IFM Chemistry Computational Chemistry 2010, 7.5 hp LAB2. Computer laboratory exercise 1 (LAB2): Quantum chemical calculations
|
|
|
- Rosanna Rice
- 6 years ago
- Views:
Transcription
1 Computer laboratory exercise 1 (LAB2): Quantum chemical calculations Introduction: The objective of the second computer laboratory exercise is to get acquainted with a program for performing quantum chemical ab initio calculations and to perform calculations on different molecules to study electronic properties and reaction mechanisms. A lab report is required for LAB2. Hand in the report by submitting it on It s learning in Computer labs > Computer lab2 > Lab report LAB2, select Submit answer and upload your file. Quantum chemical calculations using Gaussian98 In this computer laboratory exercise, the quantum chemical program Gaussian98 will be used. Gaussian98 is one version of the Gaussian series of programs, where the latest and most advanced version is called Gaussian09. 1 Gaussian is one of the most widely used quantum chemical program packages for molecular applications, and is used both in industry and in many scientific areas in academia. It is a licensed software that runs on different platforms, on single CPU systems or in parallel multiprocessor systems. The program was created by John A. Pople, which received the Nobel prize in chemistry in 1998 for his development of computational methods in quantum chemistry. 2 The prize was shared with Walter Kohn for his development of the density-functional theory, a type of calculation that can be performed using Gaussian. Practical guide The terms L-, M-, and R-click refers to clicking with the left, middle and right mouse button, respectively. The g98 input file: The basic structure of a Gaussian input file (filename.inp) consists of a series of lines in an ASCII text file, and includes several different sections. Example of the contents of an input file: Sections in input file: # RHF/6-31G** Opt Freq Route section Blank line Route section done A water molecule Title section Blank line Title done 0 1 Charge and multiplicity O H H Molecular geometry Blank line Molecular specification done (Additional sections for specific job types) (Blank line)
2 Route section: line begins with #. Specifies level of theory, calculation type, and other options, e.g. print options o Level of theory: QC method/basis set, e.g. HF/STO-3G, MP2/6-31G**, B3LYP/6-31G* o Calculation type: single point calculation no keyword, optimization Opt keyword, frequency calculation Freq keyword, scan along predefined coordinates Scan keyword o Print options: to be able to visualize molecular orbitals in Molekel, add print options GFPrint and Pop=Full Title section details about the calculation, not interpreted by program Molecular specification o Charge: first entry on fifth row specifies the systems total charge, e.g. 0, 1, -1 etc. o Multiplicity: is given by M = 2 S + 1, where S is the total spin. M = 1 for a singlet (all electrons paired), 2 for a doublet (i.e. one unpaired spin), 3 for a triplet (two unpaired spins) etc. o Molecular geometry: specifies the type and the position of the atomic nuclei of the system in Cartesian coordinates or Z-matrix Start a g98 calculation: Start the calculation by typing: g98.run filename & (Note filename and not filename.inp) in a terminal window. The calculation will generate an output file called filename.out. The g98 output file: The output file (filename.out) contains a lot of information about the calculation and the results. The content depends on what type of calculation that has been performed and on what print options that was specified. The units are usually Hartree (atomic unit) for energy and Ångström for distance. Examine the output file by e.g. reading it in a text editor, or using the more command in a terminal window. Alternatively, extract information from the output file by the grep command and appropriate text search string 3. Examples of important text strings in output files: It is good to make sure that the calculation has finished successfully by checking that the last line of the output file contains Normal termination of Gaussian 98. Use for instance the tail command. The total energy of the system is printed after SCF Done. In a single point calculation only one such line is found, in an optimization one line for each iteration is found. Use for instance the more or grep commands. Detailed information about molecular orbitals is listed after Molecular Orbital Coefficients. The number of steps required for convergence in a geometry optimization can be found by searching for Step number. The Mulliken charges are listed after Total atomic charges. The dipole moment is written after Tot=, one line below Dipole moment. To check the progress of an optimization the command grep -A4 Item filename.out can be useful. 3 grep text string filename.out Prints lines of filename.out matching the pattern text string 2
3 Visualization using Molekel4.3: Start the program by typing molekel4.3 & in a terminal window. Two windows will appear, a Molekel Main Window and a Main Interface Window. Read a molecule in xyz-format by R-click and hold in the Main Window. Choose Load > xyz, tick atomic symbol, and L-click OK. Select a file and L- click Accept. Note that Molekel4.3 does not read Gaussian input files. Load a g98 output file by R-click and hold in the Main Window. Choose Load > gaussian log. Select a file and L-click Accept. Inspect the molecule in the Main Window by rotating the molecule with the left mouse button and translating with the middle button. Zoom in or out by holding down the Shift key and using the middle button. It is possible to save the coordinates of a new molecule orientation by Write > xyz (current orient.). Change the appearance of the molecule to Ball & Stick by clicking that option in the Interface Window. Display atom labels by R-clicking the Main Window, choosing Labels and marking atom labels (unmark to remove labels). To display atomic charges (Mulliken), R-click the Main Window, choose Labels and tick atom charges ( un-tick to remove). To display dipole moment, R-click the Main Window, choose Dipole moment and read the value in Debye and/or display the dipole moment graphically by ticking the box show dipole moment. Measure atomic distances by R-clicking in the Main Window, and selecting Geometry > Distance to enter picking mode. Click on the two atoms of interest to measure the distance. The measured values are displayed in the Main Window and printed in the terminal window. Angles and dihedrals are measured similarly by Geometry > Angle and Geometry > Dihedral. Exit picking mode by M-clicking. To display molecular orbitals: o create a file (filename.macu) by R-clicking in the Main Window, choosing Compute > Orbital, selecting an orbital in the list, and clicking Accept. Type a name for the molecular orbital (filename.macu), and click Accept. o load existing macu files by R-clicking in the Main Window, and selecting Surface. Select load, choose a file from the list, and click Accept. Tick both signs, change the cutoff to e.g and click create surface. When loading an output file from a geometry optimization, the last (and hopefully converged) geometry is shown. To view the previous geometries, go to Animate > Series of coords. Click on first to view the starting structure, and then go through the geometries of the optimization, step by step, with next >. Unmark keep bonds for bonds to be formed and broken. Display vibrations with Animate > Frequency. Select a vibrational mode by clicking choose and then selecting a vibration in the list and clicking Accept. Click on animate to display the vibration and end with stop. Select another vibration in the same way. Click cancel to exit animation mode. 3
4 Exercises Save all your files for the second computer laboratory exercise in the directory Lab2 created in LAB1. Start the calculations from this directory. Download the prepared input files from the folder Computer lab 2 > Input files LAB2 on It s learning. 1. Molecular orbitals of H 2 and F 2 Calculations: Perform single point Hartree-Fock calculations using the STO-3G basis set on the diatomic molecules H 2 and F 2. Use the experimentally determined geometries (R H H =0.74 Å, and R F F =1.42 Å) and print the molecular orbitals and basis functions in the output file (keywords GFPrint and Pop=Full). Input files for these calculations are found in the files H2_MO.inp and F2_MO.inp. Analysis: Visualize the molecular orbitals using Molekel and draw molecular orbital energy diagrams for the two molecules. Verify that the molecular orbital coefficients in the output file are in agreement with the visualized molecular orbitals. Classify the molecular orbitals with symmetry labels, i.e σ or π, and indicate with an asterisk (*) if the orbital is antibonding. 2. Equilibrium geometries and reaction energy of HCN + 3 H 2 CH 4 + NH 3 Calculations: Optimize the geometries of HCN, H 2, CH 4, and NH 3 using Hartree- Fock and the 6-31G** basis set (keyword Opt). Input files are available in HCN.inp, H2.inp, CH4.inp, and NH3.inp. Analysis: Measure the covalent bond lengths of the optimized geometries using Molekel or Molden. Calculate the reaction energy (in kj/mol) for the reaction HCN + 3 H 2 CH 4 + NH 3 from the total energies of the optimized structures. Compare the computed value with the experimental U obtained from H in e.g. SI Chemical Data (remember that H = U + n RT). 3. Molecular orbitals, Mulliken charges and dipole moments of H 2 O Calculations: Perform a single point calculation at the HF/6-31G** level of theory on the experimental geometry of H 2 O, in order to study the molecular orbitals (use keywords GFPrint and Pop=Full), Mulliken charges, and dipole moment. An input file is available in H2O_MO.inp. Perform another single point calculation on the same geometry using the correlation method MP2 and the 6-31G** basis set (exchange the keyword RHF with MP2 and add the keyword Density). Analysis: In X-ray Photoelectron Spectroscopy (XPS), the ionization energy of H 2 O is about 18 ev. On the basis of the HF calculation, from which molecular orbital does the ionization occur? What does the molecular orbital look like (use Molekel to visualize)? What are the Mulliken charges of the atoms in H 2 O? How does the dipole moment calculated with the HF and MP2 methods compare to the experimental dipole moment of 1.85 Debye? Extra: Perform RHF and MP2 calculations using the near-hartree-fock-limit basis set G(3d2f,3p2d) and compare the obtained dipole moments. 4. Product and binding energy of the HF + OH reaction Calculations: Study the acid-base reaction between HF and OH by performing a geometry optimization of the geometry enclosed in the input file OH_HF.inp using Hartree-Fock and the 6-31G** basis set. With the same method, study the reaction between Be 2+ and LiF starting from Be_LiF.inp. 4
5 Analysis: Describe what happens in the reactions? What product is formed? Calculate the binding energy 4 of the product complex. 5. Reaction coordinate and activation energy of a proton transfer reaction Calculations: Proton transfer reactions occur in many catalytic and biochemical reactions. Use the model system H 3 O + + H 2 O to study the energy barrier of proton transfer. A potential energy surface (PES) scan is the mapping of the PES by calculating the energy of the system at several distinct values of for instance a bond length or bond angle. In a rigid PES scan, a series of single point energies are performed on geometries in which a certain parameter is varied and the other parameters kept fixed. In a relaxed scan, however, a certain parameter is systematically varied while all other degrees of freedom are optimized. Perform a rigid potential energy surface scan at the B3LYP/6-31G** level of theory by scanning the O-H distance in steps of 0.1 Å. A starting geometry is available in H3O_H2O.inp. Analysis: In plotting program of your choice, plot the energy as a function of the O-H distance. What is the activation energy? 6. Identify the OH vibrational modes of the formic acid monomer and dimer Calculations: Perform geometry optimization and frequency calculation on formic acid (HCOOH) at the HF/6-31G* level. The input file HCOOH.inp is available. Construct a starting geometry for a hydrogen bonded HCOOH dimer 5, optimize the geometry and calculate the vibrational frequencies. Analysis: In Molekel, examine the OH vibrational modes (vibration/displacement and frequency) of the two systems. How does the dimerization affect the OH vibrational frequency? 7. Changes in molecular orbital energies of PEDOT with increasing chain length Calculations: Perform single point calculations using the B3LYP hybrid density functional and the 6-31G** basis set for 1, 2 and 3 fused monomers of 3,4- ethylenedioxythiophene (EDOT) (input files: pedot1.inp etc). Analysis: Study how the HOMO and LUMO energies change with increasing length of the organic polymer poly(3,4-ethylenedioxythiophene) (PEDOT). Identify the molecular orbital energy of the HOMO (the highest occupied orbital) and of the LUMO (the lowest unoccupied/virtual orbital), and plot both MO energies as a function of the number of fused monomers in a diagram. In addition, plot the LUMO-HOMO energy difference versus chain length. If time permits, construct polymers consisting of 4, 5 and so on molecules, geometry optimize these structures, calculate the MO energies, and extend the diagrams. 4 Binding energy = energy of the product complex sum of the energies of the monomers (opposite sign) 5 Tip for construction of dimer: in Molekel, read the HCOOH geometry twice, orient the molecules relative to each other, write current orientation of both molecules, and combine coordinates in one file. 5
Introduction to Hartree-Fock calculations in Spartan
 EE5 in 2008 Hannes Jónsson Introduction to Hartree-Fock calculations in Spartan In this exercise, you will get to use state of the art software for carrying out calculations of wavefunctions for molecues,
EE5 in 2008 Hannes Jónsson Introduction to Hartree-Fock calculations in Spartan In this exercise, you will get to use state of the art software for carrying out calculations of wavefunctions for molecues,
计算物理作业二. Excercise 1: Illustration of the convergence of the dissociation energy for H 2 toward HF limit.
 计算物理作业二 Excercise 1: Illustration of the convergence of the dissociation energy for H 2 toward HF limit. In this exercise, basis indicates one of the following basis sets: STO-3G, cc-pvdz, cc-pvtz, cc-pvqz
计算物理作业二 Excercise 1: Illustration of the convergence of the dissociation energy for H 2 toward HF limit. In this exercise, basis indicates one of the following basis sets: STO-3G, cc-pvdz, cc-pvtz, cc-pvqz
Conformational energy analysis
 Lab 3 Conformational energy analysis Objective This computational project deals with molecular conformations the spatial arrangement of atoms of molecules. Conformations are determined by energy, so the
Lab 3 Conformational energy analysis Objective This computational project deals with molecular conformations the spatial arrangement of atoms of molecules. Conformations are determined by energy, so the
Project 2. Chemistry of Transient Species in Planetary Atmospheres: Exploring the Potential Energy Surfaces of CH 2 S
 Chemistry 362 Spring 2018 Dr. Jean M. Standard March 21, 2018 Project 2. Chemistry of Transient Species in Planetary Atmospheres: Exploring the Potential Energy Surfaces of CH 2 S In this project, you
Chemistry 362 Spring 2018 Dr. Jean M. Standard March 21, 2018 Project 2. Chemistry of Transient Species in Planetary Atmospheres: Exploring the Potential Energy Surfaces of CH 2 S In this project, you
A Computer Study of Molecular Electronic Structure
 A Computer Study of Molecular Electronic Structure The following exercises are designed to give you a brief introduction to some of the types of information that are now readily accessible from electronic
A Computer Study of Molecular Electronic Structure The following exercises are designed to give you a brief introduction to some of the types of information that are now readily accessible from electronic
LUMO + 1 LUMO. Tómas Arnar Guðmundsson Report 2 Reikniefnafræði G
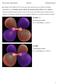 Q1: Display all the MOs for N2 in your report and classify each one of them as bonding, antibonding or non-bonding, and say whether the symmetry of the orbital is σ or π. Sketch a molecular orbital diagram
Q1: Display all the MOs for N2 in your report and classify each one of them as bonding, antibonding or non-bonding, and say whether the symmetry of the orbital is σ or π. Sketch a molecular orbital diagram
Project 3: Molecular Orbital Calculations of Diatomic Molecules. This project is worth 30 points and is due on Wednesday, May 2, 2018.
 Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Chemistry 362 Spring 2018 Dr. Jean M. Standard April 20, 2018 Project 3: Molecular Orbital Calculations of Diatomic Molecules In this project, you will investigate the molecular orbitals and molecular
Introduction to Ab Initio Quantum Chemical Computation
 c:\374-17\computation\computation17.doc for 9mar17 Prof. Patrik Callis 8mar17 Introduction to Ab Initio Quantum Chemical Computation Purpose: 1. To become acquainted with basic concepts of ab initio quantum
c:\374-17\computation\computation17.doc for 9mar17 Prof. Patrik Callis 8mar17 Introduction to Ab Initio Quantum Chemical Computation Purpose: 1. To become acquainted with basic concepts of ab initio quantum
Literature values: ΔH f, gas = % error Source: ΔH f, solid = % error. For comparison, your experimental value was ΔH f = phase:
 1 Molecular Calculations Lab: Some guideline given at the bottom of page 3. 1. Use the semi-empirical AM1 method to calculate ΔH f for the compound you used in the heat of combustion experiment. Be sure
1 Molecular Calculations Lab: Some guideline given at the bottom of page 3. 1. Use the semi-empirical AM1 method to calculate ΔH f for the compound you used in the heat of combustion experiment. Be sure
QUANTUM CHEMISTRY PROJECT 3: PARTS B AND C
 Chemistry 460 Fall 2017 Dr. Jean M. Standard November 6, 2017 QUANTUM CHEMISTRY PROJECT 3: PARTS B AND C PART B: POTENTIAL CURVE, SPECTROSCOPIC CONSTANTS, AND DISSOCIATION ENERGY OF DIATOMIC HYDROGEN (20
Chemistry 460 Fall 2017 Dr. Jean M. Standard November 6, 2017 QUANTUM CHEMISTRY PROJECT 3: PARTS B AND C PART B: POTENTIAL CURVE, SPECTROSCOPIC CONSTANTS, AND DISSOCIATION ENERGY OF DIATOMIC HYDROGEN (20
Figure 1: Transition State, Saddle Point, Reaction Pathway
 Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Potential Energy Surfaces
Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Potential Energy Surfaces
Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018)
 Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018) Abstract Students are introduced to basic features of Scigress by building molecules and performing calculations on them using semi-empirical
Learning to Use Scigress Wagner, Eugene P. (revised May 15, 2018) Abstract Students are introduced to basic features of Scigress by building molecules and performing calculations on them using semi-empirical
QUANTUM CHEMISTRY WITH GAUSSIAN : A VERY BRIEF INTRODUCTION (PART 2)
 QUANTUM CHEMISTRY WITH GAUSSIAN : A VERY BRIEF INTRODUCTION (PART 2) TARAS V. POGORELOV AND MIKE HALLOCK SCHOOL OF CHEMICAL SCIENCES, UIUC This tutorial continues introduction to Gaussian [2]. Here we
QUANTUM CHEMISTRY WITH GAUSSIAN : A VERY BRIEF INTRODUCTION (PART 2) TARAS V. POGORELOV AND MIKE HALLOCK SCHOOL OF CHEMICAL SCIENCES, UIUC This tutorial continues introduction to Gaussian [2]. Here we
Ethene. Introduction. The ethene molecule is planar (i.e. all the six atoms lie in the same plane) and has a high degree of symmetry:
 FY1006 Innføring i kvantefysikk og TFY4215 Kjemisk fysikk og kvantemekanikk Spring 2012 Chemical Physics Exercise 1 To be delivered by Friday 27.04.12 Introduction Ethene. Ethylene, C 2 H 4, or ethene,
FY1006 Innføring i kvantefysikk og TFY4215 Kjemisk fysikk og kvantemekanikk Spring 2012 Chemical Physics Exercise 1 To be delivered by Friday 27.04.12 Introduction Ethene. Ethylene, C 2 H 4, or ethene,
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Gaussian: Basic Tutorial
 Input file: # hf sto-g pop=full Water - Single Point Energy 0 H.0 H.0 H 04.5 Route Section Start with # Contains the keywords Gaussian: Basic Tutorial Route Section Title Section Charge-Multiplicity Molecule
Input file: # hf sto-g pop=full Water - Single Point Energy 0 H.0 H.0 H 04.5 Route Section Start with # Contains the keywords Gaussian: Basic Tutorial Route Section Title Section Charge-Multiplicity Molecule
MO Calculation for a Diatomic Molecule. /4 0 ) i=1 j>i (1/r ij )
 MO Calculation for a Diatomic Molecule Introduction The properties of any molecular system can in principle be found by looking at the solutions to the corresponding time independent Schrodinger equation
MO Calculation for a Diatomic Molecule Introduction The properties of any molecular system can in principle be found by looking at the solutions to the corresponding time independent Schrodinger equation
Experiment 15: Atomic Orbitals, Bond Length, and Molecular Orbitals
 Experiment 15: Atomic Orbitals, Bond Length, and Molecular Orbitals Introduction Molecular orbitals result from the mixing of atomic orbitals that overlap during the bonding process allowing the delocalization
Experiment 15: Atomic Orbitals, Bond Length, and Molecular Orbitals Introduction Molecular orbitals result from the mixing of atomic orbitals that overlap during the bonding process allowing the delocalization
Chemistry 14CL. Worksheet for the Molecular Modeling Workshop. (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell)
 Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Chemistry 14CL Worksheet for the Molecular Modeling Workshop (Revised FULL Version 2012 J.W. Pang) (Modified A. A. Russell) Structure of the Molecular Modeling Assignment The molecular modeling assignment
Hints on Using the Orca Program
 Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Hints on Using the Orca Program
Computational Chemistry Workshops West Ridge Research Building-UAF Campus 9:00am-4:00pm, Room 009 Electronic Structure - July 19-21, 2016 Molecular Dynamics - July 26-28, 2016 Hints on Using the Orca Program
Introduction to Hartree-Fock calculations using ORCA and Chemcraft
 Introduction to Hartree-Fock calculations using ORCA and Chemcraft In this exercise, you will get to use software for carrying out calculations of wavefunctions for molecules, the ORCA program. While ORCA
Introduction to Hartree-Fock calculations using ORCA and Chemcraft In this exercise, you will get to use software for carrying out calculations of wavefunctions for molecules, the ORCA program. While ORCA
Physical Chemistry II Recommended Problems Chapter 12( 23)
 Physical Chemistry II Recommended Problems Chapter 1( 3) Chapter 1(3) Problem. Overlap of two 1s orbitals recprobchap1.odt 1 Physical Chemistry II Recommended Problems, Chapter 1(3), continued Chapter
Physical Chemistry II Recommended Problems Chapter 1( 3) Chapter 1(3) Problem. Overlap of two 1s orbitals recprobchap1.odt 1 Physical Chemistry II Recommended Problems, Chapter 1(3), continued Chapter
Using Web-Based Computations in Organic Chemistry
 10/30/2017 1 Using Web-Based Computations in Organic Chemistry John Keller UAF Department of Chemistry & Biochemistry The UAF WebMO site Practical aspects of computational chemistry theory and nomenclature
10/30/2017 1 Using Web-Based Computations in Organic Chemistry John Keller UAF Department of Chemistry & Biochemistry The UAF WebMO site Practical aspects of computational chemistry theory and nomenclature
Chapter: 22. Visualization: Making INPUT File and Processing of Output Results
 Chapter: 22 Visualization: Making INPUT File and Processing of Output Results Keywords: visualization, input and output structure, molecular orbital, electron density. In the previous chapters, we have
Chapter: 22 Visualization: Making INPUT File and Processing of Output Results Keywords: visualization, input and output structure, molecular orbital, electron density. In the previous chapters, we have
General Chemistry Lab Molecular Modeling
 PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
1 Introduction to Computational Chemistry (Spartan)
 1 Introduction to Computational Chemistry (Spartan) Start Spartan by clicking Start / Programs / Spartan Then click File / New Exercise 1 Study of H-X-H Bond Angles (Suitable for general chemistry) Structure
1 Introduction to Computational Chemistry (Spartan) Start Spartan by clicking Start / Programs / Spartan Then click File / New Exercise 1 Study of H-X-H Bond Angles (Suitable for general chemistry) Structure
Jaguar DFT Optimizations and Transition State Searches
 Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Jaguar DFT Optimizations and Transition State Searches Density Functional Theory (DFT) is a quantum mechanical (QM) method that gives results superior to Hartree Fock (HF) in less computational time. A
Assignment A02: Geometry Definition: File Formats, Redundant Coordinates, PES Scans
 Assignment A02: Geometry Definition: File Formats, Redundant Coordinates, PES Scans In Assignments A00 and A01, you familiarized yourself with GaussView and G09W, you learned the basics about input (GJF)
Assignment A02: Geometry Definition: File Formats, Redundant Coordinates, PES Scans In Assignments A00 and A01, you familiarized yourself with GaussView and G09W, you learned the basics about input (GJF)
Modeling the UV-Vis Absorption of a Series of Dyes CH342L: Spectroscopy February 15, 2016
 Modeling the UV-Vis Absorption of a Series of Dyes CH342L: Spectroscopy February 15, 2016 We ll correlate the absorbance maximum of a series of dyes with structural changes between them 1. Chemicals absorb
Modeling the UV-Vis Absorption of a Series of Dyes CH342L: Spectroscopy February 15, 2016 We ll correlate the absorbance maximum of a series of dyes with structural changes between them 1. Chemicals absorb
John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks
 10/15/2016 1 WebMO & Gaussian John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Corrections and updates 9-5-2017 SCHEDULE 9-10 Intro and basic operation of WebMO and MOPAC
10/15/2016 1 WebMO & Gaussian John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Corrections and updates 9-5-2017 SCHEDULE 9-10 Intro and basic operation of WebMO and MOPAC
Name. Chem Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2
 Name Chem 322 - Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2 Click on Titan in the Start menu. When it boots, click on the right corner to make the window full-screen. icon in the
Name Chem 322 - Organic Chemistry II Laboratory Exercise Molecular Modeling Part 2 Click on Titan in the Start menu. When it boots, click on the right corner to make the window full-screen. icon in the
The Hückel Approximation Consider a conjugated molecule i.e. a molecule with alternating double and single bonds, as shown in Figure 1.
 The Hückel Approximation In this exercise you will use a program called Hückel to look at the p molecular orbitals in conjugated molecules. The program calculates the energies and shapes of p (pi) molecular
The Hückel Approximation In this exercise you will use a program called Hückel to look at the p molecular orbitals in conjugated molecules. The program calculates the energies and shapes of p (pi) molecular
Theoretical UV/VIS Spectroscopy
 Theoretical UV/VIS Spectroscopy Why is a Ruby Red When Chromium Oxide is Green? How Does a Ruby Laser Work? Goals of this Exercise: - Calculation of the energy of electronically excited states - Understanding
Theoretical UV/VIS Spectroscopy Why is a Ruby Red When Chromium Oxide is Green? How Does a Ruby Laser Work? Goals of this Exercise: - Calculation of the energy of electronically excited states - Understanding
Computational Chemistry Using the University of Alaska WebMO Site
 2/7/2017 1 Computational Chemistry Using the University of Alaska WebMO Site John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Intro and operation of WebMO and MOPAC Basic
2/7/2017 1 Computational Chemistry Using the University of Alaska WebMO Site John Keller Department of Chemistry & Biochemistry University of Alaska Fairbanks Intro and operation of WebMO and MOPAC Basic
Exercise 1: Structure and dipole moment of a small molecule
 Introduction to computational chemistry Exercise 1: Structure and dipole moment of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the dipole moment of a small
Introduction to computational chemistry Exercise 1: Structure and dipole moment of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the dipole moment of a small
Introduction to Computational Chemistry Exercise 2
 Introduction to Computational Chemistry Exercise 2 Intermolecular interactions and vibrational motion Lecturer: Antti Lignell Name Introduction In this computer exercise, we model intermolecular interactions
Introduction to Computational Chemistry Exercise 2 Intermolecular interactions and vibrational motion Lecturer: Antti Lignell Name Introduction In this computer exercise, we model intermolecular interactions
This is called a singlet or spin singlet, because the so called multiplicity, or number of possible orientations of the total spin, which is
 9. Open shell systems The derivation of Hartree-Fock equations (Chapter 7) was done for a special case of a closed shell systems. Closed shell means that each MO is occupied by two electrons with the opposite
9. Open shell systems The derivation of Hartree-Fock equations (Chapter 7) was done for a special case of a closed shell systems. Closed shell means that each MO is occupied by two electrons with the opposite
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Jack Smith. Center for Environmental, Geotechnical and Applied Science. Marshall University
 Jack Smith Center for Environmental, Geotechnical and Applied Science Marshall University -- Division of Science and Research WV Higher Education Policy Commission WVU HPC Summer Institute June 20, 2014
Jack Smith Center for Environmental, Geotechnical and Applied Science Marshall University -- Division of Science and Research WV Higher Education Policy Commission WVU HPC Summer Institute June 20, 2014
Tutorial I: IQ MOL and Basic DFT and MP2 Calculations 1 / 30
 Tutorial I: IQ MOL and Basic DFT and MP2 Calculations Q-Chem User Workshop, Denver March 21, 2015 1 / 30 2 / 30 Introduction to IQMOL DFT and MP2 Calculations 3 / 30 IQMOL and Q-CHEM IQMOL is an open-source
Tutorial I: IQ MOL and Basic DFT and MP2 Calculations Q-Chem User Workshop, Denver March 21, 2015 1 / 30 2 / 30 Introduction to IQMOL DFT and MP2 Calculations 3 / 30 IQMOL and Q-CHEM IQMOL is an open-source
Chapter 9. Covalent Bonding: Orbitals
 Chapter 9 Covalent Bonding: Orbitals EXERCISE! Draw the Lewis structure for methane, CH 4. What is the shape of a methane molecule? tetrahedral What are the bond angles? 109.5 o H H C H H Copyright Cengage
Chapter 9 Covalent Bonding: Orbitals EXERCISE! Draw the Lewis structure for methane, CH 4. What is the shape of a methane molecule? tetrahedral What are the bond angles? 109.5 o H H C H H Copyright Cengage
one ν im: transition state saddle point
 Hypothetical Potential Energy Surface Ethane conformations Hartree-Fock theory, basis set stationary points all ν s >0: minimum eclipsed one ν im: transition state saddle point multiple ν im: hilltop 1
Hypothetical Potential Energy Surface Ethane conformations Hartree-Fock theory, basis set stationary points all ν s >0: minimum eclipsed one ν im: transition state saddle point multiple ν im: hilltop 1
Instructions for Using Spartan 14
 Instructions for Using Spartan 14 Log in to the computer with your Colby ID and password. Click on the Spartan 14 icon in the dock at the bottom of your screen. I. Building Molecules Spartan has one main
Instructions for Using Spartan 14 Log in to the computer with your Colby ID and password. Click on the Spartan 14 icon in the dock at the bottom of your screen. I. Building Molecules Spartan has one main
Molecular Orbitals for Ozone
 Molecular Orbitals for Ozone Purpose: In this exercise you will do semi-empirical molecular orbital calculations on ozone with the goal of understanding the molecular orbital print out provided by Spartan
Molecular Orbitals for Ozone Purpose: In this exercise you will do semi-empirical molecular orbital calculations on ozone with the goal of understanding the molecular orbital print out provided by Spartan
Gustavus Adolphus College. Lab #5: Computational Chemistry
 CHE 372 Gustavus Adolphus College Lab #5: Computational Chemistry Introduction In this investigation we will apply the techniques of computational chemistry to several of the molecular systems that we
CHE 372 Gustavus Adolphus College Lab #5: Computational Chemistry Introduction In this investigation we will apply the techniques of computational chemistry to several of the molecular systems that we
Molecular Modeling and Conformational Analysis with PC Spartan
 Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Contents. 1. Building a molecular model 2. Additional Procedures and Options: 3. Selecting a calculation method
 Getting started with HyperChem. Program description. In this project you will create simple organic molecules and learn how to manipulate them. However, before starting you should create your home directory
Getting started with HyperChem. Program description. In this project you will create simple organic molecules and learn how to manipulate them. However, before starting you should create your home directory
Introduc)on to IQmol: Part I.!!! Shirin Faraji, Ilya Kaliman, and Anna Krylov
 Introduc)on to IQmol: Part I!!! Shirin Faraji, Ilya Kaliman, and Anna Krylov! 1 Resources! Written by Dr. Andrew Gilbert Keep yourself up to date with IQmol website: http://iqmol.org! IQmol Youtube channel:
Introduc)on to IQmol: Part I!!! Shirin Faraji, Ilya Kaliman, and Anna Krylov! 1 Resources! Written by Dr. Andrew Gilbert Keep yourself up to date with IQmol website: http://iqmol.org! IQmol Youtube channel:
( ) + ( ) + ( ) = 0.00
 Chem 3502/4502 Physical Chemistry II (Quantum Mechanics) 3 Credits Spring Semester 2006 Christopher J. Cramer Lecture 32, April 14, 2006 (Some material in this lecture has been adapted from Cramer, C.
Chem 3502/4502 Physical Chemistry II (Quantum Mechanics) 3 Credits Spring Semester 2006 Christopher J. Cramer Lecture 32, April 14, 2006 (Some material in this lecture has been adapted from Cramer, C.
Appendix D Simulating Spectroscopic Bands Using Gaussian and PGopher
 429 Appendix D Simulating Spectroscopic Bands Using Gaussian and PGopher This appendix contains methods for using Gaussian 09 121 and PGopher 120 to simulate vibrational and electronic bands of molecules.
429 Appendix D Simulating Spectroscopic Bands Using Gaussian and PGopher This appendix contains methods for using Gaussian 09 121 and PGopher 120 to simulate vibrational and electronic bands of molecules.
Transition states and reaction paths
 Transition states and reaction paths Lab 4 Theoretical background Transition state A transition structure is the molecular configuration that separates reactants and products. In a system with a single
Transition states and reaction paths Lab 4 Theoretical background Transition state A transition structure is the molecular configuration that separates reactants and products. In a system with a single
with the larger dimerization energy also exhibits the larger structural changes.
 A7. Looking at the image and table provided below, it is apparent that the monomer and dimer are structurally almost identical. Although angular and dihedral data were not included, these data are also
A7. Looking at the image and table provided below, it is apparent that the monomer and dimer are structurally almost identical. Although angular and dihedral data were not included, these data are also
BOND LENGTH WITH HYPERCHEM LITE
 BOND LENGTH WITH HYPERCHEM LITE LAB MOD2.COMP From Gannon University SIM INTRODUCTION The electron cloud surrounding the nucleus of the atom determines the size of the atom. Since this distance is somewhat
BOND LENGTH WITH HYPERCHEM LITE LAB MOD2.COMP From Gannon University SIM INTRODUCTION The electron cloud surrounding the nucleus of the atom determines the size of the atom. Since this distance is somewhat
Computational Chemistry Lab Module: Conformational Analysis of Alkanes
 Introduction Computational Chemistry Lab Module: Conformational Analysis of Alkanes In this experiment, we will use CAChe software package to model the conformations of butane, 2-methylbutane, and substituted
Introduction Computational Chemistry Lab Module: Conformational Analysis of Alkanes In this experiment, we will use CAChe software package to model the conformations of butane, 2-methylbutane, and substituted
XYZ file format Protein Data Bank (pdb) file format Z - matrix
 Chemistry block (exercise 1) In this exercise, students will be introduced how to preform simple quantum chemical calculations. Input files for Gaussian09. Output file structure. Geometry optimization,
Chemistry block (exercise 1) In this exercise, students will be introduced how to preform simple quantum chemical calculations. Input files for Gaussian09. Output file structure. Geometry optimization,
Introduction to computational chemistry Exercise I: Structure and electronic energy of a small molecule. Vesa Hänninen
 Introduction to computational chemistry Exercise I: Structure and electronic energy of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the electronic energy
Introduction to computational chemistry Exercise I: Structure and electronic energy of a small molecule Vesa Hänninen 1 Introduction In this exercise the equilibrium structure and the electronic energy
Calculating NMR Chemical Shifts for beta-ionone O
 Calculating NMR Chemical Shifts for beta-ionone O Molecular orbital calculations can be used to get good estimates for chemical shifts. In this exercise we will calculate the chemical shifts for beta-ionone.
Calculating NMR Chemical Shifts for beta-ionone O Molecular orbital calculations can be used to get good estimates for chemical shifts. In this exercise we will calculate the chemical shifts for beta-ionone.
Example: H 2 O (the car file)
 Example: H 2 O (the car file) As a practical example of DFT methods we calculate the energy and electronic properties of the water molecule. In order to carry out the DFT calculation you will need a set
Example: H 2 O (the car file) As a practical example of DFT methods we calculate the energy and electronic properties of the water molecule. In order to carry out the DFT calculation you will need a set
CHEM 344 Molecular Modeling
 CHEM 344 Molecular Modeling The Use of Computational Chemistry to Support Experimental Organic Chemistry Part 1: Molecular Orbital Theory, Hybridization, & Formal Charge * all calculation data obtained
CHEM 344 Molecular Modeling The Use of Computational Chemistry to Support Experimental Organic Chemistry Part 1: Molecular Orbital Theory, Hybridization, & Formal Charge * all calculation data obtained
NH 3 H 2 O N 2. Why do they make chemical bonds? Molecular Orbitals
 N 2 NH 3 H 2 O Why do they make chemical bonds? 5 Molecular Orbitals Why do they make chemical bonds? Stabilization Bond energy Types of Chemical Bonds Metallic Bond Ionic Bond Covalent Bond Covalent Bond
N 2 NH 3 H 2 O Why do they make chemical bonds? 5 Molecular Orbitals Why do they make chemical bonds? Stabilization Bond energy Types of Chemical Bonds Metallic Bond Ionic Bond Covalent Bond Covalent Bond
Department of Chemistry. The Intersection of Computational Chemistry and Experiment
 Department of Chemistry The Intersection of Computational Chemistry and Experiment Structure, Vibrational and Electronic Spectra of Organic Molecules Angelo R. Rossi Department of Chemistry The University
Department of Chemistry The Intersection of Computational Chemistry and Experiment Structure, Vibrational and Electronic Spectra of Organic Molecules Angelo R. Rossi Department of Chemistry The University
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
DFT and beyond: Hands-on Tutorial Workshop Tutorial 1: Basics of Electronic Structure Theory
 DFT and beyond: Hands-on Tutorial Workshop 2011 Tutorial 1: Basics of Electronic Structure Theory V. Atalla, O. T. Hofmann, S. V. Levchenko Theory Department, Fritz-Haber-Institut der MPG Berlin July 13,
DFT and beyond: Hands-on Tutorial Workshop 2011 Tutorial 1: Basics of Electronic Structure Theory V. Atalla, O. T. Hofmann, S. V. Levchenko Theory Department, Fritz-Haber-Institut der MPG Berlin July 13,
QUANTUM CHEMISTRY PROJECT 3: ATOMIC AND MOLECULAR STRUCTURE
 Chemistry 460 Fall 2017 Dr. Jean M. Standard November 1, 2017 QUANTUM CHEMISTRY PROJECT 3: ATOMIC AND MOLECULAR STRUCTURE OUTLINE In this project, you will carry out quantum mechanical calculations of
Chemistry 460 Fall 2017 Dr. Jean M. Standard November 1, 2017 QUANTUM CHEMISTRY PROJECT 3: ATOMIC AND MOLECULAR STRUCTURE OUTLINE In this project, you will carry out quantum mechanical calculations of
Be H. Delocalized Bonding. Localized Bonding. σ 2. σ 1. Two (sp-1s) Be-H σ bonds. The two σ bonding MO s in BeH 2. MO diagram for BeH 2
 The Delocalized Approach to Bonding: The localized models for bonding we have examined (Lewis and VBT) assume that all electrons are restricted to specific bonds between atoms or in lone pairs. In contrast,
The Delocalized Approach to Bonding: The localized models for bonding we have examined (Lewis and VBT) assume that all electrons are restricted to specific bonds between atoms or in lone pairs. In contrast,
Lab #3: Choice of Theoretical Method
 Lab #3: Choice of Theoretical Method These directions assume the user is familiar with the WebMO interface and can build molecules, set up calculations, etc. Exercise 1 - Determine the Proton Affinity
Lab #3: Choice of Theoretical Method These directions assume the user is familiar with the WebMO interface and can build molecules, set up calculations, etc. Exercise 1 - Determine the Proton Affinity
Theoretical Chemistry - Level II - Practical Class Molecular Orbitals in Diatomics
 Theoretical Chemistry - Level II - Practical Class Molecular Orbitals in Diatomics Problem 1 Draw molecular orbital diagrams for O 2 and O 2 +. E / ev dioxygen molecule, O 2 dioxygenyl cation, O 2 + 25
Theoretical Chemistry - Level II - Practical Class Molecular Orbitals in Diatomics Problem 1 Draw molecular orbital diagrams for O 2 and O 2 +. E / ev dioxygen molecule, O 2 dioxygenyl cation, O 2 + 25
Reikniefnafræði - Verkefni 2 Haustmisseri 2013 Kennari - Hannes Jónsson
 Háskóli Íslands, raunvísindasvið Reikniefnafræði - Verkefni 2 Haustmisseri 2013 Kennari - Hannes Jónsson Guðjón Henning 18. september 2013 1 A. Molecular orbitals of N 2 Q1: Display all the MOs for N 2
Háskóli Íslands, raunvísindasvið Reikniefnafræði - Verkefni 2 Haustmisseri 2013 Kennari - Hannes Jónsson Guðjón Henning 18. september 2013 1 A. Molecular orbitals of N 2 Q1: Display all the MOs for N 2
Quantum Chemistry. NC State University. Lecture 5. The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy
 Quantum Chemistry Lecture 5 The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy NC State University 3.5 Selective absorption and emission by atmospheric gases (source:
Quantum Chemistry Lecture 5 The electronic structure of molecules Absorption spectroscopy Fluorescence spectroscopy NC State University 3.5 Selective absorption and emission by atmospheric gases (source:
Due: since the calculation takes longer than before, we ll make it due on 02/05/2016, Friday
 Homework 3 Due: since the calculation takes longer than before, we ll make it due on 02/05/2016, Friday Email to: jqian@caltech.edu Introduction In this assignment, you will be using a commercial periodic
Homework 3 Due: since the calculation takes longer than before, we ll make it due on 02/05/2016, Friday Email to: jqian@caltech.edu Introduction In this assignment, you will be using a commercial periodic
CHEM 545 Theory and Practice of Molecular Electronic Structure. Anna I. Krylov. DON T PANIC.
 CHEM 545 Theory and Practice of Molecular Electronic Structure Anna I. Krylov http://iopenshell.usc.edu/chem545/ DON T PANIC USC Fall 2014 Things to do: 1. Install IQmol (by this Thursday). http://iqmol.org/.
CHEM 545 Theory and Practice of Molecular Electronic Structure Anna I. Krylov http://iopenshell.usc.edu/chem545/ DON T PANIC USC Fall 2014 Things to do: 1. Install IQmol (by this Thursday). http://iqmol.org/.
Lecture B6 Molecular Orbital Theory. Sometimes it's good to be alone.
 Lecture B6 Molecular Orbital Theory Sometimes it's good to be alone. Covalent Bond Theories 1. VSEPR (valence shell electron pair repulsion model). A set of empirical rules for predicting a molecular geometry
Lecture B6 Molecular Orbital Theory Sometimes it's good to be alone. Covalent Bond Theories 1. VSEPR (valence shell electron pair repulsion model). A set of empirical rules for predicting a molecular geometry
NH 3 inversion: Potential energy surfaces and transition states CH342L March 28, 2016
 N 3 inversion: Potential energy surfaces and transition states C342L March 28, 2016 Last week, we used the IR spectrum of ammonia to determine the splitting of energy levels due to inversion of the umbrella
N 3 inversion: Potential energy surfaces and transition states C342L March 28, 2016 Last week, we used the IR spectrum of ammonia to determine the splitting of energy levels due to inversion of the umbrella
Benzene Dimer: dispersion forces and electronic correlation
 Benzene Dimer: dispersion forces and electronic correlation Introduction The Benzene dimer is an ideal example of a system bound by π-π interaction, which is in several cases present in many biologically
Benzene Dimer: dispersion forces and electronic correlation Introduction The Benzene dimer is an ideal example of a system bound by π-π interaction, which is in several cases present in many biologically
Chapter 9. Covalent Bonding: Orbitals
 Chapter 9 Covalent Bonding: Orbitals Chapter 9 Table of Contents 9.1 Hybridization and the Localized Electron Model 9.2 The Molecular Orbital Model 9.3 Bonding in Homonuclear Diatomic Molecules 9.4 Bonding
Chapter 9 Covalent Bonding: Orbitals Chapter 9 Table of Contents 9.1 Hybridization and the Localized Electron Model 9.2 The Molecular Orbital Model 9.3 Bonding in Homonuclear Diatomic Molecules 9.4 Bonding
This is a very succinct primer intended as supplementary material for an undergraduate course in physical chemistry.
 1 Computational Chemistry (Quantum Chemistry) Primer This is a very succinct primer intended as supplementary material for an undergraduate course in physical chemistry. TABLE OF CONTENTS Methods...1 Basis
1 Computational Chemistry (Quantum Chemistry) Primer This is a very succinct primer intended as supplementary material for an undergraduate course in physical chemistry. TABLE OF CONTENTS Methods...1 Basis
Introductory WebMO Exercises
 Introductory WebMO Exercises These directions assume no prior knowledge of e WebMO interface and provide detailed, click by click instructions on building molecules and setting up calculations. Use your
Introductory WebMO Exercises These directions assume no prior knowledge of e WebMO interface and provide detailed, click by click instructions on building molecules and setting up calculations. Use your
Computational Material Science Part II. Ito Chao ( ) Institute of Chemistry Academia Sinica
 Computational Material Science Part II Ito Chao ( ) Institute of Chemistry Academia Sinica Ab Initio Implementations of Hartree-Fock Molecular Orbital Theory Fundamental assumption of HF theory: each electron
Computational Material Science Part II Ito Chao ( ) Institute of Chemistry Academia Sinica Ab Initio Implementations of Hartree-Fock Molecular Orbital Theory Fundamental assumption of HF theory: each electron
Atomic and Molecular Orbitals
 7 Atomic and Molecular Orbitals Chemists have developed a variety of methods for describing electrons in molecules. Lewis structures are the most familiar. These drawings assign pairs of electrons either
7 Atomic and Molecular Orbitals Chemists have developed a variety of methods for describing electrons in molecules. Lewis structures are the most familiar. These drawings assign pairs of electrons either
3: Many electrons. Orbital symmetries. l =2 1. m l
 3: Many electrons Orbital symmetries Atomic orbitals are labelled according to the principal quantum number, n, and the orbital angular momentum quantum number, l. Electrons in a diatomic molecule experience
3: Many electrons Orbital symmetries Atomic orbitals are labelled according to the principal quantum number, n, and the orbital angular momentum quantum number, l. Electrons in a diatomic molecule experience
Lecture 19: Building Atoms and Molecules
 Lecture 19: Building Atoms and Molecules +e r n = 3 n = 2 n = 1 +e +e r y even Lecture 19, p 1 Today Nuclear Magnetic Resonance Using RF photons to drive transitions between nuclear spin orientations in
Lecture 19: Building Atoms and Molecules +e r n = 3 n = 2 n = 1 +e +e r y even Lecture 19, p 1 Today Nuclear Magnetic Resonance Using RF photons to drive transitions between nuclear spin orientations in
S N 2 reaction. Br- + ClCH 3 BrCH 3 + Cl-
 FY1006 Innføring i kvantefysikk og TFY4215 Kjemisk fysikk og kvantemekanikk Spring 2012 Chemical Physics Exercise 2 To be delivered by Friday 27.04.12 S N 2 reaction. Introduction Many chemical reactions
FY1006 Innføring i kvantefysikk og TFY4215 Kjemisk fysikk og kvantemekanikk Spring 2012 Chemical Physics Exercise 2 To be delivered by Friday 27.04.12 S N 2 reaction. Introduction Many chemical reactions
Zeeman Effect Physics 481
 Zeeman Effect Introduction You are familiar with Atomic Spectra, especially the H- atom energy spectrum. Atoms emit or absorb energies in packets, or quanta which are photons. The orbital motion of electrons
Zeeman Effect Introduction You are familiar with Atomic Spectra, especially the H- atom energy spectrum. Atoms emit or absorb energies in packets, or quanta which are photons. The orbital motion of electrons
Introduction. Quantum Mechanical Computation. By Sasha Payne N. Diaz. CHEMISTRY 475-L03 1 January 2035
 Quantum Mechanical Computation By Sasha Payne N. Diaz CHEMISTRY 47-L 1 January Introduction Backgrounds for computational chemistry Understanding the behavior of materials at the atomic scale is fundamental
Quantum Mechanical Computation By Sasha Payne N. Diaz CHEMISTRY 47-L 1 January Introduction Backgrounds for computational chemistry Understanding the behavior of materials at the atomic scale is fundamental
Reikniefnafræði - Verkefni 1 Haustmisseri 2013 Kennari - Hannes Jónsson
 Háskóli Íslands, raunvísindasvið Reikniefnafræði - Verkefni 1 Haustmisseri 2013 Kennari - Hannes Jónsson Guðjón Henning 11. september 2013 A. Molecular orbitals and electron density of H 2 Q1: Present
Háskóli Íslands, raunvísindasvið Reikniefnafræði - Verkefni 1 Haustmisseri 2013 Kennari - Hannes Jónsson Guðjón Henning 11. september 2013 A. Molecular orbitals and electron density of H 2 Q1: Present
Chem 253. Tutorial for Materials Studio
 Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Chem 253 Tutorial for Materials Studio This tutorial is designed to introduce Materials Studio 7.0, which is a program used for modeling and simulating materials for predicting and rationalizing structure
Lab 1 Uniform Motion - Graphing and Analyzing Motion
 Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Lab 1 Uniform Motion - Graphing and Analyzing Motion Objectives: < To observe the distance-time relation for motion at constant velocity. < To make a straight line fit to the distance-time data. < To interpret
Organic Chemistry. Review Information for Unit 1. Atomic Structure MO Theory Chemical Bonds
 Organic Chemistry Review Information for Unit 1 Atomic Structure MO Theory Chemical Bonds Atomic Structure Atoms are the smallest representative particle of an element. Three subatomic particles: protons
Organic Chemistry Review Information for Unit 1 Atomic Structure MO Theory Chemical Bonds Atomic Structure Atoms are the smallest representative particle of an element. Three subatomic particles: protons
ABC of DFT: Hands-on session 1 Introduction into calculations on molecules
 ABC of DFT: Hands-on session 1 Introduction into calculations on molecules Tutor: Alexej Bagrets Wann? 09.11.2012, 11:30-13:00 Wo? KIT Campus Nord, Flachbau Physik, Geb. 30.22, Computerpool, Raum FE-6
ABC of DFT: Hands-on session 1 Introduction into calculations on molecules Tutor: Alexej Bagrets Wann? 09.11.2012, 11:30-13:00 Wo? KIT Campus Nord, Flachbau Physik, Geb. 30.22, Computerpool, Raum FE-6
The Hückel Approximation
 The ückel Approximation 1 In this exercise you will use a program called ückel to look at the π molecular orbitals in conjugated molecules. The program calculates the energies and shapes of π (pi) molecular
The ückel Approximation 1 In this exercise you will use a program called ückel to look at the π molecular orbitals in conjugated molecules. The program calculates the energies and shapes of π (pi) molecular
Be H. Delocalized Bonding. Localized Bonding. σ 2. σ 1. Two (sp-1s) Be-H σ bonds. The two σ bonding MO s in BeH 2. MO diagram for BeH 2
 The Delocalized Approach to Bonding: The localized models for bonding we have examined (Lewis and VBT) assume that all electrons are restricted to specific bonds between atoms or in lone pairs. In contrast,
The Delocalized Approach to Bonding: The localized models for bonding we have examined (Lewis and VBT) assume that all electrons are restricted to specific bonds between atoms or in lone pairs. In contrast,
An Introduction to Basis Sets:
 An Introduction to Basis Sets: The "Holy Grail" of computational chemistry is the calculation of the molecular orbitals (MOs) for a given molecule. IF we can calculate the MOs for a molecule, THEN we can
An Introduction to Basis Sets: The "Holy Grail" of computational chemistry is the calculation of the molecular orbitals (MOs) for a given molecule. IF we can calculate the MOs for a molecule, THEN we can
Project 1 Report File: Chem4PB3_project_1_2017-solutions last changed: 02-Feb-2017
 Project 1 Report File: Chem4PB3_project_1_2017-solutions last changed: 02-Feb-2017 1. Formaldehyde comparison of results from different methods Yellow shaded boxes are closest to experimental Method #e-
Project 1 Report File: Chem4PB3_project_1_2017-solutions last changed: 02-Feb-2017 1. Formaldehyde comparison of results from different methods Yellow shaded boxes are closest to experimental Method #e-
Application Note. U. Heat of Formation of Ethyl Alcohol and Dimethyl Ether. Introduction
 Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Application Note U. Introduction The molecular builder (Molecular Builder) is part of the MEDEA standard suite of building tools. This tutorial provides an overview of the Molecular Builder s basic functionality.
Semi-Empirical MO Methods
 Semi-Empirical MO Methods the high cost of ab initio MO calculations is largely due to the many integrals that need to be calculated (esp. two electron integrals) semi-empirical MO methods start with the
Semi-Empirical MO Methods the high cost of ab initio MO calculations is largely due to the many integrals that need to be calculated (esp. two electron integrals) semi-empirical MO methods start with the
BUILDING BASICS WITH HYPERCHEM LITE
 BUILDING BASICS WITH HYPERCHEM LITE LAB MOD1.COMP From Gannon University SIM INTRODUCTION A chemical bond is a link between atoms resulting from the mutual attraction of their nuclei for electrons. There
BUILDING BASICS WITH HYPERCHEM LITE LAB MOD1.COMP From Gannon University SIM INTRODUCTION A chemical bond is a link between atoms resulting from the mutual attraction of their nuclei for electrons. There
Molecular Orbital Theory. Molecular Orbital Theory: Electrons are located in the molecule, not held in discrete regions between two bonded atoms
 Molecular Orbital Theory Valence Bond Theory: Electrons are located in discrete pairs between specific atoms Molecular Orbital Theory: Electrons are located in the molecule, not held in discrete regions
Molecular Orbital Theory Valence Bond Theory: Electrons are located in discrete pairs between specific atoms Molecular Orbital Theory: Electrons are located in the molecule, not held in discrete regions
Quantification of JEOL XPS Spectra from SpecSurf
 Quantification of JEOL XPS Spectra from SpecSurf The quantification procedure used by the JEOL SpecSurf software involves modifying the Scofield cross-sections to account for both an energy dependency
Quantification of JEOL XPS Spectra from SpecSurf The quantification procedure used by the JEOL SpecSurf software involves modifying the Scofield cross-sections to account for both an energy dependency
Let s continue our discussion on the interaction between Fe(III) and 6,7-dihydroxynaphthalene-2- sulfonate.
 Chemistry 5995(133)-8990(013) Bioinorganic Chemistry: The Good, the Bad, and the Potential of Metals Assignment 2- Aqueous Speciation, Magnetism, Redox, UV-Vis Spectroscopy, and Pymol Let s continue our
Chemistry 5995(133)-8990(013) Bioinorganic Chemistry: The Good, the Bad, and the Potential of Metals Assignment 2- Aqueous Speciation, Magnetism, Redox, UV-Vis Spectroscopy, and Pymol Let s continue our
CHE3935. Lecture 4 Quantum Mechanical Simulation Methods Continued
 CHE3935 Lecture 4 Quantum Mechanical Simulation Methods Continued 1 OUTLINE Review Introduction to CPMD MD and ensembles The functionals of density functional theory Return to ab initio methods Binding
CHE3935 Lecture 4 Quantum Mechanical Simulation Methods Continued 1 OUTLINE Review Introduction to CPMD MD and ensembles The functionals of density functional theory Return to ab initio methods Binding
