Molecular Dynamics Simulation of Methanol-Water Mixture
|
|
|
- Dale Pearson
- 5 years ago
- Views:
Transcription
1 Molecular Dynamics Simulation of Methanol-Water Mixture Palazzo Mancini, Mara Cantoni University of Urbino Carlo Bo Abstract In this study some properties of the methanol-water mixture such as diffusivity, density, viscosity, and hydrogen bonding were calculated at different temperatures and atmospheric pressure using molecular dynamics simulations (MDS). The results were compared with the available experimental data as well as some theoretical models; overall indicating a good agreement. This shows the useful and effective application of MDS for determination of physical properties. Keywords: Molecular dynamics simulation, Einstein equation, Diffusion coefficient, Hydrogen bond - Introduction As molecular simulations are considered useful tools for the interpretation experimental data, two methods namely Monte Carlo (MC) and Molecular Dynamics (MD), can be applied. This allows us to determine macroscopic properties by evaluating exactly a theoretical model of molecular behavior using a program. [, 2] of computer Molecular simulation considers small size systems, at a typical scale of a few nanometers, and determines their behaviour from a careful computation of the interactions between their components. These computations take much more computer time than classical thermodynamic models (i. e. from a few hours to several weeks of computing time, depending on system size). As such, molecular simulation is now considered in the chemical industry as capable of filling the existing gap between experimental data and engineering models in various circumstances such as unknown chemicals, extreme conditions of temperature or pressure, and toxic compounds. The first part of this article provides a brief description of Molecular dynamic simulation
2 and how it can be analyzed for approving experimental data. In the second part, different properties of the system will be calculated by molecular dynamic simulation. Results will then be compared with available experimental data and theoretical models. 2- Molecular dynamics method 2.- What is molecular dynamics method? Molecular dynamics simulation involves integration of Newtonís equation of motion for each atom in an ensemble in order to generate information on the variation in the position, velocity, and acceleration of each particle as a function of time. The equations solved are [3]: covalently bonded atoms and the last term describes the non-bonded interactions (van der Waals and electrostatic). The constants k b, k Ë, and kˆ are force constants for the covalent bonds, bond angles, and dihedrals, respectively, r ij are the distances between atoms i and j, A ij and B ij are the Lenard Jones parameters, and q is a partial charge [3]. To calculate the properties of a real system, it is convenient to define an ensemble. An ensemble can be regarded as an imaginary collection of a very large number of systems in different quantum states with common macroscopic attributes. The ensemble average of any dynamic property (A) can be obtained from the relationship: f i m i a i () A A i p i i (4) f U i r i (2) Where f i is the force exerted on the i th atom, m i is its mass, and a i is its acceleration. The force acting on each atom is obtained from a potential function U, such as that depicted below [3]: Where A i is the value of A in quantum state i, p i represents the probability of observing the i-th state, and the angled brackets denote an ensemble average. The time averaged properties of the real system are related to the ensemble average by invoking the following assumption: A k b ij ij t A (5) k ijk ijk ij U (r r eq ) 2 ijk ( eq ) 2 2 angle 2 The above equivalence of the time ñ average k cos(n( eq and ensemble ñ average is called the Ergodic )) dihedrals hypothesis. Form of p i is determined by the A macroscopic properties that are common to ij B ij q iq i all of the systems of the ensemble. 2 6 i j r r ij ij 4 r ij (3) 2.2- Calculation of physical properties Prediction of transport properties of fluids of Where the first three terms reflect industrial interest is one of the main intramolecular interactions between motivations to perform MD simulations [4].
3 As it is not possible to observe individual atoms or molecules directly, various models are used to describe and/or predict the properties of a system with molecular V dynamic methods. On the other hand, it is k T possible to calculate macroscopic properties i Also shear viscosity can be calculated using the following equation: B xy (t) xy () dt (8) of the mixtures by calculating the interaction Where xy is the pressure tensor of the of the molecules. The partial density is one components. k B is the Boltzman constant and macroscopic property that could be calculated using molecular dynamics simulation. Also transport properties like the diffusion coefficient and shear viscosity can be computed directly from an MD simulation too [5]. In an equilibrium molecular dynamics (EMD) simulation, the self- diffusion coefficient D S α of component α is computed by taking the slope of the mean square displacement (MSD) at a long period of time: D S lim N 2dN t T is the temperature of the system GROMACS Software GROMACS is an acronym for GROningen MAchine for Chemical Simulation. It is the latest release of a versatile and very well known optimized package for molecular simulation. The package is a collection of programs and libraries for the simulation of molecular dynamics and the subsequent analysis of trajectory data. Although it is primarily targeted at biological molecules (r (t) r ()) 2 (6) with complex bonded interactions, the very i i Where N α is the number of molecules of component α, d is the spatial dimension of the system, t is the time, and r α i is the center of mass of molecule i of component α. Equivalently, D S α is given by the time integral of the velocity autocorrelation function: D S N i effective implementation of nonbonded force calculations makes GROMACS suitable for all kinds of molecular dynamics simulations based on pair potentials. Apart from normal potential functions like Lenard-Jones, Buckingham and Coulomb, it is possible to use arbitrary forms of interactions with spline-interpolated tables [7]. The GROMACS molecular dynamics code contains several algorithms that have been v (t)v () (7) developed in the Berendsen group. The leap- dn i i i Where v is the center of mass velocity of molecule i of component α. Equation (6) is known as the Einstein equation and equation (7) is often referred to as the Green-Kubo relation. [6] frog integrator is used in two versions, for molecular dynamics and stochastic dynamics [8]. 3. Result and Discussion 3.-System definition In Gromacs software, the system is defined
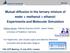
4 by its size and shape, the number and types of molecules it contains, and the coordinates and velocities of all atoms. Velocities may also be generated from a Maxwellian distribution [9]. In this study MD simulation was used to examine some physical and transport properties of the mixture of methanol ñ water. This system consists of 26 molecules of water and 26 molecules of methanol. Simulations are carried out using a fully atomistic system with the maximum size 4.6 * 2.3 *2.3 nm. The box is shown in Fig.. Gromacs software was used for the MD calculations. Simulation was performed using GROMOS 96 [, ] force field for 2 ps. MD simulations are performed at atmospheric pressure with four different temperatures 273, 283, 288, 293 k at the isothermal-isobaric ensemble (NPT). Table shows the Lenard-Jones parameters and partial charge for methanol molecule. Water molecules are modeled as SPC water model with their related parameters [] Diffusion coefficient of the methanol in the mixture As the first step, diffusion coefficient of methanol molecules in a mixture methanol-water has been studied by MD method. For this reason mean of square displacement (MSD) of methanol molecules in the mixture was calculated by Gromacs software. Fig. 2 shows MSD values versus time for methanol molecules diffusing in the methanol-water mixture at four temperatures 273, 283, 288 and 293 k. These graphs show that the behavior of MSD versus time for <t<2 ps is linear, so Einstein equation can describe the diffusion behavior of methanol. Calculated Diffusion coefficients of methanol in the mixture by Einstein equation are shown in Table and Fig. 3. Figure. Computer generated image of a methanol-water box
5 Diffusion Coefficient * ^5 (Cm^2/s) RMSD (nm^2) Table. The partial charge and Lenard-Jones parameters for methanol molecule [] Atom type Charge Lenard Jones parameters C6 C2 CH e e-6 O e e-6 H.4 Root Mean Square Deviation Displacement (RMSD) K 288 K 283 K 273 K time (ps) Figure 2. Root Mean square displacement of methanol molecules at different temperatures Calculated Diffusion Coefficient 3 MD Cal. Wilke-Chang chang Eq. Eq Temperature Figure 3. The effect of temperature on diffusion coefficient.
6 To evaluate the calculated results, the diffusion coefficient of this mixture was calculated by Wilke- Chang equation [2]. Results are shown in Table and Fig 3: D AB (7.3 8 )( M ) / 2 T 2%, while this is about % between experimental value and Eq Calculation of density In the second step, the density of methanol- water mixture was calculated at different B (9) temperatures and different points in the box. V A.6 The available experimental data for diffusion coefficient of methanol in water are shown in Table 2 [3]. The averaged error between MD results and experimental value is about The results are shown in Table 3 and Fig. 4 and 5. The comparison between experimental [3] and calculated results shows a good agreement between these data and the averaged error is less than %. Table 2. Calculated Diffusion Coefficient of Methanol in Water by MD method Diffusion coefficient Diffusion coefficient Experimental Temperature (K) by MD (cm 2 /s) by Eq. 9 (cm 2 /s) value [] * * * -5.8* * -5.42* * * -5.6* -5.8 * -5 Table 3. Calculated and experimental density in different temperatures Temperature (K) Cal. Density (kg/m^3) Exp. Density (kg/m^3) [] Error %
7 Density (kg /m^3) Density (kg/m^3) Calculated and Experimental Density MD EXP Temperature (K) 3 Figure 4. The effect of temperature on density for methanol-water mixture Local Density in the box T=273 K T=283 K T=288 K T= 293 K z Box axis (nm(nm) ).5 2 Figure 5. Calculated density in different points in the box by MD method 3.4- Shear viscosity One of the important properties of the mixtures is viscosity. It is possible to calculate this macroscopic property of the mixture by calculating the interaction of the molecules in microscopic scale. Here the viscosity of the methanol-water mixture was calculated by MD simulation and the results are compared with experimental data. As it was shown in Table 4 and Fig. 7, viscosity is decreased by increasing temperature, the same as experimental value.
8 viscosity (cp) Viscosity (cp) Mixture Viscosity K 283 K 288 K 293 K 3 K time (ps) Figure 6. Viscosity of the methanol-water mixture at different temperatures Table 4. Average calculated and experimental viscosity of the methanol-water mixture Temperature 273 K 283 K 288 K 293 K 3 K MD Calculated value Experimental value [2] Error % Experimental & Calculated viscosity of the mixture.5 MD Exp temperature (K) Figure 7. The effect of temperature on viscosity for methanol-water mixture
9 Probability 3.5- Hydrogen Bond Many thermodynamical properties (e.g., melting and boiling point) of the mixture depend on the strength of the hydrogen bonds (H-bond) formed among molecules. The H-bond is a special attractive interaction parameter that exists between an electronegative atom and a hydrogen atom bonded to another electronegative atom. Since water has two hydrogen atoms and one oxygen atom, it can form an H-bond between the Oxygen and the H atoms. Each water molecule can form up to four hydrogen bonds at the same time (two through its two lone pairs, and two through its two hydrogen atoms). The average number of H-bonds per molecule calculated in bulk water varies approximately from 2.3 to 3.8 according to the water model and the way used to define the H-bond. [4, 5] In this study the number of hydrogen atoms between water molecules and water-methanol molecules were calculated. The results, shown in Figs. 8 and 9, indicate that the number of hydrogen bonds between water molecules is from 2 to 3 but it decreases for water and methanol molecules down to. The hydrogen bond distribution is also plotted in the box, where the most probable ones form near the center, Fig. 4. Conclusion In this paper molecular dynamic theoretical method is presented to calculate density, shear viscosity, hydrogen bonding and diffusion coefficient for the mixture of methanol-water. Gromacs software was used for this purpose. The simulation was performed up to 2 ps. The results in Table and Fig. 3 clearly show that the calculated diffusion coefficient by MD method indicate a good agreement and an almost similar trend with those obtained from the Wilke-Chang equation, both of which are approved by experimental data. These values, however increase linearly with temperature (D~T). Hydrogen bond of water molecules K 283 K 288 K 293 K 3 K time (ps) Figure 8. Hydrogen bond of water molecules in the mixture
10 Probability Probability Hydrogen bond of the mixture K 283 K 288 K 293 K 3 K.5 5 time (ps) 5 2 Figure 9. Hydrogen bond of water and methanol molecules in the mixture Hydrogen Bond Distribution K 283 K 288 K 293 K z Distance axis (nm) (nm) Figure. Hydrogen bond distribution of methanol-water mixture in the box Density and viscosity similar to that of diffusion coefficient are affected by temperature. The density decreases with temperature with a decreasing deviation between MD and experimental data. This shows that MD is much more reliable at higher temperatures. In the case of viscosity, it also decreases with temperature with a decreasing deviation. After that, it decreases but with a lower deviation of MD with those of the experimental data. The reference temperature for MD calculation in the input files of the simulation is 3 K, so some of the calculated properties such as viscosity and density at the points near this temperature have the minimum deviation from the experimental data. The MD calculation for pure water in this study estimated H-bond numbering between 2 to 3 which is in a good agreement with the previous studies [4-8]. This number decreases considerably for the case of water-methanol molecules. This is due to the lower polarity of methanol molecules, compared to
11 that of water molecules. Hydrogen bond formation has the most distribution probability at the center of the box and the temperature has no effect on distribution. In the center of the box, there are water and methanol molecules in all the directions, so the hydrogen bond has the maximum distribution in this part of the system, but in the points near the sidewall of the system the number of molecules decreases so the hydrogen bonding has the minimum distribution in these points. Overall, the results show the reliability of the MD method to calculate the physical properties of the systems. One can conclude that, this method may be used to determine the macroscopic properties of the systems, instead of practical methods in the laboratory. Nomenclature T Temperature (K) M B Solvent molecular weight (kg/kmol) Solvent accumulation coefficient (-) V A Viscosity (kg/m.s) solute molar volume at boiling point (m 3 /kmol) References [] Monticella, L., Kandasamy, S. K., Periole, X., Larson, R. G., The MARTINI coarse grained force field: extension to proteins., J. Chem. Theory and Comput., 4, , (28). [2] Sadus., R. J., Molecular simulation of fluid, theory, algorithms and object-orientation., Elsevier Science B.V., (999). [3] Nobakht, A. Y., M. Shahsavan, and A. Paykani. "Numerical study of diodicity mechanism in different Tesla-type microvalves." Journal of applied research and technology.6 (23): [4] Ungerer, P., Nieto-Draghi, C., Rousseau, B., Ahunbay, G., Lachet, V., Molecular simulation of the thermophysical properties of fluids: From understanding toward quantitative predictions, Journal of Molecular Liquids, 34, 7 89, (27). [5] Krishna, R., Wesselingh, J. A., The Maxwell-Stefan approach to mass transfer, Chem. Eng. Sci., 52, 86-9, (997). [6] Dubbeldam, D., Snurr, R. Q., Recent developments in the molecular modeling of diffusion in nanoporous materials, Molecular Simulation Journal, Vol. 33, (27). [7] Lindahl, E., Hess, B. and Van der Spoel, D. GROMACS 3.: a package for molecular simulation and trajectory analysis, J Mol Model, 7:36 37, (2). [8] Van Gunsteren, WF, Berendsen, HJC, A leap frog-algorithm for stochastic dynamics, Mol Simul :73 85, (988). [9] Spoel, D.V., Lindahl, E., Hess, B., Groenhof, G., Mark, A., Berendsen, H.J.C., GROMACS: Fast, Flexible, and Free., J. Comput. Chem., 26, 7 78, (25). [] Gunsteren W.F., Krugerg, V. and Berendsen, J.C., Computer Simulation of molecular dynamics methodology, applied and perspectives in chemistry, Angew Chem Int Ed Engl, 29, , (99). [] Gunsteren W.F., Krugerg, V. and Brilleter, P., SR. and et al, The GROMOS 96 manual and user guide, Biomos and Hochschulverlag AG an der, ETH ZA/4rich, Groningen, (996). [2] Treybal., R., Mass transfer phenomena, McGraw-Hill Company, third ed., (89). [3] Perry, R. H., Perry s chemical engineer s handbook, McGraw-Hill Company, fifth ed., (984). [4] Nobakht, Ali Yousefzadi, et al. "Heat flow diversion in supported graphene nanomesh." Carbon (27). [5] Jorgensen, W. L., Madura, J. D. Optimized Intermolecular Potential Functions for liquid Hydrocarbons, Mol. Phys. 985, 56, (38). [6] Zielkiewicz, J. Structural properties of water: Comparison of the SPC, SPCE, TIP4P, and TIP5P models of water, J. Chem. Phys, 23, 45, (25). [7] Yousefzadi Nobakht, Ali, and Seungha Shin. "Anisotropic control of thermal transport in graphene/si heterostructures." Journal of Applied Physics 2.22 (26): 225.
Molecular Dynamics Simulations. Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia
 Molecular Dynamics Simulations Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia 1 An Introduction to Molecular Dynamics Simulations Macroscopic properties
Molecular Dynamics Simulations Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia 1 An Introduction to Molecular Dynamics Simulations Macroscopic properties
MD Thermodynamics. Lecture 12 3/26/18. Harvard SEAS AP 275 Atomistic Modeling of Materials Boris Kozinsky
 MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
Analysis of the simulation
 Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Lecture 18 - Covalent Bonding. Introduction. Introduction. Introduction. Introduction
 Chem 103, Section F0F Unit VII - States of Matter and Intermolecular Interactions Lecture 19 Physical states and physical changes Description of phase changes Intermolecular interactions Properties of
Chem 103, Section F0F Unit VII - States of Matter and Intermolecular Interactions Lecture 19 Physical states and physical changes Description of phase changes Intermolecular interactions Properties of
 1 Points to Remember Subject: Chemistry Class: XI Chapter: States of matter Top concepts 1. Intermolecular forces are the forces of attraction and repulsion between interacting particles (atoms and molecules).
1 Points to Remember Subject: Chemistry Class: XI Chapter: States of matter Top concepts 1. Intermolecular forces are the forces of attraction and repulsion between interacting particles (atoms and molecules).
An introduction to Molecular Dynamics. EMBO, June 2016
 An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
Introduction to molecular dynamics
 1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
Thermodynamic behaviour of mixtures containing CO 2. A molecular simulation study
 Thermodynamic behaviour of mixtures containing. A molecular simulation study V. Lachet, C. Nieto-Draghi, B. Creton (IFPEN) Å. Ervik, G. Skaugen, Ø. Wilhelmsen, M. Hammer (SINTEF) Introduction quality issues
Thermodynamic behaviour of mixtures containing. A molecular simulation study V. Lachet, C. Nieto-Draghi, B. Creton (IFPEN) Å. Ervik, G. Skaugen, Ø. Wilhelmsen, M. Hammer (SINTEF) Introduction quality issues
Molecular modeling. A fragment sequence of 24 residues encompassing the region of interest of WT-
 SUPPLEMENTARY DATA Molecular dynamics Molecular modeling. A fragment sequence of 24 residues encompassing the region of interest of WT- KISS1R, i.e. the last intracellular domain (Figure S1a), has been
SUPPLEMENTARY DATA Molecular dynamics Molecular modeling. A fragment sequence of 24 residues encompassing the region of interest of WT- KISS1R, i.e. the last intracellular domain (Figure S1a), has been
Diffusion of Water and Diatomic Oxygen in Poly(3-hexylthiophene) Melt: A Molecular Dynamics Simulation Study
 Diffusion of Water and Diatomic Oxygen in Poly(3-hexylthiophene) Melt: A Molecular Dynamics Simulation Study Julia Deitz, Yeneneh Yimer, and Mesfin Tsige Department of Polymer Science University of Akron
Diffusion of Water and Diatomic Oxygen in Poly(3-hexylthiophene) Melt: A Molecular Dynamics Simulation Study Julia Deitz, Yeneneh Yimer, and Mesfin Tsige Department of Polymer Science University of Akron
Mutual diffusion of binary liquid mixtures containing methanol, ethanol, acetone, benzene, cyclohexane, toluene and carbon tetrachloride
 - 1 - PLMMP 2016, Kyiv Mutual diffusion of binary liquid mixtures containing methanol, ethanol, acetone, benzene, cyclohexane, toluene and carbon tetrachloride Jadran Vrabec Tatjana Janzen, Gabriela Guevara-Carrión,
- 1 - PLMMP 2016, Kyiv Mutual diffusion of binary liquid mixtures containing methanol, ethanol, acetone, benzene, cyclohexane, toluene and carbon tetrachloride Jadran Vrabec Tatjana Janzen, Gabriela Guevara-Carrión,
Peptide folding in non-aqueous environments investigated with molecular dynamics simulations Soto Becerra, Patricia
 University of Groningen Peptide folding in non-aqueous environments investigated with molecular dynamics simulations Soto Becerra, Patricia IMPORTANT NOTE: You are advised to consult the publisher's version
University of Groningen Peptide folding in non-aqueous environments investigated with molecular dynamics simulations Soto Becerra, Patricia IMPORTANT NOTE: You are advised to consult the publisher's version
DIFFERENT TYPES OF INTEMOLECULAR FORCES INTERMOLECULAR FORCES
 DIFFERENT TYPES OF INTEMOLECULAR FORCES Do all the exercises in your studyguide COMPARISON OF THE THREE PHASES OF MATTER. Matter is anything that occupy space and has mass. There are three states of matter:
DIFFERENT TYPES OF INTEMOLECULAR FORCES Do all the exercises in your studyguide COMPARISON OF THE THREE PHASES OF MATTER. Matter is anything that occupy space and has mass. There are three states of matter:
Analysis of MD trajectories in GROMACS David van der Spoel
 Analysis of MD trajectories in GROMACS David van der Spoel What does MD produce? Energy terms E(t) Coordinates x(t) Velocities v(t) Forces f(t) Managing your files trjcat - merging trajectories concatenating
Analysis of MD trajectories in GROMACS David van der Spoel What does MD produce? Energy terms E(t) Coordinates x(t) Velocities v(t) Forces f(t) Managing your files trjcat - merging trajectories concatenating
Molecular Dynamic Simulation Study of the Volume Transition of PNIPAAm Hydrogels
 Molecular Dynamic Simulation Study of the Volume Transition of PNIPAAm Hydrogels Jonathan Walter 1, Jadran Vrabec 2, Hans Hasse 1 1 Laboratory of Engineering, University of Kaiserslautern, Germany 2 and
Molecular Dynamic Simulation Study of the Volume Transition of PNIPAAm Hydrogels Jonathan Walter 1, Jadran Vrabec 2, Hans Hasse 1 1 Laboratory of Engineering, University of Kaiserslautern, Germany 2 and
What is Classical Molecular Dynamics?
 What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
 A Nobel Prize for Molecular Dynamics and QM/MM What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential
A Nobel Prize for Molecular Dynamics and QM/MM What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential
States of Matter. Intermolecular Forces. The States of Matter. Intermolecular Forces. Intermolecular Forces
 Intermolecular Forces Have studied INTRAmolecular forces the forces holding atoms together to form compounds. Now turn to forces between molecules INTERmolecular forces. Forces between molecules, between
Intermolecular Forces Have studied INTRAmolecular forces the forces holding atoms together to form compounds. Now turn to forces between molecules INTERmolecular forces. Forces between molecules, between
Supporting Information
 Supporting Information Structure and Dynamics of Uranyl(VI) and Plutonyl(VI) Cations in Ionic Liquid/Water Mixtures via Molecular Dynamics Simulations Katie A. Maerzke, George S. Goff, Wolfgang H. Runde,
Supporting Information Structure and Dynamics of Uranyl(VI) and Plutonyl(VI) Cations in Ionic Liquid/Water Mixtures via Molecular Dynamics Simulations Katie A. Maerzke, George S. Goff, Wolfgang H. Runde,
Investigations of Freezing Pure Water
 Investigations of Freezing Pure Water David Meldgin Constanze Kalcher May 2, 2013 Abstract We use a the molecular simulation package LAAMPS to simulate the freezing of water. We analyze the SPC and TIP3P
Investigations of Freezing Pure Water David Meldgin Constanze Kalcher May 2, 2013 Abstract We use a the molecular simulation package LAAMPS to simulate the freezing of water. We analyze the SPC and TIP3P
Dipole-Dipole Interactions https://www.youtube.com/watch?v=cerb1d6j4-m London Dispersion Forces https://www.youtube.com/watch?
 CATALYST Lesson Plan GLE Physical Science 22. Predict the kind of bond that will form between two elements based on electronic structure and electronegativity of the elements (e.g., ionic, polar, nonpolar)
CATALYST Lesson Plan GLE Physical Science 22. Predict the kind of bond that will form between two elements based on electronic structure and electronegativity of the elements (e.g., ionic, polar, nonpolar)
Introduction to Molecular Dynamics
 Introduction to Molecular Dynamics Dr. Kasra Momeni www.knanosys.com Overview of the MD Classical Dynamics Outline Basics and Terminology Pairwise interacting objects Interatomic potentials (short-range
Introduction to Molecular Dynamics Dr. Kasra Momeni www.knanosys.com Overview of the MD Classical Dynamics Outline Basics and Terminology Pairwise interacting objects Interatomic potentials (short-range
Electonegativity, Polar Bonds, and Polar Molecules
 Electonegativity, Polar Bonds, and Polar Molecules Some Definitions Electronegativity: the ability of an atom to attract bonding electrons to itself. Intramolecular forces: the attractive force between
Electonegativity, Polar Bonds, and Polar Molecules Some Definitions Electronegativity: the ability of an atom to attract bonding electrons to itself. Intramolecular forces: the attractive force between
Lecture Presentation. Chapter 11. Liquids and Intermolecular Forces. John D. Bookstaver St. Charles Community College Cottleville, MO
 Lecture Presentation Chapter 11 Liquids and Intermolecular Forces John D. Bookstaver St. Charles Community College Cottleville, MO Properties of Gases, Liquids, and Solids State Volume Shape of State Density
Lecture Presentation Chapter 11 Liquids and Intermolecular Forces John D. Bookstaver St. Charles Community College Cottleville, MO Properties of Gases, Liquids, and Solids State Volume Shape of State Density
Intermolecular Forces of Attraction
 Name Unit Title: Covalent Bonding and Nomenclature Text Reference: Pages 189-193 Date Intermolecular Forces of Attraction Intramolecular vs. Intermolecular So far in our discussion of covalent bonding,
Name Unit Title: Covalent Bonding and Nomenclature Text Reference: Pages 189-193 Date Intermolecular Forces of Attraction Intramolecular vs. Intermolecular So far in our discussion of covalent bonding,
Chapters 11 and 12: Intermolecular Forces of Liquids and Solids
 1 Chapters 11 and 12: Intermolecular Forces of Liquids and Solids 11.1 A Molecular Comparison of Liquids and Solids The state of matter (Gas, liquid or solid) at a particular temperature and pressure depends
1 Chapters 11 and 12: Intermolecular Forces of Liquids and Solids 11.1 A Molecular Comparison of Liquids and Solids The state of matter (Gas, liquid or solid) at a particular temperature and pressure depends
Calculation of Physical Properties of the Methanol-Water Mixture Using Molecular Dynamics Simulation
 Iranan Journal of Chemcal Engneerng Vol. 6, No. 4 (Autumn), 29, IAChE Calculaton of Physcal Propertes of the Methanol-Water Mxture Usng Molecular Dynamcs Smulaton N. Farhadan, M. Sharaty-Nassar 2 - Academc
Iranan Journal of Chemcal Engneerng Vol. 6, No. 4 (Autumn), 29, IAChE Calculaton of Physcal Propertes of the Methanol-Water Mxture Usng Molecular Dynamcs Smulaton N. Farhadan, M. Sharaty-Nassar 2 - Academc
Biomolecules are dynamic no single structure is a perfect model
 Molecular Dynamics Simulations of Biomolecules References: A. R. Leach Molecular Modeling Principles and Applications Prentice Hall, 2001. M. P. Allen and D. J. Tildesley "Computer Simulation of Liquids",
Molecular Dynamics Simulations of Biomolecules References: A. R. Leach Molecular Modeling Principles and Applications Prentice Hall, 2001. M. P. Allen and D. J. Tildesley "Computer Simulation of Liquids",
Intermolecular Forces I
 I How does the arrangement of atoms differ in the 3 phases of matter (solid, liquid, gas)? Why doesn t ice just evaporate into a gas? Why does liquid water exist at all? There must be some force between
I How does the arrangement of atoms differ in the 3 phases of matter (solid, liquid, gas)? Why doesn t ice just evaporate into a gas? Why does liquid water exist at all? There must be some force between
On the calculation of solvation free energy from Kirkwood- Buff integrals: A large scale molecular dynamics study
 On the calculation of solvation free energy from Kirkwood- Buff integrals: A large scale molecular dynamics study Wynand Dednam and André E. Botha Department of Physics, University of South Africa, P.O.
On the calculation of solvation free energy from Kirkwood- Buff integrals: A large scale molecular dynamics study Wynand Dednam and André E. Botha Department of Physics, University of South Africa, P.O.
The Effect of Model Internal Flexibility Upon NEMD Simulations of Viscosity
 Draft: September 29, 1999 The Effect of Model Internal Flexibility Upon NEMD Simulations of Viscosity N. G. Fuller 1 and R. L. Rowley 1,2 Abstract The influence of model flexibility upon simulated viscosity
Draft: September 29, 1999 The Effect of Model Internal Flexibility Upon NEMD Simulations of Viscosity N. G. Fuller 1 and R. L. Rowley 1,2 Abstract The influence of model flexibility upon simulated viscosity
Water models in classical simulations
 Water models in classical simulations Maria Fyta Institut für Computerphysik, Universität Stuttgart Stuttgart, Germany Water transparent, odorless, tasteless and ubiquitous really simple: two H atoms attached
Water models in classical simulations Maria Fyta Institut für Computerphysik, Universität Stuttgart Stuttgart, Germany Water transparent, odorless, tasteless and ubiquitous really simple: two H atoms attached
Chemistry: The Central Science
 Chemistry: The Central Science Fourteenth Edition Chapter 11 Liquids and Intermolecular Forces Intermolecular Forces The attractions between molecules are not nearly as strong as the intramolecular attractions
Chemistry: The Central Science Fourteenth Edition Chapter 11 Liquids and Intermolecular Forces Intermolecular Forces The attractions between molecules are not nearly as strong as the intramolecular attractions
Mutual diffusion in the ternary mixture of water + methanol + ethanol: Experiments and Molecular Simulation
 - 1 - Mutual diffusion in the ternary mixture of water + methanol + ethanol: Experiments and Molecular Simulation Tatjana Janzen, Gabriela Guevara-Carrión, Jadran Vrabec University of Paderborn, Germany
- 1 - Mutual diffusion in the ternary mixture of water + methanol + ethanol: Experiments and Molecular Simulation Tatjana Janzen, Gabriela Guevara-Carrión, Jadran Vrabec University of Paderborn, Germany
Molecular Dynamics Simulation Study of Transport Properties of Diatomic Gases
 MD Simulation of Diatomic Gases Bull. Korean Chem. Soc. 14, Vol. 35, No. 1 357 http://dx.doi.org/1.51/bkcs.14.35.1.357 Molecular Dynamics Simulation Study of Transport Properties of Diatomic Gases Song
MD Simulation of Diatomic Gases Bull. Korean Chem. Soc. 14, Vol. 35, No. 1 357 http://dx.doi.org/1.51/bkcs.14.35.1.357 Molecular Dynamics Simulation Study of Transport Properties of Diatomic Gases Song
Why study protein dynamics?
 Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Microscopic analysis of protein oxidative damage: effect of. carbonylation on structure, dynamics and aggregability of.
 Microscopic analysis of protein oxidative damage: effect of carbonylation on structure, dynamics and aggregability of villin headpiece Drazen etrov 1,2,3 and Bojan Zagrovic 1,2,3,* Supplementary Information
Microscopic analysis of protein oxidative damage: effect of carbonylation on structure, dynamics and aggregability of villin headpiece Drazen etrov 1,2,3 and Bojan Zagrovic 1,2,3,* Supplementary Information
The micro-properties of [hmpy+] [Tf 2 N-] Ionic liquid: a simulation. study. 1. Introduction
![The micro-properties of [hmpy+] [Tf 2 N-] Ionic liquid: a simulation. study. 1. Introduction The micro-properties of [hmpy+] [Tf 2 N-] Ionic liquid: a simulation. study. 1. Introduction](/thumbs/72/66941683.jpg) ISBN 978-1-84626-081-0 Proceedings of the 2010 International Conference on Application of Mathematics and Physics Volume 1: Advances on Space Weather, Meteorology and Applied Physics Nanjing, P. R. China,
ISBN 978-1-84626-081-0 Proceedings of the 2010 International Conference on Application of Mathematics and Physics Volume 1: Advances on Space Weather, Meteorology and Applied Physics Nanjing, P. R. China,
COSMO-RS Theory. The Basics
 Theory The Basics From µ to properties Property µ 1 µ 2 activity coefficient vapor pressure Infinite dilution Gas phase Pure compound Pure bulk compound Partition coefficient Phase 1 Phase 2 Liquid-liquid
Theory The Basics From µ to properties Property µ 1 µ 2 activity coefficient vapor pressure Infinite dilution Gas phase Pure compound Pure bulk compound Partition coefficient Phase 1 Phase 2 Liquid-liquid
Liquids and Solids The Condensed States of Matter
 Liquids and Solids The Condensed States of Matter AP Chemistry Ms. Grobsky Where We Have Been And Where We Are Going In the last few chapters, we saw that atoms can form stable units called molecules by
Liquids and Solids The Condensed States of Matter AP Chemistry Ms. Grobsky Where We Have Been And Where We Are Going In the last few chapters, we saw that atoms can form stable units called molecules by
Bioengineering 215. An Introduction to Molecular Dynamics for Biomolecules
 Bioengineering 215 An Introduction to Molecular Dynamics for Biomolecules David Parker May 18, 2007 ntroduction A principal tool to study biological molecules is molecular dynamics simulations (MD). MD
Bioengineering 215 An Introduction to Molecular Dynamics for Biomolecules David Parker May 18, 2007 ntroduction A principal tool to study biological molecules is molecular dynamics simulations (MD). MD
Solids, Liquids and Gases
 WHY? Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water? Why do snowflakes have 6 sides? Why is I
WHY? Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water? Why do snowflakes have 6 sides? Why is I
Unit 10: Part 1: Polarity and Intermolecular Forces
 Unit 10: Part 1: Polarity and Intermolecular Forces Name: Block: Intermolecular Forces of Attraction and Phase Changes Intramolecular Bonding: attractive forces that occur between atoms WITHIN a molecule;
Unit 10: Part 1: Polarity and Intermolecular Forces Name: Block: Intermolecular Forces of Attraction and Phase Changes Intramolecular Bonding: attractive forces that occur between atoms WITHIN a molecule;
Advanced Molecular Dynamics
 Advanced Molecular Dynamics Introduction May 2, 2017 Who am I? I am an associate professor at Theoretical Physics Topics I work on: Algorithms for (parallel) molecular simulations including GPU acceleration
Advanced Molecular Dynamics Introduction May 2, 2017 Who am I? I am an associate professor at Theoretical Physics Topics I work on: Algorithms for (parallel) molecular simulations including GPU acceleration
= = 10.1 mol. Molar Enthalpies of Vaporization (at Boiling Point) Molar Enthalpy of Vaporization (kj/mol)
 Ch 11 (Sections 11.1 11.5) Liquid Phase Volume and Density - Liquid and solid are condensed phases and their volumes are not simple to calculate. - This is different from gases, which have volumes that
Ch 11 (Sections 11.1 11.5) Liquid Phase Volume and Density - Liquid and solid are condensed phases and their volumes are not simple to calculate. - This is different from gases, which have volumes that
Chemistry. Atomic and Molecular Structure
 Chemistry Atomic and Molecular Structure 1. The periodic table displays the elements in increasing atomic number and shows how periodicity of the physical and chemical properties of the elements relates
Chemistry Atomic and Molecular Structure 1. The periodic table displays the elements in increasing atomic number and shows how periodicity of the physical and chemical properties of the elements relates
Phase Equilibria of binary mixtures by Molecular Simulation and PR-EOS: Methane + Xenon and Xenon + Ethane
 International Journal of ChemTech Research CODEN( USA): IJCRGG ISSN : 0974-4290 Vol.5, No.6, pp 2975-2979, Oct-Dec 2013 Phase Equilibria of binary mixtures by Molecular Simulation and PR-EOS: Methane +
International Journal of ChemTech Research CODEN( USA): IJCRGG ISSN : 0974-4290 Vol.5, No.6, pp 2975-2979, Oct-Dec 2013 Phase Equilibria of binary mixtures by Molecular Simulation and PR-EOS: Methane +
(b) The measurement of pressure
 (b) The measurement of pressure The pressure of the atmosphere is measured with a barometer. The original version of a barometer was invented by Torricelli, a student of Galileo. The barometer was an inverted
(b) The measurement of pressure The pressure of the atmosphere is measured with a barometer. The original version of a barometer was invented by Torricelli, a student of Galileo. The barometer was an inverted
Chap 10 Part 4Ta.notebook December 08, 2017
 Chapter 10 Section 1 Intermolecular Forces the forces between molecules or between ions and molecules in the liquid or solid state Stronger Intermolecular forces cause higher melting points and boiling
Chapter 10 Section 1 Intermolecular Forces the forces between molecules or between ions and molecules in the liquid or solid state Stronger Intermolecular forces cause higher melting points and boiling
Chemical Physics Letters
 Chemical Physics Letters 542 (2012) 37 41 Contents lists available at SciVerse ScienceDirect Chemical Physics Letters journal homepage: www.elsevier.com/locate/cplett Thermal conductivity, shear viscosity
Chemical Physics Letters 542 (2012) 37 41 Contents lists available at SciVerse ScienceDirect Chemical Physics Letters journal homepage: www.elsevier.com/locate/cplett Thermal conductivity, shear viscosity
UB association bias algorithm applied to the simulation of hydrogen fluoride
 Fluid Phase Equilibria 194 197 (2002) 249 256 UB association bias algorithm applied to the simulation of hydrogen fluoride Scott Wierzchowski, David A. Kofke Department of Chemical Engineering, University
Fluid Phase Equilibria 194 197 (2002) 249 256 UB association bias algorithm applied to the simulation of hydrogen fluoride Scott Wierzchowski, David A. Kofke Department of Chemical Engineering, University
Chapter 12. Insert picture from First page of chapter. Intermolecular Forces and the Physical Properties of Liquids and Solids
 Chapter 12 Insert picture from First page of chapter Intermolecular Forces and the Physical Properties of Liquids and Solids Copyright McGraw-Hill 2009 1 12.1 Intermolecular Forces Intermolecular forces
Chapter 12 Insert picture from First page of chapter Intermolecular Forces and the Physical Properties of Liquids and Solids Copyright McGraw-Hill 2009 1 12.1 Intermolecular Forces Intermolecular forces
A MOLECULAR DYNAMICS STUDY OF POLYMER/GRAPHENE NANOCOMPOSITES
 A MOLECULAR DYNAMICS STUDY OF POLYMER/GRAPHENE NANOCOMPOSITES Anastassia N. Rissanou b,c*, Vagelis Harmandaris a,b,c* a Department of Applied Mathematics, University of Crete, GR-79, Heraklion, Crete,
A MOLECULAR DYNAMICS STUDY OF POLYMER/GRAPHENE NANOCOMPOSITES Anastassia N. Rissanou b,c*, Vagelis Harmandaris a,b,c* a Department of Applied Mathematics, University of Crete, GR-79, Heraklion, Crete,
Molecular Mechanics. I. Quantum mechanical treatment of molecular systems
 Molecular Mechanics I. Quantum mechanical treatment of molecular systems The first principle approach for describing the properties of molecules, including proteins, involves quantum mechanics. For example,
Molecular Mechanics I. Quantum mechanical treatment of molecular systems The first principle approach for describing the properties of molecules, including proteins, involves quantum mechanics. For example,
Contents. 1 Introduction and guide for this text 1. 2 Equilibrium and entropy 6. 3 Energy and how the microscopic world works 21
 Preface Reference tables Table A Counting and combinatorics formulae Table B Useful integrals, expansions, and approximations Table C Extensive thermodynamic potentials Table D Intensive per-particle thermodynamic
Preface Reference tables Table A Counting and combinatorics formulae Table B Useful integrals, expansions, and approximations Table C Extensive thermodynamic potentials Table D Intensive per-particle thermodynamic
Thermodynamics of Three-phase Equilibrium in Lennard Jones System with a Simplified Equation of State
 23 Bulletin of Research Center for Computing and Multimedia Studies, Hosei University, 28 (2014) Thermodynamics of Three-phase Equilibrium in Lennard Jones System with a Simplified Equation of State Yosuke
23 Bulletin of Research Center for Computing and Multimedia Studies, Hosei University, 28 (2014) Thermodynamics of Three-phase Equilibrium in Lennard Jones System with a Simplified Equation of State Yosuke
California Science Content Standards Chemistry Grades 9-12
 California Science Content Standards Chemistry Grades 9-12 Standards that all students are expected to achieve in the course of their studies are unmarked. Standards that all students should have the opportunity
California Science Content Standards Chemistry Grades 9-12 Standards that all students are expected to achieve in the course of their studies are unmarked. Standards that all students should have the opportunity
Development of a Water Cluster Evaporation Model using Molecular Dynamics
 Development of a Water Cluster Evaporation Model using Molecular Dynamics Arnaud Borner, Zheng Li, Deborah A. Levin. Department of Aerospace Engineering, The Pennsylvania State University, University Park,
Development of a Water Cluster Evaporation Model using Molecular Dynamics Arnaud Borner, Zheng Li, Deborah A. Levin. Department of Aerospace Engineering, The Pennsylvania State University, University Park,
Chapter 14. Liquids and Solids
 Chapter 14 Liquids and Solids Section 14.1 Water and Its Phase Changes Reviewing What We Know Gases Low density Highly compressible Fill container Solids High density Slightly compressible Rigid (keeps
Chapter 14 Liquids and Solids Section 14.1 Water and Its Phase Changes Reviewing What We Know Gases Low density Highly compressible Fill container Solids High density Slightly compressible Rigid (keeps
Molecular Dynamics Simulation of a Nanoconfined Water Film
 Molecular Dynamics Simulation of a Nanoconfined Water Film Kyle Lindquist, Shu-Han Chao May 7, 2013 1 Introduction The behavior of water confined in nano-scale environment is of interest in many applications.
Molecular Dynamics Simulation of a Nanoconfined Water Film Kyle Lindquist, Shu-Han Chao May 7, 2013 1 Introduction The behavior of water confined in nano-scale environment is of interest in many applications.
Supporting Information for: Physics Behind the Water Transport through. Nanoporous Graphene and Boron Nitride
 Supporting Information for: Physics Behind the Water Transport through Nanoporous Graphene and Boron Nitride Ludovic Garnier, Anthony Szymczyk, Patrice Malfreyt, and Aziz Ghoufi, Institut de Physique de
Supporting Information for: Physics Behind the Water Transport through Nanoporous Graphene and Boron Nitride Ludovic Garnier, Anthony Szymczyk, Patrice Malfreyt, and Aziz Ghoufi, Institut de Physique de
Exercise 2: Solvating the Structure Before you continue, follow these steps: Setting up Periodic Boundary Conditions
 Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Journal of Physics: Conference Series PAPER. To cite this article: Jiramate Kitjanon et al 2017 J. Phys.: Conf. Ser
 Journal of Physics: Conference Series PAPER Transferability of Polymer Chain Properties between Coarse-Grained and Atomistic Models of Natural Rubber Molecule Validated by Molecular Dynamics Simulations
Journal of Physics: Conference Series PAPER Transferability of Polymer Chain Properties between Coarse-Grained and Atomistic Models of Natural Rubber Molecule Validated by Molecular Dynamics Simulations
The Molecular Dynamics Method
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
Intermolecular Forces and Liquids and Solids
 Intermolecular Forces and Liquids and Solids Chapter 11 Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. A phase is a homogeneous part of the system in contact
Intermolecular Forces and Liquids and Solids Chapter 11 Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. A phase is a homogeneous part of the system in contact
What factors affect whether something is a solid, liquid or gas? What actually happens (breaks) when you melt various types of solids?
 States of Mattter What factors affect whether something is a solid, liquid or gas? What actually happens (breaks) when you melt various types of solids? What external factors affect whether something is
States of Mattter What factors affect whether something is a solid, liquid or gas? What actually happens (breaks) when you melt various types of solids? What external factors affect whether something is
S OF MATTER TER. Unit. I. Multiple Choice Questions (Type-I)
 Unit 5 STATE TES TE S OF MATTER MA TER I. Multiple Choice Questions (Type-I) 1. A person living in Shimla observed that cooking food without using pressure cooker takes more time. The reason for this observation
Unit 5 STATE TES TE S OF MATTER MA TER I. Multiple Choice Questions (Type-I) 1. A person living in Shimla observed that cooking food without using pressure cooker takes more time. The reason for this observation
CE 530 Molecular Simulation
 1 CE 530 Molecular Simulation Lecture 1 David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Time/s Multi-Scale Modeling Based on SDSC Blue Horizon (SP3) 1.728 Tflops
1 CE 530 Molecular Simulation Lecture 1 David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Time/s Multi-Scale Modeling Based on SDSC Blue Horizon (SP3) 1.728 Tflops
Chem 1075 Chapter 13 Liquids and Solids Lecture Outline
 Chem 1075 Chapter 13 Liquids and Solids Lecture Outline Slide 2-3 Properties of Liquids Unlike gases, liquids respond dramatically to temperature and pressure changes. We can study the liquid state and
Chem 1075 Chapter 13 Liquids and Solids Lecture Outline Slide 2-3 Properties of Liquids Unlike gases, liquids respond dramatically to temperature and pressure changes. We can study the liquid state and
Chem 112 Dr. Kevin Moore
 Chem 112 Dr. Kevin Moore Gas Liquid Solid Polar Covalent Bond Partial Separation of Charge Electronegativity: H 2.1 Cl 3.0 H Cl δ + δ - Dipole Moment measure of the net polarity in a molecule Q Q magnitude
Chem 112 Dr. Kevin Moore Gas Liquid Solid Polar Covalent Bond Partial Separation of Charge Electronegativity: H 2.1 Cl 3.0 H Cl δ + δ - Dipole Moment measure of the net polarity in a molecule Q Q magnitude
Lecture Presentation. Chapter 11. Liquids and Intermolecular Forces Pearson Education, Inc.
 Lecture Presentation Chapter 11 Liquids and States of Matter The fundamental difference between states of matter is the strength of the intermolecular forces of attraction. Stronger forces bring molecules
Lecture Presentation Chapter 11 Liquids and States of Matter The fundamental difference between states of matter is the strength of the intermolecular forces of attraction. Stronger forces bring molecules
CHEMISTRY Matter and Change. Chapter 12: States of Matter
 CHEMISTRY Matter and Change Chapter 12: States of Matter CHAPTER 12 States of Matter Section 12.1 Section 12.2 Section 12.3 Section 12.4 Gases Forces of Attraction Liquids and Solids Phase Changes Click
CHEMISTRY Matter and Change Chapter 12: States of Matter CHAPTER 12 States of Matter Section 12.1 Section 12.2 Section 12.3 Section 12.4 Gases Forces of Attraction Liquids and Solids Phase Changes Click
Supplemntary Infomation: The nanostructure of. a lithium glyme solvate ionic liquid at electrified. interfaces
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 207 Supplemntary Infomation: The nanostructure of a lithium glyme solvate ionic liquid
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 207 Supplemntary Infomation: The nanostructure of a lithium glyme solvate ionic liquid
MOLECULAR DYNAMICS SIMULATION OF THE STRUCTURE OF C6 ALKANES INTRODUCTION. A. V. Anikeenko, A. V. Kim, and N. N. Medvedev UDC 544.2: 544.
 Journal of Structural Chemistry. Vol. 51, No. 6, pp. 1090-1096, 2010 Original Russian Text Copyright 2010 by A. V. Anikeenko, A. V. Kim, and N. N. Medvedev MOLECULAR DYNAMICS SIMULATION OF THE STRUCTURE
Journal of Structural Chemistry. Vol. 51, No. 6, pp. 1090-1096, 2010 Original Russian Text Copyright 2010 by A. V. Anikeenko, A. V. Kim, and N. N. Medvedev MOLECULAR DYNAMICS SIMULATION OF THE STRUCTURE
Professor K. Intermolecular forces
 Professor K Intermolecular forces We've studied chemical bonds which are INTRAmolecular forces... We now explore the forces between molecules, or INTERmolecular forces which you might rightly assume to
Professor K Intermolecular forces We've studied chemical bonds which are INTRAmolecular forces... We now explore the forces between molecules, or INTERmolecular forces which you might rightly assume to
Brief Review of Statistical Mechanics
 Brief Review of Statistical Mechanics Introduction Statistical mechanics: a branch of physics which studies macroscopic systems from a microscopic or molecular point of view (McQuarrie,1976) Also see (Hill,1986;
Brief Review of Statistical Mechanics Introduction Statistical mechanics: a branch of physics which studies macroscopic systems from a microscopic or molecular point of view (McQuarrie,1976) Also see (Hill,1986;
Chapter 11. Liquids and Intermolecular Forces
 Chapter 11 Liquids and Intermolecular Forces States of Matter The three states of matter are 1) Solid Definite shape Definite volume 2) Liquid Indefinite shape Definite volume 3) Gas Indefinite shape Indefinite
Chapter 11 Liquids and Intermolecular Forces States of Matter The three states of matter are 1) Solid Definite shape Definite volume 2) Liquid Indefinite shape Definite volume 3) Gas Indefinite shape Indefinite
Intermolecular Forces and Liquids and Solids
 PowerPoint Lecture Presentation by J. David Robertson University of Missouri Intermolecular Forces and Liquids and Solids Chapter 11 Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction
PowerPoint Lecture Presentation by J. David Robertson University of Missouri Intermolecular Forces and Liquids and Solids Chapter 11 Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction
of its physical and chemical properties.
 8.4 Molecular Shapes VSEPR Model The shape of a molecule determines many of its physical and chemical properties. Molecular l geometry (shape) can be determined with the Valence Shell Electron Pair Repulsion
8.4 Molecular Shapes VSEPR Model The shape of a molecule determines many of its physical and chemical properties. Molecular l geometry (shape) can be determined with the Valence Shell Electron Pair Repulsion
Supplemental Material for Temperature-sensitive colloidal phase behavior induced by critical Casimir forces
 Supplemental Material for Temperature-sensitive colloidal phase behavior induced by critical Casimir forces Minh Triet Dang, 1 Ana Vila Verde, 2 Van Duc Nguyen, 1 Peter G. Bolhuis, 3 and Peter Schall 1
Supplemental Material for Temperature-sensitive colloidal phase behavior induced by critical Casimir forces Minh Triet Dang, 1 Ana Vila Verde, 2 Van Duc Nguyen, 1 Peter G. Bolhuis, 3 and Peter Schall 1
6 Hydrophobic interactions
 The Physics and Chemistry of Water 6 Hydrophobic interactions A non-polar molecule in water disrupts the H- bond structure by forcing some water molecules to give up their hydrogen bonds. As a result,
The Physics and Chemistry of Water 6 Hydrophobic interactions A non-polar molecule in water disrupts the H- bond structure by forcing some water molecules to give up their hydrogen bonds. As a result,
Introduction. Monday, January 6, 14
 Introduction 1 Introduction Why to use a simulation Some examples of questions we can address 2 Molecular Simulations Molecular dynamics: solve equations of motion Monte Carlo: importance sampling Calculate
Introduction 1 Introduction Why to use a simulation Some examples of questions we can address 2 Molecular Simulations Molecular dynamics: solve equations of motion Monte Carlo: importance sampling Calculate
University of Groningen. Characterization of oil/water interfaces van Buuren, Aldert Roelf
 University of Groningen Characterization of oil/water interfaces van Buuren, Aldert Roelf IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
University of Groningen Characterization of oil/water interfaces van Buuren, Aldert Roelf IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
fiziks Institute for NET/JRF, GATE, IIT-JAM, JEST, TIFR and GRE in PHYSICAL SCIENCES
 Content-Thermodynamics & Statistical Mechanics 1. Kinetic theory of gases..(1-13) 1.1 Basic assumption of kinetic theory 1.1.1 Pressure exerted by a gas 1.2 Gas Law for Ideal gases: 1.2.1 Boyle s Law 1.2.2
Content-Thermodynamics & Statistical Mechanics 1. Kinetic theory of gases..(1-13) 1.1 Basic assumption of kinetic theory 1.1.1 Pressure exerted by a gas 1.2 Gas Law for Ideal gases: 1.2.1 Boyle s Law 1.2.2
Gromacs Workshop Spring CSC
 Gromacs Workshop Spring 2007 @ CSC Erik Lindahl Center for Biomembrane Research Stockholm University, Sweden David van der Spoel Dept. Cell & Molecular Biology Uppsala University, Sweden Berk Hess Max-Planck-Institut
Gromacs Workshop Spring 2007 @ CSC Erik Lindahl Center for Biomembrane Research Stockholm University, Sweden David van der Spoel Dept. Cell & Molecular Biology Uppsala University, Sweden Berk Hess Max-Planck-Institut
What are covalent bonds?
 Covalent Bonds What are covalent bonds? Covalent Bonds A covalent bond is formed when neutral atoms share one or more pairs of electrons. Covalent Bonds Covalent bonds form between two or more non-metal
Covalent Bonds What are covalent bonds? Covalent Bonds A covalent bond is formed when neutral atoms share one or more pairs of electrons. Covalent Bonds Covalent bonds form between two or more non-metal
Michael W. Mahoney Department of Physics, Yale University, New Haven, Connecticut 06520
 JOURNAL OF CHEMICAL PHYSICS VOLUME 115, NUMBER 23 15 DECEMBER 2001 Quantum, intramolecular flexibility, and polarizability effects on the reproduction of the density anomaly of liquid water by simple potential
JOURNAL OF CHEMICAL PHYSICS VOLUME 115, NUMBER 23 15 DECEMBER 2001 Quantum, intramolecular flexibility, and polarizability effects on the reproduction of the density anomaly of liquid water by simple potential
Improved Adaptive Resolution Molecular Dynamics Simulation
 Improved Adaptive Resolution Molecular Dynamics Simulation Iuliana Marin, Virgil Tudose, Anton Hadar, Nicolae Goga, Andrei Doncescu To cite this version: Iuliana Marin, Virgil Tudose, Anton Hadar, Nicolae
Improved Adaptive Resolution Molecular Dynamics Simulation Iuliana Marin, Virgil Tudose, Anton Hadar, Nicolae Goga, Andrei Doncescu To cite this version: Iuliana Marin, Virgil Tudose, Anton Hadar, Nicolae
Chapter 11. Liquids and Intermolecular Forces
 Chapter 11. Liquids and Intermolecular Forces 11.1 A Molecular Comparison of Gases, Liquids, and Solids Gases are highly compressible and assume the shape and volume of their container. Gas molecules are
Chapter 11. Liquids and Intermolecular Forces 11.1 A Molecular Comparison of Gases, Liquids, and Solids Gases are highly compressible and assume the shape and volume of their container. Gas molecules are
Ch. 11: Liquids and Intermolecular Forces
 Ch. 11: Liquids and Intermolecular Forces Learning goals and key skills: Identify the intermolecular attractive interactions (dispersion, dipole-dipole, hydrogen bonding, ion-dipole) that exist between
Ch. 11: Liquids and Intermolecular Forces Learning goals and key skills: Identify the intermolecular attractive interactions (dispersion, dipole-dipole, hydrogen bonding, ion-dipole) that exist between
What determines whether a substance will be a solid, liquid, or gas? Thursday, April 24, 14
 What determines whether a substance will be a solid, liquid, or gas? Answer: The attractive forces that exists between its particles. Answer: The attractive forces that exists between its particles. For
What determines whether a substance will be a solid, liquid, or gas? Answer: The attractive forces that exists between its particles. Answer: The attractive forces that exists between its particles. For
Molecular Dynamics (MD) Model Generation for the Graphite Nanoplatelets Modified Asphalt
 Fourth International Conference on Sustainable Construction Materials and Technologies http://www.claisse.info/proceedings.htm SCMT4 Las Vegas, USA, August 7-11, 2016 Molecular Dynamics (MD) Model Generation
Fourth International Conference on Sustainable Construction Materials and Technologies http://www.claisse.info/proceedings.htm SCMT4 Las Vegas, USA, August 7-11, 2016 Molecular Dynamics (MD) Model Generation
Lecture 11: Potential Energy Functions
 Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Intermolecular Forces & Condensed Phases
 Intermolecular Forces & Condensed Phases CHEM 107 T. Hughbanks READING We will discuss some of Chapter 5 that we skipped earlier (Van der Waals equation, pp. 145-8), but this is just a segue into intermolecular
Intermolecular Forces & Condensed Phases CHEM 107 T. Hughbanks READING We will discuss some of Chapter 5 that we skipped earlier (Van der Waals equation, pp. 145-8), but this is just a segue into intermolecular
Intermolecular Forces and Liquids and Solids. Chapter 11. Copyright The McGraw Hill Companies, Inc. Permission required for
 Intermolecular Forces and Liquids and Solids Chapter 11 Copyright The McGraw Hill Companies, Inc. Permission required for 1 A phase is a homogeneous part of the system in contact with other parts of the
Intermolecular Forces and Liquids and Solids Chapter 11 Copyright The McGraw Hill Companies, Inc. Permission required for 1 A phase is a homogeneous part of the system in contact with other parts of the
1.3 Molecular Level Presentation
 1.3.1 Introduction A molecule is the smallest chemical unit of a substance that is capable of stable, independent existence. Not all substances are composed of molecules. Some substances are composed of
1.3.1 Introduction A molecule is the smallest chemical unit of a substance that is capable of stable, independent existence. Not all substances are composed of molecules. Some substances are composed of
Chapter 11 Intermolecular Forces, Liquids, and Solids
 Chemistry, The Central Science, 11th edition Theodore L. Brown, H. Eugene LeMay, Jr., and Bruce E. Bursten Chapter 11, Liquids, and Solids States of Matter The fundamental difference between states of
Chemistry, The Central Science, 11th edition Theodore L. Brown, H. Eugene LeMay, Jr., and Bruce E. Bursten Chapter 11, Liquids, and Solids States of Matter The fundamental difference between states of
Ch. 11 States of matter
 Ch. 11 States of matter States of Matter Solid Definite volume Definite shape Liquid Definite volume Indefinite shape (conforms to container) Gas Indefinite volume (fills any container) Indefinite shape
Ch. 11 States of matter States of Matter Solid Definite volume Definite shape Liquid Definite volume Indefinite shape (conforms to container) Gas Indefinite volume (fills any container) Indefinite shape
Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar.
 Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar. 25, 2002 Molecular Dynamics: Introduction At physiological conditions, the
Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar. 25, 2002 Molecular Dynamics: Introduction At physiological conditions, the
Chapter 11 Intermolecular Forces, Liquids, and Solids
 Chemistry, The Central Science, 11th edition Theodore L. Brown, H. Eugene LeMay, Jr., and Bruce E. Bursten Chapter 11, Liquids, and Solids John D. Bookstaver St. Charles Community College Cottleville,
Chemistry, The Central Science, 11th edition Theodore L. Brown, H. Eugene LeMay, Jr., and Bruce E. Bursten Chapter 11, Liquids, and Solids John D. Bookstaver St. Charles Community College Cottleville,
