Comparative Mapping of Seed Dormancy Loci Between Tropical and Temperate Ecotypes of Weedy Rice (Oryza sativa L.)
|
|
|
- Bertram Morton
- 5 years ago
- Views:
Transcription
1 INVESTIGATION Comparative Mapping of Seed Dormancy Loci Between Tropical and Temperate Ecotypes of Weedy Rice (Oryza sativa L.) Lihua Zhang,*,1 Jieqiong Lou,*,1 Michael E. Foley, and Xing-You Gu*,2 *Agronomy, Horticulture, and Plant Science Department, South Dakota State University, Brookings, South Dakota and Northern Crop Sciences Laboratory, United States Department of Agriculture-Agricultural Research Service, Fargo, North Dakota ABSTRACT Genotypic variation at multiple loci for seed dormancy (SD) contributes to plant adaptation to diverse ecosystems. Weedy rice (Oryza sativa) was used as a model to address the similarity of SD genes between distinct ecotypes. A total of 12 quantitative trait loci (QTL) for SD were identified in one primary and two advanced backcross (BC) populations derived from a temperate ecotype of weedy rice (34.3 N Lat.). Nine (75%) of the 12 loci were mapped to the same positions as those identified from a tropical ecotype of weedy rice (7.1 N Lat.). The high similarity suggested that the majority of SD genes were conserved during the ecotype differentiation. These common loci are largely those collocated/linked with the awn, hull color, pericarp color, or plant height loci. Phenotypic correlations observed in the populations support the notion that indirect selections for the wild-type morphological characteristics, together with direct selections for germination time, were major factors influencing allelic distributions of SD genes across ecotypes. Indirect selections for crop-mimic traits (e.g., plant height and flowering time) could also alter allelic frequencies for some SD genes in agroecosystems. In addition, 3 of the 12 loci were collocated with segregation distortion loci, indicating that some gametophyte development genes could also influence the genetic equilibria of SD loci in hybrid populations. The SD genes with a major effect on germination across ecotypes could be used as silencing targets to develop transgene mitigation (TM) strategies to reduce the risk of gene flow from genetically modified crops into weed/wild relatives. KEYWORDS seed dormancy weed comparative genomics quantitative trait locus segregation distortion Weeds are unwanted plants that have adapted to agroecosystems and compete with crop cultivars (Harlan 1965; Booth et al. 2003). Seed dormancy (SD) plays a critical role in the adaptation. Weed seeds, usually dormant upon maturation, may survive in the soil for months to years, depending on genotypes and environments. Presumably, the genotypic differentiation of SD in a species occurred at multiple loci during Copyright 2017 Zhang et al. doi: Manuscript received February 13, 2017; accepted for publication June 2, 2017; published Early Online June 6, This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License ( licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Supplemental material is available online at doi: /g /-/dc1. 1 These authors contributed equally to this work. 2 Corresponding author: Agronomy, Horticulture, and Plant Science Department, South Dakota State University, SNP 248B, Box 2140C, 1390 College Ave., Brookings, SD xingyou.gu@sdstate.edu evolution before specific populations adapted to agroecosystems as weeds, given the relatively short history of domestication for major crops (,9000 yr; Chopra and Prakash 2002). Thus, it is important to know about the degree of similarity in SD genes between distinct ecotypes and factors influencing their genotypic/allelic frequencies. This information may help design new weed management strategies. We selected weedy rice as a model system to address the ecological genetic issues in this research. Weedy rice refers to various forms of plants that belong to the Oryza genus and infest rice fields from tropical to temperate areas (Oka 1988; Delouche et al. 2007). The rice Oryza sativa was domesticated from the wild ancestor (O. rufipogon Griff.) and differentiated into the indica and japonica subspecies that are distributed across tropical/subtropical and temperate areas, respectively (Khush and Brar 2002). The origin of the conspecific weedy rice was associated with the domestication and subspeciation processes. For example, weedy rice populations can be indicaor japonica-like. The indica-like populations in tropical areas (tropical ecotypes) could originate from natural variants of the wild ancestor, or Volume 7 August
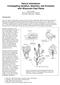
2 Figure 1 A framework linkage map and positions of seed dormancy QTL identified in this and previous research. The map was marked with RM loci segregating in the BC 1 F 1 EM93-1//EM93-1/LD population. Asterisks indicate segregation distortion loci in favor of the allele from EM93-1 ( )orld( ). Black bars indicate C.I. (equivalent to one-lod support lengths) of qsd, qan, or qhc detected in this BC 1 F 1 population (Figure 2B). Five-point stars indicate qsds detected in the BC 2 F 1 (9) (Figure 3) and/or BC 2 F 1 (139) (Figure 4), but not in this BC 1 F 1 population. Ovals indicate approximate positions of qsds previously detected in the BC 1 F 1 EM93-1//EM93-1/SS18-2 population or its advanced generations (Gu et al. 2004; Ye et al. 2010). LD and SS18-2 are temperate and tropical ecotypes of weedy rice, respectively. Ch, chromosome; LD, pure line LüDao; LOD, logarithm of the odds; qan, QTL for awn; qhc, QTL for hull color; qsd, QTL for seed dormancy; QTL, quantitative trait loci; RM, rice microsatellite. from hybrids between wild and cultivated rice. The japonica-like populations in temperate areas (temperate ecotypes) that are historically absent of wild rice may originate from old/extinct cultivars, or hybrids between indica and japonica cultivars (Suh et al. 1997; Tang and Morishima 1997). Despite the ecotype differentiation, weedy rice populations, particularly those adapted to an ecosystem for a long period, usually have strong SD and some other wild-type characteristics (Oka 1988; Delouche et al. 2007). The phenotypic similarity between distinct ecotypes could arise from the same or different sets of genes, depending on the relatedness of weed populations and the coevolutionary relationship between SD and the other adaptive or domestication-related traits in local ecosystems. We used a comparative mapping approach to infer the differentiation at QTL for SD (qsd) between temperate and tropical ecotypes of weedy rice. Wild and weedy rice are divergent from cultivated rice in SD, as evaluated under controlled environment conditions (Veasey et al. 2004; Gu et al. 2005a). Several lines of wild (O. rufipogan or O. nivara)/weedy rice were crossed with cultivars to identify QTL associated with domestication-related traits, including SD or germination capacity (Cai and Morishima 2000; Thomson et al. 2003; Gu et al. 2005b; Lee et al. 2005; Li et al. 2006; Jing et al. 2008; Subudhi et al. 2012; Mispan et al. 2013). The number of reported SD QTL varied with mapping populations or environments (years) from a few to 20. Some of them remain to be confirmed because several factors in a distant cross, such as partial sterility or low seed set, segregation distortion, and seed shattering, could have a negative impact on the QTL analysis (Cai and Morishima 2000). In the previous research, we identified 10 SD QTL in a primary and advanced BC populations derived from a tropical ecotype of weedy rice (Gu et al. 2004; Ye et al. 2010). All of these 10 loci have been confirmed, and some of them have been cloned. The objectives of this research were to: (1) identify SD QTL for a temperate ecotype of weedy rice and (2) compare these loci with those mapped for the tropical ecotype to infer shared genetic and evolutionary mechanisms underlying the adaptive trait. MATERIALS AND METHODS Parental lines and mapping populations Two ecotypes of weedy rice: The pure lines, LD and SS18-2, were selected from the previous research (Gu et al. 2005a) to represent the temperate and tropical ecotypes of weedy rice, respectively. LD was purified from LüDao (in Chinese), a population of volunteer rice historically present in the Lianyungang area ( N Lat.) of East China (Jiang et al. 1985). This population was similar to some local landraces (O. sativa ssp. japonica) in plant type and seed (spikelet) morphology (black hull and red pericarp colors, long awn, and medium grain), but different from the old landraces in seed shattering and dormancy (Jiang et al. 1985). SS18-2 was purified from SS18, a population of weedy rice from the Songkla (7.18 N Lat.) area of Southern Thailand (Tang and Morishima 1997), and is similar to LD in seed morphology and dormancy (Supplemental Material, Table S1 in File S1). Despite the phenotypic similarity, there was no direct relationship 2606 L. Zhang et al.
3 n Table 1 Summary of segregation distortion loci linked to seed dormancy QTL in the BC 1 F 1 (EM93-1//EM93-1/LD) and BC 2 F 1 populations Locus (QTL) Chr Population Number of Plants Genotypic (Allelic) Frequency Homozygote (Allele from EM93-1) Heterozygote (Allele from LD) Chi-Square Value a RM176 (qsd6-3) 6 BC 1 F BC 2 F 1 (9) BC 2 F 1 (139) RM21197 (qsd7-1) 7 BC 1 F BC 2 F 1 (9) BC 2 F 1 (139) b RM28607 (qsd12) 12 BC 1 F BC 2 F 1 (9) QTL, quantitative trait loci; Chr, chromosome; LD, pure line LüDao. a Significance of the deviation from the 1:1 expectation at probability levels of P, 0.05, P, 0.01, or P, b This population was segregating for a short segment containing qsd7-1/ RM21197 on Chr 7 (Figure 4A) that may not encompass the segregation distortion locus. in origin between these two geographically isolated weed populations. Based on diagnostic characteristics and isozyme markers, LD and SS18 were classified into the japonica-andindica-like groups of weedy rice, respectively (Tang and Morishima 1997). Recurrent parent and BC populations: EM93-1, an early maturation semidwarf indica line (Ye et al. 2013), was used as the female, recurrent parent to develop BC populations. The BC 1 F 1 EM93-1//EM93-1/LD population, which had been previously evaluated for phenotypic correlations between seed-related traits (Gu et al. 2005a), was used to scan for QTL along the LD genome. In addition, two BC 1 F 1 plants (#9 and 139), which were similar to EM93-1 in flowering time, were selected to develop the BC 2 F 1 (9) EM93-1/BC 1 F 1 plant #9 and BC 2 F 1 (139) EM93-1/BC 1 F 1 plant #139 populations. The BC 2 F 1 (9) and (139) populations were used to confirm the detected QTL and to identify additional loci whose effects on germination may have been masked by some major genes segregating in the BC 1 F 1 population (Ye et al. 2010). Plant cultivation, and seed harvesting and storage The BC 1 F 1 and BC 2 F 1 populations were grown in greenhouses in different years. To capture all available genotypes in a mapping population, hybrid seeds were air-dried to break dormancy, germinated at 30 and 100% relative humidity for 5 d, and cultured with a nutrient solution (Yoshida et al. 1976) for 2 wk. Seedlings were transplanted into pots (28 cm diameter 25 cm height), with one plant per pot, and filled with a mixture of clay soil and Sunshine #1 medium (Sun Gro Horticulture). Greenhouse temperatures were set at 29/21 for day/night, and the day-lengths were natural, except from the 6th to 8th wk when a 10-hr (8:00 18:00) short-day treatment was used to synchronize flowering. Plants were tagged for flowering dates when the first panicle in a plant emerged from the leaf sheath. Panicles were covered with white pollination bags at 10 d after flowering and the bags fixed to bamboo poles to prevent shattering due to brushing or shaking the plant. Seeds were harvested at 40 d after flowering, air-dried in the greenhouse for 3 d, and stored in a freezer (220 )tomaintainthestatus of dormancy developed on the plant (i.e., primary dormancy). Phenotypic identifications for SD and morphologies SD: The primary dormancy was evaluated by germination percentage for both seeds and caryopses from the BC 1 F 1 and for seeds from the BC 2 F 1 populations. A seed in grass species usually refers to a dispersal unit, which consists of the seed component (embryo, endosperm, and testa) and covering (pericarp and hull, or lemma and palea) tissues, whereas a caryopsis is a hull-removed seed enclosed by the pericarp. To evaluate seed germination, after-ripening (AR) treatments were used to release part of the primary dormancy to better display genotypic variation on the percentage scale. Briefly, seeds from each plant were allocated into three or four sets and stored in a lab room (24 25 ) fora series of 7 or 10 d intervals to obtain various degrees of partially AR samples. Caryopses were prepared by hand removal of the hull from non-ar seeds. About 50 seeds/caryopses were distributed in a 9 cm petri dish, which was lined with a filter paper and wetted with 8 ml deionized water. A germination experiment was replicated three times in an incubator set for 30, 100% relative humidity, and dark conditions. Germinated seeds (radicle protrusion. 3 mm) were counted at day 7. Awn: The BC 2 F 1 populations were evaluated for the morphological traits awn, hull color, and pericarp color, to confirm their correlations with SD in the BC 1 F 1 EM93-1//EM93-1/LD population (Gu et al. 2005a). An awn is a needle-like appendage extended from the terminal end of a lemma and functions in aiding seed dispersal or movement into wet soil. The awn trait varies in length with plants, as well as with seeds on a panicle, in a segregating population. Thus, the trait was quantified by the mean awn length, and the percentage of seeds with an awn, in a random sample of.50 seeds from a BC 2 F 1 plant. Hull color: This trait was measured with the ChromaMeter Minolta CR310, which transfers reflectance spectra into the L,a,andb readings to quantify blackness, redness, and yellowness, respectively. The L readings range from 0 to 100, with 0 and 100 indicating completely nonreflective (black) and perfectly reflective (white), respectively. The a readings vary from 2100 to 100, with negative and positive values indicating greenness and redness, respectively. The b readings also vary from 2100 to 100, with negative and positive values indicating blueness and yellowness, respectively. The reflectance spectra were measured using 100 seeds in a 6 cm petri dish on a dark background, and means of three independent readings for each of the spectra used for data analysis. Pericarp color: This trait was visually scored as red/brown (1) or white (0) for correlation analysis. This was partly because most BC 2 F 1 plants had an insufficient amount of seeds to prepare caryopses for the reflection spectrum measurement after the replicated germination tests. In addition, the pigment trait is controlled by the gene Rc encoding a bhlh familiar transcription factor in rice (Sweeney et al. 2006; Furukawa et al. 2007). This regulatory gene is also one of the QTL for SD (i.e., qsd7-1) and its functional alleles are present in tropical and temperate ecotypes of weedy red rice, including LD and SS18-2, to control maternal tissue-imposed dormancy (Gu et al. 2011). Volume 7 August 2017 Comparative Study on Seed Dormancy 2607
4 Figure 2 Genome-wide scan for seed dormancy QTL in the BC 1 F 1 EM93-1//EM93-1/LD population. (A) Frequency distributions of percent germination for seeds or caryopses. N was the number of plants evaluated for germination at DAR. (B) Distributions of LR along the 12 Ch. Seed dormancy QTL (qsd) were inferred by peaks of the LR distributions above the threshold. Ch, chromosome; DAR, days after ripening; LR, likelihood ratio; qsd, QTL for seed dormancy; QTL, quantitative trait loci. Marker genotyping and map construction Fresh leaves were used to prepare genomic DNA samples for marker genotyping. More than 300 rice microsatellite markers from the 12 chromosomes (Chrs) of rice (McCouch et al. 2002), including all of those used to map the SS18-2 genome (Gu et al. 2004), were screened for polymorphism between EM93-1 and LD. Information on the markers (primer sequences, repeat motifs, and genomic positions) is available in the Gramene database ( org/markers/microsat/). DNA extraction, marker amplification by polymerase chain reaction (PCR), and PCR product display by electrophoresis in a 6% nondenatured polyacrylamide gel were performed using the previously described methods (Gu et al. 2004). Marker genotypes were scored using the AlphaEaseFC (Alpha Innotech) gel imaging system. Polymorphic markers with a size difference suitable to score were used to genotype the BC 1 F 1 s to develop a linkage map covering the weedy rice genome. Markers on the Chr or Chr segments heterozygous for the BC 1 F 1 plants #9 and #139 were used to genotype the BC 2 F 1 (9) and BC 2 F 1 (139) populations, respectively, to develop partial linkage maps. Linkage maps were constructed using MAPMAKER/EXP 3.0 (Lincoln et al. 1992). Map distances in centimorgan (cm) were converted from recombination fractions using the Kosambi mapping function. Markers were grouped at the LOD score of 3.0 and the maximum distance of 50 cm (equivalent to 0.38 recombination fraction). Linkage groups were assigned to the 12 Chrs based on markers physical positions (McCouch et al. 2002). Orders of closely linked (a few cms) markers were also checked for physical positions on the Nipponbare genome sequence (International Rice Genome Sequencing Project 2005). Data and QTL analysis Germination data from the BC 1 F 1 population were used to infer correlations for the degree of dormancy between seeds and caryopses and between the AR treatments. Data from the BC 2 F 1 populations were used to estimate correlations of SD with each of the morphological traits. Linear correlation analysis was performed using the SAS CORR program. For QTL analysis, germination data (x) were transformed by sin 21 (x) 20.5 to improve the normality. The analysis was performed using Windows QTL Cartographer V2.5_011 (Wang et al. 2012). The interval mapping program was used to scan for QTL at 1 cm walking speed and 1000 permutations at 5% error rate. The composite interval mapping program was used to define QTL peak positions and C.I. (one-lod support regions), and to estimate the effects of the mapped loci and their contributions to the phenotypic variances (R 2 ). Data availability Data for the origin and differentiation of the parental lines and data for trait correlations in the BC populations are available as Tables S1 S3 in File S1. Phenotypic and genotypic datasets from the mapping populations are available upon request. RESULTS Genetic differentiation and linkage map F 1 plants from the EM93-1/LD cross had 65% seed set, which was 30% lower than the seed set rate for F 1 EM93-1/SS18-2 plants (Gu et al. 2004). Partial sterility is a characteristic of hybrid F 1 s from an intersubspecific cross in rice (Oka 1988). The common parent EM93-1 in the two crosses is an indica line. The lower seed set rate in the EM93-1/ LD cross supports that LD is japonica-like (Tang and Morishima 1997) and genetic differentiation between the parents is greater than the level for an introsubspecific cross. DNA polymorphism between EM93-1 and LD was 60%, based on 256 markers amplified for alleles of predicted molecular sizes. Based on the BC 1 F 1 EM93-1//EM93-1/LD (LD-BC 1 F 1 ) population, a linkage map (Figure 1) was constructed using 139 markers, with 0.027% missing values. This map covered a total of 1650 cm for the 12 Chrs, with 2608 L. Zhang et al.
5 n Table 2 Summary of QTL for seed dormancy (qsd) identified in the BC 1 F 1 (EM93-1//EM93-1/LD) and BC 2 F 1 populations QTL Chr Peak (cm) a LR b R 2b Effect c Germination c Population qsd1-1 1 RM220 (4) DAR BC 2 F 1 (139) qsd1-2 1 RM212 (22) DAR BC 1 F 1 RM212 (22) DAR qsd4 4 RM3839 (3) DAR BC 1 F 1 RM3839 (2) DAR RM3839 (1) DAR qsd6-1 6 RM314 (7) DAR BC 1 F 1 RM314 (8) DAR RM314 (14) DAR RM557 (0) DAR BC 2 F 1 (139) qsd6-2 6 RM587 (0) DAR BC 2 F 1 (139) qsd6-3 6 RM528 (16) DAR BC 2 F 1 (9) RM528 (13) DAR RM528 (23) DAR BC 2 F 1 (139) RM528 (6) DAR qsd7-1 7 RM21197 (0) DAR BC 1 F 1 RM21197 (0) DAR RM21197 (1) DAR RM21197 (0) DAR RM21197 (21) Caryopsis RM21197 (0) DAR BC 2 F 1 (9) RM21197 (2) DAR RM21197 (0) DAR BC 2 F 1 (139) RM21197 (21) DAR qsd7-2 7 RM505 (1) DAR BC 2 F 1 (9) RM505 (4) DAR qsd8 8 RM339 (21) DAR BC 1 F 1 RM339 (22) DAR RM339 (20) DAR RM339 (22) DAR BC 2 F 1 (9) qsd9 9 RM524 (21) DAR BC 1 F 1 RM524 (23) DAR qsd10 10 RM244 (22) Caryopsis BC 1 F 1 RM244 (3) DAR BC 2 F1 (139) qsd12 12 RM28607 (22) DAR BC 1 F 1 RM28607 (24) DAR RM28607 (3) DAR RM28607 (5) DAR BC 2 F 1 (9) QTL, quantitative trait loci; Chr, chromosome; LR, likelihood ratio; DAR, days after ripening; qsd, QTL for seed dormancy. a Number in the parentheses is the genetic distance of the peak located above (2) or below the marker on the Chr or Chr segment in Figure 1, Figure 3A, or Figure 4A. b LR and proportion of the variance explained by the QTL (R2 ). c Difference between the heterozygous and homozygous genotypes at the locus in arcsine-transformed percent germination for intact seeds at DAR or for caryopsis. the mean intermarker distance being 12.9 (6 8.9) cm. About 45% of the 139 markers were also located on the linkage map constructed using the BC 1 F 1 EM93-1//EM93-1/SS18-2 (SS18-BC 1 F 1 ) population (Gu et al. 2004). The total genetic distance was 250 cm shorter for the LD than for the SS18-2 genome in the EM93-1 background. Segregation distortion was observed for some or all of the markers on Chrs 6, 7, and 12 in the LD-BC 1 F 1 population (Figure 1). The segregation ratio was biased against heterozygotes for the markers on Chrs 7 and 12, but toward the heterozygote for RM176 located near the end of the long arm of Chr 6 (Table 1). Because the BC 1 F 1 population was developed using the F 1 EM93-1/LD plants as the male parent, the distortions must be caused by functionally differentiated genes for male gametophyte development or pollination of the hybrid (sporophyte). Such sporo-gametophytic interaction genes were associated with partial sterility of hybrids from distant crosses in the O. sativa complex (Oka 1988). In contrast, a segregation distortion was not detected for any of the loci on Chrs 6 and 7, and the markers distal to the major SD QTL qsd12, in the SS18-BC 1 F 1 population (Gu et al. 2004). QTL associated with SD in the BC 1 F 1 population The frequency distribution pattern of percent germination varied with seeds, caryopses, or days of AR (DAR) in the LD-BC 1 F 1 population (Figure 2A). Correlations (r) of seed germination between any two of the 1, 11, 21, and 31 DAR treatments were significant (Table S2 in File S1), but coefficients of determination were relatively low (R 2 = ). Similarly, the degree of dormancy between seeds and caryopses was positively correlated, but the R 2 values ( ) were even lower than the estimates for seeds at the different DAR (Table S2 in File S1). Therefore, all these measurements were used to detect qsd. A total of eight qsds were detected in the population (Figure 2B). Of them, qsd7-1 was the only one whose effect could be detected by seeds at 1, 11, 21, and 31 DAR. This major QTL contributed more to the variance in germinability for caryopses (R 2 = 0.27) than for seeds (R 2 = ), and was collocated with Rc. The collocation accounted for the phenotypic correlation between the dormancy and pericarp color traits (Gu et al. 2005a). The remaining loci were associated with one to three of the five measurements (Table 2). LD and EM93-1 contribute Volume 7 August 2017 Comparative Study on Seed Dormancy 2609
6 Figure 3 Mapping of seed dormancy QTL in the BC 2 F 1 (9) population. (A) A partial linkage map. The map was constructed with markers on 10 Ch or Ch segments segregating in the population. Black bars indicate one-lod support lengths for qsd, qan, or qhc. Ovals indicate positions of qsds previously detected in the BC 1 F 1 EM93-1//EM93-1/SS18-2 population (Gu et al. 2004). (B) Frequency distributions of percent germination. N was the number of BC 2 F 1 plants evaluated at 7, 14, or 21 DAR. (C) Distributions of LRs along the map. qsds were inferred by peaks of the LR distributions above the threshold. Ch, chromosome; DAR, days of after-ripening; LOD, logarithm of the odds; LR, likelihood ratio; qan, QTL for awn; qhc, QTL for hull color; qsd, QTL for seed dormancy; QTL, quantitative trait loci. the dormancy-enhancing allele to seven and one (qsd1-2) of the eight QTL, respectively. QTL associated with SD in the BC 2 F 1 (9) population BC 1 F 1 plant #9 is heterozygous for Chr 9 and part of the others except Chrs 5 and 10, while the remainder of the plant genome was synchronized by EM93-1. The total length of the heterozygous regions on the 10 Chrs is 600 cm, as estimated based on the BC 2 F 1 (9) population of 130 plants (Figure 3A). The heterozygous regions cover peak-containing (one-lod support) intervals for qsd7-1, 8, 9,and12 detected in the BC 1 F 1 population. Phenotypic variation for SD at 7, 14, and 21 DAR (Figure 3B), and segregating distortion for the three loci (Table 1), were observed in the BC 2 F 1 (9) population. A total of five qsds, including qsd7-1, 8, and 12, but not qsd9, were detected in the advanced BC population (Figure 3C). The new locus qsd6-3 was located on the RM interval near the end of the long arm of Chr 6, and has the dormancy-enhancing allele from LD (Table 3). The other new locus, qsd7-2, is the second QTL on Chr 7 and has the dormancyenhancing allele from EM93-1. qsd12 accounted for 67% of the variance in germination percentage at21darwheneffectsoftheotherqtlwerenotsignificant in the BC 2 F 1 (9) population (Table 2). However, qsd12 seffectwasnotsignificant at 7 and 14 DAR when the others were detectable. These results suggest that qsd12 maintained an inhibitory effect on germination longer than the other SD QTL. In addition, the severe segregation distortion for the qsd12-containing region, which greatly reduced the genotypic frequency for heterozygotes (6%) in the BC 2 F 1 population (Table 1), must alsolowerthepowertoevaluatetheqtl s effect on germination at an early stage of AR. QTL associated with SD in the BC 2 F 1 (139) population The BC 1 F 1 plant #139 is heterozygous for Chr 6 and part of the others except Chrs 9 and 12, while the remainder of the plant genome was synchronized by EM93-1. The total length of the heterozygous regions on the 10 Chrs is 550 cm, as estimated based on the BC 2 F 1 (139) population of 151 plants (Figure 4A). The heterozygous regions cover peak-containing intervals of qsd4, 6-1, 7-1, 8, and 10 detected in the BC 1 F 1,andqSD6-3 detected in the BC 2 F 1 (9) population. Phenotypic variation for SD at 7, 14, and 21 DAR (Figure 4B), and segregation distortion for RM176 on Chr 6 (Table 1), were observed in this population. A total of six qsds, including qsd6-1, 6-3, 7-1, and10 were detected (Figure 4C), but qsd4 and 8 were not significant in the advanced BC population. Two new loci (qsd1-1 and 6-2) were identified in the BC 2 F 1 population and both have the dormancy-enhancing allele from LD (Table 2). Of the three QTL on Chr 6, qsd6-3 contributed most to the phenotypic variance (Table 2). The BC 2 F 1 (139) population segregated on Chr 7 for a segment of 30 cm encompassing qsd7-1 and its flanking markers from RM481 to RM5481 (Figure 4A). However, segregation distortion was not detected for these markers, including RM21197 located within the qsd7-1 underlying gene (Gu et al. 2011). This result indicates that the gene responsible for the segregation distortion in the BC 1 F 1 and BC 2 F 1 (9) populations locates outside the 30 cm segment L. Zhang et al.
7 n Table 3 Summary of QTL for the awn and hull color traits identified in the BC 1 F 1 (EM93-1//EM93-1/LD) and BC 2 F 1 populations QTL Chr Peak (cm) a LR b R 2b Effect c Measurement d Population Awn qan4-1 4 RM185 (21) % Awned seeds BC 1 F 1 RM185 (3) % Awned seeds BC 2 F 1 (9) RM185 (0) Awn length RM5979 (1) % Awned seeds BC 2 F 1 (139) RM5979 (1) Awn length qan8 8 RM23292 (0) % Awned seeds BC 1 F 1 RM23292 (217) % Awned seeds BC 2 F 1 (9) RM23292 (219) Awn length RM23292 (28) % Awned seeds BC 2 F 1 (139) RM23292 (21) Awn length Hull color qhc2 2 RM526 (3) L (Blackness) BC 2 F 1 (9) a (Redness) qhc4 4 RM252 (0) Visual score BC 1 F 1 RM252 (0) L (Blackness) BC 2 F 1 (9) RM252 (0) a (Redness) RM252 (0) L (Blackness) BC 2 F 1 (139) RM252 (0) a (Redness) RM252 (0) b (Yellowness) qhc7 7 RM5481 (1) a (Redness) QTL, quantitative trait loci; Chr, chromosome; LR, likelihood ratio. a Number in the parentheses is the genetic distance of the peak located above (2) or below the marker on the Chr or Chr segment in Figure 1, Figure 3A, or Figure 4A. b LR and proportion of the variance explained by the QTL (R2 ). c Difference between the heterozygous and homozygous genotypes in the trait value. d The trait awn was measured by the percentage of seeds with awn and the mean awn length for seeds from a plant; and the hull color was measured by visual scores (dark vs. straw) for the BC 1 F 1 population and by reflection spectrum readings for darkness (L ), red redness (a ), and yellowness (b ) for the BC 2 F 1 population. QTL associated with the seed morphological traits Phenotypic variation for each of the morphological traits and their correlations with SD were observed in the two BC 2 F 1 populations, with the presence of awn, dark pigment on the hull, or red pigment on the pericarp tissue tending to reduce germination percentage (Table S3 in File S1). The phenotypic correlations were similar to those observed in the three BC 1 F 1 populations derived from different lines of weedy rice, including LD and SS18-2 (Gu et al. 2005a). Two awn QTL (qan4-1 and 8) were detected in each of the LD-BC 1 F 1 and two BC 2 F 1 populations (Table 3). In the BC 2 F 1 populations, the contribution of qan8 to the phenotypic variance (R 2 ) was three to four times greater for the percentage of awned seeds than for awn length, while qan4-1 contributed almost equally to the two measurements. qan4-1 and 8 were linked to but not collocated with qsd4 and 8, respectively (Figure 1). AmajorQTL(qHC4) and two modifiers (qhc 2 and 7) were associated with hull color (Table 3). qhc4 was detected in all of the three populations and contributed most to phenotypic variances in the visual score and component reflection spectra. This major QTL was collocated with qsd4 (Figure 1). The modifiers qhc2 and 7 were detected in the BC 2 F 1 #9 and #139 populations, respectively. qhc7 contributed 8% to the phenotypic variance for the red reflectance only and was collocated with qsd7-1/rc (Figure 4A). It is likely that the modifier qhc7 could be the Rc locus, which was associated with the visual score for the pericarp color in the two BC 2 F 1 populations. This is because the red pigment can be seen on intact straw hull-colored seeds. DISCUSSION Similarity of SD genes between distinct ecotypes of weedy rice A total of 12 SD QTL were identified from the primary and advanced BC populations developed using LD as the nonrecurrent parent. Two-thirds (eight) of these loci were detected from the BC 1 F 1 population, and the remaining 1/3 identified from the BC 2 F 1 populations where 65% of the genome was synchronized by the recurrent parent EM93-1. The temperate ecotype line LD has dormancy-enhancing alleles at 10 (83%) of the 12 loci. The estimate of 83% is close to the previous observation that the tropical ecotype line SS18-2 has dormancy-enhancing alleles at 80% of the 10 QTL detected in the EM93-1 background (Ye et al. 2010). The SD QTL identifiedfromthe populationswithld orss18-2 as the nonrecurrent parent represent a majority of the reported loci differentiated between wild/weedy and cultivated rice in regard to approximate map positions. For example, qsd6-1, 2,and3 are similar to those on Chr 6 reported for wild (Cai and Morishima 2000) and weedy (Jing et al. 2008) rice; qsd4, 7-1, and7-2 were located on the same marker intervals as the three QTL reported for the three accessions of weedy rice from USA (Subudhi et al. 2012; Mispan et al. 2013); and qsd1-1 (Figure 1A) and sd1 reported for wild rice (Li et al. 2006) were both mapped on the top of Chr 1. However, some loci reported by the other groups, such as qsd-2 (Jing et al. 2008), qsd3 (Subudhi et al. 2012), and sd12 (Li et al. 2006), were not detected our research. It is possible that some SD genes could have been eliminated from founders of the LD and SS18 populations, or lost during evolution of the weed ecotypes. The SS18 (7 N) and LD (34 N) populations acquired a similar level of SD in geographically isolated ecosystems because they share most genes for the adaptive trait. There are nine common loci (qsd1-1, 1-2, 4, 6-1, 7-1, 7-2, 8, 10, and 12) that are functionally differentiated for SD in both EM93-1/SS18-2 and EM93-1/LD crosses. It is estimated that the tropical and temperate ecotypes are similar in genotype for 75% of the 12 SD loci, if multiple alleles (more than two at a locus) are ignored. The estimated degree of similarity is similar to the report for the dicot model Arabidopsis thaliana (Bentsink et al. 2010). For example, of a total of 11 SD QTL identified for six ecotypes in the Landsberg erecta background, nine (82%) had an effect on delay of germination in two or Volume 7 August 2017 Comparative Study on Seed Dormancy 2611
8 Figure 4 Mapping of qsd in the BC 2 F 1 (139) population. (A) A partial linkage map. The map was constructed with markers on 10 Ch or Ch segments segregating in the population. Black bars indicate one-lod support intervals for qsd, qan, or qhc. Ovals indicate positions of qsds previously detected in the BC 1 F 1 EM93-1//EM93-1/SS18-2 population (Gu et al. 2004, 2005b; Ye et al. 2010). (B) Frequency distributions of percent germination. N was the number of BC 2 F 1 plants evaluated at 7, 14, or 21 DAR. (C) Distributions of LRs along the map. qsds were inferred by peaks of the LR distributions above the threshold. Ch, chromosome; DAR, days after ripening; LOD, logarithm of the odds; LR, likelihood ratio; qan, QTL for awn; qhc, QTL for hull color; qsd, QTL for seed dormancy; QTL, quantitative trait loci. more of the Arabidopsis populations (Bentsink et al. 2010). Thus, the comparative mapping results from the monocot and dicot models strongly suggest that naturally occurring genes controlling SD in a species were highly conserved during evolution. Evolutionary mechanisms of SD The adaptive significance of SD relies on functionally differentiated alleles at multiple loci to regulate the time of germination in local ecosystems. This and the previous research in weedy rice revealed several mechanisms involved in regulating genotypic/allelic frequencies at SD loci in both natural and agricultural ecosystems. The first mechanism is direct selection for the time of germination, which is critical for locally adapted genotypes to complete their life cycle. An extreme example is the domestication of cereal crops by artificial selection for rapid germination mutants (Harlan 1965). The second mechanism is indirect selection for wild-type characteristics correlated with SD. A phenotypic selection for the presence of awn, dark hull, or red pericarp tended to enhance SD (Table S3 in File S1). The indirect selections retained the dormancyenhancing alleles at the loci (e.g., qsd4, 7-1, and 8) linked to or collocated with the genes for the interrelated traits (Figure 1; Gu et al. 2005b; Mispan et al. 2013). Some of the linkage drags or collocations could be pleiotropic effects of single genes. For example, qsd7-1 and Rc are underlain by the same transcription factor gene (Os07g1120) regulating both abscisic acid (a dormancy-inducing hormone) and flavonoid (red pigments) biosynthesis pathways in early developing seeds (Gu et al. 2011). The indirect selections explain why SD is generally stronger in black-hulled awned red rice populations than in those without the wild-type characteristics (Delouche et al. 2007). Genome-wide phylogenetic analyses revealed that the hull color, pericarp color, and awn gene-containing regions were intensively selected during domestication and are informative for research on origins of weedy rice (Qiu et al. 2014; Kanapeckas et al. 2016; Li et al. 2017). The third mechanism is indirect selection for crop-mimic traits, such as plant height and flowering time. LD contains dormancy-reducing alleles at qsd1-2 and qsd7-2. Both loci also have an effect on plant height, when the QTL alleles were introduced from SS18-2 into the EM93-1 background (Ye et al. 2013). qsd1-2 was cloned as semidwarf1 (sd1), a major gene for plant height. The semidwarf line EM93-1 carries a dormancy-enhancing allele, while a vast majority of wild/weedy rice lines (including LD and SS18-2) have a dormancy-reducing allele at qsd1-2/sd1 (Ye et al. 2015). A high frequency of the dormancyreducing allele in the nondomesticated germplasm is indicative that the natural selection on such a pleiotropic gene has a greater impact on plant height than on dormancy. Collocation was also reported for the SD/heading date QTL on Chr 3 (Sdr1/Hd8; Takeuchi et al. 2003) and 6(qSD-6-2/qHD-6; Jing et al. 2008), with the dormancy-enhancing alleles delaying flowering. Thus, correlational selections for crop-mimic traits may not favor the retention of a dormancy-enhancing allele, but could contribute to genetic diversity in germination capacity. The other mechanism was inferred by the three segregation distortion loci (SDL) linked to (qsd7-1) or collocated with (qsd6-3 and 12) an SD locus. The segregation distortion favored a transmission of the dormancy-reducing alleles at qsd7-1 and 12, or the dormancy-enhancing allele at qsd6-3, through gametes produced by heterozygotes from 2612 L. Zhang et al.
9 the EM93-1/LD cross (Table 1). A similar pattern of segregation distortion was also observed for a qsd12-containing region, when it was heterozygous for the alleles from SS18-2 and EM93-1; this SDL had a larger effect on eliminating the dormancy-enhancing allele through the male than through the female gametes (Gu et al. 2015). Natural hybridization occurs between weedy and cultivated rice at a low rate (Oka 1988; Delouche et al. 2007). Thus, gametophyte development genes that cause segregation distortions in hybrid populations could also influence allelic frequencies for some SD loci, but the influence varies depending on crosses. Possible applications of SD genes In addition to understanding the origins of conspecific/congeneric weeds in agroecosystems (Qiu et al. 2014; Kanapeckas et al. 2016; Li et al. 2017), SD genes conserved across ecotypes could be manipulated to develop a TM strategy. The TM strategy was proposed to complement transgene containment techniques to reduce the risk of gene flow from genetically modified (GM) crops to wild relatives (Gressel 1999). The basic concept is a built-in linkage between a fitness-enhancing transgene (e.g., herbicide resistance) and a mitigating factor (e.g., reduced SD), which has no negative effect on the GM crop but could reduce the adaptability of weed/crop hybrids to lower the transgene s frequency in a weed population across generations (Gressel 1999). Silencing SD genes could promote germination uniformity (as for cereal cultivars) and make weeds relatively easy to eliminate by agronomic practices. We are using the qsd7-1 and 12 underlying genes as silencing targets and the RNA interference and genome editing techniques to prove the TM concept in weedy red rice. ACKNOWLEDGMENTS We thank T. Nilsson, Y. Wang, and B. Carsrud for technical assistance. Funding for this research was supported by grants from the Natural Science Foundation (IOS and ), the United States Department of Agriculture-National Research Initiative ( ), the National Institute of Food and Agriculture ( ), and the American Seed Research Foundation. LITERATURE CITED Bentsink, L., J. Hanson, C. J. Hanhart, H. Blankestijn-de Vries, C. Coltrane et al., 2010 Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. USA 107: Booth, B. D., S. D. Murphy, and C. J. Swanton, 2003 Weed Ecology in Natural and Agricultural Systems. Center for Agriculture and Bioscience International Publishing, Wallingford, UK. Cai, H. W., and H. Morishima, 2000 Genomic regions affecting seed shattering and seed dormancy in rice. Theor. Appl. Genet. 100: Chopra, V. L., and S. Prakash, 2002 Evolution and Adaptation of Cereal Crops. Science Publishers, Inc., Enfield, NH. Delouche, J. C., N. R. Burgos, D. R. Gealy, G. Z. de San Martin, and R. Labrada, 2007 Weedy Rices Origin, Biology, Ecology and Control. FAO, Rome. Furukawa, T., M. Maekawa, T. Oki, I. Suda, S. Iida et al., 2007 The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49: Gressel, J., 1999 Tandem constructs: preventing the rise of superweeds. Trends Biotechnol. 17: Gu, X.-Y., S. F. Kianian, and M. E. Foley, 2004 Multiple loci and epistasis control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166: Gu, X.-Y., S. F. Kianian, and M. E. Foley, 2005a Dormancy imposed by covering tissues interrelated with seed shattering and morphological characteristics in weedy rice (Oryza sativa L.). Crop Sci. 45: Gu,X.-Y.,S.F.Kianian,G.A.Hareland,B.L.Hoffer,andM.E.Foley, 2005b Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa). Theor. Appl. Genet. 110: Gu, X.-Y., M. E. Foley, D. P. Horvath, J. V. Anderson, J. Feng et al., 2011 Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189: Gu, X.-Y., J. Zhang, H. Ye, L. Zhang, and J. Feng, 2015 Genotyping of endosperms to determine seed dormancy genes regulating germination through embryonic, endospermic, or maternal tissues in rice. G3 (Bethesda) 5: Harlan, J. R., 1965 The possible role of weed races in the evolution of cultivated plants. Euphytica 14: International Rice Genome Sequencing Project, 2005 The map-based sequence of the rice genome. Nature 436: Jiang, H., J. Wu, G. Wang, and S. Wang, 1985 A study on LüDao from Liangyungang. Crop Germplasm Resources 2: 4 7 (in Chinese). Jing, W., L. Jiang, W. Zhang, H. Zhai, and J. Wan, 2008 Mapping QTL for seed dormancy in weedy rice. Acta Agron. Sin. 34: Kanapeckas, K. L., C. C. Vigueira, A. Ortiz, K. A. Gettler, N. R. Burgos et al., 2016 Escape to ferality: the endoferal origin of weedy rice from crop rice through de-domestication. PLoS One 11: e Khush, G. S., and D. S. Brar, 2002 Rice, pp in Evolution and Adaptation of Cereal Crops, edited by Chopra, V. L., and S. Prakash. Science Publishers, Inc., Enfield, NH. Lee,S.J.,C.S.Oh,J.P.Suh,S.R.McCouch,andS.N.Ahn,2005 Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa O. rufipogon BC1F7 population. Plant Breed. 124: Li, C., A. Zhou, and T. Sang, 2006 Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 170: Li, L. F., Y. L. Li, Y. Jia, A. L. Caicedo, and K. M. Olsen, 2017 Signatures of adaptation in the weedy rice genome. Nat. Genet. 49: Lincoln, S., M. Daly, and E. Lander, 1992 Constructing Genetic Maps with MAPMAKER/EXP 3.0, Ed. 3. Whitehead Institute, Cambridge, MA. McCouch, S. R., L. Teytelman, Y. B. Xu, K. B. Lobos, K. Clare et al., 2002 Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: Mispan, M. S., L. Zhang, J. Feng, and X.-Y. Gu, 2013 Quantitative trait locus and haplotype analyses of wild and crop-mimic traits in U.S. weedy rice. G3 3: Oka, H. I., 1988 Origin of Cultivated Rice. Japan Science Society Press, Tokyo. Qiu, J., J. Zhu, F. Fu, C. Y. Ye, W. Wang et al., 2014 Genome re-sequencing suggested a weedy rice origin from domesticated indica-japonica hybridization: a case study from southern China. Planta 240: Subudhi, P. K., A. Parco, P. K. Singh, T. DeLeon, R. Karan et al., 2012 Genetic architecture of seed dormancy in U.S. weedy rice in different genetics backgrounds. Crop Sci. 52: Suh, H. S., Y. I. Sato, and H. Morishima, 1997 Genetic characterization of weedy rice (Oryza sativa L.) based on morphophysiology, isozymes and RAPD markers. Theor. Appl. Genet. 94: Sweeney, M. T., M. J. Thomson, B. E. Pfeil, and S. R. McCouch, 2006 Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: Takeuchi, Y., S. Y. Lin, T. Sasaki, and M. Yano, 2003 Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theor. Appl. Genet. 107: Tang, L. H., and H. Morishima, 1997 Genetic characterization of weedy rices and the inference on their origins. Breed. Sci. 47: Thomson, M. J., T. H. Tai, A. M. McClung, X. H. Lai, M. E. Hinga et al., 2003 Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: Veasey, E. A., M. G. Karasawa, P. P. Santos, M. S. Rosa, E. Mamani et al., 2004 Variation in the loss of seed dormancy during after-ripening of wild and cultivated rice species. Ann. Bot. 94: Volume 7 August 2017 Comparative Study on Seed Dormancy 2613
10 Wang, S., C. J. Basten, and Z.-B. Zeng, 2012 Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. Ye, H., M. E. Foley, and X.-Y. Gu, 2010 New seed dormancy loci detected from weedy rice-derived advanced populations with major QTL alleles removed from the background. Plant Sci. 179: Ye, H., D. H. Beighley, J. Feng, and X.-Y. Gu, 2013 Genetic and physiological characterization of two clusters of quantitative trait loci associated with seed dormancy and plant height in rice. G3 (Bethesda) 3: Ye, H., J. Feng, L. Zhang, J. Zhang, M. S. Mispan et al., 2015 Map-based cloning of qsd1 2 identified a gibberellin synthase gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169: Yoshida, S., D. A. Forno, J. H. Cock, and K. A. Gomez, 1976 Laboratory Manual for Physiological Studies of Rice, Ed. 3. International Rice Research Institute, Manila, Philippines. Communicating editor: B. J. Andrews 2614 L. Zhang et al.
Segregation distortion in F 2 and doubled haploid populations of temperate japonica rice
 c Indian Academy of Sciences RESEARCH NOTE Segregation distortion in F 2 and doubled haploid populations of temperate japonica rice MASUMI YAMAGISHI 1,2,6, YOSHINOBU TAKEUCHI 3,7, ISAO TANAKA 4, IZUMI
c Indian Academy of Sciences RESEARCH NOTE Segregation distortion in F 2 and doubled haploid populations of temperate japonica rice MASUMI YAMAGISHI 1,2,6, YOSHINOBU TAKEUCHI 3,7, ISAO TANAKA 4, IZUMI
Classical Selection, Balancing Selection, and Neutral Mutations
 Classical Selection, Balancing Selection, and Neutral Mutations Classical Selection Perspective of the Fate of Mutations All mutations are EITHER beneficial or deleterious o Beneficial mutations are selected
Classical Selection, Balancing Selection, and Neutral Mutations Classical Selection Perspective of the Fate of Mutations All mutations are EITHER beneficial or deleterious o Beneficial mutations are selected
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Similar traits, different genes? Examining convergent evolution in related weedy rice populations
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Publications from USDA-ARS / UNL Faculty U.S. Department of Agriculture: Agricultural Research Service, Lincoln, Nebraska
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Publications from USDA-ARS / UNL Faculty U.S. Department of Agriculture: Agricultural Research Service, Lincoln, Nebraska
Genetic and physiological approach to elucidation of Cd absorption mechanism by rice plants
 Genetic and physiological approach to elucidation of Cd absorption mechanism by rice plants Satoru Ishikawa National Institute for Agro-Environmental Sciences, 3-1-3, Kannondai, Tsukuba, Ibaraki, 305-8604,
Genetic and physiological approach to elucidation of Cd absorption mechanism by rice plants Satoru Ishikawa National Institute for Agro-Environmental Sciences, 3-1-3, Kannondai, Tsukuba, Ibaraki, 305-8604,
PEST MANAGEMENT: WEEDS
 PEST MANAGEMENT: WEEDS Outcrossing Frequency and Phenotypes of Outcrosses Based on Flowering of Red Rice Accessions and Clearfield Cultivars in the Grand Prairie V.K. Shivrain, N.R. Burgos, J.A. Bullington,
PEST MANAGEMENT: WEEDS Outcrossing Frequency and Phenotypes of Outcrosses Based on Flowering of Red Rice Accessions and Clearfield Cultivars in the Grand Prairie V.K. Shivrain, N.R. Burgos, J.A. Bullington,
Principles of QTL Mapping. M.Imtiaz
 Principles of QTL Mapping M.Imtiaz Introduction Definitions of terminology Reasons for QTL mapping Principles of QTL mapping Requirements For QTL Mapping Demonstration with experimental data Merit of QTL
Principles of QTL Mapping M.Imtiaz Introduction Definitions of terminology Reasons for QTL mapping Principles of QTL mapping Requirements For QTL Mapping Demonstration with experimental data Merit of QTL
Title. Authors. Characterization of a major QTL for manganese accumulation in rice grain
 Title Characterization of a major QTL for manganese accumulation in rice grain Authors Chaolei Liu, Guang Chen, Yuanyuan Li, Youlin Peng, Anpeng Zhang, Kai Hong, Hongzhen Jiang, Banpu Ruan, Bin Zhang,
Title Characterization of a major QTL for manganese accumulation in rice grain Authors Chaolei Liu, Guang Chen, Yuanyuan Li, Youlin Peng, Anpeng Zhang, Kai Hong, Hongzhen Jiang, Banpu Ruan, Bin Zhang,
Genetic diversity and population structure in rice. S. Kresovich 1,2 and T. Tai 3,5. Plant Breeding Dept, Cornell University, Ithaca, NY
 Genetic diversity and population structure in rice S. McCouch 1, A. Garris 1,2, J. Edwards 1, H. Lu 1,3 M Redus 4, J. Coburn 1, N. Rutger 4, S. Kresovich 1,2 and T. Tai 3,5 1 Plant Breeding Dept, Cornell
Genetic diversity and population structure in rice S. McCouch 1, A. Garris 1,2, J. Edwards 1, H. Lu 1,3 M Redus 4, J. Coburn 1, N. Rutger 4, S. Kresovich 1,2 and T. Tai 3,5 1 Plant Breeding Dept, Cornell
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA
 EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
RFLP facilitated analysis of tiller and leaf angles in rice (Oryza sativa L.)
 Euphytica 109: 79 84, 1999. 1999 Kluwer Academic Publishers. Printed in the Netherlands. 79 RFLP facilitated analysis of tiller and leaf angles in rice (Oryza sativa L.) Zhikang Li 1,2,3, Andrew H. Paterson
Euphytica 109: 79 84, 1999. 1999 Kluwer Academic Publishers. Printed in the Netherlands. 79 RFLP facilitated analysis of tiller and leaf angles in rice (Oryza sativa L.) Zhikang Li 1,2,3, Andrew H. Paterson
Supplementary Methods
 Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Dormancy Genes from Weedy Rice Respond Divergently to Seed
 Genetics: Published Articles Ahead of Print, published on November 4, 2005 as 10.1534/genetics.105.049155 Dormancy Genes from Weedy Rice Respond Divergently to Seed Development Environments Xing-You Gu,
Genetics: Published Articles Ahead of Print, published on November 4, 2005 as 10.1534/genetics.105.049155 Dormancy Genes from Weedy Rice Respond Divergently to Seed Development Environments Xing-You Gu,
QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative, Oryza longistaminata
 Breeding Science 59: 441 445 (2009) Note QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative, Oryza longistaminata Zhiwei Chen 1,2), Fengyi
Breeding Science 59: 441 445 (2009) Note QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative, Oryza longistaminata Zhiwei Chen 1,2), Fengyi
New insights into the history of rice domestication
 Review TRENDS in Genetics Vol.23 No.11 New insights into the history of rice domestication Michael J. Kovach, Megan T. Sweeney and Susan R. McCouch Cornell University, Department of Plant Breeding and
Review TRENDS in Genetics Vol.23 No.11 New insights into the history of rice domestication Michael J. Kovach, Megan T. Sweeney and Susan R. McCouch Cornell University, Department of Plant Breeding and
Genetic Analysis of Two Weak Dormancy Mutants Derived from Strong Seed Dormancy Wild Type Rice N22 (Oryza sativa) F
 Journal of Integrative Plant Biology 211, 3 (): 33 346 Research Article Genetic Analysis of Two Weak Dormancy Mutants Derived from Strong Seed Dormancy Wild Type Rice (Oryza sativa) F Bingyue Lu 1, Kun
Journal of Integrative Plant Biology 211, 3 (): 33 346 Research Article Genetic Analysis of Two Weak Dormancy Mutants Derived from Strong Seed Dormancy Wild Type Rice (Oryza sativa) F Bingyue Lu 1, Kun
Exam 1 PBG430/
 1 Exam 1 PBG430/530 2014 1. You read that the genome size of maize is 2,300 Mb and that in this species 2n = 20. This means that there are 2,300 Mb of DNA in a cell that is a. n (e.g. gamete) b. 2n (e.g.
1 Exam 1 PBG430/530 2014 1. You read that the genome size of maize is 2,300 Mb and that in this species 2n = 20. This means that there are 2,300 Mb of DNA in a cell that is a. n (e.g. gamete) b. 2n (e.g.
1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms.
 Practicing Biology BIG IDEA 3.A 1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms. 2. Using at least 2 well-known experiments, describe which features of DNA and RNA
Practicing Biology BIG IDEA 3.A 1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms. 2. Using at least 2 well-known experiments, describe which features of DNA and RNA
Jie Chen 1), De-Run Huang 1), Lei Wang 1), Guang-Jie Liu 1,2) and Jie-Yun Zhuang* 1) Introduction
 Breeding Science 60: 153 159 (2010) Identification of quantitative trait loci for resistance to whitebacked planthopper, Sogatella furcifera, from an interspecific cross Oryza sativa O. rufipogon Jie Chen
Breeding Science 60: 153 159 (2010) Identification of quantitative trait loci for resistance to whitebacked planthopper, Sogatella furcifera, from an interspecific cross Oryza sativa O. rufipogon Jie Chen
Eiji Yamamoto 1,2, Hiroyoshi Iwata 3, Takanari Tanabata 4, Ritsuko Mizobuchi 1, Jun-ichi Yonemaru 1,ToshioYamamoto 1* and Masahiro Yano 5,6
 Yamamoto et al. BMC Genetics 2014, 15:50 METHODOLOGY ARTICLE Open Access Effect of advanced intercrossing on genome structure and on the power to detect linked quantitative trait loci in a multi-parent
Yamamoto et al. BMC Genetics 2014, 15:50 METHODOLOGY ARTICLE Open Access Effect of advanced intercrossing on genome structure and on the power to detect linked quantitative trait loci in a multi-parent
Research Journal of Biology Volume 1: (Published: 16 June, 2013) ISSN:
 Research Journal of Biology Volume 1: 24 30 (Published: 16 June, 2013) RESEARCH ARTICLE Molecular mapping of early vigour related QTLs in rice Jayateertha Diwan 1, Makanhally Channbyregowda 1, Vinay Shenoy
Research Journal of Biology Volume 1: 24 30 (Published: 16 June, 2013) RESEARCH ARTICLE Molecular mapping of early vigour related QTLs in rice Jayateertha Diwan 1, Makanhally Channbyregowda 1, Vinay Shenoy
BREEDING, GENETICS, AND PHYSIOLOGY. Phenotypic Analysis of the 2006 MY2 Mapping Population in Arkansas
 BREEDING, GENETICS, AND PHYSIOLOGY Phenotypic Analysis of the 2006 MY2 Mapping Population in Arkansas E.J. Boza, K.A.K. Moldenhauer, R.D. Cartwright, S. Linscombe, J.H. Oard, and M.M. Blocker ABSTRACT
BREEDING, GENETICS, AND PHYSIOLOGY Phenotypic Analysis of the 2006 MY2 Mapping Population in Arkansas E.J. Boza, K.A.K. Moldenhauer, R.D. Cartwright, S. Linscombe, J.H. Oard, and M.M. Blocker ABSTRACT
Quantitative trait loci mapping of the stigma exertion rate and spikelet number per panicle in rice (Oryza sativa L.)
 Quantitative trait loci mapping of the stigma exertion rate and spikelet number per panicle in rice (Oryza sativa L.) M.H. Rahman, P. Yu, Y.X. Zhang, L.P. Sun, W.X. Wu, X.H. Shen, X.D. Zhan, D.B. Chen,
Quantitative trait loci mapping of the stigma exertion rate and spikelet number per panicle in rice (Oryza sativa L.) M.H. Rahman, P. Yu, Y.X. Zhang, L.P. Sun, W.X. Wu, X.H. Shen, X.D. Zhan, D.B. Chen,
The Evolutionary Genetics of Seed Shattering and Flowering Time, Two Weed Adaptive Traits in US Weedy Rice
 University of Massachusetts Amherst ScholarWorks@UMass Amherst Open Access Dissertations 9-2012 The Evolutionary Genetics of Seed Shattering and Flowering Time, Two Weed Adaptive Traits in US Weedy Rice
University of Massachusetts Amherst ScholarWorks@UMass Amherst Open Access Dissertations 9-2012 The Evolutionary Genetics of Seed Shattering and Flowering Time, Two Weed Adaptive Traits in US Weedy Rice
Map-Based Cloning and Molecular Characterization of the Seed Dormancy 10 Locus in Rice (Oryza sativa L.)
 South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange Electronic Theses and Dissertations 2018 Map-Based Cloning and Molecular Characterization
South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange Electronic Theses and Dissertations 2018 Map-Based Cloning and Molecular Characterization
Genetic Analysis of Heading Date of Japonica Rice Cultivars in Southwest China
 Rice Science, 2011, 18(4): 287 296 Copyright 2011, China National Rice Research Institute Published by Elsevier BV. All rights reserved Genetic Analysis of Heading Date of Japonica Rice Cultivars in Southwest
Rice Science, 2011, 18(4): 287 296 Copyright 2011, China National Rice Research Institute Published by Elsevier BV. All rights reserved Genetic Analysis of Heading Date of Japonica Rice Cultivars in Southwest
2 Numbers in parentheses refer to literature cited.
 A Genetic Study of Monogerm and Multigerm Characters in Beets V. F. SAVITSKY 1 Introduction Monogerm beets were found in the variety Michigan Hybrid 18 in Oregon in 1948. Two of these monogerm plants,
A Genetic Study of Monogerm and Multigerm Characters in Beets V. F. SAVITSKY 1 Introduction Monogerm beets were found in the variety Michigan Hybrid 18 in Oregon in 1948. Two of these monogerm plants,
Managing segregating populations
 Managing segregating populations Aim of the module At the end of the module, we should be able to: Apply the general principles of managing segregating populations generated from parental crossing; Describe
Managing segregating populations Aim of the module At the end of the module, we should be able to: Apply the general principles of managing segregating populations generated from parental crossing; Describe
Germplasm. Introduction to Plant Breeding. Germplasm 2/12/2013. Master Gardener Training. Start with a seed
 Introduction to Plant Breeding Master Gardener Training Start with a seed Germplasm Germplasm The greatest service which can be rendered to any country is to add a useful plant to its culture -Thomas Jefferson
Introduction to Plant Breeding Master Gardener Training Start with a seed Germplasm Germplasm The greatest service which can be rendered to any country is to add a useful plant to its culture -Thomas Jefferson
Introduction to Plant Breeding. Master Gardener Training
 Introduction to Plant Breeding Master Gardener Training Start with a seed Germplasm Germplasm The greatest service which can be rendered to any country is to add a useful plant to its culture -Thomas Jefferson
Introduction to Plant Breeding Master Gardener Training Start with a seed Germplasm Germplasm The greatest service which can be rendered to any country is to add a useful plant to its culture -Thomas Jefferson
Genetic analysis of maize kernel thickness by quantitative trait locus identification
 Genetic analysis of maize kernel thickness by quantitative trait locus identification S.S. Wen 1, G.Q. Wen 2, C.M. Liao 3 and X.H. Liu 2 1 Key Laboratory of Southwest Rice Biology and Genetic Breeding,
Genetic analysis of maize kernel thickness by quantitative trait locus identification S.S. Wen 1, G.Q. Wen 2, C.M. Liao 3 and X.H. Liu 2 1 Key Laboratory of Southwest Rice Biology and Genetic Breeding,
Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa
 Euphytica (2007) 154:317 339 DOI 10.1007/s10681-006-9210-8 Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa Susan R. McCouch Æ Megan Sweeney Æ Jiming Li
Euphytica (2007) 154:317 339 DOI 10.1007/s10681-006-9210-8 Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa Susan R. McCouch Æ Megan Sweeney Æ Jiming Li
Evolution of phenotypic traits
 Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
Propagating Plants Sexually
 Lesson C5 1 Propagating Plants Sexually Unit C. Plant and Soil Science Problem Area 5. Plant Propagation Lesson 1. Propagating Plants Sexually New Mexico Content Standard: Pathway Strand: Plant Systems
Lesson C5 1 Propagating Plants Sexually Unit C. Plant and Soil Science Problem Area 5. Plant Propagation Lesson 1. Propagating Plants Sexually New Mexico Content Standard: Pathway Strand: Plant Systems
CHAPTER 23 THE EVOLUTIONS OF POPULATIONS. Section C: Genetic Variation, the Substrate for Natural Selection
 CHAPTER 23 THE EVOLUTIONS OF POPULATIONS Section C: Genetic Variation, the Substrate for Natural Selection 1. Genetic variation occurs within and between populations 2. Mutation and sexual recombination
CHAPTER 23 THE EVOLUTIONS OF POPULATIONS Section C: Genetic Variation, the Substrate for Natural Selection 1. Genetic variation occurs within and between populations 2. Mutation and sexual recombination
Climate Change and Plant Reproduction
 Quantitative Trait Loci Mapping of Reproductive Traits Involved in Heat Stress Responses in Arabidopsis : Implications for Global Climate Change and Plant Reproduction Lazar Pavlovic, Greta Chiu, Jeffrey
Quantitative Trait Loci Mapping of Reproductive Traits Involved in Heat Stress Responses in Arabidopsis : Implications for Global Climate Change and Plant Reproduction Lazar Pavlovic, Greta Chiu, Jeffrey
Combining Ability and Heterosis in Rice (Oryza sativa L.) Cultivars
 J. Agr. Sci. Tech. (2010) Vol. 12: 223-231 Combining Ability and Heterosis in Rice (Oryza sativa L.) Cultivars M. Rahimi 1, B. Rabiei 1*, H. Samizadeh 1, and A. Kafi Ghasemi 1 ABSTRACT Quantitative valuations
J. Agr. Sci. Tech. (2010) Vol. 12: 223-231 Combining Ability and Heterosis in Rice (Oryza sativa L.) Cultivars M. Rahimi 1, B. Rabiei 1*, H. Samizadeh 1, and A. Kafi Ghasemi 1 ABSTRACT Quantitative valuations
2. Der Dissertation zugrunde liegende Publikationen und Manuskripte. 2.1 Fine scale mapping in the sex locus region of the honey bee (Apis mellifera)
 2. Der Dissertation zugrunde liegende Publikationen und Manuskripte 2.1 Fine scale mapping in the sex locus region of the honey bee (Apis mellifera) M. Hasselmann 1, M. K. Fondrk², R. E. Page Jr.² und
2. Der Dissertation zugrunde liegende Publikationen und Manuskripte 2.1 Fine scale mapping in the sex locus region of the honey bee (Apis mellifera) M. Hasselmann 1, M. K. Fondrk², R. E. Page Jr.² und
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL
 GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
Mapping QTL for Seedling Root Traits in Common Wheat
 2005,38(10):1951-1957 Scientia Agricultura Sinica 1,2,3 1 1 1 2 1 / / 100081 2 050021 3 100039 DH 10 14 11 15 5A 4B 2D 6D 7D 3 2 3 3 2 2 2 3 2 1 3 1 3 DH Mapping for Seedling Root Traits in Common Wheat
2005,38(10):1951-1957 Scientia Agricultura Sinica 1,2,3 1 1 1 2 1 / / 100081 2 050021 3 100039 DH 10 14 11 15 5A 4B 2D 6D 7D 3 2 3 3 2 2 2 3 2 1 3 1 3 DH Mapping for Seedling Root Traits in Common Wheat
Identification of quantitative trait loci associated with resistance to brown planthopper in the indica rice cultivar Col.
 Hereditas 144: 48 52 (2007) Identification of quantitative trait loci associated with resistance to brown planthopper in the indica rice cultivar Col.5 Thailand LIHONG SUN 1, YUQIANG LIU 1, LING JIANG
Hereditas 144: 48 52 (2007) Identification of quantitative trait loci associated with resistance to brown planthopper in the indica rice cultivar Col.5 Thailand LIHONG SUN 1, YUQIANG LIU 1, LING JIANG
QTL Analysis of Yield-related Traits using an Advanced Backcross Population Derived from Common Wild Rice (Oryza rufipogon L)
 Research Article Molecular Plant Breeding 2010, Vol.1 No.1 Open Access QTL Analysis of Yield-related Traits using an Advanced Backcross Population Derived from Common Wild Rice (Oryza rufipogon L) Zhaobin
Research Article Molecular Plant Breeding 2010, Vol.1 No.1 Open Access QTL Analysis of Yield-related Traits using an Advanced Backcross Population Derived from Common Wild Rice (Oryza rufipogon L) Zhaobin
2013 Breeding Progress for Rust Resistance in Confection Sunflower
 2013 Breeding Progress for Rust Resistance in Confection Sunflower Guojia Ma 1,2, Yunming Long 1,3, Sam Markell 2, Tom Gulya 1 and Lili Qi 1 1 USDA, ARS, NCSL, Fargo, ND 2 NDSU Plant Pathology, Fargo,
2013 Breeding Progress for Rust Resistance in Confection Sunflower Guojia Ma 1,2, Yunming Long 1,3, Sam Markell 2, Tom Gulya 1 and Lili Qi 1 1 USDA, ARS, NCSL, Fargo, ND 2 NDSU Plant Pathology, Fargo,
Chapter 2: Extensions to Mendel: Complexities in Relating Genotype to Phenotype.
 Chapter 2: Extensions to Mendel: Complexities in Relating Genotype to Phenotype. please read pages 38-47; 49-55;57-63. Slide 1 of Chapter 2 1 Extension sot Mendelian Behavior of Genes Single gene inheritance
Chapter 2: Extensions to Mendel: Complexities in Relating Genotype to Phenotype. please read pages 38-47; 49-55;57-63. Slide 1 of Chapter 2 1 Extension sot Mendelian Behavior of Genes Single gene inheritance
Genetic dissection of chlorophyll content at different growth stages in common wheat
 c Indian Academy of Sciences RESEARCH ARTICLE Genetic dissection of chlorophyll content at different growth stages in common wheat KUNPU ZHANG 1,2, ZHIJUN FANG 3, YAN LIANG 1 and JICHUN TIAN 1 1 State
c Indian Academy of Sciences RESEARCH ARTICLE Genetic dissection of chlorophyll content at different growth stages in common wheat KUNPU ZHANG 1,2, ZHIJUN FANG 3, YAN LIANG 1 and JICHUN TIAN 1 1 State
Inheritance of plant and tuber traits in diploid potatoes
 Inheritance of plant and tuber traits in diploid potatoes Mosquera, V. 1, Mendoza, H. A. 1, Villagómez. V. 1 and Tay, D. 1 National Agrarian University Peru; International Potato Center (CIP) E-mail: roni_atenea@yahoo.com
Inheritance of plant and tuber traits in diploid potatoes Mosquera, V. 1, Mendoza, H. A. 1, Villagómez. V. 1 and Tay, D. 1 National Agrarian University Peru; International Potato Center (CIP) E-mail: roni_atenea@yahoo.com
Quantitative trait locus analysis for ear height in maize based on a recombinant inbred line population
 Quantitative trait locus analysis for ear height in maize based on a recombinant inbred line population Z.Q. Li 4,5, H.M. Zhang 1,4, X.P. Wu 1,4, Y. Sun 3,4 and X.H. Liu 2 1 Maize Research Institute, Shanxi
Quantitative trait locus analysis for ear height in maize based on a recombinant inbred line population Z.Q. Li 4,5, H.M. Zhang 1,4, X.P. Wu 1,4, Y. Sun 3,4 and X.H. Liu 2 1 Maize Research Institute, Shanxi
MOLECULAR MAPS AND MARKERS FOR DIPLOID ROSES
 MOLECULAR MAPS AND MARKERS FOR DIPLOID ROSES Patricia E Klein, Mandy Yan, Ellen Young, Jeekin Lau, Stella Kang, Natalie Patterson, Natalie Anderson and David Byrne Department of Horticultural Sciences,
MOLECULAR MAPS AND MARKERS FOR DIPLOID ROSES Patricia E Klein, Mandy Yan, Ellen Young, Jeekin Lau, Stella Kang, Natalie Patterson, Natalie Anderson and David Byrne Department of Horticultural Sciences,
Chapter 1: Mendel s breakthrough: patterns, particles and principles of heredity
 Chapter 1: Mendel s breakthrough: patterns, particles and principles of heredity please read pages 10 through 13 Slide 1 of Chapter 1 One of Mendel s express aims was to understand how first generation
Chapter 1: Mendel s breakthrough: patterns, particles and principles of heredity please read pages 10 through 13 Slide 1 of Chapter 1 One of Mendel s express aims was to understand how first generation
Development of specific simple sequence repeat (SSR) markers for non-pollen type thermo-sensitive genic male sterile gene in rice (Oryza sativa L.
 African Journal of Biotechnology Vol. 10(73), pp. 16437-16442, 21 November, 2011 Available online at http://www.academicjournals.org/ajb DOI: 10.5897/AJB11.2401 ISSN 1684 5315 2011 Academic Journals Full
African Journal of Biotechnology Vol. 10(73), pp. 16437-16442, 21 November, 2011 Available online at http://www.academicjournals.org/ajb DOI: 10.5897/AJB11.2401 ISSN 1684 5315 2011 Academic Journals Full
DNA Structure and Function
 DNA Structure and Function Nucleotide Structure 1. 5-C sugar RNA ribose DNA deoxyribose 2. Nitrogenous Base N attaches to 1 C of sugar Double or single ring Four Bases Adenine, Guanine, Thymine, Cytosine
DNA Structure and Function Nucleotide Structure 1. 5-C sugar RNA ribose DNA deoxyribose 2. Nitrogenous Base N attaches to 1 C of sugar Double or single ring Four Bases Adenine, Guanine, Thymine, Cytosine
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
Mapping of a Major Resistance Gene to the Brown Planthopper in the Rice Cultivar Rathu Heenati
 Breeding Science 55 : 391 396 (2005) Mapping of a Major Resistance Gene to the Brown Planthopper in the Rice Cultivar Rathu Heenati Lihong Sun 1), Changchao Su 1), Chunming Wang 1), Huqu Zhai 2) and Jianmin
Breeding Science 55 : 391 396 (2005) Mapping of a Major Resistance Gene to the Brown Planthopper in the Rice Cultivar Rathu Heenati Lihong Sun 1), Changchao Su 1), Chunming Wang 1), Huqu Zhai 2) and Jianmin
Hairy s Inheritance: Investigating Variation, Selection, and Evolution with Wisconsin Fast Plants
 Introduction Hairy s Inheritance: Investigating Variation, Selection, and Evolution with Wisconsin Fast Plants Daniel Lauffer Wisconsin Fast Plants Program University of Wisconsin - Madison Since the dawn
Introduction Hairy s Inheritance: Investigating Variation, Selection, and Evolution with Wisconsin Fast Plants Daniel Lauffer Wisconsin Fast Plants Program University of Wisconsin - Madison Since the dawn
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Transferring Powdery Mildew Resistance Genes from Wild Helianthus into Cultivated Sunflower. Pilar Rojas-Barros, Chao-Chien Jan, and Thomas J.
 Transferring Powdery Mildew Resistance Genes from Wild Helianthus into Cultivated Sunflower Pilar Rojas-Barros, Chao-Chien Jan, and Thomas J. Gulya USDA-ARS, Northern Crop Science Laboratory, Fargo, ND
Transferring Powdery Mildew Resistance Genes from Wild Helianthus into Cultivated Sunflower Pilar Rojas-Barros, Chao-Chien Jan, and Thomas J. Gulya USDA-ARS, Northern Crop Science Laboratory, Fargo, ND
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/331/6019/876/dc1 Supporting Online Material for Synthetic Clonal Reproduction Through Seeds Mohan P. A. Marimuthu, Sylvie Jolivet, Maruthachalam Ravi, Lucie Pereira,
www.sciencemag.org/cgi/content/full/331/6019/876/dc1 Supporting Online Material for Synthetic Clonal Reproduction Through Seeds Mohan P. A. Marimuthu, Sylvie Jolivet, Maruthachalam Ravi, Lucie Pereira,
4/26/18. Domesticated plants vs. their wild relatives. Lettuce leaf size/shape, fewer secondary compounds
 The final exam: Tuesday, May 8 at 4:05-6:05pm in Ruttan Hall B35. 75 multiple choice questions for 150 points 50 questions from Lecture 20 27 25 questions directly from the first two exams. Key for exam
The final exam: Tuesday, May 8 at 4:05-6:05pm in Ruttan Hall B35. 75 multiple choice questions for 150 points 50 questions from Lecture 20 27 25 questions directly from the first two exams. Key for exam
Mutation, Selection, Gene Flow, Genetic Drift, and Nonrandom Mating Results in Evolution
 Mutation, Selection, Gene Flow, Genetic Drift, and Nonrandom Mating Results in Evolution 15.2 Intro In biology, evolution refers specifically to changes in the genetic makeup of populations over time.
Mutation, Selection, Gene Flow, Genetic Drift, and Nonrandom Mating Results in Evolution 15.2 Intro In biology, evolution refers specifically to changes in the genetic makeup of populations over time.
You are encouraged to answer/comment on other people s questions. Domestication conversion of plants or animals to domestic uses
 The final exam: Tuesday, May 8 at 4:05-6:05pm in Ruttan Hall B35. 75 multiple choice questions for 150 points 50 questions from Lecture 20 27 25 questions directly from the first two exams. Key for exam
The final exam: Tuesday, May 8 at 4:05-6:05pm in Ruttan Hall B35. 75 multiple choice questions for 150 points 50 questions from Lecture 20 27 25 questions directly from the first two exams. Key for exam
BREEDING AND GENETICS
 The Journal of Cotton Science 6:97-103 (2002) http://journal.cotton.org, The Cotton Foundation 2002 97 BREEDING AND GENETICS Assessment of Day-Neutral Backcross Populations of Cotton Using AFLP Markers
The Journal of Cotton Science 6:97-103 (2002) http://journal.cotton.org, The Cotton Foundation 2002 97 BREEDING AND GENETICS Assessment of Day-Neutral Backcross Populations of Cotton Using AFLP Markers
Sporic life cycles involve 2 types of multicellular bodies:
 Chapter 3- Human Manipulation of Plants Sporic life cycles involve 2 types of multicellular bodies: -a diploid, spore-producing sporophyte -a haploid, gamete-producing gametophyte Sexual Reproduction in
Chapter 3- Human Manipulation of Plants Sporic life cycles involve 2 types of multicellular bodies: -a diploid, spore-producing sporophyte -a haploid, gamete-producing gametophyte Sexual Reproduction in
POSSIBILITY TO SELECT HETEROZYGOUS GENOTYPES BY POLLEN FERTILITY IN SEGREGATION HYBRID AND BACKCROSS PROGENIES CREATED based on NUCLEAR MALE STERILITY
 565 Bulgarian Journal of Agricultural Science, 18 (No 4) 2012, 565-570 Agricultural Academy POSSIBILITY TO SELECT HETEROZYGOUS GENOTYPES BY POLLEN FERTILITY IN SEGREGATION HYBRID AND BACKCROSS PROGENIES
565 Bulgarian Journal of Agricultural Science, 18 (No 4) 2012, 565-570 Agricultural Academy POSSIBILITY TO SELECT HETEROZYGOUS GENOTYPES BY POLLEN FERTILITY IN SEGREGATION HYBRID AND BACKCROSS PROGENIES
Study of Genetic Diversity in Some Newly Developed Rice Genotypes
 International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 6 Number 10 (2017) pp. 2693-2698 Journal homepage: http://www.ijcmas.com Original Research Article https://doi.org/10.20546/ijcmas.2017.610.317
International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 6 Number 10 (2017) pp. 2693-2698 Journal homepage: http://www.ijcmas.com Original Research Article https://doi.org/10.20546/ijcmas.2017.610.317
Genetic structure and eco-geographical differentiation of cultivated Hsien rice (Oryza sativa L. subsp. indica) in China revealed by microsatellites
 Article Crop Germplasm Resources January 2013 Vol.58 No.3: 344 352 doi: 10.1007/s11434-012-5396-4 Genetic structure and eco-geographical differentiation of cultivated Hsien rice (Oryza sativa L. subsp.
Article Crop Germplasm Resources January 2013 Vol.58 No.3: 344 352 doi: 10.1007/s11434-012-5396-4 Genetic structure and eco-geographical differentiation of cultivated Hsien rice (Oryza sativa L. subsp.
QUANTITATIVE ANALYSIS OF PHOTOPERIODISM OF TEXAS 86, GOSSYPIUM HIRSUTUM RACE LATIFOLIUM, IN A CROSS AMERICAN UPLAND COTTON' Received June 21, 1962
 THE GENETICS OF FLOWERING RESPONSE IN COTTON. IV. QUANTITATIVE ANALYSIS OF PHOTOPERIODISM OF TEXAS 86, GOSSYPIUM HIRSUTUM RACE LATIFOLIUM, IN A CROSS WITH AN INBRED LINE OF CULTIVATED AMERICAN UPLAND COTTON'
THE GENETICS OF FLOWERING RESPONSE IN COTTON. IV. QUANTITATIVE ANALYSIS OF PHOTOPERIODISM OF TEXAS 86, GOSSYPIUM HIRSUTUM RACE LATIFOLIUM, IN A CROSS WITH AN INBRED LINE OF CULTIVATED AMERICAN UPLAND COTTON'
Solutions to Problem Set 4
 Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
The long and the short of it: SD1 polymorphism and the evolution of growth trait divergence in U.S. weedy rice
 Molecular Ecology (20) 20, 3743 3756 doi: 0./j.365-294X.20.0526.x The long and the short of it: SD polymorphism and the evolution of growth trait divergence in U.S. weedy rice MICHAEL REAGON,* CARRIE S.
Molecular Ecology (20) 20, 3743 3756 doi: 0./j.365-294X.20.0526.x The long and the short of it: SD polymorphism and the evolution of growth trait divergence in U.S. weedy rice MICHAEL REAGON,* CARRIE S.
Identifying Wheat Growth Stages
 AGR-224 Identifying Wheat Growth Stages Carrie A. Knott, Plant and Soil Sciences University of Kentucky College of Agriculture, Food and Environment Cooperative Extension Service Identifying growth stages
AGR-224 Identifying Wheat Growth Stages Carrie A. Knott, Plant and Soil Sciences University of Kentucky College of Agriculture, Food and Environment Cooperative Extension Service Identifying growth stages
Lecture WS Evolutionary Genetics Part I 1
 Quantitative genetics Quantitative genetics is the study of the inheritance of quantitative/continuous phenotypic traits, like human height and body size, grain colour in winter wheat or beak depth in
Quantitative genetics Quantitative genetics is the study of the inheritance of quantitative/continuous phenotypic traits, like human height and body size, grain colour in winter wheat or beak depth in
Growth Regulator Effects on Flowering in Maize
 Growth Regulator Effects on Flowering in Maize Eric Bumann July 14, 2008 My Background Research Associate at Pioneer Hi-Bred in Johnston, IA Production research 5 years in greenhouse research B.S. in Horticulture
Growth Regulator Effects on Flowering in Maize Eric Bumann July 14, 2008 My Background Research Associate at Pioneer Hi-Bred in Johnston, IA Production research 5 years in greenhouse research B.S. in Horticulture
Awide range of natural variation for flowering time
 Copyright Ó 2006 by the Genetics Society of America DOI: 10.1534/genetics.105.050500 Substitution Mapping of dth1.1, a Flowering-Time Quantitative Trait Locus (QTL) Associated With Transgressive Variation
Copyright Ó 2006 by the Genetics Society of America DOI: 10.1534/genetics.105.050500 Substitution Mapping of dth1.1, a Flowering-Time Quantitative Trait Locus (QTL) Associated With Transgressive Variation
Gene Flow Between Crops and Their Wild Progenitors
 Gene Flow Between Crops and Their Wild Progenitors Roberto Papa Università Politecnica delle Marche, Ancona, Italy Paul Gepts University of California, Davis, California, U.S.A. INTRODUCTION Gene flow
Gene Flow Between Crops and Their Wild Progenitors Roberto Papa Università Politecnica delle Marche, Ancona, Italy Paul Gepts University of California, Davis, California, U.S.A. INTRODUCTION Gene flow
Little White Lies: Pericarp Color Provides Insights into the Origins and Evolution of Southeast Asian Weedy Rice
 INVESTIGATION Little White Lies: Pericarp Color Provides Insights into the Origins and Evolution of Southeast Asian Weedy Rice Yongxia Cui,*,,1 Beng Kah Song,,1,2 Lin-Feng Li,, Ya-Ling Li, Zhongyun Huang,**
INVESTIGATION Little White Lies: Pericarp Color Provides Insights into the Origins and Evolution of Southeast Asian Weedy Rice Yongxia Cui,*,,1 Beng Kah Song,,1,2 Lin-Feng Li,, Ya-Ling Li, Zhongyun Huang,**
Introduction to QTL mapping in model organisms
 Human vs mouse Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University www.biostat.jhsph.edu/~kbroman [ Teaching Miscellaneous lectures] www.daviddeen.com
Human vs mouse Introduction to QTL mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University www.biostat.jhsph.edu/~kbroman [ Teaching Miscellaneous lectures] www.daviddeen.com
ACTA AGRONOMICA SINICA SSR. Classification for Some Sterile Lines and Their Restorers of Hybrid Rice with SSR Markers
 32 2 2006 2 169 175 ACTA AGRONOMICA SINICA Vol132, No12 pp1 169-175 Feb1, 2006 SSR 1 1,2, # 1 1 1 1, 3 2 2 Ξ ( 1, 330045 ; 2,,100094) : ( Oryza sativa L. ) 12 36 SSR(simple sequence repeats), 5 7 54 300,
32 2 2006 2 169 175 ACTA AGRONOMICA SINICA Vol132, No12 pp1 169-175 Feb1, 2006 SSR 1 1,2, # 1 1 1 1, 3 2 2 Ξ ( 1, 330045 ; 2,,100094) : ( Oryza sativa L. ) 12 36 SSR(simple sequence repeats), 5 7 54 300,
Major questions of evolutionary genetics. Experimental tools of evolutionary genetics. Theoretical population genetics.
 Evolutionary Genetics (for Encyclopedia of Biodiversity) Sergey Gavrilets Departments of Ecology and Evolutionary Biology and Mathematics, University of Tennessee, Knoxville, TN 37996-6 USA Evolutionary
Evolutionary Genetics (for Encyclopedia of Biodiversity) Sergey Gavrilets Departments of Ecology and Evolutionary Biology and Mathematics, University of Tennessee, Knoxville, TN 37996-6 USA Evolutionary
The evolutionary success of flowering plants is to a certain
 An improved genetic model generates highresolution mapping of QTL for protein quality in maize endosperm Rongling Wu*, Xiang-Yang Lou*, Chang-Xing Ma*, Xuelu Wang, Brian A. Larkins, and George Casella*
An improved genetic model generates highresolution mapping of QTL for protein quality in maize endosperm Rongling Wu*, Xiang-Yang Lou*, Chang-Xing Ma*, Xuelu Wang, Brian A. Larkins, and George Casella*
Advances in M olecular Genetics of T iller ing Characters in R ice
 2006, 18 (1) : 80 84 Acta Agriculturae J iangxi 1, 2 (1., 330200; 2., 225009) : ( ) : ; ; ; ; : S511. 032 : A : 1001-8581 (2006) 01-0080 - 05 Advances in M olecular Genetics of T iller ing Characters in
2006, 18 (1) : 80 84 Acta Agriculturae J iangxi 1, 2 (1., 330200; 2., 225009) : ( ) : ; ; ; ; : S511. 032 : A : 1001-8581 (2006) 01-0080 - 05 Advances in M olecular Genetics of T iller ing Characters in
Maize Genetics Cooperation Newsletter Vol Derkach 1
 Maize Genetics Cooperation Newsletter Vol 91 2017 Derkach 1 RELATIONSHIP BETWEEN MAIZE LANCASTER INBRED LINES ACCORDING TO SNP-ANALYSIS Derkach K. V., Satarova T. M., Dzubetsky B. V., Borysova V. V., Cherchel
Maize Genetics Cooperation Newsletter Vol 91 2017 Derkach 1 RELATIONSHIP BETWEEN MAIZE LANCASTER INBRED LINES ACCORDING TO SNP-ANALYSIS Derkach K. V., Satarova T. M., Dzubetsky B. V., Borysova V. V., Cherchel
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated
 Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Gene mapping in model organisms
 Gene mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman Goal Identify genes that contribute to common human diseases. 2
Gene mapping in model organisms Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman Goal Identify genes that contribute to common human diseases. 2
Maternal control of seed development mediated by the flavonoid biosynthesis pathway
 P- END1 Maternal control of seed development mediated by the flavonoid biosynthesis pathway Maha Aljabri, James Doughty, and Rod Scott University of Bath, UK Many plants exhibit post-zygotic barriers to
P- END1 Maternal control of seed development mediated by the flavonoid biosynthesis pathway Maha Aljabri, James Doughty, and Rod Scott University of Bath, UK Many plants exhibit post-zygotic barriers to
Genetic Analysis and Mapping of Genes for Culm Length and Internode Diameter in Progeny from an Interspecific Cross in Rice
 Plant Breed. Biotech. 2018 (June) 6(2):140~146 https://doi.org/10.9787/pbb.2018.6.2.140 RESEARCH ARTICLE Online ISSN: 2287-9366 Print ISSN: 2287-9358 Genetic Analysis and Mapping of Genes for Culm Length
Plant Breed. Biotech. 2018 (June) 6(2):140~146 https://doi.org/10.9787/pbb.2018.6.2.140 RESEARCH ARTICLE Online ISSN: 2287-9366 Print ISSN: 2287-9358 Genetic Analysis and Mapping of Genes for Culm Length
Assessment of Genetic Diversity of Pawpaw Cultivars with Inter-Simple Sequence Repeat Markers
 Assessment of Genetic Diversity of Pawpaw Cultivars with Inter-Simple Sequence Repeat Markers Kirk W. Pomper*, Sheri B. Crabtree, and Snake C. Jones Kentucky State University What is Pawpaw? Asimina triloba
Assessment of Genetic Diversity of Pawpaw Cultivars with Inter-Simple Sequence Repeat Markers Kirk W. Pomper*, Sheri B. Crabtree, and Snake C. Jones Kentucky State University What is Pawpaw? Asimina triloba
unique aspect of pollen competition. A pollen mixture may Petunia hybrida
 Acta 801. Neerl. 31(1/2), February 1982, p. 97-103, The effect of delayed pollination in Petunia hybrida Gabriella+Bergamini Mulcahy* D.L. Mulcahy*, and P.L. Pfahler ** * Departmentof Botany, University
Acta 801. Neerl. 31(1/2), February 1982, p. 97-103, The effect of delayed pollination in Petunia hybrida Gabriella+Bergamini Mulcahy* D.L. Mulcahy*, and P.L. Pfahler ** * Departmentof Botany, University
PHENOTYPIC AND GENOTYPIC ANALYSIS FOR EARLY MORNING FLOWERING TRAITS AT FLOWERING STAGE IN RICE (Oryza sativa L.)
 Proceedings of the 3 rd International Conference on Agriculture and Forestry, Vol. 2, 2016, pp. 158-166 Copyright TIIKM ISSN: 2362-1036 online DOI: https://doi.org/10.17501/icoaf.2016.2114 PHENOTYPIC AND
Proceedings of the 3 rd International Conference on Agriculture and Forestry, Vol. 2, 2016, pp. 158-166 Copyright TIIKM ISSN: 2362-1036 online DOI: https://doi.org/10.17501/icoaf.2016.2114 PHENOTYPIC AND
Table S1 List of primers used for genotyping and qrt-pcr.
 Table S1 List of primers used for genotyping and qrt-pcr. genotyping! allele! ligomer*! 5'-sequence-3'! rice! d10-2! F! TTGGCTTTGCCTCGTTTC!!! R! AGCCTCCACTTGTACTGTG! Arabidopsis! max2-3, max2-4! F! ACTCTCTCCGACCTCCCTGACG!!!
Table S1 List of primers used for genotyping and qrt-pcr. genotyping! allele! ligomer*! 5'-sequence-3'! rice! d10-2! F! TTGGCTTTGCCTCGTTTC!!! R! AGCCTCCACTTGTACTGTG! Arabidopsis! max2-3, max2-4! F! ACTCTCTCCGACCTCCCTGACG!!!
The Plant Cell, November. 2017, American Society of Plant Biologists. All rights reserved
 The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field.
 Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Reinforcement Unit 3 Resource Book. Meiosis and Mendel KEY CONCEPT Gametes have half the number of chromosomes that body cells have.
 6.1 CHROMOSOMES AND MEIOSIS KEY CONCEPT Gametes have half the number of chromosomes that body cells have. Your body is made of two basic cell types. One basic type are somatic cells, also called body cells,
6.1 CHROMOSOMES AND MEIOSIS KEY CONCEPT Gametes have half the number of chromosomes that body cells have. Your body is made of two basic cell types. One basic type are somatic cells, also called body cells,
GENETICS - CLUTCH CH.1 INTRODUCTION TO GENETICS.
 !! www.clutchprep.com CONCEPT: HISTORY OF GENETICS The earliest use of genetics was through of plants and animals (8000-1000 B.C.) Selective breeding (artificial selection) is the process of breeding organisms
!! www.clutchprep.com CONCEPT: HISTORY OF GENETICS The earliest use of genetics was through of plants and animals (8000-1000 B.C.) Selective breeding (artificial selection) is the process of breeding organisms
Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.)
 Magwa et al. BMC Genetics (2016) 17:28 DOI 10.1186/s12863-016-0340-2 RESEARCH ARTICLE Open Access Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations
Magwa et al. BMC Genetics (2016) 17:28 DOI 10.1186/s12863-016-0340-2 RESEARCH ARTICLE Open Access Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations
Ch 11.Introduction to Genetics.Biology.Landis
 Nom Section 11 1 The Work of Gregor Mendel (pages 263 266) This section describes how Gregor Mendel studied the inheritance of traits in garden peas and what his conclusions were. Introduction (page 263)
Nom Section 11 1 The Work of Gregor Mendel (pages 263 266) This section describes how Gregor Mendel studied the inheritance of traits in garden peas and what his conclusions were. Introduction (page 263)
Full file at CHAPTER 2 Genetics
 CHAPTER 2 Genetics MULTIPLE CHOICE 1. Chromosomes are a. small linear bodies. b. contained in cells. c. replicated during cell division. 2. A cross between true-breeding plants bearing yellow seeds produces
CHAPTER 2 Genetics MULTIPLE CHOICE 1. Chromosomes are a. small linear bodies. b. contained in cells. c. replicated during cell division. 2. A cross between true-breeding plants bearing yellow seeds produces
The shift from perennial to annual habit was of great importance. Convergent evolution of perenniality in rice and sorghum
 Convergent evolution of perenniality in rice and sorghum F. Y. Hu*,D.Y.Tao, E. Sacks*, B. Y. Fu*, P. Xu,J.Li, Y. Yang, K. McNally*, G. S. Khush*, A. H. Paterson, and Z.-K. Li* *International Rice Research
Convergent evolution of perenniality in rice and sorghum F. Y. Hu*,D.Y.Tao, E. Sacks*, B. Y. Fu*, P. Xu,J.Li, Y. Yang, K. McNally*, G. S. Khush*, A. H. Paterson, and Z.-K. Li* *International Rice Research
Identification of quantitative trait loci that regulate Arabidopsis root system size
 Genetics: Published Articles Ahead of Print, published on September 12, 2005 as 10.1534/genetics.105.047555 Identification of quantitative trait loci that regulate Arabidopsis root system size and plasticity
Genetics: Published Articles Ahead of Print, published on September 12, 2005 as 10.1534/genetics.105.047555 Identification of quantitative trait loci that regulate Arabidopsis root system size and plasticity
Duplication of an upstream silencer of FZP increases grain yield in rice
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41477-017-0042-4 In the format provided by the authors and unedited. Duplication of an upstream silencer of FZP increases grain yield in rice
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41477-017-0042-4 In the format provided by the authors and unedited. Duplication of an upstream silencer of FZP increases grain yield in rice
(Write your name on every page. One point will be deducted for every page without your name!)
 POPULATION GENETICS AND MICROEVOLUTIONARY THEORY FINAL EXAMINATION (Write your name on every page. One point will be deducted for every page without your name!) 1. Briefly define (5 points each): a) Average
POPULATION GENETICS AND MICROEVOLUTIONARY THEORY FINAL EXAMINATION (Write your name on every page. One point will be deducted for every page without your name!) 1. Briefly define (5 points each): a) Average
Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach
 Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach ABSTRACT SUBMITTED TO JAMIA MILLIA ISLAMIA NEW DELHI IN PARTIAL FULFILMENT OF
Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach ABSTRACT SUBMITTED TO JAMIA MILLIA ISLAMIA NEW DELHI IN PARTIAL FULFILMENT OF
