Lecture 10: December 22, 2009
|
|
|
- Diana Ryan
- 5 years ago
- Views:
Transcription
1 Computational Genomics Fall Semester, 2009 Lecture 0: December 22, 2009 Lecturer: Roded Sharan Scribe: Adam Weinstock and Yaara Ben-Amram Segre 0. Context Free Grammars 0.. Introduction In lecture 6, we saw Nussinov s algorithm[6], which solves the problem of RNA folding by maximizing base-pairing. However, that solution uses an arbitrary scoring function and provides no tools for probabilistic modeling of different RNA sequences. We now wish to create profiles to describe the folding patterns of RNA families and give a likelihood score to the pertinence of an RNA sequence to a specific family. As seen in lectures 7-8, HMMs can be used to create probabilistic profiles for sequence families. Such models are limited, however, as they are unable to describe relations between characters in distant locations along the sequence. In this lecture, we shall review stochastic context free grammars and show how they may be used to overcome the latter limitation Reminder: Formal Languages A formal language, or grammar, G is defined by a quartet G = (V, Σ, S, R) Where: V is a non-terminal alphabet (e.g., {A, B, C, D, E,... } ). Σ is a terminal alphabet (e.g., {a, c, g, t} ). S V is a special start symbol. R is a set of rewriting rules called productions. Formal languages are divided into four categories: Regular grammars The simplest grammars, can be described using rules of the forms W aw, W a. They can be decided using deterministic finite automata (DFA). Note that an HMM is equivalent to a DFA with probabilities assigned to each transition. This lecture is based on Durbin et al., Biological Sequence Analysis (cambridge, 98) and slides by B. Majoros, Duke university.
2 2 Computational Genomics c Tel Aviv Univ. Context free grammars (CFGs) constructed using rules of the form W β, where β is any combination of terminals and non-terminals. They are equivalent to push down automata. Context sensitive grammars constructed using rules of the form αw β αγβ, where α, β, γ are combinations of terminals and non-terminals. They are equivalent to linear bounded automata. Unrestricted grammars constructed using rules of the form αw β γ, where α, β, γ are combinations of terminals and non-terminals. They are equivalent to Turing machines Context Free Grammars Like all formal languages, a context-free grammar is defined by a quartet G = (V, Σ, S, R). Productions in R are rules of the form: X λ, where X V, λ (V Σ). The context-freeness is imposed by the requirement that the left hand side of each production rule may contain only a single symbol, and that symbol must be a non-terminal. Thus, a CFG cannot specify context-sensitive rules (wxz wδz) where the derivation of X is dependent on X s neighbors w, z Derivations A derivation (or parse) consists of a series of applications of production rules from R, beginning with the start non-terminal S and ending with the terminal string x: S s s 2 s 3... x Where s i (V Σ), x Σ. Each step, we replace non-terminal X with the right hand side of a rule from R, whose left hand side is X. If such a derivation exists, we say that G generates x, and denote it by S x. CFGs can be ambiguous. There may be many ways to derive x. Two derivations generating x may vary only in the order in which production rules are applied, which seems somewhat redundant. Consider the following example: G = ({S, A, B}, {a, b}, S, R) R = {S AB, A a, B b} Two derivations can create the string ab :. S AB ab ab
3 Context Free Grammars 3 2. S AB Ab ab In order to avoid this redundancy, we will concentrate on leftmost derivations, where the leftmost non-terminal is always replaced first. All permutations of a certain set of productions will be represented by a single leftmost derivation. Note that we may still encounter multiple derivations of x Context-Free Versus Regular The advantage of CFGs over HMMs lies in their ability to model arbitrary runs of matching pairs of elements, such as matching pairs of parentheses:... ((((((((... ))))))))... When the number of matching pairs is unbounded, a finite-state model such as an HMM is inadequate to enforce the constraint that all left elements must have a matching right element. In contrast, in a CFG we can use rules such as X (X). A sample derivation using such a rule is: X (X) ((X)) (((X))) ((((X)))) (((((X))))) An additional rule such as X ɛ is necessary to terminate the recursion Parse Trees Figure 0.: A sample parse tree deriving the sequence on the right[4]. A parse tree is a representation of a parse of a string by a CFG. The root of the tree is the start non-terminal S. The leaves terminal symbols of the string. Internal nodes non-terminals. The children of an internal node are the productions of that non-terminal (left-to-right order). Note that:
4 4 Computational Genomics c Tel Aviv Univ.. A subtree spans a contiguous sequence segment. 2. Each leftmost derivation corresponds with a single parse tree. This relation is one to one and onto Chomsky Normal Form Any CFG which does not derive the empty string (i.e., ɛ / L(G)) can be converted into an equivalent grammar in Chomsky Normal Form (CNF). A CNF grammar is one in which all productions are of the form: X Y Z or X a for non-terminals X, Y, Z, and terminal a. Transforming a CFG into CNF can be accomplished by appropriately-ordered application of the following operations: Eliminating useless symbols (non-terminals that only derive ɛ) Eliminating null productions (X ɛ) Eliminating unit productions (X Y ) Factoring long right hand side expressions (A abc factored into A ab, B bc, C c) Factoring terminals (A cb is factored into A CB, C c) Example: NON-CNF: S ast tsa csg gsc L L NNNN N a c g t CNF: S AS T T S A CS G GS C NL S A SA S T ST S C SC S G SG L NL 2 L 2 NN N a c g t A a
5 Stochastic Context Free Grammers 5 C c G g T t CNF allows us to easily implement parsers, at the expanse of adding many non-terminals and productions. Though CNF is a less intuitive way to view the problem domain, it may be beneficial in terms of computation. 0.2 Stochastic Context Free Grammers A stochastic context-free grammar (SCFG) is a CFG plus a probability distribution on productions: G = (V, Σ, S, R, P p) Where P p : R [0, ]. P p provides a probability distribution for each non-terminal X. That is, the probabilities of all its productions sum to one: P p(x λ) = X V X λ The probability of a derivation S x is the product of the probabilities of all its productions: P P (X i λ i ) i We can sum over all possible (leftmost) derivations of a given string x to get the probability that G will generate x at random: P (x G) = P (S x G) = j P (S j x G) Note: We do not state given G ( G) on many occasions in order to avoid long equations. Example: Consider the grammar: G = (V G, Σ, S, R G, P G ) V G = {S, L, N}, Σ = {a, c, g, t} R G is the set consisting of:
6 6 Computational Genomics c Tel Aviv Univ. S ast tsa csg gsc L L NNNN N a c g t The probability of the sequence acgtacgtacgt is given by: P (acgtacgtacgt) = P (S ast acsgt acgscgt acgtsacgt acgtlacgt acgtnnnnacgt acgtannnacgt acgtacnnacgt acgtacgnacgt acgtacgtacgt) = = Note that this sequence has only one possible leftmost derivation under grammar G, and therefore the sum consists of a single term. 0.3 The Parsing Problem Given a grammar G and a string x, we would like to know: Can G derive x? If so, what series of productions would be used during the derivation? (There may be multiple answers.) When G is an SCFG, additional questions arise: What is the probability that G derives x? [P (x G)] What is the most probable derivation of x via G? [argmax j (P (S j x G))] 0.4 Solving the Parsing Problem: The CYK Algorithm (Cocke and Schwartz[2], 970; Younger[7], 967; Kasami, 965[5]) The CYK algorithm is a dynamic programming algorithm which utilizes the fact that all subtrees of a given parse tree span a contiguous subsequence segment. Given a grammar G = (V, Σ, S, R) in CNF, we build a dynamic programming matrix D, such that D i,j holds the set of non-terminals that could derive the subsequence x[i, j]. We start with subsequences of length one, and increase subsequence length by one each iteration. We initialize all cells D i,i to contain all non-terminals which produce x i. The remainder of the DP matrix is then
7 Solving the Parsing Problem: The CYK Algorithm 7 computed along diagonals parallel to the main diagonal. Computing cell D i,j, we attempt to find all partitions: x[i, k], x[k +, j] and productions such that: Note that (A BC) R (0.) B x[i, k] (0.2) C x[k +, j] (0.3) B x[i, k] iff B D i,k (0.4) C x[k +, j] iff C D k+,j (0.5) It follows that G generates x iff S D,n, which signifies S x. Figure 0.2: The CYK algorithm: D i,j contains all the non-terminals X which can derive the entire subsequence: actagctatctagcttacggtaatcgcatcgcgc. D k+,j contains only those non-terminals which can derive the red subsequence. D i,k contains only those non-terminals which can derive the green subsequence. Image by B. Majoros Pseudocode for the CYK Algorithm. Initialization: i n, D i,i = {A A x i R}
8 8 Computational Genomics c Tel Aviv Univ. 2. For each i = n- down to do 3. Termination: For each j = i+ up to n do D i,j = {A A BC R, s.t. B D i,k C D k+,j, i k < j} S x iff S D,n Complexity: We denote the number of non-terminals by m = V and the length of the sequence x by n = x. Note that in CNF the number of production rules is at most R = m 3 + m Σ = O(m 3 ) since each rule is either of the form A BC or A a. For each of the O(n 2 ) cells of the DP matrix we check all combinations of O(n) partitions and O(m 3 ) rules, hence total time complexity is O(n 3 m 3 ). For each cell we hold the set of possible non-terminals deriving the subsequence x[i, j]. As there are potentially O(m) such non-terminals, total space complexity is O(n 2 m). 0.5 Extending HMMs for SCFG SCFGs extend CFGs in much the same way HMMs extended regular grammars. We will now present the inside and outside algorithms[], which are analogous to the forward and backward algorithms used in HMMs. The SCFG model can be viewed as inferring the hidden data (the non-terminals) from the observed data (the sequence of terminals), gradually reconstructing the parse tree from the bottom up, ending at the root. 0.6 The Inside Algorithm The inside algorithm is an analog of the forward algorithm. It evaluates the probability of deriving the subsequence x[i, j] given that the root of the corresponding subtree is X. We denote this as: α(i, j, X) = P (X x[i, j] G) = P (x[i, j] X ij, G) (0.6) where X ij is the event of the non-terminal X being the root of the subtree spanning x[i, j]. Computing α(i, j, X) is similar to the computation of D i,j in the CYK algorithm (from the leaves of the parse tree towards its root). The difference is that if previously we accumulated all partitions and productions that allow for the derivation of x[i, j] from X, we now accumulate the probabilities of all such combinations. α(i, j, X) = Y,Z j P (X Y Z)P (x[i, k] Y ik, G)P (x[k +, j] Z (k+)j, G) (0.7) k=i
9 The Inside Algorithm 9 Figure 0.3: The inside and outside algorithms are analogous to the forward and backward algorithms used in HMMs. Image by B. Majoros. With probability P (X Y Z) we use the rule X Y Z, in which case we know that both Y ik and Z (k+)j occur. But from (0.7) we know that the probability P (x[i, k] Y ik, G) of Y being the root of the subtree spanning x[i, k] is exactly α(i, k, Y ), which was calculated in a previous iteration. Similarly, P (x[k +, j] Z (k+)j, G) = α(k +, j, Z) Pseudocode for the Inside Algorithm. Initialization: for i = up to n do foreach non-terminal X do α(i, i, X) = P (X x i ) 2. for i = n down to do for j = i + up to n do foreach non-terminal X do α(i, j, X) = j P (X Y Z)α(i, k, Y )α(k +, j, Z) Y,Z k=i
10 0 Computational Genomics c Tel Aviv Univ. The probability P (x G) of the full input sequence x can then be found in the final cell of the matrix: α(, n, S). Complexity: Complexity analysis for the inside algorithm is essentially the same as that of CYK, aside of the fact that we now have to save the probability corresponding with each possible non-terminal. Therefore the complexity is again O(n 3 m 3 ) time and O(m 2 n) space. 0.7 The Outside Algorithm The outside algorithm is an analog of the backward algorithm. It evaluates the probability β(i, j, X) of a complete parse tree rooted at S for the complete sequence x, excluding the parse subtree for subsequence x[i, j] rooted at X. β(i, j, X) = P (S x... x i Xx j+... x n ) = P (x[, i ], X ij, x[j +, n] G) (0.8) During computation we shall make use of α(i, j, X) values, applying the inside algorithm is therefore a precondition for outside computation. We initialize β(, n, S) to one, since the root of all parse trees is the start non-terminal S. The rest of the DP matrix is filled in diagonals from the root towards the leaves. A parse in which X ij occurs has one of two forms: X ij is the left child of a rule Y XZ. In this case S x... x i Y x k+... x n x... x i XZx k+... x n x... x i Xx j+... x k x k+... x n Note that Z x j+..x k (this means Z ik occurs). The probability of this event is therefore P (x..x i Y x k+..x n )P (Y XZ)P (Z x j+..x k ) = β(i, k, Y )P (Y XZ)α(j+, k, Z) (0.9) X ij is the right child of a rule Y ZX. In a similar fashion, the probability of this event is P (x..x k Y x j+..x n )P (Y ZX)P (Z x k..x i ) = β(k, j, Y )P (Y ZX)α(k, i, Z) (0.0)
11 The Outside Algorithm Each iteration, the length of the excluded sequence diminishes by one (j i decreases). At the end of the computation, the value β(i, i, X) represents P (x..x i Xx i+..x n ), thus we get P (x G) = X V P (X x i )β(i, i, X) (0.) Figure 0.4: Example of a parse where X ij is the left child of a rule Y XZ. The probability of this event is β(i, k, Y )P (Y XZ)α(j +, k, Z). Image by B. Majoros 0.7. Pseudocode for the Outside Algorithm. Initialization: β(, n, S) = foreach X S set β(, n, X) = 0 2. for i = up to n do for j = n down to i do foreach non-terminal X do β(i, j, X) = n P (Y XZ)α(j +, k, Z)β(i, k, Y )+ Y,Z V Y,Z V k=j+ i P (Y ZX)α(k, i, Z)β(k, j, Y ) k=
12 2 Computational Genomics c Tel Aviv Univ. 3. Termination (for any i): P (x G) = X V P (X x i )β(i, i, X) Complexity: Time and space complexity for the outside algorithm are identical to those of the inside algorithm Inferring the Probabilities X ij We now wish to infer the probabilities of all events X ij. These are informative, since they allow us to evaluate the probability of specific events leading to the final structure of the RNA molecule (in our case). For example, we could give the likelihood that a certain subsequence forms a stem-loop structure rather than a bifurcation of two smaller secondary structures. We wish to calculate P (X ij x, G). By Bayes rule: applying the chain rule we get P (X ij x, G) = P (X ij x, G) = P (x G) P (X ij, x G) (0.2) P (x G) P (x..x i Xx j+..x n G)P (x i..x j x..x i Xx j+..x n ) (0.3) Since we are dealing with a context free grammar, the probability of applying a certain rule to a non-terminal X is independent of the flanking sequences w, z. It follows that Going back to the full expression we get P (X ij x, G) = P (X Y Z) = P (wxz wy Zz) (0.4) P (x i..x j x..x i Xx j+..x n ) = P (x i..x j X ij ) (0.5) P (x G) P (x..x i Xx j+..x n G)P (x i..x j X ij ) (0.6) In summation: P (X ij x, G) = β(i, j, X)α(i, j, X) (0.7) P (x G) 0.8 Parameter Estimation Using EM We seldom know the exact probability of each production rule. The parameters of our model (probabilities of the different production rules) are usually estimated given a training set of sequences. As was the case with HMMs, the difficulty lies in the fact that even for our
13 Parameter Estimation Using EM 3 training sequences, the hidden states are unknown. We do not know which non-terminal is responsible for the emission of which character sequence. We use a version of the EM algorithm in order to estimate the model s parameters:. Give an initial estimate of the parameters. Using this initial estimate, apply the inside and outside algorithms to the sequences of the training set. 2. Summing over all parse trees deriving the training set, calculate the following expectations: Expectation of the number of times each rule is applied: E(X Y Z). Expectation of the number of times a non-terminal X appears: E(X). 3. Having calculated these values, give a new estimate of the parameters and update the model: P new (X Y Z) = E(X Y Z)/E(X) (0.8) 4. Repeat the process iteratively, until convergence is reached (the likelihood of the training set given the new estimate increases only marginally) Calculating E(X Y Z) Applying the rule X Y Z at position (i, j) is described by this derivation: S x..x i Xx j+..x n x..x i Y Zx j+..x n x..x i x i..x k Zx j+..x n x..x i x i..x k x k+..x j x j+..x n = x for some i k < j. The probability of this event is therefore: P (X ij, X Y Z x, G) = P (x G) P (X ij, X Y Z, x G) (0.9) j = β(i, j, X)P (X Y Z)α(i, k, Y )α(k +, j, Z) (0.20) P (x G) k=i Summing over all possible positions (i, j) we get the expectation of X Y Z: E(X Y Z) = P (x G) n i= j n β(i, j, X)P (X Y Z)α(i, k, Y )α(k +, j, Z) (0.2) j= k=i
14 4 Computational Genomics c Tel Aviv Univ Calculating E(X a) Applying the rule X a at position (i, i) (the non-terminal X emits a = x i ) is described by this derivation: The probability of this event is therefore: S x..x i Xx i+..x n x..x i a x i+..x n Where δ(x i, a) = P (X ii, X a x, G) = P (x G) P (X ii, X a, x G) (0.22) = P (x G) δ(x i, a)β(i, i, X)P (X a) (0.23) { if x i = a 0 otherwise Summing over all possible positions (i, i) we get the expectation of X a: E(X a) = P (x G) n δ(x i, a)β(i, i, X)P (X a) (0.24) i= Calculating E(X) The expectation of the non-terminal X is the sum of P (X ij x, G) over all positions (i, j). That is: n n E(X) = P (X ij x, G) (0.25) From (0.7) we get: E(X) = P (x G) i= n i= j= n β(i, j, X)α(i, j, X) (0.26) j=
15 Covariance Models EM Update Equations We deduce the following EM update equations: P new (X Y Z X) = E(X Y Z) E(X) = j n n β(i, j, X)P (X Y Z)α(i, k, Y )α(k +, j, Z) i= j= k=i n n β(i, j, X)α(i, j, X) i= j= (0.27) P new (X a X) = E(X a) E(X) = n δ(x i, a)β(i, i, X)P (X a) i= n i= j= n β(i, j, X)α(i, j, X) (0.28) 0.9 Covariance Models Covariance models[3] provide a general modeling scheme for RNA families, using SCFGs. The problem we shall address is creating an SCFG model for the consensus structure of an RNA family. CMs will also provide us with the means to decide whether a given RNA sequence conforms to the consensus structure, and assign that decision with a likelihood score.
16 6 Computational Genomics c Tel Aviv Univ A Basic SCFG for RNA Structure Consider an SCFG with 7 state types, as shown in figure (0.5): Figure 0.5: A basic SCFG that describes RNA folding behavior[4]. It can efficiently describe the typical behavior of RNA folding: P states emit base paired nucleotides (stems). L, R states emit unpaired nucleotides (single strands, loop segments, bulge, dangling end). B states allow for bifurcations (junctions). S states represent the start of a new structure, at the root of the tree or immediately following a bifurcation. D states allow for deletions with respect to the consensus. E states allow for the termination of a structure (note that emitting states only allow us to extend a structure) Note, however, that these state types are generic and not position specific. While this grammar represents the probabilities of transitions from one state to the other, it does not allow for different behaviors at different positions along the consensus, e.g. position 5 is more likely to be part of a stem, position 4 is more likely to contain a purine, etc.
17 Covariance Models A Simple Example Here is a simple SCFG which demonstrates how, by assigning uniform probabilities, we can favor a parse tree which maximizes base pairing. In effect, this simple grammar imitates Nussinov s algorithm. G = (V G, Σ, S, R G, P G ) V G = {S, P, L, R, B, E} Σ = {a, c, g, u} S P L R B E P asu usa csg gsc L as cs gs us R G = R Sa Sc Sg Su B SS E ɛ For each non-terminal, all productions have equal probabilities. Specifically, all emitting productions have probability. Lets examine two possible parses of the string cacgug : 4. S cacgug : S P csg cp g casug cap ug cacsgug cacegug cacgug This derivation has the probability: = = S 2 cacgug : S L cs cr csg clg casg carg casug calug cacsug cacrug cacsgug cacegug cacgug This derivation has the probability: = = The first parse is significantly more likely. All emitting productions have the general form S X wsz (where X V G and w, z Σ {ɛ}) and therefore a probability of. For 20 this reason, the most probable parse contains a minimal number of productions. The pair emitting state is thus preferred over the left/right emitting states, whenever possible. Hence the most probable parse yields maximal base pairing. Note: This is not entirely accurate, since this model puts a probability penalty on branching. The branching itself decreases the probability of the parse by a factor of 5 (S B SS). Terminating the new branch further decreases the probability by a factor of 5 (S E ɛ). Nussinov s algorithm has no such penalty Extending the Basic Grammar To allow for position specific behavior, each production along the parse tree generating the consensus is assigned a unique non-terminal. These non-terminals are called nodes. Eight
18 8 Computational Genomics c Tel Aviv Univ. node types are defined, in accordance with the basic state types: Figure 0.6: Eight different node types allow for position specific behavior[4]. Each node is given a (unique) name, comprised of a node type and a serial number, e.g. MATL 2, signifying that the 2nd production along the parse tree should be a left emission. When constructing the parse tree, we prefer MATL nodes over MATR nodes. This is rather an arbitrary choice, meant to reduce ambiguity, similar to our choice of leftmost derivations. Figure 0.7 illustrates a parse tree corresponding with a given consensus structure. Note that this kind of parse tree cannot derive sequences deviating from the consensus it contains no insertion/deletion nodes. Figure 0.7: Creating a guide (parse) tree for a consensus structure[4].
19 Covariance Models The Final CM In order to account for deviations from the consensus, each node is expanded into several states, divided into 2 sets: A split set Represents the main consensus state. A parse tree must visit exactly one state from each split set. The split set allows for deletions: a split state may emit less symbols (terminals) than indicated by the node type. An insert set Allows for insertions with respect to the consensus. A parse tree may pass through the insert states zero or more times. Figure 0.8 details the translation of node types to split and insert sets. Figure 0.8: Split and insert sets corresponding with different node types. The split set is written in square brackets[4]. Different split states signify the emission/deletion of consensus symbols. For example, a pair emitting node of type MATP may emit both symbols (MP), only the left symbol (ML), only the right symbol (MR), or none of the two (D). Only emitting nodes (MATP, MATL, MATR) may have deletions. Non-emitting nodes cannot have deletions and so their split sets are singletons. Transitions: A split state can transit to an insert state in the same node or a split state in the next node. An IL state can transit to itself, an IR in the same node or a split state in the next node. An IR can transit to itself or to a split state in the next node. Figure 0.9 depicts the resulting CM.
20 20 Computational Genomics c Tel Aviv Univ. Figure 0.9: Expanding the nodes to create the final CM. On the right: CM created by expanding the guide tree in figure 0.7. On the left: An enlarged segment of the CM, depicting the states and transitions of nodes MATP 6, MATP 7, MATR 8[4]. 0.0 Specialized Inside/Outside Algorithms for CMs Unlike CNF grammars, CMs follow a strict form. Specialized versions of the inside and outside algorithms can be made to utilize that strictness of form to compute posterior probabilities P (X ij x, G) more efficiently. Notations for this section: A CM has n states (non-terminals) W, W 2,..., W m. Each state pertains to one of the 7 state types defined earlier. W is the (root) start state for the whole CM. There may be several end states as a CM usually represents a multi-branched structure, each branch (e.g. stem-loop) having its own end state. s v {P, L, R, D, S, B, E} denotes the type of state v.
21 Specialized Inside/Outside Algorithms for CMs 2 End states can generate the null sequence ɛ. In fact, all parses end with the production W v ɛ for some v, s v = E. It is therefore necessary to assign a probability to the generation of the null sequence. Let x[j +, j] = ɛ denote the null subsequence, preceding the terminal x j. L v, R v W v. {0, } denote the number of terminals emitted to the left and right by state t v (y) denotes the probability of transition P (W v W y ). e v (x i, x j ) denotes the probability of emitting the terminal x i to the left and x j to the right, by state v. This notation will be used for all emission probabilities (regardless of whether x i, x j are emitted or not): for P states e(x i, x j ) = e(x i )e(x j ) for L states e(x i, x j ) = e(x i ) for R states e(x i, x j ) = e(x j ) for non-emitting states e(x i, x j ) = C v denotes the children of state W v. C v is an ordered list of indices y for the states W y that W v can make a transition to. P v denotes the children of state W v. P v is an ordered list of indices y for the states W y that can make a transition to W v. θ denotes the model s parameters the probabilities corresponding with each production and emission. Figure 0.0: Properties of all state types according to our notation[4].
22 22 Computational Genomics c Tel Aviv Univ. Figure 0.0 summarizes the properties of all 7 state types. Lets examine some of the qualities of CMs:. A state has 6 productions at the most A split state can transition to insert states in the same node (at most 2), or to split states in the next node (at most 4). A general SCFG state may have as many as O(m 2 ) productions. We find that in CMs, although the number of states is m = O(n), each state has only a constant number of productions. 2. B states have only one production A bifurcation (BIF) node always transits to 2 start nodes (BEGL, BEGR). All three node types are non-emitting, have singleton split sets, and no insert sets. So we get a single production (with probability ) W v W y W z, where s v = B, s y = S, s z = S. 3. A single branching production The B production is the only one to produce two non-terminals. This is important, since such productions are computationally difficult. Generating the subsequence x[i, j] from W v using the branching production W v W y W z is described by this derivation: W v W y W z x i..x k W z x i..x k x k+..x j We have to consider all partitions x[i, k],x[k +, j] such that W y x[i, k] and W z x[k +, j]. This may take O(n) operations. 4. Ordered states Transitions are only possible from a node to itself or the next. This dictates that y > v for all indices y C v, or y v for insert states. This prevents futile cycles composed of non-emitting states Inside Algorithm for CMs The specialized inside algorithm computes α(i, j, W v ) differently for each state type s v. Non-emitting states may generate the null sequence. It is therefore necessary to calculate the probability of deriving sequences of length 0, α(j +, j, W v ). Generating the null sequence ɛ: s v = E: An end state can only generate a null sequence, so P (W v ɛ s v = E) = s v S, D: Start and deletion states cannot generate a null sequence directly, but may, through non-emitting productions, derive an end state. Assume y C v and
23 Specialized Inside/Outside Algorithms for CMs 23 s z = E (possibly y = z), the derivation is then: W v W y W z ɛ The probability of this event is: P (W v W y, W y ɛ θ) = P (W v W y )P (W y ɛ) = t v (y)α(j +, j, W y ) (0.29) Summing over all possible y C v we get: α(j +, j, W v ) = t v (y)α(j +, j, W y ) (0.30) y C v s v = B: A bifurcation state may derive two empty structures through this derivation: W v W y W z ɛw z ɛɛ Since P (W v W y W z ) =, the probability of this event is: P (W v ɛ) = α(j +, j, W y )α(j +, j, W z ) (0.3) s v P, L, R: Emitting states cannot generate a null sequence, so P (W v ɛ s v {P, L, R}) = Pseudocode for the Specialized Inside Algorithm. Initialization: for j = 0 up to n do for v = m down to do s v = E : s v = S, D : t v (y)α(j +, j, W y ) α(j +, j, W v ) = y C v s v = B : α(j +, j, W y )α(j +, j, W z ) s v = P, L, R : 0 2. for i = n down to do for j = i up to n do
24 24 Computational Genomics c Tel Aviv Univ. for v = m down to do s v = E : 0 s v = P, j = i : 0 j α(i, j, W v ) = s v = B : α(i, k, W y )α(k +, j, W z ) k=i otherwise : e v (x i, x j ) t v (y)α(i + L v, j R v, W y ) y C v Complexity: For each of the O(n 2 ) cells of the DP matrix we have to compute α(i, j, W v ) for all v m. This takes O(n) time if s v = B and O() otherwise (since C v 6). Suppose the CM has b bifurcation states and a other states (a + b = m), then total time complexity is O(an 2 + bn 3 ). Space complexity is O(mn 2 ) Outside algorithm for CMs The specialized outside algorithm seems to be simpler than the general one, since most productions have a single child (non-terminal). Actually, this is true for all productions save one the branching production. Branching always yields two S type states, so these require special attention. Outside computation requires values claculated by the inside algorithm, unlike the general version, the specialized outside only requires α(i, j, W z ) for s z = S. Computing β(i, j, W v ) : s v P, L, R, B, D, E : Assume W v is generated by the pair emitting production W y x i W v x j+ for some y P v, s y = P. The event W x..x i W v x j+ corresponds with the derivation: W x..x i 2 W y x j+2..x n x..x i 2 x i W v x j+ x j+2..x n The probability of this event is: P (x..x i W v x j+..x n θ) = P (x..x i 2 W y x j+2..x n )P (W y x i W v x j+ ) (0.32) = β(i, j +, W y )t y (v)e y (x i, x j+ ) (0.33) In the general case (without asserting s v = P ) x i, x j+ may or may not have been emited by W y, depending on L y, R y. we then get: P (x..x i W v x j+..x n θ) = β(i L y, j + R y, W y )t y (v)e y (x i L y, x j+ R y ) (0.34) Summing over all possible y P v we get: β(i, j, W v ) = y P v β(i L y, j + R y, W y )t y (v)e y (x i L y, x j+ R y ) (0.35)
25 Specialized Inside/Outside Algorithms for CMs 25 s v = S, W v is the root: The root W has β(, n, W ) = and β(i, j, W ) = 0 for all other values of (i, j). s v = S, W v is a left child: A left child is generated by the branching production W y W v W z where s y = B, s z = S and W y has C y = {v, z}. The event W x..x i W v x j+ corresponds with the derivation: W x..x i W y x k+..x n x..x i W v W z x k+..x n x..x i W v x j+..x k x k+..x n for some j k n. The probability of this event is: P (x..x i W v x j+..x n θ) = (0.36) = P (x..x i W y x k+..x n )P (W y W v W z )P (W z x j+..x k ) (0.37) = β(i, k, W y ) α(j +, k, W z ) (0.38) The above derivation is not leftmost, yet the order of derivation does not affect the probability of events. Summing over all possible j k n we get: β(i, j, W v ) = n β(i, k, W y )α(j +, k, W z ) (0.39) k=j The first term of the sum (k = j) represents a case where W z generates a null sequence. s v = S, W v is a right child: A right child is generated by the branching production W y W z W v where s y = B, s z = S and W y has C y = {z, v}. Following the guidelines of the previous case we get: β(i, j, W v ) = i β(k, j, W y )α(k, i, W z ) (0.40) k= The last term of the sum (k = i) represents a case where W z generates a null sequence Pseudocode for the specialized outside algorithm. Initialization: β(, n, W ) =
26 26 Computational Genomics c Tel Aviv Univ. 2. for i = up to n + do for j = n down to i do for v = 2 up to m do for s v = S, P v = y, C y = {v, z} : n β(i, k, W y )α(j +, k, W z ) k=j for s v = S, P v = y, C y = {v, z} : β(i, j, W v ) = i β(k, j, W y )α(k, i, W z ) k= for s v P, L, R, B, D, E : β(i L y, j + R y, W y )t y (v)e(x i L y, x j+ R y ) y P v Complexity: Time and space complexity for the specialized outside algorithm are identical to those of the specialized inside algorithm. NOTE: For a specialized version of the EM algorithm, refer to Biological Sequence Analysis, Durbin et al. (Cambridge, 98). 0. Creating a Consensus Structure From an MSA In some cases, the consensus structure of a family of RNA sequences is unknown. For the purpose of creating a CM, we must first construct a consensus structure for the family (or training set). We assume that an MSA of all input sequences is given. Later we will show that any random MSA will suffice. Notations for this section: A denotes an MSA of m RNA sequences x, x 2... x m. We denote n = A. We assume that x... x m are gapped according to the MSA. A column of the MSA is considered an insertion if it contains > 50% gaps (this is a common heuristic). A i denotes the i th column of A. A[i, j] denotes columns i through j of A. P (x i = c) denotes the (empirical) probability of a sequence x A having the character c at position i. This can also be described as the relative frequency of the character c in column A i.
27 Creating a Consensus Structure From an MSA 27 A consensus structure of A is defined by a set C of index pairs (i, j), such that i < j n. A pair (i, j) C signifies that columns A i, A j are base paired. Since any base can only pair with one other base, an index i may only appear once in C. 0.. Mutual Information The mutual information of two random variables is a quantity that measures the mutual dependence of the two variables. Formally, the mutual information I(X; Y ) of two discrete random variables X and Y is defined as follows: I(X; Y ) = y Y P (x, y) P (x, y) log P (x)p 2 (y) x X (0.4) where P (x) P (X = x), P 2 (y) P (Y = y), P (x, y) P (X = x, Y = y). Examples of maximal and minimal I(X, Y ): Assume X and Y are independent, then P (x, y) = P (x)p 2 (y) for all x X, y Y. We get: I(X; Y ) = P (x, y) P (x, y) log (0.42) P (x)p 2 (y) x X y Y = P (x, y) log P (x)p 2 (y) (0.43) P (x)p 2 (y) x X y Y = P (x, y) log = 0 (0.44) x X y Y Assume X can have n possible outcomes {a... a n }, each occuring with equal probability n, Y has n outcomes {b... b n }. Assume that X, Y are completely co-dependent, meaning that for all i n, a i occurs iff b i occurs. We get: I(X; Y ) = = = n n P (a i, b j ) P (a i, b j ) log P j= (a i )P 2 (b j ) (0.45) n P (a i, b i ) P (a i, b i ) log P (a i )P 2 (b i ) (0.46) n n n log n = log n = log n n (0.47) i= i= i= n n i=
28 28 Computational Genomics c Tel Aviv Univ Mutual Information of MSA Columns In our context, we use the mutual information bit-score to determine whether two MSA columns should base pair or not. This is reasonable since base-paired columns should show a significant correlation. Adapting the general formula to our context, let M i,j denote the mutual information of columns i and j of an MSA: M i,j = Notes and observations: a,b={a,c,g,u} P (x i = a, x j = b) log 2 P (x i = a, x j = b) P (x i = a)p (x j = b) x i has 4 possible outcomes {A, C, G, T } so 0 M i,j 2 (0.48) For some sequence x k in the MSA, the character x k i or x k j (or both) may be deleted. In that case, the probability P (x k i = a, x k j = b) is undefined. Such sequences are excluded when computing M i,j. Two degenerate (constant) columns i, j have M i,j = 0. Assume k.x k i = A, k.x k j = G for example. Then the sum has a single term: M i,j = P (x i = A, x j = G ) log 2 P (x i = A, x j = G ) P (x i = A )P (x j = G ) = log 2 = 0 = 0 (0.49) 0..3 Finding Consensus Structure with Maximal M i,j We wish to find a consensus structure C which maximizes (i,j) C M i,j (the sum of mutual information values over all paired columns). To accomplish this we use a DP algorithm, similar to Nussinov s maximal base-pairing algorithm; We build a DP matrix S, such that S i,j holds the optimal (in terms of maximum M i,j ) structure for A[i, j]. General flow of the algorithm:. Initialize all cells S i,i (the leaves) to zero. 2. The matrix is then filled from leaves to root. Computing cell S i,j, we choose the best of four options: (a) Add unpaired column i at the beginning of the optimal structure for A[i +, j]. (b) Add unpaired column j at the end of the optimal structure for A[i, j ]. (c) Surround the optimal structure for A[i +, j ] with base paired columns i, j. This increases the structure s score by M i,j.
29 Creating a Consensus Structure From an MSA 29 (d) Concatenate two smaller structures A[i, k], A[k +, j] for some i < k < j. S i,j then contains the sum of the two scores. 3. When the matrix has been filled, S,n holds the optimal score for the complete MSA. 4. Tracing back the path yielding maximal score, we find the optimal structure for A. The recursion for S i,j : S i,j = max { } S i+,j, S i,j, S i+,j + M i,j, max {S i,k + S k+,j } i<k<j (0.50) Complexity: for each of the O(n 2 ) cells of the DP matrix we check all O(n) partitions. Therefore, the total time complexity is O(n 3 ). Each cell holds a single value, total space complexity is therefore O(n 2 ). Figure 0.: The covariance model training algorithm[3] 0..4 Working Without an MSA CMs provide a powerful tool for the modeling of RNA folding. The chink in the armor, so to speak, is having to rely on an initial MSA to construct the CM. MSAs are generic and do not factor the specific behavior of RNA folding in any way. We introduce an iterative approach that allows us to construct a CM from a random MSA (sequences are gapped randomly):. A consensus structure is derived from the (initially random) MSA.
30 30 Computational Genomics c Tel Aviv Univ. 2. The model s parameters are optimized by EM. 3. The CM is used to align each of the sequences to the consensus a new MSA is derived. 4. The process is repeated until convergence. It should be noted, that although the quality of the CM is retained when using a random MSA, time to convergence increases significantly (when compared to an initial MSA of good quality). 0.2 Application of CMs trna Modeling trnas provide an ideal test case for CM. It is the largest gene family in most genomes, sequences exhibit significant variation while secondary structure is widely conserved. Durbin and Eddy created two covariance models for a data set of 45 aligned trna sequences. 00 trna sequences were randomly selected to serve as a test set, an additional group of a 00 sequences was randomly chosen to serve as a training set. Two training modes were used: Mode A CM was trained using a trusted alignment of the training set, incorporating data obtained from X-ray crystalography. Mode U CM was trained using a random alignment of the training set. The A model converged rapidly and had an average bit score 2 of The U model showed a similar score of 56.7 bits, yet took greater time to converge. The results are given in the following table: Training Mode # of Iterations Bit Score Accuracy A % U % An alignment produced by an HMM trained using the same data yields 30 bits of information, about half the information achieved by the CM alignments. Note that both models achieve at least 90% accuracy 3. It should be stated that a degapped alignment of the sequences (where sequences are simply placed one on top of the other) yields only 30% accuracy. 2 P (x θ) The information bit score is defined as log 2 P (r θ), where P (r θ) is the average probability of a random sequence r with r = x being generated by the model. Intuitively, information bit means that a sequence x is twice as likely to be generated by the model than a random sequence of equal length. 3 Accuracy is defined as the percent of symbol pairs aligned according to the CM, that are also aligned according to the trusted alignment.
31 Application of CMs trna Modeling 3 Figure 0.2: trna secondary structure. Next, the A45 model (trained using all 45 aligned sequences) was used to classify a test set consisting of sequences from GenBank structural RNA database, and C. elegans genomic sequence (6.3 Mb total). Here is a partial list of the results: As seen on figure (0.3), Any cutoff score between.7 and 25.9 cleanly separates all the non-trnas from the 547 cytoplasmic trnas, giving a sensitivity of over 99.98%. All 4 C. elegans trnas were detected with a score greater than 3 (00% sensitivity). 26 out of 522 annotated trnas from GenBank were missed (95% sensitivity). All 26 missed trnas were mitochondrial, 22 of them completely lack the D-loop. Of all annotated non-trna molecules, none were mis-classified as trnas (00% specificity). In conclusion, Covariance Models supply an elegant probabilistic solution for predicting RNA secondary structure, showing remarkable results.
32 32 Computational Genomics c Tel Aviv Univ. Figure 0.3: Number of hits versus score in bits, using the A45 model to search sequences from Genbank and C.elegans genomic sequence. Non-tRNA hits are shown in white. Hits to cytoplasmic trnas are shown in black. Other trna hits, in gray, are the scores of 868 mitochondrial, chloroplast, viral, and selenocysteine trnas. Arrows indicate the gap between the highest non-trna hit (with two trna-related exceptions) and the lowest scoring cytoplasmic trna.[3]
33 Bibliography [] J. Baker. Trainable grammars for speech recognition. Speech communication papers presented at the 97th Meeting of the Acoustical Society of America, 979. [2] John Cocke and Jacob T. Schwartz. Programming languages and their compilers: Preliminary notes. Technical report, Courant Institute of Mathematical Sciences, New York University, 970. [3] Sean R. Eddy and Richard Durbin. Rna sequence analysis using covariance models. Computational Biology, 994. [4] R. Durbin et al. Biological sequence analysis. Cambridge University Press, 998. [5] Tadao Kasami. An efficient recognition and syntax-analysis algorithm for context-free languages. Scientific report AFCRL , 965. [6] Ruth Nussinov and Ann B. Jacobson. Fast algorithm for predicting the secondary structure of single-stranded rna. PNAS, 980. [7] Daniel H. Younger. Recognition and parsing of context-free languages in time n 3. Information and Control,
CS681: Advanced Topics in Computational Biology
 CS681: Advanced Topics in Computational Biology Can Alkan EA224 calkan@cs.bilkent.edu.tr Week 10 Lecture 1 http://www.cs.bilkent.edu.tr/~calkan/teaching/cs681/ RNA folding Prediction of secondary structure
CS681: Advanced Topics in Computational Biology Can Alkan EA224 calkan@cs.bilkent.edu.tr Week 10 Lecture 1 http://www.cs.bilkent.edu.tr/~calkan/teaching/cs681/ RNA folding Prediction of secondary structure
RNA Basics. RNA bases A,C,G,U Canonical Base Pairs A-U G-C G-U. Bases can only pair with one other base. wobble pairing. 23 Hydrogen Bonds more stable
 RNA STRUCTURE RNA Basics RNA bases A,C,G,U Canonical Base Pairs A-U G-C G-U wobble pairing Bases can only pair with one other base. 23 Hydrogen Bonds more stable RNA Basics transfer RNA (trna) messenger
RNA STRUCTURE RNA Basics RNA bases A,C,G,U Canonical Base Pairs A-U G-C G-U wobble pairing Bases can only pair with one other base. 23 Hydrogen Bonds more stable RNA Basics transfer RNA (trna) messenger
RNA Secondary Structure Prediction
 RNA Secondary Structure Prediction 1 RNA structure prediction methods Base-Pair Maximization Context-Free Grammar Parsing. Free Energy Methods Covariance Models 2 The Nussinov-Jacobson Algorithm q = 9
RNA Secondary Structure Prediction 1 RNA structure prediction methods Base-Pair Maximization Context-Free Grammar Parsing. Free Energy Methods Covariance Models 2 The Nussinov-Jacobson Algorithm q = 9
RNA Search and! Motif Discovery" Genome 541! Intro to Computational! Molecular Biology"
 RNA Search and! Motif Discovery" Genome 541! Intro to Computational! Molecular Biology" Day 1" Many biologically interesting roles for RNA" RNA secondary structure prediction" 3 4 Approaches to Structure
RNA Search and! Motif Discovery" Genome 541! Intro to Computational! Molecular Biology" Day 1" Many biologically interesting roles for RNA" RNA secondary structure prediction" 3 4 Approaches to Structure
Properties of Context-Free Languages
 Properties of Context-Free Languages Seungjin Choi Department of Computer Science and Engineering Pohang University of Science and Technology 77 Cheongam-ro, Nam-gu, Pohang 37673, Korea seungjin@postech.ac.kr
Properties of Context-Free Languages Seungjin Choi Department of Computer Science and Engineering Pohang University of Science and Technology 77 Cheongam-ro, Nam-gu, Pohang 37673, Korea seungjin@postech.ac.kr
CONTEXT-SENSITIVE HIDDEN MARKOV MODELS FOR MODELING LONG-RANGE DEPENDENCIES IN SYMBOL SEQUENCES
 CONTEXT-SENSITIVE HIDDEN MARKOV MODELS FOR MODELING LONG-RANGE DEPENDENCIES IN SYMBOL SEQUENCES Byung-Jun Yoon, Student Member, IEEE, and P. P. Vaidyanathan*, Fellow, IEEE January 8, 2006 Affiliation:
CONTEXT-SENSITIVE HIDDEN MARKOV MODELS FOR MODELING LONG-RANGE DEPENDENCIES IN SYMBOL SEQUENCES Byung-Jun Yoon, Student Member, IEEE, and P. P. Vaidyanathan*, Fellow, IEEE January 8, 2006 Affiliation:
Finite Automata Theory and Formal Languages TMV027/DIT321 LP4 2018
 Finite Automata Theory and Formal Languages TMV027/DIT321 LP4 2018 Lecture 14 Ana Bove May 14th 2018 Recap: Context-free Grammars Simplification of grammars: Elimination of ǫ-productions; Elimination of
Finite Automata Theory and Formal Languages TMV027/DIT321 LP4 2018 Lecture 14 Ana Bove May 14th 2018 Recap: Context-free Grammars Simplification of grammars: Elimination of ǫ-productions; Elimination of
Foundations of Informatics: a Bridging Course
 Foundations of Informatics: a Bridging Course Week 3: Formal Languages and Semantics Thomas Noll Lehrstuhl für Informatik 2 RWTH Aachen University noll@cs.rwth-aachen.de http://www.b-it-center.de/wob/en/view/class211_id948.html
Foundations of Informatics: a Bridging Course Week 3: Formal Languages and Semantics Thomas Noll Lehrstuhl für Informatik 2 RWTH Aachen University noll@cs.rwth-aachen.de http://www.b-it-center.de/wob/en/view/class211_id948.html
An Introduction to Bioinformatics Algorithms Hidden Markov Models
 Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Non-context-Free Languages. CS215, Lecture 5 c
 Non-context-Free Languages CS215, Lecture 5 c 2007 1 The Pumping Lemma Theorem. (Pumping Lemma) Let be context-free. There exists a positive integer divided into five pieces, Proof for for each, and..
Non-context-Free Languages CS215, Lecture 5 c 2007 1 The Pumping Lemma Theorem. (Pumping Lemma) Let be context-free. There exists a positive integer divided into five pieces, Proof for for each, and..
On the Sizes of Decision Diagrams Representing the Set of All Parse Trees of a Context-free Grammar
 Proceedings of Machine Learning Research vol 73:153-164, 2017 AMBN 2017 On the Sizes of Decision Diagrams Representing the Set of All Parse Trees of a Context-free Grammar Kei Amii Kyoto University Kyoto
Proceedings of Machine Learning Research vol 73:153-164, 2017 AMBN 2017 On the Sizes of Decision Diagrams Representing the Set of All Parse Trees of a Context-free Grammar Kei Amii Kyoto University Kyoto
98 Algorithms in Bioinformatics I, WS 06, ZBIT, D. Huson, December 6, 2006
 98 Algorithms in Bioinformatics I, WS 06, ZBIT, D. Huson, December 6, 2006 8.3.1 Simple energy minimization Maximizing the number of base pairs as described above does not lead to good structure predictions.
98 Algorithms in Bioinformatics I, WS 06, ZBIT, D. Huson, December 6, 2006 8.3.1 Simple energy minimization Maximizing the number of base pairs as described above does not lead to good structure predictions.
CSCI Compiler Construction
 CSCI 742 - Compiler Construction Lecture 12 Cocke-Younger-Kasami (CYK) Algorithm Instructor: Hossein Hojjat February 20, 2017 Recap: Chomsky Normal Form (CNF) A CFG is in Chomsky Normal Form if each rule
CSCI 742 - Compiler Construction Lecture 12 Cocke-Younger-Kasami (CYK) Algorithm Instructor: Hossein Hojjat February 20, 2017 Recap: Chomsky Normal Form (CNF) A CFG is in Chomsky Normal Form if each rule
Algorithms in Bioinformatics
 Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri RNA Structure Prediction Secondary
Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri RNA Structure Prediction Secondary
CYK Algorithm for Parsing General Context-Free Grammars
 CYK Algorithm for Parsing General Context-Free Grammars Why Parse General Grammars Can be difficult or impossible to make grammar unambiguous thus LL(k) and LR(k) methods cannot work, for such ambiguous
CYK Algorithm for Parsing General Context-Free Grammars Why Parse General Grammars Can be difficult or impossible to make grammar unambiguous thus LL(k) and LR(k) methods cannot work, for such ambiguous
Hidden Markov Models
 Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Hidden Markov Models Outline 1. CG-Islands 2. The Fair Bet Casino 3. Hidden Markov Model 4. Decoding Algorithm 5. Forward-Backward Algorithm 6. Profile HMMs 7. HMM Parameter Estimation 8. Viterbi Training
Even More on Dynamic Programming
 Algorithms & Models of Computation CS/ECE 374, Fall 2017 Even More on Dynamic Programming Lecture 15 Thursday, October 19, 2017 Sariel Har-Peled (UIUC) CS374 1 Fall 2017 1 / 26 Part I Longest Common Subsequence
Algorithms & Models of Computation CS/ECE 374, Fall 2017 Even More on Dynamic Programming Lecture 15 Thursday, October 19, 2017 Sariel Har-Peled (UIUC) CS374 1 Fall 2017 1 / 26 Part I Longest Common Subsequence
Hidden Markov Models. Main source: Durbin et al., Biological Sequence Alignment (Cambridge, 98)
 Hidden Markov Models Main source: Durbin et al., Biological Sequence Alignment (Cambridge, 98) 1 The occasionally dishonest casino A P A (1) = P A (2) = = 1/6 P A->B = P B->A = 1/10 B P B (1)=0.1... P
Hidden Markov Models Main source: Durbin et al., Biological Sequence Alignment (Cambridge, 98) 1 The occasionally dishonest casino A P A (1) = P A (2) = = 1/6 P A->B = P B->A = 1/10 B P B (1)=0.1... P
order is number of previous outputs
 Markov Models Lecture : Markov and Hidden Markov Models PSfrag Use past replacements as state. Next output depends on previous output(s): y t = f[y t, y t,...] order is number of previous outputs y t y
Markov Models Lecture : Markov and Hidden Markov Models PSfrag Use past replacements as state. Next output depends on previous output(s): y t = f[y t, y t,...] order is number of previous outputs y t y
To make a grammar probabilistic, we need to assign a probability to each context-free rewrite
 Notes on the Inside-Outside Algorithm To make a grammar probabilistic, we need to assign a probability to each context-free rewrite rule. But how should these probabilities be chosen? It is natural to
Notes on the Inside-Outside Algorithm To make a grammar probabilistic, we need to assign a probability to each context-free rewrite rule. But how should these probabilities be chosen? It is natural to
This lecture covers Chapter 7 of HMU: Properties of CFLs
 This lecture covers Chapter 7 of HMU: Properties of CFLs Chomsky Normal Form Pumping Lemma for CFs Closure Properties of CFLs Decision Properties of CFLs Additional Reading: Chapter 7 of HMU. Chomsky Normal
This lecture covers Chapter 7 of HMU: Properties of CFLs Chomsky Normal Form Pumping Lemma for CFs Closure Properties of CFLs Decision Properties of CFLs Additional Reading: Chapter 7 of HMU. Chomsky Normal
Today s Lecture: HMMs
 Today s Lecture: HMMs Definitions Examples Probability calculations WDAG Dynamic programming algorithms: Forward Viterbi Parameter estimation Viterbi training 1 Hidden Markov Models Probability models
Today s Lecture: HMMs Definitions Examples Probability calculations WDAG Dynamic programming algorithms: Forward Viterbi Parameter estimation Viterbi training 1 Hidden Markov Models Probability models
Context Free Grammars
 Automata and Formal Languages Context Free Grammars Sipser pages 101-111 Lecture 11 Tim Sheard 1 Formal Languages 1. Context free languages provide a convenient notation for recursive description of languages.
Automata and Formal Languages Context Free Grammars Sipser pages 101-111 Lecture 11 Tim Sheard 1 Formal Languages 1. Context free languages provide a convenient notation for recursive description of languages.
Page 1. References. Hidden Markov models and multiple sequence alignment. Markov chains. Probability review. Example. Markovian sequence
 Page Hidden Markov models and multiple sequence alignment Russ B Altman BMI 4 CS 74 Some slides borrowed from Scott C Schmidler (BMI graduate student) References Bioinformatics Classic: Krogh et al (994)
Page Hidden Markov models and multiple sequence alignment Russ B Altman BMI 4 CS 74 Some slides borrowed from Scott C Schmidler (BMI graduate student) References Bioinformatics Classic: Krogh et al (994)
THEORY OF COMPUTATION (AUBER) EXAM CRIB SHEET
 THEORY OF COMPUTATION (AUBER) EXAM CRIB SHEET Regular Languages and FA A language is a set of strings over a finite alphabet Σ. All languages are finite or countably infinite. The set of all languages
THEORY OF COMPUTATION (AUBER) EXAM CRIB SHEET Regular Languages and FA A language is a set of strings over a finite alphabet Σ. All languages are finite or countably infinite. The set of all languages
Improved TBL algorithm for learning context-free grammar
 Proceedings of the International Multiconference on ISSN 1896-7094 Computer Science and Information Technology, pp. 267 274 2007 PIPS Improved TBL algorithm for learning context-free grammar Marcin Jaworski
Proceedings of the International Multiconference on ISSN 1896-7094 Computer Science and Information Technology, pp. 267 274 2007 PIPS Improved TBL algorithm for learning context-free grammar Marcin Jaworski
Follow sets. LL(1) Parsing Table
 Follow sets. LL(1) Parsing Table Exercise Introducing Follow Sets Compute nullable, first for this grammar: stmtlist ::= ε stmt stmtlist stmt ::= assign block assign ::= ID = ID ; block ::= beginof ID
Follow sets. LL(1) Parsing Table Exercise Introducing Follow Sets Compute nullable, first for this grammar: stmtlist ::= ε stmt stmtlist stmt ::= assign block assign ::= ID = ID ; block ::= beginof ID
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 11 Project
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 11 Project
Stephen Scott.
 1 / 27 sscott@cse.unl.edu 2 / 27 Useful for modeling/making predictions on sequential data E.g., biological sequences, text, series of sounds/spoken words Will return to graphical models that are generative
1 / 27 sscott@cse.unl.edu 2 / 27 Useful for modeling/making predictions on sequential data E.g., biological sequences, text, series of sounds/spoken words Will return to graphical models that are generative
Introduction to Theory of Computing
 CSCI 2670, Fall 2012 Introduction to Theory of Computing Department of Computer Science University of Georgia Athens, GA 30602 Instructor: Liming Cai www.cs.uga.edu/ cai 0 Lecture Note 3 Context-Free Languages
CSCI 2670, Fall 2012 Introduction to Theory of Computing Department of Computer Science University of Georgia Athens, GA 30602 Instructor: Liming Cai www.cs.uga.edu/ cai 0 Lecture Note 3 Context-Free Languages
Hidden Markov Models
 Andrea Passerini passerini@disi.unitn.it Statistical relational learning The aim Modeling temporal sequences Model signals which vary over time (e.g. speech) Two alternatives: deterministic models directly
Andrea Passerini passerini@disi.unitn.it Statistical relational learning The aim Modeling temporal sequences Model signals which vary over time (e.g. speech) Two alternatives: deterministic models directly
FORMAL LANGUAGES, AUTOMATA AND COMPUTABILITY
 15-453 FORMAL LANGUAGES, AUTOMATA AND COMPUTABILITY Chomsky Normal Form and TURING MACHINES TUESDAY Feb 4 CHOMSKY NORMAL FORM A context-free grammar is in Chomsky normal form if every rule is of the form:
15-453 FORMAL LANGUAGES, AUTOMATA AND COMPUTABILITY Chomsky Normal Form and TURING MACHINES TUESDAY Feb 4 CHOMSKY NORMAL FORM A context-free grammar is in Chomsky normal form if every rule is of the form:
CSCE 471/871 Lecture 3: Markov Chains and
 and and 1 / 26 sscott@cse.unl.edu 2 / 26 Outline and chains models (s) Formal definition Finding most probable state path (Viterbi algorithm) Forward and backward algorithms State sequence known State
and and 1 / 26 sscott@cse.unl.edu 2 / 26 Outline and chains models (s) Formal definition Finding most probable state path (Viterbi algorithm) Forward and backward algorithms State sequence known State
Doctoral Course in Speech Recognition. May 2007 Kjell Elenius
 Doctoral Course in Speech Recognition May 2007 Kjell Elenius CHAPTER 12 BASIC SEARCH ALGORITHMS State-based search paradigm Triplet S, O, G S, set of initial states O, set of operators applied on a state
Doctoral Course in Speech Recognition May 2007 Kjell Elenius CHAPTER 12 BASIC SEARCH ALGORITHMS State-based search paradigm Triplet S, O, G S, set of initial states O, set of operators applied on a state
Multiple Sequence Alignment using Profile HMM
 Multiple Sequence Alignment using Profile HMM. based on Chapter 5 and Section 6.5 from Biological Sequence Analysis by R. Durbin et al., 1998 Acknowledgements: M.Sc. students Beatrice Miron, Oana Răţoi,
Multiple Sequence Alignment using Profile HMM. based on Chapter 5 and Section 6.5 from Biological Sequence Analysis by R. Durbin et al., 1998 Acknowledgements: M.Sc. students Beatrice Miron, Oana Răţoi,
Chomsky Normal Form and TURING MACHINES. TUESDAY Feb 4
 Chomsky Normal Form and TURING MACHINES TUESDAY Feb 4 CHOMSKY NORMAL FORM A context-free grammar is in Chomsky normal form if every rule is of the form: A BC A a S ε B and C aren t start variables a is
Chomsky Normal Form and TURING MACHINES TUESDAY Feb 4 CHOMSKY NORMAL FORM A context-free grammar is in Chomsky normal form if every rule is of the form: A BC A a S ε B and C aren t start variables a is
Markov Chains and Hidden Markov Models. COMP 571 Luay Nakhleh, Rice University
 Markov Chains and Hidden Markov Models COMP 571 Luay Nakhleh, Rice University Markov Chains and Hidden Markov Models Modeling the statistical properties of biological sequences and distinguishing regions
Markov Chains and Hidden Markov Models COMP 571 Luay Nakhleh, Rice University Markov Chains and Hidden Markov Models Modeling the statistical properties of biological sequences and distinguishing regions
Processing/Speech, NLP and the Web
 CS460/626 : Natural Language Processing/Speech, NLP and the Web (Lecture 25 Probabilistic Parsing) Pushpak Bhattacharyya CSE Dept., IIT Bombay 14 th March, 2011 Bracketed Structure: Treebank Corpus [ S1[
CS460/626 : Natural Language Processing/Speech, NLP and the Web (Lecture 25 Probabilistic Parsing) Pushpak Bhattacharyya CSE Dept., IIT Bombay 14 th March, 2011 Bracketed Structure: Treebank Corpus [ S1[
STA 414/2104: Machine Learning
 STA 414/2104: Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistics! rsalakhu@cs.toronto.edu! http://www.cs.toronto.edu/~rsalakhu/ Lecture 9 Sequential Data So far
STA 414/2104: Machine Learning Russ Salakhutdinov Department of Computer Science! Department of Statistics! rsalakhu@cs.toronto.edu! http://www.cs.toronto.edu/~rsalakhu/ Lecture 9 Sequential Data So far
CSCE 478/878 Lecture 9: Hidden. Markov. Models. Stephen Scott. Introduction. Outline. Markov. Chains. Hidden Markov Models. CSCE 478/878 Lecture 9:
 Useful for modeling/making predictions on sequential data E.g., biological sequences, text, series of sounds/spoken words Will return to graphical models that are generative sscott@cse.unl.edu 1 / 27 2
Useful for modeling/making predictions on sequential data E.g., biological sequences, text, series of sounds/spoken words Will return to graphical models that are generative sscott@cse.unl.edu 1 / 27 2
MA/CSSE 474 Theory of Computation
 MA/CSSE 474 Theory of Computation CFL Hierarchy CFL Decision Problems Your Questions? Previous class days' material Reading Assignments HW 12 or 13 problems Anything else I have included some slides online
MA/CSSE 474 Theory of Computation CFL Hierarchy CFL Decision Problems Your Questions? Previous class days' material Reading Assignments HW 12 or 13 problems Anything else I have included some slides online
Pair Hidden Markov Models
 Pair Hidden Markov Models Scribe: Rishi Bedi Lecturer: Serafim Batzoglou January 29, 2015 1 Recap of HMMs alphabet: Σ = {b 1,...b M } set of states: Q = {1,..., K} transition probabilities: A = [a ij ]
Pair Hidden Markov Models Scribe: Rishi Bedi Lecturer: Serafim Batzoglou January 29, 2015 1 Recap of HMMs alphabet: Σ = {b 1,...b M } set of states: Q = {1,..., K} transition probabilities: A = [a ij ]
Markov Chains and Hidden Markov Models. = stochastic, generative models
 Markov Chains and Hidden Markov Models = stochastic, generative models (Drawing heavily from Durbin et al., Biological Sequence Analysis) BCH339N Systems Biology / Bioinformatics Spring 2016 Edward Marcotte,
Markov Chains and Hidden Markov Models = stochastic, generative models (Drawing heavily from Durbin et al., Biological Sequence Analysis) BCH339N Systems Biology / Bioinformatics Spring 2016 Edward Marcotte,
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2011 1 HMM Lecture Notes Dannie Durand and Rose Hoberman October 11th 1 Hidden Markov Models In the last few lectures, we have focussed on three problems
Computational Genomics and Molecular Biology, Fall 2011 1 HMM Lecture Notes Dannie Durand and Rose Hoberman October 11th 1 Hidden Markov Models In the last few lectures, we have focussed on three problems
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 07: profile Hidden Markov Model http://bibiserv.techfak.uni-bielefeld.de/sadr2/databasesearch/hmmer/profilehmm.gif Slides adapted from Dr. Shaojie Zhang
EECS730: Introduction to Bioinformatics Lecture 07: profile Hidden Markov Model http://bibiserv.techfak.uni-bielefeld.de/sadr2/databasesearch/hmmer/profilehmm.gif Slides adapted from Dr. Shaojie Zhang
60-354, Theory of Computation Fall Asish Mukhopadhyay School of Computer Science University of Windsor
 60-354, Theory of Computation Fall 2013 Asish Mukhopadhyay School of Computer Science University of Windsor Pushdown Automata (PDA) PDA = ε-nfa + stack Acceptance ε-nfa enters a final state or Stack is
60-354, Theory of Computation Fall 2013 Asish Mukhopadhyay School of Computer Science University of Windsor Pushdown Automata (PDA) PDA = ε-nfa + stack Acceptance ε-nfa enters a final state or Stack is
Expectation Maximization (EM)
 Expectation Maximization (EM) The EM algorithm is used to train models involving latent variables using training data in which the latent variables are not observed (unlabeled data). This is to be contrasted
Expectation Maximization (EM) The EM algorithm is used to train models involving latent variables using training data in which the latent variables are not observed (unlabeled data). This is to be contrasted
Logic. proof and truth syntacs and semantics. Peter Antal
 Logic proof and truth syntacs and semantics Peter Antal antal@mit.bme.hu 10/9/2015 1 Knowledge-based agents Wumpus world Logic in general Syntacs transformational grammars Semantics Truth, meaning, models
Logic proof and truth syntacs and semantics Peter Antal antal@mit.bme.hu 10/9/2015 1 Knowledge-based agents Wumpus world Logic in general Syntacs transformational grammars Semantics Truth, meaning, models
Lecture 4: Hidden Markov Models: An Introduction to Dynamic Decision Making. November 11, 2010
 Hidden Lecture 4: Hidden : An Introduction to Dynamic Decision Making November 11, 2010 Special Meeting 1/26 Markov Model Hidden When a dynamical system is probabilistic it may be determined by the transition
Hidden Lecture 4: Hidden : An Introduction to Dynamic Decision Making November 11, 2010 Special Meeting 1/26 Markov Model Hidden When a dynamical system is probabilistic it may be determined by the transition
CS20a: summary (Oct 24, 2002)
 CS20a: summary (Oct 24, 2002) Context-free languages Grammars G = (V, T, P, S) Pushdown automata N-PDA = CFG D-PDA < CFG Today What languages are context-free? Pumping lemma (similar to pumping lemma for
CS20a: summary (Oct 24, 2002) Context-free languages Grammars G = (V, T, P, S) Pushdown automata N-PDA = CFG D-PDA < CFG Today What languages are context-free? Pumping lemma (similar to pumping lemma for
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2014 1 HMM Lecture Notes Dannie Durand and Rose Hoberman November 6th Introduction In the last few lectures, we have focused on three problems related
Computational Genomics and Molecular Biology, Fall 2014 1 HMM Lecture Notes Dannie Durand and Rose Hoberman November 6th Introduction In the last few lectures, we have focused on three problems related
CS5371 Theory of Computation. Lecture 7: Automata Theory V (CFG, CFL, CNF)
 CS5371 Theory of Computation Lecture 7: Automata Theory V (CFG, CFL, CNF) Announcement Homework 2 will be given soon (before Tue) Due date: Oct 31 (Tue), before class Midterm: Nov 3, (Fri), first hour
CS5371 Theory of Computation Lecture 7: Automata Theory V (CFG, CFL, CNF) Announcement Homework 2 will be given soon (before Tue) Due date: Oct 31 (Tue), before class Midterm: Nov 3, (Fri), first hour
Grammar formalisms Tree Adjoining Grammar: Formal Properties, Parsing. Part I. Formal Properties of TAG. Outline: Formal Properties of TAG
 Grammar formalisms Tree Adjoining Grammar: Formal Properties, Parsing Laura Kallmeyer, Timm Lichte, Wolfgang Maier Universität Tübingen Part I Formal Properties of TAG 16.05.2007 und 21.05.2007 TAG Parsing
Grammar formalisms Tree Adjoining Grammar: Formal Properties, Parsing Laura Kallmeyer, Timm Lichte, Wolfgang Maier Universität Tübingen Part I Formal Properties of TAG 16.05.2007 und 21.05.2007 TAG Parsing
O 3 O 4 O 5. q 3. q 4. Transition
 Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Advanced Natural Language Processing Syntactic Parsing
 Advanced Natural Language Processing Syntactic Parsing Alicia Ageno ageno@cs.upc.edu Universitat Politècnica de Catalunya NLP statistical parsing 1 Parsing Review Statistical Parsing SCFG Inside Algorithm
Advanced Natural Language Processing Syntactic Parsing Alicia Ageno ageno@cs.upc.edu Universitat Politècnica de Catalunya NLP statistical parsing 1 Parsing Review Statistical Parsing SCFG Inside Algorithm
Harvard CS 121 and CSCI E-207 Lecture 12: General Context-Free Recognition
 Harvard CS 121 and CSCI E-207 Lecture 12: General Context-Free Recognition Salil Vadhan October 11, 2012 Reading: Sipser, Section 2.3 and Section 2.1 (material on Chomsky Normal Form). Pumping Lemma for
Harvard CS 121 and CSCI E-207 Lecture 12: General Context-Free Recognition Salil Vadhan October 11, 2012 Reading: Sipser, Section 2.3 and Section 2.1 (material on Chomsky Normal Form). Pumping Lemma for
4 : Exact Inference: Variable Elimination
 10-708: Probabilistic Graphical Models 10-708, Spring 2014 4 : Exact Inference: Variable Elimination Lecturer: Eric P. ing Scribes: Soumya Batra, Pradeep Dasigi, Manzil Zaheer 1 Probabilistic Inference
10-708: Probabilistic Graphical Models 10-708, Spring 2014 4 : Exact Inference: Variable Elimination Lecturer: Eric P. ing Scribes: Soumya Batra, Pradeep Dasigi, Manzil Zaheer 1 Probabilistic Inference
REDUCING THE WORST CASE RUNNING TIMES OF A FAMILY OF RNA AND CFG PROBLEMS, USING VALIANT S APPROACH
 REDUCING THE WORST CASE RUNNING TIMES OF A FAMILY OF RNA AND CFG PROBLEMS, USING VALIANT S APPROACH SHAY ZAKOV, DEKEL TSUR, AND MICHAL ZIV-UKELSON Abstract. We study Valiant s classical algorithm for Context
REDUCING THE WORST CASE RUNNING TIMES OF A FAMILY OF RNA AND CFG PROBLEMS, USING VALIANT S APPROACH SHAY ZAKOV, DEKEL TSUR, AND MICHAL ZIV-UKELSON Abstract. We study Valiant s classical algorithm for Context
Context-Free Grammars (and Languages) Lecture 7
 Context-Free Grammars (and Languages) Lecture 7 1 Today Beyond regular expressions: Context-Free Grammars (CFGs) What is a CFG? What is the language associated with a CFG? Creating CFGs. Reasoning about
Context-Free Grammars (and Languages) Lecture 7 1 Today Beyond regular expressions: Context-Free Grammars (CFGs) What is a CFG? What is the language associated with a CFG? Creating CFGs. Reasoning about
Expectation Maximization (EM)
 Expectation Maximization (EM) The Expectation Maximization (EM) algorithm is one approach to unsupervised, semi-supervised, or lightly supervised learning. In this kind of learning either no labels are
Expectation Maximization (EM) The Expectation Maximization (EM) algorithm is one approach to unsupervised, semi-supervised, or lightly supervised learning. In this kind of learning either no labels are
CS5371 Theory of Computation. Lecture 9: Automata Theory VII (Pumping Lemma, Non-CFL)
 CS5371 Theory of Computation Lecture 9: Automata Theory VII (Pumping Lemma, Non-CFL) Objectives Introduce Pumping Lemma for CFL Apply Pumping Lemma to show that some languages are non-cfl Pumping Lemma
CS5371 Theory of Computation Lecture 9: Automata Theory VII (Pumping Lemma, Non-CFL) Objectives Introduce Pumping Lemma for CFL Apply Pumping Lemma to show that some languages are non-cfl Pumping Lemma
CMPT-825 Natural Language Processing. Why are parsing algorithms important?
 CMPT-825 Natural Language Processing Anoop Sarkar http://www.cs.sfu.ca/ anoop October 26, 2010 1/34 Why are parsing algorithms important? A linguistic theory is implemented in a formal system to generate
CMPT-825 Natural Language Processing Anoop Sarkar http://www.cs.sfu.ca/ anoop October 26, 2010 1/34 Why are parsing algorithms important? A linguistic theory is implemented in a formal system to generate
FORMAL LANGUAGES, AUTOMATA AND COMPUTATION
 FORMAL LANGUAGES, AUTOMATA AND COMPUTATION DECIDABILITY ( LECTURE 15) SLIDES FOR 15-453 SPRING 2011 1 / 34 TURING MACHINES-SYNOPSIS The most general model of computation Computations of a TM are described
FORMAL LANGUAGES, AUTOMATA AND COMPUTATION DECIDABILITY ( LECTURE 15) SLIDES FOR 15-453 SPRING 2011 1 / 34 TURING MACHINES-SYNOPSIS The most general model of computation Computations of a TM are described
CPS 220 Theory of Computation
 CPS 22 Theory of Computation Review - Regular Languages RL - a simple class of languages that can be represented in two ways: 1 Machine description: Finite Automata are machines with a finite number of
CPS 22 Theory of Computation Review - Regular Languages RL - a simple class of languages that can be represented in two ways: 1 Machine description: Finite Automata are machines with a finite number of
Algorithms in Bioinformatics FOUR Pairwise Sequence Alignment. Pairwise Sequence Alignment. Convention: DNA Sequences 5. Sequence Alignment
 Algorithms in Bioinformatics FOUR Sami Khuri Department of Computer Science San José State University Pairwise Sequence Alignment Homology Similarity Global string alignment Local string alignment Dot
Algorithms in Bioinformatics FOUR Sami Khuri Department of Computer Science San José State University Pairwise Sequence Alignment Homology Similarity Global string alignment Local string alignment Dot
Statistical NLP: Hidden Markov Models. Updated 12/15
 Statistical NLP: Hidden Markov Models Updated 12/15 Markov Models Markov models are statistical tools that are useful for NLP because they can be used for part-of-speech-tagging applications Their first
Statistical NLP: Hidden Markov Models Updated 12/15 Markov Models Markov models are statistical tools that are useful for NLP because they can be used for part-of-speech-tagging applications Their first
NPDA, CFG equivalence
 NPDA, CFG equivalence Theorem A language L is recognized by a NPDA iff L is described by a CFG. Must prove two directions: ( ) L is recognized by a NPDA implies L is described by a CFG. ( ) L is described
NPDA, CFG equivalence Theorem A language L is recognized by a NPDA iff L is described by a CFG. Must prove two directions: ( ) L is recognized by a NPDA implies L is described by a CFG. ( ) L is described
Computational Models: Class 5
 Computational Models: Class 5 Benny Chor School of Computer Science Tel Aviv University March 27, 2019 Based on slides by Maurice Herlihy, Brown University, and modifications by Iftach Haitner and Yishay
Computational Models: Class 5 Benny Chor School of Computer Science Tel Aviv University March 27, 2019 Based on slides by Maurice Herlihy, Brown University, and modifications by Iftach Haitner and Yishay
CMSC 330: Organization of Programming Languages. Pushdown Automata Parsing
 CMSC 330: Organization of Programming Languages Pushdown Automata Parsing Chomsky Hierarchy Categorization of various languages and grammars Each is strictly more restrictive than the previous First described
CMSC 330: Organization of Programming Languages Pushdown Automata Parsing Chomsky Hierarchy Categorization of various languages and grammars Each is strictly more restrictive than the previous First described
Statistical Machine Learning from Data
 Samy Bengio Statistical Machine Learning from Data Statistical Machine Learning from Data Samy Bengio IDIAP Research Institute, Martigny, Switzerland, and Ecole Polytechnique Fédérale de Lausanne (EPFL),
Samy Bengio Statistical Machine Learning from Data Statistical Machine Learning from Data Samy Bengio IDIAP Research Institute, Martigny, Switzerland, and Ecole Polytechnique Fédérale de Lausanne (EPFL),
Pushdown Automata: Introduction (2)
 Pushdown Automata: Introduction Pushdown automaton (PDA) M = (K, Σ, Γ,, s, A) where K is a set of states Σ is an input alphabet Γ is a set of stack symbols s K is the start state A K is a set of accepting
Pushdown Automata: Introduction Pushdown automaton (PDA) M = (K, Σ, Γ,, s, A) where K is a set of states Σ is an input alphabet Γ is a set of stack symbols s K is the start state A K is a set of accepting
Lecture 7 Sequence analysis. Hidden Markov Models
 Lecture 7 Sequence analysis. Hidden Markov Models Nicolas Lartillot may 2012 Nicolas Lartillot (Universite de Montréal) BIN6009 may 2012 1 / 60 1 Motivation 2 Examples of Hidden Markov models 3 Hidden
Lecture 7 Sequence analysis. Hidden Markov Models Nicolas Lartillot may 2012 Nicolas Lartillot (Universite de Montréal) BIN6009 may 2012 1 / 60 1 Motivation 2 Examples of Hidden Markov models 3 Hidden
Natural Language Processing : Probabilistic Context Free Grammars. Updated 5/09
 Natural Language Processing : Probabilistic Context Free Grammars Updated 5/09 Motivation N-gram models and HMM Tagging only allowed us to process sentences linearly. However, even simple sentences require
Natural Language Processing : Probabilistic Context Free Grammars Updated 5/09 Motivation N-gram models and HMM Tagging only allowed us to process sentences linearly. However, even simple sentences require
Computational Models - Lecture 5 1
 Computational Models - Lecture 5 1 Handout Mode Iftach Haitner. Tel Aviv University. November 28, 2016 1 Based on frames by Benny Chor, Tel Aviv University, modifying frames by Maurice Herlihy, Brown University.
Computational Models - Lecture 5 1 Handout Mode Iftach Haitner. Tel Aviv University. November 28, 2016 1 Based on frames by Benny Chor, Tel Aviv University, modifying frames by Maurice Herlihy, Brown University.
A gentle introduction to Hidden Markov Models
 A gentle introduction to Hidden Markov Models Mark Johnson Brown University November 2009 1 / 27 Outline What is sequence labeling? Markov models Hidden Markov models Finding the most likely state sequence
A gentle introduction to Hidden Markov Models Mark Johnson Brown University November 2009 1 / 27 Outline What is sequence labeling? Markov models Hidden Markov models Finding the most likely state sequence
Computational Models - Lecture 4 1
 Computational Models - Lecture 4 1 Handout Mode Iftach Haitner and Yishay Mansour. Tel Aviv University. April 3/8, 2013 1 Based on frames by Benny Chor, Tel Aviv University, modifying frames by Maurice
Computational Models - Lecture 4 1 Handout Mode Iftach Haitner and Yishay Mansour. Tel Aviv University. April 3/8, 2013 1 Based on frames by Benny Chor, Tel Aviv University, modifying frames by Maurice
Parsing with Context-Free Grammars
 Parsing with Context-Free Grammars Berlin Chen 2005 References: 1. Natural Language Understanding, chapter 3 (3.1~3.4, 3.6) 2. Speech and Language Processing, chapters 9, 10 NLP-Berlin Chen 1 Grammars
Parsing with Context-Free Grammars Berlin Chen 2005 References: 1. Natural Language Understanding, chapter 3 (3.1~3.4, 3.6) 2. Speech and Language Processing, chapters 9, 10 NLP-Berlin Chen 1 Grammars
11 The Max-Product Algorithm
 Massachusetts Institute of Technology Department of Electrical Engineering and Computer Science 6.438 Algorithms for Inference Fall 2014 11 The Max-Product Algorithm In the previous lecture, we introduced
Massachusetts Institute of Technology Department of Electrical Engineering and Computer Science 6.438 Algorithms for Inference Fall 2014 11 The Max-Product Algorithm In the previous lecture, we introduced
Hidden Markov Models
 Hidden Markov Models Outline CG-islands The Fair Bet Casino Hidden Markov Model Decoding Algorithm Forward-Backward Algorithm Profile HMMs HMM Parameter Estimation Viterbi training Baum-Welch algorithm
Hidden Markov Models Outline CG-islands The Fair Bet Casino Hidden Markov Model Decoding Algorithm Forward-Backward Algorithm Profile HMMs HMM Parameter Estimation Viterbi training Baum-Welch algorithm
Computing if a token can follow
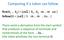 Computing if a token can follow first(b 1... B p ) = {a B 1...B p... aw } follow(x) = {a S......Xa... } There exists a derivation from the start symbol that produces a sequence of terminals and nonterminals
Computing if a token can follow first(b 1... B p ) = {a B 1...B p... aw } follow(x) = {a S......Xa... } There exists a derivation from the start symbol that produces a sequence of terminals and nonterminals
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, etworks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, etworks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2014
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Statistical Sequence Recognition and Training: An Introduction to HMMs
 Statistical Sequence Recognition and Training: An Introduction to HMMs EECS 225D Nikki Mirghafori nikki@icsi.berkeley.edu March 7, 2005 Credit: many of the HMM slides have been borrowed and adapted, with
Statistical Sequence Recognition and Training: An Introduction to HMMs EECS 225D Nikki Mirghafori nikki@icsi.berkeley.edu March 7, 2005 Credit: many of the HMM slides have been borrowed and adapted, with
Lecture Notes on Inductive Definitions
 Lecture Notes on Inductive Definitions 15-312: Foundations of Programming Languages Frank Pfenning Lecture 2 August 28, 2003 These supplementary notes review the notion of an inductive definition and give
Lecture Notes on Inductive Definitions 15-312: Foundations of Programming Languages Frank Pfenning Lecture 2 August 28, 2003 These supplementary notes review the notion of an inductive definition and give
Hidden Markov Models. Ivan Gesteira Costa Filho IZKF Research Group Bioinformatics RWTH Aachen Adapted from:
 Hidden Markov Models Ivan Gesteira Costa Filho IZKF Research Group Bioinformatics RWTH Aachen Adapted from: www.ioalgorithms.info Outline CG-islands The Fair Bet Casino Hidden Markov Model Decoding Algorithm
Hidden Markov Models Ivan Gesteira Costa Filho IZKF Research Group Bioinformatics RWTH Aachen Adapted from: www.ioalgorithms.info Outline CG-islands The Fair Bet Casino Hidden Markov Model Decoding Algorithm
Theory of Computation 8 Deterministic Membership Testing
 Theory of Computation 8 Deterministic Membership Testing Frank Stephan Department of Computer Science Department of Mathematics National University of Singapore fstephan@comp.nus.edu.sg Theory of Computation
Theory of Computation 8 Deterministic Membership Testing Frank Stephan Department of Computer Science Department of Mathematics National University of Singapore fstephan@comp.nus.edu.sg Theory of Computation
Evolutionary Models. Evolutionary Models
 Edit Operators In standard pairwise alignment, what are the allowed edit operators that transform one sequence into the other? Describe how each of these edit operations are represented on a sequence alignment
Edit Operators In standard pairwise alignment, what are the allowed edit operators that transform one sequence into the other? Describe how each of these edit operations are represented on a sequence alignment
Hidden Markov Models. Ron Shamir, CG 08
 Hidden Markov Models 1 Dr Richard Durbin is a graduate in mathematics from Cambridge University and one of the founder members of the Sanger Institute. He has also held carried out research at the Laboratory
Hidden Markov Models 1 Dr Richard Durbin is a graduate in mathematics from Cambridge University and one of the founder members of the Sanger Institute. He has also held carried out research at the Laboratory
CISC4090: Theory of Computation
 CISC4090: Theory of Computation Chapter 2 Context-Free Languages Courtesy of Prof. Arthur G. Werschulz Fordham University Department of Computer and Information Sciences Spring, 2014 Overview In Chapter
CISC4090: Theory of Computation Chapter 2 Context-Free Languages Courtesy of Prof. Arthur G. Werschulz Fordham University Department of Computer and Information Sciences Spring, 2014 Overview In Chapter
Section 1 (closed-book) Total points 30
 CS 454 Theory of Computation Fall 2011 Section 1 (closed-book) Total points 30 1. Which of the following are true? (a) a PDA can always be converted to an equivalent PDA that at each step pops or pushes
CS 454 Theory of Computation Fall 2011 Section 1 (closed-book) Total points 30 1. Which of the following are true? (a) a PDA can always be converted to an equivalent PDA that at each step pops or pushes
A parsing technique for TRG languages
 A parsing technique for TRG languages Daniele Paolo Scarpazza Politecnico di Milano October 15th, 2004 Daniele Paolo Scarpazza A parsing technique for TRG grammars [1]
A parsing technique for TRG languages Daniele Paolo Scarpazza Politecnico di Milano October 15th, 2004 Daniele Paolo Scarpazza A parsing technique for TRG grammars [1]
Hidden Markov Models
 Hidden Markov Models Slides revised and adapted to Bioinformática 55 Engª Biomédica/IST 2005 Ana Teresa Freitas Forward Algorithm For Markov chains we calculate the probability of a sequence, P(x) How
Hidden Markov Models Slides revised and adapted to Bioinformática 55 Engª Biomédica/IST 2005 Ana Teresa Freitas Forward Algorithm For Markov chains we calculate the probability of a sequence, P(x) How
Context Free Grammars: Introduction
 Context Free Grammars: Introduction Context free grammar (CFG) G = (V, Σ, R, S), where V is a set of grammar symbols Σ V is a set of terminal symbols R is a set of rules, where R (V Σ) V S is a distinguished
Context Free Grammars: Introduction Context free grammar (CFG) G = (V, Σ, R, S), where V is a set of grammar symbols Σ V is a set of terminal symbols R is a set of rules, where R (V Σ) V S is a distinguished
Hidden Markov Models. x 1 x 2 x 3 x K
 Hidden Markov Models 1 1 1 1 2 2 2 2 K K K K x 1 x 2 x 3 x K Viterbi, Forward, Backward VITERBI FORWARD BACKWARD Initialization: V 0 (0) = 1 V k (0) = 0, for all k > 0 Initialization: f 0 (0) = 1 f k (0)
Hidden Markov Models 1 1 1 1 2 2 2 2 K K K K x 1 x 2 x 3 x K Viterbi, Forward, Backward VITERBI FORWARD BACKWARD Initialization: V 0 (0) = 1 V k (0) = 0, for all k > 0 Initialization: f 0 (0) = 1 f k (0)
In this chapter, we explore the parsing problem, which encompasses several questions, including:
 Chapter 12 Parsing Algorithms 12.1 Introduction In this chapter, we explore the parsing problem, which encompasses several questions, including: Does L(G) contain w? What is the highest-weight derivation
Chapter 12 Parsing Algorithms 12.1 Introduction In this chapter, we explore the parsing problem, which encompasses several questions, including: Does L(G) contain w? What is the highest-weight derivation
Automata and Computability. Solutions to Exercises
 Automata and Computability Solutions to Exercises Spring 27 Alexis Maciel Department of Computer Science Clarkson University Copyright c 27 Alexis Maciel ii Contents Preface vii Introduction 2 Finite Automata
Automata and Computability Solutions to Exercises Spring 27 Alexis Maciel Department of Computer Science Clarkson University Copyright c 27 Alexis Maciel ii Contents Preface vii Introduction 2 Finite Automata
Predicting RNA Secondary Structure Using Profile Stochastic Context-Free Grammars and Phylogenic Analysis
 Fang XY, Luo ZG, Wang ZH. Predicting RNA secondary structure using profile stochastic context-free grammars and phylogenic analysis. JOURNAL OF COMPUTER SCIENCE AND TECHNOLOGY 23(4): 582 589 July 2008
Fang XY, Luo ZG, Wang ZH. Predicting RNA secondary structure using profile stochastic context-free grammars and phylogenic analysis. JOURNAL OF COMPUTER SCIENCE AND TECHNOLOGY 23(4): 582 589 July 2008
Statistical Methods for NLP
 Statistical Methods for NLP Sequence Models Joakim Nivre Uppsala University Department of Linguistics and Philology joakim.nivre@lingfil.uu.se Statistical Methods for NLP 1(21) Introduction Structured
Statistical Methods for NLP Sequence Models Joakim Nivre Uppsala University Department of Linguistics and Philology joakim.nivre@lingfil.uu.se Statistical Methods for NLP 1(21) Introduction Structured
CONTRAfold: RNA Secondary Structure Prediction without Physics-Based Models
 Supplementary Material for CONTRAfold: RNA Secondary Structure Prediction without Physics-Based Models Chuong B Do, Daniel A Woods, and Serafim Batzoglou Stanford University, Stanford, CA 94305, USA, {chuongdo,danwoods,serafim}@csstanfordedu,
Supplementary Material for CONTRAfold: RNA Secondary Structure Prediction without Physics-Based Models Chuong B Do, Daniel A Woods, and Serafim Batzoglou Stanford University, Stanford, CA 94305, USA, {chuongdo,danwoods,serafim}@csstanfordedu,
Computational Models - Lecture 4
 Computational Models - Lecture 4 Regular languages: The Myhill-Nerode Theorem Context-free Grammars Chomsky Normal Form Pumping Lemma for context free languages Non context-free languages: Examples Push
Computational Models - Lecture 4 Regular languages: The Myhill-Nerode Theorem Context-free Grammars Chomsky Normal Form Pumping Lemma for context free languages Non context-free languages: Examples Push
