Physical Chemistry Final Take Home Fall 2003
|
|
|
- Mae Walsh
- 6 years ago
- Views:
Transcription
1 Physical Chemistry Final Take Home Fall 2003 Do one of the following questions. These projects are worth 30 points (i.e. equivalent to about two problems on the final). Each of the computational problems below took me less than 20 minutes to do. Under no circumstances should your report be more than one written page (and perhaps a scatter plot). Answer the specific questions. Save all your molecule structures. If you use an SGI, use your own account. Give your SGI account name if you used an SGI or the PC number if you used a PC and give the names of all the files that you generated. You don't need to type your report; hand written is fine. Your reports are due at the beginning of the final. Problem 1 uses ISIS/Base pharmachophore searches for ACE inhibitors to explore the importance of several options within the cfs algorithm. Problem 2 uses ME Gibbs Free energy of solvation descriptors in a QSAR study of anticancer compounds. Problem 3 uses ME and electrostatics descriptors in a QSAR study of anticancer compounds. Problem 4 uses ME and continuum solvation calculations to look at solvent effects on tautomeric equilibria. You can use any written source, as long as that source does not involve the active efforts of any other individual, other than yourself. For example, you can use the tutorials, your book, your notes, and references in the library, but you cannot use , fax, letter, teleconference, voice, or video communications. You may of course ask questions of me (x3315, ), but I may not be available Wednesday night. If you wish to contact me before you start, so I can check in for questions- I'll be happy to do so. I will be more than happy to help you with software related issues, for example how to add or delete descriptors, use the Builder, or switching between ISIS/Base forms. See the note at the end for help with Printing issues from ME. 1. Use the pharmachophore from Figure 8 in the Henry, Güner paper to do five cfs searches that explore the importance of the options in the cfs procedure. These searches will be just like the ones we did in class except for changing the options in the cfs search (as listed below). These options concern the molecular mechanics portion of the cfs procedure. This pharmachophore is already built and is available in the "D:/ISIS/ace" folder, so you don't need to build it. Just open the q4.skc file in ISIS/Draw. Use the CMC3D database (password "isis/3d"). Remember that 3D queries need to be pasted into the Model box from the Model tab of the search form. (Pasting your query in the Structure box will give an error for a cfs search). In evaluating the hits from a pharmachophore search it is important to determine how many of the known active compounds are recovered from the database along with the new compounds. There are 362 compounds in the database that are listed as "antihypertensive." f course not all of these are ACE inhibitors, but the recovery rate of these compounds will none-the-less help us to evaluate the cfs procedure. To determine the number of "antihypertensive" compounds in your hit list do the following: 1. After your cfs search is complete, pull down the Search menu, slide right on Set Search Domain to, and choose Current List. 2. Switch to the Query mode by clicking on the Query button in the top button bar. Switch to the Structure form by clicking on the Structure tab on the "index card deck." 3. In the Class query box enter the text: like "%antihyper%"
2 4. Pull down the Search menu and choose By Form. Record the number of hits. 5. Remember to return the search to the full database by pulling down the Search menu, slide right on Set Search Domain to, and choose Entire Database. 6. Remember to do Step 5 above: choose Entire Database. Start with the cfs options set to the values we used in class. These options were chosen to ensure rapid search times: Record the results in the table below. Use the following sets of cfs options listed in the table. You may get a couple errors along the way. These won't have an effect on the final outcome. Max. no. of attempted fittings Global relaxation TDF 5 off off off off Van der Waals energy difference Total hits Antihypertensives recovered Answer the following questions: 1. Which change gave the best improvement in the recovery of known antihypertensives? 2. Which change gave the best improvement in the totals number of hits? 3. The global relaxation description in the ISIS documentation explains: You can specify that ISIS rotate single bonds in other parts of the structure to reduce nonbonded interactions throughout the molecule. When global relaxation is active during bump checking, ISIS finds structures that might be eliminated if ISIS rotated single bonds
3 solely in the region that matched the query. Global relaxation increases the size of your hit list and produces more realistic 3D structures. Global relaxation does a full molecular mechanics minimization at the chosen conformation. Does this rather time consuming step make a big difference in the hit rate and the recovery of known antihypertensives? 4. The maximum number of attempted fittings and the Torsional degress of freedom options concern mostly statistical issues rather than chemical issues. The global relaxation and Van der Waals energy difference options are more "physical chemical" issues that concern strain energy and the thermodynamic accessibility of the strained conformations that fit the query. Are statistical or thermodynamic issues more important for good cfs searches as implemented in this algorithm? 2. A group of quinones ( 2,5-bis(1-aziridinyl)-p-benzoquinones ) has been studied for their antitumor activity. You will find a selection of these in the benzo/ directory and the database file "benzo.mdb" in your SGI account or in D:/moe/ on the PC's. You will do a QSAR study similar to the study we did in lab. The base structure is: See S. P. Gupta, "Quantitative Structure- R1 Activity Relationship Studies on Anticancer Drugs," Chem. Rev. 1994,94, and R2 Table 7, for more information if you would like (but not required)(see me for a copy). Use the instructions for the ACE QSAR study in the CAMD tutorial as the basis for this assignment. However, this time we want to do a better job of considering electrostatic effects. This database includes F H2 and F oct already entered in the descriptor list (see Problem 3 below). Remove these descriptors from the database. They won't be necessary for this exercise. For this exercise use the PEE based electrostatic descriptors in the table below. These descriptors attempt to determine the effect of the electrostatic distribution in the molecule on the activity of the compounds in a more precise way than possible with "bulk" descriptors like the dipole moment. PEE_VSA+6 Sum of v i where q i is greater than 0.3. PEE_VSA+5 Sum of v i where q i is in the range [0.25,0.30). PEE_VSA+4 Sum of v i where q i is in the range [0.20,0.25). PEE_VSA+3 Sum of v i where q i is in the range [0.15,0.20). PEE_VSA+2 Sum of v i where q i is in the range [0.10,0.15). PEE_VSA+1 Sum of v i where q i is in the range [0.05,0.10). PEE_VSA+0 Sum of v i where q i is in the range [0.00,0.05). PEE_VSA-0 Sum of v i where q i is in the range [-0.05,0.00). PEE_VSA-1 Sum of v i where q i is in the range [-0.10,-0.05). PEE_VSA-2 Sum of v i where q i is in the range [-0.15,-0.10). PEE_VSA-3 Sum of v i where q i is in the range [-0.20,-0.15). PEE_VSA-4 Sum of v i where q i is in the range [-0.25,-0.20). PEE_VSA-5 Sum of v i where q i is in the range [-0.30,-0.25). PEE_VSA-6 Sum of v i where q i is less than
4 Remember to follow the guidelines concerning descriptor correlations and the maximum number of descriptors. Find a small set of descriptors that give a reasonable QSAR. Then add logp(o/w) and redo the QSAR to determine if solvation effects that are not correlated with the electrostatic distribution are important. Answer the following questions. 1. Give your best QSAR equation using the PEE_VSA descriptors. Include the R 2 and R values and the RELATIVE IMPRTACE for each of the descriptors. 2. Give your best QSAR equation using the same descriptors as in question 1 but including logp(o/w). Include the R 2 and R values and the RELATIVE IMPRTACE for each of the descriptors. 3. Give the R 2 and R values for the QSAR using logp(o/w) alone. 4. Does inclusion of the PEE_VSA descriptors markedly improve the QSAR over the classical logp(o/w) descriptor alone? 5. Does the inclusion of the logp(o/w) descriptor markedly improve the QSAR using only the PEE_VSA descriptors? What does this imply about hydrophobic effects? (ote that PEE_VSA+0 and PEE_VSA-0 should also correlate with hydrophobic effects to some extent). 6. Give the name of the computer and the database file that you generated. 3. A group of quinones ( 2,5-bis(1-aziridinyl)-p-benzoquinones ) has been studied for their antitumor activity. You will find a selection of these in the benzo/ directory and the database file "benzo.mdb" in your SGI account or in D:/moe/ on the PC's. You will do a QSAR study similar to the study we did in lab. The base structure is: See S. P. Gupta, "Quantitative Structure- R1 Activity Relationship Studies on Anticancer Drugs," Chem. Rev. 1994,94, and R2 Table 7, for more information if you would like (but not required)(see me for a copy). Use the instructions for the ACE QSAR study in the CAMD tutorial as the basis for this assignment. However, this time we want to do a better job of considering solvation effects. This database includes F H2 and F oct already entered in the descriptor list (see the CAMD Introduction). Make sure to use E_solv in the new descriptors that you add. E_solv uses the GB/SA solvation calculation for its values, so make sure to set up the MMFF94 force field with solvation in the same way as we did for the molecular mechanics solvation Gibbs Free Energy exercises (don't use the distance dependent dielectric). F H2 and E_solv should give the same values. They are both the Gibbs Free Energy of solvation in water. However, the F H2 values are derived from empirical correlations with experimental data extracted into group additive substituent constants. And, of course the GB/SA calculations are based on the first principles approach using the Born Approximation. Remember to follow the guidelines concerning descriptor correlations and the maximum number of descriptors. This dataset is fun since many different combinations of descriptors do a good job. So don't spend a lot of time trying to find the "perfect" QSAR equation. A reasonable QSAR that follows the general guidelines is fine. However, do consider that F H2, F oct, E_solv,
5 and log P(o/w) are all closely related. It is best not to use more than two of this set in your final QSAR to avoid distortion of the final QSAR results (even though they are not necessarily all strongly correlated). Answer the following questions: 1. How well correlated are F H2 and E_solv? Since both are rough approximations, it is not easy to decide which will give the best approximation to the experimental solvation free energy. 2. Give your best QSAR equation. Include the R 2 and R values and the RELATIVE IMPRTACE for each of the descriptors. 3. Give your best QSAR equation using the same descriptors as in question 2 but without F H2, F oct, or E_solv if you used them in question 2. Include the R 2 and R values and the RELATIVE IMPRTACE for each of the descriptors. 4. Do inclusion of F H2, F oct, or E_solv markedly improve the QSAR over the classical descriptors (e. g. log P(o/w), mr, vdw_vol, ASA, apol, dipole)? 5. Give the name of name of the computer and the database file that you generated. 4. Solvent effects can have a large influence on tautomeric equilibria. Polar and nonpolar molecules are differentially stabilized in polar solvents. 1 Consider the tautomeric equilibrium in the system: H H 2-Hydroxypyridine is favored in the gas phase, but the pyridone is favored in aqueous solution. The molecular mechanics steric energies of these two molecules can't be directly compared, since they don't have the same number and types of chemical bonds. However, we can compare the GB/SA solvation energies. The experimental results require that the solvation energy of the pyridone should be more favorable (larger negative) than for 2-hydroxypyridine. Do the GB/SA solvation energies calculated using ME reproduce the experimental trends? Use the procedure that we used in lab from the Molecular Mechanics tutorial to determine the solvation energy for these two compounds. Remember to consider conformational isomerism in determining the global minimum energy. In addition, why is the solvation energy of the pyridone more favorable? Create a database containing these two compounds and use the QuaSar-Descriptors application to calculate the dipole moment, ASA, and logp(o/w) for these two compounds. Use the following instructions to create the database: 1. With one of the two molecules in the ME main window, pull down the File menu, slide right on ew and choose Database. In the file librarian dialog box enter the database file name as "D:/moe/XXXpyridine.mdb", where XXX are your initials. 2. In the Database Viewer, pull down the Field menu and choose Create Field.
6 3. In the dialog bar at the top of spreadsheet window pull down the Create Field type menu and choose molecule. In the adjacent ame dialog box enter "mol". Press enter. A column label "mol" should appear. 4. Pull down the Entry menu and choose Add Entry In the ew Entry window click K. The current molecule in the ME window should be transferred into the database. 5. Close the current molecule in the ME main window. Build or Load the file for the other of the two molecules. In the Database Viewer, once again pull down the Entry menu and choose Add Entry In the ew Entry window click K. The current molecule in the ME window should be transferred into the database as the second molecule. 6. You can now add your descriptors by pulling down the Compute menu and choosing Descriptors. Answer the following questions: 1. Which molecule is better stabilized in aqueous solvent? Do the GB/SA solvation energies calculated using ME reproduce the experimental trends? 2. Using the dipole moment and the ASA values, decide whether the charge distribution or the solvent accessible surface plays a bigger role in determining the relative solvation energy of the two compounds. Remember the surface tension term used in the GB/SA approach is 8.72 cal/å Given the answer to questions 1 and 2, why is this molecule better stabilized by aqueous solvation? 4. Does the logp(o/w) value correlate as expected with the solvation energy and the dipole moment? 5. Give the name of the computer and the database file that you generated. Printing Plots from ME on a PC Use the Start menu to run the Paint program. Make the window that you want to print the active window. Press Alt-Print Screen. Switch to the Paint program, pull down the Edit menu and choose Paste. For scatter plots, the background will be black. To switch to a white background, pull down the Image menu ancd choose Invert Colors. Print the graphics from the Paint program as you would normally. Literature Cited: 1. W. J. Hehre, L. D. Burke, A. J. Shusterman, W. J. Pietro, Experiments in Computational rganic Chemistry, Wavenfunction, Inc., Irvine, CA, Experiment 6: Substituent and Solvent Effects on Tautomeric Equilibria.
Conformational Analysis of n-butane
 Conformational Analysis of n-butane In this exercise you will calculate the Molecular Mechanics (MM) single point energy of butane in various conformations with respect to internal rotation around the
Conformational Analysis of n-butane In this exercise you will calculate the Molecular Mechanics (MM) single point energy of butane in various conformations with respect to internal rotation around the
ISIS/Draw "Quick Start"
 ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
ISIS/Draw "Quick Start" Click to print, or click Drawing Molecules * Basic Strategy 5.1 * Drawing Structures with Template tools and template pages 5.2 * Drawing bonds and chains 5.3 * Drawing atoms 5.4
1 An Experimental and Computational Investigation of the Dehydration of 2-Butanol
 1 An Experimental and Computational Investigation of the Dehydration of 2-Butanol Summary. 2-Butanol will be dehydrated to a mixture of 1-butene and cis- and trans-2-butene using the method described in
1 An Experimental and Computational Investigation of the Dehydration of 2-Butanol Summary. 2-Butanol will be dehydrated to a mixture of 1-butene and cis- and trans-2-butene using the method described in
Molecular Modeling and Conformational Analysis with PC Spartan
 Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
General Chemistry Lab Molecular Modeling
 PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
Chemistry 14CL. Worksheet for the Molecular Modeling Workshop. (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell)
 Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Literature values: ΔH f, gas = % error Source: ΔH f, solid = % error. For comparison, your experimental value was ΔH f = phase:
 1 Molecular Calculations Lab: Some guideline given at the bottom of page 3. 1. Use the semi-empirical AM1 method to calculate ΔH f for the compound you used in the heat of combustion experiment. Be sure
1 Molecular Calculations Lab: Some guideline given at the bottom of page 3. 1. Use the semi-empirical AM1 method to calculate ΔH f for the compound you used in the heat of combustion experiment. Be sure
3. An Introduction to Molecular Mechanics
 3. An Introduction to Molecular Mechanics Introduction When you use Chem3D to draw molecules, the program assigns bond lengths and bond angles based on experimental data. The program does not contain real
3. An Introduction to Molecular Mechanics Introduction When you use Chem3D to draw molecules, the program assigns bond lengths and bond angles based on experimental data. The program does not contain real
Introduction to Hartree-Fock calculations in Spartan
 EE5 in 2008 Hannes Jónsson Introduction to Hartree-Fock calculations in Spartan In this exercise, you will get to use state of the art software for carrying out calculations of wavefunctions for molecues,
EE5 in 2008 Hannes Jónsson Introduction to Hartree-Fock calculations in Spartan In this exercise, you will get to use state of the art software for carrying out calculations of wavefunctions for molecues,
Assignment 1: Molecular Mechanics (PART 1 25 points)
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard August 19, 2015 Assignment 1: Molecular Mechanics (PART 1 25 points) In this assignment, you will perform some molecular mechanics calculations using the
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard August 19, 2015 Assignment 1: Molecular Mechanics (PART 1 25 points) In this assignment, you will perform some molecular mechanics calculations using the
Tutorials on Library Design E. Lounkine and J. Bajorath (University of Bonn) C. Muller and A. Varnek (University of Strasbourg)
 Tutorials on Library Design E. Lounkine and J. Bajorath (University of Bonn) C. Muller and A. Varnek (University of Strasbourg) The purpose of this tutorial is to generate a library of potential inhibitors
Tutorials on Library Design E. Lounkine and J. Bajorath (University of Bonn) C. Muller and A. Varnek (University of Strasbourg) The purpose of this tutorial is to generate a library of potential inhibitors
Calculating NMR Chemical Shifts for beta-ionone O
 Calculating NMR Chemical Shifts for beta-ionone O Molecular orbital calculations can be used to get good estimates for chemical shifts. In this exercise we will calculate the chemical shifts for beta-ionone.
Calculating NMR Chemical Shifts for beta-ionone O Molecular orbital calculations can be used to get good estimates for chemical shifts. In this exercise we will calculate the chemical shifts for beta-ionone.
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
ICM-Chemist How-To Guide. Version 3.6-1g Last Updated 12/01/2009
 ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
Ligand Scout Tutorials
 Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
APBS electrostatics in VMD - Software. APBS! >!Examples! >!Visualization! >! Contents
 Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
SeeSAR 7.1 Beginners Guide. June 2017
 SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
Assignment 1: Molecular Mechanics (PART 2 25 points)
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard September 2, 2015 Assignment 1: Molecular Mechanics (PART 2 25 points) In this assignment, you will perform some additional molecular mechanics calculations
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard September 2, 2015 Assignment 1: Molecular Mechanics (PART 2 25 points) In this assignment, you will perform some additional molecular mechanics calculations
Chem 310, Organic Chemistry Lab Molecular Modeling Using Macromodel
 Chem 310, Organic Chemistry Lab Molecular Modeling Using Macromodel This is a molecular modeling experiment, and should be written up in your lab notebook just as if it were a normal "wet-chemistry" experiment.
Chem 310, Organic Chemistry Lab Molecular Modeling Using Macromodel This is a molecular modeling experiment, and should be written up in your lab notebook just as if it were a normal "wet-chemistry" experiment.
The OptiSage module. Use the OptiSage module for the assessment of Gibbs energy data. Table of contents
 The module Use the module for the assessment of Gibbs energy data. Various types of experimental data can be utilized in order to generate optimized parameters for the Gibbs energies of stoichiometric
The module Use the module for the assessment of Gibbs energy data. Various types of experimental data can be utilized in order to generate optimized parameters for the Gibbs energies of stoichiometric
v Prerequisite Tutorials GSSHA WMS Basics Watershed Delineation using DEMs and 2D Grid Generation Time minutes
 v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
v. 10.1 WMS 10.1 Tutorial GSSHA WMS Basics Creating Feature Objects and Mapping Attributes to the 2D Grid Populate hydrologic parameters in a GSSHA model using land use and soil data Objectives This tutorial
OECD QSAR Toolbox v.4.1. Tutorial illustrating new options for grouping with metabolism
 OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
3. An Introduction to Molecular Mechanics
 3. An Introduction to Molecular Mechanics Introduction When you use Chem3D to draw molecules, the program assigns bond lengths and bond angles based on experimental data. The program does not contain real
3. An Introduction to Molecular Mechanics Introduction When you use Chem3D to draw molecules, the program assigns bond lengths and bond angles based on experimental data. The program does not contain real
Exercise 2: Solvating the Structure Before you continue, follow these steps: Setting up Periodic Boundary Conditions
 Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
1 Introduction to Computational Chemistry (Spartan)
 1 Introduction to Computational Chemistry (Spartan) Start Spartan by clicking Start / Programs / Spartan Then click File / New Exercise 1 Study of H-X-H Bond Angles (Suitable for general chemistry) Structure
1 Introduction to Computational Chemistry (Spartan) Start Spartan by clicking Start / Programs / Spartan Then click File / New Exercise 1 Study of H-X-H Bond Angles (Suitable for general chemistry) Structure
Patrick: An Introduction to Medicinal Chemistry 5e MOLECULAR MODELLING EXERCISES CHAPTER 17
 MOLECULAR MODELLING EXERCISES CHAPTER 17 Exercise 17.6 Conformational analysis of n-butane Introduction Figure 1 Butane Me Me In this exercise, we will consider the possible stable conformations of butane
MOLECULAR MODELLING EXERCISES CHAPTER 17 Exercise 17.6 Conformational analysis of n-butane Introduction Figure 1 Butane Me Me In this exercise, we will consider the possible stable conformations of butane
Tutorial 12 Excess Pore Pressure (B-bar method) Undrained loading (B-bar method) Initial pore pressure Excess pore pressure
 Tutorial 12 Excess Pore Pressure (B-bar method) Undrained loading (B-bar method) Initial pore pressure Excess pore pressure Introduction This tutorial will demonstrate the Excess Pore Pressure (Undrained
Tutorial 12 Excess Pore Pressure (B-bar method) Undrained loading (B-bar method) Initial pore pressure Excess pore pressure Introduction This tutorial will demonstrate the Excess Pore Pressure (Undrained
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Table of Contents A red arrow was used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 Swain Home
Jaguar DFT Optimizations and Transition State Searches
 Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Assignment 2: Conformation Searching (50 points)
 Chemistry 380.37 Fall 2015 Dr. Jean M. Standard September 16, 2015 Assignment 2: Conformation Searching (50 points) In this assignment, you will use the Spartan software package to investigate some conformation
Chemistry 380.37 Fall 2015 Dr. Jean M. Standard September 16, 2015 Assignment 2: Conformation Searching (50 points) In this assignment, you will use the Spartan software package to investigate some conformation
POC via CHEMnetBASE for Identifying Unknowns
 Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Table of Contents A red arrow is used to identify where buttons and functions are located in CHEMnetBASE. Figure Description Page Entering the Properties of Organic Compounds (POC) Database 1 CHEMnetBASE
Titrator 3.0 Tutorial: Calcite precipitation
 Titrator 3.0 Tutorial: Calcite precipitation November 2008 Steve Cabaniss A. Introduction This brief tutorial is intended to acquaint you with some of the features of the program Titrator. It assumes that
Titrator 3.0 Tutorial: Calcite precipitation November 2008 Steve Cabaniss A. Introduction This brief tutorial is intended to acquaint you with some of the features of the program Titrator. It assumes that
WMS 9.0 Tutorial GSSHA Modeling Basics Infiltration Learn how to add infiltration to your GSSHA model
 v. 9.0 WMS 9.0 Tutorial GSSHA Modeling Basics Infiltration Learn how to add infiltration to your GSSHA model Objectives This workshop builds on the model developed in the previous workshop and shows you
v. 9.0 WMS 9.0 Tutorial GSSHA Modeling Basics Infiltration Learn how to add infiltration to your GSSHA model Objectives This workshop builds on the model developed in the previous workshop and shows you
Expt MM 1. MOLECULAR MODELING AND PREDICTIONS OF EQUILIBRIUM CONSTANT FOR MENTHONE (trans) AND ISOMENTHONE (cis) ISOMERS (MM)
 Expt MM 1 MOLECULAR MODELING AND PREDICTIONS OF EQUILIBRIUM CONSTANT FOR MENTHONE (trans) AND ISOMENTHONE (cis) ISOMERS (MM) Important Modification Note the software in use may be changed in 2008 to Scigress.
Expt MM 1 MOLECULAR MODELING AND PREDICTIONS OF EQUILIBRIUM CONSTANT FOR MENTHONE (trans) AND ISOMENTHONE (cis) ISOMERS (MM) Important Modification Note the software in use may be changed in 2008 to Scigress.
E-EROS TUTORIAL Encyclopedia of Reagents for Organic Synthesis at the UIUC Chemistry Library
 E-EROS TUTORIAL Encyclopedia of Reagents for Organic Synthesis at the UIUC Chemistry Library For any questions about e-eros please contact Tina Chrzastowski or call (217) 333-3737. Introduction About e-eros:
E-EROS TUTORIAL Encyclopedia of Reagents for Organic Synthesis at the UIUC Chemistry Library For any questions about e-eros please contact Tina Chrzastowski or call (217) 333-3737. Introduction About e-eros:
LAB 2 - ONE DIMENSIONAL MOTION
 Name Date Partners L02-1 LAB 2 - ONE DIMENSIONAL MOTION OBJECTIVES Slow and steady wins the race. Aesop s fable: The Hare and the Tortoise To learn how to use a motion detector and gain more familiarity
Name Date Partners L02-1 LAB 2 - ONE DIMENSIONAL MOTION OBJECTIVES Slow and steady wins the race. Aesop s fable: The Hare and the Tortoise To learn how to use a motion detector and gain more familiarity
Application Note. U. Heat of Formation of Ethyl Alcohol and Dimethyl Ether. Introduction
 Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
ON SITE SYSTEMS Chemical Safety Assistant
 ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
ON SITE SYSTEMS Chemical Safety Assistant CS ASSISTANT WEB USERS MANUAL On Site Systems 23 N. Gore Ave. Suite 200 St. Louis, MO 63119 Phone 314-963-9934 Fax 314-963-9281 Table of Contents INTRODUCTION
Lab 1: Handout GULP: an Empirical energy code
 Lab 1: Handout GULP: an Empirical energy code We will be using the GULP code as our energy code. GULP is a program for performing a variety of types of simulations on 3D periodic solids, gas phase clusters,
Lab 1: Handout GULP: an Empirical energy code We will be using the GULP code as our energy code. GULP is a program for performing a variety of types of simulations on 3D periodic solids, gas phase clusters,
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build a userdefined
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build a userdefined QSAR Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build a userdefined QSAR Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed
Project 2. Chemistry of Transient Species in Planetary Atmospheres: Exploring the Potential Energy Surfaces of CH 2 S
 Chemistry 362 Spring 2018 Dr. Jean M. Standard March 21, 2018 Project 2. Chemistry of Transient Species in Planetary Atmospheres: Exploring the Potential Energy Surfaces of CH 2 S In this project, you
Chemistry 362 Spring 2018 Dr. Jean M. Standard March 21, 2018 Project 2. Chemistry of Transient Species in Planetary Atmospheres: Exploring the Potential Energy Surfaces of CH 2 S In this project, you
Chem 1 Kinetics. Objectives. Concepts
 Chem 1 Kinetics Objectives 1. Learn some basic ideas in chemical kinetics. 2. Understand how the computer visualizations can be used to benefit the learning process. 3. Understand how the computer models
Chem 1 Kinetics Objectives 1. Learn some basic ideas in chemical kinetics. 2. Understand how the computer visualizations can be used to benefit the learning process. 3. Understand how the computer models
Experiment 4: Equilibrium Thermodynamics of a Keto-Enol Tautomerism Reaction
 Experiment 4: Equilibrium Thermodynamics of a Keto-Enol Tautomerism Reaction Reading: SGN: Experiment 21 (p.256-263), Experiment 43 (p.456-459). Quanta: Nuclear magnetic resonance, Relaxation All reactions
Experiment 4: Equilibrium Thermodynamics of a Keto-Enol Tautomerism Reaction Reading: SGN: Experiment 21 (p.256-263), Experiment 43 (p.456-459). Quanta: Nuclear magnetic resonance, Relaxation All reactions
2: SIMPLE HARMONIC MOTION
 2: SIMPLE HARMONIC MOTION Motion of a mass hanging from a spring If you hang a mass from a spring, stretch it slightly, and let go, the mass will go up and down over and over again. That is, you will get
2: SIMPLE HARMONIC MOTION Motion of a mass hanging from a spring If you hang a mass from a spring, stretch it slightly, and let go, the mass will go up and down over and over again. That is, you will get
Tutorial: Structural Analysis of a Protein-Protein Complex
 Molecular Modeling Section (MMS) Department of Pharmaceutical and Pharmacological Sciences University of Padova Via Marzolo 5-35131 Padova (IT) @contact: stefano.moro@unipd.it Tutorial: Structural Analysis
Molecular Modeling Section (MMS) Department of Pharmaceutical and Pharmacological Sciences University of Padova Via Marzolo 5-35131 Padova (IT) @contact: stefano.moro@unipd.it Tutorial: Structural Analysis
Vector Analysis: Farm Land Suitability Analysis in Groton, MA
 Vector Analysis: Farm Land Suitability Analysis in Groton, MA Written by Adrienne Goldsberry, revised by Carolyn Talmadge 10/9/2018 Introduction In this assignment, you will help to identify potentially
Vector Analysis: Farm Land Suitability Analysis in Groton, MA Written by Adrienne Goldsberry, revised by Carolyn Talmadge 10/9/2018 Introduction In this assignment, you will help to identify potentially
Version 1.2 October 2017 CSD v5.39
 Mogul Geometry Check Table of Contents Introduction... 2 Example 1. Using Mogul to assess intramolecular geometry... 3 Example 2. Using Mogul to explain activity data... 5 Conclusions... 8 Further Exercises...
Mogul Geometry Check Table of Contents Introduction... 2 Example 1. Using Mogul to assess intramolecular geometry... 3 Example 2. Using Mogul to explain activity data... 5 Conclusions... 8 Further Exercises...
The EpH module. Table of Contents. Section 3
 The module Use to calculate and plot isothermal (Pourbaix) diagrams. Note: accesses compound databases, i.e. treats the aqueous phase as an ideal solution. Table of Contents Section 1 Section 2 Section
The module Use to calculate and plot isothermal (Pourbaix) diagrams. Note: accesses compound databases, i.e. treats the aqueous phase as an ideal solution. Table of Contents Section 1 Section 2 Section
ES205 Analysis and Design of Engineering Systems: Lab 1: An Introductory Tutorial: Getting Started with SIMULINK
 ES205 Analysis and Design of Engineering Systems: Lab 1: An Introductory Tutorial: Getting Started with SIMULINK What is SIMULINK? SIMULINK is a software package for modeling, simulating, and analyzing
ES205 Analysis and Design of Engineering Systems: Lab 1: An Introductory Tutorial: Getting Started with SIMULINK What is SIMULINK? SIMULINK is a software package for modeling, simulating, and analyzing
Cerius 2. Hypothesis & Receptor Models. Release 4.0 April 1999
 Cerius 2 Hypothesis & Receptor Models Release 4.0 April 1999 Molecular Simulations Inc. 9685 Scranton Road San Diego, CA 92121-3752 619/458-9990 Fax: 619/458-0136 Copyright * This document is copyright
Cerius 2 Hypothesis & Receptor Models Release 4.0 April 1999 Molecular Simulations Inc. 9685 Scranton Road San Diego, CA 92121-3752 619/458-9990 Fax: 619/458-0136 Copyright * This document is copyright
Watershed Modeling Orange County Hydrology Using GIS Data
 v. 10.0 WMS 10.0 Tutorial Watershed Modeling Orange County Hydrology Using GIS Data Learn how to delineate sub-basins and compute soil losses for Orange County (California) hydrologic modeling Objectives
v. 10.0 WMS 10.0 Tutorial Watershed Modeling Orange County Hydrology Using GIS Data Learn how to delineate sub-basins and compute soil losses for Orange County (California) hydrologic modeling Objectives
The View Data module
 The module Use to examine stored compound data (H, S, C p (T), G, etc.) in Compound type databases and list solutions and their constituents in Solution type databases. Table of contents Section 1 Section
The module Use to examine stored compound data (H, S, C p (T), G, etc.) in Compound type databases and list solutions and their constituents in Solution type databases. Table of contents Section 1 Section
Using Microsoft Excel
 Using Microsoft Excel Objective: Students will gain familiarity with using Excel to record data, display data properly, use built-in formulae to do calculations, and plot and fit data with linear functions.
Using Microsoft Excel Objective: Students will gain familiarity with using Excel to record data, display data properly, use built-in formulae to do calculations, and plot and fit data with linear functions.
Physical Chemistry II Laboratory
 Kuwata Spring 2003 Physical Chemistry II Laboratory The Rovibrational Spectra of H 35 Cl and H 37 Cl Using FTIR Write-Up Due Date: Thursday, April 17 (You may record spectra and write your reports in teams
Kuwata Spring 2003 Physical Chemistry II Laboratory The Rovibrational Spectra of H 35 Cl and H 37 Cl Using FTIR Write-Up Due Date: Thursday, April 17 (You may record spectra and write your reports in teams
Wikipedia - Stellar classification:
 Stars and Hertzprung-Russell Diagram Introductory Astronomy laboratory exercise with Stellarium Mike Chu Name Stellarium is an open source and cross-platform application from www.stellarium.org. A star
Stars and Hertzprung-Russell Diagram Introductory Astronomy laboratory exercise with Stellarium Mike Chu Name Stellarium is an open source and cross-platform application from www.stellarium.org. A star
ICM-Chemist-Pro How-To Guide. Version 3.6-1h Last Updated 12/29/2009
 ICM-Chemist-Pro How-To Guide Version 3.6-1h Last Updated 12/29/2009 ICM-Chemist-Pro ICM 3D LIGAND EDITOR: SETUP 1. Read in a ligand molecule or PDB file. How to setup the ligand in the ICM 3D Ligand Editor.
ICM-Chemist-Pro How-To Guide Version 3.6-1h Last Updated 12/29/2009 ICM-Chemist-Pro ICM 3D LIGAND EDITOR: SETUP 1. Read in a ligand molecule or PDB file. How to setup the ligand in the ICM 3D Ligand Editor.
Motion II. Goals and Introduction
 Motion II Goals and Introduction As you have probably already seen in lecture or homework, and if you ve performed the experiment Motion I, it is important to develop a strong understanding of how to model
Motion II Goals and Introduction As you have probably already seen in lecture or homework, and if you ve performed the experiment Motion I, it is important to develop a strong understanding of how to model
Protein Bioinformatics Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski
 Protein Bioinformatics 260.655 Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski Goals: Approx. Time [1] Use the Protein Data Bank PDB website. 10 minutes [2] Use the WebMol Viewer.
Protein Bioinformatics 260.655 Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski Goals: Approx. Time [1] Use the Protein Data Bank PDB website. 10 minutes [2] Use the WebMol Viewer.
NMRPredict Functional Block Diagram
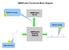 NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
NMRPredict Functional Block Diagram 2 Submit to server. NMRPredict Server NMRPredict DeskTop Software Review results. 3 1 Input structure. Starting the NMRPredict Interface Click on the NMRPredict icon.
From BASIS DD to Barista Application in Five Easy Steps
 Y The steps are: From BASIS DD to Barista Application in Five Easy Steps By Jim Douglas our current BASIS Data Dictionary is perfect raw material for your first Barista-brewed application. Barista facilitates
Y The steps are: From BASIS DD to Barista Application in Five Easy Steps By Jim Douglas our current BASIS Data Dictionary is perfect raw material for your first Barista-brewed application. Barista facilitates
Absorption Spectra of Conjugated Molecules
 Expt 1: Absorption Spectra of onjugated Molecules EM 361 Introduction Absorption Spectra of onjugated Molecules The purpose of this experiment is to measure the absorption spectra of two series of cyanine
Expt 1: Absorption Spectra of onjugated Molecules EM 361 Introduction Absorption Spectra of onjugated Molecules The purpose of this experiment is to measure the absorption spectra of two series of cyanine
Project 3: Molecular Orbital Calculations of Diatomic Molecules. This project is worth 30 points and is due on Wednesday, May 2, 2018.
 Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Introduction to Computer Tools and Uncertainties
 Experiment 1 Introduction to Computer Tools and Uncertainties 1.1 Objectives To become familiar with the computer programs and utilities that will be used throughout the semester. To become familiar with
Experiment 1 Introduction to Computer Tools and Uncertainties 1.1 Objectives To become familiar with the computer programs and utilities that will be used throughout the semester. To become familiar with
The data for this lab comes from McDonald Forest. We will be working with spatial data representing the forest boundary, streams, roads, and stands.
 GIS LAB 6 Using the Projection Utility. Converting Data to Oregon s Approved Lambert Projection. Determining Stand Size, Stand Types, Road Length, and Stream Length. This lab will ask you to work with
GIS LAB 6 Using the Projection Utility. Converting Data to Oregon s Approved Lambert Projection. Determining Stand Size, Stand Types, Road Length, and Stream Length. This lab will ask you to work with
Electric Fields and Equipotentials
 OBJECTIVE Electric Fields and Equipotentials To study and describe the two-dimensional electric field. To map the location of the equipotential surfaces around charged electrodes. To study the relationship
OBJECTIVE Electric Fields and Equipotentials To study and describe the two-dimensional electric field. To map the location of the equipotential surfaces around charged electrodes. To study the relationship
Using the Stock Hydrology Tools in ArcGIS
 Using the Stock Hydrology Tools in ArcGIS This lab exercise contains a homework assignment, detailed at the bottom, which is due Wednesday, October 6th. Several hydrology tools are part of the basic ArcGIS
Using the Stock Hydrology Tools in ArcGIS This lab exercise contains a homework assignment, detailed at the bottom, which is due Wednesday, October 6th. Several hydrology tools are part of the basic ArcGIS
PHY 123 Lab 1 - Error and Uncertainty and the Simple Pendulum
 To print higher-resolution math symbols, click the Hi-Res Fonts for Printing button on the jsmath control panel. PHY 13 Lab 1 - Error and Uncertainty and the Simple Pendulum Important: You need to print
To print higher-resolution math symbols, click the Hi-Res Fonts for Printing button on the jsmath control panel. PHY 13 Lab 1 - Error and Uncertainty and the Simple Pendulum Important: You need to print
A Computer Study of Molecular Electronic Structure
 A Computer Study of Molecular Electronic Structure The following exercises are designed to give you a brief introduction to some of the types of information that are now readily accessible from electronic
A Computer Study of Molecular Electronic Structure The following exercises are designed to give you a brief introduction to some of the types of information that are now readily accessible from electronic
Name. Chem Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2
 Name Chem 322 - Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2 Click on Titan in the Start menu. When it boots, click on the right corner to make the window full-screen. icon in the
Name Chem 322 - Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2 Click on Titan in the Start menu. When it boots, click on the right corner to make the window full-screen. icon in the
From BASIS DD to Barista Application in Five Easy Steps
 Y The steps are: From BASIS DD to Barista Application in Five Easy Steps By Jim Douglas our current BASIS Data Dictionary is perfect raw material for your first Barista-brewed application. Barista facilitates
Y The steps are: From BASIS DD to Barista Application in Five Easy Steps By Jim Douglas our current BASIS Data Dictionary is perfect raw material for your first Barista-brewed application. Barista facilitates
Free Energy Change and Activation Barrier for a Menshutkin Reaction Including Effects of the Solvent
 Proposed Exercise for the Physical Chemistry Section of the Teaching with Cache Workbook: Free Energy Change and Activation Barrier for a Menshutkin Reaction Including Effects of the Solvent Contributed
Proposed Exercise for the Physical Chemistry Section of the Teaching with Cache Workbook: Free Energy Change and Activation Barrier for a Menshutkin Reaction Including Effects of the Solvent Contributed
Building Inflation Tables and CER Libraries
 Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Building Inflation Tables and CER Libraries January 2007 Presented by James K. Johnson Tecolote Research, Inc. Copyright Tecolote Research, Inc. September 2006 Abstract Building Inflation Tables and CER
Tutorial 23 Back Analysis of Material Properties
 Tutorial 23 Back Analysis of Material Properties slope with known failure surface sensitivity analysis probabilistic analysis back analysis of material strength Introduction Model This tutorial will demonstrate
Tutorial 23 Back Analysis of Material Properties slope with known failure surface sensitivity analysis probabilistic analysis back analysis of material strength Introduction Model This tutorial will demonstrate
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.4. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
Senior astrophysics Lab 2: Evolution of a 1 M star
 Senior astrophysics Lab 2: Evolution of a 1 M star Name: Checkpoints due: Friday 13 April 2018 1 Introduction This is the rst of two computer labs using existing software to investigate the internal structure
Senior astrophysics Lab 2: Evolution of a 1 M star Name: Checkpoints due: Friday 13 April 2018 1 Introduction This is the rst of two computer labs using existing software to investigate the internal structure
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Spatial Analysis using Vector GIS THE GOAL: PREPARATION:
 PLAN 512 GIS FOR PLANNERS Department of Urban and Environmental Planning University of Virginia Fall 2006 Prof. David L. Phillips Spatial Analysis using Vector GIS THE GOAL: This tutorial explores some
PLAN 512 GIS FOR PLANNERS Department of Urban and Environmental Planning University of Virginia Fall 2006 Prof. David L. Phillips Spatial Analysis using Vector GIS THE GOAL: This tutorial explores some
Exercises for Part I: HSC
 Thermodynamic and process modelling in metallurgy and mineral processing 477415S 17 August 2018 Exercises for Part I: HSC This document contains exercises for different modules of the HSC software. It
Thermodynamic and process modelling in metallurgy and mineral processing 477415S 17 August 2018 Exercises for Part I: HSC This document contains exercises for different modules of the HSC software. It
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
Newton's 2 nd Law. . Your end results should only be interms of m
 Newton's nd Law Introduction: In today's lab you will demonstrate the validity of Newton's Laws in predicting the motion of a simple mechanical system. The system that you will investigate consists of
Newton's nd Law Introduction: In today's lab you will demonstrate the validity of Newton's Laws in predicting the motion of a simple mechanical system. The system that you will investigate consists of
BASIC TECHNOLOGY Pre K starts and shuts down computer, monitor, and printer E E D D P P P P P P P P P P
 BASIC TECHNOLOGY Pre K 1 2 3 4 5 6 7 8 9 10 11 12 starts and shuts down computer, monitor, and printer P P P P P P practices responsible use and care of technology devices P P P P P P opens and quits an
BASIC TECHNOLOGY Pre K 1 2 3 4 5 6 7 8 9 10 11 12 starts and shuts down computer, monitor, and printer P P P P P P practices responsible use and care of technology devices P P P P P P opens and quits an
BCMB/CHEM 8190 Lab Exercise Using Maple for NMR Data Processing and Pulse Sequence Design March 2012
 BCMB/CHEM 8190 Lab Exercise Using Maple for NMR Data Processing and Pulse Sequence Design March 2012 Introduction Maple is a powerful collection of routines to aid in the solution of mathematical problems
BCMB/CHEM 8190 Lab Exercise Using Maple for NMR Data Processing and Pulse Sequence Design March 2012 Introduction Maple is a powerful collection of routines to aid in the solution of mathematical problems
OECD QSAR Toolbox v.4.1. Step-by-step example for building QSAR model
 OECD QSAR Toolbox v.4.1 Step-by-step example for building QSAR model Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed to take
OECD QSAR Toolbox v.4.1 Step-by-step example for building QSAR model Background Objectives The exercise Workflow of the exercise Outlook 2 Background This is a step-by-step presentation designed to take
Rate Law Determination of the Crystal Violet Reaction. Evaluation copy
 Rate Law Determination of the Crystal Violet Reaction Computer 30 In this experiment, you will observe the reaction between crystal violet and sodium hydroxide. One objective is to study the relationship
Rate Law Determination of the Crystal Violet Reaction Computer 30 In this experiment, you will observe the reaction between crystal violet and sodium hydroxide. One objective is to study the relationship
1. Open polymath: 2. Go to Help, Contents F1 or Press F1
 Polymath Tutorial Process Fluid Transport 1. Open polymath: 2. Go to Help, Contents F1 or Press F1 1 3. Read the section titled Introduction to Polymath both getting started and Variables and expressions
Polymath Tutorial Process Fluid Transport 1. Open polymath: 2. Go to Help, Contents F1 or Press F1 1 3. Read the section titled Introduction to Polymath both getting started and Variables and expressions
Molecular modeling with InsightII
 Molecular modeling with InsightII Yuk Sham Computational Biology/Biochemistry Consultant Phone: (612) 624 7427 (Walter Library) Phone: (612) 624 0783 (VWL) Email: shamy@msi.umn.edu How to run InsightII
Molecular modeling with InsightII Yuk Sham Computational Biology/Biochemistry Consultant Phone: (612) 624 7427 (Walter Library) Phone: (612) 624 0783 (VWL) Email: shamy@msi.umn.edu How to run InsightII
Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018)
 Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018) Abstract Students are introduced to basic features of Scigress by building molecules and performing calculations on them using semi-empirical
Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018) Abstract Students are introduced to basic features of Scigress by building molecules and performing calculations on them using semi-empirical
Instructions for Using Spartan 14
 Instructions for Using Spartan 14 Log in to the computer with your Colby ID and password. Click on the Spartan 14 icon in the dock at the bottom of your screen. I. Building Molecules Spartan has one main
Instructions for Using Spartan 14 Log in to the computer with your Colby ID and password. Click on the Spartan 14 icon in the dock at the bottom of your screen. I. Building Molecules Spartan has one main
Center of Mass. Evaluation copy
 Center of Mass Experiment 19 INTRODUCTION In the most of the previous experiments you have examined the motion of a single object as it underwent a variety of motions. You learned that an object subject
Center of Mass Experiment 19 INTRODUCTION In the most of the previous experiments you have examined the motion of a single object as it underwent a variety of motions. You learned that an object subject
Agilent All Ions MS/MS
 Agilent All Ions MS/MS Workflow Overview A Determine fragment ions for LC/MS Quant method B Develop final Quant method Develop LC/MS Qualitative Analysis method Process data with Find by Formula Build
Agilent All Ions MS/MS Workflow Overview A Determine fragment ions for LC/MS Quant method B Develop final Quant method Develop LC/MS Qualitative Analysis method Process data with Find by Formula Build
2: SIMPLE HARMONIC MOTION
 2: SIMPLE HARMONIC MOTION Motion of a Mass Hanging from a Spring If you hang a mass from a spring, stretch it slightly, and let go, the mass will go up and down over and over again. That is, you will get
2: SIMPLE HARMONIC MOTION Motion of a Mass Hanging from a Spring If you hang a mass from a spring, stretch it slightly, and let go, the mass will go up and down over and over again. That is, you will get
Experiment 0 ~ Introduction to Statistics and Excel Tutorial. Introduction to Statistics, Error and Measurement
 Experiment 0 ~ Introduction to Statistics and Excel Tutorial Many of you already went through the introduction to laboratory practice and excel tutorial in Physics 1011. For that reason, we aren t going
Experiment 0 ~ Introduction to Statistics and Excel Tutorial Many of you already went through the introduction to laboratory practice and excel tutorial in Physics 1011. For that reason, we aren t going
Experiment 13. Dilutions and Data Handling in a Spreadsheet rev 1/2013
 Absorbance Experiment 13 Dilutions and Data Handling in a Spreadsheet rev 1/2013 GOAL: This lab experiment will provide practice in making dilutions using pipets and introduce basic spreadsheet skills
Absorbance Experiment 13 Dilutions and Data Handling in a Spreadsheet rev 1/2013 GOAL: This lab experiment will provide practice in making dilutions using pipets and introduce basic spreadsheet skills
Searching Substances in Reaxys
 Searching Substances in Reaxys Learning Objectives Understand that substances in Reaxys have different sources (e.g., Reaxys, PubChem) and can be found in Document, Reaction and Substance Records Recognize
Searching Substances in Reaxys Learning Objectives Understand that substances in Reaxys have different sources (e.g., Reaxys, PubChem) and can be found in Document, Reaction and Substance Records Recognize
Chemistry 5021/8021 Computational Chemistry 3/4 Credits Spring Semester 2000 ( Due 4 / 3 / 00 )
 Chemistry 5021/8021 Computational Chemistry 3/4 Credits Spring Semester 2000 ( Due 4 / 3 / 00 ) This problem set will take longer than the last one in the sense that you may need to submit some jobs, leave,
Chemistry 5021/8021 Computational Chemistry 3/4 Credits Spring Semester 2000 ( Due 4 / 3 / 00 ) This problem set will take longer than the last one in the sense that you may need to submit some jobs, leave,
NMR Predictor. Introduction
 NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
NMR Predictor This manual gives a walk-through on how to use the NMR Predictor: Introduction NMR Predictor QuickHelp NMR Predictor Overview Chemical features GUI features Usage Menu system File menu Edit
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
OECD QSAR Toolbox v.3.3. Predicting skin sensitisation potential of a chemical using skin sensitization data extracted from ECHA CHEM database
 OECD QSAR Toolbox v.3.3 Predicting skin sensitisation potential of a chemical using skin sensitization data extracted from ECHA CHEM database Outlook Background The exercise Workflow Save prediction 23.02.2015
OECD QSAR Toolbox v.3.3 Predicting skin sensitisation potential of a chemical using skin sensitization data extracted from ECHA CHEM database Outlook Background The exercise Workflow Save prediction 23.02.2015
