A Thesis Presented to The Academic Faculty. Miranda McDaniel
|
|
|
- Camilla Dickerson
- 5 years ago
- Views:
Transcription
1 Proton Coupled Electron Transfer as Explored by the Tryptophan Cation Radical Formation in Biomimetic Peptides A Thesis Presented to The Academic Faculty by Miranda McDaniel In Partial Fulfillment of the Requirements for the Degree B.S. in Biochemistry in the School of Chemistry and Biochemistry Georgia Institute of Technology May 2014
2 Proton Coupled Electron Transfer as Explored by the Tryptophan Cation Radical Formation in Biomimetic Peptides Approved by: Dr. Bridgette Barry, Advisor School of Chemistry and Biochemistry Georgia Institute of Technology Dr. Eliezer Hershkovits School of Chemistry and Biochemistry Georgia Institute of Technology Dr. William Baron School of Chemistry and Biochemistry Georgia Institute of Technology Date Approved: May 2014 ii
3 TABLE OF CONTENTS Page LIST OF FIGURES LIST OF SYMBOLS AND ABBREVIATIONS ABSTRACT vii viii iv CHAPTER 1 Introduction 1 2 Literature Review 4 Proton Coupled Electron Transfer 4 Cytochrome C Peroxidase 5 Biomimetic Peptides 7 3 Materials and Methods 9 4 Results 12 5 Discussion 14 6 Summary 16 REFERENCES 17 iii
4 LIST OF FIGURES Page Figure 1: Beta-hairpin Peptides 9 Figure 2: CD Spectrum of Peptide A 12 Figure 3: CD Spectra of Peptide M and W 13 iv
5 NOMENCLATURE PCET OEC EPR NMR CcP CD SWV DPV Proton Coupled Electron Transfer Oxygen Evolving Complex Electron Paramagnetic Resonance Nuclear Magnetic Resonance Cytochrome c Peroxidase Circular Dichroism Square Wave Voltammetry Differential Pulse Voltammetry v
6 ABSTRACT Proton coupled electron transfer (PCET) is a mechanism exploited by many biological processes including the oxygen evolving complex (OEC) of photosystem II. PCET functions largely by the formation of a stable radical species. One amino acid with the ability to form such a stable cation radical is tryptophan. In this study, biomimetic beta-hairpin peptides have been synthesized with a cross-strand interacting tryptohan residue. The peptides have been characterized by circular dichroism spectroscopy, to be further explored by electrochemistry and electron paramagnetic resonance (EPR) spectroscopy. These techniques will provide insight into the formation of the tryptophan radical, the factors influencing its stabilization, the effect of the local amino acid environment, and, ultimately, the fundamentals of proton coupled electron transfer. vi
7 CHAPTER 1 INTRODUCTION Proton coupled electron transfer (PCET) is a mechanism of many redox reactions by which a proton and an electron are transferred in a concerted manner. Many biological processes, such as those performed by the oxygen evolving complex of photosystem II in the chloroplasts of plants and those of the ribonucleotide reductase enzyme of DNA synthesis and repair, function by such a mechanism. An important process of the PCET reaction is the formation of a cation radical species from an amino acid residue. One such amino acid with the ability to facilitate the reaction is tryptophan, which contains an aromatic, hydrophobic side chain. The cation formation from tryptophan is also an essential reaction for the enzymatic activity of cytochrome c peroxidase (CcP), an enzyme responsible for the catalysis of H 2 O 2 oxidation to form water. In an attempt to better understand the ability of and mechanism by which the cation radical forms, and ultimately by which the PCET reaction occurs, the activity of CcP and similar tryptophan containing biomolecules can be monitored. Miller et al. 1 have effectively used site-directed mutagenesis to probe the protein environment that stabilizes the radical of compound I (or compound ES) of CcP. In the mutagenesis, the tryptophan in position 191 of the sequence was replaced by glycine or glutamine. The mutants were then crystallized and a monovalent cation binding site was observed in the cavity that was formerly occupied by the side chain of Trp-191. It was further determined that the cation binding site was created by partial negative charges in the cavity and that the Trp-191 side chain resides in a consensus potassium binding site. In a study by Huyett et al. 2, compound I of the CcP enzyme was characterized by 1
8 continuous wave and pulsed Q- band electron nuclear double resonance (ENDOR) spectroscopy. The results allowed for the complete characterization of the active site of compound I, which was found to contain an oxyferryl heme coupled to the Trp-191 π- cation radical by weak spin exchange. In order to develop a more detailed understanding of the mechanism by which compound I forms in heme peroxidases, Hiner et al. 3 provided a focused review stating that the intermediate stores two oxidizing equivalents from hydrogen peroxide; one equivalent stored as an oxyferryl iron center and the second as a radical on the porphyrin ring or a tryptophan residue. The mechanism of action for the heterolytic cleavage of the hydrogen peroxide has been attributed to the acid-base catalytic activity of the conserved distal histidine, with assistance from the distal arginine and asparagine. Though comprehensive in its methods, results, and discussion, the review outlines an important need for more research to further understand and define the mechanism, stabilization, and environment of the cation radical formation observed in cytochrome c peroxidase. Further investigation into this process will provide valuable insight into the complex mechanism of PCET. To propel the study of PCET by an exploration of the tryptophan cation radical formation, two water-soluble biomimetic peptides, Peptide M (IMDRYRVRNGDRIWIRLR) and Peptide W (IMDRWRVRNGDRIHIRLR) have been synthesized. These peptides were engineered to retain a hydrogen bonded beta-hairpin structure allowing for a tryptophan residue to interact with a cross-strand amino acid. The tryptophan residue interacts with the cross-strand residues of Tyrosine and Histidine in Peptides M and W, respectively. Through a series of techniques, including circular 2
9 dichroism (CD), differential pulse voltammetry, and electron paramagnetic resonance (EPR), knowledge can be gained about the formation of a tryptophan cation radical, the factors influencing its stabilization, and the effect of the local amino acid environment. Once a comprehensive characterization of the radical activity in these prototypes is completed, details of the previously unknown fundamentals of proton coupled electron transfer can be resolved and understood. 3
10 CHAPTER 2 LITERATURE REVIEW Proton Coupled Electron Transfer While the mechanism of PCET is not completely understood, it is known that the reaction is facilitated by the formation of a cation radical species from an amino acid residue. Tryptophan is one such amino acid whose cation radical formation is being studied. For example, Sjodin et al. 4 explored the coupling of electron and proton transfer by investigating model complexes containing an oxidized tyrosine or tryptophan residue. Through differential pulse voltammetry and laser flash photolysis with transient absorption detection of Ru-Tyr and Ru-Trp, the group was able to determine that PCET proceeds in a competition between a stepwise reaction and a single step reaction. During the stepwise process, electron transfer is followed by protonation of an amino acid radical (ETPT). In a concerted reaction (CEP), the electron and proton are transferred simultaneously. The group studied the effects of solution ph, the strength of the Ru III oxidant, and the identity of the amino acid on the mechanism that would dominate the reaction. Furthermore, Zhang et al. 5 established that tryptophan oxidation is an appropriate model for understanding how electrons and protons are coupled in PCET. The team employs laser flash-quench methods to determine the intramolecular oxidation kinetics of tryptophan derivatives with water as a proton acceptor. Ultimately, the group concludes that the CEP reaction is a fundamental process that is essential for the understanding of PCET. While the details comprising the order of electron and proton transfer are becoming clear, there is still a lack of understanding of the mechanism by which this 4
11 transfer occurs. Even then, if the mechanism is well understood and accepted, the application of the mechanism provides another obstacle to overcome. By generating alternative model structures and peptides for PCET, it could be possible to better understand the mechanism of the transfer through manipulation of amino acid residues. Furthermore, by conducting experimentation on a model structure, as opposed to the actual complex in vitro, the conditions of stable radical formation and PCET can be optimized and the effects of ph and the neighboring residues on the amino acid environment can be explored. Cytochrome C Peroxidase Knowing the role of tryptophan oxidation in the study of proton and electron coupling, the tryptophan radical and its formation should also be considered. Cytochrome c peroxidase (CcP) is an enzyme whose mechanism exploits the tryptophan cation radical. This enzyme is responsible for the catalysis of H 2 O 2 oxidation to form water. In the catalytic cycle, the proximal tryptophan is known to be oxidized to a cation radical. Bonagura et al. 6 used site-directed mutagenesis and electron paramagnetic resonance (EPR) spectroscopy to introduce an ascorbate peroxidase cation binding site into CcP and show that the cation-containing mutants of CcP are no longer able to form a stable tryptophan radical. The results of this experimentation led to the conclusion that the reactivity of a redox active amino acid side chain is largely controlled by long-range electrostatic effects. Moreover, the results indicated that the oxidation and reduction of the proximal tryptophan residue is essential during the oxidation of ferrocytochrome c. Similarly, Miller et al. 1 employed site-directed mutagenesis to probe the protein environment that stabilizes the radical of compound I (or compound ES) of CcP. The 5
12 tryptophan in position 191 (Trp-191) was replaced by a glycine or glutamine residue and these mutant forms were crystallized. The crystallization revealed a monovalent cation binding site in the cavity, previously occupied by a Trp-191 side chain. It was further determined that the cation binding site was created by partial negative charges in the cavity and the Trp-191 side chain resides in a consensus potassium binding site created by partial negative charges at the backbone of carbonyl oxygen atoms of residues at positions 175 and 177, carbonyl ends of a long alpha-helix, heme propionates, and the carboxylate side chain of Asp-235. Thus, the negative potential enveloping the side chain of the Trp-191 is sufficient to account for the stability of the radical. As more knowledge is gained about the stability of the radical, it is possible to optimize the conditions under which the tryptophan radical can be formed. To better understand the formation of the intermediate compound I of the CcP enzyme, Huyett et al. 2 characterized the enzyme by continuous wave and pulsed Q-band electron nuclear double resonance (ENDOR) spectroscopy. The active site of compound I was found to contain an oxyferral heme coupled to the Trp-191 π-cation radical by weak spin exchange. In congruence with the results from this study, Hiner et al. 3 provided a review in which the group outlines the methods by which the intermediate stores the two oxidizing equivalents from hydrogen peroxide. One equivalent is stored as an oxyferryl iron center and the second as a radical on the porphyrin ring or a tryptophan residue. The conserved distal histidine, with assistance from the distal arginine and asparagine, is known to perform the acid-base catalysis necessary for the heterolytic cleavage of the hydrogen peroxide. 6
13 Biomimetic Peptides With a basic understanding of the radical formation in the OEC as a foundation, biomimetic beta-hairpin peptides are being synthesized and characterized with the goal of modeling the radical formation and PCET within these proteins. Sibert et al. 7 designed a novel amino acid sequence containing a tyrosine residue. This residue can be oxidized by ultraviolet photolysis or electrochemical methods. The group characterizes the peptide with NMR spectroscopy, EPR spectroscopy, and electrochemistry to determine that the tyrosine residue couples with histidine in an interstrand α-cation interaction responsible for stabilizing the tyrosyl radical. While peptides have been used as models for the PCET mechanism, there have yet to be studies that incorporate the cation radical formation ability of tryptophan into the peptides for the understanding of the PCET reactions of photosystem II. Based on a basic understanding of PCET and cation radical formation in the oxygen-evolving complex and cytochrome c peroxidase, biomimetic peptides have been designed to contain a tryptophan residue and maintain a hydrogen bonded beta-hairpin motif. Peptide M (IMDRYRVRNGDRIWIRLR) and Peptide W (IMDRWRVRNGDRIHIRLR) have been synthesized, allowing for the tryptophan residue to interact with the cross-strand residues of Tyrosine and Histidine, respectively. Initially, the peptides will be characterized by circular dichroism spectroscopy, to provide valuable information regarding the secondary structure. A third peptide, Peptide A (IMDRYRVRNGDRIHIRLR) was designed and characterized by Sibert et al. 7 by the same CD methods employed here. The results of the CD were then confirmed with NMR spectroscopy and can now be used as a control for the identification of Peptide M and Peptide W secondary structure. Once the motif is established, other methods of 7
14 characterization can be applied. Similar to the methods employed by Sjödin et al. 4 and Sibert et al. 7, electrochemistry, specifically differential pulse voltammetry, can be used to measure the peak potential of the peptide across a range of ph environments. Finally, EPR, as used by Bonagura et al. 6 and Sibert et al. 7, can be used to study the unpaired electrons within the peptide. Ideally, the tryptophan radical would be successfully generated and probed in these model peptides, allowing for the understanding of its stabilization and formation. With a fundamental understanding of the tryptophan radical formation in these prototypes and the dependency on ph, proximal amino acid environment, and charge, the mechanism of proton coupled electron transfer can be better understood and exploited for a new means of energy conversion. 8
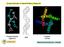
15 CHAPTER 3 MATERIALS AND METHODS Peptide Engineering Biomimetic peptides have been engineered to contain tryptophan residues and maintain hydrogen bonded beta-hairpin motifs. Peptide M (IMDRYRVRNGDRIWIRLR) and Peptide W (IMDRWRVRNGDRIHIRLR) have been synthesized, allowing for the tryptophan residue to interact with the cross-strand residues of Tyrosine and Histidine, respectively. Peptide samples were synthesized by Genscript (> 95% purity). Peptide A Peptide M Peptide W Figure 1: Beta-hairpin motifs of Peptide A, Peptide M, and Peptide W. Figures on the left show the predicted cross-strand interactions. Figures on the right show the beta-hairpin models as predicted by PEP-FOLD Structure Prediction online modeling program. 9
16 Circular Dichroism CD spectra of 100 µm solutions were obtained using a JASCO J-810 CD Polarimeter. Spectra were collected under the following conditions: sensitivity of 100 mdeg, data pitch of 1 nm, scan speed of 50 nm/min, response time of 1 s, and a bandwidth of 1 nm. The CD spectra of peptide M and peptide W have been compared to that of Peptide A at ph 6.5. Peptide A (IMDRYRVRNGDRIHIRLR) was used as a template and is known to contain the beta-hairpin motif by the NMR spectroscopy analysis performed by Sibert et at. 7 10
17 CHAPTER 4 RESULTS Peptide A (Control) Circular dichroism was employed as a method for confirming the beta hairpin secondary structure of the peptides. By passing a beam of circularly polarized light through the sample containing buffered peptide, secondary structure can be deduced by changes in ellipticity. The following spectrum was collected for Peptide A and is characteristic of a beta hairpin structure based on a minimum around 218 nm. The betahairpin structure for the peptide was confirmed by NMR spectroscopy, as designated in the previous literature. By using this peptide, and its CD spectrum, as a control, conclusions can be drawn regarding the secondary structure of the other peptides, Peptide M and W. Figure 2: The CD spectrum of 100 µm Peptide A collected at a ph of 6.5 obtained at 20 o C (bold line, pre-melt), 80 o C (dashed-line) and at 20 o C (thin-line, post-melt) showing the characteristic betahairpin peak. The tick mark is 1 mdeg. 11
18 Peptide M and W Peptide M and Peptide W were studied under the same conditions. The CD data showed the characteristic minimum of ellipticity around 218 nm. Because the secondary structure was confirmed for Peptide A by NMR, comparisons that can be made between the CD spectra for Peptide A and Peptide M or W can provide valuable information regarding the secondary structure of these peptides of interest. Based on the high level of similarity, it is possible to conclude that these two peptides, M and W, share the same beta-hairpin secondary structure as Peptide A. The CD spectra collected at ph 6.5 have been provided below (Figure 3). Figure 3: The CD spectrum of 100 µm Peptide M (top) and W (bottom) collected at a ph of 6.5 obtained at 20 o C (bold line, pre-melt), 80 o C (dashed-line) and at 20 o C (thin-line, post-melt) showing the characteristic beta-hairpin peak. The tick mark is 1 mdeg. 12
19 It is also important to analyze the effects of melting on the secondary structure of the peptides. By altering the peptide environment from 20 C to 80 C, it was possible to measure the changes in ellipticity and, ultimately, determine the robustness and reversibility of the beta-hairpin folding. The CD spectra shown in Figures 2 and 3 include the measurements at 20 C before a melt, 80 C during a melt, and 20 C after a melt. The spectra taken at 80 C has a minimum ellipticity of smaller magnitude than that measured at 20 C before the melt measured at the same wavelength. This observation is significant for two reasons. Firstly, even after melting, the peptide retains a significant amount of the beta-hairpin structure, relative to an unfolded peptide sequence. This indicates that the folding of these peptides is stable and robust, even at high temperatures. Secondly, the secondary structure that is lost to melting is reestablished upon cooling, such that the secondary structure is equivalent to that of the pre-melted conditions. The stability, rigidity, and reversibility, in congruence with the beta-hairpin conformation, make these peptides ideal candidates for further characterization. 13
20 CHAPTER 5 DISCUSSION Based on the circular dichroism spectroscopy, it can be concluded that the peptide sequences being explored in this research, M and W, are folded into the beta-haipin motif. This conclusion was based on the analysis of the spectra in regards to both the predicted CD spectrum of a beta-sheet peptide fold, as well as the comparison to the spectrum of a peptide determined to fold into a beta-hairpin by NMR spectroscopy. Betahairpin motifs are expected to have a characteristic minimum of ellipticity at around nm, which is not characteristic of either alpha helices or random coils. Both Peptide M and W spectra show this characteristic minimum, so the secondary structure of a betahairpin can be inferred. To further support this conclusion, the CD spectra of these peptides were compared to the CD spectrum of Peptide A. NMR spectroscopy was employed by Sibert et al. 7 to determine the secondary structure and support the findings from the CD data for this peptide. The NMR analysis concluded that the amino acid sequence was folded into the predicted motif and supported the analysis of the CD spectrum. As such, the CD spectra of Peptide M and W can be compared to that of Peptide A, and conclusions can be drawn about the secondary structure of the peptides. Based on a high level of similarity between the three spectra, there exists sufficient evidence to conclude that Peptide M and Peptide W are folded into the beta-hairpin motif, as predicted by the PEP- FOLD Structure Prediction online modeling program. With this conclusion, along with the high stability of the peptides, it becomes necessary and possible to further explore the peptide environment. Literature predicts that
21 the aromatic tryptophan residue will be crucial in a proton coupled electron transfer mechanism that can occur between the tryptophan and its cross-strand interacting amino acid residue. By applying analytical techniques, such as differential pulse voltammetry and electron paramagnetic resonance spectroscopy, to probe the electron environment, it is possible to determine the mechanism of radical formation and proton coupled electron transfer that occurs within the cross-strand interacting aromatic amino acids. 15
22 CHAPTER 6 SUMMARY In this study, biomimetic peptides containing cross-strand interacting tryptophan residues were characterized by circular dichroism spectroscopy. By exploiting this technique in the characterization of the peptides, it was possible to determine their secondary structure. Based on the comparison of the CD spectra with that of a known beta-hairpin peptide, the secondary structure was accurately determined to be a betahairpin with a stable conformation. Because of the rigidity of this peptide fold, the tryptophan amino acid is held in close proximity with the tyrosine or histidine in Peptides M and W, respectively. As such, this biomimetic scaffold can be explored by other analytical techniques to develop an understanding of the cross-strand interaction. If the formation of the tryptophan radical, the factors influencing its stabilization, and the effect of the local amino acid environment can be understood, then the mechanism of proton coupled electron transfer can begin to be elucidated. 16
23 REFERENCES 1. Miller, M. A., Han, G. W., and Kraut, J. (1994) A cation binding motif stabilizes the compound I radical of cytochrome c peroxidase, Proceedings of the National Academy of Sciences 91, Huyett, J. E., Doan, P. E., Gurbiel, R., Houseman, A. L. P., Sivaraja, M., Goodin, D. B., and Hoffman, B. M. (1995) Compound ES of Cytochrome c Peroxidase Contains a Trp. pi.- Cation Radical: Characterization by Continuous Wave and Pulsed Q- Band External Nuclear Double Resonance Spectroscopy, Journal of the American Chemical Society 117, Hiner, A. N. P., Raven, E. L., Thorneley, R. N. F., Garcıá- Cánovas, F., and Rodrıǵuez- López, J. N. (2002) Mechanisms of compound I formation in heme peroxidases, Journal of inorganic biochemistry 91, Sjödin, M., Styring, S., Wolpher, H., Xu, Y., Sun, L., and Hammarström, L. (2005) Switching the redox mechanism: models for proton- coupled electron transfer from tyrosine and tryptophan, Journal of the American Chemical Society 127, Zhang, M.- T., and Hammarström, L. (2011) Proton- coupled electron transfer from tryptophan: A concerted mechanism with water as proton acceptor, Journal of the American Chemical Society 133, Bonagura, C. A., Sundaramoorthy, M., Pappa, H. S., Patterson, W. R., and Poulos, T. L. (1996) An engineered cation site in cytochrome c peroxidase alters the reactivity of the redox active tryptophan, Biochemistry 35, Sibert, R., Josowicz, M., Porcelli, F., Veglia, G., Range, K., and Barry, B. A. (2007) Proton- coupled electron transfer in a biomimetic peptide as a model of enzyme regulatory mechanisms, Journal of the American Chemical Society 129,
CHAPTER 29 HW: AMINO ACIDS + PROTEINS
 CAPTER 29 W: AMI ACIDS + PRTEIS For all problems, consult the table of 20 Amino Acids provided in lecture if an amino acid structure is needed; these will be given on exams. Use natural amino acids (L)
CAPTER 29 W: AMI ACIDS + PRTEIS For all problems, consult the table of 20 Amino Acids provided in lecture if an amino acid structure is needed; these will be given on exams. Use natural amino acids (L)
Contents. xiii. Preface v
 Contents Preface Chapter 1 Biological Macromolecules 1.1 General PrincipIes 1.1.1 Macrornolecules 1.2 1.1.2 Configuration and Conformation Molecular lnteractions in Macromolecular Structures 1.2.1 Weak
Contents Preface Chapter 1 Biological Macromolecules 1.1 General PrincipIes 1.1.1 Macrornolecules 1.2 1.1.2 Configuration and Conformation Molecular lnteractions in Macromolecular Structures 1.2.1 Weak
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor
 LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
Biomolecules: lecture 10
 Biomolecules: lecture 10 - understanding in detail how protein 3D structures form - realize that protein molecules are not static wire models but instead dynamic, where in principle every atom moves (yet
Biomolecules: lecture 10 - understanding in detail how protein 3D structures form - realize that protein molecules are not static wire models but instead dynamic, where in principle every atom moves (yet
Principles of Physical Biochemistry
 Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Properties of amino acids in proteins
 Properties of amino acids in proteins one of the primary roles of DNA (but not the only one!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids repeated
Properties of amino acids in proteins one of the primary roles of DNA (but not the only one!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids repeated
Dana Alsulaibi. Jaleel G.Sweis. Mamoon Ahram
 15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
1. What is an ångstrom unit, and why is it used to describe molecular structures?
 1. What is an ångstrom unit, and why is it used to describe molecular structures? The ångstrom unit is a unit of distance suitable for measuring atomic scale objects. 1 ångstrom (Å) = 1 10-10 m. The diameter
1. What is an ångstrom unit, and why is it used to describe molecular structures? The ångstrom unit is a unit of distance suitable for measuring atomic scale objects. 1 ångstrom (Å) = 1 10-10 m. The diameter
Lecture 15: Enzymes & Kinetics. Mechanisms ROLE OF THE TRANSITION STATE. H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl. Margaret A. Daugherty.
 Lecture 15: Enzymes & Kinetics Mechanisms Margaret A. Daugherty Fall 2004 ROLE OF THE TRANSITION STATE Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products
Lecture 15: Enzymes & Kinetics Mechanisms Margaret A. Daugherty Fall 2004 ROLE OF THE TRANSITION STATE Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products
Basics of protein structure
 Today: 1. Projects a. Requirements: i. Critical review of one paper ii. At least one computational result b. Noon, Dec. 3 rd written report and oral presentation are due; submit via email to bphys101@fas.harvard.edu
Today: 1. Projects a. Requirements: i. Critical review of one paper ii. At least one computational result b. Noon, Dec. 3 rd written report and oral presentation are due; submit via email to bphys101@fas.harvard.edu
NH 2. Biochemistry I, Fall Term Sept 9, Lecture 5: Amino Acids & Peptides Assigned reading in Campbell: Chapter
 Biochemistry I, Fall Term Sept 9, 2005 Lecture 5: Amino Acids & Peptides Assigned reading in Campbell: Chapter 3.1-3.4. Key Terms: ptical Activity, Chirality Peptide bond Condensation reaction ydrolysis
Biochemistry I, Fall Term Sept 9, 2005 Lecture 5: Amino Acids & Peptides Assigned reading in Campbell: Chapter 3.1-3.4. Key Terms: ptical Activity, Chirality Peptide bond Condensation reaction ydrolysis
THE UNIVERSITY OF MANITOBA. PAPER NO: 409 LOCATION: Fr. Kennedy Gold Gym PAGE NO: 1 of 6 DEPARTMENT & COURSE NO: CHEM 4630 TIME: 3 HOURS
 PAPER NO: 409 LOCATION: Fr. Kennedy Gold Gym PAGE NO: 1 of 6 DEPARTMENT & COURSE NO: CHEM 4630 TIME: 3 HOURS EXAMINATION: Biochemistry of Proteins EXAMINER: J. O'Neil Section 1: You must answer all of
PAPER NO: 409 LOCATION: Fr. Kennedy Gold Gym PAGE NO: 1 of 6 DEPARTMENT & COURSE NO: CHEM 4630 TIME: 3 HOURS EXAMINATION: Biochemistry of Proteins EXAMINER: J. O'Neil Section 1: You must answer all of
Problems from Previous Class
 1 Problems from Previous lass 1. What is K m? What are the units of K m? 2. What is V max? What are the units of V max? 3. Write down the Michaelis-Menten equation. 4. What order of reaction is the reaction
1 Problems from Previous lass 1. What is K m? What are the units of K m? 2. What is V max? What are the units of V max? 3. Write down the Michaelis-Menten equation. 4. What order of reaction is the reaction
BCH 4053 Exam I Review Spring 2017
 BCH 4053 SI - Spring 2017 Reed BCH 4053 Exam I Review Spring 2017 Chapter 1 1. Calculate G for the reaction A + A P + Q. Assume the following equilibrium concentrations: [A] = 20mM, [Q] = [P] = 40fM. Assume
BCH 4053 SI - Spring 2017 Reed BCH 4053 Exam I Review Spring 2017 Chapter 1 1. Calculate G for the reaction A + A P + Q. Assume the following equilibrium concentrations: [A] = 20mM, [Q] = [P] = 40fM. Assume
SUPPORTING INFORMATION. Proton Coupled Electron Transfer in Tyrosine and a Beta Hairpin Maquette: Reaction Dynamics on the Picosecond Time Scale
 SUPPORTING INFORMATION Proton Coupled Electron Transfer in Tyrosine and a Beta Hairpin Maquette: Reaction Dynamics on the Picosecond Time Scale Cynthia V. Pagba, 1 San-Hui Chi, 2 Joseph Perry, 2 and Bridgette
SUPPORTING INFORMATION Proton Coupled Electron Transfer in Tyrosine and a Beta Hairpin Maquette: Reaction Dynamics on the Picosecond Time Scale Cynthia V. Pagba, 1 San-Hui Chi, 2 Joseph Perry, 2 and Bridgette
CHEM 3653 Exam # 1 (03/07/13)
 1. Using phylogeny all living organisms can be divided into the following domains: A. Bacteria, Eukarya, and Vertebrate B. Archaea and Eukarya C. Bacteria, Eukarya, and Archaea D. Eukarya and Bacteria
1. Using phylogeny all living organisms can be divided into the following domains: A. Bacteria, Eukarya, and Vertebrate B. Archaea and Eukarya C. Bacteria, Eukarya, and Archaea D. Eukarya and Bacteria
Introduction to" Protein Structure
 Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Calculate a rate given a species concentration change.
 Kinetics Define a rate for a given process. Change in concentration of a reagent with time. A rate is always positive, and is usually referred to with only magnitude (i.e. no sign) Reaction rates can be
Kinetics Define a rate for a given process. Change in concentration of a reagent with time. A rate is always positive, and is usually referred to with only magnitude (i.e. no sign) Reaction rates can be
Exam I Answer Key: Summer 2006, Semester C
 1. Which of the following tripeptides would migrate most rapidly towards the negative electrode if electrophoresis is carried out at ph 3.0? a. gly-gly-gly b. glu-glu-asp c. lys-glu-lys d. val-asn-lys
1. Which of the following tripeptides would migrate most rapidly towards the negative electrode if electrophoresis is carried out at ph 3.0? a. gly-gly-gly b. glu-glu-asp c. lys-glu-lys d. val-asn-lys
Recommended Reading: 23, 29 (3rd edition); 22, 29 (4th edition) Ch 102 Problem Set 7 Due: Thursday, June 1 Before Class. Problem 1 (1 points) Part A
 Recommended Reading: 23, 29 (3rd edition); 22, 29 (4th edition) Ch 102 Problem Set 7 Due: Thursday, June 1 Before Class Problem 1 (1 points) Part A Kinetics experiments studying the above reaction determined
Recommended Reading: 23, 29 (3rd edition); 22, 29 (4th edition) Ch 102 Problem Set 7 Due: Thursday, June 1 Before Class Problem 1 (1 points) Part A Kinetics experiments studying the above reaction determined
BSc and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes Marking
Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes Marking
Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur. Lecture - 06 Protein Structure IV
 Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur Lecture - 06 Protein Structure IV We complete our discussion on Protein Structures today. And just to recap
Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur Lecture - 06 Protein Structure IV We complete our discussion on Protein Structures today. And just to recap
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Protein Dynamics. The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron.
 Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Citation for published version (APA): Nouri-Nigjeh, E. (2011). Electrochemistry in the mimicry of oxidative drug metabolism Groningen: s.n.
 University of Groningen Electrochemistry in the mimicry of oxidative drug metabolism Nouri-Nigjeh, Eslam IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish
University of Groningen Electrochemistry in the mimicry of oxidative drug metabolism Nouri-Nigjeh, Eslam IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish
Bi 8 Midterm Review. TAs: Sarah Cohen, Doo Young Lee, Erin Isaza, and Courtney Chen
 Bi 8 Midterm Review TAs: Sarah Cohen, Doo Young Lee, Erin Isaza, and Courtney Chen The Central Dogma Biology Fundamental! Prokaryotes and Eukaryotes Nucleic Acid Components Nucleic Acid Structure DNA Base
Bi 8 Midterm Review TAs: Sarah Cohen, Doo Young Lee, Erin Isaza, and Courtney Chen The Central Dogma Biology Fundamental! Prokaryotes and Eukaryotes Nucleic Acid Components Nucleic Acid Structure DNA Base
Protein Structure. W. M. Grogan, Ph.D. OBJECTIVES
 Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
4. The Michaelis-Menten combined rate constant Km, is defined for the following kinetic mechanism as k 1 k 2 E + S ES E + P k -1
 Fall 2000 CH 595C Exam 1 Answer Key Multiple Choice 1. One of the reasons that enzymes are such efficient catalysts is that a) the energy level of the enzyme-transition state complex is much higher than
Fall 2000 CH 595C Exam 1 Answer Key Multiple Choice 1. One of the reasons that enzymes are such efficient catalysts is that a) the energy level of the enzyme-transition state complex is much higher than
Biophysics 490M Project
 Biophysics 490M Project Dan Han Department of Biochemistry Structure Exploration of aa 3 -type Cytochrome c Oxidase from Rhodobacter sphaeroides I. Introduction: All organisms need energy to live. They
Biophysics 490M Project Dan Han Department of Biochemistry Structure Exploration of aa 3 -type Cytochrome c Oxidase from Rhodobacter sphaeroides I. Introduction: All organisms need energy to live. They
Chapter 2: Chemical Basis of Life
 Chapter 2: Chemical Basis of Life Chemistry is the scientific study of the composition of matter and how composition changes. In order to understand human physiological processes, it is important to understand
Chapter 2: Chemical Basis of Life Chemistry is the scientific study of the composition of matter and how composition changes. In order to understand human physiological processes, it is important to understand
1. Amino Acids and Peptides Structures and Properties
 1. Amino Acids and Peptides Structures and Properties Chemical nature of amino acids The!-amino acids in peptides and proteins (excluding proline) consist of a carboxylic acid ( COOH) and an amino ( NH
1. Amino Acids and Peptides Structures and Properties Chemical nature of amino acids The!-amino acids in peptides and proteins (excluding proline) consist of a carboxylic acid ( COOH) and an amino ( NH
Supporting Information
 Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Dental Biochemistry EXAM I
 Dental Biochemistry EXAM I August 29, 2005 In the reaction below: CH 3 -CH 2 OH -~ ethanol CH 3 -CHO acetaldehyde A. acetoacetate is being produced B. ethanol is being oxidized to acetaldehyde C. acetaldehyde
Dental Biochemistry EXAM I August 29, 2005 In the reaction below: CH 3 -CH 2 OH -~ ethanol CH 3 -CHO acetaldehyde A. acetoacetate is being produced B. ethanol is being oxidized to acetaldehyde C. acetaldehyde
HIV protease inhibitor. Certain level of function can be found without structure. But a structure is a key to understand the detailed mechanism.
 Proteins are linear polypeptide chains (one or more) Building blocks: 20 types of amino acids. Range from a few 10s-1000s They fold into varying three-dimensional shapes structure medicine Certain level
Proteins are linear polypeptide chains (one or more) Building blocks: 20 types of amino acids. Range from a few 10s-1000s They fold into varying three-dimensional shapes structure medicine Certain level
Lecture 12. Metalloproteins - II
 Lecture 12 Metalloproteins - II Metalloenzymes Metalloproteins with one labile coordination site around the metal centre are known as metalloenzyme. As with all enzymes, the shape of the active site is
Lecture 12 Metalloproteins - II Metalloenzymes Metalloproteins with one labile coordination site around the metal centre are known as metalloenzyme. As with all enzymes, the shape of the active site is
Membrane Proteins: 1. Integral proteins: 2. Peripheral proteins: 3. Amphitropic proteins:
 Membrane Proteins: 1. Integral proteins: proteins that insert into/span the membrane bilayer; or covalently linked to membrane lipids. (Interact with the hydrophobic part of the membrane) 2. Peripheral
Membrane Proteins: 1. Integral proteins: proteins that insert into/span the membrane bilayer; or covalently linked to membrane lipids. (Interact with the hydrophobic part of the membrane) 2. Peripheral
ELECTRON PARAMAGNETIC RESONANCE
 ELECTRON PARAMAGNETIC RESONANCE = MAGNETIC RESONANCE TECHNIQUE FOR STUDYING PARAMAGNETIC SYSTEMS i.e. SYSTEMS WITH AT LEAST ONE UNPAIRED ELECTRON Examples of paramagnetic systems Transition-metal complexes
ELECTRON PARAMAGNETIC RESONANCE = MAGNETIC RESONANCE TECHNIQUE FOR STUDYING PARAMAGNETIC SYSTEMS i.e. SYSTEMS WITH AT LEAST ONE UNPAIRED ELECTRON Examples of paramagnetic systems Transition-metal complexes
Spectroscopy Chapter 13
 Spectroscopy Chapter 13 Electromagnetic Spectrum Electromagnetic spectrum in terms of wavelength, frequency and Energy c=λν c= speed of light in a vacuum 3x108 m/s v= frequency in Hertz (Hz s-1 ) λ= wavelength
Spectroscopy Chapter 13 Electromagnetic Spectrum Electromagnetic spectrum in terms of wavelength, frequency and Energy c=λν c= speed of light in a vacuum 3x108 m/s v= frequency in Hertz (Hz s-1 ) λ= wavelength
BIMS 503 Exam I. Sign Pledge Here: Questions from Robert Nakamoto (40 pts. Total)
 BIMS 503 Exam I September 24, 2007 _ /email: Sign Pledge Here: Questions from Robert Nakamoto (40 pts. Total) Questions 1-6 refer to this situation: You are able to partially purify an enzyme activity
BIMS 503 Exam I September 24, 2007 _ /email: Sign Pledge Here: Questions from Robert Nakamoto (40 pts. Total) Questions 1-6 refer to this situation: You are able to partially purify an enzyme activity
Chapter 15: Enyzmatic Catalysis
 Chapter 15: Enyzmatic Catalysis Voet & Voet: Pages 496-508 Slide 1 Catalytic Mechanisms Catalysis is a process that increases the rate at which a reaction approaches equilibrium Rate enhancement depends
Chapter 15: Enyzmatic Catalysis Voet & Voet: Pages 496-508 Slide 1 Catalytic Mechanisms Catalysis is a process that increases the rate at which a reaction approaches equilibrium Rate enhancement depends
Enzyme Catalysis & Biotechnology
 L28-1 Enzyme Catalysis & Biotechnology Bovine Pancreatic RNase A Biochemistry, Life, and all that L28-2 A brief word about biochemistry traditionally, chemical engineers used organic and inorganic chemistry
L28-1 Enzyme Catalysis & Biotechnology Bovine Pancreatic RNase A Biochemistry, Life, and all that L28-2 A brief word about biochemistry traditionally, chemical engineers used organic and inorganic chemistry
Bioinorganic Chemistry
 PRINCIPLES OF Bioinorganic Chemistry Stephen J. Lippard MASSACHUSETTS INSTITUTE OF TECHNOLOGY Jeremy M. Berg JOHNS HOPKINS SCHOOL OF MEDICINE f V University Science Books Mill Valley, California Preface
PRINCIPLES OF Bioinorganic Chemistry Stephen J. Lippard MASSACHUSETTS INSTITUTE OF TECHNOLOGY Jeremy M. Berg JOHNS HOPKINS SCHOOL OF MEDICINE f V University Science Books Mill Valley, California Preface
Protein structure. Protein structure. Amino acid residue. Cell communication channel. Bioinformatics Methods
 Cell communication channel Bioinformatics Methods Iosif Vaisman Email: ivaisman@gmu.edu SEQUENCE STRUCTURE DNA Sequence Protein Sequence Protein Structure Protein structure ATGAAATTTGGAAACTTCCTTCTCACTTATCAGCCACCT...
Cell communication channel Bioinformatics Methods Iosif Vaisman Email: ivaisman@gmu.edu SEQUENCE STRUCTURE DNA Sequence Protein Sequence Protein Structure Protein structure ATGAAATTTGGAAACTTCCTTCTCACTTATCAGCCACCT...
Keynotes in Organic Chemistry
 Keynotes in Organic Chemistry Second Edition ANDREW F. PARSONS Department of Chemistry, University of York, UK Wiley Contents Preface xi 1 Structure and bonding 1 1.1 Ionic versus covalent bonds 1 1.2
Keynotes in Organic Chemistry Second Edition ANDREW F. PARSONS Department of Chemistry, University of York, UK Wiley Contents Preface xi 1 Structure and bonding 1 1.1 Ionic versus covalent bonds 1 1.2
2: CHEMICAL COMPOSITION OF THE BODY
 1 2: CHEMICAL COMPOSITION OF THE BODY Although most students of human physiology have had at least some chemistry, this chapter serves very well as a review and as a glossary of chemical terms. In particular,
1 2: CHEMICAL COMPOSITION OF THE BODY Although most students of human physiology have had at least some chemistry, this chapter serves very well as a review and as a glossary of chemical terms. In particular,
BIOCHEMISTRY Course Outline (Fall, 2011)
 BIOCHEMISTRY 402 - Course Outline (Fall, 2011) Number OVERVIEW OF LECTURE TOPICS: of Lectures INSTRUCTOR 1. Structural Components of Proteins G. Brayer (a) Amino Acids and the Polypeptide Chain Backbone...2
BIOCHEMISTRY 402 - Course Outline (Fall, 2011) Number OVERVIEW OF LECTURE TOPICS: of Lectures INSTRUCTOR 1. Structural Components of Proteins G. Brayer (a) Amino Acids and the Polypeptide Chain Backbone...2
Dental Biochemistry Exam The total number of unique tripeptides that can be produced using all of the common 20 amino acids is
 Exam Questions for Dental Biochemistry Monday August 27, 2007 E.J. Miller 1. The compound shown below is CH 3 -CH 2 OH A. acetoacetate B. acetic acid C. acetaldehyde D. produced by reduction of acetaldehyde
Exam Questions for Dental Biochemistry Monday August 27, 2007 E.J. Miller 1. The compound shown below is CH 3 -CH 2 OH A. acetoacetate B. acetic acid C. acetaldehyde D. produced by reduction of acetaldehyde
From Amino Acids to Proteins - in 4 Easy Steps
 From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
Outline. The ensemble folding kinetics of protein G from an all-atom Monte Carlo simulation. Unfolded Folded. What is protein folding?
 The ensemble folding kinetics of protein G from an all-atom Monte Carlo simulation By Jun Shimada and Eugine Shaknovich Bill Hawse Dr. Bahar Elisa Sandvik and Mehrdad Safavian Outline Background on protein
The ensemble folding kinetics of protein G from an all-atom Monte Carlo simulation By Jun Shimada and Eugine Shaknovich Bill Hawse Dr. Bahar Elisa Sandvik and Mehrdad Safavian Outline Background on protein
Protein Structure Analysis and Verification. Course S Basics for Biosystems of the Cell exercise work. Maija Nevala, BIO, 67485U 16.1.
 Protein Structure Analysis and Verification Course S-114.2500 Basics for Biosystems of the Cell exercise work Maija Nevala, BIO, 67485U 16.1.2008 1. Preface When faced with an unknown protein, scientists
Protein Structure Analysis and Verification Course S-114.2500 Basics for Biosystems of the Cell exercise work Maija Nevala, BIO, 67485U 16.1.2008 1. Preface When faced with an unknown protein, scientists
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
BIOCHEMISTRY. František Vácha. JKU, Linz.
 BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
Enzyme function: the transition state. Enzymes & Kinetics V: Mechanisms. Catalytic Reactions. Margaret A. Daugherty A B. Lecture 16: Fall 2003
 Lecture 16: Enzymes & Kinetics V: Mechanisms Margaret A. Daugherty Fall 2003 Enzyme function: the transition state Catalytic Reactions A B Catalysts (e.g. enzymes) act by lowering the transition state
Lecture 16: Enzymes & Kinetics V: Mechanisms Margaret A. Daugherty Fall 2003 Enzyme function: the transition state Catalytic Reactions A B Catalysts (e.g. enzymes) act by lowering the transition state
Catalytic Reactions. Intermediate State in Catalysis. Lecture 16: Catalyzed reaction. Uncatalyzed reaction. Enzymes & Kinetics V: Mechanisms
 Enzyme function: the transition state Catalytic Reactions Lecture 16: Enzymes & Kinetics V: Mechanisms Margaret A. Daugherty Fall 2003 A B Catalysts (e.g. enzymes) act by lowering the transition state
Enzyme function: the transition state Catalytic Reactions Lecture 16: Enzymes & Kinetics V: Mechanisms Margaret A. Daugherty Fall 2003 A B Catalysts (e.g. enzymes) act by lowering the transition state
Protein Structure Marianne Øksnes Dalheim, PhD candidate Biopolymers, TBT4135, Autumn 2013
 Protein Structure Marianne Øksnes Dalheim, PhD candidate Biopolymers, TBT4135, Autumn 2013 The presentation is based on the presentation by Professor Alexander Dikiy, which is given in the course compedium:
Protein Structure Marianne Øksnes Dalheim, PhD candidate Biopolymers, TBT4135, Autumn 2013 The presentation is based on the presentation by Professor Alexander Dikiy, which is given in the course compedium:
4 Examples of enzymes
 Catalysis 1 4 Examples of enzymes Adding water to a substrate: Serine proteases. Carbonic anhydrase. Restrictions Endonuclease. Transfer of a Phosphoryl group from ATP to a nucleotide. Nucleoside monophosphate
Catalysis 1 4 Examples of enzymes Adding water to a substrate: Serine proteases. Carbonic anhydrase. Restrictions Endonuclease. Transfer of a Phosphoryl group from ATP to a nucleotide. Nucleoside monophosphate
Magnetic Resonance Spectroscopy EPR and NMR
 Magnetic Resonance Spectroscopy EPR and NMR A brief review of the relevant bits of quantum mechanics 1. Electrons have spin, - rotation of the charge about its axis generates a magnetic field at each electron.
Magnetic Resonance Spectroscopy EPR and NMR A brief review of the relevant bits of quantum mechanics 1. Electrons have spin, - rotation of the charge about its axis generates a magnetic field at each electron.
Organic Chemistry 112 A B C - Syllabus Addendum for Prospective Teachers
 Chapter Organic Chemistry 112 A B C - Syllabus Addendum for Prospective Teachers Ch 1-Structure and bonding Ch 2-Polar covalent bonds: Acids and bases McMurry, J. (2004) Organic Chemistry 6 th Edition
Chapter Organic Chemistry 112 A B C - Syllabus Addendum for Prospective Teachers Ch 1-Structure and bonding Ch 2-Polar covalent bonds: Acids and bases McMurry, J. (2004) Organic Chemistry 6 th Edition
1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI )
 Uses of NMR: 1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI ) 3) NMR is used as a method for determining of protein, DNA,
Uses of NMR: 1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI ) 3) NMR is used as a method for determining of protein, DNA,
Model Mélange. Physical Models of Peptides and Proteins
 Model Mélange Physical Models of Peptides and Proteins In the Model Mélange activity, you will visit four different stations each featuring a variety of different physical models of peptides or proteins.
Model Mélange Physical Models of Peptides and Proteins In the Model Mélange activity, you will visit four different stations each featuring a variety of different physical models of peptides or proteins.
Research Science Biology The study of living organisms (Study of life)
 Scientific method Why is there a hypothesis and prediction? If only prediction: then there is no way to finish the prediction and conclude whether the results support the hypothesis If surfaces are sampled
Scientific method Why is there a hypothesis and prediction? If only prediction: then there is no way to finish the prediction and conclude whether the results support the hypothesis If surfaces are sampled
Circular Dichroism & Optical Rotatory Dispersion. Proteins (KCsa) Polysaccharides (agarose) DNA CHEM 305. Many biomolecules are α-helical!
 Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
The Structure and Functions of Proteins
 Wright State University CORE Scholar Computer Science and Engineering Faculty Publications Computer Science and Engineering 2003 The Structure and Functions of Proteins Dan E. Krane Wright State University
Wright State University CORE Scholar Computer Science and Engineering Faculty Publications Computer Science and Engineering 2003 The Structure and Functions of Proteins Dan E. Krane Wright State University
THE UNIVERSITY OF MANITOBA. PAPER NO: _1_ LOCATION: 173 Robert Schultz Theatre PAGE NO: 1 of 5 DEPARTMENT & COURSE NO: CHEM / MBIO 2770 TIME: 1 HOUR
 THE UNIVERSITY OF MANITOBA 1 November 1, 2016 Mid-Term EXAMINATION PAPER NO: _1_ LOCATION: 173 Robert Schultz Theatre PAGE NO: 1 of 5 DEPARTMENT & COURSE NO: CHEM / MBIO 2770 TIME: 1 HOUR EXAMINATION:
THE UNIVERSITY OF MANITOBA 1 November 1, 2016 Mid-Term EXAMINATION PAPER NO: _1_ LOCATION: 173 Robert Schultz Theatre PAGE NO: 1 of 5 DEPARTMENT & COURSE NO: CHEM / MBIO 2770 TIME: 1 HOUR EXAMINATION:
Nanobiotechnology. Place: IOP 1 st Meeting Room Time: 9:30-12:00. Reference: Review Papers. Grade: 40% midterm, 60% final report (oral + written)
 Nanobiotechnology Place: IOP 1 st Meeting Room Time: 9:30-12:00 Reference: Review Papers Grade: 40% midterm, 60% final report (oral + written) Midterm: 5/18 Oral Presentation 1. 20 minutes each person
Nanobiotechnology Place: IOP 1 st Meeting Room Time: 9:30-12:00 Reference: Review Papers Grade: 40% midterm, 60% final report (oral + written) Midterm: 5/18 Oral Presentation 1. 20 minutes each person
Protein Folding experiments and theory
 Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
1/23/2012. Atoms. Atoms Atoms - Electron Shells. Chapter 2 Outline. Planetary Models of Elements Chemical Bonds
 Chapter 2 Outline Atoms Chemical Bonds Acids, Bases and the p Scale Organic Molecules Carbohydrates Lipids Proteins Nucleic Acids Are smallest units of the chemical elements Composed of protons, neutrons
Chapter 2 Outline Atoms Chemical Bonds Acids, Bases and the p Scale Organic Molecules Carbohydrates Lipids Proteins Nucleic Acids Are smallest units of the chemical elements Composed of protons, neutrons
Biomolecules: lecture 9
 Biomolecules: lecture 9 - understanding further why amino acids are the building block for proteins - understanding the chemical properties amino acids bring to proteins - realizing that many proteins
Biomolecules: lecture 9 - understanding further why amino acids are the building block for proteins - understanding the chemical properties amino acids bring to proteins - realizing that many proteins
Chemistry. for the life and medical sciences. Mitch Fry and Elizabeth Page. second edition
 hemistry for the life and medical sciences Mitch Fry and Elizabeth Page second edition ontents Preface to the second edition Preface to the first edition about the authors ix x xi 1 elements, atoms and
hemistry for the life and medical sciences Mitch Fry and Elizabeth Page second edition ontents Preface to the second edition Preface to the first edition about the authors ix x xi 1 elements, atoms and
Protein Structure. Role of (bio)informatics in drug discovery. Bioinformatics
 Bioinformatics Protein Structure Principles & Architecture Marjolein Thunnissen Dep. of Biochemistry & Structural Biology Lund University September 2011 Homology, pattern and 3D structure searches need
Bioinformatics Protein Structure Principles & Architecture Marjolein Thunnissen Dep. of Biochemistry & Structural Biology Lund University September 2011 Homology, pattern and 3D structure searches need
5) An amino acid that doesn't exist in proteins: - Tyrosine - Tryptophan - Cysteine - ornithine* 6) How many tripeptides can be formed from using one
 Biochemistry First 1) An amino acid that does not have L & D: - proline - serine - Glycine* - valine 2) An amino acid that exists in beta-bends and turns: - Gly* - Pro - Arg - Asp 3) An amino acid that
Biochemistry First 1) An amino acid that does not have L & D: - proline - serine - Glycine* - valine 2) An amino acid that exists in beta-bends and turns: - Gly* - Pro - Arg - Asp 3) An amino acid that
Advanced Certificate in Principles in Protein Structure. You will be given a start time with your exam instructions
 BIRKBECK COLLEGE (University of London) Advanced Certificate in Principles in Protein Structure MSc Structural Molecular Biology Date: Thursday, 1st September 2011 Time: 3 hours You will be given a start
BIRKBECK COLLEGE (University of London) Advanced Certificate in Principles in Protein Structure MSc Structural Molecular Biology Date: Thursday, 1st September 2011 Time: 3 hours You will be given a start
2. In regards to the fluid mosaic model, which of the following is TRUE?
 General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
Circular dichroism spectroscopic studies on structures formed by telomeric DNA sequences in vitro
 Circular dichroism spectroscopic studies on structures formed by telomeric DNA sequences in vitro ZHANG Xiaoyan 1, CAO Enhua 1, SUN Xueguang 1 & BAI Chunli 2 1. Institute of Biophysics, Chinese Academy
Circular dichroism spectroscopic studies on structures formed by telomeric DNA sequences in vitro ZHANG Xiaoyan 1, CAO Enhua 1, SUN Xueguang 1 & BAI Chunli 2 1. Institute of Biophysics, Chinese Academy
BIRKBECK COLLEGE (University of London)
 BIRKBECK COLLEGE (University of London) SCHOOL OF BIOLOGICAL SCIENCES M.Sc. EXAMINATION FOR INTERNAL STUDENTS ON: Postgraduate Certificate in Principles of Protein Structure MSc Structural Molecular Biology
BIRKBECK COLLEGE (University of London) SCHOOL OF BIOLOGICAL SCIENCES M.Sc. EXAMINATION FOR INTERNAL STUDENTS ON: Postgraduate Certificate in Principles of Protein Structure MSc Structural Molecular Biology
Presenter: She Zhang
 Presenter: She Zhang Introduction Dr. David Baker Introduction Why design proteins de novo? It is not clear how non-covalent interactions favor one specific native structure over many other non-native
Presenter: She Zhang Introduction Dr. David Baker Introduction Why design proteins de novo? It is not clear how non-covalent interactions favor one specific native structure over many other non-native
Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination
 Lecture 9 M230 Feigon Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination Reading resources v Roberts NMR of Macromolecules, Chap 4 by Christina Redfield
Lecture 9 M230 Feigon Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination Reading resources v Roberts NMR of Macromolecules, Chap 4 by Christina Redfield
CHEMISTRY (CHEM) CHEM 208. Introduction to Chemical Analysis II - SL
 Chemistry (CHEM) 1 CHEMISTRY (CHEM) CHEM 100. Elements of General Chemistry Prerequisite(s): Completion of general education requirement in mathematics recommended. Description: The basic concepts of general
Chemistry (CHEM) 1 CHEMISTRY (CHEM) CHEM 100. Elements of General Chemistry Prerequisite(s): Completion of general education requirement in mathematics recommended. Description: The basic concepts of general
Conformational Geometry of Peptides and Proteins:
 Conformational Geometry of Peptides and Proteins: Before discussing secondary structure, it is important to appreciate the conformational plasticity of proteins. Each residue in a polypeptide has three
Conformational Geometry of Peptides and Proteins: Before discussing secondary structure, it is important to appreciate the conformational plasticity of proteins. Each residue in a polypeptide has three
Protein folding. Today s Outline
 Protein folding Today s Outline Review of previous sessions Thermodynamics of folding and unfolding Determinants of folding Techniques for measuring folding The folding process The folding problem: Prediction
Protein folding Today s Outline Review of previous sessions Thermodynamics of folding and unfolding Determinants of folding Techniques for measuring folding The folding process The folding problem: Prediction
NAME. EXAM I I. / 36 September 25, 2000 Biochemistry I II. / 26 BICH421/621 III. / 38 TOTAL /100
 EXAM I I. / 6 September 25, 2000 Biochemistry I II. / 26 BIH421/621 III. / 8 TOTAL /100 I. MULTIPLE HOIE (6 points) hoose the BEST answer to the question by circling the appropriate letter. 1. An amino
EXAM I I. / 6 September 25, 2000 Biochemistry I II. / 26 BIH421/621 III. / 8 TOTAL /100 I. MULTIPLE HOIE (6 points) hoose the BEST answer to the question by circling the appropriate letter. 1. An amino
Translation. A ribosome, mrna, and trna.
 Translation The basic processes of translation are conserved among prokaryotes and eukaryotes. Prokaryotic Translation A ribosome, mrna, and trna. In the initiation of translation in prokaryotes, the Shine-Dalgarno
Translation The basic processes of translation are conserved among prokaryotes and eukaryotes. Prokaryotic Translation A ribosome, mrna, and trna. In the initiation of translation in prokaryotes, the Shine-Dalgarno
NMR Spectroscopy of Polymers
 UNESCO/IUPAC Course 2005/2006 Jiri Brus NMR Spectroscopy of Polymers Brus J 1. part At the very beginning the phenomenon of nuclear spin resonance was studied predominantly by physicists and the application
UNESCO/IUPAC Course 2005/2006 Jiri Brus NMR Spectroscopy of Polymers Brus J 1. part At the very beginning the phenomenon of nuclear spin resonance was studied predominantly by physicists and the application
Details of Protein Structure
 Details of Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Anne Mølgaard, Kemisk Institut, Københavns Universitet Learning Objectives
Details of Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Anne Mølgaard, Kemisk Institut, Københavns Universitet Learning Objectives
CONFOCHECK. Innovation with Integrity. Infrared Protein Analysis FT-IR
 CONFOCHECK Infrared Protein Analysis Innovation with Integrity FT-IR CONFOCHECK: FT-IR System for Protein Analytics FT-IR Protein Analysis Infrared spectroscopy measures molecular vibrations due to the
CONFOCHECK Infrared Protein Analysis Innovation with Integrity FT-IR CONFOCHECK: FT-IR System for Protein Analytics FT-IR Protein Analysis Infrared spectroscopy measures molecular vibrations due to the
Biochemistry 3100 Sample Problems Binding proteins, Kinetics & Catalysis
 (1) Draw an approximate denaturation curve for a typical blood protein (eg myoglobin) as a function of ph. (2) Myoglobin is a simple, single subunit binding protein that has an oxygen storage function
(1) Draw an approximate denaturation curve for a typical blood protein (eg myoglobin) as a function of ph. (2) Myoglobin is a simple, single subunit binding protein that has an oxygen storage function
Chemical Properties of Amino Acids
 hemical Properties of Amino Acids Protein Function Make up about 15% of the cell and have many functions in the cell 1. atalysis: enzymes 2. Structure: muscle proteins 3. Movement: myosin, actin 4. Defense:
hemical Properties of Amino Acids Protein Function Make up about 15% of the cell and have many functions in the cell 1. atalysis: enzymes 2. Structure: muscle proteins 3. Movement: myosin, actin 4. Defense:
CHEMISTRY (CHEM) CHEM 5800 Principles Of Materials Chemistry. Tutorial in selected topics in materials chemistry. S/U grading only.
 Chemistry (CHEM) 1 CHEMISTRY (CHEM) CHEM 5100 Principles of Organic and Inorganic Chemistry Study of coordination compounds with a focus on ligand bonding, electron counting, molecular orbital theory,
Chemistry (CHEM) 1 CHEMISTRY (CHEM) CHEM 5100 Principles of Organic and Inorganic Chemistry Study of coordination compounds with a focus on ligand bonding, electron counting, molecular orbital theory,
Biochemistry Quiz Review 1I. 1. Of the 20 standard amino acids, only is not optically active. The reason is that its side chain.
 Biochemistry Quiz Review 1I A general note: Short answer questions are just that, short. Writing a paragraph filled with every term you can remember from class won t improve your answer just answer clearly,
Biochemistry Quiz Review 1I A general note: Short answer questions are just that, short. Writing a paragraph filled with every term you can remember from class won t improve your answer just answer clearly,
It s the amino acids!
 Catalytic Mechanisms HOW do enzymes do their job? Reducing activation energy sure, but HOW does an enzyme catalysis reduce the energy barrier ΔG? Remember: The rate of a chemical reaction of substrate
Catalytic Mechanisms HOW do enzymes do their job? Reducing activation energy sure, but HOW does an enzyme catalysis reduce the energy barrier ΔG? Remember: The rate of a chemical reaction of substrate
BBS501 Section 1 9:00 am 10:00 am Monday thru Friday LRC 105 A & B
 BBS501 Section 1 9:00 am 10:00 am Monday thru Friday LRC 105 A & B Lecturers: Dr. Yie-Hwa Chang Room M130 Phone: #79263 E-mail:changy@slu.edu Dr. Tomasz Heyduk Room M99 Phone: #79238 E-mail: heydukt@slu.edu
BBS501 Section 1 9:00 am 10:00 am Monday thru Friday LRC 105 A & B Lecturers: Dr. Yie-Hwa Chang Room M130 Phone: #79263 E-mail:changy@slu.edu Dr. Tomasz Heyduk Room M99 Phone: #79238 E-mail: heydukt@slu.edu
Molecular Basis of K + Conduction and Selectivity
 The Structure of the Potassium Channel: Molecular Basis of K + Conduction and Selectivity -Doyle, DA, et al. The structure of the potassium channel: molecular basis of K + conduction and selectivity. Science
The Structure of the Potassium Channel: Molecular Basis of K + Conduction and Selectivity -Doyle, DA, et al. The structure of the potassium channel: molecular basis of K + conduction and selectivity. Science
REDOX ACTIVE TYROSINE RESIDUES IN BIOMIMETIC BETA HAIRPINS
 REDOX ACTIVE TYROSINE RESIDUES IN BIOMIMETIC BETA HAIRPINS A Dissertation Presented to The Academic Faculty by Robin S. Sibert In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy
REDOX ACTIVE TYROSINE RESIDUES IN BIOMIMETIC BETA HAIRPINS A Dissertation Presented to The Academic Faculty by Robin S. Sibert In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy
e 2m e c I, (7.1) = g e β B I(I +1), (7.2) = erg/gauss. (7.3)
 Chemistry 126 Molecular Spectra & Molecular Structure Week # 7 Electron Spin Resonance Spectroscopy, Supplement Like the hydrogen nucleus, an unpaired electron in a sample has a spin of I=1/2. The magnetic
Chemistry 126 Molecular Spectra & Molecular Structure Week # 7 Electron Spin Resonance Spectroscopy, Supplement Like the hydrogen nucleus, an unpaired electron in a sample has a spin of I=1/2. The magnetic
JEFFERSON COLLEGE COURSE SYLLABUS CHM201 ORGANIC CHEMISTRY II. 5 Credit Hours. Prepared by: Richard A. Pierce
 JEFFERSON COLLEGE COURSE SYLLABUS CHM201 ORGANIC CHEMISTRY II 5 Credit Hours Prepared by: Richard A. Pierce Revised Date: January 2008 by Ryan H. Groeneman Arts & Science Education Dr. Mindy Selsor, Dean
JEFFERSON COLLEGE COURSE SYLLABUS CHM201 ORGANIC CHEMISTRY II 5 Credit Hours Prepared by: Richard A. Pierce Revised Date: January 2008 by Ryan H. Groeneman Arts & Science Education Dr. Mindy Selsor, Dean
Read more about Pauling and more scientists at: Profiles in Science, The National Library of Medicine, profiles.nlm.nih.gov
 2018 Biochemistry 110 California Institute of Technology Lecture 2: Principles of Protein Structure Linus Pauling (1901-1994) began his studies at Caltech in 1922 and was directed by Arthur Amos oyes to
2018 Biochemistry 110 California Institute of Technology Lecture 2: Principles of Protein Structure Linus Pauling (1901-1994) began his studies at Caltech in 1922 and was directed by Arthur Amos oyes to
Peptides And Proteins
 Kevin Burgess, May 3, 2017 1 Peptides And Proteins from chapter(s) in the recommended text A. Introduction B. omenclature And Conventions by amide bonds. on the left, right. 2 -terminal C-terminal triglycine
Kevin Burgess, May 3, 2017 1 Peptides And Proteins from chapter(s) in the recommended text A. Introduction B. omenclature And Conventions by amide bonds. on the left, right. 2 -terminal C-terminal triglycine
Matter and Substances Section 3-1
 Matter and Substances Section 3-1 Key Idea: All matter is made up of atoms. An atom has a positively charges core surrounded by a negatively charged region. An atom is the smallest unit of matter that
Matter and Substances Section 3-1 Key Idea: All matter is made up of atoms. An atom has a positively charges core surrounded by a negatively charged region. An atom is the smallest unit of matter that
Major Types of Association of Proteins with Cell Membranes. From Alberts et al
 Major Types of Association of Proteins with Cell Membranes From Alberts et al Proteins Are Polymers of Amino Acids Peptide Bond Formation Amino Acid central carbon atom to which are attached amino group
Major Types of Association of Proteins with Cell Membranes From Alberts et al Proteins Are Polymers of Amino Acids Peptide Bond Formation Amino Acid central carbon atom to which are attached amino group
Examples of Protein Modeling. Protein Modeling. Primary Structure. Protein Structure Description. Protein Sequence Sources. Importing Sequences to MOE
 Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
