NMR Predictor. Introduction
|
|
|
- Alice Hamilton
- 6 years ago
- Views:
Transcription
1 NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit menu Options menu Examples View menu Examples Help menu Toolbar Panels Molecule View Panel Table of Chemical Shifts NMR Spectrum Preview Panel NMR Spectrum Display Panel NMR Prediction pop-up menu NMR Spectrum Viewer Menu system File menu View menu Help Toolbar Panels Spectrum View Panel Spectrum Display Panel Status bar NMR Spectrum Viewer Pop-up Menu References Introduction Predicting the NMR spectrum for a chemical compound can play an important role in structure validation and elucidation. The NMR Predictor is a standalone tool that can predict both 1 H and
2 13 C NMR spectra of organic compounds. NMR Predictor QuickHelp The picture below gives a quick overview on the capabilities of ChemAxon's NMR Predictor. Fig. 1 NMR Predictor QuickHelp NMR Predictor Overview The NMR Predictor application is able to predict NMR spectra for standard organic molecules containing the most frequent atoms (molecules containing H, C, N, O, F, Cl, Br, I, P, S, Si, Se, B, Sn, Ge, Te and As atoms). The chemical shifts are estimated by a mixed HOSE- and linear
3 model based on a topological description scheme and are in relation to the chemical shift of tetramethylsilane ((TMS)=0 ppm). 13C and 1H chemical shift training data were retrieved for training from the NMRShift Database. You can read more about the NMR chemical shift model description here. The NMR Predictor is integrated into MarvinSketch (you can reach it under the Calculations > NMR menu), and contains three components to discover NMR spectra of molecules. Fig. 2 The Calculations > NMR menu in MarvinSketch and its three components NMR prediction is also accessible via cxcalc. To get started with the NMR Predictor via cxcalc, use the cxcalc nmr -h command. Chemical features The NMR Predictor has the following basic features: Prediction of 13C and 1H NMR chemical shifts Spin-spin couplings are taken into account according to the first order approximation H-H, H-F and C-F couplings are considered during NMR spectrum calculation Diastereotopic protons are differentiated NMR Spectrum Viewer is able to display NMR spectra in JCAMP-DX format. GUI features The NMR Predictor graphical user interface (GUI) has the following features: Export predicted spectrum to MOL file Export predicted spectrum to JCAMP-DX file and/or import JCAMP-DX (*.jdx) reference spectrum Create PDF file as a report of your prediction, containing molecule structure, predicted
4 spectrum, and related tables Detached Copy to clipboard action for all predictor panels and tables is available Update molecule from MarvinSketch; Toggle between decoupled and coupled NMR spectrum Toggle between explicit and implicit hydrogen display Select NMR prediction frequency from a predetermined list Add common organic solvent peaks to predicted spectrum Add tautomer peaks to predicted spectrum Restore default NMR predictor settings, e.g. prediction frequency, display and view options Display realistic or line NMR spectra Add atom indices or chemical shift values to signals as spectrum labels Display spectrum scale in ppm or Hz units Show integral curve to assign value to NMR spectrum signals Display legend on spectrum display panel Show local maximum values of reference spectrum Personalize the color management of NMR Predictor Set chart color uniquely When you click on a peak on spectrum display panel or on an atom on molecule preview panel, selection will move to and zoom in on the selected signal Choose multiplet selection mode: individual selection in case of overlapping multiplets is available Use various modes of zoom in on spectrum Find spectrum and molecule structure related information in Atom, Multiplet, and Coupling tables Show atom indices on molecule structure corresponding to the different multiplets Atoms of the input molecule and multiplets of the NMR spectrum are linked together. Upon selection of an atom the corresponding multiplet is highlighted and vice versa. A single NMR prediction is allowed to contain more molecules. Usage You can predict 13C NMR and 1H NMR spectra of organic molecules drawn in MarvinSketch using the Calculations > NMR menu: Draw molecule in MarvinSketch. Go to Calculations > > and choose NMR CNMR Prediction to discover the predicted 13C NMR spectrum of the molecule, or HNMR Prediction to discover the predicted H NMR spectrum of the molecule The predicted spectrum will open in CNMR Prediction window if you chose CNMR
5 Prediction, and in HNMR Prediction window if you chose HNMR Prediction, respectively. You can predict both spectra of the molecule in question, which will appear in separate windows. Fig. 3 HNMR Prediction result window and its parts Both NMR Prediction windows consist of a menu, toolbar, and four panels. The name of the window is displayed at the top left corner. At the bottom left corner of the status bar general information on the NMR prediction is shown, i.e. nucleus, measurement unit and prediction frequency. At the bottom right corner the coordinates of mouse cursor position on the NMR Spectrum Display Panel are shown. Menu elements and panels of both 13C and 1H NMR Prediction windows are discussed together. Differences will be marked by the appropriate icon (
6 for CNMR Prediction, for HNMR Prediction). Menu system The menu system contains File, Edit, Options, View and Help menus. File menu The File menu can be used to export spectra to files; to import spectrum of JCAMP-DX file format and superimpose it on predicted NMR spectrum; to remove the imported spectrum and to close the NMR Prediction result window. File > Export to PDF...: exports the molecule structure, predicted spectrum (full), and related tables to PDF file. You can select Keep view settings option on the export dialog panel to keep the actual view of spectrum during export. File > Export to JCAMP-DX...: exports predicted spectrum to JCAMP-DX (jdx) file format. File > Export to Molfile...: exports predicted spectrum to MOL format. You can export the predicted spectrum data to SDF file format. The SDF file will contain structure and NMR Spectrum fields. The NMR Spectrum field contains the relevant atom number (AN), value of chemical shift (v), unit of chemical shift (u), and multiplicity of the signal (M) in the following format: v1;u1,m1;an1 v2;u2,m2;an2... vi;ui,mi;ani. File > Import from JCAMP-DX: imports a spectrum in JCAMP-DX format. The imported spectrum will be superimposed on the predicted NMR spectrum. File > Remove Imported Spectrum: removes previously imported JCAMP-DX spectrum from NMR Spectrum Display Panel. File > Exit: closes NMR Prediction window. Edit menu The Edit menu can be used to copy specific panels to clipboard and to update the molecule from MarvinSketch. You can also apply the right-click of your mouse on the proper panel to copy it to
7 the clipboard. Edit > Copy Spectrum: copies the actual view of Spectrum Display Panel to the clipboard. Edit > Copy Spectrum Preview: copies the actual view of Spectrum Preview Panel to the clipboard. Edit > Copy Molecule: copies the actual view of Molecule View Panel to the clipboard. Edit > Copy Multiplet Table: copies Multiple Table to the clipboard. Edit > Copy Atom Table: copies Atom Table to the clipboard. Edit > Copy Coupling Table: copies Coupling Table to the clipboard. Edit > Update Molecule: updates molecule on Molecule View Panel and the whole prediction at the same time. You can switch back to MarvinSketch window without closing NMR Prediction window; modify the molecule or draw a new molecule of which NMR spectrum you wish to predict. Switch back to NMR Prediction window and either select Update Molecule or click on the Update button on Molecule View Panel to refresh NMR prediction. Options menu The Options menu can be used to set optional NMR prediction settings: Options > Spin-Spin Coupling: prediction considers spin-spin coupling; the result is splitting of signals into multiplets according to the interaction between two nuclei. Options > Implicit Hydrogen Mode: hydrogens are displayed only on hetero and terminal atoms. Note If you switch off this mode: all hydrogens will be visible on Molecule View Panel; atoms will be re-numbered on all corresponding panels; coupling table will be filled in with relevant data. Options > NMR Prediction Frequency: sets the frequency of the NMR prediction. Select prediction frequency from the predetermined list. Prediction frequency influences the fine structure of the spectrum. Options > Add Solvent Peaks...: Adds NMR signal(s) of selected common organic solvent(s) to the predicted spectrum. Select solvents from the predetermined list and click OK. The signal(s) of selected solvent(s) will be added to the predicted spectrum. When spectrum labels are displayed, you can see the name of the solvent attached to the corresponding signal. We used the NMR shift data of common organic solvents in CDCl collected by. 3 Gottlieb et al Options > Select Tautomers...: opens a dialog where tautomers of the relevant molecule are displayed. The major tautomer is automatically selected. The values of tautomer distributions are obtained from MarvinSketch's Dominant tautomer distribution calculation. You can select altogether 8 tautomers to add their signal(s) to the predicted
8 spectrum. Check the upper right check box of the appropriate tautomer. The distribution of each tautomer has to be set before proceeding. When spectrum labels are displayed, the corresponding signals of the active tautomer can be seen on Spectrum Display Panel, while the tautomer peaks are signed according to their symbols. Options > Clear Tautomers: removes all selected tautomer peaks from predicted spectrum. Options > Reset Default Settings: resets zoom and the default Options, Color, and View settings of NMR predictor. This is for CNMR Predictor: NMR Prediction Frequency: 500 [125] MHz Spectrum Display: Realistic Spectrum Spectrum Labels: Chemical Shifts Measurement Unit: ppm Zoom Follows Selection: On HNMR Predictor: Spin-Spin Coupling: On Implicit Hydrogen Mode: On NMR Prediction Frequency: 500 MHz Spectrum Display: Realistic Spectrum Spectrum Labels: Chemical Shifts Measurement Unit: ppm Integral Curve: On Zoom Follows Selection: On Examples Example #1 Toggle Spin-Spin Coupling: choose Options > Spin-Spin Coupling Fig. 4 Spin-Spin coupling option effect on the prediction results Example #2 Toggle Implicit Hydrogen Mode: choose Options > Implicit Hydrogen Mode
9 Change default setting to: Spectrum Labels > Atom Numbers; Zoom in on the certain spectrum region. Fig. 5 Implicit Hydrogen Mode effect on the prediction View menu It is to select different display options related to the predicted spectrum and the molecule structure: Spectrum Display: Realistic Spectrum: displays predicted spectrum in a realistic way. Line Spectrum: predicted chemical shifts are presented by distinct lines with proper intensity. Spectrum Labels: In order to assign signals and relevant atoms more easily, you can display the atom numbers or the chemical shift values of each signal on the NMR Spectrum Display an Molecule View Panels. Select: Atom Numbers to see atoms assigned to each signal and to display atom numbers on Molecule View Panel as well. Chemical Shifts to see the exact chemical shift value of NMR signals on the NMR Spectrum Display Panel. None to remove spectrum labels. Note that you can display only one type of label at a time.
10 Measurement Unit: the chemical shift of tetramethylsilane (TMS) is set to zero, and all other chemical shifts are predicted relative to it. The unit of display can be: Hz or ppm. Integral Curve: displays integral curve on spectrum. Default setting is on. Display Legend: displays legend on Spectrum Display Panel. The legend contains information on different functions of Spectrum Display Panel. Reference Spectrum: It is an imported JCAMP-DX NMR spectrum that can be superimposed on the predicted NMR spectrum. Display Shifts: If the imported JCAMP-DX file has "PEAKTABLE" property, the chemical shifts of the imported spectrum can be displayed. None: Remove chemical shift labels of the reference spectrum. Set Colors...: you can customize the color of the predicted spectrum, reference spectrum and selection. Zoom Follows Selection: If you select an exact atom on Molecule View Panel, or a signal on NMR Spectrum Display Panel, the appropriate signal is centered and zoomed in on NMR Spectrum Display Panel. Select Individual Multiplets: In case of overlapping multiplets, this option enables highlighting individual multiplets. Horizontal Zoom In: zooms in on spectrum in X-axis direction. Horizontal Zoom Out: zooms out of spectrum in X-axis direction. Vertical Zoom In: zooms in on spectrum in Y-axis direction. Note that the bottom of the selection window is fixed. Vertical Zoom Out: zooms out of spectrum in Y-axis direction. Note that the bottom of the selection window is fixed. Reset Zoom: displays the whole spectrum in both directions.
11 Scale Up Reference: increases intensity of imported reference spectrum. Active when reference spectrum is imported. Scale Down Reference: decreases intensity of imported reference spectrum. Active when reference spectrum is imported. Examples Selecting individual multiplets Fig. 6 Selecting individual multiplets Help menu There are two menu points that can help you get started with the NMR Predictor: Help > Quick Help Help > Help Contents Toolbar You can use toolbar elements to access selected NMR Predictor menu items:
12 Export to PDF... Spectrum Display Spectrum Labels
13 Spin-Spin Coupling Integral Curve
14 Measurement Unit Zoom Follows Selection
15 Select Individual Multiplets Horizontal Zoom In
16 Horizontal Zoom Out Vertical Zoom In
17 Vertical Zoom Out Reset Zoom
18 Scale Up Reference Scale Down Reference
19 Quick Help Fig. 7 Toolbar elements of the NMR Predictor Panels NMR Prediction window contains the Molecule View Panel, the Table of Chemical Shifts, the NMR Spectrum Preview Panel and the NMR Spectrum Display Panel to present the predicted spectrum and to display selected features. Molecule View Panel The Molecule View Panel displays the molecule that has to be drawn in MarvinSketch. Using the Ctrl button while the cursor is located in this panel, the view of molecule can be controlled: Ctrl+Mouse scroll button zooms in/out on to the molecule Ctrl+Mouse dragging moves the molecule If you select Spectrum Labels > Atom Numbers, atom numbers will appear on both the Molecule View Panel and the NMR Spectrum Display Panel. If you select Spectrum Labels > Chemical Shifts, chemical shift values of predicted multiplets will appear on both the Molecule View Panel and the NMR Spectrum Display Panel. If you have added tautomers to the predicted spectrum via the Select Tautomers... option, the layout of Molecule View Panel will change: the active tautomer is displayed on the panel. To go to the next/previous tautomer, click on the arrows next to the molecule.
20 In the picture below symbols T1, T2,..., T, mark the selected tautomers. Hover over to see tautomer structure in a pop-up window. Click on the symbol to make it active. n Fig. 8 Molecule View Panel with tautomer windows Click on the Update button after you have made any modifications on molecular structure in MarvinSketch and you want to predict the NMR spectrum of the new molecule. The effect is equal to using the Edit > Update Molecule action. Table of Chemical Shifts The following tabs are available on this panel: Multiplet information, Atom information and Coupling information tabs.table on all tabs contains data of the predicted spectrum in Multiplet or Atom point of view. Coupling table contains the calculated coupling constants when the Spin-Spin coupling option is selected. Multiplet information table has six columns, namely: Atom numbers, Chemical shift, Net intensity, Intensity pattern, Multiplet information, and Quality. You can sort data according to these columns. Atom numbers are the numbers displayed on the molecule structure and are assigned automatically. Chemical shift values are displayed in the selected Measurement Unit. Net intensity is the integration value of the relevant signal. Intensity pattern describes the relative intensity of the multiplet elements. Multiplet information is the conventional one letter abbreviation of multiplicity, e.g.: s - singlet; d - doublet; t - triplet;... Quality defines the prediction quality according to our validation method. Definitions: good, medium, rough.
21 Atom information table has five columns, namely: Atom number, Chemical shift, Net intensity, Multiplet information, and Quality. Atom numbers are the numbers displayed on the molecule structure and are assigned automatically. Chemical shift values are displayed in the selected Measurement Unit. Net intensity is the integration value of the relevant signal. Multiplet information is the conventional one letter abbreviation of multiplicity, e.g.: s - singlet; d - doublet; t - triplet;... Quality defines the prediction quality according to our validation method. Definitions: good, medium, rough. Coupling information table has four columns, namely: Atom 1, Atom 2, Value, and Quality. Atom 1 and Atom 2 are the number of atoms that the coupling constant is connected to. The value of the coupling constant is displayed in Hz. Quality defines the prediction quality according to our validation method. Definitions: good, medium, rough. NMR Spectrum Preview Panel The NMR Spectrum Preview Panel displays the whole predicted spectrum. You can zoom in and out on spectrum by using your mouse, toolbar zoom items or menu items: If you want to zoom in on specific region of the spectrum, use left-click and drag on NMR Spectrum Preview Panel. The background of the selected region will turn to white, while unselected region of the spectrum will turn to grey. You can move the selection window by left-clicking into the middle of the selection window; hold mouse button while moving the selection, and release button to place it. You can resize the selection window if you grab-and-drag its yellow side frames (except bottom frame). NMR Spectrum Display Panel The NMR Spectrum Display Panel displays the appropriate zoom region of the spectrum of the molecule presented on Molecule View Panel. Move your mouse pointer over the NMR Spectrum Display Panel and use mouse-wheel to zoom in and out on NMR spectrum along the X-axis. Using Ctrl+mouse-wheel will zoom in and out on NMR spectrum along the Y-axis. If you have added tautomers to the predicted spectrum via Select Tautomers... option and Spectrum Labels are on, the inactive tautomer signals are marked according to their symbols
22 (T1, T2,...). Fig. 9 NMR Spectrum Display Panel with tautomers NMR Prediction pop-up menu Fig. 10 NMR Prediction pop-up menu Right-clicking on any panel pops up a menu with the following element: Copy to clipboard: the panel in question will be copied to the clipboard.
23 NMR Spectrum Viewer The NMR Spectrum Viewer is part of the NMR Calculation group. It is able to display Nuclear Magnetic Resonance spectra saved in JCAMP-DX format (*.jdx). The opened spectrum can be zoomed in, exported to PDF, or simply copy-pasted as an image. The NMR Spectrum Viewer window consists of a menu, toolbar, two panels, and a status bar. Fig. 11 The NMR Spectrum Viewer Menu system The Menu contains File, View, and Help items.
24 File menu File > Import from JCAMP-DX...: opens an NMR Spectrum in JCAMP-DX format to display it in NMR Spectrum Viewer. Clicking on this menu item will launch the Open dialog window. Select an NMR spectrum in JCAMP-DX format and click on Open. File > Exit: closes the application. View menu Measurement Unit: displays NMR spectrum in one of the following units: Hz or; ppm. Display Local Maximum Places: NMR Spectrum Viewer can display local maximum places as spectrum labels when the JCAMP-DX file contains PEAKTABLE information. Horizontal Zoom In: zooms in on NMR Spectrum along the X-axis. Horizontal Zoom Out: zooms out on NMR Spectrum along the X-axis. Vertical Zoom In: zooms in on the NMR Spectrum along the Y-axis. Vertical Zoom Out: zooms out on the NMR Spectrum along the Y-axis. Reset Zoom: restores the spectrum zooming to full spectrum view. Help Help > Help Contents: opens the help page in your browser. Toolbar You can use toolbar elements to access selected NMR Spectrum Viewer menu items.
25 Import from JCAMP-DX... Display Local Maximum Places
26 Measurement Unit Horizontal Zoom In
27 Horizontal Zoom Out Vertical Zoom In
28 Vertical Zoom Out Reset Zoom Fig. 12 Toolbar elements Panels Panels can be copied separately as images by right-clicking on the appropriate panel and selecting Copy to clipboard action.
29 Spectrum View Panel Fig. 13 The Spectrum View Panel The Spectrum View Panel displays the whole imported spectrum. If you want to zoom in on specific region of the spectrum, use left-click and drag or ctrl + left-click and drag on the NMR Spectrum Preview Panel. The background of the selected region will be highlighted in white, while unselected region of the spectrum will turn to grey. You can move the selection window by left-clicking into the middle of the selection; hold mouse button while moving the selection, and release button to place it. You can resize the selection window if you grab-and-drag the yellow side frame around the highlighted area. Spectrum Display Panel Fig. 14 The Spectrum Display Panel
30 The Spectrum Display Panel displays the appropriate zoom region of the spectrum. Move your mouse pointer over the NMR Spectrum Display Panel and use mouse-wheel to zoom in and out horizontally, Ctrl+mouse-wheel to zoom in and out verically on the NMR spectrum. Status bar The status bar of NMR Spectrum Viewer displays the X and Y coordinates of mouse cursor position, and the following data get stored in the opened in a JCAMP-DX file: Nucleus; Frequency, Owner; File. Fig. 15 The NMR Viewer status bar NMR Spectrum Viewer Pop-up Menu Fig. 15 Copy to clipboard action in the NMR Spectrum Viewer panel Right-clicking on any panel pops up a menu with the following element: Copy to clipboard: The panel in question will be copied to the clipboard.
31 References Gottlieb, H.E.; Kotlyar, V.; Nudelman, A. J. Org. Chem., 1997, 62, ; doi
ISIS/Draw "Quick Start"
 ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
Login -the operator screen should be in view when you first sit down at the spectrometer console:
 Lab #2 1D 1 H Double Resonance (Selective Decoupling) operation of the 400 MHz instrument using automated sample insertion (robot) and automated locking and shimming collection of 1D 1 H spectra retrieving
Lab #2 1D 1 H Double Resonance (Selective Decoupling) operation of the 400 MHz instrument using automated sample insertion (robot) and automated locking and shimming collection of 1D 1 H spectra retrieving
Tutorial. Getting started. Sample to Insight. March 31, 2016
 Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
You w i ll f ol l ow these st eps : Before opening files, the S c e n e panel is active.
 You w i ll f ol l ow these st eps : A. O pen a n i m a g e s t a c k. B. Tr a c e t h e d e n d r i t e w i t h t h e user-guided m ode. C. D e t e c t t h e s p i n e s a u t o m a t i c a l l y. D. C
You w i ll f ol l ow these st eps : A. O pen a n i m a g e s t a c k. B. Tr a c e t h e d e n d r i t e w i t h t h e user-guided m ode. C. D e t e c t t h e s p i n e s a u t o m a t i c a l l y. D. C
OneStop Map Viewer Navigation
 OneStop Map Viewer Navigation» Intended User: Industry Map Viewer users Overview The OneStop Map Viewer is an interactive map tool that helps you find and view information associated with energy development,
OneStop Map Viewer Navigation» Intended User: Industry Map Viewer users Overview The OneStop Map Viewer is an interactive map tool that helps you find and view information associated with energy development,
Introduction to ArcMap
 Introduction to ArcMap ArcMap ArcMap is a Map-centric GUI tool used to perform map-based tasks Mapping Create maps by working geographically and interactively Display and present Export or print Publish
Introduction to ArcMap ArcMap ArcMap is a Map-centric GUI tool used to perform map-based tasks Mapping Create maps by working geographically and interactively Display and present Export or print Publish
SeeSAR 7.1 Beginners Guide. June 2017
 SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Computer simulation of radioactive decay
 Computer simulation of radioactive decay y now you should have worked your way through the introduction to Maple, as well as the introduction to data analysis using Excel Now we will explore radioactive
Computer simulation of radioactive decay y now you should have worked your way through the introduction to Maple, as well as the introduction to data analysis using Excel Now we will explore radioactive
Mnova Software for Analyzing Reaction Monitoring NMR Spectra
 Mnova Software for Analyzing Reaction Monitoring NMR Spectra Version 10 Chen Peng, PhD, VP of Business Development, US & China Mestrelab Research SL San Diego, CA, USA chen.peng@mestrelab.com 858.736.4563
Mnova Software for Analyzing Reaction Monitoring NMR Spectra Version 10 Chen Peng, PhD, VP of Business Development, US & China Mestrelab Research SL San Diego, CA, USA chen.peng@mestrelab.com 858.736.4563
How to Create a Substance Answer Set
 How to Create a Substance Answer Set Select among five search techniques to find substances Since substances can be described by multiple names or other characteristics, SciFinder gives you the flexibility
How to Create a Substance Answer Set Select among five search techniques to find substances Since substances can be described by multiple names or other characteristics, SciFinder gives you the flexibility
1. Double-click the ArcMap icon on your computer s desktop. 2. When the ArcMap start-up dialog box appears, click An existing map and click OK.
 Module 2, Lesson 1 The earth moves In this activity, you will observe worldwide patterns of seismic activity (earthquakes) and volcanic activity (volcanoes). You will analyze the relationships of those
Module 2, Lesson 1 The earth moves In this activity, you will observe worldwide patterns of seismic activity (earthquakes) and volcanic activity (volcanoes). You will analyze the relationships of those
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
APPENDIX 1 Version of 8/24/05 11:01 AM Simulation of H-NMR without a structure*:
 WARNING NOTICE: The experiments described in these materials are potentially hazardous and require a high level of safety training, special facilities and equipment, and supervision by appropriate individuals.
WARNING NOTICE: The experiments described in these materials are potentially hazardous and require a high level of safety training, special facilities and equipment, and supervision by appropriate individuals.
Chemistry Department
 Chemistry Department NMR/Instrumentation Facility Users Guide - VNMRJ Prepared by Leila Maurmann The following procedures should be used to acquire one-dimensional proton and carbon NMR data on the 400MHz
Chemistry Department NMR/Instrumentation Facility Users Guide - VNMRJ Prepared by Leila Maurmann The following procedures should be used to acquire one-dimensional proton and carbon NMR data on the 400MHz
NMRPredict Functional Block Diagram
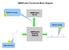 NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
GIS Workshop UCLS_Fall Forum 2014 Sowmya Selvarajan, PhD TABLE OF CONTENTS
 TABLE OF CONTENTS TITLE PAGE NO. 1. ArcGIS Basics I 2 a. Open and Save a Map Document 2 b. Work with Map Layers 2 c. Navigate in a Map Document 4 d. Measure Distances 4 2. ArcGIS Basics II 5 a. Work with
TABLE OF CONTENTS TITLE PAGE NO. 1. ArcGIS Basics I 2 a. Open and Save a Map Document 2 b. Work with Map Layers 2 c. Navigate in a Map Document 4 d. Measure Distances 4 2. ArcGIS Basics II 5 a. Work with
ICM-Chemist How-To Guide. Version 3.6-1g Last Updated 12/01/2009
 ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
Working with ArcGIS: Classification
 Working with ArcGIS: Classification 2 Abbreviations D-click R-click TOC Double Click Right Click Table of Content Introduction The benefit from the use of geographic information system (GIS) software is
Working with ArcGIS: Classification 2 Abbreviations D-click R-click TOC Double Click Right Click Table of Content Introduction The benefit from the use of geographic information system (GIS) software is
PQSMol. User s Manual
 PQSMol User s Manual c Parallel Quantum Solutions, 2006 www.pqs-chem.com Conventions used in this manual Filenames, commands typed in at the command prompt are written in typewriter font: pqsview aspirin.out
PQSMol User s Manual c Parallel Quantum Solutions, 2006 www.pqs-chem.com Conventions used in this manual Filenames, commands typed in at the command prompt are written in typewriter font: pqsview aspirin.out
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
How to Make or Plot a Graph or Chart in Excel
 This is a complete video tutorial on How to Make or Plot a Graph or Chart in Excel. To make complex chart like Gantt Chart, you have know the basic principles of making a chart. Though I have used Excel
This is a complete video tutorial on How to Make or Plot a Graph or Chart in Excel. To make complex chart like Gantt Chart, you have know the basic principles of making a chart. Though I have used Excel
3D Molecule Viewer of MOGADOC (JavaScript)
 3D Molecule Viewer of MOGADOC (JavaScript) Movement of the Molecule Rotation of the molecule: Use left mouse button to drag. Translation of the molecule: Use right mouse button to drag. Resize the molecule:
3D Molecule Viewer of MOGADOC (JavaScript) Movement of the Molecule Rotation of the molecule: Use left mouse button to drag. Translation of the molecule: Use right mouse button to drag. Resize the molecule:
How many states. Record high temperature
 Record high temperature How many states Class Midpoint Label 94.5 99.5 94.5-99.5 0 97 99.5 104.5 99.5-104.5 2 102 102 104.5 109.5 104.5-109.5 8 107 107 109.5 114.5 109.5-114.5 18 112 112 114.5 119.5 114.5-119.5
Record high temperature How many states Class Midpoint Label 94.5 99.5 94.5-99.5 0 97 99.5 104.5 99.5-104.5 2 102 102 104.5 109.5 104.5-109.5 8 107 107 109.5 114.5 109.5-114.5 18 112 112 114.5 119.5 114.5-119.5
Simulation of Second Order Spectra Using SpinWorks. CHEM/BCMB 8190 Biomolecular NMR UGA, Spring, 2005
 Simulation of Second Order Spectra Using SpinWorks CHEM/BCMB 8190 Biomolecular NMR UGA, Spring, 2005 Introduction Although we frequently assume that scalar couplings are small compared to the differences
Simulation of Second Order Spectra Using SpinWorks CHEM/BCMB 8190 Biomolecular NMR UGA, Spring, 2005 Introduction Although we frequently assume that scalar couplings are small compared to the differences
BUILDING BASICS WITH HYPERCHEM LITE
 BUILDING BASICS WITH HYPERCHEM LITE LAB MOD1.COMP From Gannon University SIM INTRODUCTION A chemical bond is a link between atoms resulting from the mutual attraction of their nuclei for electrons. There
BUILDING BASICS WITH HYPERCHEM LITE LAB MOD1.COMP From Gannon University SIM INTRODUCTION A chemical bond is a link between atoms resulting from the mutual attraction of their nuclei for electrons. There
General Chemistry Lab Molecular Modeling
 PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
4) protons experience a net magnetic field strength that is smaller than the applied magnetic field.
 1) Which of the following CANNOT be probed by an spectrometer? See sect 16.1 Chapter 16: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even number
1) Which of the following CANNOT be probed by an spectrometer? See sect 16.1 Chapter 16: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even number
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Space Objects. Section. When you finish this section, you should understand the following:
 GOLDMC02_132283433X 8/24/06 2:21 PM Page 97 Section 2 Space Objects When you finish this section, you should understand the following: How to create a 2D Space Object and label it with a Space Tag. How
GOLDMC02_132283433X 8/24/06 2:21 PM Page 97 Section 2 Space Objects When you finish this section, you should understand the following: How to create a 2D Space Object and label it with a Space Tag. How
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
Computational Study of Chemical Kinetics (GIDES)
 Computational Study of Chemical Kinetics (GIDES) Software Introduction Berkeley Madonna (http://www.berkeleymadonna.com) is a dynamic modeling program in which relational diagrams are created using a graphical
Computational Study of Chemical Kinetics (GIDES) Software Introduction Berkeley Madonna (http://www.berkeleymadonna.com) is a dynamic modeling program in which relational diagrams are created using a graphical
Cerno Application Note Extending the Limits of Mass Spectrometry
 Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Creation of Accurate Mass Library for NIST Database Search Novel MS calibration has been shown to enable accurate mass and elemental composition determination on quadrupole GC/MS systems for either molecular
Synteny Portal Documentation
 Synteny Portal Documentation Synteny Portal is a web application portal for visualizing, browsing, searching and building synteny blocks. Synteny Portal provides four main web applications: SynCircos,
Synteny Portal Documentation Synteny Portal is a web application portal for visualizing, browsing, searching and building synteny blocks. Synteny Portal provides four main web applications: SynCircos,
Ligand Scout Tutorials
 Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Application Note. U. Heat of Formation of Ethyl Alcohol and Dimethyl Ether. Introduction
 Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Chemistry 14CL. Worksheet for the Molecular Modeling Workshop. (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell)
 Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
4) protons experience a net magnetic field strength that is smaller than the applied magnetic field.
 1) Which of the following CANNOT be probed by an NMR spectrometer? See sect 15.1 Chapter 15: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even
1) Which of the following CANNOT be probed by an NMR spectrometer? See sect 15.1 Chapter 15: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even
Electric Fields and Equipotentials
 OBJECTIVE Electric Fields and Equipotentials To study and describe the two-dimensional electric field. To map the location of the equipotential surfaces around charged electrodes. To study the relationship
OBJECTIVE Electric Fields and Equipotentials To study and describe the two-dimensional electric field. To map the location of the equipotential surfaces around charged electrodes. To study the relationship
MassHunter Software Overview
 MassHunter Software Overview 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks A
MassHunter Software Overview 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks A
Preparing a PDB File
 Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
The Geodatabase Working with Spatial Analyst. Calculating Elevation and Slope Values for Forested Roads, Streams, and Stands.
 GIS LAB 7 The Geodatabase Working with Spatial Analyst. Calculating Elevation and Slope Values for Forested Roads, Streams, and Stands. This lab will ask you to work with the Spatial Analyst extension.
GIS LAB 7 The Geodatabase Working with Spatial Analyst. Calculating Elevation and Slope Values for Forested Roads, Streams, and Stands. This lab will ask you to work with the Spatial Analyst extension.
4) protons experience a net magnetic field strength that is smaller than the applied magnetic field.
 1) Which of the following CANNOT be probed by an spectrometer? See sect 16.1 Chapter 16: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even number
1) Which of the following CANNOT be probed by an spectrometer? See sect 16.1 Chapter 16: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even number
Chem 253. Tutorial for Materials Studio
 Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
McIDAS-V Tutorial Displaying Point Observations from ADDE Datasets updated July 2016 (software version 1.6)
 McIDAS-V Tutorial Displaying Point Observations from ADDE Datasets updated July 2016 (software version 1.6) McIDAS-V is a free, open source, visualization and data analysis software package that is the
McIDAS-V Tutorial Displaying Point Observations from ADDE Datasets updated July 2016 (software version 1.6) McIDAS-V is a free, open source, visualization and data analysis software package that is the
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Molecular Modeling and Conformational Analysis with PC Spartan
 Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
M E R C E R W I N WA L K T H R O U G H
 H E A L T H W E A L T H C A R E E R WA L K T H R O U G H C L I E N T S O L U T I O N S T E A M T A B L E O F C O N T E N T 1. Login to the Tool 2 2. Published reports... 7 3. Select Results Criteria...
H E A L T H W E A L T H C A R E E R WA L K T H R O U G H C L I E N T S O L U T I O N S T E A M T A B L E O F C O N T E N T 1. Login to the Tool 2 2. Published reports... 7 3. Select Results Criteria...
Quantification of JEOL XPS Spectra from SpecSurf
 Quantification of JEOL XPS Spectra from SpecSurf The quantification procedure used by the JEOL SpecSurf software involves modifying the Scofield cross-sections to account for both an energy dependency
Quantification of JEOL XPS Spectra from SpecSurf The quantification procedure used by the JEOL SpecSurf software involves modifying the Scofield cross-sections to account for both an energy dependency
3D - Structure Graphics Capabilities with PDF-4 Database Products
 3D - Structure Graphics Capabilities with PDF-4 Database Products Atomic Structure Information in the PDF-4 Databases ICDD s PDF-4 databases contain atomic structure information for a significant number
3D - Structure Graphics Capabilities with PDF-4 Database Products Atomic Structure Information in the PDF-4 Databases ICDD s PDF-4 databases contain atomic structure information for a significant number
Performing Map Cartography. using Esri Production Mapping
 AGENDA Performing Map Cartography Presentation Title using Esri Production Mapping Name of Speaker Company Name Kannan Jayaraman Agenda Introduction What s New in ArcGIS 10.1 ESRI Production Mapping Mapping
AGENDA Performing Map Cartography Presentation Title using Esri Production Mapping Name of Speaker Company Name Kannan Jayaraman Agenda Introduction What s New in ArcGIS 10.1 ESRI Production Mapping Mapping
The Kalzium Handbook. Carsten Niehaus
 Carsten Niehaus 2 Contents 1 Introduction 1 2 Kalzium quick start guide 2 3 Using Kalzium 6 3.1 Overview of Kalziums usage.................... 6 3.2 The information dialog........................ 6 3.2.1
Carsten Niehaus 2 Contents 1 Introduction 1 2 Kalzium quick start guide 2 3 Using Kalzium 6 3.1 Overview of Kalziums usage.................... 6 3.2 The information dialog........................ 6 3.2.1
2010 Autodesk, Inc. All rights reserved. NOT FOR DISTRIBUTION.
 Wastewater Profiles 2010 Autodesk, Inc. All rights reserved. NOT FOR DISTRIBUTION. The contents of this guide were created for Autodesk Topobase 2011. The contents of this guide are not intended for other
Wastewater Profiles 2010 Autodesk, Inc. All rights reserved. NOT FOR DISTRIBUTION. The contents of this guide were created for Autodesk Topobase 2011. The contents of this guide are not intended for other
This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus.
 PCI GEOMATICS GEOMATICA QUICKSTART 1. Introduction This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus. All data used in
PCI GEOMATICS GEOMATICA QUICKSTART 1. Introduction This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus. All data used in
Introduction to Spark
 1 As you become familiar or continue to explore the Cresset technology and software applications, we encourage you to look through the user manual. This is accessible from the Help menu. However, don t
1 As you become familiar or continue to explore the Cresset technology and software applications, we encourage you to look through the user manual. This is accessible from the Help menu. However, don t
Polar alignment in 5 steps based on the Sánchez Valente method
 1 Polar alignment in 5 steps based on the Sánchez Valente method Compared to the drift alignment method, this one, allows you to easily achieve a perfect polar alignment in just one step. By "perfect polar
1 Polar alignment in 5 steps based on the Sánchez Valente method Compared to the drift alignment method, this one, allows you to easily achieve a perfect polar alignment in just one step. By "perfect polar
Creation and modification of a geological model Program: Stratigraphy
 Engineering manual No. 39 Updated: 11/2018 Creation and modification of a geological model Program: Stratigraphy File: Demo_manual_39.gsg Introduction The aim of this engineering manual is to explain the
Engineering manual No. 39 Updated: 11/2018 Creation and modification of a geological model Program: Stratigraphy File: Demo_manual_39.gsg Introduction The aim of this engineering manual is to explain the
ICM-Chemist-Pro How-To Guide. Version 3.6-1h Last Updated 12/29/2009
 ICM-Chemist-Pro How-To Guide Version 3.6-1h Last Updated 12/29/2009 ICM-Chemist-Pro ICM 3D LIGAND EDITOR: SETUP 1. Read in a ligand molecule or PDB file. How to setup the ligand in the ICM 3D Ligand Editor.
ICM-Chemist-Pro How-To Guide Version 3.6-1h Last Updated 12/29/2009 ICM-Chemist-Pro ICM 3D LIGAND EDITOR: SETUP 1. Read in a ligand molecule or PDB file. How to setup the ligand in the ICM 3D Ligand Editor.
Chem 1 Kinetics. Objectives. Concepts
 Chem 1 Kinetics Objectives 1. Learn some basic ideas in chemical kinetics. 2. Understand how the computer visualizations can be used to benefit the learning process. 3. Understand how the computer models
Chem 1 Kinetics Objectives 1. Learn some basic ideas in chemical kinetics. 2. Understand how the computer visualizations can be used to benefit the learning process. 3. Understand how the computer models
(THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE)
 PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
PART 2: Analysis in ArcGIS (THIS IS AN OPTIONAL BUT WORTHWHILE EXERCISE) Step 1: Start ArcCatalog and open a geodatabase If you have a shortcut icon for ArcCatalog on your desktop, double-click it to start
Designing a Quilt with GIMP 2011
 Planning your quilt and want to see what it will look like in the fabric you just got from your LQS? You don t need to purchase a super expensive program. Try this and the best part it s FREE!!! *** Please
Planning your quilt and want to see what it will look like in the fabric you just got from your LQS? You don t need to purchase a super expensive program. Try this and the best part it s FREE!!! *** Please
Viewing and Analyzing Proteins, Ligands and their Complexes 2
 2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
CHEM Chapter 13. Nuclear Magnetic Spectroscopy (Homework) W
 CHEM 2423. Chapter 13. Nuclear Magnetic Spectroscopy (Homework) W Short Answer 1. For a nucleus to exhibit the nuclear magnetic resonance phenomenon, it must be magnetic. Magnetic nuclei include: a. all
CHEM 2423. Chapter 13. Nuclear Magnetic Spectroscopy (Homework) W Short Answer 1. For a nucleus to exhibit the nuclear magnetic resonance phenomenon, it must be magnetic. Magnetic nuclei include: a. all
2G1/3G4 GIS TUTORIAL >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
 > University of Michigan >Taubman College of Architecture > ARCH 552, Perimeter @ Work Out [T]here, Fall 2009 >September 24, 2009 2G1/3G4 GIS TUTORIAL >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
> University of Michigan >Taubman College of Architecture > ARCH 552, Perimeter @ Work Out [T]here, Fall 2009 >September 24, 2009 2G1/3G4 GIS TUTORIAL >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
The Hückel Approximation Consider a conjugated molecule i.e. a molecule with alternating double and single bonds, as shown in Figure 1.
 The Hückel Approximation In this exercise you will use a program called Hückel to look at the p molecular orbitals in conjugated molecules. The program calculates the energies and shapes of p (pi) molecular
The Hückel Approximation In this exercise you will use a program called Hückel to look at the p molecular orbitals in conjugated molecules. The program calculates the energies and shapes of p (pi) molecular
ON SITE SYSTEMS Chemical Safety Assistant
 ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
Zeeman Effect Physics 481
 Zeeman Effect Introduction You are familiar with Atomic Spectra, especially the H- atom energy spectrum. Atoms emit or absorb energies in packets, or quanta which are photons. The orbital motion of electrons
Zeeman Effect Introduction You are familiar with Atomic Spectra, especially the H- atom energy spectrum. Atoms emit or absorb energies in packets, or quanta which are photons. The orbital motion of electrons
Software BioScout-Calibrator June 2013
 SARAD GmbH BioScout -Calibrator 1 Manual Software BioScout-Calibrator June 2013 SARAD GmbH Tel.: ++49 (0)351 / 6580712 Wiesbadener Straße 10 FAX: ++49 (0)351 / 6580718 D-01159 Dresden email: support@sarad.de
SARAD GmbH BioScout -Calibrator 1 Manual Software BioScout-Calibrator June 2013 SARAD GmbH Tel.: ++49 (0)351 / 6580712 Wiesbadener Straße 10 FAX: ++49 (0)351 / 6580718 D-01159 Dresden email: support@sarad.de
4. NMR spectra. Interpreting NMR spectra. Low-resolution NMR spectra. There are two kinds: Low-resolution NMR spectra. High-resolution NMR spectra
 1 Interpreting NMR spectra There are two kinds: Low-resolution NMR spectra High-resolution NMR spectra In both cases the horizontal scale is labelled in terms of chemical shift, δ, and increases from right
1 Interpreting NMR spectra There are two kinds: Low-resolution NMR spectra High-resolution NMR spectra In both cases the horizontal scale is labelled in terms of chemical shift, δ, and increases from right
Experiment 2 - NMR Spectroscopy
 Experiment 2 - NMR Spectroscopy OBJECTIVE to understand the important role of nuclear magnetic resonance spectroscopy in the study of the structures of organic compounds to develop an understanding of
Experiment 2 - NMR Spectroscopy OBJECTIVE to understand the important role of nuclear magnetic resonance spectroscopy in the study of the structures of organic compounds to develop an understanding of
Tutorial 23 Back Analysis of Material Properties
 Tutorial 23 Back Analysis of Material Properties slope with known failure surface sensitivity analysis probabilistic analysis back analysis of material strength Introduction Model This tutorial will demonstrate
Tutorial 23 Back Analysis of Material Properties slope with known failure surface sensitivity analysis probabilistic analysis back analysis of material strength Introduction Model This tutorial will demonstrate
An area chart emphasizes the trend of each value over time. An area chart also shows the relationship of parts to a whole.
 Excel 2003 Creating a Chart Introduction Page 1 By the end of this lesson, learners should be able to: Identify the parts of a chart Identify different types of charts Create an Embedded Chart Create a
Excel 2003 Creating a Chart Introduction Page 1 By the end of this lesson, learners should be able to: Identify the parts of a chart Identify different types of charts Create an Embedded Chart Create a
Consents Resource Consents Map
 Consents Resource Consents Map Select the map from the Maps introduction page http://www.waikatoregion.govt.nz/maps/ If you have the map browser open the Resource Consents map will also display when selected
Consents Resource Consents Map Select the map from the Maps introduction page http://www.waikatoregion.govt.nz/maps/ If you have the map browser open the Resource Consents map will also display when selected
CHEMDRAW ULTRA ITEC107 - Introduction to Computing for Pharmacy. ITEC107 - Introduction to Computing for Pharmacy 1
 CHEMDRAW ULTRA 12.0 ITEC107 - Introduction to Computing for Pharmacy 1 Objectives Basic drawing skills with ChemDraw Bonds, captions, hotkeys, chains, arrows Checking and cleaning up structures Chemical
CHEMDRAW ULTRA 12.0 ITEC107 - Introduction to Computing for Pharmacy 1 Objectives Basic drawing skills with ChemDraw Bonds, captions, hotkeys, chains, arrows Checking and cleaning up structures Chemical
Determining C-H Connectivity: ghmqc and ghmbc (VnmrJ-2.2D Version: For use with the new Software)
 Determining C-H Connectivity: ghmqc and ghmbc (VnmrJ-2.2D Version: For use with the new Software) Heteronuclear Multiple Quantum Coherence (HMQC) and Heteronuclear Multiple Bond Coherence (HMBC) are 2-dimensional
Determining C-H Connectivity: ghmqc and ghmbc (VnmrJ-2.2D Version: For use with the new Software) Heteronuclear Multiple Quantum Coherence (HMQC) and Heteronuclear Multiple Bond Coherence (HMBC) are 2-dimensional
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Creating Empirical Calibrations
 030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
030.0023.01.0 Spreadsheet Manual Save Date: December 1, 2010 Table of Contents 1. Overview... 3 2. Enable S1 Calibration Macro... 4 3. Getting Ready... 4 4. Measuring the New Sample... 5 5. Adding New
Getting Started. Start ArcMap by opening up a new map.
 Start ArcMap by opening up a new map. Getting Started We now need to set up ArcMap to do some analysis using the Spatial Analyst extension. You will need to activate the Spatial Analyst extension by selecting
Start ArcMap by opening up a new map. Getting Started We now need to set up ArcMap to do some analysis using the Spatial Analyst extension. You will need to activate the Spatial Analyst extension by selecting
Astro 101 Lab #1. To advance time forward and backward, click on the arrow toolbar. From left to right, the buttons will
 Name: Astro 101 Lab #1 Lab objectives 1) Learn how to use the Stellarium planetarium program, by becoming familiar with the user interface and configuring the planetarium to your present location on Earth.
Name: Astro 101 Lab #1 Lab objectives 1) Learn how to use the Stellarium planetarium program, by becoming familiar with the user interface and configuring the planetarium to your present location on Earth.
Child Opportunity Index Mapping
 Child Opportunity Index Mapping A partnership project of How to Use and Interpret the Child Opportunity Index Maps How to Use the Child Opportunity Index Maps Getting Started: How to Find and Open a Child
Child Opportunity Index Mapping A partnership project of How to Use and Interpret the Child Opportunity Index Maps How to Use the Child Opportunity Index Maps Getting Started: How to Find and Open a Child
CHEMISTRY Organic Chemistry Laboratory II Spring 2019 Lab #5: NMR Spectroscopy
 Team Members: Unknown # CHEMISTRY 244 - Organic Chemistry Laboratory II Spring 2019 Lab #5: NMR Spectroscopy Purpose: You will learn how to predict the NMR data for organic molecules, organize this data
Team Members: Unknown # CHEMISTRY 244 - Organic Chemistry Laboratory II Spring 2019 Lab #5: NMR Spectroscopy Purpose: You will learn how to predict the NMR data for organic molecules, organize this data
BOND LENGTH WITH HYPERCHEM LITE
 BOND LENGTH WITH HYPERCHEM LITE LAB MOD2.COMP From Gannon University SIM INTRODUCTION The electron cloud surrounding the nucleus of the atom determines the size of the atom. Since this distance is somewhat
BOND LENGTH WITH HYPERCHEM LITE LAB MOD2.COMP From Gannon University SIM INTRODUCTION The electron cloud surrounding the nucleus of the atom determines the size of the atom. Since this distance is somewhat
User Guide (Windows and Macintosh)
 User Guide (Windows and Macintosh) WAVEFUNCTION Wavefunction, Inc. 18401 Von Karman Avenue, Suite 370 Irvine, CA 92612 U.S.A. www.wavefun.com Wavefunction, Inc., Japan Branch Office 3-5-2, Kouji-machi,
User Guide (Windows and Macintosh) WAVEFUNCTION Wavefunction, Inc. 18401 Von Karman Avenue, Suite 370 Irvine, CA 92612 U.S.A. www.wavefun.com Wavefunction, Inc., Japan Branch Office 3-5-2, Kouji-machi,
Preparations and Starting the program
 Preparations and Starting the program https://oldwww.abo.fi/fakultet/ookforskning 1) Create a working directory on your computer for your Chemkin work, and 2) download kinetic mechanism files AAUmech.inp
Preparations and Starting the program https://oldwww.abo.fi/fakultet/ookforskning 1) Create a working directory on your computer for your Chemkin work, and 2) download kinetic mechanism files AAUmech.inp
Introduction to ArcGIS 10.2
 Introduction to ArcGIS 10.2 Francisco Olivera, Ph.D., P.E. Srikanth Koka Lauren Walker Aishwarya Vijaykumar Keri Clary Department of Civil Engineering April 21, 2014 Contents Brief Overview of ArcGIS 10.2...
Introduction to ArcGIS 10.2 Francisco Olivera, Ph.D., P.E. Srikanth Koka Lauren Walker Aishwarya Vijaykumar Keri Clary Department of Civil Engineering April 21, 2014 Contents Brief Overview of ArcGIS 10.2...
Jasco V-670 absorption spectrometer
 Laser Spectroscopy Labs Jasco V-670 absorption spectrometer Operation instructions 1. Turn ON the power switch on the right side of the spectrophotometer. It takes about 5 minutes for the light source
Laser Spectroscopy Labs Jasco V-670 absorption spectrometer Operation instructions 1. Turn ON the power switch on the right side of the spectrophotometer. It takes about 5 minutes for the light source
HSC Chemistry 7.0 User's Guide
 HSC Chemistry 7.0 47-1 HSC Chemistry 7.0 User's Guide Sim Flowsheet Module Experimental Mode Pertti Lamberg Outotec Research Oy Information Service P.O. Box 69 FIN - 28101 PORI, FINLAND Fax: +358-20 -
HSC Chemistry 7.0 47-1 HSC Chemistry 7.0 User's Guide Sim Flowsheet Module Experimental Mode Pertti Lamberg Outotec Research Oy Information Service P.O. Box 69 FIN - 28101 PORI, FINLAND Fax: +358-20 -
4) protons experience a net magnetic field strength that is smaller than the applied magnetic field.
 1) Which of the following CANNOT be probed by an spectrometer? See sect 15.1 Chapter 15: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even number
1) Which of the following CANNOT be probed by an spectrometer? See sect 15.1 Chapter 15: 1 A) nucleus with odd number of protons & odd number of neutrons B) nucleus with odd number of protons &even number
ScienceWord and PagePlayer Chemical formulae. Dr Emile C. B. COMLAN Novoasoft Representative in Africa
 ScienceWord and PagePlayer Chemical formulae Dr Emile C. B. CMLAN Novoasoft Representative in Africa Emails: ecomlan@yahoo.com ecomlan@scienceoffice.com Web site: www.scienceoffice.com Chemical formulae
ScienceWord and PagePlayer Chemical formulae Dr Emile C. B. CMLAN Novoasoft Representative in Africa Emails: ecomlan@yahoo.com ecomlan@scienceoffice.com Web site: www.scienceoffice.com Chemical formulae
Operation of the Bruker 400 JB Stothers NMR Facility Department of Chemistry Western University
 Operation of the Bruker 400 JB Stothers NMR Facility Department of Chemistry Western University 1. INTRODUCTION...3 1.1. Overview of the Bruker 400 NMR Spectrometer...3 1.2. Overview of Software... 3 1.2.1.
Operation of the Bruker 400 JB Stothers NMR Facility Department of Chemistry Western University 1. INTRODUCTION...3 1.1. Overview of the Bruker 400 NMR Spectrometer...3 1.2. Overview of Software... 3 1.2.1.
GIS: Introductory Guide to MapPlace. Workshop Agenda. How the Maps Work. Geographic Information Systems. -The MapPlace is a web GIS application
 1 Introductory Guide to MapPlace Workshop 2007 Larry Jones MapPlace & Geoscience Databases Pat Desjardins Corporate GIS Gib McArthur Manager, Resource Information Kirk Hancock MINFILE Geologist Ward Kilby
1 Introductory Guide to MapPlace Workshop 2007 Larry Jones MapPlace & Geoscience Databases Pat Desjardins Corporate GIS Gib McArthur Manager, Resource Information Kirk Hancock MINFILE Geologist Ward Kilby
North Carolina State University Department of Chemistry Varian NMR Training Manual
 North Carolina State University Department of Chemistry Varian NMR Training Manual by J.B. Clark IV & Dr. S. Sankar 1 st Edition 05/15/2009 Section 3: Glide Program Operations for Advanced 1D & 2D Spectra
North Carolina State University Department of Chemistry Varian NMR Training Manual by J.B. Clark IV & Dr. S. Sankar 1 st Edition 05/15/2009 Section 3: Glide Program Operations for Advanced 1D & 2D Spectra
SDBS Integrated Spectral Database for Organic Compounds
 SDBS Integrated Spectral Database for Organic Compounds Sample Search for Chemistry 130 Grace Baysinger and Dr. Dave Keller SDBS URL and Description http://riodb01.ibase.aist.go.jp/sdbs/cgi-bin/cre_index.cgi?lang=eng
SDBS Integrated Spectral Database for Organic Compounds Sample Search for Chemistry 130 Grace Baysinger and Dr. Dave Keller SDBS URL and Description http://riodb01.ibase.aist.go.jp/sdbs/cgi-bin/cre_index.cgi?lang=eng
Using the Budget Features in Quicken 2008
 Using the Budget Features in Quicken 2008 Quicken budgets can be used to summarize expected income and expenses for planning purposes. The budget can later be used in comparisons to actual income and expenses
Using the Budget Features in Quicken 2008 Quicken budgets can be used to summarize expected income and expenses for planning purposes. The budget can later be used in comparisons to actual income and expenses
Carbon Structure Simulations using Crystal Viewer Tool
 Carbon Structure Simulations using Crystal Viewer Tool The Crystal Viewer tool in nanohub.org can be used to build three carbon nanostructures: graphene sheets, Buckminsterfullerene, Bucky balls (C 60
Carbon Structure Simulations using Crystal Viewer Tool The Crystal Viewer tool in nanohub.org can be used to build three carbon nanostructures: graphene sheets, Buckminsterfullerene, Bucky balls (C 60
Task 1: Open ArcMap and activate the Spatial Analyst extension.
 Exercise 10 Spatial Analyst The following steps describe the general process that you will follow to complete the exercise. Specific steps will be provided later in the step-by-step instructions component
Exercise 10 Spatial Analyst The following steps describe the general process that you will follow to complete the exercise. Specific steps will be provided later in the step-by-step instructions component
User Manuel. EurotaxForecast. Version Latest changes ( )
 User Manuel EurotaxForecast Version 1.23.0771- Latest changes (19.07.2003) Contents Preface 5 Welcome to Eurotax Forecast...5 Using this manual 6 How to use this manual?...6 Program overview 7 General
User Manuel EurotaxForecast Version 1.23.0771- Latest changes (19.07.2003) Contents Preface 5 Welcome to Eurotax Forecast...5 Using this manual 6 How to use this manual?...6 Program overview 7 General
Watershed Modeling Orange County Hydrology Using GIS Data
 v. 10.0 WMS 10.0 Tutorial Watershed Modeling Orange County Hydrology Using GIS Data Learn how to delineate sub-basins and compute soil losses for Orange County (California) hydrologic modeling Objectives
v. 10.0 WMS 10.0 Tutorial Watershed Modeling Orange County Hydrology Using GIS Data Learn how to delineate sub-basins and compute soil losses for Orange County (California) hydrologic modeling Objectives
ArcGIS 9 ArcGIS StreetMap Tutorial
 ArcGIS 9 ArcGIS StreetMap Tutorial Copyright 2001 2008 ESRI All Rights Reserved. Printed in the United States of America. The information contained in this document is the exclusive property of ESRI. This
ArcGIS 9 ArcGIS StreetMap Tutorial Copyright 2001 2008 ESRI All Rights Reserved. Printed in the United States of America. The information contained in this document is the exclusive property of ESRI. This
