Title: Robust analysis of synthetic label-free DNA junctions in solution by X-ray scattering and molecular simulation
|
|
|
- Ashley Parker
- 5 years ago
- Views:
Transcription
1 Supplementary Information Title: Robust analysis of synthetic label-free DNA junctions in solution by X-ray scattering and molecular simulation Kyuhyun Im 1,5, Daun Jeong 2,5, Jaehyun Hur 1, Sung-Jin Kim 2, Sungwoo Hwang 1, Kyeong Sik Jin 3, *, Nokyoung Park 1, *, and Kinam Kim 4 1 Frontier Research Laboratory, Samsung Advanced Institute of Technology, Samsung Electronics, Yongin, Gyeonggi-do , South Korea 2 Computational Science Group, Samsung Advanced Institute of Technology, Samsung Electronics, Yongin, Gyeonggi-do , South Korea 3 Pohang Accelerator Laboratory, Pohang University of Science and Technology, Pohang , South Korea 4 Samsung Advanced Institute of Technology, Samsung Electronics, Yongin, Gyeonggi-do , South Korea 5 These authors contributed equally to this work * Correspondence should be addressed to Nokyoung Park (n2010.park@samsung.com) and Kyeong Sik Jin (jinks@postech.ac.kr)
2 Figure S1. Gel electrophoresis analysis of branched DNA. Lane 1: ss DNA (36mer), Lane 2: 3WJ DNA, Lane 3: 4WJ DNA.
3 Table S1. Oligonucleotide sequences Strand Sequence 3WJ DNA Y1 Y2 Y3 CGA CCG ATG AAT AGC GGT CAG ATC CGT ACC TAC TCG CGA GTC GTT CGC AAT ACG ACC GCT ATT CAT CGG TCG CGA GTA GGT ACG GAT CTG CGT ATT GCG AAC HAC TCG X1 CGA CCG ATG AAT AGC GGT CAG ATC CGT ACC TAC TCG 4WJ DNA X2 X3 CGA GTA GGT ACG GAT CTG CGT ATT GCG AAC GAC TCG CGA GTC GTT CGC AAT ACG GCT GTA CGT ATG GTC TCG X4 CGA GAC CAT ACG TAC AGC ACC GCT ATT CAT CGG TCG
4 Figure S2. Guinier plots of scattering curves in response to change in temperature. Log I(q) versus q 2 for 3WJ (a) and 4WJ (b) in PBS buffer as a function of temperature. For clarity, each plot is shifted along the log I(q) axis. The straight lines were obtained from linear regression in the q 2 region.
5 Table S2. Structural parameters obtained from SAXS analysis of 3WJ and 4WJ in response to change in temperature Sample Temperature (ºC) R a g,g (nm) R b g,p(r) (nm) D c max (nm) P 1st /P d 3rd (a.u.) MD ± ± ± ± ± ± ± WJ ± ± ± ± ± ± ± ± ± ± MD ± ± ± ± ± ± ± WJ ± ± ± ± ± ± ± ± ± ± a R g,g (radius of gyration) was calculated from the Guinier analysis of SAXS data. b R g,p(r) (radius of gyration) was calculated from the p(r) function by the program GNOM. c D max (maximum dimension of particle) was obtained from the p(r) function by the program GNOM. d P 1st /P 3rd is the ratio of the 1 st peak intensity value to the 3 rd peak intensity value in the p(r) function.
6 Equilibrium structures of 3WJ and 4WJ DNA at room temperature from MD simulations a. Comparison of two force fields Here, we discuss 2 MD simulation results employing different force field parameters for nucleic acids in comparison with experimental results. The structures averaged over 100 ns for 2 models at T = 300 K, of which parameters parmbsc0 and parm99 were chosen, are displayed in Figure S3. The average structure obtained from the parmbsc0 force field is found to be a symmetric and T-shaped 3WJ DNA; in contrast, the parm99 force field yields a Y-shape. Various geometric quantities computed from the 2 models are compiled in Table S3, by using 3DNA software for structural analysis. In both cases, a few broken base pairs were observed despite the branched structure, and this is the main cause of inconsistency with experimental results that will be discussed later. The radius of gyration and helix radius of the parmbsc0 model is smaller than that of the parm99 model. It is of interest that the width of the major groove of the former is smaller than that of the latter, and the width of the minor groove is opposite. Helix radius is defined as the radial displacement of atom P in the local helix frame of each dimer. In general, the 3WJ from parmbsc0 is smaller than that from parm99. This is also revealed by displaying the averaged WAXD patterns computed from MD simulation results of the two models (Fig. S4). Differences between the 2 models can be understood as a result of the modified potential parameters for dihedral angles α and γ in the backbone, which were added in the parmbsc0 model. The dihedral potentials endow temporal stability on the double helix structure, which is consistent with recent experimental results. Enhanced stacking, originating from the potential function, is believed to induce 2 helices to form a straight double helix of 36 base pairs and a closed junction with the other arm in the resultant structure. It also reduces fluctuations of the global structure, which is revealed as smaller standard deviations of geometric quantities (Table S3).
7 We now turn to the discussion on the validity of the 2 force fields, with reference to the experimental results, by analyzing p(r). The results from SAXS experiments are considered an ensemble average of the equilibrium structures. To obtain the corresponding quantities from the simulations, we obtain the average of p(r), computed from 50 configurations separated by 2 ns in the MD simulation trajectory. P(r) from individual configurations is depicted in Figure S3 with black (thin) lines, whereas their averages are in red (thick) lines. The trajectories of 100 ns might not be sufficient to ensure the ergodicity of the system. However, we consider these trajectories as equilibrium ensembles obtained after performing a sufficiently long equilibration of 100 ns, while monitoring the stationarity of structural quantities, such as the root-mean squared deviation and number of hydrogen bonds (results not shown). The simulation results of p(r) from 2 force fields generally coincide with experimental results, with respect to the positions of main peaks and the decreasing behavior of tails at large r. However, a couple of discrepancies should be addressed in more detail. First, the difference between the heights of 2 peaks is very small for both cases compared with the experimental result. Second, the positions of the first peak in the 2 models are located at a slightly smaller r. Third, the second peak disappears in the averaged p(r) from simulation results. Finally, the tails at large r (>8 nm) do not coincide with each other, particularly in the case of the parm99 model. Considering that the reconstructed structure for 3WJ DNA from the experiments was distorted Y-shape, the parm99 model, which produced Y-shaped structures, would exhibit greater consistency with the experimental result in Figure S3; however, this is not the case. Although some individual curves have the second peak, the position of the third peaks of those curves does not match. In particular, the tail at large r exhibits a marked deviation from the experimental result. On the other hand, in the parmbsc0 model, we do not observe the second peak in p(r) from individual configurations, while the
8 positions of the third peak and tail exhibit a reasonable coincidence with the experimental result (Fig. S3). The 2 models display different level of the variance in the configurational ensemble. As pointed out earlier, the result of the parmbsc0 force field shows less structural heterogeneity compared with that of the parm99 force field. In the latter, the third peak is sometimes markedly enhanced and changes its position to, implying that one interhelical angle should decrease considerably. At this moment, we are unable to determine the proficiency of the 2 force fields because the current experimental method cannot probe the level of structural fluctuation. To summarize, both models with two force field parameters yield ensemble averaged structures, which are generally consistent with the experimental results. The parmbsc0 force field reproduces the experimental result well at large r, while the parm99 force field captures the existence of the second peak at some configurations. The relative heights of the first and third peaks are reproduced only in some instances among the structural ensemble, but not in the sense of ensemble averages for both models. b. Characterization of global structures of 3WJ DNA using p(r) The distribution of interparticle distance, p(r), is a convenient quantity to consider for validating simulation results with experimental results. Simple geometries, such as spheres, disks, and rods, are easily identified with p(r). However, some difficulties exist in specifying the complex shape of branched DNA from p(r). In solution, three helices consisting of 3WJ DNA would not be perfectly ordered as they are in a crystal, due to thermal fluctuation. Moreover, complimentary base pairs in junction regions and helical ends may be partially broken. Here, we examine how p(r) reflects various characteristics of global structures, while excluding disorder in local structures. We separately built three helices using the tool NAB, with the same sequences used in the experiment. The geometry of each helix is based on the
9 generic crystal structure of a double helix. We only considered key features of the global structure, by changing the relative positions and angles of the 3 straight helices. The 3 strands are not completely connected at the junction region in our model structures. Six representative structures are chosen: planar Y-shape, pyramid, symmetric T-shape, asymmetric T-shape, planar Y-shape with open junction, and planar Y-shape with broken base pairing at the edge. For brevity, the 6 model structures will be denoted M1 6, respectively. We analyzed and compared p(r) of model structures in terms of the relative orientation of the 3 helices and the breakage of base pairs. The former case analyzed with M1 4 is presented in Figure S5a, while the latter case with M1, M5, and M6 is shown in Figure S5b. Figure S5c displays the structures of M1 6. p(r), calculated from the model structures, is characterized by three peaks and a monotonic tail at large r (Figs. S5a, b), whereas the second peak is not pronounced and the tail decrease is not monotonic in the experimental result. The position of the first peak is related to the width of the major groove, which is found to be 1.7 nm for M1 6. The size and shape of the whole molecule determines the third peak, around r = 5.5 nm, and the tail. In Figure S5a, the third peak is lower for the planar Y structure (M1) than in the pyramid (M2), while the tail of the former exhibits a more gradual decay than the latter. Meanwhile, the symmetric T-shape (M3) yields a broader peak at the same r and heavier tail at large r (>10 nm) compared with M1, because 2 helices are aligned to form a long and straight rod. In addition, the height of the third peak becomes relatively larger. The overall symmetry also affects p(r), and the third peak for the asymmetric T-shape (M4) is enhanced further. Finally, the second peak, located around r = 3.5 nm, is related to base pairing. Figure S5b corresponds to the cases for different base pairing. The planar Y-shape with an open junction (M5), where the bases near the branch are flipped out, yields an overall broadening of three peaks compared with M1. Similarly, in the case of M6 (i.e., when the bases flip out at the edge of
10 helical arms), the second peak becomes less distinct than that of M1 (Fig. S5b). Our observations provide a valuable reference point for understanding experimental and simulation results in terms of p(r). c. Characterization of global structures in 4WJ DNA using p(r) We generated 6 basic structures representing the global geometry of the 4WJ system. We believe that the number of such structures should be more; however, at this moment our effort was focused on understanding p(r) in terms of the relative orientation of four helices. The 6 model structures denoted by M1 6 (Fig. S6) are present in a stacked, planar, planar cross, Y, tetrahedral, and pyramid shape, respectively. We display p(r) corresponding to M1 6, obtained using the programs CRYSOL and GNOM, together with the experimental results in Figure S6. In the experimental p(r), a major peak is located at, while the shoulders are observed around and. Comparing the experimental p(r) to the computed p(r) reveals a substantial variance in the relative height of the main peaks. No p(r) for M1 6, by itself, agrees with the experimental results, and M1 6 do not reproduce the position of the main peak or the second shoulder. However, it is tempting to consider a combination of appropriately weighted p(r) for M1 6 as a reasonable reproduction of the aforementioned features in the experimental results. This will be a good prototypical system for further developing the ensemble refinement of SAXS in future studies. Although not quantitative, one can assume that the contribution of M4 should be substantial, as two helices are likely to get close to each other during conformational fluctuations.
11 Table S3. Geometry of 3WJ DNA from MD trajectories of 100 ns, employing the parmbsc0 and parm99 force field parameters parmbsc0 parm99 Number of base pairs ± ± Radius of gyration (nm) ± ± Width of major groove (nm) ± ± Width of minor groove (nm) ± ± Helix radius (nm) ± ± 0.041
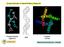
12 Figure S3. MD results: average structures and p(r) obtained from 2 force field parameters. a b c d
13 Figure S4. Averaged WAXD patterns calculated from the equilibrium structures, employing the parmbsc0 and parm99 force fields at 300 K.
14 Figure S5. Characterization of global structures of 3WJ DNA using p(r).
15 Figure S6. Characterization of global structures of 4WJ DNA using p(r). a b M1 M2 M3 M4 M5 M6
16 Figure S7. Small-angle X-ray scattering (SAXS) profiles and pair distance distribution functions from experiments and MD simulations. (a) Scattering profiles of 3WJ and 4WJ in PBS buffer at 72ºC. The solid colored lines are the experimental data, and the solid black lines are the theoretical SAXS curves for each of the 50 structural models generated from MD simulations. (b) Pair distance distribution functions for 3WJ and 4WJ, based on an analysis of the experimental SAXS data using the program GNOM. The solid colored lines are the p(r) functions from the experimental data, and the solid black lines are the p(r) functions for each of the 50 structural models obtained from MD simulations, labeled as black solid lines in a.
17 Figure S8. MD model structures of single-stranded DNA used to simulate the structure of 3WJ and 4WJ in solution at 400 K. Structural models calculated from MD simulations for a single strand of DNA using simulation conditions in Table S4. Surface rendering in the structural models was achieved using Discovery Studio 1.6 (Accelrys, Inc.). (a) M1, (b) M10, (c) M20, (d) M30, (e) M40, (f) M50
18 Table S4. Structural parameters for MD model structures of single-stranded DNA at 400 K MD R a g,p(r) (nm) D b max (nm) MD R a g,p(r) (nm) D b max (nm) M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± M ± a R g,p(r) (radius of gyration) was calculated from the p(r) function by the program GNOM. b D max (maximum dimension of particle) was obtained from the p(r) function by the program GNOM.
19 Intensity (a.u.) Fig S9. UV melting curve (UV absorption as a function of temperature). Heating runs were performed between 20 and 90 C, heating at a rate of 1 C/min, while continuously monitoring the absorbance at 260 nm.the melting temperature values are 64 and 63 o C for 3WJ and 4WJ, respectively. These were calculated from melting curves of each DNA samples in PBS buffer solutions of ph WJ DNA 3WJ DNA Temperature ( o C)
20 CD (mdeg) CD (mdeg) Fig S10. Circular dichroism spectra of 3WJ and 4WJ DNA. Both BDMs have positive and negative peaks near 280 and 250 nm, respectively, which is typical of spectra of the B- form DNA. 3WJ T20 T50 T68 T80 T30 T60 T70 T85 T40 T65 T72 T Wavelength (nm) 4WJ 10 Temperature T20 T50 T68 T80 T30 T60 T70 T85 T40 T65 T72 T Wavelength (nm)
21 MD simulation results of 3WJ and 4WJ DNA at 400 K We increased the temperature of the system up to 400 K to observe DNA thermal denaturation. As a result, the 3WJ and 4WJ DNA were shown to be stable for hundreds of nanoseconds. The number of hydrogen bonds between base pairs in 3WJ exhibited a slight decrease during a time scale of longer than 500 ns, while those in 4WJ decreased very slowly (Fig. S11). We provide 2 notes regarding this observation: the scale of the temperature and the extent of sampling in the configuration space. Although 400 K is an unphysical temperature above the boiling point of water, the TIP3P water model does not boil, and the helix arms form a stable duplex. This is primarily due to the employed interatomic potential, which has been optimized with respect to the structural and dynamical properties of nucleic acids and water at low temperature in general. The force field parameter sets chosen here for nucleic acids and water are known to enhance base stacking and hydration, respectively. Accordingly, the temperature applied in our model system does not scale consistently with the physical temperature. We believe that 400 K should correspond to an enhanced noise level whose physical temperature is still below the melting point of our model system. The second note is that the time scale reached in the simulation is hundreds of nanoseconds, which might be too short to ensure ergodicity. If the branched DNA has a very rugged landscape of potential energy in the configuration space, the system would exhibit slow relaxation. The slight decrease in the number of hydrogen bonds might correspond to either slow relaxation to a collapsed molten globule or long-term fluctuations of partially disordered states at equilibrium. In addition, the system could approach different configurations (e.g., planar or tetrahedral structures) according to the initial configuration, particularly in the case of 4WJ. The initial condition dependence of the 4WJ system at 400 K is shown in Supplementary Figure S13. The upper panel exhibits the averaged structures at 300 K and 400K starting from a tetrahedral structure, while the lower panel is for the case of a planar
22 extended structure. Figure S11 MD results: p(r) calculated from equilibrium structures of at 400 K. a. 3WJ DNA b. 4WJ DNA
23 Figure S12. MD simulation results for the stability of 3WJ and 4WJ at 400 K.
24 Figure S13. Averaged equilibrium structures at 300 K and 400 K, starting from different initial configurations.
25 Table S5. Overview of the radius of gyration (R g ) and forward scattering intensity I(0) obtained from SAXS analysis of 3WJ and 4WJ in a wide concentration range Sample Conc. (mg/ml) R g I(0) (a.u.) ± E+02 ± WJ ± E+02 ± ± E+02 ± ± E+03 ± ± E+02 ± WJ ± E+02 ± ± E+03 ± ± E+03 ±
26 Table S6. Overview of the radius of gyration (R g ), forward scattering intensity I(0) and estimation of the molecular mass obtained from SAXS analysis of 3WJ and 4WJ in solution Sample MM calculated (kda) Conc. (mg/ml) I(0) Abs R g MM SAXS (kda) 3WJ ± WJ ±
High throughput near infrared screening discovers DNA-templated silver clusters with peak fluorescence beyond 950 nm
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 High throughput near infrared screening discovers DNA-templated silver clusters with peak fluorescence
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 High throughput near infrared screening discovers DNA-templated silver clusters with peak fluorescence
Supplementary Information
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Directed self-assembly of genomic sequences into monomeric and polymeric branched DNA structures
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Directed self-assembly of genomic sequences into monomeric and polymeric branched DNA structures
Practical Bioinformatics
 5/2/2017 Dictionaries d i c t i o n a r y = { A : T, T : A, G : C, C : G } d i c t i o n a r y [ G ] d i c t i o n a r y [ N ] = N d i c t i o n a r y. h a s k e y ( C ) Dictionaries g e n e t i c C o
5/2/2017 Dictionaries d i c t i o n a r y = { A : T, T : A, G : C, C : G } d i c t i o n a r y [ G ] d i c t i o n a r y [ N ] = N d i c t i o n a r y. h a s k e y ( C ) Dictionaries g e n e t i c C o
Supporting Information for. Initial Biochemical and Functional Evaluation of Murine Calprotectin Reveals Ca(II)-
 Supporting Information for Initial Biochemical and Functional Evaluation of Murine Calprotectin Reveals Ca(II)- Dependence and Its Ability to Chelate Multiple Nutrient Transition Metal Ions Rose C. Hadley,
Supporting Information for Initial Biochemical and Functional Evaluation of Murine Calprotectin Reveals Ca(II)- Dependence and Its Ability to Chelate Multiple Nutrient Transition Metal Ions Rose C. Hadley,
Supporting Information for. Polypyrrole/Agarose based Electronically. Conductive and Reversibly Restorable Hydrogel
 Supporting Information for Polypyrrole/Agarose based Electronically Conductive and Reversibly Restorable Hydrogel Jaehyun Hur, Kyuhyun Im, Sang Won Kim, Jineun Kim, Dae-Young Chung, Tae-Ho Kim, Kyoung
Supporting Information for Polypyrrole/Agarose based Electronically Conductive and Reversibly Restorable Hydrogel Jaehyun Hur, Kyuhyun Im, Sang Won Kim, Jineun Kim, Dae-Young Chung, Tae-Ho Kim, Kyoung
SUPPORTING INFORMATION FOR. SEquence-Enabled Reassembly of β-lactamase (SEER-LAC): a Sensitive Method for the Detection of Double-Stranded DNA
 SUPPORTING INFORMATION FOR SEquence-Enabled Reassembly of β-lactamase (SEER-LAC): a Sensitive Method for the Detection of Double-Stranded DNA Aik T. Ooi, Cliff I. Stains, Indraneel Ghosh *, David J. Segal
SUPPORTING INFORMATION FOR SEquence-Enabled Reassembly of β-lactamase (SEER-LAC): a Sensitive Method for the Detection of Double-Stranded DNA Aik T. Ooi, Cliff I. Stains, Indraneel Ghosh *, David J. Segal
Number-controlled spatial arrangement of gold nanoparticles with
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Number-controlled spatial arrangement of gold nanoparticles with DNA dendrimers Ping Chen,*
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Number-controlled spatial arrangement of gold nanoparticles with DNA dendrimers Ping Chen,*
Building a Multifunctional Aptamer-Based DNA Nanoassembly for Targeted Cancer Therapy
 Supporting Information Building a Multifunctional Aptamer-Based DNA Nanoassembly for Targeted Cancer Therapy Cuichen Wu,, Da Han,, Tao Chen,, Lu Peng, Guizhi Zhu,, Mingxu You,, Liping Qiu,, Kwame Sefah,
Supporting Information Building a Multifunctional Aptamer-Based DNA Nanoassembly for Targeted Cancer Therapy Cuichen Wu,, Da Han,, Tao Chen,, Lu Peng, Guizhi Zhu,, Mingxu You,, Liping Qiu,, Kwame Sefah,
The role of the FliD C-terminal domain in pentamer formation and
 The role of the FliD C-terminal domain in pentamer formation and interaction with FliT Hee Jung Kim 1,2,*, Woongjae Yoo 3,*, Kyeong Sik Jin 4, Sangryeol Ryu 3,5 & Hyung Ho Lee 1, 1 Department of Chemistry,
The role of the FliD C-terminal domain in pentamer formation and interaction with FliT Hee Jung Kim 1,2,*, Woongjae Yoo 3,*, Kyeong Sik Jin 4, Sangryeol Ryu 3,5 & Hyung Ho Lee 1, 1 Department of Chemistry,
Supplementary Information for
 Supplementary Information for Evolutionary conservation of codon optimality reveals hidden signatures of co-translational folding Sebastian Pechmann & Judith Frydman Department of Biology and BioX, Stanford
Supplementary Information for Evolutionary conservation of codon optimality reveals hidden signatures of co-translational folding Sebastian Pechmann & Judith Frydman Department of Biology and BioX, Stanford
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Zn 2+ -binding sites in USP18. (a) The two molecules of USP18 present in the asymmetric unit are shown. Chain A is shown in blue, chain B in green. Bound Zn 2+ ions are shown as
Supplementary Figure 1 Zn 2+ -binding sites in USP18. (a) The two molecules of USP18 present in the asymmetric unit are shown. Chain A is shown in blue, chain B in green. Bound Zn 2+ ions are shown as
Crick s early Hypothesis Revisited
 Crick s early Hypothesis Revisited Or The Existence of a Universal Coding Frame Ryan Rossi, Jean-Louis Lassez and Axel Bernal UPenn Center for Bioinformatics BIOINFORMATICS The application of computer
Crick s early Hypothesis Revisited Or The Existence of a Universal Coding Frame Ryan Rossi, Jean-Louis Lassez and Axel Bernal UPenn Center for Bioinformatics BIOINFORMATICS The application of computer
Supporting Information
 Supporting Information T. Pellegrino 1,2,3,#, R. A. Sperling 1,#, A. P. Alivisatos 2, W. J. Parak 1,2,* 1 Center for Nanoscience, Ludwig Maximilians Universität München, München, Germany 2 Department of
Supporting Information T. Pellegrino 1,2,3,#, R. A. Sperling 1,#, A. P. Alivisatos 2, W. J. Parak 1,2,* 1 Center for Nanoscience, Ludwig Maximilians Universität München, München, Germany 2 Department of
NSCI Basic Properties of Life and The Biochemistry of Life on Earth
 NSCI 314 LIFE IN THE COSMOS 4 Basic Properties of Life and The Biochemistry of Life on Earth Dr. Karen Kolehmainen Department of Physics CSUSB http://physics.csusb.edu/~karen/ WHAT IS LIFE? HARD TO DEFINE,
NSCI 314 LIFE IN THE COSMOS 4 Basic Properties of Life and The Biochemistry of Life on Earth Dr. Karen Kolehmainen Department of Physics CSUSB http://physics.csusb.edu/~karen/ WHAT IS LIFE? HARD TO DEFINE,
Clay Carter. Department of Biology. QuickTime and a TIFF (Uncompressed) decompressor are needed to see this picture.
 QuickTime and a TIFF (Uncompressed) decompressor are needed to see this picture. Clay Carter Department of Biology QuickTime and a TIFF (LZW) decompressor are needed to see this picture. Ornamental tobacco
QuickTime and a TIFF (Uncompressed) decompressor are needed to see this picture. Clay Carter Department of Biology QuickTime and a TIFF (LZW) decompressor are needed to see this picture. Ornamental tobacco
SEQUENCE ALIGNMENT BACKGROUND: BIOINFORMATICS. Prokaryotes and Eukaryotes. DNA and RNA
 SEQUENCE ALIGNMENT BACKGROUND: BIOINFORMATICS 1 Prokaryotes and Eukaryotes 2 DNA and RNA 3 4 Double helix structure Codons Codons are triplets of bases from the RNA sequence. Each triplet defines an amino-acid.
SEQUENCE ALIGNMENT BACKGROUND: BIOINFORMATICS 1 Prokaryotes and Eukaryotes 2 DNA and RNA 3 4 Double helix structure Codons Codons are triplets of bases from the RNA sequence. Each triplet defines an amino-acid.
Supplemental data. Pommerrenig et al. (2011). Plant Cell /tpc
 Supplemental Figure 1. Prediction of phloem-specific MTK1 expression in Arabidopsis shoots and roots. The images and the corresponding numbers showing absolute (A) or relative expression levels (B) of
Supplemental Figure 1. Prediction of phloem-specific MTK1 expression in Arabidopsis shoots and roots. The images and the corresponding numbers showing absolute (A) or relative expression levels (B) of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI:.38/NCHEM.246 Optimizing the specificity of nucleic acid hyridization David Yu Zhang, Sherry Xi Chen, and Peng Yin. Analytic framework and proe design 3.. Concentration-adjusted
SUPPLEMENTARY INFORMATION DOI:.38/NCHEM.246 Optimizing the specificity of nucleic acid hyridization David Yu Zhang, Sherry Xi Chen, and Peng Yin. Analytic framework and proe design 3.. Concentration-adjusted
The Effect of Linker DNA on the Structure and Interaction of Nucleosome Core Particles
 Electronic Supplementary Material (ESI) for Soft Matter. This journal is The Royal Society of Chemistry 2018 SUPPLEMENTARY INFORMATION The Effect of Linker DNA on the Structure and Interaction of Nucleosome
Electronic Supplementary Material (ESI) for Soft Matter. This journal is The Royal Society of Chemistry 2018 SUPPLEMENTARY INFORMATION The Effect of Linker DNA on the Structure and Interaction of Nucleosome
Characterization of Pathogenic Genes through Condensed Matrix Method, Case Study through Bacterial Zeta Toxin
 International Journal of Genetic Engineering and Biotechnology. ISSN 0974-3073 Volume 2, Number 1 (2011), pp. 109-114 International Research Publication House http://www.irphouse.com Characterization of
International Journal of Genetic Engineering and Biotechnology. ISSN 0974-3073 Volume 2, Number 1 (2011), pp. 109-114 International Research Publication House http://www.irphouse.com Characterization of
Principles of Physical Biochemistry
 Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Advanced topics in bioinformatics
 Feinberg Graduate School of the Weizmann Institute of Science Advanced topics in bioinformatics Shmuel Pietrokovski & Eitan Rubin Spring 2003 Course WWW site: http://bioinformatics.weizmann.ac.il/courses/atib
Feinberg Graduate School of the Weizmann Institute of Science Advanced topics in bioinformatics Shmuel Pietrokovski & Eitan Rubin Spring 2003 Course WWW site: http://bioinformatics.weizmann.ac.il/courses/atib
Melting Transition of Directly-Linked Gold Nanoparticle DNA Assembly arxiv:physics/ v1 [physics.bio-ph] 10 Mar 2005
![Melting Transition of Directly-Linked Gold Nanoparticle DNA Assembly arxiv:physics/ v1 [physics.bio-ph] 10 Mar 2005 Melting Transition of Directly-Linked Gold Nanoparticle DNA Assembly arxiv:physics/ v1 [physics.bio-ph] 10 Mar 2005](/thumbs/74/69791072.jpg) Physica A (25) to appear. Melting Transition of Directly-Linked Gold Nanoparticle DNA Assembly arxiv:physics/539v [physics.bio-ph] Mar 25 Abstract Y. Sun, N. C. Harris, and C.-H. Kiang Department of Physics
Physica A (25) to appear. Melting Transition of Directly-Linked Gold Nanoparticle DNA Assembly arxiv:physics/539v [physics.bio-ph] Mar 25 Abstract Y. Sun, N. C. Harris, and C.-H. Kiang Department of Physics
Electronic supplementary material
 Applied Microbiology and Biotechnology Electronic supplementary material A family of AA9 lytic polysaccharide monooxygenases in Aspergillus nidulans is differentially regulated by multiple substrates and
Applied Microbiology and Biotechnology Electronic supplementary material A family of AA9 lytic polysaccharide monooxygenases in Aspergillus nidulans is differentially regulated by multiple substrates and
SSR ( ) Vol. 48 No ( Microsatellite marker) ( Simple sequence repeat,ssr),
 48 3 () Vol. 48 No. 3 2009 5 Journal of Xiamen University (Nat ural Science) May 2009 SSR,,,, 3 (, 361005) : SSR. 21 516,410. 60 %96. 7 %. (),(Between2groups linkage method),.,, 11 (),. 12,. (, ), : 0.
48 3 () Vol. 48 No. 3 2009 5 Journal of Xiamen University (Nat ural Science) May 2009 SSR,,,, 3 (, 361005) : SSR. 21 516,410. 60 %96. 7 %. (),(Between2groups linkage method),.,, 11 (),. 12,. (, ), : 0.
Supplemental Figure 1.
 A wt spoiiiaδ spoiiiahδ bofaδ B C D E spoiiiaδ, bofaδ Supplemental Figure 1. GFP-SpoIVFA is more mislocalized in the absence of both BofA and SpoIIIAH. Sporulation was induced by resuspension in wild-type
A wt spoiiiaδ spoiiiahδ bofaδ B C D E spoiiiaδ, bofaδ Supplemental Figure 1. GFP-SpoIVFA is more mislocalized in the absence of both BofA and SpoIIIAH. Sporulation was induced by resuspension in wild-type
Regulatory Sequence Analysis. Sequence models (Bernoulli and Markov models)
 Regulatory Sequence Analysis Sequence models (Bernoulli and Markov models) 1 Why do we need random models? Any pattern discovery relies on an underlying model to estimate the random expectation. This model
Regulatory Sequence Analysis Sequence models (Bernoulli and Markov models) 1 Why do we need random models? Any pattern discovery relies on an underlying model to estimate the random expectation. This model
SUPPLEMENTARY DATA - 1 -
 - 1 - SUPPLEMENTARY DATA Construction of B. subtilis rnpb complementation plasmids For complementation, the B. subtilis rnpb wild-type gene (rnpbwt) under control of its native rnpb promoter and terminator
- 1 - SUPPLEMENTARY DATA Construction of B. subtilis rnpb complementation plasmids For complementation, the B. subtilis rnpb wild-type gene (rnpbwt) under control of its native rnpb promoter and terminator
Supporting Information
 Supporting Information Mixed DNA-functionalized Nanoparticle Probes for Surface Enhanced Raman Scattering-based Multiplex DNA Detection Zhiliang Zhang, a, b Yongqiang Wen,* a Ying Ma, a Jia Luo, a Lei
Supporting Information Mixed DNA-functionalized Nanoparticle Probes for Surface Enhanced Raman Scattering-based Multiplex DNA Detection Zhiliang Zhang, a, b Yongqiang Wen,* a Ying Ma, a Jia Luo, a Lei
Table S1. Primers and PCR conditions used in this paper Primers Sequence (5 3 ) Thermal conditions Reference Rhizobacteria 27F 1492R
 Table S1. Primers and PCR conditions used in this paper Primers Sequence (5 3 ) Thermal conditions Reference Rhizobacteria 27F 1492R AAC MGG ATT AGA TAC CCK G GGY TAC CTT GTT ACG ACT T Detection of Candidatus
Table S1. Primers and PCR conditions used in this paper Primers Sequence (5 3 ) Thermal conditions Reference Rhizobacteria 27F 1492R AAC MGG ATT AGA TAC CCK G GGY TAC CTT GTT ACG ACT T Detection of Candidatus
Structural characterization of NiV N 0 P in solution and in crystal.
 Supplementary Figure 1 Structural characterization of NiV N 0 P in solution and in crystal. (a) SAXS analysis of the N 32-383 0 -P 50 complex. The Guinier plot for complex concentrations of 0.55, 1.1,
Supplementary Figure 1 Structural characterization of NiV N 0 P in solution and in crystal. (a) SAXS analysis of the N 32-383 0 -P 50 complex. The Guinier plot for complex concentrations of 0.55, 1.1,
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
TM1 TM2 TM3 TM4 TM5 TM6 TM bp
 a 467 bp 1 482 2 93 3 321 4 7 281 6 21 7 66 8 176 19 12 13 212 113 16 8 b ATG TCA GGA CAT GTA ATG GAG GAA TGT GTA GTT CAC GGT ACG TTA GCG GCA GTA TTG CGT TTA ATG GGC GTA GTG M S G H V M E E C V V H G T
a 467 bp 1 482 2 93 3 321 4 7 281 6 21 7 66 8 176 19 12 13 212 113 16 8 b ATG TCA GGA CAT GTA ATG GAG GAA TGT GTA GTT CAC GGT ACG TTA GCG GCA GTA TTG CGT TTA ATG GGC GTA GTG M S G H V M E E C V V H G T
The photoluminescent graphene oxide serves as an acceptor rather. than a donor in the fluorescence resonance energy transfer pair of
 Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 20XX The photoluminescent graphene oxide serves as an acceptor rather than a donor in the fluorescence
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 20XX The photoluminescent graphene oxide serves as an acceptor rather than a donor in the fluorescence
SUPPLEMENTARY INFORMATION
 DOI:.8/NCHEM. Conditionally Fluorescent Molecular Probes for Detecting Single Base Changes in Double-stranded DNA Sherry Xi Chen, David Yu Zhang, Georg Seelig. Analytic framework and probe design.. Design
DOI:.8/NCHEM. Conditionally Fluorescent Molecular Probes for Detecting Single Base Changes in Double-stranded DNA Sherry Xi Chen, David Yu Zhang, Georg Seelig. Analytic framework and probe design.. Design
Contents. xiii. Preface v
 Contents Preface Chapter 1 Biological Macromolecules 1.1 General PrincipIes 1.1.1 Macrornolecules 1.2 1.1.2 Configuration and Conformation Molecular lnteractions in Macromolecular Structures 1.2.1 Weak
Contents Preface Chapter 1 Biological Macromolecules 1.1 General PrincipIes 1.1.1 Macrornolecules 1.2 1.1.2 Configuration and Conformation Molecular lnteractions in Macromolecular Structures 1.2.1 Weak
ELECTRONIC SUPPLEMENTARY INFORMATION
 This journal is The Royal Society of Chemistry 1 ELECTRONIC SUPPLEMENTARY INFORMATION More than one non-canonical phosphodiester bond into the G-tract: formation of unusual parallel G-quadruplex structures
This journal is The Royal Society of Chemistry 1 ELECTRONIC SUPPLEMENTARY INFORMATION More than one non-canonical phosphodiester bond into the G-tract: formation of unusual parallel G-quadruplex structures
Modelling and Analysis in Bioinformatics. Lecture 1: Genomic k-mer Statistics
 582746 Modelling and Analysis in Bioinformatics Lecture 1: Genomic k-mer Statistics Juha Kärkkäinen 06.09.2016 Outline Course introduction Genomic k-mers 1-Mers 2-Mers 3-Mers k-mers for Larger k Outline
582746 Modelling and Analysis in Bioinformatics Lecture 1: Genomic k-mer Statistics Juha Kärkkäinen 06.09.2016 Outline Course introduction Genomic k-mers 1-Mers 2-Mers 3-Mers k-mers for Larger k Outline
Supplemental Table 1. Primers used for cloning and PCR amplification in this study
 Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Structural and mechanistic insight into the substrate. binding from the conformational dynamics in apo. and substrate-bound DapE enzyme
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 215 Structural and mechanistic insight into the substrate binding from the conformational
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 215 Structural and mechanistic insight into the substrate binding from the conformational
Introduction to Polymer Physics
 Introduction to Polymer Physics Enrico Carlon, KU Leuven, Belgium February-May, 2016 Enrico Carlon, KU Leuven, Belgium Introduction to Polymer Physics February-May, 2016 1 / 28 Polymers in Chemistry and
Introduction to Polymer Physics Enrico Carlon, KU Leuven, Belgium February-May, 2016 Enrico Carlon, KU Leuven, Belgium Introduction to Polymer Physics February-May, 2016 1 / 28 Polymers in Chemistry and
part 4: phenomenological load and biological inference. phenomenological load review types of models. Gαβ = 8π Tαβ. Newton.
 2017-07-29 part 4: and biological inference review types of models phenomenological Newton F= Gm1m2 r2 mechanistic Einstein Gαβ = 8π Tαβ 1 molecular evolution is process and pattern process pattern MutSel
2017-07-29 part 4: and biological inference review types of models phenomenological Newton F= Gm1m2 r2 mechanistic Einstein Gαβ = 8π Tαβ 1 molecular evolution is process and pattern process pattern MutSel
Protein Threading. Combinatorial optimization approach. Stefan Balev.
 Protein Threading Combinatorial optimization approach Stefan Balev Stefan.Balev@univ-lehavre.fr Laboratoire d informatique du Havre Université du Havre Stefan Balev Cours DEA 30/01/2004 p.1/42 Outline
Protein Threading Combinatorial optimization approach Stefan Balev Stefan.Balev@univ-lehavre.fr Laboratoire d informatique du Havre Université du Havre Stefan Balev Cours DEA 30/01/2004 p.1/42 Outline
SUPPLEMENTARY INFORMATION
 Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
Supplementary information. Porphyrin-Assisted Docking of a Thermophage Portal Protein into Lipid Bilayers: Nanopore Engineering and Characterization.
 Supplementary information Porphyrin-Assisted Docking of a Thermophage Portal Protein into Lipid Bilayers: Nanopore Engineering and Characterization. Benjamin Cressiot #, Sandra J. Greive #, Wei Si ^#,
Supplementary information Porphyrin-Assisted Docking of a Thermophage Portal Protein into Lipid Bilayers: Nanopore Engineering and Characterization. Benjamin Cressiot #, Sandra J. Greive #, Wei Si ^#,
Encoding of Amino Acids and Proteins from a Communications and Information Theoretic Perspective
 Jacobs University Bremen Encoding of Amino Acids and Proteins from a Communications and Information Theoretic Perspective Semester Project II By: Dawit Nigatu Supervisor: Prof. Dr. Werner Henkel Transmission
Jacobs University Bremen Encoding of Amino Acids and Proteins from a Communications and Information Theoretic Perspective Semester Project II By: Dawit Nigatu Supervisor: Prof. Dr. Werner Henkel Transmission
3. Evolution makes sense of homologies. 3. Evolution makes sense of homologies. 3. Evolution makes sense of homologies
 Richard Owen (1848) introduced the term Homology to refer to structural similarities among organisms. To Owen, these similarities indicated that organisms were created following a common plan or archetype.
Richard Owen (1848) introduced the term Homology to refer to structural similarities among organisms. To Owen, these similarities indicated that organisms were created following a common plan or archetype.
Supplementary Information for Topological phase transition and quantum spin Hall edge states of antimony few layers
 1 Supplementary Information for Topological phase transition and quantum spin Hall edge states of antimony few layers Sung Hwan Kim, 1, 2 Kyung-Hwan Jin, 2 Joonbum Park, 2 Jun Sung Kim, 2 Seung-Hoon Jhi,
1 Supplementary Information for Topological phase transition and quantum spin Hall edge states of antimony few layers Sung Hwan Kim, 1, 2 Kyung-Hwan Jin, 2 Joonbum Park, 2 Jun Sung Kim, 2 Seung-Hoon Jhi,
EXAM I COURSE TFY4310 MOLECULAR BIOPHYSICS December Suggested resolution
 page 1 of 7 EXAM I COURSE TFY4310 MOLECULAR BIOPHYSICS December 2013 Suggested resolution Exercise 1. [total: 25 p] a) [t: 5 p] Describe the bonding [1.5 p] and the molecular orbitals [1.5 p] of the ethylene
page 1 of 7 EXAM I COURSE TFY4310 MOLECULAR BIOPHYSICS December 2013 Suggested resolution Exercise 1. [total: 25 p] a) [t: 5 p] Describe the bonding [1.5 p] and the molecular orbitals [1.5 p] of the ethylene
Protein Dynamics. The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron.
 Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Supporting Information. An Electric Single-Molecule Hybridisation Detector for short DNA Fragments
 Supporting Information An Electric Single-Molecule Hybridisation Detector for short DNA Fragments A.Y.Y. Loh, 1 C.H. Burgess, 2 D.A. Tanase, 1 G. Ferrari, 3 M.A. Maclachlan, 2 A.E.G. Cass, 1 T. Albrecht*
Supporting Information An Electric Single-Molecule Hybridisation Detector for short DNA Fragments A.Y.Y. Loh, 1 C.H. Burgess, 2 D.A. Tanase, 1 G. Ferrari, 3 M.A. Maclachlan, 2 A.E.G. Cass, 1 T. Albrecht*
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Codon Distribution in Error-Detecting Circular Codes
 life Article Codon Distribution in Error-Detecting Circular Codes Elena Fimmel, * and Lutz Strüngmann Institute for Mathematical Biology, Faculty of Computer Science, Mannheim University of Applied Sciences,
life Article Codon Distribution in Error-Detecting Circular Codes Elena Fimmel, * and Lutz Strüngmann Institute for Mathematical Biology, Faculty of Computer Science, Mannheim University of Applied Sciences,
Introduction to SAXS at SSRL
 Everything You Ever Wanted to Know About Introduction to SAXS at SSRL SAXS But Were Afraid to Ask John A Pople Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford CA
Everything You Ever Wanted to Know About Introduction to SAXS at SSRL SAXS But Were Afraid to Ask John A Pople Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford CA
Evolvable Neural Networks for Time Series Prediction with Adaptive Learning Interval
 Evolvable Neural Networs for Time Series Prediction with Adaptive Learning Interval Dong-Woo Lee *, Seong G. Kong *, and Kwee-Bo Sim ** *Department of Electrical and Computer Engineering, The University
Evolvable Neural Networs for Time Series Prediction with Adaptive Learning Interval Dong-Woo Lee *, Seong G. Kong *, and Kwee-Bo Sim ** *Department of Electrical and Computer Engineering, The University
Circular Dichroism & Optical Rotatory Dispersion. Proteins (KCsa) Polysaccharides (agarose) DNA CHEM 305. Many biomolecules are α-helical!
 Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
Analysis of the simulation
 Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Presenter: She Zhang
 Presenter: She Zhang Introduction Dr. David Baker Introduction Why design proteins de novo? It is not clear how non-covalent interactions favor one specific native structure over many other non-native
Presenter: She Zhang Introduction Dr. David Baker Introduction Why design proteins de novo? It is not clear how non-covalent interactions favor one specific native structure over many other non-native
evoglow - express N kit distributed by Cat.#: FP product information broad host range vectors - gram negative bacteria
 evoglow - express N kit broad host range vectors - gram negative bacteria product information distributed by Cat.#: FP-21020 Content: Product Overview... 3 evoglow express N -kit... 3 The evoglow -Fluorescent
evoglow - express N kit broad host range vectors - gram negative bacteria product information distributed by Cat.#: FP-21020 Content: Product Overview... 3 evoglow express N -kit... 3 The evoglow -Fluorescent
The ideal fiber pattern exhibits 4-quadrant symmetry. In the ideal pattern the fiber axis is called the meridian, the perpendicular direction is
 Fiber diffraction is a method used to determine the structural information of a molecule by using scattering data from X-rays. Rosalind Franklin used this technique in discovering structural information
Fiber diffraction is a method used to determine the structural information of a molecule by using scattering data from X-rays. Rosalind Franklin used this technique in discovering structural information
ID14-EH3. Adam Round
 Bio-SAXS @ ID14-EH3 Adam Round Contents What can be obtained from Bio-SAXS Measurable parameters Modelling strategies How to collect data at Bio-SAXS Procedure Data collection tests Data Verification and
Bio-SAXS @ ID14-EH3 Adam Round Contents What can be obtained from Bio-SAXS Measurable parameters Modelling strategies How to collect data at Bio-SAXS Procedure Data collection tests Data Verification and
Supplementary Figures. Measuring Similarity Between Dynamic Ensembles of Biomolecules
 Supplementary Figures Measuring Similarity Between Dynamic Ensembles of Biomolecules Shan Yang, Loïc Salmon 2, and Hashim M. Al-Hashimi 3*. Department of Chemistry, University of Michigan, Ann Arbor, MI,
Supplementary Figures Measuring Similarity Between Dynamic Ensembles of Biomolecules Shan Yang, Loïc Salmon 2, and Hashim M. Al-Hashimi 3*. Department of Chemistry, University of Michigan, Ann Arbor, MI,
The 3 Genomic Numbers Discovery: How Our Genome Single-Stranded DNA Sequence Is Self-Designed as a Numerical Whole
 Applied Mathematics, 2013, 4, 37-53 http://dx.doi.org/10.4236/am.2013.410a2004 Published Online October 2013 (http://www.scirp.org/journal/am) The 3 Genomic Numbers Discovery: How Our Genome Single-Stranded
Applied Mathematics, 2013, 4, 37-53 http://dx.doi.org/10.4236/am.2013.410a2004 Published Online October 2013 (http://www.scirp.org/journal/am) The 3 Genomic Numbers Discovery: How Our Genome Single-Stranded
Pathways and Controls of N 2 O Production in Nitritation Anammox Biomass
 Supporting Information for Pathways and Controls of N 2 O Production in Nitritation Anammox Biomass Chun Ma, Marlene Mark Jensen, Barth F. Smets, Bo Thamdrup, Department of Biology, University of Southern
Supporting Information for Pathways and Controls of N 2 O Production in Nitritation Anammox Biomass Chun Ma, Marlene Mark Jensen, Barth F. Smets, Bo Thamdrup, Department of Biology, University of Southern
ydci GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC
 Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Exercise 2: Solvating the Structure Before you continue, follow these steps: Setting up Periodic Boundary Conditions
 Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
The Trigram and other Fundamental Philosophies
 The Trigram and other Fundamental Philosophies by Weimin Kwauk July 2012 The following offers a minimal introduction to the trigram and other Chinese fundamental philosophies. A trigram consists of three
The Trigram and other Fundamental Philosophies by Weimin Kwauk July 2012 The following offers a minimal introduction to the trigram and other Chinese fundamental philosophies. A trigram consists of three
evoglow - express N kit Cat. No.: product information broad host range vectors - gram negative bacteria
 evoglow - express N kit broad host range vectors - gram negative bacteria product information Cat. No.: 2.1.020 evocatal GmbH 2 Content: Product Overview... 4 evoglow express N kit... 4 The evoglow Fluorescent
evoglow - express N kit broad host range vectors - gram negative bacteria product information Cat. No.: 2.1.020 evocatal GmbH 2 Content: Product Overview... 4 evoglow express N kit... 4 The evoglow Fluorescent
ChemiScreen CaS Calcium Sensor Receptor Stable Cell Line
 PRODUCT DATASHEET ChemiScreen CaS Calcium Sensor Receptor Stable Cell Line CATALOG NUMBER: HTS137C CONTENTS: 2 vials of mycoplasma-free cells, 1 ml per vial. STORAGE: Vials are to be stored in liquid N
PRODUCT DATASHEET ChemiScreen CaS Calcium Sensor Receptor Stable Cell Line CATALOG NUMBER: HTS137C CONTENTS: 2 vials of mycoplasma-free cells, 1 ml per vial. STORAGE: Vials are to be stored in liquid N
Circular dichroism spectroscopic studies on structures formed by telomeric DNA sequences in vitro
 Circular dichroism spectroscopic studies on structures formed by telomeric DNA sequences in vitro ZHANG Xiaoyan 1, CAO Enhua 1, SUN Xueguang 1 & BAI Chunli 2 1. Institute of Biophysics, Chinese Academy
Circular dichroism spectroscopic studies on structures formed by telomeric DNA sequences in vitro ZHANG Xiaoyan 1, CAO Enhua 1, SUN Xueguang 1 & BAI Chunli 2 1. Institute of Biophysics, Chinese Academy
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Systematic Coarse-Grained Modeling of Complexation between Small Interfering RNA and Polycations Zonghui Wei 1 and Erik Luijten 1,2,3,4,a) 1 Graduate Program in Applied Physics, Northwestern
SUPPLEMENTAL MATERIAL Systematic Coarse-Grained Modeling of Complexation between Small Interfering RNA and Polycations Zonghui Wei 1 and Erik Luijten 1,2,3,4,a) 1 Graduate Program in Applied Physics, Northwestern
Supplementary Information for
 Supplementary Information for Elucidating the Solvent Effect on the Switch of the Helicity of Poly(quinoxaline-2,3-diyl)s: A Conformational Analysis by Small-Angle Neutron Scattering Yuuya Nagata, *1 Tsuyoshi
Supplementary Information for Elucidating the Solvent Effect on the Switch of the Helicity of Poly(quinoxaline-2,3-diyl)s: A Conformational Analysis by Small-Angle Neutron Scattering Yuuya Nagata, *1 Tsuyoshi
I690/B680 Structural Bioinformatics Spring Protein Structure Determination by NMR Spectroscopy
 I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
Supplementary figure 1. Comparison of unbound ogm-csf and ogm-csf as captured in the GIF:GM-CSF complex. Alignment of two copies of unbound ovine
 Supplementary figure 1. Comparison of unbound and as captured in the GIF:GM-CSF complex. Alignment of two copies of unbound ovine GM-CSF (slate) with bound GM-CSF in the GIF:GM-CSF complex (GIF: green,
Supplementary figure 1. Comparison of unbound and as captured in the GIF:GM-CSF complex. Alignment of two copies of unbound ovine GM-CSF (slate) with bound GM-CSF in the GIF:GM-CSF complex (GIF: green,
Molecular Modeling lecture 2
 Molecular Modeling 2018 -- lecture 2 Topics 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction 4. Where do protein structures come from? X-ray crystallography
Molecular Modeling 2018 -- lecture 2 Topics 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction 4. Where do protein structures come from? X-ray crystallography
SUPPORTING MATERIAL. The human DNA repair factor XPC/HR23B distinguishes stereoisomeric benzo[a]pyrenyl-dna lesions CNRS/INSERM/ULP,
![SUPPORTING MATERIAL. The human DNA repair factor XPC/HR23B distinguishes stereoisomeric benzo[a]pyrenyl-dna lesions CNRS/INSERM/ULP, SUPPORTING MATERIAL. The human DNA repair factor XPC/HR23B distinguishes stereoisomeric benzo[a]pyrenyl-dna lesions CNRS/INSERM/ULP,](/thumbs/82/86359411.jpg) SUPPORTING MATERIAL The human DNA repair factor XPC/HR23B distinguishes stereoisomeric benzo[a]pyrenyl-dna lesions V. Mocquet 1, K. Kropachev 2, M. Kolbanovskiy 2, A. Kolbanovskiy 2, A. Tapias 1, Y. Cai
SUPPORTING MATERIAL The human DNA repair factor XPC/HR23B distinguishes stereoisomeric benzo[a]pyrenyl-dna lesions V. Mocquet 1, K. Kropachev 2, M. Kolbanovskiy 2, A. Kolbanovskiy 2, A. Tapias 1, Y. Cai
Characterizing Biological Macromolecules by SAXS Detlef Beckers, Jörg Bolze, Bram Schierbeek, PANalytical B.V., Almelo, The Netherlands
 Characterizing Biological Macromolecules by SAXS Detlef Beckers, Jörg Bolze, Bram Schierbeek, PANalytical B.V., Almelo, The Netherlands This document was presented at PPXRD - Pharmaceutical Powder X-ray
Characterizing Biological Macromolecules by SAXS Detlef Beckers, Jörg Bolze, Bram Schierbeek, PANalytical B.V., Almelo, The Netherlands This document was presented at PPXRD - Pharmaceutical Powder X-ray
Hyeyoung Shin a, Tod A. Pascal ab, William A. Goddard III abc*, and Hyungjun Kim a* Korea
 The Scaled Effective Solvent Method for Predicting the Equilibrium Ensemble of Structures with Analysis of Thermodynamic Properties of Amorphous Polyethylene Glycol-Water Mixtures Hyeyoung Shin a, Tod
The Scaled Effective Solvent Method for Predicting the Equilibrium Ensemble of Structures with Analysis of Thermodynamic Properties of Amorphous Polyethylene Glycol-Water Mixtures Hyeyoung Shin a, Tod
Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study
 pubs.acs.org/jpca Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study Tomoo Miyahara, Hiroshi Nakatsuji,*, and Hiroshi Sugiyama Quantum Chemistry Research Institute, JST, CREST,
pubs.acs.org/jpca Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study Tomoo Miyahara, Hiroshi Nakatsuji,*, and Hiroshi Sugiyama Quantum Chemistry Research Institute, JST, CREST,
BM29 biosaxs data processing tutorial
 HERCULES 2014 BM29 biosaxs data processing tutorial Page 2 OUTLINE Sample Changer Primary Data Processing Model Validation HPLC-SAXS Primary Data Processing Model Validation Ab Initio Model Software in
HERCULES 2014 BM29 biosaxs data processing tutorial Page 2 OUTLINE Sample Changer Primary Data Processing Model Validation HPLC-SAXS Primary Data Processing Model Validation Ab Initio Model Software in
Supporting information for: Norovirus capsid proteins self-assemble through. biphasic kinetics via long-lived stave-like.
 Supporting information for: Norovirus capsid proteins self-assemble through biphasic kinetics via long-lived stave-like intermediates Guillaume Tresset,, Clémence Le Cœur, Jean-François Bryche, Mouna Tatou,
Supporting information for: Norovirus capsid proteins self-assemble through biphasic kinetics via long-lived stave-like intermediates Guillaume Tresset,, Clémence Le Cœur, Jean-François Bryche, Mouna Tatou,
12. Spectral diffusion
 1. Spectral diffusion 1.1. Spectral diffusion, Two-Level Systems Until now, we have supposed that the optical transition frequency of each single molecule is a constant (except when we considered its variation
1. Spectral diffusion 1.1. Spectral diffusion, Two-Level Systems Until now, we have supposed that the optical transition frequency of each single molecule is a constant (except when we considered its variation
, follows from these assumptions.
 Untangling the mechanics and topology in the frictional response of long overhand elastic knots Supplemental Information M.K. Jawed, P. Dieleman, B. Audoly, P. M. Reis S1 Experimental details In this section,
Untangling the mechanics and topology in the frictional response of long overhand elastic knots Supplemental Information M.K. Jawed, P. Dieleman, B. Audoly, P. M. Reis S1 Experimental details In this section,
Transfer of Chirality from Molecule to Phase in Self-assembled Chiral Block Copolymers
 Transfer of Chirality from Molecule to Phase in Self-assembled Chiral Block Copolymers Rong-Ming Ho,* Ming-Chia Li, Shih-Chieh Lin, Hsiao-Fang Wang, Yu-Der Lee, Hirokazu Hasegawa, and Edwin L. Thomas Supporting
Transfer of Chirality from Molecule to Phase in Self-assembled Chiral Block Copolymers Rong-Ming Ho,* Ming-Chia Li, Shih-Chieh Lin, Hsiao-Fang Wang, Yu-Der Lee, Hirokazu Hasegawa, and Edwin L. Thomas Supporting
Timing molecular motion and production with a synthetic transcriptional clock
 Timing molecular motion and production with a synthetic transcriptional clock Elisa Franco,1, Eike Friedrichs 2, Jongmin Kim 3, Ralf Jungmann 2, Richard Murray 1, Erik Winfree 3,4,5, and Friedrich C. Simmel
Timing molecular motion and production with a synthetic transcriptional clock Elisa Franco,1, Eike Friedrichs 2, Jongmin Kim 3, Ralf Jungmann 2, Richard Murray 1, Erik Winfree 3,4,5, and Friedrich C. Simmel
ion mobility spectrometry IR spectroscopy
 Debasmita Gho 29.10.2016 Introducti on Owing to its accuracy, sensitivity, and speed, mass spectrometry (MS) coupled to fragmentation techniques is the method of choice for determining the primary structure
Debasmita Gho 29.10.2016 Introducti on Owing to its accuracy, sensitivity, and speed, mass spectrometry (MS) coupled to fragmentation techniques is the method of choice for determining the primary structure
Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS)
 nature methods Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Greg L Hura, Angeli L Menon, Michal Hammel, Robert P Rambo, Farris L Poole II, Susan E Tsutakawa,
nature methods Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Greg L Hura, Angeli L Menon, Michal Hammel, Robert P Rambo, Farris L Poole II, Susan E Tsutakawa,
Supporting Information for the Effects of Electrostatic Interaction and Chirality on the
 Supporting Information for the Effects of Electrostatic Interaction and Chirality on the Friction Coefficient of Water Flow Inside Single-Walled Carbon Nanotubes and Boron Nitride Nanotubes Xingfei Wei
Supporting Information for the Effects of Electrostatic Interaction and Chirality on the Friction Coefficient of Water Flow Inside Single-Walled Carbon Nanotubes and Boron Nitride Nanotubes Xingfei Wei
The CB[n] Family: Prime Components for Self-Sorting Systems Supporting Information
![The CB[n] Family: Prime Components for Self-Sorting Systems Supporting Information The CB[n] Family: Prime Components for Self-Sorting Systems Supporting Information](/thumbs/95/124871499.jpg) The CB[n] Family: Prime Components for Self-Sorting Systems Supporting Information by Simin Liu, Christian Ruspic, Pritam Mukhopadhyay,Sriparna Chakrabarti, Peter Y. Zavalij, and Lyle Isaacs* Department
The CB[n] Family: Prime Components for Self-Sorting Systems Supporting Information by Simin Liu, Christian Ruspic, Pritam Mukhopadhyay,Sriparna Chakrabarti, Peter Y. Zavalij, and Lyle Isaacs* Department
Small-Angle X-ray Scattering (SAXS) SPring-8/JASRI Naoto Yagi
 Small-Angle X-ray Scattering (SAXS) SPring-8/JASRI Naoto Yagi 1 Wikipedia Small-angle X-ray scattering (SAXS) is a small-angle scattering (SAS) technique where the elastic scattering of X-rays (wavelength
Small-Angle X-ray Scattering (SAXS) SPring-8/JASRI Naoto Yagi 1 Wikipedia Small-angle X-ray scattering (SAXS) is a small-angle scattering (SAS) technique where the elastic scattering of X-rays (wavelength
Supplementary Figures:
 Supplementary Figures: Supplementary Figure 1: The two strings converge to two qualitatively different pathways. A) Models of active (red) and inactive (blue) states used as end points for the string calculations
Supplementary Figures: Supplementary Figure 1: The two strings converge to two qualitatively different pathways. A) Models of active (red) and inactive (blue) states used as end points for the string calculations
Supplementary Information. The Solution Structural Ensembles of RNA Kink-turn Motifs and Their Protein Complexes
 Supplementary Information The Solution Structural Ensembles of RNA Kink-turn Motifs and Their Protein Complexes Xuesong Shi, a Lin Huang, b David M. J. Lilley, b Pehr B. Harbury a,c and Daniel Herschlag
Supplementary Information The Solution Structural Ensembles of RNA Kink-turn Motifs and Their Protein Complexes Xuesong Shi, a Lin Huang, b David M. J. Lilley, b Pehr B. Harbury a,c and Daniel Herschlag
Introduction to" Protein Structure
 Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Nonadiabatic dynamics simulations of singlet fission in 2,5-bis(fluorene-9-ylidene)-2,5-dihydrothiophene crystals. Supporting Information.
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 18 Nonadiabatic dynamics simulations of singlet fission in,5-bis(fluorene-9-ylidene)-,5-dihydrothiophene
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 18 Nonadiabatic dynamics simulations of singlet fission in,5-bis(fluorene-9-ylidene)-,5-dihydrothiophene
Re- engineering cellular physiology by rewiring high- level global regulatory genes
 Re- engineering cellular physiology by rewiring high- level global regulatory genes Stephen Fitzgerald 1,2,, Shane C Dillon 1, Tzu- Chiao Chao 2, Heather L Wiencko 3, Karsten Hokamp 3, Andrew DS Cameron
Re- engineering cellular physiology by rewiring high- level global regulatory genes Stephen Fitzgerald 1,2,, Shane C Dillon 1, Tzu- Chiao Chao 2, Heather L Wiencko 3, Karsten Hokamp 3, Andrew DS Cameron
Unfolding CspB by means of biased molecular dynamics
 Chapter 4 Unfolding CspB by means of biased molecular dynamics 4.1 Introduction Understanding the mechanism of protein folding has been a major challenge for the last twenty years, as pointed out in the
Chapter 4 Unfolding CspB by means of biased molecular dynamics 4.1 Introduction Understanding the mechanism of protein folding has been a major challenge for the last twenty years, as pointed out in the
J- vs. H-type assembly: pentamethine cyanine (Cy5) as near IR chiroptical. reporter
 S1 Supporting Information: J- vs. H-type assembly: pentamethine cyanine (Cy5) as near IR chiroptical reporter Larysa I. Markova, a,b Vladimir L. Malinovskii, a Leonid D. Patsenker b and Robert Häner* a
S1 Supporting Information: J- vs. H-type assembly: pentamethine cyanine (Cy5) as near IR chiroptical reporter Larysa I. Markova, a,b Vladimir L. Malinovskii, a Leonid D. Patsenker b and Robert Häner* a
Near-instant surface-selective fluorogenic protein quantification using sulfonated
 Electronic Supplementary Material (ESI) for rganic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2014 Supplemental nline Materials for ear-instant surface-selective fluorogenic
Electronic Supplementary Material (ESI) for rganic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2014 Supplemental nline Materials for ear-instant surface-selective fluorogenic
Long Distance Radical Cation Hopping in DNA: The Effect of Thymine-Hg(II)-Thymine Base Pairs
 Long Distance Radical Cation Hopping in DNA: The Effect of Thymine-Hg(II)-Thymine Base Pairs Joshy Joseph and Gary B. Schuster* School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta,
Long Distance Radical Cation Hopping in DNA: The Effect of Thymine-Hg(II)-Thymine Base Pairs Joshy Joseph and Gary B. Schuster* School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta,
Chiral selection in wrapping, crossover, and braiding of DNA mediated by asymmetric bend-writhe elasticity
 http://www.aimspress.com/ AIMS Biophysics, 2(4): 666-694. DOI: 10.3934/biophy.2015.4.666 Received date 28 August 2015, Accepted date 29 October 2015, Published date 06 November 2015 Research article Chiral
http://www.aimspress.com/ AIMS Biophysics, 2(4): 666-694. DOI: 10.3934/biophy.2015.4.666 Received date 28 August 2015, Accepted date 29 October 2015, Published date 06 November 2015 Research article Chiral
