VT-2005 Sigma Profile Database
|
|
|
- Liliana Fleming
- 6 years ago
- Views:
Transcription
1 VT-2005 Sigma Profile Database A detailed tutorial for generating sigma profiles using Accelrys Materials Studio v3.2 software package Eric Mullins, Y.A. Liu, Richard Oldland, Shu Wang, Stanley I. Sandler, Chau-Chyun Chen, Michael Zwolak, and Kevin Seavey Honeywell Center of Excellence in Computer-Aided Design SINOPEC/FPCC/AspenTech Center of Excellence in Process Systems Engineering Department of Chemical Engineering, Virginia Tech Blacksburg, VA 24060
2 Objectives An Introduction to Sigma Profiles, P(σ) Using Accelrys Materials Studio (MS) Detailed procedure for creating a molecule Detailed procedures for optimizing molecular geometry and performing COSMO calculations Overview of the Sigma Profile averaging program
3 Sigma Profiles at a Glance Sigma Profiles, P(σ), depict the surface charge density distribution over the entire molecule. Graphically: Area vs. Charge/Area Profiles are available with our database. Each molecule s profile is unique. Profiles are sensitive 2 0 to conformation P(sigma)*Area(sigma) (A^2) Sigma (e/a^2) 1,4-dioxane water methyl acetate
4 COSMO-based models used sigma profiles to predict physical properties. 6 Solubility (SLE) Vapor-Liquid (VLE) Partition coefficients (K ow ) pka Pressure (kpa) COSMO-SAC COSMO-SAC Experimental Data Experimental Data COSMO-RS COSMO-RS T = K Phenol Mole Fraction
5 The VT-2005 sigma profile database currently contains 1266 compounds. Compounds include alkanes, alkenes, alkynes, alcohols, amines, acids, aldehydes, aromatics, epoxides, esters, ethers, fluorocarbons, chlorocarbons, ketones and others. Compounds consist of the following atoms: N, C, O, H, F, Cl, P, S, I, and Br
6 Using Accelrys Materials Studio (MS), we will construct our model molecule. We will use propane for our example. Molecules can be drawn manually or downloaded from other online sources as *.mol files and imported into MS. NIST database Scifinder National Library of Medicine
7 We begin by creating a new project in MS, and then opening a new molecule window. Select a new 3D Atomistic Document. Right click on the 3D Atomistic Document in the project window and select Rename. Type Propane and hit Enter.
8 We display the molecule with the Ball and Stick option. Right click in the molecular window and select Display Style. Select Ball and Stick in the display style window and then close the window.
9 Use the sketch tool to draw propane (C 3 H 8 ) Select the Sketch Atom tool from the toolbar. With the drop-down menu, select the Carbon atom.
10 Draw the three carbon atoms, but do not draw the hydrogen atoms for now. Left click once to place the first atom. Move your cursor and click again; it will automatically create a second atom connected with a single bond. Draw a three carbon backbone. Hitting Esc will stop adding atoms. In the instance of double bonds, simply clicking on the single bond will change its bond type.
11 The program automatically adds the required hydrogen for each molecule with the Adjust Hydrogen tool. Click the Adjust Hydrogen icon from the toolbar to add the hydrogen atoms.
12 Highlight the entire molecule and change the properties filter to molecule. Changing the filter option to molecule shows various properties like the mass, number of atoms, and the chemical formula. Quickly verify the chemical formula is correct.
13 Now we use the Clean tool to correct the bond lengths and bond angles. Left click on the Clean tool icon on the toolbar. The Clean tool does a rough geometry optimization and adjusts the bond lengths and angles to the correct values for each atom and bond type. We are now finished constructing our example molecule, Propane.
14 Now we move onto setting up the Geometry Optimization calculation. Select the DMol3 calculation from the Modules Menu or use the icon on the toolbar. The calculation dialog box will appear.
15 Select the task option in the DMol3 calculation window. Geometry optimization fixed the atomic coordinates for the molecule by minimizing the total energy of the molecule.
16 Adjust the Geometry Optimization tolerance to Fine. This can be done in the Setup or Electronic tab. This is the accuracy of the Hamiltonian matrix element convergence. This specifies the accuracy to which the SCF (Self-Consistent Field) equations are converged. The Fine setting is recommended by the software documentation for highly accurate geometry optimizations. It represents a convergence of 10-6.
17 Select DNP as the Basis Set in the Electronic tab. DNP = Double Numerical basis with Polarization functions, i.e., functions with angular momentum one higher than the highest occupied orbital in the free atom. According to the software documentation, minimal basis sets are generally inadequate for anything but qualitative results, while DNP sets are the most reliable. The DNP option instructs the program to ignore extraneous functions that eliminate certain atomic orbitals. When Quality it set to Fine, DNP is the default basis set.
18 Select GGA (local correlation) and VWN-BP (gradient-corrected functional) for the Functional option in the Setup tab. GGA = Generalized Gradient Approximation. VWN-BP = BP functional (Becke, 1986; Pedrew, 1986) with local correlation replaced by the VWN functional (Volsko, Wilk, and Nusair, 1980). Koch and Holthausen (2001) give excellent guidance about the selection of appropriate functionals.
19 Select the More button in the Job Control tab. Check the Retain server files box. This will save the files to the hard drive in the jobs directory.
20 Click the Files button at the bottom of the DMol3 calculation dialog box. Click the Save Files button. This will create the input files necessary for the calculation. They are visible in the Project window.
21 Open propane.input from the Project window. Insert the code Basis_version v4.0.0 into the input file. This statement instructs the program to use version DNP basis set instead of the default v3.5 DNP basis set. The software documentation recommends v4.0.0 for COSMO calculations.
22 Leave the Spin Unrestricted box unchecked in the Setup tab. Checking the Spin Unrestricted is necessary when working with radicals, charged molecules, and organometallics. Although it is possible to run an organic molecule with the Spin Unrestricted setting, it will be much slower computationally. Checking the Symmetry box in the setup tab can also apply to certain molecules, but not all.
23 Click the Run Files button to begin the DMol3 Geometry Optimization. The geometry optimization predicts the energy level of the molecule in the ideal gas phase. This calculation is the longest step of the procedure and will require ~75% of the time to produce a sigma profile.
24 The Project window will show the files created and tracks the progress of the calculation. The *.xsd file is the optimized geometry. All further calculations should use this file. The *.outmol file will show the ideal gas phase energy. There are additional files that do not show up in the Project window but are stored in the Jobs folder. (C:\Program Files\Accelrys\MS Modeling 3.2\Gateway\root_default\dsd\jobs)
25 We have now completed the Geometry Optimization and will proceed to the COSMO calculation. Begin by opening the geometry optimization output, vt-0003.xsd, and open another DMol3 calculation dialog box. Select the task, Energy. Leave all other options the same as the geometry optimization.
26 Click the Files button in the calculation window. Click the Save Files button to create the input files.
27 Open the propane.input file and add the COSMO calculation keywords. The COSMO keywords tell the program which parameters to use in the COSMO calculation including atomic radii, basis set version, etc.
28 Now we review the parameters and settings in the COSMO keywords. This statement turns the COSMO calculation on. Again, we will use basis set version instead of the default v3.5. The first number is the atomic number and the second term is the fitted atomic radii. Klamt (1998) fitted these parameters to several sets of data.
29 Now we will run the COSMO calculation. Click on the Run Files button from the calculation window to begin the energy calculation. Upon completion, the Project window will contain a *.cosmo and *.outmol file. There are additional files stored in the Jobs folder.
30 The *.cosmo file contains much useful information. This file contains the volume of the cavity around the molecule in the theoretical conducting medium, and it is used in the COSMO-RS/COSMO- SAC models. It also contains the condensed phase energy, the number of surface segments, and their charge.
31 We edit the *.cosmo file to make it compatible with the sigma averaging FORTRAN program. Record the number of surface segments. Delete all information above the last table, which contains the segment charges and coordinates. Then save as a *.txt text file. Delete all text above this line.
32 Our sigma averaging program uses the modified *.cosmo file to calculate the sigma profile. We average the surface charge densities from the *.cosmo file to find an effective surface charge density on a standard surface segment. σ m = r r * n av mn σ n exp n rn + rav rn + rav r r d d n av mn exp n rn + rav rn + rav
33 The averaging program creates a new text file, the sigma profile. The output file is Chemical name Sigma-profile.txt is located in the C:\Profiles\ directory. This directory must be created manually before running the averaging program. The output can be input directly into the COSMO-SAC program or can be plotted graphically.
34 We run the FORTRAN program to average the surface charges over a standardized bonding site. Run sigmaprofile.exe. Follow the prompts and enter: New COSMO output (c:/propane.txt) Chemical Name (Propane) The Number of Surface Segments
35 The sigma averaging program output is text, but can be easily converted into graphical form. Sigma values, (e/a2) [X-axis] P(σ) values, (A2) [Y-axis]
36 Using Microsoft Excel, we display the sigma profile below. Propane Sigma Profile VT-0003 Propane P(σ)*Area(σ), A Sigma, σ e/a 2
37 Acknowledgments We want to thank Alliant Techsystems (particularly Anthony Miano, Vice President), AspenTech (particularly Dustin MacNeil, Director of Worldwide University Programs, Larry Evans, Board Chairman, and Chau-Chyun Chen), China Petroleum and Chemical Corporation (particularly Wilfred Wang, President) and Honeywell Specialty Materials and Honeywell International Foundation for supporting our educational programs in computer-aided design and process systems engineering at Virginia Tech. Michael Zwolak gratefully appreciates support from an NSF Graduate Fellowship.
38 References Andzelm, Jan; Klamt, Andreas; and Kölmel, Christoph, Incorporation of solvent effects into density functional calculations of molecular energies and geometries, J. Chem. Phys., 103, (1995) Bustamante, P.; Pena, M.A.; Barra, J., The Modified Extended Hansen Method to Determine Partial Solubility Parameters of Drugs Containing a Single Hydrogen Bonding Group and Their Sodium Derivatives: Benzoic Acid/Na and Ibuprofen/Na,Int. J. of Pharmaceutics, 2000, 194, 117. Giles, Neil F. and Wilson, Grant M., Phase Equilibria on Seven Binary Mixtures, J. Chem. Eng. Data., 45, (2000). Klamt, Andreas, G. Shurmann, COSMO-A New Approach
39 References Lin, Shiang-Tai; Chang, Jaeeon; Wang, Shu; Goddard III, William A.; Sandler, Stanley I., Prediction of Vapor Pressures and Enthalpies of Vaporization Using COSMO Solvation Model, J. Phys. Chem., 108, , (2004). Lin, Shiang-Tai and Stanley I. Sandler, A Priori Phase Equilibrium Prediction from a Segment Contribution Solvation Model, Ind. Eng. Chem. Res, 41, 899(2002a). Lin, Shiang-Tai and Stanley I. Sandler, Reply to Comments on A Priori Phase Equilibrium Prediction from a Segment Contribution Solvation Model, Ind. Eng. Chem. Res., 41, 2332 (2002b). Panayiotou, Costas, Equation-of-State Models and Quantum Mechanics Calculations, Ind. Eng. Chem. Res, 42, 1495 (2003).
Sigma-Profile Database for Using COSMO-Based Thermodynamic Methods
 Ind. Eng. Chem. Res. 2006, 45, 4389-4415 4389 Sigma-Profile Database for Using COSMO-Based Thermodynamic Methods Eric Mullins, Richard Oldland, Y. A. Liu,*, Shu Wang, Stanley I. Sandler, Chau-Chyun Chen,
Ind. Eng. Chem. Res. 2006, 45, 4389-4415 4389 Sigma-Profile Database for Using COSMO-Based Thermodynamic Methods Eric Mullins, Richard Oldland, Y. A. Liu,*, Shu Wang, Stanley I. Sandler, Chau-Chyun Chen,
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Chem 253. Tutorial for Materials Studio
 Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Assignment 1: Molecular Mechanics (PART 1 25 points)
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard August 19, 2015 Assignment 1: Molecular Mechanics (PART 1 25 points) In this assignment, you will perform some molecular mechanics calculations using the
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard August 19, 2015 Assignment 1: Molecular Mechanics (PART 1 25 points) In this assignment, you will perform some molecular mechanics calculations using the
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Athena Visual Software, Inc. 1
 Athena Visual Studio Visual Kinetics Tutorial VisualKinetics is an integrated tool within the Athena Visual Studio software environment, which allows scientists and engineers to simulate the dynamic behavior
Athena Visual Studio Visual Kinetics Tutorial VisualKinetics is an integrated tool within the Athena Visual Studio software environment, which allows scientists and engineers to simulate the dynamic behavior
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Comparison of the a Priori COSMO-RS Models and Group Contribution Methods: Original UNIFAC, Modified UNIFAC(Do), and Modified UNIFAC(Do) Consortium
 pubs.acs.org/iecr Comparison of the a Priori COSMO-RS Models and Group Contribution Methods: Original UNIFAC, Modified UNIFAC(Do), and Modified UNIFAC(Do) Consortium Zhimin Xue, Tiancheng Mu,*, and Ju
pubs.acs.org/iecr Comparison of the a Priori COSMO-RS Models and Group Contribution Methods: Original UNIFAC, Modified UNIFAC(Do), and Modified UNIFAC(Do) Consortium Zhimin Xue, Tiancheng Mu,*, and Ju
Chemistry 14CL. Worksheet for the Molecular Modeling Workshop. (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell)
 Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Aspen Dr. Ziad Abuelrub
 Aspen Plus Lab Pharmaceutical Plant Design Aspen Dr. Ziad Abuelrub OUTLINE 1. Introduction 2. Getting Started 3. Thermodynamic Models & Physical Properties 4. Pressure Changers 5. Heat Exchangers 6. Flowsheet
Aspen Plus Lab Pharmaceutical Plant Design Aspen Dr. Ziad Abuelrub OUTLINE 1. Introduction 2. Getting Started 3. Thermodynamic Models & Physical Properties 4. Pressure Changers 5. Heat Exchangers 6. Flowsheet
ISIS/Draw "Quick Start"
 ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
41. Sim Reactions Example
 HSC Chemistry 7.0 41-1(6) 41. Sim Reactions Example Figure 1: Sim Reactions Example, Run mode view after calculations. General This example contains instruction how to create a simple model. The example
HSC Chemistry 7.0 41-1(6) 41. Sim Reactions Example Figure 1: Sim Reactions Example, Run mode view after calculations. General This example contains instruction how to create a simple model. The example
Stoichiometric Reactor Simulation Robert P. Hesketh and Concetta LaMarca Chemical Engineering, Rowan University (Revised 4/8/09)
 Stoichiometric Reactor Simulation Robert P. Hesketh and Concetta LaMarca Chemical Engineering, Rowan University (Revised 4/8/09) In this session you will learn how to create a stoichiometric reactor model
Stoichiometric Reactor Simulation Robert P. Hesketh and Concetta LaMarca Chemical Engineering, Rowan University (Revised 4/8/09) In this session you will learn how to create a stoichiometric reactor model
Introduction to Hartree-Fock calculations in Spartan
 EE5 in 2008 Hannes Jónsson Introduction to Hartree-Fock calculations in Spartan In this exercise, you will get to use state of the art software for carrying out calculations of wavefunctions for molecues,
EE5 in 2008 Hannes Jónsson Introduction to Hartree-Fock calculations in Spartan In this exercise, you will get to use state of the art software for carrying out calculations of wavefunctions for molecues,
COSMO-RS Theory. The Basics
 Theory The Basics From µ to properties Property µ 1 µ 2 activity coefficient vapor pressure Infinite dilution Gas phase Pure compound Pure bulk compound Partition coefficient Phase 1 Phase 2 Liquid-liquid
Theory The Basics From µ to properties Property µ 1 µ 2 activity coefficient vapor pressure Infinite dilution Gas phase Pure compound Pure bulk compound Partition coefficient Phase 1 Phase 2 Liquid-liquid
The OptiSage module. Use the OptiSage module for the assessment of Gibbs energy data. Table of contents
 The module Use the module for the assessment of Gibbs energy data. Various types of experimental data can be utilized in order to generate optimized parameters for the Gibbs energies of stoichiometric
The module Use the module for the assessment of Gibbs energy data. Various types of experimental data can be utilized in order to generate optimized parameters for the Gibbs energies of stoichiometric
Building Inflation Tables and CER Libraries
 Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
NMR Predictor. Introduction
 NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
Application Note. U. Heat of Formation of Ethyl Alcohol and Dimethyl Ether. Introduction
 Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Assignment 2: Conformation Searching (50 points)
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard September 16, 2015 Assignment 2: Conformation Searching (50 points) In this assignment, you will use the Spartan software package to investigate some conformation
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard September 16, 2015 Assignment 2: Conformation Searching (50 points) In this assignment, you will use the Spartan software package to investigate some conformation
Jaguar DFT Optimizations and Transition State Searches
 Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
v Prerequisite Tutorials GSSHA WMS Basics Watershed Delineation using DEMs and 2D Grid Generation Time minutes
 v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
Calculating NMR Chemical Shifts for beta-ionone O
 Calculating NMR Chemical Shifts for beta-ionone O Molecular orbital calculations can be used to get good estimates for chemical shifts. In this exercise we will calculate the chemical shifts for beta-ionone.
Calculating NMR Chemical Shifts for beta-ionone O Molecular orbital calculations can be used to get good estimates for chemical shifts. In this exercise we will calculate the chemical shifts for beta-ionone.
15. Studio ScaleChem Getting Started
 15. Studio ScaleChem Getting Started Terminology Analysis Brines Gases Oils Reconciliation Before we can discuss how to use Studio ScaleChem we must first discuss some terms. This will help us define some
15. Studio ScaleChem Getting Started Terminology Analysis Brines Gases Oils Reconciliation Before we can discuss how to use Studio ScaleChem we must first discuss some terms. This will help us define some
ST-Links. SpatialKit. Version 3.0.x. For ArcMap. ArcMap Extension for Directly Connecting to Spatial Databases. ST-Links Corporation.
 ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
ON SITE SYSTEMS Chemical Safety Assistant
 ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
Space Objects. Section. When you finish this section, you should understand the following:
 GOLDMC02_132283433X 8/24/06 2:21 PM Page 97 Section 2 Space Objects When you finish this section, you should understand the following: How to create a 2D Space Object and label it with a Space Tag. How
GOLDMC02_132283433X 8/24/06 2:21 PM Page 97 Section 2 Space Objects When you finish this section, you should understand the following: How to create a 2D Space Object and label it with a Space Tag. How
Automatic Watershed Delineation using ArcSWAT/Arc GIS
 Automatic Watershed Delineation using ArcSWAT/Arc GIS By: - Endager G. and Yalelet.F 1. Watershed Delineation This tool allows the user to delineate sub watersheds based on an automatic procedure using
Automatic Watershed Delineation using ArcSWAT/Arc GIS By: - Endager G. and Yalelet.F 1. Watershed Delineation This tool allows the user to delineate sub watersheds based on an automatic procedure using
Molecular Graphics. Molecular Graphics Expt. 1 1
 Molecular Graphics Expt. 1 1 Molecular Graphics The study of organic chemistry has for more than a century and a half focussed on the relationship between the structure of an organic molecule (its three-dimensional
Molecular Graphics Expt. 1 1 Molecular Graphics The study of organic chemistry has for more than a century and a half focussed on the relationship between the structure of an organic molecule (its three-dimensional
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Silica surface - Materials Studio tutorial. CREATING SiO 2 SURFACE
 Silica surface - Materials Studio tutorial CREATING SiO 2 SURFACE Our goal surface of SiO2 6.948 Ǻ Import structure The XRD experiment gives us such parameters as: lattice parameters, symmetry group and
Silica surface - Materials Studio tutorial CREATING SiO 2 SURFACE Our goal surface of SiO2 6.948 Ǻ Import structure The XRD experiment gives us such parameters as: lattice parameters, symmetry group and
Agilent MassHunter Quantitative Data Analysis
 Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch and Method Setup Outliers, Data Review, Reporting 1 MassHunter Quantitative Analysis
Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch and Method Setup Outliers, Data Review, Reporting 1 MassHunter Quantitative Analysis
LAB 2 - ONE DIMENSIONAL MOTION
 Name Date Partners L02-1 LAB 2 - ONE DIMENSIONAL MOTION OBJECTIVES Slow and steady wins the race. Aesop s fable: The Hare and the Tortoise To learn how to use a motion detector and gain more familiarity
Name Date Partners L02-1 LAB 2 - ONE DIMENSIONAL MOTION OBJECTIVES Slow and steady wins the race. Aesop s fable: The Hare and the Tortoise To learn how to use a motion detector and gain more familiarity
Getting started with BatchReactor Example : Simulation of the Chlorotoluene chlorination
 Getting started with BatchReactor Example : Simulation of the Chlorotoluene chlorination 2011 ProSim S.A. All rights reserved. Introduction This document presents the different steps to follow in order
Getting started with BatchReactor Example : Simulation of the Chlorotoluene chlorination 2011 ProSim S.A. All rights reserved. Introduction This document presents the different steps to follow in order
Prediction of Vapor/Liquid Equilibrium Behavior from Quantum Mechanical Data
 The University of Akron IdeaExchange@UAkron Honors Research Projects The Dr. Gary B. and Pamela S. Williams Honors College Spring 2016 Prediction of Vapor/Liquid Equilibrium Behavior from Quantum Mechanical
The University of Akron IdeaExchange@UAkron Honors Research Projects The Dr. Gary B. and Pamela S. Williams Honors College Spring 2016 Prediction of Vapor/Liquid Equilibrium Behavior from Quantum Mechanical
Spatial Data Analysis in Archaeology Anthropology 589b. Kriging Artifact Density Surfaces in ArcGIS
 Spatial Data Analysis in Archaeology Anthropology 589b Fraser D. Neiman University of Virginia 2.19.07 Spring 2007 Kriging Artifact Density Surfaces in ArcGIS 1. The ingredients. -A data file -- in.dbf
Spatial Data Analysis in Archaeology Anthropology 589b Fraser D. Neiman University of Virginia 2.19.07 Spring 2007 Kriging Artifact Density Surfaces in ArcGIS 1. The ingredients. -A data file -- in.dbf
APBS electrostatics in VMD - Software. APBS! >!Examples! >!Visualization! >! Contents
 Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
AAG TPoint Mapper (Version 1.40)
 AAG TPoint Mapper (Version 1.40) AAG_TPointMapper works together with Maxim DL, Pinpoint, TheSky6 and TPoint to automate the process of building a TPoint model for a GOTO telescope connected to TheSky6.
AAG TPoint Mapper (Version 1.40) AAG_TPointMapper works together with Maxim DL, Pinpoint, TheSky6 and TPoint to automate the process of building a TPoint model for a GOTO telescope connected to TheSky6.
You w i ll f ol l ow these st eps : Before opening files, the S c e n e panel is active.
 You w i ll f ol l ow these st eps : A. O pen a n i m a g e s t a c k. B. Tr a c e t h e d e n d r i t e w i t h t h e user-guided m ode. C. D e t e c t t h e s p i n e s a u t o m a t i c a l l y. D. C
You w i ll f ol l ow these st eps : A. O pen a n i m a g e s t a c k. B. Tr a c e t h e d e n d r i t e w i t h t h e user-guided m ode. C. D e t e c t t h e s p i n e s a u t o m a t i c a l l y. D. C
Physical Chemistry Final Take Home Fall 2003
 Physical Chemistry Final Take Home Fall 2003 Do one of the following questions. These projects are worth 30 points (i.e. equivalent to about two problems on the final). Each of the computational problems
Physical Chemistry Final Take Home Fall 2003 Do one of the following questions. These projects are worth 30 points (i.e. equivalent to about two problems on the final). Each of the computational problems
Tutorial. Getting started. Sample to Insight. March 31, 2016
 Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
Chem 1 Kinetics. Objectives. Concepts
 Chem 1 Kinetics Objectives 1. Learn some basic ideas in chemical kinetics. 2. Understand how the computer visualizations can be used to benefit the learning process. 3. Understand how the computer models
Chem 1 Kinetics Objectives 1. Learn some basic ideas in chemical kinetics. 2. Understand how the computer visualizations can be used to benefit the learning process. 3. Understand how the computer models
1 Introduction to Computational Chemistry (Spartan)
 1 Introduction to Computational Chemistry (Spartan) Start Spartan by clicking Start / Programs / Spartan Then click File / New Exercise 1 Study of H-X-H Bond Angles (Suitable for general chemistry) Structure
1 Introduction to Computational Chemistry (Spartan) Start Spartan by clicking Start / Programs / Spartan Then click File / New Exercise 1 Study of H-X-H Bond Angles (Suitable for general chemistry) Structure
Expt MM 1. MOLECULAR MODELING AND PREDICTIONS OF EQUILIBRIUM CONSTANT FOR MENTHONE (trans) AND ISOMENTHONE (cis) ISOMERS (MM)
 Expt MM 1 MOLECULAR MODELING AND PREDICTIONS OF EQUILIBRIUM CONSTANT FOR MENTHONE (trans) AND ISOMENTHONE (cis) ISOMERS (MM) Important Modification Note the software in use may be changed in 2008 to Scigress.
Expt MM 1 MOLECULAR MODELING AND PREDICTIONS OF EQUILIBRIUM CONSTANT FOR MENTHONE (trans) AND ISOMENTHONE (cis) ISOMERS (MM) Important Modification Note the software in use may be changed in 2008 to Scigress.
Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018)
 Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018) Abstract Students are introduced to basic features of Scigress by building molecules and performing calculations on them using semi-empirical
Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018) Abstract Students are introduced to basic features of Scigress by building molecules and performing calculations on them using semi-empirical
Preparing a PDB File
 Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Computational Chemistry Lab Module: Conformational Analysis of Alkanes
 Introduction Computational Chemistry Lab Module: Conformational Analysis of Alkanes In this experiment, we will use CAChe software package to model the conformations of butane, 2-methylbutane, and substituted
Introduction Computational Chemistry Lab Module: Conformational Analysis of Alkanes In this experiment, we will use CAChe software package to model the conformations of butane, 2-methylbutane, and substituted
PDF-4+ Tools and Searches
 PDF-4+ Tools and Searches PDF-4+ 2018 The PDF-4+ 2018 database is powered by our integrated search display software. PDF-4+ 2018 boasts 72 search selections coupled with 125 display fields resulting in
PDF-4+ Tools and Searches PDF-4+ 2018 The PDF-4+ 2018 database is powered by our integrated search display software. PDF-4+ 2018 boasts 72 search selections coupled with 125 display fields resulting in
Cerno Application Note Extending the Limits of Mass Spectrometry
 Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Example: H 2 O (the car file)
 Example: H 2 O (the car file) As a practical example of DFT methods we calculate the energy and electronic properties of the water molecule. In order to carry out the DFT calculation you will need a set
Example: H 2 O (the car file) As a practical example of DFT methods we calculate the energy and electronic properties of the water molecule. In order to carry out the DFT calculation you will need a set
Ligand Scout Tutorials
 Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
3D - Structure Graphics Capabilities with PDF-4 Database Products
 3D - Structure Graphics Capabilities with PDF-4 Database Products Atomic Structure Information in the PDF-4 Databases ICDD s PDF-4 databases contain atomic structure information for a significant number
3D - Structure Graphics Capabilities with PDF-4 Database Products Atomic Structure Information in the PDF-4 Databases ICDD s PDF-4 databases contain atomic structure information for a significant number
SeeSAR 7.1 Beginners Guide. June 2017
 SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
Electronegativity Scale F > O > Cl, N > Br > C, H
 Organic Chem Chapter 12 Alkanes Organic chemistry is the study of carbon compounds. Carbon has several properties that are worth discussing: Tetravalent Always forms 4 bonds Can form multiple bonds (double
Organic Chem Chapter 12 Alkanes Organic chemistry is the study of carbon compounds. Carbon has several properties that are worth discussing: Tetravalent Always forms 4 bonds Can form multiple bonds (double
Using a mini-gas Chromatograph (GC): Identifying Unknown Compounds
 Experiment Using a mini-gas Chromatograph (GC): Identifying Unknown Compounds There are many different types of chromatography: paper, thin layer, liquid, high-pressure liquid (HPLC) and gas (GC). Chromatography
Experiment Using a mini-gas Chromatograph (GC): Identifying Unknown Compounds There are many different types of chromatography: paper, thin layer, liquid, high-pressure liquid (HPLC) and gas (GC). Chromatography
INVESTIGATING GAS CHROMATOGRAPHY rev 8/12
 EXPERIMENT 2 INVESTIGATING GAS CHROMATOGRAPHY rev 8/12 GOAL In this experiment, you will measure and analyze the chromatogram of a mixture of seven compounds using a Vernier Mini GC. You will then vary
EXPERIMENT 2 INVESTIGATING GAS CHROMATOGRAPHY rev 8/12 GOAL In this experiment, you will measure and analyze the chromatogram of a mixture of seven compounds using a Vernier Mini GC. You will then vary
CHEMDRAW ULTRA ITEC107 - Introduction to Computing for Pharmacy. ITEC107 - Introduction to Computing for Pharmacy 1
 CHEMDRAW ULTRA 12.0 ITEC107 - Introduction to Computing for Pharmacy 1 Objectives Basic drawing skills with ChemDraw Bonds, captions, hotkeys, chains, arrows Checking and cleaning up structures Chemical
CHEMDRAW ULTRA 12.0 ITEC107 - Introduction to Computing for Pharmacy 1 Objectives Basic drawing skills with ChemDraw Bonds, captions, hotkeys, chains, arrows Checking and cleaning up structures Chemical
Creating Empirical Calibrations
 030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
Winmostar tutorial LAMMPS Melting point V X-Ability Co,. Ltd. 2017/8/17
 Winmostar tutorial LAMMPS Melting point V7.025 X-Ability Co,. Ltd. question@winmostar.com Contents Configure I. Build solid phase II. Equilibration of solid phase III. Equilibration of liquid phase IV.
Winmostar tutorial LAMMPS Melting point V7.025 X-Ability Co,. Ltd. question@winmostar.com Contents Configure I. Build solid phase II. Equilibration of solid phase III. Equilibration of liquid phase IV.
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Working with ArcGIS: Classification
 Working with ArcGIS: Classification 2 Abbreviations D-click R-click TOC Double Click Right Click Table of Content Introduction The benefit from the use of geographic information system (GIS) software is
Working with ArcGIS: Classification 2 Abbreviations D-click R-click TOC Double Click Right Click Table of Content Introduction The benefit from the use of geographic information system (GIS) software is
(THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE)
 PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
Introduction Molecular Structure Script Console External resources Advanced topics. JMol tutorial. Giovanni Morelli.
 Gen 19th, 2017 1 2 Create and edit Display and view Mesurament and labelling Surface and Orbitals 3 4 from Database Protein Enzyme Crystal Structure and Unit Cell 5 Symmetry Animation General information
Gen 19th, 2017 1 2 Create and edit Display and view Mesurament and labelling Surface and Orbitals 3 4 from Database Protein Enzyme Crystal Structure and Unit Cell 5 Symmetry Animation General information
Conformational Analysis of n-butane
 Conformational Analysis of n-butane In this exercise you will calculate the Molecular Mechanics (MM) single point energy of butane in various conformations with respect to internal rotation around the
Conformational Analysis of n-butane In this exercise you will calculate the Molecular Mechanics (MM) single point energy of butane in various conformations with respect to internal rotation around the
FROM ATOM TO POLYMER. creating and manipulating models of chemical compounds In BIOVIA Materials studio
 FROM ATOM TO POLYMER creating and manipulating models of chemical compounds In BIOVIA Materials studio What can be built in Materials Studio? Single structures What can be built in Materials Studio? Homopolymers
FROM ATOM TO POLYMER creating and manipulating models of chemical compounds In BIOVIA Materials studio What can be built in Materials Studio? Single structures What can be built in Materials Studio? Homopolymers
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Winmostar tutorial LAMMPS Polymer modeling V X-Ability Co,. Ltd. 2017/7/6
 Winmostar tutorial LAMMPS Polymer modeling V7.021 X-Ability Co,. Ltd. question@winmostar.com 2017/7/6 Contents Configure I. Register a monomer II. Define a polymer III. Build a simulation cell IV. Execute
Winmostar tutorial LAMMPS Polymer modeling V7.021 X-Ability Co,. Ltd. question@winmostar.com 2017/7/6 Contents Configure I. Register a monomer II. Define a polymer III. Build a simulation cell IV. Execute
CREATING CUSTOMIZED DATE RANGE COLLECTIONS IN PRESENTATION STUDIO
 CREATING CUSTOMIZED DATE RANGE COLLECTIONS IN PRESENTATION STUDIO Date range collections are pre-defined reporting periods for performance data. You have two options: Dynamic date ranges automatically
CREATING CUSTOMIZED DATE RANGE COLLECTIONS IN PRESENTATION STUDIO Date range collections are pre-defined reporting periods for performance data. You have two options: Dynamic date ranges automatically
Aspen Plus PFR Reactors Tutorial using Styrene with Pressure Drop Considerations By Robert P. Hesketh and Concetta LaMarca Spring 2005
 Aspen Plus PFR Reactors Tutorial using Styrene with Pressure Drop Considerations By Robert P. Hesketh and Concetta LaMarca Spring 2005 In this laboratory we will incorporate pressure-drop calculations
Aspen Plus PFR Reactors Tutorial using Styrene with Pressure Drop Considerations By Robert P. Hesketh and Concetta LaMarca Spring 2005 In this laboratory we will incorporate pressure-drop calculations
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Project 3: Molecular Orbital Calculations of Diatomic Molecules. This project is worth 30 points and is due on Wednesday, May 2, 2018.
 Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Task 1: Start ArcMap and add the county boundary data from your downloaded dataset to the data frame.
 Exercise 6 Coordinate Systems and Map Projections The following steps describe the general process that you will follow to complete the exercise. Specific steps will be provided later in the step-by-step
Exercise 6 Coordinate Systems and Map Projections The following steps describe the general process that you will follow to complete the exercise. Specific steps will be provided later in the step-by-step
7 Infrared, Thermochemistry, UV-Vis, and NMR
 7 Infrared, Thermochemistry, UV-Vis, and NMR Exercise 1 Method Dependence and Scaling for the Infrared Spectrum of Formaldehyde. Build a molecule of formaldehyde using sp 2 C and atoms. Clean up the structure
7 Infrared, Thermochemistry, UV-Vis, and NMR Exercise 1 Method Dependence and Scaling for the Infrared Spectrum of Formaldehyde. Build a molecule of formaldehyde using sp 2 C and atoms. Clean up the structure
PDF-4+ Tools and Searches
 PDF-4+ Tools and Searches PDF-4+ 2019 The PDF-4+ 2019 database is powered by our integrated search display software. PDF-4+ 2019 boasts 74 search selections coupled with 126 display fields resulting in
PDF-4+ Tools and Searches PDF-4+ 2019 The PDF-4+ 2019 database is powered by our integrated search display software. PDF-4+ 2019 boasts 74 search selections coupled with 126 display fields resulting in
Watershed Modeling Orange County Hydrology Using GIS Data
 v. 10.0 WMS 10.0 Tutorial Watershed Modeling Orange County Hydrology Using GIS Data Learn how to delineate sub-basins and compute soil losses for Orange County (California) hydrologic modeling Objectives
v. 10.0 WMS 10.0 Tutorial Watershed Modeling Orange County Hydrology Using GIS Data Learn how to delineate sub-basins and compute soil losses for Orange County (California) hydrologic modeling Objectives
Flash Point Calculation by UNIFAC
 Flash Point Calculation by UNIFAC Short Introduction and Tutorial DDBSP - Dortmund Data Bank Software Package DDBST Software & Separation Technology GmbH Marie-Curie-Straße 10 D-26129 Oldenburg Tel.: +49
Flash Point Calculation by UNIFAC Short Introduction and Tutorial DDBSP - Dortmund Data Bank Software Package DDBST Software & Separation Technology GmbH Marie-Curie-Straße 10 D-26129 Oldenburg Tel.: +49
How to Create a Substance Answer Set
 How to Create a Substance Answer Set Select among five search techniques to find substances Since substances can be described by multiple names or other characteristics, SciFinder gives you the flexibility
How to Create a Substance Answer Set Select among five search techniques to find substances Since substances can be described by multiple names or other characteristics, SciFinder gives you the flexibility
Data Mining with the PDF-4 Databases. FeO Non-stoichiometric Oxides
 Data Mining with the PDF-4 Databases FeO Non-stoichiometric Oxides This is one of three example-based tutorials for using the data mining capabilities of the PDF-4+ database and it covers the following
Data Mining with the PDF-4 Databases FeO Non-stoichiometric Oxides This is one of three example-based tutorials for using the data mining capabilities of the PDF-4+ database and it covers the following
Agilent MassHunter Quantitative Data Analysis
 Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch Table, Compound Information Setup, Calibration Curve and Globals Settings 1 MassHunter
Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch Table, Compound Information Setup, Calibration Curve and Globals Settings 1 MassHunter
MAGNETITE OXIDATION EXAMPLE
 HSC Chemistry 7.0 1 MAGNETITE OXIDATION EXAMPLE Pelletized magnetite (Fe 3 O 4 ) ore may be oxidized to hematite (Fe 2 O 3 ) in shaft furnace. Typical magnetite content in ore is some 95%. Oxidation is
HSC Chemistry 7.0 1 MAGNETITE OXIDATION EXAMPLE Pelletized magnetite (Fe 3 O 4 ) ore may be oxidized to hematite (Fe 2 O 3 ) in shaft furnace. Typical magnetite content in ore is some 95%. Oxidation is
13. Equilibrium Module - Description of Menus and Options
 15008-ORC-J 1 (57) 13. Equilibrium Module - Description of Menus and Options 15008-ORC-J 2 (57) SUMMARY HSC Equilibrium module enables user to calculate multi-component equilibrium compositions in heterogeneous
15008-ORC-J 1 (57) 13. Equilibrium Module - Description of Menus and Options 15008-ORC-J 2 (57) SUMMARY HSC Equilibrium module enables user to calculate multi-component equilibrium compositions in heterogeneous
17. E - ph (Pourbaix) Diagrams Module
 HSC - EpH 15011-ORC-J 1 (16) 17. E - ph (Pourbaix) Diagrams Module E - ph diagrams show the thermodynamic stability areas of different species in an aqueous solution. Stability areas are presented as a
HSC - EpH 15011-ORC-J 1 (16) 17. E - ph (Pourbaix) Diagrams Module E - ph diagrams show the thermodynamic stability areas of different species in an aqueous solution. Stability areas are presented as a
Transition states and reaction paths
 Transition states and reaction paths Lab 4 Theoretical background Transition state A transition structure is the molecular configuration that separates reactants and products. In a system with a single
Transition states and reaction paths Lab 4 Theoretical background Transition state A transition structure is the molecular configuration that separates reactants and products. In a system with a single
SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide
 SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide This page is intentionally left blank. SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide The ACTiSys IR Programmer and SuperCELL
SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide This page is intentionally left blank. SuperCELL Data Programmer and ACTiSys IR Programmer User s Guide The ACTiSys IR Programmer and SuperCELL
Introduction to Structure Preparation and Visualization
 Introduction to Structure Preparation and Visualization Created with: Release 2018-4 Prerequisites: Release 2018-2 or higher Access to the internet Categories: Molecular Visualization, Structure-Based
Introduction to Structure Preparation and Visualization Created with: Release 2018-4 Prerequisites: Release 2018-2 or higher Access to the internet Categories: Molecular Visualization, Structure-Based
M E R C E R W I N WA L K T H R O U G H
 H E A L T H W E A L T H C A R E E R WA L K T H R O U G H C L I E N T S O L U T I O N S T E A M T A B L E O F C O N T E N T 1. Login to the Tool 2 2. Published reports... 7 3. Select Results Criteria...
H E A L T H W E A L T H C A R E E R WA L K T H R O U G H C L I E N T S O L U T I O N S T E A M T A B L E O F C O N T E N T 1. Login to the Tool 2 2. Published reports... 7 3. Select Results Criteria...
NMRPredict Functional Block Diagram
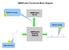 NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
Performing a Pharmacophore Search using CSD-CrossMiner
 Table of Contents Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Searching with a Pharmacophore... 4 Performing a Pharmacophore Search using CSD-CrossMiner Version 2.0
Table of Contents Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Searching with a Pharmacophore... 4 Performing a Pharmacophore Search using CSD-CrossMiner Version 2.0
Physical Properties. Alcohols can be: CH CH 2 OH CH 2 CH 3 C OH CH 3. Secondary alcohol. Primary alcohol. Tertiary alcohol
 Chapter 10: Structure and Synthesis of Alcohols 100 Physical Properties Alcohols can be: CH 3 CH 3 CH CH 2 OH * Primary alcohol CH 3 OH CH * CH 2 CH 3 Secondary alcohol CH 3 CH 3 * C OH CH 3 Tertiary alcohol
Chapter 10: Structure and Synthesis of Alcohols 100 Physical Properties Alcohols can be: CH 3 CH 3 CH CH 2 OH * Primary alcohol CH 3 OH CH * CH 2 CH 3 Secondary alcohol CH 3 CH 3 * C OH CH 3 Tertiary alcohol
1 An Experimental and Computational Investigation of the Dehydration of 2-Butanol
 1 An Experimental and Computational Investigation of the Dehydration of 2-Butanol Summary. 2-Butanol will be dehydrated to a mixture of 1-butene and cis- and trans-2-butene using the method described in
1 An Experimental and Computational Investigation of the Dehydration of 2-Butanol Summary. 2-Butanol will be dehydrated to a mixture of 1-butene and cis- and trans-2-butene using the method described in
PDF-2 Tools and Searches
 PDF-2 Tools and Searches PDF-2 2019 The PDF-2 2019 database is powered by our integrated search display software. PDF-2 2019 boasts 69 search selections coupled with 53 display fields resulting in a nearly
PDF-2 Tools and Searches PDF-2 2019 The PDF-2 2019 database is powered by our integrated search display software. PDF-2 2019 boasts 69 search selections coupled with 53 display fields resulting in a nearly
Name. Chem Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2
 Name Chem 322 - Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2 Click on Titan in the Start menu. When it boots, click on the right corner to make the window full-screen. icon in the
Name Chem 322 - Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2 Click on Titan in the Start menu. When it boots, click on the right corner to make the window full-screen. icon in the
Searching Substances in Reaxys
 Searching Substances in Reaxys Learning Objectives Understand that substances in Reaxys have different sources (e.g., Reaxys, PubChem) and can be found in Document, Reaction and Substance Records Recognize
Searching Substances in Reaxys Learning Objectives Understand that substances in Reaxys have different sources (e.g., Reaxys, PubChem) and can be found in Document, Reaction and Substance Records Recognize
mylab: Chemical Safety Module Last Updated: January 19, 2018
 : Chemical Safety Module Contents Introduction... 1 Getting started... 1 Login... 1 Receiving Items from MMP Order... 3 Inventory... 4 Show me Chemicals where... 4 Items Received on... 5 All Items... 5
: Chemical Safety Module Contents Introduction... 1 Getting started... 1 Login... 1 Receiving Items from MMP Order... 3 Inventory... 4 Show me Chemicals where... 4 Items Received on... 5 All Items... 5
Dimerization in a Tubular Reactor
 Dimerization in a Tubular Reactor Tubular reactors are very common in large-scale continuous, for example in the petroleum industry. One key design and optimization parameter is the conversion, that is
Dimerization in a Tubular Reactor Tubular reactors are very common in large-scale continuous, for example in the petroleum industry. One key design and optimization parameter is the conversion, that is
Finite Element Modules for Enhancing Undergraduate Transport Courses: Application to Fuel Cell Fundamentals
 Finite Element Modules for Enhancing Undergraduate Transport Courses: Application to Fuel Cell Fundamentals Originally published in 007 American Society for Engineering Education Conference Proceedings
Finite Element Modules for Enhancing Undergraduate Transport Courses: Application to Fuel Cell Fundamentals Originally published in 007 American Society for Engineering Education Conference Proceedings
Motion II. Goals and Introduction
 Motion II Goals and Introduction As you have probably already seen in lecture or homework, and if you ve performed the experiment Motion I, it is important to develop a strong understanding of how to model
Motion II Goals and Introduction As you have probably already seen in lecture or homework, and if you ve performed the experiment Motion I, it is important to develop a strong understanding of how to model
Computer simulation of radioactive decay
 Computer simulation of radioactive decay y now you should have worked your way through the introduction to Maple, as well as the introduction to data analysis using Excel Now we will explore radioactive
Computer simulation of radioactive decay y now you should have worked your way through the introduction to Maple, as well as the introduction to data analysis using Excel Now we will explore radioactive
Physical Chemistry Lab: The Hydrolysis of Aspirin. Each collaborative team determines the rate constant for the hydrolysis of aspirin.
 CH 348 Physical Chemistry Spring 2009 MK Bowman Adapted from: Marrs, Peter S. J. Chem. Educ. 2004 81 870. Physical Chemistry Lab: The Hydrolysis of Aspirin Each collaborative team determines the rate constant
CH 348 Physical Chemistry Spring 2009 MK Bowman Adapted from: Marrs, Peter S. J. Chem. Educ. 2004 81 870. Physical Chemistry Lab: The Hydrolysis of Aspirin Each collaborative team determines the rate constant
EXPERIMENT 1: Survival Organic Chemistry: Molecular Models
 EXPERIMENT 1: Survival Organic Chemistry: Molecular Models Introduction: The goal in this laboratory experience is for you to easily and quickly move between empirical formulas, molecular formulas, condensed
EXPERIMENT 1: Survival Organic Chemistry: Molecular Models Introduction: The goal in this laboratory experience is for you to easily and quickly move between empirical formulas, molecular formulas, condensed
