AN EVIDENCE ONTOLOGY FOR USE IN PATHWAY/GENOME DATABASES
|
|
|
- Randolph Wood
- 5 years ago
- Views:
Transcription
1 AN EVIDENCE ONTOLOGY FOR USE IN PATHWAY/GENOME DATABASES Appears in Pacific Symposium on Biocomputing 2004 Pages , World Scientific Publishers, Singapore P.D. KARP, S. PALEY, C.J. KRIEGER SRI International, 333 Ravenswood Ave., Menlo Park, CA USA P. ZHANG Carnegie Institution of Washington, Department of Plant Biology 260 Panama Street, Stanford, California Abstract. An important emerging need in Model Organism Databases (MODs) and other bioinformatics databases (DBs) is that of capturing the scientific evidence that supports the information within a DB. This need has become particularly acute as more DB content consists of computationally predicted information, such as predicted gene functions, operons, metabolic pathways, and protein properties. This paper presents an ontology for encoding the type of support and the degree of support for DB assertions, and for encoding the literature source in which that support is reported. The ontology includes a hierarchy of 35 evidence codes for modeling different types of wet-lab and computational evidence for the existence of operons and metabolic pathways, and for gene functions. We also describe an implementation of the ontology within the Pathway Tools software environment, which is used to query and update Pathway/Genome DBs such as EcoCyc, MetaCyc, and HumanCyc. 1 Introduction An important emerging need in Model Organism Databases (MODs) and other bioinformatics databases (DBs) is that of capturing the scientific evidence that supports the information within a DB. This need has become particularly acute as more DB content consists of computationally predicted information, such as predicted gene functions, operons, metabolic pathways, and protein properties. DB users want to know the type of evidence that supports assertions within a DB, and they want to know the strength of that evidence. Strength and type are in general independent parameters, although they are often related; for example, computationally generated predictions are generally held to be less reliable than are wet-lab experiments, but there are certainly unreliable types of wet-lab methods. This paper reports on an ontology for encoding the type of support and the degree of support for DB assertions, and for encoding the literature source
2 in which that support is reported. We also describe an implementation of the ontology within the Pathway Tools software environment, 1 which is used to query and update Pathway/Genome DBs (PGDBs) such as EcoCyc, 2 Meta- Cyc, 3 and HumanCyc (see URL 2 Motivations for an Evidence Ontology The evidence ontology is designed to encode information about why we believe certain assertions in a PGDB, the sources of those assertions, and the degree of confidence scientists hold in those assertions. An assertion could be the existence of a biological object described in a PGDB. For example, we would like to be able to encode the evidence supporting the existence of a gene, an operon, or a pathway that is described within a PGDB. Has the operon been predicted using a computational operon finder? Or is it supported by wet-lab experiments? If the latter, what types of experimental methods were used? It should be possible to capture multiple types of evidence: if the existence of a metabolic pathway is supported by both a computational algorithm and by two different types of wet-lab experiments, our evidence ontology should be able to capture that information, and also to capture the literature citations that are the source of that information. We also want to be able to capture two types of confidence information in the evidence ontology. If the probability of correctness of an individual piece of evidence is known, we want to capture that. For example, if we have measured the overall accuracy of an operon predictor and found that accuracy to be 80%, then computational operon predictions made by that program should be recorded to have an individual confidence of 0.8. Similarly, if we know that a wet-lab method has a probability of correctness of 0.7, we should be able to capture that information in conjunction with our evidence codes. But in addition to capturing the confidence in individual pieces of evidence, we want to capture the overall confidence in an assertion that results from synthesizing across multiple pieces of evidence. Consider a case where the existence of a metabolic pathway is supported by a computational prediction and by two wet-lab experiments. We would like a curator to be able to record his or her overall confidence level in that pathway that results from integrating those three pieces of evidence. Object existence is one class of PGDB assertion. But object properties and relationships form another important class of assertions. We should be able to encode evidence not just about object existence, but also regarding slot values stored in a PGDB, such as the type of evidence supporting the molecular weight of a protein, or the assertion that pyruvate inhibits an enzyme, or the assertion that a protein catalyzes a given reaction.
3 2.1 Related Work Gene Ontology provides an evidence ontology that satisfies some of the preceding criteria, and that formed the starting point for our work. 4 The GO evidence codes are described at URL Wherever possible we have adopted the GO evidence codes, or small variations of them, to facilitate translation between the systems. But in many cases we significantly extended or reworked the GO system because it was not designed to satisfy, and cannot satisfy, the requirements listed earlier in this section. For example: (a) The GO system does not encode specific classes of experimental methods, that is, the GO code IDA (Inferred from Direct Assay) has no subclasses to define subtypes of experimental assays (such as assays that provide evidence for the activity of an enzyme or for the presence of a promoter). (b) Strictly speaking, the GO evidence system is intended to be used to annotate the support for attachment of a GO term to a gene. It is not specifically designed for use to record evidence for the existence of a biological object, or for other types of assertions such as slot values. (c) The GO evidence system does not provide a way to associate confidence values with assertions. We are unaware of directly related work in the Artificial Intelligence community. AI work on truth maintenance systems (TMSs) is not relevant because TMSs are concerned with capturing relationships between propositions inferred by an automated reasoner, and the propositions on which those inferences depend 5. That is a different problem than trying to capture general classes of evidence that support some proposition. 3 Overview of Pathway Tools The Pathway Tools software is a reusable package for creating, querying, visualizing, and analyzing MODs. Its components include the following. PathoLogic This is a module for computationally creating a new PGDB for an organism from its annotated genome. PathoLogic includes a metabolic pathway predictor 6 and an operon predictor. Given a properly formatted Genbank entry for an annotated genome, PathoLogic can create a new PGDB within a day. Additional manual processing that is typically required before a PGDB is ready for release takes 2 3 weeks for a bacterial genome. 7 Pathway/Genome Editors This module includes a graphical interactive editing tool for every datatype managed by Pathway Tools, including genes, proteins, biochemical reactions and pathways, small molecule metabolites, and operons. Pathway/Genome Navigator This module allows users to query a PGDB and to visualize the results of a query. Visualization tools supported include visualization of chromosomes (genome browser), genes, proteins (with
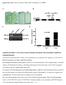
4 specialized displays for enzymes, for transporters, and for transcription factors the latter displays all operons controlled by the transcription factor), pathways, and transcription units (operons). A metabolic overview diagram is a drawing of all known metabolic pathways of an organism. Expression data for a given organism can be painted onto the metabolic overview to place expression data in a pathway context and to allow the user to discern the coordinated expression of entire pathways, or of important steps within a pathway. SRI has used Pathway Tools to develop 15 PGDBs, which are available through the BioCyc Web site at URL In addition, Pathway Tools has been licensed by 31 groups in academia and industry. PGDBs available from those groups are also listed at URL Recent enhancements to Pathway Tools include (1) ontology, visualization, and editing support for introns, exons, and alternative splicing; (2) tools for exporting PGDB information to flat files, and for importing information from flat files into PGDBs; (3) implementation of Perl and Java APIs for querying and updating PGDBs, called PerlCyc and JavaCyc, respectively (see URL 4 Pathway Tools Implementation of the Evidence Ontology This section describes how the Pathway Tools evidence ontology is presented to the user in Pathway Tools displays to give the reader an understanding of how the ontology is used. We extended Pathway Tools so that the pathway and operon predictors within PathoLogic decorate the pathway and operon PGDB objects that they create with evidence-code information to indicate computationally predicted objects as such. We extended the Editors to include functionality that allows users to interactively enter and modify evidence codes. We extended the Navigator to display evidence information. For example, the Navigator window shown in Figure 1 displays information about the transcription unit a called cbl. The flask icon at the top right of the diagram indicates that evidence from wet-lab experiments supports the existence of the transcription unit. The lower flask and computer icons adjacent to the Promoter: cbl line indicate that both wet-lab experiments and computational predictions support the existence of this promoter. Finally, the flask at the bottom right of the window indicates that experimental evidence supports the information about the activity of the transcription factor. Although our evidence system provides for more precise distinctions than simply wet-lab versus computational, we felt it best to keep our graphical a Transcription units are essentially the same as operons, although transcription units can contain single genes, whereas by definition operons must contain multiple genes.
5 Figure 1: Pathway Tools display of the EcoCyc transcription unit cbl. interface simple by displaying only a few different icons, since expecting users to learn icons for each of our 35 evidence codes would be unreasonable. Therefore, object display windows such as the transcription-unit display in Figure 1 show icons that only differentiate evidence codes at the highest level of our hierarchy, such as distinguishing experimental evidence from computational evidence. The user can click on these icons to view another screen that shows the detailed evidence codes that support the existence of an object (see Figure 2), and from what literature sources that evidence was reported. 5 The Evidence Ontology Each piece of evidence about object existence in PGDBs is recorded as a structured evidence tuple, as a value within the Citations slot of PGDB objects such
6 Figure 2: Detailed evidence report for existence of the cbl promoter. as pathways and transcription units. An evidence tuple allows us to associate several types of information within one piece of evidence. Each evidence tuple is of the form: Evidence-code : Citation : Curator : Timestamp : Probability where Evidence-code is a unique ID for the type of evidence, as provided in Table 1. Citation is an optional citation identifier such as a PubMed ID that indicates the source of the evidence. For computational evidence, the citation refers to an article describing either the general properties of the algorithm used, or its application in this case. Curator is the username of the curator who created this evidence tuple. Timestamp is an optional integer representing the time and date on which this evidence tuple was created. Probability is an optional real number that indicates the probability that the assertion supported by this evidence is correct, such as a probability provided by an algorithm. We expect that the probability portion of the evidence tuple will be used much more frequently for computational evidence than for wet-lab evidence because the accuracies of
7 computational techniques tend to be better known in general than for experimental methods. The notion of what it means for a biological object to exist varies somewhat by object type, and is difficult to define precisely. For example, what does it mean for a gene to exist? Does the existence of a gene depend only on whether some gene product is produced from a region of DNA, or on whether the exact boundaries of the gene are defined precisely? In the case of a gene, the probability of existence should not reflect whether the exact nucleotide start and stop positions of the gene are correct, but should depend only on whether a gene product is produced by an approximate region of a chromosome, due to the accuracy limits of current gene finders. In contrast, the notion of probability of existence of a transcription unit should indeed depend on where the gene boundary of the transcription unit lies, since every bacterial gene lies within some transcription unit, and the fundamental problem of predicting transcription units is defining their gene boundaries. Finally, we defined a slot Confidence that allows a PGDB to record an overall integrated probability for the existence of an object. Whereas probabilities within evidence tuples encode the probability associated with an individual piece of evidence, the Confidence slot is intended to hold the net probability that results from integrating across the potentially multiple pieces of evidence available. This integration process will in most cases be a manual process performed by a curator, that will therefore be subjective and will vary among individuals, and we are in the process of developing guidelines for that integration process. Note our implementation performs no manipulation of probability values other than permitting their entry and display. 5.1 The Hierarchy of Evidence Codes The Evidence-code components of evidence tuples denote different types of evidence. Those evidence types are arranged in a generalization specialization hierarchy as shown in Table 1. The root of that hierarchy, the node Evidence, has four direct children that define the four main evidence types: EV Comp: Inferred from Computation. The evidence for an assertion comes from a computational analysis. The assertion itself might have been made by a person or by a computer, that is, EV Comp does not specify whether manual interpretation of the computation occurred. EV Exp: Inferred from Experiment. The evidence for an assertion comes from a wet-lab experiment of some type.
8 EV Comp EV Comp HInf EV Comp HInf Positional Identification EV Comp HInf Similar To Consensus EV Comp HInf Fn From Seq EV Comp AInf EV Comp AInf Positional Identification EV Comp AInf Similar To Consensus EV Comp AInf Fn From Seq EV Comp AInf Single Directon EV Exp EV Exp IPI EV Exp IMP EV Exp IMP Site Mutation EV Exp IMP Polar Mutation EV Exp IMP Reaction Blocked EV Exp IMP Reaction Enhanced EV Exp IGI EV Exp IGI Func Complementation EV Exp IEP EV Exp IEP Gene Expression Analysis EV Exp IDA EV Exp IDA Binding Of Cellular Extracts EV Exp IDA Binding Of Purified Proteins EV Exp IDA RNA Polymerase Footprinting EV Exp IDA Transcription Init Mapping EV Exp IDA Boundaries Defined EV Exp IDA Transcript Len Determination EV Exp IDA Unpurified Protein EV Exp IDA Purified Protein Multspecies EV Exp IDA Purified Protein EV IC EV AS EV AS TAS EV AS NAS Inferred from computational analysis. Inferred by a human based on computational evidence. Human inference of promoter position. Human inf. based on similarity to consensus sequences. Human inf. of function from sequence. Inferred computationally without human oversight automated inference. Automated inf. of promoter position. Automated inf. based on similarity to consensus sequences. Automated inf. of function from sequence. Automated inf. that a single-gene directon is a transcription unit. Inferred from experiment. Inferred from physical interaction. Inferred from mutant phenotype. Site mutation. Polar mutation. Reaction blocked in mutant. Reaction enhanced in mutant. Inferred from genetic interaction. Inferred by functional complementation. Inferred from expression pattern. Gene expression analysis. Inferred from direct assay. Binding of cellular extracts. Binding of purified proteins. RNA polymerase footprinting. Transcription initiation mapping. Boundaries of transcription experimentally identified. Length of transcript experimentally determined. Assay of unpurified protein. Assay of protein purified from mixed culture. Assay of purified protein. Inferred by curator. Author statement. Traceable author statement. Non-traceable author statement. Table 1: The taxonomy of evidence types. Each row of this table defines one evidence type, giving its code and description. Indentation indicates ordering in the taxonomy, for example, EV AS TAS (fourth row) is a child of EV AS.
9 EV IC: Inferred by Curator. An assertion was inferred by a curator from relevant information such as other assertions in a database. EV AS: Author Statement. The evidence for an assertion comes from an author statement in a publication, where that publication does not provide direct experimental support for the assertion. (Ordinarily, this code will not be used directly generally one of its child codes, EV AS TAS or EV AS NAS, will be used instead.) We expect the most commonly used codes will be EV Comp and EV Exp, and their sub-codes. An HTML version of the entire evidence ontology, including detailed comments describing each evidence code, is available at URL There are several reasons why we feel this evidence system is best structured as a hierarchical ontology rather than as a flat list of controlled terms. First, we expect that the hierarchy will facilitate understanding of the evidence system by new curators because terms are grouped into logically related clusters. This aspect will be particularly important as the size and complexity of the evidence ontology grows to model additional detailed evidence types. Second, we expect the hierarchy will facilitate the curation process itself by allowing faster retrieval of relevant terms from editor menus than if retrieval was from a flat list. Third, non-leaf nodes in the evidence ontology will themselves be used in curation in cases where leaf evidence nodes do not match the evidence that is actually available, or where a publication is not specific about the type of evidence that supports some assertion, thus requiring a more general evidence code. The lower levels of the hierarchy have thus far been designed primarily for encoding the evidence for protein function, and evidence related to mechanisms of regulation of transcription initiation. There are many types of experiments and computational techniques, but our curators have made efforts to divide and group them into the categories they judge to be most meaningful. These evidence codes are not comprehensive with respect to other types of biological information. In the future, if we decide to apply evidence codes to different types of objects, we expect to extend the existing set of evidence codes to cover the types of experiments and analyses applicable to the new object types. In our implementation of the evidence codes, each code is defined as a class within the Ocelot object DBMS whose object ID is the evidence code. Slots defined for each evidence code include its name (such as Inferred by Computational Analysis ), a comment describing the code, and a slot Pertains To that lists the classes of PGDB objects to which the code can be applied. The Pathway Tools editors query this slot to determine what evidence codes
10 are applicable to a given type of objects, such as a metabolic pathway, when generating choose-lists of evidence codes for the curator. The EV AS (Author Statement) category has two subtypes: author statements that are traceable to a publication that contained direct evidence for an assertion, and statements that are not traceable in that manner. The EV Comp (Inferred by Computational Analysis) category is also divided into two subtypes: computational inferences that were made in a purely automated fashion, and inferences in which a human was involved, under the assumption that the condition of whether or not a person was involved in arbitrating among computational evidence is a factor that a scientist interpreting the evidence would consider important. An examination of the subtypes of EV Comp reveals that some of these evidence types apply only to certain PGDB object types. For example, the code EV Comp AInf Single Directon applies only to computational inference of operons (it indicates that an operon was inferred by the existence of a gene G for which the adjacent genes on both sides of G are both transcribed in the opposite direction from G, implying that those genes cannot be in the same operon as G). This property of being relevant only to specific object types applies to other evidence types in our system. 5.2 Attaching Evidence Codes to Individual Slot Values As well as using evidence tuples to record evidence about object existence, we can attach evidence tuples to individual values within a PGDB to record evidence for finer-grained assertions. For example, we could record the evidence that supports the strength of a promoter stored in a PGDB, or that supports the assertion that a given metabolite inhibits the activity of an enzyme. 5.3 Object and Relational Implementations of Evidence Tuples We implement the association of evidence tuples with individual slot values using an Ocelot mechanism called annotations. An annotation is a five-tuple of the form Frame : Slot : Value : Label : Datum that allows a labeled datum to be associated with a value within a slot of a frame. In this case, the label is the string Evidence and the datum is the evidence tuple itself. In Ocelot, annotation tuples are physically stored within the frame that they are associated with. We envision that our evidence system could be implemented in a relational DBMS by creating a table whose columns are Table-ID, Key, Column, Value, Evidence-Code, Citation, Curator, Timestamp, and Probability. Table-ID, Key, and Column are analogous to Frame and Slot in the Ocelot
11 representation they identify an object within a relational DBMS. Column and Value identify a specific relational column and value within that column to which the evidence tuple applies. When the evidence tuple applies to the entire object, Column and Value would be null. In both the Ocelot and the relational implementations, it is straightforward to compute across evidence information, such as to query all pathways with experimental evidence. 6 Use of the Evidence Ontology within EcoCyc and MetaCyc The EcoCyc DB currently records evidence codes for the existence of 341 of its 810 transcription units, for 531 of its 878 promoters, and for 952 of its 1071 DNA binding sites. The preceding evidence codes were assigned over the past few years using an earlier, more primitive evidence system. We translated codes from that system into the system described here, which was feasible because the new system was explicitly designed to be a superset of the old system. Assignment of evidence codes for pathways and protein functions by curators of the EcoCyc and MetaCyc DBs is just beginning. As an example, the anaerobic toluene degradation pathway, described in MetaCyc as it appears in Thauera aromatica, has been established by biochemical assay of the enzymes, pathway intermediates, and products. Thus, we would assign to it the evidence code EV Exp IDA (Inferred by Direct Assay), along with one or more citations to the literature. If a user were to use PathoLogic to create a PGDB for some other organism, X, and PathoLogic inferred that the anaerobic toluene degradation pathway were present in X, it would be assigned an evidence code of EV-AInf (automated inference) in the new PGDB (this assignment is done automatically by PathoLogic). A protein that is multifunctional may have the same or different evidence codes assigned to each function. In addition to containing objects for each protein and for each reaction, the Pathway Tools schema defines an object, called an enzymatic-reaction, that describes the pairing of an enzyme to a reaction. We assign the evidence code for each function of a protein to the corresponding enzymatic-reaction object rather than to the protein object so there is no ambiguity about which functional assignment each evidence code pertains to. For example, the product of the E. coli ndh gene has NADH dehydrogenase and NADH cupric reductase activities. The NADH dehydrogenase activity was established by direct assay of the purified protein. The NADH cupric reductase activity was established by observing that the reaction is blocked or enhanced in mutants with the gene missing or over-expressed, respectively. Thus, we assign the evidence code EV Exp IDA Purified Protein (along with relevant literature citations) to the enzymatic-reaction that links the ndh gene product to the NADH de-
12 hydrogenase reaction, and the codes EV-IMP Reaction Enhanced and EV Exp IMP Reaction Blocked to the enzymatic-reaction that links the protein to the NADH cupric reductase reaction. (Note that this is not intended to be a complete description of the ndh system additional codes may apply to one or both activities.) We are not restricted to using the lowest-level codes in the evidence ontology. If the enzyme was characterized by some experiment that did not precisely match one of our lowest-level codes, or the literature did not provide enough information to distinguish between them, we might assign a code of EV Exp IMP even EV Exp in place of one of the more specific codes. 7 Software Availability Pathway Tools for SUN/Solaris, Intel/Windows, and Intel/Linux is freely available to academics; a license fee applies to commercial use. See for more details. Acknowledgments Julio Collado-Vides contributed to development of evidence codes related to transcriptional regulation. This work was supported by grant R01-HG from the NIH National Human Genome Research Institute. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. 1. P.D. Karp, S. Paley, and P. Romero. The Pathway Tools Software. Bioinformatics, 18:S225 S232, P.D. Karp, M. Riley, M. Saier, I.T. Paulsen, S. Paley, and A. Pellegrini- Toole. The EcoCyc database. Nuc. Acids Res., 30(1):56 8, P.D. Karp, M. Riley, S. Paley, and A. Pellegrini-Toole. The MetaCyc database. Nuc. Acids Res., 30(1):59 61, M. Ashburner, C.A. Ball, J.A. Blake, D. Botstein, H. Butler, J.M. Cherry, A.P. Davis, K. Dolinski, S.S. Dwight, J.T. Eppig, M.A. Harris, D.P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J.C. Matese, J.E. Richardson, M. Ringwald, G.M. Rubin, and G. Sherlock. Gene Ontology: Tool for the unification of biology. Nature Genetics, 25:25 29, J. Doyle. Truth maintenance systems. Artificial Intelligence, 12(3):231 72, S. Paley and P.D. Karp. Evaluation of computational metabolic-pathway predictions for H. pylori. Bioinformatics, 18(5): , P. Romero and P.D. Karp. PseudoCyc, a Pathway/Genome Database for Pseudomonas aeruginosa. J. Mol. Microbiol. Biotech., 5(4):230 9, 2003.
The EcoCyc Database. January 25, de Nitrógeno, UNAM,Cuernavaca, A.P. 565-A, Morelos, 62100, Mexico;
 The EcoCyc Database Peter D. Karp, Monica Riley, Milton Saier,IanT.Paulsen +, Julio Collado-Vides + Suzanne M. Paley, Alida Pellegrini-Toole,César Bonavides ++, and Socorro Gama-Castro ++ January 25, 2002
The EcoCyc Database Peter D. Karp, Monica Riley, Milton Saier,IanT.Paulsen +, Julio Collado-Vides + Suzanne M. Paley, Alida Pellegrini-Toole,César Bonavides ++, and Socorro Gama-Castro ++ January 25, 2002
GOSAP: Gene Ontology Based Semantic Alignment of Biological Pathways
 GOSAP: Gene Ontology Based Semantic Alignment of Biological Pathways Jonas Gamalielsson and Björn Olsson Systems Biology Group, Skövde University, Box 407, Skövde, 54128, Sweden, [jonas.gamalielsson][bjorn.olsson]@his.se,
GOSAP: Gene Ontology Based Semantic Alignment of Biological Pathways Jonas Gamalielsson and Björn Olsson Systems Biology Group, Skövde University, Box 407, Skövde, 54128, Sweden, [jonas.gamalielsson][bjorn.olsson]@his.se,
Transitioning BioCyc to a Subscription Model
 Transitioning BioCyc to a Subscription Model Peter D. Karp SRI International ecocyc.org biocyc.org metacyc.org BioCyc.org Collection of 9,300 Pathway/Genome Databases Pathway/Genome Database (PGDB) combines
Transitioning BioCyc to a Subscription Model Peter D. Karp SRI International ecocyc.org biocyc.org metacyc.org BioCyc.org Collection of 9,300 Pathway/Genome Databases Pathway/Genome Database (PGDB) combines
Chapter 15 Active Reading Guide Regulation of Gene Expression
 Name: AP Biology Mr. Croft Chapter 15 Active Reading Guide Regulation of Gene Expression The overview for Chapter 15 introduces the idea that while all cells of an organism have all genes in the genome,
Name: AP Biology Mr. Croft Chapter 15 Active Reading Guide Regulation of Gene Expression The overview for Chapter 15 introduces the idea that while all cells of an organism have all genes in the genome,
Estimation of Identification Methods of Gene Clusters Using GO Term Annotations from a Hierarchical Cluster Tree
 Estimation of Identification Methods of Gene Clusters Using GO Term Annotations from a Hierarchical Cluster Tree YOICHI YAMADA, YUKI MIYATA, MASANORI HIGASHIHARA*, KENJI SATOU Graduate School of Natural
Estimation of Identification Methods of Gene Clusters Using GO Term Annotations from a Hierarchical Cluster Tree YOICHI YAMADA, YUKI MIYATA, MASANORI HIGASHIHARA*, KENJI SATOU Graduate School of Natural
86 Part 4 SUMMARY INTRODUCTION
 86 Part 4 Chapter # AN INTEGRATION OF THE DESCRIPTIONS OF GENE NETWORKS AND THEIR MODELS PRESENTED IN SIGMOID (CELLERATOR) AND GENENET Podkolodny N.L. *1, 2, Podkolodnaya N.N. 1, Miginsky D.S. 1, Poplavsky
86 Part 4 Chapter # AN INTEGRATION OF THE DESCRIPTIONS OF GENE NETWORKS AND THEIR MODELS PRESENTED IN SIGMOID (CELLERATOR) AND GENENET Podkolodny N.L. *1, 2, Podkolodnaya N.N. 1, Miginsky D.S. 1, Poplavsky
BMD645. Integration of Omics
 BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
V19 Metabolic Networks - Overview
 V19 Metabolic Networks - Overview There exist different levels of computational methods for describing metabolic networks: - stoichiometry/kinetics of classical biochemical pathways (glycolysis, TCA cycle,...
V19 Metabolic Networks - Overview There exist different levels of computational methods for describing metabolic networks: - stoichiometry/kinetics of classical biochemical pathways (glycolysis, TCA cycle,...
ATLAS of Biochemistry
 ATLAS of Biochemistry USER GUIDE http://lcsb-databases.epfl.ch/atlas/ CONTENT 1 2 3 GET STARTED Create your user account NAVIGATE Curated KEGG reactions ATLAS reactions Pathways Maps USE IT! Fill a gap
ATLAS of Biochemistry USER GUIDE http://lcsb-databases.epfl.ch/atlas/ CONTENT 1 2 3 GET STARTED Create your user account NAVIGATE Curated KEGG reactions ATLAS reactions Pathways Maps USE IT! Fill a gap
V14 extreme pathways
 V14 extreme pathways A torch is directed at an open door and shines into a dark room... What area is lighted? Instead of marking all lighted points individually, it would be sufficient to characterize
V14 extreme pathways A torch is directed at an open door and shines into a dark room... What area is lighted? Instead of marking all lighted points individually, it would be sufficient to characterize
Pathway Bioinformatics: Inference, Visualization, and Analysis. Peter D. Karp, Ph.D.
 Pathway Bioinformatics: Inference, Visualization, and Analysis Peter D. Karp, Ph.D. Bioinformatics Research Group SRI International pkarp@ai.sri.com BioCyc.org EcoCyc.org, MetaCyc.org, HumanCyc.org 1 SRI
Pathway Bioinformatics: Inference, Visualization, and Analysis Peter D. Karp, Ph.D. Bioinformatics Research Group SRI International pkarp@ai.sri.com BioCyc.org EcoCyc.org, MetaCyc.org, HumanCyc.org 1 SRI
OECD QSAR Toolbox v.4.1. Tutorial on how to predict Skin sensitization potential taking into account alert performance
 OECD QSAR Toolbox v.4.1 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.1 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.0. Tutorial on how to predict Skin sensitization potential taking into account alert performance
 OECD QSAR Toolbox v.4.0 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
OECD QSAR Toolbox v.4.0 Tutorial on how to predict Skin sensitization potential taking into account alert performance Outlook Background Objectives Specific Aims Read across and analogue approach The exercise
Networks & pathways. Hedi Peterson MTAT Bioinformatics
 Networks & pathways Hedi Peterson (peterson@quretec.com) MTAT.03.239 Bioinformatics 03.11.2010 Networks are graphs Nodes Edges Edges Directed, undirected, weighted Nodes Genes Proteins Metabolites Enzymes
Networks & pathways Hedi Peterson (peterson@quretec.com) MTAT.03.239 Bioinformatics 03.11.2010 Networks are graphs Nodes Edges Edges Directed, undirected, weighted Nodes Genes Proteins Metabolites Enzymes
OECD QSAR Toolbox v.4.1. Tutorial illustrating new options for grouping with metabolism
 OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
OECD QSAR Toolbox v.4.1 Tutorial illustrating new options for grouping with metabolism Outlook Background Objectives Specific Aims The exercise Workflow 2 Background Grouping with metabolism is a procedure
This document describes the process by which operons are predicted for genes within the BioHealthBase database.
 1. Purpose This document describes the process by which operons are predicted for genes within the BioHealthBase database. 2. Methods Description An operon is a coexpressed set of genes, transcribed onto
1. Purpose This document describes the process by which operons are predicted for genes within the BioHealthBase database. 2. Methods Description An operon is a coexpressed set of genes, transcribed onto
Introduction. Gene expression is the combined process of :
 1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1. In most cases, genes code for and it is that
 Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
Supplemental Materials
 JOURNAL OF MICROBIOLOGY & BIOLOGY EDUCATION, May 2013, p. 107-109 DOI: http://dx.doi.org/10.1128/jmbe.v14i1.496 Supplemental Materials for Engaging Students in a Bioinformatics Activity to Introduce Gene
JOURNAL OF MICROBIOLOGY & BIOLOGY EDUCATION, May 2013, p. 107-109 DOI: http://dx.doi.org/10.1128/jmbe.v14i1.496 Supplemental Materials for Engaging Students in a Bioinformatics Activity to Introduce Gene
Organization of Genes Differs in Prokaryotic and Eukaryotic DNA Chapter 10 p
 Organization of Genes Differs in Prokaryotic and Eukaryotic DNA Chapter 10 p.110-114 Arrangement of information in DNA----- requirements for RNA Common arrangement of protein-coding genes in prokaryotes=
Organization of Genes Differs in Prokaryotic and Eukaryotic DNA Chapter 10 p.110-114 Arrangement of information in DNA----- requirements for RNA Common arrangement of protein-coding genes in prokaryotes=
SABIO-RK Integration and Curation of Reaction Kinetics Data Ulrike Wittig
 SABIO-RK Integration and Curation of Reaction Kinetics Data http://sabio.villa-bosch.de/sabiork Ulrike Wittig Overview Introduction /Motivation Database content /User interface Data integration Curation
SABIO-RK Integration and Curation of Reaction Kinetics Data http://sabio.villa-bosch.de/sabiork Ulrike Wittig Overview Introduction /Motivation Database content /User interface Data integration Curation
Bioinformatics. Dept. of Computational Biology & Bioinformatics
 Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Computational Genomics. Systems biology. Putting it together: Data integration using graphical models
 02-710 Computational Genomics Systems biology Putting it together: Data integration using graphical models High throughput data So far in this class we discussed several different types of high throughput
02-710 Computational Genomics Systems biology Putting it together: Data integration using graphical models High throughput data So far in this class we discussed several different types of high throughput
3.B.1 Gene Regulation. Gene regulation results in differential gene expression, leading to cell specialization.
 3.B.1 Gene Regulation Gene regulation results in differential gene expression, leading to cell specialization. We will focus on gene regulation in prokaryotes first. Gene regulation accounts for some of
3.B.1 Gene Regulation Gene regulation results in differential gene expression, leading to cell specialization. We will focus on gene regulation in prokaryotes first. Gene regulation accounts for some of
Pathway Databases: A Case Study in Computational Symbolic Theories
 sponsors development of the information technology infrastructure necessary for a National Virtual Observatory (NVO). There is a close cooperation with the particle physics community through the Grid Physics
sponsors development of the information technology infrastructure necessary for a National Virtual Observatory (NVO). There is a close cooperation with the particle physics community through the Grid Physics
Biological Pathways Representation by Petri Nets and extension
 Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
GCD3033:Cell Biology. Transcription
 Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Outline. Terminologies and Ontologies. Communication and Computation. Communication. Outline. Terminologies and Vocabularies.
 Page 1 Outline 1. Why do we need terminologies and ontologies? Terminologies and Ontologies Iwei Yeh yeh@smi.stanford.edu 04/16/2002 2. Controlled Terminologies Enzyme Classification Gene Ontology 3. Ontologies
Page 1 Outline 1. Why do we need terminologies and ontologies? Terminologies and Ontologies Iwei Yeh yeh@smi.stanford.edu 04/16/2002 2. Controlled Terminologies Enzyme Classification Gene Ontology 3. Ontologies
2 GENE FUNCTIONAL SIMILARITY. 2.1 Semantic values of GO terms
 Bioinformatics Advance Access published March 7, 2007 The Author (2007). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Bioinformatics Advance Access published March 7, 2007 The Author (2007). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Integration of functional genomics data
 Integration of functional genomics data Laboratoire Bordelais de Recherche en Informatique (UMR) Centre de Bioinformatique de Bordeaux (Plateforme) Rennes Oct. 2006 1 Observations and motivations Genomics
Integration of functional genomics data Laboratoire Bordelais de Recherche en Informatique (UMR) Centre de Bioinformatique de Bordeaux (Plateforme) Rennes Oct. 2006 1 Observations and motivations Genomics
Prokaryotic Gene Expression (Learning Objectives)
 Prokaryotic Gene Expression (Learning Objectives) 1. Learn how bacteria respond to changes of metabolites in their environment: short-term and longer-term. 2. Compare and contrast transcriptional control
Prokaryotic Gene Expression (Learning Objectives) 1. Learn how bacteria respond to changes of metabolites in their environment: short-term and longer-term. 2. Compare and contrast transcriptional control
FCModeler: Dynamic Graph Display and Fuzzy Modeling of Regulatory and Metabolic Maps
 FCModeler: Dynamic Graph Display and Fuzzy Modeling of Regulatory and Metabolic Maps Julie Dickerson 1, Zach Cox 1 and Andy Fulmer 2 1 Iowa State University and 2 Proctor & Gamble. FCModeler Goals Capture
FCModeler: Dynamic Graph Display and Fuzzy Modeling of Regulatory and Metabolic Maps Julie Dickerson 1, Zach Cox 1 and Andy Fulmer 2 1 Iowa State University and 2 Proctor & Gamble. FCModeler Goals Capture
Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Chapter 18 Regulation of Gene Expression PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
AP Bio Module 16: Bacterial Genetics and Operons, Student Learning Guide
 Name: Period: Date: AP Bio Module 6: Bacterial Genetics and Operons, Student Learning Guide Getting started. Work in pairs (share a computer). Make sure that you log in for the first quiz so that you get
Name: Period: Date: AP Bio Module 6: Bacterial Genetics and Operons, Student Learning Guide Getting started. Work in pairs (share a computer). Make sure that you log in for the first quiz so that you get
INTERACTIVE CLUSTERING FOR EXPLORATION OF GENOMIC DATA
 INTERACTIVE CLUSTERING FOR EXPLORATION OF GENOMIC DATA XIUFENG WAN xw6@cs.msstate.edu Department of Computer Science Box 9637 JOHN A. BOYLE jab@ra.msstate.edu Department of Biochemistry and Molecular Biology
INTERACTIVE CLUSTERING FOR EXPLORATION OF GENOMIC DATA XIUFENG WAN xw6@cs.msstate.edu Department of Computer Science Box 9637 JOHN A. BOYLE jab@ra.msstate.edu Department of Biochemistry and Molecular Biology
Gene Ontology and overrepresentation analysis
 Gene Ontology and overrepresentation analysis Kjell Petersen J Express Microarray analysis course Oslo December 2009 Presentation adapted from Endre Anderssen and Vidar Beisvåg NMC Trondheim Overview How
Gene Ontology and overrepresentation analysis Kjell Petersen J Express Microarray analysis course Oslo December 2009 Presentation adapted from Endre Anderssen and Vidar Beisvåg NMC Trondheim Overview How
Computational methods for the analysis of bacterial gene regulation Brouwer, Rutger Wubbe Willem
 University of Groningen Computational methods for the analysis of bacterial gene regulation Brouwer, Rutger Wubbe Willem IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
University of Groningen Computational methods for the analysis of bacterial gene regulation Brouwer, Rutger Wubbe Willem IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
Topic 4 - #14 The Lactose Operon
 Topic 4 - #14 The Lactose Operon The Lactose Operon The lactose operon is an operon which is responsible for the transport and metabolism of the sugar lactose in E. coli. - Lactose is one of many organic
Topic 4 - #14 The Lactose Operon The Lactose Operon The lactose operon is an operon which is responsible for the transport and metabolism of the sugar lactose in E. coli. - Lactose is one of many organic
Introduction to Bioinformatics
 CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression Edited by Shawn Lester PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley
Chapter 18 Regulation of Gene Expression Edited by Shawn Lester PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley
Complete all warm up questions Focus on operon functioning we will be creating operon models on Monday
 Complete all warm up questions Focus on operon functioning we will be creating operon models on Monday 1. What is the Central Dogma? 2. How does prokaryotic DNA compare to eukaryotic DNA? 3. How is DNA
Complete all warm up questions Focus on operon functioning we will be creating operon models on Monday 1. What is the Central Dogma? 2. How does prokaryotic DNA compare to eukaryotic DNA? 3. How is DNA
Name Period The Control of Gene Expression in Prokaryotes Notes
 Bacterial DNA contains genes that encode for many different proteins (enzymes) so that many processes have the ability to occur -not all processes are carried out at any one time -what allows expression
Bacterial DNA contains genes that encode for many different proteins (enzymes) so that many processes have the ability to occur -not all processes are carried out at any one time -what allows expression
OECD QSAR Toolbox v.3.4. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.4 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.3. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.3 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
Name: SBI 4U. Gene Expression Quiz. Overall Expectation:
 Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
BME 5742 Biosystems Modeling and Control
 BME 5742 Biosystems Modeling and Control Lecture 24 Unregulated Gene Expression Model Dr. Zvi Roth (FAU) 1 The genetic material inside a cell, encoded in its DNA, governs the response of a cell to various
BME 5742 Biosystems Modeling and Control Lecture 24 Unregulated Gene Expression Model Dr. Zvi Roth (FAU) 1 The genetic material inside a cell, encoded in its DNA, governs the response of a cell to various
Simulation of Gene Regulatory Networks
 Simulation of Gene Regulatory Networks Overview I have been assisting Professor Jacques Cohen at Brandeis University to explore and compare the the many available representations and interpretations of
Simulation of Gene Regulatory Networks Overview I have been assisting Professor Jacques Cohen at Brandeis University to explore and compare the the many available representations and interpretations of
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Prokaryotic Gene Expression (Learning Objectives)
 Prokaryotic Gene Expression (Learning Objectives) 1. Learn how bacteria respond to changes of metabolites in their environment: short-term and longer-term. 2. Compare and contrast transcriptional control
Prokaryotic Gene Expression (Learning Objectives) 1. Learn how bacteria respond to changes of metabolites in their environment: short-term and longer-term. 2. Compare and contrast transcriptional control
Multiple Choice Review- Eukaryotic Gene Expression
 Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Lecture 4: Transcription networks basic concepts
 Lecture 4: Transcription networks basic concepts - Activators and repressors - Input functions; Logic input functions; Multidimensional input functions - Dynamics and response time 2.1 Introduction The
Lecture 4: Transcription networks basic concepts - Activators and repressors - Input functions; Logic input functions; Multidimensional input functions - Dynamics and response time 2.1 Introduction The
 Supplemental Table 1. Primers used for qrt-pcr analysis Name abf2rtf abf2rtr abf3rtf abf3rtf abf4rtf abf4rtr ANAC096RTF ANAC096RTR abf2lp abf2rp abf3lp abf3lp abf4lp abf4rp ANAC096F(XbaI) ANAC096R(BamHI)
Supplemental Table 1. Primers used for qrt-pcr analysis Name abf2rtf abf2rtr abf3rtf abf3rtf abf4rtf abf4rtr ANAC096RTF ANAC096RTR abf2lp abf2rp abf3lp abf3lp abf4lp abf4rp ANAC096F(XbaI) ANAC096R(BamHI)
RGP finder: prediction of Genomic Islands
 Training courses on MicroScope platform RGP finder: prediction of Genomic Islands Dynamics of bacterial genomes Gene gain Horizontal gene transfer Gene loss Deletion of one or several genes Duplication
Training courses on MicroScope platform RGP finder: prediction of Genomic Islands Dynamics of bacterial genomes Gene gain Horizontal gene transfer Gene loss Deletion of one or several genes Duplication
GENE ONTOLOGY (GO) Wilver Martínez Martínez Giovanny Silva Rincón
 GENE ONTOLOGY (GO) Wilver Martínez Martínez Giovanny Silva Rincón What is GO? The Gene Ontology (GO) project is a collaborative effort to address the need for consistent descriptions of gene products in
GENE ONTOLOGY (GO) Wilver Martínez Martínez Giovanny Silva Rincón What is GO? The Gene Ontology (GO) project is a collaborative effort to address the need for consistent descriptions of gene products in
REVIEW SESSION. Wednesday, September 15 5:30 PM SHANTZ 242 E
 REVIEW SESSION Wednesday, September 15 5:30 PM SHANTZ 242 E Gene Regulation Gene Regulation Gene expression can be turned on, turned off, turned up or turned down! For example, as test time approaches,
REVIEW SESSION Wednesday, September 15 5:30 PM SHANTZ 242 E Gene Regulation Gene Regulation Gene expression can be turned on, turned off, turned up or turned down! For example, as test time approaches,
UNIT 6 PART 3 *REGULATION USING OPERONS* Hillis Textbook, CH 11
 UNIT 6 PART 3 *REGULATION USING OPERONS* Hillis Textbook, CH 11 REVIEW: Signals that Start and Stop Transcription and Translation BUT, HOW DO CELLS CONTROL WHICH GENES ARE EXPRESSED AND WHEN? First of
UNIT 6 PART 3 *REGULATION USING OPERONS* Hillis Textbook, CH 11 REVIEW: Signals that Start and Stop Transcription and Translation BUT, HOW DO CELLS CONTROL WHICH GENES ARE EXPRESSED AND WHEN? First of
METABOLIC PATHWAY PREDICTION/ALIGNMENT
 COMPUTATIONAL SYSTEMIC BIOLOGY METABOLIC PATHWAY PREDICTION/ALIGNMENT Hofestaedt R*, Chen M Bioinformatics / Medical Informatics, Technische Fakultaet, Universitaet Bielefeld Postfach 10 01 31, D-33501
COMPUTATIONAL SYSTEMIC BIOLOGY METABOLIC PATHWAY PREDICTION/ALIGNMENT Hofestaedt R*, Chen M Bioinformatics / Medical Informatics, Technische Fakultaet, Universitaet Bielefeld Postfach 10 01 31, D-33501
Classification of Gene Expression Profiles: Comparison of K-means and Expectation Maximization Algorithms
 Eighth International Conference on Hybrid Intelligent Systems Classification of Gene Expression Profiles: Comparison of K-means and Expectation Maximization Algorithms Cristina Rubio-Escudero Dpto. Lenguajes
Eighth International Conference on Hybrid Intelligent Systems Classification of Gene Expression Profiles: Comparison of K-means and Expectation Maximization Algorithms Cristina Rubio-Escudero Dpto. Lenguajes
OECD QSAR Toolbox v.3.2. Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding
 OECD QSAR Toolbox v.3.2 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
OECD QSAR Toolbox v.3.2 Step-by-step example of how to build and evaluate a category based on mechanism of action with protein and DNA binding Outlook Background Objectives Specific Aims The exercise Workflow
Computational Biology: Basics & Interesting Problems
 Computational Biology: Basics & Interesting Problems Summary Sources of information Biological concepts: structure & terminology Sequencing Gene finding Protein structure prediction Sources of information
Computational Biology: Basics & Interesting Problems Summary Sources of information Biological concepts: structure & terminology Sequencing Gene finding Protein structure prediction Sources of information
CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON
 PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
OECD QSAR Toolbox v.3.3. Predicting skin sensitisation potential of a chemical using skin sensitization data extracted from ECHA CHEM database
 OECD QSAR Toolbox v.3.3 Predicting skin sensitisation potential of a chemical using skin sensitization data extracted from ECHA CHEM database Outlook Background The exercise Workflow Save prediction 23.02.2015
OECD QSAR Toolbox v.3.3 Predicting skin sensitisation potential of a chemical using skin sensitization data extracted from ECHA CHEM database Outlook Background The exercise Workflow Save prediction 23.02.2015
PROTEIN SYNTHESIS INTRO
 MR. POMERANTZ Page 1 of 6 Protein synthesis Intro. Use the text book to help properly answer the following questions 1. RNA differs from DNA in that RNA a. is single-stranded. c. contains the nitrogen
MR. POMERANTZ Page 1 of 6 Protein synthesis Intro. Use the text book to help properly answer the following questions 1. RNA differs from DNA in that RNA a. is single-stranded. c. contains the nitrogen
Grundlagen der Bioinformatik Summer semester Lecturer: Prof. Daniel Huson
 Grundlagen der Bioinformatik, SS 10, D. Huson, April 12, 2010 1 1 Introduction Grundlagen der Bioinformatik Summer semester 2010 Lecturer: Prof. Daniel Huson Office hours: Thursdays 17-18h (Sand 14, C310a)
Grundlagen der Bioinformatik, SS 10, D. Huson, April 12, 2010 1 1 Introduction Grundlagen der Bioinformatik Summer semester 2010 Lecturer: Prof. Daniel Huson Office hours: Thursdays 17-18h (Sand 14, C310a)
O 3 O 4 O 5. q 3. q 4. Transition
 Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Computational approaches for functional genomics
 Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
Intelligent GIS: Automatic generation of qualitative spatial information
 Intelligent GIS: Automatic generation of qualitative spatial information Jimmy A. Lee 1 and Jane Brennan 1 1 University of Technology, Sydney, FIT, P.O. Box 123, Broadway NSW 2007, Australia janeb@it.uts.edu.au
Intelligent GIS: Automatic generation of qualitative spatial information Jimmy A. Lee 1 and Jane Brennan 1 1 University of Technology, Sydney, FIT, P.O. Box 123, Broadway NSW 2007, Australia janeb@it.uts.edu.au
Lab 2 Worksheet. Problems. Problem 1: Geometry and Linear Equations
 Lab 2 Worksheet Problems Problem : Geometry and Linear Equations Linear algebra is, first and foremost, the study of systems of linear equations. You are going to encounter linear systems frequently in
Lab 2 Worksheet Problems Problem : Geometry and Linear Equations Linear algebra is, first and foremost, the study of systems of linear equations. You are going to encounter linear systems frequently in
GLOBEX Bioinformatics (Summer 2015) Genetic networks and gene expression data
 GLOBEX Bioinformatics (Summer 2015) Genetic networks and gene expression data 1 Gene Networks Definition: A gene network is a set of molecular components, such as genes and proteins, and interactions between
GLOBEX Bioinformatics (Summer 2015) Genetic networks and gene expression data 1 Gene Networks Definition: A gene network is a set of molecular components, such as genes and proteins, and interactions between
Biological Networks: Comparison, Conservation, and Evolution via Relative Description Length By: Tamir Tuller & Benny Chor
 Biological Networks:,, and via Relative Description Length By: Tamir Tuller & Benny Chor Presented by: Noga Grebla Content of the presentation Presenting the goals of the research Reviewing basic terms
Biological Networks:,, and via Relative Description Length By: Tamir Tuller & Benny Chor Presented by: Noga Grebla Content of the presentation Presenting the goals of the research Reviewing basic terms
Browsing Genomic Information with Ensembl Plants
 Browsing Genomic Information with Ensembl Plants Etienne de Villiers, PhD (Adapted from slides by Bert Overduin EMBL-EBI) Outline of workshop Brief introduction to Ensembl Plants History Content Tutorial
Browsing Genomic Information with Ensembl Plants Etienne de Villiers, PhD (Adapted from slides by Bert Overduin EMBL-EBI) Outline of workshop Brief introduction to Ensembl Plants History Content Tutorial
56:198:582 Biological Networks Lecture 8
 56:198:582 Biological Networks Lecture 8 Course organization Two complementary approaches to modeling and understanding biological networks Constraint-based modeling (Palsson) System-wide Metabolism Steady-state
56:198:582 Biological Networks Lecture 8 Course organization Two complementary approaches to modeling and understanding biological networks Constraint-based modeling (Palsson) System-wide Metabolism Steady-state
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Tuesday, December 27, 16
 Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Enduring understanding 3.B: Expression of genetic information involves cellular and molecular
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Enduring understanding 3.B: Expression of genetic information involves cellular and molecular
Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis
 Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis Technical Overview Introduction Metabolomics studies measure the relative abundance of metabolites
Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis Technical Overview Introduction Metabolomics studies measure the relative abundance of metabolites
Gene Switches Teacher Information
 STO-143 Gene Switches Teacher Information Summary Kit contains How do bacteria turn on and turn off genes? Students model the action of the lac operon that regulates the expression of genes essential for
STO-143 Gene Switches Teacher Information Summary Kit contains How do bacteria turn on and turn off genes? Students model the action of the lac operon that regulates the expression of genes essential for
BIOINFORMATICS LAB AP BIOLOGY
 BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
4. Why not make all enzymes all the time (even if not needed)? Enzyme synthesis uses a lot of energy.
 1 C2005/F2401 '10-- Lecture 15 -- Last Edited: 11/02/10 01:58 PM Copyright 2010 Deborah Mowshowitz and Lawrence Chasin Department of Biological Sciences Columbia University New York, NY. Handouts: 15A
1 C2005/F2401 '10-- Lecture 15 -- Last Edited: 11/02/10 01:58 PM Copyright 2010 Deborah Mowshowitz and Lawrence Chasin Department of Biological Sciences Columbia University New York, NY. Handouts: 15A
Reading Assignments. A. Genes and the Synthesis of Polypeptides. Lecture Series 7 From DNA to Protein: Genotype to Phenotype
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Boolean models of gene regulatory networks. Matthew Macauley Math 4500: Mathematical Modeling Clemson University Spring 2016
 Boolean models of gene regulatory networks Matthew Macauley Math 4500: Mathematical Modeling Clemson University Spring 2016 Gene expression Gene expression is a process that takes gene info and creates
Boolean models of gene regulatory networks Matthew Macauley Math 4500: Mathematical Modeling Clemson University Spring 2016 Gene expression Gene expression is a process that takes gene info and creates
Title: HyBrow - A Prototype System for Computer-Aided Hypothesis Evaluation
 Title: HyBrow - A Prototype System for Computer-Aided Hypothesis Evaluation Authors: Racunas S. A. #, The Department of Electrical Engineering, Pennsylvania State University, University Park, PA 16802,
Title: HyBrow - A Prototype System for Computer-Aided Hypothesis Evaluation Authors: Racunas S. A. #, The Department of Electrical Engineering, Pennsylvania State University, University Park, PA 16802,
Bioinformatics Chapter 1. Introduction
 Bioinformatics Chapter 1. Introduction Outline! Biological Data in Digital Symbol Sequences! Genomes Diversity, Size, and Structure! Proteins and Proteomes! On the Information Content of Biological Sequences!
Bioinformatics Chapter 1. Introduction Outline! Biological Data in Digital Symbol Sequences! Genomes Diversity, Size, and Structure! Proteins and Proteomes! On the Information Content of Biological Sequences!
V14 Graph connectivity Metabolic networks
 V14 Graph connectivity Metabolic networks In the first half of this lecture section, we use the theory of network flows to give constructive proofs of Menger s theorem. These proofs lead directly to algorithms
V14 Graph connectivity Metabolic networks In the first half of this lecture section, we use the theory of network flows to give constructive proofs of Menger s theorem. These proofs lead directly to algorithms
STAAR Biology Assessment
 STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
Introduction to Bioinformatics Online Course: IBT
 Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec1 Building a Multiple Sequence Alignment Learning Outcomes 1- Understanding Why multiple
Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec1 Building a Multiple Sequence Alignment Learning Outcomes 1- Understanding Why multiple
Introduction to Bioinformatics
 Systems biology Introduction to Bioinformatics Systems biology: modeling biological p Study of whole biological systems p Wholeness : Organization of dynamic interactions Different behaviour of the individual
Systems biology Introduction to Bioinformatics Systems biology: modeling biological p Study of whole biological systems p Wholeness : Organization of dynamic interactions Different behaviour of the individual
Chapter 16 Lecture. Concepts Of Genetics. Tenth Edition. Regulation of Gene Expression in Prokaryotes
 Chapter 16 Lecture Concepts Of Genetics Tenth Edition Regulation of Gene Expression in Prokaryotes Chapter Contents 16.1 Prokaryotes Regulate Gene Expression in Response to Environmental Conditions 16.2
Chapter 16 Lecture Concepts Of Genetics Tenth Edition Regulation of Gene Expression in Prokaryotes Chapter Contents 16.1 Prokaryotes Regulate Gene Expression in Response to Environmental Conditions 16.2
Chapter 17. From Gene to Protein. Biology Kevin Dees
 Chapter 17 From Gene to Protein DNA The information molecule Sequences of bases is a code DNA organized in to chromosomes Chromosomes are organized into genes What do the genes actually say??? Reflecting
Chapter 17 From Gene to Protein DNA The information molecule Sequences of bases is a code DNA organized in to chromosomes Chromosomes are organized into genes What do the genes actually say??? Reflecting
Motivating the need for optimal sequence alignments...
 1 Motivating the need for optimal sequence alignments... 2 3 Note that this actually combines two objectives of optimal sequence alignments: (i) use the score of the alignment o infer homology; (ii) use
1 Motivating the need for optimal sequence alignments... 2 3 Note that this actually combines two objectives of optimal sequence alignments: (i) use the score of the alignment o infer homology; (ii) use
Microbiome Metabolic Modeling with Pathway Tools
 Microbiome Metabolic Modeling with Pathway Tools Peter D. Karp SRI International Cost for High Quality Models of 300 Microbiome Organisms Mul/ple body sites of human, mouse, farm animals 500 species in
Microbiome Metabolic Modeling with Pathway Tools Peter D. Karp SRI International Cost for High Quality Models of 300 Microbiome Organisms Mul/ple body sites of human, mouse, farm animals 500 species in
REGULATION OF GENE EXPRESSION. Bacterial Genetics Lac and Trp Operon
 REGULATION OF GENE EXPRESSION Bacterial Genetics Lac and Trp Operon Levels of Metabolic Control The amount of cellular products can be controlled by regulating: Enzyme activity: alters protein function
REGULATION OF GENE EXPRESSION Bacterial Genetics Lac and Trp Operon Levels of Metabolic Control The amount of cellular products can be controlled by regulating: Enzyme activity: alters protein function
Preparing a PDB File
 Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
2. Cellular and Molecular Biology
 2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
A Simple Protein Synthesis Model
 A Simple Protein Synthesis Model James K. Peterson Department of Biological Sciences and Department of Mathematical Sciences Clemson University September 3, 213 Outline A Simple Protein Synthesis Model
A Simple Protein Synthesis Model James K. Peterson Department of Biological Sciences and Department of Mathematical Sciences Clemson University September 3, 213 Outline A Simple Protein Synthesis Model
Course plan Academic Year Qualification MSc on Bioinformatics for Health Sciences. Subject name: Computational Systems Biology Code: 30180
 Course plan 201-201 Academic Year Qualification MSc on Bioinformatics for Health Sciences 1. Description of the subject Subject name: Code: 30180 Total credits: 5 Workload: 125 hours Year: 1st Term: 3
Course plan 201-201 Academic Year Qualification MSc on Bioinformatics for Health Sciences 1. Description of the subject Subject name: Code: 30180 Total credits: 5 Workload: 125 hours Year: 1st Term: 3
Unit 3: Control and regulation Higher Biology
 Unit 3: Control and regulation Higher Biology To study the roles that genes play in the control of growth and development of organisms To be able to Give some examples of features which are controlled
Unit 3: Control and regulation Higher Biology To study the roles that genes play in the control of growth and development of organisms To be able to Give some examples of features which are controlled
Bi 1x Spring 2014: LacI Titration
 Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
objective functions...
 objective functions... COFFEE (Notredame et al. 1998) measures column by column similarity between pairwise and multiple sequence alignments assumes that the pairwise alignments are optimal assumes a set
objective functions... COFFEE (Notredame et al. 1998) measures column by column similarity between pairwise and multiple sequence alignments assumes that the pairwise alignments are optimal assumes a set
