minimal models for lipid membranes: local modifications around fusion objects
|
|
|
- Claude Taylor
- 5 years ago
- Views:
Transcription
1 minimal models for lipid membranes: local modifications around fusion objects Giovanni Marelli Georg August Universität, Göttingen January 21, 2013 PhD defense
2 collective phenomena structures and morphologies lipid bilayer minimal models for membranes motivations 2 / 22
3 collective phenomena structures and morphologies lipid bilayer stalk pore fusion objects transmembrane protein minimal models for membranes motivations 2 / 22
4 membrane fusion how does membrane fusion work? viral fusion K. K. Lee EMBO J. 29, 1299 (2010) minimal models for membranes motivations 3 / 22
5 membrane fusion viral fusion how does membrane fusion work? possible (heuristic) explanations? K. K. Lee EMBO J. 29, 1299 (2010) minimal models for membranes motivations 3 / 22
6 membrane fusion how does membrane fusion work? artificial pore possible (heuristic) explanations? capture morphology change and dynamics isolate metastable states minimal models for membranes motivations 3 / 22
7 membrane fusion how does membrane fusion work? artificial pore under lateral tension, low hydration identify the fusion pathways capture morphology change and dynamics isolate metastable states minimal models for membranes estimate energies and times role of the peptides motivations 3 / 22
8 contents minimal models for collective phenomena in lipid membranes lipids as diblocks atomistic martini diblock coarse-grained solvent-free soft large time scales, many lipids computational efficiency sampling from thermal fluctuations minimal models for membranes motivations 4 / 22
9 contents minimal models for collective phenomena in lipid membranes lipids as diblocks atomistic martini diblock coarse-grained solvent-free soft large time scales, many lipids computational efficiency sampling from thermal fluctuations membranes as continuum sheets minimal models for membranes motivations 4 / 22
10 contents minimal models for collective phenomena in lipid membranes lipids as diblocks atomistic martini diblock coarse-grained solvent-free soft large time scales, many lipids computational efficiency sampling from thermal fluctuations membranes as continuum sheets mechanical properties of unperturbed membranes: time and length scales energetic contributions local modifications by fusion objects thickness and density profiles in simulations and numerical calculations superposition of effects lipid mediated interactions the peptides stabilize the pore the peptide lowers the line tension of the pore minimal models for membranes motivations 4 / 22
11 overview of the thesis planar membranes reference values protein/lipid interactions superposition of effects stalk morphologies universality of structures in models and experiments minimal models for membranes hydrophobic stability conditions for stability pore/protein interactions superposition of thickness profiles motivations 5 / 22
12 model solvent-free, coarse-grained model bonded (harmonic spring, bending): U sp k B T = P N b 1 k sp i=1 2 (r i r i+1 ) 2 U b k B T = P N b 1 i=2 k b (1 cos θ i±1 ) non bonded (density functional): H nb [ρ] k B T = R «d 3 r v 2 Re 3 AA 2 ρ 2 A + v AAA 3 ρ 3 3 A corresponding eq of state: PRe 3 k B T = ρ + v AA 2 ρ 2 A v AAAρ 3 A (fluid/vapor phase coexistance) the virial coefficients are set by ρ coex and the kn b : v AA = 2 kn b +3 v ρ coex AAA = 3 kn b +2 2 ρ 2 coex mixed virial coefficient χn MF = ρ coex `vab 1 2 (v AA + v BB ) M. Hömberg and M. Müller, J. Chem. Phys. 132, (2010) minimal models for membranes model 6 / 22
13 model solvent-free, coarse-grained model bonded (harmonic spring, bending): U sp k B T = P N b 1 k sp i=1 2 (r i r i+1 ) 2 U b k B T = P N b 1 i=2 k b (1 cos θ i±1 ) non bonded (density functional): H nb [ρ] k B T = R «d 3 r v 2 Re 3 AA 2 ρ 2 A + v AAA 3 ρ 3 3 A corresponding eq of state: PRe 3 k B T = ρ + v AA 2 ρ 2 A v AAAρ 3 A (fluid/vapor phase coexistance) the virial coefficients are set by ρ coex and the kn b : v AA = 2 kn b +3 v ρ coex AAA = 3 kn b +2 2 ρ 2 coex mixed virial coefficient χn MF = ρ coex `vab 1 2 (v AA + v BB ) M. Hömberg and M. Müller, J. Chem. Phys. 132, (2010) minimal models for membranes model 6 / 22
14 model solvent-free, coarse-grained model bonded (harmonic spring, bending): U sp k B T = P N b 1 k sp i=1 2 (r i r i+1 ) 2 U b k B T = P N b 1 i=2 k b (1 cos θ i±1 ) non bonded (density functional): H nb [ρ] k B T = R «d 3 r v 2 Re 3 AA 2 ρ 2 A + v AAA 3 ρ 3 3 A corresponding eq of state: PRe 3 k B T = ρ + v AA 2 ρ 2 A v AAAρ 3 A (fluid/vapor phase coexistance) the virial coefficients are set by ρ coex and the kn b : v AA = 2 kn b +3 v ρ coex AAA = 3 kn b +2 2 ρ 2 coex mixed virial coefficient χn MF = ρ coex `vab 1 2 (v AA + v BB ) M. Hömberg and M. Müller, J. Chem. Phys. 132, (2010) minimal models for membranes model 6 / 22
15 properties of planar membranes planar lamellar membrane time scale length scale coupling moduli fluctuation spectrum minimal models for membranes planar membranes 7 / 22
16 time and length scale diffusivity D` 2E x(t) x(0) D = lim t 4t timestep: 1[ t s] 8[ps] (martini 0.04[ps], atomistic 4[fs]) K. Weiß and J. Enderlein, ChemPhysChem 13, 990 (2012) varying kn b minimal models for membranes planar membranes 8 / 22
17 time and length scale diffusivity D` 2E x(t) x(0) D = lim t 4t timestep: 1[ t s] 8[ps] (martini 0.04[ps], atomistic 4[fs]) K. Weiß and J. Enderlein, ChemPhysChem 13, 990 (2012) varying kn b thickness d = [ L] 1.08[nm] N. Kučerka et al. Biophysica Acta 1808, 2761 (2011) minimal models for membranes planar membranes 8 / 22
18 quantities fluctuations Z F s = dxdy ks (s(x, y) s) 2 2 s 2 harmonic approximation we sample from thermal fluctuations s fluctuating quantity, k s coupling constant, L patch area. minimal models for membranes planar membranes 9 / 22
19 quantities fluctuations Z F s = dxdy ks 2 harmonic approximation we sample from thermal fluctuations (s(x, y) s) 2 s 2 1 standard deviation = lim k s L L 2 (s s) 2 L s 2 minimal models for membranes planar membranes 9 / 22
20 quantities fluctuations Z F s = dxdy ks 2 harmonic approximation we sample from thermal fluctuations (s(x, y) s) 2 s 2 1 standard deviation = lim k s L L 2 (s s) 2 L s 2 σ = ρd thickness `d d 2 k el = 22.7[k B T / L 2 ] lipids/area (σ σ) 2 k com = 38.7[k B T / L 2 ] density (ρ ρ) 2 k melt = 34.6[k B T / L 2 ] σ 2 σ 2 = ρ2 ρ 2 + d2 d 2 1 = k com k melt k el minimal models for membranes planar membranes 9 / 22
21 quantities fluctuations we sample from thermal fluctuations Z F s = dxdy ks (s(x, y) s) 2 1 L 2 (s s) 2 L 2 s 2 = lim k s L s 2 harmonic approximation standard deviation compressibility σ = ρd thickness `d d 2 k el = 22.7[k B T / L 2 ] lipids/area (σ σ) 2 k com = 38.7[k B T / L 2 ] density (ρ ρ) 2 k melt = 34.6[k B T / L 2 ] σ 2 σ 2 = ρ2 ρ 2 + d2 d 2 1 = k com k melt k el minimal models for membranes planar membranes 9 / 22
22 energetic contributions bending (height), peristaltic (thickness), protrusion (lipid separation) F = 1 R 2 dxdy k 1 ben + 1 (d d) + k 2 (ρ ρ) R 1 R 2 el d 2 + k 2 melt ρ 2 Helfrich model bending: k ben bending rigidity peristaltic: k el elastic coupling c 0 spontaneous curvature a area/lipids ζ := c 0 a a c 0 protrusion: k λ protrusion rigidity γ λ microscopic surface tension minimal models for membranes planar membranes 10 / 22
23 energetic contributions bending (height), peristaltic (thickness), protrusion (lipid separation) F ben = 1 2 R dxdy kben ( 2 h(x, y)) 2 Monge representation height power spectrum h 2 (q) L 2 = 1 k ben q 4 bending: k ben bending rigidity 14.3[k B T ] peristaltic: k el elastic coupling c 0 spontaneous curvature a area/lipids ζ := c 0 a a c 0 protrusion: k λ protrusion rigidity γ λ microscopic surface tension minimal models for membranes planar membranes 10 / 22
24 energetic contributions F ben = 1 2 bending (height), peristaltic (thickness), protrusion (lipid separation) R dxdy kben ( 2 h(x, y)) 2 F per = 1 2 R dxdy kel d 2 (x,y) d 2 thickness height h 2 (q) L 2 = 1 k ben q 4 d 2 (q) L 2 = 1 k el / d 2 power spectrum bending: k ben bending rigidity 14.3[k B T ] peristaltic: k el elastic coupling 1.74/ d 2 [k B T / L 4 ] c 0 spontaneous curvature a area/lipids ζ := c 0 a a c 0 protrusion: k λ protrusion rigidity γ λ microscopic surface tension minimal models for membranes planar membranes 10 / 22
25 energetic contributions bending (height), peristaltic (thickness), protrusion (lipid separation) thickness height h 2 (q) L 2 = 1 d 2 (q) L 2 = 1 k ben q 4 + 2(k λ +γ λ q 2 ) 1 1 k ben q 4 4ζk ben q 2 +k el / d 2 + 2(k λ +γ λ q 2 ) Branningan and Brown, Biophys. J. 92, 864 (2007) power spectrum bending: k ben bending rigidity 12.5[k B T ] peristaltic: k el elastic coupling 1.96/ d 2 [k B T / L 4 ] c 0 spontaneous curvature a area/lipids ζ := c 0 a a c 0 = 0.14/ d[ L 2 ] protrusion: k λ protrusion rigidity γ λ microscopic surface tension we extract: k ben, ζ, k el minimal models for membranes planar membranes 10 / 22
26 lipid/protein interactions hydrophobic inclusion radial thickness profile weakening of the membrane continuum model superposition of effects pore stabilisation minimal models for membranes lipids/peptide 11 / 22
27 thickness profile contact angle and thinning cylindrical transmembrane protein cluster of connected monomers radius 1[ L] hydrophobic mismatch 1.3 N b /A = 38[ L 2 ] θ c = 31 o minimal models for membranes lipids/peptide 12 / 22
28 thickness profile contact angle and thinning cylindrical transmembrane protein cluster of connected monomers radius 1[ L] hydrophobic mismatch 1.3 N b /A = 38[ L 2 ] θ c = 31 o radial thickness profiles minimal models for membranes lipids/peptide 12 / 22
29 lipid frustration bond length minimal models for membranes lipids/peptide 13 / 22
30 lipid frustration oil partitions close to the peptide bond length density of the oil oil molecule: dodecane hydrocarbon tail the oil loses its translational entropy acts as a sensor to partion where most of the stress is minimal models for membranes lipids/peptide 13 / 22
31 lipid frustration oil partitions close to the peptide bond length density of the oil point to point hydrophobic density difference with and without oil oil molecule: dodecane hydrocarbon tail the oil loses its translational entropy acts as a sensor to partion where most of the stress is minimal models for membranes lipids/peptide 13 / 22
32 discrete description mean separation rescaled to the bulk value thickness from the polar/apolar density intersection Delaunay triangulation thickness mean area minimal models for membranes lipids/peptide 14 / 22
33 discrete description mean separation rescaled to the bulk value thickness from the polar/apolar density intersection bead number normalization upper and lower monolayers Delaunay triangulation thickness mean area minimal models for membranes lipids/peptide 14 / 22
34 weakening of the membrane lowering the line tension of the pore classical nucleation theory G = G0 + 2πRp λ πrp2 Pl line tension of the pore in NVT ensemble Pl = λ Rp minimal models for membranes lipids/peptide 15 / 22
35 weakening of the membrane lowering the line tension of the pore classical nucleation theory G = G0 + 2πRp λ πrp2 Pl line tension of the pore in NVT ensemble Pl = λ Rp the peptide lowers the line tension of the pore pore s radius correction N Pl = Rλp 1 αpep Rpep p minimal models for membranes lipids/peptide 15 / 22
36 continuum description F = R dxdy k ben d s(x, y) := d(x, y) d k ben 4 c + 4k ben ζ d elastic model 2 ( 2 d s) 2 + 4k ben c 0 2 d s + 2 k ben ζ d d s 2 d s + k el 2 d 2 ds 2 Euler Lagrange equations: 2 c + k el d 2 d s(x, y) = 0 2 c := 2 x 0 0 y 2 δf = 0 δd s «r/ξ sin(kr) fitting equation: Ae r ξ = 2.2[ L] k = 0.88[ L 1 ] sketch B. West, F. L. H. Brown and F. Schmid, Biophys. J. 96, 101 (2009) minimal models for membranes lipids/peptide 16 / 22
37 continuum description F = R dxdy k ben d s(x, y) := d(x, y) d k ben 4 c + 4k ben ζ d elastic model 2 ( 2 d s) 2 + 4k ben c 0 2 d s + 2 k ben ζ d d s 2 d s + k el 2 d 2 ds 2 Euler Lagrange equations: 2 c + k el d 2 d s(x, y) = 0 2 c := 2 x 0 0 y 2 δf = 0 δd s «r/ξ sin(kr) fitting equation: Ae r ξ = 2.2[ L] k = 0.88[ L 1 ] sketch B. West, F. L. H. Brown and F. Schmid, Biophys. J. 96, 101 (2009) minimal models for membranes lipids/peptide 16 / 22
38 continuum description F = R dxdy k ben d s(x, y) := d(x, y) d k ben 4 r + 4k ben ζ d elastic model 2 ( 2 d s) 2 + 4k ben c 0 2 d s + 2 k ben ζ d d s 2 d s + k el 2 d 2 ds 2 Euler Lagrange equations: 2 r + k el d 2 d s(r) = 0 2 r := 1 r (r r ) r δf δd s = 0 r/ξ sin(kr) fitting equation: Ae r ξ = 2.2[ L] k = 0.88[ L 1 ] sketch B. West, F. L. H. Brown and F. Schmid, Biophys. J. 96, 101 (2009) minimal models for membranes lipids/peptide 16 / 22
39 continuum description F = R dxdy k ben d s(x, y) := d(x, y) d k ben 4 r + 4k ben ζ d elastic model 2 ( 2 d s) 2 + 4k ben c 0 2 d s + 2 k ben ζ d d s 2 d s + k el 2 d 2 ds 2 Euler Lagrange equations: 2 r + k el d 2 d s(r) = 0 2 r := 1 r (r r ) r δf δd s = 0 r/ξ sin(kr) fitting equation: Ae r ξ = 2.2[ L] k = 0.88[ L 1 ] sketch B. West, F. L. H. Brown and F. Schmid, Biophys. J. 96, 101 (2009) minimal models for membranes lipids/peptide 16 / 22
40 fine exploration thickness characteristics the solution of the continuum model is times faster than simulations maximum thinning d max minimum distance r min minimal models for membranes lipids/peptide 17 / 22
41 superposition of effects two inclusions rmin II : minimum splits in two dmax II : superposition effect is stronger clusterisation of gramicidins: (hydrophobic matching) T. Harroun et al Biophys. J. 76, 937 (1999) minimal models for membranes lipids/peptide 18 / 22
42 superposition of effects two inclusions rmin II : minimum splits in two dmax II : superposition effect is stronger clusterisation of gramicidins: (hydrophobic matching) T. Harroun et al Biophys. J. 76, 937 (1999) minimal models for membranes lipids/peptide 18 / 22
43 many peptides thickness (simulations) minimal models for membranes thickness (calculations) lipids/peptide 19 / 22
44 many peptides thickness (simulations) minimal models for membranes thickness (calculations) lipids/peptide 19 / 22
45 many peptides thickness (simulations) minimal models for membranes thickness (calculations) lipids/peptide 19 / 22
46 summary unperturbed membranes sampling from thermal fluctuations time and length scale energetic contributions power spectrum material properties parametrisation of the continuum model minimal models for membranes conclusions 20 / 22
47 summary unperturbed membranes sampling from thermal fluctuations time and length scale energetic contributions power spectrum material properties parametrisation of the continuum model lipid/peptide thickness profile undershoot below the bulk value good agreement between simulations and calculations superposition of effects the membrane gets thinner pore stabilisation minimal models for membranes conclusions 20 / 22
48 summary unperturbed membranes sampling from thermal fluctuations time and length scale energetic contributions power spectrum material properties parametrisation of the continuum model lipid/peptide thickness profile undershoot below the bulk value good agreement between simulations and calculations superposition of effects the membrane gets thinner pore stabilisation oil partition oil partitions at the interface with the peptide (pore s rim and stalk s ends) (relaxes the lipid frustration) minimal models for membranes conclusions 20 / 22
49 summary unperturbed membranes sampling from thermal fluctuations time and length scale energetic contributions power spectrum material properties parametrisation of the continuum model oil partition oil partitions at the interface with the peptide (pore s rim and stalk s ends) (relaxes the lipid frustration) lipid/peptide thickness profile undershoot below the bulk value good agreement between simulations and calculations superposition of effects the membrane gets thinner pore stabilisation fusion pathways metastable states identify them (evolution change by lipid composition) minimal models for membranes conclusions 20 / 22
50 summary unperturbed membranes sampling from thermal fluctuations time and length scale energetic contributions power spectrum material properties parametrisation of the continuum model oil partition oil partitions at the interface with the peptide (pore s rim and stalk s ends) (relaxes the lipid frustration) lipid/peptide thickness profile undershoot below the bulk value good agreement between simulations and calculations superposition of effects the membrane gets thinner pore stabilisation fusion pathways metastable states identify them (evolution change by lipid composition) minimal models fast and fine exploration of phenomena tuned by more detailed models confirm the multiscale approach (universality of stalk s morphologies) (bilayer repulsion) minimal models for membranes conclusions 20 / 22
51 future developement lipid/peptide dynamics study the peptide diffusion by changing the peptide description study the lipid diffusivity depending on the radial distance from the peptide minimal models for membranes conclusions 21 / 22
52 future developement lipid/peptide dynamics study the peptide diffusion by changing the peptide description study the lipid diffusivity depending on the radial distance from the peptide stalk/pore/peptides competing line tensions interactions between a boundle of peptide and a stalk minimal models for membranes conclusions 21 / 22
53 acknowledgments thank you for your attention acknowledgments Marcus Müller Yuliya Smirnova Jelger Risselada Martin Hömberg Kostas Daoulas Ulrich Welling foundings: SFB project minimal models for membranes appendix 22 / 22
54 Dissipative Particle Dynamics thermostat simulation technique: MD + MDPD thermostat characteristic: the total force is the sum of the conservative, random and dissipative forces F tot = F c + F r + F d the forces are pairwise F tot i = P j Fc ij + Fr ij + Fd ij the forces are soften via a weighting function F ij = F ij w(r ij ) features of the multibody DPD: local density dependance pragmatic extension of the classical DPD that allows one to prescribe the thermodynamic behavior of a system ensembles NPT NVT NP t T tensionless membrane. P. Español and P. B. Warren Europhys. Lett (1995) S. Y. Trominov et al. J. Chem. Phys (2002) minimal models for membranes appendix 23 / 22
55 Dissipative Particle Dynamics thermostat simulation technique: MD + MDPD thermostat characteristic: the total force is the sum of the conservative, random and dissipative forces F tot = F c + F r + F d the forces are pairwise F tot i = P j Fc ij + Fr ij + Fd ij the forces are soften via a weighting function F ij = F ij w(r ij ) features of the multibody DPD: local density dependance pragmatic extension of the classical DPD that allows one to prescribe the thermodynamic behavior of a system ensembles NPT NVT NP t T tensionless membrane. P. Español and P. B. Warren Europhys. Lett (1995) S. Y. Trominov et al. J. Chem. Phys (2002) minimal models for membranes appendix 23 / 22
56 oil relaxation P αβ (r, z) = k B T ρ(r, z) + k B T ρ(r, z) + 1 V normal component Pz l (r, z) = P zz (r, z) P θθ(r,z )+P rr (r,z ) 2 no oil with oil (r,z P ) ij du ij dr x α ij x β ij x radial component P l r (r, z) = P rr (r, z) P θθ(r,z )+P zz (r,z ) 2 identification of the interface position of the relaxation by the oil minimal models for membranes appendix 24 / 22
57 oil relaxation P αβ (r, z) = k B T ρ(r, z) + k B T ρ(r, z) + 1 V normal component Pz l (r, z) = P zz (r, z) P θθ(r,z )+P rr (r,z ) 2 no oil with oil (r,z P ) ij du ij dr x α ij x β ij x radial component P l r (r, z) = P rr (r, z) P θθ(r,z )+P zz (r,z ) 2 identification of the interface position of the relaxation by the oil minimal models for membranes appendix 24 / 22
58 oil relaxation P αβ (r, z) = k B T ρ(r, z) + k B T ρ(r, z) + 1 V normal component Pz l (r, z) = P zz (r, z) P θθ(r,z )+P rr (r,z ) 2 no oil with oil (r,z P ) ij du ij dr x α ij x β ij x lateral pressure difference radial component P l r (r, z) = P rr (r, z) P θθ(r,z )+P zz (r,z ) 2 identification of the interface position of the relaxation by the oil minimal models for membranes appendix 24 / 22
59 monolayer spontaneous curvature calculation of the spontaneous curvature lateral pressure P l zz (r, z) = Pzz (r, z) Pxx (r, z) + P yy (r, z) 2 spontaneous curvature c 0 = 0.36[ L 1 ] Z L k ben c 0 = dz zpzz l (z) 0 «normal lateral pressure profile minimal models for membranes appendix 25 / 22
60 elastic model F = Z dxdy k ben 2( 2 h) 2 + k λ (z z 2 ) + γ λ `( z + ) 2 + ( z ) 2 + h z + /2 + d z ) + kcom 2 d 2 + k ben c 0 2 d + k ben ζd d 2 d + k ben 2 ( 2 d) 2 comparison with the solution of the differential equation in 2d finite elements calculation (surface evolver) finite differences calculation minimal models for membranes appendix 26 / 22
61 slab mean square displacement mean square displacement in radial slabs minimal models for membranes appendix 27 / 22
62 superposition of effects/two inclusions density plot around the inclusions division in sectors by angles minimum distance and depth the membrane is thinner between the two inclusions the position of the minimum shifts minimal models for membranes appendix 28 / 22
63 stiffness of a circular stalk we search iteratively a torus surrounding the stalk with: we count the beads inside the torus the smallest lateral radius the largest normal radius the most contact with heads the less contact with tails the center of mass of the beads is the torus position the radius of the torus is r t = A/2π, A the area occupied by the beads radial evolution diffusivity D stalk = D lipid /3 minimal models for membranes appendix 29 / 22
64 stiffness and density profile exp data PhD thesis of S. Aeffner radial density profiles - model/scale dependence Martini force field, dft, implicit solvent explicit solvent flexible chains, explicit solvent d t /d s = d t /d b = 2 d s/d b = d t /d s = 2 d t /d b = 2.5 d s /d b = 1.2 d t /d s = 2.1 d t /d b = 2.1 d s /d b = 1.0 d t /d s = 2.9 d t /d b = 2.2 d s /d b = 0.77 stiffness dft Martini minimal models for membranes appendix 30 / 22
65 oil contribution addition of small oil chains in the bilayers dft 10% oil partitioning density profile tails density difference Martini 15% minimal models for membranes appendix 31 / 22
Introduction to Computer Simulations of Soft Matter Methodologies and Applications Boulder July, 19-20, 2012
 Introduction to Computer Simulations of Soft Matter Methodologies and Applications Boulder July, 19-20, 2012 K. Kremer Max Planck Institute for Polymer Research, Mainz Overview Simulations, general considerations
Introduction to Computer Simulations of Soft Matter Methodologies and Applications Boulder July, 19-20, 2012 K. Kremer Max Planck Institute for Polymer Research, Mainz Overview Simulations, general considerations
Supplementary Information for: Controlling Cellular Uptake of Nanoparticles with ph-sensitive Polymers
 Supplementary Information for: Controlling Cellular Uptake of Nanoparticles with ph-sensitive Polymers Hong-ming Ding 1 & Yu-qiang Ma 1,2, 1 National Laboratory of Solid State Microstructures and Department
Supplementary Information for: Controlling Cellular Uptake of Nanoparticles with ph-sensitive Polymers Hong-ming Ding 1 & Yu-qiang Ma 1,2, 1 National Laboratory of Solid State Microstructures and Department
Dissipative Particle Dynamics: Foundation, Evolution and Applications
 Dissipative Particle Dynamics: Foundation, Evolution and Applications Lecture 4: DPD in soft matter and polymeric applications George Em Karniadakis Division of Applied Mathematics, Brown University &
Dissipative Particle Dynamics: Foundation, Evolution and Applications Lecture 4: DPD in soft matter and polymeric applications George Em Karniadakis Division of Applied Mathematics, Brown University &
Obtaining the Bending Modulus from a Buckled Lipid Membrane
 Obtaining the Bending Modulus from a Buckled Lipid Membrane Patrick Diggins, Mingyang Hu, Markus Deserno Department of Physics, Carnegie Mellon University, Pittsburgh PA, USA October 11, 2013 Outline Lipid
Obtaining the Bending Modulus from a Buckled Lipid Membrane Patrick Diggins, Mingyang Hu, Markus Deserno Department of Physics, Carnegie Mellon University, Pittsburgh PA, USA October 11, 2013 Outline Lipid
PHASE TRANSITIONS IN SOFT MATTER SYSTEMS
 OUTLINE: Topic D. PHASE TRANSITIONS IN SOFT MATTER SYSTEMS Definition of a phase Classification of phase transitions Thermodynamics of mixing (gases, polymers, etc.) Mean-field approaches in the spirit
OUTLINE: Topic D. PHASE TRANSITIONS IN SOFT MATTER SYSTEMS Definition of a phase Classification of phase transitions Thermodynamics of mixing (gases, polymers, etc.) Mean-field approaches in the spirit
Mesoscopic simulation for the structural change of a surfactant solution using dissipative particle dynamics
 Korean J. Chem. Eng., 26(6), 1717-1722 (2009) DOI: 10.1007/s11814-009-0235-2 RAPID COMMUNICATION Mesoscopic simulation for the structural change of a surfactant solution using dissipative particle dynamics
Korean J. Chem. Eng., 26(6), 1717-1722 (2009) DOI: 10.1007/s11814-009-0235-2 RAPID COMMUNICATION Mesoscopic simulation for the structural change of a surfactant solution using dissipative particle dynamics
arxiv: v1 [cond-mat.soft] 10 Feb 2010
![arxiv: v1 [cond-mat.soft] 10 Feb 2010 arxiv: v1 [cond-mat.soft] 10 Feb 2010](/thumbs/76/73375543.jpg) Main phase transition in lipid bilayers: phase coexistence and line tension in a soft, solvent-free, coarse-grained model Martin Hömberg and Marcus Müller Institut für Theoretische Physik, Georg-August-Universität,
Main phase transition in lipid bilayers: phase coexistence and line tension in a soft, solvent-free, coarse-grained model Martin Hömberg and Marcus Müller Institut für Theoretische Physik, Georg-August-Universität,
Origins of Mechanical and Rheological Properties of Polymer Nanocomposites. Venkat Ganesan
 Department of Chemical Engineering University of Texas@Austin Origins of Mechanical and Rheological Properties of Polymer Nanocomposites Venkat Ganesan $$$: NSF DMR, Welch Foundation Megha Surve, Victor
Department of Chemical Engineering University of Texas@Austin Origins of Mechanical and Rheological Properties of Polymer Nanocomposites Venkat Ganesan $$$: NSF DMR, Welch Foundation Megha Surve, Victor
Kobe-Brown Simulation Summer School 2015 Project: DPD Simulation of a Membrane
 Kobe-Brown Simulation Summer School 2015 Project: DPD Simulation of a Membrane Clark Bowman Karen Larson Yuji Funaki Ross Parker Tae Woo Kim Week 1: Introduction to Molecular Dynamics (MD) Computer simulation
Kobe-Brown Simulation Summer School 2015 Project: DPD Simulation of a Membrane Clark Bowman Karen Larson Yuji Funaki Ross Parker Tae Woo Kim Week 1: Introduction to Molecular Dynamics (MD) Computer simulation
Dynamics of Polymers and Membranes P. B. Sunil Kumar
 Dynamics of Polymers and Membranes P. B. Sunil Kumar Department of Physics, Indian Institute of Technology Madras, Chennai, 600036 Dissipative Particle Dynamics (DPD) - Basic model P. Espagnol and P. Warren,
Dynamics of Polymers and Membranes P. B. Sunil Kumar Department of Physics, Indian Institute of Technology Madras, Chennai, 600036 Dissipative Particle Dynamics (DPD) - Basic model P. Espagnol and P. Warren,
Analysis of the simulation
 Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
MD Thermodynamics. Lecture 12 3/26/18. Harvard SEAS AP 275 Atomistic Modeling of Materials Boris Kozinsky
 MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
Experimental Soft Matter (M. Durand, G. Foffi)
 Master 2 PCS/PTSC 2016-2017 10/01/2017 Experimental Soft Matter (M. Durand, G. Foffi) Nota Bene Exam duration : 3H ecture notes are not allowed. Electronic devices (including cell phones) are prohibited,
Master 2 PCS/PTSC 2016-2017 10/01/2017 Experimental Soft Matter (M. Durand, G. Foffi) Nota Bene Exam duration : 3H ecture notes are not allowed. Electronic devices (including cell phones) are prohibited,
Elastic Properties and Line Tension of. Self-Assembled Bilayer Membranes
 Elastic Properties and Line Tension of Self-Assembled Bilayer Membranes ELASTIC PROPERTIES AND LINE TENSION OF SELF-ASSEMBLED BILAYER MEMBRANES BY KYLE PASTOR, B.Sc. a thesis submitted to the department
Elastic Properties and Line Tension of Self-Assembled Bilayer Membranes ELASTIC PROPERTIES AND LINE TENSION OF SELF-ASSEMBLED BILAYER MEMBRANES BY KYLE PASTOR, B.Sc. a thesis submitted to the department
Structuring of hydrophobic and hydrophilic polymers at interfaces Stephen Donaldson ChE 210D Final Project Abstract
 Structuring of hydrophobic and hydrophilic polymers at interfaces Stephen Donaldson ChE 210D Final Project Abstract In this work, a simplified Lennard-Jones (LJ) sphere model is used to simulate the aggregation,
Structuring of hydrophobic and hydrophilic polymers at interfaces Stephen Donaldson ChE 210D Final Project Abstract In this work, a simplified Lennard-Jones (LJ) sphere model is used to simulate the aggregation,
Contents. Introduction List of notation
 Contents Introduction List of notation i vii 1 Geometry and variational calculus 1 1.1 Planar curves 1 1.1.1 Polar coordinate system 2 1.1.2 Curvature 3 1.1.3 Frenet-Serret equations 5 1.2 Space curves
Contents Introduction List of notation i vii 1 Geometry and variational calculus 1 1.1 Planar curves 1 1.1.1 Polar coordinate system 2 1.1.2 Curvature 3 1.1.3 Frenet-Serret equations 5 1.2 Space curves
Coarse-Grained Models!
 Coarse-Grained Models! Large and complex molecules (e.g. long polymers) can not be simulated on the all-atom level! Requires coarse-graining of the model! Coarse-grained models are usually also particles
Coarse-Grained Models! Large and complex molecules (e.g. long polymers) can not be simulated on the all-atom level! Requires coarse-graining of the model! Coarse-grained models are usually also particles
The Molecular Dynamics Method
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
Dissipative particle dynamics: Bridging the gap between atomistic and mesoscopic simulation
 Dissipative particle dynamics: Bridging the gap between atomistic and mesoscopic simulation Robert D. Groot and Patrick B. Warren Unilever Research Port Sunlight Laboratory, Quarry Road East, Bebington,
Dissipative particle dynamics: Bridging the gap between atomistic and mesoscopic simulation Robert D. Groot and Patrick B. Warren Unilever Research Port Sunlight Laboratory, Quarry Road East, Bebington,
EXAM I COURSE TFY4310 MOLECULAR BIOPHYSICS December Suggested resolution
 page 1 of 7 EXAM I COURSE TFY4310 MOLECULAR BIOPHYSICS December 2013 Suggested resolution Exercise 1. [total: 25 p] a) [t: 5 p] Describe the bonding [1.5 p] and the molecular orbitals [1.5 p] of the ethylene
page 1 of 7 EXAM I COURSE TFY4310 MOLECULAR BIOPHYSICS December 2013 Suggested resolution Exercise 1. [total: 25 p] a) [t: 5 p] Describe the bonding [1.5 p] and the molecular orbitals [1.5 p] of the ethylene
Systematic Coarse-Graining and Concurrent Multiresolution Simulation of Molecular Liquids
 Systematic Coarse-Graining and Concurrent Multiresolution Simulation of Molecular Liquids Cameron F. Abrams Department of Chemical and Biological Engineering Drexel University Philadelphia, PA USA 9 June
Systematic Coarse-Graining and Concurrent Multiresolution Simulation of Molecular Liquids Cameron F. Abrams Department of Chemical and Biological Engineering Drexel University Philadelphia, PA USA 9 June
Dissipative Particle Dynamics Simulations of Polymersomes
 17708 J. Phys. Chem. B 2005, 109, 17708-17714 Dissipative Particle Dynamics Simulations of Polymersomes Vanessa Ortiz,*,,, Steven O. Nielsen,, Dennis E. Discher,, Michael L. Klein,, Reinhard Lipowsky,
17708 J. Phys. Chem. B 2005, 109, 17708-17714 Dissipative Particle Dynamics Simulations of Polymersomes Vanessa Ortiz,*,,, Steven O. Nielsen,, Dennis E. Discher,, Michael L. Klein,, Reinhard Lipowsky,
Structure Analysis of Bottle-Brush Polymers: Simulation and Experiment
 Jacob Klein Science 323, 47, 2009 Manfred Schmidt (Mainz) Structure Analysis of Bottle-Brush Polymers: Simulation and Experiment http://www.blueberryforest.com Hsiao-Ping Hsu Institut für Physik, Johannes
Jacob Klein Science 323, 47, 2009 Manfred Schmidt (Mainz) Structure Analysis of Bottle-Brush Polymers: Simulation and Experiment http://www.blueberryforest.com Hsiao-Ping Hsu Institut für Physik, Johannes
Budding Dynamics of Individual Domains in Multicomponent Membranes Simulated by N-Varied Dissipative Particle Dynamics
 J. Phys. Chem. B 2007, 111, 5837-5849 5837 Budding Dynamics of Individual Domains in Multicomponent Membranes Simulated by N-Varied Dissipative Particle Dynamics Bingbing Hong, Feng Qiu,* Hongdong Zhang,
J. Phys. Chem. B 2007, 111, 5837-5849 5837 Budding Dynamics of Individual Domains in Multicomponent Membranes Simulated by N-Varied Dissipative Particle Dynamics Bingbing Hong, Feng Qiu,* Hongdong Zhang,
Coarse-graining, applications and mesoscopic molecular dynamics
 CS Work in progress Coarse-graining, applications and mesoscopic molecular dynamics Carsten Svaneborg Institute for Physics, Chemistry, and Pharmacy University of Southern Denmark Campusvej 55, 5320 Odense
CS Work in progress Coarse-graining, applications and mesoscopic molecular dynamics Carsten Svaneborg Institute for Physics, Chemistry, and Pharmacy University of Southern Denmark Campusvej 55, 5320 Odense
Nucleation rate (m -3 s -1 ) Radius of water nano droplet (Å) 1e+00 1e-64 1e-128 1e-192 1e-256
 Supplementary Figures Nucleation rate (m -3 s -1 ) 1e+00 1e-64 1e-128 1e-192 1e-256 Calculated R in bulk water Calculated R in droplet Modified CNT 20 30 40 50 60 70 Radius of water nano droplet (Å) Supplementary
Supplementary Figures Nucleation rate (m -3 s -1 ) 1e+00 1e-64 1e-128 1e-192 1e-256 Calculated R in bulk water Calculated R in droplet Modified CNT 20 30 40 50 60 70 Radius of water nano droplet (Å) Supplementary
Game Physics. Game and Media Technology Master Program - Utrecht University. Dr. Nicolas Pronost
 Game and Media Technology Master Program - Utrecht University Dr. Nicolas Pronost Soft body physics Soft bodies In reality, objects are not purely rigid for some it is a good approximation but if you hit
Game and Media Technology Master Program - Utrecht University Dr. Nicolas Pronost Soft body physics Soft bodies In reality, objects are not purely rigid for some it is a good approximation but if you hit
Effect of protein shape on multibody interactions between membrane inclusions
 PHYSICAL REVIEW E VOLUME 61, NUMBER 4 APRIL 000 Effect of protein shape on multibody interactions between membrane inclusions K. S. Kim, 1, * John Neu, and George Oster 3, 1 Department of Physics, Graduate
PHYSICAL REVIEW E VOLUME 61, NUMBER 4 APRIL 000 Effect of protein shape on multibody interactions between membrane inclusions K. S. Kim, 1, * John Neu, and George Oster 3, 1 Department of Physics, Graduate
Computer simulation methods (2) Dr. Vania Calandrini
 Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
Chap. 2. Polymers Introduction. - Polymers: synthetic materials <--> natural materials
 Chap. 2. Polymers 2.1. Introduction - Polymers: synthetic materials natural materials no gas phase, not simple liquid (much more viscous), not perfectly crystalline, etc 2.3. Polymer Chain Conformation
Chap. 2. Polymers 2.1. Introduction - Polymers: synthetic materials natural materials no gas phase, not simple liquid (much more viscous), not perfectly crystalline, etc 2.3. Polymer Chain Conformation
Coarse-graining limits in open and wall-bounded dissipative particle dynamics systems
 THE JOURNAL OF CHEMICAL PHYSICS 124, 184101 2006 Coarse-graining limits in open and wall-bounded dissipative particle dynamics systems Igor V. Pivkin and George E. Karniadakis a Division of Applied Mathematics,
THE JOURNAL OF CHEMICAL PHYSICS 124, 184101 2006 Coarse-graining limits in open and wall-bounded dissipative particle dynamics systems Igor V. Pivkin and George E. Karniadakis a Division of Applied Mathematics,
An efficient hybrid method for modeling lipid membranes with molecular resolution
 An efficient hybrid method for modeling lipid membranes with molecular resolution G.J.A. (Agur) Sevink, M. Charlaganov & J.G.E.M Fraaije Soft Matter Chemistry Group, Leiden University, The Netherlands
An efficient hybrid method for modeling lipid membranes with molecular resolution G.J.A. (Agur) Sevink, M. Charlaganov & J.G.E.M Fraaije Soft Matter Chemistry Group, Leiden University, The Netherlands
arxiv: v1 [cond-mat.soft] 22 Oct 2007
![arxiv: v1 [cond-mat.soft] 22 Oct 2007 arxiv: v1 [cond-mat.soft] 22 Oct 2007](/thumbs/78/78401876.jpg) Conformational Transitions of Heteropolymers arxiv:0710.4095v1 [cond-mat.soft] 22 Oct 2007 Michael Bachmann and Wolfhard Janke Institut für Theoretische Physik, Universität Leipzig, Augustusplatz 10/11,
Conformational Transitions of Heteropolymers arxiv:0710.4095v1 [cond-mat.soft] 22 Oct 2007 Michael Bachmann and Wolfhard Janke Institut für Theoretische Physik, Universität Leipzig, Augustusplatz 10/11,
Glass-Transition and Side-Chain Dynamics in Thin Films: Explaining. Dissimilar Free Surface Effects for Polystyrene and Poly(methyl methacrylate)
 Supporting Information for Glass-Transition and Side-Chain Dynamics in Thin Films: Explaining Dissimilar Free Surface Effects for Polystyrene and Poly(methyl methacrylate) David D. Hsu, Wenjie Xia, Jake
Supporting Information for Glass-Transition and Side-Chain Dynamics in Thin Films: Explaining Dissimilar Free Surface Effects for Polystyrene and Poly(methyl methacrylate) David D. Hsu, Wenjie Xia, Jake
Introduction to molecular dynamics
 1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
Effect of surfactant structure on interfacial properties
 EUROPHYSICS LETTERS 15 September 2003 Europhys. Lett., 63 (6), pp. 902 907 (2003) Effect of surfactant structure on interfacial properties L. Rekvig 1 ( ), M. Kranenburg 2, B. Hafskjold 1 and B. Smit 2
EUROPHYSICS LETTERS 15 September 2003 Europhys. Lett., 63 (6), pp. 902 907 (2003) Effect of surfactant structure on interfacial properties L. Rekvig 1 ( ), M. Kranenburg 2, B. Hafskjold 1 and B. Smit 2
Overview & Applications. T. Lezon Hands-on Workshop in Computational Biophysics Pittsburgh Supercomputing Center 04 June, 2015
 Overview & Applications T. Lezon Hands-on Workshop in Computational Biophysics Pittsburgh Supercomputing Center 4 June, 215 Simulations still take time Bakan et al. Bioinformatics 211. Coarse-grained Elastic
Overview & Applications T. Lezon Hands-on Workshop in Computational Biophysics Pittsburgh Supercomputing Center 4 June, 215 Simulations still take time Bakan et al. Bioinformatics 211. Coarse-grained Elastic
Non equilibrium thermodynamics: foundations, scope, and extension to the meso scale. Miguel Rubi
 Non equilibrium thermodynamics: foundations, scope, and extension to the meso scale Miguel Rubi References S.R. de Groot and P. Mazur, Non equilibrium Thermodynamics, Dover, New York, 1984 J.M. Vilar and
Non equilibrium thermodynamics: foundations, scope, and extension to the meso scale Miguel Rubi References S.R. de Groot and P. Mazur, Non equilibrium Thermodynamics, Dover, New York, 1984 J.M. Vilar and
Coupling the Level-Set Method with Variational Implicit Solvent Modeling of Molecular Solvation
 Coupling the Level-Set Method with Variational Implicit Solvent Modeling of Molecular Solvation Bo Li Math Dept & CTBP, UCSD Li-Tien Cheng (Math, UCSD) Zhongming Wang (Math & Biochem, UCSD) Yang Xie (MAE,
Coupling the Level-Set Method with Variational Implicit Solvent Modeling of Molecular Solvation Bo Li Math Dept & CTBP, UCSD Li-Tien Cheng (Math, UCSD) Zhongming Wang (Math & Biochem, UCSD) Yang Xie (MAE,
simulations Extracting coarse-grained elastic behavior of capsid proteins from molecular dynamics Outline Feb. 2009
 Extracting coarse-grained elastic behavior of capsid proteins from molecular dynamics simulations Stephen D. Hicks Christopher L. Henley Cornell University Gordon Conf. on Physical Virology Feb. 2009 Outline
Extracting coarse-grained elastic behavior of capsid proteins from molecular dynamics simulations Stephen D. Hicks Christopher L. Henley Cornell University Gordon Conf. on Physical Virology Feb. 2009 Outline
Perfect mixing of immiscible macromolecules at fluid interfaces
 Perfect mixing of immiscible macromolecules at fluid interfaces Sergei S. Sheiko, 1* Jing Zhou, 1 Jamie Boyce, 1 Dorota Neugebauer, 2+ Krzysztof Matyjaszewski, 2 Constantinos Tsitsilianis, 4 Vladimir V.
Perfect mixing of immiscible macromolecules at fluid interfaces Sergei S. Sheiko, 1* Jing Zhou, 1 Jamie Boyce, 1 Dorota Neugebauer, 2+ Krzysztof Matyjaszewski, 2 Constantinos Tsitsilianis, 4 Vladimir V.
Multiscale Modeling and Simulation of Soft Matter Materials
 Multiscale Modeling and Simulation of Soft Matter Materials IMA : Development and Analysis of Multiscale Methods November 2008 Paul J. Atzberger Department of Mathematics University of California Santa
Multiscale Modeling and Simulation of Soft Matter Materials IMA : Development and Analysis of Multiscale Methods November 2008 Paul J. Atzberger Department of Mathematics University of California Santa
Proteins in solution: charge-tuning, cluster formation, liquid-liquid phase separation, and crystallization
 HERCULES Specialized Course: Non-atomic resolution scattering in biology and soft matter Grenoble, September 14-19, 2014 Proteins in solution: charge-tuning, cluster formation, liquid-liquid phase separation,
HERCULES Specialized Course: Non-atomic resolution scattering in biology and soft matter Grenoble, September 14-19, 2014 Proteins in solution: charge-tuning, cluster formation, liquid-liquid phase separation,
Supplemental Information: Combined simulation and experimental study of large deformation of red blood cells in microfluidic systems
 Supplemental Materials (Not to be Published) Supplemental Information: Combined simulation and experimental study of large deformation of red blood cells in microfluidic systems David J. Quinn, Igor V.
Supplemental Materials (Not to be Published) Supplemental Information: Combined simulation and experimental study of large deformation of red blood cells in microfluidic systems David J. Quinn, Igor V.
Erythrocyte Flickering
 Author: Facultat de Física, Universitat de Barcelona, Diagonal 645, 08028 Barcelona, Spain. Advisor: Aurora Hernández-Machado In this work we will study the fluctuations of the red blood cell membrane
Author: Facultat de Física, Universitat de Barcelona, Diagonal 645, 08028 Barcelona, Spain. Advisor: Aurora Hernández-Machado In this work we will study the fluctuations of the red blood cell membrane
Computer simulation methods (1) Dr. Vania Calandrini
 Computer simulation methods (1) Dr. Vania Calandrini Why computational methods To understand and predict the properties of complex systems (many degrees of freedom): liquids, solids, adsorption of molecules
Computer simulation methods (1) Dr. Vania Calandrini Why computational methods To understand and predict the properties of complex systems (many degrees of freedom): liquids, solids, adsorption of molecules
hydrate systems Gránásy Research Institute for Solid State Physics & Optics H-1525 Budapest, POB 49, Hungary László
 Phase field theory of crystal nucleation: Application to the hard-sphere and CO 2 Bjørn Kvamme Department of Physics, University of Bergen Allégaten 55, N-5007 N Bergen, Norway László Gránásy Research
Phase field theory of crystal nucleation: Application to the hard-sphere and CO 2 Bjørn Kvamme Department of Physics, University of Bergen Allégaten 55, N-5007 N Bergen, Norway László Gránásy Research
hydrated Nafion-117 for fuel cell application
 A molecular dynamics simulation study of oxygen within hydrated Nafion-117 for fuel cell application Jeffrey P. Fuller, Giuseppe F. Brunello, Seung Soon Jang School of Materials Science and Engineering
A molecular dynamics simulation study of oxygen within hydrated Nafion-117 for fuel cell application Jeffrey P. Fuller, Giuseppe F. Brunello, Seung Soon Jang School of Materials Science and Engineering
August 27, International ANSYS Conference Nick Reynolds, Ph.D. Director, Materials Pre-Sales, US, Accelrys
 Multiscale modeling and simulation developing solutions which link the atomistic, mesoscale, and engineering scales August 27, 2008 2008 International ANSYS Conference Nick Reynolds, Ph.D. Director, Materials
Multiscale modeling and simulation developing solutions which link the atomistic, mesoscale, and engineering scales August 27, 2008 2008 International ANSYS Conference Nick Reynolds, Ph.D. Director, Materials
Shear Properties and Wrinkling Behaviors of Finite Sized Graphene
 Shear Properties and Wrinkling Behaviors of Finite Sized Graphene Kyoungmin Min, Namjung Kim and Ravi Bhadauria May 10, 2010 Abstract In this project, we investigate the shear properties of finite sized
Shear Properties and Wrinkling Behaviors of Finite Sized Graphene Kyoungmin Min, Namjung Kim and Ravi Bhadauria May 10, 2010 Abstract In this project, we investigate the shear properties of finite sized
Soft Bodies. Good approximation for hard ones. approximation breaks when objects break, or deform. Generalization: soft (deformable) bodies
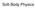 Soft-Body Physics Soft Bodies Realistic objects are not purely rigid. Good approximation for hard ones. approximation breaks when objects break, or deform. Generalization: soft (deformable) bodies Deformed
Soft-Body Physics Soft Bodies Realistic objects are not purely rigid. Good approximation for hard ones. approximation breaks when objects break, or deform. Generalization: soft (deformable) bodies Deformed
APMA 2811T. By Zhen Li. Today s topic: Lecture 2: Theoretical foundation and parameterization. Sep. 15, 2016
 Today s topic: APMA 2811T Dissipative Particle Dynamics Instructor: Professor George Karniadakis Location: 170 Hope Street, Room 118 Time: Thursday 12:00pm 2:00pm Dissipative Particle Dynamics: Foundation,
Today s topic: APMA 2811T Dissipative Particle Dynamics Instructor: Professor George Karniadakis Location: 170 Hope Street, Room 118 Time: Thursday 12:00pm 2:00pm Dissipative Particle Dynamics: Foundation,
Isotropic-Nematic Behaviour of Semiflexible Polymers in the Bulk and under Confinement
 Isotropic-Nematic Behaviour of Semiflexible Polymers in the Bulk and under Confinement S. Egorov 1, A. Milchev 2, P. Virnau 2 and K. Binder 2 1 University of Virginia 2 University of Mainz Outline Microscopic
Isotropic-Nematic Behaviour of Semiflexible Polymers in the Bulk and under Confinement S. Egorov 1, A. Milchev 2, P. Virnau 2 and K. Binder 2 1 University of Virginia 2 University of Mainz Outline Microscopic
Supporting Information for: Physics Behind the Water Transport through. Nanoporous Graphene and Boron Nitride
 Supporting Information for: Physics Behind the Water Transport through Nanoporous Graphene and Boron Nitride Ludovic Garnier, Anthony Szymczyk, Patrice Malfreyt, and Aziz Ghoufi, Institut de Physique de
Supporting Information for: Physics Behind the Water Transport through Nanoporous Graphene and Boron Nitride Ludovic Garnier, Anthony Szymczyk, Patrice Malfreyt, and Aziz Ghoufi, Institut de Physique de
Aspects of nonautonomous molecular dynamics
 Aspects of nonautonomous molecular dynamics IMA, University of Minnesota, Minneapolis January 28, 2007 Michel Cuendet Swiss Institute of Bioinformatics, Lausanne, Switzerland Introduction to the Jarzynski
Aspects of nonautonomous molecular dynamics IMA, University of Minnesota, Minneapolis January 28, 2007 Michel Cuendet Swiss Institute of Bioinformatics, Lausanne, Switzerland Introduction to the Jarzynski
Liquid-Liquid Phase Transitions and Water-Like Anomalies in Liquids
 1 Liquid-Liquid Phase Transitions and Water-Like Anomalies in Liquids Erik Lascaris Final oral examination 9 July 2014 2 Outline Anomalies in water and simple models Liquid-liquid phase transition in water
1 Liquid-Liquid Phase Transitions and Water-Like Anomalies in Liquids Erik Lascaris Final oral examination 9 July 2014 2 Outline Anomalies in water and simple models Liquid-liquid phase transition in water
Modeling of Micro-Fluidics by a Dissipative Particle Dynamics Method. Justyna Czerwinska
 Modeling of Micro-Fluidics by a Dissipative Particle Dynamics Method Justyna Czerwinska Scales and Physical Models years Time hours Engineering Design Limit Process Design minutes Continious Mechanics
Modeling of Micro-Fluidics by a Dissipative Particle Dynamics Method Justyna Czerwinska Scales and Physical Models years Time hours Engineering Design Limit Process Design minutes Continious Mechanics
File ISM02. Dynamics of Soft Matter
 File ISM02 Dynamics of Soft Matter 1 Modes of dynamics Quantum Dynamics t: fs-ps, x: 0.1 nm (seldom of importance for soft matter) Molecular Dynamics t: ps µs, x: 1 10 nm Brownian Dynamics t: ns>ps, x:
File ISM02 Dynamics of Soft Matter 1 Modes of dynamics Quantum Dynamics t: fs-ps, x: 0.1 nm (seldom of importance for soft matter) Molecular Dynamics t: ps µs, x: 1 10 nm Brownian Dynamics t: ns>ps, x:
Property Prediction with Multiscale Simulations of Silicon Containing Polymer Composites
 Silicon-Containing Polymers and Composites ACS Division of Polymer Chemistry Omni Hotel, San Diego, CA Property Prediction with Multiscale Simulations of Silicon Containing Polymer Composites Dr. Andreas
Silicon-Containing Polymers and Composites ACS Division of Polymer Chemistry Omni Hotel, San Diego, CA Property Prediction with Multiscale Simulations of Silicon Containing Polymer Composites Dr. Andreas
Martini Basics. Hands on: how to prepare a Martini
 Martini Basics Hands on: how to prepare a Martini gin and vermouth are combined at a ratio of 2:1, stirred in a mixing glass with ice cubes, then strained into a chilled cocktail glass and garnished with
Martini Basics Hands on: how to prepare a Martini gin and vermouth are combined at a ratio of 2:1, stirred in a mixing glass with ice cubes, then strained into a chilled cocktail glass and garnished with
Free Energy and Thermal Fluctuations of Neutral Lipid Bilayers
 Langmuir 001, 17, 455-463 455 Free Energy and Thermal Fluctuations of Neutral Lipid Bilayers Marian Manciu and Eli Ruckenstein* Department of Chemical Engineering, State University of New York at Buffalo,
Langmuir 001, 17, 455-463 455 Free Energy and Thermal Fluctuations of Neutral Lipid Bilayers Marian Manciu and Eli Ruckenstein* Department of Chemical Engineering, State University of New York at Buffalo,
Universal Repulsive Contribution to the. Solvent-Induced Interaction Between Sizable, Curved Hydrophobes: Supporting Information
 Universal Repulsive Contribution to the Solvent-Induced Interaction Between Sizable, Curved Hydrophobes: Supporting Information B. Shadrack Jabes, Dusan Bratko, and Alenka Luzar Department of Chemistry,
Universal Repulsive Contribution to the Solvent-Induced Interaction Between Sizable, Curved Hydrophobes: Supporting Information B. Shadrack Jabes, Dusan Bratko, and Alenka Luzar Department of Chemistry,
Supporting Information
 Supporting Information Interface-Induced Affinity Sieving in Nanoporous Graphenes for Liquid-Phase Mixtures Yanan Hou, Zhijun Xu, Xiaoning Yang * State Key Laboratory of Material-Orientated Chemical Engineering,
Supporting Information Interface-Induced Affinity Sieving in Nanoporous Graphenes for Liquid-Phase Mixtures Yanan Hou, Zhijun Xu, Xiaoning Yang * State Key Laboratory of Material-Orientated Chemical Engineering,
Sunyia Hussain 06/15/2012 ChE210D final project. Hydration Dynamics at a Hydrophobic Surface. Abstract:
 Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Randomly Triangulated Surfaces as Models for Fluid and Crystalline Membranes. G. Gompper Institut für Festkörperforschung, Forschungszentrum Jülich
 Randomly Triangulated Surfaces as Models for Fluid and Crystalline Membranes G. Gompper Institut für Festkörperforschung, Forschungszentrum Jülich Motivation: Endo- and Exocytosis Membrane transport of
Randomly Triangulated Surfaces as Models for Fluid and Crystalline Membranes G. Gompper Institut für Festkörperforschung, Forschungszentrum Jülich Motivation: Endo- and Exocytosis Membrane transport of
Dissipative Particle Dynamics
 CHAPTER 2 Dissipative Particle Dynamics Igor V. Pivkin, a,b Bruce Caswell, a and George Em Karniadakis a a Division of Applied Mathematics, Brown University, Providence, Rhode Island b Department of Materials
CHAPTER 2 Dissipative Particle Dynamics Igor V. Pivkin, a,b Bruce Caswell, a and George Em Karniadakis a a Division of Applied Mathematics, Brown University, Providence, Rhode Island b Department of Materials
Nonlinear Mechanics of Monolayer Graphene Rui Huang
 Nonlinear Mechanics of Monolayer Graphene Rui Huang Center for Mechanics of Solids, Structures and Materials Department of Aerospace Engineering and Engineering Mechanics The University of Texas at Austin
Nonlinear Mechanics of Monolayer Graphene Rui Huang Center for Mechanics of Solids, Structures and Materials Department of Aerospace Engineering and Engineering Mechanics The University of Texas at Austin
Research Statement. Shenggao Zhou. November 3, 2014
 Shenggao Zhou November 3, My research focuses on: () Scientific computing and numerical analysis (numerical PDEs, numerical optimization, computational fluid dynamics, and level-set method for interface
Shenggao Zhou November 3, My research focuses on: () Scientific computing and numerical analysis (numerical PDEs, numerical optimization, computational fluid dynamics, and level-set method for interface
Statistical thermodynamics for MD and MC simulations
 Statistical thermodynamics for MD and MC simulations knowing 2 atoms and wishing to know 10 23 of them Marcus Elstner and Tomáš Kubař 22 June 2016 Introduction Thermodynamic properties of molecular systems
Statistical thermodynamics for MD and MC simulations knowing 2 atoms and wishing to know 10 23 of them Marcus Elstner and Tomáš Kubař 22 June 2016 Introduction Thermodynamic properties of molecular systems
ACMAC s PrePrint Repository
 ACMAC s PrePrint Repository Studying multicomponent membrane tethers using numerical methods and simulations Vagelis A. Harmandaris and Markus Deserno Original Citation: Harmandaris, Vagelis A. and Deserno,
ACMAC s PrePrint Repository Studying multicomponent membrane tethers using numerical methods and simulations Vagelis A. Harmandaris and Markus Deserno Original Citation: Harmandaris, Vagelis A. and Deserno,
Mechanical properties of polymers: an overview. Suryasarathi Bose Dept. of Materials Engineering, IISc, Bangalore
 Mechanical properties of polymers: an overview Suryasarathi Bose Dept. of Materials Engineering, IISc, Bangalore UGC-NRCM Summer School on Mechanical Property Characterization- June 2012 Overview of polymer
Mechanical properties of polymers: an overview Suryasarathi Bose Dept. of Materials Engineering, IISc, Bangalore UGC-NRCM Summer School on Mechanical Property Characterization- June 2012 Overview of polymer
Journal of Physics: Conference Series PAPER. To cite this article: Jiramate Kitjanon et al 2017 J. Phys.: Conf. Ser
 Journal of Physics: Conference Series PAPER Transferability of Polymer Chain Properties between Coarse-Grained and Atomistic Models of Natural Rubber Molecule Validated by Molecular Dynamics Simulations
Journal of Physics: Conference Series PAPER Transferability of Polymer Chain Properties between Coarse-Grained and Atomistic Models of Natural Rubber Molecule Validated by Molecular Dynamics Simulations
A MOLECULAR DYNAMICS STUDY OF POLYMER/GRAPHENE NANOCOMPOSITES
 A MOLECULAR DYNAMICS STUDY OF POLYMER/GRAPHENE NANOCOMPOSITES Anastassia N. Rissanou b,c*, Vagelis Harmandaris a,b,c* a Department of Applied Mathematics, University of Crete, GR-79, Heraklion, Crete,
A MOLECULAR DYNAMICS STUDY OF POLYMER/GRAPHENE NANOCOMPOSITES Anastassia N. Rissanou b,c*, Vagelis Harmandaris a,b,c* a Department of Applied Mathematics, University of Crete, GR-79, Heraklion, Crete,
Investigation of Surfactant Efficiency Using Dissipative Particle Dynamics
 Langmuir 2003, 19, 8195-8205 8195 Investigation of Surfactant Efficiency Using Dissipative Particle Dynamics Live Rekvig,*, Marieke Kranenburg, Jocelyne Vreede, Bjørn Hafskjold, and Berend Smit Department
Langmuir 2003, 19, 8195-8205 8195 Investigation of Surfactant Efficiency Using Dissipative Particle Dynamics Live Rekvig,*, Marieke Kranenburg, Jocelyne Vreede, Bjørn Hafskjold, and Berend Smit Department
PROTEIN-PROTEIN DOCKING REFINEMENT USING RESTRAINT MOLECULAR DYNAMICS SIMULATIONS
 TASKQUARTERLYvol.20,No4,2016,pp.353 360 PROTEIN-PROTEIN DOCKING REFINEMENT USING RESTRAINT MOLECULAR DYNAMICS SIMULATIONS MARTIN ZACHARIAS Physics Department T38, Technical University of Munich James-Franck-Str.
TASKQUARTERLYvol.20,No4,2016,pp.353 360 PROTEIN-PROTEIN DOCKING REFINEMENT USING RESTRAINT MOLECULAR DYNAMICS SIMULATIONS MARTIN ZACHARIAS Physics Department T38, Technical University of Munich James-Franck-Str.
Simulation of Coarse-Grained Equilibrium Polymers
 Simulation of Coarse-Grained Equilibrium Polymers J. P. Wittmer, Institut Charles Sadron, CNRS, Strasbourg, France Collaboration with: M.E. Cates (Edinburgh), P. van der Schoot (Eindhoven) A. Milchev (Sofia),
Simulation of Coarse-Grained Equilibrium Polymers J. P. Wittmer, Institut Charles Sadron, CNRS, Strasbourg, France Collaboration with: M.E. Cates (Edinburgh), P. van der Schoot (Eindhoven) A. Milchev (Sofia),
One-particle-thick, Solvent-free, Course-grained Model for Biological and Biomimetic Fluid Membranes
 University of Pennsylvania ScholarlyCommons Departmental Papers (MSE) Department of Materials Science & Engineering 7-12-2010 One-particle-thick, Solvent-free, Course-grained Model for Biological and Biomimetic
University of Pennsylvania ScholarlyCommons Departmental Papers (MSE) Department of Materials Science & Engineering 7-12-2010 One-particle-thick, Solvent-free, Course-grained Model for Biological and Biomimetic
S(l) bl + c log l + d, with c 1.8k B. (2.71)
 2.4 DNA structure DNA molecules come in a wide range of length scales, from roughly 50,000 monomers in a λ-phage, 6 0 9 for human, to 9 0 0 nucleotides in the lily. The latter would be around thirty meters
2.4 DNA structure DNA molecules come in a wide range of length scales, from roughly 50,000 monomers in a λ-phage, 6 0 9 for human, to 9 0 0 nucleotides in the lily. The latter would be around thirty meters
Annual Report for Research Work in the fiscal year 2006
 JST Basic Research Programs C R E S T (Core Research for Evolutional Science and Technology) Annual Report for Research Work in the fiscal year 2006 Research Area : High Performance Computing for Multi-scale
JST Basic Research Programs C R E S T (Core Research for Evolutional Science and Technology) Annual Report for Research Work in the fiscal year 2006 Research Area : High Performance Computing for Multi-scale
DECRIRE LA TRANSITION VITREUSE: Simulations moléculaires et approches topologiques. Matthieu Micoulaut, LPTMC, Paris Sorbonne Universités UPMC
 DECRIRE LA TRANSITION VITREUSE: Simulations moléculaires et approches topologiques Matthieu Micoulaut, LPTMC, Paris Sorbonne Universités UPMC C. Yildrim, O. Laurent, B. Mantisi, M. Bauchy «Chasseurs d
DECRIRE LA TRANSITION VITREUSE: Simulations moléculaires et approches topologiques Matthieu Micoulaut, LPTMC, Paris Sorbonne Universités UPMC C. Yildrim, O. Laurent, B. Mantisi, M. Bauchy «Chasseurs d
Scientific Computing II
 Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
V(φ) CH 3 CH 2 CH 2 CH 3. High energy states. Low energy states. Views along the C2-C3 bond
 Example V(φ): Rotational conformations of n-butane C 3 C C C 3 Potential energy of a n-butane molecule as a function of the angle φ of bond rotation. V(φ) Potential energy/kj mol -1 0 15 10 5 eclipse gauche
Example V(φ): Rotational conformations of n-butane C 3 C C C 3 Potential energy of a n-butane molecule as a function of the angle φ of bond rotation. V(φ) Potential energy/kj mol -1 0 15 10 5 eclipse gauche
Many proteins spontaneously refold into native form in vitro with high fidelity and high speed.
 Macromolecular Processes 20. Protein Folding Composed of 50 500 amino acids linked in 1D sequence by the polypeptide backbone The amino acid physical and chemical properties of the 20 amino acids dictate
Macromolecular Processes 20. Protein Folding Composed of 50 500 amino acids linked in 1D sequence by the polypeptide backbone The amino acid physical and chemical properties of the 20 amino acids dictate
New from Ulf in Berzerkeley
 New from Ulf in Berzerkeley from crystallization to statistics of density fluctuations Ulf Rørbæk Pedersen Department of Chemistry, University of California, Berkeley, USA Roskilde, December 16th, 21 Ulf
New from Ulf in Berzerkeley from crystallization to statistics of density fluctuations Ulf Rørbæk Pedersen Department of Chemistry, University of California, Berkeley, USA Roskilde, December 16th, 21 Ulf
Multiscale Materials Modeling
 Multiscale Materials Modeling Lecture 06 Polymers These notes created by David Keffer, University of Tennessee, Knoxville, 2012. Outline Multiscale Modeling of Polymers I. Introduction II. Atomistic Simulation
Multiscale Materials Modeling Lecture 06 Polymers These notes created by David Keffer, University of Tennessee, Knoxville, 2012. Outline Multiscale Modeling of Polymers I. Introduction II. Atomistic Simulation
The effect of plasticity in crumpling of thin sheets: Supplementary Information
 The effect of plasticity in crumpling of thin sheets: Supplementary Information T. Tallinen, J. A. Åström and J. Timonen Video S1. The video shows crumpling of an elastic sheet with a width to thickness
The effect of plasticity in crumpling of thin sheets: Supplementary Information T. Tallinen, J. A. Åström and J. Timonen Video S1. The video shows crumpling of an elastic sheet with a width to thickness
High-Resolution Implementation of Self-Consistent Field Theory
 High-Resolution Implementation of Self-Consistent Field Theory Eric W. Cochran Chemical and Biological Engineering Iowa State University Carlos Garcia-Cervera Department of Mathematics University of California
High-Resolution Implementation of Self-Consistent Field Theory Eric W. Cochran Chemical and Biological Engineering Iowa State University Carlos Garcia-Cervera Department of Mathematics University of California
Degenerate ground-state lattices of membrane inclusions
 PHYSICAL REVIEW E 78, 011401 008 Degenerate ground-state lattices of membrane inclusions K. S. Kim, 1 Tom Chou,, * and Joseph Rudnick 3 1 Lawrence Livermore National Laboratory, Livermore, California 94550,
PHYSICAL REVIEW E 78, 011401 008 Degenerate ground-state lattices of membrane inclusions K. S. Kim, 1 Tom Chou,, * and Joseph Rudnick 3 1 Lawrence Livermore National Laboratory, Livermore, California 94550,
Smoothed Dissipative Particle Dynamics: theory and applications to complex fluids
 2015 DPD Workshop September 21-23, 2015, Shanghai University Smoothed Dissipative Particle Dynamics: Dynamics theory and applications to complex fluids Marco Ellero Zienkiewicz Centre for Computational
2015 DPD Workshop September 21-23, 2015, Shanghai University Smoothed Dissipative Particle Dynamics: Dynamics theory and applications to complex fluids Marco Ellero Zienkiewicz Centre for Computational
Molecular dynamics simulations of anti-aggregation effect of ibuprofen. Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov
 Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
A comparative study between dissipative particle dynamics and molecular dynamics for simple- and complex-geometry flows
 THE JOURNAL OF CHEMICAL PHYSICS 123, 104107 2005 A comparative study between dissipative particle dynamics and molecular dynamics for simple- and complex-geometry flows Eric E. Keaveny, Igor V. Pivkin,
THE JOURNAL OF CHEMICAL PHYSICS 123, 104107 2005 A comparative study between dissipative particle dynamics and molecular dynamics for simple- and complex-geometry flows Eric E. Keaveny, Igor V. Pivkin,
Thermal, electrical and mechanical simulations of field emitters using Finite Element Method
 Thermal, electrical and mechanical simulations of field emitters using Finite Element Method V. Zadin, S. Parviainen, V. Jansson, S. Vigonski, M. Veske, K. Kuppart, K. Eimre, R. Aare, A. Aabloo, F. Djurabekova
Thermal, electrical and mechanical simulations of field emitters using Finite Element Method V. Zadin, S. Parviainen, V. Jansson, S. Vigonski, M. Veske, K. Kuppart, K. Eimre, R. Aare, A. Aabloo, F. Djurabekova
Vertical Melting of a Stack of Membranes
 EPJ manuscript No. (will be inserted by the editor) Vertical Melting of a Stack of Membranes M.E.S. Borelli a, H. Kleinert b, and Adriaan M.J. Schakel c Institut für Theoretische Physik, Freie Universität
EPJ manuscript No. (will be inserted by the editor) Vertical Melting of a Stack of Membranes M.E.S. Borelli a, H. Kleinert b, and Adriaan M.J. Schakel c Institut für Theoretische Physik, Freie Universität
Curvature-Mediated Interactions Between Membrane Proteins
 74 Biophysical Journal Volume 75 November 998 74 9 Curvature-Mediated Interactions Between Membrane Proteins K. S. Kim,* John Neu, # and George Oster *Department of Physics, Graduate Group in Theoretical
74 Biophysical Journal Volume 75 November 998 74 9 Curvature-Mediated Interactions Between Membrane Proteins K. S. Kim,* John Neu, # and George Oster *Department of Physics, Graduate Group in Theoretical
Directed Assembly of Functionalized Nanoparticles with Amphiphilic Diblock Copolymers. Contents
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 Electronic Supplementary Information for Directed Assembly of Functionalized Nanoparticles
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 Electronic Supplementary Information for Directed Assembly of Functionalized Nanoparticles
7 To solve numerically the equation of motion, we use the velocity Verlet or leap frog algorithm. _ V i n = F i n m i (F.5) For time step, we approxim
 69 Appendix F Molecular Dynamics F. Introduction In this chapter, we deal with the theories and techniques used in molecular dynamics simulation. The fundamental dynamics equations of any system is the
69 Appendix F Molecular Dynamics F. Introduction In this chapter, we deal with the theories and techniques used in molecular dynamics simulation. The fundamental dynamics equations of any system is the
INTERMOLECULAR AND SURFACE FORCES
 INTERMOLECULAR AND SURFACE FORCES SECOND EDITION JACOB N. ISRAELACHVILI Department of Chemical & Nuclear Engineering and Materials Department University of California, Santa Barbara California, USA ACADEMIC
INTERMOLECULAR AND SURFACE FORCES SECOND EDITION JACOB N. ISRAELACHVILI Department of Chemical & Nuclear Engineering and Materials Department University of California, Santa Barbara California, USA ACADEMIC
Computing phase diagrams of model liquids & self-assembly of (bio)membranes
 Computing phase diagrams of model liquids & self-assembly of (bio)membranes Ulf Rørbæk Pedersen (ulf@urp.dk) Crystal structure of a simplistic model of a molecule. What is the melting temperature? A typical
Computing phase diagrams of model liquids & self-assembly of (bio)membranes Ulf Rørbæk Pedersen (ulf@urp.dk) Crystal structure of a simplistic model of a molecule. What is the melting temperature? A typical
2 Introduction to mechanics
 21 Motivation Thermodynamic bodies are being characterized by two competing opposite phenomena, energy and entropy which some researchers in thermodynamics would classify as cause and chance or determinancy
21 Motivation Thermodynamic bodies are being characterized by two competing opposite phenomena, energy and entropy which some researchers in thermodynamics would classify as cause and chance or determinancy
Introduction. Statistical physics: microscopic foundation of thermodynamics degrees of freedom 2 3 state variables!
 Introduction Thermodynamics: phenomenological description of equilibrium bulk properties of matter in terms of only a few state variables and thermodynamical laws. Statistical physics: microscopic foundation
Introduction Thermodynamics: phenomenological description of equilibrium bulk properties of matter in terms of only a few state variables and thermodynamical laws. Statistical physics: microscopic foundation
