LETTERS. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra
|
|
|
- Terence Garrison
- 5 years ago
- Views:
Transcription
1 Vol March 2008 doi: /nature06597 LETTERS Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra Yu He 1, Tao Ye 1, Min Su 2, Chuan Zhang 1, Alexander E. Ribbe 1, Wen Jiang 2 & Chengde Mao 1 DNA is renowned for its double helix structure and the base pairing that enables the recognition and highly selective binding of complementary DNA strands. These features, and the ability to create DNA strands with any desired sequence of bases, have led to the use of DNA rationally to design various nanostructures and even execute molecular computations 1 4. Of the wide range of self-assembled DNA nanostructures reported, most are one- or two-dimensional 5 9. Examples of three-dimensional DNA structures include cubes 10, truncated octahedra 11, octohedra 12 and tetrahedra 13,14, which are all comprised of many different DNA strands with unique sequences. When aiming for large structures, the need to synthesize large numbers (hundreds) of unique DNA strands poses a challenging design problem 9,15. Here, we demonstrate a simple solution to this problem: the design of basic DNA building units in such a way that many copies of identical units assemble into larger three-dimensional structures. We test this hierarchical self-assembly concept with DNA molecules that form three-point-star motifs, or tiles. By controlling the flexibility and concentration of the tiles, the one-pot assembly yields tetrahedra, dodecahedra or buckyballs that are tens of nanometres in size and comprised of four, twenty or sixty individual tiles, respectively. We expect that our assembly strategy can be adapted to allow the fabrication of a range of relatively complex three-dimensional structures. Our approach to forming DNA polyhedra is a one-pot selfassembly process illustrated in Fig. 1: individual single strands of DNA first assemble into sticky-ended, three-point-star motifs (tiles), which then further assemble into polyhedra through sticky-end association between the tiles. The three-point-star motif contains a threefold rotational symmetry and consists of seven strands: a long repetitive central strand (blue-red; strand L or L9), three identical medium strands (green; strand M), and three identical short peripheral strands (black; strand S). At the centre of the motif are three single-stranded loops (coloured red). The flexibility of the motif can be easily adjusted by varying the loop length, with increased loop length increasing tile flexibility. The termini of each branch of the tile carry two complementary, four-base-long, single-stranded overhangs, or sticky ends. Association between the sticky-ends allows the tiles to further assemble into larger structures such as the polyhedra described here. The three-point-star motif has been used for the assembly of flat two-dimensional (2D) crystals 16,17, where neighbouring units face in opposite directions of the crystal plane to cancel the intrinsic curvature of the DNA tiles. Because polyhedra are closed threedimensional (3D) objects containing a finite number of component tiles, we reasoned that three factors would promote polyhedron formation. (1) If all component DNA tiles face in the same direction, their curvatures would add up and promote the formation of closed structures. For example, some closed DNA tubular structures have been observed when all DNA tiles face the same side of the crystal plane 7. This requirement can be easily satisfied by choosing the length of each pseudo-continuous DNA duplex in the final structures to be four turns (42 bases). (2) Self-assembly is an inter-unit process. This means that higher (micromolar) DNA concentrations favour large assemblies such as flat 2D crystals, whereas lower DNA concentrations favour small assemblies such as polyhedra. This concentration-dependent kinetic effect should also provide some control over polyhedral size. (3) 2D crystal formation was found to require loops that are two to three bases long 17. Elongating the loops increases tile flexibility; this should prevent the assembly of DNA stars into large 2D crystals and instead promote the formation of smaller structures. We first tested this hypothesis by assembling a DNA tetrahedron from four three-point-star tiles. Each tile sits at a vertex, and its branches each associate with a branch from another tile to form the edges of the tetrahedron. The assembled tiles at the four vertices retain the threefold rotational symmetry of the free, individual star tiles, but are no longer planar. In fact, they are significantly bent and thus need to be quite flexible. To provide this flexibility, the loop length is designed to be five bases long. This ensures that the DNA stars will associate with each other under hybridization conditions to form highly flexible assemblies, which allows the free sticky-ends in the assemblies to meet and associate with each other to yield closed S M LL Tetrahedron Loop: 5 bases DNA: 75 nm Dodecahedron Loop: 3 bases DNA: 50 nm Buckyball Loop: 3 bases DNA: 500 nm Figure 1 Self-assembly of DNA polyhedra. Three different types of DNA single strands stepwise assemble into symmetric three-point-star motifs (tiles) and then into polyhedra in a one-pot process. There are three singlestranded loops (coloured red) in the centre of the complex. The final structures (polyhedra) are determined by the loop length (3 or 5 bases long) and the DNA concentration. 1 Department of Chemistry, 2 Markey Center for Structural Biology and Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA Nature Publishing Group
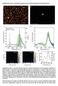
2 NATURE Vol March 2008 LETTERS structures (without any free sticky-ends). The size of the closed structures is concentration-dependent. At sufficiently low DNA concentration, we expect the formation of tetrahedra because they are the smallest closed structures that can form without deformation of the pseudo-continuous DNA duplexes. The DNA tetrahedra form when solutions containing the DNA strands are mixed and slowly cooled from 95 uc to room temperature (22 uc). Non-denaturing polyacrylamide gel electrophoresis (PAGE) of the resultant solution shows that for DNA concentrations of,100 nm, the dominant DNA complex contains four star tiles (Supplementary Fig. 1). Dynamic light scattering (DLS) provides a direct measurement of the physical sizes of the dissolved DNA complexes (Fig. 2a), yielding an apparent hydrodynamic radius of, nm (s.d.). This value agrees with the radius of the circumscribed sphere of the DNA tetrahedron model (10.9 nm), assuming 0.33 nm per base pair for the pitch and 2 nm for the diameter of a DNA duplex, respectively. To provide direct evidence for the self-assembly of DNA into tetrahedra, we imaged the samples using atomic force microscopy (AFM) and cryogenic transmission electronic microscopy (cryo- EM). DNA species appear as uniform-sized, triangular particles when imaged by AFM in air (Fig. 2b). Strong electrostatic interactions with the substrate and dehydration cause the 3D DNA tetrahedron to collapse into a 2D object with a triangle shape, which is consistent with the features observed by AFM. The most convincing experimental support for tetrahedron formation comes from cryo-em analysis (Fig. 2c, d), with the images showing that most particles have tetrahedral shapes of the expected size. The yield of correctly assembled DNA tetrahedra is,90%, as estimated from the number of particles observed by cryo-em and by gel electrophoretic analysis (Supplementary Fig. 2). We reconstructed from the experimentally observed particles a 3D structure for the DNA tetrahedron at 2.6 nm resolution, using single-particle 3D reconstruction 18. The computed projections from this 3D reconstruction match well to the individual particle images (Fig. 2d) and the class averages of raw particle images with similar views (Supplementary Fig. 4). In the reconstructed 3D tetrahedron structure (Fig. 2e), the observed edges are 16 nm long, nicely matching the designed model (16.2 nm). The thickness and width of the edge is also consistent with two DNA duplexes arranged side by side in the design. At the threefold vertices, a depression is visible, which is consistent with the central cavity in star motif. To demonstrate the versatility of the hierarchical self-assembly approach, we produced a DNA dodecahedron consisting of 20 three-point-star tiles, 12 faces (pentagons), and 30 edges. Compared to a tetrahedron, dodecahedra are less curved and the star tiles need not be nearly as flexible. Thus, the length of the central single-stranded loop is reduced to three bases long to make the star tiles much stiffer. At a low DNA concentration (50 nm), DNA star tiles readily assemble into dodecahedra (Fig. 3). DLS measurements (Fig. 3a) reveal an apparent hydrodynamic radius for the assembled objects of, nm (s.d.), which agrees with the value (23.6 nm) estimated from the designed structural model. AFM imaging in air reveals circular features with uniform sizes (Fig. 3b) and apparent diameters of,70 nm. This appearance of the assembled objects can be explained by DNA dehydration and strong electrostatic interactions between the DNA and mica substrate surface, which collapses the DNA dodecahedra into two layers. A close-up view (Fig. 3b inset) shows a component pentagon located at the centre of the top layer, which is in reasonable agreement with a collapsed 2D projection of a dodecahedron. The strongest experimental evidence for dodecahedron formation is provided by cryo-em analysis (Fig. 3c e), which reveals dodecahedron-shaped objects of the expected size (in the boxed areas in Fig. 3c and Supplementary Fig. 6). Single-particle reconstruction using experimentally observed particles yields a 3D map of the DNA dodecahedron. An icosahedral symmetry, which shares the same symmetry as a dodecahedron, was imposed during the reconstruction, resulting in a well-defined dodecahedron structure (with a resolution of 2.8 nm). The computed projections from this 3D reconstruction match well to the individual particle images (Fig. 3d) and the class averages of raw particle images with similar views (Supplementary Fig. 7). The radius of the circumscribed sphere of the reconstructed dodecahedron model is 23.5 nm, consistent with both the design and the DLS data. Each edge is 4 nm wide and 2 nm thick, c d Intensity (a. u.) 25 nm ,000 10,000 Radius (nm) 25 nm e 0 nm 3.0 nm 5 nm Figure 2 Characterization of the DNA tetrahedron by DLS, AFM and Cryo- EM. a, DLS shows the size histogram of DNA tetrahedron. b, An AFM image of DNA tetrahedra and a close-up view (inset). A height scale bar is shown at the bottom. c, A representative cryo-em image. White boxes indicate the DNA particles. For a magnified large view field, see Supplementary Fig. 3. d, Raw cryo-em images of individual particles and the corresponding projections of the DNA tetrahedron 3D structure reconstructed from the cryo-em images. These particles are selected from different image frames to represent views at different orientations. e, Three views of the reconstructed DNA tetrahedron structure Nature Publishing Group 199
3 LETTERS NATURE Vol March 2008 a b c d 24.0 ± 1.8 nm (s.d.) Intensity (a. u.) 25 nm ,000 10,000 Radius (nm) 200 nm c 50 nm d e 0 nm 3.4 nm 20 nm Figure 3 A DNA dodecahedron. a, Size histogram of the DNA dodecahedron measured by DLS. b, c, An AFM image (b) and a cryo-em image (c) of the DNA assemblies. For a magnified large view field, see Supplementary Fig. 6. d, Individual raw cryo-em images and the which are reasonable dimensions for two associated branches from two neighbouring star tiles. A truncated icosahedron structure is another type of highly symmetric polyhedron. It contains 60 vertexes, 90 edges and 32 faces (12 pentagons and 20 hexagons). Two well-known examples of such a structure are soccer balls and Buckminsterfullerene molecules (or buckyballs, C 60 ). It remains challenging to rationally design a corresponding projections of the DNA dodecahedron 3D structure reconstructed from cryo-em images. These particles are selected from different image frames to represent views at different orientations. e, Three views of the DNA dodecahedron 3D structure. molecular system that can self-assemble into such a complex structure. However, at a high DNA concentration (500 nm), the star tiles (with three-base-long, single-stranded loops at the centre) readily assemble into the buckyball structure as suggested by DLS analysis, AFM imaging and cryo-em analysis (Fig. 4). The DLS measurement indicates that the DNA assemblies have an apparent hydrodynamic radius of nm (s.d.), close to the calculated radius of the a b c d Intensity (a. u.) 25 nm ,000 10,000 Radius (nm) 200 nm 100 nm e 0 nm 3.0 nm 20 nm Figure 4 A DNA buckyball. a, Size histogram of the DNA buckyball measured by DLS. b, c, An AFM image (b) and a cryo-em image (c) of the DNA assemblies. For a magnified large view field, see Supplementary Fig. 9. d, Individual raw cryo-em images and the corresponding projections of the Nature Publishing Group DNA buckyball 3D structure reconstructed from cryo-em images. These particles are selected from different image frames to represent views at different orientations. e, Three views of the DNA buckyball structure reconstructed from cryo-em images.
4 NATURE Vol March 2008 LETTERS structural model (41.0 nm). However, the polydispersity of these assemblies is higher than that of the tetrahedron and dodecahedron assemblies. AFM imaging shows the DNA assemblies as uniformsized, round, collapsed nanostructures with a diameter of,110 nm. Once again, the most direct evidence for buckyball formation comes from cryo-em imaging (Fig. 4c and Supplementary Fig. 9). Using hand-picked particles, a buckyball 3D structure can be obtained from reconstruction. However, the size distribution and the distortion of the particles in cryo-em images look much worse than that from the AFM images. Many smaller particles and some networks are visible. These undesired particles introduce some uncertainties to the reconstruction process and lower the quality of the reconstructed structures. The 3D reconstruction merely represents the average structure of these distorted particles. Nevertheless, the resulting structure resembles a buckyball. We note that the assembly yield decreases as the size of the target structure increases: the assembly yield for the tetrahedron is, 90%, but only 76% for the larger dodecahedron and 69% for the even larger buckyball (as analysed by agarose gel electrophoresis; see Supplementary Figure S2). This trend might be rationalized by the fact that larger structures require more star tiles and are thus more difficult to assemble. However, we also note that in the cryo-em images of the DNA buckyballs, smaller objects and networks are abundant; this highlights that large structures such as buckyballs are easier to deform and break into open networks or smaller aggregates due to external disturbance (as will occur during sample preparation for imaging). The more demanding assembly and the increased lability are both likely to contribute to the apparently decreasing assembly yield for larger structures. However, the current data are not sufficient to evaluate the specific contribution of each of these factors. We have shown that DNA can be programmed to assemble into well-defined, regular polyhedra that might find use as synthetic nanocontainers or 3D structural scaffolds. The current study used only DNA three-point-star motifs as primary building blocks, but we are currently investigating whether our hierarchical assembly strategy can be applied to other DNA motifs to prepare an even wider range of 3D objects. METHODS SUMMARY Oligonucleotides. DNA sequences were adapted from previous works, which were originally designed by the SEQUIN 19 computer program: central longstrand L (blue-red): aggcaccatcgtaggtttcttgccaggcaccatcgtaggtttcttgccaggcaccatcgtaggtttcttgcc; central long strand L9 (blue-red): aggcaccatcgtaggtttaacttgccaggcaccatcgtaggtttaacttgccaggcaccatcgtaggtttaacttgcc; medium strand M (green): tagcaacctgcctggcaagcctacgatggacacggtaacgcc; short peripheral strand S (black): ttaccgtgtggttgctaggcg. Formation of DNA complexes. DNA strands were combined according to the correct molecular ratios in a tris-acetic-edta-mg 21 (TAE/Mg 21 ) buffer. Tetrahedron formation used strands L9, M and S; dodecahedron and buckyball formation used strands L, M and S. DNA assembly involved cooling solutions from 95 uc to room temperature over 48 h, using concentrations as indicated in Fig. 1 unless specifically stated otherwise. DNA samples were then directly used for characterization, without further fractionation or purification. Non-denaturing PAGE and AFM imaging and DLS. Gels containing 4% polyacrylamide were run at 4 uc. For AFM, DNA samples were imaged in tappingmode on a Multimode AFM with a Nanoscope IIIa controller (Veeco) using oxide-sharpened silicon probes in air at 22 uc. For DLS, 12 ml DNA sample solutions were measured by DynaPro 99 (Protein Solutions/Wyatt) with laser wavelength of 824 nm at 22 uc. Cryo-EM imaging. DNA sample solutions were concentrated to,3 mm, spread onto Quantifoil grids, then plunge-frozen. Data were recorded using a Gatan 4, ,080 pixel charge-coupled device (CCD) camera in a Philips CM200 transmission electron microscope with field-emission gun operating at 200 kv accelerating voltage. Single-particle reconstruction. 3D reconstructions of the DNA polyhedra used the single-particle image processing software EMAN 18. Correct symmetry for each of the polyhedra was established by processing the images, assuming different symmetries and finding the symmetry that yields a 3D reconstruction consistent with the particle images (Supplementary Figs 5 and 8). Final 3D maps were visualized using UCSF Chimera software 20. Full Methods and any associated references are available in the online version of the paper at Received 10 July; accepted 18 December Seeman, N. C. DNA in a material world. Nature 421, (2003). 2. Seeman, N. C. DNA enables nanoscale control of the structure of matter. Q. Rev. Biophys. 38, (2005). 3. Feldkamp, U. & Niemeyer, C. M. Rational design of DNA nanoarchitectures. Angew. Chem. Int. Edn Engl. 45, (2006). 4. Adleman, L. M. Molecular computation of solutions to combinatorial problems. Science 266, (1994). 5. Winfree, E., Liu, F. R., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, (1998). 6. Rothemund, P. W. K., Papadakis, N. & Winfree, E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2, (2004). 7. Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. DNA-templated selfassembly of protein arrays and highly conductive nanowires. Science 301, (2003). 8. Scheffler, M., Dorenbeck, A., Jordan, S., Wustefeld, M. & von Kiedrowski, G. Selfassembly of trisoligo-nucleotidyls: the case for nano-acetylene and nanocyclobutadiene. Angew. Chem. Int. Edn Engl. 38, (1999). 9. Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, (2006). 10. Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, (1991). 11. Zhang, Y. W. & Seeman, N. C. Construction of a DNA-truncated octahedron. J. Am. Chem. Soc. 116, (1994). 12. Shih, W. M., Quispe, J. D. & Joyce, G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, (2004). 13. Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, (2005). 14. Goodman, R. P., Berry, R. M. & Turberfield, A. J. The single-step synthesis of a DNA tetrahedron. Chem. Commun (2004). 15. Douglas, S. M., Chou, J. J. & Shih, W. M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, (2007). 16. He, Y., Chen, Y., Liu, H. P., Ribbe, A. E. & Mao, C. D. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J. Am. Chem. Soc. 127, (2005). 17. He, Y. & Mao, C. D. Balancing flexibility and stress in DNA nanostructures. Chem. Commun (2006). 18. Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: Semiautomated software for highresolution single-particle reconstructions. J. Struct. Biol. 128, (1999). 19. Seeman, N. C. Denovo design of sequences for nucleic-acid structural engineering. J. Biomol. Struct. Dyn. 8, (1990). 20. Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF chimera. J. Struct. Biol. 157, (2007). Supplementary Information is linked to the online version of the paper at Acknowledgements We thank H. Liu for help with the initial DLS experiment. This work was supported by the National Science Foundation. AFM and DLS studies were carried out in the Purdue Laboratory for Chemical Nanotechnology (PLCN). The cryo-em images were taken in the Purdue Biological Electron Microscopy Facility and the Purdue Rosen Center for Advanced Computing (RCAC) provided the computational resource for the 3D reconstructions. Author Information Reprints and permissions information is available at Correspondence and requests for materials should be addressed to C.M. (mao@purdue.edu) Nature Publishing Group 201
5 doi: /nature06597 METHODS Oligonucleotides. All oligonucleotides were purchased from IDT, Inc. and purified by 20% denaturing polyacrylamide gel electrophoresis (PAGE). Formation of DNA complexes. The TAE/Mg 21 buffer contained 40 mm Tris base (ph 8.0), 20 mm acetic acid, 2 mm EDTA and 12.5 mm magnesium acetate. At the same DNA concentrations as in Fig. 1, DNA samples were directly used after assembly for AFM imaging and DLS studies, but concentrated to,3 mm for cryo-em imaging. No fractionationation or purification was needed after assembly of DNA complexes. Non-denaturing PAGE. Gels containing 4% polyacrylamide (19:1 acrylamide/ bisacrylamide) were run on a FB-VE10-1 electrophoresis unit (FisherBiotech) at 4 uc (100 V, constant voltage). The running buffer was TAE/Mg 21 buffer. After electrophoresis, the gels were stained with Stains-All (Sigma) and scanned. PAGE is only suited for characterization of DNA tetrahedra, but not for dodecahedra and buckyballs because these are too large to migrate into the gel matrix. AFM imaging. A drop of 2 ml DNA solution was spotted onto freshly cleaved mica surface, and kept for 10 s to achieve strong adsorption. The sample drop was then washed off by 30 ml 2 mm magnesium acetate solution, and dried by compressed air. DNA samples were imaged in tapping mode on a Multimode AFM with Nanoscope IIIa controller (Veeco) using oxide-sharpened silicon probes having a resonance frequency in the range of khz (MikroMasch NSC15). The tip surface interaction was minimized by optimizing the scan set-point to the highest possible value. AFM imaging was performed at 22 uc. Cryo-EM imaging. DNA sample solutions were concentrated to,3 mm (in terms of DNA tiles) with Microcon YM-100 (100 kda) Centrifugal Filter Units. A drop of 3 ml concentrated DNA solution was pipetted onto a Quantifoil grid. Then, the grid was blotted and immediately plunge-frozen into ethane slush cooled by liquid nitrogen. The images were taken under low-dose condition to minimize radiation damages to the samples. To enhance the image contrast, under-focuses in the range of 2 4 mm were used to record the images. The calibrated magnifications used for DNA tetrahedron, dodecahedron, and buckyballs were 368,050, 352,260 and 337,760, respectively, resulting in pixel sizes of 2.21, 2.87 and 3.97 Å. Initial model. For all DNA polyhedra, initial models were built using 100 randomly selected raw particles. The initial orientation of individual particle was randomly assigned within the corresponding asymmetry unit of the polyhedron. DNA tetrahedron. 449 particles were used for the single-particle reconstruction. 110 reference projections in the tetrahedral asymmetric unit were generated at an angular interval of 4u. A projection matching algorithm was then used to determine the centre and orientation of raw particles in the iterative refinement. The tetrahedral symmetry was imposed during the reconstruction. The map resolution was determined to be at 2.6 nm using the Fourier shell correlation (0.5 threshold criterion) of two 3D maps independently built from half data sets. Control reconstructions without imposing any symmetry or with lower symmetries imposed were performed to check that particles indeed have tetrahedron symmetry (Supplementary Fig. 3). DNA dodecahedron. 725 particles were used for the single-particle reconstruction. 99 reference projections were generated in the icosahedral asymmetric unit with an angular interval of 2u. A projection matching algorithm was used to determine the particle centre and orientation. Icosahedral symmetry was imposed during reconstruction. The final resolution and map quality were evaluated as those of the DNA tetrahedron, indicating a resolution of 2.8 nm. DNA buckyball. 514 particles were used for the buckyball reconstruction. 30 reference projections resulted from an angular interval of 4u in an icosahedral asymmetric unit. Icosahedral symmetry was imposed during reconstruction Nature Publishing Group
6 "#$%$&'"&%()*#(+,-..#/0(1)2+)*1//#3$"&) *45$%/2(#&4(%$)62(1'#7$%) "#$%&'()"%&*+,-#&./#(,0/(,1&23%4(,5%6789+::%&;%,<+(,1 =./%,15%*() (6% +D(1%EO C/%D+C(3 )L :(,5E (6% E+5%E : "#$#& 89: "#$%& 89: * "#$%$#& 89:* "#$%$%& "(((((&. "(((((& ' "(((((& + 89:* "#$%$#& %'()* 89:* +,()* #,-()* %--()*.--()* "(((((&, "(((((&. "(((((& 0 "(((((&, "(((((& % "(((((& 0 "(((((& / "(((((& % "(((((& /
7 "#$%&' (+* (1(6)E% 1%3 FR8SJK (,(3BE+E E%3L? TU ( C(D%6(8 1%3 +D(1%& (EE%D:3+,1 B+%35E (6% P+@/ (, +D(1% A)3B/%56(F%EA%C+(33B5)5%C(/%56((,5:#CVB:(33EK/(N%5+LL+C#3@+%E@)A%,%@6(@%+,@)1%3 +,5+N+5#(3@+3% M>2M)5%C(/%56), M>2_#CVB:(33 "(((((&.-( "(((((& /-( "(((((& 0( 2EE%D:3BB+%35FJKY ZR8T [\8] \^8Z
8 SR,D )-.- +-,- Qa Ta ]a Ia
9 "#$%&' (.* 2 +@E C3(EE (N%6(1%5 +D(1%E C)66%EA),5+,1
10 "#$%&' )L M>2 F(& :K 6%3(4%5 FC& 5K )L +D(1%E L6)D ),3B FCK.] )6 F5K,) +DA)E%58 F(K F:K FCK F5K
11 )- +- QRR,D Qa Ta ]a Ia
12 "#$%&' (1* 2 M>2 5)5%C(/%56),Y +@E C3(EE (N%6(1%5 +D(1%E C)66%EA),5+,1
13 "#$%&' )L M>2 5)5%C(/%56), FS L)35 F:K )L +D(1%E L6)D ),3B.S +DA)E%58 /+1/%6 F(K F:K
14 +-,- )-.- TRR,D Qa Ta ]a Ia
15 "#$%&' ()4* 2 M>2 :#CVB:(33Y +@E C3(EE (N%6(1%5 +D(1%E C)66%EA),5+,1
16 Supramolecular structures : Nature 1 of 1 3/13/2008 9:36 AM Search " Advanced search Editor's Summary 13 March 2008 Supramolecular structures A variety of patterned materials and nanostructures have been made from DNA, by exploiting its programmability to control molecular interactions. But making larger, more complex three-dimensional structures with current fabrication methods would require hundreds of unique DNA strands, an impractical proposition. Help is at hand. A team from Purdue University has developed a modular approach that can be likened to a DNA equivalent of Lego bricks. A few DNA molecules are programmed to fold into a basic structural unit, with four, twenty or sixty copies of that unit then assembling according to reaction conditions into tetrahedra, dodecahedra or buckyballs, respectively. Other complex structures should also be accessible using this strategy. LETTER Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra Yu He, Tao Ye, Min Su, Chuan Zhang, Alexander E. Ribbe, Wen Jiang & Chengde Mao doi: /nature06597 First paragraph (/nature/journal/v452/n7184/abs/nature06597.html) Full Text (/nature/journal/v452/n7184 /full/nature06597.html) PDF (3,022K) (/nature/journal/v452/n7184/pdf/nature06597.pdf) Supplementary information (/nature/journal/v452/n7184/suppinfo/nature06597.html)
SUPPLEMENTARY INFORMATION
 Hierarchical Self-Assembly of Symmetric Supramolecular Polyhedra Yu He, Tao Ye, Min Su, Chuan Zhang, Alexander E. Ribbe, Wen Jiang & Chengde Mao Figure S1. Non-denaturing polyacrylamide gel electrophoresis
Hierarchical Self-Assembly of Symmetric Supramolecular Polyhedra Yu He, Tao Ye, Min Su, Chuan Zhang, Alexander E. Ribbe, Wen Jiang & Chengde Mao Figure S1. Non-denaturing polyacrylamide gel electrophoresis
Programmable DNA Self-Assemblies for Nanoscale Organization of Ligands and Proteins
 Programmable DNA Self-Assemblies for Nanoscale Organization of Ligands and Proteins NANO LETTERS 2005 Vol. 5, No. 4 729-733 Sung Ha Park, Peng Yin, Yan Liu, John H. Reif, Thomas H. LaBean,*, and Hao Yan*,
Programmable DNA Self-Assemblies for Nanoscale Organization of Ligands and Proteins NANO LETTERS 2005 Vol. 5, No. 4 729-733 Sung Ha Park, Peng Yin, Yan Liu, John H. Reif, Thomas H. LaBean,*, and Hao Yan*,
The Design and Fabrication of a Fully Addressable 8-tile DNA Lattice
 The Design and Fabrication of a Fully Addressable 8-tile DNA Lattice Chris Dwyer 1,3, Sung Ha Park 2, Thomas LaBean 3, Alvin Lebeck 3 1 Dept. of Electrical and Computer Engineering, Duke University, Durham,
The Design and Fabrication of a Fully Addressable 8-tile DNA Lattice Chris Dwyer 1,3, Sung Ha Park 2, Thomas LaBean 3, Alvin Lebeck 3 1 Dept. of Electrical and Computer Engineering, Duke University, Durham,
Nanoscale Patterning of Self-assembled Monolayers using DNA Nanostructure Templates
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Nanoscale Patterning of Self-assembled Monolayers using DNA Nanostructure Templates Sumedh P. Surwade,
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Nanoscale Patterning of Self-assembled Monolayers using DNA Nanostructure Templates Sumedh P. Surwade,
Supramolecular DNA nanotechnology. Faisal A. Aldaye
 Supramolecular DA nanotechnology Faisal A. Aldaye Department of Chemistry, McGill University Current address: Department of Systems Biology, Harvard University 200 Longwood Avenue, Boston, MA 02115, USA
Supramolecular DA nanotechnology Faisal A. Aldaye Department of Chemistry, McGill University Current address: Department of Systems Biology, Harvard University 200 Longwood Avenue, Boston, MA 02115, USA
A Study of DNA Tube Formation Mechanisms Using 4-, 8-, and 12-Helix DNA Nanostructures
 Published on Web 03/02/2006 A Study of DNA Tube Formation Mechanisms Using 4-, 8-, and 12-Helix DNA Nanostructures Yonggang Ke, Yan Liu, Junping Zhang, and Hao Yan* Contribution from the Department of
Published on Web 03/02/2006 A Study of DNA Tube Formation Mechanisms Using 4-, 8-, and 12-Helix DNA Nanostructures Yonggang Ke, Yan Liu, Junping Zhang, and Hao Yan* Contribution from the Department of
Supporting information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Supporting information Self-assembled nanopatch with peptide-organic multilayers and mechanical
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Supporting information Self-assembled nanopatch with peptide-organic multilayers and mechanical
Self-Assembly of DNA Arrays into Multilayer Stacks
 Article Subscriber access provided by HOUSTON ACADEMY OF MEDICINE Self-Assembly of DNA Arrays into Multilayer Stacks Alexey Y. Koyfman, Sergei N. Magonov, and Norbert O. Reich Langmuir, 2009, 25 (2), 1091-1096
Article Subscriber access provided by HOUSTON ACADEMY OF MEDICINE Self-Assembly of DNA Arrays into Multilayer Stacks Alexey Y. Koyfman, Sergei N. Magonov, and Norbert O. Reich Langmuir, 2009, 25 (2), 1091-1096
DNA Computing by Self Assembly. Presented by Mohammed Ashraf Ali
 DNA Computing by Self Assembly Presented by Mohammed Ashraf Ali Outline Self- Assembly Examples and behaviour of self-assembly DNA Computing Tiling Theory Nanotechnology DNA Self-assembly Visualization
DNA Computing by Self Assembly Presented by Mohammed Ashraf Ali Outline Self- Assembly Examples and behaviour of self-assembly DNA Computing Tiling Theory Nanotechnology DNA Self-assembly Visualization
Angewandte. Finite-Size, Fully Addressable DNA Tile Lattices Formed by Hierarchical Assembly Procedures**
 DNA Nanostructures DOI: 10.1002/anie.200503797 Finite-Size, Fully Addressable DNA Tile Lattices Formed by Hierarchical Assembly Procedures** Sung Ha Park, Constantin Pistol, Sang Jung Ahn, John H. Reif,
DNA Nanostructures DOI: 10.1002/anie.200503797 Finite-Size, Fully Addressable DNA Tile Lattices Formed by Hierarchical Assembly Procedures** Sung Ha Park, Constantin Pistol, Sang Jung Ahn, John H. Reif,
Interdisciplinary Nanoscience Center University of Aarhus, Denmark. Design and Imaging. Assistant Professor.
 Interdisciplinary Nanoscience Center University of Aarhus, Denmark Design and Imaging DNA Nanostructures Assistant Professor Wael Mamdouh wael@inano.dk Molecular Self-assembly Synthesis, SPM microscopy,
Interdisciplinary Nanoscience Center University of Aarhus, Denmark Design and Imaging DNA Nanostructures Assistant Professor Wael Mamdouh wael@inano.dk Molecular Self-assembly Synthesis, SPM microscopy,
Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices
 Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices Hao Yan, Thomas H. LaBean, Liping Feng, and John H. Reif* Department of Computer Science, Duke University, Durham, NC 27708
Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices Hao Yan, Thomas H. LaBean, Liping Feng, and John H. Reif* Department of Computer Science, Duke University, Durham, NC 27708
Directed Nucleation Assembly of Barcode Patterned DNA Lattices
 February 13, 2003 Directed Nucleation Assembly of Barcode Patterned DNA Lattices Hao Yan, Thomas H. LaBean, Liping Feng, John H. Reif* Department of Computer Science Duke University Durham, NC 27708 *To
February 13, 2003 Directed Nucleation Assembly of Barcode Patterned DNA Lattices Hao Yan, Thomas H. LaBean, Liping Feng, John H. Reif* Department of Computer Science Duke University Durham, NC 27708 *To
Intrinsic DNA Curvature of Double-Crossover Tiles
 Intrinsic DNA Curvature of Double-Crossover Tiles Seungjae Kim, 1 Junghoon Kim, 1 Pengfei Qian, 2 Jihoon Shin, 3 Rashid Amin, 3 Sang Jung Ahn, 4 Thomas H. LaBean, 5 Moon Ki Kim, 2,3 * and Sung Ha Park,
Intrinsic DNA Curvature of Double-Crossover Tiles Seungjae Kim, 1 Junghoon Kim, 1 Pengfei Qian, 2 Jihoon Shin, 3 Rashid Amin, 3 Sang Jung Ahn, 4 Thomas H. LaBean, 5 Moon Ki Kim, 2,3 * and Sung Ha Park,
Atomic Force Microscopy Characterization of Room- Temperature Adlayers of Small Organic Molecules through Graphene Templating
 Atomic Force icroscopy Characterization of Room- Temperature Adlayers of Small Organic olecules through Graphene Templating Peigen Cao, Ke Xu,2, Joseph O. Varghese, and James R. Heath *. Kavli Nanoscience
Atomic Force icroscopy Characterization of Room- Temperature Adlayers of Small Organic olecules through Graphene Templating Peigen Cao, Ke Xu,2, Joseph O. Varghese, and James R. Heath *. Kavli Nanoscience
Impact of multivalent charge presentation on peptide
 Supporting Information File 1 for Impact of multivalent charge presentation on peptide nanoparticle aggregation Daniel Schöne 1, Boris Schade 2, Christoph Böttcher 2 and Beate Koksch* 1 Address: 1 Institute
Supporting Information File 1 for Impact of multivalent charge presentation on peptide nanoparticle aggregation Daniel Schöne 1, Boris Schade 2, Christoph Böttcher 2 and Beate Koksch* 1 Address: 1 Institute
The Controlled Evolution of a Polymer Single Crystal
 Supporting Online Material The Controlled Evolution of a Polymer Single Crystal Xiaogang Liu, 1 Yi Zhang, 1 Dipak K. Goswami, 2 John S. Okasinski, 2 Khalid Salaita, 1 Peng Sun, 1 Michael J. Bedzyk, 2 Chad
Supporting Online Material The Controlled Evolution of a Polymer Single Crystal Xiaogang Liu, 1 Yi Zhang, 1 Dipak K. Goswami, 2 John S. Okasinski, 2 Khalid Salaita, 1 Peng Sun, 1 Michael J. Bedzyk, 2 Chad
Final Reading Assignment: Travels to the Nanoworld: pages pages pages
 Final Reading Assignment: Travels to the Nanoworld: pages 152-164 pages 201-214 pages 219-227 Bottom-up nanofabrication Can we assemble nanomachines manually? What are the components (parts)? nanoparticles
Final Reading Assignment: Travels to the Nanoworld: pages 152-164 pages 201-214 pages 219-227 Bottom-up nanofabrication Can we assemble nanomachines manually? What are the components (parts)? nanoparticles
A simple example of a polyhedral network or polynet, constructed from truncated octahedra and hexagonal prisms.
 Polyhedral Nets A simple example of a polyhedral network or polynet, constructed from truncated octahedra and hexagonal prisms. The building process indicated produces a labyrinth. The labyrinth graph
Polyhedral Nets A simple example of a polyhedral network or polynet, constructed from truncated octahedra and hexagonal prisms. The building process indicated produces a labyrinth. The labyrinth graph
History of 3D Electron Microscopy and Helical Reconstruction
 T H E U N I V E R S I T Y of T E X A S S C H O O L O F H E A L T H I N F O R M A T I O N S C I E N C E S A T H O U S T O N History of 3D Electron Microscopy and Helical Reconstruction For students of HI
T H E U N I V E R S I T Y of T E X A S S C H O O L O F H E A L T H I N F O R M A T I O N S C I E N C E S A T H O U S T O N History of 3D Electron Microscopy and Helical Reconstruction For students of HI
Fast ph-assisted functionalization of silver nanoparticles with monothiolated DNA
 Supporting Information for Fast ph-assisted functionalization of silver nanoparticles with monothiolated DNA Xu Zhang ab, Mark R. Servos b, and Juewen Liu* a a Department of Chemistry and Waterloo Institute
Supporting Information for Fast ph-assisted functionalization of silver nanoparticles with monothiolated DNA Xu Zhang ab, Mark R. Servos b, and Juewen Liu* a a Department of Chemistry and Waterloo Institute
SUPPORTING INFORMATION
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 SUPPORTING INFORMATION Materials Graphite powder (SP-1 graphite) was obtained from Bay carbon.
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 SUPPORTING INFORMATION Materials Graphite powder (SP-1 graphite) was obtained from Bay carbon.
Dumpling-Like Nanocomplex of Foldable Janus Polymer Sheet and Sphere
 Dumpling-Like Nanocomplex of Foldable Janus Polymer Sheet and Sphere Lei Gao, Ke Zhang, and Yongming Chen* Supporting Information Experimental Section Materials The triblock terpolymer, P2VP 310 -b-ptepm
Dumpling-Like Nanocomplex of Foldable Janus Polymer Sheet and Sphere Lei Gao, Ke Zhang, and Yongming Chen* Supporting Information Experimental Section Materials The triblock terpolymer, P2VP 310 -b-ptepm
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1513/dc1 Supporting Online Material for Structural Roles for Human Translation Factor eif3 in Initiation of Protein Synthesis Bunpote Siridechadilok, Christopher
www.sciencemag.org/cgi/content/full/310/5753/1513/dc1 Supporting Online Material for Structural Roles for Human Translation Factor eif3 in Initiation of Protein Synthesis Bunpote Siridechadilok, Christopher
Integer and Vector Multiplication by Using DNA
 Int. J. Open Problems Compt. Math., Vol. 1, No. 2, September 2008 Integer and Vector Multiplication by Using DNA Essam Al-Daoud 1, Belal Zaqaibeh 2, and Feras Al-Hanandeh 3 1 Computer Science Department,
Int. J. Open Problems Compt. Math., Vol. 1, No. 2, September 2008 Integer and Vector Multiplication by Using DNA Essam Al-Daoud 1, Belal Zaqaibeh 2, and Feras Al-Hanandeh 3 1 Computer Science Department,
Studies of Thermal Stability of Multivalent DNA Hybridization in a Nanostructured System
 Biophysical Journal Volume 97 July 2009 563 571 563 Studies of Thermal Stability of Multivalent DNA Hybridization in a Nanostructured System Jeanette Nangreave, Hao Yan,* and Yan Liu* Department of Chemistry
Biophysical Journal Volume 97 July 2009 563 571 563 Studies of Thermal Stability of Multivalent DNA Hybridization in a Nanostructured System Jeanette Nangreave, Hao Yan,* and Yan Liu* Department of Chemistry
Electronic Supplementary Information. Tubulation of Liposomes via the Interaction of Supramolecular Nanofibers
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Tubulation of Liposomes via the Interaction of Supramolecular
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Tubulation of Liposomes via the Interaction of Supramolecular
Active Tile Self Assembly:
 Active Tile Self Assembly: Simulating Cellular Automata at Temperature 1 Daria Karpenko Department of Mathematics and Statistics, University of South Florida Outline Introduction Overview of DNA self-assembly
Active Tile Self Assembly: Simulating Cellular Automata at Temperature 1 Daria Karpenko Department of Mathematics and Statistics, University of South Florida Outline Introduction Overview of DNA self-assembly
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry Supporting information for:
 Supporting information for: Preparation of 1-D Nanoparticle Superstructures with Tailorable Thicknesses using Gold-Binding Peptide Conjugates: New Insights into Fabrication Process and Mechanism Leekyoung
Supporting information for: Preparation of 1-D Nanoparticle Superstructures with Tailorable Thicknesses using Gold-Binding Peptide Conjugates: New Insights into Fabrication Process and Mechanism Leekyoung
Electronic supplementary information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2018 Electronic supplementary information Heterogeneous nucleation and growth of highly crystalline
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2018 Electronic supplementary information Heterogeneous nucleation and growth of highly crystalline
Supporting Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Supporting Information Self-healing supramolecular heterometallic gels based on synergistic effect
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Supporting Information Self-healing supramolecular heterometallic gels based on synergistic effect
Toward Modular Molecular Composite Nanosystems
 Toward Modular Molecular Composite Nanosystems K. Eric Drexler, PhD U.C. Berkeley 26 April 2009 Intended take-away messages: Paths are now open toward complex, self-assembled, heterogenous nanosystems
Toward Modular Molecular Composite Nanosystems K. Eric Drexler, PhD U.C. Berkeley 26 April 2009 Intended take-away messages: Paths are now open toward complex, self-assembled, heterogenous nanosystems
The sacrificial role of graphene oxide in stabilising Fenton-like catalyst GO Fe 3 O 4
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 The sacrificial role of graphene oxide in stabilising Fenton-like catalyst GO Fe 3 O 4 Nor Aida
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 The sacrificial role of graphene oxide in stabilising Fenton-like catalyst GO Fe 3 O 4 Nor Aida
Supplementary information
 Supplementary information doi: 10.1038/nchem.1025 Platinum nanocrystals selectively shaped using facet-specific peptide sequences Chin-Yi Chiu 1, Yujing Li 1, Lingyan Ruan 1, Xingchen Ye 2, Christopher
Supplementary information doi: 10.1038/nchem.1025 Platinum nanocrystals selectively shaped using facet-specific peptide sequences Chin-Yi Chiu 1, Yujing Li 1, Lingyan Ruan 1, Xingchen Ye 2, Christopher
sin" =1.22 # D "l =1.22 f# D I: In introduction to molecular electron microscopy - Imaging macromolecular assemblies
 I: In introduction to molecular electron microscopy - Imaging macromolecular assemblies Yifan Cheng Department of Biochemistry & Biophysics office: GH-S472D; email: ycheng@ucsf.edu 2/20/2015 - Introduction
I: In introduction to molecular electron microscopy - Imaging macromolecular assemblies Yifan Cheng Department of Biochemistry & Biophysics office: GH-S472D; email: ycheng@ucsf.edu 2/20/2015 - Introduction
ParM filament images were extracted and from the electron micrographs and
 Supplemental methods Outline of the EM reconstruction: ParM filament images were extracted and from the electron micrographs and straightened. The digitized images were corrected for the phase of the Contrast
Supplemental methods Outline of the EM reconstruction: ParM filament images were extracted and from the electron micrographs and straightened. The digitized images were corrected for the phase of the Contrast
Supporting Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Supporting Information High-k Polymer/Graphene Oxide Dielectrics for Low-Voltage Flexible Nonvolatile
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Supporting Information High-k Polymer/Graphene Oxide Dielectrics for Low-Voltage Flexible Nonvolatile
Nanotechnology? Source: National Science Foundation (NSF), USA
 2 2 Nanotechnology? Ability to work at the atomic, molecular and even sub-molecular levels in order to create and use material structures, devices and systems with new properties and functions Source:
2 2 Nanotechnology? Ability to work at the atomic, molecular and even sub-molecular levels in order to create and use material structures, devices and systems with new properties and functions Source:
Supporting Information
 Supporting Information Controlled Radical Polymerization and Quantification of Solid State Electrical Conductivities of Macromolecules Bearing Pendant Stable Radical Groups Lizbeth Rostro, Aditya G. Baradwaj,
Supporting Information Controlled Radical Polymerization and Quantification of Solid State Electrical Conductivities of Macromolecules Bearing Pendant Stable Radical Groups Lizbeth Rostro, Aditya G. Baradwaj,
Supporting Information
 Supporting Information Growth of Molecular Crystal Aggregates for Efficient Optical Waveguides Songhua Chen, Nan Chen, Yongli Yan, Taifeng Liu, Yanwen Yu, Yongjun Li, Huibiao Liu, Yongsheng Zhao and Yuliang
Supporting Information Growth of Molecular Crystal Aggregates for Efficient Optical Waveguides Songhua Chen, Nan Chen, Yongli Yan, Taifeng Liu, Yanwen Yu, Yongjun Li, Huibiao Liu, Yongsheng Zhao and Yuliang
Supporting Information s for
 Supporting Information s for # Self-assembling of DNA-templated Au Nanoparticles into Nanowires and their enhanced SERS and Catalytic Applications Subrata Kundu* and M. Jayachandran Electrochemical Materials
Supporting Information s for # Self-assembling of DNA-templated Au Nanoparticles into Nanowires and their enhanced SERS and Catalytic Applications Subrata Kundu* and M. Jayachandran Electrochemical Materials
The photoluminescent graphene oxide serves as an acceptor rather. than a donor in the fluorescence resonance energy transfer pair of
 Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 20XX The photoluminescent graphene oxide serves as an acceptor rather than a donor in the fluorescence
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 20XX The photoluminescent graphene oxide serves as an acceptor rather than a donor in the fluorescence
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Energy & Environmental Science. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information Self-supported formation of hierarchical
Electronic Supplementary Material (ESI) for Energy & Environmental Science. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information Self-supported formation of hierarchical
3D Motion of DNA-Au Nanoconjugates in Graphene Liquid Cell EM
 Supporting Information for 3D Motion of DNA-Au Nanoconjugates in Graphene Liquid Cell EM Qian Chen,,, Jessica Smith,, Jungwon Park,,1, Kwanpyo Kim,,2, Davy Ho, Haider I. Rasool,,!Alex Zettl,, A. Paul Alivisatos,,*!
Supporting Information for 3D Motion of DNA-Au Nanoconjugates in Graphene Liquid Cell EM Qian Chen,,, Jessica Smith,, Jungwon Park,,1, Kwanpyo Kim,,2, Davy Ho, Haider I. Rasool,,!Alex Zettl,, A. Paul Alivisatos,,*!
Reconfigurable DNA Origami Nanocapsule for ph- Controlled Encapsulation and Display of Cargo
 Supporting Information Reconfigurable DNA Origami Nanocapsule for ph- Controlled Encapsulation and Display of Cargo Heini Ijäs, Iiris Hakaste, Boxuan Shen, Mauri A. Kostiainen, and Veikko Linko* Biohybrid
Supporting Information Reconfigurable DNA Origami Nanocapsule for ph- Controlled Encapsulation and Display of Cargo Heini Ijäs, Iiris Hakaste, Boxuan Shen, Mauri A. Kostiainen, and Veikko Linko* Biohybrid
Combinatorial Algorithms and Computational Complexity for DNA Self-Assembly
 Combinatorial Algorithms and Computational Complexity for DNA Self-Assembly Ming-Yang Kao Northwestern University Evanston, Illinois USA Presented at Kyoto University on December 12, 2013 Outline of the
Combinatorial Algorithms and Computational Complexity for DNA Self-Assembly Ming-Yang Kao Northwestern University Evanston, Illinois USA Presented at Kyoto University on December 12, 2013 Outline of the
Yujuan Zhou, Kecheng Jie and Feihe Huang*
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 A redox-responsive selenium-containing pillar[5]arene-based macrocyclic amphiphile: synthesis,
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 A redox-responsive selenium-containing pillar[5]arene-based macrocyclic amphiphile: synthesis,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/NCHEM.1123 Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofibre and hydrogel Lesley E. R. O Leary, Jorge A. Fallas, Erica L. Bakota, Marci K. Kang
DOI: 10.1038/NCHEM.1123 Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofibre and hydrogel Lesley E. R. O Leary, Jorge A. Fallas, Erica L. Bakota, Marci K. Kang
DNA-Scaffolded Self-Assembling Nano-Circuitry
 DNA-Scaffolded Self-Assembling Nano-Circuitry An Ongoing Research Project with Dr. Soha Hassoun Presentation by Brandon Lucia and Laura Smith DNA-Scaffolded... DNA is special type of molecule Made of a
DNA-Scaffolded Self-Assembling Nano-Circuitry An Ongoing Research Project with Dr. Soha Hassoun Presentation by Brandon Lucia and Laura Smith DNA-Scaffolded... DNA is special type of molecule Made of a
Supporting Materials Ultra-small Sub-10 nm Near Infrared Fluorescent Mesoporous Silica Nanoparticles
 Supporting Materials Ultra-small Sub-10 nm Near Infrared Fluorescent Mesoporous Silica Nanoparticles Kai Ma, Hiroaki Sai and Ulrich Wiesner* Department of Materials Science and Engineering, Cornell University,
Supporting Materials Ultra-small Sub-10 nm Near Infrared Fluorescent Mesoporous Silica Nanoparticles Kai Ma, Hiroaki Sai and Ulrich Wiesner* Department of Materials Science and Engineering, Cornell University,
Mechanically Strong and Highly Conductive Graphene Aerogels and Its Use as. Electrodes for Electrochemical Power Sources
 Supporting Information for Mechanically Strong and Highly Conductive Graphene Aerogels and Its Use as Electrodes for Electrochemical Power Sources Xuetong Zhang, Zhuyin Sui, Bin Xu, Shufang Yue, Yunjun
Supporting Information for Mechanically Strong and Highly Conductive Graphene Aerogels and Its Use as Electrodes for Electrochemical Power Sources Xuetong Zhang, Zhuyin Sui, Bin Xu, Shufang Yue, Yunjun
SELF-ASSEMBLED DNA NANOSTRUCTURES AND DNA-TEMPLATED SILVER NANOWIRES. Sung Ha Park. Department of Physics Duke University
 SELF-ASSEMBLED DNA NANOSTRUCTURES AND DNA-TEMPLATED SILVER NANOWIRES by Sung Ha Park Department of Physics Duke University Date: Approved: Gleb Finkelstein, Supervisor Thomas H. LaBean, Co-supervisor John
SELF-ASSEMBLED DNA NANOSTRUCTURES AND DNA-TEMPLATED SILVER NANOWIRES by Sung Ha Park Department of Physics Duke University Date: Approved: Gleb Finkelstein, Supervisor Thomas H. LaBean, Co-supervisor John
Supplementary Material for. Zinc Oxide-Black Phosphorus Composites for Ultrasensitive Nitrogen
 Electronic Supplementary Material (ESI) for Nanoscale Horizons. This journal is The Royal Society of Chemistry 2018 Supplementary Material for Zinc Oxide-Black Phosphorus Composites for Ultrasensitive
Electronic Supplementary Material (ESI) for Nanoscale Horizons. This journal is The Royal Society of Chemistry 2018 Supplementary Material for Zinc Oxide-Black Phosphorus Composites for Ultrasensitive
Tuning structure and function in tetra(aniline)-based rod-coil-rod architectures
 Tuning structure and function in tetra(aniline)-based rod-coil-rod architectures Chinwe U. Udeh, a Patrice Rannou, b Benjamin P. Brown, a,c James O. Thomas, c,d and Charl F. J. Faul a,d* a School of Chemistry,
Tuning structure and function in tetra(aniline)-based rod-coil-rod architectures Chinwe U. Udeh, a Patrice Rannou, b Benjamin P. Brown, a,c James O. Thomas, c,d and Charl F. J. Faul a,d* a School of Chemistry,
Overview of New Structures for DNA-Based Nanofabrication and Computation
 Overview of New Structures for DNA-Based Nanofabrication and Computation Thomas H. LaBean, Hao Yan, Sung Ha Park, Liping Feng, Peng Yin, Hanying Li, Sang Jung Ahn, Dage Liu, Xiaoju Guan, and John H. Reif
Overview of New Structures for DNA-Based Nanofabrication and Computation Thomas H. LaBean, Hao Yan, Sung Ha Park, Liping Feng, Peng Yin, Hanying Li, Sang Jung Ahn, Dage Liu, Xiaoju Guan, and John H. Reif
Synthesis of Colloidal Au-Cu 2 S Heterodimers via Chemically Triggered Phase Segregation of AuCu Nanoparticles
 SUPPORTING INFORMATION Synthesis of Colloidal Au-Cu 2 S Heterodimers via Chemically Triggered Phase Segregation of AuCu Nanoparticles Nathan E. Motl, James F. Bondi, and Raymond E. Schaak* Department of
SUPPORTING INFORMATION Synthesis of Colloidal Au-Cu 2 S Heterodimers via Chemically Triggered Phase Segregation of AuCu Nanoparticles Nathan E. Motl, James F. Bondi, and Raymond E. Schaak* Department of
Introduction to Nanoparticle Tracking Analysis (NTA) Measurement Principle of ZetaView
 Technical Note Nanoparticle Tracking Key words: Introduction to Nanoparticle Tracking Analysis (NTA) Measurement Principle of ZetaView Particle characterization, Nanoparticle Tracking Analysis (NTA), Brownian
Technical Note Nanoparticle Tracking Key words: Introduction to Nanoparticle Tracking Analysis (NTA) Measurement Principle of ZetaView Particle characterization, Nanoparticle Tracking Analysis (NTA), Brownian
Experimental details. General
 Experimental details General TiO 2 P25 was purchased from Degussa; methyl methacrylate (MMA, 99%), 2,2 -azobis(2- methylpropionamidine) dihydrochloride (97%), titanium (IV) isopropoxide (97%), concentrated
Experimental details General TiO 2 P25 was purchased from Degussa; methyl methacrylate (MMA, 99%), 2,2 -azobis(2- methylpropionamidine) dihydrochloride (97%), titanium (IV) isopropoxide (97%), concentrated
Supporting Information for. Co-crystal Engineering: A Novel Method to Get One-dimensional (1D) Carbon
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2016 Supporting Information for Co-crystal Engineering: A Novel Method to Get One-dimensional (1D)
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2016 Supporting Information for Co-crystal Engineering: A Novel Method to Get One-dimensional (1D)
Supplementary Figure 1: Power dependence of hot-electrons reduction of 4-NTP to 4-ATP. a) SERS spectra of the hot-electron reduction reaction using
 Supplementary Figure 1: Power dependence of hot-electrons reduction of 4-NTP to 4-ATP. a) SERS spectra of the hot-electron reduction reaction using 633 nm laser excitation at different powers and b) the
Supplementary Figure 1: Power dependence of hot-electrons reduction of 4-NTP to 4-ATP. a) SERS spectra of the hot-electron reduction reaction using 633 nm laser excitation at different powers and b) the
Supplementary Information
 Supplementary Information Supramolecular polymerization of hydrogen-bonded rosettes with anthracene chromophores: regioisomeric effect on nanostructures Deepak D. Prabhu, Keisuke Aratsu, Mitsuaki Yamauchi,
Supplementary Information Supramolecular polymerization of hydrogen-bonded rosettes with anthracene chromophores: regioisomeric effect on nanostructures Deepak D. Prabhu, Keisuke Aratsu, Mitsuaki Yamauchi,
Electronic Supplementary Information
 Electronic Supplementary Information Uniform and Rich Wrinkled Electrophoretic Deposited Graphene Film: A Robust Electrochemical Platform for TNT Sensing Longhua Tang, Hongbin Feng, Jinsheng Cheng and
Electronic Supplementary Information Uniform and Rich Wrinkled Electrophoretic Deposited Graphene Film: A Robust Electrochemical Platform for TNT Sensing Longhua Tang, Hongbin Feng, Jinsheng Cheng and
- Supporting Information - Controlled Assembly of Eccentrically Encapsulated Gold Nanoparticles
 - Supporting Information - S1 Controlled Assembly of Eccentrically Encapsulated Gold Nanoparticles Tao Chen, Miaoxin Yang, Xinjiao Wang, Li Huey Tan, Hongyu Chen* Division of Chemistry and Biological Chemistry,
- Supporting Information - S1 Controlled Assembly of Eccentrically Encapsulated Gold Nanoparticles Tao Chen, Miaoxin Yang, Xinjiao Wang, Li Huey Tan, Hongyu Chen* Division of Chemistry and Biological Chemistry,
SUPPLEMENTARY INFORMATION
 SUPPLMTARY IFORMATIO a doi:10.108/nature10402 b 100 nm 100 nm c SAXS Model d ulers assigned to reference- Back-projected free class averages class averages Refinement against single particles Reconstructed
SUPPLMTARY IFORMATIO a doi:10.108/nature10402 b 100 nm 100 nm c SAXS Model d ulers assigned to reference- Back-projected free class averages class averages Refinement against single particles Reconstructed
Electronic Supplementary Information. Experimental details graphene synthesis
 Electronic Supplementary Information Experimental details graphene synthesis Graphene is commercially obtained from Graphene Supermarket (Reading, MA, USA) 1 and is produced via a substrate-free gas-phase
Electronic Supplementary Information Experimental details graphene synthesis Graphene is commercially obtained from Graphene Supermarket (Reading, MA, USA) 1 and is produced via a substrate-free gas-phase
Molecular electron microscopy
 Molecular electron microscopy - Imaging macromolecular assemblies Yifan Cheng Department of Biochemistry & Biophysics office: GH-S427B; email: ycheng@ucsf.edu 2/22/2013 - Introduction of Molecular Microscopy:
Molecular electron microscopy - Imaging macromolecular assemblies Yifan Cheng Department of Biochemistry & Biophysics office: GH-S427B; email: ycheng@ucsf.edu 2/22/2013 - Introduction of Molecular Microscopy:
Controlled Electroless Deposition of Nanostructured Precious Metal Films on Germanium Surfaces
 SUPPORTING INFORMATION. Controlled Electroless Deposition of Nanostructured Precious Metal Films on Germanium Surfaces Lon A. Porter, Jr., Hee Cheul Choi, Alexander E. Ribbe, and Jillian M. Buriak Department
SUPPORTING INFORMATION. Controlled Electroless Deposition of Nanostructured Precious Metal Films on Germanium Surfaces Lon A. Porter, Jr., Hee Cheul Choi, Alexander E. Ribbe, and Jillian M. Buriak Department
Supporting Information for
 Supporting Information for Multilayer CuO@NiO Hollow Spheres: Microwave-Assisted Metal-Organic-Framework Derivation and Highly Reversible Structure-Matched Stepwise Lithium Storage Wenxiang Guo, Weiwei
Supporting Information for Multilayer CuO@NiO Hollow Spheres: Microwave-Assisted Metal-Organic-Framework Derivation and Highly Reversible Structure-Matched Stepwise Lithium Storage Wenxiang Guo, Weiwei
Visible-light Driven Plasmonic Photocatalyst Helical Chiral TiO 2 Nanofibers
 Visible-light Driven Plasmonic Photocatalyst Ag/AgCl @ Helical Chiral TiO 2 Nanofibers Dawei Wang, Yi Li*, Gianluca Li Puma, Chao Wang, Peifang Wang, Wenlong Zhang, and Qing Wang Fig. S1. The reactor of
Visible-light Driven Plasmonic Photocatalyst Ag/AgCl @ Helical Chiral TiO 2 Nanofibers Dawei Wang, Yi Li*, Gianluca Li Puma, Chao Wang, Peifang Wang, Wenlong Zhang, and Qing Wang Fig. S1. The reactor of
University of Groningen
 University of Groningen "Giant Surfactants" Created by the Fast and Efficient Functionalization of a DNA Tetrahedron with a Temperature-Responsive Polymer Wilks, Thomas R.; Bath, Jonathan; de Vries, Jan
University of Groningen "Giant Surfactants" Created by the Fast and Efficient Functionalization of a DNA Tetrahedron with a Temperature-Responsive Polymer Wilks, Thomas R.; Bath, Jonathan; de Vries, Jan
Supporting Information
 Supporting Information Janus Hollow Spheres by Emulsion Interfacial Self-Assembled Sol-Gel Process Fuxin Liang, Jiguang Liu, Chengliang Zhang, Xiaozhong Qu, Jiaoli Li, Zhenzhong Yang* State Key Laboratory
Supporting Information Janus Hollow Spheres by Emulsion Interfacial Self-Assembled Sol-Gel Process Fuxin Liang, Jiguang Liu, Chengliang Zhang, Xiaozhong Qu, Jiaoli Li, Zhenzhong Yang* State Key Laboratory
A dual redox-responsive supramolecular amphiphile fabricated by selenium-containing pillar[6]arene-based molecular recognition
![A dual redox-responsive supramolecular amphiphile fabricated by selenium-containing pillar[6]arene-based molecular recognition A dual redox-responsive supramolecular amphiphile fabricated by selenium-containing pillar[6]arene-based molecular recognition](/thumbs/88/115999894.jpg) Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2018 A dual redox-responsive supramolecular amphiphile fabricated by selenium-containing pillar[6]arene-based
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2018 A dual redox-responsive supramolecular amphiphile fabricated by selenium-containing pillar[6]arene-based
Introduction to Nanotechnology Chapter 5 Carbon Nanostructures Lecture 1
 Introduction to Nanotechnology Chapter 5 Carbon Nanostructures Lecture 1 ChiiDong Chen Institute of Physics, Academia Sinica chiidong@phys.sinica.edu.tw 02 27896766 Section 5.2.1 Nature of the Carbon Bond
Introduction to Nanotechnology Chapter 5 Carbon Nanostructures Lecture 1 ChiiDong Chen Institute of Physics, Academia Sinica chiidong@phys.sinica.edu.tw 02 27896766 Section 5.2.1 Nature of the Carbon Bond
Supporting Information
 Supporting Information Ultrathin Spinel-Structured Nanosheets Rich in Oxygen Deficiencies for Enhanced Electrocatalytic Water Oxidation** Jian Bao, Xiaodong Zhang,* Bo Fan, Jiajia Zhang, Min Zhou, Wenlong
Supporting Information Ultrathin Spinel-Structured Nanosheets Rich in Oxygen Deficiencies for Enhanced Electrocatalytic Water Oxidation** Jian Bao, Xiaodong Zhang,* Bo Fan, Jiajia Zhang, Min Zhou, Wenlong
The Golden Section, the Pentagon and the Dodecahedron
 The Golden Section, the Pentagon and the Dodecahedron C. Godsalve email:seagods@hotmail.com July, 009 Contents Introduction The Golden Ratio 3 The Pentagon 3 4 The Dodecahedron 8 A few more details 4 Introduction
The Golden Section, the Pentagon and the Dodecahedron C. Godsalve email:seagods@hotmail.com July, 009 Contents Introduction The Golden Ratio 3 The Pentagon 3 4 The Dodecahedron 8 A few more details 4 Introduction
Amphiphilic diselenide-containing supramolecular polymers
 Electronic Supplementary Material (ESI) for Polymer Chemistry. This journal is The Royal Society of Chemistry 2014 Amphiphilic diselenide-containing supramolecular polymers Xinxin Tan, Liulin Yang, Zehuan
Electronic Supplementary Material (ESI) for Polymer Chemistry. This journal is The Royal Society of Chemistry 2014 Amphiphilic diselenide-containing supramolecular polymers Xinxin Tan, Liulin Yang, Zehuan
A Biosensor for the Detection of Single Base Mismatches in microrna
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 215 Supporting Information A Biosensor for the Detection of Single Base Mismatches in microrna Jieon
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 215 Supporting Information A Biosensor for the Detection of Single Base Mismatches in microrna Jieon
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis):
 SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
Lecture CIMST Winter School. 1. What can you see by TEM?
 Lecture CIMST Winter School Cryo-electron microscopy and tomography of biological macromolecules 20.1.2011 9:00-9:45 in Y03G91 Dr. Takashi Ishikawa OFLB/005 Tel: 056 310 4217 e-mail: takashi.ishikawa@psi.ch
Lecture CIMST Winter School Cryo-electron microscopy and tomography of biological macromolecules 20.1.2011 9:00-9:45 in Y03G91 Dr. Takashi Ishikawa OFLB/005 Tel: 056 310 4217 e-mail: takashi.ishikawa@psi.ch
Supplementary Information. Large Scale Graphene Production by RF-cCVD Method
 Supplementary Information Large Scale Graphene Production by RF-cCVD Method Enkeleda Dervishi, *a,b Zhongrui Li, b Fumiya Watanabe, b Abhijit Biswas, c Yang Xu, b Alexandru R. Biris, d Viney Saini, a,b
Supplementary Information Large Scale Graphene Production by RF-cCVD Method Enkeleda Dervishi, *a,b Zhongrui Li, b Fumiya Watanabe, b Abhijit Biswas, c Yang Xu, b Alexandru R. Biris, d Viney Saini, a,b
CHARACTERIZATION of NANOMATERIALS KHP
 CHARACTERIZATION of NANOMATERIALS Overview of the most common nanocharacterization techniques MAIN CHARACTERIZATION TECHNIQUES: 1.Transmission Electron Microscope (TEM) 2. Scanning Electron Microscope
CHARACTERIZATION of NANOMATERIALS Overview of the most common nanocharacterization techniques MAIN CHARACTERIZATION TECHNIQUES: 1.Transmission Electron Microscope (TEM) 2. Scanning Electron Microscope
A metal ion triggered shrinkable supramolecular hydrogel and controlled release by an amphiphilic peptide dendron
 Supporting Information For A metal ion triggered shrinkable supramolecular hydrogel and controlled release by an amphiphilic peptide dendron Long Qin, a Pengfei Duan, a Fan Xie, a Li Zhang a and Minghua
Supporting Information For A metal ion triggered shrinkable supramolecular hydrogel and controlled release by an amphiphilic peptide dendron Long Qin, a Pengfei Duan, a Fan Xie, a Li Zhang a and Minghua
Chapter 12. Nanometrology. Oxford University Press All rights reserved.
 Chapter 12 Nanometrology Introduction Nanometrology is the science of measurement at the nanoscale level. Figure illustrates where nanoscale stands in relation to a meter and sub divisions of meter. Nanometrology
Chapter 12 Nanometrology Introduction Nanometrology is the science of measurement at the nanoscale level. Figure illustrates where nanoscale stands in relation to a meter and sub divisions of meter. Nanometrology
Supporting Information
 Supporting Information Materials and Methods: Tris-hydroxymethylaminomethane (Tris) was purchased from USB. All the other reagents used in the experiments were purchased from Sigma. All the DNA oligonucleotides
Supporting Information Materials and Methods: Tris-hydroxymethylaminomethane (Tris) was purchased from USB. All the other reagents used in the experiments were purchased from Sigma. All the DNA oligonucleotides
Electron Microscopy of Tobacco Mosaic Virus Prepared with the Aid of Negative Staining-Carbon Film Techniques
 J. gen. Virol. (1976), :3I, 265-269 Printed in Great Britain 265 Electron Microscopy of Tobacco Mosaic Virus Prepared with the Aid of Negative Staining-Carbon Film Techniques (Accepted 13 January I976)
J. gen. Virol. (1976), :3I, 265-269 Printed in Great Britain 265 Electron Microscopy of Tobacco Mosaic Virus Prepared with the Aid of Negative Staining-Carbon Film Techniques (Accepted 13 January I976)
Chapter 10. Nanometrology. Oxford University Press All rights reserved.
 Chapter 10 Nanometrology Oxford University Press 2013. All rights reserved. 1 Introduction Nanometrology is the science of measurement at the nanoscale level. Figure illustrates where nanoscale stands
Chapter 10 Nanometrology Oxford University Press 2013. All rights reserved. 1 Introduction Nanometrology is the science of measurement at the nanoscale level. Figure illustrates where nanoscale stands
Interaction of Gold Nanoparticle with Proteins
 Chapter 7 Interaction of Gold Nanoparticle with Proteins 7.1. Introduction The interfacing of nanoparticle with biomolecules such as protein is useful for applications ranging from nano-biotechnology (molecular
Chapter 7 Interaction of Gold Nanoparticle with Proteins 7.1. Introduction The interfacing of nanoparticle with biomolecules such as protein is useful for applications ranging from nano-biotechnology (molecular
Intensity (a.u.) Intensity (a.u.) Raman Shift (cm -1 ) Oxygen plasma. 6 cm. 9 cm. 1mm. Single-layer graphene sheet. 10mm. 14 cm
 Intensity (a.u.) Intensity (a.u.) a Oxygen plasma b 6 cm 1mm 10mm Single-layer graphene sheet 14 cm 9 cm Flipped Si/SiO 2 Patterned chip Plasma-cleaned glass slides c d After 1 sec normal Oxygen plasma
Intensity (a.u.) Intensity (a.u.) a Oxygen plasma b 6 cm 1mm 10mm Single-layer graphene sheet 14 cm 9 cm Flipped Si/SiO 2 Patterned chip Plasma-cleaned glass slides c d After 1 sec normal Oxygen plasma
Supplementary Figure 1. (a-b) EDX of Mo 2 and Mo 2
 Supplementary Figure 1. (a-b) EDX of Mo 2 C@NPC/NPRGO and Mo 2 C@NPC. Supplementary Figure 2. (a) SEM image of PMo 12 2-PPy, (b) TEM, (c) HRTEM, (d) STEM image and EDX elemental mapping of C, N, P, and
Supplementary Figure 1. (a-b) EDX of Mo 2 C@NPC/NPRGO and Mo 2 C@NPC. Supplementary Figure 2. (a) SEM image of PMo 12 2-PPy, (b) TEM, (c) HRTEM, (d) STEM image and EDX elemental mapping of C, N, P, and
An inorganic-organic hybrid supramolecular nanotube as high-performance anode for lithium ion batteries
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2018 An inorganic-organic hybrid supramolecular nanotube as high-performance anode for lithium
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2018 An inorganic-organic hybrid supramolecular nanotube as high-performance anode for lithium
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. The asymmetric unit in para-iodio-phenylalanine crystal. The 50% probability ellipsoid representation was prepared using the Mercury Software. Colors are as
Supplementary Figures Supplementary Figure 1. The asymmetric unit in para-iodio-phenylalanine crystal. The 50% probability ellipsoid representation was prepared using the Mercury Software. Colors are as
SUPPORTING INFORMATION
 Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2017 SUPPORTING INFORMATION Synthesis of Circular and Triangular Gold Nanorings with
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2017 SUPPORTING INFORMATION Synthesis of Circular and Triangular Gold Nanorings with
Kavli Workshop for Journalists. June 13th, CNF Cleanroom Activities
 Kavli Workshop for Journalists June 13th, 2007 CNF Cleanroom Activities Seeing nm-sized Objects with an SEM Lab experience: Scanning Electron Microscopy Equipment: Zeiss Supra 55VP Scanning electron microscopes
Kavli Workshop for Journalists June 13th, 2007 CNF Cleanroom Activities Seeing nm-sized Objects with an SEM Lab experience: Scanning Electron Microscopy Equipment: Zeiss Supra 55VP Scanning electron microscopes
Number-controlled spatial arrangement of gold nanoparticles with
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Number-controlled spatial arrangement of gold nanoparticles with DNA dendrimers Ping Chen,*
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Number-controlled spatial arrangement of gold nanoparticles with DNA dendrimers Ping Chen,*
Debjit Roy, Saptarshi Mandal, Chayan K. De, Kaushalendra Kumar and Prasun K. Mandal*
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Nearly Suppressed Photoluminescence Blinking of Small Sized, Blue-Green-Orange-Red
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Nearly Suppressed Photoluminescence Blinking of Small Sized, Blue-Green-Orange-Red
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/NCHEM.1290 Metal-directed, chemically tunable assembly of one-, two- and threedimensional crystalline protein arrays 1 2 1 1 1,3 Jeffrey D. Brodin 1, X. I. Ambroggio 2, Chunyan Tang 1, Kristin
DOI: 10.1038/NCHEM.1290 Metal-directed, chemically tunable assembly of one-, two- and threedimensional crystalline protein arrays 1 2 1 1 1,3 Jeffrey D. Brodin 1, X. I. Ambroggio 2, Chunyan Tang 1, Kristin
Supplementary Note 1: Dark field measurements and Scattering properties of NPoM geometries
 Supplementary Note 1: Dark field measurements and Scattering properties of NPoM geometries Supplementary Figure 1: Dark field scattering properties of individual nanoparticle on mirror geometries separated
Supplementary Note 1: Dark field measurements and Scattering properties of NPoM geometries Supplementary Figure 1: Dark field scattering properties of individual nanoparticle on mirror geometries separated
Molecular Self-Assembly: Models and Algorithms
 Molecular Self-Assembly: Models and Algorithms Ashish Goel Stanford University MS&E 319/CS 369X; Research topics in optimization; Stanford University, Spring 2003-04 http://www.stanford.edu/~ashishg Self-Assembly
Molecular Self-Assembly: Models and Algorithms Ashish Goel Stanford University MS&E 319/CS 369X; Research topics in optimization; Stanford University, Spring 2003-04 http://www.stanford.edu/~ashishg Self-Assembly
DNA-based Nanomaterials
 I DNA-based Nanomaterials Nanotechnologies for the Life Sciences Vol. 2 Biological and Pharmaceutical Nanomaterials. Edited by Challa S. S. R. Kumar Copyright 8 2006 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
I DNA-based Nanomaterials Nanotechnologies for the Life Sciences Vol. 2 Biological and Pharmaceutical Nanomaterials. Edited by Challa S. S. R. Kumar Copyright 8 2006 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
Supporting Information
 Supporting Information Materials Multiwalled carbon nanotubes (MWNTs, φ = 10-30 nm) were purchased from Nanotech Port Co. Ltd. (Shenzhen, China). Tetraethylorthosilicate (TEOS, >98 %), Triton X-100 and
Supporting Information Materials Multiwalled carbon nanotubes (MWNTs, φ = 10-30 nm) were purchased from Nanotech Port Co. Ltd. (Shenzhen, China). Tetraethylorthosilicate (TEOS, >98 %), Triton X-100 and
