A FAST, ACCURATE TWO-STEP LINEAR MIXED MODEL FOR GENETIC ANALYSIS APPLIED TO REPEAT MRI MEASUREMENTS
|
|
|
- Tracy Walters
- 5 years ago
- Views:
Transcription
1 A FAST, ACCURATE TWO-STEP LINEAR MIXED MODEL FOR GENETIC ANALYSIS APPLIED TO REPEAT MRI MEASUREMENTS Qifan Yang 1,4, Gennady V. Roshchupkin 2, Wiro J. Niessen 2, Sarah E. Medland 3, Alyssa H. Zhu 1, Paul M. Thompson 1, and Neda Jahanshad 1 * 1. Imaging Genetics Center, Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Marina del Rey, California, 90292, USA; 2. Department of Medical Informatics, Radiology & Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands; 3. Queensland, Institute of Medical Research, Berghofer Medical Research Institute, Royal Brisbane Hospital, Brisbane, QLD 4029 Australia; 4. Computational Biology and Bioinformatics program, Department of Biological Sciences, University of Southern California, California, 90007, USA; *corresponding author neda.jahanshad@usc.edu Abstract: Large-scale biobanks are being collected around the world in efforts to better understand human health and risk factors for disease. They often survey hundreds of thousands of individuals, combining questionnaires with clinical, genetic, demographic, and imaging assessments; some of this data may be collected longitudinally. Genetic associations analysis of such datasets requires methods to properly handle relatedness, population structure and other types of biases introduced by confounders. Most popular and accurate approaches rely on linear mixed model (LMM) algorithms, which are iterative and computational complexity of each iteration scales by the square of the sample size, slowing the pace of discoveries (up to several days for single trait analysis), and, furthermore, limiting the use of repeat phenotypic measurements. Here, we describe our new, non-iterative, much faster and accurate Two-Step Linear Mixed Model (2sLMM) approach, that has a computational complexity that scales linearly with sample size. We show that the first step retains accurate estimates of the heritability (the proportion of the trait variance explained by additive genetic factors), even when increasingly complex genetic relationships between individuals are modeled. Second step provides a faster framework to obtain the effect sizes of covariates in regression model. We applied Two-Step LMM to real data from the UK Biobank, which recently released genotyping information and processed MRI data from 9,725 individuals. We used the left and right hippocampus volume (HV) as repeated measures, and observed increased and more accurate heritability estimation, consistent with simulations. Keywords: SNP, Heritability, Repeated Measures, Empirical Kinship, Imaging Genetics, Genome Wide Association Studies (GWAS), Big Data 1. Introduction Genome-wide association studies (GWAS) of complex human traits can enable an understanding of the trait s underlying biological architecture, for example revealing genetic risk factors for disease. However, large-scale populations on the order of tens to hundreds of thousands of individuals are often needed to reliably detect the relatively small effects of single nucleotide polymorphisms. This need for large sample sizes has launched widespread interest in biobank studies that collect data from large populations that may include siblings or other relatives; therefore, some degree of known or cryptic relatedness may exist between individuals included in the study. In order to include all participants while accurately accounting for the fact that some samples may not be fully independent, a linear mixed model (LMM) is often used to improve 1
2 association power and accuracy and avoid an inflation of results, which would occur if wrongfully assuming independence in the population. Active efforts exist for the development of LMM methods, yet current methods continue to have limitations that prevent them from being practical for GWAS of hundreds or thousands of phenotypes (traits). Factored spectrally transformed linear mixed models [2], and genome-wide efficient mixed-model association [3] are two widely used exact models for LMM. However, these iterative optimization methods are computationally intensive, on the O(N $ ) for each iteration. Bayesian mixed models [4], have been proposed to overcome some of this complexity, and each iteration takes on the order of O(N); these methods do not estimate narrow-sense heritability, and they require the sample size to be sufficiently large to attain model accuracy. Because of the iterative optimization strategy, the computation time for most existing methods increases rapidly as the sample sizes become larger, to ensure convergence. This all makes them unfeasible for scaling on large datasets or analyzing many phenotypes, which is essential requirement for such methods in this coming biobank era. We note that large-scale genetics problems have motivated the development of faster methods to be developed [7, 8], yet to the best of our knowledge, these methods assume only minimal or no genetic relationships among individuals. Here, we build on and fill a gap in existing work and develop a novel approach to avoid time-consuming iterative steps. We propose an efficient, unbiased and robust Two-Step Linear Mixed Model method (2sLMM), which not only can be used to incorporate the complex genetic relationships among individuals within a large population, but also repeated measures from the same individuals. This two-step method can be used to analyze the heritability of a trait with multiple measurements, while also being to identify genetic loci that may be significantly associated with the trait s population variance through association testing. The 2sLMM consists of two steps: 1) to estimate heritability of the trait (h $ ) and then the effect size of independent variables, β. Here, we use dimensionality reduction methods for preprocessing and momentmatching regression [8, 11] to estimate the variance components due to genetic and environmental effects. Using the genetic relationship matrix derived from genome-wide genotypes of single nucleotide polymorphisms (SNPs), this heritability is commonly referred to as SNP-based heritability; 2) in the second step, we use Generalized Least Squares (GLS) regressions. While the time complexity for dimensionality reduction is the same as Singular Value Decomposition (SVD), the time complexity for our estimation steps is simply O(N)without any iterations, scaling only linearly with sample size, offering an efficient framework to analyze repeated measurements. Consider if each subject had one repeat measurement, then the sample size would double, and the computational burden for many other algorithms would quadruple, per iteration. We show our 2sLMM, for estimating heritability and fixed effects, is capable of modeling complex genetic relationships in population studies with relatedness and repeated measurements. This framework could dramatically reduce the estimation time from hours or days [3, 4, 9], to approximately 5 minutes per trait, when analyzing large scale genetics populations with a large number of traits. Our method is motivated primarily by applications for neuroimaging genetics, where GWAS can be performed on a large number of traits, representing volume, or area of various regions, even over a million voxels within a brain image [5]. Large scale biobank initiatives such as UK Biobank, are making it possible to do such image wide analyses in approximately 10,000 individuals at once [27], are methods are currently confining such analyses to unrelated individuals and analysis of single, non-repeated measurements. Here, 2
3 we demonstrate our method on brain volumes derived from MRI measurements. As there is little evidence for genetic lateralization between left and right hemispheres [28], we further use these measures as repeated measurements from approximately 10,000 individuals scanned as part of the UK Biobank [17], to demonstrate the robustness in our method. 2. Methods 2.1 Linear Mixed Models for single measurements The common framework for modeling a trait Y according to a genetic component, g, an environmental component, e, and fixed effects X is as follows: Y = X β + g + e (1) here, if there are N individuals, Y for a single trait is an N 1 vector, X is an N k matrix of k fixed effects, such as age, sex, and in the case of MRI-based volumetric phenotypes, intracranial volume (ICV), which is commonly added as a covariate. Linear mixed models (LMM) are often used to model complex genetic relationships among individuals, including cryptic relatedness, which may be a common occurrence in large-scale population studies such as the UK Biobank. In LMM, the genetic effect g and the total error, e, are often modeled as in [4]: g ~ N(0, σ 7 $ K ) (2) e ~ N (0, σ 9 $ Σ ) (3) where σ 7 $ is the additive genetic variance. K is an N N genetic relationship matrix (GRM), and every entry K [i, j] corresponds to the degree of genetic similarity between subject i and subject j [1]. The total error, e, is modeled as a normal distribution, where σ 9 $ is the environmental variance and Σ is an N N covariance matrix for total error. The structure of Σ is often given based on experimental design or prior information [10]. In independent studies, individual subject errors are independently and identically distributed (i.i.d.) and, therefore, Σ is an N N identity matrix I. 2.2 Two-Step Linear Mixed Model for repeat measurements Suppose the number of subjects is N; now, for each subject we now have more than one measurement, or repeat measurements, that we will use collectively to model the genetic effect. For p measurements, equation (1) may now be written using matrix notation. Let Y, X and e be Np 1 vectors, representing repeated traits, fixed effects and environmental component. Y = [y BB y B$ y BD y $B y $$ y $D y EB y E$ y ED ] F (4) X = [x BB x B$ x BD x $B x $$ x $D x EB x E$ x ED ] F (5) e = [e BB e B$ e BD e $B e $$ e $D e EB e E$ e ED ] F (6) Specifically, under the notations of (4), (5) and (6), suppose g, K, Σ are Np 1 repeated genetic component, Np Np repeated GRM and Np Np covariance matrix for total error. The Linear mixed model with p measurements is similarly expressed as equation (1), (2) and (3) Y = Xβ + g + e (7) 3
4 g ~ N(0, σ 7 $ K) (8) e ~ N (0, σ $ 9 Σ) (9) the GRM for repeated measures, K is assumed to be expressed as K = K Δ, where K is an N N GRM of subjects (no repeated measurements), and Δ is a p p definite matrix with Δ [i, j] = 1, for all i and j [10]. Σ is the covariance matrix for total errors. The matrix form of repeated LMM is mathematically equivalent to y KL = x KL β + g KL + e KL ; i = 1, 2,, N; j = 1, 2,, p (10) where y KL is the j repeated measurement for i subject, x th th KL is a 1 k vector of k fixed effects, as above. e KL is the j total error for i subject. For example, e th th KL could represent the total effect of environmental effects and individual measurement errors [11]. In order to estimate heritability h $ = σ $ 7 / (σ $ 7 + σ $ $ $ 9 ), it is assumed that σ 7 and σ 9 may be estimated as σ $ 7 and σ$ 9, respectively, and therefore, h $ = σ $ 7 / (σ $ 7 + σ $ 9 ). However, going back to equations (8) and (9), we need to model K and Σ appropriately for the repeated measurements. For repeated GRM, K in (8), this requires modeling the repeated identities in the matrix K, and results in a rank deficient, noninvertible matrix. The effect Σ in (9) now also includes subject specific environmental effects, which would be shared across repeat measurements for subject i, in addition to the measurement errors that correspond to unique errors for each measurement j. In this case, Σ will not be an identity matrix. If there is a correlation between the errors of any p measures i and j, then Σ i, j = 1 ρ I + ρ I Δ, where I is an Np Np identity matrix, I is a N N identity matrix and ρ is the correlation coefficient [10]. While this Σ could be modeled as two error terms, in this paper we focus on the LMM case where the repeated measurement correlation coefficient ρ is negligible. Studies have shown that this assumption performs similarly to the more complex model which accounts for correlated errors [9]. Future work will more accurately account for the non-zero correlation; in the case of our current work, much of this shared error across intra-subject measurements may also be absorbed by the repeated measurement used as a covariate, as we will describe. Our goal is to obtain fast, accurate and robust estimators h $ and β. This may include the fixed effect of a SNP, β RES, when tested individually or when tens of millions of SNPs are tested in genome-wide association studies (GWAS). The 2sLMM method consists two steps to estimate h $ and β. In the first step, we use a dimensional reduction method to compute SVD-transformation matrices S B and U for preprocessing, and use moment matching regression to estimate σ $ $ 7, σ 9 and h $ ; in the second, we use the Generalized Least Squares (GLS) method to estimate β. We describe these steps in a diagram in Figure 1 and in the following sections. 4
5 Figure 1. Procedure of 2sLMM for repeat measurements. Here, dimensionality reduction preprocessing is necessary to obtain an efficient, unbiased and consistent and Best Linear Unbiased Estimator (BLUE) σ $ 9. σ$ 7 can then be estimated only when σ$ 9 is known; it is also efficient, unbiased and consistent. Based on the statistical properties of OLS, h $ = σ $ 7 / (σ $ 7 + σ $ 9 ) is the most appropriate estimator Step 1 part 1: SVD dimensionality reduction preprocessing First, find a SVD-transformation matrix S B, such that S F B X = 0, S F B g = 0, S F B S B = I (11, 12, 13) If the conditions stated in equations (11), (12) and (13) are satisfied, then the original LMM (7) $ is equivalent to the following new linear mixed model, where σ 9 is the only unknown parameter. S F B Y = S F B e (14) S F B e ~ N (0, σ 9 I) (15) The existence of S B is proved by Theorem 1 (Supplementary Materials) and can be computed using Fast Singular Value Decomposition (Fast SVD) [12] Step 1 part 2: Estimate σ e 2 After SVD dimensionality reduction preprocessing, we estimate σ 9 $ using moment matching regression [8, 11], which is mathematically equivalent to LD regression [13] under some constrains. The SVD-transformed phenotypes are S B F Y. To estimate σ 9 $, we regress the 2 nd moment of transformed phenotypes S B F YY F S B, onto the variance component of S B F e, the matrix I: vec(s F B YY F S B ) = σ $ 9 vec I + ε B (16) where vec() is a vectorization function -- with an input of any matrix it simply stacks the columns; ε B is the regression error. The OLS estimator of (16) is numerically solved as σ $ 9 = trace(s F B YY F S B )/trace(i) (17) The unbiased, consistent and efficient properties of σ$ 9 are summarized in Theorem 2 and Theorem 3 (Supplementary Materials). The variance of σ$ 9 is (a b c ) c E approximated by the asymptotic variance based on the consistency of σ 9 Supplementary Materials) Step 1 part 3: Estimate σ g 2 and h 2 [24], which is $ (Theorem 4, σ 7 $ and h $ can also be estimated using SVD-moment matching regression. In order to estimate σ 7 $, we compute a SVD-transformation matrix U, such that U F X = 0, U F U = I (18, 19) We then apply the matrix U to both sides of the original linear mixed model (7) 5
6 U F Y = U F g + U F e (20) where U F g ~ N (0, σ $ 7 U F KU), and U F e ~ N (0, σ $ 9 I). Given σ $ 9, we use the moment-matching regression [8, 11] and consider a simpler, but still accurate linear regression for (20), where ε $ is the regression error: vec U F YY F U σ $ 9 vec I = σ $ 7 vec U F KU + ε $ (21) Then an unbiased, consistent and efficient OLS estimator σ$ 7 is derived σ $ 7 = trace(u F KUU F YY F U σ$ 9 U F KU)/trace(U F KUU F KU) (22) The variance of σ$ 7 is (a f c ) c [24], where p is the number of repeat measurements (Theorem 4, DE Supplementary Materials). Finally, the heritability estimator is obtained through h $ = σ $ 7 / (σ $ 7 + σ $ 9 ), and variance of estimated heritability is calculated by Delta method [25] Step 2: Estimate β var h $ = (a c b ) c a c f g ac h var σ 7 b $ + (a f c ) c a c f g ac h var σ$ 9 (23) b β = (X F M kb X) kb X F M kb Y (24) cov(β) = (X F M kb X) kb (σ $ 7 + σ $ 9 ) (25) in which M is the OLS estimator of variance-covariance matrix [15]. In particular, M = h $ K + (1 h $ )I (26) β is obtained using GLS method, so β is also unbiased, consistent and efficient, which is an important consideration in large scale big data applications such as imaging genetics. 2.3 Simulation methods To determine the accuracy and efficiency of our 2sLMM method, we simulated several conditions similar the real neuroimaging genetics dataset we use in the next section (from UK Biobank), and compared our estimates to those obtained from Ge et al. s moment-matching regression for SNP-based Heritability Estimation (MMHE) [8, 11], whose application scope now includes repeated measurements, (Repeated MMHE) [11]. Suppose the total number of simulations is N RKm, and the number of subjects is N. We perform simulation analysis for the proposed LMM with repeat measurements: Y = Xβ + g + e (7) g ~ N(0, σ 7 $ K) (8) e ~ N (0, σ $ 9 Σ) (9) where, Σ is an identity matrix I and the number of repeated measurements p is 2. To make the simulated GRM (the matrix K ) close to a real GRM from a family-based population, we first randomly generated a 2-trial binomial distributed N m genotype matrix Z, where the number of SNPs, m is 100,000, and probability of success for each trial is 0.2. Here, the probability of 6
7 success for each binomial trial corresponds to the minor allele frequency (MAF). In particular, every column of Z needs to be the normalized. K is obtained by K = K Δ, K = Z F Z/m. We designated the true values for our simulation based on our real data example (see next section). Suppose fixed effects X in (7) include an intercept, age, sex, and a scaled version of intracranial volume (scaled ICV), X = [1 X p79 X q9r X qspt9uvwx ]. Motivated by the distribution of data in the UK Biobank, we sampled X p79 from a normal distribution with mean 60 and variance 10. X q9r is a binary variable, coded here as 0 for female and 1 for male. X q9r was simulated using a discrete uniform distribution, with only two values, 0 and 1. X qspt9uvwx was drawn from a normal distribution with mean 1.2 and variance 0.05, X qspt9uvwx ~ N 1.2, For robustness, we also performed a follow-up test modeling in noise: X qspt9uvwx was drawn from a contaminated normal distribution, in which 91% of simulated X qspt9uvwx were from N 1.2, 0.05 as before, while the remaining 9% are drawn from another normal distribution with a much higher variance, N 1.2, 3 such that now, X qspt9uvwx ~ 0.91N 1.2, N (1.2, 3). The design of contaminated normal distribution follows from [16]. Using random sampling with replacement, 100 simulations were conducted for 10,000 subjects. For each simulated sample, both our 2sLMM method and MMHE [11] were run. True values of estimators are assigned as h $ = 0.4, sum of errors σ $ 7 + σ $ 9 = 170,000, β } = 6,600, β p79 = -15, β q9r = 5, β qspt9uvwx = -1, Analysis with real data The UK Biobank is a large scale epidemiological study, collecting genotypes, brain MRIs, as well as numerous other variables from adults aged in the UK. Information regarding brain imaging analysis from the UK Biobank is detailed in [17]. Summary volumetric data has been made available in their database and was accessed here as part of application # As of July 2017, a sample size of 9,725 individuals had non-missing data on age at scan, sex, left and right subcortical volumes, and scaled intracranial volume (scaledicv). Raw genotypes were downloaded and used to create the GRM in the raremetalworker (RMW) software package ( which calculates the GRM according to [2]; we did not use the X chromosome in our GRM calculation. The resulting GRM matrix is full rank and invertible. 9,725 subjects in the UK Biobank were analyzed using our 2sLMM method for a repeat analysis study of hippocampal volume heritability. To date, large-scale genome-wide association initiatives from large scale consortia, including the ENIGMA consortium, use a bilaterally averaged measure of hippocampal volume as the trait of interest [18, 19, 20]. This avoids possible left/right mismatches across different cohorts, and also reduces noise compared to running each left or right volumes separately. However, if the lateralized volumes are considered as repeated measures, rather than averaged, the effective sample size may be higher, possibly leading to more stable estimates than the current averaging techniques. To demonstrate these repeat measurements, 9,725 left and 9,725 right measurements are considered separately as individual elements of trait vector Y. Fixed effects X in (7) included the scaled ICV to control for head size, age, sex and an intercept. Due to the limited exclusion criteria, many health conditions are represented in UK Biobank; for the current study, we did not eliminate or control for any conditions and performed our estimates on the more diverse phenotypic pool. 7
8 Using this data, we first estimate the heritability of hippocampal volume using bilaterally averaged measures from all 9,725 subjects, resulting in 9,725 data points. Second, we estimate the heritability of hippocampal volume using each of the lateral measures from all 9,725 subjects as unique samples as described above, resulting in 19,450 data points. 3. Results 3.1 Simulation results For a given simulated condition, we plot the resulting estimate of each of the 100 simulations. Simulation Set 1 (Figure 2; Table 1) is generated given X qspt9uvwx ~ N (1.2, 0.05), while simulation Set 2 (Figure 3; Table 2) is the test for robustness, where X qspt9uvwx ~ 0.91N 1.2, N (1.2, 0.3). For Set 1 and Set 2, the same GRM (single measure or repeated measures) were run using 2sLMM, and Repeated MMHE. Mean, bias, standard deviation (SD) and mean squared error (MSE) are reported in Table 1 and Table 2 for simulated estimators, h $, σ $ 7, σ $ 9, β p79 of 2sLMM. Repeated MMHE [11] does not provide estimates of β values, so we only report the outputs for h $, σ $ 7, and σ $ 9. Simulation Set 1: N = 1000, N RKm = 100, p = 2, X qspt9uvwx ~ N 1.2, 0.05 Figure 2. Heritability estimate simulations ( N = 1000, N RKm = 100 ). Top: Two-Step Linear Mixed Model (2sLMM); Bottom: Repeated moment-matching heritability estimation (Repeated MMHE [11]). Columns correspond to estimations of h $, σ $ 7, σ $ 9, and for our method, β p79. The x-axis reflects the number of each of the 100 simulations. The same scales are used for the y-axes for both 2sLMM and MMHE. Table 1. Simulation Statistics (N = 1000, N RKm = 100) for 2sLMM and Repeated MMHE Estimators True Mean Bias Standard Dev MSE values Repeated Repeated Repeated Repeated 2sLMM 2sLMM 2sLMM 2sLMM MMHE MMHE MMHE MMHE h k k k σ g ƒ σ2 e ƒ β age
9 Simulation Set 2: N = 1000, N RKm = 100, p = 2, X qspt9uvwx ~ 0.91N 1.2, N (1.2, 0.3) Figure 3. Heritability estimate simulations ( N = 1000, N RKm = 100, X qspt9uvwx ~0.91N 1.2, N (1.2, 0.3) Top: Two-Step Linear Mixed Model (2sLMM); Bottom: Repeated moment-matching heritability estimation (Repeated MMHE [11]). Columns correspond to estimations of h $, σ $ 7, σ $ 9, and for our method, β p79. The x-axis reflects the number of each of the 100 simulations. The same scales are used for the y-axes for both 2sLMM and MMHE. Table 2. Simulation Statistics simulations ( N = 1000, N RKm = 100, X qspt9uvwx ~ 0.91N 1.2, N (1.2, 0.3)) for 2sLMM and Repeated MMHE True Estimator Mean Bias Standard Dev MSE values Repeated Repeated Repeated Repeated 2sLMM 2sLMM 2sLMM 2sLMM MMHE MMHE MMHE MMHE h k k k 0.02 σ g ƒ σ2 e ƒ β age k Analysis with real data from the UK Biobank Y is the average of left and right hippocampal volumes from each of the 9,725 subjects. Fixed effects X in (1) included intercept, age, sex and scaled ICV to control for head size. Using MMHE [8] for a single (not-repeated) measurement, the heritability estimate for a single measurement is h $ = 0.06 ( σ 7 $ = and σ 9 $ = ), which despite MMHE being similar to LD score regression under certain conditions [8], the result is far less than the genomewide summary statistic heritability calculated using LD Score Regression [13] for hippocampal volume in [21] h $ = 0.135, using GWAS summary data from a comparable sample size (N ~ 11,600) from the ENIGMA Consortium [19]. For hippocampus repeated measurements, we ran both our method and Repeated MMHE [11], and we compare h $, σ $ 7, σ $ 9, β }, β p79, β q9r, β vwx running time (implemented with MATLAB and CPU) in Table 3, where β } is the estimated β for intercepts. The standard error of the heritability estimate se(h $ ) is estimated using block Jackknife as in [11] (block size = 266) 9
10 for Repeated MMHE and 2sLMM, we refer to this as Jackknife se(h $ ). Additionally, se(h $ ) is calculated for 2sLMM, which is approximated by the asymptotic se(h $ ). As seen in Table 3, even with MMHE, which we have shown may underestimate heritability, we see that using the repeat measurement approach is far more powerful (h $ ~ 0.32 as opposed to h $ ~0.06) and is capable of attributing more of the variance to additive genetic effects. Our 2sLMM method can capture a even greater portion of the genetic variance h $ ~ 0.44, which is closer to the hippocampal heritability estimates suggested by twin and family studies of approximately 70% [20]. Furthermore, 2sLMM provides effect size estimates for fixed effects. Table 3. Estimation results for repeated measurements of hippocampal volume in UK Biobank. The 2sLMM provides higher estimates of heritability as well as estimates for the beta coefficients of fixed effects. Estimators and Time Repeated MMHE 2sLMM σ g σ2 e h Jackknife se(h 2 ) se(h 2 ) β se(β 0 ) β age se(β age ) β sex se(β sex ) β ICV se(β ICV ) Matlab CPU Time min min 4. Discussion In this paper, we propose a fast, statistically efficient and accurate Two-Step Linear Mixed Model (2sLMM) for population studies with cryptic relatedness and repeated measurements. Speed and accuracy are arguably the most important two criteria to evaluate a statistical model. Most iterative linear mixed models [2-4] based on Restricted Maximal Likelihood (ReML) or Bayesian methods, require many iterations to obtain accurate results. While fast, non-iterative, linear mixed models exist, under certain conditions some important estimators such as h $ or β for a fixed variable, may be inefficient, underestimated or inaccurate with large sampling errors. This is due to the fact that some of these existing fast LMMs often ignore or just simplify the complex genetic relationships between individuals. In order to estimate β related to fixed effects such as age, sex or more importantly, SNPs, the Two-Step ProbAbel package [15] for example, first ignores genetic effects to estimate non-genetic β values. Non-repeated [8] or Repeated MMHE [11], a moment-matching heritability method works well for population data; this method gives fast and unbiased heritability estimates as shown in Table 3. However, for populations that include family data, the GRM deviates significantly from an 10
11 identity matrix, resulting in non-normally distributed, highly correlated and heteroscedastic regression errors in moment-matching methods such as those in [8, 11, 22]. Figure 2 and Figure 3 show that if we use these models that do not support highly related samples, MMHE tends to $ overestimate σ 9 and underestimate σ $ 7, resulting in underestimated h $. In Figure 3, our robustness test showed that when scaled ICV contains small portion of outliers, Repeated MMHE gives h $, σ $ 9, σ$ 7 with relatively larger standard deviations, which may explain why Repeated MMHE generates lower heritability than 2sLMM, since the ICV data from UK Biobank contains many outliers. If we estimate β directly using results from the momentmatching regression, the bias and standard deviations of σ$ 9 and σ$ 7 will be further magnified, and the β will be inaccurate. In contrast, our fast method projects the original data onto a lower dimensional space, but keeps all the necessary information for estimation. This dimensionality reduction method ensures that the estimated heritability and β have minimum estimation standard errors, as shown in our simulation studies (Figure 2, Figure 3). In Table 3, Jackknife standard errors are reported for Repeated LMM and MMHE as they are conducted in the original papers [8, 11], indicating both methods are stable. The asymptotic standard error of h $, is close to the simulation standard error in Table 1 and Table 2. This reflects the statistical power of our 2sLMM, when applied to largescale neuroimaging genetics datasets. Table 2 and Figure 3 show that the 2sLMM method is very robust, and insensitive to outliers, ensuring our heritability analysis is not biased by the outliers from the data. Furthermore, estimated β for age, sex, and ICV (intracranial volume) are reported in Table 3. The score test for significance of β p79, β q9r, and β vwx can be obtained with asymptotic χ $ distributed statistics [15]. As expected, significant effects of age and ICV are observed, while sex differences are not detected to be significant for the hippocampal volume in our study; this may be due to simultaneously controlling for the other factors ICV and age. This proposed 2sLMM method has some application limitations. Most notably, the method is applied in the case where we are considering independent repeat measurements, which is regarded as the simplest case to quantify the total errors from experiments and environment. In the more precise and complex repeated measurement model, the total error e could incorporate many additive effects, such as intrasubject and intersubject effects, household effects and measurement effects [23]. A fast, efficient and robust statistical method is still an open problem. Despite this limitation, other studies have shown that this assumption performs similarly to the more complex model which accounts for correlated errors in intrasubject measurements [9]. in our case, much of this shared error across the left and right volume measurements may also be absorbed by the repeated ICV measurement fitted in X. In future work, we will evaluate the performance of our model for genome-wide association results, to identify genome-wide significant SNPs for subcortical measures or other genetic related phenotypes. Compared with iterative methods, this non-iterative and accurate method will show much greater time improvement when running on GPU, given GPU optimizes the matrix multiplication far more efficiently than current CPU implementations. Our method will also be further extended to support more complex linear mixed models, to account for additional intra-subject and inter-subject relationships, including common environment, or household effects. 11
12 Acknowledgements NIH grant U54 EB from the Big Data to Knowledge (BD2K) Program. This research has been conducted using the UK Biobank Resource under Application Number References [1] Yang, J, et al. "GCTA: a tool for genome-wide complex trait analysis." The American Journal of Human Genetics 88.1 (2011): [2] Lippert, C, et al. "FaST linear mixed models for genome-wide association studies." Nature methods 8.10 (2011): [3] Zhou, X, and Stephens, M. "Genome-wide efficient mixed-model analysis for association studies." Nature genetics 44.7 (2012): [4] Loh, P., et al. "Efficient Bayesian mixed-model analysis increases association power in large cohorts." Nature genetics 47.3 (2015): [5] Medland, S. E., et al. "Whole-genome analyses of whole-brain data: working within an expanded search space." Nature neuroscience 17.6 (2014): [6] Jahanshad, N. et al. "Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA DTI working group." Neuroimage 81 (2013): [7] Roshchupkin, G. V., et al. "HASE: Framework for efficient high-dimensional association analyses." Scientific reports 6 (2016). [8] Ge, T, et al. "Phenome-wide heritability analysis of the UK Biobank." PLoS genetics 13.4 (2017): e [9] Eu-Ahsunthornwattana, J, et al. "Comparison of methods to account for relatedness in genome-wide association studies with family-based data." PLoS genetics 10.7 (2014): e [10] Naik, D. N., and Rao S. S. "Analysis of multivariate repeated measures data with a Kronecker product structured covariance matrix." Journal of Applied Statistics 28.1 (2001): [11] Ge, T, et al. "Heritability analysis with repeat measurements and its application to restingstate functional connectivity." Proceedings of the National Academy of Sciences (2017): [12] Holmes, M., et al.. "Fast SVD for large-scale matrices." Workshop on Efficient Machine Learning at NIPS. Vol [13] Bulik-Sullivan, B. K., et al. "LD Score regression distinguishes confounding from polygenicity in genome-wide association studies." Nature genetics 47.3 (2015): [14] Harville, D. "Extension of the Gauss-Markov theorem to include the estimation of random effects." The Annals of Statistics (1976): [15] Aulchenko, Y. S., et al. "ProbABEL package for genome-wide association analysis of imputed data." BMC bioinformatics 11.1 (2010): 134. [16] Maas, C. JM, and Joop J. Hox. "Robustness issues in multilevel regression analysis." Statistica Neerlandica 58.2 (2004): [17] Miller, K. L., et al. "Multimodal population brain imaging in the UK Biobank prospective epidemiological study." Nature neuroscience (2016): [18] Stein, M. B., et al. "Hippocampal volume in women victimized by childhood sexual abuse." Psychological medicine 27.4 (1997): [19] Hibar, D. P., et al. "Common genetic variants influence human subcortical brain structures." Nature (2015):
13 [20] Hibar, D. P., et al. "Novel genetic loci associated with hippocampal volume." Nature communications 8 (2017): [21] Franke, B. et al. "Genetic influences on schizophrenia and subcortical brain volumes: largescale proof-of-concept and roadmap for future studies." Nature neuroscience 19.3 (2016): 420. [22] Kleijnen, J PC. "White noise assumptions revisited: Regression metamodels and experimental designs in practice." Proceedings of the 38th conference on Winter simulation. Winter Simulation Conference, [23] Lange, K. Mathematical and statistical methods for genetic analysis. Springer Science & Business Media, [24] Shao, J. "Asymptotic distribution of the weighted least squares estimator." Annals of the Institute of Statistical Mathematics 41.2 (1989): [25] Oehlert, G. W. "A note on the delta method." The American Statistician 46.1 (1992): [26] Satizabal, C. L., et al. "Genetic Architecture of Subcortical Brain Structures in Over 40,000 Individuals Worldwide." biorxiv (2017): [27] Elliott, L., et al. "The genetic basis of human brain structure and function: 1,262 genomewide associations found from 3,144 GWAS of multimodal brain imaging phenotypes from 9,707 UK Biobank participants." BioRxiv (2017): [28] Wright, I. C., et al. "Meta-analysis of regional brain volumes in schizophrenia." American Journal of Psychiatry (2000):
14 Supplemental Materials Theorem 1. There exists a matrix U, such that U F X = 0 and U F U = I. Similarly, there exists a matrix W, such that W F (U F KU) = 0 and W F W = I. Then S B is defined as S B = UW, which satisfies conditions (11), (12) and (13). Furthermore, the dimensionality reduction results, (14) and (15) hold, without impacting the estimate accuracy for σ $ 9. Proof. U and W are related to the left null space for X and U F KU. Left null space is a subspace of its corresponding matrix. For any given singular matrix A, any vector x is in the left null space of A if it satisfies x F A = 0. Consequently, if a matrix H satisfies H F A = 0 and H F H = I, then all the columns of H are considered as all the orthogonal bases in the left null space of A. Using the concept of left null space, it is obvious that the U and W exist if and only if both X and K are singular matrices, or rank deficient. In practice, every column of X is a kind of fixed effect. In most cases, GWAS data contain hundreds or thousands of subjects, but with less than ten fixed effects to be considered, simultaneously. Thus, the column number of X is much smaller the row number Np, which is a sufficient condition to ensure that X is rank deficient. Second, the GRM for repeated measures, K, is assumed to be expressed as K = K Δ, where K is the N N GRM of subjects (no repeated measurements), and Δ is a p p definite matrix with Δ [i, j] = 1, for all i and j [10]. Therefore, K is a singular matrix. Since X is rank deficient and K is singular, U and W exist based on the definition of the left null space. Given the existence and properties of U and W, (11), (12) and (13) must be true. As S B = UW, S F B e in (15) is linear transformation of a normal random variable e, such that S F B e is also normal-distributed. Next note that S B F Y = S B F Xβ + S B F g + S B F e = S B F e (27) where S F B Xβ and S F B g are cancelled out because S F B X = 0, and S F B g = W F U F g ~ N (0, W F U F KUW) = N (0, 0) (28) In sum, (14) and (15) are true. o Theorem 2. σ$ 9 is a Best Linear Unbiased Estimator (BLUE) when Σ is an identity matrix. Proof. Note that the best only means that among all the unbiased linear estimators for σ $ 9, the best one has the lowest variance. σ$ 9 is a BLUE if only σ$ 9 is suitable for Gauss-Markov theorem [14]. The Gauss-Markov theorem confirms that in a linear regression model where errors are uncorrelated, with expectation 0 and equal variance, the BLUE of coefficients is given by the OLS estimator. Therefore, we only need to ensure the expectation and variance of ε B are both 0. Given e B = S F B e, let us define the k th value of e B is e B,, and the k th value of ε B is ε B,, and the rank of Z B is D. Z B = S BF Y = S BF e in (14) 14
15 ε B = vec Z B Z F B σ $ 9 vec I = E( ε B ) = E e B,B e B,B e B,B e B, e B,$ e B,B e B,$ e B, e B, e B,B e B, e B, E e B,B e B,B e B,B e B, e B,$ e B,B e B,$ e B, e B, e B,B e B, e B, e B,B e B,B e B,B e B, e B,$ e B,B e B,$ e B, e B, e B,B e B, e B, σ $ 9 vec I = = 0 e B,B e B,B e B,B e B, e B,$ e B,B e B,$ e B, e B, e B,B e B, e B, E e B,B e B,B e B,B e B, e B,$ e B,B e B,$ e B, e B, e B,B e B, e B, k th value of ε B is ε B,, then there exist the corresponding i(k) and j(k) to k, such that k 1 = j k 1 D + i k 1, 0 i k 1 D 1. ε B, = e B,K( ) e B,L( ) (29) This is equivalent to i k = mod k 1, D + 1, and j k = (k i(k))/d + 1, where mod k 1, D is the modulo, i.e., the remainder after the division of k 1 by D. cov ε B,, ε B, c = cov e B,K e B,L, e B,K c e B,L c = E (e B,K e B,L, e B,K c e B,L c ) Therefore, cov ε B,, ε B, c = 1 if and only if i k B = j k B = i k $ = j k $. Specifically, var ε B,K = 1 for any i. We can now conclude that σ$ 9 is suitable for the Gauss- Markov theorem and σ$ 9 is a BLUE. o Theorem 3. σ$ 9 is an unbiased, consistent and efficient estimator. Proof. In statistics, an estimator is said to be unbiased, when the mean of the sampling distribution is equal to the true parameter being estimated; it is said to be a consistent estimator if and only if as the number of data points increases indefinitely, the resulting sequences of estimators will converge to the true parameter. For a linear regression model, if errors have finite variance, and are uncorrelated with the corresponding regressor, the estimator of any regressor coefficient is unbiased and consistent. In terms of regressor errors ε B in (19) and Theorem 2, its variance is equal to 1 which is finite. And it is known that ε B are uncorrelated with vec(i), which proves the consistent and unbiased properties of σ $ 9. The efficiency of an estimator is that among all unbiased estimators, an efficient estimator has the lowest variance. Efficiency requires that the errors have finite variance and are homoscedastic. First, Theorem 2 shows that the variance of ε B are finite. Furthermore, E ( ε $ B vec I ) is independent of the regressor vec I, implying that the variance of ε B is homoscedastic. o 15
16 Theorem 4 The variance of σ$ 9 is (a b c ) c, which is approximated by asymptotic variance based on E the consistency of σ $ 9. Similarly, the variance of σ$ 7 is (a f c ) c, where p is the number of repeat measurements. Proof. For variances of σ$ 9 and σ $ 7, in general, they are derived from (17) and (22), which can be regarded as the same mixed model: T = M, M ~ N (0, σ $ œ R) (31, 32) vec T F T = vec M + ε (33) $ $ where σ œ represents genetic variance σ 7 or environmental variance σ $ 9, D D matrix R $ represents Genetic Relationship Matrix, or covariance matrix for total errors. In this paper, σ œ is solved with Ordinary Least Square, whose result is numerically close to Generalized Least $ $ Square. Equation (3.6) and (3.9) in [25] prove that the approximate rate of σ œ to true σ œ is $ o( D). After detailed calculation of sample variance of ε, the asymptotic variance of σ œ is (a ž c ) c. $ $ Back to our estimates, the reduced dimension space to estimate σ 9 and σ 7 is on the order of N and pn, leading to variance of σ$ 9 and σ$ 7 is (a b c ) c E DE and (a f c ) c, separately. DE o 16
Efficient Bayesian mixed model analysis increases association power in large cohorts
 Linear regression Existing mixed model methods New method: BOLT-LMM Time O(MM) O(MN 2 ) O MN 1.5 Corrects for confounding? Power Efficient Bayesian mixed model analysis increases association power in large
Linear regression Existing mixed model methods New method: BOLT-LMM Time O(MM) O(MN 2 ) O MN 1.5 Corrects for confounding? Power Efficient Bayesian mixed model analysis increases association power in large
HERITABILITY ESTIMATION USING A REGULARIZED REGRESSION APPROACH (HERRA)
 BIRS 016 1 HERITABILITY ESTIMATION USING A REGULARIZED REGRESSION APPROACH (HERRA) Malka Gorfine, Tel Aviv University, Israel Joint work with Li Hsu, FHCRC, Seattle, USA BIRS 016 The concept of heritability
BIRS 016 1 HERITABILITY ESTIMATION USING A REGULARIZED REGRESSION APPROACH (HERRA) Malka Gorfine, Tel Aviv University, Israel Joint work with Li Hsu, FHCRC, Seattle, USA BIRS 016 The concept of heritability
(Genome-wide) association analysis
 (Genome-wide) association analysis 1 Key concepts Mapping QTL by association relies on linkage disequilibrium in the population; LD can be caused by close linkage between a QTL and marker (= good) or by
(Genome-wide) association analysis 1 Key concepts Mapping QTL by association relies on linkage disequilibrium in the population; LD can be caused by close linkage between a QTL and marker (= good) or by
Multidimensional heritability analysis of neuroanatomical shape. Jingwei Li
 Multidimensional heritability analysis of neuroanatomical shape Jingwei Li Brain Imaging Genetics Genetic Variation Behavior Cognition Neuroanatomy Brain Imaging Genetics Genetic Variation Neuroanatomy
Multidimensional heritability analysis of neuroanatomical shape Jingwei Li Brain Imaging Genetics Genetic Variation Behavior Cognition Neuroanatomy Brain Imaging Genetics Genetic Variation Neuroanatomy
Association studies and regression
 Association studies and regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Association studies and regression 1 / 104 Administration
Association studies and regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Association studies and regression 1 / 104 Administration
MIXED MODELS THE GENERAL MIXED MODEL
 MIXED MODELS This chapter introduces best linear unbiased prediction (BLUP), a general method for predicting random effects, while Chapter 27 is concerned with the estimation of variances by restricted
MIXED MODELS This chapter introduces best linear unbiased prediction (BLUP), a general method for predicting random effects, while Chapter 27 is concerned with the estimation of variances by restricted
Restricted Maximum Likelihood in Linear Regression and Linear Mixed-Effects Model
 Restricted Maximum Likelihood in Linear Regression and Linear Mixed-Effects Model Xiuming Zhang zhangxiuming@u.nus.edu A*STAR-NUS Clinical Imaging Research Center October, 015 Summary This report derives
Restricted Maximum Likelihood in Linear Regression and Linear Mixed-Effects Model Xiuming Zhang zhangxiuming@u.nus.edu A*STAR-NUS Clinical Imaging Research Center October, 015 Summary This report derives
Nature Genetics: doi: /ng Supplementary Figure 1. Number of cases and proxy cases required to detect association at designs.
 Supplementary Figure 1 Number of cases and proxy cases required to detect association at designs. = 5 10 8 for case control and proxy case control The ratio of controls to cases (or proxy cases) is 1.
Supplementary Figure 1 Number of cases and proxy cases required to detect association at designs. = 5 10 8 for case control and proxy case control The ratio of controls to cases (or proxy cases) is 1.
Heritability estimation in modern genetics and connections to some new results for quadratic forms in statistics
 Heritability estimation in modern genetics and connections to some new results for quadratic forms in statistics Lee H. Dicker Rutgers University and Amazon, NYC Based on joint work with Ruijun Ma (Rutgers),
Heritability estimation in modern genetics and connections to some new results for quadratic forms in statistics Lee H. Dicker Rutgers University and Amazon, NYC Based on joint work with Ruijun Ma (Rutgers),
Supplementary Information
 Supplementary Information 1 Supplementary Figures (a) Statistical power (p = 2.6 10 8 ) (b) Statistical power (p = 4.0 10 6 ) Supplementary Figure 1: Statistical power comparison between GEMMA (red) and
Supplementary Information 1 Supplementary Figures (a) Statistical power (p = 2.6 10 8 ) (b) Statistical power (p = 4.0 10 6 ) Supplementary Figure 1: Statistical power comparison between GEMMA (red) and
Supplementary Materials for Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control
 Supplementary Materials for Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control Xiaoquan Wen Department of Biostatistics, University of Michigan A Model
Supplementary Materials for Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control Xiaoquan Wen Department of Biostatistics, University of Michigan A Model
FaST linear mixed models for genome-wide association studies
 Nature Methods FaS linear mixed models for genome-wide association studies Christoph Lippert, Jennifer Listgarten, Ying Liu, Carl M Kadie, Robert I Davidson & David Heckerman Supplementary Figure Supplementary
Nature Methods FaS linear mixed models for genome-wide association studies Christoph Lippert, Jennifer Listgarten, Ying Liu, Carl M Kadie, Robert I Davidson & David Heckerman Supplementary Figure Supplementary
Multiple regression. CM226: Machine Learning for Bioinformatics. Fall Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar
 Multiple regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Multiple regression 1 / 36 Previous two lectures Linear and logistic
Multiple regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Multiple regression 1 / 36 Previous two lectures Linear and logistic
EFFICIENT COMPUTATION WITH A LINEAR MIXED MODEL ON LARGE-SCALE DATA SETS WITH APPLICATIONS TO GENETIC STUDIES
 Submitted to the Annals of Applied Statistics EFFICIENT COMPUTATION WITH A LINEAR MIXED MODEL ON LARGE-SCALE DATA SETS WITH APPLICATIONS TO GENETIC STUDIES By Matti Pirinen, Peter Donnelly and Chris C.A.
Submitted to the Annals of Applied Statistics EFFICIENT COMPUTATION WITH A LINEAR MIXED MODEL ON LARGE-SCALE DATA SETS WITH APPLICATIONS TO GENETIC STUDIES By Matti Pirinen, Peter Donnelly and Chris C.A.
Lecture WS Evolutionary Genetics Part I 1
 Quantitative genetics Quantitative genetics is the study of the inheritance of quantitative/continuous phenotypic traits, like human height and body size, grain colour in winter wheat or beak depth in
Quantitative genetics Quantitative genetics is the study of the inheritance of quantitative/continuous phenotypic traits, like human height and body size, grain colour in winter wheat or beak depth in
Learning gradients: prescriptive models
 Department of Statistical Science Institute for Genome Sciences & Policy Department of Computer Science Duke University May 11, 2007 Relevant papers Learning Coordinate Covariances via Gradients. Sayan
Department of Statistical Science Institute for Genome Sciences & Policy Department of Computer Science Duke University May 11, 2007 Relevant papers Learning Coordinate Covariances via Gradients. Sayan
Association Testing with Quantitative Traits: Common and Rare Variants. Summer Institute in Statistical Genetics 2014 Module 10 Lecture 5
 Association Testing with Quantitative Traits: Common and Rare Variants Timothy Thornton and Katie Kerr Summer Institute in Statistical Genetics 2014 Module 10 Lecture 5 1 / 41 Introduction to Quantitative
Association Testing with Quantitative Traits: Common and Rare Variants Timothy Thornton and Katie Kerr Summer Institute in Statistical Genetics 2014 Module 10 Lecture 5 1 / 41 Introduction to Quantitative
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature25973 Power Simulations We performed extensive power simulations to demonstrate that the analyses carried out in our study are well powered. Our simulations indicate very high power for
doi:10.1038/nature25973 Power Simulations We performed extensive power simulations to demonstrate that the analyses carried out in our study are well powered. Our simulations indicate very high power for
Lecture 2: Linear Models. Bruce Walsh lecture notes Seattle SISG -Mixed Model Course version 23 June 2011
 Lecture 2: Linear Models Bruce Walsh lecture notes Seattle SISG -Mixed Model Course version 23 June 2011 1 Quick Review of the Major Points The general linear model can be written as y = X! + e y = vector
Lecture 2: Linear Models Bruce Walsh lecture notes Seattle SISG -Mixed Model Course version 23 June 2011 1 Quick Review of the Major Points The general linear model can be written as y = X! + e y = vector
Linear Regression (1/1/17)
 STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
Variance Component Models for Quantitative Traits. Biostatistics 666
 Variance Component Models for Quantitative Traits Biostatistics 666 Today Analysis of quantitative traits Modeling covariance for pairs of individuals estimating heritability Extending the model beyond
Variance Component Models for Quantitative Traits Biostatistics 666 Today Analysis of quantitative traits Modeling covariance for pairs of individuals estimating heritability Extending the model beyond
GLIDE: GPU-based LInear Detection of Epistasis
 GLIDE: GPU-based LInear Detection of Epistasis Chloé-Agathe Azencott with Tony Kam-Thong, Lawrence Cayton, and Karsten Borgwardt Machine Learning and Computational Biology Research Group Max Planck Institute
GLIDE: GPU-based LInear Detection of Epistasis Chloé-Agathe Azencott with Tony Kam-Thong, Lawrence Cayton, and Karsten Borgwardt Machine Learning and Computational Biology Research Group Max Planck Institute
LECTURE 2 LINEAR REGRESSION MODEL AND OLS
 SEPTEMBER 29, 2014 LECTURE 2 LINEAR REGRESSION MODEL AND OLS Definitions A common question in econometrics is to study the effect of one group of variables X i, usually called the regressors, on another
SEPTEMBER 29, 2014 LECTURE 2 LINEAR REGRESSION MODEL AND OLS Definitions A common question in econometrics is to study the effect of one group of variables X i, usually called the regressors, on another
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#7:(Mar-23-2010) Genome Wide Association Studies 1 The law of causality... is a relic of a bygone age, surviving, like the monarchy,
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#7:(Mar-23-2010) Genome Wide Association Studies 1 The law of causality... is a relic of a bygone age, surviving, like the monarchy,
Previous lecture. Single variant association. Use genome-wide SNPs to account for confounding (population substructure)
 Previous lecture Single variant association Use genome-wide SNPs to account for confounding (population substructure) Estimation of effect size and winner s curse Meta-Analysis Today s outline P-value
Previous lecture Single variant association Use genome-wide SNPs to account for confounding (population substructure) Estimation of effect size and winner s curse Meta-Analysis Today s outline P-value
Applications of Randomized Methods for Decomposing and Simulating from Large Covariance Matrices
 Applications of Randomized Methods for Decomposing and Simulating from Large Covariance Matrices Vahid Dehdari and Clayton V. Deutsch Geostatistical modeling involves many variables and many locations.
Applications of Randomized Methods for Decomposing and Simulating from Large Covariance Matrices Vahid Dehdari and Clayton V. Deutsch Geostatistical modeling involves many variables and many locations.
Bayesian Regression (1/31/13)
 STA613/CBB540: Statistical methods in computational biology Bayesian Regression (1/31/13) Lecturer: Barbara Engelhardt Scribe: Amanda Lea 1 Bayesian Paradigm Bayesian methods ask: given that I have observed
STA613/CBB540: Statistical methods in computational biology Bayesian Regression (1/31/13) Lecturer: Barbara Engelhardt Scribe: Amanda Lea 1 Bayesian Paradigm Bayesian methods ask: given that I have observed
Least Squares Estimation-Finite-Sample Properties
 Least Squares Estimation-Finite-Sample Properties Ping Yu School of Economics and Finance The University of Hong Kong Ping Yu (HKU) Finite-Sample 1 / 29 Terminology and Assumptions 1 Terminology and Assumptions
Least Squares Estimation-Finite-Sample Properties Ping Yu School of Economics and Finance The University of Hong Kong Ping Yu (HKU) Finite-Sample 1 / 29 Terminology and Assumptions 1 Terminology and Assumptions
Genotype Imputation. Biostatistics 666
 Genotype Imputation Biostatistics 666 Previously Hidden Markov Models for Relative Pairs Linkage analysis using affected sibling pairs Estimation of pairwise relationships Identity-by-Descent Relatives
Genotype Imputation Biostatistics 666 Previously Hidden Markov Models for Relative Pairs Linkage analysis using affected sibling pairs Estimation of pairwise relationships Identity-by-Descent Relatives
PCA and admixture models
 PCA and admixture models CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar, Alkes Price PCA and admixture models 1 / 57 Announcements HW1
PCA and admixture models CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar, Alkes Price PCA and admixture models 1 / 57 Announcements HW1
25 : Graphical induced structured input/output models
 10-708: Probabilistic Graphical Models 10-708, Spring 2013 25 : Graphical induced structured input/output models Lecturer: Eric P. Xing Scribes: Meghana Kshirsagar (mkshirsa), Yiwen Chen (yiwenche) 1 Graph
10-708: Probabilistic Graphical Models 10-708, Spring 2013 25 : Graphical induced structured input/output models Lecturer: Eric P. Xing Scribes: Meghana Kshirsagar (mkshirsa), Yiwen Chen (yiwenche) 1 Graph
Lecture 2: Linear and Mixed Models
 Lecture 2: Linear and Mixed Models Bruce Walsh lecture notes Introduction to Mixed Models SISG, Seattle 18 20 July 2018 1 Quick Review of the Major Points The general linear model can be written as y =
Lecture 2: Linear and Mixed Models Bruce Walsh lecture notes Introduction to Mixed Models SISG, Seattle 18 20 July 2018 1 Quick Review of the Major Points The general linear model can be written as y =
Linear Regression. In this problem sheet, we consider the problem of linear regression with p predictors and one intercept,
 Linear Regression In this problem sheet, we consider the problem of linear regression with p predictors and one intercept, y = Xβ + ɛ, where y t = (y 1,..., y n ) is the column vector of target values,
Linear Regression In this problem sheet, we consider the problem of linear regression with p predictors and one intercept, y = Xβ + ɛ, where y t = (y 1,..., y n ) is the column vector of target values,
Lecture 2: Genetic Association Testing with Quantitative Traits. Summer Institute in Statistical Genetics 2017
 Lecture 2: Genetic Association Testing with Quantitative Traits Instructors: Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2017 1 / 29 Introduction to Quantitative Trait Mapping
Lecture 2: Genetic Association Testing with Quantitative Traits Instructors: Timothy Thornton and Michael Wu Summer Institute in Statistical Genetics 2017 1 / 29 Introduction to Quantitative Trait Mapping
Methods for Cryptic Structure. Methods for Cryptic Structure
 Case-Control Association Testing Review Consider testing for association between a disease and a genetic marker Idea is to look for an association by comparing allele/genotype frequencies between the cases
Case-Control Association Testing Review Consider testing for association between a disease and a genetic marker Idea is to look for an association by comparing allele/genotype frequencies between the cases
GWAS IV: Bayesian linear (variance component) models
 GWAS IV: Bayesian linear (variance component) models Dr. Oliver Stegle Christoh Lippert Prof. Dr. Karsten Borgwardt Max-Planck-Institutes Tübingen, Germany Tübingen Summer 2011 Oliver Stegle GWAS IV: Bayesian
GWAS IV: Bayesian linear (variance component) models Dr. Oliver Stegle Christoh Lippert Prof. Dr. Karsten Borgwardt Max-Planck-Institutes Tübingen, Germany Tübingen Summer 2011 Oliver Stegle GWAS IV: Bayesian
Software for genome-wide association studies having multivariate responses: Introducing MAGWAS
 Software for genome-wide association studies having multivariate responses: Introducing MAGWAS Chad C. Brown 1 and Alison A. Motsinger-Reif 1,2 1 Department of Statistics, 2 Bioinformatics Research Center
Software for genome-wide association studies having multivariate responses: Introducing MAGWAS Chad C. Brown 1 and Alison A. Motsinger-Reif 1,2 1 Department of Statistics, 2 Bioinformatics Research Center
Extending the MR-Egger method for multivariable Mendelian randomization to correct for both measured and unmeasured pleiotropy
 Received: 20 October 2016 Revised: 15 August 2017 Accepted: 23 August 2017 DOI: 10.1002/sim.7492 RESEARCH ARTICLE Extending the MR-Egger method for multivariable Mendelian randomization to correct for
Received: 20 October 2016 Revised: 15 August 2017 Accepted: 23 August 2017 DOI: 10.1002/sim.7492 RESEARCH ARTICLE Extending the MR-Egger method for multivariable Mendelian randomization to correct for
A General Framework for Variable Selection in Linear Mixed Models with Applications to Genetic Studies with Structured Populations
 A General Framework for Variable Selection in Linear Mixed Models with Applications to Genetic Studies with Structured Populations Joint work with Karim Oualkacha (UQÀM), Yi Yang (McGill), Celia Greenwood
A General Framework for Variable Selection in Linear Mixed Models with Applications to Genetic Studies with Structured Populations Joint work with Karim Oualkacha (UQÀM), Yi Yang (McGill), Celia Greenwood
DNA polymorphisms such as SNP and familial effects (additive genetic, common environment) to
 1 1 1 1 1 1 1 1 0 SUPPLEMENTARY MATERIALS, B. BIVARIATE PEDIGREE-BASED ASSOCIATION ANALYSIS Introduction We propose here a statistical method of bivariate genetic analysis, designed to evaluate contribution
1 1 1 1 1 1 1 1 0 SUPPLEMENTARY MATERIALS, B. BIVARIATE PEDIGREE-BASED ASSOCIATION ANALYSIS Introduction We propose here a statistical method of bivariate genetic analysis, designed to evaluate contribution
Machine Learning Linear Regression. Prof. Matteo Matteucci
 Machine Learning Linear Regression Prof. Matteo Matteucci Outline 2 o Simple Linear Regression Model Least Squares Fit Measures of Fit Inference in Regression o Multi Variate Regession Model Least Squares
Machine Learning Linear Regression Prof. Matteo Matteucci Outline 2 o Simple Linear Regression Model Least Squares Fit Measures of Fit Inference in Regression o Multi Variate Regession Model Least Squares
1 Cricket chirps: an example
 Notes for 2016-09-26 1 Cricket chirps: an example Did you know that you can estimate the temperature by listening to the rate of chirps? The data set in Table 1 1. represents measurements of the number
Notes for 2016-09-26 1 Cricket chirps: an example Did you know that you can estimate the temperature by listening to the rate of chirps? The data set in Table 1 1. represents measurements of the number
Lecture 3: Linear Models. Bruce Walsh lecture notes Uppsala EQG course version 28 Jan 2012
 Lecture 3: Linear Models Bruce Walsh lecture notes Uppsala EQG course version 28 Jan 2012 1 Quick Review of the Major Points The general linear model can be written as y = X! + e y = vector of observed
Lecture 3: Linear Models Bruce Walsh lecture notes Uppsala EQG course version 28 Jan 2012 1 Quick Review of the Major Points The general linear model can be written as y = X! + e y = vector of observed
Linear Models 1. Isfahan University of Technology Fall Semester, 2014
 Linear Models 1 Isfahan University of Technology Fall Semester, 2014 References: [1] G. A. F., Seber and A. J. Lee (2003). Linear Regression Analysis (2nd ed.). Hoboken, NJ: Wiley. [2] A. C. Rencher and
Linear Models 1 Isfahan University of Technology Fall Semester, 2014 References: [1] G. A. F., Seber and A. J. Lee (2003). Linear Regression Analysis (2nd ed.). Hoboken, NJ: Wiley. [2] A. C. Rencher and
A Course in Applied Econometrics Lecture 7: Cluster Sampling. Jeff Wooldridge IRP Lectures, UW Madison, August 2008
 A Course in Applied Econometrics Lecture 7: Cluster Sampling Jeff Wooldridge IRP Lectures, UW Madison, August 2008 1. The Linear Model with Cluster Effects 2. Estimation with a Small Number of roups and
A Course in Applied Econometrics Lecture 7: Cluster Sampling Jeff Wooldridge IRP Lectures, UW Madison, August 2008 1. The Linear Model with Cluster Effects 2. Estimation with a Small Number of roups and
Empirical Bayes Moderation of Asymptotically Linear Parameters
 Empirical Bayes Moderation of Asymptotically Linear Parameters Nima Hejazi Division of Biostatistics University of California, Berkeley stat.berkeley.edu/~nhejazi nimahejazi.org twitter/@nshejazi github/nhejazi
Empirical Bayes Moderation of Asymptotically Linear Parameters Nima Hejazi Division of Biostatistics University of California, Berkeley stat.berkeley.edu/~nhejazi nimahejazi.org twitter/@nshejazi github/nhejazi
Lecture 5: BLUP (Best Linear Unbiased Predictors) of genetic values. Bruce Walsh lecture notes Tucson Winter Institute 9-11 Jan 2013
 Lecture 5: BLUP (Best Linear Unbiased Predictors) of genetic values Bruce Walsh lecture notes Tucson Winter Institute 9-11 Jan 013 1 Estimation of Var(A) and Breeding Values in General Pedigrees The classic
Lecture 5: BLUP (Best Linear Unbiased Predictors) of genetic values Bruce Walsh lecture notes Tucson Winter Institute 9-11 Jan 013 1 Estimation of Var(A) and Breeding Values in General Pedigrees The classic
Comparison of multiple imputation methods for systematically and sporadically missing multilevel data
 Comparison of multiple imputation methods for systematically and sporadically missing multilevel data V. Audigier, I. White, S. Jolani, T. Debray, M. Quartagno, J. Carpenter, S. van Buuren, M. Resche-Rigon
Comparison of multiple imputation methods for systematically and sporadically missing multilevel data V. Audigier, I. White, S. Jolani, T. Debray, M. Quartagno, J. Carpenter, S. van Buuren, M. Resche-Rigon
Multiple Linear Regression
 Multiple Linear Regression University of California, San Diego Instructor: Ery Arias-Castro http://math.ucsd.edu/~eariasca/teaching.html 1 / 42 Passenger car mileage Consider the carmpg dataset taken from
Multiple Linear Regression University of California, San Diego Instructor: Ery Arias-Castro http://math.ucsd.edu/~eariasca/teaching.html 1 / 42 Passenger car mileage Consider the carmpg dataset taken from
Basics of Multivariate Modelling and Data Analysis
 Basics of Multivariate Modelling and Data Analysis Kurt-Erik Häggblom 2. Overview of multivariate techniques 2.1 Different approaches to multivariate data analysis 2.2 Classification of multivariate techniques
Basics of Multivariate Modelling and Data Analysis Kurt-Erik Häggblom 2. Overview of multivariate techniques 2.1 Different approaches to multivariate data analysis 2.2 Classification of multivariate techniques
This model of the conditional expectation is linear in the parameters. A more practical and relaxed attitude towards linear regression is to say that
 Linear Regression For (X, Y ) a pair of random variables with values in R p R we assume that E(Y X) = β 0 + with β R p+1. p X j β j = (1, X T )β j=1 This model of the conditional expectation is linear
Linear Regression For (X, Y ) a pair of random variables with values in R p R we assume that E(Y X) = β 0 + with β R p+1. p X j β j = (1, X T )β j=1 This model of the conditional expectation is linear
Latent Variable Methods for the Analysis of Genomic Data
 John D. Storey Center for Statistics and Machine Learning & Lewis-Sigler Institute for Integrative Genomics Latent Variable Methods for the Analysis of Genomic Data http://genomine.org/talks/ Data m variables
John D. Storey Center for Statistics and Machine Learning & Lewis-Sigler Institute for Integrative Genomics Latent Variable Methods for the Analysis of Genomic Data http://genomine.org/talks/ Data m variables
Applied Statistics and Econometrics
 Applied Statistics and Econometrics Lecture 6 Saul Lach September 2017 Saul Lach () Applied Statistics and Econometrics September 2017 1 / 53 Outline of Lecture 6 1 Omitted variable bias (SW 6.1) 2 Multiple
Applied Statistics and Econometrics Lecture 6 Saul Lach September 2017 Saul Lach () Applied Statistics and Econometrics September 2017 1 / 53 Outline of Lecture 6 1 Omitted variable bias (SW 6.1) 2 Multiple
Asymptotic distribution of the largest eigenvalue with application to genetic data
 Asymptotic distribution of the largest eigenvalue with application to genetic data Chong Wu University of Minnesota September 30, 2016 T32 Journal Club Chong Wu 1 / 25 Table of Contents 1 Background Gene-gene
Asymptotic distribution of the largest eigenvalue with application to genetic data Chong Wu University of Minnesota September 30, 2016 T32 Journal Club Chong Wu 1 / 25 Table of Contents 1 Background Gene-gene
Analyzing metabolomics data for association with genotypes using two-component Gaussian mixture distributions
 Analyzing metabolomics data for association with genotypes using two-component Gaussian mixture distributions Jason Westra Department of Statistics, Iowa State University Ames, IA 50011, United States
Analyzing metabolomics data for association with genotypes using two-component Gaussian mixture distributions Jason Westra Department of Statistics, Iowa State University Ames, IA 50011, United States
Combining SEM & GREML in OpenMx. Rob Kirkpatrick 3/11/16
 Combining SEM & GREML in OpenMx Rob Kirkpatrick 3/11/16 1 Overview I. Introduction. II. mxgreml Design. III. mxgreml Implementation. IV. Applications. V. Miscellany. 2 G V A A 1 1 F E 1 VA 1 2 3 Y₁ Y₂
Combining SEM & GREML in OpenMx Rob Kirkpatrick 3/11/16 1 Overview I. Introduction. II. mxgreml Design. III. mxgreml Implementation. IV. Applications. V. Miscellany. 2 G V A A 1 1 F E 1 VA 1 2 3 Y₁ Y₂
Package MACAU2. R topics documented: April 8, Type Package. Title MACAU 2.0: Efficient Mixed Model Analysis of Count Data. Version 1.
 Package MACAU2 April 8, 2017 Type Package Title MACAU 2.0: Efficient Mixed Model Analysis of Count Data Version 1.10 Date 2017-03-31 Author Shiquan Sun, Jiaqiang Zhu, Xiang Zhou Maintainer Shiquan Sun
Package MACAU2 April 8, 2017 Type Package Title MACAU 2.0: Efficient Mixed Model Analysis of Count Data Version 1.10 Date 2017-03-31 Author Shiquan Sun, Jiaqiang Zhu, Xiang Zhou Maintainer Shiquan Sun
SUPPLEMENTARY APPENDICES FOR WAVELET-DOMAIN REGRESSION AND PREDICTIVE INFERENCE IN PSYCHIATRIC NEUROIMAGING
 Submitted to the Annals of Applied Statistics SUPPLEMENTARY APPENDICES FOR WAVELET-DOMAIN REGRESSION AND PREDICTIVE INFERENCE IN PSYCHIATRIC NEUROIMAGING By Philip T. Reiss, Lan Huo, Yihong Zhao, Clare
Submitted to the Annals of Applied Statistics SUPPLEMENTARY APPENDICES FOR WAVELET-DOMAIN REGRESSION AND PREDICTIVE INFERENCE IN PSYCHIATRIC NEUROIMAGING By Philip T. Reiss, Lan Huo, Yihong Zhao, Clare
Structure in Data. A major objective in data analysis is to identify interesting features or structure in the data.
 Structure in Data A major objective in data analysis is to identify interesting features or structure in the data. The graphical methods are very useful in discovering structure. There are basically two
Structure in Data A major objective in data analysis is to identify interesting features or structure in the data. The graphical methods are very useful in discovering structure. There are basically two
Using Genomic Structural Equation Modeling to Model Joint Genetic Architecture of Complex Traits
 Using Genomic Structural Equation Modeling to Model Joint Genetic Architecture of Complex Traits Presented by: Andrew D. Grotzinger & Elliot M. Tucker-Drob Paper: Grotzinger, A. D., Rhemtulla, M., de Vlaming,
Using Genomic Structural Equation Modeling to Model Joint Genetic Architecture of Complex Traits Presented by: Andrew D. Grotzinger & Elliot M. Tucker-Drob Paper: Grotzinger, A. D., Rhemtulla, M., de Vlaming,
ISyE 691 Data mining and analytics
 ISyE 691 Data mining and analytics Regression Instructor: Prof. Kaibo Liu Department of Industrial and Systems Engineering UW-Madison Email: kliu8@wisc.edu Office: Room 3017 (Mechanical Engineering Building)
ISyE 691 Data mining and analytics Regression Instructor: Prof. Kaibo Liu Department of Industrial and Systems Engineering UW-Madison Email: kliu8@wisc.edu Office: Room 3017 (Mechanical Engineering Building)
Sparse BLAS-3 Reduction
 Sparse BLAS-3 Reduction to Banded Upper Triangular (Spar3Bnd) Gary Howell, HPC/OIT NC State University gary howell@ncsu.edu Sparse BLAS-3 Reduction p.1/27 Acknowledgements James Demmel, Gene Golub, Franc
Sparse BLAS-3 Reduction to Banded Upper Triangular (Spar3Bnd) Gary Howell, HPC/OIT NC State University gary howell@ncsu.edu Sparse BLAS-3 Reduction p.1/27 Acknowledgements James Demmel, Gene Golub, Franc
Robust Bayesian Variable Selection for Modeling Mean Medical Costs
 Robust Bayesian Variable Selection for Modeling Mean Medical Costs Grace Yoon 1,, Wenxin Jiang 2, Lei Liu 3 and Ya-Chen T. Shih 4 1 Department of Statistics, Texas A&M University 2 Department of Statistics,
Robust Bayesian Variable Selection for Modeling Mean Medical Costs Grace Yoon 1,, Wenxin Jiang 2, Lei Liu 3 and Ya-Chen T. Shih 4 1 Department of Statistics, Texas A&M University 2 Department of Statistics,
Power and sample size calculations for designing rare variant sequencing association studies.
 Power and sample size calculations for designing rare variant sequencing association studies. Seunggeun Lee 1, Michael C. Wu 2, Tianxi Cai 1, Yun Li 2,3, Michael Boehnke 4 and Xihong Lin 1 1 Department
Power and sample size calculations for designing rare variant sequencing association studies. Seunggeun Lee 1, Michael C. Wu 2, Tianxi Cai 1, Yun Li 2,3, Michael Boehnke 4 and Xihong Lin 1 1 Department
MODEL-FREE LINKAGE AND ASSOCIATION MAPPING OF COMPLEX TRAITS USING QUANTITATIVE ENDOPHENOTYPES
 MODEL-FREE LINKAGE AND ASSOCIATION MAPPING OF COMPLEX TRAITS USING QUANTITATIVE ENDOPHENOTYPES Saurabh Ghosh Human Genetics Unit Indian Statistical Institute, Kolkata Most common diseases are caused by
MODEL-FREE LINKAGE AND ASSOCIATION MAPPING OF COMPLEX TRAITS USING QUANTITATIVE ENDOPHENOTYPES Saurabh Ghosh Human Genetics Unit Indian Statistical Institute, Kolkata Most common diseases are caused by
Biostatistics-Lecture 16 Model Selection. Ruibin Xi Peking University School of Mathematical Sciences
 Biostatistics-Lecture 16 Model Selection Ruibin Xi Peking University School of Mathematical Sciences Motivating example1 Interested in factors related to the life expectancy (50 US states,1969-71 ) Per
Biostatistics-Lecture 16 Model Selection Ruibin Xi Peking University School of Mathematical Sciences Motivating example1 Interested in factors related to the life expectancy (50 US states,1969-71 ) Per
. Also, in this case, p i = N1 ) T, (2) where. I γ C N(N 2 2 F + N1 2 Q)
 Supplementary information S7 Testing for association at imputed SPs puted SPs Score tests A Score Test needs calculations of the observed data score and information matrix only under the null hypothesis,
Supplementary information S7 Testing for association at imputed SPs puted SPs Score tests A Score Test needs calculations of the observed data score and information matrix only under the null hypothesis,
Homoskedasticity. Var (u X) = σ 2. (23)
 Homoskedasticity How big is the difference between the OLS estimator and the true parameter? To answer this question, we make an additional assumption called homoskedasticity: Var (u X) = σ 2. (23) This
Homoskedasticity How big is the difference between the OLS estimator and the true parameter? To answer this question, we make an additional assumption called homoskedasticity: Var (u X) = σ 2. (23) This
Introduction to Statistical Genetics (BST227) Lecture 6: Population Substructure in Association Studies
 Introduction to Statistical Genetics (BST227) Lecture 6: Population Substructure in Association Studies Confounding in gene+c associa+on studies q What is it? q What is the effect? q How to detect it?
Introduction to Statistical Genetics (BST227) Lecture 6: Population Substructure in Association Studies Confounding in gene+c associa+on studies q What is it? q What is the effect? q How to detect it?
Model-Free Knockoffs: High-Dimensional Variable Selection that Controls the False Discovery Rate
 Model-Free Knockoffs: High-Dimensional Variable Selection that Controls the False Discovery Rate Lucas Janson, Stanford Department of Statistics WADAPT Workshop, NIPS, December 2016 Collaborators: Emmanuel
Model-Free Knockoffs: High-Dimensional Variable Selection that Controls the False Discovery Rate Lucas Janson, Stanford Department of Statistics WADAPT Workshop, NIPS, December 2016 Collaborators: Emmanuel
Supplementary Note on Bayesian analysis
 Supplementary Note on Bayesian analysis Structured variability of muscle activations supports the minimal intervention principle of motor control Francisco J. Valero-Cuevas 1,2,3, Madhusudhan Venkadesan
Supplementary Note on Bayesian analysis Structured variability of muscle activations supports the minimal intervention principle of motor control Francisco J. Valero-Cuevas 1,2,3, Madhusudhan Venkadesan
Proportional Variance Explained by QLT and Statistical Power. Proportional Variance Explained by QTL and Statistical Power
 Proportional Variance Explained by QTL and Statistical Power Partitioning the Genetic Variance We previously focused on obtaining variance components of a quantitative trait to determine the proportion
Proportional Variance Explained by QTL and Statistical Power Partitioning the Genetic Variance We previously focused on obtaining variance components of a quantitative trait to determine the proportion
Testing Some Covariance Structures under a Growth Curve Model in High Dimension
 Department of Mathematics Testing Some Covariance Structures under a Growth Curve Model in High Dimension Muni S. Srivastava and Martin Singull LiTH-MAT-R--2015/03--SE Department of Mathematics Linköping
Department of Mathematics Testing Some Covariance Structures under a Growth Curve Model in High Dimension Muni S. Srivastava and Martin Singull LiTH-MAT-R--2015/03--SE Department of Mathematics Linköping
Introduction to Machine Learning. PCA and Spectral Clustering. Introduction to Machine Learning, Slides: Eran Halperin
 1 Introduction to Machine Learning PCA and Spectral Clustering Introduction to Machine Learning, 2013-14 Slides: Eran Halperin Singular Value Decomposition (SVD) The singular value decomposition (SVD)
1 Introduction to Machine Learning PCA and Spectral Clustering Introduction to Machine Learning, 2013-14 Slides: Eran Halperin Singular Value Decomposition (SVD) The singular value decomposition (SVD)
DEGseq: an R package for identifying differentially expressed genes from RNA-seq data
 DEGseq: an R package for identifying differentially expressed genes from RNA-seq data Likun Wang Zhixing Feng i Wang iaowo Wang * and uegong Zhang * MOE Key Laboratory of Bioinformatics and Bioinformatics
DEGseq: an R package for identifying differentially expressed genes from RNA-seq data Likun Wang Zhixing Feng i Wang iaowo Wang * and uegong Zhang * MOE Key Laboratory of Bioinformatics and Bioinformatics
Statistical Power of Model Selection Strategies for Genome-Wide Association Studies
 Statistical Power of Model Selection Strategies for Genome-Wide Association Studies Zheyang Wu 1, Hongyu Zhao 1,2 * 1 Department of Epidemiology and Public Health, Yale University School of Medicine, New
Statistical Power of Model Selection Strategies for Genome-Wide Association Studies Zheyang Wu 1, Hongyu Zhao 1,2 * 1 Department of Epidemiology and Public Health, Yale University School of Medicine, New
NIH Public Access Author Manuscript Stat Sin. Author manuscript; available in PMC 2013 August 15.
 NIH Public Access Author Manuscript Published in final edited form as: Stat Sin. 2012 ; 22: 1041 1074. ON MODEL SELECTION STRATEGIES TO IDENTIFY GENES UNDERLYING BINARY TRAITS USING GENOME-WIDE ASSOCIATION
NIH Public Access Author Manuscript Published in final edited form as: Stat Sin. 2012 ; 22: 1041 1074. ON MODEL SELECTION STRATEGIES TO IDENTIFY GENES UNDERLYING BINARY TRAITS USING GENOME-WIDE ASSOCIATION
Probability of Detecting Disease-Associated SNPs in Case-Control Genome-Wide Association Studies
 Probability of Detecting Disease-Associated SNPs in Case-Control Genome-Wide Association Studies Ruth Pfeiffer, Ph.D. Mitchell Gail Biostatistics Branch Division of Cancer Epidemiology&Genetics National
Probability of Detecting Disease-Associated SNPs in Case-Control Genome-Wide Association Studies Ruth Pfeiffer, Ph.D. Mitchell Gail Biostatistics Branch Division of Cancer Epidemiology&Genetics National
A Bootstrap Test for Causality with Endogenous Lag Length Choice. - theory and application in finance
 CESIS Electronic Working Paper Series Paper No. 223 A Bootstrap Test for Causality with Endogenous Lag Length Choice - theory and application in finance R. Scott Hacker and Abdulnasser Hatemi-J April 200
CESIS Electronic Working Paper Series Paper No. 223 A Bootstrap Test for Causality with Endogenous Lag Length Choice - theory and application in finance R. Scott Hacker and Abdulnasser Hatemi-J April 200
Regression Analysis for Data Containing Outliers and High Leverage Points
 Alabama Journal of Mathematics 39 (2015) ISSN 2373-0404 Regression Analysis for Data Containing Outliers and High Leverage Points Asim Kumer Dey Department of Mathematics Lamar University Md. Amir Hossain
Alabama Journal of Mathematics 39 (2015) ISSN 2373-0404 Regression Analysis for Data Containing Outliers and High Leverage Points Asim Kumer Dey Department of Mathematics Lamar University Md. Amir Hossain
Introduction to Machine Learning
 10-701 Introduction to Machine Learning PCA Slides based on 18-661 Fall 2018 PCA Raw data can be Complex, High-dimensional To understand a phenomenon we measure various related quantities If we knew what
10-701 Introduction to Machine Learning PCA Slides based on 18-661 Fall 2018 PCA Raw data can be Complex, High-dimensional To understand a phenomenon we measure various related quantities If we knew what
Estimation of Parameters in Random. Effect Models with Incidence Matrix. Uncertainty
 Estimation of Parameters in Random Effect Models with Incidence Matrix Uncertainty Xia Shen 1,2 and Lars Rönnegård 2,3 1 The Linnaeus Centre for Bioinformatics, Uppsala University, Uppsala, Sweden; 2 School
Estimation of Parameters in Random Effect Models with Incidence Matrix Uncertainty Xia Shen 1,2 and Lars Rönnegård 2,3 1 The Linnaeus Centre for Bioinformatics, Uppsala University, Uppsala, Sweden; 2 School
CS 4491/CS 7990 SPECIAL TOPICS IN BIOINFORMATICS
 CS 4491/CS 7990 SPECIAL TOPICS IN BIOINFORMATICS * Some contents are adapted from Dr. Hung Huang and Dr. Chengkai Li at UT Arlington Mingon Kang, Ph.D. Computer Science, Kennesaw State University Problems
CS 4491/CS 7990 SPECIAL TOPICS IN BIOINFORMATICS * Some contents are adapted from Dr. Hung Huang and Dr. Chengkai Li at UT Arlington Mingon Kang, Ph.D. Computer Science, Kennesaw State University Problems
Model Selection Tutorial 2: Problems With Using AIC to Select a Subset of Exposures in a Regression Model
 Model Selection Tutorial 2: Problems With Using AIC to Select a Subset of Exposures in a Regression Model Centre for Molecular, Environmental, Genetic & Analytic (MEGA) Epidemiology School of Population
Model Selection Tutorial 2: Problems With Using AIC to Select a Subset of Exposures in a Regression Model Centre for Molecular, Environmental, Genetic & Analytic (MEGA) Epidemiology School of Population
Simple Linear Regression Model & Introduction to. OLS Estimation
 Inside ECOOMICS Introduction to Econometrics Simple Linear Regression Model & Introduction to Introduction OLS Estimation We are interested in a model that explains a variable y in terms of other variables
Inside ECOOMICS Introduction to Econometrics Simple Linear Regression Model & Introduction to Introduction OLS Estimation We are interested in a model that explains a variable y in terms of other variables
Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization
 Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization arxiv:1606.03729v1 [stat.me] 12 Jun 2016 Stephen Burgess 1, Jack Bowden 2 Frank Dudbridge
Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization arxiv:1606.03729v1 [stat.me] 12 Jun 2016 Stephen Burgess 1, Jack Bowden 2 Frank Dudbridge
Bayesian construction of perceptrons to predict phenotypes from 584K SNP data.
 Bayesian construction of perceptrons to predict phenotypes from 584K SNP data. Luc Janss, Bert Kappen Radboud University Nijmegen Medical Centre Donders Institute for Neuroscience Introduction Genetic
Bayesian construction of perceptrons to predict phenotypes from 584K SNP data. Luc Janss, Bert Kappen Radboud University Nijmegen Medical Centre Donders Institute for Neuroscience Introduction Genetic
The STS Surgeon Composite Technical Appendix
 The STS Surgeon Composite Technical Appendix Overview Surgeon-specific risk-adjusted operative operative mortality and major complication rates were estimated using a bivariate random-effects logistic
The STS Surgeon Composite Technical Appendix Overview Surgeon-specific risk-adjusted operative operative mortality and major complication rates were estimated using a bivariate random-effects logistic
Analysis of longitudinal neuroimaging data with OLS & Sandwich Estimator of variance
 Analysis of longitudinal neuroimaging data with OLS & Sandwich Estimator of variance Bryan Guillaume Reading workshop lifespan neurobiology 27 June 2014 Supervisors: Thomas Nichols (Warwick University)
Analysis of longitudinal neuroimaging data with OLS & Sandwich Estimator of variance Bryan Guillaume Reading workshop lifespan neurobiology 27 June 2014 Supervisors: Thomas Nichols (Warwick University)
Practical Linear Algebra: A Geometry Toolbox
 Practical Linear Algebra: A Geometry Toolbox Third edition Chapter 12: Gauss for Linear Systems Gerald Farin & Dianne Hansford CRC Press, Taylor & Francis Group, An A K Peters Book www.farinhansford.com/books/pla
Practical Linear Algebra: A Geometry Toolbox Third edition Chapter 12: Gauss for Linear Systems Gerald Farin & Dianne Hansford CRC Press, Taylor & Francis Group, An A K Peters Book www.farinhansford.com/books/pla
Previous lecture. P-value based combination. Fixed vs random effects models. Meta vs. pooled- analysis. New random effects testing.
 Previous lecture P-value based combination. Fixed vs random effects models. Meta vs. pooled- analysis. New random effects testing. Interaction Outline: Definition of interaction Additive versus multiplicative
Previous lecture P-value based combination. Fixed vs random effects models. Meta vs. pooled- analysis. New random effects testing. Interaction Outline: Definition of interaction Additive versus multiplicative
COS513: FOUNDATIONS OF PROBABILISTIC MODELS LECTURE 9: LINEAR REGRESSION
 COS513: FOUNDATIONS OF PROBABILISTIC MODELS LECTURE 9: LINEAR REGRESSION SEAN GERRISH AND CHONG WANG 1. WAYS OF ORGANIZING MODELS In probabilistic modeling, there are several ways of organizing models:
COS513: FOUNDATIONS OF PROBABILISTIC MODELS LECTURE 9: LINEAR REGRESSION SEAN GERRISH AND CHONG WANG 1. WAYS OF ORGANIZING MODELS In probabilistic modeling, there are several ways of organizing models:
REPLICATION VARIANCE ESTIMATION FOR THE NATIONAL RESOURCES INVENTORY
 REPLICATION VARIANCE ESTIMATION FOR THE NATIONAL RESOURCES INVENTORY J.D. Opsomer, W.A. Fuller and X. Li Iowa State University, Ames, IA 50011, USA 1. Introduction Replication methods are often used in
REPLICATION VARIANCE ESTIMATION FOR THE NATIONAL RESOURCES INVENTORY J.D. Opsomer, W.A. Fuller and X. Li Iowa State University, Ames, IA 50011, USA 1. Introduction Replication methods are often used in
Linear models. Linear models are computationally convenient and remain widely used in. applied econometric research
 Linear models Linear models are computationally convenient and remain widely used in applied econometric research Our main focus in these lectures will be on single equation linear models of the form y
Linear models Linear models are computationally convenient and remain widely used in applied econometric research Our main focus in these lectures will be on single equation linear models of the form y
Two-Level Designs to Estimate All Main Effects and Two-Factor Interactions
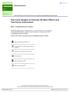 Technometrics ISSN: 0040-1706 (Print) 1537-2723 (Online) Journal homepage: http://www.tandfonline.com/loi/utch20 Two-Level Designs to Estimate All Main Effects and Two-Factor Interactions Pieter T. Eendebak
Technometrics ISSN: 0040-1706 (Print) 1537-2723 (Online) Journal homepage: http://www.tandfonline.com/loi/utch20 Two-Level Designs to Estimate All Main Effects and Two-Factor Interactions Pieter T. Eendebak
Missing Data Issues in the Studies of Neurodegenerative Disorders: the Methodology
 Missing Data Issues in the Studies of Neurodegenerative Disorders: the Methodology Sheng Luo, PhD Associate Professor Department of Biostatistics & Bioinformatics Duke University Medical Center sheng.luo@duke.edu
Missing Data Issues in the Studies of Neurodegenerative Disorders: the Methodology Sheng Luo, PhD Associate Professor Department of Biostatistics & Bioinformatics Duke University Medical Center sheng.luo@duke.edu
Supplementary Information for Efficient Bayesian mixed model analysis increases association power in large cohorts
 Supplementary Information for Efficient Bayesian mixed model analysis increases association power in large cohorts Po-Ru Loh, George Tucker, Brendan K Bulik-Sullivan, Bjarni J Vilhjálmsson, Hilary K Finucane,
Supplementary Information for Efficient Bayesian mixed model analysis increases association power in large cohorts Po-Ru Loh, George Tucker, Brendan K Bulik-Sullivan, Bjarni J Vilhjálmsson, Hilary K Finucane,
25 : Graphical induced structured input/output models
 10-708: Probabilistic Graphical Models 10-708, Spring 2016 25 : Graphical induced structured input/output models Lecturer: Eric P. Xing Scribes: Raied Aljadaany, Shi Zong, Chenchen Zhu Disclaimer: A large
10-708: Probabilistic Graphical Models 10-708, Spring 2016 25 : Graphical induced structured input/output models Lecturer: Eric P. Xing Scribes: Raied Aljadaany, Shi Zong, Chenchen Zhu Disclaimer: A large
Resampling techniques for statistical modeling
 Resampling techniques for statistical modeling Gianluca Bontempi Département d Informatique Boulevard de Triomphe - CP 212 http://www.ulb.ac.be/di Resampling techniques p.1/33 Beyond the empirical error
Resampling techniques for statistical modeling Gianluca Bontempi Département d Informatique Boulevard de Triomphe - CP 212 http://www.ulb.ac.be/di Resampling techniques p.1/33 Beyond the empirical error
Principal Component Analysis
 Principal Component Analysis Yingyu Liang yliang@cs.wisc.edu Computer Sciences Department University of Wisconsin, Madison [based on slides from Nina Balcan] slide 1 Goals for the lecture you should understand
Principal Component Analysis Yingyu Liang yliang@cs.wisc.edu Computer Sciences Department University of Wisconsin, Madison [based on slides from Nina Balcan] slide 1 Goals for the lecture you should understand
