Exploiting Tree Kernels for High Performance Chemical Induced Disease. relation extraction.
|
|
|
- Silvester Summers
- 5 years ago
- Views:
Transcription
1 Exploiting Tree Kernels for High Performance Chemical Induced Disease Relation Extraction Nagesh C. Panyam, Karin Verspoor, Trevor Cohn and Kotagiri Ramamohanarao Department of Computing and Information Systems, The University of Melbourne, Australia {karin.verspoor, t.cohn, Abstract Machine learning approaches based on supervised classification have emerged as effective methods for Biomedical relation extraction such as the Chemical-Induced- Disease (CID) task. These approaches owe their success to a rich set of features crafted from the lexical and syntactic regularities in the text. Kernel methods are an effective alternative to manual feature engineering and have been successfully used in similar tasks such as text classification. In this paper, we study the effectiveness of tree kernels for Chemical-Disease relation extraction. Our experiments demonstrate that subset tree kernels increase the F-score to 61.7% as compared to 57.9% achieved with simple feature engineering. We also describe the strengths and shortcomings of tree kernel approaches for the CID relation extraction task. 1 Introduction Scientific publications in the fields of biomedical and life sciences are vast and growing fast (Haas et al., 2014). Prior research has shown that Chemicals and Diseases and their relationships are among the most searched topics by PubMed users (Dogan et al., 2009), due to their importance in applications such as toxicology, drug discovery and safety surveillance. Efforts to manually curate and extract these important concepts such as Chemicals and Diseases and their relationships have led to the creation of structured databases such as the Comparative Toxicogenomics Database (CTD) (Davis et al., 2012). However, manual curation is unlikely to scale (Baumgartner et al., 2007) and has stimulated research interest in automated relation extraction. The recent shared task for Chemical-Induced- Disease relation extraction (CID) organized by BioCreative-V (Wei et al., 2015), has made available a large body of annotated PubMed abstracts for the valuable Chemical-Disease relations. The shared task revealed that CID relation extraction is a difficult task with best reported systems achieving an F-score of about 57%. Study of the participating teams approaches reveals that most approaches (14 out of 18) were based on Support Vector Machines (SVMs) (Burges, 1998), modeling relation extraction as a supervised classification problem. Most of these systems obtain their performance through a rich feature set that is manually crafted by studying the syntactic and lexical regularities in the text. Substantial performance boost is also drawn from custom heuristics such as postprocessing rules (Zhou et al., 2016). Designing such an effective relation extraction system involves extensive feature engineering and domain expertise. Kernel methods in NLP (Collins and Duffy, 2001) have been designed precisely to address this problem of manual feature engineering. These methods enable an efficient and comprehensive exploration of a very high dimensional feature space and to automatically adapt to the dominant patterns expressed in the training set. In our work, we show that kernel methods can be used for boosting relation extraction performance without having to manually engineer additional features. We demonstrate through experiments that combining tree kernels over constituent parses with simple lexical and syntactic features can substantially enhance the performance of the CID task. We also discuss the strengths and weaknesses of these methods which can assist in the design of better methods in the future.
2 2 Related Work Our system is developed in the context of the CID subtask described in BioCreative-V (Wei et al., 2015). Many teams, including the top scoring team (Wei et al., 2015), model the CID task as a supervised binary classification problem. In addition to the annotated PubMed abstracts, alternate sources of information such as the Chemical Toxicology Database (CTD) (Davis et al., 2012) were used. Similar biomedical relation extraction tasks that have been studied are drug-drug interaction (Bjorne et al., 2011) and protein-protein interaction (Lan et al., 2009). A subsequence kernel was presented by (Bunescu and Mooney, 2005) for protein-protein interaction extraction. Richer kernels that use constituent parses or dependency structures are studied in (Chowdhury et al., 2011; Airola et al., 2008) for the protein-protein interaction extraction. Recent approaches have focused on broadening the scope of word matching from a simple lexical match to more complex semantic matching (Saleh et al., 2014). The suitability of these methods for the CID task remains to be explored. 3 Approach Our goal is to minimize task specific and domain specific feature engineering. We therefore explore the power of domain independent techniques such as kernel methods for effective relation extraction. Kernel methods automatically explore a large feature space and can reduce the need for rich hand crafted features. In our system, we do not employ any preprocessing or custom filtering techniques. We use simple text based features and a knowledge base (CTD) look up as our primary feature set. Further knowledge extraction from text is accomplished through tree kernels. We cast the CID relation extraction as a binary classification problem. The input to the classifier is a pair of chemical and disease mentions. From the set of predicted relation mentions, we extract their normalized entity ids (MeSH Ids) and add it to the final list of Chemical-Disease relations. We built and tested two types of classifiers, namely linear classifier and tree kernel classifier. The linear classifier uses a flat list of simple features. The tree kernel classifier uses kernel methods over constituent parse trees of input sentences. The detailed steps are described below: 3.1 Linear classifier 1. Every chemical mention (C) that appears in the article is paired with every other disease mention (D) to generate an entity pair (C-D) for classification. An entity pair in which both the entity mentions are within a sentence are referred to as intrasentence pairs, and those that cross a sentence boundary are referred to as intersentence pairs. The full test data is the union of intrasentence and intersentence entity pairs. 2. Intersentence and intrasentence pairs are grouped separately for training and testing with two separate classifiers. No further filtering or post-processing of (C-D) pairs is performed. 3. At training time, we label a (C-D) pair as positive if there exists a valid CID relation between these entities, using the relation annotations. At test time, the label is inferred from the classifier output. 4. Features for intrasentence pairs include verbs, bag of words, POS tags, dependency parse and the token distance between entity mentions in the sentence. 5. Features for intersentence pairs include the POS tags and bag of words of the two sentences containing entity mentions, distance (number of sentences) between them, statistical features of the entity mentions in the document (frequency of mentions), entity frequencies and zonal information (document zone containing the mentions). 6. We use the Chemical Toxicology Database (Davis et al., 2012) to generate a binary feature I ctd (C, D) that evaluates to 1 if the (C-D) pair is known to be related in the CTD database and 0 otherwise. We used a Support Vector Machine (SVM) with linear kernel from Scikit (Pedregosa et al., 2011) to classify candidate entity pairs. The predicted (C-D) pairs from the sentence level and document level classifiers are combined to form the final list of document level CID relations. We refer to this system as Linear Classifier. 3.2 Tree Kernel Classifier Kernel methods have gained wide spread acceptance, because they allow direct computation of similarity (dot product) between two examples in an implicitly mapped high dimensional
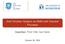
3 JJ Cyclic Pre NP VBN (a) Full constituent parse tree. dysosmia Disease S VBN induced mid VP IN by mid PP NP PZA Chemical (b) Few of its fragments (subset trees). VP PP IN by PP NP PZA dysosmia Figure 1: Illustration for the sentence Cyclic dysosmia induced by PZA (Pyrazinamide) : a) Full constituent parse tree. b) Few of its fragments (implicitly) considered by the subset tree kernel. Entity focus is incorporated by prefixing special tags such as pre, mid, post, Chemical and Disease.. space (Collins and Duffy, 2001; Zelenko et al., 2002). A tree kernel implicitly maps a given tree into a very high dimensional feature space of tree fragments, as illustrated with an example in Figure 1. The kernel score between two trees is the count of common tree fragments between them. We used tree kernels over constituent parse trees of sentences, to efficiently compute the syntactic similarity between two sentences. Different variants of the tree kernels are proposed based on what constitutes a tree fragment, such as subtrees or subsets of nodes. Efficient algorithms with linear time complexity in the average case are presented in (Moschitti, 2006b). The formal definition of the tree kernel is discussed below. Given two trees T 1 and T 2 and the set of all possible tree fragments F = {f 1, f 2,...}, an indicator function I i (n) is defined which evaluates to 1 if the fragment f i is rooted at node n and 0 otherwise. The unnormalized kernel score is given by k (T 1, T 2 ) = (n1, n2) n 2 N T2 n 1 N T1 where N T1 and N T2 are the sets of nodes of T 1 and T 2 respectively and (n1, n2) = F i=1 I i(n 1 )I i (n 2 ). The normalized kernel score is given by K(T 1, T 2 ) = k (T 1, T 2 ) k (T 1, T 1 ) k (T 2, T 2 ) We experimented with subtree and subset tree kernels over constituent parse trees and found subset tree kernels to be superior for our task. In the rest of the paper, we mean subset tree kernel when we refer to tree kernels. We used Stanford CoreNLP (Manning et al., 2014) to generate the constituent parse trees. For tree kernels we use the SVM-LIGHT-TK toolkit 1 (Moschitti, 2006a) that offers kernel implementation within SVM 2 (Joachims, 1999). For each intrasentence (C-D) pair we get a single parse tree and for each intersentence pair we get a forest of two constituent parse trees, from each sentence containing one of the two entity mentions. The contribution from flat features as defined in the section 3.1 can be combined with tree kernels by linearly combining the dot products of the flat feature vectors and the tree kernel. That is, the kernel for the new classifier (linear + tree kernel) is computed as the sum of the linear kernel over flat features and the tree kernel over the constituent parse trees. We report results for these classifiers, namely Linear, Tree Kernel and Linear + Tree Kernel classifier in Section Tree kernels with entity focus Tree kernels attempt to classify a sentence in its entirety and in its default form are unaware of the entity mentions in the sentence. This approach is suitable if our goal is to simply detect if a sentence expresses a relation or not. However, to render greater focus on the entity mentions, we can preprocess the sentence to highlight the location of a word with reference to entity mentions. We prefixed all words in the sentence with pre, mid, and post tags, based on whether they are located prior to, in between, or post entity mentions, before generating the constituent parse trees
4 4 Evaluation 4.1 Dataset and Evaluation metrics We work with the dataset provided by BioCreative-V (Wei et al., 2015). It comprises 3 subsets, referred to as training, development and test set. Each subset consists of 500 PubMed articles (Title and Abstract only), that are fully annotated with Chemical and Disease mentions and the CID relations. Our goal is to extract Chemical-Disease relations at the document (PubMed abstract) level and the metrics are standard Precision (P), Recall (R) and F1 measure ( 2P R P +R ). 4.2 Results We measure the effectiveness of our relation extraction system over the provided test data set, as set out in the CID task. We use the standard entity annotations provided with the data set. Given the limited annotated data, we decided to use both the training and development data set for classifier training with default settings and no custom parameter tuning. Results for intersentence, intrasentence and the full set of (C-D) pairs are presented for linear classifier, tree kernel classifier and their combination, in Table 1. We also present the results for the Linear classifier without the CTD feature. Finally, the table also contains the results reported in a prior work by (Zhou et al., 2016) for the CID task. A comparative study with this prior work is presented in Section 5. To summarize, our final system (linear + tree kernel) achieves an F-score of 61.7% over the CID test data. Note that the combination of linear and tree kernels outperforms the linear and tree kernel classifiers individually. The Table also reveals the substantial contribution of the CTD look up feature towards the linear classifier s performance. 5 Discussion Comparison with prior art. Previously published results in the CID BioCreative-V task used custom entity recognition tools. Therefore, their CID performance is not directly comparable without replicating their entity annotation process. A more accurate comparison can be made with (Zhou et al., 2016) who follow a similar evaluation process. Their system uses gold standard entity annotations and is trained on the CID training and development datasets and evaluated on Test Data Classifier P R F1 Intrasentence Lin - CTD Intrasentence Lin Intrasentence TK Intrasentence Lin + TK Intersentence Lin - CTD Intersentence Lin Intersentence TK Intersentence Lin + TK Full test Lin - CTD Full test Lin Full test TK Full test Lin + TK Full test (Zhou et al., 2016) Table 1: Results on CID test data for Linear classifier (Lin), Tree Kernel (TK) and their combination (Lin+TK). The performance of the linear classifier without CTD feature (Lin - CTD) is also shown. the CID test data. They report an F-score of 61.3%. Significantly, their system relies on task specific post-processing rules, without which their F1 score drops to 56.0%. Our system performs better (61.7%), reflecting a substantive advantage in precision, without using heuristics or task specific rules. Effectiveness of Tree Kernels. We note that tree kernels can significantly improve the performance of CID relation extraction as illustrated in the results. Also, this additional performance is obtained using PubMed abstracts and not external information sources. These results suggest that a greater amount of information exists in annotated text that is easier to extract with tree kernels as compared to manual feature mining for richer patterns. Further, tree kernels have an effect of increasing the precision of the classifiers, specially for intersentence cases. This is likely due to the fact that tree kernels enable stringent comparison of sentence structures (constituent parse trees) as compared to the lenient approach of bag of words matching with linear kernels. Further enhancements. Incorporating entity focus to tree kernels (section 3.2.1) produced a slight improvement (to 61.7% from 61.0%). This approach is likely to be beneficial for sentences that express multiple relations (> 1) between multiple entity pairs. In the CID dataset, we found
5 that sentences expressing multiple relations constitute around 14%, 15% and 14% of training, development and test datasets respectively. Alternate approaches that discriminate parts of the sentence based on the relation expressed are likely to further improve the performance. In the context of intersentence (C-D) pairs, we are currently using only the two sentences s i, s j that contain the entity mentions. However, the actual relationship might be collectively expressed by any subset of the sentences in the document. We attempted to model the whole of the document as a forest of parse trees of all its sentences, but did not observe any improvement in performance. For the CID task where more than 30% of the (C- D) pairs cross sentence boundaries, effective intersentence relation extraction remains a challenge. 6 Summary and Conclusion In this work, we show that tree kernels were very effective in CID relation extraction and boosted F1 score to 61.7% as compared to 57.9% achieved with a linear classifier using simple handcrafted features alone. In future work, we seek to improve intersentence relation extraction from documents. References Antti Airola, Sampo Pyysalo, Jari Björne, Tapio Pahikkala, Filip Ginter, and Tapio Salakoski A graph kernel for protein-protein interaction extraction. In Proceedings of the workshop on current trends in biomedical natural language processing, pages 1 9. Association for Computational Linguistics. William A Baumgartner, K Bretonnel Cohen, Lynne M Fox, George Acquaah-Mensah, and Lawrence Hunter Manual curation is not sufficient for annotation of genomic databases. Bioinformatics, 23(13):i41 i48. Jari Bjorne, Antti Airola, Tapio Pahikkala, and Tapio Salakoski Drug-drug interaction extraction from biomedical texts with SVM and RLS classifiers. CEUR Workshop Proceedings, 761: Razvan C. Bunescu and Raymond J. Mooney Subsequence kernels for relation extraction. In NIPS. Christopher JC Burges A tutorial on support vector machines for pattern recognition. Data mining and knowledge discovery, 2(2): Faisal Mahbub Chowdhury, Alberto Lavelli, and Alessandro Moschitti A Study on Dependency Tree Kernels for Automatic Extraction of Protein-Protein Interaction. BioNLP 2011, (2011): Michael Collins and Nigel Duffy Convolution kernels for natural language. In Advances in neural information processing systems, pages Allan Peter Davis, Cynthia Grondin Murphy, Robin Johnson, Jean M Lay, Kelley Lennon-Hopkins, Cynthia Saraceni-Richards, Daniela Sciaky, Benjamin L King, Michael C Rosenstein, Thomas C Wiegers, et al The comparative toxicogenomics database: update Nucleic acids research, page gks994. Rezarta Islamaj Dogan, G Craig Murray, Aurélie Névéol, and Zhiyong Lu Understanding pubmed R user search behavior through log analysis. Database, 2009:bap018. Laura Haas, Melissa Cefkin, Cheryl Kieliszewski, Wil Plouffe, and Mary Roth The ibm research accelerated discovery lab. SIGMOD Rec., 43(2):41 48, December. Thorsten Joachims Advances in kernel methods. chapter Making Large-scale Support Vector Machine Learning Practical, pages MIT Press, Cambridge, MA, USA. Man Lan, Chew Lim Tan, and Jian Su Feature generation and representations for protein-protein interaction classification. Journal of Biomedical Informatics, 42: Christopher D Manning, Mihai Surdeanu, John Bauer, Jenny Finkel, Steven J Bethard, and David Mc- Closky The stanford corenlp natural language processing toolkit. In Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations, pages Alessandro Moschitti. 2006a. Efficient convolution kernels for dependency and constituent syntactic trees. In Machine Learning: ECML 2006, pages Springer. Alessandro Moschitti. 2006b. Making tree kernels practical for natural language learning. In EACL. Fabian Pedregosa, Gaël Varoquaux, Alexandre Gramfort, Vincent Michel, Bertrand Thirion, Olivier Grisel, Mathieu Blondel, Peter Prettenhofer, Ron Weiss, Vincent Dubourg, et al Scikit-learn: Machine learning in python. The Journal of Machine Learning Research, 12: I Saleh, Alessandro Moschitti, Preslav Nakov, L Màrquez, and S Joty Semantic Kernels for Semantic Parsing. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages
6 Chih-Hsuan Wei, Yifan Peng, Robert Leaman, Allan Peter Davis, Carolyn J Mattingly, Jiao Li, Thomas C Wiegers, and Zhiyong Lu Overview of the biocreative v chemical disease relation (cdr) task. In Proceedings of the fifth BioCreative challenge evaluation workshop, Sevilla, Spain. Dmitry Zelenko, Chinatsu Aone, and Anthony Richardella Kernel methods for relation extraction. Journal of Machine Learning Research, 3: Huiwei Zhou, Huijie Deng, Long Chen, Yunlong Yang, Chen Jia, and Degen Huang Exploiting syntactic and semantics information for chemical disease relation extraction. Database, 2016:baw048.
CLRG Biocreative V
 CLRG ChemTMiner @ Biocreative V Sobha Lalitha Devi., Sindhuja Gopalan., Vijay Sundar Ram R., Malarkodi C.S., Lakshmi S., Pattabhi RK Rao Computational Linguistics Research Group, AU-KBC Research Centre
CLRG ChemTMiner @ Biocreative V Sobha Lalitha Devi., Sindhuja Gopalan., Vijay Sundar Ram R., Malarkodi C.S., Lakshmi S., Pattabhi RK Rao Computational Linguistics Research Group, AU-KBC Research Centre
Semantic Role Labeling via Tree Kernel Joint Inference
 Semantic Role Labeling via Tree Kernel Joint Inference Alessandro Moschitti, Daniele Pighin and Roberto Basili Department of Computer Science University of Rome Tor Vergata 00133 Rome, Italy {moschitti,basili}@info.uniroma2.it
Semantic Role Labeling via Tree Kernel Joint Inference Alessandro Moschitti, Daniele Pighin and Roberto Basili Department of Computer Science University of Rome Tor Vergata 00133 Rome, Italy {moschitti,basili}@info.uniroma2.it
ChemDataExtractor: A toolkit for automated extraction of chemical information from the scientific literature
 : A toolkit for automated extraction of chemical information from the scientific literature Callum Court Molecular Engineering, University of Cambridge Supervisor: Dr Jacqueline Cole 1 / 20 Overview 1
: A toolkit for automated extraction of chemical information from the scientific literature Callum Court Molecular Engineering, University of Cambridge Supervisor: Dr Jacqueline Cole 1 / 20 Overview 1
Sieve-Based Spatial Relation Extraction with Expanding Parse Trees
 Sieve-Based Spatial Relation Extraction with Expanding Parse Trees Jennifer D Souza and Vincent Ng Human Language Technology Research Institute University of Texas at Dallas Richardson, TX 75083-0688 {jld082000,vince}@hlt.utdallas.edu
Sieve-Based Spatial Relation Extraction with Expanding Parse Trees Jennifer D Souza and Vincent Ng Human Language Technology Research Institute University of Texas at Dallas Richardson, TX 75083-0688 {jld082000,vince}@hlt.utdallas.edu
Hierachical Name Entity Recognition
 Hierachical Name Entity Recognition Dakan Wang, Yu Wu Mentor: David Mcclosky, Mihai Surdeanu March 10, 2011 1 Introduction In this project, we investigte the hierarchical name entity recognition problem
Hierachical Name Entity Recognition Dakan Wang, Yu Wu Mentor: David Mcclosky, Mihai Surdeanu March 10, 2011 1 Introduction In this project, we investigte the hierarchical name entity recognition problem
Special Topics in Computer Science
 Special Topics in Computer Science NLP in a Nutshell CS492B Spring Semester 2009 Speaker : Hee Jin Lee p Professor : Jong C. Park Computer Science Department Korea Advanced Institute of Science and Technology
Special Topics in Computer Science NLP in a Nutshell CS492B Spring Semester 2009 Speaker : Hee Jin Lee p Professor : Jong C. Park Computer Science Department Korea Advanced Institute of Science and Technology
Temporal and spatial approaches for land cover classification.
 Temporal and spatial approaches for land cover classification. Ryabukhin Sergey sergeyryabukhin@gmail.com Abstract. This paper describes solution for Time Series Land Cover Classification Challenge (TiSeLaC).
Temporal and spatial approaches for land cover classification. Ryabukhin Sergey sergeyryabukhin@gmail.com Abstract. This paper describes solution for Time Series Land Cover Classification Challenge (TiSeLaC).
Fast Computing Grammar-driven Convolution Tree Kernel for Semantic Role Labeling
 Fast Computing Grammar-driven Convolution Tree Kernel for Semantic Role Labeling Wanxiang Che 1, Min Zhang 2, Ai Ti Aw 2, Chew Lim Tan 3, Ting Liu 1, Sheng Li 1 1 School of Computer Science and Technology
Fast Computing Grammar-driven Convolution Tree Kernel for Semantic Role Labeling Wanxiang Che 1, Min Zhang 2, Ai Ti Aw 2, Chew Lim Tan 3, Ting Liu 1, Sheng Li 1 1 School of Computer Science and Technology
Spatial Role Labeling CS365 Course Project
 Spatial Role Labeling CS365 Course Project Amit Kumar, akkumar@iitk.ac.in Chandra Sekhar, gchandra@iitk.ac.in Supervisor : Dr.Amitabha Mukerjee ABSTRACT In natural language processing one of the important
Spatial Role Labeling CS365 Course Project Amit Kumar, akkumar@iitk.ac.in Chandra Sekhar, gchandra@iitk.ac.in Supervisor : Dr.Amitabha Mukerjee ABSTRACT In natural language processing one of the important
HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH
 HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH Hoang Trang 1, Tran Hoang Loc 1 1 Ho Chi Minh City University of Technology-VNU HCM, Ho Chi
HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH Hoang Trang 1, Tran Hoang Loc 1 1 Ho Chi Minh City University of Technology-VNU HCM, Ho Chi
Support Vector Machine & Its Applications
 Support Vector Machine & Its Applications A portion (1/3) of the slides are taken from Prof. Andrew Moore s SVM tutorial at http://www.cs.cmu.edu/~awm/tutorials Mingyue Tan The University of British Columbia
Support Vector Machine & Its Applications A portion (1/3) of the slides are taken from Prof. Andrew Moore s SVM tutorial at http://www.cs.cmu.edu/~awm/tutorials Mingyue Tan The University of British Columbia
Multiword Expression Identification with Tree Substitution Grammars
 Multiword Expression Identification with Tree Substitution Grammars Spence Green, Marie-Catherine de Marneffe, John Bauer, and Christopher D. Manning Stanford University EMNLP 2011 Main Idea Use syntactic
Multiword Expression Identification with Tree Substitution Grammars Spence Green, Marie-Catherine de Marneffe, John Bauer, and Christopher D. Manning Stanford University EMNLP 2011 Main Idea Use syntactic
Bio-Medical Text Mining with Machine Learning
 Sumit Madan Department of Bioinformatics - Fraunhofer SCAI Textual Knowledge PubMed Journals, Books Patents EHRs What is Bio-Medical Text Mining? Phosphorylation of glycogen synthase kinase 3 beta at Threonine,
Sumit Madan Department of Bioinformatics - Fraunhofer SCAI Textual Knowledge PubMed Journals, Books Patents EHRs What is Bio-Medical Text Mining? Phosphorylation of glycogen synthase kinase 3 beta at Threonine,
ToxiCat: Hybrid Named Entity Recognition services to support curation of the Comparative Toxicogenomic Database
 ToxiCat: Hybrid Named Entity Recognition services to support curation of the Comparative Toxicogenomic Database Dina Vishnyakova 1,2, 4, *, Julien Gobeill 1,3,4, Emilie Pasche 1,2,3,4 and Patrick Ruch
ToxiCat: Hybrid Named Entity Recognition services to support curation of the Comparative Toxicogenomic Database Dina Vishnyakova 1,2, 4, *, Julien Gobeill 1,3,4, Emilie Pasche 1,2,3,4 and Patrick Ruch
CMU at SemEval-2016 Task 8: Graph-based AMR Parsing with Infinite Ramp Loss
 CMU at SemEval-2016 Task 8: Graph-based AMR Parsing with Infinite Ramp Loss Jeffrey Flanigan Chris Dyer Noah A. Smith Jaime Carbonell School of Computer Science, Carnegie Mellon University, Pittsburgh,
CMU at SemEval-2016 Task 8: Graph-based AMR Parsing with Infinite Ramp Loss Jeffrey Flanigan Chris Dyer Noah A. Smith Jaime Carbonell School of Computer Science, Carnegie Mellon University, Pittsburgh,
A Support Vector Method for Multivariate Performance Measures
 A Support Vector Method for Multivariate Performance Measures Thorsten Joachims Cornell University Department of Computer Science Thanks to Rich Caruana, Alexandru Niculescu-Mizil, Pierre Dupont, Jérôme
A Support Vector Method for Multivariate Performance Measures Thorsten Joachims Cornell University Department of Computer Science Thanks to Rich Caruana, Alexandru Niculescu-Mizil, Pierre Dupont, Jérôme
Annotating Derivations: A New Evaluation Strategy and Dataset for Algebra Word Problems
 Annotating Derivations: A New Evaluation Strategy and Dataset for Algebra Word Problems Shyam Upadhyay 1 Ming-Wei Chang 2 1 University of Illinois at Urbana-Champaign, IL, USA 2 Microsoft Research, Redmond,
Annotating Derivations: A New Evaluation Strategy and Dataset for Algebra Word Problems Shyam Upadhyay 1 Ming-Wei Chang 2 1 University of Illinois at Urbana-Champaign, IL, USA 2 Microsoft Research, Redmond,
Driving Semantic Parsing from the World s Response
 Driving Semantic Parsing from the World s Response James Clarke, Dan Goldwasser, Ming-Wei Chang, Dan Roth Cognitive Computation Group University of Illinois at Urbana-Champaign CoNLL 2010 Clarke, Goldwasser,
Driving Semantic Parsing from the World s Response James Clarke, Dan Goldwasser, Ming-Wei Chang, Dan Roth Cognitive Computation Group University of Illinois at Urbana-Champaign CoNLL 2010 Clarke, Goldwasser,
Tuning as Linear Regression
 Tuning as Linear Regression Marzieh Bazrafshan, Tagyoung Chung and Daniel Gildea Department of Computer Science University of Rochester Rochester, NY 14627 Abstract We propose a tuning method for statistical
Tuning as Linear Regression Marzieh Bazrafshan, Tagyoung Chung and Daniel Gildea Department of Computer Science University of Rochester Rochester, NY 14627 Abstract We propose a tuning method for statistical
Machine Learning for natural language processing
 Machine Learning for natural language processing Classification: Maximum Entropy Models Laura Kallmeyer Heinrich-Heine-Universität Düsseldorf Summer 2016 1 / 24 Introduction Classification = supervised
Machine Learning for natural language processing Classification: Maximum Entropy Models Laura Kallmeyer Heinrich-Heine-Universität Düsseldorf Summer 2016 1 / 24 Introduction Classification = supervised
TALP at GeoQuery 2007: Linguistic and Geographical Analysis for Query Parsing
 TALP at GeoQuery 2007: Linguistic and Geographical Analysis for Query Parsing Daniel Ferrés and Horacio Rodríguez TALP Research Center Software Department Universitat Politècnica de Catalunya {dferres,horacio}@lsi.upc.edu
TALP at GeoQuery 2007: Linguistic and Geographical Analysis for Query Parsing Daniel Ferrés and Horacio Rodríguez TALP Research Center Software Department Universitat Politècnica de Catalunya {dferres,horacio}@lsi.upc.edu
Quark-gluon tagging in the forward region of ATLAS at the LHC.
 CS229. Fall 2016. Final Project Report. Quark-gluon tagging in the forward region of ATLAS at the LHC. Rob Mina (robmina), Randy White (whiteran) December 15, 2016 1 Introduction In modern hadron colliders,
CS229. Fall 2016. Final Project Report. Quark-gluon tagging in the forward region of ATLAS at the LHC. Rob Mina (robmina), Randy White (whiteran) December 15, 2016 1 Introduction In modern hadron colliders,
SUPPORT VECTOR MACHINE
 SUPPORT VECTOR MACHINE Mainly based on https://nlp.stanford.edu/ir-book/pdf/15svm.pdf 1 Overview SVM is a huge topic Integration of MMDS, IIR, and Andrew Moore s slides here Our foci: Geometric intuition
SUPPORT VECTOR MACHINE Mainly based on https://nlp.stanford.edu/ir-book/pdf/15svm.pdf 1 Overview SVM is a huge topic Integration of MMDS, IIR, and Andrew Moore s slides here Our foci: Geometric intuition
Information Extraction from Text
 Information Extraction from Text Jing Jiang Chapter 2 from Mining Text Data (2012) Presented by Andrew Landgraf, September 13, 2013 1 What is Information Extraction? Goal is to discover structured information
Information Extraction from Text Jing Jiang Chapter 2 from Mining Text Data (2012) Presented by Andrew Landgraf, September 13, 2013 1 What is Information Extraction? Goal is to discover structured information
Chunking with Support Vector Machines
 NAACL2001 Chunking with Support Vector Machines Graduate School of Information Science, Nara Institute of Science and Technology, JAPAN Taku Kudo, Yuji Matsumoto {taku-ku,matsu}@is.aist-nara.ac.jp Chunking
NAACL2001 Chunking with Support Vector Machines Graduate School of Information Science, Nara Institute of Science and Technology, JAPAN Taku Kudo, Yuji Matsumoto {taku-ku,matsu}@is.aist-nara.ac.jp Chunking
Abstention Protocol for Accuracy and Speed
 Abstention Protocol for Accuracy and Speed Abstract Amirata Ghorbani EE Dept. Stanford University amiratag@stanford.edu In order to confidently rely on machines to decide and perform tasks for us, there
Abstention Protocol for Accuracy and Speed Abstract Amirata Ghorbani EE Dept. Stanford University amiratag@stanford.edu In order to confidently rely on machines to decide and perform tasks for us, there
Machine Learning Analyses of Meteor Data
 WGN, The Journal of the IMO 45:5 (2017) 1 Machine Learning Analyses of Meteor Data Viswesh Krishna Research Student, Centre for Fundamental Research and Creative Education. Email: krishnaviswesh@cfrce.in
WGN, The Journal of the IMO 45:5 (2017) 1 Machine Learning Analyses of Meteor Data Viswesh Krishna Research Student, Centre for Fundamental Research and Creative Education. Email: krishnaviswesh@cfrce.in
Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function.
 Bayesian learning: Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function. Let y be the true label and y be the predicted
Bayesian learning: Machine learning comes from Bayesian decision theory in statistics. There we want to minimize the expected value of the loss function. Let y be the true label and y be the predicted
Mining coreference relations between formulas and text using Wikipedia
 Mining coreference relations between formulas and text using Wikipedia Minh Nghiem Quoc 1, Keisuke Yokoi 2, Yuichiroh Matsubayashi 3 Akiko Aizawa 1 2 3 1 Department of Informatics, The Graduate University
Mining coreference relations between formulas and text using Wikipedia Minh Nghiem Quoc 1, Keisuke Yokoi 2, Yuichiroh Matsubayashi 3 Akiko Aizawa 1 2 3 1 Department of Informatics, The Graduate University
Machine Learning for Structured Prediction
 Machine Learning for Structured Prediction Grzegorz Chrupa la National Centre for Language Technology School of Computing Dublin City University NCLT Seminar Grzegorz Chrupa la (DCU) Machine Learning for
Machine Learning for Structured Prediction Grzegorz Chrupa la National Centre for Language Technology School of Computing Dublin City University NCLT Seminar Grzegorz Chrupa la (DCU) Machine Learning for
Statistical Machine Learning Theory. From Multi-class Classification to Structured Output Prediction. Hisashi Kashima.
 http://goo.gl/jv7vj9 Course website KYOTO UNIVERSITY Statistical Machine Learning Theory From Multi-class Classification to Structured Output Prediction Hisashi Kashima kashima@i.kyoto-u.ac.jp DEPARTMENT
http://goo.gl/jv7vj9 Course website KYOTO UNIVERSITY Statistical Machine Learning Theory From Multi-class Classification to Structured Output Prediction Hisashi Kashima kashima@i.kyoto-u.ac.jp DEPARTMENT
Natural Language Processing CS Lecture 06. Razvan C. Bunescu School of Electrical Engineering and Computer Science
 Natural Language Processing CS 6840 Lecture 06 Razvan C. Bunescu School of Electrical Engineering and Computer Science bunescu@ohio.edu Statistical Parsing Define a probabilistic model of syntax P(T S):
Natural Language Processing CS 6840 Lecture 06 Razvan C. Bunescu School of Electrical Engineering and Computer Science bunescu@ohio.edu Statistical Parsing Define a probabilistic model of syntax P(T S):
Statistical Machine Learning Theory. From Multi-class Classification to Structured Output Prediction. Hisashi Kashima.
 http://goo.gl/xilnmn Course website KYOTO UNIVERSITY Statistical Machine Learning Theory From Multi-class Classification to Structured Output Prediction Hisashi Kashima kashima@i.kyoto-u.ac.jp DEPARTMENT
http://goo.gl/xilnmn Course website KYOTO UNIVERSITY Statistical Machine Learning Theory From Multi-class Classification to Structured Output Prediction Hisashi Kashima kashima@i.kyoto-u.ac.jp DEPARTMENT
Lab 12: Structured Prediction
 December 4, 2014 Lecture plan structured perceptron application: confused messages application: dependency parsing structured SVM Class review: from modelization to classification What does learning mean?
December 4, 2014 Lecture plan structured perceptron application: confused messages application: dependency parsing structured SVM Class review: from modelization to classification What does learning mean?
Regularization Introduction to Machine Learning. Matt Gormley Lecture 10 Feb. 19, 2018
 1-61 Introduction to Machine Learning Machine Learning Department School of Computer Science Carnegie Mellon University Regularization Matt Gormley Lecture 1 Feb. 19, 218 1 Reminders Homework 4: Logistic
1-61 Introduction to Machine Learning Machine Learning Department School of Computer Science Carnegie Mellon University Regularization Matt Gormley Lecture 1 Feb. 19, 218 1 Reminders Homework 4: Logistic
Modeling Biological Processes for Reading Comprehension
 Modeling Biological Processes for Reading Comprehension Vivek Srikumar University of Utah (Previously, Stanford University) Jonathan Berant, Pei- Chun Chen, Abby Vander Linden, BriEany Harding, Brad Huang,
Modeling Biological Processes for Reading Comprehension Vivek Srikumar University of Utah (Previously, Stanford University) Jonathan Berant, Pei- Chun Chen, Abby Vander Linden, BriEany Harding, Brad Huang,
Modern Information Retrieval
 Modern Information Retrieval Chapter 8 Text Classification Introduction A Characterization of Text Classification Unsupervised Algorithms Supervised Algorithms Feature Selection or Dimensionality Reduction
Modern Information Retrieval Chapter 8 Text Classification Introduction A Characterization of Text Classification Unsupervised Algorithms Supervised Algorithms Feature Selection or Dimensionality Reduction
Empirical Risk Minimization, Model Selection, and Model Assessment
 Empirical Risk Minimization, Model Selection, and Model Assessment CS6780 Advanced Machine Learning Spring 2015 Thorsten Joachims Cornell University Reading: Murphy 5.7-5.7.2.4, 6.5-6.5.3.1 Dietterich,
Empirical Risk Minimization, Model Selection, and Model Assessment CS6780 Advanced Machine Learning Spring 2015 Thorsten Joachims Cornell University Reading: Murphy 5.7-5.7.2.4, 6.5-6.5.3.1 Dietterich,
Text Mining. Dr. Yanjun Li. Associate Professor. Department of Computer and Information Sciences Fordham University
 Text Mining Dr. Yanjun Li Associate Professor Department of Computer and Information Sciences Fordham University Outline Introduction: Data Mining Part One: Text Mining Part Two: Preprocessing Text Data
Text Mining Dr. Yanjun Li Associate Professor Department of Computer and Information Sciences Fordham University Outline Introduction: Data Mining Part One: Text Mining Part Two: Preprocessing Text Data
arxiv: v2 [cs.lg] 5 May 2015
![arxiv: v2 [cs.lg] 5 May 2015 arxiv: v2 [cs.lg] 5 May 2015](/thumbs/88/115102450.jpg) fastfm: A Library for Factorization Machines Immanuel Bayer University of Konstanz 78457 Konstanz, Germany immanuel.bayer@uni-konstanz.de arxiv:505.0064v [cs.lg] 5 May 05 Editor: Abstract Factorization
fastfm: A Library for Factorization Machines Immanuel Bayer University of Konstanz 78457 Konstanz, Germany immanuel.bayer@uni-konstanz.de arxiv:505.0064v [cs.lg] 5 May 05 Editor: Abstract Factorization
Deep Learning for Natural Language Processing. Sidharth Mudgal April 4, 2017
 Deep Learning for Natural Language Processing Sidharth Mudgal April 4, 2017 Table of contents 1. Intro 2. Word Vectors 3. Word2Vec 4. Char Level Word Embeddings 5. Application: Entity Matching 6. Conclusion
Deep Learning for Natural Language Processing Sidharth Mudgal April 4, 2017 Table of contents 1. Intro 2. Word Vectors 3. Word2Vec 4. Char Level Word Embeddings 5. Application: Entity Matching 6. Conclusion
KSU Team s System and Experience at the NTCIR-11 RITE-VAL Task
 KSU Team s System and Experience at the NTCIR-11 RITE-VAL Task Tasuku Kimura Kyoto Sangyo University, Japan i1458030@cse.kyoto-su.ac.jp Hisashi Miyamori Kyoto Sangyo University, Japan miya@cse.kyoto-su.ac.jp
KSU Team s System and Experience at the NTCIR-11 RITE-VAL Task Tasuku Kimura Kyoto Sangyo University, Japan i1458030@cse.kyoto-su.ac.jp Hisashi Miyamori Kyoto Sangyo University, Japan miya@cse.kyoto-su.ac.jp
What s Cooking? Predicting Cuisines from Recipe Ingredients
 What s Cooking? Predicting Cuisines from Recipe Ingredients Kevin K. Do Department of Computer Science Duke University Durham, NC 27708 kevin.kydat.do@gmail.com Abstract Kaggle is an online platform for
What s Cooking? Predicting Cuisines from Recipe Ingredients Kevin K. Do Department of Computer Science Duke University Durham, NC 27708 kevin.kydat.do@gmail.com Abstract Kaggle is an online platform for
Aspect Term Extraction with History Attention and Selective Transformation 1
 Aspect Term Extraction with History Attention and Selective Transformation 1 Xin Li 1, Lidong Bing 2, Piji Li 1, Wai Lam 1, Zhimou Yang 3 Presenter: Lin Ma 2 1 The Chinese University of Hong Kong 2 Tencent
Aspect Term Extraction with History Attention and Selective Transformation 1 Xin Li 1, Lidong Bing 2, Piji Li 1, Wai Lam 1, Zhimou Yang 3 Presenter: Lin Ma 2 1 The Chinese University of Hong Kong 2 Tencent
MACHINE LEARNING FOR NATURAL LANGUAGE PROCESSING
 MACHINE LEARNING FOR NATURAL LANGUAGE PROCESSING Outline Some Sample NLP Task [Noah Smith] Structured Prediction For NLP Structured Prediction Methods Conditional Random Fields Structured Perceptron Discussion
MACHINE LEARNING FOR NATURAL LANGUAGE PROCESSING Outline Some Sample NLP Task [Noah Smith] Structured Prediction For NLP Structured Prediction Methods Conditional Random Fields Structured Perceptron Discussion
Anomaly Detection for the CERN Large Hadron Collider injection magnets
 Anomaly Detection for the CERN Large Hadron Collider injection magnets Armin Halilovic KU Leuven - Department of Computer Science In cooperation with CERN 2018-07-27 0 Outline 1 Context 2 Data 3 Preprocessing
Anomaly Detection for the CERN Large Hadron Collider injection magnets Armin Halilovic KU Leuven - Department of Computer Science In cooperation with CERN 2018-07-27 0 Outline 1 Context 2 Data 3 Preprocessing
Learning Features from Co-occurrences: A Theoretical Analysis
 Learning Features from Co-occurrences: A Theoretical Analysis Yanpeng Li IBM T. J. Watson Research Center Yorktown Heights, New York 10598 liyanpeng.lyp@gmail.com Abstract Representing a word by its co-occurrences
Learning Features from Co-occurrences: A Theoretical Analysis Yanpeng Li IBM T. J. Watson Research Center Yorktown Heights, New York 10598 liyanpeng.lyp@gmail.com Abstract Representing a word by its co-occurrences
Encoding Tree Pair-based Graphs in Learning Algorithms: the Textual Entailment Recognition Case
 Encoding Tree Pair-based Graphs in Learning Algorithms: the Textual Entailment Recognition Case Alessandro Moschitti DII University of Trento Via ommarive 14 38100 POVO (T) - Italy moschitti@dit.unitn.it
Encoding Tree Pair-based Graphs in Learning Algorithms: the Textual Entailment Recognition Case Alessandro Moschitti DII University of Trento Via ommarive 14 38100 POVO (T) - Italy moschitti@dit.unitn.it
CLEF 2017: Multimodal Spatial Role Labeling (msprl) Task Overview
 CLEF 2017: Multimodal Spatial Role Labeling (msprl) Task Overview Parisa Kordjamshidi 1, Taher Rahgooy 1, Marie-Francine Moens 2, James Pustejovsky 3, Umar Manzoor 1 and Kirk Roberts 4 1 Tulane University
CLEF 2017: Multimodal Spatial Role Labeling (msprl) Task Overview Parisa Kordjamshidi 1, Taher Rahgooy 1, Marie-Francine Moens 2, James Pustejovsky 3, Umar Manzoor 1 and Kirk Roberts 4 1 Tulane University
Classification of Study Region in Environmental Science Abstracts
 Classification of Study Region in Environmental Science Abstracts Jared Willett, Timothy Baldwin, David Martinez and Angus Webb Department of Computing and Information Systems NICTA Victoria Research Laboratory
Classification of Study Region in Environmental Science Abstracts Jared Willett, Timothy Baldwin, David Martinez and Angus Webb Department of Computing and Information Systems NICTA Victoria Research Laboratory
Lecture 13: Structured Prediction
 Lecture 13: Structured Prediction Kai-Wei Chang CS @ University of Virginia kw@kwchang.net Couse webpage: http://kwchang.net/teaching/nlp16 CS6501: NLP 1 Quiz 2 v Lectures 9-13 v Lecture 12: before page
Lecture 13: Structured Prediction Kai-Wei Chang CS @ University of Virginia kw@kwchang.net Couse webpage: http://kwchang.net/teaching/nlp16 CS6501: NLP 1 Quiz 2 v Lectures 9-13 v Lecture 12: before page
Multi-Plant Photovoltaic Energy Forecasting Challenge: Second place solution
 Multi-Plant Photovoltaic Energy Forecasting Challenge: Second place solution Clément Gautrais 1, Yann Dauxais 1, and Maël Guilleme 2 1 University of Rennes 1/Inria Rennes clement.gautrais@irisa.fr 2 Energiency/University
Multi-Plant Photovoltaic Energy Forecasting Challenge: Second place solution Clément Gautrais 1, Yann Dauxais 1, and Maël Guilleme 2 1 University of Rennes 1/Inria Rennes clement.gautrais@irisa.fr 2 Energiency/University
Generalization to a zero-data task: an empirical study
 Generalization to a zero-data task: an empirical study Université de Montréal 20/03/2007 Introduction Introduction and motivation What is a zero-data task? task for which no training data are available
Generalization to a zero-data task: an empirical study Université de Montréal 20/03/2007 Introduction Introduction and motivation What is a zero-data task? task for which no training data are available
Deep Learning for NLP
 Deep Learning for NLP Instructor: Wei Xu Ohio State University CSE 5525 Many slides from Greg Durrett Outline Motivation for neural networks Feedforward neural networks Applying feedforward neural networks
Deep Learning for NLP Instructor: Wei Xu Ohio State University CSE 5525 Many slides from Greg Durrett Outline Motivation for neural networks Feedforward neural networks Applying feedforward neural networks
Planet Labels - How do we use our planet?
 Planet Labels - How do we use our planet? Timon Ruban Stanford University timon@stanford.edu 1 Introduction Rishabh Bhargava Stanford University rish93@stanford.edu Vincent Sitzmann Stanford University
Planet Labels - How do we use our planet? Timon Ruban Stanford University timon@stanford.edu 1 Introduction Rishabh Bhargava Stanford University rish93@stanford.edu Vincent Sitzmann Stanford University
NLP Homework: Dependency Parsing with Feed-Forward Neural Network
 NLP Homework: Dependency Parsing with Feed-Forward Neural Network Submission Deadline: Monday Dec. 11th, 5 pm 1 Background on Dependency Parsing Dependency trees are one of the main representations used
NLP Homework: Dependency Parsing with Feed-Forward Neural Network Submission Deadline: Monday Dec. 11th, 5 pm 1 Background on Dependency Parsing Dependency trees are one of the main representations used
hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference
 CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
DM-Group Meeting. Subhodip Biswas 10/16/2014
 DM-Group Meeting Subhodip Biswas 10/16/2014 Papers to be discussed 1. Crowdsourcing Land Use Maps via Twitter Vanessa Frias-Martinez and Enrique Frias-Martinez in KDD 2014 2. Tracking Climate Change Opinions
DM-Group Meeting Subhodip Biswas 10/16/2014 Papers to be discussed 1. Crowdsourcing Land Use Maps via Twitter Vanessa Frias-Martinez and Enrique Frias-Martinez in KDD 2014 2. Tracking Climate Change Opinions
Understanding Comments Submitted to FCC on Net Neutrality. Kevin (Junhui) Mao, Jing Xia, Dennis (Woncheol) Jeong December 12, 2014
 Understanding Comments Submitted to FCC on Net Neutrality Kevin (Junhui) Mao, Jing Xia, Dennis (Woncheol) Jeong December 12, 2014 Abstract We aim to understand and summarize themes in the 1.65 million
Understanding Comments Submitted to FCC on Net Neutrality Kevin (Junhui) Mao, Jing Xia, Dennis (Woncheol) Jeong December 12, 2014 Abstract We aim to understand and summarize themes in the 1.65 million
A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery
 AtomNet A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery Izhar Wallach, Michael Dzamba, Abraham Heifets Victor Storchan, Institute for Computational and
AtomNet A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery Izhar Wallach, Michael Dzamba, Abraham Heifets Victor Storchan, Institute for Computational and
Learning to translate with neural networks. Michael Auli
 Learning to translate with neural networks Michael Auli 1 Neural networks for text processing Similar words near each other France Spain dog cat Neural networks for text processing Similar words near each
Learning to translate with neural networks Michael Auli 1 Neural networks for text processing Similar words near each other France Spain dog cat Neural networks for text processing Similar words near each
More on HMMs and other sequence models. Intro to NLP - ETHZ - 18/03/2013
 More on HMMs and other sequence models Intro to NLP - ETHZ - 18/03/2013 Summary Parts of speech tagging HMMs: Unsupervised parameter estimation Forward Backward algorithm Bayesian variants Discriminative
More on HMMs and other sequence models Intro to NLP - ETHZ - 18/03/2013 Summary Parts of speech tagging HMMs: Unsupervised parameter estimation Forward Backward algorithm Bayesian variants Discriminative
Support Vector Machines (SVM) in bioinformatics. Day 1: Introduction to SVM
 1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
Support Vector Machines
 Support Vector Machines Reading: Ben-Hur & Weston, A User s Guide to Support Vector Machines (linked from class web page) Notation Assume a binary classification problem. Instances are represented by vector
Support Vector Machines Reading: Ben-Hur & Weston, A User s Guide to Support Vector Machines (linked from class web page) Notation Assume a binary classification problem. Instances are represented by vector
Syntactic Patterns of Spatial Relations in Text
 Syntactic Patterns of Spatial Relations in Text Shaonan Zhu, Xueying Zhang Key Laboratory of Virtual Geography Environment,Ministry of Education, Nanjing Normal University,Nanjing, China Abstract: Natural
Syntactic Patterns of Spatial Relations in Text Shaonan Zhu, Xueying Zhang Key Laboratory of Virtual Geography Environment,Ministry of Education, Nanjing Normal University,Nanjing, China Abstract: Natural
Intelligent Systems (AI-2)
 Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 23, 2015 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 23, 2015 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Penn Treebank Parsing. Advanced Topics in Language Processing Stephen Clark
 Penn Treebank Parsing Advanced Topics in Language Processing Stephen Clark 1 The Penn Treebank 40,000 sentences of WSJ newspaper text annotated with phrasestructure trees The trees contain some predicate-argument
Penn Treebank Parsing Advanced Topics in Language Processing Stephen Clark 1 The Penn Treebank 40,000 sentences of WSJ newspaper text annotated with phrasestructure trees The trees contain some predicate-argument
Identifying interactions between chemical entities in biomedical text
 Identifying interactions between chemical entities in biomedical text Andre Lamurias 1,*, João D. Ferreira 1 and Francisco M. Couto 1 1 LaSIGE, Departamento de Informática, Faculdade de Ciências, Universidade
Identifying interactions between chemical entities in biomedical text Andre Lamurias 1,*, João D. Ferreira 1 and Francisco M. Couto 1 1 LaSIGE, Departamento de Informática, Faculdade de Ciências, Universidade
PCA and Autoencoders
 PCA and Autoencoders Tyler Manning-Dahan INSE 6220 - Fall 2017 Concordia University Abstract In this paper, I compare two dimensionality reduction techniques for processing images before learning a multinomial
PCA and Autoencoders Tyler Manning-Dahan INSE 6220 - Fall 2017 Concordia University Abstract In this paper, I compare two dimensionality reduction techniques for processing images before learning a multinomial
The SUBTLE NL Parsing Pipeline: A Complete Parser for English Mitch Marcus University of Pennsylvania
 The SUBTLE NL Parsing Pipeline: A Complete Parser for English Mitch Marcus University of Pennsylvania 1 PICTURE OF ANALYSIS PIPELINE Tokenize Maximum Entropy POS tagger MXPOST Ratnaparkhi Core Parser Collins
The SUBTLE NL Parsing Pipeline: A Complete Parser for English Mitch Marcus University of Pennsylvania 1 PICTURE OF ANALYSIS PIPELINE Tokenize Maximum Entropy POS tagger MXPOST Ratnaparkhi Core Parser Collins
Information Extraction from Biomedical Text. BMI/CS 776 Mark Craven
 Information Extraction from Biomedical Text BMI/CS 776 www.biostat.wisc.edu/bmi776/ Mark Craven craven@biostat.wisc.edu Spring 2012 Goals for Lecture the key concepts to understand are the following! named-entity
Information Extraction from Biomedical Text BMI/CS 776 www.biostat.wisc.edu/bmi776/ Mark Craven craven@biostat.wisc.edu Spring 2012 Goals for Lecture the key concepts to understand are the following! named-entity
Outliers Treatment in Support Vector Regression for Financial Time Series Prediction
 Outliers Treatment in Support Vector Regression for Financial Time Series Prediction Haiqin Yang, Kaizhu Huang, Laiwan Chan, Irwin King, and Michael R. Lyu Department of Computer Science and Engineering
Outliers Treatment in Support Vector Regression for Financial Time Series Prediction Haiqin Yang, Kaizhu Huang, Laiwan Chan, Irwin King, and Michael R. Lyu Department of Computer Science and Engineering
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Introduction to the CoNLL-2004 Shared Task: Semantic Role Labeling
 Introduction to the CoNLL-2004 Shared Task: Semantic Role Labeling Xavier Carreras and Lluís Màrquez TALP Research Center Technical University of Catalonia Boston, May 7th, 2004 Outline Outline of the
Introduction to the CoNLL-2004 Shared Task: Semantic Role Labeling Xavier Carreras and Lluís Màrquez TALP Research Center Technical University of Catalonia Boston, May 7th, 2004 Outline Outline of the
Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network
 Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network Marko Tscherepanow and Franz Kummert Applied Computer Science, Faculty of Technology, Bielefeld
Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network Marko Tscherepanow and Franz Kummert Applied Computer Science, Faculty of Technology, Bielefeld
6.891: Lecture 24 (December 8th, 2003) Kernel Methods
 6.891: Lecture 24 (December 8th, 2003) Kernel Methods Overview ffl Recap: global linear models ffl New representations from old representations ffl computational trick ffl Kernels for NLP structures ffl
6.891: Lecture 24 (December 8th, 2003) Kernel Methods Overview ffl Recap: global linear models ffl New representations from old representations ffl computational trick ffl Kernels for NLP structures ffl
Intelligent Systems (AI-2)
 Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 24, 2016 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Intelligent Systems (AI-2) Computer Science cpsc422, Lecture 19 Oct, 24, 2016 Slide Sources Raymond J. Mooney University of Texas at Austin D. Koller, Stanford CS - Probabilistic Graphical Models D. Page,
Linear Classifiers IV
 Universität Potsdam Institut für Informatik Lehrstuhl Linear Classifiers IV Blaine Nelson, Tobias Scheffer Contents Classification Problem Bayesian Classifier Decision Linear Classifiers, MAP Models Logistic
Universität Potsdam Institut für Informatik Lehrstuhl Linear Classifiers IV Blaine Nelson, Tobias Scheffer Contents Classification Problem Bayesian Classifier Decision Linear Classifiers, MAP Models Logistic
The Noisy Channel Model and Markov Models
 1/24 The Noisy Channel Model and Markov Models Mark Johnson September 3, 2014 2/24 The big ideas The story so far: machine learning classifiers learn a function that maps a data item X to a label Y handle
1/24 The Noisy Channel Model and Markov Models Mark Johnson September 3, 2014 2/24 The big ideas The story so far: machine learning classifiers learn a function that maps a data item X to a label Y handle
Features of Statistical Parsers
 Features of tatistical Parsers Preliminary results Mark Johnson Brown University TTI, October 2003 Joint work with Michael Collins (MIT) upported by NF grants LI 9720368 and II0095940 1 Talk outline tatistical
Features of tatistical Parsers Preliminary results Mark Johnson Brown University TTI, October 2003 Joint work with Michael Collins (MIT) upported by NF grants LI 9720368 and II0095940 1 Talk outline tatistical
NLP Programming Tutorial 11 - The Structured Perceptron
 NLP Programming Tutorial 11 - The Structured Perceptron Graham Neubig Nara Institute of Science and Technology (NAIST) 1 Prediction Problems Given x, A book review Oh, man I love this book! This book is
NLP Programming Tutorial 11 - The Structured Perceptron Graham Neubig Nara Institute of Science and Technology (NAIST) 1 Prediction Problems Given x, A book review Oh, man I love this book! This book is
The Infinite PCFG using Hierarchical Dirichlet Processes
 S NP VP NP PRP VP VBD NP NP DT NN PRP she VBD heard DT the NN noise S NP VP NP PRP VP VBD NP NP DT NN PRP she VBD heard DT the NN noise S NP VP NP PRP VP VBD NP NP DT NN PRP she VBD heard DT the NN noise
S NP VP NP PRP VP VBD NP NP DT NN PRP she VBD heard DT the NN noise S NP VP NP PRP VP VBD NP NP DT NN PRP she VBD heard DT the NN noise S NP VP NP PRP VP VBD NP NP DT NN PRP she VBD heard DT the NN noise
Machine Learning (CS 567) Lecture 2
 Machine Learning (CS 567) Lecture 2 Time: T-Th 5:00pm - 6:20pm Location: GFS118 Instructor: Sofus A. Macskassy (macskass@usc.edu) Office: SAL 216 Office hours: by appointment Teaching assistant: Cheol
Machine Learning (CS 567) Lecture 2 Time: T-Th 5:00pm - 6:20pm Location: GFS118 Instructor: Sofus A. Macskassy (macskass@usc.edu) Office: SAL 216 Office hours: by appointment Teaching assistant: Cheol
MACHINE LEARNING. Vapnik-Chervonenkis (VC) Dimension. Alessandro Moschitti
 MACHINE LEARNING Vapnik-Chervonenkis (VC) Dimension Alessandro Moschitti Department of Information Engineering and Computer Science University of Trento Email: moschitti@disi.unitn.it Computational Learning
MACHINE LEARNING Vapnik-Chervonenkis (VC) Dimension Alessandro Moschitti Department of Information Engineering and Computer Science University of Trento Email: moschitti@disi.unitn.it Computational Learning
Random Coattention Forest for Question Answering
 Random Coattention Forest for Question Answering Jheng-Hao Chen Stanford University jhenghao@stanford.edu Ting-Po Lee Stanford University tingpo@stanford.edu Yi-Chun Chen Stanford University yichunc@stanford.edu
Random Coattention Forest for Question Answering Jheng-Hao Chen Stanford University jhenghao@stanford.edu Ting-Po Lee Stanford University tingpo@stanford.edu Yi-Chun Chen Stanford University yichunc@stanford.edu
S NP VP 0.9 S VP 0.1 VP V NP 0.5 VP V 0.1 VP V PP 0.1 NP NP NP 0.1 NP NP PP 0.2 NP N 0.7 PP P NP 1.0 VP NP PP 1.0. N people 0.
 /6/7 CS 6/CS: Natural Language Processing Instructor: Prof. Lu Wang College of Computer and Information Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang The grammar: Binary, no epsilons,.9..5
/6/7 CS 6/CS: Natural Language Processing Instructor: Prof. Lu Wang College of Computer and Information Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang The grammar: Binary, no epsilons,.9..5
Fantope Regularization in Metric Learning
 Fantope Regularization in Metric Learning CVPR 2014 Marc T. Law (LIP6, UPMC), Nicolas Thome (LIP6 - UPMC Sorbonne Universités), Matthieu Cord (LIP6 - UPMC Sorbonne Universités), Paris, France Introduction
Fantope Regularization in Metric Learning CVPR 2014 Marc T. Law (LIP6, UPMC), Nicolas Thome (LIP6 - UPMC Sorbonne Universités), Matthieu Cord (LIP6 - UPMC Sorbonne Universités), Paris, France Introduction
Extraction of Opposite Sentiments in Classified Free Format Text Reviews
 Extraction of Opposite Sentiments in Classified Free Format Text Reviews Dong (Haoyuan) Li 1, Anne Laurent 2, Mathieu Roche 2, and Pascal Poncelet 1 1 LGI2P - École des Mines d Alès, Parc Scientifique
Extraction of Opposite Sentiments in Classified Free Format Text Reviews Dong (Haoyuan) Li 1, Anne Laurent 2, Mathieu Roche 2, and Pascal Poncelet 1 1 LGI2P - École des Mines d Alès, Parc Scientifique
a) b) (Natural Language Processing; NLP) (Deep Learning) Bag of words White House RGB [1] IBM
![a) b) (Natural Language Processing; NLP) (Deep Learning) Bag of words White House RGB [1] IBM a) b) (Natural Language Processing; NLP) (Deep Learning) Bag of words White House RGB [1] IBM](/thumbs/92/107948882.jpg) c 1. (Natural Language Processing; NLP) (Deep Learning) RGB IBM 135 8511 5 6 52 yutat@jp.ibm.com a) b) 2. 1 0 2 1 Bag of words White House 2 [1] 2015 4 Copyright c by ORSJ. Unauthorized reproduction of
c 1. (Natural Language Processing; NLP) (Deep Learning) RGB IBM 135 8511 5 6 52 yutat@jp.ibm.com a) b) 2. 1 0 2 1 Bag of words White House 2 [1] 2015 4 Copyright c by ORSJ. Unauthorized reproduction of
Joint Emotion Analysis via Multi-task Gaussian Processes
 Joint Emotion Analysis via Multi-task Gaussian Processes Daniel Beck, Trevor Cohn, Lucia Specia October 28, 2014 1 Introduction 2 Multi-task Gaussian Process Regression 3 Experiments and Discussion 4 Conclusions
Joint Emotion Analysis via Multi-task Gaussian Processes Daniel Beck, Trevor Cohn, Lucia Specia October 28, 2014 1 Introduction 2 Multi-task Gaussian Process Regression 3 Experiments and Discussion 4 Conclusions
An Empirical Study of Building Compact Ensembles
 An Empirical Study of Building Compact Ensembles Huan Liu, Amit Mandvikar, and Jigar Mody Computer Science & Engineering Arizona State University Tempe, AZ 85281 {huan.liu,amitm,jigar.mody}@asu.edu Abstract.
An Empirical Study of Building Compact Ensembles Huan Liu, Amit Mandvikar, and Jigar Mody Computer Science & Engineering Arizona State University Tempe, AZ 85281 {huan.liu,amitm,jigar.mody}@asu.edu Abstract.
SUPPORT VECTOR REGRESSION WITH A GENERALIZED QUADRATIC LOSS
 SUPPORT VECTOR REGRESSION WITH A GENERALIZED QUADRATIC LOSS Filippo Portera and Alessandro Sperduti Dipartimento di Matematica Pura ed Applicata Universit a di Padova, Padova, Italy {portera,sperduti}@math.unipd.it
SUPPORT VECTOR REGRESSION WITH A GENERALIZED QUADRATIC LOSS Filippo Portera and Alessandro Sperduti Dipartimento di Matematica Pura ed Applicata Universit a di Padova, Padova, Italy {portera,sperduti}@math.unipd.it
Learning and Inference over Constrained Output
 Learning and Inference over Constrained Output Vasin Punyakanok Dan Roth Wen-tau Yih Dav Zimak Department of Computer Science University of Illinois at Urbana-Champaign {punyakan, danr, yih, davzimak}@uiuc.edu
Learning and Inference over Constrained Output Vasin Punyakanok Dan Roth Wen-tau Yih Dav Zimak Department of Computer Science University of Illinois at Urbana-Champaign {punyakan, danr, yih, davzimak}@uiuc.edu
Fast and Effective Kernels for Relational Learning from Texts
 Fast and Effective Kernels for Relational Learning from Texts Alessandro Moschitti MOCHITTI@DIT.UNITN.IT Department of Information and Communication Technology, University of Trento, 38050 Povo di Trento,
Fast and Effective Kernels for Relational Learning from Texts Alessandro Moschitti MOCHITTI@DIT.UNITN.IT Department of Information and Communication Technology, University of Trento, 38050 Povo di Trento,
Uncovering protein interaction in abstracts and text using a novel linear model and word proximity networks
 Uncovering protein interaction in abstracts and text using a novel linear model and word proximity networks Alaa Abi-Haidar 1,6, Jasleen Kaur 1, Ana Maguitman 2, Predrag Radivojac 1, Andreas Retchsteiner
Uncovering protein interaction in abstracts and text using a novel linear model and word proximity networks Alaa Abi-Haidar 1,6, Jasleen Kaur 1, Ana Maguitman 2, Predrag Radivojac 1, Andreas Retchsteiner
Motif Extraction and Protein Classification
 Motif Extraction and Protein Classification Vered Kunik 1 Zach Solan 2 Shimon Edelman 3 Eytan Ruppin 1 David Horn 2 1 School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel {kunikver,ruppin}@tau.ac.il
Motif Extraction and Protein Classification Vered Kunik 1 Zach Solan 2 Shimon Edelman 3 Eytan Ruppin 1 David Horn 2 1 School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel {kunikver,ruppin}@tau.ac.il
Introduction to Support Vector Machines
 Introduction to Support Vector Machines Andreas Maletti Technische Universität Dresden Fakultät Informatik June 15, 2006 1 The Problem 2 The Basics 3 The Proposed Solution Learning by Machines Learning
Introduction to Support Vector Machines Andreas Maletti Technische Universität Dresden Fakultät Informatik June 15, 2006 1 The Problem 2 The Basics 3 The Proposed Solution Learning by Machines Learning
SemEval-2013 Task 3: Spatial Role Labeling
 SemEval-2013 Task 3: Spatial Role Labeling Oleksandr Kolomiyets, Parisa Kordjamshidi, Steven Bethard and Marie-Francine Moens KU Leuven, Celestijnenlaan 200A, Heverlee 3001, Belgium University of Colorado,
SemEval-2013 Task 3: Spatial Role Labeling Oleksandr Kolomiyets, Parisa Kordjamshidi, Steven Bethard and Marie-Francine Moens KU Leuven, Celestijnenlaan 200A, Heverlee 3001, Belgium University of Colorado,
Personal Project: Shift-Reduce Dependency Parsing
 Personal Project: Shift-Reduce Dependency Parsing 1 Problem Statement The goal of this project is to implement a shift-reduce dependency parser. This entails two subgoals: Inference: We must have a shift-reduce
Personal Project: Shift-Reduce Dependency Parsing 1 Problem Statement The goal of this project is to implement a shift-reduce dependency parser. This entails two subgoals: Inference: We must have a shift-reduce
10/17/04. Today s Main Points
 Part-of-speech Tagging & Hidden Markov Model Intro Lecture #10 Introduction to Natural Language Processing CMPSCI 585, Fall 2004 University of Massachusetts Amherst Andrew McCallum Today s Main Points
Part-of-speech Tagging & Hidden Markov Model Intro Lecture #10 Introduction to Natural Language Processing CMPSCI 585, Fall 2004 University of Massachusetts Amherst Andrew McCallum Today s Main Points
