A 3D Statistical Fluid Registration Algorithm
|
|
|
- Clifford Campbell
- 6 years ago
- Views:
Transcription
1 A 3D Statistical Fluid Registration Algorithm Caroline Brun* 1, Natasha Leporé* 1,5, Xavier Pennec 2, Yi-Yu Chou 1, Agatha D. Lee 1, Greig de Zubicaray 3, Katie McMahon 3, Margaret J. Wright 4, and Paul M. Thompson 1 [* equal contribution] Abstract. In this paper, we further investigate a Statistically-Assisted Fluid Image Registration Algorithm (SAFIRA) that was recently developed in [2]. SAFIRA was built in a Lagrangian framework, which naturally incorporates two types of statistics on shape variations - deformation vectors and tensors - in the regularization term of the registration. This makes structural brain MRI registrations more accurate and biologically realistic. Here, we add to the understanding of the energetic behavior of the system through looking at its Hamiltonian. Furthermore, we compare the vector-statistics version to the non-statistical one and to the widely-used fluid registration, which is based on the Navier-Stokes equation. For the statistical registration, we use prior information on a training set independent from the dataset to be analyzed. Registration accuracy is measured on a pre-labeled data set for all 3 methods, and we also compute the heritability of brain structure using 46 twin pairs. SAFIRA detected genetic effects more extensively and showed improved registration accuracy compared to the other two algorithms. 1 Introduction Nonlinear registration is the warping of one object onto another, using measures of image similarity or corresponding features extracted from the images to guide the deformation. It is widely used in brain imaging for computational anatomy studies. Tensor-based morphometry (TBM), for example, identifies systematic differences in brain structure by statistically analyzing deformations that align images from many subjects to a common template. Registration methods commonly combine two terms: a similarity measure (distance or measure of agreement between two images) that drives the transformation and a regularizer that ensures its smoothness. This extra term is added to the registration function being optimized, to enforce desirable transformation properties such as smoothness, invertibility and inverse-consistency. For instance, the similarity criterion is regarded as a body force introduced into mechanical equations that govern linear elastic motion (Hooke s Law) in [1] or viscous fluid equations (Navier-Stokes equation) in [4]. Other algorithms rely on Gaussian filtering [17] or enforce particular properties of the deformation such as diffeomorphic trajectories [10]. When registering structural magnetic resonance brain images, the information available (voxel intensity, pre-defined landmarks) is rather limited and correspondence mappings are not unique. Consequently, a realistic model is needed to achieve deformations that are closer to an independently defined ground truth. This can be done for instance, as we chose to do here, by incorporating statistical information on the data set into the deformation. Some registration methods
2 have been developed to encode information on the natural variability in brain structure [7], but none of the 14 most widely used methods incorporates statistical information, except in the form of stationary Laplacian-type priors that only enforce smoothness [8]. However, in [5], the authors nonlinearly rescaled statistics on the strain tensors to use them in a demons-like registration algorithm. In [2], we implemented a Statistically-Assisted Fluid Registration Algorithm (SAFIRA). The non-statistical version of SAFIRA is the fluid equivalent of the elastic registration first introduced in [11], where the standard Euclidean elastic regularizer was replaced by one based on Riemannian metrics. The algorithm was re-formulated using the Lagrangian formalism to describe the dynamics of the system. This new formulation made it clear how to include the different types of statistics on the expected deformations [3]. The version of SAFIRA using statistics on the displacements was shown to give more accurate registrations and better detection sensitivity in a group-wise statistical analysis [3]. For those tests, the prior information was extracted from the dataset that is being analyzed. Here we build on this work in two ways. First, we look at the system in the energy domain by building the Hamiltonian of the system, to better understand the transfer of energy between the different components of the registration equation. Secondly, we present extensive tests of SAFIRA, by comparing the more advantageous statistical version of the algorithm against other fluid registration algorithms such as the standard Navier-Stokes fluid algorithm [4] [9], and the non-statistical version of SAFIRA. However, to verify the independent effect of the prior information on the deformation, we use statistics computed from another dataset comparable to the one studied. Indeed, our tests were done using two data sets: the manually-labeled LP BA40, which is one of the standard datasets used in the field to compare the accuracy and precision of different registration algorithms [8] and a 3D dataset consisted of 23 pairs of identical twins and 23 pairs of same-sex fraternal twins. As both groups are composed of healthy young adults, each of them can serve as a training set when using the statistical version of the algorithm to register the subjects from the other group to a given template. The vector-based statistical algorithm is shown to perform better in terms of accuracy and detection. We also investigate the detection power of each algorithm via heritability measures on the twins dataset. 2 Defining SAFIRA 2.1 Introduction A 3D brain image volume may be regarded as embedded in a deformable continuummechanical system for which each voxel is seen as a particle [4,?]. The system is then solved using a Newtonian mechanics equation of the form: d q(x, t) dt = q Cost α q Reg( q) β q, (1) where Cost = Sim(I, J q) = (I(x) J(q(x)) 2 dx is the similarity criterion, here the sum of squared intensity differences (SSD) and Reg is the regularizing term. q is obtained from equation (1) at each time step t and integrated over time
3 to find the displacement q. It is not immediately clear how to include statistical information into this equation, hence the need for the Lagrangian formalism. 2.2 The generalized Lagrangian When a system is subjected to conservative and non-conservative forces, the path followed by a mechanical system minimizes the action S, defined as δs = t1 t 0 ( L δq d ) L dt q + F. r dt, (2) where L(q, q) = T ( q) V (q) is the conservative Lagrangian accounting for the kinetic and the potential energies (T ( q) and V (q), respectively) and F is the nonconservative force during the virtual displacement δr, q is the displacement and q, the velocity., ( ) L d dt ( ) L + F. q ( ) r = 0 (3) A complete derivation can be found for example in [16]. This dynamic equation defines the motion of the non-conservative system at each time t. 2.3 Defining SAFIRA In this section, we use the Lagrangian theory presented in (2.2) to reformulate equation (1). Given the definition of the kinetic energy and acknowledging the conservative properties of the similarity term, the different terms of equation (1) may be defined as follows: Kinetic energy: T = 1 2 q j 2 2 Potential Energy: V = Cost(q) (4) Nonconservative energy: V F = αreg( q) β q 2 2 In this case, by deriving each term of Eq. 4, one can verify that Eqs. 1 and 3 correspond to each other with: ( ) d L = d q L dt q j dt = qcost(i, J, q) (5) ( ) r F = α q Reg( q) + β q This definition is used to find q at each time step. In the next section, we describe how statistical information is added to the non-conservative terms. This allows the system to dissipate more energy when it is moving in an expected direction (that agrees with this prior statistical information) or to keep or even inject energy when it is moving in an unlikely direction. This makes it easier to surpass local minima and keep moving towards a more plausible minimum.
4 2.4 Incorporating statistics The two non-conservative terms in Eq. 4 can be modified to incorporate the covariance of the displacements and deformation matrices. First, the regularizer in our algorithm, here labeled as Reg Riem ( q), can be modified to include statistics on the deformation tensors (see [3]). However, here we will focus only on the second non-conservative term, 1 2 β q 2 2. The Euclidean norm. 2 in this term can be replaced by a Mahalanobis distance: F ( ) r = α q Reg Riem ( q) + β q T j cov 1 q j q j (6) with cov qj = 1 N i (q i q j ) T (q i q j ), the covariance of the displacements q at a voxel j summed over the images i. 2.5 Implementation SAFIRA is a multi-resolution algorithm. In the implementation, we neglects second order terms (hence, the equation used in practice is q Cost α q Reg( q) β ˆ q = 0). Here ˆ q represents the solution to either statistical or non-statistical versions (see 2.4). For each resolution: 1. Define a grid on the template and set q(x, t = 0) = 0. Then at each time step δt: 2. Calculate q Cost 3. Solve the PDE to find q at each point in the grid, using a gradient descent (RK4). Here, q(q, t = 0) = γg F with γ = 0.3 and G a Gaussian. 4. Find the time step using the user defined maximal flow allowed. 5. Integrate q to find the displacement q, within this time step. 6. Compute the Jacobian of the displacements. If the Jacobian determinant falls below 0.5, then re-grid the template and return to Step Obtain the new displacement field once the Jacobian value is acceptable. Here, we do not use a traditional gradient descent but an explicit iterative method (RK4). This was chosen so that the resolution is less sensitive to local minima than a first-order method. 3 Hamiltonian mechanics: conservation of energy? Using a Lagrangian structure makes each term easier to interpret. However, computing the corresponding Hamiltonian H is crucial to understand and characterize the interaction between the different energy terms. While the energy of conservative systems is maintained with t, this is no longer the case when non-conservative forces are added. In the context of registration, summarizing the transfers between the different types of energies is of considerable interest, as it can allow tuning of the local speed of registration. The Hamiltonian H represents the energy of the conservative system and may be derived from the conservative Lagrangian L as
5 H = p q L with p = ( ) L q q (7) where p is the momentum of the system and q is the displacement. For nonconservative systems, we have ṗ = d ( ) ( ) ( ) L L r = + F dt q q Consequently, dh dt = d L (p q L) = ṗ q + p q dt L L q q q t As in our case, L t = 0, dh dt L = ṗ q q, we obtain : dh dt = F ( ) r q (8) For our system, H = α q Reg( q) + β ˆ q 1 2 q 2 + Cost(q), so the kinetic and cost energies lost are transferred to the regularizer and dissipation terms over time. Hence, changing the weights of the different terms in the regularizer and the dissipation can change the speed at which the registration converges at each voxel. 4 Data and Analysis 4.1 Data Acquisition and preprocessing The first dataset is composed of 3D structural brain MRI scans of 23 monozygotic (MZ) and 23 dizygotic (DZ) same-sex twin pairs (age range: years), as well as one scan from an identically scanned healthy subject used as the common target for the fluid registration. We collected 3D T 1-weighted MP-RAGE images (with parameters T R = 2500ms, T E = 3.83ms, T I = 1500ms, flip angle = 15 o, coronal orientation, FOV 230mm) on a 4 Tesla Brucker Medspec whole body scanner (Wesley Hospital, Brisbane, Australia). Non-brain tissue was deleted from the MRI images using the Brain Surface Extraction Software (BSE) [13] and linearly aligned to the Colin27 template. All scans were then aligned to the ICBM53 template using 9-parameter registration - FMRIB s Linear Image Registration Toolbox, FLIRT, prior to the nonlinear registrations described in this paper. Secondly, the LP BA40 dataset contains 40 images (20 males and 20 females - average age 29.2 ± 6.3 yrs. Details can be found at The volumes from the two datasets were intensity and space-normalized and resized to a common resolution.
6 4.2 Algorithms and data analysis All subjects 3D scans were non-linearly registered to a common template using the three algorithms (non-statistical and vector-statistics version of SAFIRA and traditional fluid). In each case, vector fields, and their corresponding Jacobian matrices J were computed at each voxel, resulting in a scalar value for the Jacobian determinant, det(j). The det(j) s express the local differences in volume (3D) or area (2D) between each subject and the target image: detj(u) > 1 indicates a local excess in the image being studied, compared to the template, while detj(u) < 1 indicates a local deficit. In the case when the statistical algorithm was used, the prior information was computed from a first round of registration on the other dataset, i.e., all the LPBA40 volumes were registered to a common template, voxelwise displacement vectors were obtained for each subject, from which a voxelwise covariance matrix resulted. This statistical information was then incorporated in the deformation process when using the statistical version of the algorithm to register the twin images to the same template (and vice versa). 4.3 Genetic influence on brain structures The detection power of the different algorithms, for use in a morphometry study, was compared by computing genetic measures from the twin dataset. To measure the resemblance between twin pairs, we first computed the intraclass correlation coefficient (ICC) for both the MZ and DZ groups in the cerebrum according σ to the equation: ICC = 2 b (σb 2 ). σ2 +σ2 b is the pooled variance between pairs and w σw 2 is the variance within pairs [12]. From there, we computed the heritability h 2 = 2(r(MZ) r(dz)), where r(mz) and r(dz) are the intraclass correlation values for the MZ and DZ groups, respectively. Heritability is an estimate of the proportion of the observed variation in a measurement that is attributable to genetic differences among individuals. 4.4 Accuracy of volume quantification As SAFIRA was primarily developed to study volume and shape differences between subjects as in TBM analyses, we estimated the accuracy of the three algorithms by measuring the volume differences in the 53 structures defined in the LP BA40 database. The differences were computed between one (randomly chosen) subject from the 40 subjects that was used as template, and each of the other 39 images registered to it. Briefly, the deformation field obtained from the registration of one subject s smri to the template is applied to the corresponding subject s labeled image and the labels of it are compared to the template s labeled image (the ground truth) (see [8] for more details). 5 Results The heritability is displayed as a set of 3D maps for the whole cerebrum in the figure (left). The anatomical pattern is consistent overall for the 3 methods:
7 subcortical structures are shown to be influenced by genetic differences across individuals. However, the vector-based algorithm performs better compared to the traditional fluid and non-statistical version of SAFIRA. This is particularly notable in the occipital lobes, which have been found to be highly genetically influenced, perhaps as they mature early in life according to a consistent genetic program (white arrow). This can also be noticed in the prefrontal areas, where a high heritability is found by the statistical code only. These regions were previously shown to be related to IQ [14], which was shown to be partly heritable, according to the American Psychological Association. The heritability maps obtained with the statistical version also show less blurriness. On the right of the figure, we show the volume quantification error for all ROIs. Incorporating vector-based statistics on the deformation field during the registration improves volumetric matching for most delineated structures, and makes volume quantification more accurate. This is the especially clear for subcortical gray matter structures, such as the caudate and putamen. 6 Conclusion Here, we thoroughly studied the vector-statistics version of SAFIRA and showed its advantages in terms of accuracy and sensitivity to genetic influences on brain structure. The results confirmed the findings of [3], where the prior information was provided by the same studied dataset. In particular, we compared it to the traditional fluid method commonly used in tensor-based morphometry, an application for which SAFIRA was primarily designed. More extensive genetic effects were detected with the statistical algorithm together with less noisy findings, maybe related to an improvement of the accuracy of the registration. We also computed the Hamiltonian of the system, which was crucial to understand the exchange of energy, and thus how the incorporation of statistics influences the deformation pattern. References 1. Bajcsy R, Kovačič S,Multiresolution elastic matching, Computer vision, graphics and image processing, 46, (1989) Anonymous, see Supplemental material Anonymous, see Supplemental material Christensen GE et al., Deformable templates using large deformation kinematics, IEEE Trans. Image Process. 5, (1996) Commowick O et al., Incorporating Statistical Measures of Anatomical Variability in Atlas-to-Subject Registration for Conformal Brain Radiotherapy, MICCAI (2005), Hulshoff Pol, HE et al., Genetic Contributions to Human Brain Morphology and Intelligence, J Neurosci 26, (2006) Gee, JC, On matching brain volumes, Pattern Recognit, (1999) 8. Klein, A et al., Evaluation of 14 nonlinear deformation algorithms applied to human brain mri registration, NeuroImage 46, (2009) Leporé N et al., Tensor based morphometry using fast fluid registration an application to deafness, SPIE (2008)
8 5. Commowick O et al., Incorporating Statistical Measures of Anatomical Variability in Atlas-to-Subject Registration for Conformal Brain Radiotherapy, MICCAI(2005), Hulshoff Pol, HE et al., Genetic Contributions to Human Brain Morphology and Intelligence, JNeurosci26,(2006) Gee, JC, On matching brain volumes, PatternRecognit,(1999) 8. Klein, A et al., Evaluation of 14 nonlinear deformation algorithms applied to human brain mri registration, NeuroImage46,(2009) Leporé N et al., Tensor based morphometry using fast fluid registration an application to deafness, SPIE(2008) 10. Miller MI, Computational anatomy: shape, growth, and atrophy comparison via diffeomorphism, Neuroimage23,(2004) Pennec, X et al., Riemannian elasticity: A statistical regularization framework for non-linear registration, MICCAI(2005) Scout PE, Fleiss JL, Intraclass correlations: Uses in assessing rater reliability, Psychol Bull 2, (1979) Shattuck, DW et al., BrainSuite: an automated cortical surface identification tool, MedIA 8, (2002) Shawn, P et al., Intellectual ability and cortical development in Children and Adolescent, Nature440,(2006) Sowell E et al., Mapping cortical change across the human life span, NatNeurosci Neurosci 6, (2003) Tveter, FT Hamilton s equations of motion for non-conservative systems, Celestial Mechanics and Dynamical Astronomy, 60 (1994) Vercauteren T et al., Symmetric Log-Domain Diffeomorphic Registration: A Demons-based Approach, MICCAI(2008) Fig. 1. Left: maps of heritability coefficients. Blue colors: h 2 = =0nogeneticinfluence 0 influence --redcolors:h h 2 = =0.75 high heritability. Right: Volume Quantification Error. Blue shows a better registration accuracy.
Multi-Atlas Tensor-Based Morphometry and its Application to a Genetic Study of 92 Twins
 Multi-Atlas Tensor-Based Morphometry and its Application to a Genetic Study of 92 Twins Natasha Leporé 1, Caroline Brun 1, Yi-Yu Chou 1, Agatha D. Lee 1, Marina Barysheva 1, Greig I. de Zubicaray 2, Matthew
Multi-Atlas Tensor-Based Morphometry and its Application to a Genetic Study of 92 Twins Natasha Leporé 1, Caroline Brun 1, Yi-Yu Chou 1, Agatha D. Lee 1, Marina Barysheva 1, Greig I. de Zubicaray 2, Matthew
Tensor-Based Analysis of Genetic Influences on Brain Integrity using DTI in 100 Twins
 Tensor-Based Analysis of Genetic Influences on Brain Integrity using DTI in 100 Twins Agatha D. Lee 1, Natasha Leporé 1, Caroline Brun 1, Yi-Yu Chou 1, Marina Barysheva 1, Ming-Chang Chiang 1, Sarah K.
Tensor-Based Analysis of Genetic Influences on Brain Integrity using DTI in 100 Twins Agatha D. Lee 1, Natasha Leporé 1, Caroline Brun 1, Yi-Yu Chou 1, Marina Barysheva 1, Ming-Chang Chiang 1, Sarah K.
Morphometrics with SPM12
 Morphometrics with SPM12 John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. What kind of differences are we looking for? Usually, we try to localise regions of difference.
Morphometrics with SPM12 John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. What kind of differences are we looking for? Usually, we try to localise regions of difference.
Neuroimage Processing
 Neuroimage Processing Instructor: Moo K. Chung mkchung@wisc.edu Lecture 10-11. Deformation-based morphometry (DBM) Tensor-based morphometry (TBM) November 13, 2009 Image Registration Process of transforming
Neuroimage Processing Instructor: Moo K. Chung mkchung@wisc.edu Lecture 10-11. Deformation-based morphometry (DBM) Tensor-based morphometry (TBM) November 13, 2009 Image Registration Process of transforming
Longitudinal growth analysis of early childhood brain using deformation based morphometry
 Longitudinal growth analysis of early childhood brain using deformation based morphometry Junki Lee 1, Yasser Ad-Dab'bagh 2, Vladimir Fonov 1, Alan C. Evans 1 and the Brain Development Cooperative Group
Longitudinal growth analysis of early childhood brain using deformation based morphometry Junki Lee 1, Yasser Ad-Dab'bagh 2, Vladimir Fonov 1, Alan C. Evans 1 and the Brain Development Cooperative Group
Quantitative Neuro-Anatomic and Functional Image Assessment Recent progress on image registration and its applications
 Quantitative Neuro-Anatomic and Functional Image Assessment Recent progress on image registration and its applications Guido Gerig Sarang Joshi Tom Fletcher Applications of image registration in neuroimaging
Quantitative Neuro-Anatomic and Functional Image Assessment Recent progress on image registration and its applications Guido Gerig Sarang Joshi Tom Fletcher Applications of image registration in neuroimaging
Morphometry. John Ashburner. Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK.
 Morphometry John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Morphometry is defined as: Measurement of the form of organisms or of their parts. The American Heritage
Morphometry John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Morphometry is defined as: Measurement of the form of organisms or of their parts. The American Heritage
IMA Preprint Series # 2298
 RELATING FIBER CROSSING IN HARDI TO INTELLECTUAL FUNCTION By Iman Aganj, Neda Jahanshad, Christophe Lenglet, Arthur W. Toga, Katie L. McMahon, Greig I. de Zubicaray, Margaret J. Wright, Nicholas G. Martin,
RELATING FIBER CROSSING IN HARDI TO INTELLECTUAL FUNCTION By Iman Aganj, Neda Jahanshad, Christophe Lenglet, Arthur W. Toga, Katie L. McMahon, Greig I. de Zubicaray, Margaret J. Wright, Nicholas G. Martin,
Morphometry. John Ashburner. Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Voxel-Based Morphometry
 Morphometry John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing followed by SPM Tissue
Morphometry John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing followed by SPM Tissue
Computational Brain Anatomy
 Computational Brain Anatomy John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing followed
Computational Brain Anatomy John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing followed
Comparison of Standard and Riemannian Fluid Registration for Tensor-Based Morphometry in HIV/AIDS
 Comparison of Standard and Riemannian Fluid Registration for Tensor-Based Morphometry in HIV/AIDS Caroline Brun 1, Natasha Lepore 1, Xavier Pennec 2, Yi-Yu Chou 1, Oscar L. Lopez 3, Howard J. Aizenstein
Comparison of Standard and Riemannian Fluid Registration for Tensor-Based Morphometry in HIV/AIDS Caroline Brun 1, Natasha Lepore 1, Xavier Pennec 2, Yi-Yu Chou 1, Oscar L. Lopez 3, Howard J. Aizenstein
Morphometrics with SPM12
 Morphometrics with SPM12 John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. What kind of differences are we looking for? Usually, we try to localise regions of difference.
Morphometrics with SPM12 John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. What kind of differences are we looking for? Usually, we try to localise regions of difference.
Multivariate Statistical Analysis of Deformation Momenta Relating Anatomical Shape to Neuropsychological Measures
 Multivariate Statistical Analysis of Deformation Momenta Relating Anatomical Shape to Neuropsychological Measures Nikhil Singh, Tom Fletcher, Sam Preston, Linh Ha, J. Stephen Marron, Michael Wiener, and
Multivariate Statistical Analysis of Deformation Momenta Relating Anatomical Shape to Neuropsychological Measures Nikhil Singh, Tom Fletcher, Sam Preston, Linh Ha, J. Stephen Marron, Michael Wiener, and
Deformation Morphometry: Basics and Applications
 Deformation Morphometry: Basics and Applications Valerie Cardenas Nicolson, Ph.D. Assistant Adjunct Professor NCIRE, UCSF, SFVA Center for Imaging of Neurodegenerative Diseases VA Challenge Clinical studies
Deformation Morphometry: Basics and Applications Valerie Cardenas Nicolson, Ph.D. Assistant Adjunct Professor NCIRE, UCSF, SFVA Center for Imaging of Neurodegenerative Diseases VA Challenge Clinical studies
Medical Image Analysis
 Medical Image Analysis Instructor: Moo K. Chung mchung@stat.wisc.edu Lecture 3. Deformation-based Morphometry (DBM) January 30, 2007 Deformation based Morphometry (DBM) It uses deformation fields obtained
Medical Image Analysis Instructor: Moo K. Chung mchung@stat.wisc.edu Lecture 3. Deformation-based Morphometry (DBM) January 30, 2007 Deformation based Morphometry (DBM) It uses deformation fields obtained
Statistical Analysis of Tensor Fields
 Statistical Analysis of Tensor Fields Yuchen Xie Baba C. Vemuri Jeffrey Ho Department of Computer and Information Sciences and Engineering University of Florida Abstract. In this paper, we propose a Riemannian
Statistical Analysis of Tensor Fields Yuchen Xie Baba C. Vemuri Jeffrey Ho Department of Computer and Information Sciences and Engineering University of Florida Abstract. In this paper, we propose a Riemannian
Brain Differences Visualized in the Blind using Tensor Manifold Statistics and Diffusion Tensor Imaging
 Brain Differences Visualized in the Blind using Tensor Manifold Statistics and Diffusion Tensor Imaging Agatha D. Lee Natasha Lepore Franco Lepore Flamine Alary Caroline Brun Yiyu Chou Marina Barysheva
Brain Differences Visualized in the Blind using Tensor Manifold Statistics and Diffusion Tensor Imaging Agatha D. Lee Natasha Lepore Franco Lepore Flamine Alary Caroline Brun Yiyu Chou Marina Barysheva
Optimized Conformal Parameterization of Cortical Surfaces Using Shape Based Matching of Landmark Curves
 Optimized Conformal Parameterization of Cortical Surfaces Using Shape Based Matching of Landmark Curves Lok Ming Lui 1, Sheshadri Thiruvenkadam 1, Yalin Wang 1,2,TonyChan 1, and Paul Thompson 2 1 Department
Optimized Conformal Parameterization of Cortical Surfaces Using Shape Based Matching of Landmark Curves Lok Ming Lui 1, Sheshadri Thiruvenkadam 1, Yalin Wang 1,2,TonyChan 1, and Paul Thompson 2 1 Department
An Anatomical Equivalence Class Based Joint Transformation-Residual Descriptor for Morphological Analysis
 An Anatomical Equivalence Class Based Joint Transformation-Residual Descriptor for Morphological Analysis Sajjad Baloch, Ragini Verma, and Christos Davatzikos University of Pennsylvania, Philadelphia,
An Anatomical Equivalence Class Based Joint Transformation-Residual Descriptor for Morphological Analysis Sajjad Baloch, Ragini Verma, and Christos Davatzikos University of Pennsylvania, Philadelphia,
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Multivariate models of inter-subject anatomical variability
 Multivariate models of inter-subject anatomical variability Wellcome Trust Centre for Neuroimaging, UCL Institute of Neurology, 12 Queen Square, London WC1N 3BG, UK. Prediction Binary Classification Curse
Multivariate models of inter-subject anatomical variability Wellcome Trust Centre for Neuroimaging, UCL Institute of Neurology, 12 Queen Square, London WC1N 3BG, UK. Prediction Binary Classification Curse
Regularized Nonrigid Registration of Lung CT Images by Preserving Tissue Volume and Vesselness Measure
 Regularized Nonrigid Registration of Lung CT Images by Preserving Tissue Volume and Vesselness Measure Kunlin Cao 1,3, Kaifang Du 2,3, Kai Ding 2,3, Joseph M. Reinhardt 2,3,, and Gary E. Christensen 1,3
Regularized Nonrigid Registration of Lung CT Images by Preserving Tissue Volume and Vesselness Measure Kunlin Cao 1,3, Kaifang Du 2,3, Kai Ding 2,3, Joseph M. Reinhardt 2,3,, and Gary E. Christensen 1,3
Registration of anatomical images using geodesic paths of diffeomorphisms parameterized with stationary vector fields
 Registration of anatomical images using geodesic paths of diffeomorphisms parameterized with stationary vector fields Monica Hernandez, Matias N. Bossa, and Salvador Olmos Communication Technologies Group
Registration of anatomical images using geodesic paths of diffeomorphisms parameterized with stationary vector fields Monica Hernandez, Matias N. Bossa, and Salvador Olmos Communication Technologies Group
Unbiased Volumetric Registration via Nonlinear Elastic Regularization
 Unbiased Volumetric Registration via Nonlinear Elastic Regularization Igor Yanovsky, Carole Le Guyader, Alex Leow, Arthur Toga, Paul Thompson, Luminita Vese To cite this version: Igor Yanovsky, Carole
Unbiased Volumetric Registration via Nonlinear Elastic Regularization Igor Yanovsky, Carole Le Guyader, Alex Leow, Arthur Toga, Paul Thompson, Luminita Vese To cite this version: Igor Yanovsky, Carole
A Riemannian Framework for Denoising Diffusion Tensor Images
 A Riemannian Framework for Denoising Diffusion Tensor Images Manasi Datar No Institute Given Abstract. Diffusion Tensor Imaging (DTI) is a relatively new imaging modality that has been extensively used
A Riemannian Framework for Denoising Diffusion Tensor Images Manasi Datar No Institute Given Abstract. Diffusion Tensor Imaging (DTI) is a relatively new imaging modality that has been extensively used
BMI/STAT 768: Lecture 04 Correlations in Metric Spaces
 BMI/STAT 768: Lecture 04 Correlations in Metric Spaces Moo K. Chung mkchung@wisc.edu February 1, 2018 The elementary statistical treatment on correlations can be found in [4]: http://www.stat.wisc.edu/
BMI/STAT 768: Lecture 04 Correlations in Metric Spaces Moo K. Chung mkchung@wisc.edu February 1, 2018 The elementary statistical treatment on correlations can be found in [4]: http://www.stat.wisc.edu/
Tract-Specific Analysis for DTI of Brain White Matter
 Tract-Specific Analysis for DTI of Brain White Matter Paul Yushkevich, Hui Zhang, James Gee Penn Image Computing & Science Lab Department of Radiology University of Pennsylvania IPAM Summer School July
Tract-Specific Analysis for DTI of Brain White Matter Paul Yushkevich, Hui Zhang, James Gee Penn Image Computing & Science Lab Department of Radiology University of Pennsylvania IPAM Summer School July
Improved Correspondence for DTI Population Studies via Unbiased Atlas Building
 Improved Correspondence for DTI Population Studies via Unbiased Atlas Building Casey Goodlett 1, Brad Davis 1,2, Remi Jean 3, John Gilmore 3, and Guido Gerig 1,3 1 Department of Computer Science, University
Improved Correspondence for DTI Population Studies via Unbiased Atlas Building Casey Goodlett 1, Brad Davis 1,2, Remi Jean 3, John Gilmore 3, and Guido Gerig 1,3 1 Department of Computer Science, University
Game Physics. Game and Media Technology Master Program - Utrecht University. Dr. Nicolas Pronost
 Game and Media Technology Master Program - Utrecht University Dr. Nicolas Pronost Soft body physics Soft bodies In reality, objects are not purely rigid for some it is a good approximation but if you hit
Game and Media Technology Master Program - Utrecht University Dr. Nicolas Pronost Soft body physics Soft bodies In reality, objects are not purely rigid for some it is a good approximation but if you hit
Spatiotemporal Anatomical Atlas Building
 Spatiotemporal Anatomical Atlas Building Population Shape Regression For Random Design Data Brad Davis 1, P. Thomas Fletcher 2, Elizabeth Bullitt 1, Sarang Joshi 2 1 The University of North Carolina at
Spatiotemporal Anatomical Atlas Building Population Shape Regression For Random Design Data Brad Davis 1, P. Thomas Fletcher 2, Elizabeth Bullitt 1, Sarang Joshi 2 1 The University of North Carolina at
Soft Bodies. Good approximation for hard ones. approximation breaks when objects break, or deform. Generalization: soft (deformable) bodies
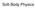 Soft-Body Physics Soft Bodies Realistic objects are not purely rigid. Good approximation for hard ones. approximation breaks when objects break, or deform. Generalization: soft (deformable) bodies Deformed
Soft-Body Physics Soft Bodies Realistic objects are not purely rigid. Good approximation for hard ones. approximation breaks when objects break, or deform. Generalization: soft (deformable) bodies Deformed
Diffeomorphic Diffusion Registration of Lung CT Images
 Diffeomorphic Diffusion Registration of Lung CT Images Alexander Schmidt-Richberg, Jan Ehrhardt, René Werner, and Heinz Handels Institute of Medical Informatics, University of Lübeck, Lübeck, Germany,
Diffeomorphic Diffusion Registration of Lung CT Images Alexander Schmidt-Richberg, Jan Ehrhardt, René Werner, and Heinz Handels Institute of Medical Informatics, University of Lübeck, Lübeck, Germany,
Sparse Scale-Space Decomposition of Volume Changes in Deformations Fields
 Sparse Scale-Space Decomposition of Volume Changes in Deformations Fields Lorenzi Marco 1, Bjoern H Menze 1,2, Marc Niethammer 3, Nicholas Ayache 1, and Xavier Pennec 1 for the Alzheimer's Disease Neuroimaging
Sparse Scale-Space Decomposition of Volume Changes in Deformations Fields Lorenzi Marco 1, Bjoern H Menze 1,2, Marc Niethammer 3, Nicholas Ayache 1, and Xavier Pennec 1 for the Alzheimer's Disease Neuroimaging
arxiv: v2 [cs.cg] 29 Oct 2018
![arxiv: v2 [cs.cg] 29 Oct 2018 arxiv: v2 [cs.cg] 29 Oct 2018](/thumbs/94/121718460.jpg) Computational Technologies for Brain Morphometry Zicong Zhou Ben Hildebrandt Xi Chen Guojun Liao October 10, 2018 arxiv:1810.04833v2 [cs.cg] 29 Oct 2018 Abstract In this paper, we described a set of computational
Computational Technologies for Brain Morphometry Zicong Zhou Ben Hildebrandt Xi Chen Guojun Liao October 10, 2018 arxiv:1810.04833v2 [cs.cg] 29 Oct 2018 Abstract In this paper, we described a set of computational
Medical Image Synthesis via Monte Carlo Simulation
 Medical Image Synthesis via Monte Carlo Simulation An Application of Statistics in Geometry & Building a Geometric Model with Correspondence James Z. Chen, Stephen M. Pizer, Edward L. Chaney, Sarang Joshi,
Medical Image Synthesis via Monte Carlo Simulation An Application of Statistics in Geometry & Building a Geometric Model with Correspondence James Z. Chen, Stephen M. Pizer, Edward L. Chaney, Sarang Joshi,
Schild s Ladder for the parallel transport of deformations in time series of images
 Schild s Ladder for the parallel transport of deformations in time series of images Lorenzi Marco 1,2, Nicholas Ayache 1, Xavier Pennec 1, and the Alzheimer s Disease Neuroimaging Initiative 1 Project
Schild s Ladder for the parallel transport of deformations in time series of images Lorenzi Marco 1,2, Nicholas Ayache 1, Xavier Pennec 1, and the Alzheimer s Disease Neuroimaging Initiative 1 Project
Template estimation form unlabeled point set data and surfaces for Computational Anatomy
 Template estimation form unlabeled point set data and surfaces for Computational Anatomy Joan Glaunès 1 and Sarang Joshi 1 Center for Imaging Science, Johns Hopkins University, joan@cis.jhu.edu SCI, University
Template estimation form unlabeled point set data and surfaces for Computational Anatomy Joan Glaunès 1 and Sarang Joshi 1 Center for Imaging Science, Johns Hopkins University, joan@cis.jhu.edu SCI, University
Fast Geodesic Regression for Population-Based Image Analysis
 Fast Geodesic Regression for Population-Based Image Analysis Yi Hong 1, Polina Golland 2, and Miaomiao Zhang 2 1 Computer Science Department, University of Georgia 2 Computer Science and Artificial Intelligence
Fast Geodesic Regression for Population-Based Image Analysis Yi Hong 1, Polina Golland 2, and Miaomiao Zhang 2 1 Computer Science Department, University of Georgia 2 Computer Science and Artificial Intelligence
Multivariate Statistics of the Jacobian Matrices in Tensor Based Morphometry and Their Application to HIV/AIDS
 Multivariate Statistics of the Jacobian Matrices in Tensor Based Morphometry and Their Application to HIV/AIDS Natasha Lepore 1, Caroline A. Brun 1, Ming-Chang Chiang 1,Yi-YuChou 1, Rebecca A. Dutton 1,KiraleeM.Hayashi
Multivariate Statistics of the Jacobian Matrices in Tensor Based Morphometry and Their Application to HIV/AIDS Natasha Lepore 1, Caroline A. Brun 1, Ming-Chang Chiang 1,Yi-YuChou 1, Rebecca A. Dutton 1,KiraleeM.Hayashi
Statistical Computing on Manifolds for Computational Anatomy. X. Pennec Computational Anatomy 1. Talairach & Tournoux, 1988
 Xavier Pennec Statistical registration: Pair-wise and Group-wise Alignment and Atlas formation, Brisbane, Australia, Friday Nov, 007. Statistical Computing on Manifolds for Computational Anatomy X. Pennec
Xavier Pennec Statistical registration: Pair-wise and Group-wise Alignment and Atlas formation, Brisbane, Australia, Friday Nov, 007. Statistical Computing on Manifolds for Computational Anatomy X. Pennec
Diffeomorphic Shape Trajectories for Improved Longitudinal Segmentation and Statistics
 Diffeomorphic Shape Trajectories for Improved Longitudinal Segmentation and Statistics Prasanna Muralidharan 1, James Fishbaugh 1, Hans J. Johnson 2, Stanley Durrleman 3, Jane S. Paulsen 2, Guido Gerig
Diffeomorphic Shape Trajectories for Improved Longitudinal Segmentation and Statistics Prasanna Muralidharan 1, James Fishbaugh 1, Hans J. Johnson 2, Stanley Durrleman 3, Jane S. Paulsen 2, Guido Gerig
Improved Correspondence for DTI Population Studies Via Unbiased Atlas Building
 Improved Correspondence for DTI Population Studies Via Unbiased Atlas Building Casey Goodlett 1,BradDavis 1,2,RemiJean 3, John Gilmore 3, and Guido Gerig 1,3 1 Department of Computer Science, University
Improved Correspondence for DTI Population Studies Via Unbiased Atlas Building Casey Goodlett 1,BradDavis 1,2,RemiJean 3, John Gilmore 3, and Guido Gerig 1,3 1 Department of Computer Science, University
Symmetric Log-Domain Diffeomorphic Registration: A Demons-Based Approach
 Symmetric Log-Domain Diffeomorphic Registration: A Demons-Based Approach Tom Vercauteren 1, Xavier Pennec 2,AymericPerchant 1, and Nicholas Ayache 2 1 Mauna Kea Technologies, France 2 Asclepios, INRIA
Symmetric Log-Domain Diffeomorphic Registration: A Demons-Based Approach Tom Vercauteren 1, Xavier Pennec 2,AymericPerchant 1, and Nicholas Ayache 2 1 Mauna Kea Technologies, France 2 Asclepios, INRIA
ACCURATE measurement of differences in brain anatomy
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 1, JANUARY 2008 129 Generalized Tensor-Based Morphometry of HIV/AIDS Using Multivariate Statistics on Deformation Tensors Natasha Lepore*, Caroline Brun,
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 1, JANUARY 2008 129 Generalized Tensor-Based Morphometry of HIV/AIDS Using Multivariate Statistics on Deformation Tensors Natasha Lepore*, Caroline Brun,
Metamorphic Geodesic Regression
 Metamorphic Geodesic Regression Yi Hong, Sarang Joshi 3, Mar Sanchez 4, Martin Styner, and Marc Niethammer,2 UNC-Chapel Hill/ 2 BRIC, 3 University of Utah, 4 Emory University Abstract. We propose a metamorphic
Metamorphic Geodesic Regression Yi Hong, Sarang Joshi 3, Mar Sanchez 4, Martin Styner, and Marc Niethammer,2 UNC-Chapel Hill/ 2 BRIC, 3 University of Utah, 4 Emory University Abstract. We propose a metamorphic
How Many Gradients are Sufficient in High-Angular Resolution Diffusion Imaging (HARDI)?
 How Many Gradients are Sufficient in High-Angular Resolution Diffusion Imaging (HARDI)? Liang Zhan 1, Ming-Chang Chiang 1, Alex D. Leow 1, Siwei Zhu 2, Marina Barysheva 1, Arthur W. Toga 1, Katie L. McMahon
How Many Gradients are Sufficient in High-Angular Resolution Diffusion Imaging (HARDI)? Liang Zhan 1, Ming-Chang Chiang 1, Alex D. Leow 1, Siwei Zhu 2, Marina Barysheva 1, Arthur W. Toga 1, Katie L. McMahon
DT-REFinD: Diffusion Tensor Registration With Exact Finite-Strain Differential
 DT-REFinD: Diffusion Tensor Registration With Exact Finite-Strain Differential The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
DT-REFinD: Diffusion Tensor Registration With Exact Finite-Strain Differential The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
Analysis of Longitudinal Shape Variability via Subject Specific Growth Modeling
 Analysis of Longitudinal Shape Variability via Subject Specific Growth Modeling James Fishbaugh 1, Marcel Prastawa 1, Stanley Durrleman 2, Joseph Piven 3 for the IBIS Network, and Guido Gerig 1 1 Scientific
Analysis of Longitudinal Shape Variability via Subject Specific Growth Modeling James Fishbaugh 1, Marcel Prastawa 1, Stanley Durrleman 2, Joseph Piven 3 for the IBIS Network, and Guido Gerig 1 1 Scientific
Population Based Analysis of Directional Information in Serial Deformation Tensor Morphometry
 Population Based Analysis of Directional Information in Serial Deformation Tensor Morphometry Colin Studholme 1,2 and Valerie Cardenas 1,2 1 Department of Radiiology, University of California San Francisco,
Population Based Analysis of Directional Information in Serial Deformation Tensor Morphometry Colin Studholme 1,2 and Valerie Cardenas 1,2 1 Department of Radiiology, University of California San Francisco,
Basics of Diffusion Tensor Imaging and DtiStudio
 Basics of Diffusion Tensor Imaging and DtiStudio DTI Basics 1 DTI reveals White matter anatomy Gray matter White matter DTI uses water diffusion as a probe for white matter anatomy Isotropic diffusion
Basics of Diffusion Tensor Imaging and DtiStudio DTI Basics 1 DTI reveals White matter anatomy Gray matter White matter DTI uses water diffusion as a probe for white matter anatomy Isotropic diffusion
NIH Public Access Author Manuscript IEEE Trans Med Imaging. Author manuscript; available in PMC 2014 May 29.
 NIH Public Access Author Manuscript Published in final edited form as: IEEE Trans Med Imaging. 2009 December ; 28(12): 1914 1928. doi:10.1109/tmi.2009.2025654. DT-REFinD: Diffusion Tensor Registration
NIH Public Access Author Manuscript Published in final edited form as: IEEE Trans Med Imaging. 2009 December ; 28(12): 1914 1928. doi:10.1109/tmi.2009.2025654. DT-REFinD: Diffusion Tensor Registration
Geodesic Refinement Using James-Stein Estimators
 Geodesic Refinement Using James-Stein Estimators Greg M. Fleishman 1,2, P. Thomas Fletcher 3, Boris A. Gutman 2, Gautam Prasad 2, Yingnian Wu 4, and Paul M. Thompson 2 1 UC Los Angeles, Department of Bioengineering
Geodesic Refinement Using James-Stein Estimators Greg M. Fleishman 1,2, P. Thomas Fletcher 3, Boris A. Gutman 2, Gautam Prasad 2, Yingnian Wu 4, and Paul M. Thompson 2 1 UC Los Angeles, Department of Bioengineering
Bayesian Principal Geodesic Analysis in Diffeomorphic Image Registration
 Bayesian Principal Geodesic Analysis in Diffeomorphic Image Registration Miaomiao Zhang and P. Thomas Fletcher School of Computing, University of Utah, Salt Lake City, USA Abstract. Computing a concise
Bayesian Principal Geodesic Analysis in Diffeomorphic Image Registration Miaomiao Zhang and P. Thomas Fletcher School of Computing, University of Utah, Salt Lake City, USA Abstract. Computing a concise
A new algorithm for the computation of the group logarithm of diffeomorphisms
 A new algorithm for the computation of the group logarithm of diffeomorphisms Matias Bossa and Salvador Olmos GTC, I3A, University of Zaragoza, Spain, {bossa,olmos}@unizar.es Abstract. There is an increasing
A new algorithm for the computation of the group logarithm of diffeomorphisms Matias Bossa and Salvador Olmos GTC, I3A, University of Zaragoza, Spain, {bossa,olmos}@unizar.es Abstract. There is an increasing
RS-fMRI analysis in healthy subjects confirms gender based differences
 RS-fMRI analysis in healthy subjects confirms gender based differences Alberto A. Vergani (aavergani@uninsubria.it) PhD student in Computer Science and Computational Mathematics University of Insubria
RS-fMRI analysis in healthy subjects confirms gender based differences Alberto A. Vergani (aavergani@uninsubria.it) PhD student in Computer Science and Computational Mathematics University of Insubria
Spatially Adaptive Log-Euclidean Polyaffine Registration Based on Sparse Matches
 Spatially Adaptive Log-Euclidean Polyaffine Registration Based on Sparse Matches Maxime Taquet 1,2, Benoît Macq 1, and Simon K. Warfield 2 1 ICTEAM Institute, Université catholique de Louvain, Louvain-La-Neuve,
Spatially Adaptive Log-Euclidean Polyaffine Registration Based on Sparse Matches Maxime Taquet 1,2, Benoît Macq 1, and Simon K. Warfield 2 1 ICTEAM Institute, Université catholique de Louvain, Louvain-La-Neuve,
Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain
 Available online at www.sciencedirect.com Medical Image Analysis 12 (2008) 26 41 www.elsevier.com/locate/media Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling
Available online at www.sciencedirect.com Medical Image Analysis 12 (2008) 26 41 www.elsevier.com/locate/media Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling
CIND Pre-Processing Pipeline For Diffusion Tensor Imaging. Overview
 CIND Pre-Processing Pipeline For Diffusion Tensor Imaging Overview The preprocessing pipeline of the Center for Imaging of Neurodegenerative Diseases (CIND) prepares diffusion weighted images (DWI) and
CIND Pre-Processing Pipeline For Diffusion Tensor Imaging Overview The preprocessing pipeline of the Center for Imaging of Neurodegenerative Diseases (CIND) prepares diffusion weighted images (DWI) and
Lecture I: Constrained Hamiltonian systems
 Lecture I: Constrained Hamiltonian systems (Courses in canonical gravity) Yaser Tavakoli December 15, 2014 1 Introduction In canonical formulation of general relativity, geometry of space-time is given
Lecture I: Constrained Hamiltonian systems (Courses in canonical gravity) Yaser Tavakoli December 15, 2014 1 Introduction In canonical formulation of general relativity, geometry of space-time is given
2800 Lyngby, Denmark, Abstract
 Comparison of three lters in the solution of the Navier-Stokes equation in registration C. Gramkow 1;3 and M. Bro-Nielsen 1;2;3 1 IMM - Department of Mathematical Modelling, Technical University of Denmark,
Comparison of three lters in the solution of the Navier-Stokes equation in registration C. Gramkow 1;3 and M. Bro-Nielsen 1;2;3 1 IMM - Department of Mathematical Modelling, Technical University of Denmark,
BMI/STAT 768: Lecture 09 Statistical Inference on Trees
 BMI/STAT 768: Lecture 09 Statistical Inference on Trees Moo K. Chung mkchung@wisc.edu March 1, 2018 This lecture follows the lecture on Trees. 1 Inference on MST In medical imaging, minimum spanning trees
BMI/STAT 768: Lecture 09 Statistical Inference on Trees Moo K. Chung mkchung@wisc.edu March 1, 2018 This lecture follows the lecture on Trees. 1 Inference on MST In medical imaging, minimum spanning trees
Nonlinear Registration of Diffusion MR Images Based on Fiber Bundles
 Nonlinear Registration of Diffusion MR Images Based on Fiber Bundles Ulas Ziyan 1, Mert R. Sabuncu 1, Lauren J. O Donnell 2,3, and Carl-Fredrik Westin 1,3 1 MIT Computer Science and Artificial Intelligence
Nonlinear Registration of Diffusion MR Images Based on Fiber Bundles Ulas Ziyan 1, Mert R. Sabuncu 1, Lauren J. O Donnell 2,3, and Carl-Fredrik Westin 1,3 1 MIT Computer Science and Artificial Intelligence
Math background. Physics. Simulation. Related phenomena. Frontiers in graphics. Rigid fluids
 Fluid dynamics Math background Physics Simulation Related phenomena Frontiers in graphics Rigid fluids Fields Domain Ω R2 Scalar field f :Ω R Vector field f : Ω R2 Types of derivatives Derivatives measure
Fluid dynamics Math background Physics Simulation Related phenomena Frontiers in graphics Rigid fluids Fields Domain Ω R2 Scalar field f :Ω R Vector field f : Ω R2 Types of derivatives Derivatives measure
Contrast Mechanisms in MRI. Michael Jay Schillaci
 Contrast Mechanisms in MRI Michael Jay Schillaci Overview Image Acquisition Basic Pulse Sequences Unwrapping K-Space Image Optimization Contrast Mechanisms Static and Motion Contrasts T1 & T2 Weighting,
Contrast Mechanisms in MRI Michael Jay Schillaci Overview Image Acquisition Basic Pulse Sequences Unwrapping K-Space Image Optimization Contrast Mechanisms Static and Motion Contrasts T1 & T2 Weighting,
Rician Noise Removal in Diffusion Tensor MRI
 Rician Noise Removal in Diffusion Tensor MRI Saurav Basu, Thomas Fletcher, and Ross Whitaker University of Utah, School of Computing, Salt Lake City, UT 84112, USA Abstract. Rician noise introduces a bias
Rician Noise Removal in Diffusion Tensor MRI Saurav Basu, Thomas Fletcher, and Ross Whitaker University of Utah, School of Computing, Salt Lake City, UT 84112, USA Abstract. Rician noise introduces a bias
Covariance Tracking Algorithm on Bilateral Filtering under Lie Group Structure Yinghong Xie 1,2,a Chengdong Wu 1,b
 Applied Mechanics and Materials Online: 014-0-06 ISSN: 166-748, Vols. 519-50, pp 684-688 doi:10.408/www.scientific.net/amm.519-50.684 014 Trans Tech Publications, Switzerland Covariance Tracking Algorithm
Applied Mechanics and Materials Online: 014-0-06 ISSN: 166-748, Vols. 519-50, pp 684-688 doi:10.408/www.scientific.net/amm.519-50.684 014 Trans Tech Publications, Switzerland Covariance Tracking Algorithm
Adaptive diffeomorphic multi-resolution demons and its application to same. modality medical image registration with large deformation 1
 Adaptive diffeomorphic multi-resolution demons and its application to same modality medical image registration with large deformation 1 Chang Wang 1 Qiongqiong Ren 1 Xin Qin 1 Yi Yu 1 1 (School of Biomedical
Adaptive diffeomorphic multi-resolution demons and its application to same modality medical image registration with large deformation 1 Chang Wang 1 Qiongqiong Ren 1 Xin Qin 1 Yi Yu 1 1 (School of Biomedical
Parametric non-rigid registration using a stationary velocity field
 Parametric non-rigid registration using a stationary velocity field Marc Modat, Pankaj Daga, M. Jorge Cardoso, Sebastien Ourselin, Centre for Medical Image Computing, University College London, WC1 6BT,
Parametric non-rigid registration using a stationary velocity field Marc Modat, Pankaj Daga, M. Jorge Cardoso, Sebastien Ourselin, Centre for Medical Image Computing, University College London, WC1 6BT,
Symmetric Image Normalization in the Diffeomorphic Space
 Symmetric Image Normalization in the Diffeomorphic Space Brian Avants, Charles Epstein, James Gee Penn Image Computing & Science Lab Departments of Radiology and Mathematics University of Pennsylvania
Symmetric Image Normalization in the Diffeomorphic Space Brian Avants, Charles Epstein, James Gee Penn Image Computing & Science Lab Departments of Radiology and Mathematics University of Pennsylvania
Computing the Geodesic Interpolating Spline
 Computing the Geodesic Interpolating Spline Anna Mills 1, Tony Shardlow 1, and Stephen Marsland 2 1 The University of Manchester, UK 2 Massey University, NZ Abstract. We examine non-rigid image registration
Computing the Geodesic Interpolating Spline Anna Mills 1, Tony Shardlow 1, and Stephen Marsland 2 1 The University of Manchester, UK 2 Massey University, NZ Abstract. We examine non-rigid image registration
Geodesics, Parallel Transport & One-parameter Subgroups for Diffeomorphic Image Registration
 Geodesics, Parallel Transport & One-parameter Subgroups for Diffeomorphic Image Registration Marco Lorenzi 1,2 and Xavier Pennec 1 1 Project Team Asclepios, INRIA Sophia Antipolis, France 2 LENITEM, IRCCS
Geodesics, Parallel Transport & One-parameter Subgroups for Diffeomorphic Image Registration Marco Lorenzi 1,2 and Xavier Pennec 1 1 Project Team Asclepios, INRIA Sophia Antipolis, France 2 LENITEM, IRCCS
Construction of a Deformable Spatiotemporal MRI Atlas of the Fetal Brain: Evaluation of Similarity Metrics and Deformation Models
 Construction of a Deformable Spatiotemporal MRI Atlas of the Fetal Brain: Evaluation of Similarity Metrics and Deformation Models Ali Gholipour 1, Catherine Limperopoulos 2, Sean Clancy 1, Cedric Clouchoux
Construction of a Deformable Spatiotemporal MRI Atlas of the Fetal Brain: Evaluation of Similarity Metrics and Deformation Models Ali Gholipour 1, Catherine Limperopoulos 2, Sean Clancy 1, Cedric Clouchoux
Spatial encoding in Magnetic Resonance Imaging. Jean-Marie BONNY
 Spatial encoding in Magnetic Resonance Imaging Jean-Marie BONNY What s Qu est an image ce qu une? image? «a reproduction of a material object by a camera or a related technique» Multi-dimensional signal
Spatial encoding in Magnetic Resonance Imaging Jean-Marie BONNY What s Qu est an image ce qu une? image? «a reproduction of a material object by a camera or a related technique» Multi-dimensional signal
Fluid Extensions for Optical Flow and Diffusion-based Image Registration
 Fluid Extensions for Optical Flow and Diffusion-based Image Registration Jens-Peer Kuska 1, Patrick Scheibe 2, Ulf-Dietrich Braumann 2 1 Interdisciplinary Centre for Bioinformatics, University Leipzig,
Fluid Extensions for Optical Flow and Diffusion-based Image Registration Jens-Peer Kuska 1, Patrick Scheibe 2, Ulf-Dietrich Braumann 2 1 Interdisciplinary Centre for Bioinformatics, University Leipzig,
Anatomical Regularization on Statistical Manifolds for the Classification of Patients with Alzheimer s Disease
 Anatomical Regularization on Statistical Manifolds for the Classification of Patients with Alzheimer s Disease Rémi Cuingnet 1,2, Joan Alexis Glaunès 1,3, Marie Chupin 1, Habib Benali 2, and Olivier Colliot
Anatomical Regularization on Statistical Manifolds for the Classification of Patients with Alzheimer s Disease Rémi Cuingnet 1,2, Joan Alexis Glaunès 1,3, Marie Chupin 1, Habib Benali 2, and Olivier Colliot
Vector Calculus. A primer
 Vector Calculus A primer Functions of Several Variables A single function of several variables: f: R $ R, f x (, x ),, x $ = y. Partial derivative vector, or gradient, is a vector: f = y,, y x ( x $ Multi-Valued
Vector Calculus A primer Functions of Several Variables A single function of several variables: f: R $ R, f x (, x ),, x $ = y. Partial derivative vector, or gradient, is a vector: f = y,, y x ( x $ Multi-Valued
On Mean Curvature Diusion in Nonlinear Image Filtering. Adel I. El-Fallah and Gary E. Ford. University of California, Davis. Davis, CA
 On Mean Curvature Diusion in Nonlinear Image Filtering Adel I. El-Fallah and Gary E. Ford CIPIC, Center for Image Processing and Integrated Computing University of California, Davis Davis, CA 95616 Abstract
On Mean Curvature Diusion in Nonlinear Image Filtering Adel I. El-Fallah and Gary E. Ford CIPIC, Center for Image Processing and Integrated Computing University of California, Davis Davis, CA 95616 Abstract
Cortical Shape Analysis using the Anisotropic Global Point Signature
 Cortical Shape Analysis using the Anisotropic Global Point Signature Anand A Joshi 1,3, Syed Ashrafulla 1, David W Shattuck 2, Hanna Damasio 3 and Richard M Leahy 1 1 Signal and Image Processing Institute,
Cortical Shape Analysis using the Anisotropic Global Point Signature Anand A Joshi 1,3, Syed Ashrafulla 1, David W Shattuck 2, Hanna Damasio 3 and Richard M Leahy 1 1 Signal and Image Processing Institute,
Biometrics: Introduction and Examples. Raymond Veldhuis
 Biometrics: Introduction and Examples Raymond Veldhuis 1 Overview Biometric recognition Face recognition Challenges Transparent face recognition Large-scale identification Watch list Anonymous biometrics
Biometrics: Introduction and Examples Raymond Veldhuis 1 Overview Biometric recognition Face recognition Challenges Transparent face recognition Large-scale identification Watch list Anonymous biometrics
Regularization of Diffusion Tensor Field Using Coupled Robust Anisotropic Diffusion Filters
 Regularization of Diffusion Tensor Field Using Coupled Robust Anisotropic Diffusion Filters Songyuan Tang a, Yong Fan a, Hongtu Zhu b, Pew-Thian Yap a Wei Gao a, Weili Lin a, and Dinggang Shen a a Department
Regularization of Diffusion Tensor Field Using Coupled Robust Anisotropic Diffusion Filters Songyuan Tang a, Yong Fan a, Hongtu Zhu b, Pew-Thian Yap a Wei Gao a, Weili Lin a, and Dinggang Shen a a Department
Uncertainty Quantification for LDDMM Using a Low-rank Hessian Approximation
 Uncertainty Quantification for LDDMM Using a Low-rank Hessian Approximation Xiao Yang, Marc Niethammer, Department of Computer Science and Biomedical Research Imaging Center University of North Carolina
Uncertainty Quantification for LDDMM Using a Low-rank Hessian Approximation Xiao Yang, Marc Niethammer, Department of Computer Science and Biomedical Research Imaging Center University of North Carolina
Application of Trace-Norm and Low-Rank Matrix Decomposition for Computational Anatomy
 Application of Trace-Norm and Low-Rank Matrix Decomposition for Computational Anatomy Nematollah Batmanghelich 1 Ali Gooya 1 Stathis Kanterakis 1 Ben Taskar 2 Christos Davatzikos 1 1 Section of Biomedical
Application of Trace-Norm and Low-Rank Matrix Decomposition for Computational Anatomy Nematollah Batmanghelich 1 Ali Gooya 1 Stathis Kanterakis 1 Ben Taskar 2 Christos Davatzikos 1 1 Section of Biomedical
Hierarchical statistical shape analysis and prediction of sub-cortical brain structures
 Medical Image Analysis xxx (2007) xxx xxx www.elsevier.com/locate/media Hierarchical statistical shape analysis and prediction of sub-cortical brain structures Anil Rao a,b, *, Paul Aljabar a, Daniel Rueckert
Medical Image Analysis xxx (2007) xxx xxx www.elsevier.com/locate/media Hierarchical statistical shape analysis and prediction of sub-cortical brain structures Anil Rao a,b, *, Paul Aljabar a, Daniel Rueckert
Introduction to General Linear Models
 Introduction to General Linear Models Moo K. Chung University of Wisconsin-Madison mkchung@wisc.edu September 27, 2014 In this chapter, we introduce general linear models (GLM) that have been widely used
Introduction to General Linear Models Moo K. Chung University of Wisconsin-Madison mkchung@wisc.edu September 27, 2014 In this chapter, we introduce general linear models (GLM) that have been widely used
Robust Tensor Splines for Approximation of Diffusion Tensor MRI Data
 Robust Tensor Splines for Approximation of Diffusion Tensor MRI Data Angelos Barmpoutis, Baba C. Vemuri, John R. Forder University of Florida Gainesville, FL 32611, USA {abarmpou, vemuri}@cise.ufl.edu,
Robust Tensor Splines for Approximation of Diffusion Tensor MRI Data Angelos Barmpoutis, Baba C. Vemuri, John R. Forder University of Florida Gainesville, FL 32611, USA {abarmpou, vemuri}@cise.ufl.edu,
Riemann Surface. David Gu. SMI 2012 Course. University of New York at Stony Brook. 1 Department of Computer Science
 Riemann Surface 1 1 Department of Computer Science University of New York at Stony Brook SMI 2012 Course Thanks Thanks for the invitation. Collaborators The work is collaborated with Shing-Tung Yau, Feng
Riemann Surface 1 1 Department of Computer Science University of New York at Stony Brook SMI 2012 Course Thanks Thanks for the invitation. Collaborators The work is collaborated with Shing-Tung Yau, Feng
Joint T1 and Brain Fiber Diffeomorphic Registration Using the Demons
 Joint T1 and Brain Fiber Diffeomorphic Registration Using the Demons Viviana Siless, Pamela Guevara, Xavier Pennec, Pierre Fillard To cite this version: Viviana Siless, Pamela Guevara, Xavier Pennec, Pierre
Joint T1 and Brain Fiber Diffeomorphic Registration Using the Demons Viviana Siless, Pamela Guevara, Xavier Pennec, Pierre Fillard To cite this version: Viviana Siless, Pamela Guevara, Xavier Pennec, Pierre
Mass Preserving Mappings and Image Registration
 Mass Preserving Mappings and Image Registration Steven Haker 1, Allen Tannenbaum 2, and Ron Kikinis 1 1 Department of Radiology, Surgical Planning Laboratory Brigham and Women s Hospital, Boston, MA 02115,
Mass Preserving Mappings and Image Registration Steven Haker 1, Allen Tannenbaum 2, and Ron Kikinis 1 1 Department of Radiology, Surgical Planning Laboratory Brigham and Women s Hospital, Boston, MA 02115,
Random walks for deformable image registration Dana Cobzas
 Random walks for deformable image registration Dana Cobzas with Abhishek Sen, Martin Jagersand, Neil Birkbeck, Karteek Popuri Computing Science, University of Alberta, Edmonton Deformable image registration?t
Random walks for deformable image registration Dana Cobzas with Abhishek Sen, Martin Jagersand, Neil Birkbeck, Karteek Popuri Computing Science, University of Alberta, Edmonton Deformable image registration?t
Non-parametric Diffeomorphic Image Registration with the Demons Algorithm
 Non-parametric Image Registration with the Demons Algorithm Tom Vercauteren, Xavier Pennec, Aymeric Perchant, Nicholas Ayache To cite this version: Tom Vercauteren, Xavier Pennec, Aymeric Perchant, Nicholas
Non-parametric Image Registration with the Demons Algorithm Tom Vercauteren, Xavier Pennec, Aymeric Perchant, Nicholas Ayache To cite this version: Tom Vercauteren, Xavier Pennec, Aymeric Perchant, Nicholas
Unsupervised Learning with Permuted Data
 Unsupervised Learning with Permuted Data Sergey Kirshner skirshne@ics.uci.edu Sridevi Parise sparise@ics.uci.edu Padhraic Smyth smyth@ics.uci.edu School of Information and Computer Science, University
Unsupervised Learning with Permuted Data Sergey Kirshner skirshne@ics.uci.edu Sridevi Parise sparise@ics.uci.edu Padhraic Smyth smyth@ics.uci.edu School of Information and Computer Science, University
Brain Lesion Segmentation: A Bayesian Weighted EM Approach
 Brain Lesion Segmentation: A Bayesian Weighted EM Approach Senan Doyle, Florence Forbes, Michel Dojat November 19, 2009 Table of contents Introduction & Background A Weighted Multi-Sequence Markov Model
Brain Lesion Segmentation: A Bayesian Weighted EM Approach Senan Doyle, Florence Forbes, Michel Dojat November 19, 2009 Table of contents Introduction & Background A Weighted Multi-Sequence Markov Model
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Lin Mei NNA54176B Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Figures: 10 # Tables: 0 # Videos: 0 This checklist is
Corresponding Author: Manuscript Number: Manuscript Type: Lin Mei NNA54176B Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Figures: 10 # Tables: 0 # Videos: 0 This checklist is
Curves in the configuration space Q or in the velocity phase space Ω satisfying the Euler-Lagrange (EL) equations,
 Physics 6010, Fall 2010 Hamiltonian Formalism: Hamilton s equations. Conservation laws. Reduction. Poisson Brackets. Relevant Sections in Text: 8.1 8.3, 9.5 The Hamiltonian Formalism We now return to formal
Physics 6010, Fall 2010 Hamiltonian Formalism: Hamilton s equations. Conservation laws. Reduction. Poisson Brackets. Relevant Sections in Text: 8.1 8.3, 9.5 The Hamiltonian Formalism We now return to formal
Consistent Linear-Elastic Transformations for Image Matching
 Consistent Linear-Elastic Transformations for Image Matching Gary E. Christensen Department of Electrical and Computer Engineering The University of Iowa, Iowa City, Iowa 52242, USA gary-christensen@uiowa.edu
Consistent Linear-Elastic Transformations for Image Matching Gary E. Christensen Department of Electrical and Computer Engineering The University of Iowa, Iowa City, Iowa 52242, USA gary-christensen@uiowa.edu
A Unified Statistical Approach to Deformation-Based Morphometry
 NeuroImage 14, 000 000 (2001) doi:10.1006/nimg.2001.0862, available online at http://www.idealibrary.com on A Unified Statistical Approach to Deformation-Based Morphometry M. K. Chung,* K. J. Worsley,*,
NeuroImage 14, 000 000 (2001) doi:10.1006/nimg.2001.0862, available online at http://www.idealibrary.com on A Unified Statistical Approach to Deformation-Based Morphometry M. K. Chung,* K. J. Worsley,*,
Dictionary Learning on Riemannian Manifolds
 Dictionary Learning on Riemannian Manifolds Yuchen Xie Baba C. Vemuri Jeffrey Ho Department of CISE, University of Florida, Gainesville FL, 32611, USA {yxie,vemuri,jho}@cise.ufl.edu Abstract. Existing
Dictionary Learning on Riemannian Manifolds Yuchen Xie Baba C. Vemuri Jeffrey Ho Department of CISE, University of Florida, Gainesville FL, 32611, USA {yxie,vemuri,jho}@cise.ufl.edu Abstract. Existing
Short Term Memory Quantifications in Input-Driven Linear Dynamical Systems
 Short Term Memory Quantifications in Input-Driven Linear Dynamical Systems Peter Tiňo and Ali Rodan School of Computer Science, The University of Birmingham Birmingham B15 2TT, United Kingdom E-mail: {P.Tino,
Short Term Memory Quantifications in Input-Driven Linear Dynamical Systems Peter Tiňo and Ali Rodan School of Computer Science, The University of Birmingham Birmingham B15 2TT, United Kingdom E-mail: {P.Tino,
NSTTUTE RESEARCH. ON POVER1YD,scWK~J~~
 340-76 NSTTUTE FOR RESEARCH ON POVER1YD,scWK~J~~ ON JENSEN"l,S METHOD FOR TWINS Arthur S. Goldberger: ON JENSEW'IS METHOD FOR TWINS Arthur S. Goldberger IYIarch 1976 I am grateful to Glen Cain, Robin Hogarth,
340-76 NSTTUTE FOR RESEARCH ON POVER1YD,scWK~J~~ ON JENSEN"l,S METHOD FOR TWINS Arthur S. Goldberger: ON JENSEW'IS METHOD FOR TWINS Arthur S. Goldberger IYIarch 1976 I am grateful to Glen Cain, Robin Hogarth,
Spatial encoding in Magnetic Resonance Imaging. Jean-Marie BONNY
 Spatial encoding in Magnetic Resonance Imaging Jean-Marie BONNY What s Qu est an image ce qu une? image? «a reproduction of a material object by a camera or a related technique» Multi-dimensional signal
Spatial encoding in Magnetic Resonance Imaging Jean-Marie BONNY What s Qu est an image ce qu une? image? «a reproduction of a material object by a camera or a related technique» Multi-dimensional signal
