Systems Analysis of Stochastic and Population Balance Models for Chemically Reacting Systems
|
|
|
- Gabriella May
- 6 years ago
- Views:
Transcription
1 Systems Analysis of Stochastic and Population Balance Models for Chemically Reacting Systems by Eric Lynn Haseltine A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY (Chemical Engineering) at the UNIVERSITY OF WISCONSIN MADISON 25
2 c Copyright by Eric Lynn Haseltine 25 All Rights Reserved
3 To Lori and Grace, for their love and support i
4 ii Systems Analysis of Stochastic and Population Balance Models for Chemically Reacting Systems Eric Lynn Haseltine Under the supervision of Professor James B. Rawlings At the University of Wisconsin Madison Chemical reaction models present one method of analyzing complex reaction pathways. Most models of chemical reaction networks employ a traditional, deterministic setting. The shortcomings of this traditional framework, namely difficulty in accounting for population heterogeneity and discrete numbers of reactants, motivate the need for more flexible modeling frameworks such as stochastic and cell population balance models. How to efficiently use models to perform systems-level tasks such as parameter estimation and feedback controller design is important in all frameworks. Consequently, this thesis focuses on three main areas: 1. improving the methods used to simulate and perform systems-level tasks using stochastic models, 2. formulating and applying cell population balance models to better account for experimental data, and 3. applying moving-horizon estimation to improve state estimates for nonlinear reaction systems. For stochastic models, we have derived and implemented techniques that improve simulation efficiency and perform systems-level tasks using these simulations. For discrete stochastic models, these systems-level tasks rely on approximate, biased sensitivities, whereas continuous models (i.e. stochastic differential equations) permit calculation of unbiased sensitivities. Numerous examples illustrate the efficiency of these methods, including an application to modeling of batch crystallization systems. We have also investigated using cell population balance models to incorporate both intracellular and extracellular levels of information in viral infections. Given experimental images of the focal infection system for vesicular stomatitis virus, we have applied these models to better understand the dynamics of multiple rounds of virus infection and the interferon (antiviral) host response. The model provides estimates of key parameters and suggests that the experimental technique may cause salient features in the data. We have also proposed an
5 efficient and accurate model decomposition that predicts population-level measurements of intracellular and extracellular species. Finally, we have assessed the capabilities of several state estimators, including movinghorizon estimation (MHE) and the extended Kalman filter (EKF). When multiple optima arise in the estimation problem, the judicious use of constraints and nonlinear optimization as employed by MHE can lead to improved state estimates and closed-loop control performance than the EKF. This improvement comes at the price of the computational expense required to solve the MHE optimization. iii
6 iv
7 v Acknowledgments Whatever you do, work at it with all your heart, as working for the Lord, not for men, since you know that you will receive an inheritance from the Lord as a reward. -Colossians 3:23-24 I first thank God, creator of heaven and earth, by whose grace I have had the opportunity to complete the work comprising this thesis. I thank my wife Lori, for her love, patience, and support. I would not have had the courage to aim so high without your encouragement. Also, the years in Madison would not have been as special without your presence. I thank my daughter Grace, who has always been able to make me smile during this past year no matter how far graduation seemed away. I am grateful to my family: my parents, Doug and Lydia, and my brother, David. Without your support and guidance through the years of my life, I would not be where I am today. I also wish to thank my in-laws, Carl and Linda Rutkowski, in particular for supporting my wife these past five years. I thank my extended church family at Mad City Church: the Billers, the Thompsons, the Smiths, the Sells, and the Konkols. In particular, I wish to acknowledge Shane and Karen Biller, who have loved, supported, and prayed for my family as if we were part of their own. There are many people in the chemical engineering department at the University of Wisconsin whom I must also acknowledge. First, I thank my advisor, Jim Rawlings, for giving me great latitude to exercise my creativity and to study interesting problems. I am always amazed by your ability to identify the important problems in a field. It has been a great honor to work with you and learn from you. I am also grateful to John Yin for first listening to my modeling ideas, then making ways for me to collaborate with his group. I am deeply indebted to Gabriele Pannocchia, who always made time to answer my questions, no matter how trivial. Since imitation is the highest form of flattery, I have tried to be as patient, kind, and understanding to my junior group members as you were to me. I could always count on either reasoning out research problems or taking a break for humor with Aswin Venkat (a.k.a. the British spy). Thank you, Matt Tenny, for your help in the office and the weight room, although perhaps I would have graduated sooner if you had not introduced me to Nethack. Brian Odelson and Daniel Patience always kept me from taking research too seriously, be it rounding everyone up for a game of darts, or getting MJ to drop by for an ice cream break. Thanks also to John Eaton for Octave and Linux support; who would have figured five years ago that I would install Linux on my laptop? It has been a pleasure getting to know Paul
8 vi Larsen, Murali Rajamani, and especially Ethan Mastny, who listened to almost all of my ideas on stochastic simulation. I also thank former Rawlings group members Jenny Wang, Scott Middlebrooks, and Chris Rao for their help during my first years in the group. Finally, I have had the great pleasure of getting to know the Yin group over the past year. In particular, I thank Vy Lam for graciously putting up with of my experimental questions. I am also grateful to Patrick Suthers and Hwijin Kim for their friendship. ERIC LYNN HASELTINE University of Wisconsin Madison February 25
9 vii Contents Abstract Acknowledgments List of Tables List of Figures ii v xiii xv Chapter 1 Introduction 1 Chapter 2 Literature Review Traditional Deterministic Reaction Models Systems Level Tasks for Deterministic Models Optimal Control State Estimation Parameter Estimation Sensitivities Stochastic Reaction Models Monte Carlo Simulation of the Stochastic Model Performing Systems Level Tasks with Stochastic Models Population Balance Models Chapter 3 Motivation Current Limitations of Stochastic Models Integration Methods Systems Level Tasks Current Limitations of Traditional Deterministic Models Current Limitations of State Estimation Techniques Chapter 4 Approximations for Stochastic Reaction Models Stochastic Partitioning Slow Reaction Subset Fast Reaction Subset The Combined System The Equilibrium Approximation
10 viii The Langevin and Deterministic Approximations Numerical Implementation of the Approximations Simulating the Equilibrium Approximation Simulating the Langevin and Deterministic Approximations: Exact Next Reaction Time Simulating the Langevin and Deterministic Approximations: Approximate Next Reaction Time Practical Implementation Examples Enzyme Kinetics Simple Crystallization Intracellular Viral Infection Critical Analysis of the Stochastic Approximations Chapter 5 Sensitivities for Stochastic Models The Chemical Master Equation Sensitivities for Stochastic Systems Approximate Methods for Generating Sensitivities Deterministic Approximation for the Sensitivity Finite Difference Sensitivities Examples Parameter Estimation With Approximate Sensitivities High-Order Rate Example Revisited Steady-State Analysis Lattice-Gas Example Conclusions Chapter 6 Sensitivity Analysis of Discrete Markov Chain Models Smoothed Perturbation Analysis Coin Flip Example State-Dependent Simulation Example Smoothing by Integration Sensitivity Calculation for Stochastic Chemical Kinetics Conclusions and Future Directions Chapter 7 Sensitivity Analysis of Stochastic Differential Equation Models The Master Equation Sensitivity Examples Simple Reversible Reaction Oregonator Applications of Parametric Sensitivities Parameter Estimation
11 7.3.2 Calculating Steady States Simple Dumbbell Model of a Polymer in Solution Conclusions Chapter 8 Stochastic Simulation of Particulate Systems Introduction Stochastic Chemical Kinetics Overview Stochastic Formulation of Isothermal Chemical Kinetics Extension of the Problem Scope Interpretation of the Simulation Output Crystallization Model Assumptions Stochastic Simulation of Batch Crystallization Isothermal Nucleation and Growth Nonisothermal Nucleation and Growth Isothermal Nucleation, Growth, and Agglomeration Parameter Estimation With Stochastic Models Trust-Region Optimization Finite Difference Sensitivities Parameter Estimation for Isothermal Nucleation, Growth, and Agglomeration Critical Analysis of Stochastic Simulation as a Modeling Tool Conclusions Chapter 9 Population Balance Models for Cellular Systems Population Balance Modeling Application of the Model to Viral Infections Intracellular Model Extracellular Events Final Model Refinements Model Solution Application to In Vitro and In Vivo Conditions In Vitro Experiment In Vivo Initial Infection In Vivo Drug Therapy Future Outlook and Impact Chapter 1 Modeling Virus Dynamics: Focal Infections Experimental System Modeling the Experiment Modeling the Measurement Analyzing and Modeling the Images Propagation of VSV on BHK-21 Cells ix
12 x Development of a Reaction-Diffusion Model Analysis of the Model Fit Propagation of VSV on DBT Cells Refinement of the Reaction-Diffusion Model Discussion Model Prediction: Infection Propagation in the Presence of Interferon Inhibitors Conclusions Appendix Chapter 11 Multi-level Dynamics of Viral Infections Modeling Framework Examples Initial Infection for a Generic Viral Infection VSV/DBT Focal Infection Model Solution Conclusions Chapter 12 Moving-Horizon State Estimation Formulation of the Estimation Problem Nonlinear Observability Extended Kalman Filtering Monte Carlo Filters Moving-Horizon Estimation Example Comparison of Results Evaluation of Arrival Cost Strategies EKF Failure Chemical Reaction Systems Example Example Computational Expense Conclusions Appendix Derivation of the MHE Smoothing Formulation Derivation of the MHE Filtering Formulation Equivalence of the Full Information and Least Squares Formulations Evolution of a Nonlinear Probability Density
13 Chapter 13 Closed Loop Performance Using Moving-Horizon Estimation Regulator Disturbance Models for Nonlinear Models Plant-model Mismatch: Exothermic CSTR Example Maximum Yield Example Conclusions Chapter 14 Conclusions 277 Bibliography 281 Vita 293 xi
14 xii
15 xiii List of Tables 2.1 Types of cell population models Model parameters and reaction extents for the enzyme kinetics example Model parameters and reaction extents for the simple crystallization example Comparison of time steps for the simple crystallization example Model parameters and reaction extents for the intracellular viral infection example Simulation time comparison for the intracellular viral infection example Parameters for the lattice-gas example Parameters for the coin flip example Parameter values for the simple reversible reaction Parameter values for the Oregonator system of reactions Parameters for the simple dumbbell model Results for the simple dumbbell model Nucleation and growth parameters for an isothermal batch crystallizer Nonisothermal nucleation and growth parameters for a batch crystallizer Nucleation, growth, and agglomeration parameters for an isothermal, batch crystallizer Parameters for the parameter estimation example Estimated parameters Model parameters for in vitro simulation Model parameters for in vivo simulation Comparison of actual and fitted parameter values for in vivo simulation of an initial infection Additional model parameters for in vivo drug therapy Parameters used to describe the experimental conditions Parameter estimates for the VSV/BHK-21 focal infection models Hessian analysis for the parameter estimates of the original VSV/BHK-21 focal infection model
16 xiv 1.4 Hessian analysis for the parameter estimates of the revised VSV/BHK-21 focal infection model Parameter estimates for the VSV/DBT focal infection models Hessian analysis for the parameter estimates of the reaction-diffusion VSV/DBT focal infection model Hessian analysis for the parameter estimates of the first segregated VSV/DBT focal infection model Hessian analysis for the parameter estimates of the second segregated VSV/DBT focal infection model Model parameters for the initial infection simulation Initial conditions and rate constants for the intracellular reactions of the VSV infection of DBT cells Initial conditions and rate constants for the reactions describing the intracellular host antiviral response of the VSV infection of DBT cells Extracellular model parameters for the infection of DBT cells by VSV Sample size required to ensure that the relative mean square error at zero is less than EKF steady-state behavior, no measurement or state noise EKF steady-state behavior, no measurement or state noise A priori initial conditions for state estimation Effects of a priori initial conditions, constraints, and horizon length on state estimation Comparison of MHE and EKF computational expense Model Steady States for a Plant with T c = 3 K, T = 35 K Maximum yield CSTR parameters
17 xv List of Figures 2.1 Microscopic volume considered in the equation of continuity for two dimensions Optimal control seeks to drive the output to set point Parameter estimation seeks to minimize the deviations between the model prediction and the data Illustration of the strong law of large numbers given a uniform distribution Illustration of the central limit theorem given a uniform distribution Computational time per simulation as a function of n Ao Extent of reaction as a function of n Ao Finite difference sensitivity for the stochastic model Cyclic nature of viral infections Comparison of the stochastic-equilibrium simulation to exact stochastic simulation Comparison of approximate tau-leap simulation to exact stochastic simulation Comparison of approximate stochastic-langevin simulation to exact stochastic simulation Comparison of exact stochastic-deterministic simulation to exact stochastic simulation Comparison of approximate stochastic-deterministic simulation to exact stochastic simulation Squared error trends for the exact and approximate stochastic-deterministic simulations Intracellular viral infections: (a) typical and (b) aborted Evolution of the template probability distribution for the (a) exact stochastic and (b) approximate stochastic-deterministic simulations Comparisons of the template probability distribution for the exact stochastic and approximate stochastic-deterministic simulations Comparison of the template mean and standard deviation for exact stochastic, approximate stochastic-deterministic, and deterministic simulations Comparison of the genome mean and standard deviation for exact stochastic, approximate stochastic-deterministic, and deterministic simulations
18 xvi 4.12 Comparison of the structural protein mean and standard deviation for exact stochastic, approximate stochastic-deterministic, and deterministic simulations Comparison of the exact, approximate, and central finite difference sensitivities for a second-order reaction Comparison of the exact and approximate sensitivities for the high-order rate example Relative error of the approximate sensitivity s with respect to the exact sensitivity s as the number of n A,o molecules increases for the high-order rate example Comparison of the exact, approximate, and finite difference sensitivity for the high-order rate example Comparison of the (a) parameter estimates per Newton-Raphson iteration and (b) model fit at iteration 2 using the approximate and finite difference sensitivities for the high-order rate example Results for the lattice-gas model Mean E[S n ] as a function of the number of coin flips n Mean sensitivity E[Sn] θ as a function of the number of coin flips n Comparison of nominal and perturbed path for SPA analysis SPA analysis of the discrete decision Illustration of the branching nature of the perturbed path for SPA analysis Mean E[n k ] as a function of the number of decisions k Mean sensitivity E[n k] θ as a function of the number of decisions k Comparison of the exact and simulated (a) mean and (b) mean integrated sensitivity for the irreversible reaction 2A B Results for the simple reversible reaction re-using the same random numbers Results for the simple reversible reaction using different random numbers Results for one trajectory of the Oregonator cyclical reactions Results for parameter estimation of the simple reversible reaction example Results for steady-state analysis of the Oregonator reaction example: estimated state per Newton iteration Method for calculating the population balance from stochastic simulation Mean of the stochastic solution for an isothermal crystallization with nucleation and growth, 1 simulation, characteristic particle size =.1, system volume V = Mean of the stochastic solution for an isothermal crystallization with nucleation and growth, average of 1 simulations, characteristic particle size =.1, system volume V = Average stochastic simulation time based on 1 simulations and V =
19 xvii 8.5 Mean of the stochastic solution for an isothermal crystallization with nucleation and growth, average of 1 simulations, characteristic particle size =.1, system volume V = Deterministic solution by orthogonal collocation for isothermal crystallization with nucleation and growth Deterministic solution by orthogonal collocation for isothermal crystallization with nucleation and growth, inclusion of the diffusivity term Total and supersaturated monomer profiles for nonisothermal crystallization Crystallizer and cooling jacket temperature profiles Mean of the exact stochastic solution for nonisothermal crystallization with nucleation and growth Mean of the approximate stochastic solution for nonisothermal crystallization with nucleation and growth, propensity of no reaction a = Deterministic solution by orthogonal collocation for nonisothermal crystallization with nucleation and growth, inclusion of the diffusivity term Zeroth moment comparisons First moment comparisons Mean of the stochastic solution for an isothermal crystallization with nucleation, growth, and agglomeration Comparison of final model prediction and measurements for the parameter estimation example Convergence of parameter estimates as a function of the optimization iteration Fit of a structured, unsegregated model to experimental results Time evolution of intracellular components and secreted virus for the intracellular model Fit of a structured, unsegregated model to experimental results Dynamic in vivo response of the cell population balance to initial infection Extracellular model fit to dynamic in vivo response of an initial infection Dynamic in vivo response to initial treatment with inhibitor drugs I 1 and I Effect of drug therapy on in vivo steady states Overview of the experimental system Measurement model Comparison of representative experimental images to model fits Comparison of the initial uninfected cell concentration for the original and revised models Comparison of representative experimental images to model fits for VSV propagation on DBT cells Comparison of intracellular production rates of virus and interferon for the segregated model of VSV propagation on DBT cells
20 xviii 1.7 Comparison of representative experimental images to model predictions for VSV propagation on DBT cells in the presence of interferon inhibitors Experimental (averaged) images obtained from the dynamic propagation of VSV on BHK-21 cells Experimental (averaged) images obtained from the dynamic propagation of VSV on DBT cells (a) Comparison of the full and decoupled model solutions for the initial infection example. (b) Percent error for the decoupled model solution, assuming the full solution is exact Schematic of modeled events for the infection of DBT cells by VSV Detailed schematic of modeled events for the up-regulation of interferon (IFN) genes Comparison of experimental data, simple segregated model fit, and the developed model Comparison of total production of virus (VSV) and interferon (IFN) per cell for the simple segregated model and intracellularly-structured, segregated model Dynamic measurement of mrna species for the focal infection system Comparison of potential point estimates (mean and mode) for (a) unimodal and (b) bimodal a posteriori distributions Example of using the kernel method to estimate the density of samples drawn from a normal distribution Example of using a histogram to estimate the density of samples drawn from a normal distribution Extended Kalman filter results Contours of P (x 1 y, y 1 ) Clipped extended Kalman filter results Moving-horizon estimation results Contours of max P (x 1, x y x, y 1 ) A posteriori density P (x 1 y, y 1 ) calculated using a Monte Carlo filter with density estimation Contours of P (x 4 y,..., y 4 ) Contours of max P (x 1,..., x 4 y x 1,...,x,..., y 4 ) with the arrival cost approximated 3 using the smoothing update Contours of max P (x 1,..., x 4 y x 1,...,x,..., y 4 ) with the arrival cost approximated 3 as a uniform prior Contours of max P (x 1,..., x 1 y x 1,...,x,..., y 1 ) with the arrival cost approximated 9 using the smoothing update Extended Kalman filter results Clipped extended Kalman filter results
21 12.16Moving-horizon estimation results Extended Kalman filter results Clipped extended Kalman filter results Moving-horizon estimation results Extended Kalman filter results Moving-horizon estimation results Extended Kalman filter results Moving-horizon estimation results Clipped extended Kalman filter results Moving-horizon estimation results Clipped extended Kalman filter results Moving-horizon estimation results General diagram of closed-loop control for the model-predictive control framework Exothermic CSTR diagram Steady states for the Exothermic CSTR example Exothermic CSTR feed disturbance Exothermic CSTR results: rejection of a feed disturbance using an output disturbance model Exothermic CSTR: Comparison of best nonlinear results to linear MPC results Maximum yield CSTR Maximum yield CSTR steady states Maximum yield CSTR: temporary output disturbance Maximum yield CSTR results xix
22 xx
23 1 Chapter 1 Introduction Chemical reaction models present one method of assimilating and interpreting complex reaction pathways. Usually a deterministic framework is employed to model these networks of chemical reactions. This framework assumes that a system evolves in a continuous, wellprescribed manner. Systems-level tasks seek to extract the maximum amount of utility from these models. Most of these tasks, such as parameter estimation and feedback control, can be posed in terms of optimization problems. For systems containing few numbers of particles, such as intracellular reaction networks, concentrations are not large enough to justify applying the usual smoothly-varying assumption made in deterministic models. Rather, there are a countably finite number of chemical species in the given system. Stochastic reaction models consider such mesoscopic phenomena in terms of discrete, molecular events that, given a cursory examination, occur in a random fashion. These stochastic simulations are merely realizations of a deterministically evolving probability distribution. Here, one must use simulation to reconstruct moments of this distribution due to the tremendous size of the probability space. The basis for these models is well established in the literature, but the methods that govern the exact simulation of these models often become computationally expensive to evaluate and hence have great room for improvement. Additionally, relatively little work has been performed in extending systems-level tasks to handling these sorts of models. Consequently, there exists a need to first formulate reasonable analogs of these traditionally deterministic tasks in a stochastic setting, and then propose methods for efficiently performing these tasks. One of the simplest, yet most intriguing biological organisms is the virus. The virus contains enough genetic information to replicate itself given the machinery of a living host. So powerful is this strategy that viral infections present one of the most potent threats to human survival and well-being. The Joint United Nations Programme on HIV/AIDS (UNAIDS) estimates that in 22, 42 million people were living with HIV/AIDS, 5 million people were newly infected with HIV, and 3.1 million people died due to AIDS related illnesses. The World Health Organization estimates that of the 17 million people currently suffering from hepatitis C, roughly one million will develop cancer of the liver during the next 1 years. In the United States alone, researchers estimate that the 5 million cases of the common cold contracted annually cost $4 billion in health care costs and lost productivity [31]. Hence there is a vi-
24 2 tal humanitarian and economic interest in systematically understanding how viral infections progress and how this progression can be controlled. Accordingly, researchers have invested significant amounts of time and money towards determining the roles that individual components such as the genome or proteins play in viral infections. As of yet, however, there exists no comprehensive picture that quantitatively incorporates and integrates data on viral infections from multiple levels. Again, models offer one manner of consolidating the vast amount of information contained across these levels, and systems-level tasks provide one method of conveniently extracting information. This dissertation considers the role of deterministic and stochastic models in assimilating dynamic data. The primary focus is on maximizing the information available from these models as well as applying such models to experimental systems. The remainder of this thesis is organized as follows: Chapter 2 reviews literature pertaining to simulation of deterministic and stochastic chemical reaction models and methods for extracting information from these simulations, such as parameter estimation and state estimation. Here, we introduce the sensitivity as a useful quantity for performing systems-level tasks. Chapter 3 provides motivation for solving the problems addressed in this thesis. Chapters 4 through 7 examine stochastic simulation with an emphasis on stochastic chemical kinetics. We present this material in the following order: In Chapter 4, we derive approximations for stochastic chemical kinetics for systems with coupled fast and slow reactions. These approximations lead to simulation strategies that result in drastic reductions of computational expense when compared to exact simulation methods. Chapter 5 considers biased approximations for calculating mean sensitivities from simulation for the stochastic chemical kinetics problem, and then applies these sensitivities to calculate steady states and estimate parameters. Chapter 6 explains how the discrete nature of the stochastic chemical kinetics formulation makes obtaining unbiased estimates of mean sensitivities difficult, then explores several techniques for calculating these unbiased estimates. Chapter 7 considers unbiased estimates for sensitivities of simulations governed by stochastic differential equations. Here, we simply differentiate the continuous samples paths to obtain the desired sensitivities, then use the sensitivities to perform useful tasks. Chapter 8 applies some of the stochastic simulation methods developed in previous chapters to solve the batch crystallization population balance. The flexibility of the simulation allows the modeler to focus on modeling the experimental system rather than the numerical methods required to solve the resulting models.
25 Chapters 9 through 11 address population balance models for viral infections. We consider the following issues: Chapter 9 derives a population balance model incorporating information from both the intracellular and extracellular levels of description. To explore the utility of this model, we compare numerical results from this model to other simpler models for experimentally relevant conditions. Chapter 1 considers modeling of experimental data from the focal infection system. This experimental system provides dynamic image data for multiple rounds of virus infection and antiviral host response. Here, we place an emphasis on determining the minimal level of modeling complexity necessary to adequately describe the experimental data. Chapter 11 proposes a decomposition technique for solving population balance models when flow of information is restricted from the extracellular to intracellular level. The goal is to efficiently and accurately solve population balance models while reconstructing population-level dynamics for intracellular and extracellular species. Chapters 12 and 13 consider one specific system-level task, namely state estimation. These chapters focus on the probabilistic formulation of the state estimation problem, in which the goal is to calculate the state estimate that maximizes the a posteriori distribution (the probability of the current state conditioned on all available experimental measurements). We examine the following topics: Chapter 12 outlines conditions for generating multiple modes in the a posteriori distribution for some relevant chemically reacting systems. We then construct examples exhibiting such conditions, and compare how several state estimators, namely the extended Kalman filter, moving-horizon estimator, and Monte Carlo filters, perform for these examples. Chapter 13 examines how multiple modes in the a posteriori distribution can affect the performance of closed-loop feedback control for different estimators. Finally, Chapter 14 presents conclusions, outlines major accomplishments, and discusses potential areas of future work. 3
26 4
27 5 Chapter 2 Literature Review Models for chemical reaction networks usually arise in a traditional, deterministic setting. Given a deterministic model, we can consider performing various systems level tasks such as parameter estimation and control. We can generally pose these tasks in terms of an optimization. In this context, a quantity known as the sensitivity becomes useful for efficient solution of the optimization. The shortcomings of the traditional deterministic framework motivate the need for alternatives that provide a more flexible foundation for chemical reaction modeling. Two such alternatives are stochastic and population balance models. This chapter presents a brief review of the modeling literature for both these subjects and the traditional models. 2.1 Traditional Deterministic Reaction Models In a deterministic setting, we perform mass balances for the reactants and products of interest using the equation of continuity. Here we define the mass of these species as a function of time (t) and the internal (y) and external (x) characteristics of the system: η(t, z)dz = mass of reactants or products (2.1) [ ] [ ] x external characteristics z = = (2.2) y internal characteristics We now consider an arbitrary, time-varying control volume V (t) spanning a space in z. This volume has a time-varying surface S(t). The normal vector n s points from the surface away from the volume, and the vector v s specifies the velocity of the surface. The vector v z specifies the velocity of material flowing through the volume. Figure 2.1 depicts a low-dimensional representation of this volume. Assuming that V (t) contains a statistically significant amount of mass, the conservation equation for the species contained in V (t) is d η(t, z)dz = R η dz dt V (t) V (t) }{{}}{{} accumulation generation S(t) F n s dω }{{} convective + diffusive flux + S(t) η(t, z)(v s n s )dω } {{ } flux due to surface motion (2.3)
28 6 z 2 S(t) v z V (t) v z n s n s v s v s z 1 Figure 2.1: Microscopic volume considered in the equation of continuity for two dimensions. in which R η refers to the production rate of the species η, F is the total flux, and dω is the differential change in the surface. Making use of the Leibniz formula permits differentiating the volume integral d η(t, z)dz = dt V (t) V (t) Substituting equation (2.4) into equation (2.3) yields V (t) η(t, z) dz + η(t, z) (v s n s ) dω (2.4) t S(t) η(t, z) dz = R η dz F n s dω (2.5) t V (t) S(t) Now apply the divergence theorem to the surface integral to obtain V (t) Combining all terms into the same integral yields η(t, z) dz = R η dz Fdz (2.6) t V (t) V (t) V (t) η(t, z) dz + F R η dz = (2.7) t Since the element V (t) is arbitrary, the argument of the integral must be zero; this result yields the microscopic equation of continuity: η(t, z) t + F = R η (2.8)
29 Equation (2.8) is the most general form of our proposed model. Both Bird, Stewart, and Lightfoot [11] and Deen [24] derive this equation without consideration of internal characteristics. We consider a time-varying control element, so our derivation is more akin to that of Deen [24]. Traditionally, one assumes that there are no internal characteristics of interest. Equation (2.8) then further reduces to: η(t, x) t + F = R η (2.9) Additionally, we can write the total flux F as the sum of convective and diffusive fluxes F = η(t, x)v x + f (2.1) We now assume that the reactor is well-stirred so that neither η nor R η depend on the external coordinates x. This assumption implies that there is no diffusive flux, i.e. f =, which yields η(t, x) t + (η(t)v x ) = R η (2.11) Next, we integrate over the time-varying reactor volume V e : η(t) + (η(t)v x ) dx = R η dx (2.12) V e t V e η(t) dx + (η(t)v x ) dx = R η dx (2.13) V e t V e V e dη V e dt + (ηv x ) dx = R η V e (2.14) V e in which we have dropped the time dependence of η for notational convenience. Applying the divergence theorem to change the volume integral to a surface integral yields dη V e dt + n e (ηv x ) dω e = R η V e (2.15) S e in which S e is the time-varying surface of the reactor volume V e, dω e is the differential change in this surface, and n e is the normal vector with respect to the surface pointing away from the reactor volume. Clearly η does not change within the reactor volume. However, changes to the surface as well as influx and outflow of material across the reactor boundary affect η as follows n e (ηv x ) dω e = n e (ηv x ) dω e,1 + n e (ηv x ) dω e,2 (2.16) S e S e,1 S e,2 }{{}}{{} flow across the reactor surface surface expansion due to reactor volume changes = q f η f qη +η dv e dt 7 (2.17)
30 8 in which q and q f are respectively the effluent and feed volumetric flow rates, and η f is the concentration of η in the feed. The resulting conservation equation is V e df dt q f η f + qη + η dv e dt = R ηv e (2.18) d(ηv e ) dt = q f η f qη + R η V e (2.19) Equation (2.19) is commonly associated with continuous stirred tank reactors (CSTR s). Alternatively, we could have derived the plug flow reactor (PFR) design equation by starting with equation (2.9) and assuming that the reactor is well mixed in only two external dimensions. 2.2 Systems Level Tasks for Deterministic Models Performing systems level tasks such as parameter estimation, model based feedback control, and process and product design requires a different set of tools than those required for pure simulation. Many systems level tasks are conveniently posed as optimization problems. We briefly review several of these tasks, namely optimal control, state estimation, and parameter estimation, and introduce the sensitivity as a useful quantity for performing these tasks Optimal Control Optimal control consists of minimizing an objective of the form min Φ = u,...,u N N (y k y sp ) T Q(y k y sp ) + (u k u sp ) T R(u k u sp ) + ( u k ) T S u k (2.2a) k= s.t. x k+1 = F (x k, u k ) (2.2b) y k = h(x k ) (2.2c) in which u k = u k u k 1, d(x k ), g(u k ) (2.2d) y k is the measurement at time t k ; u k is the input at time t k ; x k is the state at time t k ; F (x k, u k ) is the solution to a first-principles model (e.g. equation (2.19)) over the time interval [t k, t k+1 ); y sp and u sp are the measurement and input, respectively, at the desired set point; Q and R are matrices that penalize deviations of the measurement and input from set point; and S is a matrix that penalizes changes in the input.
31 In general, the optimal control problem considers an infinite number of decisions, i.e. the control horizon N is infinite. As shown in Figure 2.2, the goal of optimization (2.2) is to drive the measurements to their set points. Most control applications consist of discrete time sample, so we have formulated the model, equation (2.2b), in discrete time also. 9 u k y k k 1 Past Present k k + 1 k + 2 k Future value of control objective Figure 2.2: Optimal control seeks to drive the output to set point by minimizing deviations of both the output y and the input u from their respective set points. There is a wealth of control literature that examines the properties of the equation (2.2). For example, this formulation does not even guarantee that the controller will drive the outputs to set point. Rather, one must include additional conditions such as enforcing a terminal penalty on each optimization (i.e. y N = y sp ) or adding a terminal penalty to the final measurement y N that quantifies the cost-to-go for an infinite horizon. We refer the interested reader to the literature for additional information on this subject [119, 118, 9] State Estimation State estimation poses the problem: given a time course of experimental measurements and a dynamic model of the system, what is the most likely state of the system? This problem is usually formulated probabilistically, that is, we would like to calculate ˆx k k = arg max x k P (x k y,..., y k ) (2.21) in which x k is the state at time t k, y k is the measurement at time t k, and ˆx k k is the a posteriori state estimate of x at time t k given all measurements up to time t k. The nature of the
32 1 estimator depends greatly on the choice of dynamic model. For linear, unconstrained systems with additive Gaussian noise, the Kalman filter [144] provides a closed-form solution to equation (2.21). For constrained or nonlinear systems, solution of this equation may or may not be tractable. One computationally attractive method for addressing the nonlinear system is the extended Kalman filter, which first linearizes the nonlinear system, then applies the Kalman filter update equations to the linearized system [144]. This technique assumes that the a posteriori distribution is normally distributed (unimodal). Examples of implementations include estimation for the production of silicon/germanium alloy films [93], polymerization reactions [13], and fermentation processes [55]. However, the extended Kalman filter, or EKF, is at best an ad hoc solution to a difficult problem, and hence there exist many barriers to the practical implementation of EKFs (see, for example, Wilson et al. [163]) Parameter Estimation Parameter estimation seeks to reconcile model predictions with experimental data, as shown in Figure 2.3. In particular, we would like to maximize the probability of the mean parameter y k k 2 A + B C 25 3 Figure 2.3: Parameter estimation seeks to minimize the deviations between the model prediction (solid line) and the data (points). set θ given the measurements y k s max P Θ Y,...,Y θ N (θ y,..., y N ) (2.22) in which θ and y k are realizations of the random variables Θ and Y k, respectively. For convenience, we drop the subscript denoting the random variable unless required for clarity. We assume that the measurements y k s are generated from an underlying deterministic model
33 11 whose measurements are corrupted by noise, i.e. in which x k+1 = F (x k ; θ) (2.23) y k = h(x k ) + v k (2.24) v k N (, Π) k =,..., N (2.25) the state variables x k s are simply convenient functions of the parameters θ and the variables v k s are realizations of the normally distributed random variable ξ N (, Π) 1. Using Bayes Theorem to manipulate the joint distribution P (θ y,..., y N ) yields P (θ y,..., y N ) P (y,..., y N ) = P (y }{{},..., y N θ)p (θ) (2.26) constant P (θ y,..., y N ) P (y,..., y N θ)p (θ) (2.27) In general, P (θ) is assumed to be a noninformative prior so as not to unduly influence the estimate of the parameters. For the chosen disturbances (i.e. normally distributed), Box and Tiao show that the noninformative prior is the distribution P (θ) = constant [14]. We derive the distribution of P (y,..., y N, θ) from the known distribution P (v,..., v N, θ) in the manner described by Ross [13]. This derivation require use of the inverse function theorem from calculus [132]. First define the function mapping (v,..., v N, θ) onto (y,..., y N, θ) as We require that h(x (θ)) + v f(v,..., v N, θ) =. h(x N (θ)) + v N θ (2.28) 1. f(v,..., v N, θ) can be uniquely solved for v,..., v N and θ in terms of y,..., y N and θ. This condition is trivially true because v k = y k h(x k (θ)) k =,..., N (2.29a) θ = θ (2.29b) 2. f(v,..., v N, θ) has continuous partial derivatives at all points and the determinant of 1 The notation N (, Π) refers to a normally distributed random variable with mean and covariance Π.
34 12 its Jacobian is nonzero. The Jacobian J of equation (2.28) is J = f(v,..., v N, θ) z T = z T = I h(x (θ)) x T.... I h(x N (θ)) x T N x θ T x N θ T (2.3) I [ ] v T... v T N θt (2.31) If h(x k ) and x k are at least once continuously differentiable for all k =,..., N, then the Jacobian has continuous partial derivatives. Also, J is a block-upper triangular matrix with ones on the diagonal, so its determinant is one (nonzero). Since these conditions hold, we can calculate the distribution P (y,..., y N, θ) via Then the desired conditional is P (y,..., y N, θ) = det(j) 1 P (v,..., v N, θ) (2.32) ( N ) = P ξ (v k ) P (θ) (2.33) k= P (y,..., y N θ) = P (y,..., y N, θ) (2.34) P (θ) N = P ξ (v k ) (2.35) = k= N P ξ (y k h(x k (θ))) (2.36) k= We derive the desired optimization problem next: max θ P (θ y,..., y N ) max θ N P ξ (v k ) (2.37) k= = max log θ = max θ min θ ( N ) P ξ (v k ) k= (2.38) N log P ξ (y k h(x k (θ))) (2.39) k= N k=1 1 2 (y k h(x k )) T Π 1 (y k h(x k )) (2.4)
35 13 Therefore, this problem is equivalent to the optimization min θ Φ = 1 2 N e T k Π 1 e k k=1 e k = y k h(x k ) x k+1 = F (x k ; θ) (2.41a) (2.41b) (2.41c) We refer the reader to Box and Tiao [14] and Stewart, Caracotsios, and Sørensen [145] for a more detailed account of estimating parameters from data. Their discussion includes, for example, calculation of confidence intervals for estimated parameters Sensitivities We define the sensitivity s as s = x θ T (2.42) in which x is the state of the system and θ is a vector containing the parameters of interest for the system. This quantity is useful for efficiently performing optimization. In particular, sensitivities provide precise first-order information about the solution of the system, and this first-order information is manipulated to calculate gradients and Hessians that guide the nonlinear optimization routines. For example, consider the nonlinear optimization for parameter estimation, equation (2.41). A strict local solution to this optimization is obtained when the gradient is zero and the Hessian is positive definite. Calculating these quantities yields θ Φ = 1 θ T e T k 2 Π 1 e k (2.43) k = ( ) h(xk ) x T k x T k k θ T Π 1 e k (2.44) = ( ) h(xk ) T x T s k Π 1 e k (2.45) k k θθ Φ = θ T θφ (2.46) ( = θ T ( ) ) h(xk ) T x T s k Π 1 e k (2.47) k k = k ( h(xk ) x T k ) T s k Π 1 h(x k) x T s k + k ( h(xk ) x T k 2 x k θ k θ T k ) T Π 1 e k (2.48)
36 14 The sensitivity s clearly arises in calculation of both of these quantities. Next, we consider calculation of sensitivities for ordinary differential equations (ODE s) and differential algebraic equations (DAE s). This analysis basically summarizes the excellent work presented by Caracotsios et al. [17]. ODE Sensitivities ODE systems may be written in the following form: dx = f(x, θ) dt (2.49a) x() = x (2.49b) Accordingly, we can obtain an expression for the evolution of the sensitivity by differentiating equation (2.49a) by the parameters θ: ( ) dx θ T = f(x, θ) (2.5) dt T θ ( ) d x f(x, θ) x f(x, θ) θ dt θ T = x T + T θ θ T θ T (2.51) ds f(x, θ) f(x, θ) = dt x T s + θ T (2.52) This analysis demonstrates that the evolution equation for the sensitivity is the following ODE system: ds dt s i,j () = f(x, θ) = x T s + { 1 if x,i = θ j otherwise f(x, θ) θ T (2.53a) (2.53b) Equation (2.53) demonstrates two distinctive features about the evolution equation for the sensitivity: 1. it is linear with respect to s, and 2. it depends only on the current values of s and x. Therefore, we can solve for s by merely integrating equation (2.53) along with the ODE system (2.49). DAE Sensitivities DAE systems consider the following general form: = g(ẋ, x, θ) (2.54a) x() = x ẋ() = ẋ (2.54b) (2.54c)
37 where x is the state of the system, ẋ is the first derivative of x, and θ is a vector containing the parameters of interest for the system. Again, we define the sensitivity s by equation (2.42) and differentiate equation (2.54a) by the θ to determine an expression for the evolution of the sensitivity: = g(ẋ, x, θ) (2.55) T θ g(ẋ, x, θ) ẋ g(ẋ, x, θ) x g(ẋ, x, θ) θ = ẋ T + T θ x T + T θ θ T θ T (2.56) ( ) g(ẋ, x, θ) d x g(ẋ, x, θ) x g(ẋ, x, θ) = ẋ T dt θ T + x T + T θ θ T (2.57) g(ẋ, x, θ) ds g(ẋ, x, θ) g(ẋ, x, θ) = ẋ T + dt x T s + θ T (2.58) This analysis demonstrates that the evolution equation for the sensitivity of a DAE system yields a linear DAE system: 15 g(ẋ, x, θ) = ẋ T ṡ + { 1 if x,i = θ j s i,j () = otherwise ṡ() = ṡ g(ẋ, x, θ) g(ẋ, x, θ) x T s + θ T (2.59a) (2.59b) (2.59c) As is the case for the original DAE system (2.54), we must pick a consistent initial condition (i.e. s and ṡ must satisfy equation (2.59a)). Again, we find that we can solve for the sensitivities of the system by merely integrating equation (2.59) along with the original DAE system (2.54). 2.3 Stochastic Reaction Models When dealing with systems containing a countably finite number of molecules, deterministic models make the unrealistic assumptions that 1. mesoscopic phenomena can be treated as continuous events; and 2. identical systems given identical perturbations behave precisely the same. For example, most models of intracellular kinetics inherently examine a small number of molecules contained within a single cell (the finite number of chromosomes in the nucleus, for example), making the first assumption invalid. Additionally, identical systems given identical perturbations may elicit completely different responses. Stochastic models of chemical kinetics make no such assumptions, and hence offer one alternative to traditional deterministic models. These models have recently received an increased amount of attention from the modeling community (see, for example, [3, 91, 79]).
38 16 Stochastic models of chemical kinetics postulate a deterministic evolution equation for the probability of being in a state rather than the state itself, as is the case in the usual deterministic models. Gillespie outlines the derivation of the evolution equation for this probability distribution in depth [48]. The basis of this derivation depends on the fundamental hypothesis of the stochastic formulation of chemical kinetics, which defines the reaction parameter c µ characterizing reaction µ as: c µ dt = average probability, to first order in dt, that a particular combination of µ reactant molecules will react accordingly in the next time interval dt. We also define h µ as the number of distinct molecular reactant combinations for reaction µ at a given time, and a µ (n)dt = h µ c µ δt as the probability, first order in dt, that a µ reaction will occur in the next time interval dt. Given this fundamental hypothesis, the governing equation for this system is the chemical master equation dp (n, t) dt m = a k (n ν k )P (n ν k, t) a k (n)p (n, t) (2.6) k=1 in which n is the state of the system in terms of number of molecules (a p-vector), P (n, t) is the probability that the system is in state n at time t, a k (n)dt is the probability to order dt that reaction k occurs in the time interval [t, t + dt), and ν k is the kth column of the stoichiometric matrix ν (a p m matrix). Here, we assume that the initial condition P (n, t ) is known. The solution of equation (2.6) is computationally intractable for all but the simplest systems. Rather, Monte Carlo methods are employed to reconstruct the probability distribution and its statistics (usually the mean and variance). We consider such methods subsequently Monte Carlo Simulation of the Stochastic Model Monte Carlo methods take advantage of the fact that any statistic can be written in terms of a large sample limit of observations, i.e. h(n) 1 h(n)p (n, t)dn = lim N N N h(n i ) 1 N i=1 N h(n i ) for N sufficiently large (2.61) i=1
39 17 Mean (E[X]) Number of Samples 4 5 Figure 2.4: Illustration of the strong law of large numbers given a uniform distribution over the interval [, 1]. As the number of samples increases, the sample mean converges to the true mean of.5. in which n i is the ith Monte Carlo reconstruction of the state n. Accordingly, the desired statistic can be reconstructed to sufficient accuracy given a large enough number of observations. This statement follows as a direct result of the strong law of large numbers, which we state next. Theorem 2.1 (Strong Law of Large Numbers [13].) Let X 1, X 2,..., X n be a sequence of independent and identically distributed random variables, each having finite mean E[X i ] = m. Then, with probability 1, X X n lim = m (2.62) n n Proof: See Ross for details of the proof [13]. In this case, reconstructions of the desired statistic, i.e. h(n i ), are independent and identically distributed variables according to the common density function given by the chemical master equation (2.6). Therefore, sampling sufficiently many of these h(n i ) gives us the convergence to h(n) specified by the strong law of large numbers. We illustrate the strong law of large numbers with a simple example. Consider a uniform distribution over the interval [, 1]. This distribution has a finite mean of.5. The strong law of large numbers requires the average of samples drawn from this distribution to approach the mean with probability one. Figure 2.4 plots the average as a function of sample size; clearly this value approaches.5 as the number of samples increases. Unfortunately, the strong law of large numbers gives no indication as to the accuracy of the reconstructed statistic given a finite number of samples. An estimate for the degree of accuracy actually arises from the central limit theorem, which we state next.
40 18 Theorem 2.2 (Central Limit Theorem [13].) Let X 1, X 2,..., X n be a sequence of independent and identically distributed random variables, each having finite mean m and finite variance σ 2. Then the distribution of Z n = X X n nm σ (2.63) n tends to the standard normal as n. That is, lim P (Z n a) = 1 a e x2 /2 dx n 2π Proof: See Ross for details of the proof [13]. In this case, we now expect the reconstruction of the desired statistic, i.e. h(n), to be normally distributed assuming a large enough finite sample N. Simulating this statistic multiple times (e.g. twenty samples of h(n) reconstructed from N samples each, or 2 N total samples) permits indirect estimation of standard statistics for h(n) such as confidence intervals. How does one check whether or not the finite sample size N is large enough to justify invocation of the central limit theorem, then? Kreyszig proposes the following rule of thumb for determining this number of samples: if the skewness of the distribution is small, use at least twenty and fifty samples to reconstruct the mean and variance, respectively [75]. We can also reconstruct multiple realizations of the Z N distribution, then use statistical tests such as the Shapiro-Wilk test to test this distribution for normality [137, 131]. If these tests indicate normality, then we are free to apply the usual statistical inferences for the Z N distribution and hence obtain some measure of the accuracy of the reconstructed statistic h(n). We illustrate the central limit theorem using again the uniform density over the range [, 1]. Figure 2.5 compares the Monte Carlo reconstructed density for Z N to the standard normal distribution. For N = 1, the reconstructed density of Z N is obviously not normal; in fact, this plot merely reconstructs the underlying uniform distribution (appropriately shifted). For N = 2, the reconstructed density of Z N compares favorably to the standard normal. These statistical theorems, then, ultimately require samples to be drawn exactly from the master equation. For nontrivial examples, direct solution of the master equation is not feasible. Alternatively, one could consider an exact stochastic simulation of the fundamental hypothesis as examined by Gillespie [45]. This method examines the joint probability function, P (τ, µ)dτ, that governs when the next reaction occurs, and which reaction occurs. Here, ( ) p P (τ, µ n, t) = a µ (n) exp a k (n)τ (2.64) in which P (τ, µ n, t)dτ is the probability that the next reaction will occur in the infinitesimal time interval [t + τ, t + τ + dτ) and will be a µ reaction, given that the original state is n at time t. One can then construct numerical algorithms for simulating trajectories obeying the density (2.64). To our knowledge, no one has yet demonstrated the equivalence between the chemical master equation and stochastic simulation. The fact that these two formulas are somehow k=1
41 19 Probability Density (f(z1)) (a) Z 1 Probability Density (f(z2)) (b) 4 Z 2 Figure 2.5: Illustration of the central limit theorem given a uniform distribution over the interval [, 1]: (a) N = 1 sample and (b) N = 2 samples. Solid line plots the Monte Carlo reconstructed density. Dashed line plots the standard normal distribution. equivalent rests solely on the basis that both arise from the fundamental hypothesis. This reasoning is tantamount to the logical statements if A implies B and A implies C, then B implies C and C implies B. This reasoning is incorrect. Here, we demonstrate that one can derive equations (2.6) and (2.64) from one another. Theorem 2.3 (Equivalence of the master equation and the next reaction probability density.) Assume that P (N, t ) is known, where [ ] N = n n 1... (2.65) The probability densities generated by the chemical master equation (i.e. equation (2.6)) and the joint
42 2 density P (τ, µ n, t)dτ (i.e. equation (2.64)) are identical. Proof. If these probability densities are indeed equivalent, the evolution equations for these densities must be equivalent. Therefore we can prove this theorem by demonstrating that (1) P (τ, µ n, t)dτ gives rise to the chemical master equation and (2) the chemical master equation gives rise to P (τ, µ n, t)dτ. 1. Given P (τ, µ n, t)dτ, derive the chemical master equation. We consider propagating the marginal density P (n j, t) (dropping the conditional argument (N, t ) for convenience) from time t to the future time t + dτ. Noting that the probability of having multiple reactions occur over this time is order dτ, we have P (n j, t + dτ) =P (n j, t) + m k=1 ( 1 ) m lim P (τ, k n j, t)dτ τ k=1 (2.66) P (n j ν k, t) lim τ P (τ, k n j ν k, t)dτ + O(dτ) (2.67) Manipulating this equation gives rise to the chemical master equation: lim dτ P (n j, t + dτ) P (n j, t) = a k (n j )P (n j, t) + P (n j ν k, t)a k (n j ν k ) + O(1) (2.68) dτ P (n j, t + dτ) P (n j, t) m = lim a k (n j )P (n j, t) + P (n j ν k, t)a k (n j ν k ) + O(1) dτ dτ dp (n j, t) dt = 2. Given the chemical master equation, derive P (τ, µ n, t)dτ. k=1 (2.69) m a k (n j )P (n j, t) + P (n j ν k, t)a k (n j ν k ) (2.7) k=1 In this case, the master equation (2.6) is known. Given that the system is in state n at time t, we seek to derive the probability that the next reaction will occur at time t+τ and will be reaction µ. This statement is equivalent to specifying (a) P (n, t) = 1 and (b) no reactions occur over the interval [t, t + τ). Accordingly, the master equation reduces to the following form: d dt P (n, t ) m P (n + ν 1, t k=1 a k(n)... P (n, t ) ). = a 1 (n)... P (n + ν 1, t )...., P (n + ν m, t ) a m (n)... P (n + ν m, t ) t t t + τ (2.71)
43 in which we have now effectively conditioned each P (n, t ) on the basis that no reaction occurs over the given interval. Solving for the desired probabilities yields P (n, t ) = exp P (n + ν j, t ) = ( ) m a k (n)(t t) k=1 a j (n) m k=1 a k(n) [ 1 exp ( 21 (2.72) )] m a k (n)(t t), 1 j m (2.73) Our strategy now is to first note that P (τ, µ n, t)dτ consists of the independent probabilities k=1 P (τ, µ n, t)dτ = P (µ n, t)p (τ n, t)dτ (2.74) then solve for these marginal densities as a function of the P (n, t ) s. Conceptually, P (τ n, t)dτ is the probability that the first reaction occurs in the interval [t+τ, t+τ +dτ). We solve for this quantity by taking advantage of its relationship with P (n, t + τ) P (τ n, t)dτ = m j=1 dp (n + ν j, t ) dt t =t+τ dτ (2.75) = dp (n, t ) dt dτ (2.76) t =t+τ m = a k (n)p (n, t + τ)dτ (2.77) = k=1 ( ) m m a k (n) exp a k (n)τ dτ (2.78) k=1 As expected, P (τ n, t)dτ is independent of µ. Similarly, we express P (µ n, t) as a function of the P (n + ν j, t ) s P (µ n, t) = = = P (n + ν µ, t ) m k=1 P (n + ν k, t ) [ ( a µ (n) m k=1 a 1 exp k(n) m j=1 [ a j (n) m k=1 a 1 exp k(n) a µ (n) m k=1 a k(n) ( k=1 )] m a k (n)(t t) k=1 (2.79) )] (2.8) m a k (n)(t t) k=1 (2.81) As expected, P (µ n, t) is independent of τ.
44 22 Combining the two marginal densities, we obtain as claimed. P (τ, µ n, t)dτ = P (µ n, t)p (τ n, t)dτ (2.82) ( ) a µ (n) m m = m k=1 a a k (n) exp a k (n)τ dτ (2.83) k(n) k=1 k=1 ( ) m = a µ (n) exp a k (n)τ dτ (2.84) k=1 Theorem 2.4 (Reconstruction of the master equation density from exact simulation.) Assuming conservation of mass and a finite number of reactions, then the probability density at a single future time point t reconstructed from Monte Carlo simulations converges to the density governed by the chemical master equation almost surely over the interval [t, t]. That is, { } P lim P N(n i, t N, t ) = P (n i, t N, t ) = 1 i = 1,..., n s (2.85) N in which N is the number of exact Monte Carlo simulations, P N (n, t N, t ) is the Monte Carlo reconstruction of the probability density given N exact simulations, P (n, t N, t ) is the density governed by the master equation, and n s is the total number of possible species. Proof: We must show that { } P ψ : lim n i,n(ψ, t) = n i (ψ, t) = 1 i = 1,... n s (2.86) N in which N = [ n 1... n ns ] T, and n i,n is the Monte Carlo reconstruction of n i given N simulations. Let ɛ >. We must show that there exists an N such that if m > N, P {ψ : n i,m (ψ, t) = n i (ψ, t)} 1 < ɛ i = 1,... n s (2.87) The assumption of conservation of mass and a finite number of reactants indicates that n s is finite. Choose X i (ψ, t) = δ(ψ n i, t) (2.88) in which the random variable ψ is generated by running an exact stochastic simulation until time t. The mean of this random variable is P (n i, t N, t ). Theorem 2.3 states that any
45 simulation scheme obeying the next reaction probability density P (τ, µ n, t) generates exact trajectories from the master equation. Therefore, we can apply the strong law of large numbers, which says that there exists an N i i = 1,..., n s such that if m > N i, 23 P {ψ : X i,m (ψ, t) = P (n i, t)} 1 ɛ 2 i = 1,... n s (2.89) Let N = max i N i. Then if m > N, P {ψ : n i,m (ψ, t) = n i (ψ, t)} 1 P {ψ : n i,n (ψ, t) = n i (ψ, t)} 1 i = 1,... n s (2.9) ɛ 2 i = 1,... n s (2.91) < ɛ i = 1,... n s (2.92) Since ɛ is arbitrary, the proof is complete. In his seminal works, Gillespie proposes two simple and efficient methods for generating exact trajectories obeying the probability function P (τ, µ) [45, 46]. Theorem 2.3 proves that these trajectories obey exactly the chemical master equation (2.6). Gillespie appropriately named these algorithms the direct method and the next reaction method. We summarize these methods in algorithms 1 and 2. Algorithm 1 Direct Method. Initialize. Set the time, t, equal to zero. Set the number of species n to n. 1. Calculate: (a) the reaction rates a k (n) for k = 1,..., m; and (b) the total reaction rate, r tot = m k=1 a k(n). 2. Select two random numbers p 1, p 2 from the uniform distribution (, 1). Let τ = log(p 1 )/r tot. Choose j such that j 1 j a k (n) < p 2 r tot a k (n) k=1 k=1 3. Let t t + τ. Let n n + ν j. Go to 1. Exact algorithms such as the direct method treat microscopic phenomena as discrete, molecular events. For intracellular models, this feature is appealing because of the inherently
46 24 Algorithm 2 First Reaction Method. Initialize. Set the time, t, equal to zero. Set the number of species n to n. 1. Calculate the reaction rates a k (n) for k = 1,..., m. 2. Select m random numbers p 1,..., p m from the uniform distribution (, 1). Let τ k = log(p k )/a k (n), k = 1,..., m. Choose j such that j = arg min τ k k 3. Let t t + τ j. Let n n + ν j. Go to 1. small number of molecules contained within a single cell (the finite number of chromosomes in the nucleus, for example). As models become progressively more complex, however, these algorithms often become expensive computationally. Some recent efforts have focused upon reducing this computational load. He, Zhang, Chen, and Yang employ a deterministic equilibrium assumption on polymerization reaction kinetics [61]. Gibson and Bruck refine the first reaction method, i.e. algorithm 2, to reduce the required number of random numbers, a technique that works best for systems in which some reactions occur much more frequently than others [43]. Rao and Arkin demonstrate how to numerically simulate systems reduced by the quasi-steady-state assumption [113]. This work expands upon ideas by Janssen [69, 7] and Vlad and Pop [157] who first examined the adiabatic elimination of fast relaxing variables in stochastic chemical kinetics. Resat, Wiley, and Dixon address systems with reaction rates varying by several orders of magnitude by applying a probability-weighted Monte Carlo approach, but this method increases error in species fluctuations [126]. Gillespie examines two approximate methods, tau leaping and k α leaping, for accelerating simulations by modeling the selection of fast reactions with Poisson distributions [5]. These methods employ explicit, first-order Euler approximations that permit larger time steps to be taken than exact methods by allowing multiple firings of fast reactions by approximating the next reaction distribution. In explicit tau leaping, one chooses a fixed time step τ, then increments the state by n(t + τ) n(t) + m ν k P k (a k (n(t))τ) (2.93) k=1 in which P k (a k (n(t))τ) is a Poisson random variable with mean a k (n(t))τ. In k α leaping, one chooses a particular reaction to undergo a predetermined number of events k α, then deter-
47 mines the time τ required for these events to occur by drawing a gamma random variable Γ(a α (n), k α ). Using this value of τ, one draws Poisson random variables to determine how many events the remaining reactions undergo. A subsequent paper by Gillespie and Petzold discusses the error associated with the tau leaping approximation by using Taylor-series expansion arguments [51]. These conditions specify restrictions on the time increment τ to ensure that the error in the reconstructed mean and variance remain below a user-specified tolerance. However, this error only quantifies the effects of the reaction rate (a j (n) s) dependence upon the state n, not the effect of approximating the exact next reaction distribution with a Poisson distribution. Rathinam, Petzold, Cao, and Gillespie later present a first-order implicit version of tau leaping, i.e. 25 m m n(t + τ) n(t) + ν k a k (n(t + τ)) + ν k [P k (a k (n(t))τ) a k (n(t))] (2.94) k=1 k=1 This method has greater numerical stability than the explicit version [117] Performing Systems Level Tasks with Stochastic Models Employing kinetic Monte Carlo models for systems level tasks is an area of active research. Raimondeau, Aghalayam, Mhadeshwar, and Vlachos consider sensitivities via finite differences and parameter estimation for kinetic Monte Carlo simulations [15]. Drews, Braatz, and Alkire consider calculating the sensitivity for the mean of multiple Monte Carlo simulations via finite differences, and apply this method to copper electrodeposition to determine which parameter perturbations most significantly affect the measurements [25]. Gallivan and Murray consider model reduction techniques for the chemical master equation [39], then use the reduced models to determine optimal open-loop temperature profiles for epitaxial thin film growth [38]. Lou and Christofides consider control of growth rate and surface roughness in thin film growth [81, 82], employing proportional integral control that uses a kinetic Monte Carlo model to provide information about interactions between outputs and manipulated inputs. This simple form of feedback control does not require an optimization. Laurenzi uses a genetic algorithm to estimate parameters for a model of aggregating blood platelets and neutrophils [78]. Armaou and Kevrekidis employ a coarse time-stepper and a direct stochastic optimization method (Hooke-Jeeves) to determine an optimal control policy for a set of reactions on a catalyst surface [4]. Siettos, Armaou, Makeev and Kevrekidis use the coarse time stepper to identify the local linearization of the nonlinear stochastic model at a steady state of interest [138]. Given the local linearization of the model, standard linear quadratic control theory is then applied. Armaou, Siettos and Kevrekidis consider extending this control approach to spatially distributed processes [5]. Finally Siettos, Maroudas and Kevrekidis construct bifurcation diagrams for the mean of the stochastic models [139].
48 Population Balance Models Stochastic models of chemical kinetics pose one alternative to traditional deterministic models for modeling intracellular kinetics. Many biological systems of interest, however, consist of populations of cells influencing one another. Here, we consider the dynamic behavior of cell populations undergoing viral infections. Traditionally, mathematical models for viral infections have focused solely on events occurring in either the intracellular or extracellular level. At the intracellular level, kinetic models have been applied to examine the dynamics of how viruses harness host cells to replicate more virus [73, 27, 29, 3], and how drugs targeting specific virus components affect this replication [122, 3]. These models, however, consider only one infection cycle, whereas infections commonly consist of numerous infection cycles. At the extracellular level, researchers have considered how drug therapies affect the dynamics of populations of viruses [164, 62, 98, 13, 1]. These models, though, neglect the fact that these drugs target specific intracellular viral components. To better understand the interplay of intracellular and extracellular events, a different modeling framework is necessary. We propose cell population balances as one such framework. Mathematical models for cell population dynamics may be effectively grouped by two distinctive features: whether or not the model has structure, and whether or not the model has segregations [6]. If a model has structure, then multiple intracellular components affect the dynamics of the cell population. If a model has segregations, then some cellular characteristic can be employed to distinguish among different cells in a population. Table 2.1 summarizes the different combinations of models arising from these features. In this context, current extracellular models are equivalent to unstructured, unsegregated models because the cells in each population (uninfected and infected cells) are assumed indistinguishable from each other. Unsegregated Unstructured Most idealized case Cell population treated as one-component solute Structured Multicomponent average cell description Segregated Single component, heterogeneous individual cells Multicomponent description of cell-to-cell heterogeneity Most realistic case Table 2.1: Types of cell population models [6] The derivation of structured, segregated models stems from the equation of continuity. In particular, the derivation is identical as before up to the microscopic equation (2.8), but now considers the effect of various internal segregations upon the population behavior.
49 Fredrickson, Ramkrishna, and Tsuchiya consider the details of this derivation in their seminal contribution [36]. In recent years, this modeling framework has returned to the literature as researchers strive to adequately reconcile model predictions with the dynamics demonstrated by experimental data [8, 1, 33]. Also, new measurements such as flow cytometry offer the promise of actually differentiating between cells of a given population [1, 67], again implying the need to model distinctions between cells in a given population. 27 Notation a µ (n) c µ dt dω dω e µth reaction rate average probability to O(dt) that reaction µ will occur in the next time interval dt differential change in the control surface S(t) differential change in the reactor surface S e e k deviation between the predicted and actual measurement at time t k F total flux of the quantity η(t, z) f diffusive contribution to the total flux F h µ J m N (m, C) number of distinct molecular reactant combinations for reaction µ at a given time Jacobian mean of a probability distribution normal distribution with mean m and covariance C N matrix containing all possible molecular configurations at time t n vector of the number of molecules for each chemical species n i n e n s n s P P(m) ith Monte Carlo reconstruction of the vector n normal vector pointing from the reactor surface S e away from the volume V e normal vector pointing from the surface S(t) away from the volume V (t) total number of possible species probability random number drawn from the Poisson distribution with mean m p random number from the uniform distribution (, 1) q effluent volumetric flow rate q f R η r tot feed volumetric flow rate production rate of the species η sum of reaction rates S e S(t) s ṡ t t k V e V (t) time-varying surface of the reactor volume V e arbitrary, time-varying control volume spanning a space in z sensitivity of the state x with respect to the parameters θ first derivative of the sensitivity with respect to time time discrete sampling time time-varying reactor volume arbitrary, time-varying control volume spanning a space in z
50 28 v k v s v x v z X x x ẋ x k Y k y realization of the variable ξ at time t k velocity vector for the surface S(t) x-component of the velocity vector v z velocity vector for material flowing through the volume V (t) random variable external characteristics state first derivative of the state with respect to time state at time t k distribution for the measurement y k internal characteristics y k Z N z Γ δ η(t, z)dz Θ θ measurement at time t k random variable whose limiting distribution as N is the normal distribution internal and external characteristics random number drawn from the gamma distribution Dirac delta function mass of reactants or products distribution for the parameter set θ parameter set for a given model µ one possible reaction in the stochastic kinetics framework ν stoichiometric matrix ξ N (, Π)-distributed random variable Π covariance matrix for the random variable ξ σ standard deviation τ time of the next stochastic reaction φ objective function ψ random variable
51 29 Chapter 3 Motivation The motivation for this work is the current state of stochastic and deterministic methods used to model chemically reacting systems. For example, the rapid growth of biological measurements on the intracellular level (e.g. microarray and proteomic data) will require much more complicated models to adequately assimilate the data contained by these measurements. Therefore we seek to improve the current techniques used to evaluate and manipulate stochastic and deterministic models. In this chapter, we examine the current limitations of the existing methods for using stochastic models, traditional deterministic models, and state estimation techniques. 3.1 Current Limitations of Stochastic Models We see two primary limitations of current methods for handling stochastic models: 1. exact integration methods scale with the number of reaction events, and 2. methods for performing systems level tasks require the use of noisy finite difference techniques. We illustrate these points next Integration Methods The current options for performing exact simulation of stochastic chemical kinetics are Gillespie s direct and first reaction methods [45, 46], and the next reaction method of Gibson and Bruck [43]. Gibson and Bruck [43] analyze the computational expenditure of these methods, and find that Gillespie s methods at best scale with the number of reaction events, whereas their next reaction method scales with the log of the number of reaction events. To illustrate this point, we consider the simple reaction in which 2A k 1 k 1 B a(ɛ) = 1 2 k 1n A (n A 1) (3.1)
52 3 k 1 = 4/(3n Ao ) and k 1 =.1, ɛ is the dimensionless extent of reaction, a(ɛ) is the reaction propensity function, n A is the number of A molecules, and n Ao is the initial number of A molecules. We consider simulating this system in which there are initially zero B molecules and a variable number of A molecules. For this system, the number of possible reactions scales with n Ao. We scale rate constants for reactions with nonlinear rates so that the dimensionless extent of reaction remains constant as the variable n Ao changes. Figure 3.1 demonstrates that the computational time for one simulation scales linearly with n Ao, as expected. Time required for simulation (sec) Initial number of A molecules (n Ao ) 18 2 Figure 3.1: Computational time per simulation as a function of n Ao. Line represents the leastsquares fit of the data assuming that a simulation with n Ao = requires no computational time. The question arises, then, as to the suitability of these methods for simulating intracellular chemistry. As an example, we consider the case of a rapidly growing Escherichia coli (or E. coli) cell. For this circumstance, one E. Coli cell contains approximately four molecules of deoxyribonucleic acid (DNA), 1 molecules of messenger ribonucleic acid (mrna), and 1 6 proteins [6]. Simulating these conditions with methods that scale with the number of reaction events is clearly acceptable for modeling the DNA and mrna species, but simulating events at the protein level is not a trivial task. Now consider Figure 3.2, which plots how an intensive variable such as the extent of reaction ɛ changes as n Ao increases. This figure demonstrates that, as the number of molecules increases, the extent appears to be converging to a smoothly-varying deterministic trajectory. This simulation exhibits precisely the mathematical result proven by Kurtz: in the thermodynamic limit (n, V, n/v = constant), the master equation written for n (number
53 of molecules) collapses to the a deterministic equation for c (concentration of molecules) [76]. The appeal of the deterministic equation is that the computational time required for its solution does not scale with the simulated number of molecules. For E. Coli, such an approximation may certainly be valid for reactions among proteins, but not for those among DNA. We address this issue further in Chapter Extent of reaction Time 6 n Ao 1 n Ao 2 n Ao deterministic 8 1 Figure 3.2: Extent of reaction as a function of n Ao Systems Level Tasks A secondary issue arising from stochastic models is how to extract information from these models. Currently, most researchers merely integrate these types of models to determine the dynamic behavior of the system given a specific initial condition and inputs. As pointed out previously, this integration is potentially expensive. One recent strategy for obtaining more information from the model involves using finite difference methods to obtain estimates of the model sensitivity [15, 25], then using these sensitivities for parameter estimation and steady-state analysis. For example, we could determine the sensitivity of reaction 3.1 to the forward rate constant k 1 by evaluating the central finite difference s = n A F (k 1 + δ) F (k 1 δ) k 1 2δ (3.2) in which s is the sensitivity of the state n A with respect to the parameter k 1, F (x) yields a trajectory from a stochastic model integration given the parameter k 1 = x and n Ao initial molecules, and δ is a perturbation to the parameter k 1.
54 32 Figure 3.3 plots the perturbed trajectories and the desired sensitivity. At the smaller perturbation of δ =.2k 1, the stochastic fluctuations of the simulation dominate, yielding a noisy, poor sensitivity estimate. The larger perturbation of δ =.8k 1 yields a smoother sensitivity, but the accuracy of the central finite difference is questionable. There is obviously significant room for improvement in the methods used to calculate this quantity. We consider this issue further in Chapters 5 and 6. Additionally, little work has focused on how best to use information obtained from simulations of stochastic differential equations. Accordingly, we consider sensitivities for these types of models in Chapter 7. Finally, we apply many of the tools developed in these chapters to crystallization systems in Chapter 8. n A n A δ +δ Time δ +δ Time (a) (b) S S Figure 3.3: Finite difference sensitivity for the stochastic model: (a) small perturbation (δ =.2k 1 ) and (b) large perturbation (δ =.8k 1 ).
55 3.2 Current Limitations of Traditional Deterministic Models 33 We restrict this examination to modeling of viral infections, although the same arguments generally hold for virtually all systems involving populations of cells. Figure 3.4 generalizes the cyclic nature of viral infections. The initiation of a viral infection occurs when the virus is introduced to a host organism. The virus then targets specific uninfected host cells for infection. Once infected, these host cells become in essence factories that replicate and secrete the virus. The cycle of infection and virus production then continues. During this infection cycle, uninfected cells may continue to reproduce. This cycle is essentially the one proposed by Nowak and May [98]. Uninfected Cells Infected Cells Generation death Free Virus Figure 3.4: Cyclic nature of viral infections. These types of models usually assume that the production rate of virus is directly proportional to the concentration of infected cells. This assumption generally permits reduction of the model to a coupled set of ordinary differential equations (e.g. three ODE s to model the uninfected cell population, the infected cell population, and the virus population). This assumption is a gross simplification; in fact, many modelers have focused entirely on considering the complex chemistry required at the intracellular level to produce viral progeny [73, 27, 29, 3]. A more realistic picture of viral infections consists of a combination of the intracellular and extracellular levels. As described in Chapter 2, cell population balance models offer one means of combining these two levels. Since the literature review uncovered little active research in this area, we therefore seek to explore the utility of the cell population balance in explaining biological phenomena. We believe that refined versions of these models may lead to insights on how to best control viral propagation. We first explore the utility of the cell population balance in a numerical setting in Chapter 9, then investigate whether or not these types of models are useful in explaining actual experimental data in Chapter 1. Finally, we introduce an approximation that significantly reduces the computational expense of solving this class of models in Chapter 11.
56 Current Limitations of State Estimation Techniques It is well established that the Kalman filter is the optimal state estimator for unconstrained, linear systems subject to normally distributed state and measurement noise. Many physical systems, however, exhibit nonlinear dynamics and have states subject to hard constraints, such as nonnegative concentrations or pressures. Hence Kalman filtering is no longer directly applicable. Perhaps the most popular method for estimating the state of nonlinear systems is the extended Kalman filter, which first linearizes the nonlinear system, then applies the Kalman filter update equations to the linearized system [144]. The extended Kalman filter assumes that the a posteriori distribution is normally distributed (unimodal), hence the mean and the mode of the distribution are equivalent. Questions that arise are: how does this strategy perform when multiple modes arise in the a posteriori distribution? Also, are multiple modes even a concern for chemically reacting systems? Finally, can multiple modes in the estimator hinder closed-loop performance? We address the first two of these questions in Chapter 12, and the final question in Chapter 13. Notation a(ɛ) c k j n n A s δ ɛ reaction propensity function concentrations for all reaction species rate constant for reaction k number of molecules for all reaction species number of molecules for species A sensitivity finite difference perturbation extent of reaction
57 35 Chapter 4 Approximations for Stochastic Reaction Models 1 Exact methods are available for the simulation of isothermal, well-mixed stochastic chemical kinetics. As increasingly complex physical systems are modeled, however, these methods become difficult to solve because the computational burden scales with the number of reaction events [43]. We address one aspect of this problem: the case in which reacting species fluctuate by different orders of magnitude. We expand upon the idea of a partitioned system [113, 157] and simulation via Gillespie s direct method [45, 46] to construct approximations that reduce the computational burden for simulation of these species. In particular, we partition the system into subsets of fast and slow reactions. We make various approximations for the fast reactions (either invoking an equilibrium approximation, or treating them deterministically or as Langevin equations), and treat the slow reactions as stochastic events. Such approximations can significantly reduce computational load while accurately reconstructing at least the first two moments of the probability distribution for each species. This chapter provides a theoretical background for such approximations and outlines strategies for computing these approximations. First, we examine the theoretical underpinnings of the approximations. Next, we propose numerical algorithms for performing the simulations, review several practical implementation issues, and propose a further approximation. We then consider three motivating examples drawn from the fields of enzyme kinetics, particle technology, and biotechnology that illustrate the accuracy and computational efficiency of these approximations. Finally, we critically examine the technique and present conclusions. 4.1 Stochastic Partitioning The key ideas are to 1) model the state of the reaction system using extents of reaction as opposed to molecules of species, and 2) partition the state into subsets of fast and slow reactions. With these two modeling choices, we can exploit the structure of the chemical master equation, the governing equation for the evolution of the system probability density, by 1 Portions of this chapter appear in Haseltine and Rawlings [57].
58 36 making order of magnitude arguments. We then derive the master equations that govern the fast and slow reaction subsets. This section outlines these manipulations in greater detail. We model the state of the system, x, using an extent for each irreversible reaction 2. An extent of reaction model is consistent with a molecule balance model since n = n + ν T x (4.1) in which, assuming that there are m extents of reaction and p chemical species: x is the state of the system in terms of extents (an m-vector), n is the number of molecules (a p-vector), n is the initial number of molecules (a p-vector), and ν is the stoichiometric matrix (an m p-matrix). The upper and lower bounds of x are constrained by the limiting reactant species. We arbitrarily set the initial condition to the origin. Given assumptions outlined by Gillespie [48], the governing equation for this system is the chemical master equation dp (x; t) dt m = a k (x I k )P (x I k ; t) a k (x)p (x; t) (4.2) k=1 in which P (x; t) is the probability that the system is in state x at time t, a k (x)dt is the probability to order dt that reaction k occurs in the time interval [t, t + dt), and I k is the k th column of the (m m)-identity matrix I. The structure of I arises for this particular chemical master equation because the reactions are irreversible. Also, we have implicitly conditioned the master equation (4.2) on a specific initial condition, i.e. n. Generalizing the analysis presented in this chapter to a distribution of initial conditions (n,1,..., n,n ) is straightforward due to the relation P (x n,1,..., n,n ; t) = j P (x n,j ; t)p (n,j ) (4.3) and the fact that the values of P (n,j ) are specified in the initial condition. Now we examine the time scale over which the extents of reaction change. We must first determine a relevant time scale so that we can partition the extents into two subsets: those that have small propensity functions (a k (x) s) and occur few if any times over the time scale, and those that have large propensity functions and occur numerous times over the given time 2 Note that reversible reactions can be modeled as two irreversible reactions.
59 scale. We designate these subsets of x as the (m l)-vector y and the l-vector z, respectively. Note that [ ] [ ] y I y x = and I = z I z (4.4) in which I y and I z are (m l m l)- and (l l)-identity matrices, respectively. We also partition the reaction propensities into groups of fast (c j ) and slow (b j ) Equation (4.2) becomes a 1 (y, z; t) b 1 (y, z; t).. a m l (y, z; t) b m l (y, z; t) = a m l+1 (y, z; t) c 1 (y, z; t).. a m (y, z; t) c l (y, z; t) 37 (4.5) dp (y, z; t) dt m l = b j (y I y j, z)p (y Iy j, z; t) b j(y, z)p (y, z; t) j=1 l + c k (y, z I z k )P (y, z Iz k ; t) c k(y, z)p (y, z; t) (4.6) k=1 Ultimately, we are interested the determining an approximate governing equation for the evolution of the joint density, P (y, z; t), in regimes where fast reaction extents are much greater than slow reaction extents. Denoting the total extent space as X, we define a subspace X p X for which [ ] y c k (y, z) b j (y, z) 1 k l, 1 j m l, X p (4.7) z By defining the conditional and marginal probabilities over this subspace as [ ] y P (y, z; t) = P (z y; t)p (y; t) X p (4.8) z P (y; t) = [ ] y P (y, z; t) X p (4.9) z z we can alternatively derive evolution equations for both the marginal probability of the slow reactions, P (y; t), and the probability of the fast reactions conditioned on the slow reactions, P (z y; t). Consequently, we then know how the fast and slow reactions evolve over this time scale. Also, this partitioning is similar to that used by Rao and Arkin [113], who partition the master equation by species to treat the quasi-steady-state assumption. We partition by reaction extents to treat fast and slow reactions.
60 38 All the manipulations performed in the next two subsections apply only for fast and slow reactions in the partitioned subspace X p. To simplify the presentation of the results, we drop the implied notation from all subsequent equations. [ y z ] X p Slow Reaction Subset We first address the subset of slow reaction extents y. From the definition of the marginal density, P (y; t) = P (y, z; t) (4.1) z Differentiating equation (4.1) with respect to time yields dp (y; t) dt = z dp (y, z; t) dt (4.11) Now substitute the master equation (4.6) into equation (4.11) and manipulate to yield dp (y; t) = m l b j (y I y j dt, z)p (y Iy j, z; t) b j(y, z)p (y, z; t) z j=1 ) l + c k (y, z I z k )P (y, z Iz k ; t) c k(y, z)p (y, z; t) = = + k=1 z ( z m l b j (y I y j, z)p (y Iy j, z; t) b j(y, z)p (y, z; t) j=1 ) l c k (y, z I z k )P (y, z Iz k ; t) c k(y, z)p (y, z; t) k=1 (4.12) (4.13) } {{ } m l b j (y I y j, z)p (y Iy j, z; t) b j(y, z)p (y, z; t) (4.14) z j=1 Equation (4.14) is exact; we have made no approximations in its derivation. Also, if we rewrite the joint density in terms of the conditional density using the definition P (y, z; t) = P (z y; t)p (y; t) (4.15) then one interpretation of this analysis is that the evolution of the marginal P (y; t) depends on the conditional density P (z y; t). We consider deriving an evolution equation for this conditional density next.
61 Fast Reaction Subset We now address the evolution of the probability density for the subset of fast reactions conditioned on the subset of slow reactions, P (z y; t). For our starting point, we use order of magnitude arguments, i.e. equation (4.7), to approximate the original master equation (4.6) as dp (y, z; t) dt l c k (y, z I z k )P (y, z Iz k ; t) c k(y, z)p (y, z; t) (4.16) k=1 We define this approximate joint density as P A (y, z; t), and thus its evolution equation is dp A (y, z; t) dt l c k (y, z I z k )P A(y, z I z k ; t) c k(y, z)p A (y, z; t) (4.17) k=1 Following Rao and Arkin [113], we define the joint density P A (y, z; t) as the product of the desired conditional density P A (z y; t) and the marginal density P A (y; t): Differentiating equation (4.18) with respect to time yields dp A (y, z; t) dt P A (y, z; t) = P A (z y; t)p A (y; t) (4.18) = dp A(z y; t) dt Solving equation (4.19) for the desired conditional derivative yields dp A (z y; t) dt ( 1 dpa (y, z; t) = P A (y; t) dt P A (y; t) + dp A(y; t) P A (z y; t) (4.19) dt dp ) A(y; t) P A (z y; t) dt (4.2) Evaluating the marginal evolution equation by summing equation (4.17) over the fast extents z yields dp A (y; t) dt = z l c k (y, z I z k )P A(y, z I z k ; t) c k(y, z)p A (y, z; t) (4.21) k=1 = (4.22) Consequently, equation (4.19) becomes dp A (z y; t) dt ( l ) 1 = c k (y, z I z k P A (y; t) )P A(y, z I z k ; t) c k(y, z)p A (y, z; t) = k=1 (4.23) l c k (y, z I z k )P A(z I z k y; t) c k(y, z)p A (z y; t) (4.24) k=1 which is the desired closed-form expression for the conditional density P A (z y; t).
62 The Combined System For the slow reactions, we approximate the joint density P (y, z; t) as P (y, z; t) P A (z y; t)p (y; t) (4.25) Combining the evolution equations for the slow and fast reaction extents, i.e. equations (4.14) and (4.24) respectively, then yields the following coupled master equations dp (y; t) dt dp A (z y; t) dt ( ) ( m l b j (y I y j, z)p A(z y I y j ; t) P (y I y j ; t) z = j=1 z l c k (y, z I z k)p A (z I z k y; t) c k (y, z)p A (z y; t) k=1 ) b j (y, z)p A (z y; t) P (y; t) (4.26a) (4.26b) From these equations, using order of magnitude arguments to produce a time-scale separation has clearly had two effects: first, the coupled expressions for the marginal and conditional evolution equations in (4.26) are Markov in nature; and second, the evolution equation for the fast extents conditioned on the slow extents, P A (z y), has decoupled from the slow extent marginal, P (y). Additionally, exact solution of the coupled master equations (4.26) is at least as difficult as the original master equation (4.2) due to the fact that one must solve an individual master equation of the form of equation (4.26b) for every element of the slow conditional equation (4.26a). From a simulation perspective, equation (4.26) is also as difficult to evaluate as the original master equation (4.2) since both of the coupled master equations are discrete and time-varying. However, approximating the fast extents can significantly reduce the computational expense involved with simulating these coupled equations. Different approximations are applicable based on the characteristic relaxation times of the fast and slow extents. Next, we investigate two such approximations: an equilibrium approximation for the case in which the fast extents relax significantly faster than the slow extents, and a Langevin or deterministic approximation for the case in which both fast and slow extents relax at similar rates The Equilibrium Approximation We first consider the case in which the relaxation time for the fast extents is significantly smaller than the expected time to the first slow reaction. To illustrate this case, we consider the simple example A k 1 k2 B k 3 C (4.27)
63 We denote the extents of reaction for this example as ɛ 1, ɛ 2, and ɛ 3, and define the reaction propensities as 41 a 1 (x) = k 1 n A a 2 (x) = k 2 n B a 3 (x) = k 3 n C (4.28a) (4.28b) (4.28c) If k 1, k 2 k 3, then we can partition ɛ 1 and ɛ 2 as the fast reactions z, and ɛ 3 as the slow extent of reaction y. Additionally, we would expect the fast extents of reaction to equilibrate (relax) before the expected time to the first slow reaction. Returning to the master equation formalism, this equilibration implies that we should approximate the fast reactions, equation (4.26b), as l c k (y, z I z k )P A(z I z k y; t) c k(y, z)p A (z y; t) (4.29) k=1 The resulting coupled master equations are ( ) ( m l dp (y; t) b j (y I y j dt, z)p A(z y I y j ; t) P (y I y j ; t) z z = j=1 l c k (y, z I z k)p A (z I z k y; t) c k (y, z)p A (z y; t) k=1 b j (y, z)p A (z y; t) ) P (y; t) (4.3a) (4.3b) This coupled system, equation (4.3), is markedly similar to the governing equations for the slow-scale simulation recently proposed by Cao, Gillespie, and Petzold [16]. Their derivation deviates from ours, however, and the differences deserve some attention. First, Cao, Gillespie, and Petzold [16] partition on the basis of fast and slow species rather than extents, with fast species affected by at least one fast reaction and slow species affected by solely slow reactions. We have chosen to remain in the extent space because extents are equilibrating, not chemical species. Also, Cao, Gillespie, and Petzold [16] use the construct of a virtual fast system to arrive at an evolution equation for the slow species (similar to our evolution equation for the slow extent marginal, equation (4.14)), a choice that obviates the need for defining an evolution equation for the conditional density P (z y). In contrast to this approach, we believe that our approach has a much tighter connection to the original master equation due to the fact we derived the coupled system, equation (4.3), directly from the the original master equation, and the fact that we can obtain an approximate value of the joint density P (y, z; t) through equation (4.25). Also, all approximations arise directly from order of magnitude and relaxation time arguments The Langevin and Deterministic Approximations We now consider the case in which both fast and slow extents relax at similar time scales. Revisiting the reaction example 4.27, we consider the case in which k 1 k 2, k 3 and n Ao
64 42 n Bo, n Co in which the notation n Ao refers to the initial number of A molecules. For this example, we partition ɛ 1 as the fast extent of reaction z, and ɛ 2 and ɛ 3 as the slow extents of reaction y. Until a significant amount of A has been consumed, we would expect numerous firings of ɛ 1 interspersed with relatively few firings of ɛ 2 and ɛ 3. Clearly the system never equilibrates, but rather fast and slow reactions fire until the fast extent reaches a similar order of magnitude as one of the slow extents. Note also that, in contrast to the equilibrium approximation, we have introduced the number of molecules into the time-scale argument. For most cases, we expect this time-scale argument to involve large numbers of reacting molecules, but such involvement is not always the case as demonstrated in the viral infection example presented later in this chapter. Rather, we require that the magnitude of the fast reactions remain large relative to the magnitude of the slow reactions through the expected time of the first slow reaction. Returning to the master equation formalism, this process requires a different approximation for the conditional density P (z y). We proceed by demonstrating as outlined by Gardiner [41] how this subset can be approximated using the Langevin approximation. Define the characteristic size of the system to be Ω, and use this size to recast the master equation (4.24) in terms of intensive variables (let z z/ω). Performing a Kramers-Moyal expansion on this master equation results in a system size expansion in Ω. In the limit as z and Ω become large, the discrete master equation (4.26b) can be approximated by its first two differential moments with the continuous Fokker-Planck equation P A (z y; t) t = l i=1 z i (A i (y, z)p A (z y; t))+ 1 2 l l i=1 j=1 2 z i z j ( Bij (y, z) 2 P A (z y; t) ) (4.31) in which (noting that z consists of extents of reaction): A(y, z) = [B(y, z)] 2 = l I z i c i (y, z) = i=1 [ c 1 (y, z) c 2 (y, z) c l (y, z)] T (4.32) l I z i (I z i ) T c i (y, z) = diag (c 1 (y, z), c 2 (y, z),..., c l (y, z)) (4.33) i=1 Here, diag(a,..., z) defines a matrix with elements a,..., z on the diagonal. Equation (4.31) has Itô solution of the form dz i = A i (y, z)dt + l B ij (y, z)dw j 1 i l (4.34a) j=1 = c i (y, z)dt + c i (y, z)dw i 1 i l (4.34b) in which W is a vector of Wiener processes. Equation (4.34) is the chemical Langevin equation, whose formulation was recently readdressed by Gillespie [49]. Note the difference between equations (4.31) and (4.34). The Fokker-Planck equation (4.31) specifies the distribution of the stochastic process, whereas the stochastic differential equation (4.34) specifies how the trajectories of the state evolve. Also, bear in mind that whether or not a given Ω is large
65 enough to permit truncation of the system size expansion is relative. In this case, Ω is of sufficient magnitude to make this approximation valid for only a subset of the reactions, not the entire system. Combining the evolution equations for the slow and fast reaction extents, i.e. equations (4.26a) and (4.31) respectively, the problem of interest is the coupled set of master equations 43 dp (y; t) dt P A (z y; t) t m l ( k=1 ( = z z b k (y I y k, z k )P A(z k y Iy k ; t)dz ) P (y I y k ; t) b k (y, z )P A (z y; t)dz ) P (y; t) (4.35a) l i=1 z i (A i (y, z)p A (z y; t)) l l i=1 j=1 2 ( Bij (y, z) 2 P A (z y; t) ) z i z j (4.35b) If we can solve these equations simultaneously, then we in fact have an approximate solution to the original master equation (4.6) due to the definition of the conditional density given by equation (4.25). Note that the solution is approximate due to the fact that we have used the Fokker-Planck approximation for the master equation of the fast reactions. In the thermodynamic limit (z, Ω, z = z/ω = finite), the intensive variables for the fast subset of reactions (z s) evolve deterministically [76]. Accordingly, we propose further approximating the Langevin equation (4.34) as In this case, the coupled master equations (4.35) reduce to dp (y; t) dt m l dz i = c i (y, z)dt 1 i l (4.36) b k (y I y k, z(t))p (y Iy k ; t) b k(y, z(t))p (y; t) k=1 (4.37a) dz i =c i (y, z)dt 1 i l (4.37b) in which z(t) is the solution to the differential equation (4.36). The benefit of this assumption is that equation (4.36) can be solved rigorously using an ODE solver. Unfortunately for physical systems, the thermodynamic limit is obviously unattainable. However, knowledge of the modeled system can lead to this simplification. If the magnitude of the fluctuations in this term is small compared to the sensitivity of c i (y, z) to the subset y, then equation (4.36) is a valid approximation. This approximation is also valid if one is primarily concerned with the fluctuations in the small-numbered species as opposed to the large-numbered species, assuming that the extents approximated by equation (4.36) predominantly affect the population size of large-numbered species.
66 Numerical Implementation of the Approximations We now outline procedures for implementing the equilibrium, Langevin, and deterministic approximations presented in the previous section. We propose using simulation to reconstruct moments of the underlying master equation. For the slow reactions, Gillespie [47] outlines a general method for exact stochastic simulation that is applicable to the desired problem, equation (4.26a). This method examines the joint probability function, P (τ, µ), that governs when the next reaction occurs, and which reaction occurs. We present a brief derivation of this function. We proceed by noting that the key probabilistic questions are: when will the next reaction occur, and which reaction will it be [45]? For this end, we define { b µ (y, z; t)dt = z b µ(y, z)p A (z y; t)dt equilibrium approximation z b µ(y, z )P A (z y; t)dz dt Langevin or deterministic approximation (4.38) in which b µ (y, z; t)dt is the probability (first order in dt) that reaction µ occurs in the next time interval dt. We express the joint probability P (τ, µ)dτ as the product of the independent probabilities P (τ, µ)dτ = P (τ)p (µ)dτ (4.39) in which P (τ) is the probability that no reaction occurs within [t, t + τ), and P (µ)dτ is the probability that reaction µ takes place within [t + τ, t + τ + dτ). To determine P (τ), consider the change in this probability over the differential increment in time dt, assuming that probabilities are independent over disjoint periods of time [68]: m l P (τ + dt) = P (τ) 1 b j (y, z; t + τ)dt (4.4a) j=1 = P (τ)(1 r y tot (t)dt) (4.4b) Here, r y tot (t) is the sum of reaction rates for subset y at time t. Rearranging equation (4.4a) and taking the limit as dt yields the differential equation dp (τ) = r y tot dt (t)p (τ) (4.41) which has solution ( P (τ) = exp The joint probability function P (τ, µ) is therefore: t+τ P (τ, µ) = b µ (y, z; t + τ) exp t r y tot (t )dt ) ( t+τ t r y tot (t )dt ) (4.42) (4.43)
67 We now address our key questions by conditioning the joint probability function P (τ, µ): P (τ, µ) = P (µ τ)p (τ) (4.44) in which P (τ) is the probability that a reaction occurs in the differential instant after time t+τ, and P (µ τ) is the probability that this reaction will be µ. First note that by definition: P (τ) = 45 l P (τ, µ) (4.45) µ=1 Implicit in this equation is the assumption that a reaction occurs, and hence the probability of not having a reaction is zero. Then by rearranging equation (4.44) and incorporating (4.45), it can be deduced that: P (µ τ) = P (τ, µ) m l µ=1 P (τ, µ) (4.46) Equation (4.46) can be solved exactly by employing equation (4.43) to yield: P (µ τ) = b µ (y, z; t + τ) m l j=1 b j(y, z; t + τ) We then solve equation (4.45) by employing equation (4.43): m l ( t+τ ) P (τ) = b j (y, z; t + τ) exp r y tot (t )dt j=l t t+τ ) = r y tot ( (t + τ) exp r y tot (t )dt t (4.47) (4.48a) (4.48b) Using Monte Carlo simulation, we obtain realizations of the desired joint probability function P (τ, µ) by randomly selecting τ and µ from the probability densities defined by equations (4.48b) and (4.47). Such a method is the equivalent of the direct method for hybrid systems. Given two random numbers p 1 and p 2 uniformly distributed on (, 1), τ and µ are constrained accordingly: µ 1 k=l+1 t+τ t r y tot (t )dt + log(p 1 ) = b k (y, z; t + τ) < p 2 r y tot (t + τ) µ k=l+1 b k (y, z; t + τ) (4.49a) (4.49b) Simulating the different approximations require slightly different algorithms, which we address next.
68 Simulating the Equilibrium Approximation We first address the equilibrium approximation. For this case, b j (y, z; t) = z b j (y, z)p A (z y; t) 1 j m l (4.5) Additionally, the quantities b j (y, z; t) are actually time invariant between slow reactions. Thus, the integral constraint (4.49a) reduces to the algebraic relation τ = log(p 1) r y tot (t) (4.51) Algorithm 3 Exact solution of the partitioned stochastic system for the equilibrium approximation. Off-line. Partition the set x of m extents of reaction into fast and slow extents. Determine the partitioned stoichiometric matrices (the (m l p)-matrix ν y and the (l p)-matrix ν z ) and the reaction propensity laws (a y k (y, z) s). Also, choose a strategy for solving the distribution P A(z y) given by equation (4.3) for the fast reactions in the partitioned case. Initialize. Set the time, t, equal to zero. Set the number of species n to n. 1. Solve for the distribution P A (z y), denoting all possible combinations of z as (z(),..., z(t)). Record the initial value of z as z(i). 2. For subset y, calculate (a) the reaction propensities, b j (y, z) = z b j(y, z)p A (z y) (b) the total reaction propensity, r y tot = m l k=1 b j(y, z). j = 1,..., m l, and 3. Select three random numbers p 1, p 2, and p 3 from the uniform distribution (, 1). 4. Choose z(j) from the distribution P A (z y) such that Set ˆν z = z(j) z(i). j 1 P A (z(k) y) < p 1 k=1 5. Let τ = log(p 2 )/r y tot. Choose j such that j 1 b k (y, z) < p 3 r y tot k=1 j P A (z(k) y) k=1 j b k (y, z) k=1 6. Let n n + ( ν y j ) T + ˆνz, where ν y j is the jth row of ν y. Go to step 1. Algorithm 3 presents one method of solving this system. Note that we could draw a sample from the equilibrium distribution P A (z y) at any time to determine a current value of
69 the state, which may be desirable for sampling the system at uniform time increments. Also, this algorithm is very similar to the slow-scale stochastic simulation algorithm proposed by Cao, Gillespie, and Petzold [16], with the exception that our algorithm partitions extents as opposed to species. Solution of the equilibrated density P A (z y) deserves some further attention. If we stack probabilities for all possible values of the fast extents into a vector P, we can recast the continuous-time master equation as a vector-matrix problem, i.e. 47 dp dt = AP (equilibrium assumption) (4.52) in which A is the matrix of reaction propensities. The equilibrium distribution is then the null space of the matrix A, which we can compute numerically. In general, we expect A to be a sparse matrix. Consequently, we can efficiently solve the linear system (4.52) for P using Krylov iterative methods [153] such as the biconjugate gradient stabilized method. Cao, Gillespie, and Petzold [16] outline some alternative, approximate methods for evaluating this equilibrated density Simulating the Langevin and Deterministic Approximations: Exact Next Reaction Time We now address methods for simulating the Langevin and deterministic approximations. These approximations have time-varying reaction propensities, so we must satisfy equation (4.49a) by integrating r y tot and the fast subset of reactions z forward in time until the following condition is met: b j (y, z; t) = t+τ t z r y tot (t )dt + log(p 1 ) = (4.53) m l r y tot (t) = b j (y, z; t) (4.54) j=l b j (y, z )P A (z y; t)dz 1 j m l (4.55) For the Langevin approximation, we propose reconstructing the density P A (z y; t) by simulating the stochastic differential equation (4.34) (also known as the Langevin equation). In this case, equation (4.55) becomes b j (y, z; t) 1 N N b j (y, z k ) 1 j m l (4.56) k=1 in which z k is the kth of N simulations of equation (4.34). For the deterministic approximation, equation (4.37) indicates that we need only solve for the deterministic evolution of the fast extents. We propose using algorithm 4 to solve this partitioned reaction system, in which we
70 48 Algorithm 4 Exact solution of the partitioned stochastic system for the Langevin and deterministic approximations. Off-line. Determine the criteria for when and how the set x of m extents of reaction should be partitioned. Determine the stoichiometric matrices of the form given in equation (4.1) and reaction propensity laws for the unpartitioned (the (m p)-matrix ν and a k (x) s) and partitioned cases (the (m l p)- matrix ν y, the (l p)-matrix ν z, and a y k (y, z) s). Also, determine the necessary Langevin or deterministic equations for the fast reactions in the partitioned case. Initialize. Set the time, t, equal to zero. Set the number of species n to n. 1. If the partitioning criteria established off-line are met, go to step Calculate (a) the reaction propensities, r k = a k (x), and (b) the total reaction propensity, r tot = m k=1 r k. 3. Select two random numbers p 1, p 2 from the uniform distribution (, 1). Let τ = log(p 1 )/r tot. Choose j such that j 1 j r k < p 2 r tot k=1 4. Let t t + τ. Let n n + ν T j, where ν j is the j th row of ν. Go to step For subset y, calculate (a) the reaction propensities, r y k = by k (y, z), and (b) the total reaction propensity, r y tot = m l k=1 ry k. 6. Select two random numbers p 1, p 2 from the uniform distribution (, 1). 7. Determine ˆν z = (ν z ) T [z(t + τ) z(t)] by integrating r y tot (t) and the subset of fast reactions z until the following condition is met: k=1 r k t+τ t r y tot (t )dt + log(p 1 ) = m l s.t. : r y tot (t) = a y k (y, z; t) k=1 8. Let t t + τ. Let n n + ˆν z. 9. Choose j such that j 1 r y k < p 2r y tot (t) Current values of the r y k s and ry tot should be available from step Let n n + ( ν y ) T j, where ν y j is the jth row of ν y. Go to step 1. k=1 j k=1 r y k
71 choose only to use one simulation to evaluate equation (4.56) for the Langevin case. Using more than one simulation to evaluate equation (4.56) for the Langevin case is also possible. Over the time interval τ, implementation of this algorithm actually enforces the more stringent requirement that dp (y) = (4.57) dt Hence equation (4.22) is exact, not approximate Simulating the Langevin and Deterministic Approximations: Approximate Next Reaction Time One major difficulty in this method is satisfying the constraint t+τ t r y tot (t )dt + log(p 1 ) = (4.58) in step 7 of the algorithm 4 as opposed to the simple algebraic relation for τ used in the unmodified Gillespie algorithm (i.e. step 3 of algorithm 4). This constraint can prove to be computationally expensive. If the reaction propensities for the fast subset of extents z change insignificantly over the stochastic time step τ, the unmodified Gillespie algorithm can still provide an approximate solution. When the reaction propensities change significantly over τ, steps can be taken to reduce the error of the Gillespie algorithm. One idea is to scale the stochastic time step τ by artificially introducing a probability of no reaction into the system: Let a dt be the contrived probability, first order in dt, that no reaction occurs in the next time interval dt. This probability does not affect the number of molecules of the modeled reaction system while allowing adjustment of the stochastic time step by changing the magnitude of a. Theoretically, as the magnitude of a becomes infinite, the total reaction rate becomes infinite. As the total reaction rate approaches infinity, the error of the stochastic simulation subject to constraints approaches zero because the algorithm checks whether or not a reaction occurs at every time. Even though the method outlined by Gillespie [47] and Jansen [68] is exact, for this case there is still error associated with 1) the number of simulations performed since it is a Monte Carlo method, and 2) integration of the Langevin equations for the fast extents of reaction. Thus it is plausible that these errors may be greater than the error introduced by the approximation. Hence our approximation may often prove to be less computationally expensive than the exact simulation while generating an acceptable amount of simulation error. The approximation modifies steps 5-1 of the algorithm 4 with those given by algorithm 5.
72 5 Algorithm 5 Approximate solution of the partitioned stochastic system. 5. For subset y, calculate (a) the reaction propensities, r y k = by k (y, z), and (b) the total reaction propensity, r y tot = m l k= ry k. 6. Select two random numbers p 1, p 2 from the uniform distribution (, 1). 7. Let τ = log(p 1 )/r tot. Integrate subset z over the range [t, t + τ) to determine ˆν z = (ν z ) T [z(t + τ) z(t)]. Let t t + τ. Let n n + ˆν z. 8. Recalculate the reaction propensities r y k s and the total reaction propensity ry tot (t). Choose j such that j 1 j r y k < p 2r y tot (t) ( 9. Let n n + Go to step 1. ν y j k= ) T, where ν y j is the jth row of ν y. k= r y k 4.3 Practical Implementation Partitioning of the state x into fast and slow extents should be intuitive. We recommend maintaining at least two orders of magnitude difference between the values of the partitioned reaction propensities. It may also be helpful to generate results for a full stochastic simulation, and then identify which reactions are bottlenecks (i.e. ones occurring most frequently). Note that there may exist several regimes that require different partitioning of the state. Also, care should be exercised to maintain the validity of the order of magnitude partition between y and z. It is obviously undesirable for slow reaction extents to become the same order of magnitude of the fast extents during the time increment τ. Finally, nothing precludes one from invoking the equilibrium approximation for one subset of fast reactions, and the deterministic or Langevin approximation for another subset of reactions. We did not carry out such an analysis for notational simplicity. 4.4 Examples We now consider three motivating examples that illustrate the accuracy of the approximations. For clarity, we first briefly review the nomenclature that indicates which approximations, if any, are performed in a given simulation. We can either perform a purely stochastic simu-
73 51 Parameter Symbol Value reaction propensity 4.59a a 1 (x) k 1 n E n S reaction propensity 4.59b a 2 (x) k 2 n ES reaction propensity 4.59c a 3 (x) k 3 n ES reaction 4.59a rate constant k 1 2. reaction 4.59b rate constant k 2 2. reaction 4.59c rate constant k 3 1. initial number of E molecules n Eo 2 initial number of S molecules n So 1 initial number of ES molecules n ESo initial number of P molecules n P o Table 4.1: Model parameters and reaction extents for the enzyme kinetics example lation on the unpartitioned reaction system, or we can partition the system into fast and slow reactions. For this partitioned case, a stochastic-equilibrium simulation equilibrates the fast reactions, a stochastic-langevin simulation treats the fast reactions as Langevin equations, and a stochastic-deterministic simulation treats the fast reactions deterministically. We can then simulate this partitioned reaction system by exact simulation, in which the next reaction time exactly accounts for the time dependence of the fast reactions upon the slow reactions; or by an approximate simulation, which neglects this time dependence but scales the next reaction time with a propensity of no reaction. For comparison to other approximate techniques, we simulate the simple crystallization example using implicit tau leaping. In contrast to the partitioning techniques proposed here, tau leaping approximates the number of times every reaction fires in a fixed time interval using a rate-dependent Poisson distribution. The details of this method are presented in Chapter Enzyme Kinetics We consider the simple enzyme kinetics problem E + S k 1 ES ɛ 1 (4.59a) ES k 2 E + S ɛ 2 (4.59b) ES k 3 E + P ɛ 3 (4.59c) The model parameters and the reaction extents are given in Table 4.1. For this example, the first and second reactions equilibrate before the expected time of one third reaction. Hence we partition the extents of reaction (ɛ i s) as follows: ɛ 3 comprises the subset of slow reactions y, and ɛ 1 and ɛ 2 comprise the subset of fast reactions z.
74 52 2 Number of Molecules S ES P E 2 4 Time Figure 4.1: Comparison of the stochastic-equilibrium simulation (dashed lines) to exact stochastic simulation (solid lines) based on 5 simulations. We calculate the averages of all species using fifty simulations sampled at a time interval of.1 units. We use both the stochastic-equilibrium and exact simulations to compute these averages. For the stochastic-equilibrium simulation, solving for the equilibrium distribution in equation (4.52) is easiest if one treats the fast reactions ɛ 1 and ɛ 2 as one extent. Figure 4.1 presents the results of the comparison. The stochastic-equilibrium simulation provides an excellent reconstruction of the mean behavior. The exact simulation requires roughly twentythree times the amount of computational expense as the stochastic-equilibrium simulation. We refer the interested reader to Cao, Gillespie, and Petzold [16] for additional examples and discussion on the equilibrium approximation. While their derivation of the equilibrium approximation differs from ours, their simulation algorithm is very similar to our algorithm Simple Crystallization Consider a simplified reaction system for the crystallization of species A: 2A k 1 B ɛ 1 (4.6a) A + C k 2 D ɛ 2 (4.6b) The model parameters and the reaction extents are given in Table 4.2. For this example, the first reaction occurs many more times than the second reaction. Hence we partition the extents of reaction (ɛ i s) as follows 3 : ɛ 2 comprises the subset of slow reactions y, and 3 Reactions are partitioned on the basis of the magnitude of their extents, not their rate constants.
75 53 Parameter Symbol Value reaction propensity 4.6a a 1 (x) 1 2 k 1n A (n A 1) reaction propensity 4.6b a 2 (x) k 2 n A n C reaction 4.6a rate constant k reaction 4.6b rate constant k initial number of A molecules n Ao initial number of B molecules n Bo initial number of C molecules n Co 1 initial number of D molecules n Do Table 4.2: Model parameters and reaction extents for the simple crystallization example ɛ 1 comprises the subset of fast reactions z. We first integrate the system using the implicit tau leap method [117]. We choose a time step of.2, and generate Poisson random numbers using code from Numerical Recipes in C [14]. Figure 4.2 demonstrates that this approximation adequately reconstructs the mean and standard deviation for all species. We next perform an approximate stochastic-langevin simulation. Here we approximate the fast reaction subset using the Langevin approximation and attempt to reconstruct the first two moments of each species. The Langevin equations are integrated using the Euler-Murayama method [4] with a time increment of.1. We account for the time-varying propensity of the slow reaction by employing the approximate scheme, setting the propensity of no reaction (a ) to 1. Figure 4.3 compares these results to the exact stochastic results for ten thousand simulations. The approximation accurately reconstructs the mean and standard deviation for all species. Next, we approximate the fast reaction subset deterministically and attempt to reconstruct the first two moments of each species based upon ten thousand simulations. For this case, we consider both the exact and approximate stochastic-deterministic simulations. Figure 4.4 compares the results of exact stochastic simulation to the exact stochastic deterministic solution. This approximation does an excellent job of reconstructing all of the means as well as the standard deviations for species C and D. However, we are not able to reconstruct the standard deviations for species A and B. This phenomenon is expected because by approximating ɛ 1 deterministically, we neglect all fluctuations caused by the first reaction. Figure 4.5 compares the results of exact stochastic simulation to the approximate stochastic-deterministic solution given a small value for the propensity of no reaction, a. For this value of a, the approximation accurately reconstructs the means of species A and B, but fails to reconstruct the moments of species C and D as well as the standard deviations of species A and B. This phenomenon indicates that the value of a is too small. By examining the cumulative squared error, however, Figure 4.6 demonstrates that increasing the value of a results in comparable error for the approximate and exact stochastic-deterministic simulations. Here, the least squares error is based on the deviation of the species C trajectories between the
76 54 Number of Molecules ( 1 5 ) B A 4 6 Time 8 (a) 1 Number of Molecules A B 4 6 Time 8 (b) 1 Number of Molecules σ C σ Time 7 8 (c) 9 1 Number of Molecules σ D σ Time 7 8 (d) 9 1 Figure 4.2: Comparison of approximate tau-leap simulation (points) to exact stochastic simulation (lines) based on 1, simulations and time step of.2. (a) Comparison of the mean for species A and B. (b) Comparison of the standard deviations for species A and B. (c) Comparison of the mean (C) and standard deviation (±σ) for species C. (d) compares the mean (D) and standard deviation (±σ) for species D. approximation techniques and the exact stochastic simulation. Table 4.3 compares the order of magnitude of the limiting time step for the different methods in this example. The major improvement in the approximate methods is that the time step is now limited by the slow reaction time as opposed to the fast reaction time. Note that the solution methods for the partitioned reaction system require more computational expense per limiting time step than the exact stochastic solution method. However, we still observed an order of magnitude improvement in computational expense by employing the approximate solution methods. Also, the results indicate that the tau leap method is the fastest approximation. This result is a little misleading because we employed an implicit first-order method for tau leaping, whereas we integrated deterministic equations using stiff predictor-corrector methods. For a comparison using the same order of method, we expect the
77 55 Number of Molecules ( 1 5 ) B A 4 6 Time 8 (a) 1 Number of Molecules A B 4 6 Time 8 (b) 1 Number of Molecules σ C σ Time 7 8 (c) 9 1 Number of Molecules σ D σ Time 7 8 (d) 9 1 Figure 4.3: Comparison of approximate stochastic-langevin simulation (points) to exact stochastic simulation (lines) based on 1, simulations, propensity of no reaction a = 1, and Langevin time step of.1. (a) Comparison of the mean for species A and B. (b) Comparison of the standard deviations for species A and B. (c) Comparison of the mean (C) and standard deviation (±σ) for species C. (d) compares the mean (D) and standard deviation (±σ) for species D. stochastic-deterministic simulation to yield slightly faster results than tau leaping because the former method does not draw any Poisson random variables.
78 56 Number of Molecules ( 1 5 ) B A 4 6 Time 8 (a) 1 Number of Molecules A B 2 A B 4 6 Time 8 (b) 1 Number of Molecules σ C σ Time 7 8 (c) 9 1 Number of Molecules σ D σ Time 7 8 (d) 9 1 Figure 4.4: Comparison of exact stochastic-deterministic simulation (points) to exact stochastic simulation (lines) based on 1, simulations. (a) Comparison of the mean for species A and B. (b) Comparison of the standard deviations for species A and B. (c) Comparison of the mean (C) and standard deviation (±σ) for species C. (d) Comparison of the mean (D) and standard deviation (±σ) for species D. Solution Method System Type Limiting Time Step O(Time Step) Relative CPU Time Exact Stochastic unpartitioned fast reaction time O(1 5 ) 12.3 Tau Leap unpartitioned slow reaction time O(.25) 1. Stochastic- slow reaction time partitioned Langevin (Langevin integration) O(.1) 1.31 Stochastic- slow reaction time (ODE partitioned Deterministic solver) O(1) 1.4 Table 4.3: Comparison of time steps for the simple crystallization example
79 57 Number of Molecules ( 1 5 ) B A 4 6 Time 8 (a) 1 Number of Molecules A B 2 A B 4 6 Time 8 (b) 1 Number of Molecules σ C σ Time 7 8 (c) 9 1 Number of Molecules σ D σ Time 7 8 (d) 9 1 Figure 4.5: Comparison of approximate stochastic-deterministic simulation (points) to exact stochastic simulation (lines) based on 1, simulations and propensity of no reaction a =.1. (a) Comparison of the mean for species A and B. (b) Comparison of the standard deviations for species A and B. (c) Comparison of the mean (C) and standard deviation (±σ) for species C. (d) Comparison of the mean (D) and standard deviation (±σ) for species D.
80 58 1 (a) Squared Error 1 Approximate Exact Propensity of No Reaction, a 1 (b) Squared Error 1 Approximate.1.1 Exact.1 1 Propensity of No Reaction, a 1 Figure 4.6: Squared error trends for the exact and approximate stochastic-deterministic simulations based on 1, simulations. The squared error is calculated from the deviation of the moments for species C between the approximation techniques and the exact stochastic simulation. (a) Plot of the error in the mean of species C. (b) Plot of the error in the standard deviation of species C.
81 59 Parameter Symbol Value reaction propensity 4.61a a 1 (x) k 1 (template) reaction propensity 4.61b a 2 (x) k 2 (genome) reaction propensity 4.61c a 3 (x) k 3 (template) reaction propensity 4.61d a 4 (x) k 4 (template) reaction propensity 4.61e a 5 (x) k 5 (struct) reaction propensity 4.61f a 6 (x) k 6 (genome)(struct) reaction 4.61a rate constant k 1 1. day 1 reaction 4.61b rate constant k 2.25 day 1 reaction 4.61c rate constant k 3 1. day 1 reaction 4.61d rate constant k 4.25 day 1 reaction 4.61e rate constant k day 1 reaction 4.61f rate constant k (molecules day) 1 initial number of template molecules template o 1 initial number of genome molecules genome o initial number of struct molecules struct o Table 4.4: Model parameters and reaction extents for the intracellular viral infection example Intracellular Viral Infection We now consider a general model of an infection of a cell by a virus. A reduced system model consists of the following reaction mechanism [143]: nucleotides template genome ɛ 1 (4.61a) nucleotides + genome template ɛ 2 (4.61b) nucleotides + amino acids template struct ɛ 3 (4.61c) template degraded ɛ 4 (4.61d) struct secreted/degraded ɛ 5 (4.61e) genome + struct secreted virus ɛ 6 (4.61f) where genome and template are the genomic and template viral nucleic acids respectively, and struct is the viral structural protein. Additional assumptions include: 1. nucleotides and amino acids are available at constant concentrations, and 2. template catalyzes reactions (4.61a) and (4.61c). We are interested in the time evolution of the template, genome, and struct species. We assume that the initial infection of a cell corresponds to the insertion of one template molecule into
82 6 Number of Molecules struct genome template (a) Time (Days) 15 (b) 2 Number of Molecules template struct genome Time (Days) 4 5 Figure 4.7: Intracellular viral infections: (a) typical and (b) aborted. the cell. The model parameters and reaction extents are presented in Table 4.4. This model has two interesting features best illustrated by the two exact stochastic simulations presented in Figure 4.7. First, the three components of the model exhibit fluctuations that vary by differing orders of magnitude. For the same time scale, the struct species fluctuates by hundreds to thousands of molecules, whereas the template and genome species fluctuate by tens of molecules. Second, the model solution exhibits a bimodal distribution. In particular, a cell may exhibit either a typical infection in which all species become populated, or an aborted infection in which all species are eliminated from the cell. When the number of template and struct molecules are greater than zero and one hundred respectively, reactions 4.61c and 4.61e occur many more times than the remaining reactions. Hence when template > and struct > 1, we partition the system as follows:
83 61 Solution Method System Type Relative CPU Time Exact Stochastic unpartitioned 51.5 Stochastic-Deterministic partitioned 1 Table 4.5: Simulation time comparison for the intracellular viral infection example ɛ 1, ɛ 2, ɛ 4, and ɛ 6 comprise the subset of slow reactions y, and ɛ 3 and ɛ 5 comprise the subset of fast reactions z. Figure 4.7 indicates that the simulation should traverse between the partitioned and unpartitioned reaction systems. Since our approximation makes fast reactions continuous events as opposed to discrete ones, we round all species when transitioning from the approximate to exact stochastic simulation to prevent non-integer values. This rounding only affects the struct species, and therefore introduces negligible error into the system. We choose to approximate the fast reaction subset deterministically, so we employ the approximate stochastic-deterministic simulation with propensity of no reaction a =. We compare the approximate stochastic-deterministic simulation to the exact stochastic simulation by reconstructing the statistics for each species based upon one thousand simulations. We also compare the evolution of the mean for these two simulations to the solution of the purely deterministic model. Figures 4.8 through 4.1 compare the time evolution of the probability distribution for template, the small numbered species. These figures indicate that the approximate stochasticdeterministic simulation accurately reconstructs the entire template probability distribution. Note that the purely deterministic model, however, is unable to accurately reconstruct even the evolution of the mean. This phenomenon occurs because the deterministic model cannot describe the bimodal nature of the probability density. Figure 4.11 compares the evolution of the mean and standard deviation for the genome species. Again, the approximate simulation accurately reconstructs the time evolution of these moments. Figure 4.12 compares the evolution of the mean and standard deviation for the struct, the large numbered species. Surprisingly, the approximate stochastic-deterministic simulation accurately reconstructs the time evolution of both of these statistics. Since we approximated the fast reactions deterministically, we did not expect to accurately reconstruct moments higher than the mean for the large numbered species. For this example, though, fluctuations in the small numbered species, template, are amplified into the struct species via reaction 4.61c. Thus we are able to accurately reconstruct moments of order higher than zero. Table 4.5 compares the computational expense between the exact stochastic and approximate stochastic-deterministic solution methods. The approximate solution method results in a fifty-fold reduction in computational expense over the exact solution method.
84 62 (a) Probability Time (days) Template Molecules (b).1 Probability Time (days) Template Molecules Figure 4.8: Evolution of the template probability distribution for the (a) exact stochastic and (b) approximate stochastic-deterministic simulations. 4.5 Critical Analysis of the Stochastic Approximations The primary contribution of this work is the idea of partitioning a purely stochastic reaction system using extents of reaction into subsets of slow and fast reactions. Using order of magnitude arguments, we can derive approximate Markov evolution equations for the slow extent marginal and the fast extents conditioned on the slow extents. The evolution equation for the fast extents conditioned on the slow extents is a closed-form expression, whereas the evo-
85 (a).6 Probability Approximate Stochastic Time (Days) 15 (b) 2 Probability Template Molecules Figure 4.9: Comparisons of the (a) (template =,t) and (b) (template,t = 2 days) cross-sections of the template probability distribution for the exact stochastic (solid line) and approximate stochastic-deterministic (dashed line) simulations. lution equation for the slow extent marginal depends on this conditional probability. Using relaxation time arguments, we can propose two approximations for the fast extents: an equilibrium approximation when the fast extents relax faster than the slow extents, and a Langevin or deterministic approximation when both fast and slow extents exhibit similar relaxation times. The equilibrium assumption is similar in nature to the slow-reaction simulation recently proposed in the literature by Cao, Gillespie, and Petzold [16]. In contrast to this approach, we believe that our approach has a much tighter connection to the original master equation. By equilibrating the fast reaction subset, we can substantially reduce the computational requirement by integrating the system over a much larger time step than the exact stochastic simulation. This method requires solving for the equilibrium distribution of the fast reactions.
86 64 25 Template Molecules σ template σ Deterministic template Time (Days) 15 2 Figure 4.1: Comparison of the template mean and standard deviation (±σ) for exact stochastic (solid lines), approximate stochastic-deterministic (long dashed lines), and deterministic (short dashed lines) simulations. Genome Molecules Deterministic +σ genome genome σ Time (Days) 15 2 Figure 4.11: Comparison of the genome mean and standard deviation (±σ) for exact stochastic (solid lines), approximate stochastic-deterministic (dashed lines), and deterministic (points) simulations. If there are few fast extents or many of fast extents are independent of one another, then exactly solving for this distribution is possible as illustrated by the enzyme kinetics example. If there are a large number of coupled fast extents, then exact solution may not be computationally
87 65 14 Struct Molecules ( 1 3 ) σ struct σ 1 Time (Days) Deterministic struct 15 2 Figure 4.12: Comparison of the structural protein (struct) mean and standard deviation (±σ) for exact stochastic (solid lines), approximate stochastic-deterministic (dashed lines), and deterministic (points) simulations. feasible. For example, consider the coupled, fast reactions A + E B + E C + E D + E A minimal representation of these reactions requires three (reversible) extents of reaction, which is difficult to solve given a reasonable number of molecules for each species. By approximating the fast reaction subset using Langevin equations, we can reduce the computational requirement by integrating the system over a much larger time step than the exact stochastic simulation. However, we must now employ schemes for integrating stochastic differential equations. By approximating the fast reaction subset deterministically, we can bound the computational requirements for simulation of the system. For this case, we can employ existing and robust ordinary differential equation solvers for integration of this reaction subset. In contrast, the computational expense for exact stochastic simulation scales with the number of reaction events. For an example, reconsider simulation of the simple crystallization system presented in section Doubling the initial amount of A doubles the number of times the fast reaction must occur, and thus significantly increases the computational load of an exact stochastic simulation. On the other hand, if the fast reaction is approximated deterministically, then doubling the initial amount of A does not require stochastic simulation of any additional reaction events, and thus results in no change in the computational load. The partitioning techniques presented here sacrifice some numerical accuracy for a bound on the computational load. By equilibrating some fast reactions, one cannot expect to accurately reconstruct statistics for species affected by these fast reactions at very fine time scales. However, we are often interested in the macroscopic behavior of the system, and it
88 66 may not be possible to even observe a physical system at such fine time scales. Approximating some discrete, molecular reaction events as continuous events via the Langevin approximation loses the discrete nature of the entire system. However, as illustrated by the simple crystallization example, this approximation still accurately reconstructs at least the first two moments of each reacting species. Furthermore, approximating fast reactions deterministically eliminates all fluctuations contributed to the system by these reactions. Depending upon the system and the modeling objective, though, these sacrifices may be acceptable. In the simple crystallization example, the stochastic-deterministic simulations accurately reconstructed the means of all species as well as the standard deviations for the small numbered species. If fluctuations in the larger species are not of interest, then these results are acceptable. In the intracellular viral infection example, the approximate stochastic-deterministic simulation accurately reconstructed the evolution of the probability distribution for the small numbered species, as well as the means and standard deviations for the large numbered species. Here, amplification of fluctuations from the small to large numbered species (template to struct) led to accurate estimates of the statistics of large numbered species. A secondary contribution of this work is an approximate simulation for master equations subject to time-varying constraints. As demonstrated by the simple crystallization example, this approximate simulation approaches the accuracy of the exact simulation as the magnitude of the propensity of no reaction increases. This approximation is most useful for cases in which the total reaction rate, r tot, is not integrable analytically. For this case, we must use an ODE solver with a stopping criterion to determine the next reaction time. Since calling such an ODE solver requires some overhead computational expense, performing the approximate simulation may be computationally favorable. The work presented here reflects only a fraction of the approximations that should prove useful for simulating stochastic chemical kinetics. For example, one could simulate fast reactions using tau-leaping schemes instead of deterministic or Langevin approximations. Also, we did not address the quasi-steady state assumption (QSSA). In a deterministic setting, the QSSA equilibrates the rate of change for a given chemical species. In terms of our previous example, reaction 4.27, such an assumption would set = a 1 (x) a 2 (x) a 3 (x). For the discrete master equation, however, it is unlikely that such a situation can arise due to the integer nature of all chemical species. The most likely situation is for either ɛ 1 > and ɛ 2 = ɛ 3 =, or ɛ 2, ɛ 3 ɛ 1. In this case, we would expect to almost never find a B molecule in an exact simulation. Although Rao and Arkin [113] recently addressed this issue, they assumed a Markovian form for their governing master equation rather than derive it directly from the original master equation (4.2). A tighter connection between the original and approximate systems should be possible. We believe that the future of stochastic simulation lies in software packages that can 1. adaptively partition reactions into subsets, using appropriate approximations for each subset (i.e. exact, Poisson, Langevin, or deterministic approximations); and 2. adaptively adjust the integration time step to control the error induced at each step.
89 For reconstruction of only the mean and variance, this software should dramatically reduce the amount of computational expense required to generate approximate realizations from the underlying master equation. We envision that the primary benefit of the tools presented in this work is bridging the gap from the microscopic to the macroscopic. In particular, researchers are becoming increasingly interested in modeling nanomaterials, phenomena at interfaces, and site interactions on catalysts. In each of these problems, macroscopic interactions in the bulk influence microscopic interactions at interfaces. Although most of the action is at the interface, we cannot neglect the bulk or we lose the ability to model the effect of process design and control strategies. The techniques presented here provide one method of modeling these interactions. 67 Notation A matrix of reaction propensities a j (n) jth reaction propensity (rate) b j (y, z) jth slow reaction rate averaged over values of the fast extents b j (y, z) jth slow reaction rate c j (y, z) jth fast reaction rate I identity matrix k j N n j n jo n n n,j P P P A rate constant for reaction k number of Monte Carlo simulations number of molecules for species j initial number of molecules for species j number of molecules for all reaction species initial number of molecules for all reaction species jth initial number of molecules for all reaction species probability vector for all possible values of the extents of reaction probability approximate probability (reduced by order of magnitude arguments) p random number from the uniform distribution (, 1) r tot r y tot t W x y z z ɛ sum of reaction rates sum of reaction rates for the slow reaction partition time vector of Wiener processes state of the system in terms of extents subset of slow reaction extents subset of fast reaction extents subset of fast reaction extents scaled by Ω extent of reaction µ one possible reaction in the stochastic kinetics framework ν stoichiometric matrix
90 68 σ τ Ω standard deviation time of the next stochastic reaction characteristic system size
91 69 Chapter 5 Sensitivities for Stochastic Models Recently, models of isothermal, well-mixed stochastic chemical kinetics and Monte Carlo techniques for simulating these models have garnered significant attention from researchers in a wide variety of disciplines. This chapter considers a next logical step in applying these models: performing systems level tasks such as parameter estimation and steady-state analysis. One useful quantity in performing these tasks is the sensitivity. Various methods for calculating sensitivities of the underlying probability distribution and its moments are considered. For nontrivial models, the most computationally efficient method of evaluating the sensitivity consists of coupling an approximate evolution equation for the sensitivity with Monte Carlo reconstruction of the desired moments. Several parameter estimation and steady-state analysis examples demonstrate that, for systems level tasks, this approximation is well suited. We also show that highly-accurate sensitivities are not critical because optimization algorithms generally converge without exact gradients. This chapter is organized as follows. First we review the chemical kinetics master equation and define the sensitivity of moments of this equation with respect to model parameters. Next we propose and compare several methods for calculating approximations of the sensitivities with an eye on computational efficiency. Finally we illustrate how to use the sensitivities for (1) calculating parameter estimates for several linear and nonlinear kinetic models and (2) performing steady-state analysis. 5.1 The Chemical Master Equation The governing equation for the system of interest is again the chemical master equation. In this case, however, we consider the dependence of the master equation upon the set of parameters θ dp (n, t; θ) m = a k (n ν k, θ)p (n ν k, t; θ) a k (n, θ)p (n, t; θ) (5.1) dt in which k=1 n is the state of the system in terms of number of molecules (a p-vector), θ is a vector containing the system parameters (an l-vector),
92 7 P (n, t; θ) is the probability that the system is in state n at time t given parameters θ, a k (n, θ)dt is the probability to order dt that reaction k occurs in the time interval [t, t+dt), and ν k is the kth column of the stoichiometric matrix ν (a p m matrix). Here, we assume that the initial condition P (n, t ; θ) is known. One useful quantity in performing systems level tasks is the sensitivity. We consider in the next section the calculation of the sensitivity for stochastic systems governed by the chemical master equation. 5.2 Sensitivities for Stochastic Systems The sensitivity indicates how responsive the state is to perturbations of a given parameter. For the master equation (5.1), the state is the probability P (n, t; θ), and its sensitivity is s(n, t; θ) = P (n, t; θ) θ Here, s(n, t; θ) is an l-vector. We derive the evolution equation for this sensitivity by differentiating the master equation (5.1) with respect to the parameters θ dp (n, t; θ) = θ dt θ ds(n, t; θ) dt = (5.2) m a k (n ν k, θ)p (n ν k, t; θ) a k (n, θ)p (n, t; θ) (5.3) k=1 m a k (n ν k, θ)s(n ν k, t; θ) a k (n, θ)s(n, t; θ)+ k=1 a k (n ν k, θ) θ We make two observations about equation (5.4): 1. it is linear in the sensitivity s(n, t; θ) and P (n ν k, t; θ) a k(n, θ) P (n, t; θ) (5.4) θ 2. solution of this equation requires simultaneous solution of the master equation (5.1), but not vice versa. i.e. For engineering purposes, we are interested in moments of the probability distribution, g(n) = n g(n)p (n, t; θ) (5.5) in which g(n) and g(n) are q-vectors. For example, we might seek to implement control moves that drive the mean system behavior towards a desired set point. Such tasks require knowledge of how sensitive these moments are with respect to the parameters. The master equation (5.1)
93 indicates that the probability distribution evolves continuously with time; consequently, moments of this distribution (assuming that they are well defined) evolve continuously as well. Therefore we can simply differentiate equation (5.5) with respect to the parameters to define the sensitivity of these moments, s(g(n)), as follows: θ T g(n) = s(g(n), t; θ) = n 71 θ T g(n)p (n, t; θ) (5.6) n g(n)s(n, t; θ) T (5.7) Here, s(g(n), t; θ) is a q l matrix. Equation (5.7) indicates that these sensitivities depend upon the sensitivity of the master equation, s(n, t; θ). Therefore, the exact solution of s(g(n)) requires solving the following set of coupled equations: dp (n, t; θ) dt ds(n, t; θ) dt = = m a k (n ν k, θ)p (n ν k, t; θ) a k (n, θ)p (n, t; θ) k=1 m a k (n ν k, θ)s(n ν k, t; θ) a k (n, θ)s(n, t; θ) k=1 + a k(n ν k, θ) θ P (n ν k, t; θ) a k(n, θ) P (n, t; θ) θ (5.8a) (5.8b) s(g(n), t; θ) = n g(n)s(n, t; θ) T (5.8c) Exact solution of even just the master equation (5.1) is computationally intractable for all but the simplest systems. Consequently, exact calculation of both the master equation and its sensitivity (i.e. equation (5.8)) is also intractable in general. However, Monte Carlo methods such as those proposed by Gillespie [45, 46] and Gibson and Bruck [43] can reconstruct moments of the master equation to some degree of precision (error associated with the finite number of simulations corrupts these reconstructed quantities). In the next section, we examine methods for reconstructing the sensitivities given only information about how moments of the master equation evolve Approximate Methods for Generating Sensitivities Approximate methods of generating sensitivities for this system include 1. deriving an approximate model for the sensitivity of a desired moment and 2. applying finite difference schemes. The primary benefit of these alternatives is that they require only reconstruction of the desired moment, not necessarily via solution of the master equation (5.1). For systems level tasks
94 72 such as parameter estimation and steady-state analysis, we are particularly interested in the dynamic behavior of the mean n n = n np (n, t; θ) (5.9) and its sensitivity s = s(n, t; θ) = n ns(n, t; θ) T (5.1) in which n is a p-vector and s is a p l matrix. We consider deriving approximations for the mean sensitivity s subsequently. We note that the sensitivity for any moment could be derived and calculated similarly Deterministic Approximation for the Sensitivity Combining equations (5.4) and (5.1) yields the following evolution equation for the mean sensitivity s ds dt = θ T = θ T = θ T = θ T n m n (a k (n ν k, θ)p (n ν k, t; θ) a k (n, θ)p (n, t; θ)) (5.11) k=1 ( m na k (n ν k, θ)p (n ν k, t; θ) ) na k (n, θ)p (n, t; θ) (5.12) n n ( m (n + ν k )a k (n, θ)p (n, t; θ) ) na k (n, θ)p (n, t; θ) (5.13) n n m ν k a k (n, θ)p (n, t; θ) (5.14) k=1 k=1 k=1 n Consider a Taylor series expansion of a k (n, θ) about the mean value n a k (n, θ) = a k (n, θ) + a k(n, θ) n T (n n) + 1 n=n 2 (n n)t 2 a k (n) n n T (n n) + (5.15) n=n One approximation consists of incorporating only the first two terms of the expansion (5.15) into equation (5.14) to obtain ds dt θ T = θ T m ( ν k a k (n, θ) + a ) k(n, θ) n T (n n) P (n, θ; t) (5.16) k=1 n n=n m ν k a k (n, θ) (5.17) k=1 = νa(n, θ) (5.18) T θ ( ) a(n, θ) n a(n, θ) θ = ν n T + T θ θ T θ T (5.19)
95 in which ( ds a(n, θ) dt ν n T s + a(n, θ) = ) a(n, θ) θ T 73 (5.2) [ a 1 (n, θ) a m (n, θ)] T (5.21) Equation (5.2), then, is the first-order approximation of the sensitivity evolution equation assuming that the mean n is known. Logically, then, we must specify how we plan on calculating the mean. Clearly we can also approximate the mean evolution equation using the first two terms of the truncated Taylor series expansion (5.15) as follows: dn m dt = ν k a k (n)p (n, t; θ) (5.22) k=1 n m ( ν k a k (n, θ) + a ) k(n, θ) n T (n n) P (n, θ; t) (5.23) n=n = k=1 n m ν k a k (n, θ) (5.24) k=1 dn dt νa(n, θ) (5.25) Equation (5.25) is the usual deterministic approximation of the chemical master equation [154]. In general, the mean behavior of the chemical master equation does not obey the deterministic equation (5.25); see Arkin, Ross, and McAdams [3] and Srivastava, You, Summers, and Yin [143] for recent biological examples of this phenomenon. Therefore, we do not advise calculating both the mean and the sensitivity in this fashion. We propose to estimate the mean by averaging the results of multiple Monte Carlo simulations, and to approximate the sensitivity of the mean using equation (5.2). Since both the mean and the sensitivity are linear functions, exchanging the order of evaluation is valid. So the following strategies are equivalent: 1. Evaluate s k, n k for every simulation using equation (5.2), in which n k denotes the kth Monte Carlo simulation of n. Since the reaction rate vector a(n, θ) is constant between reaction events, equation (5.2) can be solved exactly via a matrix exponential [21]. Finally, calculate s = E[s k ], in which E[n] denotes the expectation of n. 2. Evaluate n k for every simulation, calculate E[n k ], then calculate s using E[n k ] and equation (5.2). The first option is presumably the more computationally expensive option since exact solution of equation (5.2) requires evaluation of a matrix exponential for every reaction step. The second option, however, may experience difficulties because depending on the behavior of the mean n, explicit strategies for evaluating equation (5.2) (e.g. Runge-Kutta methods) may require small time steps to ensure stability; and
96 74 random noise associated with the finite number of Monte Carlo simulations may induce inaccuracies for higher-order methods. In spite of these problems, we advocate using the second option to calculate the approximate sensitivity if performing Monte Carlo simulations is computationally expensive. We note that elementary chemical reactions are generally bimolecular. For this case, the Taylor series expansion consists exactly of the first three terms of equation (5.15), and we expect that equation (5.2) adequately approximates the true sensitivity. For unimolecular or zero-order reactions, the Taylor series expansion is exact, so equation (5.2) is exact. Finally, reducing the master equation to a series of moments truncates some of the information contained by the probability distribution of the initial state. For the remainder of this chapter, we assume that this probability distribution is a delta function at the initial mean value. Our method is not restricted to this particular choice of distribution, however. Rather, one may set this initial distribution arbitrarily via proper configuration of the Monte Carlo simulations used to reconstruct the desired moments as discussed in Chapter 4 (see equation (4.3)) Finite Difference Sensitivities For finite differences, we assume that we have some evolution equation for the mean n that depends on the system parameters θ n k+1 = F (n k ; θ, Ω) (5.26) Here, the notation n k denotes the value of the mean n at time t k. Also, Ω denotes the string of random numbers used to propagate the state. Recall that the sensitivity s indicates how sensitive the mean is to perturbations of a given parameter, i.e. s k = n k θ T (5.27) We could then approximate the jth column of the desired sensitivity using, for example, a central difference scheme: s k+1,j = F (n k; θ + δc j, Ω 1 ) F (n k ; θ δc j, Ω 2 ) 2δ + i O(δ 2 ) (5.28) Here, δ is a small positive constant, c j is the jth unit vector, and i is a vector of ones. If we use the mean of Monte Carlo simulations to determine the state propagation function F (n k ; θ, Ω) and choose Ω 1 Ω 2, then we have essentially amplified the error associated with the finite number of simulations into evaluation of equation (5.28). On the other hand, evaluating the means by using the same strings of random numbers, i.e. Ω 1 = Ω 2, eliminates this amplification. However, we now have the potential of choosing a sufficiently small, non-zero perturbation such that F (n k ; θ+δc j, Ω 1 ) = F (n k ; θ δc j, Ω 2 ). If we choose the parameter perturbation to be too large, then the O(δ 2 ) is not negligible in equation (5.28). Hence special care must be
97 taken in the selection of the perturbation δ. The subsequent chapter, Chapter 6, discusses these subtleties in greater detail. Finally, the computational expense of this method may be prohibitive if evaluating the mean is computationally intensive because calculating the sensitivity requires, in this case, two mean evaluations per parameter. In contrast, calculating the additional sensitivities using the approximate calculation of equation (5.2) does not require any additional stochastic simulations. Raimondeau, Aghalayam, Mhadeshwar, and Vlachos recently examined using finite differences to calculate sensitivities for kinetic Monte Carlo simulations [15]. However, they use only a single simulation to generate their sensitivity and require relatively large parameter perturbations to generate measurable changes in model responses (one of their examples uses a parameter perturbation of approximately 3%). These authors make no appeal to the master equation nor to the fact that the mean should be a smoothly-varying function. We interpret their approach as a mean sensitivity calculation using a poor reconstruction of the mean. Due to the large choice of parameter perturbation, we infer that the authors did not use the same strings of random numbers to evaluate equation (5.28), i.e. Ω 1 Ω 2. Drews, Braatz, and Alkire also recently examined using finite differences to calculate sensitivities for kinetic Monte Carlo code simulating copper electrodeposition [25]. These authors consider the specific case of the mean sensitivity, and derive finite differences for cases with significant finite simulation error. In these cases, the finite simulation error is greater than higher-order contributions of the finite difference expansion, so the authors derive firstorder finite differences that minimize the variance of the finite simulation error. No appeal is made to the master equation, and they implicitly assume that the mean should be a smoothlyvarying function. Their computational requirements certainly motivate the approximations made in this chapter, however. Each simulation required on average 64 hours to complete, and the total computational requirement was 92,547 hours for 22 parameters. Additionally, the authors employed perturbations of +1% and 5%, so the accuracy of the finite difference is questionable. Solving for the approximate sensitivity would require only one mean evaluation (roughly 14 hours) plus the computational time required for the sensitivity calculation, a computational savings of at least an order of magnitude. These authors have also chosen a rather large parameter perturbation, again leading us to infer that they did not use the same strings of random numbers to evaluate equation (5.28), i.e. Ω 1 Ω Examples We now illustrate these different methods of calculating the sensitivity with two simple examples. For clarity, we first briefly review the nomenclature that indicates which approximations, if any, are performed in a given simulation. We can either reconstruct the mean exactly by solving the master equation, or approximately via Monte Carlo simulation. Given a reconstruction of the mean, we can then calculate the sensitivity using the approximate equation (5.2), or by finite differences, i.e. equation (5.28). Solving the exact sensitivity of the mean requires solution of equation (5.8), namely the master equation, the desired moment, and their respective
98 s Time Exact Approximate Finite Difference Figure 5.1: Comparison of the exact, approximate, and central finite difference sensitivities for a second-order reaction. sensitivities. Second-Order Reaction Example We consider the simple second-order reaction A B a 1 = 1 2 k 1n A (n A 1) (5.29a) with initial condition n A,o = 2 and n B,o =, and k 1 =.333. For this example, we define x = [ n A n B ] T, θ = k1, s = n B / k 1 The reaction rate is nonlinear, implying that equation (5.2) is an approximation of the actual sensitivity. We solve for the exact sensitivity. We also reconstruct the mean via Monte Carlo simulation, then calculate the sensitivity by both the approximate equation (5.2) and central finite differences. Each mean evaluation is calculated by averaging fifty Monte Carlo simulations. Additionally, we perturbed k 1 by 1% to generate the finite difference sensitivity. Figure 5.1 compares the exact, approximate, and central finite difference sensitivities. For this example, the exact and approximate sensitivities are virtually identical. The central finite difference sensitivity, on the other hand, yields a very noisy and poor reconstruction at roughly twice the cost of the approximate sensitivity. Performing more Monte Carlo simulations per each mean evaluation would improve this estimate at the expense of additional computational burden.
99 Approximate Exact s n Ao = Time 8 1 Figure 5.2: Comparison of the exact and approximate sensitivities for the high-order rate example. High-Order Reaction Example We consider the simple set of reactions A B a 1 = k 1n A 1 + Kn A (5.3a) B A a 2 = k 2 n B (5.3b) with initial condition n A,o = 2 and n B,o =, and parameters k 1 = 4., k 2 =.1, and K = 2/n A,o. For this example, we define x = [ n A n B ] T, θ = K, s = na / K The first reaction rate is nonlinear, implying that equation (5.2) is an approximation of the actual sensitivity. In fact, for this case the Taylor series expansion (5.15) has an infinite number of terms. We solve for the exact sensitivity. We also reconstruct the mean exactly, then solve for the sensitivity via the approximate equation (5.2). Figure 5.2 plots this comparison, and demonstrates a large discrepancy between the exact and approximate sensitivities. As the initial number of A molecules increases, Figure 5.3 shows that the relative error between the exact and approximate sensitivities decreases. This trend is expected because, in the thermodynamic limit (i.e. x, Ω, z = x/ω constant), the chemical master equation reduces to a deterministic evolution equation for the concentrations z of the form given by the first-order approximation of the mean, equation (5.25) [76]. Next, we consider reconstructing the mean of the system via Monte Carlo simulation, and evaluate the sensitivity by both the approximate equation (5.2) and central finite differences. For this example, we set the initial condition n A,o = 2. Each mean evaluation is calculated by averaging fifty Monte Carlo simulations. Figure 5.4 compares the exact, approximate,
100 n Ao = 2 (s s)/s Time 6 n Ao = 2 n Ao = Figure 5.3: Relative error of the approximate sensitivity s with respect to the exact sensitivity s as the number of n A,o molecules increases for the high-order rate example s Time 6 Exact Approximate Finite Difference 8 1 Figure 5.4: Comparison of the exact, approximate, and finite difference sensitivity for the highorder rate example. and central finite difference sensitivities. The approximate sensitivity differs significantly from the exact sensitivity at later times but compares favorably with the approximate sensitivity obtained from using an exact reconstruction of the mean (i.e. Figure 5.2). Therefore the error in the approximate sensitivity is due to the truncation of the Taylor series expansion and not the Monte Carlo simulations. We perturbed K by 1% to generate the finite difference sensitivity. This sensitivity better approximates the exact sensitivity, but this method amplifies the error associated with the finite number of simulations. Additionally, the computational expense is roughly twice that required for the approximate sensitivity. Finally, we note that this computa-
101 tional expense results from perturbing only a single parameter. If we had required sensitivities for all parameters (k 1, k 2, and K), the computational expense would triple since the required number of simulations scales linearly with the desired number of sensitivities. In contrast, determining additional sensitivities using the approximate calculation does not require any additional stochastic simulations Parameter Estimation With Approximate Sensitivities The goal of parameter estimation is to determine the set of parameters that best reconciles the measurements with model predictions. The classical approach is to assume that measurements are corrupted by normally distributed noise. Accordingly, we calculate the optimal parameters via the least squares optimization min θ Φ = 1 2 e T k Π 1 e k s.t.: x k+1 = F (x k, θ) k e k = y k h(x k ) (5.31a) (5.31b) (5.31c) in which e k s denote the difference between the measurements y k and the model predictions h(x k ), and Π is the covariance matrix for the measurement noise. For the optimal set of parameters, the gradient θ Φ is zero. We can numerically evaluate the gradient according to θ Φ = 1 θ T e T k 2 Π 1 e k (5.32) k = ( ) h(xk ) x T k x T k k θ T Π 1 e k (5.33) = ( ) h(xk ) T x T s k Π 1 e k (5.34) k k Equation (5.34) indicates that the gradient depends upon s k, the sensitivity of the state with respect to the parameters. In general, most experiments do not include many replicates due to cost and time constraints. Therefore, the best experimental data we are likely to obtain is the average. In fitting these data to stochastic models governed by the master equation, we accordingly choose the mean n as the the state of interest. Monte Carlo simulation and evaluation of equation (5.2) provide estimates of the mean and the sensitivities. Since equation (5.2) is approximate (firstorder with respect to the mean), evaluating the gradient using this sensitivity is also approximate. For the sake of illustration, we obtain optimal parameter estimates using an optimization scheme analogous to the Newton-Raphson method. In particular, we perform a Taylor
102 8 series expansion of the gradient around the current parameter estimate θ k to generate the next estimate θ k+1 θ Φ θk+1 θ Φ θk + θθ Φ θk (θ k+1 θ k ) (5.35) Since we desire the gradient at the next iterate to be zero, ( ) 1 θ k+1 = θ k θθ Φ θk θ Φ θk (5.36) Differentiating the gradient (i.e. equation (5.34)) yields the Hessian ( θθ Φ = θ T ( ) ) h(xk ) T x T s k Π 1 e k k k (5.37) = k ( h(xk ) x T k ) T s k Π 1 h(x k) x T s k + k ( h(xk ) x T k 2 x k θ k θ T k ) T Π 1 e k (5.38) Making the usual Gauss-Newton approximation for the Hessian (i.e. e k ), we obtain θθ Φ k ( h(xk ) x T k ) T s k Π 1 h(x k) x T s k (5.39) k Finally, since we estimate both the mean and the sensitivities using Monte Carlo simulations, the finite number of simulations introduces some error into both of these estimates. Properly specifying a convergence criteria for this method must take this error into account. Raimondeau, Aghalayam, Mhadeshwar, and Vlachos argue that using kinetic Monte Carlo simulation to perform parameter estimation is too computationally expensive [15]. They claim that a model with two to three parameters requiring.5 hours per simulation needs roughly 1 5 function evaluations for direct optimization. We believe that the actual number of function evaluations required for direct optimization is significantly lower if one uses the approximate sensitivity coupled with the optimization scheme presented in this section. In the next example, we demonstrate that surprisingly few function evaluations lead to accurate parameter estimates High-Order Rate Example Revisited We consider parameter estimation for the high-order rate example reactions (5.3). Our experimental data consists of the time evolution of species A obtained from the average of fifty Monte Carlo simulations. We assume that the values of k 1 and k 2 are known, and attempt to estimate K using the Newton-Raphson method described in the previous section. Sensitivities for this method are obtained using both the approximate and central finite difference sensitivities. For each method, mean evaluations are calculated by averaging fifty Monte Carlo simulations using the same strings of random numbers (note that a different string of random numbers is used to generate the experimental data). Hence calculation of the approximate
103 81 Measurement h(e(x)) K Actual (K = 1) Iteration 2 4 Time (a) 18 2 (b) 1 Figure 5.5: Comparison of the (a) parameter estimates per Newton-Raphson iteration and (b) model fit at iteration 2 using the approximate (dashed line) and finite difference (solid line) sensitivities for the high-order rate example. Points represent the actual measurement data. sensitivity requires one mean evaluation per iteration, while the finite difference sensitivity requires three mean evaluations (K, K δ, and K + δ). We perturbed K by 1% to calculate the central finite difference sensitivity. Figure 5.5 plots the results of this parameter estimation. Both sensitivities lead to correct estimation of the parameter K in approximately the same number of Newton-Raphson iterations. Clearly the error in the approximate sensitivity does not significantly hinder the search. Additionally, neither method converges to the true parameter value. This phenomenon results due to the fact that different strings of random numbers are used to generate the experimental data and the data used to estimate the parameter K. Finally, the estimation using the central finite difference required roughly three times the computational expense of that using the approximate sensitivity.
104 Steady-State Analysis Exact determination of steady states requires solving for the stationary state of the master equation (5.1). The difficulty of this task is comparable to that of solving the dynamic response. The next logical question, then, is if we can determine steady states from simulation. Unfortunately, we can only reconstruct the entire probability distributions from an infinite number of simulations. Given a finite number of simulations, we can reconstruct only a limited number of moments. Hence we can seek to find a steady state consistent with this desired number of moments. An additional complication associated with simulation is that we only have information from integrating the model forward in time. At steady state, we know that x k+1 = x k = steady state (5.4) Thus, we propose two methods for determining steady states from simulation: 1. Run Monte Carlo simulation for a long time. 2. Guess the steady state. Check: x k+1 = x k for short simulation? If not, use a Newton-Raphson search algorithm to search for an improved estimate of the steady state: (x k+1 x k ) θj+1 (x k+1 x k ) θj + θ T (x k+1 x k ) (θ j+1 θ j ) (5.41) θj = (x k+1 x k ) θj + (s k+1 s k ) θj (θ j+1 θ j ) (5.42) θ j+1 = θ j (s k+1 s k ) 1 (x k+1 x k ) θj (5.43) Here, θ j denotes the value of the initial state at iteration j. x k denotes the value of the state x at simulation time k for a given iteration. The second method, recently employed by Makeev et al. [85] in the same capacity, uses short bursts of simulation to determine whether or not the system is at a steady state. Clearly this method may be significantly faster than the first method, which requires a lengthy simulation. Additionally, employing the second method permits use of the approximate sensitivity which can be calculated inexpensively from simulation. We consider an example of the second method in the next example.
105 83 Parameter Value Total catalyst sites 2 2 k k k 3 k k 5.36 k Table 5.1: Parameters for the lattice-gas example Lattice-Gas Example We consider the following lattice-gas reaction model [85]: A + k 1 A (5.44a) A k 2 A + (5.44b) A + B k 3 C + 2 (5.44c) B k 4 2B (5.44d) C + k 5 C (5.44e) C k 6 C + (5.44f) All reactions are elementary as written. Parameters for these reactions are given in Table 5.1. Figure 5.6 plots the results for a dynamic simulation of the lattice-gas model and the convergence of the steady-state search algorithm. For this example, the model response corresponds to a limit cycle; therefore, the eigenvalues of the state sensitivity matrix s k+1 = x k+1 / x T k should contain values with absolute value greater than unity to reflect the unstable nature of this steady state. The search algorithm finds a steady state within the region of this limit cycle with eigenvalues (calculated using the approximate sensitivity) ] T λ (s k+1 ) = [ Hence the approximate sensitivity indicates that the steady state is indeed unstable. 5.5 Conclusions We have examined various methods of calculating sensitivities for the moments of the chemical master equation, and explicitly derived methods for calculating the mean sensitivity. Exact solution of the mean sensitivity requires solving the chemical master equation and its sensitivity, a task that is infeasible for all but trivial systems. For more complex systems, the mean and
106 84 Surface Species C A B 1 Time C Surface Species Iteration A B Figure 5.6: Results for the lattice-gas model: (a) dynamic response of the model from an empty lattice initial condition and (b) convergence of the steady-state search algorithm. its sensitivity must be reconstructed from Monte Carlo simulations. If carefully implemented, finite differences can generate accurate sensitivities. However, the computational expense of this method scales linearly with the number of parameters, and is particularly burdensome for computationally intensive Monte Carlo simulations. In contrast, employing a first-order approximation of the sensitivity permits inexpensive calculation of the mean sensitivity from a reconstruction of the mean. Knowledge of model sensitivities permits execution of systems level tasks such as parameter estimation, optimal control, and steady-state analysis. In these operations, highlyaccurate sensitivities are not critical because optimization algorithms generally converge, albeit more slowly, without exact gradients. For use in an optimization context, the efficient evaluation of the approximate sensitivity proposed in this chapter seems well suited.
107 Notation 85 a(n, t) a k (n) c j e k i n n n k P s s s k t t k x y k z δ λ θ ν Π φ Ω vector of all reaction rates (a k (n) s) kth reaction rate the jth unit vector deviation between the predicted and actual measurement vectors at time t k a vector of ones vector of the number of molecules for all reaction species vector of the mean number of molecules for all reaction species kth Monte Carlo reconstruction of the vector n probability sensitivity of the state x with respect to the parameters θ sensitivity of the mean n with respect to the parameters θ kth Monte Carlo simulation reconstruction of the sensitivity s time discrete sampling time state of the system measurement vector at time t k state vector x scaled by the characteristic system size Ω finite difference perturbation eigenvalue parameter vector for a given model stoichiometric matrix covariance matrix for the measurement noise objective function value characteristic system size
108 86
109 87 Chapter 6 Sensitivity Analysis of Discrete Markov Chain Models In the previous chapter, we considered two approximations to the sensitivity equation: (1) finite differences, which offer inherently biased estimates of the sensitivity for significant computational expense, and (2) a first-order approximation to the sensitivity that required trivial computational expense. The second of these methods is analogous to the stochastic fluid models currently proposed in the field of perturbation analysis [19, 167]. For use in the context of unconstrained optimization, we demonstrated that both these approximations to the sensitivities permit efficient optimization. In this chapter, we consider methods for exactly calculating sensitivities for discrete Markov chain models from solely simulation information. In general, a discrete Markov chain model provides simple rules for propagating the discrete state n forward in time, i.e. P (n k+1 ) = P (n k+1 n k )P (n k ) (6.1) in which P ( ) denotes the probability of ( ), and n k refers to the state at time t k. Usually the accessible state space is too large to permit computation of the entire probability distribution, so we are forced to sample the distribution via Monte Carlo methods. These methods take advantage of the fact that any statistic can be written in terms of a large sample limit of observations, i.e. h(n) 1 h(n)p (n, t)dn = lim N N N h(n i ) 1 N i=1 N h(n i ) for N sufficiently large (6.2) in which n i is the ith Monte Carlo reconstruction of the state n. The desired statistic can then be reconstructed to sufficient accuracy given a large enough number of observations. Ultimately, we are interested in calculating sensitivities of expectations, i.e. i=1 s = E[h(n; θ)] θ (6.3a) E [h(n; θ + )] E [h(n; θ)] = lim N i=1 = lim lim h(ni ; θ + ) h(n i ; θ) N N (6.3b) (6.3c)
110 88 in which θ is a parameter of interest 1 For purposes of simulation, we must truncate the number of simulations N at some (hopefully) large, finite value. Then perhaps the easiest approximation to solving equation (6.3) is to fix at some nonzero value and use, for example, a forward finite difference scheme: s = N i=1 h(ni ; θ +, Ω i 1 ) h(ni ; θ, Ω i 2 ) N + O( ) + O(N.5 ) (6.4) Here, Ω i 1 and Ωi 2 refer to the string of random numbers used in the ith simulation. As noted by Fu and Hu [37] in their introduction, we can reduce the variance of this estimate by taking Ω i 1 = Ωi 2, that is, by using the same seed for multiple simulations. However, finite difference methods are inherently biased estimators due to the fact that the O( ) term does not go to zero as N. Additionally, these methods can suffer tremendously from finite simulation error. If h(n) can only be reconstructed to several significant figures, we must choose a large value for the perturbation, causing the O(N.5 )/ term to dominate the expression (6.3). Alternatively, we could seek to derive unbiased sensitivity estimates from the simulated sample paths alone. Accordingly, we would like to be able to justify the interchange of expectation and differentiation in equation (6.3). This particular problem has been wellcharacterized in the field of perturbation analysis; see, for example, Ho and Cao [63] and Cassandras and Lafortune [18]. When n is discrete, it is clear that for any finite N we can always choose a + > such that N i=1 h(ni ; θ + ) h(n i ; θ) N = if < < + (6.5) To overcome this problem, we must devise a means to make the sample paths continuous so that the exchange of expectation and differentiation is valid. In this chapter, we consider smoothing by both conditional expectation (smoothed perturbation analysis) and integration. 1 This analysis can easily be extended to multiple parameters. We choose to examine a single parameter for notational simplicity.
111 6.1 Smoothed Perturbation Analysis 89 Smoothed perturbation analysis or SPA smooths discrete sample paths by using conditional expectation. Choosing a characterization z for each simulated sample path, we see that h(n; θ) = n = n = n = n = z h(n; θ)p (n; θ) (6.6) h(n; θ)p (n, z; θ) (6.7) z h(n; θ)p (n, z; θ) P (z) (6.8) P (z) z h(n; θ)p (n z; θ)p (z) (6.9) z P (z) n h(n; θ)p (n z; θ) (6.1) = z P (z) n h(n; θ)p (n z; θ) (6.11) = z P (z)e[h(n; θ) z; θ] (6.12) E[h(n; θ) z; θ] varies continuously with θ, therefore we can evaluate the desired sensitivity by differentiating both sides of equation (6.12) θ h(n; θ) = θ P (z)e[h(n; θ) z; θ] (6.13) z s = z P (z) E[h(n; θ) z; θ] (6.14) θ = z P (z) lim E[h(n; θ) z, θ + ] E[h(n; θ) z, θ] (6.15) Because each Monte Carlo sample path determines the characterization z, equation (6.15) corresponds to first differentiating the smoothed sample paths, then averaging the results. We refer the interested reader to Fu and Hu [37] for the proofs of the unbiasedness of this estimator. The remaining questions are how to choose the characterization z, and how to evaluate the conditional expectation E[h(n; θ) z; θ]. We examine these issues further by considering two motivating examples.
112 Coin Flip Example We consider the example of flipping a coin. Define S n to be the sum of n-independent flips, in which S n = n x j (6.16) j=1 P (X = ) = θ (6.17) P (X = 1) = 1 θ (6.18) θ 1 (6.19) Here, x j is the jth realization of the random variable X. It is straightforward to show that E[S n ] = n E[x j ] (6.2) We are interested in calculating the sensitivity of S n with respect to the parameter θ. It is easy to show that E[S n ] = E[S n ] θ j=1 n 1 θ = n(1 θ) (6.21) j=1 = n (6.22) For the sake of illustration, we compute the SPA estimate for this process. We choose the characterization z i to be the ith outcome of the n flips given the nominal parameter value θ, e.g. z i = { x i 1 (θ) =,..., xi n(θ) = 1 }. Then equation (6.2) becomes E[S n ] = N P (z i )E[Sn z i i ] (6.23) i=1 = 1 N = 1 N N E[Sn z i i ] (6.24) i=1 N n E[x i j z i ] (6.25) i=1 We turn our attention towards calculating the quantity E[x i j zi ]. Suppose that the jth flip yields x j (θ) =. Because each of the n flips are independent, each x j depends only on the jth element of the characterization z. Therefore, we must calculate the conditional probabilities P (x j (θ + ) = x j (θ) = ) and P (x j (θ + ) = 1 x j (θ) = ). To do so, we use the fact that the random variable X(θ) can be written in terms of the uniform distribution U j=1 P (X(θ) = ) = P (U < θ) (6.26)
113 Assuming that the parameter perturbation > 2, we evaluate the conditional probabilities: P (x j (θ + ) = x j (θ) = ) = P (U < θ + U < θ) (6.27) = P (U < θ +, U < θ) P (U < θ) 91 (6.28) = 1 (6.29) P (x j (θ + ) = 1 x j (θ) = ) = P (U > θ + U < θ) (6.3) Then the desired conditional expectation is = (6.31) E[x j (θ) z] θ P (x j (θ + ) = x j (θ) = ) P (x j (θ) = x j (θ) = ) = lim ( ) (6.32) + P (x j(θ + ) = 1 x j (θ) = ) P (x j (θ) = x j (θ) = ) (1 ) (6.33) = (6.34) Alternatively, we consider the case in which the jth flip yields x j = 1. Then the conditional probabilities are P (x j (θ + ) = x j (θ) = 1) = P (U < θ + U > θ) (6.35) P (θ < U < θ + ) = P (U > θ) (6.36) = 1 θ (6.37) P (x j (θ + ) = 1 x j (θ) = 1) = P (U > θ + U > θ) (6.38) and the desired conditional expectation is P (U > θ +, U > θ) = P (U > θ) 1 (θ + ) = 1 θ (6.39) (6.4) E[x j (θ) z] θ P (x j (θ + ) = x j (θ) = 1) P (x j (θ) = 1 x j (θ) = 1) = lim (1 ) (6.41) + P (x j(θ + ) = 1 x j (θ) = 1) P (x j (θ) = 1 x j (θ) = 1) (1 1) (6.42) = lim (1 θ) (6.43) = 1 1 θ (6.44) 2 We can also calculate the conditional expectations assuming that <.
114 92 Parameter Symbol Value Finite difference perturbation.1 Number of simulations N 5 Coin flip probability θ.25 Table 6.1: Parameters for the coin flip example E[S n ] Number of coin flips n Exact Monte Carlo 8 1 Figure 6.1: Mean E[S n ] as a function of the number of coin flips n From this analysis, it is clear that the only trials that impact the sensitivity are those in which x j (θ) = 1. Our estimator of the sensitivity is then E[S n ] = 1 N n 1 θ N 1 θ 1 { x i j(θ) 1 } (6.45) i=1 j=1 { 1 if δ = 1 {δ} = (6.46) otherwise We now examine the numerical results for this process via simulation. We use the parameters given in Table 6.1. Figure 6.1 plots the average E[S n ] as a function of the number of flips n. Figure 6.2 plots the exact, SPA, and finite difference sensitivities s as a function of the number of flips n. This figure illustrates that the SPA estimate varies significantly less than the finite difference estimate. In fact, the SPA sensitivity appears to have roughly the same amount of error as the simulated estimate of the mean.
115 93-2 Exact SPA Finite Difference -4 E[S n ] θ Number of coin flips n 8 1 Figure 6.2: Mean sensitivity E[Sn] θ State-Dependent Simulation Example as a function of the number of coin flips n In the previous example, each of the n flips are independent, and the probability for choosing heads or tails depended solely on the parameter θ. We now consider calculating the sensitivity for a Markov chain in which the transition probabilities are state dependent, e.g. { n P (n k+1 n k ; θ) = k + ν 1 if U < H 1 (n k ; θ) (6.47) n k + ν 2 if U > H 1 (n k ; θ) [ ] T n k = A k B k (6.48) [ ] ] 1 1 ν = [ν 1 ν 2 = (6.49) 1 1 r 1 (n k ; θ) = k 1 A k 1 + θa k (6.5) r 2 (n k ; θ) = k 2 B k (6.51) H j (n k ; θ) = r j (n k ; θ) 2 k=1 r k(n k ; θ) (6.52) We consider a total of n discrete decisions. One characterization for this system is z i = {n, v i 1 (θ),..., vi n(θ)}; namely, the initial state n and the string of discrete decisions v j (θ) s for the ith simulation. We note that the simulation uses random numbers to generate the discrete decisions (v j (θ) s). Identifying the discrete decision by the v j (θ) s is more conducive for calculating sensitivities than the string of random numbers. We turn our attention towards calculating the quantity E[n i 1 zi ]. Suppose that the first
116 94 x k Nominal path Perturbed path Figure 6.3: Comparison of nominal and perturbed path for SPA analysis U H(n, θ) 1 H(n, θ + ) U H(n, θ) 1 H(n, θ + ) Figure 6.4: SPA analysis of the discrete decision. Given a positive perturbation >, H(n, θ + ) < H(n, θ). Therefore if decision v 2 is chosen given the nominal parameter θ, no perturbed parameter can change the choice to v 1. decision yields v 1 (θ) = ν 1. We again ask the same question: what if we had chosen ν 2 instead of ν 1? Figure 6.3 illustrates this question, in which the a new perturbed path deviates from the nominal path. Therefore, we must calculate the conditional probabilities P (v 1 (n, θ + ) = ν 1 v j (n, θ) = ν 1, n ) and P (v 1 (n, θ + ) = ν 2 v j (n, θ) = ν 1, n ). To do so, we again use the fact that the random variable v 1 (n, θ) can be written in terms of the uniform distribution U P (v 1 (n, θ) = ν 1 ) = P (U < H 1 (n, θ)) (6.53)
117 Assuming that the parameter perturbation > 3, we evaluate the conditional probabilities: 95 P (v (θ +, n ) = ν 1 v (θ, n ) = ν 1, n ) = P (U < H 1 (θ +, n ) U < H 1 (θ, n )) (6.54) = P (U < H 1(θ +, n ), U < H 1 (θ, n )) P (U < H 1 (θ, n )) = H 1(θ +, n ) H 1 (θ, n ) (6.55) (6.56) P (v (θ +, n ) = ν 2 v (θ, n ) = ν 1, n ) = P (U > H 1 (θ +, n ) U < H 1 (θ, n )) (6.57) = P (H 1(θ +, n ) < U < H 1 (θ, n )) P (U < H 1 (θ, n )) = H 1(θ, n ) H 1 (θ +, n ) H 1 (θ, n ) (6.58) (6.59) We note that if ν 2 is chosen, no perturbation > could change the reaction; see Figure 6.4 for an illustration of why. Defining Then the desired conditional expectation is k 1 n k = n + v j (θ, n j ) (6.6) j= k 1 ˆn k = n + v j (θ +, ˆn j ) (6.61) j= E[n 1 (θ) z] θ P (v (θ +, n ) = ν 1 v (θ, n ) = ν 1 ) = lim (n 1 n 1 ) (6.62) + P (v (θ +, n ) = ν 2 v (θ, n ) = ν 1 ) (ˆn 1 n 1 ) (6.63) H 1 (θ, n ) H 1 (θ +, n ) = lim (ˆn 1 n 1 ) H 1 (θ, n ) (6.64) = θ [H 1(θ, n )] (ν 2 ν 1 ) (6.65) H 1 (θ, n ) In general, we are interested in calculating the sensitivity of E[n j (θ) z], i.e. E[n j (θ) z] θ 1 = lim N N v j 1 i=1 v P (ˆn j (θ + ) zi (θ) (ˆn j n j ) (6.66) so we must consider the probability P (n j (θ + ) z i (θ)). Using the properties of conditional 3 We can also calculate the conditional expectations assuming that <.
118 96 x k Nominal path 1 2 Perturbed paths Figure 6.5: Illustration of the branching nature of the perturbed path for SPA analysis densities and Markov chains, we have P (n j (θ + ) z i (θ)) = P (v j,..., v 1 ; θ + z i (θ)) (6.67) v 1 v j 1 = P (v j ; θ + z i (θ), v j 1,..., v 1 ; θ + ) P (v 1 ; θ + z i (θ)) v 1 v j 1 (6.68) in which we use the notation P ( ; θ) to denote that the quantity P ( ) is a function of the parameter θ. It is clear that this process branches at every discrete decision, as shown in Figure 6.5, and that we must follow each of these branches with nonzero weight throughout the duration of the simulation. Figures 6.6 and 6.7 plot the mean and sensitivity comparison for this example. The SPA estimate demonstrates superior reconstruction of the sensitivity in comparison to finite differences, albeit at a greater computational expense. Surprisingly, though, the SPA estimate for each sample path did not require tracking perturbed paths for all possible state combinations due to the coalescing of many perturbed paths. However, we do not expect this feature to hold for more models that span larger dimensions, particularly those that include more discrete decisions of the form n k + ν 1 if U < H 1 (n k ; θ) n k + ν 2 if H 1 (n k ; θ) < U H 2 (n k ; θ) P (n k+1 n k ; θ) = (6.69). n k + ν m if U > H m 1 (n k ; θ) Accordingly, we could consider using a particle filter to track the perturbed paths.
119 E[x k ] 1 Exact Monte Carlo Number of decisions k 8 1 Figure 6.6: Mean E[n k ] as a function of the number of decisions k E[x k ] θ Number of decisions k Exact SPA Finite Difference 8 1 Figure 6.7: Mean sensitivity E[n k] θ as a function of the number of decisions k 6.2 Smoothing by Integration In some cases, the SPA estimate is not easily calculable. Consequently, we are interested in simpler means of calculating sensitivities. As noted in the previous section, conditional expectation provides one means of smoothing discrete sample paths. In some sense, expectation may be viewed as an integration over the state space. For some systems, it may be more
120 98 advantageous to integrate over a variable other than the state space. In this section, we consider the simple example of a state-dependent timing event with no discrete decisions. In particular, we address calculation of sensitivities for stochastic chemical kinetics given only one reaction. In this case, infinitesimal perturbation changes effect only when the single reaction occurs. To begin this analysis, we examine the first possible reaction. We can then define the discrete state n as n(t; θ) = { n, t t < t + τ 1 n + ν, t t + τ 1 (6.7) τ 1 = log(p 1) r tot (n ; θ) (6.71) in which t is the initial time, n is the initial state, t +τ 1 is the next reaction time, ν T is the stoichiometric matrix, ant θ is a vector of parameters. We can write equation (6.7) alternatively as n(t; θ) = n + ν t t δ ( t τ 1 (n ; θ) ) dt (6.72) The smoothing trick that we apply here is to define an integrated sensitivity s I in terms of the state integrated with respect to time, i.e. s I ( t ) θ T n(t ; θ)dt (6.73) t Integrating equation (6.72) with respect to time yields ( t t n(t ; θ)dt = t t n + ν t t δ ( t τ 1 (n ; θ) ) dt ) dt (6.74) We can differentiate equation (6.74) with respect to the parameters θ to yield τ 1 s I (t; θ) = s I + ν r tot (n ; θ) t t δ ( t τ 1 (θ) ) dt (6.75) We can similarly show for an arbitrary number of µ reactions that t t t n(t ; θ)dt t µ = n + ν δ ( t τ j (n + (j 1)ν; θ) ) dt dt (6.76) t t s I (t; θ) = s I + ν t t j=1 µ δ ( t τ j (n + (j 1)ν; θ) ) dt (6.77) j=1 Convergence of s I to the integral of the mean sensitivity follows from the law of large numbers. We consider a simple example to illustrate this technique. The single reaction is 2A k B (6.78)
121 Exact Simulation (a) x Time 6 8 (b) 1 s I Exact Simulation 2 4 Time Figure 6.8: Comparison of the exact and simulated (a) mean and (b) mean integrated sensitivity for the irreversible reaction 2A B. with the reaction elementary as written. The initial condition is n A = 2 and n B = molecules, and the parameter value is k = 1/15. We solve for the mean and its integrated sensitivity both exactly (via solution of the master equation) and by Monte Carlo reconstruction. For the latter case, we average fifty simulations to reconstruct the mean behavior and apply equation (6.77) to evaluate the integrated sensitivity. Figure 6.8 presents the results for this case, and demonstrates excellent agreement between the exact and reconstructed values for both the mean and the integrated sensitivity. In general, we require values for the sensitivity rather than the integrated sensitivity. There are numerous possibilities for deriving this quantity. For example, a polynomial can be fitted through the integrated sensitivity. The derivative of this fitted polynomial would then provide an estimate for the desired sensitivity. As seen in Figure 6.8 (b), however, the reconstructed integrated sensitivity can be noisy. Therefore, we recommend against low-order differencing of the integrated sensitivity due to the fact that such differencing amplifies noise.
122 1 6.3 Sensitivity Calculation for Stochastic Chemical Kinetics Thus far, we have considered calculation of sensitivities for first the discrete-time case (only choosing from a finite number of discrete events for the next reaction), and the time-dependent case with no discrete event selection. The stochastic chemical kinetics problem, however, is a combination of both time-dependent and discrete events. We envision that this problem could be addressed using the tools presented in previous sections, namely smoothed perturbation analysis (for the which reaction choice) and smoothing by integration (for the timing of the reaction). The problematic part for this particular problem, however, is really in implementing SPA. Because time is continuous, the selection of discrete events does not have the property of occurring at the same time for every simulation, as was the case in the discrete-time case. Hence there is no fortuitous coalescing of perturbed paths; in fact, one must nominally track every generated perturbed path to obtain the SPA estimate. Such a task seems unreasonable computationally for all but the simplest models. One potential means around this problem is to bound the computational expense by tracking only the paths that contribute the most to the SPA estimate. However, the problem of continuous time again appears because the perturbed paths are potentially at different time points in the simulation, making comparison of these paths difficult. 6.4 Conclusions and Future Directions This chapter explored methods for solving the sensitivity of moments of the master equation from simulation via smoothing. We first examined smoothing by conditional expectation, or smoothed perturbation analysis, to address the case of sensitivities for time-independent, discrete event systems. We then applied smoothing by time integration to account for the effect of parameters on the timing of continuous events. Finally, we briefly examined how one might apply these two methods to evaluate sensitivities for stochastic chemical kinetics. As of the writing of this thesis, we do not know of a satisfactory method for efficiently evaluating unbiased estimates for the sensitivity of moments of the discrete master equation. Thus we are forced to conclude that the best options for evaluating these sensitivities are either the approximation proposed previously in Chapter 5, or finite differences. We can speculate on possible methods for doing so as presented next. We speculate that directly solving the companion sensitivity equation to the master equation may offer some hope for calculation of unbiased sensitivities. Considering all the individual probabilities and their respective sensitivities, i.e. P (n ; θ) P (n; θ) = P (n 1 ; θ) (6.79) S(n; θ) =. P (n; θ) θ T (6.8)
123 We can write the evolution equations for the master equation and its sensitivity as linear systems 11 dp (n; θ) = A(θ)P (n; θ) dt (6.81) ds(n; θ) = A(θ)S(n; θ) + J(θ)P (n; θ) dt (6.82) J(θ) = A(θ) θ (6.83) Integrating equation (6.82) with respect to time yields the convolution integral S(n; θ) = e At S(n; θ) + t e A(t t ) J(θ)P (n, t ; θ)dt (6.84) The primary drawback to this method is that the sensitivity equation (6.84) has the same large dimensionality as the master equation, the same problem that forced us to use simulation to solve the master equation. We can attempt to solve the sensitivity equation using simulation, but this method also suffers several drawbacks. First, the sensitivity is not a probability distribution, so we must recast the problem into a form conducive for solution by simulation. Even after doing so, solving for the sensitivity requires knowledge of the probability distribution, which is presumably reconstructed from simulation. Hence even if we could exactly solve for the sensitivity, the result would only be as accurate as the reconstructed probability density. The primary appeal of this method, though, is that the simulations used to reconstruct the probability density can also be used to evaluate the sensitivity, i.e. the convolution integral in equation (6.84). If we can efficiently store and retrieve this information, solving the sensitivity equation would require little or no additional simulation. Notation H j n N P p j P r j S n S s I t U v j X jth transition probability discrete state vector number of simulations probability jth random number vector of probabilities jth transition rate sum of n-independent coin flips matrix of probability sensitivity vectors time-integrated sensitivity time uniform distribution jth discrete decision random variable
124 12 x j z ν τ θ θ Ω jth realization of the random variable X characterization of a simulated trajectory finite difference perturbation stoichiometric matrix next reaction time parameter vector of parameters random number string used for simulation
125 13 Chapter 7 Sensitivity Analysis of Stochastic Differential Equation Models The purpose of this chapter is to develop and present methods for using stochastic differential equation models for purposes other than pure simulation. As a simulation tool, these types of models are becoming an increasingly popular method for introducing science and engineering students and researchers to the molecular world in which random fluctuations are an important physical phenomena to be captured in the model. If we consider systems levels tasks, such as parameter estimation, model-based feedback control, and process and product design, we require a different set of tools than those required for pure simulation. Many systems level tasks are conveniently posed as optimization problems, and brute force optimization of these highly noisy simulation models either fails outright or is so time consuming that the entire exercise becomes tedious and frustrating. Simply attaching an optimization method to a stochastic simulation model is inefficient if we do not consider the engineering task that might come later when the simulation is created. We propose adding a small piece of code to the stochastic simulation that exactly computes the sensitivity of the trajectory to all model parameters of interest. These parameters may be kinetic parameters to be estimated from data or control decisions used to control the dynamic or steady-state behavior of the system. Sensitivity analysis of stochastic differential equations (SDEs) is by no means a new concept. To the best of our knowledge, Dacol and Rabitz [23] first proposed such an analysis. These authors suggested using a Green s function approach to solve for the sensitivity of moments of the underlying probability distribution. In this chapter, we propose differentiating simulated sample paths directly to calculate the same sensitivities. We first review the master equation of interest and define the sensitivity of moments of this equation with respect to model parameters. Next we propose and compare several methods for calculating these sensitivities with an eye on computational efficiency. Finally, we illustrate how to use the sensitivities for calculating parameter estimates, computing steady states, and computing quantities for polymer models.
126 The Master Equation We consider the following master (Fokker-Planck) equation: P (x, t; θ) t = l i=1 x i (A i (x; θ)p (x, t; θ)) l l i=1 j=1 2 x i x j ( Bij (x; θ) 2 P (x, t; θ) ) (7.1) in which x is the state vector for the system, θ is the vector of parameters, t is time, P (x, t; θ) is the probability distribution function, A i denotes the ith element of the vector A, and B ij denotes the (i, j)th element of the matrix B. Many different boundary conditions are possible for this system (see, for example, Gardiner [41]); for this chapter, we use reflecting boundary conditions of the form A i (x; θ)p (x, t; θ) l l i=1 j=1 x j ( Bij (x; θ) 2 P (x, t; θ) ) = (7.2) unless specified otherwise. Defining the sensitivity S(x, t; θ) as S(x, t; θ) P (x, t; θ) θ (7.3) we can differentiate equation (7.1) with respect to θ to obtain the sensitivity evolution equation P (x, t; θ) = θ t θ S(x, t; θ) t = l i=1 l i=1 x i l l i=1 j=1 x i (A i (x; θ)p (x, t; θ)) ( Ai (x; θ) 2 θ x i x j l l i=1 j=1 ) P (x, t; θ) + A i (x; θ)s(x, t; θ) ( Bij (x; θ) θ 2 ( Bij (x; θ) 2 P (x, t; θ) ) x i x j ) 2B ij (x; θ)p (x, t; θ) + B ij (x; θ) 2 S(x, t; θ) Clearly solution of equation (7.5) requires the solution of equation (7.1), but not vice-versa. In general, we are interested in moments of the probability distribution, i.e. g(x) = x (7.4) (7.5) g(ω)p (ω, t; θ)dω (7.6) in which g(x) and g(x) are vectors. For example, we might seek to implement control moves that drive the mean system behavior towards a desired set point. Such tasks require knowledge of how sensitive these moments are with respect to the parameters. The master equation (7.1) indicates that the probability distribution evolves continuously with time; consequently, moments of this distribution (assuming that they are well defined) evolve continuously as well.
127 Therefore we can simply differentiate equation (7.6) with respect to the parameters to define the sensitivity of these moments, s(g(x)), as follows: 15 θ T g(x) = θ T g(ω)p (ω, t; θ)dω (7.7) x s(g(x), t; θ) = g(x)s(x, t; θ) T (7.8) x Here, s(g(x), t; θ) is a matrix. Equation (7.8) indicates that these sensitivities depend upon the sensitivity of the master equation, S(n, t; θ). Therefore, the exact solution of s(g(x)) requires simultaneous solution of equations (7.1), (7.5), and (7.8). As opposed to exactly solving for the desired moments of the master equation, we can reconstruct these moments via simulation. The master equation (7.1) has Itô solution of the form dx i = A i (x; θ)dt + l B ij (x; θ)dw j (7.9) in which W is a vector of Wiener processes. We can simulate trajectories of equation (7.9) by using, for example, an Euler scheme [4], then tabulate this trajectory information to reconstruct the desired moments (7.6) by applying the law of large numbers j=1 g(x) = x 1 g(ω)p (ω, t; θ)dω = lim N N N g(x i ) 1 N i=1 N g(x i ) for finite N (7.1) i=1 in which N is the number of simulated trajectories and x i is the value of the state for the ith simulation. Logically, then, we could also attempt to reconstruct the sensitivities from the simulated sample paths alone. This analysis requires some care; in particular, we must justify interchanging the operators of expectation and differentiation, i.e. E[g(x i ; θ +, Ω 1 )] E[g(x i [ ; θ, Ω 2 )] lim = E lim g(x i ; θ +, Ω 1 ) g(x i ] ; θ, Ω 2 ) (7.11) in which E( ) denotes the expectation operator and Ω 1 and Ω 2 refer to the random numbers used to generate the desired expectations. Because the individual sample paths are continuous, the interchange is justifiable and we can merely differentiate equation (7.9) with respect to θ θ dx i = l A i (x; θ)dt + B ij (x; θ)dw j (7.12) θ ds i = A i(x; θ) x j=1 s i + A i(x; θ) dt + θ l ( Bij (x; θ) s i + B ) ij(x; θ) dw j (7.13) x θ j=1
128 16 in which s i is defined as s i x i (7.14) θ Consequently, we can evaluate the desired moments and sensitivities of these moments by simultaneously evaluating equations (7.9) and (7.13). Additionally, we have the choice of using either the same strings of random numbers for evaluation of equation (7.11), i.e. Ω 1 = Ω 2, or different strings of random numbers. The former case corresponds to differentiating individual sample paths and consequently using the same values for the state and Brownian increments to evaluate the desired moments and their sensitivities. This subtle distinction actually results in dramatic differences in the evaluated sensitivities as pointed out by Fu and Hu [37]. We illustrate this point in the examples. 7.2 Sensitivity Examples We now consider two motivating examples comparing parametric and finite difference sensitivities. The first example is a single, reversible reaction that demonstrates the accuracy of the parametric and finite difference sensitivities. The second example consists of the Oregonator reactions and illustrates the superiority of the parametric sensitivity over finite differences Simple Reversible Reaction We consider the reversible reaction 2A k 1 B ɛ =.5k 1 c A (c A 1) k 2 c B (7.15) k 1 in which ɛ denotes the extent of reaction. Parameter values for this example are given in Table 7.1. We solve the master equation (7.1) and its sensitivity (7.5) for the extent of reaction ɛ by using finite differences to discretizing the ɛ dimension ( ɛ = 2), then using DASKR (a variant of the package DASPK [15]) to integrate the resulting system of differential-algebraic equations. We also use simulation to evaluate the mean of the stochastic differential equation (7.9) and its sensitivity (7.13). Here, we use a first-order Euler integration with a time increment of t = 1 2. We reconstruct the mean with ten simulations. Figure 7.1 compares the mean results for the master equation, parametric sensitivity, and finite difference sensitivity. For this figure, we have chosen a central finite difference scheme with a perturbation of 1 8 of the parameter value. Figure 7.1 (a) demonstrates that ten simulations yield a reasonable approximation to the mean. Figures 7.1 (b) and (c) illustrate that the parametric and finite difference mean sensitivities yield indistinguishable results (to the scale of the graph), and that these results are similar to those of the master equation. Figure 7.2 compares the mean sensitivity of parameter k 2 for the master equation and finite difference sensitivity. Rather than use the same random numbers for evaluation of the sensitivity, we evaluate the perturbed expectations using different strings of random numbers (i.e. Ω 1 Ω 2 in equation (7.11)). Figure 7.2 presents these results for a parameter perturbation
129 of 1%. The finite difference result is substantially noisier than when using the same strings of random numbers for each perturbation (e.g. Figure 7.1 (b)). The noise directly results from the error due to the finite number of simulations used to reconstruct the mean. In fact, the finite simulation error completely swamps the sensitivity calculation when using a parameter perturbation of 1% or less. This result underscores the importance of differentiating the sample trajectories to obtain sensitivity information. 17 Parameter Value k 1 1/45 k 2 1/3 P (c A = 15, c B = 25, t = ) 1 Table 7.1: Parameter values for the simple reversible reaction Oregonator We now consider the Oregonator system of reactions [32] W + B k 1 A (7.16) A + B k 2 X (7.17) Y + A k 3 2A + C (7.18) 2A k 4 Z (7.19) C k 5 B (7.2) Reactions are elementary as written. Parameters for this system are given in Table 7.2. It is assumed that concentrations of species W and Y remain constant throughout the reaction. Additionally, we track only species A, B, and C since species X and Z are products. We use simulation to evaluate a single trajectory of the stochastic differential equation (7.9) and its sensitivity (7.13). Here, we use a first-order Euler integration with a time increment of t = 1 3. The initial condition is P (c A = 5, c B = 1, c C = 2, t ) = 1. Figure 7.3 presents the results for this example. Figure 7.3 (a) demonstrates that for the given set of parameters, these reactions yield a stable, oscillatory response. Although Figure 7.3 (b) shows good visual agreement between the parametric and finite difference sensitivities, plot (c) clearly shows that the difference between these two sensitivities is actually increasing with time even though the finite difference perturbation is small (1 8 of the parameter k 1 ).
130 18 Number of molecules A B Exact Simulation (a) 2 4 Time sk B A Exact Parametric FD (b) -2 sk Time A B 6 8 Exact Parametric FD (c) Time Figure 7.1: Results for the simple reversible reaction: (a) comparison of the exact and reconstructed mean (by simulation); (b) comparison of the exact, parametric, and finite difference (FD) sensitivities for parameter k 1 ; and (c) comparison of the exact, parametric, and finite difference (FD) sensitivities for parameter k 2. Here, the finite difference perturbation is 1 8 of each parameter.
131 19 sk B A Exact FD 2 4 Time Figure 7.2: Results for the simple reversible reaction: comparison of the exact and finite difference (FD) sensitivities for parameter k 1 using different random numbers for each finite difference expectation. Here, the finite difference perturbation is 1 1 of the parameter k 1. Parameter Value k 1 c W 2 k 2.1 k 3 c Y 14 k 4.16 k 5 26 Table 7.2: Parameter values for the Oregonator system of reactions. 7.3 Applications of Parametric Sensitivities We now turn our attention to applications of parametric sensitivities. We first consider estimating parameters for the simple, reversible reaction of section We then perform steady-state analysis for the Oregonator reactions of section Finally, we use parametric sensitivities to evaluate the viscosity of a simple dumbbell model Parameter Estimation The goal of parameter estimation is to determine the set of parameters that best reconciles the measurements with model predictions. The classical approach is to assume that measurements are corrupted by normally distributed noise. Accordingly, we calculate the optimal parameters
132 11 Concentration 1 3 Sensitivity 1 4 Sensitivity c A c B c C c A c B c C Time Time Time c A c B c C Figure 7.3: Results for one trajectory of the Oregonator cyclical reactions: (a) simulated trajectory, (b) parametric and finite difference (FD) sensitivity for parameter k 1, and (c) difference between the parametric and finite difference sensitivities. Here, the finite difference perturbation is 1 8 of the parameter.
133 via the least squares optimization min θ Φ = 1 2 e T k Π 1 e k s.t.: x k+1 = F (x k ; θ) k e k = y k h(x k ) 111 (7.21a) (7.21b) (7.21c) in which Φ is the objective function value, e k s denote the difference between the measurements y k and the model predictions h(x k ), and Π is the covariance matrix for the measurement noise. For the optimal set of parameters, the gradient θ Φ is zero. We can numerically evaluate the gradient according to θ Φ = 1 θ T e T k 2 Π 1 e k (7.22) k = ( ) h(xk ) x T k x T k k θ T Π 1 e k (7.23) = ( ) h(xk ) T x T s k Π 1 e k (7.24) k k Equation (7.24) indicates that the gradient depends upon s k, the sensitivity of the state with respect to the parameters. In general, most experiments do not include many replicates due to cost and time constraints. Therefore, the best experimental data we are likely to obtain is the average. In fitting these data to stochastic models governed by the master equation, we accordingly choose the mean x as the the state of interest. Monte Carlo simulation and parametric sensitivities provide estimates of the mean and its sensitivity. For the sake of illustration, we obtain optimal parameter estimates using an unconstrained, line-search optimization with BFGS Hessian update; for further details on this method, we refer the interested reader to Nocedal and Wright [97]. Here, we provide the optimizer with both the objective function and the gradient given in equations (7.21) and (7.24), respectively. Although the Monte Carlo reconstruction of the mean is nominally stochastic, by reusing the same string of random numbers for every optimization iteration the objective function given in equation (7.21) becomes, in a sense, deterministic. Additionally, the objective function is continuous with respect to the parameters. Some care must be taken to ensure that the string of random numbers used by the optimization gives a representative reconstruction of the mean (recall that the finite number of simulations introduces some error in the reconstructed mean). Practically, this condition can be checked by optimizing the model with several different random number strings. We reconsider the simple reversible reaction of section We assume that we can measure the average amount of c B with a sampling time of t =.2. Experimental data are generated using the parameters given in section with the exception that one hundred
134 112 Measurement nb Time k (a) (b) 1 Parameter k Iteration Figure 7.4: Results for parameter estimation of the simple reversible reaction example: (a) comparison of the experimental (points) and predicted (line) measurements and (b) convergence of the optimized parameters (dashed lines) to the true values (solid lines) for the proposed scheme. simulations are used to generate the mean behavior. For the parameter estimation, we attempt to estimate both log base ten values of k 1 and k 2 using a different seed for the random number generator than that used to generate the experimental data. We estimate log 1 values to prevent both numerical conditioning problems and negative estimates of the rate constants. Figure 7.4 presents the results of this estimation. The experimental and predicted measurements agree well, and the parameter estimates quickly converge to close to the true values. The offset between the estimated and true parameters is expected due to the finite simulation error since different seeds are used to generate the experimental and predicted measurements. To determine the accuracy of the optimization, we analyze both the gradient and the Hessian of the objective function. Differentiating the gradient (i.e. equation (7.24)) yields the
135 Hessian θθ Φ θθ Φ = θ T ( k ( ) ) h(xk ) T s k Π 1 e k x T k 113 (7.25) = k ( h(xk ) x T k ) T s k Π 1 h(x k) x T s k + k ( h(xk ) x T k 2 x k θ k θ T k ) T Π 1 e k (7.26) Making the usual Gauss-Newton approximation for the Hessian (i.e. e k ), we obtain θθ Φ k ( h(xk ) x T k ) T s k Π 1 h(x k) x T s k (7.27) k For this optimization, the values of the gradient and the approximate Hessian are θ Φ = [ ] [ ] θθ Φ = Examining the eigenvalue/eigenvector (λ/ν) decomposition of the Hessian yields [ ] λ 1 = , ν 1 = [ ] λ 2 = , ν 2 = (7.28) (7.29) (7.3) (7.31) Because the gradient is reasonably small and the Hessian is positive definite (eigenvalues are positive), we conclude that the optimizer has indeed converged to a local minimum Calculating Steady States Exact determination of steady states requires solving for the stationary state of the master equation (7.1). The difficulty of this task is comparable to that of solving the dynamic response. In this section, we use the result from section 5.4 which allows us to determine stationary points for moments of the underlying probability distribution given short bursts of simulation. The difference in the analysis presented here is that (1) the considered master equation is of the Fokker-Planck type, i.e. equation (7.1), and (2) sensitivities of the simulated moments can be determined exactly. We now apply this method to the Oregonator system of reactions previously presented in section For the steady-state calculation, we calculate the evolution of the mean using a short burst of simulation ( ss = 1 2 ). We use an Euler integration with time increment t = 1 3 to evaluate ss. One hundred simulations are used to reconstruct the mean. Figure 7.5 presents the convergence of the steady-state calculation per completed Newton iteration. The majority
136 114 x C B A Iteration Figure 7.5: Results for steady-state analysis of the Oregonator reaction example: estimated state per Newton iteration. of the convergence occurs within the first five iterations. The calculated mean and sensitivity are [ T x = ] (7.32) s = (7.33) Analyzing the eigenvalues of the mean sensitivity yields [ ] λ = i i (7.34) which indicates by linear stability analysis (see Chen [21] for further details) that the steady state is unstable, as expected Simple Dumbbell Model of a Polymer in Solution We now consider calculation of the zero-shear viscosity for a simple dumbbell model of a polymer molecule in solution. For this model, two dumbbells are connected by a Hookean spring. We track the coordinates of each dumbbell, in which ( ) H dx 1 = ζ (x 2 x 1 ) + ( v) T x 1 dt + 2DdW 1 (7.35a) dx 2 = ( Hζ ) (x 2 x 1 ) + ( v) T x 2 dt + 2DdW 2 (7.35b) v = γ (7.35c)
137 in which x 1 and x 2 are the Cartesian coordinates of each dumbbell, H is the spring constant, ζ is the friction coefficient, v is the velocity field, D is the diffusivity of each bead, and W is a vector of Wiener processes. The stress τ is defined as 115 τ =< Hqq T > nkt δ (7.36) in which < > denotes the expectation operator and q = x 1 x 2. For this system, the viscosity η is η = τ 12 γ (7.37) γ= Defining γ as the parameter of interest, the viscosity η clearly becomes a function of the sensitivities s 1 = x 1 γ s 2 = x 2 γ (7.38) (7.39) Parameter Symbol Value Friction coefficient ζ 1. Diffusivity D 1 4 Spring constant H 1. Table 7.3: Parameters for the simple dumbbell model. Quantity Symbol Value Analytical viscosity η Estimated viscosity η e ± Table 7.4: Results for the simple dumbbell model. Standard deviation calculated by grouping the simulation results into groups of ten, then determining the standard deviation of the resulting ten averages. We simulate equation (7.35) using an Euler discretization with time increment of t = 1 2. The expectation < Hqq T > is calculated by averaging the time courses of one hundred simulations with a time period of 1. Parameters for the model are given in Table 7.3. For this simple example, the viscosity can be calculated exactly as η = Dζ2 4H (7.4) Table 7.4 presents the results of this simulation. The viscosity calculated using parametric sensitivities compares favorably to the exact value.
138 Conclusions We have proposed differentiating simulated sample paths to obtain parametric sensitivities for models consisting of stochastic differential equations. The sensitivity equations are evaluated simultaneously with the model equations to yield accurate, first-order information about the simulated trajectories. Two simple examples demonstrated the accuracy of this technique in comparison to both finite differences and the solution of the underlying master equation and its sensitivity. These results underscore the importance of differencing each simulated trajectory rather than trajectories generated using different strings of random numbers. However, we observed little difference between the accuracy of parametric and finite difference sensitivities. Additionally, we have demonstrated how these sensitivities can be used to perform systems-level tasks for this class of models. The examples included using nonlinear optimization to estimate parameters, performing steady-state analysis, and evaluating derivatives for polymer models efficiently. We expect these tools to prove useful in a wide range of applications, from more complex polymer models to financial models. Notation c j D e k g(x) H h(x k ) k j N P (x, t; θ) q S(x, t; θ) s(g(x)) s t W x x i y k ɛ η η e λ ν concentration of species j diffusivity difference vector between the measurements y k and the model predictions h(x k ) at time t k moment of the probability distribution spring constant model-predicted measurement vector at time t k rate constant for the jth reaction number of simulated trajectories probability distribution function distance vector for the dumbbell model sensitivity of the probability distribution function sensitivity of a moment of the probability distribution sensitivity of x for a simulated trajectory time vector of Wiener processes state vector value of the state x for the ith simulation measurement vector at time t k extent of reaction viscosity estimated viscosity eigenvalue eigenvector
139 117 Φ Π ζ τ θ Ω objective function value covariance matrix for the measurement noise friction coefficient shear matrix vector of model parameters random number string used for simulation
140 118
141 119 Chapter 8 Stochastic Simulation of Particulate Systems 1 The stochastic chemical kinetics approach provides one method of formulating the stochastic crystallization population balance equation (PBE). In this formulation, crystal nucleation and growth are modeled as sequential additions of solubilized ions or molecules (units) to either other units or an assembly of any number of units. Monte Carlo methods provide one means of solving this problem. In this chapter, we assess the limitations of such methods by both (1) simulating models for isothermal and nonisothermal size-independent nucleation, growth and agglomeration; and (2) performing parameter estimation using these models. We also derive the macroscopic (deterministic) PBE from the stochastic formulation, and compare the numerical solutions of the stochastic and deterministic PBEs. The results demonstrate that even as we approach the thermodynamic limit, in which the deterministic model becomes valid, stochastic simulation provides a general, flexible solution technique for examining many possible mechanisms. Thus the stochastic simulation permits the user to focus more on modeling issues as opposed to solution techniques. 8.1 Introduction Both deterministic and stochastic frameworks have been used to describe the time evolution of a population of particles. The classical deterministic framework consists of coupled population, mass, and energy balances which describe crystal nucleation, growth, agglomeration, and breakage as smooth, continuous processes. Randolph and Larson [11], Hulburt and Katz [65], and Ramkrishna and Borwanker [18, 19] have extensively studied the analysis and treatment of the deterministic population balance equation (PBE) to these crystal formation mechanisms. Hulburt and Katz [65] made a seminal contribution in which they develop a population balance that includes an arbitrary number of characteristic variables. They use the method of moments to solve the PBE for a variety of applications such as modeling systems with one or two length dimensions, and modeling agglomerating systems. Ramkrishna [17] 1 Portions of this chapter to appear in Haseltine, Patience, and Rawlings [56].
142 12 provides an excellent summary of techniques used to solve the deterministic balances for models in a single distributed dimension. Ma, Braatz, and Tafti [84] apply high resolution methods to solve the deterministic balances with two characteristic length scales. If the population is large, single microscopic events such as incorporation of growth units into a crystal lattice and biparticle collisions are not significant. Microscopic events tend to occur on short time scales relative to those required to make a significant change in the macroscopic particle size density (PSD). If fluctuations about the average PSD are large, then the deterministic PBE is no longer valid. Large fluctuations about the average density occur when the population modeled is small. Examples of small populations in particulate systems in which fluctuations are significant include such varied applications as aggregation of platelets and neutrophils in flow fields, growth and aggregation of proteins, and aggregation of cell mixtures [79]. The deterministic PBE is also not valid in modeling precipitation reactions in micelles in which the micelles act as micro-scale reactors containing a small population of fine particles [86, 8]. In contrast to the deterministic framework, the stochastic framework models crystal nucleation, growth and agglomeration as random, discrete processes. Ramkrishna and Borwanker [18, 19] introduce the stochastic framework to modeling particulate processes. The authors show that the deterministic PBE is one of an infinite sequence of equations, called product densities, that describe the mean behavior and fluctuations about the mean behavior of the PSD. The deterministic PBE is, in fact, the expectation density of the infinite sequence of equations satisfied by the product density equations. As the population decreases, higher order product density equations are required to describe the time behavior and fluctuations about the expected behavior of the population. We refer the interested reader to Ramkrishna [17] for the details of this analysis. One approach to solving the stochastic model for any population of crystals is the Monte Carlo simulation method. Kendall [72] first applies the concept of exponentially distributed time intervals between birth and death events in a single-species population. Shah, Ramkrishna, and Borwanker [136] use the same approach and simulate breakage and agglomeration in a dispersed-phase system. The rates of agglomeration and breakage are proportional to the number of particles in the system and the size-dependent mechanism of breakage and agglomeration. Laurenzi and Diamond [79] apply the same technique as Shah, Ramkrishna, and Borwanker to model aggregation kinetics of platelets and neutrophils in flow fields. Gooch and Hounslow [52] apply a Monte Carlo technique similar to Shah, Ramkrishna, and Borwanker to model breakage and agglomeration. Gooch and Hounslow calculate the event time interval from the numerical solution to the zeroth moment equation with N = 1 for breakage, and N = 1 for agglomeration. Manjunath et al. [86] and Bandyopadhyaya et al. [8] use the stochastic approach to model precipitation in small micellar systems. The model specifies the minimum number of solubilized ions and molecules to form a stable nucleus. Once a particle nucleates, growth is rapid and depletes the micelle of growth units. Brownian collisions govern the interaction between micelles. Solubilized ions and molecules are transferred during collisions.
143 In the stochastic approach developed here, nucleation and growth in a large-scale batch crystallizer are considered as a sequence of bimolecular chemical reactions. In particular, solubilized ions or molecules (units) sequentially add to other units or to an assembly of any number of units. Both Gillespie [46] and Shah, Ramkrishna, and Borwanker [136] propose equivalent methods for simulating exact trajectories of this random process. The expected behavior of the system can then be evaluated by averaging over many trajectory simulations. The burden of model solution rests mainly with the computing hardware, and these Monte Carlo simulations can be time intensive depending on the number of particles and the size of the molecular unit. Currently, desktop computers can simulate systems with reasonably large particle populations and small molecular units in a matter of seconds or minutes. In this chapter we first review the stochastic formulation of chemical kinetics and summarize the exact simulation method used to solve this system. We then extend the scope of the formulation to describe nonisothermal systems. Since this extension leads to a constraint that hinders the computation, we suggest an approximation that overcomes this obstacle. We then outline assumptions for formulation of the crystallization model. We illustrate the dependence of the stochastic solution on key stochastic parameters, such as cluster size and simulation volume. We also provide an analysis showing the connection between the stochastic formulation and the deterministic PBE. Next, we solve the stochastic formulation for models incorporating isothermal and nonisothermal, size-independent nucleation, growth and agglomeration and contrast the solution to that from the deterministic framework. We then address how to estimate parameters using stochastic models, and provide an example. Finally, we assess the limitations of the Monte Carlo simulation technique Stochastic Chemical Kinetics Overview In this section, we first review the stochastic formulation of chemical kinetics and one computational method for solving this problem. We then relax key assumptions of this problem formulation in order to address other interesting physical systems, and discuss one approximate computational solution method Stochastic Formulation of Isothermal Chemical Kinetics The stochastic formulation of chemical kinetics has its physical basis in the kinetic theory of gases [48]. The modeled system consists of well-mixed, gas-phase chemical species maintained at thermal equilibrium. The key model assumptions include 1) a hard-sphere molecular model and 2) non-reactive collisions occur much more frequently than reactive collisions. It is then possible to derive a deterministic time-evolution equation not for the state, but rather for the probability of being in a given state at a specific time. This evolution equation is the chemical master equation dp (x, t) dt = m a k (x ν k )P (x ν k, t) a k (x)p (x, t) (8.1) k=1
144 122 in which x is the state of the system in terms of number of molecules (a p-vector), P (x, t) is the probability that the system is in state x at time t, a k (x)dt is the probability to order dt that reaction k occurs in the time interval [t, t + dt), and ν k is the kth column of the stoichiometric matrix ν (a p m matrix). Here, we assume that the initial condition P (x, t ) is known. The solution of equation (8.1) is computationally intractable for all but the simplest systems. Rather, Monte Carlo methods are employed to reconstruct the probability distribution and its moments (usually the mean and variance). Monte Carlo methods take advantage of the strong law of large numbers, which permits reconstruction of functions of the probability distribution g(x) by drawing exact samples from this distribution, i.e. g(x) 1 g(x)p (x, t)dx = lim N N N g(x i ) 1 N i=1 N g(x i ) for N sufficiently large (8.2) in which g(x) is the average value of g(x), N is the number of samples, and x i is the ith Monte Carlo reconstruction of x. One efficient method for generating exact trajectories from the master equation is Gillespie s direct method [45, 46]. As noted previously, this particular simulation method is equivalent to the interval of quiescence technique proposed by Shah et al. [136]. This method was previously summarized in algorithm 1. i= Extension of the Problem Scope The previous problem formulation is quite restrictive from a modeling perspective. Firstly, many systems of interest are not solely gas phase. This restriction can be overcome by judicious modeling assumptions to ensure that neither thermodynamics nor conservation laws are violated. Secondly, the reaction propensities (a k s) often change between reaction events. For example, subjecting the system to a deterministic energy balance introduces time-varying reaction propensities into the system. In such cases the problem of interest is actually the following master equation subject to constraints: dp (x; t) dt dy(t) dt = m a k (x ν k, y)p (x ν k ; t) a k (x, y)p (x; t) k=1 (8.3a) = b(p (x), y; t) (8.3b) To solve equation (8.3) exactly, we must revise algorithm 1 to account for the time dependence of the propensity functions, a k (x, y) [47]. Since r tot and r k are functions of time, they must be
145 recalculated after determination of τ in order to choose which reaction occurs next. The major difficulty in this method is that in step 2 of the algorithm 1, we must now satisfy the constraint t+τ t 123 r tot (t )dt + log(p 1 ) = (8.4) as opposed to a simple algebraic relation. This constraint often proves to be computationally expensive. If the reaction propensities do not change significantly over the stochastic time step τ, the unmodified algorithm 1 can still provide an approximate solution. When the reaction propensities change significantly over τ, steps can be taken to reduce the error of algorithm 1. One idea is to scale the stochastic time step τ by artificially introducing a probability of no reaction into the system [57]: Let a dt be the contrived probability, first order in dt, that no reaction occurs in the next time interval dt. This probability does not affect the number of molecules of the modeled reactive system while allowing adjustment of the stochastic time step by changing the magnitude of a. Theoretically, as the magnitude of a becomes infinite, the total reaction rate becomes infinite. As the total reaction rate approaches infinity, the error of the stochastic simulation subject to ODE constraints approaches zero because the algorithm checks whether or not a reaction occurs at every instant of time. Practically, the algorithm should first check the no reaction propensity at each iteration to prevent needless calculation of the entire range of actual reactions. Finally, we note that even though the method outlined by Gillespie is exact [47], there is still error associated with the finite number of simulations performed since it is a Monte Carlo method. Thus it is plausible that the inherent sampling error may be greater than the error introduced by our approximation. Hence our approximation may often prove to be less computationally expensive than the simulation by Gillespie [47] while generating an acceptable amount of simulation error. We summarize our approximation in algorithm Interpretation of the Simulation Output Stochastic simulations of population balances involve two inherent and completely different distributions. First, each particle size N j has its own probability distribution P (N j, t) dictating the likelihood that the particle size contains a prescribed number of particles. Second, the population balance encompasses the entire distribution of these N j s. For the simulation results in this chapter, we perform multiple simulations given a specific initial condition. For each particle size at a given time, we then average over all simulations to obtain the expected numbers of particles for the given size, i.e. N j (t) = N Nj(t) i (8.5) i=1
146 124 Algorithm 6 Approximate Method (time-dependent reaction propensities). Initialize. Set the time, t, equal to zero. Set x and y to x and y, respectively. 1. Calculate: (a) the reaction propensities, r k = a k (x, y), and (b) the total reaction propensity, r tot = m k= r k. 2. Select two random numbers p 1, p 2 from the uniform distribution (, 1). 3. Let τ = log(p 1 )/r tot. Integrate dy/dt = b(x, y; t) over the range [t, t + τ) to determine y(t + τ). Let t t + τ. 4. Recalculate the reaction propensities r k s and the total reaction propensity r tot. Choose j such that j 1 j r k < p 2 r tot k= k= r k 5. Let x x + ν j. Update y if necessary. Go to 1. Here, N j (t) is clearly a scalar value. Finally, we tabulate all of these N j (t) s, the expected number of particles, to yield a mean population balance. This procedure is illustrated in Figure Crystallization Model Assumptions Certain key assumptions ensure the validity of the stochastic problem formulation. These assumptions are: 1. The system of interest is a well-mixed, constant volume, batch crystallizer. The wellmixed assumption implies that the crystallizer temperature is homogeneous; that is, if any event creates a temperature change, the thermal energy is instantaneously distributed throughout the crystallizer. 2. Particles have discrete sizes and size changes occur in discrete increments. On an atomic level, this assumption is physically true since crystals are composed of a discrete number
147 125 Distributions Scalars N 1 N 1.. Sample and Average Tabulate N j. N j. N n Stochastic Realizations N n Stochastic Averages Distribution of Stochastic Averages Figure 8.1: Method for calculating the population balance from stochastic simulation. Each particle size N j has its own inherent probability distribution. Monte Carlo methods provide samples from these distributions, and the samples are averaged to yield the mean value. Tabulating the mean values yields the mean of the stochastic population balance. of molecules. 3. The degree of supersaturation acts as the thermodynamic driving force for crystallization. This assumption is necessary to account for the system thermodynamics. Otherwise we would need to employ molecular dynamics simulations using an appropriate model for the potential energy function to more accurately describe the time evolution of the population balance. The downside of that choice is that our problem of interest, the macroscopic behavior of the crystallizer, becomes computationally intractable. The additional assumptions we use to simplify the solution of the population balance and reduce computational load are: 1. Physical properties for the heat capacity, liquid and crystal densities, and the heat of crystallization remain constant. 2. Nucleation, growth, and agglomeration rate constants are independent of temperature. 3. Crystal growth occurs in integer steps of a monomer unit. 4. The number of saturated monomers is an empirical function of temperature.
148 Stochastic Simulation of Batch Crystallization To illustrate the solution of the population balance via stochastic simulation, we examine three examples: 1. isothermal nucleation and growth; 2. nonisothermal nucleation and growth; and 3. isothermal nucleation, growth, and agglomeration. The mechanisms for each of these examples are size-independent. Also, we define the following nomenclature: M tot, M sat, and M are the total number of monomers, number of saturated monomers, and number of supersaturated monomers, respectively, on a per volume basis. Hence: is the characteristic volume of one monomer unit. N n is the number of particles with size l n = (n + 1). V is the system volume. M = M tot M sat (8.6) V mon is the initial volume of monomer. For these examples, V mon = 8V. n mon is the initial number of monomer particles, and is determined by the relation: n mon = V mon (8.7) n seed is the initial number of seed particles. For these examples, n seed = 1V Isothermal Nucleation and Growth Consider the isothermal reaction system with second-order nucleation and growth and a uniformly incremented volume scale = l i l i 1 : 2M kn N 1 (8.8a) N n + M kg N n+1 (8.8b) The model parameters are given in Table 8.1. We have chosen to model the crystallization mechanism using a volume scale in order to conserve mass (recall the constant crystal density assumption). In accord with this choice, the initial number of monomers are computed based on the assigned value of. Finally, we quadratically distribute the seeds over the particle volume interval l [2, 2.5].
149 127 Parameter Symbol Value nucleation rate constant k n growth rate constant k g number of saturated monomers M sat Table 8.1: Nucleation and growth parameters for an isothermal batch crystallizer Figure 8.2: Mean of the stochastic solution for an isothermal crystallization with nucleation and growth, 1 simulation, characteristic particle size =.1, system volume V = 1 Since k g is a constant, size-independent growth exhibits the same kinetics as the secondorder reaction: A + M kg B (8.9) Here the number of species A molecules is equivalent to the zeroth moment of the particle distribution N. We can reduce computational expense by using reaction (8.9) to calculate the total reaction propensity (r tot ) in the algorithm 1, then only calculating reaction propensities as needed to determine the next reaction. Simulation Results The stochastic simulation contains two parameters, the simulation volume V and the characteristic particle size, that do not exist in deterministic population balances. In deterministic
150 128 Figure 8.3: Mean of the stochastic solution for an isothermal crystallization with nucleation and growth, average of 1 simulations, characteristic particle size =.1, system volume V = 1 Average Time for One Simulation (sec) e-5 1e Characteristic Particle Size.1 1 Figure 8.4: Average stochastic simulation time based on 1 simulations and V = 1
151 129 Crystals Volume Time Figure 8.5: Mean of the stochastic solution for an isothermal crystallization with nucleation and growth, average of 1 simulations, characteristic particle size =.1, system volume V = 1 population balances, the simulation volume is specified by the volume of the modeled crystallizer. In general, stochastic techniques cannot simulate the system volume due to excessive computational expense. To overcome this difficulty, we invoke the well-mixed assumption, choose a volume that accurately represents the system, and average the results of multiple simulations given this volume. 2 Care must be taken to ensure that the results are generated from a sufficient number of simulations. For an example, consider the case in which =.1 and V = 1. For one simulation, Figure 8.2 shows that each particle size is sparsely populated, making discrete transitions between states clearly observable. Averaging over one hundred simulations, Figure 8.3 demonstrates that the particle sizes are more densely populated, thus credibly reproducing the average system behavior. Varying the characteristic particle size varies the initial number of monomer units. As decreases, the initial number of monomer units increases. Since the computational expense scales with the number of reactant molecules, this expense increases. Figure 8.4 illustrates this point by examining the average computational expense for ten simulations as a function of. In addition, the dispersion among particle sizes associated with the stochastic simulation becomes less pronounced as decreases. The effects of manipulating are illustrated in Figures 8.3 and Rate constants of order greater than one are volume dependent in the stochastic simulation because reactions are molecular events.
152 13 Derivation of the Macroscopic Population Balance as the Limit of the Master Equation The results of the stochastic simulations lead to the belief that, under appropriate conditions, the deterministic population balance arises from the master equation system representation. We now prove this assertion. The discrete master equation is of the form given in equation (8.1). Define the characteristic size of the system to be Ω, and use this size to recast the master equation (8.1) in terms of intensive variables (let z x/ω). Performing a Kramers-Moyal expansion on this master equation results in a system size expansion in Ω. In the limit as x and Ω become large, the discrete master equation can be approximated by its first two differential moments. This approximation is the continuous Fokker-Planck equation [41]: P (z; t) t A(z) = B(z) 2 = = l i=1 z i (A i (z)p (z; t)) m ν k a k (z) k=1 m ν k ν T k a k(z) k=1 l l i=1 j=1 2 ( Bij (z) 2 P (z; t) ) z i z j (8.1a) (8.1b) (8.1c) Equation (8.1) has Itô solution of the form: dz i = A i (z)dt + l B ij (z)dw j (8.11) in which W is a vector of Wiener processes. The Fokker-Planck equation (8.1) specifies the distribution of the stochastic process, whereas the stochastic differential equation (8.11) specifies how the trajectories of the state evolve. By taking the thermodynamic limit (x, Ω, z = x/ω = finite), equation (8.11) approaches the deterministic limit [76]: j=1 dz i dt = A i(z) (8.12) The deterministic limit implies that the probability P (z; t) collapses to a delta function. Now consider the two densities N(l i, t) and f(l, t), representing the discrete and continuous population balances, respectively. These densities are functions of the characteristic particle size l and the time t. N(l i, t) has units of number of crystals per volume, and f(l, t) has units of number of crystals per volume per characteristic particle size. Define the system volume, V, as the extensive characteristic size of the system, Ω. For the kinetic mechanism (8.8), equation
153 131 (8.12) defines the the discrete population balance accordingly: dm tot dt = k n M 2 k g MN(l i, t) i=1 (8.13a) dn(l 1, t) = 1 dt 2 k nm 2 k g MN(l 1, t) (8.13b) dn(l i, t) = k g M [N(l i 1, t) N(l i, t)], dt i = 2,..., (8.13c) For small and a 1, it is apparent that the following equality should hold: N(l a, t) = la+ 2 l a 2 l a = (a + 1) f(l, t)dl (8.14a) (8.14b) Differentiating equation (8.14a) with respect to time yields: dn(l a, t) dt = d dt la+ 2 l a 2 f(l, t)dl = la+ 2 l a 2 f(l, t) dl (8.15) t For a > 1, apply the definition given by (8.13c) into equation (8.15): k g M[N(l a, t) N(l a, t)] = la+ 2 l a 2 f(l, t) dl (8.16) t Rewriting the left hand side in terms of an integral over the particle size l and regrouping yields: la+ 2 f(l, t) + k g M[f(l, t) f(l, t)]dl = (8.17) t l a 2 Since the bounds on the integral of equation (8.17) are arbitrary, i.e., they hold for any a such that a > 1, one solution is to set the integrand to zero: f(l, t) t + k g M[f(l, t) f(l, t)] = (8.18) Mccoy [92] suggests considering a Taylor series expansion to determine the difference f(l, t) f(l, t): f(l, t) f(l, t) = f(l, t) + [(l ) l] f(l, t) l 2! l 2 [(l ) l] (8.19a) f(l, t) = f(l, t) f(l, t) l 2 l (8.19b) Hence the desired difference is: f(l, t) f(l, t) f(l, t) = 2 2 f(l, t) l 2 l (8.2)
154 132 For sufficiently small, the first partial derivative of equation (8.2) adequately approximates this difference: f(l, t) t = k g M [f(l, t) f(l, t)] (8.21a) k gm f(l, t) l (8.21b) where k g = k g. Equation (8.21b) is the corresponding macroscopic population balance equation for well-mixed systems with only nucleation and growth, and is defined over the range l <. The boundary condition for equation (8.21b) at l = is: f(, t) = (8.22) The other boundary condition, f(, t), can be determined by examining the zeroth moment (µ ) of equation (8.21b) and noting that only nucleation influences the number of particles: f(l, t) dl = t dµ dt k gm f(l, t) dl l (8.23a) = k gm(f(, t) f(, t)) (8.23b) 1 2 k nm 2 = k gmf(, t) (8.23c) f(, t) = k nm 2k g Finally, conservation of monomer dictates: dm tot dt = k n M 2 k g MN(l i, t) i=1 k n M 2 k g M f(l, t)dl (8.23d) (8.24a) (8.24b) In summary, in the thermodynamic limit and as becomes small, the stochastic formulation yields the following deterministic formulation: f(l, t) = k f(l, t) t gm l dm tot = k n M 2 k g M dt f(, t) = k nm 2k g f(l, t)dl (8.25a) (8.25b) (8.25c) Using these results, we solve the deterministic population balance for =.1 using orthogonal collocation on finite elements [121, 127]. Figure 8.6 presents the resulting population balance discretized to. Note that in comparison to the mean of the stochastic solution,
155 133 Figure 8.6: Deterministic solution by orthogonal collocation for isothermal crystallization with nucleation and growth, results discretized to a characteristic particle size =.1, system volume V = 1 i.e. Figure 8.3, the deterministic solution displays no dispersion in either the seed or nucleated particle distributions. This result indicates that the simulated characteristic particle size, =.1, is large enough to merit including higher order terms of the f(l, t) f(l, t) expansion. The next correction is the diffusivity term commonly used to model growth rate dispersion. The corresponding formulation for this model is: ( f(l, t) f(l, t) = k t gm 2 ) f(l, t) l 2 l 2 dm tot = k n M 2 k g M f(l, t)dl dt f(, t) = k nm 2k g + f(l, t) 2 l l= f(l, t) = l l= (8.26a) (8.26b) (8.26c) (8.26d) Figure 8.7 presents this population balance discretized to. Comparison of this result to Figure 8.3, the mean of the stochastic solution, demonstrates excellent agreement between the two distributions. In contrast to prior modeling efforts (e.g. [111]), however, the diffusivity term is a function of the growth rate, not a constant. Hence when the growth rate is zero, growth rate dispersion ceases.
156 134 Figure 8.7: Deterministic solution by orthogonal collocation for isothermal crystallization with nucleation and growth, inclusion of the diffusivity term, results discretized to a characteristic particle size =.1, system volume V = 1 The key differences between the stochastic and deterministic population balances are somewhat subtle and deserve further attention. First, the stochastic population balance has discrete particle sizes containing an integer number of particles. The deterministic population balance, on the other hand, has continuous particle sizes, and integration over a range of particle sizes yields a real number of particles contained within this range. Second, the number of particles contained in each size class of the stochastic population balance is governed by an individual probability distribution; hence different simulations may yield different numbers of particles in a particular size class at the same time even if the initial condition is identical. Only in the large number (thermodynamic) limit do these probability distributions collapse to delta functions (single values) for the concentration of particles in a given size class. In the deterministic population balance, simulating a given initial condition multiple times yields the same number of particles over a given size range at the same simulation time. We note that Ramkrishna [16] provides a similar, but different perspective than ours on the connection between the stochastic and deterministic population balances. In his work, Ramkrishna considers continuous particle size classes, and demonstrates that the deterministic population balance can be obtained by averaging the governing master equation. Our derivation considers discrete particle sizes and derives the deterministic population balance as the large number (thermodynamic) limit of the governing master equation. We shy away from averaging because of literature examples demonstrating that this equivalence does not always
157 135 hold in the small molecule limit [143] Nonisothermal Nucleation and Growth In this example, we are interested in modeling a nonisothermal crystallizer whose temperature is regulated by a cooling jacket. We consider the reaction system: 2M N n + M kn N 1 H n rxn (8.27a) kg N n+1 H g rxn (8.27b) For the deterministic case, the energy balance should satisfy the following equation: dt dt = UA ( ) ( ρc p V (T j T ) Hn rxn 1 ρc p 2 k nm 2 Hg rxn k g M ρc p ) f(l, t)dl (8.28) Stochastically, we differentiate between enthalpy changes due to interaction with the cooling jacket and enthalpy changes due to nucleation and growth reactions. We treat enthalpy changes due to reactions stochastically in that they instantaneously release a specified heat of reaction upon completion. On the other hand, we treat enthalpy changes due to interaction with the cooling jacket continuously, giving rise to a deterministic enthalpy loss expression. This treatment of the energy balance with stochastic and deterministic contributions is discussed further by Vlachos [156]. Hence our simulation plan is as follows: 1. Upon completion of a reaction event, update the temperature due to the enthalpy of reaction. 2. Between reaction events, update the temperature using the following equation: dt dt = UA ρc p V (T j T ) (8.29) Since the monomer saturation, M sat, is a function of temperature, the monomer supersaturation, M, is also a function of temperature and we must apply an algorithm that accounts for time-dependent reaction propensities. We quadratically distribute the seeds over the crystal volume interval [2, 2.5]. The cooling temperature profile for the jacket (T j ) follows an exponentially decreasing trajectory. The solubility relationship for the number of monomer is given by: log 1 M sat = 2.25 log 1 T +.4 T (8.3) The model parameters are given in Table 8.2. The results for the mean of the exact stochastic simulation are presented in Figures 8.8 through 8.1. Figure 8.11 presents the result for the mean of the approximate stochastic simulation with propensity of no reaction a = 1. The discretized solution of the deterministic population balance including the diffusivity term is presented in Figure These figures
158 136 Parameter Symbol Value nucleation rate constant k n growth rate constant k g characteristic particle volume.1 initial crystallizer temperature T o initial cooling jacket temperature T j,o crystallizer heat transfer coefficient area UA 5 solution density heat capacity ρc p 1 simulation system volume V 1 nucleation and growth heats of reaction Hrxn n = Hrxn g.1 Table 8.2: Nonisothermal nucleation and growth parameters for a batch crystallizer Number of Monomer Molecules Time Number of Supersaturated Molecules Figure 8.8: Total and supersaturated monomer profiles for nonisothermal crystallization demonstrate agreement between the mean of the exact stochastic solution, the mean of the approximate stochastic solution, and the deterministic solution. Figures 8.13 and 8.14 compare of the zeroth and first moments of the approximate stochastic simulation to the exact stochastic simulation. Here, we define the jth moment of the stochastic simulation µ j as µ j = x x j N x (8.31) in which N n is the average number of particles in the nth size class. Varying the value of the propensity of no reaction, a, controls the stochastic time in the approximate stochastic solution. For this simulation, the value of a =.1 is clearly too small to account for the
159 Crystallizer Temperature 3 25 Jacket Time Figure 8.9: Crystallizer and cooling jacket temperature profiles Figure 8.1: Mean of the exact stochastic solution for nonisothermal crystallization with nucleation and growth, average of 5 simulations, characteristic particle size =.1, system volume V = 1
160 138 Figure 8.11: Mean of the approximate stochastic solution for nonisothermal crystallization with nucleation and growth, average of 5 simulations, characteristic particle size =.1, system volume V = 1, propensity of no reaction a = 1 time-varying reaction propensities as evidenced by the poor initial reconstruction of the moments. However, as the value of a increases, the resulting population balances tend towards the exact stochastic solution. Although accuracy increases as a increases, computational expense increases as well. Hence the value of a must be carefully selected to balance the two. Also, our implementation of the exact stochastic simulation employed an ODE solver with a stopping criteria to account for the time-varying reaction propensities, whereas the approximate solution did not require an ODE solver. As a result, the exact solution was two orders of magnitude slower than the approximate solution Isothermal Nucleation, Growth, and Agglomeration We examine the same reactions as in mechanism (8.8), but now consider particle agglomeration as well: 2M kn N 1 (8.32a) N l + M N p + N q kg N n+1 k a Np+q (8.32b) (8.32c) The model parameters are given in Table 8.3. For size-independent agglomeration, k a is a
161 139 Figure 8.12: Deterministic solution by orthogonal collocation for nonisothermal crystallization with nucleation and growth, inclusion of the diffusivity term, results discretized to a characteristic particle size =.1, system volume V = 1 Percent Error a =.1 a = 1 a = Time Figure 8.13: Zeroth moment comparisons, mean of the stochastic solution for nonisothermal crystallization with nucleation and growth, average of 5 simulations, characteristic particle size =.1, system volume V = 1
162 14 Percent Error a =.1 a = 1 a = Time Figure 8.14: First moment comparisons, mean of the stochastic solution for nonisothermal crystallization with nucleation and growth, average of 5 simulations, characteristic particle size =.1, system volume V = 1 Parameter Symbol Value nucleation rate constant k n growth rate constant k g agglomeration rate constant k a simulation system volume V 1 characteristic particle volume.1 number of saturated monomers M sat Table 8.3: Nucleation, growth, and agglomeration parameters for an isothermal, batch crystallizer constant. To make the simulation efficient, we note that this type of agglomeration exhibits the same kinetics as the second-order reaction: 2A ka C (8.33) Again, the number of species A molecules is equivalent to the zeroth moment of the particle distribution N, so we can use reaction (8.33) to calculate the propensity of all agglomeration events occurring. In steps 1 and 4 of algorithm 1, we use this value in calculation of the total reaction rate. Next, we first determine which type of reaction occurs (nucleation, growth, or agglomeration), then which specific event occurs, again calculating reaction propensities only as needed.
163 141 Figure 8.15: Mean of the stochastic solution for an isothermal crystallization with nucleation, growth, and agglomeration; average of 5 simulations; characteristic particle size =.1; system volume V = 1 The results of this simulation are presented in Figure In contrast to Figure 8.3, the equivalent reaction system without agglomeration, we see that agglomeration increases the observed particle dispersion phenomenon. 8.5 Parameter Estimation With Stochastic Models The goal of parameter estimation is to determine the set of parameters that best reconciles the experimental measurements with model predictions. The classical approach is to assume that measurements are corrupted by normally distributed noise. Accordingly, we calculate the optimal parameters via the least squares optimization min θ Φ = 1 2 e T k Re k s.t.: n k+1 = F (n k, θ) k e k = y k h(n k ) (8.34a) (8.34b) (8.34c) in which e k s denote the difference between the measurements y k s and the model predictions h(n k ) s 3. In general, most experiments do not include many replicates due to cost 3 We assume that the measurement residuals e k s are normally distributed with zero mean and R 1 covariance.
164 142 and time constraints. Therefore, the best experimental data we are likely to obtain is in the form of moments of the master equation, i.e. equation (8.2). Clearly the master equation (8.1) demonstrates that these moments are twice continuously differentiable, so standard nonlinear optimization algorithms apply to fitting these moments to data. In fitting data to stochastic models governed by the master equation, we choose the mean x as the the state of interest. Monte Carlo simulation provides an estimate of this mean, albeit to some degree of error due to the finite simulation error. In the following subsections, we present a trust-region optimization method, discuss the calculation of finite difference sensitivities, and provide an example of estimating parameters for the nucleation, growth, and agglomeration mechanism of section Trust-Region Optimization We perform optimization (8.34) using a trust-region method employing a Gauss-Newton approximation of the Hessian. This method has provable convergence to stationary points (i.e. θ Φ ) [97]. Algorithm 7 presents the basic steps of this method. Evaluation of the objective function is relatively expensive since it requires integrating the stochastic model. Therefore, we choose to accept all parameter changes that reduce the value of the objective function and solve the trust-region subproblem exactly using a quadratic programming solver. Also, we scale the optimized parameters using a log 1 transformation. The trust-region subproblem requires knowledge of both the gradient and the Hessian. We can numerically evaluate both of these quantities θ Φ = 1 θ T e T k 2 Re k (8.37) k = ( ) h(nk ) n T k n T k k θ T Re k (8.38) = ( ) h(nk ) T n T S k Re k (8.39) k k θθ Φ k ( h(nk ) n T k ) T S k R h(n k) n T S k (8.4) k which indicates dependence upon S k, the sensitivity of the state with respect to the parameters Finite Difference Sensitivities We assume that the unknown evolution equation for the mean x depends on the system parameters θ x k+1 = F (x k, θ) (8.41)
165 143 Algorithm 7 Trust Region Optimization. Given k =, >, (, ), and η [,.25). while (not converged) 1. Solve the subproblem p k = arg min p R n m k(p) = Φ θk + θ Φ T θ k p pt θθ Φ θk p s.t.: p k (8.35a) (8.35b) 2. Evaluate 3. if ρ k <.25 ρ k = Φ(θ k) Φ(θ k + p k ) m k () m k (p k ) (8.36) k+1 =.25 p k else if ρ k >.75 and p k = k else end if end if if ρ k > η k+1 = min(2 k, ) k+1 = k else θ k+1 = θ k + p k θ k+1 = θ k end if 4. k k + 1 end while
166 144 Parameter Symbol Value simulations per measurement evaluation n sim 1 finite difference perturbation δ.1θ j 1 transmittance constant k t 3 measurement inverse covariance R diag([1 8, 1]) Table 8.4: Parameters for the parameter estimation example. Here, θ j is the jth element of the vector θ. Here, the notation x k denotes the value of the mean x at time t k. The sensitivity s indicates how sensitive the mean is to perturbations of a given parameter, i.e. s k = x k θ T (8.42) We can then approximate the jth component of the desired sensitivity using, for example, a central difference scheme: s k+1,j = F (x k, θ + δe j ) F (x k, θ δe j ) 2δ + i O(δ 2 ) (8.43) in which δ is a small positive constant, e j is the jth unit vector, and i is a vector of ones. Finite difference methods have several potential problems when used in conjunction with Monte Carlo reconstructed quantities as discussed in Chapters 6 and 5. To reduce the finite simulation error, we re-seed the random number generator before each sample used to generate the mean x k. In doing so, we must take special care in the selection of the perturbation δ to ensure that its effect on the mean is sufficiently large; otherwise, the positive and negative perturbations are approximately equal (i.e. F (x k, θ + δe j ) F (x k, θ δe j )) resulting in a poor reconstruction of the sensitivity. Finally, the computational expense of this method can be prohibitive if evaluating the mean is computationally intensive because calculating the sensitivity requires, in this case, two mean evaluations per parameter. Drews, Braatz, and Alkire [25] recently examined using finite differences to calculate sensitivities for kinetic Monte Carlo code simulating copper electrodeposition. These authors consider the specific case of the mean sensitivity, and derive finite differences for cases with significant finite simulation error. In these cases, the finite simulation error is greater than higher-order contributions of the finite difference expansion, so the authors derive first-order finite differences that minimize the variance of the finite simulation error. We circumvent the need for such expressions by appealing to the law of large numbers; that is, we reduce the variance of the finite simulation error by merely increasing the number of simulations used to evaluate the mean when necessary.
167 145 Transformed Parameter Symbol Actual Value Estimated Value nucleation rate constant log 1 k n ±.3 growth rate constant log 1 k g ±.2 agglomeration rate constant log 1 k a ±.5 Table 8.5: Estimated parameters Number of Supersaturated Molecules Time Transmittance Figure 8.16: Comparison of final model prediction and measurements for the parameter estimation example Parameter Estimation for Isothermal Nucleation, Growth, and Agglomeration We reconsider the isothermal nucleation, growth, and agglomeration example given in section Traditional measurements for batch crystallizers yield moments of the PBE, so we assume that we can measure both the supersaturated monomer and transmittance, i.e. [ ] M y = exp ( k t µ 2 ) (8.44) µ 2 = x x 2 N x (8.45) in which µ 2 is the second moment of the particle distribution and M is the average amount of supersaturated monomer. Parameters for the optimization routine are given in Table 8.4. Using the kinetic mechanism (8.32), we generate results from one simulation for the experimental measurements, then attempt to fit the parameters using subsequent simulations. Table 8.5 compares the actual and estimated parameter values. We also report 95%
168 log 1 k g log 1 k a log 1 k n Iteration Figure 8.17: Convergence of parameter estimates as a function of the optimization iteration. confidence intervals for the estimated parameter values calculated by ignoring the effect of the finite simulation error. The results indicate excellent agreement between the actual and fitted parameters. The slight discrepancies in the fit most likely result from the finite simulation error (we simulated the experimental and predicted measurements using different seeds for the random number generator). Figure 8.16, which plots both the experimental and model predicted measurements, also demonstrates excellent agreement between the model and the experiment. Figure 8.17 plots the convergence of the parameter estimates as a function of the optimization iteration. This result indicates that the convergence to the optimal parameter values occurs relatively quickly (roughly five iterations). Each iteration requires seven mean evaluations (six for the finite difference calculations and one for the predicted step). Raimondeau, Aghalayam, Mhadeshwar, and Vlachos [15] argue that using kinetic Monte Carlo simulation to perform parameter estimation is too computationally expensive. They claim that a model with two to three parameters needs roughly 1 5 function (mean) evaluations for direct optimization. For this example, in contrast, the required number of mean evaluations is less than 1 2. In general, we expect that the actual number of function evaluations required for direct optimization is significantly lower than their estimate when using an appropriate optimization scheme. 8.6 Critical Analysis of Stochastic Simulation as a Modeling Tool Thus far, we have demonstrated the efficacy of stochastic simulation as a macroscopic modeling tool. Now we address the benefits and shortcomings of this technique. The primary
169 shortcoming of stochastic simulation is the computational expense. Since the computational expense of stochastic simulation scales with the number of reactant molecules, this expense increases as the modeled volume increases or the characteristic particle size decreases. Also, the computational expense is significantly greater than that required to solve the equivalent deterministic system. However, as computing power continues to increase, this discrepancy will become less of a hindrance in solving the stochastic PBE. Perhaps the greatest advantages of stochastic simulation are its flexibility and ease of implementation. The simple algorithms presented in this chapter are applicable to any reaction mechanism. For example, adding agglomeration to the preexisting isothermal nucleation and growth code required addition of the n(n 1)/2 possible agglomeration reactions between n possible particle sizes. We expect that adding more complicated mechanisms or tracking more than one crystal characteristic are straightforward extensions of this algorithm. Implementing size-dependent growth, for example, requires only making the reaction propensities functions of length (i.e., r k = a k (x, y, l k )). To track two characteristic lengths, we need only explicitly account for each particle and define mechanisms for growth of each characteristic length. The most difficult part of augmenting the reaction mechanism is deciding how to store and update the active particle sizes. To illustrate these points, we invite the interested reader to download and examine codes that simulate isothermal nucleation and growth, and isothermal nucleation, growth, and agglomeration from our web site at haseltin/stochsims.tar. Addition of agglomeration requires approximately sixty additional lines of code to the nucleation and growth code. The majority of this code updates the data structure employed to account for existing crystal sizes. In contrast, attempting to examine nucleation, growth, and agglomeration using orthogonal collocation most likely requires major revision of the solution technique, such as adaptive mesh algorithms. Stochastic simulation inherently accounts for each crystal in the simulation. Hence we see stochastic simulation as a general solution technique that allows the user to focus on key modeling issues as opposed to population balance solution methods. We also demonstrated one method of performing parameter estimation with stochastic models. By applying appropriate nonlinear optimization routines, we can obtain optimal parameter values with surprisingly few evaluations of the stochastic model. The primary drawback to the presented method is the calculation of sensitivities via finite differences. Finite difference methods quickly become expensive to evaluate as both the number of parameters and the computational burden of evaluating the stochastic model increase. Finally, refined optimization of Monte Carlo simulations requires quantifying the effects of the finite simulation error on both the model constraint (an error-in-variables formulation is more appropriate) and the termination criteria.
170 Conclusions Stochastic simulation provides one alternative to solving the deterministic crystallization population balance. For systems with small numbers of monomer and seed, the stochastic crystallization model is more realistic than the deterministic model because it inherently accounts for the system fluctuations. In the limit as the numbers of monomer and seed become large, the deterministic model becomes valid. Even for this case, stochastic simulation provides a general, flexible solution technique for examining many possible reaction mechanisms. Additionally, optimization of the stochastic model for purposes such as parameter estimation is feasible and requires relatively few evaluations of the model. Simulation results presented in this chapter illustrate these claims. Thus stochastic simulation should permit the user to focus more on modeling issues as opposed to solution techniques. Notation A crystallizer area a dt contrived probability, first order in dt, that no reaction occurs in the next time interval dt a k (x)dt probability to order dt that reaction k occurs in the time interval [t, t + dt) C p e e j f(l, t)dl g(x) h(x k ) i k a k g k n l M M M tot M sat N N j heat capacity error vector jth unit vector concentration of particles average value of the quantity g(x) model prediction of the measurement vector at time t k vector of ones agglomeration rate constant growth rate constant nucleation rate constant characteristic particle size average amount of supersaturated monomer number of supersaturated monomers total number of monomers number of saturated monomers number of Monte Carlo samples jth particle size N n number of particles with size l n = (n + 1) n mon n seed P (x, t) p k R r k initial number of monomer particles initial number of seed particles probability that the system is in state x at time t kth uniformly-distributed random number inverse covariance matrix of the measurement noise kth reaction propensity
171 149 r tot S s T T j,o T o t U V V mon W x x x i y total reaction propensity sensitivity matrix of the state sensitivity of the mean x temperature initial cooling jacket temperature initial crystallizer temperature time crystallizer heat transfer coefficient system volume initial volume of monomer the Wiener process state vector in terms of number of molecules average state vector ith Monte Carlo reconstruction of x vector of state-dependent variables y k z k Hrxn g Hrxn n δ η measurement vector at time t k state vector in terms of concentration (intensive variable) characteristic volume of one monomer unit trust-region optimization parameter at step k trust-region optimization parameter growth heat of reaction nucleation heat of reaction small positive constant trust-region optimization parameter µ j jth moment of the particle size distribution ν stoichiometric matrix Φ objective function value ρ p ρ k τ θ Ω solution density trust-region optimization parameter at step k next reaction time vector of model parameters characteristic system size
172 15
173 151 Chapter 9 Population Balance Models for Cellular Systems 1 To date, most models of viral infections have focused exclusively on modeling either the intracellular level or the extracellular level. To more realistically model these infections, we propose incorporating both levels of information into the description. One way of performing this task in a deterministic setting is to derive cell population balances from the equation of continuity. In this chapter, we first outline the basics of deriving and solving these population balance models for viral infections. Next, we construct a population balance model for a generic viral infection. We examine the behavior of this model given in vitro and in vivo conditions, and compare the results to other model candidates. Finally, we present conclusions and consider the future role of cell population balances in modeling virus dynamics. 9.1 Population Balance Modeling The general population balance equation for cell populations arises from the seminal contribution of Fredrickson, Ramkrishna, and Tsuchiya [36]. In recent years, this modeling framework has returned to the literature as researchers strive to adequately reconcile model predictions with the dynamics demonstrated by experimental data [8, 1, 33]. Also, new measurements such as flow cytometry offer the promise of actually differentiating between cells of a given population [1, 67], again implying the need to model distinctions between cells in a given population. Here, we present a brief derivation for models encompassing a population of infected cells as well as intracellular and extracellular components of interest. In a deterministic setting, we can model the infected cell population by deriving a cell population balance from the equation of continuity. Here we define the concentration of infected cells as a function of time (t) and the internal (y) and external (x) characteristics of the 1 Portions of this chapter to appear in Haseltine, Rawlings, and Yin [6].
174 152 system: η(t, z)dz = concentration of infected cells (9.1) [ ] [ ] x external characteristics z = = (9.2) y internal characteristics We can then write a conservation equation for these cells by considering an arbitrary control volume V (t) spanning a space in x and y, assuming that V (t) contains a statistically significant number of cells. Following the same arguments presented in section 2.1 results in the microscopic equation of continuity, equation (2.8). This equation is the most general form of our proposed model. We reiterate that the only assumption made thus far is that we consider a statistically significant number of cells. We now must specify segregations for the infected cell population. First, we assume that the cells are well-mixed; this assumption allows us to eliminate the spatial dimensions from equation (2.8): η(t, y) t + (η(t, y)v y ) = R η (9.3) Next, we propose differentiating among the stage of infection for infected cells by using the infected cell age. The cell age acts as a clock that starts upon initial infection of an uninfected cell and ends upon the death of this cell. Although such a parameter cannot be explicitly measured, it can nonetheless be identified experimentally through its effect upon other observable quantities such as the expression of viral products. Because the age changes with time in the usual way, the age velocity term is unity, y = τ = infected cell age (9.4) v y = 1 (9.5) Additionally, modeling the intracellular biochemical network necessitates augmenting the cell population balance with mass balances for viral components (labeled component i). Since the intracellular components are also segregated by the cell age, derivation of these mass balances follows that for the infected cell population (i.e. from equation (2.3) to (9.3)), yielding in which i j t + i j τ = R j + E j j = 1,..., n (9.6) R j is the intracellular production rate of component j. Processes such as transcription and translation of the viral genome are examples of events contributing to R j. E j accounts for the effect of extracellular events on the intracellular production rate of component j. An example of such an event includes superinfection of an infected cell, which inserts additional viral genome and proteins into the cell.
175 We model extracellular components (labeled component e) as well-mixed and unsegregated (i.e. having no τ-dependence). The production rates for extracellular components may also be a function of both extracellular (E) and intracellular (R) events. In this case, however, infected cells produce and secrete extracellular components at an age-dependent rate. The conservation equation for the extracellular component, then, includes an integration of the intracellular rate over the infected cell population: e τd k t = E k + η(t, τ)r k dτ k = 1,..., m (9.7) Here, τ d specifies the age of the oldest infected cell. Examples of processes contributing to E k and R k include regeneration of uninfected cells and secretion of virus from infected cells, respectively. The comprehensive model for this system is η(t, τ) t 153 η(t, τ) + = R η τ (9.8a) i j t + i j τ = R j + E j j = 1,..., n (9.8b) e k t = E k + τd η(t, τ)r k dτ k = 1,..., m (9.8c) 9.2 Application of the Model to Viral Infections We now consider application of this model to a generic viral infection. We first outline the basic intracellular and extracellular events occurring in such an infection, discuss further model refinements, and present the numerical technique used to solve the final model Intracellular Model At the intracellular level, we incorporate events from a simple structured model of virus growth [143]: nucleotides + gen k 1 tem ɛ 1 = k 1 i V1 i gen (9.9a) V1 amino acids nucleotides str k 2 V 2, tem str ɛ 2 = k 2 i V2 i tem (9.9b) k 3 tem gen ɛ 3 = k 3 i tem (9.9c) k 4 degraded ɛ4 = k 4 i str (9.9d) gen + str k 5 secreted virus ɛ5 = k 5 i gen i str (9.9e) Here, gen and tem are the genomic and template viral nucleic acids respectively, str is the viral structural protein, V 1 and V 2 are viral enzymes that catalyze their respective reactions,
176 154 and the reaction rates are given by the ɛ expressions. These events account for the insertion of the viral genome into the host nucleus, production of a viral template used to replicate the viral genome and mass-produce viral structural protein, and the assembly and secretion of viral progeny. We assume that host nucleotides and amino acids are available at constant concentrations. Therefore, the only intracellular components that we must track are the tem, gen, str, V 1, and V 2 components Extracellular Events At the extracellular level, we adopt a standard model [98]: virus + uninfected cell virus infected cell uninfected cell k 6 infected cell ɛ6 = k 6 e vir e unc (9.1a) k 7 degraded ɛ7 = k 7 e vir (9.1b) k 8 death ɛ8 = k 8 e inf (9.1c) k 9 death ɛ9 = k 9 e unc (9.1d) precursors k 1 uninfected cell ɛ 1 = k 1 (9.1e) These events address the intuitive notions of cell growth, death, and infection by free virus. From this point forward, we use the abbreviations unc, inf, and vir for uninfected host cells, infected host cells, and virus Final Model Refinements Further model assumptions include: Reaction rates of intracellular and extracellular events follow simple, mass-action kinetics. All reactions are elementary as written except for enzyme-catalyzed reactions, in which case the expressions result from performing model reduction on Michaelis- Menten kinetics. Infected cells are created at age zero due to interaction between uninfected cells and free virus, and infected cells die at an exponential rate until age τ d R η = k 6 e unc e vir δ(τ) η(t, τ) (k 8 + δ(τ τ d )) (9.11) Here, δ is the Dirac delta function. Also, an initial infection corresponds to insertion of 1 gen/cell, 8 V 1 /cell, and 4 V 2 /cell into an uninfected cell. No superinfection of infected cells occurs. Concentrations of intracellular enzymes remain constant throughout the life cycle of an infected cell.
177 155 Therefore, our final model is η(t, τ) η(t, τ) + t τ i tem (t, τ) + i tem(t, τ) t τ i gen (t, τ) + i gen(t, τ) t τ i str (t, τ) + i str(t, τ) t τ i V1 (t, τ) + i V 1 (t, τ) t τ i V2 (t, τ) + i V 2 (t, τ) t τ de unc dt de vir dt = k 6 e unc e vir δ(τ) (k 8 + δ(τ τ d )) η(t, τ) (9.12a) = R tem (9.12b) = R gen + δ(τ) (9.12c) = R str (9.12d) = 8δ(τ) (9.12e) = 4δ(τ) (9.12f) = k 1 k 9 e unc k 6 e unc e vir (9.12g) = k 7 e vir k 6 e unc e vir + τd η(t, τ)r vir (τ)dτ (9.12h) Model Solution To solve the model, we use orthogonal collocation on finite elements of Lagrange polynomials [155, 121, 127]. This method approximates functions of multiple coordinates, e.g. η(t, τ), by a linear combination of Lagrange interpolation polynomials: η(t, τ) n L j (τ)η(t, τ j ) (9.13) j=1 in which L j is a Lagrange interpolation polynomial of degree n, and η(t, τ j ) is the function evaluated at the point τ j. Accordingly, we can approximate the age derivative at each collocation point as η(t, τ) τ τ=τj n j=1 L j (τ) τ j η(t, τ j ) (9.14) n A ij η(t, τ j ) (9.15) j=1 in which the matrix A is the derivative weight matrix. Also, we can approximately evaluate integrals by using quadrature τd η(t, τ)dτ n q j η(t, τ j ) (9.16) j=1
178 156 where q j is the j th quadrature weight. This method is known as the global orthogonal collocation method when only one collocation element is applied to the entire domain of interest. Alternatively, one could split the domain into multiple subdomains, then apply a collocation element to each subdomain; in this case, the method is called orthogonal collocation on finite elements. Collocation on finite elements permits concentration of elements in regions where sharp gradients exist, a case that normally causes difficulties in global orthogonal collocation. At the junction of finite elements, one imposes continuity of the population η(t, τ), i.e. η(t, τ) τ = η(t, τ) τ + (9.17) in which the boundary between elements occurs at τ = τ, and τ and τ + represent the boundaries of the adjoining finite elements [127]. Note that the number of boundary conditions at the junction of elements is equal to the number of partial derivatives due to segregations (i.e. τ), and that the order of each boundary condition is one less than the order of its partial derivative. Unless otherwise specified, we use only one collocation element in our discretization. The collocation method is very sensitive to large changes of the order of magnitude for the approximating function. Since equation (9.12a) indicates that η(t, τ) changes exponentially, we use a logarithmic transformation to scale η(t, τ). Applying this method to equation (9.12) in effect discretizes the integro-partial differential equation into a system of differential algebraic equations (DAE s). We then use the software package DASPK [15] to integrate the DAE system. Orthogonal collocation on finite elements presents merely one manner of solving equation (9.12). We refer the interested reader to Mantzaris, Daoutidis, and Srienc [87, 88, 89] for an excellent overview of other numerical methods used to solve similar equations. 9.3 Application to In Vitro and In Vivo Conditions To better understand the cell population balance, we apply model (9.12) to both in vitro and in vivo conditions. We also compare the results of the model to other commonly used models In Vitro Experiment Here we construct an in silico example to simulate a laboratory experiment. Our apparatus is a well-mixed, batch reactor containing uninfected cells in which nutrients are provided to sustain cells without growth. We assume that assays are available that measure the concentration of uninfected cells, infected cells, virus, genome, template, structural protein, and V 1 and V 2 viral enzymes. With the goal of determining the intracellular kinetics, we consider performing the following experiment: infect a population of cells and measure components for a sample of cells. Although this technique has the disadvantage of introducing the population dynamics into the measurements, sampling a statistically significant number of cells has two primary advantages:
179 157 Parameter Value Units τ d 1 days k cell/(#-day) k cell/(#-day) k 3.7 day 1 k 4 2. day 1 k cell/(#-day) k host/(#-day) k day 1 k day 1 k day 1 k 1 #/(host-day) Table 9.1: Model parameters for in vitro simulation stochastic effects and cell to cell variations should average out, and we can adjust the sample size so that each component can be detected by its assay and consistency with the key assumption of the continuity equation (statistically significant number of cells) is maintained. We simulate the population balance model (9.12) with parameters given in Table 9.1 for the following initial conditions: 1. extracellular virus >> uninfected cells (all uninfected cells are infected initially), and 2. extracellular virus > uninfected cells (only a fraction of uninfected cells are infected initially). Experimental observations indicate that infected cells die [83]. Perhaps the simplest way to account for cell death is to combine the intracellular model (9.9) with a simple population balance, i.e. de unc dt de inf = k 5 e unc k 2 e unc e vir (9.18a) = k 4 e dt inf + k 2 e unc e vir (9.18b) de vir = k 3 e vir dt k 2 e unc c vir + R vir e inf (9.18c) Equation (9.18) is a structured, unsegregated model. Next, we perform parameter estimation and model reduction 2 to obtain an optimal fit of the structured, unsegregated model (9.18) to the data generated by the population balance (structured, segregated) model (9.12). For the sake of brevity, we do not report any of the fitted rate constants ( k s). Examining this optimal fit provides insight into the limitations of structured, unsegregated models. 2 Rawlings and Ekerdt [12] provide the details of this method.
180 158 Case 1: All Uninfected Cells Infected Initially Figure 9.1 presents the results for this case. These results indicate that the structured, unsegregated model provides an excellent fit to the data. Since all uninfected cells are infected within a relatively short period of time (roughly ten days), the approximation that all cells behave the same is valid; hence the good fit to the data. We contrast these results to those obtained from only simulating the intracellular events (i.e. Figure 9.2). Over the same time, the purely intracellular model predicts that all intracellular components increase monotonically throughout the experiment. We therefore infer that the phenomenon of cell death causes the maxima observed in the measured intracellular components. This observation reiterates the fact that experiments of this type introduce the population dynamics into the measurements. Case 2: A Fraction of Uninfected Cells Infected Initially Figure 9.3 presents the results for this case. Examination of these results indicate that roughly two rounds of infection initiation occur (marked by peaks in the infected cell population): the first round within the first ten days of the experiment, corresponding to the initial infection; and the second round at roughly 75 to 1 days, corresponding to infection of uninfected cells from virus produced by the first round of infected cells. Since the structured, unsegregated model assumes that all cells behave on average the same, it cannot adequately describe the phenomenon of multiple rounds of infection. As a result, this model provides a sub-par fit to the data. Also, we note that multiple rounds of infection have been observed experimentally in continuous flow reactors [66, 151, 134, 74] as opposed to the conditions simulated here which are batch experiments In Vivo Initial Infection We now consider the in vivo behavior of the cell population balance for an initial infection of a virus-free host. Here, the initial condition is the steady state of the system with no virus. For the sake of illustration, we account for the host immune response very simply: comparison of Tables 9.1 and 9.2 shows that, in contrast to the in vitro system, the in vivo system: clears extracellular virus more rapidly (faster decay due to a larger value of k 7 ), and uninfected host cells are produced at a nonzero rate (k 1 is now nonzero). Figure 9.4 demonstrates the host response for all extracellular components. The system exhibits three stages of infection: first, a period of relative dormancy for roughly two infection cycles (2 days 2τ d ); next, a cycle of rapid infection leading first to a peak in the infected cell then virus population; and finally, an approach to an infected steady state. In the first stage, both the extracellular virus and infected cell populations are actually increasing steadily. However, in contrast to the rapid rate of infection observed during the second stage, the first stage appears to be dormant on the scale of Figure 9.4.
181 159 uninfected cells ( 1 5 #/host) Time (Days) 8 1 infected cells ( 1 5 #/host) Time (Days) 8 1 virus ( 1 7 #/host) Time (Days) 8 1 tem ( 1 5 #/host) Time (Days) 8 1 gen ( 1 6 #/host) Time (Days) 8 1 struct ( 1 8 #/host) Time (Days) 8 1 Figure 9.1: Fit of a structured, unsegregated model to experimental results. Initial condition is such that all uninfected cells are quickly infected by virus. Points present the experimental data obtained by solving the population balance model (structured, segregated model). Lines present the optimal fit of the structured, unsegregated model to the experimental data.
182 16 Concentration str secreted virus gen tem Time (Days) 8 1 Figure 9.2: Time evolution of intracellular components and secreted virus for the intracellular model Parameter Value Units τ d 1 days k cell/(#-day) k cell/(#-day) k 3.7 day 1 k 4 2. day 1 k cell/(#-day) k host/(#-day) k 7 1. day 1 k day 1 k day 1 k #/(host-day) Table 9.2: Model parameters for in vivo simulation For the in vivo case, structured, unsegregated models do not offer an adequate representation of the system. Firstly, the average cell approximation ignores the cyclic nature of an infection because we must assume that the average cell reaches a steady state when intuitively we know that cells are regenerating and dying. Secondly, our intracellular model (see Figure 9.2) does not reach a steady state over the life time of an infected cell (i.e. 1 days), so making the in vivo model reach a steady state requires unphysical changes to either the intracellular or extracellular description.
183 161 uninfected cells ( 1 5 #/host) Time (Days) 15 2 infected cells ( 1 4 #/host) Time (Days) 15 2 virus ( 1 6 #/host) Time (Days) 15 2 tem ( 1 5 #/host) Time (Days) gen ( 1 5 #/host) struct ( 1 8 #/host) Time (Days) Time (Days) 15 2 Figure 9.3: Fit of a structured, unsegregated model to experimental results. Initial condition is such that not all uninfected cells are initially infected by virus. Points present the experimental data obtained by solving the population balance model (structured, segregated model). Lines present the optimal fit of the structured, unsegregated model to the experimental data.
184 162 Extracellular Components ( 1 7 #/host) uninfected cells Time (Days) virus infected cells Figure 9.4: Dynamic in vivo response of the cell population balance to initial infection Extracellular Components ( 1 7 #/host) uninfected cells Time (Days) virus infected cells Figure 9.5: Extracellular model fit to dynamic in vivo response of an initial infection Alternatively, we could incorporate only the extracellular events (9.1) in a mathematical description as so: de unc dt de inf = ˆk 1 ˆk 5 e unc ˆk 2 e unc e vir (9.19a) = ˆk 4 e dt inf + ˆk 2 e unc e vir (9.19b) de vir = ˆk 3 e vir dt ˆk 2 e unc c vir + ˆk 6 e inf (9.19c)
185 Model (9.19) Model (9.12) Parameter Fit Value 95% Confidence Interval Parameter Value Units ˆk ± k #/(host-day) ˆk ± k host/(#-day) ˆk ± k 7 1. day 1 ˆk ± k day 1 ˆk ± k day 1 ˆk 6.14 ± NA day 1 unc(t=) ± unc(t=) 1 8 #/host vir(t=) 37.2 ±9.25 vir(t=) 1 #/host Table 9.3: Comparison of actual and fitted parameter values for in vivo simulation of an initial infection 163 This model differs only from that of Wodarz and Nowak [164] in that we assume infection of an uninfected cell by a virus consumes the virus. Again, we attempt to optimally fit this model (9.19) to the cell population balance results 3. Figure 9.5 shows that this model cannot exhibit the same behavior as the cell population balance; most noticeably, the purely extracellular model cannot capture the dynamics of the initial dormant phase nor the burst of virus that follows the peak in the infected cell population. Table 9.3 illustrates that the fitted and actual parameters do not match to 95% confidence, but all fitted parameters are roughly the same order of magnitude with the exception of the virus decay parameter (k 7 and ˆk 3 ) and the initial virus concentration. This discrepancy occurs because the purely extracellular model (9.19) lumps all intracellular virus production events together. This result indicates that unstructured, lumped parameter models can supply unreliable estimates for parameters that govern individual events In Vivo Drug Therapy Now we consider in vivo response to drug therapy. In particular, we examine the extracellular effect that viral enzyme inhibitors I 1 and I 2 produce by affecting the intracellular enzymes V 1 and V 2, respectively. Thus, the extracellular events associated with the drug therapy are k I 13 1 degraded / secreted ɛ13 = k 13 e I1 (9.2a) I 1 + unc k 14 I 1 (adsorbed) + unc ɛ 14 = k 14 e I1 e unc (9.2b) k I 15 2 degraded / secreted ɛ15 = k 15 e I2 (9.2c) I 2 + unc k 16 I 2 (adsorbed) + unc ɛ 16 = k 16 e I2 e unc (9.2d) 3 Optimal fit corresponds to a least squares fit for the residual log 1 (y k + ci) log 1 (s k + ci), where log 1 is the base ten logarithm, y k is the measurement vector, s k is the model predicted measurement vector, i is a vector of ones, and c is a small constant. Also, the initial uninfected cell and virus concentrations were used as model parameters.
186 164 In equations (9.2b) and (9.2d), we use the notation (adsorbed) to designate that the extracellular drugs have been adsorbed into a cell. Intracellularly, these drugs then interact as so: For this situation, we assume that: K 1 V 1 + I 1 V I 1 (9.21a) K 2 V 2 + I 2 V I 2 (9.21b) k I 11 1 secreted ɛ11 = k 11 i I1 (9.21c) k I 12 2 secreted ɛ12 = k 12 i I2 (9.21d) 1. equilibrium holds for the intracellular reactions (9.21a) and (9.21b); 2. all other reactions in (9.21) and (9.2) are elementary as written; 3. the inhibitors interact only with uninfected cells; and 4. the extracellular drug intake can be modeled as an overdamped second-order, linear function [99] of the form ( u Ij (t) = ū Ij [1 exp ζt ) ( cosh βt + ζ )] βt sinh (9.22a) τ u τ u β τ u β = ζ (9.22b) assuming that a change in the drug intake occurs at time t =. Parameters for this model are given in Tables 9.2 and 9.4. The initial condition for this model corresponds to the steady state of the previous section (see Figure 9.4). Figure 9.6 presents the dynamic response for in vivo drug therapy. This response demonstrates the characteristic pharmacokinetic lag observed experimentally in viral treatments [13, 11]; however, this lag is directly attributable to modeled events, namely the drug intake dynamics, the assumption that the drugs interact only with uninfected cells, and the intracellular dynamics of drug interaction with virus enzymes. In contrast, purely extracellular models must lump each of these individual events into (generally) a single parameter to describe this lag, as examined by Perelson et al. [11]. Another attractive feature of the cell population balance over the purely extracellular model is the ability to examine the effects that perturbations to the intracellular model have upon the extracellular components. As an example, we consider the effect that changes in the efficacy of the viral inhibitors have upon the extracellular uninfected cell and virus concentrations. Such a change in efficacy may result, for example, by a mutation in the viral enzymes causing decreased efficiency in the viral enzyme-inhibitor interaction. Also, we assume that intracellular drug concentrations cannot exceed values of 45 and 6 #/cell for i I1 and i I2, respectively, due to adverse side-effects of the inhibitors. Plots (a) and (b) of Figure 9.7 present
187 165 Parameter Value Units K 1 1. cell/# K 2 1. cell/# k day 1 k day 1 k day 1 k host/(#-day) k day 1 k host/(#-day) ū I #/(host-day) ū I #/(host-day) ζ 1.1 unitless τ u 1. day Table 9.4: Additional model parameters for in vivo drug therapy the results for the nominal case. If the goal of the drug therapy is to maximize the uninfected cell concentration while minimizing the virus concentration, then the optimal treatment strategy is to maximize intake of both drugs. Plots (c) and (d) of Figure 9.7 present the results for a mutated virus corresponding to an 8% and 9% decrease in the binding constants K 1 and K 2, respectively. After the mutation, the optimal treatment strategy is actually to maximize I 1 intake and stop treatment with I 2.
188 166 Extracellular Inhibitor ( 1 6 #/host) I 1 I Time (Days) Cells ( 1 7 #/host) Time (Days) infected uninfected 4 5 Extracellular Virus ( 1 7 #/host) Time (Days) 4 5 Figure 9.6: Dynamic in vivo response to initial treatment with inhibitor drugs I 1 and I 2.
189 (a) increasing I Intracellular I2 (#/cell) (b) increasing I1 Uninfected Cells ( 1 6 #/host) Extracellular Virus (#/host) Intracellular I2 (#/cell) increasing I1 (c) Intracellular I2 (#/cell) 1 8 (d) increasing I1 Uninfected Cells ( 1 6 #/host) Extracellular Virus (#/host) Intracellular I2 (#/cell) Figure 9.7: Effect of drug therapy on in vivo steady states. Amount of (a) uninfected cells and (b) extracellular virus given nominal drug efficacy. Amount of (c) uninfected cells and (d) extracellular virus given reduced drug efficacy due to virus mutation.
190 Future Outlook and Impact The cell population balance offers an intuitive, flexible environment for modeling the combined intracellular and extracellular events associated with viral infections. Because this model has segregations, it can account for observed phenomena such as multiple rounds of infection and pharmacokinetic delays associated with drug treatments of infections. Because this model has structure, it can examine the effects that each intracellular component has upon the dynamics of the extracellular components. Neither structured, unsegregated models nor purely extracellular models can account for both of these phenomena. Validation of cell population balance models requires experimental measurements of both extracellular populations and intracellular viral components. Traditional assays already offer a means for measuring extracellular populations; for example, clinicians routinely measure both host CD4+ T-cells and virus titers in HIV-infected patients. Methods such as polymerase chain reaction (PCR), western blotting, and plaque assays offer quantitative intracellular measurements of the viral genome, proteins, and infectious viral progeny, respectively. Cell population balance models provide one method of adequately assimilating the data contained in these measurements. For in vitro experiments, we suspect that modifications to existing protocols may yield new information about the structure of the population balance model. For example, most studies of replication for animal viruses rely on one-step growth curves in which all cells in a culture are infected simultaneously [162]. While such experiments have supplied information on the intracellular dynamics of a single infection cycle, they offer no insight into how virus-mediated activities, such as activation of cellular antiviral responses and cell-cell communication, may influence the subsequent dynamics of viral propagation. New in vitro methods currently being developed [26, 28] allow viruses to infect cells sequentially rather than simultaneously, opening new opportunities to probe virus-host interactions at multiple levels. A good quantitative model of how viral infections propagate will lead to better understanding of how to best control this propagation. For example, steady-state analysis for in vitro drug therapy revealed that the optimal treatment strategy for one particular virus mutation requires stopping treatment with one drug. This counterintuitive result highlights a potential pitfall of current strategies that aim to thwart the emergence of drug-resistant virus mutants by employing multiple anti-viral drugs. Another intriguing possibility would be to perform sensitivity analysis for both intracellular components and rate constants to determine which ones have the greatest impact upon extracellular components such as the virus concentration. This analysis could then focus drug development towards those candidates having maximum therapeutic benefit. One could also consider tailoring therapies by characterizing both the virus and immune system for a given individual, rather than relying on general drug regimens obtained from the best average response for a given study.
191 Notation 169 A c E j e j i i j K j k j k j ˆk j L j (τ) log 1 q j R j R η s k t u j ū j V (t) v y x y y k z β δ ɛ j η(t, z)dz η(t, τ j ) τ τ d τ u ζ derivative weight matrix for orthogonal collocation small constant extracellular production rate extracellular viral component a vector of ones intracellular viral component equilibrium constant for the segregated, structured model reaction rate constant for the segregated, structured model reaction rate constant for the unsegregated, structured model reaction rate constant for the purely extracellular model Lagrange interpolation polynomial of degree n for orthogonal collocation base ten logarithm jth quadrature weight for orthogonal collocation jth intracellular production rate production rate for the infected cell population η measurement vector predicted by the model time second-order input for extracellular component j input for extracellular component j arbitrary, time-varying control volume spanning a space in z vector specifying the y-component velocity of cells flowing through the volume V external characteristics internal characteristics experimental measurement vector internal and external characteristics parameter for the second-order input function Dirac delta function jth reaction rate concentration of infected cells infected cell concentration evaluated at the point τ j infected cell age age of the oldest infected cell permitted by the model natural period of the second-order input function damping coefficient of the second-order input function
192 17
193 171 Chapter 1 Modeling Virus Dynamics: Focal Infections We consider using dynamic models to obtain a better quantitative and integrative understanding of both viral infections and cellular antiviral mechanisms. We expect this approach to provide key insights into mechanisms of viral pathogenesis and host immune responses, as well as facilitate development of effective anti-viral strategies. Our focus, however, is not to incorporate all the wealth of information already known about either of these topics; rather, we seek to identify the critical biological and experimental phenomena that give rise to the experimental observations. We consider the focal infection system described by Duca et al. [26], which permits quantification of multiple rounds of viral infection. This experimental system provides a unique platform for studying multiple rounds of the virus replication cycle as well as the innate ability of host cells to combat the invading virus. We consider the example virus/host system of vesicular stomatitis virus (VSV) propagating on either baby hamster kidney (BHK-21) cells or murine astrocytoma (DBT) cells. VSV is a member of the Rhabdoviridae family consisting of enveloped RNA viruses [129]. Its compact genome is only approximately 12 kb in length, and encodes genetic information for five proteins. Because VSV is highly infective and grows to high titer in cell culture, it is viewed as a model system for studying viral replication [64, 7]. Also, VSV infection can elicit an interferon-mediated antiviral response from host cells [129]. Thus the studied experimental system provides a platform for further probing the quantitative dynamics of this antiviral response. A great wealth of information is known about the interferon antiviral response (see, for example, [133, 54]). We seek to elucidate what level of complexity is requisite to explain the experimental data. Yin and McCaskill [165] first proposed a reaction-diffusion model to capture the dynamics of plaque formation due to viral infection. The authors derived model solutions for this formulation in several limiting cases. You and Yin [166] later refined this model and used a finite difference method to numerically solve the time progression of the resulting model. Fort [34] and Fort and Méndez [35] revised the model of You and Yin [166] to account for the delay associated with intracellular events required to replicate virus, and derived expressions for the velocity of the propagating front. These works, however, focused on explaining the
194 172 Step 1: Monolayers fixed at selected times uninfected cells Step 2: Removal of agar and washes focal infection Measurement Imaging Step 3: Antibody labeling for viral glycoprotein infection spread Key: Step 4: Detection by antibody immunofluorescence Antibody Virus Dead cell Infected cell Uninfected cell Figure 1.1: Overview of the experimental system. Initially, host cells are grown in a confluent monolayer on a plate. The cells are then covered by a layer of agar. To initiate the infection, a pipette (one mm radius) is used to carefully remove a small portion of the agar in the center of the plate. An initial inoculum of virus is then placed in the resulting hole in the agar, initiating the infection. The agar overlay serves to restrict virus propagation to nearby cells. To monitor the infection spread, monolayers are fixed at various times post-infection. The agar overlay is removed and the cells are rinsed several times, the last time with a labeled antibody that binds specifically to the viral glycoprotein coating the exterior of the virus capsid. Images of the monolayers are then acquired using an inverted epifluorescent microscope. velocity of the infection front, a quantity derived from experimentally-obtained images of the infection spread. Our goal in this chapter is to explain the infection dynamics contained within the entire images. In this chapter, we first briefly review the experimental system of interest. Next, we outline the steps taken to analyze the experimental measurements (images of the infection spread) and propose a measurement model. We then successively formulate, fit, and refine models using the analyzed images, first for VSV infection of BHK-21 cells, then for DBT cells. Finally, we analyze the results of the parameter fitting and present conclusions. 1.1 Experimental System Here we briefly review the experimental system of interest; for detailed information on the experimental procedure, we refer the interested reader to Duca et al. [26]. This system permits dynamic, spatial quantification of virus protein via antibody immunofluorescence. Figure 1.1 presents a general schematic of this experimental system along with a digital image acquired during such an infection. Initially, host cells are grown in a confluent monolayer on a plate. The cells are then covered by a layer of agar. To initiate the infection, a pipette (one mm radius) is used to carefully remove a small portion of the agar in the center of the plate. An
195 173 Parameter Symbol Value Cell volume V c ml Initial number of uninfected cells n unc, 1 6 cells Number of viruses in the initial inoculum n vir, viruses Radius of the plate r plate 1.75 cm Table 1.1: Parameters used to describe the experimental conditions. initial inoculum of virus is then placed in the resulting hole in the agar, initiating the infection. The agar overlay serves to restrict virus propagation to nearby cells. To monitor the infection spread, monolayers are fixed at various times post-infection. The agar overlay is removed and the cells are rinsed several times, the last time with a labeled antibody that binds specifically to the viral glycoprotein coating the exterior of the virus capsid. Images of the monolayers are then acquired using an inverted epiflourescent microscope Modeling the Experiment Table 1.1 presents parameters used to model the experimental conditions. We assume that cells are spherical objects, with the height of the cell monolayer equal to the resulting cell diameter. Concentrations for all species are calculated assuming that the volume of the monolayer is cylindrical. The dimensions of this cylinder are given by the height of the cell monolayer and the radius of the plate. We model the concentration of the initial virus inoculum using the piecewise linear continuous function c vir,, r <.75 cm ( c vir (t =, r) = 1 2 cm (r.75)) c vir,.75 cm r.125 cm, r > 1.25 cm (1.1) Modeling the Measurement We assume that the measurement process (steps one through four in Figure 1.1) is an equilibrium process in which virus associates indiscriminately with cells in the monolayer. Additionally, dead cells undergo a change in morphology which decreases their ability to remain bound to the plate during removal of the agar overlay. We account for this effect by estimating k wash, the fraction of dead cells that adhere to the plate after the removal of the agar overlay and the subsequent washes. Accordingly, the amount of virus bound to host cells is given by the expression c K m = vir-host (1.2) c vir (c unc + c infc + k wash c dc ) in which K m is the equilibrium constant, and c vir, c unc, c infc, c dc, and c vir-host refer to the concentrations of virus, uninfected cells, infected cells, dead cells, and virus-host complexes, respectively.
196 174 Intensity Original Image Intensity Averaged Image.. k m i bgd 1 1 v min v max v min v max Virus-Host Complex Virus-Host Complex Figure 1.2: Measurement model. The original images quantize the virus-host concentration, a continuous variable, onto the integer-valued intensity. Each pixel in the averaged images is the mean of 4 pixels from the original image, and we approximate the step-wise discontinuous intensity (incremented by 1/4) as a piece-wise, continuous function Analyzing and Modeling the Images We have reduced the amount of information in each image by partitioning the images into blocks of 2 pixels by 2 pixels, then averaging the pixels contained in each block. This averaging technique has the primary benefit of drastically reducing the total number of pixels that must be analyzed (in the case of the largest image, from roughly two million to five thousand pixels) while retaining the prominent features of the infection spread. We assume that the intensity of each pixel in the image is due to the background fluorescence of cells and linear variation in the concentration of virus-host complexes, which fluoresce due to the labeled antibody. In the original images, the intensity information quantizes this essentially continuous variable into a step-wise, discontinuous signal (integer valued from to the saturating value of 255). For the averaged images, the intensity information is step-wise, discontinuous with increments of 1/4. We approximate this signal using a piecewise continuous function. The comparison between the measurement model for the original and averaged images is illustrated in Figure 1.2. The measurement model is then: i bgd, c vir-host v min y m = k m c vir-host + i bgd, v min < c vir-host < v max (1.3) 255, c vir-host v max in which y m is the intensity measurement, k m is the conversion constant from concentration to intensity, i bgd is the background fluorescence (in intensity), and v min and v max are the minimum and maximum detectable virus-host concentrations.
197 175 Time (hours) Data Original Model + Initial Inoculation Figure 1.3: Comparison of representative experimental images to model fits. The full set of experimental images are available in the appendix. Original Model refers to the derived reaction-diffusion model. + Initial Inoculation incorporates the variation in the concentration of uninfected cells within the radius of the initial inoculation. The white scale bar in the upper left-hand corner of the experimental images is one millimeter. 1.2 Propagation of VSV on BHK-21 Cells We first consider propagation of VSV on baby hamster kidney (BHK-21) cells. The first column of images in Figure 1.3 presents representative images for the time course of the experiment; the full set of experimental images are available in the appendix. For this virus/host system, the images demonstrate two prominent features: (1) the infection propagates unimpeded outward radially and (2) the band of intensity amplifies from the first to the third measurement. We now consider models to quantitatively capture both of these features.
198 Development of a Reaction-Diffusion Model We extend the reaction-diffusion model first proposed by Yin and McCaskill [165] and later refined by You and Yin [166] to model this infection. We consider only extracellular species, namely virus, uninfected cells, infected cells, and dead cells. In this context, only the virus is allowed to diffuse, and we model the following reactions: virus + uninfected cell k 1 infected cell (1.4a) infected cell k 2 Y virus (1.4b) in which Y is the yield of virus per infected cell. We assume that the infection propagation is radially symmetric. The concentrations of all species are then segregated by both time and radial distance, giving rise to the following governing equations for the model: c vir t c unc = 1 r r ( D eff vir r c vir r ) + R vir (1.5a) t =R unc (1.5b) c infc =R t infc (1.5c) Dvir eff 1 φ vir 2 + φ (1.5d) φ = V e (c unc + c infc ) c j (t =, r) known, dc vir dr = r=,rmax (1.5e) (1.5f) in which the reaction terms (e.g. R vir ) are dictated by the stoichiometry of reaction (1.4) assuming that the reactions are elementary as written. Also, diffusivity of the virus is hindered due to the presence of uninfected and infected cells on the plate. An effective diffusivity accounts for this effect. We solve equation (1.5) by discretizing the spatial dimension using central differences with an increment of.25 cm, then solving the resulting set of differential-algebraic equations using the package DASKR, a variant of the predictor-corrector solver DASPK [15], with the banded solver option. We determine optimal parameter estimates by solving the following least squares optimization min Φ = min e T k Re k θ θ k s.t.: e k = y k h(x k ; θ) ] T x k = [c vir c unc c infc c dc Equation (1.5) which minimizes the sum of squared residuals between the vectorized images y k and the model-predicted images h(x k ; θ) in a pixel by pixel comparison by manipulating the model parameters θ. Here we use a log 1 transformation of the parameters for the optimization.
199 cunc 1 7 (#/ml) Original Model + Initial Inoculation.8 1 Radius (cm) Figure 1.4: Comparison of the initial uninfected cell concentration for the original and revised (accounting for the initial inoculation effect) models. The second column of images in Figure 1.3 presents the results for the optimal fit. In comparison to the experimental data, the results demonstrate similar radial propagation of the infection front, but do not capture the amplification of intensity observed through the first three samples. To refine the model, we propose that the resulting amplification results from an initial condition effect. In particular, we allow the initial concentration of uninfected cells to vary within the radius of the initial inoculum, and introduce the parameter c unc, in which ( c unc (t =, r) = cm ( 1 c unc, c unc, ), ) c unc, (r.75) c unc,, c unc,, r <.75 cm.75cm r.125 cm r > 1.25 cm (1.6) Performing the parameter estimation with this additional degree of freedom yields the altered initial concentration profile for uninfected cells in Figure 1.4 as well as the optimal fit presented in the third column of images in Figure 1.3. Clearly this fit captures both the outward radial propagation of the infection as well as the amplification of the intensity in the first three images of the time series data Analysis of the Model Fit Table 1.2 presents the parameter estimates for both the original and refined models. Both models predict roughly the same estimates for all parameters. Also, adding the parameter c unc, reduces the objective function Φ by about five percent. Ware et al. [16] use laser light-scattering spectroscopy to estimate the diffusivity of the VSV virion to be cm 2 /sec. Converting this value to cm 2 /hr and taking the log 1
200 178 Model 1 Model 2 Parameter Units log 1 Value log 1 Value k 1 hr k 2 cm 3 /hr D vir cm 2 /hr Y i bgd k m K m cm k wash c unc, cm 3 NA 6.25 Φ Table 1.2: Parameter estimates for the VSV/BHK-21 focal infection models. Parameters are estimated for the log 1 transformation of the parameters. NA denotes that the parameter is not applicable for the given model. Eigenvector log 1 Parameter v 1 v 2 v 3 v 4 v 5 v 6 v 7 k k D vir Y i bgd k m K m k wash Eigenvalue 1.5e7 2.23e5 6.12e5 4.88e6 3.58e7 3.21e8 1.7e9 Table 1.3: Hessian analysis for the parameter estimates of the original VSV/BHK-21 focal infection model. Parameters are estimated for the log 1 transformation of the parameters. Negative eigenvalues are likely due to error in the finite difference approximation used to calculate the Hessian. yields a value of 4.8. This value is very close to the estimated values of 3.87 and 3.94 (see Table 1.2). Table 1.3 analyzes the Hessian of the objective function for the parameter estimates of the original model. This analysis indicates that two linear combinations of parameters cannot be estimated due to negative eigenvalues (which most likely result from errors in the finite difference approximation of the Hessian). The first of these two linear combinations of parameters, i.e. v 1, is primarily constituted by the first reaction rate constant k 1 and the virus yield Y. The second rate constant k 2 accounts for virtually all of the second of these linear combinations. Table 1.4 analyzes the Hessian of the objective function for the parameter estimates of the revised model. This analysis indicates that two linear combinations of parameters cannot be estimated due to negative eigenvalues. These two linear combinations of parameters
201 Eigenvector log 1 Parameter v 1 v 2 v 3 v 4 v 5 v 6 v 7 v 8 k k D vir Y i bgd k m K m k wash c unc, Eigenvalue 1.64e7 2.92e4 7.66e3 5.31e5 7.59e6 3.6e7 3.24e8 1.24e9 Table 1.4: Hessian analysis for the parameter estimates of the revised VSV/BHK-21 focal infection model. Parameters are estimated for the log 1 transformation of the parameters. Negative eigenvalues are likely due to error in the finite difference approximation used to calculate the Hessian. 179 correspond roughly to those of the original model. The modeling process gives insight into the key biological and experimental phenomena giving rise to the observed experimental measurements. First, manipulation of the initial concentration of uninfected cells within the radius of the initial inoculum accounts for the amplification of the intensity in the first three images of the time-series data. This effect has two possible causes: either cells are damaged or removed when a hole is removed from agar at the initiation of the experiment, or uninfected cells but not infected cells continue to grow during the first portion of the experiment. Second, the infection spread is well characterized by considering only extracellular species in the model development. We could have incorporated intracellular infection events (transcription, translation, replication, and assembly of virus) into the model description, but the additional parameters necessary for this model would not be justifiable for the given experimental data. 1.3 Propagation of VSV on DBT Cells We now consider propagation of VSV on murine astrocytoma (DBT) cells. The first column of images in Figure 1.5 presents a representative time course for the experiment; the full set of experimental images are available in the appendix. For this virus/host system, the images demonstrate three prominent features: (1) the infection propagates unimpeded outward radially for the first three images, (2) the intensity of the measurement amplifies from the first to the third measurement, and (3) the infection spread is halted after the third image and the intensity of the measurement diminishes. This particular cell line is known to have an antiviral strategy, namely the interferon signaling pathway. We now consider models to quantitatively capture all of these features.
202 18 Time (hours) Data Reaction- Diffusion Model Segregated Model, Fit 1 Segregated Model, Fit Figure 1.5: Comparison of representative experimental images to model fits for VSV propagation on DBT cells. The white scale bar in the upper left-hand corner of the experimental images is one millimeter Refinement of the Reaction-Diffusion Model We refine the reaction-diffusion model proposed in the previous section to model this infection. In addition to the extracellular species considered previously (virus, uninfected cells, infected cells, and dead cells), we also model interferon (without any distinction between the types α, β, and γ) and inoculated cells. Both virus and interferon are permitted to diffuse. We
203 181 account for the following reactions: virus + uninfected cell infected cell infected cell k 1 infected cell (1.7a) k 2 Y virus + dead cell (1.7b) k 3 infected cell + interferon (1.7c) uninfected cell + interferon k4 inoculated cell inoculated cell k 5 inoculated cell + interferon inoculated cell + virus k1 inoculated cell (1.7d) (1.7e) (1.7f) infected cell + virus k1 infected cell (1.7g) This reaction mechanism makes the following assumptions: 1. interferon binds to uninfected cells to form inoculated cells that are resistant to viral infection, 2. super-infection of infected cells does not alter the yield of virus per infected cell, and 3. virus binds indiscriminately to uninfected, infected, and inoculated cells. We again assume that the infection propagation is radially symmetric. The concentrations of all species are then segregated by both time and radial distance, giving rise to the following governing equations for the model: c vir t c ifn t c unc = 1 r = 1 r ( r r Dvir eff r c vir r ( Difn eff r c ifn r c infc ) + R vir (1.8a) ) + R ifn (1.8b) = R unc, = R t t infc (1.8c) c inoc c = R inoc, dc = R t t dc (1.8d) Dvir eff = 2D 1 φ vir 2 + φ, Deff ifn = 2D 1 φ ifn (1.8e) 2 + φ φ = V e (c unc + c infc + c inoc ) dc vir dc dr =, ifn r=,rmax dr = r=,rmax c i (t =, r) known (1.8f) (1.8g) (1.8h) in which the reaction terms (e.g. R vir ) are dictated by the stoichiometry of reaction (1.7) assuming that the reactions are elementary as written. Additionally, the initial images of the infection indicate a ring-like pattern in the intensity. We account for this phenomenon by estimating two parameters, c unc,1 and c unc,2, that determine the shape of the initial radial profile
204 182 for the uninfected cell concentration, i.e. c unc,2, r <.25 cm c unc,2 2(c unc,2 c unc,1 ) cm (r.25),.25 cm r <.75 cm c unc (t =, r) = c unc,1,.75 r <.1 cm c unc, 2(c unc, c unc,1 ) cm (r.25).1 cm, r <.15 cm c unc,, r >.15 cm (1.9) We estimate the optimal parameters using the same spatial discretization and nonlinear optimization as in the previous section. The second column of images in Figure 1.5 present the optimal fits for this model. In comparison to the experimentally obtained images, this model is able to capture quantitatively the radial propagation of the infection front. However, the fit only qualitatively captures the increase and decrease in the intensity of the experimental data. To better quantitatively capture the temporal changes in this intensity, we propose incorporating the life cycle of infected cells. We therefore segregate the infected cell population by the age of infection τ, and model the intracellular production rates of virus and interferon using first-order plus time delay expressions, i.e. r vir (τ) = K vir [1 exp ( k vir (τ d vir ))] (1.1) r ifn (τ) = K ifn [1 exp ( k ifn (τ d ifn ))] (1.11) We also assume that infected cells cannot live longer than age τ d, at which point these cells die. This model requires fitting of four more parameters than the reaction-diffusion model (seven additional parameters are required for the first-order plus time delay description, but this description obviates the need for the virus yield Y and the rate constants k 2 and k 3 ). The considered reactions now become: virus + uninfected cell k 1 infected cell (1.12a) infected cell virus (age dependent) (1.12b) infected cell infected cell + interferon (age dependent) (1.12c) uninfected cell + interferon k4 inoculated cell inoculated cell k 5 inoculated cell + interferon inoculated cell + virus k1 inoculated cell (1.12d) (1.12e) (1.12f) infected cell + virus k1 infected cell (all ages) (1.12g)
205 The model equations are then the following set of coupled integro-partial differential equations c vir = 1 ( Dvir eff t r r r c ) τd vir + c r infc (τ)r vir (τ)dτ + R vir (1.13a) c ifn = 1 ( Difn eff t r r r c ) τd ifn + c r infc (τ)r ifn (τ)dτ + R ifn (1.13b) dc vir dr dc infc dτ c unc c infc 183 = R unc, + c infc = R t t τ infc (1.13c) c inoc c = R inoc, dc = R t t dc (1.13d) Dvir eff = 2D 1 φ vir 2 + φ, Deff ifn = 2D 1 φ ifn (1.13e) 2 + φ τd φ = V e (c unc + c infc dτ + c inoc ) dc =, ifn r=,rmax dr = r=,rmax dc = k 1 c vir c unc, infc τ= dτ = τ=τd c i (t =, r) known (1.13f) (1.13g) (1.13h) (1.13i) We discretize the age dimension using orthogonal collocation on Lagrange polynomials [155] with seventeen points, and use the same spatial discretization scheme as in the reactiondiffusion model. The third and fourth columns of images in Figure 1.5 present the optimal fits for this model. In comparison to the experimentally obtained images, this model is able to capture quantitatively both the radial propagation of the infection front and the changes in the intensity of the experimental data. The optimization also yields two sets of parameters with similar fits and similar values of the objective function, but different values for the parameters. Most overtly different are the estimates for the intracellular production rates of virus and interferon, which suggest two different mechanisms for up-regulation of the interferon pathway. These production rates are presented in Figure 1.6. In the first fit, the estimated maximum age of infected cells is roughly 26 hours, and the production of interferon lags significantly after the production of interferon. For the second fit, the estimated maximum age of infected cells is only roughly 17 hours, and the production of interferon closely precedes the virus production. Additionally, the production rates in the second fit are approximately an order of magnitude lower than the production rates in the first fit Discussion The models provide estimates for key parameters in the viral infection and host response. In this case, the three model fits only predict similar parameter values for the background fluo-
206 184 Production Rate (#/hour) Infection Age (hours) Virus Interferon 25 (a) 3 Production Rate (#/hour) Infection Age (hours) Virus Interferon (b) 18 Figure 1.6: Comparison of intracellular production rates of virus and interferon for the segregated model of VSV propagation on DBT cells. rescence i bgd and the viral diffusivity D vir. The remaining parameters are generally different by at least an order of magnitude. Ware et al. [16] estimate the diffusivity of the VSV virion to be cm 2 /sec. Converting this value to cm 2 /hr and taking the log 1 yields a value of 4.8. The estimated values of this diffusivity, D vir in Table 1.5, are all within an order of magnitude of this value. Porterfield et al. [12] and Nichol and Deutsch [96] estimate the diffusivity of γ-interferon to be and cm 2 /sec, respectively. Converting these values to cm 2 /hr and
207 185 Reaction-Diffusion Segregated Fit 1 Segregated Fit 2 Parameter Units log 1 Value log 1 Value log 1 Value k 1 hr k 2 cm 3 /hr NA NA k 3 cm 3 /hr NA NA k 4 cm 3 /hr k 5 cm 3 /hr D vir cm 2 /hr D ifn cm 2 /hr Y NA NA i bgd k m /K m k wash c unc,1 cm c unc,2 cm k vir hr 1 NA k ifn hr 1 NA K vir hr 1 NA K ifn hr 1 NA d vir hr 1 NA d ifn hr 1 NA τ d hr NA Φ Table 1.5: Parameter estimates for the VSV/DBT focal infection models. Parameters are estimated for the log 1 transformation of the parameters. NA denotes that the parameter is not applicable for the given model. taking the log 1 yields values of 2.83 to 2.57, respectively. These values have the same order of magnitude as the fits for the reaction-diffusion model and the first segregated fit. The second segregated fit predicts the diffusivity of interferon to be roughly two orders of magnitude greater than either of the previously reported values. The infection spread is not well characterized by considering only extracellular species in the model development. Incorporation of simple first-order plus time delay expressions for the production rates of virus and interferon leads to significantly improved quantitative prediction of the given experimental data (roughly a 17% decrease in the objective function Φ via the addition of four parameters). Additionally, the model fits suggest two different possible mechanisms for production of both virus and interferon. For VSV infection of Krebs- 2 carcinoma cells [158] and mouse L cells [161], experimental studies place the first detectable amount of interferon between four and eight hours, respectively. These results suggest that the second segregated fit is more realistic than the first segregated fit.
208 186 log 1 Eigenvector Parameter v 1 v 2 v 3 v 4 v 5 v 6 k k k k k D vir D ifn Y i bgd k m/k m k wash 1 c unc, c unc, Eigenvalue 2.83e7 6.81e6 4.8e6 2.66e6 3.37e5 1.2 log 1 Eigenvector Parameter v 7 v 8 v 9 v 1 v 11 v 12 v 13 k k k k k D vir D ifn Y i bgd k m/k m k wash c unc, c unc, Eigenvalue 1.89e6 6.81e6 2.9e7 3.61e7 1.22e8 4.15e8 1.27e9 Table 1.6: Hessian analysis for the parameter estimates of the reaction-diffusion VSV/DBT focal infection model. Parameters are estimated for the log 1 transformation of the parameters. Negative eigenvalues are likely due to error in the finite difference approximation used to calculate the Hessian. Unreported values denote that the contribution of the parameter to the eigenvector is less than Table 1.6 presents the Hessian analysis for the reaction-diffusion model. This analysis indicates that roughly five linear combinations of parameters cannot be estimated from the experimental data. However, Figure 1.5 demonstrates that this model is not capable of capturing the infection dynamics, particularly the magnitude of the intensity. Tables 1.7 and 1.8 present the Hessian analysis of the objective function Φ for the segregated model fits. Roughly five linear combinations of parameters yield negative eigenvalues for both fits, indicating that these parameter combinations cannot be estimated from the experimental data. This analysis indicates that the experimental measurements are not informative
209 187 log 1 Eigenvector Parameter v 1 v 2 v 3 v 4 v 5 v 6 v 7 v 8 v 9 k k 4.77 k D vir.77 D ifn i bgd k m/k m k wash f unc, f unc, k vir k ifn K vir K ifn d vir d ifn.77 τ d.77 Eigenvalue 2.27e14 7.8e e5 6.18e4 2.61e3 4.32e3 1.2e4 2.7e4 7.33e4 log 1 Eigenvector Parameter v 1 v 11 v 12 v 13 v 14 v 15 v 16 v 17 k k 4.77 k D vir.77 D ifn i bgd k m/k m k wash f unc, f unc, k vir k ifn K vir K ifn d vir d ifn.77 τ d.77 Eigenvalue 3.19e5 4.93e5 6.32e5 1.44e7 9.39e7 8.87e8 7.8e e14 Table 1.7: Hessian analysis for the parameter estimates of the first segregated VSV/DBT focal infection model. Parameters are estimated for the log 1 transformation of the parameters. Negative eigenvalues are likely due to error in the finite difference approximation used to calculate the Hessian. Unreported values denote that the contribution of the parameter to the eigenvector is less than
210 188 log 1 Eigenvector Parameter v 1 v 2 v 3 v 4 v 5 v 6 v 7 v 8 v 9 k k k D vir D ifn i bgd k m/k m k wash f unc, f unc, k vir k ifn K vir K ifn d vir d ifn τ d Eigenvalue 5.4e5 2.39e5 1.38e5 5.37e4 1.35e2 9.32e3 1.21e4 2.66e5 1.8e5 Eigenvector log 1 Parameter v 1 v 11 v 12 v 13 v 14 v 15 v 16 v 17 k k k D vir D ifn i bgd k m/k m k wash f unc, f unc, k vir k ifn K vir K ifn d vir d ifn τ d Eigenvalue 1.85e5 4.74e5 2.9e6 2.74e6 8.68e6 5.2e7 1.37e8 9.6e8 Table 1.8: Hessian analysis for the parameter estimates of the second segregated VSV/DBT focal infection model. Parameters are estimated for the log 1 transformation of the parameters. Negative eigenvalues are likely due to error in the finite difference approximation used to calculate the Hessian. Unreported values denote that the contribution of the parameter to the eigenvector is less than
211 189 Time (hours) Data Segregated Model, Fit 1 Segregated Model, Fit Figure 1.7: Comparison of representative experimental images to model predictions for VSV propagation on DBT cells in the presence of interferon inhibitors. The white scale bar in the lower left-hand corner of the experimental images is one millimeter. enough to distinguish between these different mechanisms Model Prediction: Infection Propagation in the Presence of Interferon Inhibitors To validate the model, we compare model predictions of the infection propagation in the presence of interferon inhibitors to experimentally-obtained images. We assume that the dosing of interferon inhibitor is sufficiently large to completely inhibit production of interferon. Accordingly, we set the constants k 5 and K 2 corresponding to interferon production from inoculated cells and the production rate of interferon in infected cells to zero. Figure 1.7 compares the results for the experimental data with the segregated model predictions. In both cases, the models over-predict the radial propagation of the infection front for the latter two time points. Additionally, the first segregated model predicts even farther propagation of the infection front than the second segregated model. The most likely explanation for the deviations between the data and predictions is that the dosing of the interferon inhibitor is not large enough to completely eliminate the host antiviral response.
Nonlinear Stochastic Modeling and State Estimation of Weakly Observable Systems: Application to Industrial Polymerization Processes
 Nonlinear Stochastic Modeling and State Estimation of Weakly Observable Systems: Application to Industrial Polymerization Processes Fernando V. Lima, James B. Rawlings and Tyler A. Soderstrom Department
Nonlinear Stochastic Modeling and State Estimation of Weakly Observable Systems: Application to Industrial Polymerization Processes Fernando V. Lima, James B. Rawlings and Tyler A. Soderstrom Department
Extending the Tools of Chemical Reaction Engineering to the Molecular Scale
 Extending the Tools of Chemical Reaction Engineering to the Molecular Scale Multiple-time-scale order reduction for stochastic kinetics James B. Rawlings Department of Chemical and Biological Engineering
Extending the Tools of Chemical Reaction Engineering to the Molecular Scale Multiple-time-scale order reduction for stochastic kinetics James B. Rawlings Department of Chemical and Biological Engineering
State Estimation using Moving Horizon Estimation and Particle Filtering
 State Estimation using Moving Horizon Estimation and Particle Filtering James B. Rawlings Department of Chemical and Biological Engineering UW Math Probability Seminar Spring 2009 Rawlings MHE & PF 1 /
State Estimation using Moving Horizon Estimation and Particle Filtering James B. Rawlings Department of Chemical and Biological Engineering UW Math Probability Seminar Spring 2009 Rawlings MHE & PF 1 /
Stochastic Modeling of Chemical Reactions
 Stochastic Modeling of Chemical Reactions Eric Haseltine Department of Chemical Engineering University of Wisconsin-Madison TWMCC 27 September Texas-Wisconsin Modeling and Control Consortium 22TWMCC Outline
Stochastic Modeling of Chemical Reactions Eric Haseltine Department of Chemical Engineering University of Wisconsin-Madison TWMCC 27 September Texas-Wisconsin Modeling and Control Consortium 22TWMCC Outline
Real-Time Feasibility of Nonlinear Predictive Control for Semi-batch Reactors
 European Symposium on Computer Arded Aided Process Engineering 15 L. Puigjaner and A. Espuña (Editors) 2005 Elsevier Science B.V. All rights reserved. Real-Time Feasibility of Nonlinear Predictive Control
European Symposium on Computer Arded Aided Process Engineering 15 L. Puigjaner and A. Espuña (Editors) 2005 Elsevier Science B.V. All rights reserved. Real-Time Feasibility of Nonlinear Predictive Control
Lessons in Estimation Theory for Signal Processing, Communications, and Control
 Lessons in Estimation Theory for Signal Processing, Communications, and Control Jerry M. Mendel Department of Electrical Engineering University of Southern California Los Angeles, California PRENTICE HALL
Lessons in Estimation Theory for Signal Processing, Communications, and Control Jerry M. Mendel Department of Electrical Engineering University of Southern California Los Angeles, California PRENTICE HALL
State Estimation of Linear and Nonlinear Dynamic Systems
 State Estimation of Linear and Nonlinear Dynamic Systems Part I: Linear Systems with Gaussian Noise James B. Rawlings and Fernando V. Lima Department of Chemical and Biological Engineering University of
State Estimation of Linear and Nonlinear Dynamic Systems Part I: Linear Systems with Gaussian Noise James B. Rawlings and Fernando V. Lima Department of Chemical and Biological Engineering University of
Constrained State Estimation Using the Unscented Kalman Filter
 16th Mediterranean Conference on Control and Automation Congress Centre, Ajaccio, France June 25-27, 28 Constrained State Estimation Using the Unscented Kalman Filter Rambabu Kandepu, Lars Imsland and
16th Mediterranean Conference on Control and Automation Congress Centre, Ajaccio, France June 25-27, 28 Constrained State Estimation Using the Unscented Kalman Filter Rambabu Kandepu, Lars Imsland and
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2014
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2016
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2016 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2016 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
natural development from this collection of knowledge: it is more reliable to predict the property
 1 Chapter 1 Introduction As the basis of all life phenomena, the interaction of biomolecules has been under the scrutiny of scientists and cataloged meticulously [2]. The recent advent of systems biology
1 Chapter 1 Introduction As the basis of all life phenomena, the interaction of biomolecules has been under the scrutiny of scientists and cataloged meticulously [2]. The recent advent of systems biology
ROBUST CONSTRAINED ESTIMATION VIA UNSCENTED TRANSFORMATION. Pramod Vachhani a, Shankar Narasimhan b and Raghunathan Rengaswamy a 1
 ROUST CONSTRINED ESTIMTION VI UNSCENTED TRNSFORMTION Pramod Vachhani a, Shankar Narasimhan b and Raghunathan Rengaswamy a a Department of Chemical Engineering, Clarkson University, Potsdam, NY -3699, US.
ROUST CONSTRINED ESTIMTION VI UNSCENTED TRNSFORMTION Pramod Vachhani a, Shankar Narasimhan b and Raghunathan Rengaswamy a a Department of Chemical Engineering, Clarkson University, Potsdam, NY -3699, US.
ECE521 week 3: 23/26 January 2017
 ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
UNIVERSITY OF CALIFORNIA. Los Angeles. Distributed Model Predictive Control of Nonlinear. and Two-Time-Scale Process Networks
 UNIVERSITY OF CALIFORNIA Los Angeles Distributed Model Predictive Control of Nonlinear and Two-Time-Scale Process Networks A dissertation submitted in partial satisfaction of the requirements for the degree
UNIVERSITY OF CALIFORNIA Los Angeles Distributed Model Predictive Control of Nonlinear and Two-Time-Scale Process Networks A dissertation submitted in partial satisfaction of the requirements for the degree
Derivations for order reduction of the chemical master equation
 2 TWMCC Texas-Wisconsin Modeling and Control Consortium 1 Technical report number 2006-02 Derivations for order reduction of the chemical master equation Ethan A. Mastny, Eric L. Haseltine, and James B.
2 TWMCC Texas-Wisconsin Modeling and Control Consortium 1 Technical report number 2006-02 Derivations for order reduction of the chemical master equation Ethan A. Mastny, Eric L. Haseltine, and James B.
Gaussian processes. Chuong B. Do (updated by Honglak Lee) November 22, 2008
 Gaussian processes Chuong B Do (updated by Honglak Lee) November 22, 2008 Many of the classical machine learning algorithms that we talked about during the first half of this course fit the following pattern:
Gaussian processes Chuong B Do (updated by Honglak Lee) November 22, 2008 Many of the classical machine learning algorithms that we talked about during the first half of this course fit the following pattern:
Lecture (9) Reactor Sizing. Figure (1). Information needed to predict what a reactor can do.
 Lecture (9) Reactor Sizing 1.Introduction Chemical kinetics is the study of chemical reaction rates and reaction mechanisms. The study of chemical reaction engineering (CRE) combines the study of chemical
Lecture (9) Reactor Sizing 1.Introduction Chemical kinetics is the study of chemical reaction rates and reaction mechanisms. The study of chemical reaction engineering (CRE) combines the study of chemical
Stochastic Spectral Approaches to Bayesian Inference
 Stochastic Spectral Approaches to Bayesian Inference Prof. Nathan L. Gibson Department of Mathematics Applied Mathematics and Computation Seminar March 4, 2011 Prof. Gibson (OSU) Spectral Approaches to
Stochastic Spectral Approaches to Bayesian Inference Prof. Nathan L. Gibson Department of Mathematics Applied Mathematics and Computation Seminar March 4, 2011 Prof. Gibson (OSU) Spectral Approaches to
Fast Probability Generating Function Method for Stochastic Chemical Reaction Networks
 MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 71 (2014) 57-69 ISSN 0340-6253 Fast Probability Generating Function Method for Stochastic Chemical Reaction
MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 71 (2014) 57-69 ISSN 0340-6253 Fast Probability Generating Function Method for Stochastic Chemical Reaction
Nonlinear Model Predictive Control Tools (NMPC Tools)
 Nonlinear Model Predictive Control Tools (NMPC Tools) Rishi Amrit, James B. Rawlings April 5, 2008 1 Formulation We consider a control system composed of three parts([2]). Estimator Target calculator Regulator
Nonlinear Model Predictive Control Tools (NMPC Tools) Rishi Amrit, James B. Rawlings April 5, 2008 1 Formulation We consider a control system composed of three parts([2]). Estimator Target calculator Regulator
BAYESIAN ESTIMATION BY SEQUENTIAL MONTE CARLO SAMPLING FOR NONLINEAR DYNAMIC SYSTEMS
 BAYESIAN ESTIMATION BY SEQUENTIAL MONTE CARLO SAMPLING FOR NONLINEAR DYNAMIC SYSTEMS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate
BAYESIAN ESTIMATION BY SEQUENTIAL MONTE CARLO SAMPLING FOR NONLINEAR DYNAMIC SYSTEMS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate
, MODELLING AND CONTROL OF SUSTAINED OSCILLATIONS IN THE CONTINUOUS EMULSION POLYMERIZATION OF VINYL ACETATE. by Mark James Pollock
 , MODELLING AND CONTROL OF SUSTAINED OSCILLATIONS IN THE CONTINUOUS EMULSION POLYMERIZATION OF VINYL ACETATE by Mark James Pollock A Thesi s Submitted to the Schoo~ of Graduate Studies in Partial Fulfilment
, MODELLING AND CONTROL OF SUSTAINED OSCILLATIONS IN THE CONTINUOUS EMULSION POLYMERIZATION OF VINYL ACETATE by Mark James Pollock A Thesi s Submitted to the Schoo~ of Graduate Studies in Partial Fulfilment
Here represents the impulse (or delta) function. is an diagonal matrix of intensities, and is an diagonal matrix of intensities.
 19 KALMAN FILTER 19.1 Introduction In the previous section, we derived the linear quadratic regulator as an optimal solution for the fullstate feedback control problem. The inherent assumption was that
19 KALMAN FILTER 19.1 Introduction In the previous section, we derived the linear quadratic regulator as an optimal solution for the fullstate feedback control problem. The inherent assumption was that
Optimizing Economic Performance using Model Predictive Control
 Optimizing Economic Performance using Model Predictive Control James B. Rawlings Department of Chemical and Biological Engineering Second Workshop on Computational Issues in Nonlinear Control Monterey,
Optimizing Economic Performance using Model Predictive Control James B. Rawlings Department of Chemical and Biological Engineering Second Workshop on Computational Issues in Nonlinear Control Monterey,
Estimating Disturbance Covariances From Data For Improved Control Performance
 Estimating Disturbance Covariances From Data For Improved Control Performance by Brian J. Odelson A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY
Estimating Disturbance Covariances From Data For Improved Control Performance by Brian J. Odelson A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY
Condensed Table of Contents for Introduction to Stochastic Search and Optimization: Estimation, Simulation, and Control by J. C.
 Condensed Table of Contents for Introduction to Stochastic Search and Optimization: Estimation, Simulation, and Control by J. C. Spall John Wiley and Sons, Inc., 2003 Preface... xiii 1. Stochastic Search
Condensed Table of Contents for Introduction to Stochastic Search and Optimization: Estimation, Simulation, and Control by J. C. Spall John Wiley and Sons, Inc., 2003 Preface... xiii 1. Stochastic Search
In search of the unreachable setpoint
 In search of the unreachable setpoint Adventures with Prof. Sten Bay Jørgensen James B. Rawlings Department of Chemical and Biological Engineering June 19, 2009 Seminar Honoring Prof. Sten Bay Jørgensen
In search of the unreachable setpoint Adventures with Prof. Sten Bay Jørgensen James B. Rawlings Department of Chemical and Biological Engineering June 19, 2009 Seminar Honoring Prof. Sten Bay Jørgensen
Pattern Recognition and Machine Learning
 Christopher M. Bishop Pattern Recognition and Machine Learning ÖSpri inger Contents Preface Mathematical notation Contents vii xi xiii 1 Introduction 1 1.1 Example: Polynomial Curve Fitting 4 1.2 Probability
Christopher M. Bishop Pattern Recognition and Machine Learning ÖSpri inger Contents Preface Mathematical notation Contents vii xi xiii 1 Introduction 1 1.1 Example: Polynomial Curve Fitting 4 1.2 Probability
Statistics: Learning models from data
 DS-GA 1002 Lecture notes 5 October 19, 2015 Statistics: Learning models from data Learning models from data that are assumed to be generated probabilistically from a certain unknown distribution is a crucial
DS-GA 1002 Lecture notes 5 October 19, 2015 Statistics: Learning models from data Learning models from data that are assumed to be generated probabilistically from a certain unknown distribution is a crucial
Experimental designs for multiple responses with different models
 Graduate Theses and Dissertations Graduate College 2015 Experimental designs for multiple responses with different models Wilmina Mary Marget Iowa State University Follow this and additional works at:
Graduate Theses and Dissertations Graduate College 2015 Experimental designs for multiple responses with different models Wilmina Mary Marget Iowa State University Follow this and additional works at:
Stochastic Analogues to Deterministic Optimizers
 Stochastic Analogues to Deterministic Optimizers ISMP 2018 Bordeaux, France Vivak Patel Presented by: Mihai Anitescu July 6, 2018 1 Apology I apologize for not being here to give this talk myself. I injured
Stochastic Analogues to Deterministic Optimizers ISMP 2018 Bordeaux, France Vivak Patel Presented by: Mihai Anitescu July 6, 2018 1 Apology I apologize for not being here to give this talk myself. I injured
 5.1 2D example 59 Figure 5.1: Parabolic velocity field in a straight two-dimensional pipe. Figure 5.2: Concentration on the input boundary of the pipe. The vertical axis corresponds to r 2 -coordinate,
5.1 2D example 59 Figure 5.1: Parabolic velocity field in a straight two-dimensional pipe. Figure 5.2: Concentration on the input boundary of the pipe. The vertical axis corresponds to r 2 -coordinate,
Online monitoring of MPC disturbance models using closed-loop data
 Online monitoring of MPC disturbance models using closed-loop data Brian J. Odelson and James B. Rawlings Department of Chemical Engineering University of Wisconsin-Madison Online Optimization Based Identification
Online monitoring of MPC disturbance models using closed-loop data Brian J. Odelson and James B. Rawlings Department of Chemical Engineering University of Wisconsin-Madison Online Optimization Based Identification
Linear-Quadratic Optimal Control: Full-State Feedback
 Chapter 4 Linear-Quadratic Optimal Control: Full-State Feedback 1 Linear quadratic optimization is a basic method for designing controllers for linear (and often nonlinear) dynamical systems and is actually
Chapter 4 Linear-Quadratic Optimal Control: Full-State Feedback 1 Linear quadratic optimization is a basic method for designing controllers for linear (and often nonlinear) dynamical systems and is actually
Dynamic System Identification using HDMR-Bayesian Technique
 Dynamic System Identification using HDMR-Bayesian Technique *Shereena O A 1) and Dr. B N Rao 2) 1), 2) Department of Civil Engineering, IIT Madras, Chennai 600036, Tamil Nadu, India 1) ce14d020@smail.iitm.ac.in
Dynamic System Identification using HDMR-Bayesian Technique *Shereena O A 1) and Dr. B N Rao 2) 1), 2) Department of Civil Engineering, IIT Madras, Chennai 600036, Tamil Nadu, India 1) ce14d020@smail.iitm.ac.in
ECE276A: Sensing & Estimation in Robotics Lecture 10: Gaussian Mixture and Particle Filtering
 ECE276A: Sensing & Estimation in Robotics Lecture 10: Gaussian Mixture and Particle Filtering Lecturer: Nikolay Atanasov: natanasov@ucsd.edu Teaching Assistants: Siwei Guo: s9guo@eng.ucsd.edu Anwesan Pal:
ECE276A: Sensing & Estimation in Robotics Lecture 10: Gaussian Mixture and Particle Filtering Lecturer: Nikolay Atanasov: natanasov@ucsd.edu Teaching Assistants: Siwei Guo: s9guo@eng.ucsd.edu Anwesan Pal:
A Note on the Particle Filter with Posterior Gaussian Resampling
 Tellus (6), 8A, 46 46 Copyright C Blackwell Munksgaard, 6 Printed in Singapore. All rights reserved TELLUS A Note on the Particle Filter with Posterior Gaussian Resampling By X. XIONG 1,I.M.NAVON 1,2 and
Tellus (6), 8A, 46 46 Copyright C Blackwell Munksgaard, 6 Printed in Singapore. All rights reserved TELLUS A Note on the Particle Filter with Posterior Gaussian Resampling By X. XIONG 1,I.M.NAVON 1,2 and
A First Course on Kinetics and Reaction Engineering Unit 30.Thermal Back-Mixing in a PFR
 Unit 30.Thermal Back-Mixing in a PFR Overview One advantage offered by a CSTR when running an exothermic reaction is that the cool feed gets heated by mixing with the contents of the reactor. As a consequence
Unit 30.Thermal Back-Mixing in a PFR Overview One advantage offered by a CSTR when running an exothermic reaction is that the cool feed gets heated by mixing with the contents of the reactor. As a consequence
Engineering. Green Chemical. S. Suresh and S. Sundaramoorthy. and Chemical Processes. An Introduction to Catalysis, Kinetics, CRC Press
 I i Green Chemical Engineering An Introduction to Catalysis, Kinetics, and Chemical Processes S. Suresh and S. Sundaramoorthy CRC Press Taylor & Francis Group Boca Raton London New York CRC Press is an
I i Green Chemical Engineering An Introduction to Catalysis, Kinetics, and Chemical Processes S. Suresh and S. Sundaramoorthy CRC Press Taylor & Francis Group Boca Raton London New York CRC Press is an
Real-Time Software Transactional Memory: Contention Managers, Time Bounds, and Implementations
 Real-Time Software Transactional Memory: Contention Managers, Time Bounds, and Implementations Mohammed El-Shambakey Dissertation Submitted to the Faculty of the Virginia Polytechnic Institute and State
Real-Time Software Transactional Memory: Contention Managers, Time Bounds, and Implementations Mohammed El-Shambakey Dissertation Submitted to the Faculty of the Virginia Polytechnic Institute and State
Theory and Practice of Data Assimilation in Ocean Modeling
 Theory and Practice of Data Assimilation in Ocean Modeling Robert N. Miller College of Oceanic and Atmospheric Sciences Oregon State University Oceanography Admin. Bldg. 104 Corvallis, OR 97331-5503 Phone:
Theory and Practice of Data Assimilation in Ocean Modeling Robert N. Miller College of Oceanic and Atmospheric Sciences Oregon State University Oceanography Admin. Bldg. 104 Corvallis, OR 97331-5503 Phone:
MOST control systems are designed under the assumption
 2076 IEEE TRANSACTIONS ON AUTOMATIC CONTROL, VOL. 53, NO. 9, OCTOBER 2008 Lyapunov-Based Model Predictive Control of Nonlinear Systems Subject to Data Losses David Muñoz de la Peña and Panagiotis D. Christofides
2076 IEEE TRANSACTIONS ON AUTOMATIC CONTROL, VOL. 53, NO. 9, OCTOBER 2008 Lyapunov-Based Model Predictive Control of Nonlinear Systems Subject to Data Losses David Muñoz de la Peña and Panagiotis D. Christofides
DISCRETE STOCHASTIC PROCESSES Draft of 2nd Edition
 DISCRETE STOCHASTIC PROCESSES Draft of 2nd Edition R. G. Gallager January 31, 2011 i ii Preface These notes are a draft of a major rewrite of a text [9] of the same name. The notes and the text are outgrowths
DISCRETE STOCHASTIC PROCESSES Draft of 2nd Edition R. G. Gallager January 31, 2011 i ii Preface These notes are a draft of a major rewrite of a text [9] of the same name. The notes and the text are outgrowths
Autonomous Navigation for Flying Robots
 Computer Vision Group Prof. Daniel Cremers Autonomous Navigation for Flying Robots Lecture 6.2: Kalman Filter Jürgen Sturm Technische Universität München Motivation Bayes filter is a useful tool for state
Computer Vision Group Prof. Daniel Cremers Autonomous Navigation for Flying Robots Lecture 6.2: Kalman Filter Jürgen Sturm Technische Universität München Motivation Bayes filter is a useful tool for state
A Robust Strategy for Joint Data Reconciliation and Parameter Estimation
 A Robust Strategy for Joint Data Reconciliation and Parameter Estimation Yen Yen Joe 1) 3), David Wang ), Chi Bun Ching 3), Arthur Tay 1), Weng Khuen Ho 1) and Jose Romagnoli ) * 1) Dept. of Electrical
A Robust Strategy for Joint Data Reconciliation and Parameter Estimation Yen Yen Joe 1) 3), David Wang ), Chi Bun Ching 3), Arthur Tay 1), Weng Khuen Ho 1) and Jose Romagnoli ) * 1) Dept. of Electrical
Quantitative Biology II Lecture 4: Variational Methods
 10 th March 2015 Quantitative Biology II Lecture 4: Variational Methods Gurinder Singh Mickey Atwal Center for Quantitative Biology Cold Spring Harbor Laboratory Image credit: Mike West Summary Approximate
10 th March 2015 Quantitative Biology II Lecture 4: Variational Methods Gurinder Singh Mickey Atwal Center for Quantitative Biology Cold Spring Harbor Laboratory Image credit: Mike West Summary Approximate
Stochastic Models, Estimation and Control Peter S. Maybeck Volumes 1, 2 & 3 Tables of Contents
 Navtech Part #s Volume 1 #1277 Volume 2 #1278 Volume 3 #1279 3 Volume Set #1280 Stochastic Models, Estimation and Control Peter S. Maybeck Volumes 1, 2 & 3 Tables of Contents Volume 1 Preface Contents
Navtech Part #s Volume 1 #1277 Volume 2 #1278 Volume 3 #1279 3 Volume Set #1280 Stochastic Models, Estimation and Control Peter S. Maybeck Volumes 1, 2 & 3 Tables of Contents Volume 1 Preface Contents
CS281 Section 4: Factor Analysis and PCA
 CS81 Section 4: Factor Analysis and PCA Scott Linderman At this point we have seen a variety of machine learning models, with a particular emphasis on models for supervised learning. In particular, we
CS81 Section 4: Factor Analysis and PCA Scott Linderman At this point we have seen a variety of machine learning models, with a particular emphasis on models for supervised learning. In particular, we
an introduction to bayesian inference
 with an application to network analysis http://jakehofman.com january 13, 2010 motivation would like models that: provide predictive and explanatory power are complex enough to describe observed phenomena
with an application to network analysis http://jakehofman.com january 13, 2010 motivation would like models that: provide predictive and explanatory power are complex enough to describe observed phenomena
OPTIMAL ESTIMATION of DYNAMIC SYSTEMS
 CHAPMAN & HALL/CRC APPLIED MATHEMATICS -. AND NONLINEAR SCIENCE SERIES OPTIMAL ESTIMATION of DYNAMIC SYSTEMS John L Crassidis and John L. Junkins CHAPMAN & HALL/CRC A CRC Press Company Boca Raton London
CHAPMAN & HALL/CRC APPLIED MATHEMATICS -. AND NONLINEAR SCIENCE SERIES OPTIMAL ESTIMATION of DYNAMIC SYSTEMS John L Crassidis and John L. Junkins CHAPMAN & HALL/CRC A CRC Press Company Boca Raton London
Inferences about Parameters of Trivariate Normal Distribution with Missing Data
 Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 7-5-3 Inferences about Parameters of Trivariate Normal Distribution with Missing
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 7-5-3 Inferences about Parameters of Trivariate Normal Distribution with Missing
Chapter 1. Introduction. 1.1 Background
 Chapter 1 Introduction Science is facts; just as houses are made of stones, so is science made of facts; but a pile of stones is not a house and a collection of facts is not necessarily science. Henri
Chapter 1 Introduction Science is facts; just as houses are made of stones, so is science made of facts; but a pile of stones is not a house and a collection of facts is not necessarily science. Henri
Partially Observable Markov Decision Processes (POMDPs)
 Partially Observable Markov Decision Processes (POMDPs) Sachin Patil Guest Lecture: CS287 Advanced Robotics Slides adapted from Pieter Abbeel, Alex Lee Outline Introduction to POMDPs Locally Optimal Solutions
Partially Observable Markov Decision Processes (POMDPs) Sachin Patil Guest Lecture: CS287 Advanced Robotics Slides adapted from Pieter Abbeel, Alex Lee Outline Introduction to POMDPs Locally Optimal Solutions
Stochastic Processes
 qmc082.tex. Version of 30 September 2010. Lecture Notes on Quantum Mechanics No. 8 R. B. Griffiths References: Stochastic Processes CQT = R. B. Griffiths, Consistent Quantum Theory (Cambridge, 2002) DeGroot
qmc082.tex. Version of 30 September 2010. Lecture Notes on Quantum Mechanics No. 8 R. B. Griffiths References: Stochastic Processes CQT = R. B. Griffiths, Consistent Quantum Theory (Cambridge, 2002) DeGroot
New Advances in Uncertainty Analysis and Estimation
 New Advances in Uncertainty Analysis and Estimation Overview: Both sensor observation data and mathematical models are used to assist in the understanding of physical dynamic systems. However, observational
New Advances in Uncertainty Analysis and Estimation Overview: Both sensor observation data and mathematical models are used to assist in the understanding of physical dynamic systems. However, observational
Mathematical Theory of Control Systems Design
 Mathematical Theory of Control Systems Design by V. N. Afarias'ev, V. B. Kolmanovskii and V. R. Nosov Moscow University of Electronics and Mathematics, Moscow, Russia KLUWER ACADEMIC PUBLISHERS DORDRECHT
Mathematical Theory of Control Systems Design by V. N. Afarias'ev, V. B. Kolmanovskii and V. R. Nosov Moscow University of Electronics and Mathematics, Moscow, Russia KLUWER ACADEMIC PUBLISHERS DORDRECHT
c 2011 JOSHUA DAVID JOHNSTON ALL RIGHTS RESERVED
 c 211 JOSHUA DAVID JOHNSTON ALL RIGHTS RESERVED ANALYTICALLY AND NUMERICALLY MODELING RESERVOIR-EXTENDED POROUS SLIDER AND JOURNAL BEARINGS INCORPORATING CAVITATION EFFECTS A Dissertation Presented to
c 211 JOSHUA DAVID JOHNSTON ALL RIGHTS RESERVED ANALYTICALLY AND NUMERICALLY MODELING RESERVOIR-EXTENDED POROUS SLIDER AND JOURNAL BEARINGS INCORPORATING CAVITATION EFFECTS A Dissertation Presented to
Coordinating multiple optimization-based controllers: new opportunities and challenges
 Coordinating multiple optimization-based controllers: new opportunities and challenges James B. Rawlings and Brett T. Stewart Department of Chemical and Biological Engineering University of Wisconsin Madison
Coordinating multiple optimization-based controllers: new opportunities and challenges James B. Rawlings and Brett T. Stewart Department of Chemical and Biological Engineering University of Wisconsin Madison
The Material Balance for Chemical Reactors
 The Material Balance for Chemical Reactors Copyright c 2015 by Nob Hill Publishing, LLC 1 General Mole Balance V R j Q 0 c j0 Q 1 c j1 Conservation of mass rate of accumulation of component j = + { rate
The Material Balance for Chemical Reactors Copyright c 2015 by Nob Hill Publishing, LLC 1 General Mole Balance V R j Q 0 c j0 Q 1 c j1 Conservation of mass rate of accumulation of component j = + { rate
The Material Balance for Chemical Reactors. Copyright c 2015 by Nob Hill Publishing, LLC
 The Material Balance for Chemical Reactors Copyright c 2015 by Nob Hill Publishing, LLC 1 General Mole Balance V R j Q 0 c j0 Q 1 c j1 Conservation of mass rate of accumulation of component j = + { rate
The Material Balance for Chemical Reactors Copyright c 2015 by Nob Hill Publishing, LLC 1 General Mole Balance V R j Q 0 c j0 Q 1 c j1 Conservation of mass rate of accumulation of component j = + { rate
Nonlinear and/or Non-normal Filtering. Jesús Fernández-Villaverde University of Pennsylvania
 Nonlinear and/or Non-normal Filtering Jesús Fernández-Villaverde University of Pennsylvania 1 Motivation Nonlinear and/or non-gaussian filtering, smoothing, and forecasting (NLGF) problems are pervasive
Nonlinear and/or Non-normal Filtering Jesús Fernández-Villaverde University of Pennsylvania 1 Motivation Nonlinear and/or non-gaussian filtering, smoothing, and forecasting (NLGF) problems are pervasive
CONTROL SYSTEMS, ROBOTICS AND AUTOMATION Vol. XI Stochastic Stability - H.J. Kushner
 STOCHASTIC STABILITY H.J. Kushner Applied Mathematics, Brown University, Providence, RI, USA. Keywords: stability, stochastic stability, random perturbations, Markov systems, robustness, perturbed systems,
STOCHASTIC STABILITY H.J. Kushner Applied Mathematics, Brown University, Providence, RI, USA. Keywords: stability, stochastic stability, random perturbations, Markov systems, robustness, perturbed systems,
Learning Model Predictive Control for Iterative Tasks: A Computationally Efficient Approach for Linear System
 Learning Model Predictive Control for Iterative Tasks: A Computationally Efficient Approach for Linear System Ugo Rosolia Francesco Borrelli University of California at Berkeley, Berkeley, CA 94701, USA
Learning Model Predictive Control for Iterative Tasks: A Computationally Efficient Approach for Linear System Ugo Rosolia Francesco Borrelli University of California at Berkeley, Berkeley, CA 94701, USA
TIME SERIES ANALYSIS. Forecasting and Control. Wiley. Fifth Edition GWILYM M. JENKINS GEORGE E. P. BOX GREGORY C. REINSEL GRETA M.
 TIME SERIES ANALYSIS Forecasting and Control Fifth Edition GEORGE E. P. BOX GWILYM M. JENKINS GREGORY C. REINSEL GRETA M. LJUNG Wiley CONTENTS PREFACE TO THE FIFTH EDITION PREFACE TO THE FOURTH EDITION
TIME SERIES ANALYSIS Forecasting and Control Fifth Edition GEORGE E. P. BOX GWILYM M. JENKINS GREGORY C. REINSEL GRETA M. LJUNG Wiley CONTENTS PREFACE TO THE FIFTH EDITION PREFACE TO THE FOURTH EDITION
Intelligent Systems Statistical Machine Learning
 Intelligent Systems Statistical Machine Learning Carsten Rother, Dmitrij Schlesinger WS2015/2016, Our model and tasks The model: two variables are usually present: - the first one is typically discrete
Intelligent Systems Statistical Machine Learning Carsten Rother, Dmitrij Schlesinger WS2015/2016, Our model and tasks The model: two variables are usually present: - the first one is typically discrete
Sequential Monte Carlo Methods for Bayesian Computation
 Sequential Monte Carlo Methods for Bayesian Computation A. Doucet Kyoto Sept. 2012 A. Doucet (MLSS Sept. 2012) Sept. 2012 1 / 136 Motivating Example 1: Generic Bayesian Model Let X be a vector parameter
Sequential Monte Carlo Methods for Bayesian Computation A. Doucet Kyoto Sept. 2012 A. Doucet (MLSS Sept. 2012) Sept. 2012 1 / 136 Motivating Example 1: Generic Bayesian Model Let X be a vector parameter
Optimal Control. McGill COMP 765 Oct 3 rd, 2017
 Optimal Control McGill COMP 765 Oct 3 rd, 2017 Classical Control Quiz Question 1: Can a PID controller be used to balance an inverted pendulum: A) That starts upright? B) That must be swung-up (perhaps
Optimal Control McGill COMP 765 Oct 3 rd, 2017 Classical Control Quiz Question 1: Can a PID controller be used to balance an inverted pendulum: A) That starts upright? B) That must be swung-up (perhaps
Nonlinear Optimization for Optimal Control Part 2. Pieter Abbeel UC Berkeley EECS. From linear to nonlinear Model-predictive control (MPC) POMDPs
 Nonlinear Optimization for Optimal Control Part 2 Pieter Abbeel UC Berkeley EECS Outline From linear to nonlinear Model-predictive control (MPC) POMDPs Page 1! From Linear to Nonlinear We know how to solve
Nonlinear Optimization for Optimal Control Part 2 Pieter Abbeel UC Berkeley EECS Outline From linear to nonlinear Model-predictive control (MPC) POMDPs Page 1! From Linear to Nonlinear We know how to solve
Preface Acknowledgments
 Contents Preface Acknowledgments xi xix Chapter 1: Brief review of static optimization methods 1 1.1. Introduction: Significance of Mathematical Models 1 1.2. Unconstrained Problems 4 1.3. Equality Constraints
Contents Preface Acknowledgments xi xix Chapter 1: Brief review of static optimization methods 1 1.1. Introduction: Significance of Mathematical Models 1 1.2. Unconstrained Problems 4 1.3. Equality Constraints
Introduction to Computational Stochastic Differential Equations
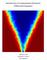 Introduction to Computational Stochastic Differential Equations Gabriel J. Lord Catherine E. Powell Tony Shardlow Preface Techniques for solving many of the differential equations traditionally used by
Introduction to Computational Stochastic Differential Equations Gabriel J. Lord Catherine E. Powell Tony Shardlow Preface Techniques for solving many of the differential equations traditionally used by
Run-to-Run MPC Tuning via Gradient Descent
 Ian David Lockhart Bogle and Michael Fairweather (Editors), Proceedings of the nd European Symposium on Computer Aided Process Engineering, 7 - June, London. c Elsevier B.V. All rights reserved. Run-to-Run
Ian David Lockhart Bogle and Michael Fairweather (Editors), Proceedings of the nd European Symposium on Computer Aided Process Engineering, 7 - June, London. c Elsevier B.V. All rights reserved. Run-to-Run
CHEMICAL ENGINEERING (CHE)
 Chemical Engineering (CHE) 1 CHEMICAL ENGINEERING (CHE) CHE 2033 Introduction to Chemical Process Engineering Prerequisites: CHEM 1515 and ENSC 2213 Description: Concurrent enrollment in MATH 2233 or 3263,
Chemical Engineering (CHE) 1 CHEMICAL ENGINEERING (CHE) CHE 2033 Introduction to Chemical Process Engineering Prerequisites: CHEM 1515 and ENSC 2213 Description: Concurrent enrollment in MATH 2233 or 3263,
Introduction. Chapter 1
 Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
Karl-Rudolf Koch Introduction to Bayesian Statistics Second Edition
 Karl-Rudolf Koch Introduction to Bayesian Statistics Second Edition Karl-Rudolf Koch Introduction to Bayesian Statistics Second, updated and enlarged Edition With 17 Figures Professor Dr.-Ing., Dr.-Ing.
Karl-Rudolf Koch Introduction to Bayesian Statistics Second Edition Karl-Rudolf Koch Introduction to Bayesian Statistics Second, updated and enlarged Edition With 17 Figures Professor Dr.-Ing., Dr.-Ing.
Estimation for state space models: quasi-likelihood and asymptotic quasi-likelihood approaches
 University of Wollongong Research Online University of Wollongong Thesis Collection 1954-2016 University of Wollongong Thesis Collections 2008 Estimation for state space models: quasi-likelihood and asymptotic
University of Wollongong Research Online University of Wollongong Thesis Collection 1954-2016 University of Wollongong Thesis Collections 2008 Estimation for state space models: quasi-likelihood and asymptotic
Computational methods for continuous time Markov chains with applications to biological processes
 Computational methods for continuous time Markov chains with applications to biological processes David F. Anderson anderson@math.wisc.edu Department of Mathematics University of Wisconsin - Madison Penn.
Computational methods for continuous time Markov chains with applications to biological processes David F. Anderson anderson@math.wisc.edu Department of Mathematics University of Wisconsin - Madison Penn.
Stochastic Processes. Theory for Applications. Robert G. Gallager CAMBRIDGE UNIVERSITY PRESS
 Stochastic Processes Theory for Applications Robert G. Gallager CAMBRIDGE UNIVERSITY PRESS Contents Preface page xv Swgg&sfzoMj ybr zmjfr%cforj owf fmdy xix Acknowledgements xxi 1 Introduction and review
Stochastic Processes Theory for Applications Robert G. Gallager CAMBRIDGE UNIVERSITY PRESS Contents Preface page xv Swgg&sfzoMj ybr zmjfr%cforj owf fmdy xix Acknowledgements xxi 1 Introduction and review
WIDE AREA CONTROL THROUGH AGGREGATION OF POWER SYSTEMS
 WIDE AREA CONTROL THROUGH AGGREGATION OF POWER SYSTEMS Arash Vahidnia B.Sc, M.Sc in Electrical Engineering A Thesis submitted in partial fulfilment of the requirements for the degree of Doctor of Philosophy
WIDE AREA CONTROL THROUGH AGGREGATION OF POWER SYSTEMS Arash Vahidnia B.Sc, M.Sc in Electrical Engineering A Thesis submitted in partial fulfilment of the requirements for the degree of Doctor of Philosophy
in this web service Cambridge University Press
 BROWNIAN RATCHETS Illustrating the development of Brownian ratchets, from their foundations, to their role in the description of life at the molecular scale and in the design of artificial nano-machinery,
BROWNIAN RATCHETS Illustrating the development of Brownian ratchets, from their foundations, to their role in the description of life at the molecular scale and in the design of artificial nano-machinery,
CONTENTS. Preface List of Symbols and Notation
 CONTENTS Preface List of Symbols and Notation xi xv 1 Introduction and Review 1 1.1 Deterministic and Stochastic Models 1 1.2 What is a Stochastic Process? 5 1.3 Monte Carlo Simulation 10 1.4 Conditional
CONTENTS Preface List of Symbols and Notation xi xv 1 Introduction and Review 1 1.1 Deterministic and Stochastic Models 1 1.2 What is a Stochastic Process? 5 1.3 Monte Carlo Simulation 10 1.4 Conditional
INTRODUCTION TO PATTERN RECOGNITION
 INTRODUCTION TO PATTERN RECOGNITION INSTRUCTOR: WEI DING 1 Pattern Recognition Automatic discovery of regularities in data through the use of computer algorithms With the use of these regularities to take
INTRODUCTION TO PATTERN RECOGNITION INSTRUCTOR: WEI DING 1 Pattern Recognition Automatic discovery of regularities in data through the use of computer algorithms With the use of these regularities to take
Modelling Under Risk and Uncertainty
 Modelling Under Risk and Uncertainty An Introduction to Statistical, Phenomenological and Computational Methods Etienne de Rocquigny Ecole Centrale Paris, Universite Paris-Saclay, France WILEY A John Wiley
Modelling Under Risk and Uncertainty An Introduction to Statistical, Phenomenological and Computational Methods Etienne de Rocquigny Ecole Centrale Paris, Universite Paris-Saclay, France WILEY A John Wiley
Parameter Estimation in Stochastic Chemical Kinetic Models. Rishi Srivastava. A dissertation submitted in partial fulfillment of
 Parameter Estimation in Stochastic Chemical Kinetic Models by Rishi Srivastava A dissertation submitted in partial fulfillment of the requirement for the degree of Doctor of Philosophy (Chemical Engineering)
Parameter Estimation in Stochastic Chemical Kinetic Models by Rishi Srivastava A dissertation submitted in partial fulfillment of the requirement for the degree of Doctor of Philosophy (Chemical Engineering)
Stability of Ensemble Kalman Filters
 Stability of Ensemble Kalman Filters Idrissa S. Amour, Zubeda Mussa, Alexander Bibov, Antti Solonen, John Bardsley, Heikki Haario and Tuomo Kauranne Lappeenranta University of Technology University of
Stability of Ensemble Kalman Filters Idrissa S. Amour, Zubeda Mussa, Alexander Bibov, Antti Solonen, John Bardsley, Heikki Haario and Tuomo Kauranne Lappeenranta University of Technology University of
Handout 1: Introduction to Dynamic Programming. 1 Dynamic Programming: Introduction and Examples
 SEEM 3470: Dynamic Optimization and Applications 2013 14 Second Term Handout 1: Introduction to Dynamic Programming Instructor: Shiqian Ma January 6, 2014 Suggested Reading: Sections 1.1 1.5 of Chapter
SEEM 3470: Dynamic Optimization and Applications 2013 14 Second Term Handout 1: Introduction to Dynamic Programming Instructor: Shiqian Ma January 6, 2014 Suggested Reading: Sections 1.1 1.5 of Chapter
Contents. Set Theory. Functions and its Applications CHAPTER 1 CHAPTER 2. Preface... (v)
 (vii) Preface... (v) CHAPTER 1 Set Theory Definition of Set... 1 Roster, Tabular or Enumeration Form... 1 Set builder Form... 2 Union of Set... 5 Intersection of Sets... 9 Distributive Laws of Unions and
(vii) Preface... (v) CHAPTER 1 Set Theory Definition of Set... 1 Roster, Tabular or Enumeration Form... 1 Set builder Form... 2 Union of Set... 5 Intersection of Sets... 9 Distributive Laws of Unions and
CHEMISTRY. Overview. Choice of topic
 CHEMISTRY Overview An extended essay in chemistry provides students with an opportunity to investigate a particular aspect of the materials of our environment. Such extended essays must be characterized
CHEMISTRY Overview An extended essay in chemistry provides students with an opportunity to investigate a particular aspect of the materials of our environment. Such extended essays must be characterized
Basic modeling approaches for biological systems. Mahesh Bule
 Basic modeling approaches for biological systems Mahesh Bule The hierarchy of life from atoms to living organisms Modeling biological processes often requires accounting for action and feedback involving
Basic modeling approaches for biological systems Mahesh Bule The hierarchy of life from atoms to living organisms Modeling biological processes often requires accounting for action and feedback involving
Chemical Reactions and Chemical Reactors
 Chemical Reactions and Chemical Reactors George W. Roberts North Carolina State University Department of Chemical and Biomolecular Engineering WILEY John Wiley & Sons, Inc. x Contents 1. Reactions and
Chemical Reactions and Chemical Reactors George W. Roberts North Carolina State University Department of Chemical and Biomolecular Engineering WILEY John Wiley & Sons, Inc. x Contents 1. Reactions and
State Estimation of Linear and Nonlinear Dynamic Systems
 State Estimation of Linear and Nonlinear Dynamic Systems Part II: Observability and Stability James B. Rawlings and Fernando V. Lima Department of Chemical and Biological Engineering University of Wisconsin
State Estimation of Linear and Nonlinear Dynamic Systems Part II: Observability and Stability James B. Rawlings and Fernando V. Lima Department of Chemical and Biological Engineering University of Wisconsin
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 7 Approximate
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 7 Approximate
STABILITY CONDITIONS AND OBSERVER DESIGN FOR A CONTINUOUS CRYSTALLIZER
 STABILITY CONDITIONS AND OBSERVER DESIGN FOR A CONTINUOUS CRYSTALLIZER Juan Du and B. Erik Ydstie* Carnegie Mellon University Pittsburgh, PA 15213 Abstract The population balance equation is used to describe
STABILITY CONDITIONS AND OBSERVER DESIGN FOR A CONTINUOUS CRYSTALLIZER Juan Du and B. Erik Ydstie* Carnegie Mellon University Pittsburgh, PA 15213 Abstract The population balance equation is used to describe
Optimal Decentralized Control of Coupled Subsystems With Control Sharing
 IEEE TRANSACTIONS ON AUTOMATIC CONTROL, VOL. 58, NO. 9, SEPTEMBER 2013 2377 Optimal Decentralized Control of Coupled Subsystems With Control Sharing Aditya Mahajan, Member, IEEE Abstract Subsystems that
IEEE TRANSACTIONS ON AUTOMATIC CONTROL, VOL. 58, NO. 9, SEPTEMBER 2013 2377 Optimal Decentralized Control of Coupled Subsystems With Control Sharing Aditya Mahajan, Member, IEEE Abstract Subsystems that
Completion Time of Fuzzy GERT-type Networks with Loops
 Completion Time of Fuzzy GERT-type Networks with Loops Sina Ghaffari 1, Seyed Saeid Hashemin 2 1 Department of Industrial Engineering, Ardabil Branch, Islamic Azad university, Ardabil, Iran 2 Department
Completion Time of Fuzzy GERT-type Networks with Loops Sina Ghaffari 1, Seyed Saeid Hashemin 2 1 Department of Industrial Engineering, Ardabil Branch, Islamic Azad university, Ardabil, Iran 2 Department
Population Games and Evolutionary Dynamics
 Population Games and Evolutionary Dynamics William H. Sandholm The MIT Press Cambridge, Massachusetts London, England in Brief Series Foreword Preface xvii xix 1 Introduction 1 1 Population Games 2 Population
Population Games and Evolutionary Dynamics William H. Sandholm The MIT Press Cambridge, Massachusetts London, England in Brief Series Foreword Preface xvii xix 1 Introduction 1 1 Population Games 2 Population
PROPERTIES OF POLYMERS
 PROPERTIES OF POLYMERS THEIR CORRELATION WITH CHEMICAL STRUCTURE; THEIR NUMERICAL ESTIMATION AND PREDICTION FROM ADDITIVE GROUP CONTRIBUTIONS Third, completely revised edition By D.W. VÄN KREVELEN Professor-Emeritus,
PROPERTIES OF POLYMERS THEIR CORRELATION WITH CHEMICAL STRUCTURE; THEIR NUMERICAL ESTIMATION AND PREDICTION FROM ADDITIVE GROUP CONTRIBUTIONS Third, completely revised edition By D.W. VÄN KREVELEN Professor-Emeritus,
Basics of Uncertainty Analysis
 Basics of Uncertainty Analysis Chapter Six Basics of Uncertainty Analysis 6.1 Introduction As shown in Fig. 6.1, analysis models are used to predict the performances or behaviors of a product under design.
Basics of Uncertainty Analysis Chapter Six Basics of Uncertainty Analysis 6.1 Introduction As shown in Fig. 6.1, analysis models are used to predict the performances or behaviors of a product under design.
Geometry in a Fréchet Context: A Projective Limit Approach
 Geometry in a Fréchet Context: A Projective Limit Approach Geometry in a Fréchet Context: A Projective Limit Approach by C.T.J. Dodson University of Manchester, Manchester, UK George Galanis Hellenic
Geometry in a Fréchet Context: A Projective Limit Approach Geometry in a Fréchet Context: A Projective Limit Approach by C.T.J. Dodson University of Manchester, Manchester, UK George Galanis Hellenic
Bayesian Methods for Machine Learning
 Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
