InSilicoVA: A Method to Automate Cause of Death Assignment for Verbal Autopsy
|
|
|
- Morris Carter
- 5 years ago
- Views:
Transcription
1 InSilicoVA: A Method to Automate Cause of Death Assignment for Verbal Autopsy Samuel J. Clark 1,4,5,6,7,*, Tyler McCormick 1,2, Zehang Li 2, and Jon Wakefield 2,3 1 Department of Sociology, University of Washington 2 Department of Statistics, University of Washington 3 Department of Biostatistics, University of Washington 4 Institute of Behavioral Science (IBS), University of Colorado at Boulder 5 MRC/Wits Rural Public Health and Health Transitions Research Unit (Agincourt), School of Public Health, Faculty of Health Sciences, University of the Witwatersrand 6 ALPHA Network, London School of Hygience and Tropical Medicine, London, UK 7 INDEPTH Network, Accra, Ghana * Correspondence to: work@samclark.net August 24, 2013 Working Paper no. 133 Center for Statistics and the Social Sciences University of Washington i
2 Abstract Verbal autopsies (VA) are widely used to provide cause-specific mortality estimates in developing world settings where vital registration does not function well. VAs assign cause(s) to a death by using information describing the events leading up to the death, provided by care givers. Typically physicians read VA interviews and assign causes using their expert knowledge. Physician coding is often slow, and individual physicians bring bias to the coding process that results in non-comparable cause assignments. These problems significantly limit the utility of physician-coded VAs. A solution to both is to use an algorithmic approach that formalizes the cause-assignment process. This ensures that assigned causes are comparable and requires many fewer person-hours so that cause assignment can be conducted quickly without disrupting the normal work of physicians. Peter Byass InterVA method (Byass et al., 2012) is the most widely used algorithmic approach to VA coding and is aligned with the WHO 2012 standard VA questionnaire (Leitao et al., 2013). The statistical model underpinning InterVA can be improved; uncertainty needs to be quantified, and the link between the population-level CSMFs and the individual-level cause assignments needs to be statistically rigorous. Addressing these theoretical concerns provides an opportunity to create new software using modern languages that can run on multiple platforms and will be widely shared. Building on the overall framework pioneered by InterVA, our work creates a statistical model for automated VA cause assignment. Acknowledgments Preparation of this manuscript was partially supported by the Bill and Melinda Gates Foundation. The authors are grateful to Peter Byass, Basia Zaba, Kathleen Kahn, Stephen Tollman, Adrian Raftery, Philip Setel and Osman Sankoh for helpful discussions. ii
3 Contents 1 Introduction 1 2 InterVA Byass Description of InterVA Our Notation for InterVA: Our Description of InterVA Data Requirements: Our Presentation of the InterVA Model and Algorithm Evaluation of InterVA Model & Algorithm InSilicoVA InSilicoVA Notation: InSilicoVA Data: InSilicoVA Algorithm Testing & Comparing InSilicoVA and InterVA Simulation Study Application to Real Data - Agincourt HDSS Results Discussion Summary of Findings Future Work iii
4 1 Introduction Verbal autopsy (VA) is a common approach for determining cause of death in regions where deaths are not recorded routinely. VAs are a standardized questionnaire administered to caregivers, family members, or others knowledgeable of the circumstances of a recent death with the goal of using these data to infer the likely causes of death (Byass et al., 2012). The VA survey instrument asks questions related to the deceased individual s medical condition (did the person have diarrhea, for example) and related to other factors surrounding the death (did the person die in an automotive accident, for example). VA has been widely used by researchers in Health and Demographic Surveillance Sites (HDSS), such as the INDEPTH Network (Sankoh and Byass, 2012), and has recently received renewed attention from the World Health Organization through the release of an update to the widely used standard VA questionnaire (see statistics/verbalautopsystandards/). The main statistical challenge with VA data is to ascertain patterns in responses that correspond to a pre-defined set of causes of death. Typically the nature of such patterns is not known a priori and measurements are subject to various types of measurement error, which are discussed in further detail below. There are two credible methods for automating the assignment of cause of death from VA data. InterVA (see for example: Byass et al., 2003, 2006, 2012) is a proprietary algorithm developed and maintained by Peter Byass, and it has been used extensively by both the AL- PHA (Maher et al., 2010) and INDEPTH networks of HDSS sites and a wide variety of other research groups. At this time InterVA appears to be the de facto standard method. The Institute for Health Metrics and Evaluation (IHME) have proposed a number of additional methods (for example: Flaxman et al., 2011; James et al., 2011; Murray et al., 2011), some of which build on earlier work by King and Lu (King et al., 2010; King and Lu, 2008). Among their methods, the Simplified Symptom Pattern Recognition (SSPR) method (Murray et al., 2011) is most directly comparable to InterVA and appears to have the best chance of becoming widely used. The SSPR and related methods require a so-called gold standard database, a database consisting of a large number of deaths where the cause has been certified by medical professionals and is considered reliable, and further, where the symptoms for those deaths are also verifiable by medical professionals. Deaths recorded in a gold standard database are typically in-hospital deaths. Given regional variation in the prevalence and familiarity of medical professionals with certain causes of death, region-specific gold standard databases are also necessary. The majority of public health and epidemiology researchers and officials do not have access to such a gold standard database, motivating development of methods to infer cause of death using VA data without access to gold standard databases. Aside from the challenges related to obtaining useful gold standard training data, using VA data to infer cause assignment is also statistically challenging because there are multiple sources of variation and error present in VA data. We identify three sources of variation and propose a novel method called InSilicoVA to address these sources of variation. First, though responses on the VA instrument may be the respondents recollection or best guess about what has happened, they are not necessarily accurate. This type of error is present in all survey data but may be especially pronounced in VA data because respondents have 1
5 varying levels of familiarity with the circumstances surrounding a death. Also, the definition of an event of interest may be different for each respondent. A question about diarrhea, for example, requires the respondent to be sufficiently involved in the decedent s medical care to know this information and to have a definition of diarrhea that maps relatively well to accepted clinical standards. A second source of variability arises from individual variation in presentation of diseases. Statistical methods must be sufficiently robust to overfitting to appreciate that two individuals with the same cause of death may have slightly different presentations, and thus, different VA reports. Third, like InterVA, InSilicoVA will use physician elicited conditional probabilities. These probabilities, representing the consensus estimate of the likelihood that a person will experience a given symptom if they died from a certain cause, are unknown. Simulation and results with data from the Agincourt MRC/Wits Rural Public Health and Heath Transitions Research Unit (Agincourt) indicate that results obtained with conditional probabilities assumed fixed and known (as is done in both InterVA and the SSPR method) can underestimate uncertainty in population cause of death distributions. We evaluate the sensitivity to these prior probabilities. InSilicoVA incorporates uncertainty from these sources and propagates the uncertainty between individual cause assignments and population distributions. Accounting for these sources of error produces confidence intervals for both individual cause assignments and population distributions. These confidence intervals reflect more realistically the complexity of cause assignment with VA data. A unified framework for both individual cause and population distribution estimation also means that additional information about individual causes, such as physician coded VAs, can easily be used in InSilicoVA, even if physician coded records are only available for a subset of cases. Further, we can exactly quantify the contribution of each VA questionnaire item in classifying a case into a specific cause, affording the possibility for item reduction by identifying which inputs are most useful for discriminating between causes. This feature could lead to a more efficient, streamlined survey mechanism and set of conditional probabilities for elicitation. In Section 2 we describe InterVA and present open challenges which we address with InSilicoVA. In Section 3 we present the InSilicoVA model. Section 4 describes applications of both methods to simulated and real data, and Section 4.3 provides the results. We conclude with a very brief discussion in Section 5. 2 InterVA 2.1 Byass Description of InterVA Below is Byass summary of the InterVA method presented in Byass et al. (2012). Bayes theorem links the probability of an event happening given a particular circumstance with the unconditional probability of the same event and the conditional probability of the circumstance given the event. If the event of interest is a particular cause of death, and the circumstance is part of the events leading to death, then Bayes theorem can be applied in terms of circumstances and causes of death. Specifically, if there are a predetermined set of possible 2
6 causes of death C 1... C m and another set of indicators I 1... I n representing various signs, symptoms and circumstances leading to death, then Bayes general theorem for any particular C i and I j can be stated as: P (C i I j ) = P (I j C i ) P (C i ) P (I j C i ) P (C i ) + P (I j!c i ) P (!C i ) (1) where P (!C i ) is (1 P (C i )). Over the whole set of causes of death C 1... C m a set of probabilities for each C i can be calculated using a normalising assumption so that the total conditional probability over all causes totals unity: P (C i I j ) = P (I j C i ) P (C i ) m i=1 P (C i) (2) Using an initial set of unconditional probabilities for causes of death C 1... C m (which can be thought of as P (C i I 0 )) and a matrix of conditional probabilities P (I j C i ) for indicators I 1... I n and causes C 1... C m, it is possible to repeatedly apply the same calculation process for each I 1... I n that applies to a particular death: P (C i I 1...n ) = P (I j C i ) P (C i I 0...n 1 ) m i=1 P (C i I 0...n 1 ) (3) This process typically results in the probabilities of most causes reducing, while a few likely causes are characterised by their increasing probabilities as successive indicators are processed. In the same article Byass describes the process of defining the conditional probabilities P (I j C i ). Apart from the mathematics, the major challenge in building a probabilistic model covering all causes of death to a reasonable level of detail lies in populating the matrix of conditional probabilities P (I j C i ). There is no overall source of data available which systematically quantifies probabilities of various signs, symptoms and circumstances leading to death in terms of their associations with particular causes. Therefore, these conditional probabilities have to be estimated from a diversity of incomplete sources (including previous InterVA models) and modulated by expert opinion. In the various versions of InterVA that have been developed, expert panels have been convened to capture clinical expertise on the relationships between indicators and causes. In this case, an expert panel convened in Geneva in December 2011 and continued to deliberate subsequently, particularly considering issues that built on previous InterVA versions. Experience has shown that gradations in levels of perceived probabilities correspond more to a logarithmic than linear scale, and in the expert consultation for InterVA-4, we used a perceived probability scale that was subsequently converted to numbers on a logarithmic scale as shown below. 3
7 Table 1: InterVA Conditional Probability Letter-Value Correspondances Label Value Interpretation I 1.0 Always A 0.8 Almost always A 0.5 Common A 0.2 B 0.1 Often B 0.05 B 0.02 C 0.01 Unusual C C D vrare D D E Hardly ever N 0.0 Never The physician-derived conditional probabilities that are supplied with the InterVA software (Byass, 2013) are coded using the letter codes in the leftmost column of Table 1. We rewrite, interpret and discuss the InterVA model below. 2.2 Our Notation for InterVA: Deaths: y j j {1,..., J}, Y = [y1,..., y J ] Signs/symptoms: s k {0, 1}, k {1,..., K}, S = [s1,..., s K ] Causes: c n n {1,..., N}, N n=1 c j,n = 1 Fraction of all deaths that are cause n, the cause-specific mortality fraction (CSMF): f n n {1,..., N}, F = [f1,..., f N ], N n=1 f n = Our Description of InterVA Data Requirements: 1. For each death y j, a VA interview produces a binary-valued vector of signs/symptoms: S j = {s j,1, s j,2,... s j,k } (4) S is the J K matrix whose rows are the S j for each death. 2. A K N matrix of conditional probabilities reflecting physicians opinions about how 4
8 likely a given sign/symptom is for a death resulting from a given cause: Pr(s 1 c 1 ) Pr(s 1 c 2 ) Pr(s 1 c N ) Pr(s 2 c 1 ) Pr(s 2 c 2 ) Pr(s 2 c N ) P = Pr(s K c 1 ) Pr(s K c 2 ) Pr(s K c N ) As supplied with the InterVA software (Byass, 2013) P does not contain internally consistent probabilities 1. This is easy to understand by noting that these probabilities are not derived from a well-defined event space that would constrain them to be consistent with one another. As described by Byass above in Section 2.1 the physicians provide a letter grade for each conditional probability, and these correspond to a ranking of perceived likelihood of a given sign/symptom if the death is due to a given cause. These letter grades are then turned into numbers in the range [0, 1] (NB: 0.0 and 1.0 are included) using Table 1. Consequently it is not possible to assume that the members of P will behave as expected when one attempts to calculate complements and use more than one in an expression in a way that should be consistent. 3. An initial guess of F, F = [f n,..., f N ] (5) 2.4 Our Presentation of the InterVA Model and Algorithm For a specific death y j we can imagine and examine the two-dimensional joint distribution (c j,n, s j,k ): where Pr(c j,n s j,k ) = P (s j,k c j,n ) Pr(c j,n ) Pr(s j,k ) Pr(s j,k c j,n ) Pr(c j,n ) = Pr(s j,k c j,n ) Pr(c j,n ) + x x = Pr(s j,k c j,n ) Pr( c j,n ) (7) Looking on the RHS of (6), we have Pr(s j,k c j,n ) from the conditional probabilities from physicians and Pr(c j,n ) f n. If the conditional probabilities P were well-behaved, then x = N n =1, n n (6) Pr(s j,k c j,n ) Pr(c j,n ) (8) 1 The P supplied with InterVA has many logical inconsistencies, for example situations where conditional probabilities should add up to equal another: Pr(fast breathing for 2 weeks or longer HIV) + Pr(fast breathing for less than 2 weeks HIV) Pr(fast breathing HIV), or where they just do not make sense: Pr(fast breathing for 2 weeks or longer sepsis) > Pr(fast breathing sepsis). The P supplied with InterVA-4 (Byass, 2013) is a matrix with 17,526 entries. We have investigated automated ways of correcting the inconsistencies, but with every attempt we discover more, so we have concluded that the entries in P need to be re-elicited from physicians using an approach that ensures that they are consistent. 5
9 However since the P supplied with the InterVA software (Byass, 2013) are not consistent with one another this calculation does not produce useful results. InterVA solves this with an arbitrary reformulation of the relationship. For each death y j and over all signs/symptoms s k associated with y j : (j, n) : Propensity(c j,n S j ) = f n K [Pr(s j,k c n )] s k (9) For each death y j these Propensities do not add to 1.0 so they need to be normalized to produce well-behaved probabilities: k=1 (j, n) : Pr(c j,n ) = Propensity(c j,n S j ) N n=1 Propensity(c j,n S j ) (10) The population-level CSMFs F are calculated by adding up the results of calculating (10) for all causes for all deaths: J n : f n = Pr(c j,n ) (11) j=1 2.5 Evaluation of InterVA Model & Algorithm In effect what InterVA does is distribute a given death among a number of predefined causes. The cause with the largest fraction is assumed to be the primary cause, followed with decreasing significance by the remaining causes in order from largest to smallest. The conceptual construct of a partial death is central to InterVA and is interchanged freely with the probability of dying from a given cause. This equivalence is not real and is at the heart of the theoretical problems with InterVA. At a high level InterVA proposes a very useful solution to the fundamental challenge that all automated VA coding algorithms face - how to characterize and use the relationship that exists between signs/symptoms and causes of death. In a perfect world we would have medically certified patient records that include the results of real autopsies, and we could use those to model this relationship and use the results of that model in our cause-assignment algorithms. But in that perfect world there is no use for VA at all. So by definition we live in the world where that type of gold standard data do not and will not exist most of the time for most of the developing world where VAs are conducted. Byass solution to this is to accept the limitations on the expert knowledge that physicians can provide to substitute for gold standard data, and further, to elicit and then organize that information in a very useful format the conditional probabilities matrix P above in (5). In sum, Byass has sorted through a variety of possible general strategies and settled on one that is both doable and produces useful results. Where we contribute is to help refine the statistical and computational methods used to conduct the cause assignments. In order to do that we have evaluated InterVA thoroughly, 6
10 and we have identified a number of weaknesses that we feel can be addressed. The brief list below is by necessity a blunt description of those weaknesses, which nevertheless do not reduce the importance of InterVA as described just above. There are several theoretical problems with the InterVA model: 1. The derivation presented in (1) through (3) is incorrect; in particular (2) does not follow from (1), and (3) does not follow from (2). As described just above, (11) requires that probabilities be equated with fractional deaths, which is conceptually difficult. 2. InterVA s statistical model is not probabilistic in the recognized sense because it does not include elements that can vary unpredictably, and hence there is no randomness. Although P contains probabilities (see discussion with (5) above), these are not allowed to vary in the estimation procedure used to assign causes of death P is effectively a fixed input to the model. 3. Because the model does not contain features that are allowed to vary unpredictably, it is not possible to quantify uncertainty to produce probabilistic error bounds. 4. If we ignore the errors in the derivation and work with (9) - (11) as if they were correct, there are additional problems. Equation (9) is at the core of InterVA and demonstrates two undesirable features: (a) For a specific individual the propensity for each cause is deterministically affected by f n, what Byass terms the prior probability of cause n, effectively a userdefined parameter of the algorithm. This means that the final individual-level cause assignments are a deterministic transformation of the so-called prior i.e. the results are not only sensitive to but depend directly on the prior. (b) The expression in (9) captures only one valance in the relationship between signs/symptoms and a cause of death it acknowledges and reacts only to the presence of a sign/symptom but not to its absence, effectively throwing away half of the information in the dataset. To include information conveyed by the absence of a sign/symptom, (9) needs a term that involves something like 1 Pr(s j,k C n ). [The components of InSilicoVA in (22) and (27) below include this term.] This is undesirable for two reasons: (1) signs/symptoms are not selecting causes that fit and de-selecting causes that don t, but rather just de-selecting causes, and (2) the final probabilities are typically the product of a large number of very small numbers, and hence their numeric values can become extremely small, small enough to interact badly and unpredictable with the numerical storage/arithmetic capacity of the software and computers used to calculate them. A simple log transformation would solve this problem. 5. Finally, (11) is a deterministic transformation of the individual-level cause assignments to produce a population-level CSMF; simply another way of stating the individual-level cause assignments. Because the individual-level cause assignments are not probabilistic, neither are the resulting CSMFs. In addition there are idiosyncrasies that affect the current implementation of InterVA (Byass, 7
11 2013) and some oddities having to do with the matrix of conditional probabilities provided with Byass InterVA. We will not describe those here. InSilicoVA is designed to overcome these problems and provide a valid statistical framework on which further refinements can be built. 3 InSilicoVA InSilicoVA is a statistical model and computational algorithm to automate assignment of cause of death from data obtained by VA interviews. Broadly the method aims to: Follow in the footsteps of InterVA building on its strengths and addressing its weaknesses. Produce consistent, comparable cause assignments and CSMFs. Be statistically and computationally valid and extensible. Provide a means to quantify uncertainty. Be able to function to assign causes to a single death. The name InSilicoVA is inspired by in-vitro studies that mimic real biology but in more controlled circumstances (often on glass petri dishes). In this case we are assigning causes to deaths using a computer that performs the required calculations using a silicon chip. Further, we owe a great debt to the InterVA (interpret VA) method that provides useful philosophical and practical frameworks on which to build the new method - so we have stuck to the structure of InterVA s name. 3.1 InSilicoVA Notation: Deaths: y j j {1,..., J}, Y = [y1,..., y J ] Signs/symptoms: s k {0, 1}, k {1,..., K}, S = [s1,..., s K ] Causes of death: c n n {1,..., N}, C = [c1,..., c N ] For individual j, probability of cause n given S j : l j,n = Pr(y j = c n S j ), j {1,..., J}, n {1,..., N}, L j = [l j,1,..., l j,n ], N n=1 l j,n = 1 Count of all deaths that are cause n, the cause-specific death count (CSDC): m n n {1,..., N}, M = [m1,..., m N ], N n=1 m n = J Fraction of all deaths that are cause n, the cause-specific mortality fraction (CSMF): f n n {1,..., N}, F = [f1,..., f N ], N n=1 f n = 1 8
12 3.2 InSilicoVA Data: 1. For each death y j, the VA interview produces a binary-valued vector of signs/symptoms: S j = {s j,1, s j,2,... s j,k } (12) S is the J K matrix whose rows are the S j for each death. The columns of S are assumed to be independent given C, i.e. there is no systematic relationship between the signs/symptoms for a given cause. This is very obviously not a justifiable assumption. Signs and symptoms come in characteristic sets depending on the cause of death, so there is some correlation between them, conditional on a given cause. Nonetheless we assume independence in order to facilitate initial construction and testing of our model, and most pragmatically, so that we can utilize the matrix of conditional probabilities supplied by Byass with the InterVA software (Byass, 2013) it is impossible to either regenerate or significantly improve upon these without significant resources with which to organize meetings of physicians with the relevant experience who can provide this information. This S j is the same as (4) used by InterVA. 2. A K N matrix of conditional probabilities reflecting physicians opinions about how likely a given sign/symptom is for a death resulting from a given cause: Pr(s 1 c 1 ) Pr(s 1 c 2 ) Pr(s 1 c N ) Pr(s 2 c 1 ) Pr(s 2 c 2 ) Pr(s 2 c N ) P =..... (13). Pr(s K c 1 ) Pr(s K c 2 ) Pr(s K c N ) InSilicoVA assumes that the components of P are consistent with one another. In the simulation study described in Section 4.1, we construct consistent values for P, but when we test the model on real data in Section 4.2, we have no option other than using the inconsistent P supplied with the InterVA software (Byass, 2013). 3. An initial guess of F, F = [f n,..., f N ] 3.3 InSilicoVA Algorithm We are interested in the joint distribution ( F, Y ) given the set of observed signs/symptoms S. The posterior distribution is: Pr( F, Y S) = Pr(S Y, F ) Pr( Y F ) Pr( F ) Pr(S) Pr(S Y, F ) Pr( Y F ) Pr( F ) (14) = J Pr(S y j, F ) Pr(y j F ) Pr( F ) (15) j=1 9
13 Because individual cause assignments are independent, individual sign/symptom vectors S j are independent from F (the CSMFs), and we have: Pr( F, Y S) J Pr(S y j ) Pr(y j F ) Pr( F ) (16) We will use a Gibbs sampler to sample from this posterior as follows: G.1 start with an initial guess of F, F G.2 sample Y F, S G.3 sample F Y, S j=1 G.4 repeat steps G.2 and G.3 until F and Y converge This algorithm is generic and allows a rich range of models. For the moment the InsilicoVA model is: s j,k c n Bernoulli(Pr(s j,k c n )) (17) y j = c n F Multinomial N (1, F ) (18) F Dirichlet( α), α is N-dimensional and constant (19) Then the posterior in (16) is: Pr( F, Y S) J j=1 k=1 K Pr(s j,k y j = c n ) Pr(y j = c n F ) Pr( F ) (20) This formulation is computationally efficient because of Multinomial/Dirichlet conjugacy, and because using Bayes rule we have, for step G.2: where the l j,n that compose L j are: l j,n = Pr(y j = c n S j ) y j = c n F, S j Multinomial N (1, L j ) (21) = Pr(y j = c n ) Pr( S j y j = c n ) Pr( S j ) substituting f n = Pr(y j = c n ) and using the data to calculate = the probability of a specific S j given the cause assignment y j = c n f n K k=1 (Pr(s ) j,k y j = c n ) s j,k [1 Pr(sj,k y j = c n )] (1 s j,k) N n =1 f n (Pr(s ) K (22) j,k y j = c n ) s j,k [1 Pr(sj,k y j = c n )] (1 s j,k) k=1 10
14 We can also derive from (20) the distribution of F conditional on Y, for step G.3: F Y, S Dirichlet( M + α) (23) where m n = J [y j = c n ], using Iverson s bracket notation:[z] = j=1 { 1 if z true 0 if z false (24) In summary, the Gibbs sampler proceeds given suitable initialization F by: G.2 sampling a cause for each death to generate a new Y F, S: y j = c n F, S j Multinomial N (1, L j ) (25) where l j,n = f n K k=1 (Pr(s ) j,k y j = c n ) s j,k [1 Pr(sj,k y j = c n )] (1 s j,k) N n =1 f n (Pr(s ) K (26) j,k y j = c n ) s j,k [1 Pr(sj,k y j = c n )] (1 s j,k) k=1 G.3 sampling a new F Y, S: F Y, S Dirichlet( M + α) (27) The resulting sample of ( F, Y ) and the L j that go with it form the output of the method. These are distributions of CSMFs at the population level and probabilities of dying from each cause at the individual level. These distributions can be summarized as required to produce point values and measures of uncertainty in F and L j. Deaths often result from more than one cause. InSilicoVA accommodates this possibility by producing a separate distribution of the probabilities of being assigned to each cause; that is N distributions, one for each cause. In contrast, InterVA reports one value for each cause, and those values sum to unity across causes for a single death. Finally, with a suitable F, InSilicoVA can be used to assign causes (and their associated l j,n ) to a single death by repeatedly drawing causes using (25). This requires no more information than InterVA to accomplish the same objective, and it produces uncertainty bounds around the probabilities of being assigned to each cause. 4 Testing & Comparing InSilicoVA and InterVA To evaluate both InSilicoVA and InterVA we fit them to simulated and real data. We have created R code that implements both methods. The R code for InterVA matches the results produced by Peter Byass implementation (Byass, 2013). 11
15 4.1 Simulation Study Our simulated data are generated using this procedure: 1. Draw a set of Pr(s k c n ) so that they have the same distribution and range as those provided with Byass InterVA software (Byass, 2013). 2. Draw a set of simulated deaths from a made up distribution of deaths by cause. 3. For each simulated death, assign a set of signs/symptoms by applying the conditional probabilities simulated in step 1. These simulated data have the same overall features as the data required for either InterVA or InSilicoVA, and we know both the real population distribution of deaths by cause and the true individual cause assignments. Our simulation study poses three questions: 1. Fair comparison of InSilicoVA and InterVA. To make this comparison we generate 100 simulated datasets and apply both methods to each dataset. We summarize the results with individual-level and population-level error measures. We refer to this as fair because we apply both methods in their simplest form to data that fulfill all the requirements of both methods. Since the data are effectively perfect we expect both methods to perform well. 2. Influence of numeric values of Pr(s k c n ). Given the structure of InterVA, we are concerned that the results of InterVA may be sensitive to the exact numerical values taken by the conditional probabilities supplied by physicians. To test this we rescale the simulated Pr(s k c n ) so that their range is restricted to [ ]. We again simulate 100 datasets and apply both methods to each dataset and summarize the resulting errors for each method. 3. Reporting errors. As we mentioned in the introduction, we are concerned about reporting errors for any algorithmic approach to VA cause assignment. To investigate this we randomly recode our simulated signs/symptoms so that a small fraction are wrong - i.e. coded 0 when the sign/symptom exists (15%) or 1 when there is no sign/symptom (10%). We do this for 100 simulated datasets and summarize errors resulting from application of both methods. 4.2 Application to Real Data - Agincourt HDSS To investigate the behavior of InSilicoVA and InterVA on real data, we apply both methods to the VA data generated by the Agincourt health and demographic surveillance system (HDSS) in South Africa (Kahn et al., 2012) from roughly 1993 to the present. The Agincourt site continuously monitors the population of 21 villages located in the Bushbuckridge District of Mpumalanga Province in northeast South Africa. This is a rural population living in what was during Apartheid a black homeland. The Agincourt HDSS was established in the early 1990s with the purpose of guiding the reorganization of South Africa s health system. 12
16 Since then the goals of the HDSS have evolved and now it contributes to evaluation of national policy at population, household and individual levels. The population covered by the Agincourt site is approximately eighty-thousand. For this test we us the physician-generated conditional probabilities P and initial guess of the CSMFs F provided by Byass with the InterVA software (Byass, 2013). 4.3 Results The results of our simulation study are summarized graphically as a set of Figures 1 3. The Agincourt results are presented in Figure 4. 5 Discussion 5.1 Summary of Findings InSilicoVA begins to solve most of the critical problems with InterVA. The results of applying both methods to simulated data indicate that InSilicoVA performs well under all circumstances except reporting errors, but even in this situation InSilicoVA performs far better than InterVA. InSilicoVA and InterVA both perform relatively well when the simulated data are perfect. InSilicoVA s performance is not affected by changing the magnitudes and ranges of the conditional probability inputs, whereas InterVA s performance suffers dramatically. With reporting errors both methods performance is negatively impacted, but InterVA becomes effectively useless. Applied to one specific real dataset, both methods produce qualitatively similar results, but InSilicoVA is far more conservative and produces confidence bounds, whereas InterVA does not. For Agincourt, Figure 4.A shows the causes with the largest difference between the In- SilicoVA and InterVA estimates of the CSMF. InSilicoVA classifies a larger portion of deaths as due to causes labeled as other. This indicates that these causes are related to either the communicable or non-communicable diseases, but there is not enough information to make a more specific classification. This feature of InSilicoVA identifies cases that are difficult to classify using available data and may, for example, be good candidates for physician review. We view this behavior as a strength of InSilicoVA because it is consistent with the fundamental weakness of the VA approach, namely that both the information obtained from a VA interview and the expert knowledge and/or gold standard used to characterize the relationship between signs/symptoms and causes are inherently weak and incomplete, and consequently it is very difficult or impossible to make highly specific cause assignments using VA. Given this, we do not want a method that is artificially precise, i.e. forces fine-tuned classification when there is insufficient information. Hence we view InSilicoVA s behavior as reasonable, honest (in that it does not over interpret the data) and useful. Useful in 13
17 the sense that it identifies where our information is particularly weak and therefore where we need to apply more effort either to data or to interpretation, like addition physician reviews. 5.2 Future Work We plan a variety of additional work on InSilicoVA: 1. Explore the possibility of replacing the Dirichlet distribution in (19), (23) and (27) with a mixture of Normals on the baseline logit transformed set of f n s. This provides additional flexible parameters to allow each CSMF to have its own mean and variance. 2. Embed InSilicoVA in a spatio-temporal model that allows F to vary smoothly through space and time. This would provide a parsimonious way of exploring spatio-temporal variation in the CSMFs while using the data as efficiently as possible. 3. Create the ability to add physician cause assignments to (22) and (26) so that information in that form can be utilized when available. The physician codes will require pre-processing to remove physician-specific bias in cause assignment, perhaps using a rater reliability method (for example: Salter-Townshend and Murphy, 2012). 4. Most importantly, address the obviously invalid assumption that the signs/symptoms are independent given a specific cause. This will require modeling of the signs/symptoms and the physician-provided conditional probabilities so that important dependencies can be accommodated. Further, this will require additional consultation with physicians and acquisition of new expert knowledge to characterize these dependencies. All of this will require a generous grant and the collaboration of a large number of experts. This will very likely greatly improve the performance and robustness of the method. 5. Critically, re-elicit the conditional probabilities P from physicians so that they are logically well-behaved, i.e. fully consistent with one another and their complements. 6. Focus and sharpen VA questionnaire. Quantify the influence of each sign/symptom to: (1) potentially eliminate low-value signs/symptoms and thereby make the VA interview more efficient, and/or (2) suggest sign/symptom types that appear particularly useful, and potentially suggest augmenting VA interviews based on that information. 7. Explore new possibilities for refining the conditional probabilities P and potentially for entirely new models. 14
18 Individual accuracy CSMF estimation Accuracy Mean Absolute Error InSilicoVA InterVA InSilicoVA InterVA A Figure 1: Simulation setup 1: Fair Comparison. A: InSilicoVA correctly assigns cause of death correctly effectively 100% of the time. InterVA is less accurate in assigning individual causes of death. B: InSilicoVA s errors in identifying CSMFs are consistently very small. InterVA s errors are also generally small, but the distribution has a long tail in the direction of large errors sometimes InterVA s errors are large. B 15
19 Individual accuracy CSMF estimation Accuracy Mean Absolute Error InSilicoVA InterVA InSilicoVA InterVA A Figure 2: Simulation setup 2: Conditional Probabilities in the range [ ]. A: InSilicoVA correctly assigns cause of death correctly effectively 100% of the time. InterVA correctly assigns cause of death correctly 80% of the time with wide variation all the way down to as low as 60% and never above about 90%. B: InSilicoVA s errors in identifying CSMFs are consistently very small. InterVA s errors in identifying the CSMFs are larger and more variable. B 16
20 Individual accuracy CSMF estimation Accuracy Mean Absolute Error InSilicoVA InterVA InSilicoVA InterVA A Figure 3: Simulation setup 3: Reporting Errors. Both methods suffer, but InterVA suffers a lot more. A: InSilicoVA correctly assigns cause of death correctly about 70% of the time. InterVA correctly assigns cause of death correctly about 40% of the time. B: InSilicoVA s errors in identifying CSMFs are still consistently small. InterVA s errors in identifying the CSMFs are larger. B 17
21 Most discrepency Within 4% Other and unspecified NCD Acute abdomen Sepsis (non obstetric) Other and unspecified external CoD Pulmonary tuberculosis Digestive neoplasms Chronic obstructive pulmonary dis Other and unspecified infect dis Stroke Diarrhoeal diseases Diabetes mellitus Other and unspecified neoplasms Reproductive neoplasms MF Other and unspecified cardiac dis Meningitis and encephalitis Assault Breast neoplasms Asthma Road traffic accident Intentional self harm 20% 15% 10% 5% 0 5% 10% 15% 20% InSilico InterVA A 4% 2% 0 2% 4% InSilico InterVA B Estimated cause specific mortality fractions Other and unspecified NCD Sepsis (non obstetric) Pulmonary tuberculosis Digestive neoplasms Chronic obstructive pulmonary dis Other and unspecified infect dis InterVA InSilicoVA Stroke Diarrhoeal diseases Diabetes mellitus Other and unspecified neoplasms Estimated CSMF C Figure 4: Agincourt Application. A: Differences in CSMFs (InSilicoVA InterVA) displaying the 10 specific causes that differ most. InSilicoVA is less willing to make highly specific classifications and thus produces larger CSMFs associated with less-specific causes and smaller CSMFs associated with more specific causes. B: Same as (A) but for the 10 largest differences in CSMFs that are still less than 4%, which as a group includes HIV even though it is not in the top 10 that are plotted here. C: The CSMFs produced by both models on their natural scale, the same 10 causes as in A for which the differences are greatest. 18
22 References Byass, P. (2013). Interva software. Byass, P., D. Chandramohan, S. J. Clark, L. D Ambruoso, E. Fottrell, W. J. Graham, A. J. Herbst, A. Hodgson, S. Hounton, K. Kahn, et al. (2012). Strengthening standardised interpretation of verbal autopsy data: the new interva-4 tool. Global health action 5. Byass, P., E. Fottrell, D. L. Huong, Y. Berhane, T. Corrah, K. Kahn, L. Muhe, et al. (2006). Refining a probabilistic model for interpreting verbal autopsy data. Scandinavian journal of public health 34 (1), Byass, P., D. L. Huong, and H. Van Minh (2003). A probabilistic approach to interpreting verbal autopsies: methodology and preliminary validation in vietnam. Scandinavian Journal of Public Health 31 (62 suppl), Flaxman, A. D., A. Vahdatpour, S. Green, S. L. James, C. J. Murray, and Consortium Population Health Metrics Research (2011). Random forests for verbal autopsy analysis: multisite validation study using clinical diagnostic gold standards. Popul Health Metr 9 (29). James, S. L., A. D. Flaxman, C. J. Murray, and Consortium Population Health Metrics Research (2011). Performance of the tariff method: validation of a simple additive algorithm for analysis of verbal autopsies. Popul Health Metr 9 (31). Kahn, K., M. A. Collinson, F. X. Gómez-Olivé, O. Mokoena, R. Twine, P. Mee, S. A. Afolabi, B. D. Clark, C. W. Kabudula, A. Khosa, et al. (2012). Profile: Agincourt health and socio-demographic surveillance system. International journal of epidemiology 41 (4), King, G. and Y. Lu (2008). Verbal autopsy methods with multiple causes of death. Statistical Science 100 (469). King, G., Y. Lu, and K. Shibuya (2010). Designing verbal autopsy studies. Popul Health Metr 8 (19). Leitao, J., D. Chandramohan, P. Byass, R. Jakob, K. Bundhamcharoen, and C. Choprapowan (2013). Revising the WHO verbal autopsy instrument to facilitate routine cause-of-death monitoring. Global Health Action 6 (21518). Maher, D., S. Biraro, V. Hosegood, R. Isingo, T. Lutalo, P. Mushati, B. Ngwira, M. Nyirenda, J. Todd, and B. Zaba (2010). Translating global health research aims into action: the example of the alpha network*. Tropical Medicine & International Health 15 (3), Murray, C. J., S. L. James, J. K. Birnbaum, M. K. Freeman, R. Lozano, A. D. Lopez, and Consortium Population Health Metrics Research (2011). Simplified symptom pattern method for verbal autopsy analysis: multisite validation study using clinical diagnostic gold standards. Popul Health Metr 9 (30). 19
23 Salter-Townshend, M. and T. B. Murphy (2012). Sentiment analysis of online media. Lausen, B., van del Poel, D. and Ultsch, A.(eds.). Algorithms from and for Nature and Life. Studies in Classification, Data Analysis, and Knowledge Organization. Sankoh, O. and P. Byass (2012). The indepth network: filling vital gaps in global epidemiology. International Journal of Epidemiology 41 (3),
How to Read 100 Million Blogs (& Classify Deaths Without Physicians)
 ary King Institute for Quantitative Social Science Harvard University () How to Read 100 Million Blogs (& Classify Deaths Without Physicians) (6/19/08 talk at Google) 1 / 30 How to Read 100 Million Blogs
ary King Institute for Quantitative Social Science Harvard University () How to Read 100 Million Blogs (& Classify Deaths Without Physicians) (6/19/08 talk at Google) 1 / 30 How to Read 100 Million Blogs
Local calibration of verbal autopsy algorithms
 Local calibration of verbal autopsy algorithms arxiv:1810.10572v1 [stat.me] 24 Oct 2018 Abhirup Datta Department of Biostatistics, Johns Hopkins University and Jacob Fiksel Department of Biostatistics,
Local calibration of verbal autopsy algorithms arxiv:1810.10572v1 [stat.me] 24 Oct 2018 Abhirup Datta Department of Biostatistics, Johns Hopkins University and Jacob Fiksel Department of Biostatistics,
How to Read 100 Million Blogs (& Classify Deaths Without Physicians)
 How to Read 100 Million Blogs (& Classify Deaths Without Physicians) Gary King Institute for Quantitative Social Science Harvard University (7/1/08 talk at IBM) () (7/1/08 talk at IBM) 1 / 30 References
How to Read 100 Million Blogs (& Classify Deaths Without Physicians) Gary King Institute for Quantitative Social Science Harvard University (7/1/08 talk at IBM) () (7/1/08 talk at IBM) 1 / 30 References
On the errors introduced by the naive Bayes independence assumption
 On the errors introduced by the naive Bayes independence assumption Author Matthijs de Wachter 3671100 Utrecht University Master Thesis Artificial Intelligence Supervisor Dr. Silja Renooij Department of
On the errors introduced by the naive Bayes independence assumption Author Matthijs de Wachter 3671100 Utrecht University Master Thesis Artificial Intelligence Supervisor Dr. Silja Renooij Department of
Machine Learning
 Machine Learning 10-601 Tom M. Mitchell Machine Learning Department Carnegie Mellon University August 30, 2017 Today: Decision trees Overfitting The Big Picture Coming soon Probabilistic learning MLE,
Machine Learning 10-601 Tom M. Mitchell Machine Learning Department Carnegie Mellon University August 30, 2017 Today: Decision trees Overfitting The Big Picture Coming soon Probabilistic learning MLE,
CS 188: Artificial Intelligence Spring Today
 CS 188: Artificial Intelligence Spring 2006 Lecture 9: Naïve Bayes 2/14/2006 Dan Klein UC Berkeley Many slides from either Stuart Russell or Andrew Moore Bayes rule Today Expectations and utilities Naïve
CS 188: Artificial Intelligence Spring 2006 Lecture 9: Naïve Bayes 2/14/2006 Dan Klein UC Berkeley Many slides from either Stuart Russell or Andrew Moore Bayes rule Today Expectations and utilities Naïve
Approach to identifying hot spots for NCDs in South Africa
 Approach to identifying hot spots for NCDs in South Africa HST Conference 6 May 2016 Noluthando Ndlovu, 1 Candy Day, 1 Benn Sartorius, 2 Karen Hofman, 3 Jens Aagaard-Hansen 3,4 1 Health Systems Trust,
Approach to identifying hot spots for NCDs in South Africa HST Conference 6 May 2016 Noluthando Ndlovu, 1 Candy Day, 1 Benn Sartorius, 2 Karen Hofman, 3 Jens Aagaard-Hansen 3,4 1 Health Systems Trust,
Two-sample Categorical data: Testing
 Two-sample Categorical data: Testing Patrick Breheny April 1 Patrick Breheny Introduction to Biostatistics (171:161) 1/28 Separate vs. paired samples Despite the fact that paired samples usually offer
Two-sample Categorical data: Testing Patrick Breheny April 1 Patrick Breheny Introduction to Biostatistics (171:161) 1/28 Separate vs. paired samples Despite the fact that paired samples usually offer
Introduction to Machine Learning
 Introduction to Machine Learning CS4375 --- Fall 2018 Bayesian a Learning Reading: Sections 13.1-13.6, 20.1-20.2, R&N Sections 6.1-6.3, 6.7, 6.9, Mitchell 1 Uncertainty Most real-world problems deal with
Introduction to Machine Learning CS4375 --- Fall 2018 Bayesian a Learning Reading: Sections 13.1-13.6, 20.1-20.2, R&N Sections 6.1-6.3, 6.7, 6.9, Mitchell 1 Uncertainty Most real-world problems deal with
Discrete Mathematics and Probability Theory Spring 2016 Rao and Walrand Note 14
 CS 70 Discrete Mathematics and Probability Theory Spring 2016 Rao and Walrand Note 14 Introduction One of the key properties of coin flips is independence: if you flip a fair coin ten times and get ten
CS 70 Discrete Mathematics and Probability Theory Spring 2016 Rao and Walrand Note 14 Introduction One of the key properties of coin flips is independence: if you flip a fair coin ten times and get ten
CS 188: Artificial Intelligence Spring Announcements
 CS 188: Artificial Intelligence Spring 2011 Lecture 12: Probability 3/2/2011 Pieter Abbeel UC Berkeley Many slides adapted from Dan Klein. 1 Announcements P3 due on Monday (3/7) at 4:59pm W3 going out
CS 188: Artificial Intelligence Spring 2011 Lecture 12: Probability 3/2/2011 Pieter Abbeel UC Berkeley Many slides adapted from Dan Klein. 1 Announcements P3 due on Monday (3/7) at 4:59pm W3 going out
Introduction to Machine Learning
 Uncertainty Introduction to Machine Learning CS4375 --- Fall 2018 a Bayesian Learning Reading: Sections 13.1-13.6, 20.1-20.2, R&N Sections 6.1-6.3, 6.7, 6.9, Mitchell Most real-world problems deal with
Uncertainty Introduction to Machine Learning CS4375 --- Fall 2018 a Bayesian Learning Reading: Sections 13.1-13.6, 20.1-20.2, R&N Sections 6.1-6.3, 6.7, 6.9, Mitchell Most real-world problems deal with
Discrete Mathematics and Probability Theory Fall 2015 Lecture 21
 CS 70 Discrete Mathematics and Probability Theory Fall 205 Lecture 2 Inference In this note we revisit the problem of inference: Given some data or observations from the world, what can we infer about
CS 70 Discrete Mathematics and Probability Theory Fall 205 Lecture 2 Inference In this note we revisit the problem of inference: Given some data or observations from the world, what can we infer about
Part 7: Glossary Overview
 Part 7: Glossary Overview In this Part This Part covers the following topic Topic See Page 7-1-1 Introduction This section provides an alphabetical list of all the terms used in a STEPS surveillance with
Part 7: Glossary Overview In this Part This Part covers the following topic Topic See Page 7-1-1 Introduction This section provides an alphabetical list of all the terms used in a STEPS surveillance with
Bayesian Inference. Introduction
 Bayesian Inference Introduction The frequentist approach to inference holds that probabilities are intrinsicially tied (unsurprisingly) to frequencies. This interpretation is actually quite natural. What,
Bayesian Inference Introduction The frequentist approach to inference holds that probabilities are intrinsicially tied (unsurprisingly) to frequencies. This interpretation is actually quite natural. What,
Probability and Statistics
 Probability and Statistics Kristel Van Steen, PhD 2 Montefiore Institute - Systems and Modeling GIGA - Bioinformatics ULg kristel.vansteen@ulg.ac.be CHAPTER 4: IT IS ALL ABOUT DATA 4a - 1 CHAPTER 4: IT
Probability and Statistics Kristel Van Steen, PhD 2 Montefiore Institute - Systems and Modeling GIGA - Bioinformatics ULg kristel.vansteen@ulg.ac.be CHAPTER 4: IT IS ALL ABOUT DATA 4a - 1 CHAPTER 4: IT
Equivalence of random-effects and conditional likelihoods for matched case-control studies
 Equivalence of random-effects and conditional likelihoods for matched case-control studies Ken Rice MRC Biostatistics Unit, Cambridge, UK January 8 th 4 Motivation Study of genetic c-erbb- exposure and
Equivalence of random-effects and conditional likelihoods for matched case-control studies Ken Rice MRC Biostatistics Unit, Cambridge, UK January 8 th 4 Motivation Study of genetic c-erbb- exposure and
Discrete Mathematics and Probability Theory Spring 2014 Anant Sahai Note 10
 EECS 70 Discrete Mathematics and Probability Theory Spring 2014 Anant Sahai Note 10 Introduction to Basic Discrete Probability In the last note we considered the probabilistic experiment where we flipped
EECS 70 Discrete Mathematics and Probability Theory Spring 2014 Anant Sahai Note 10 Introduction to Basic Discrete Probability In the last note we considered the probabilistic experiment where we flipped
Quantitative Understanding in Biology 1.7 Bayesian Methods
 Quantitative Understanding in Biology 1.7 Bayesian Methods Jason Banfelder October 25th, 2018 1 Introduction So far, most of the methods we ve looked at fall under the heading of classical, or frequentist
Quantitative Understanding in Biology 1.7 Bayesian Methods Jason Banfelder October 25th, 2018 1 Introduction So far, most of the methods we ve looked at fall under the heading of classical, or frequentist
INTRODUCTION. In March 1998, the tender for project CT.98.EP.04 was awarded to the Department of Medicines Management, Keele University, UK.
 INTRODUCTION In many areas of Europe patterns of drug use are changing. The mechanisms of diffusion are diverse: introduction of new practices by new users, tourism and migration, cross-border contact,
INTRODUCTION In many areas of Europe patterns of drug use are changing. The mechanisms of diffusion are diverse: introduction of new practices by new users, tourism and migration, cross-border contact,
Designing Information Devices and Systems I Spring 2018 Lecture Notes Note Introduction to Linear Algebra the EECS Way
 EECS 16A Designing Information Devices and Systems I Spring 018 Lecture Notes Note 1 1.1 Introduction to Linear Algebra the EECS Way In this note, we will teach the basics of linear algebra and relate
EECS 16A Designing Information Devices and Systems I Spring 018 Lecture Notes Note 1 1.1 Introduction to Linear Algebra the EECS Way In this note, we will teach the basics of linear algebra and relate
A.I. in health informatics lecture 2 clinical reasoning & probabilistic inference, I. kevin small & byron wallace
 A.I. in health informatics lecture 2 clinical reasoning & probabilistic inference, I kevin small & byron wallace today a review of probability random variables, maximum likelihood, etc. crucial for clinical
A.I. in health informatics lecture 2 clinical reasoning & probabilistic inference, I kevin small & byron wallace today a review of probability random variables, maximum likelihood, etc. crucial for clinical
Table 2.14 : Distribution of 125 subjects by laboratory and +/ Category. Test Reference Laboratory Laboratory Total
 2.5. Kappa Coefficient and the Paradoxes. - 31-2.5.1 Kappa s Dependency on Trait Prevalence On February 9, 2003 we received an e-mail from a researcher asking whether it would be possible to apply the
2.5. Kappa Coefficient and the Paradoxes. - 31-2.5.1 Kappa s Dependency on Trait Prevalence On February 9, 2003 we received an e-mail from a researcher asking whether it would be possible to apply the
The Problem. Sustainability is an abstract concept that cannot be directly measured.
 Measurement, Interpretation, and Assessment Applied Ecosystem Services, Inc. (Copyright c 2005 Applied Ecosystem Services, Inc.) The Problem is an abstract concept that cannot be directly measured. There
Measurement, Interpretation, and Assessment Applied Ecosystem Services, Inc. (Copyright c 2005 Applied Ecosystem Services, Inc.) The Problem is an abstract concept that cannot be directly measured. There
Designing Information Devices and Systems I Fall 2018 Lecture Notes Note Introduction to Linear Algebra the EECS Way
 EECS 16A Designing Information Devices and Systems I Fall 018 Lecture Notes Note 1 1.1 Introduction to Linear Algebra the EECS Way In this note, we will teach the basics of linear algebra and relate it
EECS 16A Designing Information Devices and Systems I Fall 018 Lecture Notes Note 1 1.1 Introduction to Linear Algebra the EECS Way In this note, we will teach the basics of linear algebra and relate it
Probabilistic Reasoning
 Course 16 :198 :520 : Introduction To Artificial Intelligence Lecture 7 Probabilistic Reasoning Abdeslam Boularias Monday, September 28, 2015 1 / 17 Outline We show how to reason and act under uncertainty.
Course 16 :198 :520 : Introduction To Artificial Intelligence Lecture 7 Probabilistic Reasoning Abdeslam Boularias Monday, September 28, 2015 1 / 17 Outline We show how to reason and act under uncertainty.
13.4 INDEPENDENCE. 494 Chapter 13. Quantifying Uncertainty
 494 Chapter 13. Quantifying Uncertainty table. In a realistic problem we could easily have n>100, makingo(2 n ) impractical. The full joint distribution in tabular form is just not a practical tool for
494 Chapter 13. Quantifying Uncertainty table. In a realistic problem we could easily have n>100, makingo(2 n ) impractical. The full joint distribution in tabular form is just not a practical tool for
Bayesian Methods for Machine Learning
 Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Medical Question Answering for Clinical Decision Support
 Medical Question Answering for Clinical Decision Support Travis R. Goodwin and Sanda M. Harabagiu The University of Texas at Dallas Human Language Technology Research Institute http://www.hlt.utdallas.edu
Medical Question Answering for Clinical Decision Support Travis R. Goodwin and Sanda M. Harabagiu The University of Texas at Dallas Human Language Technology Research Institute http://www.hlt.utdallas.edu
CHAPTER 2 Estimating Probabilities
 CHAPTER 2 Estimating Probabilities Machine Learning Copyright c 2017. Tom M. Mitchell. All rights reserved. *DRAFT OF September 16, 2017* *PLEASE DO NOT DISTRIBUTE WITHOUT AUTHOR S PERMISSION* This is
CHAPTER 2 Estimating Probabilities Machine Learning Copyright c 2017. Tom M. Mitchell. All rights reserved. *DRAFT OF September 16, 2017* *PLEASE DO NOT DISTRIBUTE WITHOUT AUTHOR S PERMISSION* This is
MULTIPLE REGRESSION AND ISSUES IN REGRESSION ANALYSIS
 MULTIPLE REGRESSION AND ISSUES IN REGRESSION ANALYSIS Page 1 MSR = Mean Regression Sum of Squares MSE = Mean Squared Error RSS = Regression Sum of Squares SSE = Sum of Squared Errors/Residuals α = Level
MULTIPLE REGRESSION AND ISSUES IN REGRESSION ANALYSIS Page 1 MSR = Mean Regression Sum of Squares MSE = Mean Squared Error RSS = Regression Sum of Squares SSE = Sum of Squared Errors/Residuals α = Level
The subjective worlds of Frequentist and Bayesian statistics
 Chapter 2 The subjective worlds of Frequentist and Bayesian statistics 2.1 The deterministic nature of random coin throwing Suppose that, in an idealised world, the ultimate fate of a thrown coin heads
Chapter 2 The subjective worlds of Frequentist and Bayesian statistics 2.1 The deterministic nature of random coin throwing Suppose that, in an idealised world, the ultimate fate of a thrown coin heads
A4. Methodology Annex: Sampling Design (2008) Methodology Annex: Sampling design 1
 A4. Methodology Annex: Sampling Design (2008) Methodology Annex: Sampling design 1 Introduction The evaluation strategy for the One Million Initiative is based on a panel survey. In a programme such as
A4. Methodology Annex: Sampling Design (2008) Methodology Annex: Sampling design 1 Introduction The evaluation strategy for the One Million Initiative is based on a panel survey. In a programme such as
Lecture 2. Judging the Performance of Classifiers. Nitin R. Patel
 Lecture 2 Judging the Performance of Classifiers Nitin R. Patel 1 In this note we will examine the question of how to udge the usefulness of a classifier and how to compare different classifiers. Not only
Lecture 2 Judging the Performance of Classifiers Nitin R. Patel 1 In this note we will examine the question of how to udge the usefulness of a classifier and how to compare different classifiers. Not only
2018 PAA Short Course on Bayesian Small Area Estimation using Complex Data Introduction and Overview
 2018 PAA Short Course on Bayesian Small Area Estimation using Complex Data Introduction and Overview Sam Clark 1, Zehang Richard Li 2, Jon Wakefield 2,3 1 Department of Sociology, Ohio State University,
2018 PAA Short Course on Bayesian Small Area Estimation using Complex Data Introduction and Overview Sam Clark 1, Zehang Richard Li 2, Jon Wakefield 2,3 1 Department of Sociology, Ohio State University,
Reinforcement Learning Wrap-up
 Reinforcement Learning Wrap-up Slides courtesy of Dan Klein and Pieter Abbeel University of California, Berkeley [These slides were created by Dan Klein and Pieter Abbeel for CS188 Intro to AI at UC Berkeley.
Reinforcement Learning Wrap-up Slides courtesy of Dan Klein and Pieter Abbeel University of California, Berkeley [These slides were created by Dan Klein and Pieter Abbeel for CS188 Intro to AI at UC Berkeley.
Chapter 1. Scientific Process and Themes of Biology
 Chapter 1 Scientific Process and Themes of Biology What is Science? u Scientific knowledge is acquired using a rigorous process u Science is an organized way of gathering and analyzing evidence about the
Chapter 1 Scientific Process and Themes of Biology What is Science? u Scientific knowledge is acquired using a rigorous process u Science is an organized way of gathering and analyzing evidence about the
CROSS SECTIONAL STUDY & SAMPLING METHOD
 CROSS SECTIONAL STUDY & SAMPLING METHOD Prof. Dr. Zaleha Md. Isa, BSc(Hons) Clin. Biochemistry; PhD (Public Health), Department of Community Health, Faculty of Medicine, Universiti Kebangsaan Malaysia
CROSS SECTIONAL STUDY & SAMPLING METHOD Prof. Dr. Zaleha Md. Isa, BSc(Hons) Clin. Biochemistry; PhD (Public Health), Department of Community Health, Faculty of Medicine, Universiti Kebangsaan Malaysia
Hypothesis testing. Chapter Formulating a hypothesis. 7.2 Testing if the hypothesis agrees with data
 Chapter 7 Hypothesis testing 7.1 Formulating a hypothesis Up until now we have discussed how to define a measurement in terms of a central value, uncertainties, and units, as well as how to extend these
Chapter 7 Hypothesis testing 7.1 Formulating a hypothesis Up until now we have discussed how to define a measurement in terms of a central value, uncertainties, and units, as well as how to extend these
ECE521 week 3: 23/26 January 2017
 ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
Introduction to AI Learning Bayesian networks. Vibhav Gogate
 Introduction to AI Learning Bayesian networks Vibhav Gogate Inductive Learning in a nutshell Given: Data Examples of a function (X, F(X)) Predict function F(X) for new examples X Discrete F(X): Classification
Introduction to AI Learning Bayesian networks Vibhav Gogate Inductive Learning in a nutshell Given: Data Examples of a function (X, F(X)) Predict function F(X) for new examples X Discrete F(X): Classification
Bayesian Estimation of Prediction Error and Variable Selection in Linear Regression
 Bayesian Estimation of Prediction Error and Variable Selection in Linear Regression Andrew A. Neath Department of Mathematics and Statistics; Southern Illinois University Edwardsville; Edwardsville, IL,
Bayesian Estimation of Prediction Error and Variable Selection in Linear Regression Andrew A. Neath Department of Mathematics and Statistics; Southern Illinois University Edwardsville; Edwardsville, IL,
Discrete Probability and State Estimation
 6.01, Fall Semester, 2007 Lecture 12 Notes 1 MASSACHVSETTS INSTITVTE OF TECHNOLOGY Department of Electrical Engineering and Computer Science 6.01 Introduction to EECS I Fall Semester, 2007 Lecture 12 Notes
6.01, Fall Semester, 2007 Lecture 12 Notes 1 MASSACHVSETTS INSTITVTE OF TECHNOLOGY Department of Electrical Engineering and Computer Science 6.01 Introduction to EECS I Fall Semester, 2007 Lecture 12 Notes
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2016
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2016 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2016 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Two-sample Categorical data: Testing
 Two-sample Categorical data: Testing Patrick Breheny October 29 Patrick Breheny Biostatistical Methods I (BIOS 5710) 1/22 Lister s experiment Introduction In the 1860s, Joseph Lister conducted a landmark
Two-sample Categorical data: Testing Patrick Breheny October 29 Patrick Breheny Biostatistical Methods I (BIOS 5710) 1/22 Lister s experiment Introduction In the 1860s, Joseph Lister conducted a landmark
Agreement Coefficients and Statistical Inference
 CHAPTER Agreement Coefficients and Statistical Inference OBJECTIVE This chapter describes several approaches for evaluating the precision associated with the inter-rater reliability coefficients of the
CHAPTER Agreement Coefficients and Statistical Inference OBJECTIVE This chapter describes several approaches for evaluating the precision associated with the inter-rater reliability coefficients of the
1 Using standard errors when comparing estimated values
 MLPR Assignment Part : General comments Below are comments on some recurring issues I came across when marking the second part of the assignment, which I thought it would help to explain in more detail
MLPR Assignment Part : General comments Below are comments on some recurring issues I came across when marking the second part of the assignment, which I thought it would help to explain in more detail
Food delivered. Food obtained S 3
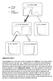 Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Weighted Majority and the Online Learning Approach
 Statistical Techniques in Robotics (16-81, F12) Lecture#9 (Wednesday September 26) Weighted Majority and the Online Learning Approach Lecturer: Drew Bagnell Scribe:Narek Melik-Barkhudarov 1 Figure 1: Drew
Statistical Techniques in Robotics (16-81, F12) Lecture#9 (Wednesday September 26) Weighted Majority and the Online Learning Approach Lecturer: Drew Bagnell Scribe:Narek Melik-Barkhudarov 1 Figure 1: Drew
FCE 3900 EDUCATIONAL RESEARCH LECTURE 8 P O P U L A T I O N A N D S A M P L I N G T E C H N I Q U E
 FCE 3900 EDUCATIONAL RESEARCH LECTURE 8 P O P U L A T I O N A N D S A M P L I N G T E C H N I Q U E OBJECTIVE COURSE Understand the concept of population and sampling in the research. Identify the type
FCE 3900 EDUCATIONAL RESEARCH LECTURE 8 P O P U L A T I O N A N D S A M P L I N G T E C H N I Q U E OBJECTIVE COURSE Understand the concept of population and sampling in the research. Identify the type
Application of Indirect Race/ Ethnicity Data in Quality Metric Analyses
 Background The fifteen wholly-owned health plans under WellPoint, Inc. (WellPoint) historically did not collect data in regard to the race/ethnicity of it members. In order to overcome this lack of data
Background The fifteen wholly-owned health plans under WellPoint, Inc. (WellPoint) historically did not collect data in regard to the race/ethnicity of it members. In order to overcome this lack of data
FORMULATION OF THE LEARNING PROBLEM
 FORMULTION OF THE LERNING PROBLEM MIM RGINSKY Now that we have seen an informal statement of the learning problem, as well as acquired some technical tools in the form of concentration inequalities, we
FORMULTION OF THE LERNING PROBLEM MIM RGINSKY Now that we have seen an informal statement of the learning problem, as well as acquired some technical tools in the form of concentration inequalities, we
Measurements and Data Analysis
 Measurements and Data Analysis 1 Introduction The central point in experimental physical science is the measurement of physical quantities. Experience has shown that all measurements, no matter how carefully
Measurements and Data Analysis 1 Introduction The central point in experimental physical science is the measurement of physical quantities. Experience has shown that all measurements, no matter how carefully
Bayesian analysis in nuclear physics
 Bayesian analysis in nuclear physics Ken Hanson T-16, Nuclear Physics; Theoretical Division Los Alamos National Laboratory Tutorials presented at LANSCE Los Alamos Neutron Scattering Center July 25 August
Bayesian analysis in nuclear physics Ken Hanson T-16, Nuclear Physics; Theoretical Division Los Alamos National Laboratory Tutorials presented at LANSCE Los Alamos Neutron Scattering Center July 25 August
Modeling conditional dependence among multiple diagnostic tests
 Received: 11 June 2017 Revised: 1 August 2017 Accepted: 6 August 2017 DOI: 10.1002/sim.7449 RESEARCH ARTICLE Modeling conditional dependence among multiple diagnostic tests Zhuoyu Wang 1 Nandini Dendukuri
Received: 11 June 2017 Revised: 1 August 2017 Accepted: 6 August 2017 DOI: 10.1002/sim.7449 RESEARCH ARTICLE Modeling conditional dependence among multiple diagnostic tests Zhuoyu Wang 1 Nandini Dendukuri
Probability Theory. Introduction to Probability Theory. Principles of Counting Examples. Principles of Counting. Probability spaces.
 Probability Theory To start out the course, we need to know something about statistics and probability Introduction to Probability Theory L645 Advanced NLP Autumn 2009 This is only an introduction; for
Probability Theory To start out the course, we need to know something about statistics and probability Introduction to Probability Theory L645 Advanced NLP Autumn 2009 This is only an introduction; for
Introduction. Chapter 1
 Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
, (1) e i = ˆσ 1 h ii. c 2016, Jeffrey S. Simonoff 1
 Regression diagnostics As is true of all statistical methodologies, linear regression analysis can be a very effective way to model data, as along as the assumptions being made are true. For the regression
Regression diagnostics As is true of all statistical methodologies, linear regression analysis can be a very effective way to model data, as along as the assumptions being made are true. For the regression
Fundamentals of Applied Sampling
 1 Chapter 5 Fundamentals of Applied Sampling Thomas Piazza 5.1 The Basic Idea of Sampling Survey sampling is really quite remarkable. In research we often want to know certain characteristics of a large
1 Chapter 5 Fundamentals of Applied Sampling Thomas Piazza 5.1 The Basic Idea of Sampling Survey sampling is really quite remarkable. In research we often want to know certain characteristics of a large
Elements of probability theory
 The role of probability theory in statistics We collect data so as to provide evidentiary support for answers we give to our many questions about the world (and in our particular case, about the business
The role of probability theory in statistics We collect data so as to provide evidentiary support for answers we give to our many questions about the world (and in our particular case, about the business
Inferring the Causal Decomposition under the Presence of Deterministic Relations.
 Inferring the Causal Decomposition under the Presence of Deterministic Relations. Jan Lemeire 1,2, Stijn Meganck 1,2, Francesco Cartella 1, Tingting Liu 1 and Alexander Statnikov 3 1-ETRO Department, Vrije
Inferring the Causal Decomposition under the Presence of Deterministic Relations. Jan Lemeire 1,2, Stijn Meganck 1,2, Francesco Cartella 1, Tingting Liu 1 and Alexander Statnikov 3 1-ETRO Department, Vrije
CS 188: Artificial Intelligence. Our Status in CS188
 CS 188: Artificial Intelligence Probability Pieter Abbeel UC Berkeley Many slides adapted from Dan Klein. 1 Our Status in CS188 We re done with Part I Search and Planning! Part II: Probabilistic Reasoning
CS 188: Artificial Intelligence Probability Pieter Abbeel UC Berkeley Many slides adapted from Dan Klein. 1 Our Status in CS188 We re done with Part I Search and Planning! Part II: Probabilistic Reasoning
Natural Language Processing. Classification. Features. Some Definitions. Classification. Feature Vectors. Classification I. Dan Klein UC Berkeley
 Natural Language Processing Classification Classification I Dan Klein UC Berkeley Classification Automatically make a decision about inputs Example: document category Example: image of digit digit Example:
Natural Language Processing Classification Classification I Dan Klein UC Berkeley Classification Automatically make a decision about inputs Example: document category Example: image of digit digit Example:
Q-Matrix Development. NCME 2009 Workshop
 Q-Matrix Development NCME 2009 Workshop Introduction We will define the Q-matrix Then we will discuss method of developing your own Q-matrix Talk about possible problems of the Q-matrix to avoid The Q-matrix
Q-Matrix Development NCME 2009 Workshop Introduction We will define the Q-matrix Then we will discuss method of developing your own Q-matrix Talk about possible problems of the Q-matrix to avoid The Q-matrix
Introduction: MLE, MAP, Bayesian reasoning (28/8/13)
 STA561: Probabilistic machine learning Introduction: MLE, MAP, Bayesian reasoning (28/8/13) Lecturer: Barbara Engelhardt Scribes: K. Ulrich, J. Subramanian, N. Raval, J. O Hollaren 1 Classifiers In this
STA561: Probabilistic machine learning Introduction: MLE, MAP, Bayesian reasoning (28/8/13) Lecturer: Barbara Engelhardt Scribes: K. Ulrich, J. Subramanian, N. Raval, J. O Hollaren 1 Classifiers In this
ECLT 5810 Linear Regression and Logistic Regression for Classification. Prof. Wai Lam
 ECLT 5810 Linear Regression and Logistic Regression for Classification Prof. Wai Lam Linear Regression Models Least Squares Input vectors is an attribute / feature / predictor (independent variable) The
ECLT 5810 Linear Regression and Logistic Regression for Classification Prof. Wai Lam Linear Regression Models Least Squares Input vectors is an attribute / feature / predictor (independent variable) The
Lecture 10: Introduction to reasoning under uncertainty. Uncertainty
 Lecture 10: Introduction to reasoning under uncertainty Introduction to reasoning under uncertainty Review of probability Axioms and inference Conditional probability Probability distributions COMP-424,
Lecture 10: Introduction to reasoning under uncertainty Introduction to reasoning under uncertainty Review of probability Axioms and inference Conditional probability Probability distributions COMP-424,
Topic Models. Brandon Malone. February 20, Latent Dirichlet Allocation Success Stories Wrap-up
 Much of this material is adapted from Blei 2003. Many of the images were taken from the Internet February 20, 2014 Suppose we have a large number of books. Each is about several unknown topics. How can
Much of this material is adapted from Blei 2003. Many of the images were taken from the Internet February 20, 2014 Suppose we have a large number of books. Each is about several unknown topics. How can
Base Rates and Bayes Theorem
 Slides to accompany Grove s handout March 8, 2016 Table of contents 1 Diagnostic and Prognostic Inference 2 Example inferences Does this patient have Alzheimer s disease, schizophrenia, depression, etc.?
Slides to accompany Grove s handout March 8, 2016 Table of contents 1 Diagnostic and Prognostic Inference 2 Example inferences Does this patient have Alzheimer s disease, schizophrenia, depression, etc.?
1 Probabilities. 1.1 Basics 1 PROBABILITIES
 1 PROBABILITIES 1 Probabilities Probability is a tricky word usually meaning the likelyhood of something occuring or how frequent something is. Obviously, if something happens frequently, then its probability
1 PROBABILITIES 1 Probabilities Probability is a tricky word usually meaning the likelyhood of something occuring or how frequent something is. Obviously, if something happens frequently, then its probability
Basic Probabilistic Reasoning SEG
 Basic Probabilistic Reasoning SEG 7450 1 Introduction Reasoning under uncertainty using probability theory Dealing with uncertainty is one of the main advantages of an expert system over a simple decision
Basic Probabilistic Reasoning SEG 7450 1 Introduction Reasoning under uncertainty using probability theory Dealing with uncertainty is one of the main advantages of an expert system over a simple decision
Probability. Lecture Notes. Adolfo J. Rumbos
 Probability Lecture Notes Adolfo J. Rumbos October 20, 204 2 Contents Introduction 5. An example from statistical inference................ 5 2 Probability Spaces 9 2. Sample Spaces and σ fields.....................
Probability Lecture Notes Adolfo J. Rumbos October 20, 204 2 Contents Introduction 5. An example from statistical inference................ 5 2 Probability Spaces 9 2. Sample Spaces and σ fields.....................
Machine Learning Linear Classification. Prof. Matteo Matteucci
 Machine Learning Linear Classification Prof. Matteo Matteucci Recall from the first lecture 2 X R p Regression Y R Continuous Output X R p Y {Ω 0, Ω 1,, Ω K } Classification Discrete Output X R p Y (X)
Machine Learning Linear Classification Prof. Matteo Matteucci Recall from the first lecture 2 X R p Regression Y R Continuous Output X R p Y {Ω 0, Ω 1,, Ω K } Classification Discrete Output X R p Y (X)
Uncertainty and Rules
 Uncertainty and Rules We have already seen that expert systems can operate within the realm of uncertainty. There are several sources of uncertainty in rules: Uncertainty related to individual rules Uncertainty
Uncertainty and Rules We have already seen that expert systems can operate within the realm of uncertainty. There are several sources of uncertainty in rules: Uncertainty related to individual rules Uncertainty
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation. Volker Tresp Summer 2014
 Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
Bayesian Networks: Construction, Inference, Learning and Causal Interpretation Volker Tresp Summer 2014 1 Introduction So far we were mostly concerned with supervised learning: we predicted one or several
ZecoByte. Panelgranskning. SQCT - Survey Quality Control Tool. technical presentation. 12 april 2012
 SQCT - Survey Quality Control Tool Panelgranskning SQCT - Survey Quality Control Tool 12 april 2012 technical presentation Innehåll Bakgrund sid. 1 Uppdrag sid. 1 SQCT sid. 1 Resultat sid. 1 Om sid. 2
SQCT - Survey Quality Control Tool Panelgranskning SQCT - Survey Quality Control Tool 12 april 2012 technical presentation Innehåll Bakgrund sid. 1 Uppdrag sid. 1 SQCT sid. 1 Resultat sid. 1 Om sid. 2
Lecture 1: Probability Fundamentals
 Lecture 1: Probability Fundamentals IB Paper 7: Probability and Statistics Carl Edward Rasmussen Department of Engineering, University of Cambridge January 22nd, 2008 Rasmussen (CUED) Lecture 1: Probability
Lecture 1: Probability Fundamentals IB Paper 7: Probability and Statistics Carl Edward Rasmussen Department of Engineering, University of Cambridge January 22nd, 2008 Rasmussen (CUED) Lecture 1: Probability
Dengue Forecasting Project
 Dengue Forecasting Project In areas where dengue is endemic, incidence follows seasonal transmission patterns punctuated every few years by much larger epidemics. Because these epidemics are currently
Dengue Forecasting Project In areas where dengue is endemic, incidence follows seasonal transmission patterns punctuated every few years by much larger epidemics. Because these epidemics are currently
9/2/2010. Wildlife Management is a very quantitative field of study. throughout this course and throughout your career.
 Introduction to Data and Analysis Wildlife Management is a very quantitative field of study Results from studies will be used throughout this course and throughout your career. Sampling design influences
Introduction to Data and Analysis Wildlife Management is a very quantitative field of study Results from studies will be used throughout this course and throughout your career. Sampling design influences
Tutorial on Approximate Bayesian Computation
 Tutorial on Approximate Bayesian Computation Michael Gutmann https://sites.google.com/site/michaelgutmann University of Helsinki Aalto University Helsinki Institute for Information Technology 16 May 2016
Tutorial on Approximate Bayesian Computation Michael Gutmann https://sites.google.com/site/michaelgutmann University of Helsinki Aalto University Helsinki Institute for Information Technology 16 May 2016
Accelerated warning of a bioterrorist event or endemic disease outbreak
 Accelerated warning of a bioterrorist event or endemic disease outbreak Luke Marsh Dstl, Porton Down, Salisbury, Wiltshire, SP4 0JQ Email: lmarsh@dstl.gov.uk Abstract - This paper discusses an alarm algorithm
Accelerated warning of a bioterrorist event or endemic disease outbreak Luke Marsh Dstl, Porton Down, Salisbury, Wiltshire, SP4 0JQ Email: lmarsh@dstl.gov.uk Abstract - This paper discusses an alarm algorithm
Computational methods are invaluable for typology, but the models must match the questions: Commentary on Dunn et al. (2011)
 Computational methods are invaluable for typology, but the models must match the questions: Commentary on Dunn et al. (2011) Roger Levy and Hal Daumé III August 1, 2011 The primary goal of Dunn et al.
Computational methods are invaluable for typology, but the models must match the questions: Commentary on Dunn et al. (2011) Roger Levy and Hal Daumé III August 1, 2011 The primary goal of Dunn et al.
MACHINE LEARNING INTRODUCTION: STRING CLASSIFICATION
 MACHINE LEARNING INTRODUCTION: STRING CLASSIFICATION THOMAS MAILUND Machine learning means different things to different people, and there is no general agreed upon core set of algorithms that must be
MACHINE LEARNING INTRODUCTION: STRING CLASSIFICATION THOMAS MAILUND Machine learning means different things to different people, and there is no general agreed upon core set of algorithms that must be
An English translation of Laplace s Théorie Analytique des Probabilités is available online at the departmental website of Richard Pulskamp (Math
 An English translation of Laplace s Théorie Analytique des Probabilités is available online at the departmental website of Richard Pulskamp (Math Dept, Xavier U, Cincinnati). Ed Jaynes would have rejoiced
An English translation of Laplace s Théorie Analytique des Probabilités is available online at the departmental website of Richard Pulskamp (Math Dept, Xavier U, Cincinnati). Ed Jaynes would have rejoiced
Empirical Bayes Moderation of Asymptotically Linear Parameters
 Empirical Bayes Moderation of Asymptotically Linear Parameters Nima Hejazi Division of Biostatistics University of California, Berkeley stat.berkeley.edu/~nhejazi nimahejazi.org twitter/@nshejazi github/nhejazi
Empirical Bayes Moderation of Asymptotically Linear Parameters Nima Hejazi Division of Biostatistics University of California, Berkeley stat.berkeley.edu/~nhejazi nimahejazi.org twitter/@nshejazi github/nhejazi
Item Response Theory and Computerized Adaptive Testing
 Item Response Theory and Computerized Adaptive Testing Richard C. Gershon, PhD Department of Medical Social Sciences Feinberg School of Medicine Northwestern University gershon@northwestern.edu May 20,
Item Response Theory and Computerized Adaptive Testing Richard C. Gershon, PhD Department of Medical Social Sciences Feinberg School of Medicine Northwestern University gershon@northwestern.edu May 20,
Introduction to Design of Experiments
 Introduction to Design of Experiments Jean-Marc Vincent and Arnaud Legrand Laboratory ID-IMAG MESCAL Project Universities of Grenoble {Jean-Marc.Vincent,Arnaud.Legrand}@imag.fr November 20, 2011 J.-M.
Introduction to Design of Experiments Jean-Marc Vincent and Arnaud Legrand Laboratory ID-IMAG MESCAL Project Universities of Grenoble {Jean-Marc.Vincent,Arnaud.Legrand}@imag.fr November 20, 2011 J.-M.
Discrete Probability and State Estimation
 6.01, Spring Semester, 2008 Week 12 Course Notes 1 MASSACHVSETTS INSTITVTE OF TECHNOLOGY Department of Electrical Engineering and Computer Science 6.01 Introduction to EECS I Spring Semester, 2008 Week
6.01, Spring Semester, 2008 Week 12 Course Notes 1 MASSACHVSETTS INSTITVTE OF TECHNOLOGY Department of Electrical Engineering and Computer Science 6.01 Introduction to EECS I Spring Semester, 2008 Week
A Bayesian Method for Guessing the Extreme Values in a Data Set
 A Bayesian Method for Guessing the Extreme Values in a Data Set Mingxi Wu University of Florida May, 2008 Mingxi Wu (University of Florida) May, 2008 1 / 74 Outline Problem Definition Example Applications
A Bayesian Method for Guessing the Extreme Values in a Data Set Mingxi Wu University of Florida May, 2008 Mingxi Wu (University of Florida) May, 2008 1 / 74 Outline Problem Definition Example Applications
Assessing Risks. Tom Carter. tom/sfi-csss. Complex Systems
 Assessing Risks Tom Carter http://cogs.csustan.edu/ tom/sfi-csss Complex Systems July, 2002 Our general topics: Assessing risks Using Bayes Theorem The doomsday argument References 2 The quotes Science
Assessing Risks Tom Carter http://cogs.csustan.edu/ tom/sfi-csss Complex Systems July, 2002 Our general topics: Assessing risks Using Bayes Theorem The doomsday argument References 2 The quotes Science
BIOL 51A - Biostatistics 1 1. Lecture 1: Intro to Biostatistics. Smoking: hazardous? FEV (l) Smoke
 BIOL 51A - Biostatistics 1 1 Lecture 1: Intro to Biostatistics Smoking: hazardous? FEV (l) 1 2 3 4 5 No Yes Smoke BIOL 51A - Biostatistics 1 2 Box Plot a.k.a box-and-whisker diagram or candlestick chart
BIOL 51A - Biostatistics 1 1 Lecture 1: Intro to Biostatistics Smoking: hazardous? FEV (l) 1 2 3 4 5 No Yes Smoke BIOL 51A - Biostatistics 1 2 Box Plot a.k.a box-and-whisker diagram or candlestick chart
Do not copy, post, or distribute
 10 ESTIMATION STUDIES Inferring the Parameters of a Population from the Statistics of a Sample 10.0 LEARNING OBJECTIVES In this chapter, we discuss the construction and interpretation of estimation studies.
10 ESTIMATION STUDIES Inferring the Parameters of a Population from the Statistics of a Sample 10.0 LEARNING OBJECTIVES In this chapter, we discuss the construction and interpretation of estimation studies.
A Note on Bayesian Inference After Multiple Imputation
 A Note on Bayesian Inference After Multiple Imputation Xiang Zhou and Jerome P. Reiter Abstract This article is aimed at practitioners who plan to use Bayesian inference on multiplyimputed datasets in
A Note on Bayesian Inference After Multiple Imputation Xiang Zhou and Jerome P. Reiter Abstract This article is aimed at practitioners who plan to use Bayesian inference on multiplyimputed datasets in
MODULE -4 BAYEIAN LEARNING
 MODULE -4 BAYEIAN LEARNING CONTENT Introduction Bayes theorem Bayes theorem and concept learning Maximum likelihood and Least Squared Error Hypothesis Maximum likelihood Hypotheses for predicting probabilities
MODULE -4 BAYEIAN LEARNING CONTENT Introduction Bayes theorem Bayes theorem and concept learning Maximum likelihood and Least Squared Error Hypothesis Maximum likelihood Hypotheses for predicting probabilities
Probability Hal Daumé III. Computer Science University of Maryland CS 421: Introduction to Artificial Intelligence 27 Mar 2012
 1 Hal Daumé III (me@hal3.name) Probability 101++ Hal Daumé III Computer Science University of Maryland me@hal3.name CS 421: Introduction to Artificial Intelligence 27 Mar 2012 Many slides courtesy of Dan
1 Hal Daumé III (me@hal3.name) Probability 101++ Hal Daumé III Computer Science University of Maryland me@hal3.name CS 421: Introduction to Artificial Intelligence 27 Mar 2012 Many slides courtesy of Dan
A Posteriori Corrections to Classification Methods.
 A Posteriori Corrections to Classification Methods. Włodzisław Duch and Łukasz Itert Department of Informatics, Nicholas Copernicus University, Grudziądzka 5, 87-100 Toruń, Poland; http://www.phys.uni.torun.pl/kmk
A Posteriori Corrections to Classification Methods. Włodzisław Duch and Łukasz Itert Department of Informatics, Nicholas Copernicus University, Grudziądzka 5, 87-100 Toruń, Poland; http://www.phys.uni.torun.pl/kmk
where Female = 0 for males, = 1 for females Age is measured in years (22, 23, ) GPA is measured in units on a four-point scale (0, 1.22, 3.45, etc.
 Notes on regression analysis 1. Basics in regression analysis key concepts (actual implementation is more complicated) A. Collect data B. Plot data on graph, draw a line through the middle of the scatter
Notes on regression analysis 1. Basics in regression analysis key concepts (actual implementation is more complicated) A. Collect data B. Plot data on graph, draw a line through the middle of the scatter
Examples of frequentist probability include games of chance, sample surveys, and randomized experiments. We will focus on frequentist probability sinc
 FPPA-Chapters 13,14 and parts of 16,17, and 18 STATISTICS 50 Richard A. Berk Spring, 1997 May 30, 1997 1 Thinking about Chance People talk about \chance" and \probability" all the time. There are many
FPPA-Chapters 13,14 and parts of 16,17, and 18 STATISTICS 50 Richard A. Berk Spring, 1997 May 30, 1997 1 Thinking about Chance People talk about \chance" and \probability" all the time. There are many
Hierarchical kernel applied to mixture model for the classification of binary predictors
 Hierarchical kernel applied to mixture model for the classification of binary predictors Seydou Sylla, Stephane Girard, Abdou Kâ Diongue, Aldiouma Diallo, Cheikh Sokhna To cite this version: Seydou Sylla,
Hierarchical kernel applied to mixture model for the classification of binary predictors Seydou Sylla, Stephane Girard, Abdou Kâ Diongue, Aldiouma Diallo, Cheikh Sokhna To cite this version: Seydou Sylla,
Regularization. CSCE 970 Lecture 3: Regularization. Stephen Scott and Vinod Variyam. Introduction. Outline
 Other Measures 1 / 52 sscott@cse.unl.edu learning can generally be distilled to an optimization problem Choose a classifier (function, hypothesis) from a set of functions that minimizes an objective function
Other Measures 1 / 52 sscott@cse.unl.edu learning can generally be distilled to an optimization problem Choose a classifier (function, hypothesis) from a set of functions that minimizes an objective function
