Evolution equation of phenotype distribution: General formulation and application to error catastrophe
|
|
|
- Blake Owens
- 5 years ago
- Views:
Transcription
1 Evolution equation of phenotype distribution: General formulation and application to error catastrophe Katsuhiko Sato 1, * and Kunihiko Kaneko 1,2, 1 Complex Systems Biology Project, ERATO, JST, Komaba, Meguro-ku, Tokyo , Japan 2 Department of Pure and Applied Sciences, University of Tokyo, Komaba, Meguro-ku, Tokyo , Japan Received 7 November 2006; published 19 June 2007 An equation describing the evolution of phenotypic distribution is derived using methods developed in statistical physics. The equation is solved by using the singular perturbation method, and assuming that the number of bases in the genetic sequence is large. Applying the equation to the mutation-selection model by Eigen provides the critical mutation rate for the error catastrophe. Phenotypic fluctuation of clones individuals sharing the same gene is introduced into this evolution equation. With this formalism, it is found that the critical mutation rate is sometimes increased by the phenotypic fluctuations, i.e., noise can enhance robustness of a fitted state to mutation. Our formalism is systematic and general, while approximations to derive more tractable evolution equations are also discussed. DOI: /PhysRevE PACS numbers: Kg, A, r I. INTRODUCTION For decades, quantitative studies of evolution in laboratories have used bacteria and other microorganisms 1 3. Changes in phenotypes, such as enzyme activity and gene expressions introduced by mutations in genes, are measured along with the changes in their population distribution in phenotypes 4 7. Following such experimental advances, it is important to analyze the evolution equation of population distribution of concerned genotypes and phenotypes. In general, fitness for reproduction is given by a phenotype, not directly by a genetic sequence. Here, we consider evolution in a fixed environment, so that the fitness is given as a fixed function of the phenotype. A phenotype is determined by mapping a genetic sequence. This phenotype is typically represented by a continuous scalar variable, such as enzyme activity, protein abundances, and body size. For studying the evolution of a phenotype, it is essential to establish a description of the distribution function for a continuous phenotypic variable, where the fitness for survival, given as a function of such a continuous variable, determines population distribution changes over generations. However, since a gene is originally encoded on a base sequence such as AGCTGCTT in DNA, it is represented by a symbol sequence of a large number of discrete elements. Mutation in a sequence is not originally represented by a continuous change. Since the fitness is given as a function of phenotype, we need to map base sequences of a large number of elements onto a continuous phenotypic variable x, where the fitness is represented as a function of x, instead of the base sequence itself. A theoretical technique and careful analysis are needed to project a discrete symbol sequence onto a continuous variable. Mutation in a nucleotide sequence is random, and is represented by a stochastic process. Thus, a method of deriving a diffusion equation from a random walk is often applied. *Electronic address: sato@complex.c.u-tokyo.ac.jp Electronic address: kaneko@complex.c.u-tokyo.ac.jp However, the selection process depends on the phenotype. If a phenotype is given as a function of a sequence, the fitness is represented by a continuous variable mapped from a base sequence. Since the population changes through the selection of fitness, the distribution of the phenotype changes accordingly. If the mapping to the phenotype variable is represented properly, the evolutionary process will be described by the dynamics of the distribution of the variable, akin to a Fokker-Planck equation. In fact, there have been several approaches to representing the gene with a continuous variable. Kimura 8 developed the population distribution of a continuous fitness. Also, for certain conditions, a Fokker-Planck type equation has been mathematically analyzed 9 11 and applied to several biological examples Generalizing these studies provides a systematic derivation of an equation describing the evolution of the distribution of the phenotypic variable. We adopt selection-mutation models describing the molecular biological evolution discussed by Eigen 16, Kauffman 17, and others, and take a continuum limit assuming that the number of bases N in the genetic sequence is large, and derive the evolution equation systematically in terms of the expansion of 1/N. In particular, we refer to Eigen s equation 16, originally introduced for the evolution of RNA, where the fitness is given as a function of a sequence. Mutation into a sequence is formulated by a master equation, which is transformed to a diffusionlike equation. With this representation, population dynamics over a large number of species is reduced to one simple integro-differential equation with one variable. Although the equation obtained is a nonlinear equation for the distribution, we can adopt techniques developed in the analysis of the linear Fokker-Planck equation, such as the eigenfunction expansion and perturbation methods. So far, we have assumed a fixed, unique mapping from a genotype to a phenotype. However, there are phenotypic fluctuations in individuals sharing the same genotype, which has recently been measured quantitatively as a stochastic gene expression Relevance of such fluctuations to evolution has also been discussed In this case, map /2007/756/ The American Physical Society
2 KATSUHIKO SATO AND KUNIHIKO KANEKO ping from a gene gives the average of the phenotype, but phenotype of each individual fluctuates around the average. In the second part of the present paper, we introduce this isogenic phenotypic fluctuation into our evolution equation. Indeed, our framework of Fokker-Planck type equations is fitted to include such fluctuations, so that one can discuss the effect of isogenic phenotypic fluctuations on the evolution. The outline of the present paper is as follows: We first establish a sequence model in Sec. II. For deriving the evolution equation from the sequence model, we postulate the assumption that the transition probability of phenotype values is uniquely determined by the original phenotype value. The assumption may appear too demanding at first sight, but we show that it is not unnatural from the viewpoint of evolutionary biology. In fact, most models studied so far satisfy this postulate. With this assumption, we derive a Fokker- Planck type equation of phenotypic distribution using the Kramers-Moyal expansion method from statistical physics 29,30, and following the standard technique in the Markov chain process We discuss the validity of this expansion method to derive the equation, also from a biological point of view. As an example of the application of our formulation, we study the Eigen s model in Sec. III, and estimate the critical mutation rates at which error catastrophe occurs, using a singular perturbation method. In Sec. V, we discuss the range of the applicability of our method and possible extensions to it. Following the formulation and application of the Fokker- Planck type equations for evolution, we study the effect of isogenic phenotypic fluctuations. While fluctuation in the mapping from a genotype to phenotype modifies the fitness function in the equation, our formulation itself is applicable. We will also discuss how this fluctuation changes the conditions for the error catastrophe, by adopting Eigen s model. For concluding the paper, we discuss generality of our formulation, and the relevance of isogenic phenotypic fluctuation to evolution. II. DERIVATION OF EVOLUTION EQUATION We consider a population of individuals having a haploid genotype, which is encoded on a sequence consisting of N sites consider, for example, DNA or RNA. The gene is represented by this symbol sequence, which is assigned from a set of numbers, such as 1,1. This set of numbers is denoted by S. By denoting the state value of the ith site by s i S, the configuration of the sequence is represented by the ordered set s=s 1,...,s N. We assume that a scalar phenotype variable x is assigned for each sequence s. This mapping from sequence to phenotype is given as function xs. Examples of the phenotype include the activity of some enzyme protein, infection rate of bacteria virus, and replication rate of RNA. In general, the function xs is a degenerate function, i.e., many different sequences are mapped onto the same phenotypic value x. Each sequence is reproduced with rate A, which is assumed to depend only on the phenotypic value x, asax; this assumption may be justified by choosing the phenotypic value x to relate to the replication. For example, if a protein is involved in the metabolism of a replicating cell, its activity may affect the replication rate of the cell and of the protein itself. In the replication of the sequence, mutation generally occurs; for simplicity, we consider only the substitution of si. With a given constant mutation rate over all sites in the sequence, the state s i of the daughter sequence is changed from s i of the mother sequence, where the value s i is assigned from the members of the set S with an equal probability. We call this type of mutation symmetric mutation 34. The mutation is represented by the transition probability Qs s, from the mother s to the daughter s sequence. The probability Q is uniquely determined from the sequence s, the mutation rate, and the number of members of S. The setup so far is essentially the same as adopted by Eigen et al. 16, where the fitness is given as a function of the RNA sequence or DNA sequence of virus. Now we assume that the transition probability depends only on the phenotypic value x, i.e., the function Q can be written in terms of a probability function W, which depends only on x, Wx x, as Qs s = W xs x. 1 ssx=xs This assumption may appear too demanding. However, most models of sequence evolution somehow adopt this assumption. For example, in Eigen s model, fitness is given as a function of the Hamming distance from a given optimal sequence. By assigning a phenotype x as the Hamming distance, the above condition is satisfied his will be discussed later. In Kauffman s NK model, if we set N1, K1, and K/N1, this assumption is also satisfied see Appendix A. For the RNA secondary structure model 35, this assumption seems to hold approximately, from statistical estimates through numerical simulations. Some simulations on a cell model with catalytic reaction networks 22,36 also support the assumption on the transition probability, in which the phenotype of a cell is given by the concentration of a specific chemical, and the mutation changes the path in the network. In fact, a similar assumption has been made in evolution theory with a gene substitution process 37,38. The validity of this assumption in experiments has to be confirmed. Consider a selection experiment to enhance some function through mutation, such as the evolution of a certain protein to enhance its activity 7. In this case, the assumption means that the activity distribution over the mutant proteins is statistically similar as long as they have the same activity, even though their mother protein sequences are different. With the above setup, we consider the population of these sequences and their dynamics, allowing for overlap between generations, by taking a continuous-time model 34. Wedo not consider the death rate of the sequence explicitly since its consideration introduces only an additional term, as will be shown later. The time-evolution equation of the probability distribution Pˆ s,t of the sequence s is given by
3 EVOLUTION EQUATION OF PHENOTYPE DISTRIBUTION: Pˆ s,t = ĀPˆ s,t + A xs Qs spˆ s,t, s 2 as specified by Eigen 16. Here the quantity Ā is the average fitness of the population at time t, defined by Ā = s A(xs)Pˆ s,t and Q is the transition probability satisfying s Qs s=1 for any s. According to the assumption 1, Eq. 2 is transformed into the equation for Px,t, which is the probability distribution of the sequences having the phenotypic value x, defined by Px,t= ssx=xs Pˆ s,t. The equation is given by = Ā + AxWx xpx,t, 3 x where the function W satisfies Wx x = 1 for any x, 4 x as shown. Since N is sufficiently large, the variable x is regarded as a continuous variable. By using the Kramers-Moyal expansion 29,30,39, with the help of property 4, we obtain = Ax Ā 1 n + n n=1 n! x nm nxax, 5 where m n x is the nth moment about the value x, defined by m n x=x x n Wx xdx. Let us discuss the conditions for the convergence of expansion 5, without mathematical rigor. For convergence, it is natural to assume that the function Wx x decays sufficiently fast as x gets far from x, by the definition of the moment. Here, the transition Wx x is a result of n point mutants of the original sequence s for n=0,1,2,...,n. Accordingly, we introduce a set of quantities, w n (xs x), as the fitness distribution of n point mutants of the original sequence s. Naturally, w 0 (xs x)=(xs x), which does not contribute to the nth moment m n n1. Next, we introduce the probability p n that a daughter sequence is an n point mutant n=0,1,2,...,n from her mother sequence, which are determined only by the mutation rate and the sequence length N. Indeed, p n s form a binomial distribution, characterized by and N. In terms of the quantities w n and p n, we are able to write down the transition probability W as N W xs x = p n w n xs x. n=0 Now, we discuss if W(xs x) decays sufficiently fast with xs x. First, we note that the width of the domain, in 6 which w n (xs x) is not close to zero, increases with n since n-point mutants involve an increasing number of changes in the phenotype with larger values of n. Hence, in order for the series on the right-hand side in Eq. 5 to converge, at least the single-point-mutant transition w 1 (xs x) has to decay sufficiently fast with xs x. In other words, the phenotypic value of a single-point mutant s of the mother sequence s must not vary much from that of the original sequence, i.e., xs xs should not be large continuity condition. In general, the domain x xs, in which w n (xs x) 0, increases with n. On the other hand, the term p n decreases with n and with the power of n. Hence, as long as the mutation rate is not large, the contribution of w n to W is expected to decay with n. Thus, if the continuity condition with regard to a single-point mutant and a sufficiently low mutation rate are satisfied, the requirement on W(xs x) should be fulfilled. Hence the convergence of the expansion is expected. Following the argument, we further restrict our study to the case with a small mutation rate 55 such that N 1 holds. The transition probability W in Eq. 6 is written as W xs x 1 N xs x + Nw 1 xs x, 7 where we have used the property that p n s form the binomial distribution characterized by and N. Introducing a new parameter, =N, that gives the average of the number of changed sites at a single-point mutant, and using the transition probability 7, we obtain = Ax Ā 1 n + n n=1 n! x nm 1 n xax, where m n 1 x is the nth moment of w 1 x x, i.e., m n 1 x =x x n w 1 x xdx. When we stop the expansion at the second order, as is often adopted in statistical physics, we obtain = Ax Ā 8 + x m 1 1 x x m 2 1 xax. 9 Equations 8 and 9 are basic equations for the evolution of the distribution function. Equation 9 is an approximation of Eq. 8; however, it is often more tractable, with the help of techniques developed for solving the Fokker-Planck equation see Appendix B and 40, while there is no established standard method for solving Eq. 8. At the boundary condition we naturally impose that there is no probability flux, which is given by
4 KATSUHIKO SATO AND KUNIHIKO KANEKO 1 n n 1 n=1 n! x n 1m 1 n xaxx=x1,x2 in the case of Eq. 8 and m 1 1 x x m 2 1 =0 xaxx=x 1,x 2 =0, in the case of Eq. 9, where x 1 and x 2 are the positions of the left and right boundaries, respectively. Next, as an example of the application of our formula, we derive the evolution equation for Eigen s model, and estimate the error threshold, with the help of a singular perturbation theory. Through this application, we can see the validity of Eq. 9 as an approximation of Eq. 8. Two additional remarks: First, introduction of the death of individuals is rather straightforward. By including the death rate Dx in the evolution equation, the first term in Eq. 8 or Eq. 9 is replaced by (Ax Dx Ā D )Px,t, where D Dxdx. Second, instead of deriving each term in Eq. 9 from microscopic models, it may be possible to adopt it as a phenomenological equation, with parameters or functions to be determined heuristically from experiments. III. APPLICATION OF ERROR THRESHOLD IN EIGEN MODEL In the Eigen model 16, the set S of the site state values is given by 1,1, and the fitness replication rate of the sequence is given as a function of its Hamming distance from the target sequence 1,,1, i.e., the fitness of an individual sequence is given as a function of the number n of the sites of the sequence having value 1. Hence it is appropriate to define a phenotypic value x in the Eigen model as a monotonic function of the number n; we determine it as x=2n N/N, in the range 1,1. Accordingly, the replication rate A of the sequence can be written as a function of x, i.e., Ax; it is natural to postulate that A is a non-negative and bounded function over the whole domain. If the sequence length N is sufficiently large, the phenotypic variable x can be regarded as a continuous variable, since the step size of x x=2/n approaches 0 as N goes to infinity. In order to derive the evolution equation of form 8 corresponding to the Eigen model, we only need to know the function w 1 in that model. Recall that in our formulation the mutation rate is assumed to be so small that only a singlepoint mutation is considered. Due to the assumption of the symmetric mutation, this distribution function is obtained as w 1 x x x=1+x/2, w 1 x x+x=1 x/2, and w 1 x x=0 for any other x. Accordingly, the nth moment is given by m n 1 x=1+x/2 x n +1 x/2x n. Now, we obtain 1 = Ax Ā + n=1 n! + 1 x 2 2 N n 1+x xn 2 n 2 N nax, 12 where =N, the mutation rate per sequence. When we ignore the moment terms higher than the second order, we have = Ax Ā N xx 1 N xax. 13 In fact, if we focus on a change near x0 o be specific xo1/ N, the truncation of the expansion up to the second order is validated (or equivalently, if we define x=2n N/ N instead of 2n N/N, and expand Eq. 3 by 1/ N instead of 1/N, terms higher than the second order are negligible, as is also discussed in 9. However, in this case, the validity is restricted to xo1 i.e., n N/2O1, which means xo1/ N in the original variable). Now we solve Eq. 13 with a standard singular perturbation method see Appendix B, and then return to Eq. 12. According to the analysis in Appendix B, the stationary solution of the equation of form 13 is given by the eigenfunction corresponding to the largest eigenvalue of the linear operator L defined by L=Ax+2/xx+/xAx with =1/N. Now we consider the eigenvalue problem AxPx +2 xx + xaxpx = Px, 14 where Px0, with to be determined. Since is very small because N is sufficiently large, a singular perturbation method, the WKB approximation 41, is applied. Let us put Px = e 1/ x0 x R,xdx, 15 where x 0 is some constant and R is a function of and x, which is expanded with respect to as R,x = R 0 x + R 1 x + 2 R 2 x Retaining only the zeroth order terms in in Eq. 14, weget Ax +2xR 0 x + R 0 2 xax =, 17 which is formally solved for R 0 as R ± 0 x= x±gx/2 where gx=x 2 +2//Ax 1. Hence the general solution of Eq. 14 up to the zeroth order in is given by Px=e 1/ x + x R0 0 xdx +e 1/ x x R0 0 xdx with and constants to be determined. Now, recall the boundary conditions 11; P has to take the two branches in R 0 as Px=e 1/ x + x R0 b xdx for xx b and Px=e 1/ x x R0 b xdx for xx b, where x b is defined as the value at which gx has the minimum value. Next, from the continuity of P at x b, = follows, while from the con
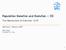
5 EVOLUTION EQUATION OF PHENOTYPE DISTRIBUTION: f(x) A(x) λ x p x b x FIG. 1. Examples of profiles of the evaluation function f for three values of. The gray, thin, and broken curves give the profiles of f for =0.31, =0.386, and =0.49, respectively, where f is defined by fx=ax1 1 1 x 2 and A is given by Ax=1+0.2x 0.25x x+0.1x 0.75; the profile of A is indicated by the black curve. This illustrates determination of x b and x p ; x b is given by the position where f takes a maximum, while x p is given as the position where the line y= fx b crosses the curve of A. For 0.386, fx has a maximum value at x=x b, and thus the critical mutation rate for the error threshold is estimated to be * = tinuity of P/x at x b, the function g has to vanish at x=x b. This requirement gx b =0 determines the value of the unknown parameter as = Ax b 1 2 x b2. 18 From the form of function P, Eq. 15, we find that P has its peak at the point x=x p, where R 0 x vanishes, i.e., Ax p = holds. Then, Px approaches x x p in the limit +0. These results are consistent with the requirement that the mean replication rate in the steady state be equal to the largest eigenvalue of the system see Appendix B. The stationary solution of Eq. 12 is obtained by following the same procedure of singular perturbation. Consider the eigenvalue problem n 1+x n! xn 2 2n + 1 x 2 2nAxPx 1 AxPx + n=1 = Px. 19 By putting Px=e 1/ x0 x R xdx 0 and taking only the zeroth order terms in, we obtain Ax + 1+x 2 e2r 0 x x 2 e 2R 0 x 1Ax =, which gives R 0 ± x = 1 2 log Ax ± ĝx 1+x with ĝx=1+1//ax x 2. By defining again the value x=x b at which ĝx takes the minimum, P is represented as Px=e 1/ x + x R0 b xdx for x x b and Px=e 1/ x x R0 b xdx for xx b. The continuity of P/x at x=x b requires ĝx b =0, which determines the value of as = Ax b xb Again, Px=x x p, in the limit +0, with x p given by the condition Ax p =. When x b 1, the form 20 approaches Eq. 18 asymptotically. This implies that the time evolution equation 8, if restricted to x1, is accurately approximated by Eq. 9 that keeps the terms only up to the second moment. Let us estimate the threshold mutation rate for error catastrophe. This error threshold is defined as the critical mutation rate * at which the peak position x p of the stationary distribution drops from x p 0tox p =0, with an increase of. We use the following procedure to obtain the critical value *. First consider an evaluation function whose form corresponds to that of eigenvalue 20 as fx = Ax1 1 1 x 2, 21 and find the position at which the function fx takes the maximum value. This procedure is equivalent to obtaining x b in the above analysis, since the relation fx= ( 2 A 2 x/ Ax x 2 )ĝx and the requirement that ĝx b =0 and dĝx/dx x=xb =0 lead to dfx/dx x=xb =0. Obviously, x b is given as a function of ; thus we denote it by x b. The position x b determines the position x p of the stationary distribution through the relation Ax p == fx b as in the above analysis. If A is flat around x=0 and higher than there in some positive region x0, x p discontinuously changes from x p 0tox p =0 at some critical mutation rate *, when increases from zero. A schematic illustration of this transition is given in Fig. 1. As a simple example of this estimate of error threshold, let us consider the case Ax =1+A 0 x x 0, 22 with A 0 0 and 0x 0 1, and as the Heaviside step function, defined as x=0 for x0 and x=1 for x0. This form is chosen for simplicity, but it should be noted that the threshold-type behavior of growth is rather common in a biological system. According to the procedure given above,
6 KATSUHIKO SATO AND KUNIHIKO KANEKO the critical mutation rate is straightforwardly obtained as * =A 0 /1+A x0 ; for *, x p =x 0 and for *, x p =0. Remark. Statistical mechanics approach to the mutationselection model has been developed recently 34,42 47.An exact transformation from the sequence model Eigen model 16 into a class of Ising models 43,45 has recently been reported. Hence the sequence model is treated analytically with methods developed in statistical physics. Rigorous estimation of the error threshold for various fitness landscapes 34,46 and relaxation times of species distribution have been obtained 47. In fact, our estimate above agrees with that given by their analysis. Their method is indeed powerful when a microscopic model is prescribed in correspondence with a spin model. However, even if such a microscopic model is not given, our formulation with a Fokker-Planck type equation will be applicable because it only requires estimation of moments in the fitness landscape. Alternatively, by giving a phenomenological model describing the fitness without microscopic process, it is possible to derive the evolution equation of population distribution. Hence our formulation has a broad range of potential applications. IV. CONSIDERATION OF PHENOTYPIC FLUCTUATION In this section, we include the fluctuation in the mapping from genetic sequence to the phenotype into our formula, and examine how it influences the error catastrophe. We first explain the term phenotypic fluctuation briefly, and show that in its presence our formulation 8 remains valid by redefining the function Ax. By applying the formulation, we study how the introduction of the phenotypic fluctuation changes the critical mutation rate * for the error catastrophe. In general, even for individuals with identical gene sequences in a fixed environment, the phenotypic values are distributed. Some examples are the activities of proteins synthesized from the identical DNA 48, the thermal fluctuation of the shapes of RNA molecules with a given identical sequence 49, and the numbers of specific proteins for isogenic bacteria. Indeed, the phenotypic fluctuation of isogenetic biological elements is rather large 19 22,25. Itis also suggested theoretically that the isogenetic fluctuation is larger than the fluctuation induced by genetic mutation 26,27. Next, the phenotype x from each individual with the sequence s is distributed, which is denoted by P phe s,x. We assume that the form of distribution P phe is characterized only in terms of its mean value, i.e., the distributions P phe s having the same mean value X take the same form; it is not unnatural to assume that those with the same mean phenotype value have a similar fluctuation value 56. By representing the mean value of the phenotype x by x s, the distribution P phe is written as P phe s,x= Pˆ phe(x s,x), where Pˆ phe is a function of x and x, which is normalized with respect to x, i.e., satisfying Pˆ phex,xdx=1. In our formulation, the replication rate A of the sequence with the phenotypic value x is given by a function of phenotypic value x, denoted by Ax. The mean replication rate  of the species s is calculated by  x s = Pˆ phe x s,x Axdx. 23 As in the case of Eq. 1, we assume that the transition probability from s to s during the replication is represented only by its mean values x s and x s, i.e., the transition probability function is written as W(x s x s). With this setup, the population dynamics of the whole sequences is represented in terms of the distribution of the mean value x only, so that we can use our formulation 8 even when the phenotypic fluctuation is taken into account; we need only replace the replication rate A in Eq. 8 by the mean replication rate  obtained from Eq. 23. Now, we can study the influence of phenotypic fluctuation on the error threshold by taking the step fitness function Ax of Eq. 22 and including the phenotypic fluctuation as given in Eq. 23. We consider a simple case where the form of Pˆ phe is given by a constant function within a given range we call this the piecewise flat case. Our aim is to illustrate the effect of the phenotypic fluctuation on the error threshold, so we evaluate the critical mutation rate * using the simpler form fx=ax1 /2x 2 from Eq. 18, while the use of the form 21 gives the same qualitative result. With this simpler evaluation function, the critical mutation rate * is given by 0 * = 2A 0 1+A 0 x 0 2, 24 in the case without phenotypic fluctuation. Here we examine if this critical value * 0 increases under isogenic phenotypic fluctuation. We make two further technical assumptions in the following analysis: First, we assume that A 0 in the form 22 is sufficiently small so that the value of critical * is not large, because our formalism is applicable only when is small. Second, we extend the range of x to, for simplicity. This does not cause problems because we have set the range of x 0 to 0, 1. Hence the stationary distribution has its peak around the range 0x1; everywhere outside this range, the distribution vanishes. In general Pˆ phe is distributed around its peak. As a typical case we study the following simple case in which distribution Pˆ phe of the phenotype of the species s is given by = 0 for x x, Pˆ phe F 1 x s,x for x x x +, for x + x, where gives the half-width of the distribution. This is an illustration of a threshold-type dependence often observed in biological systems. The letter F represents the piecewiseflat distribution case. Then,  is calculated by
7 EVOLUTION EQUATION OF PHENOTYPE DISTRIBUTION: A(x) FIG. 2. Example of profiles of the mean fitness functions without phenotypic fluctuation case solid curve, with a constant phenotypic fluctuation over a given range given by Eq. 25 broken curve, where we set Ax=1+0.1x 0.5 and = x = 1 for x x 0,  F x 1+ A 0 2 x x 0 for x 0 x x 0 +, 1+A 0 for x 0 + x. An example of  F x is shown in Fig. 2. The evaluation function f in Sec. III is given by f F x=â F x1 /2x 2. We are interested in whether or not there is a case where *F is greater than * 0 in this situation, so that we will consider only the case where the position x *F b x F b *F is within the range x 0,x 0, because from the profile of  F we can readily find that *F is smaller than * *F 0 when x b x 0. When x 0 /2+A 0 x 0, the position x *F b is within the range x 0,x 0. In that case, x *F b and *F are given by x *F b 2x 0 and *F A 0 /4x 0 to the first order of A 0, respectively. Comparing this *F with * 0 in Eq. 24, we conclude that *F * 0 for 02+ 2/4x0, and *F * 0 for 2+ 2/4x0 x 0. Hence, when the half width of the distribution P phe is within the range (2 + 2/4x0,x 0 ), the critical mutation rate for the error catastrophe threshold is increased. In other words, the isogenic phenotypic fluctuation increases the robustness of the high fitness state against mutation. We also studied the case in which Pˆ phex,x decreases linearly around its peak, i.e., with a triangular form. In this case, the phenotypic fluctuation decreases the critical mutation rate as long as A 0 is small, while it can increase for sufficiently large values of A 0, for a certain range of the values of width of phenotypic fluctuation. On the other hand, for a distribution P phe x,x=n+1/2 n+1 x x n (x x )(x + x) with a positive number n, the increase of critical mutation rate is possible if 0n1 57. We have also studied the case with Gaussian distribution, in which case the increase of the critical mutation rate is not observed. V. DISCUSSION In the present paper, we have presented a general formulation to describe the evolution of phenotype distribution. A partial differential equation describing the temporal evolution of phenotype distribution is presented with a selfconsistently determined growth term. Once a microscopic model is provided, each term in this evolution equation is explicitly determined so that one can derive the evolution of phenotype distribution straightforwardly. This Eq. 8 is obtained as a result of Kramers-Moyal expansion, which includes infinite order of derivatives. However, this expansion is often summed to a single term in the large number limit of base sequences, with the aid of singular perturbation. If the value of a phenotype variable x is much smaller than unity which is the maximal possible value giving rise to the fittest state, the terms higher than the second order can be neglected, so that a Fokker-Planck type equation with a self-consistent growth term is derived. The validity of this truncation is confirmed by putting x=2n N/ N and verifying that the third or higher order moment is negligible compared with the second-order moment. Thus the equation up to its second order, Eq. 9, is relevant to analyzing the initial stage of evolution starting from a low-fitness value. As a starting point for our formalism, we adopted Eq. 2, which is called the coupled mutation-selection equation 50. Although it is a natural and general choice for studying the evolution, a simpler and approximate form may be used if the mutation rate and the selection pressure are sufficiently small. This form given by Pˆ s,t/= ĀPˆ s,t + s Qs spˆ s,t, is called the parallel mutationselection equation 51,52. It approaches the coupled mutation-selection equation 2, in the limits of small mutation rate and selection pressure, as shown in 50. Ifwe start from this approximate, parallel mutation-selection equation, and follow the procedure presented in this paper, we obtain /=Ax Ā+/x m 1 1 x /xm 1 2 x. In general, this equation is more tractable than Eq. 9, as the techniques developed in Fokker-Planck equations are
8 KATSUHIKO SATO AND KUNIHIKO KANEKO straightforwardly applied as discussed in 40, and it is also useful in describing evolution. Setting Ax=x 2 and replacing m 1 1 and m 2 1 with some constants, the equation is reduced to that introduced by Kimura 8; while setting Ax=x, m 1 1 xx, and replacing m 2 1 with some constant derive the equation by Levine 9. Because our formalism is general, these earlier studies are derived by approximating our evolution equation suitably. Besides the generality, another merit of our formulation lies in its use of the phenotype as a variable describing the distribution, rather than the fitness as adopted by Kimura. Whereas the phenotype is an inherent variable directly mapped from the genetic sequence, the fitness is a function of the phenotype and environment, and may be more strongly influenced than the phenotype, by environmental conditions. The evaluation of the transition matrix by mutation in Eq. 8 would be more complicated if we used the fitness as a variable, due to crucial dependence of fitness values on the environmental conditions. In the formalism by phenotype distribution, environmental change is feasible by changing the growth term Ax accordingly. Our formalism does include the fitness-based equation as a special case, by setting Ax=x. Another merit in our formulation is that it easily takes isogenic phenotypic fluctuation into account without changing the form of the equation, but only by modifying Ax. By applying this equation, we obtained the influence of isogenic phenotype fluctuations on error catastrophe. The critical mutation rate for the error catastrophe increases because of the fluctuation, in a certain case. This implies that the fluctuation can enhance the robustness of a high-fitness state against mutation. In fact, the relevance of isogenic phenotypic fluctuations on evolution has been recently proposed 25 27, and change in phenotypic fluctuation through evolution has been experimentally verified 7,25. In general, phenotypic fluctuations and a mutation-selection process for artificial evolution have been extensively studied recently. The present formulation will be useful in analyzing such experimental data, as well as in elucidating the relevance of phenotypic fluctuations to evolution. ACKNOWLEDGMENTS The authors would like to thank P. Marcq, S. Sasa, and T. Yomo for useful discussion. APPENDIX A: ESTIMATION OF THE TRANSITION PROBABILITY IN THE NK MODEL In the NK model 17,53, the fitness f of a sequence s is given by fs = 1 i s, N i=1 where i is the contribution of the ith site to the fitness, which is a function of s i and the state values of other K sites. The function i takes a value chosen uniformly from 0, 1 at N random. We assume that the phenotype x of the sequence s is given by x= fs. When N1, K1, and K/N1, the phenotype distribution of mutants of a given sequence s whose phenotype is x is characterized only by the phenotype x without the need to specify the sequence s. For showing this, we first examine the one-point mutant case. We consider the number of changed sites of sites at which s are changed due to a single-point mutation. By assuming that the average number of changed sites is K, the distribution of the number of changed sites n, denoted by P site n, is approximately given by P site ne n K2 /2K, A1 with the help of the limiting form of binomial distribution. Here, we have omitted the normalization constant. Next, we study the distribution of the difference between the phenotype x of the original sequence and the phenotype x of its one-point mutant, given the number n of changed sites of the single-point mutant. We denote the distribution as P diff n;x, where X=x x. Here the average of x is xn n/n, since N n sites are unchanged. Thus, according to the central limit theorem, the distribution is estimated as P diff n;xexp x X + n 2 N 2n 2 N 2, A2 where 2 is the variance of the distribution of the value of. This variance is estimated from the probability distribution P s,i that the sequence is generated. Although the explicit form of P s,i is hard to obtain unless i and s are given, it is estimated by means of the most probable distribution, obtained as follows: Find the distribution that maximizes the evaluation function S called entropy defined by S= 0 1 Plog Pd under the conditions 0 1 Pd=1 and 0 1 Pd=x. Accordingly the variance 2 may depend on x. Combining these distributions A1 and A2 gives the distribution of X without constraint on the number of changed sites: N PX = P site np diff n;xexp n=1 x X + K 2 N 2K 2 + x 2 N 2 This result indicates that the phenotype distribution of single-point mutants from the original sequence s having the phenotype x is characterized by its phenotype x only; s is not necessary. Similarly, one can show that phenotype distribution of n-point mutants is also characterized only by x. Hence the transition probability in the NK model is described only in terms of the phenotypes of the sequences, when N1, K1, and K/N
9 EVOLUTION EQUATION OF PHENOTYPE DISTRIBUTION: APPENDIX B: MATHEMATICAL STRUCTURE OF THE EQUATION OF FORM (9) We first rewrite Eq. 9 as = Ā + Lx, B1 where L is a linear operator, defined by Lx=Ax +/xfx+ 2 /x 2 gx with fx= m 1 1 xax and gx=/2m 1 2 xax. The linear operator L of the above form is transformed to an Hermite operator using variable transformations 54 see below, so that L is represented by a complete set of eigenfunctions and corresponding eigenvalues, which are denoted by i x and i i=0,1,2,..., respectively. Eigenvalues are real and not degenerated, so that they are arranged as According to 40, Px,t is expanded as Px,t = a i i x, B2 i=0 where a i satisfies da i = a i i a j j. B3 dt j=0 The prime over the sum symbol indicates that the summation is taken except for those of noncontributing eigenfunctions as defined in 40. Stationary solutions of Eq. B3 are given by a k =1 and a i =0 for ik. Among these stationary solutions, only the solution a 0 =1 and a i =0 for i0 is stable. Hence the eigenfunction for the largest eigenvalue he largest replication rate gives the stationary distribution function. Now it is important to obtain eigenfunctions and eigenvalues of L, in particular the largest eigenvalue 0 and its corresponding eigenfunction 0. Hence we focus our attention on the eigenvalue problem Ax + 2 fx + x x 2gx Px = Px, B4 where is a constant and P is a function of x. We can transform eigenvalue problem B4 to a Schrödinger equation-type eigenvalue problem as follows: First we introduce a new variable y related to x as yx x = x0h/gxdx, where x0 and h are constants. Next, we consider a new function y related to Px as y = gx y h e y 0 fˆy/2hdy, Pxx=xy where y 0 is some constant, xy the inverse function of yx, and fˆ a function of y defined by fˆy =gxfx h + 1 dgx. 2 dx x=xy Using these new quantities y and and rewriting eigenvalue problem B4 suitably, we get Vy + h 2 y 2y = y, B5 where Vy=Ây+dfˆy/dy/2 fˆ2 y/4h with Ây =A(xy). 1 S. F. Elena and R. E. Lenski, Nat. Rev. Genet. 4, R. E. Lenski, M. R. Rose, S. C. Simpson, and S. C. Tadler, Am. Nat. 183, M. Hegreness, N. Shoresh, D. Hartl, and R. Kishony, Science 311, A. E. Mayo, Y. Setty, S. Shavit, A. Zaslaver, and U. Alon, PLoS Biol. 4, E. Dekel and U. Alon, Nature London 436, A. Kashiwagi, W. Noumachi, M. Katsuno, M. T. Alam, I. Urabe, and T. Yomo, J. Mol. Evol. 52, Y. Ito, T. Kawama, I. Urabe, and T. Yomo, J. Mol. Evol. 58, M. Kimura, Proc. Natl. Acad. Sci. U.S.A. 54, L. S. Tsimring, H. Levine, and D. A. Kessler, Phys. Rev. Lett. 76, U. Gerland and T. Hwa, J. Mol. Evol. 55, M. Lassig, F. Tria, and L. Peliti, Europhys. Lett. 62, C. Taylor, D. Fudenberg, A. Sasaki, and M. A. Nowak, Bull. Math. Biol. 66, T. Antal and I. Scheuring, Bull. Math. Biol. 68, A. Traulsen, J. C. Claussen, and C. Hauert, Phys. Rev. Lett. 95, T. Reichenbach, M. Mobilia, and E. Frey, Phys. Rev. E 74, M. Eigen, J. McCaskill, and P. Schuster, J. Phys. Chem. 92, S. Kauffman, The Origins of Order Oxford University Press, New York, J. Spudich and D. Koshland, Nature London 262, M. B. Elowitz, A. J. Levine, E. D. Siggia, and P. S. Swain, Science 297, M. Kaern, T. C. Elston, W. J. Blake, and J. J. Collins, Nat. Rev. Genet. 6, J. Hasty, J. Pradines, M. Dolnik, and J. J. Collins, Proc. Natl. Acad. Sci. U.S.A. 97, C. Furusawa, T. Suzuki, A. Kashiwagi, T. Yomo, and K. Kaneko, Biophysics 1, M. Ueda, Y. Sako, T. Tanaka, P. Devreotes, and T. Yanagida, Science 294, A. Bar-Even, J. Paulsson, N. Maheshri, M. Carmi, E. O Shea, Y. Pilpel, and N. Barkai, Nat. Genet. 38, K. Sato, Y. Ito, T. Yomo, and K. Kaneko, Proc. Natl. Acad. Sci
10 KATSUHIKO SATO AND KUNIHIKO KANEKO U.S.A. 100, K. Kaneko, Life: An Introduction to Complex Systems Biology Springer, Berlin, K. Kaneko and C. Furusawa, J. Theor. Biol. 240, L. W. Ancel, Theor Popul. Biol. 58, N. G. van Kampen, Stochastic Processes in Physics and Chemistry North-Holland, Amsterdam, R. Kubo, K. Matsuo, and K. Kitahara, J. Stat. Phys. 9, John G. Kemeny and J. Laurie Snell, Finite Markov Chains Springer, New York, 1981, Chap E. Baake, M. Baake, A. Bovier, and M. Klein, J. Math. Biol. 50, C. J. Burke and M. Rosenblatt, Ann. Math. Stat. 29, E. Baake and H. Wagner, Genet. Res. 78, M. S. Waterman, Introduction to Computational Biology: Maps, Sequences, and Genomes Chapman and Hall, London, C. Furusawa and K. Kaneko, Phys. Rev. E 73, J. H. Gillespie, Theor Popul. Biol. 23, H. A. Orr, Evolution Lawrence, Kans. 56, H. Haken, Synergetics: An Introduction Nonequilibrium Phase Transitions and Self-Organization in Physics, Chemistry, and Biology Springer-Verlag, Berlin, K. Sato and K. Kaneko, Phys. Biol. 3, P. M. Morse and H. Feshbach, Methods of Theoretical Physics McGraw-Hill, New York, 1953, pp T. Garske, Bull. Math. Biol. 68, E. Baake, M. Baake, and H. Wagner, Phys. Rev. Lett. 78, L. Peliti, Europhys. Lett. 57, I. Leuthausser, J. Stat. Phys. 48, D. B. Saakian and C. K. Hu, Proc. Natl. Acad. Sci. U.S.A. 103, D. Saakian and C. K. Hu, Phys. Rev. E 69, H. Yang, G. Luo, P. Karnchanaphanurach, T. M. Louie, I. Rech, S. Cova, L. Xun, and X. S. Xie, Science 302, L. W. Ancel and W. Fontana, J. Exp. Zool. 288, J. Hofbauer, J. Math. Biol. 23, J. F. Crow and M. Kimura, An Introduction to Population Genetics Theory Harper & Row, New York, E. Akin, The Geometry of Population Genetics Springer- Verlag, Berlin, B. Levitan and S. Kauffman, Mol. Divers. 1, H. Risken, The Fokker-Planck Equation: Methods of Solution and Applications, 2nd ed. Springer-Verlag, Berlin, 1996, pp. 103 and The value is a mutation rate per site, and is a dimensionless parameter, while Ax is the reproduction rate per time. 56 This is true if we include in the Eigen model a transcription error to protein. Also, the phenotype fluctuation of fluorescence of bacteria cells in 7 seem to support this assumption. 57 In this case, x b * =2/1 nx 0 and * =A 0 n +1 n+1 /4 n+1 1 n/x 0 1 n to the lowest order of A 0. The condition for * 0 * is given by 0n1 and (1 1 nn+1/8 1/1 n )x 0 x 0 when n is close to
Living organisms are composed of many different types of
 On the relation between fluctuation and response in biological systems Katsuhiko Sato*, Yoichiro Ito, Tetsuya Yomo*, and Kunihiko Kaneko* *Department of Pure and Applied Sciences, University of Tokyo,
On the relation between fluctuation and response in biological systems Katsuhiko Sato*, Yoichiro Ito, Tetsuya Yomo*, and Kunihiko Kaneko* *Department of Pure and Applied Sciences, University of Tokyo,
Relationship among phenotypic plasticity, phenotypic fl uctuations, robustness, and evolvability 529
 Relationship among phenotypic plasticity, phenotypic fl uctuations, robustness, and evolvability 59 Relationship among phenotypic plasticity, phenotypic fluctuations, robustness, and evolvability; Waddington
Relationship among phenotypic plasticity, phenotypic fl uctuations, robustness, and evolvability 59 Relationship among phenotypic plasticity, phenotypic fluctuations, robustness, and evolvability; Waddington
Evolution of Robustness to Noise and Mutation in Gene Expression Dynamics
 to Noise and Mutation in Gene Epression Dynamics Kunihiko Kaneko 1,2 * 1 Department of Pure and Applied Sciences, University of Tokyo, Tokyo Japan, 2 Comple Systems Biology Project, Eploratory Research
to Noise and Mutation in Gene Epression Dynamics Kunihiko Kaneko 1,2 * 1 Department of Pure and Applied Sciences, University of Tokyo, Tokyo Japan, 2 Comple Systems Biology Project, Eploratory Research
arxiv:physics/ v1 [physics.bio-ph] 27 Jun 2001
![arxiv:physics/ v1 [physics.bio-ph] 27 Jun 2001 arxiv:physics/ v1 [physics.bio-ph] 27 Jun 2001](/thumbs/72/67609644.jpg) Maternal effects in molecular evolution Claus O. Wilke Digital Life Laboratory, Mail Code 36-93, Caltech, Pasadena, CA 925 wilke@caltech.edu (Printed: May 3, 27) arxiv:physics/693v [physics.bio-ph] 27
Maternal effects in molecular evolution Claus O. Wilke Digital Life Laboratory, Mail Code 36-93, Caltech, Pasadena, CA 925 wilke@caltech.edu (Printed: May 3, 27) arxiv:physics/693v [physics.bio-ph] 27
Linear Algebra of Eigen s Quasispecies Model
 Linear Algebra of Eigen s Quasispecies Model Artem Novozhilov Department of Mathematics North Dakota State University Midwest Mathematical Biology Conference, University of Wisconsin La Crosse May 17,
Linear Algebra of Eigen s Quasispecies Model Artem Novozhilov Department of Mathematics North Dakota State University Midwest Mathematical Biology Conference, University of Wisconsin La Crosse May 17,
Evolution of Phenotype as selection of Dynamical Systems 1 Phenotypic Fluctuation (Plasticity) versus Evolution
 Evolution of Phenotype as selection of Dynamical Systems 1 Phenotypic Fluctuation (Plasticity) versus Evolution 2 Phenotypic Fluctuation versus Genetic Variation consistency between Genetic and Phenotypic
Evolution of Phenotype as selection of Dynamical Systems 1 Phenotypic Fluctuation (Plasticity) versus Evolution 2 Phenotypic Fluctuation versus Genetic Variation consistency between Genetic and Phenotypic
Linear Algebra of the Quasispecies Model
 Linear Algebra of the Quasispecies Model Artem Novozhilov Department of Mathematics North Dakota State University SIAM Conference on the Life Sciences, Charlotte, North Caroline, USA August 6, 2014 Artem
Linear Algebra of the Quasispecies Model Artem Novozhilov Department of Mathematics North Dakota State University SIAM Conference on the Life Sciences, Charlotte, North Caroline, USA August 6, 2014 Artem
Population Genetics and Evolution III
 Population Genetics and Evolution III The Mechanisms of Evolution: Drift São Paulo / January 2019 SMRI (Italy) luca@peliti.org 1 Drift 2 The Population Genetics Triad Sewall Wright Ronald A. Fisher Motoo
Population Genetics and Evolution III The Mechanisms of Evolution: Drift São Paulo / January 2019 SMRI (Italy) luca@peliti.org 1 Drift 2 The Population Genetics Triad Sewall Wright Ronald A. Fisher Motoo
Handbook of Stochastic Methods
 C. W. Gardiner Handbook of Stochastic Methods for Physics, Chemistry and the Natural Sciences Third Edition With 30 Figures Springer Contents 1. A Historical Introduction 1 1.1 Motivation I 1.2 Some Historical
C. W. Gardiner Handbook of Stochastic Methods for Physics, Chemistry and the Natural Sciences Third Edition With 30 Figures Springer Contents 1. A Historical Introduction 1 1.1 Motivation I 1.2 Some Historical
arxiv:physics/ v1 [physics.bio-ph] 30 Sep 2002
![arxiv:physics/ v1 [physics.bio-ph] 30 Sep 2002 arxiv:physics/ v1 [physics.bio-ph] 30 Sep 2002](/thumbs/73/68632814.jpg) Zipf s Law in Gene Expression Chikara Furusawa Center for Developmental Biology, The Institute of Physical arxiv:physics/0209103v1 [physics.bio-ph] 30 Sep 2002 and Chemical Research (RIKEN), Kobe 650-0047,
Zipf s Law in Gene Expression Chikara Furusawa Center for Developmental Biology, The Institute of Physical arxiv:physics/0209103v1 [physics.bio-ph] 30 Sep 2002 and Chemical Research (RIKEN), Kobe 650-0047,
56:198:582 Biological Networks Lecture 8
 56:198:582 Biological Networks Lecture 8 Course organization Two complementary approaches to modeling and understanding biological networks Constraint-based modeling (Palsson) System-wide Metabolism Steady-state
56:198:582 Biological Networks Lecture 8 Course organization Two complementary approaches to modeling and understanding biological networks Constraint-based modeling (Palsson) System-wide Metabolism Steady-state
Relaxation Property and Stability Analysis of the Quasispecies Models
 Commun. Theor. Phys. (Beijing, China) 52 (2009) pp. 726 734 c Chinese Physical Society and IOP Publishing Ltd Vol. 52, No. 4, October 15, 2009 Relaxation Property and Stability Analysis of the Quasispecies
Commun. Theor. Phys. (Beijing, China) 52 (2009) pp. 726 734 c Chinese Physical Society and IOP Publishing Ltd Vol. 52, No. 4, October 15, 2009 Relaxation Property and Stability Analysis of the Quasispecies
Testing the transition state theory in stochastic dynamics of a. genetic switch
 Testing the transition state theory in stochastic dynamics of a genetic switch Tomohiro Ushikubo 1, Wataru Inoue, Mitsumasa Yoda 1 1,, 3, and Masaki Sasai 1 Department of Computational Science and Engineering,
Testing the transition state theory in stochastic dynamics of a genetic switch Tomohiro Ushikubo 1, Wataru Inoue, Mitsumasa Yoda 1 1,, 3, and Masaki Sasai 1 Department of Computational Science and Engineering,
Statistical-mechanical study of evolution of robustness in noisy environments
 PHYSICAL REVIEW E 8, 599 29 Statistical-mechanical study of evolution of robustness in noisy environments Ayaka Sakata* and Koji Hukushima Graduate School of Arts and Sciences, The University of Tokyo,
PHYSICAL REVIEW E 8, 599 29 Statistical-mechanical study of evolution of robustness in noisy environments Ayaka Sakata* and Koji Hukushima Graduate School of Arts and Sciences, The University of Tokyo,
STOCHASTIC PROCESSES IN PHYSICS AND CHEMISTRY
 STOCHASTIC PROCESSES IN PHYSICS AND CHEMISTRY Third edition N.G. VAN KAMPEN Institute for Theoretical Physics of the University at Utrecht ELSEVIER Amsterdam Boston Heidelberg London New York Oxford Paris
STOCHASTIC PROCESSES IN PHYSICS AND CHEMISTRY Third edition N.G. VAN KAMPEN Institute for Theoretical Physics of the University at Utrecht ELSEVIER Amsterdam Boston Heidelberg London New York Oxford Paris
Handbook of Stochastic Methods
 Springer Series in Synergetics 13 Handbook of Stochastic Methods for Physics, Chemistry and the Natural Sciences von Crispin W Gardiner Neuausgabe Handbook of Stochastic Methods Gardiner schnell und portofrei
Springer Series in Synergetics 13 Handbook of Stochastic Methods for Physics, Chemistry and the Natural Sciences von Crispin W Gardiner Neuausgabe Handbook of Stochastic Methods Gardiner schnell und portofrei
arxiv: v1 [cond-mat.stat-mech] 3 Apr 2007
![arxiv: v1 [cond-mat.stat-mech] 3 Apr 2007 arxiv: v1 [cond-mat.stat-mech] 3 Apr 2007](/thumbs/74/69882095.jpg) A General Nonlinear Fokker-Planck Equation and its Associated Entropy Veit Schwämmle, Evaldo M. F. Curado, Fernando D. Nobre arxiv:0704.0465v1 [cond-mat.stat-mech] 3 Apr 2007 Centro Brasileiro de Pesquisas
A General Nonlinear Fokker-Planck Equation and its Associated Entropy Veit Schwämmle, Evaldo M. F. Curado, Fernando D. Nobre arxiv:0704.0465v1 [cond-mat.stat-mech] 3 Apr 2007 Centro Brasileiro de Pesquisas
Chapter 6 - Random Processes
 EE385 Class Notes //04 John Stensby Chapter 6 - Random Processes Recall that a random variable X is a mapping between the sample space S and the extended real line R +. That is, X : S R +. A random process
EE385 Class Notes //04 John Stensby Chapter 6 - Random Processes Recall that a random variable X is a mapping between the sample space S and the extended real line R +. That is, X : S R +. A random process
HIGH FRICTION LIMIT OF THE KRAMERS EQUATION : THE MULTIPLE TIME SCALE APPROACH. Lydéric Bocquet
 HIGH FRICTION LIMIT OF THE KRAMERS EQUATION : THE MULTIPLE TIME SCALE APPROACH Lydéric Bocquet arxiv:cond-mat/9605186v1 30 May 1996 Laboratoire de Physique, Ecole Normale Supérieure de Lyon (URA CNRS 1325),
HIGH FRICTION LIMIT OF THE KRAMERS EQUATION : THE MULTIPLE TIME SCALE APPROACH Lydéric Bocquet arxiv:cond-mat/9605186v1 30 May 1996 Laboratoire de Physique, Ecole Normale Supérieure de Lyon (URA CNRS 1325),
Molecular models of biological evolution have attracted
 Exact solution of the Eigen model with general fitness functions and degradation rates David B. Saakian* and Chin-Kun Hu* *Institute of Physics, Academia Sinica, ankang, Taipei 11529, Taiwan; and Yerevan
Exact solution of the Eigen model with general fitness functions and degradation rates David B. Saakian* and Chin-Kun Hu* *Institute of Physics, Academia Sinica, ankang, Taipei 11529, Taiwan; and Yerevan
Quasispecies distribution of Eigen model
 Vol 16 No 9, September 2007 c 2007 Chin. Phys. Soc. 1009-1963/2007/16(09)/2600-08 Chinese Physics and IOP Publishing Ltd Quasispecies distribution of Eigen model Chen Jia( ), Li Sheng( ), and Ma Hong-Ru(
Vol 16 No 9, September 2007 c 2007 Chin. Phys. Soc. 1009-1963/2007/16(09)/2600-08 Chinese Physics and IOP Publishing Ltd Quasispecies distribution of Eigen model Chen Jia( ), Li Sheng( ), and Ma Hong-Ru(
Critique of the Fox-Lu model for Hodgkin-Huxley fluctuations in neuron ion channels
 Critique of the Fox-Lu model for Hodgkin-Huxley fluctuations in neuron ion channels by Ronald F. Fox Smyrna, Georgia July 0, 018 Subtle is the Lord, but malicious He is not. Albert Einstein, Princeton,
Critique of the Fox-Lu model for Hodgkin-Huxley fluctuations in neuron ion channels by Ronald F. Fox Smyrna, Georgia July 0, 018 Subtle is the Lord, but malicious He is not. Albert Einstein, Princeton,
1.3 Forward Kolmogorov equation
 1.3 Forward Kolmogorov equation Let us again start with the Master equation, for a system where the states can be ordered along a line, such as the previous examples with population size n = 0, 1, 2,.
1.3 Forward Kolmogorov equation Let us again start with the Master equation, for a system where the states can be ordered along a line, such as the previous examples with population size n = 0, 1, 2,.
Extending the Tools of Chemical Reaction Engineering to the Molecular Scale
 Extending the Tools of Chemical Reaction Engineering to the Molecular Scale Multiple-time-scale order reduction for stochastic kinetics James B. Rawlings Department of Chemical and Biological Engineering
Extending the Tools of Chemical Reaction Engineering to the Molecular Scale Multiple-time-scale order reduction for stochastic kinetics James B. Rawlings Department of Chemical and Biological Engineering
Dynamical-Systems Perspective to Stem-cell Biology: Relevance of oscillatory gene expression dynamics and cell-cell interaction
 Dynamical-Systems Perspective to Stem-cell Biology: Relevance of oscillatory gene expression dynamics and cell-cell interaction Kunihiko Kaneko Universal Biology Inst., Center for Complex-Systems Biology,
Dynamical-Systems Perspective to Stem-cell Biology: Relevance of oscillatory gene expression dynamics and cell-cell interaction Kunihiko Kaneko Universal Biology Inst., Center for Complex-Systems Biology,
Gillespie s Algorithm and its Approximations. Des Higham Department of Mathematics and Statistics University of Strathclyde
 Gillespie s Algorithm and its Approximations Des Higham Department of Mathematics and Statistics University of Strathclyde djh@maths.strath.ac.uk The Three Lectures 1 Gillespie s algorithm and its relation
Gillespie s Algorithm and its Approximations Des Higham Department of Mathematics and Statistics University of Strathclyde djh@maths.strath.ac.uk The Three Lectures 1 Gillespie s algorithm and its relation
Escape Problems & Stochastic Resonance
 Escape Problems & Stochastic Resonance 18.S995 - L08 & 09 dunkel@mit.edu Examples Illustration by J.P. Cartailler. Copyright 2007, Symmation LLC. transport & evolution: stochastic escape problems http://evolutionarysystemsbiology.org/intro/
Escape Problems & Stochastic Resonance 18.S995 - L08 & 09 dunkel@mit.edu Examples Illustration by J.P. Cartailler. Copyright 2007, Symmation LLC. transport & evolution: stochastic escape problems http://evolutionarysystemsbiology.org/intro/
Are there species smaller than 1mm?
 Are there species smaller than 1mm? Alan McKane Theoretical Physics Division, University of Manchester Warwick, September 9, 2013 Alan McKane (Manchester) Are there species smaller than 1mm? Warwick, September
Are there species smaller than 1mm? Alan McKane Theoretical Physics Division, University of Manchester Warwick, September 9, 2013 Alan McKane (Manchester) Are there species smaller than 1mm? Warwick, September
16. Working with the Langevin and Fokker-Planck equations
 16. Working with the Langevin and Fokker-Planck equations In the preceding Lecture, we have shown that given a Langevin equation (LE), it is possible to write down an equivalent Fokker-Planck equation
16. Working with the Langevin and Fokker-Planck equations In the preceding Lecture, we have shown that given a Langevin equation (LE), it is possible to write down an equivalent Fokker-Planck equation
Lecture Notes: Markov chains
 Computational Genomics and Molecular Biology, Fall 5 Lecture Notes: Markov chains Dannie Durand At the beginning of the semester, we introduced two simple scoring functions for pairwise alignments: a similarity
Computational Genomics and Molecular Biology, Fall 5 Lecture Notes: Markov chains Dannie Durand At the beginning of the semester, we introduced two simple scoring functions for pairwise alignments: a similarity
Transport of single molecules along the periodic parallel lattices with coupling
 THE JOURNAL OF CHEMICAL PHYSICS 124 204901 2006 Transport of single molecules along the periodic parallel lattices with coupling Evgeny B. Stukalin The James Franck Institute The University of Chicago
THE JOURNAL OF CHEMICAL PHYSICS 124 204901 2006 Transport of single molecules along the periodic parallel lattices with coupling Evgeny B. Stukalin The James Franck Institute The University of Chicago
Inter Intra Molecular Dynamics as an Iterated Function System
 Journal of the Physical Society of Japan Vol 74, No 9, September, 2005, pp 2386 2390 #2005 The Physical Society of Japan LETTERS Inter Intra Molecular Dynamics as an Iterated Function System Kunihiko KANEKO
Journal of the Physical Society of Japan Vol 74, No 9, September, 2005, pp 2386 2390 #2005 The Physical Society of Japan LETTERS Inter Intra Molecular Dynamics as an Iterated Function System Kunihiko KANEKO
stochastic, evolutionary game dynamics in finite populations
 PHYSICAL REVIEW E 80, 011909 2009 Deterministic evolutionary game dynamics in finite populations Philipp M. Altrock* and Arne Traulsen Emmy-Noether Group of Evolutionary Dynamics, Department of Evolutionary
PHYSICAL REVIEW E 80, 011909 2009 Deterministic evolutionary game dynamics in finite populations Philipp M. Altrock* and Arne Traulsen Emmy-Noether Group of Evolutionary Dynamics, Department of Evolutionary
Viral evolution model with several time scales
 Viral evolution model with several time scales AA Archibasov Samara National Research University, 4 Moskovskoe Shosse, 4486, Samara, Russia Abstract In this paper a viral evolution model with specific
Viral evolution model with several time scales AA Archibasov Samara National Research University, 4 Moskovskoe Shosse, 4486, Samara, Russia Abstract In this paper a viral evolution model with specific
NATURAL SELECTION FOR WITHIN-GENERATION VARIANCE IN OFFSPRING NUMBER JOHN H. GILLESPIE. Manuscript received September 17, 1973 ABSTRACT
 NATURAL SELECTION FOR WITHIN-GENERATION VARIANCE IN OFFSPRING NUMBER JOHN H. GILLESPIE Department of Biology, University of Penmyluania, Philadelphia, Pennsyluania 19174 Manuscript received September 17,
NATURAL SELECTION FOR WITHIN-GENERATION VARIANCE IN OFFSPRING NUMBER JOHN H. GILLESPIE Department of Biology, University of Penmyluania, Philadelphia, Pennsyluania 19174 Manuscript received September 17,
Boulder 07: Modelling Stochastic Gene Expression
 Boulder 7: Modelling Stochastic Gene Expression Peter Swain, Physiology, McGill University swain@cnd.mcgill.ca The whys and wherefores of stochasticity A system evolves stochastically if its dynamics is
Boulder 7: Modelling Stochastic Gene Expression Peter Swain, Physiology, McGill University swain@cnd.mcgill.ca The whys and wherefores of stochasticity A system evolves stochastically if its dynamics is
First and Last Name: 2. Correct The Mistake Determine whether these equations are false, and if so write the correct answer.
 . Correct The Mistake Determine whether these equations are false, and if so write the correct answer. ( x ( x (a ln + ln = ln(x (b e x e y = e xy (c (d d dx cos(4x = sin(4x 0 dx xe x = (a This is an incorrect
. Correct The Mistake Determine whether these equations are false, and if so write the correct answer. ( x ( x (a ln + ln = ln(x (b e x e y = e xy (c (d d dx cos(4x = sin(4x 0 dx xe x = (a This is an incorrect
Stochastic Particle Methods for Rarefied Gases
 CCES Seminar WS 2/3 Stochastic Particle Methods for Rarefied Gases Julian Köllermeier RWTH Aachen University Supervisor: Prof. Dr. Manuel Torrilhon Center for Computational Engineering Science Mathematics
CCES Seminar WS 2/3 Stochastic Particle Methods for Rarefied Gases Julian Köllermeier RWTH Aachen University Supervisor: Prof. Dr. Manuel Torrilhon Center for Computational Engineering Science Mathematics
On state-space reduction in multi-strain pathogen models, with an application to antigenic drift in influenza A
 On state-space reduction in multi-strain pathogen models, with an application to antigenic drift in influenza A Sergey Kryazhimskiy, Ulf Dieckmann, Simon A. Levin, Jonathan Dushoff Supporting information
On state-space reduction in multi-strain pathogen models, with an application to antigenic drift in influenza A Sergey Kryazhimskiy, Ulf Dieckmann, Simon A. Levin, Jonathan Dushoff Supporting information
Nonlinear Analysis: Modelling and Control, Vilnius, IMI, 1998, No 3 KINK-EXCITATION OF N-SYSTEM UNDER SPATIO -TEMPORAL NOISE. R.
 Nonlinear Analysis: Modelling and Control Vilnius IMI 1998 No 3 KINK-EXCITATION OF N-SYSTEM UNDER SPATIO -TEMPORAL NOISE R. Bakanas Semiconductor Physics Institute Go štauto 11 6 Vilnius Lithuania Vilnius
Nonlinear Analysis: Modelling and Control Vilnius IMI 1998 No 3 KINK-EXCITATION OF N-SYSTEM UNDER SPATIO -TEMPORAL NOISE R. Bakanas Semiconductor Physics Institute Go štauto 11 6 Vilnius Lithuania Vilnius
Lecture 12: Detailed balance and Eigenfunction methods
 Miranda Holmes-Cerfon Applied Stochastic Analysis, Spring 2015 Lecture 12: Detailed balance and Eigenfunction methods Readings Recommended: Pavliotis [2014] 4.5-4.7 (eigenfunction methods and reversibility),
Miranda Holmes-Cerfon Applied Stochastic Analysis, Spring 2015 Lecture 12: Detailed balance and Eigenfunction methods Readings Recommended: Pavliotis [2014] 4.5-4.7 (eigenfunction methods and reversibility),
Lecture 4: Transcription networks basic concepts
 Lecture 4: Transcription networks basic concepts - Activators and repressors - Input functions; Logic input functions; Multidimensional input functions - Dynamics and response time 2.1 Introduction The
Lecture 4: Transcription networks basic concepts - Activators and repressors - Input functions; Logic input functions; Multidimensional input functions - Dynamics and response time 2.1 Introduction The
arxiv:cond-mat/ v2 [cond-mat.stat-mech] 23 Feb 1998
![arxiv:cond-mat/ v2 [cond-mat.stat-mech] 23 Feb 1998 arxiv:cond-mat/ v2 [cond-mat.stat-mech] 23 Feb 1998](/thumbs/78/77115469.jpg) Multiplicative processes and power laws arxiv:cond-mat/978231v2 [cond-mat.stat-mech] 23 Feb 1998 Didier Sornette 1,2 1 Laboratoire de Physique de la Matière Condensée, CNRS UMR6632 Université des Sciences,
Multiplicative processes and power laws arxiv:cond-mat/978231v2 [cond-mat.stat-mech] 23 Feb 1998 Didier Sornette 1,2 1 Laboratoire de Physique de la Matière Condensée, CNRS UMR6632 Université des Sciences,
LANGER'S METHOD FOR WEAKLY BOUND
 PFC/JA-82-5 LANGER'S METHOD FOR WEAKLY BOUND STATES OF THE HELMHOLTZ EQUATION WITH SYMMETRIC PROFILES by George L. Johnston Plasma Fusion Center Massachusetts Institute of Technology Cambridge, Massachusetts
PFC/JA-82-5 LANGER'S METHOD FOR WEAKLY BOUND STATES OF THE HELMHOLTZ EQUATION WITH SYMMETRIC PROFILES by George L. Johnston Plasma Fusion Center Massachusetts Institute of Technology Cambridge, Massachusetts
QUADRATIC MAJORIZATION 1. INTRODUCTION
 QUADRATIC MAJORIZATION JAN DE LEEUW 1. INTRODUCTION Majorization methods are used extensively to solve complicated multivariate optimizaton problems. We refer to de Leeuw [1994]; Heiser [1995]; Lange et
QUADRATIC MAJORIZATION JAN DE LEEUW 1. INTRODUCTION Majorization methods are used extensively to solve complicated multivariate optimizaton problems. We refer to de Leeuw [1994]; Heiser [1995]; Lange et
Stochastic Processes at Single-molecule and Single-cell levels
 Stochastic Processes at Single-molecule and Single-cell levels Hao Ge haoge@pu.edu.cn Beijing International Center for Mathematical Research 2 Biodynamic Optical Imaging Center Peing University, China
Stochastic Processes at Single-molecule and Single-cell levels Hao Ge haoge@pu.edu.cn Beijing International Center for Mathematical Research 2 Biodynamic Optical Imaging Center Peing University, China
Two special equations: Bessel s and Legendre s equations. p Fourier-Bessel and Fourier-Legendre series. p
 LECTURE 1 Table of Contents Two special equations: Bessel s and Legendre s equations. p. 259-268. Fourier-Bessel and Fourier-Legendre series. p. 453-460. Boundary value problems in other coordinate system.
LECTURE 1 Table of Contents Two special equations: Bessel s and Legendre s equations. p. 259-268. Fourier-Bessel and Fourier-Legendre series. p. 453-460. Boundary value problems in other coordinate system.
Series Solution of Linear Ordinary Differential Equations
 Series Solution of Linear Ordinary Differential Equations Department of Mathematics IIT Guwahati Aim: To study methods for determining series expansions for solutions to linear ODE with variable coefficients.
Series Solution of Linear Ordinary Differential Equations Department of Mathematics IIT Guwahati Aim: To study methods for determining series expansions for solutions to linear ODE with variable coefficients.
A Detailed Look at a Discrete Randomw Walk with Spatially Dependent Moments and Its Continuum Limit
 A Detailed Look at a Discrete Randomw Walk with Spatially Dependent Moments and Its Continuum Limit David Vener Department of Mathematics, MIT May 5, 3 Introduction In 8.366, we discussed the relationship
A Detailed Look at a Discrete Randomw Walk with Spatially Dependent Moments and Its Continuum Limit David Vener Department of Mathematics, MIT May 5, 3 Introduction In 8.366, we discussed the relationship
Langevin Methods. Burkhard Dünweg Max Planck Institute for Polymer Research Ackermannweg 10 D Mainz Germany
 Langevin Methods Burkhard Dünweg Max Planck Institute for Polymer Research Ackermannweg 1 D 55128 Mainz Germany Motivation Original idea: Fast and slow degrees of freedom Example: Brownian motion Replace
Langevin Methods Burkhard Dünweg Max Planck Institute for Polymer Research Ackermannweg 1 D 55128 Mainz Germany Motivation Original idea: Fast and slow degrees of freedom Example: Brownian motion Replace
From time series to superstatistics
 From time series to superstatistics Christian Beck School of Mathematical Sciences, Queen Mary, University of London, Mile End Road, London E 4NS, United Kingdom Ezechiel G. D. Cohen The Rockefeller University,
From time series to superstatistics Christian Beck School of Mathematical Sciences, Queen Mary, University of London, Mile End Road, London E 4NS, United Kingdom Ezechiel G. D. Cohen The Rockefeller University,
MATHEMATICAL MODELS - Vol. III - Mathematical Modeling and the Human Genome - Hilary S. Booth MATHEMATICAL MODELING AND THE HUMAN GENOME
 MATHEMATICAL MODELING AND THE HUMAN GENOME Hilary S. Booth Australian National University, Australia Keywords: Human genome, DNA, bioinformatics, sequence analysis, evolution. Contents 1. Introduction:
MATHEMATICAL MODELING AND THE HUMAN GENOME Hilary S. Booth Australian National University, Australia Keywords: Human genome, DNA, bioinformatics, sequence analysis, evolution. Contents 1. Introduction:
2. Mathematical descriptions. (i) the master equation (ii) Langevin theory. 3. Single cell measurements
 1. Why stochastic?. Mathematical descriptions (i) the master equation (ii) Langevin theory 3. Single cell measurements 4. Consequences Any chemical reaction is stochastic. k P d φ dp dt = k d P deterministic
1. Why stochastic?. Mathematical descriptions (i) the master equation (ii) Langevin theory 3. Single cell measurements 4. Consequences Any chemical reaction is stochastic. k P d φ dp dt = k d P deterministic
Fitness landscapes and seascapes
 Fitness landscapes and seascapes Michael Lässig Institute for Theoretical Physics University of Cologne Thanks Ville Mustonen: Cross-species analysis of bacterial promoters, Nonequilibrium evolution of
Fitness landscapes and seascapes Michael Lässig Institute for Theoretical Physics University of Cologne Thanks Ville Mustonen: Cross-species analysis of bacterial promoters, Nonequilibrium evolution of
Compartmentalization and Cell Division through Molecular Discreteness and Crowding in a Catalytic Reaction Network
 Life 214, 4, 586-597; doi:1.339/life44586 Article OPEN ACCESS life ISSN 275-1729 www.mdpi.com/journal/life Compartmentalization and Cell Division through Molecular Discreteness and Crowding in a Catalytic
Life 214, 4, 586-597; doi:1.339/life44586 Article OPEN ACCESS life ISSN 275-1729 www.mdpi.com/journal/life Compartmentalization and Cell Division through Molecular Discreteness and Crowding in a Catalytic
An Introduction to Evolutionary Game Theory: Lecture 2
 An Introduction to Evolutionary Game Theory: Lecture 2 Mauro Mobilia Lectures delivered at the Graduate School on Nonlinear and Stochastic Systems in Biology held in the Department of Applied Mathematics,
An Introduction to Evolutionary Game Theory: Lecture 2 Mauro Mobilia Lectures delivered at the Graduate School on Nonlinear and Stochastic Systems in Biology held in the Department of Applied Mathematics,
Casting Polymer Nets To Optimize Molecular Codes
 Casting Polymer Nets To Optimize Molecular Codes The physical language of molecules Mathematical Biology Forum (PRL 2007, PNAS 2008, Phys Bio 2008, J Theo Bio 2007, E J Lin Alg 2007) Biological information
Casting Polymer Nets To Optimize Molecular Codes The physical language of molecules Mathematical Biology Forum (PRL 2007, PNAS 2008, Phys Bio 2008, J Theo Bio 2007, E J Lin Alg 2007) Biological information
Molecular evolution - Part 1. Pawan Dhar BII
 Molecular evolution - Part 1 Pawan Dhar BII Theodosius Dobzhansky Nothing in biology makes sense except in the light of evolution Age of life on earth: 3.85 billion years Formation of planet: 4.5 billion
Molecular evolution - Part 1 Pawan Dhar BII Theodosius Dobzhansky Nothing in biology makes sense except in the light of evolution Age of life on earth: 3.85 billion years Formation of planet: 4.5 billion
Gene Expression as a Stochastic Process: From Gene Number Distributions to Protein Statistics and Back
 Gene Expression as a Stochastic Process: From Gene Number Distributions to Protein Statistics and Back June 19, 2007 Motivation & Basics A Stochastic Approach to Gene Expression Application to Experimental
Gene Expression as a Stochastic Process: From Gene Number Distributions to Protein Statistics and Back June 19, 2007 Motivation & Basics A Stochastic Approach to Gene Expression Application to Experimental
A Moment Closure Method for Stochastic Chemical Reaction Networks with General Kinetics
 MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 70 2013 785-800 ISSN 0340-6253 A Moment Closure Method for Stochastic Chemical Reaction Networks with General
MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 70 2013 785-800 ISSN 0340-6253 A Moment Closure Method for Stochastic Chemical Reaction Networks with General
Population Genetics: a tutorial
 : a tutorial Institute for Science and Technology Austria ThRaSh 2014 provides the basic mathematical foundation of evolutionary theory allows a better understanding of experiments allows the development
: a tutorial Institute for Science and Technology Austria ThRaSh 2014 provides the basic mathematical foundation of evolutionary theory allows a better understanding of experiments allows the development
Error threshold in simple landscapes
 arxiv:cond-mat/9610028v1 [cond-mat.dis-nn] 4 Oct 1996 Error threshold in simple landscapes Silvio Franz ICTP, Strada Costiera 10, I-34100 Trieste (Italy) and Luca Peliti Groupe de Physico-Chimie Théorique,
arxiv:cond-mat/9610028v1 [cond-mat.dis-nn] 4 Oct 1996 Error threshold in simple landscapes Silvio Franz ICTP, Strada Costiera 10, I-34100 Trieste (Italy) and Luca Peliti Groupe de Physico-Chimie Théorique,
DNA Bubble Dynamics. Hans Fogedby Aarhus University and Niels Bohr Institute Denmark
 DNA Bubble Dynamics Hans Fogedby Aarhus University and Niels Bohr Institute Denmark Principles of life Biology takes place in wet and warm environments Open driven systems - entropy decreases Self organization
DNA Bubble Dynamics Hans Fogedby Aarhus University and Niels Bohr Institute Denmark Principles of life Biology takes place in wet and warm environments Open driven systems - entropy decreases Self organization
Time fractional Schrödinger equation
 Time fractional Schrödinger equation Mark Naber a) Department of Mathematics Monroe County Community College Monroe, Michigan, 48161-9746 The Schrödinger equation is considered with the first order time
Time fractional Schrödinger equation Mark Naber a) Department of Mathematics Monroe County Community College Monroe, Michigan, 48161-9746 The Schrödinger equation is considered with the first order time
Emergence of Rules in Cell Society: Differentiation, Hierarchy, and Stability
 Emergence of Rules in Cell Society: Differentiation, Hierarchy, and Stability arxiv:adap-org/9802002v1 13 Feb 1998 Chikara Furusawa and Kunihiko Kaneko Department of Pure and Applied Sciences University
Emergence of Rules in Cell Society: Differentiation, Hierarchy, and Stability arxiv:adap-org/9802002v1 13 Feb 1998 Chikara Furusawa and Kunihiko Kaneko Department of Pure and Applied Sciences University
Comment on A BINOMIAL MOMENT APPROXIMATION SCHEME FOR EPIDEMIC SPREADING IN NETWORKS in U.P.B. Sci. Bull., Series A, Vol. 76, Iss.
 Comment on A BINOMIAL MOMENT APPROXIMATION SCHEME FOR EPIDEMIC SPREADING IN NETWORKS in U.P.B. Sci. Bull., Series A, Vol. 76, Iss. 2, 23-3, 24 Istvan Z. Kiss, & Prapanporn Rattana July 2, 24 School of
Comment on A BINOMIAL MOMENT APPROXIMATION SCHEME FOR EPIDEMIC SPREADING IN NETWORKS in U.P.B. Sci. Bull., Series A, Vol. 76, Iss. 2, 23-3, 24 Istvan Z. Kiss, & Prapanporn Rattana July 2, 24 School of
Physics 250 Green s functions for ordinary differential equations
 Physics 25 Green s functions for ordinary differential equations Peter Young November 25, 27 Homogeneous Equations We have already discussed second order linear homogeneous differential equations, which
Physics 25 Green s functions for ordinary differential equations Peter Young November 25, 27 Homogeneous Equations We have already discussed second order linear homogeneous differential equations, which
Can evolution paths be explained by chance alone? arxiv: v1 [math.pr] 12 Oct The model
![Can evolution paths be explained by chance alone? arxiv: v1 [math.pr] 12 Oct The model Can evolution paths be explained by chance alone? arxiv: v1 [math.pr] 12 Oct The model](/thumbs/89/98225676.jpg) Can evolution paths be explained by chance alone? Rinaldo B. Schinazi Department of Mathematics University of Colorado at Colorado Springs rinaldo.schinazi@uccs.edu arxiv:1810.05705v1 [math.pr] 12 Oct
Can evolution paths be explained by chance alone? Rinaldo B. Schinazi Department of Mathematics University of Colorado at Colorado Springs rinaldo.schinazi@uccs.edu arxiv:1810.05705v1 [math.pr] 12 Oct
MA22S3 Summary Sheet: Ordinary Differential Equations
 MA22S3 Summary Sheet: Ordinary Differential Equations December 14, 2017 Kreyszig s textbook is a suitable guide for this part of the module. Contents 1 Terminology 1 2 First order separable 2 2.1 Separable
MA22S3 Summary Sheet: Ordinary Differential Equations December 14, 2017 Kreyszig s textbook is a suitable guide for this part of the module. Contents 1 Terminology 1 2 First order separable 2 2.1 Separable
F n = F n 1 + F n 2. F(z) = n= z z 2. (A.3)
 Appendix A MATTERS OF TECHNIQUE A.1 Transform Methods Laplace transforms for continuum systems Generating function for discrete systems This method is demonstrated for the Fibonacci sequence F n = F n
Appendix A MATTERS OF TECHNIQUE A.1 Transform Methods Laplace transforms for continuum systems Generating function for discrete systems This method is demonstrated for the Fibonacci sequence F n = F n
Growth equations for the Wolf-Villain and Das Sarma Tamborenea models of molecular-beam epitaxy
 PHYSICAL REVIEW E VOLUME 54, NUMBER 6 DECEMBER 1996 Growth equations for the Wolf-Villain and Das Sarma Tamborenea models of molecular-beam epitaxy Zhi-Feng Huang and Bing-Lin Gu Chinese Center of Advanced
PHYSICAL REVIEW E VOLUME 54, NUMBER 6 DECEMBER 1996 Growth equations for the Wolf-Villain and Das Sarma Tamborenea models of molecular-beam epitaxy Zhi-Feng Huang and Bing-Lin Gu Chinese Center of Advanced
LIST OF MATHEMATICAL PAPERS
 LIST OF MATHEMATICAL PAPERS 1961 1999 [1] K. Sato (1961) Integration of the generalized Kolmogorov-Feller backward equations. J. Fac. Sci. Univ. Tokyo, Sect. I, Vol. 9, 13 27. [2] K. Sato, H. Tanaka (1962)
LIST OF MATHEMATICAL PAPERS 1961 1999 [1] K. Sato (1961) Integration of the generalized Kolmogorov-Feller backward equations. J. Fac. Sci. Univ. Tokyo, Sect. I, Vol. 9, 13 27. [2] K. Sato, H. Tanaka (1962)
Stochastic Processes around Central Dogma
 Stochastic Processes around Central Dogma Hao Ge haoge@pku.edu.cn Beijing International Center for Mathematical Research Biodynamic Optical Imaging Center Peking University, China http://www.bicmr.org/personal/gehao/
Stochastic Processes around Central Dogma Hao Ge haoge@pku.edu.cn Beijing International Center for Mathematical Research Biodynamic Optical Imaging Center Peking University, China http://www.bicmr.org/personal/gehao/
Measures for information propagation in Boolean networks
 Physica D 227 (2007) 100 104 www.elsevier.com/locate/physd Measures for information propagation in Boolean networks Pauli Rämö a,, Stuart Kauffman b, Juha Kesseli a, Olli Yli-Harja a a Institute of Signal
Physica D 227 (2007) 100 104 www.elsevier.com/locate/physd Measures for information propagation in Boolean networks Pauli Rämö a,, Stuart Kauffman b, Juha Kesseli a, Olli Yli-Harja a a Institute of Signal
A Method for Estimating Mean First-passage Time in Genetic Algorithms
 A Method for Estimating Mean First-passage Time in Genetic Algorithms Hiroshi Furutani Department of Information Science, Kyoto University of Education, Fushimi-ku, Kyoto, 612-8522 Japan This paper presents
A Method for Estimating Mean First-passage Time in Genetic Algorithms Hiroshi Furutani Department of Information Science, Kyoto University of Education, Fushimi-ku, Kyoto, 612-8522 Japan This paper presents
1 Review of di erential calculus
 Review of di erential calculus This chapter presents the main elements of di erential calculus needed in probability theory. Often, students taking a course on probability theory have problems with concepts
Review of di erential calculus This chapter presents the main elements of di erential calculus needed in probability theory. Often, students taking a course on probability theory have problems with concepts
Lecture 12: Detailed balance and Eigenfunction methods
 Lecture 12: Detailed balance and Eigenfunction methods Readings Recommended: Pavliotis [2014] 4.5-4.7 (eigenfunction methods and reversibility), 4.2-4.4 (explicit examples of eigenfunction methods) Gardiner
Lecture 12: Detailed balance and Eigenfunction methods Readings Recommended: Pavliotis [2014] 4.5-4.7 (eigenfunction methods and reversibility), 4.2-4.4 (explicit examples of eigenfunction methods) Gardiner
Symbiotic Sympatric Speciation: Compliance with Interaction-driven Phenotype Differentiation from a Single Genotype
 Symbiotic Sympatric Speciation: Compliance with Interaction-driven Phenotype Differentiation from a Single Genotype Kunihiko Kaneko (Univ of Tokyo, Komaba) with Tetsuya Yomo (Osaka Univ.) experiment Akiko
Symbiotic Sympatric Speciation: Compliance with Interaction-driven Phenotype Differentiation from a Single Genotype Kunihiko Kaneko (Univ of Tokyo, Komaba) with Tetsuya Yomo (Osaka Univ.) experiment Akiko
and the compositional inverse when it exists is A.
 Lecture B jacques@ucsd.edu Notation: R denotes a ring, N denotes the set of sequences of natural numbers with finite support, is a generic element of N, is the infinite zero sequence, n 0 R[[ X]] denotes
Lecture B jacques@ucsd.edu Notation: R denotes a ring, N denotes the set of sequences of natural numbers with finite support, is a generic element of N, is the infinite zero sequence, n 0 R[[ X]] denotes
Gene regulation: From biophysics to evolutionary genetics
 Gene regulation: From biophysics to evolutionary genetics Michael Lässig Institute for Theoretical Physics University of Cologne Thanks Ville Mustonen Johannes Berg Stana Willmann Curt Callan (Princeton)
Gene regulation: From biophysics to evolutionary genetics Michael Lässig Institute for Theoretical Physics University of Cologne Thanks Ville Mustonen Johannes Berg Stana Willmann Curt Callan (Princeton)
Shift in the velocity of a front due to a cutoff
 PHYSICAL REVIEW E VOLUME 56, NUMBER 3 SEPTEMBER 1997 Shift in the velocity of a front due to a cutoff Eric Brunet* and Bernard Derrida Laboratoire de Physique Statistique, ENS, 24 rue Lhomond, 75005 Paris,
PHYSICAL REVIEW E VOLUME 56, NUMBER 3 SEPTEMBER 1997 Shift in the velocity of a front due to a cutoff Eric Brunet* and Bernard Derrida Laboratoire de Physique Statistique, ENS, 24 rue Lhomond, 75005 Paris,
Normalization integrals of orthogonal Heun functions
 Normalization integrals of orthogonal Heun functions Peter A. Becker a) Center for Earth Observing and Space Research, Institute for Computational Sciences and Informatics, and Department of Physics and
Normalization integrals of orthogonal Heun functions Peter A. Becker a) Center for Earth Observing and Space Research, Institute for Computational Sciences and Informatics, and Department of Physics and
Predicting Fitness Effects of Beneficial Mutations in Digital Organisms
 Predicting Fitness Effects of Beneficial Mutations in Digital Organisms Haitao Zhang 1 and Michael Travisano 1 1 Department of Biology and Biochemistry, University of Houston, Houston TX 77204-5001 Abstract-Evolutionary
Predicting Fitness Effects of Beneficial Mutations in Digital Organisms Haitao Zhang 1 and Michael Travisano 1 1 Department of Biology and Biochemistry, University of Houston, Houston TX 77204-5001 Abstract-Evolutionary
arxiv: v2 [math-ph] 11 Jun 2015
![arxiv: v2 [math-ph] 11 Jun 2015 arxiv: v2 [math-ph] 11 Jun 2015](/thumbs/91/105961841.jpg) On the relevance of q-distribution functions: The return time distribution of restricted random walker Jaleh Zand 1,, Ugur Tirnakli 2,, and Henrik Jeldtoft Jensen 1, 1 Centre for Complexity Science and
On the relevance of q-distribution functions: The return time distribution of restricted random walker Jaleh Zand 1,, Ugur Tirnakli 2,, and Henrik Jeldtoft Jensen 1, 1 Centre for Complexity Science and
Lognormal Moment Closures for Biochemical Reactions
 Lognormal Moment Closures for Biochemical Reactions Abhyudai Singh and João Pedro Hespanha Abstract In the stochastic formulation of chemical reactions, the dynamics of the the first M -order moments of
Lognormal Moment Closures for Biochemical Reactions Abhyudai Singh and João Pedro Hespanha Abstract In the stochastic formulation of chemical reactions, the dynamics of the the first M -order moments of
Stochastic Processes around Central Dogma
 Stochastic Processes around Central Dogma Hao Ge haoge@pu.edu.cn Beijing International Center for Mathematical Research Biodynamic Optical Imaging Center Peing University, China http://www.bicmr.org/personal/gehao/
Stochastic Processes around Central Dogma Hao Ge haoge@pu.edu.cn Beijing International Center for Mathematical Research Biodynamic Optical Imaging Center Peing University, China http://www.bicmr.org/personal/gehao/
Restarting a Genetic Algorithm for Set Cover Problem Using Schnabel Census
 Restarting a Genetic Algorithm for Set Cover Problem Using Schnabel Census Anton V. Eremeev 1,2 1 Dostoevsky Omsk State University, Omsk, Russia 2 The Institute of Scientific Information for Social Sciences
Restarting a Genetic Algorithm for Set Cover Problem Using Schnabel Census Anton V. Eremeev 1,2 1 Dostoevsky Omsk State University, Omsk, Russia 2 The Institute of Scientific Information for Social Sciences
Entropy ISSN
 Entropy 2004, 6, 223 232 Full Paper Entropy ISSN 1099-4300 www.mdpi.org/entropy/ INTERNAL STRUCTURE OF ELEMENTARY PARTICLE AND POSSIBLE DETERMINISTIC MECHANISM OF BIOLOGICAL EVOLUTION Alexey V. Melkikh
Entropy 2004, 6, 223 232 Full Paper Entropy ISSN 1099-4300 www.mdpi.org/entropy/ INTERNAL STRUCTURE OF ELEMENTARY PARTICLE AND POSSIBLE DETERMINISTIC MECHANISM OF BIOLOGICAL EVOLUTION Alexey V. Melkikh
Monte Carlo and cold gases. Lode Pollet.
 Monte Carlo and cold gases Lode Pollet lpollet@physics.harvard.edu 1 Outline Classical Monte Carlo The Monte Carlo trick Markov chains Metropolis algorithm Ising model critical slowing down Quantum Monte
Monte Carlo and cold gases Lode Pollet lpollet@physics.harvard.edu 1 Outline Classical Monte Carlo The Monte Carlo trick Markov chains Metropolis algorithm Ising model critical slowing down Quantum Monte
The Evolution of Sex Chromosomes through the. Baldwin Effect
 The Evolution of Sex Chromosomes through the Baldwin Effect Larry Bull Computer Science Research Centre Department of Computer Science & Creative Technologies University of the West of England, Bristol
The Evolution of Sex Chromosomes through the Baldwin Effect Larry Bull Computer Science Research Centre Department of Computer Science & Creative Technologies University of the West of England, Bristol
Derivation of amplitude equations for nonlinear oscillators subject to arbitrary forcing
 PHYSICAL REVIEW E 69, 066141 (2004) Derivation of amplitude equations for nonlinear oscillators subject to arbitrary forcing Catalina Mayol, Raúl Toral, and Claudio R. Mirasso Department de Física, Universitat
PHYSICAL REVIEW E 69, 066141 (2004) Derivation of amplitude equations for nonlinear oscillators subject to arbitrary forcing Catalina Mayol, Raúl Toral, and Claudio R. Mirasso Department de Física, Universitat
Citation PHYSICAL REVIEW E (2005), 72(1) RightCopyright 2005 American Physical So
 TitlePhase transitions in multiplicative Author(s) Shimazaki, H; Niebur, E Citation PHYSICAL REVIEW E (2005), 72(1) Issue Date 2005-07 URL http://hdl.handle.net/2433/50002 RightCopyright 2005 American
TitlePhase transitions in multiplicative Author(s) Shimazaki, H; Niebur, E Citation PHYSICAL REVIEW E (2005), 72(1) Issue Date 2005-07 URL http://hdl.handle.net/2433/50002 RightCopyright 2005 American
Basic modeling approaches for biological systems. Mahesh Bule
 Basic modeling approaches for biological systems Mahesh Bule The hierarchy of life from atoms to living organisms Modeling biological processes often requires accounting for action and feedback involving
Basic modeling approaches for biological systems Mahesh Bule The hierarchy of life from atoms to living organisms Modeling biological processes often requires accounting for action and feedback involving
The fate of beneficial mutations in a range expansion
 The fate of beneficial mutations in a range expansion R. Lehe (Paris) O. Hallatschek (Göttingen) L. Peliti (Naples) February 19, 2015 / Rutgers Outline Introduction The model Results Analysis Range expansion
The fate of beneficial mutations in a range expansion R. Lehe (Paris) O. Hallatschek (Göttingen) L. Peliti (Naples) February 19, 2015 / Rutgers Outline Introduction The model Results Analysis Range expansion
Stochastically driven genetic circuits
 Stochastically driven genetic circuits CHAOS 16, 026103 2006 L. S. Tsimring Institute for Nonlinear Science, University of California, San Diego, La Jolla, California 92093-0402 D. Volfson Institute for
Stochastically driven genetic circuits CHAOS 16, 026103 2006 L. S. Tsimring Institute for Nonlinear Science, University of California, San Diego, La Jolla, California 92093-0402 D. Volfson Institute for
Energy spectrum for a short-range 1/r singular potential with a nonorbital barrier using the asymptotic iteration method
 Energy spectrum for a short-range 1/r singular potential with a nonorbital barrier using the asymptotic iteration method A. J. Sous 1 and A. D. Alhaidari 1 Al-Quds Open University, Tulkarm, Palestine Saudi
Energy spectrum for a short-range 1/r singular potential with a nonorbital barrier using the asymptotic iteration method A. J. Sous 1 and A. D. Alhaidari 1 Al-Quds Open University, Tulkarm, Palestine Saudi
The Moran Process as a Markov Chain on Leaf-labeled Trees
 The Moran Process as a Markov Chain on Leaf-labeled Trees David J. Aldous University of California Department of Statistics 367 Evans Hall # 3860 Berkeley CA 94720-3860 aldous@stat.berkeley.edu http://www.stat.berkeley.edu/users/aldous
The Moran Process as a Markov Chain on Leaf-labeled Trees David J. Aldous University of California Department of Statistics 367 Evans Hall # 3860 Berkeley CA 94720-3860 aldous@stat.berkeley.edu http://www.stat.berkeley.edu/users/aldous
arxiv: v1 [cond-mat.stat-mech] 6 Mar 2008
![arxiv: v1 [cond-mat.stat-mech] 6 Mar 2008 arxiv: v1 [cond-mat.stat-mech] 6 Mar 2008](/thumbs/86/93817801.jpg) CD2dBS-v2 Convergence dynamics of 2-dimensional isotropic and anisotropic Bak-Sneppen models Burhan Bakar and Ugur Tirnakli Department of Physics, Faculty of Science, Ege University, 35100 Izmir, Turkey
CD2dBS-v2 Convergence dynamics of 2-dimensional isotropic and anisotropic Bak-Sneppen models Burhan Bakar and Ugur Tirnakli Department of Physics, Faculty of Science, Ege University, 35100 Izmir, Turkey
Modeling Networks of Evolving Populations
 Modeling Networks of Evolving Populations Sean Elliott under the direction of Dominic Skinner Department of Mathematics Massachusetts Institute of Technology and the Research Science Institute Abstract
Modeling Networks of Evolving Populations Sean Elliott under the direction of Dominic Skinner Department of Mathematics Massachusetts Institute of Technology and the Research Science Institute Abstract
Chapter 2 Ensemble Theory in Statistical Physics: Free Energy Potential
 Chapter Ensemble Theory in Statistical Physics: Free Energy Potential Abstract In this chapter, we discuss the basic formalism of statistical physics Also, we consider in detail the concept of the free
Chapter Ensemble Theory in Statistical Physics: Free Energy Potential Abstract In this chapter, we discuss the basic formalism of statistical physics Also, we consider in detail the concept of the free
