Phylogenetic Relationships of Yessotoxin-Producing Dinoflagellates, Based on the Large Subunit and Internal Transcribed Spacer Ribosomal DNA Domains
|
|
|
- Sophia Marsh
- 6 years ago
- Views:
Transcription
1 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Jan. 2009, p Vol. 75, No /09/$ doi: /aem Copyright 2009, American Society for Microbiology. All Rights Reserved. Phylogenetic Relationships of Yessotoxin-Producing Dinoflagellates, Based on the Large Subunit and Internal Transcribed Spacer Ribosomal DNA Domains Meredith D. A. Howard, 1 * G. Jason Smith, 2 and Raphael M. Kudela 1 Ocean Science Department, University of California, Santa Cruz, 1156 High Street, Santa Cruz, California 95064, 1 and Moss Landing Marine Laboratories, 8272 Moss Landing Rd., Moss Landing, California Received 9 April 2008/Accepted 6 November 2008 Yessotoxin (YTX) is a globally distributed marine toxin produced by some isolates of the dinoflagellate species Protoceratium reticulatum, Lingulodinium polyedrum, and Gonyaulax spinifera within the order Gonyaulacales. The process of isolating cells and testing each isolate individually for YTX production during toxic blooms are labor intensive, and this impedes our ability to respond quickly to toxic blooms. In this study, we used molecular sequences from the large subunit and internal transcribed spacer genomic regions in the ribosomal operon of known YTX-producing dinoflagellates to determine if genetic differences exist among geographically distinct populations or between toxic and nontoxic isolates within species. In all analyses, all three YTX-producing species fell within the Gonyaulacales order in agreement with morphological taxonomy. Phylogenetic analyses of available rrna gene sequences indicate that the capacity for YTX production appears to be confined to the order Gonyaulacales. These findings indicate that Gonyaulacoloid dinoflagellate species are the most likely to produce YTX and thus should be prioritized for YTX screening during events. Dinoflagellate species that fall outside of the Gonyaulacales order are unlikely to produce YTX. Although the rrna operon offers multiple sequence domains to resolve species level diversification within this dinoflagellate order, these domains are not sufficiently variable to provide robust markers for YTX toxicity. * Corresponding author. Present address: Southern California Coastal Water Research Project and University of Southern California, 3535 Harbor Blvd., Suite 110, Costa Mesa, CA Phone: (714) Fax: (714) Mhoward@sccwrp.org. Published ahead of print on 14 November Yessotoxin (YTX) is a globally distributed toxin originally isolated from scallops, Patinopecten yessoensis, for which the toxin class was named (36). Due to the potential public health implications of ingesting contaminated shellfish, YTX is a regularly monitored marine toxin in New Zealand, Europe, and Japan. In 2002, the European Commission placed YTX into a separate phycotoxin group and established a regulatory level of 1 g g 1 of YTX equivalents in shellfish meat intended for human consumption (directive 2002/225/EC); Japan and New Zealand have similar regulatory language. There are three confirmed producers of the YTX phycotoxin: the dinoflagellates Protoceratium reticulatum (Claparhde and Lachmann) Buetschli, Lingulodinium polyedrum (Stein) Dodge, and Gonyaulax spinifera Dodge. YTX production within and among dinoflagellate species tested to date is highly variable. Previous reports have identified both nontoxic and toxic isolates of L. polyedrum from Spain, the United Kingdom, and California (3, 18, 43, 47, 58). In addition, several YTXpositive isolates of L. polyedrum have been identified in Italy (9, 62) and Ireland (J. Silke, personal communication), yet only nontoxic isolates have been observed in Norway waters (44). Several nontoxic isolates of G. spinifera have been identified from the United Kingdom (21, 58) and one from the United States (18), yet several toxic isolates from New Zealand (45) and Italy (46) have been observed. Water samples from New Zealand yielded both nontoxic and toxic isolates of G. spinifera, including one isolate that produced the highest amount of YTX per cell for any genus to date (45; L. Rhodes, personal communication). YTX-producing isolates of P. reticulatum have been described from Canada (58), Italy (5), Norway (50), Japan (10, 52), New Zealand (30, 34, 51, 52), Spain (41, 42), South Africa (12, 21), and the United Kingdom (58). Together, these studies indicate that while YTX production by P. reticulatum appears to be globally distributed, a large variation in the toxicity (0.3 to 79 pg YTX cell 1 ) is observed between isolates of this species. To add to the challenges of establishing monitoring criteria for potential YTX-producing dinoflagellates, there have been conflicting studies published on the toxicity of P. reticulatum from U.S. coastal waters, with reports indicating both the presence and absence of YTX accumulation in cultured isolates from California, Florida, and Washington (41, 42; B. Paz, personal communication). During environmental YTX events, many dinoflagellates (including species not previously identified to produce YTX) have been isolated, grown in culture, and tested for the production of YTX, particularly members of the Prorocentrum genus which frequently co-occur during these events. This process is labor intensive and dependent on the isolation of a number of cells sufficient for YTX analysis. Given the variation in the expression of toxicity among isolates of the same species and the limited morphological variability within species, we attempted to identify coarse scale genetic markers that would differentiate between spatially and temporally distinct isolates while simultaneously identifying YTX-accumulating strains of L. polyedrum, P. reticulatum, and G. spinifera. Given the worldwide distribution of potentially toxic dinoflagellates and the global observations of YTX, we speculated that phylogenetic analysis of molecular sequence information 54
2 VOL. 75, 2009 PHYLOGENY OF YESSOTOXIN-PRODUCING DINOFLAGELLATES 55 TABLE 1. List of the cultures analyzed, including the label of each, the GenBank accession number, the isolation location, the year of isolation, and the culture collection or person from which the culture was received Species Label from these YTX-producing dinoflagellates could be used as a tool to help identify other potential yet unidentified YTXproducing dinoflagellates. The resultant molecular database could also enhance the subsequent development of robust detection probes for monitoring YTX-producing species. The use of molecular phylogenetics is now widely utilized in addition to traditional methodologies (such as morphology, ultrastructure, life-cycle information, and fossil record) to understand the evolutionary history of dinoflagellates and to evaluate the relationships within genera among morphologically indistinguishable species. The rrna operon comprises the genome regions that code for the RNA components of the ribosomes and consists of the large subunit (LSU), the small subunit (SSU), and the 5.8S genes, the latter bound by internal transcribed spacer regions 1 and 2 (ITS1 and ITS2). The rrna gene has been the target of many phylogenetic studies because ribosomes are universally present in living organisms and functional constraints have resulted in high sequence conservation within these domains (13, 26, 53). The LSU consists of many structural domains (D1 to D12), and the D1, D3, and D8 domains are particularly useful to evaluate the phylogenetic relationships of closely related species (27, 32, 54) since these variable regions flank more conservative regions. The ITS regions are commonly used to analyze closely related and geographically different species (6, 24, 28, 48, 53, 56). The rrna operon has also been widely and successfully targeted for the design of primers and probes to detect or quantify harmful algal bloom species (2, 31, 32, 55). MATERIALS AND METHODS Accession no. ITS1-5.8S-ITS2 LSU D1-D2 Laboratory cultures. Cultures were obtained from commercial or individual culture collections. Table 1 lists each species, isolate label name, GenBank accession number, location of isolation, year of isolation, and person or culture collection from which the culture was received. All of the isolates labeled CCMP were received from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton. Only the extracted DNA was received for the L. polyedrum Location Yr Culture collection Lingulodinium polyedrum CCMP1931 EU EU Scripps Pier, La Jolla, CA 1998 Provasoli-Guillard National Center for Culture of Marine Phytoplankton CCMP1936 EU EU Scripps Pier, La Jolla, CA A EU EU Scripps Pier, La Jolla, CA 1998 P. Franks, Scripps Institute of Oceanography CCAP 1121/2 EU EU Loch Creran, Argyll, Scotland 1996 Culture Collection of Algae and Protozoa Protoceratium reticulatum CCMP404 EU EU Salton Sea, CA 1966 Provasoli-Guillard National Center for Culture of Marine Phytoplankton CCMP1889 EU EU Friday Harbor, San Juan 1983 Island, WA CTCC01 EU EU South Africa 2006 G. Pitcher, Marine and Coastal Management, South Africa Gonyaulax spinifera CCMP409 EU EU West Boothbay Harbor, MA 1986 Provasoli-Guillard National Center for Culture of Marine Phytoplankton Prorocentrum minimum CCMP1329 EU Great South Bay, Long Island, NY 1958 Provasoli-Guillard National Center for Culture of Marine Phytoplankton culture CCAP1121/2, obtained from the Culture Collection of Algae and Protozoa (CCAP, Scotland). The cultures were grown under nonaxenic conditions in autoclaved glass flasks with either f/2 or L1 medium (15, 16, 17) with seawater stocks obtained from Granite Canyon, California. The culture densities were grown to approximately 10 3 cells ml 1. The culture conditions were standardized with growth temperatures of 21 C ( 1 C) and 87 mol photons m 2 s 1 irradiance using Sylvania grow-lite spectrally corrected light sources on a 14:10 light/dark cycle for cultures CCMP1931, CCMP1936, 104A (all L. polyedrum), and CCMP404 (P. reticulatum). The remaining cultures CCMP1889 and CTCC01 (P. reticulatum), CCMP409 (G. spinifera), and CCMP1329 (P. minimum) were grown at 15 C ( 1 C), with 70 mol photons m 2 s 1 irradiance using Sylvania grow-lite spectrally corrected light sources on a 12:12 light/dark cycle. Duplicate samples of each culture were gently filtered onto Poretics, 5.0- m polycarbonate filters (Osmonics, Inc.), at mid-exponential growth phase. The cells were resuspended in medium and centrifuged in a Fisher Scientific Marathon 8K centrifuge for 3 min at 1,073 g (4,000 rpm). The supernatant was pipetted off, and the pellets were immediately frozen in liquid nitrogen and stored at 80 C until analysis. DNA extraction and PCR amplification. The genomic DNA was extracted from the pellets using NucleoSpin plant kits (BD Biosciences) according to the manufacturer s instructions. Approximately 50 ng of genomic DNA was amplified using 50- l PCRs containing nuclease-free water (Fisher Scientific), 1 JumpStart PCR buffer (10 mm Tris-HCl [ph 8.3], 50 mm KCl; Sigma-Aldrich), 2.5 mm MgCl, 0.2 mm deoxynucleoside triphosphate (Advantage ultrapure PCR deoxynucleotide mix; BD Biosciences), 200 nm of gene-specific oligonucleotide primers, and 0.5 U JumpStart Taq DNA polymerase (Sigma-Aldrich). The ITS1, 5.8S, and ITS2 regions (approximately 800 bp) were amplified from each culture using several primers, each of which is listed in Table 2. The Lp1F and Lp2R oligonucleotide pairs were based on the L. polyedrum ribosomal DNA (rdna) sequence AF and designed to amplify the entire ITS1-5.8S-ITS2 region from the 3 end of the SSU rdna and 5 end of the LSU rdna. This dinoflagellate-specific primer set was used successfully to amplify this domain from isolates CCMP404, CCMP1889, and CTCC01 (all P. reticulatum) and CCMP1931, CCMP1936, 104A, and CCAP1121/2 (all L. polyedrum). The more generic oligonucleotide pair 1400F and 38R targeting the same domain (29) were needed to amplify the entire ITS1-5.8S-ITS2 domain from the G. spinifera isolate CCMP409. All amplifications were carried out in duplicate using the Perkin Elmer GeneAmp PCR system 2400 and the following cycling conditions: initial template denaturation for 5 min at 94 C, followed by 33 cycles of 94 C for 30 s, 58 C for 30 s, and extension at 72 C for 1 min and 15 s, followed by a final extension at 72 C for 7 min. A 700-bp fragment of the LSU rdna, spanning the D1 and D2 regions of the
3 56 HOWARD ET AL. APPL. ENVIRON. MICROBIOL. Primer TABLE 2. List of primers used in the DNA amplification of cultures Sequence Reference (if applicable) Lp1F 5 ATTATGTCCCTGCCCTTTGTACAC3 Lp2R 5 ACTGAAGGAATCCTGTTTGGTTTC3 1400F 5 CTGCCCTTTGTACACACCGCCCGTC R 5 CGCTTATTGATATGCTTA3 29 D1R 5 ACCCGCTGAATTTAAGCATA3 26 D2C 5 CCTTGGTCCGTGTTTCAAGA3 26 hypervariable domains (26, 27, 54), was amplified using the primers D1R (forward) and D2C (reverse) for all cultures. All amplifications were carried out in duplicate using the Perkin Elmer GeneAmp PCR system 2400 as described above with the annealing temperature decreased to 55 C. The quality and specificity of all the amplification products were assessed by agarose gel electrophoresis. Individual product bands visualized by staining with ethidium bromide were purified using the Wizard SV gel and PCR clean-up system (Promega) according to the manufacturer s instructions. Products eluted from the solid-phase matrix were further cleaned by isopropanol precipitation. After verifying the purity and concentration by agarose gel electrophoresis, individual template samples were sequenced bidirectionally from the appropriate oligonucleotide primer using a commercial service (Northwoods DNA, Inc.). Consensus sequences obtained for this study were deposited in GenBank using the BankIT facility (Table 1). Alignment and phylogenetic analysis. The sequences were aligned using ClustalX, version 1.8 (60), and subsequently adjusted manually. The new ITS and LSU sequences from this study (Table 1) were aligned with existing ITS and LSU sequences available in GenBank (Tables 3 and 4). Phylogenetic relationships were inferred based on the simple pairwise differences of nucleotides (p-distance) derived from the alignment, ignoring gaps. The p-distance represents the number of nucleotide differences divided by the total number of nucleotides compared. The neighbor joining (NJ) distance method and maximum parsimony (MP) estimation of phylogenetic relationships were conducted using the Molecular Evolutionary Genetics Analysis (MEGA) program version 3.1 (23). Bootstrapping values of 1,000 replicates were used to assess the consistency of each derived topology. Due to differences in the availability of sequence datasets for the targeted domains, two outgroups were used separately in these analyses: the dinoflagellate Heterocapsa sp. (GenBank accession number AB084100) was used in the ITS analysis, and the dinoflagellate Symbiodinium microadriaticum TABLE 3. List of sequences used in the ITS region rdna phylogenetic analysis from GenBank Species Accession no. Origin Akashiwo sanguinea AY South Korea AY South Korea AY New York AB Japan Alexandrium catenella AJ Spain AJ Spain Alexandrium margalefeii AJ Italy Gonyaulax spinifera AF Heterocapsa sp. AB Japan Lingulodinium polyedrum AF Korea AM Italy Karenia brevis AF AF Karenia mikimotoi AM United Kingdom Karlodinium sp. AM Spain Karlodinium micrum AF North Carolina AJ Norway Prorocentrum micans AF Prorocentrum minimum AF North Carolina AF AF North Carolina Protoceratium reticulatum AM Italy TABLE 4. List of sequences used in the LSU region rdna phylogenetic analysis from GenBank Species Accession no. Origin Akashiwo sanguinea AF AF AY South Korea AY South Korea AY New York Alexandrium catenella AF California Alexandrium margalefeii AY Alexandrium pseudogonyaulax AY Cochlodinium polykrikoides AY South Korea AF Gonyaulax baltica AY Gonyaulax digitale AY Gonyaulax elongata AY United Kingdom Gonyaulax membranacea AY Ireland Gonyaulax polygramma DQ Korea Gonyaulax spinifera DQ New Zealand DQ New Zealand AY EF Heterocapsa triquetra AF Denmark Lingulodinium polyedrum AF Korea EF Korea Karenia brevis AY Florida AY Florida AY Florida AY Texas AY Texas Karenia mikimotoi AY Florida Karlodinium micrum AY Australia AY New Zealand DQ U92257 New Zealand AY New Zealand Karlodinium veneficum DQ United Kingdom Prorocentrum mexicanum AJ AF Prorocentrum micans AY AF Korea AF Denmark AY Protoceratium reticulatum AY South Africa AF EF Italy EF Pyrodinium bahamense var. AY The Philippines compressum Symbiodinium microadriaticum AF (GenBank accession number AF060896) was used in the LSU analysis. These outgroups were selected based on their having the largest p-distance with respect to the YTX-producing taxa. RESULTS ITS regions. Phylogenetic analysis of the ITS alignments placed all the YTX-producing species identified to date within a monophyletic clade encompassing the dinoflagellate order Gonyaulacales. The resultant topology was well supported (bootstrap values were 99% by NJ analysis [Fig. 1] and 90% by parsimony [Fig. 2]), which agrees with morphologically based taxonomic data. Differences in the intraspecific diversity within the YTX-producing taxa were also revealed by this analysis. There was very low or no diversity among isolates of L. polyedrum. Intraspecific diversity increased among P. reticulatum
4 VOL. 75, 2009 PHYLOGENY OF YESSOTOXIN-PRODUCING DINOFLAGELLATES 57 FIG. 1. NJ analysis of the ITS rdna for all species listed in Tables 1 and 3. Bootstrap values (1,000 replicates) are listed as percentages of 100, and only values greater than 50% are shown. Heterocapsa sp. (GenBank accession number AB084100) was the outgroup. Cultures of Lingulodinium polyedrum and Protoceratium reticulatum in bold are toxic, and cultures of Lingulodinium polyedrum and Gonyaulax spinifera in italics are nontoxic. Cultures of Protoceratium reticulatum for which there are conflicting studies on toxicity are indicated with double asterisks ( **) after the name. The toxicity is unknown for all of the YTX-producing species in the regular font. isolates and was highest among the G. spinifera isolates sequenced to date. For the rrna loci sequenced for this study, alignments revealed that there was no intraspecific genetic diversity (0.000 p-distance) among the known isolates of L. polyedrum isolates. However, a comparison of these sequences to a GenBank record for this species (AF377944) revealed two separate nucleotide differences (0.006 p-distance) in the ITS1 region, each representing a transition. There was no genetic divergence (0.000 p-distance) between the P. reticulatum culture isolated from Washington (EU532484, culture CCMP1889) and the toxic South Africa isolate (EU532486, culture CTCC01); however, the culture from the Salton Sea (EU532485, CCMP404), a saline lake in California, was highly divergent (0.235 p-distance) from the two coastal ocean isolates. The Washington and California cultures have been reported to be toxic (41) and nontoxic (42; B. Paz, personal communication), respectively. For the purposes of this study, we will consider the toxicity of these cultures uncertain. The GenBank sequence AM was only slightly divergent (0.003 p-distance) from the coastal ocean isolates (EU532484, culture CCMP1889, and EU532486, culture CTCC01) and more divergent (0.235 p-distance) from the saline lake isolate (EU532485, culture CCMP404). There were only two G. spinifera isolates used in the ITS analysis (due to a lack of sequence information for the ITS region in GenBank). These two isolates were genetically distinct (0.350 p-distance), and the toxicity was known for one of those sequences (EU532487, CCMP409). LSU region. In both the NJ (Fig. 3) and maximum parsimony (Fig. 4) analyses of the LSU domains D1 and D2, the Gonyaulacales order again formed a distinct clade, although with lower support (bootstrap values 80% and 56%, respectively) compared with the ITS analyses. As in the ITS analysis, molecular discrimination among geographically distinct isolates of L. polyedrum was not detected (0.000 p-distance). However, this LSU domain exhibited more variability among the strains from the GenBank database. While the Korean isolate described in GenBank (AF377944) had a similar level of divergence as that observed for the L. polyedrum cultures sequenced in this study (0.006 p-distance), this divergence was due to only 3 nucleotide differences in the sequences, likely due to sequencing errors. Another submission again from Korean waters (EF613357) exhibited greater divergence at this locus (0.041 p-distance). While there was no genetic difference (0.000 p-distance) among the coastal ocean P. reticulatum cul-
5 58 HOWARD ET AL. APPL. ENVIRON. MICROBIOL. FIG. 2. Maximum parsimony analysis of the ITS rdna for all species listed in Tables 1 and 3. Bootstrap values (1,000 replicates) are listed as percentages of 100, and only values greater than 50% are shown. Heterocapsa sp. (GenBank accession number AB084100) was the outgroup. Cultures of Lingulodinium polyedrum and Protoceratium reticulatum in bold are toxic, and cultures of Lingulodinium polyedrum and Gonyaulax spinifera in italics are nontoxic. Cultures of Protoceratium reticulatum for which there are conflicting studies on toxicity are indicated with double asterisks ( **) after the name. The toxicity is unknown for all of the YTX-producing species in the regular font. tures, there was a slight difference ( p-distance) between these isolates and the saline lake isolate (EU532476), which was consistent with the ITS analyses. The LSU sequences from GenBank were genetically identical to the coastal P. reticulatum cultures sequenced in this study. The sequence diversity was considerably higher among isolates designated as G. spinifera. Interestingly, the two toxin-producing strains of G. spinifera, DQ and DQ (45), were genetically identical to each other (0.000 p-distance) but distinct from the non-toxin-producing isolate EU (0.330 p-distance) and GenBank strain AY (0.3 p-distance), toxicity unknown. The toxic strains were less divergent from GenBank strain EF (0.130 p-distance), toxicity unknown. The GenBank submission assigned as G. membranacea (AY154965) was found to be identical to the LSU sequence derived from the G. spinifera strain (CCMP409) used in this study, indicating an incorrect species annotation either in the original submission or in the CCMP culture collection. DISCUSSION The evolution of dinoflagellates has been intensely studied, and it has become common to use traditional methodologies, such as thecal plate and whole-cell morphology, ultrastructure, life cycles, and fossil record, in combination with molecular phylogenetics (13, 59). The dinoflagellates consist of six major groups, Gymnodiniales, Gonyaulacales, Prorocentrales, Peridiniales, Dinophysiales, and Suessiales (59). While most dinoflagellates have been classified according to morphological characteristics, a range of observations indicate a consistent pattern of differences in phycotoxicity within morphologically identical species. The potential for molecular sequence information to provide additional higherresolution discriminatory characters may help our understanding of the evolutionary differences and monitoring of bloom dynamics between isolates of the same species. There are a large number of studies that have used nuclear rrna genes successfully to evaluate relationships among closely related taxa within genera and particularly within species that are morphologically indistinguishable using the variable domains within the LSU region (27, 29, 32, 54) and potentially more rapidly evolving ITS region (6, 24, 28, 48, 53, 56). While a consensus molecular phylogeny has not been accepted for the Dinophyceae as a whole, there are broad consistencies in the topological positions of some taxonomic orders within this group. For example, previous dinoflagellate
6 VOL. 75, 2009 PHYLOGENY OF YESSOTOXIN-PRODUCING DINOFLAGELLATES 59 FIG. 3. NJ analysis of the LSU rdna for all species listed in Tables 1 and 4. Bootstrap values (1,000 replicates) are listed as percentages of 100, and only values greater than 50% are shown. Symbiodinium microadriaticum (GenBank accession number AF060896) was the outgroup. Cultures of Lingulodinium polyedrum, Protoceratium reticulatum, and Gonyaulax spinifera in bold are toxic and cultures of Lingulodinium polyedrum and Gonyaulax spinifera in italics are nontoxic. Cultures of Protoceratium reticulatum for which there are conflicting studies on toxicity are indicated with double asterisks ( **) after the name. The toxicity is unknown for all of the YTX-producing species in the regular font.
7 60 HOWARD ET AL. APPL. ENVIRON. MICROBIOL. FIG. 4. Maximum parsimony analysis of the LSU rdna for all species listed in Tables 1 and 4. Bootstrap values (1,000 replicates) are listed as percentages of 100, and only values greater than 50% are shown. Symbiodinium microadriaticum (GenBank accession number AF060896) was the outgroup. Cultures of Lingulodinium polyedrum, Protoceratium reticulatum, and Gonyaulax spinifera in bold are toxic, and cultures of Lingulodinium polyedrum and Gonyaulax spinifera in italics are nontoxic. Cultures of Protoceratium reticulatum for which there are conflicting studies on toxicity are indicated with double asterisks ( **) after the name. The toxicity is unknown for all of the YTX-producing species in the regular font.
8 VOL. 75, 2009 PHYLOGENY OF YESSOTOXIN-PRODUCING DINOFLAGELLATES 61 phylogeny studies have found the Gonyaulacales order to be a monophyletic and more recently diverged group relative to other described orders within the Dinophyceae (37, 63). In all phylogenetic analyses in this study, the Gonyaulacales order formed a distinct clade, consistent with previously published studies (7, 13, 37, 49, 59, 63). The molecular sequence analysis for the two loci examined in this study support the validity of the Gonyaulacales and firmly place all known YTXproducing species within this dinoflagellate order. The Prorocentrales order was included in these analyses because Prorocentrum micans was frequently present when YTX was detected in California mussels samples (18; M. Silver, unpublished data), and several isolates from the United Kingdom have been tested for YTX production (58). The results of this study, however, suggest that non-gonyaulacaloid dinoflagellate species (such as Prorocentrum) are unlikely to produce YTX, and testing such species that fall outside of the Gonyaulacales order may be ineffective for identifying the biological origin of YTX during events. The L. polyedrum species showed very low intraspecific diversity, and at this level of genetic assessment, there did not appear to be geographically distinct populations but rather a global distribution of ribotypes within this species. Consequently, rrna operons provide no indirect genetic markers for YTX production, and lack of diversity at this level hints that variation in toxicity may be due to environmental conditions or genomic variability. Toxin production in other algae, such as Alexandrium spp., P. reticulatum, and Pseudo-nitzschia spp., has been shown to be influenced by environmental conditions, particularly nutrients (1, 4, 14, 19, 38, 39, 40), although under common culture conditions, some isolate-specific variation can be maintained (22, 57). Future studies should evaluate if there are specific genes that are turned on during toxin production (42), which would explain the differences between toxic and nontoxic isolates of the same, genetically similar species. The lack of genetic diversity indicates that overall quantification may be the most effective methodology in toxic bloom monitoring of L. polyedrum. A quantitative PCR method based on the SSU has been developed for the quantification of L. polyedrum from southern California (35). The results from this study of the LSU and ITS regions of L. polyedrum populations suggest that the SSU quantitative PCR method can be applied to other geographic regions to aid in monitoring L. polyedrum bloom dynamics. There was a large sequence divergence observed between multiple isolates in the LSU analyses of G. spinifera, which is consistent with other studies of the LSU region of the Gonyaulax genus (11, 20, 45, 46). While a limited number of LSU sequences were used in this study, distinct ribotypes of toxic G. spinifera (DQ and DQ151558) were revealed. In a similar phylogenetic analysis of the LSU region, toxic strains of G. spinifera from New Zealand (DQ and DQ151558) (45) and Italy (46) grouped together and formed a clade distinct from other strains and had a high intraspecific variability compared with GenBank strains (46). The high levels of intraspecific genetic divergence detected in our analysis (30 to 40%) as well as from Riccardi et al. (46) suggest that G. spinifera is undergoing rapid diversification. Considerable morphological variability is observed for G. spinifera (8, 61), and it is possible that cryptic species may exist within the known isolates of G. spinifera. Notwithstanding current taxonomic assignments, the members of the genus Gonyaulax may provide fruitful targets for the development of molecular probes distinguishing isolates of differing toxicity and for the examination of how the expression of toxin production may drive the evolution or diversification of harmful algal species. Additional multilocus and ultrastructural studies using a large number of unique G. spinifera isolates will be needed to thoroughly describe the genetic variability within this species. Due to the conflicting reports of toxicity for P. reticulatum cultures CCMP404 and CCMP1889, it is difficult to determine if there are genetically distinguishable isolates of this species based on toxicity. However, there are several published studies evaluating the influence of environmental conditions on the production of YTX in P. reticulatum. In isolates of P. reticulatum from Emilia-Romagna, Italy, YTX production increased within the cells when cultures were grown under higher salinity and temperature conditions and under both replete and severe phosphate-limited nutrient conditions (14). YTX released from cells into the medium was found to be higher in cultures grown under nitrogen limitation, lower temperature or lower salinity conditions. Those authors concluded that environmental conditions directly affect toxin production and that decreased temperature and salinity will decrease toxin production but will not terminate toxin production in cells (14). In culture, the stationary growth phase, as well as the addition of the micronutrient selenium but not iron or cobalt, significantly increased YTX production by P. reticulatum (33, 34). These published studies suggest that environmental conditions can influence toxicity and therefore that genetically distinct isolates based on toxicity may not exist. Future research on this species will need to evaluate the genetic diversity of strains for which a complete toxin profile has already been established, such as the isolated cultures from Spain (42). We hypothesized that the rrna genes may be useful for the evaluation of genetic variability among toxic and nontoxic isolates. However, the results of this study show that the constrained sequence variability of several rrna operons do not provide robust markers of YTX toxicity among species for most genera in the Gonyaulacales order. While this study and the results of Riccardi et al. (46) suggest that this application might be possible for G. spinifera, these results are based on small sets of isolates, and therefore, a larger number of G. spinifera isolates needs to be analyzed to characterize the true genetic and toxicity associations within this species. If Gonyaulax is indeed undergoing rapid diversification, this genus may be suitable for genomic tools, but we suggest that future studies focus on both physiological and genomic assays for YTX production in Gonyaulacales. Our results do demonstrate, however, that confirmed YTX production is currently confined to the order Gonyaulacales within the Dinophyceae and that species within this taxonomic order should be given priority for future testing and field collections associated with monitoring for YTX contamination events. Interestingly, a previous study evaluating the origin of paralytic shellfish poisoning (PSP) toxins concluded that PSP toxin-producing species are randomly distributed throughout all dinoflagellate groups and, based on this widespread toxin distribution, speculated that PSP toxins had multiple independent origins in the Gymnodiniales and Gonyaulacales orders (63). While other
9 62 HOWARD ET AL. APPL. ENVIRON. MICROBIOL. species within the Gonyaulacales order have not been documented to produce YTX congeners (e.g., Alexandrium catenella), all YTX-producing species identified so far remain within this group. Our results contrast with this hypothesis of multiple origins for PSP toxins, as the occurrence of YTX production within several distinct genera in the Gonyaulacales support the hypothesis that YTX biosynthetic capacity arose early in the divergence of this order and consequently later in the evolutionary history of the Dinophyceae. ACKNOWLEDGMENTS This work was supported by the University of California Coastal Environmental Quality Initiative Program award 05TCEQI awarded to R.M.K. and M.D.A.H. and other grants awarded to M.D.A.H., including the Ida Benson Lynn fellowship, the Marilyn C. Davis memorial scholarship, the University of California, the Santa Cruz Women s Club, and the Ocean Science Department at the University of California, Santa Cruz. G.J.S. acknowledges prior NSF support (OCE ) for molecular facilities. We thank Bethany Jenkins and Kendra Hayashi. We thank the Marine Pollution Studies Laboratory at Granite Canyon, University of California, Davis, for the seawater they provided, and Grant Pitcher and Peter Franks for providing algal strains used in this study. We also thank three anonymous reviewers who provided useful commentary on an earlier version of the manuscript. REFERENCES 1. Anderson, D. M., D. M. Kulis, J. J. Sullivan, S. Hall, and C. Lee Dynamics and physiology of saxitoxin production by the dinoflagellates Alexandrium spp. Mar. Biol. 104: Anderson, D. M Identification of harmful algal species using molecular probes: an emerging perspective, p In P. Lassus, G. Arzul, P. Gentien, and C. Marcaillou (ed.), Harmful marine algal blooms. Lavoisier, Paris, France. 3. Armstrong, M., and R. Kudela Evaluation of California isolates of Lingulodinium polyedrum for the production of yessotoxin. Afr. J. Mar. Sci. 28: Armstrong Howard, M. D., W. P. Cochlan, R. M. Kudela, N. Ladizinsky, and R. M. Kudela Nitrogenous preference of toxic Pseudo-nitzschia australis (Bacillariophyceae) from field and laboratory experiments. Harmful Algae 6: Boni, L., A. Ceredi, F. Guerrini, A. Milandri, R. Pistocchi, R. Poletti, and M. Pompei Toxic Gonyaulax Grindley Reinecke in the north-western Adriatic Sea (Italy). Abstr. 10th Int. Harmful Algal Bloom Conf., St. Petersburg, FL. 6. Coyer, J. A., G. J. Smith, and R. A. Andersen Evolution of Macrocystis spp. (Phaeophyceae) as determined by ITS1 and ITS2 sequences. J. Phycol. 37: Daugbjerg, N., G. Hansen, J. Larsen, and Ø. Moestrup Phylogeny of some of the major genera of dinoflagellates based on ultrastructure and partial LSU rdna sequence data, including the erection of three new genera of unarmored dinoflagellates. Phycologia 39: Dodge, J. D Some revisions of the family Gonyaulacaceae (Dinophyceae) based on scanning electron microscope study. Bot. Mar. 32: Draisci, R., E. Ferretti, L. Palleschi, C. Marchiafava, R. Poletti, A. Milandri, A. Ceredi, and M. Pompei High levels of yessotoxin in mussels and presence of yessotoxin and homoyessotoxin in dinoflagellates of the Adriatic Sea. Toxicon 37: Eiki, K., M. Satake, K. Koike, T. Ogata, T. Mitsuya, and Y. Oshima Confirmation of yessotoxin production by the dinoflagellate Protoceratium reticulatum in Mutsu Bay. Fish. Sci. 71: Ellegaard, M., N. Daugbjerg, A. Rochon, J. Lewis, and I. Harding Morphological and LSU rdna sequence variation within the Gonyaulax spinifera-spiniferites group (Dinophyceae) and proposal of G. elongata comb. nov. and G. membranacea comb. nov. Phycologia 42: Fawcett, A., G. C. Pitcher, S. Bernard, A. D. Cembella, and R. M. Kudela Contrasting wind patterns and toxigenic phytoplankton in the southern Benguela upwelling system. Mar. Ecol. Prog. Ser. 348: Fensome, R. A., J. F. Saldarriaga, and F. J. R. Taylor Dinoflagellate phylogeny revisited: reconciling morphological and molecular based phylogenies. Grana 38: Guerrini, F., P. Ciminiello, C. Dell Aversano, L. Tartaglione, E. Fattorusso, L. Boni, and R. Pistocchi Influence of temperature, salinity and nutrient limitation on yessotoxin production and release by the dinoflagellate Protoceratium reticulatum in batch cultures. Harmful Algae 6: Guillard, R. R. L., and J. H. Ryther Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8: Guillard, R. R. L Culture of phytoplankton for feeding marine invertebrates, p In W. L. Smith and M. H. Chanely (ed.), Culture of marine invertebrate animals. Plenum Press, New York, NY. 17. Guillard, R. R. L., and P. E. Hargraves Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32: Howard, M. D. A., M. Silver, and R. M. Kudela Yessotoxin detected in mussel (Mytilus californicus) and phytoplankton samples from the U.S. west coast. Harmful Algae 7: Hwang, D. F., and Y. H. Lu Influence of environmental and nutritional factors on growth, toxicity and toxin profile of dinoflagellate Alexandrium minutum. Toxicon 38: Kim, K.-Y., Y.-S. Kim, C.-H. Hwang, C.-K. Lee, W.-A. Lim, and C.-H. Kim Phylogenetic analysis of dinoflagellate Gonyaulax polygramma Stein responsible for harmful algal blooms based on the partial LSU rdna sequence data. Algae 21: Krock, B., T. Alpermann, U. Tilmann, G. C. Pitcher, and A. D. Cembella Yessotoxin profiles of marine dinoflagellates Protoceratium reticulatum and Gonyaulax spinifera, p In O. Moestrup (ed.), Proceedings of the XII International Conference on Harmful Algae. International Society for the Study of Harmful Algae and Intergovernmental Oceanographic Commission, Copenhagen, Denmark, 4 to 8 September Kudela, R. M., A. Roberts, and M. Armstrong Laboratory analyses of nutrient stress and toxin production in Pseudo-nitzschia spp. from Monterey Bay, California, p In K. A. Steidinger, J. H. Landsberg, C. R. Tomas, and G. A. Vargo (ed.), Harmful algae Florida and Wildlife Conservation Commission, Florida Institute of Oceanography, and Commission of UNESCO, St. Petersburg, FL. 23. Kumar, S., K. Tamura, and M. Nei MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: LaJeunesse, T. C Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a species level marker. J. Phycol. 37: Reference deleted. 26. Lenaers, G., L. Maroteaux, B. Michot, and M. Herzog Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J. Mol. Evol. 29: Lenaers, G., C. Scholin, Y. Bhaud, D. Saint-Hilaire, and M. Herzog A molecular phylogeny of dinoflagellate protests (Pyrrhophyta) inferred from the sequence of 24S rrna divergent domains D1 and D8. J. Mol. Evol. 32: Litaker, R. W., M. W. Vandersea, S. R. Kibler, K. S. Reece, N. A. Stokes, F. M. Lutzoni, B. A. Yonish, M. A. West, M. N. D. Black, and P. A. Tester Recognizing dinoflagellate species using ITS rdna sequences. J. Phycol. 43: Lundholm, N., Ø. Moestrup, G. R. Hasle, and K. Hoef-Emden A study of the Pseudo-nitzschia pseudodelicatissima/cuspidata complex (Bacillariophyceae): what is P. pseudodelicatissima? J. Phycol. 39: MacKenzie, L., P. Holland, P. McNabb, V. Beuzenberg, A. Selwood, and T. Suzuki Complex toxin profiles in phytoplankton and greenshell mussels (Perna canaliculus), revealed by LC-MS/MS analysis. Toxicon 40: Mikulski, C. M., S. L. Morton, and G. J. Doucette Development and application of LSU rrna probes for Karenia brevis in the Gulf of Mexico, USA. Harmful Algae 4: Miller, P. E., and C. A. Scholin Identification of culture Pseudonitzschia (Bacillariophyceae) using species-specific LSU rrna targeted fluorescent probes. J. Phycol. 32: Mitrovic, S. M., M. Fernández Amandi, L. MacKenzie, A. Furey, and K. J. James Effects of selenium, iron and cobalt addition to growth and yessotoxin production of the toxic marine dinoflagellate Protoceratium reticulatum in culture. J. Exp. Mar. Biol. Ecol. 313: Mitrovic, S. M., B. Hamilton, L. MacKenzie, A. Furey, and K. J. James Persistence of yessotoxin under light and dark conditions. Mar. Environ. Res. 60: Moorthi, S. D., P. D. Countway, B. A. Stauffer, and D. A. Caron Use of quantitative real-time PCR to investigate the dynamics of the red tide dinoflagellate Lingulodinium polyedrum. Microb. Ecol. 52: Murata, M., M. Kumagai, J. S. Lee, and T. Yasumoto Isolation and structure of yessotoxin, a novel polyether compound implicated in diarrhetic shellfish poisoning. Tetrahedron Lett. 28: Murray, S., M. F. Jørgensen, S. Y. W. Ho, D. J. Patterson, and L. S. Jermiin Improving the analysis of dinoflagellate phylogeny based on rdna. Protist 156: Pan, Y., D. V. Subba Rao, K. H. Mann, R. G. Brown, and R. Pocklington Effects of silicate limitation on production of domoic acid, a neurotoxin, by the diatom Pseudo-nitzschia multiseries. I. Batch culture studies. Mar. Ecol. Prog. Ser. 131: Pan, Y., D. V. Subba Rao, K. H. Mann, W. K. W. Li, and W. G. Harrison.
10 VOL. 75, 2009 PHYLOGENY OF YESSOTOXIN-PRODUCING DINOFLAGELLATES Effects of silicate limitation on production of domoic acid, a neurotoxin, by the diatom Pseudo-nitzschia multiseries. II. Continuous culture studies. Mar. Ecol. Prog. Ser. 131: Pan, Y., D. V. Subba Rao, and K. H. Mann Changes in domoic acid production and cellular chemical composition of the toxigenic diatom Pseudonitzschia multiseries under phosphate limitation. J. Phycol. 32: Paz, B., P. Riobo, M. L. Fernández, S. Fraga, and J. M. Franco Production and release of yessotoxins by the dinoflagellates Protoceratium reticulatum and Lingulodinium polyedrum in culture. Toxicon 44: Paz, B., P. Riobó, I. Ramilo, and J. M. Franco Yessotoxins profile in strains of Protoceratium reticulatum from Spain and USA. Toxicon 50: Plumley, F. G Marine algal toxins: biochemistry, genetics, and molecular biology. Limnol. Oceanogr. 42: Ramstad, H., P. Hovgaard, T. Yasumoto, S. Larsen, and T. Aune Monthly variations in diarrhetic toxins and yessotoxin in shellfish from coast to the inner part of the Sognefjord, Norway. Toxicon 39: Rhodes, L., P. McNabb, M. de Salas, L. Briggs, V. Beuzenberg, and M. Gladstone Yessotoxin production by Gonyaulax spinifera. Harmful Algae 5: Riccardi, M., F. Guerrini, F. Roncarati, A. Milandri, M. Cangini, S. Pigozzi, E. Riccardi, A. Ceredi, P. Ciminiello, C. Dell Aversano, E. Fattorusso, M. Forino, L. Tartaglione, and R. Pistocchi. Gonyaulax spinifera from the Adriatic Sea: toxin production and phylogenetic analysis. Harmful Algae, in press. 47. Riobo, P., B. Paz, M. Fernandez, S. Fraga, and J. M. Franco Lipophylic toxins of different strains of Ostreopsidaceae and Gonyaulacaceae. Abstr. 10th Int. Harmful Algal Bloom Conf., St. Petersburg, FL. 48. Saito, K., T. Drgon, J. A. F. Robledo, D. N. Krupatkina, and G. R. Vasta Characterization of the rrna locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl. Environ. Microbiol. 68: Saldarriaga, J. R., F. J. R. Taylor, T. Cavalier-Smith, S. Menden-Deuer, and P. J. Keeling Molecular data and the evolutionary history of dinoflagellates. Eur. J. Protistol. 40: Samdal, I. A., L. J. Naustvoll, C. D. Olseng, L. R. Briggs, and C. O. Miles Use of ELISA to identify Protoceratium reticulatum as a source of yessotoxin in Norway. Toxicon 44: Satake, M., K. Terawawa, Y. Kadowaki, and T. Yasumoto Relative configuration of yessotoxin and isolation of two new analogs from toxic scallops. Tetrahedron Lett. 37: Satake, M., T. Ichimura, K. Sekiguchi, S. Yoshimatsu, and Y. Oshima Confirmation of yessotoxin and 45, 46, 47-trinoryessotoxin production by Protoceratium reticulatum collected in Japan. Nat. Toxins 7: Schlötterer, C Ribosomal DNA probes and primers, p In A. Karp, P. G. Isaac, and D. S. Ingram (ed.), Molecular tools for screening biodiversity. Chapman & Hall, London, United Kingdom. 54. Scholin, C. A., M. Herzog, M. L. Sogin, and D. M. Anderson Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rrna gene. J. Phycol. 30: Scholin, C. A., R. Marin, III, and P. E. Miller DNA probe-based assays for rapid detection of toxic algal species in environmental samples. J. Phycol. 32(Suppl.): Shao, P., Y.-Q. Chen, H. Zhou, J. Yuan, L.-H. Qu, D. Zhao, and Y.-S. Lin Genetic variability in Gymnodiniaceae ITS regions: implications for species identification and phylogenetic analysis. Mar. Biol. 144: Smith, G. J., N. L. Ladizinsky, and P. E. Miller Amino acid profiles in species and strains of Pseudo-nitzschia from Monterey Bay, California: insights into the metabolic role(s) of domoic acid, p In G. M. Hallegraeff, S. I. Blackburn, C. J. Bolch, and R. J. Lewis (ed.), Harmful algal blooms IOC/UNESCO, Paris, France. 58. Stobo, L. A., J. Lewis, M. A. Quilliam, W. R. Hardstaff, S. Gallacher, L. Webster, E. A. Smith, and M. McKenzie Detection of yessotoxin in UK and Canadian isolates of phytoplankton and optimization and validation of LC-MS methods. Can. Tech. Rep. Fish. Aquat. Sci. 2498: Taylor, F. J. R Illumination of confusion? Dinoflagellate molecular phylogenetic data viewed from a primarily morphological standpoint. Phycol. Res. 52: Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: Tomas, C. R Identifying marine phytoplankton, p Academic Press, San Diego, CA. 62. Tubaro, A., L. Sidari, R. Della Loggia, and T. Yasumoto Occurrence of yessotoxin in phytoplankton and mussels from northern Adriatic Sea, p In B. Requera, J. Blanco, M. L. Fernández, and T. Wyatt (ed.), Harmful algae. Proceedings of the VIII International Conference on Harmful Algae. Xunta de Galicia and Intergovernmental Oceanographic Commission of UNESCO, Vigo, Spain. 63. Zardoya, R., E. Costas, V. López-Rodas, A. Garrido-Pertierra, and J. M. Bautista Revised dinoflagellate phylogeny inferred from molecular analysis of large-subunit ribosomal RNA gene sequences. J. Mol. Evol. 41: Downloaded from on July 7, 2018 by guest
Azaspiracids producing dinoflagellates
 Azaspiracids producing dinoflagellates Kazuya Takahashi and Mitsunori Iwataki Univ Tokyo, Japan Azadinium spp. Found in 2009 as a producer of lipophilic polyether toxin Azaspiracids (AZAs), responsible
Azaspiracids producing dinoflagellates Kazuya Takahashi and Mitsunori Iwataki Univ Tokyo, Japan Azadinium spp. Found in 2009 as a producer of lipophilic polyether toxin Azaspiracids (AZAs), responsible
State-of-the-Art Liquid Chromatography- Tandem Mass Spectrometry Detection Methods for Shellfish Toxins (ASP, DSP, PSP)
 State-of-the-Art Liquid Chromatography- Tandem Mass Spectrometry Detection Methods for Shellfish Toxins (ASP, DSP, PSP) utline 1. Introduction 2. Toxins 3. Mass spectrometry - functional principle - ionization
State-of-the-Art Liquid Chromatography- Tandem Mass Spectrometry Detection Methods for Shellfish Toxins (ASP, DSP, PSP) utline 1. Introduction 2. Toxins 3. Mass spectrometry - functional principle - ionization
Harmful Algal Blooms (HABs) 5 Applications
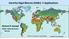 Harmful Algal Blooms (HABs) 5 Applications Richard P. Stumpf NOAA, National Ocean Service HAB occurrences worldwide Image from whoi.edu/redtide HAB applications: short term Management: Monitoring and Response
Harmful Algal Blooms (HABs) 5 Applications Richard P. Stumpf NOAA, National Ocean Service HAB occurrences worldwide Image from whoi.edu/redtide HAB applications: short term Management: Monitoring and Response
Shifting N:P ratios do not affect the competitive traits of dinoflagellates under present and future climate scenarios
 Shifting N:P ratios do not affect the competitive traits of dinoflagellates under present and future climate scenarios Maarten De Rijcke, Gabriel Orellana, Julie Vanden Bussche, Lynn Vanhaecke, Karel A.C.
Shifting N:P ratios do not affect the competitive traits of dinoflagellates under present and future climate scenarios Maarten De Rijcke, Gabriel Orellana, Julie Vanden Bussche, Lynn Vanhaecke, Karel A.C.
MOLECULAR DETECTION OF HABs SPECIES :
 MOLECULAR DETECTION OF HABs SPECIES : How it complement to HAB MONITORING PROGRAM Chui Pin LEAW PhD IOES, UM Species recognition Taxonomy is a fundamental research that ones could not neglect in any discipline
MOLECULAR DETECTION OF HABs SPECIES : How it complement to HAB MONITORING PROGRAM Chui Pin LEAW PhD IOES, UM Species recognition Taxonomy is a fundamental research that ones could not neglect in any discipline
Recent Harmful Algal blooms (HABs) Events in Indonesia
 Recent Harmful Algal blooms (HABs) Events in Indonesia By Hikmah Thoha Research Center for Oceanography, Indonesian Institute of Science Indonesia What We Know, and What We Do Not Know on HABs Outline
Recent Harmful Algal blooms (HABs) Events in Indonesia By Hikmah Thoha Research Center for Oceanography, Indonesian Institute of Science Indonesia What We Know, and What We Do Not Know on HABs Outline
FLOW: Amigos de Bolsa Chica Citizen Science Program. Plankton Collection and Identification Report
 FLOW: Amigos de Bolsa Chica Citizen Science Program Plankton Collection and Identification Report Date: 05/03/13 Time: 10:30 AM Collectors: Judy H., Dennis P., Nicole G., Joana T. Tide: ebb (going out)
FLOW: Amigos de Bolsa Chica Citizen Science Program Plankton Collection and Identification Report Date: 05/03/13 Time: 10:30 AM Collectors: Judy H., Dennis P., Nicole G., Joana T. Tide: ebb (going out)
Observation system for early warning of HAB events
 Observation system for early warning of HAB events Vera L. Trainer, NOAA Fisheries Northwest Fisheries Science Center Marine Biotoxins Program Seattle, Washington, USA Juan de Fuca eddy Regional HAB OOS
Observation system for early warning of HAB events Vera L. Trainer, NOAA Fisheries Northwest Fisheries Science Center Marine Biotoxins Program Seattle, Washington, USA Juan de Fuca eddy Regional HAB OOS
Ireland: Current Conditions
 To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
Received 21 January 2009, revised 11 February 2009, accepted 16 February 2009.
 Communications The first recorded bloom of Pseudochattonella farcimen (Dictyochophyceae, Heterokonta),(Riisberg I.,2008)intheGulfof Gdańsk* OCEANOLOGIA, 51(1), 2009. pp. 139 143. C 2009,byInstituteof Oceanology
Communications The first recorded bloom of Pseudochattonella farcimen (Dictyochophyceae, Heterokonta),(Riisberg I.,2008)intheGulfof Gdańsk* OCEANOLOGIA, 51(1), 2009. pp. 139 143. C 2009,byInstituteof Oceanology
Species composition and long-term dynamics of potentially toxic dinoflagellate species in benthic assemblages of Peter the Great Bay, Sea of Japan
 Species composition and long-term dynamics of potentially toxic dinoflagellate species in benthic assemblages of Peter the Great Bay, Sea of Japan Marina Selina & Tatiana Morozova tatiana_morozova@mail.ru
Species composition and long-term dynamics of potentially toxic dinoflagellate species in benthic assemblages of Peter the Great Bay, Sea of Japan Marina Selina & Tatiana Morozova tatiana_morozova@mail.ru
Ireland: Current Conditions
 ATLANTIC OCEAN IRISH SEA HAB Bulletin [status of harmful and toxic algae] Ireland: Current Conditions Shellfish biotoxin report (last week) Week 42:4 th 10 th October,2015 Week runs from Sunday to Saturday
ATLANTIC OCEAN IRISH SEA HAB Bulletin [status of harmful and toxic algae] Ireland: Current Conditions Shellfish biotoxin report (last week) Week 42:4 th 10 th October,2015 Week runs from Sunday to Saturday
Phylogenetic relationship of Alexandrium monilatum (Dinophyceae) to other Alexandrium species based on 18S ribosomal RNA gene sequences
 Harmful Algae 5 (2006) 275 280 A. monilatum was first identified and described from waters collected near Melbourne on the east coast of Florida (Howell, 1953). Because many of these blooms are associated
Harmful Algae 5 (2006) 275 280 A. monilatum was first identified and described from waters collected near Melbourne on the east coast of Florida (Howell, 1953). Because many of these blooms are associated
PICES-2010 Annual Meeting Portland, OR October 27, Jenny Q. Lane Peter T. Raimondi Raphael M. Kudela
 The development of toxigenic Pseudo-nitzschia bloom models in Monterey Bay, CA, and their application at a single monitoring site within the model domain PICES-2010 Annual Meeting Portland, OR October
The development of toxigenic Pseudo-nitzschia bloom models in Monterey Bay, CA, and their application at a single monitoring site within the model domain PICES-2010 Annual Meeting Portland, OR October
MiGA: The Microbial Genome Atlas
 December 12 th 2017 MiGA: The Microbial Genome Atlas Jim Cole Center for Microbial Ecology Dept. of Plant, Soil & Microbial Sciences Michigan State University East Lansing, Michigan U.S.A. Where I m From
December 12 th 2017 MiGA: The Microbial Genome Atlas Jim Cole Center for Microbial Ecology Dept. of Plant, Soil & Microbial Sciences Michigan State University East Lansing, Michigan U.S.A. Where I m From
Distribution of Dinoflagellate Cysts in the Surface Sediment of the South China Sea, Area I: Gulf of Thailand and East Coast of Peninsular Malaysia
 Distribution of Dinoflagellate Cysts in the Surface Sediment of the South China Sea, Area I: Gulf of Thailand and East Coast of Peninsular Malaysia Thaithaworn Lirdwitayaprasit Department of Marine Science,
Distribution of Dinoflagellate Cysts in the Surface Sediment of the South China Sea, Area I: Gulf of Thailand and East Coast of Peninsular Malaysia Thaithaworn Lirdwitayaprasit Department of Marine Science,
West Coast HABs affect:
 West Coast HABs affect: Fish (both wild and penned Marine Birds Marine Mammals Humans & Invertebrates Five West Coast Harmful Algal Bloom Species ϑ Dinoflagellate, Alexandrium spp. ϑ Pennate diatom, Pseudo-nitzschia
West Coast HABs affect: Fish (both wild and penned Marine Birds Marine Mammals Humans & Invertebrates Five West Coast Harmful Algal Bloom Species ϑ Dinoflagellate, Alexandrium spp. ϑ Pennate diatom, Pseudo-nitzschia
Testing for Grazer Adaptation to Toxic Algae
 Testing for Grazer Adaptation to Toxic Algae by Michael B. Finiguerra, Hans G. Dam, and David E. Avery Part I Introduction and Background Phytoplankton, microscopic single-celled algae, are natural components
Testing for Grazer Adaptation to Toxic Algae by Michael B. Finiguerra, Hans G. Dam, and David E. Avery Part I Introduction and Background Phytoplankton, microscopic single-celled algae, are natural components
Management of recreational waters in relationship with harmful microalgae blooms (HAB) in the Mediterranean Sea
 EVK3-2001- 00046 WORKSHOP Management of recreational waters in relationship with harmful microalgae blooms (HAB) in the Mediterranean Sea 25 26th October 2004 Hotel Don Antonio, Peguera- Calvià, Mallorca
EVK3-2001- 00046 WORKSHOP Management of recreational waters in relationship with harmful microalgae blooms (HAB) in the Mediterranean Sea 25 26th October 2004 Hotel Don Antonio, Peguera- Calvià, Mallorca
David W. Seaborn 1, A. Michelle Seaborn 2, William M. Dunstan 2, and Harold G. Marshall 1
 Virginia Journal of Science Volume 5, Number 4 Winter 1999 Growth and Feeding Studies on the Algal Feeding Stage of a Pfiesteria-like Dinoflagellate David W. Seaborn 1, A. Michelle Seaborn 2, William M.
Virginia Journal of Science Volume 5, Number 4 Winter 1999 Growth and Feeding Studies on the Algal Feeding Stage of a Pfiesteria-like Dinoflagellate David W. Seaborn 1, A. Michelle Seaborn 2, William M.
TRANSFORMATION OF PARALYTIC SHELLFISH TOXINS IN BENTHIC CLAM, POLYMESODA SIMILIS FED WITH TOXIC DINOFLAGELLATE, ALEXANDRIUM MINUTUM (DINOPHYCEAE)
 TRANSFORMATION OF PARALYTIC SHELLFISH TOXINS IN BENTHIC CLAM, POLYMESODA SIMILIS FED WITH TOXIC DINOFLAGELLATE, ALEXANDRIUM MINUTUM (DINOPHYCEAE) ABSTRACT P. T. Lim 1, P. S. Daniel Ngilek 1, C. P., Leaw
TRANSFORMATION OF PARALYTIC SHELLFISH TOXINS IN BENTHIC CLAM, POLYMESODA SIMILIS FED WITH TOXIC DINOFLAGELLATE, ALEXANDRIUM MINUTUM (DINOPHYCEAE) ABSTRACT P. T. Lim 1, P. S. Daniel Ngilek 1, C. P., Leaw
Ireland: Current Conditions
 ATLANTIC OCEAN IRISH SEA HAB Bulletin [status of harmful and toxic algae] Ireland: Current Conditions Shellfish biotoxin report (last week) National Monitoring Programme Designated Sampling Sites NORTH
ATLANTIC OCEAN IRISH SEA HAB Bulletin [status of harmful and toxic algae] Ireland: Current Conditions Shellfish biotoxin report (last week) National Monitoring Programme Designated Sampling Sites NORTH
Parting the Red Seas: The Optics of Red Tides
 Parting the Red Seas: The Optics of Red Tides H.M. Dierssen 1*, Kudela, R.M. 2, Ryan, J.P. 3 1 University of Connecticut, Department of Marine Science, Groton, CT 06340. 2 University of California, Ocean
Parting the Red Seas: The Optics of Red Tides H.M. Dierssen 1*, Kudela, R.M. 2, Ryan, J.P. 3 1 University of Connecticut, Department of Marine Science, Groton, CT 06340. 2 University of California, Ocean
Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
Dr. Amira A. AL-Hosary
 Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
Evaluating the Ribosomal Internal Transcribed Spacer (ITS) as a Candidate Dinoflagellate Barcode Marker
 Evaluating the Ribosomal Internal Transcribed Spacer (ITS) as a Candidate Dinoflagellate Barcode Marker Rowena F. Stern 1 *, Robert A. Andersen 2, Ian Jameson 3, Frithjof C. Küpper 4, Mary-Alice Coffroth
Evaluating the Ribosomal Internal Transcribed Spacer (ITS) as a Candidate Dinoflagellate Barcode Marker Rowena F. Stern 1 *, Robert A. Andersen 2, Ian Jameson 3, Frithjof C. Küpper 4, Mary-Alice Coffroth
Effects of environmental variables on midsummer dinoflagellate community in the Neva Estuary. Mikhail Golubkov, Vera Nikulina, Sergey Golubkov
 Effects of environmental variables on midsummer dinoflagellate community in the Neva Estuary Mikhail Golubkov, Vera Nikulina, Sergey Golubkov Introduction Dinoflagellates are single-celled eukaryotes that
Effects of environmental variables on midsummer dinoflagellate community in the Neva Estuary Mikhail Golubkov, Vera Nikulina, Sergey Golubkov Introduction Dinoflagellates are single-celled eukaryotes that
FLOW: Amigos de Bolsa Chica Citizen Science Program. Plankton Collection and Identification Report
 FLOW: Amigos de Bolsa Chica Citizen Science Program Plankton Collection and Identification Report Date: 04/26/13 Time: 2:35 PM Collectors: Judy H., Chuck D., Belen C., Joana T. (analysis also performed
FLOW: Amigos de Bolsa Chica Citizen Science Program Plankton Collection and Identification Report Date: 04/26/13 Time: 2:35 PM Collectors: Judy H., Chuck D., Belen C., Joana T. (analysis also performed
Ireland: Predictions. NMP Current closures ASP AZP DSP PSP
 HAB Bulletin [status of harmful and toxic algae] Ireland: Predictions ASP event: Low AZP event: High DSP event: High PSP event: High (site specific, moderate in general) Week 30: July 17 th - July 22 nd
HAB Bulletin [status of harmful and toxic algae] Ireland: Predictions ASP event: Low AZP event: High DSP event: High PSP event: High (site specific, moderate in general) Week 30: July 17 th - July 22 nd
Ireland: Current Conditions
 To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
Phylogenetic relationship of Alexandrium tamiyavanichii (Dinophyceae) to other Alexandrium species based on ribosomal RNA gene sequences
 Harmful Algae 1 (2002) 59 68 Phylogenetic relationship of Alexandrium tamiyavanichii (Dinophyceae) to other Alexandrium species based on ribosomal RNA gene sequences Gires Usup, Leaw C. Pin, Asmat Ahmad,
Harmful Algae 1 (2002) 59 68 Phylogenetic relationship of Alexandrium tamiyavanichii (Dinophyceae) to other Alexandrium species based on ribosomal RNA gene sequences Gires Usup, Leaw C. Pin, Asmat Ahmad,
Microbes usually have few distinguishing properties that relate them, so a hierarchical taxonomy mainly has not been possible.
 Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy. Slowly evolving molecules (e.g., rrna) used for large-scale structure; "fast- clock" molecules for fine-structure.
 Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Ireland: Current Conditions
 To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
Ireland: Current Conditions
 To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
To subscribe to receive the Irish HAB Bulletin by email: http://www.marine.ie/home/site-area/about-us/marine-institute-subscriptions?language=en Under Marine Institute Subscriptions tick the HAB bulletin
Ch 10. Classification of Microorganisms
 Ch 10 Classification of Microorganisms Student Learning Outcomes Define taxonomy, taxon, and phylogeny. List the characteristics of the Bacteria, Archaea, and Eukarya domains. Differentiate among eukaryotic,
Ch 10 Classification of Microorganisms Student Learning Outcomes Define taxonomy, taxon, and phylogeny. List the characteristics of the Bacteria, Archaea, and Eukarya domains. Differentiate among eukaryotic,
Seasonal cycle of phytoplankton community composition off Newport, Oregon, in 2009
 Seasonal cycle of phytoplankton community composition off Newport, Oregon, in 29 Xiuning Du 1, William Peterson 2 1 College of Environmental science and Engineering, Ocean University of China, Qingdao,
Seasonal cycle of phytoplankton community composition off Newport, Oregon, in 29 Xiuning Du 1, William Peterson 2 1 College of Environmental science and Engineering, Ocean University of China, Qingdao,
Taxonomy. Content. How to determine & classify a species. Phylogeny and evolution
 Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
CLASSIC BOOKS OF OCEANOGRAPHY AND MARINE BIOLOGY: AN ANNOTATED BIBLIOGRAPHY. I. MARINE BOTANY
 CLASSIC BOOKS OF OCEANOGRAPHY AND MARINE BIOLOGY: AN ANNOTATED BIBLIOGRAPHY. I. MARINE BOTANY Introduction Janet Webster Hatfield Marine Science Center Oregon State University Newport, OR 97365 janet.webster@oregonstate.edu
CLASSIC BOOKS OF OCEANOGRAPHY AND MARINE BIOLOGY: AN ANNOTATED BIBLIOGRAPHY. I. MARINE BOTANY Introduction Janet Webster Hatfield Marine Science Center Oregon State University Newport, OR 97365 janet.webster@oregonstate.edu
Microbial Taxonomy. Microbes usually have few distinguishing properties that relate them, so a hierarchical taxonomy mainly has not been possible.
 Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Production of Paralytic Shellfish Toxin (PST) by Alexandrium tamarense in response to Copepod grazing
 Production of Paralytic Shellfish Toxin (PST) by Alexandrium tamarense in response to Copepod grazing mage must be placed here, if Viviana used. Bravo Carvajal Erik Selander Size 55x55 mm. Align right
Production of Paralytic Shellfish Toxin (PST) by Alexandrium tamarense in response to Copepod grazing mage must be placed here, if Viviana used. Bravo Carvajal Erik Selander Size 55x55 mm. Align right
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut
 Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
Toxin Levels and Profiles in Microalgae from the North-Western Adriatic Sea 15 Years of Studies on Cultured Species
 Mar. Drugs 2012, 10, 140-162; doi:10.3390/md10010140 Review OPEN ACCESS Marine Drugs ISSN 1660-3397 www.mdpi.com/journal/marinedrugs Toxin Levels and Profiles in Microalgae from the North-Western Adriatic
Mar. Drugs 2012, 10, 140-162; doi:10.3390/md10010140 Review OPEN ACCESS Marine Drugs ISSN 1660-3397 www.mdpi.com/journal/marinedrugs Toxin Levels and Profiles in Microalgae from the North-Western Adriatic
Chapters 25 and 26. Searching for Homology. Phylogeny
 Chapters 25 and 26 The Origin of Life as we know it. Phylogeny traces evolutionary history of taxa Systematics- analyzes relationships (modern and past) of organisms Figure 25.1 A gallery of fossils The
Chapters 25 and 26 The Origin of Life as we know it. Phylogeny traces evolutionary history of taxa Systematics- analyzes relationships (modern and past) of organisms Figure 25.1 A gallery of fossils The
THE ARABIAN SEA: A NATURAL EXPERIMENT IN PHYTOPLANKTON BIOGEOGRAPHY
 THE ARABIAN SEA: A NATURAL EXPERIMENT IN PHYTOPLANKTON BIOGEOGRAPHY LONG TERM GOALS A. Michelle Wood Department of Biology University of Oregon Eugene, Oregon 97403 541-346-0454 email:miche@darkwing.,uoregon.edu
THE ARABIAN SEA: A NATURAL EXPERIMENT IN PHYTOPLANKTON BIOGEOGRAPHY LONG TERM GOALS A. Michelle Wood Department of Biology University of Oregon Eugene, Oregon 97403 541-346-0454 email:miche@darkwing.,uoregon.edu
INTRODUCTION MATERIALS AND METHODS. Alexandrium tamarense (strain ATKR ) used in the present study was isolated in Kure Bay,
 Nitrate Availability for the Accumulation of Shinorine, Palythine, and Porphyra-334 (Mycosporine-like Amino Acids) in Dinoflagellate Alexandrium tamarense Nobuyuki Kobashi 1, Ai Murata 1, Hitomi Taira
Nitrate Availability for the Accumulation of Shinorine, Palythine, and Porphyra-334 (Mycosporine-like Amino Acids) in Dinoflagellate Alexandrium tamarense Nobuyuki Kobashi 1, Ai Murata 1, Hitomi Taira
SHORT COMMUNICATION JOURNAL OF PLANKTON RESEARCH VOLUME 24 NUMBER 7 PAGES
 SHORT COMMUNICATION No difference found in ribosomal DNA sequences from physiologically diverse clones of Karenia brevis (Dinophyceae) from the Gulf of Mexico P. LORET 1,7, T. TENGS 2, T. A. VILLAREAL
SHORT COMMUNICATION No difference found in ribosomal DNA sequences from physiologically diverse clones of Karenia brevis (Dinophyceae) from the Gulf of Mexico P. LORET 1,7, T. TENGS 2, T. A. VILLAREAL
Shellfish biotoxin report (last week)
 Pilot HAB Bulletin [status of harmful and toxic algae] Ireland: Historic Trends, Current Conditions and Predictions Ireland HISTORIC TRENDS 2003-2012 Shellfish Toxicity: does not include winter carry over
Pilot HAB Bulletin [status of harmful and toxic algae] Ireland: Historic Trends, Current Conditions and Predictions Ireland HISTORIC TRENDS 2003-2012 Shellfish Toxicity: does not include winter carry over
Chapter 26: Phylogeny and the Tree of Life Phylogenies Show Evolutionary Relationships
 Chapter 26: Phylogeny and the Tree of Life You Must Know The taxonomic categories and how they indicate relatedness. How systematics is used to develop phylogenetic trees. How to construct a phylogenetic
Chapter 26: Phylogeny and the Tree of Life You Must Know The taxonomic categories and how they indicate relatedness. How systematics is used to develop phylogenetic trees. How to construct a phylogenetic
Microbial Taxonomy and the Evolution of Diversity
 19 Microbial Taxonomy and the Evolution of Diversity Copyright McGraw-Hill Global Education Holdings, LLC. Permission required for reproduction or display. 1 Taxonomy Introduction to Microbial Taxonomy
19 Microbial Taxonomy and the Evolution of Diversity Copyright McGraw-Hill Global Education Holdings, LLC. Permission required for reproduction or display. 1 Taxonomy Introduction to Microbial Taxonomy
RECOGNIZING DINOFLAGELLATE SPECIES USING ITS rdna SEQUENCES 1. Kimberly S. Reece, Nancy A. Stokes. François M. Lutzoni. Bryan A.
 J. Phycol. 43, 344 355 (2007) r 2007 No claim to original US government works DOI: 10.1111/j.1529-8817.2007.00320.x RECOGNIZING DINOFLAGELLATE SPECIES USING ITS rdna SEQUENCES 1 R. Wayne Litaker 2, Mark
J. Phycol. 43, 344 355 (2007) r 2007 No claim to original US government works DOI: 10.1111/j.1529-8817.2007.00320.x RECOGNIZING DINOFLAGELLATE SPECIES USING ITS rdna SEQUENCES 1 R. Wayne Litaker 2, Mark
Microbial Diversity and Assessment (II) Spring, 2007 Guangyi Wang, Ph.D. POST103B
 Microbial Diversity and Assessment (II) Spring, 007 Guangyi Wang, Ph.D. POST03B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm General introduction and overview Taxonomy [Greek
Microbial Diversity and Assessment (II) Spring, 007 Guangyi Wang, Ph.D. POST03B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm General introduction and overview Taxonomy [Greek
2018 OCB Workshop. What goes around comes around: Connectivity and microevolution in the plankton. Tatiana Rynearson
 What goes around comes around: Connectivity and microevolution in the plankton 208 OCB Workshop Tatiana Rynearson Graduate School of Oceanography, University of Rhode Island BGC and plankton Inter-specific
What goes around comes around: Connectivity and microevolution in the plankton 208 OCB Workshop Tatiana Rynearson Graduate School of Oceanography, University of Rhode Island BGC and plankton Inter-specific
Speciation mechanisms. Pavel Škaloud, Algal speciation & evolution lab Charles University, Prague Czech Republic
 Speciation mechanisms Pavel Škaloud, Algal speciation & evolution lab Charles University, Prague Czech Republic Species concepts What are the general causes of protist speciation? 1. Allopatric / sympatric
Speciation mechanisms Pavel Škaloud, Algal speciation & evolution lab Charles University, Prague Czech Republic Species concepts What are the general causes of protist speciation? 1. Allopatric / sympatric
Fluorometry Project Chlorophyll Temperature Time Series
 Fluorometry Project Ocean Institute + Scripps Institution of Oceanography Chlorophyll Temperature Time Series The California Current Long Term Ecological Research (CCE LTER) Phytoplankton Phytoplankton
Fluorometry Project Ocean Institute + Scripps Institution of Oceanography Chlorophyll Temperature Time Series The California Current Long Term Ecological Research (CCE LTER) Phytoplankton Phytoplankton
8/23/2014. Phylogeny and the Tree of Life
 Phylogeny and the Tree of Life Chapter 26 Objectives Explain the following characteristics of the Linnaean system of classification: a. binomial nomenclature b. hierarchical classification List the major
Phylogeny and the Tree of Life Chapter 26 Objectives Explain the following characteristics of the Linnaean system of classification: a. binomial nomenclature b. hierarchical classification List the major
A Study of the Moss Parasite Eocronartium muscicola By: Alicia Knudson Advisor: Dr. Elizabeth Frieders
 A Study of the Moss Parasite Eocronartium muscicola By: Alicia Knudson Advisor: Dr. Elizabeth Frieders Abstract The genus Eocronartium contains a single described species of parasitic fungus on moss plants
A Study of the Moss Parasite Eocronartium muscicola By: Alicia Knudson Advisor: Dr. Elizabeth Frieders Abstract The genus Eocronartium contains a single described species of parasitic fungus on moss plants
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics
 POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
Bioinformatics tools for phylogeny and visualization. Yanbin Yin
 Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
Exploring Microbes in the Sea. Alma Parada Postdoctoral Scholar Stanford University
 Exploring Microbes in the Sea Alma Parada Postdoctoral Scholar Stanford University Cruising the ocean to get us some microbes It s all about the Microbe! Microbes = microorganisms an organism that requires
Exploring Microbes in the Sea Alma Parada Postdoctoral Scholar Stanford University Cruising the ocean to get us some microbes It s all about the Microbe! Microbes = microorganisms an organism that requires
SEQUENCING NUCLEAR MARKERS IN FRESHWATER GREEN ALGAE: CHARA SUBSECTION WILLDENOWIA
 SEQUENCING NUCLEAR MARKERS IN FRESHWATER GREEN ALGAE: CHARA SUBSECTION WILLDENOWIA Stephen D. Gottschalk Department of Biological Sciences, Fordham University, 441 E Fordham Rd, Bronx, NY 10458, USA ABSTRACT
SEQUENCING NUCLEAR MARKERS IN FRESHWATER GREEN ALGAE: CHARA SUBSECTION WILLDENOWIA Stephen D. Gottschalk Department of Biological Sciences, Fordham University, 441 E Fordham Rd, Bronx, NY 10458, USA ABSTRACT
Pectenotoxin-2 and dinophysistoxin-1 in suspended and sedimenting organic matter in the Baltic Sea
 Limnol. Oceanogr., 51(5), 2006, 2300 2307 E 2006, by the American Society of Limnology and Oceanography, Inc. Pectenotoxin-2 and dinophysistoxin-1 in suspended and sedimenting organic matter in the Baltic
Limnol. Oceanogr., 51(5), 2006, 2300 2307 E 2006, by the American Society of Limnology and Oceanography, Inc. Pectenotoxin-2 and dinophysistoxin-1 in suspended and sedimenting organic matter in the Baltic
Amy Driskell. Laboratories of Analytical Biology National Museum of Natural History Smithsonian Institution, Wash. DC
 DNA Barcoding Amy Driskell Laboratories of Analytical Biology National Museum of Natural History Smithsonian Institution, Wash. DC 1 Outline 1. Barcoding in general 2. Uses & Examples 3. Barcoding Bocas
DNA Barcoding Amy Driskell Laboratories of Analytical Biology National Museum of Natural History Smithsonian Institution, Wash. DC 1 Outline 1. Barcoding in general 2. Uses & Examples 3. Barcoding Bocas
Parallel Measurements of Light Scattering and Characterization of Marine Particles in Water: An Evaluation of Methodology
 Parallel Measurements of Light Scattering and Characterization of Marine Particles in Water: An Evaluation of Methodology Dariusz Stramski Marine Physical Laboratory Scripps Institution of Oceanography
Parallel Measurements of Light Scattering and Characterization of Marine Particles in Water: An Evaluation of Methodology Dariusz Stramski Marine Physical Laboratory Scripps Institution of Oceanography
Forecasting inshore red tide blooms using recent past offshore conditions on the West Florida Shelf
 Forecasting inshore red tide blooms using recent past offshore conditions on the West Florida Shelf Harford 1, Bill, Rykowski 2, MB, Babcock 2 EA, Karnauskas 3, M, Sagarese 3, SR, Walter 3, JF. (1) Cooperative
Forecasting inshore red tide blooms using recent past offshore conditions on the West Florida Shelf Harford 1, Bill, Rykowski 2, MB, Babcock 2 EA, Karnauskas 3, M, Sagarese 3, SR, Walter 3, JF. (1) Cooperative
PHYLOGENY AND SYSTEMATICS
 AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 11 Chapter 26 Activity #15 NAME DATE PERIOD PHYLOGENY AND SYSTEMATICS PHYLOGENY Evolutionary history of species or group of related species SYSTEMATICS Study
AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 11 Chapter 26 Activity #15 NAME DATE PERIOD PHYLOGENY AND SYSTEMATICS PHYLOGENY Evolutionary history of species or group of related species SYSTEMATICS Study
DIFFERENTIAL GROWTH RATES OF MICRO-ALGAE IN VARIOUS CULTURE MEDIA
 DIFFERENTIAL GROWTH RATES OF MICRO-ALGAE IN VARIOUS CULTURE MEDIA C. P. GOPINATHAN Central Marine Fisheries Research Institute, Research Centre, Tuticorin. ABSTRACT Growth and multiplication of three algae,
DIFFERENTIAL GROWTH RATES OF MICRO-ALGAE IN VARIOUS CULTURE MEDIA C. P. GOPINATHAN Central Marine Fisheries Research Institute, Research Centre, Tuticorin. ABSTRACT Growth and multiplication of three algae,
Microbial 16S rdna Sequencing of Cultivable Bacteria Associated with Toxic Dinoflagellate, Pyrodinium bahamense var. compressum
 Transactions on Science and Technology Vol. 4, No. 3-3, 318-323, 2017 Microbial 16S rdna Sequencing of Cultivable Bacteria Associated with Toxic Dinoflagellate, Pyrodinium bahamense var. compressum Salley
Transactions on Science and Technology Vol. 4, No. 3-3, 318-323, 2017 Microbial 16S rdna Sequencing of Cultivable Bacteria Associated with Toxic Dinoflagellate, Pyrodinium bahamense var. compressum Salley
Harmful Algal Blooms (HABS) OCEA 101
 Harmful Algal Blooms (HABS) OCEA 101 HARMFUL ALGAL BLOOMS The definition of a HAB is not clear-cut, since it is a societal term, not a scientific term, that describes a diverse array of blooms (both microscopic
Harmful Algal Blooms (HABS) OCEA 101 HARMFUL ALGAL BLOOMS The definition of a HAB is not clear-cut, since it is a societal term, not a scientific term, that describes a diverse array of blooms (both microscopic
Ireland: Predictions. NMP Current closures ASP AZP DSP PSP
 HAB Bulletin [status of harmful and toxic algae] Ireland: Predictions ASP event: Moderate High - increasing steadily. AZP event: Highest - Ongoing DSP event: High ongoing PSP event: Low decreasing ( -
HAB Bulletin [status of harmful and toxic algae] Ireland: Predictions ASP event: Moderate High - increasing steadily. AZP event: Highest - Ongoing DSP event: High ongoing PSP event: Low decreasing ( -
Chad Burrus April 6, 2010
 Chad Burrus April 6, 2010 1 Background What is UniFrac? Materials and Methods Results Discussion Questions 2 The vast majority of microbes cannot be cultured with current methods Only half (26) out of
Chad Burrus April 6, 2010 1 Background What is UniFrac? Materials and Methods Results Discussion Questions 2 The vast majority of microbes cannot be cultured with current methods Only half (26) out of
Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be mixotrophic
 AQUATIC MICROBIAL ECOLOGY Vol. 4: 133 15, 25 Published September 6 Aquat Microb Ecol Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be
AQUATIC MICROBIAL ECOLOGY Vol. 4: 133 15, 25 Published September 6 Aquat Microb Ecol Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be
SPECIATION. REPRODUCTIVE BARRIERS PREZYGOTIC: Barriers that prevent fertilization. Habitat isolation Populations can t get together
 SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
This is a repository copy of Microbiology: Mind the gaps in cellular evolution.
 This is a repository copy of Microbiology: Mind the gaps in cellular evolution. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/114978/ Version: Accepted Version Article:
This is a repository copy of Microbiology: Mind the gaps in cellular evolution. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/114978/ Version: Accepted Version Article:
Long-term changes in microalgae communities on the Russian East coast with emphasis on toxic and bloom forming species
 Long-term changes in microalgae communities on the Russian East coast with emphasis on toxic and bloom forming species Tatiana Yu. Orlova, Polina A. Kameneva, Tatiana V. Morozova A.V. Zhirmunsky Institute
Long-term changes in microalgae communities on the Russian East coast with emphasis on toxic and bloom forming species Tatiana Yu. Orlova, Polina A. Kameneva, Tatiana V. Morozova A.V. Zhirmunsky Institute
Microbial Grazers Lab
 Microbial Grazers Lab Objective: Measure the rate at which bacteria are consumed by predators. Overview Size based food webs Microbial loop concepts acterial predators Methods to assess microbial grazing
Microbial Grazers Lab Objective: Measure the rate at which bacteria are consumed by predators. Overview Size based food webs Microbial loop concepts acterial predators Methods to assess microbial grazing
Phylogeny, biogeography, and species boundaries within the Alexandrium minutum group
 Harmful Algae 4 (2005) 1004 1020 www.elsevier.com/locate/hal Phylogeny, biogeography, and species boundaries within the Alexandrium minutum group E.L. Lilly a, *, K.M. Halanych b, D.M. Anderson c a Department
Harmful Algae 4 (2005) 1004 1020 www.elsevier.com/locate/hal Phylogeny, biogeography, and species boundaries within the Alexandrium minutum group E.L. Lilly a, *, K.M. Halanych b, D.M. Anderson c a Department
EXAMINING ALLELOPATHIC COMPETITON OF A RED TIDE ALGA WITHIN TWO DIFFERENT MARINE COMMUNITIES
 EXAMINING ALLELOPATHIC COMPETITON OF A RED TIDE ALGA WITHIN TWO DIFFERENT MARINE COMMUNITIES A Thesis Presented to The Academic Faculty by Elizabeth McMillan In Partial Fulfillment of the Requirements
EXAMINING ALLELOPATHIC COMPETITON OF A RED TIDE ALGA WITHIN TWO DIFFERENT MARINE COMMUNITIES A Thesis Presented to The Academic Faculty by Elizabeth McMillan In Partial Fulfillment of the Requirements
Bioinformatics. Dept. of Computational Biology & Bioinformatics
 Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Genomic insights into the taxonomic status of the Bacillus cereus group. Laboratory of Marine Genetic Resources, Third Institute of Oceanography,
 1 2 3 Genomic insights into the taxonomic status of the Bacillus cereus group Yang Liu 1, Qiliang Lai 1, Markus Göker 2, Jan P. Meier-Kolthoff 2, Meng Wang 3, Yamin Sun 3, Lei Wang 3 and Zongze Shao 1*
1 2 3 Genomic insights into the taxonomic status of the Bacillus cereus group Yang Liu 1, Qiliang Lai 1, Markus Göker 2, Jan P. Meier-Kolthoff 2, Meng Wang 3, Yamin Sun 3, Lei Wang 3 and Zongze Shao 1*
Chapter 19. Microbial Taxonomy
 Chapter 19 Microbial Taxonomy 12-17-2008 Taxonomy science of biological classification consists of three separate but interrelated parts classification arrangement of organisms into groups (taxa; s.,taxon)
Chapter 19 Microbial Taxonomy 12-17-2008 Taxonomy science of biological classification consists of three separate but interrelated parts classification arrangement of organisms into groups (taxa; s.,taxon)
Biology 211 (2) Week 1 KEY!
 Biology 211 (2) Week 1 KEY Chapter 1 KEY FIGURES: 1.2, 1.3, 1.4, 1.5, 1.6, 1.7 VOCABULARY: Adaptation: a trait that increases the fitness Cells: a developed, system bound with a thin outer layer made of
Biology 211 (2) Week 1 KEY Chapter 1 KEY FIGURES: 1.2, 1.3, 1.4, 1.5, 1.6, 1.7 VOCABULARY: Adaptation: a trait that increases the fitness Cells: a developed, system bound with a thin outer layer made of
Microbial DNA qpcr Multi-Assay Kit Clostridium perfringens Pathogenicity
 Microbial DNA qpcr Multi-Assay Kit Clostridium perfringens Pathogenicity Cat. no. 330043 BBID-1507ZR-3 For real-time PCR-based, application-specific microbial identification or profiling The Clostridium
Microbial DNA qpcr Multi-Assay Kit Clostridium perfringens Pathogenicity Cat. no. 330043 BBID-1507ZR-3 For real-time PCR-based, application-specific microbial identification or profiling The Clostridium
JOURNAL OF ENVIRONMENTAL SCIENCES 66 (2018) Available online at ScienceDirect.
 JOURNAL OF ENVIRONMENTAL SCIENCES 66 (2018) 246 254 Available online at www.sciencedirect.com ScienceDirect www.elsevier.com/locate/jes Identification of a marine woloszynskioid dinoflagellate Biecheleriopsis
JOURNAL OF ENVIRONMENTAL SCIENCES 66 (2018) 246 254 Available online at www.sciencedirect.com ScienceDirect www.elsevier.com/locate/jes Identification of a marine woloszynskioid dinoflagellate Biecheleriopsis
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Prokaryotes Vs. Eukaryotes
 The Microbial World Prokaryotes Vs. Eukaryotes Mircrobes of the Ocean Primary Producers Are the organisms that produce bio-mass from inorganic compounds (autotrophs). -Photosynthetic autotrophs Phytoplankton
The Microbial World Prokaryotes Vs. Eukaryotes Mircrobes of the Ocean Primary Producers Are the organisms that produce bio-mass from inorganic compounds (autotrophs). -Photosynthetic autotrophs Phytoplankton
The Microbial World. Chapter 5
 The Microbial World Chapter 5 Viruses Non-cellular infectious agents that have two basic characteristics: Not capable of reproduction without a host cell Structure: Nucleic acid core- can be DNA or RNA
The Microbial World Chapter 5 Viruses Non-cellular infectious agents that have two basic characteristics: Not capable of reproduction without a host cell Structure: Nucleic acid core- can be DNA or RNA
Regional Forecasting of California HABs
 Regional Forecasting of California HABs Chlorophyll Domoic Acid Clarissa Anderson UC Santa Cruz West Coast Regional HAB Summit February 12, 2009 Trainer et al 2000, Hickey and Banas 2003, in Kudela 2005
Regional Forecasting of California HABs Chlorophyll Domoic Acid Clarissa Anderson UC Santa Cruz West Coast Regional HAB Summit February 12, 2009 Trainer et al 2000, Hickey and Banas 2003, in Kudela 2005
Phylogenetic Analysis of Reticulitermes speratus using the Mitochondrial Cytochrome C Oxidase Subunit I Gene* 1
 Mokchae Konghak 38(2) : 135~139, 2010 Phylogenetic Analysis of Reticulitermes speratus using the Mitochondrial Cytochrome C Oxidase Subunit I Gene* 1 Moon Jung Cho* 2, Keum Shin* 2, Young-Kyoon Kim* 2,
Mokchae Konghak 38(2) : 135~139, 2010 Phylogenetic Analysis of Reticulitermes speratus using the Mitochondrial Cytochrome C Oxidase Subunit I Gene* 1 Moon Jung Cho* 2, Keum Shin* 2, Young-Kyoon Kim* 2,
Lecture 11 Friday, October 21, 2011
 Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Bio 1B Lecture Outline (please print and bring along) Fall, 2007
 Bio 1B Lecture Outline (please print and bring along) Fall, 2007 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #5 -- Molecular genetics and molecular evolution
Bio 1B Lecture Outline (please print and bring along) Fall, 2007 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #5 -- Molecular genetics and molecular evolution
DINOFLAGELLATES are an important group of unicellular
 J. Eukaryot. Microbiol., 50(6), 2003 pp. 417 421 2003 by the Society of Protozoologists Molecular Characterization of Nuclear Small Subunit (18S)-rDNA Pseudogenes in a Symbiotic Dinoflagellate (Symbiodinium,
J. Eukaryot. Microbiol., 50(6), 2003 pp. 417 421 2003 by the Society of Protozoologists Molecular Characterization of Nuclear Small Subunit (18S)-rDNA Pseudogenes in a Symbiotic Dinoflagellate (Symbiodinium,
Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition
 Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition David D. Pollock* and William J. Bruno* *Theoretical Biology and Biophysics, Los Alamos National
Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition David D. Pollock* and William J. Bruno* *Theoretical Biology and Biophysics, Los Alamos National
PLANT BIOLOGY (PBIO) Plant Biology (PBIO) 1
 Plant Biology (PBIO) 1 PLANT BIOLOGY (PBIO) PBIO 1052 How Plants Shaped Our World (LN) Description: This course is an eclectic dive into the world of plants and their influence on human society. Students
Plant Biology (PBIO) 1 PLANT BIOLOGY (PBIO) PBIO 1052 How Plants Shaped Our World (LN) Description: This course is an eclectic dive into the world of plants and their influence on human society. Students
DNA Sequencing as a Method for Larval Identification in Odonates
 DNA Sequencing as a Method for Larval Identification in Odonates Adeline Harris 121 North St Apt 3 Farmington, ME 04938 Adeline.harris@maine.edu Christopher Stevens 147 Main St Apt 1 Farmington, ME 04938
DNA Sequencing as a Method for Larval Identification in Odonates Adeline Harris 121 North St Apt 3 Farmington, ME 04938 Adeline.harris@maine.edu Christopher Stevens 147 Main St Apt 1 Farmington, ME 04938
Chapter 19: Taxonomy, Systematics, and Phylogeny
 Chapter 19: Taxonomy, Systematics, and Phylogeny AP Curriculum Alignment Chapter 19 expands on the topics of phylogenies and cladograms, which are important to Big Idea 1. In order for students to understand
Chapter 19: Taxonomy, Systematics, and Phylogeny AP Curriculum Alignment Chapter 19 expands on the topics of phylogenies and cladograms, which are important to Big Idea 1. In order for students to understand
2001 State of the Ocean: Chemical and Biological Oceanographic Conditions in the Newfoundland Region
 Stock Status Report G2-2 (2) 1 State of the Ocean: Chemical and Biological Oceanographic Conditions in the Background The Altantic Zone Monitoring Program (AZMP) was implemented in 1998 with the aim of
Stock Status Report G2-2 (2) 1 State of the Ocean: Chemical and Biological Oceanographic Conditions in the Background The Altantic Zone Monitoring Program (AZMP) was implemented in 1998 with the aim of
a,bD (modules 1 and 10 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Lack of allelopathic effects of the domoic acid-producing marine diatom Pseudo-nitzschia multiseries
 MARINE ECOLOGY PROGRESS SERIES Vol. 288: 21 33, 2005 Published March 10 Mar Ecol Prog Ser Lack of allelopathic effects of the domoic acid-producing marine diatom Pseudo-nitzschia multiseries Nina Lundholm
MARINE ECOLOGY PROGRESS SERIES Vol. 288: 21 33, 2005 Published March 10 Mar Ecol Prog Ser Lack of allelopathic effects of the domoic acid-producing marine diatom Pseudo-nitzschia multiseries Nina Lundholm
"Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky
 MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
