Estimating Recombination Rates. LRH selection test, and recombination
|
|
|
- Edmund Booth
- 5 years ago
- Views:
Transcription
1 Estimating Recombination Rates LRH selection test, and recombination Recall that LRH tests for selection by looking at frequencies of specific haplotypes. Clearly the test is dependent on the recombination rate. Higher recombination rate destroys homozygosity It turns out that recombination rates do vary a lot in the genome, and there are many regions with little or no recombination
2 Daly et al., 2001 Daly and others were looking at a 500kb region in 5q31 (Crohn disease region) 103 SNPs were genotyped in 129 trios. The direct approach is to do a case-control analysis using individual SNPs. Instead, they decided to focus on haplotypes to corect for local correlation. The study finds that large blocks (upto 100kb) show no evidence of recombination, and contain only 2-4 haplotypes There is some recombination across blocks Daly et al, 2001
3 Recombination in human chromosome 22 (Mb scale) Dawson et al. Nature 2002 Q: Can we give a direct count of the number of the recombination events? Recombination hot-spots (fine scale)
4 Recombination rates (chimp/human) Fine scale recombination rates differ between chimp and human The six hot-spots seen in human are not seen in chimp Estimating recombination rate Given population data, can you predict the scaled recombination rate! in a small region? Can you predict fine scale variation in recombination rates (across 2-3kb)?
5 Combinatorial Bounds for estimating recombination rate Recall that expected #recombinations =! log n Procedure Generate N random ARGs that results in the given sample Compute mean of the number of recombinations Alternatively, generate a summary statistic s from the population. For each!, generate many populations, and compute the mean and variance of s (This only needs to be done once). Use this to select the most likely! What is the correct summary statistic? Today, we talk about the min. number of recombination events as a possible summary statistic. It is not the most natural, but it is the most interesting computationally. The Infinite Sites Assumption & the 4 gamete condition Consider a history without recombination. No pair of sites shows all four gametes 00,01,10,11. A pair of sites with all 4 gametes implies a recombination event
6 Hudson & Kaplan Any pair of sites (i,j) containing 4 gametes must admit a recombination event. Disjoint (non-overlapping) sites must contain distinct recombination events, which can be summed! This gives a lower bound on the number of recombination events. Based on simulations, this bound is not tight. Myers and Griffiths 03: Idea 1 Let B(i,j) be a lower bound on the number of recombinations between sites i and j. Define Partition P = 1 = i 1 < i 2 < K < i k = n 1=i 1 i 2 i 3 i 4 i 5 i 6 i k =n k"1 R(P) = # B(i j,i j +1 ) is a lower bound for all P! j=1 Can we compute max P R(P) efficiently?
7 The R m bound Let R m ( j) = max Pj R(P j ), for all partitions of the first j columns Computing R m ( j) for all j is sufficient (why?) for j = 2Kn R m ( j) = max 1"k< j R m (k) + B(k, j) Improved lower bounds The R m bound also gives a general technique for combining local lower bounds into an overall lower bound. In the example, R m =2, but we cannot give any ARG with 2 recombination events. Can we improve upon Hudson and Kaplan to get better local lower bounds?
8 Myers & Griffiths: Idea 2 Consider the history of individuals. Let H t denote the number of distinct haplotypes at time t One of three things might happen at time t: Mutation: H t increase by at most 1 Recombination: H t increase by at most 1 Coalescence: H t does not increase The R H bound H " Number of extant & distinct halotypes E " Number of mutation events R " Number of Recombination events H " R + E +1 # R $ H % E %1 Infinite sites " E # S R " H # S #1 Ex: R>= 8-3-1=
9 R H bound In general, R H can be quite weak: consider the case when S>H However, it can be improved Partitioning idea: sum R H over disjoint intervals Apply to any subset of columns. Ex: Apply R H to the yellow columns Caveat : Computing max H'" H R(H') is NP - complete! (BB 05) Computing the R H bound Goal: Compute Max H R(H ) It is equivalent to the following: Find the smallest subset of columns such that every pair of rows is distinguished by at least one column For example, if we choose columns 1, 8, rows 1,2, and rows 5,6 remain identical. If choose columns 1,8,15 all rows are distinct : : : : : : : : (BB 05)
10 Computing R H A greedy heuristic: Remove all redundant rows. Set of columns, C=Ø Set S = {all pairs of rows} Iterate while (S<>Ø): Select a column c that separates maximum number of pairs P in S. C=C+{c} S=S-P Return n-1- C Computing R H How tight is R H? Clearly, by removing a haplotype, R H decreases. However, the number of recombinations needed doesn t really change
11 R s bound: Observation I! Non-informative column: If a site contains at most one 1, or one 0, then in any history, it can be obtained by adding a mutation to a branch.! EX: if a is the haplotype containing a 1, It can simply be added to the branch without increasing number of recombination events! R(M) = R(M-{s}) a s : : 1 a b c R s bound: Observation 2 Redundant rows: If two rows h 1 and h 2 are identical, then R(M) = R(M-{h 1 }) r 1 r 2 c
12 Rs bound: Observation 3 Suppose M has no noninformative columns, or redundant rows. Then, at least one of the haplotypes is a recombinant. There exists h s.t. R(M) = R(M-{h})+1 Which h should you choose? R s bound (Procedural) Procedure Compute_R s (M) If " non-informative column s return (Compute_R s (M-{s})) Else if " redundant row h return (Compute_R s (M-{h})) Else return (1 + min h (Compute_R s (M-{h}))
13 Results Additional results/problems Using dynamic programming, R s can be computed in 2^n poly(mn) time. Also, Rs can be augmented to handle intermediates. Are there poly. time lower bounds? The number of connected components in the conflict graph is a lower bound (BB 04). Fast algorithms for computing ARGs with minimum recombination. Poly. Time to get ARG with 0 recombination Poly. Time to get ARGs that are galled trees (Gusfield 03)
14 Underperforming lower bounds Sometimes, R s can be quite weak An R I lower bound that uses intermediates can help (BB 05) LPL data set 71 individuals, 9.7Kbp genomic sequence R m =22, R h =70
Integer Programming in Computational Biology. D. Gusfield University of California, Davis Presented December 12, 2016.!
 Integer Programming in Computational Biology D. Gusfield University of California, Davis Presented December 12, 2016. There are many important phylogeny problems that depart from simple tree models: Missing
Integer Programming in Computational Biology D. Gusfield University of California, Davis Presented December 12, 2016. There are many important phylogeny problems that depart from simple tree models: Missing
Integer Programming for Phylogenetic Network Problems
 Integer Programming for Phylogenetic Network Problems D. Gusfield University of California, Davis Presented at the National University of Singapore, July 27, 2015.! There are many important phylogeny problems
Integer Programming for Phylogenetic Network Problems D. Gusfield University of California, Davis Presented at the National University of Singapore, July 27, 2015.! There are many important phylogeny problems
Phylogenetic Networks with Recombination
 Phylogenetic Networks with Recombination October 17 2012 Recombination All DNA is recombinant DNA... [The] natural process of recombination and mutation have acted throughout evolution... Genetic exchange
Phylogenetic Networks with Recombination October 17 2012 Recombination All DNA is recombinant DNA... [The] natural process of recombination and mutation have acted throughout evolution... Genetic exchange
Humans have two copies of each chromosome. Inherited from mother and father. Genotyping technologies do not maintain the phase
 Humans have two copies of each chromosome Inherited from mother and father. Genotyping technologies do not maintain the phase Genotyping technologies do not maintain the phase Recall that proximal SNPs
Humans have two copies of each chromosome Inherited from mother and father. Genotyping technologies do not maintain the phase Genotyping technologies do not maintain the phase Recall that proximal SNPs
Aphylogenetic network is a generalization of a phylogenetic tree, allowing properties that are not tree-like.
 INFORMS Journal on Computing Vol. 16, No. 4, Fall 2004, pp. 459 469 issn 0899-1499 eissn 1526-5528 04 1604 0459 informs doi 10.1287/ijoc.1040.0099 2004 INFORMS The Fine Structure of Galls in Phylogenetic
INFORMS Journal on Computing Vol. 16, No. 4, Fall 2004, pp. 459 469 issn 0899-1499 eissn 1526-5528 04 1604 0459 informs doi 10.1287/ijoc.1040.0099 2004 INFORMS The Fine Structure of Galls in Phylogenetic
A Phylogenetic Network Construction due to Constrained Recombination
 A Phylogenetic Network Construction due to Constrained Recombination Mohd. Abdul Hai Zahid Research Scholar Research Supervisors: Dr. R.C. Joshi Dr. Ankush Mittal Department of Electronics and Computer
A Phylogenetic Network Construction due to Constrained Recombination Mohd. Abdul Hai Zahid Research Scholar Research Supervisors: Dr. R.C. Joshi Dr. Ankush Mittal Department of Electronics and Computer
Haplotyping as Perfect Phylogeny: A direct approach
 Haplotyping as Perfect Phylogeny: A direct approach Vineet Bafna Dan Gusfield Giuseppe Lancia Shibu Yooseph February 7, 2003 Abstract A full Haplotype Map of the human genome will prove extremely valuable
Haplotyping as Perfect Phylogeny: A direct approach Vineet Bafna Dan Gusfield Giuseppe Lancia Shibu Yooseph February 7, 2003 Abstract A full Haplotype Map of the human genome will prove extremely valuable
Finding Haplotype Block Boundaries by Using the Minimum-Description-Length Principle
 Am. J. Hum. Genet. 73:336 354, 2003 Finding Haplotype Block Boundaries by Using the Minimum-Description-Length Principle Eric C. Anderson and John Novembre Department of Integrative Biology, University
Am. J. Hum. Genet. 73:336 354, 2003 Finding Haplotype Block Boundaries by Using the Minimum-Description-Length Principle Eric C. Anderson and John Novembre Department of Integrative Biology, University
Allen Holder - Trinity University
 Haplotyping - Trinity University Population Problems - joint with Courtney Davis, University of Utah Single Individuals - joint with John Louie, Carrol College, and Lena Sherbakov, Williams University
Haplotyping - Trinity University Population Problems - joint with Courtney Davis, University of Utah Single Individuals - joint with John Louie, Carrol College, and Lena Sherbakov, Williams University
Introduction to population genetics & evolution
 Introduction to population genetics & evolution Course Organization Exam dates: Feb 19 March 1st Has everybody registered? Did you get the email with the exam schedule Summer seminar: Hot topics in Bioinformatics
Introduction to population genetics & evolution Course Organization Exam dates: Feb 19 March 1st Has everybody registered? Did you get the email with the exam schedule Summer seminar: Hot topics in Bioinformatics
One-to-one functions and onto functions
 MA 3362 Lecture 7 - One-to-one and Onto Wednesday, October 22, 2008. Objectives: Formalize definitions of one-to-one and onto One-to-one functions and onto functions At the level of set theory, there are
MA 3362 Lecture 7 - One-to-one and Onto Wednesday, October 22, 2008. Objectives: Formalize definitions of one-to-one and onto One-to-one functions and onto functions At the level of set theory, there are
Learning ancestral genetic processes using nonparametric Bayesian models
 Learning ancestral genetic processes using nonparametric Bayesian models Kyung-Ah Sohn October 31, 2011 Committee Members: Eric P. Xing, Chair Zoubin Ghahramani Russell Schwartz Kathryn Roeder Matthew
Learning ancestral genetic processes using nonparametric Bayesian models Kyung-Ah Sohn October 31, 2011 Committee Members: Eric P. Xing, Chair Zoubin Ghahramani Russell Schwartz Kathryn Roeder Matthew
Proportional Variance Explained by QLT and Statistical Power. Proportional Variance Explained by QTL and Statistical Power
 Proportional Variance Explained by QTL and Statistical Power Partitioning the Genetic Variance We previously focused on obtaining variance components of a quantitative trait to determine the proportion
Proportional Variance Explained by QTL and Statistical Power Partitioning the Genetic Variance We previously focused on obtaining variance components of a quantitative trait to determine the proportion
Mathematical Approaches to the Pure Parsimony Problem
 Mathematical Approaches to the Pure Parsimony Problem P. Blain a,, A. Holder b,, J. Silva c, and C. Vinzant d, July 29, 2005 Abstract Given the genetic information of a population, the Pure Parsimony problem
Mathematical Approaches to the Pure Parsimony Problem P. Blain a,, A. Holder b,, J. Silva c, and C. Vinzant d, July 29, 2005 Abstract Given the genetic information of a population, the Pure Parsimony problem
Random Lifts of Graphs
 27th Brazilian Math Colloquium, July 09 Plan of this talk A brief introduction to the probabilistic method. A quick review of expander graphs and their spectrum. Lifts, random lifts and their properties.
27th Brazilian Math Colloquium, July 09 Plan of this talk A brief introduction to the probabilistic method. A quick review of expander graphs and their spectrum. Lifts, random lifts and their properties.
(tree searching technique) (Boolean formulas) satisfying assignment: (X 1, X 2 )
 Algorithms Chapter 5: The Tree Searching Strategy - Examples 1 / 11 Chapter 5: The Tree Searching Strategy 1. Ex 5.1Determine the satisfiability of the following Boolean formulas by depth-first search
Algorithms Chapter 5: The Tree Searching Strategy - Examples 1 / 11 Chapter 5: The Tree Searching Strategy 1. Ex 5.1Determine the satisfiability of the following Boolean formulas by depth-first search
Population Genetics. with implications for Linkage Disequilibrium. Chiara Sabatti, Human Genetics 6357a Gonda
 1 Population Genetics with implications for Linkage Disequilibrium Chiara Sabatti, Human Genetics 6357a Gonda csabatti@mednet.ucla.edu 2 Hardy-Weinberg Hypotheses: infinite populations; no inbreeding;
1 Population Genetics with implications for Linkage Disequilibrium Chiara Sabatti, Human Genetics 6357a Gonda csabatti@mednet.ucla.edu 2 Hardy-Weinberg Hypotheses: infinite populations; no inbreeding;
The Pure Parsimony Problem
 Haplotyping and Minimum Diversity Graphs Courtney Davis - University of Utah - Trinity University Some Genetics Mother Paired Gene Representation Physical Trait ABABBA AAABBB Physical Trait ABA AAA Mother
Haplotyping and Minimum Diversity Graphs Courtney Davis - University of Utah - Trinity University Some Genetics Mother Paired Gene Representation Physical Trait ABABBA AAABBB Physical Trait ABA AAA Mother
The Tutte polynomial: sign and approximability
 : sign and approximability Mark Jerrum School of Mathematical Sciences Queen Mary, University of London Joint work with Leslie Goldberg, Department of Computer Science, University of Oxford Durham 23rd
: sign and approximability Mark Jerrum School of Mathematical Sciences Queen Mary, University of London Joint work with Leslie Goldberg, Department of Computer Science, University of Oxford Durham 23rd
Aditya Bhaskara CS 5968/6968, Lecture 1: Introduction and Review 12 January 2016
 Lecture 1: Introduction and Review We begin with a short introduction to the course, and logistics. We then survey some basics about approximation algorithms and probability. We also introduce some of
Lecture 1: Introduction and Review We begin with a short introduction to the course, and logistics. We then survey some basics about approximation algorithms and probability. We also introduce some of
ACO Comprehensive Exam October 14 and 15, 2013
 1. Computability, Complexity and Algorithms (a) Let G be the complete graph on n vertices, and let c : V (G) V (G) [0, ) be a symmetric cost function. Consider the following closest point heuristic for
1. Computability, Complexity and Algorithms (a) Let G be the complete graph on n vertices, and let c : V (G) V (G) [0, ) be a symmetric cost function. Consider the following closest point heuristic for
The Maximum Flow Problem with Disjunctive Constraints
 The Maximum Flow Problem with Disjunctive Constraints Ulrich Pferschy Joachim Schauer Abstract We study the maximum flow problem subject to binary disjunctive constraints in a directed graph: A negative
The Maximum Flow Problem with Disjunctive Constraints Ulrich Pferschy Joachim Schauer Abstract We study the maximum flow problem subject to binary disjunctive constraints in a directed graph: A negative
Spectral Graph Theory Lecture 2. The Laplacian. Daniel A. Spielman September 4, x T M x. ψ i = arg min
 Spectral Graph Theory Lecture 2 The Laplacian Daniel A. Spielman September 4, 2015 Disclaimer These notes are not necessarily an accurate representation of what happened in class. The notes written before
Spectral Graph Theory Lecture 2 The Laplacian Daniel A. Spielman September 4, 2015 Disclaimer These notes are not necessarily an accurate representation of what happened in class. The notes written before
Algorithm Design Strategies V
 Algorithm Design Strategies V Joaquim Madeira Version 0.0 October 2016 U. Aveiro, October 2016 1 Overview The 0-1 Knapsack Problem Revisited The Fractional Knapsack Problem Greedy Algorithms Example Coin
Algorithm Design Strategies V Joaquim Madeira Version 0.0 October 2016 U. Aveiro, October 2016 1 Overview The 0-1 Knapsack Problem Revisited The Fractional Knapsack Problem Greedy Algorithms Example Coin
Efficient Haplotype Inference with Boolean Satisfiability
 Efficient Haplotype Inference with Boolean Satisfiability Joao Marques-Silva 1 and Ines Lynce 2 1 School of Electronics and Computer Science University of Southampton 2 INESC-ID/IST Technical University
Efficient Haplotype Inference with Boolean Satisfiability Joao Marques-Silva 1 and Ines Lynce 2 1 School of Electronics and Computer Science University of Southampton 2 INESC-ID/IST Technical University
Partitioning Algorithms that Combine Spectral and Flow Methods
 CS369M: Algorithms for Modern Massive Data Set Analysis Lecture 15-11/11/2009 Partitioning Algorithms that Combine Spectral and Flow Methods Lecturer: Michael Mahoney Scribes: Kshipra Bhawalkar and Deyan
CS369M: Algorithms for Modern Massive Data Set Analysis Lecture 15-11/11/2009 Partitioning Algorithms that Combine Spectral and Flow Methods Lecturer: Michael Mahoney Scribes: Kshipra Bhawalkar and Deyan
1 Primals and Duals: Zero Sum Games
 CS 124 Section #11 Zero Sum Games; NP Completeness 4/15/17 1 Primals and Duals: Zero Sum Games We can represent various situations of conflict in life in terms of matrix games. For example, the game shown
CS 124 Section #11 Zero Sum Games; NP Completeness 4/15/17 1 Primals and Duals: Zero Sum Games We can represent various situations of conflict in life in terms of matrix games. For example, the game shown
( v 1 + v 2 ) + (3 v 1 ) = 4 v 1 + v 2. and ( 2 v 2 ) + ( v 1 + v 3 ) = v 1 2 v 2 + v 3, for instance.
 4.2. Linear Combinations and Linear Independence If we know that the vectors v 1, v 2,..., v k are are in a subspace W, then the Subspace Test gives us more vectors which must also be in W ; for instance,
4.2. Linear Combinations and Linear Independence If we know that the vectors v 1, v 2,..., v k are are in a subspace W, then the Subspace Test gives us more vectors which must also be in W ; for instance,
A Class Representative Model for Pure Parsimony Haplotyping
 A Class Representative Model for Pure Parsimony Haplotyping Daniele Catanzaro, Alessandra Godi, and Martine Labbé June 5, 2008 Abstract Haplotyping estimation from aligned Single Nucleotide Polymorphism
A Class Representative Model for Pure Parsimony Haplotyping Daniele Catanzaro, Alessandra Godi, and Martine Labbé June 5, 2008 Abstract Haplotyping estimation from aligned Single Nucleotide Polymorphism
Classical Complexity and Fixed-Parameter Tractability of Simultaneous Consecutive Ones Submatrix & Editing Problems
 Classical Complexity and Fixed-Parameter Tractability of Simultaneous Consecutive Ones Submatrix & Editing Problems Rani M. R, Mohith Jagalmohanan, R. Subashini Binary matrices having simultaneous consecutive
Classical Complexity and Fixed-Parameter Tractability of Simultaneous Consecutive Ones Submatrix & Editing Problems Rani M. R, Mohith Jagalmohanan, R. Subashini Binary matrices having simultaneous consecutive
Haplotyping estimation from aligned single nucleotide polymorphism fragments has attracted increasing
 INFORMS Journal on Computing Vol. 22, No. 2, Spring 2010, pp. 195 209 issn 1091-9856 eissn 1526-5528 10 2202 0195 informs doi 10.1287/ijoc.1090.0333 2010 INFORMS A Class Representative Model for Pure Parsimony
INFORMS Journal on Computing Vol. 22, No. 2, Spring 2010, pp. 195 209 issn 1091-9856 eissn 1526-5528 10 2202 0195 informs doi 10.1287/ijoc.1090.0333 2010 INFORMS A Class Representative Model for Pure Parsimony
On the Complexity of SNP Block Partitioning Under the Perfect Phylogeny Model
 On the Complexity of SNP Block Partitioning Under the Perfect Phylogeny Model Jens Gramm Wilhelm-Schickard-Institut für Informatik, Universität Tübingen, Germany. Tzvika Hartman Dept. of Computer Science,
On the Complexity of SNP Block Partitioning Under the Perfect Phylogeny Model Jens Gramm Wilhelm-Schickard-Institut für Informatik, Universität Tübingen, Germany. Tzvika Hartman Dept. of Computer Science,
12. LOCAL SEARCH. gradient descent Metropolis algorithm Hopfield neural networks maximum cut Nash equilibria
 12. LOCAL SEARCH gradient descent Metropolis algorithm Hopfield neural networks maximum cut Nash equilibria Lecture slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley h ttp://www.cs.princeton.edu/~wayne/kleinberg-tardos
12. LOCAL SEARCH gradient descent Metropolis algorithm Hopfield neural networks maximum cut Nash equilibria Lecture slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley h ttp://www.cs.princeton.edu/~wayne/kleinberg-tardos
A conjecture on the alphabet size needed to produce all correlation classes of pairs of words
 A conjecture on the alphabet size needed to produce all correlation classes of pairs of words Paul Leopardi Thanks: Jörg Arndt, Michael Barnsley, Richard Brent, Sylvain Forêt, Judy-anne Osborn. Mathematical
A conjecture on the alphabet size needed to produce all correlation classes of pairs of words Paul Leopardi Thanks: Jörg Arndt, Michael Barnsley, Richard Brent, Sylvain Forêt, Judy-anne Osborn. Mathematical
Q1) Explain how background selection and genetic hitchhiking could explain the positive correlation between genetic diversity and recombination rate.
 OEB 242 Exam Practice Problems Answer Key Q1) Explain how background selection and genetic hitchhiking could explain the positive correlation between genetic diversity and recombination rate. First, recall
OEB 242 Exam Practice Problems Answer Key Q1) Explain how background selection and genetic hitchhiking could explain the positive correlation between genetic diversity and recombination rate. First, recall
A fast estimate for the population recombination rate based on regression
 Genetics: Early Online, published on April 15, 213 as 1.1534/genetics.113.21 A fast estimate for the population recombination rate based on regression Kao Lin, Andreas Futschik, Haipeng Li CAS Key Laboratory
Genetics: Early Online, published on April 15, 213 as 1.1534/genetics.113.21 A fast estimate for the population recombination rate based on regression Kao Lin, Andreas Futschik, Haipeng Li CAS Key Laboratory
Task Assignment. Consider this very small instance: t1 t2 t3 t4 t5 p p p p p
 Task Assignment Task Assignment The Task Assignment problem starts with n persons and n tasks, and a known cost for each person/task combination. The goal is to assign each person to an unique task so
Task Assignment Task Assignment The Task Assignment problem starts with n persons and n tasks, and a known cost for each person/task combination. The goal is to assign each person to an unique task so
Lecture Notes 7 Random Processes. Markov Processes Markov Chains. Random Processes
 Lecture Notes 7 Random Processes Definition IID Processes Bernoulli Process Binomial Counting Process Interarrival Time Process Markov Processes Markov Chains Classification of States Steady State Probabilities
Lecture Notes 7 Random Processes Definition IID Processes Bernoulli Process Binomial Counting Process Interarrival Time Process Markov Processes Markov Chains Classification of States Steady State Probabilities
Zebo Peng Embedded Systems Laboratory IDA, Linköping University
 TDTS 01 Lecture 8 Optimization Heuristics for Synthesis Zebo Peng Embedded Systems Laboratory IDA, Linköping University Lecture 8 Optimization problems Heuristic techniques Simulated annealing Genetic
TDTS 01 Lecture 8 Optimization Heuristics for Synthesis Zebo Peng Embedded Systems Laboratory IDA, Linköping University Lecture 8 Optimization problems Heuristic techniques Simulated annealing Genetic
CS168: The Modern Algorithmic Toolbox Lectures #11 and #12: Spectral Graph Theory
 CS168: The Modern Algorithmic Toolbox Lectures #11 and #12: Spectral Graph Theory Tim Roughgarden & Gregory Valiant May 2, 2016 Spectral graph theory is the powerful and beautiful theory that arises from
CS168: The Modern Algorithmic Toolbox Lectures #11 and #12: Spectral Graph Theory Tim Roughgarden & Gregory Valiant May 2, 2016 Spectral graph theory is the powerful and beautiful theory that arises from
Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination)
 12/5/14 Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination) Linkage Disequilibrium Genealogical Interpretation of LD Association Mapping 1 Linkage and Recombination v linkage equilibrium ²
12/5/14 Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination) Linkage Disequilibrium Genealogical Interpretation of LD Association Mapping 1 Linkage and Recombination v linkage equilibrium ²
/633 Introduction to Algorithms Lecturer: Michael Dinitz Topic: Matroids and Greedy Algorithms Date: 10/31/16
 60.433/633 Introduction to Algorithms Lecturer: Michael Dinitz Topic: Matroids and Greedy Algorithms Date: 0/3/6 6. Introduction We talked a lot the last lecture about greedy algorithms. While both Prim
60.433/633 Introduction to Algorithms Lecturer: Michael Dinitz Topic: Matroids and Greedy Algorithms Date: 0/3/6 6. Introduction We talked a lot the last lecture about greedy algorithms. While both Prim
The genomes of recombinant inbred lines
 The genomes of recombinant inbred lines Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman C57BL/6 2 1 Recombinant inbred lines (by sibling mating)
The genomes of recombinant inbred lines Karl W Broman Department of Biostatistics Johns Hopkins University http://www.biostat.jhsph.edu/~kbroman C57BL/6 2 1 Recombinant inbred lines (by sibling mating)
Discussion of Dimension Reduction... by Dennis Cook
 Discussion of Dimension Reduction... by Dennis Cook Department of Statistics University of Chicago Anthony Atkinson 70th birthday celebration London, December 14, 2007 Outline The renaissance of PCA? State
Discussion of Dimension Reduction... by Dennis Cook Department of Statistics University of Chicago Anthony Atkinson 70th birthday celebration London, December 14, 2007 Outline The renaissance of PCA? State
Population Genetics I. Bio
 Population Genetics I. Bio5488-2018 Don Conrad dconrad@genetics.wustl.edu Why study population genetics? Functional Inference Demographic inference: History of mankind is written in our DNA. We can learn
Population Genetics I. Bio5488-2018 Don Conrad dconrad@genetics.wustl.edu Why study population genetics? Functional Inference Demographic inference: History of mankind is written in our DNA. We can learn
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin
 Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium. November 12, 2012
 Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium November 12, 2012 Last Time Sequence data and quantification of variation Infinite sites model Nucleotide diversity (π) Sequence-based
Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium November 12, 2012 Last Time Sequence data and quantification of variation Infinite sites model Nucleotide diversity (π) Sequence-based
The E-M Algorithm in Genetics. Biostatistics 666 Lecture 8
 The E-M Algorithm in Genetics Biostatistics 666 Lecture 8 Maximum Likelihood Estimation of Allele Frequencies Find parameter estimates which make observed data most likely General approach, as long as
The E-M Algorithm in Genetics Biostatistics 666 Lecture 8 Maximum Likelihood Estimation of Allele Frequencies Find parameter estimates which make observed data most likely General approach, as long as
3.4 Relaxations and bounds
 3.4 Relaxations and bounds Consider a generic Discrete Optimization problem z = min{c(x) : x X} with an optimal solution x X. In general, the algorithms generate not only a decreasing sequence of upper
3.4 Relaxations and bounds Consider a generic Discrete Optimization problem z = min{c(x) : x X} with an optimal solution x X. In general, the algorithms generate not only a decreasing sequence of upper
BIOINFORMATICS. SequenceLDhot: Detecting Recombination Hotspots. Paul Fearnhead a 1 INTRODUCTION 2 METHOD
 BIOINFORMATICS Vol. 00 no. 00 2006 Pages 1 5 SequenceLDhot: Detecting Recombination Hotspots Paul Fearnhead a a Department of Mathematics and Statistics, Lancaster University, Lancaster LA1 4YF, UK ABSTRACT
BIOINFORMATICS Vol. 00 no. 00 2006 Pages 1 5 SequenceLDhot: Detecting Recombination Hotspots Paul Fearnhead a a Department of Mathematics and Statistics, Lancaster University, Lancaster LA1 4YF, UK ABSTRACT
BP -HOMOLOGY AND AN IMPLICATION FOR SYMMETRIC POLYNOMIALS. 1. Introduction and results
 BP -HOMOLOGY AND AN IMPLICATION FOR SYMMETRIC POLYNOMIALS DONALD M. DAVIS Abstract. We determine the BP -module structure, mod higher filtration, of the main part of the BP -homology of elementary 2- groups.
BP -HOMOLOGY AND AN IMPLICATION FOR SYMMETRIC POLYNOMIALS DONALD M. DAVIS Abstract. We determine the BP -module structure, mod higher filtration, of the main part of the BP -homology of elementary 2- groups.
Complexity and Approximation of the Minimum Recombination Haplotype Configuration Problem
 Complexity and Approximation of the Minimum Recombination Haplotype Configuration Problem Lan Liu 1, Xi Chen 3, Jing Xiao 3, and Tao Jiang 1,2 1 Department of Computer Science and Engineering, University
Complexity and Approximation of the Minimum Recombination Haplotype Configuration Problem Lan Liu 1, Xi Chen 3, Jing Xiao 3, and Tao Jiang 1,2 1 Department of Computer Science and Engineering, University
Beyond Galled Trees Decomposition and Computation of Galled Networks
 Beyond Galled Trees Decomposition and Computation of Galled Networks Daniel H. Huson & Tobias H.Kloepper RECOMB 2007 1 Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or
Beyond Galled Trees Decomposition and Computation of Galled Networks Daniel H. Huson & Tobias H.Kloepper RECOMB 2007 1 Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or
Modelling Genetic Variations with Fragmentation-Coagulation Processes
 Modelling Genetic Variations with Fragmentation-Coagulation Processes Yee Whye Teh, Charles Blundell, Lloyd Elliott Gatsby Computational Neuroscience Unit, UCL Genetic Variations in Populations Inferring
Modelling Genetic Variations with Fragmentation-Coagulation Processes Yee Whye Teh, Charles Blundell, Lloyd Elliott Gatsby Computational Neuroscience Unit, UCL Genetic Variations in Populations Inferring
Inductive Learning. Inductive hypothesis h Hypothesis space H size H. Example set X. h: hypothesis that. size m Training set D
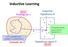 Inductive Learning size m Training set D Inductive hypothesis h - - + - + + - - - + + - + - + + - - + + - p(x): probability that example x is picked + from X Example set X + - L Hypothesis space H size
Inductive Learning size m Training set D Inductive hypothesis h - - + - + + - - - + + - + - + + - - + + - p(x): probability that example x is picked + from X Example set X + - L Hypothesis space H size
An Analysis on Recombination in Multi-Objective Evolutionary Optimization
 An Analysis on Recombination in Multi-Objective Evolutionary Optimization Chao Qian, Yang Yu, Zhi-Hua Zhou National Key Laboratory for Novel Software Technology Nanjing University, Nanjing 20023, China
An Analysis on Recombination in Multi-Objective Evolutionary Optimization Chao Qian, Yang Yu, Zhi-Hua Zhou National Key Laboratory for Novel Software Technology Nanjing University, Nanjing 20023, China
Day 12. Thursday June 7, 2012
 Day 12 Thursday June 7, 2012 1 A Word on Well-Ordering We talked about the principle of induction. Given a formula ϕ(n) then to prove n.ϕ(n) then it is sufficent to prove ϕ(0). n.ϕ(n) ϕ(n + 1) We mentioned
Day 12 Thursday June 7, 2012 1 A Word on Well-Ordering We talked about the principle of induction. Given a formula ϕ(n) then to prove n.ϕ(n) then it is sufficent to prove ϕ(0). n.ϕ(n) ϕ(n + 1) We mentioned
GWAS for Compound Heterozygous Traits: Phenotypic Distance and Integer Linear Programming Dan Gusfield, Rasmus Nielsen.
 GWAS for Compound Heterozygous Traits: Phenotypic Distance and Integer Linear Programming Dan Gusfield, Rasmus Nielsen December 11, 2016 GWAS In Genome Wide Association Studies (GWAS) we try to locate
GWAS for Compound Heterozygous Traits: Phenotypic Distance and Integer Linear Programming Dan Gusfield, Rasmus Nielsen December 11, 2016 GWAS In Genome Wide Association Studies (GWAS) we try to locate
Section 1.5. Solution Sets of Linear Systems
 Section 1.5 Solution Sets of Linear Systems Plan For Today Today we will learn to describe and draw the solution set of an arbitrary system of linear equations Ax = b, using spans. Ax = b Recall: the solution
Section 1.5 Solution Sets of Linear Systems Plan For Today Today we will learn to describe and draw the solution set of an arbitrary system of linear equations Ax = b, using spans. Ax = b Recall: the solution
Section Notes 8. Integer Programming II. Applied Math 121. Week of April 5, expand your knowledge of big M s and logical constraints.
 Section Notes 8 Integer Programming II Applied Math 121 Week of April 5, 2010 Goals for the week understand IP relaxations be able to determine the relative strength of formulations understand the branch
Section Notes 8 Integer Programming II Applied Math 121 Week of April 5, 2010 Goals for the week understand IP relaxations be able to determine the relative strength of formulations understand the branch
Counting All Possible Ancestral Configurations of Sample Sequences in Population Genetics
 1 Counting All Possible Ancestral Configurations of Sample Sequences in Population Genetics Yun S. Song, Rune Lyngsø, and Jotun Hein Abstract Given a set D of input sequences, a genealogy for D can be
1 Counting All Possible Ancestral Configurations of Sample Sequences in Population Genetics Yun S. Song, Rune Lyngsø, and Jotun Hein Abstract Given a set D of input sequences, a genealogy for D can be
Leveraging Big Data: Lecture 13
 Leveraging Big Data: Lecture 13 http://www.cohenwang.com/edith/bigdataclass2013 Instructors: Edith Cohen Amos Fiat Haim Kaplan Tova Milo What are Linear Sketches? Linear Transformations of the input vector
Leveraging Big Data: Lecture 13 http://www.cohenwang.com/edith/bigdataclass2013 Instructors: Edith Cohen Amos Fiat Haim Kaplan Tova Milo What are Linear Sketches? Linear Transformations of the input vector
17 Non-collinear alignment Motivation A B C A B C A B C A B C D A C. This exposition is based on:
 17 Non-collinear alignment This exposition is based on: 1. Darling, A.E., Mau, B., Perna, N.T. (2010) progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147.
17 Non-collinear alignment This exposition is based on: 1. Darling, A.E., Mau, B., Perna, N.T. (2010) progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147.
Graph coloring, perfect graphs
 Lecture 5 (05.04.2013) Graph coloring, perfect graphs Scribe: Tomasz Kociumaka Lecturer: Marcin Pilipczuk 1 Introduction to graph coloring Definition 1. Let G be a simple undirected graph and k a positive
Lecture 5 (05.04.2013) Graph coloring, perfect graphs Scribe: Tomasz Kociumaka Lecturer: Marcin Pilipczuk 1 Introduction to graph coloring Definition 1. Let G be a simple undirected graph and k a positive
 that of Phylotree.org, mtdna tree Build 1756 (Supplementary TableS2). is resulted in 78 individuals allocated to the hg B4a1a1 and three individuals to hg Q. e control region (nps 57372 and nps 1602416526)
that of Phylotree.org, mtdna tree Build 1756 (Supplementary TableS2). is resulted in 78 individuals allocated to the hg B4a1a1 and three individuals to hg Q. e control region (nps 57372 and nps 1602416526)
Combinatorial Optimization
 Combinatorial Optimization 2017-2018 1 Maximum matching on bipartite graphs Given a graph G = (V, E), find a maximum cardinal matching. 1.1 Direct algorithms Theorem 1.1 (Petersen, 1891) A matching M is
Combinatorial Optimization 2017-2018 1 Maximum matching on bipartite graphs Given a graph G = (V, E), find a maximum cardinal matching. 1.1 Direct algorithms Theorem 1.1 (Petersen, 1891) A matching M is
Learning Objectives. Zeroes. The Real Zeros of a Polynomial Function
 The Real Zeros of a Polynomial Function 1 Learning Objectives 1. Use the Remainder and Factor Theorems 2. Use the Rational Zeros Theorem to list the potential rational zeros of a polynomial function 3.
The Real Zeros of a Polynomial Function 1 Learning Objectives 1. Use the Remainder and Factor Theorems 2. Use the Rational Zeros Theorem to list the potential rational zeros of a polynomial function 3.
AUTHORIZATION TO LEND AND REPRODUCE THE THESIS. Date Jong Wha Joanne Joo, Author
 AUTHORIZATION TO LEND AND REPRODUCE THE THESIS As the sole author of this thesis, I authorize Brown University to lend it to other institutions or individuals for the purpose of scholarly research. Date
AUTHORIZATION TO LEND AND REPRODUCE THE THESIS As the sole author of this thesis, I authorize Brown University to lend it to other institutions or individuals for the purpose of scholarly research. Date
theta H H H H H H H H H H H K K K K K K K K K K centimorgans
 Linkage Phase Recall that the recombination fraction ρ for two loci denotes the probability of a recombination event between those two loci. For loci on different chromosomes, ρ = 1=2. For loci on the
Linkage Phase Recall that the recombination fraction ρ for two loci denotes the probability of a recombination event between those two loci. For loci on different chromosomes, ρ = 1=2. For loci on the
On the Fixed Parameter Tractability and Approximability of the Minimum Error Correction problem
 On the Fixed Parameter Tractability and Approximability of the Minimum Error Correction problem Paola Bonizzoni, Riccardo Dondi, Gunnar W. Klau, Yuri Pirola, Nadia Pisanti and Simone Zaccaria DISCo, computer
On the Fixed Parameter Tractability and Approximability of the Minimum Error Correction problem Paola Bonizzoni, Riccardo Dondi, Gunnar W. Klau, Yuri Pirola, Nadia Pisanti and Simone Zaccaria DISCo, computer
Ma/CS 6b Class 23: Eigenvalues in Regular Graphs
 Ma/CS 6b Class 3: Eigenvalues in Regular Graphs By Adam Sheffer Recall: The Spectrum of a Graph Consider a graph G = V, E and let A be the adjacency matrix of G. The eigenvalues of G are the eigenvalues
Ma/CS 6b Class 3: Eigenvalues in Regular Graphs By Adam Sheffer Recall: The Spectrum of a Graph Consider a graph G = V, E and let A be the adjacency matrix of G. The eigenvalues of G are the eigenvalues
Data Mining Prof. Pabitra Mitra Department of Computer Science & Engineering Indian Institute of Technology, Kharagpur
 Data Mining Prof. Pabitra Mitra Department of Computer Science & Engineering Indian Institute of Technology, Kharagpur Lecture 21 K - Nearest Neighbor V In this lecture we discuss; how do we evaluate the
Data Mining Prof. Pabitra Mitra Department of Computer Science & Engineering Indian Institute of Technology, Kharagpur Lecture 21 K - Nearest Neighbor V In this lecture we discuss; how do we evaluate the
An Overview of Combinatorial Methods for Haplotype Inference
 An Overview of Combinatorial Methods for Haplotype Inference Dan Gusfield 1 Department of Computer Science, University of California, Davis Davis, CA. 95616 Abstract A current high-priority phase of human
An Overview of Combinatorial Methods for Haplotype Inference Dan Gusfield 1 Department of Computer Science, University of California, Davis Davis, CA. 95616 Abstract A current high-priority phase of human
CS357: CTL Model Checking (two lectures worth) David Dill
 CS357: CTL Model Checking (two lectures worth) David Dill 1 CTL CTL = Computation Tree Logic It is a propositional temporal logic temporal logic extended to properties of events over time. CTL is a branching
CS357: CTL Model Checking (two lectures worth) David Dill 1 CTL CTL = Computation Tree Logic It is a propositional temporal logic temporal logic extended to properties of events over time. CTL is a branching
Phylogenetic Networks, Trees, and Clusters
 Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Random Variable. Pr(X = a) = Pr(s)
 Random Variable Definition A random variable X on a sample space Ω is a real-valued function on Ω; that is, X : Ω R. A discrete random variable is a random variable that takes on only a finite or countably
Random Variable Definition A random variable X on a sample space Ω is a real-valued function on Ω; that is, X : Ω R. A discrete random variable is a random variable that takes on only a finite or countably
Cosets and Lagrange s theorem
 Cosets and Lagrange s theorem These are notes on cosets and Lagrange s theorem some of which may already have been lecturer. There are some questions for you included in the text. You should write the
Cosets and Lagrange s theorem These are notes on cosets and Lagrange s theorem some of which may already have been lecturer. There are some questions for you included in the text. You should write the
Linear Regression (1/1/17)
 STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
CS325: Analysis of Algorithms, Fall Final Exam
 CS: Analysis of Algorithms, Fall 0 Final Exam I don t know policy: you may write I don t know and nothing else to answer a question and receive percent of the total points for that problem whereas a completely
CS: Analysis of Algorithms, Fall 0 Final Exam I don t know policy: you may write I don t know and nothing else to answer a question and receive percent of the total points for that problem whereas a completely
R(p, k) = the near regular complete k-partite graph of order p. Let p = sk+r, where s and r are positive integers such that 0 r < k.
 MATH3301 EXTREMAL GRAPH THEORY Definition: A near regular complete multipartite graph is a complete multipartite graph with orders of its partite sets differing by at most 1. R(p, k) = the near regular
MATH3301 EXTREMAL GRAPH THEORY Definition: A near regular complete multipartite graph is a complete multipartite graph with orders of its partite sets differing by at most 1. R(p, k) = the near regular
Integer Linear Programs
 Lecture 2: Review, Linear Programming Relaxations Today we will talk about expressing combinatorial problems as mathematical programs, specifically Integer Linear Programs (ILPs). We then see what happens
Lecture 2: Review, Linear Programming Relaxations Today we will talk about expressing combinatorial problems as mathematical programs, specifically Integer Linear Programs (ILPs). We then see what happens
Probabilistic Systems Analysis Spring 2018 Lecture 6. Random Variables: Probability Mass Function and Expectation
 EE 178 Probabilistic Systems Analysis Spring 2018 Lecture 6 Random Variables: Probability Mass Function and Expectation Probability Mass Function When we introduce the basic probability model in Note 1,
EE 178 Probabilistic Systems Analysis Spring 2018 Lecture 6 Random Variables: Probability Mass Function and Expectation Probability Mass Function When we introduce the basic probability model in Note 1,
Chapter 9: Relations Relations
 Chapter 9: Relations 9.1 - Relations Definition 1 (Relation). Let A and B be sets. A binary relation from A to B is a subset R A B, i.e., R is a set of ordered pairs where the first element from each pair
Chapter 9: Relations 9.1 - Relations Definition 1 (Relation). Let A and B be sets. A binary relation from A to B is a subset R A B, i.e., R is a set of ordered pairs where the first element from each pair
1 Measurable Functions
 36-752 Advanced Probability Overview Spring 2018 2. Measurable Functions, Random Variables, and Integration Instructor: Alessandro Rinaldo Associated reading: Sec 1.5 of Ash and Doléans-Dade; Sec 1.3 and
36-752 Advanced Probability Overview Spring 2018 2. Measurable Functions, Random Variables, and Integration Instructor: Alessandro Rinaldo Associated reading: Sec 1.5 of Ash and Doléans-Dade; Sec 1.3 and
IE 400 Principles of Engineering Management. The Simplex Algorithm-I: Set 3
 IE 4 Principles of Engineering Management The Simple Algorithm-I: Set 3 So far, we have studied how to solve two-variable LP problems graphically. However, most real life problems have more than two variables!
IE 4 Principles of Engineering Management The Simple Algorithm-I: Set 3 So far, we have studied how to solve two-variable LP problems graphically. However, most real life problems have more than two variables!
Linear equations in linear algebra
 Linear equations in linear algebra Samy Tindel Purdue University Differential equations and linear algebra - MA 262 Taken from Differential equations and linear algebra Pearson Collections Samy T. Linear
Linear equations in linear algebra Samy Tindel Purdue University Differential equations and linear algebra - MA 262 Taken from Differential equations and linear algebra Pearson Collections Samy T. Linear
18.312: Algebraic Combinatorics Lionel Levine. Lecture 22. Smith normal form of an integer matrix (linear algebra over Z).
 18.312: Algebraic Combinatorics Lionel Levine Lecture date: May 3, 2011 Lecture 22 Notes by: Lou Odette This lecture: Smith normal form of an integer matrix (linear algebra over Z). 1 Review of Abelian
18.312: Algebraic Combinatorics Lionel Levine Lecture date: May 3, 2011 Lecture 22 Notes by: Lou Odette This lecture: Smith normal form of an integer matrix (linear algebra over Z). 1 Review of Abelian
Data Mining and Matrices
 Data Mining and Matrices 08 Boolean Matrix Factorization Rainer Gemulla, Pauli Miettinen June 13, 2013 Outline 1 Warm-Up 2 What is BMF 3 BMF vs. other three-letter abbreviations 4 Binary matrices, tiles,
Data Mining and Matrices 08 Boolean Matrix Factorization Rainer Gemulla, Pauli Miettinen June 13, 2013 Outline 1 Warm-Up 2 What is BMF 3 BMF vs. other three-letter abbreviations 4 Binary matrices, tiles,
Independencies. Undirected Graphical Models 2: Independencies. Independencies (Markov networks) Independencies (Bayesian Networks)
 (Bayesian Networks) Undirected Graphical Models 2: Use d-separation to read off independencies in a Bayesian network Takes a bit of effort! 1 2 (Markov networks) Use separation to determine independencies
(Bayesian Networks) Undirected Graphical Models 2: Use d-separation to read off independencies in a Bayesian network Takes a bit of effort! 1 2 (Markov networks) Use separation to determine independencies
SAT in Bioinformatics: Making the Case with Haplotype Inference
 SAT in Bioinformatics: Making the Case with Haplotype Inference Inês Lynce 1 and João Marques-Silva 2 1 IST/INESC-ID, Technical University of Lisbon, Portugal ines@sat.inesc-id.pt 2 School of Electronics
SAT in Bioinformatics: Making the Case with Haplotype Inference Inês Lynce 1 and João Marques-Silva 2 1 IST/INESC-ID, Technical University of Lisbon, Portugal ines@sat.inesc-id.pt 2 School of Electronics
Evolution of Populations. Chapter 17
 Evolution of Populations Chapter 17 17.1 Genes and Variation i. Introduction: Remember from previous units. Genes- Units of Heredity Variation- Genetic differences among individuals in a population. New
Evolution of Populations Chapter 17 17.1 Genes and Variation i. Introduction: Remember from previous units. Genes- Units of Heredity Variation- Genetic differences among individuals in a population. New
Asymptotic Analysis. Slides by Carl Kingsford. Jan. 27, AD Chapter 2
 Asymptotic Analysis Slides by Carl Kingsford Jan. 27, 2014 AD Chapter 2 Independent Set Definition (Independent Set). Given a graph G = (V, E) an independent set is a set S V if no two nodes in S are joined
Asymptotic Analysis Slides by Carl Kingsford Jan. 27, 2014 AD Chapter 2 Independent Set Definition (Independent Set). Given a graph G = (V, E) an independent set is a set S V if no two nodes in S are joined
Combining the cycle index and the Tutte polynomial?
 Combining the cycle index and the Tutte polynomial? Peter J. Cameron University of St Andrews Combinatorics Seminar University of Vienna 23 March 2017 Selections Students often meet the following table
Combining the cycle index and the Tutte polynomial? Peter J. Cameron University of St Andrews Combinatorics Seminar University of Vienna 23 March 2017 Selections Students often meet the following table
O.K. But what if the chicken didn t have access to a teleporter.
 The intermediate value theorem, and performing algebra on its. This is a dual topic lecture. : The Intermediate value theorem First we should remember what it means to be a continuous function: A function
The intermediate value theorem, and performing algebra on its. This is a dual topic lecture. : The Intermediate value theorem First we should remember what it means to be a continuous function: A function
Knapsack and Scheduling Problems. The Greedy Method
 The Greedy Method: Knapsack and Scheduling Problems The Greedy Method 1 Outline and Reading Task Scheduling Fractional Knapsack Problem The Greedy Method 2 Elements of Greedy Strategy An greedy algorithm
The Greedy Method: Knapsack and Scheduling Problems The Greedy Method 1 Outline and Reading Task Scheduling Fractional Knapsack Problem The Greedy Method 2 Elements of Greedy Strategy An greedy algorithm
Bio nformatics. Lecture 23. Saad Mneimneh
 Bio nformatics Lecture 23 Protein folding The goal is to determine the three-dimensional structure of a protein based on its amino acid sequence Assumption: amino acid sequence completely and uniquely
Bio nformatics Lecture 23 Protein folding The goal is to determine the three-dimensional structure of a protein based on its amino acid sequence Assumption: amino acid sequence completely and uniquely
The Lefthanded Local Lemma characterizes chordal dependency graphs
 The Lefthanded Local Lemma characterizes chordal dependency graphs Wesley Pegden March 30, 2012 Abstract Shearer gave a general theorem characterizing the family L of dependency graphs labeled with probabilities
The Lefthanded Local Lemma characterizes chordal dependency graphs Wesley Pegden March 30, 2012 Abstract Shearer gave a general theorem characterizing the family L of dependency graphs labeled with probabilities
CSC 411 Lecture 3: Decision Trees
 CSC 411 Lecture 3: Decision Trees Roger Grosse, Amir-massoud Farahmand, and Juan Carrasquilla University of Toronto UofT CSC 411: 03-Decision Trees 1 / 33 Today Decision Trees Simple but powerful learning
CSC 411 Lecture 3: Decision Trees Roger Grosse, Amir-massoud Farahmand, and Juan Carrasquilla University of Toronto UofT CSC 411: 03-Decision Trees 1 / 33 Today Decision Trees Simple but powerful learning
MATH 56A: STOCHASTIC PROCESSES CHAPTER 1
 MATH 56A: STOCHASTIC PROCESSES CHAPTER. Finite Markov chains For the sake of completeness of these notes I decided to write a summary of the basic concepts of finite Markov chains. The topics in this chapter
MATH 56A: STOCHASTIC PROCESSES CHAPTER. Finite Markov chains For the sake of completeness of these notes I decided to write a summary of the basic concepts of finite Markov chains. The topics in this chapter
xy xyy 1 = ey 1 = y 1 i.e.
 Homework 2 solutions. Problem 4.4. Let g be an element of the group G. Keep g fixed and let x vary through G. Prove that the products gx are all distinct and fill out G. Do the same for the products xg.
Homework 2 solutions. Problem 4.4. Let g be an element of the group G. Keep g fixed and let x vary through G. Prove that the products gx are all distinct and fill out G. Do the same for the products xg.
