NAMD - An Exploring Tool
|
|
|
- Blaze Copeland
- 5 years ago
- Views:
Transcription
1 NAMD - An Exploring Tool 20 years of computer science innovation and collaboration 1993: HP workstation cluster 1994: Writing NAMD in C : Commodity Linux clusters 2002: Parallel on 3000 cores,10 5 atoms 2007: Graphics processors (GPUs) 2013: Petascale supercomputers Enabling ground-breaking simulations on the world s most powerful computers Integrating experimental data Scriptable steering and analysis Free energy calculations Multiple-copy algorithms Hundreds of millions of atoms Phillips et al., J. Comp. Chem. 26, 1781 (2005) HIV (2013) Cadherin (2005) GPU Serving 70,000 users on affordable hardware Evolution of computer hardware requires continual algorithmic development......over 50 method papers Multilevel Summation Method faster calculation of electrostatic forces accelerated using GPUs D. Hardy, et al., J. Chem. Theory Comput. In press. (2015)
2 : VMD A Thinking Tool Key person John Stone (UIUC) used by 100,000 scientists worldwide video: VMD: A Thinking Tool to visualize and analyze trajectories from small peptides...
3 VMD: A Thinking Tool...to extremely large biological structures VMD: A Thinking Tool and provides state-of-the-art rendering to communicate scientific results video:
4 VMD Plugins Advanced Tools developed In-House and by External Users Analysis Modeling Visualization Collaboration APBSRun CatDCD Contact Map GofRGUI HeatMapper ILSTools IRSpecGUI MultiSeq NAMD Energy NAMD Plot NetworkView NMWiz ParseFEP PBCTools PMEpot PropKa GUI RamaPlot RMSD Tool RMSD Trajectory Tool RMSD Visualizer Tool Salt Bridges Sequence Viewer Symmetry Tool Timeline VolMap AutoIonize AutoPSF Chirality Cionize Cispeptide CGTools Dowser Force Field Toolkit Inorganic Builder MDFF Membrane Merge Structs Molefacture Mutator Nanotube Paratool Psfgen RESPTool RNAView Solvate SSRestraints Topotools Clipping Plane Tool Clone Rep DemoMaster Dipole Watcher Intersurf Navigate NavFly MultiMolAnim Color Scale Bar Remote Palette Tool ViewChangeRender ViewMaster Virtual DNA Viewer VMD Movie Maker Simulation AutoIMD IMDMenu NAMD GUI NAMD Server QMTool BioCoRE Chat BioCoRE Login BioCoRE VMD Shared Views Remote Control Data Import and Plotting Data Import Multiplot PDBTool MultiTex MolFile I/O Plugins Externally Hosted Plugins Check sidechains MultiMSMS Interactive Essential Dynamics Mead Ionize Clustering Tool itrajcomp Swap RMSD Intervor SurfVol vmdice VMD Plugins Advanced Tools developed In-House and by External Users Analysis Modeling Visualization Collaboration APBSRun CatDCD Contact Map GofRGUI HeatMapper ILSTools IRSpecGUI MultiSeq NAMD Energy NAMD Plot NetworkView NMWiz ParseFEP PBCTools PMEpot PropKa GUI RamaPlot RMSD Tool RMSD Trajectory Tool RMSD Visualizer Tool Salt Bridges Sequence Viewer Symmetry Tool Timeline VolMap AutoIonize AutoPSF Chirality Cionize Cispeptide CGTools Dowser Force Field Toolkit Inorganic Builder MDFF Membrane Merge Structs Molefacture Mutator Nanotube Paratool Psfgen RESPTool RNAView Solvate SSRestraints Topotools Clipping Plane Tool Clone Rep DemoMaster Dipole Watcher Intersurf Navigate NavFly MultiMolAnim Color Scale Bar Remote Palette Tool ViewChangeRender ViewMaster Virtual DNA Viewer VMD Movie Maker Simulation AutoIMD IMDMenu NAMD GUI NAMD Server QMTool BioCoRE Chat BioCoRE Login BioCoRE VMD Shared Views Remote Control Data Import and Plotting Data Import Multiplot PDBTool MultiTex MolFile I/O Plugins Externally Hosted Plugins Check sidechains MultiMSMS Interactive Essential Dynamics Mead Ionize Clustering Tool itrajcomp Swap RMSD Intervor SurfVol vmdice
5 VMD Plugins Advanced Tools developed In-House and by External Users Analysis Modeling Visualization Collaboration APBSRun CatDCD Contact Map GofRGUI HeatMapper ILSTools IRSpecGUI MultiSeq NAMD Energy NAMD Plot NetworkView NMWiz ParseFEP PBCTools PMEpot PropKa GUI RamaPlot RMSD Tool RMSD Trajectory Tool RMSD Visualizer Tool Salt Bridges Sequence Viewer Symmetry Tool Timeline VolMap AutoIonize AutoPSF Chirality Cionize Cispeptide CGTools Dowser Force Field Toolkit Inorganic Builder MDFF Membrane Merge Structs Molefacture Mutator Nanotube Paratool Psfgen RESPTool RNAView Solvate SSRestraints Topotools Clipping Plane Tool Clone Rep DemoMaster Dipole Watcher Intersurf Navigate NavFly MultiMolAnim Color Scale Bar Remote Palette Tool ViewChangeRender ViewMaster Virtual DNA Viewer VMD Movie Maker Simulation AutoIMD IMDMenu NAMD GUI NAMD Server QMTool BioCoRE Chat BioCoRE Login BioCoRE VMD Shared Views Remote Control Data Import and Plotting Data Import Multiplot PDBTool MultiTex MolFile I/O Plugins Externally Hosted Plugins Check sidechains MultiMSMS Interactive Essential Dynamics Mead Ionize Clustering Tool itrajcomp Swap RMSD Intervor SurfVol vmdice VMD Plugins Advanced Tools developed In-House and by External Users Analysis Modeling Visualization Collaboration APBSRun CatDCD Contact Map GofRGUI HeatMapper ILSTools IRSpecGUI MultiSeq NAMD Energy NAMD Plot NetworkView NMWiz ParseFEP PBCTools PMEpot PropKa GUI RamaPlot RMSD Tool RMSD Trajectory Tool RMSD Visualizer Tool Salt Bridges Sequence Viewer Symmetry Tool Timeline VolMap AutoIonize AutoPSF Chirality Cionize Cispeptide CGTools Dowser Force Field Toolkit Inorganic Builder MDFF Membrane Merge Structs Molefacture Mutator Nanotube Paratool Psfgen RESPTool RNAView Solvate SSRestraints Topotools Clipping Plane Tool Clone Rep DemoMaster Dipole Watcher Intersurf Navigate NavFly MultiMolAnim Color Scale Bar Remote Palette Tool ViewChangeRender ViewMaster Virtual DNA Viewer VMD Movie Maker Simulation AutoIMD IMDMenu NAMD GUI NAMD Server QMTool ProDy interface BioCoRE Chat BioCoRE Login BioCoRE VMD Shared Views Remote Control Data Import and Plotting Data Import Multiplot PDBTool MultiTex MolFile I/O Plugins Externally Hosted Plugins Check sidechains MultiMSMS Interactive Essential Dynamics Mead Ionize Clustering Tool itrajcomp Swap RMSD Intervor SurfVol vmdice
6 VMD Plugins Advanced Tools developed In-House and by External Users Analysis APBSRun CatDCD Contact Map GofRGUI HeatMapper ILSTools IRSpecGUI MultiSeq NAMD Energy NAMD Plot NetworkView NMWiz ParseFEP PBCTools PMEpot PropKa GUI RamaPlot RMSD Tool RMSD Trajectory Tool RMSD Visualizer Tool Salt Bridges Sequence Viewer Symmetry Tool Timeline VolMap Modeling AutoIonize AutoPSF Chirality Cionize Cispeptide CGTools Dowser Force Field Toolkit Inorganic Builder MDFF Membrane Merge Structs Molefacture Mutator Nanotube Paratool Psfgen RESPTool RNAView Solvate SSRestraints Topotools Visualization Clipping Plane Tool Clone Rep DemoMaster Dipole Watcher Intersurf Navigate NavFly MultiMolAnim Color Scale Bar Remote Palette Tool ViewChangeRender ViewMaster Virtual DNA Viewer VMD Movie Maker Simulation AutoIMD IMDMenu NAMD GUI NAMD Server QMTool Collaboration A tool to identify events in molecular dynamics trajectories BioCoRE Chat BioCoRE Login BioCoRE VMD Shared Views Remote Control Data Import and Plotting Data Import Multiplot PDBTool MultiTex MolFile I/O Plugins Externally Hosted Plugins Check sidechains MultiMSMS Interactive Essential Dynamics Mead Ionize Clustering Tool itrajcomp Swap RMSD Intervor SurfVol vmdice Extreme Scaling Workshop, 2013, MD)simulations)can)now)fold)proteins Schulten)et)al.)Nature(Physics)6:)751,)2010;)Pande)et)al)JACS)133:664,)2011;)Shaw)et)al) Science)334:517,)2011))))) Villin)Headpiece)) (26)a.a.) But+what+do+we+learn? AtomicElevel)detail) of)folding)dynamics Schulten)et)al.)Biophys(J(94:L75,)2008,)97:)2009 Key person Peter Freddolino (now U. Michigan)
7 Folding Dynamics of Villin Headpiece Unveiled MD)simulaQons)explored)key)folding)transiQons)not)seen)before Schulten)et)al.)Biophys(J(94:L75,)2008,)97:) (μs) Key)Folding)Intermediates Folding Simulations (1.3 ms) Unravel Growth Mechanism of Disease Causative Amyloid Wei+Han)and)Schulten)JACS,)136:12450E12460,) )Tip What)do)we)learn? Similar)affinity)to) both)tips Faster)kinetics)at) + )tip EM)Image) AmyloidEβ)Fibril E )Tip + )tip)catalyzes) structural)change)of) monomer) Reconstructed)from)1.3+millisecond+ atomic/coarseegrained)simulations)) using)the)pace)force)field.
8 force (pn) Forced Unfolding of Titin Ig Domain Titin = muscle s third protein mm µm nm AFM tip exerting force on Ig domain Forced Unfolding of Titin Ig Domain rupture force rupture force SMD simulation Extension (nm) Rief et al., Science 276 (1997) AFM experiment Schulten et al., Biophysical Journal, 75 (1998) rupture force (pn) We agreed with experiment. So what 2k F did 0 ( ) 2 1 we discover? ( ) = F ( ) + 1 Schulten et al., J. Chem. Phys. 119 (2003) 2 k F 00 + O(1/k 2 ) 1998 simulation experiment Hummer and Szabo model Log(velocity(Å/ns))
9 Our simulation revealed the unfolding process in atomic level detail Key persons Barry Izralev (UIUC) Hui Lu (now UIC) Force peak at H-bond breaking in shear mode, in parallel to external force on protein Confirmed by computational + experimental (mutation) collaboration Schulten and Fernandez et al., Nature, 402 (1999) Water molecules participate in H-bond breaking From one domain to many: multi-ig elasticity Key person Jen Hsin (now Google) Hsin and Schulten et al., Annu. Rev. Biophys., 40 (2011) von Castelmur et al. (Crystallography and simulation) PNAS 105 (2008) Quasi-equilibrium Principle All degrees of freedom not constraint by forces are in equilibrium and thermodynamics can be assumed! Extension = X j hx j exp[fx j /k B T ]i Vj / hexp[fx j /k B T ]i Vj = g(f) Crystallography Titin is a muscle and simulation protein with permit 300 domains. investigation into We look at six domains. how the system functions. Force = g 1 (Extension)
10 Ultrastable Biomass Adhesion Complex Single Molecule AFM and Steered Molecular Dynamics (SMD) combined to detail Bacterium-Biomass Adhesion Complex Key person: Rafael Bernardi(UIUC) Cohesin Dockerin Carbohydrate Binding Module Challenging environments guided nature in the development of ultrastable protein complexes Strongest Measured Adhesion Bond Adhesion becomes stronger when force is applied Collaboration w/ Hermann Gaub (Munich) Nat. Commun. 5, 5635 (2014) Simulation Experiment Cohesin Dockerin As one pulls, the adhesion contact comes closer
11 From the Strongest to the Softest Ankyrin are very common protein motifs related to mechano-gating PREDICTIONS FROM SIMULATIONS Spring constant ~ 5 mn/m 340,000 atoms 20 nanoseconds M. Sotomayor, et. al., Structure 13, 669 (2005) AFM MEASUREMENTS Spring constant ~ 2.4 mn/m G. Lee, et. al., Nature 440, 246 (2006) VERY SOFT Key person: Marcos Sotomayor (now OSU) Ankyrin repeats form an extremely soft spring 1m 0.5g Ankyrin activates mechano-gating of TRPN1 channels for hearing and touch in flies A(spring(characterized(by(the( constant(of(5mn/m(is(stretched( 1m(with(by(a(0.5g(weight. Simulations Assist in the Design of Nanopore Devices for DNA Sensing Protein Nanopore Conducts ssdna Graphene Nanopore Sensing Device A cis trans ssdna α-hemolysin 3 Klaus asks Amit Meller: Can I simulate for you? Amit Meller tests Klaus: Which end threads faster, 3 or 5? trans MD simulations combined with quantum mechanics calculations I DS V DS A. Girdhar, C. Sathe, K. Schulten, J.-P. Leburton, Nanotechnology, in press (2015) A Counting nucleobases Current
12 Simulations Assist in the Design of Nanopore Devices for DNA Sensing Protein Nanopore Conducts ssdna A cis trans α-hemolysin 3 3 Key person: Aleksei Aksimentiev (UIUC) Klaus: (after 3 weeks) Faster when 3 enters first! Amit: Yes, yes! But why? But why? Graphene Nanopore Sensing Device trans MD simulations combined with Current quantum mechanics calculations 1 I DS V DS A. Girdhar, C. Sathe, K. Schulten, J.-P. Leburton, Nanotechnology, in press (2015) A Counting nucleobases Simulations Assist in the Design of Nanopore Devices for DNA Sensing J.Mathé, A. Aksimentiev, D. R. Nelson, K. Schulten, A. Meller. Proc. Natl. Acad. Sci. U.S.A. 102, (2005) Graphene Nanopore Sensing Device cis α-hemolysin 5 MD simulations combined with quantum mechanics calculations Current 1 A 3 trans I DS 2 3 trans 3 3 ssdna ssdna bases get tilted one way in narrow pore! V DS A. Girdhar, C. Sathe, K. Schulten, J.-P. Leburton, Nanotechnology, in press (2015) A Counting nucleobases 4 5
13 All-Atom Molecular Dynamics Today HIV Capsid Number of Atoms Lysozyme Aquaporin ATP Synthase Photosynthetic Chromatophore (100 nm) (2 nm) 3 Ribosome STMV Year Discover How Membrane Protein Functions - Aquaporin Schulten, R. Stroud, et al. Science, 296, 525, (2002) Key persons: Emad Tajkhorshid (UIUC), Morten Jensen (now Shaw Res.) Nobel Prize in 2003 was awarded to Peter Agre water orientation prevents proton passage + - protein dipole + water diffuses 10 times slower than in bulk -
14 Aquaporin Conducts Water, But Not Protons 20 proton passage barrier calculated by Quantum Mechanics water orientation prevents proton passage + - protein dipole Free energy (kcal/mol) kcal/mol Coordinate along the channel (A) Å + G. A. Voth, Schulten et al., Proteins: Struct. Funct., Bioinf., 55, 223 (2004) R. Stroud, Schulten, et al. Science, 296, 525, (2002) - All-Atom Molecular Dynamics Today HIV Capsid Number of Atoms Lysozyme Aquaporin ATP Synthase Photosynthetic Chromatophore (100 nm) (2 nm) 3 Ribosome STMV Year
15 Blue+Waters+Fights+AnCbioCc+Drug+Resistance+G++ Todays+Medical+Emergency+No.+One ERY ErmB protein ribosome nucleotides The erythromycin drug disturbs bacterial protein synthesis. - wider antimicrobial spectrum than penicillin - treats eyes, respiratory tract infections. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. PNAS 111: , Collaborators: Mankin (UIC),Wilson (LMUM) BlueWaters Opens New Chapter in HIV Treatment HIVE1)virion 186)hexamers) 12)pentamers 30
16 BW One-Microsecond Simulation Includes 64 Million Atoms Key person: Juan Perilla (UIUC) Structure Discovered on Blue Waters: Nature 497 (2013) Blue Waters Reveals Structure of HIV Capsid Binding of cypa Infection Structure shows: capsid tricks human cell and infects it; counterstrategy seen Key person: Juan Perilla (UIUC) Binding of E2 Premature uncoating No infection F.)DiazEGriffero,)Viruses)(2011))
17 Blue Waters Uncovers Photosynthesis 100 million atom simulation Key person Abhi Singaroy (UIUC) Sun Light Absorption Induces Elementary Chemistry
18 Finally Sun Light Produces Biological Fuel ATP
NAMD - An Exploring Tool
 Introduction to Molecular Dynamics, NAMD and VMD, and Examples Klaus Schulten, Dept. Physics, U. Illinois at Urbana-Champaign Chicago Indianapolis University$of$Illinois$$ at$urbana7champaign Thanks guys!
Introduction to Molecular Dynamics, NAMD and VMD, and Examples Klaus Schulten, Dept. Physics, U. Illinois at Urbana-Champaign Chicago Indianapolis University$of$Illinois$$ at$urbana7champaign Thanks guys!
Mechanical Proteins. Stretching imunoglobulin and fibronectin. domains of the muscle protein titin. Adhesion Proteins of the Immune System
 Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
Mechanical Proteins. Stretching imunoglobulin and fibronectin. domains of the muscle protein titin. Adhesion Proteins of the Immune System
 Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
NIH Center for Macromolecular Modeling and Bioinformatics Developer of VMD and NAMD. Beckman Institute
 NIH Center for Macromolecular Modeling and Bioinformatics Developer of VMD and NAMD 5 faculty members (2 physics, 1 chemistry, 1 biochemistry, 1 computer science); 8 developers; 1 system admin; 15 post
NIH Center for Macromolecular Modeling and Bioinformatics Developer of VMD and NAMD 5 faculty members (2 physics, 1 chemistry, 1 biochemistry, 1 computer science); 8 developers; 1 system admin; 15 post
NIH Center for Macromolecular Modeling and Bioinformatics Developer of VMD and NAMD. Beckman Institute
 NIH Center for Macromolecular Modeling and Bioinformatics Developer of VMD and NAMD 5 faculty members (2 physics, 1 chemistry, 1 biochemistry, 1 computer science); 8 developers; 1 system admin; 15 post
NIH Center for Macromolecular Modeling and Bioinformatics Developer of VMD and NAMD 5 faculty members (2 physics, 1 chemistry, 1 biochemistry, 1 computer science); 8 developers; 1 system admin; 15 post
Introduction to Protein Structures - Molecular Graphics Tool
 Lecture 1a Introduction to Protein Structures - Molecular Graphics Tool amino acid tyrosine LH2 energetics traficking Ubiquitin enzymatic control BPTI Highlights of the VMD Molecular Graphics Program >
Lecture 1a Introduction to Protein Structures - Molecular Graphics Tool amino acid tyrosine LH2 energetics traficking Ubiquitin enzymatic control BPTI Highlights of the VMD Molecular Graphics Program >
Improved Resolution of Tertiary Structure Elasticity in Muscle Protein
 Improved Resolution of Tertiary Structure Elasticity in Muscle Protein Jen Hsin and Klaus Schulten* Department of Physics and Beckman Institute, University of Illinois at Urbana-Champaign, Urbana, Illinois
Improved Resolution of Tertiary Structure Elasticity in Muscle Protein Jen Hsin and Klaus Schulten* Department of Physics and Beckman Institute, University of Illinois at Urbana-Champaign, Urbana, Illinois
Klaus Schulten Department of Physics and Theoretical and Computational Biophysics Group University of Illinois at Urbana-Champaign
 Klaus Schulten Department of Physics and Theoretical and Computational Biophysics Group University of Illinois at Urbana-Champaign GTC, San Jose Convention Center, CA Sept. 20 23, 2010 GPU and the Computational
Klaus Schulten Department of Physics and Theoretical and Computational Biophysics Group University of Illinois at Urbana-Champaign GTC, San Jose Convention Center, CA Sept. 20 23, 2010 GPU and the Computational
Introduction to Protein Structures - Molecular Graphics Tool
 Lecture 1a Introduction to Protein Structures - Molecular Graphics Tool amino acid tyrosine LH2 energetics traficking Ubiquitin enzymatic control BPTI VMD - a Tool to Think Carl Woese Atomic, CG, Particle:
Lecture 1a Introduction to Protein Structures - Molecular Graphics Tool amino acid tyrosine LH2 energetics traficking Ubiquitin enzymatic control BPTI VMD - a Tool to Think Carl Woese Atomic, CG, Particle:
Don t forget to bring your MD tutorial. Potential Energy (hyper)surface
 Don t forget to bring your MD tutorial Lab session starts at 1pm You will have to finish an MD/SMD exercise on α-conotoxin in oxidized and reduced forms Potential Energy (hyper)surface What is Force? Energy
Don t forget to bring your MD tutorial Lab session starts at 1pm You will have to finish an MD/SMD exercise on α-conotoxin in oxidized and reduced forms Potential Energy (hyper)surface What is Force? Energy
VMD: Visual Molecular Dynamics. Our Microscope is Made of...
 VMD: Visual Molecular Dynamics Computational Microscope / Tool to Think amino acid tyrosine enzymatic control BPTI VMD tutorial traficking Ubiquitin case study http://www.ks.uiuc.edu/training/casestudies/
VMD: Visual Molecular Dynamics Computational Microscope / Tool to Think amino acid tyrosine enzymatic control BPTI VMD tutorial traficking Ubiquitin case study http://www.ks.uiuc.edu/training/casestudies/
Analyzing Ion channel Simulations
 Analyzing Ion channel Simulations (Neher and Sakmann, Scientific American 1992) Single channel current (Heurteaux et al, EMBO 2004) Computational Patch Clamp (Molecular Dynamics) Atoms move according to
Analyzing Ion channel Simulations (Neher and Sakmann, Scientific American 1992) Single channel current (Heurteaux et al, EMBO 2004) Computational Patch Clamp (Molecular Dynamics) Atoms move according to
Steered Molecular Dynamics Introduction and Examples
 Steered Molecular Dynamics Introduction and Examples Klaus Schulten Justin Gullingsrud Hui Lu avidin Sergei Izrailev biotin Rosemary Braun Ferenc Molnar Dorina Kosztin Barry Isralewitz Why Steered Molecular
Steered Molecular Dynamics Introduction and Examples Klaus Schulten Justin Gullingsrud Hui Lu avidin Sergei Izrailev biotin Rosemary Braun Ferenc Molnar Dorina Kosztin Barry Isralewitz Why Steered Molecular
The living cell is a society of molecules: molecules assembling and cooperating bring about life!
 The Computational Microscope Computational microscope views the cell 100-1,000,000 processors http://micro.magnet.fsu.edu/cells/animals/images/animalcellsfigure1.jpg The living cell is a society of molecules:
The Computational Microscope Computational microscope views the cell 100-1,000,000 processors http://micro.magnet.fsu.edu/cells/animals/images/animalcellsfigure1.jpg The living cell is a society of molecules:
Molecular Dynamics Flexible Fitting
 Molecular Dynamics Flexible Fitting Ryan McGreevy Research Programmer University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Molecular Dynamics Flexible
Molecular Dynamics Flexible Fitting Ryan McGreevy Research Programmer University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Molecular Dynamics Flexible
Enhanced Sampling and Free Energy Applications in Biomolecular Modeling
 Enhanced Sampling and Free Energy Applications in Biomolecular Modeling Emad Tajkhorshid NIH Center for Macromolecular Modeling and Bioinformatics Beckman Institute for Advanced Science and Technology
Enhanced Sampling and Free Energy Applications in Biomolecular Modeling Emad Tajkhorshid NIH Center for Macromolecular Modeling and Bioinformatics Beckman Institute for Advanced Science and Technology
The Molecular Dynamics Method
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
Our Microscope is Made of...
 The Computational Microscope Computational microscope views at atomic resolution... Rs E M RER SER N RER C GA L... how living cells maintain health and battle disease John%Stone Our Microscope is Made
The Computational Microscope Computational microscope views at atomic resolution... Rs E M RER SER N RER C GA L... how living cells maintain health and battle disease John%Stone Our Microscope is Made
Folding WT villin in silico Three folding simulations reach native state within 5-8 µs
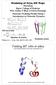 Modeling of Cryo-EM Maps Workshop Baylor College of Medicine Klaus Schulten, U. Illinois at Urbana-Champaign Molecular Modeling Flexible Fitting 1: Introduction to Molecular Dynamics MD force field Equation
Modeling of Cryo-EM Maps Workshop Baylor College of Medicine Klaus Schulten, U. Illinois at Urbana-Champaign Molecular Modeling Flexible Fitting 1: Introduction to Molecular Dynamics MD force field Equation
Hands-on Course in Computational Structural Biology and Molecular Simulation BIOP590C/MCB590C. Course Details
 Hands-on Course in Computational Structural Biology and Molecular Simulation BIOP590C/MCB590C Emad Tajkhorshid Center for Computational Biology and Biophysics Email: emad@life.uiuc.edu or tajkhors@uiuc.edu
Hands-on Course in Computational Structural Biology and Molecular Simulation BIOP590C/MCB590C Emad Tajkhorshid Center for Computational Biology and Biophysics Email: emad@life.uiuc.edu or tajkhors@uiuc.edu
Computational Structural Biology and Molecular Simulation. Introduction to VMD Molecular Visualization and Analysis
 Computational Structural Biology and Molecular Simulation Introduction to VMD Molecular Visualization and Analysis Emad Tajkhorshid Department of Biochemistry, Beckman Institute, Center for Computational
Computational Structural Biology and Molecular Simulation Introduction to VMD Molecular Visualization and Analysis Emad Tajkhorshid Department of Biochemistry, Beckman Institute, Center for Computational
The Computational Microscope
 The Computational Microscope Computational microscope views at atomic resolution... Rs SER RER C E M RER N GA L... how living cells maintain health and battle disease John Stone Our Microscope is Made
The Computational Microscope Computational microscope views at atomic resolution... Rs SER RER C E M RER N GA L... how living cells maintain health and battle disease John Stone Our Microscope is Made
Molecular dynamics simulation. CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror
 Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Structure of the HIV-1 Capsid Assembly by a hybrid approach
 Structure of the HIV-1 Capsid Assembly by a hybrid approach Juan R. Perilla juan@ks.uiuc.edu Theoretical and Computational Biophysics Group University of Illinois at Urbana-Champaign Virion HIV infective
Structure of the HIV-1 Capsid Assembly by a hybrid approach Juan R. Perilla juan@ks.uiuc.edu Theoretical and Computational Biophysics Group University of Illinois at Urbana-Champaign Virion HIV infective
GPU-Accelerated Analysis and Visualization of Large Structures Solved by Molecular Dynamics Flexible Fitting
 GPU-Accelerated Analysis and Visualization of Large Structures Solved by Molecular Dynamics Flexible Fitting John E. Stone, Ryan McGreevy, Barry Isralewitz, and Klaus Schulten Theoretical and Computational
GPU-Accelerated Analysis and Visualization of Large Structures Solved by Molecular Dynamics Flexible Fitting John E. Stone, Ryan McGreevy, Barry Isralewitz, and Klaus Schulten Theoretical and Computational
Overview & Applications. T. Lezon Hands-on Workshop in Computational Biophysics Pittsburgh Supercomputing Center 04 June, 2015
 Overview & Applications T. Lezon Hands-on Workshop in Computational Biophysics Pittsburgh Supercomputing Center 4 June, 215 Simulations still take time Bakan et al. Bioinformatics 211. Coarse-grained Elastic
Overview & Applications T. Lezon Hands-on Workshop in Computational Biophysics Pittsburgh Supercomputing Center 4 June, 215 Simulations still take time Bakan et al. Bioinformatics 211. Coarse-grained Elastic
What can be learnt from MD
 - 4.0 H-bond energy (kcal/mol) 0 What can be learnt from MD Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis ATPase, a molecular motor
- 4.0 H-bond energy (kcal/mol) 0 What can be learnt from MD Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis ATPase, a molecular motor
Atomistic Modeling of Small-Angle Scattering Data Using SASSIE-web
 Course Introduction Atomistic Modeling of Small-Angle Scattering Data Using SASSIE-web September 21-23, 2016 Advanced Photon Source Argonne National Laboratory, Argonne, IL ccpsas.org Scatters & Simulators
Course Introduction Atomistic Modeling of Small-Angle Scattering Data Using SASSIE-web September 21-23, 2016 Advanced Photon Source Argonne National Laboratory, Argonne, IL ccpsas.org Scatters & Simulators
Principles and Applications of Molecular Dynamics Simulations with NAMD
 Principles and Applications of Molecular Dynamics Simulations with NAMD Nov. 14, 2016 Computational Microscope NCSA supercomputer JC Gumbart Assistant Professor of Physics Georgia Institute of Technology
Principles and Applications of Molecular Dynamics Simulations with NAMD Nov. 14, 2016 Computational Microscope NCSA supercomputer JC Gumbart Assistant Professor of Physics Georgia Institute of Technology
TR&D3: Brownian Mover DBP4: Biosensors
 TR&D3: Brownian Mover DBP4: Biosensors Aleksei Aksimentiev Lead co-investigator of TR&D3-BD and DBP4 Associate Professor of Physics, UIUC http://bionano.physics.illinois.edu/ TR&D3 Brownian Mover Overview
TR&D3: Brownian Mover DBP4: Biosensors Aleksei Aksimentiev Lead co-investigator of TR&D3-BD and DBP4 Associate Professor of Physics, UIUC http://bionano.physics.illinois.edu/ TR&D3 Brownian Mover Overview
Anatoly B. Kolomeisky Department of Chemistry Center for Theoretical Biological Physics How to Understand Molecular Transport through Channels: The
 Anatoly B. Kolomeisy Department of Chemistry Center for Theoretical Biological Physics How to Understand Molecular Transport through Channels: The Role of Interactions Transport Through Channels Oil pumping
Anatoly B. Kolomeisy Department of Chemistry Center for Theoretical Biological Physics How to Understand Molecular Transport through Channels: The Role of Interactions Transport Through Channels Oil pumping
Retinal Proteins (Rhodopsins) Vision, Bioenergetics, Phototaxis. Bacteriorhodopsin s Photocycle. Bacteriorhodopsin s Photocycle
 Molecular chanisms of Photoactivation and Spectral Tuning in Retinal Proteins Emad Tajkhorshid Theoretical and Computational Biophysics Group Beckman Institute University of Illinois at Urbana-Champaign
Molecular chanisms of Photoactivation and Spectral Tuning in Retinal Proteins Emad Tajkhorshid Theoretical and Computational Biophysics Group Beckman Institute University of Illinois at Urbana-Champaign
BMB November 17, Single Molecule Biophysics (I)
 BMB 178 2017 November 17, 2017 14. Single Molecule Biophysics (I) Goals 1. Understand the information SM experiments can provide 2. Be acquainted with different SM approaches 3. Learn to interpret SM results
BMB 178 2017 November 17, 2017 14. Single Molecule Biophysics (I) Goals 1. Understand the information SM experiments can provide 2. Be acquainted with different SM approaches 3. Learn to interpret SM results
ATPase Synthase - A Molecular Double Motor
 ATPase Synthase - A Molecular Double Motor RC bc 1 bc 1 ATPase Photosynthesic Unit of Purple Bacteria Module that converts sun light into chemical energy (ATP) Light in hν H + Q/QH 2 /Q bc 1 ADP ATP out
ATPase Synthase - A Molecular Double Motor RC bc 1 bc 1 ATPase Photosynthesic Unit of Purple Bacteria Module that converts sun light into chemical energy (ATP) Light in hν H + Q/QH 2 /Q bc 1 ADP ATP out
Protein Dynamics. The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron.
 Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Structure Investigation of Fam20C, a Golgi Casein Kinase
 Structure Investigation of Fam20C, a Golgi Casein Kinase Sharon Grubner National Taiwan University, Dr. Jung-Hsin Lin University of California San Diego, Dr. Rommie Amaro Abstract This research project
Structure Investigation of Fam20C, a Golgi Casein Kinase Sharon Grubner National Taiwan University, Dr. Jung-Hsin Lin University of California San Diego, Dr. Rommie Amaro Abstract This research project
The Molecular Dynamics Simulation Process
 The Molecular Dynamics Simulation Process For textbooks see: M.P. Allen and D.J. Tildesley. Computer Simulation of Liquids.Oxford University Press, New York, 1987. D. Frenkel and B. Smit. Understanding
The Molecular Dynamics Simulation Process For textbooks see: M.P. Allen and D.J. Tildesley. Computer Simulation of Liquids.Oxford University Press, New York, 1987. D. Frenkel and B. Smit. Understanding
The Force Field Toolkit (fftk)
 Parameterizing Small Molecules Using: The Force Field Toolkit (fftk) Christopher G. Mayne, Emad Tajkhorshid Beckman Institute for Advanced Science and Technology University of Illinois, Urbana-Champaign
Parameterizing Small Molecules Using: The Force Field Toolkit (fftk) Christopher G. Mayne, Emad Tajkhorshid Beckman Institute for Advanced Science and Technology University of Illinois, Urbana-Champaign
Molecular Dynamics Flexible Fitting
 Molecular Dynamics Flexible Fitting Ryan McGreevy Research Programmer University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Molecular Dynamics Flexible
Molecular Dynamics Flexible Fitting Ryan McGreevy Research Programmer University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Molecular Dynamics Flexible
Molecular Dynamics Simulations of Ions Diffusion in Carbon Nanotubes Embedded in Cell Membrane
 Copyright 2014 Tech Science Press CMES, vol.98, no.3, pp.247-259, 2014 Molecular Dynamics Simulations of Ions Diffusion in Carbon Nanotubes Embedded in Cell Membrane Qing Song Tu 1, Michelle Lee 2, Samuel
Copyright 2014 Tech Science Press CMES, vol.98, no.3, pp.247-259, 2014 Molecular Dynamics Simulations of Ions Diffusion in Carbon Nanotubes Embedded in Cell Membrane Qing Song Tu 1, Michelle Lee 2, Samuel
Our Microscope is Made of...
 The Computational Microscope Computational microscope views at atomic resolution... Rs E M RER SER N RER C GA L... how living cells maintain health and battle disease John%Stone Our Microscope is Made
The Computational Microscope Computational microscope views at atomic resolution... Rs E M RER SER N RER C GA L... how living cells maintain health and battle disease John%Stone Our Microscope is Made
Signaling Proteins: Mechanical Force Generation by G-proteins G
 Signaling Proteins: Mechanical Force Generation by G-proteins G Ioan Kosztin Beckman Institute University of Illinois at Urbana-Champaign Collaborators: Robijn Bruinsma (Leiden & UCLA) Paul O Lague (UCLA)
Signaling Proteins: Mechanical Force Generation by G-proteins G Ioan Kosztin Beckman Institute University of Illinois at Urbana-Champaign Collaborators: Robijn Bruinsma (Leiden & UCLA) Paul O Lague (UCLA)
APBS electrostatics in VMD - Software. APBS! >!Examples! >!Visualization! >! Contents
 Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Announcements & Lecture Points
 Announcements & Lecture Points Homework 3 (Klaus Schulten s Lecture): Due Wednesday. Quiz returned, next homework on Wednesday. Today s Lecture: Protein Folding, Misfolding, Aggregation. Experimental Approach
Announcements & Lecture Points Homework 3 (Klaus Schulten s Lecture): Due Wednesday. Quiz returned, next homework on Wednesday. Today s Lecture: Protein Folding, Misfolding, Aggregation. Experimental Approach
Course Introduction. SASSIE CCP-SAS Workshop. January 23-25, ISIS Neutron and Muon Source Rutherford Appleton Laboratory UK
 Course Introduction SASSIE CCP-SAS Workshop January 23-25, 2017 ISIS Neutron and Muon Source Rutherford Appleton Laboratory UK ccpsas.org Scatters & Simulators Develop a community of users and developers
Course Introduction SASSIE CCP-SAS Workshop January 23-25, 2017 ISIS Neutron and Muon Source Rutherford Appleton Laboratory UK ccpsas.org Scatters & Simulators Develop a community of users and developers
Alchemical free energy calculations in OpenMM
 Alchemical free energy calculations in OpenMM Lee-Ping Wang Stanford Department of Chemistry OpenMM Workshop, Stanford University September 7, 2012 Special thanks to: John Chodera, Morgan Lawrenz Outline
Alchemical free energy calculations in OpenMM Lee-Ping Wang Stanford Department of Chemistry OpenMM Workshop, Stanford University September 7, 2012 Special thanks to: John Chodera, Morgan Lawrenz Outline
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability
 Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Steered Molecular Dynamics Studies of Titin Domains
 Steered Molecular Dynamics Studies of Titin Domains Mu Gao Department of Physics and Beckman Institute University of Illinois at Urbana-Champaign Overview Biology Background AFM experiments Modeling and
Steered Molecular Dynamics Studies of Titin Domains Mu Gao Department of Physics and Beckman Institute University of Illinois at Urbana-Champaign Overview Biology Background AFM experiments Modeling and
What can be learnt from MD. Titin Micelle Light harvesting complex Reaction Center Restriction enzyme Ubiquitin Biotin - Avidin
 What can be learnt from MD Titin Micelle Light harvesting complex Reaction Center Restriction enzyme Ubiquitin Biotin - Avidin Putting Your Hands on a Protein with Interactive Molecular Dynamics - Interactive
What can be learnt from MD Titin Micelle Light harvesting complex Reaction Center Restriction enzyme Ubiquitin Biotin - Avidin Putting Your Hands on a Protein with Interactive Molecular Dynamics - Interactive
Introduction to MD simulations of DNA systems. Aleksei Aksimentiev
 Introduction to MD simulations of DNA systems Aleksei Aksimentiev Biological Modeling at Different Scales spanning orders of magnitude in space and time The computational microscope Massive parallel computer
Introduction to MD simulations of DNA systems Aleksei Aksimentiev Biological Modeling at Different Scales spanning orders of magnitude in space and time The computational microscope Massive parallel computer
Steered Molecular Dynamics Introduction and Examples
 Steered Molecular Dynamics Introduction and Examples Klaus Schulten Justin Gullingsrud Hui Lu avidin Sergei Izrailev biotin Rosemary Braun Ferenc Molnar Dorina Kosztin Barry Isralewitz Why Steered Molecular
Steered Molecular Dynamics Introduction and Examples Klaus Schulten Justin Gullingsrud Hui Lu avidin Sergei Izrailev biotin Rosemary Braun Ferenc Molnar Dorina Kosztin Barry Isralewitz Why Steered Molecular
Supporting Information. Molecular Dynamics Simulation of. DNA Capture and Transport in. Heated Nanopores
 Supporting Information Molecular Dynamics Simulation of DNA Capture and Transport in Heated Nanopores Maxim Belkin and Aleksei Aksimentiev Department of Physics, University of Illinois at Urbana-Champaign
Supporting Information Molecular Dynamics Simulation of DNA Capture and Transport in Heated Nanopores Maxim Belkin and Aleksei Aksimentiev Department of Physics, University of Illinois at Urbana-Champaign
Effects of interaction between nanopore and polymer on translocation time
 Effects of interaction between nanopore and polymer on translocation time Mohammadreza Niknam Hamidabad and Rouhollah Haji Abdolvahab Physics Department, Iran University of Science and Technology (IUST),
Effects of interaction between nanopore and polymer on translocation time Mohammadreza Niknam Hamidabad and Rouhollah Haji Abdolvahab Physics Department, Iran University of Science and Technology (IUST),
Multimedia : Fibronectin and Titin unfolding simulation movies.
 I LECTURE 21: SINGLE CHAIN ELASTICITY OF BIOMACROMOLECULES: THE GIANT PROTEIN TITIN AND DNA Outline : REVIEW LECTURE #2 : EXTENSIBLE FJC AND WLC... 2 STRUCTURE OF MUSCLE AND TITIN... 3 SINGLE MOLECULE
I LECTURE 21: SINGLE CHAIN ELASTICITY OF BIOMACROMOLECULES: THE GIANT PROTEIN TITIN AND DNA Outline : REVIEW LECTURE #2 : EXTENSIBLE FJC AND WLC... 2 STRUCTURE OF MUSCLE AND TITIN... 3 SINGLE MOLECULE
Supporting Material for. Microscopic origin of gating current fluctuations in a potassium channel voltage sensor
 Supporting Material for Microscopic origin of gating current fluctuations in a potassium channel voltage sensor J. Alfredo Freites, * Eric V. Schow, * Stephen H. White, and Douglas J. Tobias * * Department
Supporting Material for Microscopic origin of gating current fluctuations in a potassium channel voltage sensor J. Alfredo Freites, * Eric V. Schow, * Stephen H. White, and Douglas J. Tobias * * Department
BUDE. A General Purpose Molecular Docking Program Using OpenCL. Richard B Sessions
 BUDE A General Purpose Molecular Docking Program Using OpenCL Richard B Sessions 1 The molecular docking problem receptor ligand Proteins typically O(1000) atoms Ligands typically O(100) atoms predicted
BUDE A General Purpose Molecular Docking Program Using OpenCL Richard B Sessions 1 The molecular docking problem receptor ligand Proteins typically O(1000) atoms Ligands typically O(100) atoms predicted
Adaptive Heterogeneous Computing with OpenCL: Harnessing hundreds of GPUs and CPUs
 Adaptive Heterogeneous Computing with OpenCL: Harnessing hundreds of GPUs and CPUs Simon McIntosh-Smith simonm@cs.bris.ac.uk Head of Microelectronics Research University of Bristol, UK 1 ! Collaborators
Adaptive Heterogeneous Computing with OpenCL: Harnessing hundreds of GPUs and CPUs Simon McIntosh-Smith simonm@cs.bris.ac.uk Head of Microelectronics Research University of Bristol, UK 1 ! Collaborators
NSF Summer School UIUC 2003 Molecular Machines of the Living Cell : Photosynthetic. Unit from Light Absorption to ATP Synthesis Melih Sener
 Beckman Institute NSF Summer School UIUC 2003 Molecular Machines of the Living Cell : Photosynthetic Unit from Light Absorption to ATP Synthesis Melih Sener Thorsten Ritz Ioan Kosztin Ana Damjanovic Theoretical
Beckman Institute NSF Summer School UIUC 2003 Molecular Machines of the Living Cell : Photosynthetic Unit from Light Absorption to ATP Synthesis Melih Sener Thorsten Ritz Ioan Kosztin Ana Damjanovic Theoretical
Molecular dynamics simulations of a single stranded (ss) DNA
 Molecular dynamics simulations of a single stranded (ss) DNA Subhasish Chatterjee 1, Bonnie Gersten 1, Siddarth Thakur 2, Alexander Burin 2 1 Department of Chemistry, Queens College and the Graduate Center
Molecular dynamics simulations of a single stranded (ss) DNA Subhasish Chatterjee 1, Bonnie Gersten 1, Siddarth Thakur 2, Alexander Burin 2 1 Department of Chemistry, Queens College and the Graduate Center
Interdisciplinary Nanoscience Center University of Aarhus, Denmark. Design and Imaging. Assistant Professor.
 Interdisciplinary Nanoscience Center University of Aarhus, Denmark Design and Imaging DNA Nanostructures Assistant Professor Wael Mamdouh wael@inano.dk Molecular Self-assembly Synthesis, SPM microscopy,
Interdisciplinary Nanoscience Center University of Aarhus, Denmark Design and Imaging DNA Nanostructures Assistant Professor Wael Mamdouh wael@inano.dk Molecular Self-assembly Synthesis, SPM microscopy,
20 IEEE NANOTECHNOLOGY MAGAZINE MARCH /09/$ IEEE
 REPRINT: Modeling transport through synthetic nanopores. Aleksei Aksimentiev, Robert K. Brunner, Eduardo Cruz-Chu, Jeffrey Comer, and Klaus Schulten. IEEE Nanotechnology, 3:20-28, 2009. Copyright of IEEE
REPRINT: Modeling transport through synthetic nanopores. Aleksei Aksimentiev, Robert K. Brunner, Eduardo Cruz-Chu, Jeffrey Comer, and Klaus Schulten. IEEE Nanotechnology, 3:20-28, 2009. Copyright of IEEE
The Molecular Dynamics Method
 H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
Announcements. Homework 3 (Klaus Schulten s Lecture): Due Wednesday at noon. Next homework assigned. Due Wednesday March 1.
 Announcements Homework 3 (Klaus Schulten s Lecture): Due Wednesday at noon. Next homework assigned. Due Wednesday March 1. No lecture next Monday, Feb. 27 th! (Homework is a bit longer.) Marco will have
Announcements Homework 3 (Klaus Schulten s Lecture): Due Wednesday at noon. Next homework assigned. Due Wednesday March 1. No lecture next Monday, Feb. 27 th! (Homework is a bit longer.) Marco will have
How is molecular dynamics being used in life sciences? Davide Branduardi
 How is molecular dynamics being used in life sciences? Davide Branduardi davide.branduardi@schrodinger.com Exploring molecular processes with MD Drug discovery and design Protein-protein interactions Protein-DNA
How is molecular dynamics being used in life sciences? Davide Branduardi davide.branduardi@schrodinger.com Exploring molecular processes with MD Drug discovery and design Protein-protein interactions Protein-DNA
Free energy calculation from steered molecular dynamics simulations using Jarzynski s equality
 JOURNAL OF CHEMICAL PHYSICS VOLUME 119, NUMBER 6 8 AUGUST 2003 Free energy calculation from steered molecular dynamics simulations using Jarzynski s equality Sanghyun Park and Fatemeh Khalili-Araghi Beckman
JOURNAL OF CHEMICAL PHYSICS VOLUME 119, NUMBER 6 8 AUGUST 2003 Free energy calculation from steered molecular dynamics simulations using Jarzynski s equality Sanghyun Park and Fatemeh Khalili-Araghi Beckman
Evolution of Translation: Dynamics of Recognition in RNA:Protein Complexes
 Evolution of Translation: Dynamics of Recognition in RNA:Protein Complexes Zaida (Zan) Luthey-Schulten Dept. Chemistry, Beckman Institute, Biophysics, Institute of Genomics Biology, & Physics NIH Workshop
Evolution of Translation: Dynamics of Recognition in RNA:Protein Complexes Zaida (Zan) Luthey-Schulten Dept. Chemistry, Beckman Institute, Biophysics, Institute of Genomics Biology, & Physics NIH Workshop
Evaluation of the binding energy of viral capsid proteins using a Virtual AFM
 Departament de Física de la Matèria Condensada, Facultat de Física, Universitat de Barcelona, Diagonal 645, 08028 Barcelona, Spain. Advisor: Prof. David Reguera López Abstract: Viruses are biological agents
Departament de Física de la Matèria Condensada, Facultat de Física, Universitat de Barcelona, Diagonal 645, 08028 Barcelona, Spain. Advisor: Prof. David Reguera López Abstract: Viruses are biological agents
Our Microscope is Made of...
 The Computational Microscope Computational microscope views at atomic resolution... Rs E M RER SER N RER C GA L... how living cells maintain health and battle disease John%Stone Our Microscope is Made
The Computational Microscope Computational microscope views at atomic resolution... Rs E M RER SER N RER C GA L... how living cells maintain health and battle disease John%Stone Our Microscope is Made
Modeling of Cryo-EM Maps Workshop Baylor College of Medicine Klaus Schulten, U. Illinois at Urbana-Champaign
 Modeling of Cryo-EM Maps Workshop Baylor College of Medicine Klaus Schulten, U. Illinois at Urbana-Champaign Molecular Modeling Flexible Fitting 3: Application to Ribosome Proofreading of codon recognition
Modeling of Cryo-EM Maps Workshop Baylor College of Medicine Klaus Schulten, U. Illinois at Urbana-Champaign Molecular Modeling Flexible Fitting 3: Application to Ribosome Proofreading of codon recognition
Computational Modeling of Protein Kinase A and Comparison with Nuclear Magnetic Resonance Data
 Computational Modeling of Protein Kinase A and Comparison with Nuclear Magnetic Resonance Data ABSTRACT Keyword Lei Shi 1 Advisor: Gianluigi Veglia 1,2 Department of Chemistry 1, & Biochemistry, Molecular
Computational Modeling of Protein Kinase A and Comparison with Nuclear Magnetic Resonance Data ABSTRACT Keyword Lei Shi 1 Advisor: Gianluigi Veglia 1,2 Department of Chemistry 1, & Biochemistry, Molecular
Lots of thanks for many reasons!
 Lots of thanks for many reasons! Structure and function of glutamate transporters from free energy simulations Turgut Baştuğ TOBB ETU Transporter structures Transporters have larger structures, which
Lots of thanks for many reasons! Structure and function of glutamate transporters from free energy simulations Turgut Baştuğ TOBB ETU Transporter structures Transporters have larger structures, which
Investigation of physiochemical interactions in
 Investigation of physiochemical interactions in Bulk and interfacial water Aqueous salt solutions (new endeavor) Polypeptides exhibiting a helix-coil transition Aqueous globular proteins Protein-solvent
Investigation of physiochemical interactions in Bulk and interfacial water Aqueous salt solutions (new endeavor) Polypeptides exhibiting a helix-coil transition Aqueous globular proteins Protein-solvent
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Supporting Information. for An atomistic modeling of F-actin mechanical responses and determination of. mechanical properties
 Supporting Information for An atomistic modeling of F-actin mechanical responses and determination of mechanical properties Authors: Si Li 1, Jin Zhang 2, Chengyuan Wang 1, Perumal Nithiarasu 1 1. Zienkiewicz
Supporting Information for An atomistic modeling of F-actin mechanical responses and determination of mechanical properties Authors: Si Li 1, Jin Zhang 2, Chengyuan Wang 1, Perumal Nithiarasu 1 1. Zienkiewicz
SECOND PUBLIC EXAMINATION. Honour School of Physics Part C: 4 Year Course. Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS
 2757 SECOND PUBLIC EXAMINATION Honour School of Physics Part C: 4 Year Course Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS TRINITY TERM 2013 Monday, 17 June, 2.30 pm 5.45 pm 15
2757 SECOND PUBLIC EXAMINATION Honour School of Physics Part C: 4 Year Course Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS TRINITY TERM 2013 Monday, 17 June, 2.30 pm 5.45 pm 15
Nonequilibrium thermodynamics at the microscale
 Nonequilibrium thermodynamics at the microscale Christopher Jarzynski Department of Chemistry and Biochemistry and Institute for Physical Science and Technology ~1 m ~20 nm Work and free energy: a macroscopic
Nonequilibrium thermodynamics at the microscale Christopher Jarzynski Department of Chemistry and Biochemistry and Institute for Physical Science and Technology ~1 m ~20 nm Work and free energy: a macroscopic
GPU Acceleration of Cutoff Pair Potentials for Molecular Modeling Applications
 GPU Acceleration of Cutoff Pair Potentials for Molecular Modeling Applications Christopher Rodrigues, David J. Hardy, John E. Stone, Klaus Schulten, Wen-Mei W. Hwu University of Illinois at Urbana-Champaign
GPU Acceleration of Cutoff Pair Potentials for Molecular Modeling Applications Christopher Rodrigues, David J. Hardy, John E. Stone, Klaus Schulten, Wen-Mei W. Hwu University of Illinois at Urbana-Champaign
Short Announcements. 1 st Quiz today: 15 minutes. Homework 3: Due next Wednesday.
 Short Announcements 1 st Quiz today: 15 minutes Homework 3: Due next Wednesday. Next Lecture, on Visualizing Molecular Dynamics (VMD) by Klaus Schulten Today s Lecture: Protein Folding, Misfolding, Aggregation
Short Announcements 1 st Quiz today: 15 minutes Homework 3: Due next Wednesday. Next Lecture, on Visualizing Molecular Dynamics (VMD) by Klaus Schulten Today s Lecture: Protein Folding, Misfolding, Aggregation
Cells. Structural and functional units of living organisms
 Cells Structural and functional units of living organisms Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells Proks Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells
Cells Structural and functional units of living organisms Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells Proks Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells
Exploring the Changes in the Structure of α-helical Peptides Adsorbed onto Carbon and Boron Nitride based Nanomaterials
 Exploring the Changes in the Structure of α-helical Peptides Adsorbed onto Carbon and Boron Nitride based Nanomaterials Dr. V. Subramanian Chemical Laboratory, IPC Division CSIR-Central Leather Research
Exploring the Changes in the Structure of α-helical Peptides Adsorbed onto Carbon and Boron Nitride based Nanomaterials Dr. V. Subramanian Chemical Laboratory, IPC Division CSIR-Central Leather Research
Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate
 Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate Shidi Zhao a, Laura Restrepo-Pérez b, Misha Soskine c, Giovanni Maglia c, Chirlmin Joo b, Cees Dekker b and Aleksei Aksimentiev
Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate Shidi Zhao a, Laura Restrepo-Pérez b, Misha Soskine c, Giovanni Maglia c, Chirlmin Joo b, Cees Dekker b and Aleksei Aksimentiev
The Art of Building Small from molecular switches to motors
 The Art of Building Small from molecular switches to motors Stockholm december 8 th, 2016 Ben Feringa Bioinspired Motion the early days fascinated by but no copy or mimic! 17 th of December 1903 at Kitty
The Art of Building Small from molecular switches to motors Stockholm december 8 th, 2016 Ben Feringa Bioinspired Motion the early days fascinated by but no copy or mimic! 17 th of December 1903 at Kitty
The Molecular Dynamics Method
 H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
Scanning Force Microscopy. i.e. Atomic Force Microscopy. Josef A. Käs Institute for Soft Matter Physics Physics Department. Atomic Force Microscopy
 Scanning Force Microscopy i.e. Atomic Force Microscopy Josef A. Käs Institute for Soft Matter Physics Physics Department Atomic Force Microscopy Single Molecule Force Spectroscopy Zanjan 4 Hermann E. Gaub
Scanning Force Microscopy i.e. Atomic Force Microscopy Josef A. Käs Institute for Soft Matter Physics Physics Department Atomic Force Microscopy Single Molecule Force Spectroscopy Zanjan 4 Hermann E. Gaub
NAME: PERIOD: DATE: A View of the Cell. Use Chapter 8 of your book to complete the chart of eukaryotic cell components.
 NAME: PERIOD: DATE: A View of the Cell Use Chapter 8 of your book to complete the chart of eukaryotic cell components. Cell Part Cell Wall Centriole Chloroplast Cilia Cytoplasm Cytoskeleton Endoplasmic
NAME: PERIOD: DATE: A View of the Cell Use Chapter 8 of your book to complete the chart of eukaryotic cell components. Cell Part Cell Wall Centriole Chloroplast Cilia Cytoplasm Cytoskeleton Endoplasmic
ATP hydrolysis 1 1 1
 ATP hydrolysis 1 1 1 ATP hydrolysis 2 2 2 The binding zipper 1 3 3 ATP hydrolysis/synthesis is coupled to a torque Yasuda, R., et al (1998). Cell 93:1117 1124. Abrahams, et al (1994). Nature 370:621-628.
ATP hydrolysis 1 1 1 ATP hydrolysis 2 2 2 The binding zipper 1 3 3 ATP hydrolysis/synthesis is coupled to a torque Yasuda, R., et al (1998). Cell 93:1117 1124. Abrahams, et al (1994). Nature 370:621-628.
Structural investigation of single biomolecules
 Structural investigation of single biomolecules NMR spectroscopy and X-ray crystallography are currently the most common techniques capable of determining the structures of biological macromolecules like
Structural investigation of single biomolecules NMR spectroscopy and X-ray crystallography are currently the most common techniques capable of determining the structures of biological macromolecules like
Supporting information for. DNA Origami-Graphene Hybrid Nanopore for DNA Detection
 Supporting information for DNA Origami-Graphene Hybrid Nanopore for DNA Detection Amir Barati Farimani, Payam Dibaeinia, Narayana R. Aluru Department of Mechanical Science and Engineering Beckman Institute
Supporting information for DNA Origami-Graphene Hybrid Nanopore for DNA Detection Amir Barati Farimani, Payam Dibaeinia, Narayana R. Aluru Department of Mechanical Science and Engineering Beckman Institute
SAM Teachers Guide Chemical Bonds
 SAM Teachers Guide Chemical Bonds Overview Students discover that the type of bond formed ionic, non polar covalent, or polar covalent depends on the electronegativity of the two atoms that are bonded
SAM Teachers Guide Chemical Bonds Overview Students discover that the type of bond formed ionic, non polar covalent, or polar covalent depends on the electronegativity of the two atoms that are bonded
Molecular dynamics simulation of Aquaporin-1. 4 nm
 Molecular dynamics simulation of Aquaporin-1 4 nm Molecular Dynamics Simulations Schrödinger equation i~@ t (r, R) =H (r, R) Born-Oppenheimer approximation H e e(r; R) =E e (R) e(r; R) Nucleic motion described
Molecular dynamics simulation of Aquaporin-1 4 nm Molecular Dynamics Simulations Schrödinger equation i~@ t (r, R) =H (r, R) Born-Oppenheimer approximation H e e(r; R) =E e (R) e(r; R) Nucleic motion described
5th CCPN Matt Crump. Thermodynamic quantities derived from protein dynamics
 5th CCPN 2005 -Matt Crump Thermodynamic quantities derived from protein dynamics Relaxation in Liquids (briefly!) The fluctuations of each bond vector can be described in terms of an angular correlation
5th CCPN 2005 -Matt Crump Thermodynamic quantities derived from protein dynamics Relaxation in Liquids (briefly!) The fluctuations of each bond vector can be described in terms of an angular correlation
Part III - Bioinformatics Study of Aminoacyl trna Synthetases. VMD Multiseq Tutorial Web tools. Perth, Australia 2004 Computational Biology Workshop
 Part III - Bioinformatics Study of Aminoacyl trna Synthetases VMD Multiseq Tutorial Web tools Perth, Australia 2004 Computational Biology Workshop Multiple Sequence Alignments The aminoacyl-trna synthetases,
Part III - Bioinformatics Study of Aminoacyl trna Synthetases VMD Multiseq Tutorial Web tools Perth, Australia 2004 Computational Biology Workshop Multiple Sequence Alignments The aminoacyl-trna synthetases,
Controllable Water Channel Gating of Nanometer Dimensions
 Published on Web 04/21/2005 Controllable Water Channel Gating of Nanometer Dimensions Rongzheng Wan, Jingyuan Li, Hangjun Lu, and Haiping Fang*, Contribution from Shanghai Institute of Applied Physics,
Published on Web 04/21/2005 Controllable Water Channel Gating of Nanometer Dimensions Rongzheng Wan, Jingyuan Li, Hangjun Lu, and Haiping Fang*, Contribution from Shanghai Institute of Applied Physics,
A Brief Guide to All Atom/Coarse Grain Simulations 1.1
 A Brief Guide to All Atom/Coarse Grain Simulations 1.1 Contents 1. Introduction 2. AACG simulations 2.1. Capabilities and limitations 2.2. AACG commands 2.3. Visualization and analysis 2.4. AACG simulation
A Brief Guide to All Atom/Coarse Grain Simulations 1.1 Contents 1. Introduction 2. AACG simulations 2.1. Capabilities and limitations 2.2. AACG commands 2.3. Visualization and analysis 2.4. AACG simulation
Characterizing Structural Transitions of Membrane Transport Proteins at Atomic Detail Mahmoud Moradi
 Characterizing Structural Transitions of Membrane Transport Proteins at Atomic Detail Mahmoud Moradi NCSA Blue Waters Symposium for Petascale Science and Beyond Sunriver, Oregon May 11, 2015 Outline Introduction
Characterizing Structural Transitions of Membrane Transport Proteins at Atomic Detail Mahmoud Moradi NCSA Blue Waters Symposium for Petascale Science and Beyond Sunriver, Oregon May 11, 2015 Outline Introduction
Molecular Visualization. Introduction
 Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Folding of small proteins using a single continuous potential
 JOURNAL OF CHEMICAL PHYSICS VOLUME 120, NUMBER 17 1 MAY 2004 Folding of small proteins using a single continuous potential Seung-Yeon Kim School of Computational Sciences, Korea Institute for Advanced
JOURNAL OF CHEMICAL PHYSICS VOLUME 120, NUMBER 17 1 MAY 2004 Folding of small proteins using a single continuous potential Seung-Yeon Kim School of Computational Sciences, Korea Institute for Advanced
Chapter Cells and the Flow of Energy A. Forms of Energy 1. Energy is capacity to do work; cells continually use energy to develop, grow,
 Chapter 6 6.1 Cells and the Flow of Energy A. Forms of Energy 1. Energy is capacity to do work; cells continually use energy to develop, grow, repair, reproduce, etc. 2. Kinetic energy is energy of motion;
Chapter 6 6.1 Cells and the Flow of Energy A. Forms of Energy 1. Energy is capacity to do work; cells continually use energy to develop, grow, repair, reproduce, etc. 2. Kinetic energy is energy of motion;
PTYS 214 Spring Announcements. Midterm #1 on Tuesday! Be on time! No one enters after the first person leaves! Do your homework!
 PTYS 214 Spring 2018 Announcements Midterm #1 on Tuesday! Be on time! No one enters after the first person leaves! Do your homework! 1 Last time - Properties of Life Organization, energy utilization, homeostasis,
PTYS 214 Spring 2018 Announcements Midterm #1 on Tuesday! Be on time! No one enters after the first person leaves! Do your homework! 1 Last time - Properties of Life Organization, energy utilization, homeostasis,
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis.
 Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
