A macroecological theory of microbial biodiversity. Correspondence: K Locey, Department of Biology, Indiana University, 261 Jordan Hall, 1001
|
|
|
- Mervyn Chambers
- 5 years ago
- Views:
Transcription
1 A macroecological theory of microbial biodiversity 2 William R. Shoemaker 1*, Kenneth J. Locey 1*, Jay T. Lennon Department of Biology, Indiana University, Bloomington, IN USA * Authors contributed equally to the study 6 Correspondence: K Locey, Department of Biology, Indiana University, 261 Jordan Hall, 1001 East 3rd Street, Bloomington, IN USA. kjlocey@indiana.edu
2 24 Microorganisms are the most abundant, diverse, and functionally important organisms on Earth. Over the past decade, microbial ecologists have produced the largest ever 26 community datasets. However, these data are rarely used to uncover law-like patterns of commonness and rarity, test theories of biodiversity, or explore unifying explanations for 28 the structure of microbial communities. Using a global-scale compilation of >20,000 samples from environmental, engineered, and host-related ecosystems, we test the power of 30 competing theories to predict distributions of microbial abundance and diversity- abundance scaling laws. We show that these patterns are best explained by the synergistic 32 interaction of stochastic processes that are captured by lognormal dynamics. We demonstrate that lognormal dynamics have predictive power across scales of abundance, a 34 criterion that is essential to biodiversity theory. By understanding the multiplicative and stochastic nature of ecological processes, scientists can better understand the structure and 36 dynamics of Earth s largest and most diverse ecological systems. 38 A central goal of ecology is to explain and predict patterns of biodiversity across evolutionarily distant taxa and scales abundance 1-4. Over the past century, this endeavor has 40 focused almost exclusively on macroscopic plants and animals (i.e., macroorganisms), giving little attention to the most abundant and taxonomically, functionally, and metabolically diverse 42 organisms on Earth, i.e., microorganisms 1-4. However, global-scale efforts to catalog microbial diversity across environmental, engineered, and host-related ecosystems has created an 44 opportunity to understand biodiversity using a scale of data that far surpasses the largest macrobial datasets 5. While commonness and rarity in microbial systems has become 46 increasingly studied over the past decade, such patterns are rarely investigated in the context of unified relationships that are predictable under general principles of biodiversity. 2
3 48 One of the most frequently documented patterns of microbial diversity in recent years is the rare biosphere, which describes how the majority of taxa in an environmental sample are 50 represented by few gene sequences 6, 7. While the rare biosphere has become a primary pattern of microbial ecology 6-8, it also reflects the universally uneven nature of one of ecology s 52 fundamental patterns, i.e., the species abundance distribution (SAD) 9. The SAD is among the most intensively studied patterns of commonness and rarity, and is central to biodiversity theory 54 and the study of patterns in abundance, distribution, and diversity across scales of space and time (i.e., macroecology) 9. However, microbiologists have largely overlooked the connection of the 56 SAD to theories of biodiversity and macroecology and the ability for some of those theories to predict other intensively studied patterns such as the species-area curve or distance-decay 58 relationship 10. Since the 1930 s, ecologists have developed more than 20 models that predict the SAD While some of these models are purely statistical and only predict the shape of the SAD (e.g., Gamma, Inverse Gamma), others encode the principles and mechanisms of competing theories , 9. Of all existing SAD models, none have been more successful than the distributions known as the lognormal and log-series, which often serve as standards against which other models are 64 tested 2. The lognormal is characterized by a right-skewed frequency distribution that becomes approximately normal under log-transformation; hence the name lognormal. Historically, the 66 lognormal is said to emerge from the multiplicative interactions of stochastic processes 11. Examples of these lognormal dynamics are the multiplicative nature of growth and the 68 stochastic nature of population dynamics. Another example is the stochastic nature of individual dispersal and the energetic costs that are multiplied across geographic distance. While most 70 ecological processes likely have multiplicative interactions 11, many theories of biodiversity 3
4 (e.g., neutral theory, stochastic geometry, stochastic resource limitation theory) include a 72 stochastic component 2, Lognormal dynamics should become increasingly important for large communities, a result of the central limit theorem and law of large numbers 11. Yet despite 74 being one of the most successful models of the SAD among communities of macroorganisms, the lognormal does not seem to be predicted by any general theory of biodiversity and is only 76 rarely used in microbial studies Like the lognormal, the log-series has also been successful in predicting the SAD Though commonly used since the 1940 s, the log-series is the form of the SAD that is predicted by one of the most recent, successful, and unified theories of biodiversity, i.e., the maximum 80 entropy theory of ecology (METE) 4. In ecological terms, METE states that the expected form of an ecological pattern is that which can occur in the greatest number of ways for a given set of 82 constraints, i.e., the principle of maximum entropy 4, 20. METE uses only the number of species (S) and total number of individuals (N) as its empirical inputs to predict the SAD. Using the most 84 comprehensive global-scale data compilations of macroscopic plants and animals, METE outperformed the lognormal and often explained > 90% of variation in abundance within and 86 among communities 21, 22. The success of METE has made the log-series the most highly supported model of the SAD 4. But despite its success, METE has not been tested with microbial 88 data and it is unknown whether METE can predict microbial SADs, a crucial requirement for a macroecological theory of biodiversity The lognormal, log-series, and other models of biodiversity have competed to predict the SAD for several decades. However, few studies have gone beyond the SAD to test multiple 92 models using several patterns of commonness and rarity. For example, recently discovered relationships show how aspects of commonness and rarity scale across as many as 30 orders of 4
5 94 magnitude, from the smallest sampling scales of molecular surveys to the scale of all organisms on Earth 5. Such scaling laws are among the most powerful relationships in biology, revealing 96 how one variable (e.g., S) changes in a proportional way across orders of magnitude in another variable (e.g., N). However, the mechanisms that give rise to these scaling laws were not 98 reported and it remains to be seen whether any biodiversity theory can predict and unify them. It also remains to be seen whether the model that best predicts the SAD would also best explain 100 how aspects of commonness and rarity scale with N. In this study we ask whether the lognormal and log-series can reasonably predict 102 microbial SADs and whether either model can reproduce recently discovered diversity- abundance scaling relationships 5. We used a compilation of 16S ribosomal RNA (rrna) 104 community-level surveys from over 20,000 unique locations, ranging from glaciers to 106 hydrothermal vents to hospital rooms. We contextualize the results of the lognormal and the logseries against two other well-known SAD models; one that predicts a highly uneven form, i.e., the Zipf distribution, and one that predicts a highly even form, i.e., the Broken-stick. Because 108 general theories of biodiversity should make accurate predictions regardless of the size of a 110 sample, community, or microbiome, we tested whether the performance of these four longstanding models are influenced by a primary constraint on the form of the SAD, i.e., sample abundance (N). We discuss our findings in the context of greater unification across domains of 112 life, paradigms of biodiversity theory, and in the context of how lognormal dynamics may underpin microbial ecological processes RESULTS Predicting distributions of microbial abundance 5
6 118 The lognormal explained nearly 94% of the variation within and among microbial SADs, compared to 91% for the Zipf distribution and 64% for log-series predicted by METE (Fig. 2 and 120 Table 1). The performance of the Simultaneous Broken-stick (hereafter referred to as the Broken-stick) was too poor to be evaluated. While close to the predictive power of the 122 lognormal, the Zipf distribution greatly over-predicted the abundance of the most abundant taxa (N max ). In some cases, the predicted N max was greater than the empirical value for sample 124 abundance (N). The Zipf distribution was also sensitive to the exclusion of singleton OTUs and percent cutoff in sequence similarity (Table S3 Fig. S3). In this way, the Zipf reasonably predicts 126 the abundance of intermediately abundant taxa, but often fails for the most dominant and rare taxa 22, 24 (Tables S1 and S2). In contrast to the other models, the lognormal produced unbiased 128 predictions for the abundances of dominant and rare taxa, regardless of cutoffs in percent similarity and the exclusion of singleton OTUs (Figs. S1, S2; Tables S1, S2). 130 Predictive power across scales of sample abundance (N) 132 The performance of SAD models across scales of N is rarely, if ever, examined. While the log- series has been successful among communities of macroscopic plants and animals 21, 22, N for the 134 vast majority of these samples was less than a few thousand organisms 21, 22. In contrast, the log- series predicted by METE has yet to be tested using microbial data, i.e., where N often represents 136 millions of sampled 16S rrna gene reads. We found that the lognormal performed well across all orders of magnitude in N with no 138 indication of weakening at higher orders of magnitude. The performance of METE s log-series, however, was much more variable and often provided fits to microbial SADs that were too poor 140 to interpret. As a result, the form of the SAD predicted by the most successful theory of 6
7 biodiversity for macroorganisms (i.e., METE), failed across orders of magnitude in microbial N. 142 This was the case for SADs from different systems and within SADs that were resampled to smaller N (Fig. 3, Fig. S3). While the Zipf distribution also provided reasonable fits that 144 improved with increasing N, the Broken Stick increasingly failed for greater N. This latter result supports previously documented patterns of decreasing species evenness with increasing N 5,25 ; a 146 trend that the lognormal captures without apparent bias. 148 Diversity-Abundance Scaling Laws Recently, aspects of taxonomic diversity have been shown to scale with N at rates that 150 were similar for molecular surveys of microorganisms and individual counts of macroorganisms 5. These aspects of diversity include dominance (i.e., the abundance of the most abundant OTU; 152 N max ), evenness (i.e., similarity in abundance among OTUs), and rarity (i.e., concentration of taxa at low abundances). We found that the lognormal best reproduced these diversity-abundance 154 scaling relationships 5 (Table 2 and Fig. 4). While the Zipf approximated the rate at which N max scaled with N, it greatly over-predicted the y-intercept and hence, the actual value of N max (Fig ). Additionally, neither the log-series predicted by METE nor the Broken-stick came close to reproducing the observed diversity-abundance scaling relationships (Fig. 4, Table 2). 158 DISCUSSION 160 In this study, we asked whether widely known and successful models of biodiversity could predict microbial SADs and also unify SADs with recently discovered diversity-abundance 162 scaling laws. We found that the lognormal provided the most accurate predictions for nearly all patterns in our study. This is in sharp contrast to studies of macroorganisms where the log-series 7
8 164 distribution predicted by the maximum entropy theory of ecology (METE) was overwhelmingly supported 21, 22. Such discrepancies in model performance suggest there are fundamental 166 differences between macroorganisms and microorganisms that point to the importance of lognormal dynamics. Specifically, that multiplicative processes (e.g., growth) and stochastic 168 outcomes (i.e., population fluctuations) produce a central limiting pattern within large and heterogeneous communities where species partition multiple resources 11. Instead of identifying a 170 particular process (e.g., dispersal limitation, resource competition), we propose that lognormal dynamics underpin the fundamental nature of microbial communities 11, There are fundamental differences in how ecologists study communities of microscopic and macroscopic organisms. In our study, we accounted for some of the artifacts that could 174 potentially contribute to the highly uneven microbial SADs. For example, we tested for the effects of percent similarity cutoffs that are used for defining an OTU, along with the influence 176 of singletons and sample size. However, there are other caveats that deserve attention. First, ecologists sample microbial communities at spatial scales that greatly exceed the scales of their 178 interactions 26. As a result, samples of microbial communities probably lump together many ecologically distinct taxa that do not partition the same resources or occupy the same 180 microhabitats. If microbial studies commonly lump together species that belong to different ecological communities, then this may in fact lead to the emergence of a power-law SAD (e.g., 182 the Zipf) 27. We expect that the increasing performance of the Zipf with greater N, is evidence of a power-law SAD arising from the mixture of lognormal microbial communities. While the 184 connection between the lognormal and the Zipf needs further study, a macroecological theory of microbial biodiversity should allow for this dynamic. 8
9 186 Finally, in rejecting the log-series as a model for microbial SADs, we are not rejecting METE altogether. We are instead rejecting the log-series as METE s primary form of the SAD In fact, METE appears capable of predicting both the lognormal and the Zipf 40. This is because in using METE, one tries to infer the most likely form of an ecological pattern for a particular set 190 of variables (e.g., N, S) and constraints (e.g., N/S). Consequently, the forms of ecological patterns predicted by METE could change depending on the constraints and state variables used 40. For 192 example, METE predicts that the SAD is a power law if it constrains the SAD to N/S while including a resource variable 40. However, METE has not been as fully developed to predict 194 forms of the SAD other than the log-series and it remains to be seen whether METE can predict the form of the lognormal (i.e., Poisson lognormal) used in our study. If so, and if it can 196 reconcile why a log-series SAD works best for macrobes and a lognormal works best for microbes, then METE may indeed be a unified theory of biodiversity. Until then, microbial 198 communities and microbiomes appear to be shaped by the multiplicative interactions of stochastic processes that, while highly complex, inevitably lead to predictable patterns of 200 biodiversity Data METHODS We used one of the largest compilations of microbial community and microbiome data to date, consisting of bacterial and archaeal community sequence data over 20,000 unique 208 geographic sites. These data were compiled in a previous study 5 and include 14,962 sites from the Earth Microbiome Project (EMP) 28, 4,303 sites from the Data Analysis and Coordination 9
10 210 Center (DACC) for the National Institutes of Health (NIH) Common Fund supported Human Microbiome Project (HMP) 29, as well as 1,319 non-experimental sequencing projects consisting 212 of processed 16S rrna amplicon reads from the Argonne National Laboratory metagenomics server MG-RAST 30. All sequence data were previously processed using established pipelines to 214 remove low quality sequence reads and chimeras Additional information pertaining to the datasets can be found in the supplement and in previous studies Description of SAD models 218 In this study we ask whether the lognormal, log-series, and two other classic SAD models that have some success in microbial ecology, i.e., the Simultaneous broken-stick 12 and the Zipf 220 distribution 31, 32 can reasonably predict microbial SADs (Fig. 1). We evaluated the performance of each model with and without singletons and across different percent cutoffs for sequence 222 similarity used to cluster 16S rrna reads into operational taxonomic units (OTUs). 224 Lognormal To avoid fractional abundances and to account for sampling error, we used a Poisson-based sampling model of the lognormal, i.e., the Poisson lognormal 33. We used the 226 maximum likelihood estimate of the Poisson lognormal as our species abundance model of lognormal dynamics. The likelihood estimate of the single composite parameter λ (composed of 228 the mean (μ) and standard deviation (σ)) of the Poisson lognormal is derived via numerical maximization of the likelihood surface 33. Once λ is found, the probability mass function for the 230 Poisson lognormal (hereafter lognormal) is derived using: p n =!! λ! e!! n p!"(!)!" where p!" is the lognormal probability. 10
11 232 METE The maximum entropy theory of ecology (METE) uses only two empirical inputs to 234 predict the SAD: species richness (S) and total abundance (N) of individuals (or sequence reads) in a sample. To predict the SAD, METE assumes that the expected shape of the SAD is that 236 which can occur in the highest number of ways, an assumption based on the principle of maximum entropy (MaxEnt) 20. Using METE, the shape of the SAD was predicted by calculating 238 the probability that the abundance of a species is n given S and N: Φ(n S, N) = 1 e!!" log(β!! ) n where the single fitted parameter β is defined by the equation! N S = e!!"!!!! e!!" /n!!! 240 Where N/S is the average abundance of species. This approach to predicting the MaxEnt form of the SAD yields the log-series distribution 4, Broken-stick The Broken-stick model predicts a high similarity in abundance among species 244 and hence, predicts one of the most even SADs of any model. The Broken-stick model predicts the SAD as the simultaneous breaking of a stick of length N at S - 1 randomly chosen points The Broken-stick has also has a purely statistical equivalent, i.e., the geometric distribution 34, 35 : f(k) = (1 p)!!! p The Broken-stick has no free parameters and predicts only one form of the SAD for a given 248 combination of N and S. Though rarely recognized, the geometric distribution is a maximum entropy solution when using N and S as hard constraints, i.e., the predicted SAD must have S 250 species and a sum of N individuals. 11
12 252 Zipf distribution The Zipf (i.e., the discrete Pareto distribution) distribution is a power-law model that predicts one of the most uneven forms of the SAD. This distribution is based on a 254 power-law of frequency of ranked data and is characterized by one parameter (γ), where the frequency of the k th rank is inversely proportional to k, i.e., p(k) k!, with γ often ranging 256 between -1 and -2 31, The Zipf distribution predicts the frequency of elements of rank k out of N elements with parameter γ as: f(k; γ, N) =! 1/k!!!! (1/n! ) 258 We calculated the maximum likelihood estimate of γ using numerical maximization, which was then used to generate the predicted form of the SAD. 260 Testing SAD predictions 262 Our SAD predictions were based on the rank-abundance form of the SAD, i.e., a vector of species abundances ranked from most to least abundant (Fig. 1). Because the predicted form 264 of each model preserves S (i.e., number of species), we were able to directly compare (rank-for- rank) the observed and predicted SADs using regression to find the percent of variation in 266 abundance among species that is explained by each model. We generated the predicted forms of the SAD using previously developed code 21 ( ecology) and the public repository macroecotools ( To prevent bias in our results due to the overrepresentation of a particular dataset, we 270 performed 10,000 bootstrap iterations using a sample size of 200 SADs drawn randomly from each dataset. The sample size was determined based on the number of SADs that the numerical 272 estimator used to generate the Zipf distribution was able to solve for the smallest dataset (i.e
13 SADs from MG-RAST). We then calculated the modified coefficient of determination (r 2 m) 274 around the 1:1 line (as per previous tests of METE 21, 25, 39 ) with the following equation. r!! = 1 (log!"(obs! ) log!" (pred! ))! (log!" (obs! ) log!" (obs! ))! 276 It is possible to obtain negative r 2 m values because the relationship is not fitted but instead, is 278 performed by estimating the variation around the 1:1 line with a constrained slope of 1.0 and a constrained intercept of , 25, 39. In addition, we have provided the mean, standard deviation, 280 and kernel density estimates of the log-likelihood and parameter values for all models that contain a free parameter (Tables S5, Figures S5). 282 Diversity-abundance scaling relationships 284 To determine whether the SAD models tested here can explain previously reported diversity-abundance scaling relationships 5, we first calculated the values of N max, Simpson s 286 measure of species evenness, and the log-modulo of skewness as a measure of rarity derived from predicted SADs of each model, as in ref. 5. We examined these diversity metrics against the 288 values of N in the observed SADs. We used simple linear regression on log-transformed axes to quantify the slopes of the scaling relationships, which become scaling exponents when axes are 290 arithmetically scaled, i.e., log(y) = zlog(x) is equivalent to y = x z, where z is the slope and scaling exponent. These scaling exponents were compared to the reported exponents 5. We calculated the 292 percent difference between the diversity metrics reported by each SAD model and the mean of the exponents reported for the EMP, HMP, and MG-RAST datasets. 13
14 294 We could not assess the ability of the SAD models to predict the scaling relationship of S to N, as in ref. 5. This was because all of the SAD models used in our study return SADs with the 296 same value of S as the empirical form. 298 Influence of total abundance on model performance We used ordinary least-squares regression to assess the relationship between the 300 performance of each SAD model and the number of sequences in a given sample (N). While the aim of our study was to capture the influence of sample sequence abundance (N) on SAD model 302 performance, we also rarefied within SADs. We performed bootstrapped resampling on rarefied sets of SADs to determine the influence of subsampled N on model performance. This bootstrap 304 sampling procedure consisted of sampling SADs at given fractions of sample N and then calculating the mean r!!, repeated 100 times for each model. SADs were sampled at 50%, 25%, %, 6.25%, 3.125%, and % percent of sample N. This subsampling analysis was computationally exhaustive and required SADs with N large enough to be halved 6 times and 308 still large enough to be analyzed with SAD models. Likewise, we only used SADs for which predictions from each SAD model could be obtained at each scale of subsampled N. Altogether; 310 we were able to use 10 SADs that met these criteria. 312 Computing code 314 We used open source computing code to obtain the maximum-likelihood estimates and predicted forms of the SAD for the Broken-stick, the lognormal, the prediction of METE (i.e. the 316 log-series distribution), and the Zipf distribution (github.com/weecology/macroecotools, 14
15 github.com/weecology/mete). This is the same code used in studies that showed support for 318 METE among communities of macroscopic plants and animals All analyses can be reproduced or modified for further exploration by using code, data, and following directions 320 provided here: Acknowledgements We thank Jack Gilbert and Sean Gibbons for providing EMP data and guidance on using it. We 324 also thank the researchers who collected, sequenced, and provided metagenomic data on MG- RAST as well as the individuals who maintain and provide the MG-RAST service. We also 326 acknowledge the researchers who provided the open-source code for conducting some of our analyses. Finally, we would like to thank The Human Microbiome Project Consortium for 328 providing their data on the National Institutes of Health s publically accessible Data Analysis and Coordination Center server. This work was supported by a National Science Foundation 330 Dimensions of Biodiversity Grant (# to JTL and KJL) and the U.S. Army Research Office (W911NF to JTL). 332 Author contributions WRS and KJL conceived, designed, and performed the experiments, analyzed the data, 334 and contributed materials/analysis tools. WRS, KJL, and JTL wrote the paper. 336 Competing financial interests The authors declare no conflict of interest
16 340 References 1. Brown, J.H., Mehlman, D.W. & Stevens, G.C. Spatial variation in abundance. Ecology 76, (1995). 2. Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton 344 University Press: Princeton (2001). 3. McGill, B.J. Towards a unification of unified theories of biodiversity. Ecol. Lett. 13, (2010). 4. Harte, J. Maximum entropy and ecology: a theory of abundance, distribution, and energetics. 348 (Oxford University Press, New York, USA, 2011). 5. Locey, K.J. & Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. 350 Sci. USA 113, (2016). 6. Sogin, M.L. et al. Microbial diversity in the deep sea and the underexplored rare biosphere. 352 Proc. Natl. Acad. Sci. USA 103, (2006). 7. Lynch, M.D.J. & Neufeld, J.D. Ecology and exploration of the rare biosphere. Nature Rev. 354 Microbiol. 13, (2015). 8. Reid, A. & Buckley, M. The Rare Biosphere: A report from the American Academy of 356 Microbiology. Washington, DC: American Academy of Microbiology (2011). 9. McGill, B.J. et al. Species abundance distributions: Moving beyond single prediction theories 358 to integration within an ecological framework. Ecol. Lett. 10, (2007). 10. Horner-Devine, M.C., Lage, M., Hughes, J.B. & Bohannan, B.J.M. A taxa-area relationship 360 for bacteria. Nature 432, (2004). 11. Putnam, R. Community Ecology. Chapman & Hall, London, United Kingdom (1993). 16
17 MacArthur, R. On the relative abundance of species. Amer. Nat. 94, (1960). 13. Sih, A., Englund, G. & Wooster, D. Emergent impacts of multiple predators on prey. Trends 364 Ecol. Evolut. 13, (1998). 14. Dunbar, J., Barns, S.M., Ticknor, L.O. & Kuske, C.R. Empirical and theoretical Bacterial 366 diversity in four Arizona soils. Appl. Environ. Microbiol. 68, (2002). 15. Curtis, T.P., Sloan, W.T. & Scannell, J.W. Estimating prokaryotic diversity and its limits. 368 Proc. Natl. Acad. Sci. USA 99, (2002). 16. Bohannan, B. J. M. & Hughes, J. New approaches to analyzing microbial biodiversity data. 370 Curr. Opin. Microbiol. 6, (2003). 17. Schloss, P.D. & Handelsman, J. Status of the microbial census. Microbiol. Mol. Biol. R. 68, (2004). 18. Pedrós-Alió, C. & Manrubia, S. The vast unknown microbial biosphere. Proc. Natl. Acad. 374 Sci. USA; e-pub ahead of print 3 June 2016, doi: /pnas (2016). 19. Fisher, R.A., Corbet, A.S. & Williams, C.B. The relation between the number of species and 376 the number of individuals in a random sample of an animal population. J. Anim. Ecol. 12, (1943) Jaynes, E.T. Probability Theory: The Logic of Science. Cambridge University Press: New York (2003) White, E.P., Thibault, K.M. & Xiao, X. Characterizing species abundance distributions across taxa and ecosystems using a simple maximum entropy model. Ecology 93, (2012). 22. Baldridge, E., Xiao, X. & White, E.P. An extensive comparison of species-abundance 384 distribution models. biorxiv. doi: (2015). 17
18 23. McGill, B. Strong and weak tests of macroecological theory. Oikos 102, (2003) Ulrich, W., Ollik, M., & Ugland, K. I. A meta-analysis of species-abundance distributions. Oikos 119, (2010) Locey, K.J. & White, E.P. How species richness and total abundance constrain the distribution of abundance. Ecol. Lett. 16, (2013) Fierer, N. & Lennon, J. T. The generation and maintenance of diversity in microbial communities. Am. J. Bot. 98, (2011) Allen, A.P., Li, B. & Charnov, E.L. Population fluctuations, power laws and mixtures of lognormal distributions. Ecol. Lett. 4, 1 3 (2001) Gilbert, J.A., Jansson, J.K. & Knight, R. The Earth Microbiome project: successes and aspirations. BMC Biol. 12, 69 (2014) Turnbaugh, P.J., Ley, R.E., Hamady, M., Fraser-Liggett, C.M., Knight, R. & Gordon, J.I. The human microbiome project. Nature 449, (2007) Meyer, F. et al. The metagenomics RAST server a public resource for the automatic phylo- genetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008) Gans, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, (2005) Dumbrell, A.J., Nelson, M., Helgason, T., Dytham, C. & Fitter, A.H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 4, (2010). 33. Magurran, A.E. & McGill, B.J. Biological diversity frontiers in measurement and 406 assessment. Oxford University Press: New York (2011). 18
19 34. Cohen, J.E. Alternate derivations of a species-abundance relation. Amer. Nat. 102, (1968). 35. Heip, C.H.R., Herman, P.M.J. & Soetaert, K. Indices of diversity and evenness. Oceanis 24, (1998). 36. Zipf, G.K. Human behavior and the principle of least effort. Addison-Wesley, Cambridge, 412 USA (1949). 37. Newman, M.E.J. Power laws, Pareto distributions and Zipf s law. Contemp. Phys. 46, (2005). 38. Newman, R.E.J. Power laws, Pareto distributions and Zipf s law. biorxiv. doi: /j.cities (2006). 39. Xiao, X., McGlinn, D.J. & White, E.P. A strong test of the Maximum Entropy Theory of 418 Ecology. Amer. Nat. 185, (2015). 40. Harte, J., & Newman, E. Maximum information entropy: a foundation for ecological theory. 420 Trends Ecol. Evolut. 29, (2014). 41. Ulrich, W., Ollik, M., & Ugland, K. I. A meta-analysis of species-abundance distributions. 422 Oikos 119, (2010). 19
20 424 Figure 1. Forms of predicted species abundance distributions (SAD) in rank-abundance form, i.e., ordered from the most abundant species (N max ) to least the abundant on the x-axis. The grey 426 line represents one SAD that was randomly chosen from our data. Each model was fit to the observed SAD; see Methods. The Simultaneous Broken-stick is known to produce an overly 428 even SAD. The log-series often explains SADs for plant and animal communities but has gone untested among microbes 22. The Zipf distribution is a power law model that produces one of the 430 most uneven forms of the SAD, often predicting more singletons and greater dominance (i.e., N max ) than other models. Finally, the Poisson lognormal, a lognormal model with Poisson-based 432 sampling error, tends to be similar to the unevenness of the Zipf distribution, but predicts more realistic N max. Importantly, each model used here predicts an SAD with the same richness of the 434 observed SAD, which is often not the case in other studies that fail to use maximum likelihood expectations
21 Figure 2. Relationships between predicted abundance and observed abundance for each SAD 438 model. All species of all examined SADs are plotted, with hotter colors (e.g. red) reveal a greater density of species abundances. The black diagonal line is the 1:1 line, around which a perfect 440 prediction would fall. The box within each subplot is a histogram of the per-sad modified rsquared (r2m) values from a range of zero to one, with left-skewed histograms suggesting a better 442 fit of the model to the data. The value at the top-left of each sub-plot is the mean r2m value for 10,000 bootstrapped samples (see methods). Each dot represents the observed abundance versus 444 the predicted abundance for each species in the data
22 Figure 3. The relationship of model performance (via modified r2m) to the total number of 16S 464 rrna reads (N) for each SAD. The modified r-square value r2m is the variation in the observed SAD that is explained by the predicted SAD (as in Fig. 2). The performance of the Broken-stick 466 model and of the log-series distribution predicted by the maximum entropy theory of ecology (METE) decreases for greater N. With the exception of a small group of point, the lognormal 468 provides r2m values of 0.95 or greater across scales of N. The Zipf provide better explanations of microbial SADs with increasing N. The grey dashed horizontal line is placed where the r2m 470 equals zero. The r2m can take negative values because it does not represent a fitted relationship, i.e., the y-intercept is constrained to 0 and the slope is constrained to 1. Results from the simple 472 linear regression can be found in Table S
23 Figure 4. Predictions of absolute dominance (i.e., greatest species abundance within an SAD, 476 Nmax) using the dominance scaling relationships of each model (Table 1) and the r2m of the relationship. Because of the negative r2m values for the Broken-stick and the log-series, only the 478 lognormal and the Zipf are capable of providing meaningful predictions of Nmax. This figure demonstrates the differences in Nmax produced by models (i.e., lognormal and Zipf) that perform 480 well at predicting the SAD and closely approximate the dominance scaling exponent (Table 2). Hotter colors indicate a higher density of data points, i.e., results from SADs
24 484 Table 1. Comparison of the performance of species abundance distribution (SAD) models for microbial datasets. The mean site-specific r-square (r 2 m) and standard error (σ!!! ) for each model 486 from 10,000 bootstrapped samples of 200 SADs: Broken-stick, the log-series predicted by the Maximum Entropy Theory of Ecology (METE), the lognormal, and the Zipf power law 488 distribution. The lognormal and the Zipf provide the best predictions for how abundance varies among taxa. The lognormal and the Zipf are also characterized by lower standard errors than the 490 Broken-stick and the log-series. Model rm 2 r2 m Lognormal Zipf Log-series Broken-stick
25 Table 2. In general, the lognormal comes closest to reproducing the scaling exponents of 496 diversity-abundance scaling relationships 5. These scaling relationships pertain to absolute dominance (N max ), Simpson s metric of species evenness, and skewness of the SAD. The percent 498 difference and percent error is given between the scaling exponents predicted from each SAD model and the mean of the scaling exponents for the EMP, HMP, and MG-RAST reported in 500 Table 1 of 5, i.e., where the mean for N max was 1.0, the mean for evenness was -0.48, and the mean for skewness was The p-values were < for all scaling exponents. 502 Model Diversity metric Slope % Di erence Lognormal N max Evenness Skewness Zipf N max Evenness Skewness Log-series N max Evenness Skewness Broken-stick N max Evenness Skewness
Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
Metacommunities Spatial Ecology of Communities
 Spatial Ecology of Communities Four perspectives for multiple species Patch dynamics principles of metapopulation models (patchy pops, Levins) Mass effects principles of source-sink and rescue effects
Spatial Ecology of Communities Four perspectives for multiple species Patch dynamics principles of metapopulation models (patchy pops, Levins) Mass effects principles of source-sink and rescue effects
Oikos. Appendix 1 and 2. o20751
 Oikos o20751 Rosindell, J. and Cornell, S. J. 2013. Universal scaling of species-abundance distributions across multiple scales. Oikos 122: 1101 1111. Appendix 1 and 2 Universal scaling of species-abundance
Oikos o20751 Rosindell, J. and Cornell, S. J. 2013. Universal scaling of species-abundance distributions across multiple scales. Oikos 122: 1101 1111. Appendix 1 and 2 Universal scaling of species-abundance
Mechanisms, Theory, Data, & Prediction in Macroecology
 Mechanisms, Theory, Data, & Prediction in Macroecology A Weecology Production Ethan P. White @ethanwhite ethan@weecology.org ethanwhite.org You are free to: Copy, share, adapt, or re-mix; Photograph, film,
Mechanisms, Theory, Data, & Prediction in Macroecology A Weecology Production Ethan P. White @ethanwhite ethan@weecology.org ethanwhite.org You are free to: Copy, share, adapt, or re-mix; Photograph, film,
Rank-abundance. Geometric series: found in very communities such as the
 Rank-abundance Geometric series: found in very communities such as the Log series: group of species that occur _ time are the most frequent. Useful for calculating a diversity metric (Fisher s alpha) Most
Rank-abundance Geometric series: found in very communities such as the Log series: group of species that occur _ time are the most frequent. Useful for calculating a diversity metric (Fisher s alpha) Most
Disentangling spatial structure in ecological communities. Dan McGlinn & Allen Hurlbert.
 Disentangling spatial structure in ecological communities Dan McGlinn & Allen Hurlbert http://mcglinn.web.unc.edu daniel.mcglinn@usu.edu The Unified Theories of Biodiversity 6 unified theories of diversity
Disentangling spatial structure in ecological communities Dan McGlinn & Allen Hurlbert http://mcglinn.web.unc.edu daniel.mcglinn@usu.edu The Unified Theories of Biodiversity 6 unified theories of diversity
DNA reassociation kinetics and diversity indices: richness is not rich enough
 Oikos 7: 778, 2008 doi: 0./j.2007.0030-299.63.x, # 2007 The authors. Journal compilation # Oikos 2007 Subject Editor: Heikki Setälä, Accepted 2 November 2007 DNA reassociation kinetics and diversity indices:
Oikos 7: 778, 2008 doi: 0./j.2007.0030-299.63.x, # 2007 The authors. Journal compilation # Oikos 2007 Subject Editor: Heikki Setälä, Accepted 2 November 2007 DNA reassociation kinetics and diversity indices:
Outline Classes of diversity measures. Species Divergence and the Measurement of Microbial Diversity. How do we describe and compare diversity?
 Species Divergence and the Measurement of Microbial Diversity Cathy Lozupone University of Colorado, Boulder. Washington University, St Louis. Outline Classes of diversity measures α vs β diversity Quantitative
Species Divergence and the Measurement of Microbial Diversity Cathy Lozupone University of Colorado, Boulder. Washington University, St Louis. Outline Classes of diversity measures α vs β diversity Quantitative
A meta-analysis of species abundance distributions
 Oikos 000: 001 007, 2009 doi: 10.1111/j.1600-0706.2009.18236.x 2009 The Authors. Journal compilation 2009 Oikos Subject Editor: Tim Benton. Accepted 3 November 2009 A meta-analysis of species abundance
Oikos 000: 001 007, 2009 doi: 10.1111/j.1600-0706.2009.18236.x 2009 The Authors. Journal compilation 2009 Oikos Subject Editor: Tim Benton. Accepted 3 November 2009 A meta-analysis of species abundance
Exploring Microbes in the Sea. Alma Parada Postdoctoral Scholar Stanford University
 Exploring Microbes in the Sea Alma Parada Postdoctoral Scholar Stanford University Cruising the ocean to get us some microbes It s all about the Microbe! Microbes = microorganisms an organism that requires
Exploring Microbes in the Sea Alma Parada Postdoctoral Scholar Stanford University Cruising the ocean to get us some microbes It s all about the Microbe! Microbes = microorganisms an organism that requires
How species richness and total abundance constrain the distribution of abundance
 Utah State University DigitalCommons@USU Biology Faculty Publications Biology 2013 How species richness and total abundance constrain the distribution of abundance Kenneth J. Locey Utah State University
Utah State University DigitalCommons@USU Biology Faculty Publications Biology 2013 How species richness and total abundance constrain the distribution of abundance Kenneth J. Locey Utah State University
MiGA: The Microbial Genome Atlas
 December 12 th 2017 MiGA: The Microbial Genome Atlas Jim Cole Center for Microbial Ecology Dept. of Plant, Soil & Microbial Sciences Michigan State University East Lansing, Michigan U.S.A. Where I m From
December 12 th 2017 MiGA: The Microbial Genome Atlas Jim Cole Center for Microbial Ecology Dept. of Plant, Soil & Microbial Sciences Michigan State University East Lansing, Michigan U.S.A. Where I m From
A General Unified Niche-Assembly/Dispersal-Assembly Theory of Forest Species Biodiversity
 A General Unified Niche-Assembly/Dispersal-Assembly Theory of Forest Species Biodiversity Keith Rennolls CMS, University of Greenwich, Park Row, London SE10 9LS k.rennolls@gre.ac.uk Abstract: A generalised
A General Unified Niche-Assembly/Dispersal-Assembly Theory of Forest Species Biodiversity Keith Rennolls CMS, University of Greenwich, Park Row, London SE10 9LS k.rennolls@gre.ac.uk Abstract: A generalised
Microbiome: 16S rrna Sequencing 3/30/2018
 Microbiome: 16S rrna Sequencing 3/30/2018 Skills from Previous Lectures Central Dogma of Biology Lecture 3: Genetics and Genomics Lecture 4: Microarrays Lecture 12: ChIP-Seq Phylogenetics Lecture 13: Phylogenetics
Microbiome: 16S rrna Sequencing 3/30/2018 Skills from Previous Lectures Central Dogma of Biology Lecture 3: Genetics and Genomics Lecture 4: Microarrays Lecture 12: ChIP-Seq Phylogenetics Lecture 13: Phylogenetics
Lecture: Mixture Models for Microbiome data
 Lecture: Mixture Models for Microbiome data Lecture 3: Mixture Models for Microbiome data Outline: - - Sequencing thought experiment Mixture Models (tangent) - (esp. Negative Binomial) - Differential abundance
Lecture: Mixture Models for Microbiome data Lecture 3: Mixture Models for Microbiome data Outline: - - Sequencing thought experiment Mixture Models (tangent) - (esp. Negative Binomial) - Differential abundance
Diversity partitioning without statistical independence of alpha and beta
 1964 Ecology, Vol. 91, No. 7 Ecology, 91(7), 2010, pp. 1964 1969 Ó 2010 by the Ecological Society of America Diversity partitioning without statistical independence of alpha and beta JOSEPH A. VEECH 1,3
1964 Ecology, Vol. 91, No. 7 Ecology, 91(7), 2010, pp. 1964 1969 Ó 2010 by the Ecological Society of America Diversity partitioning without statistical independence of alpha and beta JOSEPH A. VEECH 1,3
Lecture 2: Diversity, Distances, adonis. Lecture 2: Diversity, Distances, adonis. Alpha- Diversity. Alpha diversity definition(s)
 Lecture 2: Diversity, Distances, adonis Lecture 2: Diversity, Distances, adonis Diversity - alpha, beta (, gamma) Beta- Diversity in practice: Ecological Distances Unsupervised Learning: Clustering, etc
Lecture 2: Diversity, Distances, adonis Lecture 2: Diversity, Distances, adonis Diversity - alpha, beta (, gamma) Beta- Diversity in practice: Ecological Distances Unsupervised Learning: Clustering, etc
Lecture 3: Mixture Models for Microbiome data. Lecture 3: Mixture Models for Microbiome data
 Lecture 3: Mixture Models for Microbiome data 1 Lecture 3: Mixture Models for Microbiome data Outline: - Mixture Models (Negative Binomial) - DESeq2 / Don t Rarefy. Ever. 2 Hypothesis Tests - reminder
Lecture 3: Mixture Models for Microbiome data 1 Lecture 3: Mixture Models for Microbiome data Outline: - Mixture Models (Negative Binomial) - DESeq2 / Don t Rarefy. Ever. 2 Hypothesis Tests - reminder
On the Statistical Machinery of Alien Species Distribution
 On the Statistical Machinery of Alien Species Distribution M G Bowler Department of Physics, University of Oxford Keble Road, Oxford OX1 3RH, UK (m.bowler1@physics.ox.ac.uk) C K Kelly Department of Zoology,
On the Statistical Machinery of Alien Species Distribution M G Bowler Department of Physics, University of Oxford Keble Road, Oxford OX1 3RH, UK (m.bowler1@physics.ox.ac.uk) C K Kelly Department of Zoology,
NGSS Example Bundles. Page 1 of 23
 High School Conceptual Progressions Model III Bundle 2 Evolution of Life This is the second bundle of the High School Conceptual Progressions Model Course III. Each bundle has connections to the other
High School Conceptual Progressions Model III Bundle 2 Evolution of Life This is the second bundle of the High School Conceptual Progressions Model Course III. Each bundle has connections to the other
Spiral Waves Emergence in a Cyclic Predator-Prey Model
 Spiral Waves Emergence in a Cyclic Predator-Prey Model Luo-Luo Jiang 1,Wen-XuWang 2,XinHuang 3, and Bing-Hong Wang 1 1 Department of Modern Physics, University of Science and Technology of China, Hefei
Spiral Waves Emergence in a Cyclic Predator-Prey Model Luo-Luo Jiang 1,Wen-XuWang 2,XinHuang 3, and Bing-Hong Wang 1 1 Department of Modern Physics, University of Science and Technology of China, Hefei
Supplementary Figures.
 Supplementary Figures. Supplementary Figure 1 The extended compartment model. Sub-compartment C (blue) and 1-C (yellow) represent the fractions of allele carriers and non-carriers in the focal patch, respectively,
Supplementary Figures. Supplementary Figure 1 The extended compartment model. Sub-compartment C (blue) and 1-C (yellow) represent the fractions of allele carriers and non-carriers in the focal patch, respectively,
Package breakaway. R topics documented: March 30, 2016
 Title Species Richness Estimation and Modeling Version 3.0 Date 2016-03-29 Author and John Bunge Maintainer Package breakaway March 30, 2016 Species richness estimation is an important
Title Species Richness Estimation and Modeling Version 3.0 Date 2016-03-29 Author and John Bunge Maintainer Package breakaway March 30, 2016 Species richness estimation is an important
Emergence of diversity in a biological evolution model
 Journal of Physics: Conference Series PAPER OPE ACCESS Emergence of diversity in a biological evolution model To cite this article: R Wang and C Pujos 2015 J. Phys.: Conf. Ser. 604 012019 Related content
Journal of Physics: Conference Series PAPER OPE ACCESS Emergence of diversity in a biological evolution model To cite this article: R Wang and C Pujos 2015 J. Phys.: Conf. Ser. 604 012019 Related content
Chapter 54: Community Ecology
 AP Biology Guided Reading Name Chapter 54: Community Ecology Overview 1. What does community ecology explore? Concept 54.1 Community interactions are classified by whether they help, harm, or have no effect
AP Biology Guided Reading Name Chapter 54: Community Ecology Overview 1. What does community ecology explore? Concept 54.1 Community interactions are classified by whether they help, harm, or have no effect
Other resources. Greengenes (bacterial) Silva (bacteria, archaeal and eukarya)
 General QIIME resources http://qiime.org/ Blog (news, updates): http://qiime.wordpress.com/ Support/forum: https://groups.google.com/forum/#!forum/qiimeforum Citing QIIME: Caporaso, J.G. et al., QIIME
General QIIME resources http://qiime.org/ Blog (news, updates): http://qiime.wordpress.com/ Support/forum: https://groups.google.com/forum/#!forum/qiimeforum Citing QIIME: Caporaso, J.G. et al., QIIME
Frequency in % of each F-scale rating in outbreaks
 Frequency in % of each F-scale rating in outbreaks 2 1 1 1 F/EF1 F/EF2 F/EF3-F/EF5 1954 196 197 198 199 2 21 Supplementary Figure 1 Distribution of outbreak tornadoes by F/EF-scale rating. Frequency (in
Frequency in % of each F-scale rating in outbreaks 2 1 1 1 F/EF1 F/EF2 F/EF3-F/EF5 1954 196 197 198 199 2 21 Supplementary Figure 1 Distribution of outbreak tornadoes by F/EF-scale rating. Frequency (in
SUPPLEMENTARY INFORMATION
 City of origin as a confounding variable. The original study was designed such that the city where sampling was performed was perfectly confounded with where the DNA extractions and sequencing was performed.
City of origin as a confounding variable. The original study was designed such that the city where sampling was performed was perfectly confounded with where the DNA extractions and sequencing was performed.
Transitivity a FORTRAN program for the analysis of bivariate competitive interactions Version 1.1
 Transitivity 1 Transitivity a FORTRAN program for the analysis of bivariate competitive interactions Version 1.1 Werner Ulrich Nicolaus Copernicus University in Toruń Chair of Ecology and Biogeography
Transitivity 1 Transitivity a FORTRAN program for the analysis of bivariate competitive interactions Version 1.1 Werner Ulrich Nicolaus Copernicus University in Toruń Chair of Ecology and Biogeography
Microbiota: Its Evolution and Essence. Hsin-Jung Joyce Wu "Microbiota and man: the story about us
 Microbiota: Its Evolution and Essence Overview q Define microbiota q Learn the tool q Ecological and evolutionary forces in shaping gut microbiota q Gut microbiota versus free-living microbe communities
Microbiota: Its Evolution and Essence Overview q Define microbiota q Learn the tool q Ecological and evolutionary forces in shaping gut microbiota q Gut microbiota versus free-living microbe communities
The implications of neutral evolution for neutral ecology. Daniel Lawson Bioinformatics and Statistics Scotland Macaulay Institute, Aberdeen
 The implications of neutral evolution for neutral ecology Daniel Lawson Bioinformatics and Statistics Scotland Macaulay Institute, Aberdeen How is How is diversity Diversity maintained? maintained? Talk
The implications of neutral evolution for neutral ecology Daniel Lawson Bioinformatics and Statistics Scotland Macaulay Institute, Aberdeen How is How is diversity Diversity maintained? maintained? Talk
Spatial scaling of species abundance distributions
 Ecography 35: 549 556, 12 doi:.1111/j.16-587.11.7128.x 11 The Authors. Ecography 11 Nordic Society Oikos Subject Editor: Pedro Peres-Neto. Accepted 25 August 11 Spatial scaling of species abundance distributions
Ecography 35: 549 556, 12 doi:.1111/j.16-587.11.7128.x 11 The Authors. Ecography 11 Nordic Society Oikos Subject Editor: Pedro Peres-Neto. Accepted 25 August 11 Spatial scaling of species abundance distributions
Metabolic trade-offs promote diversity in a model ecosystem
 Metabolic trade-offs promote diversity in a model ecosystem Anna Posfai, Thibaud Taillefumier, Ben Weiner, Ned Wingreen Princeton University q-bio Rutgers University, July 25 2017 How can we explain species
Metabolic trade-offs promote diversity in a model ecosystem Anna Posfai, Thibaud Taillefumier, Ben Weiner, Ned Wingreen Princeton University q-bio Rutgers University, July 25 2017 How can we explain species
Test of neutral theory predic3ons for the BCI tree community informed by regional abundance data
 Test of neutral theory predic3ons for the BCI tree community informed by regional abundance data Anne%e Ostling Cody Weinberger Devin Riley Ecology and Evolu:onary Biology University of Michigan 1 Outline
Test of neutral theory predic3ons for the BCI tree community informed by regional abundance data Anne%e Ostling Cody Weinberger Devin Riley Ecology and Evolu:onary Biology University of Michigan 1 Outline
Island biogeography. Key concepts. Introduction. Island biogeography theory. Colonization-extinction balance. Island-biogeography theory
 Island biogeography Key concepts Colonization-extinction balance Island-biogeography theory Introduction At the end of the last chapter, it was suggested that another mechanism for the maintenance of α-diversity
Island biogeography Key concepts Colonization-extinction balance Island-biogeography theory Introduction At the end of the last chapter, it was suggested that another mechanism for the maintenance of α-diversity
EARTH SYSTEM: HISTORY AND NATURAL VARIABILITY Vol. III - Global Biodiversity and its Variation in Space and Time - D. Storch
 GLOBAL BIODIVERSITY AND ITS VARIATION IN SPACE AND TIME D. Storch Charles University, Center for Theoretical Study, Prague, Czech Republic Keywords: species diversity, interspecific interactions, communities,
GLOBAL BIODIVERSITY AND ITS VARIATION IN SPACE AND TIME D. Storch Charles University, Center for Theoretical Study, Prague, Czech Republic Keywords: species diversity, interspecific interactions, communities,
Supplementary File 4: Methods Summary and Supplementary Methods
 Supplementary File 4: Methods Summary and Supplementary Methods Methods Summary Constructing and implementing an SDM model requires local measurements of community composition and rasters of environmental
Supplementary File 4: Methods Summary and Supplementary Methods Methods Summary Constructing and implementing an SDM model requires local measurements of community composition and rasters of environmental
Chad Burrus April 6, 2010
 Chad Burrus April 6, 2010 1 Background What is UniFrac? Materials and Methods Results Discussion Questions 2 The vast majority of microbes cannot be cultured with current methods Only half (26) out of
Chad Burrus April 6, 2010 1 Background What is UniFrac? Materials and Methods Results Discussion Questions 2 The vast majority of microbes cannot be cultured with current methods Only half (26) out of
Microbial Taxonomy and the Evolution of Diversity
 19 Microbial Taxonomy and the Evolution of Diversity Copyright McGraw-Hill Global Education Holdings, LLC. Permission required for reproduction or display. 1 Taxonomy Introduction to Microbial Taxonomy
19 Microbial Taxonomy and the Evolution of Diversity Copyright McGraw-Hill Global Education Holdings, LLC. Permission required for reproduction or display. 1 Taxonomy Introduction to Microbial Taxonomy
Microbes and mountains: metagenetics on Mount Fuji, Japan. Jonathan Adams, Biology Department, SNU, Korea
 Microbes and mountains: metagenetics on Mount Fuji, Japan Jonathan Adams, Biology Department, SNU, Korea jonadams@snu.ac.kr Until about a decade ago, culturing could only yield 8,000 described species
Microbes and mountains: metagenetics on Mount Fuji, Japan Jonathan Adams, Biology Department, SNU, Korea jonadams@snu.ac.kr Until about a decade ago, culturing could only yield 8,000 described species
Studying Life. Lesson Overview. Lesson Overview. 1.3 Studying Life
 Lesson Overview 1.3 Characteristics of Living Things What characteristics do all living things share? Living things are made up of basic units called cells, are based on a universal genetic code, obtain
Lesson Overview 1.3 Characteristics of Living Things What characteristics do all living things share? Living things are made up of basic units called cells, are based on a universal genetic code, obtain
Chapter 6 Population and Community Ecology
 Chapter 6 Population and Community Ecology Friedland and Relyea Environmental Science for AP, second edition 2015 W.H. Freeman and Company/BFW AP is a trademark registered and/or owned by the College Board,
Chapter 6 Population and Community Ecology Friedland and Relyea Environmental Science for AP, second edition 2015 W.H. Freeman and Company/BFW AP is a trademark registered and/or owned by the College Board,
Supplementary Information
 Supplementary Information Altitudinal patterns of diversity and functional traits of metabolically active microorganisms in stream biofilms Linda Wilhelm 1, Katharina Besemer 2, Lena Fragner 3, Hannes
Supplementary Information Altitudinal patterns of diversity and functional traits of metabolically active microorganisms in stream biofilms Linda Wilhelm 1, Katharina Besemer 2, Lena Fragner 3, Hannes
Phylogenetic diversity and conservation
 Phylogenetic diversity and conservation Dan Faith The Australian Museum Applied ecology and human dimensions in biological conservation Biota Program/ FAPESP Nov. 9-10, 2009 BioGENESIS Providing an evolutionary
Phylogenetic diversity and conservation Dan Faith The Australian Museum Applied ecology and human dimensions in biological conservation Biota Program/ FAPESP Nov. 9-10, 2009 BioGENESIS Providing an evolutionary
How to quantify biological diversity: taxonomical, functional and evolutionary aspects. Hanna Tuomisto, University of Turku
 How to quantify biological diversity: taxonomical, functional and evolutionary aspects Hanna Tuomisto, University of Turku Why quantify biological diversity? understanding the structure and function of
How to quantify biological diversity: taxonomical, functional and evolutionary aspects Hanna Tuomisto, University of Turku Why quantify biological diversity? understanding the structure and function of
Community Structure. Community An assemblage of all the populations interacting in an area
 Community Structure Community An assemblage of all the populations interacting in an area Community Ecology The ecological community is the set of plant and animal species that occupy an area Questions
Community Structure Community An assemblage of all the populations interacting in an area Community Ecology The ecological community is the set of plant and animal species that occupy an area Questions
Standards A complete list of the standards covered by this lesson is included in the Appendix at the end of the lesson.
 Lesson 8: The History of Life on Earth Time: approximately 45-60 minutes, depending on length of discussion. Can be broken into 2 shorter lessons Materials: Double timeline (see below) Meter stick (to
Lesson 8: The History of Life on Earth Time: approximately 45-60 minutes, depending on length of discussion. Can be broken into 2 shorter lessons Materials: Double timeline (see below) Meter stick (to
arxiv: v1 [q-bio.qm] 7 Aug 2017
![arxiv: v1 [q-bio.qm] 7 Aug 2017 arxiv: v1 [q-bio.qm] 7 Aug 2017](/thumbs/74/70713977.jpg) HIGHER ORDER EPISTASIS AND FITNESS PEAKS KRISTINA CRONA AND MENGMING LUO arxiv:1708.02063v1 [q-bio.qm] 7 Aug 2017 ABSTRACT. We show that higher order epistasis has a substantial impact on evolutionary
HIGHER ORDER EPISTASIS AND FITNESS PEAKS KRISTINA CRONA AND MENGMING LUO arxiv:1708.02063v1 [q-bio.qm] 7 Aug 2017 ABSTRACT. We show that higher order epistasis has a substantial impact on evolutionary
Biology Assessment. Eligible Texas Essential Knowledge and Skills
 Biology Assessment Eligible Texas Essential Knowledge and Skills STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules
Biology Assessment Eligible Texas Essential Knowledge and Skills STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules
POPULATIONS and COMMUNITIES
 POPULATIONS and COMMUNITIES Ecology is the study of organisms and the nonliving world they inhabit. Central to ecology is the complex set of interactions between organisms, both intraspecific (between
POPULATIONS and COMMUNITIES Ecology is the study of organisms and the nonliving world they inhabit. Central to ecology is the complex set of interactions between organisms, both intraspecific (between
STAAR Biology Assessment
 STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
Evidence for Competition
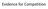 Evidence for Competition Population growth in laboratory experiments carried out by the Russian scientist Gause on growth rates in two different yeast species Each of the species has the same food e.g.,
Evidence for Competition Population growth in laboratory experiments carried out by the Russian scientist Gause on growth rates in two different yeast species Each of the species has the same food e.g.,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11226 Supplementary Discussion D1 Endemics-area relationship (EAR) and its relation to the SAR The EAR comprises the relationship between study area and the number of species that are
doi:10.1038/nature11226 Supplementary Discussion D1 Endemics-area relationship (EAR) and its relation to the SAR The EAR comprises the relationship between study area and the number of species that are
Title: Fitting and comparing competing models of the species abundance distribution: assessment and prospect
 Title: Fitting and comparing competing models of the species abundance distribution: assessment and prospect Journal Issue: Frontiers of Biogeography, 6(2) Author: Matthews, Thomas J, Conservation Biogeography
Title: Fitting and comparing competing models of the species abundance distribution: assessment and prospect Journal Issue: Frontiers of Biogeography, 6(2) Author: Matthews, Thomas J, Conservation Biogeography
Parameter Sensitivity In A Lattice Ecosystem With Intraguild Predation
 Parameter Sensitivity In A Lattice Ecosystem With Intraguild Predation N. Nakagiri a, K. Tainaka a, T. Togashi b, T. Miyazaki b and J. Yoshimura a a Department of Systems Engineering, Shizuoka University,
Parameter Sensitivity In A Lattice Ecosystem With Intraguild Predation N. Nakagiri a, K. Tainaka a, T. Togashi b, T. Miyazaki b and J. Yoshimura a a Department of Systems Engineering, Shizuoka University,
Microbial Taxonomy. Microbes usually have few distinguishing properties that relate them, so a hierarchical taxonomy mainly has not been possible.
 Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Inferring species abundance distribution across spatial scales
 Oikos 9: 780, 200 doi: 0./j.600-0706.2009.7938.x, # 2009 The Authors. Journal compilation # 2009 Oikos Subject Editor: Bradford Hawkins. Accepted 23 June 2009 Inferring species abundance distribution across
Oikos 9: 780, 200 doi: 0./j.600-0706.2009.7938.x, # 2009 The Authors. Journal compilation # 2009 Oikos Subject Editor: Bradford Hawkins. Accepted 23 June 2009 Inferring species abundance distribution across
Name Block Date. The Quadrat Study: An Introduction
 Name Block Date The Quadrat Study: An Introduction A quadrat study can almost be thought of as a snapshot of the ecosystem during a particular year and at a particular time of that year. The plant and
Name Block Date The Quadrat Study: An Introduction A quadrat study can almost be thought of as a snapshot of the ecosystem during a particular year and at a particular time of that year. The plant and
Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes. - Supplementary Information -
 Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes - Supplementary Information - Martin Bartl a, Martin Kötzing a,b, Stefan Schuster c, Pu Li a, Christoph Kaleta b a
Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes - Supplementary Information - Martin Bartl a, Martin Kötzing a,b, Stefan Schuster c, Pu Li a, Christoph Kaleta b a
Chapter 6 Population and Community Ecology. Thursday, October 19, 17
 Chapter 6 Population and Community Ecology Module 18 The Abundance and Distribution of After reading this module you should be able to explain how nature exists at several levels of complexity. discuss
Chapter 6 Population and Community Ecology Module 18 The Abundance and Distribution of After reading this module you should be able to explain how nature exists at several levels of complexity. discuss
Censusing the Sea in the 21 st Century
 Censusing the Sea in the 21 st Century Nancy Knowlton & Matthieu Leray Photo: Ove Hoegh-Guldberg Smithsonian s National Museum of Natural History Estimates of Marine/Reef Species Numbers (Millions) Marine
Censusing the Sea in the 21 st Century Nancy Knowlton & Matthieu Leray Photo: Ove Hoegh-Guldberg Smithsonian s National Museum of Natural History Estimates of Marine/Reef Species Numbers (Millions) Marine
Frontiers in Microbiology
 coloring book! 1 Frontiers in Microbiology As we are taking much better account of the unseen majority of life, unravel the biogeochemical processes that microbes facilitate, thereby making planet Earth
coloring book! 1 Frontiers in Microbiology As we are taking much better account of the unseen majority of life, unravel the biogeochemical processes that microbes facilitate, thereby making planet Earth
Campbell Essential Biology, 5e (Simon/Yeh) Chapter 1 Introduction: Biology Today. Multiple-Choice Questions
 Campbell Essential Biology, 5e (Simon/Yeh) Chapter 1 Introduction: Biology Today Multiple-Choice Questions 1) In what way(s) is the science of biology influencing and changing our culture? A) by helping
Campbell Essential Biology, 5e (Simon/Yeh) Chapter 1 Introduction: Biology Today Multiple-Choice Questions 1) In what way(s) is the science of biology influencing and changing our culture? A) by helping
Unifying theories of molecular, community and network evolution 1
 Carlos J. Melián National Center for Ecological Analysis and Synthesis, University of California, Santa Barbara Microsoft Research Ltd, Cambridge, UK. Unifying theories of molecular, community and network
Carlos J. Melián National Center for Ecological Analysis and Synthesis, University of California, Santa Barbara Microsoft Research Ltd, Cambridge, UK. Unifying theories of molecular, community and network
REPORT Does Mother Nature really prefer rare species or are log-left-skewed SADs a sampling artefact?
 Ecology Letters, (23) 6: 766 773 doi: 1.146/j.1461-248.23.491.x REPORT Does Mother Nature really prefer rare species or are log-left-skewed SADs a sampling artefact? Brian J. McGill Department of Ecology
Ecology Letters, (23) 6: 766 773 doi: 1.146/j.1461-248.23.491.x REPORT Does Mother Nature really prefer rare species or are log-left-skewed SADs a sampling artefact? Brian J. McGill Department of Ecology
Package SPECIES. R topics documented: April 23, Type Package. Title Statistical package for species richness estimation. Version 1.
 Package SPECIES April 23, 2011 Type Package Title Statistical package for species richness estimation Version 1.0 Date 2010-01-24 Author Ji-Ping Wang, Maintainer Ji-Ping Wang
Package SPECIES April 23, 2011 Type Package Title Statistical package for species richness estimation Version 1.0 Date 2010-01-24 Author Ji-Ping Wang, Maintainer Ji-Ping Wang
Network isolation and local diversity in neutral metacommunities
 Oikos 119: 1355 1363, 2010 doi: 10.1111/j.1600-0706.2010.18272.x 2009 The Authors. Journal compilation 2010 Oikos Subject Editor: José Alexandre Felizola. Accepted 14 December 2009 Network isolation and
Oikos 119: 1355 1363, 2010 doi: 10.1111/j.1600-0706.2010.18272.x 2009 The Authors. Journal compilation 2010 Oikos Subject Editor: José Alexandre Felizola. Accepted 14 December 2009 Network isolation and
of a landscape to support biodiversity and ecosystem processes and provide ecosystem services in face of various disturbances.
 L LANDSCAPE ECOLOGY JIANGUO WU Arizona State University Spatial heterogeneity is ubiquitous in all ecological systems, underlining the significance of the pattern process relationship and the scale of
L LANDSCAPE ECOLOGY JIANGUO WU Arizona State University Spatial heterogeneity is ubiquitous in all ecological systems, underlining the significance of the pattern process relationship and the scale of
Microbes usually have few distinguishing properties that relate them, so a hierarchical taxonomy mainly has not been possible.
 Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy. Slowly evolving molecules (e.g., rrna) used for large-scale structure; "fast- clock" molecules for fine-structure.
 Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
Microbial Taxonomy Traditional taxonomy or the classification through identification and nomenclature of microbes, both "prokaryote" and eukaryote, has been in a mess we were stuck with it for traditional
This is a repository copy of Microbiology: Mind the gaps in cellular evolution.
 This is a repository copy of Microbiology: Mind the gaps in cellular evolution. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/114978/ Version: Accepted Version Article:
This is a repository copy of Microbiology: Mind the gaps in cellular evolution. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/114978/ Version: Accepted Version Article:
Model Based Statistics in Biology. Part V. The Generalized Linear Model. Chapter 16 Introduction
 Model Based Statistics in Biology. Part V. The Generalized Linear Model. Chapter 16 Introduction ReCap. Parts I IV. The General Linear Model Part V. The Generalized Linear Model 16 Introduction 16.1 Analysis
Model Based Statistics in Biology. Part V. The Generalized Linear Model. Chapter 16 Introduction ReCap. Parts I IV. The General Linear Model Part V. The Generalized Linear Model 16 Introduction 16.1 Analysis
Major questions of evolutionary genetics. Experimental tools of evolutionary genetics. Theoretical population genetics.
 Evolutionary Genetics (for Encyclopedia of Biodiversity) Sergey Gavrilets Departments of Ecology and Evolutionary Biology and Mathematics, University of Tennessee, Knoxville, TN 37996-6 USA Evolutionary
Evolutionary Genetics (for Encyclopedia of Biodiversity) Sergey Gavrilets Departments of Ecology and Evolutionary Biology and Mathematics, University of Tennessee, Knoxville, TN 37996-6 USA Evolutionary
Supplementary materials Quantitative assessment of ribosome drop-off in E. coli
 Supplementary materials Quantitative assessment of ribosome drop-off in E. coli Celine Sin, Davide Chiarugi, Angelo Valleriani 1 Downstream Analysis Supplementary Figure 1: Illustration of the core steps
Supplementary materials Quantitative assessment of ribosome drop-off in E. coli Celine Sin, Davide Chiarugi, Angelo Valleriani 1 Downstream Analysis Supplementary Figure 1: Illustration of the core steps
IUCN Red List Process. Cormack Gates Keith Aune
 IUCN Red List Process Cormack Gates Keith Aune The IUCN Red List Categories and Criteria have several specific aims to provide a system that can be applied consistently by different people; to improve
IUCN Red List Process Cormack Gates Keith Aune The IUCN Red List Categories and Criteria have several specific aims to provide a system that can be applied consistently by different people; to improve
Curriculum Map. Biology, Quarter 1 Big Ideas: From Molecules to Organisms: Structures and Processes (BIO1.LS1)
 1 Biology, Quarter 1 Big Ideas: From Molecules to Organisms: Structures and Processes (BIO1.LS1) Focus Standards BIO1.LS1.2 Evaluate comparative models of various cell types with a focus on organic molecules
1 Biology, Quarter 1 Big Ideas: From Molecules to Organisms: Structures and Processes (BIO1.LS1) Focus Standards BIO1.LS1.2 Evaluate comparative models of various cell types with a focus on organic molecules
Four aspects of a sampling strategy necessary to make accurate and precise inferences about populations are:
 Why Sample? Often researchers are interested in answering questions about a particular population. They might be interested in the density, species richness, or specific life history parameters such as
Why Sample? Often researchers are interested in answering questions about a particular population. They might be interested in the density, species richness, or specific life history parameters such as
and just what is science? how about this biology stuff?
 Welcome to Life on Earth! Rob Lewis 512.775.6940 rlewis3@austincc.edu 1 The Science of Biology Themes and just what is science? how about this biology stuff? 2 1 The Process Of Science No absolute truths
Welcome to Life on Earth! Rob Lewis 512.775.6940 rlewis3@austincc.edu 1 The Science of Biology Themes and just what is science? how about this biology stuff? 2 1 The Process Of Science No absolute truths
Investigation of Possible Biases in Tau Neutrino Mass Limits
 Investigation of Possible Biases in Tau Neutrino Mass Limits Kyle Armour Departments of Physics and Mathematics, University of California, San Diego, La Jolla, CA 92093 (Dated: August 8, 2003) We study
Investigation of Possible Biases in Tau Neutrino Mass Limits Kyle Armour Departments of Physics and Mathematics, University of California, San Diego, La Jolla, CA 92093 (Dated: August 8, 2003) We study
1. The basic structural and physiological unit of all living organisms is the A) aggregate. B) organelle. C) organism. D) membrane. E) cell.
 Name: Date: Test File Questions 1. The basic structural and physiological unit of all living organisms is the A) aggregate. B) organelle. C) organism. D) membrane. E) cell. 2. A cell A) can be composed
Name: Date: Test File Questions 1. The basic structural and physiological unit of all living organisms is the A) aggregate. B) organelle. C) organism. D) membrane. E) cell. 2. A cell A) can be composed
Natal versus breeding dispersal: Evolution in a model system
 Evolutionary Ecology Research, 1999, 1: 911 921 Natal versus breeding dispersal: Evolution in a model system Karin Johst 1 * and Roland Brandl 2 1 Centre for Environmental Research Leipzig-Halle Ltd, Department
Evolutionary Ecology Research, 1999, 1: 911 921 Natal versus breeding dispersal: Evolution in a model system Karin Johst 1 * and Roland Brandl 2 1 Centre for Environmental Research Leipzig-Halle Ltd, Department
Probing diversity in a hidden world: applications of NGS in microbial ecology
 Probing diversity in a hidden world: applications of NGS in microbial ecology Guus Roeselers TNO, Microbiology & Systems Biology Group Symposium on Next Generation Sequencing October 21, 2013 Royal Museum
Probing diversity in a hidden world: applications of NGS in microbial ecology Guus Roeselers TNO, Microbiology & Systems Biology Group Symposium on Next Generation Sequencing October 21, 2013 Royal Museum
LETTER Hubbell s fundamental biodiversity parameter and the Simpson diversity index
 Ecology Letters, (25) 8: 386 39 doi: 1.1111/j.1461-248.25.729. LETTER Hubbell s fundamental biodiversity parameter and the Simpson diversity inde Fangliang He* and Xin-Sheng Hu Department of Renewable
Ecology Letters, (25) 8: 386 39 doi: 1.1111/j.1461-248.25.729. LETTER Hubbell s fundamental biodiversity parameter and the Simpson diversity inde Fangliang He* and Xin-Sheng Hu Department of Renewable
4/4/2017. Extrinsic Isolating Barriers. 1. Biological species concept: 2. Phylogenetic species concept:
 Chapter 13 The origin of species 13.1 What Is a Species? p. 414 Ways to identify species 1. Biological species concept: 1. There are many different concepts of species 2. Species are important taxonomic
Chapter 13 The origin of species 13.1 What Is a Species? p. 414 Ways to identify species 1. Biological species concept: 1. There are many different concepts of species 2. Species are important taxonomic
EVOLUTION OF COMPLEX FOOD WEB STRUCTURE BASED ON MASS EXTINCTION
 EVOLUTION OF COMPLEX FOOD WEB STRUCTURE BASED ON MASS EXTINCTION Kenichi Nakazato Nagoya University Graduate School of Human Informatics nakazato@create.human.nagoya-u.ac.jp Takaya Arita Nagoya University
EVOLUTION OF COMPLEX FOOD WEB STRUCTURE BASED ON MASS EXTINCTION Kenichi Nakazato Nagoya University Graduate School of Human Informatics nakazato@create.human.nagoya-u.ac.jp Takaya Arita Nagoya University
ANALYSIS OF CHARACTER DIVERGENCE ALONG ENVIRONMENTAL GRADIENTS AND OTHER COVARIATES
 ORIGINAL ARTICLE doi:10.1111/j.1558-5646.2007.00063.x ANALYSIS OF CHARACTER DIVERGENCE ALONG ENVIRONMENTAL GRADIENTS AND OTHER COVARIATES Dean C. Adams 1,2,3 and Michael L. Collyer 1,4 1 Department of
ORIGINAL ARTICLE doi:10.1111/j.1558-5646.2007.00063.x ANALYSIS OF CHARACTER DIVERGENCE ALONG ENVIRONMENTAL GRADIENTS AND OTHER COVARIATES Dean C. Adams 1,2,3 and Michael L. Collyer 1,4 1 Department of
Supplementary Information: The origin of bursts and heavy tails in human dynamics
 Supplementary Information: The origin of bursts and heavy tails in human dynamics Albert-László Barabási Department of Physics, University of Notre Dame, IN 46556, USA (Dated: February 7, 2005) 1 Contents
Supplementary Information: The origin of bursts and heavy tails in human dynamics Albert-László Barabási Department of Physics, University of Notre Dame, IN 46556, USA (Dated: February 7, 2005) 1 Contents
Niche Modeling. STAMPS - MBL Course Woods Hole, MA - August 9, 2016
 Niche Modeling Katie Pollard & Josh Ladau Gladstone Institutes UCSF Division of Biostatistics, Institute for Human Genetics and Institute for Computational Health Science STAMPS - MBL Course Woods Hole,
Niche Modeling Katie Pollard & Josh Ladau Gladstone Institutes UCSF Division of Biostatistics, Institute for Human Genetics and Institute for Computational Health Science STAMPS - MBL Course Woods Hole,
Do not copy, post, or distribute
 14 CORRELATION ANALYSIS AND LINEAR REGRESSION Assessing the Covariability of Two Quantitative Properties 14.0 LEARNING OBJECTIVES In this chapter, we discuss two related techniques for assessing a possible
14 CORRELATION ANALYSIS AND LINEAR REGRESSION Assessing the Covariability of Two Quantitative Properties 14.0 LEARNING OBJECTIVES In this chapter, we discuss two related techniques for assessing a possible
Culturing captures members of the soil rare biosphereemi_
 bs_bs_banner Environmental Microbiology (2012) doi:10.1111/j.1462-2920.2012.02817.x Correspondence Culturing captures members of the soil rare biosphereemi_2817 1..6 Ashley Shade, 1 Clifford S. Hogan,
bs_bs_banner Environmental Microbiology (2012) doi:10.1111/j.1462-2920.2012.02817.x Correspondence Culturing captures members of the soil rare biosphereemi_2817 1..6 Ashley Shade, 1 Clifford S. Hogan,
arxiv:cond-mat/ v2 [cond-mat.stat-mech] 13 Jan 1999
![arxiv:cond-mat/ v2 [cond-mat.stat-mech] 13 Jan 1999 arxiv:cond-mat/ v2 [cond-mat.stat-mech] 13 Jan 1999](/thumbs/92/108237926.jpg) arxiv:cond-mat/9901071v2 [cond-mat.stat-mech] 13 Jan 1999 Evolutionary Dynamics of the World Wide Web Bernardo A. Huberman and Lada A. Adamic Xerox Palo Alto Research Center Palo Alto, CA 94304 February
arxiv:cond-mat/9901071v2 [cond-mat.stat-mech] 13 Jan 1999 Evolutionary Dynamics of the World Wide Web Bernardo A. Huberman and Lada A. Adamic Xerox Palo Alto Research Center Palo Alto, CA 94304 February
Part 2: Models of Food-Web Structure
 Part 2: Models of Food-Web Structure Stochastic models of food-web structure -Two Parameters: S (species number) and C (connectance) -Randomly assign each species a niche value from to 1 -Use simple rules
Part 2: Models of Food-Web Structure Stochastic models of food-web structure -Two Parameters: S (species number) and C (connectance) -Randomly assign each species a niche value from to 1 -Use simple rules
Microbial analysis with STAMP
 Microbial analysis with STAMP Conor Meehan cmeehan@itg.be A quick aside on who I am Tangents already! Who I am A postdoc at the Institute of Tropical Medicine in Antwerp, Belgium Mycobacteria evolution
Microbial analysis with STAMP Conor Meehan cmeehan@itg.be A quick aside on who I am Tangents already! Who I am A postdoc at the Institute of Tropical Medicine in Antwerp, Belgium Mycobacteria evolution
Marine Resources Development Foundation/MarineLab Grades: 9, 10, 11, 12 States: AP Biology Course Description Subjects: Science
 Marine Resources Development Foundation/MarineLab Grades: 9, 10, 11, 12 States: AP Biology Course Description Subjects: Science Highlighted components are included in Tallahassee Museum s 2016 program
Marine Resources Development Foundation/MarineLab Grades: 9, 10, 11, 12 States: AP Biology Course Description Subjects: Science Highlighted components are included in Tallahassee Museum s 2016 program
Science Online Instructional Materials Correlation to the 2010 Biology Standards of Learning and Curriculum Framework
 and Curriculum Framework Provider York County School Divison Course Title Biology Last Updated 2010-11 Course Syllabus URL http://yorkcountyschools.org/virtuallearning/coursecatalog.aspx BIO.1 The student
and Curriculum Framework Provider York County School Divison Course Title Biology Last Updated 2010-11 Course Syllabus URL http://yorkcountyschools.org/virtuallearning/coursecatalog.aspx BIO.1 The student
ecological area-network relations: methodology Christopher Moore cs765: complex networks 16 November 2011
 ecological area-network relations: methodology Christopher Moore cs765: complex networks 16 November 2011 ecology: the study of the spatial and temporal patterns of the distribution and abundance of organisms,
ecological area-network relations: methodology Christopher Moore cs765: complex networks 16 November 2011 ecology: the study of the spatial and temporal patterns of the distribution and abundance of organisms,
Habitat loss and the disassembly of mutalistic networks
 Habitat loss and the disassembly of mutalistic networks Miguel A. Fortuna, Abhay Krishna and Jordi Bascompte M. A. Fortuna (fortuna@ebd.csic.es), A. Krishna and J. Bascompte, Integrative Ecology Group
Habitat loss and the disassembly of mutalistic networks Miguel A. Fortuna, Abhay Krishna and Jordi Bascompte M. A. Fortuna (fortuna@ebd.csic.es), A. Krishna and J. Bascompte, Integrative Ecology Group
Taxonomy and Clustering of SSU rrna Tags. Susan Huse Josephine Bay Paul Center August 5, 2013
 Taxonomy and Clustering of SSU rrna Tags Susan Huse Josephine Bay Paul Center August 5, 2013 Primary Methods of Taxonomic Assignment Bayesian Kmer Matching RDP http://rdp.cme.msu.edu Wang, et al (2007)
Taxonomy and Clustering of SSU rrna Tags Susan Huse Josephine Bay Paul Center August 5, 2013 Primary Methods of Taxonomic Assignment Bayesian Kmer Matching RDP http://rdp.cme.msu.edu Wang, et al (2007)
Package hierdiversity
 Version 0.1 Date 2015-03-11 Package hierdiversity March 20, 2015 Title Hierarchical Multiplicative Partitioning of Complex Phenotypes Author Zachary Marion, James Fordyce, and Benjamin Fitzpatrick Maintainer
Version 0.1 Date 2015-03-11 Package hierdiversity March 20, 2015 Title Hierarchical Multiplicative Partitioning of Complex Phenotypes Author Zachary Marion, James Fordyce, and Benjamin Fitzpatrick Maintainer
Community phylogenetics review/quiz
 Community phylogenetics review/quiz A. This pattern represents and is a consequent of. Most likely to observe this at phylogenetic scales. B. This pattern represents and is a consequent of. Most likely
Community phylogenetics review/quiz A. This pattern represents and is a consequent of. Most likely to observe this at phylogenetic scales. B. This pattern represents and is a consequent of. Most likely
