I (0-0) No phosphate (M)-NDPA-AMP (M)-NDPA-ADP (P)-NDPA-ATP I (0-1)
|
|
|
- Julie Bradford
- 5 years ago
- Views:
Transcription
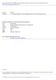
1 a) No phosphate b) CD (mdeg) (M)-NDPA-AMP (M)-NDPA-ADP (P)-NDPA-ATP Absorbance (norm.) I (0-1) I (0-0) No phosphate (M)-NDPA-AMP (M)-NDPA-ADP (P)-NDPA-ATP Supplementary Figure 1 Adenosine phosphates induced helical assembly of NDPA. a, CD and b, normalized absorption spectra of NDPA assemblies with various bound adenosine phosphates (5 x 10-5 M in aq. HEPES buffer). Normalized absorption spectra shows that binding of ATP or ADP leads to strong interchromophoric interactions among NDPA, AMP binding leads to weaker aggregates as evident from the I (0-0) /I (0-1) value AMP = 1.07, ADP = 0.95, ATP = 0.91.
2 C2 I Supplementary Figure 2 Conformations of NDPA. Two conformations of NDPA (C2 and I) shown with the NDPA plane perpendicular to the view (note that two chloride ions, linked to zinc, were added to have neutral molecules). Right: schematic representation of a stack of such molecules as viewed from the top, with a rotation angle of 60 : long axes of NDPA in black and red arrows show with which zinc atoms, zinc atom 1 has shortest distances: successively 2, 3 and 4. Two conformations of the NDPA molecule have been studied: the so-called C2 conformation, which possesses an axis of order 2, and the so-called I conformation, which possesses an inversion center (Supplementary Figure 2). The potential energy difference between both structures is negligible, around 0.1 kcal mol -1 in favor of I.
3 Supplementary Figure 3 Potential energy components. Evolution of the potential energy components per molecule in the NDPA-AMP systems during the MD run (block average over ten structures); for the four energy components, the energy scale spans 10 kcal, to easily compare the energy fluctuations. See Supplementary Note 1 and 2 for detailed explanation.
4 M P Supplementary Figure 4 MD snapshots of NDPA-AMP assemblies. Snapshots at the end of the MD run for M- helix (left) and P-helix (right) of NDPA-AMP assemblies (from C2 conformation). The Zn atoms are shown in purple balls. The difference in potential energy between these structures is around 2 kcal per mol per molecule in favor of the left-handed (M) assembly. The detailed atomistic structure analyses provided further insights into the differences in stability for the left-handed and right-handed helices. Two assemblies with opposite helicity are shown in this figure (last snapshot of MD run), both issued from C2 conformation: M corresponds to C2-L-1, and P to C2- R-2. From the potential energy estimates, these two structures possess a total energy difference of more than 2.1 kcal per mol per molecule, mostly originating from van der Waals interactions (see dark green line and light blue line in Supplementary Figure 3). As supported by the H-bond analysis, the left-handed assembly shows a higher global order than the respective right-handed assembly. On the basis of the modelled structures, several structural parameters have been computed: the average distances and the angles between neighbouring NDPA fragments and the so-called rotation strength, which is an estimate of the local chirality, see Supplementary Table 3. While the average angle between successive NDPA long axes are identical for the two structures (with opposite signs), i.e. around 56, the distances between successive NDPA differ: around 4.0 Å for M-helix while it is around 4.4 Å for the P-helix. Moreover, the deviations (deviation along the NDPA stack) are larger in the case of the P-helix, which is in line with the apparently more disordered global structure for P than for M, as depicted in this figure. The rotation strength provides direct information of the local chirality. The smaller deviation in the M-helix (Supplementary Table 3) further illustrates the differences in global order.
5 M P Supplementary Figure 5 MD snapshots of NDPA-AMP assemblies. Details of the inner structure of a snapshot at the end of MD for left-handed (left) and right-handed (right) NDPA-AMP assemblies, as extracted from a short stack in the middle of a snapshot at the end of MD. The Zn atoms are depicted in balls, with different color code depending on which side of the NDPA they are located, to show the chirality of the assembly. NDPA are depicted in thick sticks and H-bonds are shown in dashed light blue lines. The arrows guide the eye to show the chirality, the arrow pointing towards the back of the view. In the P-helix, a NDPA stacking defect appears in the middle of the stack. However, for the M-helix, the stacking of the NDPA is regular, the NDPA planes are all parallel. This difference in local order for the two types of helices can a priori be attributed to the intrinsic chirality of the ribose (D), which stabilizes more strongly the M-helix in the case of a single point (one phosphate) attachment. In the P- helix, the ribose moieties are oriented at the periphery of the stack, outside the tube formed by the Zn ions. However, in the M-helix they are within the tube formed by the Zn atoms (see Supplementary Figure 4, M versus P). They are closer to the NDPA cores, with which they can form a relatively high number of H-bonds. As the orientation of the ribose moieties in the M-helix make narrower tubes and improves the order of the molecules, partly thanks to hydrogen bonds with NDPA, the average number of - contacts per molecule is higher in M than in P (17.6 for M versus 15.3 for P). These results are in line with the more stabilizing van der Waals interactions for the M-helix compared to the P-helix.
6 Supplementary Figure 6 MD snapshots of NDPA-ADP assembly: (M)-NDPA-ADP helix displayed with a depth cue intensity (right) and without a depth cue intensity (left). The arrow indicates the location of a ribose moiety, whose hydroxyl groups point to the oxygen atoms of naphthalenediimide cores. With ADP, there are two grafting points. To evaluate which pairs of complexes graft to ADP, we started from C2 AMP-substituted helices and noticed that the closest grafting points belong to adjacent molecules (1-2 motif). An ADP substituted M-helix and P-helix were then built and submitted to MM and MD simulations. Similar observations to AMP were also made with NDPA-ADP, both in terms of energy and morphology between the left-handed and right-handed helices, although the amplitude of the differences is smaller. The M- helix is slightly more stable than the P-helix, by 0.5 kcal per mol per molecule on average in the last 10 ns of MD, and the average percentage of oxygen atoms of NDPA involved in hydrogen bonding with hydroxyl groups of adenosine is 27% in the M-helix and 21% in the P-helix. The rotation angle between successive NDPA-ADP is around 58 ± 10, similar to what is found for NDPA-AMP (57 ± 11 ), i.e. around.3 NDPA per turn for both systems. The stacking distance of NDPA is slightly reduced from 4.0 Å to 3.8 Å, from AMP to ADP, likely due to the extra constraint imposed by ADP on adjacent molecules, which form dimers. The simulated CD spectrum for M-NDPA-ADP (Figure. 2, main article) shows a negative bisignated signal with zero crossing around 380 nm, similar to that with AMP.
7 . Supplementary Figure 7 Binding modes of ATP to NDPA. Top view and side view of representative and trimers of NDPA-ATP. The arrows indicate the angle (smaller than 90 ) between neighboring molecules. See Supplementary Note 3 for detailed explanation.
8 Supplementary Figure 8 Potential energy changes. Evolution of the potentiel energy (kcal per mol per molecule) during 20 ns dynamics for representative right-handed assemblies built from and motifs based on NDPA- ATP. Energetically, the trimers seem much more stable than the trimers; the energy difference between the most stable trimers of both motifs that we built are more than 4 kcal per mol per molecule. These structures are only a starting point, as they do not imply that their stacking is favorable. The assemblies are still clearly more stable than the assembly.
9 a) b) Supplementary Figure 9 MD snapshots of NDPA-ATP assembly: a, Details of the inner structure of snapshots at the end of the MD for (P) NDPA-ATP assemblies with the (left) and (right) binding motifs. b, Schematic representing top view of two stacked trimers, showing ATP forming motif in red, and the angles between successive NDIs along the stack (the NDIs are numbered according to their location in the trimer, from 1 for the molecule at the bottom to 3 for the molecule at the top. In the second trimer, the NDIs are numbered similarly from 1 to 3 ). See Supplementary Note 4 for detailed explanation.
10 O H O N H N H O N Supplementary Figure 10 Role of Hydrogen Bonding. Hydrogen bonding between adenosine and NDPA in NDPA-AMP, NDPA-ADP, and NDPA-ATP assemblies: radial distribution function (RDF) between the hydrogen atoms of the hydroxyl groups of adenosine and the oxygen atoms of NDPA (left); RDF between the hydrogen atoms of the amino groups of adenosine and the oxygen atoms of NDPA (right). Experimentally, for NDPA-ATP, P-helices are more stable than the M-helices, which is opposite to the situation with AMP and ADP. As the helical pitches of the NDPA stacks are similar in the three systems, the change of helicity has to be found in the adenosine phosphate complexes. Their organization is indeed very different in AMP or ADP systems on one side, and ATP systems on the other side. With AMP and ADP, the adenosine moieties are able to be close enough to the naphthalenediimide cores to form H-bonds with them in the case of left-handed structures, principally via the hydroxyl groups (the amino groups of adenosine marginally establish hydrogen bonds with NDPA). This figure shows a plot of radial distribution function (RDF) between the hydrogen atoms of the hydroxyl groups of adenosine and the oxygen atoms of NDPA. There is a sharp peak at around 2 Å, a distance characteristic of hydrogen bonding, both for AMP and ADP systems. These interactions, however, no longer exist in the ATP systems, as the adenosine moieties are pointing away. The trace only appears after about 8 Å in the RDF, indicating that the ribose moieties are far from the cores. As both right-handed and left-handed helices share that characteristic, the main interactions biasing the energy balance towards right-handed helices in ATP systems are to be found in the periphery of the stacks, at the level of the chiral adenosine moieties. However, due to large constraints where ATP binds to the zinc coordinated dipicolylethylenediamine receptors, slight changes in the conformation of these rigid parts of the assemblies and associated changes in long-range interactions involving the adenosine moieties can produce many local minima with energies within a few kcal per mol per molecule span. Because this conformational space is very difficult to explore exhaustively, we prevent ourselves here in comparing the P-helix versus M-helix from an energetic point of view.
11 (P)-NDPA-ATP (P)-NDPA-ATP + 1 eq. AMP a) 8 b) 0.8 CD (mdeg) 0 CD (mdeg) 0.4 (P)-NDPA-ATP (P)-NDPA-ATP + 1 eq. AMP AMP X (M)-NDPA-AMP (P)-NDPA-ATP Supplementary Figure 11 Competitive binding between AMP and ATP. a, CD and b, absorption spectra of (P)- NDPA-ATP (5 x 10-5 M) with different equivalents of AMP. Lack of any significant change in the signal confirms inability of AMP to replace ATP competitively. Schematic on the right side depicts the same process.
12 Absorbance a) b) I (0-1) I(0-0) (M)-NDPA-ADP ATP (P)-NDPA-ATP Absorbance I (0-1) I (0-0) (M)-NDPA-AMP ATP (P)-NDPA-ATP Supplementary Figure 12 ATP based competitive replacement. Changes in the absorption spectra of a, (M)- NDPA-ADP and b, (M)-NDPA-AMP on competitive replacement with multivalent ATP (5 x 10-5 M in aq. HEPES buffer). Replacement of AMP and ADP by ATP show similar CD signal changes and thus cannot differentiate the two processes. But a closer look at the absorption spectra can distinguish the two processes. Whereas, removal of ADP by ATP in a) do not show any significant change in absorption spectra, a significant decrease in the intensity along with 2 nm red shift of absorption peak at 380 nm was observed when AMP was substituted by ATP as seen in b). This high band intensity at 380 nm, characteristic of weak NDI aggregates, are expected from AMP with one coordinating site, which lack the ability to clip chromophores for better stabilization. Whereas ADP and ATP with two and three point of attachments can clip to stabilize the chromophore aggregates, thus justifying the observed variation in absorption spectra (Supplementary Figure1b).
13 8 (P)-NDPA-ATP 4 CD (mdeg) 0-4 (M)-NDPA-AMP mole fraction of ATP Supplementary Figure 13 Chiral amplification experiment. Majority rules experiment with a mixture of AMP and ATP upon binding to NDPA (5 x 10-5 M in aq. HEPES buffer). The total concentration of phosphates is chosen such that all the binding sites are occupied and CD intensity was monitored at the CD maxima of 390 nm (positive) and 394 nm (negative). Zero CD intensity do not appear at 0.5 mole fraction of ATP due to non-racemic nature of the (M)-NDPA-AMP and (P)-NDPA-ATP helices which is reflected in their unequal maximum CD intensity (Supplementary Figure 1a).
14 a) b) o C 20 o -6 C 30 o C 40 o C CD (normalized) ln k Time (min) 40 C 30 C 20 C 10 C /T (K -1 ) c) 1.0 CD 390 nm) 0.5 Trial 1 Trial Time (min) Supplementary Figure 14 Kinetic analysis of the state-1 state-2 stereomutation process of (P)-NDPA-ATP assembly: a, Time dependent CD intensity at 390 nm monitored at different temperature (c = 5 x 10-5 M, 1 eq. ATP, with 0.28 U ml -1 CIAP) and corresponding fits (solid lines) using 1 st order kinetic model and b, the resultant Arrhenius plot. Table in c shows the increase in rate constant with temperature, which almost doubles with every 10 K rise. These data corresponds to the Figure 4d in main text. To show the reproducibility of the kinetic analysis, ATP hydrolysis was repeated multiple times and a sample data is presented in c). CD signal variation of two identical samples of (P)-NDPA-ATP with CIAP = 0.42 U ml -1 at 35 C was repeated and their respective 1 st order kinetic fit (solid line) is presented in c). We notice that the error margins in these rate constant values are within 5 % as calculated over repeated experiments (K trial1 =0.331 min -1 and K trial2 =0.333 min -1 ). All CD signals were normalized between zero and one for ease of fitting into kinetic model.
15 D ATP eq., Temp a) 15 o C b) 0, 15 o c) C o C 1, 15 o C 1, 95 o C CD (mdeg) d) 1.0 ATP eq., Temp e) 382 1, 15 o C Absorbance (norm) evident that as temperature 340 increases, the CD 380 signal vanishes Cooling along with blue shift in absorption spectra (382 nm to Absorbance Wavelenght (max, nm) , 15 o C A Temperature ( o C) CD (mdeg) f) 10 CD (mdeg) 0 Heating Cooling Temperature ( o C) ATP : CIAP 1, 95 o 1 : 0 U/ml C 1 : 0.28 U/ml 0.8 1,15 o C Supplementary Figure 15 Temperature dependent helical assembly. Temperature 5 dependent changes in a, CD Heating 381 and b, 0.5absorption spectra upon heating and cooling (P)-NDPA-ATP assembly (5 x 10-5 M in aq. HEPES buffer). c, Temperature dependent variation in CD intensity at 390 nm upon one complete cycle of heating and cooling. It is 380 nm). Upon cooling, the signal red shifts again to 382 nm along with recovery of CD signal. The vibronic features Time (sec) of the absorption spectra at high temperature do not resemble that of a monomeric NDPA. These observations indicate that the decrease in CD signal with increasing temperature at both (1) and (2) state of (P)-NDPA-ATP (Figure 4d, main text) is due to weaker ATP binding at elevated temperatures (and not their detachment as monomeric absorption features were not observed).
16 a) b) Absorbance (norm.) CIAP 0 U/ml 0.28 U/ml 0.56 U/ml 0.70 U/ml CD (mdeg) NDPA + CIAP NDPA Monomer CIAP NDPA Monomer Supplementary Figure 16 Enzyme interaction with NDPA. a, Absorption and b, CD spectral variations on addition of CIAP to NDPA. All measurements were done with 5 x 10-5 M solution in aq. HEPES buffer. Schematic on right side depicts the influence of CIAP on state of NDPA assembly. Absence of any significant change in absorption spectra and the lack of CD signal confirm that the CIAP do not have any specific interactions with NDPA.
17 a) Absorbance (norm.) 1.0 I (0-0) I (0-1) Pi 0 eq. 1 eq. 2 eq. 3 eq. NDPA Monomer Pi (PO 4 ) 3- (rac)-ndpa-pi b) I (0-0) Absorbance NDPA 1.0 I (0-1) (P)-NDPA-ATP (rac)-ndpa-pi after stereomutation 3 eq. Pi (PO 4 ) (P)-NDPA-ATP CIAP (rac)-ndpa-pi Supplementary Figure 17 Pi binding of NDPA. a, Absorption spectra of NDPA upon binding with Pi [(PO 4 ) 3- ]. Changes in the ratio of band intensity suggest significant interchromophoric interactions in NDPA on binding to Pi. b, Changes in the absorption spectra upon enzymatic hydrolysis of ATP with CIAP and its comparison with Pi bound NDPA. Schematic on right side depicts the respective processes. All experiments were at 5 x 10-5 M NDPA in aq. HEPES. Absorption spectra obtained after complete hydrolysis is significantly different from that of NDPA alone without phosphates, indicating that it is not monomeric. We see that after stereomutation, the absorption spectra show blue shift of band along with change in the ratio of band intensity indicating weakening of aggregates. As Pi is shown to strongly interact with NDPA, 3 eq. of Pi released upon complete hydrolysis of ATP can bind to the NDPA resulting in racemic assembly due to achiral nature of Pi.
18 (M)-NDPA-AMP (rac)-ndpa-pi a) 2 0 t = 20 min -2 t = 0 min CD (mdeg) (M)-NDPA-ADP (rac)-ndpa-pi b) c) CD 394 nm U/ml 0.42 U/ml 0.56 U/ml Time (min) Absorbance (norm.) 0.5 I (0-0) I (0-1) (M)-NDPA-AMP CIAP (rac)-ndpa-pi d) 4 CD (mdeg) min 0 min CD 394 nm e) f) U / ml 0.84 U / ml Time (min) Absorbance (norm.) I (0-1) I (0-0) (M)-NDPA-ADP CIAP (rac)-ndpa-pi Supplementary Figure 18 Kinetic analysis of NDPA bound AMP and ADP hydrolysis. Time dependent CD spectra showing the racemisation process of a, (M)-NDPA-AMP (c = 5 x 10-5 M) with CIAP (0.42 U ml -1 ), d, (M)- NDPA-ADP (c = 5 x 10-5 M) with CIAP (0.84 U ml -1 ) at 35 C. Schematic shows the pictorial representation of the enzymatic AMP and ADP hydrolysis. b, and e, show the corresponding CD intensity monitored at 394 nm probing the racemisation process of (M)-NDPA-AMP and (M)-NDPA-ADP respectively, with varying concentrations of CIAP at 35 C. c, and f, show changes in absorption spectra corresponding to hydrolysis of AMP/ADP prebound to NDPA with CIAP (0.70 U ml -1, 35 o C). We notice that hydrolysis leads to weakening of aggregates as seen from the change in ratio of the I (0-0) /I (0-1) but doesn t show monomeric features, suggesting Pi binding to the stacks.
19 a) b) I 1.0 (0-1) Absorbance (norm.) c) 0.5 I (0-0) CIAP NDPA (M)-NDPA-AMP (rac)-ndpa-pi after stereomutation 1 eq. Pi (PO 4 ) 3- Absorbance (norm.) 1.0 I (0-1) NDPA CIAP I (0-0) (M)-NDPA-ADP (rac)-ndpa-pi after stereomutation 2 eq. Pi (PO 4 ) 3- (M)-NDPA-AMP (rac)-ndpa-pi (M)-NDPA-ADP Supplementary Figure 19 Absorption changes during stereomutation: Changes in the absorption spectra upon enzymatic hydrolysis of a, (M)-NDPA-AMP and b, (M)-NDPA-ADP with CIAP (0.7 U ml -1, 35 C) and its comparison with completely racemized state (rac)-ndpa-pi obtained by action of CIAP as well as the Pi bound NDPA. Absorption spectra obtained after complete hydrolysis is very different from that of NDPA alone without phosphates, as seen from their I (0-0) /I (0-1) value indicating that it is not monomeric. Also the spectra after stereomutation matches well with the Pi bound NDPA stacks confirming that NDPA is indeed aggregated due to Pi binding.
20 a) 0.42 U / ml b) 0.28 U / ml c) U / ml U / ml 0.56 U / ml CD (normalized) Time (min) CD (normalized) Time (min) CD 394 nm) Time (min) Trial 1 Trial 2 Supplementary Figure 20 Kinetic analysis of NDPA bound ADP and AMP hydrolysis. Time dependent CD signal change on action of varying CIAP concentration to a, (M)-NDPA-ADP, b, (M)-NDPA-AMP at 35 C and their respective 1 st order kinetic fit (solid line). a, Correspond to kinetic decay data of Supplementary Figures 18e and b, for Supplementary Figures 18b. CD signals were normalized between zero and one for ease of fitting into kinetic model. To show the reproducibility of the kinetic analysis, AMP hydrolysis was repeated multiple times and a sample data is presented in c). CD signal variation of two identical samples of (M)-NDPA-AMP with CIAP = 0.40 U ml -1 at 35 C was repeated and their respective 1 st order kinetic fit (solid line) is presented in c). We notice that the error margins in these rate constant values are within 5 % as calculated over repeated experiments (K trial1 =0.116 min -1 and K trial2 =0.118 min -1 ).
21 a) b) Rate Constant (k, min -1 ) AMP ADP CIAP (U / ml) CD (mdeg) (M)-NDPA-AMP (M)-NDPA-ADP Time (sec) Supplementary Figure 21 Comparative Enzymatic hydrolysis. a, Plot of hydrolysis rate constants of NDPA bound AMP and ADP (k amp and k adp ) with varying concentration of CIAP at 35 C as shown in Supplementary Table 5. We see that at a given CIAP concentration, k amp is higher than k adp confirming that ADP hydrolysis is much slower than AMP. Confirming the same fact b, shows decay kinetics by plotting time dependent variation in CD signal of (M)-NDPA-AMP and (M)-NDPA-ADP (c=5 x 10-5 M)with CIAP (0.84 U ml -1 ) at 35 o C monitored at 390 nm.
22 Absorbance I (0-1) (2) I (0-0) 0.6 (1) a) b) 0.3 (1) (2) Absorbance I (0-1) I (0-0) (3) (2) (3) (2) Supplementary Figure 22 Absorption changes in stepwise hydrolysis of ATP: Variation in absorption spectra upon stepwise hydrolysis of ATP during a, 1 2 and b, 2 3 (c = 5 x 10-5 M, 35 C with CIAP (0.7 U ml -1 )). We see that the transformation from 1 2 (when the CD signal reverses in Fig. 4c) do not show any change in absorption spectra as expected of the transition from ATP ADP (Supplementary Figure 12a). However, the later process from 2 3 (when the CD signal goes to zero in Fig. 4f), we notice changes in vibronic features of NDPA absorbance (I (0-0)/I (0-1) value) as expected of the transition from ADP AMP Pi (Supplementary Figure 12b).
23 Time ATP + CIAP(t 4 ) ATP + CIAP(t 3 ) ATP ATP + CIAP(t 2 ) ADP ATP + CIAP(t 1 ) AMP ATP ppm Supplementary Figure P NMR signature of enzymatic process. Time dependent changes in the 31 P NMR spectra of ATP in presence of CIAP and its comparison with ATP without CIAP. We notice that the peak at ppm (characteristic of ATP, marked with red star symbol)vanishes soon after addition of CIAP, whereas the other two (-11.2 ppm, ppm, marked with blue triangle symbol) remain which is characteristic of ADP, which further changes with time to give only AMP signal followed by conversion to Pi and adenosine. This enzymatic ATP hydrolysis without NDPA also follows the same pathway and confirms our finding by circular dichroism in presence of NDPA.
24 a) 0.24 b) c) 0.12 A 820 nm (M)-NDPA-AMP (M)-NDPA-ADP A 820 nm ATP without NDPA (P)-NDPA-ATP A 820 nm 8 4 ADP without NDPA (M)-NDPA-ADP Time (min) Time (min) Time (min) Supplementary Figure 24 Inorganic method for Pi estimation. Pi assay results using Chen s method to determine the amount of Pi released on enzymatic hydrolysis. a) shows comparative hydrolysis kinetics of NDPA bound AMP and ADP. We see that ADP hydrolysis is indeed very slow when compared with AMP as proven previously by monitoring CD signal (Supplementary Figure11, Supplementary Figure12). These measurements were done with CIAP (0.84 U ml -1 ) and NDPA (c = 5x10-5 M) at 35 C. Comparative hydrolysis rate of b) ATP and c) ADP with and without NDPA, as probed by Pi assay. It shows that phosphate hydrolysis is slightly faster in free state compared to NDPA bound state. All measurements were done with CIAP (0.84 U ml -1 ) and NDPA (c = 5x10-5 M) at 35 C. This difference in the rate of bound and unbound phosphates could be due to easy access of unbound phosphates to enzyme. However, unlike the helix reversal measurements for kinetic analysis, Chen s assay is not an in situ measurement due to highly acidic conditions required for metal complex formation, so aliquots were taken at regular intervals for measurements. In addition, this method can only probe the net Pi released and cannot distinguish the stepwise hydrolysis of ATP via ADP and AMP to Pi.
25 5 (M)-NDPA-AMP + 1 eq. ATP (M)-NDPA-ADP + 1 eq. ATP CD (mdeg) Time (min) Supplementary Figure 25 Mechanistic insight on enzymatic action. Time dependent variation in CD signal of a) 5 x 10-5 M NDPA (aq. HEPES buffer) on competitive replacement of AMP and ADP by 1 eq. of ATP (CIAP=0.70 U ml -1 ) at 35 o C monitored at 390 nm. To understand if enzyme selectively hydrolyses the unbound or bound phosphates, CD kinetic measurements of (P)-NDPA-ATP in presence of other phosphates (one eq. of ATP followed by addition of 1 eq. of either AMP or ADP) were performed. As expected, due to competitive binding, ATP should be bound to NDPA to form (P)-NDPA-ATP stacks, whereas AMP or ADP must be free in solution. Gradual decrease in signal from time t=0 suggests that CIAP has no preferential action to unbound phosphates compared to bound ones. If enzymatic action were to be preferential to unbound phosphates, we should have obtained constant CD signal initially till all unbound phosphates are consumed and only then signal should have started to decrease.
26 Supplementary Table 1 Potential energy of NDPA-AMP: Average potential energy for the 8 types of AMP bound supramolecular assemblies during MD; the average potential energy of the most stable assembly has been set to zero, and that of other assemblies rescaled accordingly to allow direct energy comparison between the assemblies (R stands for right-handed helical assembly (P); L stands for left-handed helical assembly (M)). The standard deviation is around 1.8 kcal per mol for all types of assemblies. See Supplementary Note 1 for detailed explanation. Type of assembly Average potential energy (kcal per mol per molecule) C2-L C2-L-2 C2-R C2-R I-L I-L I-R I-R-2 2.6
27 Supplementary Table 2 Average percentage of oxygen atoms of NDPA involved in hydrogen bonding with hydroxyl groups of adenosine in AMP (C=O (NDPA)--- H-O (ribose)). Despite similar H-bonding energies between the assemblies, their hydrogen-bonding network is different, and thus the orientation of adenosine. Analysing hydrogen bonding between the hydroxyl groups of ribose and the oxygen atoms of NDPA during the last 10 ns of MD, it is found that left-handed assemblies develop up to two hydrogen bonds per molecule. These interactions can take place between molecules n and n+2 or n+3, thus contributing to long-range ordering of the assembly. Such hydrogen bonds are two to three times less numerous in the P-helix. Type of assembly Average proportion of oxygen atoms of NDPA involved in hydrogen bonding (%) C2-L-1 45 C2-L-2 39 C2-R-1 15 C2-R-2 15
28 Supplementary Table 3 Structural parameters of different types of helices in NDPA-AMP assemblies, with amplitude of deviations along the stack. See Supplementary Figure 5 for detailed explanation. The rotation strength between two neighbouring molecules is calculated as 1 x 2. r12/ 1 2, the mixed product between two successive dipoles along the stack. *The average number of - contacts is calculated from the total number of interactions at short distance (from 3.5 to 5.5 Å) between the four rings centers of adjacent NDPA molecules along the stack, divided by the total number of NDPA molecules. Structural parameter Left-handed helix (M) Right-handed helix (P) Average angle between NDPA ± ± 19.3 Average distance between NDPA 4.04 Å ± 0.42 Å 4.45 Å ± 0.92 Å Average «rotation strength» ± ± 0.21 Average number of H-bonds per NDPA-AMP in the stack Average number of - contacts per NDPA-AMP in the stack*
29 Supplementary Table 4 Kinetic analysis of the (1) (2) stereomutation process of (P)-NDPA-ATP assembly: Variation in rate constant of stereomutation in (P)-NDPA-ATP assembly upon increasing temperatures (c = 5 x 10-5 M, 1 eq. ATP, with 0.28 U ml -1 CIAP) as obtained by using 1 st order kinetic model. The rate constant increases with temperature, which almost doubles with every 10 K rise. These data corresponds to the Figure 4d in main text and Supplementary Figure 14. T (K) k ATP (min -1 )
30 Supplementary Table 5 Comparative rate constant of NDPA bound AMP and ADP hydrolysis at various CIAP concentrations and 35 C as obtained from the respective 1 st order kinetic fit shown in Supplementary Figure 20. CIAP (U ml -1 ) k AMP (min -1 ) k ADP (min -1 )
31 Supplementary Notes Supplementary Note 1 NDPA-AMP assembly: When grafting AMP, for the I conformer, the molecule loses its inversion center because of the chirality of AMP; however, the new structure obtained will still be referred to in the text as I, in reference to the symmetry of the NDPA core. For both C2 and I, two orientations of AMP are possible, and the molecules can assemble as left and right-handed helices. In total 8 systems were thus built: 2 types of NDPA conformations (I and C2) x 2 orientations of the AMP (1 and 2) with respect to the zinc atom x 2 left- or right-handed chirality of the NDPA stack (L and R). The conformers are thus named accordingly, such as C2-L-1 or I-R-2. In these assemblies, one of the two zinc atoms of a given molecule n is close to a zinc atom of molecules n+1 and n-1, but can also be close to a zinc atom of other molecules (for instance n-3 or n+3 if the rotation angle between successive molecules is 60 ; see Supplementary Figure 1). This information provides a clue about the possibilities of grafting that exist when replacing AMP with ADP or ATP, which have two or three grafting points, respectively. Molecular Dynamics is performed on these 8 types of supramolecular assemblies, each constituted of 32 NDPA molecules and 64 AMP molecules, so that each assembly is modelled in comparison to its mirror image structure, but with the ribose keeping a right-handed (D) chirality. To give insights into their relative stabilities, the average potential energy and standard deviation have been calculated after a sufficiently large relaxation time (the last 10 nanoseconds of MD for all structures, except O2-R-2 and I-L- 1, where the calculation was performed on the last 5 nanoseconds, as the assembly required more time to relax), and displayed in Supplementary Table 1. The potential energies show a clear energy segregation between the modelled stacks, on a scale of about 3.5 kcal per mol per molecule (i.e. nearly 112 kcal per mol for the 32 NDPA stack). The most striking segregation is between C2 and I families of assemblies, the least stable assemblies being those built from I conformations, where defects and stack bending appear during MD. This behavior reflects steric crowding in I systems, which explains this high level of energy segregation. Within C2 assemblies, the most stable structures are left-handed: from 1.5 up to 2.4 kcal per mol per molecule more stable than their right-handed counterparts (Supplementary Table 1). This is a large energy difference, since this potential energy is estimated per NDPA in the stack. Between the two C2 lefthanded assemblies, a small energy difference of 0.3 kcal per mol per molecule is estimated, suggesting that the orientation of AMP grafting on Zn atom does not appear very critical for left-handed (M) structure. Supplementary Note 2 (P)-NDPA-AMP vs (M)-NDPA-AMP: To evaluate the origin of the energy difference between right-handed and left-handed C2 assemblies of NDPA-AMP, we extracted the valence (bonding terms) and long-range (non-bonding) interactions from the energy profiles and displayed their evolution on a scale of around 10 kcal per mol per molecule, see Supplementary Figure 3. From the results
32 obtained, it is difficult to attribute the energy difference solely to a given type of interactions, as there are multiple cancellation effects. However, we can observe that the largest energy differences arise mainly from van der Waals interactions. Supplementary Note 3 NDPA-ATP assembly: With ATP, there are three grafting points, which give rise to much more different possibilities of grafting than with AMP. One possible type of assembly is formed of trimers interacting by long-range interactions, a trimer being composed of three stacked NDPA molecules bonded together by two ATP. Another possible type of assembly results from shifted ATP binding modes, i.e. where ATP molecules share three stacked NDPA molecules with two other ATP molecules, instead of one as in a trimer. As a result, all NDPA molecules are linked together to form a supermolecule. The assemblies with shifted binding modes that we built, however, showed a large level of disorder during MD, indicative that this type of organization is unfavorable. We thus focused on assemblies built from trimers. Despite this focus on a more limited set of conformers, there are still different possibilities to bind ATP to three NDPA molecules. The trimers are highly constrained, because the zinc complexes are rigid, and because these rigid entities are bonded together via ATP. Because of the different possibilities of binding, and the presence of strong geometric constraints, there are different local minima that are only accessible by building them systematically (this is opposite to flexible systems, where computational methods such as MD can be used to explore their potential energy surface). Due to this large number of possibilities, an exhaustive exploration was not possible. However, the simulation of about twenty trimers allowed us to distinguish two families of trimers, whose members have similar designs. The two families are characterized by the order of binding of the stacked molecules: or The x-y-z design means that molecule x is linked to molecule y, and molecule y is linked to molecule z, while x, y, and z refers to the order of the molecules along the stacking direction (Supplementary Figure 7). Supplementary Note 4 In Supplementary Figure 9, the right-handed and assemblies are displayed. The striking characteristic of assemblies is that the organization of the adenosine phosphate complexes is not helical. While the naphthalenediimide cores do form a right-handed helix, the adenosine phosphate complexes form two columns on both sides of the assembly (Supplementary Figure 9). As the pitch of the naphthalenediimide helix is similar, whatever the adenosine phosphate (6 molecules with ATP, versus 6.3 with AMP and ADP), as well as the average rotation angle between neighboring molecules (60 versus 57-58, respectively), the peculiar arrangement of the complexes is to be explained by symmetry. With AMP, the angle between the adenosine phosphate complexes is about 60, i.e. the same as the angle between the naphthalenediimide cores. With ADP, because each adenosine phosphate complex
33 involves two naphthalenediimide cores, the angle between the complexes is twice the angle between the naphthalenediimide cores, about 120 (or -60, considering that the molecules have a C 2 axis along the stacking direction). Thus, for left-handed assemblies, the adenosine phosphate complexes can be regarded as forming a left-handed helix with motifs separated by 120 (pitch of 3.1 adenosine phosphate complexes, i.e. 6.3 molecules), or a right-handed helix with motifs separated by 60 (pitch of 6.3 adenosine phosphate complexes, i.e molecules). With ATP, the angle between the adenosine phosphate complexes is three times the angle between the naphthalenediimide cores, about 180. As a result, the pitch of the assembly of adenosine phosphate complexes is of only two complexes, effectively corresponding to six NDPA molecules. But an assembly with a pitch of only two motifs can not be called an helix; the trimers are translational images from each other along the stacking direction. We then investigated in more details the R representative assembly. Ten structures extracted every nanosecond between 10 and 20 ns of MD were used to study the angle between neighboring molecules. Though the average angle is of 60, a more detailed analysis reveals that there are three population distributions: the angle between molecules 1 and 2, 1-2 is 81 ± 4. Similarly, 2-3 is 44 ± 4 (see Supplementary Figure 7). And 3-1 = 54 ± 4, 1 refering to a molecule belonging to a neigboring trimer (Figure 2, main article). The first two angles deviate from the angle found in the NDPA-AMP arrangement (~57 ), due to the constraints imposed by ATP. The third one, however, measured between molecules belonging to adjacent trimers, i.e. more free to rotate with respect to one another, is not much affected. This result indicates weak steric hindrance between trimers, which are thus able to stack while conserving the natural angle between single NDPA molecules. The sum of the three angles is 179, corresponding well to the symmetry condition necessary for having adenosine phosphate complexes forming columns instead of helices. Supplementary Methods Materials: CIAP (3 units mg -1 ) was purchased from Sisco Research Laboratory Pvt. Ltd. India. All other chemicals were purchased from the commercial sources and were used as such. Spectroscopic grade solvents were used for all optical measurements. Simulation of CD spectra: The calculation of the excitonic CD spectra involves two steps. First, the lowest 30 excited states of the 32 NDPA molecules extracted from the MD trajectories above are computed at the INDO/SCI level (using an active space of 30 occupied and 30 empty molecular orbitals). Then, an excitonic Hamiltonian encompassing a total of 32x30 basis functions (30 localized excitations per molecule) is built on the basis of INDO/SCI 1 excitation energies and exciton couplings. The latter are calculated as Coulomb interactions between transition densities, thus going beyond the usual point dipole
34 model. 2 Diagonalization of this Hamiltonian yields a set of 960 exciton states with energies ħ and wavefunctions >, for which the oscillator strength f and the rotational strength R are computed as: 3 f μ i, n 2 i, n µ i, n G 2,...Supplementary Equation 1 R h c μ i, n G G μ i, n j, n' µ i, n j, n' j, n' ( r n r n' ),.Supplementary Equation 2 where c is the speed of light, µ i,n the transition dipole moment from the ground state g> to the excited state i> of molecule n along the stack, μ ( i, n g h. c.) the corresponding dipole operator, G> the i, n µ i, n ground state of the helical stack (product state of all g>), and c i n the exciton state i, n i, n, wavefunctions expanded in terms of the c, eigenvectors. The absorption/cd response at input frequency i n is calculated on the basis of the oscillator/rotational strengths as: Abn( ) f G( ), Supplementary Equation 3 CD( ) R G( ),..Supplementary Equation 4 where G(- ) is a Gaussian function centered around with variance = 0.1 ev. The brackets denote a configurational average over the positional and energetic disorder as explored during the MD simulations. Here, a total of 8 supramolecular helical structures, each consisting of 32 (for NDPA-AMP and NDPA- ADP assemblies) or 33 (for NDPA-ATP assemblies) molecules, were used. This approach was found to yield CD spectra that are stable with respect to configurational averaging. Synthesis and Procedures: NDPA was synthesized based on the reported procedure and was accordingly characterized. 4, 5 Supplementary References: 1 Ridley, J. & Zerner, M. An intermediate neglect of differential overlap technique for spectroscopy: Pyrrole and the azines. Theor. Chim. Acta 32, (1973).
35 2 Beljonne, D., Cornil, J., Silbey, R., Millié, P., Brédas, J. L. Interchain interactions in conjugated materials: The exciton model versus the supermolecular approach, J. Chem. Phys. 112, (2000). 3 Spano, F. C., Meskers, S. C. J., Hennebicq, E. & Beljonne, D. Probing excitation delocalization in supramolecular chiral stacks by means of circularly polarized light: experiment and modeling,. J. Am. Chem. Soc. 129, (2007). 4 Kumar, M., Jonnalagadda, N. & George, S. J. Molecular recognition driven self-assembly and chiral induction in naphthalene diimide amphiphiles. Chem. Commun. 48, (2012). 5 Lee, H. N. et al. Pyrophosphate-selective fluorescent chemosensor at physiological ph: formation of a unique excimer upon addition of pyrophosphate. J. Am. Chem. Soc. 129, (2007).
i) impact of interchain interactions
 i) impact of interchain interactions multiple experimental observations: in dilute solutions or inert matrices: the photoluminescence quantum yield of a given conjugated polymers can be very large: up
i) impact of interchain interactions multiple experimental observations: in dilute solutions or inert matrices: the photoluminescence quantum yield of a given conjugated polymers can be very large: up
Quantum Chemistry. NC State University. Lecture 5. The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy
 Quantum Chemistry Lecture 5 The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy NC State University 3.5 Selective absorption and emission by atmospheric gases (source:
Quantum Chemistry Lecture 5 The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy NC State University 3.5 Selective absorption and emission by atmospheric gases (source:
in Halogen-Bonded Complexes
 9 Resonance Assistance and Cooperativity in Halogen-Bonded Complexes Previously appeared as Covalency in Resonance-Assisted Halogen Bonds Demonstrated with Cooperativity in N-Halo-Guanine Quartets L. P.
9 Resonance Assistance and Cooperativity in Halogen-Bonded Complexes Previously appeared as Covalency in Resonance-Assisted Halogen Bonds Demonstrated with Cooperativity in N-Halo-Guanine Quartets L. P.
Overview of Photosynthesis
 Overview of Photosynthesis In photosynthesis, green plants absorb energy from the sun and use the energy to drive an endothermic reaction, the reaction between carbon dioxide and water that produces glucose
Overview of Photosynthesis In photosynthesis, green plants absorb energy from the sun and use the energy to drive an endothermic reaction, the reaction between carbon dioxide and water that produces glucose
Water-methanol separation with carbon nanotubes and electric fields
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 215 Supplementary Information: Water-methanol separation with carbon nanotubes and electric fields
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 215 Supplementary Information: Water-methanol separation with carbon nanotubes and electric fields
XV 74. Flouorescence-Polarization-Circular-Dichroism- Jablonski diagram Where does the energy go?
 XV 74 Flouorescence-Polarization-Circular-Dichroism- Jablonski diagram Where does the energy go? 1) Excite system through A Absorbance S 0 S n Excite from ground excited singlet S = 0 could be any of them
XV 74 Flouorescence-Polarization-Circular-Dichroism- Jablonski diagram Where does the energy go? 1) Excite system through A Absorbance S 0 S n Excite from ground excited singlet S = 0 could be any of them
Structure and Bonding of Organic Molecules
 Chem 220 Notes Page 1 Structure and Bonding of Organic Molecules I. Types of Chemical Bonds A. Why do atoms forms bonds? Atoms want to have the same number of electrons as the nearest noble gas atom (noble
Chem 220 Notes Page 1 Structure and Bonding of Organic Molecules I. Types of Chemical Bonds A. Why do atoms forms bonds? Atoms want to have the same number of electrons as the nearest noble gas atom (noble
Calculate a rate given a species concentration change.
 Kinetics Define a rate for a given process. Change in concentration of a reagent with time. A rate is always positive, and is usually referred to with only magnitude (i.e. no sign) Reaction rates can be
Kinetics Define a rate for a given process. Change in concentration of a reagent with time. A rate is always positive, and is usually referred to with only magnitude (i.e. no sign) Reaction rates can be
March CUME Organic Chemistry
 March CUME Organic Chemistry Department of Chemistry, University of Missouri Columbia Saturday, March 21, 1998, @Nine O Clock, 221 Chemistry Dr. Rainer Glaser Announced Reading Circular Dichroism and Exciton-Coupled
March CUME Organic Chemistry Department of Chemistry, University of Missouri Columbia Saturday, March 21, 1998, @Nine O Clock, 221 Chemistry Dr. Rainer Glaser Announced Reading Circular Dichroism and Exciton-Coupled
Assessing the potential of atomistic molecular dynamics simulations to probe reversible. protein-protein recognition and binding
 Supporting information for Assessing the potential of atomistic molecular dynamics simulations to probe reversible protein-protein recognition and binding Luciano A. Abriata and Matteo Dal Peraro Laboratory
Supporting information for Assessing the potential of atomistic molecular dynamics simulations to probe reversible protein-protein recognition and binding Luciano A. Abriata and Matteo Dal Peraro Laboratory
Fluorescence 2009 update
 XV 74 Fluorescence 2009 update Jablonski diagram Where does the energy go? Can be viewed like multistep kinetic pathway 1) Excite system through A Absorbance S 0 S n Excite from ground excited singlet
XV 74 Fluorescence 2009 update Jablonski diagram Where does the energy go? Can be viewed like multistep kinetic pathway 1) Excite system through A Absorbance S 0 S n Excite from ground excited singlet
4. Circular Dichroism - Spectroscopy
 4. Circular Dichroism - Spectroscopy The optical rotatory dispersion (ORD) and the circular dichroism (CD) are special variations of absorption spectroscopy in the UV and VIS region of the spectrum. The
4. Circular Dichroism - Spectroscopy The optical rotatory dispersion (ORD) and the circular dichroism (CD) are special variations of absorption spectroscopy in the UV and VIS region of the spectrum. The
Chapter 6- An Introduction to Metabolism*
 Chapter 6- An Introduction to Metabolism* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Energy of Life
Chapter 6- An Introduction to Metabolism* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Energy of Life
Chapter 6 Chemical Reactivity and Mechanisms
 Chapter 6 Chemical Reactivity and Mechanisms 6.1 Enthalpy Enthalpy (ΔH or q) is the heat energy exchange between the reaction and its surroundings at constant pressure Breaking a bond requires the system
Chapter 6 Chemical Reactivity and Mechanisms 6.1 Enthalpy Enthalpy (ΔH or q) is the heat energy exchange between the reaction and its surroundings at constant pressure Breaking a bond requires the system
SUPPLEMENTARY INFORMATION
 Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
2: CHEMICAL COMPOSITION OF THE BODY
 1 2: CHEMICAL COMPOSITION OF THE BODY Although most students of human physiology have had at least some chemistry, this chapter serves very well as a review and as a glossary of chemical terms. In particular,
1 2: CHEMICAL COMPOSITION OF THE BODY Although most students of human physiology have had at least some chemistry, this chapter serves very well as a review and as a glossary of chemical terms. In particular,
Conjugated Systems, Orbital Symmetry and UV Spectroscopy
 Conjugated Systems, Orbital Symmetry and UV Spectroscopy Introduction There are several possible arrangements for a molecule which contains two double bonds (diene): Isolated: (two or more single bonds
Conjugated Systems, Orbital Symmetry and UV Spectroscopy Introduction There are several possible arrangements for a molecule which contains two double bonds (diene): Isolated: (two or more single bonds
Chapter 4 Spectroscopy
 Chapter 4 Spectroscopy The beautiful visible spectrum of the star Procyon is shown here from red to blue, interrupted by hundreds of dark lines caused by the absorption of light in the hot star s cooler
Chapter 4 Spectroscopy The beautiful visible spectrum of the star Procyon is shown here from red to blue, interrupted by hundreds of dark lines caused by the absorption of light in the hot star s cooler
Bulk behaviour. Alanine. FIG. 1. Chemical structure of the RKLPDA peptide. Numbers on the left mark alpha carbons.
 Bulk behaviour To characterise the conformational behaviour of the peptide, first we looked at the statistics of alpha carbons and the torsion angles. Then they were correlated with positions of positively
Bulk behaviour To characterise the conformational behaviour of the peptide, first we looked at the statistics of alpha carbons and the torsion angles. Then they were correlated with positions of positively
Downloaded from UvA-DARE, the institutional repository of the University of Amsterdam (UvA)
 Downloaded from UvA-DARE, the institutional repository of the University of Amsterdam (UvA) http://dare.uva.nl/document/351205 File ID 351205 Filename 5: Vibrational dynamics of the bending mode of water
Downloaded from UvA-DARE, the institutional repository of the University of Amsterdam (UvA) http://dare.uva.nl/document/351205 File ID 351205 Filename 5: Vibrational dynamics of the bending mode of water
(Excerpt from S. Ji, Molecular Theory of the Living Cell: Concepts, Molecular Mechanisms, and Biomedical Applications, Springer, New York, 2012)
 2.2 The Franck-Condon Principle (FCP) 2.2.1 FCP and Born-Oppenheimer Approximation The Franck-Condon Principle originated in molecular spectroscopy in 1925 when J. Franck proposed (and later Condon provided
2.2 The Franck-Condon Principle (FCP) 2.2.1 FCP and Born-Oppenheimer Approximation The Franck-Condon Principle originated in molecular spectroscopy in 1925 when J. Franck proposed (and later Condon provided
Luminescence. Photoluminescence (PL) is luminescence that results from optically exciting a sample.
 Luminescence Topics Radiative transitions between electronic states Absorption and Light emission (spontaneous, stimulated) Excitons (singlets and triplets) Franck-Condon shift(stokes shift) and vibrational
Luminescence Topics Radiative transitions between electronic states Absorption and Light emission (spontaneous, stimulated) Excitons (singlets and triplets) Franck-Condon shift(stokes shift) and vibrational
Structural and mechanistic insight into the substrate. binding from the conformational dynamics in apo. and substrate-bound DapE enzyme
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 215 Structural and mechanistic insight into the substrate binding from the conformational
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 215 Structural and mechanistic insight into the substrate binding from the conformational
Chapter 6 Cyclic urea - a new central unit in bent-core compounds
 82 Chapter 6 Cyclic urea - a new central unit in bent-core compounds A new class of five-ring bent-core molecules with a cyclic urea group as a central unit was synthesized [94]. A significant difference
82 Chapter 6 Cyclic urea - a new central unit in bent-core compounds A new class of five-ring bent-core molecules with a cyclic urea group as a central unit was synthesized [94]. A significant difference
A very brief history of the study of light
 1. Sir Isaac Newton 1672: A very brief history of the study of light Showed that the component colors of the visible portion of white light can be separated through a prism, which acts to bend the light
1. Sir Isaac Newton 1672: A very brief history of the study of light Showed that the component colors of the visible portion of white light can be separated through a prism, which acts to bend the light
Supplementary information for the paper
 Supplementary information for the paper Structural correlations in the generation of polaron pairs in lowbandgap polymers for photovoltaics Supplementary figures Chemically induced OD 0,1 0,0-0,1 0,1 0,0-0,1
Supplementary information for the paper Structural correlations in the generation of polaron pairs in lowbandgap polymers for photovoltaics Supplementary figures Chemically induced OD 0,1 0,0-0,1 0,1 0,0-0,1
NPTEL/IITM. Molecular Spectroscopy Lectures 1 & 2. Prof.K. Mangala Sunder Page 1 of 15. Topics. Part I : Introductory concepts Topics
 Molecular Spectroscopy Lectures 1 & 2 Part I : Introductory concepts Topics Why spectroscopy? Introduction to electromagnetic radiation Interaction of radiation with matter What are spectra? Beer-Lambert
Molecular Spectroscopy Lectures 1 & 2 Part I : Introductory concepts Topics Why spectroscopy? Introduction to electromagnetic radiation Interaction of radiation with matter What are spectra? Beer-Lambert
The Study of Chemical Reactions. Mechanism: The complete, step by step description of exactly which bonds are broken, formed, and in which order.
 The Study of Chemical Reactions Mechanism: The complete, step by step description of exactly which bonds are broken, formed, and in which order. Thermodynamics: The study of the energy changes that accompany
The Study of Chemical Reactions Mechanism: The complete, step by step description of exactly which bonds are broken, formed, and in which order. Thermodynamics: The study of the energy changes that accompany
CN NC. dha-7. dha-6 R' R. E-vhf (s-trans) E-vhf (s-cis) R CN. Z-vhf (s-cis) Z-vhf (s-trans) R = AcS R' = AcS
 R' R R' R dha-6 dha-7 R' R R' R E-vhf (s-cis) E-vhf (s-trans) R R' R R' Z-vhf (s-cis) Z-vhf (s-trans) R = R' = Supplementary Figure 1 Nomenclature of compounds. Supplementary Figure 2 500 MHz 1 H NMR spectrum
R' R R' R dha-6 dha-7 R' R R' R E-vhf (s-cis) E-vhf (s-trans) R R' R R' Z-vhf (s-cis) Z-vhf (s-trans) R = R' = Supplementary Figure 1 Nomenclature of compounds. Supplementary Figure 2 500 MHz 1 H NMR spectrum
Spectroscopy and the Particle in a Box
 Spectroscopy and the Particle in a Box Introduction The majority of colors that we see result from transitions between electronic states that occur as a result of selective photon absorption. For a molecule
Spectroscopy and the Particle in a Box Introduction The majority of colors that we see result from transitions between electronic states that occur as a result of selective photon absorption. For a molecule
( ) x10 8 m. The energy in a mole of 400 nm photons is calculated by: ' & sec( ) ( & % ) 6.022x10 23 photons' E = h! = hc & 6.
 Introduction to Spectroscopy Spectroscopic techniques are widely used to detect molecules, to measure the concentration of a species in solution, and to determine molecular structure. For proteins, most
Introduction to Spectroscopy Spectroscopic techniques are widely used to detect molecules, to measure the concentration of a species in solution, and to determine molecular structure. For proteins, most
Lecture #8 9/21/01 Dr. Hirsh
 Lecture #8 9/21/01 Dr. Hirsh Types of Energy Kinetic = energy of motion - force x distance Potential = stored energy In bonds, concentration gradients, electrical potential gradients, torsional tension
Lecture #8 9/21/01 Dr. Hirsh Types of Energy Kinetic = energy of motion - force x distance Potential = stored energy In bonds, concentration gradients, electrical potential gradients, torsional tension
Handout 13 Interpreting your results. What to make of your atomic coordinates, bond distances and angles
 Handout 13 Interpreting your results What to make of your atomic coordinates, bond distances and angles 1 What to make of the outcome of your refinement There are several ways of judging whether the outcome
Handout 13 Interpreting your results What to make of your atomic coordinates, bond distances and angles 1 What to make of the outcome of your refinement There are several ways of judging whether the outcome
Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study
 pubs.acs.org/jpca Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study Tomoo Miyahara, Hiroshi Nakatsuji,*, and Hiroshi Sugiyama Quantum Chemistry Research Institute, JST, CREST,
pubs.acs.org/jpca Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study Tomoo Miyahara, Hiroshi Nakatsuji,*, and Hiroshi Sugiyama Quantum Chemistry Research Institute, JST, CREST,
Supporting Information
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2016 Supporting Information Lipid molecules can induce an opening of membrane-facing
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2016 Supporting Information Lipid molecules can induce an opening of membrane-facing
16.1 Introduction to NMR Spectroscopy. Spectroscopy. Spectroscopy. Spectroscopy. Spectroscopy. Spectroscopy 4/11/2013
 What is spectroscopy? NUCLEAR MAGNETIC RESONANCE (NMR) spectroscopy may be the most powerful method of gaining structural information about organic compounds. NMR involves an interaction between electromagnetic
What is spectroscopy? NUCLEAR MAGNETIC RESONANCE (NMR) spectroscopy may be the most powerful method of gaining structural information about organic compounds. NMR involves an interaction between electromagnetic
Chapter 11. Intermolecular Forces and Liquids & Solids
 Chapter 11 Intermolecular Forces and Liquids & Solids The Kinetic Molecular Theory of Liquids & Solids Gases vs. Liquids & Solids difference is distance between molecules Liquids Molecules close together;
Chapter 11 Intermolecular Forces and Liquids & Solids The Kinetic Molecular Theory of Liquids & Solids Gases vs. Liquids & Solids difference is distance between molecules Liquids Molecules close together;
Analysis of the simulation
 Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase
 SUPPLEMENTARY INFORMATION Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase Louise C Briggs, Geoff S Baldwin, Non Miyata, Hisao Kondo, Xiaodong Zhang, Paul S
SUPPLEMENTARY INFORMATION Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase Louise C Briggs, Geoff S Baldwin, Non Miyata, Hisao Kondo, Xiaodong Zhang, Paul S
Cation exchange induced transformation of InP magic-sized clusters
 Supplementary Information to Accompany: Cation exchange induced transformation of InP magic-sized clusters Jennifer L. Stein, Molly I. Steimle Maxwell W. Terban, Alessio Petrone, Simon J. L. Billinge,
Supplementary Information to Accompany: Cation exchange induced transformation of InP magic-sized clusters Jennifer L. Stein, Molly I. Steimle Maxwell W. Terban, Alessio Petrone, Simon J. L. Billinge,
Basic Organic Chemistry Course code : CHEM (Pre-requisites : CHEM 11122)
 Basic Organic Chemistry Course code : CHEM 12162 (Pre-requisites : CHEM 11122) Chapter 01 Mechanistic Aspects of S N2,S N1, E 2 & E 1 Reactions Dr. Dinesh R. Pandithavidana Office: B1 222/3 Phone: (+94)777-745-720
Basic Organic Chemistry Course code : CHEM 12162 (Pre-requisites : CHEM 11122) Chapter 01 Mechanistic Aspects of S N2,S N1, E 2 & E 1 Reactions Dr. Dinesh R. Pandithavidana Office: B1 222/3 Phone: (+94)777-745-720
Ch. 9- Molecular Geometry and Bonding Theories
 Ch. 9- Molecular Geometry and Bonding Theories 9.0 Introduction A. Lewis structures do not show one of the most important aspects of molecules- their overall shapes B. The shape and size of molecules-
Ch. 9- Molecular Geometry and Bonding Theories 9.0 Introduction A. Lewis structures do not show one of the most important aspects of molecules- their overall shapes B. The shape and size of molecules-
Energy. Position, x 0 L. Spectroscopy and the Particle-in-a-Box. Introduction
 Spectroscopy and the Particle-in-a-Box Introduction The majority of colors that we see result from transitions between electronic states that occur as a result of selective photon absorption. For a molecule
Spectroscopy and the Particle-in-a-Box Introduction The majority of colors that we see result from transitions between electronic states that occur as a result of selective photon absorption. For a molecule
CD Basis Set of Spectra that is used is that derived from comparing the spectra of globular proteins whose secondary structures are known from X-ray
 CD Basis Set of Spectra that is used is that derived from comparing the spectra of globular proteins whose secondary structures are known from X-ray crystallography An example of the use of CD Modeling
CD Basis Set of Spectra that is used is that derived from comparing the spectra of globular proteins whose secondary structures are known from X-ray crystallography An example of the use of CD Modeling
Protein Dynamics. The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron.
 Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Isothermal experiments characterize time-dependent aggregation and unfolding
 1 Energy Isothermal experiments characterize time-dependent aggregation and unfolding Technical ote Introduction Kinetic measurements have, for decades, given protein scientists insight into the mechanisms
1 Energy Isothermal experiments characterize time-dependent aggregation and unfolding Technical ote Introduction Kinetic measurements have, for decades, given protein scientists insight into the mechanisms
Department of Chemistry and Biochemistry University of Lethbridge. Biochemistry II. Bioenergetics
 Department of Chemistry and Biochemistry University of Lethbridge II. Bioenergetics Slide 1 Bioenergetics Bioenergetics is the quantitative study of energy relationships and energy conversion in biological
Department of Chemistry and Biochemistry University of Lethbridge II. Bioenergetics Slide 1 Bioenergetics Bioenergetics is the quantitative study of energy relationships and energy conversion in biological
Supplementary Figure 1 Interlayer exciton PL peak position and heterostructure twisting angle. a, Photoluminescence from the interlayer exciton for
 Supplementary Figure 1 Interlayer exciton PL peak position and heterostructure twisting angle. a, Photoluminescence from the interlayer exciton for six WSe 2 -MoSe 2 heterostructures under cw laser excitation
Supplementary Figure 1 Interlayer exciton PL peak position and heterostructure twisting angle. a, Photoluminescence from the interlayer exciton for six WSe 2 -MoSe 2 heterostructures under cw laser excitation
and Ultraviolet Spectroscopy
 Organic Chemistry, 7 th Edition L. G. Wade, Jr. Chapter 15 Conjugated Systems, Orbital Symmetry, and Ultraviolet Spectroscopy 2010, Prentice all Conjugated Systems Conjugated double bonds are separated
Organic Chemistry, 7 th Edition L. G. Wade, Jr. Chapter 15 Conjugated Systems, Orbital Symmetry, and Ultraviolet Spectroscopy 2010, Prentice all Conjugated Systems Conjugated double bonds are separated
Fluorescence (Notes 16)
 Fluorescence - 2014 (Notes 16) XV 74 Jablonski diagram Where does the energy go? Can be viewed like multistep kinetic pathway 1) Excite system through A Absorbance S 0 S n Excite from ground excited singlet
Fluorescence - 2014 (Notes 16) XV 74 Jablonski diagram Where does the energy go? Can be viewed like multistep kinetic pathway 1) Excite system through A Absorbance S 0 S n Excite from ground excited singlet
Circular Dichroism & Optical Rotatory Dispersion. Proteins (KCsa) Polysaccharides (agarose) DNA CHEM 305. Many biomolecules are α-helical!
 Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability
 Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Chapter 5. Nucleophilic aliphatic substitution mechanism. by G.DEEPA
 Chapter 5 Nucleophilic aliphatic substitution mechanism by G.DEEPA 1 Introduction The polarity of a carbon halogen bond leads to the carbon having a partial positive charge In alkyl halides this polarity
Chapter 5 Nucleophilic aliphatic substitution mechanism by G.DEEPA 1 Introduction The polarity of a carbon halogen bond leads to the carbon having a partial positive charge In alkyl halides this polarity
Chapter 4. Glutamic Acid in Solution - Correlations
 Chapter 4 Glutamic Acid in Solution - Correlations 4. Introduction Glutamic acid crystallises from aqueous solution, therefore the study of these molecules in an aqueous environment is necessary to understand
Chapter 4 Glutamic Acid in Solution - Correlations 4. Introduction Glutamic acid crystallises from aqueous solution, therefore the study of these molecules in an aqueous environment is necessary to understand
two slits and 5 slits
 Electronic Spectroscopy 2015January19 1 1. UV-vis spectrometer 1.1. Grating spectrometer 1.2. Single slit: 1.2.1. I diffracted intensity at relative to un-diffracted beam 1.2.2. I - intensity of light
Electronic Spectroscopy 2015January19 1 1. UV-vis spectrometer 1.1. Grating spectrometer 1.2. Single slit: 1.2.1. I diffracted intensity at relative to un-diffracted beam 1.2.2. I - intensity of light
Chapter 6. Chemical Bonding
 Chapter 6 Chemical Bonding Section 6.1 Intro to Chemical Bonding 6.1 Objectives Define chemical bond. Explain why most atoms form chemical bonds. Describe ionic and covalent bonding. Explain why most chemical
Chapter 6 Chemical Bonding Section 6.1 Intro to Chemical Bonding 6.1 Objectives Define chemical bond. Explain why most atoms form chemical bonds. Describe ionic and covalent bonding. Explain why most chemical
Kirkwood-Buff Integrals for Aqueous Urea Solutions Based upon the Quantum Chemical Electrostatic Potential and Interaction Energies
 Supporting Information for Kirkwood-Buff Integrals for Aqueous Urea Solutions Based upon the Quantum Chemical Electrostatic Potential and Interaction Energies Shuntaro Chiba, 1* Tadaomi Furuta, 2 and Seishi
Supporting Information for Kirkwood-Buff Integrals for Aqueous Urea Solutions Based upon the Quantum Chemical Electrostatic Potential and Interaction Energies Shuntaro Chiba, 1* Tadaomi Furuta, 2 and Seishi
12. Spectral diffusion
 1. Spectral diffusion 1.1. Spectral diffusion, Two-Level Systems Until now, we have supposed that the optical transition frequency of each single molecule is a constant (except when we considered its variation
1. Spectral diffusion 1.1. Spectral diffusion, Two-Level Systems Until now, we have supposed that the optical transition frequency of each single molecule is a constant (except when we considered its variation
Experiment Section Fig. S1 Fig. S2
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2018 Supplementary Materials Experiment Section The STM experiments were carried out in an ultrahigh
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2018 Supplementary Materials Experiment Section The STM experiments were carried out in an ultrahigh
Of Electrons, Energy, and Excitons
 Of,, and Gert van der Zwan July 17, 2014 1 / 35 Bacterial 2 / 35 Bacterial Bacterial LH1, LH2: excitonic interaction and energy transfer. RC, cytochromes: electron transfer reactions. Q, UQ: proton transfer
Of,, and Gert van der Zwan July 17, 2014 1 / 35 Bacterial 2 / 35 Bacterial Bacterial LH1, LH2: excitonic interaction and energy transfer. RC, cytochromes: electron transfer reactions. Q, UQ: proton transfer
I690/B680 Structural Bioinformatics Spring Protein Structure Determination by NMR Spectroscopy
 I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
The biomolecules of terrestrial life
 Functional groups in biomolecules Groups of atoms that are responsible for the chemical properties of biomolecules The biomolecules of terrestrial life Planets and Astrobiology (2017-2018) G. Vladilo 1
Functional groups in biomolecules Groups of atoms that are responsible for the chemical properties of biomolecules The biomolecules of terrestrial life Planets and Astrobiology (2017-2018) G. Vladilo 1
Chapter 13: Phenomena
 Chapter 13: Phenomena Phenomena: Scientists measured the bond angles of some common molecules. In the pictures below each line represents a bond that contains 2 electrons. If multiple lines are drawn together
Chapter 13: Phenomena Phenomena: Scientists measured the bond angles of some common molecules. In the pictures below each line represents a bond that contains 2 electrons. If multiple lines are drawn together
Solutions and Non-Covalent Binding Forces
 Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supporting Information Kinetic Resolution of Constitutional Isomers Controlled by Selective Protection inside a Supramolecular Nanocapsule Simin Liu, Haiying Gan, Andrew T. Hermann,
SUPPLEMENTARY INFORMATION Supporting Information Kinetic Resolution of Constitutional Isomers Controlled by Selective Protection inside a Supramolecular Nanocapsule Simin Liu, Haiying Gan, Andrew T. Hermann,
Effect of Electric Field on Condensed-Phase Molecular Systems. II. Stark Effect on the Hydroxyl Stretch Vibration of Ice
 Effect of Electric Field on Condensed-Phase Molecular Systems. II. Stark Effect on the Hydroxyl Stretch Vibration of Ice Sunghwan Shin, Hani Kang, Daeheum Cho, Jin Yong Lee, *, and Heon Kang *, Department
Effect of Electric Field on Condensed-Phase Molecular Systems. II. Stark Effect on the Hydroxyl Stretch Vibration of Ice Sunghwan Shin, Hani Kang, Daeheum Cho, Jin Yong Lee, *, and Heon Kang *, Department
Investigation of Cation Binding and Sensing by new Crown Ether core substituted Naphthalene Diimide systems
 Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 218 Supplementary Material Investigation
Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 218 Supplementary Material Investigation
pyridoxal phosphate synthase
 Supplementary Information 13 C-NMR snapshots of the complex reaction coordinate of pyridoxal phosphate synthase Jeremiah W. Hanes, Ivan Keresztes, and Tadhg P. Begley * Department of Chemistry and Chemical
Supplementary Information 13 C-NMR snapshots of the complex reaction coordinate of pyridoxal phosphate synthase Jeremiah W. Hanes, Ivan Keresztes, and Tadhg P. Begley * Department of Chemistry and Chemical
Vibronic Coupling in Quantum Wires: Applications to Polydiacetylene
 Vibronic Coupling in Quantum Wires: Applications to Polydiacetylene An Exhaustively Researched Report by Will Bassett and Cole Johnson Overall Goal In order to elucidate the absorbance spectra of different
Vibronic Coupling in Quantum Wires: Applications to Polydiacetylene An Exhaustively Researched Report by Will Bassett and Cole Johnson Overall Goal In order to elucidate the absorbance spectra of different
What dictates the rate of radiative or nonradiative excited state decay?
 What dictates the rate of radiative or nonradiative excited state decay? Transitions are faster when there is minimum quantum mechanical reorganization of wavefunctions. This reorganization energy includes
What dictates the rate of radiative or nonradiative excited state decay? Transitions are faster when there is minimum quantum mechanical reorganization of wavefunctions. This reorganization energy includes
EXAM INFORMATION. Radial Distribution Function: B is the normalization constant. d dx. p 2 Operator: Heisenberg Uncertainty Principle:
 EXAM INFORMATION Radial Distribution Function: P() r RDF() r Br R() r B is the normalization constant., p Operator: p ^ d dx Heisenberg Uncertainty Principle: n ax n! Integrals: xe dx n1 a x p Particle
EXAM INFORMATION Radial Distribution Function: P() r RDF() r Br R() r B is the normalization constant., p Operator: p ^ d dx Heisenberg Uncertainty Principle: n ax n! Integrals: xe dx n1 a x p Particle
What Is Organic Chemistry?
 What Is Organic Chemistry? EQ: What is Organic Chemistry? Read: pages 1-3 Answer the questions in your packet Basics of Organic Chem 1 Chapter 1: Structure and Bonding Key terms Organic Chemistry Inorganic
What Is Organic Chemistry? EQ: What is Organic Chemistry? Read: pages 1-3 Answer the questions in your packet Basics of Organic Chem 1 Chapter 1: Structure and Bonding Key terms Organic Chemistry Inorganic
Dave S. Walker and Geraldine L. Richmond*
 J. Phys. Chem. C 2007, 111, 8321-8330 8321 Understanding the Effects of Hydrogen Bonding at the Vapor-Water Interface: Vibrational Sum Frequency Spectroscopy of H 2 O/HOD/D 2 O Mixtures Studied Using Molecular
J. Phys. Chem. C 2007, 111, 8321-8330 8321 Understanding the Effects of Hydrogen Bonding at the Vapor-Water Interface: Vibrational Sum Frequency Spectroscopy of H 2 O/HOD/D 2 O Mixtures Studied Using Molecular
An Introduction to Metabolism
 An Introduction to Metabolism I. All of an organism=s chemical reactions taken together is called metabolism. A. Metabolic pathways begin with a specific molecule, which is then altered in a series of
An Introduction to Metabolism I. All of an organism=s chemical reactions taken together is called metabolism. A. Metabolic pathways begin with a specific molecule, which is then altered in a series of
Organic Chemistry Peer Tutoring Department University of California, Irvine
 Organic Chemistry Peer Tutoring Department University of California, Irvine Arash Khangholi (akhangho@uci.edu) Cassandra Amezquita (camezqu1@uci.edu) Jiana Machhor (jmachhor@uci.edu) OCHEM 51A Professor
Organic Chemistry Peer Tutoring Department University of California, Irvine Arash Khangholi (akhangho@uci.edu) Cassandra Amezquita (camezqu1@uci.edu) Jiana Machhor (jmachhor@uci.edu) OCHEM 51A Professor
Lecture 11: Protein Folding & Stability
 Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall Protein Folding: What we know. Protein Folding
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 3
 ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 3 ENZYMES AS BIOCATALYSTS * CATALYTIC EFFICIENCY *SPECIFICITY Having discussed
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 3 ENZYMES AS BIOCATALYSTS * CATALYTIC EFFICIENCY *SPECIFICITY Having discussed
Section 10/5/06. Junaid Malek, M.D.
 Section 10/5/06 Junaid Malek, M.D. Equilibrium Occurs when two interconverting states are stable This is represented using stacked arrows: State A State B, where State A and State B can represent any number
Section 10/5/06 Junaid Malek, M.D. Equilibrium Occurs when two interconverting states are stable This is represented using stacked arrows: State A State B, where State A and State B can represent any number
with the larger dimerization energy also exhibits the larger structural changes.
 A7. Looking at the image and table provided below, it is apparent that the monomer and dimer are structurally almost identical. Although angular and dihedral data were not included, these data are also
A7. Looking at the image and table provided below, it is apparent that the monomer and dimer are structurally almost identical. Although angular and dihedral data were not included, these data are also
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
Zn(II) and Cd(II) based complexes for probing the enzymatic hydrolysis of Na 4 P 2 O 7 by Alkaline phosphatase in physiological condition
 Supplementary information Zn(II) and Cd(II) based complexes for probing the enzymatic hydrolysis of Na 4 P 2 O 7 by Alkaline phosphatase in physiological condition Priyadip Das, Sourish Bhattacharya, Sandhya
Supplementary information Zn(II) and Cd(II) based complexes for probing the enzymatic hydrolysis of Na 4 P 2 O 7 by Alkaline phosphatase in physiological condition Priyadip Das, Sourish Bhattacharya, Sandhya
Chapter 15 Dienes, Resonance, and Aromaticity
 Instructor Supplemental Solutions to Problems 2010 Roberts and Company Publishers Chapter 15 Dienes, Resonance, and Aromaticity Solutions to In-Text Problems 15.2 The delocalization energy is the energy
Instructor Supplemental Solutions to Problems 2010 Roberts and Company Publishers Chapter 15 Dienes, Resonance, and Aromaticity Solutions to In-Text Problems 15.2 The delocalization energy is the energy
Bio10 Cell and Molecular Lecture Notes SRJC
 Basic Chemistry Atoms Smallest particles that retain properties of an element Made up of subatomic particles: Protons (+) Electrons (-) Neutrons (no charge) Isotopes Atoms of an element with different
Basic Chemistry Atoms Smallest particles that retain properties of an element Made up of subatomic particles: Protons (+) Electrons (-) Neutrons (no charge) Isotopes Atoms of an element with different
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Information Figure S1: (a) Initial configuration of hydroxyl and epoxy groups used in the MD calculations based on the observations of Cai et al. [Ref 27 in the
SUPPLEMENTARY INFORMATION Supplementary Information Figure S1: (a) Initial configuration of hydroxyl and epoxy groups used in the MD calculations based on the observations of Cai et al. [Ref 27 in the
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
Chapter 13. Conjugated Unsaturated Systems. +,., - Allyl. What is a conjugated system? AllylicChlorination (High Temperature)
 What is a conjugated system? Chapter 13 Conjugated Unsaturated Systems Conjugated unsaturated systems have a p orbital on a carbon adjacent to a double bond The p orbital may be empty (a carbocation The
What is a conjugated system? Chapter 13 Conjugated Unsaturated Systems Conjugated unsaturated systems have a p orbital on a carbon adjacent to a double bond The p orbital may be empty (a carbocation The
Supplementary Figure 1: Power dependence of hot-electrons reduction of 4-NTP to 4-ATP. a) SERS spectra of the hot-electron reduction reaction using
 Supplementary Figure 1: Power dependence of hot-electrons reduction of 4-NTP to 4-ATP. a) SERS spectra of the hot-electron reduction reaction using 633 nm laser excitation at different powers and b) the
Supplementary Figure 1: Power dependence of hot-electrons reduction of 4-NTP to 4-ATP. a) SERS spectra of the hot-electron reduction reaction using 633 nm laser excitation at different powers and b) the
Chem 3502/4502 Physical Chemistry II (Quantum Mechanics) 3 Credits Spring Semester Christopher J. Cramer. Lecture 30, April 10, 2006
 Chem 3502/4502 Physical Chemistry II (Quantum Mechanics) 3 Credits Spring Semester 20056 Christopher J. Cramer Lecture 30, April 10, 2006 Solved Homework The guess MO occupied coefficients were Occupied
Chem 3502/4502 Physical Chemistry II (Quantum Mechanics) 3 Credits Spring Semester 20056 Christopher J. Cramer Lecture 30, April 10, 2006 Solved Homework The guess MO occupied coefficients were Occupied
Chapter 02 Chemical Basis of Life. Multiple Choice Questions
 Seeleys Essentials of Anatomy and Physiology 8th Edition VanPutte Test Bank Full Download: http://testbanklive.com/download/seeleys-essentials-of-anatomy-and-physiology-8th-edition-vanputte-test-bank/
Seeleys Essentials of Anatomy and Physiology 8th Edition VanPutte Test Bank Full Download: http://testbanklive.com/download/seeleys-essentials-of-anatomy-and-physiology-8th-edition-vanputte-test-bank/
Benzene a remarkable compound. Chapter 14 Aromatic Compounds. Some proposed structures for C 6 H 6. Dimethyl substituted benzenes are called xylenes
 Benzene a remarkable compound Chapter 14 Aromatic Compounds Discovered by Faraday 1825 Formula C 6 H 6 Highly unsaturated, but remarkably stable Whole new class of benzene derivatives called aromatic compounds
Benzene a remarkable compound Chapter 14 Aromatic Compounds Discovered by Faraday 1825 Formula C 6 H 6 Highly unsaturated, but remarkably stable Whole new class of benzene derivatives called aromatic compounds
Chemistry Review. Structure of an Atom. The six most abundant elements of life. Types of chemical bonds. U n i t 2 - B i o c h e m i s t r y
 Chemistry Review Structure of an Atom are organized into shells or levels around the nucleus. Atoms are most stable when their outer or valence shell is. The six most abundant elements of life Types of
Chemistry Review Structure of an Atom are organized into shells or levels around the nucleus. Atoms are most stable when their outer or valence shell is. The six most abundant elements of life Types of
Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013
 Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013 Lecture 10. Biochemical Transformations II. Phosphoryl transfer and the kinetics and thermodynamics of energy currency in the cell: ATP and GTP.
Chemistry 5.07SC Biological Chemistry I Fall Semester, 2013 Lecture 10. Biochemical Transformations II. Phosphoryl transfer and the kinetics and thermodynamics of energy currency in the cell: ATP and GTP.
Using Molecular Dynamics to Compute Properties CHEM 430
 Using Molecular Dynamics to Compute Properties CHEM 43 Heat Capacity and Energy Fluctuations Running an MD Simulation Equilibration Phase Before data-collection and results can be analyzed the system
Using Molecular Dynamics to Compute Properties CHEM 43 Heat Capacity and Energy Fluctuations Running an MD Simulation Equilibration Phase Before data-collection and results can be analyzed the system
Homework Problem Set 4 Solutions
 Chemistry 380.37 Dr. Jean M. Standard omework Problem Set 4 Solutions 1. A conformation search is carried out on a system and four low energy stable conformers are obtained. Using the MMFF force field,
Chemistry 380.37 Dr. Jean M. Standard omework Problem Set 4 Solutions 1. A conformation search is carried out on a system and four low energy stable conformers are obtained. Using the MMFF force field,
Photoelectron Spectroscopy of the Hydroxymethoxide Anion, H 2 C(OH)O
 Supplementary Material for: Photoelectron Spectroscopy of the Hydroxymethoxide Anion, H 2 C(OH)O Allan M. Oliveira, Julia H. Lehman, Anne B. McCoy 2 and W. Carl Lineberger JILA and Department of Chemistry
Supplementary Material for: Photoelectron Spectroscopy of the Hydroxymethoxide Anion, H 2 C(OH)O Allan M. Oliveira, Julia H. Lehman, Anne B. McCoy 2 and W. Carl Lineberger JILA and Department of Chemistry
Protein Structure. W. M. Grogan, Ph.D. OBJECTIVES
 Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
CHAPTER 6 STUDY GUIDE. phosphate work. energy adenosine In order for organisms to carry out life processes their cells need (1).
 CHAPTER 6 STUDY GUIDE THE FLOW OF ENERGY Section 6.1 Energy for Cells In your textbook, read about ATP. Use each of the terms below just once to complete the passage: released exergonic endergonic ATP
CHAPTER 6 STUDY GUIDE THE FLOW OF ENERGY Section 6.1 Energy for Cells In your textbook, read about ATP. Use each of the terms below just once to complete the passage: released exergonic endergonic ATP
2. Polar Covalent Bonds: Acids and Bases
 2. Polar Covalent Bonds: Acids and Bases Based on McMurry s Organic Chemistry, 6 th edition, Chapter 2 2003 Ronald Kluger Department of Chemistry University of Toronto 2.1 Polar Covalent Bonds: Electronegativity!
2. Polar Covalent Bonds: Acids and Bases Based on McMurry s Organic Chemistry, 6 th edition, Chapter 2 2003 Ronald Kluger Department of Chemistry University of Toronto 2.1 Polar Covalent Bonds: Electronegativity!
Section 3 Electronic Configurations, Term Symbols, and States
 Section 3 Electronic Configurations, Term Symbols, and States Introductory Remarks- The Orbital, Configuration, and State Pictures of Electronic Structure One of the goals of quantum chemistry is to allow
Section 3 Electronic Configurations, Term Symbols, and States Introductory Remarks- The Orbital, Configuration, and State Pictures of Electronic Structure One of the goals of quantum chemistry is to allow
