Data Mining in Protein Binding Cavities
|
|
|
- Rosaline Palmer
- 5 years ago
- Views:
Transcription
1 In Proc. GfKl 2004, Dortmund: Data Mining in Protein Binding Cavities Katrin Kupas and Alfred Ultsch Data Bionics Research Group, University of Marburg, D Marburg, Germany Abstract. The molecular function of a protein is coupled to the binding of a substrate or an endogenous ligand to a well defined binding cavity. To detect functional relationships among proteins, their binding-site exposed physicochemical characteristics were described by assigning generic pseudocenters to the functional groups of the amino acids flanking a particular active site. These pseudocenters were assembled into small substructures and their spatial similarity with appropriate chemical properties was examined. If two substructures of two binding cavities are found to be similar, they form the basis for an expanded comparison of the complete cavities. Preliminary tests indicate the benefit of this method and motivate further studies. 1 Introduction In a biological system multiple biochemical pathways are proceeded and regulated via the complementary recognition properties of proteins and their substrates. The ligand accommodates the binding cavity of the protein according to the lock-and-key principle. Two fold requirements are given: on the one hand, the ligand needs to fit sterically into the binding cavity of the protein. On the other hand, the spatial arrangement of ligand and receptor must correspond to a complementary physicochemical pattern. The shape and function of a protein, e.g. of an enzyme together with its active site is not exclusively represented by a unique amino acid sequence. Accordingly, proteins with deviating amino acid sequence, even adopting a different folding pattern, can nevertheless exhibit related binding cavities to accommodate a ligand. Low sequence homology does not imply any conclusions on binding site differences or similarities. For this reason one has to regard the three-dimensional structure as a prerequisite for a reliable comparison of proteins. Such structures are available for many examples from X-ray crystallography. In literature, different methods based on the description of the spatial protein structures in terms of a reduced set of appropriate descriptors have been reported. In addition to the shape, it is required to Copyright by Springer-Verlag, all rights are reserved.
2 2 Kupas and Ultsch code correctly the exposed physicochemical properties in a geometrical and also chemical sense. In this paper, we describe a new algorithm to compare protein binding sites by the use of common local regions. These local regions form the basis for the further comparison of two binding cavities. Similar local regions among sets of spatially arranged descriptors of two binding cavities provide a coordinate system which will be used in the next step to perform other substructure searches for related cavities. Once a convincing match is detected, it can be assumed that the two active centers are capable to bind similar ligands and thus exhibit related function. The paper is organized as follows: In Chapter 2, other approaches to classify binding cavities are reviewed. Chapter 3 describes the underlying theory and concept of our algorithm used for cavity matching. The local region in descriptor space is defined. In Chapter 4, some preliminary results of a binding cavity matching are presented. Conclusions are given in Chapter 5. 2 Other approaches Previously reported approaches to classify binding cavities can be assigned to three categories, according to the information they use for the classification: 1. Sequence alignments. 2. Comparison of folding patterns and secondary structure elements. 3. Comparison of 3D substructural epitopes. Sequence alignments If two proteins show high sequence identity one can assume structural and most likely also functional similarity among them. The mostly applied procedures were presented by Needleman and Wunsch (1970) and by Waterman (1984). Nevertheless, they are computationally and memory-wise quite demanding, so that often heuristic methods are used such as FASTA(Pearson and Lipman (1988),Pearson (1990)) and BLAST (Altschul et al. (1990)). These procedures do not find an optimal solution in all cases, but generally reveal good approximative results. Comparison of folding patterns and secondary structure elements In general, sequence alignment methods are only capable to detect relationships among proteins if sequence identity exceeds beyond 35%. To classify more distant proteins, information about their three-dimensional structure has to be incorporated to the comparison. Many methods, that establish classification and assignment of proteins to structural families, exploit global fold similarities. Hierarchical procedures have been developed which classify proteins according to their folding, their evolutionary ancestors, or according to their functional role. These systems operate either automatically or are
3 Data Mining in Protein Binding Cavities 3 dependent on manual intervention. Many of the classification schemes treat proteins as being composed by domains and classify them in terms of the properties of their individual domains (Ponting and Russell (2002)). Important approaches for such classifications are: SCOP(Murzin et al. (1995), Lo Conte et al. (2002)), CATH (Orengo et al. (1997, 2000)), FSSP/DALI (Holm and Sander (1996), Holm (1998)), MMDB (Gibrat et al. (1996)). The EN- ZYME (Bairoch (2000)) and BRENDA database (Schomburg et al. (2002)) annotate proteins with respect to the catalyzed reaction. Comparison of 3D substructural epitopes Beyond these relationships, proteins can possess a similar function even if they do not have any sequence and/or folding homology in common. Accordingly, methods that compare proteins only with respect to their folding pattern cannot detect such similarities. Procedures which seek for similar substructures in proteins are better adapted to discover similarities in such cases. The first group of algorithms comprises methods that scan protein structural databases in terms of pre-calculated or automatically generated templates. A typical example of such a template is the catalytic triad in serine proteases. A substantial advantage to restrict to relatively small templates is due to the fact that even large data collections can be scanned efficiently. Some of the best known procedures based on templates are ASSAM introduced by Artymiuk et al. (1993,2003), TESS/PROCAT by Wallace et al. (1996, 1997), PINTS by Stark and Russell (2003), DRESPAT by Wangikar et al. (2003) as well as the methods of Hamelryck (2003) and Kleywegt (1999). The second group includes approaches to compare substructural epitopes of proteins which operate independent of any template definition. For the similarity search the whole proteins or substructures are used. The group of Ruth Nussinov and Haim Wolfson developed many approaches to compare entire receptor structures or substructures. The individual methods essentially differ whether the protein structure is represented by their C α -atoms or grid points on their solvent-accessible surface, or by so-called sparse critical points, a compressed description of the solvent-accessible surface. In each case, the different procedures use geometric hashing (Bachar et al. (1993)) for common substructure detection. They perform completely independent of sequence or fold homology. The approach of Rosen (1998) permits an automatic comparison of binding cavities. Kinoshita et al. (2003) use a graph-based algorithm to compare the surfaces of two proteins. Other methods, such as GENE FIT of Lehtonen et al. (1999) and the approach of Poirrette et al. (1997) use genetic algorithms to optimally superimpose proteins in identified substructure ranges.
4 4 Kupas and Ultsch 3 Theory and Algorithm The algorithm builds on the approach of Schmitt et al. (2002). The physicochemical properties of the cavity-flanking residues are condensed into a restricted set of generic pseudocenters corresponding to five properties essential for molecular recognition: hydrogen-bond donor (DO), hydrogen-bond acceptor (AC), mixed donor/acceptor (DA), hydrophobic aliphatic (AL) and aromatic (PI). The pseudocenters express the features of the 20 different amino acids in terms of five well-placed physicochemical properties. Fig. 1. Surface (left) and pseudocenters (right) of a binding cavity with bound ligand The idea for this algorithm resides on the concept that common substructures of two binding cavities have an arrangement of these pseudocenters in common. Therefore local regions are regarded. They are composed by a center under consideration and the three nearest neighboring centers forming a pyramid. The pyramid was chosen as similarity measure because it corresponds to the smallest spatial unit spanned by these four centers. Systematically every pseudocenter in a cavity is selected as the curent center and forms a local region with its three nearest neighbors. Following the procedure, the binding cavities are partitioned by all local regions to be possibly inscribed. Accordingly, the mutual comparison of binding cavities is reduced to a multiple comparison of the different local regions. Therefore, the spatial and physicochemical characteristics of the local regions are considered separately. The spatial features of the local regions were chosen under the aspects of using a minimum number of descriptors to identify local regions and producing minimal measuring errors. A set of six spatial descriptors was tested. Three of them, the height of the pyramid, the area of the triangle spanned by the three neighbor centers and the distance between the root point of the height and the barycenter of this triangle, have been chosen.
5 Data Mining in Protein Binding Cavities 5 Every pair of local regions of two cavities with the same physicochemical and spatial properties forms the basis for the comparison of the two cavities. These two local regions are matched and the score of the appropriate overlay of the cavities is calculated. 1. Two pyramids with appropriate chemical and spatial characteristics of different binding cavities give rise to a coordinate transformation, which optimally superimposes both pyramids. That means if pyramid A consists of the points (A1, A2, A3, A4) and pyramid B of the points (B1, B2, B3, B4), then a rotation/translation has to be found, so that the sum of the squares of the distances from A i to B i adopts a minimal value (Prokrustes analysis). 2. Subsequently this coordinate transformation is applied to the whole cavities. Then, every pair of pseudocenters of the two cavities, which mutually match chemically and fall close to each other beyond a threshold of 1 Å is counted. This number of successful matches is the score for this pair of pyramids. 3. The superpositioning is done for all pairs of pyramids with the same physicochemical and sterical properties of these two cavities. The maximum of the resulting scores is determined. 4. Relating this maximum score to the maximally achievable score, i.e. the number of pseudocenters in the smaller of the two cavities, gives an estimate of the maximally achievable score. This algorithm has a set of advantages contrary to a consideration only of the individual pseudocenters. With a Prokrustes analysis concerning the individual pseudocenters the coordinate transformation must be accomplished for all pairs of chemically identical pseudocenters and the best match has to be calculated. By consideration of local regions four suitable pseudocenters are given, which have to be matched. Thus the number of computations for the superpositioning of the two cavities is reduced. Only those overlaps with a match of all four pseudocenters have to be computed. A local region composed of four centers gives a good initialization for the Prokrustes analysis. Fewer degrees of freedom exist for the coordinate transformation. A further advantage is that not all binding cavities of a data base have to be considered. Only those cavities containing a suitable local region come into consideration for the surface overlay of the cavities. The remaining cavities without a suitable local region are not consulted. 4 First Results The approach based on local regions for the comparison of protein active sites has been tested with four pairs of binding cavities with well known common substructures (Siemon (2001)). Other similarities between the proteins than these pairs were not expected. The proteins from where the binding cavities
6 6 Kupas and Ultsch had been extracted are the following: an Adenylate Kinase (1ake.2), an allosteric Chorismate Mutase (1csm.3), the Chorismate Mutase of E. Coli (1ecm.5), a Bovine-Actin-Profilin Complex (1hlu.1), a heat shock cognate Protein (1kay.1), Trypsin (1tpo.1), the Uridylate Kinase (1ukz.1) and Proteinase K (2prk.2) (Protein Data Base code (PDB)). The pairs of proteins with well-known common substructures in their binding cavities are 1ake.2/1ukz.1 (Kinases), 1csm.3/1ecm.5 (Isomerases), 1hlu.1/1kay.1 (Hydrolases) and 1tpo.1/2prk.1 (Serine Proteinases). The resulting scores after mutual match are shown in Figure 1. 1ake.2 1csm.3 1ecm.5 1hlu.1 1kay.1 1tpo.1 1ukz.1 2prk.1 1ake csm ecm hlu kay tpo ukz prk Table 1. Resulting scores of a mutual comparison of four pairs of binding cavities with well-known common substructures The numbers are given as percentage with respect to the maximally achievable score (see section 3.3). The table shows that those cavities which are known to possess common substructures also achieve the best scores, whereas the best fit found for the other cavities reveals in most of the cases significantly smaller values. The results have been examined by an expert. The coordinate transformations and the matching pseudocenters of the known pairs of binding cavities were identical with the estimated analogy. 5 Conclusions We presented a new algorithm to find common substructures and compare protein binding cavities. The cavities are partitioned into small local regions with spatial and physicochemical properties. They are formed by pseudocenters assigned to five different physicochemical qualities. The local region exists of a center under consideration and its three nearest neighbors. The physicochemical characteristics of the local regions are the combination of the physicochemical attributes assigned to each pseudocenter. The spatial characteristics were described by the height of the pyramid, the area of the
7 Data Mining in Protein Binding Cavities 7 triangle spanned by the three neighbor centers and the distance between the root point of the height and the barycenter of this triangle. The advantage of this algorithm is, that only those cavities are observed, that share a common local region, expressed in terms of the pyramid. Such substructures, which are represented by pseudocenters widely distributed over the cavity and so aren t biologically relevant, are a priori excluded from the consideration. An advantage of the use of an ESOM for classifying protein binding cavities is the fast comparison of one individual cavity with the entire database. All candidates of proteins of the whole database sharing in common similar local regions are identified in one step. The comparison of four pairs of binding cavities with well-known common substructures led to promising results. It can be assumed that the approach of dividing protein binding cavities into local regions and comparing them is capable to detect similar substructures in the cavities. References ALTSCHUL, S. F., GISH, W., MILLER, W., MYERS, E. W. and LIPMAN, D. J. (1990). Basic local alignment search tool. J. Mol. Biol., 215, ARTYMIUK, P. J., GRINDLEY, H. M., RICE, D. W. and WILLETT, P. (1993). Identification of tertiary structure resemblance in proteins using a maximal common subgraph isomorphism algorithm. J. Mol. Biol., 229, ARTYMIUK, P. J., SPRIGGS, R. V. and WILLETT, P. (2003). Searching for patterns of amino acids in 3D protein structures. J. Chem. Inf. Comput. Sci., 43, BACHAR, O., FISCHER, D., NUSSINOV, R. and WOLFSON, H. (1993). A computer vision based technique for 3-D sequence-independent structural comparison of proteins. Protein Eng., 6, BAIROCH, A. (2000). The ENZYME database in Nucleic Acids Res., 28, GIBRAT, J. F., MADEJ, T. and BRYANT, S. H. (1996). Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, HAMELRYCK, T. (2003). Efficient identification of side-chain patterns using a multidimensional index tree. Proteins, 51, HOLM, L. and SANDER, C. (1996). The FSSP database: fold classification based on structure-structure alignment of proteins. Nucleic Acids Res., 24, HOLM, S. (1998). Touring protein fold space with Dali/FSSP. KINOSHITA, K. and NAKAMURA, H. (2003). Identification of protein biochemical functions by similarity search using the molecular surface database ef-site. Protein Sci., 12, KLEYWEYGT, G. J. (1999). Recognition of spatial motifs in protein structures. J. Mol. Biol., 285, LEHTONEN, J. V., DENESSIOUK, K., MAY, A. C. and JOHNSON, M. S. (1999). Finding local structural similarities among families of unrelated protein structures: a generic nonlinear alignment algorithm. Proteins, 34, LO CONTE, L., BRENNER, S. E., HUBBARD, T. J., CHOTHIA, C. and MURZIN, A. G. (2002). SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30,
8 8 Kupas and Ultsch MURZIN, A. G., BRENNER, S. E., HUBHARD, T. and CHOTHIA, C. (1995). SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, NEEDLEMAN, S.B. and WUNSCH, C.D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, ORENGO, C. A., MICHIE, A. D., JONES, S., JONES, D. T., SWINDELLS, M. B. and THORNTON, J. M., (1997). CATH a hierarchic classification of protein domain structures. Structure, 5, ORENGO, C. A., PEARL, F. M., LEE, D., BRAY, J. E., SILLITOe, I., TODD, A. E., HARRISON, A. P. and THORNTON, J. M. (2000). Assigning genomic sequences to CATH. Nucleic Acids Res 28, PEARSON, W. R. and LIPMAN, D. J. (1988). Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A, 85, PEARSON, W. R. (1990). Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol, 183, POIRETTE, A. R., ARTYMIUK, P. J., RICE, D. W. and WILLETT, P. (1997). Comparison of protein surfaces using a genetic algorithm. J. Comput. Aided Mol. Des., 11, PONTING, C. P. and RUSSELL, R. R. (2002). The natural history of protein domains. Annu. Rev. Biophys. Biomol. Struct., 31, ROSEN, M., LIN, S. L., WOLFSON, H. and NUSSINOV, R. (1998). Molecular shape comparisons in searches for active sites and functional similarity. Protein Eng., 11, RUSSELL, R. B. (1998). Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J. Mol. Biol., 279, SCHMITT, S., KUHN, D. and KLEBE, G. (2002). A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol., 323, SCHOMBURG, I., CHANG, A. and SCHOMBURG, D. (2002). BRENDA, enzyme data and metabolic information. Nucleic Acids. Res., 30, SIEMON, R. (2001). Einige Werkzeuge zum Einsatz von selbstorganisierenden Neuronalen Netzen zur Strukturanalyse von Wirkstoff-Rezeptoren.Diplomarbeit, , FB Mathematik u. Informatik STARK, A., SUNYAEV, S. and RUSSELL, R. B. (2003). A model for statistical significance of local similarities in structure. J. Mol. Biol., 326, STARK, A. and RUSSELL, R. B. (2003). Annotation in three dimensions. PINTS: Patterns in Non-homologous Tertiary Structures. Nucleic Acids Res., 31, WALLACE, A. C., LASKOWSKI, R. A. and THORNTON, J. M. (1996). Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci., 5, WALLACE, A. C., BORKAKOTI, N. and THORNTON, J. M. (1997). TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci., 6, WANGIKAR, P. P., TENDULKAR, A. V., RAMYA, S., MALI, D. N. and SARAWAGI, S. (2003). Functional sites in protein families uncovered via an objective and automated graph theoretic approach. J. Mol. Biol., 326,
9 Data Mining in Protein Binding Cavities 9 WATERMAN, M.S. (1984). General methods for sequence comparison. Bull. Math. Biol., 46,
Number sequence representation of protein structures based on the second derivative of a folded tetrahedron sequence
 Number sequence representation of protein structures based on the second derivative of a folded tetrahedron sequence Naoto Morikawa (nmorika@genocript.com) October 7, 2006. Abstract A protein is a sequence
Number sequence representation of protein structures based on the second derivative of a folded tetrahedron sequence Naoto Morikawa (nmorika@genocript.com) October 7, 2006. Abstract A protein is a sequence
Homology Modeling. Roberto Lins EPFL - summer semester 2005
 Homology Modeling Roberto Lins EPFL - summer semester 2005 Disclaimer: course material is mainly taken from: P.E. Bourne & H Weissig, Structural Bioinformatics; C.A. Orengo, D.T. Jones & J.M. Thornton,
Homology Modeling Roberto Lins EPFL - summer semester 2005 Disclaimer: course material is mainly taken from: P.E. Bourne & H Weissig, Structural Bioinformatics; C.A. Orengo, D.T. Jones & J.M. Thornton,
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Protein Structure & Motifs
 & Motifs Biochemistry 201 Molecular Biology January 12, 2000 Doug Brutlag Introduction Proteins are more flexible than nucleic acids in structure because of both the larger number of types of residues
& Motifs Biochemistry 201 Molecular Biology January 12, 2000 Doug Brutlag Introduction Proteins are more flexible than nucleic acids in structure because of both the larger number of types of residues
Protein Structure: Data Bases and Classification Ingo Ruczinski
 Protein Structure: Data Bases and Classification Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Reference Bourne and Weissig Structural Bioinformatics Wiley, 2003 More References
Protein Structure: Data Bases and Classification Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Reference Bourne and Weissig Structural Bioinformatics Wiley, 2003 More References
In-Depth Assessment of Local Sequence Alignment
 2012 International Conference on Environment Science and Engieering IPCBEE vol.3 2(2012) (2012)IACSIT Press, Singapoore In-Depth Assessment of Local Sequence Alignment Atoosa Ghahremani and Mahmood A.
2012 International Conference on Environment Science and Engieering IPCBEE vol.3 2(2012) (2012)IACSIT Press, Singapoore In-Depth Assessment of Local Sequence Alignment Atoosa Ghahremani and Mahmood A.
2 Dean C. Adams and Gavin J. P. Naylor the best three-dimensional ordination of the structure space is found through an eigen-decomposition (correspon
 A Comparison of Methods for Assessing the Structural Similarity of Proteins Dean C. Adams and Gavin J. P. Naylor? Dept. Zoology and Genetics, Iowa State University, Ames, IA 50011, U.S.A. 1 Introduction
A Comparison of Methods for Assessing the Structural Similarity of Proteins Dean C. Adams and Gavin J. P. Naylor? Dept. Zoology and Genetics, Iowa State University, Ames, IA 50011, U.S.A. 1 Introduction
CSCE555 Bioinformatics. Protein Function Annotation
 CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
Introduction to Comparative Protein Modeling. Chapter 4 Part I
 Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
A General Model for Amino Acid Interaction Networks
 Author manuscript, published in "N/P" A General Model for Amino Acid Interaction Networks Omar GACI and Stefan BALEV hal-43269, version - Nov 29 Abstract In this paper we introduce the notion of protein
Author manuscript, published in "N/P" A General Model for Amino Acid Interaction Networks Omar GACI and Stefan BALEV hal-43269, version - Nov 29 Abstract In this paper we introduce the notion of protein
Chemogenomic: Approaches to Rational Drug Design. Jonas Skjødt Møller
 Chemogenomic: Approaches to Rational Drug Design Jonas Skjødt Møller Chemogenomic Chemistry Biology Chemical biology Medical chemistry Chemical genetics Chemoinformatics Bioinformatics Chemoproteomics
Chemogenomic: Approaches to Rational Drug Design Jonas Skjødt Møller Chemogenomic Chemistry Biology Chemical biology Medical chemistry Chemical genetics Chemoinformatics Bioinformatics Chemoproteomics
Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche
 Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche The molecular structure of a protein can be broken down hierarchically. The primary structure of a protein is simply its
Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche The molecular structure of a protein can be broken down hierarchically. The primary structure of a protein is simply its
Procheck output. Bond angles (Procheck) Structure verification and validation Bond lengths (Procheck) Introduction to Bioinformatics.
 Structure verification and validation Bond lengths (Procheck) Introduction to Bioinformatics Iosif Vaisman Email: ivaisman@gmu.edu ----------------------------------------------------------------- Bond
Structure verification and validation Bond lengths (Procheck) Introduction to Bioinformatics Iosif Vaisman Email: ivaisman@gmu.edu ----------------------------------------------------------------- Bond
2MHR. Protein structure classification is important because it organizes the protein structure universe that is independent of sequence similarity.
 Protein structure classification is important because it organizes the protein structure universe that is independent of sequence similarity. A global picture of the protein universe will help us to understand
Protein structure classification is important because it organizes the protein structure universe that is independent of sequence similarity. A global picture of the protein universe will help us to understand
A Tool for Structure Alignment of Molecules
 A Tool for Structure Alignment of Molecules Pei-Ken Chang, Chien-Cheng Chen and Ming Ouhyoung Department of Computer Science and Information Engineering, National Taiwan University {zick, ccchen}@cmlab.csie.ntu.edu.tw,
A Tool for Structure Alignment of Molecules Pei-Ken Chang, Chien-Cheng Chen and Ming Ouhyoung Department of Computer Science and Information Engineering, National Taiwan University {zick, ccchen}@cmlab.csie.ntu.edu.tw,
Single alignment: Substitution Matrix. 16 march 2017
 Single alignment: Substitution Matrix 16 march 2017 BLOSUM Matrix BLOSUM Matrix [2] (Blocks Amino Acid Substitution Matrices ) It is based on the amino acids substitutions observed in ~2000 conserved block
Single alignment: Substitution Matrix 16 march 2017 BLOSUM Matrix BLOSUM Matrix [2] (Blocks Amino Acid Substitution Matrices ) It is based on the amino acids substitutions observed in ~2000 conserved block
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/309/5742/1868/dc1 Supporting Online Material for Toward High-Resolution de Novo Structure Prediction for Small Proteins Philip Bradley, Kira M. S. Misura, David Baker*
www.sciencemag.org/cgi/content/full/309/5742/1868/dc1 Supporting Online Material for Toward High-Resolution de Novo Structure Prediction for Small Proteins Philip Bradley, Kira M. S. Misura, David Baker*
Large-Scale Genomic Surveys
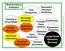 Bioinformatics Subtopics Fold Recognition Secondary Structure Prediction Docking & Drug Design Protein Geometry Structural Informatics Homology Modeling Sequence Alignment Structure Classification Gene
Bioinformatics Subtopics Fold Recognition Secondary Structure Prediction Docking & Drug Design Protein Geometry Structural Informatics Homology Modeling Sequence Alignment Structure Classification Gene
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics. ECS 254; Phone: x3748
 CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 2/15/07 CAP5510 1 EM Algorithm Goal: Find θ, Z that maximize Pr
CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 2/15/07 CAP5510 1 EM Algorithm Goal: Find θ, Z that maximize Pr
Automated Identification of Protein Structural Features
 Automated Identification of Protein Structural Features Chandrasekhar Mamidipally 1, Santosh B. Noronha 1, and Sumantra Dutta Roy 2 1 Dept. of Chemical Engg., IIT Bombay, Powai, Mumbai - 400 076, India
Automated Identification of Protein Structural Features Chandrasekhar Mamidipally 1, Santosh B. Noronha 1, and Sumantra Dutta Roy 2 1 Dept. of Chemical Engg., IIT Bombay, Powai, Mumbai - 400 076, India
Automated Identification of Protein Structural Features
 Automated Identification of Protein Structural Features Chandrasekhar Mamidipally 1, Santosh B. Noronha 1, Sumantra Dutta Roy 2 1 Dept. of Chemical Engg., IIT Bombay, Powai, Mumbai - 400 076, INDIA. chandra
Automated Identification of Protein Structural Features Chandrasekhar Mamidipally 1, Santosh B. Noronha 1, Sumantra Dutta Roy 2 1 Dept. of Chemical Engg., IIT Bombay, Powai, Mumbai - 400 076, INDIA. chandra
Structural Alignment of Proteins
 Goal Align protein structures Structural Alignment of Proteins 1 2 3 4 5 6 7 8 9 10 11 12 13 14 PHE ASP ILE CYS ARG LEU PRO GLY SER ALA GLU ALA VAL CYS PHE ASN VAL CYS ARG THR PRO --- --- --- GLU ALA ILE
Goal Align protein structures Structural Alignment of Proteins 1 2 3 4 5 6 7 8 9 10 11 12 13 14 PHE ASP ILE CYS ARG LEU PRO GLY SER ALA GLU ALA VAL CYS PHE ASN VAL CYS ARG THR PRO --- --- --- GLU ALA ILE
Sequence analysis and comparison
 The aim with sequence identification: Sequence analysis and comparison Marjolein Thunnissen Lund September 2012 Is there any known protein sequence that is homologous to mine? Are there any other species
The aim with sequence identification: Sequence analysis and comparison Marjolein Thunnissen Lund September 2012 Is there any known protein sequence that is homologous to mine? Are there any other species
A profile-based protein sequence alignment algorithm for a domain clustering database
 A profile-based protein sequence alignment algorithm for a domain clustering database Lin Xu,2 Fa Zhang and Zhiyong Liu 3, Key Laboratory of Computer System and architecture, the Institute of Computing
A profile-based protein sequence alignment algorithm for a domain clustering database Lin Xu,2 Fa Zhang and Zhiyong Liu 3, Key Laboratory of Computer System and architecture, the Institute of Computing
Heteropolymer. Mostly in regular secondary structure
 Heteropolymer - + + - Mostly in regular secondary structure 1 2 3 4 C >N trace how you go around the helix C >N C2 >N6 C1 >N5 What s the pattern? Ci>Ni+? 5 6 move around not quite 120 "#$%&'!()*(+2!3/'!4#5'!1/,#64!#6!,6!
Heteropolymer - + + - Mostly in regular secondary structure 1 2 3 4 C >N trace how you go around the helix C >N C2 >N6 C1 >N5 What s the pattern? Ci>Ni+? 5 6 move around not quite 120 "#$%&'!()*(+2!3/'!4#5'!1/,#64!#6!,6!
Do Aligned Sequences Share the Same Fold?
 J. Mol. Biol. (1997) 273, 355±368 Do Aligned Sequences Share the Same Fold? Ruben A. Abagyan* and Serge Batalov The Skirball Institute of Biomolecular Medicine Biochemistry Department NYU Medical Center
J. Mol. Biol. (1997) 273, 355±368 Do Aligned Sequences Share the Same Fold? Ruben A. Abagyan* and Serge Batalov The Skirball Institute of Biomolecular Medicine Biochemistry Department NYU Medical Center
Algorithms in Bioinformatics FOUR Pairwise Sequence Alignment. Pairwise Sequence Alignment. Convention: DNA Sequences 5. Sequence Alignment
 Algorithms in Bioinformatics FOUR Sami Khuri Department of Computer Science San José State University Pairwise Sequence Alignment Homology Similarity Global string alignment Local string alignment Dot
Algorithms in Bioinformatics FOUR Sami Khuri Department of Computer Science San José State University Pairwise Sequence Alignment Homology Similarity Global string alignment Local string alignment Dot
Grouping of amino acids and recognition of protein structurally conserved regions by reduced alphabets of amino acids
 Science in China Series C: Life Sciences 2007 Science in China Press Springer-Verlag Grouping of amino acids and recognition of protein structurally conserved regions by reduced alphabets of amino acids
Science in China Series C: Life Sciences 2007 Science in China Press Springer-Verlag Grouping of amino acids and recognition of protein structurally conserved regions by reduced alphabets of amino acids
1. Protein Data Bank (PDB) 1. Protein Data Bank (PDB)
 Protein structure databases; visualization; and classifications 1. Introduction to Protein Data Bank (PDB) 2. Free graphic software for 3D structure visualization 3. Hierarchical classification of protein
Protein structure databases; visualization; and classifications 1. Introduction to Protein Data Bank (PDB) 2. Free graphic software for 3D structure visualization 3. Hierarchical classification of protein
CMPS 6630: Introduction to Computational Biology and Bioinformatics. Structure Comparison
 CMPS 6630: Introduction to Computational Biology and Bioinformatics Structure Comparison Protein Structure Comparison Motivation Understand sequence and structure variability Understand Domain architecture
CMPS 6630: Introduction to Computational Biology and Bioinformatics Structure Comparison Protein Structure Comparison Motivation Understand sequence and structure variability Understand Domain architecture
Chapter 5. Proteomics and the analysis of protein sequence Ⅱ
 Proteomics Chapter 5. Proteomics and the analysis of protein sequence Ⅱ 1 Pairwise similarity searching (1) Figure 5.5: manual alignment One of the amino acids in the top sequence has no equivalent and
Proteomics Chapter 5. Proteomics and the analysis of protein sequence Ⅱ 1 Pairwise similarity searching (1) Figure 5.5: manual alignment One of the amino acids in the top sequence has no equivalent and
CS612 - Algorithms in Bioinformatics
 Fall 2017 Databases and Protein Structure Representation October 2, 2017 Molecular Biology as Information Science > 12, 000 genomes sequenced, mostly bacterial (2013) > 5x10 6 unique sequences available
Fall 2017 Databases and Protein Structure Representation October 2, 2017 Molecular Biology as Information Science > 12, 000 genomes sequenced, mostly bacterial (2013) > 5x10 6 unique sequences available
The PDB is a Covering Set of Small Protein Structures
 doi:10.1016/j.jmb.2003.10.027 J. Mol. Biol. (2003) 334, 793 802 The PDB is a Covering Set of Small Protein Structures Daisuke Kihara and Jeffrey Skolnick* Center of Excellence in Bioinformatics, University
doi:10.1016/j.jmb.2003.10.027 J. Mol. Biol. (2003) 334, 793 802 The PDB is a Covering Set of Small Protein Structures Daisuke Kihara and Jeffrey Skolnick* Center of Excellence in Bioinformatics, University
Bridging protein local structures and protein functions
 DOI 10.1007/s00726-008-0088-8 REVIEW ARTICLE Bridging protein local structures and protein functions Zhi-Ping Liu Æ Ling-Yun Wu Æ Yong Wang Æ Xiang-Sun Zhang Æ Luonan Chen Received: 21 February 2008 /
DOI 10.1007/s00726-008-0088-8 REVIEW ARTICLE Bridging protein local structures and protein functions Zhi-Ping Liu Æ Ling-Yun Wu Æ Yong Wang Æ Xiang-Sun Zhang Æ Luonan Chen Received: 21 February 2008 /
Introduction to sequence alignment. Local alignment the Smith-Waterman algorithm
 Lecture 2, 12/3/2003: Introduction to sequence alignment The Needleman-Wunsch algorithm for global sequence alignment: description and properties Local alignment the Smith-Waterman algorithm 1 Computational
Lecture 2, 12/3/2003: Introduction to sequence alignment The Needleman-Wunsch algorithm for global sequence alignment: description and properties Local alignment the Smith-Waterman algorithm 1 Computational
Lecture 2, 5/12/2001: Local alignment the Smith-Waterman algorithm. Alignment scoring schemes and theory: substitution matrices and gap models
 Lecture 2, 5/12/2001: Local alignment the Smith-Waterman algorithm Alignment scoring schemes and theory: substitution matrices and gap models 1 Local sequence alignments Local sequence alignments are necessary
Lecture 2, 5/12/2001: Local alignment the Smith-Waterman algorithm Alignment scoring schemes and theory: substitution matrices and gap models 1 Local sequence alignments Local sequence alignments are necessary
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Genome Databases The CATH database
 Genome Databases The CATH database Michael Knudsen 1 and Carsten Wiuf 1,2* 1 Bioinformatics Research Centre, Aarhus University, DK-8000 Aarhus C, Denmark 2 Centre for Membrane Pumps in Cells and Disease
Genome Databases The CATH database Michael Knudsen 1 and Carsten Wiuf 1,2* 1 Bioinformatics Research Centre, Aarhus University, DK-8000 Aarhus C, Denmark 2 Centre for Membrane Pumps in Cells and Disease
The CATH Database provides insights into protein structure/function relationships
 1999 Oxford University Press Nucleic Acids Research, 1999, Vol. 27, No. 1 275 279 The CATH Database provides insights into protein structure/function relationships C. A. Orengo, F. M. G. Pearl, J. E. Bray,
1999 Oxford University Press Nucleic Acids Research, 1999, Vol. 27, No. 1 275 279 The CATH Database provides insights into protein structure/function relationships C. A. Orengo, F. M. G. Pearl, J. E. Bray,
Predicting Protein Function and Binding Profile via Matching of Local Evolutionary and Geometric Surface Patterns
 doi:10.1016/j.jmb.2008.12.072 J. Mol. Biol. (2009) 387, 451 464 Available online at www.sciencedirect.com Predicting Protein Function and Binding Profile via Matching of Local Evolutionary and Geometric
doi:10.1016/j.jmb.2008.12.072 J. Mol. Biol. (2009) 387, 451 464 Available online at www.sciencedirect.com Predicting Protein Function and Binding Profile via Matching of Local Evolutionary and Geometric
Sequence Alignment Techniques and Their Uses
 Sequence Alignment Techniques and Their Uses Sarah Fiorentino Since rapid sequencing technology and whole genomes sequencing, the amount of sequence information has grown exponentially. With all of this
Sequence Alignment Techniques and Their Uses Sarah Fiorentino Since rapid sequencing technology and whole genomes sequencing, the amount of sequence information has grown exponentially. With all of this
Functional diversity within protein superfamilies
 Functional diversity within protein superfamilies James Casbon and Mansoor Saqi * Bioinformatics Group, The Genome Centre, Barts and The London, Queen Mary s School of Medicine and Dentistry, Charterhouse
Functional diversity within protein superfamilies James Casbon and Mansoor Saqi * Bioinformatics Group, The Genome Centre, Barts and The London, Queen Mary s School of Medicine and Dentistry, Charterhouse
Protein Science (1997), 6: Cambridge University Press. Printed in the USA. Copyright 1997 The Protein Society
 1 of 5 1/30/00 8:08 PM Protein Science (1997), 6: 246-248. Cambridge University Press. Printed in the USA. Copyright 1997 The Protein Society FOR THE RECORD LPFC: An Internet library of protein family
1 of 5 1/30/00 8:08 PM Protein Science (1997), 6: 246-248. Cambridge University Press. Printed in the USA. Copyright 1997 The Protein Society FOR THE RECORD LPFC: An Internet library of protein family
MSAT a Multiple Sequence Alignment tool based on TOPS
 MSAT a Multiple Sequence Alignment tool based on TOPS Te Ren, Mallika Veeramalai, Aik Choon Tan and David Gilbert Bioinformatics Research Centre Department of Computer Science University of Glasgow Glasgow,
MSAT a Multiple Sequence Alignment tool based on TOPS Te Ren, Mallika Veeramalai, Aik Choon Tan and David Gilbert Bioinformatics Research Centre Department of Computer Science University of Glasgow Glasgow,
INTERPLAY BETWEEN MULTIPLE ALIGNMENT AND STRUCTURAL ANALYSIS OF PROTEINS: SEQUENCE VERSUS STRUCTURE SIMILARITY. (SHORT REVIEW)
 INTERPLAY BETWEEN MULTIPLE ALIGNMENT AND STRUCTURAL ANALYSIS OF PROTEINS: SEQUENCE VERSUS STRUCTURE SIMILARITY. (SHORT REVIEW) P. RAJARAJESWARI 1 Dr.ALLAM APPARAO 2 1. RESEARCH SCHOLAR, COMPUTER SCIENCE
INTERPLAY BETWEEN MULTIPLE ALIGNMENT AND STRUCTURAL ANALYSIS OF PROTEINS: SEQUENCE VERSUS STRUCTURE SIMILARITY. (SHORT REVIEW) P. RAJARAJESWARI 1 Dr.ALLAM APPARAO 2 1. RESEARCH SCHOLAR, COMPUTER SCIENCE
Introduction to Evolutionary Concepts
 Introduction to Evolutionary Concepts and VMD/MultiSeq - Part I Zaida (Zan) Luthey-Schulten Dept. Chemistry, Beckman Institute, Biophysics, Institute of Genomics Biology, & Physics NIH Workshop 2009 VMD/MultiSeq
Introduction to Evolutionary Concepts and VMD/MultiSeq - Part I Zaida (Zan) Luthey-Schulten Dept. Chemistry, Beckman Institute, Biophysics, Institute of Genomics Biology, & Physics NIH Workshop 2009 VMD/MultiSeq
Protein structure similarities Patrice Koehl
 348 Protein structure similarities Patrice Koehl Comparison of protein structures can reveal distant evolutionary relationships that would not be detected by sequence information alone. This helps to infer
348 Protein structure similarities Patrice Koehl Comparison of protein structures can reveal distant evolutionary relationships that would not be detected by sequence information alone. This helps to infer
Building 3D models of proteins
 Building 3D models of proteins Why make a structural model for your protein? The structure can provide clues to the function through structural similarity with other proteins With a structure it is easier
Building 3D models of proteins Why make a structural model for your protein? The structure can provide clues to the function through structural similarity with other proteins With a structure it is easier
Protein Folds, Functions and Evolution
 Article No. jmbi.1999.3054 available online at http://www.idealibrary.com on J. Mol. Biol. (1999) 293, 333±342 Protein Folds, Functions and Evolution Janet M. Thornton 1,2 *, Christine A. Orengo 1, Annabel
Article No. jmbi.1999.3054 available online at http://www.idealibrary.com on J. Mol. Biol. (1999) 293, 333±342 Protein Folds, Functions and Evolution Janet M. Thornton 1,2 *, Christine A. Orengo 1, Annabel
InDel 3-5. InDel 8-9. InDel 3-5. InDel 8-9. InDel InDel 8-9
 Lecture 5 Alignment I. Introduction. For sequence data, the process of generating an alignment establishes positional homologies; that is, alignment provides the identification of homologous phylogenetic
Lecture 5 Alignment I. Introduction. For sequence data, the process of generating an alignment establishes positional homologies; that is, alignment provides the identification of homologous phylogenetic
ALL LECTURES IN SB Introduction
 1. Introduction 2. Molecular Architecture I 3. Molecular Architecture II 4. Molecular Simulation I 5. Molecular Simulation II 6. Bioinformatics I 7. Bioinformatics II 8. Prediction I 9. Prediction II ALL
1. Introduction 2. Molecular Architecture I 3. Molecular Architecture II 4. Molecular Simulation I 5. Molecular Simulation II 6. Bioinformatics I 7. Bioinformatics II 8. Prediction I 9. Prediction II ALL
Identification of Representative Protein Sequence and Secondary Structure Prediction Using SVM Approach
 Identification of Representative Protein Sequence and Secondary Structure Prediction Using SVM Approach Prof. Dr. M. A. Mottalib, Md. Rahat Hossain Department of Computer Science and Information Technology
Identification of Representative Protein Sequence and Secondary Structure Prediction Using SVM Approach Prof. Dr. M. A. Mottalib, Md. Rahat Hossain Department of Computer Science and Information Technology
As of December 30, 2003, 23,000 solved protein structures
 The protein structure prediction problem could be solved using the current PDB library Yang Zhang and Jeffrey Skolnick* Center of Excellence in Bioinformatics, University at Buffalo, 901 Washington Street,
The protein structure prediction problem could be solved using the current PDB library Yang Zhang and Jeffrey Skolnick* Center of Excellence in Bioinformatics, University at Buffalo, 901 Washington Street,
Analysis on sliding helices and strands in protein structural comparisons: A case study with protein kinases
 Sliding helices and strands in structural comparisons 921 Analysis on sliding helices and strands in protein structural comparisons: A case study with protein kinases V S GOWRI, K ANAMIKA, S GORE 1 and
Sliding helices and strands in structural comparisons 921 Analysis on sliding helices and strands in protein structural comparisons: A case study with protein kinases V S GOWRI, K ANAMIKA, S GORE 1 and
proteins SHORT COMMUNICATION MALIDUP: A database of manually constructed structure alignments for duplicated domain pairs
 J_ID: Z7E Customer A_ID: 21783 Cadmus Art: PROT21783 Date: 25-SEPTEMBER-07 Stage: I Page: 1 proteins STRUCTURE O FUNCTION O BIOINFORMATICS SHORT COMMUNICATION MALIDUP: A database of manually constructed
J_ID: Z7E Customer A_ID: 21783 Cadmus Art: PROT21783 Date: 25-SEPTEMBER-07 Stage: I Page: 1 proteins STRUCTURE O FUNCTION O BIOINFORMATICS SHORT COMMUNICATION MALIDUP: A database of manually constructed
Alignment & BLAST. By: Hadi Mozafari KUMS
 Alignment & BLAST By: Hadi Mozafari KUMS SIMILARITY - ALIGNMENT Comparison of primary DNA or protein sequences to other primary or secondary sequences Expecting that the function of the similar sequence
Alignment & BLAST By: Hadi Mozafari KUMS SIMILARITY - ALIGNMENT Comparison of primary DNA or protein sequences to other primary or secondary sequences Expecting that the function of the similar sequence
Study of Mining Protein Structural Properties and its Application
 Study of Mining Protein Structural Properties and its Application A Dissertation Proposal Presented to the Department of Computer Science and Information Engineering College of Electrical Engineering and
Study of Mining Protein Structural Properties and its Application A Dissertation Proposal Presented to the Department of Computer Science and Information Engineering College of Electrical Engineering and
Bioinformatics. Dept. of Computational Biology & Bioinformatics
 Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
CMPS 6630: Introduction to Computational Biology and Bioinformatics. Tertiary Structure Prediction
 CMPS 6630: Introduction to Computational Biology and Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the
CMPS 6630: Introduction to Computational Biology and Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the
CMPS 3110: Bioinformatics. Tertiary Structure Prediction
 CMPS 3110: Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the laws of physics! Conformation space is finite
CMPS 3110: Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the laws of physics! Conformation space is finite
Analysis and Prediction of Protein Structure (I)
 Analysis and Prediction of Protein Structure (I) Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 2006 Free for academic use. Copyright @ Jianlin Cheng
Analysis and Prediction of Protein Structure (I) Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 2006 Free for academic use. Copyright @ Jianlin Cheng
fconv Tutorial Part 2
 fconv Tutorial Part 2 Gerd Neudert Introduction to some basic fconv features based on version 1.08 Introduction Definitions Welcome to the second part of fconv tutorials! If you have not already done part
fconv Tutorial Part 2 Gerd Neudert Introduction to some basic fconv features based on version 1.08 Introduction Definitions Welcome to the second part of fconv tutorials! If you have not already done part
Neural Networks for Protein Structure Prediction Brown, JMB CS 466 Saurabh Sinha
 Neural Networks for Protein Structure Prediction Brown, JMB 1999 CS 466 Saurabh Sinha Outline Goal is to predict secondary structure of a protein from its sequence Artificial Neural Network used for this
Neural Networks for Protein Structure Prediction Brown, JMB 1999 CS 466 Saurabh Sinha Outline Goal is to predict secondary structure of a protein from its sequence Artificial Neural Network used for this
Motif Prediction in Amino Acid Interaction Networks
 Motif Prediction in Amino Acid Interaction Networks Omar GACI and Stefan BALEV Abstract In this paper we represent a protein as a graph where the vertices are amino acids and the edges are interactions
Motif Prediction in Amino Acid Interaction Networks Omar GACI and Stefan BALEV Abstract In this paper we represent a protein as a graph where the vertices are amino acids and the edges are interactions
Computational approaches for functional genomics
 Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
From fold to function John Moult* and Eugene Melamud
 384 From fold to function John Moult* and Eugene Melamud A number of recent advances have been made in deriving function information from protein structure. A fold relationship to an already characterized
384 From fold to function John Moult* and Eugene Melamud A number of recent advances have been made in deriving function information from protein structure. A fold relationship to an already characterized
Biophysics 101: Genomics & Computational Biology. Section 8: Protein Structure S T R U C T U R E P R O C E S S. Outline.
 Biophysics 101: Genomics & Computational Biology Section 8: Protein Structure Faisal Reza Nov. 11 th, 2003 B101.pdb from PS5 shown at left with: animated ball and stick model, colored CPK H-bonds on, colored
Biophysics 101: Genomics & Computational Biology Section 8: Protein Structure Faisal Reza Nov. 11 th, 2003 B101.pdb from PS5 shown at left with: animated ball and stick model, colored CPK H-bonds on, colored
Optimization of a New Score Function for the Detection of Remote Homologs
 PROTEINS: Structure, Function, and Genetics 41:498 503 (2000) Optimization of a New Score Function for the Detection of Remote Homologs Maricel Kann, 1 Bin Qian, 2 and Richard A. Goldstein 1,2 * 1 Department
PROTEINS: Structure, Function, and Genetics 41:498 503 (2000) Optimization of a New Score Function for the Detection of Remote Homologs Maricel Kann, 1 Bin Qian, 2 and Richard A. Goldstein 1,2 * 1 Department
Measuring quaternary structure similarity using global versus local measures.
 Supplementary Figure 1 Measuring quaternary structure similarity using global versus local measures. (a) Structural similarity of two protein complexes can be inferred from a global superposition, which
Supplementary Figure 1 Measuring quaternary structure similarity using global versus local measures. (a) Structural similarity of two protein complexes can be inferred from a global superposition, which
Protein Structure Prediction
 Page 1 Protein Structure Prediction Russ B. Altman BMI 214 CS 274 Protein Folding is different from structure prediction --Folding is concerned with the process of taking the 3D shape, usually based on
Page 1 Protein Structure Prediction Russ B. Altman BMI 214 CS 274 Protein Folding is different from structure prediction --Folding is concerned with the process of taking the 3D shape, usually based on
Introduction to" Protein Structure
 Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
The molecular functions of a protein can be inferred from
 Global mapping of the protein structure space and application in structure-based inference of protein function Jingtong Hou*, Se-Ran Jun, Chao Zhang, and Sung-Hou Kim* Department of Chemistry and *Graduate
Global mapping of the protein structure space and application in structure-based inference of protein function Jingtong Hou*, Se-Ran Jun, Chao Zhang, and Sung-Hou Kim* Department of Chemistry and *Graduate
Bioinformatics. Macromolecular structure
 Bioinformatics Macromolecular structure Contents Determination of protein structure Structure databases Secondary structure elements (SSE) Tertiary structure Structure analysis Structure alignment Domain
Bioinformatics Macromolecular structure Contents Determination of protein structure Structure databases Secondary structure elements (SSE) Tertiary structure Structure analysis Structure alignment Domain
Amino Acid Structures from Klug & Cummings. 10/7/2003 CAP/CGS 5991: Lecture 7 1
 Amino Acid Structures from Klug & Cummings 10/7/2003 CAP/CGS 5991: Lecture 7 1 Amino Acid Structures from Klug & Cummings 10/7/2003 CAP/CGS 5991: Lecture 7 2 Amino Acid Structures from Klug & Cummings
Amino Acid Structures from Klug & Cummings 10/7/2003 CAP/CGS 5991: Lecture 7 1 Amino Acid Structures from Klug & Cummings 10/7/2003 CAP/CGS 5991: Lecture 7 2 Amino Acid Structures from Klug & Cummings
Syllabus of BIOINF 528 (2017 Fall, Bioinformatics Program)
 Syllabus of BIOINF 528 (2017 Fall, Bioinformatics Program) Course Name: Structural Bioinformatics Course Description: Instructor: This course introduces fundamental concepts and methods for structural
Syllabus of BIOINF 528 (2017 Fall, Bioinformatics Program) Course Name: Structural Bioinformatics Course Description: Instructor: This course introduces fundamental concepts and methods for structural
Structural biology and drug design: An overview
 Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Automatic Epitope Recognition in Proteins Oriented to the System for Macromolecular Interaction Assessment MIAX
 Genome Informatics 12: 113 122 (2001) 113 Automatic Epitope Recognition in Proteins Oriented to the System for Macromolecular Interaction Assessment MIAX Atsushi Yoshimori Carlos A. Del Carpio yosimori@translell.eco.tut.ac.jp
Genome Informatics 12: 113 122 (2001) 113 Automatic Epitope Recognition in Proteins Oriented to the System for Macromolecular Interaction Assessment MIAX Atsushi Yoshimori Carlos A. Del Carpio yosimori@translell.eco.tut.ac.jp
Enzyme Catalysis & Biotechnology
 L28-1 Enzyme Catalysis & Biotechnology Bovine Pancreatic RNase A Biochemistry, Life, and all that L28-2 A brief word about biochemistry traditionally, chemical engineers used organic and inorganic chemistry
L28-1 Enzyme Catalysis & Biotechnology Bovine Pancreatic RNase A Biochemistry, Life, and all that L28-2 A brief word about biochemistry traditionally, chemical engineers used organic and inorganic chemistry
Protein surface descriptors for binding sites comparison and ligand prediction
 Protein surface descriptors for binding sites comparison and ligand prediction Rayan Chikhi Internship report, Kihara Bioinformatics Laboratory, Purdue University, 2007 Abstract. Proteins molecular recognition
Protein surface descriptors for binding sites comparison and ligand prediction Rayan Chikhi Internship report, Kihara Bioinformatics Laboratory, Purdue University, 2007 Abstract. Proteins molecular recognition
CISC 889 Bioinformatics (Spring 2004) Sequence pairwise alignment (I)
 CISC 889 Bioinformatics (Spring 2004) Sequence pairwise alignment (I) Contents Alignment algorithms Needleman-Wunsch (global alignment) Smith-Waterman (local alignment) Heuristic algorithms FASTA BLAST
CISC 889 Bioinformatics (Spring 2004) Sequence pairwise alignment (I) Contents Alignment algorithms Needleman-Wunsch (global alignment) Smith-Waterman (local alignment) Heuristic algorithms FASTA BLAST
Protein Structure. W. M. Grogan, Ph.D. OBJECTIVES
 Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Homology modeling of Ferredoxin-nitrite reductase from Arabidopsis thaliana
 www.bioinformation.net Hypothesis Volume 6(3) Homology modeling of Ferredoxin-nitrite reductase from Arabidopsis thaliana Karim Kherraz*, Khaled Kherraz, Abdelkrim Kameli Biology department, Ecole Normale
www.bioinformation.net Hypothesis Volume 6(3) Homology modeling of Ferredoxin-nitrite reductase from Arabidopsis thaliana Karim Kherraz*, Khaled Kherraz, Abdelkrim Kameli Biology department, Ecole Normale
Sequence Alignments. Dynamic programming approaches, scoring, and significance. Lucy Skrabanek ICB, WMC January 31, 2013
 Sequence Alignments Dynamic programming approaches, scoring, and significance Lucy Skrabanek ICB, WMC January 31, 213 Sequence alignment Compare two (or more) sequences to: Find regions of conservation
Sequence Alignments Dynamic programming approaches, scoring, and significance Lucy Skrabanek ICB, WMC January 31, 213 Sequence alignment Compare two (or more) sequences to: Find regions of conservation
MATHEMATICAL MODELS - Vol. III - Mathematical Modeling and the Human Genome - Hilary S. Booth MATHEMATICAL MODELING AND THE HUMAN GENOME
 MATHEMATICAL MODELING AND THE HUMAN GENOME Hilary S. Booth Australian National University, Australia Keywords: Human genome, DNA, bioinformatics, sequence analysis, evolution. Contents 1. Introduction:
MATHEMATICAL MODELING AND THE HUMAN GENOME Hilary S. Booth Australian National University, Australia Keywords: Human genome, DNA, bioinformatics, sequence analysis, evolution. Contents 1. Introduction:
MAPPIS: Multiple 3D Alignment of Protein-Protein Interfaces
 MAPPIS: Multiple 3D Alignment of Protein-Protein Interfaces Alexandra Shulman-Peleg 1, Maxim Shatsky 1, Ruth Nussinov 2,3 and Haim J. Wolfson 1 1 School of Computer Science, Raymond and Beverly Sackler
MAPPIS: Multiple 3D Alignment of Protein-Protein Interfaces Alexandra Shulman-Peleg 1, Maxim Shatsky 1, Ruth Nussinov 2,3 and Haim J. Wolfson 1 1 School of Computer Science, Raymond and Beverly Sackler
CAP 5510 Lecture 3 Protein Structures
 CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
The typical end scenario for those who try to predict protein
 A method for evaluating the structural quality of protein models by using higher-order pairs scoring Gregory E. Sims and Sung-Hou Kim Berkeley Structural Genomics Center, Lawrence Berkeley National Laboratory,
A method for evaluating the structural quality of protein models by using higher-order pairs scoring Gregory E. Sims and Sung-Hou Kim Berkeley Structural Genomics Center, Lawrence Berkeley National Laboratory,
Prediction of protein function from sequence analysis
 Prediction of protein function from sequence analysis Rita Casadio BIOCOMPUTING GROUP University of Bologna, Italy The omic era Genome Sequencing Projects: Archaea: 74 species In Progress:52 Bacteria:
Prediction of protein function from sequence analysis Rita Casadio BIOCOMPUTING GROUP University of Bologna, Italy The omic era Genome Sequencing Projects: Archaea: 74 species In Progress:52 Bacteria:
Structure to Function. Molecular Bioinformatics, X3, 2006
 Structure to Function Molecular Bioinformatics, X3, 2006 Structural GeNOMICS Structural Genomics project aims at determination of 3D structures of all proteins: - organize known proteins into families
Structure to Function Molecular Bioinformatics, X3, 2006 Structural GeNOMICS Structural Genomics project aims at determination of 3D structures of all proteins: - organize known proteins into families
Secondary Structure. Bioch/BIMS 503 Lecture 2. Structure and Function of Proteins. Further Reading. Φ, Ψ angles alone determine protein structure
 Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Detection of Protein Binding Sites II
 Detection of Protein Binding Sites II Goal: Given a protein structure, predict where a ligand might bind Thomas Funkhouser Princeton University CS597A, Fall 2007 1hld Geometric, chemical, evolutionary
Detection of Protein Binding Sites II Goal: Given a protein structure, predict where a ligand might bind Thomas Funkhouser Princeton University CS597A, Fall 2007 1hld Geometric, chemical, evolutionary
An Introduction to Sequence Similarity ( Homology ) Searching
 An Introduction to Sequence Similarity ( Homology ) Searching Gary D. Stormo 1 UNIT 3.1 1 Washington University, School of Medicine, St. Louis, Missouri ABSTRACT Homologous sequences usually have the same,
An Introduction to Sequence Similarity ( Homology ) Searching Gary D. Stormo 1 UNIT 3.1 1 Washington University, School of Medicine, St. Louis, Missouri ABSTRACT Homologous sequences usually have the same,
Homology. and. Information Gathering and Domain Annotation for Proteins
 Homology and Information Gathering and Domain Annotation for Proteins Outline WHAT IS HOMOLOGY? HOW TO GATHER KNOWN PROTEIN INFORMATION? HOW TO ANNOTATE PROTEIN DOMAINS? EXAMPLES AND EXERCISES Homology
Homology and Information Gathering and Domain Annotation for Proteins Outline WHAT IS HOMOLOGY? HOW TO GATHER KNOWN PROTEIN INFORMATION? HOW TO ANNOTATE PROTEIN DOMAINS? EXAMPLES AND EXERCISES Homology
Sequence and Structure Alignment Z. Luthey-Schulten, UIUC Pittsburgh, 2006 VMD 1.8.5
 Sequence and Structure Alignment Z. Luthey-Schulten, UIUC Pittsburgh, 2006 VMD 1.8.5 Why Look at More Than One Sequence? 1. Multiple Sequence Alignment shows patterns of conservation 2. What and how many
Sequence and Structure Alignment Z. Luthey-Schulten, UIUC Pittsburgh, 2006 VMD 1.8.5 Why Look at More Than One Sequence? 1. Multiple Sequence Alignment shows patterns of conservation 2. What and how many
Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
 Design of a Novel Globular Protein Fold with Atomic-Level Accuracy Brian Kuhlman, Gautam Dantas, Gregory C. Ireton, Gabriele Varani, Barry L. Stoddard, David Baker Presented by Kate Stafford 4 May 05 Protein
Design of a Novel Globular Protein Fold with Atomic-Level Accuracy Brian Kuhlman, Gautam Dantas, Gregory C. Ireton, Gabriele Varani, Barry L. Stoddard, David Baker Presented by Kate Stafford 4 May 05 Protein
Week 10: Homology Modelling (II) - HHpred
 Week 10: Homology Modelling (II) - HHpred Course: Tools for Structural Biology Fabian Glaser BKU - Technion 1 2 Identify and align related structures by sequence methods is not an easy task All comparative
Week 10: Homology Modelling (II) - HHpred Course: Tools for Structural Biology Fabian Glaser BKU - Technion 1 2 Identify and align related structures by sequence methods is not an easy task All comparative
Christian Sigrist. November 14 Protein Bioinformatics: Sequence-Structure-Function 2018 Basel
 Christian Sigrist General Definition on Conserved Regions Conserved regions in proteins can be classified into 5 different groups: Domains: specific combination of secondary structures organized into a
Christian Sigrist General Definition on Conserved Regions Conserved regions in proteins can be classified into 5 different groups: Domains: specific combination of secondary structures organized into a
Protein Structures. 11/19/2002 Lecture 24 1
 Protein Structures 11/19/2002 Lecture 24 1 All 3 figures are cartoons of an amino acid residue. 11/19/2002 Lecture 24 2 Peptide bonds in chains of residues 11/19/2002 Lecture 24 3 Angles φ and ψ in the
Protein Structures 11/19/2002 Lecture 24 1 All 3 figures are cartoons of an amino acid residue. 11/19/2002 Lecture 24 2 Peptide bonds in chains of residues 11/19/2002 Lecture 24 3 Angles φ and ψ in the
Introduction to Bioinformatics
 Introduction to Bioinformatics Jianlin Cheng, PhD Department of Computer Science Informatics Institute 2011 Topics Introduction Biological Sequence Alignment and Database Search Analysis of gene expression
Introduction to Bioinformatics Jianlin Cheng, PhD Department of Computer Science Informatics Institute 2011 Topics Introduction Biological Sequence Alignment and Database Search Analysis of gene expression
09/06/25. Computergestützte Strukturbiologie (Strukturelle Bioinformatik) Non-uniform distribution of folds. Scheme of protein structure predicition
 Sequence identity Structural similarity Computergestützte Strukturbiologie (Strukturelle Bioinformatik) Fold recognition Sommersemester 2009 Peter Güntert Structural similarity X Sequence identity Non-uniform
Sequence identity Structural similarity Computergestützte Strukturbiologie (Strukturelle Bioinformatik) Fold recognition Sommersemester 2009 Peter Güntert Structural similarity X Sequence identity Non-uniform
