni-Co, and Co Clusters on Methane. Activation
 Supporting Information for: Consequences of Surface Oxophilicity of Ni, Ni-Co, and Co Clusters on Methane Activation Weifeng Tu, 1 Mireille Ghoussoub, Chandra Veer Singh,,3** and Ya-Huei (Cathy) Chin 1,*
Supporting Information for: Consequences of Surface Oxophilicity of Ni, Ni-Co, and Co Clusters on Methane Activation Weifeng Tu, 1 Mireille Ghoussoub, Chandra Veer Singh,,3** and Ya-Huei (Cathy) Chin 1,*
Sunyia Hussain 06/15/2012 ChE210D final project. Hydration Dynamics at a Hydrophobic Surface. Abstract:
 Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Computing free energy: Thermodynamic perturbation and beyond
 Computing free energy: Thermodynamic perturbation and beyond Extending the scale Length (m) 1 10 3 Potential Energy Surface: {Ri} 10 6 (3N+1) dimensional 10 9 E Thermodynamics: p, T, V, N continuum ls
Computing free energy: Thermodynamic perturbation and beyond Extending the scale Length (m) 1 10 3 Potential Energy Surface: {Ri} 10 6 (3N+1) dimensional 10 9 E Thermodynamics: p, T, V, N continuum ls
Accelerated Quantum Molecular Dynamics
 Accelerated Quantum Molecular Dynamics Enrique Martinez, Christian Negre, Marc J. Cawkwell, Danny Perez, Arthur F. Voter and Anders M. N. Niklasson Outline Quantum MD Current approaches Challenges Extended
Accelerated Quantum Molecular Dynamics Enrique Martinez, Christian Negre, Marc J. Cawkwell, Danny Perez, Arthur F. Voter and Anders M. N. Niklasson Outline Quantum MD Current approaches Challenges Extended
Supporting Information for. Dynamics Study"
 Supporting Information for "CO 2 Adsorption and Reactivity on Rutile TiO 2 (110) in Water: An Ab Initio Molecular Dynamics Study" Konstantin Klyukin and Vitaly Alexandrov,, Department of Chemical and Biomolecular
Supporting Information for "CO 2 Adsorption and Reactivity on Rutile TiO 2 (110) in Water: An Ab Initio Molecular Dynamics Study" Konstantin Klyukin and Vitaly Alexandrov,, Department of Chemical and Biomolecular
Advanced sampling. fluids of strongly orientation-dependent interactions (e.g., dipoles, hydrogen bonds)
 Advanced sampling ChE210D Today's lecture: methods for facilitating equilibration and sampling in complex, frustrated, or slow-evolving systems Difficult-to-simulate systems Practically speaking, one is
Advanced sampling ChE210D Today's lecture: methods for facilitating equilibration and sampling in complex, frustrated, or slow-evolving systems Difficult-to-simulate systems Practically speaking, one is
Ab Ini'o Molecular Dynamics (MD) Simula?ons
 Ab Ini'o Molecular Dynamics (MD) Simula?ons Rick Remsing ICMS, CCDM, Temple University, Philadelphia, PA What are Molecular Dynamics (MD) Simulations? Technique to compute statistical and transport properties
Ab Ini'o Molecular Dynamics (MD) Simula?ons Rick Remsing ICMS, CCDM, Temple University, Philadelphia, PA What are Molecular Dynamics (MD) Simulations? Technique to compute statistical and transport properties
Monte Carlo. Lecture 15 4/9/18. Harvard SEAS AP 275 Atomistic Modeling of Materials Boris Kozinsky
 Monte Carlo Lecture 15 4/9/18 1 Sampling with dynamics In Molecular Dynamics we simulate evolution of a system over time according to Newton s equations, conserving energy Averages (thermodynamic properties)
Monte Carlo Lecture 15 4/9/18 1 Sampling with dynamics In Molecular Dynamics we simulate evolution of a system over time according to Newton s equations, conserving energy Averages (thermodynamic properties)
10 Reaction rates and equilibrium Answers to practice questions. OCR Chemistry A. number 1 (a) 1: The enthalpy change, H;
 1 (a) 1: The enthalpy change, H; 2: The activation energy, E a 1 (b) H is unaffected as it is the difference between the reactants and products E a decreases as a catalyst allows an alternative route of
1 (a) 1: The enthalpy change, H; 2: The activation energy, E a 1 (b) H is unaffected as it is the difference between the reactants and products E a decreases as a catalyst allows an alternative route of
Molecular Dynamics Simulations of Fusion Materials: Challenges and Opportunities (Recent Developments)
 Molecular Dynamics Simulations of Fusion Materials: Challenges and Opportunities (Recent Developments) Fei Gao gaofeium@umich.edu Limitations of MD Time scales Length scales (PBC help a lot) Accuracy of
Molecular Dynamics Simulations of Fusion Materials: Challenges and Opportunities (Recent Developments) Fei Gao gaofeium@umich.edu Limitations of MD Time scales Length scales (PBC help a lot) Accuracy of
Energy Barriers and Rates - Transition State Theory for Physicists
 Energy Barriers and Rates - Transition State Theory for Physicists Daniel C. Elton October 12, 2013 Useful relations 1 cal = 4.184 J 1 kcal mole 1 = 0.0434 ev per particle 1 kj mole 1 = 0.0104 ev per particle
Energy Barriers and Rates - Transition State Theory for Physicists Daniel C. Elton October 12, 2013 Useful relations 1 cal = 4.184 J 1 kcal mole 1 = 0.0434 ev per particle 1 kj mole 1 = 0.0104 ev per particle
Atomic Layer Deposition of Zinc Oxide: Study on the Water Pulse Reactions from First-Principles
 This is an open access article published under a Creative Commons Attribution (CC-BY) License, which permits unrestricted use, distribution and reproduction in any medium, provided the author and source
This is an open access article published under a Creative Commons Attribution (CC-BY) License, which permits unrestricted use, distribution and reproduction in any medium, provided the author and source
Supporting Information for. Ab Initio Metadynamics Study of VO + 2 /VO2+ Redox Reaction Mechanism at the Graphite. Edge Water Interface
 Supporting Information for Ab Initio Metadynamics Study of VO + 2 /VO2+ Redox Reaction Mechanism at the Graphite Edge Water Interface Zhen Jiang, Konstantin Klyukin, and Vitaly Alexandrov,, Department
Supporting Information for Ab Initio Metadynamics Study of VO + 2 /VO2+ Redox Reaction Mechanism at the Graphite Edge Water Interface Zhen Jiang, Konstantin Klyukin, and Vitaly Alexandrov,, Department
Oxygen Reduction Reaction
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Oxygen Reduction Reaction Oxygen is the most common oxidant for most fuel cell cathodes simply
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Oxygen Reduction Reaction Oxygen is the most common oxidant for most fuel cell cathodes simply
Supporting Information
 1 Supporting Information 2 3 4 Hydrothermal Decomposition of Amino Acids and Origins of Prebiotic Meteoritic Organic Compounds 5 6 7 Fabio Pietrucci 1, José C. Aponte 2,3,*, Richard Starr 2,4, Andrea Pérez-Villa
1 Supporting Information 2 3 4 Hydrothermal Decomposition of Amino Acids and Origins of Prebiotic Meteoritic Organic Compounds 5 6 7 Fabio Pietrucci 1, José C. Aponte 2,3,*, Richard Starr 2,4, Andrea Pérez-Villa
Supporting Information
 Supporting Information The effect of strong acid functional groups on electrode rise potential in capacitive mixing by double layer expansion Marta C. Hatzell, a Muralikrishna Raju, b Valerie J. Watson,
Supporting Information The effect of strong acid functional groups on electrode rise potential in capacitive mixing by double layer expansion Marta C. Hatzell, a Muralikrishna Raju, b Valerie J. Watson,
1. (i) 2H 2 O 2 2H 2 O + O 2 ALLOW any correct multiple including fractions IGNORE state symbols 1
 1. (i) 2H 2 O 2 2H 2 O + O 2 ALLOW any correct multiple including fractions IGNORE state symbols 1 More crowded particles OR more particles per (unit) volume ALLOW particles are closer together DO NOT
1. (i) 2H 2 O 2 2H 2 O + O 2 ALLOW any correct multiple including fractions IGNORE state symbols 1 More crowded particles OR more particles per (unit) volume ALLOW particles are closer together DO NOT
( ) ( ) ( ) + ( ) Ä ( ) Langmuir Adsorption Isotherms. dt k : rates of ad/desorption, N : totally number of adsorption sites.
 Langmuir Adsorption sotherms... 1 Summarizing Remarks... Langmuir Adsorption sotherms Assumptions: o Adsorp at most one monolayer o Surace is uniorm o No interaction between adsorbates All o these assumptions
Langmuir Adsorption sotherms... 1 Summarizing Remarks... Langmuir Adsorption sotherms Assumptions: o Adsorp at most one monolayer o Surace is uniorm o No interaction between adsorbates All o these assumptions
AP Chemistry - Notes - Chapter 12 - Kinetics Page 1 of 7 Chapter 12 outline : Chemical kinetics
 AP Chemistry - Notes - Chapter 12 - Kinetics Page 1 of 7 Chapter 12 outline : Chemical kinetics A. Chemical Kinetics - chemistry of reaction rates 1. Reaction Rates a. Reaction rate- the change in concentration
AP Chemistry - Notes - Chapter 12 - Kinetics Page 1 of 7 Chapter 12 outline : Chemical kinetics A. Chemical Kinetics - chemistry of reaction rates 1. Reaction Rates a. Reaction rate- the change in concentration
Supporting Online Material (1)
 Supporting Online Material The density functional theory (DFT) calculations were carried out using the dacapo code (http://www.fysik.dtu.dk/campos), and the RPBE (1) generalized gradient correction (GGA)
Supporting Online Material The density functional theory (DFT) calculations were carried out using the dacapo code (http://www.fysik.dtu.dk/campos), and the RPBE (1) generalized gradient correction (GGA)
Adsorption of gases on solids (focus on physisorption)
 Adsorption of gases on solids (focus on physisorption) Adsorption Solid surfaces show strong affinity towards gas molecules that it comes in contact with and some amt of them are trapped on the surface
Adsorption of gases on solids (focus on physisorption) Adsorption Solid surfaces show strong affinity towards gas molecules that it comes in contact with and some amt of them are trapped on the surface
CHEMICAL KINETICS (RATES OF REACTION)
 Kinetics F322 1 CHEMICAL KINETICS (RATES OF REACTION) Introduction Chemical kinetics is concerned with the dynamics of chemical reactions such as the way reactions take place and the rate (speed) of the
Kinetics F322 1 CHEMICAL KINETICS (RATES OF REACTION) Introduction Chemical kinetics is concerned with the dynamics of chemical reactions such as the way reactions take place and the rate (speed) of the
Monte Carlo Simulation of Long-Range Self-Diffusion in Model Porous Membranes and Catalysts
 Monte Carlo Simulation of Long-Range Self-Diffusion in Model Porous Membranes and Catalysts Brian DeCost and Dr. Sergey Vasenkov College of Engineering, University of Florida Industrial processes involving
Monte Carlo Simulation of Long-Range Self-Diffusion in Model Porous Membranes and Catalysts Brian DeCost and Dr. Sergey Vasenkov College of Engineering, University of Florida Industrial processes involving
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/NCHEM.1680 On the nature and origin of dicationic, charge-separated species formed in liquid water on X-ray irradiation Stephan Thürmer, 1 Milan Ončák, 2 Niklas Ottosson, 3 Robert Seidel,
DOI: 10.1038/NCHEM.1680 On the nature and origin of dicationic, charge-separated species formed in liquid water on X-ray irradiation Stephan Thürmer, 1 Milan Ončák, 2 Niklas Ottosson, 3 Robert Seidel,
Exercise 2: Solvating the Structure Before you continue, follow these steps: Setting up Periodic Boundary Conditions
 Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Kinetics. Chapter 14. Chemical Kinetics
 Lecture Presentation Chapter 14 Yonsei University In kinetics we study the rate at which a chemical process occurs. Besides information about the speed at which reactions occur, kinetics also sheds light
Lecture Presentation Chapter 14 Yonsei University In kinetics we study the rate at which a chemical process occurs. Besides information about the speed at which reactions occur, kinetics also sheds light
What is Classical Molecular Dynamics?
 What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
2 Reaction kinetics in gases
 2 Reaction kinetics in gases October 8, 2014 In a reaction between two species, for example a fuel and an oxidizer, bonds are broken up and new are established in the collision between the species. In
2 Reaction kinetics in gases October 8, 2014 In a reaction between two species, for example a fuel and an oxidizer, bonds are broken up and new are established in the collision between the species. In
Ab Initio Study of Hydrogen Storage on CNT
 Ab Initio Study of Hydrogen Storage on CNT Zhiyong Zhang, Henry Liu, and KJ Cho Stanford University Presented at the ICNT 2005, San Francisco Financial Support: GCEP (Global Climate and Energy Project)
Ab Initio Study of Hydrogen Storage on CNT Zhiyong Zhang, Henry Liu, and KJ Cho Stanford University Presented at the ICNT 2005, San Francisco Financial Support: GCEP (Global Climate and Energy Project)
Random Walks A&T and F&S 3.1.2
 Random Walks A&T 110-123 and F&S 3.1.2 As we explained last time, it is very difficult to sample directly a general probability distribution. - If we sample from another distribution, the overlap will
Random Walks A&T 110-123 and F&S 3.1.2 As we explained last time, it is very difficult to sample directly a general probability distribution. - If we sample from another distribution, the overlap will
Chemical Kinetics. Kinetics is the study of how fast chemical reactions occur. There are 4 important factors which affect rates of reactions:
 Chemical Kinetics Kinetics is the study of how fast chemical reactions occur. There are 4 important factors which affect rates of reactions: reactant concentration temperature action of catalysts surface
Chemical Kinetics Kinetics is the study of how fast chemical reactions occur. There are 4 important factors which affect rates of reactions: reactant concentration temperature action of catalysts surface
Photoinduced Water Oxidation at the Aqueous. GaN Interface: Deprotonation Kinetics of. the First Proton-Coupled Electron-Transfer Step
 Supporting Information Photoinduced Water Oxidation at the Aqueous Interface: Deprotonation Kinetics of the First Proton-Coupled Electron-Transfer Step Mehmed Z. Ertem,,,* eerav Kharche,,* Victor S. Batista,
Supporting Information Photoinduced Water Oxidation at the Aqueous Interface: Deprotonation Kinetics of the First Proton-Coupled Electron-Transfer Step Mehmed Z. Ertem,,,* eerav Kharche,,* Victor S. Batista,
Supplemental Information
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics Supplemental Information Hydrogen reactivity on highly-hydroxylated TiO 2 (110) surface prepared via carboxylic acid adsorption
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics Supplemental Information Hydrogen reactivity on highly-hydroxylated TiO 2 (110) surface prepared via carboxylic acid adsorption
ADSORPTION ON SURFACES. Kinetics of small molecule binding to solid surfaces
 ADSORPTION ON SURFACES Kinetics of small molecule binding to solid surfaces When the reactants arrive at the catalyst surface, reactions are accelerated Physisorption and Chemisorption 1- diffusion to
ADSORPTION ON SURFACES Kinetics of small molecule binding to solid surfaces When the reactants arrive at the catalyst surface, reactions are accelerated Physisorption and Chemisorption 1- diffusion to
Module 5: "Adsoption" Lecture 25: The Lecture Contains: Definition. Applications. How does Adsorption occur? Physisorption Chemisorption.
 The Lecture Contains: Definition Applications How does Adsorption occur? Physisorption Chemisorption Energetics Adsorption Isotherms Different Adsorption Isotherms Langmuir Adsorption Isotherm file:///e
The Lecture Contains: Definition Applications How does Adsorption occur? Physisorption Chemisorption Energetics Adsorption Isotherms Different Adsorption Isotherms Langmuir Adsorption Isotherm file:///e
Mechanisms of H- and OH-assisted CO activation as well as C-C coupling on the flat Co(0001) surface Revisited
 Electronic Supplementary Material (ESI) for Catalysis Science & Technology. This journal is The Royal Society of Chemistry 2016 Mechanisms of H- and OH-assisted CO activation as well as C-C coupling on
Electronic Supplementary Material (ESI) for Catalysis Science & Technology. This journal is The Royal Society of Chemistry 2016 Mechanisms of H- and OH-assisted CO activation as well as C-C coupling on
Current address: Department of Chemistry, Hong Kong Baptist University, Kowloon Tong, Hong Kong,
 Hydrolysis of Cisplatin - A Metadynamics Study Supporting Information Justin Kai-Chi Lau a and Bernd Ensing* b Department of Chemistry and Applied Bioscience, ETH Zurich, USI Campus, Computational Science,
Hydrolysis of Cisplatin - A Metadynamics Study Supporting Information Justin Kai-Chi Lau a and Bernd Ensing* b Department of Chemistry and Applied Bioscience, ETH Zurich, USI Campus, Computational Science,
Supporting Information for PbTiO 3
 Supporting Information for PbTiO 3 (001) Capped with ZnO(11 20): An Ab-Initio Study of Effect of Substrate Polarization on Interface Composition and CO 2 Dissociation Babatunde O. Alawode and Alexie M.
Supporting Information for PbTiO 3 (001) Capped with ZnO(11 20): An Ab-Initio Study of Effect of Substrate Polarization on Interface Composition and CO 2 Dissociation Babatunde O. Alawode and Alexie M.
Section 8.1 The Covalent Bond
 Section 8.1 The Covalent Bond Apply the octet rule to atoms that form covalent bonds. Describe the formation of single, double, and triple covalent bonds. Contrast sigma and pi bonds. Relate the strength
Section 8.1 The Covalent Bond Apply the octet rule to atoms that form covalent bonds. Describe the formation of single, double, and triple covalent bonds. Contrast sigma and pi bonds. Relate the strength
Lecture 11: Potential Energy Functions
 Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Kinetic Monte Carlo (KMC)
 Kinetic Monte Carlo (KMC) Molecular Dynamics (MD): high-frequency motion dictate the time-step (e.g., vibrations). Time step is short: pico-seconds. Direct Monte Carlo (MC): stochastic (non-deterministic)
Kinetic Monte Carlo (KMC) Molecular Dynamics (MD): high-frequency motion dictate the time-step (e.g., vibrations). Time step is short: pico-seconds. Direct Monte Carlo (MC): stochastic (non-deterministic)
Electrochemical Properties of Materials for Electrical Energy Storage Applications
 Electrochemical Properties of Materials for Electrical Energy Storage Applications Lecture Note 3 October 11, 2013 Kwang Kim Yonsei Univ., KOREA kbkim@yonsei.ac.kr 39 Y 88.91 8 O 16.00 7 N 14.01 34 Se
Electrochemical Properties of Materials for Electrical Energy Storage Applications Lecture Note 3 October 11, 2013 Kwang Kim Yonsei Univ., KOREA kbkim@yonsei.ac.kr 39 Y 88.91 8 O 16.00 7 N 14.01 34 Se
An Introduction to Chemical Kinetics
 An Introduction to Chemical Kinetics Michel Soustelle WWILEY Table of Contents Preface xvii PART 1. BASIC CONCEPTS OF CHEMICAL KINETICS 1 Chapter 1. Chemical Reaction and Kinetic Quantities 3 1.1. The
An Introduction to Chemical Kinetics Michel Soustelle WWILEY Table of Contents Preface xvii PART 1. BASIC CONCEPTS OF CHEMICAL KINETICS 1 Chapter 1. Chemical Reaction and Kinetic Quantities 3 1.1. The
Modeling the Free Energy Landscape for Janus Particle Self-Assembly in the Gas Phase. Andy Long Kridsanaphong Limtragool
 Modeling the Free Energy Landscape for Janus Particle Self-Assembly in the Gas Phase Andy Long Kridsanaphong Limtragool Motivation We want to study the spontaneous formation of micelles and vesicles Applications
Modeling the Free Energy Landscape for Janus Particle Self-Assembly in the Gas Phase Andy Long Kridsanaphong Limtragool Motivation We want to study the spontaneous formation of micelles and vesicles Applications
Free energy simulations
 Free energy simulations Marcus Elstner and Tomáš Kubař January 14, 2013 Motivation a physical quantity that is of most interest in chemistry? free energies Helmholtz F or Gibbs G holy grail of computational
Free energy simulations Marcus Elstner and Tomáš Kubař January 14, 2013 Motivation a physical quantity that is of most interest in chemistry? free energies Helmholtz F or Gibbs G holy grail of computational
Selectivity in the initial C-H bond cleavage of n-butane on PdO(101)
 Supporting Information for Selectivity in the initial C-H bond cleavage of n-butane on PdO(101) Can Hakanoglu (a), Feng Zhang (a), Abbin Antony (a), Aravind Asthagiri (b) and Jason F. Weaver (a) * (a)
Supporting Information for Selectivity in the initial C-H bond cleavage of n-butane on PdO(101) Can Hakanoglu (a), Feng Zhang (a), Abbin Antony (a), Aravind Asthagiri (b) and Jason F. Weaver (a) * (a)
Electroreduction of N 2 to ammonia at ambient conditions on mononitrides of Zr, Nb, Cr, and V A DFT guide for experiments
 Electronic Supplementary Information Electroreduction of N to ammonia at ambient conditions on mononitrides of Zr, Nb, r, and V DFT guide for experiments Younes bghoui a, nna L. Garden b, Jakob G. Howalt
Electronic Supplementary Information Electroreduction of N to ammonia at ambient conditions on mononitrides of Zr, Nb, r, and V DFT guide for experiments Younes bghoui a, nna L. Garden b, Jakob G. Howalt
Supplementary Information
 Electronic Supplementary Material (ESI) for Catalysis Science & Technology. This journal is The Royal Society of Chemistry 2015 Supplementary Information Insights into the Synergistic Role of Metal-Lattice
Electronic Supplementary Material (ESI) for Catalysis Science & Technology. This journal is The Royal Society of Chemistry 2015 Supplementary Information Insights into the Synergistic Role of Metal-Lattice
Joint ICTP-IAEA Workshop on Fusion Plasma Modelling using Atomic and Molecular Data January 2012
 2327-3 Joint ICTP-IAEA Workshop on Fusion Plasma Modelling using Atomic and Molecular Data 23-27 January 2012 Qunatum Methods for Plasma-Facing Materials Alain ALLOUCHE Univ.de Provence, Lab.de la Phys.
2327-3 Joint ICTP-IAEA Workshop on Fusion Plasma Modelling using Atomic and Molecular Data 23-27 January 2012 Qunatum Methods for Plasma-Facing Materials Alain ALLOUCHE Univ.de Provence, Lab.de la Phys.
Electronic Circuits for Mechatronics ELCT 609 Lecture 2: PN Junctions (1)
 Electronic Circuits for Mechatronics ELCT 609 Lecture 2: PN Junctions (1) Assistant Professor Office: C3.315 E-mail: eman.azab@guc.edu.eg 1 Electronic (Semiconductor) Devices P-N Junctions (Diodes): Physical
Electronic Circuits for Mechatronics ELCT 609 Lecture 2: PN Junctions (1) Assistant Professor Office: C3.315 E-mail: eman.azab@guc.edu.eg 1 Electronic (Semiconductor) Devices P-N Junctions (Diodes): Physical
Exploring the anomalous behavior of metal nanocatalysts with finite temperature AIMD and x-ray spectra
 Exploring the anomalous behavior of metal nanocatalysts with finite temperature AIMD and x-ray spectra F.D. Vila DOE grant DE-FG02-03ER15476 With computer support from DOE - NERSC. Importance of Theoretical
Exploring the anomalous behavior of metal nanocatalysts with finite temperature AIMD and x-ray spectra F.D. Vila DOE grant DE-FG02-03ER15476 With computer support from DOE - NERSC. Importance of Theoretical
Self assembly of Carbon Atoms in Interstellar Space and Formation Mechanism of Naphthalene from Small Precursors: A Molecular Dynamics Study
 Self assembly of Carbon Atoms in Interstellar Space and Formation Mechanism of Naphthalene from Small Precursors: A Molecular Dynamics Study Niladri Patra UIC Advisor: Dr. H. R. Sadeghpour ITAMP Large
Self assembly of Carbon Atoms in Interstellar Space and Formation Mechanism of Naphthalene from Small Precursors: A Molecular Dynamics Study Niladri Patra UIC Advisor: Dr. H. R. Sadeghpour ITAMP Large
Computing free energy: Replica exchange
 Computing free energy: Replica exchange Extending the scale Length (m) 1 10 3 Potential Energy Surface: {Ri} 10 6 (3N+1) dimensional 10 9 E Thermodynamics: p, T, V, N continuum ls Macroscopic i a t e regime
Computing free energy: Replica exchange Extending the scale Length (m) 1 10 3 Potential Energy Surface: {Ri} 10 6 (3N+1) dimensional 10 9 E Thermodynamics: p, T, V, N continuum ls Macroscopic i a t e regime
Supporting Information
 Supporting Information Indirect Four-Electron Oxygen Reduction Reaction on Carbon Materials Catalysts in Acidic Solutions Guo-Liang Chai* 1, Mauro Boero 2, Zhufeng Hou 3, Kiyoyuki Terakura 3,4 and Wendan
Supporting Information Indirect Four-Electron Oxygen Reduction Reaction on Carbon Materials Catalysts in Acidic Solutions Guo-Liang Chai* 1, Mauro Boero 2, Zhufeng Hou 3, Kiyoyuki Terakura 3,4 and Wendan
Theoretical Models for Chemical Kinetics
 Theoretical Models for Chemical Kinetics Thus far we have calculated rate laws, rate constants, reaction orders, etc. based on observations of macroscopic properties, but what is happening at the molecular
Theoretical Models for Chemical Kinetics Thus far we have calculated rate laws, rate constants, reaction orders, etc. based on observations of macroscopic properties, but what is happening at the molecular
Bacterial Outer Membrane Porins as Electrostatic Nanosieves: Exploring Transport Rules of Small Polar Molecules
 Bacterial Outer Membrane Porins as Electrostatic Nanosieves: Exploring Transport Rules of Small Polar Molecules Harsha Bajaj, Silvia Acosta Gutiérrez, Igor Bodrenko, Giuliano Malloci, Mariano Andrea Scorciapino,
Bacterial Outer Membrane Porins as Electrostatic Nanosieves: Exploring Transport Rules of Small Polar Molecules Harsha Bajaj, Silvia Acosta Gutiérrez, Igor Bodrenko, Giuliano Malloci, Mariano Andrea Scorciapino,
Unit 12: Chemical Kinetics
 Unit 12: Chemical Kinetics Author: S. Michalek Introductory Resources: Zumdahl v. 5 Chapter 12 Main Ideas: Integrated rate laws Half life reactions Reaction Mechanisms Model for chemical kinetics Catalysis
Unit 12: Chemical Kinetics Author: S. Michalek Introductory Resources: Zumdahl v. 5 Chapter 12 Main Ideas: Integrated rate laws Half life reactions Reaction Mechanisms Model for chemical kinetics Catalysis
a micro PLUMED2.0 Tutorial for ESPResSo users PLUMED2.0: portable plugin for free-energy calculations with molecular dynamics
 a micro PLUMED2.0 Tutorial for ESPResSo users PLUMED2.0: portable plugin for free-energy calculations with molecular dynamics ESPResSo summer school 2013 Organized by CECAM, SimTech, ICP and University
a micro PLUMED2.0 Tutorial for ESPResSo users PLUMED2.0: portable plugin for free-energy calculations with molecular dynamics ESPResSo summer school 2013 Organized by CECAM, SimTech, ICP and University
!n[a] =!n[a] o. " kt. Half lives. Half Life of a First Order Reaction! Pressure of methyl isonitrile as a function of time!
![!n[a] =!n[a] o. kt. Half lives. Half Life of a First Order Reaction! Pressure of methyl isonitrile as a function of time! !n[a] =!n[a] o. kt. Half lives. Half Life of a First Order Reaction! Pressure of methyl isonitrile as a function of time!](/thumbs/77/76481346.jpg) Half lives Half life: t 1/2 t 1/2 is the time it takes for the concentration of a reactant to drop to half of its initial value. For the reaction A! products Half Life of a First Order Reaction! Pressure
Half lives Half life: t 1/2 t 1/2 is the time it takes for the concentration of a reactant to drop to half of its initial value. For the reaction A! products Half Life of a First Order Reaction! Pressure
Introduction Statistical Thermodynamics. Monday, January 6, 14
 Introduction Statistical Thermodynamics 1 Molecular Simulations Molecular dynamics: solve equations of motion Monte Carlo: importance sampling r 1 r 2 r n MD MC r 1 r 2 2 r n 2 3 3 4 4 Questions How can
Introduction Statistical Thermodynamics 1 Molecular Simulations Molecular dynamics: solve equations of motion Monte Carlo: importance sampling r 1 r 2 r n MD MC r 1 r 2 2 r n 2 3 3 4 4 Questions How can
Chapter 14 Chemical Kinetics
 Chapter 14 Chemical Kinetics Factors that Affect Reaction rates Reaction Rates Concentration and Rate The Change of Concentration with Time Temperature and Rate Reactions Mechanisms Catalysis Chemical
Chapter 14 Chemical Kinetics Factors that Affect Reaction rates Reaction Rates Concentration and Rate The Change of Concentration with Time Temperature and Rate Reactions Mechanisms Catalysis Chemical
Supporting information. Realizing Two-Dimensional Magnetic Semiconductors with. Enhanced Curie Temperature by Antiaromatic Ring Based
 Supporting information Realizing Two-Dimensional Magnetic Semiconductors with Enhanced Curie Temperature by Antiaromatic Ring Based Organometallic Frameworks Xingxing Li and Jinlong Yang* Department of
Supporting information Realizing Two-Dimensional Magnetic Semiconductors with Enhanced Curie Temperature by Antiaromatic Ring Based Organometallic Frameworks Xingxing Li and Jinlong Yang* Department of
a micro PLUMED Tutorial for FHI-aims users PLUMED: portable plugin for free-energy calculations with molecular dynamics
 a micro PLUMED Tutorial for FHI-aims users PLUMED: portable plugin for free-energy calculations with molecular dynamics Density functional theory and beyond: Computational materials science for real materials
a micro PLUMED Tutorial for FHI-aims users PLUMED: portable plugin for free-energy calculations with molecular dynamics Density functional theory and beyond: Computational materials science for real materials
Figure 1: Transition State, Saddle Point, Reaction Pathway
 Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Potential Energy Surfaces
Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Potential Energy Surfaces
Collision Theory. Reaction Rates A little review from our thermodynamics unit. 2. Collision with Incorrect Orientation. 1. Reactants Must Collide
 Reaction Rates A little review from our thermodynamics unit Collision Theory Higher Temp. Higher Speeds More high-energy collisions More collisions that break bonds Faster Reaction In order for a reaction
Reaction Rates A little review from our thermodynamics unit Collision Theory Higher Temp. Higher Speeds More high-energy collisions More collisions that break bonds Faster Reaction In order for a reaction
The Potential Energy Surface
 The Potential Energy Surface In this section we will explore the information that can be obtained by solving the Schrödinger equation for a molecule, or series of molecules. Of course, the accuracy of
The Potential Energy Surface In this section we will explore the information that can be obtained by solving the Schrödinger equation for a molecule, or series of molecules. Of course, the accuracy of
BAE 820 Physical Principles of Environmental Systems
 BAE 820 Physical Principles of Environmental Systems Catalysis of environmental reactions Dr. Zifei Liu Catalysis and catalysts Catalysis is the increase in the rate of a chemical reaction due to the participation
BAE 820 Physical Principles of Environmental Systems Catalysis of environmental reactions Dr. Zifei Liu Catalysis and catalysts Catalysis is the increase in the rate of a chemical reaction due to the participation
N independent electrons in a volume V (assuming periodic boundary conditions) I] The system 3 V = ( ) k ( ) i k k k 1/2
![N independent electrons in a volume V (assuming periodic boundary conditions) I] The system 3 V = ( ) k ( ) i k k k 1/2 N independent electrons in a volume V (assuming periodic boundary conditions) I] The system 3 V = ( ) k ( ) i k k k 1/2](/thumbs/90/101212070.jpg) Lecture #6. Understanding the properties of metals: the free electron model and the role of Pauli s exclusion principle.. Counting the states in the E model.. ermi energy, and momentum. 4. DOS 5. ermi-dirac
Lecture #6. Understanding the properties of metals: the free electron model and the role of Pauli s exclusion principle.. Counting the states in the E model.. ermi energy, and momentum. 4. DOS 5. ermi-dirac
7/19/2011. Models of Solution. State of Equilibrium. State of Equilibrium Chemical Reaction
 Models of Solution Chemistry- I State of Equilibrium A covered cup of coffee will not be colder than or warmer than the room temperature Heat is defined as a form of energy that flows from a high temperature
Models of Solution Chemistry- I State of Equilibrium A covered cup of coffee will not be colder than or warmer than the room temperature Heat is defined as a form of energy that flows from a high temperature
Supplementary Figure 1 Morpholigical properties of TiO 2-x SCs. The statistical particle size distribution (a) of the defective {001}-TiO 2-x SCs and
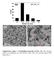 Supplementary Figure 1 Morpholigical properties of TiO 2-x s. The statistical particle size distribution (a) of the defective {1}-TiO 2-x s and their typical TEM images (b, c). Quantity Adsorbed (cm 3
Supplementary Figure 1 Morpholigical properties of TiO 2-x s. The statistical particle size distribution (a) of the defective {1}-TiO 2-x s and their typical TEM images (b, c). Quantity Adsorbed (cm 3
Energy Landscapes and Accelerated Molecular- Dynamical Techniques for the Study of Protein Folding
 Energy Landscapes and Accelerated Molecular- Dynamical Techniques for the Study of Protein Folding John K. Prentice Boulder, CO BioMed Seminar University of New Mexico Physics and Astronomy Department
Energy Landscapes and Accelerated Molecular- Dynamical Techniques for the Study of Protein Folding John K. Prentice Boulder, CO BioMed Seminar University of New Mexico Physics and Astronomy Department
Chemical Kinetics. Goal. Objectives
 4 Chemical Kinetics To understand the physical Goal factors that determine reaction rates. bjectives After this chapter, you should be able to: describe the factors that determine reaction rates. identify
4 Chemical Kinetics To understand the physical Goal factors that determine reaction rates. bjectives After this chapter, you should be able to: describe the factors that determine reaction rates. identify
Hydrides and Dihydrogen as Ligands: Hydrogenation Catalysis
 Hydrides and Dihydrogen as Ligands: Hydrogenation Catalysis Synthesis of Organometallic Complex Hydrides Reaction of MCO with OH -, H -, or CH 2 CHR 2 M(CO) n + OH - = M(CO) n-1 (COOH) - = HM(CO) n-1 -
Hydrides and Dihydrogen as Ligands: Hydrogenation Catalysis Synthesis of Organometallic Complex Hydrides Reaction of MCO with OH -, H -, or CH 2 CHR 2 M(CO) n + OH - = M(CO) n-1 (COOH) - = HM(CO) n-1 -
Brown et al, Chemistry, 2nd ed (AUS), Ch. 12:
 Kinetics: Contents Brown et al, Chemistry, 2 nd ed (AUS), Ch. 12: Why kinetics? What is kinetics? Factors that Affect Reaction Rates Reaction Rates Concentration and Reaction Rate The Change of Concentration
Kinetics: Contents Brown et al, Chemistry, 2 nd ed (AUS), Ch. 12: Why kinetics? What is kinetics? Factors that Affect Reaction Rates Reaction Rates Concentration and Reaction Rate The Change of Concentration
AP CHEMISTRY 2009 SCORING GUIDELINES
 2009 SCORING GUIDELINES Question 1 (10 points) Answer the following questions that relate to the chemistry of halogen oxoacids. (a) Use the information in the table below to answer part (a)(i). Acid HOCl
2009 SCORING GUIDELINES Question 1 (10 points) Answer the following questions that relate to the chemistry of halogen oxoacids. (a) Use the information in the table below to answer part (a)(i). Acid HOCl
Supporting Information
 Prebiotic NH 3 formation: Insights from simulations András Stirling 1, Tamás Rozgonyi 2, Matthias Krack 3, Marco Bernasconi 4 1 Institute of Organic Chemistry, Research Centre for Natural Sciences of the
Prebiotic NH 3 formation: Insights from simulations András Stirling 1, Tamás Rozgonyi 2, Matthias Krack 3, Marco Bernasconi 4 1 Institute of Organic Chemistry, Research Centre for Natural Sciences of the
Acidic Water Monolayer on Ruthenium(0001)
 Acidic Water Monolayer on Ruthenium(0001) Youngsoon Kim, Eui-seong Moon, Sunghwan Shin, and Heon Kang Department of Chemistry, Seoul National University, 1 Gwanak-ro, Seoul 151-747, Republic of Korea.
Acidic Water Monolayer on Ruthenium(0001) Youngsoon Kim, Eui-seong Moon, Sunghwan Shin, and Heon Kang Department of Chemistry, Seoul National University, 1 Gwanak-ro, Seoul 151-747, Republic of Korea.
Design of Efficient Catalysts with Double Transition Metal. Atoms on C 2 N Layer
 Supporting Information Design of Efficient Catalysts with Double Transition Metal Atoms on C 2 N Layer Xiyu Li, 1, Wenhui Zhong, 2, Peng Cui, 1 Jun Li, 1 Jun Jiang 1, * 1 Hefei National Laboratory for
Supporting Information Design of Efficient Catalysts with Double Transition Metal Atoms on C 2 N Layer Xiyu Li, 1, Wenhui Zhong, 2, Peng Cui, 1 Jun Li, 1 Jun Jiang 1, * 1 Hefei National Laboratory for
Chemistry 2000 Lecture 1: Introduction to the molecular orbital theory
 Chemistry 2000 Lecture 1: Introduction to the molecular orbital theory Marc R. Roussel January 5, 2018 Marc R. Roussel Introduction to molecular orbitals January 5, 2018 1 / 24 Review: quantum mechanics
Chemistry 2000 Lecture 1: Introduction to the molecular orbital theory Marc R. Roussel January 5, 2018 Marc R. Roussel Introduction to molecular orbitals January 5, 2018 1 / 24 Review: quantum mechanics
Metadynamics. Day 2, Lecture 3 James Dama
 Metadynamics Day 2, Lecture 3 James Dama Metadynamics The bare bones of metadynamics Bias away from previously visited configuraaons In a reduced space of collecave variables At a sequenaally decreasing
Metadynamics Day 2, Lecture 3 James Dama Metadynamics The bare bones of metadynamics Bias away from previously visited configuraaons In a reduced space of collecave variables At a sequenaally decreasing
Available online at ScienceDirect. Physics Procedia 71 (2015 ) 30 34
 Available online at www.sciencedirect.com ScienceDirect Physics Procedia 71 (2015 ) 30 34 18th Conference on Plasma-Surface Interactions, PSI 2015, 5-6 February 2015, Moscow, Russian Federation and the
Available online at www.sciencedirect.com ScienceDirect Physics Procedia 71 (2015 ) 30 34 18th Conference on Plasma-Surface Interactions, PSI 2015, 5-6 February 2015, Moscow, Russian Federation and the
Molecular dynamics simulations of fluorine molecules interacting with a Siˆ100 (2 1) surface at 1000 K
 Molecular dynamics simulations of fluorine molecules interacting with a Siˆ100 (2 1) surface at 1000 K T. A. Schoolcraft a) and A. M. Diehl Department of Chemistry, Shippensburg University, Shippensburg,
Molecular dynamics simulations of fluorine molecules interacting with a Siˆ100 (2 1) surface at 1000 K T. A. Schoolcraft a) and A. M. Diehl Department of Chemistry, Shippensburg University, Shippensburg,
A Theoretical Study of Oxidation of Phenoxy and Benzyl Radicals by HO 2
 A Theoretical Study of xidation of Phenoxy and Benzyl adicals by 2 S. Skokov, A. Kazakov, and F. L. Dryer Department of Mechanical and Aerospace Engineering Princeton University, Princeton, NJ 08544 We
A Theoretical Study of xidation of Phenoxy and Benzyl adicals by 2 S. Skokov, A. Kazakov, and F. L. Dryer Department of Mechanical and Aerospace Engineering Princeton University, Princeton, NJ 08544 We
Explanation: They do this by providing an alternative route or mechanism with a lower activation energy
 Catalysts Definition: Catalysts increase reaction rates without getting used up. Explanation: They do this by providing an alternative route or mechanism with a lower Comparison of the activation energies
Catalysts Definition: Catalysts increase reaction rates without getting used up. Explanation: They do this by providing an alternative route or mechanism with a lower Comparison of the activation energies
OCN 623: Thermodynamic Laws & Gibbs Free Energy. or how to predict chemical reactions without doing experiments
 OCN 623: Thermodynamic Laws & Gibbs Free Energy or how to predict chemical reactions without doing experiments Definitions Extensive properties Depend on the amount of material e.g. # of moles, mass or
OCN 623: Thermodynamic Laws & Gibbs Free Energy or how to predict chemical reactions without doing experiments Definitions Extensive properties Depend on the amount of material e.g. # of moles, mass or
Insights into Different Products of Nitrosobenzene and Nitrobenzene. Hydrogenation on Pd(111) under the Realistic Reaction Condition
 Insights into Different Products of Nitrosobenzene and Nitrobenzene Hydrogenation on Pd(111) under the Realistic Reaction Condition Lidong Zhang a, Zheng-Jiang Shao a, Xiao-Ming Cao a,*, and P. Hu a,b,*
Insights into Different Products of Nitrosobenzene and Nitrobenzene Hydrogenation on Pd(111) under the Realistic Reaction Condition Lidong Zhang a, Zheng-Jiang Shao a, Xiao-Ming Cao a,*, and P. Hu a,b,*
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Information Figure S1: (a) Initial configuration of hydroxyl and epoxy groups used in the MD calculations based on the observations of Cai et al. [Ref 27 in the
SUPPLEMENTARY INFORMATION Supplementary Information Figure S1: (a) Initial configuration of hydroxyl and epoxy groups used in the MD calculations based on the observations of Cai et al. [Ref 27 in the
Introduction to Geometry Optimization. Computational Chemistry lab 2009
 Introduction to Geometry Optimization Computational Chemistry lab 9 Determination of the molecule configuration H H Diatomic molecule determine the interatomic distance H O H Triatomic molecule determine
Introduction to Geometry Optimization Computational Chemistry lab 9 Determination of the molecule configuration H H Diatomic molecule determine the interatomic distance H O H Triatomic molecule determine
An Introduction to Quantum Chemistry and Potential Energy Surfaces. Benjamin G. Levine
 An Introduction to Quantum Chemistry and Potential Energy Surfaces Benjamin G. Levine This Week s Lecture Potential energy surfaces What are they? What are they good for? How do we use them to solve chemical
An Introduction to Quantum Chemistry and Potential Energy Surfaces Benjamin G. Levine This Week s Lecture Potential energy surfaces What are they? What are they good for? How do we use them to solve chemical
CHAPTER 11: CHROMATOGRAPHY FROM A MOLECULAR VIEWPOINT
 CHAPTER 11: CHROMATOGRAPHY FROM A MOLECULAR VIEWPOINT Contrasting approaches 1. bulk transport (e.g., c c = W ) t x + D c x goal: track concentration changes advantage: mathematical rigor (for simple models).
CHAPTER 11: CHROMATOGRAPHY FROM A MOLECULAR VIEWPOINT Contrasting approaches 1. bulk transport (e.g., c c = W ) t x + D c x goal: track concentration changes advantage: mathematical rigor (for simple models).
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland
 Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Sampling the free energy surfaces of collective variables
 Sampling the free energy surfaces of collective variables Jérôme Hénin Enhanced Sampling and Free-Energy Calculations Urbana, 12 September 2018 Please interrupt! struct bioinform phys chem theoretical
Sampling the free energy surfaces of collective variables Jérôme Hénin Enhanced Sampling and Free-Energy Calculations Urbana, 12 September 2018 Please interrupt! struct bioinform phys chem theoretical
Estimating Relationship Development Spreadsheet and Unit-as-an-Independent Variable Regressions
 Estimating Relationship Development Spreadsheet and Unit-as-an-Independent Variable Regressions Raymond P. Covert and Noah L. Wright MCR, LLC MCR, LLC rcovert@mcri.com nwright@mcri.com ABSTRACT MCR has
Estimating Relationship Development Spreadsheet and Unit-as-an-Independent Variable Regressions Raymond P. Covert and Noah L. Wright MCR, LLC MCR, LLC rcovert@mcri.com nwright@mcri.com ABSTRACT MCR has
Study of Ozone in Tribhuvan University, Kathmandu, Nepal. Prof. S. Gurung Central Department of Physics, Tribhuvan University, Kathmandu, Nepal
 Study of Ozone in Tribhuvan University, Kathmandu, Nepal Prof. S. Gurung Central Department of Physics, Tribhuvan University, Kathmandu, Nepal 1 Country of the Mt Everest 2 View of the Mt Everest 3 4 5
Study of Ozone in Tribhuvan University, Kathmandu, Nepal Prof. S. Gurung Central Department of Physics, Tribhuvan University, Kathmandu, Nepal 1 Country of the Mt Everest 2 View of the Mt Everest 3 4 5
= 16! = 16! W A = 3 = 3 N = = W B 3!3!10! = ΔS = nrln V. = ln ( 3 ) V 1 = 27.4 J.
 Answer key: Q1A Both configurations are equally likely because the particles are non-interacting (i.e., the energy does not favor one configuration over another). For A M = 16 N = 6 W A = 16! 0.9 101 =
Answer key: Q1A Both configurations are equally likely because the particles are non-interacting (i.e., the energy does not favor one configuration over another). For A M = 16 N = 6 W A = 16! 0.9 101 =
