The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity
|
|
|
- Andra Barker
- 5 years ago
- Views:
Transcription
1 Development 124, (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity Claudia Wittmann 1, Olaf Bossinger 2, Bob Goldstein 3,, Martin Fleischmann 1, Reto Kohler 1, Karin Brunschwig 1, Heinz Tobler 1 and Fritz Müller 1, * 1 Institute of Zoology, University of Fribourg, Pérolles, 1700 Fribourg, Switzerland 2 Institut für Genetik, Heinrich-Heine Universität Düsseldorf, Universitätsstr. 1, Düsseldorf, Germany 3 MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK *Author for correspondence ( fritz.mueller@unifr.ch) Present address: Department of Biology, Indiana University, Jordan Hall. Bloomington, Indiana 47405, USA Present address: Department of Molecular and Cell Biology, 385 LSA, University of California, Berkeley, California , USA SUMMARY Clusters of homeobox-containing HOM-C/hox genes determine the morphology of animal body plans and body parts and are thought to mediate positional information. Here, we describe the onset of embryonic expression of ceh- 13, the Caenorhabditis elegans orthologue of the Drosophila labial gene, which is the earliest gene of the C. elegans Hox gene cluster to be activated in C. elegans development. At the beginning of gastrulation, ceh-13 is asymmetrically expressed in posterior daughters of anteroposterior divisions, first in the posterior daughter of the intestinal precursor cell E and then in all posterior daughters of the AB descendants ABxxx. In this paper, we present evidence that supports position-independent activation of ceh-13 during early C. elegans embryogenesis, which integrates cell fate determinants and cell polarity cues. Our findings imply that mechanisms other than cell-extrinsic anteroposterior positional signals play an important role in the activation and regulation of the C. elegans Hox gene ceh- 13. Key words: homeobox, asymmetry, Caenorhabditis elegans, polarity, intestine INTRODUCTION The underlying principle of every developmental program is that single cells divide to give rise to daughter cells with distinct developmental fates. Differences between daughter cells can originate from differences in the daughter cells environment or can be generated by unequal segregation of cell fate determinants during oriented cell division. The development of the nematode Caenorhabditis elegans follows a remarkably reproducible lineage pattern (Sulston et al., 1983; Sulston and Horvitz, 1977), whereby oriented asymmetric cleavages play essential roles (reviewed by Guo and Kemphues, 1996). Oriented asymmetric divisions produce cells in appropriate spatial relationships that differ in size, cell cycle timing, cell fate and cleavage spindle orientation (Sulston et al., 1983). Many of these asymmetric cleavages are externally triggered. At the 4-cell stage, for example, the P 2 cell induces the EMS cell both to become polarized to give rise to distinct daughters MS and E (intestinal precursor) and to direct its division axis for a correct partitioning of developmental potential (Schierenberg, 1987; Goldstein, 1992, 1993, 1995a,b; Bossinger and Schierenberg, 1996). C. elegans has a repertoire of developmental regulatory genes that is remarkably similar to that found in other animals with less determinant cell lineages. These genes include the homeobox-containing HOM-C/Hox gene family that are found in a wide variety of organisms. They determine the animal body plan and are thought to mediate positional information. C. elegans has a cluster of four evolutionarily conserved HOM- C/Hox genes (Bürglin et al., 1991; Bürglin and Ruvkun, 1993; Kenyon and Wang, 1991; Wang et al., 1993; Wilson et al., 1994; Hunter and Kenyon, 1995) which, during development, are expressed in consecutive domains along the anteroposterior body axis (Wang et al., 1993; Clark et al., 1993; Wittmann et al., unpublished data). In this paper, we describe the early expression of ceh-13, the orthologue of the Drosophila labial gene (Schaller et al., 1990; Sharkey et al., 1997; Wittmann et al., unpublished data). ceh- 13 is the earliest gene of the C. elegans Hox gene cluster to be activated in development. Its protein product first appears in the 26-cell-stage embryos in the Ep cell and soon afterwards also in the posterior daughters of anteroposterior cell divisions in the AB lineage. ceh-13 has a dynamic expression pattern, which shows the typical characteristics of a labial gene (Wittmann et al., unpublished data). Using skn-1 and pop-1 mutant backgrounds that change cell fates, as well as by performing trypan blue experiments to block cell-cell interactions, we show that ceh-13 expression in Ep relies on the intestinal fate and is independent of the position of the cell in the embryo. This finding is in agreement with the recent data on mab-5 (Cowing and Kenyon, 1996) and suggests that the expression pattern of the C. elegans Hox genes must be established by a
2 4194 C. Wittmann and others complex lineage-based molecular mechanism that differs from that operating in Drosophila and vertebrates, where HOM- C/Hox gene expression is strongly influenced by the position of a nucleus or a cell in the embryo. Furthermore, we show that the onset of early ceh-13 expression depends on asymmetric cell divisions along the anteroposterior axis, thus providing strong evidence for an animal-wide system directing anteroposterior cellular polarity during embryonic development. MATERIALS AND METHODS Worm strains and culture methods C. elegans strains were cultured using standard conditions (Brenner, 1974). The following mutations and rearrangements were used in this analysis: LG I: pop-1(zu189), lin-17(n671), ht1(i;v) LG II: rol-6(n1276e187), swis1[rol-6(su1006) + ceh-13::gfp] LG IV: skn-1(zn67), nt1(iv;v) LG V: him-5(e1490) The pop-1(zu189) mutation was given to us by Dr. J. Priess, Seattle (USA) and the other strains used in this study were provided by the Caenorhabdidtis Genetics Center at the University of Minnesota, USA. The strain FR307 is of the genotype swex307[rol-6(su1006) + ceh-13::gfp]; rol-6(n1276e187)ii. Immunolocalization Anti-CEH-13 was used as a primary antibody at a working dilution of 1:40 to 1:100 (Wittmann et al., unpublished data). The secondary antibodies, anti-rabbit IgG FITC conjugate (Sigma F-0382), were incubated overnight at 4 C with fixed worms in the following proportions: 150 µl of antibody for 200 µl of compact worms and 200 µl of antibody buffer A (AbA; Finney and Ruvkun, 1990). After this purification step, they were used at a working dilution of 1:10. Embryos were immunostained on poly-lysine slides as described (Bowerman et al., 1993; Okkema and Fire, 1994) or, for better results, by a non-freeze-crack large-scale method. For this, embryos were collected by hypochlorite treatment (Lewis and Fleming, 1995), rinsed at least twice in M9 and pure water. They were resuspended in a fixation solution containing freshly solubilized 1-2% paraformaldehyde, 80 mm KCl, 20 mm NaCl, 10 mm EGTA, 5 mm spermidine HCl, 30 mm sodium Pipes ph 7.4 and 50% methanol, immediately frozen in dry-ice/ethanol or liquid nitrogen. Embryos were thawed on ice, incubated on ice for 1 hour, washed once with 100 mm Tris-HCl ph 7.4, 1% Triton X-100, 1 mm EDTA and blocked for half an hour in AbB (Antibody buffer B; Finney and Ruvkun, 1990). Incubation with primary antibodies was conducted overnight at room temperature in AbA + goat serum (DAKO X907; at least in a 1:2000 dilution). Embryos were washed in 3-4 changes of AbB (Finney and Ruvkun, 1990) at room temperature before incubation with the secondary antibodies. Embryos were mounted on 0.5% poly-lysine slides or on agarose pads (2% agarose in 50mM Tris-HCl ph 9.5, 50 mm MgCl 2) in a 1:1 ratio of 100 mm Tris ph 9.5 and Vectashield (Vector Inc. Burlingame, CA; H-1000) + 2 mg/ml DAPI. Fluorescence was enhanced using 50 to 100 W mercury bulbs and specific filters: nm for FITC and 365 nm for DAPI. C. elegans germline transformation and in vivo observation The translational ceh-13::gfp fusion gene (Chalfie et al., 1994; Heim et al., 1995) used in this study was constructed by inserting the PstI fragment derived from ppdpst37 (Wittmann et al., unpublished data) into the vector ppd95.69 (provided by A. Fire). This construct encodes a CEH-13::GFP fusion protein. Injection procedure, genome integration and transgenic homozygote selection were performed as described for the β-galactosidase fusion (Mello and Fire, 1995; Wittmann et al., unpublished data). Of the stable lines, FR301 was chosen for the integration of the transforming DNA into a chromosomal locus. swex301[rol-6(su1006) + ceh-13::gfp]; rol- 6(n1276e187)II worms were treated with 1500 rads of gamma radiation and their F 3 progeny screened for the presence of 100% Rol- 6 individuals, resulting in the isolation of the integration swis1[rol- 6(su1006) + ceh-13::gfp] in the strain FR308. FR308 hermaphrodites were out-crossed to N2 males 5 times (FR317) and the site of integration was mapped to LG II. To visualize the CEH-13::GFP, worms were excited with a nm light source (50 to 100 W mercury bulb). Whole-mount in situ hybridization mrna in situ hybridization was executed as described (Seydoux and Fire, 1995) with a prolongation of the detection step to overnight. The two 356 bp and 839 bp long antisense probes were generated on gelpurified EcoRI inserts from the ceh-13 cdna clone Bar23C with the primers 5002 (5 -TTTCGTCTATCACCTTC-3 ) and 5006 (5 -GATG- GAGGTGGAAGAGTG-3 ), respectively. As control, the sense probe was produced with the primer 3270 (5 -GCTCATCCATCCAACTAC- 3 ) and a 650 bp long SacI fragment from Bar23C (length of the probe: 536 b). Another negative control was performed with a pbs plasmid probe, which was produced by amplifying NaeI linearized plasmid with a T7 primer. Blastomere culture and analysis AB and P 1 blastomeres were isolated from the N2 strain and from FR307 (swex307[rol-6(su1006) + ceh-13::gfp]; rol-6(n1276e187)ii): 2/6AB blastomeres and 6/8 P 1 blastomeres were from N2 and were examined using the anti-ceh-13 antibody; the rest were examined for GFP as above. Methods for removing eggshells and vitelline membranes and for culturing C. elegans cells have been described (Edgar, 1995). Isolated cells were identified on the basis of size; for example, in the 2-cell-stage AB is larger than P 1. Fixation and immunostaining was performed according to Goldstein (1995b). Blastomere identification and lineage analysis Ep and ABxxxp (see Results and Fig. 1) were identified as ceh-13- expressing cells by reconstructing immunolocalized 28-cell-stage embryos. Embryos were photographed (see below for image processing) and comparison of these pictures with the original embryos and hand-made models (Sulston et al., 1983) allowed the final reconstruction. For detailed cell lineage analysis in wild-type embryos and in mutant backgrounds, CEH-13::GFP-expressing embryos were mounted in egg salts on 2% agarose pads, sealed with nail polish and submitted to manual 4-D recording. Specific cells were traced until selected time-points. Trypan blue incubation, blastomere removal, cytochalasin D block and blastomere removal Eggs were dissected out of gravid adults with a scalpel in a drop of distilled water on a microscope slide. Uncleaved zygotes were identified under the dissecting microscope and transferred with a drawnout glass mouth pipette to a second microscope slide, carrying either a thin layer of 3-5% agar in H 2O as a cushion (blastomere removal) or a poly-lysine layer (cytochalasin D blocks, blastomere removal). The eggs were covered with a coverslip and the edges sealed with Vaseline (Sulston and Horvitz, 1977). Manipulations were performed under Nomarski optics using a 100 oil immersion objective. The impermeable eggshell and underlying vitelline membrane were perforated with brief pulses of a N2-pumped, microscope coupled dye laser (Lambda Physics, Göttingen; Schierenberg, 1984) by using the laser dye Rhodamine 6G. Penetration was performed in cell culture media containing 15% fetal calf serum (Edgar, 1995) with 14 mg/ml trypan blue (Sigma Nr 961). This concentration reliably blocked early cellular interactions without showing toxic effects (Bossinger and
3 Early embryonic Hox gene activation 4195 Schierenberg, 1996). To neutralize the inhibitive effect of trypan blue in cleavage-blocking experiments and after blastomere isolation, 40% instead of 15% FCS was used (Bossinger and Schierenberg, 1996). Cell division of 1-, 2- and 4-cell stages were inhibited by adding 5 µg/ml cytochalasin D to the cell culture medium (Bossinger and Schierenberg, 1992). For the blastomere isolation experiments, cells were extruded through a laser-induced hole in the eggshell as described in Schierenberg and Wood (1985). Image processing Figs 1A-D and 3 are the result of Fuji PROVIA 400 slides taken with an Olympus OM 4-Ti camera mounted on a Zeiss Axioplan microscope. These pictures were either digitalized (Fig. 1A-D) or exposed on photography paper using the Cibachrome procedure (Fig. 3). Fig. 4 was assembled from 6-10 second-integrated fluorescent signals. The image analysis system consisted of a Hamamatsu chilled CCD C5985 camera mounted on the Zeiss Axioplan microscope ( pixels/cm), a Hamamatsu chilled CCD driver C6391 and a Hamamatsu image processor Argus-20. Figs 1E, 5 and 7 are framegrabbed pictures ( pixels/cm) from video recordings, using a Panasonic WV-BC700 video camera (without infrared filter) and a time-lapse video recorder (Panasonic, AG-6720-E), linked to a Leica DM IRBE inverted microscope. The sensitivity of the Panasonic camera was virtually enhanced (up to 32) with the Panasonic WV- CU204 camera control unit (Bossinger and Schierenberg, 1992) and a Hamamatsu image processor Argus-10. The frame-grabbing was realized using the Movie Machine II (Fast Multimedia AG) videocard and the Movie TV II (release 1.02, version 2.02) software in the Windows environment. Final image processing was performed in the MacIntosh environment, using the Adobe Photoshop 3.0 software (US patent ). A final resolution of 300 dpi was chosen. Manipulations, such as contrast enhancement, color balance refinement, color curve adjustment or filtering were always applied to the total surface of the picture for avoiding any modifications of the original signal(s). Confocal microscopy was performed on a Biorad confocal microscope and images were processed in Adobe Photoshop as above. RESULTS Early embryonic ceh-13 activation occurs asymmetrically in different cell lineages By using specific anti-ceh-13 antibodies, weak CEH-13 expression was first detected in 26-cell-stage embryos in the intestinal precursor cell Ep. At the 28-cell stage, the nucleus of Ep was brightly stained (Fig. 1A,C). Nuclear CEH-13 also appeared in posterior-ventral ABxxxp cells and cytoplasmatic staining was detectable in anterior-dorsal ABxxxp cells (Fig. 1A-D). ABxxpp cells showed higher CEH-13 levels than ABxxap cells. During the next round of division, the CEH-13 protein was inherited by the daughters of Ep and ABxxxp cells, and became nuclear in all cells. The antibody staining pattern matched exactly that of a ceh-13::lacz reporter gene and of the identically regulated ceh-13::gfp reporter gene in strain FR317 (Fig. 1E). Furthermore, confocal microscopical analysis of 50- cell-stage embryos containing the ceh-13::gfp marker gene revealed low levels of ceh-13 expression in the Da and Dp cells (Fig. 1F). A summary of the first ceh-13 activation in the AB, D and E lineages is shown in Fig. 2. In situ hybridization experiments revealed that ceh-13 mrna was not present at early embryonic stages. It appeared first in the Ep cell of 26-cell-stage embryos and soon afterwards in all ABxxxp cells, only shortly before CEH-13 (Fig. 3 and data not shown). This suggested that ceh-13 is an early zygotic gene, whose expression pattern is determined at the level of transcription. Therefore, and since the anti-ceh-13 and the CEH-13::LACZ or CEH-13::GFP patterns overlapped exactly, we conclude that the strain containing the integrated ceh-13::gfp reporter gene (FR317) can be used to analyse the establishment of the CEH-13 expression pattern during early embryonic development in more detail. Correct ceh-13 expression in the Ep cell depends on its intestinal fate but not on the position of the cell in the embryo The fact that early ceh-13 expression occurs in the AB, D and E lineage, but not in the MS and C lineages (see Fig. 2), suggested that the onset of ceh-13 expression may depend on specific lineage and cell fate cues. To address this hypothesis, we have tested whether the Ep cell fate is necessary and sufficient for ceh-13 expression. First, we analysed the ceh-13::gfp expression pattern in a pop-1(zu189) mutant background. pop- 1(zu189) is a maternal-effect lethal mutation and in embryos isolated from pop-1 homozygous mothers, the anterior daughter of EMS, MS, adopts the fate of its posterior sister cell E (Lin et al., 1995). The genomic ceh-13::gfp insertion (swis1) was crossed into the pop-1 lin-17/ht1(i:v)i; him-5/ht1(i;v)v strain. Embryos from pop-1 lin-17 I; swis1[rol-6(su1006) ceh- 13::gfp]II homozygotes were lineaged until we could detect fluorescence, i.e., until the Epl and Epr cells were generated (we have looked at Epl and Epr, since they usually show stronger fluorescence than the Ep cell). We then looked for ectopic ceh-13 expression in the MS descendants. Out of 18 lineaged embryos, 4 showed bright and 8 showed weak signals in the MSpa and MSpp cells (Fig. 4A). No signal was observed in any of the MSa daughters. Furthermore, MSa and MSp daughters in lin-17 I; swis1[rol-6(su1006) ceh-13::gfp]ii control embryos (n=6) did not show fluorescence at all (Fig. 4B). Given these results, we conclude that the E fate is sufficient for ectopic expression of ceh-13 in the MSp cell. To determine whether gut determination is necessary for ceh-13 expression in Ep, we have incubated FR307 4-cell-stage embryos (swex307[rol-6(su1006) + ceh-13::gfp]; rol- 6(n1276e187)II) with trypan blue. This polysulfated hydrocarbon dye binds strongly to the cell membranes and inhibits the interaction between P 2 and EMS and thus prevents gut induction, resulting in two EMS daughters that both have MSlike characteristics (Bossinger and Schierenberg, 1996). The treated embryos (n=12) show several signs of normal cell differentiation, e.g. muscle twitching and programmed cell death, indicating that trypan blue in the applied concentration is not generally toxic (Bossinger and Schierenberg, 1996). However, using two different markers (birefringency and binding of the gut-specific antibody ICB4; Laufer et al., 1980; Okamoto and Thomson, 1985), no signs of gut differentiation could be found (12/12). Furthermore, in trypan-blue-treated FR307 embryos, the 4-EMS descendants behaved like MS cells and did not show any CEH-13::GFP fluorescence (data not shown), suggesting that gut induction is necessary for normal CEH-13 expression in Ep. To further investigte this question, we have analyzed the distribution of CEH-13::GFP in the skn-1(zu67) background. Mutations in the maternal-effect gene skn-1 prevent EMS from producing both pharyngeal and intestinal cells (Bowerman et al., 1992). Instead, its daughters E and MS both make body
4 4196 C. Wittmann and others of 21 lineaged skn-1 animals that do not form gut granules showed normal ceh-13 expression in the Ep daughter cells. Again, this may be caused by incomplete penetrance of the skn- 1 allele zu67. Weak ceh-13 in skn-1 animals without gut differentiation contrasts the situation in trypan-blue-treated embryos where, in absence of gut induction, no ceh-13 expression occurs. This difference may be explained by the fact that ceh-13 regulation is only partially SKN-1 dependent. SKN-1 is required for specifying the identity of the EMS blastomere and both of its descendants E and MS, but MS does not express ceh-13. Thus, SKN-1 is necessary but not sufficient to specify the E fate and additional factors must act to establish the gut founder cell. It has been suggested that P2 is the source of a polarizing signal that orients the EMS cleavage axis and causes EMS to divide asymmetrically and to produce MS and E fates (Goldstein, 1992, 1993). If this induction is inhibited by treating 4-cellstage embryos with trypan blue, no gut is formed and no ceh- 13 expression is observed in the Ep cell, indicating that P 2 - EMS interaction may be required for ceh-13 activation. In skn-1 embryos, this interaction may still occur and provide the cue for the activation of low levels of ceh-13 expression even in the absence of differentiated gut. Altogether, our data with pop-1 and skn-1 mutants and the results with trypan-bluetreated embryos demonstrate that normal ceh-13 expression in Ep correlates with the intestinal cell fate, but does not depend on the position of the cell in the embryo. Fig. 1. Expression of ceh-13 in C. elegans early embryos. (A-D) 28- cell-stage embryo (this embryo has16 AB-derived cells, we call it a 16-AB stage). (A,C) Ventral and (B,D) dorsal views. (A,B) Immunofluorescent staining of Ep and ABxxxp cells with anti- CEH-13 antibodies. The ABarap cell stains very weakly. (C,D) The same embryo stained with the DNA-binding dye DAPI. The names of the blastomeres are indicated (see Materials and Methods for blastomere identification); lower cases are used for AB-derivatives. (E) 32-AB-stage embryo expressing the ceh-13::gfp transgene. All fluorescent nuclei are visible. CEH-13::GFP is present in Epl and Epr (arrowheads) and in 14 out of 16 daughters of ABxxxp (arrows). The image has been generated by adding fluorescent pictures of two different focal planes. (F) Confocal microscope section of an about 50-cell-stage embryo showing CEH-13::GFP in Da and Dp (arrows). All embryos are oriented with the anterior end to the left. Embryos measure approximately 50 µm along their anteroposterior axis. wall muscles and hypodermal cells similar to the C blastomere, the anterior daughter of P 2 (Bowermann et al., 1992). We have lineaged embryos from skn-1(iv); swis1[rol-6(su1006)+ceh- 13::gfp] II (FR525) homozygous mothers until the 2 daughters of Ep were born and scored for the presence of CEH-13::GFP in these cells. Since the zu67 allele shows incomplete penetrance (Bowerman et al., 1992, 1993), we also scored gut fluorescence in our lineaged embryos after a few hours of further development. The Ep daughters of skn-1 embryos with differentiated gut (n=10) always showed wild-type levels of CEH- 13 (equal levels of GFP fluorescence in Ep daughters and AB descendants, Fig. 4D). Out of 21 lineaged embryos that did not develop gut granules, however, 15 showed a significantly reduced CEH-13::GFP signal in the two daughter cells of Ep (Fig. 4C,E,F). This demonstrates that mutations in skn-1 can interfere with the normal expression of ceh-13, but only in Ep cells that do not differentiate gut granules. Surprisingly, 6 out Asymmetric ceh-13 activation does not depend on cell-cell contacts An important finding is that ceh-13 activation in the E and soon afterwards in the AB lineages always occurs in the posterior daughters of anteroposterior cell division (cf. Fig. 2). In the D lineage, weak expression was observed in the Da and Dp cells, and it is tempting to speculate that onset of ceh-13 expression below the detection level may already occur in the D cell, the posterior daughter of the P 3 cell. From our data, thus, an apparent rule emerges; the potential to produce CEH-13 is present in the posterior daughters of the 4th anteroposterior division in the E and possibly the D lineage and of the 5th division in the AB lineage (which corresponds also to the 4th anteroposterior cleavage, since the 3rd cleavage in the AB lineage occurs along the left-right axis) and is distributed to the daughters of these cells. This asymmetric ceh-13 activation pattern may be the result of asymmetric segregation of regulatory factors or of cell-cell interactions, or both. Because pop-1 mutants express ceh-13 ectopically in their MSp cells (Fig. 4A), it is unlikely that asymmetric activation depends on specific cell-cell interactions of Ep with its neighboring cells. To confirm this assumption, we have removed the late P 2 -blastomere after the division of the AB cells from 2 to 4 through a laser-induced hole in the eggshell (n=6, Fig. 5). In these embryos, gut induction had already occurred and ceh-13::gfp is correctly expressed in Ep. This result demonstrates that after gut induction cell-cell contacts between E and C, Cp and P 3, as well as between Ep and D, Cp and P 4, are not required for correct ceh-13::gfp activation in Ep. Furthermore, removal of P 2 had no influence on the ceh-13 expression pattern in the ABxxxp cells of the 32- AB cell stage (14±2 fluorescent nuclei, n=6; data not shown). These results suggest that ceh-13 activation in Ep and in the
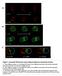
5 Early embryonic Hox gene activation ZYGOTE = P0 Fig. 2. Summary of the initial ceh- 13 expression in the AB, E and D lineages of C. elegans. Numbers indicate the number of anteroposterior divisions from the beginning of development. a stands for anterior, p for posterior, l for left and r for right. The 3rd division in the AB lineage is a left-right division, whose daughters are indicated as ABxl/r. Filled circles, cells expressing ceh-13; open circles, cells not expressing ceh AB (P0a) P1 (P0p) ABa ABp ABal/r ABpl/r 3 ABal/ra ABal/rp ABpl/ra ABpl/rp ABal/raa ABal/rap 28-cell stage 2 ABal/rpa ABal/rpp 5 5 ABpl/raa ABpl/rap ABpl/rpa ABpl/rpp EMS P2 3 MS E 4 Ea Ep Epl Epr 4 P3 P4 D 5 5 C ABxxxp cells does not depend on interactions with any of the P 2 descendants. In order to investigate a possible role of postdivision cellcell interactions in ceh-13 activation, ceh-13::gfp-carrying embryos were incubated with cytochalasin D. Cytochalasin D disrupts the C. elegans actin cytoskeleton within 1 minute of exposure time and blocks cell divisions upon continuous incubation without stopping nuclear divisions (Hill and Strome, 1988). The embryos were treated with cytochalasin D at different timepoints in development, followed under the microscope until the number of their nuclei reached the equivalent of 28- or 46-cell-stage embryos and assayed for ceh-13::gfp expression. Some embryos were further incubated for later scoring of the intestinal-specific rhabditin granules by fluorescence (Cowan and McIntosh, 1985; Laufer et al., 1980; Goldstein, 1995a,b). No GFP expression could be detected in arrested zygotes (n=8). 2-cell-stage embryos also showed no GFP expression, even if gut granules could later be scored (n=10, Fig. 6A,B). 4-cell-stage-blocked embryos, however, could be classified into three categories: non-fluorescing embryos (n=4), embryos with ceh-13::gfp expression in all ABp nuclei only (n=6, Fig. 6D), and embryos with expression in all ABp and EMS nuclei (n=6, Fig. 6F; in these embryos, EMS expression was always correlated with gut granule formation). From these results, we conclude that, after the 4- cell stage, neither normal cell-cell contacts nor an intact actin cytoskeleton are required for the onset of ceh-13 expression in the ABp and EMS lineages, which therefore differ from the ABa and P2 lineages. Alternatively, however, it is also possible that normal cell-cell interactions are required, but that inductions occur between non-normal cells in the blocked embryos. Contact between AB and P1 is not required for ceh- 13 expression It has been proposed that a polarising induction of P 1 participates in specifying anteroposterior differences within the AB lineage (Hutter and Schnabel, 1994). To test whether an AB- P 1 interaction is necessary for ceh-13 activation, we isolated AB and P 1 blastomeres and cultured them separately. When the Fig. 3. Localization of ceh-13 mrna in a 26-cell-stage C. elegans embryo by whole-mount in situ hybridization. Nomarski picture showing staining in Ep (arrowhead). Fig. 4. ceh-13::gfp expression in pop-1 and skn-1 mutant backgrounds. (A) 50-cell-stage lin-17 pop-1 embryo showing expression of the ceh-13::gfp reporter in the daughter cells of Ep (arrowheads), and in the ectopic intestinal cells MSpa and MSpp (arrows). (B) Lin-17 control embryos show normal CEH-13::GFP expression in the Ep cells (arrowhead), but no expression in MSp daughters (arrow). (C) Skn-1 embryo showing weaker expression than normal in the Ep cell (arrowhead). (D) Skn-1 escaper (with gut fluorescence) showing normal expression in the Ep cell (arrowhead). (E) Nomarski picture of C after a few hours of development. (F) Same embryo as in E, UV excited. No gut autofluorescence was detected.
6 4198 C. Wittmann and others Fig. 5. CEH-13::GFP expression in an embryo in which P 2 has been removed. (A) Laser removal of P 2 was performed at the time when the two anterior AB cells started to divide. (B) The same embryo at the equivalent of the 28-cell stage. Ep is indicated with an arrowhead. (C) Same embryo as in B: the CEH-13::GFP fusion protein was expressed in Ep (arrowhead). In Drosophila, position-specific signals located along the anteroposterior axis establish the pattern of HOM-C gene expression (Lawrence and Morata, 1994). In the nematode C. elegans, it is not yet clear how the pattern of HOM-C/Hox gene expression is regulated. Our data suggest that cell fate determinants rather than positional information play a fundamental role in ceh-13 activation. Indeed, ceh-13 is ectopically expressed in the MS lineage of pop-1 embryos, indicating that the activation of this gene in the intestinal precursor cell Ep is independent on the position of this cell in the embryo. Furthermore, cytochalasin D and P 2 -removal experiments suggest that ceh-13 activation does not depend on the interaction of Ep with its neighbouring cells. Rather, the Ep cell fate itself is necessary and sufficient for correct activation of ceh-13. This is also supported by strongly reduced levels of CEH-13::GFP in skn-1 embroys, which fail to produce intestinal cells (Bowermann et al., 1992), and by the complete absence of ceh- 13 expression in embryos where gut induction was inhibited by trypan blue treatment (Bossinger and Schierenberg, 1996). Furthermore, incubation of 4-cell-stage embryos with cytochalasin D blocks the onset of ceh-13 expression in the ABa cell, but not in the ABp cell, suggesting that the regulation of ceh-13 expression is modulated by cell-specific cues. Altogether, our results indicate that the onset of ceh-13 expression during early embryonic development is cell fate dependent. isolates reached the equivalent of the 26-cell stage (16 ABa like cells, 10 P 1 -derived cells), each isolate was examined for CEH-13 expression (see Materials and Methods). Five of six AB isolates contained CEH-13 in up to 8 of the total 16 cells (Fig. 7A). All of eight P 1 isolates were positive for CEH-13 in the Ep cell as demonstrated by cell lineaging analysis (Fig. 7B). Even cell isolations carried out very early in the 2-cell stage (5 minutes after the start of cytokinesis in P 0 for AB isolates, and 7 minutes for P 1 isolates) were positive for CEH- 13. These results were confirmed by removing either the AB or the P 1 cell from 2-cell embryos through a laser-introduced hole in the eggshell (results not shown). Altogether, our data suggest that contact between P 1 and AB is not likely to be required for ceh-13 activation. DISCUSSION ceh-13 activation depends on cell fate and on anteroposterior cell polarity The homolog of the Drosophila HOM-C gene labial, ceh-13, is the first gene of the C. elegans Hox gene cluster to be activated during development (Wittmann et al., unpublished data). The early expression of ceh-13 begins at the 26-cell stage, shortly before the onset of gastrulation in the intestinal precursor cell Ep, and soon afterwards in all ABxxxp cells. One division round later, the ceh-13::gfp reporter gene becomes activated in the Da and Dp cells, however, no ceh-13 expression is seen in the MS and C lineages at this stage. Fig. 6. CEH-13::GFP expression in cytochalasin-d-blocked embryos. (A) Nomarski picture of an embryo blocked at the 2-cell stage and incubated in cytochalasin D until the embryo reached the equivalent of the 28-cell stage. (B) Fluorescent picture of the same embryo as in A showing rhabditin granules in P 1 but no ceh-13::gfp expression. (C) Nomarski picture of a 4-cell-stage-blocked embryo. (D) Fluorescent picture of the embryo in C with CEH-13::GFP expression in the ABp cell. (E) Nomarski picture of another 4-cellstage-blocked embryo. (F) Same embryo as in E. CEH-13::GFP fluorescence was detected in the ABp and EMS cells. No expression was present in the ABa cell.
7 Early embryonic Hox gene activation 4199 Fig. 7. Confocal microscope sections of the results of 2-cell-stage blastomere isolations, processed for anti-ceh-13 fluorescence at the equivalent of the 26-cell stage. (A) 8 of the 16 progeny of an AB isolate showed nuclear CEH-13; the identity of these 8 cells was not determined. 5 of the positive nuclei are shown in this section. (B) One cell of the 10 progeny of P 1 showed nuclear CEH-13. Each cell was identified by following the cell lineage of the isolate and also by DAPI-staining of the fixed isolate. The stained cell is Ep. Our findings are consistent with recent data of another member of the C. elegans Hox gene cluster, mab-5. Cowing and Kenyon (1996) demonstrated that the expression of mab-5 in mesodermal and ectodermal tissues during embryonic and postembryonic development is highly correlated with cell fate and does not depend on cell position. These findings imply that mechanisms other than cell-extrinsic A-P positional signals play an important role in the activation and regulation of the C. elegans Hox genes. In addition to cell fate determinants, polarity cues are also important in regulating early ceh-13 expression, an interpretation supported by the observation that the first activation of ceh-13 in the E, AB and probably also D lineages always occurs in the posterior daughters of the 4th embryonic anteroposterior division (corresponding to the 5th division in the AB lineage). This suggests that these cells contain information regarding their position along the last division axis and that this information is passed along to the ceh-13 regulatory pathway. The polarity of the E cell, e.g., does not depend on its position in the embryo, since in pop-1 mutants the two daughters of the MS cell, which ectopically adopts the fate of its posterior sister cell E (Lin et al., 1995), show the same difference (i.e., only the daughters of the MSp cell express ceh-13). In summary, our data demonstrate that the cells first expressing ceh-13 in early embryos are always the posterior daughters of anteroposterior cell division, regardless of where they are born, and suggest the existence of an anteroposterior cellular polarity system that is involved in the activation of ceh-13. This cellular polarity system may act cell-autonomously and be present in all daugthers of the 4th anteroposterior divisions of the embryo, irrespective of whether these cells express ceh-13 or not. Alternatively, however, there could be a global, cell-extrinsic polarity system acting in the embryo. The existence of an animal-wide system directing anteroposterior cellular polarity has already been proposed (Way et al., 1994; Priess, 1994). Such a system may also apply in later developmental stages and for other genes. For example, a MEC-3-expressing cell is always the anterior daughter of a mechanoreceptor-generating asymmetric division (reviewed in Way et al., 1994) and MAB-5::LACZ first appears in the posterior daughters of the ABpxppp cells (Cowing and Kenyon, 1992). VAB-7, another homeodomain-containing protein is first present in the posterior daughters of Cxx cells (Ahringer, 1996). This suggests that similar polarity cues acting in ceh-13 activation may be involved in the regulation of these genes. The control of HOM-C/hox gene expression by cellular rather than global embryonic polarity cues does not seem to be restricted to C. elegans. In Drosophila, higher levels of LAB have been observed in the delaminating procephalic neuroblasts compared to the protein levels observed in the ectodermal sisters of these cells during the development of the nervous system (Diederich et al., 1989). The cell division that generates these neuroblasts, is asymmetric and controlled by the INSCUTEABLE protein (Kraut et al., 1996), suggesting that labial expression is also correlated with cellular polarity. Therefore, the mechanisms integrating cellular polarity and homeobox gene expression, particularly labial-like gene activity, may have been conserved during evolution. Molecular mechanisms of ceh-13 regulation The fact that ceh-13 activation is dependent on cell fate and cell polarity strongly suggests that the regulatory information, or at least a part of it, is segregated asymmetrically to the ceh- 13-expressing cells. During the first few embryonic divisions, differentially localized maternal gene products account for the various developmental potentials of the first few embryonic cells (Priess, 1994). Our data show that at least two of these proteins, SKN-1 and POP-1, influence ceh-13 expression in Ep and may function as cell-specific or cell-lineage-specific modulators of ceh-13 activation. It remains to be determined whether these two regulatory proteins directly bind to the promoter region of ceh-13. The activation of ceh-13 expression may depend on other activator and inhibitor molecules that are differently distributed during early embryonic development. 1- and 2-cell embryos blocked by cytochalasin D did not express GFP::CEH-13, suggesting that early embryos may contain one or several inhibitors of ceh-13 expression. After the 4-cell stage, the cytochalasin D sensitivity is released in ABp and EMS, but not in ABa and P2. The finding that 4-cell-stageblocked embryos can be classified into non-fluorescing embryos, embryos with ceh-13::gfp expression in all ABp nuclei only, and embryos with expression in all ABp and EMS nuclei, suggest that these inhibitors could be inactivated in the ABp and EMS lineages as the result of inducing events. In the ABa lineage, the inhibitor of ceh-13 expression may then be segregated away from the ceh-13-expressing ABaxxp cells. Alternatively, the ABa descendants may require a positional signal or a cell-cell interaction. In the ABp and E lineages, ceh-13 activation may depend on an activator molecule that is distributed to the ceh-13-expressing Ep and ABpxxp cells. The correct distribution of both, the inhibitor in the ABa
8 4200 C. Wittmann and others lineage and the activator in the ABp and E lineages, may depend on the same polarity system that is active during the 4th embryonic anteroposterior cell division. Further analysis will be needed to reveal the mechanisms of early ceh-13 activation and to understand the role of asymmetric cell divisions in ceh-13 expression and in embryonic patterning in general. We thank Drs J. R. Priess (Fred Hutchinson Cancer Research Center, Seattle), C. C. Mello (University of Massachusetts, Worcester) and the C. elegans genetic stock center for providing strains, Dr A. Fire (Carnegie Institution of Washington, Baltimore) for the vectors, Dr Monique Zetka for critical reading of the manuscript, M. Zbinden for excellent technical help and Ilford for image printing. This research was supported by Swiss National Science Foundation Grants and /1. REFERENCES Ahringer, J. (1996). Posterior patterning by the Caenorhabditis elegans evenskipped homolog vab-7. Genes Dev. 10, Bossinger, O., and Schierenberg, E. (1996). Early embryonic induction in C. elegans can be inhibited with polysulfated hydrocarbon dyes. Dev. Biol. 176, Bossinger, O. and Schierenberg, E. (1992). Transfer and tissue-specific accumulation of cytoplasmic components in embryos of Caenorhabditis elegans and Rhabditis dolichura. Development 114, Bowerman, B., Eaton, B. A., and Priess, J. R. (1992). skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68, Bowerman, B., Draper, B. W., Mello, C. C., and Priess, J. R. (1993). The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74, Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, Bürglin, T.R., Ruvkun, G., Coulson, A., Hawkins, N.C., McGhee, J.D., Schaller, D., Wittmann, C., Müller, F., Waterston, R.H. (1991). Nematode homeobox cluster. Nature 351, 703. Bürglin, T.R. and Ruvkun, G. (1993). The Caenorhabditis elegans homeobox gene cluster. Curr. Opin. Genet. Dev. 3, Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W., Prasher, D.C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, Clark, S.G., Chisholm, A.D. and Horvitz, H.R. (1993). Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell 74, Cowan, A.E. and McIntosh, J.R. (1985). Mapping the distribution of differentiation potential for intestine, muscle, and hypodermis during early development in Caenorhabditis elegans. Cell 41, Cowing, D. W., and Kenyon, C. (1992). Expression of the homeotic gene mab- 5 during Caenorhabditis elegans embryogenesis. Development 116, Cowing, D., and Kenyon, D. (1996). Correct Hox gene expression established indpendently of position in Caenorhabditis elegans. Nature 382, Diederich, R. J., Merrill, V. K., Pultz, M. A., and Kaufman, T. C. (1989). Isolation, structure, and expression of labial, a homeotic gene of the Antennapedia Complex involved in Drosophila head development. Genes Dev. 3, Edgar, L.G. (1995). Blastomere culture and analysis. In Caenorhabditis elegans: Modern Biological Analysis of an Organism (ed. H. F. Epstein and D. C. Shakes), pp New York: Academic Press. Finney, M. and Ruvkun, G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, Goldstein, B. (1992). Induction of gut in Caenorhabditis elegans embryos. Nature 357, Goldstein, B. (1993). Establishment of gut fate in the E lineage of C. elegans: the roles of lineage-dependent mechanisms and cell interactions. Development 118, Goldstein, B. (1995a). An analysis of the response to gut induction in the C. elegans embryo. Development 121, Goldstein, B. (1995b). Cell contacts orient some cell division axes in the Caenorhabditis elegans embryo. J. Cell Biol. 129, Guo, S., and Kemphues, K. J. (1996). Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr. Opin. Gen. Dev. 6, Heim, R., Cubitt, A., Tsien, R. (1995). Improved green fluorescence. Nature 373, Hill, D.P. and Strome, S. (1988). An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 125, Hunter, C.P. and Kenyon, C. (1995). Specification of anteroposterior cell fates in Caenorhabditis elegans by Drosophila Hox proteins. Nature 377, Hutter, H., and Schnabel, R. (1994). glp-1 and inductions establishing embryonic axis in C. elegans. Development 120, Kenyon C. and Wang, B.B. (1991). A cluster of Antennapedia-class homeobox genes in a nonsegmented animal. Science 253, Kraut, R., Chia, W., Yeh Jan, L., Nung Jan, Y., and Knoblich, J. A. (1996). Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, Laufer, J.S., Bazzicalupo, P., Wood, W.B. (1980). Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 19, Lawrence, P.A. and Morata, G. (1994). Homeobox genes: Their function in Drosophila segmentation and pattern formation. Cell 78, Lewis, J.A. and Fleming, J.T. (1995). Basic culture methods. In Caenorhabditis elegans: Modern Biological Analysis of an Organism (ed. H. F. Epstein and D. C. Shakes), pp New York: Academic Press. Lin, R., Thompson, S., Priess, J.R. (1995). pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83, Mello, C. C., and Fire, A. (1995). DNA transformation. In In Caenorhabditis elegans: Modern Biological Analysis of an Organism (ed. H. F. Epstein and D. C. Shakes), pp New York: Academic Press. Okamoto, H. and Thomson, J.N. (1985). Monoclonal antibodies which distinguish certain classes of neuronal and supporting cells in the nervous tissue of the nematode Caenorhabditis elegans. J. Neurosci. 5, Okkema, P.G. and Fire, A. (1994). The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development 120, Priess, J. R. (1994). Establishment of initial asymmetry in early Caenorhabditis elegans embryos. Curr. Opin. Genet. Dev. 4, Schaller, D., Wittmann, C., Spicher, A., Müller, F., and Tobler, H. (1990). Cloning and analysis of three new homeobox genes from the nematode Caenorhabditis elegans. Nucl. Acids Res. 18, Schierenberg, E. (1984). Altered cell division rates after laser-induced cell fusion in nematode embryos. Dev. Biol. 101, Schierenberg, E. and Wood, W.B. (1985). Control of cell-cycle timing in early embryos of Caenorhabditis elegans. Dev. Biol. 107, Schierenberg, E. (1987). Reversal of cellular polarity and early cell-cell interactions in the embryo of Caenorhabditis elegans. Dev. Biol. 122, Seydoux, G. and Fire, A. (1995). In In Caenorhabditis elegans: Modern Biological Analysis of an Organism (ed. H. F. Epstein and D. C. Shakes), pp New York: Academic Press. Sharkey, M., Graba, Y. and Scott, M.P. (1997). Hox genes in evolution: protein surfaces and paralog groups. Trends Genet. 13, Sulston, J. E., and Horvitz, H. R. (1977). Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol. 56, Sulston, J. E., Schierenberg, E., White, J. G., and Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, Wang, B., Müller-Immerglück, M.M., Austin, J., Robinson, N.T., Chisholm, A. and Kenyon, C (1993). A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell 74, Way, J. C., Wang, L., Run, J.-Q., and Hung, M.-S. (1994). Cell polarity and the mechanism of asymmetric cell division. BioEssays 16, Wilson, R. et al. (1994). 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature 368, (Accepted 11 August 1997)
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Caenorhabditis elegans
 Caenorhabditis elegans Why C. elegans? Sea urchins have told us much about embryogenesis. They are suited well for study in the lab; however, they do not tell us much about the genetics involved in embryogenesis.
Caenorhabditis elegans Why C. elegans? Sea urchins have told us much about embryogenesis. They are suited well for study in the lab; however, they do not tell us much about the genetics involved in embryogenesis.
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 11. Development: Differentiation and Determination
 KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
Combinatorial specification of blastomere identity by glp-1-dependent cellular interactions in the nematode Caenorhabditis elegans
 Development 20, 3325-3338 (994) Printed in Great Britain The Company of Biologists Limited 994 3325 Combinatorial specification of blastomere identity by glp--dependent cellular interactions in the nematode
Development 20, 3325-3338 (994) Printed in Great Britain The Company of Biologists Limited 994 3325 Combinatorial specification of blastomere identity by glp--dependent cellular interactions in the nematode
Exam 1 ID#: October 4, 2007
 Biology 4361 Name: KEY Exam 1 ID#: October 4, 2007 Multiple choice (one point each) (1-25) 1. The process of cells forming tissues and organs is called a. morphogenesis. b. differentiation. c. allometry.
Biology 4361 Name: KEY Exam 1 ID#: October 4, 2007 Multiple choice (one point each) (1-25) 1. The process of cells forming tissues and organs is called a. morphogenesis. b. differentiation. c. allometry.
Homeotic genes in flies. Sem 9.3.B.6 Animal Science
 Homeotic genes in flies Sem 9.3.B.6 Animal Science So far We have seen that identities of each segment is determined by various regulators of segment polarity genes In arthopods, and in flies, each segment
Homeotic genes in flies Sem 9.3.B.6 Animal Science So far We have seen that identities of each segment is determined by various regulators of segment polarity genes In arthopods, and in flies, each segment
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
Green Fluorescent Protein (GFP) Today s Nobel Prize in Chemistry
 In the news: High-throughput sequencing using Solexa/Illumina technology The copy number of each fetal chromosome can be determined by direct sequencing of DNA in cell-free plasma from pregnant women Confession:
In the news: High-throughput sequencing using Solexa/Illumina technology The copy number of each fetal chromosome can be determined by direct sequencing of DNA in cell-free plasma from pregnant women Confession:
MBios 401/501: Lecture 14.2 Cell Differentiation I. Slide #1. Cell Differentiation
 MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
Regulative Development in a Nematode Embryo: A Hierarchy of Cell Fate Transformations
 Developmental Biology 215, 1 12 (1999) Article ID dbio.1999.9423, available online at http://www.idealibrary.com on Regulative Development in a Nematode Embryo: A Hierarchy of Cell Fate Transformations
Developmental Biology 215, 1 12 (1999) Article ID dbio.1999.9423, available online at http://www.idealibrary.com on Regulative Development in a Nematode Embryo: A Hierarchy of Cell Fate Transformations
Developmental genetics: finding the genes that regulate development
 Developmental Biology BY1101 P. Murphy Lecture 9 Developmental genetics: finding the genes that regulate development Introduction The application of genetic analysis and DNA technology to the study of
Developmental Biology BY1101 P. Murphy Lecture 9 Developmental genetics: finding the genes that regulate development Introduction The application of genetic analysis and DNA technology to the study of
Midterm 1. Average score: 74.4 Median score: 77
 Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
interactions and are regulated by maternal GLP-1 signaling in Caenorhabditis
 Development 122, 4105-4117 (1996) Printed in Great ritain The Company of iologists Limited 1996 DEV3495 4105 lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are
Development 122, 4105-4117 (1996) Printed in Great ritain The Company of iologists Limited 1996 DEV3495 4105 lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans
 Development 127, 4587-4598 (2) Printed in Great Britain The Company of Biologists Limited 2 DEV4424 4587 Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans
Development 127, 4587-4598 (2) Printed in Great Britain The Company of Biologists Limited 2 DEV4424 4587 Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation and cell fate patterning in early C.
 Development 128, 4301-4314 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV3466 4301 The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation
Development 128, 4301-4314 (2001) Printed in Great Britain The Company of Biologists Limited 2001 DEV3466 4301 The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation
Genes, Development, and Evolution
 14 Genes, Development, and Evolution Chapter 14 Genes, Development, and Evolution Key Concepts 14.1 Development Involves Distinct but Overlapping Processes 14.2 Changes in Gene Expression Underlie Cell
14 Genes, Development, and Evolution Chapter 14 Genes, Development, and Evolution Key Concepts 14.1 Development Involves Distinct but Overlapping Processes 14.2 Changes in Gene Expression Underlie Cell
18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis
 18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis An organism arises from a fertilized egg cell as the result of three interrelated processes: cell division, cell
18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis An organism arises from a fertilized egg cell as the result of three interrelated processes: cell division, cell
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Establishment of POP-1 asymmetry in early C. elegans embryos
 Development 130, 3547-3556 2003 The Company of Biologists Ltd doi:10.1242/dev.00563 3547 Establishment of POP-1 asymmetry in early C. elegans embryos Frederick D. Park and James R. Priess Division of Basic
Development 130, 3547-3556 2003 The Company of Biologists Ltd doi:10.1242/dev.00563 3547 Establishment of POP-1 asymmetry in early C. elegans embryos Frederick D. Park and James R. Priess Division of Basic
MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION
 MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION Drosophila is the best understood of all developmental systems, especially at the genetic level, and although it is an invertebrate it has had an enormous
MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION Drosophila is the best understood of all developmental systems, especially at the genetic level, and although it is an invertebrate it has had an enormous
C. elegans L1 cell adhesion molecule functions in axon guidance
 C. elegans L1 cell adhesion molecule functions in axon guidance Biorad Lihsia Chen Dept. of Genetics, Cell Biology & Development Developmental Biology Center C. elegans embryogenesis Goldstein lab, UNC-Chapel
C. elegans L1 cell adhesion molecule functions in axon guidance Biorad Lihsia Chen Dept. of Genetics, Cell Biology & Development Developmental Biology Center C. elegans embryogenesis Goldstein lab, UNC-Chapel
Unicellular: Cells change function in response to a temporal plan, such as the cell cycle.
 Spatial organization is a key difference between unicellular organisms and metazoans Unicellular: Cells change function in response to a temporal plan, such as the cell cycle. Cells differentiate as a
Spatial organization is a key difference between unicellular organisms and metazoans Unicellular: Cells change function in response to a temporal plan, such as the cell cycle. Cells differentiate as a
Developmental Biology Lecture Outlines
 Developmental Biology Lecture Outlines Lecture 01: Introduction Course content Developmental Biology Obsolete hypotheses Current theory Lecture 02: Gametogenesis Spermatozoa Spermatozoon function Spermatozoon
Developmental Biology Lecture Outlines Lecture 01: Introduction Course content Developmental Biology Obsolete hypotheses Current theory Lecture 02: Gametogenesis Spermatozoa Spermatozoon function Spermatozoon
Lecture 7. Development of the Fruit Fly Drosophila
 BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
Question Set # 4 Answer Key 7.22 Nov. 2002
 Question Set # 4 Answer Key 7.22 Nov. 2002 1) A variety of reagents and approaches are frequently used by developmental biologists to understand the tissue interactions and molecular signaling pathways
Question Set # 4 Answer Key 7.22 Nov. 2002 1) A variety of reagents and approaches are frequently used by developmental biologists to understand the tissue interactions and molecular signaling pathways
Asymmetric movements of cytoplasmic components in Caenorhabditis elegans zygotes
 J. Embryol. exp. Morph. 97 Supplement, 15-29 (1986) 15 Printed in Great Britain The Company of Biologists Limited 1986 Asymmetric movements of cytoplasmic components in Caenorhabditis elegans zygotes SUSAN
J. Embryol. exp. Morph. 97 Supplement, 15-29 (1986) 15 Printed in Great Britain The Company of Biologists Limited 1986 Asymmetric movements of cytoplasmic components in Caenorhabditis elegans zygotes SUSAN
Development of Drosophila
 Development of Drosophila Hand-out CBT Chapter 2 Wolpert, 5 th edition March 2018 Introduction 6. Introduction Drosophila melanogaster, the fruit fly, is found in all warm countries. In cooler regions,
Development of Drosophila Hand-out CBT Chapter 2 Wolpert, 5 th edition March 2018 Introduction 6. Introduction Drosophila melanogaster, the fruit fly, is found in all warm countries. In cooler regions,
Supplementary Information
 Supplementary Information Supplementary figures % Occupancy 90 80 70 60 50 40 30 20 Wt tol-1(nr2033) Figure S1. Avoidance behavior to S. enterica was not observed in wild-type or tol-1(nr2033) mutant nematodes.
Supplementary Information Supplementary figures % Occupancy 90 80 70 60 50 40 30 20 Wt tol-1(nr2033) Figure S1. Avoidance behavior to S. enterica was not observed in wild-type or tol-1(nr2033) mutant nematodes.
POP-1 and Anterior Posterior Fate Decisions in C. elegans Embryos
 Cell, Vol. 92, 229 239, January 23, 1998, Copyright 1998 by Cell Press POP-1 and Anterior Posterior Fate Decisions in C. elegans Embryos Rueyling Lin, Russell J. Hill, and James R. Priess* Division of
Cell, Vol. 92, 229 239, January 23, 1998, Copyright 1998 by Cell Press POP-1 and Anterior Posterior Fate Decisions in C. elegans Embryos Rueyling Lin, Russell J. Hill, and James R. Priess* Division of
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila July 9, 2008 Drosophila Development Overview Fertilization Cleavage Gastrulation Drosophila body plan Oocyte formation Genetic control
Developmental Biology Biology 4361 Axis Specification in Drosophila July 9, 2008 Drosophila Development Overview Fertilization Cleavage Gastrulation Drosophila body plan Oocyte formation Genetic control
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Upstream Elements Regulating mir-241 and mir-48 Abstract Introduction
 Upstream Elements Regulating mir-241 and mir-48 Hanna Vollbrecht, Tamar Resnick, and Ann Rougvie University of Minnesota: Twin Cities Undergraduate Research Scholarship 2012-2013 Abstract Caenorhabditis
Upstream Elements Regulating mir-241 and mir-48 Hanna Vollbrecht, Tamar Resnick, and Ann Rougvie University of Minnesota: Twin Cities Undergraduate Research Scholarship 2012-2013 Abstract Caenorhabditis
Developmental switches and gene regulatory networks in the most completely described animal, the nematode C. elegans
 Developmental switches and gene regulatory networks in the most completely described animal, the nematode. elegans Joel Rothman U Santa Barbara Generating developmental diversity: Symmetry breaking Dr.
Developmental switches and gene regulatory networks in the most completely described animal, the nematode. elegans Joel Rothman U Santa Barbara Generating developmental diversity: Symmetry breaking Dr.
Mesoderm Induction CBT, 2018 Hand-out CBT March 2018
 Mesoderm Induction CBT, 2018 Hand-out CBT March 2018 Introduction 3. Books This module is based on the following books: - 'Principles of Developement', Lewis Wolpert, et al., fifth edition, 2015 - 'Developmental
Mesoderm Induction CBT, 2018 Hand-out CBT March 2018 Introduction 3. Books This module is based on the following books: - 'Principles of Developement', Lewis Wolpert, et al., fifth edition, 2015 - 'Developmental
A complementation test would be done by crossing the haploid strains and scoring the phenotype in the diploids.
 Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
A Genetic Mosaic Screen of Essential Zygotic Genes in Caenorhabditis elegans
 Copyright 0 1991 by the Genetics Society of America A Genetic Mosaic Screen of Essential Zygotic Genes in Caenorhabditis elegans Elizabeth A. Bucher and Iva Greenwald Department of Molecular Biology, Princeton
Copyright 0 1991 by the Genetics Society of America A Genetic Mosaic Screen of Essential Zygotic Genes in Caenorhabditis elegans Elizabeth A. Bucher and Iva Greenwald Department of Molecular Biology, Princeton
Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases
 Development 130, 1255-1265 2003 The Company of Biologists Ltd doi:10.1242/dev.00284 1255 Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases Adrian A. Cuenca
Development 130, 1255-1265 2003 The Company of Biologists Ltd doi:10.1242/dev.00284 1255 Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases Adrian A. Cuenca
The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development
 Developmental Biology 285 (2005) 510 523 Genomes & Developmental Control The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development Morris
Developmental Biology 285 (2005) 510 523 Genomes & Developmental Control The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development Morris
Wan-Ju Liu 1,2, John S Reece-Hoyes 3, Albertha JM Walhout 3 and David M Eisenmann 1*
 Liu et al. BMC Developmental Biology 2014, 14:17 RESEARCH ARTICLE Open Access Multiple transcription factors directly regulate Hox gene lin-39 expression in ventral hypodermal cells of the C. elegans embryo
Liu et al. BMC Developmental Biology 2014, 14:17 RESEARCH ARTICLE Open Access Multiple transcription factors directly regulate Hox gene lin-39 expression in ventral hypodermal cells of the C. elegans embryo
Drosophila Life Cycle
 Drosophila Life Cycle 1 Early Drosophila Cleavage Nuclei migrate to periphery after 10 nuclear divisions. Cellularization occurs when plasma membrane folds in to divide nuclei into cells. Drosophila Superficial
Drosophila Life Cycle 1 Early Drosophila Cleavage Nuclei migrate to periphery after 10 nuclear divisions. Cellularization occurs when plasma membrane folds in to divide nuclei into cells. Drosophila Superficial
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila
 Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
GENETIC CONTROL OF CELL INTERACTIONS IN NEMATODE DEVELOPMENT
 Annu. Rev. Genet. 1991. 25. 411-36 Copyright 199l by Annual Reviews Inc. All rights reserved GENETIC CONTROL OF CELL INTERACTIONS IN NEMATODE DEVELOPMENT E. J. Lambie and Judith Kimble Laboratory of Molecular
Annu. Rev. Genet. 1991. 25. 411-36 Copyright 199l by Annual Reviews Inc. All rights reserved GENETIC CONTROL OF CELL INTERACTIONS IN NEMATODE DEVELOPMENT E. J. Lambie and Judith Kimble Laboratory of Molecular
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1178343/dc1 Supporting Online Material for Starvation Protects Germline Stem Cells and Extends Reproductive Longevity in C. elegans This PDF file includes: Giana Angelo
www.sciencemag.org/cgi/content/full/1178343/dc1 Supporting Online Material for Starvation Protects Germline Stem Cells and Extends Reproductive Longevity in C. elegans This PDF file includes: Giana Angelo
Reading: Chapter 5, pp ; Reference chapter D, pp Problem set F
 Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Early Development in Invertebrates
 Developmental Biology Biology 4361 Early Development in Invertebrates October 25, 2006 Early Development Overview Cleavage rapid cell divisions divisions of fertilized egg into many cells Gastrulation
Developmental Biology Biology 4361 Early Development in Invertebrates October 25, 2006 Early Development Overview Cleavage rapid cell divisions divisions of fertilized egg into many cells Gastrulation
Investigating C. elegans development through mosaic analysis
 Primer 4761 Investigating C. elegans development through mosaic analysis John Yochem and Robert K. Herman Department of Genetics, Cell Biology and Development, University of Minnesota, 6-160 Jackson Hall,
Primer 4761 Investigating C. elegans development through mosaic analysis John Yochem and Robert K. Herman Department of Genetics, Cell Biology and Development, University of Minnesota, 6-160 Jackson Hall,
AP Biology Gene Regulation and Development Review
 AP Biology Gene Regulation and Development Review 1. What does the regulatory gene code for? 2. Is the repressor by default active/inactive? 3. What changes the repressor activity? 4. What does repressor
AP Biology Gene Regulation and Development Review 1. What does the regulatory gene code for? 2. Is the repressor by default active/inactive? 3. What changes the repressor activity? 4. What does repressor
Figure 1. Automated 4D detection and tracking of endosomes and polarity markers.
 A t = 0 z t = 90 B t = 0 z t = 90 C t = 0-90 t = 0-180 t = 0-300 Figure 1. Automated 4D detection and tracking of endosomes and polarity markers. A: Three different z planes of a dividing SOP shown at
A t = 0 z t = 90 B t = 0 z t = 90 C t = 0-90 t = 0-180 t = 0-300 Figure 1. Automated 4D detection and tracking of endosomes and polarity markers. A: Three different z planes of a dividing SOP shown at
purpose of this Chapter is to highlight some problems that will likely provide new
 119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
Chapter 10 Development and Differentiation
 Part III Organization of Cell Populations Chapter Since ancient times, people have wondered how organisms are formed during the developmental process, and many researchers have worked tirelessly in search
Part III Organization of Cell Populations Chapter Since ancient times, people have wondered how organisms are formed during the developmental process, and many researchers have worked tirelessly in search
BIOLOGY 111. CHAPTER 5: Chromosomes and Inheritance
 BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
Axis determination in flies. Sem 9.3.B.5 Animal Science
 Axis determination in flies Sem 9.3.B.5 Animal Science All embryos are in lateral view (anterior to the left). Endoderm, midgut; mesoderm; central nervous system; foregut, hindgut and pole cells in yellow.
Axis determination in flies Sem 9.3.B.5 Animal Science All embryos are in lateral view (anterior to the left). Endoderm, midgut; mesoderm; central nervous system; foregut, hindgut and pole cells in yellow.
University of Massachusetts Medical School Wan-Ju Liu University of Maryland
 University of Massachusetts Medical School escholarship@umms Program in Systems Biology Publications and Presentations Program in Systems Biology 5-13-2014 Multiple transcription factors directly regulate
University of Massachusetts Medical School escholarship@umms Program in Systems Biology Publications and Presentations Program in Systems Biology 5-13-2014 Multiple transcription factors directly regulate
Patterning of Caenorhabditis elegans Posterior Structures by the Abdominal-B Homolog, egl-5
 Developmental Biology 207, 215 228 (1999) Article ID dbio.1998.9124, available online at http://www.idealibrary.com on Patterning of Caenorhabditis elegans Posterior Structures by the Abdominal-B Homolog,
Developmental Biology 207, 215 228 (1999) Article ID dbio.1998.9124, available online at http://www.idealibrary.com on Patterning of Caenorhabditis elegans Posterior Structures by the Abdominal-B Homolog,
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Principles of Experimental Embryology
 Biology 4361 Developmental Biology Principles of Experimental Embryology June 16, 2008 Overview What forces affect embryonic development? The embryonic environment: external and internal How do forces
Biology 4361 Developmental Biology Principles of Experimental Embryology June 16, 2008 Overview What forces affect embryonic development? The embryonic environment: external and internal How do forces
Principles of Experimental Embryology
 Biology 4361 Developmental Biology Principles of Experimental Embryology September 19, 2006 Major Research Questions How do forces outside the embryo affect its development? (Environmental Developmental
Biology 4361 Developmental Biology Principles of Experimental Embryology September 19, 2006 Major Research Questions How do forces outside the embryo affect its development? (Environmental Developmental
Bypass and interaction suppressors; pathway analysis
 Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
Supplementary Figure 1. Nature Genetics: doi: /ng.3848
 Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Identifying cells using Immunofluorscence Rachel Scalzo Bio-300, 001 Professor Bentley 9/24/2014
 Identifying cells using Immunofluorscence Rachel Scalzo Bio-300, 001 Professor Bentley 9/24/2014 2 Abstract In this experiment, we used immunofluorescence to isolate certain organelles and see where they
Identifying cells using Immunofluorscence Rachel Scalzo Bio-300, 001 Professor Bentley 9/24/2014 2 Abstract In this experiment, we used immunofluorescence to isolate certain organelles and see where they
Lesson Overview. Gene Regulation and Expression. Lesson Overview Gene Regulation and Expression
 13.4 Gene Regulation and Expression THINK ABOUT IT Think of a library filled with how-to books. Would you ever need to use all of those books at the same time? Of course not. Now picture a tiny bacterium
13.4 Gene Regulation and Expression THINK ABOUT IT Think of a library filled with how-to books. Would you ever need to use all of those books at the same time? Of course not. Now picture a tiny bacterium
10/03/2014. Eukaryotic Development. + Differentiation vs. Development. Differentiation. Development
 Differentiation vs. Development What comes to mind when you think of differentiation? Eukaryotic Development What about development? Presented by: Sean, Daria, Emily, and Maggie Example: Human Development
Differentiation vs. Development What comes to mind when you think of differentiation? Eukaryotic Development What about development? Presented by: Sean, Daria, Emily, and Maggie Example: Human Development
Specification of the C. elegans MS blastomere by the T-box factor TBX-35
 Development epress online publication date 10 July 2006 RESEARCH ARTICLE 3097 Development 133, 3097-3106 (2006) doi:10.1242/dev.02475 Specification of the C. elegans MS blastomere by the T-box factor TBX-35
Development epress online publication date 10 July 2006 RESEARCH ARTICLE 3097 Development 133, 3097-3106 (2006) doi:10.1242/dev.02475 Specification of the C. elegans MS blastomere by the T-box factor TBX-35
Honors Biology Reading Guide Chapter 11
 Honors Biology Reading Guide Chapter 11 v Promoter a specific nucleotide sequence in DNA located near the start of a gene that is the binding site for RNA polymerase and the place where transcription begins
Honors Biology Reading Guide Chapter 11 v Promoter a specific nucleotide sequence in DNA located near the start of a gene that is the binding site for RNA polymerase and the place where transcription begins
The Caenorhabditis elegans par-5 Gene Encodes a Protein Required for Cellular Asymmetry in the Early Embryo
 Developmental Biology 241, 47 58 (2002) doi:10.1006/dbio.2001.0489, available online at http://www.idealibrary.com on The Caenorhabditis elegans par-5 Gene Encodes a 14-3-3 Protein Required for Cellular
Developmental Biology 241, 47 58 (2002) doi:10.1006/dbio.2001.0489, available online at http://www.idealibrary.com on The Caenorhabditis elegans par-5 Gene Encodes a 14-3-3 Protein Required for Cellular
16 The Cell Cycle. Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization
 The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
Chromosome Chr Duplica Duplic t a ion Pixley
 Chromosome Duplication Pixley Figure 4-6 Molecular Biology of the Cell ( Garland Science 2008) Figure 4-72 Molecular Biology of the Cell ( Garland Science 2008) Interphase During mitosis (cell division),
Chromosome Duplication Pixley Figure 4-6 Molecular Biology of the Cell ( Garland Science 2008) Figure 4-72 Molecular Biology of the Cell ( Garland Science 2008) Interphase During mitosis (cell division),
Lesson Overview Meiosis
 11.4 THINK ABOUT IT As geneticists in the early 1900s applied Mendel s laws, they wondered where genes might be located. They expected genes to be carried on structures inside the cell, but which structures?
11.4 THINK ABOUT IT As geneticists in the early 1900s applied Mendel s laws, they wondered where genes might be located. They expected genes to be carried on structures inside the cell, but which structures?
Three different fusions led to three basic ideas: 1) If one fuses a cell in mitosis with a cell in any other stage of the cell cycle, the chromosomes
 Section Notes The cell division cycle presents an interesting system to study because growth and division must be carefully coordinated. For many cells it is important that it reaches the correct size
Section Notes The cell division cycle presents an interesting system to study because growth and division must be carefully coordinated. For many cells it is important that it reaches the correct size
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
1. What are the three general areas of the developing vertebrate limb? 2. What embryonic regions contribute to the developing limb bud?
 Study Questions - Lecture 17 & 18 1. What are the three general areas of the developing vertebrate limb? The three general areas of the developing vertebrate limb are the proximal stylopod, zeugopod, and
Study Questions - Lecture 17 & 18 1. What are the three general areas of the developing vertebrate limb? The three general areas of the developing vertebrate limb are the proximal stylopod, zeugopod, and
MCB 141 Midterm I Feb. 14, 2012
 Write your name and student ID# on EVERY PAGE of your exam MCB 141 Midterm I Feb. 14, 2012 Question #1 Question #2 Question #3 Question #4 BONUS / 28 pts / 27 pts / 25 pts / 20 pts / 1 pt TOTAL / 100 pts
Write your name and student ID# on EVERY PAGE of your exam MCB 141 Midterm I Feb. 14, 2012 Question #1 Question #2 Question #3 Question #4 BONUS / 28 pts / 27 pts / 25 pts / 20 pts / 1 pt TOTAL / 100 pts
Cell lineage in marine nematode Enoplus brevis
 Development 125, 143-150 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV7605 143 Cell lineage in marine nematode Enoplus brevis Dmitrii A. Voronov* and Yuri V. Panchin Institute
Development 125, 143-150 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV7605 143 Cell lineage in marine nematode Enoplus brevis Dmitrii A. Voronov* and Yuri V. Panchin Institute
Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct
 SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
BIS &003 Answers to Assigned Problems May 23, Week /18.6 How would you distinguish between an enhancer and a promoter?
 Week 9 Study Questions from the textbook: 6 th Edition: Chapter 19-19.6, 19.7, 19.15, 19.17 OR 7 th Edition: Chapter 18-18.6 18.7, 18.15, 18.17 19.6/18.6 How would you distinguish between an enhancer and
Week 9 Study Questions from the textbook: 6 th Edition: Chapter 19-19.6, 19.7, 19.15, 19.17 OR 7 th Edition: Chapter 18-18.6 18.7, 18.15, 18.17 19.6/18.6 How would you distinguish between an enhancer and
Biology 218, practise Exam 2, 2011
 Figure 3 The long-range effect of Sqt does not depend on the induction of the endogenous cyc or sqt genes. a, Design and predictions for the experiments shown in b-e. b-e, Single-cell injection of 4 pg
Figure 3 The long-range effect of Sqt does not depend on the induction of the endogenous cyc or sqt genes. a, Design and predictions for the experiments shown in b-e. b-e, Single-cell injection of 4 pg
An intracellular phase transition drives nucleolar assembly and size control
 An intracellular phase transition drives nucleolar assembly and size control Steph Weber Department of Biology McGill University Membranes spatially organize eukaryotic cells Cells also contain membraneless
An intracellular phase transition drives nucleolar assembly and size control Steph Weber Department of Biology McGill University Membranes spatially organize eukaryotic cells Cells also contain membraneless
Structure and Function Analysis of LIN-14, a Temporal Regulator of Postembryonic Developmental Events in Caenorhabditis elegans
 MOLECULAR AND CELLULAR BIOLOGY, Mar. 2000, p. 2285 2295 Vol. 20, No. 6 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Structure and Function Analysis of LIN-14,
MOLECULAR AND CELLULAR BIOLOGY, Mar. 2000, p. 2285 2295 Vol. 20, No. 6 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Structure and Function Analysis of LIN-14,
MIR-237 is Likely a Developmental Timing Gene that Regulates the L2-to-L3 Transition in C. Elegans
 Marquette University e-publications@marquette Master's Theses (2009 -) Dissertations, Theses, and Professional Projects MIR-237 is Likely a Developmental Timing Gene that Regulates the L2-to-L3 Transition
Marquette University e-publications@marquette Master's Theses (2009 -) Dissertations, Theses, and Professional Projects MIR-237 is Likely a Developmental Timing Gene that Regulates the L2-to-L3 Transition
Cell Fate Specification in the C. elegans Embryo
 a DEVELOPMENTAL DYNAMICS 239:1315 1329, 2010 SPECIAL ISSUE REVIEWS A PEER REVIEWED FORUM Cell Fate Specification in the C. elegans Embryo Morris F. Maduro* Cell specification requires that particular subsets
a DEVELOPMENTAL DYNAMICS 239:1315 1329, 2010 SPECIAL ISSUE REVIEWS A PEER REVIEWED FORUM Cell Fate Specification in the C. elegans Embryo Morris F. Maduro* Cell specification requires that particular subsets
178 Part 3.2 SUMMARY INTRODUCTION
 178 Part 3.2 Chapter # DYNAMIC FILTRATION OF VARIABILITY WITHIN EXPRESSION PATTERNS OF ZYGOTIC SEGMENTATION GENES IN DROSOPHILA Surkova S.Yu. *, Samsonova M.G. St. Petersburg State Polytechnical University,
178 Part 3.2 Chapter # DYNAMIC FILTRATION OF VARIABILITY WITHIN EXPRESSION PATTERNS OF ZYGOTIC SEGMENTATION GENES IN DROSOPHILA Surkova S.Yu. *, Samsonova M.G. St. Petersburg State Polytechnical University,
Drosophila Somatic Anterior-Posterior Axis (A-P Axis) Formation
 Home Biol 4241 Luria-Delbruck 1943 Hershey-Chase 1952 Meselson-Stahl 1958 Garapin et al. 1978 McClintock 1953 King-Wilson 1975 Sanger et al. 1977 Rothberg et al. 2011 Jeffreys et al. 1985 Bacterial Genetics
Home Biol 4241 Luria-Delbruck 1943 Hershey-Chase 1952 Meselson-Stahl 1958 Garapin et al. 1978 McClintock 1953 King-Wilson 1975 Sanger et al. 1977 Rothberg et al. 2011 Jeffreys et al. 1985 Bacterial Genetics
5/4/05 Biol 473 lecture
 5/4/05 Biol 473 lecture animals shown: anomalocaris and hallucigenia 1 The Cambrian Explosion - 550 MYA THE BIG BANG OF ANIMAL EVOLUTION Cambrian explosion was characterized by the sudden and roughly simultaneous
5/4/05 Biol 473 lecture animals shown: anomalocaris and hallucigenia 1 The Cambrian Explosion - 550 MYA THE BIG BANG OF ANIMAL EVOLUTION Cambrian explosion was characterized by the sudden and roughly simultaneous
Developmental Biology 3230 Midterm Exam 1 March 2006
 Name Developmental Biology 3230 Midterm Exam 1 March 2006 1. (20pts) Regeneration occurs to some degree to most metazoans. When you remove the head of a hydra a new one regenerates. Graph the inhibitor
Name Developmental Biology 3230 Midterm Exam 1 March 2006 1. (20pts) Regeneration occurs to some degree to most metazoans. When you remove the head of a hydra a new one regenerates. Graph the inhibitor
First posted online on 21 April 2004 as /dev.01110
 First posted online on 21 April 2004 as 10.1242/dev.01110 Access the Development most recent version epress at http://dev.biologists.org/lookup/doi/10.1242/dev.01110 online publication date 21 April 2004
First posted online on 21 April 2004 as 10.1242/dev.01110 Access the Development most recent version epress at http://dev.biologists.org/lookup/doi/10.1242/dev.01110 online publication date 21 April 2004
9/4/2015 INDUCTION CHAPTER 1. Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology. Fig 1.
 INDUCTION CHAPTER 1 Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology Fig 1.1 1 EVOLUTION OF METAZOAN BRAINS GASTRULATION MAKING THE 3 RD GERM LAYER
INDUCTION CHAPTER 1 Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology Fig 1.1 1 EVOLUTION OF METAZOAN BRAINS GASTRULATION MAKING THE 3 RD GERM LAYER
The Timing of lin-4 RNA Accumulation Controls the Timing of Postembryonic Developmental Events in Caenorhabditis elegans
 Developmental Biology 210, 87 95 (1999) Article ID dbio.1999.9272, available online at http://www.idealibrary.com on The Timing of lin-4 RNA Accumulation Controls the Timing of Postembryonic Developmental
Developmental Biology 210, 87 95 (1999) Article ID dbio.1999.9272, available online at http://www.idealibrary.com on The Timing of lin-4 RNA Accumulation Controls the Timing of Postembryonic Developmental
Bi 1x Spring 2014: LacI Titration
 Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
Patterning the. moving beyond the cell lineage
 atterning the C. elegans embryo erspective atterning the C. elegans embryo moving beyond the lineage The Caenorhabditis elegans embryo undergoes a series of stereotyped cleavages that generates the organs
atterning the C. elegans embryo erspective atterning the C. elegans embryo moving beyond the lineage The Caenorhabditis elegans embryo undergoes a series of stereotyped cleavages that generates the organs
A diploid somatic cell from a rat has a total of 42 chromosomes (2n = 42). As in humans, sex chromosomes determine sex: XX in females and XY in males.
 Multiple Choice Use the following information for questions 1-3. A diploid somatic cell from a rat has a total of 42 chromosomes (2n = 42). As in humans, sex chromosomes determine sex: XX in females and
Multiple Choice Use the following information for questions 1-3. A diploid somatic cell from a rat has a total of 42 chromosomes (2n = 42). As in humans, sex chromosomes determine sex: XX in females and
Is Molecular Genetics Becoming Less Reductionistic?
 Is Molecular Genetics Becoming Less Reductionistic? Notes from recent case studies on mapping C. elegans and the discovery of microrna Richard M. Burian Virginia Tech rmburian@vt.edu Outline Introduction
Is Molecular Genetics Becoming Less Reductionistic? Notes from recent case studies on mapping C. elegans and the discovery of microrna Richard M. Burian Virginia Tech rmburian@vt.edu Outline Introduction
FGF signaling functions in the hypodermis to regulate fluid balance in C. elegans
 Research article 2595 FGF signaling functions in the hypodermis to regulate fluid balance in C. elegans Peng Huang and Michael J. Stern* Yale University School of Medicine, Department of Genetics, I-354
Research article 2595 FGF signaling functions in the hypodermis to regulate fluid balance in C. elegans Peng Huang and Michael J. Stern* Yale University School of Medicine, Department of Genetics, I-354
Both the apoptotic suicide pathway and phagocytosis are required for a programmed cell death in Caenorhabditis elegans
 Johnsen and Horvitz BMC Biology (2016) 14:39 DOI 10.1186/s12915-016-0262-5 RESEARCH ARTICLE Open Access Both the apoptotic suicide pathway and phagocytosis are required for a programmed cell death in Caenorhabditis
Johnsen and Horvitz BMC Biology (2016) 14:39 DOI 10.1186/s12915-016-0262-5 RESEARCH ARTICLE Open Access Both the apoptotic suicide pathway and phagocytosis are required for a programmed cell death in Caenorhabditis
NucView TM 488 Caspase-3 Assay Kit for Live Cells
 NucView TM 488 Caspase-3 Assay Kit for Live Cells Catalog Number: 30029 (100-500 assays) Contact Information Address: Biotium, Inc. 3423 Investment Blvd. Suite 8 Hayward, CA 94545 USA Telephone: (510)
NucView TM 488 Caspase-3 Assay Kit for Live Cells Catalog Number: 30029 (100-500 assays) Contact Information Address: Biotium, Inc. 3423 Investment Blvd. Suite 8 Hayward, CA 94545 USA Telephone: (510)
Supplementary Information for. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria
 Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
DEVELOPMENT. Shuyi Huang, Premnath Shetty, Scott M. Robertson and Rueyling Lin*
 RESEARCH ARTICLE 2685 Development 134, 2685-2695 (2007) doi:10.1242/dev.008268 Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and
RESEARCH ARTICLE 2685 Development 134, 2685-2695 (2007) doi:10.1242/dev.008268 Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and
C. elegans Dynamin Mediates the Signaling of Phagocytic Receptor CED-1 for the Engulfment and Degradation of Apoptotic Cells
 Developmental Cell 10, 743 757, June, 2006 ª2006 Elsevier Inc. DOI 10.1016/j.devcel.2006.04.007 C. elegans Dynamin Mediates the Signaling of Phagocytic Receptor CED-1 for the Engulfment and Degradation
Developmental Cell 10, 743 757, June, 2006 ª2006 Elsevier Inc. DOI 10.1016/j.devcel.2006.04.007 C. elegans Dynamin Mediates the Signaling of Phagocytic Receptor CED-1 for the Engulfment and Degradation
