THE ACCESSIBILITY OF TYPE I CU(II) CENTERS IN LACCASE, AZURIN, AND STELLACYANIN TO EXCHANGEABLE HYDROGEN AND AMBIENT WATER
|
|
|
- Ethel Floyd
- 6 years ago
- Views:
Transcription
1 THE ACCESSIBILITY OF TYPE I CU(II) CENTERS IN LACCASE, AZURIN, AND STELLACYANIN TO EXCHANGEABLE HYDROGEN AND AMBIENT WATER W. B. MIMS AND J. L. DAVIS Bell Laboratories, Murray Hill, New Jersey J. PEISACH Albert Einstein College ofmedicine, Bronx, New York and Bell Laboratories, Murray Hill, New Jersey Type 1 Cu(II) centers are found in a number of low molecular weight electron-transporting proteins, including azurin, plastocyanin, and stellacyanin, and in larger, copper-containing oxidases, such as laccase, ascorbate oxidase, and ceruloplasmin (1, 2). (The last three also contain other types of CU[II].) The outstanding physical characteristic of Type 1 Cu(II) is an intense optical absorption near 600 nm, with a molar extinction coefficient ranging from 3,000-5,000. This arises from a metal-to-ligand charge transfer (3-5). Another property is a small nuclear hyperfine interaction in the EPR of 3-9 x 10-3 cm-', which is believed to be due to the nontetragonal, and likely near tetrahedral, symmetry of the metal site (6, 7). The structures of two low molecular weight Type 1 Cu(II) proteins have been determined by x-ray crystallographic analysis. The first, poplar leaf plastocyanin (8), has been analyzed to 1.6-A resolution. The ligands to Cu(II) consist of a cysteinyl sulfur, believed responsible for the charge transfer described above (3, 4), two imidazole nitrogens, and at a distance of 1.9 A, which is rather remote for the formation of a chemical bond, a methionyl sulfur. A similar arrangement of ligands is found for azurin, based on x-ray data refined to 3-A resolution (9). From various spectroscopic studies, it is believed that the cysteinyl sulfur and at least one imidazole nitrogen are bound to the metal ion in all Type 1 Cu(II) proteins (10-17). The methionyl sulfur, however, is not conserved in the structure, since stellacyanin lacks methionine (7, 18). From calculations of the partial specific volumes for plastocyanin, it was determined that a significant portion of the crystalline protein, 36%, is occupied by water (8). Most of this water is probably lodged between protein ABSTRACT The characteristic deuterium modulation pattern was observed in the electron spin-echo envelopes for laccase, decupro laccase (from which Type 2 copper had been removed), stellacyanin, and azurin that had been exchanged against D20. From the decay rate of the modulation pattern and from a quantitative analysis of the modulation depth, we conclude that the Cu(II) sites in these proteins are directly accessible to solvent. Similar results were obtained for laccase and decupro laccase. molecules but some within the protein structure. X-ray crystallographic analysis is of insufficient resolution, however, to determine its location. Here we present evidence that water is found close to Type 1 Cu(II) in azurin, stellacyanin, and laccase. Further, we show that stellacyanin is more open to solvent than is azurin. These conclusions are based on detailed comparisons of the electron spin-echo decay envelopes observed when these proteins are prepared in D20 and H20 solutions. The nuclear modulation pattern due to coupling between the paramagnetic center and the deuterons in D20 is extracted from the modulation patterns due to other nuclei, such as '4N (14) and 'H, by dividing the envelopes one into the other (19, 20). The depth of the deuteron modulation pattern can then be related to the number and distance of the interacting 2H nuclei (21). Since the depth of the deuteron modulation is far greater than can be accounted for from interaction with exchangeable peptide and side-chain deuterons, we conclude that it arises from D20 close to Cu(II). MATERIALS AND METHODS Laccase and stellacyanin were purified according to established procedures from lac acetone powders (Saito and Company, Tokyo, Japan), derived from Rhus vernicifera, the Japanese lac tree (13, 22). Decupro laccase from which Type 2 Cu(II) had been removed, was prepared according to the method of Graziani et al. (23) modified by Avigliano et al. (17). Pseudomonas aeruginosa azurin was purchased from Public Health Laboratory Service, Porton Down, England. Laccase in 0.05 M phosphate buffer, ph 5.8, decupro laccase in 0.01 M phosphate buffer, ph 7.0, stellacyanin in 0.01 M phosphate buffer, ph 6, and azurin in 0.01 M phosphate buffer, ph 7, were each separated from adventitious Cu(II) by passage through a Chelex 100 column that had been previously equilibrated with the buffers in which the individual proteins were BIoPHYs. J. e BiophysicalSociety /84/04/755/12 $1.00 Volume45 April
2 dissolved. The removal of free copper as well as the integrity of Type 1 Cu(II) centers was monitored by EPR. Exchange against D20 at 50 was achieved by diluting protein samples with five volumes of D20, and then concentrating by pressure filtration (Amicon Corp., Scientific Sys. Div., Danvers, MA) to the original volume. This procedure was repeated three times for each protein solution that, after concentration, was frozen in liquid N2. The total time for D20 exchange varied from 2 to 3 h, a time period comparable with that required for exchange of the majority of peptidic protons in azurin, but far less than that required to exchange the C2 proton of metal-ligated imidazole (24). The concentration of proteins used for electron spin-echo (ESE) studies varied from 0.4 mm for stellacyanin to 2 mm for laccase. It was particularly important to study azurin in dilute solution as it has the tendency to aggregate in the absence of glycerol' (25). X-band ESE experiments were carried out at liquid He temperatures on an instrument described elsewhere using a tunable cavity (25). This cavity is the same as that shown in reference 13 with the addition of a screw mounted in the side of the cavity block and projecting near one end of the dipole resonator. The screw can be rotated from the top of the helium Dewar flask, thus varying the capacitive load on the resonator and changing its resonance frequency. This cavity was adjusted to the same resonance frequency for all samples. When performing three-pulse experiments, the microwave phase of the first two transmitter pulses was reversed in alternate spin-echo cycles to eliminate glitches in the envelope caused by the passage of unwanted two-pulse echoes (26) through the boxcar gate.2 Data from individual scans were digitized on a Nicolet Computer (1074; Nicolet Instrument Corp., Madison, WI) and transferred to a VAX computer for further processing and Fourier transformation (27). Two recordings of the electron spin-echo envelope were made, one with a sample prepared in aqueous medium consisting of normal water (99.98% H20) and the other on a sample prepared in a D20 medium. The recorded envelope for the second (D20-exchanged) sample was then divided by the envelope for the first (H20) sample to detect the changes brought about by the H20/D20 exchange. The rationale for the dividing procedure stems from a theorem that states that the envelope modulating function when a number of nuclei are all coupled to the same electron spin is given by the product of the modulation functions due to each individual nucleus (28, 29). Thus Vmod (II I212. IN) = Vmjd(II) X VmOd (I2) X... X VmOd (IN)* (1) At first it might seem from this theorem that the envelope dividing procedure would succeed in eliminating completely all contributions other than those due to the deuterium nuclei and the replaced protons. Thus, by Eq. I... V.Od(II, I2 ID IN)/ VmOd VI1, I2 IH ***IN) = V. (ID)/Vmjd (IH) (2) The product theorem (Eq. 1) is not, however, strictly applicable to the frozen solution materials, which we are studying, since any observed modulation function is really the average of many modulation functions, each one corresponding to a different orientation of the paramagnetic complex. The theorem would be rigorously true only if it could be shown that, for the system concerned, the average of the product was the same as 'See reference 25, Fig. 6. 2The "deglitching" procedure was suggested by Dr. M. K. Bowman. 3The product formula assumes a somewhat less simple form in the case of three-pulse echoes (29). Because the product formula is in either case not rigorously true for frozen solution samples, this difference need not concern us. the product of the averages.4 The envelope dividing procedure is, nevertheless, a useful first step, and largely eliminates information that is irrelevant to the present measurement, such as modulation due to '4N nuclei (13, 14, 16, 17). Residual 'H modulation in the quotient (as predicted by Eq. 2), and other unwanted effects, such as those arising from small differences in the phase memory times of the two samples compared, can be removed by high- and low-pass filtering to leave a quotient function that clearly displays the effect of 2H coupling to the paramagnetic center. The depth of the deuterium modulation pattern was determined by fitting a cosine function of the appropriate period to a selected cycle in the quotient curve and by calculating the quantity d = 1/2-1/4 [y(trough A) + y (trough B)]/y(peak), (3) where y is the ordinate of the fitted function, y (trough A) and y (trough B) are its values at adjacent minima, and y(peak) is the value of the function at the maximum between. Alternatively, one may calculate the quantity d = 1/2- y (trough)/ [y (peak A) + y (peak B)], (4) where y(peak A) and y(peak B) are values at adjacent maxima and y (trough) is the value at the minimum between. The parameter d is half the peak-to-trough amplitude of the cosine modulating function divided by the peak value (or mean peak value) of the echo envelope quotient for the selected cycle. The depth measurements obtained in Eqs. 3 and 4 might, in principle, be interpreted by comparing them with depth measurements made on simulated electron spin envelopes. However, because of the complexity of the protein-aquo system, and because of the additional difficulty introduced by the deuterium quadrupole moment, this approach would require numerous elaborate computer simulations that would be expensive and possibly unconvincing. We have, therefore, tried the following simplified procedure. Measurements on the protein samples were followed by measurements of exactly the same type on hydrated metal ion complexes that were designed to serve as magnetic model systems. The deuterated forms of the complexes were prepared with a mixture of D20 and H20 so that they gave deuterium modulation depths of the same order as those found in the proteins. By considering the modulation depth as the sum of depths due to many deuterons, those near and those far away, we were thus able to make a series of direct comparisons between the proximity of water molecules in the proteins and the hydrated complexes. To go beyond this point and assign numerical values to the water distances in the proteins, it is necessary to know the structure of the hydrated metal ion complex, including the outer-sphere structure, in some detail. Here again, one has the option of using echo envelope data to solve the structural problem by fitting observed data to computer simulations, and in this case we have done so, because the situation is much simpler for hydrated complexes than for proteins. Furthermore, we have been able to avoid the difficulty of the deuteron quadrupole moment by taking the distance information from an earlier study made on hydrated complexes containing no D20 (21). There remains, of course, a question as to whether the water molecules are oriented towards the metal ion in the protein active site in the same way as they are towards the metal ion in a hydrated complex. If they are, then the hydrogen nuclei in the protein are t0.8 A further away from the metal ion than the centers of the associated water molecules. The general conclusions reached in this paper can, to a large degree, be inferred from a study of the figures, in particular Figs. I and 2, which show that according to some appropriate weighted average, deuterium 4A further complication arises in three-pulse experiments, where the product formula contains two terms. See reference BIOPHYSICAL JOURNAL VOLUME
3 s C STELLACYANIN 3-pulse LD2O A W~~~ ~B H20 rl146ns * S S ; ~~~~~'l4j; i., s Aqm.... I. A.Ii.' a-;e 1 45l :,! i. - - lxb -: J {k, b;!1 -, w- i i 0--.n-;:.^3< :i "1 i ^ '*,l..s 4'* 5 3 *.:. vs.4,!, f : t; *-.qll I **---v T S -~~~~~~~~~J l' d vl-o.4 *,.,_..I ~~4. - :s ~ s ~ z~ n F*^ - ; ^ s ',. -14, T+r(As) T r (ps) FIGURE 1 (Upper left) Two-pulse electron spin-echo envelopes for stellacyanin. (Upper right) Three-pulse electron spin-echo envelopes for stellacyanin obtained with r = 146 ns. (Lower left) Three-pulse electron spin-echo decay envelopes for stellacyanin obtained with r = 220 ns. (Lower right) Three-pulse electron spin-echo decay envelopes for laccase obtained with T = 220 ns. Curves A show the ESE envelopes for copper proteins after exchange against D20. Curves B show envelopes obtained for the same samples before D20 exchange. The modulation component due to exchanged deuterium can be seen in the C curves, which were obtained by dividing a B curve into an A curve. Measurements of the peak-to-trough amplitudes in the deuterium modulation pattern are used to estimate how close the exchanged D20 molecules and deuterons are to the Cu(II) active site. All data was taken at liquid helium temperatures with a spectrometer frequency of 9,507 MHz and a magnetic-field setting of 3,293 G. atoms are about as near to the active site in copper proteins, which have been exchanged against D20, as they are to the metal ions in aquo complexes where every sixth hydrogen is a deuteron. However, a much more careful analysis is needed to extract numbers and to provide a basis for discussing the active site geometry. In the next section we give a detailed description of this analysis. Because this is the first study of its type and since certain results disagree with those obtained in single crystal x-ray crystallographic measurements (8), we have discussed at some length the assumptions and approximations on which our argument is founded. A-*- r4--=.- U T - t FIGURE 2 (Upper eft) Two-pulse electron spin-echo decay envelopes for Cu(II)-aquo complex. (Upper right) Three-pulse electron spin-echo decay envelopes for Cu(II)-aquo obtained with r = 220 ns. (Lower left) Two-pulse electron spin-echo decay envelopes for Nd(III)-aquo. (Lower right) Three-pulse electron spin-echo decay envelope for Nd(III)-aquo obtained with r = 220 ns. Curves A show the ESE envelopes for partially deuterated aquo complexes of 2.5 mm Cu(II) and Nd(III) in which the H20/D20 ratio was 5:1. Curves B show envelopes obtained for wholly protonated complexes. The modulation component due to deuterium can be seen in the C curves, which were obtained by dividing a B curve into an A curve. All data was taken at liquid helium temperatures with a spectrometer frequency, 9,507 MHz and a magnetic-field setting of 3,293 G. RESULTS AND ANALYSIS Echo Envelope Data for Copper Proteins Fig. 1 upper left shows two-pulse echo envelope data obtained for stellacyanin; the upper right and lower left sections of Fig. I show the corresponding three-pulse data with the pulse I to pulse II times T set at 146 and 220 ns, respectively. The lower right section of Fig. 1 shows the three-pulse data for laccase, with r = 220 ns. Curves A in MIMS ET AL. Accessibility of Type I Cu(II) Centers 757
4 Fig. 1 were obtained with the D20 exchanged sample. Curves B were obtained with the sample in a normal H20 medium and curves C show the function obtained by dividing the B curves into the A curves. Similar results were obtained for decupro laccase and for azurin. In Fig. 1 the data have been processed by a low-pass filter with a cut off at 12 MHz. The low-pass filter eliminates some noise and also most of the 'H modulation that remains after the dividing procedure. It had little effect on the three-pulse data because the signal-to-noise ratio of the raw data was good, and most of the 'H modulation had already been eliminated by setting r equal to a whole number of 'H nuclear cycles (frequency suppression effect).5 The C curves in Fig. I have also been processed by a high-pass filter with cut off below 0.2 MHz, the direct current (dc) level being subsequently restored to show the relative magnitude of the deuterium modulation. High-pass filtering is useful when preparing the figures because it removes upward or downward drifts in the quotient function that occur whenever there are small differences between the phase memory decay of the A and B envelopes. However, the procedure of high-pass filtering and dc restoration can lead to errors and was not used when making the numerical determinations of the modulation depth, d, shown later in Table I. The problem of fitting a cosine wave in the presence of an upward or downward slope in the quotient function was solved in this case by adding a linear term to the fitting model function. All data throughout this study were taken with the magnetic field, Ho, equaling 3,293 G and the microwave frequency, f, equaling 9,507 MHz. Other instrumental settings (for example the microwave pulse recurrence rate) were, as far as possible, kept the same to facilitate comparisons between different curves and to minimize errors. Note that the C curves in Fig. 1 all show clearly the characteristic 2H-modulation pattern (f = 2.14 MHz, period = 465 ns) thus indicating the presence of exchangeable water or hydrogen near the Cu(II) site in the proteins. Also note that the r = 220 ns three-pulse stellacyanin data in the lower left-hand corner of Fig. 1 show the 2H modulation more clearly and with less interference than the two-pulse data in the upper left-hand section or the three-pulse data taken at a different T setting in the upper right. The same was true for laccase, decupro laccase, and azurin. The reasons why these data are best are easily identified. Three-pulse data tend to yield better results because there is less phase memory decay. Moreover, at the particular setting, r = 220 ns, there is a fortunate conjunction of numbers involving the frequency suppression effect (see footnote 5). The 'H component is largely eliminated because 220 ns is approximately a whole number of proton periods. (At 3,293 G, the 'H-NMR frequency is 14 MHz and the period 71.3 ns.) Also the 1.4-MHz component due to '4N in an imidazole ligand of Cu(II) (30) is largely 'See reference 25, pp suppressed.6 The latter is an important advantage because the 1.4-MHz frequency lies close to the 2H frequency and would otherwise be a source of interference in the measurement (see, for example, the spectra in Fig. 8). Finally, 220 ns is close to one-half period of the 2H NMR frequency, a setting at which one would expect the 2H modulation to be maximized. The experimental curves in Fig. 1 demonstrate qualitatively the presence of exchangeable hydrogen near the Cu(II) site in the proteins, but they do not directly convey information regarding the number of hydrogen atoms and their location. A more detailed interpretation might be attempted by computing a number of envelope functions for trial geometries (31). Indeed, because the theory of the modulation effect involves very few approximations and is, for practical purposes, an exact theory, one could in principle compute any given envelope function if all the parameters of the problem were known, including the distances and orientations of all water molecules and of other hydrogen containing species. There are, however, too many unknows for the method of trial simulation to appear promising, and we shall look for a simpler, if more approximate, analytical framework. As an aid in developing a suitable analysis and in order to have a set of standard curves for comparison, we have made echo-envelope measurements on two well characterized aquo complexes in H20, and in mixed H20/D20 solutions, using the same instrumental settings as those used in the protein experiments. Echo-Envelope Data for Cu(II)/Aquo and Nd(III)/Aquo Complexes Our first choice as a standard of comparison was the Cu(II) aquo complex formed by dissolving hydrated CuS04 in H20 or in a mixture of H20 and D20. An equal quantity of glycerol was added to the solution to promote the formation of a glass when frozen. Some results obtained with Cu(II)/aquo samples are shown in Fig. 2. As in Fig. 1 the A curves are echo envelopes recorded for a deuterated sample, prepared here by mixing H20 and D20 in a 5:1 proportion. The B curves show equivalent data obtained for a sample prepared with H20 only, and the C curves show the function obtained by dividing an A curve by a B curve. High-pass and low-pass filtering was applied as in the case of curves C in Fig. 1. When comparing the upper left and upper right sections of Fig. 2 with the corresponding protein data in Fig. 1, note that the Cu(II)/aquo sample gives results that are somewhat different from those obtained with the copper proteins. The decay of the modulation pattern is too rapid, and in the two-pulse case, there is a prominent secondharmonic component. The reasons for this rapid decay will be discussed shortly. Meanwhile, we present in the lower 6See reference 25, Fig. 14, p BIOPHYSICAL JOURNAL VOLUME
5 left and lower right sections of Fig. 2 the equivalent data obtained for the Nd(III)/aquo complex formed by dissolving hydrated NdCl3 in H20 or in a H20/D20 mixture. Here again an equal volume of glycerol was added to promote glass formation, and the ratio of H20/D20 in the deuterated sample was 5:1. The results obtained with this sample resembled the protein data more closely and the Nd(III)/aquo complex will, therefore, be used in the analysis as the preferred standard of comparison. The Nd(III)/aquo complex is not, of course, proposed as a chemical model but only as a magnetic model in which the interactions between deuterons and the unpaired electron spin are of a similar order of magnitude. Modulation Depth as a Source of Information A rough idea as to the nearness of the exchangeable hydrogen nuclei in the copper proteins can be obtained by comparing the ratio plots (C curves of Fig. 1 for these proteins with the corresponding ratio plots for the standards (C curves of Fig. 2). Thus, for example, the deuterium modulation depth observed for laccase is about the same as that observed for Cu/aquo, H20/D20 = 5:1, (Upper right of Fig. 2) and is somewhat deeper than that observed for the Nd/aquo standard sample. We might, therefore, conclude that the active site in deuteriumexchanged laccase is coordinated by one D20 molecule, just as is the Cu(II) center in the Cu/aquo standard (in which one molecule out of six is D20). A rudimentary interpretation along these lines cannot, however, be easily extended to include the stellacyanin or azurin data, which show a modulation pattern appreciably shallower than that obtained for either of the standards. Moreover, as we shall see, the conclusion reached in this way for laccase is inconsistent with the observed modulation decay rates. We must, therefore, consider the origin of the modulation patterns for frozen solution samples in greater detail, both to extract the information contained in the decay rate of the deuterium pattern and to see how ambient water can contribute to the observed result. The envelope modulation patterns due to coupling between the electron spin and one of the surrounding nuclei have the form V2p = 1-3/4d2 + d272(t)cos w- - 1/4d2n2'(r) cos 2 wi (5) V3P = 1 - d3n3(r')(1 - cos WT'). (6) Eq. 5 refers to two-pulse experiments and Eq. 6 to threepulse experiments. The time T' = T + T is the total time between the second pulse and the stimulated echo, and w/2 ir is the nuclear frequency. The depths d2 and d3 in two-pulse and three-pulse experiments are not the same because the frequency suppression effect (see footnote 5) enters into d3 and the effects of averaging over many orientations of the complex may differ in the two cases. According to theory, the depth of modulation for weakly dipolar-coupled nuclei (i.e., those for which the nuclear Zeeman energy exceeds the coupling energy) is proportional to the inverse sixth power of the radial distance.7 Thus, for one nucleus we can write d = al/r, (7) where a is a constant depending on the spin and magnetic moment of the nucleus, the g-value of the electron to which it is coupled, and the Zeeman field setting. The constant a also includes the effect of averaging over various orientations of the complex with respect to the Zeeman field, and in a three-pulse experiment it depends on the setting r (which determines the magnitude of the frequency suppression effects [see footnote 5]). Let us suppose that we have two deuterons coupled to the same electron at distances r, and r2. Then according to the product theorem (see Eq. 1), the resulting deuterium modulation pattern would have the form V12 = I - (d, + d2)(i - cos wir') + d1d2(1 - cos wr')2, (8) where d, = a/r,6 and d2 = al/r26. However, in the experimental data the modulation amplitude due to all of the deuterium nuclei is only of the order of 10-20% (dtotal 0.1), which suggests that higher order product terms, such as the term in d1d2 above must make a relatively small contribution. We shall, therefore, drop terms in higher powers of d, and assume that, for the present purpose, modulation depths can simply be added. (A precise accounting for small terms is, in any case, not likely to be useful here because as pointed out earlier, the product formula is not accurate for averages over many orientations of a complex.) This approximation makes it easy to take into account those cases in which a number of like nuclei interact with the same electron spin. Thus, for example, if a paramagnetic ion is surrounded by n nuclei all at the same distance, then the resulting modulation depth, d, is given by (see footnote 7) d = an/r6. (9) The contribution to the modulation depth due to the deuterons in ambient water can be found by integrating a/r6 over a suitable geometric domain. Consider, for example, the geometry in Fig. 3 a and suppose that deuterons are distributed uniformly, with density p, throughout a volume extending from a sphere of closest approach, with radius rmn, out to infinity. Integrating Eq. 7 we obtain the result d(rmin, oo) = 4 irpa/3r.,in (10) for the total modulation depth. This formula might be used to describe a case in which a paramagnetic ion was 7See reference 21, Eqs MIMS ET AL. Accessibility of Type I Cu(II) Centers 759
6 -- FIGURE 3 Schematic representations of a paramagnetic ion in aqueous medium, partially exposed to solvent or totally embedded in a protein. a represents a hydrated ion in an aqueous medium. The depth of the hydrogen modulation in the ESE envelope can be calculated by integrating contributions due to nuclei lying in the volume that extends from a minimum radius r.jn to r = o. A correction is needed to account for the orientation of water molecules in the first coordination sphere. b, c, and d are schematic representations for an ion partially exposed to aqueous solvent. These geometries can be used to model a protein in which the metal ion is (b) in a crevice, or (c) on the surface, or (d) below the surface of the protein. See Eq immersed in a uniformly deuterated medium, rmin being determined by the radius of the ion and by the geometry of the molecules constituting the deuterated medium. (A correction for nonuniformity of the deuteron distribution is considered later.) The result for the geometry shown in Fig. 3 b, in which the deuterated medium is excluded from all but a solid angle,,6, follows immediately. Thus d(rmin, 0, 0) = 4.pal3rmin (11) The Fig. 3 b geometry might be used to model a protein containing a crevice that exposes the active site to the surrounding (deuterated) solvent. Other simple geometries are shown in Fig. 3 c and d. In c and in d d(rmin, a, 27r) = 2 7rpa/ 3r3,j (12) d(z) = 7rpa/6z3. (13) The geometries in Fig. 3 c and d might be used to model a protein in which the active site lies on or near the surface. If the top of the sphere of closest approach is level with the surface, then z = rmin, and d(z = rmin) = rpa/6rm3 (14) Results for many other simple geometries can be derived 760 similarly. Individual deuterons at a distance, rd, can also be taken into account by adding contributions d(rd) = a/rd6 to the overall depth. Expressions such as Eqs enable one to compare the depths of modulation expected for different hypothetical geometries of the protein active site without making a detailed simulation in each case. They also enable one to relate measurements made on protein samples to equivalent measurements made on the standard, and, hence, if the geometrical arrangement of deuterons about the paramagnetic ion is known for the standard, to estimate the distribution of deuterons in the protein. Essentially what one does here is to find the ratio between the modulation depth observed for the protein and the modulation depth observed in the same modulation cycle for the standard. This is most conveniently done by deriving a value of the proportionality constant a from the measurements on the standard and then using this value of a to interpret the protein data. To proceed with this calculation of a we require a formula giving the modulation depth for the Nd(III) aquo complex. Unfortunately, we cannot use Eq. 9, with rmin as the ionic radius, because the hydrogen nuclei are not uniformly distributed in the first coordination sphere, the water molecules being oriented with the oxygen atoms closest to the Nd(III) ion and the oxygen to hydrogen bonds directed away from it. There is also the problem that first sphere coordination is by water, whereas the outer matrix consists of an equal mixture of water and glycerol. The distribution of hydrogen nuclei in the Nd(III)/aquo complex is, in fact, more closely approximated by the geometrical arrangement shown in Fig. 4 b than by the simple uniform distribution in Fig. 4 a. By comparing the two figures it can be seen that the effect of water molecule a FIGURE 4 Two models for a metal ion in an aqueous solvent. (a) Uniform distribution of hydrogen atoms beyond an effective inner radius rmin. (b) First coordination sphere considered separately to account for the orientation of water molecules in the electrostatic field of the ion. The first sphere for the Nd(III) solvated ion contains nine water molecules. Comparisons between observed ESE envelopes and computer simulations b 0.8 A greater than have shown that r. = 3.0 A for this ion. This radius is the value of rmin obtained by fitting ESE data to model a by Eq. 10. The possibility of water molecule orientation is allowed for in the present work by adding 0.8 A to the values of rmin derived by Eqs The adjusted radii are taken as the distance of the nearest hydrogen atoms. BIOPHYSICAL JOURNAL VOLUME
7 orientation in b is to "compress" a portion of the uniformly hydrogenated medium shown in a into a thin spherical shell with radius rz,. The radius, rcs, in b is larger than the - rmin effective inner radius rn in a, and the difference r,, may be regarded as a correction applied to rmin in order to allow for water-molecule orientation. The difficulty of making a reliable analysis for this more complicated model can be bypassed by using the results of some earlier work in which the experimentally observed proton-modulation pattern for Nd(III)/aquo with the complex embedded in a glycerol/water matrix was simulated for a variety of Nd-H distances (32). In this earlier work it was shown that the echo-envelope data can be matched by assuming that in the first coordination sphere, 18 hydrogen nuclei are distributed about the Nd(III) ion at a distance of 3.0 A. Substituting r = r, = 3.0 A in Eq. 9 and allowing for the fact that in our standard sample one in six protons are replaced by deuterium (i.e., setting n = 3), we find that the modulation depth due to the first coordination sphere d(first sphere) = 4.12 x 1O-3 a A-6 units. (15) To find where the first sphere ends and the outer water/glycerol matrix begins, i.e., to find the radius rmat in Fig. 4 b, we assume that the volume between the ion (radius = ri) and the matrix is equal to the volume of nine water molecules. Substituting r, = 1.04 A (33) for Nd(III) and assuming a density of 1.0 for water, we obtain an inner radius rmat = 4.02 A for the matrix. The density p of deuterium nuclei in the 5:1 H20/D20 solution, mixed 1:1 with glycerol, is 5.61 x 10-3 per cm-24. Assuming a uniform distribution of deuterons in the matrix and applying Eq. 10 with rma, = 4.02 A, we find that the modulation depth due to the outer matrix d(outer matrix) = x 10-3 a A-6 units. (16) Summing Eq. 15 and Eq. 16 we have d[(nd(iii)/aquo standard)] = 4.48 x 1O-3a A-6 units (17) (By comparing Eq. 15 and Eq. 16 it can be seen that the very approximate assumptions regarding the structure of the outer matrix of water and glycerol do not have a large effect on the result.) If one assumed that the outer matrix were wholly water and not 1:1 water/glycerol (but still with H20/D20 = 5:1), then the depth computed in Eq. 17 would have been 4.84 x 10-3 aa-6 units. The same number could also have been obtained by using the simpler model in Fig. 4 a and by substituting rmin = 2.16 A in Eq. 10. The difference of =0.8 A between the radius of the first coordination sphere, rc,, = 3.0 A, in the Fig. 4 b model and the corresponding inner radius, rmin = 2.16 A, according to the Fig. 10 a model is, therefore, the correction that must be added to rmin to arrive at the distance of the nearest hydrogen nuclei when one wishes to allow for water molecular orientation. Modulation Decay as a Further Source of Information Although we shall not attempt here to make any kind of numerical analysis based on the modulation decay rate, the decay function is, nevertheless, a valuable qualitative source of information. First we review the manner in which the decay factors %2(r), n'2(r) and m(r') originate. The factors v start from unity at time zero. They correspond to the time transform of a lineshape function, which describes the broadening of the NMR line due to the proximity of the electron, and they would be calculable if one knew the detailed geometrical arrangement in advance.8 (The lineshape calculation must here also include weighting factors specifying the depth of modulation for each orientation of the complex.) The broadening for nuclei lying some distance away from the electron is quite small and the decay for these nuclei is relatively slow, whereas for close lying nuclei the broadening is considerable and the modulation depth falls off substantially in one or two cycles. For any particular set of experimental data, the line-broadening function associated with the decay of modulation depth can be obtained by taking the Fourier transforms of the functions in curves C of Figs. 1 and 2. (See for example the frequency spectra in Fig. 5.) From the above it can be seen that the modulation decay function offers a potentially useful means of discriminating between near nuclei and remote nuclei. However, there is an additional factor to be considered in the case of deuterium. The 2H nucleus has a small nuclear quadrupole moment that interacts with the electrostatic fields in water and in other molecular systems commonly found in biological materials. In zero field, the quadrupolar interaction for D20 corresponds to a line splitting of = 160 KHz (34). This is too small a perturbation to split the deuterium lines observed here into components, but it contributes to the broadening in a way that is independent of the electronnuclear distance. Detailed calculations, taking the quadrupolar interaction into account, have been made by Shubin and Dikanov (35). From the present point of view the main effect of the quadrupolar interaction is to obscure the distinction between remote and near nuclei and to make it more difficult to use the decay rate to distinguish between them. In spite of this difficulty some useful conclusions can be drawn from a comparison between the protein data and the Cu(II)/aquo data, the most striking difference being in the two-pulse decay envelope, which clearly shows a second harmonic in the Cu(II)/aquo case. This harmonic is generated by the sum of the two nuclear frequencies associated with the electron spin-up and spin-down orientations. It decays much more slowly than the fundamental at w because the two nuclear transitions undergo opposite and approximately equal shifts in the field of the electron, 8See reference 32, Fig. 6. MIMS ET AL. Accessibility of Type I Cu(II) Centers 761
8 so that the resultant broadening of the 2 w harmonic is small in comparison with the broadening of the fundamental (see footnote 8). It is, therefore, often easy to see this harmonic in the later part of the two-pulse time waveform. For 2H, the difference between the two-pulse decay functions, 72(T) and p2(r), is much less marked since the quadrupolar shift is independent of the electron-spin orientation and actually contributes twice as much broadening to the second harmonic as it does to the fundamental. As a result, the second harmonic is not always easy to see in the two-pulse time waveform for deuterium. However, it is clearly visible when the electron-nuclear coupling is significantly larger than the quadrupolar shift. Curve C in the upper left section of Fig. 2 shows that this is the case for the Cu(II)/aquo complex. It is, on the other hand, not true for stellacyanin (Curves C in Fig. 1) nor for laccase, decupro laccase, and azurin. We, therefore, conclude that the electron-nuclear coupling is weaker and the hydrogen nuclei are further from the Cu(II) ion in the proteins than they are in the Cu(II)/aquo complex. The three-pulse data tell a similar story, although the distinction is less immediately obvious than in the two-pulse case. The function p3(r') decays less rapidly for the Cu(II) proteins than for the Cu(II)/aquo complex. (The line broadening associated with the three-pulse decay factors can be seen in Fig. 5.) For the Nd(III)/aquo complex the decay rates also differ from those found in the copper proteins. The discrepancy is smaller, however, suggesting that the deuteron distances in the protein approximate more closely to those found in Nd(III)/aquo than to those found in the Cu(II)/ aquo complex. Derivation of the Modulation Depth from Envelope Data Although the decay functions can be useful as a source of additional information, they constitute a major practical difficulty when attempting to measure the depth of modulation because (a) we are only able to record the echo envelope after a certain dead time has elapsed and cannot see all of the first deuterium modulation cycle, and (b) we do not know the exact forms of the decay function for the materials concerned. It is, therefore, not possible to measure the true depth directly nor to make a reliable extrapolation to zero time. Approximate extrapolations can be attempted as, for example, when deriving Fourier cosine transforms9 (Fig. 5). However, in the present experiments we avoided the uncertainties of data extrapolation when determining the modulation depth and based the measurement solely on the observed portion of the envelope. To minimize errors due to decay, we made the measurement on the first available complete cycle containing the neces- 9See for example reference 25, pp Azurin lr *l46 ns MHz Stellocyanin r * 146 ns MHz MHz 6 Azurin r-220ns MHz Stellacyanin r * 220ns M Hz Nd (M1)-aquo Tr 220ns MHz FIGURE 5 Cosine Fourier transforms of curves obtained by taking the ratio of three-pulse electron spin-echo envelopes for deuterated and nondeuterated samples. The ratio curves (see curves C in Figs. 1 and 2) were reconstructed through the dead-time region before being transformed. The r = 146 data show some residual contamination due to "N. The r = 220 data were therefore used in all measurements of the modulation depth. The deuterium peak (2.14 MHz) is narrower for the Cu(II) proteins than for the Cu(II)/aquo and Nd(III)/aquo models. This indicates that the deuterium nuclei are somewhat further away from the metal ion in the proteins than in the models. sary maximum and minimum points. In the case of the = 220 ns three-pulse data, this measurement covers the time range between the two peaks at r' = 465 ns and at T' = 930 ns, Eq. 4 being used to find the depth, d. Results are given in Table I. Measurements made one-half a deuterium cycle later, between the two troughs at r' = 697 ns and ' = 1,162 ns are also shown in Table I for comparison (depth computation by Eq. 3). The errors that arise as a result of decay are partially eliminated by referring the protein data to the data obtained with the standard sample, and they would cancel if the decay rates for the two were identical. However, note that from the last column of Table I in which the ' = ns and the T' = 697-1,162 ns measurements are compared, the decay rates do in fact differ to some extent. Fortunately, the nuclear distances vary only as the cube root (Eqs ) or sixth root (Eq. 9) of the modulation depth, thus reducing the significance of any errors. Substituting the observed depth parameter da = (Table I) for the Nd(III)/aquo complex in Eq. 17, we BIOPHYSICAL JOURNAL VOLUME
9 TABLE I DEPTH PARAMETERS FOR THE DEUTERIUM MODULATION PATTERNS OBSERVED IN THREE-PULSE ECHO EXPERIMENTS FOR FOUR PROTEIN SAMPLES AND FOR TWO STANDARD INORGANIC COMPLEXES Depth Depth Ratio Sample parameter parameter da/db da db Stellacyanin Azurin Laccase Decupro laccase Nd(III)/aquo Cu(II)/aquo The Nd(III)/aquo and Cu(II)/aquo samples were prepared with H20/ D20 = 5:1, and with a 1:1 water/glycerol ratio. All samples were measured at a magnetic field H. = 3,293 G, a microwave frequencyf = 9507 MHz, and X = 220 ns. The depth parameters da and db are half of the peak-to-trough amplitude divided by the peak amplitude. The parameter da is measured by fitting the deuterium modulation pattern between 465 and 929 ns (see Eq. 4), and the parameter db by fitting the pattern one-half cycle later between 697 and 1,162 nsec (see Eq. 3). The ratio da/db shows the decay of the modulation pattern between these two measurements. obtain the value a = 24.0 A6 units (18) for the proportionality constant occurring in Eqs With this value the numerical relationships between the modulation depth and distances, rmin, z, for the geometries in Fig. 9 become for a to c in Fig. 3 and da(r = rmin to r = oo) = t/rin (1 9) da(z) = 0.85/z3 (20) for d in Fig. 3. For curve A in Fig. 1 = 1, for curve B r = i,/4ir (i.e., the fraction of the sphere corresponding to the crevice), and for curve C r = 0.5. The parameter p, i.e., the number of hydrogen nuclei per unit volume in water, was taken to be 6.34 x Io-2 per (A)3 when deriving Eqs. 19 and 20 from Eqs The distances rmin and z in Eqs. 19 and 20 are in angstrom units, and the depth parameters refer to the modulation cycle extending from 465 to 929 ns, which was the cycle used to find a. Note that our procedure for measuring the modulation depth is not the only possible one, nor is it the best one in all cases. Measurements of the area under the MHz peak in the Fourier transform ratio plot (see Fig. 5) and comparison with a standard might be a more practical approach in cases where it is difficult to isolate the deuterium modulation pattern from other spectral components (for example, if it were necessary to make the measurements at the -r = 146 setting rather than the r = 220 setting [see Fig. 5]). Comparison of the areas under the peak requires the introduction of some questionable steps in the analysis, however. If a cosine Fourier transform is used to derive the line spectrum, then a zero time extrapolation must first be made, and the accuracy of the result depends on the success of the extrapolation. If Fourier power transform methods are used (36), then differences in the decay function must be allowed for, and the height of the time waveforms above the baseline has to be determined to arrive at the relative intensity of the line. Because we had a good signal-to-noise ratio and could (by using the r equals 220 ns three-pulse setting) avoid serious spectral contamination, we preferred to make the measurement directly on the time waveform. The effects of decay could then be minimized by selecting an early modulation cycle, and the measurements could be made on a function that was related in a clear and obvious manner to the experimental data. DISCUSSION Modulation depths; da, for the deuterium cycle between 465 and 929 ns (with a trough at 697 ns), as observed in three-pulse experiments with r = 220 ns, are shown in Table I and will be used in this discussion. Note that (a) laccase and decupro laccase, where Type 2 Cu(II) had been reversibly removed, give approximately the same result (b) the modulation depth for laccase and decupro laccase is almost twice that for the other two copper proteins, and (c) the modulation depth for laccase is approximately the same as that for the 5:1 H20/D20 Nd(III)/aquo complex. As pointed out earlier, the slow decay rate of the modulation pattern, and the lack of a strong second harmonic in the two-pulse curve suggests that the large modulation depths observed for laccase and the other copper proteins are not due to a single, close lying exchangeable D20 molecule in the primary coordination sphere of the Cu(II) ion (nor, for the same reason, to a close lying deuteron belonging to the protein). We must, therefore, turn to other models to explain the observed modulation and consider, by the methods developed in the preceding section, the contributions due to deuterons in the ambient D20 medium. Let us first consider the hemispherical geometry of Fig. 3 c, i.e., let us set = 0.5 in Eq. 19. Substituting the observed modulation depth d = for laccase, we find that the effective distance of closest approach rmin is 2.85 A. This effective radius is calculated by assuming a uniform distribution of deuterons and by ignoring possible orientations of the oxygen atoms in D20 towards the Cu(II) ion. With orientation taken into account, the situation would tend to resemble that which is assumed for the Nd(III)/ aquo complex and the distance of the nearest deuterons would be larger than rmin. A precise calculation is not possible unless we know the actual orientations of the D20 molecules, which are seen by the Cu(II) active site in laccase. However, assuming that the correction for orientation of the nearest water molecules is 0.8 A as in Nd(III)/ MIMS ET AL. Accessibility of Type I Cu(II) Centers 763
10 aquo (see Discussion following Eq. 17), then the nearest deuterons in laccase, according to the hemispherical model, are estimated to lie at 3.7 A. (A recalculation of the orientation correction for a hypothetical complex with radii larger than those in Nd(III)/aquo would result in a smaller correction, and hence a distance slightly less than 3.7 A.) For the crevice geometry of Fig. 3 b, r < 0.5 and the resulting estimates for distance would be shorter. Thus, for example, if the solid angle +V = ir (D = 0.25), then by Eq. 19 the estimate of rmin for laccase becomes 2.26 A. Adding 0.8 A to allow for orientation of the water molecules, we estimate that for this model, the nearest deuterons are at 3.1 A. This distance, which is comparable with the distance in the Nd(III)/aquo complex would also be consistent with the observed modulation decay rate. Obviously there is a trade off between distance and the solid angle of exposure to the environment. However, certain models would be difficult or impossible to reconcile with the data. The embedded model of Fig. 3 d would, by Eq. 20, require an effective minimum distance z = 1.80 A. Adding an extra 0.8 A for the water-molecule orientation effect would yield an estimated distance 2.6 A for the nearest deuterons. For deuterons at this distance, the decay rate of the modulation pattern would be too fast and the echo envelope would resemble that obtained for the Cu(II)/aquo complex, in contradiction to the observed result. If the Cu(II) center were buried more deeply in the protein, the decay rate would be slower and would resemble the observed decay rate more closely, but the modulation would be too shallow. A substantially larger value for v is not compatible with the experimental results at all. Thus, for instance, a 1I-A increase in the estimated distance of the nearest deuterons (corresponding to a I-A increase in z) would reduce the modulation depth by a factor 3.8: 1; a 2-A increase would reduce the depth by 9.4:1. Note also that the exchange of deuterium for 'H on the remote nitrogen atoms of imidazole ligands to Cu(II) ( 1017) would produce a relatively minor effect. Substituting R = 4.16 A and a = 24.0 (A)9 (Eq. 18) in Eq. 7, we see that the contribution to the depth da, due to one such deuterated imidazole is The exchange of 2H for 'H on two such imidazole ligands would contribute to da, a quantity that is more than one order of magnitude less than the observed depth. This substitution would have only a very small effect on the overall depth and could not, in any circumstances, explain our observations. It is perhaps no surprise to find that the Cu(II) site in laccase is accessible to water because accessibility to small molecules is a normal requirement for catalytic action. More interesting is the observation that a very similar situation exists for the electron-transfer proteins, stellacyanin and azurin, where the depth parameters are, respectively, 1.62:1 and 1.90:1 less than for laccase. The results for these proteins suggest that (a) the crevice is narrower, or (b) the Cu(II) is more deeply buried, or (c) some combination of the two. Deuteron exchange on the remote nitrogen of imidazole ligands is again excluded as the primary source of the observed modulation pattern because the effect would be an order of magnitude too small. Note that even if we ascribe the whole of the difference between laccase and the electron transfer proteins to increased distance from the surface, the required increase in distance is quite small, as one can see from the cube law dependence in the Eqs. 19 and 20. Applying the cube law we find that the distance for the Cu(II) ion would be 18% greater in stellacyanin and 24% greater in azurin than in laccase. The distance between the active site and the solvent must, therefore, be much the same in all three proteins. The interesting overall result emerging from the analysis is that the metal centers in the Cu(II) proteins examined do not seem buried. They are either in the surface of the protein or in a shallow crevice with access to the surrounding medium. This conclusion is to be compared with measurements based on x-ray crystallographic data for two different Type 1 Cu(II) proteins, plastocyanin and azurin (8, 9). For the former, the point of closest approach of the Cu(II) to external solvent is the edge of the His-87 imidazole ligand, a distance of -6 A from the metal ion to ambient water. For azurin, the distance from Cu(II) to solvent is 7.8 A. This distance has been confirmed by measurements based on proton relaxation data on the same protein in aqueous solution (37). However, substituting the latter value for z in Eq. 20, we find that the corresponding depth, da, for azurin would be , i.e., 43 times less than the observed depth of ESE envelope modulation. How might these points of disagreement be reconciled? First, as regards the x-ray data, it has been demonstrated in numerous cases that protein structure in the crystalline state may not be the same as protein structure in solution. Our measurements suggest that in aqueous medium, the active site opens up and becomes more accessible to its environment than in the crystalline state. Second, in regard to the disagreement with the relaxation data, this could occur because the waters observed in the electron spin-echo experiment are in slow exchange with solvent on the NMR time scale and do not show up in the relaxation experiment. The data also suggest that copper in stellacyanin is more accessible to solvent than the copper in azurin. This is in accord with the findings of Mauk et al. (38) based on their kinetic estimate of distance of electron transfer from metalloprotein redox sites to the electron transfer agent, Fe(EDTA)2-. From their studies, they conclude that the electron transfer distance is -3.7 A for stellacyanin and -5.5 A for azurin. Note that the latter distance, though larger than the distance from Cu(II) to water-molecule hydrogen atoms, as estimated from our data, is significantly less than the 7.8-A measurement from the Cu(II) to the outside of the protein, which is based on x-ray crystallographic analysis. 764 BIOPHYSICAL JOURNAL VOLUME
11 In summary, we conclude from the electron spin-echo measurements that the protein structure in azurin and in other certain copper proteins is more open to solvent than originally believed. Because similar access may be available for other small molecules as well, any discussion of electron-transfer reactions of proteins and small molecules should be reconsidered in the light of these results. That portion of this work carried out at the Albert Einstein College of Medicine was supported in part by the U.S. Public Health Service grant HL to J. Peisach. Receivedfor publication 14 April 1983 and in final form 28 September REFERENCES 1. Malkin, R., and B. G. Malmstrom The state and function of copper in biological systems. Adv. Enzymol. Relat. Areas Mol. Biol. 33: Fee, J. A Copper proteins: systems containing the "blue" copper center. Struct. Bonding. 23: McMillan, D. R., R. C. Rosenberg, and H. B. Gray Preparation and spectroscopic studies of cobalt(ii) derivatives of blue copper proteins. Proc. Natl. Acad. Sci. USA. 71: Solomon, E. I., J. W. Hare, and H. B. Gray Spectroscopic studies and a structural model for blue copper centers in proteins. Proc. Natl. Acad. Sci. USA. 73: Peisach, J., and W. B. Mims The linear electric field effect in stellacyanin, azurin and in some simple copper model compounds. Eur. J. Biochem. 84: Blumberg, W. E Some aspects of models of copper complexes. In The Biochemistry of Copper. J. Peisach, P. Aisen, and W. E. Blumberg, editors. Academic Press, Inc., New York Peisach, J., W. G. Levine, and W. E. Blumberg Structural properties of stellacyanin, a copper mucoprotein from Rhus vernicifera, the Japanese lac tree. J. Biol. Chem. 242: Colman, P. M., H. C. Freeman, J. M. Guss, M. Murata, V. A. Norris, J. A. M. Ramshaw, and M. P. Venkatappa X-ray crystal structure analysis of plastocyanin at 2.7 A resolution. Nature (Lond.). 272: Adman, E. T., R. E. Stenkamp, L. C. Sieker, and L. H. Jensen A crytallographic model for azurin at 3 A resolution. J. Mol. Biol. 123: Markley, J. L., E. L. Ulrich, S. P. Berg, and D. W. Krogman Nuclear magnetic resonance studies of copper binding sites of blue copper proteins: oxidized, reduced and apoplastocyanin. Biochemistry. 14: Ugurbil, K., A. Norton, A. Allerhand, and R. Bersohn Studies of individual carbon sites of Azurin from Pseudomonas aeruginosa by natural abundance carbon- 13 nuclear magnetic resonance spectroscopy. Biochemistry. 16: Hill, H. A. O., and L. Wing-Kai Investigation of the structure of the blue copper protein from Rhus vernicifera, stellacyanin, by proton magnetic resonance spectroscopy. J. Inorg. Biochem. 11: Mims, W. B., and J. Peisach Assignment of a ligand in stellacyanin by a pulsed electron paramagnetic resonance method. Biochemistry. 15: Mims, W. B., and J. Peisach Measurement of '4N superhyperfine frequencies in stellacyanin by an electron spin echo method. J. Biol. Chem. 254: Roberts, J. E., T. G. Brown, B. M. Hoffman, and J. Peisach Electron-nuclear double resonance of stellacyanin, a blue-copper protein. J. Am. Chem. Soc. 102: Mondovi, B., M. T. Graziani, W. B. Mims, R. Oltzik, and J. Peisach Pulsed electron paramagnetic resonance studies of Types I and II copper of Rhus Vernicifera laccase and porcine ceruloplasmin. Biochemistry. 16: Avigliano, L., J. L. Davis, M. T. Graziani, A. Marchesini, W. B. Mims, B. Mondovi, and J. Peisach Electron spin echo spectroscopic studies of Type 1 and Type 2 copper in Rhus Vernicifera laccase and in Cucurbita Pepo Medullosa ascorbate oxidate. FE,BS (Fed. Eur. Biochem. Soc.) Lett. 56: Bergman, C., E. K. Gandvik, P. 0. Nyman, and L. Strid The amino acid sequence of stellacyanin from the lacquer tree. Biochem. Biophys. Res. Commun. 77: Zweier, J., P. Aisen, J. Peisach, and W. B. Mims Pulsed EPR studies of copper complexes of transferrin. J. Biol. Chem. 254: Zweier, J., J. Peisach, and W. B. Mims Electron spin echo studies of the copper complexes of conalbumin. J. Biol. Chem. 257: Mims, W. B., J. Peisach, and J. L. Davis Nuclear modulation of the electron spin echo envelope in glassy materials. J. Chem. Phys. 66: Reinhammer, B Purification and properties of laccase and stellacyanin from Rhus Vernicifera. Biochim. Biophys. Acta. 205: Graziani, M. T., L. Morpurgo, G. Rotilio, and B. Mondovi Selective removal of Type 2 copper from Rhus Vernicifera laccase. FEBS (Fed. Eur. Biochem. Soc.) Lett. 70: Hill, H. A. O., and B. E. Smith Characteristics of azurin from Pseudomonas aeruginosa via 270-MHz 'H nuclear magnetic resonance spectroscopy. J. Inorg. Biochem. 11: Mims, W. B., and J. Peisach Electron spin echo spectroscopy and the study of metalloproteins. In Biological Magnetic Resonance. L. J. Berliner and J. Reuben, editors. Plenum Press, New York. 3: Peisach, J., W. B. Mims, and J. L. Davis Studies of the electron-nuclear coupling between Fe(III) and '4N in cytochrome P-450 and in a series of low spin heme compounds. J. Biol. Chem. 254: Shimizu, T., W. B. Mims, J. Peisach, and J. L. Davis Analysis of the electron spin echo decay envelope for Nd3+:ATP complexes. J. Chem. Phys. 70: Mims, W. B Envelope modulation in spin-echo experiments. Phys. Rev. B5: Dikanov, S. A., A. A. Shubin, and V. N. Parmon Modulation effects in the electron spin echo resulting from hyperfine interaction with a nucleus of an arbitrary spin. J. Magn. Reson. 42: Mims, W. B., and J. Peisach The nuclear modulation effect in electron spin echoes for complexes of Cu2" and imidazole with '4N and '5N. J. Chem. Phys. 69: Kevan, L Modulation of electron spin-echo decay in solids. In Time Domain Electron Spin Resonance. L. Kevan and R. N. Schwartz, editors. Interscience Publishers, Inc., New York Mims, W. B., and J. L. Davis Proton modulation of the electron spin echo envelope in an Nd3+:aquo glass. J. Chem. Phys. 64: Shannon, R. D., and C. T. Prewitt Effective ionic radii in oxides and fluorides. Acta Crystallogr. Sect. B. Struct. Crystallogr. Cryst. Chem. 25: Edmonds, D. T Nuclear quadrupole double resonance. Phys. Rep. Phys. Lett. C: Shubin, A. A., and S. A. Dikanov, The Influence of nuclear quadrupole interactions upon electron spin echo modulation MIMS ET AL. Accessibility of Type I Cu(II) Centers 765
e 2m e c I, (7.1) = g e β B I(I +1), (7.2) = erg/gauss. (7.3)
 Chemistry 126 Molecular Spectra & Molecular Structure Week # 7 Electron Spin Resonance Spectroscopy, Supplement Like the hydrogen nucleus, an unpaired electron in a sample has a spin of I=1/2. The magnetic
Chemistry 126 Molecular Spectra & Molecular Structure Week # 7 Electron Spin Resonance Spectroscopy, Supplement Like the hydrogen nucleus, an unpaired electron in a sample has a spin of I=1/2. The magnetic
An Introduction to Hyperfine Structure and Its G-factor
 An Introduction to Hyperfine Structure and Its G-factor Xiqiao Wang East Tennessee State University April 25, 2012 1 1. Introduction In a book chapter entitled Model Calculations of Radiation Induced Damage
An Introduction to Hyperfine Structure and Its G-factor Xiqiao Wang East Tennessee State University April 25, 2012 1 1. Introduction In a book chapter entitled Model Calculations of Radiation Induced Damage
( ) x10 8 m. The energy in a mole of 400 nm photons is calculated by: ' & sec( ) ( & % ) 6.022x10 23 photons' E = h! = hc & 6.
 Introduction to Spectroscopy Spectroscopic techniques are widely used to detect molecules, to measure the concentration of a species in solution, and to determine molecular structure. For proteins, most
Introduction to Spectroscopy Spectroscopic techniques are widely used to detect molecules, to measure the concentration of a species in solution, and to determine molecular structure. For proteins, most
Electron Spin Resonance, Basic principle of NMR, Application of NMR in the study of Biomolecules, NMR imaging and in vivo NMR spectromicroscopy
 Electron Spin Resonance, Basic principle of NMR, Application of NMR in the study of Biomolecules, NMR imaging and in vivo NMR spectromicroscopy Mitesh Shrestha Electron Spin Resonance Electron paramagnetic
Electron Spin Resonance, Basic principle of NMR, Application of NMR in the study of Biomolecules, NMR imaging and in vivo NMR spectromicroscopy Mitesh Shrestha Electron Spin Resonance Electron paramagnetic
PRACTICAL ASPECTS OF NMR RELAXATION STUDIES OF BIOMOLECULAR DYNAMICS
 PRACTICAL ASPECTS OF MR RELAXATIO STUDIES OF BIOMOLECULAR DYAMICS Further reading: Can be downloaded from my web page Korzhnev D.E., Billeter M., Arseniev A.S., and Orekhov V. Y., MR Studies of Brownian
PRACTICAL ASPECTS OF MR RELAXATIO STUDIES OF BIOMOLECULAR DYAMICS Further reading: Can be downloaded from my web page Korzhnev D.E., Billeter M., Arseniev A.S., and Orekhov V. Y., MR Studies of Brownian
Inorganic Spectroscopic and Structural Methods
 Inorganic Spectroscopic and Structural Methods Electromagnetic spectrum has enormous range of energies. Wide variety of techniques based on absorption of energy e.g. ESR and NMR: radiowaves (MHz) IR vibrations
Inorganic Spectroscopic and Structural Methods Electromagnetic spectrum has enormous range of energies. Wide variety of techniques based on absorption of energy e.g. ESR and NMR: radiowaves (MHz) IR vibrations
ELECTRON PARAMAGNETIC RESONANCE
 ELECTRON PARAMAGNETIC RESONANCE = MAGNETIC RESONANCE TECHNIQUE FOR STUDYING PARAMAGNETIC SYSTEMS i.e. SYSTEMS WITH AT LEAST ONE UNPAIRED ELECTRON Examples of paramagnetic systems Transition-metal complexes
ELECTRON PARAMAGNETIC RESONANCE = MAGNETIC RESONANCE TECHNIQUE FOR STUDYING PARAMAGNETIC SYSTEMS i.e. SYSTEMS WITH AT LEAST ONE UNPAIRED ELECTRON Examples of paramagnetic systems Transition-metal complexes
Solid-state NMR and proteins : basic concepts (a pictorial introduction) Barth van Rossum,
 Solid-state NMR and proteins : basic concepts (a pictorial introduction) Barth van Rossum, 16.02.2009 Solid-state and solution NMR spectroscopy have many things in common Several concepts have been/will
Solid-state NMR and proteins : basic concepts (a pictorial introduction) Barth van Rossum, 16.02.2009 Solid-state and solution NMR spectroscopy have many things in common Several concepts have been/will
Module 13: Chemical Shift and Its Measurement
 Subject Chemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy CHE_P12_M13_e-Text TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction 3. Shielding and deshielding
Subject Chemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy CHE_P12_M13_e-Text TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction 3. Shielding and deshielding
NUCLEAR MAGNETIC RESONANCE. The phenomenon of nuclear magnetic resonance will be used to study magnetic moments of nuclei.
 14 Sep 11 NMR.1 NUCLEAR MAGNETIC RESONANCE The phenomenon of nuclear magnetic resonance will be used to study magnetic moments of nuclei. Theory: In addition to its well-known properties of mass, charge,
14 Sep 11 NMR.1 NUCLEAR MAGNETIC RESONANCE The phenomenon of nuclear magnetic resonance will be used to study magnetic moments of nuclei. Theory: In addition to its well-known properties of mass, charge,
Polarised Nucleon Targets for Europe, 2nd meeting, Bochum 2005
 Polarised Nucleon Targets for Europe, nd meeting, Bochum Temperature dependence of nuclear spin-lattice relaxations in liquid ethanol with dissolved TEMPO radicals H. Štěpánková, J. Englich, J. Kohout,
Polarised Nucleon Targets for Europe, nd meeting, Bochum Temperature dependence of nuclear spin-lattice relaxations in liquid ethanol with dissolved TEMPO radicals H. Štěpánková, J. Englich, J. Kohout,
Ferdowsi University of Mashhad
 Spectroscopy in Inorganic Chemistry Nuclear Magnetic Resonance Spectroscopy spin deuterium 2 helium 3 The neutron has 2 quarks with a -e/3 charge and one quark with a +2e/3 charge resulting in a total
Spectroscopy in Inorganic Chemistry Nuclear Magnetic Resonance Spectroscopy spin deuterium 2 helium 3 The neutron has 2 quarks with a -e/3 charge and one quark with a +2e/3 charge resulting in a total
Chapter 8 Magnetic Resonance
 Chapter 8 Magnetic Resonance 9.1 Electron paramagnetic resonance 9.2 Ferromagnetic resonance 9.3 Nuclear magnetic resonance 9.4 Other resonance methods TCD March 2007 1 A resonance experiment involves
Chapter 8 Magnetic Resonance 9.1 Electron paramagnetic resonance 9.2 Ferromagnetic resonance 9.3 Nuclear magnetic resonance 9.4 Other resonance methods TCD March 2007 1 A resonance experiment involves
Determining Protein Structure BIBC 100
 Determining Protein Structure BIBC 100 Determining Protein Structure X-Ray Diffraction Interactions of x-rays with electrons in molecules in a crystal NMR- Nuclear Magnetic Resonance Interactions of magnetic
Determining Protein Structure BIBC 100 Determining Protein Structure X-Ray Diffraction Interactions of x-rays with electrons in molecules in a crystal NMR- Nuclear Magnetic Resonance Interactions of magnetic
Scalar (contact) vs dipolar (pseudocontact) contributions to isotropic shifts.
 Scalar (contact) vs dipolar (pseudocontact) contributions to isotropic shifts. Types of paramagnetic species: organic radicals, and complexes of transition metals, lanthanides, and actinides. Simplest
Scalar (contact) vs dipolar (pseudocontact) contributions to isotropic shifts. Types of paramagnetic species: organic radicals, and complexes of transition metals, lanthanides, and actinides. Simplest
Introduction to Electron Paramagnetic Resonance Spectroscopy
 Introduction to Electron Paramagnetic Resonance Spectroscopy Art van der Est, Department of Chemistry, Brock University St. Catharines, Ontario, Canada 1 EPR Spectroscopy EPR is magnetic resonance on unpaired
Introduction to Electron Paramagnetic Resonance Spectroscopy Art van der Est, Department of Chemistry, Brock University St. Catharines, Ontario, Canada 1 EPR Spectroscopy EPR is magnetic resonance on unpaired
Nuclear Magnetic Resonance (NMR) Spectroscopy Introduction:
 Nuclear Magnetic Resonance (NMR) Spectroscopy Introduction: Nuclear magnetic resonance spectroscopy (NMR) is the most powerful tool available for organic structure determination. Like IR spectroscopy,
Nuclear Magnetic Resonance (NMR) Spectroscopy Introduction: Nuclear magnetic resonance spectroscopy (NMR) is the most powerful tool available for organic structure determination. Like IR spectroscopy,
ATOMIC STRUCTURE, ELECTRONS, AND PERIODICITY
 ATOMIC STRUCTURE, ELECTRONS, AND PERIODICITY All matter is made of atoms. There are a limited number of types of atoms; these are the elements. (EU 1.A) Development of Atomic Theory Atoms are so small
ATOMIC STRUCTURE, ELECTRONS, AND PERIODICITY All matter is made of atoms. There are a limited number of types of atoms; these are the elements. (EU 1.A) Development of Atomic Theory Atoms are so small
NMR in Structural Biology
 NMR in Structural Biology Exercise session 2 1. a. List 3 NMR observables that report on structure. b. Also indicate whether the information they give is short/medium or long-range, or perhaps all three?
NMR in Structural Biology Exercise session 2 1. a. List 3 NMR observables that report on structure. b. Also indicate whether the information they give is short/medium or long-range, or perhaps all three?
Magnetic Resonance Spectroscopy EPR and NMR
 Magnetic Resonance Spectroscopy EPR and NMR A brief review of the relevant bits of quantum mechanics 1. Electrons have spin, - rotation of the charge about its axis generates a magnetic field at each electron.
Magnetic Resonance Spectroscopy EPR and NMR A brief review of the relevant bits of quantum mechanics 1. Electrons have spin, - rotation of the charge about its axis generates a magnetic field at each electron.
Assignment 3 Due Tuesday, March 31, 2009
 Assignment 3 Due Tuesday, March 31, 2009 Download and read the Math_techniques.pdf file from the Handouts section of the class web page. Do problems 1, 2, and 4 following section C (for problem 1, you
Assignment 3 Due Tuesday, March 31, 2009 Download and read the Math_techniques.pdf file from the Handouts section of the class web page. Do problems 1, 2, and 4 following section C (for problem 1, you
BMB/Bi/Ch 173 Winter 2018
 BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4:00-5:00pm and SFL 229, Monday 3/5 4:00-5:30pm.
BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4:00-5:00pm and SFL 229, Monday 3/5 4:00-5:30pm.
Biophysical Chemistry: NMR Spectroscopy
 Relaxation & Multidimensional Spectrocopy Vrije Universiteit Brussel 9th December 2011 Outline 1 Relaxation 2 Principles 3 Outline 1 Relaxation 2 Principles 3 Establishment of Thermal Equilibrium As previously
Relaxation & Multidimensional Spectrocopy Vrije Universiteit Brussel 9th December 2011 Outline 1 Relaxation 2 Principles 3 Outline 1 Relaxation 2 Principles 3 Establishment of Thermal Equilibrium As previously
SUPPLEMENTARY NOTE 1: ADDITIONAL CHARACTERIZATION OF NANODIAMOND SOLUTIONS AND THE OVERHAUSER EFFECT
 1 SUPPLEMENTARY NOTE 1: ADDITIONAL CHARACTERIZATION OF NANODIAMOND SOLUTIONS AND THE OVERHAUSER EFFECT Nanodiamond (ND) solutions were prepared using high power probe sonication and analyzed by dynamic
1 SUPPLEMENTARY NOTE 1: ADDITIONAL CHARACTERIZATION OF NANODIAMOND SOLUTIONS AND THE OVERHAUSER EFFECT Nanodiamond (ND) solutions were prepared using high power probe sonication and analyzed by dynamic
DETECTION OF UNPAIRED ELECTRONS
 DETECTION OF UNPAIRED ELECTRONS There are experimental methods for the detection of unpaired electrons. One of the hallmarks of unpaired electrons in materials is interaction with a magnetic field. That
DETECTION OF UNPAIRED ELECTRONS There are experimental methods for the detection of unpaired electrons. One of the hallmarks of unpaired electrons in materials is interaction with a magnetic field. That
All measurement has a limit of precision and accuracy, and this must be taken into account when evaluating experimental results.
 Chapter 11: Measurement and data processing and analysis 11.1 Uncertainty and error in measurement and results All measurement has a limit of precision and accuracy, and this must be taken into account
Chapter 11: Measurement and data processing and analysis 11.1 Uncertainty and error in measurement and results All measurement has a limit of precision and accuracy, and this must be taken into account
ATOMIC STRUCTURE, ELECTRONS, AND PERIODICITY
 ATOMIC STRUCTURE, ELECTRONS, AND PERIODICITY All matter is made of atoms. There are a limited number of types of atoms; these are the elements. (EU 1.A) Development of Atomic Theory Atoms are so small
ATOMIC STRUCTURE, ELECTRONS, AND PERIODICITY All matter is made of atoms. There are a limited number of types of atoms; these are the elements. (EU 1.A) Development of Atomic Theory Atoms are so small
Chapter 7. Nuclear Magnetic Resonance Spectroscopy
 Chapter 7 Nuclear Magnetic Resonance Spectroscopy I. Introduction 1924, W. Pauli proposed that certain atomic nuclei have spin and magnetic moment and exposure to magnetic field would lead to energy level
Chapter 7 Nuclear Magnetic Resonance Spectroscopy I. Introduction 1924, W. Pauli proposed that certain atomic nuclei have spin and magnetic moment and exposure to magnetic field would lead to energy level
16. Reactions of the Radical Pairs. The I P Step of the Paradigm
 16. Reactions of the Radical Pairs. The I P Step of the Paradigm Let us consider the product forming step in the general photochemical paradigm, i.e., the I P process shown in Figure 1. From the discussion
16. Reactions of the Radical Pairs. The I P Step of the Paradigm Let us consider the product forming step in the general photochemical paradigm, i.e., the I P process shown in Figure 1. From the discussion
Slow symmetric exchange
 Slow symmetric exchange ϕ A k k B t A B There are three things you should notice compared with the Figure on the previous slide: 1) The lines are broader, 2) the intensities are reduced and 3) the peaks
Slow symmetric exchange ϕ A k k B t A B There are three things you should notice compared with the Figure on the previous slide: 1) The lines are broader, 2) the intensities are reduced and 3) the peaks
Chapter 10: Multi- Electron Atoms Optical Excitations
 Chapter 10: Multi- Electron Atoms Optical Excitations To describe the energy levels in multi-electron atoms, we need to include all forces. The strongest forces are the forces we already discussed in Chapter
Chapter 10: Multi- Electron Atoms Optical Excitations To describe the energy levels in multi-electron atoms, we need to include all forces. The strongest forces are the forces we already discussed in Chapter
16.1 Introduction to NMR Spectroscopy. Spectroscopy. Spectroscopy. Spectroscopy. Spectroscopy. Spectroscopy 4/11/2013
 What is spectroscopy? NUCLEAR MAGNETIC RESONANCE (NMR) spectroscopy may be the most powerful method of gaining structural information about organic compounds. NMR involves an interaction between electromagnetic
What is spectroscopy? NUCLEAR MAGNETIC RESONANCE (NMR) spectroscopy may be the most powerful method of gaining structural information about organic compounds. NMR involves an interaction between electromagnetic
CONTENTS. 2 CLASSICAL DESCRIPTION 2.1 The resonance phenomenon 2.2 The vector picture for pulse EPR experiments 2.3 Relaxation and the Bloch equations
 CONTENTS Preface Acknowledgements Symbols Abbreviations 1 INTRODUCTION 1.1 Scope of pulse EPR 1.2 A short history of pulse EPR 1.3 Examples of Applications 2 CLASSICAL DESCRIPTION 2.1 The resonance phenomenon
CONTENTS Preface Acknowledgements Symbols Abbreviations 1 INTRODUCTION 1.1 Scope of pulse EPR 1.2 A short history of pulse EPR 1.3 Examples of Applications 2 CLASSICAL DESCRIPTION 2.1 The resonance phenomenon
ATOMIC HYDROGEN REACTION RATES IN AQUEOUS SOLUTI-ON
 ATOMC HYDROGEN REACTON RATES N AQUEOUS SOLUT-ON.. e VA FREE-NDUCTON DECAY ATTENUATON David M. Bartels Chemistry Division, Argonne National Laboratory Development of a new pulsed-epr method - free induction
ATOMC HYDROGEN REACTON RATES N AQUEOUS SOLUT-ON.. e VA FREE-NDUCTON DECAY ATTENUATON David M. Bartels Chemistry Division, Argonne National Laboratory Development of a new pulsed-epr method - free induction
Zeeman Effect. Alex Povilus Physics 441- Fall 2003 December 20, 2003
 Zeeman Effect Alex Povilus Physics 441- Fall 2003 December 20, 2003 Abstract The Zeeman Effect is observed by application of a strong magnetic field to a mercury vapor cell and exciting transitions by
Zeeman Effect Alex Povilus Physics 441- Fall 2003 December 20, 2003 Abstract The Zeeman Effect is observed by application of a strong magnetic field to a mercury vapor cell and exciting transitions by
Spectroscopy. Practical Handbook of. J. W. Robinson, Ph.D., D.Sc, F.R.C.S. Department of Chemistry Louisiana State University Baton Rouge, Louisiana
 Practical Handbook of Spectroscopy Edited by J. W. Robinson, Ph.D., D.Sc, F.R.C.S. Department of Chemistry Louisiana State University Baton Rouge, Louisiana CRC Press Boca Raton Ann Arbor Boston TABLE
Practical Handbook of Spectroscopy Edited by J. W. Robinson, Ph.D., D.Sc, F.R.C.S. Department of Chemistry Louisiana State University Baton Rouge, Louisiana CRC Press Boca Raton Ann Arbor Boston TABLE
Principles of Nuclear Magnetic Resonance Microscopy
 Principles of Nuclear Magnetic Resonance Microscopy Paul T. Callaghan Department of Physics and Biophysics Massey University New Zealand CLARENDON PRESS OXFORD CONTENTS 1 PRINCIPLES OF IMAGING 1 1.1 Introduction
Principles of Nuclear Magnetic Resonance Microscopy Paul T. Callaghan Department of Physics and Biophysics Massey University New Zealand CLARENDON PRESS OXFORD CONTENTS 1 PRINCIPLES OF IMAGING 1 1.1 Introduction
Molecular spectroscopy
 Molecular spectroscopy Origin of spectral lines = absorption, emission and scattering of a photon when the energy of a molecule changes: rad( ) M M * rad( ' ) ' v' 0 0 absorption( ) emission ( ) scattering
Molecular spectroscopy Origin of spectral lines = absorption, emission and scattering of a photon when the energy of a molecule changes: rad( ) M M * rad( ' ) ' v' 0 0 absorption( ) emission ( ) scattering
The Use of NMR Spectroscopy
 Spektroskopi Molekul Organik (SMO): Nuclear Magnetic Resonance (NMR) Spectroscopy All is adopted from McMurry s Organic Chemistry The Use of NMR Spectroscopy Used to determine relative location of atoms
Spektroskopi Molekul Organik (SMO): Nuclear Magnetic Resonance (NMR) Spectroscopy All is adopted from McMurry s Organic Chemistry The Use of NMR Spectroscopy Used to determine relative location of atoms
NMR Spectroscopy. Guangjin Hou
 NMR Spectroscopy Guangjin Hou 22-04-2009 NMR History 1 H NMR spectra of water H NMR spectra of water (First NMR Spectra on Water, 1946) 1 H NMR spectra ethanol (First bservation of the Chemical Shift,
NMR Spectroscopy Guangjin Hou 22-04-2009 NMR History 1 H NMR spectra of water H NMR spectra of water (First NMR Spectra on Water, 1946) 1 H NMR spectra ethanol (First bservation of the Chemical Shift,
Lecture- 08 Emission and absorption spectra
 Atomic and Molecular Absorption Spectrometry for Pollution Monitoring Dr. J R Mudakavi Department of Chemical Engineering Indian Institute of Science, Bangalore Lecture- 08 Emission and absorption spectra
Atomic and Molecular Absorption Spectrometry for Pollution Monitoring Dr. J R Mudakavi Department of Chemical Engineering Indian Institute of Science, Bangalore Lecture- 08 Emission and absorption spectra
Chapter 13 An Introduction to Ultraviolet/Visible Molecular Absorption Spectrometry
 Chapter 13 An Introduction to Ultraviolet/Visible Molecular Absorption Spectrometry 13A Measurement Of Transmittance and Absorbance Absorption measurements based upon ultraviolet and visible radiation
Chapter 13 An Introduction to Ultraviolet/Visible Molecular Absorption Spectrometry 13A Measurement Of Transmittance and Absorbance Absorption measurements based upon ultraviolet and visible radiation
A very brief history of the study of light
 1. Sir Isaac Newton 1672: A very brief history of the study of light Showed that the component colors of the visible portion of white light can be separated through a prism, which acts to bend the light
1. Sir Isaac Newton 1672: A very brief history of the study of light Showed that the component colors of the visible portion of white light can be separated through a prism, which acts to bend the light
Chapter 4. Electrostatic Fields in Matter
 Chapter 4. Electrostatic Fields in Matter 4.1. Polarization 4.2. The Field of a Polarized Object 4.3. The Electric Displacement 4.4. Linear Dielectrics 4.5. Energy in dielectric systems 4.6. Forces on
Chapter 4. Electrostatic Fields in Matter 4.1. Polarization 4.2. The Field of a Polarized Object 4.3. The Electric Displacement 4.4. Linear Dielectrics 4.5. Energy in dielectric systems 4.6. Forces on
High Frequency Electron Paramagnetic Resonance Studies of Mn 12 Wheels
 High Frequency Electron Paramagnetic Resonance Studies of Mn 12 Wheels Gage Redler and Stephen Hill Department of Physics, University of Florida Abstract High Frequency Electron Paramagnetic Resonance
High Frequency Electron Paramagnetic Resonance Studies of Mn 12 Wheels Gage Redler and Stephen Hill Department of Physics, University of Florida Abstract High Frequency Electron Paramagnetic Resonance
Magnetic Resonance Spectroscopy
 INTRODUCTION TO Magnetic Resonance Spectroscopy ESR, NMR, NQR D. N. SATHYANARAYANA Formerly, Chairman Department of Inorganic and Physical Chemistry Indian Institute of Science, Bangalore % I.K. International
INTRODUCTION TO Magnetic Resonance Spectroscopy ESR, NMR, NQR D. N. SATHYANARAYANA Formerly, Chairman Department of Inorganic and Physical Chemistry Indian Institute of Science, Bangalore % I.K. International
Theoretical simulation of nonlinear spectroscopy in the liquid phase La Cour Jansen, Thomas
 University of Groningen Theoretical simulation of nonlinear spectroscopy in the liquid phase La Cour Jansen, Thomas IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF)
University of Groningen Theoretical simulation of nonlinear spectroscopy in the liquid phase La Cour Jansen, Thomas IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF)
Atomic Structure and Processes
 Chapter 5 Atomic Structure and Processes 5.1 Elementary atomic structure Bohr Orbits correspond to principal quantum number n. Hydrogen atom energy levels where the Rydberg energy is R y = m e ( e E n
Chapter 5 Atomic Structure and Processes 5.1 Elementary atomic structure Bohr Orbits correspond to principal quantum number n. Hydrogen atom energy levels where the Rydberg energy is R y = m e ( e E n
Structure Determination: Nuclear Magnetic Resonance Spectroscopy
 Structure Determination: Nuclear Magnetic Resonance Spectroscopy Why This Chapter? NMR is the most valuable spectroscopic technique used for structure determination More advanced NMR techniques are used
Structure Determination: Nuclear Magnetic Resonance Spectroscopy Why This Chapter? NMR is the most valuable spectroscopic technique used for structure determination More advanced NMR techniques are used
Simulation of the NMR Second Moment as a Function of Temperature in the Presence of Molecular Motion. Application to (CH 3
 Simulation of the NMR Second Moment as a Function of Temperature in the Presence of Molecular Motion. Application to (CH 3 ) 3 NBH 3 Roman Goc Institute of Physics, A. Mickiewicz University, Umultowska
Simulation of the NMR Second Moment as a Function of Temperature in the Presence of Molecular Motion. Application to (CH 3 ) 3 NBH 3 Roman Goc Institute of Physics, A. Mickiewicz University, Umultowska
CHAPTER 6--- CHEMISTRY IN BIOLOGY. Miss Queen
 CHAPTER 6--- CHEMISTRY IN BIOLOGY Miss Queen SECTION 6.1 Atoms, Elements, Compounds COMPOSITION OF MATTER Matter - Everything in universe is composed of matter Matter is anything that occupies space or
CHAPTER 6--- CHEMISTRY IN BIOLOGY Miss Queen SECTION 6.1 Atoms, Elements, Compounds COMPOSITION OF MATTER Matter - Everything in universe is composed of matter Matter is anything that occupies space or
Long-lived spin echoes in magnetically diluted system: an NMR study of the Ge single crystals Alexander M. Panich,
 Long-lived spin echoes in magnetically diluted system: an NMR study of the Ge single crystals Alexander M. Panich, Department of Physics, Ben-Gurion University of the Negev, Beer Sheva, Israel N. A. Sergeev,
Long-lived spin echoes in magnetically diluted system: an NMR study of the Ge single crystals Alexander M. Panich, Department of Physics, Ben-Gurion University of the Negev, Beer Sheva, Israel N. A. Sergeev,
NMR = Nuclear Magnetic Resonance
 NMR = Nuclear Magnetic Resonance NMR spectroscopy is the most powerful technique available to organic chemists for determining molecular structures. Looks at nuclei with odd mass numbers or odd number
NMR = Nuclear Magnetic Resonance NMR spectroscopy is the most powerful technique available to organic chemists for determining molecular structures. Looks at nuclei with odd mass numbers or odd number
I690/B680 Structural Bioinformatics Spring Protein Structure Determination by NMR Spectroscopy
 I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
Solid state 13 Cand 1 H MAS NMR investigations of C 60 (ferrocene-d 10 ) 2 complex
 Spectroscopy 17 (2003) 39 44 39 IOS Press Solid state 13 Cand 1 H MAS NMR investigations of C 60 (ferrocene-d 10 ) 2 complex E. Shabanova, K. Schaumburg and F.S. Kamounah CISMI, Department of Chemistry,
Spectroscopy 17 (2003) 39 44 39 IOS Press Solid state 13 Cand 1 H MAS NMR investigations of C 60 (ferrocene-d 10 ) 2 complex E. Shabanova, K. Schaumburg and F.S. Kamounah CISMI, Department of Chemistry,
Center for Sustainable Environmental Technologies, Iowa State University
 NMR Characterization of Biochars By Catherine Brewer Center for Sustainable Environmental Technologies, Iowa State University Introduction Nuclear magnetic resonance spectroscopy (NMR) uses a very strong
NMR Characterization of Biochars By Catherine Brewer Center for Sustainable Environmental Technologies, Iowa State University Introduction Nuclear magnetic resonance spectroscopy (NMR) uses a very strong
Name: BCMB/CHEM 8190, BIOMOLECULAR NMR FINAL EXAM-5/5/10
 Name: BCMB/CHEM 8190, BIOMOLECULAR NMR FINAL EXAM-5/5/10 Instructions: This is an open book, limited time, exam. You may use notes you have from class and any text book you find useful. You may also use
Name: BCMB/CHEM 8190, BIOMOLECULAR NMR FINAL EXAM-5/5/10 Instructions: This is an open book, limited time, exam. You may use notes you have from class and any text book you find useful. You may also use
The Nuclear Force and Limitations to the Lorentz Electrostatic Force Equation
 The Nuclear Force and Limitations to the Lorentz Electrostatic Force Equation Author: Singer, Michael Date: 1 st May 2017 3 rd July 2018 Revision Abstract In Electromagnetic Field Theory it is the interaction
The Nuclear Force and Limitations to the Lorentz Electrostatic Force Equation Author: Singer, Michael Date: 1 st May 2017 3 rd July 2018 Revision Abstract In Electromagnetic Field Theory it is the interaction
16.1 Introduction to NMR. Spectroscopy
 16.1 Introduction to NMR What is spectroscopy? Spectroscopy NUCLEAR MAGNETIC RESNANCE (NMR) spectroscopy may be the most powerful method of gaining structural information about organic compounds. NMR involves
16.1 Introduction to NMR What is spectroscopy? Spectroscopy NUCLEAR MAGNETIC RESNANCE (NMR) spectroscopy may be the most powerful method of gaining structural information about organic compounds. NMR involves
Disordered Solids. real crystals spin glass. glasses. Grenoble
 Disordered Solids real crystals spin glass glasses Grenoble 21.09.11-1 Tunneling of Atoms in Solids Grenoble 21.09.11-2 Tunneln Grenoble 21.09.11-3 KCl:Li Specific Heat specific heat roughly a factor of
Disordered Solids real crystals spin glass glasses Grenoble 21.09.11-1 Tunneling of Atoms in Solids Grenoble 21.09.11-2 Tunneln Grenoble 21.09.11-3 KCl:Li Specific Heat specific heat roughly a factor of
Nuclear Magnetic Resonance
 Nuclear Magnetic Resonance PRINCIPLES OF NMR SPECTROSCOPY Contents Principles of nuclear magnetic resonance The nmr spectrometer Basic principles in nmr application NMR tools used to obtain information
Nuclear Magnetic Resonance PRINCIPLES OF NMR SPECTROSCOPY Contents Principles of nuclear magnetic resonance The nmr spectrometer Basic principles in nmr application NMR tools used to obtain information
The Basics of Magnetic Resonance Imaging
 The Basics of Magnetic Resonance Imaging Nathalie JUST, PhD nathalie.just@epfl.ch CIBM-AIT, EPFL Course 2013-2014-Chemistry 1 Course 2013-2014-Chemistry 2 MRI: Many different contrasts Proton density T1
The Basics of Magnetic Resonance Imaging Nathalie JUST, PhD nathalie.just@epfl.ch CIBM-AIT, EPFL Course 2013-2014-Chemistry 1 Course 2013-2014-Chemistry 2 MRI: Many different contrasts Proton density T1
How to Tell Diamagnetic Materials from Paramagnetic
 How to Tell Diamagnetic Materials from Paramagnetic I wrote these notes as a followup to the Material Physics web pages of Dr. Rudolf Winter s from Aberystwyth University (Wales, UK): diamagmetism and
How to Tell Diamagnetic Materials from Paramagnetic I wrote these notes as a followup to the Material Physics web pages of Dr. Rudolf Winter s from Aberystwyth University (Wales, UK): diamagmetism and
2.1 Experimental and theoretical studies
 Chapter 2 NiO As stated before, the first-row transition-metal oxides are among the most interesting series of materials, exhibiting wide variations in physical properties related to electronic structure.
Chapter 2 NiO As stated before, the first-row transition-metal oxides are among the most interesting series of materials, exhibiting wide variations in physical properties related to electronic structure.
NMRis the most valuable spectroscopic technique for organic chemists because it maps the carbon-hydrogen framework of a molecule.
 Chapter 13: Nuclear magnetic resonance spectroscopy NMRis the most valuable spectroscopic technique for organic chemists because it maps the carbon-hydrogen framework of a molecule. 13.2 The nature of
Chapter 13: Nuclear magnetic resonance spectroscopy NMRis the most valuable spectroscopic technique for organic chemists because it maps the carbon-hydrogen framework of a molecule. 13.2 The nature of
Spin Relaxation and NOEs BCMB/CHEM 8190
 Spin Relaxation and NOEs BCMB/CHEM 8190 T 1, T 2 (reminder), NOE T 1 is the time constant for longitudinal relaxation - the process of re-establishing the Boltzmann distribution of the energy level populations
Spin Relaxation and NOEs BCMB/CHEM 8190 T 1, T 2 (reminder), NOE T 1 is the time constant for longitudinal relaxation - the process of re-establishing the Boltzmann distribution of the energy level populations
Spin Interactions. Giuseppe Pileio 24/10/2006
 Spin Interactions Giuseppe Pileio 24/10/2006 Magnetic moment µ = " I ˆ µ = " h I(I +1) " = g# h Spin interactions overview Zeeman Interaction Zeeman interaction Interaction with the static magnetic field
Spin Interactions Giuseppe Pileio 24/10/2006 Magnetic moment µ = " I ˆ µ = " h I(I +1) " = g# h Spin interactions overview Zeeman Interaction Zeeman interaction Interaction with the static magnetic field
Basic Chemistry for Biology. Honors Biology
 Basic Chemistry for Biology Honors Biology 2013-2014 Composition of Matter Matter - Everything in universe is composed of matter Matter is anything that occupies space or has mass Mass quantity of matter
Basic Chemistry for Biology Honors Biology 2013-2014 Composition of Matter Matter - Everything in universe is composed of matter Matter is anything that occupies space or has mass Mass quantity of matter
Multi-Electron Atoms II
 Multi-Electron Atoms II LS Coupling The basic idea of LS coupling or Russell-Saunders coupling is to assume that spin-orbit effects are small, and can be neglected to a first approximation. If there is
Multi-Electron Atoms II LS Coupling The basic idea of LS coupling or Russell-Saunders coupling is to assume that spin-orbit effects are small, and can be neglected to a first approximation. If there is
Supplementary Information Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes
 Supplementary Information Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes Elka R. Georgieva 1,2*, Peter P. Borbat 1,2, Haley D. Norman 3 and Jack H. Freed 1,2* 1 Department
Supplementary Information Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes Elka R. Georgieva 1,2*, Peter P. Borbat 1,2, Haley D. Norman 3 and Jack H. Freed 1,2* 1 Department
Scattering by Groups of Particles
 Scattering by Groups of Particles 2 Overview The interactions between light and groups of particles in paint films are highly complex. While we can accurately calculate the scattering of an individual
Scattering by Groups of Particles 2 Overview The interactions between light and groups of particles in paint films are highly complex. While we can accurately calculate the scattering of an individual
PAPER No. : 8 (PHYSICAL SPECTROSCOPY) MODULE No. : 5 (TRANSITION PROBABILITIES AND TRANSITION DIPOLE MOMENT. OVERVIEW OF SELECTION RULES)
 Subject Chemistry Paper No and Title Module No and Title Module Tag 8 and Physical Spectroscopy 5 and Transition probabilities and transition dipole moment, Overview of selection rules CHE_P8_M5 TABLE
Subject Chemistry Paper No and Title Module No and Title Module Tag 8 and Physical Spectroscopy 5 and Transition probabilities and transition dipole moment, Overview of selection rules CHE_P8_M5 TABLE
Chapter 9: Molecular Geometry and Bonding Theories
 Chapter 9: Molecular Geometry and Bonding Theories 9.1 Molecular Geometries -Bond angles: angles made by the lines joining the nuclei of the atoms in a molecule -Bond angles determine overall shape of
Chapter 9: Molecular Geometry and Bonding Theories 9.1 Molecular Geometries -Bond angles: angles made by the lines joining the nuclei of the atoms in a molecule -Bond angles determine overall shape of
Principles of Nuclear Magnetic Resonance in One and Two Dimensions
 Principles of Nuclear Magnetic Resonance in One and Two Dimensions Richard R. Ernst, Geoffrey Bodenhausen, and Alexander Wokaun Laboratorium für Physikalische Chemie Eidgenössische Technische Hochschule
Principles of Nuclear Magnetic Resonance in One and Two Dimensions Richard R. Ernst, Geoffrey Bodenhausen, and Alexander Wokaun Laboratorium für Physikalische Chemie Eidgenössische Technische Hochschule
Model Answer (Paper code: AR-7112) M. Sc. (Physics) IV Semester Paper I: Laser Physics and Spectroscopy
 Model Answer (Paper code: AR-7112) M. Sc. (Physics) IV Semester Paper I: Laser Physics and Spectroscopy Section I Q1. Answer (i) (b) (ii) (d) (iii) (c) (iv) (c) (v) (a) (vi) (b) (vii) (b) (viii) (a) (ix)
Model Answer (Paper code: AR-7112) M. Sc. (Physics) IV Semester Paper I: Laser Physics and Spectroscopy Section I Q1. Answer (i) (b) (ii) (d) (iii) (c) (iv) (c) (v) (a) (vi) (b) (vii) (b) (viii) (a) (ix)
Introduction to Relaxation Theory James Keeler
 EUROMAR Zürich, 24 Introduction to Relaxation Theory James Keeler University of Cambridge Department of Chemistry What is relaxation? Why might it be interesting? relaxation is the process which drives
EUROMAR Zürich, 24 Introduction to Relaxation Theory James Keeler University of Cambridge Department of Chemistry What is relaxation? Why might it be interesting? relaxation is the process which drives
Technical University of Denmark
 Technical University of Denmark Page 1 of 10 pages Written test, 12 December 2012 Course name: Introduction to medical imaging Course no. 31540 Aids allowed: None. Pocket calculator not allowed "Weighting":
Technical University of Denmark Page 1 of 10 pages Written test, 12 December 2012 Course name: Introduction to medical imaging Course no. 31540 Aids allowed: None. Pocket calculator not allowed "Weighting":
An introduction to Solid State NMR and its Interactions
 An introduction to Solid State NMR and its Interactions From tensor to NMR spectra CECAM Tutorial September 9 Calculation of Solid-State NMR Parameters Using the GIPAW Method Thibault Charpentier - CEA
An introduction to Solid State NMR and its Interactions From tensor to NMR spectra CECAM Tutorial September 9 Calculation of Solid-State NMR Parameters Using the GIPAW Method Thibault Charpentier - CEA
Internal Structures of Electron, Neutron, Proton and Nuclei particle masses are not arbitrary
 Internal Structures of Electron, Neutron, Proton and Nuclei particle masses are not arbitrary Jose P Koshy josepkoshy@gmail.com Abstract: The mass of neutron is slightly greater than1838 electrons. So
Internal Structures of Electron, Neutron, Proton and Nuclei particle masses are not arbitrary Jose P Koshy josepkoshy@gmail.com Abstract: The mass of neutron is slightly greater than1838 electrons. So
SUPPLEMENTARY INFORMATION
 Fast spin information transfer between distant quantum dots using individual electrons B. Bertrand, S. Hermelin, S. Takada, M. Yamamoto, S. Tarucha, A. Ludwig, A. D. Wieck, C. Bäuerle, T. Meunier* Content
Fast spin information transfer between distant quantum dots using individual electrons B. Bertrand, S. Hermelin, S. Takada, M. Yamamoto, S. Tarucha, A. Ludwig, A. D. Wieck, C. Bäuerle, T. Meunier* Content
Spin Dynamics Basics of Nuclear Magnetic Resonance. Malcolm H. Levitt
 Spin Dynamics Basics of Nuclear Magnetic Resonance Second edition Malcolm H. Levitt The University of Southampton, UK John Wiley &. Sons, Ltd Preface xxi Preface to the First Edition xxiii Introduction
Spin Dynamics Basics of Nuclear Magnetic Resonance Second edition Malcolm H. Levitt The University of Southampton, UK John Wiley &. Sons, Ltd Preface xxi Preface to the First Edition xxiii Introduction
Conclusion. 109m Ag isomer showed that there is no such broadening. Because one can hardly
 Conclusion This small book presents a description of the results of studies performed over many years by our research group, which, in the best period, included 15 physicists and laboratory assistants
Conclusion This small book presents a description of the results of studies performed over many years by our research group, which, in the best period, included 15 physicists and laboratory assistants
Physics 221B Spring 2012 Notes 42 Scattering of Radiation by Matter
 Physics 221B Spring 2012 Notes 42 Scattering of Radiation by Matter 1. Introduction In the previous set of Notes we treated the emission and absorption of radiation by matter. In these Notes we turn to
Physics 221B Spring 2012 Notes 42 Scattering of Radiation by Matter 1. Introduction In the previous set of Notes we treated the emission and absorption of radiation by matter. In these Notes we turn to
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
PRACTICAL ASPECTS OF NMR RELAXATION STUDIES OF BIOMOLECULAR DYNAMICS
 PRACTICAL ASPECTS OF MR RELAXATIO STUDIES OF BIOMOLECULAR DYAMICS Further reading: (Can be downloaded from my web page Korzhnev D.E., Billeter M., Arseniev A.S., and Orekhov V. Y., MR Studies of Brownian
PRACTICAL ASPECTS OF MR RELAXATIO STUDIES OF BIOMOLECULAR DYAMICS Further reading: (Can be downloaded from my web page Korzhnev D.E., Billeter M., Arseniev A.S., and Orekhov V. Y., MR Studies of Brownian
Matter mass space atoms solid, a liquid, a gas, or plasm elements compounds mixtures atoms Compounds chemically combined Mixtures not chemically
 SOL PS.2 THE NATURE OF MATTER Matter is anything that has mass and occupies space. All matter is made up of small particles called atoms. Matter can exist as a solid, a liquid, a gas, or plasma. Matter
SOL PS.2 THE NATURE OF MATTER Matter is anything that has mass and occupies space. All matter is made up of small particles called atoms. Matter can exist as a solid, a liquid, a gas, or plasma. Matter
PAPER No. 12: ORGANIC SPECTROSCOPY. Module 19: NMR Spectroscopy of N, P and F-atoms
 Subject Chemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy CHE_P12_M19_e-Text TABLE OF CONTENTS 1. Learning Outcomes 2. 15 N NMR spectroscopy 3. 19 F NMR spectroscopy
Subject Chemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy CHE_P12_M19_e-Text TABLE OF CONTENTS 1. Learning Outcomes 2. 15 N NMR spectroscopy 3. 19 F NMR spectroscopy
Chapters 9&10 Structure and Bonding Theories
 Chapters 9&10 Structure and Bonding Theories Ionic Radii Ions, just like atoms, follow a periodic trend in their radii. The metal ions in a given period are smaller than the non-metal ions in the same
Chapters 9&10 Structure and Bonding Theories Ionic Radii Ions, just like atoms, follow a periodic trend in their radii. The metal ions in a given period are smaller than the non-metal ions in the same
Chapter 8: Electrons in Atoms Electromagnetic Radiation
 Chapter 8: Electrons in Atoms Electromagnetic Radiation Electromagnetic (EM) radiation is a form of energy transmission modeled as waves moving through space. (see below left) Electromagnetic Radiation
Chapter 8: Electrons in Atoms Electromagnetic Radiation Electromagnetic (EM) radiation is a form of energy transmission modeled as waves moving through space. (see below left) Electromagnetic Radiation
T 1, T 2, NOE (reminder)
 T 1, T 2, NOE (reminder) T 1 is the time constant for longitudinal relaxation - the process of re-establishing the Boltzmann distribution of the energy level populations of the system following perturbation
T 1, T 2, NOE (reminder) T 1 is the time constant for longitudinal relaxation - the process of re-establishing the Boltzmann distribution of the energy level populations of the system following perturbation
Introduction to Modern Physics
 SECOND EDITION Introduction to Modern Physics John D. McGervey Case Western Reserve University Academic Press A Subsidiary of Harcourt Brace Jovanovich Orlando San Diego San Francisco New York London Toronto
SECOND EDITION Introduction to Modern Physics John D. McGervey Case Western Reserve University Academic Press A Subsidiary of Harcourt Brace Jovanovich Orlando San Diego San Francisco New York London Toronto
Circular Dichroism & Optical Rotatory Dispersion. Proteins (KCsa) Polysaccharides (agarose) DNA CHEM 305. Many biomolecules are α-helical!
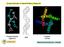 Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
Circular Dichroism & Optical Rotatory Dispersion Polysaccharides (agarose) DNA Proteins (KCsa) Many biomolecules are α-helical! How can we measure the amount and changes in amount of helical structure
4 Spin-echo, Spin-echo Double Resonance (SEDOR) and Rotational-echo Double Resonance (REDOR) applied on polymer blends
 4 Spin-echo, Spin-echo ouble Resonance (SEOR and Rotational-echo ouble Resonance (REOR applied on polymer blends The next logical step after analyzing and concluding upon the results of proton transversal
4 Spin-echo, Spin-echo ouble Resonance (SEOR and Rotational-echo ouble Resonance (REOR applied on polymer blends The next logical step after analyzing and concluding upon the results of proton transversal
Using Molecular Dynamics to Compute Properties CHEM 430
 Using Molecular Dynamics to Compute Properties CHEM 43 Heat Capacity and Energy Fluctuations Running an MD Simulation Equilibration Phase Before data-collection and results can be analyzed the system
Using Molecular Dynamics to Compute Properties CHEM 43 Heat Capacity and Energy Fluctuations Running an MD Simulation Equilibration Phase Before data-collection and results can be analyzed the system
Copyright Mark Brandt, Ph.D A third method, cryogenic electron microscopy has seen increasing use over the past few years.
 Structure Determination and Sequence Analysis The vast majority of the experimentally determined three-dimensional protein structures have been solved by one of two methods: X-ray diffraction and Nuclear
Structure Determination and Sequence Analysis The vast majority of the experimentally determined three-dimensional protein structures have been solved by one of two methods: X-ray diffraction and Nuclear
Fundamentals of Spectroscopy for Optical Remote Sensing. Course Outline 2009
 Fundamentals of Spectroscopy for Optical Remote Sensing Course Outline 2009 Part I. Fundamentals of Quantum Mechanics Chapter 1. Concepts of Quantum and Experimental Facts 1.1. Blackbody Radiation and
Fundamentals of Spectroscopy for Optical Remote Sensing Course Outline 2009 Part I. Fundamentals of Quantum Mechanics Chapter 1. Concepts of Quantum and Experimental Facts 1.1. Blackbody Radiation and
Lecture #6 Chemical Exchange
 Lecture #6 Chemical Exchange Topics Introduction Effects on longitudinal magnetization Effects on transverse magnetization Examples Handouts and Reading assignments Kowalewski, Chapter 13 Levitt, sections
Lecture #6 Chemical Exchange Topics Introduction Effects on longitudinal magnetization Effects on transverse magnetization Examples Handouts and Reading assignments Kowalewski, Chapter 13 Levitt, sections
Last Updated:
 Last Updated: 2014 07 30 Generation of the EPR ignal MR and EPR are similar in a way that the amount of absorption energy required to for a transition between atomic or molecular states. pectroscopy generally
Last Updated: 2014 07 30 Generation of the EPR ignal MR and EPR are similar in a way that the amount of absorption energy required to for a transition between atomic or molecular states. pectroscopy generally
Worksheet 2.1. Chapter 2: Atomic structure glossary
 Worksheet 2.1 Chapter 2: Atomic structure glossary Acceleration (in a mass spectrometer) The stage where the positive ions are attracted to negatively charged plates. Alpha decay The emission of an alpha
Worksheet 2.1 Chapter 2: Atomic structure glossary Acceleration (in a mass spectrometer) The stage where the positive ions are attracted to negatively charged plates. Alpha decay The emission of an alpha
V27: RF Spectroscopy
 Martin-Luther-Universität Halle-Wittenberg FB Physik Advanced Lab Course V27: RF Spectroscopy ) Electron spin resonance (ESR) Investigate the resonance behaviour of two coupled LC circuits (an active rf
Martin-Luther-Universität Halle-Wittenberg FB Physik Advanced Lab Course V27: RF Spectroscopy ) Electron spin resonance (ESR) Investigate the resonance behaviour of two coupled LC circuits (an active rf
