Missing Data in Longitudinal Studies: Mixed-effects Pattern-Mixture and Selection Models
|
|
|
- Jessie Nash
- 6 years ago
- Views:
Transcription
1 Missing Data in Longitudinal Studies: Mixed-effects Pattern-Mixture and Selection Models Hedeker D & Gibbons RD (1997). Application of random-effects pattern-mixture models for missing data in longitudinal studies. Psychological Methods, 2, Chapter 14 in Hedeker & Gibbons (2006), Longitudinal Data Analysis, Wiley. 1
2 Example: Treatment-Related Change Across Time Data from the NIMH Schizophrenia collaborative study on treatment related changes in overall severity. IMPS item 79, Severity of Illness, was scored as: 1 = normal 2 = borderline mentally ill 3 = mildly ill 4 = moderately ill 5 = markedly ill 6 = severely ill 7 = among the most extremely ill Sample size at Week Group completers PLC (n=108) % DRUG (n=329) % Drug = Chlorpromazine, Fluphenazine, or Thioridazine 2
3 Descriptive Statistics - NIMH Schizophrenia study Observed IMPS79 Means, n, and sd week 0 week 1 week 3 week 6 placebo n drug n pooled sd Correlations: n = 313 and 321 n 424 week 0 week 1 week 3 week 6 week week week week
4 4
5 Mixed-effects regression model (MRM) - Schizophrenia study IMP S79 = Drug + T ime + (Drug T ime) + Subj + (Subj T ime) + Error IMPS79 ij = β 0 +β 1 Drug i +β 2 SWeek j +β 3 (Drug i SWeek j ) + υ 0i +υ 1i SWeek j +ε ij i = 1,..., N subjects j = 1,..., n i obs Drug = 0 for placebo, 1 for drug SWeek = 0, 1 = 1, 2 = 1.41, 3 = 1.73, 4 = 2, 5 = 2.24, 6 =
6 NIMH Schizophrenia Study - IMPS79 across Time: ML Estimates (se) Completers All Subjects N = 335 N = 437 est. se p < est. se p < intercept Drug (0 = plc; 1 = drug) Time (sqrt week) Drug by Time Intercept variance sd =.63 sd =.61 Int-Time covariance r = -.04 r =.07 Time variance sd =.45 sd =.49 6
7 Fitted and Obs. Means across Time by Condition ˆ IMP S ij = Drug i.34 T ime j.64(d i T j ) 7
8 Missing Data and Incomplete Data Models y y (O) R = 0 (observed) y (M) R = 1 (missing) GEE assumes Missing Completely at Random (MCAR) P (R y, X) = P (R X) for all y conditional on covariates, R is independent of y (O) and y (M) covariate-dependent missingness Likelihood-based MRM assumes Missing at Random (MAR) P (R y, X) = P (R X, y (O) ) for all y (M) conditional on covariates and observed values of the dependent variable, R is independent of y (M) ignorable non-response 8
9 Missing Not At Random (MNAR) Models When the data are nonignorable (i.e., MNAR), standard statistical models can yield badly biased results The observed data provide no information to either confirm or refute ignorability cannot test MAR versus MNAR Two general classes of MNAR models Pattern mixture models - use missing data pattern information in the longitudinal modeling Selection models - modeling of both the longitudinal and missingness processes will be illustrated in terms of MRMs, however they can be more broadly defined and utilized 9
10 Comments on MNAR models Ordinary MRM (and other full-likelihood models) assume MAR, these extended models do not Use of nonignorable models can be helpful in conducting a sensitivity analysis; to see how the conclusions might vary as a function of what is assumed about the missing data Not necessarily a good idea to rely on a single MNAR model, because the assumptions about the missing data are impossible to assess with the observed data One should use MNAR models sensibly, possibly examining several types of such models for a given dataset 10
11 Pattern-mixture models for missing data Little (1993, 1994, 1995); Hedeker & Gibbons (1997) divide subjects into groups depending on their missing data pattern the missing data pattern is a between-subjects variable to be used in longitudinal data analysis For 3 timepoints, there are eight (2 3 ) possible missing data patterns: pattern group time1 time2 time3 1 O O O 2 O O M 3 O M O 4 M O O where, O=observed and M=missing 5 M M O 6 O M M 7 M O M 8 M M M Since MMM provides no data, it is ignored in the analysis 11
12 Representing patterns with dummy-coded variables pattern D1 D2 D3 D4 D5 D6 OOO OOM OMO MOO MMO OMM MOM these dummy-codes represent deviations from pattern OOO Other coding schemes can be used ( effect or sequential coding) these variables are used as main effects and interactions 12
13 Classification of Subjects based on missing-data Drop i = 0 subject measured at week 6 (last timepoint) 1 subject missing at week 6 (last timepoint) Drug Drop group group completer dropout total placebo (.65) (.35) drug (.81) (.19) total Dropout not independent of Drug χ 2 1 = 11.25, p <.001 Is dropout related to severity of illness? Does dropout moderate the influence of other variables effects on severity of illness? 13
14 Mixed-effects pattern mixture model: Schiz data augment the basic MRM of IMPS79 over time: IMPS79 ij = β 0 +β 1 Drug i +β 2 SWeek j +β 3 (Drug i SWeek j )+υ 0i +υ 1i SWeek j +ε ij, with variables based on the missing data patterns e.g., completers (N = 335) vs non-completers (N = 102) Drop = 0 or 1 for those that did not or did dropout from the trial (i.e., were not measured at the final study timepoint) IMPS79 ij = β 0 + β 1 Drug i + β 2 SWeek j + β 3 (Drug i SWeek j ) + β0 D Drop i + βd 1 (Drop i Drug i ) + βd 2 (Drop i Sweek j) + β3 D (Drop i Drug i Sweek j ) + υ 0i + υ 1i SWeek j + ε ij β 0, β 1, β 2, and β 3 are for the completer subsample β D 0, β D 1, β D 2, and β D 3 how dropouts differ from completers three-way interaction is of particular interest - indicates how the drug by time interaction varies with study completion 14
15 15
16 Less Restrictive Pattern Mixture Model use week of dropout variable D i in forming missing data patterns six missing data patterns: five dropout weeks and completers Let D m = D 1,..., D 5 denote dummy-variables which contrast each dropout pattern to the completers IMPS79 ij = β 0 + β 1 Drug i + β 2 SWeek j + β 3 (Drug i SWeek j ) + 5 m=1 β m 0 D m + β m 1 (D m Drug i ) + β m 2 (D m Sweek j ) + β3 m (D m Drug i Sweek j ) + υ 0i + υ 1i SWeek j + ε ij β 0, β 1, β 2, β 3 are for completers β0 m, βm 1, βm 2, βm 3 completers the β m 3 indicate how dropout group m differs from parameters are of great interest 16
17 parameter est se p < est se p < est se p < Int β Drug β SWeek β Drug SWeek β Dropout Drop = 1 (N = 102) D = 1 (N = 37) Int β Drug β SWeek β Drug SWeek β D = 2 (N = 10) Int β Drug β SWeek β Drug SWeek β D = 3 (N = 42) Int β Drug β SWeek β Drug SWeek β D = 4 (N = 5) Int β Drug β SWeek β Drug SWeek β D = 5 (N = 8) Int β Drug β SWeek β Drug SWeek β Deviance
18 18
19 Pattern-mixture averaged results (Little, 1995) Obtained averaging over missing-data patterns e.g., completers and dropouts Uses sample proportions as estimates of missing-data pattern proportions Depends on model for missing-data patterns e.g., completer versus dropout status varies by tx Completer Dropout placebo 70/108 placebo 38/108 drug 265/329 drug 64/ / /437 19
20 Pattern-mixture averaged results ˆ β = ˆπ c ˆβc + ˆπ d ˆβd = ˆβ c + ˆπ d ˆβD ˆβ c correspond to the coefficients in the current model formulation not involving dropout (i.e., intercept, drug, time, drug by time) (ˆβ d ˆβ c ) = ˆβ D correspond to the dropout-related coefficients in the current model formulation (i.e., dropout, dropout by drug, dropout by time, dropout by drug by time) ˆπ d is the sample proportion of dropouts averaged estimates are linear combinations of model estimates (obtained by simple arithmetic or via ESTIMATE statement in SAS) 20
21 Placebo Intercept (5.22) + ( ) = (0.32) = Completers Dropouts Placebo Time effect ( 0.39) + ( ) = (0.25) = Completers Dropouts Drug Intercept difference (0.20) + ( ) = ( 0.40) = Completers Dropouts Drug Time difference ( 0.54) + ( ) = ( 0.64) = Completers Dropouts 21
22 SAS example using ESTIMATE /* pattern-mixture random intercept and trend model /* using marginal dropout proportion to estimate averaged results */ PROC MIXED METHOD=ML COVTEST; CLASS id; MODEL imps79 = sweek drug sweek*drug dropout dropout*sweek dropout*drug dropout*drug*sweek / SOLUTION; RANDOM INTERCEPT sweek /SUB=id TYPE=UN G GCORR; ESTIMATE avg int INTERCEPT 1 sweek 0 drug 0 sweek*drug 0 dropout.2334 dropout*sweek 0 dropout*drug 0 dropout*drug*sweek 0; ESTIMATE avg sweek INTERCEPT 0 sweek 1 drug 0 sweek*drug 0 dropout 0 dropout*sweek.2334 dropout*drug 0 dropout*drug*sweek 0; ESTIMATE avg drug INTERCEPT 0 sweek 0 drug 1 sweek*drug 0 dropout 0 dropout*sweek 0 dropout*drug.2334 dropout*drug*sweek 0; ESTIMATE avg sweek*drug INTERCEPT 0 sweek 0 drug 0 sweek*drug 1 dropout 0 dropout*sweek 0 dropout*drug 0 dropout*drug*sweek.2334; RUN; where dropout =0 (completers) or =1 (dropouts) and 102/437 =.2334 (i.e., dropout proportion) 22
23 Don t have (or like) SAS? (i.e., can I do pattern-mixture modeling with SPSS?... YES!) Weighted Effect Coding (Darlington, 1990, Regression and Linear Models, pp ): in ANOVA context yields comparisons of each cell mean, except the reference cell, with a weighted average of cell means Effect coding level D becomes Weighted effect coding level D weight of level 1 weight of level 2 Y = β 0 + β 1 D + e β 0 = weighted average of Y means across the two levels β 1 = difference between level 1 mean and the weighted average 23
24 SPSS example using weighted effect coding Step 1 - do some data management to create the necessary variables SCHIZREP.SAV ID subject s ID IMPS79 severity of illness (dependent variable) Week study week (= 0, 1, 2, 3, 4, 5, 6) Drug =0 (placebo) or =1 (drug) SexM =0 (female) or =1 (male) SWeek DrugSwk sqrt of Week product of Drug by Sweek Week max subject s maximum value of Week Dropout =0 (Week max = 6) or =1 (Week max < 6) DropW DropWDrug DropWSweek DropWDrugSwk = ˆπ D ˆπ C = (Dropout = 0) or =1 (Dropout = 1) product of DropW by Drug product of DropW by SWeek product of DropW by Drug by SWeek 24
25 Model with weighted effect coding IMPS79 ij = β 0 + β 1 Drug i + β 2 SWeek j + β 3 (Drug i SWeek j ) + β DW 0 DropW i + β DW 1 (DropW i Drug i ) + β DW 2 (DropW i Sweek j ) + β DW 3 (DropW i Drug i Sweek j ) + υ 0i + υ 1i SWeek j + ε ij β 0, β 1, β 2, and β 3 are weighted averages over completers and dropouts (i.e., exactly what we want!) β0 DW, β1 DW, β2 DW, and β3 DW how dropouts differ from the weighted averages 25
26 SPSS syntax for model with weighted effect coding Schizpm.sps MIXED Imps79 WITH Drug Sweek DrugSwk DropW DropWDrug DropWSweek DropWDrugSwk /FIXED = Drug Sweek DrugSwk DropW DropWDrug DropWSweek DropWDrugSwk /METHOD = ML /PRINT = SOLUTION TESTCOV /RANDOM INTERCEPT Sweek SUBJECT(ID) COVTYPE(UN). 26
27 Pattern-mixture averaged results using either approach (ESTIMATE in SAS or weighted effect coding in SPSS), we get: parameter estimate std error p-value Intercept Drug SWeek DrugSwk Great! We re done, right? 27
28 I ve got some good news, and some not-so-good news averaged estimates are fine standard errors are not exactly correct 28
29 Standard Errors for Averaged Estimates ˆV (ˆ β) = ˆV (ˆ β)f + n d n c N 3 (ˆβ D ) 2 where, ˆV (ˆ β)f is the variance treating the sample proportions as known, i.e., the square of the standard error one gets using ˆ β = ˆβ c + ˆπ d ˆβD and not taking into account the fact that π d is estimated (i.e., this is obtained using methods that yield linear combinations of estimates and their associated standard errors) simple augmentation of ˆV (ˆ β)f to get correct standard errors 29
30 Calculation of ˆV (ˆ β) = ˆV (ˆ β)f + n d n c N 3 ( ˆβ D ) 2 parameter ˆ β ˆV (ˆ β)f ˆβD Augment ˆV (ˆ β) SE intercept (.0898) 2 = time (.0670) 2 = drug.1086 (.1029) 2 = drug time (.0776) 2 = Augment = n d n c N 3 ( ˆβ D ) 2, here, n d n c N 3 = (437) 3 =
31 Pattern-mixture averaged results - using drug-stratified proportions Placebo Intercept (5.22) + ( ) = (0.32) = Completers Dropouts Placebo Time effect ( 0.39) + ( ) = (0.25) = Completers Dropouts Drug Intercept difference (0.20) + ( ) = ( 0.40) = Completers Dropouts Drug Time difference ( 0.54) + ( ) = ( 0.64) = Completers Dropouts 31
32 Calculation of ˆV (ˆ β) = ˆV (ˆ β)f + n d n c N 3 ( ˆβ D ) 2 parameter ˆ β ˆV (ˆ β)f ˆβD Augment ˆV (ˆ β) SE intercept (.0879) 2 = time (.0698) 2 = drug.1241 (.1043) 2 = drug time (.0772) 2 = Augment = n d n c N 3 ( ˆβ D ) 2, where n d n c N 3 = (108) 3 = for placebo n d n c N 3 = (329) 3 = for drug 32
33 NIMH Schizophrenia Study - IMPS79 across Time: MRM models Ordinary Pattern mixture PM averaged parameter est se p < est se p < est se p < Int β Drug β SWeek β Drug SWeek β Int β0 D Drug β1 D SWeek β2 D Drug SWeek β3 D Deviance
34 Mixed-effects selection models These models have also been called random-coefficient selection models (Little, 95) random-effects-dependent models (Hogan & Laird, 97) shared parameter models (Wu & Carroll, 88; Ten Have et al., 98) One specifies both a model for the longitudinal outcome and a model for the dropout (or missingness) Both models depend on random subject effects, most or all of which are shared by both models 34
35 Longitudinal model - ordinary MRM of y i y i = X i β + Z i υ i + ε i Dropout model - grouped/discrete time survival analysis of D i log( log(1 P(D i = j D i j))) = W i α + υ i α W i includes dropout predictors, some or all may be in X i To the extent that α are nonzero, this is a nonignorable model because missingness, here characterized simply as dropout time, is dependent on both y O i and y M i (via υ i ) 35
36 Treatment (denoted Drug) by last wave (denoted Maxweek) Maxweek Drug Total placebo (.12) (.05) (.15) (.02) (.02) (.65) drug (.07) (.02) (.08) (.01) (.02) (.81) dropout is more common among the placebo group Pearson χ 2 test yields p <.025; Mantel-Haenszel χ 2 test for trend yields p <
37 Mixed-effects selection model - Schiz study Longitudinal model: IMPS79 ij = β 0 +β 1 Drug i +β 2 SWeek j +β 3 (Drug i SWeek j ) + υ 0i +υ 1i SWeek j +ε ij or, after orthogonalizing the random effects (υ i = Sθ i, where Σ υ = SS, Cholesky factorization) IMPS79 ij = β 0 + β 1 Drug i + β 2 SWeek j + β 3 (Drug i SWeek j ) + (σ υ0 + σ υ 01 SWeek j ) θ 0i + σ υ 1 SWeek j θ 1i + ε ij with σ υ 01 = σ υ01 /σ υ0 and σ υ 1 = σ 2 υ 1 σ 2 υ 01 /σ 2 υ 0 37
38 Dropout model: log( log(1 P(D i = j D i j))) = α 0j + α 1 Drug i + α 2 θ 0i + α 3 θ 1i + α 4 (Drug i θ 0i ) + α 5 (Drug i θ 1i ) or as P(D i j) = 1 exp( exp(α 0j + α 1 Drug i + α 2 θ 0i + α 3 θ 1i + α 4 (Drug i θ 0i ) + α 5 (Drug i θ 1i ))) random effects are summaries of a person s observed and unobserved y data this shared parameter model is a nonignorable model if α 2 = α 3 = α 4 = α 5 = 0 is rejected test of whether a particular model of ignorability is reasonable vs a particular model of nonignorability (i.e., not a general test of ignorability) 38
39 Separate and shared parameter models Separate (deviance = ) Shared (deviance = ) parameter ML est std error p-value ML est std error p-value Outcome intercept β Drug β SWeek β Drug Sweek β Dropout Drug α Random intercept α Random slope α Drug intercept α Drug slope α separate parameter model yields identical results as running these two models, one for y i and one for D i, separately shared parameter model fits better, χ 2 4 = 30.1, p <.0001 for longitudinal component, conclusions are same as MAR model marginally significant slope: for the placebo group there is a tendency to dropout as the slope increases significant negative Drug slope: the slope effect is opposite for the drug group; drug patients with more negative slopes (i.e., greater improvement) are more likely to drop out 39
40 NIMH Schizophrenia Study: Severity across Time ML Estimates (se) random intercept and slope models Shared Pattern Completers All cases Parameter Mixture N = 335 N = 437 N = 437 N = 437 intercept (.109) (.088) (.088) (.089) Drug (0=P; 1=D) (.123) (.101) (.102) (.105) Time (sqrt wk) (.073) (.068) (.073) (.071) Drug by Time (.083) (.078) (.083) (.078) 40
41 Conclusions Mixed-effects regression models (MRMs) useful for incomplete longitudinal data can handle subjects measured incompletely or at different timepoints missing data assumed MAR dependent on covariates and available data on dependent variable 41
42 Mixed-effects pattern-mixture and selection (i.e., shared parameter) models augment MRM Pattern-mixture adds missing-data pattern as between-subjects factor assesses degree to which missingness influences outcomes assesses degree to which missingness interacts with model terms (i.e., intervention group, intervention group by time) Selection missingness in terms of important covariates missingness in terms of (shared) random subject effects Does not invent data, attempts to maximize information obtained from available data 42
43 SAS MIXED code - SCHIZPM5b.SAS DATA one; INFILE c:\mixdemo\schizrep.dat ; INPUT id imps79 week drug sex ; /* the coding for the variables is as follows: id = subject id number imps79 = overall severity (1=normal,..., 7=among the most extremely ill) week = 0,1,2,3,4,5,6 (most of the obs. are at weeks 0,1,3, and 6) drug 0=placebo 1=drug (chlorpromazine, fluphenazine, or thioridazine) sex 0=female 1=male */ /* compute the square root of week to linearize relationship */ sweek = SQRT(week); /* calculate the maximum value of week for each subject (suppress the printing of the output for this procedure) */ PROC MEANS NOPRINT; CLASS id; VAR week; OUTPUT OUT=two MAX=maxweek; RUN; /* determine if a subject has data at week 6 dropout = 0 (for completers) or = 1 (for dropouts) */ DATA three; SET two; dropout=0; IF maxweek LT 6 THEN dropout=1; 43
44 /* dataset with all subjects (adding the dropout variable) */ DATA four; MERGE one three; BY id; IF id NE.; /* random intercept and trend model */ PROC MIXED DATA=four METHOD=ML COVTEST; CLASS id; MODEL imps79 = sweek drug sweek*drug / SOLUTION; RANDOM INTERCEPT sweek /SUB=id TYPE=UN G GCORR; RUN; /* pattern-mixture random intercept and trend model /* using marginal dropout proportion to estimate averaged results */ PROC MIXED DATA=four METHOD=ML COVTEST; CLASS id; MODEL imps79 = sweek drug sweek*drug dropout dropout*sweek dropout*drug dropout*drug*sweek / SOLUTION; RANDOM INTERCEPT sweek /SUB=id TYPE=UN G GCORR; ESTIMATE avg int INTERCEPT 1 sweek 0 drug 0 sweek*drug 0 dropout.2334 dropout*sweek 0 dropout*drug 0 dropout*drug*sweek 0; ESTIMATE avg sweek INTERCEPT 0 sweek 1 drug 0 sweek*drug 0 dropout 0 dropout*sweek.2334 dropout*drug 0 dropout*drug*sweek 0; ESTIMATE avg drug INTERCEPT 0 sweek 0 drug 1 sweek*drug 0 dropout 0 dropout*sweek 0 dropout*drug.2334 dropout*drug*sweek 0; ESTIMATE avg sweek*drug INTERCEPT 0 sweek 0 drug 0 sweek*drug 1 dropout 0 dropout*sweek 0 dropout*drug 0 dropout*drug*sweek.2334; RUN; 44
45 /* pattern-mixture random intercept and trend model /* using drug-specific dropout proportions to estimate averaged results */ PROC MIXED DATA=four METHOD=ML COVTEST; CLASS id; MODEL imps79 = sweek drug sweek*drug dropout dropout*sweek dropout*drug dropout*drug*sweek / SOLUTION; RANDOM INTERCEPT sweek /SUB=id TYPE=UN G GCORR; ESTIMATE avg int INTERCEPT 1 sweek 0 drug 0 sweek*drug 0 dropout dropout*sweek 0 dropout*drug 0 dropout*drug*sweek 0; ESTIMATE avg sweek INTERCEPT 0 sweek 1 drug 0 sweek*drug 0 dropout 0 dropout*sweek dropout*drug 0 dropout*drug*sweek 0; ESTIMATE avg drug INTERCEPT 0 sweek 0 drug 1 sweek*drug 0 dropout 0 dropout*sweek 0 dropout*drug dropout*drug*sweek 0; ESTIMATE avg sweek*drug INTERCEPT 0 sweek 0 drug 0 sweek*drug 1 dropout 0 dropout*sweek 0 dropout*drug 0 dropout*drug*sweek.19453; RUN; 45
46 SAS code - SCHZ2MODS.SAS TITLE1 shared parameter model of time to dropout and imps79 across time ; DATA one; INFILE c:\mixdemo\schizrep.dat ; INPUT id imps79 week drug sex ; /* The coding for the variables is as follows: id = subject id number imps79 = overall severity (1=normal,..., 7=most extremely ill) week = 0,1,2,3,4,5,6 (most of the obs. are at weeks 0,1,3, and 6) drug 0=placebo 1=drug (chlorpromazine, fluphenazine, or thioridazine) sex 0=female 1=male */ /* compute the square root of week to linearize relationship */ sweek = SQRT(week); /* calculate the maximum value of week for each subject and get drug in this aggregated dataset too */ PROC MEANS NOPRINT; CLASS id; VAR week drug; OUTPUT OUT=two MAX(week drug)=maxweek drug; /* setting up imps79 across time and maxweek as one outcome vector */ DATA daty; SET one; outcome = imps79; ind = 0; DATA datr; SET two; outcome = maxweek; ind = 1; IF id NE.; DATA all; SET daty datr; BY id; uppercase represents specific SAS syntax; lowercase represents user-defined the SAS dataset all includes the (n i + 1) 1 outcome vector y i, named outcome, which contains y i as its first n i elements and D i as its final element ind with values of 0 or 1, is also defined; this variable will be used to distinguish between the y i and D i elements 46
47 /* random trend model for imps79 */ PROC MIXED DATA=daty METHOD=ML COVTEST; CLASS id; MODEL outcome = drug sweek sweek*drug / SOLUTION; RANDOM INTERCEPT sweek /SUB=id TYPE=UN G GCORR; /* model for time to dropout - grouped-time proportional hazards model */ PROC LOGISTIC DATA=datr; MODEL outcome = drug / LINK = CLOGLOG; 47
48 /* separate modeling of imps79 and time to dropout */ PROC NLMIXED DATA=all; PARMS b0=5.35 b1=.05 b2=-.34 b3=-.64 sde=.76 v0=.61 v01=.02 v1=.49 a1=-.69 i1=-1.95 i2=-1.70 i3=-.99 i4=-.84 i5=-.69; IF (ind = 0) THEN DO; z = (outcome - (b0 + b1*drug + b2*sweek + b3*drug*sweek + (v0 + v01*sweek/v0)*u1 + SQRT(v1*v1 - (v01*v01)/(v0*v0))*sweek*u2)); p = (1 / SQRT(2* *sde*sde)) * EXP(-.5 * (z*z) / (sde*sde)); END; IF (ind = 1) THEN DO; z = a1*drug; IF (outcome=1) THEN p = 1 - EXP(0 - EXP(i1+z)); ELSE IF (outcome=2) THEN p = (1 - EXP(0 - EXP(i2+z))) - (1 - EXP(0 - EXP(i1+z))); ELSE IF (outcome=3) THEN p = (1 - EXP(0 - EXP(i3+z))) - (1 - EXP(0 - EXP(i2+z))); ELSE IF (outcome=4) THEN p = (1 - EXP(0 - EXP(i4+z))) - (1 - EXP(0 - EXP(i3+z))); ELSE IF (outcome=5) THEN p = (1 - EXP(0 - EXP(i5+z))) - (1 - EXP(0 - EXP(i4+z))); ELSE IF (outcome=6) THEN p = 1 - (1 - EXP(0 - EXP(i5+z))); END; IF (p > 1e-8) THEN ll = LOG(p); else ll = -1e100; MODEL outcome GENERAL(ll); RANDOM u1 u2 NORMAL([0,0], [1,0,1]) SUBJECT=id; 48
49 /* shared parameter model of imps79 and time to dropout */ PROC NLMIXED DATA=all; PARMS b0=5.35 b1=.05 b2=-.34 b3=-.64 sde=.76 v0=.61 v01=.02 v1=.49 a1=-.69 a2=0 a3=0 a4=0 a5=0 i1=-1.95 i2=-1.70 i3=-.99 i4=-.84 i5=-.69; IF (ind = 0) THEN DO; z = (outcome - (b0 + b1*drug + b2*sweek + b3*drug*sweek + (v0 + v01*sweek/v0)*u1 + SQRT(v1*v1 - (v01*v01)/(v0*v0))*sweek*u2)); p = (1 / SQRT(2* *sde*sde)) * EXP(-.5 * (z*z) / (sde*sde)); END; IF (ind = 1) THEN DO; z = a1*drug + a2*u1 + a3*u2 + a4*u1*drug + a5*u2*drug; IF (outcome=1) THEN p = 1 - EXP(0 - EXP(i1+z)); ELSE IF (outcome=2) THEN p = (1 - EXP(0 - EXP(i2+z))) - (1 - EXP(0 - EXP(i1+z))); ELSE IF (outcome=3) THEN p = (1 - EXP(0 - EXP(i3+z))) - (1 - EXP(0 - EXP(i2+z))); ELSE IF (outcome=4) THEN p = (1 - EXP(0 - EXP(i4+z))) - (1 - EXP(0 - EXP(i3+z))); ELSE IF (outcome=5) THEN p = (1 - EXP(0 - EXP(i5+z))) - (1 - EXP(0 - EXP(i4+z))); ELSE IF (outcome=6) THEN p = 1 - (1 - EXP(0 - EXP(i5+z))); END; IF (p > 1e-8) THEN ll = LOG(p); else ll = -1e100; MODEL outcome GENERAL(ll); RANDOM u1 u2 NORMAL([0,0], [1,0,1]) SUBJECT=id; 49
Mixed-Effects Pattern-Mixture Models for Incomplete Longitudinal Data. Don Hedeker University of Illinois at Chicago
 Mixed-Effects Pattern-Mixture Models for Incomplete Longitudinal Data Don Hedeker University of Illinois at Chicago This work was supported by National Institute of Mental Health Contract N44MH32056. 1
Mixed-Effects Pattern-Mixture Models for Incomplete Longitudinal Data Don Hedeker University of Illinois at Chicago This work was supported by National Institute of Mental Health Contract N44MH32056. 1
Mixed Models for Longitudinal Binary Outcomes. Don Hedeker Department of Public Health Sciences University of Chicago.
 Mixed Models for Longitudinal Binary Outcomes Don Hedeker Department of Public Health Sciences University of Chicago hedeker@uchicago.edu https://hedeker-sites.uchicago.edu/ Hedeker, D. (2005). Generalized
Mixed Models for Longitudinal Binary Outcomes Don Hedeker Department of Public Health Sciences University of Chicago hedeker@uchicago.edu https://hedeker-sites.uchicago.edu/ Hedeker, D. (2005). Generalized
Why analyze as ordinal? Mixed Models for Longitudinal Ordinal Data Don Hedeker University of Illinois at Chicago
 Why analyze as ordinal? Mixed Models for Longitudinal Ordinal Data Don Hedeker University of Illinois at Chicago hedeker@uic.edu www.uic.edu/ hedeker/long.html Efficiency: Armstrong & Sloan (1989, Amer
Why analyze as ordinal? Mixed Models for Longitudinal Ordinal Data Don Hedeker University of Illinois at Chicago hedeker@uic.edu www.uic.edu/ hedeker/long.html Efficiency: Armstrong & Sloan (1989, Amer
Mixed Models for Longitudinal Ordinal and Nominal Outcomes
 Mixed Models for Longitudinal Ordinal and Nominal Outcomes Don Hedeker Department of Public Health Sciences Biological Sciences Division University of Chicago hedeker@uchicago.edu Hedeker, D. (2008). Multilevel
Mixed Models for Longitudinal Ordinal and Nominal Outcomes Don Hedeker Department of Public Health Sciences Biological Sciences Division University of Chicago hedeker@uchicago.edu Hedeker, D. (2008). Multilevel
Advantages of Mixed-effects Regression Models (MRM; aka multilevel, hierarchical linear, linear mixed models) 1. MRM explicitly models individual
 Advantages of Mixed-effects Regression Models (MRM; aka multilevel, hierarchical linear, linear mixed models) 1. MRM explicitly models individual change across time 2. MRM more flexible in terms of repeated
Advantages of Mixed-effects Regression Models (MRM; aka multilevel, hierarchical linear, linear mixed models) 1. MRM explicitly models individual change across time 2. MRM more flexible in terms of repeated
Mixed Models for Longitudinal Ordinal and Nominal Outcomes. Don Hedeker Department of Public Health Sciences University of Chicago
 Mixed Models for Longitudinal Ordinal and Nominal Outcomes Don Hedeker Department of Public Health Sciences University of Chicago hedeker@uchicago.edu https://hedeker-sites.uchicago.edu/ Hedeker, D. (2008).
Mixed Models for Longitudinal Ordinal and Nominal Outcomes Don Hedeker Department of Public Health Sciences University of Chicago hedeker@uchicago.edu https://hedeker-sites.uchicago.edu/ Hedeker, D. (2008).
Application of Item Response Theory Models for Intensive Longitudinal Data
 Application of Item Response Theory Models for Intensive Longitudinal Data Don Hedeker, Robin Mermelstein, & Brian Flay University of Illinois at Chicago hedeker@uic.edu Models for Intensive Longitudinal
Application of Item Response Theory Models for Intensive Longitudinal Data Don Hedeker, Robin Mermelstein, & Brian Flay University of Illinois at Chicago hedeker@uic.edu Models for Intensive Longitudinal
GEE for Longitudinal Data - Chapter 8
 GEE for Longitudinal Data - Chapter 8 GEE: generalized estimating equations (Liang & Zeger, 1986; Zeger & Liang, 1986) extension of GLM to longitudinal data analysis using quasi-likelihood estimation method
GEE for Longitudinal Data - Chapter 8 GEE: generalized estimating equations (Liang & Zeger, 1986; Zeger & Liang, 1986) extension of GLM to longitudinal data analysis using quasi-likelihood estimation method
Mixed Models for Longitudinal Ordinal and Nominal Data
 Mixed Models for Longitudinal Ordinal and Nominal Data Hedeker, D. (2008). Multilevel models for ordinal and nominal variables. In J. de Leeuw & E. Meijer (Eds.), Handbook of Multilevel Analysis. Springer,
Mixed Models for Longitudinal Ordinal and Nominal Data Hedeker, D. (2008). Multilevel models for ordinal and nominal variables. In J. de Leeuw & E. Meijer (Eds.), Handbook of Multilevel Analysis. Springer,
Lab 11. Multilevel Models. Description of Data
 Lab 11 Multilevel Models Henian Chen, M.D., Ph.D. Description of Data MULTILEVEL.TXT is clustered data for 386 women distributed across 40 groups. ID: 386 women, id from 1 to 386, individual level (level
Lab 11 Multilevel Models Henian Chen, M.D., Ph.D. Description of Data MULTILEVEL.TXT is clustered data for 386 women distributed across 40 groups. ID: 386 women, id from 1 to 386, individual level (level
Contrasting Marginal and Mixed Effects Models Recall: two approaches to handling dependence in Generalized Linear Models:
 Contrasting Marginal and Mixed Effects Models Recall: two approaches to handling dependence in Generalized Linear Models: Marginal models: based on the consequences of dependence on estimating model parameters.
Contrasting Marginal and Mixed Effects Models Recall: two approaches to handling dependence in Generalized Linear Models: Marginal models: based on the consequences of dependence on estimating model parameters.
over Time line for the means). Specifically, & covariances) just a fixed variance instead. PROC MIXED: to 1000 is default) list models with TYPE=VC */
 CLP 944 Example 4 page 1 Within-Personn Fluctuation in Symptom Severity over Time These data come from a study of weekly fluctuation in psoriasis severity. There was no intervention and no real reason
CLP 944 Example 4 page 1 Within-Personn Fluctuation in Symptom Severity over Time These data come from a study of weekly fluctuation in psoriasis severity. There was no intervention and no real reason
An Application of a Mixed-Effects Location Scale Model for Analysis of Ecological Momentary Assessment (EMA) Data
 An Application of a Mixed-Effects Location Scale Model for Analysis of Ecological Momentary Assessment (EMA) Data Don Hedeker, Robin Mermelstein, & Hakan Demirtas University of Illinois at Chicago hedeker@uic.edu
An Application of a Mixed-Effects Location Scale Model for Analysis of Ecological Momentary Assessment (EMA) Data Don Hedeker, Robin Mermelstein, & Hakan Demirtas University of Illinois at Chicago hedeker@uic.edu
SAS Code for Data Manipulation: SPSS Code for Data Manipulation: STATA Code for Data Manipulation: Psyc 945 Example 1 page 1
 Psyc 945 Example page Example : Unconditional Models for Change in Number Match 3 Response Time (complete data, syntax, and output available for SAS, SPSS, and STATA electronically) These data come from
Psyc 945 Example page Example : Unconditional Models for Change in Number Match 3 Response Time (complete data, syntax, and output available for SAS, SPSS, and STATA electronically) These data come from
Analyzing Group by Time Effects in Longitudinal Two-Group Randomized Trial Designs With Missing Data
 Journal of Modern Applied Statistical Methods Volume Issue 1 Article 6 5-1-003 Analyzing Group by Time Effects in Longitudinal Two-Group Randomized Trial Designs With Missing Data James Algina University
Journal of Modern Applied Statistical Methods Volume Issue 1 Article 6 5-1-003 Analyzing Group by Time Effects in Longitudinal Two-Group Randomized Trial Designs With Missing Data James Algina University
ST3241 Categorical Data Analysis I Multicategory Logit Models. Logit Models For Nominal Responses
 ST3241 Categorical Data Analysis I Multicategory Logit Models Logit Models For Nominal Responses 1 Models For Nominal Responses Y is nominal with J categories. Let {π 1,, π J } denote the response probabilities
ST3241 Categorical Data Analysis I Multicategory Logit Models Logit Models For Nominal Responses 1 Models For Nominal Responses Y is nominal with J categories. Let {π 1,, π J } denote the response probabilities
General Linear Model (Chapter 4)
 General Linear Model (Chapter 4) Outcome variable is considered continuous Simple linear regression Scatterplots OLS is BLUE under basic assumptions MSE estimates residual variance testing regression coefficients
General Linear Model (Chapter 4) Outcome variable is considered continuous Simple linear regression Scatterplots OLS is BLUE under basic assumptions MSE estimates residual variance testing regression coefficients
STAT 705 Generalized linear mixed models
 STAT 705 Generalized linear mixed models Timothy Hanson Department of Statistics, University of South Carolina Stat 705: Data Analysis II 1 / 24 Generalized Linear Mixed Models We have considered random
STAT 705 Generalized linear mixed models Timothy Hanson Department of Statistics, University of South Carolina Stat 705: Data Analysis II 1 / 24 Generalized Linear Mixed Models We have considered random
Time-Invariant Predictors in Longitudinal Models
 Time-Invariant Predictors in Longitudinal Models Today s Class (or 3): Summary of steps in building unconditional models for time What happens to missing predictors Effects of time-invariant predictors
Time-Invariant Predictors in Longitudinal Models Today s Class (or 3): Summary of steps in building unconditional models for time What happens to missing predictors Effects of time-invariant predictors
Growth Mixture Model
 Growth Mixture Model Latent Variable Modeling and Measurement Biostatistics Program Harvard Catalyst The Harvard Clinical & Translational Science Center Short course, October 28, 2016 Slides contributed
Growth Mixture Model Latent Variable Modeling and Measurement Biostatistics Program Harvard Catalyst The Harvard Clinical & Translational Science Center Short course, October 28, 2016 Slides contributed
A Bayesian Nonparametric Approach to Monotone Missing Data in Longitudinal Studies with Informative Missingness
 A Bayesian Nonparametric Approach to Monotone Missing Data in Longitudinal Studies with Informative Missingness A. Linero and M. Daniels UF, UT-Austin SRC 2014, Galveston, TX 1 Background 2 Working model
A Bayesian Nonparametric Approach to Monotone Missing Data in Longitudinal Studies with Informative Missingness A. Linero and M. Daniels UF, UT-Austin SRC 2014, Galveston, TX 1 Background 2 Working model
Introduction to lnmle: An R Package for Marginally Specified Logistic-Normal Models for Longitudinal Binary Data
 Introduction to lnmle: An R Package for Marginally Specified Logistic-Normal Models for Longitudinal Binary Data Bryan A. Comstock and Patrick J. Heagerty Department of Biostatistics University of Washington
Introduction to lnmle: An R Package for Marginally Specified Logistic-Normal Models for Longitudinal Binary Data Bryan A. Comstock and Patrick J. Heagerty Department of Biostatistics University of Washington
SAS Syntax and Output for Data Manipulation:
 CLP 944 Example 5 page 1 Practice with Fixed and Random Effects of Time in Modeling Within-Person Change The models for this example come from Hoffman (2015) chapter 5. We will be examining the extent
CLP 944 Example 5 page 1 Practice with Fixed and Random Effects of Time in Modeling Within-Person Change The models for this example come from Hoffman (2015) chapter 5. We will be examining the extent
Mixed-effects Polynomial Regression Models chapter 5
 Mixed-effects Polynomial Regression Models chapter 5 1 Figure 5.1 Various curvilinear models: (a) decelerating positive slope; (b) accelerating positive slope; (c) decelerating negative slope; (d) accelerating
Mixed-effects Polynomial Regression Models chapter 5 1 Figure 5.1 Various curvilinear models: (a) decelerating positive slope; (b) accelerating positive slope; (c) decelerating negative slope; (d) accelerating
ANOVA Longitudinal Models for the Practice Effects Data: via GLM
 Psyc 943 Lecture 25 page 1 ANOVA Longitudinal Models for the Practice Effects Data: via GLM Model 1. Saturated Means Model for Session, E-only Variances Model (BP) Variances Model: NO correlation, EQUAL
Psyc 943 Lecture 25 page 1 ANOVA Longitudinal Models for the Practice Effects Data: via GLM Model 1. Saturated Means Model for Session, E-only Variances Model (BP) Variances Model: NO correlation, EQUAL
STAT 7030: Categorical Data Analysis
 STAT 7030: Categorical Data Analysis 5. Logistic Regression Peng Zeng Department of Mathematics and Statistics Auburn University Fall 2012 Peng Zeng (Auburn University) STAT 7030 Lecture Notes Fall 2012
STAT 7030: Categorical Data Analysis 5. Logistic Regression Peng Zeng Department of Mathematics and Statistics Auburn University Fall 2012 Peng Zeng (Auburn University) STAT 7030 Lecture Notes Fall 2012
Joint Modeling of Longitudinal Item Response Data and Survival
 Joint Modeling of Longitudinal Item Response Data and Survival Jean-Paul Fox University of Twente Department of Research Methodology, Measurement and Data Analysis Faculty of Behavioural Sciences Enschede,
Joint Modeling of Longitudinal Item Response Data and Survival Jean-Paul Fox University of Twente Department of Research Methodology, Measurement and Data Analysis Faculty of Behavioural Sciences Enschede,
Time-Invariant Predictors in Longitudinal Models
 Time-Invariant Predictors in Longitudinal Models Topics: What happens to missing predictors Effects of time-invariant predictors Fixed vs. systematically varying vs. random effects Model building strategies
Time-Invariant Predictors in Longitudinal Models Topics: What happens to missing predictors Effects of time-invariant predictors Fixed vs. systematically varying vs. random effects Model building strategies
Bayesian methods for missing data: part 1. Key Concepts. Nicky Best and Alexina Mason. Imperial College London
 Bayesian methods for missing data: part 1 Key Concepts Nicky Best and Alexina Mason Imperial College London BAYES 2013, May 21-23, Erasmus University Rotterdam Missing Data: Part 1 BAYES2013 1 / 68 Outline
Bayesian methods for missing data: part 1 Key Concepts Nicky Best and Alexina Mason Imperial College London BAYES 2013, May 21-23, Erasmus University Rotterdam Missing Data: Part 1 BAYES2013 1 / 68 Outline
Introduction to mtm: An R Package for Marginalized Transition Models
 Introduction to mtm: An R Package for Marginalized Transition Models Bryan A. Comstock and Patrick J. Heagerty Department of Biostatistics University of Washington 1 Introduction Marginalized transition
Introduction to mtm: An R Package for Marginalized Transition Models Bryan A. Comstock and Patrick J. Heagerty Department of Biostatistics University of Washington 1 Introduction Marginalized transition
Longitudinal analysis of ordinal data
 Longitudinal analysis of ordinal data A report on the external research project with ULg Anne-Françoise Donneau, Murielle Mauer June 30 th 2009 Generalized Estimating Equations (Liang and Zeger, 1986)
Longitudinal analysis of ordinal data A report on the external research project with ULg Anne-Françoise Donneau, Murielle Mauer June 30 th 2009 Generalized Estimating Equations (Liang and Zeger, 1986)
Introduction to Random Effects of Time and Model Estimation
 Introduction to Random Effects of Time and Model Estimation Today s Class: The Big Picture Multilevel model notation Fixed vs. random effects of time Random intercept vs. random slope models How MLM =
Introduction to Random Effects of Time and Model Estimation Today s Class: The Big Picture Multilevel model notation Fixed vs. random effects of time Random intercept vs. random slope models How MLM =
Hierarchical Generalized Linear Models. ERSH 8990 REMS Seminar on HLM Last Lecture!
 Hierarchical Generalized Linear Models ERSH 8990 REMS Seminar on HLM Last Lecture! Hierarchical Generalized Linear Models Introduction to generalized models Models for binary outcomes Interpreting parameter
Hierarchical Generalized Linear Models ERSH 8990 REMS Seminar on HLM Last Lecture! Hierarchical Generalized Linear Models Introduction to generalized models Models for binary outcomes Interpreting parameter
UNIVERSITY OF TORONTO Faculty of Arts and Science
 UNIVERSITY OF TORONTO Faculty of Arts and Science December 2013 Final Examination STA442H1F/2101HF Methods of Applied Statistics Jerry Brunner Duration - 3 hours Aids: Calculator Model(s): Any calculator
UNIVERSITY OF TORONTO Faculty of Arts and Science December 2013 Final Examination STA442H1F/2101HF Methods of Applied Statistics Jerry Brunner Duration - 3 hours Aids: Calculator Model(s): Any calculator
Categorical Predictor Variables
 Categorical Predictor Variables We often wish to use categorical (or qualitative) variables as covariates in a regression model. For binary variables (taking on only 2 values, e.g. sex), it is relatively
Categorical Predictor Variables We often wish to use categorical (or qualitative) variables as covariates in a regression model. For binary variables (taking on only 2 values, e.g. sex), it is relatively
Shared Random Parameter Models for Informative Missing Data
 Shared Random Parameter Models for Informative Missing Data Dean Follmann NIAID NIAID/NIH p. A General Set-up Longitudinal data (Y ij,x ij,r ij ) Y ij = outcome for person i on visit j R ij = 1 if observed
Shared Random Parameter Models for Informative Missing Data Dean Follmann NIAID NIAID/NIH p. A General Set-up Longitudinal data (Y ij,x ij,r ij ) Y ij = outcome for person i on visit j R ij = 1 if observed
Estimation: Problems & Solutions
 Estimation: Problems & Solutions Edps/Psych/Stat 587 Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Fall 2017 Outline 1. Introduction: Estimation of
Estimation: Problems & Solutions Edps/Psych/Stat 587 Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Fall 2017 Outline 1. Introduction: Estimation of
SAS Syntax and Output for Data Manipulation: CLDP 944 Example 3a page 1
 CLDP 944 Example 3a page 1 From Between-Person to Within-Person Models for Longitudinal Data The models for this example come from Hoffman (2015) chapter 3 example 3a. We will be examining the extent to
CLDP 944 Example 3a page 1 From Between-Person to Within-Person Models for Longitudinal Data The models for this example come from Hoffman (2015) chapter 3 example 3a. We will be examining the extent to
Topic 28: Unequal Replication in Two-Way ANOVA
 Topic 28: Unequal Replication in Two-Way ANOVA Outline Two-way ANOVA with unequal numbers of observations in the cells Data and model Regression approach Parameter estimates Previous analyses with constant
Topic 28: Unequal Replication in Two-Way ANOVA Outline Two-way ANOVA with unequal numbers of observations in the cells Data and model Regression approach Parameter estimates Previous analyses with constant
Example 7b: Generalized Models for Ordinal Longitudinal Data using SAS GLIMMIX, STATA MEOLOGIT, and MPLUS (last proportional odds model only)
 CLDP945 Example 7b page 1 Example 7b: Generalized Models for Ordinal Longitudinal Data using SAS GLIMMIX, STATA MEOLOGIT, and MPLUS (last proportional odds model only) This example comes from real data
CLDP945 Example 7b page 1 Example 7b: Generalized Models for Ordinal Longitudinal Data using SAS GLIMMIX, STATA MEOLOGIT, and MPLUS (last proportional odds model only) This example comes from real data
Designing Multilevel Models Using SPSS 11.5 Mixed Model. John Painter, Ph.D.
 Designing Multilevel Models Using SPSS 11.5 Mixed Model John Painter, Ph.D. Jordan Institute for Families School of Social Work University of North Carolina at Chapel Hill 1 Creating Multilevel Models
Designing Multilevel Models Using SPSS 11.5 Mixed Model John Painter, Ph.D. Jordan Institute for Families School of Social Work University of North Carolina at Chapel Hill 1 Creating Multilevel Models
Course Introduction and Overview Descriptive Statistics Conceptualizations of Variance Review of the General Linear Model
 Course Introduction and Overview Descriptive Statistics Conceptualizations of Variance Review of the General Linear Model PSYC 943 (930): Fundamentals of Multivariate Modeling Lecture 1: August 22, 2012
Course Introduction and Overview Descriptive Statistics Conceptualizations of Variance Review of the General Linear Model PSYC 943 (930): Fundamentals of Multivariate Modeling Lecture 1: August 22, 2012
Beyond GLM and likelihood
 Stat 6620: Applied Linear Models Department of Statistics Western Michigan University Statistics curriculum Core knowledge (modeling and estimation) Math stat 1 (probability, distributions, convergence
Stat 6620: Applied Linear Models Department of Statistics Western Michigan University Statistics curriculum Core knowledge (modeling and estimation) Math stat 1 (probability, distributions, convergence
Lecture 10: Introduction to Logistic Regression
 Lecture 10: Introduction to Logistic Regression Ani Manichaikul amanicha@jhsph.edu 2 May 2007 Logistic Regression Regression for a response variable that follows a binomial distribution Recall the binomial
Lecture 10: Introduction to Logistic Regression Ani Manichaikul amanicha@jhsph.edu 2 May 2007 Logistic Regression Regression for a response variable that follows a binomial distribution Recall the binomial
Lecture 3.1 Basic Logistic LDA
 y Lecture.1 Basic Logistic LDA 0.2.4.6.8 1 Outline Quick Refresher on Ordinary Logistic Regression and Stata Women s employment example Cross-Over Trial LDA Example -100-50 0 50 100 -- Longitudinal Data
y Lecture.1 Basic Logistic LDA 0.2.4.6.8 1 Outline Quick Refresher on Ordinary Logistic Regression and Stata Women s employment example Cross-Over Trial LDA Example -100-50 0 50 100 -- Longitudinal Data
Model Assumptions; Predicting Heterogeneity of Variance
 Model Assumptions; Predicting Heterogeneity of Variance Today s topics: Model assumptions Normality Constant variance Predicting heterogeneity of variance CLP 945: Lecture 6 1 Checking for Violations of
Model Assumptions; Predicting Heterogeneity of Variance Today s topics: Model assumptions Normality Constant variance Predicting heterogeneity of variance CLP 945: Lecture 6 1 Checking for Violations of
STAT 5500/6500 Conditional Logistic Regression for Matched Pairs
 STAT 5500/6500 Conditional Logistic Regression for Matched Pairs Motivating Example: The data we will be using comes from a subset of data taken from the Los Angeles Study of the Endometrial Cancer Data
STAT 5500/6500 Conditional Logistic Regression for Matched Pairs Motivating Example: The data we will be using comes from a subset of data taken from the Los Angeles Study of the Endometrial Cancer Data
Statistical Methods. Missing Data snijders/sm.htm. Tom A.B. Snijders. November, University of Oxford 1 / 23
 1 / 23 Statistical Methods Missing Data http://www.stats.ox.ac.uk/ snijders/sm.htm Tom A.B. Snijders University of Oxford November, 2011 2 / 23 Literature: Joseph L. Schafer and John W. Graham, Missing
1 / 23 Statistical Methods Missing Data http://www.stats.ox.ac.uk/ snijders/sm.htm Tom A.B. Snijders University of Oxford November, 2011 2 / 23 Literature: Joseph L. Schafer and John W. Graham, Missing
De-mystifying random effects models
 De-mystifying random effects models Peter J Diggle Lecture 4, Leahurst, October 2012 Linear regression input variable x factor, covariate, explanatory variable,... output variable y response, end-point,
De-mystifying random effects models Peter J Diggle Lecture 4, Leahurst, October 2012 Linear regression input variable x factor, covariate, explanatory variable,... output variable y response, end-point,
Introduction to Within-Person Analysis and RM ANOVA
 Introduction to Within-Person Analysis and RM ANOVA Today s Class: From between-person to within-person ANOVAs for longitudinal data Variance model comparisons using 2 LL CLP 944: Lecture 3 1 The Two Sides
Introduction to Within-Person Analysis and RM ANOVA Today s Class: From between-person to within-person ANOVAs for longitudinal data Variance model comparisons using 2 LL CLP 944: Lecture 3 1 The Two Sides
Estimation: Problems & Solutions
 Estimation: Problems & Solutions Edps/Psych/Soc 587 Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Spring 2019 Outline 1. Introduction: Estimation
Estimation: Problems & Solutions Edps/Psych/Soc 587 Carolyn J. Anderson Department of Educational Psychology c Board of Trustees, University of Illinois Spring 2019 Outline 1. Introduction: Estimation
Time Invariant Predictors in Longitudinal Models
 Time Invariant Predictors in Longitudinal Models Longitudinal Data Analysis Workshop Section 9 University of Georgia: Institute for Interdisciplinary Research in Education and Human Development Section
Time Invariant Predictors in Longitudinal Models Longitudinal Data Analysis Workshop Section 9 University of Georgia: Institute for Interdisciplinary Research in Education and Human Development Section
Analysis of Incomplete Non-Normal Longitudinal Lipid Data
 Analysis of Incomplete Non-Normal Longitudinal Lipid Data Jiajun Liu*, Devan V. Mehrotra, Xiaoming Li, and Kaifeng Lu 2 Merck Research Laboratories, PA/NJ 2 Forrest Laboratories, NY *jiajun_liu@merck.com
Analysis of Incomplete Non-Normal Longitudinal Lipid Data Jiajun Liu*, Devan V. Mehrotra, Xiaoming Li, and Kaifeng Lu 2 Merck Research Laboratories, PA/NJ 2 Forrest Laboratories, NY *jiajun_liu@merck.com
Basics of Modern Missing Data Analysis
 Basics of Modern Missing Data Analysis Kyle M. Lang Center for Research Methods and Data Analysis University of Kansas March 8, 2013 Topics to be Covered An introduction to the missing data problem Missing
Basics of Modern Missing Data Analysis Kyle M. Lang Center for Research Methods and Data Analysis University of Kansas March 8, 2013 Topics to be Covered An introduction to the missing data problem Missing
Research Design: Topic 18 Hierarchical Linear Modeling (Measures within Persons) 2010 R.C. Gardner, Ph.d.
 Research Design: Topic 8 Hierarchical Linear Modeling (Measures within Persons) R.C. Gardner, Ph.d. General Rationale, Purpose, and Applications Linear Growth Models HLM can also be used with repeated
Research Design: Topic 8 Hierarchical Linear Modeling (Measures within Persons) R.C. Gardner, Ph.d. General Rationale, Purpose, and Applications Linear Growth Models HLM can also be used with repeated
Review of CLDP 944: Multilevel Models for Longitudinal Data
 Review of CLDP 944: Multilevel Models for Longitudinal Data Topics: Review of general MLM concepts and terminology Model comparisons and significance testing Fixed and random effects of time Significance
Review of CLDP 944: Multilevel Models for Longitudinal Data Topics: Review of general MLM concepts and terminology Model comparisons and significance testing Fixed and random effects of time Significance
Time-Invariant Predictors in Longitudinal Models
 Time-Invariant Predictors in Longitudinal Models Today s Topics: What happens to missing predictors Effects of time-invariant predictors Fixed vs. systematically varying vs. random effects Model building
Time-Invariant Predictors in Longitudinal Models Today s Topics: What happens to missing predictors Effects of time-invariant predictors Fixed vs. systematically varying vs. random effects Model building
TMA 4275 Lifetime Analysis June 2004 Solution
 TMA 4275 Lifetime Analysis June 2004 Solution Problem 1 a) Observation of the outcome is censored, if the time of the outcome is not known exactly and only the last time when it was observed being intact,
TMA 4275 Lifetime Analysis June 2004 Solution Problem 1 a) Observation of the outcome is censored, if the time of the outcome is not known exactly and only the last time when it was observed being intact,
Testing Indirect Effects for Lower Level Mediation Models in SAS PROC MIXED
 Testing Indirect Effects for Lower Level Mediation Models in SAS PROC MIXED Here we provide syntax for fitting the lower-level mediation model using the MIXED procedure in SAS as well as a sas macro, IndTest.sas
Testing Indirect Effects for Lower Level Mediation Models in SAS PROC MIXED Here we provide syntax for fitting the lower-level mediation model using the MIXED procedure in SAS as well as a sas macro, IndTest.sas
Binary Logistic Regression
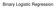 The coefficients of the multiple regression model are estimated using sample data with k independent variables Estimated (or predicted) value of Y Estimated intercept Estimated slope coefficients Ŷ = b
The coefficients of the multiple regression model are estimated using sample data with k independent variables Estimated (or predicted) value of Y Estimated intercept Estimated slope coefficients Ŷ = b
Random Coefficient Model (a.k.a. multilevel model) (Adapted from UCLA Statistical Computing Seminars)
 STAT:5201 Applied Statistic II Random Coefficient Model (a.k.a. multilevel model) (Adapted from UCLA Statistical Computing Seminars) School math achievement scores The data file consists of 7185 students
STAT:5201 Applied Statistic II Random Coefficient Model (a.k.a. multilevel model) (Adapted from UCLA Statistical Computing Seminars) School math achievement scores The data file consists of 7185 students
Maximum Likelihood Estimation; Robust Maximum Likelihood; Missing Data with Maximum Likelihood
 Maximum Likelihood Estimation; Robust Maximum Likelihood; Missing Data with Maximum Likelihood PRE 906: Structural Equation Modeling Lecture #3 February 4, 2015 PRE 906, SEM: Estimation Today s Class An
Maximum Likelihood Estimation; Robust Maximum Likelihood; Missing Data with Maximum Likelihood PRE 906: Structural Equation Modeling Lecture #3 February 4, 2015 PRE 906, SEM: Estimation Today s Class An
Multilevel Models in Matrix Form. Lecture 7 July 27, 2011 Advanced Multivariate Statistical Methods ICPSR Summer Session #2
 Multilevel Models in Matrix Form Lecture 7 July 27, 2011 Advanced Multivariate Statistical Methods ICPSR Summer Session #2 Today s Lecture Linear models from a matrix perspective An example of how to do
Multilevel Models in Matrix Form Lecture 7 July 27, 2011 Advanced Multivariate Statistical Methods ICPSR Summer Session #2 Today s Lecture Linear models from a matrix perspective An example of how to do
More Statistics tutorial at Logistic Regression and the new:
 Logistic Regression and the new: Residual Logistic Regression 1 Outline 1. Logistic Regression 2. Confounding Variables 3. Controlling for Confounding Variables 4. Residual Linear Regression 5. Residual
Logistic Regression and the new: Residual Logistic Regression 1 Outline 1. Logistic Regression 2. Confounding Variables 3. Controlling for Confounding Variables 4. Residual Linear Regression 5. Residual
Known unknowns : using multiple imputation to fill in the blanks for missing data
 Known unknowns : using multiple imputation to fill in the blanks for missing data James Stanley Department of Public Health University of Otago, Wellington james.stanley@otago.ac.nz Acknowledgments Cancer
Known unknowns : using multiple imputation to fill in the blanks for missing data James Stanley Department of Public Health University of Otago, Wellington james.stanley@otago.ac.nz Acknowledgments Cancer
" M A #M B. Standard deviation of the population (Greek lowercase letter sigma) σ 2
 Notation and Equations for Final Exam Symbol Definition X The variable we measure in a scientific study n The size of the sample N The size of the population M The mean of the sample µ The mean of the
Notation and Equations for Final Exam Symbol Definition X The variable we measure in a scientific study n The size of the sample N The size of the population M The mean of the sample µ The mean of the
Some methods for handling missing values in outcome variables. Roderick J. Little
 Some methods for handling missing values in outcome variables Roderick J. Little Missing data principles Likelihood methods Outline ML, Bayes, Multiple Imputation (MI) Robust MAR methods Predictive mean
Some methods for handling missing values in outcome variables Roderick J. Little Missing data principles Likelihood methods Outline ML, Bayes, Multiple Imputation (MI) Robust MAR methods Predictive mean
Lecture 4 Multiple linear regression
 Lecture 4 Multiple linear regression BIOST 515 January 15, 2004 Outline 1 Motivation for the multiple regression model Multiple regression in matrix notation Least squares estimation of model parameters
Lecture 4 Multiple linear regression BIOST 515 January 15, 2004 Outline 1 Motivation for the multiple regression model Multiple regression in matrix notation Least squares estimation of model parameters
Time-Invariant Predictors in Longitudinal Models
 Time-Invariant Predictors in Longitudinal Models Topics: Summary of building unconditional models for time Missing predictors in MLM Effects of time-invariant predictors Fixed, systematically varying,
Time-Invariant Predictors in Longitudinal Models Topics: Summary of building unconditional models for time Missing predictors in MLM Effects of time-invariant predictors Fixed, systematically varying,
Supplemental Materials. In the main text, we recommend graphing physiological values for individual dyad
 1 Supplemental Materials Graphing Values for Individual Dyad Members over Time In the main text, we recommend graphing physiological values for individual dyad members over time to aid in the decision
1 Supplemental Materials Graphing Values for Individual Dyad Members over Time In the main text, we recommend graphing physiological values for individual dyad members over time to aid in the decision
NATIONAL UNIVERSITY OF SINGAPORE EXAMINATION (SOLUTIONS) ST3241 Categorical Data Analysis. (Semester II: )
 NATIONAL UNIVERSITY OF SINGAPORE EXAMINATION (SOLUTIONS) Categorical Data Analysis (Semester II: 2010 2011) April/May, 2011 Time Allowed : 2 Hours Matriculation No: Seat No: Grade Table Question 1 2 3
NATIONAL UNIVERSITY OF SINGAPORE EXAMINATION (SOLUTIONS) Categorical Data Analysis (Semester II: 2010 2011) April/May, 2011 Time Allowed : 2 Hours Matriculation No: Seat No: Grade Table Question 1 2 3
Binomial Model. Lecture 10: Introduction to Logistic Regression. Logistic Regression. Binomial Distribution. n independent trials
 Lecture : Introduction to Logistic Regression Ani Manichaikul amanicha@jhsph.edu 2 May 27 Binomial Model n independent trials (e.g., coin tosses) p = probability of success on each trial (e.g., p =! =
Lecture : Introduction to Logistic Regression Ani Manichaikul amanicha@jhsph.edu 2 May 27 Binomial Model n independent trials (e.g., coin tosses) p = probability of success on each trial (e.g., p =! =
Introduction to Bayesian Statistics and Markov Chain Monte Carlo Estimation. EPSY 905: Multivariate Analysis Spring 2016 Lecture #10: April 6, 2016
 Introduction to Bayesian Statistics and Markov Chain Monte Carlo Estimation EPSY 905: Multivariate Analysis Spring 2016 Lecture #10: April 6, 2016 EPSY 905: Intro to Bayesian and MCMC Today s Class An
Introduction to Bayesian Statistics and Markov Chain Monte Carlo Estimation EPSY 905: Multivariate Analysis Spring 2016 Lecture #10: April 6, 2016 EPSY 905: Intro to Bayesian and MCMC Today s Class An
Lecture 12: Effect modification, and confounding in logistic regression
 Lecture 12: Effect modification, and confounding in logistic regression Ani Manichaikul amanicha@jhsph.edu 4 May 2007 Today Categorical predictor create dummy variables just like for linear regression
Lecture 12: Effect modification, and confounding in logistic regression Ani Manichaikul amanicha@jhsph.edu 4 May 2007 Today Categorical predictor create dummy variables just like for linear regression
Statistical Methods III Statistics 212. Problem Set 2 - Answer Key
 Statistical Methods III Statistics 212 Problem Set 2 - Answer Key 1. (Analysis to be turned in and discussed on Tuesday, April 24th) The data for this problem are taken from long-term followup of 1423
Statistical Methods III Statistics 212 Problem Set 2 - Answer Key 1. (Analysis to be turned in and discussed on Tuesday, April 24th) The data for this problem are taken from long-term followup of 1423
SAS Code: Joint Models for Continuous and Discrete Longitudinal Data
 CHAPTER 14 SAS Code: Joint Models for Continuous and Discrete Longitudinal Data We show how models of a mixed type can be analyzed using standard statistical software. We mainly focus on the SAS procedures
CHAPTER 14 SAS Code: Joint Models for Continuous and Discrete Longitudinal Data We show how models of a mixed type can be analyzed using standard statistical software. We mainly focus on the SAS procedures
Implementation of Pairwise Fitting Technique for Analyzing Multivariate Longitudinal Data in SAS
 PharmaSUG2011 - Paper SP09 Implementation of Pairwise Fitting Technique for Analyzing Multivariate Longitudinal Data in SAS Madan Gopal Kundu, Indiana University Purdue University at Indianapolis, Indianapolis,
PharmaSUG2011 - Paper SP09 Implementation of Pairwise Fitting Technique for Analyzing Multivariate Longitudinal Data in SAS Madan Gopal Kundu, Indiana University Purdue University at Indianapolis, Indianapolis,
DIAGNOSTICS FOR STRATIFIED CLINICAL TRIALS IN PROPORTIONAL ODDS MODELS
 DIAGNOSTICS FOR STRATIFIED CLINICAL TRIALS IN PROPORTIONAL ODDS MODELS Ivy Liu and Dong Q. Wang School of Mathematics, Statistics and Computer Science Victoria University of Wellington New Zealand Corresponding
DIAGNOSTICS FOR STRATIFIED CLINICAL TRIALS IN PROPORTIONAL ODDS MODELS Ivy Liu and Dong Q. Wang School of Mathematics, Statistics and Computer Science Victoria University of Wellington New Zealand Corresponding
Missing Covariate Data in Matched Case-Control Studies
 Missing Covariate Data in Matched Case-Control Studies Department of Statistics North Carolina State University Paul Rathouz Dept. of Health Studies U. of Chicago prathouz@health.bsd.uchicago.edu with
Missing Covariate Data in Matched Case-Control Studies Department of Statistics North Carolina State University Paul Rathouz Dept. of Health Studies U. of Chicago prathouz@health.bsd.uchicago.edu with
Exam Applied Statistical Regression. Good Luck!
 Dr. M. Dettling Summer 2011 Exam Applied Statistical Regression Approved: Tables: Note: Any written material, calculator (without communication facility). Attached. All tests have to be done at the 5%-level.
Dr. M. Dettling Summer 2011 Exam Applied Statistical Regression Approved: Tables: Note: Any written material, calculator (without communication facility). Attached. All tests have to be done at the 5%-level.
Faculty of Health Sciences. Correlated data. Count variables. Lene Theil Skovgaard & Julie Lyng Forman. December 6, 2016
 Faculty of Health Sciences Correlated data Count variables Lene Theil Skovgaard & Julie Lyng Forman December 6, 2016 1 / 76 Modeling count outcomes Outline The Poisson distribution for counts Poisson models,
Faculty of Health Sciences Correlated data Count variables Lene Theil Skovgaard & Julie Lyng Forman December 6, 2016 1 / 76 Modeling count outcomes Outline The Poisson distribution for counts Poisson models,
STA 4504/5503 Sample Exam 1 Spring 2011 Categorical Data Analysis. 1. Indicate whether each of the following is true (T) or false (F).
 STA 4504/5503 Sample Exam 1 Spring 2011 Categorical Data Analysis 1. Indicate whether each of the following is true (T) or false (F). (a) T In 2 2 tables, statistical independence is equivalent to a population
STA 4504/5503 Sample Exam 1 Spring 2011 Categorical Data Analysis 1. Indicate whether each of the following is true (T) or false (F). (a) T In 2 2 tables, statistical independence is equivalent to a population
Multinomial Logistic Regression Models
 Stat 544, Lecture 19 1 Multinomial Logistic Regression Models Polytomous responses. Logistic regression can be extended to handle responses that are polytomous, i.e. taking r>2 categories. (Note: The word
Stat 544, Lecture 19 1 Multinomial Logistic Regression Models Polytomous responses. Logistic regression can be extended to handle responses that are polytomous, i.e. taking r>2 categories. (Note: The word
STA 303 H1S / 1002 HS Winter 2011 Test March 7, ab 1cde 2abcde 2fghij 3
 STA 303 H1S / 1002 HS Winter 2011 Test March 7, 2011 LAST NAME: FIRST NAME: STUDENT NUMBER: ENROLLED IN: (circle one) STA 303 STA 1002 INSTRUCTIONS: Time: 90 minutes Aids allowed: calculator. Some formulae
STA 303 H1S / 1002 HS Winter 2011 Test March 7, 2011 LAST NAME: FIRST NAME: STUDENT NUMBER: ENROLLED IN: (circle one) STA 303 STA 1002 INSTRUCTIONS: Time: 90 minutes Aids allowed: calculator. Some formulae
Lecture 3 Linear random intercept models
 Lecture 3 Linear random intercept models Example: Weight of Guinea Pigs Body weights of 48 pigs in 9 successive weeks of follow-up (Table 3.1 DLZ) The response is measures at n different times, or under
Lecture 3 Linear random intercept models Example: Weight of Guinea Pigs Body weights of 48 pigs in 9 successive weeks of follow-up (Table 3.1 DLZ) The response is measures at n different times, or under
Advanced Quantitative Data Analysis
 Chapter 24 Advanced Quantitative Data Analysis Daniel Muijs Doing Regression Analysis in SPSS When we want to do regression analysis in SPSS, we have to go through the following steps: 1 As usual, we choose
Chapter 24 Advanced Quantitative Data Analysis Daniel Muijs Doing Regression Analysis in SPSS When we want to do regression analysis in SPSS, we have to go through the following steps: 1 As usual, we choose
Logistic Regression - problem 6.14
 Logistic Regression - problem 6.14 Let x 1, x 2,, x m be given values of an input variable x and let Y 1,, Y m be independent binomial random variables whose distributions depend on the corresponding values
Logistic Regression - problem 6.14 Let x 1, x 2,, x m be given values of an input variable x and let Y 1,, Y m be independent binomial random variables whose distributions depend on the corresponding values
Statistical tables are attached Two Hours UNIVERSITY OF MANCHESTER. May 2007 Final Draft
 Statistical tables are attached wo Hours UNIVERSIY OF MNHESER Ma 7 Final Draft Medical Statistics M377 Electronic calculators ma be used provided that the cannot store tet nswer LL si questions in SEION
Statistical tables are attached wo Hours UNIVERSIY OF MNHESER Ma 7 Final Draft Medical Statistics M377 Electronic calculators ma be used provided that the cannot store tet nswer LL si questions in SEION
Acknowledgements. Outline. Marie Diener-West. ICTR Leadership / Team INTRODUCTION TO CLINICAL RESEARCH. Introduction to Linear Regression
 INTRODUCTION TO CLINICAL RESEARCH Introduction to Linear Regression Karen Bandeen-Roche, Ph.D. July 17, 2012 Acknowledgements Marie Diener-West Rick Thompson ICTR Leadership / Team JHU Intro to Clinical
INTRODUCTION TO CLINICAL RESEARCH Introduction to Linear Regression Karen Bandeen-Roche, Ph.D. July 17, 2012 Acknowledgements Marie Diener-West Rick Thompson ICTR Leadership / Team JHU Intro to Clinical
Outline. Linear OLS Models vs: Linear Marginal Models Linear Conditional Models. Random Intercepts Random Intercepts & Slopes
 Lecture 2.1 Basic Linear LDA 1 Outline Linear OLS Models vs: Linear Marginal Models Linear Conditional Models Random Intercepts Random Intercepts & Slopes Cond l & Marginal Connections Empirical Bayes
Lecture 2.1 Basic Linear LDA 1 Outline Linear OLS Models vs: Linear Marginal Models Linear Conditional Models Random Intercepts Random Intercepts & Slopes Cond l & Marginal Connections Empirical Bayes
Biostatistics Workshop Longitudinal Data Analysis. Session 4 GARRETT FITZMAURICE
 Biostatistics Workshop 2008 Longitudinal Data Analysis Session 4 GARRETT FITZMAURICE Harvard University 1 LINEAR MIXED EFFECTS MODELS Motivating Example: Influence of Menarche on Changes in Body Fat Prospective
Biostatistics Workshop 2008 Longitudinal Data Analysis Session 4 GARRETT FITZMAURICE Harvard University 1 LINEAR MIXED EFFECTS MODELS Motivating Example: Influence of Menarche on Changes in Body Fat Prospective
Describing Within-Person Change over Time
 Describing Within-Person Change over Time Topics: Multilevel modeling notation and terminology Fixed and random effects of linear time Predicted variances and covariances from random slopes Dependency
Describing Within-Person Change over Time Topics: Multilevel modeling notation and terminology Fixed and random effects of linear time Predicted variances and covariances from random slopes Dependency
More about linear mixed models
 Faculty of Health Sciences Contents More about linear mixed models Analysis of repeated measurements, NFA 2016 Julie Lyng Forman & Lene Theil Skovgaard Department of Biostatistics, University of Copenhagen
Faculty of Health Sciences Contents More about linear mixed models Analysis of repeated measurements, NFA 2016 Julie Lyng Forman & Lene Theil Skovgaard Department of Biostatistics, University of Copenhagen
Solutions for Examination Categorical Data Analysis, March 21, 2013
 STOCKHOLMS UNIVERSITET MATEMATISKA INSTITUTIONEN Avd. Matematisk statistik, Frank Miller MT 5006 LÖSNINGAR 21 mars 2013 Solutions for Examination Categorical Data Analysis, March 21, 2013 Problem 1 a.
STOCKHOLMS UNIVERSITET MATEMATISKA INSTITUTIONEN Avd. Matematisk statistik, Frank Miller MT 5006 LÖSNINGAR 21 mars 2013 Solutions for Examination Categorical Data Analysis, March 21, 2013 Problem 1 a.
Monday 7 th Febraury 2005
 Monday 7 th Febraury 2 Analysis of Pigs data Data: Body weights of 48 pigs at 9 successive follow-up visits. This is an equally spaced data. It is always a good habit to reshape the data, so we can easily
Monday 7 th Febraury 2 Analysis of Pigs data Data: Body weights of 48 pigs at 9 successive follow-up visits. This is an equally spaced data. It is always a good habit to reshape the data, so we can easily
STAT 5500/6500 Conditional Logistic Regression for Matched Pairs
 STAT 5500/6500 Conditional Logistic Regression for Matched Pairs The data for the tutorial came from support.sas.com, The LOGISTIC Procedure: Conditional Logistic Regression for Matched Pairs Data :: SAS/STAT(R)
STAT 5500/6500 Conditional Logistic Regression for Matched Pairs The data for the tutorial came from support.sas.com, The LOGISTIC Procedure: Conditional Logistic Regression for Matched Pairs Data :: SAS/STAT(R)
multilevel modeling: concepts, applications and interpretations
 multilevel modeling: concepts, applications and interpretations lynne c. messer 27 october 2010 warning social and reproductive / perinatal epidemiologist concepts why context matters multilevel models
multilevel modeling: concepts, applications and interpretations lynne c. messer 27 october 2010 warning social and reproductive / perinatal epidemiologist concepts why context matters multilevel models
Longitudinal Modeling with Logistic Regression
 Newsom 1 Longitudinal Modeling with Logistic Regression Longitudinal designs involve repeated measurements of the same individuals over time There are two general classes of analyses that correspond to
Newsom 1 Longitudinal Modeling with Logistic Regression Longitudinal designs involve repeated measurements of the same individuals over time There are two general classes of analyses that correspond to
Describing Change over Time: Adding Linear Trends
 Describing Change over Time: Adding Linear Trends Longitudinal Data Analysis Workshop Section 7 University of Georgia: Institute for Interdisciplinary Research in Education and Human Development Section
Describing Change over Time: Adding Linear Trends Longitudinal Data Analysis Workshop Section 7 University of Georgia: Institute for Interdisciplinary Research in Education and Human Development Section
ISQS 5349 Final Exam, Spring 2017.
 ISQS 5349 Final Exam, Spring 7. Instructions: Put all answers on paper other than this exam. If you do not have paper, some will be provided to you. The exam is OPEN BOOKS, OPEN NOTES, but NO ELECTRONIC
ISQS 5349 Final Exam, Spring 7. Instructions: Put all answers on paper other than this exam. If you do not have paper, some will be provided to you. The exam is OPEN BOOKS, OPEN NOTES, but NO ELECTRONIC
